Ethylene inhibits the production of jasmonic acid to facilitate the emergence of rice seedlings from the soil.
Abstract
Elongation of the mesocotyl and coleoptile facilitates the emergence of rice (Oryza sativa) seedlings from soil and is affected by various genetic and environment factors. The regulatory mechanism underlying this process remains largely unclear. Here, we examined the regulation of mesocotyl and coleoptile growth by characterizing a gaoyao1 (gy1) mutant that exhibits a longer mesocotyl and longer coleoptile than its original variety of rice. GY1 was identified through map-based cloning and encodes a PLA1-type phospholipase that localizes in chloroplasts. GY1 functions at the initial step of jasmonic acid (JA) biosynthesis to repress mesocotyl and coleoptile elongation in etiolated rice seedlings. Ethylene inhibits the expression of GY1 and other genes in the JA biosynthesis pathway to reduce JA levels and enhance mesocotyl and coleoptile growth by promoting cell elongation. Genetically, GY1 acts downstream of the OsEIN2-mediated ethylene signaling pathway to regulate mesocotyl/coleoptile growth. Through analysis of the resequencing data from 3000 rice accessions, we identified a single natural variation of the GY1 gene, GY1376T, which contributes to mesocotyl elongation in rice varieties. Our study reveals novel insights into the regulatory mechanism of mesocotyl/coleoptile elongation and should have practical applications in rice breeding programs.
INTRODUCTION
Flowering plants begin their life cycle as a seed beneath the soil surface. With adequate water and proper temperature, the seeds germinate and seedlings develop. The seedlings withstand complex environments to emerge from the soil and initiate their growth in light. For rice (Oryza sativa), the coleoptile is an essential structure of the seedlings that protects the plumule as it moves through the soil layers. Below the coleoptile, a short stem, also called the mesocotyl, develops in the dark, which pushes the base of the coleoptile toward the soil surface. The elongation of the mesocotyl and coleoptile is regulated by various genetic and environmental factors, which stimulate the emergence of rice seedlings (Takahashi, 1978; Helms et al., 1989; Dilday et al., 1990).
Plant hormones and other related factors affect the growth of the mesocotyl and coleoptile. For instance, abscisic acid promotes mesocotyl growth through increasing the cell division activity of the meristem (Watanabe et al., 2001). Strigolactones inhibit mesocotyl growth through inhibiting cell division, and cytokinin antagonizes strigolactones to regulate mesocotyl elongation (Hu et al., 2010, 2014). The photoreceptor mutant osphyA-1 exhibits longer mesocotyls and coleoptiles under continuous far-red light (Takano et al., 2001). Furthermore, light inhibits mesocotyl and coleoptile growth by regulating jasmonic acid (JA) biosynthesis (Svyatyna and Riemann, 2012). Mutants of OsAOS1/CPM1, OsAOC/CPM2, and OsJAR1/OsGH3.5, which are involved in JA biosynthesis (Svyatyna and Riemann, 2012), showed a longer coleoptile phenotype under red light (Biswas et al., 2003; Riemann et al., 2003, 2008; Svyatyna and Riemann, 2012). Expansins are cell wall-loosening proteins that induce wall stress relaxation and volumetric extension of plant cells (Cho and Cosgrove, 2000; Li et al., 2003). Expansins are encoded by substantial gene families and have classically been divided into two subfamilies, α- and β-expansins (Li et al., 2003). Overexpression of AtEXP4 in transgenic Arabidopsis thaliana plants led to slightly longer petioles, larger leaf blades, and larger cells than controls (Cho and Cosgrove, 2000). Overexpressing the expansin gene OsEXP4 promoted elongation of the mesocotyl and coleoptile in rice (Choi et al., 2003).
Recent studies of seedling emergence in Arabidopsis suggest that ethylene regulates etiolated growth and the seedlings’ response to soil through interacting with photomorphogenesis factors (Zhong et al., 2014; Shi et al., 2016a, 2016b). Ethylene is an important gaseous plant hormone that regulates various processes in plant growth and development (Bleecker and Kende, 2000; Guo and Ecker, 2004; Rzewuski and Sauter, 2008; Ma et al., 2010). In Arabidopsis, the perception of ethylene is based on the binding of ethylene to its five membrane receptors (Hua and Meyerowitz, 1998). After perception, a negative regulator, CONSTITUTIVE TRIPLE RESPONSE1, is repressed (Kieber et al., 1993), and the central signal transducer ETHYLENE-INSENSITIVE2 (EIN2) is cleaved at the C-terminal end. This C-terminal end is further translocated into the nucleus for activation of transcription factors EIN3/EIN3-Like 1 (EIN3/EIL1) (Chao et al., 1997; Alonso et al., 1999; Ju et al., 2012; Qiao et al., 2012; Wen et al., 2012). EIN2 also enters the P-body, where it represses the translation of EIN3 Binding F-Box 1/2, which in turn stabilizes EIN3/EIL1 and thereby activates diverse ethylene responses (Li et al., 2015). Our previous studies demonstrated that MaoHuZi7 (MHZ7/OsEIN2), MHZ6/OsEIL1, and OsEIL2 regulate root and coleoptile growth in rice etiolated seedlings (Ma et al., 2013; Yang et al., 2015b).
Ethylene and JA regulate plant growth, development, and defense responses, both synergistically and antagonistically (Song et al., 2014a; Liu et al., 2015; Yang et al., 2015a). The induction of PLANT DEFENSIN 1.2 and ETHYLENE RESPONSE FACTOR1 relies on both ethylene and JA (Penninckx et al., 1998; Lorenzo et al., 2003). Ethylene and JA both promote root hair growth (Zhu et al., 2006). Furthermore, JA promotes the degradation of Jasmonate-ZIM domain protein (JAZ), which interacts with and represses EIN3/EIL1, suggesting a synergistic regulation between ethylene and JA (Zhu et al., 2011). Recently, EIN3 and MYC2 have been found to interact with each other at different levels to regulate the antagonism between ethylene and JA in Arabidopsis (Song et al., 2014a, 2014b; Zhang et al., 2014).
The biosynthesis of JA initiates with phospholipases in chloroplasts, which release α-linolenic acid from the chloroplast membrane (Wang et al., 2000; Hyun et al., 2008; Ellinger et al., 2010; Liu et al., 2015). The lipoxygenases (LOXs) then oxidate α-linolenic acid to produce 13-hydroperoxy-octadecatrienoic acid (13-HPOT) and 9-HPOT (Feussner and Wasternack, 2002; Bannenberg et al., 2009; Chauvin et al., 2013). After allene oxide synthase converts hydroperoxides to an unstable 12,13 (S)-epoxylinolenic acid, the allene oxide cyclase catalyzes the conversion of the product to (9S,13S)-12-oxo-phytodienoic acid (Ziegler et al., 2000), which is later transferred into peroxisomes and reduced to OPC-8:0 by 12-oxophytodienoic acid reductase (OPR) (Schaller et al., 2000). The OPC-8:0 is esterified with CoA by OPC-8:CoA ligase 1 (OPCL1) and then subjected to three cycles of β-oxidation, which involves the action of acyl-CoA oxidase, the multifunctional protein, and 3-ketoacyl-CoA thiolase to generate JA (Hayashi et al., 1999; Eastmond and Graham, 2000; Eastmond et al., 2000; Schaller and Stintzi, 2009). The active form of JA, jasmonoyl-isoleucine, is synthesized by JAR1 through activating the amide-linked conjugation of the carboxyl group to Ile (Staswick et al., 2002).
Although interactions between ethylene and JA affect various responses, their roles in mesocotyl/coleoptile elongation and the emergence of rice seedlings are not known. In this study, we identified and characterized the rice mutant gaoyao1 (gy1), which exhibits long mesocotyls and long coleoptiles. Through map-based cloning, we found that GY1 encodes a phospholipase that functions at the initial step of the JA biosynthesis pathway. JA inhibits mesocotyl/coleoptile elongation, and ethylene suppresses JA biosynthesis to promote mesocotyl/coleoptile growth. We further identified a single elite allele of GY1 from 3000 rice accessions and found that this allele is associated with long mesocotyls of rice seedlings. Our study reveals a mechanism by which the mesocotyl and coleoptile elongate when rice seedlings emerge from the soil and should facilitate breeding for new rice cultivars that are adapted to growing in dry soil.
RESULTS
The gy1 Mutant Has Longer Mesocotyls and Coleoptiles
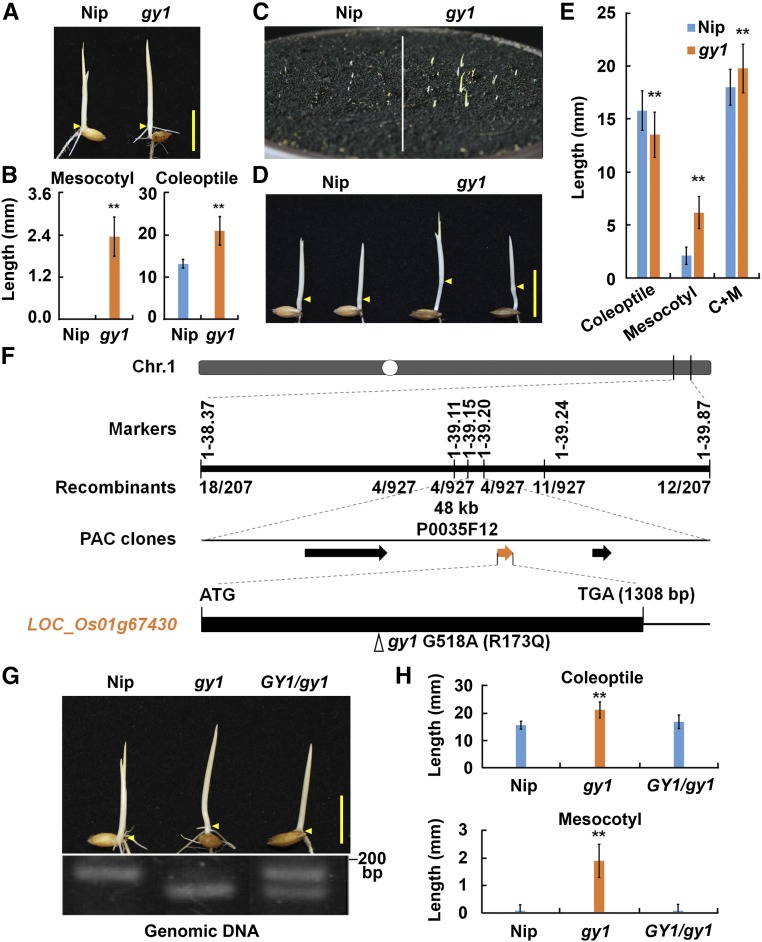
In screening for ethylene response mutants in rice (O. sativa cv Nipponbare), a mutant exhibiting longer mesocotyl and longer coleoptile than the wild-type Nipponbare (Nip) in etiolated seedlings was identified and named gaoyao1 (a Chinese name meaning high waist) (Figures 1A and 1B). In a backcross of the gy1 mutant with Nip, all members of the F1 generation exhibited a wild-type phenotype, and the F2 generation segregated at a ratio of 3:1 (wild-type phenotype:mutant phenotype) (Supplemental Table 1), indicating that the mutant harbors a recessive mutation in a single gene. However, the gy1 mutant seemed to exhibit normal ethylene response and had similar ethylene production in comparison to Nip (Supplemental Figures 1A to 1C).
Figure 1.
Phenotype of the gy1 Mutant and Map-Based Cloning of GY1.
(A) Comparison of Nip and gy1 etiolated seedlings grown in water for 3 d after germination in darkness. Arrowheads indicate positions of coleoptilar nodes between the mesocotyl and coleoptile. Bars = 10 mm.
(B) Mesocotyl and coleoptile length of etiolated seedlings represented in (A). The values are means ± sd of 20 to 30 seedlings per sample. The asterisks indicate significant difference compared with Nip (**P < 0.01, Student’s t test).
(C) Emergence of Nip and gy1 from soil. Germinated seeds were sown at a depth of 2 cm and seedlings were grown for 3 d under a 14-h-light/10-h-dark photoperiod.
(D) Etiolated Nip and gy1 seedlings grown in soil as in (C). Arrowheads indicate positions of coleoptilar nodes between the mesocotyl and coleoptile. Bar = 10 mm.
(E) Coleoptile and mesocotyl length of etiolated seedlings represented in (D). The values are means ± sd of 20 to 30 seedlings per sample. The asterisks indicate significant difference compared with Nip (**P < 0.01, Student’s t test).
(F) Map-based cloning of GY1. The GY1 locus was mapped to a 48-kb region between marker 1-39.15 and marker 1-39.20 of chromosome 1. The numbers below the markers indicate the number of recombinant individuals among the examined individuals with the mutant phenotype from the F2 segregated population. The mutation site of gy1 (G518A for nucleotide change and R173Q for amino acid change) is indicated by an arrowhead in the schematic diagram of GY1. The black box represents the coding region of GY1.
(G) The GY1 genomic sequence complemented the gy1 mutant phenotype. Etiolated seedlings from Nip, gy1, and the GY1-complemented line of gy1 grown for 3 d after germination are shown. Arrowheads indicate positions of nodes. The lower panel is the confirmation of the transgenic line by cleaving PCR products with Aor51H I. The PCR products were amplified with the dCAPS primers listed in Supplemental Table 7. Bar = 10 mm.
(H) Coleoptile and mesocotyl length of etiolated seedlings represented in (G). The values are means ± sd of 20 to 30 seedlings per sample. The asterisks indicate significant difference compared with Nip (**P < 0.01, Student’s t test).
Long mesocotyls and coleoptiles may facilitate the emergence of rice seedlings from the soil (Dilday et al., 1990). We then measured the emergence rate of the gy1 mutant and Nip. The germinated seeds were sown in potted soil at a depth of 2 cm at 28°C. Three days later, about half of the gy1 mutant seedlings had emerged from the soil, while only a few Nip seedlings had emerged (Figure 1C). At a later stage, all the Nip and gy1 mutants emerged and the gy1 mutant seedlings were significantly taller than Nip seedlings at all stages examined (Supplemental Figure 2). Three-day-old seedlings were removed from the soil and compared. The gy1 mutant has much longer mesocotyls than Nip, and the difference in mesocotyl length was greater in soil-grown etiolated seedlings than that in water-grown etiolated seedlings (Figures 1A, 1B, 1D, and 1E). These results indicate that the longer mesocotyl and coleoptile in the gy1 mutant may facilitate early emergence of seedlings from soil.
Map-Based Cloning of GY1
Although we used a rice T-DNA-tagged population for mutant screening, most of our identified mutants harbored point and/or short InDel mutations (Ma et al., 2013, 2014; Yang et al., 2015b; Yin et al., 2015), which are most likely generated during mass production of rice embryogenic calli and longer callus growth periods (Wei et al., 2016). The tagging efficiency of T-DNA-tagged populations is only 5 to 10% in rice, and the relationship between flanking sequence and phenotype is very weak (Wei et al., 2016). Based on our previous studies (Ma et al., 2013, 2014; Yang et al., 2015b; Yin et al., 2015), map-based cloning was used to identify the gene responsible for the gy1 phenotype. Using 927 individuals with the gy1 phenotype segregated in the F2 population derived from a cross between the gy1 mutant and an indica cultivar TN1, the GY1 locus was located to a 48-kb region of chromosome 1. This region contained three predicted genes, according to information in the rice genome annotation project (http://rice.plantbiology.msu.edu/). None of these genes was differentially expressed between Nip and the gy1 mutant (Supplemental Figure 1D). Genome sequencing indicated that the gy1 mutant had a single base substitution at 518 bp in a predicted putative lipase gene LOC_Os01g67430. This single base substitution of G to A in gy1 resulted in mutation of GY1 at 173th amino acid, from arginine (R) to glutamine (Q) (R173Q) (Figure 1F).
To test whether this putative lipase gene could rescue the phenotype of the gy1 mutant, a plasmid (pC-GY1) harboring the genomic sequence of the candidate gene, including the coding region and the 2022-bp upstream and 1476-bp downstream regions, was transformed into the gy1 mutant plants. Transgenic plants showed complete complementation of the gy1 phenotype at the seedling and mature stages (Figures 1G and 1H; Supplemental Figure 3). These results indicate that GY1 is located at the LOC_Os01g67430 locus and encodes a putative lipase.
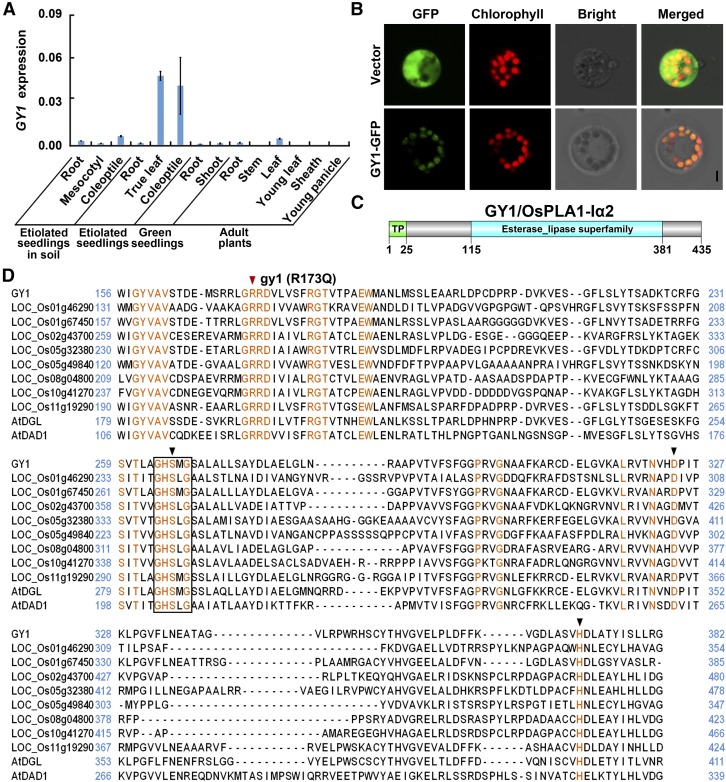
Spatial Expression of GY1 and Protein Subcellular Localization
The expression pattern of GY1 was analyzed in organs of Nip plants from the vegetative to reproductive stages using qPCR. GY1 was highly expressed in the true leaf and coleoptile of etiolated seedlings, and exhibited much lower expression levels in the other organs examined (Figure 2A). In soil-grown etiolated seedlings, GY1 expression was also higher in coleoptiles than in the other tested organs (Figure 2A). This result suggests that GY1 plays important roles in coleoptiles.
Figure 2.
GY1 Expression, Protein Subcellular Localization, and Alignment Analysis of GY1 with Its Homologs.
(A) GY1 expression level in different organs of Nip detected by qPCR relative to OsActin2 expression. “Etiolated seedlings in soil” indicate that the germinated seeds were sown in soil at a depth of 4 cm. “Etiolated seedlings” and “Green seedlings” represent seedlings cultured under regular water conditions in darkness and in 14 h light/10 h dark, respectively. “Adult plants” represent plants grown in the field. The values are means ± sd of three biological replicates (independent pools of tissue) per sample.
(B) GY1 is localized in chloroplasts of rice protoplasts as revealed by the merged green fluorescence of GY1-GFP fusion protein and the red autofluorescence of chlorophyll. Bar = 5 μm.
(C) A schematic diagram of GY1 protein. A transit peptide (TP) and an esterase/lipase domain are indicated.
(D) Alignment of GY1 and its homologs. Multiple alignments of amino acid sequences of GY1 and its homologs in rice and Arabidopsis were conducted using MEGA6. Identical residues are shaded in orange. The black box indicates the highly conserved GXSXG motif of PLA1. The black arrowheads indicate a triad of amino acids in the catalytic center of PLA1. The red arrowhead indicates the mutation site (R173Q) in gy1.
GY1 protein was predicted to localize in the chloroplasts using LocTree3 (Supplemental Figure 4A). To examine the cellular localization of GY1, a rice protoplast transient assay was performed using the pBI-GY1-GFP construct in which the GY1 coding sequence was fused in frame with GFP to generate a GY1:GFP fusion protein. The fluorescence signals of the GY1:GFP fusion protein colocalized with the chlorophyll red autofluorescence in the chloroplasts of rice and Arabidopsis protoplasts (Figure 2B; Supplemental Figure 4B). These results suggest that GY1 localizes in the chloroplast. While the materials used for rice protoplast preparation were etiolated seedlings, and the protoplasts were only cultured in light for a few hours during plasmid transformation, we deduce that GY1 may localize in the etioplasts of etiolated seedlings.
The Lipid Substrates of Phospholipase A1 Accumulated in the gy1 Mutant
GY1 has no intron and encodes a putative phospholipase composed of 425-amino acid residues according to information from the NCBI database (http://www.ncbi.nlm.nih.gov/) (Figure 2C). Homology searches with GY1 amino acid sequences revealed that GY1 was similar to two Arabidopsis lipases, DONGLE (DGL) and DEFECTIVE IN ANTHER DEHISCENCE1 (DAD1). Both of these lipases belong to the phospholipase A1 (PLA1) family (Figure 2D) (Hyun et al., 2008), whose members are characterized by the presence of a highly conserved GXSXG motif and a catalytic triad composed of a serine (S), an aspartic acid (D), and a histidine (H) residue (Singh et al., 2012). The GY1 protein contained a GHSMG motif and a putative catalytic triad S, D, and H that fit the definition of a PLA1, suggesting that GY1 belongs to the PLA1 family. Through multiple alignment of GY1 and its homologs, we found that the mutation site in the gy1 mutant at the 173th amino acid was a highly conserved residue in all of the compared proteins from rice and Arabidopsis (Figure 2D).
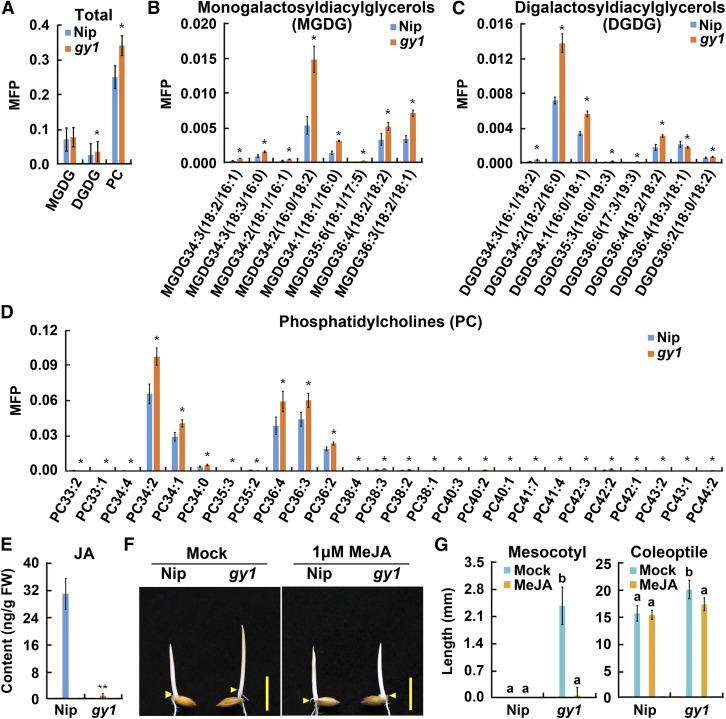
Because GY1 is a member of the PLA1 family, its mutation should affect the lipid profile. Lipid analysis of the gy1 mutant and Nip showed that the levels of different species of phosphatidylcholine (PC), digalactosyldiacylglycerol, and monogalactosyldiacylglycerol, the main substrates of PLA1 (Singh et al., 2012), were significantly higher in the gy1 mutant than in Nip (Figures 3A to 3D). These results imply that the point mutation in the gy1 mutant affects the PLA1 activity of the GY1.
Figure 3.
Lipid Contents and JA Contents in Rice Seedlings and MeJA Rescue of gy1 Phenotype.
(A) Total monogalactosyldiacylglycerols (MGDG), digalactosyldiacylglycerols (DGDG), and PC levels expressed in molar fractions normalized to total polar lipids (MFP) in etiolated seedlings from Nip and gy1 grown for 3 d after germination in dark. The values are means ± sd of four biological replicates (independent pools of tissue) per sample. The asterisks indicate significant difference compared with the corresponding values in Nip (*P < 0.05, Student’s t test).
(B) Levels of monogalactosyldiacylglycerol species in etiolated seedlings of Nip and gy1. Other indications are as in (A).
(C) Levels of digalactosyldiacylglycerol species in etiolated seedlings of Nip and gy1. Other indications are as in (A).
(D) Levels of PC species in etiolated seedlings from Nip and gy1. Other indications are as in (A).
(E) JA content in etiolated seedlings from Nip and gy1 grown for 3 d after germination in dark. The values are means ± sd of three biological replicates (independent pools of tissue) per sample. The asterisks indicate significant difference compared with Nip (**P < 0.01, Student’s t test).
(F) MeJA (1 μM) treatment rescued gy1 phenotype. The etiolated seedlings were grown for 3 d after germination in dark. Arrowheads indicate positions of the coleoptilar nodes between mesocotyl and coleoptile. Bar = 10 mm.
(G) Mesocotyl and coleoptile length of etiolated seedlings represented in (F). The values are means ± sd of 20 to 30 seedlings per sample. Different letters above each column indicate significant difference between the compared pairs (P < 0.05, Student’s t test).
GY1 Mediates the Initial Step of JA Biosynthesis
Since GY1 clusters with AtDGL (Supplemental Figure 5 and Supplemental Data Set 1), which is the first enzyme in the JA biosynthesis pathway, we predict that GY1 also initiates JA biosynthesis in rice. The JA contents in the etiolated seedlings of Nip and the gy1 mutant were then measured to examine whether the GY1 mutation hampered JA accumulation. The JA content was significantly lower in the shoots of gy1 etiolated seedlings than in those of Nip etiolated seedlings, indicating that mutation in GY1 blocked JA production (Figure 3E). osaoc, which harbors a mutation in the gene encoding a downstream JA biosynthesis enzyme OsAOC (Riemann et al., 2013), also exhibited longer mesocotyls and coleoptiles in etiolated seedlings than its wild-type Nihonmasari (Supplemental Figure 6). These results indicate that disruption of the JA biosynthesis pathway underlies the long mesocotyl and long coleoptile phenotype of etiolated seedlings.
To further examine the role of JA in mesocotyl/coleoptile growth regulation, we investigated whether JA application could rescue the gy1 mutant phenotype. A concentration of 1 μΜ ΜeJA was selected because this level did not affect coleoptile growth of Nip seedlings (Supplemental Figure 7). With 1 μΜ ΜeJA treatment, the gy1 mutant phenotype was almost completely recovered and resembled Nip seedlings (Figure 3F), and the lengths of gy1 mesocotyls and coleoptiles were similar to those of Nip mesocotyls and coleoptiles of seedlings of the same age under normal growth conditions (Figure 3G). These results indicate that GY1 is a PLA1 family protein mediating the initial step of the JA biosynthesis pathway, and JA represses mesocotyl and coleoptile elongation in etiolated rice seedlings.
Emergence of Rice Seedlings Correlates with Reduced Jasmonic Acid Content but Increased Ethylene Production
Considering that the longer mesocotyl and coleoptile of the gy1 mutant facilitate seedling emergence from soil (Figures 1C and 1D; Supplemental Figure 2), disruption of JA biosynthesis leads to mesocotyl and coleoptile elongation (Figures 1A and 1B and Figure 3E; Supplemental Figure 6), JA application rescued the gy1 mutant phenotype (Figures 3F and 3G), and soil-grown Nip seedlings have longer mesocotyls than the water-grown Nip seedlings (Figures 1A, 1B, 1D, and 1E), we speculated that JA content may be related to the emergence process of rice seedlings from under the soil.
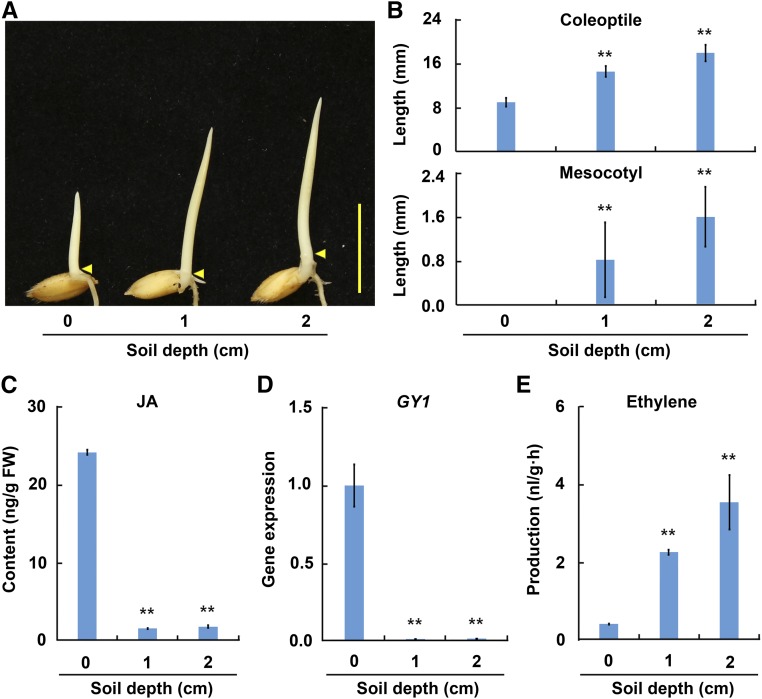
The germinated Nip seeds were sown in pots covered with different depths of soil (0, 1, and 2 cm) and grown for 3 d, and then the seedlings were removed for evaluation. The length of the mesocotyl and coleoptile increased with increasing seed-sowing depth (Figures 4A and 4B). The JA content and GY1 expression level were also measured in this process, and values from both parameters decreased drastically at greater soil depths (Figures 4C and 4D). These results suggest that less JA promotes the emergence of rice seedlings from thicker soil cover.
Figure 4.
Phenotypic and Hormone Changes in Rice Seedlings after Seed-Sowing at Different Soil Depths.
(A) Comparison of Nip etiolated seedlings sown at different depths in soil. The germinated seeds were covered with 0, 1, and 2 cm soil and the seedlings were grown for 3 d in darkness. Arrowheads indicate positions of coleoptilar nodes between the mesocotyl and coleoptile. Bar = 10 mm.
(B) Coleoptile and mesocotyl length of Nip etiolated seedlings represented in (A). The values are means ± sd of 20 to 30 seedlings per sample. The asterisks indicate significant difference compared with the “0 cm” value (**P < 0.01, Student’s t test).
(C) JA contents in shoots of Nip seedlings sown at different depths. The values are means ± sd of three biological replicates (independent pools of tissue) per sample. The asterisks indicate significant difference compared with the “0 cm” value (**P < 0.01, Student’s t test).
(D) GY1 relative expression in shoots of Nip seedlings sown at different depths. The expression was detected by qPCR relative to OsActin2. The values are means ± sd of three biological replicates (independent pools of tissue) per sample. The asterisks indicate significant difference compared with the “0 cm” value (**P < 0.01, Student’s t test).
(E) Ethylene production in Nip seedlings sown at different depths. The values are means ± sd of three biological replicates (independent pools of tissue) per sample. The asterisks indicate significant difference compared with the “0 cm” value (**P < 0.01, Student’s t test).
We previously revealed that MHZ7/OsEIN2-overexpressing rice seedlings exhibited the long mesocotyl and long coleoptile phenotype upon ethylene treatment (Ma et al., 2013), resembling the gy1 mutant (Figure 1A). In Arabidopsis seedlings, ethylene production was activated by soil cover (Zhong et al., 2014). Based on these findings, we propose that the GY1-mediated JA biosynthesis that inhibits mesocotyl and coleoptile elongation may be related to the ethylene pathway. We then examined ethylene production during rice seedling emergence when seedlings were sown at the same soil depths, from 0 to 2 cm. Ethylene emission increased in the rice seedlings with increasing seed-sowing depth (Figure 4E).
All these results indicate that, when rice seedlings are covered with soil, ethylene production is induced, whereas JA biosynthesis is inhibited, and that these changes in hormone production promote the elongation of mesocotyls and coleoptiles of rice seedlings, facilitating their emergence from soil.
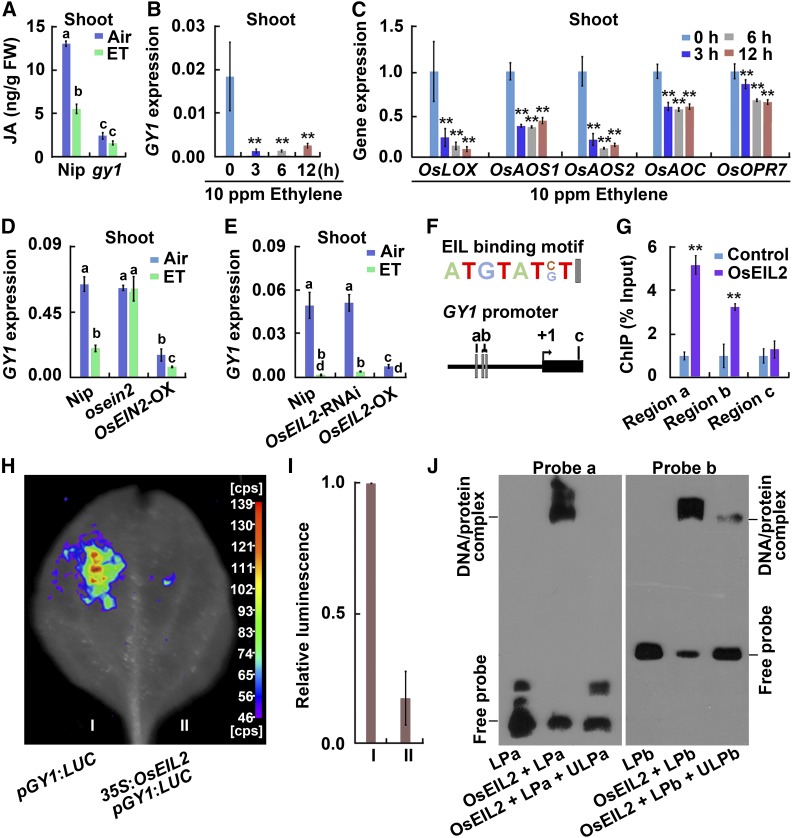
JA Biosynthesis Is Repressed by the Ethylene Pathway
To further explore the relationship between JA and ethylene during elongation of etiolated rice seedlings, we measured the JA content in the presence or absence of ethylene treatment. Under 10 ppm ethylene, the JA content in Nip etiolated seedlings decreased in comparison with that of the non-treatment control, whereas the JA content in gy1 etiolated seedlings did not differ in the presence or absence of ethylene treatment (Figure 5A). GY1 expression in 2-d-old etiolated seedlings was also significantly inhibited by ethylene after 3 to 12 h of treatment (Figure 5B). We further examined the expression of other genes in the JA biosynthesis pathway and all the tested genes were significantly inhibited by ethylene at different levels (Figure 5C). We also subjected deetiolated seedlings (etiolated seedlings exposed to light for 1 h) to the ethylene treatment. In these deetiolated seedlings, the JA content and expression of GY1 and most other JA biosynthesis genes were higher than those in etiolated seedlings (Supplemental Figures 8A and 8B). After ethylene treatment, clear reductions in JA content and the expression of GY1 and other JA biosynthesis genes were still observed in the deetiolated seedlings (Supplemental Figures 8A and 8B). All these results indicate that ethylene inhibits JA accumulation through suppressing the expression of GY1 and other genes in the JA biosynthesis pathway.
Figure 5.
Ethylene Inhibits GY1 Expression through OsEIL2.
(A) JA content in etiolated seedlings from Nip and gy1 grown for 3 d after germination in darkness. ET indicates 10 ppm ethylene treatment for 8 h. The values are means ± sd of three biological replicates (independent pools of tissue) per sample. Different letters above each column indicate significant difference between the compared pairs (P < 0.05, Student’s t test).
(B) GY1 expression in shoots of Nip etiolated seedlings after 10 ppm ethylene treatment for 8 h. The quantitation was performed by qPCR relative to OsActin2 expression. The values are means ± sd of three biological replicates (independent pools of tissue) per sample. The asterisks indicate significant difference compared with the “0 h” value (**P < 0.01, Student’s t test).
(C) Expression of OsLOX, OsAOS1, OsAOS2, OsAOC, and OsOPR7 from the JA biosynthesis pathway in shoots of etiolated Nip seedlings. The seedlings were treated with 10 ppm ethylene for 8 h, and the quantitation was performed by qPCR relative to OsActin2 expression. The expression level of each gene at the “0 h” treatment was set to 1 and other values were compared with it. The values are means ± sd of three biological replicates (independent pools of tissue) per sample. The asterisks indicate significant difference compared with the “0 h” value (**P < 0.01, Student’s t test).
(D) GY1 expression level in the shoots of Nip, mhz7/osein2, and MHZ7/OsEIN2-OX etiolated seedlings after treatment with 10 ppm ethylene (ET) for 8 h. The quantitation was performed by qPCR relative to OsActin2 expression. The values are means ± sd of three biological replicates (independent pools of tissue) per sample. Different letters above each column indicate significant difference between the compared pairs (P < 0.05, Student’s t test).
(E) GY1 expression in etiolated seedlings from Nip, OsEIL2-RNAi, and OsEIL2-OX after 10 ppm ethylene treatment for 8 h. Other indications are as in (D).
(F) The EIL binding motif in the GY1 promoter region. a, b, and c indicate regions examined by ChIP-PCR in (G). a and b also indicate probe a and probe b used in (J), respectively.
(G) Enrichment fold of the ChIP-PCR signals from the regions shown in (F). The ChIP analysis was performed with the wild-type Nip (Control) and 35S:OsEIL2:GFP (OsEIL2) transgenic plants. Error bars indicate ± sd from three biological repeats (independent pools of tissue). Asterisks indicate that the difference between OsEIL2 value and control value is significant (P < 0.01, Student’s t test).
(H) OsEIL2 represses the promoter activity of GY1 in a transient expression assay in tobacco leaves. 35S:OsEIL2 was used as an effector plasmid and pGY1:LUC as a reporter plasmid harboring the GY1 promoter-driven LUC. More than five biological replicates (independent pools of tissue) were performed with similar results.
(I) Quantitative analysis of luminescence intensity in samples from (G). The values are means ± sd of four biological replicates (independent pools of tissue). The asterisks indicate significant difference compared with the control (**P < 0.01, Student’s t test).
(J) OsEIL2 protein binds to the promoter region of GY1 containing the EIL binding site. GST-tagged OsEIL2 N terminus fusion protein (OsEIL2) was incubated with biotin-labeled DNA fragments (probe a and probe b), respectively. An excess of unlabeled probe (competitor) was also added to compete with the biotin-labeled promoter region for OsEIL2 binding. LPa indicates labeled probe a. ULPa indicates unlabeled probe a. LPb indicates labeled probe b. ULPb indicates unlabeled probe b. The upper bands are labeled probes bound with OsEIL2 protein.
To determine whether the inhibition of GY1 by ethylene depends on the ethylene signaling pathway, we examined GY1 expression in ethylene response mutants/RNAi plants and in plants overexpressing MHZ7/OsEIN2 and OsEIL2, which were established in our previous studies (Ma et al., 2013; Yang et al., 2015b). MHZ7/OsEIN2 is homologous to EIN2, a central membrane protein in Arabidopsis ethylene signaling (Ma et al., 2013). OsEIL2 is homologous to the transcription factor EIN3 of the Arabidopsis ethylene signaling pathway and specifically regulates the coleoptile ethylene response of etiolated rice seedlings (Yang et al., 2015b). The ethylene-mediated inhibition of GY1 expression was blocked in the mhz7/osein2 mutant, but not in the OsEIL2-RNAi etiolated seedlings (Figures 5D and 5E). Plants overexpressing MHZ7/OsEIN2 and OsEIL2 had low expression levels of GY1 compared with Nip under normal conditions, and the gene expression was further reduced with ethylene treatment (Figures 5D and 5E). These results indicate that ethylene signaling represses GY1 expression in a MHZ7/OsEIN2-dependent manner.
As OsEIL2 overexpression repressed GY1 expression, we analyzed whether this transcription factor can bind to the GY1 promoter and inhibit the promoter activity. In the promoter region of GY1, three EIL consensus sequences were identified (Figure 5F) (Zhong et al., 2009; Boutrot et al., 2010; Yang et al., 2015b). To assess whether OsEIL2 could bind to the EIL binding motif in the promoter of GY1 in vivo, we performed chromatin immunoprecipitation (ChIP) analysis with the 35S:OsEIL2-GFP transgenic rice plants. The ChIP-PCR signals were enriched at the OsEIL2 binding region containing the EIL binding motif in the 35S:OsEIL2-GFP transgenic plants (Figure 5G). The 2022-bp fragment upstream of the ATG start codon of GY1 was further cloned into the reporter plasmid that carried a LUCIFERASE (LUC) gene. The effector plasmid 35S:OsEIL2, together with the pGY1:LUC reporter plasmid, were coinjected into tobacco (Nicotiana tabacum) leaves for a transient expression assay. OsEIL2 significantly decreased the LUC activity driven by the GY1 promoter compared with the control (Figures 5H and 5I). These results suggest that OsEIL2 inhibits the GY1 promoter activity. An electrophoresis mobility shift assay (EMSA) was further performed to test whether OsEIL2 could directly bind to the GY1 promoter, and OsEIL2 was found to specifically bind to the two probes containing the EIL binding motif (Figure 5J). Taken together, these results suggest that OsEIL2 inhibits GY1 expression through direct binding to the GY1 promoter and suppression of GY1 promoter activity.
Genetic Relationship between GY1 and the Ethylene Signaling Gene MHZ7/OsEIN2 in Rice
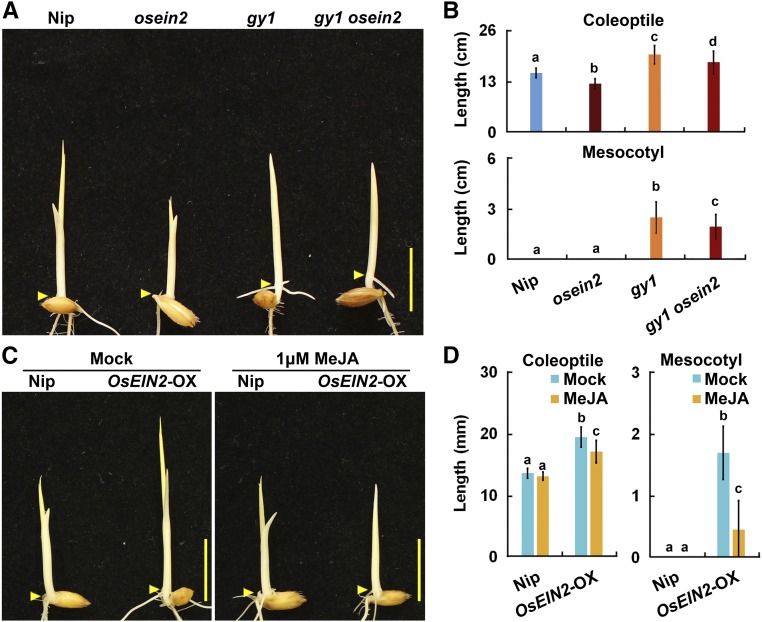
To further examine the genetic relationship between GY1 and the ethylene signaling pathway, we generated the double mutant gy1 osein2 through a genetic cross of gy1 with mhz7/osein2, which was identified in our previous study (Ma et al., 2013). The etiolated seedlings of the double mutant gy1 osein2 exhibited a long mesocotyl and long coleoptile, very similar to the gy1 phenotype (Figures 6A and 6B). This result suggests that GY1 acts downstream of MHZ7/OsEIN2 to regulate the elongation of the mesocotyl and coleoptile in etiolated rice seedlings.
Figure 6.
Genetic Interaction of GY1 with OsEIN2.
(A) Etiolated Nip, gy1, osein2, and the gy1 osein2 double mutant seedlings grown for 3 d after germination in darkness. Arrowheads indicate the coleoptilar nodes between the mesocotyl and coleoptile. Bar = 10 mm.
(B) Coleoptile and mesocotyl length of etiolated Nip, gy1, osein2, and gy1 osein2 double mutant seedlings grown for 3 d after germination in darkness. The values are means ± sd of 20 to 30 seedlings per sample. Different letters above each column indicate significant difference between the compared pairs (P < 0.05, Student’s t test).
(C) Etiolated seedlings from Nip and MHZ7/OsEIN2-OX without (mock) or with 1 μM MeJA treatment grown for 3 d after germination in darkness. Arrowheads indicate the coleoptilar nodes between the mesocotyl and coleoptile. Bar = 10 mm.
(D) Coleoptile and mesocotyl length of etiolated seedlings from Nip and MHZ7/OsEIN2-OX without (mock) or with 1 μM MeJA treatment grown for 3 d after germination in darkness. The values are means ± sd of 20 to 30 seedlings per sample. Different letters above each column indicate significant difference between the compared pairs (P < 0.05, Student’s t test).
We further applied JA to the MHZ7/OsEIN2-overexpressing plants to test whether JA has any effects on MHZ7/OsEIN2 function. With 1 μΜ MeJA treatment, the long mesocotyl and long coleoptile of MHZ7/OsEIN2-OX seedlings were largely suppressed (Figures 6C and 6D), indicating that ethylene signaling-promoted mesocotyl and coleoptile growth at least partially requires a reduction in JA levels. These results support the notion that the JA pathway acts downstream of the ethylene pathway to regulate mesocotyl and coleoptile growth in etiolated rice seedlings.
Altered Gene Expression and Cell Elongation Correlate with the Long Mesocotyl and Coleoptile of gy1
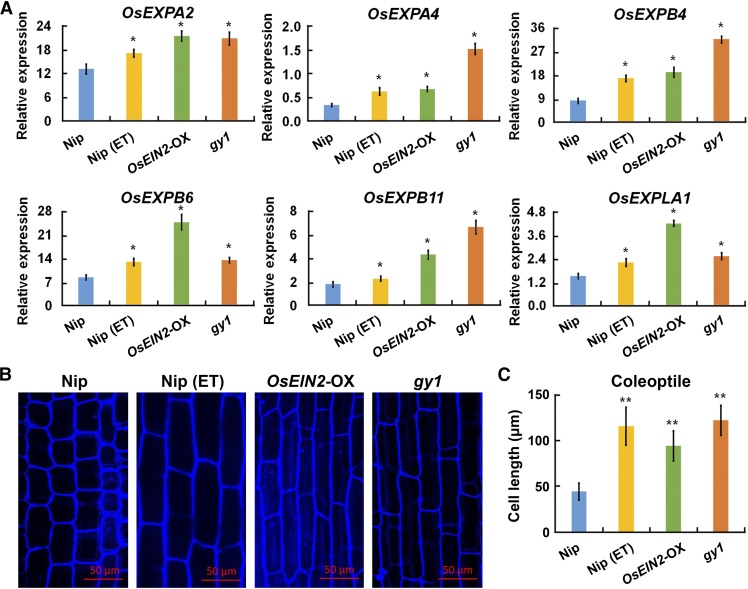
To understand the molecular aspects of mesocotyl/coleoptile elongation, the mesocotyls of 3-d-old etiolated Nip and gy1 mutant seedlings, which had been sown under 4 cm of soil, were harvested for mRNA library preparation, sequencing, and further bioinformatics analysis. The differentially expressed genes between Nip and the gy1 mutant were then analyzed. Through enrichment analysis for Gene Ontology terms, differentially expressed genes were significantly enriched in these categories: external encapsulating structure, extracellular region, cell wall, cell cycle, DNA metabolic process, response to endogenous stimulus, and response to stimulus (Supplemental Figure 9A). Expression of the JA biosynthesis-related gene OsJMT and JA-induced genes OsJIP-1, OsJIP-2, OsJIP-3, and OsJIP-4 in the term “response to endogenous stimulus” was significantly lower in the gy1 mutant than in Nip (Supplemental Figure 9B). By contrast, the expression of the expansin family of genes OsEXPA2, OsEXPA4, OsEXPB4, OsEXPB6, OsEXPB11, and OsEXPLA1 in the term “cell wall” was significantly higher in the gy1 mutant than in Nip (Supplemental Figure 9C). We further analyzed the expression of expansin family genes in the shoots of etiolated Nip, Nip subjected to 10 ppm ethylene treatment, MHZ7/OsEIN2-OX (Ma et al., 2013), and gy1 mutant seedlings by qPCR. The latter three materials had long mesocotyls/coleoptiles. The expression of OsEXPA2, OsEXPA4, OsEXPB4, OsEXPB6, OsEXPB11, and OsEXPLA1 was significantly increased in Nip subjected to ethylene treatment, MHZ7/OsEIN2-OX, and the gy1 mutant compared with Nip (Figure 7A). These results suggest that a reduction in the expression of JA-responsive genes and upregulation of expansin genes are related to the elongation of the mesocotyl/coleoptile in etiolated rice seedlings.
Figure 7.
Expression of Expansin Family Genes and Cell Length of Coleoptiles in Different Seedlings.
(A) The expression level of OsEXPA2, OsEXPA4, OsEXPB4, OsEXPB6, OsEXPB11, and OsEXPLA1 (expansin-like A1) in the shoots of etiolated Nip, Nip with 10 ppm ethylene (ET) treatment, MHZ7/OsEIN2-OX, and gy1 seedlings, detected by qPCR relative to OsActin2 expression. The values are means ± sd of three biological replicates (independent pools of tissue) per sample. The asterisks indicate significant difference compared with Nip (P < 0.05, Student’s t test).
(B) Coleoptile cells of etiolated Nip, Nip with 10 ppm ethylene treatment, MHZ7/OsEIN2-OX, and gy1 seedlings. Bar = 50 μm.
(C) Cell lengths of the coleoptiles of etiolated Nip, Nip with 10 ppm ethylene treatment, MHZ7/OsEIN2-OX, and gy1 seedlings. The values are means ± sd of 20 to 30 cells per sample. The asterisks indicate significant difference compared with Nip (**P < 0.01, Student’s t test).
Since the expansin family of genes mainly promote cell length (Huang et al., 2000; Choi et al., 2003; Li et al., 2003; Sharova, 2007), we examined the cell length of the etiolated seedlings from the above plants. The coleoptile cells of MHZ7/OsEIN2-OX and the gy1 mutant were significantly longer than those of Nip under normal conditions, and ethylene treatment also increased the cell length of Nip (Figures 7B and 7C). These results indicate that the ethylene signaling pathway, as well as the GY1 mutation, facilitates cell elongation to promote mesocotyl and coleoptile growth in rice seedlings.
A Natural Allelic Variation of GY1 Contributes to Mesocotyl Elongation
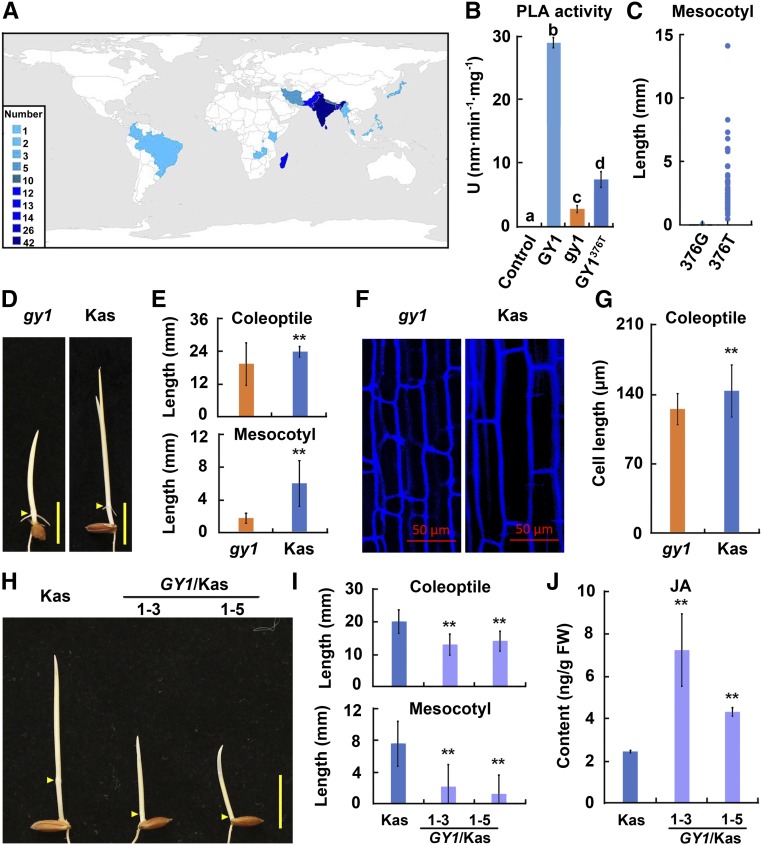
Although GY1 mutation-mediated mesocotyl elongation promotes emergence of seedlings, other yield-related agronomic traits were affected in the gy1 mutant under field growth condition (Supplemental Table 2). We therefore investigated whether any natural variations of GY1 are present, which are relevant to the long mesocotyl phenotype in etiolated seedlings of rice accessions. We meta-analyzed the coding sequence of GY1 using the public resequencing data of 3000 rice accessions with representative diversity (Li et al., 2014). Compared with the Nip reference GY1 sequence, we found that a single G376T single nucleotide polymorphism (SNP) existed in 141 rice accessions, which belonged to different rice subgroups and originated from different countries/regions in Asia, Africa, and South America (Figure 8A; Supplemental Table 3). This G376T SNP caused the 126th amino acid to change from glycine to cysteine. To assess whether the variation could influence the enzymatic activity, we measured the PLA activity of GY1, gy1 (GY1518A), and GY1376T. These results indicated that the PLA activity of gy1 and GY1376T was drastically lower than that of GY1. However, the PLA activity of GY1376T was significantly higher than that of gy1 (Figure 8B).
Figure 8.
Natural Variation of GY1 Correlates with Mesocotyl Elongation in Rice Accessions and GY1 Suppresses the Long Mesocotyl and Coleoptile Phenotypes in Kasalath Etiolated Seedlings.
(A) Distribution of the rice accessions harboring the 376T SNP in GY1. Countries are labeled with different shades of blue according to the number of varieties distributed in each country. The countries include Asian countries (India, Bangladesh, Sri Lanka, Pakistan, Nepal, Iran, Japan, Malaysia, Myanmar, and Philippines), African countries (Madagascar, Burundi, Kenya, Liberia, and Zambia), South American countries (Brazil and Colombia), and an Oceania country (Fiji).
(B) PLA activity of GY1, gy1 (GY1518A), and GY1376T. PLA activity was measured in vitro with dye-labeled PC as a substrate at 30°C. GY1, gy1 (GY1518A), and GY1376T were expressed as fusion proteins with the SUMO peptide. SUMO peptide was used as a control. Values are means ± sd for four independent replicates. Different letters above each column indicate significant difference between the compared pairs (P < 0.05, LSD and S-N-K test).
(C) The 376T SNP of GY1 correlates with long mesocotyls in rice accessions. The 376G SNP of GY1 is related to short/no mesocotyl.
(D) Etiolated seedlings of gy1 and Kasalath (Kas) grown for 3 d after germination in darkness. Arrowheads indicate the coleoptilar nodes between mesocotyl and coleoptile. Bar = 10 mm.
(E) Coleoptile and mesocotyl length of etiolated seedlings from (C). The values are means ± sd of 20 to 30 seedlings per sample. The asterisks indicate significant difference compared with gy1 (**P < 0.01, Student’s t test).
(F) Coleoptile cells of etiolated seedlings from gy1 and Kas. Bar = 50 μm.
(G) Cell length of coleoptile as shown in (E). The values are means ± sd of 20 to 30 cells per sample. The asterisks indicate significant difference compared with gy1 (**P < 0.01, Student’s t test).
(H) GY1376G expression inhibits the long mesocotyl and coleoptile phenotype in Kas with GY1376T. Phenotype of etiolated seedlings from Kas and two transgenic lines transformed with GY1376G grown for 3 d after germination in darkness. Arrowheads indicate the coleoptilar nodes between the mesocotyl and coleoptile. The dCAPS marker was used to genotype GY1 and GY1376T by cleaving PCR products with FspI. dCAPS primers are listed in Supplemental Table 7. Bar = 10 mm.
(I) Coleoptile and mesocotyl length of etiolated seedlings from (G). The values are means ± sd of 20 to 30 seedlings per sample. The asterisks indicate significant difference compared with Kas (**P < 0.01, Student’s t test).
(J) JA contents in the shoots of etiolated seedlings from Kas (with GY1376T) and two transgenic lines transformed with GY1376G grown for 3 d after germination in darkness. The values are means ± sd of three biological replicates (independent pools of tissue) per sample. The asterisks indicate significant difference compared with Kas (**P < 0.01, Student’s t test).
To elucidate whether the natural variant of 376T SNP in the GY1 gene is associated with any change in mesocotyl phenotype in etiolated seedlings, we collected 51 available rice accessions from the above 141 rice accessions harboring the 376T SNP according to the public resequencing data and conducted an association analysis. These accessions belonged to different rice subgroups. We found that 44 rice accessions with 376T SNP exhibited a longer mesocotyl phenotype compared with Nip with 376G in GY1, whereas the other seven rice accessions exhibited a short mesocotyl phenotype like Nip. After resequencing the GY1 gene in all 51 rice accessions, we found that the seven rice accessions exhibiting the short mesocotyl phenotype actually had the 376G SNP in their GY1 gene (Figure 8C; Supplemental Figure 10 and Supplemental Table 4). These results indicate that the 376T SNP of the GY1 gene correlates with the relatively long mesocotyls of etiolated rice seedlings.
Previously, five quantitative trait loci (QTL) for mesocotyl elongation were detected from backcross inbred lines developed from Kasalath and Nipponbare (Lee et al., 2012). The present GY1 locus overlapped with one of them. Kasalath (with GY1376T) etiolated seedlings also had a long mesocotyl and coleoptile, which were even longer than those of gy1 probably due to increased cell elongation (Figures 8D to 8G; Supplemental Table 4).
To further demonstrate the role of GY1 in mesocotyl regulation, the pC-GY1 plasmid used to complement the gy1 mutant was transformed into Kasalath plants with the GY1376T genotype. The generated GY1-transgenic plants in the Kasalath background exhibited shorter mesocotyls and coleoptiles than Kasalath (Figures 8H and 8I). These transgenic plants also had higher levels of JA contents than Kasalath (Figure 8J). All the results suggest that the natural variant allele GY1376T of the GY1 gene in Kasalath and other rice varieties is responsible for the long mesocotyl phenotype in etiolated seedlings.
DISCUSSION
In this study, we characterized the rice mutant gy1 and revealed that JA inhibits the elongation of mesocotyls and coleoptiles of rice seedlings. During the emergence of rice seedlings from soil cover, ethylene production increases and GY1-mediated JA biosynthesis is then inhibited through the direct binding of OsEIL2 to the promoter region of GY1, ultimately promoting mesocotyl and coleoptile elongation of the rice seedlings. We found that an elite natural allele of GY1 with the 376T SNP is associated with long mesocotyls and this finding may be used in breeding programs to select cultivars that can grow in dry land.
GY1-Mediated JA Biosynthesis Inhibits Mesocotyl and Coleoptile Elongation
Our study demonstrates that GY1-mediated JA biosynthesis inhibits mesocotyl and coleoptile elongation in etiolated rice seedlings. The following pieces of evidence support our conclusion. First, the gy1 mutant exhibited longer mesocotyls and longer coleoptiles than its wild-type Nip, and GY1 could completely complement this phenotype (Figure 1). Second, JA content significantly decreased in the gy1 mutant compared with that of Nip (Figure 3E). Third, the cpm2/osaoc mutant, which is also a JA biosynthesis pathway mutant, had a long mesocotyl and long coleoptile phenotype that resembled that of gy1 (Supplemental Figure 6). Fourth, 1 µM MeJA could rescue the long mesocotyl and long coleoptile phenotype of the gy1 mutant (Figures 3F and 3G). Fifth, a natural variation of 376T SNP in GY1, which resulted in an amino acid change from glycine to cysteine at position 126 of GY1, was identified from 3000 rice accessions. The presence of the 376T SNP was positively correlated with the long mesocotyl phenotype (Figure 8C; Supplemental Tables 3 and 4). Sixth, transforming the Kasalath variety harboring GY1376T with the wild-type GY1376G gene from Nip could largely suppress the long mesocotyl and long coleoptile phenotype of Kasalath (Figures 8H and 8I), and the transgenic lines had higher JA content than Kasalath (Figure 8J). Seventh, GY1 has PLA activity, and gy1 and GY1376T have much lower PLA activity than GY1. However, GY1376T has higher PLA activity than gy1. From all the evidence above, we conclude that the phospholipase GY1-mediated JA biosynthesis inhibits elongation of the mesocotyl and coleoptile in etiolated rice seedlings.
While mutation of GY1 promotes mesocotyl and coleoptile elongation in the gy1 mutant, the roots were not significantly affected (Supplemental Figure 1A), indicating the specificity of GY1 function in seedlings. When analyzing the spatial expression of GY1 in mesocotyls of Nip seedlings, the germinated Nip seeds were covered with soil to a 4 cm depth for three days, and under this condition, mesocotyl growth was promoted. In these mesocotyls, the expression of GY1 turned out to be very low (Figure 2A), consistent with the negative relationship between GY1 expression and mesocotyl elongation.
It should be noted that the etiolated seedlings of gy1 exhibit both longer mesocotyls and longer coleoptiles than Nip. In a natural population, the 376T SNP of GY1 is only associated with the long mesocotyl phenotype in etiolated seedlings, and the coleoptile length of seedlings was distributed over a large range due to background differences (Supplemental Table 4). Since there is no easy way to analyze the genetic effect of 376T SNP on the coleoptile phenotype among plants in a natural population by association analysis, we performed a complementation test in the Kasalath variety, which harbors GY1376T, using the wild-type GY1376G from Nip. We found that GY1376T regulated the long coleoptile and the long mesocotyl phenotypes in etiolated seedlings.
This study reveals that GY1-mediated JA biosynthesis inhibits mesocotyl/coleoptile elongation of etiolated rice seedlings, affecting seedling emergence from soil cover. This finding is consistent with previous reports that show that JA inhibits seed germination in various plants, e.g., Arabidopsis, alfalfa (Medicago sativa), cornflower (Centaurea cyanus), rice, cress (Lepidium sativum), maize (Zea mays) (Wilen et al., 1994; Liu et al., 2015). However, the genes underlying the inhibition are still largely unknown. (9S,13S)-12-oxo-phytodienoic acid, a key metabolic intermediate during JA biosynthesis, acts synergistically with abscisic acid to inhibit germination in Arabidopsis (Dave et al., 2011). Arabidopsis DAD1, DGL, and DLAH are homologs of GY1 and exhibit PLA activity (Ishiguro et al., 2001; Hyun et al., 2008; Ellinger et al., 2010; Seo et al., 2011). DLAH-overexpressing transgenic seeds were tolerant to higher temperature and had higher germination percentages than did wild-type seeds (Seo et al., 2011). However, it is not clear whether Arabidopsis DAD1 and DGL are also involved in the seed germination process, although they both regulate floral development through JA synthesis (Ishiguro et al., 2001; Hyun et al., 2008).
In addition to its role in mesocotyl/coleoptile elongation and emergence from soil, GY1 affects floral development, as judged from the gy1 mutant phenotype at maturity (Supplemental Figure 3). Floral development in plants is controlled by multiple signals and JA is one of them (Yuan and Zhang, 2015). The Arabidopsis homolog DAD1 and DGL-mediated JA biosynthesis regulates floral development (Ishiguro et al., 2001; Hyun et al., 2008). A gy1 allelic mutant extra glume1 (eg1) also exhibits an abnormal number of pistils and an extra glume-like organ in rice (Li et al., 2009; Zhang et al., 2016). The abnormal flower development of the gy1 mutant may cause the reduction of yield-related traits (Supplemental Table 2). EG1/GY1 exhibited lipase activity in vitro (Zhang et al., 2016) and regulated lipid metabolism (Zhang et al., 2016) and JA biosynthesis (Cai et al., 2014) during rice floral development. Our analysis further showed that GY1 was an etioplast-localized PLA in etiolated seedlings (Figures 2B to 2D and 8B), preferred PC as a substrate (Figures 3A to 3D), and participated in JA biosynthesis to regulate skotomorphogenesis and floral development (Figures 3E and 3F; Supplemental Figure 3). Furthermore, the JA signaling repressor EG2/OsJAZ1 not only interacts with the JA receptor OsCOI1b to trigger OsJAZ1’s degradation during spikelet development, but also interacts with OsMYC2 to repress an E-class gene OsMADS1 that is crucial for spikelet development (Cai et al., 2014). These studies indicate that JA synthesis and signaling control the development of the inflorescence and flower.
Ethylene Signaling Inhibits JA Biosynthesis to Promote Mesocotyl/Coleoptile Elongation
In Arabidopsis, the ethylene pathway plays an important role in the growth of etiolated seedlings under soil environments (Zhong et al., 2014; Shi et al., 2016a, 2016b). Here, we find that the ethylene signaling pathway inhibits JA biosynthesis and eventually promotes mesocotyl and coleoptile elongation in etiolated rice seedlings to facilitate their emergence from soil. The following evidence supports this conclusion. First, ethylene production in rice seedlings increases along with the seed-sowing depth in soil (Figure 4E). By contrast, GY1 expression and JA contents are reduced as the sowing depth increases (Figures 4C and 4D). Mesocotyl and coleoptile growth are promoted as the sowing depth increases (Figures 4A and 4B). Second, ethylene inhibits the expression of GY1 and other genes in the JA biosynthesis pathway and further reduces the JA content in etiolated rice seedlings (Figures 5A to 5C). Third, overexpression of ethylene signaling components MHZ7/OsEIN2 and OsEIL2 inhibits GY1 expression (Figures 5D and 5E). Fourth, mutation of MHZ7/OsEIN2 in the mhz7/osein2 mutant disrupts the ethylene-mediated inhibition of GY1 (Figure 5D). Fifth, OsEIL2 directly binds to the promoter region of GY1, as demonstrated by EMSA and ChIP-PCR assays, and represses GY1 promoter activity in a tobacco leaf transient expression assay (Figures 5G to 5J). Sixth, double mutant analysis reveals that GY1 acts downstream of MHZ7/OsEIN2 to affect mesocotyl/coleoptile growth (Figures 6A and 6B). Seventh, JA application to the MHZ7/OsEIN2-overexpressing plants reduces mesocotyl/coleoptile elongation, suggesting that JA acts downstream of ethylene signaling for functional regulation (Figures 6C and 6D). All of this evidence demonstrates that ethylene interacts with JA to control mesocotyl/coleoptile elongation during the early emergence of rice seedlings.
It should be noted that although the gy1 mutant already has a relatively long mesocotyl/coleoptile, ethylene treatment further promotes mesocotyl/coleoptile elongation (Supplemental Figures 1A to 1C). Additionally, it seems that the gy1 mutant was more sensitive to ethylene than Nip during seedling emergence under soil-grown conditions. These facts suggest that, in addition to the inhibition of GY1-mediated JA biosynthesis, ethylene may regulate mesocotyl/coleoptile growth through other downstream mechanisms/factors. It has been reported that ethylene induces cell wall acidification and increases the extensibility of cells in Rumex palustris (Vreeburg et al., 2005). This is largely attributed to the acidic pH optimum for expansin activity (McQueen-Mason et al., 1992). Furthermore, ethylene regulates the stability of intracellular microtubules through WAVE-DAMPENED2-LIKE5 to mediate etiolated hypocotyl cell elongation in Arabidopsis (Sun et al., 2015). In addition, many loci have been identified that may regulate mesocotyl growth. Using a population of backcross inbred lines, five QTL have been found to control mesocotyl length in rice (Lee et al., 2012). GY1 overlaps with one of these QTL. Through genome-wide association analysis of 270 rice accessions, Wu et al. (2015) identified 16 loci that control mesocotyl growth in rice seedlings grown in water or in sand. Recently, using the same genome-wide association analysis method, Lu et al. (2016) analyzed 469 rice accessions and discovered 23 loci involved in the regulation of mesocotyl/shoot growth. Dissection of all these loci may reveal the complete mechanisms involved in mesocotyl regulation.
When analyzing the role of ethylene signaling in GY1 inhibition, MHZ7/OsEIN2 appeared to be fully required for ethylene-mediated GY1 inhibition, whereas knocking down of OsEIL2 seemed not to significantly affect the ethylene-induced GY1 inhibition in shoots (Figures 5D and 5E). However, in our previous study, we found that coleoptile growth of the OsEIL2-RNAi seedlings was insensitive to ethylene (Yang et al., 2015b). The discrepancy then existed that the same OsEIL2-RNAi plants are insensitive to ethylene with respect to ethylene-promoted coleoptile elongation but sensitive to ethylene with respect to ethylene-induced GY1 inhibition. This phenomenon may be explained by the fact that although OsEIL2 expression was reduced in the OsEIL2-RNAi plants, significant levels of residual OsEIL2 transcripts remained (Yang et al., 2015b) and that the resulting OsEIL2 activity may be enough to inhibit GY1 expression, but not reach the threshold to promote coleoptile elongation in response to ethylene. Alternatively, differential sensitivity to ethylene may be one of the reasons for different responses. Other explanations/possibilities cannot be excluded.
Natural Variation of 376T SNP in GY1 Correlates with Mesocotyl Elongation in Rice Accessions
Although GY1 mutation-mediated mesocotyl elongation may facilitate seedling emergence from soil, this mutation affects flowering development and yield-related traits (Supplemental Table 2). Such a defect prevents the use of the GY1 mutation in breeding efforts. We thus examined whether elite natural alleles are present in different rice accessions. After analyzing the resequencing data of 3000 rice accessions (Li et al., 2014), only one natural variant of GY1 was discovered, which harbors a 376T instead of the original 376G present in Nip. This elite GY1376T allele is positively correlated with mesocotyl elongation in rice. Introducing a wild-type GY1376G into the Kasalath accession, which harbors the GY1376T and has a long mesocotyl and coleoptile phenotype, increases the JA content and largely inhibits mesocotyl and coleoptile growth, indicating that GY1 may be the major factor regulating mesocotyl and coleoptile growth. Indeed, GY1 is located within a major QTL for mesocotyl regulation, which was identified from backcross inbred lines developed from Kasalath and Nipponbare (Lee et al., 2012). From these analyses, we identified an elite natural variation of GY1376T, which promotes mesocotyl elongation and has no yield penalty. This allele may be introduced into accessions lacking it by breeding to produce new rice cultivars, and these cultivars should be suitable for growing in dry land due to their high emergence rate.
The 141 rice varieties harboring 376T in GY1 mainly belong to the subgroups Aus/boro, Indica type, and Aromatic type (basmati/sandri type) (Supplemental Table 3). These accessions mainly originated from Asian countries with tropical monsoon climates, including Bangladesh, India, Pakistan, and Nepal. Long-term cultivation and domestication may have selected the GY1376T variant, which enables the plant to withstand such climates. It should be noted that only a small proportion of Japonica rice has this allele (Supplemental Table 3). The elite allele should be introduced into Japonica varieties to test the effect of this allele on mesocotyl and coleoptile elongation and rice adaptation to dry upland.
Taken together, our study reveals that ethylene-inhibited GY1-mediated JA biosynthesis promotes mesocotyl/coleoptile elongation in etiolated rice seedlings. We further associate an elite natural allele GY1376T with mesocotyl elongation in different rice accessions. Our study provides insight into the mechanism underlying mesocotyl/coleoptile elongation, and this knowledge should facilitate the practical use of the elite allele in rice breeding programs.
METHODS
Plant Materials and Growth Conditions
The gy1 mutant used in this study was in the Japonica background (Oryza sativa cv Nipponbare) and isolated from a T-DNA insertion population (Ma et al., 2013). The cpm2 mutant (O. sativa cv Nihonmasari) was kindly provided by Moritoshi Iino (Graduate School of Science, Osaka City University). The seeds of various rice accessions were kindly provided by Zhi-Kang Li (Institute of Crop Sciences/National Key Facilities for Crop Gene Resources and Genetic Improvement, Chinese Academy of Agricultural Sciences). For material propagation, crossing, and investigation of agronomic traits, rice plants were cultivated at the Experimental Station of the Institute of Genetics and Developmental Biology in Beijing during the natural growing seasons from May to October.
Seeds were germinated in water at 37°C in darkness for 2 d as previously described (Ma et al., 2013). To measure the mesocotyls and coleoptiles, the germinated seeds were incubated in darkness at 28°C and grown for 3 to 4 d as indicated in each experiment. For ethylene treatments, the germinated seeds were incubated in darkness at 28°C for 2 to 3 d and then treated with 10 ppm ethylene (Ma et al., 2013). For the seedling emergence experiment, germinated seeds were sown at a 2-cm depth in pots containing soil. The pots were placed under 20,000 lux white fluorescent light (14 h light/10 h dark) at 28°C and with 75% relative humidity.
Map-Based Cloning
To determine the GY1 locus, the mutant gy1 was crossed with an Indica variety, cv TN1. The genomic DNA of etiolated seedlings from members of the F2 population exhibiting the mutant phenotype was extracted using the SDS method (Dellaporta et al., 1983). For rough mapping, a DNA pool generated from 15 individuals with the mutant phenotype was used. A total of 927 individuals with the mutant phenotype selected from the F2 population were used for fine mapping. The insertion-deletion (Idl) markers and simple sequence repeat markers were the same as previously described (Ma et al., 2013). The GY1 locus was mapped to chromosome 1 between Idl1-39.15 (5′-TTGTTGTGCATACTACAGCTAG-3′ and 5′-CTAAGCTAATTTCAAGACTGCC-3′) and Idl1-39.20 (5′-TGATGCAGTCAACTGGCTTG-3′ and 5′-ACAATCAACGGGTCGACATC-3′), which is a 48-kb region containing three genes. The candidate gene was finally determined by sequencing all three genes in this region. The gy1 mutation was confirmed via the dCAPS (derived cleaved amplified polymorphic sequence) marker. The restriction enzyme site was introduced by reverse primer 5′-acgagacgagcacgtAGCGC-3′ (519th nucleotide). When amplifying GY1 (GY1518G) and gy1 (GY1518A), we could generate the Aor51HI restriction site (AGCGCT) in the product of gy1 but not in the product of GY1 (see Supplemental Table 7). The PCR product was then cleaved by Aor51HI, and the resulting product of gy1 was shorter than that of GY1.
Vector Construction and Rice Transformation
For construction of the complementation plasmid pC-GY1, a fragment of GY1 was inserted into the pCAMBIA2300 vector with SmaI and Sse8387 I sites to generate pC-GY1. This insertion fragment of 4806 bp was a GY1 genomic sequence containing the 1308-bp GY1 coding sequence, the 2022-bp promoter sequence upstream of the ATG start codon of GY1, and the 1476-bp sequence downstream of the TGA stop codon of GY1.
For subcellular localization, the pBI-GY1-GFP plasmid was produced. The coding sequence of GY1 was digested with BamHI/HindIII and cloned into the pBI221-GFP vector that had also been digested with BamHI/HindIII. The GFP coding sequence was fused in frame to the 3′ end of GY1.
For the transcriptional regulation assay in tobacco leaves, the coding sequence (2500 bp) of OsEIL2 was inserted into the pCAMBIA2300-35S-ocs vector to generate the effector plasmid 35S:OsEIL2. The GY1 promoter fragment (2022 bp) upstream from the ATG start codon of GY1 was cloned into pGWB435 (Invitrogen) to generate the reporter plasmid pGY1:LUC.
The primers used to generate the constructs are listed in Supplemental Table 5.
The pC-GY1 plasmid was transfected into Agrobacterium tumefaciens strain EHA105 through electroporation, and the strain harboring the construct was then transformed into the gy1 mutant or the Kasalath variety for complementation tests. Rice transformation was performed as described previously (Wuriyanghan et al., 2009). The complemented gy1 mutant and Kasalath variety were genotyped by dCAPS marker analysis using the primers listed in Supplemental Table 7. The PCR products from gy1 and Kasalath were digested with the restriction enzymes Aor51HI and FspI, respectively.
Subcellular Localization Assays
To determine the subcellular localization of GY1, the pBI-GY1-GFP construct was transformed into rice protoplasts and Arabidopsis protoplasts by polyethylene glycol-mediated transfection as described (Bart et al., 2006; Yang et al., 2015b). After 12 to 16 h of incubation at 28°C under darkness, GFP fluorescence was observed with a confocal laser scanning microscope (Leica TCS SP5).
Phylogenetic Analysis
The protein sequences of GY1 homologs in Arabidopsis thaliana and rice were downloaded from Phytozome v.11 and then aligned with ClustalX v2.1 (Supplemental Data Set 1). The neighbor-joining method was used to construct a phylogenetic tree with 1000 replicates in Phylip v3.69. The tree was viewed in MEGA v.7.
Lipid Analysis
Lipid extractions for rice tissues were performed using a modified version of the Bligh and Dyer’s protocol as previously described (Lam et al., 2014b). Extracted organic fractions were pooled and dried in miVac (Genevac). Samples were stored at −80°C until further analysis.
Polar lipids were analyzed using an Agilent 1260 HPLC system coupled with a triple quadrupole/ion trap mass spectrometer (5500Qtrap; SCIEX) as described previously (Lam et al., 2014a). Individual lipid species were quantified by referencing to spiked internal standards. Galactolipids (MGDGs and DGDGs) were separated using a Phenomenex Kinetex 2.6µ-C18 column as previously described (Cheong et al., 2014).
Protein Expression and PLA Activity Assay
The codons of GY1, gy1 (GY1518A), and GY1376T were optimized for expression in Escherichia coli (http://www.jcat.de) and synthesized. The fragments were cloned into pET-SUMO (Invitrogen) by TA cloning. The His6-SUMO-tagged fusion proteins were expressed in E. coli BL21 (DE3), purified using Ni-NTA (Novagen), and eluted with buffer containing 25 mM Tris-HCl (pH 7.4), 150 mM NaCl, and 250 mM imidazole. Imidazole in protein solution was removed via desalting columns (Thermo Scientific) before activity analysis.
The PLA activity was determined as described (Ishiguro et al., 2001) with some modifications. Dye-labeled 1-BODIPY FL C11-2-BODIPY FL C11-PC (Molecular Probes) was used as substrate for measuring PLA activity. In brief, 30 μL of 2 mM PLA substrate was mixed with 30 μL of 10 mM dioleoyl-phosphatidylcholine and 30 μL of 10 mM dioleoyl-phosphatidylglycerol in 5 mL reaction buffer (50 mM Tris-HCl, 140 mM NaCl, and 1 mM CaCl2, pH 7.4) to generate substrate-liposome mix. Then, ∼20 μg purified proteins and 50 μL substrate-liposome mix in total 100 μL reaction buffer were incubated at room temperature for 30 min. The fluorescence was measured at multiple time points to monitor the kinetics using a microplate reader equipped for excitation at 450 nm and fluorescence emission at 515 nm. The known activity unit of Lecitase Ultra (Novozymes) was used as a positive control to obtain the standard curve. SUMO protein was used as no-PLA control to correct for background fluorescence.
Measurement of JA Content and Ethylene Production
For JA measurement in water-grown seedlings, 2-d-old etiolated seedlings of Nip and gy1 were treated with or without 10 ppm ethylene for 24 h, and the shoots, which contain the coleoptiles and the true leaves, were harvested. Each sample was ground in liquid nitrogen, weighed (∼200 mg for each sample), and stored at −80°C until extraction.
For JA measurement during seedling emergence from soil, different volumes of 0.6% Agar A were poured into 45-mL vials to generate supporting matrix with different heights. Germinated seeds of Nip were then placed on the surface of solidified 0.6% Agar A and covered with different depths of sterilized soil (0, 1, and 2 cm) with water saturation. Finally, the agar matrix (0 cm), and the agar matrix plus the soil (1 and 2 cm) have the same height. The 3-d-old seedlings were ground in liquid nitrogen, weighed (∼200 mg for each sample), and stored at −80°C until extraction.
JA was purified and measured as previously described with some changes in detection conditions (Fu et al., 2012).
For ethylene production in water-grown seedlings, the germinated seeds were cultured in darkness in a 45-mL uncapped vial for 7 d at 28°C, and then the vials were sealed with a rubber cap for 15 h. After this treatment, ethylene emission was measured.
For ethylene production during seedling emergence from soil, a similar culturing method was used as was used to measure JA content during seedling emergence from soil in uncapped vials, and then the vials were sealed with a rubber cap for 20 h. After the treatment, ethylene emission was measured.
Ethylene production was determined by gas chromatography (GC2014; Shimadzu) using an instrument equipped with a flame ionization detector.
JA and ethylene measurements were repeated three times, and each sample had three biological and technical replicates. The averages and standard deviations are shown.
MeJA Treatment to Rescue the Mutant Phenotype
To rescue the long mesocotyl and coleoptile phenotype of gy1 with MeJA (Sigma-Aldrich), 0.01, 0.1, 1, and 10 μM MeJA were used to obtain a dose-response curve for the Nip coleoptile. The MeJA solutions were dissolved in ethanol, and equivalent volumes of ethanol were added for the mock treatment as for the control. A concentration of 1 μM MeJA was used to rescue the mutant phenotype. Germinated seeds were placed on stainless steel sieves in 5.5-liter air-tight plastic boxes, and 1.5 liters of MeJA solution and mock solution were added to these plastic boxes separately. After growth for 3 d at 28°C in darkness, the lengths of the coleoptiles and mesocotyls were measured.
Gene Expression Analysis by qPCR
Total RNA was extracted with TRIzol reagent (Invitrogen). Using a Turbo DNA-free Kit (Invitrogen) and a cDNA synthesis kit (TaKaRa), the acquired cDNA samples were used to perform qPCR with the LightCycler 480II (Roche). OsActin2 was used as the internal control. The primers used are listed in Supplemental Table 5.
ChIP-PCR Analysis
For the ChIP assay, the OsEIL2-GFP cassette was cloned into the pCAMBIA2300-35S-ocs vector to generate the 35S:OsEIL2:GFP plasmid, which was then transformed into Nip by Agrobacterium-mediated transfection. Shoots (1.5 g) of 3-d-old etiolated seedlings of the Nip and OsEIL2-GFP line were prepared for ChIP assays. In brief, each sample was cross-linked in 37 mL cross-link buffer (1% formaldehyde, 0.1 M sucrose, 10 mM sodium phosphate, and 50 mM NaCl, pH 8.0) under a vacuum in an exicator for 30 min, and then 2.5 mL of 2 M glycine was added to quench cross-linking under a vacuum. Chromatin was isolated and resuspended in 75 μL lysis buffer (1% SDS, 50 mM Tris-HCl, pH 8.0, and 10 mM EDTA). Then, 625 μL of dilution buffer (1.1% Triton X-100, 50 mM Tris-HCl, pH 8.0, 167 mM NaCl, 1.2 mM EDTA, 1 mM PMSF, and one tablet of protease inhibitor cocktail) was added and the sample was sonicated with a Bioruptor for 10 min on ice (10 s on/45 s off cycles). The chromatin sample was then incubated at 4°C overnight with GFP antibody (A6455; Invitrogen). Immunoprecipitated complexes were collected using magnetic protein G/A beads (Invitrogen) and washed with low salt (150 mM NaCl, 0.1% SDS, 1% Triton X-100, 2 mM EDTA, and 20 mM Tris-HCl, pH 8.0), high salt (500 mM NaCl, 0.1% SDS, 1% Triton X-100, 2 mM EDTA, and 20 mM Tris-HCl, pH 8.0), and LiCl washing (0.25 M LiCl, 1% Nonidet P-40, 1% sodium deoxycholate, 1 mM EDTA, and 10 mM Tris-HCl, pH 8.0) buffer successively. The immunoprecipitated complexes were incubated in lysis buffer at 65°C for 4 h. DNA was recovered with a QIAquick PCR purification kit (Qiagen) and analyzed by real-time PCR using primers as described (Supplemental Table 5). The chromatin isolated before precipitation was used as an input control, and the chromatin precipitated without antibody was used as a negative control. The “% input” value of each ChIP-PCR fragment was calculated first by normalizing the fragment amount against the input value and then by normalizing the value from the transgenic plants against the value from the control plants.
Transcriptional Regulatory Activity of OsEIL2 on the GY1 Promoter in Tobacco Leaves
The reporter plasmid (pGY1:LUC) and the effector plasmids (35S:OsEIL2) were transformed into Agrobacterium strain EHA105. The strains were later resuspended in infiltration buffer (10 mM MES, 0.2 mM acetosyringone, and 10 mM MgCl2) to an ultimate concentration of optical density (600 nm = 1). Equal amounts of different combined suspensions were infiltrated into the young leaves of 5-week-old tobacco (Nicotiana tabacum) plants with syringes without needles. After growth in darkness for 12 h, the infiltrated plants were cultivated under 16 h light/8 h darkness for 48 h at 24°C. Before observation, the leaves were sprayed with 100 mM luciferin (Promega) and placed in darkness for 5 min. A low-light cooled CCD imaging apparatus (iXon; Andor Technology) was then used to observe the LUC activity of each experiment.
EMSA
Plasmid construction and protein purification of OsEIL2 (amino acids 1–354) followed previous descriptions (Yang et al., 2015b). Single-stranded complementary oligonucleotide fragments containing the EIN3 binding elements from the GY1 promoter were synthesized (Invitrogen) and biotinylated using the Biotin 3′-end DNA Labeling Kit (Thermo Fisher Scientific). Biotin end-labeled and unlabeled oligonucleotide pairs were annealed to obtain double-stranded biotin-labeled and unlabeled probes by mixing equal amounts of each single-stranded complementary oligonucleotide fragment, incubating the fragments at 95°C for 5 min, and cooling them down to room temperature slowly overnight. The EMSA reaction solutions were prepared using the LightShift Chemiluminescent EMSA Kit (Thermo Fisher Scientific). After the reactions, the protein-probe mixtures were separated on a 6% nondenaturing neutral polyacrylamide gel in 0.5× TBE and transferred to a nylon membrane (GE). After UV light cross-linking, the DNA on the membrane was detected with the Chemiluminescent Nucleic Acid Detection Module (Thermo Fisher Scientific).
The oligonucleotide sequences used are listed in Supplemental Table 6.
RNA-Seq Library Preparation and Bioinformatics Analysis
Seeds of Nip and the gy1 mutant were germinated at 37°C in darkness for 2 d as previously described (Ma et al., 2013). The germinated seeds were sown at a 4-cm depth in pots containing soil, and the seedlings were grown under 14 h light/10 h dark at 28°C for 3 d. The etiolated seedlings were removed from the pots, and the mesocotyls were harvested. The total RNA was extracted with CTAB buffer according to previous descriptions (Fu et al., 2004; Yin et al., 2016). The library preparation and bioinformatics analysis followed previous descriptions (Yang et al., 2015b).
Agronomic Trait Analysis
After harvest, Nip and gy1 were evaluated for agronomic traits and 25 plants from each material were used for measurements. For each plant, plant height and yield-related traits were measured. Seed-setting rate was calculated as filled grain number/total grain number. Other traits were also measured, and the average from 25 plants was provided for each genotype in Supplemental Table 2. All the data were analyzed by one-way ANOVA (LSD and S-N-K) for test groups with SPSS 18.0.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: LOC_Os01g67430 (GY1), LOC_Os02g10120 (OsLOX), LOC_Os03g55800 (OsAOS1), LOC_Os03g12500 (OsAOS2), LOC_Os03g32314 (OsAOC), LOC_Os08g35740 (OsOPR7), LOC_Os01g60770 (OsEXPA2), LOC_Os05g39990 (OsEXPA4), LOC_Os10g40730 (OsEXPB4), LOC_Os10g40700 (OsEXPB6), LOC_Os02g44108 (OsEXPB11), LOC_Os03g04020 (OsEXPLA1), and LOC_Os10g36650 (OsActin2).
Supplemental Data
Supplemental Figure 1. Ethylene Response and Ethylene Production of gy1.
Supplemental Figure 2. Emergence Rate of gy1 Compared with the Nip Control.
Supplemental Figure 3. Phenotype of gy1 and GY1-Complemental Plants at Maturity Stage.
Supplemental Figure 4. GY1 Subcellular Localization in Arabidopsis Protoplasts.
Supplemental Figure 5. Phylogenetic Analysis of GY1 and Its Homologs.
Supplemental Figure 6. Phenotype Analysis of Another JA Biosynthesis Mutant.
Supplemental Figure 7. MeJA Dose-Response Curve for Coleoptile Length of Etiolated Seedlings.
Supplemental Figure 8. Ethylene Inhibits JA Level and GY1 Expression in Shoots of Deetiolated Seedlings.
Supplemental Figure 9. Identification of Differentially Expressed Genes and GO Enrichment Analysis.
Supplemental Figure 10. Phenotype of Etiolated Seedlings from Different Rice Accessions.
Supplemental Table 1. Dominant/Recessive Analysis of Mutant Phenotype.
Supplemental Table 2. Agronomic Traits of Field-Grown Nip and gy1 in 2012 and 2014.
Supplemental Table 3. The Number of Allelic Variation of GY1 in Rice Accessions.
Supplemental Table 4. The Mesocotyl and Coleoptile Length in Different Genotypes of GY1 in Etiolated Seedlings of Different Varieties.
Supplemental Table 5. Primers Used for Gene Expression Analysis and Vector Construction.
Supplemental Table 6. Oligonucleotides Used for EMSA.
Supplemental Table 7. Primers Used for gy1 Allelic Mutant Analysis.
Supplemental Data Set 1. Alignments Used to Generate the Phylogeny Presented in Supplemental Figure 5.
Supplementary Material
Acknowledgments
This work was supported by the 973 project (2015CB755702), the National Natural Science Foundation of China (31530004, 31670274, and 31600980), the Transgenic Research Projects (2016ZX08009003-002), the Postdoctoral Research Fellowship (2015M581200 to C.-C.Y.), and the State Key Lab of Plant Genomics. We thank Shuang Fang and Jin-Fang Chu (National Centre for Plant Gene Research [Beijing], Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing) for determining the JA contents. We also thank Sin-Man Lam and Guang-Hou Shui (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing) for determining the lipid contents.
AUTHOR CONTRIBUTIONS
Q.X., B.M., J.-S.Z., and S.-Y.C. conceived and designed the experiments. Q.X. performed the experiments. B.M. isolated the gy1 mutant. X.L. performed the bioinformatics analysis and some of the experiments. S.-J.H. carried out the rice transformation. Y.-H.H., C.Y., C.-C.Y., H.Z., Y.Z., and W.-K.Z. prepared some of the materials. W.-S.W. and Z.-K.L. provided rice variety seeds. Q.X., J.-S.Z., and B.M. wrote the article.
Glossary
- JA
jasmonic acid
- PC
phosphatidylcholine
- EMSA
electrophoresis mobility shift assay
- QTL
quantitative trait loci
- dCAPS
derived cleaved amplified polymorphic sequence
- ChIP
chromatin immunoprecipitation
Footnotes
Articles can be viewed without a subscription.
References
- Alonso J.M., Hirayama T., Roman G., Nourizadeh S., Ecker J.R. (1999). EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284: 2148–2152. [DOI] [PubMed] [Google Scholar]
- Bannenberg G., Martínez M., Hamberg M., Castresana C. (2009). Diversity of the enzymatic activity in the lipoxygenase gene family of Arabidopsis thaliana. Lipids 44: 85–95. [DOI] [PubMed] [Google Scholar]
- Bart R., Chern M., Park C.J., Bartley L., Ronald P.C. (2006). A novel system for gene silencing using siRNAs in rice leaf and stem-derived protoplasts. Plant Methods 2: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas K.K., Neumann R., Haga K., Yatoh O., Iino M. (2003). Photomorphogenesis of rice seedlings: a mutant impaired in phytochrome-mediated inhibition of coleoptile growth. Plant Cell Physiol. 44: 242–254. [DOI] [PubMed] [Google Scholar]
- Bleecker A.B., Kende H. (2000). Ethylene: a gaseous signal molecule in plants. Annu. Rev. Cell Dev. Biol. 16: 1–18. [DOI] [PubMed] [Google Scholar]
- Boutrot F., Segonzac C., Chang K.N., Qiao H., Ecker J.R., Zipfel C., Rathjen J.P. (2010). Direct transcriptional control of the Arabidopsis immune receptor FLS2 by the ethylene-dependent transcription factors EIN3 and EIL1. Proc. Natl. Acad. Sci. USA 107: 14502–14507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q., Yuan Z., Chen M., Yin C., Luo Z., Zhao X., Liang W., Hu J., Zhang D. (2014). Jasmonic acid regulates spikelet development in rice. Nat. Commun. 5: 3476. [DOI] [PubMed] [Google Scholar]
- Chao Q., Rothenberg M., Solano R., Roman G., Terzaghi W., Ecker J.R. (1997). Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 89: 1133–1144. [DOI] [PubMed] [Google Scholar]
- Chauvin A., Caldelari D., Wolfender J.L., Farmer E.E. (2013). Four 13-lipoxygenases contribute to rapid jasmonate synthesis in wounded Arabidopsis thaliana leaves: a role for lipoxygenase 6 in responses to long-distance wound signals. New Phytol. 197: 566–575. [DOI] [PubMed] [Google Scholar]
- Cheong W.F., Wenk M.R., Shui G. (2014). Comprehensive analysis of lipid composition in crude palm oil using multiple lipidomic approaches. J. Genet. Genomics 41: 293–304. [DOI] [PubMed] [Google Scholar]
- Cho H.T., Cosgrove D.J. (2000). Altered expression of expansin modulates leaf growth and pedicel abscission in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 97: 9783–9788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi D., Lee Y., Cho H.T., Kende H. (2003). Regulation of expansin gene expression affects growth and development in transgenic rice plants. Plant Cell 15: 1386–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dave A., Hernández M.L., He Z., Andriotis V.M.E., Vaistij F.E., Larson T.R., Graham I.A. (2011). 12-Oxo-phytodienoic acid accumulation during seed development represses seed germination in Arabidopsis. Plant Cell 23: 583–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaporta S.L., Wood J., Hicks J.B. (1983). A plant DNA minipreparation: Version II. Plant Mol. Biol. Report. 1: 19–21. [Google Scholar]
- Dilday R.H., Mgonja M.A., Amonsilpa S.A., Collins F.C., Wells B.R. (1990). Plant height vs. mesocotyl and coleoptile elongation in rice: linkage or pleiotropism. Crop Sci. 30: 815–818. [Google Scholar]
- Eastmond P.J., Graham I.A. (2000). The multifunctional protein AtMFP2 is co-ordinately expressed with other genes of fatty acid beta-oxidation during seed germination in Arabidopsis thaliana (L.) Heynh. Biochem. Soc. Trans. 28: 95–99. [DOI] [PubMed] [Google Scholar]
- Eastmond P.J., Hooks M.A., Williams D., Lange P., Bechtold N., Sarrobert C., Nussaume L., Graham I.A. (2000). Promoter trapping of a novel medium-chain acyl-CoA oxidase, which is induced transcriptionally during Arabidopsis seed germination. J. Biol. Chem. 275: 34375–34381. [DOI] [PubMed] [Google Scholar]
- Ellinger D., Stingl N., Kubigsteltig I.I., Bals T., Juenger M., Pollmann S., Berger S., Schuenemann D., Mueller M.J. (2010). DONGLE and DEFECTIVE IN ANTHER DEHISCENCE1 lipases are not essential for wound- and pathogen-induced jasmonate biosynthesis: redundant lipases contribute to jasmonate formation. Plant Physiol. 153: 114–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feussner I., Wasternack C. (2002). The lipoxygenase pathway. Annu. Rev. Plant Biol. 53: 275–297. [DOI] [PubMed] [Google Scholar]
- Fu J., Chu J., Sun X., Wang J., Yan C. (2012). Simple, rapid, and simultaneous assay of multiple carboxyl containing phytohormones in wounded tomatoes by UPLC-MS/MS using single SPE purification and isotope dilution. Anal. Sci. 28: 1081–1087. [DOI] [PubMed] [Google Scholar]
- Fu X.H., Deng S.L., Su G.H., Shi S.H. (2004). Isolating high-quality RNA from mangroves without liquid nitrogen. Plant Mol. Biol. Report. 22: 197. [Google Scholar]
- Guo H., Ecker J.R. (2004). The ethylene signaling pathway: new insights. Curr. Opin. Plant Biol. 7: 40–49. [DOI] [PubMed] [Google Scholar]
- Hayashi H., De Bellis L., Ciurli A., Kondo M., Hayashi M., Nishimura M. (1999). A novel acyl-CoA oxidase that can oxidize short-chain acyl-CoA in plant peroxisomes. J. Biol. Chem. 274: 12715–12721. [DOI] [PubMed] [Google Scholar]
- Helms R.S., Dilday R.H., Mgonja M.A., Amonsilpa S. (1989). Genetic and plant growth regulator manipulation of rice (Oryza sativa L.) mesocotyl and coleoptile lengths. Proc. Ark. Acad. Sci 43: 42–45. [Google Scholar]
- Hu Z., Yan H., Yang J., Yamaguchi S., Maekawa M., Takamure I., Tsutsumi N., Kyozuka J., Nakazono M. (2010). Strigolactones negatively regulate mesocotyl elongation in rice during germination and growth in darkness. Plant Cell Physiol. 51: 1136–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z., Yamauchi T., Yang J., Jikumaru Y., Tsuchida-Mayama T., Ichikawa H., Takamure I., Nagamura Y., Tsutsumi N., Yamaguchi S., Kyozuka J., Nakazono M. (2014). Strigolactone and cytokinin act antagonistically in regulating rice mesocotyl elongation in darkness. Plant Cell Physiol. 55: 30–41. [DOI] [PubMed] [Google Scholar]
- Hua J., Meyerowitz E.M. (1998). Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell 94: 261–271. [DOI] [PubMed] [Google Scholar]
- Huang J., Takano T., Akita S. (2000). Expression of α-expansin genes in young seedlings of rice (Oryza sativa L.). Planta 211: 467–473. [DOI] [PubMed] [Google Scholar]
- Hyun Y., et al. (2008). Cooperation and functional diversification of two closely related galactolipase genes for jasmonate biosynthesis. Dev. Cell 14: 183–192. [DOI] [PubMed] [Google Scholar]
- Ishiguro S., Kawai-Oda A., Ueda J., Nishida I., Okada K. (2001). The DEFECTIVE IN ANTHER DEHISCENCE1 gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 13: 2191–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju C., et al. (2012). CTR1 phosphorylates the central regulator EIN2 to control ethylene hormone signaling from the ER membrane to the nucleus in Arabidopsis. Proc. Natl. Acad. Sci. USA 109: 19486–19491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieber J.J., Rothenberg M., Roman G., Feldmann K.A., Ecker J.R. (1993). CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 72: 427–441. [DOI] [PubMed] [Google Scholar]
- Lam S.M., Tong L., Duan X., Petznick A., Wenk M.R., Shui G. (2014a). Extensive characterization of human tear fluid collected using different techniques unravels the presence of novel lipid amphiphiles. J. Lipid Res. 55: 289–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam S.M., Wang Y., Duan X., Wenk M.R., Kalaria R.N., Chen C.P., Lai M.K.P., Shui G. (2014b). Brain lipidomes of subcortical ischemic vascular dementia and mixed dementia. Neurobiol. Aging 35: 2369–2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.S., Sasaki K., Higashitani A., Ahn S.N., Sato T. (2012). Mapping and characterization of quantitative trait loci for mesocotyl elongation in rice (Oryza sativa L.). Rice (N. Y.) 5: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Xue D., Gao Z., Yan M., Xu W., Xing Z., Huang D., Qian Q., Xue Y. (2009). A putative lipase gene EXTRA GLUME1 regulates both empty-glume fate and spikelet development in rice. Plant J. 57: 593–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Ma M., Feng Y., Li H., Wang Y., Ma Y., Li M., An F., Guo H. (2015). EIN2-directed translational regulation of ethylene signaling in Arabidopsis. Cell 163: 670–683. [DOI] [PubMed] [Google Scholar]
- Li Y., Jones L., McQueen-Mason S. (2003). Expansins and cell growth. Curr. Opin. Plant Biol. 6: 603–610. [DOI] [PubMed] [Google Scholar]
- Li Z.K., et al.; 3,000 Rice Genomes Project (2014). The 3,000 rice genomes project. Gigascience 3: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Zhang S., Sun N., Liu H., Zhao Y., Liang Y., Zhang L., Han Y. (2015). Functional diversity of jasmonates in rice. Rice (NY) 8: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O., Piqueras R., Sánchez-Serrano J.J., Solano R. (2003). ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15: 165–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q., Zhang M., Niu X., Wang C., Xu Q., Feng Y., Wang S., Yuan X., Yu H., Wang Y., Wei X. (2016). Uncovering novel loci for mesocotyl elongation and shoot length in indica rice through genome-wide association mapping. Planta 243: 645–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma B., Chen S.Y., Zhang J.S. (2010). Ethylene signaling in rice. Chin. Sci. Bull. 55: 2204–2210. [Google Scholar]
- Ma B., Yin C.C., He S.J., Lu X., Zhang W.K., Lu T.G., Chen S.Y., Zhang J.S. (2014). Ethylene-induced inhibition of root growth requires abscisic acid function in rice (Oryza sativa L.) seedlings. PLoS Genet. 10: e1004701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma B., et al. (2013). Identification of rice ethylene-response mutants and characterization of MHZ7/OsEIN2 in distinct ethylene response and yield trait regulation. Mol. Plant 6: 1830–1848. [DOI] [PubMed] [Google Scholar]
- McQueen-Mason S., Durachko D.M., Cosgrove D.J. (1992). Two endogenous proteins that induce cell wall extension in plants. Plant Cell 4: 1425–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx I.A.M.A., Thomma B.P.H.J., Buchala A., Métraux J.P., Broekaert W.F. (1998). Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 10: 2103–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao H., Shen Z., Huang S.S.C., Schmitz R.J., Urich M.A., Briggs S.P., Ecker J.R. (2012). Processing and subcellular trafficking of ER-tethered EIN2 control response to ethylene gas. Science 338: 390–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riemann M., Riemann M., Takano M. (2008). Rice JASMONATE RESISTANT 1 is involved in phytochrome and jasmonate signalling. Plant Cell Environ. 31: 783–792. [DOI] [PubMed] [Google Scholar]
- Riemann M., Muller A., Korte A., Furuya M., Weiler E.W., Nick P. (2003). Impaired induction of the jasmonate pathway in the rice mutant hebiba. Plant Physiol. 133: 1820–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riemann M., et al. (2013). Identification of rice Allene Oxide Cyclase mutants and the function of jasmonate for defence against Magnaporthe oryzae. Plant J. 74: 226–238. [DOI] [PubMed] [Google Scholar]
- Rzewuski G., Sauter M. (2008). Ethylene biosynthesis and signaling in rice. Plant Sci. 175: 32–42. [Google Scholar]
- Schaller A., Stintzi A. (2009). Enzymes in jasmonate biosynthesis - structure, function, regulation. Phytochemistry 70: 1532–1538. [DOI] [PubMed] [Google Scholar]
- Schaller F., Biesgen C., Müssig C., Altmann T., Weiler E.W. (2000). 12-Oxophytodienoate reductase 3 (OPR3) is the isoenzyme involved in jasmonate biosynthesis. Planta 210: 979–984. [DOI] [PubMed] [Google Scholar]
- Seo Y.S., Kim E.Y., Kim W.T. (2011). The Arabidopsis sn-1-specific mitochondrial acylhydrolase AtDLAH is positively correlated with seed viability. J. Exp. Bot. 62: 5683–5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharova E.I. (2007). Expansins: proteins involved in cell wall softening during plant growth and morphogenesis. Russ. J. Plant Physiol. 54: 713–727. [Google Scholar]
- Shi H., Liu R., Xue C., Shen X., Wei N., Deng X.W., Zhong S. (2016a). Seedlings transduce the depth and mechanical pressure of covering soil using COP1 and ethylene to regulate EBF1/EBF2 for soil emergence. Curr. Biol. 26: 139–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Shen X., Liu R., Xue C., Wei N., Deng X.W., Zhong S. (2016b). The red light receptor Phytochrome B directly enhances Substrate-E3 Ligase interactions to attenuate ethylene responses. Dev. Cell 39: 597–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A., Baranwal V., Shankar A., Kanwar P., Ranjan R., Yadav S., Pandey A., Kapoor S., Pandey G.K. (2012). Rice phospholipase A superfamily: organization, phylogenetic and expression analysis during abiotic stresses and development. PLoS One 7: e30947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S., Qi T., Wasternack C., Xie D. (2014a). Jasmonate signaling and crosstalk with gibberellin and ethylene. Curr. Opin. Plant Biol. 21: 112–119. [DOI] [PubMed] [Google Scholar]
- Song S., Huang H., Gao H., Wang J., Wu D., Liu X., Yang S., Zhai Q., Li C., Qi T., Xie D. (2014b). Interaction between MYC2 and ETHYLENE INSENSITIVE3 modulates antagonism between jasmonate and ethylene signaling in Arabidopsis. Plant Cell 26: 263–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick P.E., Tiryaki I., Rowe M.L. (2002). Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation. Plant Cell 14: 1405–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Ma Q., Mao T. (2015). Ethylene regulates the Arabidopsis microtubule-associated protein WAVE-DAMPENED2-LIKE5 in etiolated hypocotyl elongation. Plant Physiol. 169: 325–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svyatyna K., Riemann M. (2012). Light-dependent regulation of the jasmonate pathway. Protoplasma 249 (Suppl 2): S137–S145. [DOI] [PubMed] [Google Scholar]
- Takahashi N. (1978). Adaptive importance of mesocotyl and coleoptile growth in rice under different moisture regimes. Aust. J. Plant Physiol. 5: 511–517. [Google Scholar]
- Takano M., Kanegae H., Shinomura T., Miyao A., Hirochika H., Furuya M. (2001). Isolation and characterization of rice phytochrome A mutants. Plant Cell 13: 521–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vreeburg R.A., Benschop J.J., Peeters A.J., Colmer T.D., Ammerlaan A.H., Staal M., Elzenga T.M., Staals R.H., Darley C.P., McQueen-Mason S.J., Voesenek L.A. (2005). Ethylene regulates fast apoplastic acidification and expansin A transcription during submergence-induced petiole elongation in Rumex palustris. Plant J. 43: 597–610. [DOI] [PubMed] [Google Scholar]
- Wang C., Zien C.A., Afitlhile M., Welti R., Hildebrand D.F., Wang X. (2000). Involvement of phospholipase D in wound-induced accumulation of jasmonic acid in Arabidopsis. Plant Cell 12: 2237–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe H., Takahashi K., Saigusa M. (2001). Morphological and anatomical effects of abscisic acid (ABA) and fluridone (FLU) on the growth of rice mesocotyls. Plant Growth Regul. 34: 273–275. [Google Scholar]
- Wei F.J., et al. (2016). Somaclonal variation does not preclude the use of rice transformants for genetic screening. Plant J. 85: 648–659. [DOI] [PubMed] [Google Scholar]
- Wen X., Zhang C., Ji Y., Zhao Q., He W., An F., Jiang L., Guo H. (2012). Activation of ethylene signaling is mediated by nuclear translocation of the cleaved EIN2 carboxyl terminus. Cell Res. 22: 1613–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilen R.W., Ewan B.E., Gusta L.V. (1994). Interaction of abscisic acid and jasmonic acid on the inhibition of seed germination and the induction of freezing tolerance. Can. J. Bot. 72: 1009–1017. [Google Scholar]
- Wu J., et al. (2015). Genome-wide Association Study (GWAS) of mesocotyl elongation based on re-sequencing approach in rice. BMC Plant Biol. 15: 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuriyanghan H., et al. (2009). The ethylene receptor ETR2 delays floral transition and affects starch accumulation in rice. Plant Cell 21: 1473–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Lu X., Ma B., Chen S.Y., Zhang J.S. (2015a). Ethylene signaling in rice and Arabidopsis: conserved and diverged aspects. Mol. Plant 8: 495–505. [DOI] [PubMed] [Google Scholar]
- Yang C., Ma B., He S.J., Xiong Q., Duan K.X., Yin C.C., Chen H., Lu X., Chen S.Y., Zhang J.S. (2015b). MAOHUZI6/ETHYLENE INSENSITIVE3-LIKE1 and ETHYLENE INSENSITIVE3-LIKE2 regulate ethylene response of roots and coleoptiles and negatively affect salt tolerance in rice. Plant Physiol. 169: 148–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin C.C., Ma B., Wang W., Xiong Q., Zhao H., Chen S.Y., Zhang J.S. (2016). RNA extraction and preparation in rice (Oryza sativa). Curr. Protoc. Plant Biol. 1: 411–418. [DOI] [PubMed] [Google Scholar]
- Yin C.C., et al. (2015). Ethylene responses in rice roots and coleoptiles are differentially regulated by a carotenoid isomerase-mediated abscisic acid pathway. Plant Cell 27: 1061–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z., Zhang D. (2015). Roles of jasmonate signalling in plant inflorescence and flower development. Curr. Opin. Plant Biol. 27: 44–51. [DOI] [PubMed] [Google Scholar]
- Zhang B., Wu S., Zhang Y., Xu T., Guo F., Tang H., Li X., Wang P., Qian W., Xue Y. (2016). A high temperature-dependent mitochondrial lipase EXTRA GLUME1 promotes floral phenotypic robustness against temperature fluctuation in rice (Oryza sativa L.). PLoS Genet. 12: e1006152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Zhu Z., An F., Hao D., Li P., Song J., Yi C., Guo H. (2014). Jasmonate-activated MYC2 represses ETHYLENE INSENSITIVE3 activity to antagonize ethylene-promoted apical hook formation in Arabidopsis. Plant Cell 26: 1105–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S., Zhao M., Shi T., Shi H., An F., Zhao Q., Guo H. (2009). EIN3/EIL1 cooperate with PIF1 to prevent photo-oxidation and to promote greening of Arabidopsis seedlings. Proc. Natl. Acad. Sci. USA 106: 21431–21436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S., Shi H., Xue C., Wei N., Guo H., Deng X.W. (2014). Ethylene-orchestrated circuitry coordinates a seedling’s response to soil cover and etiolated growth. Proc. Natl. Acad. Sci. USA 111: 3913–3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C., Gan L., Shen Z., Xia K. (2006). Interactions between jasmonates and ethylene in the regulation of root hair development in Arabidopsis. J. Exp. Bot. 57: 1299–1308. [DOI] [PubMed] [Google Scholar]
- Zhu Z., et al. (2011). Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 12539–12544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler J., Stenzel I., Hause B., Maucher H., Hamberg M., Grimm R., Ganal M., Wasternack C. (2000). Molecular cloning of allene oxide cyclase. The enzyme establishing the stereochemistry of octadecanoids and jasmonates. J. Biol. Chem. 275: 19132–19138. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.