Abstract
Type-III secreted effectors (T3Es) play critical roles during bacterial pathogenesis in plants. Plant recognition of certain T3Es can trigger defence, often accompanied by macroscopic cell death, termed the hypersensitive response (HR). Economically important species of kiwifruit are susceptible to Pseudomonas syringae pv. actinidiae (Psa), the causal agent of kiwifruit bacterial canker. Although Psa is non-pathogenic in Arabidopsis thaliana, we observed that a T3E, HopZ5 that is unique to a global outbreak clade of Psa, triggers HR and defence in Arabidopsis accession Ct-1. Ws-2 and Col-0 accessions are unable to produce an HR in response to Pseudomonas-delivered HopZ5. While Ws-2 is susceptible to virulent bacterial strain Pseudomonas syringae pv. tomato DC3000 carrying HopZ5, Col-0 is resistant despite the lack of an HR. We show that HopZ5, like other members of the YopJ superfamily of acetyltransferases that it belongs to, autoacetylates lysine residues. Through comparisons to other family members, we identified an acetyltransferase catalytic activity and demonstrate its requirement for triggering defence in Arabidopsis and Nicotiana species. Collectively, data herein indicate that HopZ5 is a plasma membrane-localized acetyltransferase with autoacetylation activity required for avirulence.
Introduction
Plants resist microbial invasion through innate defence systems including a first layer of inducible immunity that recognizes conserved pathogen-associated molecular patterns (PAMPs). Plant cell surface-bound pattern recognition receptors (PRRs) bind to corresponding PAMPs and initiate PAMP-triggered immunity (PTI)1. Successful bacterial pathogens can suppress PTI through the use of a type III secretion system (T3SS) to inject proteinaceous virulence factors, termed effectors (T3E), directly into plant cells2. In order to suppress PTI, bacterial T3Es possess a wide variety of enzymatic functions that target multiple components of the PTI signalling pathway3. On the other hand, plants carry intracellular nucleotide-binding and leucine-rich repeat resistance proteins (NLR) that can sense the presence of one or more effectors to activate a second layer of defence, effector-triggered immunity (ETI)4, that restricts pathogen growth. A recognized effector that triggers ETI is thus termed an avirulence protein (Avr). ETI is often accompanied by rapid transcriptional reprogramming of defence genes, production of antimicrobial compounds and the hypersensitive response (HR), a localized programmed cell death at the site of infection that serves to restrict pathogen spread4.
NLR proteins consist of a central nucleotide-binding domain, a carboxy-terminal leucine-rich repeat domain and a variable amino-terminal domain, often a toll-interleukin receptor (TIR)-type or coiled-coil (CC)-type, hereafter referred to as TNLs and CNLs, respectively5. NLRs characteristically require common downstream signalling components, including the lipase-like protein ENHANCED DISEASE SUSCEPTIBILITY 1 (EDS1) or the integrin-like protein NON-RACE SPECIFIC DISEASE RESISTANCE 1 (NDR1) involved in cell wall-plasma membrane adhesion for some TNL and CNL classes, respectively6–8. In addition to these two branches of NLR signalling, a shared component involves the SUPPRESSOR OF THE G2 ALLELE OF SKP1 (SGT1) gene. SGT1 is a highly conserved protein in plants that participates in a chaperone complex with HEAT SHOCK PROTEIN 90 (HSP90) and REQUIRED FOR MLA12 RESISTANCE 1 (RAR1) to maintain NLR homeostasis and was shown to be integral to defence mediated by various NLRs9, 10. NLRs have also shown a requirement for the phytohormone salicylic acid (SA) for the proper induction of systemic acquired resistance (SAR) following NLR activation, a system requiring the SA biosynthesis gene SID2 11, 12.
Pseudomonas syringae pv. actinidiae (Psa) causes a bacterial canker disease in kiwifruit and has been a global threat to the kiwifruit industry since the 1980s and consists of several distinct clades known as biovars. The source of a recent global outbreak of bacterial canker disease in kiwifruit orchards in major exporters like Italy (2008), Chile (2010) and New Zealand (2010) has been identified as biovar 3 strains of Psa which includes the NZV-13 (ICMP 18884, CP011972–3) strain from New Zealand13. The Psa NZV-13 genome was recently sequenced in order to understand its origin and virulence mechanisms14, 15. Similar to other P. syringae strains, Psa NZV-13 carries the genes encoding the T3SS and a repertoire of 34 predicted T3Es. Putative biochemical activities of Psa NZV-13 T3Es include ribosyltransferase, glutamidase, phospholyase, cysteine protease, and acetyltransferase. Interestingly, HopZ5 (AKT29515.1), a putative acetyltransferase, and HopH1 (AKT29516.1) are uniquely present among the global outbreak strains of biovar 3 including Psa NZV-1314.
The YopJ (Yersinia outer protein J) effector protein from the human gastroenteritis pathogen Yersinia pseudotuberculosis was shown to have an acetyltransferase activity. YopJ acetylates multiple MITOGEN ACTIVATED PROTEIN KINASE KINASE (MAPKK) proteins, key components of host defense, that results in inhibition of their phosphorylation and suppression of NFκB mammalian defense signalling pathways16, 17. Interestingly, plant pathogenic bacteria carry a number of YopJ-class T3Es including Ralstonia PopP2 and the Pseudomonas HopZ family of effectors18, 19. HopZ1a from P. syringae pv. syringae A2 autoacetylates a specific threonine residue (T346) with the involvement of two serine residues (S349 and S351) following in planta activation of HopZ1a by the plant cofactor IP620, 21. In addition, HopZ1a acetylates jasmonate ZIM-domain (JAZ) repressors of jasmonic acid (JA) signalling leading to their proteosomal degradation, thereby activating the JA pathway and antagonizing salicylic acid-mediated disease resistance22. Additionally, HopZ1a targets tubulin to block defence at the cell wall while simultaneously interfering with the host’s secretory pathway that has been linked to systemic immunity23–25. However, in the presence of a functional CNL gene, ZAR1, HopZ1a activates ETI in Arabidopsis thaliana (hereafter referred to as Arabidopsis)26. HopZ1a-triggered immunity requires Arabidopsis pseudokinase ZED1 that is monitored by ZAR127. The downstream genetic components required for HopZ1a-triggered immunity remains elusive26, 27. The Ralstonia solanacearum GMI1000 effector PopP2 is a nucleus-localized acetyltransferase, that autoacetylates a lysine residue (K383) required for its trans-acetylation activity18 and targets WRKY transcription factors involved in defence activation28, 29. The recognition of PopP2 in Arabidopsis is conferred by the TNL protein pair RRS1 and RPS4. PopP2 acetylates the WRKY DNA-binding domain of RRS1 resulting in destabilized RRS1 binding to its target DNA28–30. It is hypothesized that PopP2 targets other WRKY transcription factors to enhance bacterial virulence yet a detailed mechanism is unknown. Based on these findings, despite their conserved enzymatic activity, different members of the YopJ acetyltransferase family may have been specialized for targeting distinct plant defence components.
In this study we sought to examine the genetic basis of the Psa NZV-13-triggered HR in Arabidopsis. By using T3SS delivery of Psa effectors from P. fluorescens Pf0-1 (T3S), a non-pathogenic strain engineered to carry a functional T3SS31, we found that HopZ5 triggers ETI in Arabidopsis. By analogy with other HopZ members, HopZ5 encodes a putative acetyltransferase and we show that this enzymatic activity is required for its avirulence activity. In addition, HopZ5-induced cell death in Nicotiana benthamiana requires SGT1 but not EDS1 or NDR1. Furthermore, we discovered that HopZ5 triggers ETI that is not associated with HR development in an Arabidopsis accession-specific manner.
Results
HopZ5PsaNZV-13 triggers accession-specific immunity in Arabidopsis
Several P. syringae type-III secreted effectors (T3E) were shown to trigger NLR-dependent immunity in Arabidopsis26, 32–34. To examine if P. syringae pv. actinidiae (Psa) NZV-13 can also trigger immunity in Arabidopsis, we screened several accessions for Psa NZV-13-triggered hypersensitive response (HR) which is typically associated with effector-triggered immunity (ETI)35. Psa NZV-13 triggered a strong HR in the Catania-1 (Ct-1) accession but not in Col-0 and Wassilewskija-2 (Ws-2) (Fig. 1a). In order to identify the Psa NZV-13 effector that triggers HR in Ct-1, we cloned all 34 predicted T3Es (based on tBLASTx annotation followed by manual confirmation as done previously14) from Psa NZV-13 into a broad host-range vector (pBBR 1MCS-5) with expression driven by the previously characterized avrRps4 promoter33, 36. Individual T3E delivery by Pseudomonas fluorescens (Pf) Pf0-1(T3S) (hereafter, Pf0-1(T3S))31 identified HopZ5 as a cause of the HR in Ct-1 (Fig. 1a). Col-0 and Ws-2 did not develop HR in response to Pf0-1(T3S)-delivered HopZ5 or empty vector (EV). However, as expected, Pf0-1(T3S)-delivered AvrRpm1 (from P. syringae pv. maculicola M6) triggered HR in all three Arabidopsis accessions tested, indicating that our experimental conditions were optimal. To quantify HopZ5-triggered HR, we measured the leakage of ions caused during HR development in the infected leaves. As expected, Pf0-1(T3S)-delivered AvrRpm1 induced significant increase in electrolyte leakage levels in all three Arabidopsis accessions tested whereas EV did not cause a notable change. Consistent with HR data, Pf0-1(T3S)-delivered HopZ5-triggered significantly increased electrolyte leakage level as compared to EV in Ct-1 but not in Ws-2 nor Col-0 (Fig. 1b).
Figure 1.
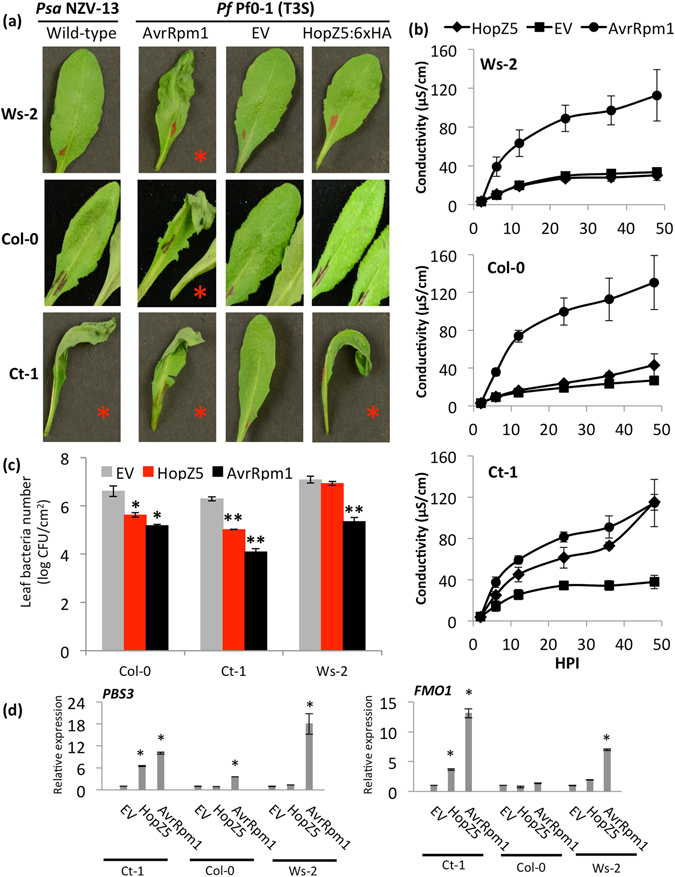
Pseudomonas syringae pv. actinidiae type III effector HopZ5 triggers accession-specific immunity in Arabidopsis. (a) Arabidopsis accession-dependent development of a hypersensitive response to P. syringae pv. actinidiae and its type-III secreted effector HopZ5. Leaves of Arabidopsis plants were infiltrated with P. syringae pv. actinidiae NZV-13 or P. fluorescens Pf0-1 (T3S) carrying an empty vector pBBR1MCS-5 (EV), hopZ5 with a C-terminal 6xHA tag under the control of the avrRps4 promoter in pBBR1MCS-5 or untagged avrRpm1 under its native promoter in pVSP61. Bacterial suspensions (1 × 108 CFU/mL) were blunt-syringe infiltrated into the leaves and photographs were taken 20 hours after infiltration (1 dpi). Development of the hypersensitive response is indicated by a red asterisk. (b) Electrolyte leakage from Arabidopsis leaf discs after infiltration with P. fluorescens Pf0-1 (T3S) carrying the indicated constructs, as in (a). Bacteria (1 × 108 CFU/mL) were blunt syringe-infiltrated into the leaves. The error bars indicate the standard error from four technical replicates. The experiment was conducted three times with similar results. (c) P. syringae pv. tomato DC3000 (Pto DC3000) growth in Arabidopsis. Pto DC3000 carrying EV, hopZ5-HA or avrRpm1 was blunt syringe-infiltrated at 5 × 105 CFU/mL into Arabidopsis leaves, and bacterial growth was determined 4 days post-infection (4 dpi). Error bars represent standard error from six technical replicates. Asterisks indicate results of Student’s t-test between selected sample and EV for that accession; *(P < 0.05), **(P < 0.01). The experiment was conducted six times with similar results. (d) Defence gene expression in Arabidopsis in response to EV, HopZ5-HA or AvrRpm1. P. fluorescens Pf0-1 (T3S) (1 × 108 CFU/mL) carrying EV, hopZ5-HA or avrRpm1 was blunt syringe-infiltrated into leaves, samples taken at 8 hours post infiltration and defence gene expression determined from extracted RNA by quantitative polymerase chain reaction. Expression for each defence gene is relative to internal EF1α expression and defence gene expression for EV samples indicated. Error bars indicate standard error from three technical replicates. Asterisks indicate results of Student’s t-test between selected sample and EV for that accession; *(P < 0.01). The experiment was conducted three times with similar results.
Next, we assessed the effect of HopZ5 on in planta growth of virulent P. syringae pv. tomato (Pto) DC3000. Pto DC3000 strains carrying hopZ5 or avrRpm1 grew significantly less (a reduction of one log) than EV in Ct-1 (Fig. 1c). Interestingly, although neither Col-0 nor Ws-2 accessions show HR in response to Pf0-1(T3S)-delivered HopZ5, we consistently observed a small but significant growth restriction in Col-0 (a reduction of almost one log) but not in Ws-2. To further examine the HR-deficient immunity in Col-0, we assessed HopZ5-triggered defence gene expression by Pf0-1(T3S)-delivery. Surprisingly, Pf0-1(T3S)-delivered HopZ5 upregulated transcript accumulation of two previously characterized defence genes PBS3 and FMO1 in Ct-1 but not in Col-0 and Ws-2 (Fig. 1d). Since HopZ5 restricts in planta Pto DC3000 growth in both Ct-1 and Col-0, we tested whether Pto DC3000 delivery of HopZ5 induces defence marker gene expression. As expected, Col-0 showed upregulated PBS3 and FMO1 transcript accumulation in response to Pto DC3000-delivered HopZ5 and AvrRpm1 compared to EV (Supplementary Fig. S1). These data suggest that HopZ5 triggers defence response in Ct-1 and Col-0.
HopZ5 triggers an NbSGT1-dependent hypersensitive response-like cell death in Nicotiana benthamiana
Agrobacterium-mediated transient expression (hereafter, agroinfiltration) of pathogen effectors in Nicotiana spp. often induces HR-like cell death (HCD)37, 38. In order to test if HopZ5 triggers HCD in Nicotiana spp., we transiently expressed C-terminally YFP-tagged HopZ5 under the control of constitutive cauliflower mosaic virus 35 S promoter (35 S promoter) in Nicotiana benthamiana via agroinfiltration. Interestingly, HopZ5 triggered a robust HCD within 2 dpi (Fig. 2a). We used a well-established virus-induced gene silencing (VIGS) system to examine the genetic components required for HopZ5-triggered HCD in N. benthamiana. We generated VIGS constructs for NbEDS1, NbNDR1 and NbSGT1 as they have been previously identified as critical components in rendering HCD39, 40. While silencing enabled significant reduction in transcriptional expression of NbEDS1, NbNDR1 and NbSGT1 compared to the empty vector silencing construct, only NbSGT1-silenced plants lost the ability to mount HCD in response to HopZ5 expression (Fig. 2a,b). This result suggests that the HopZ5-triggered activation of defence signalling, at least in N. benthamiana, requires SGT1 but not EDS1 or NDR1.
Figure 2.
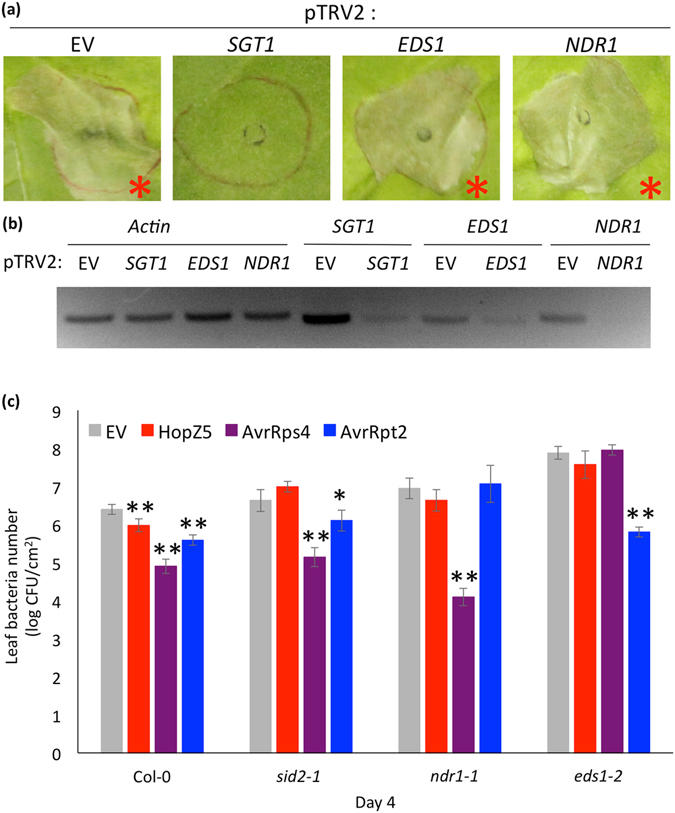
HopZ5 recognition occurs in Nicotiana benthamiana and requires SGT1 but not NDR1 or EDS1. (a) SGT1-silenced N. benthamiana loses HopZ5-triggered hypersensitive response. Two-week-old N. benthamiana seedlings were infiltrated with Agrobacterium tumefaciens AGL1 carrying pTRV2 targeting SGT1, EDS1 or NDR1 for silencing, or the empty pTRV2 vector alone. 4–5 weeks post VIGS treatment, YFP-tagged variants of HopZ5 was transiently expressed and photographs taken at 3 days post infiltration. Red asterisks indicate development of cell death symptoms. The experiment was conducted three times with similar results. (b) Silencing of target genes was confirmed by RNA extraction from multiple leaves followed by semi-quantitative polymerase chain reaction. Actin was used as control for RNA. Labels above indicate the gene tested for expression by semi-qPCR while labels below indicate the silencing construct used. (c) Pto DC3000 carrying EV, hopZ5-HA, avRps4 or avrRpt2 was blunt syringe-infiltrated at 5 × 105 CFU/mL into leaves of Arabidopsis wildtype Col-0, sid2-1, ndr1-1, or eds1-2 mutant lines. Bacterial growth was determined 4 days post-infection (4 dpi). Error bars represent standard error from six technical replicates. Asterisks indicate results of Student’s t-test between selected sample and EV for that accession; *(P < 0.05), **(P < 0.01). The experiment was conducted three times with similar results.
To test if this lack of requirement for NDR1 and EDS1 is reproduced in Arabidopsis, we tested if Col-0 mutants, eds1-2, ndr1-1 and sid2-1, were able to restrict growth of Pto DC3000 delivering HopZ5 or two different avirulence effectors, AvrRps4 or AvrRpt26, 11, 12. Surprisingly, in contrast to wild-type Col-0, all three mutants (sid2-1, ndr1-1 and eds1-2) were affected in their ability to restrict growth of Pto DC3000 delivering HopZ5 (Fig. 2c). As expected, resistance to Pto DC3000 expressing AvrRpt2 or AvrRps4 was abolished in ndr1-1 or eds1-2, respectively6.
Plasma membrane localization and autoacetylation are required for HopZ5 avirulence activity
In silico analysis of HopZ5 suggests that it belongs to the YopJ superfamily of acetyltransferases and cysteine proteases (Supplementary Fig. S2). Several members of this family including Pseudomonas syringae HopZ1, Xanthomonas campestris AvrBsT and R. solanacearum PopP2 have been characterized previously18, 26, 41. HopZ5, similar to other YopJ family members, possesses several conserved key residues including a catalytic triad consisting of histidine (H150), glutamate (E169) and cysteine (C218) and a lysine residue putatively required for auto-/trans-acetylation (K278) (Fig. 3a; Supplementary Fig. S3). Additionally, HopZ5 possesses a glycine (G2) that is conserved in most members of the HopZ family of T3Es and required for myristoylation-dependent plasma membrane localization42. The phylogenetic relationships and predicted conserved residues suggest that HopZ5 may be a plasma membrane-localized acetyltransferase.
Figure 3.
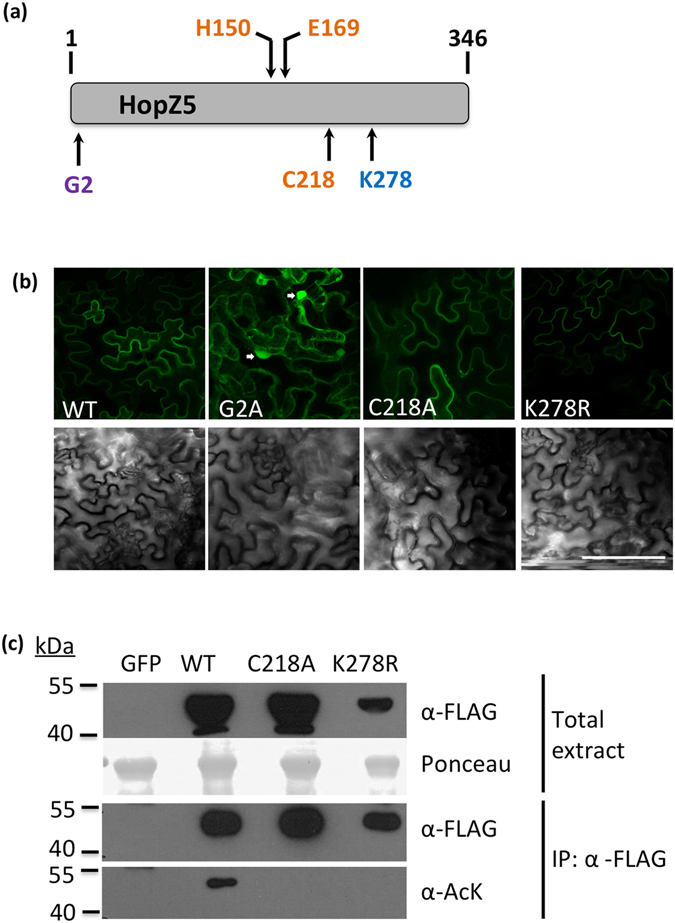
Plasma membrane localization and autoacetylation are affected in HopZ5 variants. (a) Key residues for HopZ5 function predicted through conservation between YopJ family members. Line model for HopZ5 peptide with putative myristoylation residue (glycine, G2), acetyltransferase catalytic core residues (histidine, H150; glutamate, E169; cysteine, C218) and autoacetylation-required site (lysine, K278) is shown. Mutants generated in this study include a glycine to alanine mutation (G2A), cysteine to alanine (C218A) and a lysine to arginine (K278R). (b) In planta subcellular localization of HopZ5 variants was determined in SGT1-silenced N. benthamiana. Leaves were infiltrated with Agrobacterium tumefaciens AGL1 carrying YFP-tagged hopZ5 (WT), hopZ5 (G2A), hopZ5 (C218A) or hopZ5 (K278R) under a constitutive CaMV 35 S promoter. Leaf discs were taken at 2 days post infiltration and visualized under confocal laser-scanning microscopy for localization of the YFP tag. Top and bottom panels are confocal images and corresponding bright-field images, respectively. White arrows in G2A sample indicate plant cell nuclei. Scale bar represents 50 μm. (c) Detection of acetylated wild-type HopZ5 (WT), but not HopZ5-C218A and HopZ5-K278R from transient expression in N. benthamiana SGT1-silenced plants. Protein was extracted from leaf samples 2 days post infiltration. Protein presence in total extract was tested by α-FLAG antibody and acetylation status tested by immunoblot with an α-AcK antibody after α- FLAG immunoprecipitation. Ponceau Red staining of total protein extract shows sample loading.
Similar to the work carried out for PopP218 and HopZ1a42, 43 for which critical amino acid residues were mutagenized to analyse biochemical functions, we utilized site-directed mutagenesis to generate HopZ5 variants carrying a C218A (cysteine to alanine) or K278R (lysine to arginine) mutation that would eliminate the predicted acetyltransferase activity or autoacetylation, respectively. We also generated a G2A (glycine to alanine) HopZ5 variant to alter the predicted subcellular localization. To confirm the biochemical requirement for these key residues, we then used NbSGT1-silenced N. benthamiana since HopZ5-triggered HCD was reduced in these plants, promoting HopZ5 protein accumulation and detection. Agroinfiltration of C-terminally YFP-tagged HopZ5 (HopZ5-YFP) variants allowed detection of subcellular localization of HopZ5 by confocal microscopy. We found that while wild-type HopZ5 and both C218A and K278R variants localized to the cell periphery, the G2A variant localized in the cytoplasm and nuclei of Agrobacteria-infected cells (Fig. 3b, Supplementary Fig. S4). The presence of YFP signal in the cytoplasmic strands in G2A infiltrated plants as well as in nuclei (white arrows in Fig. 3b, and red arrows colocalized with DAPI stain in Supplementary Fig. S4) suggests that wild-type HopZ5 normally localizes at the plant plasma membrane. Several R. solanacearum effector PopP2 was shown to autoacetylate lysine residues18. The C218A and K278R variants were thus further examined for their ability to acetylate internal lysine residues, as an indicator of autoacetylation ability. Transiently expressed FLAG-tagged HopZ5 variants were immunoprecipitated and probed with an antibody that detects acetylated lysine residues. We found that wild-type HopZ5 but not the C218A variant was lysine-acetylated, suggesting that C218A has lost its acetyltransferase activity and that it is autoacetylated (Fig. 3c). In addition, although catalytic residues (H150, E169 and C218) are present, lysine-acetylation of the K278R variant was not detectable, indicating either that it may be the sole acetylated lysine residue and is autoacetylated in HopZ5, or that it is required for autoacetylation in general.
To determine if plasma membrane localization and autoacetylation of HopZ5 are required for avirulence, we examined the defence responses triggered by HopZ5 variants in the resistant Arabidopsis accession Ct-1. Pf0-1(T3S)-delivered HopZ5 C218A or K278R variants did not trigger HR in Ct-1 leaves (Fig. 4a). Interestingly, the G2A variant only showed a partial loss of HR as observed through a qualitative assessment of leaf appearance (Fig. 4a). Electrolyte leakage quantitation showed a significant but not complete attenuation of ion leakage for the G2A variant while both C218A and K278R variants induced a much reduced level, similar to EV (Fig. 4b). Next, these HopZ5 variants were assessed for their ability to restrict growth of a virulent bacterial strain Pto DC3000 in Ct-1. Although the G2A variant induced significantly reduced ion leakage as compared to wild-type HopZ5 (Fig. 4b), in planta growth of Pto DC3000 carrying G2A variant was comparable to Pto DC3000 carrying wild-type hopZ5 (Fig. 4c). In addition, both C218A and K278A variants did not significantly restrict Pto DC3000 growth compared to EV. To further elucidate the early defence responses triggered by HopZ5 variants, the expression of several defence marker genes was determined. Pf0-1(T3S)-delivered wild-type HopZ5 but not G2A, C218A or K278A variants induced significant accumulation of defence marker gene expression as compared to EV (Fig. 4d). All three variants of HopZ5 were unaffected in protein expression and stability in Pto DC3000, indicating that the differences seen between wild-type HopZ5 and the variants in their ability to trigger immunity was not a result of inconsistent protein expression due to the mutations (Supplementary Fig. S5). Taken together, these data suggest that all three predicted functional residues are at least partially required for a full induction of ETI.
Figure 4.
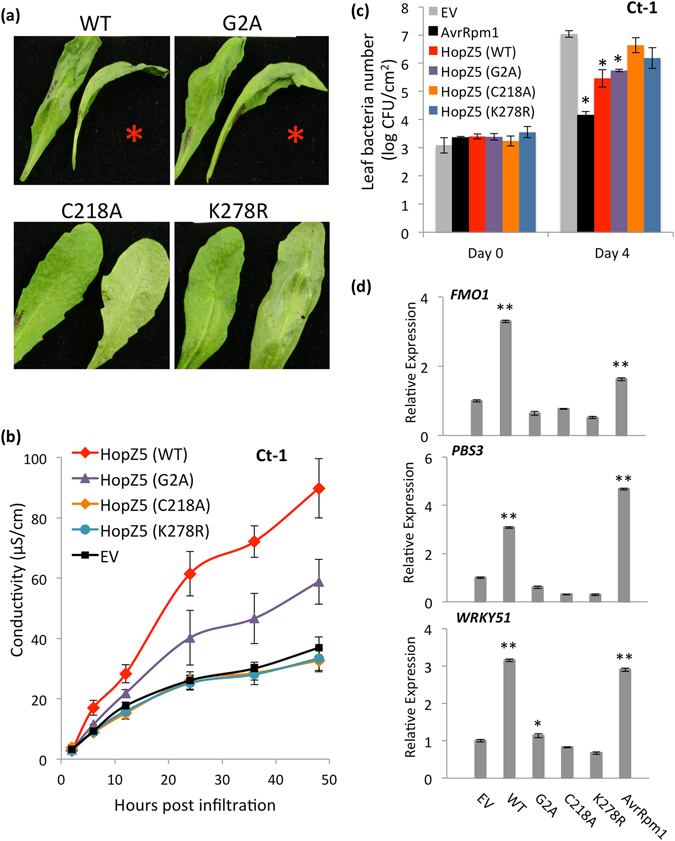
Putative acetyltransferase activity and myristoylation of HopZ5 are required for avirulence. (a) Development of the hypersensitive response in the Arabidopsis accession Ct-1 against HopZ5 variants. P. fluorescens Pf0-1 (T3S) (1 × 108 CFU/mL) carrying hopZ5-HA (WT), hopZ5(G2A)-HA, hopZ5(C218A)-HA or hopZ5(K278R)-HA was blunt syringe-infiltrated into leaves and photographs taken 20 hours after infiltration. The red asterisk indicates development of the hypersensitive response. (b) Electrolyte leakage from Arabidopsis accession Ct-1 leaf discs after infiltration with P. fluorescens Pf0-1 (T3S) expressing HopZ5 variants, as in (a). Bacteria (1 × 108 CFU/mL) were blunt syringe-infiltrated into the leaves. The error bars indicate the standard error from four technical replicates. The experiment was conducted five times with similar results. (c) Pto DC3000 growth in Arabidopsis accession Ct-1. Pto DC3000 carrying hopZ5 variants, as in (a), was blunt syringe-infiltrated at 5 × 105 CFU/mL into Arabidopsis leaves, and bacterial growth was determined 4 days post-infection (4 dpi). Error bars represent standard error from six technical replicates. Asterisks indicate results of Student’s t-test between selected sample and EV for that accession; *(P < 0.05), **(P < 0.01). The experiment was conducted nine times with similar results. (d) Defence gene expression in Arabidopsis Ct-1 in response to HopZ5 variants, as in (a). P. fluorescens Pf0-1 (T3S) (1 × 108 CFU/mL) carrying the indicated construct was blunt syringe-infiltrated into leaves and defence gene expression determined from extracted RNA by quantitative polymerase chain reaction. Expression for each defence gene is relative to internal EF1α expression and defence gene expression for EV samples indicated. Error bars indicate standard error from three technical replicates. Asterisks indicate results of Student’s t-test between selected sample and EV for that accession; *(P < 0.05), **(P < 0.01). The experiment was conducted five times with similar results.
In order to test if HopZ5-triggered defence responses in other plant species require similar properties of HopZ5, HopZ5-YFP variants were transiently expressed in N. benthamiana and N. tabacum. Agroinfiltration of wild-type HopZ5 but not GFP control induced rapid development of HCD in both N. benthamiana (Fig. 5a) and N. tabacum (Supplementary Fig. S6). Consistent with Pseudomonas-delivered HopZ5 results in Arabidopsis, the C218A variant did not induce any visible cell death. Interestingly, G2A and K278R variants induced delayed cell death development, indicating that avirulence activity of these variants was significantly reduced. To quantify the effect of the mutations on HopZ5-triggered HCD, we carried out an electrolyte leakage assay after agroinfiltration of G2A, C218A and K278R variants in N. benthamiana (Fig. 5b, Supplementary Fig. S7). Electrolyte leakage measurement at 2 days post-inoculation (dpi), when HCD symptoms started to appear prior to leaf collapse, demonstrated a statistically significant reduction for both the G2A and K278R variants and a complete loss of HCD-associated leakage for the C218A variant. These results demonstrate that the partial loss of avirulence function for HopZ5 G2A and K278A variants applies in more than one plant host (summarized in Supplementary Table 1).
Figure 5.
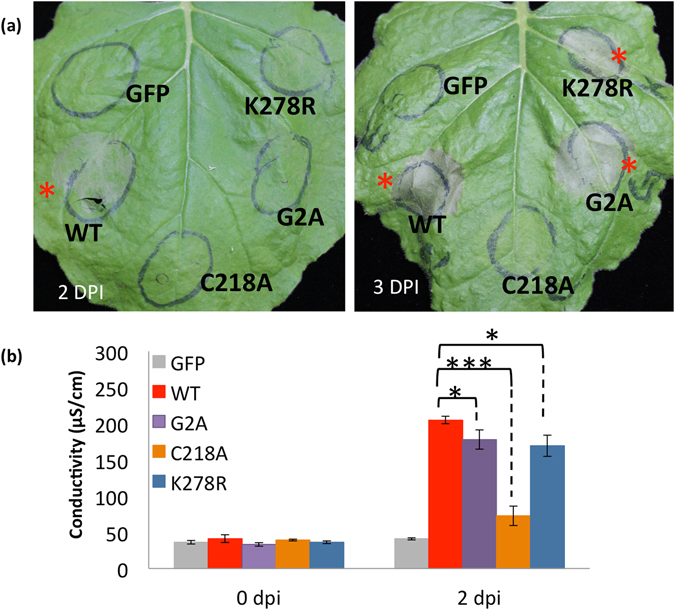
HopZ5 recognition in Nicotiana benthamiana requires acetyltransferase activity. (a) HopZ5 (WT) or mutated myristoylation (G2A), catalytic core (C218A) and autoacetylation-required (K278R) variants tagged with 3xFLAG were transiently expressed under the CaMV 35 S promoter in N. benthamiana Photographs were taken at 2 or 3 days post infiltration (dpi). Red asterisks indicate development of hypersensitive response-like cell death symptoms. This experiment was conducted six times with identical results. (b) Electrolyte leakage from N. benthamiana leaf discs triggered by Agrobacterium-mediated transient expression of HopZ5 or variants, as in (a), was determined at 0 and 2 days post infiltration (dpi). Infiltrations were carried out by blunt syringe at OD600 = 0.4 for all samples. Error bars represent standard error from four technical replicates. Asterisks indicate results of Student’s t-test between selected samples; *(P < 0.05), ***(P < 0.001). This experiment was conducted three times with identical results.
HopZ5 triggers defence that is not associated with HR in Col-0
To clarify the HopZ5-triggered bacterial immunity without HR in Col-0 plants, we generated stable transgenic Col-0 lines expressing HopZ5-YFP variants under the control of the 35 S promoter. Using multiple independent lines for wild-type HopZ5 (Z-1, Z-4, Z-5), C218A (C-1, C-4), K278R (K-2, K-6) and G2A (G-2, G-3), we first tested for expression of the transgene in T2 plants. Notably, we were only able to identify low-expression lines for wild-type HopZ5 (Z) and G2A (G) variants while C218A (C) and K278R (K) variant lines had significantly higher transgene expression levels (Fig. 6a). Additionally, T2 plants for the Z lines alone showed some stunted growth and early flowering phenotypes compared to wild-type Col-0 and other mutant variants (Fig. 6b). Importantly, we found that PR1 expression, a late defence response marker, was significantly upregulated in the Z lines despite the low HopZ5 transgene expression level (Fig. 6c). G plant lines showed PR1 transcript accumulation levels intermediate between Z lines and wild-type Col-0, while in C plant lines, PR1 expression was indistinguishable from Col-0. The K plant lines showed only slightly elevated PR1 transcript accumulation compared to Col-0 (approximately 2-fold), indicating a minor upregulation of defence responses. Next, we assessed whether the transgenic expression of HopZ5 variants could restrict Pto DC3000 growth as a result of upregulated defence response. Consistent with PR1 expression analysis, Z plant lines showed significant reduction in Pto DC3000 growth compared to wild-type Col-0 (Fig. 6d). Unexpectedly, the G plant lines did not show significant reduction of Pto DC3000 growth compared to Col-0 which could be due to the low expression levels of the G2A variant.
Figure 6.
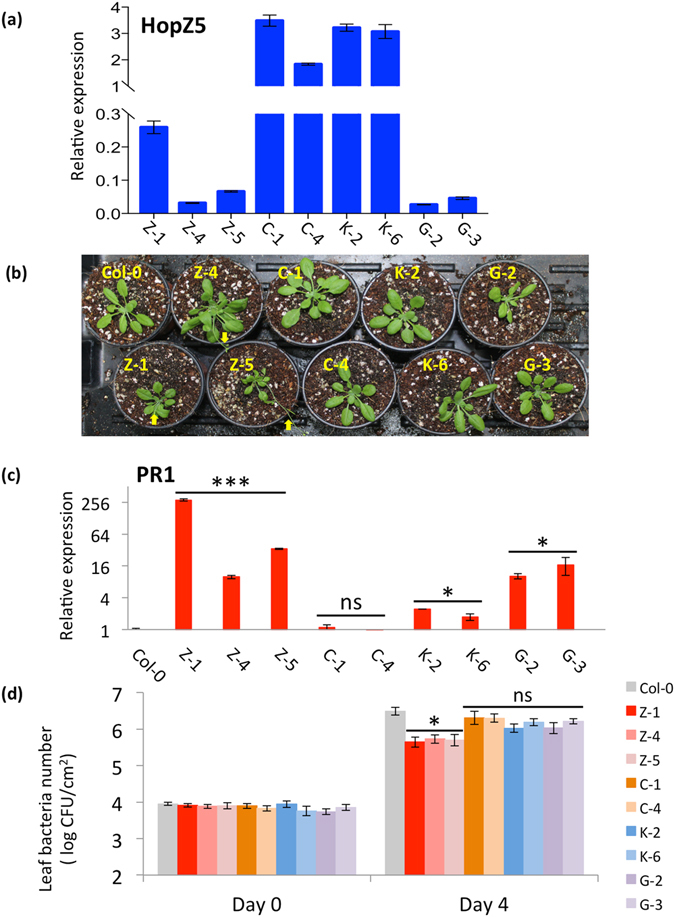
HopZ5 variants are compromised in their ability to trigger defence when overexpressed in Arabidopsis thaliana Col-0. (a) hopZ5-transgene expression was determined in hopZ5 stable expression lines in the Col-0 background. 15–20 T2 seedlings from two or three independent lines per hopZ5 variant (hopZ5-wild-type: Z-1, Z-4, Z-5; C218A: C-1, C-4; K278R: K-2, K-6; G2A: G-2, G-3) were harvested after selection for three weeks on solid media containing kanamycin. RNA was extracted and cDNA synthesized and used for quantitative polymerase chain reaction (qPCR) for the hopZ5-YFP transgene. Expression is relative to internal EF1α expression only. Error bars indicate standard error from three technical replicates. Asterisks indicate results of Student’s t-test between selected sample and Col-0 wild-type also grown on plates but without kanamycin; ***(P < 0.0001). This experiment was conducted three times with similar results. (b) Stable transgenic lines expressing wild-type HopZ5 show altered morphology. 2-week-old T2 plants as in (a) were transferred to soil after selection on kanamycin. Photographs are of representative 5-week-old plants that showed a differential phenotype only. Typically between 1-in-3 (Z-1) to 1-in-4 plants (Z-4 and Z-5) showed a stunted/early flowering phenotype, putatively representative of Mendelian segregation and individuals homozygous for the transgene. Yellow arrows indicate the inflorescence in the early-flowering plants. (c) PR1 marker gene expression was determined in hopZ5 stable expression lines as in (a). Expression is relative to internal EF1α expression and PR1 expression for wildtype. Error bars indicate standard error from three technical replicates. Asterisks indicate results of Student’s t-test between selected sample and Col-0 wild-type also grown on plates but without kanamycin; ***(P < 0.0001). This experiment was conducted three times with similar results. (d) Pto DC3000 growth in hopZ5 stable expression lines in Col-0. Pto DC3000 was blunt syringe-infiltrated at 5 × 105 CFU/mL into leaves of each stable expression line from (a) and bacterial growth was determined 4 days post-infection (4 dpi). Error bars represent standard error from six technical replicates. Asterisks indicate results of Student’s t-test between selected sample and Col-0 wild-type; ns (P > 0.05), *(P < 0.05), **(P < 0.01). The experiment was conducted three times with similar results.
Taken together, the elevated PR1 expression and reduced Pto DC3000 growth relative to wild-type Col-0 in HopZ5 Z lines (Fig. 6c,d), coupled to upregulation of ETI marker genes in response to HopZ5 delivery by Pto DC3000 (Supplementary Fig. S1) and virulent pathogen growth restriction (Fig. 1c) suggest that HopZ5 triggers defence that is not associated with HR in Col-0.
Discussion
We have identified HopZ5 from P. syringae pv. actinidiae NZV-13 as an avirulence effector in Arabidopsis accessions Ct-1 and Col-0. HopZ5 delivery by Pseudomonas T3S resulted in HR and bacterial growth restriction in Ct-1. Interestingly, in Col-0, neither HR nor elevated electrolyte leakage was observed but Pto DC3000 growth was significantly restricted in the presence of HopZ5. Enhanced defence marker gene expression when HopZ5 was delivered from Pseudomonas or transgenically expressed in Col-0 indicates the presence of a functional recognition system in Col-0. The difference between Pf0-1(T3S) and Pto DC3000 delivery of HopZ5 is unknown but likely to be an issue of delivery efficiency since in our Pf0-1(T3S) delivery experiments defence marker gene upregulation in Col-0 was poor even for the positive control AvrRpm1 (Fig. 1d). In addition, acetyltransferase activity and plasma membrane localization are required for HopZ5-triggered immunity in both Arabidopsis and Nicotiana benthamiana.
Separation of HR from pathogen growth restriction was demonstrated in ndr1 mutant plants that showed HR in response to Pto DC3000 delivering AvrRpm1, AvrB or AvrPphB but lost the ability to restrict growth of these strains44. Similarly, Turnip crinkle virus (TCV)-triggered HR is mediated by the CNL gene HRT while the full disease resistance is mediated in conjunction with a second recessive determinant rrt 45. Moreover, TCV resistance has a non-canonical requirement for EDS1 as well as RAR1 and SGT1 but these genes are not required for HR46. The converse, HR-independent resistance, was recently shown in Arabidopsis metacaspase mutants, mc1 and mc2. mc1 and mc2 mutants show reduced and increased cell death responses to Pto DC3000 delivering AvrRpm1, respectively, but have no effect on bacterial growth47. Similarly, immunity triggered by unrelated bacterial type III effectors HopA1 (previously HopPsyA) and AvrRps4 specifically in the Col-0 accession are HR-independent48, 49. The locus that confers suppression of AvrRps4- or HopA1-triggered HR in Col-0, HR REGULATOR IN EDS1 PATHWAY (HED1), was previously mapped to the bottom arm of chromosome 5 in Arabidopsis but the exact genetic determinant of this phenotype remains to be elucidated48. Furthermore, Heidrich and colleagues have elegantly demonstrated that the cell compartment-specific localization of EDS1 is linked to pathogen growth restriction (nucleus alone) or HR (nucleo-cytoplasmic) in response to Pto DC3000 carrying avrRps4 50.
Yet another example of HR uncoupled from defence is the defense, no death1 (dnd1) mutant that shows constitutively elevated defence responses but is unable to mount HR in response to avirulent bacterial strains51. dnd1 carries a null mutation in a cyclic nucleotide-gated ion channel AtCNGC2 predicted to be involved in Ca2+ signalling during defence responses52. Interestingly, a similar gain of constitutive defence phenotype was observed in dnd2/AtCNGC4 and cpr22/AtCNGC11-AtCNGC12 although the latter is able to show HR52–54. The involvement of different AtCNGCs in HR-independent defence suggests that they are negative regulators of defence. Notably, it is possible due to its membrane-localization that HopZ5 targets an ion channel in Col-0 triggering HR-independent immunity.
The closest homolog of HopZ5 is AvrBsT from X. campestris pv. vesicatoria (Xcv; also called X. euvesicatoria), sharing 58% amino acid identity. AvrBsT triggers HR and immunity (as measured by virulent bacterial growth) in Arabidopsis accession Pitzal-0 (Pi-0) but not in Col-0 and Landsberg erecta-0 (Ler-0)55. Notably, Pi-0 was the only accession out of 71 total tested that showed AvrBsT-triggered HR. Suppression of AvrBsT-triggered immunity/HR is conferred by the SUPPRESSOR OF AVRBST ELICITED RESISTANCE1 (SOBER1) gene encoding a carboxylesterase. In contrast to AvrBsT, Col-0 showed disease resistance to virulent bacterial pathogen carrying hopZ5 (Figs 1c and 6). Thus, it is likely that the genetic basis of suppression of HopZ5-triggered HR is different from AvrBsT. However, it remains to be seen if SOBER1 plays a role in HopZ5-mediated immunity in Col-0.
The separation of HR incidence from immunity was originally ascribed to a matter of amplitude, particularly because ETI was perceived as an amplified PTI response56. However, as in the aforementioned examples, the resolution of HR from immunity can be complex. In the case of HopZ5, it is conceivable that a dominant suppressor or negative regulator of HR may exist in Col-0. This suppressor would specifically interfere with HR signalling triggered by HopZ5 or reduce the general immunity level below the threshold that is required for HR development but not immunity. Further investigation into the dynamics behind HR-associated and HR-independent HopZ5-triggered immunity could serve to illustrate a larger role of HR in relation to immunity. Indeed, the observation that while NDR1- and EDS1-silencing did not affect HCD in N. benthamiana, the loss of either of these genes in Arabidopsis affected immunity against HopZ5, is suggestive of a differentiation between HR (or HCD) from growth restriction. Identification of the putative suppressor of HR in Col-0 plants will enable us to elucidate the point where HR and immunity diverge for HopZ5.
The YopJ family members are present in various plant pathogenic bacteria including Pseudomonas, Xanthomonas, Acidovorax and Ralstonia (Supplementary Fig. S2)19. Xcv, the causal agent of tomato bacterial spot disease, carries four different members of the YopJ family, namely avrXv4, avrRxv, avrBsT and xopJ 57–59. AvrXv4 is a cytoplasmic effector with small ubiquitin modifier (SUMO)-protease activity that modifies a large number of proteins in planta and triggers defence responses in wild tomato relative Solanum pennellii and N. benthamiana 60. AvrRxv recruits a plant 14-3-3 protein for its virulence and avirulence activities61. AvrBsT, originally shown to trigger disease resistance in pepper plants62, is a cytoplasm-localized acetyltransferase that binds to and suppresses cytoplasmic SNF1-related kinase (SnRK1) that is required for AvrBs1-triggered HR63. Furthermore, AvrBsT acetylates microtubule-associated proteins to disrupt the microtubule network that is required for plant defence41. XopJ, much like its closest homolog, HopZ4, binds and proteolytically degrades proteasomal subunit RPT6 to suppress plant immunity at the plant cell membrane64. R. solanacearum GMI1000 carries three YopJ family members avrA, popP1 and popP2. AvrA and PopP1 trigger acetyltransferase activity-dependent HR in N. tabacum and N. glutinosa, respectively, that is involved in restriction of host range28, 65. The HopZ family of effectors are found in many Pseudomonas syringae strains. HopZ1a was shown to target both, JAZ repressors affecting salicylic acid-mediated resistance via activation of jasmonate signalling, and tubulin that results in disruption of the microtubule network and associated protein secretion and defence at the cell wall22, 24. HopZ2 from pea pathogen P. syringae pv. pisi 895 A physically associates with an Arabidopsis transmembrane domain containing protein MILDEW RESISTANCE LOCUS2 (MLO2) that is required for resistance against necrotrophic pathogens Botrytis cinerea and Magnaporthe oryzae 66. Cytoplasmic HopZ3 from bean pathogen P. syringae pv. syringae B728a has recently been shown to target Arabidopsis RIN4, various receptor-like cytoplasmic kinases (RLCKs) and the MAP kinase MPK4, all involved in defence signalling in planta 67. HopZ4 from cucumber pathogen P. syringae pv. lachrymans MAFF301315 binds to proteasomal subunit RPT6 and inhibits proteasome activity that is required for disease resistance68. The majority of HopZ effectors have been shown to be localized at the plant cell membrane, likely via myristoylation, and their subcellular localization is required for function42, 68. Based on these findings and our results (Figs 3 and 4), it is expected that HopZ5 targets a plasma membrane-localized defence component(s).
Plant resistance against YopJ family members has been genetically characterized so far for HopZ1a, AvrBsT and PopP2. HopZ1a targets pseudokinase ZED1 and this event is recognized by ZAR126, 27. PopP2 directly acetylates WRKY DNA-binding domain of RRS1 and activates RPS4-dependent immunity28–30. AvrBsT-mediated immunity in pepper involves both CaSGT1 and CaPIK169. CaPIK1 is a pepper receptor-like cytoplasmic kinase that forms a complex with CaSGT1 and triggers phosphorylation of CaSGT1, subsequent monomerization and nuclear localization. AvrBsT appears to interact with this complex and interferes with this process, leading to a cell death response in an AvrBsT catalytic activity-independent manner but dependent on CaPIK1 phosphorylation of CaSGT1 through specific accumulation of AvrBsT-CaSGT1 in the cytoplasm69. Furthermore, two other interactors have been found to contribute to the cell death response triggered by AvrBsT in pepper, CaHSP70a and CaALDH170, 71. As yet it remains unclear how these multiple targets integrate to trigger a cell death response to AvrBsT.
As mentioned previously, few of the characterized homologs of HopZ5 share an in planta target while some have a large number of targets that could all additively contribute to its virulence function67. Additionally, distinct NLRs recognize corresponding YopJ family T3Es to activate plant immunity. However, certain cues can be taken from previous studies on YopJ family T3Es to predict circumstances for HopZ5. Based on these findings, we hypothesize that HopZ5 could target a host protein(s) that has not been implicated in other HopZ effector-triggered immunity or susceptibility studies. Furthermore, we predict that the in planta target and the corresponding NLR of HopZ5 are likely to be plasma membrane localized.
A lysine residue that was shown to be autoacetylated in PopP2 is conserved among the majority of the YopJ family of effectors. In the case of PopP2, the autoacetylated lysine is required for its avirulence activity18. However, it was shown that HopZ1a does not require this conserved lysine for its function in a native promoter context21, 24, 72. Some members of the YopJ family of effectors were shown to acetylate serine and threonine residues, including YopJ17 and HopZ1a21. We have shown here that the conserved lysine residue (K278) is required for HopZ5 autoacetylation of lysine residues in planta (Fig. 3c). Lysine 278 of HopZ5 seems to either be the only lysine residue that is autoacetylated since the mutation of lysine 278 to arginine (K278R) abolished HopZ5 lysine autoacetylation, or is required for autoacetylation activity. Since the antibody (α-AcK) we used for testing HopZ5 autoacetylation did not allow us to detect non-lysine acetylation, additional autoacetylation of other residues in HopZ5 may still occur in both C218A and K278R variants. The crystal structure of HopZ1a revealed that the analogous residue to K278 in HopZ5, K211 in HopZ1a, is involved in coordination of the plant-derived cofactor IP620. This suggests that HopZ5 K278 could also be involved in coordination of IP6 and thus loss of this residue in the K278R variant results in loss of acetylation activity. Further analysis of HopZ5 autoacetylation and in planta target(s) will help unveil the mechanisms involved here. Nevertheless, it is conceivable that K278 could be the only lysine residue critical for HopZ5 avirulence. However, agroinfiltration of K278R variant of HopZ5 in Nicotiana spp. could still induce delayed HCD. This partial loss of avirulence activity of the K278R variant of HopZ5 in Nicotiana spp. suggests that the requirement of K278 for HopZ5 function might be dose-dependent.
We also observed a similar partial loss of function of the G2A variant which lost plasma membrane localization, presumably through lack of myristoylation (Fig. 3, Supplementary Table 1). This partial loss of function could be due to the cytoplasm-localized HopZ5 G2A variant that could still interact with its target protein and induce HCD. Identification of the host in planta target(s) of HopZ5 will be essential in better understanding the requirement for subcellular localization in initiation of HopZ5-triggered immunity in the future.
We show here that SGT1 is required for HopZ5-mediated HCD in N. benthamiana. Analysis of the genetic requirement for AvrBsT-triggered immunity in Arabidopsis indicated that NDR1 (fully), EDS1 (partially), and SID2 (partially) were required55. A similar study of ZAR1-mediated immunity against HopZ1a in Arabidopsis could not identify requirement for SID2, NDR1, EDS1 or SGT1 26. HopZ5-triggered immunity required SID2, NDR1 and EDS1. It must be noted, however, that due to the weak immunity triggered by HopZ5 in the Col-0 background, the full requirements for SID2 and EDS1 seen in our experiments may mirror the partial requirements for these two genes for AvrBsT-triggered immunity. Despite this, our results strongly suggest that the recognition of HopZ5 (and possibly AvrBsT) is likely mediated by an as yet uncharacterized NLR. The identification that both NDR1 and EDS1 are required for immunity against HopZ5 in Arabidopsis was initially surprising and appeared to contradict our expectations from observations of HCD development in N. benthamiana. However, due to the uncoupling of HR (or HCD) from immunity in Col-0 plants, we were able to explain the requirement for both NDR1 and EDS1 in immunity mounted in response to HopZ5 irrespective of the requirement for HCD (or HR).
Nevertheless, that HopZ5 does not require NDR1 nor EDS1 to activate HCD in N. benthamiana, can be explained by a redundancy between these two canonical pathways in N. benthamiana, or that the EDS1 or NDR1 levels that do accumulate in the silenced plants are not rate-limiting. Redundancy between EDS1 and NDR1 occurs in Arabidopsis RPP7- and RPP8-mediated immunity against the oomycete pathogen Hyaloperonospora arabidopsidis that was not abolished in the absence of EDS1 or NDR1 alone but was attenuated in the eds1 ndr1 double mutant73. Therefore, it is plausible that EDS1 and NDR1 play a functionally redundant role in HopZ5-triggered immunity. Alternatively, HopZ5-triggered immunity may require an as yet undiscovered signalling component of ETI.
In addition to SGT1, a non-canonical CNL, NRG1, is a required genetic component for immunity triggered by the TNL, N, against tobacco mosaic virus in Nicotiana 74. NRG1 and its homologs, including Arabidopsis ADR1, are conserved across all plant families75. Interestingly, ADR1 and ADR1-L2 were shown to be critical for several CNLs and TNLs76, 77. Consequently, it has been suggested that NRG1/ADR1-type helper NLRs could be critical downstream genetic components of a large number of canonical NLR-mediated resistances75–77. Any involvement of NRG1 or ADR1 in HopZ5-triggered immunity in Nicotiana or Arabidopsis, respectively, remains to be explored.
Elucidation of the genetic components required for HopZ5-triggered immunity is of particular interest in the interaction between Psa and kiwifruit given that HopZ5 is unique to the global outbreak strains of Psa. The gain of effectors HopZ5 and HopH1 was proposed as a transposon-mediated lateral transfer event14. HopZ5 is unique to Psa, while HopH1 is present in multiple pathogenic Pseudomonas strains and is believed to be a helper protein78. We do not yet know whether this lateral transfer event played a significant role in the rapid global spread of this disease, but if it did this research may help us develop potent options for application of resistance to Psa.
Methods
Construction of broad-host range vector
The multiple cloning site (MCS) of the broad host range vector, pBBR1MCS-579, was modified by inserting a fusion construct containing 128 bp upstream sequence from the start codon of avrRps4 (avrRps4 promoter) and 596 bp of the golden gate cloning system compatible lacZ gene containing flanking BsaI restriction enzyme sites from binary vector pICH86988. This fusion was generated by standard PCR using primers 5′-CGCAGATCTTTCCCCGAAGATTAGGAACT-3′ and 5′-GCCAGCTGCGGTCTCCCATTGGGAAGCCTCTTTGTCAAAG-3′ for the avrRps4 promoter and 5′-CTTTGACAAAGAGGCTTCCCAATGGGAGACCGCAGCTG-3′ and 5′-TATCGATAAGCTGAGACCGTCACAG-3′ for lacZ followed by overlap PCR to create a DNA fragment containing 5′-BglII-avrRps4 pro-lacZ-ClaI-3′. The amplified DNA fragment was digested with BglII and ClaI, purified from a 1.0% agarose gel, and ligated into BglII- and ClaI-treated pBBR1MCS-5. The sequence of the final pBBR1MCS-5:avrRps4 pro construct was confirmed by sequencing with M13F and M13R primers.
Bacterial type-III effector library construction
Type-III secreted effectors were predicted from the genomic sequence of Pseudomonas syringae pv. actinidiae NZV-13 by homology to known Pseudomonas effectors and annotated using Geneious software [Biomatters, NZ]. All predicted effectors with or without a discernible Hrp box were cloned using the golden gate cloning system80 into a binary vector (pICH86988) under the control of a 35 S CaMV promoter and TMV Ω leader for transient expression screens or into the broad host-range vector (pBBR1MCS-5:avrRps 4 pro) for type-III secretion system delivery.
Bacterial strains
Pseudomonas syringae pv. actinidiae NZV-13 (ICMP18884) was obtained from Plant & Food Research (Mt Albert, NZ). Cloning of broad-host range and binary vectors was carried out in Escherichia coli DH5α. Bacterial type-III effector delivery from the broad host-range vector was achieved using Pseudomonas fluorescens Pf0-1 carrying pLN18 (Pf0-1(T3S)). Bacterial pathogen growth curves were carried out using P. syringae pv. tomato DC3000 (NCPPB4369). Transient expression assays in Nicotiana tabacum or Nicotiana benthamiana was achieved using Agrobacterium tumefaciens AGL1. Bacteria were grown on agar solidified L-medium (E. coli or Agrobacterium) or King’s B medium (Pseudomonas), with antibiotics appropriate for genomic resistance and/or plasmid vector carried. Plasmids were mobilized from E. coli DH5α to Pf Pf0-1(T3S) or Pto DC3000 strains by triparental mating using E. coli HB101 (pRK2013) as a helper strain.
Plant material
Arabidopsis accessions used in this research were obtained from The Sainsbury Laboratory (Norwich, UK) or Plant & Food Research (Mt Albert, NZ). Seeds were sown on Dalton’s Premium Seed raising mix [Fruitfed, NZ] mixed with coarse grain vermiculite, transferred to individual cell trays two weeks after germination and grown to 4–5 weeks old before use. N. benthamiana and N. tabacum Wisconsin 38 seeds were sown on Dalton’s Premium Seed raising mix [Fruitfed, NZ], transferred to individual pots one week after germination and grown for 4–6 weeks before use. Plants were grown in short day conditions with 11 hours light at 22 °C.
Bacterial infiltrations for HR assays in Arabidopsis
Psa or Pf Pf0-1(T3S) were streaked from glycerol stocks onto King’s B plates with antibiotic selection and grown for two days at 28 °C. Bacteria were then harvested from plates, resuspended in 10 mM MgCl2 and diluted to OD600 = 0.2 (1 × 108 CFU/mL) for HR assays. Infiltrations were carried out on fully expanded leaves of 4- to 5-week-old Arabidopsis with a blunt 1 mL syringe. HR was assayed visually at 18–22 hours post infiltration (hpi).
Electrolyte leakage assays
Pf Pf0-1(T3S) strains were grown and infiltrated as for HR assays. Leaf discs were taken with a cork borer from infiltrated leaves when dry, washed in deionized water for half an hour before being floated on deionized water (n = 4). Electrolyte leakage was measured on a Horiba B-173 Twin Cond conductivity meter at indicated time points.
N. benthamiana electrolyte leakage
Electrolyte leakage measurements were carried out as described previously81. Briefly, Agro-infiltration was carried out as described above and two leaf discs taken per sample (n = 6) at 0 and 2 dpi, floated on 2 mL deionized water with shaking at 160 rpm for 2 hours. Leaf discs were removed before electrolyte measurements were taken.
In planta bacterial growth assays
Pto DC3000 strains were streaked and resuspended as for HR assays and diluted to OD600 = 0.001 (5 × 105 CFU/mL) before blunt syringe infiltrations into fully expanded leaves of 4- to 5-week-old Arabidopsis plants. At 4 dpi, leaf discs were taken and ground in sterile 10 mM MgCl2. Each sample was then serially diluted in sterile 10 mM MgCl2 and 20 μL spots of each sample (n = 6) and dilution were plated on King’s B plates with appropriate antibiotics. After 2 days incubation at 28 °C the CFU were counted for the least dilute sample possible.
Agrobacterium tumefaciens transient infiltration
Agrobacterium tumefaciens AGL1 strains were streaked onto L-medium plates from glycerol stocks and grown for two days at 28 °C and further inoculated in liquid L-medium with antibiotics and grown overnight before centrifugation and resuspension in Agro-infiltration buffer (10 mM MgCl2, 10 mM MES pH 5,6). Suspensions were adjusted to OD600 = 0.4 for blunt syringe infiltrations into N. benthamiana or N. tabacum leaves. Hypersensitive response-like cell death was assayed at 2–3 days post infiltration (dpi) for tobacco and 3–4 dpi for N. benthamiana.
Gene expression by qPCR
Total RNA was extracted from untreated, MgCl2 mock-treated or Pf Pf0-1(T3S)-infiltrated or Pto DC3000-infiltrated leaves from 4- to 6-week-old Arabidopsis plants via Tri-Reagent [Sigma Aldrich, NZ] and BCP [Sigma Aldrich, NZ] extraction. cDNA was synthesized from RNA using the Maxima First strand synthesis kit following manufacturer’s instructions [Thermo Fisher, NZ]. Quantitative PCR was carried out on a Roche Lightcycler™ LCII using SYBR Green mastermix [Thermo Fisher, NZ]. Primers used for qPCR are listed in Supplementary Table 2.
SDS-PAGE and Western blot
N. benthamiana leaves were infiltrated with A. tumefaciens AGL1 carrying binary vectors for tagged proteins. Leaf tissue was harvested at 2 dpi and snap frozen before grinding and total protein extraction. Samples were immunoprecipitated (IP) using anti-FLAG affinity gel [Sigma Aldrich, NZ]. Total protein and IP samples were boiled in loading buffer containing DTT [Sigma Aldrich, NZ] to denature proteins. SDS-polyacrylamide gel electrophoresis was performed, subsequently blotted onto PVDF membranes [Sigma Aldrich, NZ] and probed with horseradish peroxidase-conjugated antibody against the epitope tag (α-FLAG [Sigma Aldrich, NZ] or α-GFP [Santa Cruz Biotechnology, USA]) or against acetylation (α-AcK) [Cell Signaling, USA]. Visualization was achieved using Pierce Pico and Femto reagents [Thermo Fischer, NZ]. Protein loading was visualized using Ponceau staining [Thermo Fischer, NZ].
Virus-induced gene silencing
Two-week-old N. benthamiana seedlings were infiltrated with pTRV1 and pTRV2 (carrying the target gene sequence) at total OD600 = 1 (Mixture of OD600 = 0.5 of each pTRV1 and pTRV2), into 2 cotyledons. Infiltrated plants were grown 4–5 weeks before infiltration with A. tumefaciens AGL1 carrying YFP-tagged hopZ5. Samples were also taken from infiltrated plants to confirm silencing by RNA extraction and qPCR.
Confocal microscopy
SGT1-silenced N. benthamiana plants were transiently infiltrated with A. tumefaciens AGL1 carrying YFP-tagged HopZ5 variants. Leaf discs were taken at 2 dpi from randomly selected infiltrated area, avoiding leaf veins, and mounted in water for confocal laser scanning microscopy. A Carl Zeiss LSM700 with excitation at 488 nm with a 20 mW Argon laser with emission filters set between 500 and 530 nm was used for imaging.
Stable transgenic line generation
A. tumefaciens AGL1 carrying C-terminally YFP-tagged HopZ5 constructs in binary vectors were transformed into Arabidopsis plants (Col-0) by floral dip transformation as described previously82. Transgenic plants were selected on agar-solidified Murashige and Skoog (MS) [Duchefa, Total Labs, NZ] plates containing 50 μg/mL kanamycin and confirmed by qPCR for transgene expression. T2 seeds harvested from each confirmed T1 parent line were used for experiments after selection on agar-solidified MS medium supplemented with kanamycin.
Electronic supplementary material
Acknowledgements
This work was carried out with the support of ‘Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ011913)’ Rural Development Administration, Republic of Korea, and Bioprotection Centre of Research Excellence, New Zealand. J.J and S.C are the recipients of PhD scholarships from Plant & Food Research Institute and Zespri, respectively, in New Zealand.
Author Contributions
J.J., S.C. and K.H.S. designed the experiments. J.J., S.C., M.P., D.S.C., A.S. and C.S. conducted the experiments and prepared figures. J.J., S.C., E.H.R., M.D.T. and K.H.S. wrote the manuscript. All authors reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Jay Jayaraman and Sera Choi contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-03704-x
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Zipfel C, Robatzek S. Pathogen-associated molecular pattern-triggered immunity: Veni, Vidi…? Plant Physiol. 2010;154:551–554. doi: 10.1104/pp.110.161547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Macho AP, Zipfel C. Targeting of plant pattern recognition receptor-triggered immunity by bacterial type-III secretion system effectors. Curr. Opin. Microbiol. 2015;23:14–22. doi: 10.1016/j.mib.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 3.Toruño TY, Stergiopoulos I, Coaker G. Plant pathogen effectors: cellular probes interfering with plant defenses in spatial and temporal manners. Annu. Rev. Phytopathol. 2016;54:419–441. doi: 10.1146/annurev-phyto-080615-100204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cui H, Tsuda K, Parker JE. Effector-triggered immunity: From pathogen perception to robust defense. Annu. Rev. Plant Biol. 2015;66:487–511. doi: 10.1146/annurev-arplant-050213-040012. [DOI] [PubMed] [Google Scholar]
- 5.Qi D, Innes RW. Recent advances in plant NLR structure, function, localization, and signaling. Front. Immunol. 2013;4:348. doi: 10.3389/fimmu.2013.00348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aarts N, et al. Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. USA. 1998;95:10306–10311. doi: 10.1073/pnas.95.17.10306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Falk A, et al. EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc. Natl. Acad. Sci. USA. 1999;96:3292–3297. doi: 10.1073/pnas.96.6.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knepper C, Savory EA, Day B. Arabidopsis NDR1 is an integrin-like protein with a role in fluid loss and plasma membrane-cell wall adhesion. Plant Physiol. 2011;156:286–300. doi: 10.1104/pp.110.169656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shirasu K. The HSP90-SGT1 chaperone complex for NLR immune sensors. Annu. Rev. Plant Biol. 2009;60:139–164. doi: 10.1146/annurev.arplant.59.032607.092906. [DOI] [PubMed] [Google Scholar]
- 10.Zhang M, Kadota Y, Prodromou C, Shirasu K, Pearl LH. Structural basis for assembly of Hsp90-Sgt1-CHORD protein complexes: Implications for chaperoning of NLR innate immunity receptors. Mol. Cell. 2010;39:269–281. doi: 10.1016/j.molcel.2010.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nawrath C, Metraux J-P. Salicylic acid induction–deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell. 1999;11:1393–1404. doi: 10.1105/tpc.11.8.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wildermuth MC, et al. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature. 2001;414:562–565. doi: 10.1038/35107108. [DOI] [PubMed] [Google Scholar]
- 13.Chapman JR, et al. Phylogenetic relationships among global populations of Pseudomonas syringae pv. actinidiae. Phytopathology. 2012;102:1034–1044. doi: 10.1094/PHYTO-03-12-0064-R. [DOI] [PubMed] [Google Scholar]
- 14.McCann HC, et al. Genomic analysis of the kiwifruit pathogen Pseudomonas syringae pv. actinidiae provides insight into the origins of an emergent plant disease. PLoS Pathog. 2013;9:e1003503. doi: 10.1371/journal.ppat.1003503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Templeton MD, Warren BA, Andersen MT, Rikkerink EHA, Fineran PC. Complete DNA sequence of Pseudomonas syringae pv. actinidiae, the causal agent of kiwifruit canker disease. Genome Announc. 2015;3:e01054–15. doi: 10.1128/genomeA.01054-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orth K, et al. Inhibition of the mitogen-activated protein kinase kinase superfamily by a Yersinia effector. Science. 1999;285:1920–1923. doi: 10.1126/science.285.5435.1920. [DOI] [PubMed] [Google Scholar]
- 17.Mukherjee S, et al. Yersinia YopJ acetylates and inhibits kinase activation by blocking phosphorylation. Science. 2006;312:1211–1214. doi: 10.1126/science.1126867. [DOI] [PubMed] [Google Scholar]
- 18.Tasset C, et al. Autoacetylation of the Ralstonia solanacearum effector PopP2 targets a lysine residue essential for RRS1-R-mediated immunity in Arabidopsis. PLoS Pathog. 2010;6:e1001202. doi: 10.1371/journal.ppat.1001202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lewis JD, et al. The YopJ superfamily in plant-associated bacteria. Mol. Plant Pathol. 2011;12:928–937. doi: 10.1111/j.1364-3703.2011.00719.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Z-M, et al. Structure of a pathogen effector reveals the enzymatic mechanism of a novel acetyltransferase family. Nat. Struct. Mol. Biol. 2016;23:847–852. doi: 10.1038/nsmb.3279. [DOI] [PubMed] [Google Scholar]
- 21.Ma K-W, et al. Two serine residues in Pseudomonas syringae effector HopZ1a are required for acetyltransferase activity and association with the host co-factor. New Phytol. 2015;208:1157–1168. doi: 10.1111/nph.13528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang S, et al. Bacterial effector activates jasmonate signaling by directly targeting JAZ transcriptional repressors. PLoS Pathog. 2013;9:e1003715. doi: 10.1371/journal.ppat.1003715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Macho AP, Guevara CM, Tornero P, Ruiz-Albert J, Beuzón CR. The Pseudomonas syringae effector protein HopZ1a suppresses effector-triggered immunity. New Phytol. 2010;187:1018–1033. doi: 10.1111/j.1469-8137.2010.03381.x. [DOI] [PubMed] [Google Scholar]
- 24.Lee AH-Y, et al. A bacterial acetyltransferase destroys plant microtubule networks and blocks secretion. PLoS Pathog. 2012;8:e1002523. doi: 10.1371/journal.ppat.1002523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang D, Weaver ND, Kesarwani M, Dong X. Induction of protein secretory pathway is required for systemic acquired resistance. Science. 2005;308:1036–1040. doi: 10.1126/science.1108791. [DOI] [PubMed] [Google Scholar]
- 26.Lewis JD, Wu R, Guttman DS, Desveaux D. Allele-specific virulence attenuation of the Pseudomonas syringae HopZ1a Type III effector via the Arabidopsis ZAR1 resistance protein. PLoS Genet. 2010;6:e1000894. doi: 10.1371/journal.pgen.1000894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lewis JD, et al. The Arabidopsis ZED1 pseudokinase is required for ZAR1-mediated immunity induced by the Pseudomonas syringae type III effector HopZ1a. Proc. Natl. Acad. Sci. 2013;110:18722–18727. doi: 10.1073/pnas.1315520110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le Roux C, et al. A receptor pair with an integrated decoy converts pathogen disabling of transcription factors to immunity. Cell. 2015;161:1074–1088. doi: 10.1016/j.cell.2015.04.025. [DOI] [PubMed] [Google Scholar]
- 29.Sarris PF, et al. A plant immune receptor detects pathogen effectors that target WRKY transcription factors. Cell. 2015;161:1089–1100. doi: 10.1016/j.cell.2015.04.024. [DOI] [PubMed] [Google Scholar]
- 30.Deslandes L, et al. Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive RRS1-R gene, a member of a novel family of resistance genes. Proc. Natl. Acad. Sci. USA. 2002;99:2404–2409. doi: 10.1073/pnas.032485099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thomas WJ, Thireault CA, Kimbrel JA, Chang JH. Recombineering and stable integration of the Pseudomonas syringae pv. syringae 61 hrp/hrc cluster into the genome of the soil bacterium Pseudomonas fluorescens Pf0-1: Stable integration of a T3SS-locus into Pf0-1. Plant J. 2009;60:919–928. doi: 10.1111/j.1365-313X.2009.03998.x. [DOI] [PubMed] [Google Scholar]
- 32.Bisgrove SR, Simonich MT, Smith NM, Sattler A, Innes RW. A disease resistance gene in Arabidopsis with specificity for two different pathogen avirulence genes. Plant Cell. 1994;6:927–933. doi: 10.1105/tpc.6.7.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hinsch M, Staskawicz B. Identification of a new Arabidopsis disease resistance locus, RPs4, and cloning of the corresponding avirulence gene, avrRps4, from Pseudomonas syringae pv. pisi. Mol. Plant-Microbe Interact. 1996;9:55–61. doi: 10.1094/MPMI-9-0055. [DOI] [PubMed] [Google Scholar]
- 34.Kunkel BN, Bent AF, Dahlbeck D, Innes RW, Staskawicz BJ. RPS2, an Arabidopsis disease resistance locus specifying recognition of Pseudomonas syringae strains expressing the avirulence gene avrRpt2. Plant Cell. 1993;5:865–875. doi: 10.1105/tpc.5.8.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johansson ON, et al. A quick and robust method for quantification of the hypersensitive response in plants. PeerJ. 2015;3:e1469. doi: 10.7717/peerj.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sohn KH, Lei R, Nemri A, Jones JDG. The downy mildew effector proteins ATR1 and ATR13 promote disease susceptibility in Arabidopsis thaliana. Plant Cell. 2007;19:4077–4090. doi: 10.1105/tpc.107.054262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wei C-F, et al. A Pseudomonas syringae pv. tomato DC3000 mutant lacking the type III effector HopQ1-1 is able to cause disease in the model plant Nicotiana benthamiana. Plant J. 2007;51:32–46. doi: 10.1111/j.1365-313X.2007.03126.x. [DOI] [PubMed] [Google Scholar]
- 38.Wroblewski T, et al. Comparative large-scale analysis of interactions between several crop species and the effector repertoires from multiple pathovars of Pseudomonas and Ralstonia. Plant Physiol. 2009;150:1733–1749. doi: 10.1104/pp.109.140251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cai X-Z, Xu Q-F, Wang C-C, Zheng Z. Development of a virus-induced gene-silencing system for functional analysis of the RPS2-dependent resistance signalling pathways in Arabidopsis. Plant Mol. Biol. 2006;62:223–232. doi: 10.1007/s11103-006-9016-z. [DOI] [PubMed] [Google Scholar]
- 40.Liu Y, Schiff M, Marathe R, Dinesh-Kumar SP. Tobacco RAR1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J. 2002;30:415–429. doi: 10.1046/j.1365-313X.2002.01297.x. [DOI] [PubMed] [Google Scholar]
- 41.Cheong MS, et al. AvrBsT acetylates Arabidopsis ACIP1, a protein that associates with microtubules and is required for immunity. PLoS Pathog. 2014;10:e1003952. doi: 10.1371/journal.ppat.1003952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lewis JD, Abada W, Ma W, Guttman DS, Desveaux D. The HopZ family of Pseudomonas syringae type III effectors require myristoylation for virulence and avirulence functions in Arabidopsis thaliana. J. Bacteriol. 2008;190:2880–2891. doi: 10.1128/JB.01702-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma W, Dong FFT, Stavrinides J, Guttman DS. Type III effector diversification via both pathoadaptation and horizontal transfer in response to a coevolutionary arms race. PLoS Genet. 2006;2:e209. doi: 10.1371/journal.pgen.0020209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Century KS, Holub EB, Staskawicz BJ. NDR1, a locus of Arabidopsis thaliana that is required for disease resistance to both a bacterial and a fungal pathogen. Proc. Natl. Acad. Sci. USA. 1995;92:6597–6601. doi: 10.1073/pnas.92.14.6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cooley MB, Pathirana S, Wu HJ, Kachroo P, Klessig DF. Members of the Arabidopsis HRT/RPP8 family of resistance genes confer resistance to both viral and oomycete pathogens. Plant Cell. 2000;12:663–676. doi: 10.1105/tpc.12.5.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chandra-Shekara AC, et al. Signaling requirements and role of salicylic acid in HRT- and rrt-mediated resistance to turnip crinkle virus in Arabidopsis. Plant J. 2004;40:647–659. doi: 10.1111/j.1365-313X.2004.02241.x. [DOI] [PubMed] [Google Scholar]
- 47.Coll NS, et al. Arabidopsis type I metacaspases control cell death. Science. 2010;330:1393–1397. doi: 10.1126/science.1194980. [DOI] [PubMed] [Google Scholar]
- 48.Gassmann W. Natural variation in the Arabidopsis response to the avirulence gene hopPsyA uncouples the hypersensitive response from disease resistance. Mol. Plant-Microbe Interact. 2005;18:1054–1060. doi: 10.1094/MPMI-18-1054. [DOI] [PubMed] [Google Scholar]
- 49.Gassmann W, Hinsch ME, Staskawicz BJ. The Arabidopsis RPS4 bacterial-resistance gene is a member of the TIR-NBS-LRR family of disease-resistance genes. Plant J. 1999;20:265–277. doi: 10.1046/j.1365-313X.1999.t01-1-00600.x. [DOI] [PubMed] [Google Scholar]
- 50.Heidrich K, et al. Arabidopsis EDS1 connects pathogen effector recognition to cell compartment-specific immune responses. Science. 2011;334:1401–1404. doi: 10.1126/science.1211641. [DOI] [PubMed] [Google Scholar]
- 51.Yu IC, Parker J, Bent AF. Gene-for-gene disease resistance without the hypersensitive response in Arabidopsis dnd1 mutant. Proc. Natl. Acad. Sci. USA. 1998;95:7819–7824. doi: 10.1073/pnas.95.13.7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Clough SJ, et al. The Arabidopsis dnd1 ‘defense, no death’ gene encodes a mutated cyclic nucleotide-gated ion channel. Proc. Natl. Acad. Sci. USA. 2000;97:9323–9328. doi: 10.1073/pnas.150005697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Balagué C, et al. HLM1, an essential signaling component in the hypersensitive response, is a member of the cyclic nucleotide-gated channel ion channel family. Plant Cell. 2003;15:365–379. doi: 10.1105/tpc.006999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yoshioka K, et al. The chimeric Arabidopsis cyclic nucleotide-gated ion channel11/12 activates multiple pathogen resistance responses. Plant Cell. 2006;18:747–763. doi: 10.1105/tpc.105.038786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cunnac S, et al. A conserved carboxylesterase is a suppressor of avrbst-elicited resistance in Arabidopsis. Plant Cell. 2007;19:688–705. doi: 10.1105/tpc.106.048710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Coll NS, Epple P, Dangl JL. Programmed cell death in the plant immune system. Cell Death Differ. 2011;18:1247–1256. doi: 10.1038/cdd.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ciesiolka LD, et al. Regulation of expression of avirulence gene avrRxv and identification of a family of host interaction factors by sequence analysis of avrBsT. Mol. Plant-Microbe Interact. 1999;12:35–44. doi: 10.1094/MPMI.1999.12.1.35. [DOI] [PubMed] [Google Scholar]
- 58.Astua-Monge G, et al. Xv4-vrxv4: a new gene-for-gene interaction identified between Xanthomonas campestris pv. vesicatoria race T3 and wild tomato relative Lycopersicon pennellii. Mol. Plant-Microbe Interact. 2000;13:1346–1355. doi: 10.1094/MPMI.2000.13.12.1346. [DOI] [PubMed] [Google Scholar]
- 59.Noël L, Thieme F, Gäbler J, Büttner D, Bonas U. XopC and XopJ, two novel type III effector proteins from Xanthomonas campestris pv. vesicatoria. J. Bacteriol. 2003;185:7092–7102. doi: 10.1128/JB.185.24.7092-7102.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roden J, Eardley L, Hotson A, Cao Y, Mudgett MB. Characterization of the Xanthomonas AvrXv4 effector, a SUMO protease translocated into plant cells. Mol. Plant-Microbe Interact. 2004;17:633–643. doi: 10.1094/MPMI.2004.17.6.633. [DOI] [PubMed] [Google Scholar]
- 61.Whalen MC, et al. Identification of a host 14-3-3 protein that interacts with Xanthomonas effector AvrRxv. Physiol. Mol. Plant Pathol. 2008;72:46–55. doi: 10.1016/j.pmpp.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Minsavage GV. Gene-For-gene relationships specifying disease resistance in Xanthomonas campestris pv. vesicatoria - pepper interactions. Mol. Plant. Microbe Interact. 1990;3:41–47. doi: 10.1094/MPMI-3-041. [DOI] [Google Scholar]
- 63.Szczesny R, et al. Suppression of the AvrBs1-specific hypersensitive response by the YopJ effector homolog AvrBsT from Xanthomonas depends on a SNF1-related kinase. New Phytol. 2010;187:1058–1074. doi: 10.1111/j.1469-8137.2010.03346.x. [DOI] [PubMed] [Google Scholar]
- 64.Üstün S, Börnke F. The Xanthomonas campestris type III effector XopJ proteolytically degrades proteasome subunit RPT6. Plant Physiol. 2015;168:107–119. doi: 10.1104/pp.15.00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Poueymiro M, et al. Two type III secretion system effectors from Ralstonia solanacearum GMI1000 determine host-range specificity on tobacco. Mol. Plant-Microbe Interact. 2009;22:538–550. doi: 10.1094/MPMI-22-5-0538. [DOI] [PubMed] [Google Scholar]
- 66.Lewis JD, et al. Quantitative Interactor Screening with next-generation Sequencing (QIS-Seq) identifies Arabidopsis thaliana MLO2 as a target of the Pseudomonas syringae type III effector HopZ2. BMC Genomics. 2012;13:8. doi: 10.1186/1471-2164-13-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee J, et al. Acetylation of an NB-LRR plant immune-effector complex suppresses immunity. Cell Rep. 2015;13:1670–1682. doi: 10.1016/j.celrep.2015.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Üstün S, König P, Guttman DS, Börnke F. HopZ4 from Pseudomonas syringae, a member of the HopZ type III effector family from the YopJ superfamily, inhibits the proteasome in plants. Mol. Plant. Microbe Interact. 2014;27:611–623. doi: 10.1094/MPMI-12-13-0363-R. [DOI] [PubMed] [Google Scholar]
- 69.Kim NH, Kim DS, Chung EH, Hwang BK. Pepper suppressor of the G2 allele of skp1 interacts with the receptor-like cytoplasmic kinase1 and type III effector AvrBsT and promotes the hypersensitive cell death response in a phosphorylation-dependent manner. Plant Physiol. 2014;165:76–91. doi: 10.1104/pp.114.238840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kim NH, Hwang BK. Pepper aldehyde dehydrogenase CaALDH1 interacts with Xanthomonas effector AvrBsT and promotes effector-triggered cell death and defence responses. J. Exp. Bot. 2015;66:3367–3380. doi: 10.1093/jxb/erv147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim NH, Hwang BK. Pepper Heat shock protein 70a Interacts with the type III effector AvrBsT and triggers plant cell death and immunity. Plant Physiol. 2015;167:307–322. doi: 10.1104/pp.114.253898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rufián JS, et al. Auto-acetylation on K289 is not essential for HopZ1a-mediated plant defense suppression. Front. Microbiol. 2015;6:e684. doi: 10.3389/fmicb.2015.00684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McDowell JM, et al. Downy mildew (Peronospora parasitica) resistance genes in Arabidopsis vary in functional requirements for NDR1, EDS1, NPR1 and salicylic acid accumulation. Plant J. 2000;22:523–529. doi: 10.1046/j.1365-313x.2000.00771.x. [DOI] [PubMed] [Google Scholar]
- 74.Peart JR, Mestre P, Lu R, Malcuit I, Baulcombe DC. NRG1, a CC-NB-LRR protein, together with N, a TIR-NB-LRR protein, mediates resistance against tobacco mosaic virus. Curr. Biol. 2005;15:968–973. doi: 10.1016/j.cub.2005.04.053. [DOI] [PubMed] [Google Scholar]
- 75.Collier SM, Hamel L-P, Moffett P. Cell death mediated by the N-terminal domains of a unique and highly conserved class of NB-LRR protein. Mol. Plant-Microbe Interact. 2011;24:918–931. doi: 10.1094/MPMI-03-11-0050. [DOI] [PubMed] [Google Scholar]
- 76.Bonardi V, et al. Expanded functions for a family of plant intracellular immune receptors beyond specific recognition of pathogen effectors. Proc. Natl. Acad. Sci. USA. 2011;108:16463–16468. doi: 10.1073/pnas.1113726108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dong OX, et al. TNL-mediated immunity in Arabidopsis requires complex regulation of the redundant ADR1 gene family. New Phytol. 2016;210:960–973. doi: 10.1111/nph.13821. [DOI] [PubMed] [Google Scholar]
- 78.Mohr TJ, et al. Naturally occurring nonpathogenic isolates of the plant pathogen Pseudomonas syringae lack a type III secretion system and effector gene orthologues. J. Bacteriol. 2008;190:2858–2870. doi: 10.1128/JB.01757-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kovach ME, et al. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 80.Engler C, Kandzia R, Marillonnet S. A one pot, one step, precision cloning method with high throughput capability. PLoS ONE. 2008;3:e3647. doi: 10.1371/journal.pone.0003647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Oh C-S, Pedley KF, Martin GB. Tomato 14-3-3 protein 7 positively regulates immunity-associated programmed cell death by enhancing protein abundance and signaling ability of MAPKKKα. Plant Cell. 2010;22:260–272. doi: 10.1105/tpc.109.070664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


