Abstract
Background
Hundreds of genetic variants are thought to contribute to variation in asthma risk by modulating gene expression. Methods that increase the power of genome wide association studies (GWAS) to identify risk-associated variants are needed.
Objective
To develop a method that aggregates the evidence for association with disease risk across expression quantitative trait loci (eQTLs) of a gene and use this approach to identify asthma risk genes.
Methods
We developed a gene-based test and software package called EUGENE that (1) is applicable to GWAS summary statistics; (2) considers both cis- and trans-eQTLs; (3) incorporates eQTLs identified in different tissues; and (4) uses simulations to account for multiple testing. We applied this approach to two published asthma GWAS (combined N=46,044) and used mouse studies to provide initial functional insights into two genes with novel genetic associations.
Results
We tested the association between asthma and 17,190 genes which were found to have cis-and/or trans-eQTLs across 16 published eQTL studies. At an empirical false discovery rate of 5%, 48 genes were associated with asthma risk. Of these, for 37 the association was driven by eQTLs located in established risk loci for allergic disease, including six genes not previously implicated in disease aetiology (eg. LIMS1, TINF2 and SAFB). The remaining 11 significant genes represent potential novel genetic associations with asthma. The association with four of these replicated in an independent GWAS: B4GALT3, USMG5, P2RY13 and P2RY14, which are genes involved in nucleotide synthesis or nucleotide-dependent cell activation. In mouse studies, P2ry13 and P2ry14 – purinergic receptors activated by ADP and UDP-sugars, respectively – were up-regulated after allergen challenge, notably in airway epithelial cells, eosinophils and neutrophils. Intranasal exposure with the receptor agonists induced the release of IL-33 and subsequent eosinophil infiltration into the lungs.
Conclusion
We identified novel associations between asthma and eQTLs for four genes related to nucleotide synthesis/signaling, and demonstrate the power of gene-based analyses of GWAS.
Keywords: Inflammation, eQTL, transcriptome, predisposition, obesity, EUGENE, VEGAS, PrediXcan, TWAS, ZNF707, AOAH, CLK3, UDP-glucose
INTRODUCTION
Asthma is a highly polygenic disease, with potentially hundreds or thousands of risk variants with small effects contributing to variation in disease risk1. A small number of risk-associated variants has been identified through genome-wide association studies (GWAS), but the majority remain to be mapped. Identifying risk-associated variants is important because these could point to genes that were not previously suspected to be involved in disease pathophysiology (eg.2, 3) or that could represent drug targets with greater probability of clinical success4, 5.
Several approaches have been proposed to increase the power of GWAS to identify variants with a modest but reproducible association with disease risk. These include larger sample sizes, the analysis of more refined phenotypes6, 7, multivariate association analysis of related phenotypes8, gene-based association analyses9, 10, and association analyses restricted to functional variants, such as those that regulate gene expression levels11. The aim of this study was to develop a method that combined the two latter approaches and apply it to results from a published asthma GWAS to help identify new genes whose expression was associated with genotype and related to disease risk.
Specifically, we hypothesized that if the expression of a gene is causally related to asthma, and gene expression is regulated by multiple independent expression quantitative trait loci (eQTLs), then a gene-based approach that captures the aggregate signals from these eQTLs would be expected to improve power over the alternative approach of testing each variant individually. Recently, Gamazon et al.12 described a gene-based association method based on the same concept, called PrediXcan. Briefly, this approach includes three steps: first, for a given gene, eQTLs are identified from transcriptome data sets. Second, a model that can be used to predict gene expression levels based on the aggregate effect of those eQTLs is trained on a reference transcriptome data set. And third, this model is used to infer expression levels for a target GWAS data set that includes individuals genotyped for those eQTLs but for whom actual gene expression levels might not be available. The genetically-inferred gene expression levels can then tested for association with the phenotype of interest (eg. asthma).
As highlighted by Gamazon et al.12, PrediXcan has several advantages over other gene-based tests, such as VEGAS9. However, in our view, it has one major limitation: unlike VEGAS, it is not applicable to GWAS summary statistics, which are typically more readily available, and therefore can be applied to a larger sample size than available GWAS data sets with individual-level genetic data. The TWAS approach developed by Gusev et al.13 addresses this caveat, but in its current release is applicable only to a relatively small number of genes (4,284 from two blood eQTL studies), cis- but not trans-acting eQTLs (eg. those located >1 Mb from the target gene), and to a single reference transcriptome dataset at a time.
In this study, we developed a gene-based association approach, called EUGENE, that combines the biological focus of PrediXcan and TWAS, and the versatility of VEGAS. Our approach also considers eQTL evidence across different tissues and estimates empirical false discovery rates (FDR), while accounting for the LD between variants. We applied this new approach to a published asthma GWAS14 to try to identify novel genes whose genetic component of gene expression is associated with asthma risk. Finally, we investigated whether results from mouse models of experimental acute allergic asthma are consistent with a contribution of two selected genes to disease pathophysiology.
METHODS
EUGENE approach
The proposed gene-based approach is described in detail in the Online Repository. Briefly, for a given gene, our approach includes four steps. First, we identify a set of variants that influence gene expression in any cell type or tissue relevant to the disease or trait of interest, based on results from published eQTL studies. Including eQTLs identified in tissues not thought to be relevant for the disease of interest might improve power, but this is something we did not consider in our study. We include in this list eQTLs located in cis (< 1 Mb from the target gene) or trans (> 1 Mb away or in a different chromosome). This list is then reduced to a sub-set of eQTLs with linkage disequilibrium (LD) r2<0.1; we refer to these as “independent eQTLs” for a given gene (see Figure E1 in this article’s Online Repository at www.jacionline.org). Second, we extract association results for these independent eQTLs from a disease or trait GWAS of interest and then calculate a gene-based statistic Q, as the sum of the 1-df chi-squares for the individual eQTLs. This represents the aggregate evidence for association in that GWAS across the independent eQTLs of that gene. Third, we perform simulations using individual-level genetic data to estimate the statistical significance of Q, while accounting for the residual LD between eQTLs. Fourth, false-discovery rate (FDR) thresholds are also estimated empirically to account for multiple testing. Simulations show that the type-I error rate of EUGENE is close to the nominal expectation (Table E1 in the Online Repository). The software and input files required to run EUGENE are freely available at https://genepi.qimr.edu.au/staff/manuelF.
Application of EUGENE to published GWAS of asthma
We applied EUGENE to a published asthma GWAS14 to illustrate the utility of the proposed approach. This GWAS included 6,685 individuals with both asthma and hayfever and 14,091 asthma- and hayfever-free controls, all of European descent, tested for association with 4.9 million SNPs with a minor allele frequency >1%. In the original analysis of individual SNPs, eleven independent variants were found to be associated with disease risk at a genome-wide significance level of P<3×10−8. We used EUGENE to identify genes with an association with disease risk in the Ferreira et al. study14 at an empirical FDR of 0.05 (corresponding to a P-value threshold of 1.9×10−4). At this FDR level, 5% of genes called significant (ie. with a P<1.9×10−4) are expected to be false-positive associations due to multiple testing. To confirm putative novel associations, we then applied EUGENE to an independent asthma GWAS, the GABRIEL study15, for which summary statistics are publicly available. After excluding overlapping samples (the Busselton study), results from the GABRIEL study were based on 9,967 asthmatics and 15,301 controls.
Predicted direction of effect of gene expression on asthma risk
EUGENE can be used to identify a set of genes with expression levels determined by eQTLs, and for which the eQTLs are collectively associated with disease risk. However, unlike PrediXcan12 or TWAS13, EUGENE does not directly provide the predicted direction of effect of gene expression on disease risk. To understand whether a genetically determined increase in gene expression levels was predicted to increase or decrease disease risk, we compared the direction of effect of each eQTL on gene expression reported on the transcriptome GWAS with the effect on asthma risk reported in the Ferreira et al.14 asthma GWAS. Based on this information, for each eQTL we report whether the allele associated with increased gene expression is associated with an increased or decreased asthma risk.
Functional studies in the mouse
We selected two putative novel asthma risk genes for preliminary functional studies in the mouse: P2RY13 and P2RY14. The criteria used to select these genes for functional follow-up were as follows: (1) significant gene-based association with asthma in the discovery GWAS at an empirical FDR of 5%; (2) the eQTLs that contribute to the significant gene-based association were in low LD (r2<0.1) with established asthma risk variants (those with a P<5×10−8 in published GWAS of asthma, hayfever, eczema and/or allergies); and (3) the gene-based association replicated (P<0.05) in an independent GWAS. Four genes satisfied all three criteria: P2RY13, P2RY14, USMG5 and B4GALT3. We prioritized the former two for follow-up because functional experiments were feasible with available tools/reagents (both are cell-surface receptors). We performed two sets of experiments, which are described in detail in the Online Repository and were performed in accordance with the Animal Care and Ethics Committees of the University of Queensland (Brisbane, Australia).
First, we used an established mouse model of acute allergic asthma16 to identify the cell types in the lung that express P2ry13 and P2ry14 in the context of allergen-induced airway inflammation. Two groups of wild-type C57Bl/6 mice were anesthetized and sensitized intranasally (i.n.) with either saline solution (group 1) or 100 μg of HDM extract on day 0. Subsequently, mice were challenged with either saline (group 1) or 5 μg of HDM (group 2) at day 14, 15, 16 and 17 and sacrificed 3 hours later. Total RNA was isolated from the left lung and quantitative real-time PCR performed to measure overall gene expression. To identify individual cell types in the lung expressing P2ry13 and P2ry14, bronchoalveolar lavage fluid (BALF) was collected and cells stained with anti-P2ry13 or anti-P2ry14antibodies. Cells were then stained with cell-type specific fluorescently labeled antibodies and enumerated using a BD LSR Fortessa cytometer. To assess expression in airway epithelial cells, paraffin-embedded lung sections were prepared as previously described17 and probed with anti-P2ry13 or anti-P2ry14 antibodies. Photomicrographs were taken at 400× and 1000× magnification at room temperature and acquired using Olympus Image Analysis Software.
We performed a second set of experiments to test the hypothesis that P2ry13 or P2ry14 receptor activation could influence the release of alarmins, such as IL-33, and contribute to airway inflammation. Naïve mice were inoculated i.n. with saline, 10 nM 2-methyl-ADP (P2ry13 agonist), 10 nM UDP-glucose (P2ry14 agonist) or 10 nM ATP (agonist for all P2ry receptors, except P2ry6 and P2ry14), all in 50 uL. For comparison, three additional groups of mice were inoculated with 100 ug of HDM, 100 ug of cockroach extract (Blattella germanica) or 25 ug of Alternaria alternata extract. Two hours post-challenge, BALF was collected as described above and IL-33 levels measured by ELISA. Seventy-two hours post-challenge, BALF was again collected to obtain immune cell counts and stained for flow cytometry as described above.
RESULTS
Application of EUGENE to results from a published asthma GWAS
We applied our proposed gene-based test of association to a published asthma GWAS14 including 6,685 cases and 14,091 controls to identify genes with eQTLs collectively associated with disease risk. We tested the association with 17,190 genes (Figure 1) which were found to have cis-eQTLs (N=13,557), trans-eQTLs (N=315) or both (N=3,318), across 16 published eQTL studies, representing 12 different cell types or tissues relevant to asthma (Table E2 in the Online Repository).
Figure 1.
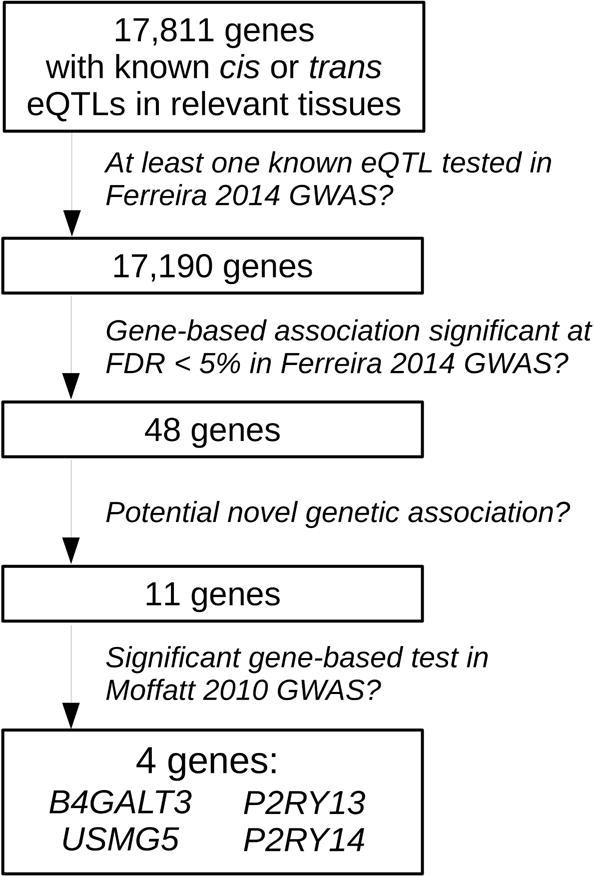
Outline of analytical procedure
Of the 17,190 genes tested, 48 genes were associated with asthma at an empirical FDR of 0.05 (Table 1 and Figure 2). Of these, 31 (65%) were located within 1 Mb (or on the MHC region) of established risk variants for allergic disease (highlighted with a ‘+’ in Figure 2 and listed in Table E3 in the Online Repository). For example, for TSLP18 (gene-based P=7×10−6), we identified six independent cis-eQTLs in five tissues, of which four were individually associated with asthma risk at P<0.05 (Table E4 in the Online Repository). Multiple genes within the same risk locus had significant associations with asthma: 12 in the MHC15; 11 on 17q122; three on 2q1215; and two on 16p1314. Some of these associations resulted from eQTLs being shared between neighboring genes, as observed for ORMDL3, GSDMB and ZPBP2 on 17q12 (Table E5 in the Online Repository), and for CLEC16A and SOCS1 on 16p13 (r2 between rs35441874 and rs7184491 [cf. Table E3] is 0.64). eQTL sharing could arise, for example, if an underlying causal variant disrupts the activity of a regulatory element that controls the expression of multiple genes. But that was not always the case; in the MHC region, the individual LTA eQTL that was most strongly associated with asthma (P=2×10−5) was in low LD (r2≤0.02) with the eQTLs for the other 11 significant MHC genes (Table E6 in the Online Repository). Similar results were observed for NEU1. Therefore, at least in the MHC region, the multiple significant associations observed were not entirely explained by eQTLs shared between genes.
Table 1.
Forty eight genes with a significant (FDR < 5%) association with asthma risk in the Ferreira et al.14 GWAS.
| Gene | Position | N eQTLs | N eQTLs tested | N eQTLs with P<0.05 | Best individual eQTL
|
EUGENE P-value | Potential novel associationa | |
|---|---|---|---|---|---|---|---|---|
| SNP | P-value | |||||||
| HLA-DQB1 | 6:32627244 | 78 | 26 | 15 | rs1063355 | 1.8 × 10−13 | <10−6 | No |
| GSDMB | 17:38060848 | 15 | 11 | 5 | rs2952140 | 1.2 × 10−8 | < 10−6 | No |
| LIMS1 | 2:109150857 | 15 | 14 | 4 | rs1063355 | 1.8 × 10−13 | < 10−6 | No* |
| TLR1 | 4:38792298 | 9 | 6 | 3 | rs12233670 | 1.4 × 10−11 | <10−6 | No |
| ORMDL3 | 17:38077294 | 19 | 14 | 5 | rs2952140 | 1.2 × 10−8 | <10−6 | No |
| IKZF3 | 17:37921198 | 9 | 7 | 3 | rs7207600 | 4.5 × 10−7 | < 10−6 | No |
| IL18RAP | 2:103035149 | 18 | 16 | 6 | rs13018263 | 5.0 × 10−6 | <10−6 | No |
| CLEC16A | 16:11038345 | 4 | 4 | 2 | rs35441874 | 2.9 × 10−8 | <10−6 | No |
| ZPBP2 | 17:38024417 | 2 | 2 | 1 | rs9916765 | 1.9 × 10−9 | <10−6 | No |
| GRB7 | 17:37894180 | 2 | 1 | 1 | rs14050 | 1.4 × 10−7 | <10−6 | No |
| TINF2 | 14:24708849 | 4 | 4 | 2 | rs3135006 | 1.7 × 10−6 | < 10−6 | No* |
| TAP2 | 6:32789610 | 48 | 36 | 11 | rs2858312 | 1.9 × 10−5 | 2.0 × 10−6 | No |
| TAP1 | 6:32812986 | 13 | 12 | 4 | rs6928482 | 2.0 × 10−8 | 2.0 × 10−6 | No |
| HSPA1B | 6:31795512 | 13 | 13 | 6 | rs13215091 | 4.7 × 10−4 | 2.0 × 10−6 | No |
| TSLP | 5:110405760 | 6 | 6 | 4 | rs17132582 | 3.2 × 10−4 | 2.0 × 10−6 | No |
|
| ||||||||
| DYNC1H1 | 14:102430865 | 6 | 5 | 2 | rs4906262 | 1.1 × 10−5 | 2.0 × 10−6 | Yes |
|
| ||||||||
| HLA-DRB1 | 6:32546546 | 97 | 46 | 18 | rs3806156 | 1.4 × 10−4 | 4.0 × 10−6 | No |
| IL18R1 | 2:102927989 | 11 | 11 | 5 | rs6751967 | 3.2 × 10−6 | 4.0 × 10−6 | No |
| SOCS1 | 16:11348262 | 7 | 6 | 5 | rs7184491 | 3.5 × 10−6 | 4.0 × 10−6 | No |
| CISD3 | 17:36886488 | 4 | 4 | 2 | rs2941503 | 1.6 × 10−7 | 5.0 × 10−6 | No |
| PGAP3 | 17:37827375 | 2 | 2 | 1 | rs903502 | 1.5 × 10−6 | 6.0 × 10−6 | No |
| IL1RL2 | 2:102803433 | 2 | 2 | 1 | rs9646944 | 6.7 × 10−7 | 9.0 × 10−6 | No |
| CLK3 | 15:74890841 | 2 | 2 | 1 | rs9268853 | 1.8 × 10−6 | 9.0 × 10−6 | No* |
| SMAD3 | 15:67356101 | 7 | 7 | 2 | rs17293632 | 2.0 × 10−7 | 1.1 × 10−5 | No |
| SAFB | 19:5623046 | 2 | 2 | 1 | rs9268853 | 1.8 × 10−6 | 1.1 × 10−5 | No* |
|
| ||||||||
| P2RY13 | 3:151044100 | 8 | 8 | 5 | rs9877416 | 1.2 × 10−4 | 1.2 × 10−5 | Yes |
|
| ||||||||
| AOAH | 7:36552456 | 26 | 21 | 3 | rs9268853 | 1.8 × 10−6 | 1.3 × 10−5 | No* |
| SLC44A4 | 6:31830969 | 2 | 2 | 1 | rs9275141 | 1.1 × 10−6 | 1.3 × 10−5 | No |
| STARD3 | 17:37793318 | 7 | 3 | 1 | rs2941503 | 1.6 × 10−7 | 1.5 × 10−5 | No |
| LTA | 6:31539831 | 16 | 13 | 5 | rs2442752 | 1.5 × 10−5 | 1.6 × 10−5 | No |
| MED24 | 17:38175350 | 5 | 5 | 2 | rs7502514 | 4.8 × 10−5 | 1.7 × 10−5 | No |
|
| ||||||||
| HIBADH | 7:27565061 | 14 | 12 | 6 | rs6951856 | 9.6 × 10−5 | 2.9 × 10−5 | Yes |
| P2RY12 | 3:151055168 | 6 | 6 | 4 | rs17282940 | 7.0 × 10−5 | 3.0 × 10−5 | Yes |
|
| ||||||||
| NR1D1 | 17:38249040 | 5 | 4 | 2 | rs12150298 | 2.8 × 10−6 | 3.0 × 10−5 | No |
| ZNF707 | 8:144766622 | 9 | 8 | 3 | rs17609240 | 1.5 × 10−6 | 4.2 × 10−5 | No* |
| TOP2A | 17:38544768 | 1 | 1 | 1 | rs2102928 | 4.1 × 10−5 | 4.9 × 10−5 | No |
| HLA-DRB6 | 6:32520490 | 60 | 13 | 6 | rs522254 | 6.3 × 10−4 | 5.4 × 10−5 | No |
|
| ||||||||
| REEP3 | 10:65281123 | 6 | 4 | 2 | rs7898489 | 9.1 × 10−6 | 6.1 × 10−5 | Yes |
| PTCSC3 | 14:36605314 | 2 | 2 | 2 | rs7148603 | 1.8 × 10−4 | 6.5 × 10−5 | Yes |
| P2RY14 | 3:150929905 | 13 | 12 | 5 | rs10513393 | 1.1 × 10−4 | 7.2 × 10−5 | Yes |
|
| ||||||||
| HLA-DQA1 | 6:32595956 | 79 | 29 | 9 | rs504594 | 1.7 × 10−5 | 8.1 × 10−5 | No |
|
| ||||||||
| ACO2 | 22:41865129 | 3 | 3 | 2 | rs960596 | 1.3 × 10−4 | 9.4 × 10−5 | Yes |
|
| ||||||||
| HCP5 | 6:31368479 | 23 | 22 | 4 | rs2071595 | 6.7 × 10−6 | 9.6 × 10−5 | No |
| NEU1 | 6:31825436 | 8 | 8 | 5 | rs9267901 | 9.1 × 10−4 | 9.8 × 10−5 | No |
| MICB | 6:31462658 | 41 | 30 | 9 | rs9268764 | 3.3 × 10−5 | 1.2 × 10−4 | No |
|
| ||||||||
| B4GALT3 | 1:161141100 | 10 | 9 | 3 | rs1668873 | 1.5 × 10−3 | 1.2 × 10−4 | Yes |
| USMG5 | 10:105148798 | 16 | 14 | 4 | rs1163073 | 4.9 × 10−4 | 1.5 × 10−4 | Yes |
| F12 | 5:176829141 | 3 | 3 | 2 | rs4976765 | 1.7 × 10−3 | 1.5 × 10−4 | Yes |
Potential novel genetic associations with asthma (highlighted in grey) are those for which the gene-based association was not driven by eQTLs located in known allergy risk loci. Known allergy loci are defined as those that contain a variant reported to be associated with asthma or other allergic diseases with P<5×10−8 in published GWAS.
Genes not located in an asthma risk locus but for which the gene-based association was driven by trans-eQTLs in LD with allergy risk variants (see Table E7 for more details).
Figure 2. Summary of association results obtained for 17,190 genes by applying the proposed gene-based test of association to a published asthma GWAS14.
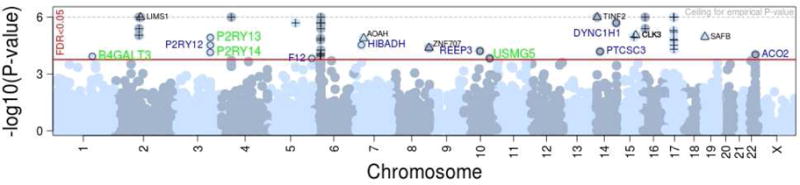
The red horizontal line shows the P-value threshold corresponding to an empirical FDR of 5% (P=1.9×10−4). Forty eight genes exceeded this threshold, including (1) 31 genes located in established risk loci for allergic disease (denoted by ‘+’); (2) six genes located in new risk loci but with a gene-based association that was driven by trans-eQTLs located in the MHC or near ORMDL3 (denoted by ‘Δ’;gene name shown in black font) and (3) 11 genes with a gene-based association that was not driven by eQTLs located in established allergy risk loci (denoted by ‘○’), including four (green font) for which the association replicated in an independent GWAS15. The y-axis represents the −log10 of the simulation-derived gene-based P-value, which accounts for the residual LD between eQTLs of a given gene. The P-value was based on up to 1 million simulations, and so it could not exceed a P=10−6 (dashed grey line).
On the other hand, six (12%; LIMS1, AOAH, ZNF707, CLK3, SAFB and TINF2; ‘Δ’ in Figure 2) of the 48 genes significant at an FDR of 0.05 were not located in established risk loci for asthma but the significant gene-based associations were (in most cases, entirely) driven by trans-eQTLs located in the MHC region or near ORMDL3 (Table E7 in the Online Repository). These include for example variant rs9268853, which is a trans-eQTL for CLK3 (P=7×10−17) in PBMCs19, SAFB (P=3×10−6) in whole-blood20 and AOAH in three tissues (best P=10−61,19–21) (Table E8 in the Online Repository). This variant has also been found to be a cis-eQTL (P<5×10−8) for HLA-DQ and HLA-DR genes across multiple tissues (not shown). These results suggest that MHC and 17q12 variants might contribute to asthma risk not only by directly modulating the expression of nearby genes, but also by indirectly influencing the expression of genes in different chromosomes (eg. through cis-mediation22).
Of potential greater interest, 11 (23%) of the 48 significant genes were located in potential novel asthma risk loci and the gene-based associations were not driven by established allergy risk variants (‘○’ in Figure 2 and Table 1). As some of these genes might represent false-positive findings, we studied their association with asthma in an independent GWAS.
Replication of the putative novel gene-based associations in an independent asthma GWAS
To confirm the putative novel associations, we applied EUGENE to an independent GWAS of asthma with publicly available summary statistics15. Based on results for 9,967 asthmatics and 15,301 controls, four of the 11 genes selected for replication had a significant gene-based association (P<0.05; Table E9 in the Online Repository), when simulations show that on average the expected number of genes significant at this threshold by chance alone given multiple testing was 0.53 (SD=0.77).
We then explored whether the discovery and replication associations for those four genes were consistent by comparing the direction of effect on disease risk for individual eQTLs. Overall, the direction of effect for most eQTLs of a given gene was the same between the two independent GWAS (Table E10 in the Online Repository). For example, of the seven eQTLs for USMG5 that were individually associated with asthma risk in either study, for six the allele that increased asthma risk was the same (or was on the same haplotype) in both studies; one eQTL was not tested in the replication GWAS, and so the direction of effect could not be compared. Therefore, the association between asthma risk and these four genes is generally consistent at the individual eQTL level between the two independent GWAS. Henceforth, we refer to these four genes with a reproducible gene-based association with asthma as “putative novel asthma risk genes”.
Contribution of cis- and trans-eQTLs to significant gene-based associations
For three (P2RY13, P2RY14, USMG5) of the four putative novel risk genes, the gene-based association with asthma was entirely driven by cis-eQTLs. Most of these eQTLs were identified by eQTL studies of whole-blood expression levels (Table E11 in the Online Repository). For the fourth gene, B4GALT3, three cis (in neutrophils, blood and fibroblasts) and one trans (in blood) eQTL contributed to the association with asthma (Table E11 in the Online Repository). The latter (rs1668873) was located 44 Mb away on chromosome 1 and was previously reported to associate with mean platelet volume and count23, 24. This variant is also a cis-eQTL for NUAK220, a nuclear transcriptional modulator that has been shown to induce the expression of B4GALT525, a galactosyltransferase related to B4GALT326. Therefore, these results suggest that both direct (cis-eQTLs) and indirect (via transcriptional modulators such as NUAK2) genetic effects on B4GALT3 expression might contribute to asthma risk.
Genetically predicted direction of effect of gene expression on asthma risk
To assess the direction of effect of gene expression on disease risk, we focused on the independent eQTLs for each gene that were individually associated with asthma in the discovery and/or replication GWAS. These variants had the greatest contribution to the significant gene-based tests. When we compared the direction of effect for each eQTL between asthma risk and expression levels, we found that the allele associated with increased gene expression was also associated with increased asthma risk for all independent eQTLs of P2RY13 and P2RY14 (Table E11 in the Online Repository). The same pattern of results was observed for six of the seven eQTLs of USMG5; for example, the rs1163073:C allele that was associated with asthma risk (OR=1.09, P=0.0005), was associated with increased USMG5 expression in five different cell types or tissues (neutrophils, LCLs, skin, PBMCs and blood). These results suggest that in the tissues or cell types considered in our analysis, a genetically determined increase in gene expression for each of these four genes is associated with increased disease risk. For B4GALT3, there was no clear pattern across multiple eQTLs: of the four alleles associated with increased gene expression, two (in neutrophils and whole-blood) were associated with increased and two (in fibroblasts and whole-blood) with decreased disease risk. Such differences between eQTLs could arise, for example, if B4GALT3 has opposing functional effects on different cell types relevant to asthma (eg. activation in one, inhibition in another). Further studies are required to test this possibility.
Functional studies in the mouse
The four putative novel asthma risk genes identified in our genetic association analysis are involved in nucleotide synthesis (B4GALT3, USMG5) and nucleotide-dependent cell activation (P2RY13, P2RY14). Based on this observation, we hypothesise that genetic dysregulation of nucleotide signaling contributes to asthma risk. In depth functional experiments that comprehensively test this hypothesis were beyond the scope of this study. Nonetheless, we carried out two sets of experiments in the mouse to provide preliminary functional support for the involvement in allergic asthma for two of these four nucleotide-related genes: P2RY13 and P2RY14. Both are cell-surface receptors with known agonists, and so were well suited for functional studies.
First, we used an established experimental model of acute allergic asthma16 to study P2ry13 and P2ry14 expression in the lungs of C57Bl/6 mice sensitized and subsequently challenged with house dust mite (HDM) allergen. In this model, mice develop granulocytic airway inflammation that has a predominant eosinophil contribution16. When considering overall lung expression, HDM challenge resulted in a significant increase in P2ry13 and P2ry14 expression, relative to control mice challenged with a saline solution (Figure 3A). To understand which lung cell types contributed to this increase in gene expression, we used flow cytometry to measure protein expression in airway epithelial cells (AECs) and major immune cell types collected through bronchoalveolar lavage. There was widespread expression of both receptors in AECs, both at baseline and after HDM challenge (Figure 3B and 3C). Most eosinophils collected in BALF after HDM challenge stained positive for both receptors (Figure 3D); expression in neutrophils was also high (Figure 3E). Lymphocytes and dendritic cells had low expression of both receptors (Figure E2 in the Online Repository).
Figure 3. Expression levels of P2ry13 and P2ry14 in lung of C57Bl/6 mice sensitized and then challenged with a saline solution or a house dust mite extract.
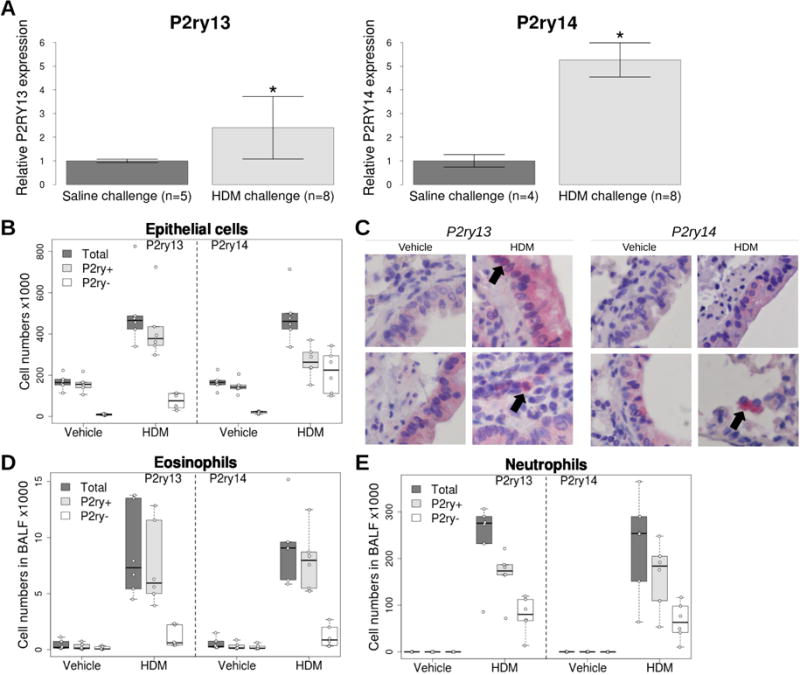
(A) Overall gene expression in lung. Expression levels were normalized to Hprt and are expressed as fold-change over saline challenge group. Results show mean +/− SD in each group. * Wilcoxon rank sum test P < 0.005 when comparing HDM and saline groups. (B, D and E) Expression of P2ry13 and P2ry14 based on flow cytometry analysis in lung epithelial cells, or eosinophils and neutrophils collected in BALF after saline or HDM challenge. (C) Expression of P2ry13 and P2ry14 in lung sections of mice challenged with saline or HDM.
Secondly, given the high level of P2ry13 and P2ry14 expression observed in AECs at baseline (ie. in the absence of allergen challenge), and the previously reported pro-inflammatory effect of the respective agonists (eg. 27, 28), we postulated that receptor activation could promote airway inflammation by inducing the release of alarmins. To test this possibility, we collected BALF from naïve mice 2 and 72 hours after intra-nasal challenge with saline, ADP (selective P2ry13 agonist) or UDP-glucose (selective P2ry14 agonist). At 2 hours post challenge, BALF levels of the alarmin IL-33 were significantly greater in mice exposed to the receptor agonists than in control mice (Figure 4A). Of note, nucleotide-induced IL-33 levels were comparable to allergen-induced IL-33 levels, indicating that both ADP and UDP-glucose are sufficient to induce IL-33 release. Furthermore, 72 hours after challenge, the number of BALF eosinophils and lymphocytes were significantly higher in agonist-treated mice (Figure 4B and 4C), but this was not the case for neutrophils, dendritic cells and monocytes (Figure E3 in the Online Repository). These results demonstrate that selective agonists of P2ry13 and P2ry14 can promote airway inflammation, even in the absence of allergen stimulation.
Figure 4. In vivo exposure to P2ry13 and P2ry14 receptor agonists in naïve C57Bl/6 mice.
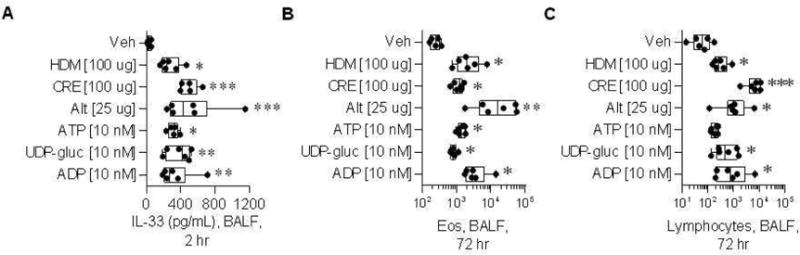
Mice were challenged with either vehicle, one of three allergens (HDM, CRE, Alt) or one of three nucleotides (ATP, UPD-glucose, ADP), and euthanized 2 and 72 hours after challenge. (A) IL-33 expression in bronchoalveolar lavage fluid (BALF) collected 2 hours post challenge. Total number of eosinophils (B) and lymphocytes (C) recruited to the BALF at 72 hours post challenge, based on flow cytometry analysis. Veh: vehicle. HDM: house dust mite. CRE: cockroach. Alt: alternaria.
DISCUSSION
Dysregulation of gene expression is thought to be a common mechanism by which genetic variants can influence cellular function and, ultimately, variation in human traits and disease risk. This proposition is supported by the observation that eQTLs are more likely to be trait-associated when compared to random variants18 and was the motivation for the gene-based approach developed in this study.
EUGENE has some advantages when compared to other gene-based association approaches, of which we highlight three: VEGAS9, PrediXcan12 and TWAS13. When compared to VEGAS, EUGENE avoids the requirement to use an arbitrary distance (same for all genes) from the known gene boundaries to define which SNPs to include in the gene-based analysis. If the distance is too small (eg. +/−5 kb), then the contribution of important more distantly located eQTLs might be missed, while a large distance (eg. +/− 1 Mb) could result in testing a large number of variants, many of which are likely to be unrelated to gene expression/function; in both cases, the power to detect a significant gene-based association is reduced. Also, because the analysis in EUGENE is restricted to variants previously shown to influence gene expression, whether in cis or in trans, a significant gene-based trait association directly implies that genetically-determined differences in gene expression contribute to trait variation. On the other hand, when compared to the recently described PrediXcan approach12, the main advantage of EUGENE is that it is applicable to GWAS summary statistics, which are typically easier to share than data sets with individual level genetic data. TWAS13, which is conceptually very similar to PrediXcan, is applicable to summary statistics but its current release only includes weights for a relatively small number of genes obtained from three eQTL studies, although this is likely to increase in the future. Another major difference when compared to both of these approaches is that with EUGENE, eQTLs identified in transcriptome studies of different cell types (and/or upon cell stimulation) can be included in the same association analysis, all contributing with equal weight to the gene-based statistic. This might be important for traits or diseases for which multiple cell types or tissues are known to play a role in the underlying pathophysiology. In the PrediXcan and TWAS approaches, the weights assigned to different eQTLs in the model used to predict gene expression levels are based on the effect (eg. regression coefficient) of those variants on expression levels measured in a single reference transcriptome data set. The extent to which those weights remain appropriate (ie. yield good prediction) if the reference transcriptome and target data sets are very different (eg. in age composition) is unclear. A disadvantage of EUGENE is that the direction of effect between gene expression and disease risk (or trait variation) is not directly inferred. To do so with the EUGENE approach, the effect of individual eQTLs that contribute to the gene-based test needs to be compared ad-hoc between the transcriptome and trait GWAS. These eQTLs also provide a specific small group of variants to test in validation studies. Lastly, EUGENE (but not the other three approaches) estimates FDR thresholds empirically, taking into account the LD between eQTLs of the same or different genes. This is important to account for multiple testing.
When we applied EUGENE to a published GWAS of asthma, we identified 48 genes with a significant gene-based association at an FDR of 0.05, including 11 associations that were not driven by established genetic risk variants for allergic disease. For four of these genes (B4GALT3, USMG5, P2RY13 and P2RY14), the association was nominally significant in an independent asthma GWAS and so we refer to these as putative novel asthma risk genes.
B4GALT3 encodes the widely-expressed enzyme β-1,4-galactosyltransferase III that catalyzes the transfer of galactose from UDP-galactose to N-acetylglucosamine, to form N-acetyllactosamine and UDP26, 29. How variation in B4GALT3 expression might contribute to asthma risk is unclear, but potential mechanisms include activation of β1 integrin30, which is important in the initiation of T-cell inflammatory responses31, or by influencing extracellular release of UDP-galactose32, a P2RY14 agonist.
USMG5 encodes a small subunit of ATP synthase33, an enzyme responsible for ATP synthesis in the mitochondria. USMG5 knockdown in HeLa cells causes the loss of ATP synthase, resulting in lower ATP synthesis and slower cell growth34. In CD4+ T-cells, mitochondria produce the ATP that is rapidly released into the extracellular space upon cell stimulation35. In turn, this ATP establishes an autocrine feedback through purinergic receptors that is essential for proper T-cell activation36. Given these observations, we speculate that genetically-determined increased USMG5 expression results in increased mitochondrial production of ATP, increased extracellular ATP release and increased T-cell activation. In turn, this would translate into an increased risk of asthma. ATP synthase has also been detected at the surface of different cell types37, where it is thought to play different physiological roles, for example, HDL endocytosis in hepatocytes via P2RY13 activation38 and non-conventional T-cell activation39. Whether USMG5 associates with membrane ATP synthase, and so could potentially influence its ectopic roles, remains to be determined.
P2RY13, also known as GPR86 or GPR94, is a purinergic receptor highly expressed in the immune system, lung and skin, but also in the brain40–42; it displays a significant homology with the nearby P2RY12 and P2RY14 genes, sharing 48 and 45% amino acid identity40. P2RY13 is strongly activated by ADP40, a degradation product of ATP. Airway epithelial goblet cells are a major source of extracellular ADP, which is released as a co-cargo molecule from mucin-containing granules43. In turn, ADP has been shown to enhance antigen-induced degranulation in mast cells, through a P2RY13-dependent mechanism27. ADP has also been reported to promote IL-6 release from keratinocytes42, inhibit TNF-alpha and IL-12 production by mature DCs44 and promote chemotaxis of immature DCs45; however, these studies did not specifically test if the observed ADP effects were mediated by P2RY13. Results from our genetic association analyses indicate that a genetically-determined increase in P2RY13 expression increases asthma risk, which is consistent with the pro-inflammatory effect suggested for ADP and P2RY13 by these functional studies.
P2RY14, also known as GPR105, encodes a G protein-coupled receptor that is potently and selectively activated by UDP-sugars, especially UDP-glucose46. UDP-glucose is thought to be an extracellular pro-inflammatory mediator47, constitutively released by different cell types including airway epithelial cells48. Importantly, infection with respiratory syncytial virus (RSV) or treatment with IL-13 significantly increases UDP-glucose release by airway epithelial cells49, and this coincides with increased mucus secretion50. Known pro-inflammatory effects of UDP-glucose acting through P2RY14 include inhibition of TLR9-dependent IFN-alpha production51, increased chemotaxis of neutrophils52, induction of mast cell degranulation53, production of IL-828, 54 and STAT3-dependent epidermal inflammation55. A small molecule antagonist for P2RY14 was recently developed and shown to effectively block chemotaxis of freshly isolated human neutrophils56. Of note, plasma UDP-glucose levels are elevated in mice fed a high-fat diet57, which raises the possibility that obesity might contribute to chronic P2RY14 activation and, in that way, increase asthma risk and/or severity. Studies that investigate this possibility are underway.
When we studied the expression of both P2ry13 and P2ry14 in mice, we found that both genes were highly expressed in AECs, both at baseline and after allergen challenge. High expression was also observed in infiltrating eosinophils after challenge and, to a smaller extent, neutrophils and monocytes. High expression in AECs suggested that receptor activation could contribute to airway inflammation even in the absence of allergen stimulation. This was indeed what we observed when naïve mice were challenged intra-nasally with either ADP or UDP-glucose: 72 hours after challenge, the numbers of BALF eosinophils were significantly increased when compared to control mice. Interestingly, eosinophil influx into the airways was preceded by a significant increase in the levels of the alarmin IL-33. The effect of both receptor agonists on IL-33 release was comparable in magnitude to that observed with allergens known to have a potent effect on IL-33 production, namely the fungus Alternaria alternata58. These results demonstrate that activation of P2ry13 and P2ry14, in addition to P2y258, can strongly induce IL-33 release in mice. Interestingly, P2ry13 expression was observed in the nuclei of AECs after allergen challenge. Given that IL-33 is constitutively stored in the nuclei of AECs59, it is possible that intracellular activation of P2ry13 expressed on the nuclear membrane plays a role in allergen-induced IL-33 release.
In conclusion, our genetic findings establish an association between asthma risk and genes involved in nucleotide synthesis (B4GALT3, USMG5) and nucleotide-dependent cell activation (P2RY13, P2RY14). In mice, in vivo activation of P2ry13 and P2ry14 induced IL-33 release and subsequent eosinophilic airway infiltration. These observations suggest that genetic dysregulation of nucleotide signaling contributes to the risk of asthma (allergic and, potentially, also non-allergic) and other related conditions; studies that test this possibility are now warranted. Our results also show that re-analysis of published GWAS with a gene-based test that exclusively focuses on documented eQTLs has the potential to identify novel associations.
Supplementary Material
KEY MESSAGES.
In humans, asthma risk is associated with genetically-determined expression of four genes related to nucleotide synthesis (B4GALT3, USMG5) and nucleotide-dependent cell activation (P2RY13 and P2RY14).
In mice, intranasal exposure with selective agonists for P2ry13 (ADP) or P2ry14 (UDP-glucose) induced the release of IL-33 and eosinophil infiltration into the lungs, in the absence of allergen stimulation.
CAPSULE SUMMARY.
Using a new method for gene-based analysis of GWAS results, we identified a genetic association between asthma risk and eQTLs for B4GALT3 and USMG5, which are involved in the production of UDP-galactose and ATP respectively, and P2RY13 and P2RY14, two G protein-coupled receptors activated respectively by ADP and UDP-sugars. Functional studies in the mouse show that activation of P2ry13 or P2ry14 induces the release of IL-33 and eosinophil infiltration into the lungs, in the absence of allergen stimulation. Functional studies that characterize in depth the contribution of these four genes to asthma pathophysiology are warranted.
Acknowledgments
We thank all study participants, including customers of 23andMe who answered surveys, as well as the employees of 23andMe, who together made this research possible.
Sources of funding: This work was supported in part by the National Human Genome Research Institute of the National Institutes of Health (R44HG006981) and the National Health and Medical Research Council of Australia (613627 and APP1036550).
ABBREVIATIONS
- AECs
airway epithelial cells
- ADP
adenosine 5-diphosphate
- Alt
Alternaria allergen
- ATP
adenosine 5-triphosphate
- BALF
bronchoalveolar lavage fluid
- CRE
cockroach allergen
- DC
dendritic cells
- eQTL
expression quantitative trait locus
- FDR
false discovery rate
- GWAS
Genome Wide Association Study
- HDM
house dust mite. IL: interleukin
- LCLs
lymphoblastoid cell lines
- LD
linkage disequilibrium
- SNP
single nucleotide polymorphism
- UDP
uridine-diphosphoglucose (UDP-glucose)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ferreira MA, Matheson MC, Duffy DL, Marks GB, Hui J, Le Souef P, et al. Identification of IL6R and chromosome 11q13.5 as risk loci for asthma. Lancet. 2011;378:1006–14. doi: 10.1016/S0140-6736(11)60874-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–3. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 3.Vicente CT, Edwards SL, Hillman KM, Kaufmann S, Mitchell H, Bain L, et al. Long-Range Modulation of PAG1 Expression by 8q21 Allergy Risk Variants. Am J Hum Genet. 2015 doi: 10.1016/j.ajhg.2015.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Okada Y, Wu D, Trynka G, Raj T, Terao C, Ikari K, et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506:376–81. doi: 10.1038/nature12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nelson MR, Tipney H, Painter JL, Shen J, Nicoletti P, Shen Y, et al. The support of human genetic evidence for approved drug indications. Nat Genet. 2015;47:856–60. doi: 10.1038/ng.3314. [DOI] [PubMed] [Google Scholar]
- 6.Ferreira MA. Improving the power to detect risk variants for allergic disease by defining case-control status based on both asthma and hay Fever. Twin Res Hum Genet. 2014;17:505–11. doi: 10.1017/thg.2014.59. [DOI] [PubMed] [Google Scholar]
- 7.Bonnelykke K, Sleiman P, Nielsen K, Kreiner-Moller E, Mercader JM, Belgrave D, et al. A genome-wide association study identifies CDHR3 as a susceptibility locus for early childhood asthma with severe exacerbations. Nat Genet. 2014;46:51–5. doi: 10.1038/ng.2830. [DOI] [PubMed] [Google Scholar]
- 8.Galesloot TE, van Steen K, Kiemeney LA, Janss LL, Vermeulen SH. A comparison of multivariate genome-wide association methods. PLoS One. 2014;9:e95923. doi: 10.1371/journal.pone.0095923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu JZ, McRae AF, Nyholt DR, Medland SE, Wray NR, Brown KM, et al. A versatile gene-based test for genome-wide association studies. Am J Hum Genet. 2010;87:139–45. doi: 10.1016/j.ajhg.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li MX, Gui HS, Kwan JS, Sham PC. GATES: a rapid and powerful gene-based association test using extended Simes procedure. Am J Hum Genet. 2011;88:283–93. doi: 10.1016/j.ajhg.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nicolae DL, Gamazon E, Zhang W, Duan S, Dolan ME, Cox NJ. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gamazon ER, Wheeler HE, Shah KP, Mozaffari SV, Aquino-Michaels K, Carroll RJ, et al. A gene-based association method for mapping traits using reference transcriptome data. Nat Genet. 2015;47:1091–8. doi: 10.1038/ng.3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gusev A, Ko A, Shi H, Bhatia G, Chung W, Penninx BW, et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat Genet. 2016;48:245–52. doi: 10.1038/ng.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferreira MA, Matheson MC, Tang CS, Granell R, Ang W, Hui J, et al. Genome-wide association analysis identifies 11 risk variants associated with the asthma with hay fever phenotype. J Allergy Clin Immunol. 2014;133:1564–71. doi: 10.1016/j.jaci.2013.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moffatt MF, Gut IG, Demenais F, Strachan DP, Bouzigon E, Heath S, et al. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010;363:1211–21. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ullah MA, Revez JA, Loh Z, Simpson J, Zhang V, Bain L, et al. Allergen-induced IL-6 trans-signaling activates gammadelta T cells to promote type 2 and type 17 airway inflammation. J Allergy Clin Immunol. 2015;136:1065–73. doi: 10.1016/j.jaci.2015.02.032. [DOI] [PubMed] [Google Scholar]
- 17.Davidson S, Kaiko G, Loh Z, Lalwani A, Zhang V, Spann K, et al. Plasmacytoid dendritic cells promote host defense against acute pneumovirus infection via the TLR7-MyD88-dependent signaling pathway. J Immunol. 2011;186:5938–48. doi: 10.4049/jimmunol.1002635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Torgerson DG, Ampleford EJ, Chiu GY, Gauderman WJ, Gignoux CR, Graves PE, et al. Meta-analysis of genome-wide association studies of asthma in ethnically diverse North American populations. Nat Genet. 2011;43:887–92. doi: 10.1038/ng.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeller T, Wild P, Szymczak S, Rotival M, Schillert A, Castagne R, et al. Genetics and beyond–the transcriptome of human monocytes and disease susceptibility. PLoS One. 2010;5:e10693. doi: 10.1371/journal.pone.0010693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Westra HJ, Peters MJ, Esko T, Yaghootkar H, Schurmann C, Kettunen J, et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet. 2013;45:1238–43. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim S, Becker J, Bechheim M, Kaiser V, Noursadeghi M, Fricker N, et al. Characterizing the genetic basis of innate immune response in TLR4-activated human monocytes. Nat Commun. 2014;5:5236. doi: 10.1038/ncomms6236. [DOI] [PubMed] [Google Scholar]
- 22.Pierce BL, Tong L, Chen LS, Rahaman R, Argos M, Jasmine F, et al. Mediation analysis demonstrates that trans-eQTLs are often explained by cis-mediation: a genome-wide analysis among 1,800 South Asians. PLoS Genet. 2014;10:e1004818. doi: 10.1371/journal.pgen.1004818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berndt SI, Gustafsson S, Magi R, Ganna A, Wheeler E, Feitosa MF, et al. Genome-wide meta-analysis identifies 11 new loci for anthropometric traits and provides insights into genetic architecture. Nat Genet. 2013;45:501–12. doi: 10.1038/ng.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soranzo N, Spector TD, Mangino M, Kuhnel B, Rendon A, Teumer A, et al. A genome-wide meta-analysis identifies 22 loci associated with eight hematological parameters in the HaemGen consortium. Nat Genet. 2009;41:1182–90. doi: 10.1038/ng.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuga W, Tsuchihara K, Ogura T, Kanehara S, Saito M, Suzuki A, et al. Nuclear localization of SNARK; its impact on gene expression. Biochem Biophys Res Commun. 2008;377:1062–6. doi: 10.1016/j.bbrc.2008.10.143. [DOI] [PubMed] [Google Scholar]
- 26.Lo NW, Shaper JH, Pevsner J, Shaper NL. The expanding beta 4-galactosyltransferase gene family: messages from the databanks. Glycobiology. 1998;8:517–26. doi: 10.1093/glycob/8.5.517. [DOI] [PubMed] [Google Scholar]
- 27.Gao ZG, Ding Y, Jacobson KA. P2Y(13) receptor is responsible for ADP-mediated degranulation in RBL-2H3 rat mast cells. Pharmacol Res. 2010;62:500–5. doi: 10.1016/j.phrs.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muller T, Bayer H, Myrtek D, Ferrari D, Sorichter S, Ziegenhagen MW, et al. The P2Y14 receptor of airway epithelial cells: coupling to intracellular Ca2+ and IL-8 secretion. Am J Respir Cell Mol Biol. 2005;33:601–9. doi: 10.1165/rcmb.2005-0181OC. [DOI] [PubMed] [Google Scholar]
- 29.Guo S, Sato T, Shirane K, Furukawa K. Galactosylation of N-linked oligosaccharides by human beta-1,4-galactosyltransferases I, II, III, IV, V, and VI expressed in Sf-9 cells. Glycobiology. 2001 Oct;11(10):813–20. doi: 10.1093/glycob/11.10.813. [DOI] [PubMed] [Google Scholar]
- 30.Liao WC, Liu CH, Chen CH, Hsu WM, Liao YY, Chang HM, et al. beta-1,4-Galactosyltransferase III suppresses extravillous trophoblast invasion through modifying beta1-integrin glycosylation. Placenta. 2015;36:357–64. doi: 10.1016/j.placenta.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 31.Suzuki K, Okuno T, Yamamoto M, Pasterkamp RJ, Takegahara N, Takamatsu H, et al. Semaphorin 7A initiates T-cell-mediated inflammatory responses through alpha1beta1 integrin. Nature. 2007;446:680–4. doi: 10.1038/nature05652. [DOI] [PubMed] [Google Scholar]
- 32.Lazarowski ER. Quantification of extracellular UDP-galactose. Anal Biochem. 2010;396:23–9. doi: 10.1016/j.ab.2009.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meyer B, Wittig I, Trifilieff E, Karas M, Schagger H. Identification of two proteins associated with mammalian ATP synthase. Mol Cell Proteomics. 2007;6:1690–9. doi: 10.1074/mcp.M700097-MCP200. [DOI] [PubMed] [Google Scholar]
- 34.Ohsakaya S, Fujikawa M, Hisabori T, Yoshida M. Knockdown of DAPIT (diabetes-associated protein in insulin-sensitive tissue) results in loss of ATP synthase in mitochondria. J Biol Chem. 2011;286:20292–6. doi: 10.1074/jbc.M110.198523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ledderose C, Bao Y, Lidicky M, Zipperle J, Li L, Strasser K, et al. Mitochondria are gate-keepers of T cell function by producing the ATP that drives purinergic signaling. J Biol Chem. 2014;289:25936–45. doi: 10.1074/jbc.M114.575308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yip L, Woehrle T, Corriden R, Hirsh M, Chen Y, Inoue Y, et al. Autocrine regulation of T-cell activation by ATP release and P2X7 receptors. FASEB J. 2009;23:1685–93. doi: 10.1096/fj.08-126458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vantourout P, Radojkovic C, Lichtenstein L, Pons V, Champagne E, Martinez LO. Ecto-F(1)- ATPase: a moonlighting protein complex and an unexpected apoA-I receptor. World J Gastroenterol. 2010;16:5925–35. doi: 10.3748/wjg.v16.i47.5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jacquet S, Malaval C, Martinez LO, Sak K, Rolland C, Perez C, et al. The nucleotide receptor P2Y13 is a key regulator of hepatic high-density lipoprotein (HDL) endocytosis. Cell Mol Life Sci. 2005;62:2508–15. doi: 10.1007/s00018-005-5194-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scotet E, Martinez LO, Grant E, Barbaras R, Jeno P, Guiraud M, et al. Tumor recognition following Vgamma9Vdelta2 T cell receptor interactions with a surface F1-ATPase-related structure and apolipoprotein A-I. Immunity. 2005;22:71–80. doi: 10.1016/j.immuni.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 40.Communi D, Gonzalez NS, Detheux M, Brezillon S, Lannoy V, Parmentier M, et al. Identification of a novel human ADP receptor coupled to G(i) J Biol Chem. 2001;276:41479–85. doi: 10.1074/jbc.M105912200. [DOI] [PubMed] [Google Scholar]
- 41.Zhang FL, Luo L, Gustafson E, Palmer K, Qiao X, Fan X, et al. P2Y(13): identification and characterization of a novel Galphai-coupled ADP receptor from human and mouse. J Pharmacol Exp Ther. 2002;301:705–13. doi: 10.1124/jpet.301.2.705. [DOI] [PubMed] [Google Scholar]
- 42.Inoue K, Hosoi J, Denda M. Extracellular ATP has stimulatory effects on the expression and release of IL-6 via purinergic receptors in normal human epidermal keratinocytes. J Invest Dermatol. 2007;127:362–71. doi: 10.1038/sj.jid.5700526. [DOI] [PubMed] [Google Scholar]
- 43.Kreda SM, Seminario-Vidal L, van Heusden CA, O’Neal W, Jones L, Boucher RC, et al. Receptor-promoted exocytosis of airway epithelial mucin granules containing a spectrum of adenine nucleotides. J Physiol. 2010;588:2255–67. doi: 10.1113/jphysiol.2009.186643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.la Sala A, Ferrari D, Corinti S, Cavani A, Di Virgilio F, Girolomoni G. Extracellular ATP induces a distorted maturation of dendritic cells and inhibits their capacity to initiate Th1 responses. J Immunol. 2001;166:1611–7. doi: 10.4049/jimmunol.166.3.1611. [DOI] [PubMed] [Google Scholar]
- 45.Idzko M, Dichmann S, Ferrari D, Di Virgilio F, la Sala A, Girolomoni G, et al. Nucleotides induce chemotaxis and actin polymerization in immature but not mature human dendritic cells via activation of pertussis toxin-sensitive P2y receptors. Blood. 2002;100:925–32. doi: 10.1182/blood.v100.3.925. [DOI] [PubMed] [Google Scholar]
- 46.Chambers JK, Macdonald LE, Sarau HM, Ames RS, Freeman K, Foley JJ, et al. A G protein-coupled receptor for UDP-glucose. J Biol Chem. 2000;275:10767–71. doi: 10.1074/jbc.275.15.10767. [DOI] [PubMed] [Google Scholar]
- 47.Harden TK, Sesma JI, Fricks IP, Lazarowski ER. Signalling and pharmacological properties of the P2Y receptor. Acta Physiol (Oxf) 2010;199:149–60. doi: 10.1111/j.1748-1716.2010.02116.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lazarowski ER, Shea DA, Boucher RC, Harden TK. Release of cellular UDP-glucose as a potential extracellular signaling molecule. Mol Pharmacol. 2003;63:1190–7. doi: 10.1124/mol.63.5.1190. [DOI] [PubMed] [Google Scholar]
- 49.Okada SF, Zhang L, Kreda SM, Abdullah LH, Davis CW, Pickles RJ, et al. Coupled nucleotide and mucin hypersecretion from goblet-cell metaplastic human airway epithelium. Am J Respir Cell Mol Biol. 2011;45:253–60. doi: 10.1165/rcmb.2010-0253OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kreda SM, Okada SF, van Heusden CA, O’Neal W, Gabriel S, Abdullah L, et al. Coordinated release of nucleotides and mucin from human airway epithelial Calu-3 cells. J Physiol. 2007;584:245–59. doi: 10.1113/jphysiol.2007.139840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shin A, Toy T, Rothenfusser S, Robson N, Vorac J, Dauer M, et al. P2Y receptor signaling regulates phenotype and IFN-alpha secretion of human plasmacytoid dendritic cells. Blood. 2008;111:3062–9. doi: 10.1182/blood-2007-02-071910. [DOI] [PubMed] [Google Scholar]
- 52.Sesma JI, Kreda SM, Steinckwich-Besancon N, Dang H, Garcia-Mata R, Harden TK, et al. The UDP-sugar-sensing P2Y(14) receptor promotes Rho-mediated signaling and chemotaxis in human neutrophils. Am J Physiol Cell Physiol. 2012;303:C490–8. doi: 10.1152/ajpcell.00138.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gao ZG, Ding Y, Jacobson KA. UDP-glucose acting at P2Y14 receptors is a mediator of mast cell degranulation. Biochem Pharmacol. 2010;79:873–9. doi: 10.1016/j.bcp.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Arase T, Uchida H, Kajitani T, Ono M, Tamaki K, Oda H, et al. The UDP-glucose receptor P2RY14 triggers innate mucosal immunity in the female reproductive tract by inducing IL-8. J Immunol. 2009;182:7074–84. doi: 10.4049/jimmunol.0900001. [DOI] [PubMed] [Google Scholar]
- 55.Jokela TA, Karna R, Makkonen KM, Laitinen JT, Tammi RH, Tammi MI. Extracellular UDP-glucose activates P2Y14 Receptor and Induces Signal Transducer and Activator of Transcription 3 (STAT3) Tyr705 phosphorylation and binding to hyaluronan synthase 2 (HAS2) promoter, stimulating hyaluronan synthesis of keratinocytes. J Biol Chem. 2014;289:18569–81. doi: 10.1074/jbc.M114.551804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barrett MO, Sesma JI, Ball CB, Jayasekara PS, Jacobson KA, Lazarowski ER, et al. A selective high-affinity antagonist of the P2Y14 receptor inhibits UDP-glucose-stimulated chemotaxis of human neutrophils. Mol Pharmacol. 2013;84:41–9. doi: 10.1124/mol.113.085654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu J, Morinaga H, Oh D, Li P, Chen A, Talukdar S, et al. GPR105 ablation prevents inflammation and improves insulin sensitivity in mice with diet-induced obesity. J Immunol. 2012;189:1992–9. doi: 10.4049/jimmunol.1103207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kouzaki H, Iijima K, Kobayashi T, O’Grady SM, Kita H. The danger signal, extracellular ATP, is a sensor for an airborne allergen and triggers IL-33 release and innate Th2-type responses. J Immunol. 2011;186:4375–87. doi: 10.4049/jimmunol.1003020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moussion C, Ortega N, Girard JP. The IL-1-like cytokine IL-33 is constitutively expressed in the nucleus of endothelial cells and epithelial cells in vivo: a novel ‘alarmin’? PLoS One. 2008;3:e3331. doi: 10.1371/journal.pone.0003331. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


