The removal of repressive histone marks in a cytokinin-rich environment primes de novo activation of WUSCHEL by B-type ARABIDOPSIS RESPONSE REGULATORs and HD-ZIP III transcription factors.
Abstract
Plant cells are totipotent and competent to regenerate from differentiated organs. It has been known for six decades that cytokinin-rich medium induces shoot regeneration from callus cells. However, the underlying molecular mechanism remains elusive. The homeodomain transcription factor WUSCHEL (WUS) is essential for de novo establishment of the shoot stem cell niche in Arabidopsis thaliana. We found that WUS-positive (WUS+) cells mark the shoot progenitor region during regeneration. A cytokinin-rich environment initially promotes the removal of the repressive histone mark H3K27me3 at the WUS locus in a cell cycle-dependent manner. Subsequently, the B-type ARABIDOPSIS RESPONSE REGULATORs (ARRs) ARR1, ARR2, ARR10, and ARR12, which function as transcriptional activators in the cytokinin signaling pathway, spatially activate WUS expression through binding with microRNA165/6-targeted HD-ZIP III transcription factors. Thus, our results provide important insights into the molecular framework for cytokinin-directed shoot regeneration and reveal a two-step mechanism for de novo activation of WUS.
INTRODUCTION
Plants can be regenerated through somatic embryogenesis or de novo organogenesis (Birnbaum and Sánchez Alvarado, 2008; Duclercq et al., 2011; Sugimoto et al., 2011; Su and Zhang, 2014; Gaillochet and Lohmann, 2015). De novo organogenesis refers to the in vitro formation of shoots or roots from cultured explants. In the model plant Arabidopsis thaliana, callus, a pluripotent cell mass, forms from explants on auxin-rich callus-inducing medium (CIM). Subsequently, culturing the callus on cytokinin-rich shoot-inducing medium (SIM) induces the differentiation of the callus into shoots (Skoog and Miller, 1957; Duclercq et al., 2011; Cheng et al., 2013). It is well known that de novo shoot regeneration is dependent on cell division progression (Che et al., 2007), the acquisition of organogenetic competence on CIM (Che et al., 2007; Atta et al., 2009; Kareem et al., 2015), the partition of auxin and cytokinin responses (Gordon et al., 2007), cytokinin signal perception (Ishida et al., 2008; Cheng et al., 2013), and stem cell commitment (Che et al., 2002; Cheng et al., 2013; Ikeuchi et al., 2016).
Plant stem cells reside in stem cell niches, which produce signals that regulate the balance between self-renewal and the generation of daughter cells that differentiate into new tissues (Barton, 2010; Ha et al., 2010; Aichinger et al., 2012; Heidstra and Sabatini, 2014; Holt et al., 2014; ten Hove et al., 2015). In the shoot apical meristem (SAM), the expression of the WUS gene defines the organizing center (OC), and the WUS protein acts as a nonautonomous signal to maintain stem cells by activating CLAVATA3 (CLV3) (Schoof et al., 2000; Yadav et al., 2011; Daum et al., 2014; Bustamante et al., 2016). The cytokinin response is mediated by B-type ARABIDOPSIS RESPONSE REGULATORs (ARRs), which activate a number of cytokinin-responsive genes including A-type ARRs (To and Kieber, 2008). There is evidence demonstrating that cytokinin plays important roles in the maintenance of the SAM and WUS expression (Gaillochet et al., 2015). For example, a cytokinin response maximum and expression of the cytokinin receptor His Kinase4 both localize to the OC (Chickarmane et al., 2012). This local cytokinin biosynthesis acts as a positional cue for OC and WUS expression within the stem cell niche (Kurakawa et al., 2007; Gordon et al., 2009; Chickarmane et al., 2012; Adibi et al., 2016). Moreover, WUS represses A-type ARRs, thereby reinforcing WUS expression by increasing cytokinin responses (Leibfried et al., 2005).
In Arabidopsis, four B-type ARR transcription factors, ARR1, ARR2, ARR10, and ARR12, play essential roles in shoot regeneration. Compared with the wild type, the shoot regenerative capacity is impaired in the arr1 arr10 arr12 mutant (Mason et al., 2005; Ishida et al., 2008; Zhang et al., 2015). The A-type ARRs play opposite roles in shoot regeneration. The regeneration capacity is markedly reduced when A-type ARRs such as ARR7 or ARR15 are overexpressed (Buechel et al., 2010). However, how WUS is de novo activated by cytokinin is largely elusive.
Our knowledge of de novo shoot regeneration is mostly derived from snapshot images of fixed samples. Studies that use fixed samples lack real-time temporal information such as the length of the cell cycle in specific cell types; therefore, how SAM fate is spatiotemporally specified remains unclear at both the cellular and molecular levels. In this study, using time-lapsed imaging and genetic analyses, we showed that WUS-positive (WUS+) cells mark the shoot progenitor region during shoot regeneration in Arabidopsis. We further demonstrated that the WUS locus is subjected to epigenetic reprogramming upon transfer to cytokinin-rich SIM. Moreover, we found that B-type ARRs (ARR1, ARR2, ARR10, and ARR12) interact with microRNA165/6 (miR165/6)-targeted HD-ZIP III transcription factors and that this complex spatially induces WUS expression within the callus.
RESULTS
WUS Is Essential for Establishing the Stem Cell Niche during Regeneration
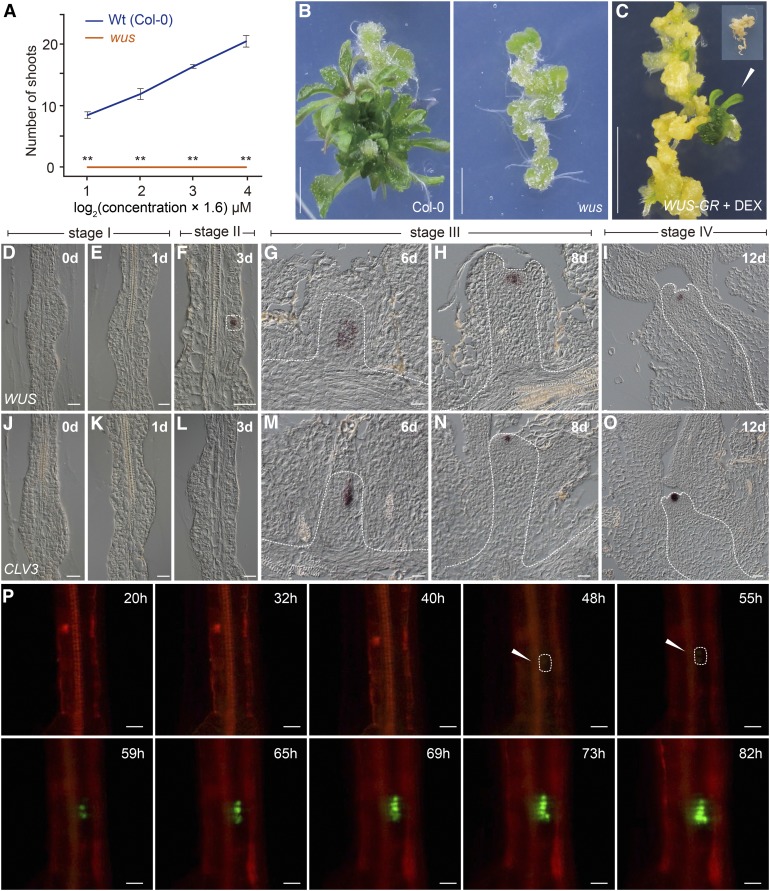
We performed de novo shoot regeneration using hypocotyls from various Arabidopsis lines as explants. After 25 d on SIM, the regenerative capacity was calculated as the number of regenerated shoots in a given number of explants. Consistent with a previous report (Chatfield et al., 2013), the wus mutant failed to regenerate shoots (0%, n = 32) (Figures 1A and 1B), whereas overexpression of dexamethasone (DEX)-inducible WUS (Pro35S:WUS-GR [Rat glucocorticoid receptor]) led to shoot regeneration on hormone-free medium supplemented with DEX (12% for +DEX, n = 40; 0% for −DEX, n = 40) (Figure 1C). In comparison, other SAM-defective mutants such as clv3, shoot meristemless (stm), and the cup-shaped cotyledon2 (cuc2) cuc3 double mutant exhibited normal or reduced shoot regeneration rates (Supplemental Figure 1). Hence, these results indicate that WUS is essential for establishing the stem cell niche during shoot regeneration.
Figure 1.
Expression of WUS during Shoot Regeneration.
(A) and (B) Shoot regeneration in the wild type and the wus mutant. Hypocotyls were used as explants for the regeneration assays. The regenerative capacity in (A) was calculated as the number of shooted explants/total number of explants. Data are expressed as mean ± sd, n = 32; **P < 0.01, Student’s t test. Bar = 0.5 cm.
(C) Shoot regeneration assay of Pro35S:WUS-GR. The hypocotyls were cultured on CIM and transferred to hormone-free MS medium supplemented with or without 10 μM DEX, n = 40. Inset: No shoots were regenerated in the absence of DEX. Bar = 0.5 cm.
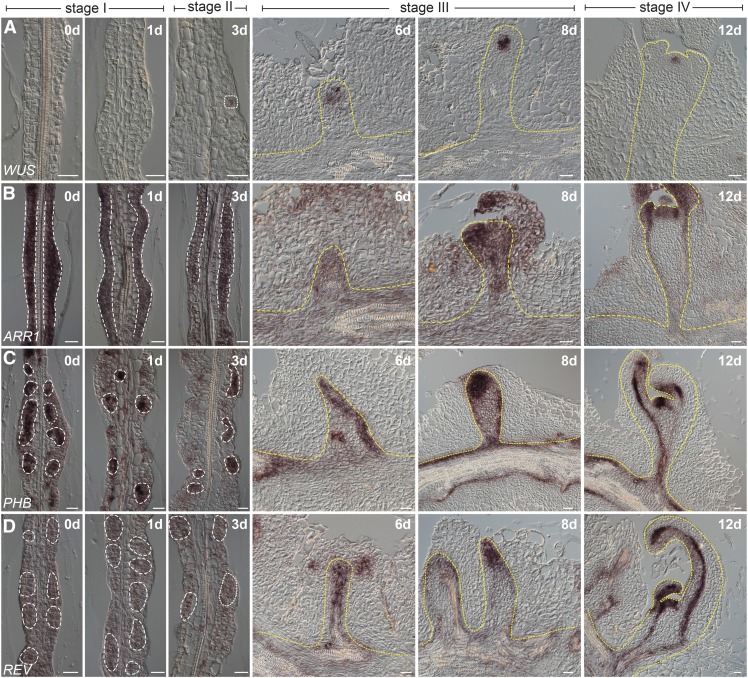
(D) to (O) Expression of WUS ([D] to [I]) and CLV3 ([J] to [O]) in wild-type (Col-0) explants. WUS+ cells and the developing SAM are marked by dash lines. Bar = 20 µm.
(P) Live imaging of WUS+ cells (green) and their progeny in the ProWUS:3xVENUS-N7 reporter line (Col-0). Cell outlines were stained by propidium iodide (red). Bar = 20 µm.
WUS+ Cells Mark the Shoot Progenitor Region during Shoot Regeneration
Using live imaging analyses with fluorescent reporters, CUC2+ cells were shown to contribute to shoot progenitor formation, whereas WUS+ cells label nonprogenitor cell regions (Gordon et al., 2007): WUS is broadly expressed throughout callus tissue by 5 d of induction on SIM, declines after 10 d culture, and is later upregulated within the center of phyllotactic shoot meristems. Unexpectedly, we found that the expression pattern of this WUS reporter construct did not mimic that of its endogenous mRNA during shoot regeneration (Supplemental Figure 2). We harvested the explants on SIM on days 0, 3, 6, 8, and 12. RNA in situ hybridization assays revealed that WUS mRNA accumulated in single cells dispersed throughout the explant on day 3 (Figures 1D to 1F). At 6 d of induction, a few clusters of WUS+ cells were observed (Figure 1G).
To visualize the real-time dynamics of WUS expression, we constructed a ProWUS:3xVENUS-N7 reporter, which showed the same expression pattern as WUS mRNA (Supplemental Figure 3). This WUS reporter was functional, as ProWUS:WUS was able to rescue the wus phenotype (87%, n = 38, wus −/−). We performed time-lapse recordings of the early phase during shoot regeneration using a live cell imaging system and divided the shoot regeneration process into four stages according to dynamic WUS expression patterns and SAM morphology (Figure 1; Supplemental Figure 3 and Supplemental Movie 1). At stage I (day 0 to 2 after transfer to SIM), the callus cells underwent cell division (Supplemental Movie 2). At stage II (day 2 to 4 after transfer to SIM), disperse WUS expression was observed. At stage III (day 5 to 10 after transfer to SIM), the stem cell niche was established: The WUS+ cells continued to proliferate, giving rise to a population of WUS+ cells that eventually resided in newly formed SAMs where CLV3+ cells were observed (Figures 1J to 1O). At stage IV (day 10 and thereafter), the regenerated shoots could be visibly identified and the first leaf primordium was differentiated. Together, these results demonstrate that WUS+ cells mark the shoot progenitor region during shoot regeneration.
STM, along with WUS, plays important roles in the maintenance of undifferentiated cells in the SAM (Gallois et al., 2002). The phenotype of the stm mutant is similar to that of the wus mutant (Long et al., 1996). We found that STM was expressed in the developing SAM beginning at stage III (6 d after transfer to SIM) (Supplemental Figure 4), implying that STM plays a minor role in de novo establishment of the SAM.
Recently, it has been shown that plant root regeneration follows the developmental stages of embryonic patterning and is guided by spatial information provided by complementary hormone domains (Efroni et al., 2016). To probe whether a similar developmental program mediates shoot regeneration, we examined the expression of WUSCHEL-RELATED HOMEOBOX2 (WOX2), which is initially expressed in the zygote and acts as a cell fate regulator in the apical/shoot lineage (Breuninger et al., 2008) (Supplemental Figure 4). While WUS was activated during embryogenesis and shoot regeneration (Gallois et al., 2002) (Figures 1D to 1I), we did not detect WOX2 transcripts in wild-type explants on SIM (Supplemental Figure 4), suggesting that embryogenesis and de novo shoot regeneration deploy different genetic programs to establish the SAM.
ARR1 and ARR2 Directly Activate WUS
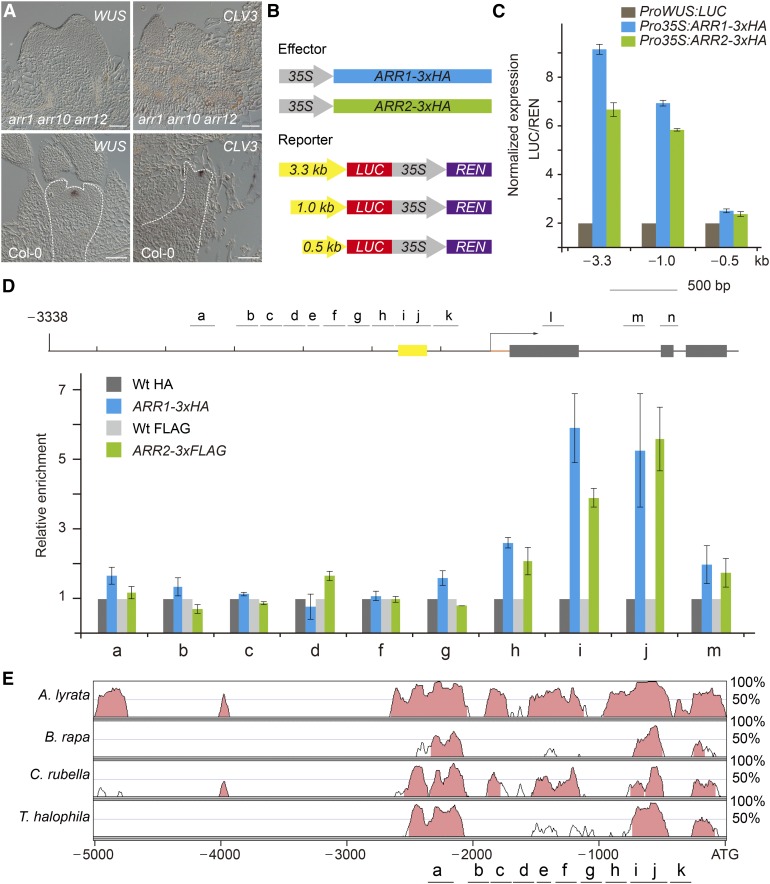
In Arabidopsis, four B-type ARR transcription factors, ARR1, ARR2, ARR10, and ARR12, are essential for shoot regeneration. Compared with the wild type, the shoot regenerative capacity, but not the root regenerative capacity, was impaired in the arr1 arr10 arr12 and arr2 arr12 mutants (Supplemental Figure 5) (Mason et al., 2005; Ishida et al., 2008; Zhang et al., 2015). WUS and CLV3 transcripts were never observed in the explants of the arr1 arr10 arr12 mutant (Figure 2A; Supplemental Figure 5). These results, together with the previous finding that cytokinin signaling activates WUS expression through both CLV-dependent and CLV-independent pathways in the inflorescence SAM (Gordon et al., 2009), suggest that these B-type ARRs are required for de novo activation of WUS.
Figure 2.
ARR1 and ARR2 Bind to the WUS Promoter.
(A) Expression of WUS and CLV3 in the wild type and the B-type arr mutant. Bar = 50 µm.
(B) Diagrams of the B-type ARR effector and WUS reporter constructs used for transient expression analyses in (C).
(C) Transient activation assays in Arabidopsis protoplasts. Error bars represent se (n = 3 biological replicates [independent experiments]).
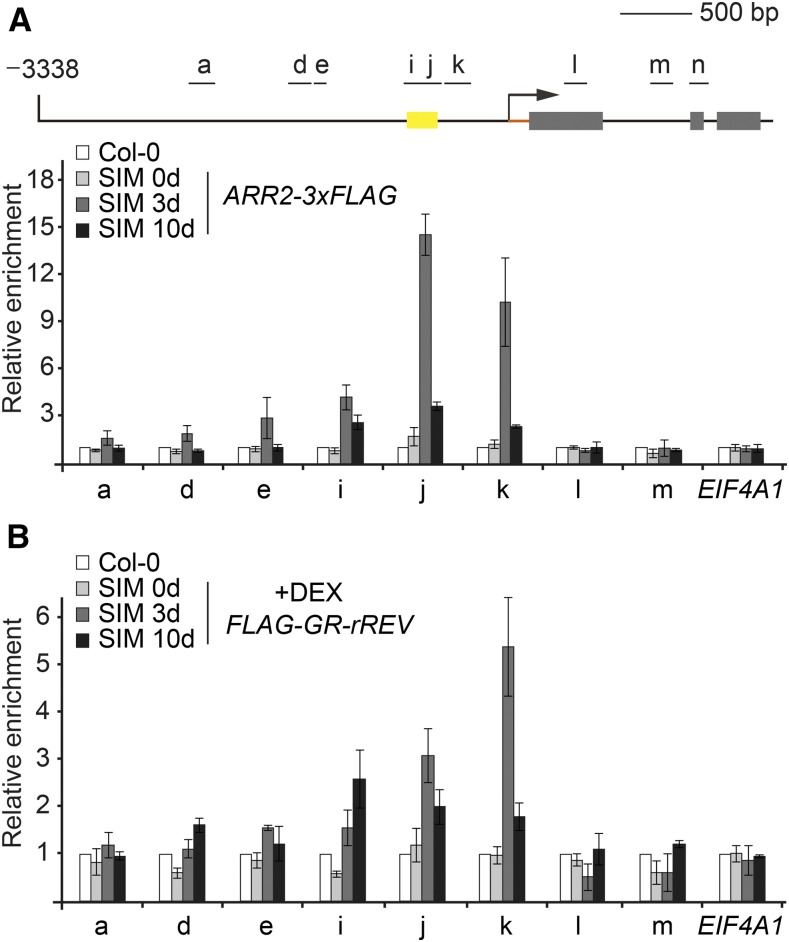
(D) ChIP analyses using Pro35S:ARR1-3xHA and Pro35S:ARR2-3xFLAG explants grown on SIM. A diagram of the WUS genomic region is shown. Black lines and gray boxes indicate promoter and coding regions, respectively. Yellow box indicates the promoter region (−541 to −726 bp), which is absolutely necessary for WUS expression in the stem cell niche (Bäurle and Laux, 2005). The chromatin extract was immunoprecipitated with anti-HA and anti-FLAG beads. Error bars represent se (n = 3 biological replicates).
(E) Pairwise alignment using mVISTA (Mayor et al., 2000) of the genomic region of WUS from four Brassicaceae that are very closely related to Arabidopsis (Bailey et al., 2006). y axis: the percentage of identity.
To determine whether B-type ARRs (ARR1, ARR2, ARR10, and ARR12) activate WUS, we performed transient assays in Arabidopsis protoplasts with ProWUS:LUC (luciferase) as a reporter. Overexpression of ARR1-3xHA or ARR2-3xHA resulted in an ∼6.0- to 8.0-fold increase in LUC activity in wild-type protoplasts (Figures 2B and 2C). This enhancement was abolished when the region from −500 to −1000 bp in the WUS promoter was removed (Figures 2B and 2C). To test whether B-type ARR binds to the regulatory sequence of WUS in vivo, chromatin extracts from wild-type, Pro35S:ARR1-3xHA, or Pro35S:ARR2-3xFLAG explants on SIM (7 d after transfer to SIM) were immunoprecipitated with anti-HA or anti-FLAG antibody. There was no apparent enrichment of the genomic fragments in wild-type samples. In contrast, the region approximately −500 to −750 bp from the translational start codon was readily amplified in Pro35S:ARR1-3xHA and Pro35S:ARR2-3xFLAG samples (Figure 2D). Notably, this region is present in two highly conserved cis-regulatory regions in the WUS promoter in four Brassicaceae that are closely related to Arabidopsis, as revealed by phylogenetic shadowing (Figure 2E). Moreover, this region overlaps with the region (−541 to −726 bp) that is absolutely necessary for WUS expression in the inflorescence meristem stem cell niche (Figure 2D; Supplemental Figure 6) (Bäurle and Laux, 2005). Using electrophoretic mobility shift assay (EMSA), we identified two regions (−550 to −620 bp and −700 to −760 bp) as the core binding sites for ARR2 (Supplemental Figure 6). Thus, these results together indicate that ARR1 and ARR2 activate WUS directly.
Division-Dependent Removal of a Repressive Epigenetic Mark Is Required for the Induction of WUS by Cytokinin
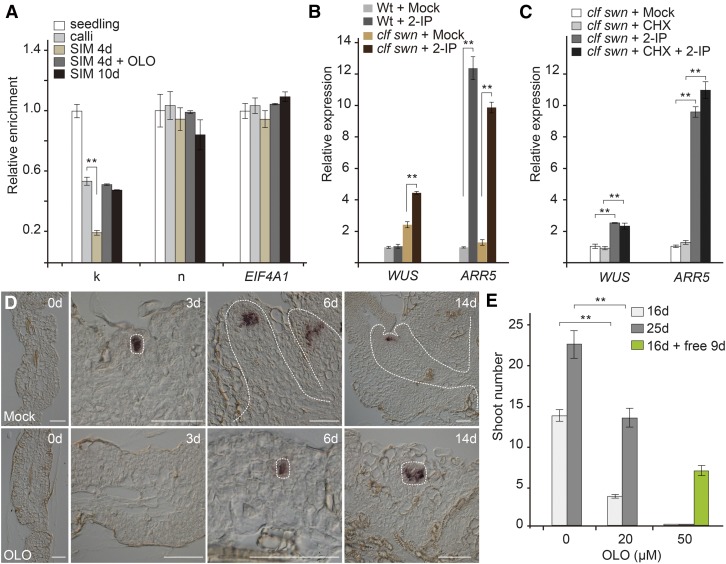
The above results suggest that B-type ARRs (ARR1, ARR2, ARR10, and ARR12) activate WUS directly. However, the induction of WUS by cytokinin required ∼2 to 3 d of culturing on SIM (Figures 1F and 1P; Supplemental Figure 3), implying that the WUS locus is initially incompetent for transcriptional activation by B-type ARRs. The transcribed region of WUS carries the repressive histone modification trimethylation of lysine 27 of histone 3 (H3K27me3), which is established and maintained by Polycomb group (PcG) proteins (Sawarkar and Paro, 2010; Li et al., 2011; Liu et al., 2011; Simon and Kingston, 2013; Xiao and Wagner, 2015). We found that the H3K27me3 level at the WUS locus was gradually reduced after transfer to cytokinin-rich SIM (Figure 3A; Supplemental Figure 7A). In Arabidopsis, CURLY LEAF (CLF) and SWINGER (SWN) encode H3K27me3 methyltransferases. The clf swn double mutant displays a global reduction in H3K27me3 levels (Goodrich et al., 1997; Chanvivattana et al., 2004). We found that the expression of WUS was rapidly induced in clf swn within 2 h of cytokinin treatment (Figure 3B). Importantly, this induction was direct, because the addition of cycloheximide (CHX; a translational inhibitor) did not interfere with WUS induction by cytokinin (Figure 3C). Consistently, clf +/− swn explants developed shoots earlier than wild-type explants on SIM (Supplemental Figure 7B).
Figure 3.
Direct Activation of WUS by Cytokinin.
(A) Temporal accumulation of H3K27me3 at the WUS locus. Wild-type explants were harvested 0, 4, and 10 d after transfer to SIM. The chromatin extract was immunoprecipitated with anti-H3K27me3 antibody. EUKARYOTIC TRANSLATION INITIATION FACTOR 4A1 (EIF4A1) was used as a negative control. **P < 0.01, Student’s t test. The results for the WUS promoter region k and n (Figure 2C) are shown. Error bars represent se (n = 3 biological replicates).
(B) Induction of WUS by cytokinin. Explants from wild-type or clf swn plants were treated with water (mock), 2-IP (a cytokinin) for 2 h. The expression of WUS was normalized to that of TUB. The expression of WUS in the mock wild-type sample was set to 1. **P < 0.01, Student’s t test.
(C) Cytokinin directly induces WUS expression. Wild-type and clf swn explants were treated with ethanol (mock), 2-IP, CHX, and 2-IP + CHX for 2 h. **P < 0.01, Student’s t test.
(D) and (E) Olo treatment (20 µM) delays WUS activation (D) and shoot regeneration (E). n = 24; error bars represent sd; **P < 0.01. Bar = 50 µm.
Olomoucine (a cyclin-dependent kinase inhibitor) blocks cell cycle progression at the G1-S and G2-M phases (Supplemental Movie 3) (Planchais et al., 2000; Sun et al., 2014). Whereas WUS was induced ∼2 to 3 d after cytokinin treatment, the addition of olomoucine prevented WUS induction on day 3 (Figure 3D; Supplemental Figure 8). Cytokinin also induced A-type ARR expression. However, A-type ARR expression was unaffected by olomoucine treatment (which had the strongest effect on WUS) (Supplemental Figure 9). In agreement with this result, the addition of olomoucine caused a delay in the decrease in H3K27me3 levels at the WUS locus, as well as delayed shoot regeneration (Figures 3A and 3E; Supplemental Figure 7A). Subsequent removal of olomoucine restored shoot regeneration (Figure 3E; Supplemental Figure 10). Thus, division-dependent removal of H3K27me3 is required for the induction of WUS by cytokinin.
B-Type ARRs Interact with HD-ZIP III Transcription Factors
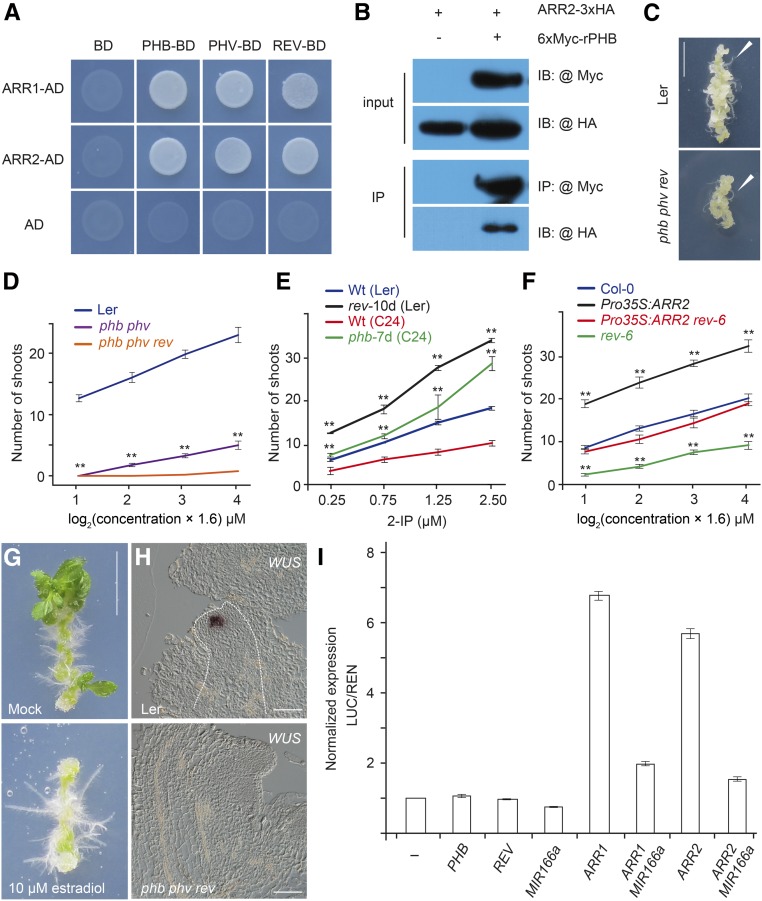
Next, we sought to determine where B-type ARRs (ARR1, ARR2, ARR10, and ARR12) activate WUS within the callus. Analyses of the synthetic cytokinin reporter ProTCSn:GFP (Müller and Sheen, 2008) revealed that the transcriptional activities of B-type ARRs were evenly distributed within calli after transfer to SIM (Supplemental Figure 11). This led us to propose that WUS+ cells are spatially specified by B-type ARRs with the help of other factors. To test this idea, we screened for ARR1/2-interacting transcription cofactors using yeast two-hybrid assays with proteins implicated in the commitment to form SAMs (Supplemental Table 1) and found that miR165/6-targeted HD-ZIP III transcription factors (PHABULOSA [PHB], PHAVOLUTA [PHV], and REVOLUTA [REV]) strongly interacted with ARR1 and ARR2 (Figure 4A). The interaction between B-type ARRs and PHB or REV was further confirmed by in vitro pull-down, coimmunoprecipitation (CoIP), and bimolecular luminescence complementation (BiLC) assays (Figure 4B; Supplemental Figure 12).
Figure 4.
The B-Type ARR-HD ZIP III Complex Activates WUS.
(A) Yeast two-hybrid assay. ARR1/2 was fused to the GAL4 activation domain (AD), and PHB/PHV/REV was fused to the GAL4 DNA binding domain (BD). Interaction assays were performed on SD-Ade-Leu-Trp-His plates with 15 mM 3-amino-1,2,4,-triazole.
(B) CoIP assay. ARR2-3xHA and 6xMyc-rPHB (miR165/6-resistant form of PHB) fusion proteins were transiently expressed in Nicotiana benthamiana leaves. The protein extract was immunoprecipitated with anti-Myc agarose beads, blotted, and probed with anti-HA or anti-Myc antibody.
(C) Root regeneration in wild-type and phb phv rev plants. Arrows indicate regenerated roots. Bar = 0.5 cm.
(D) and (E) Shoot regeneration in the wild type, phb phv, phb phv rev, rev-10d, and phb-7d. n = 24; error bars represent sd; **P < 0.01.
(F) Shoot regeneration in Col-0, Pro35S:ARR2, rev-6, and Pro35S:ARR2 rev-6, n = 24; error bars represent sd; **P < 0.01.
(G) Shoot regeneration assay of XVE-MIR166A on SIM supplemented with or without 10 μM 17-β-estradiol. Bar = 0.5 cm.
(H) Expression of WUS in wild-type and phb phv rev explants. Bar = 50 µm.
(I) Transient activation assays in protoplasts. Error bars represent se (n = 3 biological replicates).
B-Type ARR and HD-ZIP III Function as Partners in Shoot Regeneration
Simultaneous inactivation of miR165/6-targeted HD-ZIP III genes impairs the commitment to form SAMs, leading to the production of a radial symmetric structure in the apical region (Prigge et al., 2005). On the contrary, expressing PHB under the control of the regulatory sequence of PLETHORA2, a root meristem-specific gene, converts root apical meristems into shoots (Smith and Long, 2010). To explore the role of HD-ZIP III transcription factors in cytokinin-induced shoot regeneration, we performed regeneration assays using the loss-of-function mutants, phb phv and phb phv rev, and gain-of-function mutants, phb-7d and rev-10d, in which PHB or REV is highly expressed (Emery et al., 2003; Carlsbecker et al., 2010). Explants of phb phv and phb phv rev exhibited normal callus formation rates and root regenerative capacity (Figure 4C). Compared with the wild type, the shoot regenerative capacity was compromised in phb phv and abolished in phb phv rev triple mutants (Figure 4D; Supplemental Figure 13). In contrast, both phb-7d and rev-10d mutants exhibited higher regenerative capacity than the wild type (Figure 4E; Supplemental Figure 13). Explants of the inducible miR165/6 transgenic line exhibited reduced shoot regeneration on SIM in the presence of 17-β-estradiol (20.8% for +17-β-estradiol, n = 24; 87.5% for mock, n = 24; Figure 4G). Similarly, the elevated shoot regenerative capacity conferred by ARR2 overexpression was suppressed by the rev mutant (Figure 4F; Supplemental Figure 13). Furthermore, the increased level of cytokinin was not sufficient for inducing shoot regeneration in phb phv rev triple mutants (Figure 4D). Thus, the above results collectively suggest that B-type ARR and HD-ZIP III function as partners in shoot regeneration.
Expression analyses revealed that PHB was expressed properly in arr1 arr10 arr12 (Supplemental Figure 14A). Likewise, the expression of A-type ARRs in response to cytokinin was normal in explants of the phb phv rev triple mutant (Supplemental Figure 14B). WUS was never activated in the calli of phb phv rev on SIM (Figure 4H). Transient assays with ProWUS:LUC revealed that HD-ZIP III proteins alone could not activate WUS expression (Figure 4I). However, the activation of WUS by B-type ARRs was substantially suppressed in protoplasts overexpressing miR165/6, indicating that miR165/6-targeted HD-ZIPIII proteins are required for the induction of WUS by B-type ARRs.
To probe when the B-type ARR-HD-ZIP III complex binds to the WUS locus during shoot regeneration, we performed time-course chromatin immunoprecipitation (ChIP) experiments using Pro35S:ARR2-3xFLAG and the DEX-treated Pro35S:FLAG-GR-rREV explants. While both ARR2-3xFLAG and FLAG-GR-rREV highly accumulated in the explants, the occupancy of ARR2-3xFLAG at the WUS promoter was low on SIM on day 0 and became evident after 3 d (Figure 5). FLAG-GR-rREV bound to the same region as ARR2-3xFLAG, and its enrichment also peaked on day 3 after transfer to SIM. Thus, these results reveal that the timing for recruiting the B-type ARR-HD-ZIP III transcriptional complex coincides with the timing for de novo activation of WUS.
Figure 5.
ARR2 and REV Bind to the WUS Promoter.
ChIP analyses using explants from wild-type (Col-0), Pro35S:ARR2-3xFLAG, and the DEX-treated Pro35S:FLAG-GR-rREV plants on SIM on days 0, 3, and 10. For Pro35S:FLAG-GR-rREV, 10 µM DEX was added to the SIM. A diagram of the WUS genomic region is shown. Black lines and gray boxes indicate the promoter and coding region, respectively. Yellow box indicates the promoter region (−541 to −726 bp), which is absolutely necessary for WUS expression in the stem cell niche (Bäurle and Laux, 2005). The chromatin extract was immunoprecipitated with anti-FLAG beads. Error bars represent se (n = 3 biological replicates). The enrichment in the wild-type samples was set to 1.0. EIF4A1 was used as a control.
The B-Type ARR-HD-ZIP III Complex Spatially Induces WUS within the Callus
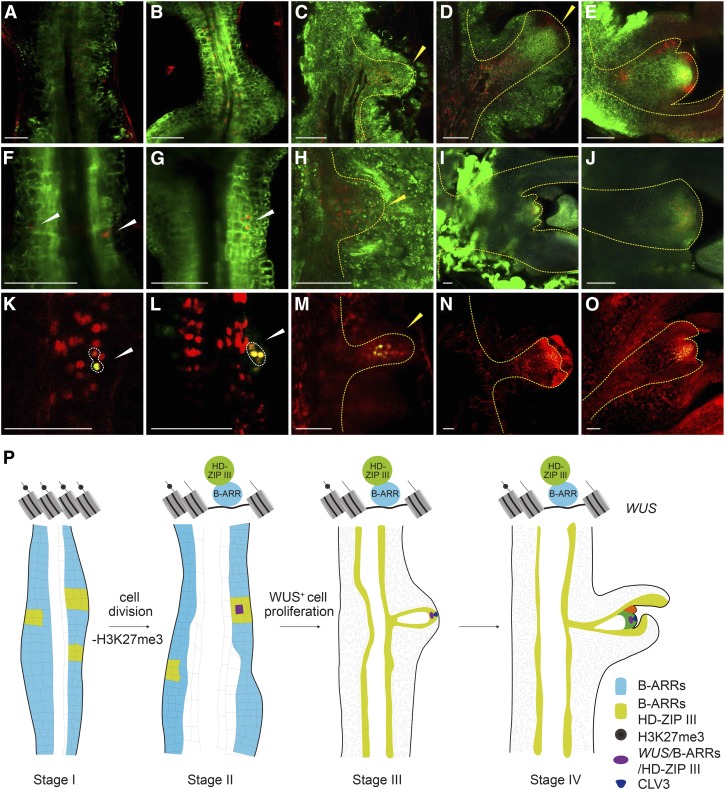
Using in situ hybridization and time-revolved imaging, we examined the temporal expression pattern of WUS, B-type ARRs, and HD-ZIP III genes during shoot regeneration. Although qRT-PCR showed that ARR2 was highly expressed in calli, we did not detect ARR2 mRNA by in situ hybridization for an unknown reason (Supplemental Figure 15). At stage I, widespread accumulation of ARR1 mRNA was observed in rapidly dividing callus cells (Figure 6B; Supplemental Figure 11). Consistently, TCSn reporter activity was evenly distributed within calli (Figures 7A to 7E). PHB and REV exhibited distinct expression patterns, with their mRNAs enriched in small patches of cells (Figures 6C and 6D; Supplemental Figure 16B). At stages II and III, ProTCSn:GFP and ProREV:DsRED-N7 mirS reporter analyses demonstrated that the expression domains of TCSn and REV overlapped (Figures 7A to 7E; Supplemental Figure 11). Notably, WUS was induced in cells where HD-ZIP III and ARR1 were coexpressed (Figures 6 and 7F to 7O; Supplemental Figure 16A), in agreement with the role of the B-type ARR-HD-ZIP III transcriptional complex in shoot regeneration. Beginning at stage IV, PHB and REV were expressed in the SAM and the adaxial side of the leaf primordium, whereas ARR1 was expressed broadly in the SAM (Figures 6B to 6D). Consistent with these findings, the local application of DEX onto Pro35S:rREV-GR phb phv rev explants induced onsite shoot formation (Supplemental Figure 17).
Figure 6.
Expression of ARR1, HD-ZIP III, and WUS during Shoot Regeneration.
Expression of WUS (A), ARR1 (B), PHB (C), and REV (D) during shoot regeneration. Bar = 20 µm.
Figure 7.
The B-Type ARR-HD-ZIP III Complex Spatially Activates WUS within the Callus.
(A) to (O) Expression of ProTCSn:GFP ProREV:DsRED-N7 mirS ([A] to [E]), ProTCSn:GFP ProWUS:DsRED-N7 ([F] to [J]), and ProREV:DsRED-N7 mirS ProWUS:3xVENUS-N7 ([K] to [O]). The WUS+ cell and the developing SAM are indicated by white and yellow dashed lines, respectively. Bar = 50 µm.
(P) A two-step mechanism for cytokinin-directed shoot regeneration. Four stages (stage I to IV) are shown. Different colors indicate the expression patterns of B-type ARRs, CLV3, WUS, and HD-ZIP III, respectively.
B-Type ARR and HD-ZIP III Cooperatively Regulate Shoot Development in Seedlings
To investigate the role of B-type ARRs and HD-ZIP III in the SAM during normal plant development, we generated a plant in which a chimeric ARR2 protein fused with the repressor motif SRDX was expressed from the promoter of REV (ProREV:ARR2-SRDX). ARR2-SRDX has a dominant-negative effect on the cytokinin response (Heyl et al., 2008). While rev-6 and ProREV:ARR2-SRDX plants were phenotypically indistinguishable from the wild type, the ProREV:ARR2-SRDX rev-6 double mutant exhibited strong defects in the SAM. Out of 26 F2 seedlings, nine produced a single cotyledon and two developed a radial symmetric structure, which resembles the phb phv rev triple mutant (Supplemental Figure 18A). In situ hybridization analyses revealed that WUS transcripts were barely detectable in both phv phv rev and ProREV:ARR2-SRDX rev-6 seedlings, which did not have SAMs (Supplemental Figures 18B to 18D). Thus, these results suggest that B-ARRs and HD-ZIP III cooperatively activate or maintain WUS expression during normal development.
DISCUSSION
In vitro shoot regeneration is crucial for both plant developmental biology investigations and plant biotechnology. Although a key molecular pathway that maintains the stem cell niche in the SAM and the signaling cascade that tranduces cytokinin signals have been identified, less is known about how SAM is de novo committed by cytokinin. Our results provide some important insights into the molecular framework for cytokinin-directed de novo shoot regeneration.
Epigenetic Reprogramming at the WUS Locus during Shoot Regeneration
While WUS is not absolutely required for vegetative growth, it plays an essential role in shoot regeneration. Using in situ hybridization and real-time imaging, we found that WUS+ cells mark the shoot progenitor region during shoot regeneration. Our results further show that clearance of epigenetic marks at the WUS locus drives shoot regeneration. Upon transfer to cytokinin-rich SIM, the repressive histone marker H3K27me3 at the WUS locus is gradually removed in conjunction with cell proliferation. Subsequently, the B-type ARR-HD-ZIP III transcriptional complex directly induces WUS expression, thereby promoting the commitment to form the SAM (Figure 7P). An interesting future research direction would be to uncover the molecular mechanism by which cytokinin-rich SIM directs epigenetic reprogramming at the WUS locus in a cell cycle-dependent manner. We envision two possible molecular mechanisms. First, the gradual removal of H3K27me3 at the WUS locus might be caused by positive histone demethylation catalyzed by histone demethylases including RELATIVE OF EARLY FLOWERING6 (REF6) and EARLY FLOWERING6 in Arabidopsis (Lu et al., 2011; Crevillén et al., 2014). Recently, two studies demonstrated that the recruitment of REF6 to a specific genomic locus is mediated by its four Cys2His2 zinc fingers, which directly recognize the CTCTGYTY motif (Cui et al., 2016; Li et al., 2016). Although the regenerative capacity of the ref6 and elf6 mutants has not been explored, sequence analyses revealed that there are three CTCTG motifs between −566 and −445 bp in the WUS promoter region. Another possible molecular mechanism is Polycomb eviction, which is responsible for the reduction in H3K27me3 levels with cell cycle progression (Sun et al., 2014). In this scenario, the binding of the B-type ARR or B-type ARR-HD-ZIP III transcriptional complex displaces PcG protein, leading to the failure to maintain the repressive histone methylation at the WUS locus during cell division.
Diverse Regulatory Regions in WUS Promoter
It is currently unknown whether the same cis-elements or regulatory regions are responsible for de novo activation or maintenance of WUS expression in the SAM. Through promoter deletion analyses, Bäurle and Laux (2005) revealed that the D5 region (−541 to −726 bp) is absolutely necessary for WUS expression in the stem cell niche. The genomic fragment carrying the D5 deletion did not rescue the wus phenotype and abolished WUS expression in the inflorescence meristem. This result is in good agreement with our finding that ARR1 and ARR2 activate WUS through binding to two fragments (−550 to −620 bp and −700 to −760 bp) within the D5 region (Supplemental Figure 6). Using a series of synthetic reporters that carry tetrameric tandem repeats of WUS promoter fragments, Bäurle and Laux identified a 57-bp regulatory region (−712 to −655 bp) that confers WUS transcription in the SAM stem cell niche (Bäurle and Laux, 2005). Surprisingly, this 57-bp regulatory region does not overlap with the ARR1/2 binding regions. Does this 57-bp regulatory region also play a role in de novo activation of WUS during shoot regeneration? Similarly, are the two ARR1/2 binding regions involved in the maintenance of WUS expression in the SAM during normal plant development? To address these questions, we should dissect whether the 57-bp region is sufficient to drive WUS expression during shoot regeneration. In addition, saturating mutagenesis of the regulatory sequences using the clustered regularly interspaced short palindromic repeat/Cas9 (CRISPR/cas9) approach in the native genomic context will reveal how the diverse cis-elements in the D5 region cooperatively regulate WUS expression (Canver et al., 2015).
The Conserved Role of B-Type ARR and HD-ZIP III in Distinct Meristematic Populations
It is poorly understood how WUS is de novo activated during embryogenesis, shoot regeneration, and axillary meristem initiation. Whether the proposed two-step model and the corresponding B-type ARR/HD-ZIP III-WUS cascade are conserved within these distinct meristematic populations is still unknown. Analyses using TCS reporter revealed a high cytokinin response in shoot meristem progenitor cells during shoot regeneration and axillary meristem initiation (Cheng et al., 2013; Q. Wang et al., 2014; Y. Wang et al., 2014). However, TCS activity is barely detected in the apical region at the early embryonic stages (Müller and Sheen, 2008; Zürcher et al., 2013). These results imply that WUS is activated in the same way during shoot regeneration and axillary meristem initiation, whereas embryogenesis utilizes another, unknown mechanism.
Our regeneration assays showed that miR165/6-targeted HD-ZIP III genes are essential for shoot regeneration. Similarly, the rev mutant lacks axillary buds (Otsuga et al., 2001; Shi et al., 2016). Moreover, in the absence of these genes, the apical portion of the embryo lacks central tissue and forms a single, radially symmetric cotyledon (Prigge et al., 2005). Therefore, although previous studies have revealed complicated, redundant, and antagonistic functions of HD-ZIP III transcription factors in meristem development (Emery et al., 2003; Prigge et al., 2005; Lee and Clark, 2015), their function in de novo activation of WUS is likely to be conserved.
Spatial Activation of WUS during Shoot Regeneration
Another interesting question raised by our study is how WUS is spatially activated within callus. Using TCS and ARR1 reporters, the cytokinin response was found to be evenly distributed within the callus at early stages of shoot regeneration. Interestingly, ARR1 reporter activities were restricted to discrete regions and enriched in the regenerated meristem at the late stages. This observation suggests that miR165/6-targeted HD-ZIP III transcription factors play roles in restricting the regionalization of ARR1. In this scenario, the colocalization of B-type ARRs and HD-ZIP III might contribute to the maintenance of WUS expression in the developing SAM.
Our genetic data and expression analyses demonstrated that the B-type ARR-HD-ZIP III transcriptional complex spatially activates WUS. Given the observation that B-type ARRs and HD-ZIP III are expressed in a broader domain than WUS, dissecting other factors that induce WUS expression represents another central challenge. Identification of the progenitor of WUS+ cells using cell lineage assays and dissection of the additional cofactor of the B-type ARR-HD-ZIP III transcriptional complex might enable us to localize the cell where WUS is activated within the callus at a more precise resolution.
In conclusion, our results illustrate the molecular mechanism determining “when” and “where” de novo shoot regeneration is going to happen in the callus and suggest a two-step model for de novo activation of WUS.
METHODS
Plant Materials
Arabidopsis thaliana (ecotypes Col-0, Ler, and C24) and Nicotiana benthamiana were grown at 21°C (day)/19°C (night) in long days (16 h light/8 h dark, with a light intensity of 80 μmol/m2/s using Philips TLD 36W/865 and 36W/830 bulbs). phb-7d (C24 ecotype), phb-6 phv-5 (Ler ecotype), rev-6 (Col-0 ecotype), phb-6 phv-5 rev-9 (Ler ecotype), rev-10d (Ler ecotype), Pro35S:FLAG-GR-rREV (Col-0 ecotype), ProTCSn:GFP (Col-0 ecotype), clf-29 swn-21 (Col-0 ecotype), Pro35S:WUS-GR (Ler ecotype), cuc2-3 cuc3-105 (Col-0 ecotype), clv3-7 (Col-0 ecotype), stm-7 (Col-0 ecotype, GABI_100F11), and wus (Col-0 ecotype, CS349353) were described previously (Prigge et al., 2005; Hibara et al., 2006; Carlsbecker et al., 2010; Brandt et al., 2012; He et al., 2012; Yadav et al., 2013; Zürcher et al., 2013; Schuster et al., 2014).
To produce transgenic Arabidopsis plants, the binary constructs were delivered into Agrobacterium tumefaciens strain GV3101 (pMP90) by the freeze-thaw method. Transgenic plants were generated by the floral dip method (Clough and Bent, 1998) and screened with 0.05% glufosinate (Basta) in soil or 40 µg/mL hygromycin or 50 µg/mL kanamycin on half-strength Murashige and Skoog (MS) plates.
Constructs
The oligonucleotide primers for all constructs are given in Supplemental Data Set 1. The map and DNA sequence for each construct is available upon request. For the yeast two-hybrid constructs, the cDNAs of PHB, PHV, REV, CUC2, CUC3, TCP FAMILY TRANSCRIPTION FACTOR4 (TCP4), ENHANCER OF SHOOT REGENERATION1 (ESR1), MONOPTEROS (MP), BRANCHED1 (BRC1), TERMINAL FLOWER2 (TFL2), and LEAF CURLING RESPONSIVENESS (LCR) were amplified and cloned into pGBKT7 (Clontech). The pGADT7 vectors for ARR1 and ARR2 have been described (Zhang et al., 2015).
To generate ProWUS:3xVENUS-N7 and ProWUS:DsRED-N7, 3xVENUS-N7 and DsRED-N7 were introduced into the vector TQ391 containing 5.7-kb upstream and 1.4-kb downstream fragments of WUS. To generate ProCLV3:GFP-ER, a GFP-ER fragment was introduced into the vector containing 1.6-kb upstream and 1.3-kb downstream fragments of CLV3. To generate the fluorescence reporter of REV/PHB, the backbone vector (TQ237 and TQ098) containing the 3.7-kb upstream region of REV or the 3.7-kb upstream region of PHB was constructed. Because REV/PHB is targeted by miR165/6, we generated miRNA-sensitive fluorescence reporters of REV/PHB. The fragments of sGFP-N7mirS and DsRED-N7mirS contained 27 bp of miR165/6 target sequence (CCTGGGATGAAGCCTGGTCCGGATTCG) from REV. sGFP-N7mirS and DsRED-N7mirS were then cloned into TQ237. To generate the ARR1 reporter, the sGFP-N7 fragment was introduced into the vector containing the 3.7-kb upstream fragment of ARR1. For the inducible line of MIR166A, the pri-MIR166A fragment was introduced into pER8 (Zuo et al., 2000). For ProREV:ARR2-SRDX, the cDNA fragment of ARR2 fused with the SRDX motif was cloned into the vector TQ237 behind the REV promoter.
BiLC constructs were generated as described (Gou et al., 2011). The cDNAs of rPHB, rPHV, and rREV were amplified and cloned into the vector JW772 behind LUCc under the control of the 35S promoter. ARR1, ARR2, ARR10, and ARR12 coding regions were cloned into the vector JW771 in front of LUCn under the control of the 35S promoter.
To generate the CoIP constructs, ARR1, ARR2, and rPHB were cloned into the binary constructs with 3xHA (JW819 for 3xHA C-terminal fusion) or 6xMyc (JW1016 for 6xMyc N-terminal fusion) tag. For the ChIP constructs, ARR1 and ARR2 were cloned into the binary constructs with 3xHA (JW819 for 3xHA carboxyl-terminal fusion) or 3xFLAG (TQ354 for 3xFLAG C-terminal fusion) tag. For the protoplast assay, the WUS promoter was cloned in front of LUC in TQ379, which harbors the Pro35S:REN cassette. Pro35S:MIR166a was generated by cloning pri-MIR166A into JW807 behind the 35S promoter.
Regeneration Experiments
Arabidopsis seeds were sterilized with 15% bleach and germinated on half-strength MS plates (2.21 g MS basal medium with vitamin powder, 0.5 g/L methylester sulfonate, 20 g/L sucrose, and 8 g/L agar, pH 5.7) in the dark. Hypocotyls or roots were excised and transferred to CIM (4.4 g MS basal medium with vitamin powder, 0.5 g/L methylester sulfonate, 20 g/L sucrose, 2.2 µM 2,4-D, 0.2 µM kinetin, and 8 g/L agar, pH 5.7) for 7 d. For shoot regeneration, the calli were then transferred to SIM (4.4 g MS basal medium with vitamin powder, 0.5 g/L methylester sulfonate, 20 g/L sucrose, 0.9 µM indole-3-acetic acid, and 8 g/L agar, pH 5.7) with different concentrations of 2-isopentenyladenine (2-IP) and incubated at 22°C under long-day conditions. For root regeneration, the calli were transferred to MS medium. The number of explants and regenerated shoots was scored. The regenerative capacity was calculated as the number of regenerated shoots in a given number of explants. Three independent experiments (biological triplicates) were performed.
Plant Treatment
2-IP, CHX, DEX, and olomoucine were dissolved in water, ethanol, or DMSO, respectively. For cytokinin treatment, wild-type or clf swn explants were treated with water (mock) or 20 μM 2-IP (for Figure 3B) and with ethanol (mock), 20 μM 2-IP, 10 μM CHX, or 20 μM 2-IP + 10 μM CHX (for Figure 3C). For olomoucine treatment, DMSO (mock) or 20 or 50 μM olomoucine was added to SIM. For DEX treatment, 10 μM DEX or ethanol (mock) was used.
Expression Analysis
Total RNA was extracted with Trizol reagent (Invitrogen). One microgram of total RNA was DNase I treated and used for cDNA synthesis with oligo(dT) primer (Fermentas). The average expression levels were calculated from 2−ΔΔCt values. Biological triplicates (three independent experiments) with technical triplicates were performed. The qRT-PCR primers for TUBULIN (TUB) have been described (Wang et al., 2009). The oligonucleotide primers for all of the genes are given in Supplemental Data Set 1.
In Situ Hybridization
RNA in situ hybridization was performed as described (Wang et al., 2009; Lian et al., 2013). cDNA fragments of PHB, REV, ARR1, ARR2, ARR5, WUS, CLV3, STM, and WOX2 were amplified and cloned into T-vector, respectively. In vitro transcription was performed with T3 or T7 RNA polymerase (Roche) in which linearized vectors were added as templates. All primers used for preparing probes are listed in Supplemental Data Set 1.
Microscopy
For RNA in situ imaging, slides were mounted with water and observed under an Olympus BX63 microscope equipped with a DP73 digital camera and differential interference contrast modules. For confocal imaging, the explants were manually sectioned into thin slices. Prepared specimens were observed and scanned under an Olympus FV1000 confocal microscope. Proper filter sets and lasers were selected for fluorescence signal scanning. For GFP, excitation light wavelength was 488 nm and emission was 510 to 550 nm; for VENUS excitation, 515 nm, emission 540 to 580 nm; for dsRED excitation, 561 nm, emission 569 to 700 nm; for propidium iodide excitation, 514 nm, emission 631 to 690 nm.
For live cell imaging, after culturing on CIM for 7 days and propidium iodide staining, the explants were transferred into a cell imaging dish (Eppendorf, 0,030,740.017) containing SIM. After marking positions, the explants were scanned with a Leica DMi8 (for Supplemental Movie 1) and Zeiss 880 inverted microscope (for Supplemental Movie 2 and 3) every 1-3 h. The live cell imaging system was operated at 22°C in long days.
Yeast Two-Hybrid Assay
Plasmids were transformed into yeast strain AH109 (Clontech) by the LiCl-PEG method. The transformants were selected on SD-Leu-Trp plates. The interactions were tested on SD-Leu-Trp-His (SD-LWH) or SD-Ade-Leu-Trp-His (SD-ALWH) plates with 3-amino-1,2,4,-triazole. At least 10 individual clones were analyzed.
CoIP and Immunoblot Analyses
Agrobacteria-infiltrated N. benthamiana leaves were used for CoIP analyses. The soluble proteins were extracted in extraction buffer (50 mM HEPES, 10 mM EDTA, 50 mM NaCl, 10% glycerol, 1% PVPP, 2 mM DTT, 1 mM PMSF, 10 µM MG-132, and 1× protease inhibitor cocktail, pH 7.5). Immunoprecipitation was performed with anti-Myc beads (Sigma-Aldrich; E6654) for 1 h at 4°C. The beads were washed three times with wash buffer (50 mM HEPES, 150 mM NaCl, 10 mM EDTA, 0.1% Triton X-100, 10% glycerol, and 1 mM PMSF, pH 7.5). 3xHA- or 6xMyc-fusion proteins were detected by immunoblot with anti-HA-peroxidase (Roche; 12013819001; 1;1000) or anti-Myc (Millipore; 05-724; 1:1000) antibody.
Pull-Down Assay
Full-length PHB was cloned into pGEX-4T-1 (Amersham Biosciences) to express GST-PHB fusion proteins. Full-length ARR1 and ARR2 were amplified and cloned into pRSF-Duet (Novagen) to express 6xHis-ARR1/ARR2 fusion proteins. The constructs were then transformed into Escherichia coli (strain Rosetta), and expression of the fusion protein was induced by 0.1 mM isopropyl-β-d-thiogalactoside. The induced cells were lysed by sonication in purification buffer (20 mM Tris-HCl and 100 mM NaCl, pH 8.0) containing 2 mM PMSF. One milliliter of lysate and 600 ng purified ARR1 or ARR2 proteins were incubated with 30 μL glutathione Sepharose 4B resins (Amersham/GE Biosciences) in 500 mL binding buffer (20 mM Tris-HCl, 50 mM NaCl, and 1 mM PMSF, pH 7.4) at 4°C for 3 h. The beads were then washed six times with 1 mL wash buffer (20 mM Tris-HCl, 150 mM NaCl, and 1 mM PMSF, pH 7.4). The washed pellet was resuspended in 2× SDS sample buffer and boiled for 5 min before loading onto SDS-PAGE gels. 6xHis- or GST-fusion proteins were detected by immunoblot with anti-His or anti-GST antibody. The membrane was exposed to a CCD-equipped camera (Tanon 5200S).
EMSA Assay
To construct plasmids for the expression of recombinant ARR2 protein in E. coli, the full-length coding sequence of ARR2 was amplified and cloned into pRSF-Duet. The ARR2-6xHis proteins were purified using a Ni-NTA purification system (Qiagen). Double-stranded oligonucleotide probes were synthesized and labeled with biotin at the 5′ end. EMSA was performed using a LightShift Chemiluminescent EMSA kit (Thermo Scientific). Briefly, biotin-labeled probes were incubated in 1× binding buffer, 2.5% glycerol, 5 mM MgCl2, and 50 ng/μL poly(dI dC) with or without purified ARR2 proteins at room temperature for 20 min. For unlabeled probe competition, unlabeled probes were added to the reactions. For the supershift assay, ARR2 proteins were mixed with different concentrations of anti-His antibody (Abmart; M30111). The probe sequences are listed in Supplemental Data Set 1.
BiLC Analysis and N. benthamiana Transient Assay
For the N. benthamiana transient assay, Agrobacterium was resuspended in infiltration buffer (10 mM methylester sulfonate, 10 mM MgCl2, and 150 µM acetosyringone, pH 5.7) at OD600 = 0.8. Pro35S:P19-HA (Papp et al., 2003) was coinfiltrated to inhibit gene silencing. The plants were incubated at 22°C for 3 d. BiLC assay was performed as described (Gou et al., 2011). Agrobacterium was resuspended in infiltration buffer at OD600 = 0.8.
For the dual-luciferase reporter assay, Arabidopsis protoplasts were prepared using 3-week-old Arabidopsis (Col-0) leaves according to a published protocol (Yoo et al., 2007). After transfection, the protoplasts were cultured on 22°C for ∼13 h. The protoplasts were then lysed with passive lysis buffer (Promega; E1910). LUC and REN activities were quantified and measured with a luminometer (Promega 20/20). LUC activity was calculated by normalizing to that of REN. Three independent experiments (biological triplicates) were performed.
ChIP Analysis
Briefly, wild-type, Pro35S:ARR1-3xHA, Pro35S:ARR2-3xFLAG, or the DEX-treated Pro35S:FLAG-GR-rREV explants on SIM were fixed according to a published protocol (Yu et al., 2013). The chromatin extract was immunoprecipitated with anti-HA beads (Sigma-Aldrich; E6779), anti-FLAG beads (Sigma-Aldrich; F2426), or anti-H3K27me3 (Millipore; 07-449). ChIP DNA was reverse cross-linked and purified with a PCR purification kit (Qiagen). One microliter of DNA was used for qRT-PCR analyses. The relative enrichment of ARR1-3xHA, ARR2-3xFLAG, and FLAG-GR-rREV on the WUS promoter was calculated by normalizing the amount of each immunoprecipitated fragment to input DNA and then by normalizing the value for transgenic plants against the value for the wild type as a negative control. For H3K27me3 enrichment, the EIF4A1 locus was used as a negative control. The enrichment level in wild-type seedlings was set to 1. For all of the ChIP experiments, three biological experiments were performed.
Phylogenetic Shadowing
Phylogenetic shadowing was performed using publicly available genomic sequences. The WUS promoters were aligned by mVISTA with default settings (Mayor et al., 2000). The conserved regions of the WUS promoters were exported to MacVector 10 and aligned using ClustalW (Larkin et al., 2007) and BoxShade (http://www.ch.embnet.org/software/BOX_form.html). Accession numbers for the WUS sequences from Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html) are as follows: Arabidopsis lyrata 931663; Brassica rapa 024485; Capsella rubella 10016192; Thellungiella halophila 10023091.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: ARR1 (At3g16857), ARR2 (At4g16110), ARR10 (At4g31920), ARR12 (At2g25180), ARR5 (At3g48100), STM (At1g62360), CUC2 (At5g53950), CUC3 (At1g76420), WUS (At2g17950), CLV3 (At2g27250), WOX2 (AT5G59340), CLF (At2g23380), SWN (At4g02020), PHB (At2g34710), PHV (At1g30490), REV (At5g60690), EIF4A1 (At3g13920), and TUB (At5g62690).
Supplemental Data
Supplemental Figure 1. Shoot Regeneration in clv3, stm, and cuc2 cuc3.
Supplemental Figure 2. Comparison of the Expression Patterns of WUS Reporters.
Supplemental Figure 3. Dynamic Expression Patterns of WUS and CLV3 during Shoot Regeneration.
Supplemental Figure 4. Expression Patterns of STM and WOX2.
Supplemental Figure 5. Regeneration Assay in the arr Mutant.
Supplemental Figure 6. EMSA Assays.
Supplemental Figure 7. The Progressive Decrease in H3K27me3 Marks at the WUS Locus Is Delayed by OLO Treatment.
Supplemental Figure 8. The Induction of WUS by Cytokinin Is Delayed by OLO Treatment.
Supplemental Figure 9. The Induction of ARR5 by Cytokinin Is Not Delayed by OLO Treatment.
Supplemental Figure 10. The Induction of Shoot Regeneration by Cytokinin Is Delayed by OLO Treatment.
Supplemental Figure 11. Dynamic Expression Patterns of REV, TCS, and ARR1 during Shoot Regeneration.
Supplemental Figure 12. ARR1, ARR2, ARR10, and ARR12 Bind to HD-ZIP III Proteins.
Supplemental Figure 13. Shoot Regeneration Assay of HD-ZIP III Mutants.
Supplemental Figure 14. Expression of HD-ZIP III Transcription Factors and B-Type ARRs.
Supplemental Figure 15. Expression of ARR1 and ARR2 in Wild-Type Explants.
Supplemental Figure 16. Spatial Activation of WUS by REV and PHB.
Supplemental Figure 17. Local Induction of HD-ZIP III Promotes Shoot Regeneration on SIM.
Supplemental Figure 18. Genetic Interaction between ARR2 and REV during Shoot Development.
Supplemental Table 1. Yeast Two-Hybrid Assay Using ARR2 as Bait.
Supplemental Data Set 1. Oligonucleotide Primer Sequences.
Supplemental Movie 1. Live Imaging of ProWUS:3xVENUS-N7 Explants.
Supplemental Movie 2. Live Imaging of ProRIBO:sGFP-N7 Explants without OLO Treatment.
Supplemental Movie 3. Live Imaging of ProRIBO:sGFP-N7 Explants Treated with OLO.
Supplementary Material
Acknowledgments
We thank the Arabidopsis Biological Resource Center, B. Müller, S. Wenkel, J. Fletcher, Y. Helariutta, V. Reddy, and J. Bowman for seeds; Yan Xiong, Xiao-Su Gao, Yun-Xiao He, Kun Zhang, and Hui Han for skillful technical assistance; Xian Sheng Zhang for exchanging unpublished results; and Hongtao Liu and members of the J.-W. Wang lab for discussion and comments on the manuscript. This work was supported by grants from National Key Research and Development Program (2016YFA0500800), the National Natural Science Foundation of China (31430013, 31222029, 912173023, and 31525004), the Chinese Academy of Sciences (QYZDB-SSW-SMC002), the State Key Basic Research Program of China (2013CB127000), the National Postdoctoral Program for Innovative Talents (BX201600178), the Shanghai Outstanding Academic Leader Program (15XD1504100), and the NKLPMG Key Research Program.
AUTHOR CONTRIBUTIONS
T.-Q.Z. and J.-W.W. designed the research. T.-Q.Z., H.L., and C.-M.Z. performed research. Y.J. contributed materials. T.-Q.Z., L.X., and J.-W. W. analyzed the data. J.-W.W. wrote the article.
NOTE ADDED IN PROOF
While this article was under revision, Laux’s laboratory reported that WOX2 promotes the establishment of the SAM by promoting the expression of HD-ZIP III transcription factors and ensuring the appropriate ratio of the phytohormones cytokinin and auxin during embryogenesis (Zhang et al., Developmental Cell, 2017).
Zhang, Z., Tucker, E., Hermann, M., and Laux, T. (2017). A molecular framework for the embryonic initiation of shoot meristem stem cells. Dev. Cell 40: 264–277.
Glossary
- CIM
callus-inducing medium
- SIM
shoot-inducing medium
- SAM
shoot apical meristem
- OC
organizing center
- DEX
dexamethasone
- CHX
cycloheximide
- CoIP
coimmunoprecipitation
- BiLC
bimolecular luminescence complementation
- ChIP
chromatin immunoprecipitation
- MS
Murashige and Skoog
- 2-IP
2-isopentenyladenine
- EMSA
electrophoretic mobility shift assay
References
- Adibi M., Yoshida S., Weijers D., Fleck C. (2016). Centering the organizing center in the Arabidopsis thaliana shoot apical meristem by a combination of cytokinin signaling and self-organization. PLoS One 11: e0147830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aichinger E., Kornet N., Friedrich T., Laux T. (2012). Plant stem cell niches. Annu. Rev. Plant Biol. 63: 615–636. [DOI] [PubMed] [Google Scholar]
- Atta R., Laurens L., Boucheron-Dubuisson E., Guivarc’h A., Carnero E., Giraudat-Pautot V., Rech P., Chriqui D. (2009). Pluripotency of Arabidopsis xylem pericycle underlies shoot regeneration from root and hypocotyl explants grown in vitro. Plant J. 57: 626–644. [DOI] [PubMed] [Google Scholar]
- Bailey C.D., Koch M.A., Mayer M., Mummenhoff K., O’Kane S.L. Jr., Warwick S.I., Windham M.D., Al-Shehbaz I.A. (2006). Toward a global phylogeny of the Brassicaceae. Mol. Biol. Evol. 23: 2142–2160. [DOI] [PubMed] [Google Scholar]
- Barton M.K. (2010). Twenty years on: the inner workings of the shoot apical meristem, a developmental dynamo. Dev. Biol. 341: 95–113. [DOI] [PubMed] [Google Scholar]
- Bäurle I., Laux T. (2005). Regulation of WUSCHEL transcription in the stem cell niche of the Arabidopsis shoot meristem. Plant Cell 17: 2271–2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum K.D., Sánchez Alvarado A. (2008). Slicing across kingdoms: regeneration in plants and animals. Cell 132: 697–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt R., et al. (2012). Genome-wide binding-site analysis of REVOLUTA reveals a link between leaf patterning and light-mediated growth responses. Plant J. 72: 31–42. [DOI] [PubMed] [Google Scholar]
- Breuninger H., Rikirsch E., Hermann M., Ueda M., Laux T. (2008). Differential expression of WOX genes mediates apical-basal axis formation in the Arabidopsis embryo. Dev. Cell 14: 867–876. [DOI] [PubMed] [Google Scholar]
- Buechel S., Leibfried A., To J.P., Zhao Z., Andersen S.U., Kieber J.J., Lohmann J.U. (2010). Role of A-type ARABIDOPSIS RESPONSE REGULATORS in meristem maintenance and regeneration. Eur. J. Cell Biol. 89: 279–284. [DOI] [PubMed] [Google Scholar]
- Bustamante M., Matus J.T., Riechmann J.L. (2016). Genome-wide analyses for dissecting gene regulatory networks in the shoot apical meristem. J. Exp. Bot. 67: 1639–1648. [DOI] [PubMed] [Google Scholar]
- Canver M.C., et al. (2015). BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature 527: 192–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsbecker A., et al. (2010). Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 465: 316–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanvivattana Y., Bishopp A., Schubert D., Stock C., Moon Y.H., Sung Z.R., Goodrich J. (2004). Interaction of Polycomb-group proteins controlling flowering in Arabidopsis. Development 131: 5263–5276. [DOI] [PubMed] [Google Scholar]
- Chatfield S.P., Capron R., Severino A., Penttila P.A., Alfred S., Nahal H., Provart N.J. (2013). Incipient stem cell niche conversion in tissue culture: using a systems approach to probe early events in WUSCHEL-dependent conversion of lateral root primordia into shoot meristems. Plant J. 73: 798–813. [DOI] [PubMed] [Google Scholar]
- Che P., Lall S., Howell S.H. (2007). Developmental steps in acquiring competence for shoot development in Arabidopsis tissue culture. Planta 226: 1183–1194. [DOI] [PubMed] [Google Scholar]
- Che P., Gingerich D.J., Lall S., Howell S.H. (2002). Global and hormone-induced gene expression changes during shoot development in Arabidopsis. Plant Cell 14: 2771–2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z.J., et al. (2013). Pattern of auxin and cytokinin responses for shoot meristem induction results from the regulation of cytokinin biosynthesis by AUXIN RESPONSE FACTOR3. Plant Physiol. 161: 240–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chickarmane V.S., Gordon S.P., Tarr P.T., Heisler M.G., Meyerowitz E.M. (2012). Cytokinin signaling as a positional cue for patterning the apical-basal axis of the growing Arabidopsis shoot meristem. Proc. Natl. Acad. Sci. USA 109: 4002–4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Crevillén P., Yang H., Cui X., Greeff C., Trick M., Qiu Q., Cao X., Dean C. (2014). Epigenetic reprogramming that prevents transgenerational inheritance of the vernalized state. Nature 515: 587–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X., et al. (2016). REF6 recognizes a specific DNA sequence to demethylate H3K27me3 and regulate organ boundary formation in Arabidopsis. Nat. Genet. 48: 694–699. [DOI] [PubMed] [Google Scholar]
- Daum G., Medzihradszky A., Suzaki T., Lohmann J.U. (2014). A mechanistic framework for noncell autonomous stem cell induction in Arabidopsis. Proc. Natl. Acad. Sci. USA 111: 14619–14624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duclercq J., Sangwan-Norreel B., Catterou M., Sangwan R.S. (2011). De novo shoot organogenesis: from art to science. Trends Plant Sci. 16: 597–606. [DOI] [PubMed] [Google Scholar]
- Efroni I., Mello A., Nawy T., Ip P.L., Rahni R., DelRose N., Powers A., Satija R., Birnbaum K.D. (2016). Root regeneration triggers an embryo-like sequence guided by hormonal interactions. Cell 165: 1721–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery J.F., Floyd S.K., Alvarez J., Eshed Y., Hawker N.P., Izhaki A., Baum S.F., Bowman J.L. (2003). Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 13: 1768–1774. [DOI] [PubMed] [Google Scholar]
- Gaillochet C., Lohmann J.U. (2015). The never-ending story: from pluripotency to plant developmental plasticity. Development 142: 2237–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillochet C., Daum G., Lohmann J.U. (2015). O cell, where art thou? The mechanisms of shoot meristem patterning. Curr. Opin. Plant Biol. 23: 91–97. [DOI] [PubMed] [Google Scholar]
- Gallois J.L., Woodward C., Reddy G.V., Sablowski R. (2002). Combined SHOOT MERISTEMLESS and WUSCHEL trigger ectopic organogenesis in Arabidopsis. Development 129: 3207–3217. [DOI] [PubMed] [Google Scholar]
- Goodrich J., Puangsomlee P., Martin M., Long D., Meyerowitz E.M., Coupland G. (1997). A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 386: 44–51. [DOI] [PubMed] [Google Scholar]
- Gordon S.P., Chickarmane V.S., Ohno C., Meyerowitz E.M. (2009). Multiple feedback loops through cytokinin signaling control stem cell number within the Arabidopsis shoot meristem. Proc. Natl. Acad. Sci. USA 106: 16529–16534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon S.P., Heisler M.G., Reddy G.V., Ohno C., Das P., Meyerowitz E.M. (2007). Pattern formation during de novo assembly of the Arabidopsis shoot meristem. Development 134: 3539–3548. [DOI] [PubMed] [Google Scholar]
- Gou J.Y., Felippes F.F., Liu C.J., Weigel D., Wang J.W. (2011). Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell 23: 1512–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha C.M., Jun J.H., Fletcher J.C. (2010). Shoot apical meristem form and function. Curr. Top. Dev. Biol. 91: 103–140. [DOI] [PubMed] [Google Scholar]
- He C., Chen X., Huang H., Xu L. (2012). Reprogramming of H3K27me3 is critical for acquisition of pluripotency from cultured Arabidopsis tissues. PLoS Genet. 8: e1002911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidstra R., Sabatini S. (2014). Plant and animal stem cells: similar yet different. Nat. Rev. Mol. Cell Biol. 15: 301–312. [DOI] [PubMed] [Google Scholar]
- Heyl A., Ramireddy E., Brenner W.G., Riefler M., Allemeersch J., Schmülling T. (2008). The transcriptional repressor ARR1-SRDX suppresses pleiotropic cytokinin activities in Arabidopsis. Plant Physiol. 147: 1380–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibara K., Karim M.R., Takada S., Taoka K., Furutani M., Aida M., Tasaka M. (2006). Arabidopsis CUP-SHAPED COTYLEDON3 regulates postembryonic shoot meristem and organ boundary formation. Plant Cell 18: 2946–2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt A.L., van Haperen J.M., Groot E.P., Laux T. (2014). Signaling in shoot and flower meristems of Arabidopsis thaliana. Curr. Opin. Plant Biol. 17: 96–102. [DOI] [PubMed] [Google Scholar]
- Ikeuchi M., Ogawa Y., Iwase A., Sugimoto K. (2016). Plant regeneration: cellular origins and molecular mechanisms. Development 143: 1442–1451. [DOI] [PubMed] [Google Scholar]
- Ishida K., Yamashino T., Yokoyama A., Mizuno T. (2008). Three type-B response regulators, ARR1, ARR10 and ARR12, play essential but redundant roles in cytokinin signal transduction throughout the life cycle of Arabidopsis thaliana. Plant Cell Physiol. 49: 47–57. [DOI] [PubMed] [Google Scholar]
- Kareem A., Durgaprasad K., Sugimoto K., Du Y., Pulianmackal A.J., Trivedi Z.B., Abhayadev P.V., Pinon V., Meyerowitz E.M., Scheres B., Prasad K. (2015). PLETHORA genes control regeneration by a two-step mechanism. Curr. Biol. 25: 1017–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurakawa T., Ueda N., Maekawa M., Kobayashi K., Kojima M., Nagato Y., Sakakibara H., Kyozuka J. (2007). Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445: 652–655. [DOI] [PubMed] [Google Scholar]
- Larkin M.A., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- Lee C., Clark S.E. (2015). A WUSCHEL-independent stem cell specification pathway is repressed by PHB, PHV and CNA in Arabidopsis. PLoS One 10: e0126006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibfried A., To J.P., Busch W., Stehling S., Kehle A., Demar M., Kieber J.J., Lohmann J.U. (2005). WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 438: 1172–1175. [DOI] [PubMed] [Google Scholar]
- Li C., et al. (2016). Concerted genomic targeting of H3K27 demethylase REF6 and chromatin-remodeling ATPase BRM in Arabidopsis. Nat. Genet. 48: 687–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Liu H., Cheng Z.J., Su Y.H., Han H.N., Zhang Y., Zhang X.S. (2011). DNA methylation and histone modifications regulate de novo shoot regeneration in Arabidopsis by modulating WUSCHEL expression and auxin signaling. PLoS Genet. 7: e1002243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian H., Li X., Liu Z., He Y. (2013). HYL1 is required for establishment of stamen architecture with four microsporangia in Arabidopsis. J. Exp. Bot. 64: 3397–3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Kim Y.J., Müller R., Yumul R.E., Liu C., Pan Y., Cao X., Goodrich J., Chen X. (2011). AGAMOUS terminates floral stem cell maintenance in Arabidopsis by directly repressing WUSCHEL through recruitment of Polycomb Group proteins. Plant Cell 23: 3654–3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J.A., Moan E.I., Medford J.I., Barton M.K. (1996). A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379: 66–69. [DOI] [PubMed] [Google Scholar]
- Lu F., Cui X., Zhang S., Jenuwein T., Cao X. (2011). Arabidopsis REF6 is a histone H3 lysine 27 demethylase. Nat. Genet. 43: 715–719. [DOI] [PubMed] [Google Scholar]
- Müller B., Sheen J. (2008). Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature 453: 1094–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason M.G., Mathews D.E., Argyros D.A., Maxwell B.B., Kieber J.J., Alonso J.M., Ecker J.R., Schaller G.E. (2005). Multiple type-B response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell 17: 3007–3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor C., Brudno M., Schwartz J.R., Poliakov A., Rubin E.M., Frazer K.A., Pachter L.S., Dubchak I. (2000). VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 16: 1046–1047. [DOI] [PubMed] [Google Scholar]
- Otsuga D., DeGuzman B., Prigge M.J., Drews G.N., Clark S.E. (2001). REVOLUTA regulates meristem initiation at lateral positions. Plant J. 25: 223–236. [DOI] [PubMed] [Google Scholar]
- Papp I., Mette M.F., Aufsatz W., Daxinger L., Schauer S.E., Ray A., van der Winden J., Matzke M., Matzke A.J. (2003). Evidence for nuclear processing of plant micro RNA and short interfering RNA precursors. Plant Physiol. 132: 1382–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planchais S., Glab N., Inzé D., Bergounioux C. (2000). Chemical inhibitors: a tool for plant cell cycle studies. FEBS Lett. 476: 78–83. [DOI] [PubMed] [Google Scholar]
- Prigge M.J., Otsuga D., Alonso J.M., Ecker J.R., Drews G.N., Clark S.E. (2005). Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell 17: 61–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawarkar R., Paro R. (2010). Interpretation of developmental signaling at chromatin: the Polycomb perspective. Dev. Cell 19: 651–661. [DOI] [PubMed] [Google Scholar]
- Schoof H., Lenhard M., Haecker A., Mayer K.F., Jürgens G., Laux T. (2000). The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100: 635–644. [DOI] [PubMed] [Google Scholar]
- Schuster C., Gaillochet C., Medzihradszky A., Busch W., Daum G., Krebs M., Kehle A., Lohmann J.U. (2014). A regulatory framework for shoot stem cell control integrating metabolic, transcriptional, and phytohormone signals. Dev. Cell 28: 438–449. [DOI] [PubMed] [Google Scholar]
- Shi B., et al. (2016). Two-step regulation of a meristematic cell population acting in shoot branching in Arabidopsis. PLoS Genet. 12: e1006168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon J.A., Kingston R.E. (2013). Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol. Cell 49: 808–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoog F., Miller C.O. (1957). Chemical regulation of growth and organ formation in plant tissues cultured in vitro. Symp. Soc. Exp. Biol. 11: 118–130. [PubMed] [Google Scholar]
- Smith Z.R., Long J.A. (2010). Control of Arabidopsis apical-basal embryo polarity by antagonistic transcription factors. Nature 464: 423–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y.H., Zhang X.S. (2014). The hormonal control of regeneration in plants. Curr. Top. Dev. Biol. 108: 35–69. [DOI] [PubMed] [Google Scholar]
- Sugimoto K., Gordon S.P., Meyerowitz E.M. (2011). Regeneration in plants and animals: dedifferentiation, transdifferentiation, or just differentiation? Trends Cell Biol. 21: 212–218. [DOI] [PubMed] [Google Scholar]
- Sun B., Looi L.S., Guo S., He Z., Gan E.S., Huang J., Xu Y., Wee W.Y., Ito T. (2014). Timing mechanism dependent on cell division is invoked by Polycomb eviction in plant stem cells. Science 343: 1248559. [DOI] [PubMed] [Google Scholar]
- ten Hove C.A., Lu K.J., Weijers D. (2015). Building a plant: cell fate specification in the early Arabidopsis embryo. Development 142: 420–430. [DOI] [PubMed] [Google Scholar]
- To J.P., Kieber J.J. (2008). Cytokinin signaling: two-components and more. Trends Plant Sci. 13: 85–92. [DOI] [PubMed] [Google Scholar]
- Wang J.W., Czech B., Weigel D. (2009). miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138: 738–749. [DOI] [PubMed] [Google Scholar]
- Wang Q., Kohlen W., Rossmann S., Vernoux T., Theres K. (2014a). Auxin depletion from the leaf axil conditions competence for axillary meristem formation in Arabidopsis and tomato. Plant Cell 26: 2068–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Wang J., Shi B., Yu T., Qi J., Meyerowitz E.M., Jiao Y. (2014b). The stem cell niche in leaf axils is established by auxin and cytokinin in Arabidopsis. Plant Cell 26: 2055–2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J., Wagner D. (2015). Polycomb repression in the regulation of growth and development in Arabidopsis. Curr. Opin. Plant Biol. 23: 15–24. [DOI] [PubMed] [Google Scholar]
- Yadav R.K., Perales M., Gruel J., Girke T., Jönsson H., Reddy G.V. (2011). WUSCHEL protein movement mediates stem cell homeostasis in the Arabidopsis shoot apex. Genes Dev. 25: 2025–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav R.K., Perales M., Gruel J., Ohno C., Heisler M., Girke T., Jönsson H., Reddy G.V. (2013). Plant stem cell maintenance involves direct transcriptional repression of differentiation program. Mol. Syst. Biol. 9: 654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Yu S., Cao L., Zhou C.M., Zhang T.Q., Lian H., Sun Y., Wu J., Huang J., Wang G., Wang J.W. (2013). Sugar is an endogenous cue for juvenile-to-adult phase transition in plants. eLife 2: e00269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T.Q., Lian H., Tang H., Dolezal K., Zhou C.M., Yu S., Chen J.H., Chen Q., Liu H., Ljung K., Wang J.W. (2015). An intrinsic microRNA timer regulates progressive decline in shoot regenerative capacity in plants. Plant Cell 27: 349–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J., Niu Q.W., Chua N.H. (2000). Technical advance: An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 24: 265–273. [DOI] [PubMed] [Google Scholar]
- Zürcher E., Tavor-Deslex D., Lituiev D., Enkerli K., Tarr P.T., Müller B. (2013). A robust and sensitive synthetic sensor to monitor the transcriptional output of the cytokinin signaling network in planta. Plant Physiol. 161: 1066–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.