ABSTRACT
We have recently demonstrated that an mRNA capping enzyme, Cet1, impairs promoter-proximal accumulation/pausing of RNA polymerase II (Pol II) independently of its capping activity in Saccharomyces cerevisiae to control transcription. However, it is still unknown how Pol II pausing is regulated by Cet1. Here, we show that Cet1's N-terminal domain (NTD) promotes the recruitment of FACT (facilitates chromatin transcription that enhances the engagement of Pol II into transcriptional elongation) to the coding sequence of an active gene, ADH1, independently of mRNA-capping activity. Absence of Cet1's NTD decreases FACT targeting to ADH1 and consequently reduces the engagement of Pol II in transcriptional elongation, leading to promoter-proximal accumulation of Pol II. Similar results were also observed at other genes. Consistently, Cet1 interacts with FACT. Collectively, our results support the notion that Cet1's NTD promotes FACT targeting to the active gene independently of mRNA-capping activity in facilitating Pol II's engagement in transcriptional elongation, thus deciphering a novel regulatory pathway of gene expression.
KEYWORDS: Cet1, FACT, RNA polymerase II, transcription, mRNA capping
INTRODUCTION
RNA polymerase II (Pol II) pauses at the promoter-proximal regions in Drosophila and mammals to provide an additional layer of regulation of transcription. NELF (negative elongation factor) and DSIF (DRB sensitivity factor) play important roles in such pausing of Pol II (1, 2). DSIF alone does not pause Pol II (1, 2). Rather, it targets NELF to associate with Pol II for pausing (1, 2). DSIF is present in all eukaryotes and archaea and shares homology with a bacterial transcription factor, NusG (1, 3). However, NELF is conserved only in higher eukaryotes (1), and thus, the promoter-proximal pausing of Pol II is observed in higher eukaryotes. Such pausing of Pol II has emerged as an important regulatory step of transcription (1, 2). The dissociation of NELF releases paused Pol II for productive transcriptional elongation. P-TEFb, a kinase, triggers the dissociation of NELF via phosphorylation (1, 2). In addition, P-TEFb phosphorylates DSIF and serine-2 at the carboxy-terminal domain (CTD) of the Rpb1 subunit of Pol II (1, 2). Such phosphorylation of DSIF and Pol II has stimulatory effects on transcriptional elongation (1). Thus, P-TEFb performs a crucial function in releasing paused Pol II and enhances transcriptional elongation. The recruitment of P-TEFb to the gene may occur in several ways, such as interaction with DNA-binding proteins, like c-Myc and NF-κB, or association with mediator or Brd4 (which is, in turn, bound to the acetylated tail of histone H4) (1, 2, 4–7). Further, the amount and availability of active P-TEFb are regulated via sequestering of P-TEFb into an inactive complex with 7SK RNA and HEXIM protein (1, 2, 8, 9). Thus, various factors, including signaling molecules and chromatin structure/modification, play crucial roles in controlling P-TEFb and, hence, the release of promoter-proximally paused Pol II and subsequent productive transcriptional elongation.
As in Drosophila and mammals, the pausing of Pol II has been reported in Caenorhabditis elegans (10). However, homologues of the components of the NELF complex have not been identified in C. elegans (1, 10). Thus, the pausing of Pol II in C. elegans raises the possibility that C. elegans employs a distinct mechanism to cause Pol II pausing at the promoter-proximal site. Like C. elegans, the yeast Saccharomyces cerevisiae also does not have discernible NELF homologues (1). Nechaev and Adelman mentioned in a review article (1) that promoter-proximal enrichment of Pol II was not detected in yeast, favoring the idea that the promoter-proximal pausing of Pol II is absent in yeast. Using the chromatin immunoprecipitation (ChIP) assay, we analyzed the association of Pol II with several genes in yeast (S. cerevisiae) and found a relatively high level of Pol II at the promoter-proximal site (11). Likewise, genome-wide localization analysis of Pol II using genomic tiling array (60-bp probes every ∼250 bp)-based hybridization in yeast revealed a set of genes with high accumulation of Pol II toward the 5′ end (12). However, there is much less promoter-proximal accumulation of Pol II in yeast than in Drosophila and mammals. Interestingly, we recently found that the accumulation of Pol II at the promoter-proximal site in yeast was significantly enhanced in the absence of a capping enzyme, Cet1 (an mRNA triphosphatase), indicating the role of an mRNA-capping triphosphatase in regulation of promoter-proximal pausing/accumulation of Pol II (11). Consistently, inhibition of serine-5 phosphorylation of the CTD of the largest subunit (Rpb1) of Pol II (which is essential for cotranscriptional recruitment of Cet1 via Ceg1 [13]) leads to promoter-proximal accumulation/pausing of Pol II (14). Further, we found that the deletion of the N-terminal domain (NTD) (amino acids [aa] 1 to 204) of Cet1 led to the accumulation of Pol II at the promoter-proximal site and reduced transcription (11). The aa 1 to 204 domain of Cet1 does not regulate mRNA triphosphatase or capping activity (15, 16). Thus, our previous results (11) demonstrated that the NTD of Cet1 regulates promoter-proximal accumulation of Pol II and transcription independently of mRNA-capping activity. However, it remains unknown how Cet1's NTD impairs promoter-proximal accumulation of Pol II and enhances transcription. We present evidence that Cet1's NTD targets FACT (facilitates chromatin transcription) to the active gene to promote Pol II's engagement in productive transcriptional elongation, thus impairing promoter-proximal accumulation/pausing of Pol II.
RESULTS AND DISCUSSION
Cet1 facilitates the targeting of FACT to the active gene ADH1 via its NTD.
We recently demonstrated enhanced accumulation of Pol II at the ADH1 promoter-proximal site in the cet1(Ts) (temperature-sensitive) mutant in comparison to the wild-type (WT) equivalent at the nonpermissive (11), but not the permissive, temperature (Fig. 1A). Similar promoter-proximal accumulation of Pol II was also observed at other genes (11). Intriguingly, we found that Cet1's NTD facilitated the release of paused Pol II from the promoter-proximal site (11) and hence transcription (11). To determine the mechanism of Cet1's regulation of promoter-proximal accumulation of Pol II and transcription, we hypothesized that the NTD of Cet1 might target FACT (a histone chaperone or chromatin-remodeling/regulatory factor that promotes transcriptional elongation by altering chromatin structure [17–19]) to enhance the engagement of Pol II in elongation, leading to decreased accumulation/pausing of Pol II at the promoter-proximal site. To test this, we analyzed the association of FACT with the promoter and coding sequence of ADH1 in the presence and absence of the NTD of Cet1. For this, we tagged the Spt16 subunit of the heterodimeric FACT chromatin-remodeling factor in the cet1Δ204 (which does not have an NTD) and wild-type strains and then performed the ChIP assay. An inactive region within chromosome V (Chr V) was analyzed as a nonspecific DNA control (or background signal). We found that the association of Spt16 with the coding sequence of ADH1 was decreased in the cet1Δ204 strain in comparison to the wild-type equivalent (Fig. 1B; see Fig. S1A and B in the supplemental material). However, the global level of Spt16 was not altered in the absence of Cet1's NTD (Fig. 1C). These results demonstrate that the association of FACT with ADH1 is decreased in the absence of Cet1's NTD. Further, as mentioned above, Cet1's NTD does not have mRNA-capping activity (15, 16). Thus, our results support the notion that the NTD of Cet1 facilitates the targeting of FACT to ADH1 independently of the mRNA-capping activity.
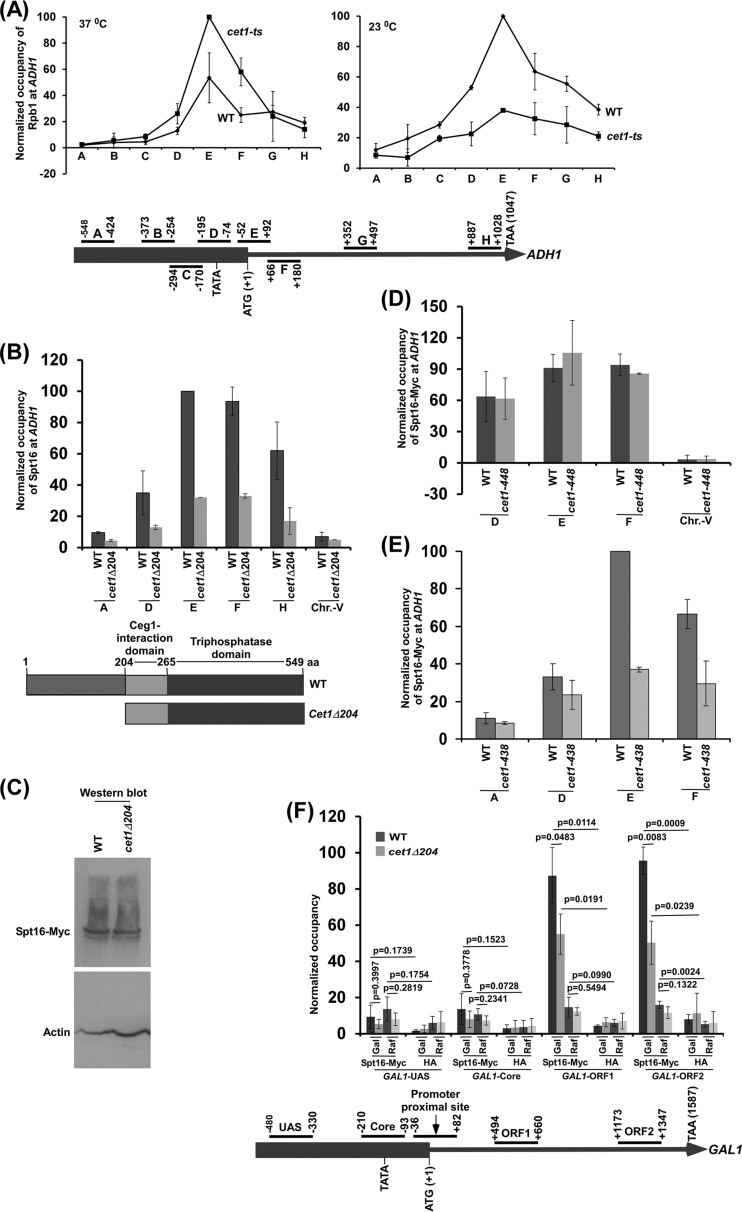
FIG 1.
The NTD (aa 1 to 204) of Cet1 targets FACT to ADH1 independently of mRNA-capping activity. (A) (Top) Analysis of Pol II association with ADH1 in the cet1(Ts) (cet1-438) mutant (YSB717) and wild-type (YSB540) strains at the nonpermissive (left) and permissive (right) temperatures. Immunoprecipitations were performed using mouse monoclonal antibody 8WG16 (Covance) against the CTD of Rpb1. The ratio of immunoprecipitate to input in the autoradiogram was measured. The maximum ratio was set to 100, and other ratios were normalized with respect to 100. The normalized ratio (represented as normalized occupancy) is plotted in the form of a histogram. (Bottom) Schematic diagram showing the locations of different primer pairs at ADH1 for ChIP analysis. The numbers are presented with respect to the position of the first nucleotide of the initiation codon (+1). (B) Analysis of FACT (Spt16-Myc) association with ADH1 in the presence and absence of the NTD of Cet1. (Top) Yeast cells were grown in YPD medium at 30°C to an OD600 of 1.0 prior to formaldehyde-based in vivo cross-linking. Immunoprecipitated DNA was analyzed by PCR using primer pairs targeted to different locations in ADH1. (Bottom) Schematic diagram of the different domains of Cet1. (C) Western blot analysis of Spt16 in the wild-type and cet1Δ204 strains. (D) ChIP analysis of FACT at ADH1 in the cet1(Ts) (cet1-448) mutant and its isogenic wild-type equivalent. Both wild-type and mutant cells were grown in YPD at 30°C to an OD600 of 0.85 and then switched to 37°C for 90 min prior to cross-linking. (E) ChIP analysis of FACT at ADH1 in the cet1(Ts) (cet1-438) mutant and its isogenic wild-type equivalent. Yeast cells were grown as for panel D. (F) ChIP analysis of Spt16 at GAL1 in the wild-type and cet1Δ204 strains in galactose (Gal)- or raffinose (Raf)-containing growth medium. The error bars indicate SD.
To complement the above-mentioned results, we next analyzed the recruitment of FACT to ADH1 in the Cet1 wild-type (YSB540) (20, 21) and temperature-sensitive mutant (cet1-448; YSB718) (20, 21) strains. The cet1(Ts) (cet1-448) mutant has a point mutation (W247A) at the Ceg1 (an enzyme with guanylyltransferase activity required for mRNA capping) interaction domain (Fig. 1B, bottom) and is stable at the nonpermissive temperature (20, 21). Thus, the cet1(Ts) mutant has an intact NTD but impaired capping activity at the nonpermissive temperature (11, 21). Using the cet1(Ts) mutant, the effect of an intact NTD of Cet1 was analyzed for recruitment of FACT to ADH1. We found that the recruitment of FACT to ADH1 is not altered in the cet1(Ts) (cet1-448) mutant strain in comparison to the wild-type equivalent at the nonpermissive temperature (Fig. 1D; see Fig. S1C in the supplemental material). These results support the idea that the stable Cet1 mutant with impaired capping activity does not alter FACT recruitment. On the other hand, the absence of Cet1's NTD impairs FACT targeting to ADH1 (Fig. 1B). Hence, the NTD, but not capping activity, of Cet1 facilitates the targeting of FACT to ADH1. Further, we found that the degradation of Cet1 in a different cet1(Ts) strain (cet1-438; YSB717) (20, 21) (with a C330W mutation in the triphosphatase domain) (Fig. 1B, bottom) at the nonpermissive temperature (20, 21) impairs FACT targeting to ADH1 (Fig. 1E). Taken together, our results support the idea that Cet1's NTD facilitates the targeting of FACT to ADH1 independently of its capping activity.
Previous studies implied that histones interact with FACT (17–19, 22, 23) and thus contribute to FACT recruitment to the gene in the absence of active transcription or Cet1's NTD. To test this, we analyzed the association of the Spt16 component of FACT with GAL1 under transcriptionally active (galactose) and inactive (raffinose) conditions in the presence and absence of Cet1's NTD. We found that Spt16 is predominantly associated with the GAL1 coding sequence under transcriptionally active conditions in galactose-containing growth medium (Fig. 1F). However, a significant level of Spt16 was not observed at GAL1 under inactive conditions in raffinose-containing growth medium (Fig. 1F). Further, we found that the predominant association of Spt16 with the GAL1 coding sequence under transcriptionally active conditions was decreased in the cet1Δ204 strain (Fig. 1F). However, Cet1's NTD is not solely required for association of FACT with the active GAL1 coding sequence, as we observed a significantly high level of Spt16 at the GAL1 coding sequence in the absence of Cet1's NTD under transcriptionally active conditions (Fig. 1F). These results support the idea that Cet1's NTD facilitates the targeting of FACT to the active GAL1 coding sequence, similar to the results at ADH1.
Cet1's NTD is involved in interaction with FACT to promote FACT recruitment to ADH1 and hence transcription.
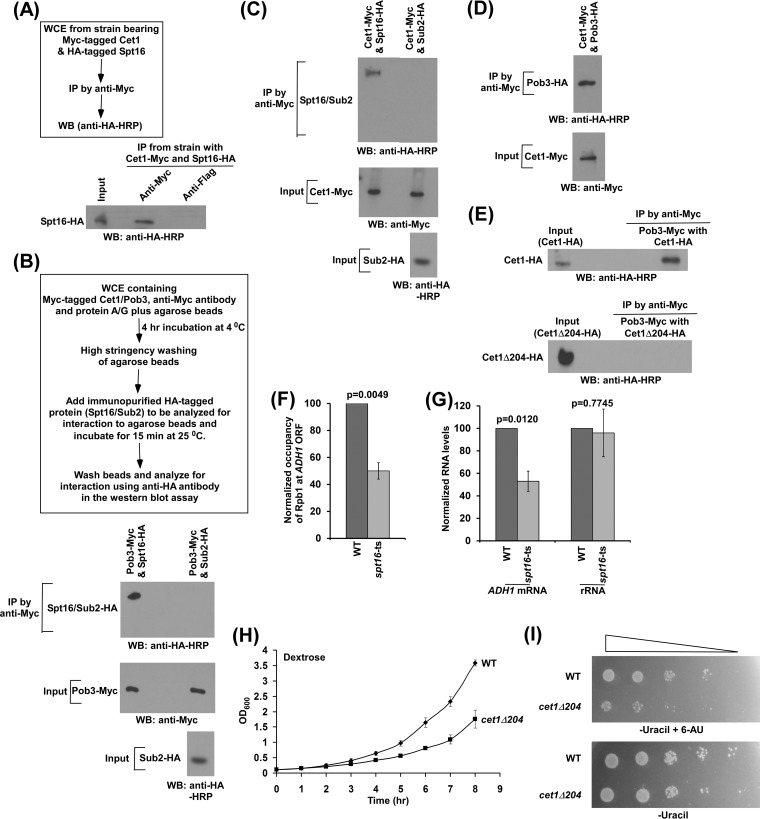
Since Cet1 facilitates the targeting of FACT to ADH1 independently of its mRNA-capping activity, it is likely to interact with FACT. To test this, we analyzed the interaction of Cet1 with FACT in vivo, using a formaldehyde-based cross-linking and coimmunoprecipitation (co-IP) assay (24, 25). For this, we tagged the CET1 and SPT16 genes with Myc and hemagglutinin (HA) epitopes, respectively, in their chromosomal loci. Using this strain, we performed a formaldehyde-based co-IP assay that revealed the interaction of Cet1 with Spt16 in vivo (Fig. 2A). Next, we analyzed their interactions in vitro. For this, we immunopurified the Spt16 and Pob3 components of FACT by Myc/HA epitope tagging and then analyzed their interaction as a positive control. The experimental strategy for this interaction analysis is shown schematically in Fig. 2B, as was done previously (24, 26). We found that Spt16 interacts with Pob3 (Fig. 2B, bottom). As a negative control, we showed that Pob3 does not interact with Sub2 (which is not known to interact with Pob3) under identical experimental conditions (Fig. 2B). Similar levels of Pob3 were present in analysis of interactions of Pob3 with Spt16 and Sub2 (Fig. 2B, input). Using identical experimental conditions, we analyzed the interaction of Cet1 with Spt16. We found that Cet1 interacts with Spt16 but not Sub2 (which is not known to interact with Cet1) (Fig. 2C). Similar levels of Cet1 were present in both interaction analyses (Fig. 2C, input). Next, we analyzed whether Cet1 can also interact with the Pob3 component of FACT. For this, we immunopurified HA-tagged Pob3 and then analyzed its interaction with Myc-tagged Cet1 following the above-described experimental protocol. We found that Cet1 interacts with Pob3 (Fig. 2D), which is required for recruitment of Spt16 (see Fig. 5J and K), and hence FACT. Collectively, our results reveal that Cet1 interacts with FACT and thus facilitates FACT targeting to the active gene.
FIG 2.
Cet1 interacts with FACT in facilitating transcription. (A) Formaldehyde-based in vivo cross-linking and co-IP assay. Schematic outline of the experimental strategy (top) and IP (bottom). WB, Western blotting. (B) (Bottom) Analysis of interaction of Pob3 with Spt16. (Top) Schematic outline of the experimental strategy to analyze protein-protein interactions in vitro. (C and D) Analysis of interaction of Cet1 with FACT. (E) Cet1 does not interact with FACT in the absence of its NTD. (F) ChIP analysis of Rpb1 association with the coding sequence of ADH1 in the wild-type and spt16(Ts) mutant strains at the nonpermissive temperature. (G) RT-PCR analysis of ADH1 mRNA levels in the wild-type and spt16(Ts) mutant strains at the nonpermissive temperature. (H) Growth analysis of wild-type and cet1Δ204 strains in liquid medium (YPD). (I) Growth analysis of wild-type and cet1Δ204 strains in solid medium with or without 6-AU. The error bars indicate SD.
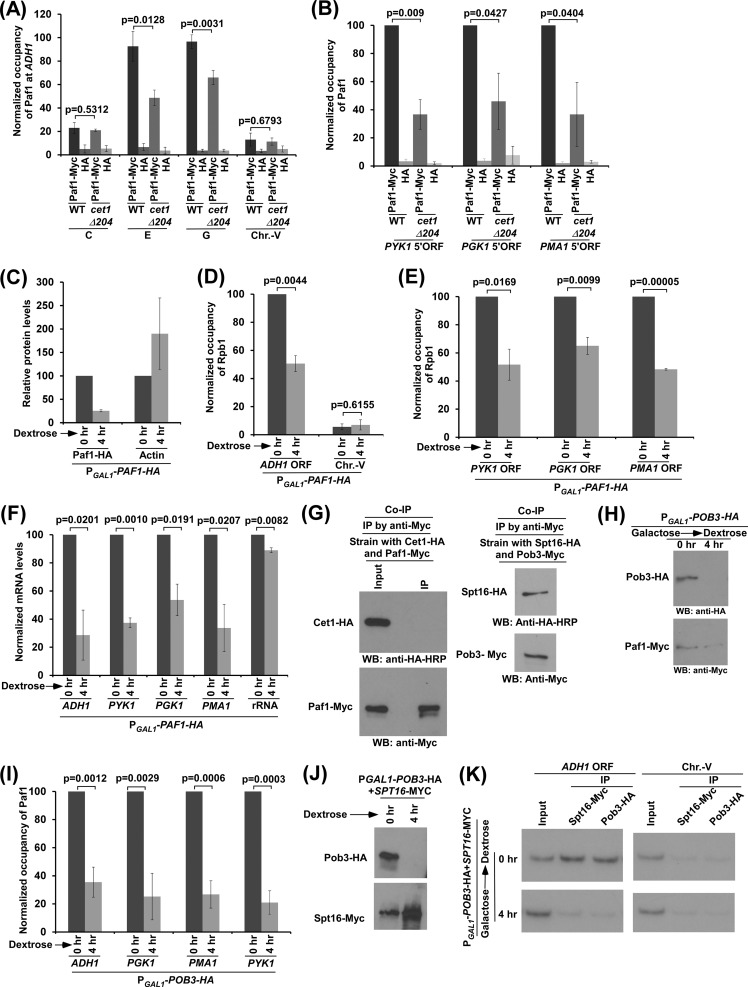
FIG 5.
Cet1's NTD recruits the Paf1C via FACT to promote transcription. (A and B) Cet1's NTD enhances recruitment of the Paf1C to the promoter-proximal sites (or 5′ ORFs) of ADH1, PGK1, PMA1, and PYK1. (C) Analysis of Paf1 and actin levels in strains expressing PAF1 under the control of the GAL1 promoter (PGAL1-PAF1-HA) after switching the carbon source in the growth medium from galactose (when total OD600 was 0.6) to dextrose. (D and E) Analysis of Rpb1 association with the ADH1, PGK1, PYK1, and PMA1 coding sequences in the strains expressing PAF1 under the control of the GAL1 promoter (PGAL1-PAF1-HA) after switching the carbon source in the growth medium from galactose to dextrose. (F) RT-PCR analysis of ADH1, PGK1, PYK1, and PMA1 mRNAs in strains expressing PAF1 under the control of the GAL1 promoter (PGAL1-PAF1-HA) after switching the carbon source in the growth medium from galactose to dextrose. (G) Co-IP analysis between Cet1 and Paf1 without in vivo cross-linking by formaldehyde. (H) Analysis of Pob3 and Paf1 levels in strains expressing HA-tagged Pob3 under the control of the GAL1 promoter (PGAL1-POB3-HA) after switching the carbon source in the growth medium from galactose to dextrose. (I) FACT enhances the recruitment of Paf1C to the promoter-proximal sites (or 5′ ORFs) of ADH1, PGK1, PMA1, and PYK1. (J and K) Pob3 is required for recruitment of Spt16 to the active gene. (J) Levels of Pob3 and Spt16 (by Western blot analysis) in the strain expressing HA-tagged Pob3 under the control of the GAL1 promoter (PGAL1-POB3-HA) after switching the carbon source in the growth medium from galactose to dextrose. (K) ChIP analysis of Spt16 and Pob3 at the ADH1 coding sequence following shutdown of Pob3 expression in dextrose-containing growth medium for 4 h. The error bars indicate SD.
Further, our ChIP results revealed that the NTD of Cet1 promotes FACT's targeting to the active gene (Fig. 1B to F). These results indicate the requirement for Cet1's NTD for interaction with FACT. To test this, we immunopurified HA-tagged Cet1 and Cet1Δ204 and then analyzed their interactions with the Pob3 component of FACT. We found that cet1Δ204 does not interact with Pob3, while Cet1 interacts with Pob3 (Fig. 2E). These results support the idea that Cet1's NTD is involved in interaction with FACT, and hence, FACT recruitment to the active gene is impaired in the absence of the NTD of Cet1 (Fig. 1B to F). In agreement with these results, the deletion of Cet1's NTD enhances promoter-proximal accumulation of Pol II at ADH1 and impairs transcription (11), as FACT promotes the association of Pol II with ADH1 and hence transcription (Fig. 2F and G), consistent with previous studies (27). Since the NTD of Cet1 facilitates transcription (11), the cellular growth of the cet1Δ204 strain is likely to be altered in comparison to that of the wild-type equivalent. Indeed, we found that the growth of yeast cells is significantly decreased in the absence of Cet1's NTD (Fig. 2H). Further, we found that the growth of the Cet1Δ204 strain is decreased in the presence of 6-azauracil (6-AU) (Fig. 2I). 6-AU decreases nucleotide pools, resulting in slow (or impaired) cellular growth upon deletion (or mutation) of the factors involved in transcriptional elongation (28). Thus, the 6-AU sensitivity of the cet1Δ204 strain supports the role of Cet1's NTD in transcriptional elongation (Fig. 2I). Similarly, the loss of targeting of FACT is likely to impair the growth of yeast cells in the presence of 6-AU. Indeed, previous studies demonstrated decreased growth of the spt16 mutant in the presence of 6-AU (29). Thus, our results support the idea that the NTD of Cet1 facilitates the targeting of FACT and, hence, transcription elongation and cellular growth.
Cet1's NTD promotes the targeting of FACT to PMA1, PGK1, and PYK1 to facilitate transcription.
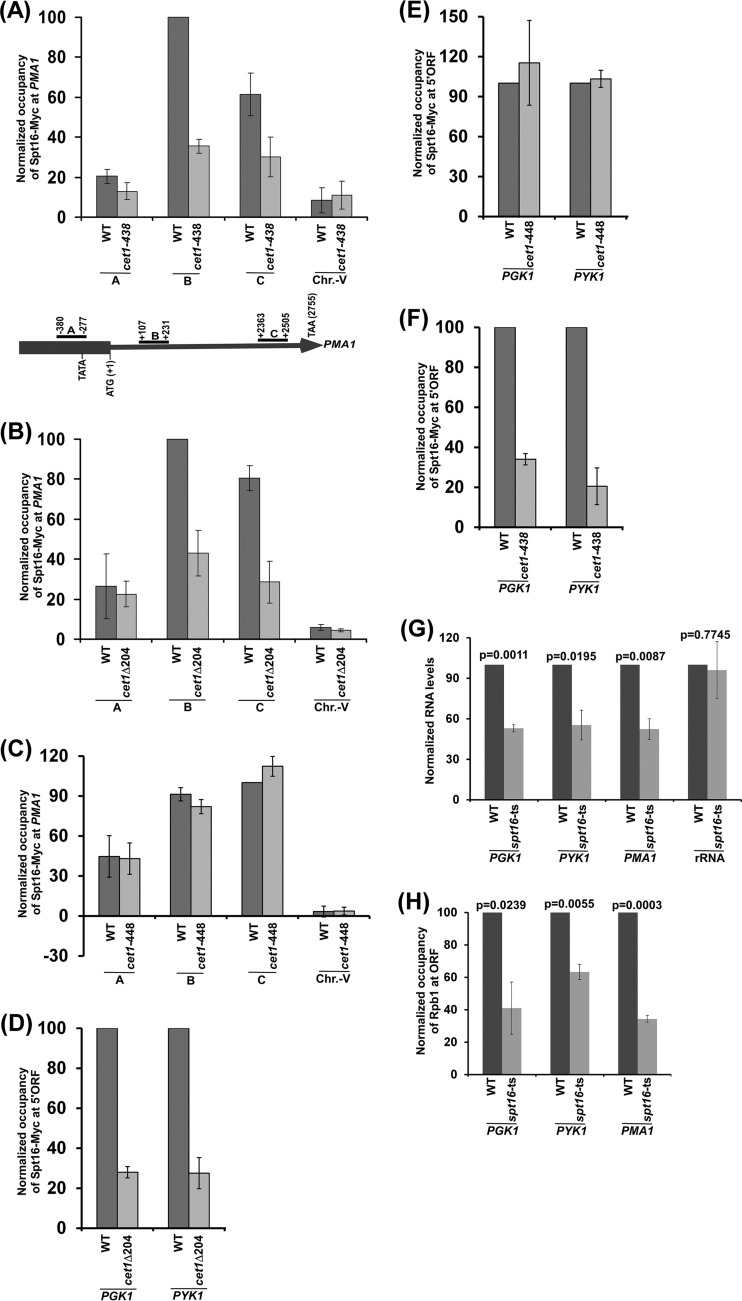
Similar to the results at ADH1, FACT is likely to be targeted to other active genes by Cet1. To test this, we analyzed the recruitment of FACT to a constitutively active gene, PMA1, in the cet1(Ts) (cet1-438, which is degraded at the nonpermissive temperature) (20, 21) and wild-type strains. We found that FACT recruitment to the PMA1 gene is decreased in the cet1(Ts) mutant strain in comparison to the wild-type equivalent (Fig. 3A). Further, we observed that the NTD of Cet1 facilitates the targeting of FACT to PMA1 (Fig. 3B). Moreover, the cet1(Ts) (cet1-448) mutant (which does not have mRNA-capping activity and is not degraded at the nonpermissive temperature) (20, 21) does not alter FACT recruitment to PMA1 (Fig. 3C). These results support the idea that Cet1 facilitates the targeting of FACT to PMA1 independently of its capping activity but via its NTD, similar to the results at ADH1. Likewise, FACT recruitment to other genes, such as PGK1 and PYK1, is also promoted by Cet1's NTD (Fig. 3D to F). Such recruitment of FACT enhances the association of Pol II with these genes and hence transcription (Fig. 3G and H). Together, our results support the role of Cet1 in facilitating FACT recruitment to the active genes independently of its capping activity, but rather, via its NTD to promote transcription, thus providing a new mechanism of FACT targeting to the active gene in transcriptional regulation.
FIG 3.
The NTD of Cet1 targets FACT to PMA1, PGK1, and PYK1. (A) Analysis of FACT (Spt16-Myc) association with PMA1 in the cet1(Ts) (cet1-438) mutant and its wild-type equivalent. (B) Analysis of FACT (Spt16-Myc) association with PMA1 in the presence and absence of the NTD of Cet1. (C) Analysis of FACT (Spt16-Myc) association with PMA1 in the cet1(Ts) (cet1-448) mutant and its wild-type equivalent. (D) ChIP analysis of FACT at the 5′ ORFs of PGK1 and PYK1 in the presence and absence of the NTD of Cet1. (E) ChIP analysis of FACT at the 5′ ORFs of PGK1 and PYK1 in the cet1(Ts) (cet1-448) mutant and its wild-type equivalent. (F) ChIP analysis of FACT at the 5′ ORFs of PGK1 and PYK1 in the cet1(Ts) (cet1-438) mutant and its wild-type equivalent. (G) RT-PCR analysis of PGK1, PYK1, and PMA1 mRNAs in the spt16(Ts) mutant and its wild-type equivalent. (H) ChIP analysis of Rpb1 at the PGK1, PYK1, and PMA1 coding sequences in the spt16(Ts) mutant and its wild-type equivalent. The error bars indicate SD.
Facilitated recruitment of FACT by Cet1's NTD does not enhance PIC formation, but rather, promotes transcriptional elongation.
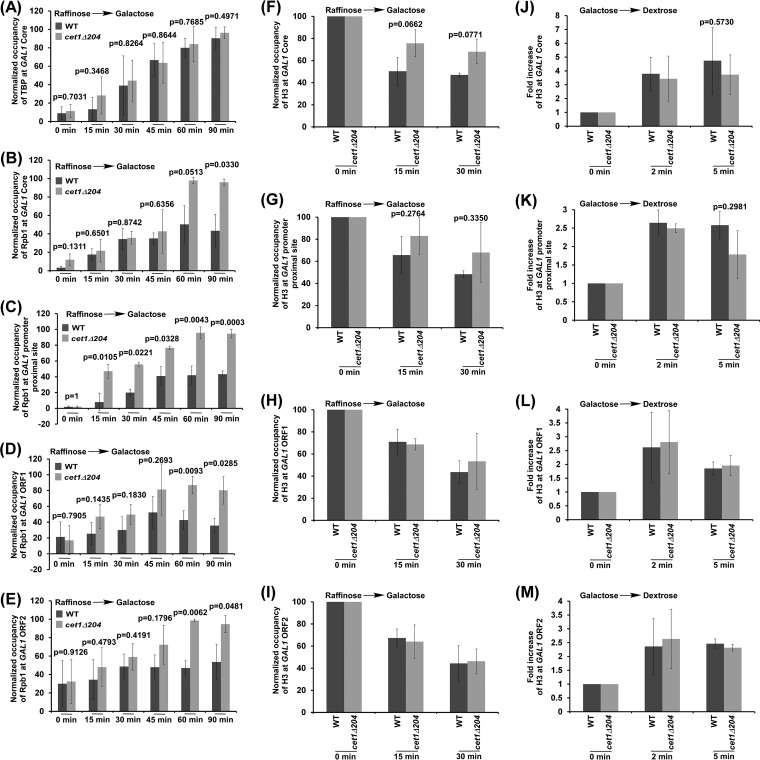
In addition to its role in transcriptional elongation, FACT is also involved in transcriptional initiation (17–19, 23, 27, 29, 30). Thus, global loss of Spt16 or its temperature-sensitive inactivation impairs both transcriptional initiation and elongation (17–19, 27, 29–31). However, localized impairment of FACT recruitment in the absence of Cet1's NTD at the beginning of transcriptional elongation would reduce transcriptional elongation but not initiation. Indeed, growth analysis in the presence of 6-AU revealed the role of Cet1's NTD in transcriptional elongation (Fig. 2I), consistent with the transcriptional defect in the cet1Δ204 strain (11). To complement this observation further, we analyzed preinitiation complex (PIC) formation and Pol II association with GAL1 in the cet1Δ204 and wild-type strains following transcriptional induction after switching the carbon source in the growth medium from raffinose to galactose. For this, we analyzed the recruitment of TATA box-binding protein (TBP) (which nucleates the assembly of the general transcription factors at the promoter to form the PIC) at the GAL1 core promoter following transcriptional induction. We found that the recruitment of TBP to the GAL1 core promoter was not altered in the cet1Δ204 strain following transcriptional induction (Fig. 4A; see Fig. S2A in the supplemental material). Likewise, the recruitment of Pol II (which joins the PIC toward the end) to the GAL1 promoter was not altered in the cet1Δ204 strain in comparison to the wild-type equivalent within 45 min of transcriptional induction (Fig. 4B). However, at the later transcriptional induction time points, accumulation of Pol II was observed at the promoter due to accumulation of Pol II at the promoter-proximal site in the cet1Δ204 strain (Fig. 4C), consistent with previous studies (11). Thus, targeted impairment of FACT recruitment in the absence of Cet1's NTD does not alter PIC formation. However, the global inactivation or loss of Spt16 impairs PIC formation (27, 29, 30). Intriguingly, we observed increased accumulation of Pol II at the GAL1 promoter-proximal site within 15 min of transcription induction in the cet1Δ204 strain (Fig. 4C). Such early accumulation of Pol II in the cet1Δ204 strain was not observed at the GAL1 core promoter or downstream coding sequence (Fig. 4B to E). Similar results were also observed at 30 or 45 transcriptional-induction time points (Fig. 4B to E; see Fig. S2B to D in the supplemental material). Thus, our results support the idea that targeted impairment of FACT recruitment in the absence of Cet1's NTD is associated with the accumulation of Pol II at the GAL1 promoter-proximal site immediately following transcriptional induction (Fig. 4B to E). Likewise, the loss of Cet1's NTD enhances accumulation of Pol II at the ADH1 promoter-proximal site (11).
FIG 4.
Analysis of Cet1's NTD function in regulation of the levels of TBP, Pol II, and histone H3 at GAL1. (A and B) ChIP analysis of TBP and Pol II at the GAL1 core promoter after switching on GAL1 transcription in galactose-containing growth medium. (C to E) ChIP analysis of Rpb1 association with the promoter-proximal site and downstream coding regions (i.e., ORF1 and ORF2) of GAL1 after switching on GAL1 transcription. (F to I) Analysis of the Cet1 NTD's role in evicting histone H3 from GAL1 after switching on transcription. (J to M) Analysis of the Cet1 NTD's role in depositing histone H3 at GAL1 after switching off GAL1 transcription in dextrose-containing growth medium. The error bars indicate SD.
Cet1's NTD targets FACT, which enhances recruitment of Paf1C to promote transcription.
Since Cet1's NTD facilitates the targeting of FACT to the active gene, chromatin assembly/disassembly at the active gene would likely be altered in the cet1Δ204 strain, as FACT is involved in regulating transcription via alteration of nucleosomal occupancy (17–19, 23, 27, 29–31). To test this, we analyzed chromatin disassembly/reassembly at the transcriptionally inducible GAL1 gene in the wild-type and cet1Δ204 strains following the experimental strategy used previously (26, 32). Briefly, we analyzed the eviction of histone H3 from the GAL1 promoter, promoter-proximal site, and downstream coding sequence following transcriptional induction in galactose-containing growth medium in the wild-type and cet1Δ204 strains to determine the role of Cet1's NTD in chromatin disassembly. We found that histone H3 eviction from the GAL1 coding sequence, promoter, or promoter-proximal site was not significantly altered in the cet1Δ204 strain (Fig. 4F to I). Similarly, we analyzed chromatin reassembly following deposition of histone H3 at the GAL1 promoter, promoter-proximal site, and coding sequence after switching the carbon source in the growth medium from galactose to dextrose in the wild-type and cet1Δ204 strains. We found that histone H3 deposition at the GAL1 coding sequence, promoter, or promoter-proximal site was not significantly altered in the cet1Δ204 strain (Fig. 4J to M). However, global inactivation of Spt16 impaired histone H3 deposition at GAL1 (31). Thus, ∼2.5-fold-impaired recruitment of FACT in the cet1Δ204 strain does not appear to have significant effects on nucleosomal occupancy (Fig. 4F to M).
Since previous studies (33–39) implicated the interaction of FACT with the Paf1 (RNA polymerase II-associated factor 1) complex (Paf1C), which promotes transcriptional elongation (33, 40), it was likely that impaired targeting of FACT in the cet1Δ204 strain would decrease the recruitment of the Paf1C to the promoter-proximal site or the 5′ end of the coding sequence to reduce transcription. To test this, we analyzed the recruitment of the Paf1C to the 5′ ends of the ADH1, PGK1, PYK1, and PMA1 coding sequences in the wild-type and cet1Δ204 strains. For this, we tagged the Paf1 component of the Paf1C with a Myc epitope in the wild-type and cet1Δ204 strains and then performed the ChIP assay at the 5′ ends of the coding sequences of these genes. Paf1 was predominantly recruited to the coding sequence (Fig. 5A), consistent with previous studies and its function in transcriptional elongation (33, 40). Further, we found that the recruitment of Paf1 to these genes was significantly impaired in the cet1Δ204 strain in comparison to the wild-type equivalent (Fig. 5A and B). Next, we analyzed the role of Paf1 in recruitment of Pol II to these genes. For this, we replaced the endogenous promoter of PAF1 with the galactose-inducible GAL1 promoter and then analyzed Pol II association with these genes after shutting down PAF1 transcription in dextrose-containing growth medium (Fig. 5C). We found that the association of Pol II with the coding sequences of these genes was decreased when the expression of Paf1 was shut down in dextrose-containing growth medium (Fig. 5C to E). Likewise, transcription of the genes was also impaired upon switching off Paf1 transcription (Fig. 5F). These results support the idea that Paf1 promotes transcription, consistent with the Paf1C's function in transcriptional elongation (33, 40). However, Paf1 does not interact with Cet1, as revealed by the co-IP assay (Fig. 5G, left). As a positive control, we showed, under the same co-IP conditions, that both Spt16 and Pob3 components of FACT interact (Fig. 5G, right). Consistently, interaction of the Paf1C with Cet1 was not found in the protein interaction database (BIOGRID). Thus, Cet1 is responsible for recruitment of the Paf1C via FACT. This is consistent with the interactions between FACT and the Paf1C (33–39) and between Cet1 and FACT (Fig. 2B to D). In agreement with these interactions, we found that FACT promotes recruitment of the Paf1C (Fig. 5H and I). Briefly, we impaired the expression of the Pob3 subunit of FACT by replacing its endogenous promoter with the galactose-inducible GAL1 promoter in dextrose-containing growth medium (Fig. 5H) and then analyzed the effect of such loss of Pob3 on recruitment of Paf1. We found that Paf1 recruitment to the 5′ ends of the ADH1, PGK1, PYK1, and PMA1 coding sequences was significantly impaired in the absence of Pob3 (Fig. 5I). Further, we found that Pob3 is required for recruitment of the Spt16 subunit of FACT (Fig. 5J and K). Thus, FACT is not recruited to the active gene (Fig. 5K) when Pob3 is not expressed (Fig. 5J), and Paf1 is not recruited in the absence of Pob3 (Fig. 5H and I). Therefore, our results reveal that FACT is required for recruitment of the Paf1C. Taken together, our results support the idea that Cet1's NTD targets FACT, which enhances recruitment of the Paf1C to promote transcription.
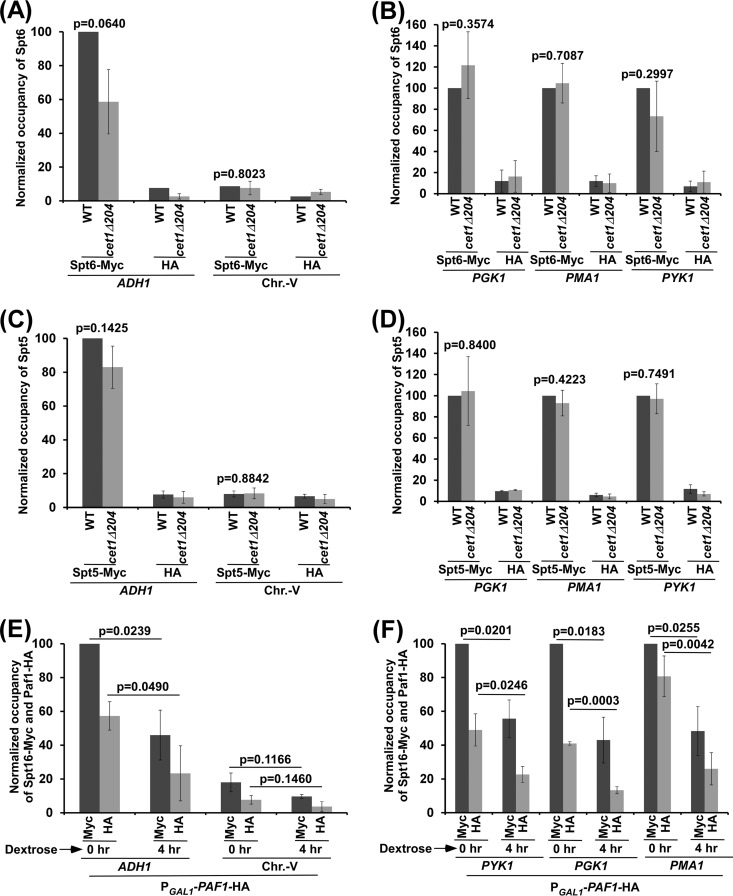
Further, Paf1C recruitment can be regulated by Spt6 (which is a histone chaperone involved in chromatin assembly and promotes transcriptional elongation) (23, 41–43) via the interaction of Cet1's NTD with Spt6, as the Paf1C interacts with Spt6 (40). To test this, we analyzed the role of Cet1's NTD in recruitment of Spt6 to ADH1, PGK1, PYK1, and PMA1. For this, we tagged Spt6 with a Myc epitope in its chromosomal locus in the wild-type and cet1Δ204 strains and then performed the ChIP assay at ADH1, PGK1, PYK1, and PMA1. We found that the absence of Cet1's NTD did not significantly alter the recruitment of Spt6 to the 5′ ends of the ADH1, PGK1, PYK1, and PMA1 coding sequences (Fig. 6A and B). Further, Spt5 (a component of the DSIF complex that is phosphorylated by Bur1 kinase) facilitates recruitment of the Paf1C (33, 44). However, we found that Cet1's NTD does not regulate Paf1C recruitment via Spt5, as the recruitment of Spt5 to the 5′ ends of the ADH1, PGK1, PMA1, and PYK1 coding sequences was not altered in the cet1Δ204 strain in comparison to the wild-type equivalent (Fig. 6C and D). Thus, Cet1's NTD enhances Paf1C recruitment via FACT but not Spt5 or Spt6. Therefore, Cet1's NTD promotes the targeting of FACT, which subsequently facilitates recruitment of the Paf1C to the active gene to enhance transcription. In addition to FACT, other factors, such as Spt5 and Pol II, also facilitate recruitment of the Paf1C to the active gene. Such recruitment of the Paf1C also promotes the targeting of FACT to the active gene, as we observed reduced association of Spt16 and Paf1 with the ADH1, PGK1, PYK1, and PMA1 coding sequences following shutdown of the expression of Paf1 (which is under the control of the GAL1 promoter) in dextrose-containing growth medium (Fig. 5C and 6E and F). In agreement with this, the Paf1C has also been previously implicated in recruitment of FACT at the active gene (45). Hence, FACT recruitment to the active gene is not completely abolished in the absence of Cet1's NTD (Fig. 1B and F and 3B and D).
FIG 6.
Cet1's NTD does not regulate recruitment of Spt5 or Spt6 to the promoter-proximal sites (or 5′ ORFs) of ADH1, PGK1, PYK1, and PMA1. (A and B) ChIP analysis of Spt6 at the 5′ ORFs of ADH1, PGK1, PYK1, and PMA1. (C and D) ChIP analysis of Spt5 at the 5′ ORFs of ADH1, PGK1, PYK1, and PMA1. (E and F) Analysis of Myc-tagged Spt16 and HA-tagged Paf1 association with the ADH1, PGK1, PYK1, and PMA1 coding sequences in the strain expressing HA-tagged Paf1 under the control of the GAL1 promoter (PGAL1-PAF1-HA) and Myc-tagged Spt16 after switching the carbon source in the growth medium from galactose to dextrose for 4 h. The error bars indicate SD.
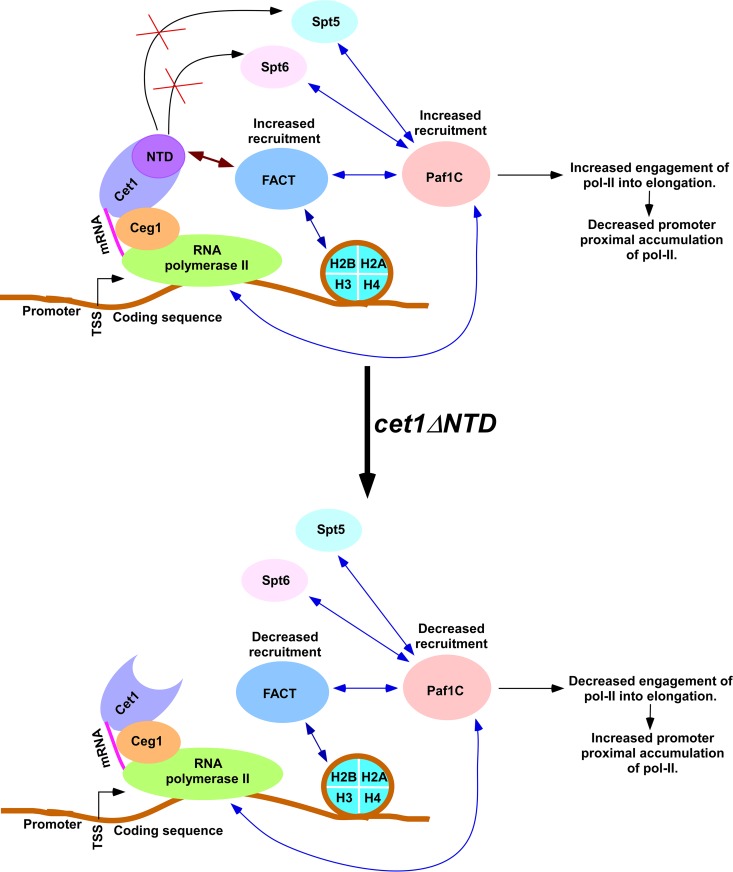
In summary, we found here that Cet1 promotes the targeting of FACT to the active genes independently of its mRNA-capping activity via its NTD (Fig. 7). FACT, in turn, facilitates recruitment of the Paf1C, which promotes transcriptional elongation (Fig. 7). Therefore, Cet1 facilitates the engagement of Pol II in elongation via enhanced recruitment of the Paf1C, impairing the accumulation of Pol II at the promoter-proximal site (Fig. 7). Consistently, Paf1 knockdown has recently been shown to enhance promoter-proximal Pol II pausing in metazoan cells (46). Thus, our results provide the basis for explaining how Cet1's NTD impairs promoter-proximal accumulation of Pol II and promotes transcription in yeast. Cet1's NTD is not evolutionarily conserved in higher eukaryotes. The absence of such a domain, therefore, is likely to contribute to promoter-proximal accumulation of Pol II in higher eukaryotes. Further, our results imply that Cet1's NTD can be specifically targeted to impair transcription in developing therapeutic agents to treat fungal infections, as Cet1's NTD is absent in humans and other higher eukaryotes.
FIG 7.
Schematic diagram showing how Cet1's NTD impairs promoter-proximal accumulation/pausing of Pol II. Cet1, which associates with Pol II via Ceg1 (35) following PIC formation and transcriptional initiation (61, 62), interacts with FACT via the NTD (Fig. 2A to E) and facilitates its targeting to the active gene in an mRNA-capping activity-independent manner (Fig. 1B to F and 3A to F). In addition, FACT also interacts with histones, and such interaction is responsible for chromatin association with FACT (15–17, 22, 23). Thus, Cet1's NTD and histones synergistically target FACT to the active gene. In the absence of Cet1's NTD, FACT targeting to the active gene is significantly decreased (Fig. 1B and 3B and D). Decreased association of FACT reduces Paf1C recruitment (Fig. 5H to K). Thus, Cet1's NTD facilitates targeting of the Paf1C via FACT (Fig. 5H to K) but not Spt5 and Spt6 (Fig. 6A to D). Paf1 promotes transcriptional elongation (33, 40) (Fig. 5D to F). Hence, loss of Cet1's NTD reduces recruitment of the Paf1C via FACT, leading to increased accumulation/pausing of Pol II at the promoter-proximal site. Like FACT, Bre1 (an E3 ubiquitin ligase for histone H2B ubiquitylation) is also involved in Paf1 recruitment, independently of Pol II (63–65). The double-headed arrows indicate interactions.
MATERIALS AND METHODS
Plasmids.
The plasmids pFA6a-13Myc-KanMX6 and pFA6a-3HA-His3MX6 (47) were used for genomic Myc and HA epitope tagging of the proteins of interest, respectively. The plasmid pRS416 was used in the 6-AU assay. The plasmid pFA6a-KanMX6-PGAL1-3HA (47) was used to delete the NTD of Cet1 with replacement of its endogenous promoter by the GAL1 promoter. The plasmid pFA6a-KanMX6-PGAL1-3HA was also used to replace the endogenous promoters of POB3 and PAF1 with the GAL1 promoter.
Yeast strains.
A yeast (S. cerevisiae) strain expressing the Cet1 mutant without the first 204 aa at the N terminus (YSB710) and its isogenic wild-type equivalent (YSB709) were obtained from the Buratowski laboratory (21). The cet1(Ts) (cet1-438 [YSB717] and cet1-448 [YSB718]) mutants and their isogenic wild-type equivalent (YSB540) were also obtained from the Buratowski laboratory (20, 21). The wild-type and spt16(Ts) mutant strains were obtained from the Stillman laboratory (29). Multiple Myc epitope tags were added at the original chromosomal locus of SPT16 in the cet1-438 (YSB717), cet1-448 (YSB718), and cet1Δ204 (YSB710) mutant and isogenic wild-type strains to generate SLY31 (Spt16-Myc in cet1-438), RSY48 (Spt16-Myc in cet1-448), SLY52 (Spt16-Myc in cet1Δ204), SLY51 (Spt16-Myc in YSB709), and SLY30 (Spt16-Myc in YSB540). Multiple Myc epitope tags were added at the original chromosomal locus of POB3 in strain W303a to generate RSY55 (Pob3-Myc in W303a). Likewise, multiple HA epitope tags were added at the C terminus at the original chromosomal locus of SUB2 in strain W303a to generate GDY75 (Sub2-HA in W303a). Myc and HA epitope tags were added at the original chromosomal loci of SPT16 and CET1, respectively, in W303a to generate SLY68 (Spt16-Myc and Cet1-HA in W303a). The endogenous promoter of POB3 in NSY18 (Paf1-Myc in W303a using the plasmid pFA6a-13Myc-KanMX6) was replaced by the galactose-inducible GAL1 promoter to generate AKY20a (using the plasmid pFA6a-His3MX6-PGAL1-3HA), which expresses Pob3 with an HA tag at the N terminus. Myc epitope tags were added at the original chromosomal locus of SPT6 in YSB709 and YSB710 to generate SLY49 (Spt6-Myc in YSB709) and SLY50 (Spt6-Myc in YSB710), respectively, using the plasmid pFA6a-13Myc-KanMX6. The endogenous promoter of CET1 in W303a was replaced by the galactose-inducible GAL1 promoter to generate AKY7a and AKY8a (using plasmid pFA6a-KanMX6-PGAL1-3HA), which express Cet1 and Cet1Δ204 with HA tags at the N-terminal ends, respectively. Strain AKY21a was generated by adding an HA epitope tag at the C-terminal end of Cet1 in its chromosomal locus in strain NSY18, which expresses Myc-tagged Paf1 in W303a. Myc epitope tags were added at the C-terminal end of Spt5 in the Cet1 wild-type (YSB709) and cet1Δ204 (YSB710) strains to generate SLY47 and SLY48, respectively. The strain AKY11a was generated by adding Myc epitope tags at the C-terminal end of Spt16 in strain RSY56, which expresses HA-tagged endogenous Pob3 under the control of the GAL1 promoter. Myc epitope tags were added at the C-terminal end of Spt16 in W303a to generate NSY16. Myc epitope tagging did not alter cellular growth (see Fig. S3 in the supplemental material).
Growth media.
The wild-type and cet1Δ204 strains were grown in YPD (yeast extract-peptone plus 2% dextrose) medium to an optical density at 600 nm (OD600) of 1.0 at 30°C prior to formaldehyde-based in vivo cross-linking. For experiments with cet1(Ts) (cet1-438 and cet1-448) mutant and wild-type strains, yeast cells were grown in YPD medium at 30°C to an OD600 of 0.85 and then transferred to 37°C for 90 min prior to cross-linking. For experiments with the spt16(Ts) mutant and its isogenic wild-type equivalent, yeast cells were grown in YPD medium at 23°C to an OD600 of 0.85 and then transferred to 37°C for 1 h prior to cross-linking or harvesting for RNA analysis. Yeast strains expressing Cet1 or Cet1Δ204 with an HA tag at the N terminus under the control of the GAL1 promoter were grown in YPG (yeast extract-peptone plus 2% galactose) medium to an OD600 of 1.0 prior to harvesting for immunopurification. For histone H3 eviction analysis at GAL1, yeast cells were grown in YPR (yeast extract-peptone plus 2% raffinose) medium to an OD600 of 0.9 and then switched to YPG medium prior to cross-linking at 0, 15, and 30 min. For histone H3 deposition analysis at GAL1, yeast cells were grown in YPG medium at an OD600 of 0.9 and then switched to YPD medium prior to cross-linking at 0, 2, and 5 min. For kinetic analysis of TBP and Pol II association with GAL1, yeast cells were grown in YPR medium to an OD600 of 0.9 and then switched to YPG medium prior to cross-linking at 0, 15, 30, 45, 60, and 90 min. For analysis of Spt16 association with GAL1, yeast strains (SLY51 and SLY52) expressing Myc-tagged Spt16 were grown in YPR or YPG medium to an OD600 of 1.0 before cross-linking.
ChIP assay.
The ChIP assay was performed as described previously (11, 48–54). Briefly, yeast cells were treated with 1% formaldehyde, collected, and resuspended in lysis buffer. Following sonication, cell lysate (400 μl lysate from 50 ml of yeast culture) was precleared by centrifugation, and then 100 μl lysate was used for each immunoprecipitation. The immunoprecipitated protein-DNA complexes were treated with proteinase K, the cross-links were reversed, and DNA was purified. The immunoprecipitated DNA was dissolved in 20 μl Tris-EDTA (TE) 8.0 (10 mM Tris HCl, pH 8.0, and 1 mM EDTA), and 1 μl of the immunoprecipitated DNA was analyzed by PCR (a total of 23 cycles). The PCR mixtures contained [α-32P]dATP (2.5 μCi for each 25-μl reaction mixture), and the PCR products were detected by autoradiography after separation on a 6% polyacrylamide gel. As a control, input DNA was isolated from 5 μl of lysate without going through the immunoprecipitation step and suspended in 100 μl of TE 8.0. To compare the PCR signal arising from the immunoprecipitated DNA with that from the input DNA, 1 μl of input DNA was used for PCR analysis. Serial dilutions of input and immunoprecipitated DNA samples were used to assess the linear range of PCR amplification, as was done previously (50, 53–55).
For the ChIP analysis of Spt16, Paf1, Spt5, and Spt6, the above-described ChIP protocol was modified as described previously (48, 49, 52, 56). Briefly, a total of 800 μl lysate was prepared from 100 ml of yeast culture. Following sonication, 400 μl lysate was used for each immunoprecipitation (using 10 μl of anti-HA or anti-Myc antibody and 100 μl of protein A/G Plus-agarose beads from Santa Cruz Biotechnology, Inc.), and the immunoprecipitated DNA sample was dissolved in 10 μl TE 8.0, 1 μl of which was used for PCR analysis (a total of 23 cycles). In parallel, PCR analysis for input DNA was performed using 1 μl DNA that was prepared by dissolving purified DNA from 5 μl lysate in 100 μl TE 8.0. The primer pairs used for PCR analysis are listed in Table 1.
TABLE 1.
Primer pairs used for PCR analysis in the ChIP assay
| Targeta | Primer pair sequences |
|---|---|
| ADH1 (A) | 5′-CTACACCAATTACACTGCCTCATT-3′, 5′-ACTTCAATAGATGGCAAATGGA-3′ |
| ADH1 (B) | 5′-GGTATACGGCCTTCCTTCCAGTTAC-3′, 5′-GAACGAGAACAATGACGAGGAAACAAAAG-3′ |
| ADH1 (C) | 5′-CTGCACAATATTTCAAGCTATACCAAGC-3′, 5′-GCCTTTGGCTTTGGAACTGGAATATC-3′ |
| ADH1 (D) | 5′-GATATTCCAGTTCCAAAGCCAAAGGC-3′, 5′-CTTAACTGGCAATGGCCAGTCAC-3′ |
| ADH1 (E) | 5′-CGGTAACAGAGCTGACACCAGAGA-3′, 5′-ACGTATCTACCAACGATTTGACCC-3′ |
| ADH1 (F) | 5′-GATATTCCAGTTCCAAAGCCAAAGGC-3′, 5′-CTTAACTGGCAATGGCCAGTCAC-3′ |
| ADH1 (G) | 5′-CTGGTTACACCCACGACGGTTCTT-3′, 5′-GCAGACTTCAAAGCCTTGTAGACG-3′ |
| ADHI (H) | 5′-CGGTAACAGAGCTGACACCAGAGA-3′, 5′-ACGTATCTACCAACGATTTGACCC-3′ |
| PYK1 (5′ ORF) | 5′-CTTGTTTCTATTTACAAGACACCAATC-3′, 5′-CTTTGGACCGATGGTACCAATG-3′ |
| PYK1 (ORF or 3′ ORF) | 5′-AAGTTTCCGATGTCGGTAACGCTAT-3′, 5′-TTGGCAAGTAAGCGATAGCTTGTTC-3′ |
| PGK1 (5′ ORF) | 5′-CAGATCATCAAGGAAGTAATTATC-3′, 5′-GTCAACTCTGATGAAGACACGC-3′ |
| PGK1 (ORF or 3′ ORF) | 5′-AGACGAAGTTGTCAAGAGCTCTGC-3′, 5′-GAAAGCAACACCTGGCAATTCCT-3′ |
| PMA1 (A) | 5′-TCGATGGTGGGTACCGCTTAT-3′, 5′-GATGTTAGACGATAATGATAGGAC-3′ |
| PMA1 (B or 5′ ORF) | 5′-ATCTTCTGACGATGACGATATCG-3′, 5′-TTCTGGAACTGGTCTAGCTTCA-3′ |
| PMA1 (C or 3′ ORF) | 5′-TCGGTGCTATGAACGGTATTATG-3′, 5′-ACATGGTAGCGATGATGTCGACAG-3′ |
| GAL1 (UAS) | 5′-CGCTTAACTGCTCATTGCTATATTG-3′, 5′-TTGTTCGGAGCAGTGCGGCGC-3′ |
| GAL1 (Core) | 5′-ATAGGATGATAATGCGATTAGTTTTTTAGCCTT-3′, 5′-GAAAATGTTGAAAGTATTAGTTAAAGTGGTTATGCA-3′ |
| GAL1 (promoter-proximal site) | 5′-ACCTCTATACTTTAACGTCAAGGAG-3′, 5′-TTTCGGCCAATGGTCTTGGTAA-3′ |
| GAL1 (ORF1) | 5′-GTGAGGAAGATCATGCTCTATACG-3′, 5′-GGCGGTTTCAAACTTGTTAGATAC-3′ |
| GAL1 (ORF2) | 5′-CAGAGGGCTAAGCATGTGTATTCT-3′, 5′-GTCAATCTCTGGACAAGAACATTC-3′ |
| Chr V | 5′-GGCTGTCAGAATATGGGGCCGTAGTA-3′, 5′-CACCCCGAAGCTGCTTTCACAATAC-3′ |
UAS, upstream activating sequence; Core, core promoter; ORF1 and ORF2, two different locations in the ORF; 5′ ORF, toward the 5′ end of the ORF; 3′ ORF, toward the 3′ end of the ORF. Different locations at a genomic locus are designated by different letters.
Autoradiograms were scanned and quantitated with the National Institutes of Health Image 1.62 program. Immunoprecipitated DNAs were quantitated as the ratio of immunoprecipitate to input in the autoradiogram. The average ChIP signal of the biologically independent experiments is reported with standard deviation (SD) (Microsoft Excel). The Student t test of Microsoft Excel (tail = 2 and types = 3) was used to determine the P values for the statistical significance of the change in the ChIP signals. The changes were considered to be statistically significant at a P value of <0.05.
WCE preparation and Western blot analysis.
To analyze the global level, yeast strains expressing HA- or Myc epitope-tagged protein were grown in 25 ml YPD medium to an OD600 of 1.0. Yeast cells were then harvested, lysed, and sonicated to prepare whole-cell extract (WCE) with solubilized chromatin following the protocol described previously for the ChIP assay (11, 48–54). The WCE was run on an SDS-polyacrylamide gel and then analyzed by Western blot assay. An anti-Myc (9E10; Santa Cruz Biotechnology, Inc.), antiactin (A2066; Sigma), anti-Flag (F3165; Sigma), anti-HA–peroxidase (H6533-1VL; Sigma), or anti-HA (F-7; Santa Cruz Biotechnology, Inc.) antibody was used for Western blot analysis.
Total RNA preparation.
Total RNA was prepared from yeast cell culture following the standard protocol (11, 50, 57, 58). Briefly, 10 ml yeast culture at a total OD600 of 1.0 in YPD medium was harvested and then suspended in 100 μl RNA preparation buffer (500 mM NaCl, 200 mM Tris-HCl, 100 mM Na2-EDTA, and 1% SDS), along with 100 μl phenol-chloroform-isoamyl alcohol and a 100-μl volume equivalent of glass beads (acid washed; Sigma). Subsequently, the yeast cell suspension was vortexed at maximum speed (10 in a VWR minivortexer; catalog no. 58816-121) five times (30 s each time). The cell suspension was placed in ice for 30 s between pulses. After vortexing, 150 μl RNA preparation buffer and 150 μl phenol-chloroform-isoamyl alcohol were added to the yeast cell suspension, followed by vortexing for 15 s at maximum speed on a VWR minivortexer. The aqueous phase was collected following 5 min of centrifugation at maximum speed in a microcentrifuge. The total RNA was isolated from the aqueous phase by precipitation with ethanol.
RT-PCR analysis.
Reverse transcriptase (RT) PCR analysis was performed according to the standard protocols (11, 59, 60). Briefly, total RNA was prepared from 10 ml of yeast culture. Ten micrograms of total RNA was used in the reverse transcription assay for both wild-type and mutant strains. The RNA was treated with RNase-free DNase (M610A; Promega) and then reverse transcribed into cDNA using oligo(dT) as described in the protocol supplied by Promega (A3800; Promega). PCR was performed using a synthesized first strand as the template and the primer pairs targeted to the ADH1, PGK1, PYK1, and PMA1 open reading frames (ORFs), as well as 18S rRNA. The RT-PCR products were separated by 2.2% agarose gel electrophoresis and visualized by ethidium bromide staining. The RT-PCR experiments were carried out three times. The experiments were biologically independent. The average signal of these biologically independent experiments is reported with SD (Microsoft Excel). The Student t test of Microsoft Excel (tail = 2 and types = 3) was used to determine the P values for the statistical significance of the change in the RT-PCR signals. The changes were considered to be statistically significant at a P value of <0.05. The primer pairs used in the PCR analysis are listed in Table 2.
TABLE 2.
Primer pairs used in RT-PCR analysis
| Target | Primer pair sequences |
|---|---|
| ADH1 | 5′-CGGTAACAGAGCTGACACCAGAGA-3′, 5′-ACGTATCTACCAACGATTTGACCC-3′ |
| PYK1 | 5′-AAGTTTCCGATGTCGGTAACGCTAT-3′, 5′-TTGGCAAGTAAGCGATAGCTTGTTC-3′ |
| PGK1 | 5′-AGACGAAGTTGTCAAGAGCTCTGC-3′, 5′-GAAAGCAACACCTGGCAATTCCT-3′ |
| PMA1 | 5′-TCGGTGCTATGAACGGTATTATG-3′, 5′-ACATGGTAGCGATGATGTCGACAG-3′ |
| 18S rRNA | 5′-GAGTCCTTGTGGCTCTTGGC-3′, 5′-AATACTGATGCCCCCGACC-3′ |
Formaldehyde-based in vivo cross-linking and co-IP assay.
The co-IP assay was performed as described previously (24, 25). Briefly, a yeast strain carrying Myc-tagged Cet1 and HA-tagged Spt16 was grown in YPD medium to an OD600 of 1.0 and then cross-linked by formaldehyde. WCE was prepared by lysing and sonicating the cross-linked yeast cells. Immunoprecipitation was performed using an anti-Myc antibody and protein A/G Plus-agarose beads. Anti-Flag was used as a nonspecific antibody. After immunoprecipitation, the agarose beads were washed as in the ChIP assay. The washed A/G Plus-agarose beads were boiled in SDS-PAGE loading buffer, and the supernatant was analyzed by SDS-PAGE and Western blotting. An anti-HA–horseradish peroxidase (HRP) antibody was used in the Western blot analysis.
Co-IP assay without in vivo cross-linking.
WCE of the yeast strain expressing Myc-tagged Paf1 and HA-tagged Cet1 (or a yeast strain expressing Myc-tagged Pob3 and HA-tagged Spt16) was prepared in radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris-Cl, pH 7.5, 0.1 mM EDTA, 1 mM MgCl2). Four hundred microliters of WCE was used for immunoprecipitation, with 10 μl (2 μg) of an anti-Myc antibody. Following 3-h incubation of WCE with antibody at 4°C in a rotor, 100 μl protein A/G Plus-agarose beads (Santa Cruz Biotechnology, Inc.) was added for 1 h at 4°C. Subsequently, the agarose beads were washed with RIPA buffer and resuspended in protein-loading dye for Western blot analysis, using an anti-Myc or anti-HA–HRP antibody as the primary antibody.
Immunopurification of Spt16, Pob3, Cet1, Cet1Δ204, and Sub2.
Immunopurification was performed as described previously (24, 26). Briefly, the yeast strains expressing HA-tagged Spt16, Sub2, Pob3, Cet1, or Cet1Δ204 were grown in 100 ml YPD-YPG medium to an OD600 of 1.0 and then harvested. Subsequently, 800 μl WCE was prepared from the culture of each strain. Immunoprecipitation was performed using anti-HA antibody and protein A/G Plus-agarose beads for 4 h at 4°C; 400 μl WCE, 100 μl protein A/G Plus-agarose beads (25% slurry), and 10 μl (2 μg) anti-HA antibody were used for each immunoprecipitation. Following immunoprecipitation, the agarose beads were washed under high-stringency washing conditions, as in the ChIP assay, but 0.5 M NaCl instead of 1 M NaCl was used for washing the beads. Then, the beads were equilibrated with buffer E (50 mM Tris base, 250 mM NaCl, 1% NP-40, 1 mM EDTA, pH 8.5) before elution of HA-tagged protein with HA peptide. The immobilized protein (HA-tagged Spt16, Sub2, Pob3, Cet1, or Cet1Δ204) on A/G Plus-agarose beads was eluted by incubating the beads in 2 bed volumes (100 μl) of buffer E containing HA peptide at a final concentration of 1 mg/ml. The beads were incubated for 30 min at 25°C on a rotor. Elution was performed three times. Buffer E (100 μl) with HA peptide was used for each elution.
Protein interaction assay in vitro.
Protein interaction analysis in vitro was performed as described previously (24, 26). Briefly, to analyze the interaction of Spt16 with Cet1, the yeast strain expressing Myc-tagged Cet1 was grown in 100 ml YPD to an OD600 of 1.0, and subsequently, 800 μl WCE was prepared. The WCE (400 μl) was used for immunoprecipitation as in the ChIP assay, using 10 μl anti-Myc antibody and 100 μl protein A/G Plus-agarose beads. The immobilized Cet1 on the beads was thoroughly washed under high-stringency washing conditions as in the ChIP assay, and then the washed beads with immobilized Cet1 were incubated with immunopurified Spt16 in buffer E containing HA peptide and aprotinin for 15 min at 25°C. Subsequently, the beads were washed with buffer W (50 mM Tris base, 2 mg/ml bovine serum albumin [BSA], 250 mM NaCl, 1% NP-40, 1 mM EDTA, pH 8.5) containing aprotinin four times (1 ml each time). The last wash was performed using buffer E containing aprotinin. The washed beads were then boiled in SDS-PAGE loading buffer, and the supernatant was subsequently analyzed by SDS-PAGE and Western blotting to determine the interaction between Spt16 and Cet1. An anti-HA–HRP antibody was used in the Western blot analysis. Likewise, the interactions of Pob3 with Spt16, Sub2, Cet1, and Cet1Δ204 were analyzed.
Growth analysis in the presence or absence of 6-AU.
The growth of the cet1Δ204 and wild-type cells was analyzed on plates containing solid synthetic complete medium lacking uracil (SC-uracil medium) (plus 2% dextrose) with or without 100 μg/ml 6-AU. Both the wild-type and cet1Δ204 strains were transformed with a low-copy-number plasmid (pRS416) expressing the URA3 gene, inoculated in liquid SC-uracil medium (with 2% dextrose), and grown to an OD600 of 0.2 at 30°C. Subsequently, the yeast cells were suspended in fresh liquid SC-uracil medium (with 2% dextrose) and grown to an OD600 of 0.4 prior to spotting (3 μl) on solid SC-uracil medium (plus 2% dextrose) with or without 100 μg/ml 6-AU. The yeast cells were spotted in serial dilutions, grown at 30°C, and photographed after 2 or 3 days. Growth analysis was carried out in biological triplicates, and consistent results were obtained. One representative set is shown in Fig. 2I.
Supplementary Material
ACKNOWLEDGMENTS
We thank Stephen Buratowski and David Stillman for the yeast strains and Michael R. Green for an anti-TBP antibody.
The work in the Bhaumik laboratory was supported by grants from the National Institutes of Health (1R15GM088798-01 and 2R15GM088798-02), the American Heart Association (15GRNT25700298), and Southern Illinois University School of Medicine.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00029-17.
REFERENCES
- 1.Nechaev S, Adelman K. 2011. Pol II waiting in the starting gates: regulating the transition from transcription initiation into productive elongation. Biochim Biophys Acta 1809:34–45. doi: 10.1016/j.bbagrm.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li J, Gilmour DS. 2011. Promoter proximal pausing and the control of gene expression. Curr Opin Genet Dev 21:231–235. doi: 10.1016/j.gde.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hirtreiter A, Damsma GE, Cheung AC, Klose D, Grohmann D, Vojnic E, Martin AC, Cramer P, Werner F. 2010. Spt4/5 stimulates transcription elongation through the RNA polymerase clamp coiled-coil motif. Nucleic Acids Res 38:4040–4051. doi: 10.1093/nar/gkq135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. 2010. c-Myc regulates transcriptional pause release. Cell 141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barboric M, Nissen RM, Kanazawa S, Jabrane-Ferrat N, Peterlin BM. 2001. NF-kappaB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol Cell 8:327–337. doi: 10.1016/S1097-2765(01)00314-8. [DOI] [PubMed] [Google Scholar]
- 6.Hargreaves DC, Horng T, Medzhitov R. 2009. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell 138:129–145. doi: 10.1016/j.cell.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donner AJ, Ebmeier CC, Taatjes DJ, Espinosa JM. 2010. CDK8 is a positive regulator of transcriptional elongation within the serum response network. Nat Struct Mol Biol 17:194–201. doi: 10.1038/nsmb.1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diribarne G, Bensaude O. 2009. 7SK RNA, a non-coding RNA regulating P-TEFb, a general transcription factor. RNA Biol 6:122–128. doi: 10.4161/rna.6.2.8115. [DOI] [PubMed] [Google Scholar]
- 9.Peterlin BM, Price DH. 2006. Controlling the elongation phase of transcription with P-TEFb. Mol Cell 23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 10.Baugh LR, Demodena J, Sternberg PW. 2009. RNA Pol II accumulates at promoters of growth genes during developmental arrest. Science 324:92–94. doi: 10.1126/science.1169628. [DOI] [PubMed] [Google Scholar]
- 11.Lahudkar S, Durairaj G, Uprety B, Bhaumik SR. 2014. A novel role for Cet1p mRNA 5′-triphosphatase in promoter proximal accumulation of RNA polymerase II in Saccharomyces cerevisiae. Genetics 196:161–176. doi: 10.1534/genetics.113.158535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim TS, Liu CL, Yassour M, Holik J, Friedman N, Buratowski S, Rando O. 2010. RNA polymerase mapping during stress responses reveals widespread nonproductive transcription in yeast. Genome Biol 11:R75. doi: 10.1186/gb-2010-11-7-r75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCracken S, Rosonina E, Fong N, Sikes M, Beyer A, O'Hare K, Shuman S, Bentley D. 1998. Role of RNA polymerase II carboxy-terminal domain in coordinating transcription with RNA processing. Cold Spring Harbor Symp Quant Biol 63:301–309. doi: 10.1101/sqb.1998.63.301. [DOI] [PubMed] [Google Scholar]
- 14.Rodríguez-Molina JB, Tseng SC, Simonett SP, Taunton J, Ansari AZ. 2016. Engineered covalent inactivation of TFIIH-kinase reveals an elongation checkpoint and results in widespread mRNA stabilization. Mol Cell 63:433–444. doi: 10.1016/j.molcel.2016.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cho EJ, Rodriguez CR, Takagi T, Buratowski S. 1998. Allosteric interactions between capping enzyme subunits and the RNA polymerase II carboxy-terminal domain. Genes Dev 12:3482–3487. doi: 10.1101/gad.12.22.3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rodriguez CR, Takagi T, Cho EJ, Buratowski S. 1999. A Saccharomyces cerevisiae RNA 5′-triphosphatase related to mRNA capping enzyme. Nucleic Acids Res 27:2181–2188. doi: 10.1093/nar/27.10.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Winkler DD, Luger K. 2011. The histone chaperone FACT: structural insights and mechanisms for nucleosome reorganization. J Biol Chem 286:18369–18374. doi: 10.1074/jbc.R110.180778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Formosa T. 2013. The role of FACT in making and breaking nucleosomes. Biochim Biophys Acta 1819:247–255. doi: 10.1016/j.bbagrm.2011.07.009. [DOI] [PubMed] [Google Scholar]
- 19.Reinberg D, Sims RJ III. 2006. De facto nucleosome dynamics. J Biol Chem 281 23297–23301. [DOI] [PubMed] [Google Scholar]
- 20.Fresco LD, Buratowski S. 1996. Conditional mutants of the yeast mRNA capping enzyme show that the cap enhances, but is not required for, mRNA splicing. RNA 2:584–596. [PMC free article] [PubMed] [Google Scholar]
- 21.Takase Y, Takagi T, Komarnitsky PB, Buratowski S. 2000. The essential interaction between yeast mRNA capping enzyme subunits is not required for triphosphatase function in vivo. Mol Cell Biol 20:9307–9316. doi: 10.1128/MCB.20.24.9307-9316.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng S, Crickard JB, Srikanth A, Reese JC. 2014. A highly conserved region within H2B is important for FACT to act on nucleosomes. Mol Cell Biol 34:303–314. doi: 10.1128/MCB.00478-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duina AA. 2011. Histone chaperones Spt6 and FACT: similarities and differences in modes of action at transcribed genes. Genet Res Int 2011:625210. doi: 10.4061/2011/625210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lahudkar S, Shukla A, Bajwa P, Durairaj G, Stanojevic N, Bhaumik SR. 2011. The mRNA cap-binding complex stimulates the formation of pre-initiation complex at the promoter via its interaction with Mot1p in vivo. Nucleic Acids Res 39:2188–2209. doi: 10.1093/nar/gkq1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hall DB, Struhl K. 2002. The VP16 activation domain interacts with multiple transcriptional components as determined by protein-protein cross-linking in vivo. J Biol Chem 277:46043–46050. doi: 10.1074/jbc.M208911200. [DOI] [PubMed] [Google Scholar]
- 26.Sen R, Ferdoush J, Kaja A, Bhaumik SR. 2016. Fine-tuning of FACT by the ubiquitin proteasome system in regulation of transcriptional elongation. Mol Cell Biol 36:1691–1703. doi: 10.1128/MCB.01112-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mason PB, Struhl K. 2003. The FACT complex travels with elongating RNA polymerase II and is important for the fidelity of transcriptional initiation in vivo. Mol Cell Biol 23:8323–8333. doi: 10.1128/MCB.23.22.8323-8333.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wyce A, Xiao T, Whelan KA, Kosman C, Walter W, Eick D, Hughes TR, Krogan NJ, Strahl BD, Berger SL. 2007. H2B ubiquitylation acts as a barrier to Ctk1 nucleosomal recruitment prior to removal by Ubp8 within a SAGArelated complex. Mol Cell 27:275–288. doi: 10.1016/j.molcel.2007.01.035. [DOI] [PubMed] [Google Scholar]
- 29.Biswas D, Dutta-Biswas R, Mitra D, Shibata Y, Strahl BD, Formosa T, Stillman DJ. 2006. Opposing roles for Set2 and yFACT in regulating TBP binding at promoters. EMBO J 25:4479–4489. doi: 10.1038/sj.emboj.7601333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ransom M, Williams SK, Dechassa ML, Das C, Linger J, Adkins M, Liu C, Bartholomew B, Tyler JK. 2009. FACT and the proteasome promote promoter chromatin disassembly and transcriptional initiation. J Biol Chem 284:23461–23471. doi: 10.1074/jbc.M109.019562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fleming AB, Kao CF, Hillyer C, Pikaart M, Osley MA. 2008. H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol Cell 31:57–66. doi: 10.1016/j.molcel.2008.04.025. [DOI] [PubMed] [Google Scholar]
- 32.Durairaj G, Chaurasia P, Lahudkar S, Malik S, Shukla A, Bhaumik SR. 2010. Regulation of chromatin assembly/disassembly by Rtt109p, a histone H3 Lys56-specific acetyltransferase, in vivo. J Biol Chem 285:30472–30479. doi: 10.1074/jbc.M110.113225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jaehning JA. 2010. The Paf1 complex: platform or player in RNA polymerase II transcription? Biochim Biophys Acta 1799:379–388. doi: 10.1016/j.bbagrm.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Squazzo SL, Costa PJ, Lindstrom DL, Kumer KE, Simic R, Jennings JL, Link AJ, Arndt KM, Hartzog GA. 2002. The Paf1 complex physically and functionally associates with transcription elongation factors in vivo. EMBO J 21:1764–1774. doi: 10.1093/emboj/21.7.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krogan NJ, Peng WT, Cagney G, Robinson MD, Haw R, Zhong G, Guo X, Zhang X, Canadien V, Richards DP, Beattie BK, Lalev A, Zhang W, Davierwala AP, Mnaimneh S, Starostine A, Tikuisis AP, Grigull J, Datta N, Bray JE, Hughes TR, Emili A, Greenblatt JF. 2004. High-definition macromolecular composition of yeast RNA-processing complexes. Mol Cell 13:225–239. doi: 10.1016/S1097-2765(04)00003-6. [DOI] [PubMed] [Google Scholar]
- 36.Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, Punna T, Peregrín-Alvarez JM, Shales M, Zhang X, Davey M, Robinson MD, Paccanaro A, Bray JE, Sheung A, Beattie B, Richards DP, Canadien V, Lalev A, Mena F, Wong P, Starostine A, Canete MM, Vlasblom J, Wu S, Orsi C, Collins SR, Chandran S, Haw R, Rilstone JJ, Gandi K, Thompson NJ, Musso G, St Onge P, Ghanny S, Lam MH, Butland G, Altaf-Ul AM, Kanaya S, Shilatifard A, O'Shea E, Weissman JS, Ingles CJ, Hughes TR, Parkinson J, Gerstein M, Wodak SJ, Emili A, Greenblatt JF. 2006. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 37.Krogan NJ, Kim M, Ahn SH, Zhong G, Kobor MS, Cagney G, Emili A, Shilatifard A, Buratowski S, Greenblatt JF. 2002. RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol Cell Biol 22:6979–6992. doi: 10.1128/MCB.22.20.6979-6992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dümpelfeld B, Edelmann A, Heurtier MA, Hoffman V, Hoefert C, Klein K, Hudak M, Michon AM, Schelder M, Schirle M, Remor M, Rudi T, Hooper S, Bauer A, Bouwmeester T, Casari G, Drewes G, Neubauer G, Rick JM, Kuster B, Bork P, Russell RB, Superti-Furga G. 2006. Proteome survey reveals modularity of the yeast cell machinery. Nature 440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 39.Gavin AC, Bösche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, Remor M, Höfert C, Schelder M, Brajenovic M, Ruffner H, Merino A, Klein K, Hudak M, Dickson D, Rudi T, Gnau V, Bauch A, Bastuck S, Huhse B, Leutwein C, Heurtier MA, Copley RR, Edelmann A, Querfurth E, Rybin V, Drewes G, Raida M, Bouwmeester T, Bork P, Seraphin B, Kuster B, Neubauer G, Superti-Furga G. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 40.Dronamraju R, Strahl BD. 2014. A feed forward circuit comprising Spt6, Ctk1 and PAF regulates Pol II CTD phosphorylation and transcription elongation. Nucleic Acids Res 42:870–881. doi: 10.1093/nar/gkt1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaplan CD, Laprade L, Winston F. 2003. Transcription elongation factors repress transcription initiation from cryptic sites. Science 301:1096–1099. doi: 10.1126/science.1087374. [DOI] [PubMed] [Google Scholar]
- 42.DeGennaro CM, Alver BH, Marguerat S, Stepanova E, Davis CP, Bähler J, Park PJ, Winston F. 2013. Spt6 regulates intragenic and antisense transcription, nucleosome positioning, and histone modifications genome-wide in fission yeast. Mol Cell Biol 33:4779–4792. doi: 10.1128/MCB.01068-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McCullough L, Connell Z, Petersen C, Formosa T. 2015. The abundant histone chaperones Spt6 and FACT collaborate to assemble, inspect, and maintain chromatin structure in Saccharomyces cerevisiae. Genetics 201:1031–1045. doi: 10.1534/genetics.115.180794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu Y, Warfield L, Zhang C, Luo J, Allen J, Lang WH, Ranish J, Shokat KM, Hahn S. 2009. Phosphorylation of the transcription elongation factor Spt5 by yeast Bur1 kinase stimulates recruitment of the PAF complex. Mol Cell Biol 29:4852–4863. doi: 10.1128/MCB.00609-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pruneski JA, Hainer SJ, Petrov KO, Martens JA. 2011. The Paf1 complex represses SER3 transcription in Saccharomyces cerevisiae by facilitating intergenic transcription-dependent nucleosome occupancy of the SER3 promoter. Eukaryot Cell 10:1283–1294. doi: 10.1128/EC.05141-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu M, Yang W, Ni T, Tang Z, Nakadai T, Zhu J, Roeder RG. 2015. RNA polymerase II-associated factor 1 regulates the release and phosphorylation of paused RNA polymerase II. Science 350:1383–1386. doi: 10.1126/science.aad2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Longtine MS, McKenzie A, Demarini DJ III, Shah NG, Wach A, Brachat A, Philippsen P, Pingle JR. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953–961. [DOI] [PubMed] [Google Scholar]
- 48.Uprety B, Lahudkar S, Malik S, Bhaumik SR. 2012. The 19S proteasome subcomplex promotes the targeting of NuA4 HAT to the promoters of ribosomal protein genes to facilitate the recruitment of TFIID for transcriptional initiation in vivo. Nucleic Acids Res 40:1969–1983. doi: 10.1093/nar/gkr977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Malik S, Shukla A, Sen P, Bhaumik SR. 2009. The 19S proteasome subcomplex establishes a specific protein interaction network at the promoter for stimulated transcriptional initiation in vivo. J Biol Chem 284:35714–35724. doi: 10.1074/jbc.M109.035709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Durairaj G, Sen R, Uprety B, Shukla A, Bhaumik SR. 2014. Sus1p facilitates pre-initiation complex formation at the SAGA-regulated genes independently of histone H2B de-ubiquitylation. J Mol Biol 426:2928–2941. doi: 10.1016/j.jmb.2014.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bhaumik SR, Green MR. 2003. Interaction of Gal4p with components of transcription machinery in vivo. Methods Enzymol 370:445–454. doi: 10.1016/S0076-6879(03)70038-X. [DOI] [PubMed] [Google Scholar]
- 52.Shukla A, Stanojevic N, Duan Z, Sen P, Bhaumik SR. 2006. Ubp8p, a histone deubiquitinase whose association with SAGA is mediated by Sgf11p, differentially regulates lysine 4 methylation of histone H3 in vivo. Mol Cell Biol 26:3339–3352. doi: 10.1128/MCB.26.9.3339-3352.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shukla A, Stanojevic N, Duan Z, Shadle T, Bhaumik SR. 2006. Functional analysis of H2B-Lys-123 ubiquitination in regulation of H3-Lys-4 methylation and recruitment of RNA polymerase II at the coding sequences of several active genes in vivo. J Biol Chem 281:19045–19054. doi: 10.1074/jbc.M513533200. [DOI] [PubMed] [Google Scholar]
- 54.Bhaumik SR, Green MR. 2002. Differential requirement of SAGA components for recruitment of TATA-box-binding protein to promoters in vivo. Mol Cell Biol 22:7365–7371. doi: 10.1128/MCB.22.21.7365-7371.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Uprety B, Sen R, Bhaumik SR. 2015. Eaf1p is required for recruitment of NuA4 in targeting TFIID to the promoters of the ribosomal protein genes for transcriptional initiation in vivo. Mol Cell Biol 35:2947–2964. doi: 10.1128/MCB.01524-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sen R, Malik S, Frankland-Searby S, Uprety B, Lahudkar S, Bhaumik SR. 2014. Rrd1p, an RNA polymerase II-specific prolyl isomerase and activator of phosphoprotein phosphatase, promotes transcription independently of rapamycin response. Nucleic Acids Res 42:9892–9907. doi: 10.1093/nar/gku703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Durairaj G, Lahudkar S, Bhaumik SR. 2014. A new regulatory pathway of mRNA export by an F-box protein, Mdm30. RNA 20:133–142. doi: 10.1261/rna.042325.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Peterson CL, Kruger W, Herskowitz I. 1991. A functional interaction between the C-terminal domain of RNA polymerase II and the negative regulator SIN1. Cell 64:1135–1143. doi: 10.1016/0092-8674(91)90268-4. [DOI] [PubMed] [Google Scholar]
- 59.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Struhl K. 2001. Current protocols in molecular biology. Wiley, New York, NY. [Google Scholar]
- 60.Malik S, Durairaj G, Bhaumik SR. 2013. Mechanisms of antisense transcription initiation from the 3-end of the GAL10 coding sequence in vivo. Mol Cell Biol 33:3549–3567. doi: 10.1128/MCB.01715-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bhaumik SR. 2011. Distinct regulatory mechanisms of eukaryotic transcriptional activation by SAGA and TFIID. Biochim Biophys Acta 1809:97–108. doi: 10.1016/j.bbagrm.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bhaumik SR, Malik S. 2008. Diverse regulatory mechanisms of eukaryotic transcriptional activation by the proteasome complex. Crit Rev Biochem Mol Biol 43:419–433. doi: 10.1080/10409230802605914. [DOI] [PubMed] [Google Scholar]
- 63.Sen R, Lahudkar S, Durairaj G, Bhaumik SR. 2013. Functional analysis of Bre1p, an E3 ligase for histone H2B ubiquitylation, in regulation of RNA polymerase II association with active genes and transcription in vivo. J Biol Chem 288:9619–9633. doi: 10.1074/jbc.M113.450403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sen R, Bhaumik SR. 2013. Transcriptional stimulatory and repressive functions of histone H2B ubiquitin ligase. Transcription 4:221–226. doi: 10.4161/trns.26623. [DOI] [PubMed] [Google Scholar]
- 65.Kim J, Roeder RG. 2009. Direct Bre1-Paf1 complex interactions and RING finger-independent Bre1-Rad6 interactions mediate histone H2B ubiquitylation in yeast. J Biol Chem 284:20582–20592. doi: 10.1074/jbc.M109.017442. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.