ABSTRACT
Viral protein R (Vpr) is an HIV-1 accessory protein whose function remains poorly understood. In this report, we sought to determine the requirement of Vpr for facilitating HIV-1 infection of monocyte-derived dendritic cells (MDDCs), one of the first cell types to encounter virus in the peripheral mucosal tissues. In this report, we characterize a significant restriction of Vpr-deficient virus replication and spread in MDDCs alone and in cell-to-cell spread in MDDC-CD4+ T cell cocultures. This restriction of HIV-1 replication in MDDCs was observed in a single round of virus replication and was rescued by the expression of Vpr in trans in the incoming virion. Interestingly, infections of MDDCs with viruses that encode Vpr mutants unable to interact with either the DCAF1/DDB1 E3 ubiquitin ligase complex or a host factor hypothesized to be targeted for degradation by Vpr also displayed a significant replication defect. While the extent of proviral integration in HIV-1-infected MDDCs was unaffected by the absence of Vpr, the transcriptional activity of the viral long terminal repeat (LTR) from Vpr-deficient proviruses was significantly reduced. Together, these results characterize a novel postintegration restriction of HIV-1 replication in MDDCs and show that the interaction of Vpr with the DCAF1/DDB1 E3 ubiquitin ligase complex and the yet-to-be-identified host factor might alleviate this restriction by inducing transcription from the viral LTR. Taken together, these findings identify a robust in vitro cell culture system that is amenable to addressing mechanisms underlying Vpr-mediated enhancement of HIV-1 replication.
IMPORTANCE Despite decades of work, the function of the HIV-1 protein Vpr remains poorly understood, primarily due to the lack of an in vitro cell culture system that demonstrates a deficit in replication upon infection with viruses in the absence of Vpr. In this report, we describe a novel cell infection system that utilizes primary human dendritic cells, which display a robust decrease in viral replication upon infection with Vpr-deficient HIV-1. We show that this replication difference occurs in a single round of infection and is due to decreased transcriptional output from the integrated viral genome. Viral transcription could be rescued by virion-associated Vpr. Using mutational analysis, we show that domains of Vpr involved in binding to the DCAF1/DDB1/E3 ubiquitin ligase complex and prevention of cell cycle progression into mitosis are required for LTR-mediated viral expression, suggesting that the evolutionarily conserved G2 cell cycle arrest function of Vpr is essential for HIV-1 replication.
KEYWORDS: Vpr, dendritic cells, human immunodeficiency virus, transcription
INTRODUCTION
Human immunodeficiency virus type 1 (HIV-1) encodes a number of proteins that allow entry and replication in human cells. In addition to the structural or enzymatic proteins that have well-defined functions in the replication cycle, there are also a number of small, accessory proteins. Accessory proteins encoded by HIV-1 are not always necessary for replication in vitro but are absolutely essential for replication in vivo (1). These proteins serve to counteract host restriction factors that would normally limit HIV-1 infection (1, 2). Of the accessory proteins encoded by HIV-1, Vpr is the only one whose function remains relatively unclear.
Vpr is a small, 96-amino-acid, 14-kDa protein that is packaged into the budding virion through associations with the p6 region of Gag (3–10). This association allows Vpr to be present in the cell at a relatively high quantity (∼200 to 300 molecules/virion) upon initial infection (11). Previous studies have extensively characterized the outcome of Vpr expression in various cell types. In cycling cells, Vpr expression results in G2/M cell cycle arrest, which culminates in the induction of apoptosis (12–14). It is well established that Vpr-mediated G2/M cell cycle arrest is mediated through its association with the Cul4A/DCAF/DDB1 E3 (CRL4DCAF1) ubiquitin ligase complex (15–17). In addition, HIV-1 Vpr has been shown to recruit and degrade a number of DNA damage response (DDR) proteins, including the SLX4-SLX1/MUS81-EME1 structure-specific endonuclease complex (SLX4com), uracil DNA glycosylase 2 (UNG2), and helicase-like transcription factor (HLTF) (18–21), via the CRL4DCAF1 complex, resulting in G2/M cell cycle arrest, although the role that this process plays during HIV-1 infection still remains unclear.
Although a number of previous studies have examined the requirement of Vpr for HIV-1 replication in various cell types, including primary CD4+ T cells and monocyte-derived macrophages (MDMs), differences in virus replication have not been consistently observed (18, 20, 22–26). Vpr expression is dispensable for infection in activated CD4+ T cells in vitro (22–25, 27), presumably due to the well-characterized cytostatic and cytopathic functions of Vpr in cycling cells (12). In contrast, recent studies with MDMs suggest that Vpr is necessary for HIV-1 envelope (Env) expression, and the purported consequence of infection of MDMs with Vpr-deficient viruses was reported to be decreased viral production and reduced cell-to-cell spread to CD4+ T cells (22, 28). Notably, there has been considerable heterogeneity in replication differences between wild-type (WT) and Vpr-deficient viruses and host responses to virus infection in MDMs, presumably due to donor and experimental variability between studies (12, 29, 30). Additionally, it has also been reported that Vpr expression in macrophages can both inhibit and induce type I interferon (IFN) responses (18, 28, 31–34).
Dendritic cells (DCs) are sentinel cells that bridge innate and adaptive immunity (35). They actively patrol peripheral tissues, including mucosal sites of HIV-1 transmission, in search of foreign pathogens. Because of this, monocyte-derived DCs (MDDCs) are among the first cells to interact with HIV-1 upon sexual transmission of the virus (36–40). While MDDCs are less susceptible to infection than activated CD4+ T cells and macrophages, they are still able to be infected ex vivo at a low but consistent level (41–44). In contrast to work with MDMs and CD4+ T cells, there have been isolated descriptions of the effects of Vpr on the HIV-1 replicative capacity in MDDCs (41, 44), with no consensus on the mechanisms accounting for the Vpr-mediated enhancement of virus replication. In this study, we use MDDCs as a model system to investigate the role of Vpr during infection. We find a robust replication defect of Vpr-deficient HIV-1 in MDDCs, and contrary to previous studies (44), the replication defect was not due to decreased Env expression in Vpr-deficient HIV-1-infected cells. Rather, the block of ΔVpr virus infection was at the step of viral transcription and could be rescued by the addition of Vpr in trans into the virion in a single-round-infection analysis. We found that mutations Vpr-Q65R and Vpr-H71R, which ablate the association of Vpr with the CRL4DCAF1 complex, or Vpr-R90K, which does not induce G2 cell cycle arrest (17, 45–50), displayed similar decreases in replication and viral transcription in a single round of infection. Together, these data show a novel postintegration block to HIV-1 replication in MDDCs at the point of viral transcription that is alleviated by virion-associated Vpr.
RESULTS
Vpr-deficient viruses display a replication defect in DCs.
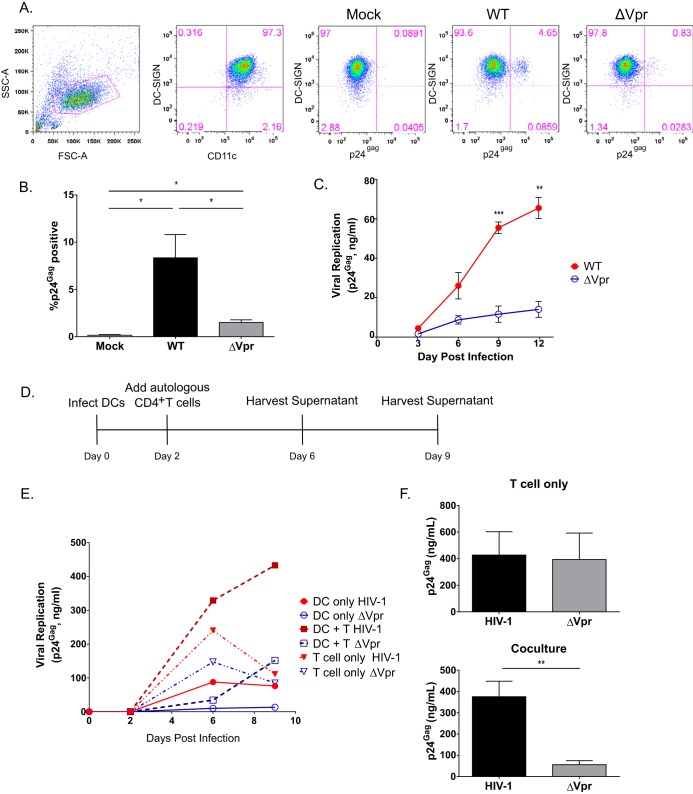
HIV-1 replication in MDDCs is restricted at the reverse transcription (RT) step by SAMHD1, which controls the size of cytosolic deoxynucleoside triphosphate (dNTP) pools (51, 52). Despite the presence of SAMHD1, MDDCs remain susceptible to HIV-1 infection in vitro at a low but measurable level (44, 53–55). We infected MDDCs with the replication-competent WT or Vpr-deficient (ΔVpr) CCR5-tropic Lai-YU2 virus and harvested cells for the determination of intracellular p24Gag expression levels by flow cytometry analysis at 3 days postinfection. The input for these infections was normalized based on the infectious titer of the viruses on TZM-bl cells. As expected, CD11c+ DC-SIGN+ MDDCs were susceptible to viral infection albeit at low levels (Fig. 1A and B). Interestingly, Lai-YU2/ΔVpr failed to establish robust infection in MDDCs (Fig. 1A and B), and there was a reproducible 3- to 5-fold decrease in the percentage of p24Gag+ cells in ΔVpr virus infections compared to WT virus infections (Fig. 1B). To determine the functional consequences of Vpr deficiency on virus spread, DCs and phytohemagglutinin (PHA)/interleukin-2 (IL-2)-activated CD4+ T cells were infected with infectious viruses (multiplicity of infection [MOI] of 1), and cell-free culture supernatants were harvested every 3 days and analyzed for p24Gag content by an enzyme-linked immunosorbent assay (ELISA). While there was some donor variability, Lai-YU2/ΔVpr infection of MDDCs derived from 3 independent donors consistently resulted in significantly lower levels of replication than did wild-type Lai-YU2 infection (Fig. 1C). In contrast to the substantial attenuation of virus spread in Lai-YU2/ΔVpr-infected DCs, both viruses replicated to similar extents in activated CD4+ T cells (Fig. 1E), in agreement with data from previously reported studies (22–25, 27). These results suggest that Vpr plays an important role in facilitating HIV-1 infection of DCs.
FIG 1.
Infection with Vpr-deficient HIV-1 results in attenuated virus replication in MDDCs and MDDC-T cell cocultures. (A) FACS profiles of mock-infected MDDCs or MDDCs infected with Lai-YU2 or Lai-YU2/ΔVpr (MOI = 1) at day 3 postinfection. Cells were stained for CD11c, DC-SIGN, and p24Gag. From left to right, the plots shown depict the gating strategy for the flow cytometry analysis and include plots of forward scatter (FSC)/side scatter (SSC) to exclude cellular debris, anti-CD11c/anti-DCSIGN staining to identify the MDDC population, and DC-SIGN/p24Gag staining to identify productively infected MDDCs in mock-infected or WT (Lai-YU2)- and ΔVpr-infected DCs. (B) Mean percentages (± standard errors of the means) of DC-SIGN+ intracellular p24Gag-positive MDDCs determined from infections of cells derived from three donors as described above for panel A. (C) Replication kinetics of Lai-YU2 and Lai-YU2/ΔVpr in MDDCs infected at an MOI of 1. MDDC supernatants were harvested every 3 days and analyzed for p24Gag content by an ELISA. Data shown are the means (± standard errors of the means) for three independent experiments with MDDCs derived from three independent donors. (D) Schematic of the DC-T cell coculture setup. MDDCs were infected with Lai-YU2 or Lai-YU2/ΔVpr (MOI = 1). At 2 days postinfection, autologous CD4+ T cells (PHA/IL-2 treated) were added at a 2:1 ratio to MDDCs or infected with cell-free virus in parallel (MOI = 1). Supernatants were harvested on day 6 and day 9 postinfection (day 3 or 6 for cell-free CD4+ T cell infection), and the p24Gag content in the culture supernatants was determined by an ELISA. (E) Kinetics of p24Gag production in cell culture supernatants from a representative infection of MDDCs only, CD4+ T cells only, or MDDC-CD4+ T cell cocultures. (F) Mean p24Gag contents (± standard errors of the means) present in the supernatants from infections of CD4+ T cells only or MDDC-CD4+ T cell cocultures at day 6 postinfection (day 3 postinfection for cell-free CD4+ T cell infections). Data are representative of infections of five independent donors. Significance was calculated by using paired Student's t tests (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Numerous studies have demonstrated robust HIV-1 replication in DC-T cell cocultures at levels higher than those observed in infections of either cell type alone and is dependent on the rapid, highly efficient transmission of DC-derived progeny virions to CD4+ T cells across infectious synapses (42, 43, 54–60). We sought to determine the effect, if any, of Vpr deficiency on DC-mediated virus spread to CD4+ T cells. MDDCs were first infected with wild-type Lai-YU2 or Lai-YU2/ΔVpr and cultured for 2 days, prior to the initiation of coculture with autologous activated CD4+ T cells (Fig. 1D). There was a substantial enhancement of virus replication in cocultures infected with the WT virus, compared to ΔVpr virus infections (∼7-fold increase) (Fig. 1E and F). Interestingly, the difference between WT and ΔVpr virus replication in DC-T cell cocultures was greater than that observed in infections of MDDCs or CD4+ T cells alone (Fig. 1E and F). Together, these results suggest that the replication defect observed in MDDCs infected with HIV-1/ΔVpr translates to CD4+ T cells during cell-to-cell contact and transmission.
Defects in Vpr infection are independent of viral glycoprotein expression.
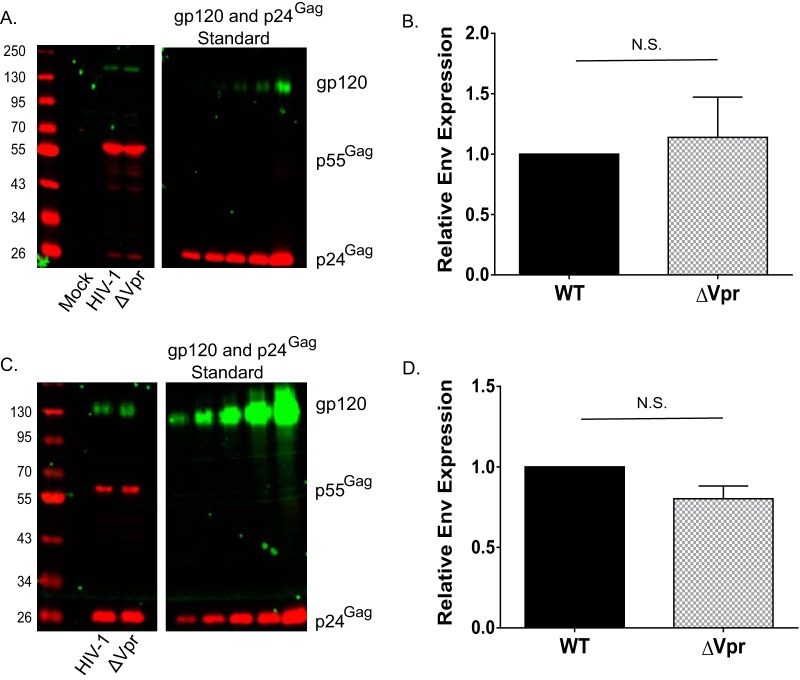
Previous studies suggested a requirement for Vpr in maintaining robust HIV-1 gp120 expression in MDMs and MDDCs by counteracting a myeloid cell-intrinsic mechanism of Env degradation (22, 28, 44). To begin to understand the underlying mechanism accounting for the replication defect of HIV-1/ΔVpr in DCs, we examined viral protein expression in MDDCs infected with wild-type Lai-YU2 or Lai-YU2/ΔVpr (MOI = 3). Infected cells were lysed at 6 days postinfection for quantitative Western blot analysis. We did not observe any steady-state differences in gp120 expression levels when values were normalized to Gag (p55 and p24) levels in MDDCs infected with the WT or ΔVpr virus (Fig. 2A). Quantification of immunoblots from infected MDDC lysates derived from four independent donors showed no significant differences in gp120 expression (Fig. 2B). We next sought to determine if Vpr deficiency might result in decreased gp120 incorporation into virus particles derived from productively infected DCs. MDDC culture supernatants were harvested on multiple days postinfection, and pooled supernatants were concentrated over a sucrose cushion prior to Western blot analysis. We again failed to observe any significant differences in the levels of gp120 incorporation between virus particles derived from WT-infected MDDCs and those derived from ΔVpr-infected MDDCs (Fig. 2C and D). The consistency of the replication defect of HIV-1/ΔVpr in MDDCs in the absence of any significant differences in gp120 expression suggests that the previously hypothesized Vpr-dependent enhancement of gp120 production is unlikely to account for the observed replication defect in the present study (28, 44).
FIG 2.
Vpr does not regulate Env expression in infected MDDCs or incorporation of Env into MDDC-derived virions. (A) Western blot analysis of mock-infected, Lai-YU2 (WT)-infected, or Lai-YU2/ΔVpr-infected MDDCs (MOI = 3) for p55Gag and gp120 expression at day 6 postinfection. (B) Quantification of Western blots for p55Gag and gp120 in infected MDDCs, as described above for panel A, from four independent experiments. The gp120 band intensity was quantified and normalized to the p55Gag values from experiments with infected MDDCs derived from 4 donors. Data shown are means (± standard errors of the means). (C) Western blot analysis of p24Gag and gp120 expression in mock-infected, Lai-YU2 (WT)-infected, or Lai-YU2/ΔVpr-infected MDDCs (MOI = 5). MDDC culture supernatants were harvested at days 3, 6, and 9 postinfection; pooled; and concentrated over a 20% sucrose cushion, and virus pellets were lysed for Western blot analysis. (D) Quantification of data from Western blot analysis of MDDC-derived virions from three independent donors. The band intensity for gp120 was quantified and normalized to the p24Gag band intensity. Data shown are means ± (standard errors of the means). Significance was calculated by using a one-sample t test (N.S., not significant [P > 0.05]).
Infection with Vpr-deficient HIV-1 does not induce type I IFN.
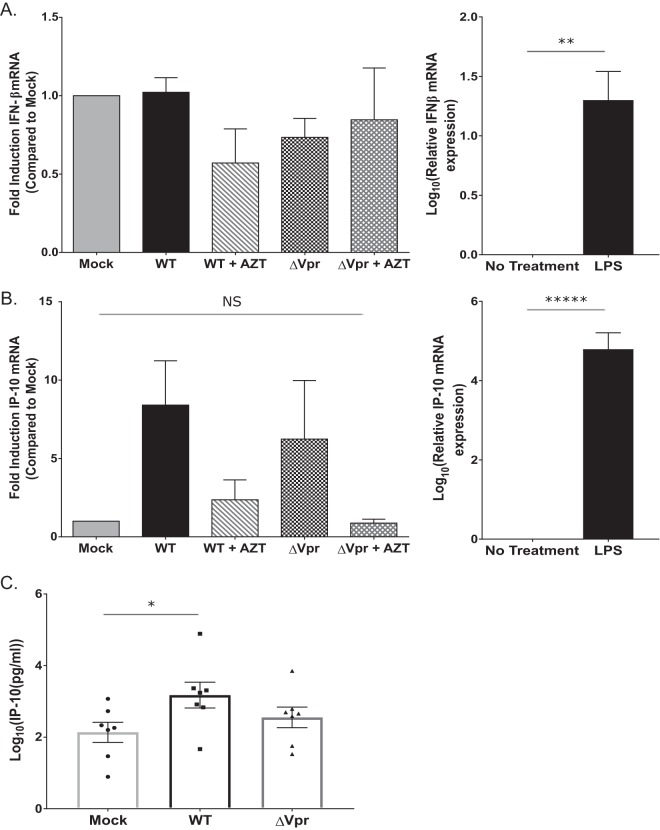
Exposure of target cells to type I IFN potently restricts HIV-1 replication in vitro (61–69). In addition, recent studies suggested that infection with ΔVpr virus induces type I IFN (18, 28, 31–34). Hence, we sought to determine if the induction of an early type I IFN response in HIV-1/ΔVpr infections of MDDCs accounts for the restricted virus replication and spread. MDDCs infected with the wild-type Lai-YU2 or Lai-YU2/ΔVpr virus were harvested at 48 h postinfection, and the mRNA expression levels of IFN-β and the type I IFN-inducible protein interferon gamma-inducible protein 10 (IP-10) were quantified by RT-quantitative PCR (qRT-PCR). At 48 h post-virus exposure, we did not detect significant increases in IFN-β mRNA levels in wild-type- or ΔVpr-infected cells compared to mock-infected cells (Fig. 3A). While the expression of the interferon stimulated gene (ISG) IP-10 was robustly induced by the establishment of productive HIV-1 infection of DCs, differences in IP-10 mRNA levels between WT and ΔVpr virus infections were not statistically significant (Fig. 3B). Note that pretreatment of cells with zidovudine (AZT) reduced the induction of IP-10 mRNA levels to those observed in mock-infected cells, suggesting that the induction of IP-10 expression in virus-exposed cells was dependent on de novo reverse transcription. In contrast, lipopolysaccharide (LPS) treatment of MDDCs for 4 h resulted in robust increases of both IFN-β and IP-10 mRNA levels (Fig. 3A and B). The inability to detect differences in the mRNA expression levels of IFN-β in MDDCs infected with the WT and ΔVpr viruses was also mirrored by the absence of differences in protein levels in infected MDDC culture supernatants (data not shown). We used a sensitive bioassay to measure type I IFN production in infected MDDC supernatants and failed to detect any type I IFN production in HIV-1-infected MDDCs over mock-infected controls (data not shown) (70). In contrast, IP-10 was robustly secreted in both Lai-YU2 (WT)- and Lai-YU2/ΔVpr-infected MDDC culture supernatants at day 3 postinfection, although the magnitude of IP-10 induction was donor dependent (Fig. 3C). Furthermore, we observed a significant increase in IP-10 production upon WT virus infection of MDDCs compared to mock-infected cells (Fig. 3C). Again, AZT pretreatment reduced the secretion of IP-10, indicating that IP-10 production is dependent on the completion of reverse transcription (Fig. 3C). Taken together, these results suggest that Vpr deficiency does not result in the induction of type I IFNs during the establishment of productive HIV-1 infection of MDDCs and is unlikely to play a role in the restriction of HIV-1/ΔVpr in DCs.
FIG 3.
Vpr deficiency does not result in enhanced type I IFN production in productively infected MDDCs. (A and B) qRT-PCR for IFN-β (A) and IP-10 (B) transcript levels in infected MDDCs at 48 h postinfection. MDDCs were mock infected or infected with Lai-YU2 or Lai-YU2/ΔVpr (MOI = 2) in the presence or absence of AZT (10 μM). The amount of IFN-β or IP-10 transcripts in infected MDDCs was normalized to the number of cells by using a GAPDH control and is reported relative to that in mock-infected MDDCs (set at a value of 1) from four independent donors. LPS treatment for 4 h was used as a positive control for IFN-β and IP-10 production. Data are the log-transformed means (±standard errors of the means) of results for seven donors. (C) The secreted level of IP-10 in MDDC culture supernatants infected with Lai-YU2 or Lai-YU2/ΔVpr (MOI = 1) at day 3 postinfection was measured by an ELISA. The data shown are the log-transformed means (± standard errors of the means) of results from independent experiments with MDDCs derived from four donors for panels A and B and from six donors for panel C. Significance was calculated by using paired Student's t test or a one-value t test (comparing normalized data) (NS, not significant [P > 0.05]; *, P < 0.5; **, P < 0.1; *****, P < 0.0001).
Infection with ΔVpr viruses results in decreased infection in a single round of replication and is rescued by virion-associated Vpr.
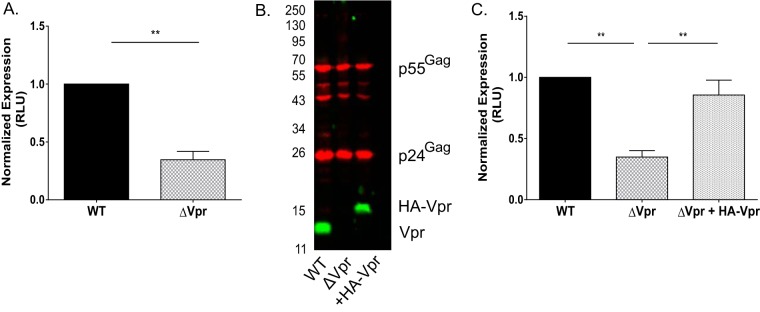
To identify the step of the virus replication cycle in MDDCs that is affected by Vpr, we next performed single-cycle-infection analysis. MDDCs were infected with HIV-1 reporter viruses pseudotyped with vesicular stomatitis virus G (VSV-G) and expressing luciferase upon the establishment of infection that express (Lai-luc Δenv/G or the WT) or do not express (Lai-luc Δenv/G ΔVpr or ΔVpr) Vpr. Infection with the ΔVpr virus resulted in a 3- to 5-fold decrease in luciferase expression compared to that with infection with the WT virus (Fig. 4A), suggesting that Vpr acts early in the HIV-1 replication cycle in MDDCs at steps preceding virion assembly and maturation. Since Vpr is a virion-associated protein, we next sought to determine whether incoming virion-associated Vpr was sufficient or if de novo-synthesized Vpr was required for the enhancement of virus replication in DCs. We produced Lai-luc Δenv/G ΔVpr complemented with hemagglutinin (HA) epitope-tagged Vpr in trans (Lai-luc Δenv/G Vpr-trans) via cotransfection of HEK293T cells with a functional HA-Vpr expression plasmid and the Lai-luc Δenv/G ΔVpr proviral plasmid. HA-Vpr was efficiently incorporated into ΔVpr virus particles to levels similar to those observed for WT virus particles (Fig. 4B). We then infected MDDCs with Lai-luc Δenv/G-WT, ΔVpr, or Vpr-trans viruses and lysed the cells on day 3 postinfection. The incorporation of Vpr in trans within incoming virus particles rescued ΔVpr virus infection in a single-round assay (Fig. 4C), suggesting that virion-incorporated Vpr is sufficient for overcoming cell-intrinsic blocks to early steps in HIV-1 replication in DCs.
FIG 4.
Infection of MDDCs with Vpr-deficient viruses results in a block to HIV-1 replication in single-round-infection analyses. (A) MDDCs infected with 40 ng p24Gag equivalents of VSV-G-pseudotyped Lai-luc Δenv (WT or ΔVpr) were lysed at 3 days postinfection, and viral replication was quantified by measuring luciferase activity in cell lysates. The luciferase activity in ΔVpr-infected cell lysates was normalized to that of WT virus-infected MDDC lysates and is reported as the mean (± standard error of the mean) of data from four independent experiments with MDDCs derived from four independent donors. (B) Western blot analysis of Vpr incorporation into virus particles (Lai-luc Δenv/G, Lai-luc Δenv/G ΔVpr, or Lai-luc Δenv/G Vpr-trans) derived from transient transfection of HEK293T cells. (C) MDDCs were infected with 40 ng p24Gag equivalents of viruses (Lai-luc Δenv, Lai-luc Δenv ΔVpr, or Lai-luc Δenv ΔVpr plus HA-Vpr) and lysed at 3 days postinfection. Cell lysates were analyzed for luciferase activity, and the data shown are normalized to values for WT virus infection and are means (±standard errors of the means) from 4 independent experiments. Significance was calculated by using paired Student's t test or a one-value t test (comparing normalized data) (**, P < 0.01). RLU, relative light units.
Proviral LTR-mediated transcriptional activity is attenuated in Vpr-deficient virus infection of DCs.
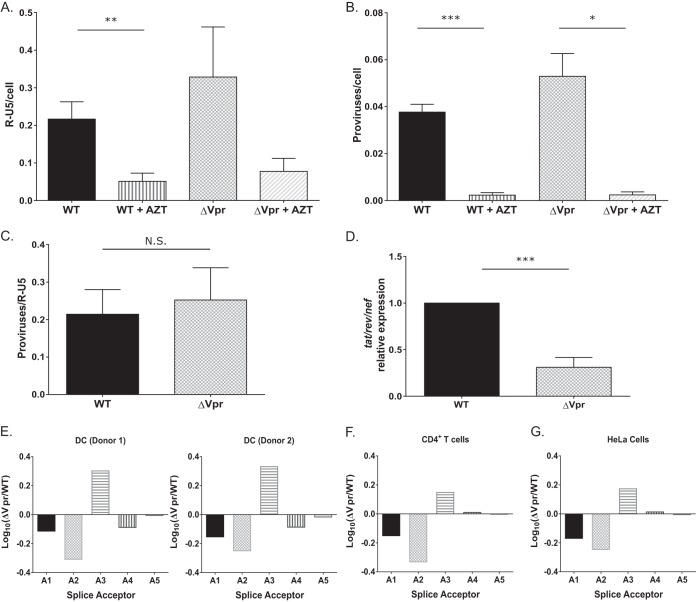
Since the block to HIV-1/ΔVpr infection in MDDCs is evident within a single round of replication and is independent of the mode of virus entry (VSV-G-pseudotyped virus infection was also restricted) (Fig. 4A), we assessed the effect of Vpr deficiency on HIV-1 RT and integration efficiency in DCs. We used quantitative PCR (qPCR) to measure RT products and the number of proviruses at day 3 postinfection using the R-U5 and Alu-Gag primer pairs, respectively (71, 72). Infections were also performed in the presence of AZT to control for contaminating input plasmid DNA. In contrast to previously reported findings (41), we saw no decrease in the number of RT products (Fig. 5A) or integrants (Fig. 5B and C) upon infection with the ΔVpr virus compared to WT virus infections (Fig. 5A to C). Previous studies suggested that Vpr can modulate HIV-1 long terminal repeat (LTR) transcriptional activity (29, 46, 73–75). We therefore asked if the block to HIV-1/ΔVpr infection occurs at the stage of viral transcript production. To determine the effect of Vpr on LTR-mediated transcription from proviruses, we used qRT-PCR to measure multiply spliced tat-rev-nef transcripts at 48 h postinfection (Fig. 5D). Similar to our findings with luciferase reporter expression in infected DCs, we observed a 4-fold decrease in the number of multiply spliced HIV-1 transcripts in HIV-1/ΔVpr-infected cells, suggesting that Vpr deficiency results in the inhibition of proviral LTR-mediated transcription in DCs.
FIG 5.
Viral transcription is attenuated in ΔVpr virus-infected MDDCs. (A to C) MDDCs infected with WT or ΔVpr viruses (MOI = 2) in the presence or absence of AZT (10 μM) were lysed at 72 h postinfection and processed for DNA isolation. Note that infected cells were cultured in the presence of indinavir (1 μM) to prevent viral spread. qPCR was used to detect early RT products (A) and integrated proviruses (B) by using the R-U5 and Alu-PCR primer sets, and the number of integrated proviruses was normalized to early RT products for each infection (C). The data reported are the means (± standard errors of the means) of results from three independent experiments. (D) The number of multiply spliced viral transcripts (tat-rev-nef) in MDDCs infected with Lai-YU2 or Lai-YU2/ΔVpr (MOI = 1) was determined at 48 h postinfection by qRT-PCR. Viral transcripts were measured by using primers specific for tat-rev-nef multiply spliced transcripts. Data shown are means (± standard errors of the means) of results from four independent experiments with MDDCs derived from four donors. (E) Quantification of the 4-kb class of splice variants for MDDCs infected with Lai-YU2 or Lai-YU2/ΔVpr (MOI = 2) for 72 h. The data were normalized, log10 transformed, and then graphed according to slice acceptor usage. (E to G) Histograms show fold changes in splicing from D1 to each of the 5 viral splice acceptor sites A1 through A5 relative to a WT control. Splicing was quantified by using a Primer ID splicing assay for MDDCs from two independent infections (E) and productively infected CD4+ T cells (F) and HeLa cells (G), and data are from a single deep-sequencing experiment. Significance was calculated by using unpaired Student's t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
We next sought to determine if the decrease in multiply spliced viral mRNA levels in HIV-1/ΔVpr-infected MDDCs was driven by changes in the pattern of viral mRNA splicing. We used a novel Primer ID-tagged deep-sequencing assay (76, 77) to determine the relative abundances of different splice variants in WT- and ΔVpr-infected DCs and compared the viral splice site usage to those observed in WT- or ΔVpr-infected CD4+ T cells and HeLa cells (Fig. 5E to G). The data depict the relative quantity of 4-kb singly spliced mRNAs for each splice acceptor and are reflective of the changes observed in the 1.8-kb multiply spliced mRNA (data not shown). We detected minor differences in splice acceptor usage between WT and ΔVpr infections of MDDCs. We observed small decreases in the use of the Vif (A1) and Vpr (A2) splice acceptors and a small increase in the use of the Tat (A3) splice acceptor, but these differences were well within the normal range of splicing variation seen in productive viral infections (77). These small differences in splice site usage were consistently observed in infections of CD4+ T cells and HeLa cells. Since the differences in splicing are both relatively small and observed in two cell types (primary activated CD4+ T cells and HeLa cells) that do not restrict ΔVpr virus replication, it is unlikely that the efficiency of viral mRNA splicing or the choice of mRNA splice acceptor sites is a factor that contributes to the restricted replication of the ΔVpr virus in MDDCs.
Mutations in the C-terminal end of Vpr or those that disrupt binding to the CRL4DCAF1 ubiquitin ligase attenuate viral replication in DCs.
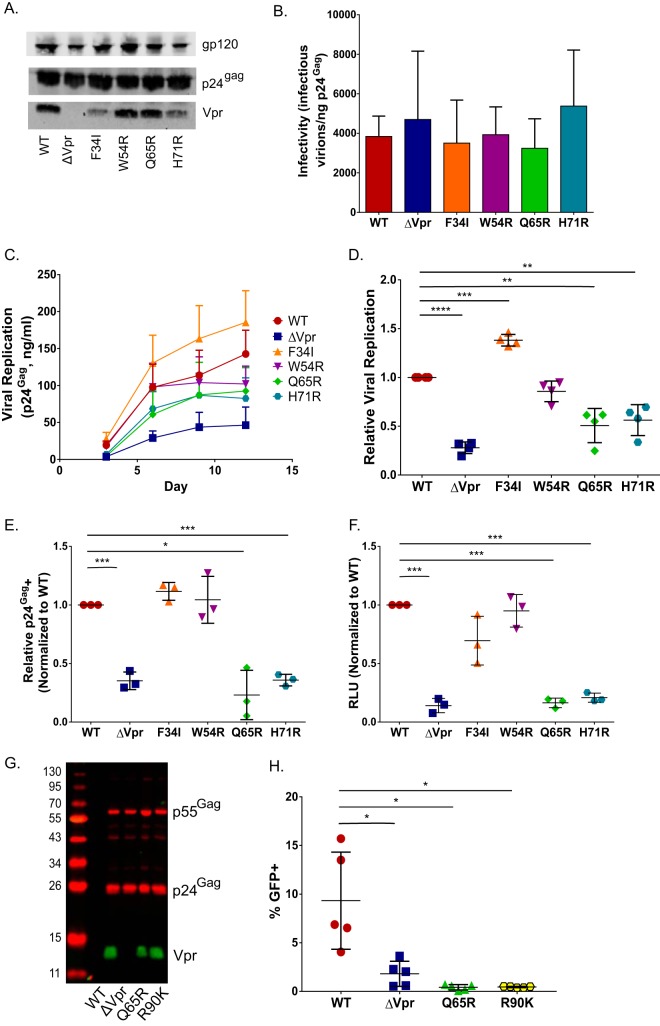
A range of functions have been attributed to Vpr, including G2/M cell cycle arrest, enhancement of the fidelity of reverse transcription, nuclear import and/or nuclear tethering of the preintegration complex, and induction of apoptosis (12–14, 30, 41, 46, 78). To clarify which of the known functions of Vpr are important for enhancing HIV-1 replication in DCs, a panel of mutations was introduced into a Vpr open reading frame (ORF) with previously characterized effects on Vpr functions. HEK293T-derived virus particles were analyzed by quantitative Western blotting to assess the incorporation of mutant Vpr proteins into viral particles (Fig. 6A). While all viral mutants expressed and incorporated Vpr in virus particles, the Vpr-F34I and Vpr-H71R mutants had slightly decreased levels of incorporation of Vpr compared to those of wild-type viruses (Fig. 6A), although both the wild-type and Vpr mutant viruses were equally infectious to TZM-bl cells on a per-particle basis (Fig. 6B). MDDCs were infected with replication-competent HIV-1 (WT or Vpr mutants) at equal MOIs, and the extent of viral replication was measured by periodic quantification of p24Gag levels in cell-free culture supernatants by an ELISA (Fig. 6C). Since there was donor-to-donor variability in the kinetics and extent of virus replication in DCs, we calculated the area under the curve of replication kinetics obtained from four independent infections (Fig. 6D). As depicted in Fig. 6C and D, infection with both the Vpr-Q65R and Vpr-H71R mutant viruses resulted in significantly attenuated virus replication and spread, similar to what was observed for ΔVpr virus replication in MDDCs (Fig. 6C and D). In contrast, the replication of both the Vpr-F34I and Vpr-W54R mutants was not significantly different from that observed with wild-type virus infections (Fig. 6C and D). Cumulative analysis revealed that the replication of the Vpr-Q65R and Vpr-H71R mutants, which lack the ability to associate with the CRL4DCAF1 complex (16–18, 30, 45, 46, 49, 79, 80), was significantly reduced (P < 0.01), similar to what was observed for ΔVpr virus infection (Fig. 6D). Interestingly, the replication of the Vpr-F34I mutant, which incorporates reduced levels of Vpr in virions (Fig. 6A) and displays a reduced association with the nuclear envelope (30, 45, 50), was slightly enhanced over that observed for wild-type virus replication (P < 0.01) (Fig. 6D), suggesting that a threshold amount of functional Vpr that is still present in the incoming virus particle is sufficient for the establishment of productive infections in DCs. Mutation Vpr-W54R, which ablates the binding of Vpr to UNG2 (19, 45, 80–82), had a negligible effect on viral replication in DCs.
FIG 6.
Vpr mutants deficient for interaction with the DCAF1/DDB1/E3 ubiquitin ligase and inducing G2 cell cycle arrest are attenuated in a single cycle of replication in MDDCs. (A) Representative Western blot analysis of HEK293T-derived Lai-YU2 (WT) and the indicated Vpr mutant viruses used for MDDC infections. Blots were probed with anti-p24Gag, anti-Vpr, and anti-gp120 antibodies. (B) Infectivity of Lai-YU2 and the corresponding Vpr mutants in TZM-bl cells, reported as the number of infectious units (blue cells) per nanogram of p24Gag equivalent, and results are the means (± standard errors of the means) of data from three independent viral preparations. (C) Viral growth curves for four independent infections of MDDCs with Lai-YU2 and the indicated Vpr mutants in DCs. Viral growth was determined by analyzing p24Gag release into cell culture supernatants at days 3, 6, 9, and 12 postinfection by an ELISA. (D) Areas under the curve compiled for four independent MDDC infections represented in panel C, normalized to the value for WT virus infection, which was set to 1 (means ± standard errors of the means). (E) Percentage of p24Gag-positive MDDCs at day 3 postinfection as measured by intracellular p24Gag staining and FACS analysis. Cells were treated with indinavir (1 μM) post-virus exposure to prevent viral spread. The data were normalized to the value for WT virus infection, which was set to 1, and depict the means (± standard errors of the means) of results from three independent infections of MDDCs from three donors. (F) MDDCs infected with 40 ng p24Gag equivalents of Lai-luc Δenv/G (WT or Vpr mutants) were lysed at 3 days postinfection, and viral replication was quantified by measuring luciferase activity in cell lysates. The luciferase activity in Vpr mutant infections was normalized to that of WT virus infections, which was set to 1, and the data shown are the means (± standard errors of the means) for three independent experiments. (G) Western blot analysis of HEK293T-derived Lai-GFP Δenv/G (WT) virus particles or the indicated Vpr mutant virus particles. (H) MDDCs infected with Lai-GFP Δenv/G (WT) or the indicated Vpr mutants (MOI = 3) were harvested at day 3 postinfection and processed for FACS analysis. The data shown are the mean percentages of GFP-positive cells (± standard errors of the means) from five independent experiments with cells derived from five independent donors. Significance was calculated by using paired Student's t test or a one-value t test (comparing normalized data) (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
We next sought to determine which of these Vpr mutants could recapitulate the single-cycle replication defect observed for HIV-1/ΔVpr infection of MDDCs (Fig. 4A). We infected MDDCs with either replication-competent viruses (Lai-YU2 WT or Vpr mutants) (MOI = 1) in the presence of a protease inhibitor (indinavir) or equal amounts of p24Gag equivalents of Lai-luc Δenv/G encoding the various Vpr mutations. Similar to the results observed with replication-competent viruses, both the number of p24Gag-postitive cells (Fig. 6E) and the level of luciferase production (Fig. 6F) from infections with the Vpr-Q65R and Vpr-H71R mutants were significantly attenuated in a single round of infection compared to isogenic WT viruses. While the host protein targeted by HIV-1 Vpr to induce G2 cell cycle arrest has not been identified, the C-terminal tail of this protein has been proposed to bind the unknown host factor, and mutations in the C-terminal tail of Vpr abrogate the ability of Vpr to induce G2 cell cycle arrest (45). To determine the role of Vpr-mediated G2 cell cycle arrest in virus infection enhancement in DCs, an additional mutation, Vpr-R90K, was introduced into a green fluorescent protein (GFP)-expressing single-cycle virus (Lai-GFP Δenv/G). The Vpr-R90K mutant can bind the CRL4DCAF1 complex but fails to induce G2 arrest in cycling cells (45, 50, 80). Despite equivalent incorporation into the virion as WT Vpr (Fig. 6G), infection of MDDCs with the Vpr-R90K mutant resulted in a significant infection defect in single-round analyses (Fig. 6H), similar to what was observed in infections with ΔVpr or Vpr-Q65R viruses, suggesting that an interaction with a putative host factor whose degradation is critical for the induction of G2 cell cycle arrest is required to enhance HIV-1 infection of DCs. Together, our data suggest that there is a novel block to HIV-1 infection in MDDCs in the absence of Vpr that is present in a single round of infection and manifests at the stage of viral transcription. Further studies are under way to determine the exact mechanism by which Vpr alleviates the DC-intrinsic block to HIV-1 replication.
DISCUSSION
In the work presented here, we examined the role of Vpr in establishing productive HIV-1 infection of DCs. Previous work in this field suggests that Vpr likely regulates a complex network of host interactions that may vary depending on the cell type infected. We find that, unlike what has been previously observed in activated CD4+ T cells and monocyte-derived macrophages (20, 22–25, 29), infection of MDDCs with ΔVpr viruses was significantly attenuated compared to WT HIV-1 infections (Fig. 1), similar to the findings reported previously by de Silva et al. (41). Interestingly, Vpr-mediated enhancement was observed for both single-round viral infection as well as spreading infections, contrary to what was reported previously (28, 41). Furthermore, the single-round replication defect could be rescued by complementing back Vpr in trans in the incoming virion (Fig. 4), indicating that incoming virion-associated Vpr is necessary for the establishment of efficient HIV-1 infection of DCs. Initiating infections with the Vpr mutants Vpr-Q65R, Vpr-H71R, and Vpr-R90K, which lack the ability to either engage the CRL4DACF1 complex or bind the yet-to-be-identified host factor(s) necessary for inducing G2 cell cycle arrest, displayed replication deficits similar to those observed for the ΔVpr virus in both spreading infections and single-round infections (Fig. 6).
Surprisingly, the block to ΔVpr virus replication in MDDCs was evident at a postintegration step and resulted in reduced numbers of viral mRNAs, suggesting that Vpr is acting either directly or indirectly to enhance transcription from the viral LTR (Fig. 5). It was reported previously that Vpr can transactivate the viral LTR in a number of cell types and that this function correlates with the ability of Vpr to induce G2 cell cycle arrest (26, 29, 46, 73, 75). Previous studies have also shown that Vpr of both simian immunodeficiency viruses (SIVs) SIVmac and SIVagm can also transactivate their respective LTRs (74, 83, 84), suggesting that this is a conserved function among nonhuman primate lentiviral Vpr proteins. While it is possible that Vpr-mediated transactivation could be more robust in DCs than in CD4+ T cells (Fig. 1E), another hypothesis is that Vpr indirectly activates transcription to promote infection in cells that have a higher barrier to infection.
Unlike most of the other lentiviral accessory proteins, Vpr is actively packaged into the budding virion through associations with the p6 region of Gag (3–6, 10). Our work with MDDCs suggests that there may be a novel role for virion-associated Vpr to enhance viral transcription and increase infection of DCs. These findings are at odds with data from recently reported studies on the role of Vpr in modulating de novo HIV-1 Env production in productively infected macrophages and MDDCs (28, 44). While we occasionally see a decrease in viral Env production during infection with the ΔVpr virus in MDDCs (one out of four donors tested), infection of MDDCs from most of the donors revealed no differences in Env expression or virion incorporation (Fig. 2). It is possible that the use of different viral clones, primary cell variation derived from multiple donors, or different infection conditions might play a role in the differences between our results and those described previously. Since we observed infection differences in a single-round-infection assay, putative effects of Vpr on Env expression are unlikely to play a role in the establishment and spread of virus infection in MDDCs and DC-T cell cocultures.
HIV-1 is not unique among primate lentiviruses in expressing a protein that functionally allows infection of DCs. HIV-2 and certain SIV lineages express Vpx, another small accessory protein that targets the host restriction factor SAMHD1 for proteosomal degradation by recruiting it to the CRL4DACF1 complex and facilitates infection of MDDCs (51, 52, 85). Interestingly, Vpr-mediated replication enhancement in MDDCs was substantially attenuated upon infection with Vpr mutants (Q65R or H71R) (Fig. 6F) that lack the ability to interact with the CRL4DCAF1 complex or upon infection with the Vpr-R90K mutant (Fig. 6H) that fails to interact with the host factor(s) hypothesized to be recruited to the CRL4DCAF1 complex for proteosomal degradation. Since Vpr is introduced into target cells along with the incoming virion because of its association with the viral capsid, we hypothesize that early interactions of Vpr with a host factor and the recruitment of this protein to the CRL4DCAF1 complex for proteosomal degradation are essential for promoting HIV-1 replication in DCs, similar to the ability of Vpx from SIVmac/SIVsmm/HIV-2 lineages to promote infection of DCs.
Across primate lentiviral Vpr evolution, the induction of the DDR and G2 cell cycle arrest are conserved functions, and Vpr proteins from diverse primate lentiviruses have been shown to associate with and degrade many DDR regulatory proteins, including SLX4com, HLTF, and UNG2 (18–21, 80, 81, 86–89). While DDR activation may represent a cell-intrinsic antiviral response, it has been suggested that both RNA and DNA viruses induce DDR signaling to promote cellular conditions that are favorable for viral replication (83, 90–93). For instance, the induction of DDR signaling activates ataxia telangiectasia mutated (ATM) kinase, which results in the activation of nuclear factor kappa light chain enhancer of activated B cells (NF-κB) (94, 95). Additionally, the DDR pathway also directly activates proinflammatory responses through the induction of interferon regulatory factors (IRFs) or through the recruitment of coactivators and chromatin-modifying complexes, such as ten-eleven translocation methylcytosine (TET) dioxygenases, which we hypothesize might also activate viral transcription (96). Since the barrier to the successful establishment of infection in noncycling, metabolically quiescent cells like MDDCs is higher than that in activated CD4+ T cells or MDMs, the Vpr-mediated activation of NF-κB and coactivator recruitment to the viral LTR might be a viral strategy for overcoming the restrictive cellular environment and for the optimal production of progeny virions. In line with this hypothesis, numerous studies have documented that Vpr is able to modulate NF-κB activity in different cell lines and primary cells, although those studies rarely agree on the mechanism of regulation or direction of modulation (26, 75, 97–102). Recent work by Höhne et al. shows similar effects of Vpr on viral replication in nonactivated primary CD4+ T cells (26), which have barriers to infection similar to those of MDDCs, including increased expression of SAMHD1 and low baseline NF-κB activity (103, 104). Some studies have shown that virion-associated Vpr-dependent activation of NF-κB occurs via a transforming growth factor β-activated kinase 1 (TAK1) signaling cascade, while other studies have shown that secreted or synthetic Vpr stimulates NF-κB signaling through a Toll-like receptor 4 (TLR4)-dependent mechanism (75, 99, 100, 102). Our data also demonstrate the upregulation of IP-10 upon HIV-1 (WT) infection (Fig. 3C), which is also dependent on NF-κB activation (105–107).These results suggest a link between Vpr-mediated NF-κB activation in MDDCs and enhanced viral gene expression and proinflammatory cytokine secretion, which may act in vivo to enhance recruitment, activation, and infection of CD4+ T cells, resulting in increased viral dissemination (Fig. 1E) (27, 75, 99).
Previous studies with peripheral blood myeloid MDDCs and MDDCs from HIV-1 elite controllers have shown that these cells may be critical for viral control, acting to capture the virus and enhance T cell-specific immunity to HIV-1 while being less susceptible to HIV-1 infection than MDDCs from healthy controls (108, 109). Understanding the mechanisms that control HIV-1 replication in MDDCs, which are overcome by Vpr, might lead to new insights into viral dissemination and persistence in vivo and the development of novel anti-HIV-1 therapeutics.
MATERIALS AND METHODS
Plasmids.
HIV-1 proviral plasmids Lai-YU2 env, Lai/Bal, Lai-luc Δenv (Env-deficient HIV-1 containing a luciferase reporter gene in place of nef), and Lai-GFP Δenv (Env-deficient HIV-1 containing GFP in place of nef) and the HA-Vpr expression plasmid were previously described (30, 110). Proviral Lai (CXCR4-tropic) clones containing Vpr mutations F34I, W54R, and H71R and a frameshift mutation in Vpr (ΔVpr) were described previously (30, 46, 80, 111). These Vpr mutations were transferred to the Lai-YU2 env, Lai-luc Δenv, or Lai-GFP Δenv proviral plasmids using ApaI and SalI restriction sites. To create proviral clones encoding the Vpr-Q65R mutation, the ApaI-SalI fragment of Lai-YU2 env was subcloned into the pSL1180 cloning vector (Stratagene), and site-directed mutagenesis was performed by using a kit (QuikChange II; Agilent Technologies) and the following primers: 5′-GCCATAATAAGAATTCTGCGACAACTGCTGTTTATCCATTTC-3′ and 5′-GAAATGGATAAACAGCAGTTGTCGCAGAATTCTTATTATGGC-3′. The mutated fragment was ligated back into Lai-YU2 env, Lai-luc Δenv, or Lai-GFP Δenv using ApaI-SalI restriction sites. The point mutation Vpr-R90K was derived by subcloning the SalI-BamHI fragment into pSL1180 and via site-directed mutagenesis (QuikChange II; Agilent Technologies) using the following primers: 5′-CGTTACTCAACAGAGGAGAGCAAAAAATGGAGCCAGTAGATCCTAGAC-3′ and 5′-GTCTAGGATCTACTGGCTCCATTTTTTGCTCTCCTCTGTTGAGTAACG-3′. The mutated fragment was cloned back into Lai-GFP Δenv.
Cells and viruses.
MDDCs were derived from monocytes isolated from peripheral blood mononuclear cells as previously described (112). TZM-bl and HEK293T cells were described previously (112–114). Replication-competent Lai-YU2 env viruses were derived by using calcium phosphate-mediated transient transfection of HEK293T cells, as described previously (115). HIV-1 vectors were generated from HEK293T cells via the cotransfection of Lai-luc Δenv or Lai-GFP Δenv with a VSV-G expression plasmid. Virus-containing cell supernatants were harvested at 2 days posttransfection, passed through 0.45-μm filters, and stored at −80°C until further use. For some experiments, virus particles were concentrated by ultracentrifugation on a 20% sucrose cushion (24,000 rpm at 4°C for 2 h with an SW28 rotor [Beckman Coulter]) (116). The virus pellets were resuspended in phosphate-buffered saline (PBS), aliquoted, and stored at −80°C until use. The capsid content of HIV-1 was determined by a p24Gag ELISA, while the infectious titer was determined via infection of TZM-bl cells (114, 117). Viral replication in MDDCs and DC-T cell cocultures was determined by measuring the p24Gag content in cell culture supernatants at the indicated days postinfection by an ELISA (117). Infection of MDDCs using a luciferase reporter virus was analyzed by using the Bright-Glo luciferase system (Promega), as previously described (118).
Drug treatments.
For the indicated experiments, cells were pretreated with AZT (10 μM; NIH AIDS Research and Reference Reagent Program) for 30 min at 37°C prior to infection, and the AZT concentration was maintained for the duration of the cultures, or cells were treated with indinavir (1 μM; NIH AIDS Research and Reference Reagent Program) post-virus exposure.
Quantitative Western blotting.
To detect Gag and Env in cell and virus particle lysates, cell lysates (normalized to equivalent amounts of cell-associated Gag) or 100 ng p24Gag virus equivalents (as determined by a quantitative ELISA) was run through SDS-PAGE gels and transferred onto nitrocellulose membranes by using a semidry transfer apparatus, as previously described (116). Blots were blocked and probed with rabbit anti-gp120 (a gift from Nancy Haigwood) and mouse anti-p24Gag (clone p24-2; NIH AIDS Research and Reference Reagent Program), followed by goat anti-mouse IgG DyLight 680 (Pierce) and goat anti-rabbit IgG DyLight 800 (Pierce). To determine Vpr incorporation, a polyclonal rabbit anti-Vpr antibody (clone 1-50; NIH AIDS Research and Reference Reagent Program) was used, followed by goat anti-rabbit IgG DyLight 700. The membranes were scanned with an Odyssey scanner (Li-Cor).
qRT-PCR.
For the quantitation of IFN-β and IP-10 mRNA levels, MDDCs (2 × 106 to 4 × 106 cells) were mock infected or infected with Lai-YU2 or Lai-YU2/ΔVpr (MOI = 2). At 48 h postinfection, cells were harvested for RNA isolation by using RNeasy RNA isolation kits (Qiagen), and cDNA was synthesized by using oligo(dT) primers and Superscript III RT (Invitrogen). cDNA corresponding to 200 ng of RNA was analyzed by qRT-PCR using SYBR green (Invitrogen) to quantify mRNA levels for IFN-β (forward primer 5′-ATTCTAACTGCAACCTTTCG-3′ and reverse primer 5′-GTTGTAGCTCATGGAAAGAG-3′), IP-10 (forward primer 5′-TCATTGGTCACCTTTTAGTG-3′ and reverse primer 5′-AAAGCAGTTAGCAAGGAAAG-3′), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (forward primer 5′-AGGGATGATGTTCTGGAGAG-3′ and reverse primer 5′-CAAGATCATCAGCAATGCCT-3′). The ΔΔCT value relative to GAPDH in the mock-infected cultures was set to 1, and data from the infected cultures are reported as fold enhancements. To determine the extent of de novo viral transcription, the number of tat-rev-nef multiply spliced transcripts was determined by qRT-PCR using SYBR green, as described previously (58), with the following primer set: forward primer 5′-GCGACGAAGACCTCCTCAG-3′ and reverse primer 5′-GAGGTGGGTTGCTTTGATAGAGA-3′. The data were normalized to GAPDH levels. As a control, MDDCs were treated with AZT (10 μM) for 30 min prior to infection, and drug levels were maintained during the course of infection.
Quantification of viral integration.
To determine the number of proviral integrants, MDDCs (3 × 106 cells) were infected with the virus (MOI = 3) for 2 h at 37°C, washed with PBS twice, and cultured for 72 h before cells were lysed for DNA extraction with a DNeasy kit (Qiagen). As a background control, MDDCs were treated with 10 μM AZT for at least 30 min prior to infection. Quantitative Alu PCR was performed by using 20 ng of DNA with the following primer sets, as described previously (72). For the first step, the following primers were used: Alu-forward (5′-GCCTCCCAAACTGCTGGGATTACAG-3′) and Gag-reverse (5′-GCTCTCGCACCCATCTCTCTCC-3′). For the second step, the following primers and probe were used: primers R-U5-F (5′-GCCTCAATAAAGCTTGCCTTGA-3′) and R-U5-R (5′-TCCACACTGACTAAAAGGGTCTGA-3′) and probe R-U5-Probe (5′–6-carboxyfluorescein [FAM]–CCAGAGTCACACAACAGACG–6-carboxytetramethylrhodamine [TAMRA]–3′). The data were normalized to a standard curve generated from infected HEK293 cell DNA (71, 72).
Splicing assay.
The splicing assay was described in detail previously (77). Briefly, cDNA primers with an internal random sequence block, as denoted by Ns in their sequences (Primer ID) (76), were designed to be located within the env intron to measure the 4-kb size class of spliced viral RNAs or to span the D4/A7 splice junction to measure the 1.8-kb size class. The reverse primer for the 4-kb size class was 5′-GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTNNNNNNNNNNNNNNGTACGCTAATACTTGTAAAGATTGCAGTACATGTACTACTT-3′, and the reverse primer for the 1.8-kb size class was 5′-GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTNNNNNNNNNNNNNNCAGTCTGAGCTGGGAGGTGGGTTGC-3′. Whole-cell RNA from infected cells was purified and used in a cDNA reaction. After the removal of the cDNA primers, PCR was carried out by using a downstream primer encoded in the cDNA primer tail and a forward primer placed just upstream of the D1 major donor site in the 5′ noncoding region, 5′-GCCTCCCTCGCGCCATCAGAGATGTGTATAAGAGACAGNNNNTGCTGAAGCGCGCACGGCAAG-3′. PCR products were sequenced by using the MiSeq platform, and sequence reads with the same Primer ID were collapsed into a single read (to correct for skewing during PCR, since each unique Primer ID tag represents a separate viral mRNA template). Data were processed by using customized scripts that are available upon request. The number of unique Primer IDs for each spliced product was used to determine the relative level of splicing from each splice donor to each splice acceptor in the viral genome, with the exception of splicing events for the nef splice acceptor A7.
Cytokine measurements.
The secreted level of IP-10 in MDDC culture supernatants was measured by using a commercially available ELISA kit (Becton Dickinson). Secreted levels of bioactive type I IFN in infected MDDC supernatants were measured by using a HEK293 ISRE-luc cell line, which expresses luciferase under the control of an IFN-inducible promoter carrying the IFN-stimulated response element (70). Briefly, HEK293 ISRE-luc cells (8 × 104) were incubated with MDDC culture supernatants for 21 h. Cells were lysed, and luciferase activity in the cell lysates was analyzed with the Bright-Glo luciferase system (Promega), as described above. Serial dilutions of recombinant interferon alpha ranging from 200 to 0.39 U/ml (PBL Interferon Source) were added to cells in each experiment for generating a standard curve.
FACS analysis.
Intracellular fluorescence-activated cell sorter (FACS) analysis for p24Gag was done by using fluorescein isothiocyanate (FITC)-conjugated anti-p24Gag monoclonal antibody (KC57; Coulter), as previously described (117). Surface staining for CD11c was done by using allophycocyanin (APC)-conjugated anti-CD11c (clone B-ly6; Becton Dickinson). Cells were analyzed by using either an LSRII or a FACSCalibur (Becton Dickinson) instrument.
Ethics statement.
This work has been deemed exempt by the Institutional Review Board of Boston University School of Medicine, since it does not meet the definition of human subjects research. All human samples used were collected in an anonymous fashion and no identifiable private information was collected.
ACKNOWLEDGMENTS
We thank Nancy Haigwood (Oregon National Primate Research Institute) for the anti-gp120 antiserum, Junzhi Wang (National Institute for the Control of Pharmaceutical and Biological Products, China) and Xuguang Li (University of Ottawa, Canada) for the HEK293 ISRE cell line, and Michael Emerman (Fred Hutchinson Cancer Research Center) for the following plasmids: Lai-YU2, Lai-luc Δenv, Bru3ori Vpr-F34I, Bru3ori Vpr-W54R, Bru3ori Vpr-H71R, Lai ΔVpr, and HA-Vpr. The following reagents were obtained from the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: anti-HIV-1 Vpr 1-50 aa polyclonal antibody from Jeffrey Kopp (11836), HIV-1 p24 Gag monoclonal antibody (24-2) from Michael H. Malim, HIV-1 p24 hybridoma (183-H12-5C) from Bruce Chesebro, zidovudine, and indinavir sulfate. We thank the Boston University Flow Cytometry Core for instrumentation and technical support. We gratefully acknowledge the assistance of the UNC High Throughput Sequencing Facility.
This work was supported by NIH grants R01 AI064099 (S.G.), T32 AI007309 (C.M.M.), and R56 AI118682 (A.J.H. and S.G.). This work was also supported by an amfAR Mathilde Krim fellowship in basic biomedical research (L.M.A.) and NIH grants R01 AI097117 (A.J.H.), P50 GM103297 (Center for HIV RNA Studies), F31 AI116406 (A.E.), and R37 AI44667 (R.I.S.). Infrastructure support was provided by the UNC Center for AIDS Research (NIH award P30 AI50410) and the UNC Lineberger Comprehensive Cancer Center (NIH award P30 CA16068).
REFERENCES
- 1.Malim MH, Emerman M. 2008. HIV-1 accessory proteins—ensuring viral survival in a hostile environment. Cell Host Microbe 3:388–398. doi: 10.1016/j.chom.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 2.Strebel K. 2013. HIV accessory proteins versus host restriction factors. Curr Opin Virol 3:692–699. doi: 10.1016/j.coviro.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Selig L, Pages JC, Tanchou V, Prévéral S, Berlioz-Torrent C, Liu LX, Erdtmann L, Darlix J, Benarous R, Benichou S. 1999. Interaction with the p6 domain of the gag precursor mediates incorporation into virions of Vpr and Vpx proteins from primate lentiviruses. J Virol 73:592–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bachand F, Yao XJ, Hrimech M, Rougeau N, Cohen EA. 1999. Incorporation of Vpr into human immunodeficiency virus type 1 requires a direct interaction with the p6 domain of the p55 gag precursor. J Biol Chem 274:9083–9091. doi: 10.1074/jbc.274.13.9083. [DOI] [PubMed] [Google Scholar]
- 5.Cohen EA, Terwilliger EF, Jalinoos Y, Proulx J, Sodroski JG, Haseltine WA. 1990. Identification of HIV-1 vpr product and function. J Acquir Immune Defic Syndr 3:11–18. [PubMed] [Google Scholar]
- 6.Yuan X, Matsuda Z, Matsuda M, Essex M, Lee TH. 1990. Human immunodeficiency virus vpr gene encodes a virion-associated protein. AIDS Res Hum Retroviruses 6:1265–1271. [DOI] [PubMed] [Google Scholar]
- 7.Lu YL, Bennett RP, Wills JW, Gorelick R, Ratner L. 1995. A leucine triplet repeat sequence (LXX)4 in p6gag is important for Vpr incorporation into human immunodeficiency virus type 1 particles. J Virol 69:6873–6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lavallée C, Yao XJ, Ladha A, Göttlinger H, Haseltine WA, Cohen EA. 1994. Requirement of the Pr55gag precursor for incorporation of the Vpr product into human immunodeficiency virus type 1 viral particles. J Virol 68:1926–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kondo E, Mammano F, Cohen EA, Göttlinger HG. 1995. The p6gag domain of human immunodeficiency virus type 1 is sufficient for the incorporation of Vpr into heterologous viral particles. J Virol 69:2759–2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Accola MA, Bukovsky AA, Jones MS, Göttlinger HG. 1999. A conserved dileucine-containing motif in p6(gag) governs the particle association of Vpx and Vpr of simian immunodeficiency viruses SIV(mac) and SIV(agm). J Virol 73:9992–9999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Müller B, Tessmer U, Schubert U, Kräusslich HG. 2000. Human immunodeficiency virus type 1 Vpr protein is incorporated into the virion in significantly smaller amounts than Gag and is phosphorylated in infected cells. J Virol 74:9727–9731. doi: 10.1128/JVI.74.20.9727-9731.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rogel ME, Wu LI, Emerman M. 1995. The human immunodeficiency virus type 1 vpr gene prevents cell proliferation during chronic infection. J Virol 69:882–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He J, Choe S, Walker R, Di Marzio P, Morgan DO, Landau NR. 1995. Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J Virol 69:6705–6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jowett JB, Planelles V, Poon B, Shah NP, Chen ML, Chen IS. 1995. The human immunodeficiency virus type 1 vpr gene arrests infected T cells in the G2 + M phase of the cell cycle. J Virol 69:6304–6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Belzile J-P, Duisit G, Rougeau N, Mercier J, Finzi A, Cohen EA. 2007. HIV-1 Vpr-mediated G2 arrest involves the DDB1-CUL4AVPRBP E3 ubiquitin ligase. PLoS Pathog 3:e85. doi: 10.1371/journal.ppat.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeHart JL, Zimmerman ES, Ardon O, Monteiro-Filho CMR, Argañaraz ER, Planelles V. 2007. HIV-1 Vpr activates the G2 checkpoint through manipulation of the ubiquitin proteasome system. Virol J 4:57. doi: 10.1186/1743-422X-4-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hrecka K, Gierszewska M, Srivastava S, Kozaczkiewicz L, Swanson SK, Florens L, Washburn MP, Skowronski J. 2007. Lentiviral Vpr usurps Cul4-DDB1[VprBP] E3 ubiquitin ligase to modulate cell cycle. Proc Natl Acad Sci U S A 104:11778–11783. doi: 10.1073/pnas.0702102104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laguette N, Brégnard C, Hue P, Basbous J, Yatim A, Larroque M, Kirchhoff F, Constantinou A, Sobhian B, Benkirane M. 2014. Premature activation of the SLX4 complex by Vpr promotes G2/M arrest and escape from innate immune sensing. Cell 156:134–145. doi: 10.1016/j.cell.2013.12.011. [DOI] [PubMed] [Google Scholar]
- 19.Bouhamdan M, Benichou S, Rey F, Navarro JM, Agostini I, Spire B, Camonis J, Slupphaug G, Vigne R, Benarous R, Sire J. 1996. Human immunodeficiency virus type 1 Vpr protein binds to the uracil DNA glycosylase DNA repair enzyme. J Virol 70:697–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lahouassa H, Blondot M-L, Chauveau L, Chougui G, Morel M, Leduc M, Guillonneau F, Ramirez BC, Schwartz O, Margottin-Goguet F. 2016. HIV-1 Vpr degrades the HLTF DNA translocase in T cells and macrophages. Proc Natl Acad Sci U S A 113:5311–5316. doi: 10.1073/pnas.1600485113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hrecka K, Hao C, Shun M-C, Kaur S, Swanson SK, Florens L, Washburn MP, Skowronski J. 2016. HIV-1 and HIV-2 exhibit divergent interactions with HLTF and UNG2 DNA repair proteins. Proc Natl Acad Sci U S A 113:E3921–E3930. doi: 10.1073/pnas.1605023113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collins DR, Lubow J, Lukic Z, Mashiba M, Collins KL. 2015. Vpr promotes macrophage-dependent HIV-1 infection of CD4+ T lymphocytes. PLoS Pathog 11:e1005054. doi: 10.1371/journal.ppat.1005054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dedera D, Hu W, Vander Heyden N, Ratner L. 1989. Viral protein R of human immunodeficiency virus types 1 and 2 is dispensable for replication and cytopathogenicity in lymphoid cells. J Virol 63:3205–3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eckstein DA, Sherman MP, Penn ML, Chin PS, De Noronha CM, Greene WC, Goldsmith MA. 2001. HIV-1 Vpr enhances viral burden by facilitating infection of tissue macrophages but not nondividing CD4+ T cells. J Exp Med 194:1407–1419. doi: 10.1084/jem.194.10.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Balliet JW, Kolson DL, Eiger G, Kim FM, McGann KA, Srinivasan A, Collman R. 1994. Distinct effects in primary macrophages and lymphocytes of the human immunodeficiency virus type 1 accessory genes vpr, vpu, and nef: mutational analysis of a primary HIV-1 isolate. Virology 200:623–631. doi: 10.1006/viro.1994.1225. [DOI] [PubMed] [Google Scholar]
- 26.Höhne K, Businger R, van Nuffel A, Bolduan S, Koppensteiner H, Baeyens A, Vermeire J, Malatinkova E, Verhasselt B, Schindler M. 2016. Virion encapsidated HIV-1 Vpr induces NFAT to prime non-activated T cells for productive infection. Open Biol 6:160046. doi: 10.1098/rsob.160046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roesch F, Richard L, Rua R, Porrot F, Casartelli N, Schwartz O. 2015. Vpr enhances tumor necrosis factor production by HIV-1-infected T cells. J Virol 89:12118–12130. doi: 10.1128/JVI.02098-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mashiba M, Collins DR, Terry VH, Collins KL. 2014. Vpr overcomes macrophage-specific restriction of HIV-1 Env expression and virion production. Cell Host Microbe 16:722–735. doi: 10.1016/j.chom.2014.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Connor RI, Chen BK, Choe S, Landau NR. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 30.Vodicka MA, Koepp DM, Silver PA, Emerman M. 1998. HIV-1 Vpr interacts with the nuclear transport pathway to promote macrophage infection. Genes Dev 12:175–185. doi: 10.1101/gad.12.2.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harman AN, Lai J, Turville S, Samarajiwa S, Gray L, Marsden V, Mercier SK, Mercier SK, Jones K, Nasr N, Rustagi A, Cumming H, Donaghy H, Mak J, Gale M, Churchill M, Hertzog P, Cunningham AL. 2011. HIV infection of dendritic cells subverts the IFN induction pathway via IRF-1 and inhibits type 1 IFN production. Blood 118:298–308. doi: 10.1182/blood-2010-07-297721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zahoor MA, Xue G, Sato H, Aida Y. 2015. Genome-wide transcriptional profiling reveals that HIV-1 Vpr differentially regulates interferon-stimulated genes in human monocyte-derived dendritic cells. Virus Res 208:156–163. doi: 10.1016/j.virusres.2015.06.017. [DOI] [PubMed] [Google Scholar]
- 33.Zahoor MA, Xue G, Sato H, Murakami T, Takeshima S, Aida Y. 2014. HIV-1 Vpr induces interferon-stimulated genes in human monocyte-derived macrophages. PLoS One 9:e106418. doi: 10.1371/journal.pone.0106418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hong HS, Bhatnagar N, Ballmaier M, Schubert U, Henklein P, Volgmann T, Heiken H, Schmidt RE, Meyer-Olson D. 2009. Exogenous HIV-1 Vpr disrupts IFN-alpha response by plasmacytoid dendritic cells (pDCs) and subsequent pDC/NK interplay. Immunol Lett 125:100–104. doi: 10.1016/j.imlet.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 35.Banchereau J, Steinman RM. 1998. Dendritic cells and the control of immunity. Nature 392:245–252. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- 36.Hladik F, McElrath MJ. 2008. Setting the stage: host invasion by HIV. Nat Rev Immunol 8:447–457. doi: 10.1038/nri2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stieh DJ, Matias E, Xu H, Fought AJ, Blanchard JL, Marx PA, Veazey RS, Hope TJ. 2016. Th17 cells are preferentially infected very early after vaginal transmission of SIV in macaques. Cell Host Microbe 19:529–540. doi: 10.1016/j.chom.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hladik F, Sakchalathorn P, Ballweber L, Lentz G, Fialkow M, Eschenbach D, McElrath MJ. 2007. Initial events in establishing vaginal entry and infection by human immunodeficiency virus type-1. Immunity 26:257–270. doi: 10.1016/j.immuni.2007.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu J, Gardner MB, Miller CJ. 2000. Simian immunodeficiency virus rapidly penetrates the cervicovaginal mucosa after intravaginal inoculation and infects intraepithelial dendritic cells. J Virol 74:6087–6095. doi: 10.1128/JVI.74.13.6087-6095.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spira AI, Marx PA, Patterson BK, Mahoney J, Koup RA, Wolinsky SM, Ho DD. 1996. Cellular targets of infection and route of viral dissemination after an intravaginal inoculation of simian immunodeficiency virus into rhesus macaques. J Exp Med 183:215–225. doi: 10.1084/jem.183.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Silva S, Planelles V, Wu L. 2012. Differential effects of Vpr on single-cycle and spreading HIV-1 infections in CD4+ T-cells and dendritic cells. PLoS One 7:e35385. doi: 10.1371/journal.pone.0035385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nobile C, Petit C, Moris A, Skrabal K, Abastado J-P, Mammano F, Schwartz O. 2005. Covert human immunodeficiency virus replication in dendritic cells and in DC-SIGN-expressing cells promotes long-term transmission to lymphocytes. J Virol 79:5386–5399. doi: 10.1128/JVI.79.9.5386-5399.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Burleigh L, Lozach P-Y, Schiffer C, Staropoli I, Pezo V, Porrot F, Canque B, Virelizier J-L, Arenzana-Seisdedos F, Amara A. 2006. Infection of dendritic cells (DCs), not DC-SIGN-mediated internalization of human immunodeficiency virus, is required for long-term transfer of virus to T cells. J Virol 80:2949–2957. doi: 10.1128/JVI.80.6.2949-2957.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang X, Zhou T, Frabutt DA, Zheng Y-H. 2016. HIV-1 Vpr increases Env expression by preventing Env from endoplasmic reticulum-associated protein degradation (ERAD). Virology 496:194–202. doi: 10.1016/j.virol.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen M, Elder RT, Yu M, O'Gorman MG, Selig L, Benarous R, Yamamoto A, Zhao Y. 1999. Mutational analysis of Vpr-induced G2 arrest, nuclear localization, and cell death in fission yeast. J Virol 73:3236–3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gummuluru S, Emerman M. 1999. Cell cycle- and Vpr-mediated regulation of human immunodeficiency virus type 1 expression in primary and transformed T-cell lines. J Virol 73:5422–5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Belzile J-P, Abrahamyan LG, Gérard FCA, Rougeau N, Cohen EA. 2010. Formation of mobile chromatin-associated nuclear foci containing HIV-1 Vpr and VPRBP is critical for the induction of G2 cell cycle arrest. PLoS Pathog 6:e1001080. doi: 10.1371/journal.ppat.1001080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wen X, Duus KM, Friedrich TD, de Noronha CMC. 2007. The HIV1 protein Vpr acts to promote G2 cell cycle arrest by engaging a DDB1 and Cullin4A-containing ubiquitin ligase complex using VprBP/DCAF1 as an adaptor. J Biol Chem 282:27046–27057. doi: 10.1074/jbc.M703955200. [DOI] [PubMed] [Google Scholar]
- 49.Tan L, Ehrlich E, Yu X-F. 2007. DDB1 and Cul4A are required for human immunodeficiency virus type 1 Vpr-induced G2 arrest. J Virol 81:10822–10830. doi: 10.1128/JVI.01380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jacquot G, Le Rouzic E, David A, Mazzolini J, Bouchet J, Bouaziz S, Niedergang F, Pancino G, Benichou S. 2007. Localization of HIV-1 Vpr to the nuclear envelope: impact on Vpr functions and virus replication in macrophages. Retrovirology 4:84. doi: 10.1186/1742-4690-4-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Laguette N, Sobhian B, Casartelli N, Ringeard M, Chable-Bessia C, Ségéral E, Yatim A, Emiliani S, Schwartz O, Benkirane M. 2011. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474:654–657. doi: 10.1038/nature10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hrecka K, Hao C, Gierszewska M, Swanson SK, Kesik-Brodacka M, Srivastava S, Florens L, Washburn MP, Skowronski J. 2011. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 474:658–661. doi: 10.1038/nature10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gringhuis SI, van der Vlist M, van den Berg LM, den Dunnen J, Litjens M, Geijtenbeek TBH. 2010. HIV-1 exploits innate signaling by TLR8 and DC-SIGN for productive infection of dendritic cells. Nat Immunol 11:419–426. doi: 10.1038/ni.1858. [DOI] [PubMed] [Google Scholar]
- 54.Turville SG, Santos JJ, Frank I, Cameron PU, Wilkinson J, Miranda-Saksena M, Dable J, Stössel H, Romani N, Piatak M, Lifson JD, Pope M, Cunningham AL. 2004. Immunodeficiency virus uptake, turnover, and 2-phase transfer in human dendritic cells. Blood 103:2170–2179. doi: 10.1182/blood-2003-09-3129. [DOI] [PubMed] [Google Scholar]
- 55.Aggarwal A, Iemma TL, Shih I, Newsome TP, McAllery S, Cunningham AL, Turville SG. 2012. Mobilization of HIV spread by diaphanous 2 dependent filopodia in infected dendritic cells. PLoS Pathog 8:e1002762. doi: 10.1371/journal.ppat.1002762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Su B, Biedma ME, Lederle A, Peressin M, Lambotin M, Proust A, Decoville T, Schmidt S, Laumond G, Moog C. 2014. Dendritic cell-lymphocyte cross talk downregulates host restriction factor SAMHD1 and stimulates HIV-1 replication in dendritic cells. J Virol 88:5109–5121. doi: 10.1128/JVI.03057-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Holl V, Xu K, Peressin M, Lederle A, Biedma ME, Delaporte M, Decoville T, Schmidt S, Laumond G, Aubertin A-M, Moog C. 2010. Stimulation of HIV-1 replication in immature dendritic cells in contact with primary CD4 T or B lymphocytes. J Virol 84:4172–4182. doi: 10.1128/JVI.01567-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gummuluru S, KewalRamani VN, Emerman M. 2002. Dendritic cell-mediated viral transfer to T cells is required for human immunodeficiency virus type 1 persistence in the face of rapid cell turnover. J Virol 76:10692–10701. doi: 10.1128/JVI.76.21.10692-10701.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pope M, Betjes MG, Romani N, Hirmand H, Cameron PU, Hoffman L, Gezelter S, Schuler G, Steinman RM. 1994. Conjugates of dendritic cells and memory T lymphocytes from skin facilitate productive infection with HIV-1. Cell 78:389–398. doi: 10.1016/0092-8674(94)90418-9. [DOI] [PubMed] [Google Scholar]
- 60.Cameron PU, Freudenthal PS, Barker JM, Gezelter S, Inaba K, Steinman RM. 1992. Dendritic cells exposed to human immunodeficiency virus type-1 transmit a vigorous cytopathic infection to CD4+ T cells. Science 257:383–387. doi: 10.1126/science.1352913. [DOI] [PubMed] [Google Scholar]
- 61.Gendelman HE, Baca LM, Turpin J, Kalter DC, Hansen B, Orenstein JM, Dieffenbach CW, Friedman RM, Meltzer MS. 1990. Regulation of HIV replication in infected monocytes by IFN-alpha. Mechanisms for viral restriction. J Immunol 145:2669–2676. [PubMed] [Google Scholar]
- 62.Dianzani F, Castilletti C, Gentile M, Gelderblom HR, Frezza F, Capobianchi MR. 1998. Effects of IFN alpha on late stages of HIV-1 replication cycle. Biochimie 80:745–754. doi: 10.1016/S0300-9084(99)80028-5. [DOI] [PubMed] [Google Scholar]
- 63.Cheney KM, McKnight Á. 2010. Interferon-alpha mediates restriction of human immunodeficiency virus type-1 replication in primary human macrophages at an early stage of replication. PLoS One 5:e13521. doi: 10.1371/journal.pone.0013521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shirazi Y, Pitha PM. 1992. Alpha interferon inhibits early stages of the human immunodeficiency virus type 1 replication cycle. J Virol 66:1321–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Baca-Regen L, Heinzinger N, Stevenson M, Gendelman HE. 1994. Alpha interferon-induced antiretroviral activities: restriction of viral nucleic acid synthesis and progeny virion production in human immunodeficiency virus type 1-infected monocytes. J Virol 68:7559–7565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Goujon C, Malim MH. 2010. Characterization of the alpha interferon-induced postentry block to HIV-1 infection in primary human macrophages and T cells. J Virol 84:9254–9266. doi: 10.1128/JVI.00854-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shirazi Y, Pitha PM. 1993. Interferon alpha-mediated inhibition of human immunodeficiency virus type 1 provirus synthesis in T-cells. Virology 193:303–312. doi: 10.1006/viro.1993.1126. [DOI] [PubMed] [Google Scholar]
- 68.Hartshorn KL, Neumeyer D, Vogt MW, Schooley RT, Hirsch MS. 1987. Activity of interferons alpha, beta, and gamma against human immunodeficiency virus replication in vitro. AIDS Res Hum Retroviruses 3:125–133. doi: 10.1089/aid.1987.3.125. [DOI] [PubMed] [Google Scholar]
- 69.Macé K, Duc Dodon M, Gazzolo L. 1989. Restriction of HIV-1 replication in promonocytic cells: a role for IFN-alpha. Virology 168:399–405. doi: 10.1016/0042-6822(89)90282-1. [DOI] [PubMed] [Google Scholar]
- 70.Larocque L, Bliu A, Xu R, Diress A, Wang J, Lin R, He R, Girard M, Li X. 2011. Bioactivity determination of native and variant forms of therapeutic interferons. J Biomed Biotechnol 2011:174615. doi: 10.1155/2011/174615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.O'Doherty U, Swiggard WJ, Jeyakumar D, McGain D, Malim MH. 2002. A sensitive, quantitative assay for human immunodeficiency virus type 1 integration. J Virol 76:10942–10950. doi: 10.1128/JVI.76.21.10942-10950.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Agosto LM, Yu JJ, Dai J, Kaletsky R, Monie D, O'Doherty U. 2007. HIV-1 integrates into resting CD4+ T cells even at low inoculums as demonstrated with an improved assay for HIV-1 integration. Virology 368:60–72. doi: 10.1016/j.virol.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Forget J, Yao XJ, Mercier J, Cohen EA. 1998. Human immunodeficiency virus type 1 vpr protein transactivation function: mechanism and identification of domains involved. J Mol Biol 284:915–923. doi: 10.1006/jmbi.1998.2206. [DOI] [PubMed] [Google Scholar]
- 74.Philippon V, Matsuda Z, Essex M. 1999. Transactivation is a conserved function among primate lentivirus Vpr proteins but is not shared by Vpx. J Hum Virol 2:167–174. [PubMed] [Google Scholar]
- 75.Varin A, Decrion A-Z, Sabbah E, Quivy V, Sire J, Van Lint C, Roques BP, Aggarwal BB, Herbein G. 2005. Synthetic Vpr protein activates activator protein-1, c-Jun N-terminal kinase, and NF-kappaB and stimulates HIV-1 transcription in promonocytic cells and primary macrophages. J Biol Chem 280:42557–42567. doi: 10.1074/jbc.M502211200. [DOI] [PubMed] [Google Scholar]
- 76.Jabara CB, Jones CD, Roach J, Anderson JA, Swanstrom R. 2011. Accurate sampling and deep sequencing of the HIV-1 protease gene using a Primer ID. Proc Natl Acad Sci U S A 108:20166–20171. doi: 10.1073/pnas.1110064108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Emery A, Zhou S, Pollom E, Swanstrom R. 2017. Characterizing HIV-1 splicing by using next-generation sequencing. J Virol 91:e02515-16. doi: 10.1128/JVI.02515-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bartz SR, Rogel ME, Emerman M. 1996. Human immunodeficiency virus type 1 cell cycle control: Vpr is cytostatic and mediates G2 accumulation by a mechanism which differs from DNA damage checkpoint control. J Virol 70:2324–2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Le Rouzic E, Belaïdouni N, Estrabaud E, Morel M, Rain J-C, Transy C, Margottin-Goguet F. 2007. HIV1 Vpr arrests the cell cycle by recruiting DCAF1/VprBP, a receptor of the Cul4-DDB1 ubiquitin ligase. Cell Cycle 6:182–188. doi: 10.4161/cc.6.2.3732. [DOI] [PubMed] [Google Scholar]
- 80.Selig L, Benichou S, Rogel ME, Wu LI, Vodicka MA, Sire J, Benarous R, Emerman M. 1997. Uracil DNA glycosylase specifically interacts with Vpr of both human immunodeficiency virus type 1 and simian immunodeficiency virus of sooty mangabeys, but binding does not correlate with cell cycle arrest. J Virol 71:4842–4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mansky LM, Preveral S, Selig L, Benarous R, Benichou S. 2000. The interaction of vpr with uracil DNA glycosylase modulates the human immunodeficiency virus type 1 in vivo mutation rate. J Virol 74:7039–7047. doi: 10.1128/JVI.74.15.7039-7047.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schröfelbauer B, Yu Q, Zeitlin SG, Landau NR. 2005. Human immunodeficiency virus type 1 Vpr induces the degradation of the UNG and SMUG uracil-DNA glycosylases. J Virol 79:10978–10987. doi: 10.1128/JVI.79.17.10978-10987.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhu Y, Gelbard HA, Roshal M, Pursell S, Jamieson BD, Planelles V. 2001. Comparison of cell cycle arrest, transactivation, and apoptosis induced by the simian immunodeficiency virus SIVagm and human immunodeficiency virus type 1 vpr genes. J Virol 75:3791–3801. doi: 10.1128/JVI.75.8.3791-3801.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mueller SM, Lang SM. 2002. The first HxRxG motif in simian immunodeficiency virus mac239 Vpr is crucial for G(2)/M cell cycle arrest. J Virol 76:11704–11709. doi: 10.1128/JVI.76.22.11704-11709.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Goujon C, Rivière L, Jarrosson-Wuilleme L, Bernaud J, Rigal D, Darlix J-L, Cimarelli A. 2007. SIVSM/HIV-2 Vpx proteins promote retroviral escape from a proteasome-dependent restriction pathway present in human dendritic cells. Retrovirology 4:2. doi: 10.1186/1742-4690-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ahn J, Vu T, Novince Z, Guerrero-Santoro J, Rapic-Otrin V, Gronenborn AM. 2010. HIV-1 Vpr loads uracil DNA glycosylase-2 onto DCAF1, a substrate recognition subunit of a cullin 4A-ring E3 ubiquitin ligase for proteasome-dependent degradation. J Biol Chem 285:37333–37341. doi: 10.1074/jbc.M110.133181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Eldin P, Chazal N, Fenard D, Bernard E, Guichou J-F, Briant L. 2014. Vpr expression abolishes the capacity of HIV-1 infected cells to repair uracilated DNA. Nucleic Acids Res 42:1698–1710. doi: 10.1093/nar/gkt974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fregoso OI, Emerman M. 2016. Activation of the DNA damage response is a conserved function of HIV-1 and HIV-2 Vpr that is independent of SLX4 recruitment. mBio 7:e01433-16. doi: 10.1128/mBio.01433-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Berger G, Lawrence M, Hué S, Neil SJD. 2014. G2/M cell cycle arrest correlates with primate lentiviral Vpr interaction with the SLX4 complex. J Virol 89:230–240. doi: 10.1128/JVI.02307-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Daniel R, Kao G, Taganov K, Greger JG, Favorova O, Merkel G, Yen TJ, Katz RA, Skalka AM. 2003. Evidence that the retroviral DNA integration process triggers an ATR-dependent DNA damage response. Proc Natl Acad Sci U S A 100:4778–4783. doi: 10.1073/pnas.0730887100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Stracker TH, Carson CT, Weitzman MD. 2002. Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature 418:348–352. doi: 10.1038/nature00863. [DOI] [PubMed] [Google Scholar]
- 92.Planelles V, Jowett JB, Li QX, Xie Y, Hahn B, Chen IS. 1996. Vpr-induced cell cycle arrest is conserved among primate lentiviruses. J Virol 70:2516–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Stivahtis GL, Soares MA, Vodicka MA, Hahn BH, Emerman M. 1997. Conservation and host specificity of Vpr-mediated cell cycle arrest suggest a fundamental role in primate lentivirus evolution and biology. J Virol 71:4331–4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nakai-Murakami C, Shimura M, Kinomoto M, Takizawa Y, Tokunaga K, Taguchi T, Hoshino S, Miyagawa K, Sata T, Kurumizaka H, Yuo A, Ishizaka Y. 2007. HIV-1 Vpr induces ATM-dependent cellular signal with enhanced homologous recombination. Oncogene 26:477–486. doi: 10.1038/sj.onc.1209831. [DOI] [PubMed] [Google Scholar]
- 95.Roshal M, Kim B, Zhu Y, Nghiem P, Planelles V. 2003. Activation of the ATR-mediated DNA damage response by the HIV-1 viral protein R. J Biol Chem 278:25879–25886. doi: 10.1074/jbc.M303948200. [DOI] [PubMed] [Google Scholar]
- 96.Nakagawa T, Lv L, Nakagawa M, Yu Y, Yu C, D'Alessio AC, Nakayama K, Fan H-Y, Chen X, Xiong Y. 2015. CRL4VprBP E3 ligase promotes monoubiquitylation and chromatin binding of TET dioxygenases. Mol Cell 57:247–260. doi: 10.1016/j.molcel.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ayyavoo V, Mahboubi A, Mahalingam S, Ramalingam R, Kudchodkar S, Williams WV, Green DR, Weiner DB. 1997. HIV-1 Vpr suppresses immune activation and apoptosis through regulation of nuclear factor κB. Nat Med 3:1117–1123. doi: 10.1038/nm1097-1117. [DOI] [PubMed] [Google Scholar]
- 98.Kogan M, Deshmane S, Sawaya BE, Gracely EJ, Khalili K, Rappaport J. 2013. Inhibition of NF-κB activity by HIV-1 Vpr is dependent on Vpr binding protein. J Cell Physiol 228:781–790. doi: 10.1002/jcp.24226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hoshino S, Konishi M, Mori M, Shimura M, Nishitani C, Kuroki Y, Koyanagi Y, Kano S, Itabe H, Ishizaka Y. 2010. HIV-1 Vpr induces TLR4/MyD88-mediated IL-6 production and reactivates viral production from latency. J Leukoc Biol 87:1133–1143. doi: 10.1189/jlb.0809547. [DOI] [PubMed] [Google Scholar]
- 100.Liu R, Tan J, Lin Y, Jia R, Yang W, Liang C, Geng Y, Qiao W. 2013. HIV-1 Vpr activates both canonical and noncanonical NF-κB pathway by enhancing the phosphorylation of IKKα/β. Virology 439:47–56. doi: 10.1016/j.virol.2013.01.020. [DOI] [PubMed] [Google Scholar]
- 101.Roux P, Alfieri C, Hrimech M, Cohen EA, Tanner JE. 2000. Activation of transcription factors NF-κB and NF-IL-6 by human immunodeficiency virus type 1 protein R (Vpr) induces interleukin-8 expression. J Virol 74:4658–4665. doi: 10.1128/JVI.74.10.4658-4665.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liu R, Lin Y, Jia R, Geng Y, Liang C, Tan J, Qiao W. 2014. HIV-1 Vpr stimulates NF-κB and AP-1 signaling by activating TAK1. Retrovirology 11:45. doi: 10.1186/1742-4690-11-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yan N, Lieberman J. 2012. SAMHD1 does it again, now in resting T cells. Nat Med 18:1611–1612. doi: 10.1038/nm.2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Baldauf H-M, Pan X, Erikson E, Schmidt S, Daddacha W, Burggraf M, Schenkova K, Ambiel I, Wabnitz G, Gramberg T, Panitz S, Flory E, Landau NR, Sertel S, Rutsch F, Lasitschka F, Kim B, König R, Fackler OT, Keppler OT. 2012. SAMHD1 restricts HIV-1 infection in resting CD4(+) T cells. Nat Med 18:1682–1687. doi: 10.1038/nm.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Brownell J, Bruckner J, Wagoner J, Thomas E, Loo Y-M, Gale M, Liang TJ, Polyak SJ, Polyak SJ. 2014. Direct, interferon-independent activation of the CXCL10 promoter by NF-κB and interferon regulatory factor 3 during hepatitis C virus infection. J Virol 88:1582–1590. doi: 10.1128/JVI.02007-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yeruva S, Ramadori G, Raddatz D. 2008. NF-kappaB-dependent synergistic regulation of CXCL10 gene expression by IL-1beta and IFN-gamma in human intestinal epithelial cell lines. Int J Colorectal Dis 23:305–317. doi: 10.1007/s00384-007-0396-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Oslund KL, Zhou X, Lee B, Zhu L, Duong T, Shih R, Baumgarth N, Hung L-Y, Wu R, Chen Y. 2014. Synergistic up-regulation of CXCL10 by virus and IFN γ in human airway epithelial cells. PLoS One 9:e100978. doi: 10.1371/journal.pone.0100978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Martin-Gayo E, Buzon MJ, Ouyang Z, Hickman T, Cronin J, Pimenova D, Walker BD, Lichterfeld M, Yu XG. 2015. Potent cell-intrinsic immune responses in dendritic cells facilitate HIV-1-specific T cell immunity in HIV-1 elite controllers. PLoS Pathog 11:e1004930. doi: 10.1371/journal.ppat.1004930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hamimi C, David A, Versmisse P, Weiss L, Bruel T, Zucman D, Appay V, Moris A, Ungeheuer M-N, Lascoux-Combe C, Barré-Sinoussi F, Muller-Trutwin M, Boufassa F, Lambotte O, Pancino G, Sáez-Cirión A, ANRS CO21 CODEX Cohort. 2016. Dendritic cells from HIV controllers have low susceptibility to HIV-1 infection in vitro but high capacity to capture HIV-1 particles. PLoS One 11:e0160251. doi: 10.1371/journal.pone.0160251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yamashita M, Emerman M. 2004. Capsid is a dominant determinant of retrovirus infectivity in nondividing cells. J Virol 78:5670–5678. doi: 10.1128/JVI.78.11.5670-5678.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Voronin Y, Holte S, Overbaugh J, Emerman M. 2009. Genetic drift of HIV populations in culture. PLoS Genet 5:e1000431. doi: 10.1371/journal.pgen.1000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Puryear WB, Akiyama H, Geer SD, Ramirez NP, Yu X, Reinhard BM, Gummuluru S. 2013. Interferon-inducible mechanism of dendritic cell-mediated HIV-1 dissemination is dependent on Siglec-1/CD169. PLoS Pathog 9:e1003291. doi: 10.1371/journal.ppat.1003291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Puryear WB, Gummuluru S. 2013. Role of glycosphingolipids in dendritic cell-mediated HIV-1 trans-infection. Adv Exp Med Biol 762:131–153. doi: 10.1007/978-1-4614-4433-6_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Vodicka MA, Goh WC, Wu LI, Rogel ME, Bartz SR, Schweickart VL, Raport CJ, Emerman M. 1997. Indicator cell lines for detection of primary strains of human and simian immunodeficiency viruses. Virology 233:193–198. doi: 10.1006/viro.1997.8606. [DOI] [PubMed] [Google Scholar]
- 115.Puryear WB, Yu X, Ramirez NP, Reinhard BM, Gummuluru S. 2012. HIV-1 incorporation of host-cell-derived glycosphingolipid GM3 allows for capture by mature dendritic cells. Proc Natl Acad Sci U S A 109:7475–7480. doi: 10.1073/pnas.1201104109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Akiyama H, Miller C, Patel HV, Hatch SC, Archer J, Ramirez N-GP, Gummuluru S. 2014. Virus particle release from glycosphingolipid-enriched microdomains is essential for dendritic cell-mediated capture and transfer of HIV-1 and henipavirus. J Virol 88:8813–8825. doi: 10.1128/JVI.00992-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hatch SC, Archer J, Gummuluru S. 2009. Glycosphingolipid composition of human immunodeficiency virus type 1 (HIV-1) particles is a crucial determinant for dendritic cell-mediated HIV-1 trans-infection. J Virol 83:3496–3506. doi: 10.1128/JVI.02249-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Akiyama H, Ramirez N-GP, Gudheti MV, Gummuluru S. 2015. CD169-mediated trafficking of HIV to plasma membrane invaginations in dendritic cells attenuates efficacy of anti-gp120 broadly neutralizing antibodies. PLoS Pathog 11:e1004751. doi: 10.1371/journal.ppat.1004751. [DOI] [PMC free article] [PubMed] [Google Scholar]