Abstract
Intracellular membrane fusion is mediated in most cases by membrane-bridging complexes of soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs). Yet in vitro, the assembly of such complexes is inefficient, and their uncatalyzed disassembly is undetectably slow. Here, we focus on the cellular machinery that orchestrates SNARE complex assembly and disassembly, thereby regulating processes ranging from vesicle trafficking to organelle fusion to neurotransmitter release. Rapid progress is being made on many fronts, including the development of more realistic cell-free reconstitutions, the application of single-molecule biophysics, and the elucidation of x-ray and high-resolution electron microscopy structures of the SNARE assembly and disassembly machineries ‘in action’.
Graphical abstract
Recent structural, biochemical and single-molecule biophysical studies have elucidated the molecular mechanisms underlying the control of SNARE complex assembly and disassembly by chaperones.
Vesicular transport in eukaryotic cells is driven by the protein nanomachines that mediate vesicle budding, vesicle movement, and vesicle docking and fusion. The culminating event, membrane fusion, delivers the cargo contained within a vesicle to a cellular compartment such as the Golgi or, in the case of exocytosis, releases it from the cell. Vesicle–target membrane fusion depends on the assembly of membrane-bridging complexes formed by soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs) [G] 1-5 (Box 1). SNAREs are also essential for some forms of ‘homotypic’ fusion, such as that between vacuoles in yeast or between early endosomes in neurosecretory cells; other homotypic fusion events (for example, between ER membranes or mitochondria) require dynamin-related proteins6. The ability of SNAREs to mediate membrane fusion thus enables a broad array of crucial cell biological processes. With great power, however, comes great risks – uncontrolled membrane fusion could quickly destroy the interior architecture of eukaryotic cells. Accordingly, the assembly and disassembly of SNARE complexes is carefully controlled7,8.
Box 1. SNARE architecture and nomenclature.
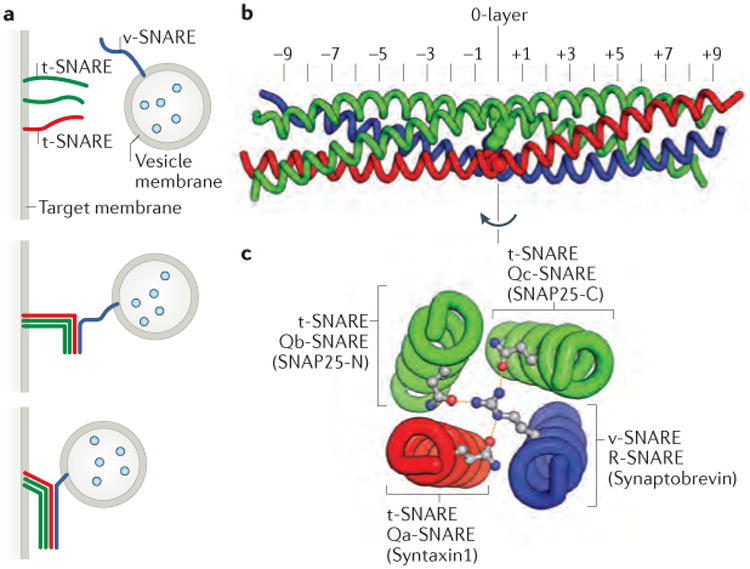
Soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs) are defined by a 60- to 70-residue ‘SNARE motif’ that can fold into an amphipathic α-helix4,5. Most but not all SNAREs contain, adjacent to the SNARE motif, a C-terminal transmembrane helix anchor. Many SNAREs also have N-terminal domains that regulate SNARE complex assembly and/or interact with other elements of the vesicle docking and fusion machinery. The formation of a productive SNARE complex entails the coupled folding and assembly of an intermolecular four-helix bundle containing four different SNARE motifs (see the figure). This bundle, at its longitudinal midpoint, contains a ‘zero layer’ comprising four large, hydrophilic residues — usually arginine or glutamine3. The zero layer residues, which are buried within the otherwise hydrophobic core of the SNARE complex, hydrogen bond with one another, presumably to ensure that the four SNARE motif α-helices assemble in the proper register5. SNAREs can be defined as vesicle-associated SNAREs (v-SNAREs) or target membrane-associated SNAREs (t-SNAREs) (see the figure, part a). Alternatively, they can be defined by the residue they contribute to the zero layer and their position within the four-helix bundle: R-SNAREs contribute an arginine to the zero layer, whereas Qa-, Qb-, and Qc-SNAREs each contribute a glutamine3 (see the figure, parts b and c). A few SNARE proteins, such as SNAP25, contain both Qb- and Qc-SNARE motifs within a single polypeptide chain. Although the original v-SNARE and t-SNARE terminology is still widely used, it is neither as specific (it does not distinguish among t-SNAREs) nor as broadly applicable (it does not apply to homotypic fusion) as the R-SNARE and Q-SNARE nomenclature. However, it is generally easy to translate between the two nomenclatures, as most v-SNAREs contain zero-layer arginines and are therefore R-SNAREs, whereas most t-SNAREs contain zero-layer glutamines and are therefore Q-SNAREs.
This Review focuses on recent progress towards elucidating the molecular mechanisms underlying SNARE complex assembly and disassembly and their control by chaperones including membrane tethering factors, Sec1-Munc18 proteins (SM proteins) [G], and N-ethylmaleimide-sensitive fusion factor (NSF) [G]. Membrane tethering factors8-10 not only mediate the initial contact between a vesicle and its target membrane but also bind SNAREs to regulate their assembly. Tethering factors work together with SM proteins1,11, which can function as templates for SNARE complex formation12. These and other findings shed new light on the mechanisms underlying synaptic vesicle fusion and its regulation by the Ca2+ sensor protein synaptotagmin13,14. The SNARE disassembly machinery consists of NSF and soluble NSF attachment proteins (SNAPs) [G]; recent structural and single-molecule biophysical studies have markedly enhanced our understanding of how stable SNARE complexes are dismantled so that the constituent SNAREs can be re-cycled and re-used15. Overall, it seems increasingly plausible that vesicle docking, SNARE complex assembly, membrane fusion, and SNARE complex disassembly and recycling may be viewed as an integrated set of reactions mediated by overlapping sets of molecular components.
SNARE complex assembly and disassembly
SNARE complex assembly, which generally involves four different SNAREs anchored in two different membranes, is topologically complex. We therefore begin the discussion with some general considerations regarding assembly–disassembly cycles and off-pathway intermediates.
SNARE complexes mediate fusion
The initial discovery of SNAREs, which were affinity purified from a bovine brain extract, resulted in the identification of three SNAREs that are essential for neurotransmitter release16. One of them, synaptobrevin, resides on synaptic vesicles, whereas the other two, syntaxin 1 and SNAP25 (synaptosomal-associated protein, 25 kDa), reside on the presynaptic plasma membrane. Synaptobrevin and syntaxin 1, similarly to most SNAREs, contain a single SNARE motif adjacent to a C-terminal transmembrane helix. SNAP25 is atypical in that it contains two SNARE motifs (SNAP25-N and SNAP25-C; see Box 1) and lacks a transmembrane anchor. Instead, SNAP25 is associated with the presynaptic membrane through palmitoyl groups that are attached to cysteine residues within the linker that joins the two SNARE motifs.
The segregation of synaptobrevin, syntaxin 1 and SNAP25 between the two membranes inspired the idea that vesicle-associated SNAREs (v-SNAREs) pair with target-membrane-associated SNAREs (t-SNARES) to generate membrane-bridging trans-SNARE complexes [G] that are capable of driving membrane fusion (FIG. 1a)17,18. Synthetic liposomes containing synaptobrevin indeed undergo lipid mixing (a proxy for fusion) with synthetic liposomes containing syntaxin 1 and SNAP25, which strongly supports the idea that trans-SNARE complexes represent a minimal machinery for membrane fusion19. More recently, content mixing assays have largely supplanted lipid mixing assays; these studies have verified the central role of SNAREs in membrane fusion and have also pointed to the key importance of other factors (discussed later)20-22.
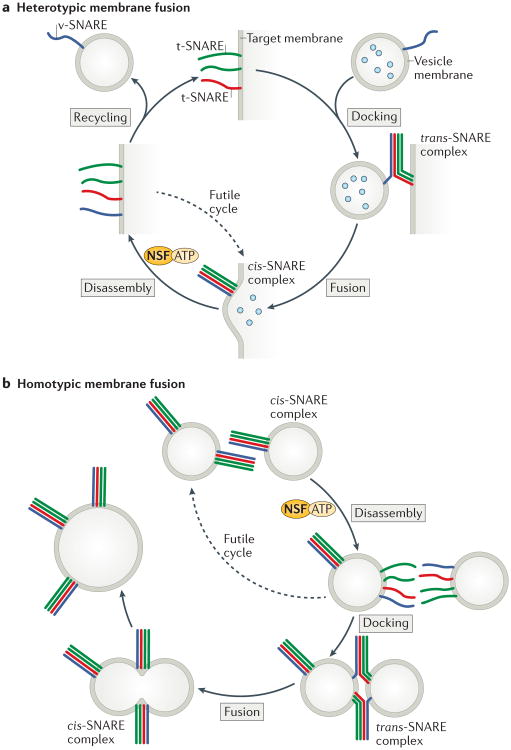
Figure 1. Cycles of SNARE assembly and disassembly.
a | Heterotypic membrane fusion is thought to begin with a v-SNARE in a vesicle and three t-SNAREs in a target membrane. Assembly into membrane-bridging trans-SNARE complexes drives membrane fusion and cargo delivery. The resulting cis-SNARE complex is disassembled by the ATPase NSF (working together with the adaptor protein SNAP; not shown), which releases the SNAREs for subsequent cycles of assembly and disassembly. If reassembly of the SNARE complex occurs before the v-SNARE is removed for recycling back to its donor (vesicle) compartment, the resulting cis-SNARE complex must be disassembled without having catalyzed membrane fusion (a futile cycle). b | In homotypic membrane fusion, each of the two membranes contains all four SNAREs. The resulting cis-SNARE complexes on each membrane must be disassembled prior to trans-SNARE complex assembly and membrane fusion. As in the case of heterotypic fusion, this cycle must operate in the continual presence of NSF and SNAP. To avoid futile cycling, chaperones are required to privilege trans-SNARE complex assembly over cis-SNARE complex assembly.
Even the simplest eukaryotes have twenty or more SNAREs, many of which are specific for particular intracellular trafficking pathways (for example, ER to Golgi or Golgi to plasma membrane)2. Extensive liposome reconstitution experiments using various combinations of SNAREs showed that lipid mixing requires cognate v-SNAREs and t-SNAREs – that is, v-SNAREs and t-SNAREs that function together in a particular trafficking pathway23. This and other evidence strongly suggests that substantial specificity, in terms of which vesicles fuse with which target membranes, is conferred by the SNAREs themselves. Later, we discuss membrane tethering factors, which also have important roles in determining the selection of target membranes for vesicle fusion.
ATP-driven SNARE disassembly
Vesicle docking is accompanied by trans-SNARE complex assembly, but membrane fusion converts the trans-SNARE complex into a cis-SNARE complex [G] in which all SNAREs are associated with the same fused membrane (FIG. 1a). cis-SNARE complexes are disassembled by the cytoplasmic proteins NSF24 and SNAPs25. Disassembled t-SNAREs are immediately available to participate in subsequent vesicle docking and fusion reactions, whereas v-SNAREs must first be recycled to the donor membrane before engaging in productive SNARE complex assembly26,27. The energy fueling this cycle of SNARE complex assembly and disassembly is expended by the ATPase NSF in breaking apart the highly stable cis-SNARE complex. The remaining steps in the cycle, including SNARE complex assembly and membrane fusion, are spontaneous – that is, energetically downhill28-31.
Topology-sensitive SNARE chaperones
The cycle of SNARE complex assembly and disassembly, as shown in FIG. 1a, raises obvious questions. For example, what prevents a freshly disassembled cis-SNARE complex from simply assembling again, creating a futile cycle? Less obviously, what prevents t-SNAREs from engaging in non-productive side reactions? In vitro, for example, syntaxin 1 and SNAP25 form non-productive 2:1 complexes in which a second copy of syntaxin 1 occupies the space that is normally reserved for the v-SNARE synaptobrevin32,33. Another non-productive side reaction involves syntaxin 1 alone, which can form dimers and tetramers34. The NSF- and SNAP-mediated disassembly of off-pathway complexes such as these would open the door to additional futile cycling.
Futile cycling could be mitigated by the involvement of additional factors that function as chaperones, by binding individual SNAREs and/or on-pathway assembly intermediates and protecting them from taking ‘wrong turns’ that would require the action of NSF (and ATP hydrolysis) to restore. A second reason to propose the involvement of SNARE chaperones is that the assembly of productive, membrane-bridging trans-SNARE complexes is topologically complex, involving multiple SNARE proteins associated with two different membranes. Chaperones might, for example, prevent the assembly of anti-parallel SNARE complexes and stable, non-cognate SNARE complexes, both of which can form in vitro35,36. Third, trans-SNARE complex assembly takes place in the presence of the SNARE disassembly machinery, and chaperones may therefore be required to provide a protected pathway for the assembly of productive trans-SNARE complexes in the presence of NSF21,37 (although it has also been suggested that NSF and SNAPs are themselves capable of distinguishing cis-SNARE complexes from trans-SNARE complexes38,39). Taken together, these considerations imply a need for ‘topology-sensitive’ chaperones to protect disassembled SNAREs from off-pathway assembly reactions, to promote the geometrically complex assembly of trans-SNARE complexes, and to protect partly or fully assembled trans-SNARE complexes from the disassembly mediated by NSF until fusion has occurred, all while allowing cis-SNARE complexes (and those off-pathway intermediates that do form) to be disassembled.
Before concluding these general considerations, it is also relevant to mention the case of homotypic fusion (FIG. 1b), in which all four SNAREs are present on each of the two membranes to be fused. The resulting cis-SNARE complexes on each membrane must first be disassembled by NSF and SNAPs, so that productive trans-SNARE complexes can then assemble. Thus, the need for chaperones that selectively promote trans-SNARE complex assembly, even in the continued presence of NSF and SNAPs, is clear. The proteins that are most clearly implicated as SNARE chaperones are membrane tethering factors and SM proteins, both of which are essential for vesicle docking and membrane fusion in vivo.
Membrane tethering factors
The close approach of two membranes bearing complementary SNAREs is a prerequisite for the formation of trans-SNARE complexes. At greater distances, and prior to SNARE engagement, membranes that are destined to fuse become tethered to one another by much larger membrane tethering factors8-10. There are two classes of membrane tethering factor: homodimers containing long stretches of predicted coiled coil and hetero-oligomers known as multisubunit tethering complexes (MTCs) [G]. The coiled-coil tethers are highly elongated, with the largest of these potentially capable of extending 300 nm or more. The MTCs are also large (>250 kDa) but are more compact than the coiled-coil homodimers, with maximum dimensions of 20-40 nm (FIG. 2a). Our understanding of membrane tethering factors is far from complete: conclusive demonstrations of membrane attachment have largely been lacking40, and high-resolution structures are in many cases missing or fragmentary. However, evidence has continued to accumulate that membrane tethering factors, by binding to SNAREs and other vesicle and target membrane proteins, function as SNARE chaperones and, more generally, as organizers of vesicle docking and fusion.
Figure 2. Membrane tethering and SNARE assembly.
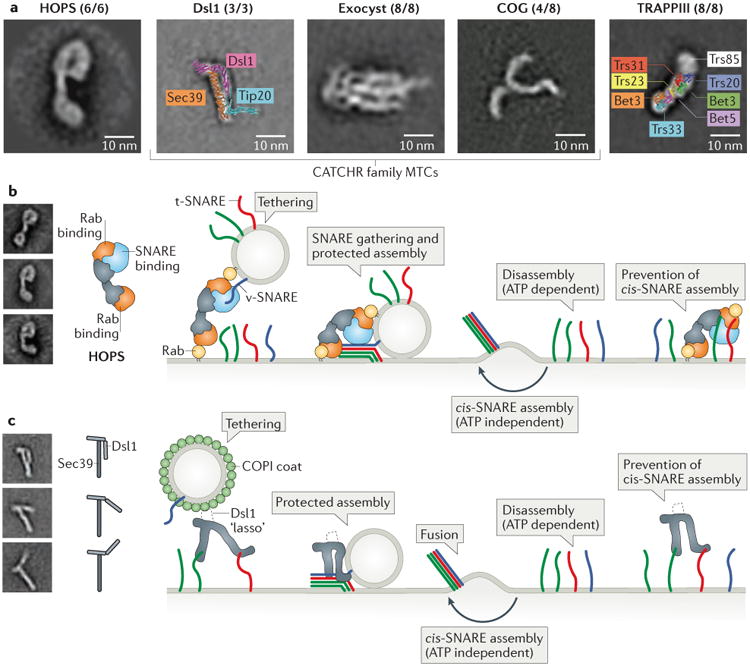
a | Membrane tethering factors include coiled-coil homodimers (not shown) and multisubunit tethering complexes (MTCs). Shown for the MTCs are representative class averages, derived from negative-stain electron micrographs, for the HOPS63, Dsl1 (Dsl1 and Sec39 subunits only)71, exocyst76, COG (Cog1-4 subunits only)77 and TRAPPIII142 complexes, as well as x-ray structure-based models for Dsl171 and TRAPPI84 complexes. The numbers in parentheses indicate the number of subunits imaged (present/total) by electron microscopy (or, in the case of Dsl1, by x-ray crystallography). Class averages reproduced with permission from REFS. 63, 71, 76, 77 and 142. b | Simplified model for HOPS complex-dependent membrane tethering and fusion. In this model, HOPS functions first as a tether, by binding to Rab proteins on two different membranes, and then as a chaperone for SNARE complex assembly. HOPS may also block the premature disassembly of trans-SNARE complexes and the futile reassembly of cis-SNARE complexes. The structure of the HOPS complex is based on negative-stain electron microscopy63. For clarity, the soluble Qc-SNARE Vam7 is shown with a membrane anchor. Electron microscopy-based class averages reproduced with permission from REF. 63. c | Model for Dsl1 complex-dependent membrane tethering and fusion. In this model, the Dsl1 complex mediates tethering by binding to SNARE proteins on the target membrane and — via an unstructured ‘lasso’ within the Dsl1 subunit — to the COPI coat on cargo-carrying vesicles. Next, the Dsl1 complex chaperones the proper assembly of trans-SNARE complexes. A hinge region within the Dsl1 subunit of the complex is evident in class averages of Dsl1–Sec39 complexes (reproduced with permission from REF. 71).
Coiled-coil tethering factors
A recent report provided the most convincing demonstration so far that coiled-coil tethering factors connect trafficking vesicles with their target membrane41. Golgins, which are the largest class of coiled-coil tethering factors, are anchored through their C-termini to the Golgi42. When individual golgins were systematically mislocalized to the outer membrane of mitochondria, they recruited the appropriate cargo-laden vesicles to mitochondria41,43,44. For example, golgin 245, which normally resides on the trans side of the Golgi and recruits endosome-derived vesicles, selectively redirected to the mitochondria vesicles carrying endosome-to-Golgi cargo, but not vesicles carrying ER-to-Golgi or intra-Golgi cargo. Overall, a panel of golgins was able to recruit vesicles in a cargo-specific manner that correlates well with their localization within Golgi stacks41.
How do golgins capture a transport vesicle? Golgins contain a large number of binding sites for Rab GTPases45,46, which in turn have long been known to have central roles in regulating vesicle transport by associating, in their activated GTP-bound states, with membranes and recruiting effector proteins such as MTCs47,48. Thus, golgins might capture vesicles by binding vesicle-associated Rab proteins. A second possibility is that golgins capture vesicles by binding directly to vesicle membranes. The golgin GMAP210 (also known as TRIP11), for example, contains an N-terminal amphipathic lipid packing sensor (ALPS) domain that binds selectively to highly curved membranes, including vesicles49,50. A third possibility is that golgins might capture vesicles by binding to their v-SNAREs, although evidence for direct golgin–v-SNARE binding is somewhat limited51,52. In addition to identifying the elements that are required for vesicle capture, it will be important to address how golgin-mediated tethering ultimately facilitates SNARE engagement and membrane fusion. Exciting progress is being made; for example, a recent study used proximity ligation and atomic force microscopy to show the functional importance of flexibility of coiled-coil tethering factors for vesicle tethering at the Golgi53.
Multisubunit tethering complexes
In contrast to the coiled-coil tethering factors, the MTCs are composed of three or more different subunits (FIG. 2a). The known MTCs can be divided into three families, each conserved from yeast to mammals: the homotypic fusion and vacuolar protein sorting (HOPS) family54,55, the complexes associated with tethering containing helical rods (CATCHR) family10,56, and the transport protein particle (TRAPP) family57. The HOPS and CATCHR families, although structurally unrelated, are particularly notable for their large number of direct interactions with other proteins that are implicated in vesicle docking and fusion, such as vesicle coat proteins, Rab GTPases, SNAREs and SM proteins (TABLE 1). This array of interactions indicates that MTCs may function as organizers of membrane docking, SNARE complex assembly and membrane fusion.
Table 1. Interactions between MTCs and other trafficking proteins.
| MTCs | Trafficking proteins | |||||||
|---|---|---|---|---|---|---|---|---|
| R-SNARE | Qa-SNARE | Qb-SNARE | Qc-SNARE | SNARE complex | Coat protein | SM protein | Rab protein | |
| HOPS | Xray12 | Xray12/PD65,66 | PD65,67 | PD65 | PDx143 | Xray144,145 | PD146 | |
| Dsl1 | GF72 | GF72 | GF71 | GF74 | ||||
| Exocyst | PD82 | GF147 | GF147 | PD81 | GF148 | PDx149 | ||
| COG | PD80,150 | PD80,150 | PDx151 | PDx152,153 | PD79,80 | PD153 | ||
| GARP | PD83 | PD83,154 | PD83 | PDx83,154 | PDx154 | |||
| TRAPP | PD155 | Xray86 | ||||||
GF: Gel filtration (purified proteins)
PD: Pull-down experiment (purified proteins)
PDx: Pull-down experiment (extract)
Xray: X-ray crystal structure of complex
HOPS: homotypic fusion and vacuolar protein sorting complex
GARP: Golgi-associated retrograde protein complex
TRAPP: transport protein particle complex
HOPS family
The HOPS family comprises both the HOPS complex [G] itself and the closely related class C core vacuole/endosome tethering (CORVET) complex. The six-subunit yeast HOPS complex, which is required for homotypic and heterotypic membrane docking and fusion in the endo-lysosomal system, is one of the best characterized MTCs (FIG. 2b)9,58. The in vitro reconstitution of yeast homotypic vacuolar fusion, using synthetic liposomes and purified proteins, has enabled an in-depth investigation of the mechanism of action of the HOPS complex59,60. For example, the HOPS complex was shown to tether membranes through its interactions with the membrane-associated Rab GTPase Ypt7, acidic phospholipids and SNAREs61,62. Negative-stain electron microscopy of the HOPS complex revealed Ypt7-binding subunits at both ends of a bilobed, flexible structure (FIG. 2a and b)63. The clear implication is that the HOPS complex tethers membranes, at least in part, by binding at both ends to membrane-associated Rab proteins.
As noted above, homotypic fusion involves two identical membranes, each carrying a full complement of four SNAREs (FIG. 1b). In order to liberate individual SNAREs so that they can then assemble into membrane-bridging trans-SNARE complexes, it is first necessary to disassemble cis-SNARE complexes on each of the participating membranes. This disassembly of the cis-complex and reassembly of the trans-complex — and therefore membrane fusion in the in vitro reconstituted system — requires Sec17 and Sec18 (the yeast SNAP and NSF protein, respectively) in addition to the HOPS complex. An attractive hypothesis is that SNAREs, having been disassembled by Sec17 and Sec18, are organized for assembly of trans-SNARE complexes by the HOPS complex. Consistent with this model, the HOPS complex binds both individual SNAREs and SNARE complexes64-67. The HOPS complex also seems to protect assembling trans-SNARE complexes from premature disassembly by Sec17 and Sec1837,68. Thus, the HOPS complex exhibits at least some of the properties that one would predict for a topology-sensitive chaperone. We discuss later a particular subunit of the HOPS complex (the SM protein Vps33) that seems to directly catalyze SNARE complex assembly.
CATCHR family
The CATCHR family comprises the exocyst, Dsl1, conserved oligomeric Golgi (COG), Golgi-associated retrograde protein (GARP) and endosome-associated recycling protein (EARP) complexes. The CATCHR-family complexes, similarly to the HOPS-family complexes, bind Rab GTPases, SNAREs, SM proteins and/or vesicle coat proteins (TABLE 1). The CATCHR complexes also seem to function both as tethers and, in collaboration with SM proteins, as chaperones for SNARE complex assembly. From a structural standpoint, the best characterized of the CATCHR complexes is the yeast Dsl1 complex, which contains the three subunits Dsl1, Tip20 and Sec39 (also known as Dsl3)69,70 (FIG. 2c). Each of these subunits is essential for the trafficking of COPI-coated vesicles from the Golgi to the ER. X-ray crystallography and negative-stain electron microscopy studies of the Dsl1 complex have shown that it has a two-legged structure containing a hinge in the middle that allows both closed and open conformations (FIG. 2c)71,72. In the closed conformation, the Dsl1 complex forms a 20-nm tower. This tower is anchored at its base, through interactions between the legs of the complex and t-SNAREs, to the ER membrane73. At the apex of the Dsl1 tower is a disordered region known as the ‘lasso’, which contains multiple binding sites for COPI74,75. The structure thus suggests that the Dsl1 complex can function as a tether, connecting COPI-coated vesicles to their target organelle. In vitro, the Dsl1 complex accelerates (albeit modestly) the assembly of cognate SNARE complexes71, which hints that the Dsl1 complex might also function as a chaperone for SNARE complex assembly.
Structural studies of subunits of the larger CATCHR complexes — GARP, EARP, COG and exocyst — point to them having a shared subunit fold, which indicates common descent from a single evolutionary progenitor, but little obvious homology at the quaternary structural level (FIG. 2a)9,10. Negative-stain electron microscopy analysis of the complete, hetero-octameric exocyst complex showed that it forms a complex bundle of rod-like elements that probably represent single subunits76. An earlier study of a COG subcomplex (comprising four of the eight subunits) revealed a rather different, three-legged structure77. Notwithstanding their architectural differences, substantial evidence indicates that each of these complexes participates in the assembly of SNARE complexes, probably with the assistance of SM proteins78-83. A more detailed understanding of the mechanics of this process awaits structural studies of MTC–SNARE (and/or MTC–SNARE–SM protein) complexes.
TRAPP family
The TRAPP family comprises several distinct TRAPP complexes: for example, TRAPPI, TRAPPII and TRAPIII in yeast. These complexes are more compact and less flexible than the HOPS or CATCHR-family complexes (FIG. 2a)84,85 and, unlike those complexes, the TRAPP family complexes function as guanine nucleotide exchange factors86 but are not known to bind SNAREs directly. The involvement of the TRAPP complexes in tethering may therefore be indirect, through their activation of Rab proteins and/or their binding to coiled-coil tethering factors or vesicle coat proteins40,87.
Sec1-Munc18 proteins and SNARE assembly
Multiple lines of evidence have emerged to suggest that MTCs work together with SM proteins, which themselves seem to be universally required for SNARE-mediated membrane fusion8.
Model of SM-catalyzed SNARE assembly
There are four major families of SM protein, and all eukaryotes seem to have at least one member of each family88. Different SM proteins, similarly to different SNAREs, are specific for different sets of intracellular trafficking pathways1. However, unlike the SNAREs, whose role in membrane fusion seems clear, the essential functional role of SM proteins has been difficult to decipher11,89,90. Progress has been hampered by, among other things, a relative dearth of high-resolution structures showing how SM proteins interact with SNAREs. However, there are now more than a dozen x-ray structures of SM proteins, representing three of the four major families, in the Protein Data Bank. Several of these structures include a bound peptide derived from the extreme N-terminus of a Qa-SNARE91-93, and two other structures contain the intact Qa-SNARE cytoplasmic domain (missing only the C-terminal transmembrane anchor)94-96. Most recently, we reported two structures of a single SM protein, bound in one case to the SNARE motif of a Qa-SNARE and in the other case to the SNARE motif of an R-SNARE12. We believe it is time to throw caution to the wind by placing these known structures into a speculative sequence (FIG. 3).
Figure 3. A structure-based mechanism for SM protein-mediated SNARE complex assembly.
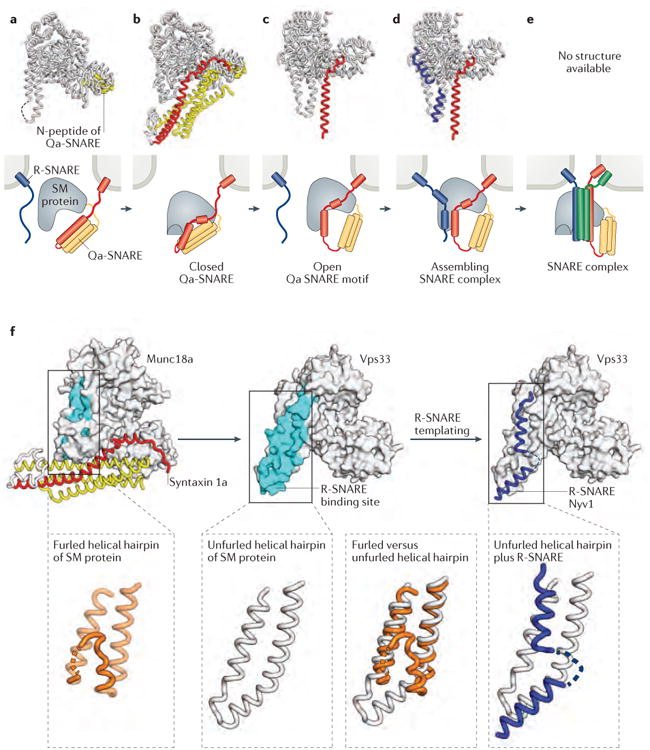
a-e | X-ray crystal structures (first row) and schematic representations (second row) of SM protein–SNARE complexes. The structures and schematics are placed in a speculative order to suggest a potential pathway for SM protein-templated SNARE complex assembly. a | Many SM proteins bind to the N-peptide lying at the extreme N-terminus of Qa- (or syntaxin-like) SNAREs. Shown is the structure of the SM protein Munc18a (grey) bound to the N-peptide of syntaxin 4 (yellow) (PDB code 3PUJ)92. b | Some Qa-SNAREs adopt an auto-inhibited conformation that binds to, and is stabilized by, the cognate SM protein. This binding mode requires a furled conformation for the helical hairpin of the SM protein. Shown is the structure of Munc18a bound to the cytoplasmic portion of syntaxin 1a (yellow and red) (PDB code 3C98)94. c | The opening of the Qa-SNARE is accompanied by the unfurling of the SM protein's helical hairpin, which exposes the R-SNARE binding site. Shown is the structure of the SM protein Vps33 (grey) bound to the SNARE motif of the Qa-SNARE Vam3 (red) (PDB code 5BUZ)12. d | Binding of the R-SNARE to the SM protein leads to a half-zippered SNARE complex representing a potential early intermediate in SNARE complex assembly. Shown is a model that combines the structures of Vps33 bound to the R-SNARE Nyv1 (blue) (PDB code 5BV0) and Vp33 bound to the Qa-SNARE Vam3 (red and yellow) (PDB code 5BUZ)12. e | After formation of the trans-SNARE complex, SM proteins may bind directly to the resulting four-helix bundle. The resulting complex has not, however, been structurally defined. f | Unfurling of the domain 3a helical hairpin of the SM protein reveals the R-SNARE binding site.
Some Qa-SNAREs contain a short N-terminal region — the N-peptide — which binds to a site on the ‘back’ of the cognate SM protein91-94,97 (Fig. 3a). The N-peptide of the neuronal Qa-SNARE syntaxin 1 has been extensively studied and is required both for synaptic vesicle fusion and for the stimulation of liposome fusion in vitro by the SM protein Munc18a 98-100. Strikingly, the syntaxin 1 N-peptide still supports liposome fusion when transplanted onto SNAP25 or when anchored, separately from the rest of syntaxin 1, in the target membrane101. These and other data indicate that the essential role of the N-peptide is to recruit the SM protein to the site of SNARE complex assembly. Other Qa-SNAREs, including those that lack an N-peptide, probably recruit SM proteins using alternative or additional strategies66,93.
The first reported structure of an SM protein, Munc18a, also included syntaxin 194,96 (Fig. 3b). Between the N-peptide of syntaxin 1 and its SNARE motif is a three-helix bundle102. The SNARE motif and the three-helix bundle interact, forming a four-helix bundle that inhibits SNARE complex assembly103,104. This autoinhibited conformation of syntaxin 1, which is marginally stable on its own, is stabilized by binding to the cleft of the SM protein94,105,106. Thus, SM proteins can clamp Qa-SNAREs in autoinhibited conformations and prevent or postpone their assembly into cis-, trans- or off-pathway (non-stoichiometric) SNARE complexes. Munc18a also stabilizes syntaxin 1 (and vice versa) in vivo107,108.
We recently reported two x-ray structures of the SM protein Vps33 bound to the SNARE motifs of the Qa-SNARE Vam3 and the R-SNARE Nyv112. All three proteins were from the thermophilic fungus Chaetomium thermophilum. In the first of these structures (FIG. 3c), the bound SNARE motif of Vam3 strongly resembles the SNARE motif of syntaxin 1 in its complex with Munc18a. The second structure reveals that the SNARE motif of an R-SNARE can bind to a highly conserved groove on the surface of the SM protein (FIG. 3d). The R-SNARE binding groove is formed by a conserved ‘helical hairpin’ — a finger-like extension emerging from domain 3a of the SM protein and consisting of two long, antiparallel α-helices connected by a loop. In the Munc18a–syntaxin 1 structure (FIG. 3b), the helical hairpin is bent back upon itself (‘furled’)92,94-96. Furling avoids a steric clash with the three-helix bundle of syntaxin 1 but also covers the R-SNARE binding groove of Munc18a (FIG. 3f). Thus, the R-SNARE binding site on the SM protein is revealed only when the helical hairpin is unfurled, which is in turn possible only when the SM protein is either uncomplexed or, as illustrated in FIG. 3c, bound to an ‘open’ Qa-SNARE.
Vps33 can bind simultaneously to the SNARE motifs of both Vam3 and Nyv112. The x-ray structure of the ternary Vps33–Vam3–Nyv1 complex has not been reported, but the crystal structures of the Vps33–Vam3 and Vps33–Nyv1 complexes place the Qa- and R-SNAREs in the proper orientation (parallel) and alignment (with zero-layer residues adjacent) for subsequent assembly (modeled in FIG. 3d). These results, together with functional studies of Vps3312 and Munc18109,110, imply that SM proteins use their helical hairpins to facilitate SNARE complex formation. At least in the case of Vps33, the SM protein functions as a template upon which an early SNARE complex assembly intermediate, resembling a half-zippered complex between the Qa- and R-SNAREs, can form.
Finally, SM proteins bind the four-helix bundles formed by assembled SNARE complexes68,111-114. The binding mode for SNARE complexes might resemble the binding mode for autoinhibited Qa-SNAREs1, but direct evidence of this has not been reported, neither has the mechanistic role of SM protein binding to SNARE complexes been definitively elucidated (Fig. 3e). Binding of SM proteins might protect trans-SNARE complexes from premature disassembly mediated by NSF and SNAPs and/or might stimulate fusion directly, perhaps by inducing membrane bending115.
The preceding discussion places the known SM protein–SNARE structures (FIG. 3a-d) into a speculative order of events. First, the SM protein is recruited to a Qa-SNARE, in either an open or an autoinhibited conformation, by binding to its N-peptide. Second, the SM protein functions as a clamp to hold the Qa-SNARE shut and prevent it from binding other SNAREs. Third, opening of the SM-bound Qa-SNARE — presumably catalyzed by another factor such as Munc13 (discussed below) — leaves the SNARE motif bound to the SM protein but enables the unfurling of the helical hairpin of the SM protein to reveal the R-SNARE binding site. Fourth, an R-SNARE embedded in the apposed membrane binds to the SM protein template, creating a half-zippered, R-SNARE–Qa-SNARE complex. Fifth, the SNARE complex assembly process continues, via a pathway that has not been elucidated, to generate a complete, fusion-competent trans-SNARE complex. This proposed sequence of events is unlikely to be universal, but we hope that it will function as a useful jumping-off point for further investigation.
Model implications
In this model, the initial contact between SNAREs is predicted to involve R- and Qa-SNAREs anchored in opposite membranes (FIG. 3). By contrast, it has been nearly axiomatic in the field that ‘acceptor’ complexes of Qa-, Qb- and Qc-SNAREs form first; indeed, the QaQbQc-SNARE complex has itself sometimes been called a t-SNARE23. However, the x-ray structures depicted in FIG. 3c and 3d hint that SM proteins function as assembly templates by binding first to R- and Qa-SNAREs. Qb- and Qc-SNAREs would join the complex subsequently. We note that what distinguishes a cis-SNARE complex from a trans-SNARE complex is whether the R- and Qa-SNAREs are in the same or different membranes. Therefore SM proteins, by somehow choosing an R-SNARE located in trans, could impose a topological filter on the first step of the overall assembly process.
The pathway we propose (FIG. 3) is consistent with a recent reconstitution of synaptic vesicle fusion21. A novel feature of this reconstitution is that it begins with liposomes containing syntaxin 1 bound to Munc18a rather than to SNAP25. The fusion of these liposomes faithfully recapitulates the fusion of synaptic vesicles in that it depends on the Ca2+ sensor protein synaptotagmin 1 (discussed in the following section), resists the SNARE complex disassembly activity of NSF and SNAPs, and depends on both Munc18a and Munc13. Munc13, a ‘priming factor’ that is essential for synaptic vesicle fusion in vivo116, opens Munc18-bound syntaxin for assembly with other SNAREs117-119. Thus, Munc13 seems to be capable of mediating the conversion between the states depicted in FIG. 3b and 3c. Munc13 and Munc18a may collaborate to provide a protected pathway for the assembly of trans-SNARE complexes in the presence of NSF and SNAPs. Intriguingly, Munc13 is structurally homologous to subunits of CATCHR-family MTCs120, which indicates that the latter may also modulate SNARE–SM protein interactions. However, it is not understood how Munc13 or MTCs might accomplish this.
Although there are several examples of the binding of SNARE motifs to SM proteins, the apparent binding affinities are low12,65,113,121. This makes sense in as much as SM proteins function as catalysts for SNARE motif assembly, and tight binding to individual SNARE motifs would slow the assembly reaction. To compensate for this relatively weak binding, the SNAREs may be gathered together by membrane tethering factors for assembly promoted by SM proteins. The potential for collaboration between MTCs and SM proteins is clearest in the case of the HOPS complex, which is unique in containing an SM protein, Vps33, as a stable subunit (FIG. 2b). The other subunits of the HOPS complex bind to the N-terminal regions of two out of the four SNAREs that are required for vacuolar fusion (Vam3 and Vam7)64,66,67. Other MTCs, which are not as tightly associated with an SM protein, could nonetheless create an ‘assembly zone’ in which the SNARE motifs are presented to the cognate SM protein. Elucidating the architecture of SNARE assembly zones will be a challenging but important goal for future investigations.
Synaptotagmin and regulated exocytosis
The most intensively studied membrane fusion reaction in biology is that between neurotransmitter-containing synaptic vesicles and the pre-synaptic plasma membrane, and indeed the SNAREs that are required for evoked neurotransmitter release have featured prominently in this Review so far. Neurotransmitter release and its physiological regulation are complex and well-reviewed topics13,122-124, with a great deal of recent study having been focused on the regulatory factors complexin and synaptotagmin 1. Complexin is generally thought to function as a clamp to prevent the premature fusion of synaptic vesicles124-127. Here, we focus primarily on the recent structural characterization of complexes between SNAREs and the Ca2+ sensor protein synaptotagmin 1128,129, a transmembrane protein that resides on synaptic vesicles. Its C2 domains, C2A and C2B, bind Ca2+, acidic phospholipids, phosphatidylinositol 4,5-bisphosphate and synaptic SNARE complexes123. Crucially, synaptotagmin 1 couples the action-potential-triggered influx of Ca2+ at the axon terminal to the SNARE-mediated fusion of synaptic vesicles with the pre-synaptic plasma membrane and the consequent release of neurotransmitters130,131. This coupling probably involves the ability of synaptotagmin 1 to bind to SNARE complexes (see however REF. 132), but it has been challenging to characterize this interaction at high resolution.
A recent crystal structure of synaptotagmin 1 and a SNARE complex, generated by connecting them using a flexible linker, revealed a conserved interface between the C2B domain of synaptotagmin 1 and a face of the SNARE complex formed by SNAP25 and syntaxin 1 (FIG. 4)129. In addition to this ‘primary’ interface, the crystal structures also revealed secondary and tertiary interfaces between synaptotagmin 1 and the SNARE complex. The residues of the SNARE complex and synaptotagmin 1 that form the primary interface are conserved among the synaptotagmins, SNAP25 homologues and syntaxin homologues that are involved in fast, Ca2+-triggered exocytosis. Moreover, the primary interface is Ca2+ independent, suggesting that it forms prior to the arrival of an action potential at the axon terminal. Mutations of two, three or five of the conserved residues disrupted binding of synaptotagmin 1 to the SNARE complex, synaptotagmin 1-dependent membrane fusion in a cell-free assay, and Ca2+-triggered exocytosis in vivo129.
Figure 4. Structural characterization of the synaptotagmin–SNARE complex interaction.
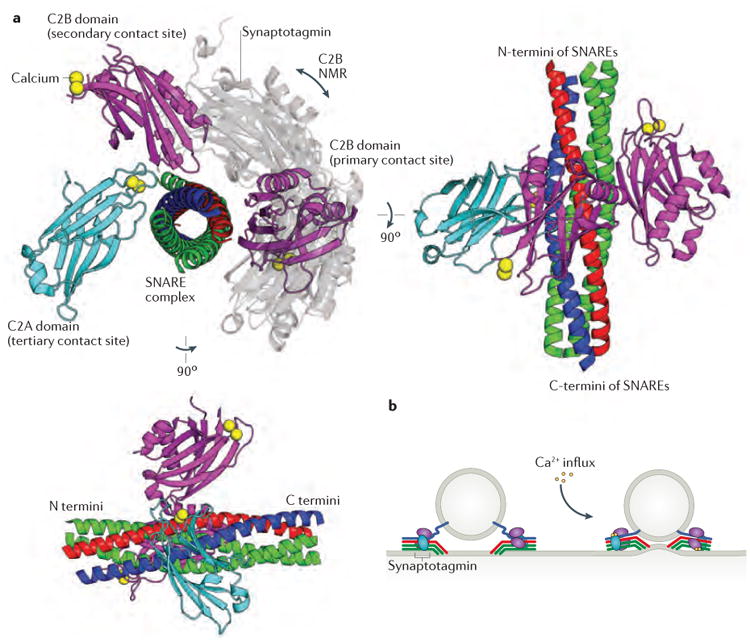
a | Superimposed structures of synaptotagmin 1 bound to the neuronal SNARE complex, determined by x-ray crystallography (PDB code 5CCH)129 and NMR (PDB code 2N1T)128, implicate the same face of the SNARE complex as being the primary contact site for the C2B domain of synaptotagmin (shown in purple for the crystal structure and in grey for the NMR structure). The x-ray crystal structure also defines secondary and tertiary binding sites on the SNARE complex for the C2A (cyan) and C2B domains of synaptotagmin. b | The structural data suggest a model in which multiple synaptotagmins and SNARE complexes form a super-complex around a docked vesicle. Calcium influx is proposed to promote interactions between synaptotagmin 1 and the plasma membrane that deform the plasma membrane, bringing it into juxtaposition with the vesicle membrane and facilitating membrane fusion129.
A second recent study, this one using NMR, identified an overlapping binding site for synaptotagmin 1 on the SNARE complex, but in this case binding involved a different face of the synaptotagmin 1 C2B domain (FIG. 4)128. Mutations targeting this interface, but different from those tested based on the crystal structure, resulted in reduced binding of synaptotagmin 1 to the SNARE complex as well as impaired function of synaptotagmin 1 in cultured neurons.
Further complicating comparison of the NMR and crystallographic studies, the NMR study used a double mutant of synaptotagmin 1 (R398Q, R399Q) that alters key residues in the primary interface as defined by the crystallographic studies. Nonetheless, both studies support the same general conclusion — that synaptotagmin 1 and the SNARE complex have multiple modes of interaction that may enable the assembly of a superstructure around the docking site between a synaptic vesicle and the presynaptic membrane128,129. Multiple synaptotagmin 1-binding sites on each SNARE complex would allow synaptotagmin 1 to bridge multiple SNARE complexes, creating a large assembly of SNAREs and Ca2+ triggers. Upon Ca2+ influx, Ca2+-driven association between synaptotagmin 1 and the plasma membrane (FIG. 4b) could reorganize and/or strain this array, inducing membrane curvature and/or closer proximity between the apposed membranes, full SNARE zippering and membrane fusion.
SNARE disassembly
So far, we have emphasized the elaborate machinery responsible for membrane docking, SNARE complex assembly and regulated membrane fusion. Following fusion, SNARE recycling — and the energization of the SNARE cycle — requires disassembly of the SNARE complex. SNARE complex disassembly is also required to resolve off-pathway SNARE complexes and to prepare membranes for homotypic fusion. The SNARE disassembly machinery comprises NSF and accessory factors known as SNAPs (no relation to the neuronal SNARE protein SNAP25). A major breakthrough in our understanding of this machinery was the recent structural characterization, using cryoEM, of intact NSF in both ADP- and ATP-bound states, as well as of SNARE–NSF–SNAP complexes39.
CryoEM structures of NSF
Each monomer of NSF comprises three domains: the N domain, which mediates SNAP–SNARE binding; the D1 domain, which mediates ATP hydrolysis; and the D2 domain, which binds stably to ATP and mediates hexamerization of NSF (FIG. 5a)15. Previous electron microscopy analysis showed that NSF forms stacked hexameric rings133,134. The quality of the new data were sufficient to determine and refine 3D reconstructions without imposing symmetry, which was instrumental in resolving nucleotide-dependent differences between the ATP- and ADP-bound states39. ATP-bound NSF adopts a ‘split-washer’ conformation, in which the D1 domains are approximately six-fold symmetric but have a slight right-handed helical pitch (FIG. 5b). By contrast, ADP-bound NSF adopts an ‘open flat washer’ conformation, in which the sixfold symmetry is broken but all six D1 domains remain essentially co-planar. The ring is more compact in the ATP-bound state, which suggests that a spring-like transition occurs from a ‘loaded’ split washer to a ‘relaxed’ open flat washer. In general agreement with previous studies133, large variations were observed in the orientations of the N domains relative to the D1 and D2 rings.
Figure 5. SNARE complex disassembly by NSF and SNAPs.
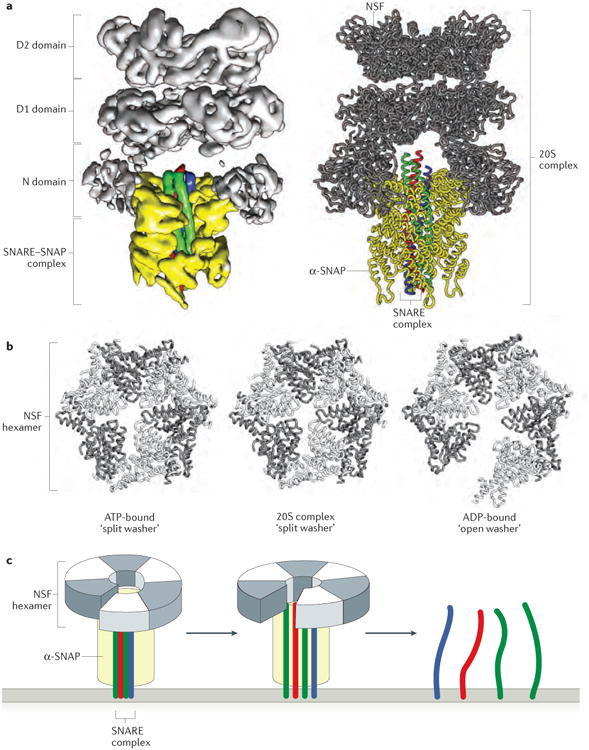
a | The near atomic-resolution cryoEM structure of the 20S complex comprising six copies of NSF, four copies of α-SNAP and the neuronal SNARE complex (PDB code 3J96)39. A ribbon model of the complex is shown next to the electron density from one of four single particle reconstructions. b | The D1 domains of the NSF hexamer are shown for three separate structures of NSF: the 20S complex, ATP-bound NSF (PDB code 3J94) and ADP-bound NSF (PDB code 3J95) (lacking α-SNAP and the SNARE complex)39. Comparison of these structures shows that NSF adopts a split washer orientation when bound to ATP and within the 20S complex, and an open washer orientation when bound to ADP. c | A simplified schematic of NSF and its interaction with the SNARE complex (only the D1 domain of NSF is shown for clarity). When bound to ATP and the SNARE complex, NSF adopts a compact, split-washer orientation. Upon ATP hydrolysis and release of inorganic phosphate, NSF undergoes a conformational change to the open-washer orientation, thereby applying rotational and shear forces to the SNARE complex. This spring-like transition unwinds the SNARE complex in a single round of ATP hydrolysis, which releases the individual SNAREs for participation in further rounds of membrane fusion39,135.
The same study also reported approximately 8-Å resolution cryo-EM reconstructions of the ‘20S complex’ that is formed by NSF (in the presence of the non-hydrolyzable ATP analog AMP-PNP), α-SNAP and the assembled SNARE motifs of syntaxin 1, SNAP25 and synaptobrevin (FIG. 5a). Four different reconstructions, potentially representing four unique molecular states, were generated39. Four molecules of α-SNAP ensheath the SNARE complex, and the N-terminal domains of NSF (now better defined than in the NSF-only structures discussed above) interact with sites on the outer surface of individual α-SNAP monomers. The symmetry mismatch (six NSF monomers interacting with four α-SNAP molecules) is such that the individual α-SNAP monomers are bound to either one or two NSF N-terminal domains. There is also a ‘handedness’ mismatch, with the four SNARE helices exhibiting a left-handed twist (which is the standard for coiled-coil interactions) whereas the four α-SNAP molecules (similarly to the D1 domains in the ATP-bound NSF hexamer) have a right-handed twist. The NSF ring is even more compact in these 20S structures than in ATP-bound NSF, which may indicate that the NSF spring is further tightened upon interaction with the α-SNAP–SNARE complex (FIG. 5b). Together, these observations support a model in which the 20S particle, as it cycles between the observed conformations (and possibly others), exerts a torque to destabilize the interactions between the four SNAREs. Additionally, the transition between the ATP-bound split washer conformation and the ADP-bound flat washer conformation of NSF may exert a shear force on the SNARE complex. Torque and shear force, applied in combination or in series, provide an appealing mechanistic explanation for SNARE complex disassembly (FIG. 5c).
Single-molecule analysis
Single molecule fluorescence experiments corroborate the idea that NSF uses a spring-loaded mechanism to unwind SNARE complexes135. In experiments using TIRF microscopy to image single SNARE complexes being disassembled by a single NSF hexamer, one round of ATP hydrolysis (per NSF monomer) was sufficient for robust disassembly. This finding contrasts with other recent reports, which concluded that multiple rounds of ATP hydrolysis are required for the disassembly of a single SNARE complexe136,137. Measurements of individual NSF hexamers indicated that SNARE complex disassembly occurs in a single step after an initial dwell period135. Strikingly, ATP hydrolysis seemed to cause a large tension that was released in a single conformational switch, reminiscent of a loaded spring being released. The addition of phosphate analogues that, in conjunction with ADP, mimic the ATP hydrolysis transition state of NSF was found to inhibit SNARE complex disassembly, suggesting that disassembly requires not only ATP hydrolysis but also the release of inorganic phosphate. This interpretation implies that NSF remains in the ATP-bound conformation — but with ADP bound — during the latent period prior to SNARE disassembly. The subsequent release of this spring-loaded state in a single step could cause gross conformational changes within the 20S particle, thereby triggering the disassembly of the bound SNARE complex and subsequent release of the monomeric SNAREs.
New roles for SNAPs?
We conclude by discussing several intriguing studies that suggest an unexpected role for SNAP proteins — which have been described as adaptors for SNARE complex disassembly — in stabilizing trans-SNARE complexes and thereby promoting membrane fusion. In the first study, Sec17 (the yeast SNAP) markedly improved binding between SNARE complexes and their cognate SM proteins68. The Sec17–SM protein–SNARE complex partially, though not completely, blocked disassembly of the SNARE complex by Sec18 (the yeast NSF protein). Based on these and other findings, the authors proposed a timer model whereby SM proteins, together with Sec17, function to delay the action of Sec18. Fusion-incompetent SNARE complexes, by contrast, would not be recognized by the SM protein and would therefore be disassembled. The disassembly delay could be important in giving trans-SNARE complexes time to mediate membrane fusion.
It also came as a surprise, given its canonical role in SNARE complex disassembly, that Sec17 was able to rescue the vacuolar fusion defect caused by short C-terminal truncations in the Qc-SNARE Vam7138. Building on this finding, a recent study showed that Sec17 can strongly stimulate the fusion of liposomes in which trans-SNARE complexes have formed but fusion pores have not yet opened139. This fusion-stimulating activity of Sec17 depended on a pair of hydrophobic loops, which in a SNAP–SNARE complex would be well-positioned for membrane insertion39. The results suggest that Sec17 can bind partially formed SNARE complexes, perhaps helping to drive SNARE zippering towards completion, and insert hydrophobic loops into the membrane, which potentially perturb the lipid bilayer and promote fusion. By contrast, another recent report suggested that α-SNAP could interfere with the full zippering of some, but not other, SNARE complexes140, which is in line with previous observations that excess Sec17 can inhibit membrane fusion141.
Together, these findings suggest an intriguing multiplicity of roles for Sec17 and possibly SNAPs in general. On the one hand, Sec17 functions as an adaptor that recruits Sec18 to cis-or off-pathway SNARE complexes for disassembly. On the other hand, Sec17 may also be recruited to trans-SNARE complexes as they assemble on SM protein templates, potentially helping to drive SNARE zippering to completion and/or perturbing the lipid bilayer, both of which would stimulate membrane fusion. At some stage the SM protein would disengage, allowing Sec17 to recruit Sec18 for SNARE complex disassembly.
Perspectives
Recent advances in the field of membrane trafficking have brought us several steps closer to a molecular framework for understanding membrane docking and fusion and its orchestration by chaperones mediating SNARE complex assembly and disassembly. Docking and fusion are likely coordinated by membrane tethering factors functioning together with small GTPases and SM proteins, but only a few relevant structures are currently available as blueprints for detailed mechanistic biochemistry. Recent advances in cryoEM provide a promising path forward, provided that strategies can be developed for coping with the intrinsic flexibility that so far seems to characterize many of the relevant complexes. It will be particularly interesting to see, as the field develops further, to what extent different SNARE complex assembly reactions (such as those mediated by HOPS complexes versus those mediated by CATCHR complexes) resemble one another mechanistically. Elucidating the shared mechanistic features implemented by different chaperone machineries should help to unlock the fundamental principles underlying intracellular membrane fusion. Finally, understanding the role of SNAPs, not only in SNARE complex disassembly but also possibly in assembly and fusion, will lead to a more holistic understanding of SNARE complex function and its regulation.
Key points.
Fusion of eukaryotic transport vesicles with target organelles requires membrane-bridging complexes of soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs).
Productive assembly of SNARE complexes, involving four different SNAREs anchored in two different membranes, is topologically complex. Both assembly and disassembly chaperones are required to ensure the temporal and spatial integrity of the intracellular trafficking network.
The initial contact of a transport vesicle with a target membrane is mediated by homo-dimeric and hetero-oligomeric membrane tethering factors. In addition to their role in tethering, these factors may also function as chaperones for SNARE complex assembly.
Sec1-Munc18 (SM) proteins interact directly with SNAREs and are required in vivo for SNARE-mediated membrane fusion. They may accelerate SNARE complex formation by functioning as templates to stabilize early assembly intermediates.
Recent x-ray crystal and NMR structures of the calcium sensor protein synaptotagmin bound to the SNARE complex reveal the molecular details of an interaction that probably underlies the exquisite sensitivity of neurotransmitter release to calcium levels.
Recent cryoEM structures and single particle FRET studies both indicate that the chaperone N-ethylmaleimide-sensitive factor (NSF) may use a ‘spring-loaded’ mechanism to disassemble SNARE complexes in a single step.
Glossary
- soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs)
Integral or peripheral membrane proteins that mediate membrane fusion by forming parallel four-helix bundles
- Sec1-Munc18 proteins
(SM proteins). Cytoplasmic proteins that bind to SNAREs and seem to be universally required for SNARE-mediated membrane fusion
- N-ethylmaleimide-sensitive fusion factor
(NSF). Homo-hexameric protein of the AAA+ ATPase family that drives the disassembly of SNARE complexes; known as Sec18 in yeast
- Soluble NSF attachment proteins
(SNAPs). Adaptor proteins that allow NSF to recognize and disassemble SNARE complexes; Sec17 is the yeast SNAP
- trans-SNARE complex
Complex containing at least one SNARE embedded within each of two apposed membranes; required for membrane fusion
- cis-SNARE complex
Non-fusogenic SNARE complex in which all SNAREs are associated with the same membrane; created from a trans-SNARE complex when membranes fuse
- Multisubunit tethering complexes
(MTCs). Large, hetero-oligomeric complexes that orchestrate vesicle docking and fusion through interactions with SNAREs, Rab proteins, SM proteins and/or vesicle coat proteins
- HOPS complex
A hetero-hexameric MTC — comprising Vps11, Vps16, Vps18, Vps33, Vps39 and Vps41 in yeast — that functions in endo-lysosomal fusion reactions (for example, the homotypic fusion of yeast vacuoles)
Biographies
Richard W. Baker is a postdoctoral researcher in the laboratory of Andres Leschziner at the University of California San Diego. His graduate work in the laboratory of Fred Hughson at Princeton University used x-ray crystallography to develop structural models of SNARE complex assembly.
Frederick M. Hughson is a professor of Molecular Biology at Princeton University. The research in his group focuses on understanding the protein machinery that generates the interior architecture of cells by guiding the movement and fusion of intracellular transport vesicles. Frederick M. Hughson's laboratory web page: http://molbio.princeton.edu/labs/hughson.
Footnotes
Competing interests statement: The authors declare no competing interests.
Subject categories: Biological sciences / Cell biology / Membrane trafficking / Membrane fusion
[URI /631/80/313/2378]
Biological sciences / Cell biology / Membrane trafficking / SNARE
[URI /631/80/313/2104]
Biological sciences / Structural biology / Electron microscopy / Cryoelectron microscopy
[URI /631/535/1258/1259]
Biological sciences / Biological techniques / Structure determination / X-ray crystallography
[URI /631/1647/2258/1266]
Biological sciences / Biophysics
[URI /631/57]
References
- 1.Südhof TC, Rothman JE. Membrane fusion: grappling with SNARE and SM proteins. Science. 2009;323:474–477. doi: 10.1126/science.1161748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jahn R, Scheller RH. SNAREs--engines for membrane fusion. Nat Rev Mol Cell Biol. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- 3.Fasshauer D, Sutton RB, Brunger AT, Jahn R. Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc Natl Acad Sci U S A. 1998;95:15781–15786. doi: 10.1073/pnas.95.26.15781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stein A, Weber G, Wahl MC, Jahn R. Helical extension of the neuronal SNARE complex into the membrane. Nature. 2009;460:525–528. doi: 10.1038/nature08156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sutton RB, Fasshauer D, Jahn R, Brunger AT. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 angstrom resolution. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- 6.McNew JA, Sondermann H, Lee T, Stern M, Brandizzi F. GTP-dependent membrane fusion. Annu Rev Cell Dev Biol. 2013;29:529–550. doi: 10.1146/annurev-cellbio-101512-122328. [DOI] [PubMed] [Google Scholar]
- 7.Bombardier JP, Munson M. Three steps forward, two steps back: mechanistic insights into the assembly and disassembly of the SNARE complex. Curr Opin Chem Biol. 2015;29:66–71. doi: 10.1016/j.cbpa.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hong W, Lev S. Tethering the assembly of SNARE complexes. Trends Cell Biol. 2014;24:35–43. doi: 10.1016/j.tcb.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 9.Kuhlee A, Raunser S, Ungermann C. Functional homologies in vesicle tethering. FEBS Lett. 2015;589:2487–2497. doi: 10.1016/j.febslet.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 10.Yu IM, Hughson FM. Tethering factors as organizers of intracellular vesicular traffic. Annu Rev Cell Dev Biol. 2010;26:137–156. doi: 10.1146/annurev.cellbio.042308.113327. [DOI] [PubMed] [Google Scholar]
- 11.Archbold JK, Whitten AE, Hu SH, Collins BM, Martin JL. SNARE-ing the structures of Sec1/Munc18 proteins. Curr Opin Struct Biol. 2014;29:44–51. doi: 10.1016/j.sbi.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 12.Baker RW, et al. A direct role for the Sec1/Munc18-family protein Vps33 as a template for SNARE assembly. Science. 2015;349:1111–1114. doi: 10.1126/science.aac7906. X-ray crystal structures of SM protein–SNARE complexes indicate that SM proteins can function as templates to stabilize an early intermediate in SNARE complex assembly. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rizo J, Xu J. The synaptic vesicle release machinery. Annu Rev Biophys. 2015;44:339–367. doi: 10.1146/annurev-biophys-060414-034057. [DOI] [PubMed] [Google Scholar]
- 14.Sudhof TC. The molecular machinery of neurotransmitter release (Nobel lecture) Angew Chem Int Ed Engl. 2014;53:12696–12717. doi: 10.1002/anie.201406359. [DOI] [PubMed] [Google Scholar]
- 15.Zhao M, Brunger AT. Recent advances in deciphering the structure and molecular mechanism of the AAA+ ATPase N-Ethylmaleimide-Sensitive Factor (NSF) J Mol Biol. 2015 doi: 10.1016/j.jmb.2015.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Söllner T, et al. SNAP receptors implicated in vesicle targeting and fusion. Nature. 1993;362:318–324. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- 17.Hanson PI, Roth R, Morisaki H, Jahn R, Heuser JE. Structure and conformational changes in NSF and its membrane receptor complexes visualized by quick-freeze/deep-etch electron microscopy. Cell. 1997;90:523–535. doi: 10.1016/s0092-8674(00)80512-7. [DOI] [PubMed] [Google Scholar]
- 18.Söllner T, Bennett MK, Whiteheart SW, Scheller RH, Rothman JE. A protein assembly-disassembly pathway in vitro that may correspond to sequential steps of synaptic vesicle docking, activation, and fusion. Cell. 1993;75:409–418. doi: 10.1016/0092-8674(93)90376-2. [DOI] [PubMed] [Google Scholar]
- 19.Weber T, et al. SNAREpins: minimal machinery for membrane fusion. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- 20.Brunger AT, Cipriano DJ, Diao J. Towards reconstitution of membrane fusion mediated by SNAREs and other synaptic proteins. Crit Rev Biochem Mol Biol. 2015;50:231–241. doi: 10.3109/10409238.2015.1023252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma C, Su L, Seven AB, Xu Y, Rizo J. Reconstitution of the vital functions of Munc18 and Munc13 in neurotransmitter release. Science. 2013;339:421–425. doi: 10.1126/science.1230473. Using liposomes bearing syntaxin 1–Munc18 (rather than syntaxin 1–SNAP25) complexes, this new reconstitution of synaptic vesicle fusion seems to faithfully recapitulate the crucial roles of Munc18, Munc13 and synaptotagmin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zick M, Wickner WT. A distinct tethering step is vital for vacuole membrane fusion. eLife. 2014;3:e03251. doi: 10.7554/eLife.03251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McNew JA, et al. Compartmental specificity of cellular membrane fusion encoded in SNARE proteins. Nature. 2000;407:153–159. doi: 10.1038/35025000. [DOI] [PubMed] [Google Scholar]
- 24.Block MR, Glick BS, Wilcox CA, Wieland FT, Rothman JE. Purification of an N-ethylmaleimide-sensitive protein catalyzing vesicular transport. Proc Natl Acad Sci U S A. 1988;85:7852–7856. doi: 10.1073/pnas.85.21.7852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clary DO, Griff IC, Rothman JE. SNAPs, a family of NSF attachment proteins involved in intracellular membrane fusion in animals and yeast. Cell. 1990;61:709–721. doi: 10.1016/0092-8674(90)90482-t. [DOI] [PubMed] [Google Scholar]
- 26.Miller SE, et al. The molecular basis for the endocytosis of small R-SNAREs by the clathrin adaptor CALM. Cell. 2011;147:1118–1131. doi: 10.1016/j.cell.2011.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pryor PR, et al. Molecular basis for the sorting of the SNARE VAMP7 into endocytic clathrin-coated vesicles by the ArfGAP Hrb. Cell. 2008;134:817–827. doi: 10.1016/j.cell.2008.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fasshauer D, Antonin W, Subramaniam V, Jahn R. SNARE assembly and disassembly exhibit a pronounced hysteresis. Nat Struct Biol. 2002;9:144–151. doi: 10.1038/nsb750. [DOI] [PubMed] [Google Scholar]
- 29.Gao Y, et al. Single reconstituted neuronal SNARE complexes zipper in three distinct stages. Science. 2012;337:1340–1343. doi: 10.1126/science.1224492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Min D, et al. Mechanical unzipping and rezipping of a single SNARE complex reveals hysteresis as a force-generating mechanism. Nature Commun. 2013;4:1705. doi: 10.1038/ncomms2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zorman S, et al. Common intermediates and kinetics, but different energetics, in the assembly of SNARE proteins. eLife. 2014;3:e03348. doi: 10.7554/eLife.03348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Margittai M, Fasshauer D, Pabst S, Jahn R, Langen R. Homo- and heterooligomeric SNARE complexes studied by site-directed spin labeling. J Biol Chem. 2001;276:13169–13177. doi: 10.1074/jbc.M010653200. [DOI] [PubMed] [Google Scholar]
- 33.Xiao W, Poirier MA, Bennett MK, Shin YK. The neuronal t-SNARE complex is a parallel four-helix bundle. Nat Struct Biol. 2001;8:308–311. doi: 10.1038/86174. [DOI] [PubMed] [Google Scholar]
- 34.Lerman JC, Robblee J, Fairman R, Hughson FM. Structural analysis of the neuronal SNARE protein syntaxin-1A. Biochemistry. 2000;39:8470–8479. doi: 10.1021/bi0003994. [DOI] [PubMed] [Google Scholar]
- 35.Weninger K, Bowen ME, Chu S, Brunger AT. Single-molecule studies of SNARE complex assembly reveal parallel and antiparallel configurations. Proc Natl Acad Sci U S A. 2003;100:14800–14805. doi: 10.1073/pnas.2036428100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Furukawa N, Mima J. Multiple and distinct strategies of yeast SNAREs to confer the specificity of membrane fusion. Sci Rep. 2014;4:4277. doi: 10.1038/srep04277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu H, Jun Y, Thompson J, Yates J, Wickner W. HOPS prevents the disassembly of trans-SNARE complexes by Sec17p/Sec18p during membrane fusion. EMBO J. 2010;29:1948–1960. doi: 10.1038/emboj.2010.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weber T, et al. SNAREpins are functionally resistant to disruption by NSF and alphaSNAP. J Cell Biol. 2000;149:1063–1072. doi: 10.1083/jcb.149.5.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao M, et al. Mechanistic insights into the recycling machine of the SNARE complex. Nature. 2015;518:61–67. doi: 10.1038/nature14148. A tour-de-force cryoEM study of NSF, SNAPs and SNAREs reveals the probable mechanism of ATP-dependent SNARE disassembly. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brunet S, Sacher M. Are all multisubunit tethering complexes bona fide tethers? Traffic. 2014;15:1282–1287. doi: 10.1111/tra.12200. [DOI] [PubMed] [Google Scholar]
- 41.Wong M, Munro S. Membrane trafficking. The specificity of vesicle traffic to the Golgi is encoded in the golgin coiled-coil proteins. Science. 2014;346:1256898. doi: 10.1126/science.1256898. The authors provide convincing evidence that golgins are bona fide vesicle tethers by relocalizing them to mitochondria and showing that appropriate vesicles are thereby recruited to this ectopic location. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Munro S. The golgin coiled-coil proteins of the Golgi apparatus. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a005256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bachert C, Linstedt AD. Dual anchoring of the GRASP membrane tether promotes trans pairing. J Biol Chem. 2010;285:16294–16301. doi: 10.1074/jbc.M110.116129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sengupta D, Truschel S, Bachert C, Linstedt AD. Organelle tethering by a homotypic PDZ interaction underlies formation of the Golgi membrane network. J Cell Biol. 2009;186:41–55. doi: 10.1083/jcb.200902110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hayes GL, et al. Multiple Rab GTPase binding sites in GCC185 suggest a model for vesicle tethering at the trans-Golgi. Mol Biol Cell. 2009;20:209–217. doi: 10.1091/mbc.E08-07-0740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sinka R, Gillingham AK, Kondylis V, Munro S. Golgi coiled-coil proteins contain multiple binding sites for Rab family G proteins. J Cell Biol. 2008;183:607–615. doi: 10.1083/jcb.200808018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barr FA. Rab GTPases and membrane identity: causal or inconsequential? J Cell Biol. 2013;202:191–199. doi: 10.1083/jcb.201306010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- 49.Drin G, Morello V, Casella JF, Gounon P, Antonny B. Asymmetric tethering of flat and curved lipid membranes by a golgin. Science. 2008;320:670–673. doi: 10.1126/science.1155821. [DOI] [PubMed] [Google Scholar]
- 50.Sato K, Roboti P, Mironov AA, Lowe M. Coupling of vesicle tethering and Rab binding is required for in vivo functionality of the golgin GMAP-210. Mol Biol Cell. 2015;26:537–553. doi: 10.1091/mbc.E14-10-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang T, Grabski R, Sztul E, Hay J. C p115-SNARE Interactions: A dynamic cycle of p115 binding monomeric SNARE motifs and releasing assembled bundles Traffic. 2015;16:148–171. doi: 10.1111/tra.12242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shorter J, Beard MB, Seemann J, Dirac-Svejstrup AB, Warren G. Sequential tethering of golgins and SNAREpin assembly by the vesicle tethering protein p115. J Cell Biol. 2002;157:45–62. doi: 10.1083/jcb.200112127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cheung PY, Limouse C, Mabuchi H, Pfeffer SR. Protein flexibility is required for vesicle tethering at the Golgi. eLife. 2015;4 doi: 10.7554/eLife.12790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Balderhaar HJ, Ungermann C. CORVET and HOPS tethering complexes -coordinators of endosome and lysosome fusion. J Cell Sci. 2013;126:1307–1316. doi: 10.1242/jcs.107805. [DOI] [PubMed] [Google Scholar]
- 55.Solinger JA, Spang A. Tethering complexes in the endocytic pathway: CORVET and HOPS. FEBS J. 2013;280:2743–2757. doi: 10.1111/febs.12151. [DOI] [PubMed] [Google Scholar]
- 56.Whyte JR, Munro S. Vesicle tethering complexes in membrane traffic. J Cell Sci. 2002;115:2627–2637. doi: 10.1242/jcs.115.13.2627. [DOI] [PubMed] [Google Scholar]
- 57.Kim JJ, Lipatova Z, Segev N. TRAPP complexes in secretion and autophagy. Front Cell Dev Biol. 2016;4:20. doi: 10.3389/fcell.2016.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wickner W. Membrane fusion: Five lipids, four SNAREs, three chaperones, two nucleotides, and a Rab, all dancing in a ring on yeast vacuoles. Annu Rev Cell Dev Biol. 2010;26:115–136. doi: 10.1146/annurev-cellbio-100109-104131. [DOI] [PubMed] [Google Scholar]
- 59.Mima J, Hickey CM, Xu H, Jun Y, Wickner W. Reconstituted membrane fusion requires regulatory lipids, SNAREs and synergistic SNARE chaperones. EMBO J. 2008;27:2031–2042. doi: 10.1038/emboj.2008.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zick M, Stroupe C, Orr A, Douville D, Wickner WT. Membranes linked by trans-SNARE complexes require lipids prone to non-bilayer structure for progression to fusion. eLife. 2014;3:e01879. doi: 10.7554/eLife.01879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hickey CM, Wickner W. HOPS initiates vacuole docking by tethering membranes before trans-SNARE complex assembly. Mol Biol Cell. 2010;21:2297–2305. doi: 10.1091/mbc.E10-01-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stroupe C, Hickey CM, Mima J, Burfeind AS, Wickner W. Minimal membrane docking requirements revealed by reconstitution of Rab GTPase-dependent membrane fusion from purified components. Proc Natl Acad Sci U S A. 2009;106:17626–17633. doi: 10.1073/pnas.0903801106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bröcker C, et al. Molecular architecture of the multisubunit homotypic fusion and vacuole protein sorting (HOPS) tethering complex. Proc Natl Acad Sci U S A. 2012;109:1991–1996. doi: 10.1073/pnas.1117797109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Krämer L, Ungermann C. HOPS drives vacuole fusion by binding the vacuolar SNARE complex and the Vam7 PX domain via two distinct sites. Mol Biol Cell. 2011;22:2601–2611. doi: 10.1091/mbc.E11-02-0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lobingier BT, Merz AJ. Sec1/Munc18 protein Vps33 binds to SNARE domains and the quaternary SNARE complex. Mol Biol Cell. 2012;23:4611–4622. doi: 10.1091/mbc.E12-05-0343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lürick A, et al. The Habc domain of the SNARE Vam3 interacts with the HOPS tethering complex to facilitate vacuole fusion. J Biol Chem. 2015;290:5405–5413. doi: 10.1074/jbc.M114.631465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stroupe C, Collins KM, Fratti RA, Wickner W. Purification of active HOPS complex reveals its affinities for phosphoinositides and the SNARE Vam7p. EMBO J. 2006;25:1579–1589. doi: 10.1038/sj.emboj.7601051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lobingier BT, Nickerson DP, Lo SY, Merz AJ. SM proteins Sly1 and Vps33 co-assemble with Sec17 and SNARE complexes to oppose SNARE disassembly by Sec18. eLife. 2014;3:e02272. doi: 10.7554/eLife.02272. Sec17 (the yeast SNAP) is shown to help load SM proteins onto SNARE complexes, thereby protecting them from disassembly. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Andag U, Neumann T, Schmitt HD. The coatomer-interacting protein Dsl1p is required for Golgi-to-endoplasmic reticulum retrieval in yeast. J Biol Chem. 2001;276:39150–39160. doi: 10.1074/jbc.M105833200. [DOI] [PubMed] [Google Scholar]
- 70.Zink S, Wenzel D, Wurm CA, Schmitt HD. A link between ER tethering and COP-I vesicle uncoating. Dev Cell. 2009;17:403–416. doi: 10.1016/j.devcel.2009.07.012. [DOI] [PubMed] [Google Scholar]
- 71.Ren Y, et al. A structure-based mechanism for vesicle capture by the multisubunit tethering complex Dsl1. Cell. 2009;139:1119–1129. doi: 10.1016/j.cell.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tripathi A, Ren Y, Jeffrey PD, Hughson FM. Structural characterization of Tip20p and Dsl1p, subunits of the Dsl1p vesicle tethering complex. Nat Struct Mol Biol. 2009;16:114–123. doi: 10.1038/nsmb.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kraynack BA, et al. Dsl1p, Tip20p, and the novel Dsl3(Sec39) protein are required for the stability of the Q/t-SNARE complex at the endoplasmic reticulum in yeast. Mol Biol Cell. 2005;16:3963–3977. doi: 10.1091/mbc.E05-01-0056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Andag U, Schmitt HD. Dsl1p, an essential component of the Golgi-endoplasmic reticulum retrieval system in yeast, uses the same sequence motif to interact with different subunits of the COPI vesicle coat. J Biol Chem. 2003;278:51722–51734. doi: 10.1074/jbc.M308740200. [DOI] [PubMed] [Google Scholar]
- 75.Suckling RJ, et al. Structural basis for the binding of tryptophan-based motifs by delta-COP. Proc Natl Acad Sci U S A. 2015;112:14242–14247. doi: 10.1073/pnas.1506186112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Heider MR, et al. Subunit connectivity, assembly determinants and architecture of the yeast exocyst complex. Nat Struct Mol Biol. 2016;23:59–66. doi: 10.1038/nsmb.3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lees JA, Yip CK, Walz T, Hughson FM. Molecular organization of the COG vesicle tethering complex. Nat Struct Mol Biol. 2010;17:1292–1297. doi: 10.1038/nsmb.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shestakova A, Suvorova E, Pavliv O, Khaidakova G, Lupashin V. Interaction of the conserved oligomeric Golgi complex with t-SNARE Syntaxin5a/Sed5 enhances intra-Golgi SNARE complex stability. J Cell Biol. 2007;179:1179–1192. doi: 10.1083/jcb.200705145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Laufman O, Kedan A, Hong W, Lev S. Direct interaction between the COG complex and the SM protein, Sly1, is required for Golgi SNARE pairing. EMBO J. 2009;28:2006–2017. doi: 10.1038/emboj.2009.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Laufman O, Hong W, Lev S. The COG complex interacts with multiple Golgi SNAREs and enhances fusogenic assembly of SNARE complexes. J Cell Sci. 2013;126:1506–1516. doi: 10.1242/jcs.122101. [DOI] [PubMed] [Google Scholar]
- 81.Dubuke ML, Maniatis S, Shaffer SA, Munson M. The exocyst subunit Sec6 interacts with assembled exocytic SNARE complexes. J Biol Chem. 2015;290:28245–28256. doi: 10.1074/jbc.M115.673806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shen D, et al. The synaptobrevin homologue Snc2p recruits the exocyst to secretory vesicles by binding to Sec6p. J Cell Biol. 2013;202:509–526. doi: 10.1083/jcb.201211148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Perez-Victoria FJ, Bonifacino JS. Dual roles of the mammalian GARP complex in tethering and SNARE complex assembly at the trans-golgi network. Mol Cell Biol. 2009;29:5251–5263. doi: 10.1128/MCB.00495-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kim YG, et al. The architecture of the multisubunit TRAPP I complex suggests a model for vesicle tethering. Cell. 2006;127:817–830. doi: 10.1016/j.cell.2006.09.029. [DOI] [PubMed] [Google Scholar]
- 85.Yip CK, Berscheminski J, Walz T. Molecular architecture of the TRAPPII complex and implications for vesicle tethering. Nat Struct Mol Biol. 2010;17:1298–1304. doi: 10.1038/nsmb.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cai Y, et al. The structural basis for activation of the Rab Ypt1p by the TRAPP membrane-tethering complexes. Cell. 2008;133:1202–1213. doi: 10.1016/j.cell.2008.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lord C, et al. Sequential interactions with Sec23 control the direction of vesicle traffic. Nature. 2011;473:181–186. doi: 10.1038/nature09969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Koumandou VL, Dacks JB, Coulson RM, Field MC. Control systems for membrane fusion in the ancestral eukaryote; evolution of tethering complexes and SM proteins. BMC Evol Biol. 2007;7:29. doi: 10.1186/1471-2148-7-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rizo J, Sudhof TC. The membrane fusion enigma: SNAREs, Sec1/Munc18 proteins, and their accomplices--guilty as charged? Ann Rev Cell Dev Biol. 2012;28:279–308. doi: 10.1146/annurev-cellbio-101011-155818. [DOI] [PubMed] [Google Scholar]
- 90.Zhang Y, et al. Munc18a does not alter fusion rates mediated by neuronal SNAREs, synaptotagmin, and complexin. J Biol Chem. 2015;290:10518–10534. doi: 10.1074/jbc.M114.630772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bracher A, Weissenhorn W. Structural basis for the Golgi membrane recruitment of Sly1p by Sed5p. EMBO J. 2002;21:6114–6124. doi: 10.1093/emboj/cdf608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hu SH, et al. Possible roles for Munc18-1 domain 3a and Syntaxin1 N-peptide and C-terminal anchor in SNARE complex formation. Proc Natl Acad Sci U S A. 2011;108:1040–1045. doi: 10.1073/pnas.0914906108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hu SH, Latham CF, Gee CL, James DE, Martin JL. Structure of the Munc18c/Syntaxin4 N-peptide complex defines universal features of the N-peptide binding mode of Sec1/Munc18 proteins. Proc Natl Acad Sci U S A. 2007;104:8773–8778. doi: 10.1073/pnas.0701124104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Burkhardt P, Hattendorf DA, Weis WI, Fasshauer D. Munc18a controls SNARE assembly through its interaction with the syntaxin N-peptide. EMBO J. 2008;27:923–933. doi: 10.1038/emboj.2008.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Burkhardt P, et al. Primordial neurosecretory apparatus identified in the choanoflagellate Monosiga brevicollis. Proc Natl Acad Sci U S A. 2011;108:15264–15269. doi: 10.1073/pnas.1106189108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Misura KM, Scheller RH, Weis WI. Three-dimensional structure of the neuronal-Sec1-syntaxin complex. Nature. 2000;404:355–362. doi: 10.1038/35006120. [DOI] [PubMed] [Google Scholar]
- 97.Dulubova I, et al. How Tlg2p/syntaxin 16 ‘snares’ Vps45. EMBO J. 2002;21:3620–3631. doi: 10.1093/emboj/cdf381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shen J, Tareste DC, Paumet F, Rothman JE, Melia TJ. Selective activation of cognate SNAREpins by Sec1/Munc18 proteins. Cell. 2007;128:183–195. doi: 10.1016/j.cell.2006.12.016. [DOI] [PubMed] [Google Scholar]
- 99.Zhou P, et al. Syntaxin-1 N-peptide and Habc-domain perform distinct essential functions in synaptic vesicle fusion. EMBO J. 2013;32:159–171. doi: 10.1038/emboj.2012.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shen J, Rathore SS, Khandan L, Rothman JE. SNARE bundle and syntaxin N-peptide constitute a minimal complement for Munc18-1 activation of membrane fusion. J Cell Biol. 2010;190:55–63. doi: 10.1083/jcb.201003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rathore SS, et al. Syntaxin N-terminal peptide motif is an initiation factor for the assembly of the SNARE-Sec1/Munc18 membrane fusion complex. Proc Natl Acad Sci U S A. 2010;107:22399–22406. doi: 10.1073/pnas.1012997108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Fernandez I, et al. Three-dimensional structure of an evolutionarily conserved N-terminal domain of syntaxin 1A. Cell. 1998;94:841–849. doi: 10.1016/s0092-8674(00)81742-0. [DOI] [PubMed] [Google Scholar]
- 103.Chen X, Lu J, Dulubova I, Rizo J. NMR analysis of the closed conformation of syntaxin-1. J Biomol NMR. 2008;41:43–54. doi: 10.1007/s10858-008-9239-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Munson M, Chen X, Cocina AE, Schultz SM, Hughson FM. Interactions within the yeast t-SNARE Sso1p that control SNARE complex assembly. Nat Struct Biol. 2000;7:894–902. doi: 10.1038/79659. [DOI] [PubMed] [Google Scholar]
- 105.Demircioglu FE, Burkhardt P, Fasshauer D. The SM protein Sly1 accelerates assembly of the ER-Golgi SNARE complex. Proc Natl Acad Sci U S A. 2014;111:13828–13833. doi: 10.1073/pnas.1408254111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Furgason ML, et al. The N-terminal peptide of the syntaxin Tlg2p modulates binding of its closed conformation to Vps45p. Proc Natl Acad Sci U S A. 2009;106:14303–14308. doi: 10.1073/pnas.0902976106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gerber SH, et al. Conformational switch of syntaxin-1 controls synaptic vesicle fusion. Science. 2008;321:1507–1510. doi: 10.1126/science.1163174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Verhage M, et al. Synaptic assembly of the brain in the absence of neurotransmitter secretion. Science. 2000;287:864–869. doi: 10.1126/science.287.5454.864. [DOI] [PubMed] [Google Scholar]
- 109.Parisotto D, et al. An extended helical conformation in domain 3a of Munc18-1 provides a template for SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) complex assembly. J Biol Chem. 2014;289:9639–9650. doi: 10.1074/jbc.M113.514273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Han GA, et al. A pivotal role for Pro-335 in balancing the dual functions of Munc18-1 domain-3a in regulated exocytosis. J Biol Chem. 2014;289:33617–33628. doi: 10.1074/jbc.M114.584805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Carpp LN, Ciufo LF, Shanks SG, Boyd A, Bryant NJ. The Sec1p/Munc18 protein Vps45p binds its cognate SNARE proteins via two distinct modes. J Cell Biol. 2006;173:927–936. doi: 10.1083/jcb.200512024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Togneri J, Cheng YS, Munson M, Hughson FM, Carr CM. Specific SNARE complex binding mode of the Sec1/Munc-18 protein, Sec1p. Proc Natl Acad Sci U S A. 2006;103:17730–17735. doi: 10.1073/pnas.0605448103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Xu Y, Su L, Rizo J. Binding of Munc18-1 to synaptobrevin and to the SNARE four-helix bundle. Biochemistry. 2010;49:1568–1576. doi: 10.1021/bi9021878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dulubova I, et al. Munc18-1 binds directly to the neuronal SNARE complex. Proc Natl Acad Sci U S A. 2007;104:2697–2702. doi: 10.1073/pnas.0611318104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Carr CM, Rizo J. At the junction of SNARE and SM protein function. Curr Opin Cell Biol. 2010;22:488–495. doi: 10.1016/j.ceb.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Augustin I, Rosenmund C, Sudhof TC, Brose N. Munc13-1 is essential for fusion competence of glutamatergic synaptic vesicles. Nature. 1999;400:457–461. doi: 10.1038/22768. [DOI] [PubMed] [Google Scholar]
- 117.Ma C, Li W, Xu Y, Rizo J. Munc13 mediates the transition from the closed syntaxin-Munc18 complex to the SNARE complex. Nat Struct Mol Biol. 2011;18:542–549. doi: 10.1038/nsmb.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yang X, et al. Syntaxin opening by the MUN domain underlies the function of Munc13 in synaptic-vesicle priming. Nat Struct Mol Biol. 2015;22:547–554. doi: 10.1038/nsmb.3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Richmond JE, Weimer RM, Jorgensen EM. An open form of syntaxin bypasses the requirement for UNC-13 in vesicle priming. Nature. 2001;412:338–341. doi: 10.1038/35085583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Li W, et al. The crystal structure of a Munc13 C-terminal module exhibits a remarkable similarity to vesicle tethering factors. Structure. 2011;19:1443–1455. doi: 10.1016/j.str.2011.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Dulubova I, Yamaguchi T, Wang Y, Sudhof TC, Rizo J. Vam3p structure reveals conserved and divergent properties of syntaxins. Nat Struct Biol. 2001;8:258–264. doi: 10.1038/85012. [DOI] [PubMed] [Google Scholar]
- 122.Jahn R, Fasshauer D. Molecular machines governing exocytosis of synaptic vesicles. Nature. 2012;490:201–207. doi: 10.1038/nature11320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sudhof TC. Neurotransmitter release: the last millisecond in the life of a synaptic vesicle. Neuron. 2013;80:675–690. doi: 10.1016/j.neuron.2013.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mohrmann R, Dhara M, Bruns D. Complexins: small but capable. Cell Mol Life Sci. 2015;72:4221–4235. doi: 10.1007/s00018-015-1998-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Krishnakumar SS, et al. Re-visiting the trans insertion model for complexin clamping. eLife. 2015;4 doi: 10.7554/eLife.04463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kümmel D, et al. Complexin cross-links prefusion SNAREs into a zigzag array. Nat Struct Mol Biol. 2011;18:927–933. doi: 10.1038/nsmb.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Trimbuch T, et al. Re-examining how complexin inhibits neurotransmitter release. eLife. 2014;3:e02391. doi: 10.7554/eLife.02391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Brewer KD, et al. Dynamic binding mode of a Synaptotagmin-1-SNARE complex in solution. Nat Struct Mol Biol. 2015;22:555–564. doi: 10.1038/nsmb.3035. NMR characterization of the interaction between neuronal SNAREs and synaptotagmin reveals a dynamic binding mode; mutagenic disruption of this binding was correlated with functional deficits. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhou Q, et al. Architecture of the synaptotagmin-SNARE machinery for neuronal exocytosis. Nature. 2015;525:62–67. doi: 10.1038/nature14975. The x-ray crystal structure of a complex between synaptotagmin and the neuronal SNARE complex reveals interfaces that are crucial for Ca2+-triggered neurotransmitter release. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Fernandez-Chacon R, et al. Synaptotagmin I functions as a calcium regulator of release probability. Nature. 2001;410:41–49. doi: 10.1038/35065004. [DOI] [PubMed] [Google Scholar]
- 131.Geppert M, et al. Synaptotagmin I: a major Ca2+ sensor for transmitter release at a central synapse. Cell. 1994;79:717–727. doi: 10.1016/0092-8674(94)90556-8. [DOI] [PubMed] [Google Scholar]
- 132.Park Y, et al. Synaptotagmin-1 binds to PIP2-containing membrane but not to SNAREs at physiological ionic strength. Nat Struct Mol Biol. 2015;22:815–823. doi: 10.1038/nsmb.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chang LF, et al. Structural characterization of full-length NSF and 20S particles. Nat Struct Mol Biol. 2012;19:268–275. doi: 10.1038/nsmb.2237. [DOI] [PubMed] [Google Scholar]
- 134.Furst J, Sutton RB, Chen J, Brunger AT, Grigorieff N. Electron cryomicroscopy structure of N-ethyl maleimide sensitive factor at 11 A resolution. EMBO J. 2003;22:4365–4374. doi: 10.1093/emboj/cdg420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ryu JK, et al. Spring-loaded unraveling of a single SNARE complex by NSF in one round of ATP turnover. Science. 2015;347:1485–1489. doi: 10.1126/science.aaa5267. Experiments using single-molecule fluorescence microscopy and magnetic tweezers provide strong support for a ‘spring-loaded’ mechanism for NSF-driven SNARE complex disassembly that is consistent with recent structural studies (REF. 39) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Cipriano DJ, et al. Processive ATP-driven substrate disassembly by the N-ethylmaleimide-sensitive factor (NSF) molecular machine. J Biol Chem. 2013;288:23436–23445. doi: 10.1074/jbc.M113.476705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Shah N, Colbert KN, Enos MD, Herschlag D, Weis WI. Three alphaSNAP and 10 ATP molecules are used in SNARE complex disassembly by N-ethylmaleimide-sensitive factor (NSF) J Biol Chem. 2015;290:2175–2188. doi: 10.1074/jbc.M114.620849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Schwartz ML, Merz AJ. Capture and release of partially zipped trans-SNARE complexes on intact organelles. J Cell Biol. 2009;185:535–549. doi: 10.1083/jcb.200811082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zick M, Orr A, Schwartz ML, Merz AJ, Wickner WT. Sec17 can trigger fusion of trans-SNARE paired membranes without Sec18. Proc Natl Acad Sci U S A. 2015;112:E2290–2297. doi: 10.1073/pnas.1506409112. Proteoliposome experiments unexpectedly indicate that Sec17 (the yeast SNAP) can, when bound to SNARE complexes, facilitate membrane fusion. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Park Y, et al. alpha-SNAP interferes with the zippering of the SNARE protein membranefusion machinery. J Biol Chem. 2014;289:16326–16335. doi: 10.1074/jbc.M114.556803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wang L, Ungermann C, Wickner W. The docking of primed vacuoles can be reversibly arrested by excess Sec17p (alpha-SNAP) J Biol Chem. 2000;275:22862–22867. doi: 10.1074/jbc.M001447200. [DOI] [PubMed] [Google Scholar]
- 142.Tan D, et al. The EM structure of the TRAPPIII complex leads to the identification of a requirement for COPII vesicles on the macroautophagy pathway. Proc Natl Acad Sci U S A. 2013;110:19432–19437. doi: 10.1073/pnas.1316356110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Darsow T, Katzmann DJ, Cowles CR, Emr SD. Vps41p function in the alkaline phosphatase pathway requires homo-oligomerization and interaction with AP-3 through two distinct domains. Mol Biol Cell. 2001;12:37–51. doi: 10.1091/mbc.12.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Baker RW, Jeffrey PD, Hughson FM. Crystal structures of the Sec1/Munc18 (SM) protein Vps33, alone and bound to the homotypic fusion and vacuolar protein sorting (HOPS) subunit Vps16. PLoS One. 2013;8:e67409. doi: 10.1371/journal.pone.0067409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Graham SC, et al. Structural basis of Vps33A recruitment to the human HOPS complex by Vps16. Proc Natl Acad Sci U S A. 2013;110:13345–13350. doi: 10.1073/pnas.1307074110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Seals DF, Eitzen G, Margolis N, Wickner WT, Price A. A Ypt/Rab effector complex containing the Sec1 homolog Vps33p is required for homotypic vacuole fusion. Proc Natl Acad Sci U S A. 2000;97:9402–9407. doi: 10.1073/pnas.97.17.9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sivaram MV, Saporita JA, Furgason ML, Boettcher AJ, Munson M. Dimerization of the exocyst protein Sec6p and its interaction with the t-SNARE Sec9p. Biochemistry. 2005;44:6302–6311. doi: 10.1021/bi048008z. [DOI] [PubMed] [Google Scholar]
- 148.Morgera F, et al. Regulation of exocytosis by the exocyst subunit Sec6 and the SM protein Sec1. Mol Biol Cell. 2012;23:337–346. doi: 10.1091/mbc.E11-08-0670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Guo W, Roth D, Walch-Solimena C, Novick P. The exocyst is an effector for Sec4p, targeting secretory vesicles to sites of exocytosis. EMBO J. 1999;18:1071–1080. doi: 10.1093/emboj/18.4.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Willett R, et al. COG complexes form spatial landmarks for distinct SNARE complexes. Nature Commun. 2013;4:1553. doi: 10.1038/ncomms2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Laufman O, Hong W, Lev S. The COG complex interacts directly with Syntaxin 6 and positively regulates endosome-to-TGN retrograde transport. J Cell Biol. 2011;194:459–472. doi: 10.1083/jcb.201102045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Miller VJ, et al. Molecular insights into vesicle tethering at the Golgi by the conserved oligomeric Golgi (COG) complex and the golgin TATA element modulatory factor (TMF) J Biol Chem. 2013;288:4229–4240. doi: 10.1074/jbc.M112.426767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Suvorova ES, Duden R, Lupashin VV. The Sec34/Sec35p complex, a Ypt1p effector required for retrograde intra-Golgi trafficking, interacts with Golgi SNAREs and COPI vesicle coat proteins. J Cell Biol. 2002;157:631–643. doi: 10.1083/jcb.200111081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Siniossoglou S, Pelham HR. Vps51p links the VFT complex to the SNARE Tlg1p. J Biol Chem. 2002;277:48318–48324. doi: 10.1074/jbc.M209428200. [DOI] [PubMed] [Google Scholar]
- 155.Cai H, et al. TRAPPI tethers COPII vesicles by binding the coat subunit Sec23. Nature. 2007;445:941–944. doi: 10.1038/nature05527. [DOI] [PubMed] [Google Scholar]