Abstract
Gene duplication is a major driver of organismal evolution. Gene retroposition is a mechanism of gene duplication whereby a gene’s transcript is used as a template to generate retroposed gene copies, or retrocopies. Intriguingly, the formation of retrocopies depends upon the enzymatic machinery encoded by retrotransposable elements, genomic parasites occurring in the majority of eukaryotes. Most retrocopies are depleted of the regulatory regions found upstream of their parental genes; therefore, they were initially considered transcriptionally incompetent gene copies, or retropseudogenes. However, examples of functional retrocopies, or retrogenes, have accumulated since the 1980s. Here, we review what we have learned about retrocopies in animals, plants and other eukaryotic organisms, with a particular emphasis on comparative and population genomic analyses complemented with transcriptomic datasets. In addition, these data have provided information about the dynamics of the different “life cycle” stages of retrocopies (i.e., polymorphic retrocopy number variants, fixed retropseudogenes and retrogenes) and have provided key insights into the retroduplication mechanisms, the patterns and evolutionary forces at work during the fixation process and the biological function of retrogenes. Functional genomic and transcriptomic data have also revealed that many retropseudogenes are transcriptionally active and a biological role has been experimentally determined for many. Finally, we have learned that not only non-long terminal repeat retroelements but also long terminal repeat retroelements play a role in the emergence of retrocopies across eukaryotes. This body of work has shown that mRNA-mediated duplication represents a widespread phenomenon that produces an array of new genes that contribute to organismal diversity and adaptation.
Keywords: retrocopy, retrogene, retropseudogene, retroCNV, new functions, testis expression, pollen expression, regulatory element
Introduction
In the past few years, we have witnessed an acceleration of eukaryotic whole genome sequencing projects, along with analyses of gene duplication events (Box 1). These data have strengthened the argument that gene duplication is an important mechanism for new gene origination and adaptation (Kaessmann 2010; Dennis and Eichler 2016; Panchy et al. 2016). In addition, population genomic data have been accumulating for several species, leading to the discovery of gene copy number variants (CNVs; Box 1) that provide additional insights into the gene duplication mechanisms, patterns and the evolutionary forces influencing whether new gene copies are fixed in or eliminated from a population (Schrider et al. 2011, 2013; Richardson et al. 2014; Zarrei et al. 2015; Cardoso-Moreira et al. 2016; Dennis and Eichler 2016; Zhu et al. 2016). One particular mechanism of gene duplication that was initially disregarded as having potential to impact the genomes and their functions is the RNA-mediated gene duplication (i.e., gene duplication involving retrotranscription of an RNA and insertion in the genome; Kaessmann et al. 2009).
Previous reviews on RNA-mediated gene duplications mainly focused on observations made in a few well-studied lineages, primarily mammals and fruit flies (Kaessmann et al. 2009) or in only one group of organisms (Richardson et al. 2014). In this review, we present and discuss a wide range of comparative and population genomic studies and highlight general trends across eukaryotes. These studies have provided information about different stages of the “life cycle” of retrocopies (retroCNVs, retropseudogenes and retrogenes; Box 1) in multiple lineages, revealing that gene retroduplication depends upon the activity of both long terminal repeat (LTR) and non-LTR retrotransposable elements, which in turn affects the number of retrocopies and their function/retention in the genomes.
While the rate of mRNA-mediated duplications can vary significantly across organisms, it is also important to consider any deletion biases in the genomes to predict the amount of retrocopies that will be found in a genome. Furthermore, the evolutionary trajectories of retrogenes appear to differ significantly to those of other types of gene duplicates, including a higher frequency of retrogenes expressed in the male germline (i.e., in testis and pollen).
mRNA-Mediated Gene Duplication
mRNA-mediated gene duplication requires the reverse transcription of a transcript and its integration in the genome. Transposable elements belonging to both major superfamilies of LTR and non-LTR retrotransposons encode a reverse transcriptase (RT) and an endonuclease/integrase (EN/INT) domain mediating the insertion of the retrocopy in the genome (Levin and Moran 2011). Using comparative genomic and population genomic data, gene copies at any of the stages of their “life cycle” can be observed (Box 1). This has aided the understanding of the mechanisms of retroduplication and of the features that need to be considered for retrocopy identification. Box 1 also includes definitions for terms and their most general use that we will employ in this review.
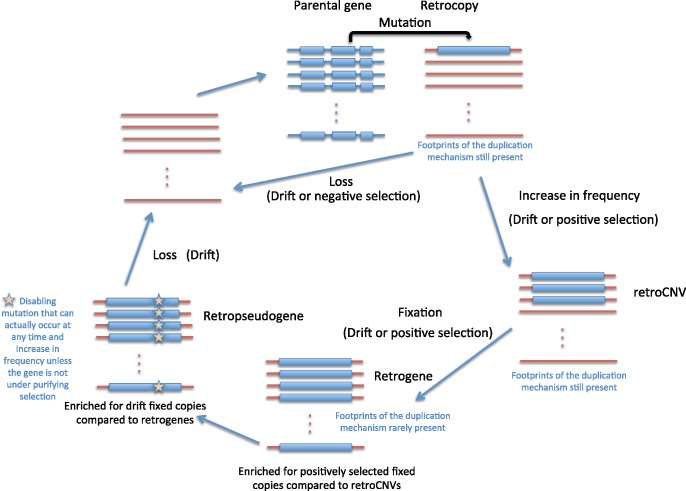
Box 1: The “Life Cycle” of Retrocopies: Nomenclature and Inferences Made from Comparisons
Gene duplication is the outcome of molecular processes that give rise to additional gene copies in genomes. This phenomenon includes both DNA-mediated and RNA-mediated duplications. When these processes take place in the germline the novel gene duplicates have the chance to reach fixation in the population. Research has often focused on the duplication of protein-coding genes, but RNA genes (rRNA, tRNA, etc.). duplicate in analogous ways. After a mutational event, a novelgene copymay increase in frequency in the population by the action of selection or drift, or it may be lost (Innan and Kondrashov 2010). The termcopy number variant(CNV) is used to refer to the fact that the products of gene duplication or deletion events might be found as polymorphic/segregating variants in the population (i.e., a gene copy is found in genomes of some but not other individuals; Feuk et al. 2006). The presence of CNVs in genomes can be assessed using population genomic data and its origin (i.e., duplication or deletion) determined from inferring the ancestral state. We often use the term gene duplication to specifically refer to duplications that are fixed in a population. Gene duplicates that show disablements (i.e., in-frame stop codons or insertions/deletions that are not a multiple of three base pairs) are consideredpseudogenes(Mighell et al. 2000; Zheng and Gerstein 2007) and they might at most have non-coding RNA regulatory roles.
If gene duplication occurs by means of reverse transcription of an mRNA and insertion in the genome, it is referred to as aretrocopy(i.e., an mRNA-mediated gene duplication; Emerson et al. 2004; Kaessmann et al. 2009). We refer to the fixed functional retrocopies asretrogenesand to those with disablements asretropseudogenes(Emerson et al. 2004; Kaessmann et al. 2009). However, some retropseudogenes are transcribed and a fraction of them are known to be involved in regulatory functions and could be considered non-protein coding retrogenes (see the “Retrocopies with regulatory functions” section).RetroCNVsrepresent retrocopies that segregate in a species and therefore can be detected through population genomic analyses (Schrider et al. 2011).
Until relatively recently, all retrocopies were referred to asprocessed pseudogenes, under the assumption that after retroposition these gene duplicates could not be expressed given the absence of regulatory regions in mRNAs (Vanin 1985; Weiner et al. 1986; Lander et al. 2001). While this terminology is still correct when referring to retropseudogenes, the term “retrocopies” is currently used more often to identify both pseudogenes and novel genes derived from retrotransposition (Kaessmann et al. 2009).
Evolutionary insights are gained from comparing retrocopies from different periods of their “life cycle” (i.e., retrocopies of different ages or fates can be compared;Figure Box 1). For example, the comparisons between the location and patterns of expression of retrogenes (functional fixed retrocopies) with retropseudogenes (disabled fixed retrocopies) can reveal the effects of both purifying and positive selection on the former (Emerson et al. 2004; Vinckenbosch et al. 2006). RetroCNVs due to gene duplication are for the most part young new gene copies that reflect more closely both insertional biases and mutational patterns than retrogenes and fixed retropseudogenes, primarily because selection has had less time to act on them (Schrider et al. 2011, 2013). Young retrogenes or retroCNVs might show molecular signatures of the mechanism by which they originated because of the limited time for mutations to accumulate. The region around the retroCNV might show the footprints of selection, if the new gene copy is adaptive and quickly increasing in frequency in the population (Long et al. 2003; Tan et al. 2016; Zhu et al. 2016).
Mechanisms of Retrocopy Formation
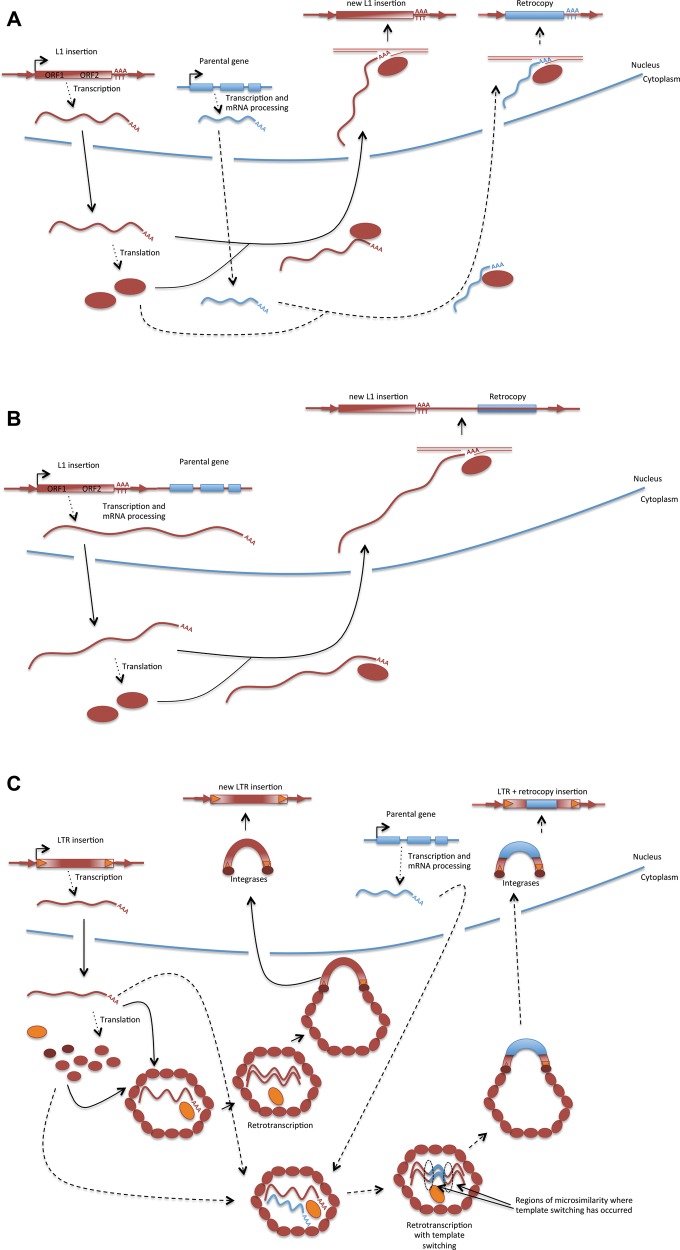
It has been recognized for decades that the abundance of retrocopies (retrogenes and retropseudogenes) in genomes should depend on the existence of a particular machinery—reverse transcriptase and endonuclease/integrase—in germ cells or in germ cell precursors (reviewed in Vanin 1985). Initially, the primary mechanism implicated was the non-LTR transposable element machinery. This stems from a biased focus in early studies on retrocopies that are extremely abundant in mammals (including humans) as a consequence of the L1 elements’ prolonged activity in their lineages (Ostertag and Kazazian 2001). L1s are long interspersed elements (LINEs) belonging to the non-LTR superfamily of retroelements. The proteins encoded by L1 elements are responsible for the transposition of non-autonomous Alu elements, as well as host gene mRNAs whose poly(A) sequences are mistakenly recognized by the L1 machinery (Wei et al. 2001). The L1 retrotransposition mechanism has been well characterized in vitro and in vivo and involves retrotranscription of the L1 in the nucleus and insertion of DNA by means of a target-primed reverse transcription (TPRT) reaction (fig. 1A and Cost et al. 2002). There is direct evidence from tissue culture that L1 is implicated in the generation of retrocopies (Tchenio et al. 1993; Maestre et al. 1995; Esnault et al. 2000). In addition, the expected hallmarks of L1 retrotransposition (i.e., a short remnant of the poly(A) tail at the 3′ end and target site duplications) have been observed in the DNA flanking retrocopies (fig. 1A andTchenio et al. 1993; Maestre et al. 1995; Esnault et al. 2000; Kaessmann et al. 2009). Thus, the generation of intronless gene copies by the L1’s enzymatic machinery is well supported (Kaessmann 2010). It has also been observed that L1 elements can produce the retrotransposition of downstream regions (fig. 1B andGoodier et al. 2000; Pickeral et al. 2000).
Fig. 1.
—(A) LINEs are autonomous non-LTR retrotransposable elements. They are transcribed by RNA polymerase II and their transcript encodes for a retrotranscriptase (RT) and additional activities (RNA binding and endonuclease activity within others; some not well characterized) in two open reading frames (Cost et al. 2002). The poly(A) of the L1 transcripts is bound by its proteins in the cytoplasm after the transcript is translated and carried to the nucleus. In the nucleus, the transcript undergoes target-primed reverse transcription (TPRT) after the endonuclease nicks the DNA and a 3′ end is available to prime the RT reaction (Cost et al. 2002). It is still unclear how all the reactions occur (Mandal and Kazazian 2016) but there are some clear hallmarks of L1-mediated retrotransposition: a short remnant of the poly(A) tail at the 3′ and target side duplications (TSDs; Vanin 1985). TPRT often produces 5′ truncated copies but well preserved 3′ end of the element including the short remnant poly(A) tail (Zingler et al. 2005). L1 elements can mediate the retrotransposition of mRNAs in cells. The poly(A) of cytoplasmic mRNAs might be recognized by L1 proteins, carried to the nucleus, undergo TPRT and be inserted in the genome. The hallmarks of this process are going to be the presence of an intronless copy of a gene with a remnant of a poly(A) tail at the 3′ end and TSDs flanking the insertion. 3′-UTRs will often be complete while there might be 5′ truncated copies from the onset (often the 5′-UTR can be shorter and sometimes the CDS can be affected by this truncation likely producing a retropseudogene). (B) L1 can produce the transduction of downstream regions when the transcript produced by the element is unusually long (i.e., it is not polyadenylated at the typical polyadenylation site) and includes the downstream region that could potentially encompass a gene (Goodier et al. 2000; Pickeral et al. 2000). (C) Autonomous LTR retrotransposable elements are transcribed and translated and the proteins encoded assemble a viral-like particle where reverse transcription of the LTR RNA occurs. The product will be a double-stranded LTR retroelement flanked by LTRs that proteins bind, bring to the nucleus and integrate in the genome (Levin and Moran 2011). When the reverse transcriptase switches transcripts might start retrotranscribing templates of cellular mRNAs and as it switches again to retrotranscribed the end of the LTR transcript produces a retrocopy that will often be an incomplete CDS but can potentially contain a whole retrocopy of cellular genes (Derr et al. 1991; Tan et al. 2016). The template switching will occur if there are by chance regions of sequence similarity between the mRNA and the nascent cDNA (Derr et al. 1991). When the template switching occurs more than once, chimeric retrocopies can be produced (Wang et al. 2006; Tan et al. 2016). These retrocopies or more often partial retrocopies will be flanked by LTR sequences on both sides.
Not all non-LTR retrotransposable elements are competent to retroduplicate genes’ transcripts, or other RNAs (e.g., they do not act in trans on other RNAs generating SINEs, i.e., short interspersed elements, or retrocopies of genes) and it does appear that transcript recognition is the limiting factor. If proteins encoded by non-LTR retroelements recognize a particular sequence of its own mRNA, as it seems to be the case for the CR1 element, this limits the types of cellular RNAs that can hitchhike and be retrotransposed (International Chicken Genome Sequencing 2004; Suh 2015).
Interestingly, the presence of LINEs and SINEs that are moved by the same autonomous machinery might reveal that either a poly(A) is recognized or there is a low stringency transcript recognition and competency to retrotranspose a broad array of transcripts (Ohshima 2013). This observation (Ohshima 2013) has prompted additional studies including a recent work in Arabidopsis thaliana and cassava (Zhu et al. 2016), two dicots harboring both LINE and SINE sequences in their genome. Zhu and colleagues found that some retroCNVs maintain the footprints of reverse transcription and insertion mechanisms (see section “Evolutionary insights from retroCNVs”) and that a few young retrocopies were likely generated by LINE retroelements. Further work is needed to determine if the association between LINE/SINE pairs and LINE-mediated gene retroposition represent a general trend in plants and other eukaryotes.
While evidence of the capacity of LTR elements to produce retrocopies has existed for several decades (Derr et al. 1991), how much these retroelements contribute to the generation of retroposed gene copies in all lineages of life is only recently beginning to be appreciated. Discoveries of LTR retroelement sequences flanking single genes in maize (Bureau et al. 1994; Jin and Bennetzen 1994) have been followed by genome-wide investigations of LTR elements involved in retrogenes formation in rice (Wang et al. 2006), Arabidopsis (Zhu et al. 2016) and more recently of retroCNVs in animals (Tan et al. 2016). It does seem that, as initially observed for the Ty1 element in yeast (fig. 1C and Derr et al. 1991), the LTR retrotranscriptase can switch template during cDNA synthesis in the cytoplasm and incorporate cellular transcripts within the new LTR copy (Tan et al. 2016). These retrocopies are flanked by LTR sequences that will eventually degenerate, but the occurrence of complete or partial long terminal direct repeats and open reading frames, in some cases, is still recognizable (Derr et al. 1991; Tan et al. 2016). LTR-mediated retrocopies have now been described in lineages where there is significant LTR activity, such as angiosperms and various metazoans (Wang et al. 2006; Tan et al. 2016). These retrocopies are flanked by LTR sequences, which can function as donors of promoter regions for a novel gene copy (fig. 1C). When the template switching occurs more than once, chimeric retrocopies of more than one transcript can be produced (Wang et al. 2006; Elrouby and Bureau 2010; Tan et al. 2016).
Although some non-LTR and LTR retrotransposable elements can generate gene retrocopies, not all retroelements included in these superfamilies might be competent to do so, as noticed above with regard to CR1 elements. How many and what kind of retrocopies are observed in a particular lineage might depend on what types of retroelements are active in that lineage at a given time (e.g., L1s in mammals or CR1s in birds). The developmental and spatial expression patterns of retroelements (i.e., what cells in the germline or pre-germline express the RT and EN/INT enzymes), the level of retroelement transcription and the stability of host mRNAs all affect the pattern of retrocopy insertions revealed by retroCNVs (Pavlicek et al. 2006; Kaessmann et al. 2009; Ohshima 2013; Richardson et al. 2014).
Retrocopy Identification
The most remarkable feature of retrocopies is the lack of introns compared with their parental genes (i.e., the genes that provide the mRNA from which retrocopies originated) due to the reverse transcription following splicing (fig. 1). This hallmark of gene retroposition has been used to identify retrocopies since the early studies on gene retroposition in the 1980s and early 1990s (Soares et al. 1985; Vanin 1985; McCarrey and Thomas 1987; Ashworth et al. 1990; Dahl et al. 1990; Fitzgerald et al. 1993; Long and Langley 1993). In these and following studies, coding and non-coding regions of the two gene copies are compared to assess whether introns occur in only one of them (Vanin 1985). However, searches of retrocopies that rely on the lack of introns engender several major caveats. First, retrocopies may inherit introns from their parent genes or they may acquire novel introns throughout intronization of their original coding sequences or via recruitment of de novo exons from flanking genomic DNA (Catania and Lynch 2008; Zhu et al. 2009; Szczesniak et al. 2011; Kang et al. 2012; Zhang et al. 2014). They may also acquire novel introns by the formation of fusion (chimeric) transcripts that include exons from nearby genes (see the “Chimeric retrogenes” section below). Importantly, intron-containing retrocopies might be overlooked, depending on the computational approach employed to search the genome. Second, intronless gene copies may also originate via DNA-mediated duplications of intronless parental genes (Zhang et al. 2011). Phylogenetic analyses to assign parental genes in gene families with both intron-containing and intronless genes should be used to assess whether any of the intronless genes were generated by another intronless copy. Several gene families, for example all groups of mammalian odorant receptor genes, exclusively contain intronless genes, thus making any inference on their duplication mechanism (RNA- vs. DNA-based) particularly challenging. Additionally, errors in the annotation of exon–intron boundaries, and the occurrence of gene isoforms with or without introns in the same section of the coding region may affect the estimates of the number of retrocopies.
Other signatures of gene retroposition may determine the duplication mechanism of putative retrocopies. Such signatures include a poly(A) tail at the 3′ end of the retrocopy and target site duplications (TSDs) flanking the insertion site in LINE-mediated gene retrotranspositions. Retrocopies that originated through an LTR-mediated mechanism may instead maintain flanking LTR sequences (fig. 1). Poly(A) tails, TSDs and LTR sequences are the only types of evidence available to infer whether a gene copy was generated through retrotransposition when the putative parental gene is intronless. Given that these signatures of retrotransposition are evolutionarily more labile than the lack of introns, they tend to be mainly useful in the assessment of young putative retrocopies.
Overall, computational strategies for genome-wide surveys of retrogenes fall into two main groups. One widely used approach relies on the comparison between annotated paralogs with similar sequences, but different gene structures (e.g., intron-containing vs. intronless genes; Betran et al. 2002; Emerson et al. 2004). This method is tailored to the discovery of functional retrogenes, particularly if the initial comparison of annotated genes includes only protein-coding sequences. A second approach consists in broad sequence similarity searches, wherein whole-genome assemblies are surveyed using protein or protein-coding DNA sequences, usually obtained from intron-containing genes. These analyses allow researchers to identify both retrogenes and retropseudogenes (Baertsch et al. 2008; Kabza et al. 2014).
In principle, studies of retrocopies based on similar methods and type of sequences should provide largely overlapping results in terms of the number of insertions. However, results tend to vary significantly among publications regarding retrocopies within the same species, mainly because of different sequence identity and alignment coverage thresholds used in the paralogous identification step, or as a result of different gene datasets. For example, estimates of retrocopies and retrogenes in human vary up to 6-fold (table 1 and supplementary table S1, Supplementary Material online).
Table 1.
Number of Retrocopies, Transcribed Retrocopies, and Retrogenes
| Taxonomic Group | Species | Retrocopiesa,b | Transcribed Retrocopiesb | Retrogenesb |
|---|---|---|---|---|
| Eutherian mammals | Human | 3,771–18,700 | 358–1,304 | ∼120–692 |
| Chimpanzee | 1,889–7,478 | 491–1,500 | 141–476 | |
| Gorilla | 4,638–7,706 | 491–1,461 | 215 | |
| Orangutan | 5,127–6,873 | 420–846 | 194 | |
| Macaque | 4,923–7,502 | 528–1,324 | 198 | |
| Common marmoset | 10,465 | |||
| Squirrel monkey | 9,320 | |||
| Mouse | 2,969–20,360 | 420 | 83–663 | |
| Rat | 3,298–7,364 | 389 | 83–567 | |
| Cow | 1,996 | 790 | ||
| Dog | 19,13–3,505 | 409 | ||
| Marsupials | Opossum | 1,992–3,036 | 421 | 256 |
| Monotremes | Platypus | 260–542 | 67 | 88–92 |
| Birds | Chicken | 70–720 | 30 | 36–321 |
| Reptiles | Anolis carolinensis | 404 | 136 | |
| Amphibians | Xenopus tropicalis | 398 | 140 | |
| Teleosts | Zebrafish | 195–652 | 119–127 | |
| Tetraodon nigroviridis | 90–644 | 60–227 | ||
| Fugu rubripes | 182 | 142 | ||
| Stickleback | 132 | 111 | ||
| Medaka | 218 | 131 | ||
| Coelacanth | 472 | 85 | ||
| Tunicates | Ciona | 110 | 96 | |
| Cephalochordates | Lancelet | 337 | 176 | |
| Dipterans | Drosophila melanogaster | 94–102 | ||
| Anopheles gambiae | 190 | |||
| Aedes aegypti | 133c | |||
| Lepidoptera | Silkworm | 27–68 | ||
| Nematodes | Caenorhabditisd | 9–48 | ||
| Dicots | Arabidopsis thaliana | 69–83 | 47–251 | |
| Poplar | 106 | 95 | ||
| Monocots | Rice | 150–1,235 | 495 | |
| Algae | Chlamydomonas | 60 | ||
| Volvox | 81 |
Retrocopies include both retropseudogenes and retrogenes except some references.
Minimum and maximum numbers of retrocopies, transcribed retrocopies and retrogenes are shown for taxa with multiple estimates.
A minimum of 133 retrogenes were annotated in A. aegypti.
Retrogenes from five Caenorhabditis species are shown.
Distinguishing between retropseudogenes and retrogenes is a primary goal of most genome-wide studies on gene retroposition. Often the retrocopies are inspected for disablements to identify retropseudogenes (Box 1) and separate them from functional or potentially functional retrocopies (e.g., Harrison et al. 2002; Emerson et al. 2004; Vinckenbosch et al. 2006; Wang et al. 2006). Another important approach to separate retropseudogenes from retrogenes is the analysis of the evolutionary dynamics of their coding sequences. Most retrogene codons should evolve under a purifying selection regime. Therefore, the ratio of nonsynonymous substitutions per site over synonymous substitutions per site, or dN/dS, should be well below one. Retropseudogenes are expected to evolve neutrally and to exhibit dN/dS values close to one. A widely used test of retrocopy functionality consists in the measure of dN/dS ratio in the retrocopy-parental gene pair, which represents the dN/dS ratio average of the lineages leading to the two genes. A combined dN/dS ratio <0.5 in the pair is considered to be evidence of evolutionary constraint on the retrocopy; if the retrocopy is nonfunctional (dN/dS ∼1), the average dN/dS should exceed 0.5 regardless of how small the dN/dS of the parental gene (Betran et al. 2002).
While this is likely to be true in many instances, a better inference of dN/dS, particularly on the retrogene lineage, should be performed by incorporating at least an additional homologous gene sequence from a different species to tests if the retrocopy lineage has evolved under negative selection, that is, if the dN/dS in this lineage is significantly lower than one. Such analyses also provide information about the different evolutionary dynamics between parent genes and retrogenes and might reveal signatures of positive selection or relaxed selection during the history of both genes (see also “Retrocopies evolutionary dynamics and pathways to biological functions”).
Finally, functional retrogenes show evidence of transcription. However, many retrogenes, particularly young ones, tend to have lower transcription levels and narrow expression breadth (see for instance Carelli et al. 2016). Indeed, a significant fraction of retrogenes is expressed primarily, if not exclusively, in the male germline. High-throughput RNA-sequencing experiments may, therefore, fail to detect transcripts of retrogenes if only a few tissues are examined. The concomitant occurrence of an intact open reading frame with length comparable to the parental gene coding sequence (CDS), a dN/dS ≪ 1, and expression in one or more tissues is generally considered a combination of traits that strongly support the functionality of a given retrogene.
The identification of functional retrocopies is a first important step in retrogene annotation. These analyses are further refined to detect other important features of putative retrogenes; for instance, the detection of their possible chimeric architecture due to fusion with exons of nearby genes, the recruitment of de novo exons (Wang et al. 2006; Kaessmann et al. 2009), or the presence of partially processed retrogenes (Soares et al. 1985; Baertsch et al. 2008; Zhang et al. 2014). Overall, analysis aimed at identifying functional retrogenes is often based on more stringent criteria in order to avoid false positives, whereas authors seeking to characterize both full-length and partial retrocopies may apply more loose thresholds, especially regarding the minimum length of retrocopy-parental gene alignments.
Surveys encompassing multiple genomes have the advantage of applying the same method to a range of species, providing a comparative framework for the analysis of the evolutionary dynamics of retrocopies (Zhu et al. 2009; Chen et al. 2011; Carelli et al. 2016). Furthermore, repositories have been established to collect retrocopy information across many species, including the RetrogeneDB database for 62 animal genomes (Kabza et al. 2014) and the RCPedia database dedicated to primate retrocopies (Navarro and Galante 2013). These are important efforts in the direction of a standardized approach to identifying and functionally characterizing retrogenes in multiple species, given that previously there has clearly been little consistency between authors in the way retrogenes are detected and analyzed.
Detection of retroCNVs
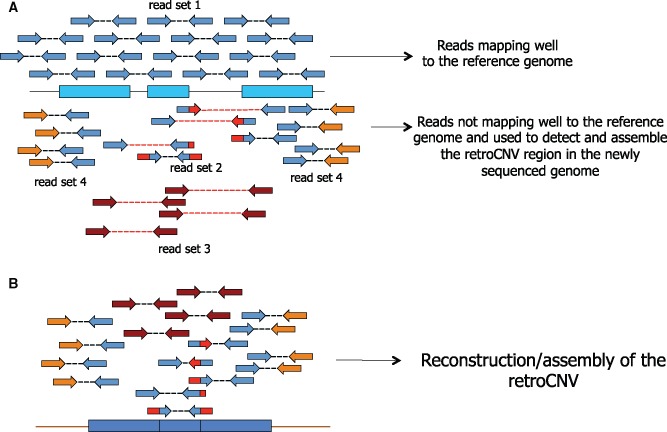
Some of the most intimate features of retrocopies have finally become accessible, even in Drosophila and other species with a rapid DNA turnover, by the onset of population genomic investigations and the discovery of retrocopy polymorphisms, or retroCNVs (Box 1). Strategies to detect retroCNVs are based on mapping short reads or paired reads from one or multiple specimens to a reference genome. These analyses rely on identifying the presence of reads that span exon–exon junctions in some individuals/strains as the primary evidence for retroCNV calls (Schrider et al. 2011, 2013; Richardson et al. 2014; Tan et al. 2016; Zhu et al. 2016). These are reads that reveal the presence of an intronless copy of an intron-containing gene in the reference genome (fig. 2). Read depth has been used to confirm the presence of an additional copy in the genome where a retroCNV is putatively identified, whereas discordant paired reads or long reads allow to fully assemble the retroCNV and the insertion site (Schrider et al. 2011, 2013; Richardson et al. 2014; Tan et al. 2016; Zhu et al. 2016). The presence of flanking repetitive regions may hamper the identification of the insertion site (Richardson et al. 2014), a problem that might be circumvented by using longer reads or mate-pair reads (Tan et al. 2016). It has been easier to assemble the insertion sites of retroCNVs in humans (Schrider et al. 2013), than in fruit flies (Tan et al. 2016), a consequence of either the mechanism of retrotransposition (non-LTR-mediated or LTR mediated; fig. 1) or the insertion site (repetitive vs. non-repetitive insertion). More detail is provided about what we have learned from retroCNVs in the section “Evolutionary insights from retroCNVs”.
Fig. 2.
—(A) Reads from the newly sequenced genome are mapped to the reference genome. Reads from the parental gene will map well (Set 1). Read from the retroCNV will not map well (Sets 2–4). They will hit the region of the parental region but there will be problems: reads spanning exon–exon junctions (Set 2), pair reads mapping farther apart than expected from de sequencing protocol (Set 3), and discordant reads (i.e., reads mapping to different regions in the genome; Set 4). (B) The reads that do not map well can be used to assemble the retroCNV.
Distribution of Retropseudogenes and Retrogenes in Eukaryotes
As for many other facets of gene and genome evolution, our knowledge of retrogenes and retropseudogenes origin and evolutionary impact is based on studies that have initially encompassed a few model species from a handful of eukaryotic lineages. Single retrogenes have been reported in several organisms; nevertheless, a simple literature search highlights the narrow taxonomic focus of such studies. As of September 2016, there were 407 papers in PubMed containing the keywords “retrogene”, “retroposed gene” or “gene retroposition”. About 90% of these articles contained the words “human”, “mouse”, “mammal”, “Drosophila” or “Arabidopsis”. Noticeably, “plant” papers in this database were outnumbered by “animal” papers 6.6 to 1 (295 vs. 45 papers). In fungi, the paucity of introns in Saccharomyces cerevisiae and most other hemiascomycetes appears to have discouraged extensive research on gene retroposition, although this mechanism has been shown to be involved in the duplication of 5S ribosomal genes in Aspergillus (Rooney and Ward 2005). As pointed out in the seminal work by Mourier and Jeffares (2003), a variety of other eukaryotes share, along with hemiascomycetes, a very low intron content and an asymmetric intron retention toward the 5′ end of genes. This appears to be the result of recombination, or gene conversion, between ancestral genes with their mRNA, which is mediated by the RT enzyme of retrotransposons (Derr et al. 1991; Pyne et al. 2005). However, several studies indicate that the common ancestor of all eukaryotes was intron-rich, and they suggest that intron paucity represents the exception rather than the rule across eukaryotes (Csuros et al. 2011; Irimia and Roy 2014). These findings, and the activity of retroelements in many genomes, warrant the scrutiny of retrocopies in eukaryotic lineages not yet assessed for this duplication mechanism.
Retrocopies have been extensively studied in mammals, in particular, in primates and rodents. Retroposed gene copies were discovered in the early 1980s (reviewed in Vanin 1985), while the first reported functional retrogene concerned the preproinsulin I gene in the rat was described in 1985 (Soares et al. 1985). Several individual retrogenes in rodents and other mammalian genomes were subsequently discovered (Andersen et al. 1986; Boer et al. 1987; McCarrey and Thomas 1987; Dyer et al. 1989). These findings were followed by genome-wide studies showing that therian mammals (placentals and marsupials) contain thousands of retropseudogenes (Goncalves et al. 2000; Ohshima et al. 2003; Torrents et al. 2003; Zhang et al. 2003; Zhang, Carriero et al. 2004; Khelifi et al. 2005; Cusack and Wolfe 2007; Terai et al. 2010; Pei et al. 2012). Other work in the past decade has revealed that up to several hundred retrocopies represent functional retrogenes in humans and other therians (Emerson et al. 2004; Marques et al. 2005; Suyama et al. 2006; Vinckenbosch et al. 2006; Sakai et al. 2007; Yu et al. 2007; Baertsch et al. 2008; Potrzebowski et al. 2008; Wang et al. 2010; Ewing et al. 2013; Zhang 2013; Navarro and Galante 2015; Carelli et al. 2016). Significantly fewer retrocopies and retrogenes were found in platypus, a monotreme, and in chicken, compared with therians (International Chicken Genome Sequencing 2004; Potrzebowski et al. 2008; Warren et al. 2008; Carelli et al. 2016). In agreement with this result, monotreme and bird genomes are devoid of L1 retroelements, the lineage of transposable elements responsible for the high rates of gene retroposition in therians (Esnault et al. 2000).
In primates, age distribution of retrocopies based on dS comparison between retrogenes and their parental genes suggests that retrocopy formation peaked ∼40 Mya, a time corresponding to an increase in activity of L1 retroelements that led to a burst in SINE/Alu repeat amplification (Ohshima et al. 2003; Marques et al. 2005). Other recent work based on phylogenetic analysis of multiple primate genomes suggests a more continuous generation rate for retrocopies, and possibly a slowing down of retroposition along the Old World Monkeys (OWMs) lineage (Zhang 2013; Navarro and Galante 2015). Navarro and Galante estimated retrocopy formation rates up to ∼140 copies per million years in the common ancestors of rhesus macaque and apes compared with rates of ∼21 copies per million years in the human lineage after the split from its common ancestor with chimpanzees (Navarro and Galante 2015). Furthermore, their investigation confirmed that New World Monkeys (NWMs) exhibit a significantly higher rate of retrocopy generation compared with OWMs, as suggested by preliminary analyses of the common marmoset genome (Marmoset Genome and Analysis 2014). Elevated copy numbers of L1 families L1PA3 and L1PA7 in NWMs might be associated with the burst of gene retroposition in these primates (Navarro and Galante 2015). These comparative genomic studies showcased how different retroelement landscapes shaped the dynamics of retrocopy formation across multiple evolutionary scales—from within-order to between-subclasses—in mammals.
More recently, retrocopy formation and evolutionary patterns have been examined across multiple chordate groups (table 1). The results of this work indicate that the lower rate of retrocopy generation found in monotremes and birds also extends to most non-mammalian vertebrates, as well as to the tunicate Ciona and the cephalochordate Amphioxus. However, the number of putative retrogenes is similar across chordate genomes, except in birds, when applying a dN/dS threshold of <0.5 in retrogene-parental gene pairs (Yu et al. 2007; Fu et al. 2010; Chen et al. 2011). These results also point to a faster turnover of retropseudogenes in non-mammalian chordates compared with mammals, with a prevalence of functional retrogenes among non-mammalian chordate retrocopies. As previously mentioned, rapid decay or deletion of retropseudogenes is shared across animals with small genomes and large effective population sizes, including Drosophila (Harrison et al. 2003).
Among non-chordate invertebrates, research on gene retroposition has historically focused on Drosophila. The first retrogene found outside mammals was the jingwei chimeric gene in D. melanogaster (Long and Langley 1993). Further analyses identified dozens of functional retrogenes, and a paucity of pseudogenes, in Drosophila, including retropseudogenes (Betran et al. 2002; Harrison et al. 2003; Dai et al. 2006; Bai et al. 2007; Zhou et al. 2008; Meisel et al. 2009; Vibranovski, Zhang et al. 2009). Genome-wide investigations of retrogene evolution have also been carried out in mosquitoes (Toups and Hahn 2010), silkworm (Toups et al. 2011; Wang et al. 2012), and Caenorhabditis nematodes (Zou et al. 2012; Zhou et al. 2015). Overall, the number of retrogenes ranges between less than 50 in nematodes and up to almost 200 in Anopheles gambiae, indicating variation in the rate of retrogene formation/retention among these invertebrate genomes (table 1 and supplementary table S1, Supplementary Material online). More work is required to determine whether different methods that have been applied to detect retrogene formation introduced variation.
In comparison to mammals and Drosophila, there has been a paucity of retrogene genome-wide studies in plants. Depending on the study, between ∼70 and ∼250 retrogenes have been found in Arabidopsis thaliana (Zhang et al. 2005; Zhu et al. 2009; Abdelsamad and Pecinka 2014). Higher estimates, however, relied only on expression to determine the functionality of retrogenes, without assessing the selective regime operating on the identified retrocopies through comparison of the evolution at nonsynonymous and synonymous sites (Abdelsamad and Pecinka 2014). Estimates of the number of retrogenes also vary significantly in rice; however, it generally appears that both dicot and monocot genomes harbor at most a few hundred retrogenes (Zhang et al. 2005; Wang et al. 2006; Zhu et al. 2009; Sakai et al. 2011; Abdelsamad and Pecinka 2014). The only study carried out in trees reported less than 100 retrogenes in poplar (Zhu et al. 2009). A comparable number of retrocopies has also been found in both unicellular (Chlamydomonas) and multicellular algae (Volvox; Jakalski et al. 2016).
Despite the methodological differences between these studies, it appears that all eukaryotes examined so far, with the possible exception of lineages with very few introns, harbor at least a few retrocopies per genome. A uniquely high rate of retrocopy accumulation is currently known only in therian mammals. This is likely the result of two independent processes: 1) a high rate of cellular mRNAs retroposition by some families of therian L1 retroelements, and 2) the slow decay of retropseudogenes in the therian lineages compared with other species where gene retroposition has been investigated, which generally share a much smaller genome size than therian mammals. Drosophila, Arabidopsis and other taxonomic groups with compact genomes often exhibit a rapid turnover of both transposable elements and pseudogenes, possibly associated with a large effective population size and the efficiency of selection against deleterious TE insertions (Lynch and Conery 2003). The analysis of more genomes in species with different effective population sizes, including plants, invertebrates and other taxa, will help determine whether the slowdown in retrocopy decay is shared across species with high nuclear DNA content, or whether it represents a unique feature of therian mammals.
While rates of retrocopy formation and number of retrogenes can vary significantly across taxa, data on gene retroposition in model organisms show that retrogenes form at a very similar pace across plants and animals. Marques and co-authors (2005) found a rate of ∼1 retrogene per million years in primates. In Drosophila melanogaster, a similar rate of ∼0.5 retrogenes per million years was observed (Bai et al. 2007). In plants, an approximate rate of 0.6 retrogenes per million years has been observed in A. thaliana (Zhang et al. 2005), whereas 0.1, 0.2 and 1 chimeric retrogenes per million years are fixed in poplar, A. thaliana and rice, respectively (Wang et al. 2006; Zhu et al. 2009).
These rates are lower than rates of DNA-based gene duplication, either tandem or interspersed, in Drosophila (Zhou et al. 2008; Zhang et al. 2010) and mammals, where segmental duplications account for a large fraction of new genes in the human and mouse genomes (She et al. 2008; Dennis and Eichler 2016). Additionally, retroCNVs appear to form at a lower rate than CNVs generated through DNA duplication in humans (Abyzov et al. 2013).
Retrocopy Evolutionary Dynamics and Pathways to Biological Functions
The long-term evolutionary trajectories of retrocopies include a range of possible outcomes that blur the blunt division between retropseudogenes and retrogenes. As pointed out from research on mammalian genomes, pseudogenes may be classified in a variety of categories depending on their disabling mutations, transcriptional activity, and potential functional role (Zheng and Gerstein 2007). In this section, we will discuss the evolutionary outcomes and consequences regarding retropseudogenes and then examine the evolution of retrogenes.
With their rich retrocopy “fossil record”, mammalian genomes undoubtedly represent a key source of information on retropseudogene origin and evolution. The wealth of genome-wide data available for humans, particularly as a result of the GENCODE project together with extensive comparative transcriptomic datasets have facilitated the dissection of retrocopy properties at an unprecedented level of detail (Pei et al. 2012; Carelli et al. 2016). All genome-wide studies on retroposed genes in mammals have shown that the majority of retrocopies present in these genomes are formed by retropseudogenes (Goncalves et al. 2000; Torrents et al. 2003; Zhang et al. 2003; Zhang, Carriero et al. 2004; Khelifi et al. 2005; Cusack and Wolfe 2007; Baertsch et al. 2008; Terai et al. 2010; Pei et al. 2012). Most retropseudogenes likely represent dead-on-arrival gene copies that have lost both protein-coding capability and transcriptional activity. However, even these gene copies may influence the evolutionary trajectory and functionality of their cognate genes. For instance, one of us found that gene conversion between a retropseudogene and a functional gene in humans led to a disease-associated mutation (Casola et al. 2012). Hundreds of historical gene conversion events involving pseudogenes and functional genes have also occurred in the human genome, with some events involving retropseudogenes (Casola C, et al., in preparation).
Retrocopies with Regulatory Functions
Transcriptome analyses have surprisingly revealed that hundreds of human and mouse retrocopies generate transcripts (Harrison et al. 2005; Frith et al. 2006; Shemesh et al. 2006; Huang et al. 2008; Pei et al. 2012). Although many retrocopy-derived transcripts represent a mere outcome of transcriptional noise, increasing evidence suggests that some of these transcripts have a biological role. In a recent study relying on human GENCODE data, a variety of signatures of biological activity, including sequence conservation, open chromatin state, DNaseI hypersensitivity sites and presence of upstream regulatory elements have been found in transcribed pseudogenes and retropseudogenes, which should perhaps be considered non-protein coding gene copies (Pei et al. 2012).
Experimental analyses of retrocopy transcripts elucidated their impact on cellular processes. For example, in DU145 prostate cancer cells, the retropseudogene PTENP1 mRNA acts as a decoy for miRNAs that also bind its parental gene PTEN, a gene related to cell growth suppression (Poliseno et al. 2010). In mouse oocytes, antisense small-interfering RNAs (siRNAs) derived from non-protein coding gene copies, including retrocopies, downregulate the expression of cognate genes (Tam et al. 2008; Watanabe et al. 2008), whereas other retrocopies mediate the degradation of both cognate genes and long noncoding RNAs (lncRNAs) during mouse spermatogenesis (Watanabe et al. 2015). Repression of parental gene expression via siRNAs derived from retrocopies and other non-protein gene copies has been suggested in rice (Guo et al. 2009).
Translation of retropseudogene RNAs, although probably a rare occurrence, may also have a role in regulating the expression of cognate genes. A recent discovery involves the truncated protein encoded by retrocopies of the tumor suppressor gene TP53. Two studies showed that the elephant’s genome contains an unusually high number of TP53 retrocopies, some of which are expressed to form truncated versions of TP53. These studies point to a possible role of peptides encoded by TP53 retrocopies in higher resistance to DNA damage and lower incidence of cancer in elephants (Abegglen et al. 2015; Sulak et al. 2016).
Protein-Coding Retrogenes
The most substantial contribution of retrocopies to protein diversity and novel phenotypes is derived from retrogenes that encode full-length proteins. While some retrogenes include protein-coding exons that have been co-opted from nearby genes, the majority of retrogenes contain the coding region inherited from their parental genes. After duplication, novel gene copies, including retrocopies, encounter different evolutionary phases that determine their possible retention and biological function. Initial fixation of retrocopies may follow one of three evolutionary scenarios, leading to the retention of their protein-coding potential: neofunctionalization, subfunctionalization, and conservation of the parental gene function (Ohno 1970; Force et al. 1999). There are several models included in each of these scenarios, which have been extensively discussed elsewhere (Hahn 2009; Innan and Kondrashov 2010). In this section, we will refer to neofunctionalization, subfunctionalization and functional conservation in a broader sense, focusing on studies that have mainly addressed the pattern of coding sequence and gene expression divergence between retrogenes and parental genes.
Neofunctionalization, or the evolution of a new biological function via the adaptive accumulation of coding or non-coding substitutions, has been argued to represent a common outcome of gene duplication since Ohno’s (1970) landmark book on gene duplication. In line with the neofunctionalization hypothesis, several papers have shown retrogenes that experienced an accelerated substitution rate in their coding sequence, compared with their cognate genes, in the early stage of their evolution (Betran and Long 2003; Jones and Begun 2005; Gayral et al. 2007; Rosso, Marques, Reichert et al. 2008; Jun et al. 2009; Matsuno et al. 2009; Quezada-Diaz et al. 2010; Tracy et al. 2010; Pegueroles et al. 2013). These findings support Ohno’s view that after duplication, one of the two gene copies is free from selective constrains and therefore accumulates “forbidden” substitutions that can initiate the path to a novel function. Following this phase, the gene copies that provide a fitness advantage begin to evolve under purifying selection regime. Some studies have also provided experimental evidence of retrogene neofunctionalization. For example, in African apes, neofunctionalization of the retrogene CDC14Bretro appears to have occurred via subcellular relocalization, a consequence of the retrogene’s protein acquiring a signal peptide specific to a different cellular compartment than the parent’s protein (Rosso, Marques, Weier et al. 2008). Direct evidence of neofunctionalization has also been found in the retrogene CYP98A8 and its paralog CYP98A9, which determined the evolution of a novel phenolic pathway in Brassicaceae (Matsuno et al. 2009). In Drosophila and mammals, novel protein functions, or expression patterns, have been observed in many retrogenes (Betran and Long 2003; Dai et al. 2008; see also Table 1 in Kaessmann et al. 2009; Carelli et al. 2016), including genes involved in the evolution of functions specific to male germline (see Box 2 and the section “Retrogene regulatory regions”). Neofunctionalization involving targeting of a novel cellular localization has also been described, for instance in the hominoid-specific Glutamate dehydrogenase retrogene GLUD2, which shows an enhanced localization in mitochondria compared with the cognate gene GLUD1 that is primarily expressed in the cytoplasm (Rosso et al. 2008). Two positively selected amino acid replacements in mitochondrial targeting sequence of GLUD2 that occurred in a common ancestor of hominoids led to the mitochondrial-specific targeting of this protein, whereas other substitutions altered the biochemical properties of this enzyme (Burki and Kaessmann 2004). Interestingly, mice transfected with the human genomic region containing GLUD2 showed transcriptomic and metabolomic changes during brain development that partially mirrored the differences observed between apes and rhesus macaque (Li et al. 2016). Furthermore, a study on cultured astrocytes of transgenic mice expressing human GLUD2 suggests a role of this gene in the uptake of glutamate and maintenance of energy homeostasis under high levels of glutamatergic signaling (Nissen et al. 2017).
The second major potential outcome of retrogenes’ evolution is subfunctionalization, which consists of the partitioning of biological functions between a gene copy and its parental gene, often via a subdivision of the original expression pattern or cellular localization between the two paralogs (Force et al. 1999; Hahn 2009; Innan and Kondrashov 2010; Kaessmann 2010). Strong evidence of subfunctionalization, however, can be hard to obtain. The frequent out-of-X movement and male germline expression pattern of retrogenes has been suggested to lead to the functional replacement of the X-linked parental genes during meiotic sex chromosome inactivation (Emerson et al. 2004; Carelli et al. 2016) and could potentially be classified as subfunctionalization, under the assumption that the parental gene was expressed during the inactivation phase, possibly as a long-lived transcript or a protein. However, the autosomal retrogenes may accumulate substitutions that change their function compared with the ancestral role of the parental X-linked genes, thus evolving under a neofunctionalization trajectory. We argue that, at least in Drosophila and some mammalian examples, many of out-of-X genes are examples of neofunctionalization (Box 2).
With regard to the categorization of evolutionary pathways in sub- or neofunctionalization, we think that two significant remarks are necessary. First, retrogenes and other gene duplicates likely experience both types of dynamics during different stages of their evolution (He and Zhang 2005). Second, assessing the evolutionary dynamics of a retrogene and its cognate gene requires in-depth comparative and functional analyses of the expression patterns (and possibly of the encoded protein) of both genes in species containing the retrogene and in several outgroup taxa. Even extensive transcriptomic datasets from multiple species do not necessarily provide information about the ancestral expression breadth of the parental gene, because gene expression might have changed independently across multiple lineages. In addition, the presence of other recent duplicates of the parental gene and the retrogenes, which are often overlooked in retrogene studies, might affect the evolutionary trajectory of all gene copies, as in the aforementioned CYP98A8/CYP98A9 retroposition event (Liu et al. 2016). Nevertheless, experimental analysis of ancestral retrogene proteins can provide information about the functional trajectory of these genes. Rosso and collaborators “resurrected” ancestral versions of the hominoid-specific retrogene CDC14Bretro and determined that the substitutions that accumulated during a phase of adaptive CDC14Bretro evolution in the common ancestor of great apes led to the adaptive relocation of this retrogene’s protein from microtubules to the endoplasmic reticulum (Rosso et al. 2008).
In the third evolutionary scenario, leading to a conserved function between a retrogene and its cognate gene, the duplicated copy is maintained because of the fitness advantage derived from having increased protein or transcript expression. While this evolutionary pathway has not been directly tested in retrogenes, it has been noticed that many retrogenes maintain the parental gene expression pattern in rice (Sakai et al. 2011) and zebrafish (Zhong et al. 2016), a possible result of selection for increase dosage of the cognate gene product. This hypothesis could be further tested by determining whether retrogene-parental gene pairs with overlapping expression show a higher combined transcription level compared with parental genes in multiple close outgroup species lacking those retrogenes.
An outcome that is not contemplated in any of the three evolutionary scenarios described above is the replacement of the original parental gene by its own retroposed copy. Ciomborowska et al. (2013) identified 25 such “orphan” retrogenes in humans, and 10 were found in a later study, with five genes overlapping in the two datasets (Carelli et al. 2016). Orphan retrogenes showed higher sequence conservation than the parental genes still present in outgroup species. Notably, two human orphan retrogenes were shown to rescue the original phenotype in Drosophila knockout mutants of the parental gene’ orthologs (Bayat et al. 2012; Carney et al. 2013).
Chimeric Retrogenes
The contribution of retrocopies to the proteome is further revealed by the wide array of gene architectures including partial or full-length coding sequences of retroposed genes merged with other transcribed sequences from genic or nongenic regions. These cases are often grouped under the broad categories of “chimeric genes”, “chimeric transcripts” or “gene fusions”. Gene architectures wherein a significant portion of the coding sequence is derived from retrocopies are usually referred to as “chimeric retrogenes”.
Following the discovery of several chimeric retrogenes in D. melanogaster (Long and Langley 1993; Wang et al. 2002; Zhang, Dean, et al. 2004; Nozawa et al. 2005; Dai et al. 2008), dozens of new cases have been described in primates (Nisole et al. 2004; Vinckenbosch et al. 2006; Baertsch et al. 2008; Ohshima and Igarashi 2010), zebrafish (Fu et al. 2010) and plants (Zhang et al. 2005; Wang et al. 2006; Zhu et al. 2009; Elrouby and Bureau 2010; see also Kaessmann et al. 2009). Additionally, many retrogenes have recruited exons that originated de novo from non-genic regions. For instance, Baertsch et al. (2008) recognized several chimeric retrogenes with de novo exons, as well as other types of gene chimerism, although overall they reported that >90% of human retrogenes are non-chimeric. However, a recent study based on extensive transcriptomic data has shown that between 28% and 52% retrogenes in nine mammals and in chicken contain a chimeric gene structure, with older retrogenes exhibiting an increase in the number of novel exons and expression breadth (Carelli et al. 2016). Most multi-exonic retrogenes in these species contain novel 5′ exons nearby existing or de novo promoters and enhancers. Some plants also show a high proportion of chimeric retrogenes, many of which appear to have rapidly formed after retroposition (Wang et al. 2006). Transcribed chimeric retroCNVs have been observed by Schrider et al. (2013) in the human genome supporting the possibility of chimeric transcription of a retrocopy immediately upon insertion. Detecting chimeric retrogenes requires tailored computational approaches that are seldom integrated in the genome-wide strategies adopted to find retrocopies, as mentioned above. Similarly, high-throughput transcriptomic data have only recently been integrated in studies focusing on retrocopies’ transcriptional activity. Thus, the frequency of chimeric retrogenes might be significantly higher than previously assumed in systems that have been less well characterized than mammals and Drosophila. Importantly, retrogene chimeras could be common in genomes with a high proportion of LTR retroelements, such as seed plants. Although retrogenes likely derived from template switching involving LTR retroelements have been reported in Arabidopsis, rice and maize (Wang et al. 2006; Elrouby and Bureau 2010; Zhu et al. 2016), the extent of such process remains largely unknown in plants.
The unique architecture of chimeric retrogenes derived from the fusion of a retrocopy with a nearby gene has often triggered the onset of novel phenotypes. For instance, in New World monkeys (genus Aotus), the Trim5-cyclophilin A chimeric retrogene is responsible for the resistance to HIV-1 (Nisole et al. 2004; Sayah et al. 2004). Interestingly, a similar fusion occurred independently in two macaque species (Brennan et al. 2008; Virgen et al. 2008). Further notable examples have been discovered in the genus Drosophila, including the first reported functional chimeric retrogene jingwei, which derived from the insertion of an Adh gene copy into the yande gene and has evolved a new substrate specificity (Long and Langley 1993; Zhang, Dean, et al. 2004). Intriguingly, three chimeric retrogenes containing an Adh retrocopy occur in three different assemblages of Drosophila species, all of them showing signatures of accelerated protein evolution after their duplication (Jones and Begun 2005).
Finally, changes in the structure of retrogenes can also arise as a result of intronization. While not ubiquitous, novel introns within the original retroposed sequence have been found in ∼10% of A. thaliana and Populus trichocarpa retrogenes (Zhu et al. 2009), in some rice retrogenes (Sakai et al. 2011) and in few retrogenes from several mammals and chicken (Fablet et al. 2009; Szczesniak et al. 2011; Carelli et al. 2016).
Box 2: Retrogenes and Sex Chromosomes
After the first set of whole genome studies of retrogenes (Betran et al. 2002; Emerson et al. 2004), it was reported that there was a very strong and intriguing pattern of retrogene duplication in Diptera and mammalian genomes with respect to sex chromosomes. A statistically significant excess of broadly transcribed (i.e., housekeeping) parental genes on the X chromosomes was found to have produced retrogenes with quite specific transcription in male germline (Betran et al. 2002; Emerson et al. 2004). This is what was termed the “out-of-the-X pattern” although it should be clarified that there is no movement of the parental gene but just gene retroduplication to an autosome. This pattern was in addition to a generally high proportion of retrogenes expressed in male germline (Betran et al. 2002; Emerson et al. 2004). The observations made initially in D. melanogaster, human and mouse have been confirmed in these species by recent analyses with more data and extended to other Drosophila species (Bai et al. 2007; Meisel et al. 2009; Vibranovski, Lopes, et al. 2009; Han and Hahn 2012), mosquito (Toups and Hahn 2010; Baker and Russell 2011), and other mammals (Potrzebowski et al. 2008; Carelli et al. 2016). Interestingly, retrogenes in poplar did not show the “out-of-the-X pattern” (Zhu et al. 2009).
It is currently well supported that this pattern is not due to mechanistic biases (e.g., mutational biases) but to the preferential fixation and preservation of the retrocopies derived from X-linked genes in these genomes. Several lines of evidence reveal this. First, it was shown in the original work in mammals and confirmed afterward that there is no excess of autosomal retropseudogenes (i.e., disabled copies that should reveal the mutational patterns; Box 1) derived from X chromosome parental genes (Emerson et al. 2004; Potrzebowski et al. 2008). In addition, in both flies and humans, retroCNVs (i.e., recent, still polymorphic retrocopies of genes; Box 1) show no out-of-the-X pattern (Schrider et al. 2011, 2013; Tan et al. 2016). These results imply that the out-of-the-X pattern is not due to insertional biases of retrocopies. Studies in fruit flies (Fontanillas et al. 2007) and humans (Gu et al. 2000) also revealed that both LTR and non-LTR retroelements lack an X-to-autosomes transposition preference. Actually in both species there is an excess of retroelement insertions on the X, although in humans there might be selective reasons for this (Bailey et al. 2000). A similar, albeit less pronounced, out-of-the-X pattern was found for DNA-mediated duplications, in spite of their different duplication mechanism compared with retroelements (Meisel et al. 2009; Vibranovski, Lopes, et al. 2009; Gallach et al. 2010; Han and Hahn 2012). Intriguingly, many of DNA-mediated duplications show a male germline-biased expression as well.
What feature/s of the new retrocopies may have been under selection in order to promote the out-of-the-X pattern? It was initially suggested that retrocopies from X-linked genes might be preserved to compensate for the repression of parental genes in the male germline due to the meiotic sex chromosome inactivation, or MSCI (Betran et al. 2002; Emerson et al. 2004). MSCI is well known to occur in eutherian and metatherian mammals (Richler et al. 1992; Turner 2007); it has been proposed that this process evolved to promote the packaging of unpaired chromatin (Turner 2007). The timing of retroduplication follows the evolutionary steps that generated strata of nonrecombining X–Y regions and Y chromosome degeneration in mammals (McLysaght 2008; Potrzebowski et al. 2008). In addition, there appears to be complementarity between the expression of X-linked parental genes and their retrogenes, as well as stronger purifying selection acting on the out-of-the-X retrogenes compared with retrogenes derived from autosomal parental genes (Potrzebowski et al. 2008; Carelli et al. 2016). These findings support the hypothesis that the out-of-the-X retrogenes might replace the parental transcript in some male germline cells. In Drosophila, the expression of X-linked genes is also regulated in the male germline and out-of-the-X duplications might be selected for (Vibranovski, Lopes, et al. 2009; Landeen et al. 2016). However, this selection towards X-to-autosome gene duplication is unlikely to represent the only factor determining the pattern and the testis-biased expression of out-of-the-X retrogenes. When the functions and mode of evolution of retrogenes are investigated in more detail, it becomes clear that there are likely additional strong selective pressures at work in the male germline. Yes, the X chromosome appears not to be a good location for testis expression in male germline in mammals and flies but it remains unclear why we observe so many testis-specific retrogenes on autosomes. Furthermore, why are many retrogenes recurrently duplicated for testis-specific functions and evolving under a positive selection regime? It has been shown that there is a phase during spermatogenesis in mammals when there is widespread spurious expression of the genome (Soumillon et al. 2013), which could influence retrogenes expression; however, retrogenes expressed specifically in testis are not a random sample of duplications from broadly expressed genes, inserted at random in autosomes, and lacking regulatory regions (see also section on retrogene regulatory regions). There are quite a few features that reveal that strong selection for new spermatogenesis or sperm functions is at work. We observe these features strongly in Drosophila but also for some genes in mammals. The selective pressures that lead to the preservation of these retrocopies have been very specific because the new genes have a limited array of functions (i.e., mitochondria energy function, glycolysis function, chromosome segregation, meiosis function, nuclear transport, transcription factors, among others; Rohozinski and Bishop 2004; Betran et al. 2006; Rohozinski et al. 2006; Gallach et al. 2010; Tracy et al. 2010; Vemuganti et al. 2010; Han and Hahn 2012; Phadnis et al. 2012). Many of these genes appear to have evolved recurrently in different lineages and under positive selection or to have neofunctionalized (i.e., acquire a different function that the parental gene did not have; e.g., some glycolysis genes have now a domain to attach to the primary segment of the sperm tail in mammals; Rohozinski and Bishop 2004; Betran et al. 2006; Rohozinski et al. 2006; Gallach et al. 2010; Tracy et al. 2010; Vemuganti et al. 2010; Han and Hahn 2012; Phadnis et al. 2012). In addition, in Drosophila, some of the retrogenes that retroduplicate recurrently are lost after evolving under strong positive selection for a period of time (i.e., they show high turnover; Tracy et al. 2010; Han and Hahn 2012; Phadnis et al. 2012). Those features are better explained by strong selective pressures in the male germline, including male–male competition, sexual antagonism and arm races with selfish elements (Betran et al. 2006; Gallach et al. 2010; Meiklejohn and Tao 2010; Tracy et al. 2010; Gallach and Betran 2011; Han and Hahn 2012; Phadnis et al. 2012). These effects might be better observed in genomes such as Drosophila where most retropseudogenes are quickly removed from the genomes as some of these retrogenes are quite young but it can also be that selection is stronger in flies due to an elevated effective population size (see above).
A fraction of retrogenes might also be retained to compensate for the dosage effects of losing a broadly expressed Y copy during Y chromosome degeneration (Hughes et al. 2015). There might be also a benefit of expressing intronless genes late in spermatogenesis given that there is no intron splicing and specific nuclear transport of these genes (Caporilli et al. 2013).
Retrogenes have also been observed to insert in the Y chromosome (Saxena et al. 1996), and to retrotranspose to the X chromosome in some lineages producing mostly female–biased genes (Emerson et al. 2004; Potrzebowski et al. 2010). Retrogene expression patterns have begun to be studied in ZW systems as well (Wang et al. 2012); however, more data need to be gathered to understand the gene movement related to sex chromosome inactivation and potential selective pressure associated with these retrogenes.
Evolutionary Insights from retroCNVs
Population genomic data have allowed the study of retrocopies that are still polymorphic in the populations (i.e., retroCNVs), leading to novel relevant insights on the evolution of retrocopies in recent years (Box 1 and text above). These retroCNVs have been key to provide information about the mechanisms of retrogene formation, the insertional biases of retrocopies, the origin of expression patterns of those genes and the evolutionary forces underlying the fixation of retrocopies in natural populations.
RetroCNVs have been studied in mammals, including humans (Abyzov et al. 2013; Ewing et al. 2013; Schrider et al. 2013; Richardson et al. 2014; Tan et al. 2016), Drosophila (Schrider et al. 2011; Cardoso-Moreira et al. 2016; Tan et al. 2016), mosquito, zebrafish, and chicken (Tan et al. 2016). They have also been recently analyzed in Arabidopsis and cassava (Zhu et al. 2016). As mentioned above, the identification of retroCNVs depends on mapping short reads or paired reads from multiple individuals onto an available reference genome. Reads that span exon–exon junctions of a gene, occur in some but not all individuals and do no map to other retrocopies in the genome are indicative of a polymorphic intronless gene copy. Discordant paired reads or long reads have been used to assemble the retroCNV and insertion site (Schrider et al. 2011, 2013; Richardson et al. 2014; Tan et al. 2016; Zhu et al. 2016) but only a small fraction of retroCNV insertion sites are usually determined (Richardson et al. 2014). Two problems contribute to this low sensitivity: the mechanism of transposition and the insertion site itself. If the retrogene is produced by an LTR element it will be flanked by the sequence of that LTR retroelement (fig. 1) that is repeated many times in the genome; standard paired reads might be of little help to locate the insertion site of that retroCNV because the discordant reads will map in the transposable element. Both longer reads and mate-pair reads could help in the event of insertions in euchromatin (Tan et al. 2016) by providing sequences farther away from the retrocopy, thus helping to anchor the retroCNV to a unique genomic locus. Determining the insertion sites of retroCNVs can increase the power to detect the changes in the allele frequency spectrum that are expected under selection because the flanking regions can be included in the analyses (see below). For retroCNVs inserted in repetitive regions (i.e., heterochromatin), we will only be able to detect selection if the allele frequency spectrum differs dramatically within the retroCNV in population samples and they will offer less information on their origin and population dynamics.
Analyses of human and Drosophila population genomic data have shown that retroCNVs do not follow the X-to-autosome pattern observed in fixed retrogenes, further supporting the view that natural selection, rather than insertional biases, drives the retention of many out-of-the-X retrocopies (Schrider et al. 2011, 2013). These studies have also shown that retrotransposition likely occurs during cell division given that many retroCNV cognate genes are expressed during this phase of the cell cycle (Abyzov et al. 2013). Importantly, the recent origin of most retroCNVs has led to the identification of molecular signatures specific of either LINEs or LTR retroelements in plants and animals, expanding on the limited perspective offered by studies in mammals wherein L1 elements have been primarily involved in the formation of retrocopies. For instance, we have learnt that non-LTR elements have produced retrocopies in plants, in spite of the limited number of these retroelements compared with LTRs in plant genomes (Zhu et al. 2016). We have also learnt that LTR elements have produced most, if not all, young retroCNVs in Drosophila, and have produced several retrocopies in mammals, mosquito, zebrafish and chicken. Therefore, LTR retroelements can now be considered important sources of retrocopies in animals (Tan et al. 2016). Far from being a mere mechanistic detail, the LTR-retroCNVs association has the important “side-effect” of providing young retrocopies with regulatory motifs embedded in the retroelements sequence; as shown for example in Drosophila, retroCNVs derived from LTR retroelements share a broad expression pattern (Tan et al. 2016).
Since retroCNV analyses are population genomic analyses, the number of retroCNVs described depends strongly on how many genomes are studied, how divergent they are and the depth at which those genomes are sequenced (Schrider et al. 2011; Cardoso-Moreira et al. 2016; Tan et al. 2016). Richardson and collaborators compared three studies on retroCNVs in humans that used the 1,000 Genomes data and suggested that the stringency of the criteria applied and the validation approach had strong influence on which retroCNVs were retrieved (Richardson et al. 2014).
RetroCNVs are also a great source of information to address questions about the evolutionary processes during the fixation of a retrocopy and discern between the models that have been proposed to explain gene duplicate retention (Hahn 2009; Innan and Kondrashov 2010). Population genetic analyses of the frequency spectrum and the variation in DNA regions flanking retroCNVs with high frequency in a population can be performed to disentangle if the retroCNV is increasing in frequency under positive selection (Schrider et al. 2013). This test requires considering the expectation for a neutral copy at the same population frequency. In addition, initial PCR validation of presence and absence of the retrogene in different strains or individuals should be performed before those analyses to provide estimates of false positives and false negatives. Schrider and collaborators were able to perform such test for 46 retroCNVs in human populations and discovered two retroCNVs that show evidence of positive selection, DHFR and GNG10, supporting evolutionary models in which natural selection drives the new duplicated genes to fixation (Schrider et al. 2013).
Retrogene Regulatory Regions
In the sections above, we have introduced two rather general and important features of retrogenes: (1) they are often stripped-down copies of genes yet (2) they are often expressed and evolve new functions. Here, we focus on reviewing the different scenarios that have been proposed to explain how retrogenes acquire their regulatory regions (i.e., promoters, binding sites for the RNA polymerase, and/or enhancers, sequences that will drive expression in particular tissues). We will go over mechanisms, some examples and the available genomic data. A major characteristic of retrogenes is their bias toward a biological function in male germline (testis or pollen; Betran et al. 2002; Emerson et al. 2004; Bai et al. 2007; Kaessmann et al. 2009; Abdelsamad and Pecinka 2014; Carelli et al. 2016). In the next paragraphs we will focus on data showing how some retrogenes acquired a testis-specific expression pattern, with an emphasis on the evidence of positive selection for male germline expression (Box 2).
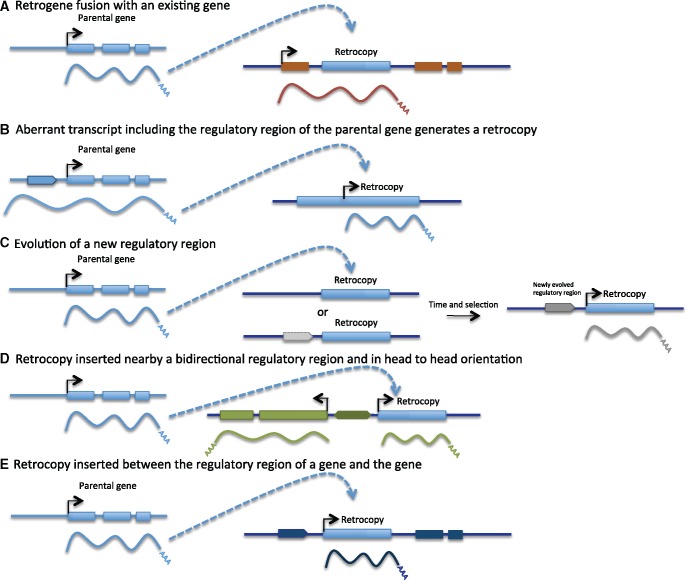
It has been known for some time that there are several manners in which a retrocopy can be transcribed right after being generated or can quickly evolve transcription (fig. 3). For some of these processes there are particular retrocopy examples, of which we will mention a few. For other processes, the evidence is indirect (i.e., derived from significant trends in the genomic data). New retrocopies can immediately (or after only a few additional mutations) acquire regulatory regions if there is a fusion with a preexisting gene upon insertion (fig. 3A). Such an outcome of retroposition is likely to often be deleterious except when gene fusions occur between a retrocopy and a recently duplicated gene or whenever it involves insertions in an intron followed by alternative splicing. Some of these retrogenes might produce chimeras (e.g., jingwei; Long and Langley 1993; Wang et al. 2000). It is also possible that some mutations that would be assumed to be deleterious might end up not being as deleterious as anticipated. We go over one such example in Drosophila below.
Figure Box 1.
—Illustration of the different fates and steps in the retrogene “life cycle” that can be compared to make evolutionary inferences. See text for more details.
Fig. 3.
—Mechanisms for retrocopy transcription. (A) The retrocopy might acquire regulatory regions from an existing gene after inserting within that gene. (B) Regulatory regions might be carried ouver from the parental gene, if an aberrant/longer transcript of the parental gene is produced. (C) A regulatory region might evolve from a proto-regulatory region or from a region with no regulatory function. (D) A retrocopy might be expressed from a bidirectional regulatory region, if inserted in head-to-head orientation. (E) A retrocopy might express if inserted between a gene and its regulatory region. See text for more details.
Retrogenes might retain regulatory regions upstream of the parental gene, particularly when they are derived from an aberrant transcript (i.e., longer at the 5′ end; fig. 3B). An example of this is Pgk2, a mammalian retrogene. The regulatory region from the parental gene likely provided initial expression of Pgk2 and testis-specific transcription evolved possibly under positive selection as the gene evolved a sperm-specific function (McCarrey and Thomas 1987; McCarrey 1990; Zhang et al. 1999; Danshina et al. 2010).
Retrogenes could also acquire expression from the genomic DNA flanking the insertion sites. They might benefit from an open chromatin state that can provide epigenetic context for the evolution of regulatory sequences either de novo or from proto-regulatory regions (fig. 3C). Alternately, retrogenes can benefit from being in head-to-head orientation with a nearby gene (fig. 3D), or by recruiting the regulatory region directly from the insertion site (i.e., inserting between a regulatory regions and a gene; fig. 3E).
There are particular examples and genome-wide analyses that support these mechanisms. In mouse and humans, detailed genomic studies have revealed that only ∼3% of retrocopies inherit regulatory regions from their parental genes, whereas 11% are transcribed from bidirectional regulatory regions of upstream genes in head-to-head orientation and 86% appear to be transcribed from newly evolved regulatory regions (Carelli et al. 2016), with the largest proportion of retrocopies becoming expressed in the testis. Interestingly, it has been shown that twice as many retrogenes are expressed in the testis compared with retropseudogenes, indicating a role for natural selection in the evolution of a male germline-related function of these genes in not only X-to-autosome duplications but in other directions as well (Vinckenbosch et al. 2006). In Drosophila, there are examples of testis-specific retrogenes for which the testis expression might be facilitated by the head-to-head orientation with a testis-specific gene or by insertion in regions where there is an abundance of testis-specific genes (Loppin et al. 2005; Bai et al. 2008). However, given that transposable elements in flies insert near female germline genes more often than near male germline genes (Fontanillas et al. 2007), these patterns have been interpreted as being due to the biased retention of testis-specific retrogenes (Bai et al. 2008). The analysis of the details of how retrogenes acquire testis expression has revealed that relocation is correlated with testis-specific expression. As mentioned in Box 2, the X chromosome appears to be a bad location to express testis-specific genes, but even in instances of autosome-to-autosome duplication, relocation appears to be correlated with the evolution of testis-specific expression or the retention of testis-specific genes (Gallach et al. 2010). When the details of some of these events are studied we encounter striking examples such as the Drosophila retrogene Dntf-2r. The molecular dissection of the Dntf-2r regulatory region pointed to a 14 bp-long motif that was present in the genome before the retrogene insertion and was driving testis-specific expression of a non-coding RNA (Sorourian et al. 2014). Thus, interestingly, it does appear that Dntf-2r acquired testis expression immediately upon insertion and that this insertion was not deleterious given that the non-coding RNA is still transcribed in the same pattern in Drosophila species with and without Dntf-2r.
Through time, regulatory regions can change. There are examples where retrogenes have functionally replaced their parental genes, with the latter acquiring a testis-specific expression. This reveals that the selective pressure for the evolution of testis-specific expression is not limited to the novel copy of a pair of paralogs (Krasnov et al. 2005).
In Arabidopsis, it has been recently shown that retrogenes are often expressed in pollen and tend to be expressed in a pattern different from the parental gene (Abdelsamad and Pecinka 2014). Additionally, these retrogenes show no expression correlation with nearby head-to-head oriented genes or other flanking genes. These authors propose that both the removal of epigenetic marks known to drive broad expression and the evolution of new regulatory regions due to selection allowed retrogenes to gain new male-specific functions (Abdelsamad and Pecinka 2014). In one instance, the de novo evolution of binding sites for the transcription factor DUO1 drives high expression of the PCR11 gene in pollen (Abdelsamad and Pecinka 2014). The lack of sex chromosomes in Arabidopsis (Giraut et al. 2011) reveals that this pattern is not always related to the presence of sex-linked genes. Indeed, selection for young testis-biased duplicated genes has also been observed in species with haplo-diploid sex determination (Wang et al. 2015).
It has been proposed that a “promiscuous/spurious” transcription phase in male germline is facilitated by a permissive chromatin state due to the transition between standard histones to testis-specific histone variants, which are then replaced by “transition proteins” (Soumillon et al. 2013). This chromatin state shift leads to the expression of many young gene duplicates that tend to be expressed in other tissues as they “age”. This model, termed the “out of the testis” hypothesis (Kaessmann 2010), is supported by the finding that young retrogenes are more often testis specific than old ones (Vinckenbosch et al. 2006; Carelli et al. 2016). This would imply that these particular genes change their expression through time. However, this age effect can also be a consequence of a high turnover (high rate of gains and losses) of testis-specific genes as opposed to broadly expressed genes that might be retained over longer evolutionary times in the genome. A high turnover is expected under transient selective pressure for those functions, i.e., in an arm race (Betran 2015). As mentioned in Box 2, there is evidence of a high rate of retrogene turnover in Drosophila. Therefore, more evidence is needed to validate the out-of-the-testis hypothesis. Carelli and collaborators made the first attempt to disentangle how often examples of changes in retrogenes’ expression patterns from testis specific to multi tissues are found and how often gene losses (i.e., turnover) of testis expressed genes occur. These authors did not find support for gene losses but they did not find evidence for changes in expression from testis to broader expression patterns of young retrogenes either (Carelli et al. 2016). However these authors analyzed a relatively limited sample size (33 functional testis-expressed retrogenes) in species that diverged long time ago. Future work relying on larger sets of retrogenes and more closely related taxa will allow to test the two hypotheses more thoroughly. Both effects likely contribute to the observation that young retrogenes are expressed in testis more often than older genes.
Concluding Remarks
Retrogenes and retropseudogenes have a profound impact on the evolution of genomes and the onset of novel gene architectures and phenotypic traits across eukaryotes. A rapidly growing body of literature on retrocopies, fueled by population genomic studies, functional genomics and transcriptomic data and comparative analyses of new genomes are revealing unanticipated levels of complexity in the formation, evolutionary trajectories and biological functions of retrogenes.
Evidence of a role of LTR retroelements in generating retrocopies is beginning to accumulate in plants and animals, providing a widespread alternative mechanism to the LINE-mediated gene retroposition so well characterized in mammals. Strikingly, both LTR and non-LTR retroelements appear to promote gene retroposition in the same species, although the former group might also contribute a “kick-start” to retrogenes evolution by providing regulatory regions to the promoter-lacking retrocopies. The recruitment of promoters and enhancers is indeed a major determinant of retrocopy long-term evolutionary dynamics. Transcriptomic studies have shown a certain promiscuity of retrogenes when it comes to recruit a promoter region: nearby genes, de novo evolved or proto-regulatory regions, even parental gene DNA motifs can be part of the regulatory suite of young and old retrogenes. Three important outcomes have emerged from these studies: first, many expressed retrocopies form chimeric genes with either genes nearby their insertion site, or with novel exons. Second, many retrogenes, particularly young ones, become expressed in the male germline. The debate on which model better explains this pattern is still ongoing and will certainly propel more research. Third, many non-protein coding retrocopies are expressed and potentially influence the activity of other genes as RNA- or peptide-mediated regulatory effectors. Several examples of such activity have been documented in the past decade and many more will likely be unveiled in the future.
Protein-coding retrogenes have been extensively studied for their potential phenotypic impact. Examples on phenotypic innovations due to retrogenes have been found in multiple lineages and might be a universal trait of these gene duplicates. They have been found in virtually every genome analyzed thus far, from tunicates, birds and fish to algae and rice, and they appear to mainly evolve following a neofunctionalization pathway, although some share their expression pattern with parental genes, suggesting that dosage effects or functional subfunctionalization may also influence their evolution.
Most of our knowledge on retrogenes and retropseudogenes derives from studies in humans, mouse, Drosophila and a few angiosperms. Although a wider taxonomic range of species has been analyzed in recent years, there is a clear gap in the functional assessment of retrocopies between these model species and other lineages, as many groups of eukaryotes have been largely neglected as far as gene retroposition goes. The widespread use of RNA-seq techniques, combined with thousands of currently available genome assemblies and re-sequencing projects, represent invaluable resources to begin closing this gap. Large-scale analyses of retrocopies relying on available as well as novel data will help answer lingering questions in this field. Are LTR retroelements responsible for the formation of most retrocopies in plants and other organisms with few non-LTR elements? Is there a correlation between LTR and non-LTR retroelements content and retrocopies number in a genome? Do non-mammalian species with large genomes contain as many retrocopies as mammals? How many non-protein coding retrocopies have a biological function? Are promoters recruited in similar ways across animals, plants, and other eukaryotes? How can we better test and compare models proposed to explain the bias of retrogenes toward male germline expression? Is the out-of-the-X process occurring outside animals? The answer to these and other pressing questions on retrocopy formation and evolution are now within reach thanks to ongoing remarkable progress in sequencing technology and genome-wide analyses of population genetic data. Given the many unexpected findings about retrocopies that have been made since their discovery nearly 40 years ago, it is safe to predict that a new wave of intriguing observations will come from future studies on retroposed gene copies.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
The authors would like to thank three anonymous reviewers and the Associate Editor for their comments and suggestions. They are also thankful to Michelle Lawing and Tomasz Koralewski for reading and commenting on this manuscript. E.B. would like to acknowledge the support from the National Institute of General Medical Sciences of the National Institutes of Health (R01GM071813). The content of this work is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. C.C. is supported by the National Institute of Food and Agriculture, U.S. Department of Agriculture, under award number TEX0-1-9599, the Texas A&M AgriLife Research, and the Texas A&M Forest Service.
Literature Cited
- Abdelsamad A, Pecinka A.. 2014. Pollen-specific activation of Arabidopsis retrogenes is associated with global transcriptional reprogramming. Plant Cell 26:3299–3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abegglen LM, et al. 2015. Potential mechanisms for cancer resistance in elephants and comparative cellular response to DNA damage in humans. JAMA 314:1850–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abyzov A, et al. 2013. Analysis of variable retroduplications in human populations suggests coupling of retrotransposition to cell division. Genome Res. 23:2042–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen RD, Birren BW, Taplitz SJ, Herschman HR.. 1986. Rat metallothionein-1 structural gene and three pseudogenes, one of which contains 5′-regulatory sequences. Mol Cell Biol. 6:302–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashworth A, Skene B, Swift S, Lovell-Badge R.. 1990. Zfa is an expressed retroposon derived from an alternative transcript of the Zfx gene. EMBO J. 9:1529–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baertsch R, Diekhans M, Kent WJ, Haussler D, Brosius J.. 2008. Retrocopy contributions to the evolution of the human genome. BMC Genomics 9:466.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Casola C, Betran E.. 2008. Evolutionary origin of regulatory regions of retrogenes in Drosophila. BMC Genomics 9:241.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Casola C, Feschotte C, Betran E.. 2007. Comparative genomics reveals a constant rate of origination and convergent acquisition of functional retrogenes in Drosophila. Genome Biol. 8:R11.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JA, Carrel L, Chakravarti A, Eichler EE.. 2000. Molecular evidence for a relationship between LINE-1 elements and X chromosome inactivation: the Lyon repeat hypothesis. Proc Natl Acad Sci U S A. 97:6634–6639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker DA, Russell S.. 2011. Role of testis-specific gene expression in sex-chromosome evolution of Anopheles gambiae. Genetics 189:1117–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayat V, et al. 2012. Mutations in the mitochondrial methionyl-tRNA synthetase cause a neurodegenerative phenotype in flies and a recessive ataxia (ARSAL) in humans. PLoS Biol. 10:e1001288.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betran E. 2015. The “life histories” of genes. J Mol Evol. 80:186–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betran E, Bai Y, Motiwale M.. 2006. Fast protein evolution and germ line expression of a Drosophila parental gene and its young retroposed paralog. Mol Biol Evol. 23:2191–2202. [DOI] [PubMed] [Google Scholar]
- Betran E, Long M.. 2003. Dntf-2r, a young Drosophila retroposed gene with specific male expression under positive Darwinian selection. Genetics 164:977–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betran E, Thornton K, Long M.. 2002. Retroposed new genes out of the X in Drosophila. Genome Res. 12:1854–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boer PH, Adra CN, Lau YF, McBurney MW.. 1987. The testis-specific phosphoglycerate kinase gene pgk-2 is a recruited retroposon. Mol Cell Biol. 7:3107–3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan G, Kozyrev Y, Hu SL.. 2008. TRIMCyp expression in old world primates Macaca nemestrina and Macaca fascicularis. Proc Natl Acad Sci U S A. 105:3569–3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bureau TE, White SE, Wessler SR.. 1994. Transduction of a cellular gene by a plant retroelement. Cell 77:479–480. [DOI] [PubMed] [Google Scholar]
- Burki F, Kaessmann H.. 2004. Birth and adaptive evolution of a hominoid gene that supports high neurotransmitter flux. Nat Genet. 36:1061–1063. [DOI] [PubMed] [Google Scholar]
- Caporilli S, Yu Y, Jiang J, White-Cooper H.. 2013. The RNA export factor, Nxt1, is required for tissue specific transcriptional regulation. PLoS Genet. 9:e1003526.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso-Moreira M, et al. 2016. Evidence for the fixation of gene duplications by positive selection in Drosophila. Genome Res. 26:787–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carelli FN, et al. 2016. The life history of retrocopies illuminates the evolution of new mammalian genes. Genome Res. 26:301–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carney TD, Struck AJ, Doe CQ.. 2013. Midlife crisis encodes a conserved zinc-finger protein required to maintain neuronal differentiation in Drosophila. Development 140:4155–4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casola C, Zekonyte U, Phillips AD, Cooper DN, Hahn MW.. 2012. Interlocus gene conversion events introduce deleterious mutations into at least 1% of human genes associated with inherited disease. Genome Res. 22:429–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catania F, Lynch M.. 2008. Where do introns come from? PLoS Biol. 6:e283.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, et al. 2011. Evolutionary patterns of RNA-based duplication in non-mammalian chordates. PLoS One 6:e21466.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciomborowska J, Rosikiewicz W, Szklarczyk D, Makalowski W, Makalowska I.. 2013. “Orphan” retrogenes in the human genome. Mol Biol Evol. 30:384–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cost GJ, Feng Q, Jacquier A, Boeke JD.. 2002. Human L1 element target-primed reverse transcription in vitro. EMBO J. 21:5899–5910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csuros M, Rogozin IB, Koonin EV.. 2011. A detailed history of intron-rich eukaryotic ancestors inferred from a global survey of 100 complete genomes. PLoS Comput Biol. 7:e1002150.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusack BP, Wolfe KH.. 2007. Not born equal: increased rate asymmetry in relocated and retrotransposed rodent gene duplicates. Mol Biol Evol. 24:679–686. [DOI] [PubMed] [Google Scholar]
- Dahl HH, Brown RM, Hutchison WM, Maragos C, Brown GK.. 1990. A testis-specific form of the human pyruvate dehydrogenase E1 alpha subunit is coded for by an intronless gene on chromosome 4. Genomics 8:225–232. [DOI] [PubMed] [Google Scholar]
- Dai H, et al. 2008. The evolution of courtship behaviors through the origination of a new gene in Drosophila. Proc Natl Acad Sci U S A. 105:7478–7483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai H, Yoshimatsu TF, Long M.. 2006. Retrogene movement within- and between-chromosomes in the evolution of Drosophila genomes. Gene 385:96–102. [DOI] [PubMed] [Google Scholar]
- Danshina PV, et al. 2010. Phosphoglycerate kinase 2 (PGK2) is essential for sperm function and male fertility in mice. Biol Reprod. 82:136–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis MY, Eichler EE.. 2016. Human adaptation and evolution by segmental duplication. Curr Opin Genet Dev. 41:44–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derr LK, Strathern JN, Garfinkel DJ.. 1991. RNA-mediated recombination in S. cerevisiae. Cell 67:355–364. [DOI] [PubMed] [Google Scholar]
- Dyer MR, Gay NJ, Walker JE.. 1989. DNA sequences of a bovine gene and of two related pseudogenes for the proteolipid subunit of mitochondrial ATP synthase. Biochem J. 260:249–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elrouby N, Bureau TE.. 2010. Bs1, a new chimeric gene formed by retrotransposon-mediated exon shuffling in maize. Plant Physiol. 153:1413–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson JJ, Kaessmann H, Betran E, Long M.. 2004. Extensive gene traffic on the mammalian X chromosome. Science 303:537–540. [DOI] [PubMed] [Google Scholar]
- Esnault C, Maestre J, Heidmann T.. 2000. Human LINE retrotransposons generate processed pseudogenes. Nat Genet. 24:363–367. [DOI] [PubMed] [Google Scholar]
- Ewing AD, et al. 2013. Retrotransposition of gene transcripts leads to structural variation in mammalian genomes. Genome Biol. 14:R22.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fablet M, Bueno M, Potrzebowski L, Kaessmann H.. 2009. Evolutionary origin and functions of retrogene introns. Mol Biol Evol. 26:2147–2156. [DOI] [PubMed] [Google Scholar]
- Feuk L, Carson AR, Scherer SW.. 2006. Structural variation in the human genome. Nat Rev Genet. 7:85–97. [DOI] [PubMed] [Google Scholar]
- Fitzgerald J, Wilcox SA, Graves JA, Dahl HH.. 1993. A eutherian X-linked gene, PDHA1, is autosomal in marsupials: a model for the evolution of a second, testis-specific variant in eutherian mammals. Genomics 18:636–642. [DOI] [PubMed] [Google Scholar]
- Fontanillas P, Hartl DL, Reuter M.. 2007. Genome organization and gene expression shape the transposable element distribution in the Drosophila melanogaster euchromatin. PLoS Genet. 3:e210.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Force A, et al. 1999. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151:1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frith MC, et al. 2006. Pseudo-messenger RNA: phantoms of the transcriptome. PLoS Genet. 2:e23.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu B, Chen M, Zou M, Long M, He S.. 2010. The rapid generation of chimerical genes expanding protein diversity in zebrafish. BMC Genomics 11:657.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallach M, Betran E.. 2011. Intralocus sexual conflict resolved through gene duplication. Trends Ecol Evol. 26:222–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallach M, Chandrasekaran C, Betran E.. 2010. Analyses of nuclearly encoded mitochondrial genes suggest gene duplication as a mechanism for resolving intralocus sexually antagonistic conflict in Drosophila. Genome Biol Evol. 2:835–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayral P, Caminade P, Boursot P, Galtier N.. 2007. The evolutionary fate of recently duplicated retrogenes in mice. J Evol Biol. 20:617–626. [DOI] [PubMed] [Google Scholar]
- Giraut L, et al. 2011. Genome-wide crossover distribution in Arabidopsis thaliana meiosis reveals sex-specific patterns along chromosomes. PLoS Genet. 7:e1002354.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves I, Duret L, Mouchiroud D.. 2000. Nature and structure of human genes that generate retropseudogenes. Genome Res. 10:672–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodier JL, Ostertag EM, Kazazian HH. Jr.. 2000. Transduction of 3′-flanking sequences is common in L1 retrotransposition. Hum Mol Genet. 9:653–657. [DOI] [PubMed] [Google Scholar]
- Gu Z, Wang H, Nekrutenko A, Li WH.. 2000. Densities, length proportions, and other distributional features of repetitive sequences in the human genome estimated from 430 megabases of genomic sequence. Gene 259:81–88. [DOI] [PubMed] [Google Scholar]
- Guo X, Zhang Z, Gerstein MB, Zheng D.. 2009. Small RNAs originated from pseudogenes: cis- or trans-acting? PLoS Comput Biol. 5:e1000449.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW. 2009. Distinguishing among evolutionary models for the maintenance of gene duplicates. J Hered. 100:605–617. [DOI] [PubMed] [Google Scholar]
- Han MV, Hahn MW.. 2012. Inferring the history of interchromosomal gene transposition in Drosophila using n-dimensional parsimony. Genetics 190:813–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison PM, et al. 2002. Molecular fossils in the human genome: identification and analysis of the pseudogenes in chromosomes 21 and 22. Genome Res. 12:272–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison PM, Milburn D, Zhang Z, Bertone P, Gerstein M.. 2003. Identification of pseudogenes in the Drosophila melanogaster genome. Nucleic Acids Res. 31:1033–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison PM, Zheng D, Zhang Z, Carriero N, Gerstein M.. 2005. Transcribed processed pseudogenes in the human genome: an intermediate form of expressed retrosequence lacking protein-coding ability. Nucleic Acids Res. 33:2374–2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Zhang J.. 2005. Rapid subfunctionalization accompanied by prolonged and substantial neofunctionalization in duplicate gene evolution. Genetics 169:1157–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YT, Chen FC, Chen CJ, Chen HL, Chuang TJ.. 2008. Identification and analysis of ancestral hominoid transcriptome inferred from cross-species transcript and processed pseudogene comparisons. Genome Res. 18:1163–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JF, Skaletsky H, Koutseva N, Pyntikova T, Page DC.. 2015. Sex chromosome-to-autosome transposition events counter Y-chromosome gene loss in mammals. Genome Biol. 16:104.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan H, Kondrashov F.. 2010. The evolution of gene duplications: classifying and distinguishing between models. Nat Rev Genet. 11:97–108. [DOI] [PubMed] [Google Scholar]
- International Chicken Genome Sequencing C. 2004. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432:695–716. [DOI] [PubMed] [Google Scholar]
- Irimia M, Roy SW.. 2014. Origin of spliceosomal introns and alternative splicing. Cold Spring Harb Perspect Biol. 6:1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakalski M, et al. 2016. Comparative genomic analysis of retrogene repertoire in two green algae Volvox carteri and Chlamydomonas reinhardtii. Biol Direct. 11:35.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin YK, Bennetzen JL.. 1994. Integration and nonrandom mutation of a plasma membrane proton ATPase gene fragment within the Bs1 retroelement of maize. Plant Cell 6:1177–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CD, Begun DJ.. 2005. Parallel evolution of chimeric fusion genes. Proc Natl Acad Sci U S A. 102:11373–11378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun J, Ryvkin P, Hemphill E, Mandoiu I, Nelson C.. 2009. The birth of new genes by RNA- and DNA-mediated duplication during mammalian evolution. J Comput Biol. 16:1429–1444. [DOI] [PubMed] [Google Scholar]
- Kabza M, Ciomborowska J, Makalowska I.. 2014. RetrogeneDB–a database of animal retrogenes. Mol Biol Evol. 31:1646–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaessmann H. 2010. Origins, evolution, and phenotypic impact of new genes. Genome Res. 20:1313–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaessmann H, Vinckenbosch N, Long M.. 2009. RNA-based gene duplication: mechanistic and evolutionary insights. Nat Rev Genet. 10:19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang LF, Zhu ZL, Zhao Q, Chen LY, Zhang Z.. 2012. Newly evolved introns in human retrogenes provide novel insights into their evolutionary roles. BMC Evol Biol. 12:128.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khelifi A, Duret L, Mouchiroud D.. 2005. HOPPSIGEN: a database of human and mouse processed pseudogenes. Nucleic Acids Res. 33:D59–D66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasnov AN, et al. 2005. A retrocopy of a gene can functionally displace the source gene in evolution. Nucleic Acids Res. 33:6654–6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landeen EL, Muirhead CA, Wright L, Meiklejohn CD, Presgraves DC.. 2016. Sex chromosome-wide transcriptional suppression and compensatory cis-regulatory evolution mediate gene expression in the Drosophila male germline. PLoS Biol. 14:e1002499.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409:860–921. [DOI] [PubMed] [Google Scholar]
- Levin HL, Moran JV.. 2011. Dynamic interactions between transposable elements and their hosts. Nat Rev Genet. 12:615–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, et al. 2016. Mice carrying a human GLUD2 gene recapitulate aspects of human transcriptome and metabolome development. Proc Natl Acad Sci U S A. 113:5358–5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, et al. 2016. Evolutionary interplay between sister cytochrome P450 genes shapes plasticity in plant metabolism. Nat Commun. 7:13026.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M, Betran E, Thornton K, Wang W.. 2003. The origin of new genes: glimpses from the young and old. Nat Rev Genet. 4:865–875. [DOI] [PubMed] [Google Scholar]
- Long M, Langley CH.. 1993. Natural selection and the origin of jingwei, a chimeric processed functional gene in Drosophila. Science 260:91–95. [DOI] [PubMed] [Google Scholar]
- Loppin B, Lepetit D, Dorus S, Couble P, Karr TL.. 2005. Origin and neofunctionalization of a Drosophila paternal effect gene essential for zygote viability. Curr Biol. 15:87–93. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS.. 2003. The origins of genome complexity. Science 302:1401–1404. [DOI] [PubMed] [Google Scholar]
- Maestre J, Tchenio T, Dhellin O, Heidmann T.. 1995. mRNA retroposition in human cells: processed pseudogene formation. EMBO J. 14:6333–6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal PK, Kazazian HH. Jr.. 2016. Purification of L1-ribonucleoprotein particles (L1-RNPs) from cultured human cells. Methods Mol Biol. 1400:299–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmoset Genome S, Analysis C.. 2014. The common marmoset genome provides insight into primate biology and evolution. Nat Genet. 46:850–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques AC, Dupanloup I, Vinckenbosch N, Reymond A, Kaessmann H.. 2005. Emergence of young human genes after a burst of retroposition in primates. PLoS Biol. 3:e357.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuno M, et al. 2009. Evolution of a novel phenolic pathway for pollen development. Science 325:1688–1692. [DOI] [PubMed] [Google Scholar]
- McCarrey JR. 1990. Molecular evolution of the human Pgk-2 retroposon. Nucleic Acids Res. 18:949–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarrey JR, Thomas K.. 1987. Human testis-specific PGK gene lacks introns and possesses characteristics of a processed gene. Nature 326:501–505. [DOI] [PubMed] [Google Scholar]
- McLysaght A. 2008. Evolutionary steps of sex chromosomes are reflected in retrogenes. Trends Genet. 24:478–481. [DOI] [PubMed] [Google Scholar]
- Meiklejohn CD, Tao Y.. 2010. Genetic conflict and sex chromosome evolution. Trends Ecol Evol. 25:215–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel RP, Han MV, Hahn MW.. 2009. A complex suite of forces drives gene traffic from Drosophila X chromosomes. Genome Biol Evol. 1:176–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mighell AJ, Smith NR, Robinson PA, Markham AF.. 2000. Vertebrate pseudogenes. FEBS Lett. 468:109–114. [DOI] [PubMed] [Google Scholar]
- Mourier T, Jeffares DC.. 2003. Eukaryotic intron loss. Science 300:1393.. [DOI] [PubMed] [Google Scholar]
- Navarro FC, Galante PA.. 2015. A genome-wide landscape of retrocopies in primate genomes. Genome Biol Evol. 7:2265–2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro FC, Galante PA.. 2013. RCPedia: a database of retrocopied genes. Bioinformatics 29:1235–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisole S, Lynch C, Stoye JP, Yap MW.. 2004. A Trim5-cyclophilin A fusion protein found in owl monkey kidney cells can restrict HIV-1. Proc Natl Acad Sci U S A. 101:13324–13328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen JD, et al. 2017. Expression of the human isoform of glutamate dehydrogenase, hGDH2, augments TCA cycle capacity and oxidative metabolism of glutamate during glucose deprivation in astrocytes. Glia 65:474–488. [DOI] [PubMed] [Google Scholar]
- Nozawa M, Aotsuka T, Tamura K.. 2005. A novel chimeric gene, siren, with retroposed promoter sequence in the Drosophila bipectinata complex. Genetics 171:1719–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. 1970. Evolution by gene duplication. New York: Springer. [Google Scholar]
- Ohshima K. 2013. RNA-mediated gene duplication and retroposons: retrogenes, LINEs, SINEs, and sequence specificity. Int J Evol Biol. 2013:424726.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima K, et al. 2003. Whole-genome screening indicates a possible burst of formation of processed pseudogenes and Alu repeats by particular L1 subfamilies in ancestral primates. Genome Biol. 4:R74.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima K, Igarashi K.. 2010. Inference for the initial stage of domain shuffling: tracing the evolutionary fate of the PIPSL retrogene in hominoids. Mol Biol Evol. 27:2522–2533. [DOI] [PubMed] [Google Scholar]
- Ostertag EM, Kazazian HH. Jr.. 2001. Biology of mammalian L1 retrotransposons. Annu Rev Genet. 35:501–538. [DOI] [PubMed] [Google Scholar]
- Panchy N, Lehti-Shiu M, Shiu SH.. 2016. Evolution of gene duplication in plants. Plant Physiol. 171:2294–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlicek A, Gentles AJ, Paces J, Paces V, Jurka J.. 2006. Retroposition of processed pseudogenes: the impact of RNA stability and translational control. Trends Genet. 22:69–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegueroles C, Laurie S, Alba MM.. 2013. Accelerated evolution after gene duplication: a time-dependent process affecting just one copy. Mol Biol Evol. 30:1830–1842. [DOI] [PubMed] [Google Scholar]
- Pei B, et al. 2012. The GENCODE pseudogene resource. Genome Biol. 13:R51.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phadnis N, Hsieh E, Malik HS.. 2012. Birth, death, and replacement of karyopherins in Drosophila. Mol Biol Evol. 29:1429–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickeral OK, Makalowski W, Boguski MS, Boeke JD.. 2000. Frequent human genomic DNA transduction driven by LINE-1 retrotransposition. Genome Res. 10:411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliseno L, et al. 2010. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 465:1033–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potrzebowski L, Vinckenbosch N, Kaessmann H.. 2010. The emergence of new genes on the young therian X. Trends Genet. 26:1–4. [DOI] [PubMed] [Google Scholar]
- Potrzebowski L, et al. 2008. Chromosomal gene movements reflect the recent origin and biology of therian sex chromosomes. PLoS Biol. 6:e80.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyne S, Skiena S, Futcher B.. 2005. Copy correction and concerted evolution in the conservation of yeast genes. Genetics 170:1501–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quezada-Diaz JE, Muliyil T, Rio J, Betran E.. 2010. Drcd-1 related: a positively selected spermatogenesis retrogene in Drosophila. Genetica 138:925–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson SR, Salvador-Palomeque C, Faulkner GJ.. 2014. Diversity through duplication: whole-genome sequencing reveals novel gene retrocopies in the human population. Bioessays 36:475–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richler C, Soreq H, Wahrman J.. 1992. X inactivation in mammalian testis is correlated with inactive X-specific transcription. Nat Genet. 2:192–195. [DOI] [PubMed] [Google Scholar]
- Rohozinski J, Bishop CE.. 2004. The mouse juvenile spermatogonial depletion (jsd) phenotype is due to a mutation in the X-derived retrogene, mUtp14b. Proc Natl Acad Sci U S A. 101:11695–11700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohozinski J, Lamb DJ, Bishop CE.. 2006. UTP14c is a recently acquired retrogene associated with spermatogenesis and fertility in man. Biol Reprod. 74:644–651. [DOI] [PubMed] [Google Scholar]
- Rooney AP, Ward TJ.. 2005. Evolution of a large ribosomal RNA multigene family in filamentous fungi: birth and death of a concerted evolution paradigm. Proc Natl Acad Sci U S A. 102:5084–5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso L, Marques AC, Reichert AS, Kaessmann H.. 2008. Mitochondrial targeting adaptation of the hominoid-specific glutamate dehydrogenase driven by positive Darwinian selection. PLoS Genet. 4:e1000150.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso L, Marques AC, Weier M, Lambert N, Lambot MA, Vanderhaeghen P, Kaessmann H.. 2008. Birth and rapid subcellular adaptation of a hominoid-specific CDC14 protein. PLoS Genet. 6:e140.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso L, et al. 2008. Birth and rapid subcellular adaptation of a hominoid-specific CDC14 protein. PLoS Biol. 6:e140.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai H, Koyanagi KO, Imanishi T, Itoh T, Gojobori T.. 2007. Frequent emergence and functional resurrection of processed pseudogenes in the human and mouse genomes. Gene 389:196–203. [DOI] [PubMed] [Google Scholar]
- Sakai H, et al. 2011. Retrogenes in rice (Oryza sativa L. ssp. japonica) exhibit correlated expression with their source genes. Genome Biol Evol. 3:1357–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena R, et al. 1996. The DAZ gene cluster on the human Y chromosome arose from an autosomal gene that was transposed, repeatedly amplified and pruned. Nat Genet. 14:292–299. [DOI] [PubMed] [Google Scholar]
- Sayah DM, Sokolskaja E, Berthoux L, Luban J.. 2004. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature 430:569–573. [DOI] [PubMed] [Google Scholar]
- Schrider DR, et al. 2013. Gene copy-number polymorphism caused by retrotransposition in humans. PLoS Genet. 9:e1003242.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrider DR, Stevens K, Cardeno CM, Langley CH, Hahn MW.. 2011. Genome-wide analysis of retrogene polymorphisms in Drosophila melanogaster. Genome Res. 21:2087–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She X, Cheng Z, Zollner S, Church DM, Eichler EE.. 2008. Mouse segmental duplication and copy number variation. Nat Genet. 40:909–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemesh R, Novik A, Edelheit S, Sorek R.. 2006. Genomic fossils as a snapshot of the human transcriptome. Proc Natl Acad Sci U S A. 103:1364–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares MB, et al. 1985. RNA-mediated gene duplication: the rat preproinsulin I gene is a functional retroposon. Mol Cell Biol. 5:2090–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorourian M, et al. 2014. Relocation facilitates the acquisition of short cis-regulatory regions that drive the expression of retrogenes during spermatogenesis in Drosophila. Mol Biol Evol. 31:2170–2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soumillon M, et al. 2013. Cellular source and mechanisms of high transcriptome complexity in the mammalian testis. Cell Rep. 3:2179–2190. [DOI] [PubMed] [Google Scholar]
- Suh A. 2015. The specific requirements for CR1 retrotransposition explain the scarcity of retrogenes in birds. J Mol Evol. 81:18–20. [DOI] [PubMed] [Google Scholar]
- Sulak M, et al. 2016. TP53 copy number expansion is associated with the evolution of increased body size and an enhanced DNA damage response in elephants. Elife 5:1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama M, Harrington E, Bork P, Torrents D.. 2006. Identification and analysis of genes and pseudogenes within duplicated regions in the human and mouse genomes. PLoS Comput Biol. 2:e76.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczesniak MW, Ciomborowska J, Nowak W, Rogozin IB, Makalowska I.. 2011. Primate and rodent specific intron gains and the origin of retrogenes with splice variants. Mol Biol Evol. 28:33–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam OH, et al. 2008. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature 453:534–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S, et al. 2016. LTR-mediated retroposition as a mechanism of RNA-based duplication in metazoans. Genome Res. 26:13–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchenio T, Segal-Bendirdjian E, Heidmann T.. 1993. Generation of processed pseudogenes in murine cells. EMBO J. 12:1487–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terai G, Yoshizawa A, Okida H, Asai K, Mituyama T.. 2010. Discovery of short pseudogenes derived from messenger RNAs. Nucleic Acids Res. 38:1163–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrents D, Suyama M, Zdobnov E, Bork P.. 2003. A genome-wide survey of human pseudogenes. Genome Res. 13:2559–2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toups MA, Hahn MW.. 2010. Retrogenes reveal the direction of sex-chromosome evolution in mosquitoes. Genetics 186:763–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toups MA, Pease JB, Hahn MW.. 2011. No excess gene movement is detected off the avian or lepidopteran Z chromosome. Genome Biol Evol. 3:1381–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tracy C, Rio J, Motiwale M, Christensen SM, Betran E.. 2010. Convergently recruited nuclear transport retrogenes are male biased in expression and evolving under positive selection in Drosophila. Genetics 184:1067–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JM. 2007. Meiotic sex chromosome inactivation. Development 134:1823–1831. [DOI] [PubMed] [Google Scholar]
- Vanin EF. 1985. Processed pseudogenes: characteristics and evolution. Annu Rev Genet. 19:253–272. [DOI] [PubMed] [Google Scholar]
- Vemuganti SA, de Villena FP, O’Brien DA.. 2010. Frequent and recent retrotransposition of orthologous genes plays a role in the evolution of sperm glycolytic enzymes. BMC Genomics 11:285.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vibranovski MD, Lopes HF, Karr TL, Long M.. 2009. Stage-specific expression profiling of Drosophila spermatogenesis suggests that meiotic sex chromosome inactivation drives genomic relocation of testis-expressed genes. PLoS Genet. 5:e1000731.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vibranovski MD, Zhang Y, Long M.. 2009. General gene movement off the X chromosome in the Drosophila genus. Genome Res. 19:897–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinckenbosch N, Dupanloup I, Kaessmann H.. 2006. Evolutionary fate of retroposed gene copies in the human genome. Proc Natl Acad Sci U S A. 103:3220–3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgen CA, Kratovac Z, Bieniasz PD, Hatziioannou T.. 2008. Independent genesis of chimeric TRIM5-cyclophilin proteins in two primate species. Proc Natl Acad Sci U S A. 105:3563–3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Long M, Vibranovski MD.. 2012. Retrogenes moved out of the z chromosome in the silkworm. J Mol Evol. 74:113–126. [DOI] [PubMed] [Google Scholar]
- Wang W, Brunet FG, Nevo E, Long M.. 2002. Origin of sphinx, a young chimeric RNA gene in Drosophila melanogaster. Proc Natl Acad Sci U S A. 99:4448–4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Zhang J, Alvarez C, Llopart A, Long M.. 2000. The origin of the Jingwei gene and the complex modular structure of its parental gene, yellow emperor, in Drosophila melanogaster. Mol Biol Evol. 17:1294–1301. [DOI] [PubMed] [Google Scholar]
- Wang W, et al. 2006. High rate of chimeric gene origination by retroposition in plant genomes. Plant Cell 18:1791–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Werren JH, Clark AG.. 2015. Genetic and epigenetic architecture of sex-biased expression in the jewel wasps Nasonia vitripennis and giraulti. Proc Natl Acad Sci U S A. 112:E3545–E3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Dong X, Ding G, Li Y.. 2010. Comparing the retention mechanisms of tandem duplicates and retrogenes in human and mouse genomes. Genet Sel Evol. 42:24.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren WC, et al. 2008. Genome analysis of the platypus reveals unique signatures of evolution. Nature 453:175–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Cheng EC, Zhong M, Lin H.. 2015. Retrotransposons and pseudogenes regulate mRNAs and lncRNAs via the piRNA pathway in the germline. Genome Res. 25:368–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, et al. 2008. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature 453:539–543. [DOI] [PubMed] [Google Scholar]
- Wei W, et al. 2001. Human L1 retrotransposition: cis preference versus trans complementation. Mol Cell Biol. 21:1429–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner AM, Deininger PL, Efstratiadis A.. 1986. Nonviral retroposons: genes, pseudogenes, and transposable elements generated by the reverse flow of genetic information. Annu Rev Biochem. 55:631–661. [DOI] [PubMed] [Google Scholar]
- Yu Z, Morais D, Ivanga M, Harrison PM.. 2007. Analysis of the role of retrotransposition in gene evolution in vertebrates. BMC Bioinformatics 8:308.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrei M, MacDonald JR, Merico D, Scherer SW.. 2015. A copy number variation map of the human genome. Nat Rev Genet. 16:172–183. [DOI] [PubMed] [Google Scholar]
- Zhang C, Gschwend AR, Ouyang Y, Long M.. 2014. Evolution of gene structural complexity: an alternative-splicing-based model accounts for intron-containing retrogenes. Plant Physiol. 165:412–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Dean AM, Brunet F, Long M.. 2004. Evolving protein functional diversity in new genes of Drosophila. Proc Natl Acad Sci U S A. 101:16246–16250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang LP, Stroud J, Eddy CA, Walter CA, McCarrey JR.. 1999. Multiple elements influence transcriptional regulation from the human testis-specific PGK2 promoter in transgenic mice. Biol Reprod. 60:1329–1337. [DOI] [PubMed] [Google Scholar]
- Zhang Q. 2013. The role of mRNA-based duplication in the evolution of the primate genome. FEBS Lett. 587:3500–3507. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wu Y, Liu Y, Han B.. 2005. Computational identification of 69 retroposons in Arabidopsis. Plant Physiol. 138:935–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YE, Vibranovski MD, Krinsky BH, Long M.. 2010. Age-dependent chromosomal distribution of male-biased genes in Drosophila. Genome Res. 20:1526–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YE, Vibranovski MD, Krinsky BH, Long M.. 2011. A cautionary note for retrocopy identification: DNA-based duplication of intron-containing genes significantly contributes to the origination of single exon genes. Bioinformatics 27:1749–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Carriero N, Gerstein M.. 2004. Comparative analysis of processed pseudogenes in the mouse and human genomes. Trends Genet. 20:62–67. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Harrison PM, Liu Y, Gerstein M.. 2003. Millions of years of evolution preserved: a comprehensive catalog of the processed pseudogenes in the human genome. Genome Res. 13:2541–2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D, Gerstein MB.. 2007. The ambiguous boundary between genes and pseudogenes: the dead rise up, or do they? Trends Genet. 23:219–224. [DOI] [PubMed] [Google Scholar]
- Zhong Z, Yang L, Zhang YE, Xue Y, He S.. 2016. Correlated expression of retrocopies and parental genes in zebrafish. Mol Genet Genomics 291:723–737. [DOI] [PubMed] [Google Scholar]
- Zhou K, Zou M, Duan M, He S, Wang G.. 2015. Identification and analysis of retrogenes in the East Asian nematode Caenorhabditis sp. 5 genome. Genome 58:349–355. [DOI] [PubMed] [Google Scholar]
- Zhou Q, et al. 2008. On the origin of new genes in Drosophila. Genome Res. 18:1446–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Tan S, Zhang Y, Zhang YE.. 2016. LINE-1-like retrotransposons contribute to RNA-based gene duplication in dicots. Sci Rep. 6:24755.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Zhang Y, Long M.. 2009. Extensive structural renovation of retrogenes in the evolution of the Populus genome. Plant Physiol. 151:1943–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zingler N, et al. 2005. Analysis of 5′ junctions of human LINE-1 and Alu retrotransposons suggests an alternative model for 5′-end attachment requiring microhomology-mediated end-joining. Genome Res. 15:780–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou M, Wang G, He S.. 2012. Evolutionary patterns of RNA-based gene duplicates in Caenorhabditis nematodes coincide with their genomic features. BMC Res Notes 5:398.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.