Abstract
Members of the genus Lactobacillus are common inhabitants of the gut, yet little is known about the traits that contribute to their ecological performance in gastrointestinal ecosystems. Lactobacillus reuteri 100-23 persists in the gut of the reconstituted Lactobacillus-free mouse after a single oral inoculation. Recently, three genes of this strain that were specifically induced (in vivo induced) in the murine gut were identified (38). We report here the detection of a gene of L. reuteri 100-23 that encodes a high-molecular-mass surface protein (Lsp) that shows homology to proteins involved in the adherence of other bacteria to epithelial cells and in biofilm formation. The three in vivo-induced genes and lsp of L. reuteri 100-23 were inactivated by insertional mutagenesis in order to study their biological importance in the murine gastrointestinal tract. Competition experiments showed that mutation of lsp and a gene encoding methionine sulfoxide reductase (MsrB) reduced ecological performance. Mutation of lsp impaired the adherence of the bacteria to the epithelium of the mouse forestomach and altered colonization dynamics. Homologues of lsp and msrB are present in the genomes of several strains of Lactobacillus and may play an important role in the maintenance of these bacteria in gut ecosystems.
The guts of mammals are colonized by a complex collection of microorganisms (the gut microbiota) that influence biochemical, physiological, immunological, and nonspecific disease resistance characteristics of the host (10). The bacterial composition of this ecosystem has been shown to be remarkably stable over time (43). Members of the gut microbiota have presumably coevolved with their host species and must possess traits that enable them to establish and maintain themselves in a lotic (flowing) and competitive environment. On the other hand, there are microbes present in the gastrointestinal tract that have a transient existence (allochthonous components), and this situation raises questions about what factors distinguish resident members of the microbiota from allochthonous members (27).
Lactobacillus reuteri strain 100-23 is a true (autochthonous) resident of the murine gut because it adheres to the nonsecretory epithelium of the forestomach and persists at constant population levels in particular regions of the gut throughout the life of the murine host (13, 39).
We maintain a colony of mice whose gut microbiota is devoid of lactobacilli but is otherwise equivalent to that of conventional mice (32). These reconstituted Lactobacillus-free (RLF) mice permit the study of Lactobacillus traits against a standardized bacteriological background. L. reuteri 100-23 and the RLF mouse gut therefore provide an appropriate model system with which to study the molecular basis of Lactobacillus autochthony.
While L. reuteri 100-23 inhabits the murine gut, genes encoding methionine sulfoxide reductase (msrB), xylose isomerase (xylA), and a protein with low similarity to methionine synthase II (met) are specifically induced (38). Additionally, this strain produces a number of surface proteins that can be extracted from the bacterial cell surface (3). We describe the identification and characterization of a gene encoding a large surface protein (Lsp) and the testing of insertional mutants of the lsp gene and of the three in vivo-induced genes for ecological performance in the RLF mouse gut. Additionally, the abilities of the lsp mutant strain and the wild type to adhere to the epithelium of the murine forestomach were compared by scanning electron microscopic (SEM) observations.
MATERIALS AND METHODS
Bacterial strains and media. The bacterial strains used in this study were L reuteri 100-23C (13), which is a plasmid-free derivative of L. reuteri 100-23 (39), and insertional mutants of this strain. Cultures were propagated anaerobically at 37°C in Lactobacilli MRS medium (Difco) unless otherwise stated. When required, antibiotics were added to the medium (erythromycin, 5 μg/ml; chloramphenicol, 7.5 μg/ml).
Characterization of genes potentially affecting ecological performance.
The genes for methionine sulfoxide reductase B (msrB) and xylose isomerase (xylA) and a gene encoding a protein with low similarity to methionine synthase II (met) were shown previously to be induced in vivo by L. reuteri 100-23C in the murine gut (38). The complete sequences of xylA and msrB were obtained by chromosomal walking using the Vectorette system (Sigma Genosys, The Woodlands, Tex.) according to the manufacturer's instructions.
Surface proteins of L. reuteri 100-23 were extracted from bacterial cells and analyzed as described previously (3). In order to identify the gene encoding a 185-kDa surface protein, a genomic library of L. reuteri 100-23 DNA was screened by colony hybridization (Western analysis) using an antiserum raised against the purified protein. To prepare the antiserum, the proteins were separated using preparative sodium dodecyl sulfate (SDS)-8% polyacrylamide gel electrophoresis (PAGE), and the polypeptides were visualized by soaking the gel in 4 M sodium acetate solution. The band corresponding to the protein of ca. 185 kDa was excised, and the polypeptide was electroeluted (Bio-Rad model 422 Electro-Eluter), dialyzed for 36 h against 5 liters of distilled water at 4°C, and freeze-dried. Antibodies were raised in New Zealand White rabbits by intramuscular injection of 20 μg of protein, followed by two boosts of 5 μg at 2 and 3 months. The antiserum was used in a 1:500 dilution in combination with 125I-labeled protein A to screen a library of L. reuteri 100-23 DNA in Escherichia coli XL1-Blue. To prepare the library, chromosomal DNA from L. reuteri was partially digested with Sau3AI and partly filled, and the DNA was ligated into λZAP II (Stratagene) and packaged in vitro according to the manufacturer's instructions. The phagemid p-Bluescript SK(−) of each positive clone was excised in vitro according to the manufacturer's instructions. One clone, which contained an insert of ca. 6 kb, was sequenced by primer walking starting with the m13 universal and m13 reverse primers located on the multiple cloning site of the vector.
Homology searches were performed against the GenBank database using the BLASTX and BLASTN algorithms (http://www.ncbi.nlm.nih.gov/BLAST). Computer-assisted analysis of nucleotide sequence data was done using the GeneJockey program (BioSoft, Cambridge, United Kingdom). The signal sequences of the putative proteins were predicted using the SignalP program (19). The number of copies of lsp was determined by Southern hybridization of genomic DNA. DNA of L. reuteri 100-23C was digested with the restriction enzymes EcoRI, AluI, and TacI (Roche), and the DNA fragments were separated by standard agarose electrophoresis. The DNA was transferred to positively charged nylon membranes by vacuum plotting using a Bio-Rad model 785 vacuum plotter according to the manufacturer's instructions. Primers LSPprobeForw and LSPprobeRev (Table 1) were used to amplify the terminal 208 bp of the known sequence of the lsp gene. The PCR product was purified, radioactively labeled, and used as a probe in hybridization experiments using standard methods (24).
TABLE 1.
Oligonucleotide primers used in this study
| Target gene | Primer | Sequence (5′-3′)a | Application |
|---|---|---|---|
| msrB | 130forward1 | TGACTGGATCCTAATATGATCGATTTGATGAACC | Insert primer |
| 130reverse1 | TGACTGAATTCCCTTACTTCTTACCTCAGTCC | Insert primer | |
| 130kon1 | GTAACGAAAACGAGGAGGATGC | Test primer | |
| 130kon2 | GTCGTCCTTCGCTTTATGAC | Test primer | |
| xylA | 139Pr1 | TGACTGGATCCTAAGTTCTTTGGAATACTTCAAACC | Insert primer |
| 139Pr2 | TGACTGAATTCGTTCAAGAAGCATTTGAGCGTC | Insert primer | |
| 139ivet2 | TGAAGGATTGGCTTCGTTTCTC | Test primer | |
| 139kon2 | ATATTGGTGAGTAGTTGGTTCC | Test primer | |
| met | 146vor1 | TGACTTCTAGATAATTTGCAAACGGCGAAATAACAC | Insert primer |
| 146rück1 | TGACTGGATCCTTCGGGACGAAGAAATTCTTC | Insert primer | |
| 146ivet4 | TTATTGCTTCAATTCTTGAACC | Test primer | |
| 146test1 | AAGCAGCGGCAACGTCATGATC | Test primer | |
| lsp | Lspvor1 | TGACTGGATCCTAAAATAGTGAAGCTGGTAGTCAACC | Insert primer |
| Lsprück1 | TGACTGAATTCTATTAGCAACGGTAATATCAC | Insert primer | |
| LspPr1 | GCAAGAGCCGAAAAAGCAAC | Test primer | |
| Lsptest1 | CTGGATTAGGTCGACGAATTCC | Test primer | |
| LSPprobeForw | CCAGATTCGAGCGATATTAC | Probe | |
| LSPprobeRev | GATCTTGGTAACCTGCGAC | Probe |
Restriction sites are italic; translation stop codons are printed in boldface.
Gene inactivation.
Gene inactivation was achieved by site-specific integration of plasmid pORI28 into the L. reuteri 100-23C chromosome as described by Russell and Klaenhammer (23) but using the temperature-sensitive plasmid pVE6007 (18) as the helper plasmid instead of pTRK669. Internal fragments of the target genes were amplified by PCR (using the insert primers in Table 1) and inserted into pORI28 (an Emr repA-negative derivative of pWV01) by directional cloning using E. coli EC1000 (which contains a copy of the pVW01 repA gene in the chromosome) as the host (15, 23). The plasmids were purified and used to electrotransform L. reuteri 100-23C containing pVE6007 (a Cmr repA-positive temperature-sensitive derivative of pWV01) using 35°C for phenotypic expression and incubation. Lactobacilli carrying both plasmids were propagated overnight at 35°C in the presence of erythromycin and chloramphenicol. Then, 50 μl of this culture was used to inoculate 10 ml of prewarmed (44°C) MRS broth without antibiotics. After incubation for 8 h at 44°C, 100 μl was plated on prewarmed MRS agar plates containing erythromycin and incubated at 44°C overnight. The transformants were recovered and subcultured at 44°C to verify purity. The absence of plasmids was confirmed by plasmid extractions and agarose gel electrophoresis (38). The integration of the pORI28-based plasmids into target genes was checked by PCR using primers flanking the target region (test primers in Table 1). To determine the stability of the integrations, the strains were propagated in MRS broth in the absence of antibiotic for ca. 90 generations and then plated on MRS agar without antibiotic. Loss of the integrated plasmid was determined by replica plating colonies on MRS agar plates with or without erythromycin.
Mutant phenotype analysis.
Xylose utilization was tested by culture in MRS broth in which glucose was replaced by 1% xylose. For analysis of cell surface proteins, bacterial cells were recovered from 16-h cultures in MRS broth by centrifugation (6,500 × g; 10 min; 4°C) and washed once with TES buffer (10 mM Tris-HCl, 1 mM EDTA, 25% sucrose, pH 8.0). After resuspension of the cells in 2 ml of TES buffer containing 5 mg of lysozyme (Sigma) and 0.1 mg of mutanolysin (Sigma) per ml, the preparation was incubated at 37°C for 30 min, cooled on ice (15 min), and centrifuged for 10 min at 2,500 × g at 0°C. The supernatant was removed, 225 μl of 10 mM phenylmethylsulfonyl fluoride in isopropanol was added, and the preparation was kept on ice for 1 h. The solution was dialyzed against 500 ml of TE1/1 solution (1 mM Tris-HCl, 1 mM EDTA, pH 8.0) for 3 h using dialysis tubing with a molecular mass cutoff of 12,000 to 14,000 kDa (Sigma). The volume was reduced to 100 μl using an Eppendorf 5531 concentrator. The resulting protein preparations were examined by standard SDS-PAGE using a precast gradient gel (4 to 20%) and the SilverQuest Silver Staining kit (Invitrogen).
Testing the ecological performance of mutants.
Cells grown in MRS broth were centrifuged (see above), washed in 0.9% sodium chloride solution, and then resuspended in an appropriate volume of the same solution to achieve the desired inoculation dose. Mutants, or 1:1 mixtures of mutant and wild type, were administered by intragastric gavage to anesthetized mice obtained from our colony of RLF mice and maintained in isolators (32). All animal experiments were approved by the Otago University Animal Ethics Committee (approval number 104/02). The mice were killed 7 days after inoculation, and lactobacilli were cultured quantitatively by previously described methods (13) from the forestomach, jejunum, and cecum. To determine the proportion of the mutant strain, the samples were plated on agar plates with and without erythromycin. Colonization dynamics of the lsp mutant strain and the wild type were determined by culture of fecal pellets collected on days 2, 4, and 6 after inoculation with ca. 105 bacteria. In additional groups of mice, the forestomach was removed and investigated by electron microscopy (see below).
Comparison of in vivo and ex vivo adherence of lsp mutant and wild type to the forestomach epithelium.
The forestomachs of mice inoculated with either the lsp mutant or the wild type were fixed in 0.1 M sodium cacodylate buffer containing 2% glutaraldehyde (pH 7.4) for 24 h at 4°C. The tissue was then washed in 0.1 M sodium cacodylate buffer (pH 7.4) and placed in 1% osmium tetroxide in 0.1 M sodium cacodylate buffer at room temperature for 1 h. The samples were washed, first with cacodylate buffer and then with distilled water. The tissue was dehydrated in an ethanol series and then critical point dried using a Baltec CPD-030 critical-point dryer. Samples were mounted on stubs and sputter coated with gold-palladium using a Bio-Rad coating unit. The forestomach epithelial surface was viewed using a Cambridge S360 scanning electron microscope operated at an accelerating voltage of 10 kV.
To study adherence of lactobacilli to the forestomach epithelium ex vivo, cells harvested from overnight cultures of the mutant and wild type were washed with phosphate-buffered saline (PBS; pH 7.4) and resuspended in PBS. The epithelia of forestomachs dissected from RLF mice were exposed to the bacterial suspensions for 5 min at room temperature. After being washed with PBS, the forestomach samples were processed as described above and viewed by SEM.
Nucleotide sequence accession numbers.
The sequences reported in this paper have been deposited in the GenBank database (accession no. AY602215 to AY602218).
RESULTS
Characterization of in vivo-induced genes and lsp.
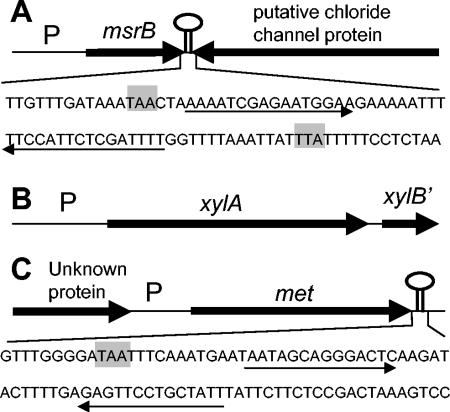
The organization of the three in vivo-induced loci is shown in Fig. 1. BlastX analysis (Table 2) of the complete gene sequences confirmed identifications that had previously been based on partial sequences (38). A sequence with a potential stem-loop structure followed by an AT-rich region was present downstream of msrB and met. These structures probably represent rho-independent terminators (Fig. 1). A gene encoding a putative chloride channel protein was found adjacent to msrB but was oriented in the opposite direction (Fig. 1). An open reading frame with homology to the gene for xylose kinase (XylB) was located downstream of xylA in the same orientation, indicating that the genes for xylose utilization by L. reuteri 100-23 were organized in an operon, as described for several other gram-positive bacteria (9). Sequence analysis of the promoter region of met revealed that gene expression is likely to be transcriptionally regulated by the T box mechanism (12). The predicted RNA sequence contains an RNA leader with a methionine specifier sequence typical of lactic acid bacteria, and expression of met may respond to methionine limitation by binding of uncharged tRNA-Met, resulting in stabilization of the antiterminator (T. M. Henkin, personal communication). This observation and the results of the sequence analysis (Table 2) suggest that met encodes a methionine synthase.
FIG. 1.
Maps of L. reuteri 100-23 genes selectively induced in the murine gastrointestinal tract. (A) msrB; (B) xylA; (C) met. P, promoter regions identified by in vivo expression technology. The sequences downstream of msrB and met are depicted, including stop codons (grey), and stem-loop structures possibly representing rho-independent terminators are indicated by arrows.
TABLE 2.
L. reuteri 100-23 genes mutated in this study with examples of matches in database, similarity, and predicted function
| Gene (bp) | Similar protein(s)a | Organism | % Identity/no. of amino acidsa | Function |
|---|---|---|---|---|
| msrB (429) | Peptide methionine sulfoxide reductase | Several organisms | 71/140 (L. johnsonii) | Reduction of oxidized methionine residues in proteins |
| xylA (1,350) | Xylose isomerase | Several organisms | 81/447 (Lactobacillus brevis) | Xylose utilization |
| met (1,137) | Hypothetical protein | Several organisms | 48/368 (L. johnsonii) | Unknown; low similarity (<25%) to methionine synthase II (cobalamin independent) of several organisms |
| lsp (5,905) | LJ0621 | L. johnsonii | 83/1,073 | Unknown |
| COG2931 | L. gasseri | 25/1,687 | Unknown | |
| Rlp | L. fermentum | 25/811 | Unknown | |
| R28 | Streptococcus pyogenes | 25/550 | Adhesion to epithelial cells | |
| Esp | Enterococcus faecalis | 24/611 | Biofilm formation | |
| Bap | S. aureus | 20/1,611 | Biofilm formation |
BlastX results.
The investigation of cell surface-associated proteins of lactobacilli isolated from the guts of different animals and humans revealed that some of these bacteria produce proteins with a high molecular mass (3). As the constitutive expression of these large proteins is associated with a high energy burden for the organism, it is likely that these large molecules are of ecological importance. L. reuteri 100-23 expressed several cell surface proteins, with the largest being ∼185 kDa (Fig. 2A). Antibodies raised against this protein detected a phagemid that contained the start of an open reading frame (5,905 bp) encoding a high-molecular-mass protein (Lsp) that had considerable nucleotide base sequence homology (86% identity over 1,899 bp) to the putative surface protein LJ0621 of Lactobacillus johnsonii NCC 533 (20). Sequence analysis using the BlastX algorithm (translated query vs. protein database) revealed similarity to several hypothetical proteins in the L. johnsonii NCC 533 genome and the unpublished Lactobacillus gasseri ATCC 33323T genome. Additionally, Lsp showed sequence similarities to regions within surface proteins of other gram-positive bacteria, including proteins involved in biofilm formation or adherence to epithelial cells (4, 28, 34, 35). Examples are shown in Table 2.
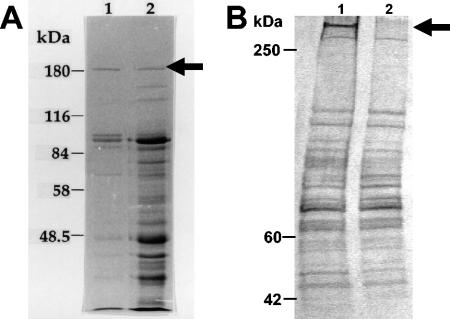
FIG. 2.
SDS-PAGE of cell surface proteins. (A) Surface proteins of L. reuteri 100-23 extracted using a buffer containing SDS and 3-mercaptoethanol without (lane 1) and with (lane 2) sonication. The protein that was used for detection of the lsp gene is marked with an arrow. (B) Surface proteins of the wild type (lane 1) and the lsp mutant (lane 2) extracted with a buffer containing lysozyme and mutanolysin. Lsp is marked with an arrow.
Lsp contained the same YSIRK-type 46-amino-acid signal peptide as the putative extracellular protein Ire0021 of L. reuteri DSM 20016T (36), indicating that the protein was transported to the cell surface. Surprisingly, the cloned partial lsp gene encoded a protein of >208 kDa, which is larger than the protein that was eluted from the SDS-PAGE gel. PCR-assisted gene walking and inverse PCR did not result in PCR products that could be used to detect the end of lsp, so the total size and sequence of the gene is not known. The Lsp homologues were predicted to be high-molecular-mass surface proteins containing a typical C-terminal gram-positive bacterial cell wall sorting signal containing the sequence LPXTG. The closest homologue, LJ0621 from L. johnsonii, has a size of 296 kDa and contains an LPQTG motif. It is therefore likely that Lsp is attached to the cell wall. Southern analysis revealed that lsp is present in one copy in the genome of L. reuteri 100-23C (data not shown).
Gene inactivation and mutant phenotype analysis.
Four different mutants were derived by integrating pORI28-based plasmids into target genes (Table 2), leading to the lsp, msrB, xylA, and met mutant strains. Insertion of the plasmids did not affect the growth of the bacteria in vitro. The xylA mutant strain did not grow in MRS broth containing xylose instead of glucose. After 90 generations of growth in the absence of antibiotic, 96.3 (xylA), 98.9 (msrB), 97.5 (met), and 99.8% (lsp) of the Lactobacillus populations retained erythromycin resistance. This was a level of in vitro stability similar to that described for insertional mutants of L. gasseri and Lactobacillus acidophilus (23).
The SDS-PAGE profiles of proteins extracted as described by Chagnaud et al. (3) showed that the 185-kDa protein that was initially used to prepare the antiserum was still produced by the lsp mutant strain (data not shown). The SDS-PAGE profile of the proteins extracted from the wild type using a buffer containing cell wall-degrading enzymes revealed the presence of a protein with a high molecular mass (ca. 300 kDa). This protein was absent from the profile of the lsp mutant (Fig. 2B).
Ecological performance of mutants in the murine gut.
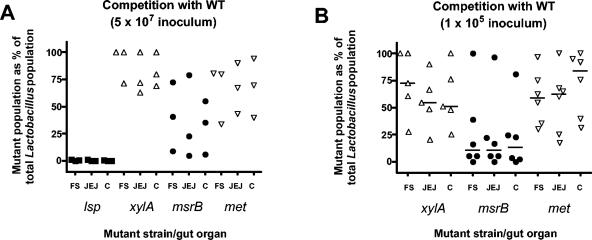
When administered as pure cultures to mice, the four mutants persisted in the gut and could be recovered from the forestomach, jejunum, and cecum at population levels similar to that of the wild type (Table 3) (13, 38). The percentages of revertants in vivo were <10% for the xylA and msrB mutants but slightly higher for the met and lsp mutants. When the mutants were used as an inoculum together with the wild type in a 1:1 ratio (ca. 5 × 107 bacteria per inoculum), the lsp mutant was recovered at rates of <1.5% of the total Lactobacillus population (Fig. 3A). In contrast, the xylA and met mutants were not impaired in colonization ability. The ability of the msrB mutant to compete with the wild type varied from mouse to mouse (Fig. 3A). Additional mice were inoculated with a smaller inoculum (ca. 105 bacteria). Populations of the msrB mutant were reduced relative to the wild type in three out of six mice (Fig. 3B). xylA and met mutant populations were not different from that of the wild type (Fig. 3B).
TABLE 3.
Populations of mutants in the murine gut 7 days after inoculation
| Mutant strain | Forestomach Lactobacillus populationa | % Emrb | Jejunum Lactobacillus population | % Emr | Cecum Lactobacillus population | % Emr |
|---|---|---|---|---|---|---|
| msrB | 8.9 (0.1) | 114.9 (2.6) | 7.4 (0.5) | 96.2 (9.6) | 7.1 (0.6) | 103.4 (6.4) |
| xylA | 8.3 (0.4) | 92.9 (6.8) | 6.2 (0.7) | 109.1 (12.1) | 7.8 (0.6) | 92.0 (7.2) |
| met | 8.7 (0.2) | 78.9 (20.1) | 7.2 (0.2) | 73.8 (11.1) | 8.2 (0.2) | 94.7 (15.2) |
| lsp | 8.0 (0.4) | 67.2 (5.3) | 6.5 (0.7) | 78.9 (4.4) | 7.2 (0.5) | 89.2 (8.3) |
Expressed as mean (three mice) log10 CFU/gram of organ; standard error of the mean in parentheses.
Mean percentage of colonies recovered on media containing erythromycin; standard error of the mean in parentheses.
FIG. 3.
Competition between wild-type and mutant strains. Mixtures of mutant and wild type (1:1) were used to inoculate RLF mice, and the percentage of mutants in the total Lactobacillus population was determined after 7 days. lsp (solid squares), xylA (open triangles up), msrB (solid circles), met (open triangles down). (F/S, forestomach; JEJ, jejunum; C, cecum). (A) Dose ca. 5 × 107 bacteria per mouse. (B) Dose ca. 105 bacteria per mouse. Values for individual mice are shown. Bars indicate medians.
Colonization dynamics and adherence of the lsp mutant.
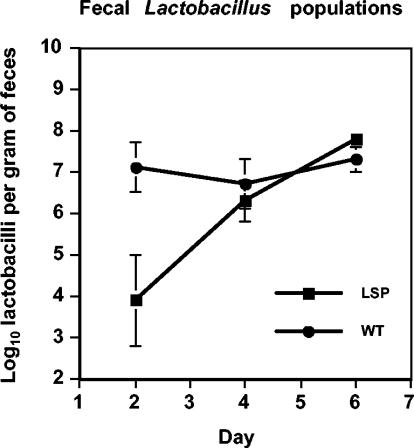
Monitoring fecal populations of lactobacilli showed that attainment of maximal population size by the lsp mutant was delayed compared to the wild type (Fig. 4). The counts of the lsp mutant were comparable on plates with and without erythromycin (data not shown).
FIG. 4.
Enumeration of lactobacilli (on MRS without erythromycin) in the feces of mice inoculated with the wild type (three mice) or the lsp mutant (four mice). Means and standard errors of the mean (bars) are shown.
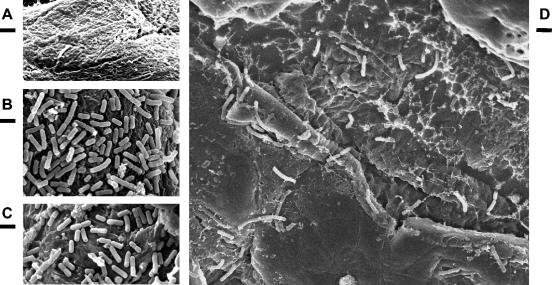
Scanning electron microscopic observations of forestomachs of mice showed that bacterial cells were absent from the forestomach epithelial surfaces of RLF mice (Fig. 5A). The forestomach epithelia of mice that had been inoculated with the wild-type strain 7 days previously were covered by a layer of adherent bacilli morphologically consistent with lactobacilli (Fig. 5B). Examination of samples from mice inoculated with the mutant also showed association of bacterial cells with the epithelium, but the coverage appeared to be slightly less than that of the wild-type strain (Fig. 5C). Examination by scanning electron microscopy of forestomachs inoculated ex vivo with wild-type cells revealed moderate numbers of rod-shaped bacterial cells associated with the epithelial surface (Fig. 5D). In contrast, extensive searches of forestomachs were required to detect any bacterial cells associated with samples that had been inoculated with the lsp mutant.
FIG. 5.
In vivo and ex vivo adherence of L. reuteri 100-23C and the lsp mutant to the forestomach epithelium assessed by SEM. The bars represent one micrometer. Forestomach epithelium of an uninoculated RLF mouse (A) and of mice inoculated with wild-type L. reuteri 100-23C (B) and the lsp mutant strain (C). (D) Ex vivo adherence of wild-type L. reuteri 100-23C to epithelium.
DISCUSSION
In contrast to humans, the gastric region of the guts of mice, rats, pigs, horses, and birds harbors relatively large populations of lactobacilli (reviewed in references 30, 31, and 42). Unlike the human stomach, which is lined with a glandular mucosa, the gastric region of these animals is lined partly with a nonglandular squamous stratified epithelium. These epithelial surfaces are colonized by lactobacilli adhering directly to the epithelium, forming a layer of bacterial cells. The mechanism by which Lactobacillus strains adhere to these epithelia has not been determined, but preliminary in vitro investigations have shown that both carbohydrate and protein molecules are likely involved (reviewed in reference 31). We have shown that a high-molecular-mass cell surface protein (Lsp) of L. reuteri 100-23 contributes to ecological performance in the murine gut. The lsp mutant could still attach to the forestomach epithelium when introduced into the gut in pure culture and given time to become established. However, attainment of a maximal population in the gut was delayed and ex vivo adherence to the forestomach epithelium was impaired. These results suggest that Lsp has a role in initiating adherence to the epithelium but that other, as yet unknown, mechanisms also mediate attachment.
Lsp is most likely attached to the bacterial cell wall, because recovery of the protein required the use of cell wall-degrading enzymes. SDS-PAGE revealed that Lsp was ca. 300 kDa and therefore much larger than the 185-kDa protein that was initially used to raise antibodies for detection of the clone containing lsp. At that time, extraction of surface proteins used a solution containing SDS that failed to recover complete Lsp but likely generated a range of peptide fragments comigrating with smaller surface proteins in SDS-PAGE. Fragments of Lsp were probably eluted from the gel together with the 185-kDA protein, accounting for antibody detection of the Lsp-producing clone.
Homologues of lsp are present in the genomes of several lactobacilli, including the human fecal isolates L. johnsonii and L. gasseri and the vaginal isolate Lactobacillus fermentum BR11 (20, 35). These proteins may have important roles in the establishment and maintenance of Lactobacillus populations as part of the normal microbiota of humans and other animals.
Methionine-sulfoxide reductase (Msr) reverses the loss of biological activity of proteins caused by oxidation of methionine to methionine sulfoxide (2). Most bacteria contain at least two genes encoding Msr activity, namely, msrA (msrA1, msrA2, and msrA3 in the case of Staphylococcus aureus) and msrB (11, 26). MsrA and msrB show substrate specificity for the two diastereomeric forms of methionine sulfoxide (MetO): R-MetO and S-MetO (17, 26). Homologues to msrA and msrB are present in the genomes of virtually all bacteria (2, 11), including Lactobacillus spp. (L. johnsonii, L. gasseri, and Lactobacillus plantarum). MsrA contributes to the protection of bacteria against oxidative damage caused by reactive nitrogen and oxygen intermediates (29). Nitric oxide (NO) is produced by epithelial cells in the gut and may act as an oxidative barrier to reduce bacterial translocation and provide a defense against pathogenic bacteria (14, 22). NO production by human bronchial epithelial cells has been reported to reduce Pseudomonas aeruginosa adherence to a cultured cell line (6). Furthermore, MsrA mutants have been reported to have reduced ability to adhere to epithelial cells compared to the wild type, and it was concluded that oxidation of methionine might affect the production and maintenance of adhesins associated with the bacterial cells (40). The ecological performance of the msrB mutant was impaired in about half of the mice used in our experiments. Therefore, the ability of the msrB mutant to inhabit the gut might have been lessened due to NO produced by epithelial cells, and the variation in mutant population levels from mouse to mouse might have been due to different levels of expression of inducible nitric oxide synthetase by murine cells. Several studies indicate that Msr activity has an important role in the virulence of pathogenic bacteria (7, 8). The importance of msrB for the colonization of the gut by L. reuteri suggests that members of the gut microbiota and pathogens apply similar strategies to cope with antimicrobial factors generated by the mammalian host.
The ecological performance of the xylA and met mutants was not impaired. This was not surprising in the case of the xylA mutant, because rodent diets contain a variety of carbohydrates. The mutant was therefore not reliant solely on xylose as a carbon and energy source. Xylose utilization might be essential for L. reuteri in gut ecosystems if the host's diet was low in easily fermentable carbohydrates but contained plant fibers rich in xylans. The in vivo induction of met may indicate methionine limitation in the murine gut. Assuming that met encodes a methionine synthase, the bacteria might compensate for the inactivation of the gene by the uptake of peptides containing methionine. L. reuteri might also possess an independent gene encoding a cobalamin-independent methionine synthase II. The genomes of several organisms, including L. plantarum, contain homologues of both genes, while the genomes of L. gasseri and L. johnsonii contain only a met homologue.
The L. reuteri genes were inactivated by insertion of a replication-defective plasmid in a single homologous recombination event. This procedure has been effective in inactivating other Lactobacillus genes (1, 16, 23). Excision of the plasmid restores the genotype and phenotype of the wild type, but the stability of the integration has generally been shown to be very good (16, 23). The rate of reversion to wild type in the case of our mutants ranged between 0.002 (lsp) and 0.04% (xylA) revertants per generation in vitro. Nevertheless, revertants will be selected and enriched in an environment where the gene is critical to ecological performance. In accordance with this, the lsp mutant, whose performance was greatly impaired in competition experiments, gave the highest percentage of revertants when colonizing the gut alone, although it was the most stable mutant in vitro (Table 3).
L. reuteri 100-23 bacteria are numerous in the forestomachs of mice (108 to 109 bacteria per g) and can be recovered from the jejunum, cecum, and feces at ∼107, 108, and 108 bacteria per g, respectively (13, 38). Interestingly, our data show that the inactivation of lsp affected the population size of the mutant throughout the gut. Adhesion of lactobacilli to the columnar epithelia lining the intestinal tract has not been described, so it is unlikely that impaired attachment was responsible for reduced population levels in regions distal to the stomach. It can therefore be assumed that L. reuteri 100-23 is not a true resident of the murine intestine but that the lactobacilli detected there are remnants of the Lactobacillus population of the forestomach, which is therefore its habitat. It is apparent from recent studies that even bacteria that are relatively numerous in a gut region may not be true residents but instead transients from a more proximal or exogenous source (5, 21, 33, 37).
The sequencing of bacterial genomes continues apace, and this information will allow insight into the qualities and characteristics of organisms encoded by genomes in relation to adaptation to life in particular environments to be gained (25, 41). The ecological importance of certain genetic traits, however, can be determined only by in vivo observations. A combination of comparative genomics, specific bacterial mutagenesis, and an appropriate experimental animal system will be required to understand the ecological performance of gut bacteria. The experimental approaches described in this paper have great potential to reveal the mechanisms by which lactobacilli function and therefore persist in the murine gut. Continuing use of this model system will doubtless improve our understanding of bacterial life in the gut.
Acknowledgments
We thank Todd Klaenhammer for providing pORI28 and E. coli EC1000 and Emmanuelle Maguin for providing pVE6007. We thank Richard Easingwood and the Otago Centre for Electron Microscopy for excellent SEM support. We thank Ralf Jack for assisting with SDS-PAGE and Markus Kranz for excellent technical assistance.
This work was financed in part by the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Barrangou, R., E. Altermann, R. Hutkins, R. Cano, and T. R. Klaenhammer. 2003. Functional and comparative genomic analyses of an operon involved in fructooligosaccharide utilization by Lactobacillus acidophilus. Proc. Natl. Acad. Sci. USA 100:8957-8962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brot, N., and H. Weissbach. 2000. Peptide methionine sulfoxide reductase: biochemistry and physiological role. Biopolymers 55:288-296. [DOI] [PubMed] [Google Scholar]
- 3.Chagnaud, P., H. F. Jenkinson, and G. W. Tannock. 1992. Cell surface-associated proteins of gastrointestinal strains of lactobacilli. Microb. Ecol. Health Dis. 5:121-131. [Google Scholar]
- 4.Cucarella, C., C. Soano, J. Valle, B. Amorena, I. Lasa, and J. R. Penades. 2001. Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J. Bacteriol. 183:2888-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dal Bello, F., J. Walter, W. P. Hammes, and C. Hertel. 2003. Increased complexity of the species composition of lactic acid bacteria in human feces revealed by alternative incubation condition. Microb. Ecol. 45:455-463. [DOI] [PubMed] [Google Scholar]
- 6.Darling, K. E., and T. J. Evans. 2003. Effects of nitric oxide on Pseudomonas aeruginosa infection of epithelial cells from a human respiratory cell line derived from a patient with cystic fibrosis. Infect. Immun. 71:2341-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dhandayuthapani, S., M. W. Blaylock, C. M. Bebear, W. G. Rasmussen, and J. B. Baseman. 2001. Peptide methionine sulfoxide reductase (MsrA) is a virulence determinant in Mycoplasma genitalium. J. Bacteriol. 183:5645-5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Douglas, T., D. S. Daniel, B. K. Parida, C. Jagannath, and S. Dhandayuthapani. 2004. Methionine sulfoxide reductase A (MsrA) deficiency affects the survival of Mycobacterium smegmatis within macrophages. J. Bacteriol. 186:3590-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Erlandson, K. A., J.-H. Park, W. El Khal, H.-H. Kao, P. Basaran, S. Brydges, and C. A. Batt. 2000. Dissolution of xylose metabolism in Lactococcus lactis. Appl. Environ. Microbiol. 66:3974-3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gordon, H. A., and L. Pesti. 1971. The gnotobiotic animal as a tool in the study of host-microbial relationships. Bacteriol. Rev. 35:390-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grimaud, R., B. Ezraty, J. K. Mitchell, D. Lafitte, C. Briand, P. J. Derrick, and F. Barras. 2001. Repair of oxidized proteins. J. Biol. Chem. 276:48915-48920. [DOI] [PubMed] [Google Scholar]
- 12.Grundy, F. J., and T. M. Henkin. 2004. Regulation of gene expression by effectors that bind to RNA. Curr. Opin. Microbiol. 7:126-131. [DOI] [PubMed] [Google Scholar]
- 13.Heng, N. C. K., J. M., Bateup, D. M. Loach, X. Wu, H. F. Jenkinson, M. Morrison, and G. W. Tannock. 1999. Influence of different functional elements of plasmid pGT232 on maintenance of recombinant plasmids in Lactobacillus reuteri populations in vitro and in vivo. Appl. Environ. Microbiol. 65:5378-5385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffman, R. A., G. Zhang, N. C. Nussler, S. L. Gleixner, H. R. Ford, R. L. Simmons, and S. C. Watkins. 1997. Constitutive expression of inducible nitric oxide synthase in the mouse ileal mucosa. Am. J. Physiol. 272:G383-G392. [DOI] [PubMed] [Google Scholar]
- 15.Leenhouts, K., G. Buist, A. Bolhuis, A. Ten Berge, J. Kiel, I. Mierau, M. Dabrowska, G. Venema, and J. Kok. 1996. A general system for generating unlabelled gene replacements in bacterial chromosomes. Mol. Gen. Genet. 253:217-224. [DOI] [PubMed] [Google Scholar]
- 16.Leloup, L., S. D. Ehrlich, M. Zagorec, and F. Morel-Deville. 1997. Single-crossover integration in the Lactobacillus sake chromosome and insertional inactivation of the ptsI and lacL genes. Appl. Environ. Microbiol. 63:2117-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lowther, W. T., H. Weissbach, F. Etienne, N. Brot, and B. W. Matthews. 2002. The mirrored methionine sulfoxide reductases of Neisseria gonorrhoeae pilB. Nat. Struct. Biol. 9:348-352. [DOI] [PubMed] [Google Scholar]
- 18.Maguin, E., P. Duwat, T. Hege, D. Ehrlich, and A. Gruss. 1992. New thermosensitive plasmid for gram-positive bacteria. J. Bacteriol. 174:5633-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 20.Pridmore, R. D., B. Berger, F. Desiere, D. Vilanova, C. Barretto, A. C. Pittet, M. C. Zwahlen, M. Rouvet, E. Altermann, R. Barrangou, B. Mollet, A. Mercenier, T. Klaenhammer, F. Arigoni, and M. A. Schell. 2004. The genome sequence of the probiotic intestinal bacterium Lactobacillus johnsonii NCC 533. Proc. Natl. Acad. Sci. USA 101:2512-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reuter, G. 2001. The Lactobacillus and Bifidobacterium microflora of the human intestine: composition and succession. Curr. Issues Intest. Microbiol. 2:43-53. [PubMed] [Google Scholar]
- 22.Roberts, P. J., G. P. Riley, K. Morgan, R. Miller, J. O. Hunter, and S. J. Middleton. 2001. The physiological expression of inducible nitric oxide synthase (iNOS) in the human colon. J. Clin. Pathol. 54:293-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Russell, W. M., and T. R. Klaenhammer. 2001. Efficient system for directed integration into the Lactobacillus acidophilus and Lactobacillus gasseri chromosomes via homologous recombination. Appl. Environ. Microbiol. 67:4361-4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Schell, M. A., M. Karmirantzou, B. Snel, D. Vilanova, B. Berger, G. Pessi, M. C. Zwahlen, F. Desiere, P. Bork, M. Delley, R. D. Pridmore, and F. Arigoni. 2002. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc. Natl. Acad. Sci. USA 99:14422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh, V. K., and J. Moskovitz. 2003. Multiple methionine sulfoxide reductase genes in Staphylococcus aureus: expression of activity and roles in tolerance of oxidative stress. Microbiology 149:2739-2747. [DOI] [PubMed] [Google Scholar]
- 27.Sonnenburg, J. L., L. T. Angenent, and J. I. Gordon. 2004. Getting a grip on things: how do communities of bacterial symbionts become established in our intestine. Nat. Immunol. 5:569-573. [DOI] [PubMed] [Google Scholar]
- 28.Stalhammar-Carlemalm, M., T. Areschoug, C. Larsson, and P. Lindahl. 1999. The R28 protein of Streptococcus pyogenes is related to several group B streptococcal surface proteins, confers protective immunity and promotes binding to human epithelial cells. Mol. Microbiol. 33:208-219. [DOI] [PubMed] [Google Scholar]
- 29.St. John, G., N. Brot, J. Ruan, H. Erdjument-Bromage, P. Tempst, H. Weissbach, and C. Nathan. 2001. Peptide methionine sulfoxide reductase from Escherichia coli and Mycobacterium tuberculosis protects bacteria against oxidative damage from reactive nitrogen intermediates. Proc. Natl. Acad. Sci. USA 98:9901-9906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tannock, G. W. 1992. Lactic microflora of pigs, mice and rats, p. 21-48. In B. J. B. Wood (ed.), The lactic acid bacteria, vol. 1. The lactic acid bacteria in health and disease. Elsevier Applied Science, London, United Kingdom.
- 31.Tannock, G. W. 1997. Normal microbiota of the gastrointestinal tract of rodents, p. 187-215. In R. I. Mackie, B. A. White, and R. E. Isaacson (ed.), Gastrointestinal microbiology, vol II. Chapman and Hall, London, United Kingdom.
- 32.Tannock, G. W., C. Crichton, G. W. Welling, J. P. Koopman, and T. Midtvedt. 1988. Reconstitution of the gastrointestinal microflora of lactobacillus-free mice. Appl. Environ. Microbiol. 54:2971-2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tannock, G. W., K. Munro, H. J. M. Harmsen, G. W. Welling, J. Smart, and P. K. Gobal. 2000. Analyses of the fecal microflora of human subjects consuming a probiotic product containing Lactobacillus rhamnosus DR20. Appl. Environ. Microbiol. 66:2578-2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Toledo-Arana, A., J. Valle, C. Solano, M. J. Arrizubieta, C. Cucarella, M. Lamata, B. Amorena, J. Leiva, J. R. Penades, and I. Lasa. 2001. The enterococcal surface protein, Esp, is involved in Enterococcus faecalis biofilm formation. Appl. Environ. Microbiol. 67:4538-4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Turner, M. S., L. M. Hafner, T. Walsh, and P. M. Giffard. 2003. Peptide surface display and secretion using two LPXTG-containing surface proteins from Lactobacillus fermentum BR11. Appl. Environ. Microbiol. 69:5855-5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wall, T., S. Roos, K. Jacobsson, A. Rosander, and H. Jonsson. 2003. Phage display reveals 52 novel extracellular and transmembrane proteins from Lactobacillus reuteri DSM 20016T. Microbiology 149:3493-3505. [DOI] [PubMed] [Google Scholar]
- 37.Walter, J., C. Hertel, G. W. Tannock, C. M. Lis, K. Munro, and W. P. Hammes. 2001. Detection of Lactobacillus, Pediococcus, Leuconostoc, and Weissella species in human feces by using group-specific PCR primers and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 67:2578-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walter, J., N. C. K. Heng, W. P. Hammes, D. M. Loach, G. W. Tannock, and C. Hertel. 2003. Identification of Lactobacillus reuteri genes specifically induced in the mouse gastrointestinal tract. Appl. Environ. Microbiol. 69:2044-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wesney, E., and G. W. Tannock. 1979. Association of rat, pig and fowl biotypes of lactobacilli with the stomach of gnotobiotic mice. Microb. Ecol. 5:35-42. [DOI] [PubMed] [Google Scholar]
- 40.Wizemann, T. M., J. Moskovitz, B. J. Pearce, D. Cundell, C. G. Arvidson, M. So, H. Weissbach, N. Brot, and H. R. Masure. 1996. Peptide methionine sulfoxide reductase contributes to the maintenance of adhesins in three major pathogens. Proc. Natl. Acad. Sci. USA 93:7985-7990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu, J., and J. I. Gordon. 2003. Inaugural article: honor thy symbionts. Proc. Natl. Acad. Sci. USA 100:10452-10459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yuki, N., T. Shimazaki, A. Kishiro, K. Watanabe, K. Uchida, T. Yuyama, and M. Morotomi. 2000. Colonization of the stratified squamous epithelium of the nonsecreting area of horse stomach by lactobacilli. Appl. Environ. Microbiol. 66:5030-5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zoetendal, E. G., A. D. L. Akkermans, and W. M. de Vos. 1998. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl. Environ. Microbiol. 64:3854-3859. [DOI] [PMC free article] [PubMed] [Google Scholar]