Summary
Pluripotent stem cells have been proposed as an unlimited source of pancreatic β cells for studying and treating diabetes. However, the long, multi-step differentiation protocols used to generate functional β cells inevitably exhibit considerable variability, particularly when applied to pluripotent cells from diverse genetic backgrounds. We have developed culture conditions that support long-term self-renewal of human multipotent pancreatic progenitors, which are developmentally more proximal to the specialized cells of the adult pancreas. These cultured pancreatic progenitor (cPP) cells express key pancreatic transcription factors, including PDX1 and SOX9, and exhibit transcriptomes closely related to their in vivo counterparts. Upon exposure to differentiation cues, cPP cells give rise to pancreatic endocrine, acinar, and ductal lineages, indicating multilineage potency. Furthermore, cPP cells generate insulin+ β-like cells in vitro and in vivo, suggesting that they offer a convenient alternative to pluripotent cells as a source of adult cell types for modeling pancreatic development and diabetes.
Keywords: pancreatic progenitors, culture conditions, pancreatic development, tissue stem cells, self-renewal, directed differentiation, β cell differentiation
Highlights
-
•
Culture on 3T3 cells enables long-term self-renewal of human pancreatic progenitors
-
•
Proliferation requires EGF, FGF10, retinoic acid, and inhibition of Notch and TGF-β
-
•
Cultured progenitors upregulate genes required for mitosis and telomere maintenance
-
•
Pancreatic duct and β-like cells are generated in vitro and in vivo
In this article, Trott and colleagues describe conditions that enable long-term self-renewal of pancreatic progenitors derived from human pluripotent stem cells. These cultured pancreatic progenitors can be expanded for at least 20 passages and are capable of differentiation into multiple pancreatic lineages, including β-like cells, both in vitro and in vivo.
Introduction
The adult pancreas comprises three major lineages: endocrine, acinar, and ductal (Pan and Wright, 2011). The endocrine compartment resides in the islets of Langerhans and consists of cells that secrete hormones required for the maintenance of euglycemia, including α cells that secrete glucagon and β cells that secrete insulin and whose failure leads to diabetes. Acinar cells produce digestive enzymes and, together with duct cells, form the exocrine pancreas. Development of the human pancreas begins with the emergence of the dorsal and ventral pancreatic buds from the posterior foregut at Carnegie stage (CS) 12 (Jennings et al., 2015, Shih et al., 2013). These rudimentary structures consist of multipotent pancreatic progenitors that proliferate extensively and undergo branching morphogenesis before fusing to form the pancreatic anlage. Each of the three major pancreatic lineages is derived from these progenitor cells following a series of cell-fate decisions and morphological changes.
In the developing foregut, transcription factor expression patterns demarcate regions that give rise to specific organs, such as the pancreas and liver. In the mouse, the transcription factor PDX1 is expressed in the emerging pancreatic buds and the neighboring antral stomach and rostral intestine (McCracken et al., 2014), and is absolutely required for pancreatic development (Jonsson et al., 1994, Offield et al., 1996). Expression of SOX9, which marks proliferative cells in a variety of tissues, distinguishes PDX1+ cells that will form the pancreas from those that give rise to other tissues (Shih et al., 2015). Expression of NKX6-1 follows that of PDX1 and SOX9 and, in humans, is required prior to transient activation of NGN3 for the generation of mature, functional β cells (Nostro et al., 2015, Russ et al., 2015). Finally, RFX6, FOXA2, and members of the GATA and HNF transcription factor families are expressed dynamically following specification of the definitive endoderm and throughout development of the pancreas (Conrad et al., 2014). Immunohistochemistry of human embryos at sequential stages during early pancreatic development suggests that the tissue specificity of these transcription factors is similar between mice and humans, although there appear to be differences in when they are expressed (Jennings et al., 2013, Jennings et al., 2015).
A series of genetic studies in mice led to the identification of numerous signaling pathways that regulate pancreatic development (Shih et al., 2013), thereby inspiring the development of protocols for the generation of pancreatic progenitors (Kroon et al., 2008) and subsequently β-like cells from human pluripotent stem cells (Pagliuca et al., 2014, Rezania et al., 2014, Russ et al., 2015). The ultimate goal of these studies is to generate functional β cells capable of maintaining euglycemia and alleviating diabetes. However, these protocols are technically challenging and expensive to conduct, often resulting in low differentiation efficiencies, partly due to the variability inherent in long, multi-step differentiation protocols that seek to recapitulate the entire developmental history of a β cell. These issues are exacerbated when such protocols are applied to genetically diverse human embryonic stem cells (hESCs) and induced pluripotent stem cells (hiPSCs). A potential solution is to initiate differentiation from an alternative cell type that is developmentally more proximal to the β cell. The obvious candidate is the PDX1+SOX9+ progenitor cell population of the emerging pancreatic bud that is capable of extensive proliferation and gives rise to all of the mature functional cells of the pancreas.
We describe a cell culture platform that enables hESC- and hiPSC-derived pancreatic progenitors to be captured and expanded in vitro. These cultured pancreatic progenitor (cPP) cells express key pancreatic transcription factors, including PDX1 and SOX9, and are stable for >25 passages. Crucially, cPP cells are closely related to their in vivo counterparts at the transcriptome level and can be differentiated into cells of the endocrine, acinar, and ductal lineages. Therefore, cPP cultures represent a convenient alternative system for studying pancreatic development that circumvents the need to repeatedly generate progenitors from pluripotent cells by directed differentiation. Finally, replacing heterochronic differentiation cultures with comparatively stable and homogeneous cPP cultures will facilitate the development of more robust protocols for the generation of pancreatic cell types from genetically diverse patient-specific hiPSC lines.
Results
Maintenance and Expansion of cPP Cells Derived from hESCs and hiPSCs
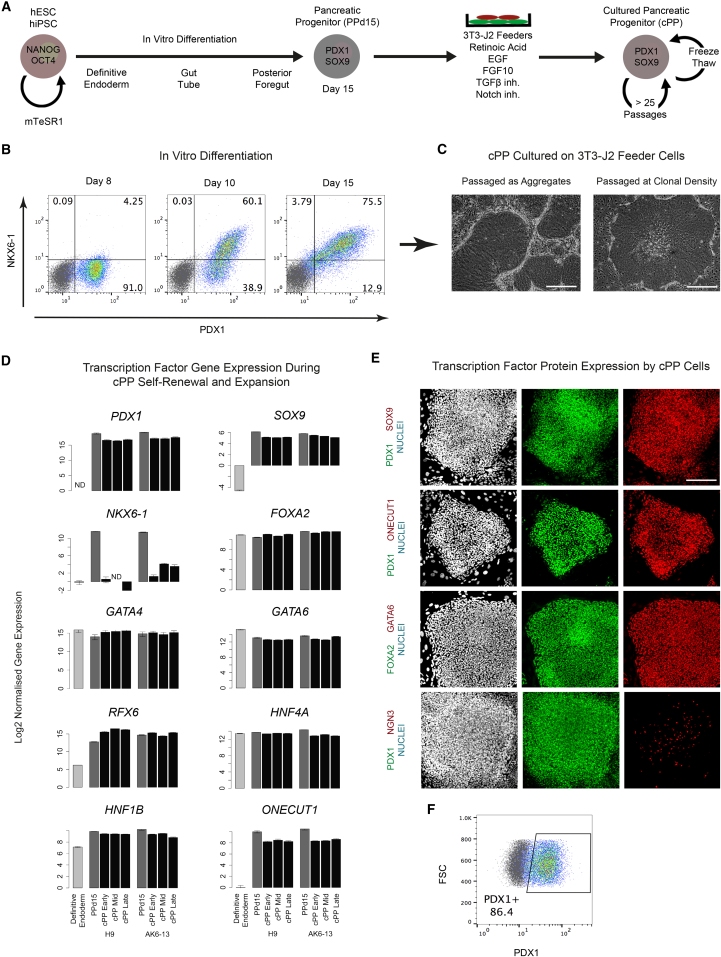
Directed differentiation guided by growth factors and small molecules facilitates the generation of diverse cell types from pluripotent stem cells. We chose to produce pancreatic progenitors from hESCs and hiPSCs (Figure S1) using reagents based on the early stages of a protocol designed to generate mature β cells (Figure S2A; Rezania et al., 2014). This differentiation strategy induced the sequential expression of PDX1 followed by NKX6-1 and yielded a median of 80% PDX1+NKX6-1+ cells after 15 days (PPd15 cells; Figures 1B and S2C). However, as is often observed during directed differentiation from pluripotent cells, the kinetics of PDX1 and NKX6-1 expression varied between cell lines (Figure S2B) (Cahan and Daley, 2013). Therefore, we sought to capture, synchronize, and expand PPd15 cells in culture.
Figure 1.
Derivation of cPP Cell Lines from hESC and hiPSC
(A) Pancreatic progenitors generated after 15 days of differentiation using the STEMdiff directed differentiation kit (PPd15 cells) were plated and expanded on a layer of 3T3-J2 feeder cells in medium supplemented with the indicated growth factors and signaling inhibitors.
(B) Intracellular flow cytometric analysis for PDX1 and NKX6-1 at days 8, 10, and 15 of differentiation using H9 hESCs.
(C) Phase-contrast images of cPP cells passaged as aggregates (left) and as single cells (right). Scale bar, 100 μm.
(D) Gene expression measured by qRT-PCR using samples harvested from PPd15 cells and cPP cells at early (6–8), middle (11–13), and late (14–18) passages. Cells were derived from both AK6-13 hiPSC and H9 hESC. Gene expression in definitive endoderm (H9 hESCs after 4 days STEMdiff differentiation) is shown for comparison. Values are plotted on a log2 scale and error bars represent the SE of three technical replicates. ND, not detected.
(E) Immunofluorescence staining of cPP cells for key pancreatic transcription factors. Scale bar, 100 μm.
(F) Intracellular flow cytometric analysis of cPP cells for PDX1. Gray dots represent control cells stained with isotype control antibodies.
The 3T3-J2 mouse embryonic fibroblast cell line has been used to culture progenitor cells derived from a variety of human tissues, including endoderm-derived intestinal stem cells (Rheinwald and Green, 1975a, Rheinwald and Green, 1975b, Wang et al., 2015). We therefore determined whether pancreatic progenitor cells could be similarly expanded, if provided with appropriate stimuli. We tested a series of signaling agonists and inhibitors previously shown to regulate pancreatic development, including EGFL7, BMP4, nicotinamide, LIF, WNT3A, R-Spondin-1, Forskolin (cAMP agonist), GSK3β inhibition (CHIR99021), and inhibitors of BMP (LDN-193189) and SHH (KAAD-cyclopamine) signaling. Ultimately, a combination of EGF, retinoic acid, and inhibitors of transforming growth factor β (TGF-β, SB431542) and Notch signaling (DAPT) was found to support long-term self-renewal of pancreatic progenitors (Figure 1A). To establish stable cPP cell lines, PPd15 cells were replated on a layer of 3T3-J2 feeder cells in the presence of these factors. Thereafter, cPP cells were routinely passaged once weekly as aggregates at an average split ratio of 1:6, although they were also capable of forming colonies at clonal density (Figure 1C). This suggests a doubling time of ∼65 hr in culture, similar to the 61 hr we routinely observe for hESCs when cultured on a layer of mouse embryonic fibroblasts.
We were able to generate self-renewing cPP cell lines from four different genetic backgrounds using two hESC (H9 and HES3) and three hiPSC cell lines (AK5-11, AK6-8, and AK6-13 derived in house); these diverse cPP cells expressed comparable levels of genes encoding key pancreatic transcription factors, including PDX1 and SOX9 (Figure S2D). Two cPP cell lines selected for further analysis (H9#1 and AK6-13) have been maintained in culture for >20 passages to date enabling >1018-fold expansion over 20 weeks. Crucially, cPP cells can be frozen and thawed with no apparent loss of proliferation or viability, suggesting cPP cells could replace pluripotent cells as a starting point for further differentiation to mature pancreatic cell types such as insulin-secreting β cells.
To determine whether cPP cultures consist of a stable and homogeneous population of cells, we measured the expression of key pancreatic transcription factors at the mRNA and protein levels. Gene expression of numerous markers of pancreatic bud cells, including PDX1 and SOX9, remained constant over extended periods in culture, indicating that our culture conditions maintain a stable population of pancreatic progenitors (Figure 1D). To determine whether cPP cultures represent a homogeneous population, we carried out immunostaining for a selection of pancreatic markers and found these to be expressed near ubiquitously at the protein level (Figure 1E). Furthermore, flow cytometric analysis showed that approximately 85% of cPP cells were PDX1+ (Figure 1F).
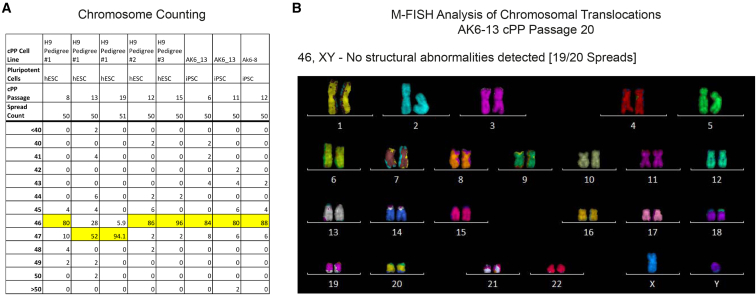
However, NKX6-1 expression was rapidly downregulated in culture, and NKX6-1 protein was not detected by immunostaining. Furthermore, we were able to establish cPP cell lines from day 7, 10, and 15 differentiation cultures (data not shown), the earliest time point being prior to expression of NKX6-1 and suggesting that cPP culture conditions stabilize pancreatic progenitors in a developmental state that precedes NKX6-1 activation. Very few cells were NGN3+, which marks early endocrine progenitors, indicating that differentiation was blocked at the progenitor stage under our culture conditions. Finally, chromosome counting showed that five out of six cPP cells carried 46 chromosomes without signs of structural changes, such as presence of fragments or dicentric chromosomes (Figure 2A). Multiplex fluorescence in situ hydridization (M-FISH) analysis on the AK6-13 line at passage 20 confirmed the absence of karyotypic abnormalities (Figure 2B). Collectively, these data demonstrate that our cPP culture conditions capture pancreatic progenitors as a near homogeneous population that is maintained stably over extended periods of time and is capable of extensive expansion.
Figure 2.
Chromosome Counting and M-FISH Analysis Reveals cPP Cells Are Genetically Stable
(A) Chromosome counting of cPP cells from diverse genetic backgrounds at different passage numbers. Values shown are the percentage of spreads with a given number of chromosomes, with the modal chromosome count for each cPP line highlighted. A modal (shared by >80% of cells) chromosome number of 46 is indicative of a normal karyotype and of karyotypic stability. Five out of six cPP cell lines analyzed exhibited a modal chromosome count of 46 after >6 passages, without evidence of fragments or dicentric chromosomes, and are considered karyotypically stable. In H9 pedigree #1, cells gradually acquired an additional isochromosome upon passaging. Traditional G-band karyotyping (data not shown) subsequently found this to be i(12) (p10)[20], an isochromosome commonly observed in hESC cultures.
(B) Multicolor fluorescence in situ hybridization (M-FISH) enables the detection of chromosomal structural abnormalities at significantly higher resolution than chromosome counting alone. M-FISH of passage 20 AK6-13 cPP cells failed to detect aneuploidy, translocations or deletions in 19/20 spreads analyzed. A representative image of a single chromosome spread is shown.
Transcriptome Analysis Demonstrates cPP Cells Are Closely Related to Their In Vivo Counterparts
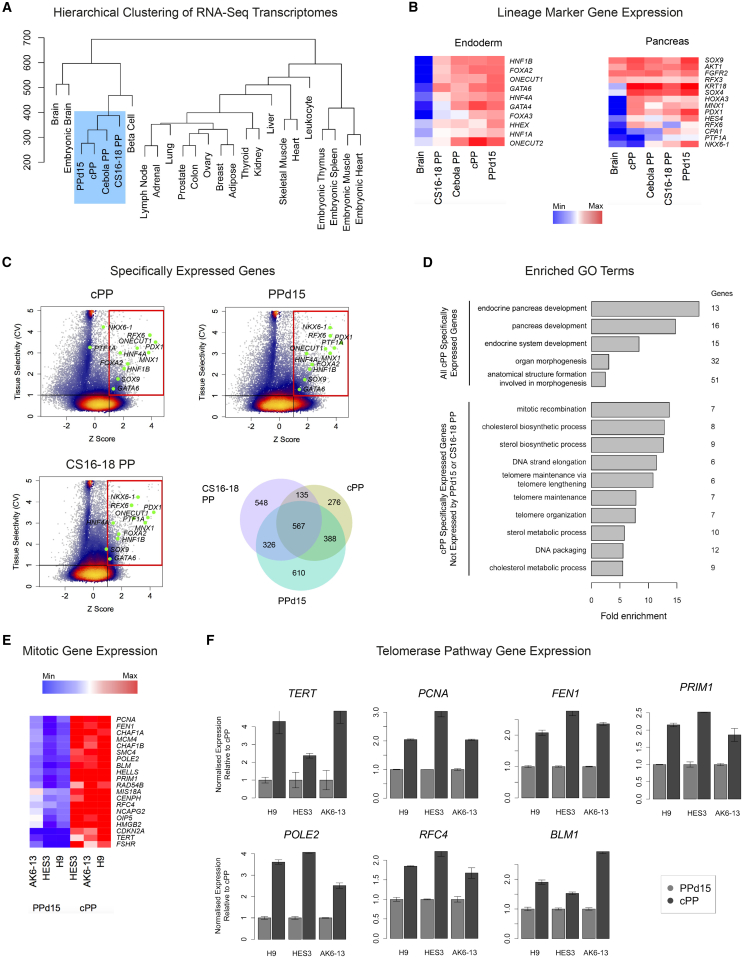
We next determined the transcriptome-wide gene counts by RNA-seq for cPP lines from three different genetic backgrounds and the PPd15 differentiation cultures from which they were established. Samples for RNA-seq were also taken from cPP cells at early, mid, and late passages. Gene expression levels correlated strongly between different cPP samples, indicating that neither genetic background nor time in culture significantly affect the cPP transcriptome (Figure S3A). However, to completely eliminate donor-specific effects on gene expression, the following analysis used mean gene counts for cPP (early passage) and PPd15 cells derived from H9 and HES3 hESCs and AK6-13 hiPSCs.
To determine how similar cPP cells are to their in vitro and in vivo counterparts, we compared the cPP transcriptome with the published transcriptomes of pancreatic progenitors differentiated in vitro (Cebola PP) and from CS16-18 human embryos (CS16-18 PP), as previously described (Cebola et al., 2015), and a diverse collection of adult and embryonic tissues (Bernstein et al., 2010, Petryszak et al., 2013). Relative to non-pancreatic tissues, cPP cells exhibited similar patterns of gene expression to both PPd15 and Cebola PP cells (Figure 3A). Furthermore, cPP, PPd15, and Cebola PP cells closely resembled in vivo pancreatic progenitors at CS16-18, and all four cell populations expressed similar levels of genes associated with endodermal and pancreatic development (Figure 3B). However, as expected, cPP cells do not express the late-stage pancreatic progenitor markers NKX6-1, PTF1A, and CPA1. When taken together, these data demonstrate that the culture conditions described here maintain cPP cells in a developmental state closely related to both the embryonic human pancreas and pancreatic progenitors generated by directed differentiation.
Figure 3.
Transcriptome Analysis of cPP Cells by RNA-Seq
(A) Hierarchical clustering of Euclidean distances between transcriptomes of diverse adult and embryonic tissues shows that in vitro and in vivo pancreatic progenitors exhibit similar patterns of gene expression. Log2-transformed gene count values were used to calculate Euclidian distances. For detailed information on the sources of data used here, see Table S1.
(B) Heatmaps showing log2-transformed gene expression levels of key endodermal and pancreatic markers by in vitro and in vivo pancreatic progenitors. Levels in brain are shown for comparison.
(C) Genes specifically expressed by cPP, PPd15, and CS16-18 pancreatic progenitors. The coefficient of variance (CV) for each protein-coding gene across the 25 tissues shown in (A) was plotted against the corresponding Z score (see Supplemental Experimental Procedures). Specifically expressed genes are located in the upper right-hand quadrant (CV >1 and Z score >1) and include genes with well-characterized roles in early pancreatic development (labeled). The color scale denotes the number of genes. The Venn diagram shows overlap between genes specifically expressed by cPP, PPd15, and CS16-18 pancreatic progenitors.
(D) Biological process Gene Ontology (GO) terms associated with all genes specifically expressed by cPP cells (above) or genes specifically expressed by cPP cells but not PPd15 or CS16-18 pancreatic progenitors (below). Only GO terms associated with >5 genes and/or an adjusted p value <0.01 are shown.
(E) Heatmap of expression levels of genes associated with the enriched GO terms mitotic recombination, DNA strand elongation, telomere maintenance, and DNA packaging. Levels are shown for individual cPP and PPd15 populations derived from three different genetic backgrounds (H9, AK6-13, and HES3) relative to the maximum detected value across the 25 different tissues shown in (A).
(F) Expression of selected telomerase pathway genes as measured by qRT-PCR in cPP and PPd15 cells. Error bars represent the SE of three technical replicates.
To further characterize the transcriptional identity of cPP cells, we sought to identify genes that distinguish them from other lineages. Specifically expressed genes were defined as those that are variably expressed across the aforementioned panel of 25 tissues (coefficient of variance >1) and whose expression is upregulated in cPP cells (Z score >1), as previously described (Cebola et al., 2015). In total 1,366 genes were identified, including numerous well-characterized markers of pancreatic progenitor cells, such as PDX1, SOX9, MNX1, and RFX6 (Figure 3C). To confirm the validity of this method, we demonstrated that these genes are not expressed by other endodermal derivatives, including liver, colon, and lung (Figure S3B). Encouragingly, around 80% of genes specifically expressed by cPP cells were shared with CS16-18 pancreatic progenitors and/or PPd15 cells. Furthermore, gene Z scores were highly correlated between these three pancreatic cell types but not with liver (Figure S3C), further demonstrating the transcriptional similarities between cPP cells and other pancreatic progenitors.
To determine the functional roles of cPP-specific genes, we analyzed associated Gene Ontology (GO) terms. The most enriched terms were those associated with endocrine pancreas development (Figure 3D, above). In order to determine how our culture conditions affect the behavior of cPP cells, we analyzed GO terms associated with genes expressed by cPP cells but not PPd15 or CS16-18 pancreatic progenitor cells (Figure 3D, below). Interestingly, the most enriched terms were those associated with aspects of cell division and telomere maintenance. Indeed, genes associated with these enriched terms, such as those encoding telomerase reverse transcriptase (TERT) and proliferating cell nuclear antigen (PCNA), were consistently upregulated in cPP cells from different genetic backgrounds, compared with the PPd15 populations from which they were derived (Figures 3E and 3F). We conclude that our feeder-based culture system maintains pancreatic progenitors as a stable population while upregulating genes required for long-term self-renewal.
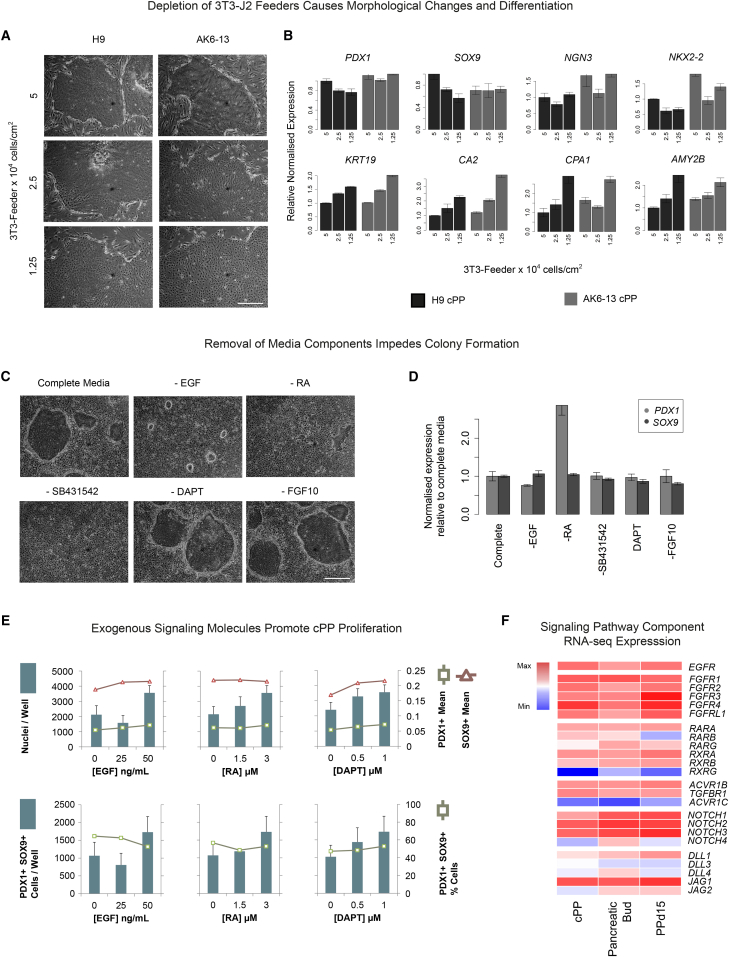
A Feeder Layer of 3T3-J2 Cells Prevents cPP Differentiation while Exogenous Signals Promote Proliferation
We next investigated the roles played by the individual components of our culture system, specifically the layer of irradiated 3T3-J2 feeder cells, stimulation with EGF, FGF10, and retinoic acid (RA), and inhibition of the TGFβ and Notch signaling pathways. To assess the importance of the feeder layer, cPP cells were subcultured onto a layer of 3T3-J2 cells plated at decreasing densities and maintained in complete cPP culture media for 7 days. At reduced feeder densities, cPP cells continued to proliferate rapidly but quickly altered their morphology and could not be serially passaged (Figure 4A). The levels of PDX1 and SOX9 remained stable, indicating cPP cells are committed to the pancreatic lineage, while markers of duct (KRT19 and CA2) and acinar (CPA1 and AMY2B) differentiation were upregulated (Figure 4B). However, we did not observe upregulation of endocrine markers (NGN3 and NKX2-2), suggesting that 3T3-J2 feeder cells are required to block further differentiation toward the ductal and acinar linages.
Figure 4.
A Layer of 3T3-Feeder Cells and Exogenous Signaling Molecules Are Required for the Maintenance and Expansion of cPP Cells
(A) Phase-contrast images of H9 and AK6-13 cPP cells after 7 days culture in complete medium on 3T3-feeder cells plated at densities of 5 × 104, 2.5 × 104, and 1.25 × 104 cells/cm2. Scale bar, 100 μm.
(B) Gene expression measured by qRT-PCR for samples harvested from cultures in (A) for endocrine (NGN3 and NKX2-2), ductal (KRT19 and CA2), and acinar (CPA1 and AMY2B) marker genes. Error bars represent the SE of three technical replicates.
(C) Phase-contrast images of cPP cells cultured for 6 days in complete medium with individual components omitted. Scale bar, 100 μm.
(D) PDX1 and SOX9 expression measured by qRT-PCR for samples harvested in (C). Error bars represent the SE of three technical replicates.
(E) Microbioreactor array (MBA) screening of factors required to propagate PDX1+SOX9+ cPP cells. Effects of reducing or removing selected factors (EGF, RA, DAPT) from complete medium containing all factors at the following levels: EGF (50 ng/mL), RA (3 μM), DAPT (1 μM), SB431542 (10 μM), and FGF10 (50 ng/mL). Top panels: effects on total nuclei per chamber, and PDX1 and SOX9 mean nuclear intensity. Lower panels: effects on the total number of PDX1+SOX9+ cells per chamber and percentage of PDX1+SOX9+ cells. Data represent the mean of ten chambers within a column treated with the given condition ± the SE.
(F) Heatmap showing RNA-seq expression levels of components of signaling pathways that regulate cPP proliferation: EGF (EGFR), FGF10 (FGFR1-4, 6 and FGFRL2), RA (RARA, RARB, RARG, RXRA, RXRB, and RXRG), SB431542 (ACVR1B [ALK4], TGFBR1 [ALK5], and ACVR1C [ALK7]), and DAPT (NOTCH1-4 and its ligands DLL1,3,4 and JAG1,2). Levels are shown relative to those observed across all 25 tissues shown in Figure 3A.
To establish the roles played by the growth factors and small molecules in our culture media, we removed each individually and assessed the effect on differentiation and proliferation. Exclusion of EGF or RA prevented cPP expansion, while removal of the TGF-β inhibitor SB431542 caused colonies to detach from the feeder layer (Figure 4C). Removal of either FGF10 or the γ-secretase inhibitor DAPT did not significantly affect colony size or morphology in the short term but, when removed from the culture media over multiple passages, led to a noticeable loss of viability. Interestingly, none of the growth factors or signaling inhibitors was individually required for maintenance of PDX1 or SOX9 expression (Figure 4D). Indeed, removal of RA actually increased PDX1 expression. These results suggest that the growth factors and inhibitors present in our culture media are primarily required to drive proliferation of cPP cells rather than maintain their developmental state.
To quantify the effect of exogenous signaling molecules on the maintenance and expansion of cPP cells, we used a microbioreactor array (MBA) screening platform to measure differentiation and proliferation (Titmarsh et al., 2012). Single cPP cells were seeded in Matrigel-coated culture chambers in the absence of feeders and exposed for 3 days to complete cPP culture media in which the levels of EGF, RA, and DAPT were varied (Figure S4). We then used an image-segmentation algorithm to identify individual nuclei and quantify immunofluorescence staining for PDX1 and SOX9, thereby enabling us to determine the percentage of double-positive cells following exposure to different growth factor regimes. Reducing the levels of any of the three factors led to a reduction in both the total number of cells and the number of PDX1+SOX9+ cells (Figure 4E). However, neither the mean levels of PDX1/SOX9 nor the percentage of PDX1+SOX9+ cells were dependent on the levels of these factors, suggesting they act primarily as mitogens. Interestingly, we noticed an increase in the number and percentage of PDX1+SOX9+ cells, but no change in the overall proliferation rate, when cells were exposed to higher concentrations of autocrine signals, particularly when provided with maximal levels of EGF, RA, and DAPT (Figure S4D). Exposure to endogenous soluble signaling molecules is therefore required to maintain PDX1 and SOX9 independently of proliferation.
When taken together, these observations demonstrate that self-renewal of cPP cells is dependent on activation of the EGF, FGF10, and RA pathways and inhibition of Notch signaling. Indeed, cPP cells and their in vitro (PPd15) and in vivo (CS16-18 pancreatic progenitor) equivalents expressed high levels of multiple receptors of EGF, FGF, RA, and Notch signaling, as well as the TGFβ receptors ALK4 and ALK5 (encoded by ACVR1B and TGFBR1, respectively) that are inhibited by SB431542 (Figure 4F). Consistent with our observations, production of FGF10 and RA by the surrounding mesenchyme is essential for expansion of the murine pancreatic bud (Bhushan et al., 2001, Martín et al., 2005, Ye et al., 2005), while EGFR is expressed throughout the pancreas and regulates islet development (Miettinen et al., 2000). Intracellular Notch signaling promotes expansion of pancreatic progenitors and prevents their further differentiation into endocrine cells (Hald et al., 2003, Murtaugh et al., 2003). Therefore, our observation that the γ-secretase inhibitor DAPT promotes proliferation of cPP cells is somewhat surprising. However, FGF10 has been shown to promote Notch activity in the developing pancreatic epithelium (Hart et al., 2003), and cPP cells express intermediate levels of the Notch effector HES1 relative to the 23 tissues described in Figure 3A (data not shown). Therefore, the relatively low concentration of DAPT added to cPP cultures most likely serves to temper Notch activity, and exceptionally high levels of Notch activity might actually suppress proliferation.
Differentiation of cPP Cells into Pancreatic Cell Types In Vitro and In Vivo
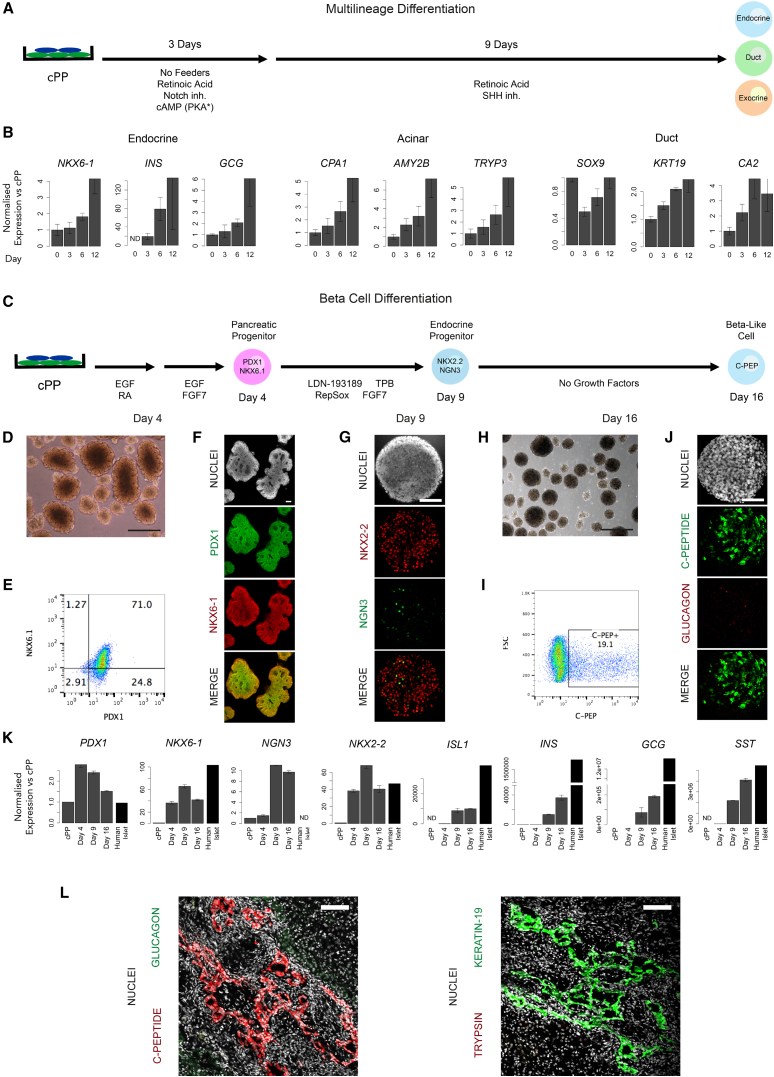
The canonical property of pancreatic progenitors is their ability to differentiate into each of the three lineages that constitute the pancreas as well as their functional derivatives. Initially, we sought to determine whether cPP cells are capable of commitment to the endocrine, duct, and acinar lineages in vitro. Since robust protocols for the directed differentiation of pancreatic duct and acinar cells have yet to be developed, cPP cells were replated in the absence of feeders and exposed to a minimal signaling regime that promotes multilineage differentiation (Figure 5A). Over the course of 12 days, we observed upregulation of endocrine (NKX6-1, INS, and GCG), acinar (CPA1, AMY2B, and TRYP3), and duct (SOX9, KRT19, and CA2) markers, demonstrating that cPP cells retain multilineage potency in vitro (Figure 5B).
Figure 5.
Testing cPP Potency In Vitro and In Vivo
(A) Feeder-depleted passage 15 H9 cPP cells were replated on Matrigel and exposed to the indicated factors that promote multilineage differentiation toward the endocrine, duct, and acinar lineages.
(B) Endocrine, exocrine, and ductal gene expression analysis in (A) after 3, 6, and 12 days. Values are shown relative to levels in undifferentiated cPP cells (day 0). Error bars represent the SE of three technical replicates.
(C) Directed differentiation of passage 10 AK6-13 cPP cells to insulin+ β-like cells using a modified version of Russ et al. (2015).
(D) Phase-contrast image of differentiating spheres undergoing branching morphogenesis after 4 days. Scale bar, 100 μm.
(E) Intracellular flow cytometric analysis of day 4 cells shows approximately 70% reactivate NKX6-1 and maintain PDX1.
(F) PDX1 and NKX6-1 immunostaining on day 4. Scale bar, 100 μm.
(G) On day 9, the majority of cells are NKX2-2+ with a proportion of these transiently NGN3+. Scale bar, 100 μm.
(H) Phase contrast image of day 16 spheres. Scale bar, 100 μm.
(I) Approximately 20% of cells are C-peptide+ on day 16.
(J) Day 16 C-peptide+ cells do not coexpress glucagon. Scale bar, 100 μm.
(K) Gene expression measured by qRT-PCR of cPP cells on days 4, 9, and 16 harvested from the differentiation protocol in (C). Levels are shown relative to those in undifferentiated cPP cells and human islets for comparison. Error bars represent the SE of three technical replicates.
(L) Immunostaining of transplanted cPP cells for markers of endocrine (C-peptide and glucagon), duct (keratin-19), and acinar (trypsin) lineages. Scale bar, 100 μm.
Of particular interest is the ability to generate β-like cells capable of secreting insulin in response to elevated glucose levels. Several groups recently published protocols that describe the differentiation of particular hESC and hiPSC cell lines into β-like cells. Activation of NKX6-1 prior to expression of NGN3 is thought to be essential for the formation of mature, functional β cells (Nostro et al., 2015, Russ et al., 2015). Therefore, we selected the four most promising protocols and assessed their ability to induce NKX6-1 expression while maintaining low levels of NGN3 (Pagliuca et al., 2014, Rezania et al., 2014, Russ et al., 2015, Zhang et al., 2009). Specifically, cPP cells were cultured as monolayers or aggregates, then exposed to the section of each differentiation protocol shown to induce NKX6-1 expression (Figure S5A). The protocol described by Russ et al. (2015) produced the highest levels of NKX6-1 expression and minimal activation of NGN3, with monolayer and suspension cultures yielding a very similar response (Figure S5B). Since the original protocol demonstrated the generation of insulin-secreting β-like cells when cells were differentiated as aggregates, we chose to use the 3D suspension platform for subsequent experiments.
Using the Russ et al. (2015) protocol, we found that around 40% of cPP cells reactivate NKX6-1. However, doubling the length of each of the first two treatments enabled the generation of nearly 70% double-positive cells, similar to the number originally reported (Figures 5E, S5C, and S5D). Interestingly, these PDX1+NKX6-1+ cells generated convoluted structures reminiscent of the branching morphogenesis of the embryonic pancreas (Figures 5D and 5F). Further differentiation induced expression of the endocrine markers NKX2-2 and NGN3, the latter in a smaller subset of cells, reflecting its transient expression during endocrine commitment (Schwitzgebel et al., 2000; Figure 5G). Finally, after 16 days, 20% of cells contained C-peptide, a proxy for insulin production, similar to the 25% reported by Russ et al. (2015). Crucially, C-peptide+ cells did not co-express the α cell hormone glucagon, suggesting that these cells are unlike the polyhormonal cells produced by earlier generations of protocols, which are unable to secrete insulin in response to elevated glucose levels. However, NGN3 levels remained high at the end of the protocol and INS mRNA levels were significantly lower than in isolated human islets, suggesting that further optimization of the protocol is required (Figure 5K).
The most stringent test of developmental potency is whether a progenitor can differentiate into a particular lineage in vivo. To assess the potency of cPP cells, we injected these cells under the renal capsules of immunodeficient mice and immunostained for markers of the three major pancreatic lineages after >23 weeks. We were able to identify large areas of cells expressing the β-cell marker C-peptide as well as the duct marker keratin 19 (KRT19), but we were unable to find trypsin+ acinar cells or glucagon+ endocrine cells (Figure 5L). However, trypsin+ cells were also observed rarely by Rezania et al. (2014) following transplantation of pancreatic progenitors, possibly because acinar cells cannot survive in the absence of ducts to carry away the digestive enzymes they secrete. The absence of cells expressing glucagon was surprising, but likely reflects generation of C-peptide+ cells by default in the absence of inductive signals required to form glucagon+ α cells.
The C-peptide+ cells did not form classical islet-like structures, but instead formed a series of interconnected cystic structures, as others have observed previously (Rostovskaya et al., 2015). Furthermore, we did not observe expansion of the progenitor population once transplanted, suggesting cPP cells differentiate rapidly into less proliferative cells in vivo. Accordingly, none of the 12 mice we assessed exhibited teratoma formation, despite transplanting >3 million cells into each mouse. These observations demonstrate that cPP cells retain the ability to differentiate into endocrine and duct cells in vivo, although it remains to be seen whether they are capable of forming acinar cells. Furthermore, the absence of teratoma formation suggests cPP cells may represent a safer alternative for transplantations than cells differentiated directly from pluripotent stem cells.
Discussion
Pluripotent stem cells have been proposed as an unlimited source of β cells for modeling and treating diabetes. However, the routine generation of functional β cells from diverse patient-derived hiPSC remains a challenge, partly because of the variability inherent in long, multi-step-directed differentiation protocols. Here, we describe a platform for long-term culture of self-renewing pancreatic progenitor cells derived from human pluripotent stem cells. These cPP cells are capable of rapid and prolonged expansion, thereby offering a convenient alternative source of β cells. Furthermore, cPP cells can be stored and transported as frozen stocks, and to date we have cultured cPP cells for up to 25 passages with no loss of proliferation. We observed that cPP cells express markers of pancreatic endocrine, duct, and acinar cells when differentiated in vitro, thereby demonstrating their multipotency, and we were able to generate up to ∼20% C-peptide+ cells using a modified version of the β cell differentiation protocol described by Russ et al. (2015). The definitive test of developmental potency is whether a cell can differentiate into a particular lineage in vivo, and cPP cells indeed generate significant numbers of keratin-19+ duct cells and C-peptide+ β-like cells when transplanted under the renal capsule of an immunodeficient mouse, although it is unclear whether they retain the ability to form acinar cells in vivo.
Cells differentiating in vitro typically do so in an unsynchronized manner, causing cultures to become progressively more heterochronic with time and reducing the efficiency with which cells can be directed toward particular lineages. Therefore, the ability to capture and synchronize differentiating progenitors is essential for developing robust protocols for generating functional β cells from diverse genetic backgrounds. Extensive molecular characterization revealed that cPP cultures generated from both hESC and hiPSC represent stable populations of cells that express early pancreatic transcription factors consistently over time. The cPP transcriptome is closely related to that of the progenitor cells of the CS16-18 pancreas. However, comparison with human embryos at different stages of development suggests that cPP cells most closely resemble cells of the pancreatic bud between CS12 and CS13, based on robust expression of PDX1, SOX9, FOXA2, and GATA4/6 and the absence of NKX6-1 and SOX17 (Jennings et al., 2013, Jennings et al., 2015).
In recent years, several groups reported methods for culturing human endodermal derivatives. Two separate reports demonstrated that hESC-derived definitive endoderm can be serially passaged and expanded if cultured on a feeder layer in the presence of appropriate mitogenic signals (Cheng et al., 2012, Sneddon et al., 2012). Subsequently, another group showed that foregut progenitor cells can be cultured in feeder-free conditions (Hannan et al., 2013). However, slow growth and variable gene expression between different lines have limited their utility. More recently, it was shown that pancreatic progenitors derived from reprogrammed endodermal cells could be expanded and passaged (Zhu et al., 2015). However, these cultures are highly heterogeneous, and it is not clear whether the minimal combination of signaling molecules and inhibitors used is sufficient to culture cells from different genetic backgrounds. Therefore, the culture system described here is the first to enable long-term self-renewal of multipotent pancreatic progenitors derived from genetically diverse hESC and hiPSC.
Intriguingly, 3T3-J2 feeders have been used to culture diverse cells types, including epidermal keratinocytes (Rheinwald and Green, 1975a, Rheinwald and Green, 1975b), corneal epithelium (Osei-Bempong et al., 2009, Rama et al., 2010), and intestinal stem cells (Wang et al., 2015), among others. Nonetheless, the mechanism(s) by which 3T3-J2 feeders stabilize cultured progenitors are unknown. An obvious candidate is signaling through the WNT/LGR5 pathway, which is required for the maintenance of organoid cultures generated from a variety of endoderm-derived adult tissues, including the pancreas (Barker et al., 2007, Barker et al., 2010, Boj et al., 2015, Huch et al., 2013). However, cPP cells do not express LGR5 and are unaffected by inhibition of endogenous WNT secretion (data not shown). Therefore, identification of the factor(s) produced by 3T3-J2 cells could lead to the discovery of a common signaling axis required to support self-renewal of progenitors from a wide variety of tissues. Finally, it will be interesting to investigate whether adapted versions of the culture system described here are capable of capturing progenitors from other endoderm-derived tissues, such as the liver, stomach, and lung, or progenitors resident in the adult pancreas.
Experimental Procedures
Human Pluripotent Stem Cell Culture and Differentiation
Human pluripotent cell lines were obtained as described in the Supplemental Experimental Procedures. Pluripotent stem cells were maintained on tissue culture plastic coated with Matrigel in mTeSR1 medium as described previously (Ludwig et al., 2006), and differentiated into pancreatic progenitors using the STEMdiff Pancreatic Progenitor kit (STEMCELL Technologies, 05120) according to the manufacturer's instructions with the following modifications: (1) cells were initially seeded into 12-well plates (Corning, 353043) at a density of 106 cells/well, and (2) stage 1 was extended to 3 days by repeating the final day's treatment. All tissue culture was carried out in 5% CO2 at 37°C.
Passaging and Maintenance of cPP Cells
Gentle cell dissociation reagent (STEMCELL Technologies, 07174) was used to passage cPP cells as aggregates that were then seeded at a 1:6 split ratio onto a layer of 3T3-J2 feeders (0.5 × 106 to 1 × 106 cells/cm2) in medium composed of advanced DMEM/F12 (Thermo Fisher Scientific, 21634010), 2 mM L-glutamine (Thermo Fisher Scientific, 25030), 100 U/mL penicillin/streptomycin (Thermo Fisher Scientific, 15140122), 1× N2 supplement (Thermo Fisher Scientific, 17502-048), 1× B27 supplement (Thermo Fisher Scientific, 17504-044), 30 nM dexamethasone (STEMCELL Technologies, 72092), 50 ng/mL EGF (R&D Systems, 236-EG-200), 50 ng/mL FGF10 (Source Bioscience, ABC144), 3 μM RA (Sigma, R2625), 10 μM SB431542 (Calbiochem, 616464), and 1 μM DAPT (Sigma, D5942). If plating single cPP cells, complete medium was supplemented with 10 μM Y27632 for the first 48 hr (Sigma, Y0503). Medium was completely replenished every 2–3 days. See Supplemental Experimental Procedures for details of 3T3-J2 cell culture.
RNA-Seq Analysis of Gene Expression
RNA was isolated from samples harvested from cPP and PPd15 cultures using an RNeasy mini kit (QIAGEN, cat. no. 74104). Feeder removal microbeads (Miltenyi Biotec, 130-095-531) were used to deplete cPP cells of 3T3 feeders prior to RNA extraction. All RNA samples had an RNA integrity number >9. RNA-seq libraries were generated using the NEBNext Ultra RNA Library Prep Kit (NEB, E7530L) and sequenced on an Illumina HiSeq 2500 system generating single-end reads of 100 bp. Table S1 contains metadata for these and public datasets used for the RNA-seq gene expression analysis. Full details of how RNA-seq reads were aligned and analyzed can be found in the Supplemental Experimental Procedures.
Multilineage Differentiation
Monolayer differentiation cultures were established as described in Supplemental Experimental Procedures. Basal differentiation medium consists of advanced DMEM/F12 (Thermo Fisher Scientific, 21634010), 2.5 g/30 mL BSA (Sigma, A9418), 2 mM L-glutamine (Thermo Fisher Scientific, 25030), 100 U/mL penicillin/streptomycin (Thermo Fisher Scientific, 15140122), and 1× B27 supplement (Thermo Fisher Scientific, 17504-044). Supplements were added as follows: days 1–3 (3 μM RA [Sigma, R2625], 1 μM DAPT [Sigma, D5942], 100 μM BNZ [Sigma, B4560]) and days 4, 7, and 10 (3 μM RA, 167 ng/mL KAAD-cyclopamine [Calbiochem, 239807]).
β Cell Differentiation
Differentiation sphere cultures were established as described in Supplemental Experimental Procedures. Basal differentiation medium consists of DMEM high glucose, 2 mM L-glutamine, and 100 U/mL penicillin/streptomycin. Supplements were added as follows: days 1–4 (1× B27 supplement, 50 ng/mL EGF, 1 μM RA [days 1–2 only], 50 ng/mL FGF7 [days 3–4 only]); days 5–10 (1× B27 supplement, 500 nM LDN-193189 [STEMCELL Technologies, 72142], 30 nM TPB [EMD Millipore, 565740], 1 μM RepSox [STEMCELL Technologies, 72392], 25 ng/mL FGF7); and days 11-17 (DMEM low glucose [Thermo Fisher Scientific, 12320-032], 2 mM L-glutamine, 1× MEM non-essential amino acids [Thermo Fisher Scientific, 11140-050]).
Transplantation Assays
cPP cells were grown to confluency to displace and eliminate feeder cells, then treated with gentle cell dissociation reagent to generate single cells. Approximately 3 × 106 to 5 × 106 cells were resuspended in 50 μL of undiluted Matrigel and injected under the kidney capsule of 8- to 12-week-old immunocompromised (NOD/SCID) mice. After 23–27 weeks, transplanted mice were euthanized and their kidneys cryopreserved prior to sectioning and immunostaining. The study protocol was approved by the National University of Singapore Institutional Review Board (NUS IRB 12–181) and Biomedical Research Council IACUC committee (151040).
Author Contributions
Conceptualization and Methodology, J.T. and N.R.D.; Investigation, J.T., E.K.T., S.O., J.W., D.M.T., and M.L.; Formal Analysis, J.T., S.L.I.J.D., M.L., and D.M.T.; Resources, M.S., J.C.W., and B.R.; Writing – Original Draft, J.T. and N.R.D.; Funding Acquisition, N.R.D., J.C.W., L.S., G.R., B.R., and S.C.
Acknowledgments
We thank Dr. Aya Wada and Prof. Huck Hui Ng for advice and guidance on cell culture techniques. We would also like to thank Dr. Michael Riedel, Dr. Jenna Moccia, and Dr. Charis Segeritz-Walko of STEMCELL Technologies for providing differentiation reagents, and Dr. Kim Robinson, Prof. Birgit Lane, and Prof. Yann Barrandon for supplying 3T3-J2 feeder cells. This work was funded by an EDB Singapore Childhood Undiagnosed Diseases Program grant and an A∗STAR Strategic Positioning Fund (SPF) Genetic Orphan Diseases Adopted: Fostering Innovation Therapy (GODAFIT) grant.
Published: June 6, 2017
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, five figures, and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2017.05.019.
Contributor Information
Jamie Trott, Email: jamie.trott@imb.a-star.edu.sg.
N. Ray Dunn, Email: ray.dunn@imb.a-star.edu.sg.
Accession Numbers
Primary RNA-seq datasets generated here are available at ArrayExpress under accession number ArrayExpress: E-MTAB-5731.
Supplemental Information
References
- Barker N., van Es J.H., Kuipers J., Kujala P., van den Born M., Cozijnsen M., Haegebarth A., Korving J., Begthel H., Peters P.J., Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- Barker N., Huch M., Kujala P., van de Wetering M., Snippert H.J., van Es J.H., Sato T., Stange D.E., Begthel H., van den Born M. LGR5+ve stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell. 2010;6:25–36. doi: 10.1016/j.stem.2009.11.013. [DOI] [PubMed] [Google Scholar]
- Bernstein B.E., Stamatoyannopoulos J.A., Costello J.F., Ren B., Milosavljevic A., Meissner A., Kellis M., Marra M.A., Beaudet A.L., Ecker J.R. The NIH roadmap epigenomics mapping consortium. Nat. Biotechnol. 2010;28:1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhushan A., Itoh N., Kato S., Thiery J.P., Czernichow P., Bellusci S., Scharfmann R. Fgf10 is essential for maintaining the proliferative capacity of epithelial progenitor cells during early pancreatic organogenesis. Development. 2001;128:5109–5117. doi: 10.1242/dev.128.24.5109. [DOI] [PubMed] [Google Scholar]
- Boj S.F., Hwang C.I., Baker L.A., Chio I.I.C., Engle D.D., Corbo V., Jager M., Ponz-Sarvise M., Tiriac H., Spector M.S. Organoid models of human and mouse ductal pancreatic cancer. Cell. 2015;160:324–338. doi: 10.1016/j.cell.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahan P., Daley G.Q. Origins and implications of pluripotent stem cell variability and heterogeneity. Nat. Rev. Mol. Cell Biol. 2013;14:357–368. doi: 10.1038/nrm3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebola I., Rodríguez-Seguí S.A., Cho C.H.H., Bessa J., Rovira M., Luengo M., Chhatriwala M., Berry A., Ponsa-Cobas J., Maestro M.A. TEAD and YAP regulate the enhancer network of human embryonic pancreatic progenitors. Nat. Cell Biol. 2015;17:615–626. doi: 10.1038/ncb3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X., Ying L., Lu L., Galvão A.M., Mills J.A., Lin H.C., Kotton D.N., Shen S.S., Nostro M.C., Choi J.K. Self-renewing endodermal progenitor lines generated from human pluripotent stem cells. Cell Stem Cell. 2012;10:371–384. doi: 10.1016/j.stem.2012.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad E., Stein R., Hunter C.S. Revealing transcription factors during human pancreatic β cell development. Trends Endocrinol. Metab. 2014;25:407–414. doi: 10.1016/j.tem.2014.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hald J., Hjorth J.P., German M.S., Madsen O.D., Serup P., Jensen J. Activated Notch1 prevents differentiation of pancreatic acinar cells and attenuate endocrine development. Dev. Biol. 2003;260:426–437. doi: 10.1016/s0012-1606(03)00326-9. [DOI] [PubMed] [Google Scholar]
- Hannan N.R.F., Fordham R.P., Syed Y.A., Moignard V., Berry A., Bautista R., Hanley N.A., Jensen K.B., Vallier L. Generation of multipotent foregut stem cells from human pluripotent stem cells. Stem Cell Reports. 2013;1:293–306. doi: 10.1016/j.stemcr.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart A., Papadopoulou S., Edlund H. Fgf10 maintains notch activation, stimulates proliferation, and blocks differentiation of pancreatic epithelial cells. Dev. Dyn. 2003;228:185–193. doi: 10.1002/dvdy.10368. [DOI] [PubMed] [Google Scholar]
- Huch M., Bonfanti P., Boj S.F., Sato T., Loomans C.J.M., van de Wetering M., Sojoodi M., Li V.S.W., Schuijers J., Gracanin A. Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/R-spondin axis. EMBO J. 2013;32:2708–2721. doi: 10.1038/emboj.2013.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings R.E., Berry A.A., Kirkwood-Wilson R., Roberts N.A., Hearn T., Salisbury R.J., Blaylock J., Piper Hanley K., Hanley N.A. Development of the human pancreas from foregut to endocrine commitment. Diabetes. 2013;62:3514–3522. doi: 10.2337/db12-1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings R.E., Berry A.A., Strutt J.P., Gerrard D.T., Hanley N.A. Human pancreas development. Development. 2015;142:3126–3137. doi: 10.1242/dev.120063. [DOI] [PubMed] [Google Scholar]
- Jonsson J., Carlsson L., Edlund T., Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature. 1994;371:606–609. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- Kroon E., Martinson L.A., Kadoya K., Bang A.G., Kelly O.G., Eliazer S., Young H., Richardson M., Smart N.G., Cunningham J. Pancreatic endoderm derived from human embryonic stem cells generates glucose-responsive insulin-secreting cells in vivo. Nat. Biotechnol. 2008;26:443–452. doi: 10.1038/nbt1393. [DOI] [PubMed] [Google Scholar]
- Ludwig T.E., Bergendahl V., Levenstein M.E., Yu J., Probasco M.D., Thomson J.A. Feeder-independent culture of human embryonic stem cells. Nat. Methods. 2006;3:637–646. doi: 10.1038/nmeth902. [DOI] [PubMed] [Google Scholar]
- Martín M., Gallego-Llamas J., Ribes V., Kedinger M., Niederreither K., Chambon P., Dollé P., Gradwohl G. Dorsal pancreas agenesis in retinoic acid-deficient Raldh2 mutant mice. Dev. Biol. 2005;284:399–411. doi: 10.1016/j.ydbio.2005.05.035. [DOI] [PubMed] [Google Scholar]
- McCracken K.W., Catá E.M., Crawford C.M., Sinagoga K.L., Schumacher M., Rockich B.E., Tsai Y.-H., Mayhew C.N., Spence J.R., Zavros Y., Wells J.M. Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature. 2014;516:400–404. doi: 10.1038/nature13863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miettinen P.J., Huotari M., Koivisto T., Ustinov J., Palgi J., Rasilainen S., Lehtonen E., Keski-Oja J., Otonkoski T. Impaired migration and delayed differentiation of pancreatic islet cells in mice lacking EGF-receptors. Development. 2000;127:2617–2627. doi: 10.1242/dev.127.12.2617. [DOI] [PubMed] [Google Scholar]
- Murtaugh L.C., Stanger B.Z., Kwan K.M., Melton D.A. Notch signaling controls multiple steps of pancreatic differentiation. Proc. Natl. Acad. Sci. USA. 2003;100:14920–14925. doi: 10.1073/pnas.2436557100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nostro M.C., Sarangi F., Yang C., Holland A., Elefanty A.G., Stanley E.G., Greiner D.L., Keller G. Efficient generation of NKX6-1+ pancreatic progenitors from multiple human pluripotent stem cell lines. Stem Cell Reports. 2015;4:591–604. doi: 10.1016/j.stemcr.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offield M.F., Jetton T.L., Labosky P.A., Ray M., Stein R.W., Magnuson M.A., Hogan B.L., Wright C.V. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development. 1996;122:983–995. doi: 10.1242/dev.122.3.983. [DOI] [PubMed] [Google Scholar]
- Osei-Bempong C., Henein C., Ahmad S. Culture conditions for primary human limbal epithelial cells. Regen. Med. 2009;4:461–470. doi: 10.2217/rme.09.7. [DOI] [PubMed] [Google Scholar]
- Pagliuca F.W., Millman J.R., Gürtler M., Segel M., Van Dervort A., Ryu J.H., Peterson Q.P., Greiner D., Melton D.A. Generation of functional human pancreatic β cells in vitro. Cell. 2014;159:428–439. doi: 10.1016/j.cell.2014.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan F.C., Wright C. Pancreas organogenesis: from bud to plexus to gland. Dev. Dyn. 2011;240:530–565. doi: 10.1002/dvdy.22584. [DOI] [PubMed] [Google Scholar]
- Petryszak R., Burdett T., Fiorelli B., Fonseca N.A., Gonzalez-Porta M., Hastings E., Huber W., Jupp S., Keays M., Kryvych N. Expression Atlas update—a database of gene and transcript expression from microarray- and sequencing-based functional genomics experiments. Nucleic Acids Res. 2013;42:D926–D932. doi: 10.1093/nar/gkt1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rama P., Matuska S., Paganoni G., Spinelli A., De Luca M., Pellegrini G. Limbal stem-cell therapy and long-term corneal regeneration. N. Engl. J. Med. 2010;363:147–155. doi: 10.1056/NEJMoa0905955. [DOI] [PubMed] [Google Scholar]
- Rezania A., Bruin J.E., Arora P., Rubin A., Batushansky I., Asadi A., O'Dwyer S., Quiskamp N., Mojibian M., Albrecht T. Reversal of diabetes with insulin-producing cells derived in vitro from human pluripotent stem cells. Nat. Biotechnol. 2014;32:1121–1133. doi: 10.1038/nbt.3033. [DOI] [PubMed] [Google Scholar]
- Rheinwald J.G., Green H. Formation of a keratinizing epithelium in culture by a cloned cell line derived from a teratoma. Cell. 1975;6:317–330. doi: 10.1016/0092-8674(75)90183-x. [DOI] [PubMed] [Google Scholar]
- Rheinwald J.G., Green H. Serial cultivation of strains of human epidermal keratinocytes: the formation of keratinizing colonies from single cells. Cell. 1975;6:331–343. doi: 10.1016/s0092-8674(75)80001-8. [DOI] [PubMed] [Google Scholar]
- Rostovskaya M., Bredenkamp N., Smith A. Towards consistent generation of pancreatic lineage progenitors from human pluripotent stem cells. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015;370:20140365. doi: 10.1098/rstb.2014.0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russ H.A., Parent A.V., Ringler J.J., Hennings T.G., Nair G.G., Shveygert M., Guo T., Puri S., Haataja L., Cirulli V. Controlled induction of human pancreatic progenitors produces functional beta-like cells in vitro. EMBO J. 2015;34:1759–1772. doi: 10.15252/embj.201591058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwitzgebel V.M., Scheel D.W., Conners J.R., Kalamaras J., Lee J.E., Anderson D.J., Sussel L., Johnson J.D., German M.S. Expression of neurogenin3 reveals an islet cell precursor population in the pancreas. Development. 2000;127:3533–3542. doi: 10.1242/dev.127.16.3533. [DOI] [PubMed] [Google Scholar]
- Shih H.P., Wang A., Sander M. Pancreas organogenesis: from lineage determination to morphogenesis. Annu. Rev. Cell Dev. Biol. 2013;29:81–105. doi: 10.1146/annurev-cellbio-101512-122405. [DOI] [PubMed] [Google Scholar]
- Shih H.P., Seymour P.A., Patel N.A., Xie R., Wang A., Liu P.P., Yeo G.W., Magnuson M.A., Sander M. A gene regulatory network cooperatively controlled by Pdx1 and Sox9 governs lineage allocation of foregut progenitor cells. Cell Rep. 2015;13:326–336. doi: 10.1016/j.celrep.2015.08.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneddon J.B., Borowiak M., Melton D.A. Self-renewal of embryonic-stem-cell-derived progenitors by organ-matched mesenchyme. Nature. 2012;491:765–768. doi: 10.1038/nature11463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titmarsh D.M., Hudson J.E., Hidalgo A., Elefanty A.G., Stanley E.G., Wolvetang E.J., Cooper-White J.J. Microbioreactor arrays for full factorial screening of exogenous and paracrine factors in human embryonic stem cell differentiation. PLoS One. 2012;7:e52405. doi: 10.1371/journal.pone.0052405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Yamamoto Y., Wilson L.H., Zhang T., Howitt B.E., Farrow M.A., Kern F., Ning G., Hong Y., Khor C.C. Cloning and variation of ground state intestinal stem cells. Nature. 2015;522:173–178. doi: 10.1038/nature14484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye F., Duvilli B., Scharfmann R. Fibroblast growth factors 7 and 10 are expressed in the human embryonic pancreatic mesenchyme and promote the proliferation of embryonic pancreatic epithelial cells. Diabetologia. 2005;48:277–281. doi: 10.1007/s00125-004-1638-6. [DOI] [PubMed] [Google Scholar]
- Zhang D., Jiang W., Liu M., Sui X., Yin X., Chen S., Shi Y., Deng H. Highly efficient differentiation of human ES cells and iPS cells into mature pancreatic insulin-producing cells. Cell Res. 2009;19:429–438. doi: 10.1038/cr.2009.28. [DOI] [PubMed] [Google Scholar]
- Zhu S., Russ H.A., Wang X., Zhang M., Ma T., Xu T., Tang S., Hebrok M., Ding S. Human pancreatic beta-like cells converted from fibroblasts. Nat. Commun. 2015;7:1–13. doi: 10.1038/ncomms10080. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.