Abstract
Mechanical damage at the time of joint injury and the ensuing inflammatory response associated with elevated levels of pro-inflammatory cytokines in the synovial fluid, are reported to contribute to the progression to osteoarthritis after injury. In this exploratory study, we used a targeted proteomics approach to follow the progression of matrix degradation in response to mechanical damage and cytokine treatment of human knee cartilage explants, and thereby to study potential molecular biomarkers. This proteomics approach allowed us to unambiguously identify and quantify multiple peptides and proteins in the cartilage medium and explants upon treatment with +/− injurious compression +/−cytokines, treatments that mimic the earliest events in post-traumatic OA. We followed degradation of different protein domains, e.g., G1/G2/G3 of aggrecan, by measuring representative peptides of matrix proteins released into the medium at 7 time points throughout the 21-day culture period. COMP neo epitopes, which were previously identified in the synovial fluid of knee injury/OA patients, were also released by these human cartilage explants treated with cyt and cyt+inj. The absence of collagen pro-peptides and elevated levels of specific COMP and COL3A1 neo-epitopes after human knee trauma may be relevant as potential biomarkers for post-traumatic OA. This model system thereby enables study of the kinetics of cartilage degradation and the identification of biomarkers within cartilage explants and those released to culture medium. Discovery proteomics revealed that candidate proteases were identified after specific treatment conditions, including MMP1, MMP-3, MMP-10 and MMP-13.
Keywords: Post-traumatic osteoarthritis, mass spectrometry, cartilage matrix, cytokines, proteomics
1. Introduction
Traumatic joint injuries can result in damage to tissues including cartilage, ligaments, meniscus, tendons, synovium and bone. In cases that involve rupture of the anterior cruciate ligament (ACL) and/or tears of the menisci, there may be no visual damage to cartilage surfaces at the time of arthroscopy, but there may be micro-damage to cartilage matrix and cell death especially in the superficial regions [1]. Direct impact mechanical damage to cartilage can also occur, ranging from surface fibrillation to cracking down to bone [2]. ACL injuries are known to trigger an immediate inflammatory response [3], which acts in conjunction with continued long-term abnormal mechanical loading of the injured knee [4] to contribute to the development of post-traumatic osteoarthritis (PTOA). Cytokines including IL-1, IL-6, TNFα, IL-8 and others [5–8] are found at elevated levels in the synovial fluid after such joint injuries. These cytokines are known to upregulate chondrocyte gene expression of various aggrecanases, collagenases and other MMPs, which are hypothesized to contribute to the degradation of cartilage matrix. In complementary in vitro models of acute joint injury [9–11], injurious mechanical compression has been shown to cause chondrocyte apoptosis and necrosis, and to potentiate the effects of cytokine treatment on cartilage matrix degradation.
The continued degeneration of cartilage after injury may take as long as 10-15 years before radiographic evidence and clinical symptoms begin to emerge [12]. During the past decade, there has been a tremendous push to discover and develop both imaging and molecular biomarkers to detect the earliest stages of PTOA and track progression of the disease [13]. An understanding of the processes that occur before clinical symptoms lead to joint deterioration depends on the discovery of such biomarkers. In particular, molecular biomarkers may also provide important knowledge of targets for therapeutic development. In this regard, synovial fluid analyses of patient samples have already provided valuable information on the progression of macromolecular degradation of cartilage ECM after injury [14]. However, reliance solely on synovial fluid aspirates has several drawbacks: (a) these patient samples can only be taken at selected intermittent intervals given patient compliance, and (b) the ongoing degradative processes occurring inside cartilage itself are not obtained.
In the present study, we have used an in vitro model of joint trauma involving the combination of a single impact injury to human cartilage explants that are immediately co-cultured in the presence of inflammatory cytokines (IL-6 and TNFα). Controlled studies using cartilage explants provide a unique opportunity to characterize the kinetics of specific cartilage degradation events via proteomic analyses of the various proteins released to the culture medium and those remaining in the cartilage. The culture medium from such studies (involving treatment of explants with various inflammatory, injurious, or therapeutic factors) has been used as a surrogate for synovial fluid in order to reduce complexity and to identify the response of cartilage to specific treatments. Recent proteomics studies of the cartilage secretome in response to specific cytokines [15–20] have enabled the discovery of new macromolecules and novel cleavage products. This advantage is now further enhanced by obtaining proteomic data to track a broad array of matrix molecules and fragments secreted by explants following mechanical injury and cytokine co-culture.
Previous proteomic studies of the secretome using explant models have generally focused on degradation products released into the medium at a single end point [21,22]. In addition, they have not included the combined effects of impact mechanical damage and inflammatory cytokine challenge that may exacerbate cartilage destruction and induce novel temporal effects on degradation processes. In the present study, we incorporated treatments involving mechanical impact injury, cytokine challenge and their combinations, and quantified the kinetics of proteins released from human cartilage explants with time over a three week period using targeted proteomics.
2. Results
2.1 Kinetics of aggrecan degradation
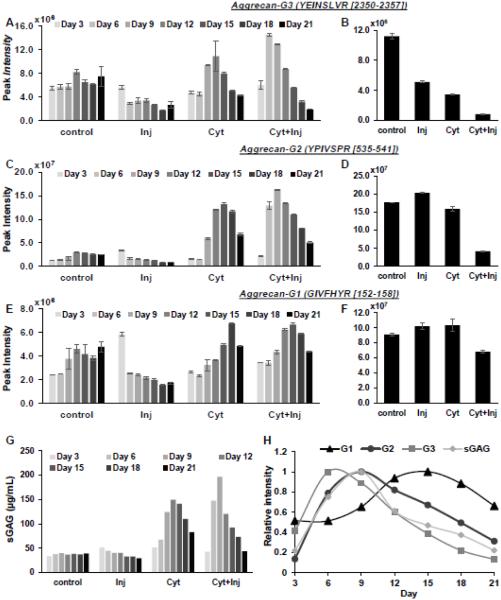
Using the experimental timeline for explant mechanical injury followed by cytokine co-culture shown schematically in Fig. 1, peptides within the G1, G2, and G3 aggrecan globular domains (Fig. 1B) that were released to the medium were measured quantitatively using the targeted mass spectrometry method, multiple reaction monitoring (MRM). Representative results from one cartilage explant in a treatment groups are presented here when at least three out of the four explants in that group had a similar response. In the injury-alone condition, the G3 peptide (2350YEINSLVR2357) showed a slightly increased release on Day 3 but reduced release thereafter (Fig. 2A). However, cytokine treatment increased the release of G3 peptide significantly, which peaked at day 12 with cyt-alone treatment and day 6 with cyt + inj treatment. In addition to increased release, we observed that the amount of G3 peptide remaining in the explant was dramatically decreased in all treatments compared to control (Fig 2B). Similarly, G2 and G1 peptides showed increased release on Day 3 in the injury-alone condition, but decreased release at subsequent time points (Fig 2C,E). Cytokine treatment also triggered increased release of G2 and G3 peptides, with the combination of cyt +inj treatment further accelerating this catabolic response (Fig 2C,E). Taken together, we observed that G3 peptides were the earliest to be released to the medium upon cyt or inj+cyt treatments, followed by G2 and lastly G1 peptides (Fig 2H). Interestingly, the measured release kinetics of total sGAG (Fig 2G) resembled most closely that of G2 peptides (Fig 2H), which is consistent with the fact that most of the GAG chains are located between the G2-G3 domains.
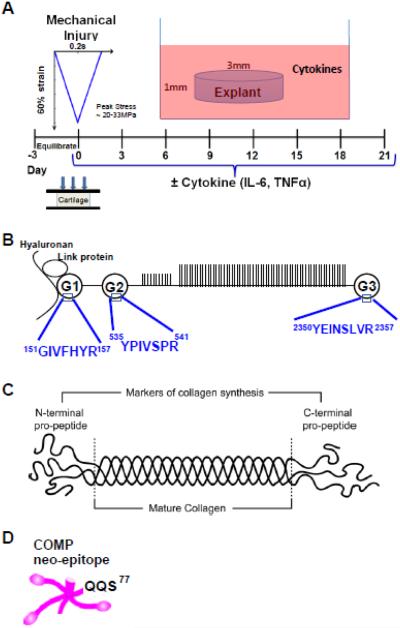
Figure 1. experimental design.
A. Human cartilage explants were subjected to unconfined mechanical compression to 60% strain (at a strain rate of 300%/s, followed by immediate release at the same rate) +/− cytokine treatment (IL-6 + TNFα) after three days of pre-equilibration. Medium changes were carried out every three days between Day 3-Day 21. B. Peptides in the G1, G2, and G3 globular domains of aggrecan were quantified using the MRM approach. C. N- and C-terminal pro-peptides are markers of collagen synthesis, and are cleaved away in mature collagen molecules. D. COMP neo-epitope QQS77, which has also been identified in the synovial fluid of patients with acute joint injury [15].
Figure 2. Aggrecan degradation kinetics.
Degradation kinetics of aggrecan G3, G2, and G1 peptides released into the medium (A, C, E) and remained in the explant (B, D, F) measured by MRM mass spectrometry. Values are average peak intensities of two technical replicates +/− standard deviation. G. GAG released into the medium for the 21 day incubation period measured by DMMB assay. H. Plot of release of G1, G2, G3 and sGAG into the medium over a 21 day time period for the cyt + inj treatment condition. Values are normalized to the maximum value.
2.2 Collagen synthesis and degradation
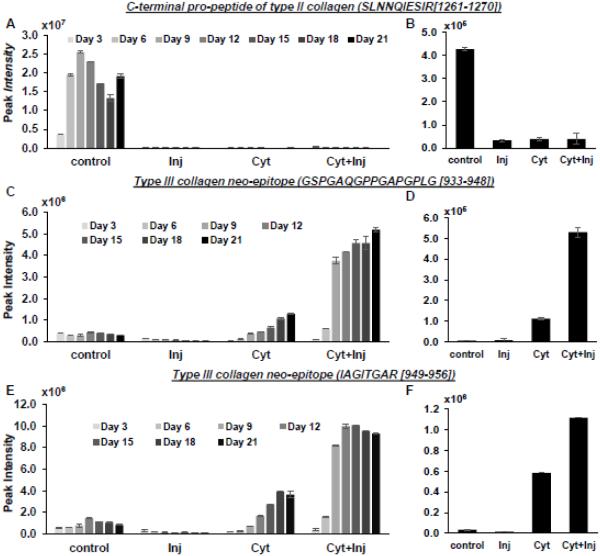
Collagen Type II c-terminal pro-peptide (1261SLNNQIESIR1270), an indicator of newly synthesized type II Collagen, was measured using the MRM method. In the medium samples, collagen c-pro-peptides were only found for the control condition (Fig 3A), indicating that injury as well as cyt-alone and cyt+inj could completely prevent synthesis of type II collagen. This result was further confirmed by the observation that much lower levels of c-pro-peptide were found in residual cartilage explants that had been subjected to inj, cyt or inj+cyt treatments (Fig 3B).
Figure 3. Collagen peptides release kinetics.
Peak intensity of release of type II collagen C-terminal pro-peptide into the medium (A) and retained in the explant (B). Release of Collagen Type III cleavage neo-epitope into the medium (C, E) and remaining in the explant (D, F). Values are average peak intensities of two technical replicates +/− standard deviation.
As an indication of collagen cleavage and degradation, we observed that two distinct neo-epitopes of collagen III were found at elevated levels in both the release medium and residual explants treated with cyt and cyt+inj (Fig 3C-F). Although injury alone did not increase collagen III degradation, the addition of injury to cytokine treatment triggered even higher levels of collagen III degradation and release compared to cytokine alone. Thus, similar to aggrecan, injury again potentiates the catabolic effect of cytokine treatment on collagen degradation. Importantly, these neo-epitopes associated with Collagen III degradation could potentially be new biomarkers for post-traumatic cartilage breakdown.
2.3 COMP neo-epitope release is elevated upon cytokine treatment
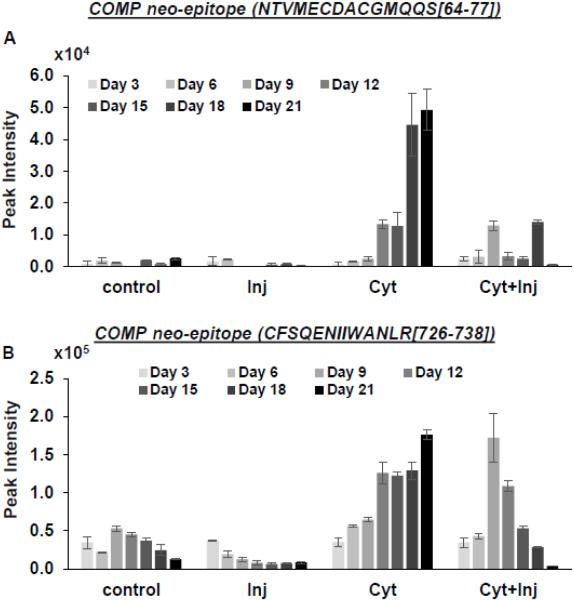
The COMP neo-epitope, NTVMECDACGMQQS (QQS77), which was found in the synovial fluid of patients with acute joint injury, was also detected in the medium samples of explants treated especially with cyt-alone (Fig 4A). Thus, our ex vivo human explant model closely resembles that of human cartilage response to injury in vivo in terms of the release of novel COMP fragments into the medium [15]. In addition, we identified another COMP neo-epitope (726CFSQENIIWANLR738) previously reported in equine tendon experiments [23] with increased release upon cytokine and cyt+inj treatments (Fig 4B). This novel neo-epitope may also be a potential biomarker for cartilage injury in PTOA.
Figure 4. Release of two COMP neo-epitopes into the medium (A, B).
Values are average peak intensities of two technical replicates +/− standard deviation.
2.4 MMPs were detected in cytokine treated conditions
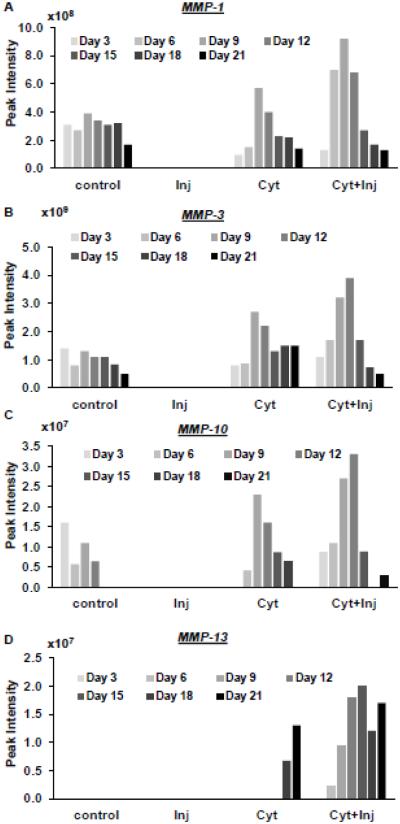
We observed in discovery experiments (run on medium from 2 explants series) that MMP-1, -3, and 10 were found in the medium samples of control (1 of 2 explants), cyt (2 of 2), and cyt + inj (2 of 2) conditions but not detected in the injury alone condition (Fig 5A-C) (2 of 2 2). MMP-13 was only found in the medium sample of cyt and cyt +inj treated conditions (Fig 4D). Thus, these results indicate that cytokine treatment stimulates the production of catabolic proteases in cartilage.
Figure 5. Peak intensities of matrix metalloproteinases.
(MMPs) released into the medium over the 21 day incubation time period. Values are peak intensities of top 3 peptides for each protein.
3. Discussion
In this study, we employed a mass spectrometry approach to characterize the kinetics of aggrecan and collagen degradation in human cartilage explants challenged with cytokines, mechanical injury, or their combination. We found that the kinetics of release of specific globular domains of aggrecan may constitute the earliest sequence of cartilage degenerative events. In addition, we discovered Type III collagen and COMP neo-epitopes which may be important potential biomarkers for cartilage degradation associated with traumatic cartilage injury.
Aggrecan degradation, a hallmark of cartilage degeneration, occurs mainly through proteolysis, with cleavage in the IGD being the most detrimental for cartilage function, as it results in a loss of aggrecan fragments containing the negatively charged sGAG chains. Electrostatic and osmotic swelling forces between these GAGs account for a significant fraction of cartilage’s equilibrium stiffness [24,25]. Additionally, the resistance to intra-tissue fluid flow provided by these closely spaced GAGs in situ results in a dramatic increase in poroelastic stiffness that normal cartilage presents to dynamic compressive loads at higher loading rates [26]. Cytokines are known to stimulate the upregulation of aggrecanases in cartilage that result in cleavage of aggrecan [27]. The NITEGE neoepitope, which is located in the IGD, was detected in the medium of bovine cartilage explants treated with IL-6 and TNFα [9]. Evidence of the kinetics of aggrecan degradation and the release of aggrecan fragments associated with specific aggrecanase cleavage sites (e.g., via Western analyses following IL-1 treatment of cartilage explants [28] ) has demonstrated that cleavage in the CS-2 and CS-1 domains occurs before that in the IGD upon cytokine treatment of cartilage explants. In the present study, using the MRM LC-MS approach, we have been able to delineate a precise timeline for release of globular domain fragments, showing that cleavage in the CS1 and CS2 domain of aggrecan (which results in G3 release) precedes cleavage in the IGD by 6 days (release of G2 and G3, Fig, 2H). Additionally, the release of sGAG (via DMMB analysis) closely resembled the time line of G2 domain peptide release, further validating our peptide measurements, as most of the GAG chains are found between G2 and G3 domain of the aggrecan molecule. The release of G1 domain occurs after the peak release of G2 domain and ~9 days later than the release of G3 peptide. These results indicate that the detection of globular domains upon inflammatory cytokine challenge to cartilage could be another useful marker in characterizing the progression of cartilage degradation.
Type II collagen is the most abundant cartilage protein, representing 80–95% of the total collagen content. In previous studies, C-propeptide of type II procollagen (CPII) has been shown to be directly related to the synthesis of collagen in cartilage [29]. Collagen type II synthesis has been shown to be increased in OA cartilage compared to healthy cartilage [29–31]. However, the serum level of CPII was found to be reduced in OA samples compared to healthy control [29]. Genomics studies have shown that both the zone of cartilage and the stage of OA progression affected the expression of the COL2 gene [32,33]; in particular, collagen II gene expression was found to be upregulated in late-stage OA cartilage [34]. Treatment of cartilage with pro-inflammatory cytokines, such as IL-1 and TNFα, can inhibit the synthesis of collagen type II by suppressing gene expression on a transcriptional level [35]. Based on our MRM targeted proteomics approach, we found that cytokine challenge inhibited collagen synthesis. In addition to the results presented here, this trend was consistent with that in three other explant replicates that we studied (unpublished results). The reason for this response could be due to the stage of OA of the tissue. We observed a decrease in collagen type II synthesis upon cytokine and injury treatment, as indicated by the reduced detection of C and N-terminal pro-peptide of collagen II in the medium as well as in the explants.
Additionally, we discovered a new neo-epitope of Type III collagen degradation, which is specifically released into the medium upon treatment with cytokines. Type III collagen molecules are present in the extracellular matrix of adult human and bovine articular cartilages extensively cross-linked to Type II collagen [36]. It has been hypothesized that Type III collagen may act as a covalent modifier that adds cohesion to a weakened, existing collagen type II fibril network as part of a chondrocyte healing response to matrix damage [36]. The putative cleavage site of collagen III is not known, but the site we discovered is in a region containing repeats of the motif GXP, and the cleavage occurred at the Gly-Ile bond, which is similar to the 1/4 -3/4 cleavage site for collagen II [37]. The increase in collagen III cleavage could indicate a further weakening of the collagen fibril network in cartilage. In addition, this new neo-epitope could be a potential biomarker for cartilage degradation in vivo.
Finally, using a discovery based approach, we investigated the release of MMPs into the medium and found that MMP-1, 3, 10, and 13 levels are elevated in cytokine treated explants. MMP-1 and 13 are involved in the degradation of collagen II. MMP-13 was previously shown to generate a collagen IX cleavage in the NC4 domain [18] . However, the role of MMP-10 in cytokine induced cartilage degradation is not yet characterized. Our results highlight the importance of MMP-10 in addition to the other MMPs that are known to play an important role. The specific function of these MMPs in matrix degradation is an important topic for future studies. The influence of age, gender, and stage of OA of donor tissue is another important aspect to address in future studies.
In summary, we have shown that a human ex vivo model of joint injury incorporating mechanical injury to cartilage and subsequent co-culture of these explants with inflammatory cytokines shows cartilage degradation profiles relevant to cartilage degradation in vivo. Using a proteomics approach, we were able to show that our model is consistent with known literature and at the same time, discover new biological insights and potential biomarkers. This makes it a valuable and feasible system to determine the efficacy of potential therapeutics.
4. Experimental procedures
4.1 Adult human cartilage harvest and explant culture
Tibial plateau (Collins Grade 0) from a 19-year old male donor was obtained postmortem from the Gift of Hope Organ and Tissue Donor Network (Itasca, IL). All procedures were approved by the Rush University Medical Center Institutional Review Board (ORA Number: 08082803-IRB01-AM01) and the Committee on the Use of Humans as Experimental Subjects at MIT. The joint surface was scored by an experienced forensic pathologist using the modified Collins grading system [38] at the time of tissue harvest. Full-thickness (~1.5mm) cartilage disks cored with a 3-mm punch were harvested within 24 hours after death of donor. A total of 16 explants, taken from both the lateral and medial side of the right tibial plateau were used for this experiment (Suppl Fig 1). Explants were pre-equilibrated for 2 days in high-glucose DMEM (4.5 g/L; Corning Cellgro, Manassas, VA) supplemented with 10mM HEPES (Gibco, Grand Island, NY), 1mM sodium pyruvate (Gibco), 0.1mM nonessential amino acids (Sigma Aldrich, St. Louis, MO), 1% insulin-transferrin-selenium (10μg/ml, 5.5μg/ml, and 5 ng/ml, respectively; Sigma), 0.4mM proline (Sigma), 20μg/ml ascorbic acid (Sigma), 100units/ml penicillin G, 100μg/ml streptomycin, and 0.25μg/ml amphotericin B (Sigma) for 2 days (5% CO2; 37°C).
4.2 Injurious compression and explant culture with exogenous cytokines
After pre-equilibration, cartilage disks were injuriously compressed in a custom-designed, incubator-housed loading apparatus [39,40]. As described previously [41], each individual cartilage disk was placed in a polysulfone chamber and subjected to radially unconfined compression to 60% final strain at a strain rate of 300%/s, followed by immediate release at the same rate (Fig. 1A). After injury, disks were immediately placed in the same equilibration medium as above for 21 days in the presence or absence of 25 ng/mL rhTNF-α, 50 ng/mL rhIL-6, and 250 ng/mL sIL-6R (R&D Systems, Minneapolis, MN). Previous studies showed that this combination of cytokines caused significantly greater sGAG loss than either cytokine alone[9,42]. There were 4 explants used for each treatment condition and they were incubated individually. Medium changes were carried out every 3 days, and used medium (300uL) was collected and stored at −80°C until analysis at each time point between day 3 and day 21 during the 21-day culture. At the end of the 21-day culture, cartilage explants were flash-frozen in liquid nitrogen.
4.3 Sample processing and mass spectrometry
4.3.1 Culture medium processing
Culture medium (25μL) from different time points was reduced by 4mM dithiothreitol for 30 minutes at 56°C, alkylated by 16mM iodoacetamide for 60 minutes in the dark at room temperature, precipitated twice with ethanol and digested by 0.25μg trypsin gold (Promega, Madison, WI) in 0.1M ammonium bicarbonate (AMBIC) buffer pH 7.8 for 16 hours on a shaker at 37°C. After drying, samples were re-suspended in 100μl 0.5M AMBIC, run through 30kDa filter (PALL Life Sciences, Westborough, MA) and desalted with reversed-phase C18 columns (SEM SS04V-SS18V) according to the manufacturer’s instructions (Nest Group, Southborough, MA).
4.3.2 Cartilage explant processing post injury
The cartilage explant disks were mounted on a cryostat via a PBS ice block, which was then mounted onto the holder using Tissue-Tek (Sakura, The Netherlands) and cryosectioned into 20μm tissue sections. These sections from the entire explant disk were combined and extracted for 24 h under gentle shaking at 4 °C using 15 volumes 4M GuHCl chaotropic extraction buffer (4M guanidine hydrochloride, 50mM sodium acetate, 100mM 6-aminocaproic acid, 5mM benzamidine, 5mM N-ethylmaleimide; pH 5.8). The tissue extracts (50μl) were then treated according to the same procedure as the explant culture medium described above.
4.3.3 Multiple reaction monitoring (MRM) mass spectrometry
We used a targeted approach to examine a predefined set of ECM proteins due to its high specificity and sensitivity for accurate quantification. The TSQ Vantage triple quadrupole mass spectrometer (ThermoFisher Scientific, Waltham MA), equipped with an Easy nano-LC system (ThermoFisher Scientific) was used for these MRM analyses. Aliquots of the desalted peptide samples were injected and quantified using a LC-MS approach and each sample was run in technical duplicates. Three to six transitions for each peptide were measured in a scheduled MRM method according to previously published work [43]. The mass spectrometer was operated in MRM mode, with both Q1 and Q3 settings at 0.7Da resolution. A spray voltage of +1700V was used with a heated ion transfer setting of 270°C for desolvation. Data were acquired using the Xcalibur version 2.1 software (ThermoFisher Scientific ). Mobile phases used were buffer A: 0.1% formic acid in water and buffer B: 0.1% formic acid in 100% acetonitrile. Separation was performed on PicoTip™ emitter 75μm×15cm capillary columns with 10μm diameter tip (New Objective, Woburn, MA), and packed with 3μm Reprosil-Pur C18-AQ resin (Dr. Maich GmbH, Ammerbuch, Germany). The on-line reversed-phase separation was performed using a flow rate of 300nl/min and a linear binary gradient from 3% B for 5min to 15% B in 3min, then to 35% B in 32min and finally to 90% B in 3min followed by a wash for 3min with 90% B, and reconditioned for 10min between each run. A standard mixture of tryptic peptides was run to check the system performance of the LC-MS setup [44].
4.3.4 Discovery proteomics
For two sets of the tissue explants, we also ran the medium samples (56 samples) using discovery mode proteomics in order to explore the medium proteome in greater depth. Discovery mode experiments (non-targeted mass spectrometry), were performed on a quadrupole Orbitrap benchtop mass spectrometer, QExactive, (Thermo Scientific) equipped with an Easy nano-LC 1000 system (ThermoFisher Scientific). Separation was performed on 75μm × 25cm, Acclaim Pepmap™ RSLC C18 capillary columns packed with 2μm particles. (ThermoFisher Scientific). A spray voltage of +2000 V was used with a heated ion transfer setting of 275°C for desolvation. The on-line reversed-phase separation was performed on an Easy nano-LC 1000 system using a flow rate of 300 nl/min and a linear binary gradient from 3% solvent B for 60 min to 35% solvent B, then to 90% solvent B for 5 min and finally isocratic flow with 90% solvent B for 5 min. An MS scan (400–1200 m/z) was recorded in the Orbitrap mass analyzer set at a resolution of 70,000 at 200 m/z, 1×106 automatic gain control (AGC) target and 100 ms maximum ion injection time. The MS was followed by data-dependent collision-induced dissociation MS/MS scans at a resolution of 15,000 on the 15 most intense multiply charged ions at 2 × 104 intensity threshold, 2 m/z isolation width and dynamic exclusion enabled for 30 seconds. Multiple peptides were measured for each protein using discovery based proteomics and the peak intensity for each protein was determined by taking the average peak area intensities of the top 3 peptides. Averaging of the top 3 peptides is based on the report that the average MS signal response for the three most intense tryptic peptides signals per mole of protein is constant [45]. The peak intensity reported represents the abundance of the protein in the medium samples.
4.4 Data Analysis
The targeted MRM data were analyzed using the Skyline 2.6 software (MacCoss Lab Software, University of Washington [46]). The relative signal from the individual transitions in combination with the expected retention time of the peak ensured the identity of the peak as measured by synthetic peptides during optimization. For simplicity, the summed areas of the individual transitions are presented in the figures throughout this paper.
Identification from discovery data was performed using the Homo sapiens taxonomy (20,200 sequences) setting of the Swiss-Prot database (SwissProt_2015_06) with Proteome Discoverer 2.0 software (ThermoFisher Scientific). The processing workflow consisted of the following nodes: Spectrum Selector for spectra pre-processing (precursor mass range: 350– 5000 Da; S/N Threshold: 1.5), Sequest-HT search engine (Protein Database: see above; Enzyme: Trypsin; Max. missed cleavage sites: 2; Peptide length range 6–144 amino acids; Precursor mass tolerance: 10 ppm; Fragment mass tolerance: 0.02 Da; Static modification: cysteine carbamidomethylation; Dynamic modification: methionine oxidation, hydroxyproline and pyro-glutamic acid (N-terminal Glu to pyroglutamic acid), and Percolator for peptide validation (FDR<1% based on peptide q-value). Results were filtered to keep only the Master protein with at least one unique peptide, and protein grouping was allowed according to the parsimony principle. For non-targeted proteomics data, the sum of the top 3 peptides for each protein was taken to reflect the intensity of the protein. Peptide intensities were quantified using a proprietary algorithm developed in Proteome Discoverer 2.0 (ThermoFisher Scientific).
Supplementary Material
Manuscript Highlights.
Proteomics analysis revealed kinetics of matrix degradation in cytokine challenged and mechanically injured human cartilage explants
Release of aggrecan G3 domain may constitute the earliest sequence of cartilage degenerative events
Collagen type II synthesis was decreased upon cytokine treatment
Collagen type III and COMP neo-epitopes may be important potential biomarkers for cartilage degradation
Acknowledgements
Supported by grants NIH-NIAMS AR060331 (AJG), A*STAR (Singapore) Fellowship and Poitras Fellowship and Poitras Pre-doctoral Fellowship (YW), Rush Ciba-Geigy Endowed Chair (SC), and the Swedish Research Council (2014−3303), the Royal Psysiographic Society, the Swedish Rheumatism Association, the Alfred Österlund Foundation, the Greta & Johan Kock and Crafoord Foundations, and the Magnus Bergvall Foundation (all PÖ). We would like to acknowledge Gift of Hope Organ and Tissue Donor Network and donors’ families for human cartilage and Dr. Arkady Margulis for tissue procurement.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
All were involved in the conception and design of the study as well as interpretation of the data. YW and YL conducted the human explant experimental work and data analysis. AK and PO conducted the mass spectrometry experimental work and data analysis. All authors critically revised the manuscript and gave final approval of the article.
Conflict of interest
The authors do not have conflict of interest.
References
- [1].Johnson DL, Urban WP, Caborn DN, Vanarthos WJ, Carlson CS. Articular cartilage changes seen with magnetic resonance imaging-detected bone bruises associated with acute anterior cruciate ligament rupture. Am. J. Sports Med. 1998;26:409–14. doi: 10.1177/03635465980260031101. [DOI] [PubMed] [Google Scholar]
- [2].Buckwalter JA. Articular cartilage injuries. Clin. Orthop. Relat. Res. 2002:21–37. doi: 10.1097/00003086-200209000-00004. [DOI] [PubMed] [Google Scholar]
- [3].Lieberthal J, Sambamurthy N, Scanzello CR. Inflammation in joint injury and post-traumatic osteoarthritis. Osteoarthritis Cartilage. 2015;23:1825–34. doi: 10.1016/j.joca.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Hosseini A, Van de Velde S, Gill TJ, Li G. Tibiofemoral cartilage contact biomechanics in patients after reconstruction of a ruptured anterior cruciate ligament. J. Orthop. Res. 2012;30:1781–8. doi: 10.1002/jor.22122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Swärd P, Frobell R, Englund M, Roos H, Struglics A. Cartilage and bone markers and inflammatory cytokines are increased in synovial fluid in the acute phase of knee injury (hemarthrosis) - a cross-sectional analysis. Osteoarthritis Cartilage. 2012;20:1302–8. doi: 10.1016/j.joca.2012.07.021. [DOI] [PubMed] [Google Scholar]
- [6].Irie K, Uchiyama E, Iwaso H. Intraarticular inflammatory cytokines in acute anterior cruciate ligament injured knee. Knee. 2003;10:93–6. doi: 10.1016/s0968-0160(02)00083-2. [DOI] [PubMed] [Google Scholar]
- [7].Higuchi H, Shirakura K, Kimura M, Terauchi M, Shinozaki T, Watanabe H, Takagishi K. Changes in biochemical parameters after anterior cruciate ligament injury. Int. Orthop. 2006;30:43–7. doi: 10.1007/s00264-005-0023-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Cameron ML, Fu FH, Paessler HH, Schneider M, Evans CH. Synovial fluid cytokine concentrations as possible prognostic indicators in the ACL-deficient knee. Knee Surg. Sports Traumatol. Arthrosc. 1994;2:38–44. doi: 10.1007/BF01552652. [DOI] [PubMed] [Google Scholar]
- [9].Sui Y, Lee JH, DiMicco MA, Vanderploeg EJ, Blake SM, Hung H-H, Plaas AHK, James IE, Song X-Y, Lark MW, Grodzinsky AJ. Mechanical injury potentiates proteoglycan catabolism induced by interleukin-6 with soluble interleukin-6 receptor and tumor necrosis factor alpha in immature bovine and adult human articular cartilage. Arthritis Rheum. 2009;60:2985–96. doi: 10.1002/art.24857. [DOI] [PubMed] [Google Scholar]
- [10].Lu YCS, Evans CH, Grodzinsky AJ. Effects of short-term glucocorticoid treatment on changes in cartilage matrix degradation and chondrocyte gene expression induced by mechanical injury and inflammatory cytokines. Arthritis Res. Ther. 2011;13:R142. doi: 10.1186/ar3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Patwari P, Lin SN, Kurz B, Cole AA, Kumar S, Grodzinsky AJ. Potent inhibition of cartilage biosynthesis by coincubation with joint capsule through an IL-1-independent pathway. Scand. J. Med. Sci. Sports. 2009;19:528–35. doi: 10.1111/j.1600-0838.2009.00911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Anderson DD, Chubinskaya S, Guilak F, Martin JA, Oegema TR, Olson SA, Buckwalter JA. Post-traumatic osteoarthritis: improved understanding and opportunities for early intervention. J. Orthop. Res. 2011;29:802–9. doi: 10.1002/jor.21359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Golightly YM, Adams SB, Kraus VB. Biomarkers of PTA. In: Olson S, Guilak F, editors. Post-Traumatic Arthritis. Springer; US: 2015. pp. 317–330. [Google Scholar]
- [14].Lohmander LS, Englund PM, Dahl LL, Roos EM. The long-term consequence of anterior cruciate ligament and meniscus injuries: osteoarthritis. Am. J. Sports Med. 2007;35:1756–69. doi: 10.1177/0363546507307396. [DOI] [PubMed] [Google Scholar]
- [15].Åhrman E, Lorenzo P, Holmgren K, Grodzinsky AJ, Dahlberg LE, Saxne T, Heinegård D, Önnerfjord P. Novel cartilage oligomeric matrix protein (COMP) neoepitopes identified in synovial fluids from patients with joint diseases using affinity chromatography and mass spectrometry. J. Biol. Chem. 2014;289:20908–20916. doi: 10.1074/jbc.M114.554683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Williams A, Smith JR, Allaway D, Harris P, Liddell S, Mobasheri A. Carprofen inhibits the release of matrix metalloproteinases 1, 3, and 13 in the secretome of an explant model of articular cartilage stimulated with interleukin 1β. Arthritis Res. Ther. 2013;15:R223. doi: 10.1186/ar4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lourido LL, Calamia V, Mateos JJ, Fernandez-Puente P, Fernandez-Tajes J, Blanco FJ, Ruiz-Romero C. Quantitative proteomic profiling of human articular cartilage degradation in osteoarthritis. J. Proteome Res. 2014;13:6096–106. doi: 10.1021/pr501024p. [DOI] [PubMed] [Google Scholar]
- [18].Danfelter M, Önnerfjord P, Heinegård D. Fragmentation of proteins in cartilage treated with interleukin-1: specific cleavage of type IX collagen by matrix metalloproteinase 13 releases the NC4 domain. J. Biol. Chem. 2007;282:36933–41. doi: 10.1074/jbc.M702491200. [DOI] [PubMed] [Google Scholar]
- [19].Wilson R, Golub SB, Rowley L, Angelucci C, Karpievitch YV, Bateman JF, Fosang AJ. Novel Elements of the Chondrocyte Stress Response Identified Using an in Vitro Model of Mouse Cartilage Degradation. J. Proteome Res. 2016;15:1033–1050. doi: 10.1021/acs.jproteome.5b01115. [DOI] [PubMed] [Google Scholar]
- [20].Lourido L, Calamia V, Fernandez-Puente P, Mateos J, Oreiro N, Blanco FJ, Ruiz-Romero C. Secretome analysis of human articular chondrocytes unravels catabolic effects of nicotine on the joint. Proteomics Clin Appl. 2015:671–680. doi: 10.1002/prca.201400186. [DOI] [PubMed] [Google Scholar]
- [21].Stevens AL, Wishnok JS, White FM, Grodzinsky AJ, Tannenbaum SR. Mechanical injury and cytokines cause loss of cartilage integrity and upregulate proteins associated with catabolism, immunity, inflammation, and repair. Mol. Cell. Proteomics. 2009;8:1475–89. doi: 10.1074/mcp.M800181-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Stenberg J, Rüetschi U, Skiöldebrand E, Kärrholm J, Lindahl A. Quantitative proteomics reveals regulatory differences in the chondrocyte secretome from human medial and lateral femoral condyles in osteoarthritic patients. Proteome Sci. 2013;11:43. doi: 10.1186/1477-5956-11-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Dakin SG, Smith RKW, Heinegård D, Önnerfjord P, Khabut A, Dudhia J. Proteomic analysis of tendon extracellular matrix reveals disease stage-specific fragmentation and differential cleavage of COMP (cartilage oligomeric matrix protein) J. Biol. Chem. 2014;289:4919–27. doi: 10.1074/jbc.M113.511972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Maroudas A. Physicochemical properties of articular cartilage. In: Freedman M, editor. Adult Articul. Cartil. Pitman Medical; Tunbridge Wells: 1979. [Google Scholar]
- [25].Buschmann MD, Grodzinsky AJ. A Molecular Model of Proteoglycan-Associated Electrostatic Forces in Cartilage Mechanics. J. Biomech. Eng. 1995;117:179–192. doi: 10.1115/1.2796000. [DOI] [PubMed] [Google Scholar]
- [26].Tavakoli Nia H, Han L, Soltani Bozchalooi I, Roughley P, Youcef-Toumi K, Grodzinsky AJ, Ortiz C. Aggrecan Nanoscale Solid–Fluid Interactions Are a Primary Determinant of Cartilage Dynamic Mechanical Properties. ACS Nano. 2015;9:2614–2625. doi: 10.1021/nn5062707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lotz MK, Kraus V. Posttraumatic osteoarthritis: pathogenesis and pharmacological treatment options. Arthritis Res. Ther. 2010;12:211. doi: 10.1186/ar3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Patwari P, Gao G, Lee JH, Grodzinsky AJ, Sandy JD. Analysis of ADAMTS4 and MT4-MMP indicates that both are involved in aggrecanolysis in interleukin-1-treated bovine cartilage. Osteoarthritis Cartilage. 2005;13:269–77. doi: 10.1016/j.joca.2004.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Nelson F, Dahlberg L, Laverty S, Reiner A, Pidoux I, Ionescu M, Fraser GL, Brooks E, Tanzer M, Rosenberg LC, Dieppe P, Robin Poole A. Evidence for altered synthesis of type II collagen in patients with osteoarthritis. J. Clin. Invest. 1998;102:2115–2125. doi: 10.1172/JCI4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Lippiello L, Hall D, Mankin HJ. Collagen synthesis in normal and osteoarthritic human cartilage. J. Clin. Invest. 1977;59:593–600. doi: 10.1172/JCI108676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Hermansson M, Sawaji Y, Bolton M, Alexander S, Wallace A, Begum S, Wait R, Saklatvala J. Proteomic Analysis of Articular Cartilage Shows Increased Type II Collagen Synthesis in Osteoarthritis and Expression of Inhibin βA (Activin A), a Regulatory Molecule for Chondrocytes. J. Biol. Chem. 2004;279:43514–43521. doi: 10.1074/jbc.M407041200. [DOI] [PubMed] [Google Scholar]
- [32].Aigner T, Zien A, Gehrsitz A, Gebhard PM, McKenna L. Anabolic and catabolic gene expression pattern analysis in normal versus osteoarthritic cartilage using complementary DNA-array technology. Arthritis Rheum. 2001;44:2777–89. doi: 10.1002/1529-0131(200112)44:12<2777::aid-art465>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- [33].Aigner T, Zhu Y, Chansky HH, Matsen FA, Maloney WJ, Sandell LJ. Reexpression of type IIA procollagen by adult articular chondrocytes in osteoarthritic cartilage. Arthritis Rheum. 1999;42:1443–50. doi: 10.1002/1529-0131(199907)42:7<1443::AID-ANR18>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- [34].Ijiri K, Zerbini LF, Peng H, Otu HH, Tsuchimochi K, Otero M, Dragomir C, Walsh N, Bierbaum BE, Mattingly D, van Flandern G, Komiya S, Aigner T, Libermann TA, Goldring MB. Differential expression of GADD45β in normal and osteoarthritic cartilage: Potential role in homeostasis of articular chondrocytes. Arthritis Rheum. 2008;58:2075–2087. doi: 10.1002/art.23504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Goldring MB, Birkhead J, Sandell LJ, Kimura T, Krane SM. Interleukin 1 suppresses expression of cartilage-specific types II and IX collagens and increases types I and III collagens in human chondrocytes. J. Clin. Invest. 1988;82:2026–2037. doi: 10.1172/JCI113823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Wu J-J, Weis MA, Kim LS, Eyre DR. Type III collagen, a fibril network modifier in articular cartilage. J. Biol. Chem. 2010;285:18537–44. doi: 10.1074/jbc.M110.112904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Howes J-M, Bihan D, Slatter DA, Hamaia SW, Packman LC, Knauper V, Visse R, Farndale RW. The recognition of collagen and triple-helical toolkit peptides by MMP-13: sequence specificity for binding and cleavage. J. Biol. Chem. 2014;289:24091–101. doi: 10.1074/jbc.M114.583443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Muehleman C, Bareither D, Huch K, Cole AA, Kuettner KE. Prevalence of degenerative morphological changes in the joints of the lower extremity. Osteoarthritis Cartilage. 1997;5:23–37. doi: 10.1016/s1063-4584(97)80029-5. [DOI] [PubMed] [Google Scholar]
- [39].Li Y, Frank EH, Wang Y, Chubinskaya S, Huang H-H, Grodzinsky AJ. Moderate dynamic compression inhibits pro-catabolic response of cartilage to mechanical injury, tumor necrosis factor-α and interleukin-6, but accentuates degradation above a strain threshold. Osteoarthr. Cartil. 2013;21:1933–41. doi: 10.1016/j.joca.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Frank EH, Jin M, Loening AM, Levenston ME, Grodzinsky AJ. A versatile shear and compression apparatus for mechanical stimulation of tissue culture explants. J. Biomech. 2000;33:1523–7. doi: 10.1016/s0021-9290(00)00100-7. [DOI] [PubMed] [Google Scholar]
- [41].Patwari P, Cook MN, DiMicco MA, Blake SM, James IE, Kumar S, Cole AA, Lark MW, Grodzinsky AJ. Proteoglycan degradation after injurious compression of bovine and human articular cartilage in vitro: interaction with exogenous cytokines. Arthritis Rheum. 2003;48:1292–301. doi: 10.1002/art.10892. [DOI] [PubMed] [Google Scholar]
- [42].Lu YCS, Evans CH, Grodzinsky AJ. Effects of short-term glucocorticoid treatment on changes in cartilage matrix degradation and chondrocyte gene expression induced by mechanical injury and inflammatory cytokines. Arthritis Res. Ther. 2011;13:R142. doi: 10.1186/ar3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Müller C, Khabut A, Dudhia J, Reinholt FP, Aspberg A, Heinegård D, Önnerfjord P. Quantitative proteomics at different depths in human articular cartilage reveals unique patterns of protein distribution. Matrix Biol. 2014;40:34–45. doi: 10.1016/j.matbio.2014.08.013. [DOI] [PubMed] [Google Scholar]
- [44].Teleman J, Karlsson C, Waldemarson S, Hansson K, James P, Malmström J, Levander F. Automated selected reaction monitoring software for accurate label-free protein quantification. J. Proteome Res. 2012;11:3766–73. doi: 10.1021/pr300256x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Silva JC, Gorenstein MV, Li G-Z, Vissers JPC, Geromanos SJ. Absolute Quantification of Proteins by LCMSE: A Virtue of Parallel ms Acquisition. Mol. Cell. Proteomics. 2005;5:144–156. doi: 10.1074/mcp.M500230-MCP200. [DOI] [PubMed] [Google Scholar]
- [46].MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–8. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.