Alternative splicing in chromatin-modifying genes is associated with temperature-dependent sex in divergent reptile lineages.
Keywords: retained intron, JARID2, JMJD3, PRC2 complex, POMC-mediated stress, sex reversal
Abstract
In many vertebrates, sex of offspring is determined by external environmental cues rather than by sex chromosomes. In reptiles, for instance, temperature-dependent sex determination (TSD) is common. Despite decades of work, the mechanism by which temperature is converted into a sex-determining signal remains mysterious. This is partly because it is difficult to distinguish the primary molecular events of TSD from the confounding downstream signatures of sexual differentiation. We use the Australian central bearded dragon, in which chromosomal sex determination is overridden at high temperatures to produce sex-reversed female offspring, as a unique model to identify TSD-specific features of the transcriptome. We show that an intron is retained in mature transcripts from each of two Jumonji family genes, JARID2 and JMJD3, in female dragons that have been sex-reversed by temperature but not in normal chromosomal females or males. JARID2 is a component of the master chromatin modifier Polycomb Repressive Complex 2, and the mammalian sex-determining factor SRY is directly regulated by an independent but closely related Jumonji family member. We propose that the perturbation of JARID2/JMJD3 function by intron retention alters the epigenetic landscape to override chromosomal sex-determining cues, triggering sex reversal at extreme temperatures. Sex reversal may then facilitate a transition from genetic sex determination to TSD, with JARID2/JMJD3 intron retention preserved as the decisive regulatory signal. Significantly, we also observe sex-associated differential retention of the equivalent introns in JARID2/JMJD3 transcripts expressed in embryonic gonads from TSD alligators and turtles, indicative of a reptile-wide mechanism controlling TSD.
INTRODUCTION
Since the initial discovery of vertebrate temperature-dependent sex determination (TSD) in 1966 (1), the mechanism by which temperature exerts its influence on sexual development has been extensively investigated (1–6). Because males and females in a TSD species have the same chromosome complement, sexual differentiation must be accomplished by differential regulation of common genes, rather than by the presence, absence, or dosage of a master sex-determining gene (7). Temperature may exert its influence at any point in the developmental cascade during which the embryo remains sexually labile (7, 8).
The Australian central bearded dragon (Pogona vitticeps) is a uniquely powerful model with which to disentangle primary from emergent molecular events associated with TSD. Dragons are exceptional for the fact that genotype (a ZW/ZZ sex chromosome system) interacts with environmental temperature to determine the fate of sexual differentiation (9, 10). ZW embryos develop as female, and ZZ embryos develop as male (11). However, at high incubation temperatures, this system is overridden so that both ZZ and ZW embryos develop as female (Fig. 1A) (9, 10). The mating of a normal ZZ male dragon with a sex-reversed ZZ female necessarily yields ZZ offspring, whose sex is determined solely by incubation temperature (9). Because female development can be specified independently by temperature or sex chromosome complement, it is possible to distinguish the effects of temperature and genotype on sex determination (Fig. 1A).
Fig. 1. Comparison of global gene expression profiles for normal and sex-reversed dragons.
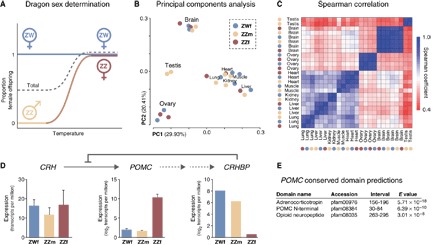
(A) In dragons, ZW embryos develop as female (ZWf; blue) regardless of environmental temperature. ZZ embryos develop as male (ZZm; yellow) at low temperatures and as sex-reversed females (ZZf; red) at high temperatures. Because female development can be specified independently by temperature or sex chromosome complement, it is possible to distinguish the effects of temperature and genotype on sex determination. (B) Principal components analysis and (C) Spearman ranked correlation clustering of global gene expression profiles for adult tissues from ZWf, ZZm, and ZZf individuals. (D) Top: Expression of POMC is activated by CRH (13), and CRH is inhibited by CRHBP (15). Bottom: Normalized expression (transcripts per million; mean ± SD; n = 2) for CRH (brain), POMC (brain), and CRHBP (liver) in ZWf, ZZm, and ZZf tissues. (E) Conserved domain prediction (protein BLAST) for the expressed sequence of POMC in dragon.
Here, we show that sex-reversed female dragons are distinguished transcriptionally from normal chromosomal females and males by (i) up-regulation of the Proopiomelanocortin (POMC)–mediated environmental stress response and (ii) the preferential retention of introns in mRNA transcribed from each of two Jumonji family genes, JARID2 and JMJD3. The retained intron sequences contain premature stop codons and are very likely to alter or abolish the function of these important epigenetic regulators. The same Jumonji introns are also differentially retained between embryonic gonads developing at either female-producing temperature (FPT) or male-producing temperature (MPT) in both alligator and turtle, which is suggestive of an ancient mechanism that may control reptile TSD.
RESULTS
To identify changes in gene expression and/or alternative splicing associated with sex reversal and thus TSD, we analyzed RNA sequencing data from adult gonad, brain, and other somatic tissues (liver, kidney, lung, heart, and muscle) in normal female (ZWf), male (ZZm), and sex-reversed female (ZZf) dragons (table S1).
We first generated global gene expression profiles and clustered these according to principal components analysis (Fig. 1B) and Spearman ranked correlation (Fig. 1C). Both methods grouped samples according to the tissue of origin, rather than sex or genotype. ZWf and ZZf ovary samples clustered separately from testis or somatic tissues but were as similar to each other as replicate samples from within each genotype (Fig. 1, B and C). Hence, all tissues, including ovaries, in sex-reversed females appear transcriptionally normal.
To find individual genes whose expression deviated reproducibly from these otherwise ordinary profiles, we performed differential gene expression analyses. By first comparing normal ZZm and ZWf dragons, we observed a previously unreported male bias in the expression of canonical muscle genes, including Actin (ACTA1; 44-fold) and Myosin (MYH1; 155-fold), in the brain (fig. S1, A to E). However, ZZf individuals exhibited normal female expression of these male-biased genes (fig. S1D). This indicates that, although they exhibit some “male-like” morphological and behavioral traits (12), sex-reversed females are not necessarily male-like in their molecular phenotypes.
Of greater relevance, ZZf brain displayed unique transcriptomic features that could be involved in TSD. Seventeen genes were classified as differentially expressed between ZZf and ZWf brains (fig. S2, A to D). Prominent among these were predicted orthologs for POMC, which is an environmental stress gene (13), and chromatin modifiers from the Jumonji family (14).
POMC was substantially overexpressed (327-fold) in the brain of ZZf, relative to ZWf or ZZm (Fig. 1D). POMC is a pituitary-expressed gene that encodes the precursor of the peptide hormone adrenocorticotropin (ACTH) (13), which is conserved in dragon (E < 1 × 10−17; Fig. 1E). ACTH stimulates synthesis and secretion of glucocorticoids from the adrenal cortex, including cortisol (corticosterone in birds and reptiles), which is the central effector hormone of the vertebrate stress response (13). POMC expression is activated by corticotropin-releasing hormone (CRH) (13) and modulated by negative feedback from inhibitory CRH-binding protein (CRHBP) (15). Although there was no difference in the expression of CRH between ZZf and ZWf dragons, CRHBP expression was reduced (262-fold) in ZZf liver, the major site of its synthesis (Fig. 1D) (15). This is consistent with a reduction in negative feedback on POMC activation, leading to its overexpression in sex-reversed females.
Because the immune and circadian systems are known to be intertwined with stress (16), it was notable that expression levels of prominent immune genes, such as IgM and IRF1, were significantly lower in ZZf dragons than ZWf or ZZm, whereas the circadian regulator, CIART, was overexpressed (fig. S2D). Moreover, canonical stress–related gene ontology terms, such as “defense response” and “response to biotic stimuli,” were enriched among the top 500 genes whose expression varied between ZZf and ZWf dragons (fig. S2F).
There is considerable evidence connecting stress and sex in vertebrates. Cortisol is a regulator of natural sex change in sequential hermaphrodite fish, which change sex in response to environmental and/or social stress (17). In birds, elevated maternal corticosterone skews the sex ratio of offspring (18), as does its presence in the yolk of two Australian TSD lizard species (19). An influence of maternal ACTH levels on sex ratio has been observed in rodents (20), and there is even evidence that maternal stress biases human sex ratios (21). Thus, the strong, potentially constitutive, activation of POMC-mediated stress observed in sex-reversed females may represent a mechanism by which environmental temperature interacts with sexual fate.
The most interesting gene identified in our differential expression analysis was the predicted ortholog for JARID2, which was expressed more highly in ZZf than ZWf tissues (fig. S2D and Fig. 2A). JARID2 belongs to the Jumonji family, whose members have JmjN, JmjC histone demethylase, and AT-rich DNA binding (ARID) domains (14), all of which are conserved in dragon. Jumonji genes are a family of epigenetic regulators with diverse developmental roles (22).
Fig. 2. Association of JARID2/JMJD3 differential IR with temperature-dependent sex.
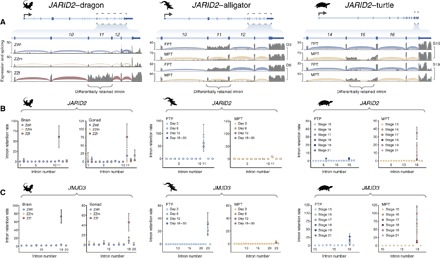
(A) Annotated gene models for the predicted ortholog of JARID2 in dragon (left), alligator (middle), and turtle (right). For dragon, normalized coverage by mapped RNA sequencing reads (gray) and density of spliced read junctions spanning annotated introns are shown for a single replicate of ZWf (blue), ZZm (yellow), and ZZf brain (red). For alligator, a single replicate of embryonic gonad after 3 and 6 days of incubation at FPT (blue) or MPT (yellow) is shown. For turtle, a single replicate of embryonic gonad at developmental stages 16 and 19 at FPT (blue) or MPT (yellow) is shown. Note that the section of zero coverage in the center of the retained intron for turtle is a string of undefined (N) bases in the genome, to which reads cannot be mapped. (B and C) Normalized rates of IR (retained transcript fragments/kilobase (kb)/spliced intron junctions; mean ± SD) are shown for individual introns in JARID2 (B) and JMJD3 (C) in ZWf, ZZm, and ZWf dragons (brain and gonad; n = 2), alligator embryonic gonad (FPT and MPT; days 3, 6, 12, and 18 to 30; n = 3), and turtle embryonic gonad (FPT and MPT; stages 15 to 19 and 21; n = 2).
In addition to its overexpression, the JARID2 locus produced a unique alternative transcript in ZZf dragons. The 11th intron of JARID2 was retained in nearly all transcripts from ZZf tissues, including the brain and gonad. In contrast, JARID2 transcripts from ZWf or ZZm dragons were spliced normally, with no sign of retention of the 11th intron (Fig. 2, A and B, and fig. S3).
Differential intron retention (IR) has recently been established as an important mode of gene regulation in eukaryotes (23). Premature stop codons within a retained intron sequence trigger mRNA degradation, inhibiting or abolishing protein expression independently of transcription levels (23). Multiple stop codons occur within the 11th intron of JARID2 (Fig. 3, A and B), so its retention should lead to transcript degradation. However, its high relative abundance indicates that degradation does not occur. If translated, the intron-retaining isoforms would produce truncated products, lacking the zinc finger DNA binding domain that is encoded downstream of intron 11 (Fig. 3C). Alternatively, specific detention of intron-retaining isoforms in the nucleus might prevent translation but protect them from degradation in the cytoplasm (24, 25).
Fig. 3. JARID2 retained intron creates premature stop.
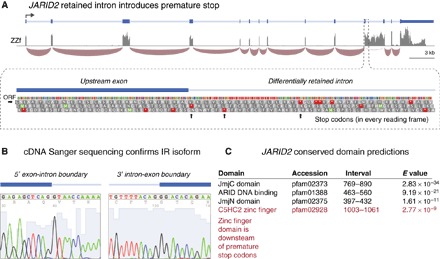
(A) Differential IR may regulate gene expression/function via the inclusion of premature stop codons within the retained intron sequence. The browser graphic shows multiple stop codons (in every reading frame) encoded within the sequence of JARID2 intron 11, which is retained in ZZf but not ZWf or ZZm dragons. ORF, open reading frame. (B) Intron-retaining isoform was validated by Sanger sequencing on complementary DNA (cDNA) using primer pairs that spanned each exon-intron boundary. (C) Conserved domain predictions (BLASTp) for the expressed JARID2 transcript from dragon included JmjN, JmjC histone demethylase, and ARID domains that characterize Jumonji family genes. The predicted zinc finger DNA binding domain is encoded downstream of intron 11 and will be deleted from a possible protein product.
The association of JARID2 differential IR with sex reversal is particularly interesting because JARID2 is a component of the master epigenetic regulator Polycomb Repressive Complex 2 (PRC2) (26, 27), in which it fulfills essential roles in tissue transdifferentiation (27) and mammalian X-chromosome inactivation (28). JARID2 appears to encourage efficient binding of PRC2 to target loci, promoting their silencing (26–28).
Wider comparisons suggest that differential IR in JARID2/JMJD3 transcripts might be associated with TSD in all reptiles. JARID2 was recently found to be differentially expressed in embryonic gonad developing at either FPT or MPT in alligator (Alligator mississippiensis) (4) and turtle (Trachemys scripta; both are TSD species) (5). We therefore interrogated RNA sequencing data from these studies (tables S2 and S3).
At FPT in alligator bipotential gonad, we found that the orthologous JARID2 intron was predominantly retained in mature transcripts, as we observed in sex-reversed ZZf dragons (Fig. 2, A and B). The same intron was predominantly spliced at MPT, similarly to normal ZWf or ZZm dragons (Fig. 2, A and B). This temperature-dependent distinction was apparent from day 3 of the experimental regime, wherein a subset of eggs was switched from FPT to MPT at day 0 (fig. S4A), and persisted throughout the entire 30-day period of comparison (Fig. 2B). JARID2 IR was also predominant at FPT at day 0, which corresponds to Ferguson developmental stage 19 (fig. S4, B and C). A change in splicing of JARID2 therefore occurred within 3 days of the temperature shift from FPT to MPT, roughly coinciding with the start of the temperature-sensitive period of development for alligator (29). JARID2 was also more highly expressed at FPT than MPT throughout the period of comparison (fig. S4, B and C).
Similarly, the orthologous JARID2 intron was differentially retained between FPT and MPT in turtle bipotential gonad. However, the directionality of the association was reversed; the intron was predominantly retained at MPT and excluded at FPT (Fig. 2, A and B). Differential IR was apparent from developmental stages 15 to 21 (Fig. 2B), encompassing the temperature-sensitive period for turtle (5). JARID2 was also more highly expressed at turtle MPT than FPT (fig. S4, B and C), again the opposite relationship to that observed in alligator and dragon. Whole embryos sampled at developmental stage 12, before the initiation of gonad development (fig. S4A), showed no difference in JARID2 splicing or expression between FPT and MPT (fig. S4, B and C).
Another member of the Jumonji family, JMJD3 (KDM6B) (14), was also reported to be up-regulated at alligator FPT relative to MPT (4) and vice versa in turtle (5). JMJD3 is an active histone demethylase and positively regulates gene expression (14). Strikingly, we found that JMJD3, just like JARID2, contained an intron that was preferentially retained at FPT in alligator and MPT in turtle (Fig. 2C). We therefore examined the predicted JMJD3 ortholog in dragon, revealing that this gene also exhibits sex reversal–specific IR (Fig. 2C) and is more highly expressed in ZZf individuals than ZWf or ZZm (fig. S5, A to C). The introns retained in JARID2 and JMJD3 are not paralogous to each other (fig. S6), implying that their capacity for differential IR does not derive from a shared sequence element.
We have therefore identified two sex reversal–specific and TSD-associated differential IR events that are expected to reduce, alter, or abolish the functionality of JARID2 and JMJD3 in at least three divergent reptile lineages (Fig. 4A). Although neither gene has known roles in sex determination, a closely related Jumonji family member, JMJD1A, is a direct regulator of the mammalian sex-determining factor SRY, and JMJD1A dysfunction causes male-to-female sex reversal in mice (30). Thus, we have identified two compelling candidate genes with the capacity to influence sex gene dosage and fulfill a sex-determining role.
Fig. 4. Model for role of JARID2/JMJD3 differential IR in temperature-dependent sex.
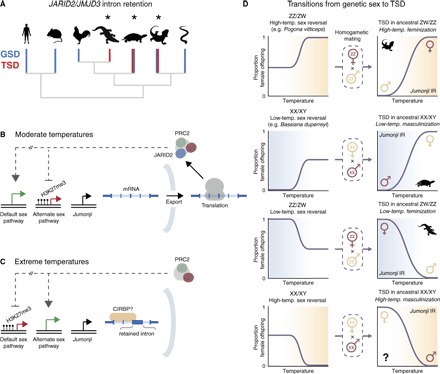
(A) Phylogenetic relationships of reptile lineages, coded for genetic sex determination (GSD; blue) and/or TSD (red), and occurrence of sex-associated JARID2/JMJD3 IR (asterisk). (B to D) The sex-determining action of JARID2/JMJD3 IR is to divert sexual differentiation from the ancestral homogametic state (the default path) to the alternate developmental pathway. (B) At moderate temperatures, JARID2/JMJD3 mRNA is spliced, exported, and translated. JARID2 facilitates recruitment of PRC2 to target genes, which are repressed via H3K27 trimethylation (26–28). JARID2 and/or JMJD3 may also directly activate genes via histone demethylation (14). (C) At extreme temperatures, introns are retained in JARID2/JMJD3 mRNA. IR may be induced by binding of CIRBP (6, 37) to JARID2/JMJD3 transcripts. IR isoforms are not exported or translated (24, 25). JARID2 is depleted from PRC2, reducing recruitment to target genes. Direct activation of genes by JARID2/JMJD3-catalyzed histone demethylation is also perturbed. The epigenetic landscape is thereby altered, repressing genes that maintain the default sex pathway and/or activating sex reversal genes. This can lead to feminization or masculinization, depending on the ancestral GSD system in a given species (8, 31, 32). (D) Four possible sex-associated patterns of IR may occur. Left: Ancestral GSD states (ZZ/ZW or XX/XY), with sex reversal at either high (beige) or low (blue) temperatures. Mating of sex-reversed and wild-type homogametic individuals causes transition to TSD (9, 30), with JARID2/JMJD3 IR maintained as the regulatory signal controlling differentiation. Right: Four TSD patterns emerge: female-specific IR at high temperatures, male-specific IR at low temperatures, female-specific IR at low temperatures, and male-specific IR at high temperatures.
DISCUSSION
Here, we propose a unified model for the molecular basis and evolution of TSD. We suggest that JARID2/JMJD3 differential IR forms the mechanistic link between environmental temperature and epigenetic state underpinning sex reversal and reptile TSD.
Because retention was favored at (i) FPT in alligator (in contrast to MPT in turtle) and (ii) high temperatures in dragon (in contrast to low temperatures in alligator/turtle), JARID2/JMJD3 IR is not simply associated with high or low incubation temperatures or with either the male or female phenotype. What occurs instead must be a complex interaction with the genetic background of each evolutionarily independent system of sex determination. As postulated previously (8, 31), we argue that some TSD species have evolved from female heterogametic sex determination systems (ZZ/ZW) and others have evolved from male heterogametic systems (XX/XY). Sex reversal can rapidly trigger the loss of the heterogametic sex chromosome and, thereby, a shift from genetic sex to TSD (9). This phenomenon has been observed in wild populations for lizard species with both ZZ/ZW (P. vitticeps) (9) and XX/XY (Bassiana duperreyi) (32) systems, triggered by high and low sex-reversing temperatures, respectively.
We suggest that the sex-determining action of JARID2/JMJD3 IR is to divert sexual differentiation from the ancestral homogametic state to the alternate developmental path, in response to extreme temperatures (Fig. 4, B and C). This model explains the variable relationships we observed between JARID2/JMJD3 IR, temperature, and sex among turtle, alligator, and dragon (Fig. 4D).
In our model (Fig. 4, B and C), JARID2/JMJD3 IR is favored at extreme temperatures. JARID2 is depleted from PRC2, reducing its recruitment to target genes. Direct activation of genes by JARID2/JMJD3-catalyzed histone demethylation (33, 34) is also perturbed. Under these conditions, we propose that the epigenetic landscape is modified, disturbing developmental canalization toward the ancestral homogametic sex (the “default” state) and thereby enabling sex reversal. This can lead to feminization or masculinization of the developing gonad, depending on the ancestral homogametic sex in a given species (Fig. 4D).
How might JARID2/JMJD3 differential IR relate to stress? Given the strong up-regulation of POMC-mediated stress that we observed in sex-reversed dragons, it is noteworthy that, in two other Australian TSD lizards, B. duperreyi and Amphibolurus muricatus, it is possible to interfere with sex ratio by manipulating yolk corticosterone levels (emulating maternal stress) during embryonic development (18). For these species, which have evolved from XX/XY and ZW/ZZ systems, respectively, corticosterone skewed sex ratio in opposing directions, consistent with the predictions of our model that accounts for different genetic sex backgrounds (Fig. 4D).
We therefore propose that POMC-mediated stress provides the temperature-sensitive signal that regulates JMJD3/JARID2 differential IR and thereby sex reversal. Consistent with this, expression of JARID2 and JMJD3 is regulated by stress in mammalian systems (34, 35). Cellular stresses, including heat shock, also directly modulate the efficiency of transcript splicing and can cause general or specific IR (24, 36). It is possible that stress-activated JARID2/JMJD3 IR, which appears to occur systemically, could have additional consequences, unrelated to sex determination, in other tissues or, alternatively, that this phenomenon simply lacks functional relevance outside the developing gonad. The response may also involve a gene previously implicated in TSD in turtles, CIRBP (6), which encodes a temperature-inducible RNA binding protein with imputed function in mRNA stability and translational regulation (37). It is plausible that IR could be induced by the binding of CIRBP to JARID2/JMJD3 mRNA during thermal stress.
Our identification of differential IR in genes critical for epigenetic regulation therefore provides another compelling link between the environmental stress and sex determination pathways. Definitive causal evidence for our proposed model will require the development of techniques for targeted gene manipulation and/or transcriptional perturbation, such as Tol2 constructs or CRISPR (clustered regularly interspaced short palindromic repeats)–Cas9 (CRISPR-associated protein 9), which have yet to be established in reptiles. Nevertheless, our observations open up new directions for investigations into the molecular mechanisms by which genes and environment interact to control reptile sex determination.
MATERIALS AND METHODS
Bearded dragon RNA sequencing libraries
We used paired-end RNA sequencing libraries from P. vitticeps, as published previously (38). Polyadenlyation-selected RNA libraries were derived from adult male (ZZm), female (ZWf), and sex-reversed female (ZZf) dragon tissues, including gonad (n = 2), brain (n = 2), and additional somatic tissues (liver, kidney, lung, heart, and muscle; n = 1). Raw sequencing reads were trimmed using TrimGalore with default parameters. After trimming and quality control (QC), a total of 386 million read pairs were retrieved (table S1). Raw libraries can be accessed under the European Nucleotide Archive study accession number PRJEB5206.
Alligator RNA sequencing libraries
We used paired-end RNA sequencing libraries from A. mississippiensis, as published previously (4). Total RNA libraries were derived from alligator embryonic gonad developing at either FPT (30°C) or MPT (33.5°C) over a 30-day time course (n = 3) (4). Raw sequencing reads were trimmed using TrimGalore with default parameters. After trimming and QC, a total of 324 million read pairs were retrieved (table S2). Raw libraries can be accessed using the DRA (DNA Databank of Japan Sequence Read Archive) accession number DRA004128-41.
Turtle RNA sequencing libraries
We used paired-end RNA sequencing libraries from T. scripta, as published previously (5). Total RNA libraries were derived from turtle embryonic gonad developing at either FPT (31°C) or MPT (26°C) at developmental stages 15 to 19 and 21 (n = 2) (5). Raw sequencing reads were trimmed using TrimGalore with default parameters. After trimming and QC, a total of 697 million sequenced read pairs were retrieved (table S3). Raw libraries can be accessed at the Sequence Read Archive under BioProject PRJNA331105.
Gene expression profiling
We used the recently assembled P. vitticeps genome (38) and accompanying gene annotation (http://gigadb.org/dataset/100166) to perform gene expression profiling, with expression values derived using Kallisto (39) alignment-free quantification over annotated coding sequences. Kallisto was used with a fixed k-mer length of 30 nucleotides. We excluded genes from downstream analyses that did not have a minimum measured abundance of ≥3 transcripts per million in multiple replicates from at least one tissue. A total of 17,818 genes fulfilled these criteria.
Comparisons of global expression profiles were performed in R. Spearman correlation matrices were generated with the corrplot package (https://cran.r-project.org/web/packages/corrplot/) and principal components analysis with the EDASeq package from Bioconductor (http://bioconductor.org/).
Differential gene expression analysis
The edgeR package from Bioconductor (http://bioconductor.org/) was used for differential gene expression analysis. A threshold of false discovery rate (FDR) < 0.01 was required for candidacy as a differentially expressed gene.
To verify that these candidates were robust to bioinformatics variables, we repeated gene quantification using a traditional alignment-based approach (as opposed to Kallisto alignment-free quantification; see above). We used a STAR-RSEM (40, 41) workflow with the following STAR-mapping parameters: --outSAMstrandField intronMotif, --outFilterIntronMotifs RemoveNoncanonicalUnannotated, --outReadsUnmapped None, --alignMatesGapMax 200000, and alignIntronMax 5000000. We also removed reads with a mapping quality score <10, as these represent potential multi-mappers.
Only genes that were concordant in their differential expression (FDR < 0.01) between both independent methods of quantification (Kallisto/STAR-RSEM) were reported as differentially expressed.
Intron retention
Normalized IR rates were assessed for all introns within JARID2 and JMJD3 using custom scripts. Our approach was analogous to that reported by Braunschweig et al. (42). Specifically, for a given intron in a given sample, the rate of retention was calculated as follows. (i) All split-mapped reads spanning the intron, with ends mapping exactly to its annotated splice junctions, were retrieved (spliced_junctions). (ii) Unspliced read fragments mapping within or overlapping (by at least 5 base pairs) the intron sequence were retrieved (retained_fragments). (iii) The IR rate was calculated as retained_fragments/intron_length (kb)/spliced_junctions. Retention rates were averaged between replicate samples and are reported as mean ± SD.
For alligator (A. mississippiensis) and turtle (T. scripta), IR was assessed in the same manner as described above. However, because no reference genome was available for T. scripta, we used the closely related Chrysemys picta draft genome and accompanying gene annotation (43).
Supplementary Material
Acknowledgments
We thank our colleagues J. Rasko and J. Wong for helpful discussions. We also thank W. Ruscoe for assisting with animal husbandry, A. Drew for providing technical support, and H. Johnston for providing aesthetic guidance. Funding: I.W.D. is supported by an Australian Postgraduate Award. Project funding was from Australian Research Council Discovery Grants DP110104377 and DP170101147 led by A.G. This research was conducted under appropriate approvals from the Victorian, New South Wales, and Queensland authorities and with approvals from the Animal Ethics Committee of the University of Canberra. Author contributions: I.W.D., C.E.H., and A.G. conceived the project. I.W.D. performed the analyses and prepared the figures. C.E.H. performed the breeding experiments, egg incubation, and molecular identification of sex reversal. J.B. performed the sequence validation of retained intron. I.W.D. conceived the molecular model. C.E.H. and A.G. conceived the evolutionary model. I.W.D. prepared the manuscript with contributions from C.E.H., J.B., J.A.M.G., P.D.W., J.S.M., and A.G. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors. All data have been published previously and are publicly available (tables S1 to S3).
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/6/e1700731/DC1
fig. S1. Comparison of gene expression in male and female dragons.
fig. S2. Comparison of gene expression in normal and sex-reversed female dragons.
fig. S3. Differential JARID2 IR in normal and sex-reversed dragons.
fig. S4. Temporal dynamics of JARID2/JMJD3 expression and splicing in alligator and turtle embryo.
fig. S5. Expression and splicing of JMJD3 in the brain and gonad from normal and sex-reversed dragons.
fig. S6. Differentially retained introns in JARID2/JMJD3 are nonparalogous.
table S1. RNA sequencing libraries for P. vitticeps.
table S2. RNA sequencing libraries for A. mississippiensis.
table S3. RNA sequencing libraries for T. scripta.
REFERENCES AND NOTES
- 1.Charnier M., Action of temperature on the sex ratio in the Agama agama (Agamidae, Lacertilia) embryo. C. R. Seances Soc. Biol. Fil. 160, 620–622 (1966). [PubMed] [Google Scholar]
- 2.Bull J. J., Vogt R. C., Temperature-dependent sex determination in turtles. Science 206, 1186–1188 (1979). [DOI] [PubMed] [Google Scholar]
- 3.Ferguson M. W., Joanen T., Temperature of egg incubation determines sex in Alligator mississippiensis. Nature 296, 850–853 (1982). [DOI] [PubMed] [Google Scholar]
- 4.Yatsu R., Miyagawa S., Kohno S., Parrott B. B., Yamaguchi K., Ogino Y., Miyakawa H., Lowers R. H., Shigenobu S., Guillette L. J. Jr., Iguchi T., RNA-seq analysis of the gonadal transcriptome during Alligator mississippiensis temperature-dependent sex determination and differentiation. BMC Genomics 17, 77 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Czerwinski M., Natarajan A., Barske L., Looger L. L., Capel B., A timecourse analysis of systemic and gonadal effects of temperature on sexual development of the red-eared slider turtle Trachemys scripta elegans. Dev. Biol. 421, 166–177 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Schroeder A. L., Metzger K. J., Miller A., Rhen T., A novel candidate gene for temperature-dependent sex determination in the common snapping turtle. Genetics 203, 557–571 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rhen T., Schroeder A., Molecular mechanisms of sex determination in reptiles. Sex. Dev. 4, 16–28 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Georges A., Ezaz T., Quinn A. E., Sarre S. D., Are reptiles predisposed to temperature-dependent sex determination? Sex. Dev. 4, 7–15 (2010). [DOI] [PubMed] [Google Scholar]
- 9.Holleley C. E., O’Meally D., Sarre S. D., Graves J. A. M., Ezaz T., Matsubara K., Azad B., Zhang X., Georges A., Sex reversal triggers the rapid transition from genetic to temperature-dependent sex. Nature 523, 79–82 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Quinn A. E., Georges A., Sarre S. D., Guarino F., Ezaz T., Graves J. A. M., Temperature sex reversal implies sex gene dosage in a reptile. Science 316, 411 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Ezaz T., Quinn A. E., Miura I., Sarre S. D., Georges A., Graves J. A. M., The dragon lizard Pogona vitticeps has ZZ/ZW micro-sex chromosomes. Chromosome Res. 13, 763–776 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Li H., Holleley C. E., Elphick M., Georges A., Shine R., The behavioural consequences of sex reversal in dragons. Proc. R. Soc. B Biol. Sci. 283, 1–7 (2016). [Google Scholar]
- 13.Cawley N. X., Li Z., Loh Y. P., 60 Years of POMC: Biosynthesis, trafficking, and secretion of pro-opiomelanocortin-derived peptides. J. Mol. Endocrinol. 56, T77–T97 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klose R. J., Kallin E. M., Zhang Y., JmjC-domain-containing proteins and histone demethylation. Nat. Rev. Genet. 7, 715–727 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Westphal N. J., Seasholtz A. F., CRH-BP: The regulation and function of a phylogenetically conserved binding protein. Front. Biosci. 11, 1878–1891 (2006). [DOI] [PubMed] [Google Scholar]
- 16.McLean M., Endocrinology, 5th edition - by L. J. DeGroot, J. L. Jameson (section authors: D. de Kretser, A. B. Grossman, J. C. Marshall, S. Melmed, J. T. Potts, and G. C. Weir). Intern. Med. J. 38, 682 (2008). [Google Scholar]
- 17.Todd E. V., Liu H., Muncaster S., Gemmell N. J., Bending Genders: The biology of natural sex change in fish. Sex. Dev. 10, 223–241 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Navara K. J., The role of steroid hormones in the adjustment of primary sex ratio in birds: Compiling the pieces of the puzzle. Integr. Comp. Biol. 53, 923–937 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Warner D. A., Radder R. S., Shine R., Corticosterone exposure during embryonic development affects offspring growth and sex ratios in opposing directions in two lizard species with environmental sex determination. Physiol. Biochem. Zool. 82, 363–371 (2009). [DOI] [PubMed] [Google Scholar]
- 20.Geiringer E., Effect of ACTH on sex ratio of the albino rat. Proc. Soc. Exp. Biol. Med. 106, 752–754 (1961). [DOI] [PubMed] [Google Scholar]
- 21.Obel C., Henriksen T. B., Secher N. J., Eskenazi B., Hedegaard M., Psychological distress during early gestation and offspring sex ratio. Hum. Reprod. 22, 3009–3012 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Landeira D., Fisher A. G., Inactive yet indispensable: The tale of Jarid2. Trends Cell Biol. 21, 74–80 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wong J. J.-L., Au A. Y. M., Ritchie W., Rasko J. E. J., Intron retention in mRNA: No longer nonsense: Known and putative roles of intron retention in normal and disease biology. BioEssays 38, 41–49 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Boutz P. L., Bhutkar A., Sharp P. A., Detained introns are a novel, widespread class of post-transcriptionally spliced introns. Genes Dev. 29, 63–80 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Graindorge A., Carre C., Gebauer F., Sex-lethal promotes nuclear retention of msl2 mRNA via interactions with the STAR protein HOW. Genes Dev. 27, 1421–1433 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaneko S., Bonasio R., Saldaña-Meyer R., Yoshida T., Son J., Nishino K., Umezawa A., Reinberg D., Interactions between JARID2 and noncoding RNAs regulate PRC2 recruitment to chromatin. Mol. Cell 53, 290–300 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanulli S., Justin N., Teissandier A., Ancelin K., Portoso M., Caron M., Michaud A., Lombard B., da Rocha S. T., Offer J., Loew D., Servant N., Wassef M., Burlina F., Gamblin S. J., Heard E., Margueron R., Jarid2 methylation via the PRC2 complex regulates H3K27me3 deposition during cell differentiation. Mol. Cell 57, 769–783 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.da Rocha S. T., Boeva V., Escamilla-Del-Arenal M., Ancelin K., Granier C., Matias N. R., Sanulli S., Chow J., Schulz E., Picard C., Kaneko S., Helin K., Reinberg D., Stewart A. F., Wutz A., Margueron R., Heard E., Jarid2 is implicated in the initial Xist-induced targeting of PRC2 to the inactive X chromosome. Mol. Cell 53, 301–316 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Lang J. W., Andrews H. V., Temperature-dependent sex determination in crocodilians. J. Exp. Zool. 270, 28–44 (1994). [Google Scholar]
- 30.Kuroki S., Matoba S., Akiyoshi M., Matsumura Y., Miyachi H., Mise N., Abe K., Ogura A., Wilhelm D., Koopman P., Nozaki M., Kanai Y., Shinkai Y., Tachibana M., Epigenetic regulation of mouse sex determination by the histone demethylase Jmjd1a. Science 341, 1106–1109 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Quinn A. E., Sarre S. D., Ezaz T., Marshall Graves J. A., Georges A., Evolutionary transitions between mechanisms of sex determination in vertebrates. Biol. Lett. 7, 443–448 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holleley C. E., Sarre S. D., O’Meally D., Georges A., Sex reversal in reptiles: Reproductive oddity or powerful driver of evolutionary change? Sex. Dev. 10, 279–287 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Herz H.-M., Shilatifard A., The JARID2–PRC2 duality. Genes Dev. 24, 857–861 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Williams K., Christensen J., Rappsilber J., Nielsen A. L., Johansen J. V., Helin K., The histone lysine demethylase JMJD3/KDM6B is recruited to p53 bound promoters and enhancer elements in a p53 dependent manner. PLOS ONE 9, e96545 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bovill E., Westaby S., Reji S., Sayeed R., Crisp A., Shaw T., Induction by left ventricular overload and left ventricular failure of the human Jumonji gene (JARID2) encoding a protein that regulates transcription and reexpression of a protective fetal program. J. Thorac. Cardiovasc. Surg. 136, 709–716 (2008). [DOI] [PubMed] [Google Scholar]
- 36.Shin C., Feng Y., Manley J. L., Dephosphorylated SRp38 acts as a splicing repressor in response to heat shock. Nature 427, 553–558 (2004). [DOI] [PubMed] [Google Scholar]
- 37.Nishiyama H., Danno S., Yokoi H., Fukumoto M., Decreased expression of cold-Inducible RNA-binding protein (CIRBP) in male germ cells at elevated temperature. Am. J. Pathol. 152, 289–296 (1998). [PMC free article] [PubMed] [Google Scholar]
- 38.Georges A., Li Q., Lian J., O’Meally D., Deakin J., Wang Z., Zhang P., Fujita M., Patel H. R., Holleley C. E., Zhou Y., Zhang X., Matsubara K., Waters P., Graves J. A. M., Sarre S. D., Zhang G., High-coverage sequencing and annotated assembly of the genome of the Australian dragon lizard Pogona vitticeps. GigaScience 4, 45 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bray N. L., Pimentel H., Melsted P., Pachter L., Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. R., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li B., Dewey C. N., RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Braunschweig U., Barbosa-Morais N. L., Pan Q., Nachman E. N., Alipanahi B., Gonatopoulos-Pournatzis T., Frey B., Irimia M., Blencowe B. J., Widespread intron retention in mammals functionally tunes transcriptomes. Genome Res. 24, 1774–1786 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shaffer H. B., Minx P., Warren D. E., Shedlock A. M., Thomson R. C., Valenzuela N., Abramyan J., Amemiya C. T., Badenhorst D., Biggar K. K., Borchert G. M., Botka C. W., Bowden R. M., Braun E. L., Bronikowski A. M., Bruneau B. G., Buck L. T., Capel B., Castoe T. A., Czerwinski M., Delehaunty K. D., Edwards S. V., Fronick C. C., Fujita M. K., Fulton L., Graves T. A., Green R. E., Haerty W., Hariharan R., Hernandez O., Hillier L. D. W., Holloway A. K., Janes D., Janzen F. J., Kandoth C., Kong L., de Koning A. P. J., Li Y., Literman R., McGaugh S. E., Mork L., O’Laughlin M., Paitz R. T., Pollock D. D., Ponting C. P., Radhakrishnan S., Raney B. J., Richman J. M., St John J., Schwartz T., Sethuraman A., Spinks P. Q., Storey K. B., Thane N., Vinar T., Zimmerman L. M., Warren W. C., Mardis E. R., Wilson R. K., The western painted turtle genome, a model for the evolution of extreme physiological adaptations in a slowly evolving lineage. Genome Biol. 14, R28 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/6/e1700731/DC1
fig. S1. Comparison of gene expression in male and female dragons.
fig. S2. Comparison of gene expression in normal and sex-reversed female dragons.
fig. S3. Differential JARID2 IR in normal and sex-reversed dragons.
fig. S4. Temporal dynamics of JARID2/JMJD3 expression and splicing in alligator and turtle embryo.
fig. S5. Expression and splicing of JMJD3 in the brain and gonad from normal and sex-reversed dragons.
fig. S6. Differentially retained introns in JARID2/JMJD3 are nonparalogous.
table S1. RNA sequencing libraries for P. vitticeps.
table S2. RNA sequencing libraries for A. mississippiensis.
table S3. RNA sequencing libraries for T. scripta.


