Abstract
Microbial rhodopsins are membrane proteins found widely in archaea, eubacteria and eukaryotes (fungal and algal species). They have various functions, such as light-driven ion pumps, light-gated ion channels, light sensors and light-activated enzymes. A light-driven proton pump bacteriorhodopsin (BR) contains a DTD motif at positions 85, 89, and 96, which is unique to archaeal proton pumps. Recently, channelrhodopsins (ChRs) containing the DTD motif, whose sequential identity is ~20% similar to BR and to cation ChRs in Chlamydomonas reinhardtii (CrCCRs), were found. While extensive studies on ChRs have been performed with CrCCR2, the molecular properties of DTD ChRs remain an intrigue. In this paper, we studied a DTD rhodopsin from G. theta (GtCCR4) using electrophysiological measurements, flash photolysis, and low-temperature difference FTIR spectroscopy. Electrophysiological measurements clearly showed that GtCCR4 functions as a light-gated cation channel, similar to other G. theta DTD ChRs (GtCCR1-3). Light-driven proton pump activity was also suggested for GtCCR4. Both electrophysiological and flash photolysis experiments showed that channel closing occurs upon reprotonation of the Schiff base, suggesting that the dynamics of retinal and channels are tightly coupled in GtCCR4. From Fourier transform infrared (FTIR) spectroscopy at 77 K, we found that the primary reaction is an all-trans to a 13-cis photoisomerization, like other microbial rhodopsins, although perturbations in the secondary structure were much smaller in GtCCR4 than in CrCCR2.
Keywords: microbial rhodopsin, light-gated channel, patch clamp, flash photolysis, light-induced difference FTIR spectra
Light-driven ion-transporting rhodopsins are important molecular machines in microbes [1–3]. They are also important tools in optogenetics [4–6]. The first-discovered protein was a light-driven outward proton pump bacteriorhodopsin (BR) in 1971 [7]. The chromophore of BR is an all-trans retinal that binds to a lysine residue through a protonated Schiff base linkage, and the Schiff base proton is transported in proton-pumping rhodopsins. For intramolecular proton transport, BR has a proton acceptor and a donor attached to the protonated Schiff base, which are D85 and D96 located at the C-helix, respectively (Table 1) [1]. T89 forms a hydrogen bond with D85 in BR [8], and these three residues are highly conserved among archaeal light-driven proton-pumping rhodopsins.
Table 1.
Amino acid sequences of the C-helix in various microbial rhodopsins
| 82 | 83 | 84 | 85 | 86 | 87 | 88 | 89 | 90 | 91 | 92 | 93 | 94 | 95 | 96 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BR | H+ pump | R | Y | A | D | W | L | F | T | T | P | L | L | L | L | D |
| PR | H+ pump | R | Y | I | D | W | L | L | T | V | P | L | L | I | C | E |
| KR2 | Na+ pump | R | Y | L | N | W | S | I | D | V | P | M | L | L | F | Q |
| FR | Cl− pump | R | Y | G | N | W | T | I | T | V | P | I | L | L | T | Q |
| pHR | Cl− pump | R | Y | L | T | W | A | L | S | T | P | M | I | L | L | A |
| CrCCR1 | cation channel | R | Y | A | E | W | L | L | T | C | R | V | I | L | I | H |
| CrCCR2 | cation channel | R | Y | A | E | W | L | L | T | C | P | V | I | L | I | H |
| CaCCR1 | cation channel | R | Y | S | E | W | L | L | C | C | P | V | I | L | I | H |
| MvCCR1 | cation channel | R | Y | M | E | W | L | M | T | C | P | V | I | L | I | A |
| GtCCR1 | cation channel | P | Y | L | D | Y | A | T | T | C | P | L | L | T | L | D |
| GtCCR2 | cation channel | P | Y | V | D | Y | C | T | T | C | P | L | L | T | L | D |
| GtCCR3 | cation channel | K | Y | L | D | Y | L | F | T | C | P | L | L | T | I | D |
| GtCCR4 | this study | K | Y | L | D | Y | I | F | T | C | P | I | L | T | L | D |
| GtACR1 | anion channel | R | M | A | S | W | L | C | T | C | P | I | M | L | G | L |
| GtACR2 | anion channel | R | M | A | S | W | L | C | T | C | P | I | M | L | G | Q |
Abbreviations: BR, bacteriorhodopsin; PR, proteorhodopsin; KR2, Krokinobacter eikastus rhodopsin 2; FR, Fulvimarina rhodopsin; pHR, Natronomonas pharaonis halorhodopsin; CrCCR1 and 2, cation channelrhodopsin (ChR) 1 and 2, respectively from Chlamydomonas reinhardtii; CaCCR1, cation ChR1 from Chlamydomonas augustae; MvCCR1, cation ChR1 from Mesostigma viride; GtCCR1, 2, 3 and 4, cation ChR1, 2, 3 and 4, respectively from Guillardia theta; GtACR1 and 2, anion ChR1 and 2, respectively from G. theta. The numbering scheme corresponds to the position of BR.
Recently, light-driven Na+ and Cl− pumping rhodopsins were found from marine bacteria [9–11]. These contain of NDQ and NTQ residues at D85, T89 and D96 positions in BR, respectively (Table 1). Therefore, the DTD, NDQ, and NTQ motifs are characteristic of a light-driven archaeal proton pump, an eubacterial Na+ pump, and an eubacterial Cl− pump, respectively [12,13]. An eubacterial proton-pump proteorhodopsin (PR) possesses the DTE motif, while an archaeal Cl− pump halorhodopsin (HR) possesses the TSA motif (Table 1). These motifs substantially characterize the function of ion-pump rhodopsins, namely, a proton pump exhibits a DTD or DTE motif, in which the first and third residues are carboxylates that act during proton transfer [13]. In contrast, the Na+ pump has a NDQ motif in which only the second residue is carboxylate, while the Cl− pump contains only neutral residues such as NTQ and TSA [13].
Compared to ion pumps, motifs are more divergent in light-gated ion channel channelrhodopsins (ChRs) (Table 1). In fact, ChR1 (CrCCR1) and 2 (CrCCR2) from Chlamydomonas reinhardtii have an ETH motif, while ChR1 from Chlamydomonas augustae (CaCCR1) and Mesostigma viride (MvCCR1) contain ECH and ETA motifs, respectively [2,14,15]. Anion ChR1 (GtACR1) and 2 (GtACR2) from Guillardia theta contain STL and STQ motifs, respectively [16]. Interestingly, Govorunova et al. reported that microbial rhodopsins possessing the DTD motif act as light-gated ion channel [17], though DTD is generally unique to the light-driven archaeal proton pump (Table 1). The cryptophyte G. theta [18] encodes more than 40 microbial rhodopsins, but two new anion-channels (GtACR1 and GtACR2) were recently found [16]. Patch-clamp measurements showed that three DTD rhodopsins with a truncated C-terminus from G. theta (GtCCR1, GtCCR2, GtCCR3) function as cation channels, suggesting that channel function in rhodopsins has evolved via multiple routes [17]. CrCCR2 has been extensively studied, revealing the opening/closing dynamics both spectroscopically and electrophysiologically [2,19–21]. The crystal structure of a chimeric protein of CrCCR1 and CrCCR2 (C1C2) was determined [22], and light-induced difference infrared spectra provided structural changes in the function of ChR [23–38]. After the emergence of the DTD ChR, its molecular properties are intriguing in comparison with other ChRs.
We have also studied microbial rhodopsins from G. theta. In this paper, we report on another DTD rhodopsin from G. theta (GtCCR4) (Table 1). GtCCR4 is homologous to GtCCR1, GtCCR2, and GtCCR3 (33, 34, and 39% identity, respectively), while the number of amino acids in GtCCR4 (367) is less than in GtCCR1 (402), GtCCR2 (433), and GtCCR3 (383) (Supplementary Fig. S1). We prepared a full-length protein of GtCCR4, and applied electrophysiological measurements, flash photolysis, and low-temperature difference FTIR spectroscopy. Electrophysiological measurements clearly showed that GtCCR4 functions as a light-gated cation channel, like other G. theta DTD rhodopsins. Flash photolysis experiments identified photocycling intermediates during the channel function of GtCCR4, and the kinetic coincidence between M decay and channel closing suggests that the dynamics of retinal and channel are tightly coupled in GtCCR4. Low-temperature FTIR spectroscopy at 77 K provides the first structural features of DTD ChR, demonstrating that the primary reaction is an all-trans to a 13-cis photoisomerization, similar to other microbial rhodopsins, although GtCCR4 has fewer secondary structural perturbations than CrCCR2. Common and unique molecular properties of DTD ChR are presented based on the present experimental results with GtCCR4.
Materials and Methods
Cloning and Expression Plasmid Construction of GtCCR4 Gene
Total RNA from G. theta CCMP2712 was obtained from the Provasoli-Guillard National Center for Marine Algae and Microbiota. Total RNA was reverse-transcribed using a SMARTerTM RACE cDNA Amplification Kit (Clontech, CA, USA). An oligo-dT primer was used for first strand synthesis. The full length of the G. theta gene 164280 (accession number MF039475) was amplified using gene-specific forward (gatcatATGACGACGTCTGCCCCTTC; “catATG” indicates the NdeI site) and reverse (gatcctcgagAACGGCCTCGGAC TCCTGC; “ctcgag” indicates the XhoI site) primers designed according to the sequence on the NCBI database and Advantage GC2 polymerase (Clontech, CA, USA). We named the gene GtCCR4. The polymerase chain reaction (PCR) product was gel-purified, sub-cloned into pGEM-T vector (Promega, WI, USA), and sequenced. For protein expression in Pichia pastoris, the full-length Gt_164280 gene was cloned between the EcoRI and XbaI sites of the pPICZB vector. The EcoRI site was created at the 5′ end of the gene while the XbaI site was created at the 3′ end by performing PCR with the forward (cgaggaattcacgATGACGACGTCTGCC; “gaattc” indicates the EcoRI site) and reverse (ctctagatcaatgatgatgat gatgatgAACGGCCTCGGACTC; “tctaga” and “atgatgatgat gatgatg” are indicate the XbaI site and His-tag encoding sequence, respectively) primers. The sequence of the gene insertion site was confirmed by DNA sequencing.
Protein Expression in P. pastoris and Purification
His-tagged GtCCR4 was expressed in P. pastoris. The cells were harvested 48–60 h after expression was induced in BMMY medium when 10 mM of all-trans-retinal (Sigma-Aldrich, MO, USA) was supplemented in the culture to a final concentration of 30 μM. Additionally, 100% filtered methanol was added to the growth medium every 24 h of induction to a final concentration of 0.5%. Membranes containing GtCCR4 were isolated as described elsewhere [39] with the following modifications. Washed P. pastoris cells were resuspended in buffer A (7 mM NaH2PO4, 7 mM EDTA, 7 mM DTT, and 1 mM phenylmethylsulfonyl fluoride (PMSF), pH 6.5) and slowly shaken with all-trans-retinal (added to a final concentration of 25 μM) in the dark at room temperature for 3–4 h in the presence of 0.5% of Westase (TaKaRa, Shiga, Japan) to digest the cell wall. The cells were vortexed several times with a 50% volume of glass beads (Φ=500 μm) and centrifuged at low speed (700×g). The supernatants were centrifuged for 30 min at 40,000×g in a fixed-angle rotor, and the GtCCR4 membrane pellets were resuspended in solubilization buffer (20 mM KH2PO4, 1% n-dodecyl-β-D-maltoside (DDM), 1 mM PMSF, pH 7.5) and stirred overnight at 4°C. The solubilization mixture was centrifuged for 30 min at 40,000×g in a fixed-angle rotor. The solubilized protein was incubated with Ni-NTA agarose (QIAGEN, Hilden, Germany) for several hours. The resin with bound GtCCR4 was washed with solubilization buffer and then treated with elution buffer (0.05 M KH2PO4, 0.4 M NaCl, 0.25% DDM, 0–1 M imidazole, pH 7.5).
Heterologous Expression in ND7/23 Cells
The GtCCR4 gene was cloned into peGFP vector between HindIII and BamHI sites in such a way that the eGFP is tagged at the C-terminus of GtCCR4. ND7/23 cells were purchased from DS Pharma Biomedical (Osaka, Japan) and cultured in high-glucose DMEM media (Wako, Osaka, Japan) in a 37°C, 5% CO2 incubator. Transfection of ND7/23 cells was performed by Lipofectamine 2000 (Invitrogen, CA, USA). Cells were supplemented with 1 μM all-trans-retinal (Sigma-Aldrich) after transfection. Expression of GtCCR4 was confirmed by eGFP fluorescence for a whole-cell patch clamp recording.
Electrophysiology
Whole-cell patch clamp recordings on ND7/23 cells were performed at room temperature with an Axopatch 200B amplifier (Molecular Devices, CA, USA). Continuous light was illuminated by OSG L12194-00-39070 (Hamamatsu Photonics, Shizuoka, Japan) via a light guide into an inverted microscope, IMT-2 (Olympus, Tokyo, Japan). The maximum light intensity was 0.33 mW/mm2 and the focus area was ~0.5 mm2. Illumination was controlled by a mechanical shutter LS6S (Vincent Associates, NY, USA). Glass pipettes were made with a micropipette puller, P-97 (Sutter Instrument, CA, USA) and fire-polished with a micro forge, MF-830 (Narishige, Tokyo, Japan). Pipette resistance was between 1.5 and 2.5 MΩ. The pipette electrode was controlled by a micro manipulator, PCS-5000 (Burleigh instruments, NY, USA). Current traces were recorded at 10 kHz and filtered to 2 kHz by an internal circuit of the amplifier. Data acquisition and shutter triggering were performed by pClamp 10 software via a Digidata 1550 (Molecular Devices). Data were analyzed by Clampfit and Origin software. The standard external solution contained 140 mM NaCl, 2 mM MgCl2, 2 mM CaCl2, 2 mM KCl, and 10 mM Hepes-NaOH (pH 7.2). The standard internal solution contained 110 mM NaCl, 2 mM MgCl2, 1 mM CaCl2, 5 mM KCl, 10 mM EGTA, and 10 mM Hepes-NaOH (pH 7.2). Osmolality of the solutions was adjusted to 300 mOsm by adding an appropriate amount of sucrose.
Flash Photolysis
The transient absorption spectra of GtCCR4 were monitored by flash photolysis measurements using a multichannel detector (Hamamatsu Photonics, Shizuoka, Japan) [40] at room temperature. The sample solution was placed in a quartz cuvette (0.7 OD with 10 mm path length) and was illuminated with a beam of second harmonics of a nano-second pulsed Nd3+-YAG laser (λ=530 nm, INDI40, Spectra-Physics, CA, USA). The laser power was 300 mJ per pulse, and repetition rate (every 200 ms) was sufficiently slower than the rate of photocycle of GtCCR4 to avoid photo-excitation of transient intermediates. The intensity of the transmitted probe light from an Xe arc lamp (L8004, Hamamatsu Photonics) was measured before and after laser excitation, and transient absorption spectra were obtained by calculating the ratio between them. Twenty spectra were averaged.
Low-Temperature Difference FTIR Spectroscopy
For FTIR spectroscopy, GtCCR4 was reconstituted into a mixture of POPE and POPG (molar ratio=3 : 1) with a protein-to-lipid molar ratio of 1 : 50 by removing DDM with Bio-Beads (SM-2, Bio-Rad, CA, USA). The reconstituted samples were washed three times with 2 mM KH2PO4 and 2 mM NaCl (pH 7.5). The pellet was resuspended in the same buffer, but the concentration was adjusted to 1.7 mg/mL. A 58 μl aliquot was placed onto a BaF2 window and dried at 4°C. Low-temperature FTIR spectroscopy was applied to the films hydrated with H2O and D2O at 77 K, as described previously [30]. To form the K intermediate, samples were illuminated with 540 nm light for 2 min. The K intermediate was photo-reversed with >600 nm light for 1 min. For each measurement, 128 interferograms were accumulated, and 50 recordings were averaged.
Results
GtCCR4 is a Light-Gated Cation Channel
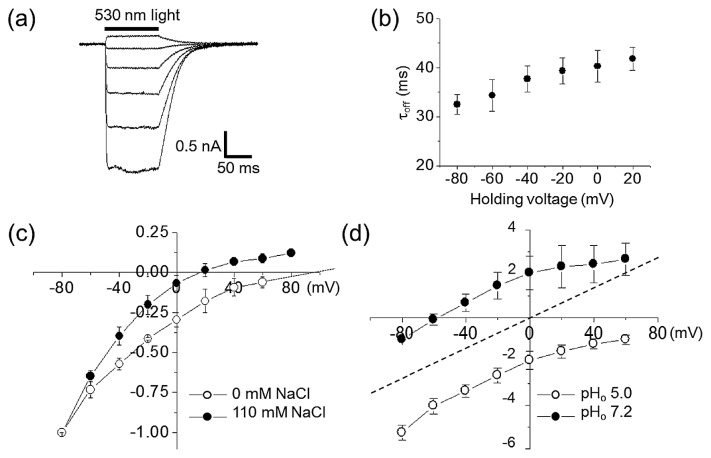
Here we cloned a new gene encoding DTD ChRs from G. theta (GtCCR4) (Table. 1 and Supplementary Fig. S1). We expressed GtCCR4 in ND7/23 cells to perform whole-cell patch-clamp recordings. Photocurrents were measured upon 300 ms illumination at a holding voltage ranging from −60 mV to +40 mV. Upon shifting the holding potential (Eh) to more positive values, the photocurrents generated by GtCCR4 reversed their direction (Fig. 1a). The current reached a plateau within 10–20 ms without significant inactivation during illumination, which was also observed in GtCCR1-3 [17]. After shutting off light, photocurrent decayed to zero with a time constant of 30–40 ms (Fig. 1b).
Figure 1.
Ion transport activity of GtCCR4 on ND7/23 cells recorded by a whole-cell patch clamp. (a) Representative photocurrents recorded at membrane potentials from −60 mV to +40 mV in 20 mV steps in standard solutions (See materials and methods). (b) Channel closing kinetics estimated by decay of photocurrents in standard solutions (N=8). Data represent the mean±SE. (c) I–V curves of stationary photocurrents with 110 mM (solid circle) or 0 mM (open circle) intercellular NaCl concentrations. The solution contained 110 mM NaCl, 2 mM CaCl2, 2 mM MgCl2, 5 mM KCl 10 mM EGTA-NaOH and 10 mM Hepes-NaOH (pH 7.2). A solution without Na+ contained 110 mM NMG-Cl, 2 mM CaCl2, 2 mM MgCl2, 5 mM KCl 10 mM EGTA-NMG and 10 mM Hepes-NMG (pH 7.2). Extracellular solution contained 140 mM NaCl, 2 mM CaCl2, 2 mM MgCl2, 2 mM KCl and Hepes-NaOH (pH 7.2). The photocurrent amplitudes were normalized at −80 mV as −1.0. Data represent the mean±SE. (d) I–V curves of stationary photocurrents with extracellular pH 5.0 (open circle) or pH 7.2 (solid circle) in the absence of NaCl. Extracellular solution contained 140 mM NMG-Cl, 2 mM CaCl2, 2 mM MgCl2 and Hepes-NMG (pH 7.2) or citric acid (pH 5.0). Intercellular solution contained 110 mM NMG-Cl, 2 mM CaCl2, 2 mM MgCl2, 5 mM KCl 10 mM EGTA-NMG and 10 mM Hepes-NMG (pH 7.2). The x- and y-axis represent the membrane potential (mV) and normalized current, respectively. The currents were first measured at pHo 7.2 (solid circle), followed by change in solution to pH 5.0 (open circle) (N=3). The photocurrent amplitudes were normalized at −80 mV at pHo 7.2 (solid circle) as −1.0. The dashed line was the theoretical curve.
To identify the nature of the transported ions, we determined reversal potentials (Er) after varying the ionic composition of the solution. When intracellular Na+ concentration was reduced from 110 mM to 0 mM by replacing it with n-methyl-D-glucamine (NMG+), Er shifted to more positive values (Fig. 1c), indicating that it passively transports Na+ across the membrane. There was an approximately 100-fold reduction in the extracellular H+ concentration (from pH 5.0 to pH 7.2) that led to a shift in Er to negative values, although Er did not correspond with theoretical Er in the ion channel when intracellular and extracellular H+ concentration were the same (Fig. 1d). Therefore, we concluded that GtCCR4 is a light-gated channel that transports both Na+ and H+. It should be noted that we observed a positive current at 0 mV in the absence of Na+ under the same intracellular and extracellular pH (Fig. 1d), suggesting the light-driven H+ pump activity for GtCCR4.
Absorption Spectra and Photocycle of GtCCR4
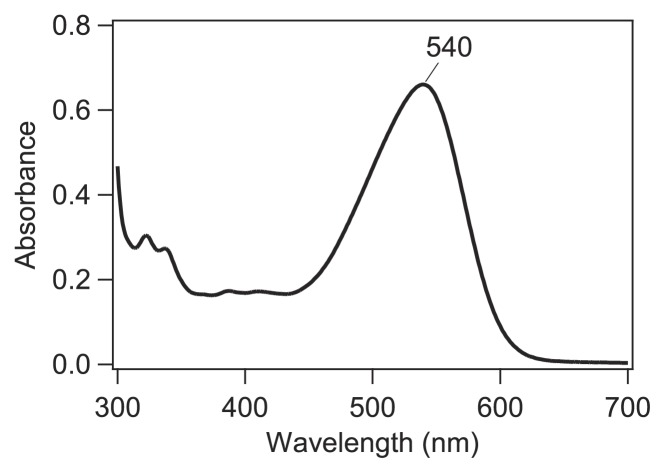
To further study the molecular properties of GtCCR4, we then attempted to purify it. His-tagged full-length GtCCR4 was expressed in P. pastoris and solubilized by 1% DDM after the addition of all-trans-retinal, and purified by Ni2+-NTA column. The elution buffer (pH 7.5) was used for the measurements of absorption spectra and flash photolysis. The absorption spectrum of GtCCR4 thus obtained exhibits a maximum at 540 nm (Fig. 2). The level of purification resulting from the expression by P. pastoris in this study was much lower than other microbial rhodopsins expressed by E. coli, and the small peaks at 300–410 nm in Figure 2 presumably originate from impurities, and not from GtCCR4. Nevertheless, we were able to monitor photocycle intermediates by flash photolysis, and light-induced difference FTIR spectra, as described next.
Figure 2.
Absorption spectrum of GtCCR4.
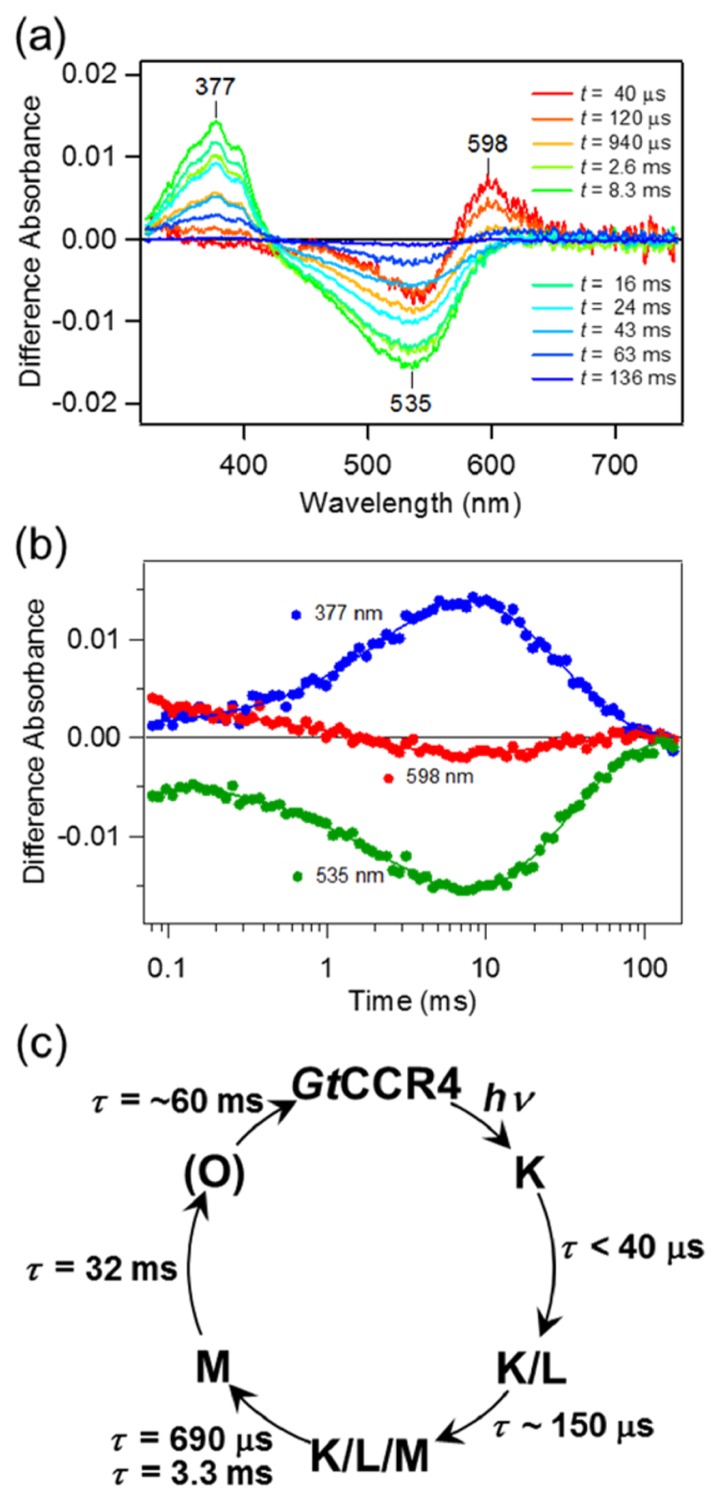
We next studied the photocycle of GtCCR4 by flash photolysis, in which the GtCCR4 protein was solubilized in detergent (DDM). Figure 3a shows the transient change in absorption of GtCCR4 upon excitation at λ=532 nm. At t=40 μs, we observed the accumulation of the red-shifted K intermediate at 598 nm which decayed in a 0.1–10 ms timeframe. Simultaneously, the bleaching signal at λ=~535 nm enlarged. This implies that another photo-intermediate, probably L, whose absorption wavelength is similar to that of its initial state, exists in the photocycle and is almost at equilibrium with K. The decay of K and L is represented by the decrease in absorption at 535 and 598 nm, respectively between 0.1 and 10 ms (Fig. 3b). The double exponential decay in K and L had a lifetime of τ=690 μs and 3.3 ms, respectively, and the accumulation of blue-shifted M was observed at 377 nm (Fig. 3a). The decay of M was highly reproducible with a single exponential function and a lifetime of τ=32 ms. At the final stage of the photocycle, we observed a slight accumulation of the red-shifted O-intermediate at t=50–100 ms at 598 nm (Fig. 3b, red circles) with an estimated lifetime of τ=~60 ms. The absorption spectra of each quasi-stable state were calculated (Supplementary Fig. S2) according to the method described previously [9]. The spectra of first component showed λmax close to that of the initial state and wide band width (FWHM=~135 nm). This suggests, even in this faster time scale (t=40 μs), the K and L are equilibrated (the K/L) with each other. Then, the absorption spectrum of second component indicated that the equilibrium more shifted toward L. In addition, a small absorption increase was observed at λ=350–410 nm which represent faster accumulation of the M. That is, the K, L and M are equilibrated in this component (the K/L/M). While the third spectrum indicates only the M was included, it was difficult to calculate the absorption spectra of the O because of its small amount of accumulation. Based on these insights, we consider that the photo-excited GtCCR4 undergoes a photocyclic reaction, as shown in Figure 3c. In this reaction, we assumed that the primary K-state is present before the accumulation of the K/L, a trend that is generally observed for most microbial rhodopsins [1,11,41], and which in fact was observed by low-temperature FTIR spectroscopy (see below).
Figure 3.
Photocycle of GtCCR4. (a) Transient absorption spectra of GtCCR4. (b) Time traces of absorption changes of GtCCR4 at 377 (blue), 535 (green) and 598 (red) nm probe wavelengths. Solid lines indicate the fitted lines based on the sequential kinetic model shown in c. (c) Photocycle scheme of GtCCR4 determined by flash photolysis measurement.
The retinal Schiff-base (RSB) is considered to be deprotonated on the M-intermediate because of blue-shifted absorption. CrCCR2 shows a similar blue-shifted state referred to as P2390 which also represents the deprotonated state of RSB [28]. The time constant of the formation of P2390 of CrCCR2 is τ=10 μs, which is much faster than that of the deprotonation of RSB of GtCCR4 (τ=690 μs and 3.3 ms). The time constant of the decay of P2390 of CrCCR2 is τ=2 ms which is also faster than the decay of M of GtCCR4 (τ=32 ms). Thus, both processes involving proton transfer are relatively slower for GtCCR4 than for CrCCR2.
The channel closing kinetics measured by the patch clamp method was about 35–40 ms (Fig. 1b) and this is close to the time-constant of M decay (τ=32 ms). This suggests that the opened channel of GtCCR4 probably closed after M decay. The channel closing of CrCCR2 occurs at 10–20 ms [28]. Thus, despite a large difference in the rates of deprotonation/protonation events of RSB, channel closing occurs on a similar time-scale between GtCCR4 and CrCCR2.
Structural Analysis of GtCCR4 by Means of Light-Induced Difference FTIR Spectroscopy at 77 K
To gain initial structural information, we then applied low-temperature FTIR spectroscopy to GtCCR4, in which the GtCCR4 protein is reconstituted into lipids. Rhodopsins generally undergo retinal photoisomerization even at 77 K, forming a red-shifted K intermediate, which can be photoreversible by illumination at a longer wavelength. We established sample illumination conditions of GtCCR4 at 77 K based on those for the C1C2 chimera of CrCCR in which 540 nm light converted dark-adapted GtCCR4 into the K intermediate, and >600 nm light reverted the K intermediate into an unphotolyzed state.
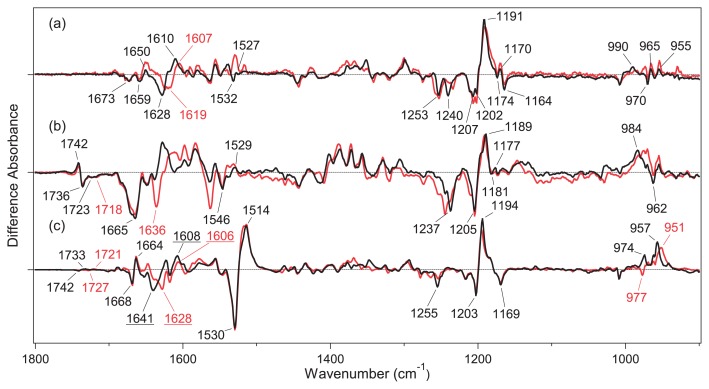
Figure 4a shows the light-induced difference FTIR spectra of GtCCR4 in the 1800–900 cm−1 region, which were measured at 77 K after the hydration with H2O (black) and D2O (red). The obtained spectra were compared with those of the C1C2 chimera of CrCCR (b) [30] and BR (c) [42]. The difference spectra were roughly normalized by use of C-C stretch of the retinal chromophore at 1210–1190 cm−1, suggesting that similar amounts of GtCCR4, C1C2 and BR are photoconverted into the K intermediate. It is well known that ethylenic C=C stretching vibration of retinal chromophore shows a linear correlation with the absorption maximum, where the C=C stretching frequency is lowered as absorption becomes red-shifted [43]. The peak pair at 1530 (−)/1514 (+) cm−1 for BR (Fig. 4c) is a characteristic feature of the formation of a red-shifted K intermediate (1514 cm−1) from BR (1530 cm−1). This is much less clear for ChRs, as seen for GtCCR4 (Fig. 4a) and C1C2 (Fig. 4b). Ethylenic C=C stretching vibration of retinal chromophore may be located at 1532 (−)/1527 (+) cm−1 for GtCCR4 (Fig. 4a), and at 1546 (−)/1529 (+) cm−1 for C1C2 (Fig. 4b). Unlike the H-D insensitive negative peaks for BR (1530 cm−1) and C1C2 (1546 cm−1), the negative peak for GtCCR4 (1532 cm−1) disappears upon hydration with D2O. We infer that other H-D sensitive vibrations mask the H-D insensitive C=C stretch for GtCCR4.
Figure 4.
Difference FTIR spectra of GtCCR4 (a), C1C2 (b) and BR (c) between the K intermediate and the unphotolyzed state in the 1800–900 cm−1 region measured at 77 K. The sample films were hydrated with H2O (black) and D2O (red). One division of the y-axis corresponds to 0.002 absorbance units. The black spectrum in (a) was multiplied by 2.6. The spectra in (b) and (c) were multiplied by 0.95 and 0.081, respectively. The data in (b) and (c) are reproduced from Ito et al. [30] and Kandori et al. [42].
Although the peaks are unclear, mirror-imaged difference spectra by alternative illumination at 540 nm and at >600 nm strongly suggest formation of the red-shifted K intermediate for GtCCR4, as well as C1C2. In fact, the C–C stretching region at 1250-1150 cm−1 is very similar among the three proteins. GtCCR4 exhibits peaks at 1253 (−), 1240 (−), 1207 (−), 1202 (−), 1191 (+), 1174 (−), 1170 (+), and 1164 (−) cm−1, among which the negative 1202 cm−1 (or 1207 cm−1) and positive 1191 cm−1 bands are assignable to the C14–C15 stretching vibration of all-trans/15-anti retinal, and the C10–C11/C14–C15 stretching vibrations of 13-cis/15-anti retinal from the literature for BR [44,45]. This indicates that photo-isomerization from the all-trans- to the 13-cis-form is the primary reaction of GtCCR4. Multiple negative peaks at 1202 and 1207 cm−1, observed only for GtCCR4, may originate from structural heterogeneity of the retinal chromophore. The negative 1255 cm−1 band in BR is composed of a mixture of D2O-insensitive C12–C13 stretching and D2O-sensitive N-H in-plane bending vibrations [46], and the former and latter seem to appear at 1253 and 1240 cm−1, respectively in GtCCR4. The bands at 1164 cm−1 were also observed at 1169 cm−1 in BR (c), but not in C1C2 (b). Since this was assigned to C10–C11 stretching vibration in BR [44], structural changes in the middle of retinal may occur similarly for GtCCR4, but not for C1C2. It should be noted that a small peak pair at 1174 (−)/1170 (+) cm−1 may originate from the photoreaction of the 13-cis/15-syn form.
Hydrogen-out-of-plane (HOOP), N-D in-plane bending, and methyl rocking vibrations appeared in the 1110–900 cm−1 region, and the presence of strong HOOP modes represent the distortion of the retinal molecule at the corresponding position. The amplitude of the positive signal is smaller for GtCCR4 than for C1C2 and BR, while presenting multiple positive peaks. Figure 4a (Supplementary Fig. S3a in more detail) shows positive peaks at 990, 965, and 955 cm−1, among which only the 990 cm−1 band is H-D exchangeable. The corresponding band for C1C2 is presumably at 984 cm−1, showing a much greater amplitude. Therefore, local chromophore distortion near the Schiff base region is much smaller in GtCCR4 than in C1C2 and BR.
C=N stretching vibrations of the protonated retinal Schiff base that appeared in the 1650–1600 cm−1 region are sensitive to H-D exchange, and the difference in frequency between C=NH and C=ND has been regarded as a probe of hydrogen-bonding strength [47–49]. The C=NH and C=ND frequencies are assignable at 1628 cm−1 and 1619 cm−1 for GtCCR4 (Fig. 4a and Supplementary Fig. S3a), at 1665 cm−1 and 1636 cm−1 for C1C2 (Fig. 4b and Supplementary Fig. S3b), and at 1641 cm−1 and 1628 cm−1 for BR (Fig. 4c and Supplementary Fig. S3c), respectively. The difference for GtCCR4 (9 cm−1) is much smaller than for C1C2 (29 cm−1), and even smaller than for BR (13 cm−1). The hydrogen-bonding acceptor of the Schiff base is a negatively charged counterion for C1C2, and a water molecule for BR. It is thus likely that the hydrogen-bonding acceptor of the Schiff base in GtCCR4 and in BR is a water molecule. Regarding the C=N stretch after retinal photoisomerization, the C=NH and C=ND stretches of K-intermediate of GtCCR4 are likely to be at 1610 and 1607 cm−1, respectively. Retinal isomerization weakens the Schiff base hydrogen bond, presumably with water.
Unlike the H-D exchangeable C=N stretch of the retinal Schiff base, H-D unexchangeable bands at 1700–1650 cm−1 in Figure 4 (Supplementary Fig. S3 in more detail) probably originate from amide-I vibrations. The peak pair at 1668 (−)/1664 (+) cm−1 in BR was assigned as amide-I vibration of α-helix, while a much larger negative peak at 1665 cm−1 in C1C2 is a signature of large structural changes of the peptide backbone in ChR [30]. This view is common for CrCCR2 [23,24,29], the most studied ChR. In contrast, a smaller change of amide-I was reported for CaCCR1 at 80 K [33], and a time-resolved FTIR study of CaCCR1 reported large changes of amide-I occurring in later intermediates [32]. Thus, limited protein structural changes to GtCCR4 upon retinal photoisomerization are similar to CaCCR1, but not to CrCCR2.
A clear spectral difference between GtCCR4 and C1C2 can be seen at ~1740 cm−1, a characteristic frequency region of protonated carboxylic acids. While there is no band at 1760–1710 cm−1 for GtCCR4 (Fig. 4a and Supplementary Fig. S3a), C1C2 shows a peak pair at 1742 (+)/1736 (−) cm−1, which is ascribable for D195, and constitutes a DC gate with C167. A similar spectral change was observed for BR at 1742 (−)/1733 (+) cm−1 (Fig. 4c and Supplementary Fig. S3c), which was assigned as D115. The corresponding residue is Thr in GtCCR4, which is consistent with the fact that spectral changes were observed for C1C2 and BR, but not for GtCCR4. Ogren et al. observed a peak pair in the difference FTIR spectra of CaCCR1 at 80 K, and proposed that one of two carboxylates in the Schiff base region (D85 and D212 for BR) is protonated [33]. This suggests that either carboxylate possesses high pKa in CaCCR1. No peaks at 1760–1710 cm−1 for GtCCR4 (Fig. 4a and Supplementary Fig. S3a) strongly suggest that two carboxylates in the Schiff base region are deprotonated, as for BR. This feature of GtCCR4 is more representative of BR than of CaCCR1.
Discussion
GtCCR4 is a DTD rhodopsin in G. theta as are GtCCR1, GtCCR2, and GtCCR3 (Table 1 and Supplementary Fig. S1) [17]. DTD is a characteristic motif of the light-driven archaeal proton pump in which the first D is the proton acceptor from the protonated Schiff base (D85 for BR), and the third D is the proton donor for the Schiff base reprotonation (D96 for BR) [13]. Therefore, the molecular properties of DTD rhodopsins are intriguing if they function as lightgated channels. The present electrophysiological experiments clearly demonstrated a cation channel function for full-length GtCCR4 (Fig. 1), as well as C-terminus truncated GtCCR1, GtCCR2, and GtCCR3 [17]. Feldbauer et al. reported that when ChR2 is expressed in electrofused giant HEK293 cells or reconstituted on planar lipid membranes, ChR2 acts as an outwardly driven proton pump [50]. Observation of a positive current under no membrane potential, no Na+ and the same intracellular and extracellular pH (Fig. 1d) suggests that GtCCR4 has the light-driven proton pump activity similarly.
Purified GtCCR4 protein absorbs maximally at 540 nm, and is considerably red-shifted from CrCCR1 and 2. The color tuning mechanism has been extensively studied in microbial rhodopsins, and several mutation studies have experimentally revealed key residues [51,52]. One of the color determinant residues is A215 in BR, and introduction of a polar amino acid such as serine or threonine causes a spectral blue-shift. The corresponding amino acid in CrCCR1 and 2 is serine, while GtCCR4 and BR contain alanine (Supplementary Fig. S1). Therefore, alanine at this position must contribute to the red-shifted absorption. In fact, GtCCR1, GtCCR2, and GtCCR3 contain alanine, alanine, and serine, whose λmax are 520 nm, 505 nm, and 460 nm, respectively [17]. This is fully consistent with the idea that the polarity at the position is the color determinant.
Microbial rhodopsins accommodate two isomeric states in the dark: all-trans, 15-anti and 13-cis, 15-syn forms [1]. Many microbial rhodopsins, except for BR, contain the all-trans, 15-anti form in the dark, while light-adaptation increases the population of the 13-cis, 15-syn form. Although the all-trans, 15-anti form is functional in light-driven ion pumps such as BR, involvement of the 13-cis, 15-syn form in the functioning of light-gated ion channels and the presence of parallel photocycles have been debated. As the amount of sample was limited, we did not perform HPLC analysis, and the isomeric ratio was unknown for GtCCR4. Nevertheless, light-induced difference FTIR spectra (Fig. 4) clearly show that the all-trans, 15-anti form is the major component in the dark, possibly with some 13-cis, 15-syn form as seen from the bands at 1174 (−)/1170 (+) cm−1. Improvement of the expression of GtCCR4 by use of P. pastoris is in progress, and a study on the 13-cis, 15-syn form is our future focus.
From FTIR spectroscopy at 77 K, it was found that the primary reaction of GtCCR4 is all-trans to 13-cis photo-isomerization, as occurs in other microbial rhodopsins. In contrast, chromophore distortion was smaller in GtCCR4 than in C1C2 and BR. Protein structural changes are much smaller in GtCCR4 than in C1C2 and CrCCR2, while this property is common to CaCCR1, which has the ECH motif. Supplementary Figure S4 shows 27 residues within 5 Å from the chromophore (retinal and side chain of lysine) in the structure of BR [53]. Among the 27 residues, 14 are identical between GtCCR4 and BR, while 9 are identical between GtCCR4 and C1C2. This suggests that the residues surrounding the retinal chromophore of GtCCR4 are more BR-like, which is consistent with the DTD motif. Further efforts by use of mutant proteins will elucidate a specific color tuning mechanism. It should be also noted that FTIR spectra at >2000 cm−1 provide useful information on the hydrogen-bonding network by monitoring X-H and X-D stretches [54,55]. Although the sample quality was not sufficient, FTIR spectroscopy of high frequency region is our future focus.
In a flash photolysis study of GtCCR4, the formation of K, L, M, and O intermediates was observed (Fig. 3), which is also common among microbial rhodopsins. Both electrophysiology and flash photolysis experiments showed that channel closing occurs upon reprotonation of the Schiff base. Although sample conditions differ between the two measurements, the results suggest that the dynamics of retinal and channels are tightly coupled in GtCCR4. In the case of CrCCR2, the best studied ChR, the time constant of reprotonation of the Schiff base (2 ms) is faster than the channel closing time (10–20 ms) [28]. Therefore, channel closing dynamics are not coupled to those of the retinal chromophore. In CrCCR2, E90 constitutes the central gate, and plays a key role in channel function [56,57]. In fact, a point mutation of E90 to arginine converted ChR2 into a chloride channel [58], and changes to the protonation of E90 have been hotly debated [27,28,35,36,38]. E90 of CrCCR2 is not conserved in GtCCR4, and there must be another mechanism for gating in DTD ChRs, where channel closing is more strictly controlled by Schiff base reprotonation. Further studies will surely reveal the detailed molecular mechanism of DTD ChR.
Conclusion
GtCCR4 contains a DTD motif like a light-driven proton pump BR. Present electrophysiological measurements clearly showed that GtCCR4 functions as a light-gated cation channel, similar to other G. theta DTD ChRs (GtCCR1-3). It was suggested that GtCCR4 has a light-driven proton pump activity. Both electrophysiological and flash photolysis experiments showed that channel closing occurs upon reprotonation of the Schiff base, which was not the case for CrCCR2. This fact suggests that the dynamics of retinal and channels are tightly coupled in GtCCR4, a DTD ChR. FTIR spectroscopy at 77 K monitored an all-trans to a 13-cis photoisomerization as the primary reaction, although perturbations in the secondary structure were much smaller in GtCCR4 than in CrCCR2. These unique properties characterize the DTD ChR.
Significance.
Functions of microbial rhodopsins are characterized by the motif in C-helix. The DTD motif is unique to an archaeal light-driven proton pump such as bacteriorhodopsin. Here we report that a DTD rhodopsin from G. theta (GtCCR4) functions as a light-gated cation channel, which also pumps protons outwardly. The primary reaction is a photoisomerization from an all-trans form to a 13-cis form of retinal, like other microbial rhodopsins, but protein structural changes are much smaller in GtCCR4 than in channelrhodopsin 2 from Chlamydomonas reinhardtii. The structural dynamics of retinal and channels are tightly coupled in the late intermediates, which are unique to GtCCR4.
Supplementary Information
Acknowledgements
This work was financially supported by grants from the Japanese Ministry of Education, Culture, Sports, Science and Technology to K.I. (26708001, 26115706, 26620005) and to H.K. (25104009, 15H02391).
Footnotes
Conflicts of Interest
All authors declare that they have no conflict of interest.
Author Contributions
H. K. directed the research, and wrote the manuscript. Y. Y. prepared samples and performed all experiments with the help of M. K. FTIR experiments were performed by Y. Y. and S. I. S. T. performed patch clamp experiments with Y. Y. K. I. performed flash photolysis experiments with Y. Y. All authors discussed and commented on the manuscript.
References
- 1.Ernst OP, Lodowski DT, Elstner M, Hegemann P, Brown LS, Kandori H. Microbial and animal rhodopsins: structures, functions, and molecular mechanisms. Chem Rev. 2014;114:126–163. doi: 10.1021/cr4003769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grote M, Engelhard M, Hegemann P. Of ion pumps, sensors and channels—perspectives on microbial rhodopsins between science and history. Biochim Biophys Acta. 2014;1837:533–545. doi: 10.1016/j.bbabio.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 3.Spudich JL, Sineshchekov OA, Govorunova EG. Mechanism divergence in microbial rhodopsins. Biochim Biophys Acta. 2014;1837:546–552. doi: 10.1016/j.bbabio.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deisseroth K. Optogenetics. Nat Methods. 2011;8:26–29. doi: 10.1038/nmeth.f.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang F, Vierock J, Yizhar O, Fenno LE, Tsunoda S, Kianianmomeni A, et al. The microbial opsin family of optogenetic tools. Cell. 2011;147:1446–1457. doi: 10.1016/j.cell.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chow BY, Boyden ES. Optogenetics and translational medicine. Sci Transl Med. 2013;5:177ps5. doi: 10.1126/scitranslmed.3003101. [DOI] [PubMed] [Google Scholar]
- 7.Oesterhelt D, Stoeckenius W. Rhodopsin-like protein from the purple membrane of Halobacterium halobium. Nat New Biol. 1971;233:149–152. doi: 10.1038/newbio233149a0. [DOI] [PubMed] [Google Scholar]
- 8.Kandori H, Yamazaki Y, Shichida Y, Raap J, Lugtenburg J, Belenky M, et al. Tight Asp-85–Thr-89 association during the pump switch of bacteriorhodopsin. Proc Natl Acad Sci USA. 2001;98:1571–1576. doi: 10.1073/pnas.98.4.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inoue K, Ono H, Abe-Yoshizumi R, Yoshizawa S, Ito H, Kogure K, et al. A light-driven sodium ion pump in marine bacteria. Nat Commun. 2013;4:1678. doi: 10.1038/ncomms2689. [DOI] [PubMed] [Google Scholar]
- 10.Yoshizawa S, Kumagai Y, Kim H, Ogura Y, Hayashi T, Iwasaki W, et al. Functional characterization of flavobacteria rhodopsins reveals a unique class of light-driven chloride pump in bacteria. Proc Natl Acad Sci USA. 2014;111:6732–6737. doi: 10.1073/pnas.1403051111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inoue K, Koua FH, Kato Y, Abe-Yoshizumi R, Kandori H. Spectroscopic study of a light-driven chloride ion pump from marine bacteria. J Phys Chem B. 2014;118:11190–11199. doi: 10.1021/jp507219q. [DOI] [PubMed] [Google Scholar]
- 12.Inoue K, Kato Y, Kandori H. Light-driven ion-translocating rhodopsins in marine bacteria. Trends Microbiol. 2015;23:91–98. doi: 10.1016/j.tim.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 13.Kandori H. Ion-pumping microbial rhodopsins. Front Mol Biosci. 2015;2:52. doi: 10.3389/fmolb.2015.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nagel G, Ollig D, Fuhrmann M, Kateriya S, Musti AM, Bamberg E, et al. Channelrhodopsin-1: a light-gated proton channel in green algae. Science. 2002;296:2395–2398. doi: 10.1126/science.1072068. [DOI] [PubMed] [Google Scholar]
- 15.Nagel G, Szellas T, Huhn W, Kateriya S, Adeishvili N, Berthold P, et al. Channelrhodopsin-2, a directly cation-selective light-gated membrane channel. Proc Natl Acid Sci USA. 2003;24:13940–13945. doi: 10.1073/pnas.1936192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Govorunova E, Sineshchekov O, Janz R, Liu X, Spudich JL. Natural light-gated anion channels: A family of microbial rhodopsins for advanced optogenetics. Science. 2015;349:647–650. doi: 10.1126/science.aaa7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Govorunova EG, Sineshchekov OA, Spudich JL. Structurally distinct cation channelrhodopsins from cryptophyte algae. Biophys J. 2016;110:2302–2304. doi: 10.1016/j.bpj.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Curtis BA, Tanifuji G, Burki F, Gruber A, Irimia M, Maruyama S, et al. Algal genomes reveal evolutionary mosaicism and the fate of nucleomorphs. Nature. 2012;492:59–65. doi: 10.1038/nature11681. [DOI] [PubMed] [Google Scholar]
- 19.Nagel G, Szellas T, Kateriya S, Adeishvili N, Hegemann P, Bamberg E. Channelrhodopsins: directly light-gated cation channels. Biochem Soc Trans. 2005;33:863–866. doi: 10.1042/BST0330863. [DOI] [PubMed] [Google Scholar]
- 20.Lórenz-Fonfría VA, Heberle J. Channelrhodopsin unchained: Structure and mechanism of a light-gated cation channel. Biochim Biophys Acta. 2014;1837:626–642. doi: 10.1016/j.bbabio.2013.10.014. [DOI] [PubMed] [Google Scholar]
- 21.Schneider F, Grimm C, Hegemann P. Biophysics of channelrhodopsin. Annu Rev Biophys. 2015;44:167–186. doi: 10.1146/annurev-biophys-060414-034014. [DOI] [PubMed] [Google Scholar]
- 22.Kato HE, Zhang F, Yizhar O, Ramakrishnan C, Nishizawa T, Hirata K, et al. Crystal structure of the channelrhodopsin light-gated cation channel. Nature. 2012;482:369–374. doi: 10.1038/nature10870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ritter E, Stehfest K, Berndt A, Hegemann P, Bartl FJ. Monitoring light-induced structural changes of Channelrhodopsin-2 by UV-visible and Fourier transform infrared spectroscopy. J Biol Chem. 2008;283:35033–35041. doi: 10.1074/jbc.M806353200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Radu I, Bamann C, Nack M, Nagel G, Bamberg E, Heberle J. Conformational changes of channelrhodopsin-2. J Am Chem Soc. 2009;131:7313–7319. doi: 10.1021/ja8084274. [DOI] [PubMed] [Google Scholar]
- 25.Nack M, Radu I, Gossing M, Bamann C, Bamberg E, von Mollard GF, et al. The DC gate in Channelrhodopsin-2: crucial hydrogen bonding interaction between C128 and D156. Photochem Photobiol Sci. 2010;9:194–198. doi: 10.1039/b9pp00157c. [DOI] [PubMed] [Google Scholar]
- 26.Stehfest K, Ritter E, Berndt A, Bartl F, Hegemann P. The branched photocycle of the slow-cycling channelrhodopsin-2 mutant C128T. J Mol Biol. 2010;398:690–702. doi: 10.1016/j.jmb.2010.03.031. [DOI] [PubMed] [Google Scholar]
- 27.Eisenhauer K, Kuhne J, Ritter E, Berndt A, Wolf S, Freier E, et al. In channelrhodopsin-2 Glu-90 is crucial for ion selectivity and is deprotonated during the photocycle. J Biol Chem. 2012;287:6904–6911. doi: 10.1074/jbc.M111.327700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lórenz-Fonfría VA, Resler T, Krause N, Nack M, Gossing M, von Mollard GF, et al. Transient protonation changes in channelrhodopsin-2 and their relevance to channel gating. Proc Natl Acad Sci USA. 2013;110:1273–1281. doi: 10.1073/pnas.1219502110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neumann-Verhoefen MK, Neumann K, Bamann C, Radu I, Heberle J, Bamberg E, et al. Ultrafast infrared spectroscopy on channelrhodopsin-2 reveals efficient energy transfer from the retinal chromophore to the protein. J Am Chem Soc. 2013;135:6968–6976. doi: 10.1021/ja400554y. [DOI] [PubMed] [Google Scholar]
- 30.Ito S, Kato HE, Taniguchi R, Iwata T, Nureki O, Kandori H. Water-containing hydrogen-bonding network in the active center of channelrhodopsin. J Am Chem Soc. 2014;136:3475–3482. doi: 10.1021/ja410836g. [DOI] [PubMed] [Google Scholar]
- 31.Muders V, Kerruth S, Lórenz-Fonfría VA, Bamann C, Heberle J, Schlesinger R. Resonance Raman and FTIR spectroscopic characterization of the closed and open states of channelrhodopsin-1. FEBS Lett. 2014;588:2301–2306. doi: 10.1016/j.febslet.2014.05.019. [DOI] [PubMed] [Google Scholar]
- 32.Lórenz-Fonfría VA, Muders V, Schlesinger R, Heberle J.Changes in the hydrogen-bonding strength of internal water molecules and cysteine residues in the conductive state of channelrhodopsin-1 J Chem Phys 14122D5072014 [DOI] [PubMed] [Google Scholar]
- 33.Ogren JI, Yi A, Mamaev S, Li H, Lugtenburg J, DeGrip WJ, et al. Comparison of the structural changes occurring during the primary phototransition of two different channelrhodopsins from Chlamydomonas algae. Biochemistry. 2015;54:377–388. doi: 10.1021/bi501243y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lórenz-Fonfría VA, Schultz BJ, Resler T, Schlesinger R, Bamann C, Bamberg E, et al. Pre-gating conformational changes in the ChETA variant of channelrhodopsin-2 monitored by nanosecond IR spectroscopy. J Am Chem Soc. 2015;137:1850–1861. doi: 10.1021/ja5108595. [DOI] [PubMed] [Google Scholar]
- 35.Kuhne J, Eisenhauer K, Ritter E, Hegemann P, Gerwert K, Bartl F. Early formation of the ion-conducting pore in channelrhodopsin-2. Angew Chem Int Ed Engl. 2015;54:4953–4957. doi: 10.1002/anie.201410180. [DOI] [PubMed] [Google Scholar]
- 36.Inaguma A, Tsukamoto H, Kato HE, Kimura T, Ishizuka T, Oishi S, et al. Chimeras of channelrhodopsin-1 and -2 from Chlamydomonas reinhardtii exhibit distinctive light-induced structural changes from channelrhodopsin-2. J Biol Chem. 2015;290:11623–11634. doi: 10.1074/jbc.M115.642256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ogren JI, Yi A, Mamaev S, Li H, Spudich JL, Rothschild KJ. Proton transfers in a channelrhodopsin-1 studied by Fourier transform infrared (FTIR) difference spectroscopy and site-directed mutagenesis. J Biol Chem. 2015;290:12719–12730. doi: 10.1074/jbc.M114.634840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Resler T, Schultz BJ, Lórenz-Fonfría VA, Schlesinger R, Heberle J. Kinetic and vibrational isotope effects of proton transfer reactions in channelrhodopsin-2. Biophys J. 2015;109:287–297. doi: 10.1016/j.bpj.2015.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Waschuk SA, Bezerra AG, Shi JrL, Brown LS. Leptosphaeria rhodopsin: Bacteriorhodopsin-like proton pump from a eukaryote. Proc Natl Acad Sci USA. 2005;102:6879–6883. doi: 10.1073/pnas.0409659102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Inoue K, Ito S, Kato Y, Nomura Y, Shibata M, Uchihashi T, et al. A natural light-driven inward proton pump. Nat Commun. 2016;7:13415. doi: 10.1038/ncomms13415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Váró G, Brown LS, Sasaki J, Kandori H, Maeda A, Needleman R, et al. Light-driven chloride ion transport by halorhodopsin from Natronobacterium pharaonis. 1. The photo chemical cycle. Biochemistry. 1995;34:14490–14499. doi: 10.1021/bi00044a027. [DOI] [PubMed] [Google Scholar]
- 42.Kandori H, Kinoshita N, Shichida Y, Maeda A. Protein structural changes in bacteriorhodopsin upon photoisomerization as revealed by polarized FTIR spectroscopy. J Phys Chem B. 1998;102:7899–7905. [Google Scholar]
- 43.Aton B, Doukas AG, Callender RH, Becher B, Ebrey TG. Resonance Raman studies of the purple membrane. Biochemistry. 1977;16:2995–2999. doi: 10.1021/bi00632a029. [DOI] [PubMed] [Google Scholar]
- 44.Smith SO, Lugtenburg J, Mathies RA. Determination of retinal chromophore structure in bacteriorhodopsin with resonance Raman spectroscopy. J Membr Biol. 1985;85:95–109. doi: 10.1007/BF01871263. [DOI] [PubMed] [Google Scholar]
- 45.Gerwert K, Siebert F. Evidence for light-induced 13-cis, 14-s-cis isomerization in bacteriorhodopsin obtained by FTIR difference spectroscopy using isotopically labelled retinals. EMBO J. 1986;5:805–811. doi: 10.1002/j.1460-2075.1986.tb04285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maeda A, Sasaki J, Pfefferle JM, Shichida Y, Yoshizawa T. Fourier transform infrared spectral studies on the Schiff base mode of all-trans bacteriorhodopsin and its photointermediates, K and L. Photochem Photobiol. 1991;54:911–921. [Google Scholar]
- 47.Aton B, Doukas AG, Narva D, Callender RH, Dinur U, Honig B. Resonance Raman studies of the primary photochemical event in visual pigments. Biophys J. 1980;29:79–94. doi: 10.1016/S0006-3495(80)85119-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baasov T, Friedman N, Sheves M. Factors affecting the C=N stretching in protonated retinal Schiff base: a model study for bacteriorhodopsin and visual pigments. Biochemistry. 1987;26:3210–3217. doi: 10.1021/bi00385a041. [DOI] [PubMed] [Google Scholar]
- 49.Rodman Gilson HS, Honig BH, Croteau A, Zarrilli G, Nakanishi K. Analysis of the factors that influence the C=N stretching frequency of polyene Schiff bases. Biophys J. 1988;53:261–269. doi: 10.1016/S0006-3495(88)83087-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feldbauer K, Zimmermann D, Pintschovius V, Spitz J, Bamann C, Bamberg E. Channelrhodopsin-2 is a leaky proton pump. Proc Natl Acad Sci USA. 2009;106:12317–12322. doi: 10.1073/pnas.0905852106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shimono K, Ikeura Y, Sudo Y, Iwamoto M, Kamo N. Environment around the chromophore in pharaonis phoborhodopsin: mutation analysis of the retinal binding site. Biochim Biophys Acta. 2001;1515:92–100. doi: 10.1016/s0005-2736(01)00394-7. [DOI] [PubMed] [Google Scholar]
- 52.Sudo Y, Okazaki A, Ono H, Yagasaki J, Sugo S, Kamiya M, et al. A blue-shifted light-driven proton pump for neural silencing. J Biol Chem. 2013;288:20624–20632. doi: 10.1074/jbc.M113.475533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Luecke H, Schobert B, Richter HT, Cartailler JP, Lanyi JK. Structure of bacteriorhodopsin at 1.55 Å resolution. J Mol Biol. 1999;291:899–911. doi: 10.1006/jmbi.1999.3027. [DOI] [PubMed] [Google Scholar]
- 54.Kandori H. Role of internal water molecules in bacteriorhodopsin. Biochim Biophys Acta. 2000;1460:177–191. doi: 10.1016/s0005-2728(00)00138-9. [DOI] [PubMed] [Google Scholar]
- 55.Furutani Y, Kandori H. Hydrogen-bonding changes of internal water molecules upon the actions of microbial rhodopsins studied by FTIR spectroscopy. Biochim Biophys Acta. 2014;1837:598–605. doi: 10.1016/j.bbabio.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 56.Ruffert K, Himmel B, Lall D, Bamann C, Bamberg E, Betz H, et al. Glutamate residue 90 in the predicted transmembrane domain 2 is crucial for cation flux through channelrhodopsin 2. Biochem Biophys Res Commun. 2011;410:737–743. doi: 10.1016/j.bbrc.2011.06.024. [DOI] [PubMed] [Google Scholar]
- 57.Gradmann D, Berndt A, Schneider F, Hegemann P. Rectification of the channelrhodopsin early conductance. Biophys J. 2011;101:1057–1068. doi: 10.1016/j.bpj.2011.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wietek J, Wiegert JS, Adeishvili N, Schneider F, Watanabe H, Tsunoda SP, et al. Conversion of channelrhodopsin into a light-gated chloride channel. Science. 2014;344:409–412. doi: 10.1126/science.1249375. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.