Abstract
Our increasing comprehension of neural crest cell development has reciprocally advanced our understanding of cadherin expression, regulation, and function. As a transient population of multipotent stem cells that significantly contribute to the vertebrate body plan, neural crest cells undergo a variety of transformative processes and exhibit many cellular behaviors, including epithelial-to-mesenchymal-transition (EMT), motility, collective cell migration, and differentiation. Multiple studies have elucidated regulatory and mechanistic details of specific cadherins during neural crest cell development in a highly contextual manner. Collectively, these results reveal that gradual changes within neural crest cells are accompanied by often times subtle, yet important, alterations in cadherin expression and function. The primary focus of this review is to coalesce recent data on cadherins in neural crest cells, from their specification to their emergence as motile cells soon after EMT, and to highlight the complexities of cadherin expression beyond our current perceptions, including the hypothesis that the neural crest EMT is a transition involving a predominantly singular cadherin switch. Further advancements in genetic approaches and molecular techniques will provide greater opportunities to integrate data from various model systems in order to distinguish unique or overlapping functions of cadherins expressed at any point throughout the ontogeny of the neural crest.
A. Formation of the neural crest
Neural crest cells are a hallmark of vertebrate embryos. Endowed with a necessary function in embryonic patterning, these cells are all the more remarkable given their transient existence in the embryo, coupled with this distinct and important function that must be achieved within a defined developmental window. Premigratory neural crest cells are specified early in development at gastrulation ((Aybar and Mayor, 2002; Basch et al., 2006; Basch et al., 2004; Ezin et al., 2009); reviewed in (Duband et al., 2015)) through the specification of the neural plate border at the lateral edges of the neural plate; these precursors later reside within the dorsal neural folds of the forming neural tube (Simoes-Costa and Bronner, 2015). As a pseudo-stratified epithelium, the neural tube (and premigratory neural crest cells) possesses specific junctional complexes to maintain this epithelial state. These include both tight and adherens junctions ((Fishwick et al., 2012; Wu et al., 2011); reviewed in (Duband et al., 2015; Taneyhill and Schiffmacher, 2013)), along with the appropriate intracellular molecules, such as Rho family GTPases, that render these cells immotile (reviewed in (Kalcheim, 2015; Taneyhill and Padmanabhan, 2013)).
To execute their important role in vertebrate patterning, premigratory neural crest cells must acquire the capacity to migrate, which typically involves delamination of these cells coupled with either a partial or complete epithelial-to-mesenchymal transition (EMT). The temporal order of these events, however, is organism- and axial level-dependent. Delamination in cranial neural cells occurs through an en masse process across vertebrate species. EMT and delamination occur concomitantly from the dorsal neural tube in the chick cranial neural crest (Duband and Thiery, 1982), and zebrafish cranial neural crest cells also simultaneously initiate EMT and emerge from the dorsal region of the neural tube (Halloran and Berndt, 2003). EMT occurs first in the neural folds of the mouse head and is later followed by delamination (Nichols, 1981, 1987; Theveneau and Mayor, 2012b), while in Xenopus, neural crest cells delaminate prior to neural tube closure, maintaining their intercellular interactions, and EMT and migration commence just before the neural tube closes. This is often referred to as a partial EMT because EMT is not completed until after the onset of migration (Alfandari et al., 2010; Sadaghiani and Thiebaud, 1987; Theveneau et al., 2010; Theveneau and Mayor, 2012c, 2013). Recent work in the cranial neural crest field has highlighted that EMT is a process that consists of at least two broad phases: 1) Delamination and separation of cells from the neuroepithelium (e.g., through de-epithelialization and decreased cell-adhesion) and 2) cell dispersal (e.g., through the adoption of mesenchymal properties and morphology changes that ultimately allow the cell to become migratory) (reviewed in (Simoes-Costa and Bronner, 2015)). At the trunk axial level in all of these vertebrates, however, neural crest cells delaminate continuously but as single cells, often in what is referred to as a “drip-like” fashion (Duband, 2010). Once neural crest cells complete their delamination and EMT, the dorsal neural tube later gives rise to the roof plate, which plays important roles in sending BMP signals to the ectoderm as well as differentiating into distinct neuronal subtypes of the central nervous system (Cohen et al., 2013; Tozer et al., 2013).
Although the timing of delamination and EMT differs across species and axial levels, the end result in each case is the generation of a migratory neural crest cell. Once motile, neural crest cells traverse the embryo but do not do so in isolation. Components of the extracellular matrix (ECM) can function to impede or promote migration, as demonstrated in vivo and ex vivo using explants of neural crest tissue, a system that recapitulates EMT and migration (see (Cousin et al., 2012; Garcia-Castro et al., 2002; Liu and Jessell, 1998; Taneyhill and Bronner-Fraser, 2005)). For example, some collagens (Duband and Thiery, 1987), fibronectin (Newgreen and Thiery, 1980), hyaluronan (Casini et al., 2012; Ori et al., 2006), and vitronectin (Delannet et al., 1994) support or promote neural crest migration, while other molecules such as versican (Perissinotto et al., 2000; Perris et al., 1996; Szabo et al., 2016), aggrecan (Perissinotto et al., 2000; Perris et al., 1996), laminin (Coles et al., 2006; Krotoski et al., 1986), other collagens (Duband and Thiery, 1987), and tenascin/cytotactin (Tan et al., 1987) limit or restrict migration of neural crest cells, in a variety of species. Neural crest cells must also intermingle with other cells to give rise to a plethora of new cell types and other structures in vertebrates. Together with neurogenic placode cells, neural crest cells assemble the cranial ganglia (Saint-Jeannet and Moody, 2014; Steventon et al., 2014), while the entire craniofacial skeleton and cartilages are derived from both neural crest and mesodermal cells (Jheon and Schneider, 2009; Noden and Trainor, 2005). Neural crest cells also form the enteric nervous system, portions of the heart, sympathetic ganglia, and pigment cells (Bronner and LeDouarin, 2012). In short, the generation of migratory neural crest cells is crucial for the proper formation and function of vertebrates.
B. Cadherin function in neural crest cell ontogeny
Transmembrane cadherin proteins are key to the formation and function of multiple organisms (Brieher & Yap, 2013; Gumbiner, 2005; Niessen & Gottardi, 2008; Niessen, Leckband, & Yap, 2011). Cadherin proteins form homodimers in cis within a cell and then primarily associate in trans with cadherin ectodomains of a similar type on adjacent cells. The cadherin cytoplasmic tail is then directly bound by members of the catenin family, such as β- and δ (p120)-catenin, and indirectly bound to α-catenin, which provides a link to the actin cytoskeleton (McCrea & Gottardi, 2016; Niessen & Gottardi, 2008) (Figure 1a). This entire complex forms the zonular adherens or cellular adherens junction, which is situated apically and laterally within epithelial cells and functions to maintain cell–cell interactions as well as facilitate the establishment of polarity. Herein, we will focus exclusively on the known expression pattern and function for cadherins, and their regulators, in neural crest cells undergoing EMT/delaminating and during their early emigration and migration (Table 1). The reader is referred to other excellent publications discussing cadherin expression and function in fully migratory neural crest cells, particularly with respect to roles in the contact inhibition of locomotion (CIL) exhibited by neural crest cells in their interactions with one another and with other cell types (Barriga & Mayor, 2015; Dady, Blavet, & Duband, 2012; Lee et al., 2013; Roycroft & Mayor, 2016; Theveneau & Mayor, 2012a; Theveneau et al., 2013).
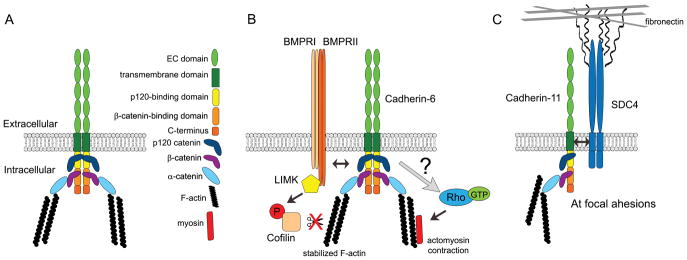
Figure 1. Full-length cadherins are multifunctional proteins in the neural crest.
(A) Cadherins are single-pass membrane proteins that dimerize in cis, interact with cadherin dimers on neighboring cells via their ectodomains in trans, and anchor the F-actin network to the cell membrane through multiple intracellular domain/catenin interactions. (B) Cadherins modulate neural crest cell contractility and apical detachment during EMT through multiple mechanisms. In zebrafish, Cadherin-6 is required for the accumulation of F-actin bundles and functions as an upstream effector of GTPase-mediated actomysoin contraction, which are both required for neural crest detachment during EMT (Clay and Halloran, 2014). In chick premigratory trunk neural crest cells, Cadherin-6B functions upstream of the BMPRII-mediated activity of LIMK, which inactivates cofilin via phosphorylation. As a result, cofilin cannot sever actin filaments, which in turn positively regulates F-actin dynamics and de-epithelialization during EMT (Park and Gumbiner, 2010, 2012). (C) In Xenopus migratory neural crest cells, Cadherin-11, but not N-cadherin, physically associates with Syndecan-4 (SDC4) at focal adhesions to bridge the extracellular matrix to the F-actin network and promote cell-substrate adhesion during migration (Langhe et al., 2016). Double arrows indicate physical interaction.
Table 1.
Functions of cadherins expressed in premigratory and early migratory neural crest cells.
| Cadherin | Species | Neural Crest Type | Axial Level | Function | References |
|---|---|---|---|---|---|
| E-cadherin | Frog Zebrafish | Premigratory | Cranial | Suppression of CIL | Scarpa et al., 2015 |
| Frog | Migratory | Cranial | Positive role in NCC migration and membrane protrusion | Huang et al., 2016 | |
|
| |||||
| N-cadherin | Chicken | Premigratory | Trunk | Negative role in EMT | Nakagawa and Takeichi 1998 |
| Premigratory | Cranial/Trunk | Cell adhesion/maintenance of neural tube pseudostratfied epithelium integrity | Bronner-Fraser et al. 1992; Nakagawa and Takeichi 1998 | ||
| Premigratory | Trunk | Pro-delamination role via indirect transcriptional regulation through N-cadherin CTF2 activity | Shoval et al., 2007 | ||
| Premigratory | Trunk | Cell adhesion/regulation of Wnt signaling | Shoval et al., 2007 | ||
| Premigratory | Trunk | Positive roles in delamination and migration following proteolysis by MMP9 | Monsonego-Ornan et al., 2012 | ||
|
| |||||
| Zebrafish | Migratory | Cranial | Positive role in migration | Piloto and Schilling, 2010 | |
| Early Migratory | Trunk | Positive role in migration | Powell et al., 2015 | ||
|
| |||||
| Frog | Migratory | Cranial | Positive role in directed migration and CIL | Theveneau et al., 2010; Theveneau et al., 2013 | |
|
| |||||
| Cadherin-6 | Chicken (Cad6B) | Premigratory | Cranial/Trunk | Establishment/segregation of premigratory NCC domain; Facilitator of neural fold fusion | Nakagawa and Takeichi 1995 |
| Early migratory | Trunk | Promotion of de-epithelialization during EMT via BMPRII/LIMK/cofilin pathway | Park and Gumbiner 2010; 2012 | ||
| Premigratory | Midbrain | Cell adhesion/maintenance of premigratory NCC epithelium | Coles et al., 2007; Schiffmacher et al., 2013 | ||
| Premigratory | Cranial | Pro-EMT role via indirect transcriptional regulation through Cadherin-6B CTF2 activity | Schiffmacher et al., 2016 | ||
|
| |||||
| Mouse | Premigratory/migratory | Cranial/Trunk | Positive role in EMT and migration | Inoue et al., 1997 | |
|
| |||||
| Zebrafish | Premigratory/migratory | Hindbrain | Positive role in EMT via regulation of F-actin assembly, Rho activity, and actomyosin, to promote detachment | Clay and Halloran, 2014 | |
|
| |||||
| Cadherin-7 | Chicken | Migratory | Cranial/Trunk | Positive role in directional migration | Nakagawa and Takeichi 1995 |
|
| |||||
| Rat | Migratory | Cranial | Positive role in migration | Takahashi and Osumi 2008 | |
|
| |||||
| Cadherin-11 | Frog | Migratory | Cranial/Trunk | Positive role in migration | Vallin et al., 1998 |
| Migratory | Cranial | Modulation of NCC specifier gene expression through regulation of Wnt/β-catenin signaling | Borchers et al., 2001 | ||
| Migratory | Cranial | Initiation of protrusive activity in filopodia and lamellipodia via interaction with GEF Trio and GTPases | Kashef et al., 2012 | ||
| Migratory | Cranial | Positive role in migration via the activity of the ADAM13-processed Cadherin-11 NTF | McCusker et al., 2009; Abbruzzese et al., 2016 | ||
| Migratory | Cranial | Positive role for full-length Cadherin-11 in CIL and directional migration | Becker et al., 2013; Abbruzzese et al., 2016 | ||
| Migratory | Cranial | Promotion of cell-substrate adhesion via interaction with Syndecan-4 at focal adhesions | Langhe et al., 2016 | ||
| Migratory | Cranial | Indirect negative role in CIL via the activity of the shed Cadherin-11 NTF | Abbruzzese et al., 2016 | ||
|
| |||||
| Chicken | Migratory | Trunk | Positive role in migration | Chalpe et al., 2010 | |
This table represents a subset of selected neural crest cadherin studies within the following models: chicken (Gallus gallus), mouse (Mus musculus), rat (Rattus norvegicus), frog (Xenopus laevis), and zebrafish (Danio rerio). Italicized functions are inferred from correlative neural crest spatio-temporal expression profiles or neural crest-specific upstream effector perturbation data. Abbreviations: contact inhibition of locomotion (CIL), neural crest cell (NCC), epithelial-to-mesenchymal transition (EMT), C-terminal fragment 2 (CTF2), N-terminal fragment (NTF).
While E-cadherin is initially present in the ectoderm, the segregation of the ectoderm into neural and non-neural domains had been previously tied to the expression of N-cadherin (and loss of E-cadherin) and maintenance of E-cadherin, respectively (Edelman et al., 1983; Ezin et al., 2009; Kemler et al., 1977; Schuh et al., 1986; Thiery et al., 1984). Premigratory neural crest cells will come to reside in the border region between these domains (neural plate border). During neurulation, the elevation of the cranial neural folds, and their eventual fusion, is accompanied by a reduction in N-cadherin therein, around Hamburger Hamilton (HH) 8 (4 somite stage (ss)) in chick (Duband et al., 1988; Hatta and Takeichi, 1986) and occurs prior to EMT in chick, frogs, and mice (Akitaya and Bronner-Fraser, 1992; Davidson and Keller, 1999; Duband et al., 1988; Hatta et al., 1987; Nakagawa and Takeichi, 1998; Radice et al., 1997), with N-cadherin expression persisting within other regions of the neural tube along the vertebrate body axis (Akitaya and Bronner-Fraser, 1992; Nakagawa and Takeichi, 1998). N-cadherin function in the neural crest has been well documented through gene perturbation assays. Overexpression of N-cadherin in the chick blocks neural crest cell emigration but does not affect the ability of neural crest cells to differentiate into the melanocyte lineage (Nakagawa and Takeichi, 1998; Shoval et al., 2007). These results are corroborated by injection of an N-cadherin function-blocking antibody that binds to the N-cadherin ectodomain, which leads to precocious emigration of neural crest cells and perturbs neural tube architecture (Bronner-Fraser et al., 1992).
Recent studies in chick, mouse, and frog, however, have revealed a more complex and dynamic expression pattern for E-cadherin and N-cadherin during neurulation (Breau et al., 2008; Dady et al., 2012; Huang et al., 2016; Lee et al., 2013). As neurulation begins at HH3 in the chick (prior to somite formation), E-cadherin transcripts and protein are found throughout the ectoderm, with N-cadherin notably absent at the transcript and protein level. With the formation of the head folds (HH7), N-cadherin transcripts are present anteriorly but the protein is conspicuously absent, whereas E-cadherin transcripts and protein are still present, and are, in fact, maintained in the neural tube until their downregulation at HH11 (Dady et al., 2012). At HH8- (3ss) in the chick, E-cadherin protein is present throughout the neural plate and ectoderm in the midbrain and anterior hindbrain, but N-cadherin protein is absent. Both E-cadherin and N-cadherin transcripts, however, are apparent in the chick head at HH8, and at this point N-cadherin protein is first detected in the apical region of the neural plate as the neural folds are elevating. At HH8+ and 9, E-cadherin and N-cadherin proteins still perdure in the neural tube, but N-cadherin protein is not detected in the dorsal neural folds/tube where the premigratory neural crest cells reside. Interestingly, as neural crest cells are actively emigrating out of the neural tube and migrating at HH10, the entire neural tube is double-positive for both N-cadherin and E-cadherin, with migratory neural crest cells also immunoreactive for E-cadherin, but not N-cadherin protein, as observed previously (Dady et al., 2012; Rogers et al., 2013) (our unpublished data). Furthermore, neural crest cell delamination occurs in two waves, with those non-neural ectoderm-derived neural crest cells delaminating first, followed by those neural crest cells originating in the neural ectoderm (Lee et al., 2013), both in chick and mouse. At the end of EMT/delamination and following neural tube closure (HH11), neural tube and migratory neural crest cells no longer express E-cadherin protein or transcripts, with the entire neural tube N-cadherin-positive at both the protein and RNA level (Dady et al., 2012).
The chick posterior hindbrain and anterior trunk possess a complex, yet slightly different, pattern of E-cadherin and N-cadherin expression (Dady et al., 2012). At HH8, E-cadherin and N-cadherin protein are expressed in a complementary pattern within the neural plate. This pattern is maintained through HH10–, with E-cadherin protein enriched in a subpopulation of premigratory neural crest cells within the dorsal neural folds, while N-cadherin is found throughout the rest of the neural tube but is absent from the premigratory neural crest, as observed in the midbrain and anterior hindbrain. At HH11 the neural tube is devoid of E-cadherin while N-cadherin protein persists, and neural crest cells delaminating and undergoing EMT at HH12 lack both N-cadherin and E-cadherin proteins. Interestingly, chick migratory neural crest cells from the rhombomere 4 (r4) stream express N-cadherin transcripts (McLennan et al., 2012), while N-cadherin and E-cadherin proteins are both observed in early migratory neural crest cells at HH11 in more anterior regions of the chick hindbrain, with E-cadherin also noted in later migratory neural crest cells (our unpublished data). Taken together, these results highlight the dynamic changes in cadherin expression patterns in different subpopulations of neural crest cells.
E-cadherin function in the premigratory and migratory cranial neural crest has been recently documented in Xenopus (Huang et al., 2016; Scarpa et al., 2015). Using explants of cranial neural crest cells, Scarpa et al. (2015) showed a switch, or change in cadherin expression, from E-cadherin in premigratory neural crest to predominantly N-cadherin in migratory neural crest during EMT, with only some E-cadherin present in this migratory population (Scarpa et al., 2015). This group also observed this switch for zebrafish neural crest cells, and, in both organisms, showed that this change is important for CIL. Overexpression of E-cadherin impedes neural crest cell migration in a cell autonomous fashion by impairing CIL through loss of protrusion formation, but does not lead to changes in N-cadherin expression upon E-cadherin expression in migratory neural crest cells. The authors then show that inhibition of CIL is dependent upon p120-catenin binding to the E-cadherin cytoplasmic tail. Conversely, E-cadherin knockdown (via a morpholino (MO) or function-blocking antibody to E-cadherin) in premigratory neural crest cells “reverts” neural crest cell polarity, with protrusions reduced at sites of cell–cell contact and increased at the free edge, and results in explanted neural crest cells exhibiting decreased intermixing. These results are in keeping with prior work indicating that N-cadherin loss of function promotes dispersion (Theveneau et al 2010; Kuriyama et al. 2014), with N-cadherin-mediated intercellular adhesion being essential for neural crest migration.
A more recent study on E-cadherin used quantitative PCR (QPCR), immunohistochemistry, and perturbation assays to localize E-cadherin in premigratory, early emigrating, and migratory cranial neural crest cells in Xenopus (Huang et al., 2016). This report reveals that several cadherin transcripts are co-expressed at these different stages of cranial neural crest cell ontogeny, including E-cadherin, N-cadherin, XB/C-cadherin, and PCNS, with Cadherin-11 observed in early emigratory and migratory neural crest cells. Furthermore, E-cadherin protein is also present at these different stages, as noted in the preceding study. MOmediated depletion of E-cadherin impeded cranial neural crest cell migration in a cell autonomous fashion in vivo with cells exhibiting reduced dispersal and remaining attached to each other. Importantly, coexpression of another cadherin after E-cadherin knockdown could not rescue this phenotype, underscoring its specificity. Finally, the extracellular domain of E-cadherin is required for appropriate cranial neural crest cell migration and in fact can rescue migration after E-cadherin knockdown, in keeping with the above study due to loss of the p120-catenin binding domain. Further experiments are necessary to determine whether E-cadherin may be playing an adhesion-independent role during neural crest migration, given its continued presence in this population of cells.
Cadherin-6B transcripts are first detected at HH6 in the forming neural folds at all axial levels in the chick (Nakagawa and Takeichi, 1995), but transcripts and protein are not noted in the cranial premigratory neural crest cell population until HH8 (Fairchild and Gammill, 2013; Nakagawa and Takeichi, 1995; Schiffmacher et al., 2014; Taneyhill et al., 2007), after the initial appearance of E-cadherin. Trunk premigratory neural crest cells also express Cadherin-6B transcripts and protein (Nakagawa and Takeichi, 1995; Taneyhill et al., 2007). Multiple studies have shown that chick midbrain and trunk migratory neural crest cells are devoid of Cadherin-6B transcripts and protein (Fairchild and Gammill, 2013; Nakagawa and Takeichi, 1995, 1998; Schiffmacher et al., 2014; Taneyhill et al., 2007); however, one report does contend that Cadherin-6B protein is present in the early trunk migratory neural crest cell population (Park and Gumbiner, 2010). Interestingly, repression of Cadherin-6B transcripts is detected earlier than loss of membrane-bound Cadherin-6B protein in the chick head (see Section D for additional details). This loss of Cadherin-6B is associated with the timing of the delamination and EMT of cranial neural crest cells (Fairchild and Gammill, 2013; Schiffmacher et al., 2014; Taneyhill et al., 2007), as MO-mediated depletion or overexpression of Cadherin-6B promotes or inhibits delamination/EMT and migration, respectively (Coles et al., 2007). Later, Cadherin-6B protein is observed in the apical region of future roof plate cells (Duband, 2010) and in the floor plate (our unpublished data), while migratory neural crest cells in the chick head and trunk express Cadherin-7 (Nakagawa and Takeichi, 1995, 1998) (see below).
In the mouse, premigratory and migratory cranial neural crest cells express Cadherin-6 (Inoue et al., 1997), suggesting an additional function for Cadherin-6 during EMT and migration that is not present in chick, likely due to differences in transcriptional regulation by species-specific cis-regulatory elements and the presence of upstream effectors. This difference in expression pattern for Cadherin-6 between chick and mouse could also perhaps reflect a change in placental mammals, as observed for Snail family members in the mouse, which, unlike in chick and Xenopus, are “swapped” with respect to their expression pattern (Sefton et al., 1998) and appear to be indispensable for neural crest cell formation (Murray and Gridley, 2006). Studies on zebrafish Cadherin-6 revealed that while reduced in the midbrain during EMT (as seen for chick Cadherin-6B), Cadherin-6 is present at low levels in hindbrain neural crest cells both prior to and during their delamination/EMT, a pattern also noted for chick Cadherin-6B (our unpublished data). After EMT, however, Cadherin-6 is not observed in migratory neural crest cells but is instead localized to neuroepithelial cells (Clay and Halloran, 2014). This group also went on to demonstrate, through live imaging in Cadherin-6 morphant embryos, that knockdown of Cadherin-6 precludes neural crest cell EMT by inhibiting the apical detachment of neural crest cells from the neuroepithelium (Figure 1B). This occurs due to a lack of accumulation of F-actin at this apical end along with an expansion in Rho activity, suggesting that, in the zebrafish hindbrain, Cadherin-6 plays a positive role in promoting EMT. This function for Cadherin-6 in augmenting EMT in neural crest cells has also been reported in chick, challenging prior findings showing the absence of Cadherin-6B in chick trunk migratory neural crest cells (Park and Gumbiner, 2010). In this report, Cadherin-6B protein was detected in early trunk migratory neural crest cells at HH10–11, on the membrane and potentially in the cytosol, and was shown to promote EMT through de-epithelialization of premigratory neural crest cells, a step that occurs prior to their delamination and EMT. This de-epithelialization step invokes non-canonical BMP signaling through the BMP type II receptor, LIM kinase/cofilin signaling, and subsequent control of the actin cytoskeleton (Park and Gumbiner, 2012) (Figure 1B). These collective results reveal that the presence, and potential functions, for cadherins in neural crest cells are axial level- and organism-dependent.
It is worth noting that the discrepancy in the preceding results for the function of Cadherin-6B during neural crest cell EMT in the chick midbrain and trunk could be attributed to the stage and/or axial level examined as well as the methods of analysis of the experimental results. The timing of the experiments during chick development (3–8 hours post-electroporation in (Coles et al., 2007), just prior to delamination/EMT, versus 20–24 hours post-electroporation in (Park and Gumbiner, 2010), well before EMT when neural crest cells are being specified), however, may help reconcile the two experiments by suggesting distinct functions for Cadherin-6B during neural crest cell specification and later during delamination/EMT (Duband et al., 2015). Given that the first group perturbed Cadherin-6B expression right before the onset of EMT in the chick head, the de-epithelialization and reduction in adhesion between premigratory neural crest cells may have already occurred. Therefore, loss of Cadherin-6B after this initial de-epithelialization would promote EMT due to a further reduction in cadherin-mediated adhesion and/or the acquisition of other features important for EMT, such as loss of apicobasal polarity and the ability to migrate. Conversely, overexpression of Cadherin-6B just prior to EMT would follow this initial Cadherin-6B-induced de-epithelialization (or early function of Cadherin-6B), leading to enhanced cell-cell adhesion and an inability for neural crest cells to undergo EMT and migrate. With respect to the trunk studies, overexpressing Cadherin-6B long before delamination and EMT would promote de-epithelialization while neural crest cells are getting specified, triggering further de-epithelialization and acquisition of motility later in development. Conversely, early Cadherin-6B knockdown would preclude all of these events and therefore inhibit delamination and EMT. This hypothesis, however, remains to be rigorously tested. Alternatively, it is possible that the method of neural crest cell egress from the neural tube (en masse in the midbrain versus a more “drip-like” fashion in the trunk) favors the loss of Cadherin-6B in the former setting, but the retention of Cadherin-6B in the latter to allow these cells to initially adhere to each other during EMT and emigration.
Migratory neural crest cells of the chick head and trunk, as well as the rat head, express Cadherin-7 (Nakagawa and Takeichi, 1995, 1998; Takahashi and Osumi, 2008), but it is not observed in comparable crest populations in Xenopus, mouse, and zebrafish (Espeseth et al., 1995; Espeseth et al., 1998; Liu et al., 2007; Liu et al., 2008; Moore et al., 2004). Cadherin-7 transcripts are present in the chick migratory neural crest cell population as early as HH10 in the midbrain, with protein detected later at HH11 in the head (our unpublished data), while “trailing” neural crest cells in the r4 stream of the chick hindbrain express Cadherin-7 (McLennan et al., 2012; McLennan et al., 2015). In the chick trunk, Cadherin-7 transcripts and protein are observed after neural crest cells have delaminated. Here, Cadherin-7 overexpression recapitulates the phenotype observed upon N-cadherin overexpression, with newly emigrating neural crest cells aggregating close to the neural tube (Nakagawa and Takeichi, 1998).
Although chick trunk migratory neural crest cells express Cadherin-11 (Chalpe, Prasad, Henke, & Paulson, 2010), to our knowledge, Cadherin-11 distribution in the chick has not been examined during cranial neural crest cell EMT and delamination. Cadherin-11 function has been well characterized in the Xenopus neural crest, however, with expression noted in premigratory and migratory cranial neural crest cell populations as well as at later stages of trunk neural crest cell migration (Hadeball, Borchers, & Wedlich, 1998; Vallin, Girault, Thiery, & Broders, 1998). Knockdown or overexpression of Xenopus Cadherin-11 leads to defects in cranial neural crest cell migration and differentiation (Borchers, David, & Wedlich, 2001; Kashef et al., 2009). Intriguingly, this overexpression phenotype can be rescued by coinjection of ADAM13 mRNA, as ADAM13 protein cleaves Cadherin-11 to release an N-terminal (NTF) that possesses a novel function (McCusker, Cousin, Neuner, & Alfandari, 2009) (discussed in Section 4). Cadherin-11-mediated adhesion is important for CIL in Xenopus migratory cranial neural crest cells (Becker, Mayor, & Kashef, 2013), and migration is promoted by the Cadherin-11 cytoplasmic tail (Borchers et al., 2001; Kashef et al., 2009).
Depletion of Cadherin-11 within neural crest cells abolishes filopodia and lamellipodia formation, resulting in membrane blebbing (Kashef et al., 2009). This observed loss of cellular protrusions is indicative of a potential defect in small GTPase function. To investigate this further, Kashef et al. (2009) attempted to rescue Cadherin-11 neural crest migration by introducing either constitutively active or dominant negative forms of Cdc42, Rac1, and RhoA (Kashef et al., 2009). The constitutively active mutants achieved rescue while the dominant negative mutants did not, but what was missing was a molecule that could serve to bridge Cadherin-11 to these small GTPases, since Cadherin-11 does not bind them directly. The guanine exchange factor (GEF) Trio had been shown previously to interact with Cadherin-11 (Backer et al., 2007), and when it, or its GEF domains, was introduced into Cadherin-11-depleted embryos, neural crest cell migration was rescued to varying extents. In addition, the domain between the p120- and β-catenin binding sites on the Cadherin-11 cytoplasmic tail could immunoprecipitate Trio, and loss of this Trio site also inhibited neural crest migration, demonstrating its importance.
In addition to promoting migration, full-length Cadherin-11 plays a role in the specification of the neural crest domain through its indirect regulation of the Wnt signaling pathway. Through loss-of-function assays, Koehler et al. (2013) found that reduced full-length Cadherin-11 pools lead to an increase in free β-catenin, which allowed for enhanced nuclear import and subsequent upregulation of Wnt target genes including CyclinD1 and c-myc (Koehler et al., 2013). These Wnt effectors then augment proliferation and upregulation of other neural crest specifiers, including Sox10 and AP-2. Moreover, Langhe et al. (2016) found that Cadherin-11 in migratory neural crest cells localizes with β1-integrin and paxillin at focal adhesions and promotes focal adhesion binding to the fibronectin-based ECM (Langhe et al., 2016). This is accomplished through the binding of Cadherin-11 to the proteoglycan Syndecan-4, which directly binds to fibronectin (Figure 1c). This complex formation is exclusive to Cadherin-11, as N-cadherin and Ccadherin were not observed in focal adhesions. Importantly, this complex also forms and functions at focal adhesion sites in other cell types, including human foreskin fibroblasts.
Taken together, the preceding studies suggest that cadherin expression in the neural crest during delamination and EMT is quite dynamic and further underscore that EMT does not encompass a simple switch from one cadherin to another (Duband et al., 2015; Rogers et al., 2013; Shook and Keller, 2003; Taneyhill and Schiffmacher, 2013; Theveneau and Mayor, 2012a, b). For instance, the repertoire of cadherins expressed in chick midbrain premigratory neural crest cells changes dramatically, both prior to and during EMT. These cells first lose N-cadherin while maintaining both Cadherin-6B and E-cadherin, leading up to closure of the neural tube. Later, a reduction in Cadherin-6B occurs just prior to EMT, while E-cadherin is still expressed and maintained in the neural tube and in premigratory and migratory neural crest cells throughout EMT and early migration. Later, E-cadherin is downregulated, and migratory neural crest cells express Cadherin-7 (our unpublished data). The situation is even more complex in the Xenopus cranial neural crest, with many cadherins observed in both premigratory and migratory neural crest cells (Huang et al., 2016), lending further credence to the idea that EMT involves simultaneous modulation of multiple cadherins.
Given the variety of cadherins that are expressed concurrently at specific axial levels, it is tempting to speculate that it is not the composition of coexpressed cadherins that regulate neural crest cell EMT and migration, but rather the overall level of the cadherins that are present. This concept of precisely controlled cadherin levels would further argue against a cadherin switch per se, in the sense that one cadherin is not completely turned off and another turned on, but rather that cadherin levels are modulated relative to one another within a neural crest cell. Evidence for this actually exists within the neural crest itself due to the presence of multiple cadherins within the premigratory and migratory neural crest cell populations. As such, changes in cadherin levels are critical, along with integration of other signaling pathways, to allow a neural crest cell to undergo EMT and migrate. While evidence from this can be gleaned partially from immunohistochemistry assays, quantitative immunoblotting experiments conducted on isolated premigratory and migratory neural crest cells will be essential to validate this hypothesis.
While regulating collective levels of coexpressed cadherins exerts control over conserved functions that additively modulate neural crest development (cell adhesion), it is likely that each cadherin, and its proteolytic fragments, possess their own unique functions. In this regard, the composition of cadherins within a given neural crest cell also plays an important role in dictating neural crest identity and behavior. At the whole protein level, cadherins share remarkable amino acid conservation, especially within motifs that compose catenin-binding domains and extracellular repeat (EC) domains. Single amino acid substitutions between coexpressed cadherins, however, can result in differential posttranslational modifications that convey a particular function, alter affinities to other protein interactors, or result in a distinct subcellular localization. In addition, minor amino acid differences between metalloproteinase cleavage sites within cadherins can alter cleavage kinetics (Caescu, Jeschke, & Turk, 2009), leading to different levels of unique N-terminal fragments generated from two cadherins processed by the same metalloproteinase (discussed in detail in Section 4). Furthermore, differences within the cadherin C-terminus dictate binding specificity and affinity to other proteins. For example, the highly conserved Cadherin-11 C-terminus (GSKDTFDDDS in chick, humans, and rodents), which is not found in E-cadherin, N-cadherin, Cadherin-7, or Cadherin-6 proteins, exclusively binds Angiomotin, a multifunctional protein involved in migration and polarity (Ortiz et al., 2015). As premigratory neural crest cells undergo EMT and become motile, or as migratory neural crest cells terminally differentiate, a change in the cadherin composition may be required to exert unique functions of newly expressed cadherins while abolishing functions of preexisting cadherins.
From an evolutionary perspective, many of the same cadherins have been retained in different organisms and are expressed in a comparable spatiotemporal pattern at a given axial level, arguing for some degree of evolutionary constraint. For example, Xenopus, chick, and mice all express E-cadherin, to some degree, within the premigratory neural crest cell population and as neural crest cells undergo EMT and migrate. It is perhaps the concentration of E-cadherin within each organism’s neural crest cells, in conjunction with the amount of other cadherins expressed, however, which facilitates the formation and/or motility of migratory neural crest cells. Other cadherins, though, do not show conservation in their expression patterns, particularly within the migratory neural crest cell population. For instance, Cadherin-7 is observed in chick and rat migratory neural crest cells, but it is not noted in Xenopus, mouse, and zebrafish, which instead express N-cadherin, Cadherin-11, and/or Cadherin-6. While these results could be interpreted to imply that distinct migratory neural crest cell cadherins have evolved for specific organisms, that is unlikely to be the case given that multiple cadherins are expressed within the migratory neural crest cell population of all of the species cited above. Altogether, the current literature provides evidence for some degree of evolutionary constraint, coupled with meticulous control over the levels of multiple cadherins, to collectively coordinate neural crest cell EMT and migration.
REGULATION OF CADHERIN LEVELS BY TRANSCRIPTION
Transcriptional repression provides one mechanism by which premigratory neural crest cells indirectly reduce cell surface cadherin levels in order to delaminate, undergo EMT, and acquire the capacity to migrate. Work from multiple labs has yielded a neural crest gene regulatory network (Simoes-Costa and Bronner, 2015; Theveneau and Mayor, 2011) that provides insight into the different transcriptional inputs functioning to reduce cadherin expression during delamination and EMT. These studies have been conducted primarily in chick and Xenopus and will be highlighted here.
The transcription factor Sip1/Zeb2 may directly or indirectly repress E-cadherin expression in the chick cranial neural crest. Sip1 is expressed in premigratory and migratory cranial neural crest cells, and loss of Sip1 through MO-mediated knockdown reveals modest increase in E-cadherin transcripts. Moreover, early migratory neural crest cells appear to possess more membrane-bound E-cadherin protein and, in turn, remain closely associated with one another near the neural tube (Rogers et al., 2013). Since E-cadherin is expressed in chick premigratory and early migratory cranial neural crest cells (Dady et al., 2012; Lee et al., 2013), these results suggest that Sip1 controls E-cadherin levels to ensure that cranial neural crest cells can disperse and properly migrate away from the neural tube. Sip1 does not, however, impact the ability of these cells to delaminate and undergo EMT, and this is consistent with reports of E-cadherin expression in the cranial neural crest during these processes (Dady et al., 2012; Huang et al., 2016; Lee et al., 2013). Sip1-depleted cells also exhibit a reduction in N-cadherin transcripts and protein. Given that N-cadherin is decreased in premigratory neural crest cells prior to their delamination and EMT (and is also not present in early migratory neural crest cells), and the persistence of Sip1 in both the premigratory and migratory cranial neural crest, it is unlikely that Sip1 directly modulates N-cadherin expression, unless Sip1 protein is modified such that it can no longer transcriptionally activate N-cadherin during the embryonic stages in which N-cadherin is downregulated. Interestingly, Cadherin-6B transcripts and protein are unaffected.
Appropriate levels of E-cadherin in controlling cranial neural crest migration may in fact be conserved in Xenopus and zebrafish. Delaminating cranial neural crest cells reduce E-cadherin transcripts through the activity of the transcriptional repressor Twist, which is positively regulated by hypoxia-inducible transcription factor 1α (Hif-1α) (Barriga, Maxwell, Reyes, & Mayor, 2013). Although Hif1α is expressed ubiquitously, it is required for neural crest migration in the head (in a cell autonomous fashion) and trunk. Depletion of Hif-1α in Xenopus leads to loss of Twist, which encodes a transcription factor critical for neural crest development (Bildsoe et al., 2009; Chen & Behringer, 1995; Hopwood, Pluck, & Gurdon, 1989; Ishii et al., 2003; Ota et al., 2004; Soo et al., 2002). This occurs in both premigratory and migratory cranial neural crest cells and results in cell aggregation that can be rescued by introduction of Twist mRNA. Conversely, Hif-1α overexpression increases cell dispersal. The authors demonstrate that loss of Hif-1α and Twist leads to increased membrane E-cadherin protein between aggregated neural crest cells in ex vivo explants. These results suggest that Hif-1α activates expression of Twist, which in turn represses E-cadherin transcription, to permit delamination of the neural crest. Interestingly, in vivo studies examining E-cadherin distribution in cranial neural crest cells possessing the Hif-1α MO reveal that, in many instances, neural crest cells still delaminate and begin their migration while still expressing membrane E-cadherin. These results indicate that other signals in vivo may maintain E-cadherin in newly delaminated neural crest cells, and, as shown recently, that E-cadherin may play a positive role during neural crest cell motility (Huang et al., 2016). As such, a role for modulating E-cadherin expression in Xenopus appears to rely on Twist, while amniotes, which lack Twist expression in the neural crest proper, use other proteins, such as Sip1/Zeb2.
Moreover, Xenopus E-cadherin is subjected to transcriptional repression through the activity of Snail proteins in combination with other co-factors. Ajuba LIM proteins are present at adherens junctions but also interact with Snail proteins to mediate repression of E-cadherin (albeit in vitro) (Langer et al., 2008). Importantly, Xenopus orthologs for Ajuba LIM (XLIMD1 and XWTIP) are expressed in a similar pattern to Xenopus Slug (Snail2) and Snail1 in premigratory and migratory neural crest cells. Furthermore, both proteins potentiate Snail repressive activity in vivo and, as such, impact neural crest development. In addition, the Polycomb Repressor Complex 2 (PRC2), which methylates histone H3 at lysine 27, influences Xenopus neural crest development through an interaction with Snail2 (Tien et al., 2015). PRC2 consists of the protein subunits EED, EZH2, and SUZ12, and EZH2 directly interacts with Snail2 in vivo. This binding is required for the ability of Snail2 to expand the neural crest domain. Notably, tri-methylation at lysine 27 on histone H3 is typically associated with transcriptionally inactive chromatin. To explore this further, the authors demonstrated that knockdown of either Snail2 or EZH2 leads to increased E-cadherin expression; this effect was in fact more robust for EZH2, indicating that it may also utilize other factors, such as Twist ((Barriga et al., 2013); see preceding paragraph), to regulate E-cadherin expression. In addition, Snail2 chromatin immunoprecipitation (ChIP) experiments revealed Snail2 association to two E-boxes, putative Snail2 binding sites, during neural crest EMT. Additional ChIP assays demonstrated that Snail2 recruits EZH2/PRC2 to chromatin regions surrounding the E-cadherin transcriptional start site, leading to tri-methylation of lysine 27 on histone H3 and transcriptional repression.
Chick Cadherin-6B is also a well-characterized cadherin in terms of its transcriptional regulation. Wnt and BMP signaling affect Cadherin-6B transcript levels, with the presence of ectopic Noggin, and, in turn, inhibition of BMP signaling, causing a reduction in Cadherin-6B transcripts in premigratory neural crest cells in the head and trunk (Sela-Donenfeld and Kalcheim, 1999). In addition, Wnt and BMP activate Cadherin-6B expression in trunk intermediate neural plate explants ex vivo (Liu and Jessell, 1998; Taneyhill and Bronner-Fraser, 2005). Myc interacting zinc finger protein-1 (Miz1) also influences Cadherin-6B expression (Kerosuo and Bronner, 2014), as Miz1 is observed in the neural tube and migratory cranial neural crest cells. Depletion of Miz1 coupled with QPCR revealed a decrease in Cadherin-6B transcripts but no change was noted for E-cadherin and N-cadherin, although Miz1 knockdown in the neural tube led to an increase or decrease in E-cadherin or N-cadherin protein, respectively.
Like Xenopus E-cadherin, Snail2 represses Cadherin-6B expression in the chick midbrain and trunk (Taneyhill et al., 2007). Depletion of Snail2 in premigratory neural crest cells leads to an upregulation of Cadherin-6B transcripts as early as 30 minutes postintroduction of a Snail2 MO. Analysis of the sequence up- and downstream of the Cadherin-6B translational start site revealed the presence of six E-boxes. Through a series of biochemical experiments, the authors revealed that Snail2 associates and binds with these E-boxes in order to repress Cadherin-6B transcription. The molecular mechanism underlying this interaction was further elaborated in a later study, which showed that Snail2 interacts with a complex of proteins that ultimately leads to Cadherin-6B repression during cranial neural crest EMT (Strobl-Mazzulla & Bronner, 2012). Depletion of either Snail2 or the adapter protein PHD12 inhibits EMT and results in loss of Cadherin-6B promoter de-acetylation. Both Snail2 and PHD12 interact directly with Sin3A (but not with each other), and Sin3A in turn recruits histone deacetylase (HDAC). The authors propose a model in which binding of Snail2 to E-boxes in the Cadherin-6B promoter, coupled with PHD12 binding to the Cadherin-6B transcriptional start site and the formation of a Sin3A/HDAC-repressive complex, results in the de-acetylation of the Cadherin-6B promoter and the repression of Cadherin-6B transcription during cranial neural crest EMT. A recent study has now shown that Snail2-mediated repression of Cadherin-6B during neural crest cell EMT is in part regulated through proteolysis of Cadherin-6B itself (Schiffmacher, Xie, & Taneyhill, 2016). The intracellular Cadherin-6B C-terminal fragment (CTF2) that arises after Cadherin-6B proteolysis associates with the endogenous Snail2 promoter in cranial neural crest cells and upregulates the expression of Snail2 and other EMT effector genes (see below for additional details).
REGULATION OF CADHERIN LEVELS THROUGH POSTTRANSLATIONAL MECHANISMS
Neural crest cells use post-translational mechanisms to modulate cadherin levels and accomplish delamination and EMT. In this section, we will examine mechanisms by which cadherin levels are reduced, including proteolysis (and its regulation), internalization/localization, and mechanical force.
1. Proteolysis and its regulation
While the expression patterns of a variety of A Distintegrin and Metalloproteinases (ADAMs) and Matrix Metalloproteases (MMPs) have been known for almost twenty years, the functional role of these proteases in controlling neural crest cell delamination, EMT, and migration through reduction in cadherin levels has only recently been explored. These proteases, and their cadherin targets, are discussed below.
Metalloproteinases
A role for ADAM10 in the neural crest is not surprising given its early embryonic lethality in the mouse and effects on neural tube integrity and morphogenesis (Hartmann et al., 2002). The expression of ADAM10 in late stage chick embryos in the trunk (>HH13) is noted along the basal lamina of the neural tube and ectoderm as well as in cultured neural crest cells in vitro (Hall & Erickson, 2003). ADAM10 possesses a similar distribution in the chick head and is also observed in premigratory cranial neural crest cells (Schiffmacher et al., 2014). Knockdown or overexpression of ADAM10 (or ADAM19, discussed below) leads to the maintenance or premature loss of Cadherin-6B in premigratory cranial neural crest cells, respectively. Chemical inhibition of ADAM10 also impairs trunk neural crest cell delamination and EMT in neural crest cell explants ex vivo (Shoval et al., 2007). At least three ADAMs (ADAM9, ADAM13, and ADAM19) localize to Xenopus cranial neural crest cells and function during neural crest cell induction and migration (Alfandari et al., 2001, 2010; Alfandari, McCusker, & Cousin, 2009; Alfandari, Wolfsberg, White, & DeSimone, 1997; Cousin, Abbruzzese, Kerdavid, Gaultier, & Alfandari, 2011; Cousin et al., 2012; McCusker et al., 2009; Neuner, Cousin, McCusker, Coyne, & Alfandari, 2009), as shown through MO-mediated depletion assays and assessment of neural crest cell markers and migration patterns. ADAM19 is also expressed by chick premigratory and migratory cranial neural crest cells and functions in cadherin proteolysis during EMT (Schiffmacher et al., 2014; see below). Moreover, presenilin-1, the catalytic component of the γ-secretase complex that liberates the intracellular cadherin tail from the plasma membrane (after cleavage of the extracellular domain), localizes to chick premigratory and migratory cranial neural crest cells (Schiffmacher et al., 2014). Thus, neural crest cells possess the correct machinery to proteolytically process cadherins (and other substrates) (Table 2).
Table 2.
Confirmed metalloproteinases and their cadherin substrates within neural crest cells.
| Cadherin | Species | Metalloproteinase | Axial Level | References |
|---|---|---|---|---|
| N-cadherin | Chicken | ADAM10 | Trunk | Shoval et al., 2007 |
| MMP-9 | Cranial/trunk | Monsonego-Ornan et al., 2012 | ||
|
| ||||
| Cadherin-6 | Chicken | ADAM10 | Cranial | Schiffmacher et al., 2014 |
| ADAM19 | Cranial | Schiffmacher et al., 2014 | ||
|
| ||||
| Cadherin-11 | Xenopus | ADAM13 | Cranial | McCusker et al., 2009 |
| ADAM9 | Cranial | McCusker et al., 2009 | ||
This table documents metalloproteinases and their known cadherin substrates, to date, within the neural crest at both cranial and trunk axial levels.
MMP expression has been documented throughout neural crest development in a variety of species. An early study did not note MMP2 expression in chick premigratory or early migratory neural crest cells; however, it was observed in later migratory cranial neural crest cells within the paraxial and branchial arch mesenchyme (Cai, Vollberg, Hahn-Dantona, Quigley, & Brauer, 2000). Later, Duong and Erickson (2004) demonstrated MMP2 expression in a “patchy” pattern in the cranial neural tube and in early emigrating cranial and trunk neural crest cells detaching from the neural tube (Duong & Erickson, 2004). Once neural crest cells migrate a short distance from the neural tube, though, they are devoid of MMP2. Given this localization pattern, functional studies using an MMP2 inhibitor and MOs were carried out and revealed a requirement for MMP2 in neural crest cell EMT but not in motility. MMP8 is likely also expressed in the mouse in early migratory cranial neural crest cells, as well as in regions through which neural crest cells migrate, and in neural crest derivatives (Giambernardi et al., 2001). MMP9 is a secreted protease that localizes to both delaminating and trunk migratory neural crest cells in the chick head and trunk (Monsonego-Ornan et al., 2012), while Xenopus cranial neural crest cells express MMP-14 (XMT1-MMP) (Harrison et al., 2004; Tomlinson et al., 2009). MMP15/MT2-MMP, an activator of pro-MMP2, is expressed in the neural folds of the very anterior chick head and open neural plate region in the trunk, while its protein product is observed along the basolateral surface/basement membrane of the neural tube (Patterson, Cavanaugh, Cantemir, Brauer, & Reedy, 2013). In zebrafish, the GPI-linked MMP17b is expressed in crestin-positive cranial neural crest cells, and knockdown of MMP17b function through MOs or chemical inhibitors leads to aberrant migration and patterning of crestin-positive trunk neural crest cells (Leigh et al., 2013).
Cadherin substrates of metalloproteinases
During neural crest cell delamination and EMT, cadherin levels are reduced through the activity of ADAMs and MMPs. To date, experimental evidence exists for not only the proteolysis of N-cadherin, Cadherin-6B, and Cadherin-11 during these processes, but also the acquisition of novel and independent functions for their respective cadherin cleavage products. These studies are highlighted in this section.
N-cadherin
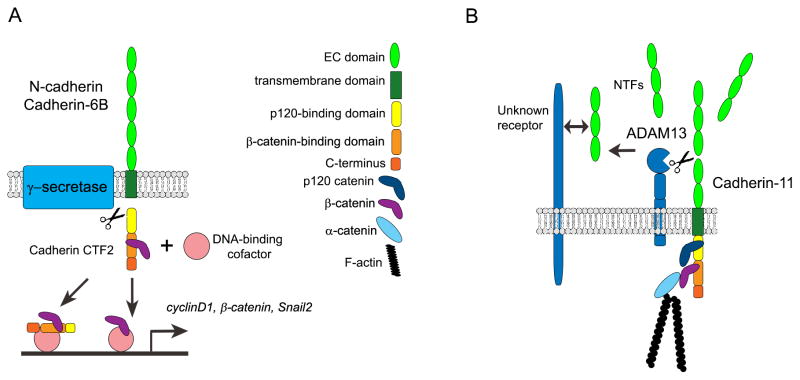
An initial role for proteolysis of N-cadherin was first suggested almost ten years ago, when Shoval et al. (2007) revealed a function for ADAM10 in mediating cleavage of N-cadherin to promote quail trunk neural crest cell EMT (Shoval et al., 2007). While the BMP4 inhibitor Noggin plays a key role in regulating the onset of delamination and EMT in the chick trunk (Sela-Donenfeld & Kalcheim, 1999, 2000, 2002), the molecular basis of this was not known. To this end, Shoval et al. (2007) showed that downregulation of noggin transcripts, and the subsequent onset of BMP4 activity, allows for the activation of ADAM10, which functions to process and deplete N-cadherin protein in the premigratory neural crest (Shoval et al., 2007). Using an ex vivo explant culture system, the authors observed retention of membrane N-cadherin in the presence of a chemical inhibitor of ADAM10, while control-treated explants were immunoreactive to antibodies raised to the N-cadherin extracellular and intracellular domains but possessed little to no membrane Ncadherin. While overexpression of N-cadherin CTF2 led to only a modest increase in the number of delaminating neural crest cells, CyclinD1 and β-catenin transcripts were increased in electroporated neural tubes as shown by in situ hybridization (Figure 2a). N-cadherin CTF2 was then identified in the nuclei of trunk neural crest cell explants, further suggesting a role in transcription. Moreover, N-cadherin CTF2 overexpression rescues the loss of delamination and EMT observed in the presence of the ADAM10 inhibitor by reducing membrane-bound N-cadherin. Although these studies did not establish a direct transcriptional role for N-cadherin CTF2 due to the long incubation time postelectroporation and the use of a nonquantitative readout for gene expression, the results are consistent with a role for N-cadherin CTF2 in gene regulation, as has been recently demonstrated for Cadherin-6B CTF2 (see below). Moreover, N-cadherin CTF2 overexpression rescues the loss of delamination and EMT observed in the presence of the ADAM10 inhibitor by reducing membrane-bound N-cadherin.
Figure 2. Proteolysis of full-length cadherins generates functional peptides that promote neural crest development.
(A) The intracellular C-termini of Cadherin-6B (in chick premigratory cranial neural crest) and N-cadherin (in chick premigratory trunk neural crest) are subjected to γ-secretase-mediated proteolysis during EMT. Newly generated soluble C-terminal fragments, or CTF2s, remain complexed with β-catenin, co-import into the nucleus, and transcriptionally regulate β-catenin/EMT effector genes including CyclinD1, β-catenin, and Snail2 (Cadherin-6B) (Schiffmacher et al., 2016; Shoval et al., 2007). (B) The ectodomain, or N-terminal fragment (NTF), of Cadherin-11 in Xenopus migratory neural crest cells is cleaved by ADAM13 and shed into the extracellular matrix, where it possibly promotes migration by binding unknown receptors (Abbruzzese et al., 2016). Double arrows indicate physical interaction.
Proteolytic processing of N-cadherin in the chick head and trunk also occurs by MMP9 and promotes neural crest cell delamination and migration (Monsonego-Ornan et al., 2012). Using a chemical inhibitor, the authors revealed that cultured cranial and trunk premigratory neural crest cells exhibit a decrease in delamination and migration and thus an overall reduction in the area occupied by migratory neural crest cells. This experiment was also performed in vivo through injection of the inhibitor into the neural tube, which corroborated the ex vivo data and also showed no effects on neural crest cell specification. As an additional means to test the involvement of MMP9 in neural crest cell EMT, an MMP9 MO was introduced unilaterally into the premigratory neural crest cell population, which led to a reduction in the number of migratory neural crest cells without affecting cell death or proliferation. Conversely, neural crest cell migration was augmented in ex vivo explants treated with MMP9 using a variety of methods. Interestingly, earlier N-cadherin loss is observed in these explants, and this was further substantiated by additional ex vivo experiments, pointing to a role for MMP9 in N-cadherin proteolysis. In embryos, however, similar assays revealed that MMP9 is not the primary protease for processing N-cadherin in the premigratory neural crest cell population/dorsal neural tube, as N-cadherin loss was still evident here in the presence of the MMP9 inhibitor. This is perhaps not surprising given the role of ADAM10, as outlined above. MMP9 does act on laminin, though, and this loss of laminin is necessary for neural crest cells to emerge from the neural tube. These results collectively suggest that MMP9 is important for the detachment and migration of neural crest cells away from the neural tube.
Cadherin-6B
In chick premigratory cranial neural crest cells, at least two ADAMs, ADAM10 and 19, and γ-secretase, proteolytically processs Cadherin-6B as these cells undergo EMT (Schiffmacher et al., 2014). Evidence for a posttranslational mechanism to reduce membrane Cadherin-6B levels during EMT was hypothesized due to the rapid loss of Cadherin-6B protein during EMT (over a 90-minute window) compared to the half-life Cadherin-6B possesses (~5.5 hours), which is similar to that observed for other cadherins. Following the onset of EMT, full-length Cadherin-6B protein decreases in cranial neural crest cells, with a concomitant increase in the Cadherin-6B shed extracellular domain NTF, as well as C-terminal fragments (CTF1 and CTF2). Importantly, the appearance of CTF1/2 is abrogated upon incubation of embryos with a broad-spectrum protease inhibitor of ADAMs and MMPs, demonstrating that intracellular processing by γ-secretase depends upon prior cleavage of the Cadherin-6B extracellular domain.
To ascertain the protease(s) responsible for cleaving Cadherin-6B, an in vitro protease assay was performed, which identified both ADAM10 and ADAM19 as putative candidate proteases for Cadherin-6B processing. These results were confirmed by demonstrating that “protease-dead” ADAM10 and ADAM19 could not process Cadherin-6B in vitro. Interestingly, neither ADAMs 12, 33, or 17 were shown to cleave Cadherin-6B in vitro, suggesting some inherent specificity in the ability of proteases to process cadherin substrates. After revealing the presence of these proteases and γ-secretase in premigratory and migratory cranial neural crest cells, functional perturbation experiments were then carried out. MO-mediated depletion of ADAM10 or ADAM19, either singly or in combination, leads to the retention of Cadherin-6B in premigratory cranial neural crest cells. Conversely, protease overexpression results in precocious loss of Cadherin-6B. Intriguingly, overexpression of the intracellular cleavage product of Cadherin-6B, CTF2, prematurely reduces membrane Cadherin-6B levels. This data support the notion of a potential independent, unique function for this Cadherin-6B cleavage product, specifically with respect to promoting EMT (Schiffmacher et al., 2014).
The molecular mechanism behind this pro-EMT function for Cadherin-6B has recently been elucidated (Schiffmacher et al., 2016). Building on their prior findings, Schiffmacher et al. (2016) discovered that, like Cadherin-6B NTFs, Cadherin-6B CTFs are generated both prior to and during EMT. As cadherin CTF2s in cultured cells have been shown to interact with catenin proteins (Ferber et al., 2008; Marambaud et al., 2002; Sadot, Simcha, Shtutman, Ben-Ze’ev, & Geiger, 1998; Uemura et al., 2006), this possibility was also tested for Cadherin-6B CTF2. Interestingly, CTF2 was shown to directly interact with β-catenin but not p120-catenin, both in vitro and in vivo in cranial neural crest cells, suggesting a function for this Cadherin-6B CTF2/ β-catenin complex in vivo. To this end, the authors revealed that the formation of a Cadherin-6B CTF2/β-catenin complex was mutually beneficial to both proteins, and dependent upon the β-catenin binding site in CTF2, leading to their respective stabilization. Furthermore, β-catenin redistributes and accumulates in the nuclei of cranial neural crest cells expressing wild-type CTF2 but not a mutant version of CTF2 deficient in β-catenin binding (MUT9). Importantly, overexpression of wild-type or MUT9 CTF2 does not change membrane β-catenin localization, suggesting that these exogenous CTF2s are not competing with endogenous cadherin CTF2s.
The nuclear co-localization and interaction of both Cadherin-6B CTF2 and β-catenin was demonstrated in vitro and ex vivo in cranial neural crest cell explant cultures. In these latter experiments, wildtype or MUT9 CTF2 was introduced into premigratory cranial neural crest cells, and quantification of β-catenin levels revealed an increase in nuclear β-catenin expression in the presence of wildtype CTF2 versus MUT9, with MUT9 and unelectroporated neural crest cells possessing low/baseline levels of nuclear β-catenin. To attribute functionality to this nuclear-localized Cadherin-6B CTF2/β-catenin complex, the authors expressed wildtype CTF2 or MUT9 for only 5 hours followed by QPCR for known EMT effector genes. Notably, CyclinD1, β-catenin, and Snail2, which encodes a transcriptional repressor of Cadherin-6B (Taneyhill et al., 2007), were all upregulated after this short incubation time in the presence of wildtype Cadherin-6B CTF2 but not MUT9, demonstrating a dependence upon an intact β-catenin binding domain in CTF2 (Figure 2A). The increase in Snail2 expression was particularly intriguing and suggested that Cadherin-6B proteolysis not only reduces cell-cell adhesion during EMT but also liberates a cleavage product that can provide input into the neural crest gene regulatory network. To further define a transcriptional role for Cadherin-6B CTF2, a Snail2-GFP reporter (Sakai et al., 2005) was co-electroporated with either wildtype or MUT9 CTF2, followed by QPCR for GFP. Consistent with the preceding QPCR results, wildtype CTF2, but not MUT9, gave a statistically significant increase in GFP transcripts. Taken together, these results reveal that Cadherin-6B CTF2 can feedback directly on Cadherin-6B transcriptional repression during cranial neural crest cell EMT.
Although cadherin CTF2 molecules have been noted in the nucleus, a direct association with chromatin had not been demonstrated. To determine whether Cadherin-6B CTF2 interacts with chromatin, a wildtype CTF2 (or MUT9) ChIP-QPCR experiment was performed in order to amplify chromatin within the endogenous Snail2 promoter that was shown to be responsive to CTF2 in the prior experiment. Two regions of chromatin within this Snail2 promoter showed association of wildtype CTF2, while MUT9 was not enriched on any chromatin region tested. These results now reveal that the presence of the β-catenin binding domain in Cadherin-6B CTF2 is critical for mediating its interaction at these specific chromatin regions.
Altogether, these data suggest that continual proteolysis of Cadherin-6B, both prior to and during EMT, is key to allow sufficient levels of a Cadherin-6B CTF2/β-catenin complex to accumulate, translocate to the nucleus, and activate genes important for cranial neural crest EMT (Schiffmacher et al., 2016). Arguably one important function for CTF2 is to protect β-catenin from degradation, allowing it to accumulate in the cytosol, followed by co-import of both proteins to the nucleus, wherein β-catenin can then modulate transcription through interactions with TCF/LEF or other proteins. These results also demonstrate, however, that CTF2/β-catenin complexes are likely interacting with other factors, such as DNA binding proteins, to mediate transactivation of target genes, which thus ascribe a scaffolding function (or other roles outside of β-catenin binding) to CTF2. Therefore, Cadherin-6B CTF2 may be controlling transcription through both indirect and direct mechanisms. While demonstrated only for Snail2, this mode of transcriptional regulation may be universally translatable to other EMT effector genes, both in the neural crest and in the context of other developmental- and disease-related EMTs.
Cadherin-11
In Xenopus, Cadherin-11 serves as a substrate for cleavage by ADAM13, which is expressed in the premigratory and migratory cranial neural crest (Alfandari et al., 1997). Overexpression of wildtype ADAM13 does not affect neural crest cell specification but does perturb the ability of neural crest cells to migrate, with cells remaining near the neural tube and failing to segregate into the three cranial streams (hyoid, mandibular, branchial) (Alfandari et al., 2001). This phenotype was later attributed to the ectopic expression of ADAM13 in other tissues, such as the ectoderm, that cranial neural crest cells encounter, as the grafting of cranial neural crest cells expressing wildtype ADAM13 into a wildtype embryo did not affect migration. Interestingly, ADAM13 protease activity is also dispensable for specification but is required for the proper migration of neural crest cells along the hyoid and branchial pathways. In support of this, grafting of ADAM13 protease-dead cranial neural crest cells into wildtype embryos, impedes, and often abolishes, the ability of these grafted cells to migrate. Migrating neural crest cells were at times observed, however, in the mandibular stream. This protease-dead ADAM13 mutant was later shown to function as a dominant negative, inhibiting endogenous ADAM13 protease activity. Additional studies with animal cap explants revealed that cells expressing wildtype, but not the protease-dead version of ADAM13, exhibit decreased adhesion with the ECM, suggesting a role for ADAM13 in ECM remodeling. In keeping with this, cultured cells expressing ADAM13 can remodel fibronectin, which depends upon the proteolytic function of ADAM13, and fibronectin is cleaved by purified ADAM13 in vitro.
A later study revealed that one means by which ADAM13 regulates neural crest cell migration is through proteolytic processing of Xenopus Cadherin-11 (McCusker et al., 2009). While full-length Cadherin-11 protein increases at stages when neural crest cells are migrating, so does a proteolytic product that corresponds to the cadherin transmembrane and cytoplasmic domains, likely generated through cleavage between EC domains 3 and 4 of Cadherin-11. Cell culture assays revealed Cadherin-11 processing by both ADAMs 9 and 13 (but not ADAM19), and further work in Xenopus was carried out to investigate the role of ADAM13. Cadherin-11 and ADAM13 biochemically interact in the embryo at stages when neural crest cells are migrating, correlating with Cadherin-11 cleavage. Importantly, overexpression of either ADAM9 or 13 (but not ADAM19) with Cadherin-11 rescues the migration defect observed upon overexpression of Cadherin-11 alone (McCusker et al., 2009), and this is dependent upon ADAM protease function. Consequently, Cadherin-11 cleavage and cranial neural crest migration is blocked in vivo upon MO-mediated depletion of ADAMs 9 and 13, as expected, but ADAM19 also showed similar results, revealing a possible indirect role that is only ascertained upon its knockdown in vivo.
The cleaved Cadherin-11 NTF (EC domains 1–3) was then examined to determine whether it might possess a unique role during migration. To this end, the Cadherin-11 NTF was first shown to bind to tissue culture cells expressing full-length Cadherin-11, and later to restore normal neural crest cell migration when co-expressed with Cadherin-11. These data suggested that the Cadherin-11 NTF may in fact compete with full-length Cadherin-11 molecules and therefore help reduce adhesion and promote migration. To further explore this role, the Cadherin-11 NTF was expressed in embryos that had been depleted for ADAM13 and ADAM19 earlier in development, which led to a rescue of the migration defect observed in these ADAM-depleted embryos. Intriguingly, rescue of this migration phenotype was only achieved in the presence of the ADAM13 MO (in combination with ADAM9 and 19 MOs) and not when MOs to other ADAMs were used in combination (e.g., MOs to ADAM9 and 19), implying that the effect is specific to ADAM13. Interestingly, introduction of MOs to ADAM9 and ADAM19 also impedes neural crest cell migration, and embryos possessing these MOs show reduced levels of ADAM13, providing a possible explanation for this observation. Alternatively, these ADAMs may affect neural crest cell specification, as seen in the heart (Komatsu et al., 2007), therefore impacting later events like migration. Moreover, knockdown of ADAM13 or ADAM19 leads to an increase in ADAM9, demonstrating that there could be some degree of compensation in the Xenopus neural crest. Finally, the Cadherin-11 NTF plays a non-cell autonomous role in promoting neural crest migration in cells lacking ADAM9, ADAM13, and ADAM19, and does not affect interactions with β-catenin nor modulate the expression of canonical Wnt target genes. Collectively, these results imply a promigratory role for the Cadherin-11 NTF in the Xenopus cranial neural crest, either through interactions with other full-length Cadherin-11 molecules to decrease cell–cell adhesion and/or by serving as a “ligand” for receptor-mediated signaling, as noted with other adhesion molecules, including cadherins (Lyon, Johnson, Williams, Sala-Newby, & George, 2009; Mechtersheimer et al., 2001; Najy, Day, & Day, 2008; Paradies & Grunwald, 1993; Utton, Eickholt, Howell, Wallis, & Doherty, 2001).
While Cadherin-11 has been shown to play a role in regulating neural crest migration (Borchers et al., 2001), a more recent study revealed that it also functions during CIL, which underlies Xenopus neural crest cell migration (Becker et al., 2013). Defects in CIL are noted in cranial neural crest cells lacking Cadherin-11 or expressing a Cadherin-11 mutant that lacks the extracellular (EC) domain and thus unable to mediate homophilic binding (Becker et al., 2013; Kashef et al., 2009). To define this pro-migratory function for the Cadherin-11 NTF in the context of CIL, Abbruzzese, Becker, Kashef, and Alfandari (2016) showed that Cadherin-11 cleavage is required for neural crest cell migration (Abbruzzese et al., 2016). To this end, a noncleavable form of full-length Cadherin-11 was generated, and it does not rescue the migration defects observed upon loss of endogenous Cadherin-11. Enhanced neural crest cell invasiveness (or inhibition of CIL) was noted in explants when neural crest cells overexpress ADAM13 or the Cadherin-11 NTF. After mutating the homophilic binding site on the Cadherin-11 NTF and expressing this version in neural crest cells, however, no change in invasiveness/CIL was noted, implying that the homophilic binding site on the Cadherin-11 NTF is correlated with this loss of CIL. As such, ADAM13 likely reduces CIL through processing of plasma membrane Cadherin-11 and production of the Cadherin-11 NTF.
To further address the role of the homophilic binding site in the Cadherin-11 NTF, deletion mutants of the NTF were generated to include just EC1 or EC1–2. EC1 was sufficient to rescue migration deficits due to full-length Cadherin-11 overexpression, and EC1–2 gave similar results to the entire Cadherin-11 NTF (EC1–3). The Cadherin-11 NTF homophilic binding mutant was also able to rescue migration in the same assay. When the non-cleavable, full-length Cadherin-11 mutant was introduced into embryos depleted for wildtype Cadherin-11, followed by injections of various Cadherin-11 NTF constructs, normal neural crest cell migration was observed in the presence of the wildtype Cadherin-11 NTF and the NTF homophilic binding mutant, demonstrating that this binding site is not required once Cadherin-11 is cleaved. Conversely, the same experiment utilizing the introduction of a full-length, homophilic binding mutant Cadherin-11 into embryos depleted for endogenous Cadherin-11 does not rescue migration, irrespective of later introduction of the Cadherin-11 NTF. Altogether, these results demonstrate that the presence of the homophilic binding site in the full-length, membrane-bound Cadherin-11 molecule is important for neural crest cell migration in vivo prior to cleavage by ADAM13. After processing, however, the Cadherin-11 NTF does not require homophilic binding for its biological function, providing evidence that the putative target for this NTF is not likely a full-length Cadherin-11 molecule but perhaps another protein/receptor (Figure 2B). Moreover, the decrease in CIL observed in the presence of ADAM13 (and likely due to ADAM13-mediated cleavage of Cadherin-11 and production of the Cadherin-11 NTF) indicates that a balance must be struck between proteolytic processing of Cadherin-11 to permit CIL to occur normally and some degree of cell-cell adhesion between migrating neural crest cells.
As discussed in Section 2, within Xenopus migratory cranial neural crest cells, Cadherin-11 is observed in cell protrusions, such that loss of Cadherin-11 leads to the absence of filopodia and lamellipodia and instead causes membrane blebbing (Kashef et al., 2009). This change in protrusion type in turn is associated with an “amoeboid-like” cell shape and subsequent mechanism of movement. Intriguingly, these morphological and migrating defects can be rescued upon introduction of a Cadherin-11 mutant lacking the extracellular domain but possessing the transmembrane and cytoplasmic domains (in theory a Cadherin-11 CTF1), while a mutant without the cytoplasmic tail (the NTF tethered to the plasma membrane) cannot rescue migration. This rescue could be attributed to the presence of the β-catenin, but not the p120-catenin, binding domain of Cadherin-11, with loss of the β-catenin binding domain yielding cells without protrusions and blebbing membranes. In addition, a mutant encoding the intracellular Cadherin-11 CTF2 did not rescue the migration defects, suggesting that the transmembrane domain also plays a critical role.
2. Internalization and localization
Neural crest cells employ other post-translational mechanisms to reduce cadherin levels, including internalizing cadherins and/or whole adherens junctions, controlling the localization/trafficking and stability of cadherins, and generating force to physically initiate delamination and EMT. These processes are discussed in detail here.
Internalization
Cadherin-6B
The internalization of intact cadherins (and/or potentially their proteolytic products) through mechanisms such as endocytosis and macropinocytosis has been demonstrated in cultured cells as a means to modulate cadherin levels (Bryant et al., 2007; Kowalczyk & Nanes, 2012; Paterson, Parton, Ferguson, Stow, & Yap, 2003; Sharma & Henderson, 2007; Solis et al., 2012; Ulrich & Heisenberg, 2009). In vivo evidence for this process during EMT, however, was lacking, until recent experiments revealed the internalization of membrane Cadherin-6B during chick cranial neural crest cell EMT (Padmanabhan & Taneyhill, 2015). Using Na1K1 ATPase as a proxy for early endosomes, this group demonstrated the presence of Cadherin-6B-positive cytoplasmic, endocytic puncta in a stable cell line expressing Cadherin-6B and in neural crest cells undergoing EMT and emigrating, both in in vivo and ex vivo explants. Further examination of the Cadherin-6B cytoplasmic tail revealed the presence of two conserved motifs that could potentially influence Cadherin-6B internalization based upon their roles in other cadherins: a dileucine (LI in Cadherin-6B) motif that positively regulates endocytosis, and a p120-catenin binding motif (EED), which serves to mask additional binding sites for the endocytic machinery when p120-catenin is bound, thereby precluding endocytosis. Stable cells lines expressing mutants of these sites revealed their biochemical functionality, with mutation of the LI motif having no effect on Cadherin-6B internalization, whereas the EED mutant showed enhanced Cadherin-6B cytosolic distribution, as predicted. In keeping with these findings, the EED mutant was more rapidly internalized, possessed a shorter half-life, and was not anchored to the actin cytoskeleton when compared to wild-type Cadherin-6B or the LI mutant. Taken together, these data establish the EED motif as a bona fide endocytic motif that regulates Cadherin-6B endocytosis. This endocytosis also occurs through a predominantly clathrin-mediated process due to co-localization of Cadherin-6B and clathrin in premigratory cranial neural crest cells undergoing EMT in vivo.
Although Cadherin-6B-positive cytosolic puncta were often positive for clathrin, in some instances these puncta were negative for clathrin but instead immunoreactive for p120-catenin. Because endocytosis relies upon the removal of p120-catenin to unmask binding sites for the endocytic machinery, these data suggested that perhaps internalization of whole adherens junctions was occurring through macropinocytosis. To address the involvement of clathrin-mediated endocytosis and macropinocytosis, pharmacological inhibitors of each were utilized in ex vivo neural crest explant cultures. Through these assays, the authors demonstrated that the appearance of internalized puncta was severely abrogated, with Cadherin-6B instead accumulating on the membrane or in large, immature macropinosomes. Consequently, inhibitor treatment also led to a loss of neural crest cell EMT. Moreover, both of these internalization processes are dynamin-dependent, as macropinosomes and membrane-bound vesicles accumulate in explanted neural crest cells treated with a dynamin-inhibitor, along with a concomitant reduction in EMT and migration (Padmanabhan and Taneyhill, 2015).
Localization, trafficking, and stability
N-cadherin
Several studies in zebrafish have examined the importance of cadherin localization and trafficking during neural crest cell migration. The Wnt target and transcription factor Ovo1 influences neural crest cell migration by controlling N-cadherin intracellular trafficking (Piloto & Schilling, 2010). Ovo1 is expressed by premigratory and a subset of migratory neural crest cells in zebrafish. MO-mediated knockdown of Ovo1 leads to the accumulation of neural crest cells dorsally that are specific to the pigment lineage. Interestingly, the phenotype of Ovo1 morphants is similar to N-cadherin-deficient mutants. A reduction in N-cadherin transcripts was noted in Ovo1-depleted embryos, but strikingly these embryos exhibit N-cadherin-positive puncta in enveloping layer cells. Microarray analysis carried out on Ovo1 morphants revealed the upregulation of genes encoding proteins involved in vesicular transport, including members of the Rab family of GTPases (rab3c, rab12, rab11-fip2, and sec6), many of which are expressed in premigratory neural crest cells. Given the presence of this trafficking gene network, the authors showed that the N-cadherin localization defects in Ovo1 morphants could be rescued, to some extent, by treating embryos with a broad-spectrum inhibitor of endo- and exocytoses. In keeping with this finding, overexpression of Rab11fip2 also leads to the presence of neural crest cell aggregates along the midline. These effects are cell autonomous and are exacerbated when Rab11fip2 is introduced into Ovo1 MO-treated embryos. Finally, a genetic interaction between Rab11fip2 and N-cadherin was also noted. Taken together, these data indicate that Ovo1 likely regulates transcription of genes involved in N-cadherin trafficking to and from neural crest cell membranes to permit migration.
The significance of proper N-cadherin localization during zebrafish neural crest migration was further underscored by data revealing a functional role for Rabconnectin-3a (Rbc3a), a protein that associates with subunits of the vacuolar-ATPase (v-ATPase) proton pump that regulates lysosomal acidification, in the neural crest (Tuttle, Hoffman, & Schilling, 2014). MO-mediated knockdown of Rbc3a causes neural crest cells to aggregate at the dorsal midline, with cells possessing a reduced velocity and directionality. Moreover, only markers of the pigment cell lineage were present, revealing a loss of neural crest cell populations that will differentiate into glial or skeletal cells. Rbc3a is also required cell autonomously in the neural crest based upon results from grafting experiments. Assays were then conducted to evaluate vesicle formation, acidification, and trafficking in Rbc3a-depleted embryos. Rbc3a MO-treated embryos accumulate early, acidified endosomes of increased size and, consequently, show a decrease in the number and size of late endosomes/lysosomes; however, many of these latter vesicles are not acidified. Rbc3a and ATP6V0A1, a v-ATPase subunit isoform, were later shown to act together in regulating endocytosis during neural crest cell migration, as knockdown of the latter by MOs phenocopies the Rbc3a MO-treated embryos. Depletion of Rbc3a also impacts Wnt target genes, with reduced expression of genes that normally promote EMT in premigratory neural crest cells. This expression is later regained in these depleted embryos after neural crest cell migration has commenced. This finding appears to correlate with β-catenin distribution (e.g., remaining on the membrane during EMT versus nuclear after migration). Loss of Rbc3a leads to enhancedWnt signaling due to the inability to efficiently remove the Wnt receptor Frizzled 7 from the neural crest cell plasma membrane. Intriguingly, Rbc3a depletion causes a decrease in N-cadherin transcripts and precocious increase in Cadherin-11 transcripts, along with loss of membrane N-cadherin protein. These results suggest an important role for Rbc3a in controlling signaling pathways that impinge upon general cadherin transcription and N-cadherin membrane localization. This may occur through the inability for Rbc3a-depleted neural crest cells to assemble signalosomes and promote Wnt signaling. Finally, the LPA receptor LPAR2 is required cell autonomously in Xenopus neural crest and functions to control neural crest migration by regulating the endocytosis of N-cadherin (Kuriyama et al., 2014).
Cell-adhesion molecule-related/downstream by oncogenes (Cdon) also regulates N-cadherin levels in the zebrafish neural crest (Powell et al., 2015). Cdon is expressed in premigratory neural crest cells and at the beginning of migration in both the head and trunk but is absent from fully migratory crestin-positive neural crest cells. MO-mediated knockdown of Cdon reveals no change in neural crest cell specification; however, trunk, but not cranial, neural crest cell migration is abrogated, with cells remaining dorsally, forming clusters, and not migrating as far ventrally as wild-type neural crest cells. Although Cdon-depleted neural crest cells exhibit an increase in velocity, they form multiple, unstable protrusions, thereby altering their ability to migrate directionally (ventrally). Cdon is required cell autonomously in migratory trunk neural crest cells, and its knockdown leads to mislocalization of N-cadherin to the cytosol of migratory neural crest cells, and specifically in leading edge membrane protrusions that mediate migration. As such, Cdon likely functions to maintain N-cadherin membrane expression to facilitate trunk neural crest cell migration. This hypothesis will require testing in future experiments, as only Cdon transcript levels were examined and Cdon is not observed in migratory neural crest cells. Therefore, Cdon protein must be maintained on the surface of neural crest cells to perform this function.
Cadherin-6B
Tetraspanin18 (Tspan18) was recently shown to stabilize Cadherin-6B levels in cranial neural crest cells, with loss of Tspan18 mediated by FoxD3 and required for neural crest cells to undergo EMT and migrate (Fairchild and Gammill, 2013). Tspan18 expression is unique to premigratory cranial neural crest cells, and MO-mediated knockdown of Tspan18 leads to loss of Cadherin-6B protein. Interestingly, Tspan18 depletion affects Cadherin-6B transcripts in the forebrain, a region from which neural crest cells do not arise. The reduction in Cadherin-6B protein in the midbrain, which possesses premigratory cranial neural crest cells, however, cannot be explained solely by a decrease in Cadherin-6B transcription (the noted reduction there was not statistically significant). Moreover, Tspan18 knockdown reduces Snail2 but augments nuclear β-catenin protein levels, as observed in vitro (Huber et al., 1996; Jamora et al., 2003), with the increase in the latter possibly mediating Cadherin-6B repression. Tspan18 depletion does not enhance neural crest cell migration, as observed for Cadherin-6B knockdown (Coles et al., 2007), and does not alter distribution of laminin or acquisition of Cadherin-7 expression, suggesting that it specifically impacts Cadherin-6B rather than delamination and mesenchymalization. Overexpression of Tspan18 impedes neural crest cell migration and results in maintenance of Cadherin-6B protein, even though Cadherin-6B transcripts are normally downregulated. These results imply that Tspan18 acts on Cadherin-6B post-translationally to control its levels during EMT. Additional experiments revealed that FoxD3 represses Tspan18 transcription during neural crest cell EMT. Collectively, these results suggest that Tspan18 protects Cadherin-6B from initial processing and/or degradation until EMT, after which time Cadherin-6B levels are reduced.
Mechanical Force
Time-lapse confocal imaging of slices taken through the chick trunk (slice cultures) have revealed that, in most cases, neural crest cells retract their apical tails while undergoing EMT and emigrating out of the neural tube (Ahlstrom & Erickson, 2009). During this process, adherens junction components are downregulated. In some instances (~11%), however, the tail physically “ruptures” from the cell body to allow neural crest cells to emerge from the neural tube. This likely occurs by surmounting the force generated by adherens junctions that are still present and mediating adhesion between premigratory neural crest cells. These data indicate that dismantling of adherens junctions to allow for neural crest cell emigration can also occur mechanically by “ripping” the neural crest cell tail containing these components away from the rest of the cell body. In other experiments, the authors note that tail retraction occurs even in the presence of adherens junction components (~36% of examined cases). It is unclear whether some degree of adherens junction disassembly occurred in this latter instance, though, given that these cells were visualized using an α-catenin or actin reporter, and cadherin expression was not directly examined.
E. Summary and Open Questions
The collection of data embodied in this review demonstrates that within neural crest cells, cadherin function goes well beyond the role of promoting cell–cell adhesion. Recent studies, especially those investigating Cadherin-6B in chick premigratory cranial neural crest cells or Cadherin-11 in Xenopus migratory cranial neural crest cells, reveal that cadherins have distinct functions at the molecular, cellular, intercellular, and tissue levels. In order to ascertain all of the functions of a cadherin within a neural crest domain at any given time, specific regulatory mechanisms, protein domains, spatiotemporal expression, and subcellular distribution must be considered. At the molecular level, subtle differences in primary sequences of protein subdomains impart novel functions on one cadherin type that are different from other coexpressed cadherins. For example, in Xenopus migratory cranial neural crest cells, transmembrane and C-terminal domains of Cadherin-11 dictate that this cadherin, but not N- or C-cadherin, associate with Syndecan-4 at focal adhesions and thus become a positive regulator of cell–fibronectin adhesion (Langhe et al., 2016). The Cadherin-11 C-terminal domain is also important for filipodial and lamellipodial extension formation, but this function is restricted to the subcellular leading edge of the cell, where the C-terminal domain can interact with Trio GEFs (Kashef et al., 2009). On the extracellular side, the Cadherin-11 homophilic binding domain located within EC1–3 of the ectodomain are required for proper neural crest cell migration (McCusker et al., 2009). Meanwhile, Cadherin-11 proteins are being proteolytically processed by ADAM13, which not only modulates the available membrane pools of full-length Cadherin-11, but also is necessary for proper migration in vivo. ADAM13-mediated proteolysis generates shed Cadherin-11 NTFs, which regulate collective cell migration via an intact homophilic binding domain. In contrast, other subdomains of the Cadherin-11 NTF impart additional functions in regulating intercellular interactions by modulating CIL (Abbruzzese et al., 2016). Moreover, Cadherin-11 proteolysis directly regulates the availability of cytosolic β-catenin and thus modulates Wnt signal transduction sensitivity (Koehler et al., 2013).
Further investigation into the primary sequences of co-expressed cadherins will yield more information on species-specific cadherin functions, and may help explain why more than one cadherin is expressed in a neural crest cell. Unique posttranslational modifications may also contribute to differential function, dictate binding affinities with other protein interactors, or direct subcellular cadherin localization. Chimeric cadherin experiments and mutational analyses will continue to aid in identifying key amino acids that distinguish roles among coexpressed cadherins.
Aside from primary sequence, there is little information regarding how one cadherin type interacts or regulates other co-expressed cadherins. Are heterophilic interactions occurring, either between full-length cadherins or cadherin fragments? It remains to be determined whether other functions are shared outside the adherens junction. Do all cadherin CTF2s regulate gene expression in the same manner? Multiple cadherin CTF2s, including those generated from E-cadherin, N-cadherin, and Cadherin-6B, upregulate Wnt effector genes including β-catenin and CyclinD1 (Gottardi et al., 2001; Klinke et al., 2015; Maretzky et al., 2005; Reiss et al., 2005; Sadot et al., 1998; Schiffmacher et al., 2016; Shoval et al., 2007; Uemura et al., 2006). Do these CTF2s also regulate their own unique subset of downstream targets? These questions warrant the need for further investigation, especially since phenotypes from cadherin gain- and loss-of-function assays can no longer be ascribed to changes in only full-length protein levels. Indeed, interpretation of phenotypes from these types of assays should not rule out the roles of cadherin NTFs and CTFs.
As more focus is placed on identifying differential functions of cadherins in neural crest cells, research on cadherin function in other systems and tissues will also benefit from these new findings, and vice versa. For example, studies conducted on other migratory cell types, such as Drosophila border and muscle founder cells, zebrafish lateral line and primordial germ cells, mouse mesodermal cells during gastrulation, and invasive cancer cells can all potentially shed light on the impact of cadherin-mediated adhesion, and its regulation, in neural crest cells as they undergo EMT and becoming migratory. In turn, data from the neural crest field will provide insight into the molecules and pathways that control the EMT and migration of cell types in a variety of species and developmental- and disease-related contexts. An excellent example of this convergence has been a better understanding of the role of Snail proteins, and the cadherins they regulate, during the EMT underlying mouse gastrulation (Carver, Jiang, Lan, Oram, & Gridley, 2001), migratory neural crest cell formation in the chick (Taneyhill et al., 2007), and cancer cell invasiveness (Batlle et al., 2000). Undoubtedly, additional findings in different species and cell types will allow for the translation of results across these connected research fields, building a blueprint for EMT and migration that will, to some degree, likely be conserved.
Interestingly, some neurocristopathies, or disorders arising from abnormalities in neural crest cell formation (including EMT and migration), can lead to dysregulated cadherin expression. For instance, Mowat-Wilson and Hirschsprung’s disease arise due to defects in Sip1 (Van de Putte et al., 2003; Wakamatsu et al., 2001), which is known to impact the levels of E-cadherin. Furthermore, mutations in Snail2, a known regulator of multiple cadherins, have been identified in humans with Waardenburg syndrome type 2D (Sanchez-Martin et al., 2002). Finally, CHARGE syndrome can be recapitulated in Xenopus embryos upon MO-mediated depletion of chromodomain helicase DNA binding domain-7, as this leads to a decrease in EMT effector genes such as Snail2 and Twist (Bajpai et al., 2010). Perhaps not surprising, to our knowledge there are no neurocristopathies that arise strictly due to mutations in a specific cadherin. This is likely due to the importance of cadherin expression throughout early development, with cadherin loss leading to embryonic lethality. Moreover, neuroblastomas and melanomas, both neural crest cell-derived tumors, exhibit reduced levels of N-cadherin, thereby promoting invasion. Melanoma growth and invasion are also evident in tumors possessing MMP-1, 2, and 9 (reviewed in (Zhang et al., 2014)). As such, understanding how cadherins are regulated will provide critical input into many of these neurocristopathies and cancers, the symptoms of which could be attributed to aberrant cadherin expression. Ultimately, this accumulation of information from multiple species and cell types will lead to achievements in preventing, correcting, or curing neurocristopathies, cancers, and other pathologies.
Acknowledgments
The authors would like to thank Drs. Dominique Alfandari and Eric Theveneau for their feedback on this review. This work was supported by NIH R01DE024217 (LAT) and a Research Scholar Grant (RSG-15-023-01-CSM) from the American Cancer Society (LAT).
References
- Abbruzzese G, Becker SF, Kashef J, Alfandari D. ADAM13 cleavage of cadherin-11 promotes CNC migration independently of the homophilic binding site. Dev Biol. 2016;415:383–390. doi: 10.1016/j.ydbio.2015.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlstrom JD, Erickson CA. The neural crest epithelial-mesenchymal transition in 4D: a ;tail’ of multiple non-obligatory cellular mechanisms. Development. 2009;136:1801–1812. doi: 10.1242/dev.034785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akitaya T, Bronner-Fraser M. Expression of cell adhesion molecules during initiation and cessation of neural crest cell migration. Dev Dyn. 1992;194:12–20. doi: 10.1002/aja.1001940103. [DOI] [PubMed] [Google Scholar]
- Alfandari D, Cousin H, Gaultier A, Smith K, White JM, Darribere T, DeSimone DW. Xenopus ADAM 13 is a metalloprotease required for cranial neural crest-cell migration. Curr Biol. 2001;11:918–930. doi: 10.1016/s0960-9822(01)00263-9. [DOI] [PubMed] [Google Scholar]
- Alfandari D, Cousin H, Marsden M. Mechanism of Xenopus cranial neural crest cell migration. Cell adhesion & migration. 2010;4:553–560. doi: 10.4161/cam.4.4.12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfandari D, McCusker C, Cousin H. ADAM function in embryogenesis. Semin Cell Dev Biol. 2009;20:153–163. doi: 10.1016/j.semcdb.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfandari D, Wolfsberg TG, White JM, DeSimone DW. ADAM 13: a novel ADAM expressed in somitic mesoderm and neural crest cells during Xenopus laevis development. Dev Biol. 1997;182:314–330. doi: 10.1006/dbio.1996.8458. [DOI] [PubMed] [Google Scholar]
- Aybar MJ, Mayor R. Early induction of neural crest cells: lessons learned from frog, fish and chick. Curr Opin Genet Dev. 2002;12:452–458. doi: 10.1016/s0959-437x(02)00325-8. [DOI] [PubMed] [Google Scholar]
- Backer S, Hidalgo-Sanchez M, Offner N, Portales-Casamar E, Debant A, Fort P, Gauthier-Rouviere C, Bloch-Gallego E. Trio controls the mature organization of neuronal clusters in the hindbrain. J Neurosci. 2007;27:10323–10332. doi: 10.1523/JNEUROSCI.1102-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajpai R, Chen DA, Rada-Iglesias A, Zhang J, Xiong Y, Helms J, Chang CP, Zhao Y, Swigut T, Wysocka J. CHD7 cooperates with PBAF to control multipotent neural crest formation. Nature. 2010;463:958–962. doi: 10.1038/nature08733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barriga EH, Maxwell PH, Reyes AE, Mayor R. The hypoxia factor Hif-1alpha controls neural crest chemotaxis and epithelial to mesenchymal transition. The Journal of cell biology. 2013;201:759–776. doi: 10.1083/jcb.201212100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barriga EH, Mayor R. Embryonic cell-cell adhesion: a key player in collective neural crest migration. Curr Top Dev Biol. 2015;112:301–323. doi: 10.1016/bs.ctdb.2014.11.023. [DOI] [PubMed] [Google Scholar]
- Basch ML, Bronner-Fraser M, Garcia-Castro MI. Specification of the neural crest occurs during gastrulation and requires Pax7. Nature. 2006;441:218–222. doi: 10.1038/nature04684. [DOI] [PubMed] [Google Scholar]
- Basch ML, Garcia-Castro MI, Bronner-Fraser M. Molecular mechanisms of neural crest induction. Birth Defects Res C Embryo Today. 2004;72:109–123. doi: 10.1002/bdrc.20015. [DOI] [PubMed] [Google Scholar]
- Batlle E, Sancho E, Franci C, Dominguez D, Monfar M, Baulida J, Garcia De Herreros A. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol. 2000;2:84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- Becker SF, Mayor R, Kashef J. Cadherin-11 mediates contact inhibition of locomotion during Xenopus neural crest cell migration. PLoS ONE. 2013;8:e85717. doi: 10.1371/journal.pone.0085717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bildsoe H, Loebel DA, Jones VJ, Chen YT, Behringer RR, Tam PP. Requirement for Twist1 in frontonasal and skull vault development in the mouse embryo. Dev Biol. 2009;331:176–188. doi: 10.1016/j.ydbio.2009.04.034. [DOI] [PubMed] [Google Scholar]
- Borchers A, David R, Wedlich D. Xenopus cadherin-11 restrains cranial neural crest migration and influences neural crest specification. Development. 2001;128:3049–3060. doi: 10.1242/dev.128.16.3049. [DOI] [PubMed] [Google Scholar]
- Breau MA, Pietri T, Stemmler MP, Thiery JP, Weston JA. A nonneural epithelial domain of embryonic cranial neural folds gives rise to ectomesenchyme. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:7750–7755. doi: 10.1073/pnas.0711344105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brieher WM, Yap AS. Cadherin junctions and their cytoskeleton(s) Current opinion in cell biology. 2013;25:39–46. doi: 10.1016/j.ceb.2012.10.010. [DOI] [PubMed] [Google Scholar]
- Bronner ME, LeDouarin NM. Development and evolution of the neural crest: an overview. Dev Biol. 2012;366:2–9. doi: 10.1016/j.ydbio.2011.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronner-Fraser M, Wolf JJ, Murray BA. Effects of antibodies against N-cadherin and N-CAM on the cranial neural crest and neural tube. Dev Biol. 1992;153:291–301. doi: 10.1016/0012-1606(92)90114-v. [DOI] [PubMed] [Google Scholar]
- Bryant DM, Kerr MC, Hammond LA, Joseph SR, Mostov KE, Teasdale RD, Stow JL. EGF induces macropinocytosis and SNX1-modulated recycling of E-cadherin. Journal of cell science. 2007;120:1818–1828. doi: 10.1242/jcs.000653. [DOI] [PubMed] [Google Scholar]
- Caescu CI, Jeschke GR, Turk BE. Active-site determinants of substrate recognition by the metalloproteinases TACE and ADAM10. The Biochemical journal. 2009;424:79–88. doi: 10.1042/BJ20090549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai DH, Vollberg TM, Sr, Hahn-Dantona E, Quigley JP, Brauer PR. MMP-2 expression during early avian cardiac and neural crest morphogenesis. Anat Rec. 2000;259:168–179. doi: 10.1002/(SICI)1097-0185(20000601)259:2<168::AID-AR7>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Carver EA, Jiang R, Lan Y, Oram KF, Gridley T. The mouse snail gene encodes a key regulator of the epithelial-mesenchymal transition. Mol Cell Biol. 2001;21:8184–8188. doi: 10.1128/MCB.21.23.8184-8188.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casini P, Nardi I, Ori M. Hyaluronan is required for cranial neural crest cells migration and craniofacial development. Dev Dyn. 2012;241:294–302. doi: 10.1002/dvdy.23715. [DOI] [PubMed] [Google Scholar]
- Chalpe AJ, Prasad M, Henke AJ, Paulson AF. Regulation of cadherin expression in the chicken neural crest by the Wnt/beta-catenin signaling pathway. Cell adhesion & migration. 2010;4:431–438. doi: 10.4161/cam.4.3.12138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZF, Behringer RR. twist is required in head mesenchyme for cranial neural tube morphogenesis. Genes Dev. 1995;9:686–699. doi: 10.1101/gad.9.6.686. [DOI] [PubMed] [Google Scholar]
- Clay MR, Halloran MC. Cadherin 6 promotes neural crest cell detachment via F-actin regulation and influences active Rho distribution during epithelial-to-mesenchymal transition. Development. 2014;141:2506–2515. doi: 10.1242/dev.105551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen M, Briscoe J, Blassberg R. Morphogen interpretation: the transcriptional logic of neural tube patterning. Curr Opin Genet Dev. 2013;23:423–428. doi: 10.1016/j.gde.2013.04.003. [DOI] [PubMed] [Google Scholar]
- Coles EG, Gammill LS, Miner JH, Bronner-Fraser M. Abnormalities in neural crest cell migration in laminin alpha5 mutant mice. Dev Biol. 2006;289:218–228. doi: 10.1016/j.ydbio.2005.10.031. [DOI] [PubMed] [Google Scholar]
- Coles EG, Taneyhill LA, Bronner-Fraser M. A critical role for Cadherin6B in regulating avian neural crest emigration. Dev Biol. 2007;312:533–544. doi: 10.1016/j.ydbio.2007.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousin H, Abbruzzese G, Kerdavid E, Gaultier A, Alfandari D. Translocation of the cytoplasmic domain of ADAM13 to the nucleus is essential for Calpain8-a expression and cranial neural crest cell migration. Dev Cell. 2011;20:256–263. doi: 10.1016/j.devcel.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousin H, Abbruzzese G, McCusker C, Alfandari D. ADAM13 function is required in the 3 dimensional context of the embryo during cranial neural crest cell migration in Xenopus laevis. Dev Biol. 2012;368:335–344. doi: 10.1016/j.ydbio.2012.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dady A, Blavet C, Duband JL. Timing and kinetics of E- to N-cadherin switch during neurulation in the avian embryo. Dev Dyn. 2012;241:1333–1349. doi: 10.1002/dvdy.23813. [DOI] [PubMed] [Google Scholar]
- Davidson LA, Keller RE. Neural tube closure in Xenopus laevis involves medial migration, directed protrusive activity, cell intercalation and convergent extension. Development. 1999;126:4547–4556. doi: 10.1242/dev.126.20.4547. [DOI] [PubMed] [Google Scholar]
- Delannet M, Martin F, Bossy B, Cheresh DA, Reichardt LF, Duband JL. Specific roles of the alpha V beta 1, alpha V beta 3 and alpha V beta 5 integrins in avian neural crest cell adhesion and migration on vitronectin. Development. 1994;120:2687–2702. doi: 10.1242/dev.120.9.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duband JL. Diversity in the molecular and cellular strategies of epithelium-to-mesenchyme transitions: Insights from the neural crest. Cell adhesion & migration. 2010;4:458–482. doi: 10.4161/cam.4.3.12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duband JL, Dady A, Fleury V. Resolving time and space constraints during neural crest formation and delamination. Curr Top Dev Biol. 2015;111:27–67. doi: 10.1016/bs.ctdb.2014.11.002. [DOI] [PubMed] [Google Scholar]
- Duband JL, Thiery JP. Distribution of fibronectin in the early phase of avian cephalic neural crest cell migration. Dev Biol. 1982;93:308–323. doi: 10.1016/0012-1606(82)90120-8. [DOI] [PubMed] [Google Scholar]
- Duband JL, Thiery JP. Distribution of laminin and collagens during avian neural crest development. Development. 1987;101:461–478. doi: 10.1242/dev.101.3.461. [DOI] [PubMed] [Google Scholar]
- Duband JL, Volberg T, Sabanay I, Thiery JP, Geiger B. Spatial and temporal distribution of the adherens-junction-associated adhesion molecule A-CAM during avian embryogenesis. Development. 1988;103:325–344. doi: 10.1242/dev.103.2.325. [DOI] [PubMed] [Google Scholar]
- Duong TD, Erickson CA. MMP-2 plays an essential role in producing epithelial-mesenchymal transformations in the avian embryo. Dev Dyn. 2004;229:42–53. doi: 10.1002/dvdy.10465. [DOI] [PubMed] [Google Scholar]
- Edelman GM, Gallin WJ, Delouvee A, Cunningham BA, Thiery JP. Early epochal maps of two different cell adhesion molecules. Proceedings of the National Academy of Sciences of the United States of America. 1983;80:4384–4388. doi: 10.1073/pnas.80.14.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espeseth A, Johnson E, Kintner C. Xenopus F-cadherin, a novel member of the cadherin family of cell adhesion molecules, is expressed at boundaries in the neural tube. Mol Cell Neurosci. 1995;6:199–211. doi: 10.1006/mcne.1995.1017. [DOI] [PubMed] [Google Scholar]
- Espeseth A, Marnellos G, Kintner C. The role of F-cadherin in localizing cells during neural tube formation in Xenopus embryos. Development. 1998;125:301–312. doi: 10.1242/dev.125.2.301. [DOI] [PubMed] [Google Scholar]
- Ezin AM, Fraser SE, Bronner-Fraser M. Fate map and morphogenesis of presumptive neural crest and dorsal neural tube. Dev Biol. 2009;330:221–236. doi: 10.1016/j.ydbio.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairchild CL, Gammill LS. Tetraspanin18 is a FoxD3-responsive antagonist of cranial neural crest epithelial-to-mesenchymal transition that maintains cadherin-6B protein. Journal of cell science. 2013;126:1464–1476. doi: 10.1242/jcs.120915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferber EC, Kajita M, Wadlow A, Tobiansky L, Niessen C, Ariga H, Daniel J, Fujita Y. A role for the cleaved cytoplasmic domain of E-cadherin in the nucleus. J Biol Chem. 2008;283:12691–12700. doi: 10.1074/jbc.M708887200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishwick KJ, Neiderer TE, Jhingory S, Bronner ME, Taneyhill LA. The tight junction protein claudin-1 influences cranial neural crest cell emigration. Mechanisms of development. 2012;129:275–283. doi: 10.1016/j.mod.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Castro MI, Marcelle C, Bronner-Fraser M. Ectodermal Wnt function as a neural crest inducer. Science. 2002;297:848–851. doi: 10.1126/science.1070824. [DOI] [PubMed] [Google Scholar]
- Giambernardi TA, Sakaguchi AY, Gluhak J, Pavlin D, Troyer DA, Das G, Rodeck U, Klebe RJ. Neutrophil collagenase (MMP-8) is expressed during early development in neural crest cells as well as in adult melanoma cells. Matrix Biol. 2001;20:577–587. doi: 10.1016/s0945-053x(01)00166-4. [DOI] [PubMed] [Google Scholar]
- Gottardi CJ, Wong E, Gumbiner BM. E-cadherin suppresses cellular transformation by inhibiting beta-catenin signaling in an adhesion-independent manner. The Journal of cell biology. 2001;153:1049–1060. doi: 10.1083/jcb.153.5.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumbiner BM. Regulation of cadherin-mediated adhesion in morphogenesis. Nature reviews. 2005;6:622–634. doi: 10.1038/nrm1699. [DOI] [PubMed] [Google Scholar]
- Hadeball B, Borchers A, Wedlich D. Xenopus cadherin-11 (Xcadherin-11) expression requires the Wg/Wnt signal. Mechanisms of development. 1998;72:101–113. doi: 10.1016/s0925-4773(98)00022-7. [DOI] [PubMed] [Google Scholar]
- Hall RJ, Erickson CA. ADAM 10: an active metalloprotease expressed during avian epithelial morphogenesis. Dev Biol. 2003;256:146–159. doi: 10.1016/s0012-1606(02)00133-1. [DOI] [PubMed] [Google Scholar]
- Halloran MC, Berndt JD. Current progress in neural crest cell motility and migration and future prospects for the zebrafish model system. Dev Dyn. 2003;228:497–513. doi: 10.1002/dvdy.10374. [DOI] [PubMed] [Google Scholar]
- Harrison M, Abu-Elmagd M, Grocott T, Yates C, Gavrilovic J, Wheeler GN. Matrix metalloproteinase genes in Xenopus development. Dev Dyn. 2004;231:214–220. doi: 10.1002/dvdy.20113. [DOI] [PubMed] [Google Scholar]
- Hartmann D, de Strooper B, Serneels L, Craessaerts K, Herreman A, Annaert W, Umans L, Lubke T, Lena Illert A, von Figura K, Saftig P. The disintegrin/metalloprotease ADAM 10 is essential for Notch signalling but not for alpha-secretase activity in fibroblasts. Human molecular genetics. 2002;11:2615–2624. doi: 10.1093/hmg/11.21.2615. [DOI] [PubMed] [Google Scholar]
- Hatta K, Takagi S, Fujisawa H, Takeichi M. Spatial and temporal expression pattern of N-cadherin cell adhesion molecules correlated with morphogenetic processes of chicken embryos. Dev Biol. 1987;120:215–227. doi: 10.1016/0012-1606(87)90119-9. [DOI] [PubMed] [Google Scholar]
- Hatta K, Takeichi M. Expression of N-cadherin adhesion molecules associated with early morphogenetic events in chick development. Nature. 1986;320:447–449. doi: 10.1038/320447a0. [DOI] [PubMed] [Google Scholar]
- Hopwood ND, Pluck A, Gurdon JB. A Xenopus mRNA related to Drosophila twist is expressed in response to induction in the mesoderm and the neural crest. Cell. 1989;59:893–903. doi: 10.1016/0092-8674(89)90612-0. [DOI] [PubMed] [Google Scholar]
- Huang C, Kratzer MC, Wedlich D, Kashef J. E-cadherin is required for cranial neural crest migration in Xenopus laevis. Dev Biol. 2016;411:159–171. doi: 10.1016/j.ydbio.2016.02.007. [DOI] [PubMed] [Google Scholar]
- Huber O, Korn R, McLaughlin J, Ohsugi M, Herrmann BG, Kemler R. Nuclear localization of beta-catenin by interaction with transcription factor LEF-1. Mechanisms of development. 1996;59:3–10. doi: 10.1016/0925-4773(96)00597-7. [DOI] [PubMed] [Google Scholar]
- Inoue T, Chisaka O, Matsunami H, Takeichi M. Cadherin-6 expression transiently delineates specific rhombomeres, other neural tube subdivisions, and neural crest subpopulations in mouse embryos. Dev Biol. 1997;183:183–194. doi: 10.1006/dbio.1996.8501. [DOI] [PubMed] [Google Scholar]
- Ishii M, Merrill AE, Chan YS, Gitelman I, Rice DP, Sucov HM, Maxson RE., Jr Msx2 and Twist cooperatively control the development of the neural crest-derived skeletogenic mesenchyme of the murine skull vault. Development. 2003;130:6131–6142. doi: 10.1242/dev.00793. [DOI] [PubMed] [Google Scholar]
- Jamora C, DasGupta R, Kocieniewski P, Fuchs E. Links between signal transduction, transcription and adhesion in epithelial bud development. Nature. 2003;422:317–322. doi: 10.1038/nature01458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jheon AH, Schneider RA. The cells that fill the bill: neural crest and the evolution of craniofacial development. J Dent Res. 2009;88:12–21. doi: 10.1177/0022034508327757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalcheim C. Epithelial-Mesenchymal Transitions during Neural Crest and Somite Development. J Clin Med. 2015:5. doi: 10.3390/jcm5010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashef J, Kohler A, Kuriyama S, Alfandari D, Mayor R, Wedlich D. Cadherin-11 regulates protrusive activity in Xenopus cranial neural crest cells upstream of Trio and the small GTPases. Genes Dev. 2009;23:1393–1398. doi: 10.1101/gad.519409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemler R, Babinet C, Eisen H, Jacob F. Surface antigen in early differentiation. Proceedings of the National Academy of Sciences of the United States of America. 1977;74:4449–4452. doi: 10.1073/pnas.74.10.4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerosuo L, Bronner ME. Biphasic influence of Miz1 on neural crest development by regulating cell survival and apical adhesion complex formation in the developing neural tube. Molecular biology of the cell. 2014;25:347–355. doi: 10.1091/mbc.E13-06-0327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinke DJ, 2nd, Horvath N, Cuppett V, Wu Y, Deng W, Kanj R. Interlocked positive and negative feedback network motifs regulate beta-catenin activity in the adherens junction pathway. Molecular biology of the cell. 2015;26:4135–4148. doi: 10.1091/mbc.E15-02-0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler A, Schlupf J, Schneider M, Kraft B, Winter C, Kashef J. Loss of Xenopus cadherin-11 leads to increased Wnt/beta-catenin signaling and up-regulation of target genes c-myc and cyclin D1 in neural crest. Dev Biol. 2013;383:132–145. doi: 10.1016/j.ydbio.2013.08.007. [DOI] [PubMed] [Google Scholar]
- Komatsu K, Wakatsuki S, Yamada S, Yamamura K, Miyazaki J, Sehara-Fujisawa A. Meltrin beta expressed in cardiac neural crest cells is required for ventricular septum formation of the heart. Dev Biol. 2007;303:82–92. doi: 10.1016/j.ydbio.2006.10.037. [DOI] [PubMed] [Google Scholar]
- Kowalczyk AP, Nanes BA. Adherens junction turnover: regulating adhesion through cadherin endocytosis, degradation, and recycling. Subcell Biochem. 2012;60:197–222. doi: 10.1007/978-94-007-4186-7_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krotoski DM, Domingo C, Bronner-Fraser M. Distribution of a putative cell surface receptor for fibronectin and laminin in the avian embryo. The Journal of cell biology. 1986;103:1061–1071. doi: 10.1083/jcb.103.3.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer EM, Feng Y, Zhaoyuan H, Rauscher FJ, 3rd, Kroll KL, Longmore GD. Ajuba LIM proteins are snail/slug corepressors required for neural crest development in Xenopus. Dev Cell. 2008;14:424–436. doi: 10.1016/j.devcel.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langhe RP, Gudzenko T, Bachmann M, Becker SF, Gonnermann C, Winter C, Abbruzzese G, Alfandari D, Kratzer MC, Franz CM, Kashef J. Cadherin-11 localizes to focal adhesions and promotes cell-substrate adhesion. Nat Commun. 2016;7:10909. doi: 10.1038/ncomms10909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RT, Nagai H, Nakaya Y, Sheng G, Trainor PA, Weston JA, Thiery JP. Cell delamination in the mesencephalic neural fold and its implication for the origin of ectomesenchyme. Development. 2013;140:4890–4902. doi: 10.1242/dev.094680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh NR, Schupp MO, Li K, Padmanabhan V, Gastonguay A, Wang L, Chun CZ, Wilkinson GA, Ramchandran R. Mmp17b is essential for proper neural crest cell migration in vivo. PLoS ONE. 2013;8:e76484. doi: 10.1371/journal.pone.0076484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Joel Duff R, Londraville RL, Marrs JA, Liu Q. Cloning and expression analysis of cadherin7 in the central nervous system of the embryonic zebrafish. Gene Expr Patterns. 2007;7:15–22. doi: 10.1016/j.modgep.2006.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JP, Jessell TM. A role for rhoB in the delamination of neural crest cells from the dorsal neural tube. Development. 1998;125:5055–5067. doi: 10.1242/dev.125.24.5055. [DOI] [PubMed] [Google Scholar]
- Liu Q, Marrs JA, Londraville RL, Wilson AL. Cadherin-7 function in zebrafish development. Cell Tissue Res. 2008;334:37–45. doi: 10.1007/s00441-008-0664-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon CA, Johnson JL, Williams H, Sala-Newby GB, George SJ. Soluble N-cadherin overexpression reduces features of atherosclerotic plaque instability. Arterioscler Thromb Vasc Biol. 2009;29:195–201. doi: 10.1161/ATVBAHA.108.178087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marambaud P, Shioi J, Serban G, Georgakopoulos A, Sarner S, Nagy V, Baki L, Wen P, Efthimiopoulos S, Shao Z, Wisniewski T, Robakis NK. A presenilin-1/gamma-secretase cleavage releases the E-cadherin intracellular domain and regulates disassembly of adherens junctions. EMBO J. 2002;21:1948–1956. doi: 10.1093/emboj/21.8.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maretzky T, Reiss K, Ludwig A, Buchholz J, Scholz F, Proksch E, de Strooper B, Hartmann D, Saftig P. ADAM10 mediates E-cadherin shedding and regulates epithelial cell-cell adhesion, migration, and beta-catenin translocation. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:9182–9187. doi: 10.1073/pnas.0500918102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCrea PD, Gottardi CJ. Beyond beta-catenin: prospects for a larger catenin network in the nucleus. Nature reviews. 2016;17:55–64. doi: 10.1038/nrm.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCusker C, Cousin H, Neuner R, Alfandari D. Extracellular cleavage of cadherin-11 by ADAM metalloproteases is essential for Xenopus cranial neural crest cell migration. Molecular biology of the cell. 2009;20:78–89. doi: 10.1091/mbc.E08-05-0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLennan R, Dyson L, Prather KW, Morrison JA, Baker RE, Maini PK, Kulesa PM. Multiscale mechanisms of cell migration during development: theory and experiment. Development. 2012;139:2935–2944. doi: 10.1242/dev.081471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLennan R, Schumacher LJ, Morrison JA, Teddy JM, Ridenour DA, Box AC, Semerad CL, Li H, McDowell W, Kay D, Maini PK, Baker RE, Kulesa PM. Neural crest migration is driven by a few trailblazer cells with a unique molecular signature narrowly confined to the invasive front. Development. 2015;142:2014–2025. doi: 10.1242/dev.117507. [DOI] [PubMed] [Google Scholar]
- Mechtersheimer S, Gutwein P, Agmon-Levin N, Stoeck A, Oleszewski M, Riedle S, Postina R, Fahrenholz F, Fogel M, Lemmon V, Altevogt P. Ectodomain shedding of L1 adhesion molecule promotes cell migration by autocrine binding to integrins. The Journal of cell biology. 2001;155:661–673. doi: 10.1083/jcb.200101099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsonego-Ornan E, Kosonovsky J, Bar A, Roth L, Fraggi-Rankis V, Simsa S, Kohl A, Sela-Donenfeld D. Matrix metalloproteinase 9/gelatinase B is required for neural crest cell migration. Dev Biol. 2012;364:162–177. doi: 10.1016/j.ydbio.2012.01.028. [DOI] [PubMed] [Google Scholar]
- Moore R, Champeval D, Denat L, Tan SS, Faure F, Julien-Grille S, Larue L. Involvement of cadherins 7 and 20 in mouse embryogenesis and melanocyte transformation. Oncogene. 2004;23:6726–6735. doi: 10.1038/sj.onc.1207675. [DOI] [PubMed] [Google Scholar]
- Murray SA, Gridley T. Snail family genes are required for left-right asymmetry determination, but not neural crest formation, in mice. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:10300–10304. doi: 10.1073/pnas.0602234103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najy AJ, Day KC, Day ML. The ectodomain shedding of E-cadherin by ADAM15 supports ErbB receptor activation. J Biol Chem. 2008;283:18393–18401. doi: 10.1074/jbc.M801329200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S, Takeichi M. Neural crest cell-cell adhesion controlled by sequential and subpopulation-specific expression of novel cadherins. Development. 1995;121:1321–1332. doi: 10.1242/dev.121.5.1321. [DOI] [PubMed] [Google Scholar]
- Nakagawa S, Takeichi M. Neural crest emigration from the neural tube depends on regulated cadherin expression. Development. 1998;125:2963–2971. doi: 10.1242/dev.125.15.2963. [DOI] [PubMed] [Google Scholar]
- Neuner R, Cousin H, McCusker C, Coyne M, Alfandari D. Xenopus ADAM19 is involved in neural, neural crest and muscle development. Mechanisms of development. 2009;126:240–255. doi: 10.1016/j.mod.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newgreen D, Thiery JP. Fibronectin in early avian embryos: synthesis and distribution along the migration pathways of neural crest cells. Cell Tissue Res. 1980;211:269–291. doi: 10.1007/BF00236449. [DOI] [PubMed] [Google Scholar]
- Nichols DH. Neural crest formation in the head of the mouse embryo as observed using a new histological technique. J Embryol Exp Morphol. 1981;64:105–120. [PubMed] [Google Scholar]
- Nichols DH. Ultrastructure of neural crest formation in the midbrain/rostral hindbrain and preotic hindbrain regions of the mouse embryo. Am J Anat. 1987;179:143–154. doi: 10.1002/aja.1001790207. [DOI] [PubMed] [Google Scholar]
- Niessen CM, Gottardi CJ. Molecular components of the adherens junction. Biochimica et biophysica acta. 2008;1778:562–571. doi: 10.1016/j.bbamem.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niessen CM, Leckband D, Yap AS. Tissue organization by cadherin adhesion molecules: dynamic molecular and cellular mechanisms of morphogenetic regulation. Physiol Rev. 2011;91:691–731. doi: 10.1152/physrev.00004.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noden DM, Trainor PA. Relations and interactions between cranial mesoderm and neural crest populations. J Anat. 2005;207:575–601. doi: 10.1111/j.1469-7580.2005.00473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori M, Nardini M, Casini P, Perris R, Nardi I. XHas2 activity is required during somitogenesis and precursor cell migration in Xenopus development. Development. 2006;133:631–640. doi: 10.1242/dev.02225. [DOI] [PubMed] [Google Scholar]
- Ortiz A, Lee YC, Yu G, Liu HC, Lin SC, Bilen MA, Cho H, Yu-Lee LY, Lin SH. Angiomotin is a novel component of cadherin-11/beta-catenin/p120 complex and is critical for cadherin-11-mediated cell migration. FASEB J. 2015;29:1080–1091. doi: 10.1096/fj.14-261594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota MS, Loebel DA, O’Rourke MP, Wong N, Tsoi B, Tam PP. Twist is required for patterning the cranial nerves and maintaining the viability of mesodermal cells. Dev Dyn. 2004;230:216–228. doi: 10.1002/dvdy.20047. [DOI] [PubMed] [Google Scholar]
- Padmanabhan R, Taneyhill LA. Cadherin-6B undergoes macropinocytosis and clathrin-mediated endocytosis during cranial neural crest cell EMT. Journal of cell science. 2015;128:1773–1786. doi: 10.1242/jcs.164426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradies NE, Grunwald GB. Purification and characterization of NCAD90, a soluble endogenous form of N-cadherin, which is generated by proteolysis during retinal development and retains adhesive and neurite-promoting function. J Neurosci Res. 1993;36:33–45. doi: 10.1002/jnr.490360105. [DOI] [PubMed] [Google Scholar]
- Park KS, Gumbiner BM. Cadherin 6B induces BMP signaling and de-epithelialization during the epithelial mesenchymal transition of the neural crest. Development. 2010;137:2691–2701. doi: 10.1242/dev.050096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KS, Gumbiner BM. Cadherin-6B stimulates an epithelial mesenchymal transition and the delamination of cells from the neural ectoderm via LIMK/cofilin mediated non-canonical BMP receptor signaling. Dev Biol. 2012;366:232–243. doi: 10.1016/j.ydbio.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AD, Parton RG, Ferguson C, Stow JL, Yap AS. Characterization of E-cadherin endocytosis in isolated MCF-7 and chinese hamster ovary cells: the initial fate of unbound E-cadherin. J Biol Chem. 2003;278:21050–21057. doi: 10.1074/jbc.M300082200. [DOI] [PubMed] [Google Scholar]
- Patterson RA, Cavanaugh AM, Cantemir V, Brauer PR, Reedy MV. MT2-MMP expression during early avian morphogenesis. Anat Rec (Hoboken) 2013;296:64–70. doi: 10.1002/ar.22618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perissinotto D, Iacopetti P, Bellina I, Doliana R, Colombatti A, Pettway Z, Bronner-Fraser M, Shinomura T, Kimata K, Morgelin M, Lofberg J, Perris R. Avian neural crest cell migration is diversely regulated by the two major hyaluronan-binding proteoglycans PG-M/versican and aggrecan. Development. 2000;127:2823–2842. doi: 10.1242/dev.127.13.2823. [DOI] [PubMed] [Google Scholar]
- Perris R, Perissinotto D, Pettway Z, Bronner-Fraser M, Morgelin M, Kimata K. Inhibitory effects of PG-H/aggrecan and PG-M/versican on avian neural crest cell migration. Faseb J. 1996;10:293–301. doi: 10.1096/fasebj.10.2.8641562. [DOI] [PubMed] [Google Scholar]
- Piloto S, Schilling TF. Ovo1 links Wnt signaling with N-cadherin localization during neural crest migration. Development. 2010;137:1981–1990. doi: 10.1242/dev.048439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell DR, Williams JS, Hernandez-Lagunas L, Salcedo E, O’Brien JH, Artinger KB. Cdon promotes neural crest migration by regulating N-cadherin localization. Dev Biol. 2015;407:289–299. doi: 10.1016/j.ydbio.2015.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radice GL, Rayburn H, Matsunami H, Knudsen KA, Takeichi M, Hynes RO. Developmental defects in mouse embryos lacking N-cadherin. Dev Biol. 1997;181:64–78. doi: 10.1006/dbio.1996.8443. [DOI] [PubMed] [Google Scholar]
- Reiss K, Maretzky T, Ludwig A, Tousseyn T, de Strooper B, Hartmann D, Saftig P. ADAM10 cleavage of N-cadherin and regulation of cell-cell adhesion and beta-catenin nuclear signalling. EMBO J. 2005;24:742–752. doi: 10.1038/sj.emboj.7600548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers CD, Saxena A, Bronner ME. Sip1 mediates an E-cadherin-to-N-cadherin switch during cranial neural crest EMT. The Journal of cell biology. 2013;203:835–847. doi: 10.1083/jcb.201305050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roycroft A, Mayor R. Molecular basis of contact inhibition of locomotion. Cell Mol Life Sci. 2016;73:1119–1130. doi: 10.1007/s00018-015-2090-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadaghiani B, Thiebaud CH. Neural crest development in the Xenopus laevis embryo, studied by interspecific transplantation and scanning electron microscopy. Dev Biol. 1987;124:91–110. doi: 10.1016/0012-1606(87)90463-5. [DOI] [PubMed] [Google Scholar]
- Sadot E, Simcha I, Shtutman M, Ben-Ze’ev A, Geiger B. Inhibition of beta-catenin-mediated transactivation by cadherin derivatives. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:15339–15344. doi: 10.1073/pnas.95.26.15339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Jeannet JP, Moody SA. Establishing the pre-placodal region and breaking it into placodes with distinct identities. Dev Biol. 2014;389:13–27. doi: 10.1016/j.ydbio.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai D, Tanaka Y, Endo Y, Osumi N, Okamoto H, Wakamatsu Y. Regulation of Slug transcription in embryonic ectoderm by b-catenin-Lef/TCF and BMP-Smad signaling. Develop Growth Differ. 2005;47:471–482. doi: 10.1111/j.1440-169X.2005.00821.x. [DOI] [PubMed] [Google Scholar]
- Sanchez-Martin M, Rodriguez-Garcia A, Perez-Losada J, Sagrera A, Read AP, Sanchez-Garcia I. SLUG (SNAI2) deletions in patients with Waardenburg disease. Human molecular genetics. 2002;11:3231–3236. doi: 10.1093/hmg/11.25.3231. [DOI] [PubMed] [Google Scholar]
- Scarpa E, Szabo A, Bibonne A, Theveneau E, Parsons M, Mayor R. Cadherin Switch during EMT in Neural Crest Cells Leads to Contact Inhibition of Locomotion via Repolarization of Forces. Dev Cell. 2015;34:421–434. doi: 10.1016/j.devcel.2015.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffmacher AT, Padmanabhan R, Jhingory S, Taneyhill LA. Cadherin-6B is proteolytically processed during epithelial-to-mesenchymal transitions of the cranial neural crest. Molecular biology of the cell. 2014;25:41–54. doi: 10.1091/mbc.E13-08-0459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffmacher AT, Xie V, Taneyhill LA. Cadherin-6B proteolysis promotes the neural crest cell epithelial-to-mesenchymal transition through transcriptional regulation. The Journal of cell biology. 2016;215:735–747. doi: 10.1083/jcb.201604006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh R, Vestweber D, Riede I, Ringwald M, Rosenberg UB, Jackle H, Kemler R. Molecular cloning of the mouse cell adhesion molecule uvomorulin: cDNA contains a B1-related sequence. Proceedings of the National Academy of Sciences of the United States of America. 1986;83:1364–1368. doi: 10.1073/pnas.83.5.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sefton M, Sanchez S, Nieto MA. Conserved and divergent roles for members of the Snail family of transcription factors in the chick and mouse embryo. Development. 1998;125:3111–3121. doi: 10.1242/dev.125.16.3111. [DOI] [PubMed] [Google Scholar]
- Sela-Donenfeld D, Kalcheim C. Regulation of the onset of neural crest migration by coordinated activity of BMP4 and Noggin in the dorsal neural tube. Development. 1999;126:4749–4762. doi: 10.1242/dev.126.21.4749. [DOI] [PubMed] [Google Scholar]
- Sela-Donenfeld D, Kalcheim C. Inhibition of noggin expression in the dorsal neural tube by somitogenesis: a mechanism for coordinating the timing of neural crest emigration. Development. 2000;127:4845–4854. doi: 10.1242/dev.127.22.4845. [DOI] [PubMed] [Google Scholar]
- Sela-Donenfeld D, Kalcheim C. Localized BMP4-noggin interactions generate the dynamic patterning of noggin expression in somites. Dev Biol. 2002;246:311–328. doi: 10.1006/dbio.2002.0672. [DOI] [PubMed] [Google Scholar]
- Sharma M, Henderson BR. IQ-domain GTPase-activating protein 1 regulates beta-catenin at membrane ruffles and its role in macropinocytosis of N-cadherin and adenomatous polyposis coli. J Biol Chem. 2007;282:8545–8556. doi: 10.1074/jbc.M610272200. [DOI] [PubMed] [Google Scholar]
- Shook D, Keller R. Mechanisms, mechanics and function of epithelial-mesenchymal transitions in early development. Mechanisms of development. 2003;120:1351–1383. doi: 10.1016/j.mod.2003.06.005. [DOI] [PubMed] [Google Scholar]
- Shoval I, Ludwig A, Kalcheim C. Antagonistic roles of full-length N-cadherin and its soluble BMP cleavage product in neural crest delamination. Development. 2007;134:491–501. doi: 10.1242/dev.02742. [DOI] [PubMed] [Google Scholar]
- Simoes-Costa M, Bronner ME. Establishing neural crest identity: a gene regulatory recipe. Development. 2015;142:242–257. doi: 10.1242/dev.105445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solis GP, Schrock Y, Hulsbusch N, Wiechers M, Plattner H, Stuermer CA. Reggies/flotillins regulate E-cadherin-mediated cell contact formation by affecting EGFR trafficking. Molecular biology of the cell. 2012;23:1812–1825. doi: 10.1091/mbc.E11-12-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soo K, O’Rourke MP, Khoo PL, Steiner KA, Wong N, Behringer RR, Tam PP. Twist function is required for the morphogenesis of the cephalic neural tube and the differentiation of the cranial neural crest cells in the mouse embryo. Dev Biol. 2002;247:251–270. doi: 10.1006/dbio.2002.0699. [DOI] [PubMed] [Google Scholar]
- Steventon B, Mayor R, Streit A. Neural crest and placode interaction during the development of the cranial sensory system. Dev Biol. 2014;389:28–38. doi: 10.1016/j.ydbio.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobl-Mazzulla PH, Bronner ME. A PHD12-Snail2 repressive complex epigenetically mediates neural crest epithelial-to-mesenchymal transition. The Journal of cell biology. 2012;198:999–1010. doi: 10.1083/jcb.201203098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo A, Melchionda M, Nastasi G, Woods ML, Campo S, Perris R, Mayor R. In vivo confinement promotes collective migration of neural crest cells. The Journal of cell biology. 2016;213:543–555. doi: 10.1083/jcb.201602083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M, Osumi N. Expression study of cadherin7 and cadherin20 in the embryonic and adult rat central nervous system. BMC developmental biology. 2008;8:87. doi: 10.1186/1471-213X-8-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan SS, Crossin KL, Hoffman S, Edelman GM. Asymmetric expression in somites of cytotactin and its proteoglycan ligand is correlated with neural crest cell distribution. Proceedings of the National Academy of Sciences of the United States of America. 1987;84:7977–7981. doi: 10.1073/pnas.84.22.7977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneyhill LA, Bronner-Fraser M. Dynamic alterations in gene expression after Wnt-mediated induction of avian neural crest. Molecular biology of the cell. 2005;16:5283–5293. doi: 10.1091/mbc.E05-03-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneyhill LA, Coles EG, Bronner-Fraser M. Snail2 directly represses cadherin6B during epithelial-to-mesenchymal transitions of the neural crest. Development. 2007;134:1481–1490. doi: 10.1242/dev.02834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneyhill LA, Padmanabhan R. Chapter 3: The Cell Biology of Neural Crest Cell Delamination and EMT. Elsevier; Chennai, India: 2013. p. 1. [Google Scholar]
- Taneyhill LA, Schiffmacher AT. Cadherin dynamics during neural crest cell ontogeny. Prog Mol Biol Transl Sci. 2013;116:291–315. doi: 10.1016/B978-0-12-394311-8.00013-3. [DOI] [PubMed] [Google Scholar]
- Theveneau E, Marchant L, Kuriyama S, Gull M, Moepps B, Parsons M, Mayor R. Collective chemotaxis requires contact-dependent cell polarity. Dev Cell. 2010;19:39–53. doi: 10.1016/j.devcel.2010.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theveneau E, Mayor R. Collective cell migration of the cephalic neural crest: the art of integrating information. Genesis. 2011;49:164–176. doi: 10.1002/dvg.20700. [DOI] [PubMed] [Google Scholar]
- Theveneau E, Mayor R. Cadherins in collective cell migration of mesenchymal cells. Current opinion in cell biology. 2012a;24:677–684. doi: 10.1016/j.ceb.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theveneau E, Mayor R. Neural crest delamination and migration: From epithelium-to-mesenchyme transition to collective cell migration. Dev Biol. 2012b doi: 10.1016/j.ydbio.2011.12.041. [DOI] [PubMed] [Google Scholar]
- Theveneau E, Mayor R. Neural crest migration: interplay between chemorepellents, chemoattractants, contact inhibition, epithelial-mesenchymal transition, and collective cell migration. Wiley Interdiscip Rev Dev Biol. 2012c;1:435–445. doi: 10.1002/wdev.28. [DOI] [PubMed] [Google Scholar]
- Theveneau E, Mayor R. Collective cell migration of epithelial and mesenchymal cells. Cell Mol Life Sci. 2013;70:3481–3492. doi: 10.1007/s00018-012-1251-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theveneau E, Steventon B, Scarpa E, Garcia S, Trepat X, Streit A, Mayor R. Chase-and-run between adjacent cell populations promotes directional collective migration. Nat Cell Biol. 2013;15:763–772. doi: 10.1038/ncb2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery JP, Delouvee A, Gallin WJ, Cunningham BA, Edelman GM. Ontogenetic expression of cell adhesion molecules: L-CAM is found in epithelia derived from the three primary germ layers. Dev Biol. 1984;102:61–78. doi: 10.1016/0012-1606(84)90175-1. [DOI] [PubMed] [Google Scholar]
- Tien CL, Jones A, Wang H, Gerigk M, Nozell S, Chang C. Snail2/Slug cooperates with Polycomb repressive complex 2 (PRC2) to regulate neural crest development. Development. 2015;142:722–731. doi: 10.1242/dev.111997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson ML, Guan P, Morris RJ, Fidock MD, Rejzek M, Garcia-Morales C, Field RA, Wheeler GN. A chemical genomic approach identifies matrix metalloproteinases as playing an essential and specific role in Xenopus melanophore migration. Chem Biol. 2009;16:93–104. doi: 10.1016/j.chembiol.2008.12.005. [DOI] [PubMed] [Google Scholar]
- Tozer S, Le Dreau G, Marti E, Briscoe J. Temporal control of BMP signalling determines neuronal subtype identity in the dorsal neural tube. Development. 2013;140:1467–1474. doi: 10.1242/dev.090118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuttle AM, Hoffman TL, Schilling TF. Rabconnectin-3a regulates vesicle endocytosis and canonical Wnt signaling in zebrafish neural crest migration. PLoS Biol. 2014;12:e1001852. doi: 10.1371/journal.pbio.1001852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura K, Kihara T, Kuzuya A, Okawa K, Nishimoto T, Bito H, Ninomiya H, Sugimoto H, Kinoshita A, Shimohama S. Activity-dependent regulation of beta-catenin via epsilon-cleavage of N-cadherin. Biochemical and biophysical research communications. 2006;345:951–958. doi: 10.1016/j.bbrc.2006.04.157. [DOI] [PubMed] [Google Scholar]
- Ulrich F, Heisenberg CP. Trafficking and cell migration. Traffic. 2009;10:811–818. doi: 10.1111/j.1600-0854.2009.00929.x. [DOI] [PubMed] [Google Scholar]
- Utton MA, Eickholt B, Howell FV, Wallis J, Doherty P. Soluble N-cadherin stimulates fibroblast growth factor receptor dependent neurite outgrowth and N-cadherin and the fibroblast growth factor receptor co-cluster in cells. J Neurochem. 2001;76:1421–1430. doi: 10.1046/j.1471-4159.2001.00140.x. [DOI] [PubMed] [Google Scholar]
- Vallin J, Girault JM, Thiery JP, Broders F. Xenopus cadherin-11 is expressed in different populations of migrating neural crest cells. Mechanisms of development. 1998;75:171–174. doi: 10.1016/s0925-4773(98)00099-9. [DOI] [PubMed] [Google Scholar]
- Van de Putte T, Maruhashi M, Francis A, Nelles L, Kondoh H, Huylebroeck D, Higashi Y. Mice lacking ZFHX1B, the gene that codes for Smad-interacting protein-1, reveal a role for multiple neural crest cell defects in the etiology of Hirschsprung disease-mental retardation syndrome. Am J Hum Genet. 2003;72:465–470. doi: 10.1086/346092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakamatsu N, Yamada Y, Yamada K, Ono T, Nomura N, Taniguchi H, Kitoh H, Mutoh N, Yamanaka T, Mushiake K, Kato K, Sonta S, Nagaya M. Mutations in SIP1, encoding Smad interacting protein-1, cause a form of Hirschsprung disease. Nature genetics. 2001;27:369–370. doi: 10.1038/86860. [DOI] [PubMed] [Google Scholar]
- Zhang D, Ighaniyan S, Stathopoulos L, Rollo B, Landman K, Hutson J, Newgreen D. The neural crest: a versatile organ system. Birth Defects Res C Embryo Today. 2014;102:275–298. doi: 10.1002/bdrc.21081. [DOI] [PubMed] [Google Scholar]