Significance
Duchenne muscular dystrophy (DMD) is a genetic X-linked neuromuscular disease characterized by severe muscle degeneration caused by absence of the protein dystrophin. In the golden retriever muscular dystrophy dog model of DMD, two atypical dogs exhibited significantly milder phenotypes compared with their severely affected littermates despite lacking dystrophin. These two notable dogs were found to have decreased expression of phosphatidylinositol transfer protein-α (PITPNA) compared with severely affected dogs. Decreased expression of PITPNA in dystrophin-deficient zebrafish and in human DMD myogenic cells ameliorated several aspects of the dystrophic phenotype, improving muscle structure, increasing survival, and increasing levels of phosphorylated Akt. Our findings present PITPNA as a genetic modifier of DMD and potential target for future therapies.
Keywords: Duchenne muscular dystrophy, genetic modifier, phosphatidylinositol transfer protein-α, skeletal muscle
Abstract
Duchenne muscular dystrophy (DMD) is a progressive muscle wasting disease caused by X-linked inherited mutations in the DYSTROPHIN (DMD) gene. Absence of dystrophin protein from the sarcolemma causes severe muscle degeneration, fibrosis, and inflammation, ultimately leading to cardiorespiratory failure and premature death. Although there are several promising strategies under investigation to restore dystrophin protein expression, there is currently no cure for DMD, and identification of genetic modifiers as potential targets represents an alternative therapeutic strategy. In a Brazilian golden retriever muscular dystrophy (GRMD) dog colony, two related dogs demonstrated strikingly mild dystrophic phenotypes compared with those typically observed in severely affected GRMD dogs despite lacking dystrophin. Microarray analysis of these “escaper” dogs revealed reduced expression of phosphatidylinositol transfer protein-α (PITPNA) in escaper versus severely affected GRMD dogs. Based on these findings, we decided to pursue investigation of modulation of PITPNA expression on dystrophic pathology in GRMD dogs, dystrophin-deficient sapje zebrafish, and human DMD myogenic cells. In GRMD dogs, decreased expression of Pitpna was associated with increased phosphorylated Akt (pAkt) expression and decreased PTEN levels. PITPNA knockdown by injection of morpholino oligonucleotides in sapje zebrafish also increased pAkt, rescued the abnormal muscle phenotype, and improved long-term sapje mutant survival. In DMD myotubes, PITPNA knockdown by lentiviral shRNA increased pAkt and increased myoblast fusion index. Overall, our findings suggest PIPTNA as a disease modifier that accords benefits to the abnormal signaling, morphology, and function of dystrophic skeletal muscle, and may be a target for DMD and related neuromuscular diseases.
Duchenne muscular dystrophy (DMD) is a progressive, X-linked muscle wasting disease caused by mutations in the DYSTROPHIN gene (1, 2). Absence of dystrophin protein from the muscle sarcolemma disrupts the link between the cytoskeleton and extracellular matrix, causing a multitude of pathological effects on muscle mechanics, signaling, and metabolic pathways. These consequences render myofibers susceptible to contraction-induced injury and cause severe muscle degeneration, fibrosis, and inflammation. Patients with DMD typically lose ambulation by age 12, and cardiorespiratory failure leads to premature death by the third decade of life (3). Despite advances in palliative support and ongoing efforts to restore dystrophin expression, there is no cure for DMD. Therefore, identification of potential genetic modifiers, which could be targets for disease therapy and discovery, are of significant interest.
Identification of genetic modifiers that reduce the pathogenic features of DMD is an emerging gateway to new therapeutic targets. Modifiers identified include osteopontin, encoded by the SPP1 gene, which is highly up-regulated in dystrophic human and mouse muscle (4, 5), and LTBP4, which regulates the availability of latent TGFβ and may mediate dilated cardiomyopathy in patients with DMD (6). Genetic ablation of osteopontin in mdx mice results in dramatic reduction of fibrosis and improvement of strength and pathophysiology of dystrophic muscle (4). Polymorphisms in both the human SPP1 and LTBP4 genes have been shown to correlate with outcomes in DMD, including rate of disease progression, loss of ambulation, and grip strength (7–10). Most recently, a common null polymorphism (R577X) in ACTN3 was found to result in significantly reduced muscle strength in young, ambulant patients with DMD, but protect from stretch-induced eccentric damage with age in α-actinin-3/mdx double knockout mice (11). In addition to these modifiers, previous work in our laboratory revealed a spontaneously occurring mutation in the promoter of Jagged1 that increased its expression in “escaper” golden retriever muscular dystrophy (GRMD) dogs, which presented with remarkably mild symptoms despite being dystrophin deficient (12). Normally, GRMD dogs exhibit severe pathology similar to patients with DMD, including early progressive muscle degeneration, fibrosis, and elevated serum creatine kinase (CK) (13). However, the two escaper dogs identified in a Brazilian GRMD colony remained ambulatory and had normal lifespans (14). In vitro analysis showed that escaper dog muscle cells had increased proliferation, and overexpression of jagged1 in dystrophin-deficient zebrafish rescued the muscle phenotype (15). Further microarray analysis of GRMD dog muscle biopsies also revealed phosphatidylinositol transfer protein-α (PITPNA) as differentially expressed in the escaper dogs compared with severely affected dogs, posing it as a potential disease modifier (15). By identifying and further investigating the mechanistic links between these genetic modifiers and dystrophic pathology, we reveal avenues for potential therapeutics for DMD.
PITPs mediate the transfer of phosphatidylinositol between two membrane compartments, thereby regulating lipid metabolism, membrane trafficking, and signaling in eukaryotic cells (16). PITPNA has largely been studied in neurons, where it is an essential component of PLC signaling and neurite outgrowth, and morpholino-mediated pitpnaa knockdown in zebrafish embryos leads to dose-dependent defects in motor neuron axons (17). Pitpnaa is the originally identified gene encoding Pitpna in zebrafish, and a second duplicate copy has just recently been discovered, termed pitpnab. In mice, loss-of-function mutations of the gene encoding PITPNA cause dose-sensitive phenotypes, including neurological dysfunction, spinocerebellar neurodegeneration, and premature death (18). PITPNA has also been shown to control extension of laminin-dependent axonal processes by regulating phosphatidylinositol 3-kinase (PI3K)-dependent signaling events during neurite remodeling (19). The PI3K complex catalyzes the production of lipid molecules that trigger the attachment of Akt to the plasma membrane, where it subsequently becomes fully activated by phosphorylation at Ser473 (20). PITPNA modulation of PI3K/Akt signaling is of interest, given the known central role of Akt in cell growth, metabolism, and apoptosis in addition to previous studies showing Akt1 to induce muscle hypertrophy (21) and differentiation of myoblasts into fused myofibers (22). In mdx mice, overexpression of constitutively active Akt results in improved muscle force generation (23, 24) and Akt activation associated with overexpression of the transmembrane protein sarcospan has also been shown to ameliorate dystrophic pathology (25).
In the present study, we investigated PITPNA as a modulator of dystrophic pathology and associated aberrant signaling in three DMD models: the GRMD dog, dystrophin-deficient sapje zebrafish (Danio rerio), and primary human DMD patient myogenic cells. We report that PITPNA repression is associated with decreased PTEN and increased phosphorylated-Akt (pAkt) expression levels. In addition, morpholino-mediated pitpnaa knockdown rescues the abnormal muscle structure normally present in homozygous-null sapje zebrafish and improves long-term survival. Finally, PITPNA knockdown by lentiviral shRNA in human DMD cells increases myoblast fusion index. These data suggest that decreased expression of PITPNA ameliorates the pathological consequences of dystrophin deficiency and may be a therapeutic entry point for the treatment of DMD.
Results
GRMD Escaper Dogs Have Decreased PITPNA and PTEN Expression.
In a Brazilian GRMD colony, two escaper dogs that eluded many of the pathological consequences of dystrophin deficiency and exhibited a very mild phenotype were identified. To identify potential compensatory mechanisms in these dogs, we compared gene expression by Agilent mRNA SurePrint Canine arrays between RNA isolated from the muscles of the two escaper dogs, four related severely affected GRMD dogs, and three normal dogs at 2 y of age. We have previously described Jagged1 as differentially expressed with 2.5-fold increased expression in the two escaper dogs (15). Among the other differentially expressed genes, Pitpna was decreased in the escaper dogs relative to severely affected dogs and noted as a potential modifier (Fig. 1A). Further qRT-PCR analysis of these samples confirmed that the escaper dogs had significantly lower Pitpna expression than severely affected dogs (P = 0.0003) and normal dogs (P = 0.043), and that severely affected dogs had significantly higher Pitpna expression compared with normal dogs (P = 0.0088) (Fig. 1B). Protein analysis of Pitpna in muscle tissue confirmed the mRNA findings and showed increased PTEN and decreased pAkt in severely affected GRMD muscle, which has been shown previously (26). Escaper dogs showed the opposite expression pattern (Fig. 1C), further suggesting PITPNA as a potentially beneficial signaling modifier.
Fig. 1.
Mildly affected escaper GRMD dogs exhibit decreased Pitpna expression and increased pAKT. (A) Venn diagram showing the number of genes differentially expressed in normal, escaper, and severely affected GRMD dogs. Pitpna was identified as the one gene differentially expressed in the mildly affected escaper dogs versus the severely affected dogs in the microarray. FDR was 5%. (B) Quantification of Pitpna mRNA expression in normal, escaper, and severely affected GRMD dogs. Data are represented as means ± SDM. *P < 0.005; **P < 0.05 by Student’s t test. (C) Western blot of protein isolated from normal, escaper, and severely affected GRMD dogs showing down-regulation of PITPNA and PTEN and up-regulation of pAkt in escaper dogs. n = 3, normal; n = 2, escaper; and n = 4, severely affected.
Pitpnaa Knockdown in sapje Zebrafish by Morpholino Injection.
We evaluated the effect of Pitpna knockdown in the dystrophin-deficient sapje zebrafish model of DMD by injection of antisense morpholino oligonucleotides (MOs) targeting pitpnaa mRNA (27), in parallel with control MOs. Given that loss-of-function mutations of the gene encoding Pitpnaa result in a range of dosage-sensitive phenotypes and premature death in zebrafish and mice (17, 18), we injected a range of pitpnaa morpholino doses into one-cell-stage sapje embryos. Injection of pitpnaa MO elicited dose-dependent down-regulation of Pitpna protein expression, with 3 ng of MO rendering Pitpna undetectable by Western blot and causing severe morphological defects and premature death as anticipated (Fig. 2 A and B). Given the observed signaling profile observed in the escaper GRMD dogs and previous evidence that the manipulation of Pitpna causes dysregulation of the PTEN/Akt pathway (19), we assayed this pathway in injected sapje fish. As observed in mildly affected GRMD dogs, sapje fish injected with pitpnaa morpholino showed significantly decreased PTEN expression and modestly increased pAkt expression (Fig. 2B), further suggesting a role for Pitpna in modifying the PI3K/Akt pathway.
Fig. 2.
Morpholino-mediated pitpnaa knockdown in zebrafish elicits dose-dependent changes in morphology and signaling. (A) Phase contrast images of sapje larvae injected with pitpnaa MO or control MO at 1 and 4 dpf. Embryos injected with 3 ng of pitpnaa MO at the one-cell stage showed abnormal development and were not viable, whereas 1 ng of pitpnaa MO did not negatively impact development. (B) Western blot showing decreased Pitpna expression with increasing doses of pitpnaa MO injected into sapje embryos. Protein lysates were harvested from sapje larvae at 4 dpf. Experiment performed in triplicate, n = 300 per replicate.
In vivo pitpnaa Knockdown Rescues the sapje Muscle Phenotype.
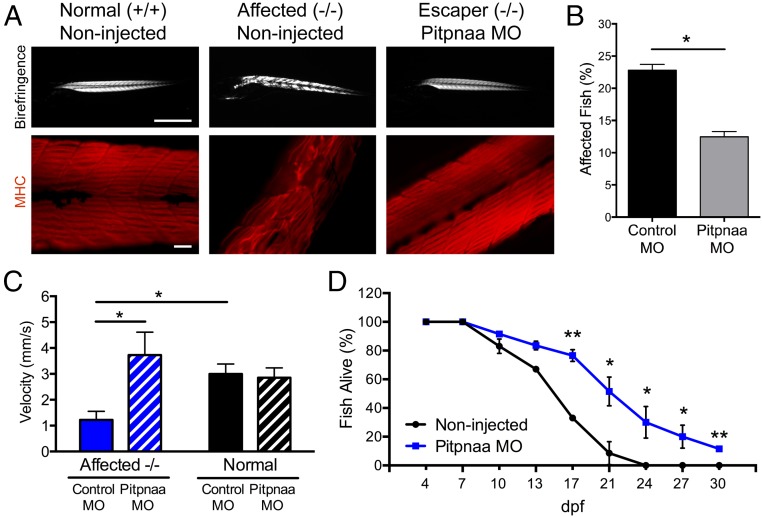
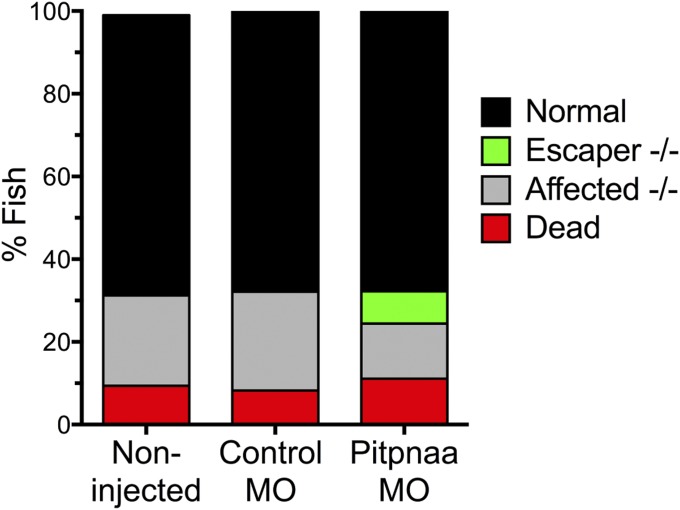
Dystrophin-deficient homozygous null sapje zebrafish exhibit abnormal muscle structure (termed hereon as “affected” fish) on 4 d postfertilization (dpf) observable by birefringence assay (28, 29), in which the transparent larvae are visualized under polarized light (Fig. 3A). Therefore, we used this characteristic to assess whether pitpnaa down-regulation could prevent manifestation of the muscle phenotype. Because homozygous null sapje fish do not survive to sexual maturity, we mated heterozygous sapje fish, which normally produce a Mendelian ratio of ∼25% affected offspring, and compared cohorts injected with 1 ng of either control MO or pitpnaa MO. The birefringence assay showed that fish injected with pitpnaa MO had a significantly lower percent of affected fish (12.5%) compared with control MO (23%) (Fig. 3B). The presence or absence of the muscle phenotype was also noted by immunofluorescent staining of larvae bodies at 4 dpf for myosin heavy chain (Fig. 3A), and subsequent genotyping confirmed that a subset of pitpnaa MO-injected fish with normal muscle structure were indeed homozygous null escaper fish. In addition, injection with the control MO did not alter expected genotype ratios or death rates compared with noninjected controls (Fig. S1).
Fig. 3.
Pitpnaa knockdown rescues sapje zebrafish muscle phenotype, improves swim velocity, and increases long-term survival. (A) Birefringence assay (Upper) and immunofluorescent staining for myosin heavy chain (Lower) showing the typical abnormal muscle phenotype of affected homozygous-null sapje fish at 4 dpf. Injection of pitpnaa MO at the one-cell stage prevented manifestation of the muscle phenotype in some homozygous-null fish, termed escaper fish, which showed healthy muscle morphology comparable to normal fish. [Scale bars: 1 mm (Upper), 100 μm (Lower).] (B) Percent of affected sapje fish as determined by birefringence assay at 4 dpf. Mating of heterozygous sapje fish normally yields ∼25% of affected embryos. Cohorts injected with 1 ng of pitpnaa MO at the one-cell stage showed significantly lower percentages of affected fish compared with control MO. Ten replicate experiments were performed, n = 200–300 per replicate. Bars represent means ± SDM. *P < 0.0001 by Student’s t test. (C) Swim velocity of sapje fish tracked on the DanioVision system. Injection with 1 ng of pitpnaa MO increased swim velocity of affected fish during a 15-min tracking period performed at 4 dpf. Bars represent means ± SEM; *P < 0.05 by two-way ANOVA and Bonferroni post hoc test. (D) Long-term survival assay showing increased survival of affected fish injected with 1 ng of pitpnaa MO at the one-cell stage. Affected fish were identified by birefringence assay at 4 dpf and followed until 30 dpf. Data represent means ± SDM. *P < 0.05, **P < 0.002 versus noninjected by Student’s t test.
Fig. S1.
Genotyping of sapje zebrafish injected with pitpnaa morpholino oligonucleotides confirms homozygous-null escaper sapje population. The genotypes of noninjected, control MO-injected, and pitpnaa MO-injected fish were determined, and it was confirmed that a subset of phenotypically normal fish were in fact homozygous null fish. Injection of either control MO or pitpnaa MO did not increase fish death or expected genotype ratios.
In Vivo pitpnaa Knockdown Improves sapje Swim Performance and Survival.
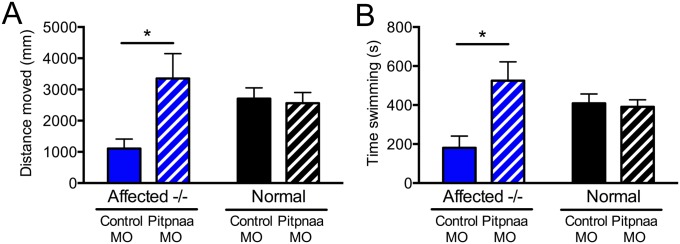
To determine the effect of decreased Pitpna expression on muscle function, swimming performance of control MO-injected versus pitpnaa MO-injected sapje fish was assessed on the Noldus DanioVision swim tracking apparatus. Individual fish were tracked in wells of a 24-well plate for a 15-min period at 4 dpf. The pitpnaa MO-injected homozygous null fish had significantly greater swim time, distance, and velocity compared with control MO-injected null fish (Fig. 3C and Fig. S2 A and B). The absence of dystrophin in homozygous null sapje fish causes premature death beginning around 10–12 dpf. To assess the effect of Pitpna down-regulation in rescuing the early death phenotype, we tracked the survival of pitpnaa MO-injected and noninjected affected sapje fish until 30 dpf. Embryos injected with pitpnaa MO exhibited significantly increased survival from 17 dpf onward compared with noninjected controls (Fig. 3D). Taken together, these results suggest that decreased Pitpna expression improves the phenotype of dystrophin-deficient zebrafish.
Fig. S2.
Pitpnaa knockdown by morpholino injection improves swim distance and time of homozygous-null sapje zebrafish. (A) Swim distance and (B) swim time of sapje fish were tracked on the DanioVision system. Injection with 1 ng of pitpnaa MO increased swim velocity of affected fish during a 15-min tracking period performed at 4 dpf. Bars represent means ± SEM; *P < 0.05 by two-way ANOVA and Bonferroni post hoc test.
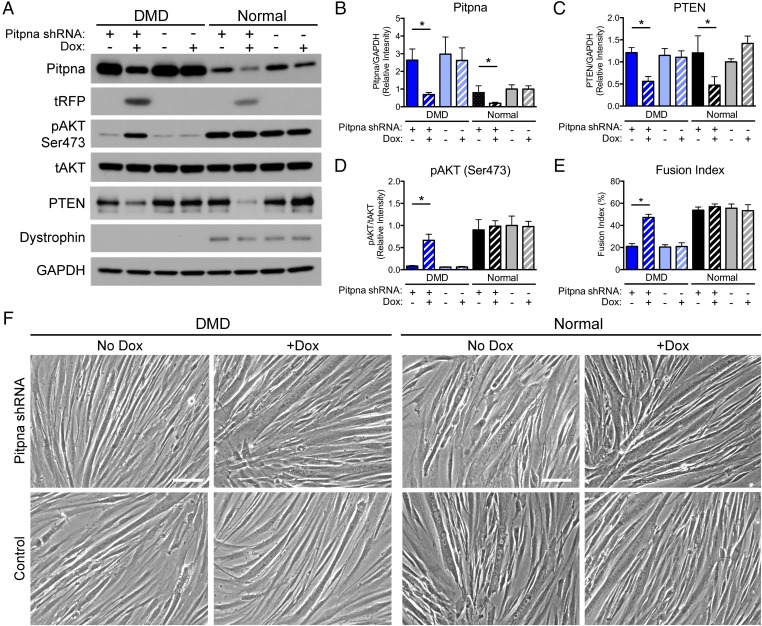
PITPNA Knockdown in Human DMD Myoblasts Increases pAkt and Fusion Index.
To determine the effect of decreased PITPNA expression in human muscle, we generated a human PITPNA doxycycline (Dox)-inducible shRNAi lentivirus (pINDUCER10 backbone) (30) and transduced the construct into primary myoblasts derived from human patients with DMD and from normal muscle biopsies to create stable muscle cell lines. PITPNA shRNA and control myoblasts were induced to differentiate into myotubes in media with or without 0.25 μg/mL of doxycycline to induce shRNA hairpin expression. Expression of the shRNA hairpin was confirmed by Western blotting, which showed significant knockdown of PITPNA and expression of the tRFP tag (Fig. 4 A and B). Similar to our observations in the escaper GRMD dogs and pitpnaa MO-injected sapje fish, PITPNA knockdown significantly decreased PTEN and increased pAKT (Ser473) in DMD myotubes (Fig. 4 C and D). Doxycycline-treated DMD myotubes also showed significantly increased fusion index approaching normal myotube levels (Fig. 4 E and F). Taken together, these results demonstrate that targeted inhibition of PITPNA in dystrophic muscle cells can modulate pAkt expression and improve muscle cell differentiation.
Fig. 4.
PITPNA knockdown by lentiviral shRNA in DMD human myotubes increases pAkt and fusion index. Normal and DMD patient-derived myoblasts were transduced with doxycycline-inducible lentivirus containing shRNA targeting PITPNA mRNA. Myoblasts were induced to differentiate into myotubes in differentiation medium with (+) and without (−) 0.25 μg/mL Dox for 5 d. (A) Western blot showing decreased PITPNA and PTEN expression and increased active pAkt (Ser473) expression with Dox treatment in transduced DMD cells. tRFP expression confirmed expression of the lentiviral construct. (B–D) Western blot quantification showing decreased PITPNA and PTEN, and increased pAKT in DMD myotubes expressing the lentiviral PITPNA shRNA. (E) Quantification of fusion index showing significant increase in DMD cells (+) Dox expressing the PITPNA shRNA compared with (−) Dox. Nuclei were counted from 10 random fields per sample, and fusion index was calculated as the percentage of nuclei within fused myotubes per total nuclei. Representative phase contrast images are shown from which fusion index was calculated. (F) Representative phase contrast images of myotubes taken at 20× magnification. Bars represent means ± SEM; *P < 0.05 by two-way ANOVA and Bonferroni post hoc test, n = 3 normal, three DMD biopsy cell lines (Scale bar: 20 μm.)
Discussion
Identification of genetic modifiers of Duchenne muscular dystrophy is an important component of understanding the disease process and identifying potential therapeutic targets. In our previous work, Pitpna was identified as differentially expressed in two escaper GRMD dogs, which exhibited a markedly milder phenotype compared with that normally associated with dystrophin deficiency. However, the role of PITPNA in skeletal muscle development and disease progression is unknown. In the present study, we have investigated PITPNA as a disease modifier in three models of DMD. Our results position PITPNA as an intriguing mediator of signaling and disease pathogenesis.
We were initially drawn to PITPNA due to its being the one gene only differentially expressed in escaper versus severe GRMD dogs that showed the same expression level on escaper versus normal dogs. However, we did not find a genetic association with the Pitpna allele in the linkage analysis, which instead led us to first investigate Jagged1 (15), one of the 65 genes that were up-regulated or down-regulated when comparing escapers to both severe and normal dogs. Despite a lack of genetic association at the Pitpna locus, we decided to further analyze its role in dystrophic muscle, which is likely via effects on secondary genes involved in the escaper phenotype. Pitpna was also noted because of its involvement in PI3K/Akt signaling. PITPs such as PITPNA perform the critical function of transferring phosphatidylinositol between membrane compartments to mediate membrane trafficking and signaling in eukaryotic cells (16, 31). Specifically, PITPNA has been shown to regulate PI3K/Akt signaling during axonal extension in neurons (19). We found that in escaper GRMD dogs, sapje zebrafish, and DMD muscle cells, decreased PITPNA expression was associated with decreased PTEN and increased Akt signaling. These results were especially interesting, given that knockdown of PTEN in muscle has been shown to prevent muscle wasting by stimulating muscle hypertrophy mediated through pAkt/mTOR signaling (32–34). In addition, induction of pAkt is known to play a significant role in muscle growth and metabolism, impacting several downstream target genes that promote muscle hypertrophy (35, 36), vascularization (37), and inhibit apoptosis (38). Conversely, aberrant PTEN signaling has been attributed to PI3K/Akt signaling dysregulation in severely affected GRMD dogs, which exhibit depressed Akt activation (26).
Increased Akt activity has been shown to ameliorate pathogenesis specifically within the context of muscular dystrophy. In mdx mice, overexpression of constitutively active Akt results in improved muscle force generation (23) and promotes sarcolemma stability by increasing expression of integrin and the dystrophin homolog utrophin (24). Further, Akt activation associated with overexpression of sarcospan has also been shown to ameliorate dystrophic pathology (25). Given the many benefits of increased Akt activity on dystrophic muscle, we were not surprised that pitpna down-regulation prevented manifestation of the muscle phenotype in our sapje zebrafish. Normally, these fish present with disorganized muscle structure on 4 dpf and begin to die precipitously around 10 dpf. The pitpnaa MO injection also improved the percentage of sapje fish that survived through 30 dpf, demonstrating that decreased Pitpna expression was beneficial to performance even when it did not prevent development of the muscle phenotype.
We repressed PITPNA expression in primary human muscle cells via doxycycline-inducible lentiviral delivery of a PITPNA shRNA and induced the stable myoblasts to differentiate into myotubes. Induction of the PITPNA shRNA with doxycycline effectively decreased PITPNA protein and PTEN expression and increased pAkt in DMD myotubes. Several previous studies have demonstrated that enhancing Akt activation promotes muscle hypertrophy (21, 23, 24, 39) and differentiation of myoblasts into fused myotubes (22). Accordingly, we found that myotubes expressing the PITPNA shRNA had significantly increased fusion index compared with controls. Given these findings, we believe that PITPNA is working, at least in part, through modulation of Akt signaling to ameliorate the dystrophic phenotype.
Despite our positive effects observed with decreased PITPNA expression, potential negative consequences of its repression must be carefully considered and monitored due to developmental defects associated with its strong knockdown and complete knockout. For example, loss-of-function mutations of the gene encoding Pitpna in mice causes dose-sensitive phenotypes, including neurological dysfunction, spinocerebellar neurodegeneration, and premature death (18). Hence, it was not surprising that injection of high doses of pitpnaa morpholino elicited severe morphological deformities and death in the sapje zebrafish. If PITPNA modulation is pursued as a therapeutic target, care must be taken not to compromise its potentially essential functions outside of skeletal muscle, and muscle-specific PITPNA modulation should be explored in future studies. Although PITPNA has been shown to be critical in neurons, it was not found to have an essential housekeeping function in murine embryonic stem cells (40), suggesting that the benefits elicited by its down-regulation may not be mediated by its role in muscle stem cells. Finally, given our observed impact of PITPNA modulation on Akt activity, the possibility for inducing unregulated cell growth must also be considered.
DMD is a multifaceted disease that will likely require a multifaceted approach to address the many features of its pathology. Although there are several promising therapeutic approaches under investigation for DMD involving the restoration of dystrophin expression at the sarcolemma, identification of disease modifiers enhances our understanding of the disease process and elevates our repertoire of potential therapeutic targets. Here, we present PITPNA as a genetic modulator of the dystrophic process, which when repressed, elicits positive effects on DMD pathogenesis via the Akt signaling pathway. These data position PITPNA as a potential DMD therapeutic target that should be carefully considered as part of a combinatorial treatment strategy.
Methods
GRMD Dogs.
GRMD dogs were housed and cared for at the University of São Paulo as previously described (41) in accordance with the animal research ethics committee of the Biosciences Institute, University of São Paulo (034/2005). Total mRNA was extracted from muscle biopsies of two escaper, four severely affected GRMD dogs, and three age-matched wild-type normal dogs. The Two-Color Microarray-Based Gene Expression Analysis-Low Input Quick Amp Labeling protocol (Agilent Technologies) was used with the SurePrint Canine 4 × 44K (Agilent Technologies) Microarray (GEO Platform GPL11351). Arrays were processed with standard procedures as previously described (15). Genes differentially expressed between normal, escaper, and severely affected animals were identified with the significance analysis of microarray (SAM) statistical approach (42). False discovery rate (FDR) was 5%. Quantitative real-time PCR was performed using 50 ng of cDNA with β-actin as an internal standard. GRMD muscle sample proteins were extracted using RIPA buffer (Boston BioProducts) with proteinase inhibitors (Roche). Soluble proteins were resolved using electrophoresis and transferred to nitrocellulose membranes (Hybond, Amersham Biosciences).
Zebrafish.
Zebrafish were housed in the Boston Children’s Hospital Aquatics Facility under the animal protocol number 09–10-1534R and maintained as previously described (43) in accordance with the Institutional Animal Care and Use Committee. Genomic DNA extracted from fish was used as the PCR template with the following primer set: forward primer 5′-CTGGTTACATTCTGAGAGACTTTC-3′ and reverse primer 5′-AGCCAGCTGAACCAATTAACTCAC-3′; as described previously (44). To knockdown pitpnaa, sapje heterozygous fish were mated, and the resulting fertilized one-cell-stage embryos were injected with a pitpnaa MO (5′-CATGTTATCTCCTTTGCCGCCCCGT-3′) (17). Knockdown of zebrafish Pitpna was confirmed by Western blot. Approximately 200–300 embryos were injected in a minimum of three replicate experiments per assay. Zebrafish proteins were extracted from a total of 50 4-dpf fish using RIPA buffer (Boston BioProducts) with proteinase inhibitors (Roche). Soluble proteins were resolved using electrophoresis and transferred to nitrocellulose membranes. The sapje dystrophic muscle phenotype was detected by using a birefringence assay as described previously (45). Immunostaining was performed in 4-dpf embryos with anti-slow muscle myosin heavy chain antibody (F59, Developmental Studies Hybridoma Bank; 1:50). The embryos were placed in 3% methyl cellulose or mounted on a glass slide and observed with fluorescent microscopes. To analyze the motor function of the zebrafish, we used the Noldus DanioVision swim tracking apparatus (46). The movement data from each larva were collected using the EthoVision XT8 software, and the detection threshold was set to detect moving red pixels. For the long-term survival assay, control and pitpnaa MO-injected sapje zebrafish were screened by birefringence on 4 dpf, and affected fish were tracked over a 30-d period. The number of surviving fish was evaluated every 3 d.
Human Samples.
Human tissue was collected and deidentified under the protocol 03–12-205R approved by the Committee of Clinical Investigation at Boston Children’s Hospital. All patients gave their written and oral consent before surgery, and all protocols were approved by the Boston Children’s Hospital human subjects internal review board (protocol 03–12-205R). Tissue from three normal and three patients with DMD was snap frozen and primary muscle cells were dissociated and frozen as described previously (47, 48). To knockdown PITPNA, we infected primary myoblasts with a doxycycline-inducible shRNAi lentivirus (49) targeting human PITPNA mRNA (5′-AATGCTTACCCCTACTGCAGA-3′) packaged into the pINDUCER10 lentivector (30). PITPNA knockdown and tRFP expression was confirmed by Western blot. Human myoblasts (48) were cultured on plates coated with 0.1% gelatin in proliferation medium, then switched to differentiation medium [2% horse serum (Gibco)/1% anti-anti/DMEM] for 7 d. Nuclei were counted from 10 random fields per sample, and fusion index was calculated as the percentage of nuclei within fused myotubes per total nuclei. Human myotubes were collected and lysed in RIPA buffer with protease inhibitor mixture. Lysates were separated by SDS/PAGE and transferred to PVDF membranes (Life Technologies).
Statistical Analysis.
All results are shown as means ± SEM and sampling distribution of the mean (SDM) as stated. Statistical analyses of the data were performed using Microsoft Excel and Prism GraphPad to implement Student’s t test and one- and two-way ANOVA followed by Bonferroni post hoc test. P values of <0.05 were considered to be statistically significant.
SI Methods are also available.
SI Methods
GRMD Dog Colony.
GRMD dogs were housed and cared for at the University of São Paulo and genotyped at birth as previously described (41). GRMD dogs were identified by microchips. Animal care and experiments were performed in accordance with the animal research ethics committee of the Biosciences Institute, University of São Paulo (034/2005).
GRMD Gene Expression Profiles.
Total mRNA was extracted from muscle biopsies of two escaper, four severely affected GRMD dogs, and four age-matched wild-type normal dogs. The Two-Color Microarray-Based Gene Expression Analysis-Low Input Quick Amp Labeling protocol (Agilent Technologies) was used with the SurePrint Canine 4 × 44K (Agilent Technologies) Microarray (GEO Platform GPL11351). Samples were labeled with Cy5 and a single RNA from a normal individual was labeled with Cy3 and used as a common reference on all arrays. Subsequently, the samples were labeled with Cy3 and the reference RNA with Cy5 to generate dye-swap technical replicates. Arrays were processed with standard procedures, as previously described (15). Averaged values of dye-swap technical replicates were used for further analysis. Genes differentially expressed between normal, escaper, and severely affected animals were identified with the significance analysis of microarray (SAM) statistical approach (42) using the following parameters: one-class unpaired responses, t statistics, 100 permutations. False discovery rate (FDR) was 5%. Gene Expression data were deposited at GEO database under accession no. GSE69040.
GRMD Quantitative RT-PCR.
Total RNA was harvested from GRMD dogs’ muscle biopsies using TRIzol (Invitrogen) following the manufacturer’s instructions. The RNA was treated with DNase (Invitrogen). A total of 1 μg of total RNA was reverse transcribed with the SuperScript III First-Strand Synthesis System (Invitrogen). Quantitative real-time PCR was performed using 50 ng of cDNA and SYBR Green PCR master mix in an ABI Prism 7100 system (Applied BioSystems). The PCR conditions were 94 °C for 15 s, 58 °C for 30 s, and 72 °C for 30 s, for 40 cycles. Primers were designed to amplify dog’s PITPNΑ and β-actin. Primers sequences were as follows: PITPNAa: sense: GCATGGAATGCCTACCCTTA and antisense: GATCTGGTTTGTGCCAGGTT; β-actin: sense: AAGATGACCCAGATCATGTTCG and antisense: GGA GTC CAT CAC GAT GCC AGT. Samples were run in triplicates, and the threshold suggested by the instrument software was used to calculate Ct. To normalize the readings, we used Ct values from the β-actin as internal standard in each run, obtaining a ΔCt value for Pitpna.
GRMD Western Blot Analysis.
GRMD muscle sample proteins were extracted using RIPA buffer (Boston BioProducts) with proteinase inhibitors (Roche). Samples were centrifuged at 13,000 × g for 10 min to remove insoluble debris. Protein concentration was measured using a BCA protein assay kit (Bio-Rad). Soluble proteins were resolved using electrophoresis with Novex 4–20% Tris-glycine gels (Life Technologies) and transferred to nitrocellulose membranes (Hybond, Amersham Biosciences). All membranes were stained with Ponceau (Sigma-Aldrich) to evaluate the amount of loaded proteins. Blots were blocked for 1 h in Tris-buffered saline Tween (TBST) containing 5% powdered skim milk and reacted overnight with the following primary antibodies: anti-PITPNA (Sigma, SAB1400211; 1:1,000), anti-pAKT (Abcam, S4273; 1:500), anti-PTEN (Cell Signaling, 9188; 1:1,000). Horseradish peroxidase (HRP)-conjugated secondary antibody (Santa Cruz Biotechnology; 1:1,000) was used to detect immunoreactive bands with Pierce ECL 2 (Thermo). Anti-beta actin antibody (anti–β-actin–HRP, Abcam; 1:10,000) was used as loading control.
Zebrafish Maintenance and Genotyping.
Zebrafish were housed in the Boston Children’s Hospital Aquatics Facility (Director Christian Lawrence) under the animal protocol number: 09–10-1534R and maintained as breeding stocks as previously described (43) in accordance with the Institutional Animal Care and Use Committee. Genomic DNA extracted from fish was used as the PCR template with the following primer set: forward primer 5′-CTGGTTACATTCTGAGAGACTTTC-3′ and reverse primer 5′-AGCCAGCTGAACCAATTAACTCAC-3′, as described previously (44).
Zebrafish pitpnaa Knockdown and Morphologic Analysis.
Sapje heterozygous fish were mated, and the resulting fertilized one-cell-stage embryos were injected with a pitpnaa MO (5′-CATGTTATCTCCTTTGCCGCCCCGT-3′) used previously (17) encompassing the PITPNAa translation initiation codon. Knockdown of zebrafish Pitpna was confirmed by Western blot. Approximately 200–300 embryos were injected in a minimum of three replicate experiments per assay. Breeding and staging of embryos was performed according to standard protocols. Embryos were imaged using a Nikon SMZ1500 microscope fitted with a QImaging Retiga 2000R camera and OpenLab software. During preparation of this manuscript, we discovered that another isoform of PITPNA in zebrafish was now known, termed pitpnab (XM_002663555.4). Our pitpna morpholino targeted the 3′-UTR of pitpnaa not homologous to the pitpnab such that pitpnab expression was likely unaffected by our morpholino injections.
Zebrafish Western Blot Analysis.
Zebrafish proteins were extracted from a total of 50 4-dpf fish using RIPA buffer (Boston BioProducts) with proteinase inhibitors (Roche). Samples were centrifuged at 13,000 × g for 10 min to remove insoluble debris. Protein concentration was measured using a BCA protein assay kit (Bio-Rad). Soluble proteins were resolved using electrophoresis with Novex 4–20% Tris-glycine gels (Life Technologies), and transferred to nitrocellulose membranes (Hybond, Amersham Biosciences). All membranes were stained with Ponceau (Sigma-Aldrich) to evaluate the amount of loaded proteins. Blots were blocked for 1 h in TBST containing 5% powdered skim milk and reacted overnight with the following primary antibodies: anti-PITPNA (Sigma, SAB1400211; 1:1,000), anti-phophoAKT (S4273, Abcam, 4060; 1:500), and anti-AKT 1,2,3 (Abcam; 1:500). HRP-conjugated secondary antibody (Santa Cruz Biotechnology; 1:1,000) was used to detect immunoreactive bands with Pierce ECL 2 (Thermo). Anti–β-actin antibody (anti–β-actin–HRP, Abcam; 1:10,000) was used as loading control.
Zebrafish Birefringence Assay.
The sapje dystrophic muscle phenotype was detected by using a birefringence assay, a technique used to analyze myofiber integrity using polarized light performed as described previously (45). Polarizing filters were placed on a bottom-lit dissection scope, and images were acquired with a QImaging Retiga 2000R camera fitted to a Nikon SMZ1500 microscope using OpenLab software. Larvae were anesthetized with tricaine and positioned relative to the polarized light to produce maximal birefringence illumination.
Zebrafish Immunostaining.
Immunostaining was performed in 4-dpf embryos. Embryos were fixed in 4% paraformaldehyde (PFA) in PBS at 4 °C overnight and dehydrated in 100% methanol. After rehydration, 4-dpf embryos were incubated in 0.1% collagenase (Sigma) in PBS for 60 min. Blocking solution containing 0.2% saponin was used for 4-dpf embryos. Anti-slow muscle myosin heavy chain antibody (F59, Developmental Studies Hybridoma Bank; 1:50) was used. The embryos were placed in 3% methyl cellulose or mounted on a glass slide and observed with fluorescent microscopes (Nikon Eclipse E1000 and Zeiss Axioplan2).
Zebrafish Motor Function Assay.
To analyze the motor function of the zebrafish, we used the Noldus DanioVision swim tracking apparatus (46). To acclimate the larvae to the new environment, 4-dpf larvae were placed individually into each well of a 24-well plate filled with 2 mL of water at 28 ± 0.5 °C 3 h before the experiment. The movement data from each larva were collected using the EthoVision XT8 software (Noldus Information Technology), and the detection threshold was set to detect moving red pixels.
Zebrafish Survival Assay.
Control and pitpnaa MO-injected sapje zebrafish were screened by birefringence on 4 dpf, and affected fish were tracked over a 30-d period. The number of surviving fish was evaluated every 3 d.
Human Tissue Collection and Processing.
Human tissue was collected and deidentified under the protocol 03–12-205R approved by the Committee of Clinical Investigation at Boston Children’s Hospital. Tissue was snap frozen and primary muscle cells were dissociated and frozen as described previously (47, 48). Three normal and three DMD biopsies were used for the following experiments.
PITPNA shRNAi Knockdown.
A doxycycline-inducible shRNAi lentivirus (49) targeting human PITPNA mRNA (5′-AATGCTTACCCCTACTGCAGA-3′) was packaged into the pINDUCER10 lentivector (30). We then infected myoblasts derived from normal and DMD patient biopsies with the lentivirus. Infected cells were selected for by addition of 1 μg/mL puromycin until all untransfected cells were dead. Addition of 0.25 μg/mL Dox hyclate initiated transcription of the turboRFP (tRFP)-shRNA cassette. PITPNA knockdown and tRFP expression was confirmed by Western blot.
In Vitro Fusion Assay.
Human myoblasts (48) were cultured on plates coated with 0.1% gelatin in proliferation medium [20% FBS (Gibco,)/1% anti-anti (Gibco)/DMEM (Gibco)]. At ∼80% confluence, the cells were switched to differentiation medium [2% horse serum (Gibco)/1% anti-anti/DMEM]. Medium was changed every other day until day 7, and images were taken on a Nikon Eclipse TS100 microscope fitted with a Spot RT3 camera. Nuclei were counted from 10 random fields per sample, and fusion index was calculated as the percentage of nuclei within fused myotubes per total nuclei.
Myotube Western Blot Analysis.
Human myotubes were collected and lysed in RIPA buffer (Boston BioProducts) with protease inhibitor mixture (Roche). Lysates were incubated on ice for 1 h, vortexing halfway through, centrifuged at 16,000 × g for 30 min at 4 °C and the supernatants retained. Protein concentration was measured using a BCA protein assay kit (Bio-Rad). Lysates were separated by SDS/PAGE on Novex 4–20% Tris⋅HCl polyacrylamide gels (Thermo Scientific) and transferred to PVDF membranes (Life Technologies). Membranes were blocked in 5% milk in TTBS and then probed with antibodies to the following: anti-PITPNA (Sigma, SAB1400211; 1:1,000), anti-tRFP (Evrogen, AB231; 1:10,000), anti–phospho-Akt Ser473 (Cell Signaling, 4060; 1:1,000), anti–total-Akt (Cell Signaling, 4691; 1:1,000), anti-PTEN (Cell Signaling 9188; 1:1,000), anti-dystrophin (Abcam 15277; 1:200), anti-GAPDH (Santa Cruz, sc-25778; 1:4,000), HRP-linked rabbit secondary (Cell Signaling, 7074), and HRP-linked mouse secondary (Cell Signaling, 7076). Blots were developed with Pierce ECL Plus (Thermo Scientific, 32132) on Blue Devil autoradiography film (Genesee Scientific, 30-101). Western blots were analyzed by ImageJ software.
Acknowledgments
We thank Victor Dubowitz and Ronal Cohn for their encouragement and support regarding our exceptional GRMD projects, as well as Jeffrey Widrick for his assistance with experiments. Funding for this work was generously provided by the Duchenne Research Fund (M.Z.), Fundação de Amparo à Pesquisa do Estado de São Paulo (M.Z. and N.M.V.), Conselho Nacional de Desenvolvimento Científico e Tecnológico (M.Z.), Institutos Nacionais de Ciência e Tecnologia (M.Z.), Associação de Assistência à Criança Deficiente (M.Z.), the Bernard F. and Alva B. Gimbel Foundation (L.M.K.), the National Institutes of Health Grant R01AR064300-01A1 (to L.M.K.), and the Boston Children’s Hospital Intellectual and Developmental Disabilities Research Center Grant NIH P30 HD-18655 (to L.M.K.). N.M.V. is supported by Muscular Dystrophy Association Development Grant MDA352465, and experiments were also supported by a grant from Pfizer, Inc. (to L.M.K.).
Footnotes
Conflict of interest statement: L.M.K. is a consultant for Pfizer, Inc., Summit Corporation PLC, and Sarepta Therapeutics for muscle disease drug therapies.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE69040).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1703556114/-/DCSupplemental.
References
- 1.Hoffman EP, Brown RH, Jr, Kunkel LM. Dystrophin: The protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51:919–928. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- 2.Monaco AP, et al. Isolation of candidate cDNAs for portions of the Duchenne muscular dystrophy gene. Nature. 1986;323:646–650. doi: 10.1038/323646a0. [DOI] [PubMed] [Google Scholar]
- 3.Emery AE, Muntoni F, Quinlivan RC. Duchenne Muscular Dystrophy. Oxford Univ Press; Oxford: 2015. [Google Scholar]
- 4.Vetrone SA, et al. Osteopontin promotes fibrosis in dystrophic mouse muscle by modulating immune cell subsets and intramuscular TGF-beta. J Clin Invest. 2009;119:1583–1594. doi: 10.1172/JCI37662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haslett JN, et al. Gene expression comparison of biopsies from Duchenne muscular dystrophy (DMD) and normal skeletal muscle. Proc Natl Acad Sci USA. 2002;99:15000–15005. doi: 10.1073/pnas.192571199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barp A, et al. Genetic modifiers of Duchenne muscular dystrophy and dilated cardiomyopathy. PLoS One. 2015;10:e0141240. doi: 10.1371/journal.pone.0141240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pegoraro E, et al. Cooperative International Neuromuscular Research Group SPP1 genotype is a determinant of disease severity in Duchenne muscular dystrophy. Neurology. 2011;76:219–226. doi: 10.1212/WNL.0b013e318207afeb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van den Bergen JC, et al. Validation of genetic modifiers for Duchenne muscular dystrophy: A multicentre study assessing SPP1 and LTBP4 variants. J Neurol Neurosurg Psychiatry. 2015;86:1060–1065. doi: 10.1136/jnnp-2014-308409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bello L, et al. Cooperative International Neuromuscular Research Group Investigators Genetic modifiers of ambulation in the cooperative international Neuromuscular research group Duchenne natural history study. Ann Neurol. 2015;77:684–696. doi: 10.1002/ana.24370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flanigan KM, et al. United Dystrophinopathy Project LTBP4 genotype predicts age of ambulatory loss in Duchenne muscular dystrophy. Ann Neurol. 2013;73:481–488. doi: 10.1002/ana.23819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hogarth MW, et al. Cooperative International Neuromuscular Research Group (CINRG) Evidence for ACTN3 as a genetic modifier of Duchenne muscular dystrophy. Nat Commun. 2017;8:14143. doi: 10.1038/ncomms14143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zucconi E, et al. Ringo: Discordance between the molecular and clinical manifestation in a golden retriever muscular dystrophy dog. Neuromuscul Disord. 2010;20:64–70. doi: 10.1016/j.nmd.2009.10.011. [DOI] [PubMed] [Google Scholar]
- 13.Kornegay JN, Tuler SM, Miller DM, Levesque DC. Muscular dystrophy in a litter of golden retriever dogs. Muscle Nerve. 1988;11:1056–1064. doi: 10.1002/mus.880111008. [DOI] [PubMed] [Google Scholar]
- 14.Zatz M, et al. A normal life without muscle dystrophin. Neuromuscul Disord. 2015;25:371–374. doi: 10.1016/j.nmd.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 15.Vieira NM, et al. Jagged 1 rescues the Duchenne muscular dystrophy phenotype. Cell. 2015;163:1204–1213. doi: 10.1016/j.cell.2015.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghosh R, Bankaitis VA. Phosphatidylinositol transfer proteins: Negotiating the regulatory interface between lipid metabolism and lipid signaling in diverse cellular processes. Biofactors. 2011;37:290–308. doi: 10.1002/biof.180. [DOI] [PubMed] [Google Scholar]
- 17.Xie Y, et al. Phosphatidylinositol transfer protein-alpha in netrin-1-induced PLC signalling and neurite outgrowth. Nat Cell Biol. 2005;7:1124–1132. doi: 10.1038/ncb1321. [DOI] [PubMed] [Google Scholar]
- 18.Alb JG, Jr, et al. Mice lacking phosphatidylinositol transfer protein-α exhibit spinocerebellar degeneration, intestinal and hepatic steatosis, and hypoglycemia. J Biol Chem. 2003;278:33501–33518. doi: 10.1074/jbc.M303591200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cosker KE, et al. Regulation of PI3K signalling by the phosphatidylinositol transfer protein PITPalpha during axonal extension in hippocampal neurons. J Cell Sci. 2008;121:796–803. doi: 10.1242/jcs.019166. [DOI] [PubMed] [Google Scholar]
- 20.Alessi DR, et al. Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J. 1996;15:6541–6551. [PMC free article] [PubMed] [Google Scholar]
- 21.Rommel C, et al. Mediation of IGF-1-induced skeletal myotube hypertrophy by PI(3)K/Akt/mTOR and PI(3)K/Akt/GSK3 pathways. Nat Cell Biol. 2001;3:1009–1013. doi: 10.1038/ncb1101-1009. [DOI] [PubMed] [Google Scholar]
- 22.Rotwein P, Wilson EM. Distinct actions of Akt1 and Akt2 in skeletal muscle differentiation. J Cell Physiol. 2009;219:503–511. doi: 10.1002/jcp.21692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim MH, et al. Myogenic Akt signaling attenuates muscular degeneration, promotes myofiber regeneration and improves muscle function in dystrophin-deficient mdx mice. Hum Mol Genet. 2011;20:1324–1338. doi: 10.1093/hmg/ddr015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peter AK, et al. Myogenic Akt signaling upregulates the utrophin-glycoprotein complex and promotes sarcolemma stability in muscular dystrophy. Hum Mol Genet. 2009;18:318–327. doi: 10.1093/hmg/ddn358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marshall JL, et al. Sarcospan-dependent Akt activation is required for utrophin expression and muscle regeneration. J Cell Biol. 2012;197:1009–1027. doi: 10.1083/jcb.201110032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feron M, et al. PTEN contributes to profound PI3K/Akt signaling pathway deregulation in dystrophin-deficient dog muscle. Am J Pathol. 2009;174:1459–1470. doi: 10.2353/ajpath.2009.080460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ile KE, et al. Zebrafish class 1 phosphatidylinositol transfer proteins: PITPbeta and double cone cell outer segment integrity in retina. Traffic. 2010;11:1151–1167. doi: 10.1111/j.1600-0854.2010.01085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bassett D, Currie PD. Identification of a zebrafish model of muscular dystrophy. Clin Exp Pharmacol Physiol. 2004;31:537–540. doi: 10.1111/j.1440-1681.2004.04030.x. [DOI] [PubMed] [Google Scholar]
- 29.Bassett DI, et al. Dystrophin is required for the formation of stable muscle attachments in the zebrafish embryo. Development. 2003;130:5851–5860. doi: 10.1242/dev.00799. [DOI] [PubMed] [Google Scholar]
- 30.Meerbrey KL, et al. The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proc Natl Acad Sci USA. 2011;108:3665–3670. doi: 10.1073/pnas.1019736108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Phillips SE, et al. The diverse biological functions of phosphatidylinositol transfer proteins in eukaryotes. Crit Rev Biochem Mol Biol. 2006;41:21–49. doi: 10.1080/10409230500519573. [DOI] [PubMed] [Google Scholar]
- 32.Latres E, et al. Insulin-like growth factor-1 (IGF-1) inversely regulates atrophy-induced genes via the phosphatidylinositol 3-kinase/Akt/mammalian target of rapamycin (PI3K/Akt/mTOR) pathway. J Biol Chem. 2005;280:2737–2744. doi: 10.1074/jbc.M407517200. [DOI] [PubMed] [Google Scholar]
- 33.Hu Z, et al. PTEN inhibition improves muscle regeneration in mice fed a high-fat diet. Diabetes. 2010;59:1312–1320. doi: 10.2337/db09-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yue F, et al. Conditional loss of Pten in myogenic progenitors leads to postnatal skeletal muscle hypertrophy but age-dependent exhaustion of satellite cells. Cell Reports. 2016;17:2340–2353. doi: 10.1016/j.celrep.2016.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peter AK, Crosbie RH. Hypertrophic response of Duchenne and limb-girdle muscular dystrophies is associated with activation of Akt pathway. Exp Cell Res. 2006;312:2580–2591. doi: 10.1016/j.yexcr.2006.04.024. [DOI] [PubMed] [Google Scholar]
- 36.Blaauw B, et al. Akt activation prevents the force drop induced by eccentric contractions in dystrophin-deficient skeletal muscle. Hum Mol Genet. 2008;17:3686–3696. doi: 10.1093/hmg/ddn264. [DOI] [PubMed] [Google Scholar]
- 37.Takahashi A, et al. Myogenic Akt signaling regulates blood vessel recruitment during myofiber growth. Mol Cell Biol. 2002;22:4803–4814. doi: 10.1128/MCB.22.13.4803-4814.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gurpur PB, Liu J, Burkin DJ, Kaufman SJ. Valproic acid activates the PI3K/Akt/mTOR pathway in muscle and ameliorates pathology in a mouse model of Duchenne muscular dystrophy. Am J Pathol. 2009;174:999–1008. doi: 10.2353/ajpath.2009.080537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lai KM, et al. Conditional activation of akt in adult skeletal muscle induces rapid hypertrophy. Mol Cell Biol. 2004;24:9295–9304. doi: 10.1128/MCB.24.21.9295-9304.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alb JG, Jr, et al. Genetic ablation of phosphatidylinositol transfer protein function in murine embryonic stem cells. Mol Biol Cell. 2002;13:739–754. doi: 10.1091/mbc.01-09-0457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Honeyman K, Carville KS, Howell JM, Fletcher S, Wilton SD. Development of a snapback method of single-strand conformation polymorphism analysis for genotyping Golden Retrievers for the X-linked muscular dystrophy allele. Am J Vet Res. 1999;60:734–737. [PubMed] [Google Scholar]
- 42.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lawrence C, Mason T. Zebrafish housing systems: A review of basic operating principles and considerations for design and functionality. ILAR J. 2012;53:179–191. doi: 10.1093/ilar.53.2.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kawahara G, Kunkel LM. Zebrafish based small molecule screens for novel DMD drugs. Drug Disc Tech. 2013;10:e91–e96. doi: 10.1016/j.ddtec.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Telfer WR, Busta AS, Bonnemann CG, Feldman EL, Dowling JJ. Zebrafish models of collagen VI-related myopathies. Hum Mol Genet. 2010;19:2433–2444. doi: 10.1093/hmg/ddq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dowling JJ, Low SE, Busta AS, Feldman EL. Zebrafish MTMR14 is required for excitation-contraction coupling, developmental motor function and the regulation of autophagy. Hum Mol Genet. 2010;19:2668–2681. doi: 10.1093/hmg/ddq153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lapan AD, Gussoni E. Isolation and characterization of human fetal myoblasts. Methods Mol Biol. 2012;798:3–19. doi: 10.1007/978-1-61779-343-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alexander MS, et al. Regulation of DMD pathology by an ankyrin-encoded miRNA. Skelet Muscle. 2011;1:27. doi: 10.1186/2044-5040-1-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fellmann C, et al. An optimized microRNA backbone for effective single-copy RNAi. Cell Reports. 2013;5:1704–1713. doi: 10.1016/j.celrep.2013.11.020. [DOI] [PubMed] [Google Scholar]