Abstract
Understanding the gene-specific risks for development of breast cancer will lead to improved clinical care for those carrying germline mutations in cancer predisposition genes. We sought to detail the spectrum of mutations and refine risk estimates for known and proposed breast cancer susceptibility genes. Targeted massively-parallel sequencing was performed to identify mutations and copy number variants in 26 known or proposed breast cancer susceptibility genes in 2134 BRCA1/2-negative women with familial breast cancer (proband with breast cancer and a family history of breast or ovarian cancer) from a largely European–Caucasian multi-institutional cohort. Case–control analysis was performed comparing the frequency of internally classified mutations identified in familial breast cancer women to Exome Aggregation Consortium controls. Mutations were identified in 8.2% of familial breast cancer women, including mutations in high-risk (odds ratio > 5) (1.4%) and moderate-risk genes (2 < odds ratio < 5) (2.9%). The remaining familial breast cancer women had mutations in proposed breast cancer genes (1.7%), Lynch syndrome genes (0.5%), and six cases had two mutations (0.3%). Case–control analysis demonstrated associations with familial breast cancer for ATM, PALB2, and TP53 mutations (odds ratio > 3.0, p < 10−4), BARD1 mutations (odds ratio = 3.2, p = 0.012), and CHEK2 truncating mutations (odds ratio = 1.6, p = 0.041). Our results demonstrate that approximately 4.7% of BRCA1/2 negative familial breast cancer women have mutations in genes statistically associated with breast cancer. We classified PALB2 and TP53 as high-risk, ATM and BARD1 as moderate risk, and CHEK2 truncating mutations as low risk breast cancer predisposition genes. This study demonstrates that large case–control studies are needed to fully evaluate the breast cancer risks associated with mutations in moderate-risk and proposed susceptibility genes.
Familial breast cancer: Pinning down susceptibility genes beyond BRCA
Women with the heritable form of breast cancer often harbor mutations in cancer-linked genes other than the usual suspects, BRCA1 and BRCA2. Slavin, Maxwell, Lilyquist, Joseph, and colleagues from major national and international cancer centers studied 2134 women with familial breast cancer who tested negative for BRCA1/2 gene mutations. The researchers sequenced 26 known or proposed breast cancer susceptibility genes and found mutations in approximately 1 in every 12 of the study subjects. They then further broke down the susceptibility genes into those that confer high-, moderate- or low-risk—although not all the proposed breast cancer genes reached statistical significance and, as such, their clinical importance remains unclear. The results support adding some of the high- and moderate-risk genes to multi-panel diagnostic tests that aim to determine the likelihood of a women developing heritable breast cancer.
Introduction
Breast cancer is the leading cause of cancer in women in the United States and the third leading cause of cancer death.1 Morbidity and mortality can be reduced by identification and clinical management of patients at high-risk.2 One major factor underlying breast cancer risk is genetic predisposition. It is currently thought that up to 10% of all breast cancers are due to an autosomal dominant susceptibility allele.3, 4 Mutant alleles in BRCA1 or BRCA2 (BRCA) account for the majority of hereditary breast cancer susceptibility.5 However, many other risk-associated alleles in other genes have now been identified through the use of multigene panels in clinical and research applications.6 An improved understanding of the spectrum of genetic susceptibility and clarification of gene-specific risks for breast cancer may result in enhanced screening, prevention, and therapeutic strategies for patients and their families.2, 4
There are few studies focusing on mutations in breast cancer susceptibility genes in affected individuals who have two or more close relatives with breast or ovarian cancer but are BRCA1/2 mutation negative.7–14 The purpose of this study was to detail the spectrum of known and proposed breast cancer susceptibility genes, highlight genotype–phenotype correlations, and refine risk estimates for mutations in these genes by examining BRCA negative familial breast cancer (FBC; proband with breast cancer and a family history of breast or ovarian cancer; see methods) women compared to the non-Finnish European controls from the Exome Aggregation Consortium (ExAC).15
Results
Phenotypic information and an overview of family history information for the 2134 individuals in the high-risk FBC is provided in Table 1. Participants were enrolled from multiple centers (see Methods). There were 2425 breast cancers in the 2134 women, with two breast cancers reported in 14% of these women. Full pathological information was available for 1055 tumors, and 62% of tumors were ER+HER2−, 16% were ER+HER2+, 9% ER−HER2+, and 14% were ER−HER2−. The majority of the cases (81%) were of European–Caucasian descent (Table 1). All FBC women had a family history of cancer, and 65% of women had at least one first-degree relative with breast cancer. A family history of ovarian cancer in at least one first or second degree relative was reported for 18% of women.
Table 1.
Phenotypic and pathological features of 2134 women with familial breast cancer
| n | % | |
|---|---|---|
| Racea | ||
| White | 1722 | 80.7 |
| Hispanic | 136 | 6.4 |
| Asian | 69 | 3.2 |
| Other | 69 | 3.2 |
| African American | 49 | 2.3 |
| Unknown | 89 | 4.2 |
| Personal history of cancerb | ||
| Second breast cancer | 291 | 13.6 |
| Ovarian cancer | 33 | 1.5 |
| Other cancer | 167 | 7.8 |
| Avg age 1st breast cancer | 47.9 ± 9.4 | |
| Avg age 2nd breast cancer | 52.9 ± 10.5 | |
| Family history of cancer | ||
| No FDR/SDR w/breast cancer | 166 | 7.8 |
| 1 FDR/SDR w/breast cancer | 419 | 19.6 |
| 2 FDR/SDR w/breast cancer | 729 | 34.2 |
| 3+ FDR/SDR w/breast cancer | 820 | 38.4 |
| FDR w/breast cancer | 1377 | 64.5 |
| FDR/SDR w/ovarian cancer | 374 | 17.5 |
| FDR/SDR w/colon cancer | 394 | 18.5 |
| Breast cancer pathology (n = 2425 cancers) | ||
| Behavior | ||
| Invasive | 1727 | 71.2 |
| In situ | 205 | 8.5 |
| Unknown | 493 | 20.3 |
| Histologyc | ||
| Ductal | 1210 | 49.9 |
| Lobular | 164 | 6.8 |
| Mixed | 132 | 5.4 |
| Other | 183 | 7.5 |
| Unknown | 736 | 30.4 |
| Grade | ||
| Low | 232 | 10.9 |
| Intermediate | 581 | 27.2 |
| High | 581 | 27.2 |
| Unknown | 1031 | 42.5 |
| ER status | ||
| Positive | 1203 | 49.6 |
| Negative | 349 | 14.4 |
| Indeterminate | 5 | 0.2 |
| Unknown | 868 | 35.8 |
| HER2 status | ||
| Negative | 888 | 36.6 |
| Positive | 289 | 11.9 |
| Indeterminate | 47 | 1.9 |
| Unknown | 1201 | 49.5 |
| Breast cancer full HR status known (n = 1055) | ||
| ER+Her2− | 651 | 61.7 |
| ER+Her2+ | 163 | 15.5 |
| ER−Her2+ | 93 | 8.8 |
| ER−Her2− | 148 | 14.0 |
FDR first degree relative, SDR second degree relative
a For race, other refers to American Indian, Alaskan Native, Native Hawaiian, Other Pacific Islander, or mixed race
b Age of BC diagnosis was unknown for seven cases
c For histology, other refers to medullary, mucinous, tubular
From the 2134 women, 2859 heterozygous single nucleotide or insertion/deletion variants were identified in 26 known or proposed breast cancer susceptibility genes (Fig. 1). Variants were classified using a modified American College of Medical Genetics and Genomics variant classification pipeline (Methods).16 Variants of uncertain significance (VUS) and likely benign/benign variants accounted for 96% of identified variants. We classified 114 (4%) of the variants as pathogenic or likely pathogenic mutations (further addressed as “mutations” throughout). In addition, analysis of next generation sequencing (NGS) data by a combination of the exome hidden Markov model17 and COpy number Detection by EXome sequencing18 algorithms, identified 14 germline copy number variants, 12 of which were classified as likely pathogenic (Supplementary Table 1). Therefore, counting single nucleotide variants, indels and copy number variants, 126 unique mutations were identified. As some mutations were recurrent, 183 total mutations were identified in FBC patients. The distribution of mutations by mutation type for each of the 26 analyzed genes are shown in Table 2. The same variant classification pipeline was then applied to the 9647 single nucleotide or insertion/deletion mutations identified in the 26 genes in ExAC data from non-Finnish European individuals (excluding samples from The Cancer Genome Atlas). As was seen in FBC women, 95% of the analyzed variants were classified as VUS or likely benign/benign and 5% of variants were classified as mutations (Fig. 1). Analysis of published exome hidden Markov model data for ExAC revealed 118 copy number variants in these genes.19 The majority of mutations identified were truncating variants in both FBC women and ExAC controls (Table 2).
Fig. 1.
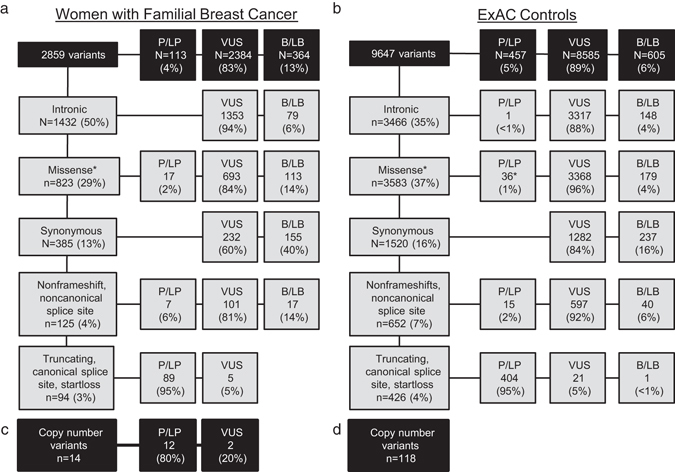
Variant classes identified in familial breast cancer women and Exome Aggregation Consortium (ExAC) controls. All single nucleotide and insertion/deletion variants in the 26 studied genes passing sequencing quality filters and with alternate allele frequencies (AAF) between 30 and 70% were classified by the described variant classification methodology. Variants are divided by general type (noncoding, missense, synonymous, loss of function and other) followed by the breakdown of pathogenic/likely pathogenic (P/LP) variants, variants of uncertain significance (VUS), and benign/likely benign (B/LB) variants in (a) familial breast cancer cases and (b) ExAC (The Cancer Genome Atlas excluded) non-Finnish European controls. Asterisks: Includes CHEK2 low-risk alleles p.I157T and p.S428F and MUTYH heterozygous pathogenic mutations in both data sets. The Exome hidden Markov model and COpy number Detection by EXome sequencing algorithms were used to identify copy number variants (CNVs) in the cases (c) and analysis of ExAC genome browser data was analyzed to identify CNVs in the controls (d)
Table 2.
Breakdown of types of pathogenic and likely pathogenic mutations identified in familial breast cancer women and ExAC controls
| Breast cancer casesa | EXAC controls | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Rates | Mutation typeb | Rates | Mutation typeb | ||||||||||
| Gene | n | % of 2134 | I/N | S | M | C | n | % of 26,375 | I/N | S | M | C | Other |
| # of unique mutations | 126 | 457 | |||||||||||
| Total # of mutations identified | 181 | 1612 | |||||||||||
| Total # of mutation carriersa | 175 | 8.2 | 1612 | 6.1 | |||||||||
| # of BC-associated mutation carriersc | 100 | 4.7 | 441 | 1.7 | |||||||||
| High risk breast cancera | 30 | 1.41 | 57 | 0.22 | |||||||||
| PALB2 a* | 19 | 0.89 | 19 | 0 | 0 | 0 | 33 | 0.13 | 28 | 2 | 0 | 2 | 1 |
| TP53* | 11 | 0.52 | 2 | 1 | 8 | 0 | 18 | 0.07 | 1 | 1 | 15 | 1 | 0 |
| CDH1 | 0 | 0.00 | 0 | 0 | 0 | 0 | 4 | 0.02 | 2 | 1 | 0 | 1 | 0 |
| PTEN | 0 | 0.00 | 0 | 0 | 0 | 0 | 1 | 0.00 | 1 | 0 | 0 | 0 | 0 |
| STK11 | 0 | 0.00 | 0 | 0 | 0 | 0 | 1 | 0.00 | 0 | 0 | 0 | 1 | 0 |
| Moderate risk breast cancera | 68 | 3.19 | 403 | 1.53 | |||||||||
| CHEK2 a* | 35 | 1.64 | 25 | 2 | 3 | 5 | 257 | 0.97 | 156 | 13 | 61 | 27 | 0 |
| ATM a* | 32 | 1.50 | 19 | 6 | 2 | 5 | 100 | 0.38 | 58 | 22 | 5 | 8 | 7 |
| NBN | 1 | 0.05 | 1 | 0 | 0 | 0 | 46 | 0.17 | 34 | 7 | 0 | 5 | 0 |
| Proposed breast cancera | 39 | 1.83 | 621 | 2.35 | |||||||||
| BARD1* | 7 | 0.33 | 6 | 1 | 0 | 0 | 33 | 0.13 | 25 | 2 | 0 | 6 | 0 |
| FANCM | 7 | 0.33 | 7 | 0 | 0 | 0 | 185 | 0.70 | 181 | 3 | 0 | 1 | 0 |
| BLM | 6 | 0.28 | 4 | 2 | 0 | 0 | 64 | 0.24 | 51 | 8 | 2 | 3 | 0 |
| RAD50 a | 4 | 0.19 | 4 | 0 | 0 | 0 | 79 | 0.30 | 75 | 4 | 0 | 0 | 0 |
| RAD51D | 4 | 0.19 | 3 | 1 | 0 | 0 | 7 | 0.03 | 6 | 0 | 0 | 1 | 0 |
| RAD51C a | 3 | 0.14 | 2 | 0 | 0 | 1 | 35 | 0.13 | 26 | 6 | 0 | 3 | 0 |
| BRIP1 | 2 | 0.09 | 2 | 0 | 0 | 0 | 46 | 0.17 | 39 | 3 | 0 | 4 | 0 |
| PPM1D | 2 | 0.09 | 2 | 0 | 0 | 0 | 27 | 0.10 | 25 | 0 | 0 | 2 | 0 |
| FANCC | 1 | 0.05 | 1 | 0 | 0 | 0 | 53 | 0.20 | 23 | 26 | 0 | 4 | 0 |
| MRE11A | 1 | 0.05 | 1 | 0 | 0 | 0 | 28 | 0.11 | 19 | 4 | 0 | 5 | 0 |
| RINT1 | 1 | 0.05 | 1 | 0 | 0 | 0 | 39 | 0.15 | 21 | 16 | 0 | 2 | 0 |
| XRCC2 | 1 | 0.05 | 1 | 0 | 0 | 0 | 13 | 0.05 | 8 | 2 | 0 | 3 | 0 |
| BAP1 | 0 | 0.00 | 0 | 0 | 0 | 0 | 12 | 0.05 | 10 | 0 | 0 | 2 | 0 |
| Lynch syndromea | 11 | 0.52 | 134 | 0.51 | |||||||||
| MSH6 a | 6 | 0.28 | 6 | 0 | 0 | 0 | 35 | 0.13 | 27 | 3 | 2 | 3 | 0 |
| MSH2 | 3 | 0.14 | 0 | 1 | 1 | 1 | 8 | 0.03 | 3 | 1 | 1 | 3 | 0 |
| PMS2 | 2 | 0.09 | 2 | 0 | 0 | 0 | 77 | 0.29 | 25 | 6 | 18 | 26 | 2 |
| MLH1 | 0 | 0.00 | 0 | 0 | 0 | 0 | 14 | 0.05 | 2 | 3 | 5 | 4 | 0 |
| MUTYH monoallelic | 33 | 1.55 | 3 | 4 | 26 | 0 | 397 | 1.51 | 21 | 9 | 366 | 1 | 0 |
a Double mutation carriers are counted once in the total number of mutation carriers. In the individual gene counts, cases with two pathogenic mutations are counted in the totals for both genes. Mutations are in CHEK2(4), ATM(3), PALB2(2), RAD50(2), MSH6(1), and RAD51C(1)
b Mutation type I/N indels and nonsense mutations, S splicing, M missense, C copy number variant, O nonconsensus splice site or other noncoding mutations
c Counting mutation carriers of genes with statisically significant odds ratios in case–control analyses (*)
From the FBC women, 183 mutations were identified in 176 women (8.2%), or 6.6% when excluding 33 women with monoallelic MUTYH mutation carriers (1.6% of cases) (Fig. 2; Table 2). The majority of women had only a single mutation identified. However, six women (0.3%) were double mutation carriers, with mutations found in two different genes, including two in ATM/CHEK2, two in PALB2/CHEK2, and one each of the following: ATM/RAD50 and MSH6/RAD50. Evaluation of the ExAC data for mutations in the 26 genes used in the study identified 1612 mutation carriers (6.1%), or 4.6% when excluding 397 monoallelic MUTYH mutation carriers (1.5% of controls) (Fig. 2; Table 2). Mutations were enriched in known high (odds ratio (OR) > 5) and moderate-risk (OR 2–5) genes among FBC women compared to ExAC controls (1.5 vs. 0.2% and 3.2 vs. 1.5%, respectively, p values <0.05) (Table 2). In contrast, the frequency of mutations in proposed breast cancer genes was greater in ExAC controls than among women with FBC (2.4 vs. 1.8%, p = 0.03). The proportions of individuals with heterozygous MUTYH and Lynch syndrome mutations were nearly identical between FBC women and ExAC controls (~1.5 and 0.5% respectively in both groups, p > 0.05).
Fig. 2.
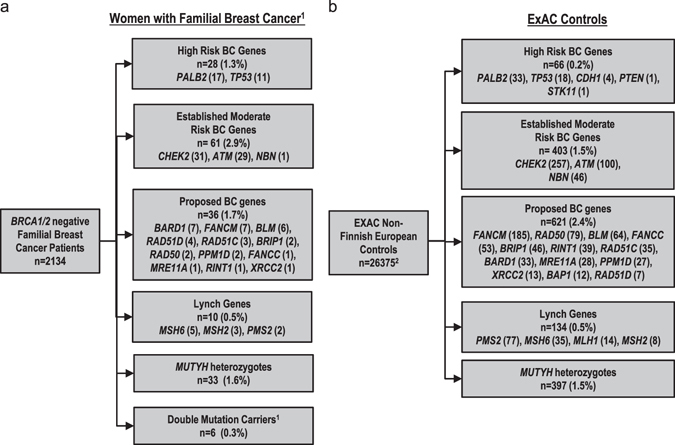
Spectrum of mutations in 26 genes in familial breast cancer women compared to Exome Aggregation Consortium (ExAC) controls. Rates of truncating, known pathogenic missense mutations, and copy number variants in (a) BRCA1/2 negative familial breast cancer women analyzed by targeted sequencing and (b) analysis of ExAC data (excluded copy number variation). Sequencing data was analyzed for mutations in high risk breast cancer genes, moderate risk breast cancer genes, proposed breast cancer genes, the Lynch syndrome genes, and MUTYH. 1Women with two pathogenic mutations were removed from single gene counts and include two cases with ATM/CHEK2 mutations, two with CHEK2/PALB2 mutations, and one patient each with ATM/RAD50, and MSH6/RAD50 mutations. 2Denominator for ExAC non-Finnish European controls (The Cancer Genome Atlas excluded) was determined by averaging the allele number for each identified variant and dividing by two.
Mutations in high-risk breast cancer genes or genes associated with syndromic diagnoses (CDH1, PALB2, PTEN, STK11, and TP53 6, 20, 21) were identified in 1.4% of the FBC cohort (Table 2). Excluding carriers of copy number variants and double mutation carriers, comparing women with FBC to ExAC controls, TP53 and PALB2 showed significant ORs of 8.17 (95% CI 3.74–18.26, p = 2.23 × 10−6) and 6.95 (CI 3.71–12.70, p = 2.33 × 10−8), respectively (Table 3). The average age at breast cancer diagnosis for TP53 mutation carriers was significantly younger than for mutation-negative individuals (Table 4) (age at diagnosis 39 years vs. 48 years, p = 8 × 10−5), and TP53 mutation carriers were more likely to have bilateral breast cancer than mutation-negative individuals (36 vs. 13%, p = 0.05). Compared to mutation-negative women, TP53 mutation carriers were not enriched for a family history of ovarian cancer (9% in mutation carriers vs. 17% in non-carriers, p > 0.05) or for first degree relatives with breast cancer (p > 0.05). Importantly, only five out of 11 (45%) TP53 mutation carriers met classic Li Fraumeni syndrome (LFS) or Chompret criteria, or had breast cancer diagnosed under the age of 31.20 TP53 mutation carriers also had a significantly higher proportion of HER2+ breast cancer compared to mutation-negative cases (83 vs. 24% in non-carriers, p = 0.004). In contrast, PALB2 mutation carriers were more likely to have ER−HER2− breast cancer (46% in mutation carriers vs. 14% in non-carriers, p = 0.006) and less likely to have ER+HER2− breast cancer (31% in mutation carriers vs. 62% in non-carriers, p = 0.04) compared to mutation-negative women. Of note, two women carrying the same CDH1 VUS, currently discordant in Clinvar as either a VUS or likely pathogenic (c.1118C>T, p.Pro373Leu), had no family history of gastric cancer and therefore did not meet criteria for hereditary diffuse gastric cancer syndrome. Both women had a history of invasive ductal (at ages 34 and 36) rather than lobular carcinoma.
Table 3.
Case–control analysis
| Gene | Case ACa | Case AN | Case AF | Control AC | Control AN | Control AF | OR | 95% CI | p-value |
|---|---|---|---|---|---|---|---|---|---|
| Gene (p < 0.05 and case number >5)b | |||||||||
| ATM | 24 | 4254 | 0.00564 | 92 | 53288 | 0.00173 | 3.28 | 2.06–5.21 | 3.64E−06 |
| BARD1 | 7 | 4254 | 0.00165 | 27 | 52157 | 0.00052 | 3.18 | 1.34–7.36 | 1.22E−02 |
| PALB2 | 17 | 4254 | 0.004 | 31 | 53738 | 0.00058 | 6.95 | 3.71–12.70 | 2.33E−08 |
| TP53 | 11 | 4254 | 0.00259 | 17 | 53579 | 0.00032 | 8.17 | 3.74–18.26 | 2.23E−06 |
| CHEK2 trunc | 23 | 4254 | 0.00541 | 169 | 50448 | 0.00335 | 1.617 | 1.03–2.51 | 4.11E−02 |
| Gene (p > 0.05 or case number < 5) | |||||||||
| RAD51D | 4 | 4254 | 0.00094 | 6 | 53110 | 0.00011 | 8.33 | 2.20–30.48 | 4.41E−03 |
| BLM | 6 | 4254 | 0.00141 | 61 | 53468 | 0.00114 | 1.24 | 0.52–2.85 | 6.36E−01 |
| BRIP1 | 2 | 4254 | 0.00047 | 42 | 53681 | 0.00078 | 0.60 | 0.10–2.33 | 7.70E−01 |
| FANCC | 1 | 4254 | 0.00024 | 49 | 52870 | 0.00093 | 0.25 | 0.01–1.48 | 1.81E−01 |
| FANCM | 7 | 4254 | 0.00165 | 184 | 53071 | 0.00347 | 0.47 | 0.22–1.01 | 5.12E−02 |
| MRE11A | 1 | 4254 | 0.00024 | 23 | 53534 | 0.00043 | 0.55 | 0.03–3.22 | 1.00E+00 |
| MSH2 | 2 | 4254 | 0.00047 | 5 | 50210 | 0.0001 | 4.72 | 0.67–22.80 | 9.84E−02 |
| MSH6 | 5 | 4254 | 0.00118 | 32 | 52301 | 0.00061 | 1.92 | 0.71–4.83 | 1.97E−01 |
| MUTYH | 33 | 4254 | 0.00776 | 396 | 53063 | 0.00746 | 1.04 | 0.72–1.48 | 7.82E−01 |
| NBN | 1 | 4254 | 0.00024 | 41 | 52529 | 0.00078 | 0.30 | 0.02–1.79 | 3.71E−01 |
| PMS2 | 2 | 4254 | 0.00047 | 51 | 49235 | 0.00104 | 0.45 | 0.08–1.72 | 4.40E−01 |
| PPM1D | 2 | 4254 | 0.00047 | 25 | 47537 | 0.00053 | 0.89 | 0.15–3.47 | 1.00E+00 |
| RAD50 | 2 | 4254 | 0.00047 | 79 | 52897 | 0.00149 | 0.31 | 0.06–1.15 | 9.14E−02 |
| RAD51C | 1 | 4254 | 0.00024 | 32 | 53293 | 0.0006 | 0.39 | 0.02–2.41 | 5.13E−01 |
| RINT1 | 1 | 4254 | 0.00024 | 37 | 53662 | 0.00069 | 0.34 | 0.02–2.06 | 5.25E−01 |
| XRCC2 | 1 | 4254 | 0.00024 | 10 | 53987 | 0.00019 | 1.27 | 0.06–8.63 | 5.66E−01 |
AC allele count, AN allele number, AF allele frequency, OR odds ratio, CI confidence interval, CHEK2trunc CHEK2 truncating mutations
a Cases with two mutations and CNVs were excluded from Case AC
b CHEK2 variants in aggregate did not reach statistical significance (See text)
Table 4.
Cancer history and breast cancer pathology characteristics of mutation carriers compared to non-mutation carriers
| No mutation | Mutation carrier | ATM | CHEK2 | PALB2 | TP53 | Candidate breasta | Ovariana | Lyncha | Double carriersa | |
|---|---|---|---|---|---|---|---|---|---|---|
| Total # of cases | 1936 | 143 | 29 | 31 | 17 | 11 | 35 | 9 | 10 | 6 |
| Average age of onset | 48 | 45 | 47 | 44 | 46 | 39 | 46 | 44 | 49 | 44 |
| p = 8e−5 | p = 0.02 | p = 1e−3 | ||||||||
| Personal cancer history | ||||||||||
| Bilateral breast cancer | 252 | 26 | 6 | 7 | 2 | 4 | 2 | 0 | 1 | 4 |
| 13% | 18% | 21% | 23% | 12% | 36% p = 0.05 | 6% | 0% | 9% | 67% p = 0.004 | |
| Ovarian cancer | 27 | 5 | 0 | 1 | 1 | 0 | 1 | 1 | 2 | 0 |
| 1% | 3% | 0% | 3% | 6% | 0% | 3% | 11% | 22% | 0% | |
| Any other cancer | 153 | 12 | 3 | 4 | 0 | 1 | 1 | 0 | 2 | 1 |
| 8% | 8% | 10% | 15% | 0% | 10% | 3% | 0% | 18% | 17% | |
| Family cancer history | ||||||||||
| FDR w/Breast cancer | 1247 | 93 | 18 | 27 | 10 | 7 | 21 | 6 | 7 | 6 |
| 64% | 65% | 62% | 87% p = 0.007 | 59% | 64% | 60% | 67% | 64% | 100% | |
| FDR/SDR w/ Ovarian cancer | 327 | 31 | 4 | 6 | 1 | 1 | 10 | 4 | 3 | 1 |
| 17% | 22% | 14% | 19% | 6% | 9% | 29% | 44% | 27% | 17% | |
| Any relative w/ Colon cancer | 348 | 34 | 8 | 9 | 2 | 3 | 7 | 3 | 2 | 2 |
| 19% | 24% | 28% | 29% | 12% | 27% | 20% | 38% | 18% | 33% | |
| Breast cancer pathology | ||||||||||
| Total # w/ER Status | 1400 | 112 | 25 | 20 | 16 | 6 | 27 | 6 | 8 | 5 |
| ER+BC | 1083 | 88 | 22 | 18 | 10 | 3 | 18 | 3 | 8 | 4 |
| 77% | 79% | 88% | 90% | 63% | 50% | 64% | 50% | 100% | 80% | |
| Total # w/Her2 Status | 1063 | 85 | 18 | 12 | 13 | 6 | 26 | 7 | 4 | 5 |
| Her2+BC | 259 | 22 | 5 | 5 | 2 | 5 | 3 | 2 | 0 | 0 |
| 24% | 26% | 28% | 42% | 15% | 83% p = 0.004 | 12% | 29% | 0% | 0% | |
| Total # w/ER +Her2 status | 1055 | 85 | 18 | 12 | 13 | 6 | 24 | 6 | 4 | 4 |
| ER+Her2− | 651 | 48 | 13 | 7 | 4 | 1 | 15 | 2 | 4 | 3 |
| 62% | 56% | 72% | 58% | 31% p = 0.04 | 17% | 63% | 33% | 100% | 75% | |
| ER–Her2− | 148 | 15 | 0 | 0 | 6 | 0 | 8 | 3 | 0 | 1 |
| 14% | 18% | 0% | 0% | 46% p = 0.006 | 0% | 33% | 50% | 0% | 20% | |
| ER+Her2+ | 163 | 16 | 4 | 4 | 3 | 2 | 2 | 1 | 0 | 0 |
| 15% | 19% | 22% | 33% | 23% | 33% | 8% | 17% | 0% | 0% | |
| ER–Her2+ | 93 | 6 | 1 | 1 | 0 | 3 | 1 | 0 | 0 | 0 |
| 9% | 7% | 6% | 8% | 0% | 50% p = 0.01 | 4% | 0% | 0% | 0% | |
a Candidate Breast: candidate breast cancer susceptibility genes (BARD1, BLM, FANCC, FANCM, MRE11A, PPM1D, RAD50, RINT1, XRCC2); Ovarian: Ovarian Cancer Genes (BRIP1, RAD51C, RAD51D); Lynch: Lynch syndrome genes (MLH1, MSH2, MSH6, PMS2); Double carriers: two women with ATM/CHEK2 mutations, two with CHEK2/PALB2 mutations, and one woman each with ATM/RAD50 and MSH6/RAD50 mutations
The ATM, CHEK2, and NBN 6 moderate-risk breast cancer genes6, 22 had the highest mutation rate in the FBC cohort at 3.19%, including five of the double mutation carriers. In the case–control analysis, excluding copy number variants and double mutation carriers, mutations in ATM were significantly enriched in FBC women compared to ExAC controls, OR = 3.28, 95% CI 2.06–5.21, p = 3.64 × 10−6 (Table 3). CHEK2 truncating mutations (OR = 1.62, 95% CI 1.03–2.51, p = 0.041) (Table 3) and CHEK2, p.1100delC alone (OR = 1.85, 95% CI 1.13–3, p = 0.017) were also enriched in FBC.23, 24 The personal history of bilateral breast cancer and other cancers were not statistically significantly higher for ATM or CHEK2 mutation carriers compared to mutation-negative women (Table 4). Cancer family history was also similar among these groups when compared to mutation-negative women, except that CHEK2 mutation carriers had a higher proportion of first-degree relatives with breast cancer (87% in mutation carriers vs. 64% in non-carriers, p = 0.007). None of the breast cancers arising in ATM and CHEK2 mutation carriers, for which hormonal status was available, were histologically classified as ER−HER2–, consistent with previous reports25 (Table 4). Only one case was identified with the common Slavic pathogenic variant NBN c.657del56 (Fig. 2); therefore precluding a case–control analysis.
Mutations in candidate breast cancer genes were only observed in 1.7% of women in this cohort (Fig. 2). Of these, only FANCM, BLM, and BARD1 had mutations in over five FBC women. Only BARD1 mutations were significantly associated with an increased risk of breast cancer (OR = 3.18, 95% CI 1.34–7.36, p = 0.012) in case–control analysis. The rates of bilateral breast cancer, ovarian cancer, family history of breast cancer in a first degree relative, family history of ovarian cancer, or pathological characteristics of the breast cancer were not significantly different in these women compared to mutation-negative women (Table 4). In addition, there were nine women with mutations in recently described ovarian cancer susceptibility genes, BRIP1, RAD51C, and RAD51D.22 A personal history of ovarian cancer only was reported for only one case; furthermore, these mutation carriers were not statistically more likely to have a first-degree or second-degree relative with ovarian cancer compared to mutation-negative individuals (Table 4).
Mutations in genes classically predisposing to colorectal cancer were infrequently observed. Mutations in Lynch syndrome (LS) genes (MLH1, MSH2, PMS2, MSH6) were identified in 11 cases, including one case with mutations in both MSH6 and RAD50. Six MSH6 mutations were detected among FBC cases, but this rate was not significantly higher than in ExAC control individuals (0.28 vs. 0.13%). Mutations in other LS genes also were not significantly associated with FBC. The age of onset of breast cancer in women with LS mutations and mutation-negative individuals was similar (age of onset 49 vs. 48, p-value not significant), and there were no differences in the rates of bilateral breast cancer or family history of breast or ovarian cancer (Table 4) or colon cancer (data not shown). Three of the ten LS gene mutation carriers had a personal history of ovarian or endometrial cancer. However, none had a personal history of colon cancer and only two had a family history of colon cancer (data not shown). All breast cancers for which information was available were classified as ER+. There were no phenotypic features identified specific to MUTYH mutation carriers; specifically, the rate of colon cancer family history was not higher in MUTYH mutation carriers compared to mutation-negative individuals (data not shown). No mutations were identified in BAP1, MLH1, PTEN, or STK11.
The frequency of copy number variants in genes associated with breast cancer in women with FBC has been largely understudied. Using COpy number Detection by EXome sequencing18 and the exome hidden Markov model,17 we identified copy number variants in 14 cases (0.7%), and classified 12 as mutations (Table 2, Supplementary Table 1). Two copy number variants, a whole gene duplication of STK11 and a single exon in-frame deletion in PTEN were classified as VUS. Five individuals had copy number variants in CHEK2, five in ATM, one in RAD51C and one in MSH2. Copy number variants in ATM, CHEK2, and RAD51C were confirmed by multiplex ligation-dependent probe amplification (MLPA) in the 11 individuals for which DNA was available (Supplementary Fig. 1). Interestingly, the MSH2 deletion was from an individual meeting Amsterdam I/II criteria for LS testing, and the individual with a RAD51C deletion had a personal history of both breast and ovarian cancer. As there were five or more individuals with copy number variants in ATM and CHEK2 each, a case–control analysis was performed individually for copy number variants identified in these genes in the FBC cohort compared to copy number variants identified in the ExAC cohort. Copy number variants in CHEK2 (OR = 3.2, CI 95% 1.64–5.85, p = 4.5 × 10−6) and ATM (OR = 7.7, 95% CI 3.13–18.1, p = 9.64 × 10−6) were enriched in the FBC women.
Discussion
We have described the spectrum of mutations in known or proposed breast cancer susceptibility genes in a large cohort of 2134 FBC patients. Overall, 8.2% of patients were found to carry a mutation in the genes analyzed. Mutations in ATM, CHEK2, and PALB2 accounted for the majority of mutations (4%), consistent with other studies showing that these are the most commonly mutated genes other than BRCA1 and BRCA2 that are associated with an increased risk of FBC.9, 10, 14, 26 While the assayed genes differ slightly between previously reported studies, our results showing a 4.3% mutation rate (Fig. 2) in high and moderate risk breast cancer predisposition genes among BRCA-negative FBC women, are also consistent with these reports.
In the present FBC cohort, only 1.3% of women carried a mutation in a high-risk breast cancer gene (Fig. 2). This risk estimate for TP53 mutations (OR = 8.17, 95% CI 3.74–18.26) has substantially tighter confidence intervals than previously reported risk estimates (OR = 11, 95% CI 0.6−201).14 However, further studies providing functional and risk associated annotation of variants, and accounting for contamination of true germline TP53 mutations with mosaic mutations that arise due to clonal hematopoiesis,27 are needed to define accurate risk estimates for breast cancer associated with TP53 inactivating mutations. PALB2 mutations were also associated with a high-risk of breast cancer (OR = 6.95, 95% CI 3.71–12.70), consistent with prior data.14 Our results support the rarity of CDH1, PTEN, and STK11 mutations in FBC cohorts and the classification of TP53 and PALB2 as high risk genes (OR > 5) for FBC. Adding samples with unexpected BRCA1/2 findings to the cohort of samples passing quality filters (n = 2211), it is important to point out that more BRCA1/2 mutations were incidentally identified in a cohort of reportedly BRCA1/2 negative individuals (3.5%) than mutations in the other high risk genes combined (1.3%). These results confirm that mutations in high risk genes outside of BRCA1/2 are exceedingly rare in FBC patients.
Regarding published moderate-risk genes (OR 2–5), this is the first large multigene case–control study that was able to estimate risks of FBC associated with mutations in ATM (OR = 3.28, 95% CI 2.06–5.21). CHEK2 truncating mutations, and c.1100delC alone were also associated with increased risks of breast cancer; however below an OR of 2, thus classifying CHEK2 as a low risk gene for FBC. Larger studies will be needed to accurately define the risk of breast cancer associated with missense mutations in these genes.
In the proposed breast cancer gene group, mutations in BARD1 were found significantly more frequently in individuals with FBC than in ExAC controls (OR = 3.18, 95% CI 1.34–7.36, p = 0.012) (Table 3). This is the first large study to show that BARD1 is a moderate risk breast cancer predisposition gene for FBC. Similarly, mutations in RAD51D were associated with increased risks of breast cancer (OR = 8.33, 95% CI 2.2−30.48, p = 0.0044). However, these findings should be interpreted with caution because the risks were based on four mutation carriers and are likely unstable. Mutation rates overall in LS genes were almost identical in FBC women and ExAC controls. However, mutations in MSH6 and MSH2 were substantially enriched (Table 2) and merit further consideration as FBC predisposition genes.21 Heterozygous mutations in MUTYH were not associated with increased breast cancer risk. Mutations in the proposed breast cancer gene groups (FANCM, BLM, RAD50, RAD51C, BRIP1, PPM1D, FANCC, MRE11A, RINT1, XRCC2, and BAP1) were rare. Therefore, the contribution of these genes to breast cancer risk could not be assessed.
Our results showing a significant association of FBC with mutations in ATM, BARD1, CHEK2, PALB2 validate a recent study investigating the association of breast cancer in a cohort of individuals who underwent genetic testing at a large testing company.28 The results here, albeit on a much smaller sample size, are derived from a cohort with well-curated family and personal cancer histories (instead of information only obtained from clinical testing report forms) and therefore provide needed validation of the results from a broad sample of individuals for whom less clinical information is available.
Consistent with other reports regarding genotype–phenotype correlations (Table 4), TP53 mutation carriers had a significantly higher portion of HER2+ breast cancer than those without mutations.29 Forty-five percent of TP53 mutation-positive women met LFS criteria. Since many women and families included in the current study were ascertained prior to multigene panel testing, emergence of well-defined medical management recommendations for LFS, and recent revisions in LFS criteria, it is likely that many of these cases would have been offered genetic testing under current standards of clinical care.4 PALB2 mutation carriers were more likely to have ER−HER2− breast cancer, whereas ATM and CHEK2 mutations were never associated with ER−HER2− breast tumors.25 BRIP1, RAD51C, and RAD51D mutation carriers were not more likely to have a first-degree or second-degree relative with ovarian cancer compared to mutation-negative FBC women, inconsistent with prior studies associating mutations in these genes with high-risks of ovarian cancer.22
This study is one of the first to thoroughly evaluate breast cancer predisposition genes included in typical multigene panels in BRCA-negative FBC and to estimate risks of cancer by comparison with the ExAC reference control dataset. Assuming one predisposition gene mutation on average per ExAC control individual, approximately 1.7% of controls had a high or moderate-risk breast cancer gene mutation and 4.4% had mutations in the 26 genes under evaluation (Fig. 2). The high mutation carrier frequency adds to previous data from control populations and clarifies the likelihood of finding unexpected or incidental hereditary cancer predisposition gene mutation carriers in the unaffected population.14 This also strengthens the justification for counseling about the potentially broad spectrum of genetic findings for individuals undergoing genetic risk assessment.2, 30 However, these results should be evaluated with caution as different ascertainment and sequencing techniques were used in the ExAC controls compared to the FBC cohort. In addition, the ethnic makeup of the FBC cohort and ExAC controls likely differs and it is possible that ORs may be over or underestimated based on differences in ethnic makeup or number of individuals of Ashkenazi Jewish status. Finally, the associations of FBC with BARD1 and CHEK2 truncating mutations are no longer significant when accounting for multiple testing (p-value threshold of <0.002), and therefore require validation in larger case–control studies.
Twelve individuals with breast cancer were identified as having copy number variants classified as likely pathogenic in these genes. Copy number variant analysis using NGS is promising, but is still in development. This is the first case–control analysis of this kind to include copy number variant evaluation. Copy number variants in CHEK2 and ATM showed significant associations. Of note, ATM showed a significantly elevated OR of 7.7 (95% CI 3.13–18.1, p = 9.64 × 10−6). Copy number variants were particularly enriched in the FBC women and will require further study. It is possible the FBC women with identified copy number variants represents an underestimate of the total number of copy number variant findings. However, it is unlikely that unidentified copy number variants account for the majority of the genetic etiology in mutation-negative FBC women. Additional research will be needed on this topic given the increasing use of NGS copy number variant calling for copy number variant detection by commercial genetic testing laboratories.
If we consider only mutations in genes that reached statistical significance in our analysis, (TP53, ATM, PALB2, BARD1, and truncating mutations in CHEK2), approximately 4.7% of BRCA negative FBC women have mutations in breast cancer susceptibility genes. Given that BRCA mutations are responsible for another 5–10% of attributable risk,7, 31 our results suggest that 86–91% of FBC women have no underlying genetic etiology that can be identified by mutational analysis of the exonic regions of genes on the majority of current breast cancer multigene panels, supporting the concept of missing heritability in breast cancer.14, 26 In this study over 90% of variants identified in these categories among our 26 selected genes were determined to be VUS (Fig. 1). Thus, additional research is needed to study the contribution of intronic, missense, and synonymous variants in the current panel genes and other candidate genes to FBC.
As shown above, the risk estimates for mutations in many proposed breast cancer genes did not reach statistical significance. Based on these data, clinical actionability of genes with low or undefined-risk remains unclear. Results from this study support targeting the known clinically-actionable high and moderate-risk genes with multigene panels as a first-tier approach to understanding breast cancer susceptibility.
Methods
The simplexo targeted resequencing
Case and control selection
2266 BRCA-negative women with FBC defined as a proband with breast cancer and at least two first-degree to third-degree relatives with breast or ovarian cancers under age 70 years were selected from multiple centers (City of Hope, University of Pennsylvania, Mayo Clinic, Memorial Sloan Kettering, British Columbia Cancer Agency, the FIRC Institute of Molecular Oncology, European Institute of Oncology, Stanford, Dana Farber Cancer Institute, and the National Institutes of Health) and their germline DNA (from blood or saliva) was sequenced for 26 known or proposed breast cancer susceptibility genes at Mayo Clinic and at the University of Pennsylvania. Germline DNA from two-hundred non-cancer controls from one of the contributing centers was used to estimate systematic sequencing artifacts of NGS. All individuals were consented and enrolled into the study through center specific Institutional Review Board approved protocols. Sequencing was completed once per sample.
Target selection and library preparation
The target region for sequencing of the candidate breast cancer susceptibility genes were selected based on coding exons. For established genes, intragenic regions were also included. Baits for solution based hybrid selection capture32 were designed using the Agilent SureSelect design tool (https://earray.chem.agilent.com/suredesign/). 500 nanograms of high molecular weight DNA was fragmented using a Covaris E-220. Fragmentation was verified by bioanalyzer. Library preparation was performed using the NEBNext Ultra DNA Library Prep kit (E7370L), and NEB Dual indexed adapters. Fragmented DNA (mean fragment size 300 ± 20 bp) was end repaired, adapter ligated, and size selected using Ampure XP /SPRI (A63881) and was PCR amplified for eight cycles. A post-PCR clean-up was performed and enrichment for targets was performed using the Agilent SureSelect protocol. Yields were assessed using BioAnalyzer. The mean library size was 300 ± 10 bp. NGS was performed on an Illumina HiSeq 2000 to an estimated 100× mean coverage for the target region to yield paired-end reads of 100 bp per sample, using 24 samples/lane.
Computational methods
FASTQ files were de-multiplexed to individual forward and reverse files. Initial QC was performed using the FASTQC33 tool kit. Read alignment was performed using BWA34 and statistics were generated using bamstats.35 The human reference GRCh37 with the addition of decoy regions V5 for improving alignment accuracy and speed was used in all cases.36 Sixty-one samples with low quality were removed using the specific criteria of: (1) samples with <1 M reads, (2) low mean bait coverage (<20) over target bases, (3) ±3 S.D. mean heterozygosity, or (4) first-degree relationship from calculated kinship coefficient values (>0.177). Similarly a variant was dropped if the site-specific call rate was less than 50%. Variant calling was performed using the Genome Analysis Toolkit (GATK) v2.6–4. For each sample, the BAM was sorted, duplicates were marked for deletion using Picard,37 indels were realigned and base recalibrated. A gVCF was created per sample using the HaplotypeCaller in GATK and all samples were recalled using the Genotype GVCF module. Variants were annotated using SnpEFF, CAVA, and ANNOVAR.38
Sample and variant level QC/QA
Pre-established criteria were used to identify ineligible individuals. Expected vs. Observed homozygosity using single nucleotide polymorphisms was computed using Plink39 and VCFtools40 for all samples to identify individual samples from males for removal (Supplementary Figure 2). Similarly, relatedness and ethnicity were estimated using kinship coefficients for all pairwise relationships using identity by state metrics. Individuals with a first-degree relative in the study or suspected duplicate samples were removed. Finally, 77 cases were found to carry BRCA mutations (including copy number variants) and were removed. A total of 2134 samples were available for subsequent analyses (Supplementary Figure 1).
In the FBC women sequenced, allelic ratios between 20 and 80% were required for variant inclusion. Allele ratios were not available for ExAC controls. Variants with allele frequencies (AF) > 0.03 in ExAC, ESP6500 and 1000 genomes were excluded (except for the known common pathogenic variant CHEK2 c.1100delC). The gene set was restricted to genes known (ATM, CDH1, CHEK2, NBN, PALB2, PTEN, STK11, and TP53) or presumed (otherwise in the list below) to have clinically relevant mutations associated with germline breast cancer susceptibility.20 The list included evaluation of introns and exons for ATM, BARD1, BLM, BRIP1, CDH1, CHEK2, MLH1, MRE11A, MSH2, MSH6, MUTYH, NBN, PALB2, PMS2, PTEN, RAD50, RAD51C, RAD51D, STK11, TP53, and XRCC2. Exon-only analysis was available for BAP1, FANCC, FANCM, PPM1D, and RINT1.
Variant classification methodology
Variants were classified by two independent methodologies and then subjected to an expert review process for final classification. Researchers classifying the variants were blinded as to the phenotype of the sample from which the variant was derived. In the first methodology, all variants with AF > 0.003 were annotated as VUS, likely benign, or benign. Variants were called pathogenic/likely pathogenic mutations when meeting one of two criteria: (1) Loss-of-function (nonsense, frameshift, splicing +/−1 or 2, whole gene deletion, truncating copy number variants (unless after a known truncating benign variant) and (2) Missense, splicing +/−3+ position, intronic or synonymous variants that were classified as pathogenic or likely pathogenic in ClinVar41 by two or more of the following clinical genetics group: Ambry, Sharing Clinical Reports Project, InVitae, GeneDX, Emory, or InSiGHT. One exception to this rule was for TP53 missense variants, due to the rarity of these variants, where only one group was required. In the second methodology of variant classification, variants were subjected to Ingenuity Variant Analysis (Qiagen Inc, Alameda, CA). The American College of Medical Genetics and Genomics and the Association for Molecular Pathology standards and guidelines for the interpretation of sequence variants were applied to obtain pathogenic/likely pathogenic/VUS/likely benign/benign calls.42 In the expert review, results were compared and all concordant calls accepted as final classifications based on the above guidelines.42 Discrepancies were evaluated based on literature and database sources and discussed by the variant classification team for consensus on a mutation or VUS/likely benign/benign final classification. Known low-risk alleles were classified in a separate low-risk allele group.
Case–control analysis
Allele counts for mutations were summed for each gene in FBC cases and non-Finnish European ExAC controls excluding samples from The Cancer Genome Atlas (TCGA) (ExAC v0.3.1 (non-TCGA), http://exac.broadinstitute.org/, accessed 29 December 2014). The ExAC dataset has been used for identifying genes associated with disease and estimating risks.15, 19, 43, 44 The allele number for ExAC controls was calculated by averaging the number of the highest quality allele calls across exonic regions. Associations with breast cancer for each gene were generated using a two-sided Fisher’s exact test using minimum likelihood method.
Identification of copy number variants
Potential copy number variants (one or greater exonic deletions and duplications) were identified with the exome hidden Markov model17 and COpy number Detection by EXome sequencing18 algorithms from NGS data. Copy number variants identified by both algorithms (n = 12) were examined in the Integrated Genome Viewer to confirm the visual presence of a low read count area spanning at least one exon. Copy number variants called were used as independent confirmation of exon deletions/duplications. Separate DNA aliquots than those used for sequencing was available from 12 of the 14 individuals for analysis by MLPA (Supplementary Table 1; Supplementary Figure 2). The ExAC genome browser (http://exac.broadinstitute.org/) data were downloaded and analyzed in the UCSC genome browser (http://genome.ucsc.edu/) for identification of copy number variants.19 All gene deletion and duplications spanning large genomic regions beyond a single gene were excluded as were deletions and duplications in repetitive regions, regions corresponding to pseudogenes, and spanning only single exons.
Clinical and statistical analysis
Clinical data was obtained for the patients in the local tumor set by IRB approved chart review after informed consent. Comparisons of rates in different groups were determined using a two-sided Fisher’s exact test of significance. Using a sample set of 2134 FBC individuals vs. 26,375 controls, this study had >80% power to detect a significant difference in breast cancer risk for genes with a >0.1% control carrier rate leading to relative risk of >4 and for genes with a >0.6% control carrier rate leading to relative risk of >2.
Data sharing statement
The mutational datasets generated during and/and/or analyzed during the current study are available from the corresponding author on reasonable request.
Electronic supplementary material
Acknowledgements
We would like to acknowledge the individuals and families that participated in the research, as well as, the funding agencies that made this work possible. We would like to thank Paolo Radice of the National Cancer Institute, Milan, Italy and Monica Barile of the European Institute of Oncology, in Milan, Italy. We would also like to thank the clinical investigators across the 39 sites in the United States contributing cases to the City of Hope Clinical Cancer Genomics Community Research Network. This work is supported by the Breast Cancer Research Foundation (F.J.C., K.L.N., K.O., J.N.W., S.M.D., M.E.R., J.M.F., J.E.G.), National Institutes of Health grants CA176785 (F.J.C), CA116167 (F.J.C and K.L.N), CA192393 (F.J.C, K.L.N., and K.O), CA92049 (J.L.), City of Hope Clinical Cancer Genomics Community Research Network and the Hereditary Cancer Research Registry was supported in part by RC4CA153828 (J.N.W.), National Cancer Institute Specialized Program of Research Excellence (SPORE) in Breast Cancer P50CA116201 (F.J.C.); Abramson Cancer Center Core grant CA016520 (K.L.N.); 3P30CA008748-4 (K.O.); CA178800 (J.V.), the U.S. Dept of Defense W81XWH-13-1-0338 (K.N.M.), the American Society of Clinical Oncology (K.N.M.), the Rooney Family Foundation (K.L.N.), the Commonwealth of Pennsylvania (K.L.N.), the Sharon Levine Corzine Cancer Research Fund (K.O.), the Robert and Kate Niehaus Clinical Cancer Initiative (K.O.), the Filomen M. D’Agostino Foundation (K.O.), the Andrew Sabin Family Fund (K.O.), STOP CANCER (T.P.S.), Oxnard Foundation (T.P.S., J.N.W.), American Cancer Society (J.N.W.), Avon Foundation (02-2013-044 to J.N. W.), Basser Center for BRCA (K.L.N., K.N.M. and S.M.D.) and the Morris and Horowitz Families Endowed Professorship (S.L.N.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Views and opinions of, and endorsements by the authors do not reflect those of the US Army or the Department of Defense. The Pennsylvania Department of Health specifically disclaims responsibility for any analyses, interpretations or conclusions.
Author contributions
T.P.S., K.N.M., J.L., J.V., K.L.N., S.N., F.J.C., J.N.W. wrote the manuscript. T.P.S., K.L.N., K.N.M., F.J.C., J.N.W., K.L.N., J.V., S.N.H., S.M.D., S.N., K.O., contributed to study design. Sample collection was completed by J.N.W., K.L.N., K.S., S.M.D., M.R., P.P., B.B., J.F., J.G., C.S., K.O., and F.J.C.. Next generation sequencing library preparation and sequencing completed by K.N.M., B.M.W.,C.H., J.L., A.M., J.V.. Informatics was completed by J.V., B.W.,V.R., T.T, S.N.H., and R.M.. Copy number variants analysis and validation completed by K.N.M, B.W., B.M.W., and K.D’A. Final mutation allele determination was completed by T.P.S., K.N.M., V.J., V.R., J.L., S.N., T.T., F.J.C., and J.N.W.
Competing interests
The authors declare that they have no competing financial interests.
Footnotes
Thomas P. Slavin, Kara N. Maxwell, Jenna Lilyquist and Joseph Vijai contributed equally to this work.
Kenneth Offit, Katherine L. Nathanson and Fergus J. Couch jointly supervised this work.
A correction to this article is available online at https://doi.org/10.1038/s41523-017-0046-2.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change History: A correction to this article has been published and is linked from the HTML version of this article.
Electronic supplementary material
Supplementary Information accompanies the paper on the npj Breast Cancer website (doi:10.1038/s41523-017-0024-8).
References
- 1.SEER. Cancer Stat Fact Sheets. http://seer.cancer.gov/statfacts/. Accessed 15 July 2016.
- 2.Weitzel JN, et al. Genetics, genomics and cancer risk assessment: state of the art and future directions in the era of personalized medicine. Cancer J. Clin. 2011;61:327–359. doi: 10.3322/caac.20128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blackwood MA, Weber BL. BRCA1 and BRCA2: from molecular genetics to clinical medicine. J. Clin. Oncol. 1998;16:1969–1977. doi: 10.1200/JCO.1998.16.5.1969. [DOI] [PubMed] [Google Scholar]
- 4.NCCN. NCCN Guidelines genetic/familial high-risk assessment: breast and ovarian V.2.2016 (2016).
- 5.Venkitaraman AR. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell. 2002;108:171–182. doi: 10.1016/S0092-8674(02)00615-3. [DOI] [PubMed] [Google Scholar]
- 6.Easton DF, et al. Gene-panel sequencing and the prediction of breast-cancer risk. N. Engl. J. Med. 2015;372:2243–2257. doi: 10.1056/NEJMsr1501341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tung N, et al. Frequency of mutations in individuals with breast cancer referred for BRCA1 and BRCA2 testing using next-generation sequencing with a 25-gene panel. Cancer. 2015;121:25–33. doi: 10.1002/cncr.29010. [DOI] [PubMed] [Google Scholar]
- 8.LaDuca H, et al. Utilization of multigene panels in hereditary cancer predisposition testing: analysis of more than 2000 patients. Genet. Med. 2014;16:830–837. doi: 10.1038/gim.2014.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cybulski C, et al. Mutations predisposing to breast cancer in 12 candidate genes in breast cancer patients from Poland. Clin. Genet. 2015;88:366–370. doi: 10.1111/cge.12524. [DOI] [PubMed] [Google Scholar]
- 10.Li J, et al. Targeted massively parallel sequencing of a panel of putative breast cancer susceptibility genes in a large cohort of multiple-case breast and ovarian cancer families. J. Med. Genet. 2016;53:34–42. doi: 10.1136/jmedgenet-2015-103452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aloraifi F, et al. Detection of novel germline mutations for breast cancer in non-BRCA1/2 families. FEBS J. 2015;282:3424–3437. doi: 10.1111/febs.13352. [DOI] [PubMed] [Google Scholar]
- 12.Kraus C, et al. Gene panel sequencing in familial breast/ovarian cancer patients identifies multiple novel mutations also in genes others than BRCA1/2. Int. J. Cancer. 2016;140:95–102. doi: 10.1002/ijc.30428. [DOI] [PubMed] [Google Scholar]
- 13.Tung N, Garber JE, Lincoln A, Domchek SM. Frequency of triple-negative breast cancer in BRCA1 mutation carriers: comparison between common Ashkenazi Jewish and other mutations. J. Clin. Oncol. 2012;30:4447–4448. doi: 10.1200/JCO.2012.44.5635. [DOI] [PubMed] [Google Scholar]
- 14.Thompson ER, et al. Panel testing for familial breast cancer: calibrating the tension between research and clinical care. J. Clin. Oncol. 2016;34:1455–1459. doi: 10.1200/JCO.2015.63.7454. [DOI] [PubMed] [Google Scholar]
- 15.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maxwell KN, et al. Evaluation of ACMG-guideline-based variant classification of cancer susceptibility and non-cancer-associated genes in families affected by breast cancer. Am. J. Hum. Genet. 2016;98:801–817. doi: 10.1016/j.ajhg.2016.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fromer M, et al. Discovery and statistical genotyping of copy-number variation from whole-exome sequencing depth. Am. J. Hum. Genet. 2012;91:597–607. doi: 10.1016/j.ajhg.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang Y, Oldridge DA, Diskin SJ, Zhang NR. CODEX: a normalization and copy number variation detection method for whole exome sequencing. Nucleic Acids Res. 2015;43:e39. doi: 10.1093/nar/gku1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruderfer DM, et al. Patterns of genic intolerance of rare copy number variation in 59,898 human exomes. Nat. Genet. 2016;48:1107–1111. doi: 10.1038/ng.3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.NCCN. NCCN genetic/familial high-risk assessment: breast and ovarian version 2.2017 (2017).
- 21.Provenzale D, et al. Genetic/familial high-risk assessment: colorectal version 1.2016, NCCN clinical practice guidelines in Oncology. J. Natl Compr. Cancer Netw. 2016;14:1010–1030. doi: 10.6004/jnccn.2016.0108. [DOI] [PubMed] [Google Scholar]
- 22.Tung N, et al. Counselling framework for moderate-penetrance cancer-susceptibility mutations. Nat. Rev. Clin. Oncol. 2016;13:581–588. doi: 10.1038/nrclinonc.2016.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shaag A, et al. Functional and genomic approaches reveal an ancient CHEK2 allele associated with breast cancer in the Ashkenazi Jewish population. Hum. Mol. Genet. 2005;14:555–563. doi: 10.1093/hmg/ddi052. [DOI] [PubMed] [Google Scholar]
- 24.Leedom TP, et al. Breast cancer risk is similar for CHEK2 founder and non-founder mutation carriers. Cancer Genet. 2016;209:403–407. doi: 10.1016/j.cancergen.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 25.Couch FJ, et al. Inherited mutations in 17 breast cancer susceptibility genes among a large triple-negative breast cancer cohort unselected for family history of breast cancer. J. Clin. Oncol. 2015;33:304–311. doi: 10.1200/JCO.2014.57.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tung N, et al. Frequency of germline mutations in 25 cancer susceptibility genes in a sequential series of patients with breast cancer. J. Clin. Oncol. 2016;34:1460–1468. doi: 10.1200/JCO.2015.65.0747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jaiswal S, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N. Engl. J. Med. 2014;371:2488–2498. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Couch, F. J. et al. Associations between cancer predisposition testing panel genes and breast cancer. JAMA Oncol. doi:10.1001/jamaoncol.2017.0424 (2017). [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- 29.Masciari S, et al. Breast cancer phenotype in women with TP53 germline mutations: a Li-Fraumeni syndrome consortium effort. Breast Cancer Res. Treat. 2012;133:1125–1130. doi: 10.1007/s10549-012-1993-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Slavin T, et al. Clinical application of multigene panels: challenges of next-generation counseling and cancer risk management. Front. Oncol. 2015;5:271. doi: 10.3389/fonc.2015.00271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bonadona V, et al. Contribution of BRCA1 and BRCA2 germ-line mutations to the incidence of breast cancer in young women: results from a prospective population-based study in France. Genes Chromosomes Cancer. 2005;43:404–413. doi: 10.1002/gcc.20199. [DOI] [PubMed] [Google Scholar]
- 32.Gumpel M, et al. Myelination and remyelination in the central nervous system by transplanted oligodendrocytes using the shiverer model. Discussion on the remyelinating cell population in adult mammals. Dev. Neurosci. 1989;11:132–139. doi: 10.1159/000111894. [DOI] [PubMed] [Google Scholar]
- 33.Andrews, S. FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/. Accessed 15 December 2016.
- 34.Li H, Durbin R. Fast and accurate long-read alignment with burrows-wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H, et al. The sequence alignment/map format and SAM tools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.1000 Genomes 1000 genomes technical reference. ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/technical/reference/phase2_reference_assembly_sequence/. Accessed 2016 (2011).
- 37.Wysoker, A., Tibbetts, K. & Fennell, T. Picard tools version 1.90. 2013. http://picard.sourceforge.net. Accessesd 22 April 2013.
- 38.Yang H, Wang K. Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat. Protoc. 2015;10:1556–1566. doi: 10.1038/nprot.2015.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chang CC, et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Danecek P, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Landrum MJ, et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42:D980–D985. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Richards S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American college of medical genetics and genomics and the association for molecular pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walsh, R. et al. Reassessment of Mendelian gene pathogenicity using 7855 cardiomyopathy cases and 60,706 reference samples. Genet. Med. 19, 192–203 (2017). [DOI] [PMC free article] [PubMed]
- 44.Nouhravesh N, et al. Analyses of more than 60,000 exomes questions the role of numerous genes previously associated with dilated cardiomyopathy. Mol. Genet. Genom Med. 2016;4:617–623. doi: 10.1002/mgg3.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


