Mechanisms that drive DNA damage-induced chromosome mobility include relaxation of external tethers to the nuclear envelope and internal chromatin–chromatin tethers. Together with microtubule dynamics, these can mobilize the genome in response to DNA damage.
Abstract
Chromatin exhibits increased mobility on DNA damage, but the biophysical basis for this behavior remains unknown. To explore the mechanisms that drive DNA damage–induced chromosome mobility, we use single-particle tracking of tagged chromosomal loci during interphase in live yeast cells together with polymer models of chromatin chains. Telomeres become mobilized from sites on the nuclear envelope and the pericentromere expands after exposure to DNA-damaging agents. The magnitude of chromatin mobility induced by a single double-strand break requires active microtubule function. These findings reveal how relaxation of external tethers to the nuclear envelope and internal chromatin–chromatin tethers, together with microtubule dynamics, can mobilize the genome in response to DNA damage.
INTRODUCTION
The structural architecture of chromosomes and their tendency to occupy specific territories within the yeast nucleus have a profound effect on their behavior. Centromeres are persistently attached to microtubule plus ends and remain tethered to the nuclear envelope via the spindle pole body (SPB) throughout the cell cycle (Winey and Bloom, 2012). Telomeres attach to the nuclear envelope through the Ku70/80/Sir4/Esc1/ protein complexes (Hediger et al., 2002; Taddei et al., 2004, 2006) and are distributed among five or six foci (Marcomini and Gasser, 2015). This organization is known as Rabl orientation, indicative of the distal position of telomeres and chromosome arms relative to the centromere. Chromatin motion is subdiffusive—a consequence of tethering, collisions, and repulsion between chains in the crowded nucleoplasm, as well as of confinement by the nuclear envelope. Understanding mechanisms that dictate chromosome behavior requires integrating chromosome cell biology with physical principles that govern fluctuating polymer chains (Zimmer and Fabre, 2011; Vasquez and Bloom, 2014; Marshall and Fung, 2016).
Using a fluorescent repressor–operator system to visualize individual genetic loci in living yeast cells, it has been shown that the cellular response to DNA damage further mobilizes the genome. In the case of DNA double-strand breaks (DSBs), loci adjacent to the DSB or loci on different chromosomes increase their range of motion (Mine-Hattab and Rothstein, 2012; Dion and Gasser, 2013). From the perspective of the polymer properties of the DNA chain, there are at least three drivers of chromatin motion: nonthermal or random, ATP-dependent forces imposed by chromatin remodeling complexes and chaperones; external tethers that link the chromatin chain to cellular structures (i.e., centromeres and telomeres); and internal tethers that cross-link chains (e.g., cohesin). Forces and tethers are not mutually exclusive and together can contribute to a particular cellular response.
Evidence for exclusively thermal fluctuations (Marshall et al., 1997; Pliss et al., 2013), as well as of ATP-dependent forces driving chain mobility, can be found in different systems (Heun et al., 2001; Weber et al., 2012). Chromosome tethers present different challenges. Tethers have specific moduli (e.g., centromere to a stiff microtubule, telomere to a mobile nuclear pore), and relaxing the modulus or complete detachment of the tether may be difficult to distinguish. In yeast, release of tethers that constrain centromeres to the spindle pole (using a conditionally functional centromere) drives a large increase in chromosome motion (Verdaasdonk et al., 2013; Strecker et al., 2016). Simulation of chromosome movement indicates that complete release of the centromere from the spindle pole is sufficient to account for the increase in mobility (Verdaasdonk et al., 2013). A different mechanism is likely to account for relaxation of centromere tethers through phosphorylation of kinetochore proteins in response to DNA damage (Strecker et al., 2016). In this case, constraints are relaxed, leading to increased mobility nonetheless. Finally, internal tethers that link sister chains (cohesin) or promote chain–chain interactions (centromeric cohesin) confine chain mobility as well (Dion et al., 2013; Ge et al., 2016; Goloborodko et al., 2016a, b; Lawrimore et al., 2016). Cohesin is recruited to sites of damage, where it is very likely to have significant consequences for chain mobility (Strom et al., 2004; Lawrimore et al., 2016).
In this study, we quantitate the magnitude of response of the major sites of chromosome tethering at telomere- and centromere-linked foci, as well as of the cohesin-rich chromatin domain surrounding the mitotic spindle, to perturbation by single DSBs and the DNA-damaging agent phleomycin. Together with using bead–spring polymer models, we explore the contribution of these tethers and the microtubule cytoskeleton as drivers of chromatin motion in live cells. Telomere mobilization within the nuclear envelope is a major driver of increased motion of a site proximal to a single DSB. The microtubule cytoskeleton is required for the full extent of chromatin response to double-strand DNA damage.
RESULTS
Chromatin mobility is increased in the presence of DNA damage
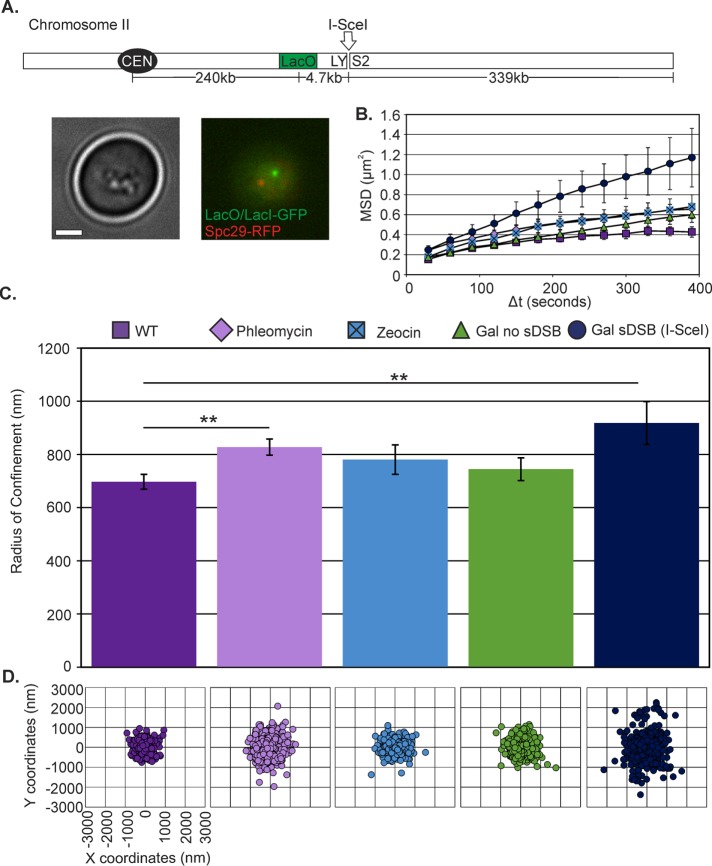
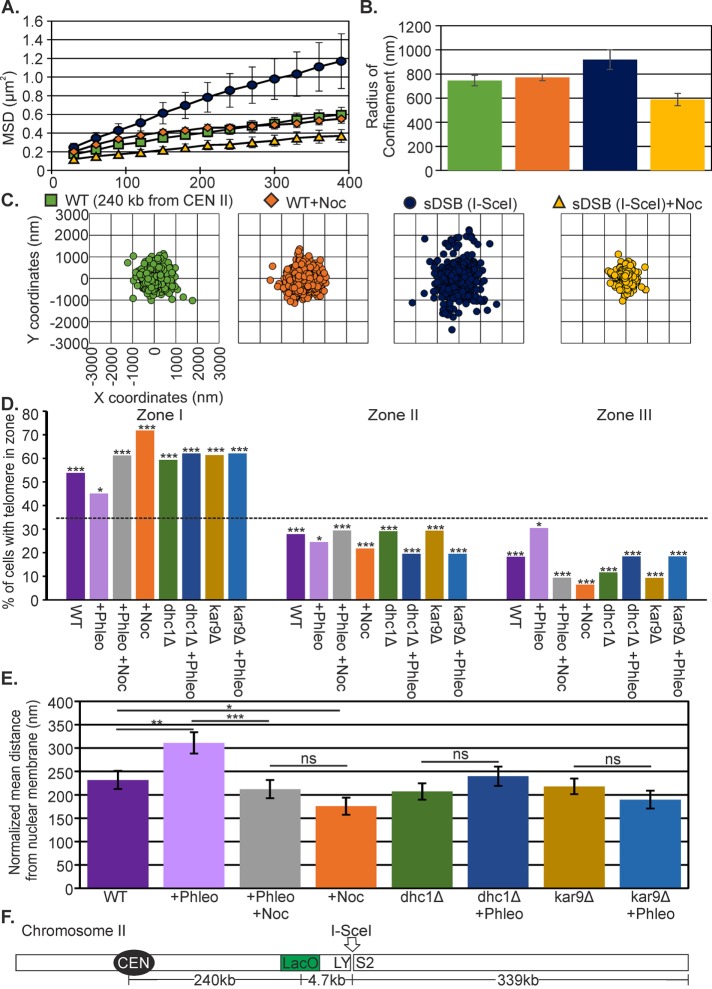
We investigated chromatin dynamics during interphase in response to two different DNA damage conditions: 1) multiple random breaks (phleomycin, Zeocin; Chen et al., 2008) and 2) a single site-specific DSB (I-SceI or HO endonuclease). To measure chromosome arm movement in vivo, we used single-particle tracking of a green fluorescent protein (GFP)–labeled chromatin array (Lac operator [LacO]/Lac repressor [LacI]–GFP) relative to the SPB (Spc29–red fluorescent protein [RFP]) at a biologically relevant time scale of 10 min at 30-s intervals. We achieve nanometer localization accuracy by Gaussian fitting the LacO/LacI-GFP chromatin array and determining the centroid of the distribution. To quantify subnuclear confinement, we calculated the radius of confinement (Rc) of the array from the SD of spot positions, σ, and the average squared deviation from the mean position, r02, and spot variance relative to the SPB, as previously described (Verdaasdonk et al., 2013).
In G1, an undamaged arm locus 240 kb from the centromere explores an area with Rc = 697 ± 27 nm (mean ± SEM; Figure 1 and Supplemental Table S1). This is consistent with measurements in G1 of an arm locus of comparable distance from the centromere (Heun et al., 2001). At low concentrations of phleomycin (3 µg/ml; see Materials and Methods; Supplemental Figure S1), mobility significantly increased to Rc = 828 ± 30 nm (Student’s t test, p = 0.003; Figure 1, B–D, and Supplemental Table S1). Zeocin, another DNA-damaging agent in the same chemical family but less effective in yeast (Supplemental Figure S1) and other fungi (Mora-Lugo et al., 2014), results in a modest increase of mobility to Rc = 781 ± 55 nm, which is not statistically different from wild type (WT; Student’s t test, p = 0.2). By comparison, DSBs due to I-SceI induction resulted in increased mobility to Rc = 918 ± 81 nm (Figure 1, B–D, Supplemental Figure S2, and Supplemental Table S1), comparable to previous observations at arm loci (Dion et al., 2012; Mine-Hattab and Rothstein, 2012).
FIGURE 1:
Chromatin response to DNA damage. (A) Top, schematic of a LacO/LacI-GFP–labeled arm locus 240 kb from CEN2 and 4.7 kb from the I-SceI cut site. I-SceI endonuclease is induced on galactose from its site of integration on chromosome VIII (Lobachev et al., 2004). Bottom, representative images in G1 unbudded cells. Green, 240-kb LacO/LacI-GFP; red, SPB labeled with Spc29-RFP. Scale bar, 1 μm. (B) MSD curves of LacO at 240 kb from CEN2 under phleomycin and Zeocin treatment and at a single DSB adjacent to LacO. (C) Rc values with SEM for WT (n = 25), phleomycin-treated (n = 68), Zeocin (n = 23), galactose (no DSB; n = 36), and I-SceI (n = 48) cells. Phleomycin and galactose sDSB (I-SceI) cells have a significant increase over WT (Student’s t test, p = 0.003 and 0.01, respectively). (D) Scatterplots illustrating variance in LacO/LacI position relative to the SPB.
Centromere-linked loci remain tethered to the spindle pole after DNA damage
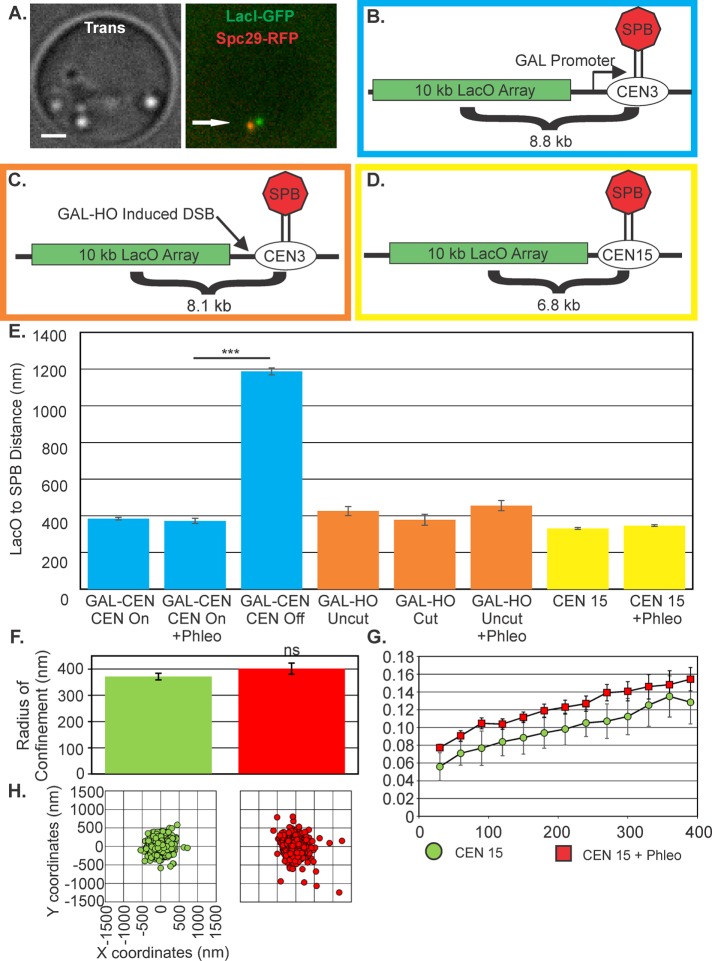
It has been reported that centromere release from the microtubule attachment is a driving mechanism for increased mobility in response to DNA damage (Strecker et al., 2016). The centromere can be experimentally detached from its microtubule attachment via activation of a proximal transcriptional promoter (Hill and Bloom, 1987; Verdaasdonk et al., 2013; Figure 2, A and B). We previously reported an increase in Rc of a centromere-linked locus from 274 to 745 nm on detachment (Verdaasdonk et al., 2013). The increased mobility is also reflected in the distance of the LacO array (8.8 kb from the GALCEN3) to the spindle pole, from 385 ± 7 to 1187 ± 19 nm in G1 cells (Figure 2E), confirming observations of increased motion of detached GALCEN chromosomes (Verdaasdonk et al., 2013).
FIGURE 2:
Centromeres are not released after DNA damage. (A) Image of centromere-proximal LacO cells in G1. Bar, 1 µm. Arrow points to SPB. (B) Schematic of LacO array 8.8 kb from galactose promoter–regulated CEN3(SGD10.2) (blue border). (C) Schematic of the HOcut-CEN3 chromosome. The chromosome contains an HO cut site adjacent to CEN3 on chromosome III (KBY8227; orange border). (D) Schematic of LacO array at 6.8 kb from CENXV (KBY8065; yellow border). (E) The average SPB to CEN-linked LacO distance in G1. Average distances were calculated from either time-lapse images at 30 s for 10 min or from single-cell populations. For GALCEN3 strain: glucose grown, 1008 cells; plus phleomycin, 154 cells; galactose grown, 609 cells. Inactivation of CEN3 on galactose (CEN Off) was significantly different from CEN3 on glucose (Student’s t test, p = 8 × 10−149). HOcut-CEN3: on glucose, 59 cells; on glucose plus phleomycin, 115 cells; and on galactose, 41 cells. CENXV untreated, 844 cells; and plus phleomycin, 1113 cells. Radius of confinement (F), MSD displacement curves (G), and scatterplots (H) of CENXV-linked lacO/LacI-GFP in untreated and phleomycin-treated cells. ns, not significantly different (Student’s t test, p = 0.2). Error bars are SEM.
An alternative method to detach the centromere is to cut a site-specific HO endonuclease recognition site that resides between a centromere-linked LacO array (8.1 kb from CEN3) and the centromere (Figure 2C and Supplemental Figure S2; Dotiwala et al., 2010; Tsabar et al., 2016). In the absence of the HO cut, LacO-LacI-GFP can be visualized as a single focus (unreplicated DNA, G1) or as a focus or two foci in metaphase, depending on the state of sister centromere separation. At this position relative to the centromere, the spots appear separated in metaphase 12% of the time, indicative of tension between sister kinetochores during metaphase (Tsabar et al., 2016). After induction of the HO endonuclease, the centromere is excised, leading to cell inviability (Dotiwala et al., 2010; Tsabar et al., 2016). LacO spots were examined 3 h postinduction, when the fraction of separated spots in metaphase was reduced to 3%, commensurate with a DSB and loss of tension (Tsabar et al., 2016). In G1, the distance from the LacO array to the spindle pole is comparable on glucose or galactose (Figure 2E). A locus adjacent to the centromere would experience increased motion or distance from the SPB only if the kinetochores detached from the microtubule as broken ends are held together by the Rad50/Mre11/Xrs2 complex (Lobachev et al., 2004).
Finally, addition of phleomycin at concentrations sufficient to elicit increase in arm motion in cells containing the GAL-CEN (Figure 2B) and WT cells containing a 10-kb LacO/LacI array 6.8 kb from CENXV does not increase the distance of the CEN-proximal array to the SPB, in contrast to the release of a centromere by transcriptional inactivation. There is no significant difference (Student’s t test, p = 0.2) in the Rc of a CEN-proximal LacO array in cells treated with phleomycin (untreated, 372 ± 12 nm; phleomycin-treated cells, 402 ± 21 nm; KBY 8065; Figure 2, D–H, and Supplemental Table S1). DNA damage either proximal to the centromere or randomly throughout the genome does not release chromosomes from their kinetochore–microtubule attachment. Thus centromere DNA is not displaced from the spindle axis after double-strand breakage.
The centromere response to DNA damage is evidenced by expansion of the pericentromere and kinetochore declustering
The pericentromere is the ∼30 kb of chromatin DNA flanking each of the 16 centromeres enriched threefold in cohesin relative to chromosome arms (Blat and Kleckner, 1999; Megee et al., 1999; D’Amours and Amon, 2004). Cohesin in the pericentromere functions as a spring together with DNA and condensin to balance force at the kinetochore from spindle microtubules (Lawrimore et al., 2015, 2016).
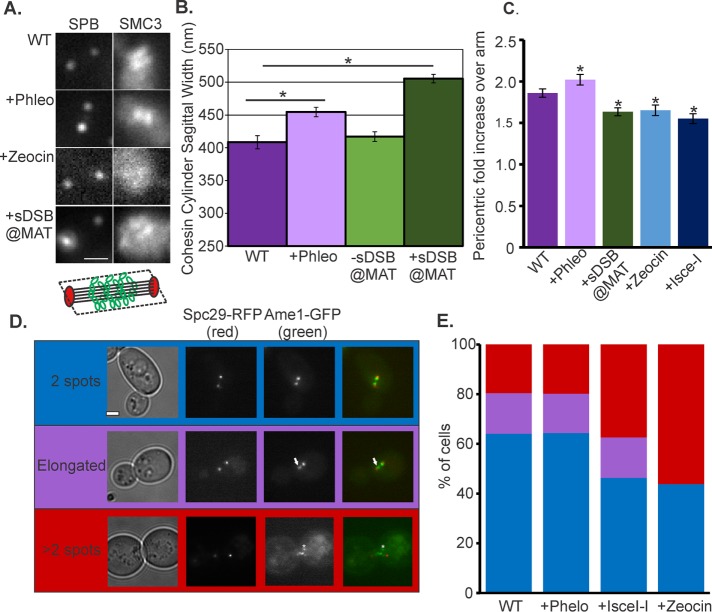
Pericentric cohesin can be visualized through the arrangement of cohesin into a barrel structure surrounding the mitotic spindle (Yeh et al., 2008; Stephens et al., 2011). On exposure to low doses of the spindle poison benomyl, the cohesin barrel expands in dimension in a spindle checkpoint–dependent mechanism (Haase et al., 2012). We treated cells with low doses of phleomycin (3 µg/ml) and examined the dimensions of pericentric cohesin tagged with Smc3-GFP. The cohesin barrel and pericentric chromatin were found to expand in diameter from 408 to 455 nm (Figure 3, A and B). In contrast, treatment with high doses (250 µg/ml) of Zeocin resulted in disruption of pericentric Smc3-GFP (Figure 3A and Supplemental Figure S3). To examine the consequences of a single DSB, we induced a DSB at the MAT locus on chromosome III via an HO endonuclease (Shroff et al., 2004). The cohesin cylinder expanded to 505 nm (Figure 3, A and B). The concentration of pericentric cohesin increased to 2.02 ± 0.40 from 1.86 ± 0.35 upon phleomycin treatment relative to WT (Figure 3C). Disruption in pericentric organization was evidenced by the reduction in Smc3-GFP fluorescence for a single DSB or exposure to Zeocin (1.55 ± 0.32 and 1.65 ± 0.42, respectively; Figure 3C).
FIGURE 3:
Pericentric cohesin responds to DNA damage. (A) Pericentric cohesin in metaphase with Smc3-GFP and Spc29-RFP in untreated, phleomycin, Zeocin, and a sDSB induced by HO endonuclease from the GAL10 promoter, target site at the MAT locus (KBY6050). Bar, 1 μm. (B) Cohesin sagittal width (schematic below A; red, spindle poles; green, cohesin; black, spindle microtubules) under DNA damage. Untreated (n = 24), plus phleomycin (n = 70), GALHO no induction (n = 38), and GALHO induction (n = 58) caused significant expansion (asterisks) of cohesin (Student’s t test, p < 0.05) compared with untreated (n = 50) and no GAL-HO (n = 20). (C) Cohesin barrel fluorescence intensity over arm untreated (n = 50), plus phleomycin (n = 40), plus Zeocin (n = 43), GALHO (n = 40), and I-SceI (n = 29). Student’s t test showed a difference between untreated and damaged cells (p < 0.05). (D) Metaphase cells showing three different kinetochore protein (Ame1-GFP) phenotypes: two spots (blue), one elongated (purple), and more than two spots (red). Asterisk denotes LacO/LacI in KBY 6047. Bar, 1 μm. (E) Bar graph showing the distribution of Ame1-GFP phenotypes in metaphase for WT cells (n = 92) plus phleomycin (n = 101), plus Zeocin (n = 153), and plus I-SceI (n = 123). Error bars are SEM.
The expansion or disruption of pericentric chromatin after DNA damage depends on the Rad9 DNA damage checkpoint, as well as on phosphorylation of histone H2A (S129A and S121A). In these mutants (rad9Δ, S129A, S121A), the cohesin barrel is not responsive to phleomycin treatment (Supplemental Figure S4). Similarly, induction of a DSB via HO does not result in expansion of the barrel in rad9Δ (Supplemental Figure S4). These findings are diagnostic of chromatin modification in the pericentromere as a consequence of DNA damage. Expansion of the cohesin barrel reflects the change in state of the chromatin network between the two spindle poles (Stephens et al., 2013).
A consequence of perturbations in pericentric cohesin is the loss of maintenance of kinetochore clustering (Anderson et al., 2009; Ng et al., 2009). Kinetochore declustering has also been reported after exposure to high concentration of DNA-damaging agents (Strecker et al., 2016). Whereas kinetochore declustering is not apparent at low doses of phleomycin, there is a marked loss of clustering after induction of I-SceI (50% cell > two clusters of sister kinetochores) and a high concentration of Zeocin (Figure 3, C–E; the latter as reported by Strecker et al., 2016). The increase in kinetochore declustering is consistent with both the decrease in pericentric cohesin (I-SceI, Figure 3C) and the loss of organization (Zeocin, Figure 3E). Taking the results together, we conclude that the increase in arm motion in response to DNA damage is not a consequence of kinetochore declustering. Kinetochore declustering reflects the change in state of the pericentromere on DNA damage, commensurate with a genome-wide response to genotoxic agents.
Telomere disruption and DNA damage result in similar increase of midarm chromatin motion
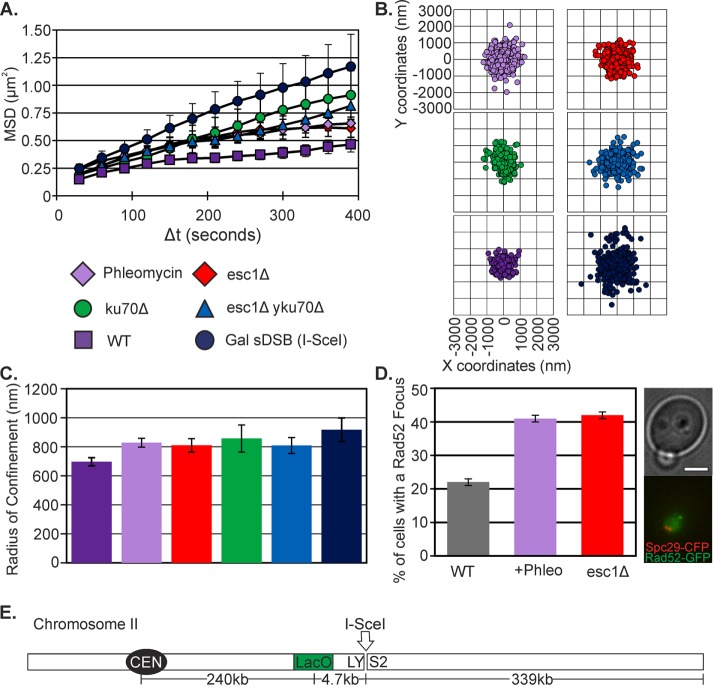
Tethering of chromosomes at their centromere and telomeres dictates their mobility (Verdaasdonk et al., 2013). To determine whether telomere detachment might account for the increase in motion on DNA damage, we examined chromosome motion in telomere-clustering mutants. The inner membrane–binding protein Esc1 and the telomere-binding complex yKu70/80 contribute to the clustering of telomeres and attachment to the nuclear envelope (Hediger et al., 2002; Taddei et al., 2006). Damage induced with phleomycin and mutants that mobilize telomeres from the nuclear envelope (yku70Δ, esc1Δ, and esc1Δ, ykuΔ double mutants) have similar mean squared displacement (MSD) curves and spot position variance and statistically similar Rc values (one-way analysis of variance [ANOVA], p = 0.9; Supplemental Table S1 and Figure 4, A–C). Increases in telomere mobility can influence the behavior of a locus 339 kb from the end of the chromosome. Rad52 focus formation is an indication of the cellular response to replication-induced or exogenous DNA damage (Lisby et al., 2001). Loss of telomere tethers in esc1Δ mutants, as well as other Esc1 functions, elicited an equivalent percentage of Rad52 foci as in cells treated with phleomycin (Figure 4D and Supplemental Figure S5).
FIGURE 4:
Telomere detachment increases arm mobility. (A) MSD curves of LacO at 240 kb from CEN2, WT (as in Figure 1A), plus phleomycin (n = 68), Gal sDSB (I-SceI; same as Figure 1A), esc1Δ (n = 39), yku70Δ (n = 17), and esc1Δ yku70Δ (n = 47). (B) Scatterplots illustrating variance in LacO/LacI position relative to the SPB. WT and Gal sDSB (I-SceI) are the same as in Figure 1D. (C) Rc values. Error bars are SEM. WT and Gal sDSB (I-SceI) are the same as in Figure 1C. There is no significant difference between plus phleomycin, esc1Δ, yku70Δ, and esc1Δ yku70Δ (one-way ANOVA, p = 0.9). (D) Rad52 focus formation in untreated (n = 358), plus phleomycin (n = 304), and esc1Δ (n = 246) G1/S cells. Error bars are SEM. (E) Schematic of LacO at 240 kb from CEN2.
Telomere-proximal regions of chromosomes are mobilized on DNA double-stranded breaks
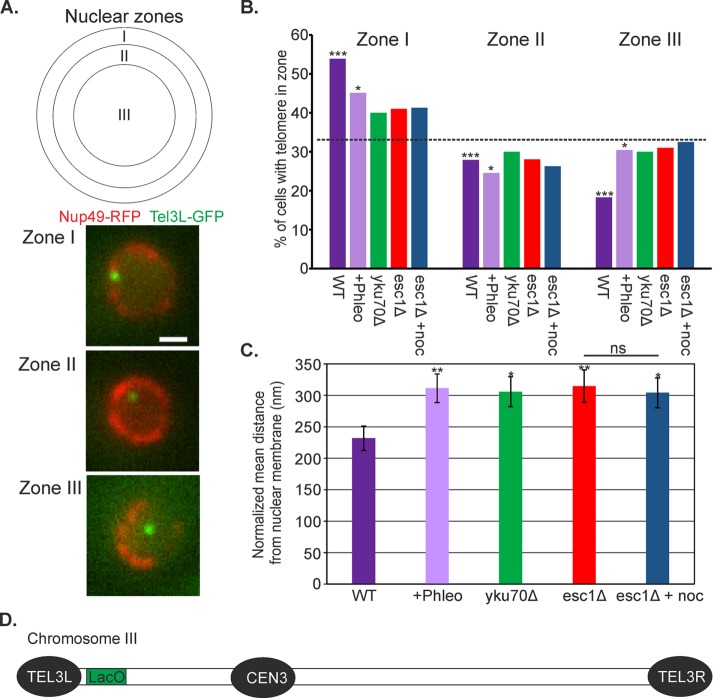
Given the similarity of chromosome motion and frequency of Rad52 foci due to DNA damage and telomere disruption, we compared the distribution of telomere positions of phleomycin-treated cells and esc1Δ and yku70Δ mutants. The nuclear envelope was marked by Nup49-RFP and the LacO array at TEL3L visualized with LacI-GFP (Figure 5). Cells in G1/early S phase were identified as unbudded or small-budded cells. The position of TEL3L from the nuclear envelope was measured and normalized by the radius of the nucleus. The mean position of the TEL3L array was 232 ± 19 nm (Figure 5C) and binned to one of three zones of equal area (Figure 5A). TEL3L is more frequently associated (within 184 nm) with the nuclear envelope (Figure 5B, zone I). We confirmed that the deletion of esc1 or yku70 alters the nuclear distribution of a LacO array integrated near the telomere on the left arm of chromosome III (TEL3L; Hediger et al., 2002; Bystricky et al., 2009; Figure 5B). Exposure to phleomycin resulted in the displacement of TEL3L from the nuclear envelope–proximal zone I to the distal zone III (>422 nm from the nuclear envelope in 40% of cells), comparable to esc1Δ and yku70Δ mutants and significantly different from the WT distribution (χ2 test, p = 0.004; Figure 5B). As previously reported (Taddei et al., 2004), telomere positions in esc1Δ and yku70Δ mutants were not significantly different from a random distribution (χ2 test, p > 0.05; random 33% occupation of each zone; dashed line in Figure 5B). In addition, the normalized mean distance of the TEL3L array from the nuclear membrane is significantly increased upon low doses of phleomycin, similar to the esc1Δ and yku70Δ mutants (Figure 5C). The addition of the microtubule-depolymerizing drug nocodazole did not alter the telomere location in esc1Δ mutants (Figure 5C; Student’s t test, p = 0.8).
FIGURE 5:
DNA damage alters telomere positioning in a way quantitatively similar to esc1Δ and yku70Δ. (A) The nucleus in 2D image stacks was partitioned into three zones of equal area. Nuclear envelope is labeled in red (Nup49-RFP) and TEL3L in green (LacO/LacI-GFP). Bar, 1 μm. (B) Distribution of LacO at TEL3L relative to the nuclear envelope in G1/S cells. The p values for the χ2 test were calculated for each data set compared with a random distribution (33% for each zone; dashed line: WT (p = 2 × 10−6, n = 104), plus phleomycin (p = 0.03, n = 102), yku70Δ (p = 0.4, n = 100), esc1Δ (p = 0.3, n = 100), and esc1Δ plus nocodazole (p = 0.4, n = 81). (C) Bar chart of the mean normalized distance of TEL3L array from the nuclear membrane. Distances were normalized to the radius of the nucleus. Error bars are SEM. Data are the same as in B. Asterisks indicate a significant difference from WT (Student’s t test; plus phleomycin, p = 0.009; yku70Δ, p = 0.02; esc1Δ, p = 0.01; esc1Δ plus nocodazole, p = 0.02). ns, not significantly different (Student’s t test, p = 0.8). (D) Schematic of the position of TEL3L-proximal LacO on chromosome III.
Polymer simulations predict that neither centromere nor telomere release is sufficient to increase mobility of midarm chromatin
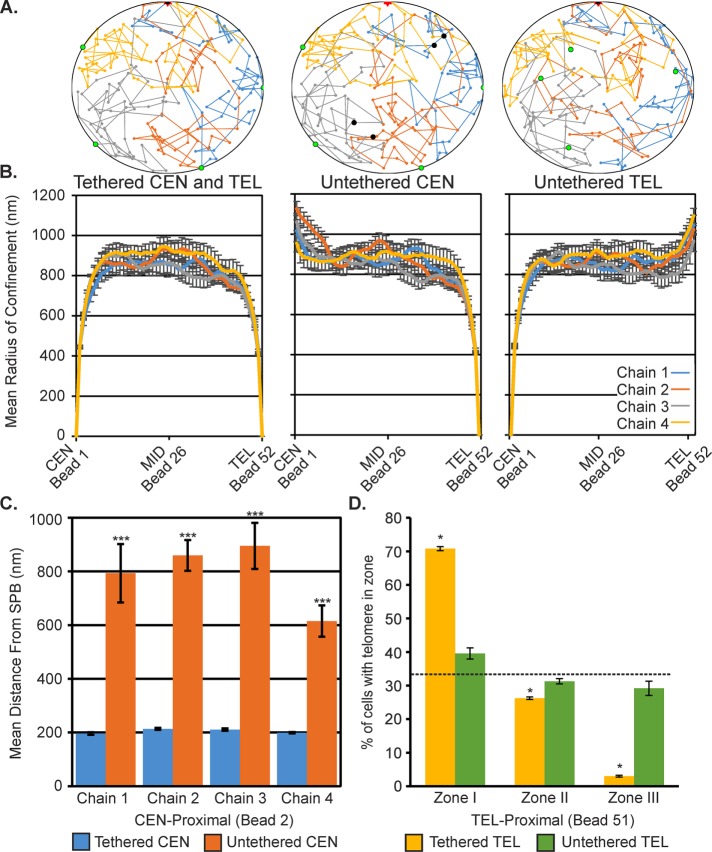
We used two-dimensional (2D) polymer simulations of four chains fluctuating within a circular “nuclear membrane” to gain insight into the physical consequences of centromere and telomere release on differing regions of a chromosome arm (Figure 6A). Each simulation was repeated 10 times with different random seeds, and the data were averaged across all simulations. Strikingly, the release of either centromeres or telomeres only increased the motion of the chains proximal to the untethered end (Figure 6B). There was no significant difference in the radii of confinement at the midpoint (bead 26) of the chains (Student’s t test, all p > 0.6) between the tethered simulations and either the centromere-untethered or telomere-untethered simulations (Figure 6B; Supplemental Figures S6 and S7). Despite the lack of a global increase in motion, the distance between the centromere-proximal region and the SPB increased in a similar manner to the GALCEN experiment (Figure 6C vs. Figure 2E). Moreover, simulated telomere release resulted in a similar shift in telomere-proximal chromosome regions toward the center of the nucleus as seen on telomere disruption and phleomycin treatment (Figure 6D vs. Figure 5B). The ensemble mean distance from the nuclear membrane across all 10 simulations of a telomere-proximal region (bead 51) increased from 138 ± 2 to 304 ± 13 nm (Student’s t test, p = 3 × 10−7). Thus additional forces may be required for the global increase in chromosome motion in the midregion of chromosome arms.
FIGURE 6:
Polymer simulations predict altered centromere and telomere localizations on release of tethering. (A) A four-chain polymer simulation in a Rabl-configuration confined within a 2000-nm diameter. Nuclear membrane (black circle) with centromeres (black dots) tethered to the SPB (red star), and telomeres (green dots) tethered to the membrane (left), centromeres untethered (middle), or telomeres untethered (right). Each simulation was repeated 10 times with different random seeds. (B) Mean radius of confinement of each bead within the simulation for tethered chains, untethered centromeres, and untethered telomeres. Error bars are SEM. (C) Mean distance from the simulation SPB for centromere-proximal bead 2 in the tethered centromere and untethered centromere simulations. All chains were significantly different with untethered centromere (Student’s t test, all p < 6 × 10−4). (D) Mean percentage of cells with telomere-proximal bead 51 within 184 nm of the simulated nuclear membrane (zone I), between 184 and 422 nm (zone II), and >422 nm (zone III). Means were calculated across 10 simulations. Tethered telomeres simulations were significantly different from random distribution (χ2 test, p = 0.03), whereas free telomeres were not significantly different (χ2 test, p = 0.9).
Cytoskeletal components are required for increase in midarm chromosome mobility and the alteration of telomere-proximal localization on DNA damage
The cytoskeleton is responsible for nuclear positioning in the vegetative cell cycle (Shaw et al., 1998; Yeh et al., 2000), chromosome pairing in the meiotic cell cycle (Chikashige et al., 1994), and the DNA damage response (Swartz et al., 2014; Chung et al., 2015; Lottersberger et al., 2015). Nuclear positioning in G1 through G2 involves the Kar9, Bim1 pathway, which links microtubules to the actin cytoskeleton via myosin (Beach et al., 2000; Yin et al., 2000), whereas positioning in metaphase/anaphase along the mother–bud axis involves cytoplasmic dynein (Yeh et al., 1995, 2000). We quantitated the motion of the 240-kb LacO/LacI-GFP (Figure 1A) under conditions that disrupt microtubule or actin cytoskeletal dynamics and the linkage between the two in the cytoplasm (Figure 7). The increase in motion of the midarm region of chromosome II after induction of a single DSB (I-SceI) depended on an active microtubule cytoskeleton (nocodazole treatment; Figure 7, A–C). The Rc was not significantly changed on nocodazole treatment in WT cells grown in glucose (Student’s t test, p = 0.07; Supplemental Table S1). The Rc for kar9Δ was significantly lower than for WT upon a single double-strand break (sDSB; I-SceI; Student’s t test, p = 0.02), whereas dhc1Δ and cells with reduced actin dynamics (latrunculin A treated) had reduced Rc values that were not significantly different from those for WT with a sDSB (I-SceI; Student’s t test, p >0.05; Supplemental Table S1; Spichal et al., 2016). Unlike telomere location in esc1Δ mutants, which was insensitive to nocodazole treatment (Figure 5, B and C), telomere redistribution on treatment with phleomycin was prevented on nocodazole treatment and Kar9 and dynein-null mutants, with TEL3L remaining primarily associated with the nuclear periphery (Figure 7, D and E). Thus DNA damage–induced redistribution of telomeres, but not the esc1Δ-induced redistribution, depends on microtubules, Kar9, and dynein (Figure 5B vs. Figure 7D).
FIGURE 7:
Damage-induced mobility and telomere position depend on the cytoskeleton. (A) MSD curves of LacO at 240 kb from CEN2 in WT (green), plus nocodazole (orange), I-SceI induced (blue), and I-SceI induced plus nocodazole (yellow) cells. (B) Rc values for WT (n = 25), plus nocodazole (n = 61), I-SceI (n = 48), and I SceI plus nocodazole (n = 20) cells. Error bars are SEM. (C) Scatterplots illustrating variance in LacO/LacI position relative to the SPB. (D) Distribution of LacO at TEL3L relative to the nuclear envelope in G1/S cells. The p values for the χ2 test were calculated for each data set to a random distribution (dashed line, 33% for each zone). WT (p = 2 × 10−6, n = 104), plus phleomycin (p = 0.03, n = 102); plus phleomycin plus nocodazole (p = 7 × 10−5, n = 86), plus nocodazole (p = 3 × 10−13, n = 78), dhc1Δ (p = 8.21 × 10−8, n = 86), dhc1Δ plus phleomycin (p = 1.43 × 10−15, n = 87), kar9 (p = 2.78 × 10−6, n = 75), and kar9 plus phleomycin (p = 7.85 × 10−9, n = 65). (E) Bar chart of the mean normalized distance of TEL3L array from the nuclear membrane. Distances were normalized to the radius of the nucleus. Error bars are SEM. Data are the same as in D. Asterisks indicate a significant difference between data sets. Students t test; WT vs. WT plus phleomycin, p = 0.009; WT vs. nocodazole, p = 0.04; WT plus phleomycin vs. WT plus phleomycin plus nocodazole, p = 0.001). ns, significant (Student’s t test, p > 0.05). (F) Schematic of LacO at 240 kb from CEN2.
DISCUSSION
The cell biology of DNA repair reveals aspects of chromosome dynamics that cannot be accounted for by the biochemistry of DNA repair or simple models of chromosome fluctuations. The mechanisms that account for increased mobilization range from activation of nucleosome remodelers (INO80) and kinesins (kinesin-14) to centromere and telomere release (Horigome et al., 2014; Chung et al., 2015; Strecker et al., 2016). None of these account for all behaviors, including 1) mobility of the damage site, 2) mobility of the genome, and 3) increased volume of the pericentromere in metaphase. The detachment of centromeres from the SPB using a GALCEN system has been shown to result in a large increase in chromosome mobility (Verdaasdonk et al., 2013; Strecker et al., 2016). To test centromere release directly, we examined the position of centromere-linked LacO arrays. As shown in Figure 2, there is no change in the position of LacO arrays after a DSB proximal to the centromere. In addition, induction of multiple lesions with phleomycin did not result in a significant increase (Student’s t test, p = 0.2) in the motion of a 6.8-kb Cen linked LacO array (Figure 2, F–H, and Supplemental Table S1). Instead, there is a significant expansion in the area occupied by pericentric chromatin within the mitotic spindle (Figure 3, A and B). The increase in area is indicative of relaxation of the constraints on the pericentromere. These data, together with the observation that the Cep3 kinetochore component is modified on exposure to damage (Strecker et al., 2016), are evidence for relaxation of the pericentromere as a means to enable chromosomes to explore more space.
A second means to mobilize the genome can be elicited through release of telomere constraints to the nuclear envelope. In budding yeast, the 32 telomeres from 16 chromosomes are clustered into four to six nuclear envelope–bound loci. These clusters constrain motion by reducing degrees of motion of the telomere and chromosome arm loci. Bead–spring polymer models indicate that the magnitude of motion depends on bead position relative to the tether (centromere or telomere; Verdaasdonk et al., 2013; Vasquez and Bloom, 2014). In vivo, release of telomeric loci from the nuclear envelope (Figure 6) or from lamin A and nuclear membrane in mammalian cells has profound effects on telomere and general chromatin mobility (Bronshtein et al., 2015). Concomitant with telomere release is the relocalization of bound Ku70/Ku80 to sites of DSBs as part of the DNA damage response (Martin et al., 1999). The Ku70/80 proteins play an essential role in genome stability and nonhomologous end-joining repair mechanisms (Martin et al., 1999). The release of telomeric constraints is translated into increased chromatin mobility along the chromosome arm. Modification of chromosome attachments to tether points reflects a polymer-based mechanism for regulating chromosome motion and enhanced homology search and repair after DNA damage.
In living cells, broken DNA ends (DSB) are not physically separated from each other on the chromosome scale (Kaye et al., 2004; Lobachev et al., 2004). Loss of essential nonhomologous repair enzymes leads to physical chromosome breaks, but even in these cases, the majority of DSBs remain in proximity (80%; Lobachev et al., 2004). Thus, even on damage, DNA ends are not “free,” but instead both ends remain proximal to one another while they explore more space. Mechanisms that shake up intact chromatin are analogous to mechanisms for untangling a ball of yarn. Shaking the ball is more effective than pulling individual strands, which might lead to knots and tighter tangles. The ways of shaking the nucleus are varied. Release from sites of tethering result in increased motion by increasing the number of available configurations of state (increase in entropy). Energetic processes are also engaged in response to DNA damage. Cytoplasmic dynein is responsible for pulling the nucleus through the neck of a budded cell, a process activated in checkpoint-deficient cells (Dotiwala et al., 2007). In addition, actin promotes nuclear dynamics in meiosis, when homology search is key (Trelles-Sticken et al., 2005; Scherthan et al., 2007; Koszul et al., 2008). Reducing actin dynamics with latrunculin A reduces the Rc of chromatin in WT or after DSB induction (Supplemental Table S1). Both actin and microtubule cytoskeletons participate in nuclear positioning, and thus it is not unexpected that both would affect chromatin behavior. The chromosomal response to DNA damage is reminiscent of genome-wide chromosome realignment seen in meiosis (Chikashige et al., 1994; Koszul et al., 2008), reflecting common mechanistic strategies for genome mobilization in cases of exogenous (DNA damage) or developmental (meiotic) perturbations.
MATERIALS AND METHODS
Nuclease cutting efficiency
We monitored cutting efficiency using quantitative PCR to measure the decrease in intensity of a fragment on introduction of a DNA DSB. For I-SceI, the cut site lies in the LYS2 gene (Figure 1). I-SceI is under control of the GAL1 promoter. Cells were grown to mid logarithmic phase in lactose-containing medium to prevent catabolite repression of the promoter in the presence of glucose. Aliquots were removed, and the remainder of the culture was switched to galactose-containing medium. Aliquots were removed at 3 and 6 h, and DNA was isolated. PCRs were performed (20 cycles) with oligonucleotides AATATGTACGGTACCACTGAAACAC (1672–1696) and GAATAGCTTTCGATTTCGACATCG (3815–3792) relative to LYS2 start (position 1) to generate a 2.1-kb fragment that flanks the I-SceI site imbedded in the open reading frame. To control for DNA input, oligonucleotides GTGCTGCTATCGATGCTACA (143–162) and AATGCGGTGTTCTTGTCTGG (842–823) relative to LEU2 start (position 1) were used to generate a 0.7-kb fragment. The resulting fragments were quantitated, and DNA fragments were analyzed according to the box-in-box method described by Hoffman et al. (2001). There is an 85% reduction in the fragment containing the LYS2 by 6 h following I-SceI induction (Supplemental Figure S2). This value is consistent with recent reports (Seeber et al., 2016). For GAL-HO, the cut site lies adjacent to CEN3, as described in Dotiwala et al. (2010) and shown schematically in Supplemental Figure S2. Cells were grown as described for I-SceI. PCRs were performed (20 cycles) with oligonucleotides TTCGTCAACTTAAAGATGACC (1kbCEN3US) and ACGCTGCAAGCTATTGAAAC (1kbCEN3DS). 1kbCEN3US is at position 113065 and 1kbCEN3DS is at position 114156 relative to CEN3 (114385–114501). In strain KBY8227, the oligonucleotides flank a 2-kb fragment containing GAL-HO and the hygromycin resistance gene (Supplemental Figure S2), yielding a 3.1-kb PCR product. On induction of GAL-HO, the cut fragment is resected, leading to deletion of the fragment, as well as CEN3, as the DSB is repaired by single-strand annealing between flanking homologous DNA segments (Supplemental Figure S2). DNA fragments were quantitated as described and normalized to uncut LEU2 PCR product.
Microscopy
Population and time-lapse images were acquired at room temperature (24°C) using a Nikon Eclipse Ti wide-field inverted microscope with a 100× Apo total internal reflection fluorescence 1.49 numerical aperture objective (Nikon, Melville, NY) and Andor Clara charge-coupled device camera (Andor, South Windsor, CT). Time-lapse stacks, each of seven 200-nm z-planes, were acquired every 30 s over a 10-min period (147 total planes per time lapse) with Nikon NIS Elements imaging software. Images were taken in transilluminated light, GFP and RFP fluorescence illumination. We chose the 30-s intervals as the best compromise between rate of acquisition and duration of time lapse due to photobleaching. The 30-s intervals were used in Verdaasdonk et al. (2013), where we report the effect of tethering on chromosome motion. Of importance, the MSD slopes of a 10-kb lacO/LacI-GFP array, 6.8 from CEN XV (KBY8065), taken at 3- and 30-s intervals are overlapping (Supplemental Figure S4). The 3-s interval allows one to estimate the slope of the MSD on a log-log plot, and the 30-s interval over a longer time scale provides a better estimate of the plateau value, which provides a better measurement of Rc.
Cohesin cylinder width
Three-dimensional stacks of images through sagittal planes were obtained as described. Line scans were drawn through the oblongate lobes of Smc3-GFP fluorescence in the sagittal (side-on) view. The width of the barrel was determined from the maximal fluorescence of each peak (Yeh et al., 2008; Haase et al., 2012).
Telomere localization
The position of the GFP spot was determined as previously described (Hediger et al., 2002). We limited our analysis to images in which the GFP foci appeared within an in-focus, circular Nup49-RFP signal. GFP foci that were in a distal z-plane from the circular Nup49-RFP signal were not measured. The distance from the spot to the nuclear envelope and the nuclear diameter were determined from a single z-stack image. Dividing the spot-to-periphery distance by the radius localized each spot to one of three zones of equal surface. Zone I has a width of 0.184 µm × nuclear radius (r), zone II has a width of 0.184–0.422 µm × r, and zone III a width of >0.422 µm × r. Confidence values for the χ2 were calculated for each data set and random and test distributions. All data except for yku70Δ were pools for two or more biological replicates. Student’s t test comparing the mean distance of the TEL3L array from the nuclear membrane assumed unequal variance and was performed using Excel.
Calculating Rc and MSD from experimental data
The GFP (LacO) and RFP (SPB) foci of the brightest planes per time point were tracked using Speckle Tracker, a custom MATLAB program (Wan et al., 2009, 2012). These coordinates were further analyzed using a custom PERL script to convert the pixels to nanometers and subtract the coordinates of the SPB from the LacO/LacI-GFP coordinates to eliminate cell and nuclear movement, subtract the means position of the resulting GFP coordinates, calculate the MSD of each time lapse, and export the MSDs and coordinates on Excel spreadsheets (Verdaasdonk et al., 2013). For MSD analysis, the images were identically analyzed using MetaMorph (Molecular Devices, Sunnyvale, CA) and MATLAB (MathWorks, Natick, MA). MATLAB was used to fit the spot positions as [μx, σx] = normfit (x – xmean) and [μy, σy] = normfit (y – ymean). The variance of the distribution of spot positions was then calculated as σ2 = mean (σ2x, σ2y). The average squared deviation from the mean position is 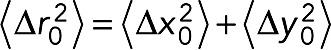 . Using a2 and
. Using a2 and 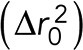 , we calculate Rc as
, we calculate Rc as 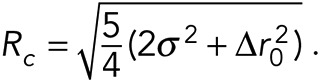
The radius of confinement was calculated for each cell to provide a distribution of values for each strain and treatment type.
Statistical analysis
For statistical comparison of reported Rc values, we used Student’s t test assuming unequal variances. To compare cohesin sagittal width measurements, we used a Student’s t test assuming equal variances (Yeh et al., 2008). One-way ANOVA and Student’s t test were performed using Excel.
Modeling description
We model a chromosome arm as a polymer chain constrained to lie within the nuclear envelope (idealized as a circle) and tethered to the boundary at either one or both ends. Following polymer physics, each chain is represented by a series of beads connected via springs. Confinement is modeled explicitly by not allowing beads to leave the circular domain. Thermal motion is introduced through a normal distribution with mean zero and variance 2KBTζ, where ζ is the drag coefficient. Thermal noise and the drag force exerted on a bead as it moves in the medium are related to each other through the fluctuation-dissipation theorem. In addition, we consider all attractive forces to be described by a wormlike chain potential (springs joining the beads) and all repulsive forces by an excluded-volume potential. The resulting equations of motion for each bead are given elsewhere (Verdaasdonk et al., 2013; Vasquez and Bloom, 2014). We use four chains each consisting of 52 beads (bead 1 = centromere; bead 52 = telomere). Assuming a molecular discretization of 5 kb/spring, this corresponds to chains that are ∼250 kb long. Once a telomere is released from the boundary, its movement is governed by the same forces acting on all other interior beads. Student’s t tests on simulation data were performed using MATLAB.
Cell preparations
Genotypes are given in Supplemental Table S2. Cells were grown to log phase at 24°C, the temperature for imaging. To induce multiple DSBs, cells were treated with 3 µg/ml phleomycin for 30–60 min or 250 µg/ml Zeocin for 1–2 h. To induce a single DSB at a known locus, cells were grown overnight in YPL (lactate) and transferred to YPG (galactose) for ∼1–5 h before imaging to induce the I-SceI or HO endonuclease (Lobachev et al., 2004; Shroff et al., 2004). To depolymerize the cytoskeletal elements, cells were treated with 20 µg/ml nocodazole (microtubules) or 0.5 µg/ml latrunculin A (actin) for 30 min to 1 h before imaging. For imaging, cells were washed and resuspended in yeast extract plus casamino acids (YC) complete medium with filter-sterilized 2% glucose or 2% galactose for activation of a Gal promoter. Cells treated with phleomycin, Zeocin, and/or nocodazole or latrunculin A were resuspended in YC complete medium solutions containing the respective concentration of each drug.
Supplementary Material
Acknowledgments
This work was supported by Grants R37 GM32238 to K.B. and T32 GM007092-39 to J.L. from the National Institutes of Health.
Abbreviations used:
- DSB
double-strand break
- GFP
green fluorescent protein
- LacI
Lac repressor
- LacO
Lac operator
- RFP
red fluorescent protein
- sDSB
single double-strand break
- SPB
spindle pole body.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-12-0846) on April 27, 2017.
REFERENCES
- Anderson M, Haase J, Yeh E, Bloom K. Function and assembly of DNA looping, clustering, and microtubule attachment complexes within a eukaryotic kinetochore. Mol Biol Cell. 2009;20:4131–4139. doi: 10.1091/mbc.E09-05-0359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach DL, Thibodeaux J, Maddox P, Yeh E, Bloom K. The role of the proteins Kar9 and Myo2 in orienting the mitotic spindle of budding yeast. Curr Biol. 2000;10:1497–1506. doi: 10.1016/s0960-9822(00)00837-x. [DOI] [PubMed] [Google Scholar]
- Blat Y, Kleckner N. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell. 1999;98:249–259. doi: 10.1016/s0092-8674(00)81019-3. [DOI] [PubMed] [Google Scholar]
- Bronshtein I, Kepten E, Kanter I, Berezin S, Lindner M, Redwood AB, Mai S, Gonzalo S, Foisner R, Shav-Tal Y, Garini Y. Loss of lamin A function increases chromatin dynamics in the nuclear interior. Nat Commun. 2015;6:8044. doi: 10.1038/ncomms9044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bystricky K, Van Attikum H, Montiel MD, Dion V, Gehlen L, Gasser SM. Regulation of nuclear positioning and dynamics of the silent mating type loci by the yeast Ku70/Ku80 complex. Mol Cell Biol. 2009;29:835–848. doi: 10.1128/MCB.01009-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Ghorai MK, Kenney G, Stubbe J. Mechanistic studies on bleomycin-mediated DNA damage: multiple binding modes can result in double-stranded DNA cleavage. Nucleic Acids Res. 2008;36:3781–3790. doi: 10.1093/nar/gkn302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikashige Y, Ding DQ, Funabiki H, Haraguchi T, Mashiko S, Yanagida M, Hiraoka Y. Telomere-led premeiotic chromosome movement in fission yeast. Science. 1994;264:270–273. doi: 10.1126/science.8146661. [DOI] [PubMed] [Google Scholar]
- Chung DK, Chan JN, Strecker J, Zhang W, Ebrahimi-Ardebili S, Lu T, Abraham KJ, Durocher D, Mekhail K. Perinuclear tethers license telomeric DSBs for a broad kinesin- and NPC-dependent DNA repair process. Nat Commun. 2015;6:7742. doi: 10.1038/ncomms8742. [DOI] [PubMed] [Google Scholar]
- D’Amours D, Amon A. At the interface between signaling and executing anaphase–Cdc14 and the FEAR network. Genes Dev. 2004;18:2581–2595. doi: 10.1101/gad.1247304. [DOI] [PubMed] [Google Scholar]
- Dion V, Gasser SM. Chromatin movement in the maintenance of genome stability. Cell. 2013;152:1355–1364. doi: 10.1016/j.cell.2013.02.010. [DOI] [PubMed] [Google Scholar]
- Dion V, Kalck V, Horigome C, Towbin BD, Gasser SM. Increased mobility of double-strand breaks requires Mec1, Rad9 and the homologous recombination machinery. Nat Cell Biol. 2012;14:502–509. doi: 10.1038/ncb2465. [DOI] [PubMed] [Google Scholar]
- Dion V, Kalck V, Seeber A, Schleker T, Gasser SM. Cohesin and the nucleolus constrain the mobility of spontaneous repair foci. EMBO Rep. 2013;14:984–991. doi: 10.1038/embor.2013.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotiwala F, Haase J, Arbel-Eden A, Bloom K, Haber JE. The yeast DNA damage checkpoint proteins control a cytoplasmic response to DNA damage. Proc Natl Acad Sci USA. 2007;104:11358–11363. doi: 10.1073/pnas.0609636104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotiwala F, Harrison JC, Jain S, Sugawara N, Haber JE. Mad2 prolongs DNA damage checkpoint arrest caused by a double-strand break via a centromere-dependent mechanism. Curr Biol. 2010;20:328–332. doi: 10.1016/j.cub.2009.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge T, Panyukov S, Rubinstein M. Self-similar conformations and dynamics in entangled melts and solutions of nonconcatenated ring polymers. Macromolecules. 2016;49:708–722. doi: 10.1021/acs.macromol.5b02319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goloborodko A, Imakaev MV, Marko JF, Mirny L. Compaction and segregation of sister chromatids via active loop extrusion. Elife. 2016a;5:e14864. doi: 10.7554/eLife.14864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goloborodko A, Marko JF, Mirny LA. Chromosome compaction by active loop extrusion. Biophys J. 2016b;110:2162–2168. doi: 10.1016/j.bpj.2016.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase J, Stephens A, Verdaasdonk J, Yeh E, Bloom K. Bub1 kinase and Sgo1 modulate pericentric chromatin in response to altered microtubule dynamics. Curr Biol. 2012;22:471–481. doi: 10.1016/j.cub.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hediger F, Neumann FR, Van Houwe G, Dubrana K, Gasser SM. Live imaging of telomeres: yKu and Sir proteins define redundant telomere-anchoring pathways in yeast. Curr Biol. 2002;12:2076–2089. doi: 10.1016/s0960-9822(02)01338-6. [DOI] [PubMed] [Google Scholar]
- Heun P, Laroche T, Shimada K, Furrer P, Gasser SM. Chromosome dynamics in the yeast interphase nucleus. Science. 2001;294:2181–2186. doi: 10.1126/science.1065366. [DOI] [PubMed] [Google Scholar]
- Hill A, Bloom K. Genetic manipulation of centromere function. Mol Cell Biol. 1987;7:2397–2405. doi: 10.1128/mcb.7.7.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman DB, Pearson CG, Yen TJ, Howell BJ, Salmon ED. Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores. Mol Biol Cell. 2001;12:1995–2009. doi: 10.1091/mbc.12.7.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horigome C, Oma Y, Konishi T, Schmid R, Marcomini I, Hauer MH, Dion V, Harata M, Gasser SM. SWR1 and INO80 chromatin remodelers contribute to DNA double-strand break perinuclear anchorage site choice. Mol Cell. 2014;55:626–639. doi: 10.1016/j.molcel.2014.06.027. [DOI] [PubMed] [Google Scholar]
- Kaye JA, Melo JA, Cheung SK, Vaze MB, Haber JE, Toczyski DP. DNA breaks promote genomic instability by impeding proper chromosome segregation. Curr Biol. 2004;14:2096–2106. doi: 10.1016/j.cub.2004.10.051. [DOI] [PubMed] [Google Scholar]
- Koszul R, Kim KP, Prentiss M, Kleckner N, Kameoka S. Meiotic chromosomes move by linkage to dynamic actin cables with transduction of force through the nuclear envelope. Cell. 2008;133:1188–1201. doi: 10.1016/j.cell.2008.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrimore J, Aicher JK, Hahn P, Fulp A, Kompa B, Vicci L, Falvo M, Taylor RM, 2nd, Bloom K. ChromoShake: a chromosome dynamics simulator reveals that chromatin loops stiffen centromeric chromatin. Mol Biol Cell. 2016;27:153–166. doi: 10.1091/mbc.E15-08-0575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrimore J, Vasquez PA, Falvo MR, Taylor RM, 2nd Vicci L, Yeh E, Forest MG, Bloom K. DNA loops generate intracentromere tension in mitosis. J Cell Biol. 2015;210:553–564. doi: 10.1083/jcb.201502046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M, Rothstein R, Mortensen UH. Rad52 forms DNA repair and recombination centers during S phase. Proc Natl Acad Sci USA. 2001;98:8276–8282. doi: 10.1073/pnas.121006298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobachev K, Vitriol E, Stemple J, Resnick MA, Bloom K. Chromosome fragmentation after induction of a double-strand break is an active process prevented by the RMX repair complex. Curr Biol. 2004;14:2107–2112. doi: 10.1016/j.cub.2004.11.051. [DOI] [PubMed] [Google Scholar]
- Lottersberger F, Karssemeijer RA, Dimitrova N, de Lange T. 53BP1 and the LINC complex promote microtubule-dependent DSB mobility and DNA repair. Cell. 2015;163:880–893. doi: 10.1016/j.cell.2015.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcomini I, Gasser SM. Nuclear organization in DNA end processing: telomeres vs double-strand breaks. DNA Repair (Amst) 2015;32:134–140. doi: 10.1016/j.dnarep.2015.04.024. [DOI] [PubMed] [Google Scholar]
- Marshall WF, Fung JC. Modeling meiotic chromosome pairing: nuclear envelope attachment, telomere-led active random motion, and anomalous diffusion. Phys Biol. 2016;13:026003. doi: 10.1088/1478-3975/13/2/026003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall WF, Straight A, Marko JF, Swedlow J, Dernburg A, Belmont A, Murray AW, Agard DA, Sedat JW. Interphase chromosomes undergo constrained diffusional motion in living cells. Curr Biol. 1997;7:930–939. doi: 10.1016/s0960-9822(06)00412-x. [DOI] [PubMed] [Google Scholar]
- Martin SG, Laroche T, Suka N, Grunstein M, Gasser SM. Relocalization of telomeric Ku and SIR proteins in response to DNA strand breaks in yeast. Cell. 1999;97:621–633. doi: 10.1016/s0092-8674(00)80773-4. [DOI] [PubMed] [Google Scholar]
- Megee PC, Mistrot C, Guacci V, Koshland D. The centromeric sister chromatid cohesion site directs Mcd1p binding to adjacent sequences. Mol Cell. 1999;4:445–450. doi: 10.1016/s1097-2765(00)80347-0. [DOI] [PubMed] [Google Scholar]
- Mine-Hattab J, Rothstein R. Increased chromosome mobility facilitates homology search during recombination. Nat Cell Biol. 2012;14:510–517. doi: 10.1038/ncb2472. [DOI] [PubMed] [Google Scholar]
- Mora-Lugo R, Zimmermann J, Rizk AM, Fernandez-Lahore M. Development of a transformation system for Aspergillus sojae based on the Agrobacterium tumefaciens-mediated approach. BMC Microbiol. 2014;14:247. doi: 10.1186/s12866-014-0247-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng TM, Waples WG, Lavoie BD, Biggins S. Pericentromeric sister chromatid cohesion promotes kinetochore biorientation. Mol Biol Cell. 2009;20:3818–3827. doi: 10.1091/mbc.E09-04-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pliss A, Malyavantham KS, Bhattacharya S, Berezney R. Chromatin dynamics in living cells: identification of oscillatory motion. J Cell Physiol. 2013;228:609–616. doi: 10.1002/jcp.24169. [DOI] [PubMed] [Google Scholar]
- Scherthan H, Wang H, Adelfalk C, White EJ, Cowan C, Cande WZ, Kaback DB. Chromosome mobility during meiotic prophase in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2007;104:16934–16939. doi: 10.1073/pnas.0704860104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeber A, Hegnauer AM, Hustedt N, Deshpande I, Poli J, Eglinger J, Pasero P, Gut H, Shinohara M, Hopfner KP, et al. RPA mediates recruitment of MRX to forks and double-strand breaks to hold sister chromatids together. Mol Cell. 2016;64:951–966. doi: 10.1016/j.molcel.2016.10.032. [DOI] [PubMed] [Google Scholar]
- Shaw SL, Maddox P, Skibbens RV, Yeh E, Salmon ED, Bloom K. Nuclear and spindle dynamics in budding yeast. Mol Biol Cell. 1998;9:1627–1631. doi: 10.1091/mbc.9.7.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shroff R, Arbel-Eden A, Pilch D, Ira G, Bonner WM, Petrini JH, Haber JE, Lichten M. Distribution and dynamics of chromatin modification induced by a defined DNA double-strand break. Curr Biol. 2004;14:1703–1711. doi: 10.1016/j.cub.2004.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spichal M, Brion A, Herbert S, Cournac A, Marbouty M, Zimmer C, Koszul R, Fabre E. Evidence for a dual role of actin in regulating chromosome organization and dynamics in yeast. J Cell Sci. 2016;129:681–692. doi: 10.1242/jcs.175745. [DOI] [PubMed] [Google Scholar]
- Stephens AD, Haase J, Vicci L, Taylor RM, 2nd, Bloom K. Cohesin, condensin, and the intramolecular centromere loop together generate the mitotic chromatin spring. J Cell Biol. 2011;193:1167–1180. doi: 10.1083/jcb.201103138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens AD, Snider CE, Haase J, Haggerty RA, Vasquez PA, Forest MG, Bloom K. Individual pericentromeres display coordinated motion and stretching in the yeast spindle. J Cell Biol. 2013;203:407–416. doi: 10.1083/jcb.201307104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strecker J, Gupta GD, Zhang W, Bashkurov M, Landry MC, Pelletier L, Durocher D. DNA damage signalling targets the kinetochore to promote chromatin mobility. Nat Cell Biol. 2016;18:281–290. doi: 10.1038/ncb3308. [DOI] [PubMed] [Google Scholar]
- Strom L, Lindroos HB, Shirahige K, Sjogren C. Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol Cell. 2004;16:1003–1015. doi: 10.1016/j.molcel.2004.11.026. [DOI] [PubMed] [Google Scholar]
- Swartz RK, Rodriguez EC, King MC. A role for nuclear envelope-bridging complexes in homology-directed repair. Mol Biol Cell. 2014;25:2461–2471. doi: 10.1091/mbc.E13-10-0569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A, Hediger F, Neumann FR, Bauer C, Gasser SM. Separation of silencing from perinuclear anchoring functions in yeast Ku80, Sir4 and Esc1 proteins. EMBO J. 2004;23:1301–1312. doi: 10.1038/sj.emboj.7600144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A, Van Houwe G, Hediger F, Kalck V, Cubizolles F, Schober H, Gasser SM. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature. 2006;441:774–778. doi: 10.1038/nature04845. [DOI] [PubMed] [Google Scholar]
- Trelles-Sticken E, Adelfalk C, Loidl J, Scherthan H. Meiotic telomere clustering requires actin for its formation and cohesin for its resolution. J Cell Biol. 2005;170:213–223. doi: 10.1083/jcb.200501042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsabar M, Haase J, Harrison B, Snider CE, Eldridge B, Kaminsky L, Hine RM, Haber JE, Bloom K. A cohesin-based partitioning mechanism revealed upon transcriptional inactivation of centromere. PLoS Genet. 2016;12:e1006021. doi: 10.1371/journal.pgen.1006021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasquez PA, Bloom K. Polymer models of interphase chromosomes. Nucleus. 2014;5:376–390. doi: 10.4161/nucl.36275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdaasdonk JS, Vasquez PA, Barry RM, Barry T, Goodwin S, Forest MG, Bloom K. Centromere tethering confines chromosome domains. Mol Cell. 2013;52:819–831. doi: 10.1016/j.molcel.2013.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan X, Cimini D, Cameron LA, Salmon ED. The coupling between sister kinetochore directional instability and oscillations in centromere stretch in metaphase PtK1 cells. Mol Biol Cell. 2012;23:1035–1046. doi: 10.1091/mbc.E11-09-0767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan X, O’Quinn RP, Pierce HL, Joglekar AP, Gall WE, DeLuca JG, Carroll CW, Liu ST, Yen TJ, McEwen BF, et al. Protein architecture of the human kinetochore microtubule attachment site. Cell. 2009;137:672–684. doi: 10.1016/j.cell.2009.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber SC, Spakowitz AJ, Theriot JA. Nonthermal ATP-dependent fluctuations contribute to the in vivo motion of chromosomal loci. Proc Natl Acad Sci USA. 2012;109:7338–7343. doi: 10.1073/pnas.1119505109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M, Bloom K. Mitotic spindle form and function. Genetics. 2012;190:1197–1224. doi: 10.1534/genetics.111.128710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E, Haase J, Paliulis LV, Joglekar A, Bond L, Bouck D, Salmon ED, Bloom KS. Pericentric chromatin is organized into an intramolecular loop in mitosis. Curr Biol. 2008;18:81–90. doi: 10.1016/j.cub.2007.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E, Skibbens RV, Cheng JW, Salmon ED, Bloom K. Spindle dynamics and cell cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae. J Cell Biol. 1995;130:687–700. doi: 10.1083/jcb.130.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E, Yang C, Chin E, Maddox P, Salmon ED, Lew DJ, Bloom K. Dynamic positioning of mitotic spindles in yeast: role of microtubule motors and cortical determinants. Mol Biol Cell. 2000;11:3949–3961. doi: 10.1091/mbc.11.11.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Pruyne D, Huffaker TC, Bretscher A. Myosin V orientates the mitotic spindle in yeast. Nature. 2000;406:1013–1015. doi: 10.1038/35023024. [DOI] [PubMed] [Google Scholar]
- Zimmer C, Fabre E. Principles of chromosomal organization: lessons from yeast. J Cell Biol. 2011;192:723–733. doi: 10.1083/jcb.201010058. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.