Abstract
Nisin, a bacteriocin, is a potential alternative to antibiotics to modulate rumen fermentation. However, little is known about its impacts on rumen microbes. This study evaluated the effects of nisin (1 and 5 μM) on in vitro rumen fermentation characteristics, microbiota, and select groups of rumen microbes in comparison with monensin (5 μM), one of the most commonly used ionophores in ruminants. Nisin had greater effects than monensin in inhibiting methane production and decreasing acetate/propionate ratio. Unlike monensin, nisin had no adverse effect on dry matter digestibility. Real-time PCR analysis showed that both monensin and nisin reduced the populations of total bacteria, fungi, and methanogens, while the population of protozoa was reduced only by monensin. Principal component analysis of bacterial 16S rRNA gene amplicons showed a clear separation between the microbiota shaped by monensin and by nisin. Comparative analysis also revealed a significant difference in relative abundance of some bacteria in different taxa between monensin and nisin. The different effects of monensin and nisin on microbial populations and bacterial communities are probably responsible for the discrepancy in their effects on rumen fermentation. Nisin may have advantages over monensin in modulating ruminal microbial ecology and reducing ruminant methane production without adversely affecting feed digestion, and thus it may be used as a potential alternative to monensin fed to ruminants.
Keywords: bacterial community, feed digestion, methane, microbial population, microbiota, monensin, nisin, rumen fermentation
Introduction
The livestock industry nowadays faces three major challenges: feed shortage, environmental pollution, and food safety, for its sustainable development (Thornton, 2010). With ruminants, due to the rumen metabolism, approximately 2 to 12% of the feed energy is wasted as methane (Johnson and Johnson, 1995), which is 23 times more potent as a greenhouse gas than CO2 (FAO, 2006). Equally challenging if not more, much of the dietary nitrogen is converted to ammonia by rumen fermentation, and eventually more than 60% of the dietary nitrogen is excreted as urea via urine, which leads to serious groundwater pollution (Firkins et al., 2007). Various antimicrobials have been extensively used in animal production in the past 60 years to promote animal growth and health. Although antibiotics have been banned for non-therapeutic purposes in the European Union since the beginning of 2006 (Franz et al., 2010), monensin (MON) is still one of the most commonly used ionophores in ruminants in other countries. Benefits of feeding MON to ruminants include improved feed digestibility, a shift of volatile fatty acids (VFA) profiles toward increased propionate but decreased acetate and an associated decrease in methanogenesis, and decreased amino acid fermentation and ruminal ammonia concentration (Russell and Houlihan, 2003). However, MON lowers dry matter intake (DMI) and often suppresses milk fat in dairy cattle (Duffield et al., 2008), and its inhibitory effect on methane emissions can be transient (Johnson and Johnson, 1995; Guan et al., 2006). Moreover, the routine use of antibiotics in animal production has also been blamed for contributing to the emergence and widespread of antimicrobial resistance and occurrence of antibiotic residues in the environment and animal products (Molina et al., 2003). Thus, antibiotic use in animal production is restricted, and non-antibiotic alternatives are highly sought after to mitigate methane emission and nitrogen excretion (Jeyanathan et al., 2014; Patra and Yu, 2014).
Bacteriocins, which are low molecular weight antibacterial peptides, have been proposed as potential alternatives to antibiotics (Cotter et al., 2013; Paiva and Breukink, 2013). Nisin is a bacteriocin produced by certain strains of Lactococcus lactis ssp. lactis, and it has been widely used in the food industry to control pathogenic bacteria (Delves-Broughton et al., 1996). In recent years, nisin has also shown its potential in other fields, including biomedical application (Shin et al., 2016) and livestock husbandry (Jüzefiak et al., 2013). Several in vitro experiments using rumen microbiota have shown that nisin can suppress amino acid deamination and methanogenesis without negative impact on DM digestibility (Shen et al., 2016) or VFA production (Callaway et al., 1997; Sar et al., 2005). Some researchers cautioned that nisin could be susceptible to rumen proteases, which may limit its utilization in vivo (Russell and Mantovani, 2002). However, Lee et al. (2002) demonstrated that nisin binding to ruminal bacteria was faster than its degradation, and it can be used to modify ruminal fermentation. One in vivo study also showed a significant 10% decrease of methane emission in sheep supplemented with nisin (Santoso et al., 2004). Thus, it is important to understand the roles of nisin in modulating rumen microbiota and its fermentation and methanogenesis.
Both nisin and MON inhibit bacteria by primarily increasing the permeability of their cell membrane, but their mechanisms of inhibition are different. Nisin inhibits susceptible bacteria by creating pores that do not have selective permeabilities and by inhibiting cell wall synthesis (Cotter et al., 2013), while MON, which is an ionophore, functions as a Na+/H+ antiporter upon inserting itself into the cell membrane (Bergen and Bates, 1984). Therefore, we hypothesized that nisin and MON might impact the rumen microbiota composition differently, though they can achieve some similar effects on rumen function. To test this hypothesis, the present study, by integrating quantitative real-time PCR and Illumina sequencing of 16S rRNA gene amplicons, investigated the changes of rumen microbial composition and their fermentation profiles in response to MON and nisin using an in vitro model.
Materials and Methods
Experimental Design
Four treatments were used in this study. One treatment received sodium monensin (Solarbio, Beijing, China) at 5 μM (MON), which is equivalent to the widely used in vivo dose in ruminants (Callaway et al., 1997). Two treatments received nisin (1200 IU/mg of solid, about 3.0% of the solid; Jianglaibio, Shanghai, China) at 1 μM (NI1) and 5 μM (NI5). The negative control (NC) received neither additive. The doses of nisin were selected based on a previous study (Callaway et al., 1997). Each treatment had four replicates.
Ruminal Inoculum and In Vitro Incubations
Three cannulated Holstein dairy cattle served as ruminal fluid donors for this in vitro study. The diet fed to these cattle contained (% DM basis) 20% corn silage, 40% grass hay (Chinese wild rye), and 40% concentrate mixture. The cows were fed twice daily at 06:00 and 18:00, and they had free access to feed and water. All animal protocols were approved by the Animal Care and Use Committee of Nanjing Agricultural University. Fresh ruminal fluid was collected through rumen cannula from the three cattle before morning feeding, mixed equally, and then poured into a sterilized bottle (1500 mL) leaving no headspace in the bottle, which was brought to the laboratory within 30 min. The mixed rumen sample was then squeezed through four layers of cheesecloth into a flask under CO2 in a water bath kept at 39°C until use.
The in vitro batch fermentation was carried out in 180 mL serum bottles. The fermentation substrate was a ground mixture of forage (25% corn silage, 17% alfalfa hay, and 8% Chinese wild rye) and concentrate (27% ground corn, 9% soybean meal, 6% cottonseed meal, 4% wheat bran, and 4% premix) at a 50:50 ratio. The buffered medium for the in vitro fermentation was prepared anaerobically as described by Theodorou et al. (1994). The anaerobic buffer medium and strained rumen fluid inoculum were combined in each bottle in a 9:1 (v/v) ratio under anaerobic conditions. A 100-mL mixture was immediately dispensed into each incubation bottle containing 1 g of ground feed substrate and respective additive. To prevent exposure to air, the headspace of the bottles was continuously flushed with CO2. Because monensin was not soluble in water, one concentrated stock solution (100×) was prepared using absolute ethanol. An equal volume of ethanol was also added to NC, NI1, and NI5. The final ethanol concentration was less than 1.0% (vol/vol). The serum bottles were each sealed with a butyl rubber stopper and secured with an aluminum crimp seal and incubated at 39°C for 24 h in a water bath with intermittent shaking by hand.
Sampling and Chemical Analysis
Gas production was measured at 3, 6, 9, 12, and 24 h using a pressure transducer (Theodorou et al., 1994). After gas measurement at each time point, 30 μL of gas sample was drawn out immediately from each bottle using a gastight syringe to determine methane concentrations using gas chromatography (GC-2014, Shimadzu, Japan) (Yang et al., 2012). At the end of the 24 h of incubation, the pH value of each in vitro culture was measured immediately using a portable pH-meter (Ecoscan pH 5, Eutech Instruments, Singapore). Then, 1 mL of culture each was preserved by adding 0.2 mL of 25% HPO3 for VFA analysis using gas chromatography (7890A, Agilent, United Kingdom) according to the method described by Mao et al. (2008). Another 1 mL of each culture was collected for subsequent analysis for ammonia-N using a colorimetric method (Chaney and Marbach, 1962). Also, 1 mL of culture each was collected for DNA extraction and subsequent microbial analysis. All the samples were stored at –20°C until analyses. The remaining content of each culture was filtered through a filter bag (ANKOM Technology, United States) to analyze apparent dry matter digestibility (DMD) gravimetrically (Blümmel et al., 1997).
DNA Extraction
Total metagenomic DNA was extracted using the bead-beating and phenol-chloroform extraction method as previously described (Dai et al., 2010). DNA was precipitated with ethanol and resuspended in 50 μL of Tris-HCl/EDTA buffer. The quality of the DNA extracts was visually checked using electrophoresis on 1.2% agarose gel (w/v) containing GoldviewTM (SaiBaiSheng, Shanghai, China), and the DNA concentration of each sample was determined using a Nanodrop 2000 (Thermo Fisher Scientific, Inc., Madison, WI, United States). The DNA samples were stored in –20°C until analyses.
Quantitative Real-Time PCR Analysis
The PCR primers used for real-time PCR quantification of total bacteria, fungi, protozoa, methanogen, Clostridium sticklandii, and Clostridium aminophilum are listed in Supplementary Table S1. Among these microbial groups, C. aminophilum and C. sticklandii are two of the main hyper-ammonia-producing bacteria (HAB) isolated from the rumen contributing to elevated deamination therein (Russell et al., 1988). Plasmid DNA containing each cloned respective target sequence was obtained by PCR and cloning (Koike et al., 2007), and the resultant recombinant plasmids were used as the standard DNA in real-time PCR. Real-time PCR was performed on a StepOnePlus platform (Applied Biosystems, Foster City, CA, United States) using SYBR Premix Ex Taq dye (Takara). Quantification of copies of 16S rRNA gene (total bacteria, C. sticklandii and C. aminophilum), 18S rRNA gene (fungi and protozoa), and methyl coenzyme-M reductase gene (mcrA, for methanogens) in each sample was performed in triplicate, and the mean value was calculated. Standard curves were generated using 10-fold serial dilutions of each standard DNA containing the target gene sequences of the respective microbial group. The absolute abundance of each microbial population was expressed as copies of the target gene/mL of culture samples.
Illumina Sequencing of 16S rRNA Gene Amplicons and Data Analysis
The V3-V4 hypervariable region of the 16S rRNA gene was amplified using primers 338 F (5′-ACTCCTACGGGAGGCAGCA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′). Unique barcodes were added to the 5′ end of both primers for multiplexing. PCR products were examined on a 2% (w/v) agarose gel, and the expected bands were each extracted and purified using the AxyPrepDNA Gel Extraction Kit (Axygen Biosciences, Foster City, CA, United States). The concentrations of the purified DNA amplicons were each quantified using a QuantiFluor® dsDNA kit (Promega, Madison, WI, United States). Amplicons from different samples were mixed in equal ratio and sequenced using the 2 × 300 pared-end kit on an Illumina MiSeq platform. The raw sequence reads were deposited into the NCBI Sequence Read Archive (SRA) database under the accession number SRP100539.
Raw fastq files were de-multiplexed, quality-filtered, and analyzed using QIIME 1.8.0 (Caporaso et al., 2010b) with the criteria as described by Mao et al. (2015b). Operational taxonomic units (OTUs) were de novo clustered with a 97% sequence similarity cutoff using UPARSE1 (version 7.1), and chimeric sequences were identified and removed using UCHIME (Edgar, 2010). The most abundant sequence within each OTU was designated as the ‘representative sequence’, and all the representative sequences were aligned against the core set of Greengenes 13.5 (DeSantis et al., 2006) using PYNAST (Caporaso et al., 2010a) with the default parameters set by QIIME. A PH Lane mask supplied by QIIME was used to remove the hypervariable regions from the aligned sequences. FASTTREE (Price et al., 2009) was used to create a phylogenetic tree of the representative sequences. Sequences were classified using the Ribosomal Database Project classifier with a standard minimum support threshold of 80% (Wang et al., 2007). Sequences identified as chloroplasts or mitochondria were removed before further analysis. Community alpha diversity measurements were estimated using the ACE, Chao1, Shannon, and Simpson indices. The interrelationships between the bacterial communities of different samples of the four treatments were visualized using principal component analysis (PCA), which was conducted using R to group the bacterial communities of different samples.
Statistical Analysis
The real-time PCR data were log transformed to improve normality. All data (rumen fermentation characteristics, absolute abundance of the microbial groups quantified by qPCR, bacterial alpha diversity indices, and the relative abundances of microbial populations at the phylum, genus and OTU levels) were analyzed using the general linear model (GLM) procedure of SAS version 9.2 (SAS Institute Inc., Cary, NC, United States), and Duncan’s multiple comparison tests were used to assess differences between the means. Differences were considered statistically significant at P ≤ 0.05. Pearson correlation coefficients were calculated using SAS version 9.2 to examine the correlation between relative abundances of bacterial genera and each of the major fermentation data. Significant correlation was considered at P ≤ 0.05.
Results
Effects of Nisin and Monensin on Rumen Fermentation Characteristics
The fermentation characteristics of different treatments are summarized in Table 1. Both nisin and MON dramatically reduced gas and methane production compared with NC (P < 0.05), with MON resulting in the lowest gas production while NI5 leading to the lowest methane production. MON also significantly reduced DMD compared with NC (P < 0.05). However, nisin at either concentration had no adverse effect on DMD (P > 0.05), though NI5 had numerically lower DMD than NC. Nisin significantly increased ammonia concentration (P < 0.05) in a dose-dependent manner, whereas MON had no effect.
Table 1.
Effects of nisin and monensin addition on gas and methane production, dry matter (DM) digestibility, and ammonia concentration in the in vitro rumen mixed cultures at 24 h.
| Item | NC | MON | NI1 | NI5 | SEM | P-value |
|---|---|---|---|---|---|---|
| Total gas (mL) | 181.9a | 131.0d | 162.2b | 148.4c | 1.48 | <0.01 |
| Methane (mL) | 23.5a | 14.5b | 14.4b | 10.6c | 0.46 | <0.01 |
| DM digestibility (%) | 58.4a | 53.1b | 59.1a | 57.4a | 0.53 | <0.01 |
| NH3-N (mM) | 10.5c | 10.7bc | 11.2b | 12.2a | 0.19 | <0.01 |
| pH value | 6.54b | 6.60a | 6.58a | 6.59a | 0.008 | <0.01 |
| Total VFA (mM) | 80.3a | 66.5c | 69.3b | 67.3bc | 0.73 | <0.01 |
| Acetate (mM) | 57.3a | 43.2b | 42.1b | 36.6c | 0.96 | <0.01 |
| Propionate (mM) | 16.4d | 20.9c | 22.2b | 26.2a | 0.35 | <0.01 |
| Acetate/Propionate | 3.50a | 2.07b | 1.90b | 1.40c | 0.086 | <0.01 |
| Butyrate (mM) | 5.67a | 2.05d | 4.06b | 3.51c | 0.131 | <0.01 |
| Valerate (mM) | 0.27a | 0.08c | 0.15b | 0.14b | 0.001 | <0.01 |
| Isobutyrate (mM) | 0.29a | 0.15b | 0.31a | 0.28a | 0.011 | <0.01 |
| Isovalerate (mM) | 0.34c | 0.16d | 0.41b | 0.52a | 0.016 | <0.01 |
| Total BCVFA (mM) | 0.62c | 0.30d | 0.72b | 0.81a | 0.017 | <0.01 |
NC = negative control (no additives); MON = monensin, 5 μM; NI1 = nisin, 1 μM; NI5 = nisin, 5 μM.
a-dMeans within a row with different superscripts differ (P < 0.05).
Compared with NC, both MON and nisin significantly increased culture pH. Both MON and nisin reduced concentrations of total VFA, acetate, butyrate, valerate, and acetate/propionate ratio (P < 0.05), but increased propionate concentration, with MON resulting in a greater than nisin. The NI5 had the lowest acetate concentration and acetate/propionate ratio compared with other treatments. With branched-chain VFA (BCVFA), however, MON and nisin had different effects. Compared with NC, concentrations of isovalerate and total BCVFA were reduced by MON but were increased by nisin (P < 0.05). MON, but not nisin, decreased the concentration of isobutyrate (P < 0.05). Overall, the results indicate that MON and nisin showed different impacts on dry matter digestibility, ammonia production, and VFA profiles, especially BCVFA profiles.
Effects of Nisin and Monensin on Rumen Microbial Populations
Quantitative real-time PCR showed that both MON and nisin significantly reduced (P < 0.05) the population of total bacteria and methanogens compared with NC (Figure 1). Both MON and nisin also significantly reduced the population of fungi (P < 0.05), with the greatest reduction observed for MON followed by NI5 and NI1. Compared with NC, the population of protozoa was reduced by MON (P < 0.05) but not by nisin (P > 0.05). The HAB quantified, C. aminophilum and C. sticklandii, which have been shown previously to be sensitive to MON and nisin, responded differently to MON and nisin. Compared to that of NC, the population of C. aminophilum was increased (P < 0.05) by both MON and nisin (Figure 2), while that of C. sticklandii was decreased by MON but increased by nisin (P < 0.05). The results indicate that MON and nisin showed different effects on the populations of some major rumen microbes.
FIGURE 1.
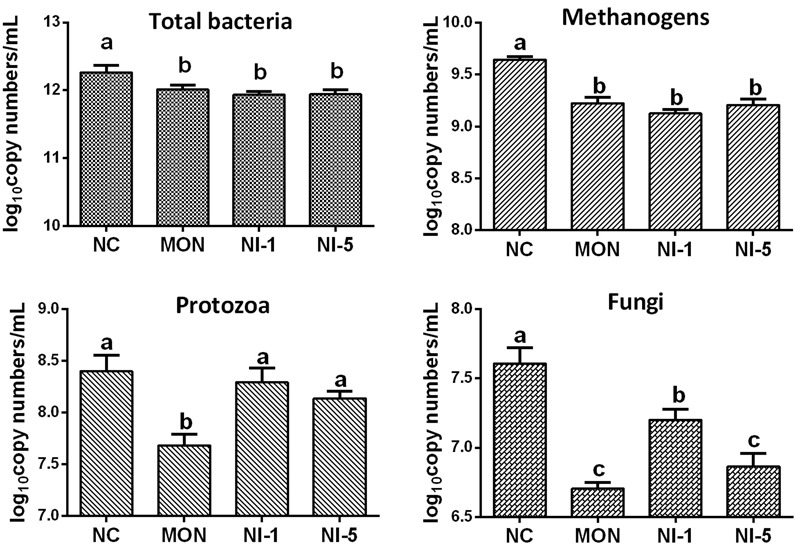
Effects of nisin and monensin addition on the population of total bacteria, methanogens, protozoa and fungi (log10 copy number of the target genes/mL) in the in vitro rumen mixed cultures. Values are means ± SE (n = 4). NC = negative control (no additives); MON = monensin, 5 μM; NI1 = nisin, 1 μM; NI5 = nisin, 5 μM. Bars with different letters (a, b, c) represent different means (P < 0.05).
FIGURE 2.
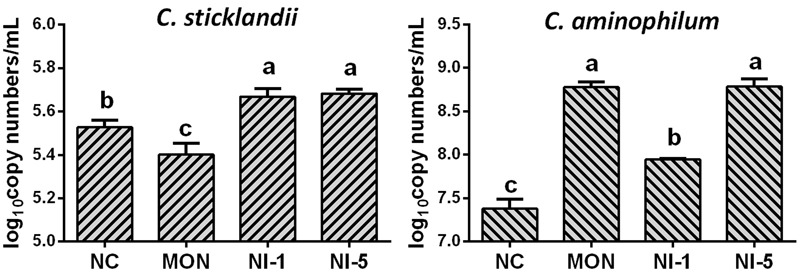
Effects of nisin and monensin addition on the population of Clostridium sticklandii and C. aminophilum (log10 copy number of 16S rRNA genes/mL) in the in vitro rumen mixed cultures. Values are means ± SE (n = 4). NC = negative control (no additives); MON = monensin, 5 μM; NI1 = nisin, 1 μM; NI5 = nisin, 5 μM. Bars with different letters (a, b, c) represent different means (P < 0.05).
Effects of Nisin and Monensin on Rumen Bacterial Communities
The effects of nisin and MON addition on the alpha diversity measurements of the ruminal bacterial community are summarized in Table 2. Across all 16 samples from the four treatments, 470,988 quality-checked sequences were classified as being bacterial. On average, at least 26,999 sequences per sample were obtained for all the treatments. The average length of the sequences was 448 bp. Greater than 99% depth coverage was achieved for all the samples. Both MON and nisin significantly reduced Shannon diversity index and increased the Simpson index compared with NC (P < 0.05), but the reduction or increase magnitudes were greater for MON than for nisin (P < 0.05). The numbers of OTUs were reduced by MON and NI5 (P < 0.05) but unaffected by NI1 (P > 0.05). The ACE and Chao 1 estimates of richness were not influenced by MON or nisin (P > 0.05).
Table 2.
Effects of nisin and monensin addition on the alpha diversity measurements of ruminal bacteria at 3% dissimilarity level.
| Item | NC | MON | NI1 | NI5 | SEM | P-value |
|---|---|---|---|---|---|---|
| # of sequences | 26,999 | 29,585 | 29,205 | 31,958 | 2,255 | 0.51 |
| Coverage (%) | 99.6 | 99.6 | 99.6 | 99.5 | 0.04 | 0.57 |
| OTU | 665a | 612b | 644ab | 613b | 13.8 | 0.05 |
| ACE | 747 | 716 | 722 | 725 | 9.2 | 0.15 |
| Chao 1 | 748 | 712 | 726 | 721 | 10.7 | 0.16 |
| Shannon index | 4.40a | 3.71d | 4.27b | 4.03c | 0.036 | <0.01 |
| Simpson index | 0.039c | 0.067a | 0.041c | 0.050b | 0.002 | <0.01 |
NC = negative control (no additives); MON = monensin, 5 μM; NI1 = nisin, 1 μM; NI5 = nisin, 5 μM; OTU = operational taxonomic units; ACE = abundance-based coverage estimator.
a-dMeans within a row with different superscripts differ (P < 0.05).
In order to understand the impacts of MON and nisin on the overall rumen bacterial community, PCA analysis was performed (Figure 3). A clear separation was seen between NC and MON along PC1, which explains >67% of total variation, while NI1 and NI5 were separated from NC and MON along PC2, which explains >21% of total variation. The separation between NI1 and NI5 was minimal.
FIGURE 3.
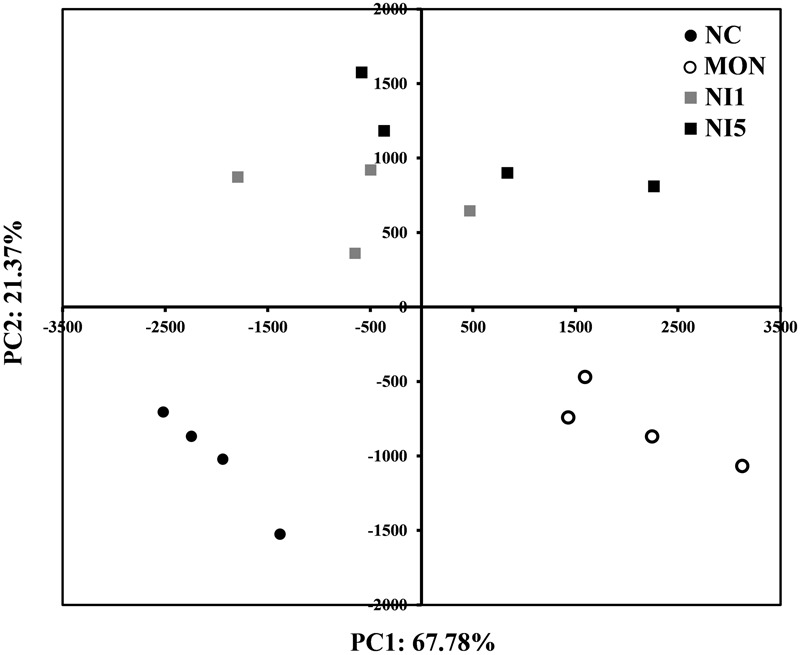
Principal component analysis (PCA) analysis of all samples based on the composition of bacterial communities. NC = negative control (no additives); MON = monensin, 5 μM; NI1 = nisin, 1 μM; NI5 = nisin, 5 μM.
There were 13 bacteria phyla identified among all the treatments, with Proteobacteria, Firmicutes, and Bacteroidetes being the most predominant phyla, representing 39.3–56.8%, 25.9–30.2%, and 14.3–25.2% of all the sequences, respectively (Table 3). The relative abundance of the bacteria phyla was altered differently by the treatments (Table 3). Both MON and nisin decreased the relative abundance of Firmicutes compared with NC (P < 0.05), with MON corresponding to a greater decrease than nisin (P < 0.05). The relative abundance of Bacteroidetes and Tenericutes was significantly lower for MON and NI5 than for NC (P < 0.05), with the lowest observed for MON. On the contrary, the relative abundance of Proteobacteria was significantly higher for MON and NI5 than for NC (P < 0.05), with the highest observed for MON. The relative abundance of Spirochaetae was decreased by MON (P < 0.05) but not influenced by nisin (P > 0.05). The relative abundance of Fibrobacteres was decreased by MON and NI5 (P < 0.05) but increased by NI1 (P < 0.05).
Table 3.
Effects of nisin and monensin addition on relative abundance of ruminal bacteria at phylum and genus level that each accounted for ≥0.5% of total sequences in at least one treatment.
| Phylum | Genus/other | Relative abundance (%) |
SEM | P-value | |||
|---|---|---|---|---|---|---|---|
| NC | MON | NI1 | NI5 | ||||
| Proteobacteria | Total | 39.3c | 56.8a | 41.1c | 46.4b | 0.84 | <0.01 |
| Ruminobacter | 30.2b | 35.1a | 25.6c | 25.3c | 0.83 | <0.01 | |
| Succinivibrio | 6.02d | 13.66b | 11.38c | 14.56a | 0.248 | <0.01 | |
| Succinimonas | 1.42c | 1.95a | 1.58bc | 1.80ab | 0.079 | <0.01 | |
| Unclassified Succinivibrionaceae | 1.47c | 5.49a | 2.09c | 4.18b | 0.245 | <0.01 | |
| Firmicutes | Total | 30.2a | 25.9c | 27.0c | 28.5b | 0.41 | <0.01 |
| Pseudobutyrivibrio | 3.82c | 1.30d | 6.44a | 4.98b | 0.109 | <0.01 | |
| Ruminococcus | 2.56a | 0.54b | 0.43b | 0.38b | 0.053 | <0.01 | |
| Oribacterium | 2.81c | 9.46a | 5.78b | 9.11a | 0.220 | <0.01 | |
| Butyrivibrio | 1.83b | 0.79d | 2.44a | 1.35c | 0.064 | <0.01 | |
| Streptococcus | 1.61a | 0.83bc | 0.68c | 0.75b | 0.033 | <0.01 | |
| Selenomonas | 1.14c | 3.68a | 2.45b | 3.96a | 0.126 | <0.01 | |
| Roseburia | 0.69b | 0.22d | 1.00a | 0.47c | 0.027 | <0.01 | |
| Anaerosporobacter | 0.71a | 0.16b | 0.17b | 0.17b | 0.021 | <0.01 | |
| Succiniclasticum | 0.69b | 1.84a | 0.96b | 1.05b | 0.157 | <0.01 | |
| Unclassified Lachnospiraceae | 7.40a | 3.19b | 3.08b | 3.09b | 0.063 | <0.01 | |
| Unclassified Christensenellaceae | 4.08a | 1.35b | 1.53b | 1.35b | 0.099 | <0.01 | |
| Unclassified Ruminococcaceae | 1.24a | 0.58c | 0.88b | 0.56c | 0.039 | <0.01 | |
| Bacteroidetes | Total | 25.2a | 14.3c | 27.0a | 21.1b | 0.68 | <0.01 |
| Prevotella | 16.30a | 9.73b | 17.30a | 11.00b | 0.806 | <0.01 | |
| Bacteroides | 0.34c | 0.99a | 0.39c | 0.63b | 0.028 | <0.01 | |
| Unclassified Rikenellaceae | 5.93b | 2.29c | 6.40ab | 7.04a | 0.322 | <0.01 | |
| Unclassified Bacteroidales | 1.80a | 0.61b | 2.14a | 1.73a | 0.127 | <0.01 | |
| Unclassified Prevotellaceae | 0.84 | 0.68 | 0.76 | 0.68 | 0.047 | 0.09 | |
| Spirochaetae | Total | 2.41a | 0.59b | 1.75a | 2.00a | 0.210 | <0.01 |
| Treponema | 2.25a | 0.58c | 1.61b | 1.93ab | 0.192 | <0.01 | |
| Fibrobacteres | Total | 0.64b | 0.07d | 1.18a | 0.43c | 0.048 | <0.01 |
| Fibrobacter | 0.64b | 0.07d | 1.18a | 0.43c | 0.048 | <0.01 | |
| Lentisphaerae | Total | 0.78 | 0.79 | 0.57 | 0.64 | 0.097 | 0.34 |
| Unclassified Lentisphaerae | 0.67 | 0.68 | 0.47 | 0.54 | 0.087 | 0.28 | |
| Tenericutes | Total | 0.85a | 0.13c | 0.96a | 0.40b | 0.086 | <0.01 |
| Anaeroplasma | 0.76a | 0.09c | 0.80a | 0.32b | 0.068 | <0.01 | |
NC = negative control (no additives); MON = monensin, 5 μM; NI1 = nisin, 1 μM; NI5 = nisin, 5 μM.
a-dMeans within a row with different superscripts differ (P < 0.05).
The bacteria that were affected differently by monensin and nisin are bolded.
A total of 69 genera of bacteria were identified, and these genera together accounted for 73.6–84.7 % of all the sequences. Only 17 of these genera were each represented by more than 0.50 % of the total sequences in at least one treatment (Table 3), and they were regarded as the “major genera”. Among the major genera, Ruminobacter was the most predominant accounting for 25.3–35.1 % of the total sequences, followed by Prevotella (9.73–17.30 %), Succinivibrio (6.02–14.56 %), and Pseudobutyrivibrio (1.30–6.44 %). In addition, a large portion (15.3–26.4 %) of the sequences could not be classified to a known genus. Unclassified Lachnospiraceae (3.08–7.40 %) and unclassified Rikenellaceae (2.29–7.04 %) were the first and the second most predominant unclassified groups, respectively. At the genus level, MON and nisin showed different effects on some bacterial genera. Compared with NC, the relative abundance of Ruminobacter was increased by MON but decreased by nisin irrespective of concentration (P < 0.05). On the contrary, the relative abundance of Pseudobutyrivibrio and unclassified Rikenellaceae was decreased by MON but increased by nisin at both concentrations (P < 0.05). The relative abundance of unclassified Bacteroidales was decreased by MON (P < 0.05) but not influenced by nisin (P > 0.05). The relative abundance of Butyrivibrio, Roseburia, and Fibrobacter was decreased by both MON and NI5 but increased by NI1 (P < 0.05). In contrast, the relative abundance of Prevotella and Anaeroplasma was decreased by MON and NI5 (P < 0.05) but not influenced by NI1 (P > 0.05). Besides, MON and nisin also had a parallel influence on some bacterial genera. The relative abundance of Succinivibrio, Succinimonas, Oribacterium, Selenomonas, and unclassified Succinivibrionaceae was increased, whereas that of Ruminococcus, Streptococcus, Anaerospoobacter, Treponema, unclassified Lachnospiraceae, unclassified Christensenellaceae, and unclassified Ruminococcaceae was decreased by MON and nisin at both concentrations (P < 0.05).
A total of 838 OTUs were clustered at a 0.03 dissimilarity cut-off across all the samples, and 46 OTUs were represented by more than 0.50 % of the total sequences in at least one treatment (Supplementary Table S2). Among these, eight OTUs were decreased (P < 0.05), six OTUs were increased (P < 0.05), and two OTUs were not influenced (P > 0.05) by MON and nisin at both concentrations. However, MON and nisin had different effects on the relative abundance of most OTUs. Compared with NC, four OTUs were decreased by MON (P < 0.05) but increased by nisin irrespective of concentration (P < 0.05). Five OTUs were increased by MON (P < 0.05) but decreased (P < 0.05) or unchanged (P > 0.05) by nisin at both concentrations. One OTU was unaffected by MON (P > 0.05) but decreased by NI1 and NI5 (P < 0.05). Furthermore, nisin also showed significant dosage effect on 20 OTUs compared with MON.
Correlations Between the Relative Abundance of Rumen Bacteria and Fermentation Parameters
The relative abundance of some of the identified rumen bacterial genera or equivalent taxa (referred to as genera) appeared to be correlated with fermentation characteristics (Figure 4). Seven genera were positively and five taxa were negatively correlated with methane production; 11 taxa were positively and seven taxa were negatively correlated with DMD; three taxa were positively and five taxa were negatively correlated with ammonia concentrations; 10 taxa were positively and seven taxa were negatively correlated with total VFA concentrations; seven taxa were positively and five taxa were negatively correlated with acetate concentrations; four taxa were positively and seven taxa were negatively correlated with propionate concentrations; 14 taxa were positively and seven taxa were negatively correlated with butyrate concentrations; and seven taxa were positively and four taxa were negatively correlated with total BCVFA concentration.
FIGURE 4.
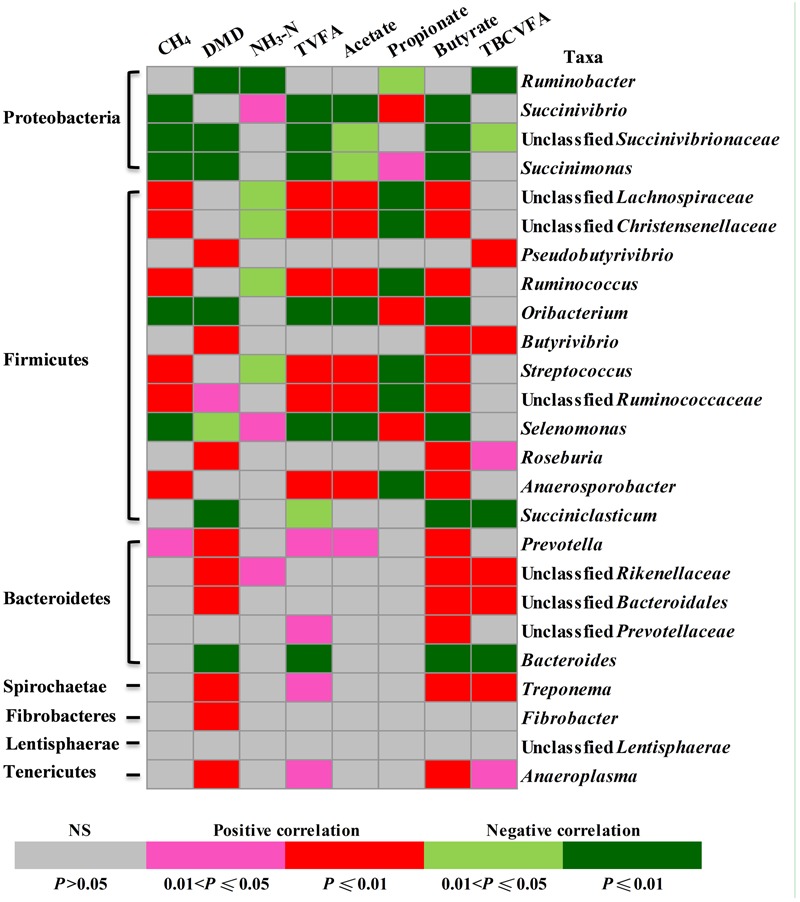
Correlations between the relative abundance of rumen bacteria (at genus level) and fermentation parameters. Cells are colored based on Pearson correlation coefficient.
Discussion
Microbes are solely responsible for feed digestion and the production of methane, VFA, and ammonia in the rumen (Patra and Yu, 2014). Previous studies have demonstrated that MON, an ionophore, and nisin, a bacteriocin, could achieve similar impacts on methane and VFA production (Callaway et al., 1997; Shen et al., 2016) but showed different effects on feed digestion (Shen et al., 2016). Therefore, comprehensive characterization of microbial populations and communities are essential to understand the mode of effects of nisin and monensin on feed digestion and rumen fermentation. This study, combining high-throughput sequencing and quantitative real-time PCR using an in vitro fermentation system, for the first time revealed the different effects on bacterial groups. Moreover, this study also shines new light on the potency of nisin as an alternative to monensin in modulating rumen fermentation.
Effects of Nisin and MON on the Major Microbial Groups and Bacterial Community Involved in Feed Digestion
As expected, monensin decreased feed digestion, consistent with previous studies (Russell and Strobel, 1988; Narvaez et al., 2013). Cellulolytic bacteria in the rumen are the major contributors to fiber degradation (Jeyanathan et al., 2014). The adverse effect of monensin on feed digestion is mainly attributed to its inhibition of cellulolytic bacteria (Russell and Strobel, 1989; Narvaez et al., 2013). F. Succinogenes, R. flavefaciens and R. albus are considered the major cellulolytic bacterial species cultured because of their high cellulose digestion ability (Krause et al., 2003). Butyrivibrio, Pseudobutyrivibrio, Oscillibacter, and Eubacterium are also recognized as fibrolytic bacterial genera (Thoetkiattikul et al., 2013). However, a meta-analysis by Koike and Kobayashi (2009) revealed that the so far recognized fibrolytic species might represent only a small proportion of the total fibrolytic population in the rumen. Indeed, a recent study reported that high abundance of some unclassified groups, including those assigned to Lachnospiraceae, Christensenellaceae, Ruminococcaceae, Rikenellaceae, Prevotellaceae, and Bacteroidales, have been found tightly adhering to forages after incubation in the rumen, indicating that these new taxa may play an important role in forage degradation in the rumen (Liu et al., 2016). In the present study, the reduced abundance of fibrolytic bacterial genera (e.g., Ruminococcus, Butyrivibrio, Pseudobutyrivibrio, and Fibrobacter) and potential fibrolytic bacteria taxa (e.g., unclassified Lachnospiraceae, unclassified Christensenellaceae, unclassified Ruminococcaceae, unclassified Rikenellaceae, and unclassified Bacteroidales) might have resulted in the decreased feed digestion in the monensin treatment. In addition, fungi and protozoa are known to contribute to fiber degradation (Martinez-Fernandez et al., 2016). Hence, the reduced fungi and protozoa populations in the MON treatment may be another reason for the decreased feed digestion therein.
The monensin addition decreased feed digestion with a concomitant reduction in methane production, which is consistent with the notion that inhibition of methanogenesis generally results in decreased feed digestion and fermentation in batch cultures (Ungerfeld, 2015). However, unaffected fiber digestibility coupled with reduced methane production in several in vitro experiments has also been reported (Morgavi et al., 2010), and the authors believed that the increased Fibrobacter population observed in these studies might have compensated fiber degradation. This phenomenon has also been found in the nisin treatments in the present study, in which methane was decreased greatly, while feed digestion was unaffected. This may be explained by shifts in microbial populations. In the nisin treatments, the relative abundance of some Gram-positive fibrolytic bacterial genera, such as Ruminococcus, unclassified Lachnospiraceae, unclassified Christensenellaceae, and unclassified Ruminococcaceae was decreased. However, besides Fibrobacter as mentioned by Morgavi et al. (2010), nisin treatment increased or maintained high relative abundance of some other fibrolytic bacterial genera, such as Pseudobutyrivibrio, Butyrivibrio, unclassified Rikenellaceae, and unclassified Bacteroidales, all of which were decreased by MON. These functionally not yet well recognized minor groups may also be important fibrolytic bacteria and contribute to the different impacts between nisin and MON. Moreover, the population of protozoa was also unaffected by nisin. Therefore, the increase or maintenance of these fibrolytic microbes in the nisin treatments might have allowed persistent efficient fiber degradation. These data suggested that the different effects of monensin and nisin on microbial community, particularly fibrolytic bacterial composition might have resulted in their discrepancy on feed digestion.
Effects of Nisin and MON on Methane Production and Related Changes in Major Microbial Groups and Bacterial Community
Corroborating the findings of several previous studies (Callaway et al., 1997; Shen et al., 2016), the present study also showed reduced methane production by nisin and monensin, with reduction by 38.7%, 54.9%, and 38.1% in the NI1, NI5, and MON treatments, respectively. However, the mechanism of action on methane production is probably different between MON and nisin. In the present study, both MON and nisin significantly reduced the population of methanogens. Narvaez et al. (2013) also found reduced methanogens population with monensin addition. However, it has been demonstrated that monensin does not directly inhibit methanogen, and it can directly inhibit H2-producing bacteria, thereby indirectly decreasing methane production (Russell and Houlihan, 2003). Supporting this notion, the present study also showed a decrease in H2-producing microorganisms, including protozoa, fungi, and Gram-positive taxa of Firmicutes, to which the primary ruminal H2-producing bacteria belong. Different with our in vitro result, one recently published in vivo study showed no effect of monensin on archaea population (Schären et al., 2017). Undoubtedly, their results further confirmed that the inhibition of methanogenesis by monensin is most likely caused by a decrease in substrate availability, rather than by direct inhibition to methanogens. In contrast, nisin has no influence on protozoa, which play an important role contributing to methane production (Morgavi et al., 2010). It should be noted that nisin also inhibited fungi and some Gram-positive taxa of Firmicutes, but to a less extent than monensin. Hence, the observed decrease in methane production by nisin is probably partly due to indirect inhibition to H2-producers. Besides, as proposed but not substantiated by Santoso et al. (2004), nisin probably reduced methane production by direct inhibition to methanogens. However, this premise can not be confirmed directly by the present study either. Therefore, future studies using pure cultures of methanogens are warranted to directly and definitively approve if nisin can directly inhibit methane production.
Effects of Nisin and MON on VFA Production and Related Changes in Rumen Microbial Composition
In the present study, both monensin and nisin additions caused an increase of propionate concentration but a decrease of acetate concentration, resulting in a decrease in the ratio of acetate to propionate. These results are consistent with those of previous in vitro studies (Callaway et al., 1997; Shen et al., 2016). In the rumen, propionate is produced through two pathways: the succinate pathway and the acrylate pathway, and the succinate pathway is the major pathway (Jeyanathan et al., 2014). Succinate is formed as an intermediate but not as an end product of the rumen fermentation (Scheifinger and Wolin, 1973). Fibrobacter succinogenes (Jeyanathan et al., 2014) and members of the Succinivibrio (Pope et al., 2011) produce succinate as their principal fermentation end product, while members of Selenomonas (e.g., Selenomonas ruminantium; Scheifinger and Wolin, 1973) produce propionate via the succinate pathway. In the present study, the abundance of Fibrobacter was much less than that of Succininivibrio. Therefore, the greatly increased relative abundance of Succinivibrio (>11.38%) and Selenomonas (>2.45%) in the MON and the nisin treatments probably contributed to increased propionate via the succinate pathway. Similar to our results, an in vivo study also found an increase in ruminal propionate proportions after monensin addition, which was caused by an increase in abundance of succinate and propionate producers and a decrease in non-producers (Schären et al., 2017). In addition, the acrylate pathway is also an important propionate-producing pathway in the rumen, in which lactate-producing bacteria such as Streptococcus bovis play a key regulatory role (Jeyanathan et al., 2014). However, the relative abundance of Streptococcus in the MON and the nisin treatments was reduced significantly. Therefore, propionate production through the acrylate pathway was probably weakened by both MON and nisin. These results suggest that monensin and nisin may increase propionate production through the succinate pathway.
The reduced acetate production in the MON and the nisin treatments can be explained by the decrease of some Gram-positive fibrolytic bacteria, such as Ruminococcus spp., which are major acetate-producing bacteria (Jeyanathan et al., 2014). In addition, the present study also revealed that some unclassified Gram-positive bacteria, such as unclassified bacteria in Lachnospiraceae, Christensenellaceae, and Ruminococcaceae, were positively correlated with acetate concentration. Thus, decreased populations of these three unclassified groups might have also contributed to the reduction in acetate production in the MON and the nisin treatments. However, nisin increased propionate production and reduced acetate production to a greater magnitude than monensin. Nisin also increased butyrate concentration to a greater extent than monensin. Protozoa were among the major butyrate producers in the rumen (Williams and Coleman, 1997), and they were positively associated with butyrate production (Mao et al., 2015a). In the present study, protozoa were decreased by monensin but not affected by nisin. Hence, the different effect on protozoal population may partly explain the higher butyrate concentration in the nisin treatment than in the MON treatment. This study observed that the relative abundance of Butyrivibrio and Pseudobutyrivibrio in the nisin treatments was significantly higher than in the MON treatment. B. fibrisolvens (Stewart et al., 1997) and Pseudobutyrivibrio xylanivorans (Kopecny et al., 2003) are important butyrate-producing species in the rumen. Therefore, the higher relative abundance of butyrate-producing bacteria in the nisin treatments may further explain the higher butyrate concentration therein than in MON treatment.
Effects of Nisin and MON on Ammonia Production and Related Changes in Some Important Microbial Groups
Ammonia concentration in ruminal in vitro batch cultures only depends on the balance between the rate of formation and utilization of ammonia by microbes. In the present study, microbial crude protein (MCP) was not measured, but the decreased bacterial populations in the nisin and the MON treatments suggested reduced ammonia utilization by microbes. Therefore, increased ammonia concentration should be expected if the formation rate was not decreased. However, the MON treatment had similar ammonia concentration as the control. It is speculative, but this may result from reduced amino acid deamination by microbes, which has been reported in earlier studies (Yang and Russell, 1993; Callaway et al., 1997). This is substantiated by the lower concentrations of BCVFA and valerate, both of which primarily arise from deamination of amino acids (Patra and Yu, 2014). Besides, we also quantified the population of protozoa, which are known protein degraders and net ammonia-producer (Firkins et al., 2007), and that of C. aminophilum and C. sticklandii, both of which are HAB (Russell et al., 1988). The reduced population of protozoa and C. sticklandii by monensin corroborates the reduced total amino acid deamination further. However, C. aminophilum increased in response to monensin, a finding contradictory to a report by Callaway et al. (1997), who found that C. aminophilum in pure cultures was inhibited by monensin though it has greater resistance than C. sticklandii. In contrast, the nisin treatments had similar total BCVFA and valerate concentrations as the control, suggesting enhanced amino acid deamination per unit of bacterial biomass. This was substantiated by the increased C. sticklandii and C. aminophilum populations, though protozoa were unaffected. Hence, the reduced ammonia utilization and enhanced amino acid deamination per unit of bacterial biomass might have contributed to the increased ammonia concentration in the nisin treatments. In an in vitro study, Sar et al. (2005) also found increased ammonia concentration after nisin addition. Contradictory to our results, Callaway et al. (1997) reported that nisin inhibited amino acid degradation by the above two HAB, and nisin was more effective than monensin in inhibiting the growth of C. aminophilum. The discrepancy in the effects of monensin and nisin on HAB populations between pure cultures and mixed cultures cannot be easily explained, but these (and possibly other) HAB may respond to them differently in mixed cultures than in pure cultures.
Alteration in Rumen Microbial Fermentation Profiles Correlated with Changes in Bacterial Community
Feed degradability, methane production, VFA profiles, and ammonia production in the rumen are some of the most important parameters indicative of microbial metabolism therein (Patra and Yu, 2015). To explore the correlations between rumen bacterial shifts (at genus level) and fermentation characteristics, Pearson’s correlation analysis was performed. This study observed a large portion of unclassified bacteria, including those unclassified within Lachnospiraceae, Rikenellaceae, Christensenellaceae, Ruminococcaceae, and Bacteroidales, which have not yet been functionally characterized. The correlation analysis revealed that the dynamic changes of these unclassified groups in response to nisin and monensin are probably responsible for or caused by, at least partially, the shifts in these fermentation characteristics. However, correlation is not causation, and a particular taxon found in association with a parameter may be merely a bystander (Hanage, 2014). It should also be noted that the correlation analysis in this study was based on the combined datasets, and lack of significant correlation between some bacterial taxa and fermentation characteristics do not necessarily mean those bacterial taxa are not important. Furthermore, as revealed by Shabat et al. (2016), the functional characteristics of a small number of species can also have a large impact on community structure and ecosystem functioning. Therefore, as recommended by other researchers (Hanage, 2014; Patra and Yu, 2015), much work is still needed to confirm these correlations and to determine their causality.
Conclusion
Nisin had greater effects than monensin in inhibiting methane production and decreasing acetate-propionate ratio. Most importantly, nisin decreased methane production without decreasing feed digestion, which is inhibited by monensin. The microbiota analysis revealed that nisin and monensin caused different alterations in the rumen microbiota and fermentation characteristics. These findings suggest that nisin can be more effective and practical than monensin in modulating rumen fermentation and mitigating methane emission, and nisin may be a potential alternative to monensin. However, future in vivo studies are needed to further validate nisin’s usefulness and efficacy in modifying rumen fermentation.
Author Contributions
WZ and JS conceived and designed the experiments. JS and ZL performed the experiments. JS and ZY analyzed the data. JS wrote the paper. ZY and WZ revised the paper. All authors agree to be accountable for all aspects of the work.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National Natural Science Foundation of China for Young Investigators (31402101), Jiangsu Provincial Natural Science Foundation for Young Investigators (BK20140696), and Fundamental Research Funds for the Central Universities (Y020150023).
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01111/full#supplementary-material
References
- Bergen W. G., Bates D. B. (1984). Ionophores: their effect on production efficiency and mode of action. J. Anim. Sci. 58 1465–1483. 10.2527/jas1984.5861465x [DOI] [PubMed] [Google Scholar]
- Blümmel M., Steingab H., Becker K. (1997). The relationship between in vitro gas production, in vitro microbial biomass yield and N-15 incorporation and its implications for the prediction of voluntary feed intake of roughages. Br. J. Nutr. 77 911–921. 10.1079/Bjn19970089 [DOI] [PubMed] [Google Scholar]
- Callaway T. R., Carneiro De Melo A. M., Russell J. B. (1997). The effect of nisin and monensin on ruminal fermentations in vitro. Curr. Microbiol. 35 90–96. 10.1007/s002849900218 [DOI] [PubMed] [Google Scholar]
- Caporaso J. G., Bittinger K., Bushman F. D., DeSantis T. Z., Andersen G. L., Knight R. (2010a). PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26 266–267. 10.1093/bioinformatics/btp636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., et al. (2010b). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7 335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaney A. L., Marbach E. P. (1962). Modified reagents for determination of urea and ammonia. Clin. Chem. 8 130–132. [PubMed] [Google Scholar]
- Cotter P. D., Ross R. P., Hill C. (2013). Bacteriocins - a viable alternative to antibiotics? Nat. Rev. Microbiol. 11 95–105. 10.1038/nrmicro2937 [DOI] [PubMed] [Google Scholar]
- Dai Z. L., Zhang J., Wu G. Y., Zhu W. Y. (2010). Utilization of amino acids by bacteria from the pig small intestine. Amino Acids 39 1201–1215. 10.1007/s00726-010-0556-9 [DOI] [PubMed] [Google Scholar]
- Delves-Broughton J., Blackburn P., Evans R. J., Hugenholtz J. (1996). Applications of the bacteriocin, nisin. Antonie Van Leeuwenhoek 69 193–202. 10.1007/Bf00399424 [DOI] [PubMed] [Google Scholar]
- Denman S. E., Mcsweeney C. S. (2006). Development of a real-time PCR assay for monitoring anaerobic fungal and cellulolytic bacterial populations within the rumen. FEMS Microbiol. Ecol. 58 572–582. 10.1111/j.1574-6941.2006.00190.x [DOI] [PubMed] [Google Scholar]
- Denman S. E., Tomkins N., Mcsweeney C. S. (2007). Quantitation and diversity analysis of ruminal methanogenic populations in response to the antimethanogenic compound bromochloromethane. FEMS Microbiol. Ecol. 62 313–322. 10.1111/j.1574-6941.2007.00394.x [DOI] [PubMed] [Google Scholar]
- DeSantis T. Z., Hugenholtz P., Larsen N., Rojas M., Brodie E. L., Keller K., et al. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72 5069–5072. 10.1128/AEM.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffield T. F., Rabiee A. R., Lean I. J. (2008). A meta-analysis of the impact of monensin in lactating dairy cattle. Part 2. Production effects. J. Dairy Sci. 91 1347–1360. 10.3168/jds.2007-0608 [DOI] [PubMed] [Google Scholar]
- Edgar R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26 2460–2461. 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- FAO (2006). Livestock’s Long Shadow: Environmental Issues and Options. Rome: Food & Agriculture Organization. [Google Scholar]
- Firkins J. L., Yu Z., Morrison M. (2007). Ruminal nitrogen metabolism: perspectives for integration of microbiology and nutrition for dairy. J. Dairy Sci. 90 E1–E16. 10.3168/jds.2006-518 [DOI] [PubMed] [Google Scholar]
- Franz C., Baser K. H. C., Windisch W. (2010). Essential oils and aromatic plants in animal feeding – a European perspective. A review. Flavour Fragr. J. 25 327–340. 10.1002/ffj.1967 [DOI] [Google Scholar]
- Guan H., Wittenberg K. M., Ominski K. H., Krause D. O. (2006). Efficacy of ionophores in cattle diets for mitigation of enteric methane. J. Anim. Sci. 84 1896–1906. 10.2527/jas.2005-652 [DOI] [PubMed] [Google Scholar]
- Hanage W. P. (2014). Microbiome science needs a healthy dose of scepticism. Nature 512 247–248. 10.1038/512247a [DOI] [PubMed] [Google Scholar]
- Jeyanathan J., Martin C., Morgavi D. P. (2014). The use of direct-fed microbials for mitigation of ruminant methane emissions: a review. Animal 8 250–261. 10.1017/S1751731113002085 [DOI] [PubMed] [Google Scholar]
- Johnson K. A., Johnson D. E. (1995). Methane Emissions from Cattle. J. Anim. Sci. 73 2483–2492. 10.2527/1995.7382483x [DOI] [PubMed] [Google Scholar]
- Jüzefiak D., Kieronczyk B., Juskiewicz J., Zdunczyk Z., Rawski M., Dlugosz J., et al. (2013). Dietary nisin modulates the gastrointestinal microbial ecology and enhances growth performance of the broiler chickens. PLoS ONE 8:e85347 10.1371/journal.pone.0085347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike S., Kobayashi Y. (2009). Fibrolytic rumen bacteria: their ecology and functions. Asian Australas. J. Anim. 22 131–138. 10.5713/ajas.2009.r.01 [DOI] [Google Scholar]
- Koike S., Yabuki H., Kobayashi Y. (2007). Validation and application of real-time polymerase chain reaction assays for representative rumen bacteria. Anim. Sci. J. 78 135–141. 10.1111/j.1740-0929.2007.00417.x [DOI] [Google Scholar]
- Kopecny J., Zorec M., Mrazek J., Kobayashi Y., Marinsek-Logar R. (2003). Butyrivibrio hungatei sp nov and Pseudobutyrivibrio xylanivorans sp nov., butyrate-producing bacteria from the rumen. Int. J. Syst. Evol. Micrbiol. 53 201–209. 10.1099/ijs.0.02345-0 [DOI] [PubMed] [Google Scholar]
- Krause D. O., Denman S. E., Mackie R. I., Morrison M., Rae A. L., Attwood G. T., et al. (2003). Opportunities to improve fiber degradation in the rumen: microbiology, ecology, and genomics. FEMS Microbiol. Rev. 27 663–693. 10.1016/S0168-6445(03)00072-X [DOI] [PubMed] [Google Scholar]
- Lee S. S., Mantovani H. C., Russell J. B. (2002). The binding and degradation of nisin by mixed ruminal bacteria. FEMS Microbiol. Ecol. 42 339–345. 10.1111/J.1574-6941.2002.Tb01023.X [DOI] [PubMed] [Google Scholar]
- Liu J. H., Zhang M. L., Xue C. X., Zhu W. Y., Mao S. Y. (2016). Characterization and comparison of the temporal dynamics of ruminal bacterial microbiota colonizing rice straw and alfalfa hay within ruminants. J. Dairy Sci. 99 9668–9681. 10.3168/jds.2016-11398 [DOI] [PubMed] [Google Scholar]
- Mao S. Y., Huo W. J., Zhu W. Y. (2015a). Microbiome-metabolome analysis reveals unhealthy alterations in the composition and metabolism of ruminal microbiota with increasing dietary grain in a goat model. Environ. Microbiol. 18 525–541. 10.1111/1462-2920.12724 [DOI] [PubMed] [Google Scholar]
- Mao S. Y., Zhang M. L., Liu J. H., Zhu W. Y. (2015b). Characterising the bacterial microbiota across the gastrointestinal tracts of dairy cattle: membership and potential function. Sci. Rep. 5:15116 10.1038/srep16116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao S. Y., Zhang G., Zhu W. Y. (2008). Effect of disodium fumarate on ruminal metabolism and rumen bacterial communities as revealed by denaturing gradient gel electrophoresis analysis of 16S ribosomal DNA. Anim. Feed Sci. Technol. 140 293–306. 10.1016/j.anifeedsci.2007.04.001 [DOI] [Google Scholar]
- Martinez-Fernandez G., Denman S. E., Yang C., Cheung J., Mitsumori M., Mcsweeney C. S. (2016). Methane inhibition alters the microbial community, hydrogen flow, and fermentation response in the rumen of cattle. Front. Microbiol. 7:1122 10.3389/fmicb.2016.01122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina A., Molina M. P., Althaus R. L., Gallego L. (2003). Residue persistence in sheep milk following antibiotic therapy. Vet. J. 165 84–89. 10.1016/S1090-0233(02)00173-9 [DOI] [PubMed] [Google Scholar]
- Morgavi D. P., Forano E., Martin C., Newbold C. J. (2010). Microbial ecosystem and methanogenesis in ruminants. Animal 4 1024–1036. 10.1017/S1751731110000546 [DOI] [PubMed] [Google Scholar]
- Narvaez N., Wang Y., Mcallister T. (2013). Effects of extracts of Humulus lupulus (hops) and Yucca schidigera applied alone or in combination with monensin on rumen fermentation and microbial populations in vitro. J. Sci. Food Agric. 93 2517–2522. 10.1002/jsfa.6068 [DOI] [PubMed] [Google Scholar]
- Paiva A. D., Breukink E. (2013). Antimicrobial Peptides and Innate Immunity. Basel: Springer Basel. [Google Scholar]
- Patra A. K., Yu Z. (2014). Effects of vanillin, quillaja saponin, and essential oils on in vitro fermentation and protein-degrading microorganisms of the rumen. Appl. Microbiol. Biotechnol. 98 897–905. 10.1007/s00253-013-4930-x [DOI] [PubMed] [Google Scholar]
- Patra A. K., Yu Z. T. (2015). Essential oils affect populations of some rumen bacteria in vitro as revealed by microarray (RumenBactArray) analysis. Front. Microbiol. 6:297 10.3389/fmicb.2015.00297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope P. B., Smith W., Denman S. E., Tringe S. G., Barry K., Hugenholtz P., et al. (2011). Isolation of Succinivibrionaceae implicated in low methane emissions from Tammar Wallabies. Science 333 646–648. 10.1126/science.1205760 [DOI] [PubMed] [Google Scholar]
- Price M. N., Dehal P. S., Arkin A. P. (2009). FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26 1641–1650. 10.1093/molbev/msp077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell J. B., Houlihan A. J. (2003). Ionophore resistance of ruminal bacteria and its potential impact on human health. FEMS Microbiol. Rev. 27 65–74. 10.1016/S0168-6445(03)00019-6 [DOI] [PubMed] [Google Scholar]
- Russell J. B., Mantovani H. C. (2002). The bacteriocins of ruminal bacteria and their potential as an alternative to antibiotics. J. Mol. Microbiol. Biotechnol. 4 347–355. [PubMed] [Google Scholar]
- Russell J. B., Strobel H. J. (1988). Effects of additives on in vitro ruminal fermentation: a comparison of monensin and bacitracin, another gram-positive antibiotic. J. Anim. Sci. 66 552–558. 10.2527/jas1988.662552x [DOI] [PubMed] [Google Scholar]
- Russell J. B., Strobel H. J. (1989). Effect of ionophores on ruminal fermentation. Appl. Environ. Microbiol. 55 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell J. B., Strobel H. J., Chen G. J. (1988). Enrichment and isolation of a ruminal bacterium with a very high specific activity of ammonia production. Appl. Environ. Microbiol. 54 872–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoso B., Mwenya B., Sar C., Gamo Y., Kobayashi T., Morikawa R., et al. (2004). Effects of supplementing galacto-oligosaccharides, Yucca schidigera or nisin on rumen methanogenesis, nitrogen and energy metabolism in sheep. Livest. Prod. Sci. 91 209–217. 10.1016/j.livprodsci.2004.08.004 [DOI] [Google Scholar]
- Sar C., Mwenya B., Pen B., Morikawa R., Takaura K., Kobayashi T., et al. (2005). Effect of nisin on ruminal methane production and nitrate/nitrite reduction in vitro. Aust. J. Agric. Res. 56 803–810. 10.1071/AR04294 [DOI] [Google Scholar]
- Schären M., Drong C., Kiri K., Riede S., Gardener M., Meyer U., et al. (2017). Differential effects of monensin and a blend of essential oils on rumen microbiota composition of transition dairy cows. J. Dairy Sci. 100 2765–2783. 10.3168/jds.2016-11994 [DOI] [PubMed] [Google Scholar]
- Scheifinger C. C., Wolin M. J. (1973). Propionate formation from cellulose and soluble sugars by combined cultures of Bacteroides succinogenes and Selenomonas ruminantium. Appl. Microbiol. 26 789–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabat S. K., Sasson G., Doron-Faigenboim A., Durman T., Yaacoby S., Berg Miller M. E., et al. (2016). Specific microbiome-dependent mechanisms underlie the energy harvest efficiency of ruminants. ISME J. 10 2958–2972. 10.1038/ismej.2016.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J. S., Liu Z., Chen Y. Y., Lv P. A., Zhu W. Y. (2016). Effects of nisin on in vitro fermentation, methanogenesis and functional microbial populations of the rumen. Acta Microbiol. Sin. 56 1348–1357. 10.13343/j.cnki.wsxb.20150559 [DOI] [PubMed] [Google Scholar]
- Shin J. M., Gwak J. W., Kamarajan P., Fenno J. C., Rickard A. H., Kapila Y. L. (2016). Biomedical applications of nisin. J. Appl. Microbiol. 120 1449–1465. 10.1111/jam.13033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart C. S., Flint H. J., Bryant M. P. (1997). “The rumen bacteria,” in The Rumen Microbial Ecosystem 2 Edn eds Hobson P. N., Stewart C. S. (New York, NY: Springer; ) 10–72. 10.1007/978-94-009-1453-7_2 [DOI] [Google Scholar]
- Sylvester J. T., Karnati S. K., Yu Z., Morrison M., Firkins J. L. (2004). Development of an assay to quantify rumen ciliate protozoal biomass in cows using real-time PCR. J. Nutr. 134 3378–3384. [DOI] [PubMed] [Google Scholar]
- Theodorou M. K., Williams B. A., Dhanoa M. S., Mcallan A. B., France J. (1994). A simple gas-production method using a pressure transducer to determine the fermentation kinetics of ruminant feeds. Anim. Feed Sci. Technol. 48 185–197. 10.1016/0377-8401(94)90171-6 [DOI] [Google Scholar]
- Thoetkiattikul H., Mhuantong W., Laothanachareon T., Tangphatsornruang S., Pattarajinda V., Eurwilaichitr L., et al. (2013). Comparative analysis of microbial profiles in cow rumen fed with different dietary fiber by tagged 16S rRNA gene pyrosequencing. Curr. Microbiol. 67 130–137. 10.1007/s00284-013-0336-3 [DOI] [PubMed] [Google Scholar]
- Thornton P. K. (2010). Livestock production: recent trends, future prospects. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365 2853–2867. 10.1098/rstb.2010.0134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungerfeld E. M. (2015). Shifts in metabolic hydrogen sinks in the methanogenesis-inhibited ruminal fermentation: a meta-analysis. Front. Microbiol. 6:37 10.3389/fmicb.2015.00037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Garrity G. M., Tiedje J. M., Cole J. R. (2007). Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73 5261–5267. 10.1128/AEM.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams A. G., Coleman G. S. (1997). “The rumen protozoa,” in The Rumen Microbial Ecosystem eds Hobson P. N., Stewart C. S. (Dordrecht: Springer; ) 73–139. 10.1007/978-94-009-1453-7_3 [DOI] [Google Scholar]
- Yang C. J., Mao S. Y., Long L. M., Zhu W. Y. (2012). Effect of disodium fumarate on microbial abundance, ruminal fermentation and methane emission in goats under different forage: concentrate ratios. Animal 6 1788–1794. 10.1017/S1751731112000857 [DOI] [PubMed] [Google Scholar]
- Yang C. M. J., Russell J. B. (1993). The effect of monensin supplementation on ruminal ammonia accumulation in vivo and the numbers of amino acid-fermenting bacteria. J. Anim. Sci. 71 3470–3476. 10.2527/1993.71123470x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


