Significance
Human exonuclease 1 (hExo1) is a 5′-structure–specific nuclease and a member of the RAD2/XPG superfamily that plays important roles in many aspects of genome maintenance. The means by which individual family members process multiple, structurally disparate substrates has been a long-standing question. The reaction intermediate structures reported here reveal that this remarkable feat is achieved by a series of orchestrated conformational changes that guide disparate substrates into a common, catalytically competent conformation, where they are cleaved by an enhanced variant of the two-metal, in-line hydrolysis mechanism. The observed motions not only enable exo- and endonucleolytic cleavage of gapped and 5′-flap substrates, respectively, but also encode unanticipated features, including mechanisms that enhance processing fidelity and account for processivity.
Keywords: Rad2/XPG superfamily, DNA repair, flap endonuclease, crystallography, exonuclease
Abstract
Human exonuclease 1 (hExo1) is a member of the RAD2/XPG structure-specific 5′-nuclease superfamily. Its dominant, processive 5′–3′ exonuclease and secondary 5′-flap endonuclease activities participate in various DNA repair, recombination, and replication processes. A single active site processes both recessed ends and 5′-flap substrates. By initiating enzyme reactions in crystals, we have trapped hExo1 reaction intermediates that reveal structures of these substrates before and after their exo- and endonucleolytic cleavage, as well as structures of uncleaved, unthreaded, and partially threaded 5′ flaps. Their distinctive 5′ ends are accommodated by a small, mobile arch in the active site that binds recessed ends at its base and threads 5′ flaps through a narrow aperture within its interior. A sequence of successive, interlocking conformational changes guides the two substrate types into a shared reaction mechanism that catalyzes their cleavage by an elaborated variant of the two-metal, in-line hydrolysis mechanism. Coupling of substrate-dependent arch motions to transition-state stabilization suppresses inappropriate or premature cleavage, enhancing processing fidelity. The striking reduction in flap conformational entropy is catalyzed, in part, by arch motions and transient binding interactions between the flap and unprocessed DNA strand. At the end of the observed reaction sequence, hExo1 resets without relinquishing DNA binding, suggesting a structural basis for its processivity.
Human exonuclease 1 (hExo1) is a member of the RAD2/XPG 5′-structure–specific nuclease superfamily (Exo1, FEN1, XPG, and GEN1) that plays essential roles in three central aspects of genome maintenance (1–4): replication, repair, and recombination. The common catalytic core of this superfamily processes a variety of disparate DNA substrates (5–8), including nicked, gapped, single-stranded 5′ flaps; Holliday junctions; and bubbles. The specificity of substrate recognition and efficiency of subsequent cleavage is tailored to the biological function of each family member (1, 3–5). hExo1 has dominant 5′–3′ exonuclease activity on nicked, gapped, or recessed-end DNA, but also cleaves 5′-flap substrates endonucleolytically (7, 8). Its primary exonucleolytic activity is important in mismatch repair (1), double-stranded break repair (2), and the DNA damage response (9). The secondary endonucleolytic activity of hExo1 is involved in processing immunoglobin class-switching recombination intermediates (10), trinucleotide repeats (11), DNA-RNA hybrids such as Okazaki fragments (12), and ribonucleotide excision repair (13). hExo1 and the predominantly endonucleolytic FEN1 family member exhibit partially overlapping functions, complicating assignment of their individual biological roles (4, 11–13). Exo1 knockout mice exhibit reduced survival, increased susceptibility to developing lymphomas (14), and sterility in both males and females (14). Defects in hExo1 are implicated in human cancers (15).
Structural studies of hExo1 (16) and FEN1 (17) have shown that a single active site cleaves both 5′ recessed ends and 5′ flaps. The different termini are accommodated by a process that “threads” 5′ flaps, but not recessed ends, through the protein (18–20). Structures of recessed substrates revealed that their 5′ phosphates are bound at the base of a conformationally diverse structural element in the active site, the mobile arch (16, 17). The scissile bonds in these complexes are located near two catalytic Mg2+ ions in the active site, or appear to move toward them. A 5′-flap substrate, determined in a bacteriophage T5 FEN complex, is threaded through an aperture in the interior of the mobile arch, also placing its scissile bond in close proximity to the two metals (21). Taken together, these observations suggest that both substrate classes could adopt a common transition state within their shared active site (16, 17).
This hypothesis has been difficult to test, because it is challenging to capture substrate or product complexes in conformations that correspond to intermediates just before or after the cleavage step without using inhibitory metals that perturb coordination geometries (16, 17) or mutations that destroy a metal-binding site (21). Accordingly, the nature of the transition state(s) has not been established definitively (16, 17, 21). Furthermore, conformational heterogeneity of the recessed or flap complexes observed in different experiments also prevented the direct structural comparisons required to test this hypothesis. Finally, no structures have been captured of threading intermediates, which might shed light on the mechanisms by which disparate starting structures are guided into a common transition state. We report on time-resolved trapping of 12 structures, representing intermediates in the hExo1 reaction cycle, which enabled us to address these questions.
Results
hExo1 Structure.
Previously, we have shown that the catalytically active N-terminal domain (residues 1–352) of hExo1 (16) forms a bean-shaped structure (Fig. 1A). Its common core region is structurally similar to FEN1 (16, 17). The duplex portion of the DNA substrate is bound on the protein surface, tethered to a surface region that includes a K+/Na+-binding site [the helix–two-turn–helix tether (H2TH)] (16). The active site is located near the point where the double-stranded DNA bifurcates into 5′ and 3′ single strands (the dsDNA “junction”), the former of which is processed (Fig. 1B). The two-helix (α4 and α5) mobile arch and an immobile two-helix (α2 and α3) wedge domain that contains a hydrophobic region are situated at this junction (16).
Fig. 1.
hExo1 catalytic domain bound to DNA. (A) Protein–DNA complex: gray, catalytic domain; cyan (circled), conserved active-site carboxylates; blue spheres, catalytic metal ions; green, mobile arch; purple, H2TH; purple sphere, K+/Na+ site; dark red, helix-turn-helix (HTH); pink, C-terminal region; gold, unprocessed strand; yellow, processed strand; red, scissile bond. This color coding scheme is used throughout, unless indicated otherwise. (B) Nucleotide naming system.
Time-Resolved Trapping of Reaction Intermediates.
We have trapped DNA complexes at various stages of both exo- and endonucleolytic cleavage (Fig. 2A and Tables S1 and S2). Substrates (Table S3) can be captured in initial binding events by cocrystallization in the absence of metals. Under favorable circumstances, various substrate intermediates and products can be obtained by initializing the reaction in crystallo with addition of Mg2+ (or Mn2+, which slows down the reaction rates and provides an anomalous signal that defines bound metal positions) and trapping intermediates by freezing the crystals in liquid nitrogen after different incubation time intervals. Mutations that substantially lowered catalytic rates, but retained metal binding, have aided trapping.
Fig. 2.
Exonucleolytic reaction cycle. (A) Assignment of structures observed in the experimental timeline (Named structures described in the main text are indicated at the bottom: r prefix, recessed-end substrates; f prefix, 5′-flap substrates) to stages along the proposed reaction coordinate (indicated at the top, numbered). At each stage, the states of major structural elements (left column) are described. Arrows indicate major motions. (B) Reaction cycle summary. The tethered DNA (Top Left; stage 1, rI) is seated at the dsDNA junction (Top Center; stage 2, rII). The scissile bond is then guided into the fully assembled active site (Top Right; stage 3, rIV) and cleaved (Bottom Right; stage 4, rVIII). The protein and DNA reset for the next cleavage cycle (Bottom Center; stage 5, rIX). (C) Structures (names indicated) illustrating the motion of the DNA along the reaction coordinate, relative to the position of the scissile bond (black square) in stage 3 (rIV). The thick dotted line indicates distance to scissile bond of the processed strand.
Table S1.
Crystallographic statistics (part 1)
| Complex | rI | rII | rIII | rIV | rV | rVI |
| PDB ID code | 5UZV | 5V04 | 5V05 | 5V06 | 5V07 | 5V08 |
| Beamline | ALS 12.3.1 SIBYLS | APS SER-CAT 22ID | ALS 12.3.1 SIBYLS | APS SER-CAT 22ID | APS SER-CAT 22ID | APS SER-CAT 22ID |
| Resolution range, Å (Highest resolution bin) | 50.00–2.45 (2.49–2.45) | 50.00–2.65 (2.70–2.65) | 50.00–2.90 (2.95–2.90) | 50.00–2.75 (2.80–2.75) | 50.00–2.15 (2.19–2.15) | 50.00–2.81 (2.86–2.81) |
| Space group | P43212 | P43212 | P43212 | P43212 | P43212 | P43212 |
| Unit cell, Å | 72.49, 72.49, 182.53 | 73.82, 73.82, 180.95 | 73.91, 73.91, 183.51 | 74.28, 74.28, 179.89 | 73.84, 73.84, 179.07 | 74.26, 74.26, 179.39 |
| Multiplicity | 6.3 (6.4) | 8.9 (6.6) | 5.3 (5.4) | 6.0 (6.5) | 7.2 (2.2) | 7.4 (7.5) |
| Completeness, % | 98.9 (100.0) | 99.6 (98.5) | 97.8 (99.8) | 99.2 (100) | 80.3 (25.5) | 99.8 (100.0) |
| Mean I/σI | 32.69 (2.36) | 26.65 (1.82) | 17.52 (2.69) | 19.19 (2.56) | 23.70 (2.83) | 25.47 (2.31) |
| Rmerge | 5.9 (65.6) | 8.1 (65.5) | 9.9 (68.5) | 9.2 (69.5) | 8.4 (30.8) | 7.7 (68.4) |
| Rmeas | 6.4 (71.6) | 8.6 (70.8) | 11.0 (75.7) | 10.1 (75.6) | 9.0 (38.0) | 8.4 (73.5) |
| CC1/2 | 0.830 | 0.870 | 0.705 | 0.698 | 0.892 | 0.888 |
| Rwork | 0.2133 (0.2857) | 0.2292 (0.3558) | 0.1992 (0.3165) | 0.2185 (0.3449) | 0.1758 (0.2251) | 0.2140 (0.3117) |
| Rfree | 0.2540 (0.3228) | 0.2558 (0.3635) | 0.2484 (0.3551) | 0.2655 (0.3507) | 0.2197 (0.2729) | 0.2623 (0.3558) |
| No. of nonhydrogen atoms | 3,309 | 3,259 | 3,253 | 3,232 | 3,311 | 3,215 |
| Protein residues | 348 | 348 | 346 | 348 | 344 | 343 |
| Nucleotides | 21 | 23 | 23 | 23 | 23 | 23 |
| Solvent/metals | 126 | 52 | 57 | 25 | 137 | 45 |
| Rms bonds, Å | 0.002 | 0.003 | 0.003 | 0.002 | 0.008 | 0.003 |
| Rms angles, ° | 0.48 | 0.54 | 0.61 | 0.46 | 1.04 | 0.49 |
| Clash score | 3.85 | 3.67 | 3.52 | 3.19 | 4.03 | 4.84 |
| Ramachandran favored, % | 97.4 | 96.5 | 96.8 | 96.8 | 96.5 | 97.9 |
| Ramachandran allowed, % | 2.6 | 3.5 | 3.2 | 3.2 | 3.5 | 2.1 |
| Ramachandran outliers, % | 0 | 0 | 0 | 0 | 0 | 0 |
Numbers in parentheses represent statistics from the highest resolution bin. All data were collected at a wavelength of 1.0 Å.
Table S2.
Crystallographic statistics (part 2)
| Complex | rVII | rVIII | rIX | f2I | f2II | f5I |
| PDB ID code | 5V09 | 5V0A | 5V0B | 5V0C | 5V0D | 5V0E |
| Beamline | ALS 12.3.1 SIBYLS | ALS 12.3.1 SIBYLS | ALS 12.3.1 SIBYLS | ALS 12.3.1 SIBYLS | APS SER-CAT 22ID | APS SER-CAT 22ID |
| Resolution range, Å (Highest resolution bin) | 50.00–2.75 (2.80–2.75) | 50.00–2.37 (2.41–2.37) | 50.00–2.63 (2.68–2.63) | 50.00–2.58 (2.73–2.58) | 50.00–2.63 (2.79–2.63) | 50.00–2.74 (2.91–2.74) |
| Space group | P43212 | P43212 | P43212 | P43212 | P43212 | P43212 |
| Unit cell, Å | 73.90 73.90 180.08 | 73.74 73.74 179.71 | 73.76 73.76 182.80 | 71.86 71.86 181.83 | 72.98 72.98 180.61 | 72.78 72.78 181.20 |
| Multiplicity | 6.3 (6.5) | 9.8 (7.0) | 5.9 (5.7) | 3.74 (3.74) | 4.73 (4.72) | 5.14 (5.19) |
| Completeness, % | 99.8 (99.7) | 99.6 (99.2) | 96.6 (99.5) | 99.7 (99.5) | 98.9 (99.7) | 99.5 (97.6) |
| Mean I/σI | 36.46 (3.27) | 33.45 (2.71) | 18.09 (3.09) | 19.82 (2.30) | 16.08 (2.17) | 16.19 (2.33) |
| Rmerge | 5.2 (68.8) | 7.4 (63.5) | 9.9 (67.4) | 4.2 (56.5) | 5.4 (59.3) | 6.0 (57.1) |
| Rmeas | 5.6 (75.0) | 7.8 (68.5) | 10.8 (74.0) | 4.9 (66.2) | 6.0 (66.1) | 6.7 (63.5) |
| CC1/2 | 0.858 | 0.896 | 0.793 | 0.877 | 0.999 (0.820) | 0.999 (0.887) |
| Rwork | 0.2168 (0.2959) | 0.2108 (0.2920) | 0.2072 (0.3232) | 0.2290 (0.3265) | 0.2053 (0.3299) | 0.2038 (0.2726) |
| Rfree | 0.2531 (0.3664) | 0.2469 (0.3532) | 0.2511 (0.3883) | 0.2725 (0.3781) | 0.2368 (0.3416) | 0.2467 (0.3520) |
| No. of nonhydrogen atoms | 3,310 | 3,304 | 3,255 | 3,101 | 3,280 | 3,286 |
| Protein residues | 347 | 346 | 344 | 347 | 348 | 348 |
| Nucleotides | 23 | 23 | 22 | 15 | 23 | 24 |
| Solvent/metals | 105 | 103 | 86 | 56 | 73 | 55 |
| Rms bonds, Å | 0.007 | 0.009 | 0.003 | 0.002 | 0.002 | 0.003 |
| Rms angles, ° | 0.75 | 0.99 | 0.54 | 0.45 | 0.51 | 0.53 |
| Clash score | 4.79 | 3.67 | 3.87 | 5.32 | 3.03 | 3.33 |
| Ramachandran favored, % | 96.5 | 98.5 | 96.2 | 95.9 | 96.5 | 95.3 |
| Ramachandran allowed, % | 3.5 | 1.5 | 3.8 | 4.1 | 3.5 | 4.7 |
| Ramachandran outliers, % | 0 | 0 | 0 | 0 | 0 | 0 |
Numbers in parentheses represent statistics from the highest resolution bin. All data collected at a wavelength of 1.0 Å.
Table S3.
DNA substrates for crystallization
| No. | Complex | Processed strand | Unprocessed strand |
| 1 | rI–rVI, rIX | 5′-(p)-TCG ACT AGC-thio-G-3′ | 5′-C-thio-GC TAG TCG ACA-thio-T-3′ |
| 2 | rVII, rVIII | 5′-(p)-ACG ACT AGC-thio-G-3′ | 5′-C-thio-GC TAG TCG TCA-thio-T-3′ |
| 3 | f2I, f2II | 5′-(p)-CTA GTA CTA GC-thio-G-3′ | 5′-C-thio-GC TAG TAC TCA-thio-T-3′ |
| 4 | f5I | 5′-(p)-TCT CGT CAC TAG C-thio-G-3′ | 5′-C-thio-GC TAG TGA TAC A-thio-T-3′ |
Recessed-End Cleavage.
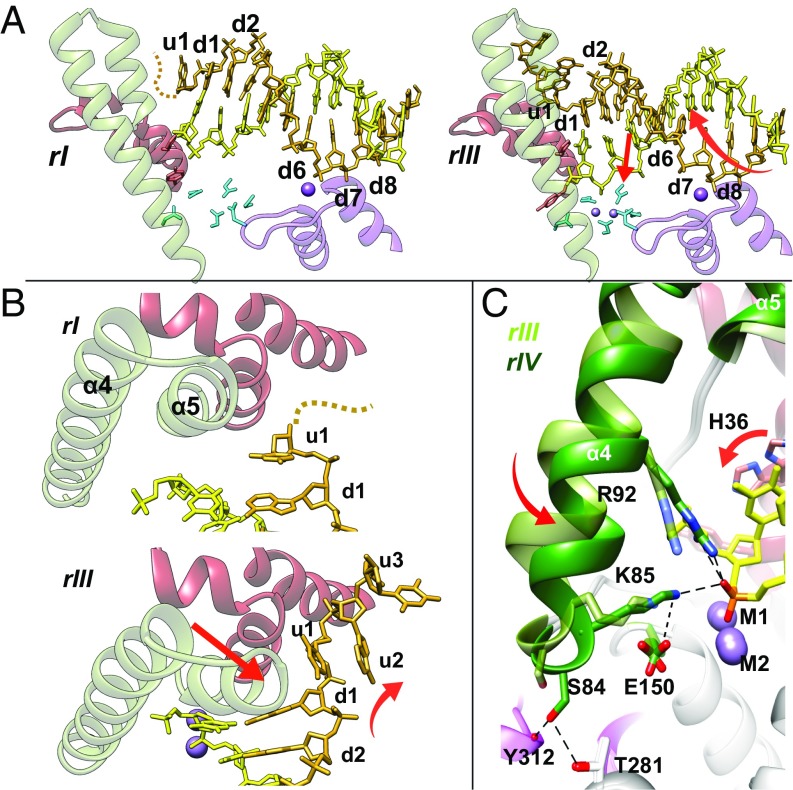
Using a substrate with a 5′ recessed end, we have identified five major stages along the exonucleolytic reaction coordinate (Fig. 2 and Movie S1), each defined by one or more experimental structures with distinct features. Stages 1 and 2 of the reaction were both obtained before initiation by Mg2+ or Mn2+ addition, and are presumed to be in dynamic equilibrium with each other. In stage 1 (rI; Figs. 2 B and C and 3A and Fig. S1), the dsDNA portion of the substrate at the d6 base pair is bound to the H2TH tether, the u1 base contacts α2, but the junction does not form contacts with the protein. Stage 2 (rII structure; Fig. 3) is characterized by two important motions relative to stage 1. First, a combined translation and rotation (screw) of the DNA moves the H2TH contact to the adjacent base (d7) in the dsDNA region (Fig. 3A). This motion “engages” the junction at the d1 base pair with the junction guide residue H36, located on the α2-α3 wedge domain (Fig. 3A). It also organizes the unprocessed ssDNA strand to bend sharply around the wedge domain (16, 17), moving u1 into the “hydrophobic wedge pocket” located between the α2- and α3-helices (Fig. 3 A and B). In the second motion, the mobile arch undergoes the first of two rotations (A), thereby adopting “open” and “closed” conformations in stages 1 and 2, respectively (Fig. 3B and Fig. S1C).
Fig. 3.
Mobile arch and DNA substrate motions before catalysis. (A) DNA motion (red arrows indicate screw motion) transitioning from stage 1 (Left, rI) to 2 (Right, rIII). The double-stranded section of the DNA translocates, shifting the contact to H2TH from d6 to d7. In rIII, the dsDNA-ssDNA junction at the d1 base pair contacts the wedge domain. The unprocessed single strand (u1–u3) is disordered in rI (dashed line) and moves in rIII to place u1 between the α2- and α3-helices, organizing u2-u3 to form an ordered, sharp bend around the α2-α3 wedge. (B) Mobile arch transitions (red arrows, rotation A) from the open (Top; stage 1, rI) to closed (Bottom; stage 2, rIII) conformation. (C) Mobile arch transitions (red arrows, rotation B) from the closed (stage 2, rIII, light green) to clamped (stage 3, rIV, dark green) conformation, placing the scissile bond next to metals M1 and M2 in a catalytically competent geometry. R92 and K85 contribute to transition-state stabilization. E150 always orients K85 (only shown in clamped form for simplicity). The clamped conformation is “latched” by S84, T281, and the Y312 main chain carbonyl.
Fig. S1.
Overview of global coupled motions of hExo1 catalytic domain and DNA to place the scissile bond for catalysis. (A) Superposition of stage 1 (rI, blue) and stage 3 (rIV, orange). Protein clamps (black arrows) toward the DNA while DNA translocates (red arrows) and the scissile bond is seated into the active site. (B) DNA screwing (red arrows) and seating (black arrows) from stage 1 (rI, blue) to stage 3 (rIV, orange). The protein is omitted for clarity except for helix-turn-helix (HTH) and H2TH, which serve as anchor points during this process. (C) Rotation A and B of the α4-α5 mobile arch from stage 1 to stage 3 (red, rI; yellow, rII; cyan, rIV; the helix axes are illustrated as arrows). Rotation A is around the bottom part of α4, whereas rotation B is around the middle part of the helix.
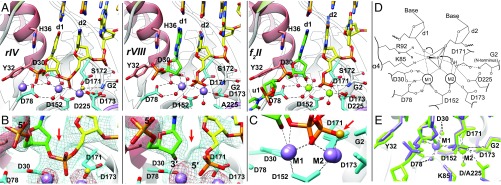
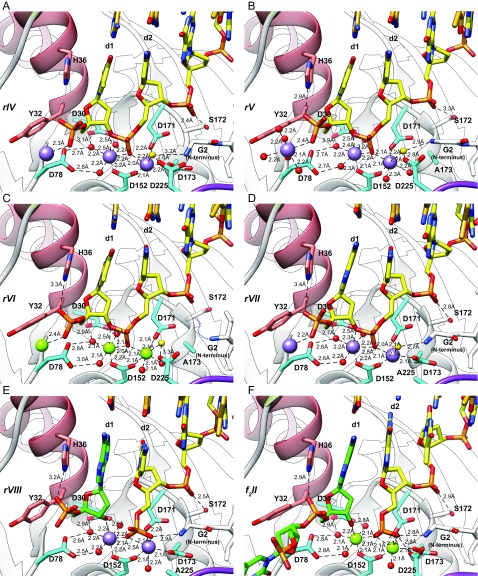
Following initiation by metal addition, we captured a third stage (structures rIV–rVII; Fig. 4 and Figs. S2 and S3) with four approaches that slow down the reaction rate: in the wild-type enzyme with Mn2+ (rIV; Fig. 4 A–C and Fig. S2A), in a D173A mutant with cognate Mn2+ ions (rV; Figs. S2B and S3) or Mg2+ (rVI; Fig. S2C), and in a D225A mutant with Mn2+ (rVII; Fig. S2D). At this stage, the DNA has twisted relative to stage 2. This motion “seats” the scissile phosphodiester bond in the active site next to two fully assembled metal-binding sites and positions a water molecule nearby (Fig. 4 and Figs. S1 A and B and S3). We interpret this stage to represent a DNA substrate just before cleavage.
Fig. 4.
Uncleaved and cleaved substrate complexes. (A) Comparison of uncleaved recessed-end DNA substrate (Left; rIV, wild-type enzyme with Mn2+) with its exonucleolytically cleaved product (Center; rVIII, D225A mutant with Mn2+) and the endonucleolytically cleaved 5′-flap product (Right; f2II, wild-type with Mg2+). Purple spheres, Mn2+; green spheres, Mg2+; gold sphere, attacking water (also Fig. S3); red spheres, waters coordinated to metals; green, cleaved nucleotide or flap products. With exception of the cleaved bond, the active-site structures remain similar before and after cleavage, consistent with states that bracket a cleavage step: Note that the d1 base remains in contact with H36 and all bases remain paired. (B) Simulated annealing omit map (blue, 1σ contour) and anomalous difference maps (red, 3σ contour) unambiguously identify uncleaved (Left, rIV) and cleaved (Right, rVIII) scissile bonds. The red arrow indicates the cleaved bond. (C) Superposition of the phosphate in the uncleaved (rIV) and cleaved (rVIII) scissile bonds shows incorporation of the attacking water (gold, red arrow) into newly formed 5′ phosphate after catalysis. (D) Proposed transition state for an enhanced, in-line, two-metal mechanism. The oxygen atoms of the phosphate in the scissile bond form a trigonal plane, stabilized by the two metals (M1 and M2), K85, and R92. Two bipyramidal apices are formed by the attacking water and 3′ oxygen of the leaving group. M2 and the protein N terminus (G2) orient the attacking water. The invariant carboxylates organize the transition geometry by direct and water-mediated (W) interactions to the metals, K85, N terminus, and attacking water. (E) Two nucleotides flanking the scissile bond of the exonucleolytic (rVIII, purple) and endonucleolytic (f2II, green) product complexes are almost perfectly superimposed.
Fig. S2.
Comparison of substrate and product complexes (same color code as in Fig. 5A, with distances labeled). (A) Active-site view of rIV (wild-type substrate complex with Mn2+ and recessed DNA). Note that the d1 base remains in contact with H36 and all bases remain paired. (B) Active-site view of rV (D173A substrate complex with Mn2+ and recessed DNA). The N terminus, S172, A173, and the attacking water are in different positions compared with rIV. (C) Active-site view of rVI (D173A substrate complex with Mg2+ and recessed DNA). The N terminus is disordered. S172, A173, and the attacking water are in different positions compared with rIV. (D) Active-site view of rVII (D225A substrate complex with Mn2+ and recessed DNA). The N terminus and the attacking water are in slightly different positions compared with rIV. (E) Active-site view of rVIII (D225A product complex with Mn2+ and recessed DNA). The attacking water is incorporated. Note that the d1 base remains in contact with H36 and all bases remain paired. (F) Active-site view of f2II (wild-type product complex with Mg2+ and 5′-flap DNA). The attacking water is incorporated. Note that the geometries are almost identical to rVIII, although the DNA substrate is different.
Fig. S3.
Position of the attacking water observed at 2.15 Å (rV complex, D173A mutant with Mn2+). Purple spheres, Mn2+; gold sphere, attacking water. Simulated annealing omit map (blue, 1σ contour) and anomalous difference maps (red, 5σ contour) identify the attacking water (red arrow) and the two active metals, respectively.
In this catalytically competent DNA substrate complex, the two catalytic Mg2+ cations (or their Mn2+ substitutions), M1 and M2, are ∼3.6 Å apart. M1 is coordinated directly by D152, and through water-mediated interactions with D30 and D78. M2 is coordinated by D152, D171, and D173 (Fig. 4 and Fig. S2). The scissile phosphodiester forms bridging interactions with both metals (Fig. 4 C–E). The water molecule is positioned appropriately for an in-line hydrolytic attack and is held in place by interactions with the N terminus, D225, and M2.
Stage 3 formation is accompanied by a second rotation of the mobile arch that “clamps” the scissile bond into place (B; Fig. 3C and Fig. S1C). The clamped arch is stabilized by hydrogen bonds that “lock” a “latch” region at its base (Fig. 3C) and Fig. S4A). The clamping motion engages two catalytically important residues, R92 and K85, to form ion pairs with a scissile bond oxygen, thereby further stabilizing the developing transition state (Figs. 3C and 4D).
Fig. S4.
Residue interactions and conformational changes around the active site. (A) Interaction of residues coordinating E150 and K85 (yellow, rIII; cyan, rIV). When the α4-α5 mobile arch moves from the closed form (stage 2) to the clamped form (stage 3), K85 points toward the scissile bond and E150 follows this motion. These two residues are stabilized by interaction with Y226 and Q285. (B) Interactions and conformational changes of active-site residues before (yellow, rII) and after (cyan, rIII) the binding of active-site metal ions. Without metals, D173 forms a hydrogen bond with the main chain of I3. After the binding of metals, this bond is broken and D173 moves and coordinates M2. The N terminus (G2) and D152 also alternate conformations with metal binding.
In stage 3, the 5′ phosphate of the processed strand is positioned at the base of the mobile arch, where it is held in place by three interactions. First, it forms a hydrogen bond with the “junction guide” residue H36, which has flipped into a down conformation, where it remains stacked with d1 (Fig. 4A and Fig. S2). Second, the “arch guide” residue, Y32, flips into a down conformation, permitting steric access of the 5′-terminal phosphate to the active site (Fig. 4A and Fig. S2). Finally, a third metal binds the 5′ phosphate. The coordination sphere of this metal is completed by additional water-mediated interactions at the mobile arch base (Fig. 4A and Fig. S2).
We have also captured an intermediate along the path of stage 3 formation (rIII; Fig. 2 A and C). In this structure, the mobile arch position is open, not clamped, and the latch is unlocked. The two metals are both present, and the arch guide residue, Y32, has rotated down (Fig. 2C and Fig. S4B). However, the scissile bond is not positioned for cleavage: The metal that binds the 5′ phosphate is absent, and the two catalytic residues, R92 and K85, remain disengaged. Although the hydrolytic water molecule is present, it is not yet positioned for in-line attack.
Following stage 3, we captured a fourth stage that corresponds to the cleaved substrate with both products still in place (rVIII; Fig. 4 A–C and Fig. S2E). This complex was captured using Mn2+ in the D225A mutant. With wild-type enzyme and Mg2+ or Mn2+, we obtain mixtures of both substrate and product in our experimental time series. In stage 4, neither the protein nor DNA has moved, but changes in the electron density around the scissile bond are consistent with its cleavage and incorporation of the attacking water into the newly formed 5′ phosphate of the resulting product (Fig. 4B). The M1 and M2 metals coordinate the termini of the two products: M2 binds the third oxygen (formerly the attacking water) of the newly formed 5′ phosphate, and M1 binds the 3′ hydroxyl of the single-nucleotide product.
Product Release.
In the final stage that we were able to capture (structure rIX; Fig. 2), we observed nucleotide monophosphate product release while retaining binding of the rest of the DNA substrate, accompanied by a partial reversal of the mobile arch motions. This state is challenging to obtain, because in prolonged experimental timelines, crystals become disordered as synchrony is lost and eventually dissolve. In state 5, the mobile arch has undergone the inverse B rotation and is now in its closed conformation. Consequently, the two catalytic residues, R92 and K85, are disengaged from the DNA. The Y32 arch guide has flipped into its up position, reinstating the steric block to the active site. The dsDNA junction, newly formed at d2, is disengaged from the H36 guide.
Flap Threading.
Complexes with 5-nt (f5I) and 2-nt (f2I) flap substrates were captured in the absence of Mg2+. In both, the dsDNA is loosely bound to the H2TH tether. In f2I, neither processed nor unprocessed DNA single strands were observed, and the junction is not engaged. The protein conformation and dsDNA position therefore closely resemble the stage 1 recessed substrate (rI).
In f5I, four of the five 5′-flap nucleotides (f2–f4) are threaded through an aperture within the mobile arch (Figs. 5 A, B, and D and Fig. S5) in which the helices have parted slightly to form a small aperture. Both guide residues, Y32 and H36, are in their up conformation. The f2-f3 nucleotides are positioned inside the arch. Their bases are flipped relative to the duplex region, pointing to the bottom of the arch. Their phosphates point toward a cluster of arginines in α5. The f4 nucleotide is positioned beyond the arch and is not flipped. The f5 base is disordered. The f1 nucleotide forms a G-T mismatch with the u1 base of the unprocessed strand. The dsDNA junction, formed here by the f1-u1 mismatch, is disengaged. The expected scissile bond (between d1 and d2) is not seated in the active site. Taken together, these results suggest that f5I corresponds to an intermediate in which the flap base proximal to the junction (f1) remains to be threaded.
Fig. 5.
Threading of uncleaved and cleaved 5′ flaps through an aperture formed by the mobile arch (green) and α2 (red). (A and B) Two views of the uncleaved, partially threaded, five-residue 5′ flap (blue). (A) View through the aperture into the active site. Four positively charged residues (R92, R93, R95, and R96; only R96 is shown) are located on the face of α4 pointing to the ssDNA and may stabilize threaded flap nucleotides. (B) Side view tracing the path of the flap out of the double-stranded region through the aperture. The red arrow indicates the expected scissile bond (d1-d2). The unprocessed strand is partially disordered (dashed line). Both Y32 and H36 are in their up conformations and disengaged. The f2 and f3 bases straddle either side of the aperture. At the entrance of the arch, u1 and f1 form a mispair. The f2 and f3 bases point in the opposite direction (“flipped”) relative to f1. The f4 base does not form extensive contacts with the protein, and f5 is disordered. R93 can interact with poly(ADP ribose), and is located adjacent to the threaded 5′-flap path. (C) Side view of the cleaved 5′-flap f2II. The f1 base has threaded through the aperture and is flipped relative to d1 (part of the cleaved flap), and f2 is disordered. The aperture straddles f1 and d1. (D) Threading and endonucleolytic cleavage. (Left) Initially, the dsDNA region is loosely tethered by H2TH (f2I). (Center) Next, the processed strand threads through the mobile arch (f5). (Right) Following dsDNA junction seating, the scissile bond is guided into the assembled active site and cleaved (f2II).
Fig. S5.
Side view tracing the path of the flap through the narrow space under arch in f5I (protein surface is shown, but the bottom part of α4 is omitted for clarity). The f2 base is flipped and contacts the active-site residues. The f3 base is also flipped and is in contact with the hydrophobic pocket formed between α2, α6, and β3. The C-terminal region forms a steric block at the arch exit. The f4 base does not form extensive contacts with the protein and is pointing to the normal orientation.
Flap Cleavage.
We captured a threaded, endonucleolytically cleaved, 2-nt flap complex, f2II, with its scissile bond placed in the active site (Fig. 5 C and D and Fig. S2F). In this structure, the mobile arch is clamped. We calculated electron density maps for both uncleaved and cleaved models. In the former, a large (5.7-sigma contour level) negative difference electron density peak was observed at the scissile bond, which was absent in the latter, consistent with a cleaved product complex containing a 5′-phosphate. This threaded complex has therefore been trapped at stage 4, as defined by the recessed-end cleavage cycle analysis. As with recessed-ended substrates, cleavage occurred 1 bp inside the duplex region (Fig. 5B). Accordingly, the product is a three-base, single-stranded 5′ flap (d1-f1-f2): d1 remains paired with the unprocessed strand, d1 and f1 are inside the arch aperture, and the f2 base is disordered. The two nucleotides flanking scissile bonds in this complex and the exonucleolytic cleavage product of stage 4 (rIV) are near-perfectly superimposed (Fig. 4E and Fig. S2 E and F). The environment of the scissile bond therefore is identical in 5′-flap cleavage and recessed-end substrates.
Conclusions
Shared Catalytic Mechanism.
We trapped structures of DNA complexes representing the exonucleolytic reaction just before and directly after the chemical cleavage step, and also captured an endonucleolytic cleavage product. In both recessed-end and 5′-flap substrates the scissile phosphodiester bond is located 1 bp in from the double-stranded junction (Fig. 4 and Fig. S2), consistent with the dominant cleavage site identified in solution (7, 8, 17). This bond is positioned between two metals in the active site that are ∼3.6 Å apart, which is the correct distance for catalysis by a two-metal mechanism, as described originally for the 3′-5′ exonucleolytic reaction of Klenow fragment (KF) DNA polymerase (22, 23). In substrate complexes, a water molecule is positioned for in-line attack of the phosphodiester bond, whereas it is absent in products, having been incorporated into the newly formed 5′ phosphate (Fig. 4 A–C). The structural similarities to KF enabled us to construct a model of the transition state (Fig. 4D), which revealed the clamped mobile arch transiently positions two key catalytic residues (16, 24), K85 and R92, to stabilize the negatively charged trigonal plane formed by three oxygens in the transition state. The two bases flanking the scissile bond in the recessed-end and 5′-flap products are placed identically in the active site (Fig. 4E and Fig. S2 E and F). These observations therefore clearly establish that exo- and endonucleolytic cleavage share a common reaction mechanism in which nucleolysis is catalyzed by the well-known two-metal, in-line hydrolysis mechanism (22, 23), but is elaborated by additional features that couple mobile arch motions with transition-state stabilization.
Substrate Guidance.
The processed strands of recessed ends and 5′ flaps are positioned in the active site by a sequence of successive, interlocking conformational changes of the mobile arch, the two guide residues, and the DNA substrate (Fig. 2). In the fully assembled active site, the mobile arch adopts two different forms: recessed-end 5′ phosphates bind to a third Mg2+ coordinated to its bottom, and 5′ flaps thread through an aperture that opens up in its interior. Together, this structural plasticity and the controlled motions enable the enzyme to guide topologically disparate substrate termini into a common reaction mechanism.
Conformational Entropy Reduction.
Decrease in the substrate DNA conformational diversity is a striking feature of the reaction as it proceeds toward its unique transition state. In stage 1 of the observed timeline, recessed ends and unthreaded 5′ flaps are both loosely held by the H2TH anchor. Their dsDNA junctions are disengaged from the protein, and their processed and unprocessed strands are disordered (e.g., rI, f2I). This disengaged state also accommodates partially threaded flaps (e.g., f5I), consistent with single-molecule studies of cleavage kinetics in FEN1 (18), which indicate that the threading reaction is catalyzed at this stage. The base pair mismatch observed in f5I between the unprocessed and partially threaded flap strands at the entry of the mobile arch aperture suggests that threading is catalyzed, in part, by transient binding interactions between the two strands, thereby reducing flap conformational entropy.
Subsequent engagement of the dsDNA junction with the protein and positioning of the processed strand in the active site dramatically reduce substrate conformational entropy. Unprocessed single strands become ordered and are placed in the wedge domain hydrophobic pocket. At stage 3 (precleavage) and stage 4 (postcleavage), the processed ends of both recessed-end and 5′-flap substrates adopt a single conformation within the active site.
Flap threading is an example of a “fly-casting” mechanism that enhances the kinetics of searching for states with low conformational entropy. An initial binding event establishes a search volume without restricting internal motions. Subsequent formation of intermediates progressively reduces conformational degrees of freedom (25). The loosely tethered, conformationally diverse complexes (rI and f2I) represent the initial, high-entropy encounter complex. Mismatch formation in f5I may represent a postulated intermediate that narrows the “binding funnel” of available conformations as threading proceeds. These structures therefore present a remarkable experimental manifestation of intermediates proposed in the fly-casting model for entropy reduction.
Mobile Arch Motions.
We identified three mobile arch conformational states: open, closed, and clamped (the open and closed states are ensembles around an average structure, and the clamped state is near unique). Each is associated with different substrate conformational degrees of freedom. In the open state, the DNA is held loosely. In the closed state, the dsDNA junction is engaged and the two single strands are ordered. In the clamped state, the active site is fully assembled, positioning the scissile bond, attacking water, two metals, and mobile catalytic residues in a catalytically competent geometry. Mobile arch motions therefore progressively restrict substrate conformational degrees of freedom as the processed strand is guided into the active site.
In other family members, the mobile arch also is important for substrate recognition and processing, and undergoes disorder/order transitions in FEN1 and in GEN1 (17, 21, 26). Arch mobility enables hExo1 activity to be manipulated allosterically. For instance, poly(ADP ribose)-binding of R93 within the arch aperture is an important known control point for hExo1 (27, 28).
Guide Residues.
The side chains of H36 and Y32 switch between “up” and “down” conformations. These residues are located on the α2-helix of the immobile wedge domain, adjacent to the mobile arch. H36 is a principal attachment point for seated dsDNA junctions, interacting with the d1 base of the processed strand, and H36A is 150-fold reduced in catalytic activity (16). The Y32 conformation controls 5′-end binding, and Y32A is 20-fold reduced in activity (16). As the mobile arch transitions from the closed state to the clamped state and the seated substrate rotates into the active site, H36 flips from the up conformation to the down conformation. This motion maintains the contact with d1, and therefore is a key contribution to the proper seating of the scissile bond, which is located between d1 and d2. Formation of the clamped mobile arch conformation also requires that Y32 flips to its down position either to maintain flap threading or to enable steric accessibility of the recessed-end 5′ phosphate-binding site. Both H36 and Y32 therefore function as steric guides that couple mobile arch motions to seating of substrates.
Processing Fidelity.
The clamped mobile arch conformation positions the two key catalytic residues K85 and R92, which are important for catalysis (16, 24). This state cannot form with incorrectly or incompletely bound substrates such as partially threaded 5′ flaps, in which the dsDNA junction is not seated and the guide residues are not triggered. Coupling transition stabilization to arch motions therefore enhances processing fidelity by suppressing inappropriate or premature substrate cleavage.
Processivity.
Nucleotide monophosphate product release is accompanied by partial reversal of the conformational sequence without relinquishing bound DNA. The mobile arch unclamps, and the dsDNA junction disengages, resulting in a loosely held substrate tethered at H2TH. This resetting motion suggests a structural basis for the observed processivity of hExo1 exonuclease activity (29, 30). Reseating would translocate the protein along the DNA by 1 bp, analogous to the motion of the substrate engagement step that followed initiation of the experimental timeline (Movie S2).
Evolution of Substrate Guidance Mechanisms.
The residues encoding the enhanced two-metal mechanism are highly conserved in the RAD2/XPG family. These residues include seven carboxylates that coordinate the two metals, either directly (D152, D171, and D173) or through a bound water (D30 and D78); D225, which orients the attacking water; and E150, which undergoes a conformational change in the clamped conformation, orienting K85. Where tested, their mutagenesis results in loss of activity (16, 31). The K85 and R92 that encode transient stabilization of the transition state are highly conserved, whereas the rest of the mobile arch sequence is not. This pattern of conservation might be expected if catalytic and processing fidelity mechanisms are common to all members, whereas the arch encodes substrate guidance specializations.
Materials and Methods
Crystallization and in Crystallo Enzyme Reaction.
The wild-type and mutant proteins were prepared as described previously (16). Oligonucleotides (Integrated DNA Technologies, Inc.; Table S3) were prepared as previously described (16). The 5′ ends of processed strands were terminated with a phosphate; all other 5′ and 3′ termini were protected with a phosphorothioate linkage. Complexes were prepared by combining 150 μM with 200 μM DNA substrate and incubating at 4 °C for 30 min. Crystals were obtained by sitting-drop vapor diffusion at 17 °C, mixing 100 μL of complex with 100 μL of precipitant solution [100 mM NaOAc (pH 7.0), 10 mM KCl, 2–4% PEG 4000]. Most crystals with recessed-end DNA appeared within 1 h after setting up trays, and crystals with 5′-flap substrates appeared after 2–4 d. Sixteen hours after their appearance, crystals were transferred into cryoprotectant solution (mother liquor with 35% ethylene glycol) for 10 min. For time-resolved experiments, the reaction was initiated by transfer into cryoprotectant supplemented with 10–20 mM MgCl2 or MnCl2. Crystals were flash-frozen in liquid nitrogen to stop the reaction at varying time intervals (∼1–600 s).
Data Collection, Structure Determination, and Refinement.
Diffraction data were collected at 100 K at the Advanced Photon Source (Argonne National Laboratory; beamlines 22-ID, SER-CAT) or the Advanced Light Source (Lawrence Berkeley National Laboratory; beamline 12.3.1, SIBYLS); all crystals diffracted to 2.2–2.9 Å resolution. Structures were determined as described previously (SI Materials and Methods and Tables S1 and S2).
SI Materials and Methods
Protein Expression and Purification.
The wild-type and D173A mutant hExo1 catalytic domain (residues 1–352) have been described previously (16). The D225A mutation was introduced as described elsewhere (16). The D173A mutant was expressed and purified as previously described (16). The wild-type and D225A mutant were expressed in Escherichia coli BL21(DE3)RIL, transformed with expression constructs. Cells were grown in 10 L of Luria broth (100 μg/mL ampicillin) for 16 h at 16 °C after induction with 0.1 mM isopropyl-β-d-thiogalactopyranoside, harvested by centrifugation (4,000 × g, 4 °C, 30 min), resuspended in lysis buffer [50 mM Hepes-NaOH (pH 7.5), 100 mM NaCl, 10 mM KCl, 10 mM MgCl2, 5 mM 2-Mercaptoethanol, 1 mM EDTA, 5% glycerol], and lysed with a cell cracker (Microfluidics). A cleared lysate was prepared by centrifugation (18,500 × g, 4 °C, 30 min), loaded onto two tandem 5-mL HisTrap HP Ni Sepharose columns (GE Healthcare Life Sciences) preequilibrated with wash buffer (lysis buffer with 300 mM NaCl and 20 mM imidazole), and eluted with 300 mM imidazole. Fractions containing pure protein were pooled and cleaved with tobacco etch virus protease (4 °C, 12 h). The cleaved mixture was loaded onto a Ni Sepharose column again to remove the hexahistidine purification tag. The eluate was loaded onto a 5-mL HiTrap SP FF column (GE Healthcare Life Sciences) equilibrated in low-ion buffer [50 mM Hepes (pH 7.5), 100 mM NaCl, 10 mM KCl, 1 mM MgCl2, 5 mM DTT, 1 mM EDTA, 5% glycerol]. Fractions containing pure protein were then eluted with high-ion buffer (low-ion buffer plus 500 mM NaCl). The purified protein in storage buffer [25 mM Hepes-NaOH (pH 7.0), 125 mM NaCl, 10 mM KCl, 2 mM TCEP, 0.1 mM EDTA, 0.1% glycerol] was concentrated to ∼15 mg/mL, flash-frozen in liquid nitrogen, and stored at −80 °C.
Oligonucleotide Substrates.
Oligonucleotides (Integrated DNA Technologies, Inc.) (Table S3) were prepared as previously described (16). The 5′ ends of processed strands were terminated with a phosphate, and all other 5′ and 3′ termini were protected with a phosphorothioate linkage to prevent nuclease digestion of the ends.
Crystallization and in Crystallo Enzyme Reaction.
Complexes were prepared by combining 150 μM with 200 μM DNA substrate and incubating at 4 °C for 30 min. Crystals were obtained by sitting-drop vapor diffusion at 17 °C, mixing 100 μL of complex with 100 μL of precipitant solution [100 mM NaOAc (pH 7.0), 10 mM KCl, 2–4% PEG 4000]. Most crystals with recessed-end DNA appeared within 1 h after setting up trays, and crystals with 5′-flap substrates appeared after 2–4 d. Sixteen hours after appearance, crystals were transferred into cryoprotectant solution (mother liquor with 35% ethylene glycol) for 10 min. For time-resolved experiments, the reaction was initiated by transfer into cryoprotectant supplemented with 10–20 mM MgCl2 or MnCl2. Crystals were flash-frozen in liquid nitrogen to stop the reaction at varying time intervals (rIII, ∼1 s; rIV, ∼5 s; rV, 30 s; rVI, 30 s; rVII, 1 min; rVIII, 6 h; rIX, 3 min; and f2II, 3 min).
Data Collection, Structure Determination, and Refinement.
Diffraction data were collected at 100 K at the Advanced Photon Source (Argonne National Laboratory; beamlines 22-ID, SER-CAT), or the Advanced Light Source (Lawrence Berkeley National Laboratory; beamline 12.3.1, SIBYLS); all crystals diffracted to 2.2–2.9 Å resolution. Data were scaled in the P43212 space group using HKL2000 (rI–rIX) or XDS (f2I-f2II, f5I) (32, 33). Data for manganese-containing complexes were collected at λ = 1.0 Å. Previously published structures [Protein Data Bank (PDB) ID code 3QEA] were used as models for molecular replacement phasing (16). Initial low-resolution refinement was carried out using deformable elastic network restraints (34). PDB ID code 3QEA served as a reference model. After initial refinement, DNA was built manually into the electron density maps. Manganese positions were identified using phased anomalous difference maps. Structures were refined using PHENIX with a maximum likelihood target and phase probability distribution (34). Movies S1 and S2 were generated using UCSF Chimera from complexes rI–rIV, rVIII, and rIX.
Supplementary Material
Acknowledgments
This work was supported, in part, by NIH Grants R01 GM091487 and P01 CA092584.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 5UZV, 5V04, 5V05, 5V06, 5V07, 5V08, 5V09, 5V0A, 5V0B, 5V0C, 5V0D, and 5V0E).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1704845114/-/DCSupplemental.
References
- 1.Genschel J, Bazemore LR, Modrich P. Human exonuclease I is required for 5′ and 3′ mismatch repair. J Biol Chem. 2002;277:13302–13311. doi: 10.1074/jbc.M111854200. [DOI] [PubMed] [Google Scholar]
- 2.Nimonkar AV, Ozsoy AZ, Genschel J, Modrich P, Kowalczykowski SC. Human exonuclease 1 and BLM helicase interact to resect DNA and initiate DNA repair. Proc Natl Acad Sci USA. 2008;105:16906–16911. doi: 10.1073/pnas.0809380105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Potenski CJ, Niu H, Sung P, Klein HL. Avoidance of ribonucleotide-induced mutations by RNase H2 and Srs2-Exo1 mechanisms. Nature. 2014;511:251–254. doi: 10.1038/nature13292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qiu J, Qian Y, Chen V, Guan MX, Shen B. Human exonuclease 1 functionally complements its yeast homologues in DNA recombination, RNA primer removal, and mutation avoidance. J Biol Chem. 1999;274:17893–17900. doi: 10.1074/jbc.274.25.17893. [DOI] [PubMed] [Google Scholar]
- 5.Ip SC, et al. Identification of Holliday junction resolvases from humans and yeast. Nature. 2008;456:357–361. doi: 10.1038/nature07470. [DOI] [PubMed] [Google Scholar]
- 6.Evans E, Fellows J, Coffer A, Wood RD. Open complex formation around a lesion during nucleotide excision repair provides a structure for cleavage by human XPG protein. EMBO J. 1997;16:625–638. doi: 10.1093/emboj/16.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee BI, Wilson DM., 3rd The RAD2 domain of human exonuclease 1 exhibits 5′ to 3′ exonuclease and flap structure-specific endonuclease activities. J Biol Chem. 1999;274:37763–37769. doi: 10.1074/jbc.274.53.37763. [DOI] [PubMed] [Google Scholar]
- 8.Keijzers G, Bohr VA, Rasmussen LJ. Human exonuclease 1 (EXO1) activity characterization and its function on flap structures. Biosci Rep. 2015;35:e00206. doi: 10.1042/BSR20150058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tomimatsu N, et al. Exo1 plays a major role in DNA end resection in humans and influences double-strand break repair and damage signaling decisions. DNA Repair (Amst) 2012;11:441–448. doi: 10.1016/j.dnarep.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vallur AC, Maizels N. Activities of human exonuclease 1 that promote cleavage of transcribed immunoglobulin switch regions. Proc Natl Acad Sci USA. 2008;105:16508–16512. doi: 10.1073/pnas.0805327105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vallur AC, Maizels N. Complementary roles for exonuclease 1 and Flap endonuclease 1 in maintenance of triplet repeats. J Biol Chem. 2010;285:28514–28519. doi: 10.1074/jbc.M110.132738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levikova M, Cejka P. The Saccharomyces cerevisiae Dna2 can function as a sole nuclease in the processing of Okazaki fragments in DNA replication. Nucleic Acids Res. 2015;43:7888–7897. doi: 10.1093/nar/gkv710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sparks JL, et al. RNase H2-initiated ribonucleotide excision repair. Mol Cell. 2012;47:980–986. doi: 10.1016/j.molcel.2012.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wei K, et al. Inactivation of Exonuclease 1 in mice results in DNA mismatch repair defects, increased cancer susceptibility, and male and female sterility. Genes Dev. 2003;17:603–614. doi: 10.1101/gad.1060603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bayram S. The exonuclease 1 Glu589Lys gene polymorphism and cancer susceptibility: Evidence based on a meta-analysis. Asian Pac J Cancer Prev. 2014;15:2571–2576. doi: 10.7314/apjcp.2014.15.6.2571. [DOI] [PubMed] [Google Scholar]
- 16.Orans J, et al. Structures of human exonuclease 1 DNA complexes suggest a unified mechanism for nuclease family. Cell. 2011;145:212–223. doi: 10.1016/j.cell.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsutakawa SE, et al. Human flap endonuclease structures, DNA double-base flipping, and a unified understanding of the FEN1 superfamily. Cell. 2011;145:198–211. doi: 10.1016/j.cell.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sobhy MA, Joudeh LI, Huang X, Takahashi M, Hamdan SM. Sequential and multistep substrate interrogation provides the scaffold for specificity in human flap endonuclease 1. Cell Reports. 2013;3:1785–1794. doi: 10.1016/j.celrep.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 19.Patel N, et al. Flap endonucleases pass 5′-flaps through a flexible arch using a disorder-thread-order mechanism to confer specificity for free 5′-ends. Nucleic Acids Res. 2012;40:4507–4519. doi: 10.1093/nar/gks051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dervan JJ, et al. Interactions of mutant and wild-type flap endonucleases with oligonucleotide substrates suggest an alternative model of DNA binding. Proc Natl Acad Sci USA. 2002;99:8542–8547. doi: 10.1073/pnas.082241699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.AlMalki FA, et al. Direct observation of DNA threading in flap endonuclease complexes. Nat Struct Mol Biol. 2016;23:640–646. doi: 10.1038/nsmb.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beese LS, Steitz TA. Structural basis for the 3′-5′ exonuclease activity of Escherichia coli DNA polymerase I: A two metal ion mechanism. EMBO J. 1991;10:25–33. doi: 10.1002/j.1460-2075.1991.tb07917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Steitz TA, Steitz JA. A general two-metal-ion mechanism for catalytic RNA. Proc Natl Acad Sci USA. 1993;90:6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pickering TJ, Garforth SJ, Thorpe SJ, Sayers JR, Grasby JA. A single cleavage assay for T5 5′–>3′ exonuclease: Determination of the catalytic parameters for wild-type and mutant proteins. Nucleic Acids Res. 1999;27:730–735. doi: 10.1093/nar/27.3.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shoemaker BA, Portman JJ, Wolynes PG. Speeding molecular recognition by using the folding funnel: The fly-casting mechanism. Proc Natl Acad Sci USA. 2000;97:8868–8873. doi: 10.1073/pnas.160259697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y, et al. Crystal structure of a eukaryotic GEN1 resolving enzyme bound to DNA. Cell Reports. 2015;13:2565–2575. doi: 10.1016/j.celrep.2015.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang F, Shi J, Chen SH, Bian C, Yu X. The PIN domain of EXO1 recognizes poly(ADP-ribose) in DNA damage response. Nucleic Acids Res. 2015;43:10782–10794. doi: 10.1093/nar/gkv939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheruiyot A, et al. Poly(ADP-ribose)-binding promotes Exo1 damage recruitment and suppresses its nuclease activities. DNA Repair (Amst) 2015;35:106–115. doi: 10.1016/j.dnarep.2015.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Myler LR, et al. Single-molecule imaging reveals the mechanism of Exo1 regulation by single-stranded DNA binding proteins. Proc Natl Acad Sci USA. 2016;113:E1170–E1179. doi: 10.1073/pnas.1516674113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Genschel J, Modrich P. Mechanism of 5′-directed excision in human mismatch repair. Mol Cell. 2003;12:1077–1086. doi: 10.1016/s1097-2765(03)00428-3. [DOI] [PubMed] [Google Scholar]
- 31.Lee Bi BI, Nguyen LH, Barsky D, Fernandes M, Wilson DM., 3rd Molecular interactions of human Exo1 with DNA. Nucleic Acids Res. 2002;30:942–949. doi: 10.1093/nar/30.4.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 34.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.