Abstract
The central step in eukaryotic homologous recombination (HR) is ATP-dependent DNA strand exchange mediated by the Rad51 recombinase. In this process, Rad51 assembles on single-stranded DNA (ssDNA) to yield a helical filament that is able to search for and invade a homologous double-stranded DNA (dsDNA), leading to strand separation in the dsDNA and formation of new base pairs between the initiating ssDNA and the complementary strand in the duplex partner. Here we have used cryo-electron microscopy (cryo-EM) to solve the structures of human RAD51 in complex with DNA molecules in their presynaptic and post-synaptic states at near atomic resolution. Our structures revealed both conserved and distinct structural features of the RAD51-DNA complexes in comparison with its prokaryotic counterpart. Importantly, we have also captured the structure of an arrested synaptic complex. The results provide insights into the molecular mechanism of DNA homology search and strand exchange processes.
Introduction
Homologous recombination (HR) plays essential roles in genetic integrity maintenance by eliminating DNA lesions such as double stranded breaks, inter-strand crosslinks, and collapsed replication forks1-5. HR is also indispensable for meiosis wherein it helps ensure the proper segregation of homologous chromosomes, and it also contributes to the generation of genetic diversity6,7. HR is a highly intricate and precisely regulated process involving a large number of proteins and complexes8,9. Among these HR factors are a family of conserved recombinases that catalyze ATP-dependent pairing and strand exchange between homologous DNA molecules.
The most studied recombinases are RecA in bactera, RadA in archaea, and Rad51 in eukaryotic cells9-14. Upon ATP binding, recombinase protomers assemble into a right-handed helical filament on ssDNA (the invading strand) called the presynaptic filament15-17. The presynaptic complex engages and samples duplex DNA to search for a homologous region in the latter. Successful homology search is followed by limited base pairing between the invading strand and the complementary strand in the dsDNA14,18-21. This HR stage has been termed as synaptic complex formation22,23. Then, more extensive DNA strand exchange occurs, and the newly formed DNA joint is bound by the recombinase filament in the post-synaptic complex. This complex is subsequently resolved with the disassembly of the recombinase filament and the recruitment of a DNA polymerase to extend the 3′ end of the invading DNA strand8,24,25. The HR reaction mediated by the conserved recombinases has been studied using biochemical, biophysical and structural approaches19,21,22,26.
X-ray crystal structures of E. coli RecA, yeast Rad51, archaea RadA in their DNA-free state have been solved, which reveal homologous core domains responsible for ATP binding and protomer-protomer interactions27-30. Electron microscopy at low resolution has also been applied to examine the structures of RecA and Rad51 in complex with ssDNA or dsDNA at different nucleotide state31-33. An atomic resolution view of RecA in complex with ssDNA and/or dsDNA has been provided by X-ray crystallography34. These structures illustrate an extended conformation of the invading DNA along the central axis of the RecA filament assembly and pairing with the complementary strand in triplet clusters. Whether or not these attributes of the presynaptic and post-synaptic complexes of RecA are shared by the eukaryotic recombinases has not been addressed. Moreover, owing to a lack of structural information of intermediate states, it remains unknown how the DNA strands in the homologous dsDNA become separated in the synaptic phase of the DNA strand exchange reaction.
In this study, we have carried out cryo-EM analyses of human RAD51 in different states of the DNA strand exchange process. These endeavors have enabled us to determine the structures of presynaptic and post-synaptic complexes of RAD51 at near atomic resolution. Furthermore, we have captured an intermediate state in the strand exchange process that allows us to visualize strand separation of the incoming duplex partner within the synaptic complex.
Results
Cryo-EM structures of presynaptic complex and post-synaptic complex
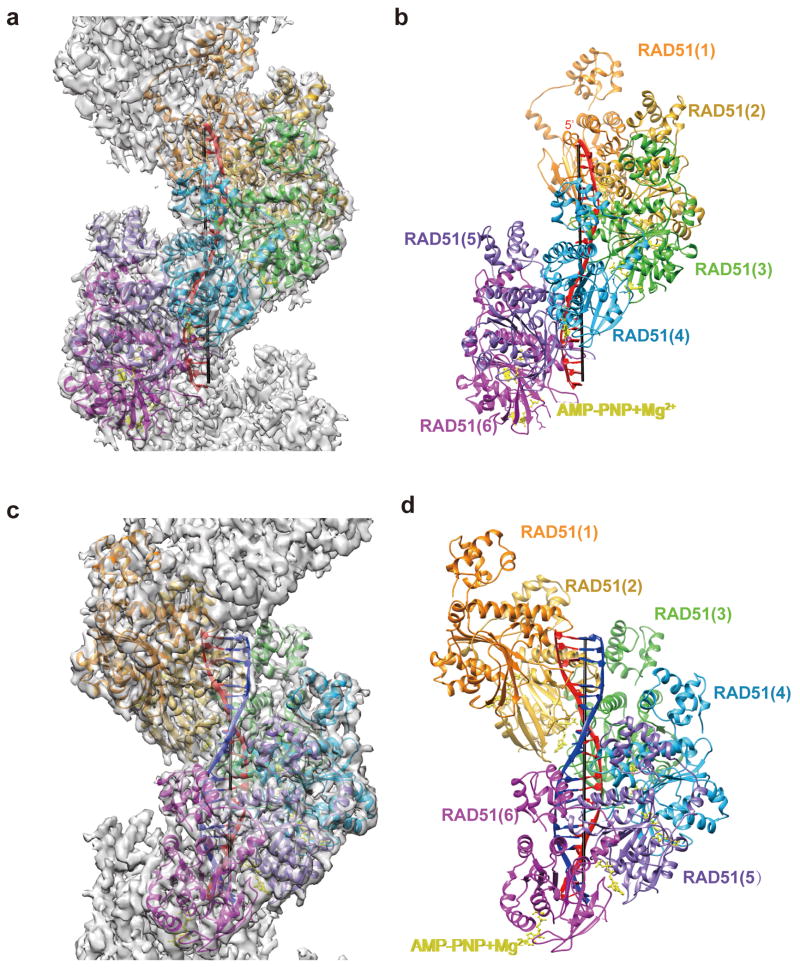
Purified human RAD51 protein was incubated with ssDNA in the presence of AMP-PNP to form the presynaptic filament (Online Methods, Supplementary Table 1 and Fig. S1). As we showed previously35,36, the ability of the presynaptic filament to conduct DNA strand exchange with a homologous duplex DNA partner is significantly enhanced by the recombinase co-factor Hop2-Mnd1 complex (Fig. S1). The presynaptic filament and post-synaptic complex generated with the presence of Hop2-Mnd1 adopt a well-ordered helical structure under conditions of cryo-EM, allowing us to use the IHRSR (Iterative Helical Real Space Reconstruction) algorithm37 for solving their 3D structures at 4.4 Å and 4.5 Å resolution, respectively (Online Methods, Table 1, Fig. 1, Fig. S2, Video S1 and S2). Both structures possess similar helical parameters of rise 15.8 Å and twist 56.77°, corresponding to a helical assembly of 6.3 RAD51 protomers per turn with about 100 Å pitch. While the overall structures of the presynaptic and post-synaptic complexes are similar, a distinct difference occurs along the helical axis of these complexes, where the latter harbors extra density (Fig. 1a and 1c). Our reconstruction of the RAD51 presynaptic complex share a very similar overall shape of the protomer with the 3D reconstruction of HsRAD51 presynaptic complex reported in a latest research although the helical symmetry deviates slightly between the two.38
Table 1. Data processing statistics for different complex states and model refinement.
| Presynaptic | Post-synaptic | Synaptic | |
|---|---|---|---|
| Data collection | |||
| TEM | Titan Krios | Titan Krios | Titan Krios |
| Camera | K2-Summit | K2-Summit | FalconII |
| Pixel size (Å) | 1.306 | 1.306 | 1.05 |
| Micrograph number | 540 | 528 | 202 |
| Image processing | |||
| Boxsize for S and R | 256 | 256 | 256 |
| Total particle number for S and R | 40,404 | 41,146 | 28,455 |
| Resolution (Å) | 4.4 | 4.5 | 5 |
| Boxsize for F | - | 320 | 400 |
| Total particle number for F | - | 2318 form 356 micrographs | 1998 |
| Good particle for F | - | 1072 | 552 |
| Resolution for F (Å) | - | 10 | 14 |
| Rise (Å) | 15.88 | 15.80 | 15.27 |
| Twist | 56.77° | 56.18° | 56.4° |
| Model building | |||
| Rms deviations (bonds) (Å2) | 0.0074 | 0.0098 | - |
| Rms deviations (angles) (Å2) | 1.03 | 1.13 | - |
| Molprobity score | 3.10 | 2.93 | - |
| Clashscore all atoms | 31.5 | 35.71 | - |
| Poor rotamers (%) | 4.59 | 3.49 | - |
| Ramachandran plot favored (%) | 91.89 | 92.07 | - |
| Ramachandran plot outliers (%) | 2.30 | 2.17 | - |
S stands for SPIDER, R stands for RELION and F stands for FREALIGN.
Figure 1. Structures of RAD51 presynaptic and post-synaptic complexes.
(a) Cryo-EM density map superimposed with the (b) atomic model of the presynaptic complex. (c) Cryo-EM density map superimposed with the (d) atomic model of the post-synaptic complex. The invading ssDNA is in red and the complementary strand from the homologous dsDNA is in blue. RAD51 protomers are in orange, golden, lime green, cyan, medium purple, magenta as the order and AMP-PNP-Mg2+ is in yellow.
We have built atomic models of the presynaptic and post-synaptic complexes from high resolution EM maps, which show key features of secondary structural elements and major side chains of the most rigid part in RAD51 as well as the DNA backbone and bases (Table 1, Fig. 1b,1d and Fig. S3). The atomic models include DNA and most of the residues of RAD51 except for the N-terminal 1-21 residues and a few residues in flexible loop regions. It is worth noting that the density of DNA in both the presynaptic and post-synaptic complex reconstructions appear as triplet clusters within asymmetric units of the helical RAD51 assembly. The current resolution does not allow us to resolve individual bases within the DNA density but the density of DNA is sufficiently clear to reveal the bound bases and base pairs arranged in distinct triplets (Fig. S3). We noticed that the density corresponding to the complementary strand was relatively weaker than that of the invading strand in the post-synaptic complex reconstruction, possibly due to the partial completion of the strand-exchange reaction. Our results thus provide evidence for a conserved mechanism of DNA engagement by RecA34 and RAD51.
Although we added Hop2-Mnd1 complex, with ∼12% of the RAD51, in our system to enhance the strand-exchange reaction, we didn't detect any prominent density corresponding to this complex in our reconstruction of the post-synaptic assembly. This could be due to the low occupancy or high flexibility of the complex within the filament so its signal was averaged out in our reconstruction process. This was confirmed by the control experiment that the reconstruction of RAD51 presynaptic complex incubated with the Hop2-Mnd1 complex appeared very similar to the presynaptic complex free of Hop2-Mnd1 (Fig. S1d), although it has been shown that Hop2-Mnd1 stably binds to RAD51 presynaptic complex35. Our observation, however, didn't rule out the possibility that Hop2-Mnd1 complex may dissociate from the RAD51 filament after the strand exchange reaction.
RAD51 structure and protomer-protomer interactions
The overall protein conformation and protomer-protomer interactions are very similar in the presynaptic and post-synaptic complexes (Fig.S4a). Consistent with the high homology among different recombinases27, the tertiary structure of human RAD51 in its functional states is similar to that described for yeast Rad51 and archaea RadA27,29 (Fig S4b, S4c and S4d). The most prominent conserved features include an N-terminal domain comprised of five α-helices, a β-strand responsible for protomer-protomer interactions, and a classical α/β ATPase core domain with both nucleotide binding Walker motifs and DNA interacting loops27 (Fig S5).
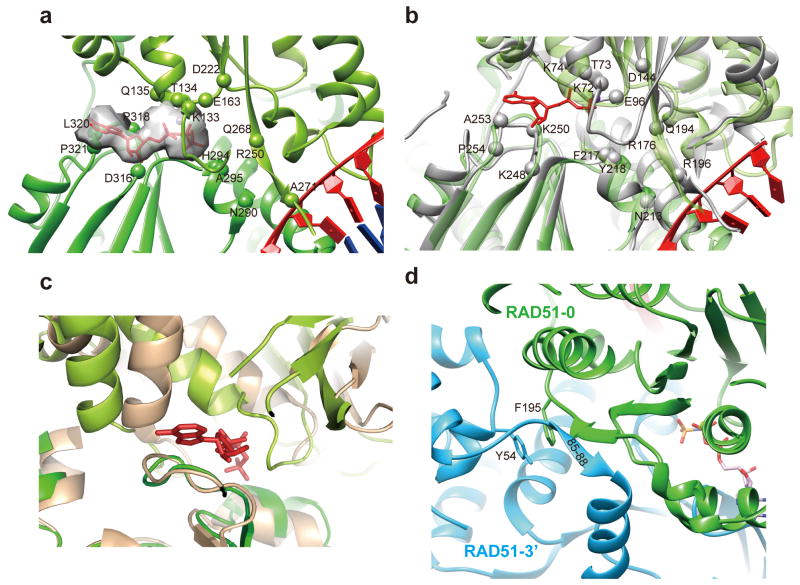
RAD51 protomers form the helical assembly mainly via three interfaces. The first is mediated by a sandwiched ATP molecule (Fig. 2a) between two adjacent protomers at a conserved interface among RecA, RadA, and Rad51. In the EM map, we can clearly identify a density corresponding to AMP-PNP bound by two neighboring RAD51 protomers. The Walker A motif spanning residues 127-GEFRTGKT134 stabilizes AMP-PNP with conserved residues K133, T134 surrounding the phosphates (Fig. 2a and Fig. S5), which is in agreement with the known role of these residues in ATP-mediated binding and hydrolysis39,40. E163, being equivalent to E96 in RecA, and D316, being critical for RAD51's recombinase activity41, surround AMP-PNP. Furthermore, the residues that are responsible for coupling nucleotide binding and nucleic acid interaction are conserved between RAD51 and RecA (Fig. 2a and 2b). In comparison with the available crystal structures of other recombinases, the nucleotide-bridged interaction interface in human RAD51 resembles that described for RadA and RecA filaments but differs from the yeast Rad51 filament, of which a ∼20° rotation and ∼10 Å translation of the protomers are needed in order to align with filaments of human RAD51 (Fig. 2c and Fig. S4b-S4d). This reflects the longer pitch of 130 Å of the yeast Rad51 filament in its crystal structure29.
Figure 2. Structure analysis of RAD51 protomer-protomer interface.
(a) AMP-PNP is buried in two neighboring RAD51 protomers coupling with ssDNA binding. Key residues involved in nucleotide binding and nucleic acid coupling are labeled in spheres. (b) The comparison of RAD51 and RecA in their nucleotide binding core. RAD51 is in limegreen and green for two protomers, respectively. RecA is in grey. Key residues involved in nucleotide binding and nucleic acid coupling are labeled in spheres for RecA. (c) The comparison of HsRAD51 and ScRad51 protomer interface. Green and salmon are used to present HsRAD51 and ScRad51, respectively, and AMP-PNP in the HsRAD51 presynaptic filament is shown in red. (d) The other two conserved protomer-protomer interfaces in eukaryotic Rad51 orthologs.
The second interaction interface is formed by a short β–strand of the linker region (residues 85-GFTT-88) between the N-terminal domain and ATPase domain of one protomer packing against the central β-sheet of the ATPase domain in the adjacent protomer (Fig. 2d). This is a unique feature of Rad51 as revealed in the crystal structures of yeast Rad51 and human RAD51 bound to the BRC peptide derived from the tumor suppressor BRCA2 28,29. The third interaction interface, which is conserved in the yeast Rad51 filament29, is an aromatic packing between residue Y54 in the N-terminal domain of one protomer and residue F195 in the ATPase domain of the adjacent protomer (Fig. 2d).
Among the protomer-protomer interactions, the ATP-mediated interface is mostly responsible to define the helical parameter of RAD51 assembly within the presynaptic and post-synaptic complexes while the other two interfaces further stabilize the assembly. Such a protomer-protomer interaction configuration also defines the DNA conformation within the complex.
Interaction of RAD51 with DNA in the presynaptic and post-synaptic complexes
As in the case of RecA, human RAD51 engages ssDNA and dsDNA in nucleotide triplet clusters. In both the presynaptic and post-synaptic complexes, two loop regions, Loop 1 and Loop 2, of RAD51 are involved in DNA interactions and play important roles in DNA strand exchange.
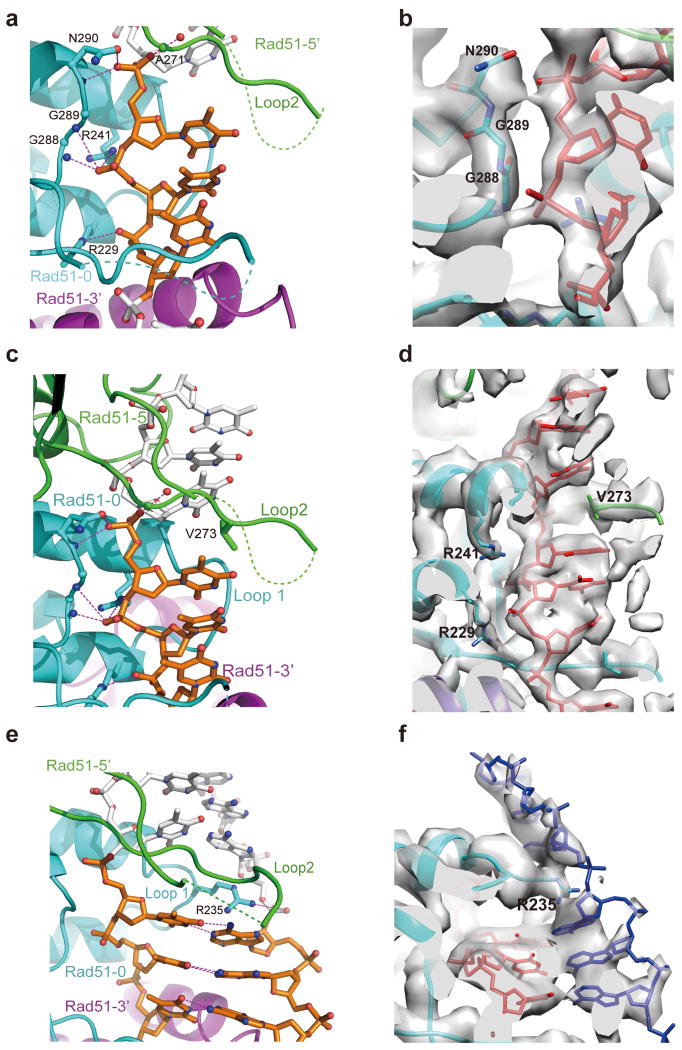
In the presynaptic complex, each triplet residues are held by three RAD51 protomers via the phosphate backbone of the DNA, with stacked bases being exposed and pointing towards the axial direction (Fig. 3a-3d, Video S1). We designate the three consecutive RAD51 protomers interacting with a nucleotide triplet from the 5′ to 3′ as RAD51-5′, RAD51-0 and RAD51-3′ (Fig. 3a). The nucleotide triplet is sandwiched by Loop 2 of RAD51-5′ and RAD51-0 in the axial direction and Loop 1 and Loop 2 of RAD51-0 in the tangential direction. From the 5′ end of a triplet, its first phosphate group interacts with residue A271 of RAD51-5′ and N290 of RAD51-0, which are equivalent to M197 and N213 in RecA. The second phosphate is bound by residues of RAD51-0 including G288 and G289 and R241, with the former two residues being the equivalents of G211 and G212 in RecA (Fig. 3b, 3d and S5). The third phosphate interacts with R229 of RAD51-0. V273 of Loop 2 in RAD51-5′, the equivalent of I199 in RecA, inserts into the space between the third base of the RAD51-5′ bound triplet and the first base of the RAD51-0 bound triplet, thus stabilizing the inter-triplet base separation with the help of other Loop 2 residues (Fig. 3d and S5). With these interactions, the RAD51 filament extends the ssDNA to ∼1.5 times the length of B-form DNA, a feature that is conserved in the RecA filament (Video S1)34.
Figure 3. Interaction of RAD51 with DNA in the presynaptic and post-synaptic complexes.
(a) In the presynaptic complex, each nucleotide triplet connects with three RAD51 protomers that are shown in green, cyan and purple following the direction from 5′ to 3′. (b) The EM density in corresponding view of (a) showing that ssDNA interacts with three RAD51 residues that are conserved in RecA. DNA is in red color. (c) The adjacent triplets in a presynaptic complex are separated and stabilized by V273 in Loop 2 of the middle RAD51. RAD51 mainly interacts with the backbone of ssDNA from the front view. (d) The EM density in corresponding view of (c) showing the interaction between V273 and ssDNA. DNA is in red color. (e) The complementary strand in the post-synaptic complex is observed in a similar extended state with ssDNA. R235 in Loop 1 facilitates the separation of the neighboring triplets from the back view of (b). Key residues are labeled with the potential hydrogen bonds highlighted. (f) The EM density in the corresponding view of (e) showing the interaction of R235 and the complementary strand in blue color. The EM density is shown at a high threshold so the complementary strand phosphate backbone is relatively weak. The invading strand is in red color.
Compared to the presynaptic complex, the interaction of RAD51 with the invading strand in the post-synaptic complex is nearly identical. Both the Loop 1 and Loop 2 regions are better resolved in the post-synaptic complex, indicating additional protein-DNA interactions to stabilize the exchanged product (Video S2, Fig. 3e and 3f). The complementary strand forms Watson-Crick base pairs with the invading strand also in triplet clusters (Video S2). We found that a specific residue R235, not available in RecA, in Loop 1 of RAD51-0 inserts into the space between two adjacent nucleotide triplets and interacts with the phosphate backbone of the complementary strand (Fig. 3e, 3f and S5). R235 also appears to undergo an electrostatic interaction with D274 in Loop 2 of the adjacent RAD51-5′ protomer (Video S2). The above observation is consistent with the finding that R235, when mutated to glutamate, abrogates DNA strand exchange even though it has only a slight impact on ssDNA binding by RAD51 42,43. The corresponding residue in E. coli RecA as identified by sequence homology alignment is D161, which appears to be important for the ssDNA binding specificity of RecA44. It is possible that R235 helps capture the DNA substrate and lower the energy state of the complementary strand after base-pairing with the invading strand has occurred.
Capture of an intermediate state of strand-exchange during synaptic complex formation
We have endeavored to visualize the displaced DNA strand in the post-synaptic complex but didn't detect any additional density in the final reconstruction. This suggests that the displaced strand either had already dissociated from the post-synaptic complex or was in a highly disordered configuration.
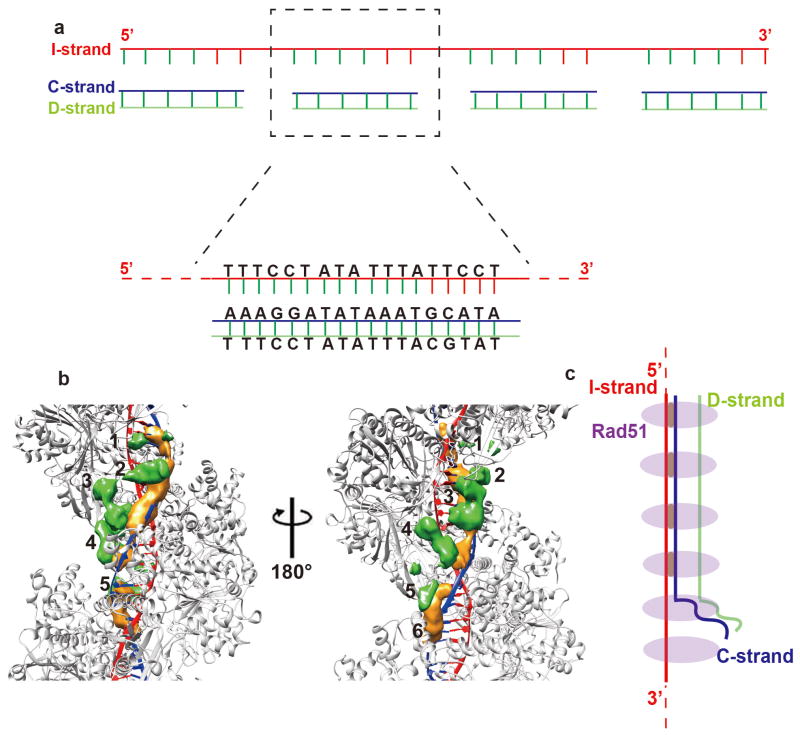
A recent study has shown that dsDNA with an identical region of 8 nucleotides can be captured by the presynaptic filament and undergoes strand exchange with the latter45. For capturing the synaptic complex by cryo-EM, we used as substrates a 72-nt ssDNA (Oligo 4 in Supplementary Table 1) with 4 identical 18-base repeats and an 18-bp dsDNA (Oligo 5/Oligo 6 in Supplementary Table 1) that bears a 5-base heterologous sequence (Fig. 4a). The use of these partially homologous DNA substrates yielded abundant helical RAD51 complexes (Fig. S6a), allowing us to apply the same helical reconstruction procedure to solve the structure of this assembly to ∼5 Å resolution. Interestingly, this structure largely resembles the post-synaptic complex with two DNA strands bound within the RAD51 helical assembly, indicating that strand exchange has occurred (Fig. S6b). Importantly, a close comparison with the post-synaptic complex formed by perfectly identical DNA substrates revealed that the complementary strand density is weaker in the reconstruction, suggesting that full strand exchange is prevented by the heterologous region in the substrates (Fig. S6b).
Figure 4. Capture of the arrested state in Rad51-mediated DNA strand exchange.
(a) Substrate design. Shown are the 72-mer invading strand that bears four 18-nt repeats and the 18-bp duplex that bears 13 nucleotides of homology to each of the repeats in the invading strand. The non-homologous nucleotides are in red. (b) The difference maps calculated by subtracting the presynaptic complex atomic model from the arrested synaptic complex 3D reconstruction with 15σ (orange color) and by subtracting the post-synaptic complex atomic model from the arrested synaptic complex 3D reconstruction with 9σ (green color) are superimposed on the atomic model of a post-synaptic complex, in which RAD51 protomers are colored grey, the invading strand is in red, and the complementary strand is in blue. Two orthogonal views are shown, with six consecutive protomers labeled by numbers. (c) A schematic interpretation of the arrested synaptic state in one hexameric RAD51 repeat. In (a) and (c), I-strand, C-strand, and D-strand stand for the invading strand, complementary strand, and displaced strand, respectively.
Because we were using the helical parameters with each RAD51 protomer as an asymmetric unit in the above reconstruction, any difference among the protomers became an average in the final reconstruction. But our uniquely designed DNA substrates may introduce structural variance within every 18 nucleotides or each helical repeat of 6 RAD51 protomers. In order to reveal the structural detail of the helical assembly, we developed an algorithm to align and reconstruct the helical assembly with every 6 contiguous RAD51 protomers as an asymmetric unit (Online Methods, Fig. S6c). This algorithm also aligns all the particle segments in register of the 18 nucleotides repeat thus allowing the capture of structural variations within each RAD51 hexameric repeat. By this method, we obtained a reconstruction of the complex at ∼12 Å resolution in which we can depict differences between the RAD51 protomers, among which the most significant one is an extra density between the ATPase domain and the DNA (Fig. S7a). These differences become more prominent upon subtracting the atomic models of post-synaptic or presynaptic complex separately from the EM map (Fig. S7b, S7c and Video S3). Within the RAD51 hexameric repeat units, every four consecutive protomers possess an extra density, whereas the last two protomers of these repeats lack this feature. Interestingly, the latter two RAD51 protomers also have a relatively weak complementary strand density, indicative of a lack of DNA strand exchange product within the confines of these protomers. Using the same image processing procedure, no extra density could be observed in the post-synaptic complex generated from the perfectly matched DNA substrates (Fig. S7d, S7e, and S7f).
Taken together, the results above reveal a synaptic reaction where strand exchange proceeds normally but becomes arrested upon encountering the heterologous region in the duplex substrate. In this arrested state, the displaced strand remains within the vicinity of the synaptic complex, likely accounting for the extra densities in the 3D reconstruction (Fig. 4c).
Discussion
Cryo-EM allowed us to visualize the different states of human RAD51-DNA complex during homologous recombination process at high resolution. The near-atomic resolution structure of presynaptic complex and post-synaptic complex has revealed similar interaction fashion but distinct features of RAD51 and bacterial RecA with their ssDNA and dsDNA substrates.
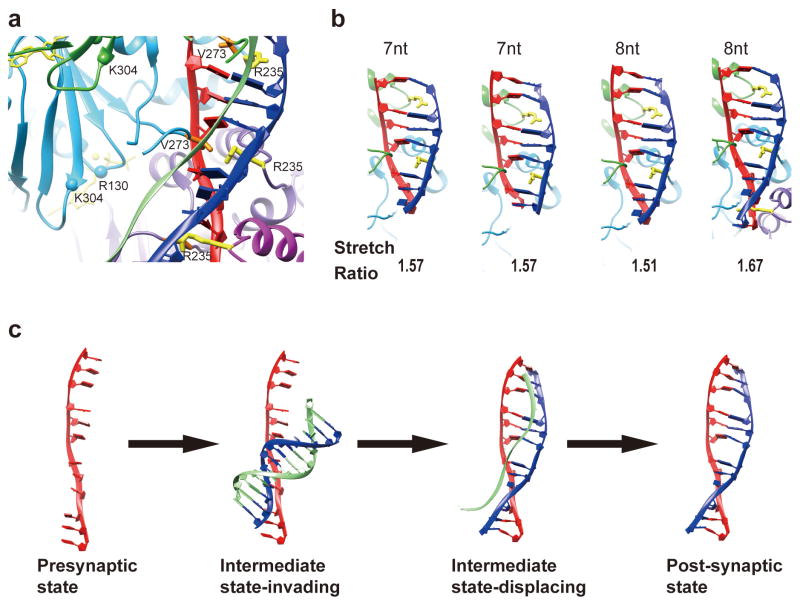
Using a specially designed DNA substrate combination, we are able to capture the structure of an arrested strand-exchange synaptic complex, in which the extra densities are proximal to the C-terminal region of the ATPase domain of RAD51. If they are from the displaced DNA strand, this would be consistent with the premise that a “secondary DNA binding site”, which encompasses R130 and K304 46, facilitates strand separation in the homologous dsDNA partner (Fig. 5a). Arg235 likely helps stabilize the complementary strand in its stretched conformation. Whether the same secondary DNA binding site is responsible for dsDNA recruitment remains to be addressed.
Figure 5. Model of RAD51-mediated DNA strand exchange with depiction of the intermediate state.
(a) The interaction of RAD51 and three DNA strands in the intermediate state. The displaced strand is located in proximity to the C-terminal part of RAD51's potential secondary DNA binding site. The invading strand (red) and the complementary strand (blue) are stabilized by V273 and R235, respectively. (b) Interaction of R235 with the complementary strand and the various stretch ratios in different length of DNA compared to the B-form DNA. (c) A hypothetical model of DNA molecule transition during the synaptic reaction.
The stabilization of the complementary strand within the post-synaptic complex in its extended state by R235 could provide a mechanistic explanation for the recent finding that a region of 8 contiguous homologous nucleotides represents the baseline for successful pairing between recombining DNA molecules. In the post-synaptic complex, the complementary strand is both under-twisted and extended as compared to B-form DNA. While the average strand extension ratio is ∼1.5, we have noticed that over a short range, e.g. 8 nucleotides, the extension ratio can reach 1.67 for a complementary strand engaged by four consecutive RAD51 protomers (the fourth configuration in Fig. 5b). We surmise that when a strand harboring 8 nucleotides of contiguous homology pairs with the invading strand, it becomes more stabilized by three R235 residues in an extended conformation, resulting in an energy state that disfavors conversion to B-form DNA. Other factors could also be involved to further stabilize the synaptic ensemble, e.g. the secondary binding site of RAD51 may help maintain the displaced strand in the unwound state (Fig. 5c). When the pairing sequence has fewer than 8 homologous nucleotides, the DNA joint is inherently labile, as, in this case, no more than two R235 residues are engaged in stabilizing the nascent DNA joint (the first and second configurations in Fig. 5b). Our model predicts the presence of other configurations of 8-nt based micro-homology search in which the DNA extension ratio is smaller (1.51) and the extended strand is being held by fewer R235 residues (for example, the third configuration in Fig. 5b). Such configurations may be less stable. The existence of heterogeneous configurations would also apply to search steps involving more than 8 nucleotides of homology. It will be interesting to investigate in the future whether there are multiple species with different stability states in the homologous recombination reaction. These species may be the target of regulation by accessory factors of the recombinase. The cryo-EM structures capturing different stages of the DNA strand exchange process and the conceptual models (Fig. 5c) that we have presented above should provide the frameworks to delineate how various factors regulate RAD51-dependent homologous recombination and chromosome damage repair.
Online Methods
DNA substrates and protein
The DNA substrates with their sequences are listed in Supplementary Table 1. All the DNA substrates were prepared and purified by gel electrophoresis, as described47. The dsDNA substrates were prepared by annealing Oligo 2 with Oligo 3, Oligo 5 with Oligo 6 or Oligo7 with Oligo 8. (Supplementary Table 1).
Human RAD51 protein was expressed in E. coli and purified using our published procedure48. The mouse Hop2-Mnd1 complex was expressed in E. coli and purified as described before49.
Homologous DNA pairing assay
The reaction was assembled in buffer (25 mM Tris-HCl, pH 7.5, 50 mM KCl, and 1 mM dithiothreitol) that contained 1 mM AMP-PNP and 2 mM MgCl2 in a final volume of 12.5 μl. The 150-mer oligo 2 (6 μM nucleotides) was incubated with RAD51 (2 μM) at 37°C for 5 min, followed by the addition of Hop2-Mnd1 (100, 200 or 400 nM) for 5 min's incubation. Then, 32P-labeled homologous dsDNA (Oligo7/Oligo8, 6 μM base pairs) was added for initiating the pairing reaction. After 15 min of incubation, the reactions were mixed with an equal volume of 1% SDS containing 1 mg/ml proteinase K. Following a 5-min incubation, the deproteinized reaction mixtures were resolved in 10% non-denaturing polyacrylamide gels in TAE buffer (30 mM Tris-acetate, pH 7.4, 0.5 mM EDTA) at 4°C. The gels were dried, and the products were visualized by autoradiography and quantified using the Quantity One software in the Personal Molecular ImagerTM (Bio-Rad)49.
Presynaptic and post-synaptic complex assembling
All the reaction steps were carried out at 37°C. For assembling pre-synaptic filaments, 72-mer poly(dT) (Oligo 1) (6 μM nucleotides), was incubated with RAD51 (2 μM) in the reaction buffer comprising of 25 mM HEPES, pH 7.5, 25 mM KCl, 4 mM MgCl2 and 4 mM AMP-PNP (Sigma) at 37°C for 60 min. As a control, the complex was also incubated with Hop2-Mnd1 (250 nM) for 25 minutes before making EM samples.
For DNA strand exchange reactions, the 150-mer ssDNA Oligo 2 (6 μM nucleotide) was incubated with RAD51 (2 μM) at 37°C for 60 min. Following the incubation with Hop2-Mnd1 (250 nM) for 10 minutes, the reaction was initiated by adding the homologous dsDNA (Oligo 2/Oligo 3; 5.1 μM base pairs) and incubated for additional 15 minutes.
Assembly of arrested synaptic complex
The 72-mer Oligo 4 (6 μM nucleotide) was used to assemble RAD51 2 μM presynaptic filaments as described above. The filaments were incubated with Hop2-Mnd1 for 10 min before the 18-bp dsDNA (Oligo 5/Oligo 6; 6 μM base pairs) was added for the synaptic reaction. The reaction was incubated for additional 20 min before cryo-EM sample preparation.
Cryo-EM specimen preparation and electron microscopy data acquisition
To prepare the cryo-EM specimen, 4 μl of the reaction mixture containing presynaptic, post-synaptic or arrested synaptic complex was placed on a glow-discharged holey carbon grid (Quantifoil Cu R1.2/1.3) and flash plunged into liquid ethane cooled at liquid nitrogen temperature using an FEI Vitrobot Mark IV with a blotting time of 3.5 s and blotting force of -2 in an environment with 100 % humidity and 20°C temperature. The grids of presynaptic and post-synaptic complexes were transferred to a Titan Krios electron microscope (FEI company) operated at the accelerating voltage of 300 kV equipped with K2 Summit direct electron counting camera (Gatan company) (Table 1). Movies of the specimens were recorded manually at a nominal magnification of 22,500 with K2 super-resolution mode yielding a pixel size of 0.653 Å using the UCSF-Image4 data collection user interface50. The specimen was exposed with a dose rate of approximately 8 counts/sec/physical-pixel (∼6.2 e-/sec/Å2) and total exposure time of 8 s with 0.25 s per frame (total 32 frames) with a defocus value ranging from -1.5 to -2.5 μm. For the arrested synaptic complex, the cryo grids were transferred to a Titan Krios microscope operated at 300 kV accelerating voltage and equipped with a Falcon II direct electron detecting device (FEI company) (Table 1). Movies of the specimen were collected on the microscope with defocus ranging from -1.5 to -2.3 μm at a nominal magnification of 75,000 with a pixel size of 1.05 Å at a dose rate of about 30 e-/sec/Å2 and total exposure time of 1.7 s with 27 frames per exposure.
Image processing of presynaptic and post-synaptic complexes
The 32 frames of dose-fractionated movie images were first aligned and summed with whole frame motion correction using dosef_gpudriftcorr51 and binned 2 fold to generate a drift-corrected summation of each movie frames. Further processing used the motion-corrected summation micrographs. The CTF parameters of each micrograph were determined using CTFFIND3 52. For each helical filament in a micrograph, we used EMAN2 subroutine e2helixboxer.py to extract the helical particle segment coordinates with a box size of 256×256 pixels and 90% overlap between adjacent particles (Table 1). Three dimensional reconstruction were executed independently in a SPIDER version of IHRSR and a modified RELION1.2 version of IHRSR (Figure S2)53-55.
In the SPIDER reconstruction approach, particles were extracted from CTF phase-flipped micrographs and submitted for 2D classification in the IMAGIC-4D package56 to exclude bad particle images. Only the particles in 2D class averages showing clear features were retained for 3D reconstruction using IHRSR in SPIDER57. The particles were aligned against a cylindrical initial model using initial helical parameters of Δφ=56.8° and Δz=15.7 Å (Table 1). After about 50 iterations, both the helical parameters and the 3D reconstruction converged to a stable solution. At the final step, we corrected the 3D map for the CTF amplitude by weighting the amplitudes calculated and averaged using the defocus values from all the images used for the back projections and for the Gaussian envelop half-width using a value of 0.25. The resolutions of the reconstruction estimated from the Fourier shell correlation (FSC) between reconstructions derived from two randomly selected halves of the data set with 0.5 criterion were about 3.8 and 4.0 Angstroms for presynaptic and post-synaptic complexes, respectively.
In the RELION reconstruction approach, the overall procedure is similar to general single particle analysis but uses a modified version of RELION with IHRSR implementation54. Briefly, reference-free 2D classification was performed to remove bad particle images. The initial model for 3D analysis was a low-pass filtered model at 60 Å resolution from the reconstruction by SPIDER approach. We performed 3D classification and refinement as in regular single particle analysis but imposed the converged helical parameters from IHRSR at the end of each iteration. The final converged reconstructions had exactly the same helical parameters as those calculated by the SPIDER approach. In order to estimate a “gold-standard” FSC resolution, we masked three neighboring protomers from two independently RELION helical reconstructed volumes from randomly separated half dataset (Fig. S2b and S2d). Based on the 0.143 criteria, these gave resolution estimation of 4.4 and 4.5 Angstrom of the presynaptic complex and post-synaptic complex, respectively. The maps were post-processed in RELION 1.2 to manually filter to the final resolution and a B-factor of -250 Å2 was applied to sharpen the maps.
Image processing of the arrested synaptic complex
For the arrested synaptic complex, the 27 movie frames were first motion-corrected using the dosef_gpudriftcorrr. We only used the motion-corrected summation of the movies for further image processing. In order to account for the potential structural difference with every 18-nt DNA repeat or 6 consecutive Rad51 protomers, we developed an algorithm to perform reconstruction of the complex in hexameric repeat (Fig. S6c). First, we picked the helical particle segments from each filament in the motion-corrected micrographs with a box size of 400×400 pixels (Table 1). The adjacent segments from the same filament have a non-overlap length of 90 pixels along the helical axis, corresponding to the rise of about one repeat of RAD51 hexamer. We firstly used the same reconstruction method as the presynaptic and post-synaptic complexes in SPIDER and searched for the converged helical parameters for RAD51 protomers.
In parallel, we used FREALIGN to perform the 3D reconstruction58. We firstly performed global parameter search using the classesbymra program of EMAN against the reference from SPIDER reconstruction. This was followed by two rounds of local refinement in FREALIGN to generate a reconstruction with RAD51 monomer as asymmetric unit in the helical assembly. Subsequently, for each filament, we re-assign the azimuthal angle of the consecutive segments to a unified set of values to follow the helical order with hexameric RAD51 repeat. We call this step “unify phi”, after which Euler angles of all the segments from the same helical filament are linked together with a unified relationship. From the longest filament image that we obtained, we thus could calculate a 3D reconstruction with hexameric RAD51 repeat as the asymmetric unit of the helical assembly. Such a model is used as reference to align all the other filaments. Because the 18-bp dsDNA has unique homologous pattern on the ssDNA, we needed to find the register of the starting RAD51 protomer of a consecutive hexameric unit that binds each 18 nucleotide repeat among the different helical assemblies. After the “unify phi” step, there are 6 possibilities for all the segments in a particular filament to match with the reference, each with an overall cross-correlation score. We calculated the value for each filament for the 6 possibilities against the reference and picked the highest score value as the correct option59. After this “register search” step, we calculated a new 3D reconstruction and updated the reference with the new map. After a few iterations of the register search, the Euler angles for all the segments stabilized and the 3D reconstruction converged to a final solution by FREALIGN.
Model building and refinement
The initial atomic model of human RAD51 protomers in both the presynaptic and post-synaptic complexes was generated from the crystal structure of yeast Rad51 (PDB ID: 1SZP) by CHAINSAW60 and rigid-body docked into the electron density map in UCSF-Chimera61. The two loops L1 (232-YSGRGEL-238) and L2 (270-VAQVDGAAMFAADPKKPI-287) involved in DNA binding are highly flexible and therefore absent in the crystal structure. We manually built the model of these two portions based on our EM map using COOT62. We also appended DNA and AMP-PNP manually in COOT, with Mg2+ added as a rule of thumb. The structure was further refined in real space in PHENIX with secondary structure restraint63. The models built were duplicated multiple times and fitted into the EM maps of the presynaptic and post-synaptic complexes in UCSF-Chimera following their corresponding helical symmetries to generate the atomic models of longer assemblies. We then manually refined the atomic models of RAD51 and DNA strands at the protomer interface in each assembly structure in COOT. Finally, the models of presynaptic and post-synaptic assemblies were refined in real-space in PHENIX with secondary structure restraint (Table 1)63.
The atomic models were cross-validated with the same method as previously described64. Briefly, each model was firstly refined against the best reconstruction. To monitor the refinement of the model and avoid over-fitting, the final model was refined against one half map (working map) and tested against the other half map (testing map) by calculating the FSC curves (Fig. S2c and S2e), which indicated that the refinement of the atomic coordinates did not suffer from over-fitting.
Data Accessibility
The EM reconstructions and their corresponding atomic coordinates are deposited in the Electron Microscopy Data Bank and the Protein Data Bank under accession codes EMD-9566 (presynaptic complex), EMD-9567 (post-synaptic complex), EMD-9568 (arrested synaptic complex), PDB 5H1B (presynaptic complex), PDB 5H1C (post-synaptic complex). Source data files and construct materials related to this work are available upon request.
Supplementary Material
Acknowledgments
We thank J.L. Lei, Y.J. Xu, T. Yang for their support in cryo-EM and high performance computation facility at the National Protein Science Facility (Beijing) at Tsinghua University. We thank X. Li for help in data collection, E. Egelman for his advice in setting up the IHRSR algorithm, T. Baker for distributing the IHRSR scripts in the SPIDER package, P. Ge and Z.H. Zhou for distributing the IHRSR-incorporated version of RELION1.2. Special thanks to C. Yan in helping building the atomic models. This work is supported by the National Science Foundation of China grant 31270765, the Key Research and Development Program of MOST grant 2016YFA0501100, the Beijing Municipal Science & Technology Commission grant Z161100000116034 to H.-W.W. and US National Institutes of Health grants CA168635, ES007061 and ES015252 to P.S.
Footnotes
Author Contributions: H.-W.W. conceived the project. H.-W.W. and P.S. designed the experiments. J.X., L.Z., and Y.X. performed EM and structural determination. W.Z. generated key research materials and performed biochemical assays. J.X., L.Z., P.S., and H.-W.W. wrote the manuscript.
Accession Codes: The 4.4 Å, 4.5 Å resolution EM structures of presynaptic and post-synaptic complex have been deposited in the Electron Microscopy Data Bank under accession codes EMD-9566 and EMD-9567 respectively. The atomic models of the presynaptic and post-synaptic complex have been deposited in the Protein Data Bank under accession codes PDB 5H1B and 5H1C. The EM density map of arrested synaptic complex have also been deposited in the Electron Microscopy Data Bank under accession code EMD-9568.
References
- 1.Wang AT, et al. A Dominant Mutation in Human RAD51 Reveals Its Function in DNA Interstrand Crosslink Repair Independent of Homologous Recombination. Molecular Cell. 2015;59:478–490. doi: 10.1016/j.molcel.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mazon G, Mimitou EP, Symington LS. SnapShot: Homologous Recombination in DNA Double-Strand Break Repair. Cell. 2010;142 doi: 10.1016/j.cell.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 3.Lambert S, et al. Homologous Recombination Restarts Blocked Replication Forks at the Expense of Genome Rearrangements by Template Exchange. Molecular Cell. 2010;39:346–359. doi: 10.1016/j.molcel.2010.07.015. [DOI] [PubMed] [Google Scholar]
- 4.van den Bosch M, Lohman PHM, Pastink A. DNA double-strand break repair by homologous recombination. Biological Chemistry. 2002;383:873–892. doi: 10.1515/BC.2002.095. [DOI] [PubMed] [Google Scholar]
- 5.Takata M, et al. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. Embo Journal. 1998;17:5497–5508. doi: 10.1093/emboj/17.18.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Da Ines O, Gallego ME, White CI. Recombination-independent mechanisms and pairing of homologous chromosomes during meiosis in plants. Mol Plant. 2014;7:492–501. doi: 10.1093/mp/sst172. [DOI] [PubMed] [Google Scholar]
- 7.Habu T, Taki T, West A, Nishimune Y, Morita T. The mouse and human homologs of DMC1, the yeast meiosis-specific homologous recombination gene, have a common unique form of exon-skipped transcript in meiosis. Nucleic Acids Res. 1996;24:470–7. doi: 10.1093/nar/24.3.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krejci L, Altmannova V, Spirek M, Zhao X. Homologous recombination and its regulation. Nucleic Acids Res. 2012;40:5795–818. doi: 10.1093/nar/gks270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sung P, Klein H. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat Rev Mol Cell Biol. 2006;7:739–50. doi: 10.1038/nrm2008. [DOI] [PubMed] [Google Scholar]
- 10.Wright WD, Heyer WD. Rad54 Functions as a Heteroduplex DNA Pump Modulated by Its DNA Substrates and Rad51 during D Loop Formation. Molecular Cell. 2014;53:420–432. doi: 10.1016/j.molcel.2013.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galkin VE, et al. The Rad51/RadA N-terminal domain activates nucleoprotein filament ATPase activity. Structure. 2006;14:983–92. doi: 10.1016/j.str.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 12.Gupta RC, Bazemore LR, Golub EI, Radding CM. Activities of human recombination protein Rad51. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:463–468. doi: 10.1073/pnas.94.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benson FE, Stasiak A, West SC. Purification and characterization of the human Rad51 protein, an analogue of E. coli RecA. EMBO J. 1994;13:5764–71. doi: 10.1002/j.1460-2075.1994.tb06914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seitz EM, Brockman JP, Sandler SJ, Clark AJ, Kowalczykowski SC. RadA protein is an archaeal RecA protein homolog that catalyzes DNA strand exchange. Genes & Development. 1998;12:1248–1253. doi: 10.1101/gad.12.9.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang SX, Yu X, Seitz EM, Kowalczykowski SC, Egelman EH. Archaeal RadA protein binds DNA as both helical filaments and octameric rings. Journal of Molecular Biology. 2001;314:1077–1085. doi: 10.1006/jmbi.2000.5213. [DOI] [PubMed] [Google Scholar]
- 16.Hilario J, Amitani I, Baskin RJ, Kowalczykowski SC. Direct imaging of human Rad51 nucleoprotein dynamics on individual DNA molecules. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:361–368. doi: 10.1073/pnas.0811965106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galletto R, Amitani I, Baskin RJ, Kowalczykowski SC. Direct observation of individual RecA filaments assembling on single DNA molecules. Nature. 2006;443:875–878. doi: 10.1038/nature05197. [DOI] [PubMed] [Google Scholar]
- 18.Register JC, Christiansen G, Griffith J. Electron-Microscopic Visualization of the Reca Protein-Mediated Pairing and Branch Migration Phases of DNA Strand Exchange. Journal of Biological Chemistry. 1987;262:12812–12820. [PubMed] [Google Scholar]
- 19.Ragunathan K, Joo C, Ha T. Real-Time Observation of Strand Exchange Reaction with High Spatiotemporal Resolution. Structure. 2011;19:1064–1073. doi: 10.1016/j.str.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mani A, Braslavsky I, Arbel-Goren R, Stavans J. Caught in the act: the lifetime of synaptic intermediates during the search for homology on DNA. Nucleic Acids Research. 2010;38:2036–2043. doi: 10.1093/nar/gkp1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Folta-Stogniew E, O'Malley S, Gupta R, Anderson KS, Radding CM. Exchange of DNA base pairs that coincides with recognition of homology promoted by E-coli RecA protein. Molecular Cell. 2004;15:965–975. doi: 10.1016/j.molcel.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 22.Danilowicz C, et al. The differential extension in dsDNA bound to Rad51 filaments may play important roles in homology recognition and strand exchange. Nucleic Acids Research. 2014;42:526–533. doi: 10.1093/nar/gkt867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Danilowicz C, et al. RecA homology search is promoted by mechanical stress along the scanned duplex DNA. Nucleic Acids Research. 2012;40:1717–1727. doi: 10.1093/nar/gkr855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li X, et al. Rad51 and Rad54 ATPase activities are both required to modulate Rad51-dsDNA filament dynamics. Nucleic Acids Research. 2007;35:4124–4140. doi: 10.1093/nar/gkm412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanchez H, Kertokalio A, van Rossum-Fikkert S, Kanaar R, Wyman C. Combined optical and topographic imaging reveals different arrangements of human RAD54 with presynaptic and postsynaptic RAD51-DNA filaments. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:11385–11390. doi: 10.1073/pnas.1306467110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van der Heijden T, et al. Homologous recombination in real time: DNA strand exchange by RecA. Molecular Cell. 2008;30:530–538. doi: 10.1016/j.molcel.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 27.Wu Y, He Y, Moya IA, Qian XG, Luo Y. Crystal structure of archaeal recombinase RadA: A snapshot of its extended conformation. Molecular Cell. 2004;15:423–435. doi: 10.1016/j.molcel.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 28.Pellegrini L, et al. Insights into DNA recombination from the structure of a RAD51-BRCA2 complex. Nature. 2002;420:287–293. doi: 10.1038/nature01230. [DOI] [PubMed] [Google Scholar]
- 29.Conway AB, et al. Crystal structure of a Rad51 filament. Nature Structural & Molecular Biology. 2004;11:791–796. doi: 10.1038/nsmb795. [DOI] [PubMed] [Google Scholar]
- 30.Story RM, Weber IT, Steitz TA. The structure of the E. coli recA protein monomer and polymer. Nature. 1992;355:318–25. doi: 10.1038/355318a0. [DOI] [PubMed] [Google Scholar]
- 31.Yu X, Jacobs SA, West SC, Ogawa T, Egelman EH. Domain structure and dynamics in the helical filaments formed by RecA and Rad51 on DNA. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:8419–8424. doi: 10.1073/pnas.111005398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.VanLoock MS, et al. ATP-Mediated conformational changes in the RecA filament. Structure. 2003;11:187–196. doi: 10.1016/s0969-2126(03)00003-0. [DOI] [PubMed] [Google Scholar]
- 33.Egelman EH. Does a stretched DNA structure dictate the helical geometry of RecA-like filaments? Journal of Molecular Biology. 2001;309:539–542. doi: 10.1006/jmbi.2001.4686. [DOI] [PubMed] [Google Scholar]
- 34.Chen ZC, Yang HJ, Pavletich NP. Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature. 2008;453:489–U3. doi: 10.1038/nature06971. [DOI] [PubMed] [Google Scholar]
- 35.Zhao W, Sung P. Significance of ligand interactions involving Hop2-Mnd1 and the RAD51 and DMC1 recombinases in homologous DNA repair and XX ovarian dysgenesis. Nucleic Acids Res. 2015;43:4055–66. doi: 10.1093/nar/gkv259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cho NW, Dilley RL, Lampson MA, Greenberg RA. Interchromosomal homology searches drive directional ALT telomere movement and synapsis. Cell. 2014;159:108–21. doi: 10.1016/j.cell.2014.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Egelman EH. The iterative helical real space reconstruction method: surmounting the problems posed by real polymers. J Struct Biol. 2007;157:83–94. doi: 10.1016/j.jsb.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 38.Short JM, et al. High-resolution structure of the presynaptic RAD51 filament on single-stranded DNA by electron cryo-microscopy. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Forget AL, Loftus MS, McGrew DA, Bennett BT, Knight KL. The human Rad51 K133A mutant is functional for DNA double-strand break repair in human cells. Biochemistry. 2007;46:3566–75. doi: 10.1021/bi062128k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Flott S, et al. Regulation of Rad51 function by phosphorylation. EMBO Rep. 2011;12:833–9. doi: 10.1038/embor.2011.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Amunugama R, et al. RAD51 protein ATP cap regulates nucleoprotein filament stability. J Biol Chem. 2012;287:8724–36. doi: 10.1074/jbc.M111.239426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reymer A, Frykholm K, Morimatsu K, Takahashi M, Norden B. Structure of human Rad51 protein filament from molecular modeling and site-specific linear dichroism spectroscopy. Proc Natl Acad Sci U S A. 2009;106:13248–53. doi: 10.1073/pnas.0902723106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prasad TK, Yeykal CC, Greene EC. Visualizing the assembly of human Rad51 filaments on double-stranded DNA. J Mol Biol. 2006;363:713–28. doi: 10.1016/j.jmb.2006.08.046. [DOI] [PubMed] [Google Scholar]
- 44.Shinohara T, et al. Loop L1 governs the DNA-binding specificity and order for RecA-catalyzed reactions in homologous recombination and DNA repair. Nucleic Acids Res. 2015;43:973–86. doi: 10.1093/nar/gku1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qi Z, et al. DNA sequence alignment by microhomology sampling during homologous recombination. Cell. 2015;160:856–69. doi: 10.1016/j.cell.2015.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cloud V, Chan YL, Grubb J, Budke B, Bishop DK. Rad51 is an accessory factor for Dmc1-mediated joint molecule formation during meiosis. Science. 2012;337:1222–5. doi: 10.1126/science.1219379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chi P, Van Komen S, Sehorn MG, Sigurdsson S, Sung P. Roles of ATP binding and ATP hydrolysis in human Rad51 recombinase function. DNA Repair. 2006;5:381–391. doi: 10.1016/j.dnarep.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 48.Sigurdsson S, Trujillo K, Song B, Stratton S, Sung P. Basis for avid homologous DNA strand exchange by human Rad51 and RPA. J Biol Chem. 2001;276:8798–806. doi: 10.1074/jbc.M010011200. [DOI] [PubMed] [Google Scholar]
- 49.Chi P, San Filippo J, Sehorn MG, Petukhova GV, Sung P. Bipartite stimulatory action of the Hop2-Mnd1 complex on the Rad51 recombinase. Genes Dev. 2007;21:1747–57. doi: 10.1101/gad.1563007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abrishami V, et al. Alignment of direct detection device micrographs using a robust Optical Flow approach. J Struct Biol. 2015;189:163–76. doi: 10.1016/j.jsb.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 51.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10:584–90. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–47. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 53.Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–30. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Clemens DL, Ge P, Lee BY, Horwitz MA, Zhou ZH. Atomic structure of T6SS reveals interlaced array essential to function. Cell. 2015;160:940–51. doi: 10.1016/j.cell.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Frank J, et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–9. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 56.van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. J Struct Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- 57.Ge P, Zhou ZH. Hydrogen-bonding networks and RNA bases revealed by cryo electron microscopy suggest a triggering mechanism for calcium switches. Proc Natl Acad Sci U S A. 2011;108:9637–42. doi: 10.1073/pnas.1018104108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grigorieff N. FREALIGN: high-resolution refinement of single particle structures. J Struct Biol. 2007;157:117–25. doi: 10.1016/j.jsb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 59.Zhang R, Nogales E. A new protocol to accurately determine microtubule lattice seam location. J Struct Biol. 2015;192:245–54. doi: 10.1016/j.jsb.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stein N. CHAINSAW: a program for mutating pdb files used as templates in molecular replacement. Journal of Applied Crystallography. 2008;41:641–643. [Google Scholar]
- 61.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 62.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 63.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–21. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yan C, et al. Structure of a yeast spliceosome at 3.6-angstrom resolution. Science. 2015;349:1182–91. doi: 10.1126/science.aac7629. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The EM reconstructions and their corresponding atomic coordinates are deposited in the Electron Microscopy Data Bank and the Protein Data Bank under accession codes EMD-9566 (presynaptic complex), EMD-9567 (post-synaptic complex), EMD-9568 (arrested synaptic complex), PDB 5H1B (presynaptic complex), PDB 5H1C (post-synaptic complex). Source data files and construct materials related to this work are available upon request.