Publisher's Note: There is an Inside Blood Commentary on this article in this issue.
Key Points
Cultured erythroid cells from DBA patients show impaired growth kinetics and altered transcriptional profiles.
Our data predict a link between GATA1 and RP mutations in the regulation of translation in erythroid differentiation.
Abstract
Diamond-Blackfan anemia (DBA) is a congenital bone marrow failure syndrome characterized by erythroid hypoplasia, usually without perturbation of other hematopoietic lineages. Approximately 65% of DBA patients with autosomal dominant inheritance have heterozygous mutations or deletions in ribosomal protein (RP) genes while <1% of patients with X-linked inheritance have been identified with mutations in the transcription factor GATA1. Erythroid cells from patients with DBA have not been well characterized, and the mechanisms underlying the erythroid specific effects of either RP or GATA1 associated DBA remain unclear. We have developed an ex vivo culture system to expand peripheral blood CD34+ progenitor cells from patients with DBA and differentiate them into erythroid cells. Cells from patients with RP or GATA1 mutations showed decreased proliferation and delayed erythroid differentiation in comparison with controls. RNA transcript analyses of erythroid cells from controls and patients with RP or GATA1 mutations showed distinctive differences, with upregulation of heme biosynthesis genes prominently in RP-mediated DBA and failure to upregulate components of the translational apparatus in GATA1-mediated DBA. Our data show that dysregulation of translation is a common feature of DBA caused by both RP and GATA1 mutations. This trial was registered at www.clinicaltrials.gov as #NCT00106015.
Introduction
Diamond-Blackfan anemia (DBA) is an inherited bone marrow failure syndrome characterized by severe macrocytic anemia, usually without perturbation of other hematopoietic lineages. Patients with DBA are generally diagnosed during infancy or early childhood, have a high frequency of congenital anomalies, and have a predisposition to cancer. The primary treatment of the anemia of DBA is corticosteroids, and although 80% of patients respond initially, approximately 40% of patients lose the steroid responsiveness, ultimately requiring chronic red cell transfusion.1,2 Approximately 65% of patients have heterozygous mutations or deletions in ribosomal protein (RP) genes, including RPS7, RPS10, RPS15, RPS17, RPS19, RPS24, RPS26, RPS27, and RPS29, as well as RPL5, RPL11, RPL15, RPL26, RPL27, RPL31, RPL35A, and RPL36.3-12 A mutation in TSR2, which has been reported as a required cofactor for 40S ribosomal subunit assembly in yeast, has been identified in one family.13 Additionally, mutations in GATA1, which encodes an erythroid transcription factor, have been demonstrated in a few patients diagnosed with DBA. The genetic causes of DBA in the remaining ∼35% of patients are as yet undetermined.
Previous studies have used a variety of approaches to model DBA, including shRNA knockdown,14-16 induced pluripotent stem cells,17 fibroblasts,18 transgenic mice,19 and zebrafish.20,21 Because of the difficulty of obtaining patient bone marrow samples, few studies have used primary DBA cells,14,22-25 and the representation of varying DBA genotypes in existing studies is quite limited. It is clear that the defect in DBA is associated with poor differentiation and, in some instances, a paucity of bone marrow–derived erythroid burst-forming units (the primitive erythroid progenitor).22 The erythroid defect in DBA has been hypothesized to result from disparate factors ranging from abnormal erythroid p53 activation to free heme toxicity resulting from delayed expression of the globin genes.25 With treatment, steroid-responsive patients develop increased levels of bone marrow–derived erythroid burst-forming units in association with clinical observations of reticulocyte response and improvement of the anemia.22,23 Although it remains unclear whether RP and GATA1-associated DBA impede erythropoiesis by parallel or convergent mechanisms, one study suggested that reduced GATA1 protein translation resulted from RP-associated DBA,26 placing GATA1 abnormalities immediately downstream of ribosomal protein insufficiency in the pathophysiology of RP and GATA1-mediated DBA.
To characterize the erythroid differentiation, steroid responsiveness, and transcriptional profiles of patients with RP and GATA1 mutations, we developed an ex vivo system to evaluate primary erythroid cells from peripheral blood CD34+ hematopoietic stem and progenitor cells, an easily accessible source that allows repeated and relatively noninvasive sampling in comparison with bone marrow–derived progenitors. Using this system, we demonstrate that CD34+ cells from patients with RP and GATA1 mutations show reduced proliferation and delayed expression of erythroid cell surface markers. CD34+ cells from corticosteroid-responsive patients with DBA show improved proliferation and differentiation in this system. This system allowed interrogation of gene expression profiles in patient samples, demonstrating differences that suggest a unifying model for the relationship of DBA caused by RP and GATA1 mutations.
Materials and methods
Cell culture
CD34+ cells were collected in acid citrate dextrose from the peripheral blood of healthy donors and patients with DBA after informed consent was obtained. Mononuclear cells were separated by centrifugation on lymphocyte separation media (MP Biomedicals), and CD34+ cells were isolated by using a CD34 microbead kit (Miltenyi Biotec) and an autoMACS Separator (Miltenyi Biotec). CD34+ cells were cultured according to the 2-step culture described by Ebert et al.27 Briefly, cells were cultured for 7 days (37°C with 5% CO2) in 12-well plates (1.5 × 105 cells/well) in proliferation media consisting of StemSpan Serum-Free Expansion Medium (Stemcell Technologies), supplemented with 100 U/mL of penicillin/streptomycin (Invitrogen), 2 mM of glutamine (Invitrogen), 40 μg/mL of human low-density lipoprotein (Stemcell Technologies), and the following cytokines: 100 ng/mL of stem cell factor, 50 ng/mL of thrombopoietin, 10 ng/mL of interleukin-3 (IL-3; Peprotech), 10 ng/mL of IL-6 (Peprotech), and 0.5 U/mL of erythropoietin (Stemcell Technologies). After 7 days, cells were cultured in proliferation media containing 3 U/mL of erythropoietin for an additional 7 days. Cultures were expanded when confluent to maintain a concentration of 1.5 × 105 cells/well.
Flow cytometric analysis
For analysis of erythroid differentiation, cells were collected at day 14 (or earlier) of in vitro culture and stained with anti-CD235–fluorescein isothiocyanate, anti-CD44-PerCP Cy5.5, and anti-CD41-phycoerythrin antibodies (BD Biosciences) for 30 minutes on ice. After washing, cells were analyzed by fluorescence-activated cell sorter Calibur 1 flow cytometer (BD Biosciences), and CD235+/CD44+/CD41− and CD235−/CD44+/CD41− cells were collected. FlowJo (version 10.0.6) software was used for analysis.
Western blot
Cell pellets were lysed by addition of radioimmunoprecipitation assay buffer and boiled in sample buffer. Proteins were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis on a 4% to 15% polyacrylamide gel (Bio-Rad) and transferred to polyvinylidene difluoride membranes (Bio-Rad). The membranes were immunoblotted with an anti-GATA1 antibody (Abcam). Anti-α-tubulin (Sigma-Aldrich) antibody was used as loading control. Proteins were visualized with chemiluminescence reagents (Pierce).
Microarray hybridization and analyses
RNA was extracted using the Purelink RNA kit (Life Technologies) with complementary DNA (cDNA) prepared by WT Plus Expression Kit (Affymetrix) from 200 ng of RNA. Labeled cDNA was hybridized to Human Gene 2.0 ST arrays (Affymetrix) according to the manufacturer’s instructions. Gene-level expression data were summarized after background correction and RMA normalization by using the Oligo Bioconductor package.28 Unsupervised hierarchical clustering was used to discover the relationship between expression patterns and disease/differentiation state. Hierarchical clustering was performed using a Euclidean distance clustering metric on expression of the most variably expressed genes across all samples (coefficient of variation was >30%; n = 109) using the ComplexHeatmap Bioconductor package.29 Gene set enrichment analysis (GSEA) was used to identify pathways altered between DBA patient and normal control cells. GSEA was performed as previously described with the Molecular Signatures Database (H, C2 curated pathways, and the RPS14 knockdown studies from C5),27,29,30 sets of previously reported GATA1 target genes,26 and a small nucleolar RNA (snoRNA) set defined as all snoRNA genes annotated on the array. Sets with false discovery rates less than 10% were considered significant.
Statistics
Statistical analyses were carried out using Prism software (Graph Pad Software). A paired t test was used for proliferation assay/growth curves. Data are presented as the mean ± standard error of the mean.
Study approval
Blood samples were collected from patients with DBA and their families in accordance with the principles of the Declaration of Helsinki, with written informed consent obtained from participants prior to inclusion in the study. This study is registered at www.clinicaltrials.gov (NCT00106015) and overseen by the Northwell Health institutional review board.
Results
Reduced proliferation of CD34+ progenitor cells from patients with DBA
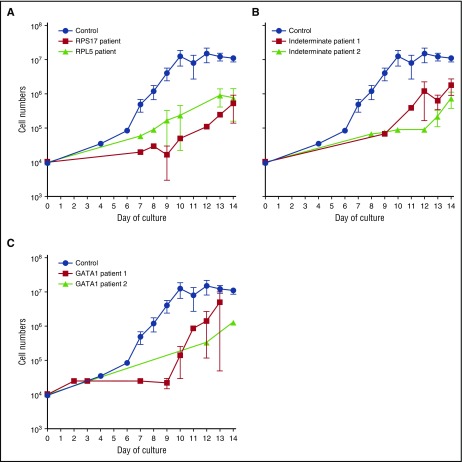
We developed a 2-phase liquid culture system to enrich and expand erythroid cells in vitro from patients with DBA and normal healthy adult donors. CD34+ progenitor cells isolated from 10–20 mL of peripheral blood were expanded for 7 days in cytokines (including 0.5 U/mL of erythropoietin) and differentiated for 7 days in the same cytokines with erythropoietin (3 U/mL).27 We characterized patients with alterations in RPL5 (n = 1), RPL35A (n = 1), RPS17 (n = 2), RPS24 (n = 1), and GATA1 (n = 2) as indeterminate DBA genotype(s) (ie, diagnosed clinically using established criteria1,2 and assessed for but lacking any currently identified genetic lesion; n = 4), with 24 healthy control donors. Each proband was analyzed in triplicate (except for one GATA1 proband because only a single sample was available; Table 1). Cell numbers were quantified throughout the culture period. At the end of the culture period, we routinely obtained ∼1 × 107 CD235+ (glycophorin A) erythroid cells from an initial population of ∼1 × 104 control CD34+ progenitor cells (Figure 1; blue lines). In contrast, all patient-derived cells, regardless of the genotype, exhibited a 10-fold or greater reduction in the number of CD235+ cells (Figure 1A-C). Nevertheless, despite these reduced growth rates, we were able to generate patient-derived erythroid cells in sufficient numbers for additional studies. Cytospin preparations of day-14 cultures demonstrated the presence of immature cells (CD41−/CD44−/CD235−), CD41−/CD44+/CD235− cells that resemble erythroid precursors, and smaller CD41−/CD44+/CD235+ cells that resemble basophilic erythroblasts in both control and DBA cultures (supplemental Figure 1, available on the Blood Web site). We did not find orthochromatic erythroblasts or reticulocytes at the culture endpoint.
Table 1.
Clinical data of DBA patients
| Patient ID | Mutation | Age, y | Age at diagnosis, mo | eADA, U/g Hb | Congenital anomalies | Current status |
|---|---|---|---|---|---|---|
| R-1 | RPS17 (c.189-190delAG) | 28 | 12 | NA | Flat thenar eminence | TD |
| R-2 | RPL5 (c.189+1G>T) | 41 | 0 | NA | ASD | TD |
| R-3 | RPL35A (deletion 3q29) | 4 | 30 | 2.4 | None | TD/SD* |
| R-4 | RPS24 (c.371A>G) | 27 | 6 | NA | None | TD |
| R-5 | RPS17 (5′UTR_3′UTRdel) | 3 | 8 | 1.7 | None | SD |
| G-1 | GATA1 (c.332G>C) | 24 | 10 | NA | None | TD/SD† |
| G-2 | GATA1 (c.332G>C) | 29 | 0 | NA | None | TD |
| I-1 | Indeterminate | 13 | 2 | NA | ASD, short stature, triphalangeal thumbs | SD |
| I-2 | Indeterminate | 9 | 22 | 1.1 | None | TD |
| I-3 | Indeterminate | 5 | 2 | 2.17 | Possible ureter duplication anomaly | TD |
| I-4 | Indeterminate | 22 | 11 | 1.48 | None | Remission |
“NA” was used if the patient was too old or already transfusion dependent at the time of the study.
ASD, atrial septal defect; eADA, erythrocyte adenosine deaminase activity; ID, identification; NA, not applicable; SD, steroid dependent; TD, transfusion dependent.
Assayed initially when transfusion dependent and later after response to corticosteroids.
Transfusion dependent but treated with corticosteroids during the study period to increase the intertransfusion interval.
Figure 1.
Reduced proliferation of erythroid precursors of patients with DBA. Growth curves of CD34+ cells from control (n = 24) and DBA patients with RP gene mutations: RPS17 (red; n = 4) or RPL5 (black; n = 3) (A); control (n = 24) and DBA patients with unidentified mutations: unknown patient 1 (red; n = 3) and unknown patient 2 (black; n = 3) (B); and control (n = 24) and DBA patients with a GATA1 mutation: GATA1 patient 1 (red; n = 3) or GATA1 patient 2 (black; n = 1) (C). Data are presented as the mean ± standard error of the mean.
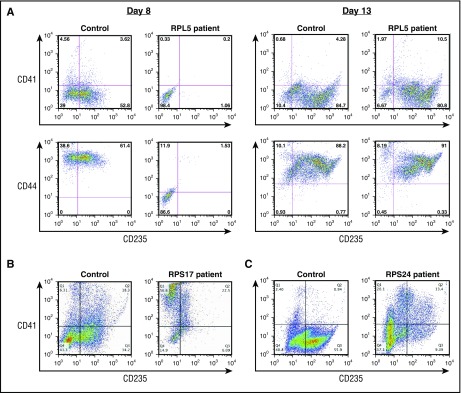
Abnormal erythroid differentiation of CD34+ progenitor cells from patients with DBA
Flow cytometric analyses revealed the presence of CD41−/CD44+/CD235+ cells in cultures of control cells harvested at day 8 but significantly reduced numbers of CD41−/CD44+/CD235+ cells in cultures of patient-derived cells, indicating that the expression of CD235 is delayed in DBA patient cultures (Figure 2A). We observed interpatient variability in the fraction of CD235+ cells at day 14 of culture. The majority of cells from DBA patients expressed CD235 by day 13 or day14 of culture (Figure 2A); however, defects in erythropoiesis persisted at day 14 in cells from 1 patient with a nonsense RPS17 mutation (Figure 2B) and from 1 patient with a missense RPS24 mutation (Figure 2C). Both of these patients’ cells showed a significant reduction in the fraction of CD41−/CD44+/CD235+ cells and a relative increase in CD41+/CD235− cells in comparison with controls, indicating a perturbation in the balance of megakaryocyte–erythroid differentiation (Figure 2B-C).
Figure 2.
Characterization of erythroid differentiation during in vitro culture. Flow cytometric analysis of cultured CD34+ cells labeled with CD235, CD44, and CD41 antibodies. (A) Control donor and a patient with an RPL5 mutation at day 8 and day 13 of terminal erythroid differentiation. (B) Control donor and a patient with a RPS17 mutation at day 14 of culture. (C) Control donor and a patient with a RPS24 mutation at day 14 of culture.
Increased proliferation of cultured CD34+ cells from a patient with DBA after steroid treatment
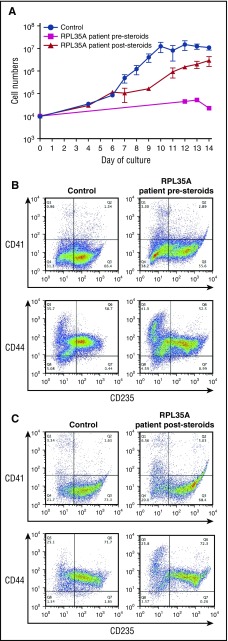
We cultured CD34+ cells from a patient with a deletion involving RPL35A (Table 1) before and after corticosteroid treatment. Culture of cells from the initial sample (prior to steroid treatment) demonstrated a significant defect in erythroid cell proliferation (Figure 3A), with production of markedly reduced numbers of CD235+ erythroblasts by day 14 of culture (Figure 3B). After these studies, the patient began a therapeutic trial of prednisolone, with peripheral blood samples collected at ∼10, 12, and 22 weeks after the initiation of treatment. No corticosteroids were added to the ex vivo cultures. However, the poststeroid samples showed significant improvement in proliferation (Figure 3A) and generation of CD235+ cells (Figure 3C). The improved proliferation we observe ex vivo correlates with a clinical response with increased reticulocytes in the peripheral blood, an increase in the hemoglobin levels, and achievement of transfusion independence by the patient. These findings are analogous to prior observations of favorable shifts in the erythropoietin dose response of bone marrow–derived erythroid progenitors ex vivo after a clinical steroid response.22,31-33 We propose that ex vivo differentiation of peripheral blood CD34+ cells is a similar proxy for steroid responsiveness in patients with DBA.
Figure 3.
Improved proliferation and erythroid differentiation in a steroid-treated patient. (A) Growth curves of CD34+ cells from control donors (n = 24) and a DBA patient with a deletion in chromosome 3q29 containing the RPL35A gene before (magenta; n = 2) and after (red; n = 3) treatment with prednisolone. The data are presented as the mean ± standard error of the mean. (B, C) Flow cytometric analysis of terminally differentiated erythroid cells from a control donor and from a patient with a deletion in chromosome 3q29 using antibodies against CD41, CD44, and CD235 before steroid treatment (B) and after steroid treatment (C).
Reduced proliferation of CD34+ progenitor cells from DBA patients with GATA1 mutations
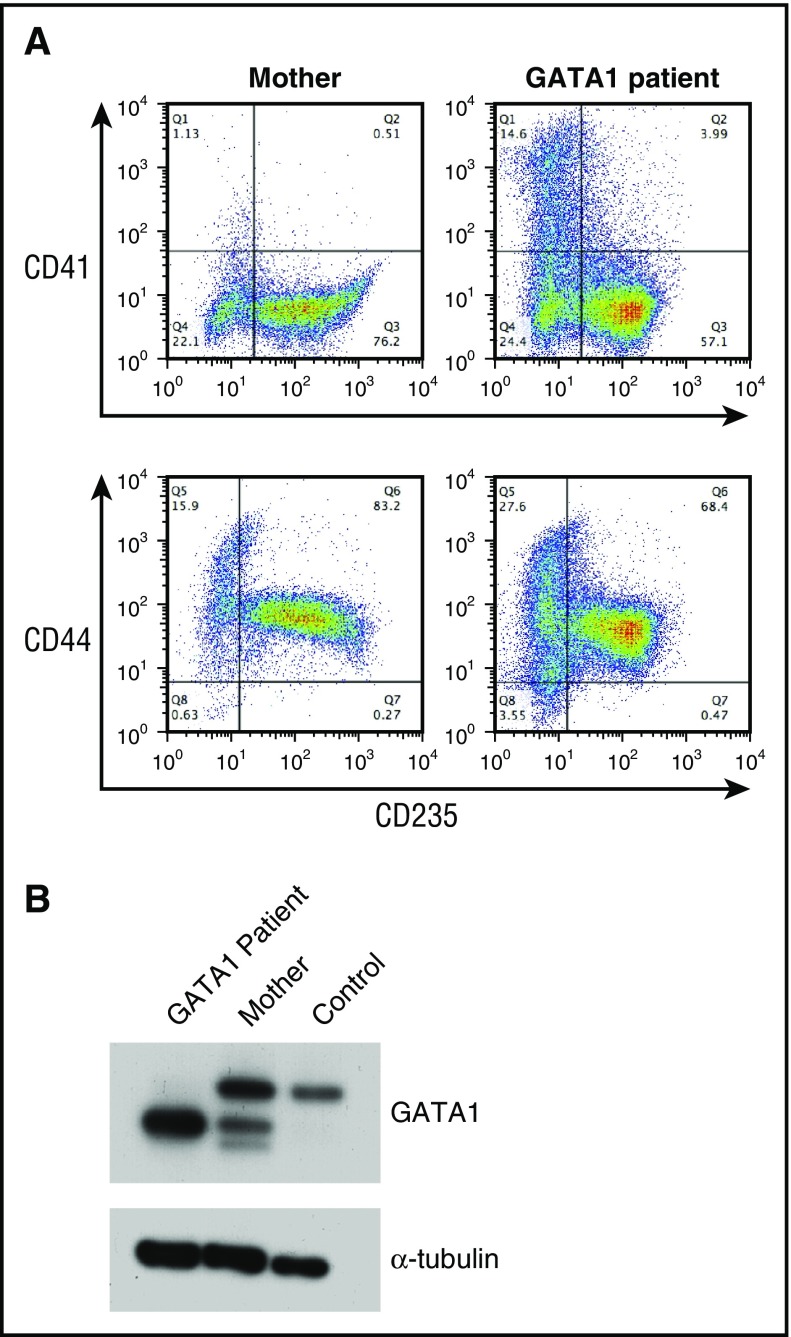
Rare DBA patients with mutations in GATA1 have been reported.34-36 We obtained CD34+ cells from the peripheral blood of 2 related patients with a splice donor mutation that results in the exclusive expression of the RNA encoding the short form of the GATA1 protein (GATA1S), which lacks the transactivation domain.34 In comparison with control CD34+ cells, the proliferation of CD34+ cells from patients with GATA1 mutations was reduced (Figure 1C). Similarly to those of other patients with DBA, cultures of cells from the GATA1 patients had an increase in the proportion of CD41+ cells, indicating a perturbation in the megakaryocyte–erythroid differentiation profile (Figure 4A).
Figure 4.
Analysis of erythroid cells from patients with a mutation in GATA1. (A) Flow cytometric analysis of CD34+ cells from the carrier mother and a DBA patient with a GATA1 mutation cultured for 14 days. The cells were labeled with CD235, CD44, and CD41 antibodies. (B) Lysates from differentiating erythroid cells from a patient with a GATA1 mutation, the carrier mother, and a control donor were immunoblotted with antibodies against GATA1 and α-tubulin (loading control).
Reverse transcription polymerase chain reaction of RNA from cells of patients with mutations in GATA1 has shown reduced levels of GATA1L messenger RNA (mRNA) in comparison with control.34 To directly assay the levels of GATA1 protein, we performed Western blotting of lysates of unfractionated day 14 cultures from 1 patient with a hemizygous GATA1 mutation, his carrier mother, and a control donor by using an antibody against GATA1 (Figure 4B). The cells from the control donor show expression of GATA1L, whereas the carrier mother showed expression of both GATA1L and GATA1S. In the cells from the patient with the GATA1 mutation, only GATA1S protein was detected, at higher levels (Figure 4B), with GATA1S protein levels exceeding the combined level of both GATA1 protein isoforms observed in the controls (Figure 4B).
Analysis of transcriptome profiles of DBA patient erythroid cells
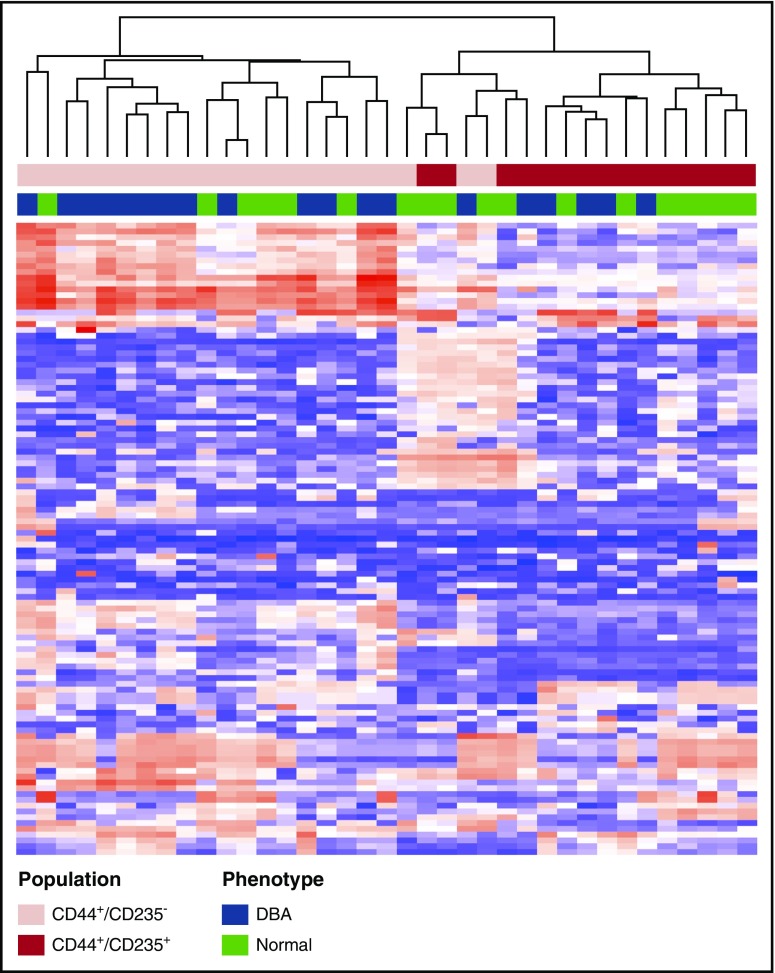
To identify genes and pathways that are perturbed in patients with DBA, we performed transcriptome analyses of RNA isolated from sorted erythroid populations at the end of our 14-day culture from control donors (n = 10) or patients with abnormalities in GATA1 (n = 3 samples from 2 probands), RPS17 (n = 2 samples from 2 probands), RPL5 (n = 1), RPS24 (n = 1), RPL35A (n = 2 samples from 1 proband), or indeterminate genotype(s) (n = 5 samples from 4 probands) using RNA microarray. Unsupervised hierarchical clustering of the most variably expressed genes (supplemental Data files) partitioned specimens into 2 groups on the basis of differentiation stage (CD41−/CD44+/CD235− vs CD41−/CD44+/CD235+; 2-tailed χ2 P < .0001) rather than by disease state (χ2 P = .071), demonstrating that gene expression among the control and DBA populations at each stage were comparable (Figure 5). These results were consistent with the similar morphology and cell surface phenotype of these populations.
Figure 5.
Unsupervised hierarchical clustering of normal and DBA erythroid microarray profiles. Unsupervised hierarchical clustering of specimens by expression levels of the most variant features (coefficient of variation is ≥30% across all DBA [blue] and normal [green] control samples) shows partitioning (top dendrogram split) predominantly by differentiation stage rather than by disease status. The cell populations assayed are CD41−/CD44+/CD235− (pink) and CD41−/CD44+/CD235+ (burgundy).
To identify functional pathways responsible for RP or GATA1-mediated erythroid failure, we used GSEA.31 These comparisons were stratified on the basis of results from the unsupervised analysis into CD41−/CD44+/CD235− and CD41−/CD44+/CD235+ cells. We compared normal CD41−/CD44+/CD235− to normal CD41−/CD44+/CD235+, all DBA samples to normal, and DBA-by-genotype to normal for both cell populations.
Comparison of control CD41−/CD44+/CD235− (N = 10) to control CD41−/CD44+/CD235+ cells (N = 8) showed marked upregulation of genes involved in heme metabolism, targets of GATA1 and E2F transcription, and evidence of increased cell cycling (supplemental Figure 2) in CD41−/CD44+/CD235+ cells. In addition, CD41−/CD44+/CD235+ cells showed marked upregulation of the translational machinery, including increased expression of genes associated with RNA polymerase I (Pol I) and Pol III, the ribosomal protein genes, and snoRNA transcription. In contrast, the predominantly upregulated gene sets in normal control CD41−/CD44+/CD235− cells were involved in cellular signaling (JAK, STAT, interleukins, tumor necrosis factor, etc).
Comparison of normal CD41−/CD44+/CD235+ cells (n = 10) to DBA CD41−/CD44+/CD235+ cells (n = 5, including 1 GATA1, 2 RP, and 2 unknown genotypes; not all cultures generated sufficient numbers of these cells [Figure 2B]) demonstrated that DBA CD41−/CD44+/CD235+ cells, despite morphologic and general mRNA expression pattern similarities to control CD41−/CD44+/CD235+, had a transcriptional profile for specific pathways that resembled that of earlier control CD41−/CD44+/CD235− (supplemental Figure 3).
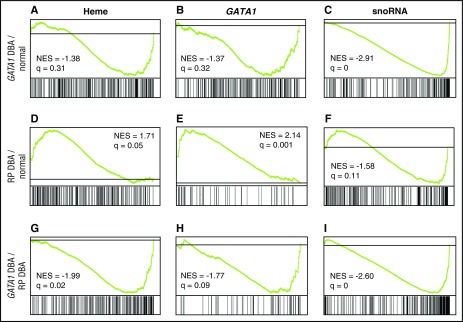
On the basis of these results, we focused further comparisons on the more homogeneous CD41−/CD44+/CD235− population. We observed distinct differences between the transcriptional profiles of normal CD41−/CD44+/CD235− cells (n = 8) to those of RP mutated (n = 6 samples from 5 probands) or GATA1 mutated (n = 3 samples from 2 probands) DBA CD41−/CD44+/CD235− cells, as well as differences in direct comparison of RP and GATA1 samples (Figure 6A-I; supplemental Data). In GSEA, genes are ranked according to differential expression, with the position of genes comprising each set (eg, heme, GATA1, snoRNA) on the x-axis. Overrepresentation of genes from a set at the top (left) or bottom (right) of the ranked list indicates over- and underexpression and is illustrated by a peak (positive) or valley (negative; green line) in relation to the gray horizontal line that represents no change. The most striking finding in the evaluation of GATA1-mutated DBA transcriptomes was that many components of the translational machinery, including snoRNAs, RNA Pol I subunits, and ribosomal protein genes, were in the “valley,” indicating statistically significant underexpression (false discovery rate [FDR] q-value = 0; Figure 6C; supplemental Data). GATA1-mutated transcriptomes showed a trend toward reduced levels of the heme biosynthetic genes compared with normal controls, but this was not statistically significant with an FDR q-value of 0.31 (Figure 6A; supplemental Data). The GATA1-mutated transcriptomes also did not demonstrate statistically significant downregulation of any of the 3 tested GATA1 gene sets (Figure 6B; supplemental Data) when compared with normal controls, a finding that likely reflects the overexpression of GATA1S in these samples along with limitations of the gene sets to distinguish targets differentially responsive to GATA1 and GATA1S. The findings in GATA1 mutated specimens suggest an important role for full-length GATA1 to generate an increased translational capacity during early erythroid differentiation, most likely at the progenitor stages.
Figure 6.
GSEA of CD235− cells. (A-I) Comparison of the transcriptome of normal CD41−/CD44+/CD235− cells to those from GATA1- or RP-mutated DBA patient CD41−/CD44+/CD235− cells, as well as differences in direct comparison of RP and GATA1 specimens, by GSEA. Genes are ranked by signal-to-noise ratio according to differential expression, with the position of genes comprising each set (heme, GATA1, snoRNA) illustrated by the vertical black bars below the plot in each comparison. Overrepresentation of genes from a set at the top (left) or bottom (right) of the ranked list indicates over- and underexpression and is illustrated by a peak (positive) or valley (negative) in the running enrichment score (green) shifted to the left or right, respectively. The gray line represents no change. The normalized enrichment score and false discovery rate q-value are shown for each comparison illustrated. Evaluation of heme synthesis genes (from the MSigDB hallmark set) demonstrated (A) modest downregulation in GATA1-mutated samples when compared with normal controls that was not statistically significant but showed significant overexpression in RP-mediated DBA in comparison with normal controls (D) and GATA1-mutated patients (G). Patients with GATA1 mutations did not show underexpression of any tested GATA1 gene set (B; GATA1 TRANSFAC set from Ref. 26 illustrated), whereas patients with RP mutations showed marked and statistically significant GATA1 target gene overexpression in comparison with both normal controls and patients harboring GATA1 mutations (E, H; GATA1 GSE628 set from Ref. 26 illustrated). Similarly, evaluation of small nucleolar RNA transcription (gene set derived from all snoRNA annotated on the array) showed highly significant downregulation in patients with GATA1 mutations in comparison with normal controls (A) and DBA patients with RP mutations (I), but no significant changes in expression of RP-mediated DBA patients when compared with normal controls (F). NES, normalized enrichment score.
Comparison of the CD41−/CD44+/CD235− transcriptional profiles of controls to patients with RP gene mutations showed statistically significant upregulation of heme synthesis genes (q = 0.05; Figure 6D) and GATA1 target genes in 2 of the 3 GATA1 gene sets tested (q = 0.001: Figure 6E; and 0.02: supplemental Data). There were no significant defects in expression of components of the translational machinery. Specifically, RP gene mRNA and snoRNA levels are similar to those of normal controls (FDR q = 0.134 and 0.114; supplemental Data and Figure 6F, respectively). Direct comparison of the transcriptional profiles of CD41−/CD44+/CD235− from patients with GATA1 and RP mutations highlighted these differences. The GATA1-mutated CD41−/CD44+/CD235− cells demonstrated significant downregulation of heme synthesis genes (q = 0.02; Figure 6G), GATA1 targets (q = 0.09; Figure 6H), and snoRNAs (q = 0; Figure 6I) in comparison with RP-mutated specimens. IPA and Metacore analyses, which are not as robust as GSEA, both identified the heme biosynthesis pathway as significantly increased in the RP samples and GATA1 as the top upstream regulator in both the RP and GATA1 datasets.
Discussion
Determining the molecular pathology that causes DBA is an essential step toward identifying novel therapies. Several models for DBA have been established, but few have used primary erythroid cells from multiple patients with DBA, including patients with mutations in at least 4 different RP genes as well as patients with GATA1 mutations. Our peripheral blood-derived ex vivo culture system represents an important advancement that should accelerate future studies of DBA pathophysiology. First, because of the ease of obtaining peripheral blood in comparison with bone marrow, we can obtain samples from many different patients, giving us a much wider survey of genotypes. Second, our system allows us to repeatedly sample patients, allowing for longitudinal studies without invasive bone marrow aspiration, which is infrequently medically indicated. Although we have currently been able to study only an isolated case, the rescue of the proliferative defects in the patient with a RPL35A mutation treated with corticosteroids agrees with the previous reports.22,31-33 Our ex vivo culture system enables future studies to better understand the determinants of corticosteroid response (and nonresponse) in these patients.
The finding that the transcriptomes of cells from RP- and GATA1-mutated patients with DBA have significant overlap makes it clear that GATA1 target genes, and not genes regulated by other known erythroid transcription factors such as KLF1, are responsible for the pathology of DBA. Consistent with the recent findings of Yang et al,25 we observe an increase in the levels of the RNA encoding the heme synthetic enzymes in RP-mutated patient cells but not in GATA1-mutated patient cells. These data suggest, but do not prove, that heme toxicity may be a feature of only RP-associated DBA as previously proposed.25 Additional biochemical studies will be required to evaluate this fully.
Erythroid cells from patients with mutations in the GATA1 gene have no detectable protein expression of the GATA1L isoform. Despite the increased level of GATA1S protein, the GATA1 patients have erythroid hypoplasia, suggesting that the GATA1S isoform cannot functionally compensate for GATA1L in erythroid development. CD41−/CD44+/CD235+ cells from patients with RP mutations showed marked downregulation of some GATA1 targets, including heme synthesis genes, and increased expression of gene sets typical of normal CD41−/CD44+/CD235− progenitors. We interpret these data to reflect an overall delay in differentiation in DBA cells rather than an indicator of proximal effect. Consistent with these results, a recent study demonstrated that GATA1S failed to properly induce erythroid differentiation in GATA1-deficient megakaryocyte–erythroid progenitor cells, which may be due to altered chromatin binding and failure of GATA1S to occupy erythroid-specific gene-regulatory elements.37 Complete elucidation of the functions of the different GATA1 isoforms in human erythroid cells is needed to fully understand erythroid development and DBA.
Ludwig et al reported selectively reduced GATA1 protein levels in differentiated erythroid cells derived from control CD34+ cells after knockdown of several RP genes, along with a signature of reduced GATA1 target gene expression in 3 RPS19-mutated DBA patients. They proposed that ribosomal protein haploinsufficiency led to preferentially decreased GATA1 mRNA translation and suggested that GATA1 acted as a master regulator of DBA.26
Our data from DBA patient cells show similarities to this work as well as important differences. We do not see evidence of downregulated GATA1 target gene expression in RP-mutated CD41−/CD44+/CD235− progenitors as opposed to the more primitive CD34+/CD45RALO/CD71+ cells analyzed by Ludwig et al.26 One possible explanation for these disparate findings is that the cells were analyzed at different stages of differentiation, Ludwig et al of more primitive cells where the levels of GATA1 would be expected to be lower, and our study of more mature erythroid cells where GATA1 levels would be expected to be higher. A second possibility is that the initial pool of CD34+ cells is deficient in “erythroid-primed” cells, perhaps due to GATA1 deficiency as suggested by Ludwig et al.26 This hypothesis can be evaluated only at the single-cell level. A third possibility is that erythroid differentiation in patient cells is blocked, but over time a small population of these cells “escape” from the restrictions of the DBA mutation, allowing the rescued clone to proliferate and differentiate normally. This escape could be due to epigenetic rescue, a possible mechanism for clinical remission.
On the basis of our results, we favor the hypothesis that the differences between our study and that of Ludwig et al are due to the analysis of cells at different stages of differentiation and maturation. We do not favor the rescue hypothesis because the transcriptome in DBA CD41−/CD44+/CD235− and CD41−/CD44+/CD235+ cells differs in specific ways from that of control samples, indicating that a complete rescue has not occurred. Second, steroid-responsive patients retain features of the DBA–erythroid phenotype including macrocytosis, elevated fetal hemoglobin levels, and elevated erythroid adenosine deaminase activity,1 suggesting that steroid-induced rescue is not complete.
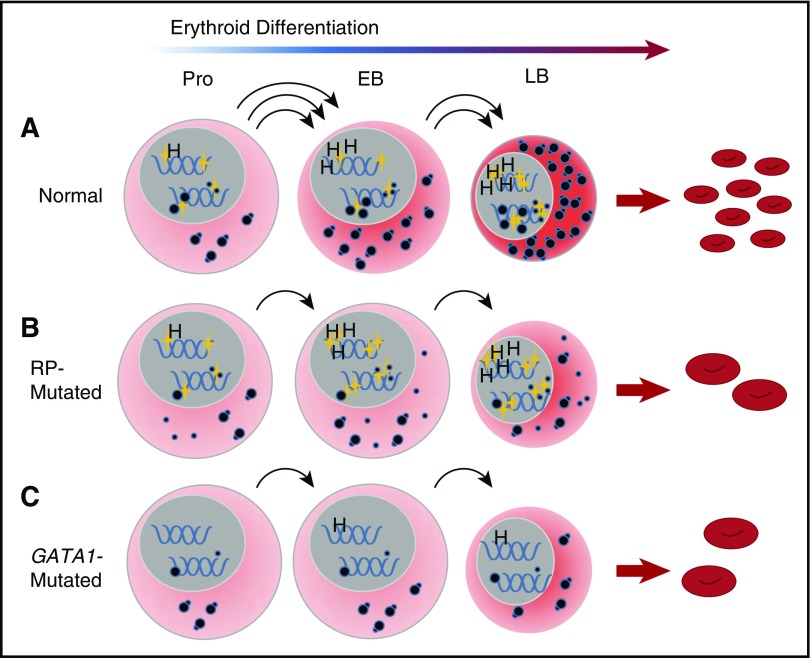
It is becoming clear that RP and GATA1 DBA represent distinct subclasses of DBA in terms of the underlying pathophysiologic mechanism of erythroid failure.38 The most striking difference in expression pattern between RP- and GATA1-mutated patient samples is the failure of mutant GATA1 cells to upregulate components of the translational apparatus, including RNA Pol I components, rRNA, snoRNA, and RP gene transcription in CD41−/CD44+/CD235− erythroblasts during differentiation. These data are consistent with the concept of GATA1 as a master regulator of translation in erythroid cells, but acting upstream rather than downstream from the insufficient complement of ribosomes caused by RP mutations. This hypothesis is supported by recent chromatin immunoprecipitation studies demonstrating GATA1 occupancy at RP promoters during erythroid differentiation.39 We propose that in DBA patients with RP mutations, mRNA translation is compromised by insufficient translational competence resulting from abnormal ribosome assembly due to haploinsufficiency of a single ribosomal protein, despite a normal GATA1-induced translation program. Insufficient translation may result in faulty translation-associated events contributing to erythroid failure. In patients with GATA1 mutations, global mRNA translation is compromised because of an inability of erythroid progenitors to upregulate the translational apparatus, including the synthesis of ribosomes (Figure 7).
Figure 7.
A convergent model for the effect of RP and GATA1 mutations on erythroid differentiation in DBA. (A) Under conditions of normal hematopoiesis, rising GATA1L levels (gold stars, nuclear) in CD41−/CD44+/CD235− cells promote the erythroid program by stimulating the expression of canonical GATA1 targets, such as heme synthesis genes, as well as enhancing expression of components of the translational apparatus, including ribosomal RNA, snoRNA, and ribosomal proteins. The net effect of these activities is the accumulation of sufficient translational competence (cytoplasmic, illustrated as mature 80S ribosomes) to support normal differentiation and proliferation from proerythroblasts into early and late basophilic erythroblasts. (B) In RP-mutated erythropoiesis, GATA1L is present to promote heme synthesis as well as upregulation of the translational apparatus; however, abortive ribosomal assembly (illustrated as a large-subunit defect with accumulation of free cytoplasmic 40S ribosomes) limits accretion of translational competence, with reduced proliferation of erythroid progenitors, delays in the maturational program, and anemia. (C) In GATA1-mediated DBA, the absence of GATA1L limits upregulation of the translational apparatus without abortive ribosome assembly, but with similar consequences to proliferation and differentiation of erythroid progenitors. This model places GATA1 as a master regulator in DBA that operates upstream from RP gene alterations. EB, early basophilic erythroblasts; H, heme synthesis genes; LB, late basophilic erythroblasts; Pro, proerythroblasts.
Acknowledgments
This work was supported by the National Institutes of Health, National Human Genome Research Institute Intramural funds (D.M.B.), grant 5R01HL079571 (J.M.L. and A.V.), and grant 7K08HL092224 (J.E.F.); the Diamond Blackfan Anemia Foundation (D.M.B., K.A.O., S.R.E., and C.A.T.), and the Arkansas Biosciences Institute (J.E.F.).
Footnotes
The data reported in this article have been deposited in the Gene Expression Omnibus database (accession number GSE89540).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: K.A.O., J.E.F., A.V., J.M.L., S.R.E., and D.M.B. designed the research study; K.A.O., S.M.A., C.A.T., D.M.B., L.B., J.L., E.A., A.E., J.E.F., and X.A. acquired the data; K.A.O., J.E.F., and D.M.B. analyzed data and wrote the manuscript; K.A.O., A.V., J.E.F., L.B., J.M.L., and D.M.B. edited the manuscript; and all authors approved the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: David M. Bodine, National Human Genome Research Institute, National Institutes of Health, 49 Convent Dr, Building 49, Room 4A04, Bethesda, MD 20892-4442; e-mail: tedyaz@mail.nih.gov.
References
- 1.Vlachos A, Blanc L, Lipton JM. Diamond Blackfan anemia: a model for the translational approach to understanding human disease. Expert Rev Hematol. 2014;7(3):359-372. [DOI] [PubMed] [Google Scholar]
- 2.Vlachos A, Muir E. How I treat Diamond-Blackfan anemia. Blood. 2010;116(19):3715-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cmejla R, Cmejlova J, Handrkova H, Petrak J, Pospisilova D. Ribosomal protein S17 gene (RPS17) is mutated in Diamond-Blackfan anemia. Hum Mutat. 2007;28(12):1178-1182. [DOI] [PubMed] [Google Scholar]
- 4.Doherty L, Sheen MR, Vlachos A, et al. . Ribosomal protein genes RPS10 and RPS26 are commonly mutated in Diamond-Blackfan anemia. Am J Hum Genet. 2010;86(2):222-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Draptchinskaia N, Gustavsson P, Andersson B, et al. . The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat Genet. 1999;21(2):169-175. [DOI] [PubMed] [Google Scholar]
- 6.Farrar JE, Nater M, Caywood E, et al. . Abnormalities of the large ribosomal subunit protein, Rpl35a, in Diamond-Blackfan anemia. Blood. 2008;112(5):1582-1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farrar JE, Vlachos A, Atsidaftos E, et al. . Ribosomal protein gene deletions in Diamond-Blackfan anemia. Blood. 2011;118(26):6943-6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gazda HT, Grabowska A, Merida-Long LB, et al. . Ribosomal protein S24 gene is mutated in Diamond-Blackfan anemia. Am J Hum Genet. 2006;79(6):1110-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gazda HT, Sheen MR, Vlachos A, et al. . Ribosomal protein L5 and L11 mutations are associated with cleft palate and abnormal thumbs in Diamond-Blackfan anemia patients. Am J Hum Genet. 2008;83(6):769-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Landowski M, O’Donohue MF, Buros C, et al. . Novel deletion of RPL15 identified by array-comparative genomic hybridization in Diamond-Blackfan anemia. Hum Genet. 2013;132(11):1265-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mirabello L, Macari ER, Jessop L, et al. . Whole-exome sequencing and functional studies identify RPS29 as a novel gene mutated in multicase Diamond-Blackfan anemia families. Blood. 2014;124(1):24-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang R, Yoshida K, Toki T, et al. . Loss of function mutations in RPL27 and RPS27 identified by whole-exome sequencing in Diamond-Blackfan anaemia. Br J Haematol. 2015;168(6):854-864. [DOI] [PubMed] [Google Scholar]
- 13.Gripp KW, Curry C, Olney AH, et al. ; UW Center for Mendelian Genomics. Diamond-Blackfan anemia with mandibulofacial dystostosis is heterogeneous, including the novel DBA genes TSR2 and RPS28. Am J Med Genet A. 2014;164(9):2240-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moniz H, Gastou M, Leblanc T, et al. ; DBA Group of Société d’Hématologie et d’Immunologie Pédiatrique-SHIP. Primary hematopoietic cells from DBA patients with mutations in RPL11 and RPS19 genes exhibit distinct erythroid phenotype in vitro. Cell Death Dis. 2012;3:e356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horos R, Ijspeert H, Pospisilova D, et al. . Ribosomal deficiencies in Diamond-Blackfan anemia impair translation of transcripts essential for differentiation of murine and human erythroblasts. Blood. 2012;119(1):262-272. [DOI] [PubMed] [Google Scholar]
- 16.Farrar JE, Quarello P, Fisher R, et al. . Exploiting pre-rRNA processing in Diamond Blackfan anemia gene discovery and diagnosis. Am J Hematol. 2014;89(10):985-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garçon L, Ge J, Manjunath SH, et al. . Ribosomal and hematopoietic defects in induced pluripotent stem cells derived from Diamond Blackfan anemia patients. Blood. 2013;122(6):912-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Avondo F, Roncaglia P, Crescenzio N, et al. . Fibroblasts from patients with Diamond-Blackfan anaemia show abnormal expression of genes involved in protein synthesis, amino acid metabolism and cancer. BMC Genomics. 2009;10:442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Devlin EE, Dacosta L, Mohandas N, Elliott G, Bodine DM. A transgenic mouse model demonstrates a dominant negative effect of a point mutation in the RPS19 gene associated with Diamond-Blackfan anemia. Blood. 2010;116(15):2826-2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Danilova N, Sakamoto KM, Lin S. Ribosomal protein S19 deficiency in zebrafish leads to developmental abnormalities and defective erythropoiesis through activation of p53 protein family. Blood. 2008;112(13):5228-5237. [DOI] [PubMed] [Google Scholar]
- 21.Uechi T, Nakajima Y, Chakraborty A, Torihara H, Higa S, Kenmochi N. Deficiency of ribosomal protein S19 during early embryogenesis leads to reduction of erythrocytes in a zebrafish model of Diamond-Blackfan anemia. Hum Mol Genet. 2008;17(20):3204-3211. [DOI] [PubMed] [Google Scholar]
- 22.Lipton JM, Kudisch M, Gross R, Nathan DG. Defective erythroid progenitor differentiation system in congenital hypoplastic (Diamond-Blackfan) anemia. Blood. 1986;67(4):962-968. [PubMed] [Google Scholar]
- 23.Iskander D, Psaila B, Gerrard G, et al. . Elucidation of the EP defect in Diamond-Blackfan anemia by characterization and prospective isolation of human EPs. Blood. 2015;125(16):2553-2557. [DOI] [PubMed] [Google Scholar]
- 24.Ohene-Abuakwa Y, Orfali KA, Marius C, Ball SE. Two-phase culture in Diamond Blackfan anemia: localization of erythroid defect. Blood. 2005;105(2):838-846. [DOI] [PubMed] [Google Scholar]
- 25.Yang Z, Keel SB, Shimamura A, et al. . Delayed globin synthesis leads to excess heme and the macrocytic anemia of Diamond Blackfan anemia and del(5q) myelodysplastic syndrome. Sci Transl Med. 2016;8(338):338ra67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ludwig LS, Gazda HT, Eng JC, et al. . Altered translation of GATA1 in Diamond-Blackfan anemia. Nat Med. 2014;20(7):748-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ebert BL, Pretz J, Bosco J, et al. . Identification of RPS14 as a 5q- syndrome gene by RNA interference screen. Nature. 2008;451(7176):335-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carvalho BS, Irizarry RA. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 2010;26(19):2363-2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32(18):2847-2849. [DOI] [PubMed] [Google Scholar]
- 30.Subramanian A, Tamayo P, Mootha VK, et al. . Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545-15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan HS, Saunders EF, Freedman MH. Diamond-Blackfan syndrome: II. In vitro corticosteroid effect on erythropoiesis. Pediatr Res. 1982;16(6):477-478. [DOI] [PubMed] [Google Scholar]
- 32.Chan HS, Saunders EF, Freedman MH. Diamond-Blackfan syndrome: I. Erythropoiesis in prednisone responsive and resistant disease. Pediatr Res. 1982;16(6):474-476. [DOI] [PubMed] [Google Scholar]
- 33.Nathan DG, Clarke BJ, Hillman DG, Alter BP, Housman DE. Erythroid precursors in congenital hypoplastic (Diamond-Blackfan) anemia. J Clin Invest. 1978;61(2):489-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sankaran VG, Ghazvinian R, Do R, et al. . Exome sequencing identifies GATA1 mutations resulting in Diamond-Blackfan anemia. J Clin Invest. 2012;122(7):2439-2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parrella S, Aspesi A, Quarello P, et al. . Loss of GATA-1 full length as a cause of Diamond-Blackfan anemia phenotype. Pediatr Blood Cancer. 2014;61(7):1319-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klar J, Khalfallah A, Arzoo PS, Gazda HT, Dahl N. Recurrent GATA1 mutations in Diamond-Blackfan anaemia. Br J Haematol. 2014;166(6):949-951. [DOI] [PubMed] [Google Scholar]
- 37.Chlon TM, McNulty M, Goldenson B, Rosinski A, Crispino JD. Global transcriptome and chromatin occupancy analysis reveal the short isoform of GATA1 is deficient for erythroid specification and gene expression. Haematologica. 2015;100(5):575-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weiss MJ, Mason PJ, Bessler M. What’s in a name? J Clin Invest. 2012;122(7):2346-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Amanatiadou EP, Papadopoulos GL, Strouboulis J, Vizirianakis IS. GATA1 and PU.1 bind to ribosomal protein genes in erythroid cells: implications for ribosomopathies. PLoS One. 2015;10(10):e0140077. [DOI] [PMC free article] [PubMed] [Google Scholar]