Abstract
Brown adipose tissue dissipates energy as heat, a process that relies on a high abundance of mitochondria and high levels of electron transport chain (ETC) complexes within these mitochondria. Two regulators of mitochondrial respiration and heat production in brown adipocytes are the transcriptional coactivator PGC-1α and its splicing isoform NT-PGC-1α, which control mitochondrial gene expression in the nucleus. Surprisingly, we found that, in brown adipocytes, some NT-PGC-1α localizes to mitochondria, whereas PGC-1α resides in the nucleus. Here we sought to investigate the role of NT-PGC-1α in brown adipocyte mitochondria. Immunocytochemistry, immunotransmission electron microscopy, and biochemical analyses indicated that NT-PGC-1α was located in the mitochondrial matrix in brown adipocytes. NT-PGC-1α was specifically enriched at the D-loop region of the mtDNA, which contains the promoters for several essential ETC complex genes, and was associated with LRP130, an activator of mitochondrial transcription. Selective expression of NT-PGC-1α and PGC-1α in PGC-1α−/− brown adipocytes similarly induced expression of nuclear DNA-encoded mitochondrial ETC genes, including the key mitochondrial transcription factor A (TFAM). Despite having comparable levels of TFAM expression, PGC-1α−/− brown adipocytes expressing NT-PGC-1α had higher expression of mtDNA-encoded ETC genes than PGC-1α−/− brown adipocytes expressing PGC-1α, suggesting a direct effect of NT-PGC-1α on mtDNA transcription. Moreover, this increase in mtDNA-encoded ETC gene expression was associated with enhanced respiration in NT-PGC-1α-expressing PGC-1α−/− brown adipocytes. Our findings reveal a previously unappreciated and isoform-specific role for NT-PGC-1α in the regulation of mitochondrial transcription in brown adipocytes and provide new insight into the transcriptional control of mitochondrial respiration.
Keywords: adipocyte, adipose tissue metabolism, gene expression, mtDNA, mitochondrial respiratory chain complex, peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α)
Introduction
Brown adipose tissue (BAT)2 dissipates energy in the form of heat. This thermogenic process relies on a high abundance of mitochondria as well as high levels of electron transport chain (ETC) complexes and uncoupling protein 1 (UCP1) within those mitochondria (1–3). The ETC system creates a proton gradient (energy) across the inner mitochondrial membrane by oxidation of glucose and fatty acids. UCP1 decreases this proton gradient by allowing protons to return to the mitochondrial matrix, resulting in heat production. Accordingly, a cold-induced increase in mitochondrial ETC activity is critically important for UCP1-mediated heat production in BAT during cold adaptation. The ETC is composed of four large multisubunit complexes (I-IV). The majority of ETC complex subunits are encoded by nuclear DNA and are imported into mitochondria, whereas 11 essential subunits of ETC complexes I, III, and IV are encoded by mtDNA (4, 5). Although the basal mitochondrial transcription machinery consisting of mitochondrial RNA polymerase (POLRMT) and mitochondrial transcription factor A (TFAM) and mitochondrial transcription factor B2 (TFB2M) has been characterized (6–8), very little is known about how mitochondrial DNA transcription is regulated in response to various physiological stimuli.
Peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α) and its splicing isoform NT-PGC-1α play crucial roles in mitochondrial biogenesis and respiration in BAT during cold adaptation (1, 3, 9, 10). Mechanistically, PGC-1α and NT-PGC-1α are highly induced by cold and coactivate a number of nuclear receptors and transcription factors, such as peroxisome proliferator-activated receptors (PPARs), estrogen-related receptors, and nuclear respiratory factors (NRFs) to activate expression of a broad set of mitochondrial genes, including electron transport chain and fatty acid oxidation genes (3, 9–12). Especially a complex with NRF-1 induces the expression of TFAM that is a key activator of mitochondrial DNA transcription as well as a regulator of mitochondrial genome replication, thus leading to a concurrent increase in mtDNA-encoded ETC gene expression (1). Several recent studies have shown additional regulatory proteins involved in mitochondrial DNA transcription in cell type- and stimulus-specific manners (13–20). Here we report the unexpected presence of NT-PGC-1α in brown adipocyte mitochondria and address its role, which is unique from PGC-1α, in the regulation of mtDNA-encoded ETC gene expression.
Results
A portion of NT-PGC-1α localizes to mitochondria in brown adipocytes
We have demonstrated that NT-PGC-1α is predominantly cytoplasmic and that its nuclear accumulation is increased by cold/β-adrenergic receptor/cAMP/PKA-mediated phosphorylation (21). Subcellular fractionation of BAT and Western blot analysis with a highly specific anti- PGC-1α antibody (Fig. 1B, bottom left panel) further revealed that, although PGC-1α was only detected in the nuclear fraction, cold-induced NT-PGC-1α was present in the highly purified mitochondrial fraction as well (Fig. 1B, bottom right panel). The cytosolic and nuclear markers were only detected in their respective fractions, indicating no contamination of the mitochondrial fraction by other subcellular fractions. To determine the mitochondrial localization of NT-PGC-1α in brown adipocytes, NT-PGC-1α was stably expressed in PGC-1α−/− brown adipocytes as described previously (21–23). It should be noted that NT-PGC-1α protein levels in PGC-1α−/− brown adipocytes were comparable with those of endogenous NT-PGC-1α induced by cAMP in wild-type brown adipocytes (Fig. 1C). Subcellular fractionation of brown adipocytes also showed that a portion of NT-PGC-1α was present in the mitochondrial fraction (Fig. 1D). We next evaluated whether mitochondrial targeting of NT-PGC-1α is regulated by cAMP-induced signaling. No change in mitochondrial content of NT-PGC-1α was observed after treatment of brown adipocytes with cAMP, indicating that mitochondrial targeting of NT-PGC-1α is independent of cAMP signaling (Fig. 1E). Consistent with our Western blot findings, immunocytochemical studies showed co-localization of NT-PGC-1α with mitochondria in PGC-1α−/− brown adipocytes (Fig. 1F). In contrast, PGC-1α was only localized in the nucleus in PGC-1α−/− brown adipocytes (Fig. 1F).
Figure 1.
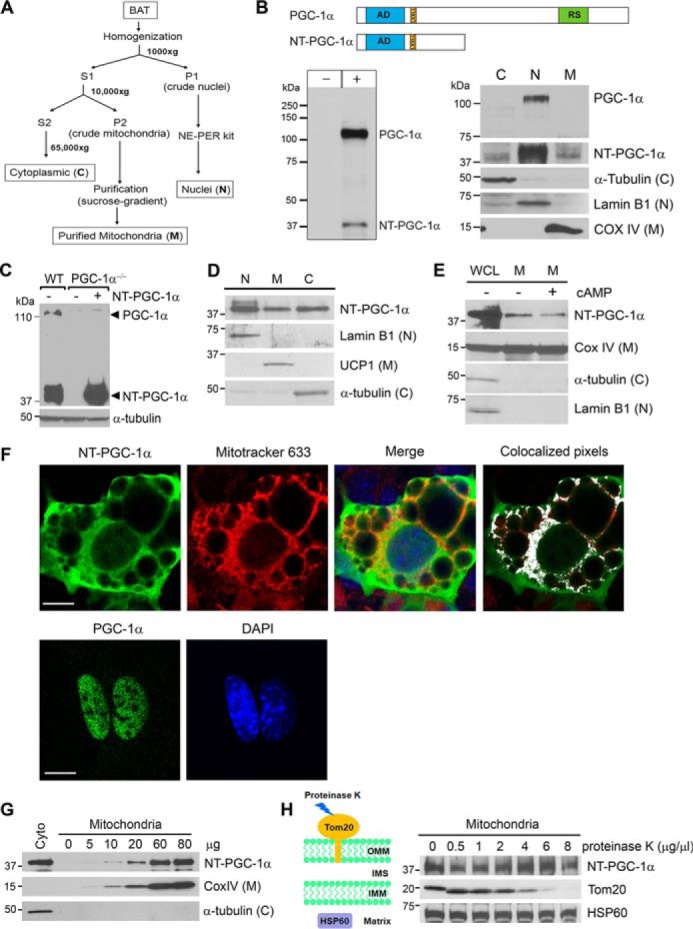
NT-PGC-1α is present in brown adipocyte mitochondria. A, schematic of subcellular fractionation. Brown adipose tissues were isolated from C57BL/6J mice exposed to 4 °C for 5 h and subjected to subcellular fractionation. B, presence of NT-PGC-1α in mitochondria. Top panel, schematic of the PGC-1α and NT-PGC-1α proteins. AD, transcription activation domain; LxxLL, nuclear receptor interaction motif; RS, arginine/serine-rich domain. Bottom left panel, PGC-1α-HA and NT-PGC-1α-HA were expressed in COS-1 cells and immunoblotted with a highly specific anti-PGC-1α monoclonal antibody (9). Bottom right panel, endogenous PGC-1α and NT-PGC-1α in BAT isolated from cold-exposed mice were immunoblotted with the same anti-PGC-1α antibody. The cytosolic (C), nuclear (N), and mitochondrial (M) markers were detected in their respective fractions. C, expression of NT-PGC-1α in PGC-1α−/− brown adipocytes. WT brown adipocytes expressing an empty vector and PGC-1α−/− brown adipocytes stably expressing NT-PGC-1α or an empty vector by retrovirus-mediated gene transfer were treated with 0.5 mm dibutyryl cAMP for 4 h. D, presence of NT-PGC-1α in brown adipocyte mitochondria. PGC-1α−/− brown adipocytes stably expressing NT-PGC-1α were treated with dibutyryl cAMP and subjected to subcellular fractionation. E, cAMP-induced signaling does not regulate mitochondrial targeting of NT-PGC-1α. PGC-1α−/− brown adipocytes stably expressing NT-PGC-1α were treated with vehicle or dibutyryl cAMP for 4 h. WGL, whole cell lysates. F, localization of NT-PGC-1α and PGC-1α in PGC-1α−/− brown adipocytes. NT-PGC-1α-HA and PGC-1α-HA were expressed in PGC-1α−/− brown adipocytes by adenovirus-mediated gene transfer and analyzed for cellular localization by indirect immunofluorescence with anti-HA antibody. Colocalized pixels were analyzed using an ImageJ analysis tool and are displayed in white on an RGB overlay image. Scale bars = 23 μm. G, correlation of the amounts of mitochondria and the protein levels of NT-PGC-1α. Mitochondrial enrichment of NT-PGC-1α was analyzed in increasing amounts of purified brown adipocyte mitochondria. H, mitochondrial NT-PGC-1α is protected from proteinase K digestion. Purified mitochondria (60 μg) were treated with increasing amounts of proteinase K.
To further confirm that mitochondrial NT-PGC-1α did not result from contamination from the cytoplasm, NT-PGC-1α was immunoblotted from increasing amounts of purified mitochondria. The levels of NT-PGC-1α closely correlated with the amounts of mitochondria that were represented by a mitochondrial protein COX IV, whereas a cytoplasmic protein, α-tubulin, was not detected regardless of mitochondria loaded (Fig. 1G). Next, to evaluate whether NT-PGC-1α localizes within the mitochondria, isolated mitochondria were treated with increasing concentrations of proteinase K, which cannot penetrate the mitochondrial membranes. Translocase of outer mitochondrial membrane 20 (Tom20) was completely degraded at high concentrations of proteinase K. On the other hand, mitochondrial matrix heat shock protein 60 (HSP60) was resistant to protease K digestion. Similarly, NT-PGC-1α was protected from proteinase K digestion (Fig. 1H), indicating that NT-PGC-1α does not adhere to the outer mitochondrial membrane in a nonspecific manner and is probably localized within the mitochondria.
NT-PGC-1α is enriched in the mitochondrial matrix and recruited to the D-loop region of mitochondrial DNA
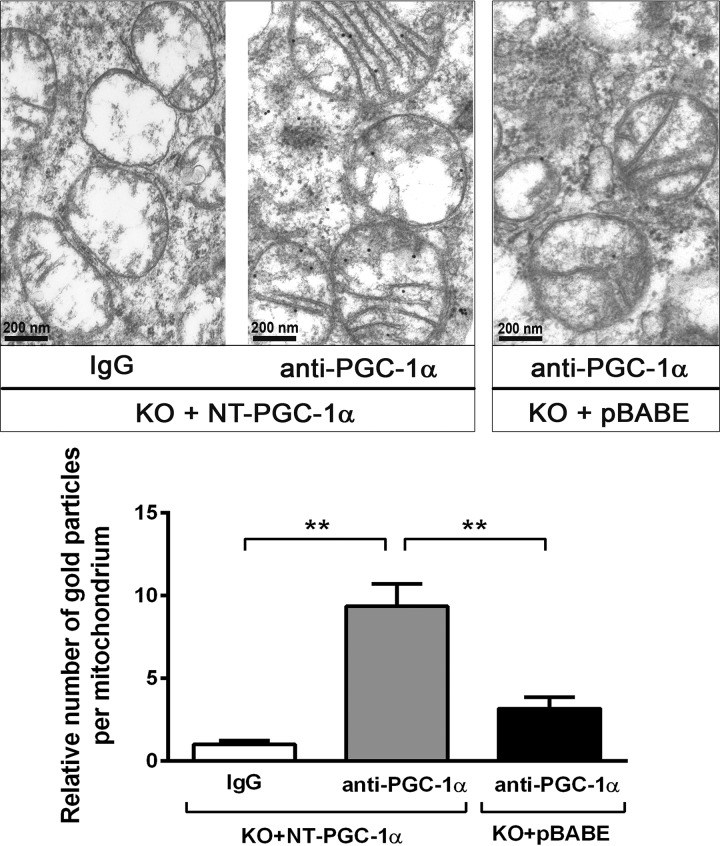
To determine the submitochondrial localization of NT-PGC-1α, immunotransmission electron microscopy was carried out using a highly specific anti-PGC-1α monoclonal antibody in PGC-1α−/− (KO) brown adipocytes expressing NT-PGC-1α or an empty vector (pBABE). NT-PGC-1α-specific immunogold particles were found in the mitochondrial matrix of PGC-1α−/− brown adipocytes expressing NT-PGC-1α (KO + NT-PGC-1α), whereas the number of these immunogold particles was greatly reduced in PGC-1α−/− brown adipocytes expressing the empty vector (KO + pBABE) (Fig. 2). In addition, this immunogold particle distribution was not observed in KO + NT-PGC-1α cells when the primary PGC-1α antibody was omitted.
Figure 2.
NT-PGC-1α localizes in the mitochondrial matrix. Immunotransmission electron microscopic analysis of NT-PGC-1α in brown adipocytes. Black dots represent immunogold particles reacted with PGC-1α antibody in PGC-1α−/− brown adipocytes (KO) expressing NT-PGC-1α or an empty vector (pBABE). Mitochondrial localization of immunogold particles was examined in 6–8 grids/group (20–30 mitochondria/grid), and the relative number of immunogold particles localized in the mitochondria in each group is shown in the bottom panel. Data are presented as the mean ± S.E. One-way ANOVA was used to compare the difference between groups: **, p < 0.01.
The finding that NT-PGC-1α was enriched in the mitochondrial matrix prompted us to ask whether NT-PGC-1α regulates mitochondrial DNA transcription. mtDNA encodes 11 essential subunits of ETC complexes I, III, and IV and two subunits of ATP synthase. The D-loop region of mtDNA contains the origin of replication and the promoters for transcription (24). Thus, to test whether mitochondrial NT-PGC-1α is recruited to the D-loop region of mtDNA, we isolated mitochondria from wild-type brown adipocytes treated with cAMP for 4 h and performed mitochondrial ChIP (mtChIP) assays using an anti-PGC-1α polyclonal antibody that has been confirmed for its specificity to immunoprecipitate NT-PGC-1α (9, 10, 22). The mtChIP showed that endogenous mitochondrial NT-PGC-1α was enriched at the D-loop region of mtDNA (Fig. 3B, top panel). No binding of NT-PGC-1α was detected at the coding region of the ND1 gene, strengthening its specific recruitment to the D-loop region. In addition, no specific protein-DNA complexes were detected at the D-loop region by immunoprecipitation with IgG. To further validate our mtChIP conditions, we examined the binding of TFAM to the D-loop region of mtDNA. TFAM was highly enriched at the D-loop region and also bound to the ND1 gene to a lesser extent (Fig. 3B, bottom panel). The latter finding is consistent with previous reports that TFAM binds to the entire mtDNA and enhances mtDNA stability (25, 26).
Figure 3.
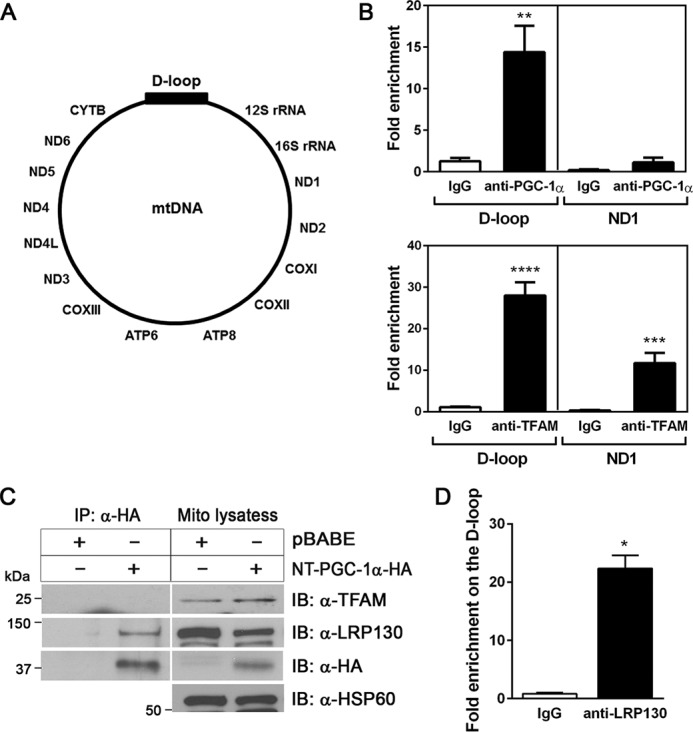
Mitochondrial NT-PGC-1α is enriched at the D-loop region of mitochondrial DNA. A, schematic of mtDNA. B, enrichment of mitochondrial NT-PGC-1α at the D-loop region of mtDNA in brown adipocytes. Mitochondria were isolated from wild-type brown adipocytes treated with 0.5 mm dibutyryl cAMP for 4 h and subjected to chromatin immunoprecipitation assays. The relative amounts of mtDNA immunoprecipitated with IgG, PGC-1α, and TFAM antibodies were analyzed by quantitative real-time PCR analysis (n = 6). Data represent mean ± S.E. **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. C, mitochondrial NT-PGC-1α does not interact with TFAM but with LRP130. Mitochondria were isolated from PGC-1α−/− brown adipocytes expressing NT-PGC-1α-HA or pBABE and subjected to immunoprecipitation (IP) using anti-HA antibody. D, enrichment of LRP130 at the mitochondrial D-loop region. Brown adipocyte mitochondria were isolated from wild-type brown adipocytes treated with 0.5 mm dibutyryl cAMP for 4 h, and mitochondrial ChIP was carried out with anti-LRP130 antibody. Data represent mean ± S.E. *, p < 0.05.
Next we examined whether mitochondrial NT-PGC-1α interacts with TFAM in brown adipocyte mitochondria. Immunoprecipitation of NT-PGC-1α-HA with anti-HA antibody did not pull down endogenous TFAM from the brown adipocyte mitochondrial lysates (Fig. 3C). In contrast, NT-PGC-1α co-precipitated a leucine-rich PPR motif-containing protein (LRP130) that is an activator of mitochondrial DNA transcription (Fig. 3C). LRP130 is localized in the mitochondrial matrix and associated with POLRMT, promoting the expression of mtDNA-encoded ETC genes (27). Consistent with the interaction between two proteins, LRP130 was enriched at the same D-loop region recognized by NT-PGC-1α (Fig. 3D). However, no interaction of mitochondrial NT-PGC-1α with POLRMT was observed (data not shown). Collectively, these results demonstrate the enrichment of NT-PGC-1α at the D-loop region and the interaction with LRP130 in the mitochondrial matrix, suggesting that NT-PGC-1α is recruited to mitochondrial transcription complexes through the interaction with LRP130.
NT-PGC-1α increases mtDNA-encoded ETC gene expression in brown adipocytes
In the nucleus, PGC-1α and NT-PGC-1α activate the expression of a broad set of mitochondrial genes, including TFAM, which is, in turn, imported to mitochondria and activates mtDNA transcription (1). Our findings further suggest that mitochondrial NT-PGC-1α may be an additional regulator of mtDNA transcription that is unique from PGC-1α. To test this possibility, we selectively expressed NT-PGC-1α and PGC-1α in PGC-1α−/− brown adipocytes and compared their effects on mtDNA-encoded ETC gene expression. NT-PGC-1α and PGC-1α produced a similar increase in gene expression of known target genes (UCP1, CIDEA, CPT1β, and PPARα) and many mitochondrial genes (TFAM, CytC, ATP5b, COX7a1, Cycs, and COX8b) encoded by nuclear DNA (nucDNA) (Fig. 4A). Consistent with this, protein levels of TFAM induced by NT-PGC-1α were comparable with those induced by PGC-1α (Fig. 4B). However, despite the same levels of TFAM, mtDNA-encoded ETC gene expression (COX1, COX2, COX3, ND1, and ND5) was significantly higher in NT-PGC-1α-expressing brown adipocytes compared with PGC-1α-expressing brown adipocytes (Fig. 4A). Moreover, higher expression levels of mtDNA-encoded ETC genes were associated with higher rates of basal and cAMP-stimulated oxygen consumption in NT-PGC-1α-expressing brown adipocytes (Fig. 4C). These data suggest that NT-PGC-1α-mediated regulation of mtDNA transcription in mitochondria is important for maximal expression of mtDNA-encoded ETC genes in brown adipocytes.
Figure 4.
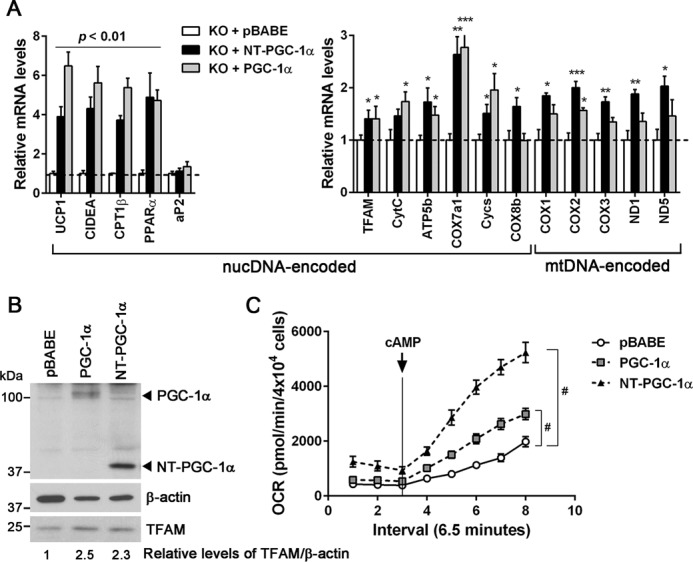
NT-PGC-1α increases mitochondrial DNA-encoded gene expression. A, differential effects of NT-PGC-1α and PGC-1α on mtDNA-encoded gene expression. PGC-1α−/− brown adipocytes stably expressing pBABE, NT-PGC-1α, and PGC-1α were assessed for nucDNA- and mtDNA-encoded gene expression (n = 6). Data represent mean ± S.E. *, p < 0.05; **, p < 0.01; ***, p < 0.001. B, Western blot analysis of TFAM in PGC-1α−/− brown adipocytes stably expressing pBABE, NT-PGC-1α, and PGC-1α. Relative protein levels of TFAM normalized to β-actin were determined by densitometric analysis using ImageJ as described under “Experimental Procedures.” C, effects of NT-PGC-1α and PGC-1α on mitochondrial respiration in PGC-1α−/− brown adipocytes. OCRs were measured at baseline and after stimulation with cAMP (n = 4) as described under “Experimental Procedures.” Representative results from three independent experiments are shown and presented as the mean ± S.E. Two-way ANOVA was used to compare the difference between groups: #, p < 0.0001.
Mitochondrially targeted MLS-NT-PGC-1α enhances mtDNA-encoded ETC gene expression and mitochondrial respiration
To assess the function of NT-PGC-1α specifically in mitochondria without its effect in the nucleus, we constructed an NT-PGC-1α that contained the mitochondrial matrix-localizing sequence (MLS) fused to the N terminus of the protein. Transiently expressed MLS-NT-PGC-1α was clearly colocalized with mitochondria (Fig. 5A). Subcellular fractionation further demonstrated that the relative mitochondrial:cytosolic ratio of MLS-NT-PGC-1α was increased by 2.3-fold compared with that of NT-PGC-1α (Fig. 5B), indicating increased targeting of NT-PGC-1α from the cytosol to the mitochondria by addition of MLS. However, nuclear localization of NT-PGC-1α was not completely prevented by addition of MLS, probably because of its ability to pass through the nuclear pores (21). A transcriptional co-activation assay using a luciferase reporter containing the PPAR response element (PPRE) elements showed that co-expression of NT-PGC-1α with PPARγ and retinoid X receptor α (RXRα) increased reporter gene expression by 10.3-fold compared with control cells, whereas MLS-NT-PGC-1α only led to a 3.7-fold increase in reporter gene expression (Fig. 5C). This result may suggest that the nuclear content of MLS-NT-PGC-1α is in part reduced because of increased mitochondrial targeting. When the MLS was added to the N terminus of PGC-1α, no change in localization was observed (supplemental Fig. S1), suggesting that multiple nuclear localization signals present at the C-terminal domain of PGC-1α might be dominant over the MLS.
Figure 5.
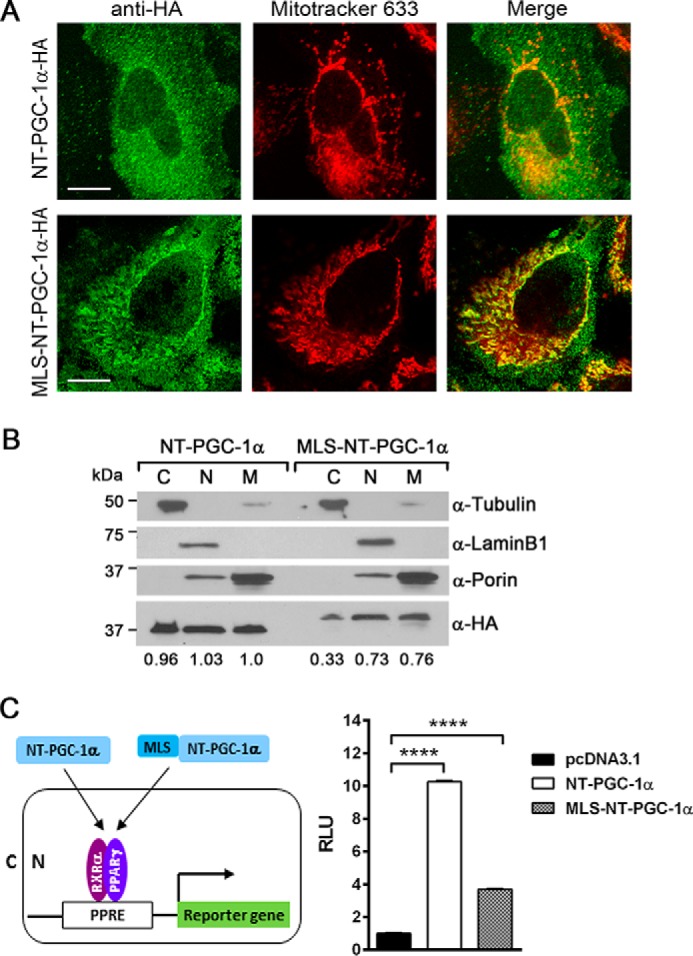
Increased mitochondrial targeting of NT-PGC-1α by addition of MLS. A, cellular localization of NT-PGC-1α and MLS-NT-PGC-1α. HeLa cells were transfected with NT-PGC-1α-HA and MLS-NT-PGC-1α-HA using FuGENE 6. Immunofluorescence was carried out using anti-HA antibody and MitoTracker Deep Red 633. Scale bars = 23 μm. B, subcellular distribution of NT-PGC-1α-HA and MLS-NT-PGC-1α-HA in HeLa cells. Relative protein levels of NT-PGC-1α-HA and MLS-NT-PGC-1α-HA in cytoplasmic (C), nuclear (N), and mitochondrial (M) fractions were determined by densitometric analysis using ImageJ as described under “Experimental Procedures.” C, effect of NT-PGC-1α and MLS-NT-PGC-1α on PPARγ/RXRα-mediated reporter gene expression in the nucleus. NT-PGC-1α or MLS-NT-PGC-1α was co-transfected with a luciferase reporter gene containing three copies of PPAR-binding sites, PPARγ, RXRα, and a Renilla luciferase reporter gene in HeLa cells. Luciferase activity was determined after 48-h transfection and normalized with Renilla luciferase activity. Data represent the mean ± S.E. of three independent experiments. One-way ANOVA was used to compare the difference between groups: ****, p < 0.0001. RLU, relative light units.
To more definitively evaluate the effect of MLS-NT-PGC-1α on mtDNA-encoded ETC gene expression in brown adipocytes, MLS-NT-PGC-1α was stably expressed in PGC-1α−/− brown adipocytes by retrovirus-mediated gene transfer. We found that MLS-NT-PGC-1α significantly elevated the expression of mtDNA-encoded ETC genes in PGC-1α−/− brown adipocytes (Fig. 6A). Expression of nuclear DNA-encoded genes (COX7a1, COX8b, ATP5b, COXIV, Cycs, UCP1, PPARα, NRF1, and TFAM) was also induced by MLS-NT-PGC-1α to some extent (Fig. 6A). However, MLS-NT-PGC-1α had no effect on mitochondrial biogenesis in PGC-1α−/− brown adipocytes (Fig. 6B). The changes in mitochondrial ETC gene expression were closely associated with enhanced mitochondrial respiration (Fig. 6C). Basal mitochondrial respiration, ATP synthase-independent proton leak, and FCCP-stimulated maximal respiration were higher in MLS-NT-PGC-1α-expressing brown adipocytes compared with control brown adipocytes.
Figure 6.
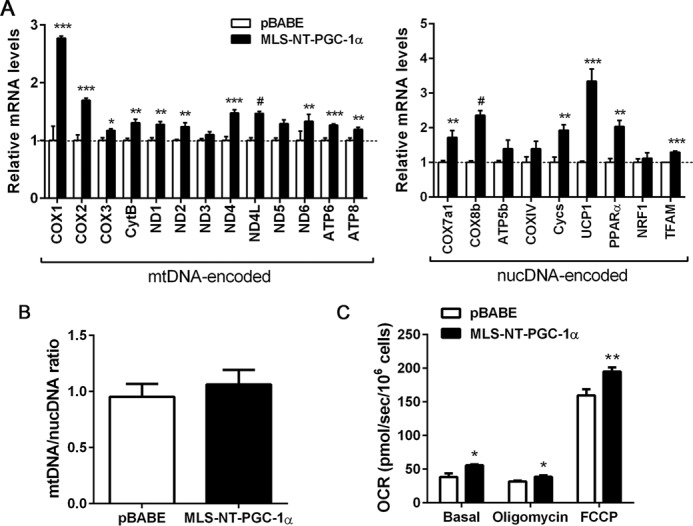
MLS-NT-PGC-1α increases mitochondrial DNA-encoded gene expression and mitochondrial respiration. A, MLS-NT-PGC-1α increases mtDNA- and nucDNA-encoded gene expression. Quantitative real-time PCR was carried out in PGC-1α−/− brown adipocytes expressing pBABE and MLS-NT-PGC-1α (n = 5). Data represent mean ± S.E. *, p < 0.05; **, p < 0.01; ***, p < 0.001; #, p < 0.0001. B, quantitative analysis of mitochondrial biogenesis. The ratio of mtDNA relative to nucDNA was analyzed by quantitative real-time PCR (n = 6/group). Data represent mean ± S.E. C, MLS-NT-PGC-1α enhances mitochondrial respiration in PGC-1α−/− brown adipocytes. OCRs were measured at baseline and after injection of oligomycin and FCCP (n = 6) as described under “Experimental Procedures.” Representative results from four independent experiments are shown and presented as the mean ± S.E. *, p < 0.05; **, p < 0.01.
Discussion
Biogenesis of functional ETC complexes requires coordinated expression of mitochondrial ETC genes from nuclear and mitochondrial genomes (4, 5). During cold adaptation, cold-inducible PGC-1α and NT-PGC-1α regulate mitochondrial respiratory capacity in BAT by two mechanisms: direct modulation of nucDNA-encoded mitochondrial ETC gene expression and indirect regulation of mtDNA-encoded ETC gene expression through TFAM (1, 10, 23). In this study, we provide the first evidence showing that NT-PGC-1α is present in brown adipocyte mitochondria, where it is enriched at the D-loop region of mtDNA, associated with LRP130, and involved in mtDNA transcription. Thus, our findings point to an additional mechanism by which NT-PGC-1α regulates mitochondrial respiration by directly modulating mtDNA-encoded ETC gene expression in mitochondria (Fig. 7). Our findings identify a previously unappreciated and isoform-specific role for NT-PGC-1α and raise a series of interesting questions, such as why NT-PGC-1α is targeted to the mitochondria, how NT-PGC-1α is imported to the mitochondria without a classical mitochondrial localizing sequence, and what components of the mitochondrial transcriptional machinery NT-PGC-1α interacts with.
Figure 7.
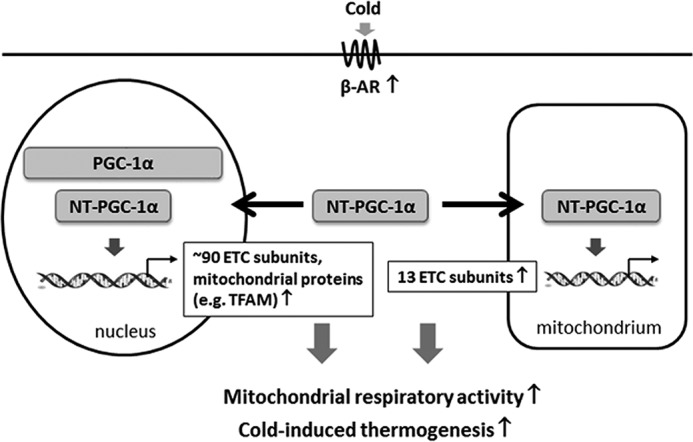
A proposed scheme of the transcriptional control of mitochondrial respiratory function by NT-PGC-1α in brown adipocytes. Cold-induced PGC-1α and NT-PGC-1α transcriptionally regulate mitochondrial respiratory function in brown adipocytes by stimulating the expression of nucDNA-encoded ETC genes and other mitochondrial genes, including TFAM. TFAM is subsequently imported to mitochondria and regulates mitochondrial DNA replication and transcription, leading to an increase in mtDNA-encoded ETC gene expression. Our findings suggest an additional mechanism by which NT-PGC-1α regulates mtDNA transcription within brown adipocyte mitochondria. Simultaneous localization of NT-PGC-1α in the nucleus and mitochondria may contribute to the coordinated regulation of nucDNA- and mtDNA-encoded ETC gene expression in response to cold.
The expression and activity of PGC-1α and NT-PGC-1α are regulated by environmental temperature in brown adipocytes. We thus speculate that this added layer of regulation of mtDNA transcription by NT-PGC-1α is in place to ensure that mtDNA-encoded ETC gene expression is regulated accordingly in response to changes in environmental temperature because an adequate supply of mitochondrially encoded subunits is required for cold-induced biogenesis of mitochondrial ETC complexes.
cAMP/PKA-mediated phosphorylation increases nuclear accumulation of NT-PGC-1α by inhibiting CRM1-mediated nuclear export (21). However, our data showed that mitochondrial targeting of NT-PGC-1α was not affected by cAMP-induced signaling. Instead, we found that sumoylation of NT-PGC-1α at Lys-183 produced an increase in mitochondrial NT-PGC-1α content,3 suggesting that modification by the small ubiquitin-like modifier may direct cytosolic NT-PGC-1α to mitochondria. Several recent studies have found that nuclear transcription factors such as the TRα1 isoform (p43), MEF2D, STAT3, and cAMP-response element-binding protein translocate to mitochondria despite the absence of a mitochondrial localizing sequence and directly modulate mtDNA transcription in a signal- and tissue-specific manner (13–20). The mitochondrial import of these nuclear transcription factors is in part assisted by chaperones such as the mitochondrial heat shock protein 70 (17, 18, 28). In addition, VDAC, a voltage-dependent anion channel, has been suggested to serve as a transport route for small proteins that lack a classical mitochondrial localizing sequence (29). Thus, it would be interesting to examine whether mtHSP70 and VDAC are involved in mitochondrial import of NT-PGC-1α.
Our data suggest that NT-PGC-1α is recruited to the mitochondrial transcriptional complexes formed on the D-loop region of mtDNA by interaction with LRP130. LRP130 has been shown to activate mitochondrial DNA transcription (27). It interacts with POLRMT but not with TFAM and increases the expression of mtDNA-encoded ETC genes, leading to enhanced mitochondrial respiration (27). In addition to its major role in mitochondria, LRP130 has been shown to interact with PGC-1α in the nucleus and increase PGC-1α-dependent transcription of nuclear genes (30). Thus, these studies further support the idea that NT-PGC-1α and LRP130 are present in the same mitochondrial transcriptional complexes and cooperate for mitochondrial DNA transcription. LRP130 is abundantly expressed in brown adipose tissue compared with white adipose tissue (31). Functional interactions between PGC-1α and LRP130 in the nucleus and between NT-PGC-1α and LRP130 in the mitochondria may be important for the coordinated regulation of mitochondrial electron transport chain proteins encoded by nuclear and mitochondrial genomes. Many nuclearly encoded ETC subunits assemble as subcomplexes in the matrix, and their redistribution to the inner mitochondrial membrane is accelerated by mitochondrially encoded ETC subunits (32, 33). Thus, the mechanism by which mitochondrial NT-PGC-1α regulates mtDNA-encoded gene expression may contribute to the efficiency of membrane-bound ETC complex production in brown adipocytes.
Two studies previously reported that PGC-1α protein is present as a prominent band at ∼90 kDa in mitochondria isolated from mouse brain, liver, and muscle and several cell lines (34, 35). However, we clearly demonstrated that PGC-1α protein expressed in COS-1 cells was detected at ∼110 kDa (Fig. 1B, bottom left panel), which is also consistent with previous reports (9, 36–38). In brown adipose tissue, endogenous PGC-1α was only detected at ∼110 kDa in the nucleus, and neither the ∼90- nor the ∼110-kDa band was detected in mitochondria (Fig. 1B, bottom right panel). Collectively, our findings reveal a novel and isoform-specific role for NT-PGC-1α in the regulation of mtDNA-encoded gene expression, providing new insight into how brown adipocytes modulate their mitochondrial respiratory capacity in response to cold.
Experimental procedures
Mice
All animal handling and experiments were conducted according to procedures reviewed and approved by the Pennington Biomedical Research Center Institutional Animal Care and Use Committee. For cold exposure experiments, C57BL/6J mice were individually housed and exposed to 4 °C for 5 h. The mice were then sacrificed to extract brown adipose tissue from the interscapular region.
Cell culture and adipocyte differentiation
HeLa cells were maintained in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin and transfected using FuGENE 6 (Roche Applied Science). Immortalized PGC-1α−/− brown preadipocytes expressing NT-PGC-1α, PGC-1α, or pBABE empty vector (21) were maintained in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin and induced for differentiation as described previously (9, 21).
Plasmid construction
The pCMV/myc/mito plasmid was used to direct NT-PGC-1α to the mitochondria by addition of the MLS to the N terminus of NT-PGC-1α. NT-PGC-1α was amplified from pcDNA3.1-NT-PGC-1α-HA (9) using primers containing PstI and NotI sites and subcloned into the PstI/NotI sites of pCMV/myc/mito. To construct a retroviral plasmid of pBABE-MLS-NT-PGC-1α-HA, MLS-NT-PGC-1α-HA was amplified from pCMV/myc/mito-NT-PGC-1α-HA using primers containing BamHI and SalI sites and subcloned into the BamHI/SalI sites of pBABE-neo. The generated plasmids were sequenced to rule out any mutations.
Retroviral and adenoviral infection
For retroviral infection, immortalized brown preadipocytes were infected in retrovirus-containing medium supplemented with 8 μg/ml of Polybrene for 8 h as described previously (21). After 48 h, neomycin-resistant clones were selected and pooled. For adenoviral infection of brown adipocytes, adenovirus was mixed and preincubated with poly-l-lysine (Mr 30,000–70,000) in Opti-MEM for 1.5 h at room temperature (21, 39). Cells were washed and infected with the adenoviral mixture/Opti-MEM for 2 h, followed by addition of DMEM complete medium and incubation overnight.
Subcellular fractionation
To obtain nuclear, cytosolic, and mitochondrial fractions, tissues or cells were suspended in ice-cold isotonic buffer (20 mm Hepes-KOH (pH 7.5), 10 mm KCl, 1.5 mm MgCl2, 1 mm EDTA, 1 mm EGTA, 250 mm sucrose, and protease inhibitors) and homogenized using a Potter-Elvehjem glass-Teflon homogenizer. After centrifugation at 1000 × g for 10 min, the pellets were washed and incubated with NE-PER reagents (Pierce) to isolate nuclei. The supernatant was centrifuged at 10,000 × g for 30 min to collect the mitochondrial pellet. Crude mitochondria were resuspended in mitochondrial isolation buffer (0.21 m mannitol, 0.07 m sucrose, 5 mm Tris, 1 mm EDTA, and protease inhibitors), overlaid on a discontinuous sucrose gradient (1.0–1.5 m), and purified by centrifugation at 64,000 × g for 30 min. The post-mitochondrial supernatant obtained after centrifugation at 10,000 × g was further centrifuged at 100,000 × g for 1 h to obtain the cytosolic protein fraction.
Proteinase K digestion assay
Purified mitochondria (60 μg) were resuspended in 100 μl of SE buffer (250 mm sucrose, 1 mm EDTA, 10 mm MOPS, and protease inhibitors) and incubated with vehicle or increasing amounts of proteinase K for 15 min on ice. After addition of 2 mm PMSF, mitochondria were spun down by centrifugation at 12,000 × g for 5 min. The pellets were rinsed with SE buffer containing PMSF and resuspended in 2× Laemmli sample buffer.
Transmission electron microscopy and immunolabeling
Differentiated brown adipocytes were collected and centrifuged at 500 rpm to form a soft pellet. The cell pellet was fixed at 4 °C in a fixative solution containing 2% glutaraldehyde and 1% paraformaldehyde and post-fixed with 1% osmium tetroxide. After dehydration using an ethanol gradient, the pellet was embedded in resin and polymerized. Thin sections from the resin blocks were mounted on nickel grids. For immunolabeling, the grids were pretreated with 2.5% sodium-meta-periodate for 20 min, washed with ddH2O and placed on droplets of 4% BSA in PBS (pH 7.2) supplemented with 0.05% Tween 20 (PBST) for 1 h. All grids were then incubated with monoclonal PGC-1α antibody or mouse IgG in 2% BSA/PBST for 1.5 h, followed by washing with PBST five times. The grids were blocked again with 2% BSA/PBST for 15 min and incubated with protein G conjugated with 15–20 nm colloidal gold particles diluted 1:10 in 1% BSA/PBST for 1 h. After successive washes with PBST and ddH2O, grids were post-fixed with 1% osmium tetroxide for 5 min, washed with ddH2O, and stained with 2% uranyl acetate for 5 min and lead acetate for 1 min. The grids were examined using a JEOL JEM 2011 transmission electron microscope at the Louisiana State University Socolofsky Microscopy Center.
Oxygen consumption assay
Immortalized brown preadipocytes were seeded at a density of 40,000 cells/well in XF24 cell culture microplates for the oxygen consumption assay. After differentiation for 7 days, brown adipocytes were washed and allowed to equilibrate in XF medium (Seahorse Biosciences) for 1 h at 37 °C without CO2. The oxygen consumption rates (OCRs) were measured at baseline and after injection of 8-CPT-cAMP (200 μm) using the XF24 analyzer (Seahorse Biosciences) as described previously (23). For oxygen consumption assays using the Oxygraph-2k (Oroboros Instruments, Innsbruck, Austria), 106 brown adipocytes were placed in a magnetically stirred respirometric chamber as described previously (22). OCR measurements were obtained at baseline and after injection of oligomycin (an inhibitor of ATP synthase) and FCCP (a chemical uncoupler) followed by antimycin A (a mitochondrial electron transport inhibitor). The values of basal respiration, oligomycin-independent proton leak, and FCCP-stimulated maximal mitochondrial respiration were determined by subtracting antimycin A-independent non-mitochondrial respiration as described in the Oroboros operator manual.
Mitochondrial chromatin immunoprecipitation assay
Brown adipocytes (6 × 150-mm dishes) were homogenized using a Potter-Elvehjem glass-Teflon homogenizer in homogenization buffer (10 mm HEPES-KOH (pH 7.4), 250 mm sucrose, and 1 mm EDTA) supplemented with protease inhibitor mixture. After centrifugation at 1000 × g for 30 min, the supernatant was collected and centrifuged at 10,000 × g for 10 min to spin down mitochondria. The pelleted mitochondria were homogenized again with two more strokes and centrifuged at 1000 × g for 10 min to remove residual nuclei. Approximately 2.5 mg of mitochondria were cross-linked with 1% formaldehyde for 30 min in cross-linking buffer (20 mm HEPES-KOH (pH 7.4), 250 mm sucrose, 2 mm EDTA, and 25 mm NaCl). The mitochondria were then suspended in PBS containing a protease inhibitor mixture and sheared using a Branson Sonifier 450 for 20 × 10 s to obtain chromatin fragments ranging from 400 bp to 1 kbp. After centrifugation at 10,000 × g for 10 min, the supernatant was diluted and precleared with BSA-blocked protein G-agarose beads for 1 h at 4 °C in IP buffer (10 mm Tris-HCl (pH 7.5), 150 mm NaCl, 0.5% BSA, and 0.5% Nonidet P-40) and incubated with IgG, PGC-1α, TFAM, or LRP130 antibody overnight at 4 °C. The immunocomplexes were precipitated with BSA-blocked protein G-agarose beads for 1 h at 4 °C and washed with a series of wash buffers. The cross-linked DNA-protein complexes were released from the beads by incubation at 65 °C overnight. The DNA samples were purified and used for real-time quantitative PCR analysis with a pair of primers specific for the D-loop region and for ND1: D-loop forward, 5′-gtggtgtcatgcatttggtatct-3′; D-loop reverse, 5′-catgaataattagccttaggtgat-3′; ND1 forward, 5′-cccattcgcgttattctt-3′; and ND1 reverse 5′-aagttgatcgtaaggaagc-3′.
Western blot and immunoprecipitation
Cells were lysed in radioimmune precipitation assay buffer and subjected to Western blot analysis as described previously (21). For immunoprecipitation, isolated mitochondria were lysed in 20 mm HEPES (pH 7.0), 150 mm NaCl, and 0.2% Nonidet P-40 supplemented with a protease inhibitor mixture. After preclearing with protein A/G-agarose beads, the lysates were incubated with IgG or primary antibody overnight at 4 °C and precipitated with protein A/G-agarose beads for 3 h at 4 °C. After washings, immunoprecipitates were subjected to Western blot analysis. Antibodies used were as follows: monoclonal and polyclonal anti-PGC-1α (9), anti-UCP1 (40), anti-Tom20, anti-Lamin B1, anti-TFAM, and anti-LRP130 from Santa Cruz Biotechnology and anti-HA, anti-α-tubulin, anti-CoxIV, and anti-HSP60 from Abcam. For detection of PGC-1α and NT-PGC-1α in brown adipose tissue and brown adipocytes, an ultra-sensitive ECL substrate (Pierce) was used. Densitometric analysis was performed to assess relative protein levels in Western blots. The integrated density of immunoreactive bands was measured using NIH ImageJ and normalized with the integrated density of the respective loading control used for Western blots.
Immunofluorescence
Cells were grown on glass coverslips and incubated with MitoTracker Deep Red 633 for 45 min to stain mitochondria. After washing with PBS, cells were fixed in cold methanol for 15 min and permeabilized with 0.2% Triton X-100/PBS for 10 min. The fixed cells were then subjected to indirect immunofluorescence using anti-HA antibody as described previously (9, 21). The cells were mounted with Vectashield mounting reagent containing 4,6-diamidino-2-phenylindole and examined with a Plan-Neofluar ×40/0.85 numerical aperture objective on a Zeiss LSM510 Meta confocal microscope. Subcellular colocalization analysis was performed using the ImageJ colocalization analysis tool following automatic thresholding parameters.
Luciferase reporter assay
HeLa cells were transiently transfected using FuGENE 6 (Roche) with (PPRE)3-TK-luc, pSV Sport-RXRα, pSV Sport-PPAPγ, pRL-SV40, and equal amounts of plasmid expressing NT-PGC-1α-HA, MLS-NTPGC-1α-HA, or pcDNA3.1 empty vector. After 48 h of transfection, cells were harvested for luciferase activity assay using a Promega Dual-Luciferase assay kit. Firefly luciferase activity was normalized using Renilla luciferase activity. Data represent mean ± S.D. of at least three independent experiments.
Quantitative real-time PCR analysis
Total RNA was isolated from cells using the RNeasy mini kit with DNase I treatment (Qiagen). cDNA synthesis and quantitative real-time PCR analysis were performed as described previously (9, 21, 23). Relative mRNA expression of the genes of interest was determined after normalization to that of cyclophilin using the ΔΔCt method. For quantitative analysis of mitochondrial biogenesis, the ratio of mitochondrial to nuclear DNA was assessed using quantitative real-time PCR with primers for NADH dehydrogenase subunit 1 (ND1) and lipoprotein lipase as described previously (10).
Statistical analysis
All data are presented as mean ± S.E. Student t tests and one-way or two-way analysis of variance (ANOVA) tests were used to compare the differences between groups using GraphPad Prism 6 software. Values of p < 0.05 were considered statistically significant: *, p < 0.05; **, p < 0.01; ***, p < 0.001; #, p < 0.0001.
Author contributions
J. S. C. conceived and coordinated the study, performed and analyzed the experiments, and wrote the manuscript. K. H. performed and analyzed the experiments. Both authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank Dr. Tom Gettys (Pennington Biomedical Research Center) for kindly providing anti-PGC-1α and anti-UCP1 antibodies. We also thank Ying Xiao (LSU Socolofsky Microscopy Center) for technical assistance and Cindi Tramonte for administrative support. We used Cell Biology and Bioimaging and Genomics Core facilities that are supported in part by COBRE (NIH8 1P30GM118430-01) and NORC (National Institutes of Health 1P30-DK072476) center grants from the National Institutes of Health.
This work was supported by National Institutes of Health Grant R01DK104748 (to J. S. C.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supplemental Fig. S1.
J. S. Chang and K. Ha, unpublished data.
- BAT
- brown adipose tissue
- ETC
- electron transport chain
- TFAM
- mitochondrial transcription factor A
- PPAR
- peroxisome proliferator-activated receptor
- NRF
- nuclear respiratory factor
- mtChIP
- mitochondrial ChIP
- POLRMT
- mitochondrial RNA polymerase
- nucDNA
- nuclear DNA
- MLS
- matrix-localizing sequence
- ddH2O
- double-distilled H2O
- OCR
- oxygen consumption rate
- FCCP
- carbonyl cyanide p-trifluoromethoxyphenylhydrazone
- ANOVA
- analysis of variance
- 8-CPT-cAMP
- 8-(4-chlorophenylthio)adenosine 3′,5′-cyclic monophosphate sodium salt.
References
- 1. Wu Z., Puigserver P., Andersson U., Zhang C., Adelmant G., Mootha V., Troy A., Cinti S., Lowell B., Scarpulla R. C., and Spiegelman B. M. (1999) Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 98, 115–124 [DOI] [PubMed] [Google Scholar]
- 2. Lowell B. B., and Spiegelman B. M. (2000) Towards a molecular understanding of adaptive thermogenesis. Nature 404, 652–660 [DOI] [PubMed] [Google Scholar]
- 3. Puigserver P., Wu Z., Park C. W., Graves R., Wright M., and Spiegelman B. M. (1998) A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 92, 829–839 [DOI] [PubMed] [Google Scholar]
- 4. Anderson S., Bankier A. T., Barrell B. G., de Bruijn M. H., Coulson A. R., Drouin J., Eperon I. C., Nierlich D. P., Roe B. A., Sanger F., Schreier P. H., Smith A. J., Staden R., and Young I. G. (1981) Sequence and organization of the human mitochondrial genome. Nature 290, 457–465 [DOI] [PubMed] [Google Scholar]
- 5. Chomyn A., Mariottini P., Cleeter M. W., Ragan C. I., Matsuno-Yagi A., Hatefi Y., Doolittle R. F., and Attardi G. (1985) Six unidentified reading frames of human mitochondrial DNA encode components of the respiratory-chain NADH dehydrogenase. Nature 314, 592–597 [DOI] [PubMed] [Google Scholar]
- 6. Montoya J., Perez-Martos A., Garstka H. L., and Wiesner R. J. (1997) Regulation of mitochondrial transcription by mitochondrial transcription factor A. Mol. Cell Biochem. 174, 227–230 [PubMed] [Google Scholar]
- 7. Cotney J., Wang Z., and Shadel G. S. (2007) Relative abundance of the human mitochondrial transcription system and distinct roles for h-mtTFB1 and h-mtTFB2 in mitochondrial biogenesis and gene expression. Nucleic Acids Res. 35, 4042–4054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tiranti V., Savoia A., Forti F., D'Apolito M. F., Centra M., Rocchi M., and Zeviani M. (1997) Identification of the gene encoding the human mitochondrial RNA polymerase (h-mtRPOL) by cyberscreening of the Expressed Sequence Tags database. Hum. Mol. Genet. 6, 615–625 [DOI] [PubMed] [Google Scholar]
- 9. Zhang Y., Huypens P., Adamson A. W., Chang J. S., Henagan T. M., Boudreau A., Lenard N. R., Burk D., Klein J., Perwitz N., Shin J., Fasshauer M., Kralli A., and Gettys T. W. (2009) Alternative mRNA splicing produces a novel biologically active short isoform of PGC-1α. J. Biol. Chem. 284, 32813–32826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chang J. S., Fernand V., Zhang Y., Shin J., Jun H. J., Joshi Y., and Gettys T. W. (2012) NT-PGC-1α protein is sufficient to link β3-adrenergic receptor activation to transcriptional and physiological components of adaptive thermogenesis. J. Biol. Chem. 287, 9100–9111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Scarpulla R. C. (2006) Nuclear control of respiratory gene expression in mammalian cells. J. Cell. Biochem. 97, 673–683 [DOI] [PubMed] [Google Scholar]
- 12. Villena J. A., Hock M. B., Chang W. Y., Barcas J. E., Giguère V., and Kralli A. (2007) Orphan nuclear receptor estrogen-related receptor α is essential for adaptive thermogenesis. Proc. Natl. Acad. Sci. U.S.A. 104, 1418–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wrutniak C., Cassar-Malek I., Marchal S., Rascle A., Heusser S., Keller J. M., Fléchon J., Dauça M., Samarut J., and Ghysdael J. (1995) A 43-kDa protein related to c-Erb A α 1 is located in the mitochondrial matrix of rat liver. J. Biol. Chem. 270, 16347–16354 [DOI] [PubMed] [Google Scholar]
- 14. Casas F., Rochard P., Rodier A., Cassar-Malek I., Marchal-Victorion S., Wiesner R. J., Cabello G., and Wrutniak C. (1999) A variant form of the nuclear triiodothyronine receptor c-ErbAα1 plays a direct role in regulation of mitochondrial RNA synthesis. Mol. Cell. Biol. 19, 7913–7924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Casas F., Pessemesse L., Grandemange S., Seyer P., Gueguen N., Baris O., Lepourry L., Cabello G., and Wrutniak-Cabello C. (2008) Overexpression of the mitochondrial T3 receptor p43 induces a shift in skeletal muscle fiber types. PLoS ONE 3, e2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pessemesse L., Schlernitzauer A., Sar C., Levin J., Grandemange S., Seyer P., Favier F. B., Kaminski S., Cabello G., Wrutniak-Cabello C., and Casas F. (2012) Depletion of the p43 mitochondrial T3 receptor in mice affects skeletal muscle development and activity. FASEB J. 26, 748–756 [DOI] [PubMed] [Google Scholar]
- 17. She H., Yang Q., Shepherd K., Smith Y., Miller G., Testa C., and Mao Z. (2011) Direct regulation of complex I by mitochondrial MEF2D is disrupted in a mouse model of Parkinson disease and in human patients. J. Clin. Invest. 121, 930–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. De Rasmo D., Signorile A., Roca E., and Papa S. (2009) cAMP response element-binding protein (CREB) is imported into mitochondria and promotes protein synthesis. FEBS J. 276, 4325–4333 [DOI] [PubMed] [Google Scholar]
- 19. Lee J., Kim C. H., Simon D. K., Aminova L. R., Andreyev A. Y., Kushnareva Y. E., Murphy A. N., Lonze B. E., Kim K. S., Ginty D. D., Ferrante R. J., Ryu H., and Ratan R. R. (2005) Mitochondrial cyclic AMP response element-binding protein (CREB) mediates mitochondrial gene expression and neuronal survival. J. Biol. Chem. 280, 40398–40401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Macias E., Rao D., Carbajal S., Kiguchi K., and DiGiovanni J. (2014) Stat3 Binds to mtDNA and regulates mitochondrial gene expression in keratinocytes. J. Invest. Dermatol. 134, 1971–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chang J. S., Huypens P., Zhang Y., Black C., Kralli A., and Gettys T. W. (2010) Regulation of NT-PGC-1α subcellular localization and function by protein kinase A-dependent modulation of nuclear export by CRM1. J. Biol. Chem. 285, 18039–18050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jun H. J., Joshi Y., Patil Y., Noland R. C., and Chang J. S. (2014) NT-PGC-1α activation attenuates high-fat diet-induced obesity by enhancing brown fat thermogenesis and adipose tissue oxidative metabolism. Diabetes 63, 3615–3625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kim J., Fernand V. E., Henagan T. M., Shin J., Huypens P., Newman S., Gettys T. W., and Chang J. S. (2016) Regulation of brown and white adipocyte transcriptome by the transcriptional coactivator NT-PGC-1α. PLoS ONE 11, e0159990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Taanman J. W. (1999) The mitochondrial genome: structure, transcription, translation and replication. Biochim. Biophys. Acta 1410, 103–123 [DOI] [PubMed] [Google Scholar]
- 25. Takamatsu C., Umeda S., Ohsato T., Ohno T., Abe Y., Fukuoh A., Shinagawa H., Hamasaki N., and Kang D. (2002) Regulation of mitochondrial D-loops by transcription factor A and single-stranded DNA-binding protein. EMBO Rep. 3, 451–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Alam T. I., Kanki T., Muta T., Ukaji K., Abe Y., Nakayama H., Takio K., Hamasaki N., and Kang D. (2003) Human mitochondrial DNA is packaged with TFAM. Nucleic Acids Res. 31, 1640–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu L., Sanosaka M., Lei S., Bestwick M. L., Frey J. H. Jr, Surovtseva Y. V., Shadel G. S., and Cooper M. P. (2011) LRP130 protein remodels mitochondria and stimulates fatty acid oxidation. J. Biol. Chem. 286, 41253–41264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Neupert W., and Herrmann J. M. (2007) Translocation of proteins into mitochondria. Annu. Rev. Biochem. 76, 723–749 [DOI] [PubMed] [Google Scholar]
- 29. Zalk R., Israelson A., Garty E. S., Azoulay-Zohar H., and Shoshan-Barmatz V. (2005) Oligomeric states of the voltage-dependent anion channel and cytochrome c release from mitochondria. Biochem. J. 386, 73–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cooper M. P., Qu L., Rohas L. M., Lin J., Yang W., Erdjument-Bromage H., Tempst P., and Spiegelman B. M. (2006) Defects in energy homeostasis in Leigh syndrome French Canadian variant through PGC-1α/LRP130 complex. Genes Dev. 20, 2996–3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cooper M. P., Uldry M., Kajimura S., Arany Z., and Spiegelman B. M. (2008) Modulation of PGC-1 coactivator pathways in brown fat differentiation through LRP130. J. Biol. Chem. 283, 31960–31967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bourges I., Ramus C., Mousson de Camaret B., Beugnot R., Remacle C., Cardol P., Hofhaus G., and Issartel J. P. (2004) Structural organization of mitochondrial human complex I: role of the ND4 and ND5 mitochondria-encoded subunits and interaction with prohibitin. Biochem. J. 383, 491–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Antonicka H., Ogilvie I., Taivassalo T., Anitori R. P., Haller R. G., Vissing J., Kennaway N. G., and Shoubridge E. A. (2003) Identification and characterization of a common set of complex I assembly intermediates in mitochondria from patients with complex I deficiency. J. Biol. Chem. 278, 43081–43088 [DOI] [PubMed] [Google Scholar]
- 34. Aquilano K., Vigilanza P., Baldelli S., Pagliei B., Rotilio G., and Ciriolo M. R. (2010) Peroxisome proliferator-activated receptor γ co-activator 1γ (PGC-1α) and sirtuin 1 (SIRT1) reside in mitochondria: possible direct function in mitochondrial biogenesis. J. Biol. Chem. 285, 21590–21599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Safdar A., Little J. P., Stokl A. J., Hettinga B. P., Akhtar M., and Tarnopolsky M. A. (2011) Exercise increases mitochondrial PGC-1α content and promotes nuclear-mitochondrial cross-talk to coordinate mitochondrial biogenesis. J. Biol. Chem. 286, 10605–10617 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36. Ruas J. L., White J. P., Rao R. R., Kleiner S., Brannan K. T., Harrison B. C., Greene N. P., Wu J., Estall J. L., Irving B. A., Lanza I. R., Rasbach K. A., Okutsu M., Nair K. S., Yan Z., et al. (2012) A PGC-1α isoform induced by resistance training regulates skeletal muscle hypertrophy. Cell 151, 1319–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chang J. S., Jun H. J., and Park M. (2016) Transcriptional coactivator NT-PGC-1α promotes gluconeogenic gene expression and enhances hepatic gluconeogenesis. Physiol. Rep. 4, e13013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shin J. H., Ko H. S., Kang H., Lee Y., Lee Y. I., Pletinkova O., Troconso J. C., Dawson V. L., and Dawson T. M. (2011) PARIS (ZNF746) repression of PGC-1α contributes to neurodegeneration in Parkinson's disease. Cell 144, 689–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Béréziat V., Moritz S., Klonjkowski B., Decaudain A., Auclair M., Eloit M., Capeau J., and Vigouroux C. (2005) Efficient adenoviral transduction of 3T3-F442A preadipocytes without affecting adipocyte differentiation. Biochimie 87, 951–958 [DOI] [PubMed] [Google Scholar]
- 40. Commins S. P., Watson P. M., Padgett M. A., Dudley A., Argyropoulos G., and Gettys T. W. (1999) Induction of uncoupling protein expression in brown and white adipose tissue by leptin. Endocrinology 140, 292–300 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.