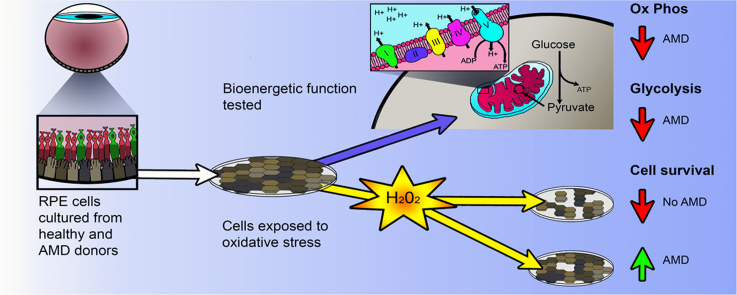
Abstract
Age-related macular degeneration (AMD) is the leading cause of blindness among older adults. It has been suggested that mitochondrial defects in the retinal pigment epithelium (RPE) underlies AMD pathology. To test this idea, we developed primary cultures of RPE to ask whether RPE from donors with AMD differ in their metabolic profile compared with healthy age-matched donors. Analysis of gene expression, protein content, and RPE function showed that these cultured cells replicated many of the cardinal features of RPE in vivo. Using the Seahorse Extracellular Flux Analyzer to measure bioenergetics, we observed RPE from donors with AMD exhibited reduced mitochondrial and glycolytic function compared with healthy donors. RPE from AMD donors were also more resistant to oxidative inactivation of these two energy-producing pathways and were less susceptible to oxidation-induced cell death compared with cells from healthy donors. Investigation of the potential mechanism responsible for differences in bioenergetics and resistance to oxidative stress showed RPE from AMD donors had increased PGC1α protein as well as differential expression of multiple genes in response to an oxidative challenge. Based on our data, we propose that cultured RPE from donors phenotyped for the presence or absence of AMD provides an excellent model system for studying “AMD in a dish”. Our results are consistent with the ideas that (i) a bioenergetics crisis in the RPE contributes to AMD pathology, and (ii) the diseased environment in vivo causes changes in the cellular profile that are retained in vitro.
Abbreviations: AMD, Age Related Macular Degeneration; ANOVA, Analysis of Variance; ARBP, 60S Acidic Ribosomal Protein P0; BEST1, Bestrophin; CAT, Catalase; cDNA, Complementary DNA; CMST, Cell Mito Stress Test; CRABP, Cellular Retinoic Acid Binding Protein; CRALBP, Cellular Retinaldehyde Binding Protein; Cyt b, Cytochrome B; CYTC, Cytochrome C; ECAR, Extra Cellular Acidification Rate; FBS, Fetal Bovine Serum; FCCP, Carbonyl Cyanide-4-(trifluoromethoxy)phenylhydrazone; FITC, Fluorescein-5-Isothiocyanate Isomer I; GAPDH, Glyceraldehyde 3-phosphate dehydrogenase; GPX1, Glutathion peroxidase 1; GSH, Glutathione; GST, Glycolytic Stress Test; GSTπ, Glutathion-S-Transferase pi; HO-1, Heme-Oxygenase; MCT3, Monocarboxylate Transporter 3; MGS, Minnesota Grading System; miRNA, Micro-RNA; MITF, Microphthalmia-associated Transcription Factor; mRNA, Messenger RNA; mtDNA, Mitochondrial DNA; NQO-1, NAD(P)H Quinone Dehydrogenase; NRF2, Nuclear Factor E2-Related Factor; OCR, Oxygen Consumption Rate; OS, Outer Segments; OxPhos, Oxidative Phosphorylation; PEDF, Pigment Epithelium-Derived Factor; PGC-1α, Peroxisome Proliferator-Activated Receptor-gamma Coactivator 1α; PMEL17, Pre-melanosome Protein 17; PPARα, Peroxisome Proliferator-Activated Receptor Alpha; PPARγ, Peroxisome Proliferator-Activated Receptor Gamma; PRDX3, Peroxiredoxin; qRT-PCR, Quantitative Reverse Transcriptase Polymerase Chain Reaction; RBP1, Retinol Binding Protein 1; RDH11, Retinal Dehydrogenase; RPE, Retinal Pigment Epithelium; rRNA, Ribosomal RNA; SOD1, Cytosolic Superoxide Dismutase; SOD2, Mitochondrial Superoxide Dismutase; SRXN1, Sulfiredoxin 1; TRYP1, Tyrosinase Related Protein
Keywords: 6 max) Age-related macular degeneration, Retinal pigment epithelium, Mitochondrial function, Glycolytic function, Antioxidants, Oxidative stress
Graphical abstract
Highlights
-
•
Primary cultures of retinal pigment epithelium (RPE) from adult donors replicate many features of RPE in vivo.
-
•
Decreased mitochondrial and glycolytic function in AMD donor RPE suggests a bioenergetic crisis contributes to AMD pathology.
-
•
The diseased environment in vivo caused changes in the cellular profile that are retained in vitro.
-
•
Altered bioenergetics and oxidation in AMD donor RPE suggest these cultures are a good model for studying "AMD in a dish".
1. Introduction
Age-related macular degeneration (AMD) is the leading cause of blindness in elderly individuals, affecting ~28% of individuals 75–85 years [29] It is predicted that 196 million people will be living with macular degeneration by 2020. As the generation of “baby boomers” grow older, this number will rise dramatically to 288 million by 2040 [46]. The number of individuals suffering from macular degeneration and other visual impairments comes with a cost. In 2013, it was estimated that the cost of visual impairment due to retinal disorders in the U.S. alone was $8.7 billion [1]. The combination of personal and public costs, as well as the large number of individuals afflicted, creates an urgent need to develop effective treatments.
AMD destroys the macula, a part of the retina supplying high acuity central vision. Patients that develop this disease lose their ability to read, drive, and recognize faces as AMD progresses. There are two forms of AMD, “wet” and “dry”, with about 10% of cases being “wet” AMD. Currently, there are treatments available for those suffering from “wet” AMD, which occurs when abnormal blood vessels grow into the retina, leading to rapid vision loss [41]. There are currently no effective treatments for “dry” AMD, characterized by loss of the retinal pigment epithelium (RPE). The RPE forms the outer blood-retinal barrier and has several key functions. RPE transport nutrients to the outer retina, absorb light and protect against photo-oxidation, regenerate the visual pigment in rhodopsin, phagocytose the tips of photoreceptors, and secrete factors required for preserving the structural integrity of the retina [44]. Because the RPE perform functions that are essential for maintaining retinal homeostasis, the loss of RPE results in photoreceptor death and blindness. A better understanding of how the environment of the diseased retina affects RPE function will provide valuable insight into disease mechanism and drive discovery of new clinical treatments that either prevent AMD or stop its progression.
The central dogma of AMD pathology has included a role for oxidative stress and oxidative damage in the retinal degeneration associated with AMD [6]. This idea has been supported by later studies in mouse models that include either global knockout of the antioxidant CuZn superoxide dismutase (SOD1) or the RPE-specific elimination of the mitochondrial manganese superoxide dismutase (SOD2) [20], [22]. The elevated retinal oxidative stress in these mouse models had detrimental effects on retinal function and caused retinal degeneration that was reminiscent of AMD. An emerging hypothesis, evolved from the idea that oxidative stress contributes to AMD pathology, involves mitochondrial dysfunction in the RPE as a prominent player in AMD pathogenesis. Strong supporting evidence from studies of human donors with AMD include the reported decrease in mitochondrial mass with disruptions in mitochondrial architecture, and an altered mitochondrial proteome evidenced by lower content of mitochondrial electron transport chain proteins [10], [33], [34] Additionally, enhanced mitochondrial DNA (mtDNA) damage has been reported in human donor RPE at stages of AMD preceding macular degeneration and vision loss [24], [45]. The ramifications of mitochondrial dysfunction include a reduced capacity for energy production, as well as detrimental effects on redox signaling and subsequent changes in gene expression.
A major limitation in understanding AMD pathology is the complexity of this multifactorial disease, which manifests in individuals over 60 years and is influenced by both environmental and genetic factors. While there are a number of good animal models available for studying specific pathways that are predicted to be involved in AMD, none of these models fully replicate all of the key features of AMD. Thus, additional model systems for studying AMD disease mechanism are needed. In this study, primary cultures of RPE from human donors with and without AMD were used to test the hypothesis that mitochondrial defects in the RPE underlie AMD pathology. We compared the bioenergetic profiles of two major energy pathways, mitochondrial oxidative phosphorylation and glycolysis. We also tested the response of these cells to an oxidative challenge. Our results show major differences in bioenergetics and oxidative stress response comparing cells from donors with or without AMD.
2. Materials and methods
2.1. Human eye procurement and grading for AMD
De-identified donor eyes were obtained from the Minnesota Lions Eye Bank (Saint Paul, MN). Eyes are obtained with the written consent of the donor or donor's family for use in medical research in accordance with the Declaration of Helsinki. The Minnesota Lions Eye Bank is licensed by the Eye Bank Association of America (accreditation #0015204) and accredited by the FDA (FDA Established Identifier 3000718538). Donor tissue is exempt from the process of Institutional Review Board approval.
Tissue handling, storage and donor exclusion criteria are as outlined previously [24], [45]. Evaluation of the presence or absence of AMD was determined by a Board Certified Ophthalmologist (Sandra R. Montezuma) from stereoscopic fundus photographs of the RPE using the criteria (RPE pigment changes and the presence, size and location of drusen) established by the Minnesota Grading System [7], [35]. Records from the Minnesota Lions Eye Bank provided demographics (age, gender, time and cause of death) for the donors used to generate RPE primary cultures (Supplement Table 1).
2.2. Cell culturing
Eyes obtained within 24 h of death were dissected and processed as described [4]. RPE cells were isolated from human donor eyecups by gently dislodging cells from Bruch's membrane with a glass rod following incubation (15 min) with 0.125% trypsin pre-heated to 37 °C. Cells were suspended in MEM and 15% serum for transport to the Ferrington laboratory, then processed for cultures as described [42] using a series of filters (70 and 40 µm) to reduce contamination of cultures with cell debris. Cells were placed in one Primaria T25 Flask and cultured in Minimum Essential Medium Eagle alpha medium (MEM-α; Sigma-Aldrich) supplemented with 15% Fetal Bovine Serum (FBS)(Atlanta Biologicals), 1 mM sodium pyruvate (Gibco), 1% nonessential amino acid (Cellgro), 50 U/mL penicillin and 50 µg/mL streptomycin (Gibco). The serum was gradually reduced to 10% FBS on day 2% and 5% FBS on day 7. Media was changed twice per week. Cells were passaged using trypsin when they reached confluence. All cell cultures were maintained in a humidified atmosphere of 95% air containing 5% CO2 at 37 °C.
Cells in passage 2 or 3 were used for characterization and functional assays. For functional assays, pigmented cells were grown to confluence in a T75 flask for approximately three months, then transferred to either 96 or 6-well plates and allowed to grow for 2–7 days. Optimal cell number and timing for procedures was experimentally determined for each assay. Cell number and condition are indicated under each experimental protocol.
2.3. Western immunoblotting
Cells (2.5×105) were grown in 6-well plates and washed with PBS and lysates were extracted on ice with either RPE lysis buffer (20 mM KCl, 50 mM Tris pH 7.8, 5 mM EDTA, 1% NP-40, 20% Glycerol) or RIPA lysis buffer (Thermo Fisher). Protein concentration was determined using a BCA assay kit (Thermo Fisher). Western blots were performed as described [31]. Proteins were resolved on SDS-PAGE gels and transferred to PVDF membranes (Millipore). Membranes were incubated overnight with primary antibodies (Supplement Table 2). The optimal protein load for each antibody was determined from initial experiments where the linear range of detection was determined. Appropriate secondary antibodies conjugated to horseradish peroxidase were used to visualize immune reactions using chemiluminescence with Super Signal West Dura Extended Duration substrate (Thermo Fisher). Immune reactions were imaged using a ChemiDoc XRS (Bio-Rad) and quantified using Quantity One software (Bio-Rad).
2.4. Immunofluorescence
Cells were grown on either fibronectin-coated chamber slides (Nunc) or transwell filters (Costar). Paraformaldehyde-fixed cells were blocked for one hour in 10% normal donkey serum and then incubated in primary antibody (Supplement Table 2) overnight. The reaction was visualized using appropriate secondary antibody. Cells were cover slipped with mounting medium containing DAPI (Vector Laboratories) and imaged with either a fluorescence microscope (Leica DM4000B) or an inverted confocal microscope (Olympus FluoView 1000). To confirm specificity of the primary antibody, control slides without primary antibody were included. H&E stained slides were imaged with an EVOS XL core microscope.
2.5. RNA isolation and qRT-PCR for RPE characterization and oxidative stress response
For RPE characterization, total RNA was prepared with the RNeasy Micro kit (Qiagen). RNA (300 ng) was used to synthesize cDNA with SuperScript III First-strand Synthesis System (ThermoFisher). Quantitative PCR analysis was performed using 2 μL of cDNA and SYBR Green (Roche) with an Eppendorf Mastercycler (Realplex2) using primers listed (Supplement Table 3). ΔCT was calculated using the housekeeping gene glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Relative expression was calculated using ΔCT values normalized to mean expression of donors without AMD.
For the oxidative stress response assay, cells were grown for 2–7 days in 6-well plates at 250,000 cells/well before treatment. Total RNA and cDNA were prepared as stated above. Expression of oxidative stress response genes was determined using quantitative reverse transcription PCR (qRT-PCR) using a BioRad iQ5 multicolor real time PCR detection system as described previously [24]. Triplicate wells of 25 μL reactions contained 1 ng cDNA, 0.2 μM Forward and Reverse primers, and 13.5 μL BioRad iQ SYBR Green Supermix. Gene information, amplicon size, annealing temperature, and primer sequences can be found in Supplement Table 3. A standard curve was included with every gene to determine efficiency.
The housekeeping gene 60 S acidic ribosomal protein P0 (ARBP) was used to calculate ΔCt of each gene of interest. To determine fold change relative to untreated controls, ΔΔCt values were calculated by subtracting the ΔCt of untreated cells from their respective hydrogen peroxide treatments at 6 or 24 h. To determine fold change relative to No AMD, ΔΔCt of each AMD donor was calculated by subtracting the mean ΔCt of No AMD cells from the ΔCt of each AMD sample for corresponding treatments. A modified Livak method was used to calculate relative expression using the efficiency for each primer.
2.6. Phagocytosis of outer segments (OS)
Sucrose gradient-purified porcine rod outer segments were obtained from Dr. Aparna Lakaraju (U Wisconsin). OS were labeled with Fluorescein-5-Isothiocyanate Isomer I (FITC) (Thermo Fisher) as described previously [37]. Labeled OS were pelleted, washed, and added to confluent RPE cultures (100,000 cells/well grown at least one month in a 4.8 cm2 well, coated with Matrigel, Corning) at a concentration of 40 OS/RPE cell. After 24-h of incubation, cells were washed with PBS, dissociated with trypsin, and collected in PBS for analysis. Flow cytometry analysis was performed using LSRII H1010 (BD Biosiences) and the data analyzed with FlowJo software.
2.7. Phagocytosis of beads
RPE cells were seeded at 1.0×105 cells / 4.8 cm2 well coated with Matrigel and grown for 24 h. One micron fluoresbrite YG Microspheres (Polyscience) were added (1×107 beads/well) and the cells cultured for an additional 24hr. To ensure that only internalized beads would be included in the measurement, cells were dissociated with Trypsin and re-seeded into a new well that was coated with Matrigel (Corning). The next day, cells were washed 3 times with DPBS (ThermoFisher) before dissociation with Trypsin and collected in DPBS. Flow cytometry analysis was performed using FALSRII H1010 (BD Biosiences) and the data analyzed with FlowJo software.
2.8. Measurement of glycolytic function using the Glycolytic Stress Test (GST)
Analysis of glycolytic function was performed on live cells using an XFe96 Extracellular Flux Analyzer (Seahorse Bioscience). Cells (4×104 cells/well) were seeded in a 96 well plate and grown for 24 h. Cells were incubated with or without 500 μM hydrogen peroxide for 24 h, washed (2X) with the GST assay medium (XF base medium DMEM supplemented with 2 mM glutamine, pH 7.4), and incubated in the GST media for 1 h at 37 °C in a non-CO2 incubator. The assay was performed using the GST assay protocol as suggested by the manufacturer (Seahorse Bioscience). Extra cellular acidification rate (ECAR) was measured under basal conditions followed by the sequential addition of 10 mM glucose, 2 µM oligomycin, and 100 mM 2-DG. A graphical description of the assay and parameters measured are provided in Supplement Fig. 1.
Fig. 1.
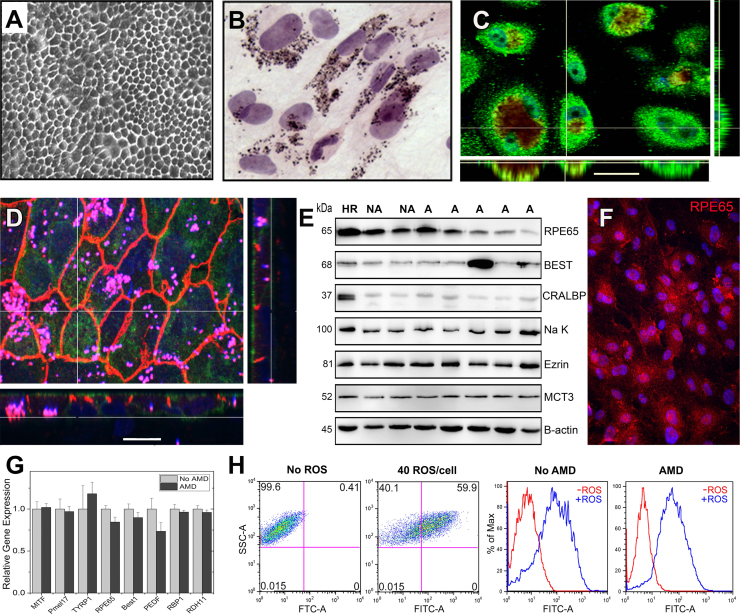
Characterization of primary human RPE cultures derived from adult donors. (A) Phase microscopy image shows confluent RPE form a monolayer with a cobblestone appearance. (B) H&E stained RPE cells visualize the pigment granules inside the cytoplasm (C,D) Confocal microscopy images of RPE cultured on transwell inserts for one month are shown. En face views of the RPE monolayer shown as maximum intensity projections through the z-axis. Also shown are cross-sections (locations shown by the white line) through the z-plane of multiple optical slices. (C) Bestrophin (green) is expressed on the basal surface. Pigment granules seen due to auto fluorescence (red) Nuclei are stained using DAPI (blue). (Scale bar 30 µm) (D) The Na-K ATPase (green) is expressed on the apical surface. ZO-1 staining (orange) marks cell borders; punctate pigment granules appear pink. Nuclei are stained using DAPI (blue). (Scale bar 15 µm) (E) RPE cultures from non-diseased (No AMD) and AMD donors contain many prototypic RPE proteins as demonstrated on Western immunoblots. Molecular mass for each protein is shown on the left. HR is RPE homogenate from a human donor. β-actin is the loading control. CRALBP, cellular retinaldehyde-binding protein; MCT3, monocarboxylate transporter 3. (F) Immunohistochemistry and fluorescent microcopy was used to detect the expression of the prototypic RPE protein, the 65 kDa Retinal Pigment Epithelium-Specific Protein RPE65. (G) RT-qPCR analysis was performed on RPE cultures from non-diseased (No AMD) (n=4) and AMD (n=7) donors. Graph shows AMD delta CT relative to the mean delta CT of the non-diseased group. Data are the mean (±SEM) normalized values. MITF, microphthalmia-associated transcription factor; Pmel17, pre-melanosome protein 1; TYRP1, tyrosinase related protein 1; Best1, Bestrophin; PEDF, pigment epithelial derived factor; RBP1,Retinol binding protein 1; RDH11, Retinal dehydrogenase. (H) FACs analysis measuring the phagocytosis of FITC-labeled rod OS by RPE from a healthy (No AMD) and an AMD donor. Dot plots (left) and histograms for cells without and with the addition of OS are shown. Data are mean (± SEM). (* denotes p<0.05).
2.9. Measurement of mitochondrial function using the Cell Mito Stress Test (CMST)
Analysis of mitochondrial function was performed on live cells using XFe96 Extracellular Flux Analyzer (Seahorse Bioscience) using the CMST assay conditions. Cells (4×104 cells/well) were seeded in a 96 well plate and grown for 24 h. Cells were incubated with or without 500 μM hydrogen peroxide for 24 h, washed (2X) with CMST assay medium (XF base medium DMEM supplemented with 2 mM glutamine, 5.5 mM glucose, and 1 mM sodium pyruvate, pH 7.4), and then incubated in CMST medium for 1 h at 37 °C in a non-CO2 incubator. The CMST assay protocol was performed as suggested by the manufacturer (Seahorse Bioscience). Oxygen consumption rate (OCR) was detected under basal conditions followed by the sequential addition of oligomycin (2 µm), FCCP (1 µm), as well as rotenone (1 μM) and antimycin A (1 μM). This allowed for measurement of the following parameters: basal respiration, OCR, ATP production, maximal respiration, spare respiratory capacity, and non-mitochondrial respiration. A graphical description of the assay and parameters measured are provided (Supplement Fig. 1).
2.10. Determining mitochondrial DNA content
DNA was prepared from confluent cells grown in a T25 using a DNeasy Mini-Prep Kit (Qiagen). The content of mtDNA was determined using quantitative real-time PCR (iQ5 Multicolor Real-Time PCR Detection System; Bio-Rad) as described [24]. Three sets of primers were used in the RT-qPCR analysis (Supplement Table 3). Total mtDNA copies were quantified by amplifying two regions of the mitochondrial genome, Cytochrome b (Cyt b) (222 bp) and the 16 S rRNA (197 bp) normalized to the invariable β-globin nuclear gene (147 bp).
2.11. Pigment Epithelium-Derived Factor (PEDF) analysis
RPE cells were seeded (1×105 cells/well) onto polyester inserts (6.5 mm diameter, 0.4 µm pores; Corning) coated with Matrigel (Corning). Cell monolayers were cultured for approximately 2 months with media changes twice per week. Samples were collected from the apical chambers 24-h after a media change. ELISA for PEDF (R & D Systems) was conducted according to the manufacturer's protocols. Growth factor concentration was derived from standard curves and normalized to chamber volume.
2.12. Cell viability
RPE cells were seeded (5×103 cells/well) in a 96-well microtiter plate and grown for 48 h in RPE media containing 1% FBS and no sodium pyruvate. RPE cells were incubated with different concentrations of hydrogen peroxide (150, 200 and 250 μM) for 24 h. Cell viability was determined using the Cyquant Direct Cell Proliferation Assay Kit (ThermoFisher). The percent of viable cells were determined by normalizing to the untreated control group.
2.13. Measures of ATP content
The cellular ATP production was assayed using ATPlite- luminescence ATP detection assay system (Perkin-Elmer). RPE cells were seeded (5×103 cells/well) in a 96-well microtiter plate and grown for 48 h in RPE media containing 1% FBS and no phenol red or sodium pyruvate. Cells were treated with hydrogen peroxide (150, 200 and 250 μM) for 24 h. ATP content was quantified using a luminescence microplate reader (Biotek, Synergy 2).
2.14. Glutathione (GSH) analysis
Intracellular GSH levels were measured using a GSH-Glo Glutathione assay kit (Promega). Cells (5×103 cells/well) were seeded in a 96-well microtiter plate and grown for 48 h in RPE media containing 1% FBS and without phenol red or sodium pyruvate. RPE cells were incubated with different concentrations of hydrogen peroxide (150, 200 and 250 μM) for 24 h. GSH content was determined with a luminescence microplate reader (BioTek, Synergy 2).
2.15. Statistical analysis
A Grubb's test was first run on each data set to remove outliers. An unpaired Student's t-test was performed to compare data from donors without AMD (No AMD) to donors with AMD (Figs. 2C, 3C, 4A-C, and 5D). A paired t-test was used to determine if there was a significant change in bioenergetics parameters after incubation with hydrogen peroxide (Fig. 2D,E and Fig. 3D,E). Two-way Analysis of Variance (ANOVA) was used to compare the effect of disease state (No AMD vs AMD) and hydrogen peroxide concentration, with a Tukey's post-hoc test when required (Fig. 5A-C). For oxidative stress response RT-qPCR and western blots, donor's fold change relative to untreated data were analyzed using One-Way ANOVA with repeated measures and a multiple comparison Dunnet's test (Fig. 5E+G). AMD fold change relative to average No AMD was tested for normal distribution (Fig. 5F+H). If data fit a normal distribution, a one-sample t-test was used. If data did not fit a normal distribution, a Wilcoxon signed-rank test was used. For both one-sample t-tests and Wilcoxon signed-rank tests, data was tested against a hypothetical mean of one. The hypothetical mean of one was chosen as fold change isrelative to a value of one. Analyses were performed using the statistical software in Origin 9.1 (Originlab Corp) or GraphPad Prism 7 (GraphPad Software). Data are reported as mean ± SEM for each group. p≤0.05 was considered statistically significant and a p-value between 0.05 and 0.10 was considered a trend.
Fig. 2.
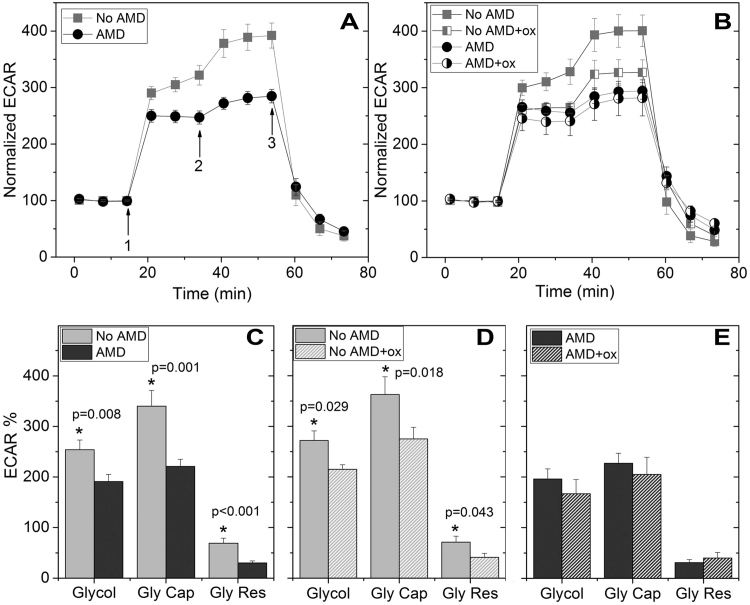
RPE from donors with AMD show reduced glycolytic function and resistance to oxidative inactivation. (A,B) Trace shows extracellular acidification rate (ECAR) normalized to baseline for cells from non-diseased and AMD donors with no treatment (A, No AMD n=9; AMD n=9) and following 24 h incubation with 500 µM hydrogen peroxide (B, No AMD n=7; AMD n=6). Arrows indicate injection of glucose (1), oligomycin (2), and 2-deoxyglucose (2-DG,3). (C,D,E) Parameters of glycolytic function calculated from data shown in A or B. Probability values for significant differences, as determined by t-test comparing No AMD with AMD (C) or paired t-test comparing glycolytic function with and without oxidation for individual samples (D,E), is provided on the graphs. See Supplement Fig. 1A for the method of calculation. All data are mean (± SEM). (* denotes p<0.05).
Fig. 3.
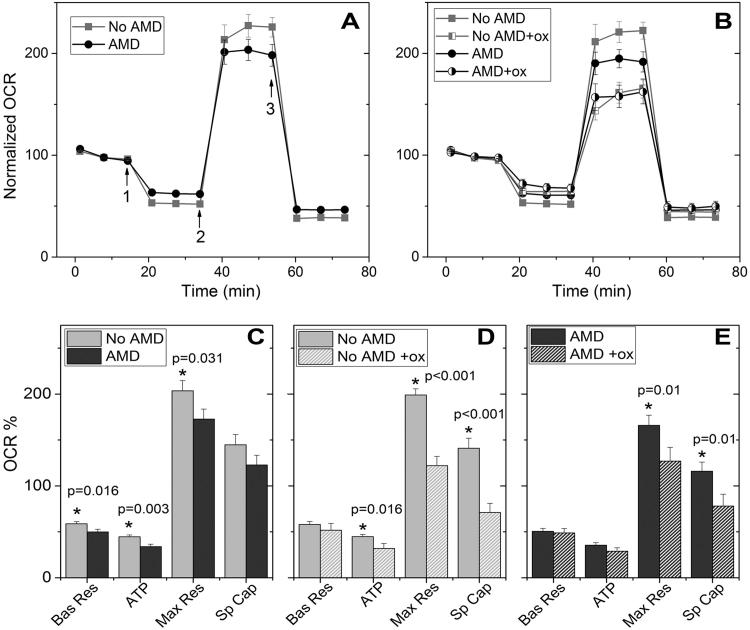
RPE from donors with AMD show reduced mitochondrial function. (A,B) Trace from an XF96 Extracellular Flux Analyzer shows the oxygen consumption rate (OCR) normalized to baseline for cells from non-diseased and AMD donors with no treatment (A, No AMD n=14; AMD n=19) and following 24 h incubation with 500 µM hydrogen peroxide (B, No AMD n=10; AMD n=15). Arrows indicate injection of oligomycin (1), FCCP (2) and antimycin and rotenone (3) to perturb mitochondrial function. (C,D,E) Parameters of mitochondrial function were calculated from data shown in A or B. Probability values for significant differences, as determined by t-test comparing No AMD with AMD (C) or paired t-test comparing values with and without oxidation for individual samples (D,E), is provided on the graphs. Bas Res=basal respiration; Max Res=maximal respiration; Sp Cap=spare capacity. See Supplement Fig. 1B for the method of calculation. All data are mean (± SEM). (* denotes p<0.05).
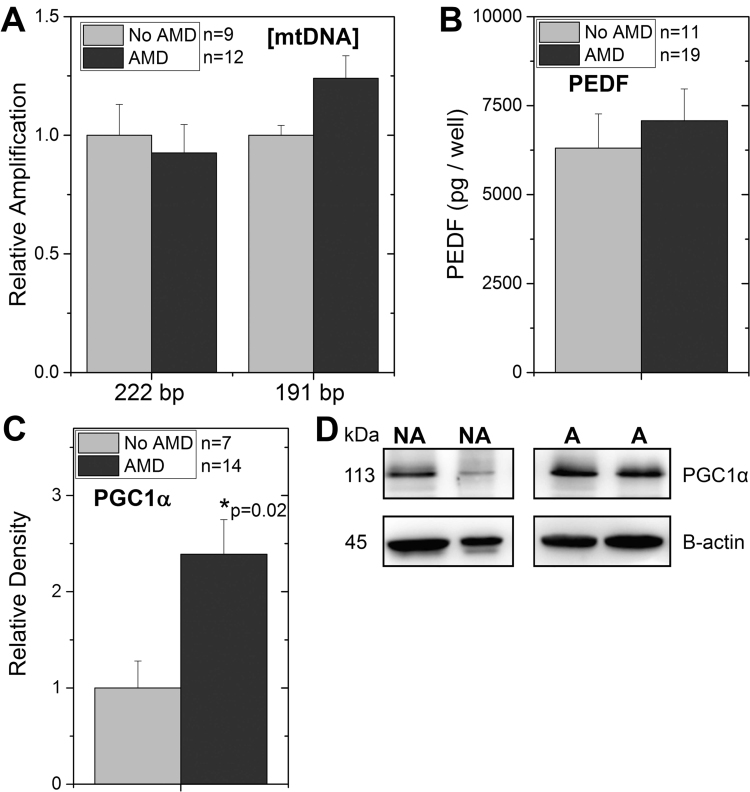
Fig. 4.
Investigating the loss in mitochondrial function for RPE from AMD donors. (A) Mitochondrial content was estimated from the real-time PCR amplification of the mitochondrial genome (Cyt b (222 bp) and16S rRNA (197 bp)) relative to amplification of the β-globin nuclear gene (a measure of total RPE). Ratios (mtDNA/ β-globin) were normalized to the mean ratio for non-diseased (No AMD) donors. (B) Graph shows results of ELISA measuring pigment epithelium derived factor (PEDF) secreted by RPE for donors without AMD (n=11) or with AMD (n=19). (C,D) Western blots were used to evaluate the content of PGC1α in cultures from donors with or without AMD. Results of densitometry (C) and representative western blot (D) are shown. * p=0.02, RPE from AMD donors were significantly higher than cells from donors without AMD. All data are mean (± SEM). (* denotes p<0.05).
Fig. 5.
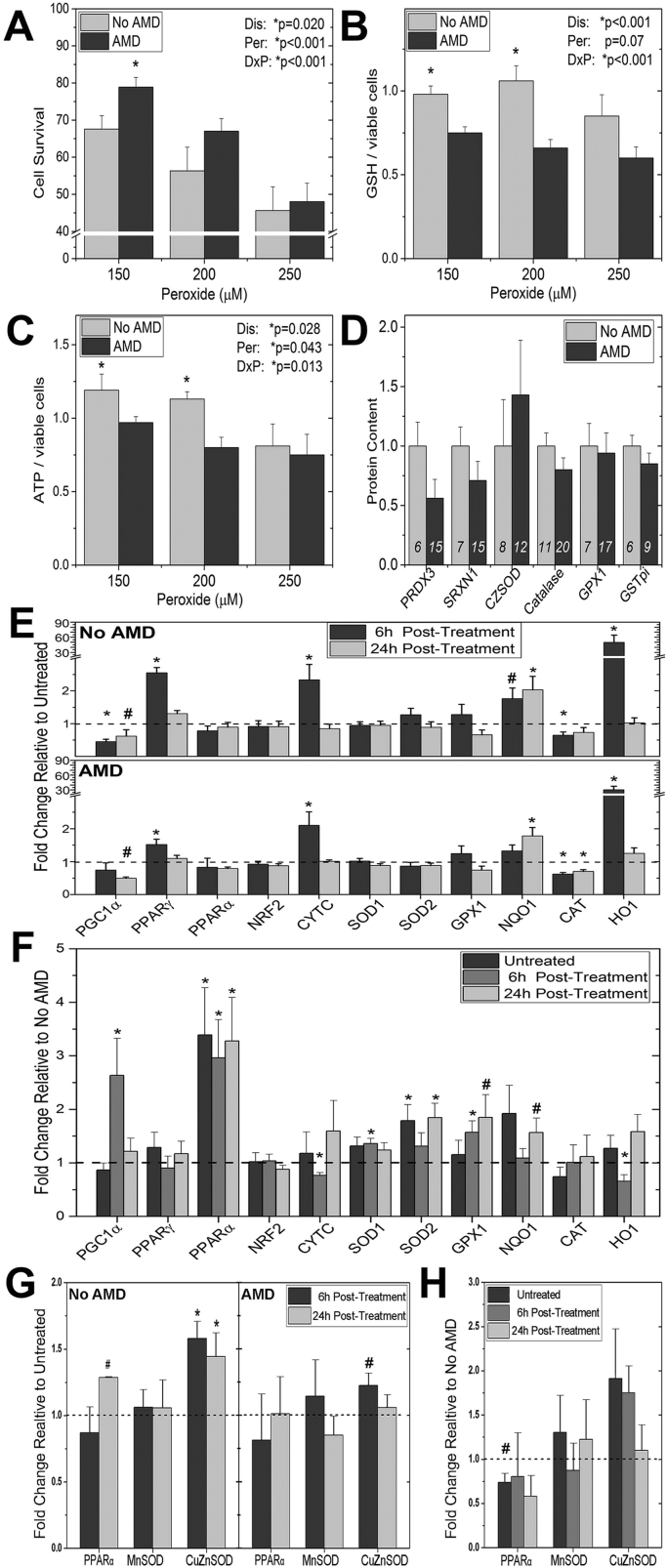
Investigating the resistance to oxidation stress in RPE from AMD donors. (A,B,C) RPE cultures were incubated with 0, 150 µm, 200 µm, or 250 µm hydrogen peroxide for 24 h. (A) Cell survival (No AMD n=9; AMD n=16) was measured using the CyQuant Direct Cell Proliferation Assay. (B) ATP content (No AMD n=8; AMD n=14) was determined using the ATPlite luminescence ATP detection assay. (C) GSH content (No AMD n=9; AMD n=16) was determined using the GSH-Glo Glutathione assay. Data are hydrogen peroxide treated relative to untreated controls. Results from the two-way ANOVA (main effects were disease and hydrogen peroxide) are shown on each graph. (D) Data are from densitometry of Western immunoblots using antibodies specific for multiple antioxidants. Results are normalized to the mean density for donors with No AMD. The number of donors for each analysis is shown within the bars. (E) mRNA was isolated from untreated controls and cells exposed to 300 µm hydrogen peroxide for either 6 h or 24 h. Results are the fold change in expression relative to untreated controls (dashed line). (F) The same Ct values used in (E) were used to calculate fold change in expression of AMD relative to the average for No AMD samples (dashed line). (G) Proteins were isolated using RIPA buffer from untreated controls and cells exposed to 300 µm hydrogen peroxide at 6 h or 24hrs. Results are the fold change of densitometry relative to untreated controls (dashed line). (H) The same densitometry values were used to calculate the fold change relative to No AMD donors. See Materials and Methods for fold change calculations. All data are mean (± SEM). ( * denotes p<0.05 and # denotes 0.10 > p > 0.05).
3. Results
3.1. Characterization of Primary RPE cultures from human donor eyes
A summary of demographics and clinical information for donors used in the experiments is provided in Supplement Table 1. The average time from death to cell harvesting was 19.5±4 h (mean ± SD). Donors were graded for the presence or absence of AMD using the criteria established for eye bank eyes [7], [35]. Donors with no clinically obvious eye disease were designated as “No AMD” (MGS1). Donors exhibiting RPE pigmentary changes and drusen were included in the “AMD” group. The majority of cells (~90%) were from donors at early (MGS2) or intermediate (MGS3) AMD. Cells in passage 2 or 3 were used for characterization and functional assays.
Confluent primary RPE cultures from donors with or without AMD developed pigmented cell monolayers with a cobblestone appearance (Fig. 1A,B). Based on anti-ZO1 staining, they also develop tight junctions and when grown on transwells, become polarized as evidenced by the apical and basal localization of the Na-K ATPase and Bestrophin, respectively (Fig. 1C,D). Results from Western immunoblotting, immunofluorescent staining, and qRT-PCR analysis show these primary cultures express prototypic RPE proteins (RPE65; Bestrophin (Best1); monocarboxylate transporter 3 (MCT3)), enzymes associated with pigment development (pre-melanosome protein 17 (Pmel17); tyrosinase related protein 1 (Tryp1)), proteins involved in the visual cycle (retinol binding protein 1 (RBP1); retinal dehydrogenase (RDH11); cellular retinoic acid binding protein (CRABP); cellular retinaldehyde binding protein (CRALBP)), and the transcription factor associated with differentiated RPE (microphthalmia-associated transcription factor (MITF) (Fig. 1E-G and Supplement Fig. 2A-C). Notably, similar levels of gene expression were observed in cells from both non-diseased and AMD donors (Fig. 1G). These primary RPE cultures also contained no measurable contamination by endothelial cells, immune cells, or Mast cells as determined by Western blot probing with anti-CD31, anti-CD45 and anti-TPSAB1 antibodies (Supplement Fig. 2D). Cultures of RPE from donors with or without AMD also retained in vivo function, as demonstrated by their ability to phagocytose photoreceptor OS (Fig. 1H) and latex beads (Supplement Fig. 2E). These results show primary RPE cultures share many of the cardinal features of RPE in vivo.
3.2. Assessment of bioenergetics and resistance to oxidative stress
To compare the bioenergetic profile of primary RPE cultures from non-diseased donors (No AMD) and donors with AMD, we measured glycolytic and mitochondrial function using an XFe96 Extracellular Flux Analyzer (Seahorse Bioscience). This analysis allows for real-time measurements of ECAR and OCR, which are indicators of glycolysis and mitochondrial respiration, respectively. The effect of hydrogen peroxide-induced oxidative stress on cell energetics was also assessed.
3.2.1. Lower glycolytic function and resistance to oxidative damage in RPE from AMD Donors
Glycolysis was assessed utilizing the glycolytic stress test and monitoring the ECAR. Fig. 2 shows a trace of the average ECAR (normalized to baseline) for cells from non-diseased and AMD donors with no treatment (A) and following 24-h incubation with 500 µM hydrogen peroxide (B). ECAR measured after sequential injections of glycolytic stressors (glucose, oligomycin, and 2-deoxyglucose) was used to calculate glycolysis, glycolytic capacity and glycolytic reserve (Supplement Fig. 1A).
All measures of glycolysis were significantly lower in RPE from donors with AMD compared with healthy donors. Glycolysis, glycolytic capacity, and glycolytic reserve were decreased 25%, 35% and 57%, respectively (Fig. 2C). Hydrogen peroxide treatment significantly decreased all measures of glycolysis in donors without AMD (Fig. 2D) but not in cells from AMD donors (Fig. 2E). In cells from donors without AMD, hydrogen peroxide treatment decreased glycolysis 21%, glycolytic capacity 24% and glycolytic reserve 42% (Fig. 2D). Cells from AMD donors were resistant to oxidative inactivation.
3.2.2. Decreased mitochondrial function and resistance to oxidative damage in RPE from AMD Donors
OCR was measured in primary RPE from donors with and without AMD using the Cell Mito Stress Test. Fig. 3 shows a trace of the average OCR (normalized to baseline) for untreated cells from non-diseased and AMD donors (A) and following 24-h incubation with 500 µM hydrogen peroxide (B). OCR measured after sequential injections of mitochondrial stressors (oligomycin, carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone (FCCP) and antimycin/rotenone) was used to calculate basal respiration, ATP production, maximal respiration, and spare capacity (Supplement Fig. 1B).
RPE from donors with AMD had significantly lower basal respiration, ATP production, and maximum respiration compared with RPE from healthy donors (Fig. 3C) Hydrogen peroxide treatment significantly decreased ATP production 29% in donors without AMD (Fig. 3D) but not in cells from AMD donors (Fig. 3E). Significant declines in maximal respiration and spare capacity were observed in both groups. Cells from donors without AMD exhibited a more substantial decrease in both maximal respiration (39% vs 23%) and spare capacity (50% vs 33%) compared with AMD donors.
3.3. Investigating potential mechanisms linked to reduced bioenergetics in RPE from AMD donors
Factors that can influence oxidative capacity include the cellular content of mitochondria, the production of growth factor PEDF by RPE [13], and the content of the transcriptional coactivator peroxisome proliferator- activated receptor-gamma coactivator 1α (PGC-1α) [19]. Mitochondrial content was estimated from qRT-PCR amplification of small segments of the mitochondrial genome localized within the region for Cyt b (222 bp) and the 16 S rRNA (197 bp). These two regions were selected because they are located in very different but stable regions of the mitochondrial genome. Amplification of the β-globin nuclear gene (147 bp), which has two copies per diploid cell, was used to estimate the number of RPE cells in each sample. Amplification of the two regions of the mitochondrial genome and β-globin nuclear gene provides an estimate of the total mtDNA copies per RPE cell. Our results show no difference in mtDNA content when comparing healthy and AMD donors (Fig. 4A).
PEDF has been shown to stabilize mitochondrial networks and improve RPE mitochondrial function [13]. To determine if differences in PEDF production could help explain the disease-related difference in mitochondrial function, we measured the content of PEDF secreted by RPE cultures using an ELISA assay (Fig. 4B). Our results show RPE from healthy and AMD donors secreted approximately the same amount of PEDF to the apical side of RPE cells grown on transwells.
Previous work has shown that PGC-1α has a positive effect on both mitochondrial metabolism and antioxidant capacity [19]. To test whether differences in content of this transcriptional coactivator could explain the reduced oxidative phosphorylation observed in RPE from AMD donors, we measured the cellular content of PGC-1α protein by Western immunoblots (Fig. 4C,D). We found that cultured RPE from donors with AMD had significantly higher levels of PGC1α compared with donors without AMD. With higher PGC1α, the prediction is that mitochondrial function would improve, which was not observed for RPE cultured from AMD donors.
3.4. Investigating potential mechanisms responsible for differential oxidative stress resistance
Analysis of the bioenergetic profile of primary RPE cultures showed RPE from donors with AMD were more resistant to hydrogen peroxide-induced decrements in both mitochondrial and glycolytic function. To further investigate these initial findings, we performed an orthogonal assay of cell death to confirm that cells from AMD donors were more resistant to oxidative stress. In both healthy and AMD donor cells, we observed a dose-dependent decrease in cell survival (p<0.001) (Fig. 5A). However, cells from AMD donors had significantly better survival (p=0.02), especially at low levels of oxidative stress.
To investigate the potential mechanistic basis for the ability of RPE from AMD donors to withstand an oxidative insult, we measured the content of ATP and GSH under the same conditions as the cell death assay. GSH is a highly abundant tripeptide composed of glycine, cysteine, and glutamic acid, with multiple roles in helping to protect the cell from an oxidative challenge, such as reducing oxidized proteins and reversibly binding to protein sulfhydryl groups to protect them during an oxidative challenge. Following incubation with increasing amounts of hydrogen peroxide, both GSH (p=0.07) and ATP (p=0.04) exhibited a dose-dependent decrease in all cells (Fig. 5B and C). Consistent with our Seahorse analysis of ATP content, cells from donors with AMD had lower levels of ATP (p=0.03). GSH content was also lower in AMD donor cells (p>0.001).
The cellular antioxidant capacity has a significant impact on how the cell responds to an oxidative challenge. We have already shown that RPE from AMD donors have significantly elevated levels of PGC-1α protein, which may contribute to their higher resistance to an oxidative insult (Fig. 4C). To determine if altered levels of antioxidants aid in the ability of RPE from AMD donors to withstand an oxidative challenge, Western immunoblotting was used to measure the content of several major antioxidants (Fig. 5D and Supplement Fig. 2F). Antioxidants were selected based on their function and localization within the cell. Peroxiredoxin (PRDX3, mitochondria), catalase (CAT, peroxisomes), and glutathione peroxidase (GPX1, cytosolic) are the major enzymes involved in protecting cells against hydrogen peroxide [36]. Following an oxidative stress, sulfiredoxin 1 (SRXN1, cytosolic) translocates into the mitochondria and preserves the antioxidant function of mitochondrial PRDX3 [32]. Glutathione-S-transferase pi (GSTπ) helps to detoxify specific products of oxidative damage, such as 4-hydroxynonenal. The cytosolic superoxide dismutase (SOD1), one of the most abundant antioxidants, reduces cellular content of superoxide by transforming it into hydrogen peroxide. Our analysis showed that under basal conditions, the relative content of these enzymes was not different for RPE from non-diseased or AMD donors.
The ability to upregulate redox sensitive genes is important in maintaining a healthy cell environment following an oxidative challenge. Therefore, we analyzed the change in gene expression of redox sensitive transcription factors and downstream genes in RPE from donors with and without AMD after treatment with hydrogen peroxide. A select number of genes exhibited a significant change in expression in response to treatment. Peroxisome proliferator-activated receptor gamma (PPARγ), cytochrome c (CYTC), NAD(P)H quinone dehydrogenase (NQO-1), and heme-oxygenase 1 (HO-1) increased in both AMD and No AMD cells after hydrogen peroxide treatment (Fig. 5E). Expression of glutathione peroxidase 1 (GPX1), superoxide dismutase 1 (SOD1), and superoxide dismutase 2 (SOD2) remained unchanged in response to treatment (Fig. 5E). CAT and PGC1α showed similar decreases in both AMD and No AMD cells after hydrogen peroxide treatment (Fig. 5E).
To determine if there were differences in gene expression between cells from healthy and AMD donors, we calculated the fold change of mRNA content in AMD cells relative to the mean content of No AMD cells (Fig. 5F). Baseline levels of gene expression were higher in AMD RPE relative to mean No AMD cells for peroxisome proliferator-activated receptor alpha (PPARα) and SOD2. At six hours post hydrogen peroxide, PGC1α, PPARα, SOD1, and GPX1 displayed higher levels of expression in AMD RPE. Expression of PPARα and SOD2 was significantly higher at 24 h post treatment. Conversely, reduced expression was observed in CYTC and HO1 in AMD RPE six hours after hydrogen peroxide exposure. These data show substantial changes in expression of redox sensitive genes in response to an oxidative challenge.
To determine if the changes seen in mRNA led to changes in protein content, western immunoblotting was performed under identical conditions used for gene expression analysis. The content of PPARα was examined based on the significant difference in mRNA levels between cells with AMD or No AMD. Additional analysis was performed on the gene products of SOD1 and SOD2 (CuZnSOD and MnSOD, respectively) due to their major role in the oxidative stress response. In cells from donors with No AMD, PPARα protein was increased approximately 25% at 24 h post-peroxide compared with untreated controls (p=0.08). Additionally, CuZnSOD protein increased significantly relative to untreated at both 6 and 24 h (Fig. 5G). In cells from donors with AMD, there was approximately a 20% increase in CuZnSOD (p=0.059) at 6 h (Fig. 5G). Comparing protein content in AMD versus No AMD RPE we observed approximately a 25% decrease in PPARα (p=0.073) in untreated controls (Fig. 5H).
4. Discussion
In this study, we showed that RPE isolated and cultured from human donor eyes retain the ability to phagocytose OS and have morphological characteristics that resemble native RPE in vivo. Functional analysis of the two major energy producing pathways, oxidative phosphorylation (OxPhos) and glycolysis, revealed that RPE from AMD donors had a reduced bioenergetic profile compared with RPE from non-diseased donors. Despite the defects in bioenergetics, RPE from AMD donors were more resistant to mitochondrial and glycolytic oxidative inactivation and also oxidation-induced cell death. Investigation of potential mechanisms responsible for the greater protection from oxidation revealed upregulation of PGC1α protein as well as differential induction of redox-sensitive transcription factors and their downstream products in the RPE cultured from AMD donors. Based on these results, we speculate that the microenvironment of the diseased retina modifies the RPE and that at least some of these changes are retained in vitro.
Data showing functional impairment in both mitochondrial OxPhos and glycolysis in RPE from AMD donors suggest AMD is associated with a bioenergetic crisis in the RPE. This idea has been previously suggested [24], [21] but had not been experimentally tested.
Evidence reported in studies using RPE directly harvested from donors with AMD found defects in mitochondrial morphology and increased mtDNA damage, which is consistent with a predicted loss in mitochondrial function in vivo [10], [24], [45]. This study, as well as a recently published comparison of primary RPE cultures from AMD and healthy donors [11], provides strong experimental evidence that altered bioenergetics is part of AMD pathology. Mitochondria are at the heart of RPE energy production since it is the site of the tricarboxylic acid (TCA) cycle, β-oxidation, and OxPhos. Substrate for the TCA cycle is also produced from the glycolytic reduction of glucose, so a deficiency in glycolysis could ultimately affect OxPhos. Thus, while OxPhos supplies the majority of ATP, the entire bioenergetic system works together to meet the energy requirements of the cell [18].
Of note, we find that the protein content of PGC1α, a transcription factor co-activator that regulates mitochondrial biogenesis, was higher in cells from AMD donors (Fig. 4C). However, the elevated PGC1α content was not sufficient to improve mitochondrial function or increase mtDNA content. Therefore, other factors are contributing to the loss in bioenergetics in AMD donor cells. For example, we had previously reported significant mtDNA damage in RPE from AMD donors [45]. Extensive damage was present in the D-loop region, where transcription factors initiate mtDNA biogenesis. This disruption in mtDNA replication would inhibit mitochondrial biogenesis.
The second major finding is that RPE cells from AMD donors were more resistant to an oxidative challenge. These data are in contrast to Golestaneh and colleagues [11], who found no change in oxidative resistance at 24 h post-hydrogen peroxide in AMD versus healthy donors. A potential explanation for the discrepant results could be the low number of donors in their study (n=5 per group) or differences in culture conditions.
Our previous proteome analysis of RPE showed AMD is associated with substantial upregulation of antioxidant enzymes in cells that were directly harvested from donors with AMD [7]. These results suggest the cell has upregulated the antioxidant capacity in vivo to accommodate the oxidative environment of the diseased retina. In the current study, we evaluated whether increased antioxidant enzymes could explain the greater resistance to oxidative stress exhibited by RPE from AMD donors. We found that levels of antioxidant enzymes were essentially equivalent irrespective of the donor's health status (Fig. 5D). These cells, which were cultured for 3–4 months prior to use, may have adapted their proteome to the new in vitro environment that had homogenous temperature, oxygen, and nutrients. In addition to its role in mitochondrial biogenesis, PGC1α also contributes to the antioxidant capacity of the cell [19] by regulating multiple transcription factors, including nuclear factor E2-related factor (NRF2), PPARα, and PPARγ [30]. PGC1α is required for induction of many reactive oxygen species detoxifying enzymes, including CuZnSOD, MnSOD, GPX1, and CAT [43]. Therefore, the increase in PGC1α content could play a role in the observed resistance of RPE from AMD donors to an oxidative insult.
It has previously been shown that the type of oxidant and extent of oxidative stress determines which pathways are activated [9]. For example, intermediate oxidative stress activates NF-κB, AP1, and MAP kinase pathways, while more elevated stress upregulates NRF2. A mild stress with minimal cell death was used for measuring changes in gene expression and protein content under an oxidative challenge (Fig. 5F-H), which may explain the observed changes in only a select number of genes relative to untreated controls. The degree of confluence, density of the cells, and the development of differentiated polarized RPE can also influence cultured RPE's response to oxidative stress [16]. The different conditions used for our oxidative stress response, cell death experiments, and measurements of bioenergetic function led to varying degrees of cell death (5–10%, 20–55%, and 40–47% respectively, data not shown). None-the-less, these individual assays were able to reveal differences between cells from non-diseased and AMD donors. As a caveat with all cultured cells, experiments performed in vitro may not be a genuine reflection of their response in vivo. More extensive investigation using different oxidizing conditions are required to define the mechanism responsible for oxidative resistance of AMD donor cells.
The altered bioenergetics and differential response to oxidative stress (Fig. 2, Fig. 3, Fig. 5) in RPE from donors with AMD suggest these cells possess a “metabolic memory” in vitro that may be linked to their in vivo environment. This idea is consistent with data from diabetic patients and animal models, where the effect of the diseased environment is sustained in cultured cells [28], [48], [8], [39]. Of note, there are many parallels between diabetic retinopathy and AMD, the most important being the increase in retinal oxidative stress and mitochondrial dysfunction [26], [27]. The metabolic memory observed in the current study and associated with cells derived from diabetic animals could result from epigenetic mechanisms that adjust gene expression to accommodate the changing cellular environment. Epigenetic modifications, which are reversible but can also persist long after the stimulus is removed, regulate gene expression at either the level of transcription or translation by altering chromatin structure (via DNA methylation and post-translational modification of histones) or by blocking mRNA translation due to binding of micro-RNAs (miRNA) to the 3′ untranslated region. DICER, an endonuclease that is involved in miRNA maturation, regulates the cellular content of miRNAs. Because the majority of the enzymes responsible for maintaining DNA methylation and histone methylation/acetylation are redox sensitive, these epigenetic modifications provide a footprint documenting the cell's exposure to an oxidative environment.
While we did not directly investigate whether epigenetic modifications in our cultured cells could explain the apparent “metabolic memory” retained by RPE from AMD donors, the importance of epigenetic modifications in AMD has started to emerge [14]. For example, disease-specific differences in DNA methylation patterns and downregulation of DICER1 have been reported in RPE/choroid of human AMD donors [17], [23], [47]. Future experiments will focus on determining if epigenetic changes have occurred in cells from AMD donors as a potential explanation for the differential response to an oxidative challenge.
This study provides insight into the molecular mechanism and underlying differences between RPE cells from AMD and No AMD donors in their bioenergetics and ability to handle an oxidative challenge. Strong evidence from human donor RPE [24], [10] and reduced OxPhos measured in primary RPE cultures suggest mitochondrial dysfunction plays a central role in AMD. We propose mitochondrial dysfunction initiates a cascade of events resulting in increased ROS production as well as activation of PGC1α and other redox sensitive transcription factors (Fig. 6). If not properly removed from the cell the increased ROS exacerbates mitochondrial dysfunction. This dysfunction is also communicated to the nucleus through retrograde signaling [25]. Mitochondrial retrograde signaling is stimulated by defects in OxPhos, mtDNA damage, decreased ATP, increased ROS, and release of Ca2+ from the mitochondria [38]. These pathways, stemming from mitochondrial dysfunction, cause changes in gene expression and protein content leading to altered bioenergetics and increased resistance to oxidative stress. In addition, signaling resulting from mitochondrial dysfunction may lead to changes in the epigenetic landscape, further altering gene expression. These adjustments allow the cell to survive. However, this system fails under conditions of chronic assault in the diseased retina.
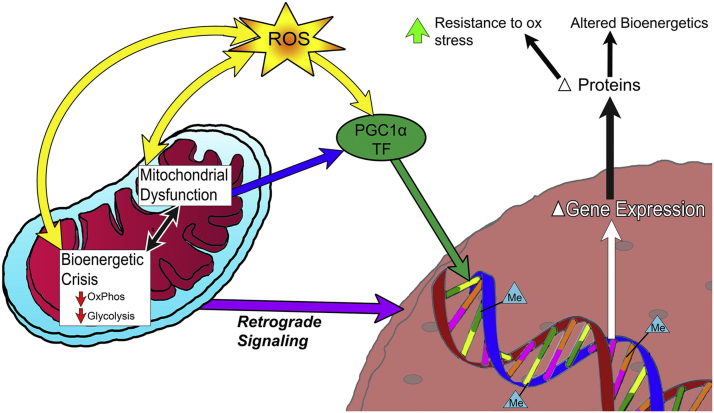
Fig. 6.
Model for AMD related changes in RPE. AMD is associated with mitochondrial dysfunction and a bioenergetic crisis, documented by decreased OxPhos and glycolysis. Mitochondrial dysfunction leads to increased ROS production. PGC1a and redox sensitive transcription factors (TF) respond to environmental changes caused by mitochondrial dysfunction. In combination with mitochondrial retrograde signaling, these pathways alter gene expression and protein content of the cell. The epigenetic landscape, denoted by CpG methylation (Me) adds another layer of complexity to gene expression, and is altered due to an oxidative challenge. These changes lead to an increased resistance to oxidative stress and an altered bioenergetics profile.
The use of primary cultures of RPE has provided a valuable model system for studying RPE physiology and function under a number of experimental conditions. Pioneer work in primary cultures led to the discovery of polarized secretion of VEGF, IL6, and IL8 by RPE [15], [3] and that VEGF secretion increased under conditions of hypoxia [3]. Primary RPE cultures from donors of different ages have also revealed age-related changes in bioenergetics, antioxidant capacity, and cell signaling [12], [2], [40]. They have also been useful for establishing drug efficacy, as demonstrated from RPE protection by compounds such as PEDF and N-acetylcysteine amide [13,5]. The current study showed that some effects of the diseased retinal environment appear to be retained in vitro, and therefore it appears that the “metabolic memory” is linked to the donor's health status. These results suggest that primary RPE cultures from human donors graded for the presence or absence of AMD provide an excellent model system to study the effects of AMD on RPE function and gene expression.
Funding
This work was supported by the Elaine and Robert Larson Endowed Vision Research Chair (to DAF); National Institutes of Health/ National Institute of Aging (T32-AG29796 to MRT) and National Eye Institute (R01 EY026012 to DAF); Beckman Initiative for Macular Research (#1303); Foundation Fighting Blindness (TA-NMT-0613-0620-UMN); Regenerative Medicine Minnesota (2015-5337);an anonymous benefactor for Macular Degeneration Research; the Lindsay Family Foundation; Minnesota Lions Vision Foundation; an unrestricted grant from Research to Prevent Blindness to the Department of Ophthalmology and Visual Neurosciences. None of the funding agencies had a role in study design, in the collection, analysis and interpretation of data, in writing the manuscript, or in the decision to submit the manuscript for publication.
Acknowledgements
The authors wish to acknowledge the contribution of the Minnesota Lions Eye Bank personnel for their assistance in procuring eyes, and to Kathy Goode and Sung Lee for photographing and processing eye tissue. Dr. T. Michael Redman (NEI) provided the antibody for RPE-65. Dr. Aparna Lakaraju (U Wisconsin) provided procine outer segments for the phagocytosis experiments.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.redox.2017.05.015.
Contributor Information
Deborah A. Ferrington, Email: Ferri013@umn.edu.
Mara C. Ebeling, Email: ebeli017@umn.edu.
Rebecca J. Kapphahn, Email: kapph001@umn.edu.
Marcia R. Terluk, Email: mrterluk@umn.edu.
Cody R. Fisher, Email: fishe765@umn.edu.
Jorge R. Polanco, Email: polan070@umn.edu.
Heidi Roehrich, Email: rohri002@umn.edu.
Michaela M. Leary, Email: leary070@umn.edu.
Zhaohui Geng, Email: gengx027@umn.edu.
James R. Dutton, Email: dutto015@umn.edu.
Sandra R. Montezuma, Email: smontezu@umn.edu.
Appendix A. Supplementary material
Supplementary material
.
Supplementary material
.
Supplementary material
.
References
- 1.Alliance for Eye and Vision Research, 20th Anniversary Special Report. October. 〈http://www.eyeresearch.org/pdf/ValueBrochure/Value_of_Vision_Research_Brochure_Page_1.pdf〉, 2013.
- 2.Bhattacharya S., Chaum E., Johnson D.A., Johnson L.R. Age-related susceptibility to apoptosis in human retinal pigment epithelial cells is triggered by disruption of p53-Mdm2 association. Invest Ophthalmol. Vis. Sci. 2012;53:8350–8366. doi: 10.1167/iovs.12-10495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blaauwgeers H.G.T., Holtkamp G.M., Rutten H. Polarized vascular endothelial growth factor secretion by human retinal pigment epithelium and localization of vascular endothelial growth factor receptors on the inner choriocapillaris. Am. J. Pathol. 1999;155:421–428. doi: 10.1016/S0002-9440(10)65138-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blenkinsop T.A., Salero E., Stern J.H., Temple S. The culture and maintenance of functional retinal pigment epithelial monolayers from adult human eyes. Methods Mol. Biol. 2016;945:45–55. doi: 10.1007/978-1-62703-125-7_4. [DOI] [PubMed] [Google Scholar]
- 5.Carver K.A., Yang D. N-Acetylcysteine amide protects against oxidative stress- induced microparticle release from human retinal pigment epithelial cells. Invest Ophthalmol. Vis. Sci. 2016;57:360–371. doi: 10.1167/iovs.15-17117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beatty S., Koh H., Phil M., Henson D., Boulton M. The role of oxidative stress in the pathogenesis of age-related macular degeneration. Surv. Opthalmol. 2000;45(2):115–134. doi: 10.1016/s0039-6257(00)00140-5. [DOI] [PubMed] [Google Scholar]
- 7.Decanini A., Nordgaard C.L., Feng X., Ferrington D.A., Olsen T.W. Changes in select redox protein of the retinal pigment epithelium in age-related macular degeneration. Am. J. Ophthalmol. 2007;143:607–615. doi: 10.1016/j.ajo.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Durham J.R., Dulmovits B.M., Cronk S.M. Pericyte chemomechanics and the angiogenic switch: insights into the pathogenesis of proliferative diabetic retinopathy? Invest Ophthalmol. Vis. Sci. 2015;56:3441–3459. doi: 10.1167/iovs.14-13945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Espinosa-Diez C., Miguel V., Mennerich D., Kietzmann T., Sanchez-Periz P., Cadenas S., Lamas S. Antioxidant responses and cellular adjustments to oxidative stress. Redox Biol. 2015;6:183–197. doi: 10.1016/j.redox.2015.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feher J., Kovacs I., Artico M., Cavallotti C., Papale A., Balacco Gabrieli C. Mitochondrial alterations of retinal pigment epithelium in age-related macular degeneration. Neurobiol. Aging. 2006;27:983–993. doi: 10.1016/j.neurobiolaging.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 11.Golestaneh N., Chu Y., Xiao Y.Y., Stoleru G.L., Theos A.C. Dysfunctional autophagy in RPE, a contributing factor in age-related macular degeneration. Cell Death Dis. 2017;8:e2537. doi: 10.1038/cddis.2016.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He Y., Ge J., Burke J.M., Myers R.L., Dong Z.Z., Tombran-Tink J. Mitochondria impairment correlates with increased sensitivity of aging RPE cells to oxidative stress. J. Ocul. Biol. Dis. Inform. 2010;3:92–108. doi: 10.1007/s12177-011-9061-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He Y., Leung K.W., Ren Y., Pei J., Ge J., Tombran-Tink J. PEDF improves mitochondrial function in RPE cells during oxidative stress. Invest Ophthalmol. Vis. Sci. 2014;55:6742–6755. doi: 10.1167/iovs.14-14696. [DOI] [PubMed] [Google Scholar]
- 14.He S., Li X., Chan N., Hinton D.R. Review: epigenetic mechanisms in ocular disease. Mol. Vis. 2013;19:665–674. [PMC free article] [PubMed] [Google Scholar]
- 15.Holtkamp G.M., Van Rossem M., De Vos A.F. Polarized secretion of IL-6 and IL-8 by human retinal pigment epithelial cells. Clin. Exp. Immunol. 1998;112:34–43. doi: 10.1046/j.1365-2249.1998.00560.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsiung J., Zhu D., Hinton D.R. Polarized human embryonic stem cell-derived retinal pigment epithelial cell monolayers have higher resistance to oxidative stress-induced cell death than nonpolarized cultures. Stem Cells Transl. Med. 2015;4(1):10–20. doi: 10.5966/sctm.2014-0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hunter A., Spechler P.A., Swanger A. DNA methylation is associated with altered gene expression in AMD. Invest Ophthalmol. Vis. Sci. 2012;53:2089–2105. doi: 10.1167/iovs.11-8449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hurley J.B., Lindsay K.J., Du J. Glucose, lactate, and the shuttling of metabolites in vertebrate retinas. J. Neurosci. Res. 2015;93:1079–1092. doi: 10.1002/jnr.23583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iacovelli J., Rowe G.C., Khadka A., Diaz-Aguilar D., Spencer C., Arany Z., Saint-Geniez M. PGC1α induces human RPE oxidative metabolism and antioxidant capacity. Invest Ophthalmol. Vis. Sci. 2016;57:1038–1051. doi: 10.1167/iovs.15-17758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Imamura Y., Noda S., Hashizume K., Shinoda K., Yamaguchi M., Uchiyama S., Shimizu T., Mizushima Y., Shirasawa T., Drusen Tsubota K. choroidal neovascularization, and retinal pigment epithelium dysfunction in SOD1-deficient mice: a model of age-related macular degeneration. Proc. Natl. Acad. Sci. USA. 2006;103(30):11282–11287. doi: 10.1073/pnas.0602131103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jarrett S.G., Lin H., Godley B.F., Boulton M.E. Mitochondrial DNA damage and it potential role in retinal degeneration. Prog. Ret Eye Res. 2008;27:596–607. doi: 10.1016/j.preteyeres.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 22.Justilien V., Pang J., Renganathan K., Zhan X., Crabb J., Kim S., Sparrow J., Hauswirth W., Lewin A. SOD2 knockdown mouse model of early AMD. Invest Opthalmol Vis. Sci. 2007;48(10):4407–4420. doi: 10.1167/iovs.07-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaneko H., Dridi S., Tarallo V. DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature. 2011;471 doi: 10.1038/nature09830. (325-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karunadharma P.P., Nordgaard C.L., Olsen T.W., Ferrington D.A. Mitochondrial DNA damage as a potential mechanism for age-related macular degeneration. Invest Ophthalmol. Vis. Sci. 2010;11:5470–5479. doi: 10.1167/iovs.10-5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kleine T., Leister D. Retrograde signaling: organelles go networking. Biochim Biophys. Acta. 2016;1857(8):1313–1325. doi: 10.1016/j.bbabio.2016.03.017. [DOI] [PubMed] [Google Scholar]
- 26.Kowluru R.A. Mitochondria damage in the pathogenesis of diabetic retinopathy and in the metabolic memory associated with its continued progress. Curr. Med. Chem. 2013;20:3226–3233. doi: 10.2174/09298673113209990029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kowluru R.A., Kowluru A., Mishra M., Kumar B. Oxidative stress and epigenetic modifications in the pathogenesis of diabetic retinopathy. Prog. Retin Eye Res. 2015;48:40–61. doi: 10.1016/j.preteyeres.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar S., Pamulapati H., Tikoo K. Fatty acid induced metabolic memory involves alterations in renal histone H3K36me2 and H3K27me3. Mol. Cell Endocrinol. 2016;422:233–242. doi: 10.1016/j.mce.2015.12.019. [DOI] [PubMed] [Google Scholar]
- 29.Leibowitz H.M., Krueger D.E., Maunder L.R., Milton R.C., Kini M.M., Kahn H.A., Nickerson R.J., Pool J., Colton T.L., Ganley J.F., Loewenstein J.I., Dawber T.R. The Framingham eye study monograph: an opthalmological and epidemiological study of cataract, glaucoma, diabetic retinopathy, and macular degeneration, and visual acuity in the general population of 2631 adults. Suv Ophthalmol. 1980;24:335–610. [PubMed] [Google Scholar]
- 30.Liang Huiyun, Ward Walter F. PGC-1α: a key regulator of energy metabolism. Adv. Physiol. Educ. 2006;30:145–151. doi: 10.1152/advan.00052.2006. [DOI] [PubMed] [Google Scholar]
- 31.Maldonado M., Kapphahn R.J., Terluk M.R., Heuss N.D., Yuan C., Gregerson D.S., Ferrington D.A. Immunoproteasome deficiency modifies the alternative pathway of NFκB signaling. PLoS One. 2013;8:e56187. doi: 10.1371/journal.pone.0056187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Noh Y.H., Baek J.Y., Joeng W., Rhee S.G., Chang T.S. Sufiredoxin translocation into the mitochondria plays a crucial role in reducing hyperoxidized peroxiredoxin III. J. Biol. Chem. 2009;284:8470–8477. doi: 10.1074/jbc.M808981200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nordgaard C.L., Berg K.M., Kapphahn R.J., Reilly C., Feng X., Olsen T.W., Ferrington D.A. Proteomics of the retinal pigment epithelium reveals altered protein expression at progressive stages of age-related macular degeneration. Invest. Ophthalmol. Vis. Sci. 2006;47(3):815–822. doi: 10.1167/iovs.05-0976. [DOI] [PubMed] [Google Scholar]
- 34.Nordgaard C.L., Karunadharma P.P., Feng X., Olsen T.W., Ferrington D.A. Mitochondrial proteomics of the retinal pigment epithelium at progressive stages of age-related macular degeneration. Invest. Ophthalmol. Vis. Sci. 2008;49(7):2848–2855. doi: 10.1167/iovs.07-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Olsen T.W., Feng X. The Minnesota Grading System of eye bank eyes for age-related macular degeneration. Invest Ophthalmol. Vis. Sci. 2004;45:4484–4490. doi: 10.1167/iovs.04-0342. [DOI] [PubMed] [Google Scholar]
- 36.Poyton R.A., Hampton M.B. Peroxiredoxins as biomarkers of oxidative stress. Biochem Biophys. Acta. 2014;(1840):906–912. doi: 10.1016/j.bbagen.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 37.Parinot C., Rieu Q., Chatagnon J., Finnemann S.C., Nandrot E. Large scale purification of porcine or bovine photoreceptor outer segments for phagocytosis assays on retinal pigment epithelial cells. JOVE. 2014;94:e52100. doi: 10.3791/52100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quiros P.M., Mottis A., Auwerx J. Mitonuclear communication in homeostasis and stress. Nat. Rev. Mol. Cell Biol. 2016;17(4):213–226. doi: 10.1038/nrm.2016.23. [DOI] [PubMed] [Google Scholar]
- 39.Reddy M.A., Das S., Zhuo C. Regulation of vascular smooth muscle cell dysfunction under diabetic conditions by miR-504. Arterioscler. Thromb. Vasc. Biol. 2016;36:864–873. doi: 10.1161/ATVBAHA.115.306770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rohrer B., Bandyopadhyay M., Beeson C. Reduced metabolic capacity in aged primary retinal pigment epithelium (RPE) is correlated with increased susceptibility to oxidative stress. Adv. Exp. Med. Biol. 2016;854:793–798. doi: 10.1007/978-3-319-17121-0_106. [DOI] [PubMed] [Google Scholar]
- 41.Seth R., Sigler E., Adelman R. Age-related macular degeneration – Review and current concepts. US Ophthalmic Rev. 2011;4(1):96–100. [Google Scholar]
- 42.Sonoda S., Spee C., Barron E., Ryan S.J., Kannan R., Hinton D.R. A protocol for the culture and differentiation of highly polarized human retinal pigment epithelial cells. Nat. Protoc. 2009;4:662–673. doi: 10.1038/nprot.2009.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pierre St, Julie Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell. 2006;127:397–408. doi: 10.1016/j.cell.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 44.Strauss O. The retinal pigment epithelium in visual function. Physiol. Rev. 2005;85:825–881. doi: 10.1152/physrev.00021.2004. [DOI] [PubMed] [Google Scholar]
- 45.Terluk M.R., Kapphahn R.J., Soukup L.M., Gong H., Gallardo C., Montezuma S.R. Investigating mitochondria as a target for treating age-related macular degeneration. J. Neurosci. 2015;35(18):7304–7311. doi: 10.1523/JNEUROSCI.0190-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wong W., Su X., Li X., Cheung C., Klein R., Cheng C., Wong T. Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: a systematic review and meta-analysis. Lancet Glob. Health. 2014;2:e106–e116. doi: 10.1016/S2214-109X(13)70145-1. [DOI] [PubMed] [Google Scholar]
- 47.Wei L., Liu B., Tuo J. Hypomethylation of the IL-17RC promotor associates with age-related macular degeneration. Cell Rep. 2012;2:1151–1158. doi: 10.1016/j.celrep.2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu X.Y., Geng Y.J., Lian J.L. High levels of glucose induce “metabolic memory” in cardiomyocyte via epigenetic histone H3 lysine 9 methylation. Mol. Biol. Rep. 2012;39:8891–8898. doi: 10.1007/s11033-012-1756-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material