Abstract
Paris is famous in China for its medicinal value and has been included in the Chinese Pharmacopoeia. Inaccurate identification of these species could confound their effective exploration, conservation, and domestication. Due to the plasticity of the morphological characteristics, correct identification among Paris species remains problematic. In this regard, we report the complete chloroplast genome of P. thibetica and P. rugosa to develop highly variable molecular markers. Comparing three chloroplast genomes, we sought out the most variable regions to develop the best cpDNA barcodes for Paris. The size of Paris chloroplast genome ranged from 162,708 to 163,200 bp. A total of 134 genes comprising 81 protein coding genes, 45 tRNA genes and 8 rRNA genes were observed in all three chloroplast genomes. Eight rapidly evolving regions were detected, as well as the difference of simple sequence repeats (SSR) and repeat sequence. Two regions of the coding gene ycf1, ycf1a and ycf1b, evolved the quickest and were proposed as core barcodes for Paris. The complete chloroplast genome sequences provide more integrated and adequate information for better understanding the phylogenetic pattern and improving efficient discrimination during species identification.
Introduction
The chloroplasts are photosynthetic organelles that provide energy to green plants. In angiosperms, most chloroplast genomes are circular, double-stranded DNA, containing a pair of inverted repeats (IRs), one large single-copy region (LSC) and one small single copy region (SSC)1, 2. Most chloroplast genomes are ranging from 120–160 kb in length and highly conserved in gene content and order3, 4. Owing to being haploid, maternal inheritance, and highly conservation in gene content and genome structure, the chloroplast genomes are valuable sources for exploring useful DNA markers for species identification, evolutionary studies and phylogenetic relationships among plant species5–7. The advance of high-throughput sequencing technologies has facilitated rapid progress of chloroplast genomics due to time-saving and low-cost advantages8. The number of chloroplast genomes of land plants released in the National Center for Biotechnology Information (NCBI) has risen to 1,011 (accessed at October 31, 2016).
The genus Paris (Melanthiaceae: Parideae)9, 10 consists of about 24 species of perennial herbs distributed in the temperate regions from Europe to eastern Asia, 22 species (12 endemic) were chiefly in China. The rhizomes of many Paris species are used in traditional Chinese medicine for more than 2000 years in China, owing to their analgesic and anti-coagulant properties, most notably as an ingredient of Yunnan Baiyao11. However, over-exploitation for economic purposes is pushing these species to the brink of extinction. The Paris genus is listed as exit-prohibited species by Environmental Protection Agency. So there is an urgent need to develop conservation strategies to prevent losses of species resources through the characterization of its genomic information and genetic structure.
Because of their medicine value, Paris species has been the subject of taxonomic studies and, particularly, species identification12, 13. However, so far, there are no efficient methods for identifying the species of Paris. Traditionally, the taxonomy and species identification of the genus Paris are based on the morphological traits, but the plasticity of its morphological characteristics made the classification of Paris very complicated, most Paris species have abundant intraspecific variations in morphology and chemical composition12, 14, 15.
Molecular methods, such as molecular marker techniques and DNA barcoding, provide effective information for taxonomy and species identification. In the past decades, the applications of diverse molecular techniques have gained increasing importance in resolving taxonomy and species identification questions. However, at the species level, the reported candidate barcoding sequences still have difficulties in the identification of Paris species. Analysis based on plastid genomic markers (psbA-tmH, rpoB, rpoCl, rbcL, matK) and nuclear gene ITS2 suggested that ITS2 can only discriminate P. polyphylla var. yunnanensis from P. polyphylla var. chinensis 16–19. Ji et al. tested the generic and infrageneric circumscription of Paris with nuclear ITS and plastid psbA-trnH, trnL-trnF DNA sequence data and supported the classification of Paris as a single genus, but the delimitation of species still remained unresolved12. All these studies have provided valuable insights for an initial molecular-based identification of Paris, but there were too little variations in those chloroplast genomic markers to address the issues of species discrimination.
Here, we sequenced and analyzed the chloroplast genome of P. rugosa and P. thibetica using the next-generation sequencing platform. Our aim was to retrieve valuable chloroplast molecular markers by comparing the chloroplast genomes among these two and recently published chloroplast genomes of Paris. Our second objective was to investigate global structural patterns of Paris chloroplast genomes and to examine variations of simple sequence repeat (SSRs) and repeat sequences among Paris chloroplast genomes. We believe that these types of resources will be useful for species-level discrimination and avoid confounding effective exploration, conservation, and domestication for Paris species.
Results
Genome Assembly and Features
We sequenced the complete chloroplast genome of two Paris species, P. rugosa and P. thibetica (Fig. 1). In total, 10,380,007 (P. rugosa) and 26,745,248 (P. thibetica) raw data reads were generated. Out of those, 401,240 and 297,202 reads were identified as the chloroplast genome sequences for P. rugosa and P. thibetica, respectively (Table 1). Chloroplast genomes showed a typical quadripartite structure, consisting of a pair of IRs (32,884–33,144 bp) separated by the LSC (84,010–84,108 bp) and SSC (12,854–12,984 bp) regions (Fig. 1 and Table 1). The chloroplast genome of P. rugosa (GenBank accession no. KY247142), with a length of 163,200 bp, was 492 bp larger than that of P. thibetica (GenBank accession no. KY247143), 210 bp larger than that of P. polyphylla var. yunnanensis (GenBank accession no. KT805945) published in our previous paper.
Figure 1.
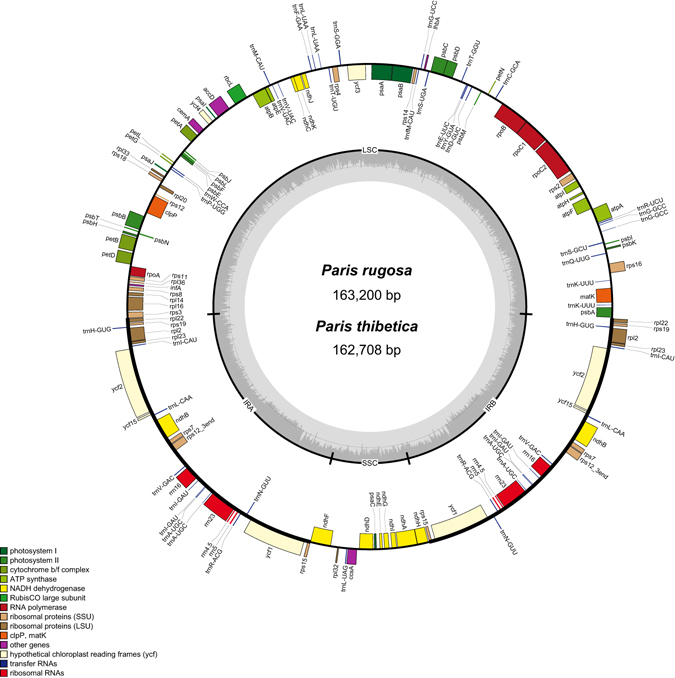
Gene map of Paris Chloroplast genome. The genes inside and outside of the circle are transcribed in the clockwise and counterclockwise directions, respectively. Genes belonging to different functional groups are shown in different colors. The thick lines indicate the extent of the inverted repeats (IRa and IRb) that separate the genomes into small single-copy (SSC) and large single-copy (LSC) regions.
Table 1.
Comparison of feature of Paris rugose, Paris thibetica, Paris polyphylla var. yunnanensis.
| Species | Paris rugosa | Paris thibetica | Paris polyphylla var. yunnanensis |
|---|---|---|---|
| Accession number | KY247142 | KY247143 | KT805945 |
| Genome size (bp) | 163200 | 162708 | 162990 |
| LSC (bp) | 84058 | 84010 | 84108 |
| SSC (bp) | 12854 | 12930 | 12984 |
| IRs (bp) | 33144 | 32884 | 32949 |
| Number of protein-coding genes1 | 81(9) | 81(9) | 81(9) |
| Number of tRNAs genes1 | 45(8) | 45(8) | 45(8) |
| Nubmer of rRNAs genes1 | 8(4) | 8(4) | 8(4) |
| GC content (%) | 37.1 | 37.2 | 37.1 |
| Raw data read number | 10380007 | 26745248 | / |
| Mapped read number | 401240 | 297202 | / |
| Chloroplast coverage(X) | 368 | 373 | / |
1The numbers in parenthesis indicate the genes duplicated in the IR regions.
Three Paris genomes identically harbored 113 different genes arranged in the same order, including 72 protein-coding genes, 37 tRNA genes and 4 rRNA genes. All these three genomes have rich AT content with an overall purine content ranging from 62. 8% to 62.9% (Table 1).
SSR and Repetitive Sequence Statistics
SSRs are repeated DNA sequences consisting of tandem repeats 1–10 bp in length per unit distributed throughout the genome (Fig. 2A). The total number of SSRs was 127 in P. polyphylla var. yunnanensis, 124 in P. rugosa and 131 in P. thibetica (Supplementary Table S3). The majority type of SSR in all species was mononucleotide, with 57 in P. polyphylla var. yunnanensis, 61 in P. rugosa and 64 in P. thibetica (Supplementary Table S1).
Figure 2.
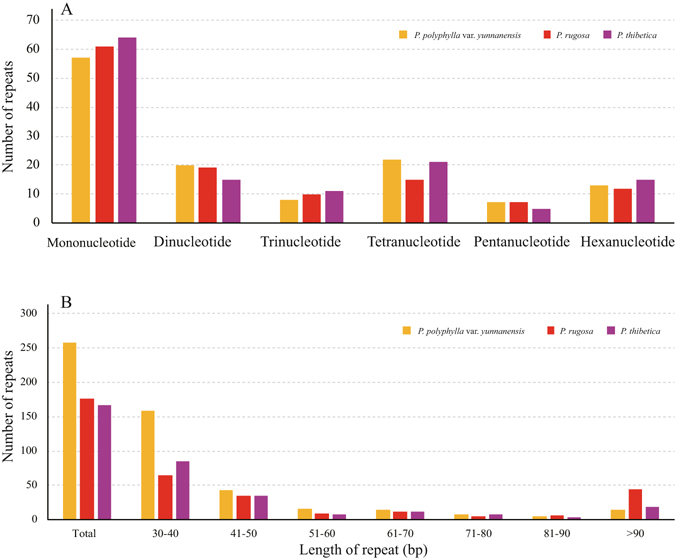
Analysis of repeated sequences in the three Paris chloroplast genomes. (A) Frequency of simple sequence repeats (SSRs) by MISA. (B) Frequency of repeat sequences determined by REPuter.
Repeat sequences with repeat unit longer than 30 bp and sequence identity greater than 90% were analyzed (Fig. 2B). P. polyphylla var. yunnanensis contained 258 repeats, of these, 159 repeats were 30–40 bp long, 85 repeats were 40–90 bp long, and 14 repeats were longer than 90 bp. P. rugosa contained 176 repeats, of these, 65 repeats were 30–40 bp long, 67 repeats were 40–90 bp long, and 44 repeats were longer than 90 bp. P. thibetica contained 167 repeats, of these, 85 repeats were 30–40 bp long, 64 repeats were 40–90 bp long, and 18 repeats were longer than 90 bp (Fig. 2B, Supplementary Table S4).
Divergent Hotspots in Paris Chloroplast Genome
A total of 902 SNPs were detected among three Paris species. To clarify the sequence divergence level, the nucleotide variability values within 600 bp in all three chloroplast genomes were calculated with DnaSP 5.0 software. The values ranged from 0 to 0.02056 with a mean of 0.00375, revealing the slight differences among the genomes. However, eight highly variable loci with higher Pi values (Pi > 0.0087) were precisely located (Fig. 3). These regions included trnS-trnG, rpoC1, psbC-trnS-psbZ, ycf2, ycf1a, trnN-ycf1, ycf1b, rpl32-trnL, of which three loci lie in the LSC region, four in the IR region, and one in the SSC region (Fig. 3).
Figure 3.
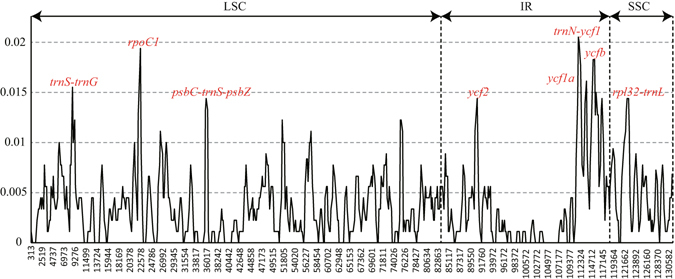
Sliding window analysis of the whole chloroplast genome of three Paris species. (window length: 600 bp, step size: 200 bp). X-axis: position of the midpoint of a window, Y-axis: nucleotide diversity of each window.
DNA barcoding of Paris
TrnN-ycf1 had some more indels and poly structure and the primers did not work well, so we gave it up in the following analyses. The variability of seven developed regions were tested together with three conventional candidate DNA barcodes (matK, rbcL and trnH-psbA) using 19 samples of Paris species. Features of ten barcode data set were shown in Table 2. There are only six variable sites of the trnH-psbA region, showing the lowest level of variability (0.68%). The variability of the ycf1a region was the highest (7.72%), followed by ycf1b region (6.47%), trnS-trnG region (6.25%), and rpl32-trnL (5.25%).
Table 2.
Variability of the seven new markers and universal chloroplast DNA barcode in Paris.
| Markers | Region | Length | Variable sites | Information sites | Nucleotide diversity | Discrimination success (%) based on Distance method | ||
|---|---|---|---|---|---|---|---|---|
| Numbers | % | Numbers | % | |||||
| trnS-trnG | LSC | 1279 | 80 | 6.25% | 38 | 2.97% | 0.00602 | 47.37% |
| rpoC1 | LSC | 999 | 24 | 2.40% | 17 | 1.70% | 0.00589 | 15.79% |
| psbC-trnS-psbZ | LSC | 1035 | 12 | 1.16% | 8 | 0.77% | 0.00226 | 42.11% |
| ycf2 | IR | 940 | 29 | 3.09% | 21 | 2.23% | 0.00846 | 36.84% |
| ycf1a | IR | 1140 | 88 | 7.72% | 57 | 5.00% | 0.01826 | 52.63% |
| ycfb | IR | 556 | 36 | 6.47% | 23 | 4.14% | 0.01442 | 31.58% |
| rpl32-trnL | SSC | 1010 | 53 | 5.25% | 31 | 3.07% | 0.00226 | 47.37% |
| ycf1a + ycf1b | IR | 1696 | 124 | 7.31% | 80 | 4.72% | 0.01737 | 89.47% |
| matK | LSC | 734 | 7 | 0.95% | 4 | 0.54% | 0.00197 | 21.05% |
| rbcL | LSC | 602 | 10 | 1.66% | 8 | 1.33% | 0.00456 | 26.32% |
| trnH-psbA | LSC | 876 | 6 | 0.68% | 5 | 0.57% | 0.00172 | 5.26% |
| matK + rbcL + trnH-psbA | LSC | 2212 | 23 | 1.04% | 17 | 0.77% | 0.00256 | 42.11% |
In the single-barcode analysis using distance method, the lowest discriminatory power was found for trnH-psbA (5.26%), followed by rpoC1(15.79%) and matK (21.05%), while ycf1a (52.63%) provided the highest discrimination rate. Combining matK + rbcL + trnH-psbA, the discrimination rate was still relatively low (42.11%). According to the single barcode discrimination power, the combination of ycf1a + ycf1b presented a higher discrimination rate (89.47%). The tree based method had the same results (Fig. 4 and Supplementary Fig. 1).
Figure 4.

NJ tree for Paris using the rbcL + matK + trnH-psbA (A) and ycf1a + ycf1b (B) DNA barcode combination. NJ topology shown with NJ/MP/ML bootstrap support values were listed at each node.
Discussion
Chloroplast Genome of Paris
Recently, more and more taxonomists have focused on chloroplast genome to investigate phylogeny relationship of related species. For example, the chloroplast genome of three species of Veroniceae20 and four species of Tila 21 were used for plant phylogenetic analysis. In this study, the complete plastid genome sequences of three Paris species were compared and the results showed that the gene structures, contents and arrangement were conserved. The size of P. thibetica, P. rugosa and P. polyphylla var. yunnanensis chloroplast genome ranged from 162,708 to 163,200 bp, nevertheless, the three Paris species had the same protein-coding genes, tRNAs and rRNAs. The length variations among Paris chloroplast genomes may result from the length of spacer and intron.
Compared with other Melanthiaceae chloroplast genomes, IR regions extended into rps15 gene in Paris and genome size is ~7 kb longer than Trillium 22. The IR/SC junction position changes may be caused by contraction or expansion of IR region, which is a common evolutionary phenomenon in plants23.
Larger and more complex repeat sequences may play an important role in the rearrangement of chloroplast genomes and sequence divergence23. In the three Paris chloroplast genomes, we found numerous repeated sequences particularly in the intergenic spacer regions and the length of repeated sequences ranged from 30 to 284 bp, similar to those reported in other angiosperm linages24, 25.
Previously, SSRs have been described as a major tool to unravel genome polymorphism across species, ecological and evolutionary studies4, 26. In three Paris chloroplast genomes, the most abundant SSR pattern was found to be stretches of mononucleotides (A/T) (Fig. 2A). More interestingly, the cpSSRs were only observed in the non-coding region27, 28. Because the chloroplast genome sequences are highly conserved among Paris, microsatellite sites for chloroplast genomes are transferable across species. The cpSSRs of three Paris species in our study are expected to be useful for the analysis of genetic diversity in Paris.
DNA barcode for Paris
DNA barcoding has been largely used as a new biological tool to facilitate accurate species identification29. The ideal DNA barcode would be a single locus that could be universally amplified and sequenced for a broad range of taxa, be easily aligned over large phylogenetic distances, and provide sufficient variation to reliably distinguish closely related species30. Unfortunately, the candidate barcodes matK and rbcL, as a “core” plant barcode, often have limited resolutions at species level. In this study, combining matK, rbcL and trnH-psbA only less than half of samples were successfully identified (Table 2). Therefore, searching for an effective barcode with high evolutionary rates is very important for specific group, such as Paris.
Chloroplast genome is endemic to plants. Therefore, chloroplast DNA barcodes are of primary choices. The “hotspot” regions which cluster more SNP and indel mutations create the highly variable regions in the chloroplast genome. In this study, we identified eight highly variable barcode including trnS-trnG, rpoC1, psbC-trnS-psbZ, ycf2, ycf1a, trnN-ycf1, ycf1b, rpl32-trnL (Fig. 3). The coding gene ycf1, trnS-trnG, rpoC1 and rpl32-trnL were the focus in previous studies to investigate sequence variation and phylogenetic analysis in angiosperms4, 31, 32.
The poor performance of three commonly used barcodes rbcL, trnH-psbA, and matK in resolving Paris species indicates that additional barcodes should be exploited for this complex group. The ycf1a and ycf1b regions can be used as a starting point to identify Paris and relative species because they are certainly the most promising sequences to accomplish DNA barcode objectives in closely related species up to now. ycf1 encodes a protein of approximately 1,800 amino acids, as the second largest gene in the chloroplast genome33. Because ycf1 is too long and too variable to permit the design of universal primers31, it has received little attention for DNA barcoding at low taxonomy, but ycf1, especially ycf1a and ycf1b may be the best barcodes at present as specific barcodes for Paris (Fig. 4 and Table 2).
The chloroplast genomes provide sufficient genetic information for species identification. In this study, we identified variable markers in the chloroplast genome for accurate Paris species identification and developed SSRs for further evolutionary studies. Such strategy to invent species-specific molecular markers was an effective approach that it will increase the efficiency and feasibility of species identification and population-based studies of Paris considering the characteristics of the chloroplast genomes.
Materials and Methods
Chloroplast Genome Sequencing
Fresh leaves were collected from Lushui, Yunnan province in South China and were identified based on morphology. Total genomic DNA was isolated from fresh leaves using the DNeasy Plant MiniKit (Qiagen, CA, USA). DNA and voucher specimens of sampled species were deposited in the herbarium of Chinese Academy of Inspection and Quarantine. DNA was sheared by nebulization with compressed nitrogen gas, yielding fragments of 500 bp in length. Paired-end libraries were prepared with the Mate Pair Library Preparation Kit (Illumina, San Diego, California, USA) in accordance with the manufacturer’s instructions. Whole genome sequences were executed using Illumina Hiseq 4000 Genome Analyzer.
Chloroplast Genome Assemblage and Annotation
For both two species, the high-throughput sequencing data were quality-controlled and assembled using SPAdes 3.6.134. The assembled sequences of the chloroplast genome were selected using the Blast program35. The contigs of the chloroplast genome were assembled using Sequencher 4.10 with default parameters and the gaps between contigs were linked by amplification with PCR-based conventional Sanger sequencing using ABI 3730. The specific primers were designed based on the flanking sequences to bridge the gaps. After that, all reads were mapped to the assembled chloroplast genome sequence using Geneious 8.136 to avoid assembly errors and proofread the contig is correct. Finally, we obtained two Paris high quality complete chloroplasts genome sequences. The assembled genomes were annotated using the Dual Organellar Genome Annotator (DOGMA)37. The circle maps of the two species were drawn using GenomeVx38.
Repeat Sequence Analysis
Perl script MISA (MIcroSAtellite identification tool, http://pgrc.ipk-gatersleben.de/misa/) was used to search for simple sequence repeat (SSRs or microsatellites) loci in the chloroplast genomes. Tandem repeats of 1–6 nucleotides were considered as microsatellites. The minimum numbers of repeats were set to 10, 6, 5, 5, 5 and 5 for mono-, di-, tri-, tetra-, penta-, and hexa-nucleotides, respectively. REPuter was used to find tandem, dispersed, and palindromic repeats, with a minimum repeat size of 30 bp and a sequence identity greater than 90%39.
Divergent Hotspots Identification
The three completed chloroplast genome sequences (P. polyphylla var. yunnanensis, P. rugose, P. thibetica) were aligned using MAFFT40 and were manually adjusted using Se-Al 2.041. To analyze nucleotide diversity (Pi), we conducted a sliding window analysis using DnaSP version 5 software42. The window length was set to 600 base pairs and the step size was set as 200 base pairs.
Highly Variable Barcode Acquisition
We collected 6 Paris species to test the barcodes designed in this study (Supplementary Table S1). The primers for amplifying the highly variable regions were designed using FastPCR (Supplementary Table S2). The primers for amplifying and sequencing the control markers of rbcL, matK and trnH-psbA were the same as previous studies33. The same DNA sequences of another 11 Paris species were downloaded from GenBank43.
The PCR amplifications were performed in a final volume of 25 μL containing 1× PCR buffer (with Mg2+), 0.25 mmol/L each dNTP, 0.25 μmol/L each primer, 1.25 U Taq polymerase, and 20–30 ng DNA. The PCR program started at 94 °C for 4 min, followed by 34 cycles of 30 s at 94 °C, 40 s at 52 °C, and 1 min at 72 °C, and ended with a final extension of 10 min at 72 °C. Both of the strands were sequenced on ABI Prism 3730xl (Applied Biosystems, Foster City, U.S.A.) following the manufacturer’s protocols.
DNA Barcoding Analysis
We evaluated the hypervariable barcodes and compared with the chloroplast genes rbcL, matK and trnH-psbA using two different methods. Firstly, the distance method was applied via the function nearNeighbour of SPIDER44. Species discrimination was considered successful if the closest K2P distance for all of the individuals of a given species belonged to only one conspecific individual. Secondly, a tree-based method was used to assess whether sequences in our data sets form species specific clusters. Neighbour-joining (NJ) trees were constructed for each individual barcode and their combinations by MEGA 6 based on a K2P distance45. Maximum likelihood (ML) analyses were performed using RAxML 8.0 with the GTR+G model46. Maximum parsimony (MP) trees were analyzed with PAUP* v4b10 program47. Relative support for the branches of the NJ, ML and MP trees were assessed via 1000 bootstrap replicates.
Electronic supplementary material
Acknowledgements
This work was supported by grants from the Specialized Funds for Inspection and Quarantine Scientific Research on Germplasm Resources from General Administration of Quality Supervision, Inspection and Quarantine of the People’s Republic of China (AQSIQ), the Basic Scientific Research Foundation of the Chinese Academy of Inspection and Quarantine (2016JK011).
Author Contributions
Y.S., S.-J.W. and N.-Z.C. designed the experiment, drafted and made revisions to the manuscript; Y.-M.D. collected samples and performed the experiment; Y.S. and J.X. analyzed the data. M.-F.L. and S.-F.Z. contributed reagents and analysis tools. All of the authors have read and approved the final manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-02083-7
Accession Codes: P. rugosa, P. thibetica and P. polyphylla var. yunnanensis chloroplast genome are available in GenBank database (accession number: KY247142, KY247143, KT805945). The Accession no. of other sequences are from KY851328 to KY851377 (Table S1).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Jansen RK, et al. Methods for obtaining and analyzing whole chloroplast genome sequences. Methods Enzymol. 2005;395:348–84. doi: 10.1016/S0076-6879(05)95020-9. [DOI] [PubMed] [Google Scholar]
- 2.Bendich AJ. Circular chloroplast chromosomes: the grand illusion. Plant Cell. 2004;16(7):1661–1666. doi: 10.1105/tpc.160771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dong W, et al. A chloroplast genomic strategy for designing taxon specific DNA mini-barcodes: A case study on ginsengs. BMC Genetics. 2014;15(1):138. doi: 10.1186/s12863-014-0138-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu C, et al. Comparative Analysis of Six Lagerstroemia Complete Chloroplast Genomes. Front Plant Sci. 2017;8(15):15. doi: 10.3389/fpls.2017.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jansen RK, et al. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci USA. 2007;104(49):19369–19374. doi: 10.1073/pnas.0709121104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang YJ, Ma PF, Li DZ. High-throughput sequencing of six bamboo chloroplast genomes: phylogenetic implications for temperate woody bamboos (Poaceae: Bambusoideae) PLOS ONE. 2011;6(5):e20596. doi: 10.1371/journal.pone.0020596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Awasthi P, Ahmad I, Gandhi SG, Bedi YS. Development of chloroplast microsatellite markers for phylogenetic analysis in Brassicaceae. Acta Biol Hung. 2012;63(4):463–473. doi: 10.1556/ABiol.63.2012.4.5. [DOI] [PubMed] [Google Scholar]
- 8.Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008;26(10):1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 9.Bremer B, et al. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Bot. J. Linn. Soc. 2009;161:105–121. doi: 10.1111/j.1095-8339.2009.00996.x. [DOI] [Google Scholar]
- 10.Zomlefer WB, Judd WS, Whitten WM, Williams NH. A synopsis of Melanthiaceae (Liliales) with focus on character evolution in tribe Melanthieae. Aliso. 2006;22:566–578. [Google Scholar]
- 11.Long CL, et al. Strategies for agrobiodiversity conservation and promotion: a case from Yunnan, China. Biodiversity & Conservation. 2003;12(6):1145–1156. doi: 10.1023/A:1023085922265. [DOI] [Google Scholar]
- 12.Ji Y, Fritsch PW, Li H, Xiao T, Zhou Z. Phylogeny and classification of Paris (Melanthiaceae) inferred from DNA sequence data. Annals of botany. 2006;98(1):245–256. doi: 10.1093/aob/mcl095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.China Plant BOLG et al. Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proc Nat Acad Sci USA108(49), 19641–19646 (2011). [DOI] [PMC free article] [PubMed]
- 14.Kato H, Terauchi R, Utech FH, Kawano S. Molecular systematics of the Trilliaceae sensu lato as inferred from rbcL sequence data. Mol phylogen evol. 1995;4(2):184–193. doi: 10.1006/mpev.1995.1018. [DOI] [PubMed] [Google Scholar]
- 15.Osaloo SK, Kawano S. Molecular systematics of Trilliaceae II. Phylogenetic analyses of Trillium and its allies using sequences of rbcL and matK genes of cpDNA and internal transcribed spacers of 18S–26S nrDNA. Plant Species Biology. 1999;14(1):75–94. doi: 10.1046/j.1442-1984.1999.00009.x. [DOI] [Google Scholar]
- 16.Zhu YJ, Chen SL, Yao H, Tan R. DNA barcoding the medicinal plants of the genus. Paris. Acta pharmaceutica Sinica. 2010;45(3):376–382. [PubMed] [Google Scholar]
- 17.Li XJ, Yang ZY, Huang YL, Ji YH. Complete Chloroplast Genome of the Medicinal Plant Paris polyphylla var. chinensis (Melanthiaceae) J Trop Subtrop Bot. 2015;23(6):601–613. [Google Scholar]
- 18.Kim JS, Kim JH. Comparative Genome Analysis and Phylogenetic Relationship of Order Liliales Insight from the Complete Plastid Genome Sequences of Two Lilies (Lilium longiflorum and Alstroemeria aurea) PLOS ONE. 2013;8(6):e68180. doi: 10.1371/journal.pone.0068180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Do HD, Kim JS, Kim JH. A trnI_CAU triplication event in the complete chloroplast genome of Paris verticillata M.Bieb. (Melanthiaceae, Liliales) Genome Biol Evol. 2014;6(7):1699–1706. doi: 10.1093/gbe/evu138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi KS, Chung MG, Park S. The complete chloroplast genome sequences of three Veroniceae species (Plantaginaceae): comparative analysis and highly divergent regions. Front plant sci. 2016;7:355. doi: 10.3389/fpls.2016.00355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cai J, Ma P-F, Li H-T, Li D-Z. Complete plastid genome sequencing of four Tilia species (Malvaceae): a comparative analysis and phylogenetic implications. PLoS One. 2015;10(11):e0142705. doi: 10.1371/journal.pone.0142705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim SC, Kim JS, Kim JH. Insight into infrageneric circumscription through complete chloroplast genome sequences of two Trillium species. Aob Plants. 2016;8:plw015. doi: 10.1093/aobpla/plw015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dong W, et al. Comparative analysis of the complete chloroplast genome sequences in psammophytic Haloxylon species (Amaranthaceae) PeerJ. 2016;4:e2699. doi: 10.7717/peerj.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greiner S, et al. The complete nucleotide sequences of the five genetically distinct plastid genomes of Oenothera, subsection Oenothera: I. Sequence evaluation and plastome evolution. Nucleic Acids Res. 2008;36(7):2366–2378. doi: 10.1093/nar/gkn081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng W, Chen J, Hao Z, Shi J. Comparative Analysis of the Chloroplast Genomic Information of Cunninghamia lanceolata (Lamb.) Hook with Sibling Species from the Genera Cryptomeria D. Don, Taiwania Hayata, and Calocedrus Kurz. Int J Mol Sci. 2016;17(7):1084. doi: 10.3390/ijms17071084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He Y, et al. The Complete Chloroplast Genome Sequences of the Medicinal Plant Pogostemon cablin. Int J Mol Sci. 2016;17(6):820. doi: 10.3390/ijms17060820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raveendar S, et al. The complete chloroplast genome of Capsicum annuum var. glabriusculum using Illumina sequencing. Molecules. 2015;20(7):13080–13088. doi: 10.3390/molecules200713080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gichira AW, et al. The complete chloroplast genome sequence of an endemic monotypic genus Hagenia (Rosaceae): structural comparative analysis, gene content and microsatellite detection. PeerJ. 2017;5:e2846. doi: 10.7717/peerj.2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hebert PDN, Cywinska A, Ball SL, DeWaard JR. Biological identifications through DNA barcodes. Proc Biol sci. 2003;270(1512):313–321. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clement WL, Donoghue MJ. Barcoding success as a function of phylogenetic relatedness in Viburnum, a clade of woody angiosperms. BMC Evol Biol. 2012;12(1):73. doi: 10.1186/1471-2148-12-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dong W, Liu J, Yu J, Wang L, Zhou S. Highly variable chloroplast markers for evaluating plant phylogeny at low taxonomic levels and for DNA barcoding. PLOS ONE. 2012;7(4):e35071. doi: 10.1371/journal.pone.0035071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Särkinen T, George M. Predicting plastid marker variation: can complete plastid genomes from closely related species help? PLoS One. 2013;8(11):e82266. doi: 10.1371/journal.pone.0082266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dong W, et al. ycf1, the most promising plastid DNA barcode of land plants. Sci rep. 2015;5:8348. doi: 10.1038/srep08348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bankevich A, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–77. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brozynska M, Furtado A, Henry RJ. Direct chloroplast sequencing: comparison of sequencing platforms and analysis tools for whole chloroplast barcoding. PLoS One. 2014;9(10):e110387. doi: 10.1371/journal.pone.0110387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kearse M, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28(12):1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wyman SK, Jansen RK, Boore JL. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2004;20(17):3252–3255. doi: 10.1093/bioinformatics/bth352. [DOI] [PubMed] [Google Scholar]
- 38.Conant GC, Wolfe KH. GenomeVx: simple web-based creation of editable circular chromosome maps. Bioinformatics. 2008;24(6):861–862. doi: 10.1093/bioinformatics/btm598. [DOI] [PubMed] [Google Scholar]
- 39.Kurtz S, et al. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001;29(22):4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rambaut, A. Sequence alignment editor. Version 2.0. Department of Zoology, University of Oxford: Oxford (2002).
- 42.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25(11):1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 43.Huang, Y. et al. Analysis of Complete Chloroplast Genome Sequences Improves Phylogenetic Resolution in Paris (Melanthiaceae). Frontiers in Plant Science 7, doi:10.3389/fpls.2016.01797 (2016). [DOI] [PMC free article] [PubMed]
- 44.Brown SD, et al. Spider: an R package for the analysis of species identity and evolution, with particular reference to DNA barcoding. Mol Ecol Resour. 2012;12(3):562–565. doi: 10.1111/j.1755-0998.2011.03108.x. [DOI] [PubMed] [Google Scholar]
- 45.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22(21):2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 47.Swofford, D. L. PAUP*. Phylogenetic analysis using parsimony (* and other methods). Version 4 (2003).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


