Abstract
The front-line antituberculosis drug isoniazid (INH) and the related drug ethionamide (ETH) are prodrugs that upon activation inhibit the synthesis of mycolic acids, leading to bactericidal activity. Coresistance to INH and ETH can be mediated by dominant mutations in the target gene inhA, encoding an enoyl-ACP reductase, or by recessive mutations in ndh, encoding a type II NADH dehydrogenase (NdhII). To address the mechanism of resistance mediated by the latter, we have isolated novel ndh mutants of Mycobacterium smegmatis and Mycobacterium bovis BCG. The M. smegmatis ndh mutants were highly resistant to INH and ETH, while the M. bovis BCG mutants had low-level resistance to INH and ETH. All mutants had defects in NdhII activity resulting in an increase in intracellular NADH/NAD+ ratios. Increasing NADH levels were shown to protect InhA against inhibition by the INH-NAD adduct formed upon INH activation. We conclude that ndh mutations mediate a novel mechanism of resistance by increasing the NADH cellular concentration, which competitively inhibits the binding of INH-NAD or ETH-NAD adduct to InhA.
Despite the declaration by the World Health Organization 10 years ago that tuberculosis (TB) was a global health emergency, the problem has worsened primarily due to the growing human immunodeficiency virus epidemic and the emergence of drug resistance (9, 12). Although TB can be cured by a regimen of several drugs for at least 6 months, the emergence of drug-resistant and multidrug-resistant TB has created new challenges to control and defeat the disease. According to the World Health Organization, drug-resistant Mycobacterium tuberculosis strains are found in at least 72 countries at a rate ranging from 3 to 41% (53). Understanding the mechanisms by which drug resistance occurs is important in order to quickly identify drug-resistant strains to treat the patients adequately. Moreover, knowledge of resistance mechanisms leads to the understanding of the mode of drug action and to the development of strategies to overcome drug resistance.
Isoniazid (INH) is one of the most effective drugs used to treat TB. INH was introduced as an antituberculosis drug in 1952 (4, 13) and, soon after, the first INH-resistant (INHr) M. tuberculosis strains were isolated (35). It is estimated that worldwide up to 28% of the M. tuberculosis strains are INHr, with a median of 6.2% in new TB cases (53). In previously treated TB cases, the percentage of INHr strains can reach up to 60% (53).
Numerous genes have been found to be associated with INH resistance in clinical isolates (43). However, to date only two genes, katG encoding a catalase peroxidase and inhA encoding an NADH-dependent enoyl-ACP reductase, have been shown to confer greater than fivefold increased susceptibility or resistance to M. tuberculosis upon gene transfer (26, 56). Mutations in katG are recessive, resulting in a loss or altered catalase peroxidase function (and subsequent INH resistance) in both INHr laboratory-isolated mutants and INHr clinical isolates (17, 22, 27, 41, 43, 57). The recessive nature of the katG mutations, i.e., the restoration of catalase peroxidase activity and INH susceptibility when replaced or complemented with the wild-type gene, is consistent with the fact that KatG activates INH (20, 56, 57) to generate a hypothetical isonicotinic acyl radical that reacts with NAD and forms an INH-NAD adduct (45). This adduct binds to and inhibits InhA (29, 40, 44, 45, 52), resulting in mycolic acid biosynthesis inhibition (49) and cell death (49, 51). In contrast, overexpressed inhA alleles or alleles causing amino acid substitutions within the structural gene have been shown to confer INH resistance in a dominant fashion, i.e., conferring INH resistance when the mutant alleles replace or complement the wild-type gene (1, 26, 51). Mutations in INHr clinical isolates of M. tuberculosis have been mapped to the promoter region or the structural gene of inhA (1, 2, 17, 22, 27, 31, 38, 41, 43). The dominant nature of these mutations was consistent with the hypothesis that inhA encodes the target of INH (1, 23, 26).
The structural analog of INH, ethionamide (ETH), another InhA inhibitor, has provided important insights into INH action. Early studies of clinical strains resistant to INH revealed that some of the strains were also coresistant to ETH and/or thiosemicarbazone, even though the patients had never been treated with those drugs (6, 7, 18, 28). A recent study of 41 ETH-resistant (ETHr) clinical isolates of M. tuberculosis demonstrated that most of the strains were also coresistant to INH and that, in 51% of these strains, the ETHr and INHr phenotypes were solely due to mutations in the inhA gene and/or its promoter region (38). Mutations in inhA or overexpression of inhA have previously been shown to confer resistance in a dominant fashion to ETH and INH (1, 26). Like INH, ETH appears to be a prodrug that is activated by the monooxygenase EthA (3, 10, 14, 50). An ETH-NAD adduct had been hypothesized to be the inhibitor of InhA (45).
Coresistance to INH and ETH has also been shown to be mediated in a recessive fashion in Mycobacterium smegmatis by mutations in ndh, a gene encoding a type II NADH dehydrogenase (NdhII) (36). The orthologue NdhII protein has been characterized in Escherichia coli as a membrane-bound, monomeric, non-proton-translocating flavoprotein that oxidizes NADH, reduces quinone, and catalyzes the transfer of electrons from the reduced flavin to quinones (15, 19, 21, 32, 54). The M. tuberculosis genome contains genes encoding both NdhII (ndh; Rv1584c) and the type I NADH dehydrogenase (NdhI) (nuoA-N; Rv3145 to -3158). The orthologue NdhI protein has been characterized in E. coli as a membrane-associated, multimeric, energy-coupling, proton-translocating enzyme that oxidizes NADH and pumps protons across the membrane using the energy generated by the redox reaction (15, 21, 54, 55). Mutations in ndh have been identified in INHr clinical isolates of M. tuberculosis (27), but no complementation experiments were done to show that the INH resistance phenotype was due to the ndh mutation in those strains and the level of ETH resistance was not measured. Therefore, to demonstrate that ndh mutations can cause INH and ETH resistance in slow-growing mycobacteria and to elucidate the mechanism of INH and ETH resistance due to ndh mutations in mycobacteria, we sought to isolate spontaneous ndh mutants in fast- and slow-growing mycobacteria. In this report, we describe the isolation and genetic characterization of spontaneous ndh mutants in M. smegmatis and Mycobacterium bovis BCG. In addition, we analyze the NdhII activity and the resulting NADH/NAD+ ratios in the INH and ETH coresistant mutants and propose a new mechanism for INH and ETH resistance in mycobacteria.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The M. smegmatis, M. bovis BCG, and M. tuberculosis H37Rv mutants were obtained from laboratory stocks. The plasmids used in this study were described previously (26, 36). The M. smegmatis strains were grown in Middlebrook 7H9 medium (Difco) supplemented with 10% (vol/vol) ADS enrichment (50 g of albumin, 20 g of dextrose, 8.5 g of sodium chloride in 1 liter water), 0.2% (vol/vol) glycerol, and 0.5% (vol/vol) Tween 80. The M. bovis BCG and M. tuberculosis H37Rv strains were grown in Middlebrook 7H9 medium (Difco) supplemented with 10% (vol/vol) OADC enrichment (Difco), 0.2% (vol/vol) glycerol, and 0.05% (vol/vol) Tween 80. The solid medium used was the same as that described above with the addition of 1.5% (wt/vol) agar.
Isolation of spontaneous mutants.
Mutants were isolated from nonmutagenized cultures grown in the media described above. The cultures were incubated with shaking at 30°C for M. smegmatis and at 37°C for M. bovis BCG and M. tuberculosis H37Rv until they reached an optical density at 600 nm (OD600) of at least 1.0. Tenfold serial dilutions were then plated on agar plates (media described above) containing either INH (25 μg/ml) for M. smegmatis mutant isolation or INH (0.25 μg/ml) and ETH (5 or 10 μg/ml) for M. bovis BCG and M. tuberculosis H37Rv mutant isolation. The plates were then incubated at 30°C for 7 days for M. smegmatis mutant isolation or at 37°C for 4 to 6 weeks for M. bovis BCG and M. tuberculosis H37Rv mutant isolation.
Assessment of Ts lethality phenotype.
The M. smegmatis ndh mutants were grown to stationary phase at 30°C. Tenfold serial dilutions were plated onto two sets of plates: one set was incubated at 30°C for 7 days (for titration of the cultures) and the other set was incubated at 42°C for 5 days. The plates incubated at 42°C were then shifted to 30°C, and the incubation continued for 7 days. A strain was considered to have a temperature-sensitive (Ts) lethal phenotype if less than 10% of its population could survive the incubation at 42°C (measured by the number of CFU per milliliter obtained after 5 days at 42°C followed by 7 days at 30°C divided by the titer).
MIC determination.
Mycobacterial cultures were grown to an OD600 of ≈1.0. Tenfold serial dilutions were plated on plates containing the following drug concentrations: for M. smegmatis strains, INH (0, 5, 10, 25, 50, 100 μg/ml), ETH (0, 5, 10, 25, 50, 100 μg/ml), triclosan (TRC; 0, 5, 10, 15, 25, 50 μg/ml), streptomycin (0.1, 0.25, 0.5, 1 μg/ml), ethambutol (0.1, 0.25, 0.5, 1, 5 μg/ml), and rifampin (5, 10, 25, 50, 75, 100 μg/ml); for M. bovis BCG strains, INH (0, 0.1, 0.2, 0.25, 0.3, 0.4, 0.5, 0.6, 0.8, 1 μg/ml), ETH (0, 2.5, 5, 10, 12.5, 15, 20 μg/ml), and TRC (0, 5, 10, 12.5, 15, 20 μg/ml). The MIC was determined as the concentration of drug that reduced the number of CFU per milliliter by 99%.
PCR amplification.
The ndh gene was amplified from chromosomal DNA using the following primers: for M. smegmatis, TW96 (5′-CGAGGAGCATCAATGAGCCA-3′) and TW97 (5′-CTCGACCGAACCGGCTAGGA-3′); for M. bovis BCG, TW539 (5′-CTGACCGGTTGGCTGGTAA-3′) and NDHB2 (5′-CGGATCCAGCAGCGGAACATGAG-3′). The products were directly sequenced bidirectionally, and the sequences were aligned against each other and the wild-type sequence.
Transformation experiments.
M. smegmatis strains (10-ml cultures) were grown at 30°C to mid-log phase (OD600 ≈ 0.7), washed twice with 10% cold glycerol, and resuspended in cold 10% glycerol (0.5 ml). To perform transformations, the cold cell suspension (150 μl) was added to the plasmid (1.5 μl) and then electroporated using the following parameters: 2.5 V, 25 μF, and 1,000 Ω. Medium (1 ml) was added to the suspension, which was incubated at 30°C for 5 h, and then plated. For M. bovis BCG strains, the same protocol was followed, but cell preparation and electroporation were performed at room temperature and incubation was done at 37°C (25).
NADH dehydrogenase assays.
Cultures (500 ml) were grown to mid-log phase (OD600 ≈ 0.7), spun down, and washed twice in cold phosphate buffer (50 mM K2HPO4 [pH 7.5], 5 mM MgSO4). The cell pellet was weighed and resuspended in cold phosphate buffer (1 ml/g of pellet). DNase and proteinase inhibitors were added to the cell suspension, and the cells were broken using a French press (four passages; 1,000 lb/in2/cm2). The cell debris was removed by centrifugation (12,000 × g, 20 min), and the membrane fraction was isolated by ultracentrifugation (100,000 × g, 90 min). The membrane fraction was then resuspended in phosphate buffer, and protein concentration was measured using the Bio-Rad protein assay. NADH dehydrogenase activities (type I and II) were assayed spectrophotometrically, in the membrane fractions, at room temperature, by measuring the rate of NHDH (reduced nicotinamide hypoxanthine dinucleotide) oxidation at 340 nm (type I) or the rate of NADH oxidation in the presence of menadione (2 mM) at 340 nm (type II), or by measuring the rate of 2,6-dichloroindophenol (DCIP) reduction at 610 nm in the presence of NHDH (type I) or NADH (type II) (36).
Determination of NADH and NAD+ cellular concentrations.
Mycobacterial cultures were grown to an OD600 ranging from 0.8 to 1.2. A sample of the cultures (1 ml) was spun, and the cell pellets were resuspended in 0.2 M HCl (0.3 ml, NAD+ extraction) or 0.2 M NaOH (0.3 ml, NADH extraction). After 10 min at 55°C, the suspensions were cooled to 0°C and neutralized by adding 0.1 M NaOH (0.3 ml, NAD+ extraction) or 0.1 M HCl (0.3 ml, NADH extraction) while vortexing at high speed. After centrifugation, the supernatants were collected and transferred to a new tube and used immediately. The concentration of NAD+ (or NADH) was obtained by measuring spectrophotometrically the rate of 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide reduction by the yeast type II alcohol dehydrogenase in the presence of phenazine ethosulfate at 570 nm (30, 46). The rate of 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide reduction is proportional to the concentration of nucleotide.
Quantification of the inhA expression levels.
Total RNA extractions, cDNA synthesis, and quantitative PCR with molecular beacons were performed as described previously (26).
Western blot analysis.
M. bovis BCG mid-log-phase cultures (10 ml) were resuspended in 0.8 ml of phosphate buffer (phosphate-buffered saline; 20 mM K2HPO4 [pH 7.5], 0.15 M NaCl) and disrupted during 10 min using a Branson Sonifier 450. Equal amounts of protein (25 μg/lane) were then separated on a sodium dodecyl sulfate-12% polyacrylamide gel as described previously (24). After electrophoresis, the proteins were transferred onto a Hybond-C Extra membrane (Amersham). Membranes were then saturated with 2% dry milk in phosphate-buffered saline-0.1% Tween 20 and incubated overnight with rabbit anti-InhA antibodies raised against the M. tuberculosis InhA protein (23) (dilution, 1/30,000). Membranes were then washed and incubated with anti-rabbit antibodies conjugated to alkaline phosphatase (1/7,000 dilution; Promega).
InhA enzymatic activity assay.
All assays were carried out on a Cary 100 Bio Spectrophotometer by monitoring oxidation of NADH at 340 nm. The M. tuberculosis InhA protein was expressed and purified as previously described (42). Three different procedures were used to assay the activity of M. tuberculosis InhA with increasing concentration of NADH. In the first experiment, the INH-NAD adduct was presynthesized by adding Mn(III) pyrophosphate (4 mM) into a mixture consisting of INH (2 mM) and NAD+ (2 mM) in 100 mM potassium phosphate buffer solution, pH 7.5 (40). The assay was performed by incubating InhA (60 nM) with increasing concentrations of NADH (10, 100, or 500 μM or 1 mM) and the INH-NAD adduct solution (10 μM) for 20 min in 100 mM potassium phosphate buffer solution, pH 7.5 (40). The reaction was initiated by adding 2-trans-dodecenoyl-coenzyme A (CoA) (50 μM). In the second assay, InhA (60 nM) was mixed together with NADH (10, 100, or 500 μM or 1 mM), INH (10 μM), and Mn(III) pyrophosphate (20 μM) in 100 mM potassium phosphate buffer solution, pH 7.5, and the reaction was initiated by adding 2-trans-dodecenoyl-CoA (50 μM). In the third experiment, the INH-NAD adduct was presynthesized (as described for the first experiment) and incubated with InhA (5 μM) for 3 h, followed by separation using a Hi-Trap (5-ml) desalting column. The InhA-inhibitor complex was collected and assayed as in the first experiment.
RESULTS
Isolation of novel ndh mutants in M. smegmatis
We have previously shown that spontaneous M. smegmatis ndh mutants could be isolated by plating nonmutagenized M. smegmatis cultures onto rich medium (Mueller-Hinton) plates containing INH (25 μg/ml; five times the MIC), incubating the plates at 30°C, and then screening for the mutants that failed to grow at 42°C (36). These mutants were found to have pleiotropic mutations in ndh conferring four independent phenotypes: INH resistance, ETH resistance, temperature sensitivity, and auxotrophy. In an attempt to identify novel alleles mapping to ndh involved in INH resistance, we isolated spontaneous INHr mutants by plating 10-fold serial dilutions of 10 independent, nonmutagenized M. smegmatis cultures onto minimal medium Middlebrook 7H10 plates containing 25 μg of INH/ml. INHr mutants were isolated at a frequency ranging from 8 × 10−7 to 9 × 10−8. When screening for thermosensitivity, 6 to 18% of these mutants were temperature sensitive for growth at 42°C. We isolated 26 mutants that were INHr Ts and prototrophs.
The mutants were highly resistant to INH (MIC ≥ 50 μg/ml, with most of the mutants being resistant to more than 100 μg/ml) (Table 1) but were susceptible to other antimycobacterial drugs, like streptomycin, ethambutol, and rifampin (data not shown). Two other antimycobacterial agents, ETH and TRC, both targeting InhA (1, 34), were also tested. As shown in Table 1, the M. smegmatis mutants were highly resistant to ETH but had a low resistance to TRC.
TABLE 1.
Characterization of M. smegmatis ndh mutants
| Strain | ndh allele | Mutation
|
MIC (μg/ml)
|
% Viability at 42°Ca | |||
|---|---|---|---|---|---|---|---|
| DNA | Amino acid | INH | ETH | TRC | |||
| mc2155 | ndh | Wild type | Wild type | 5 | 10 | 10 | 100 |
| mc22374 | ndh-64 | T50C | I17T | >100 | >100 | 25 | 55 |
| mc22375 | ndh-52 | A85C | T29P | >100 | 50 | 10 | 31 |
| mc22376 | ndh-101 | A137C | H46P | >100 | >100 | 25 | 99 |
| mc22377g | ndh-61 | G251A | G84D | >100 | >100 | 25 | 1 |
| mc22378 | ndh-11 | T299C | L100P | 100 | >100 | 10 | 1 |
| mc22379 | ndh-16 | G343Ab | A115T | >100 | >100 | 25 | 10 |
| mc22380 | ndh-13 | T364A | Y122N | >100 | >100 | 25 | 10 |
| mc22381 | ndh-32 | C433Tc | R145C | >100 | >100 | 25 | 1 |
| mc22382 | ndh-41 | T509C | F170S | >100 | >100 | 10 | 45 |
| mc22383 | ndh-23 | G559Cd | A187P | 50 | 50 | 25 | 12 |
| mc22384 | ndh-71 | T737C | V246A | >100 | >100 | 10 | 30 |
| mc22385 | ndh-17 | T815A | V272E | >100 | >100 | 10 | 46 |
| mc22386 | ndh-22 | T899Ge | V300G | >100 | >100 | 25 | 30 |
| mc22387 | ndh-31 | G1005T | Q335H | 100 | 100 | 10 | 35 |
| mc22388 | ndh-84 | T1081Cf | Y361H | >100 | 100 | 10 | 50 |
Viability at 42°C was defined as the percentage of cells that survived incubation at 42°C. Ten- fold serial dilutions of mycobacterial cultures were plated at the nonpermissive temperature (42°C). After incubation at 42°C for 4 days, the plates were incubated at the permissive temperature (30°C) for 6 days. The number of CFU per milliliter obtained after the temperature shift was divided by the titer of the culture to give the percent viability at 42°C.
Also found in mutant ndh-18. This was the only mutation previously isolated (36).
Also found in mutants ndh-63 and ndh-93.
Also found in mutant ndh-12.
Also found in mutants ndh-24, ndh-51, and ndh-83.
Also found in mutants ndh-62, ndh-81, ndh-82, and ndh-85.
The M. smegmatis ndh Ts lethal mutants are shown in bold.
To test if the mutations mapped to ndh, the INHr Ts mutants were transformed with pYUB803, an extrachromosomal plasmid (pMV261 [48]) containing the M. smegmatis ndh gene, and the resulting transformants were screened for loss of thermosensitivity and INH resistance. The pYUB803 construct restored INH sensitivity and temperature resistance phenotypes to all the INHr Ts mutants (data not shown). Furthermore, pYUB808, a replicative plasmid (pMV261) containing the M. bovis BCG mdh gene encoding the NADH-dependent malate dehydrogenase, also restored thermoresistance in all but one mutant (mc22380) (data not shown). Although M. smegmatis does not possess the NADH-specific malate dehydrogenase, Miesel et al. (36) had shown that the M. bovis BCG mdh gene was able to restore wild-type phenotypes to M. smegmatis ndh mutants. Interestingly, Molenaar et al. (37) postulated that Mdh allows for the restoration of the Ndh enzymatic activity by combining with the M. smegmatis malate:quinone oxidoreductase (Mqo) enzyme. Taken together, these transformation studies were consistent with the hypothesis that all the mutants had acquired mutations in the M. smegmatis ndh gene.
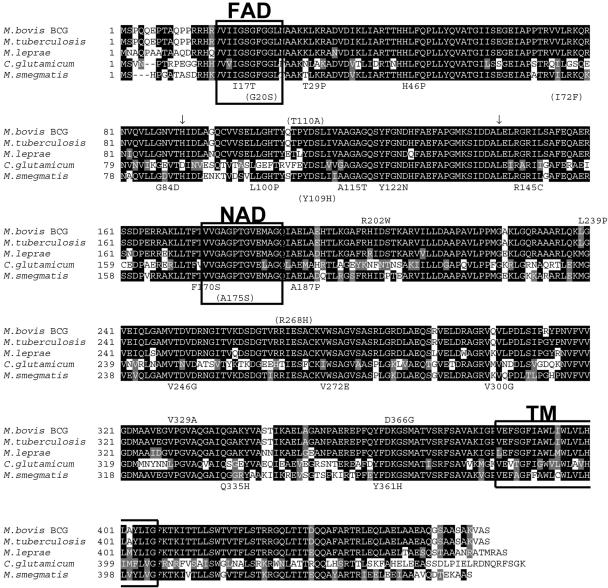
Sequence analysis revealed that all 26 mutants contained single point mutations in the ndh gene resulting in amino acid substitutions (Table 1). Eleven of the mutations were found more than once. The G559C (Ala187Pro) and T1081C (Tyr361His) mutations were found in two independent cultures, whereas the C433T (Arg145Cys) and T899G (Val300Gly) mutations were found in three independent cultures. Thus, of the 15 unique mutations identified, only 1, the Ala115Thr amino acid change, had been isolated previously and it, too, was found to be a prototroph (36). The locations of the amino acid substitutions caused by the mutations are depicted on the protein sequence (Fig. 1). The M. smegmatis ndh gene is 1,374 bp in length, encoding an open reading frame of 458 amino acids. All of the mutations caused amino acid substitutions from amino acid 17 to amino acid 361 (Fig. 1). None of the mutations was positioned in the NAD binding site and only one (Ile17Thr) was located in the FAD binding site, but most of the mutations were found in conserved regions of Ndh from M. tuberculosis, Mycobacterium leprae, and Corynebacterium glutamicum (Fig. 1).
FIG. 1.
Sequence alignment of actinomyces NdHII proteins and mutation descriptions. The NdhII proteins from M. bovis BCG, M. tuberculosis, M. leprae, Corynebacterium glutamicum, and M. smegmatis were aligned using BOXSHADE 3.21 (http://www.ch.embnet.org/software/BOX_form.html). The putative FAD and NAD binding motifs (33) and transmembrane domain (TM) (http://www.cbs.dtu.dk/services/TMHMM/) (47) are boxed, and the conserved regions have a black background. The positions and the amino acid substitutions found in M. smegmatis ndh mutants isolated in this study and in previous studies (in parentheses) (36) are shown below the M. smegmatis alignment. The positions and the amino acid substitutions found in M. bovis BCG ndh mutants isolated in this study are shown above the M. bovis BCG alignment (the base pair insertions are indicated with arrows), while the positions and the amino acid changes previously isolated in M. tuberculosis clinical isolates (27) are shown in parentheses above the M. bovis BCG alignment.
Coresistance to INH and ETH can be mediated by ndh mutations in M. bovis BCG.
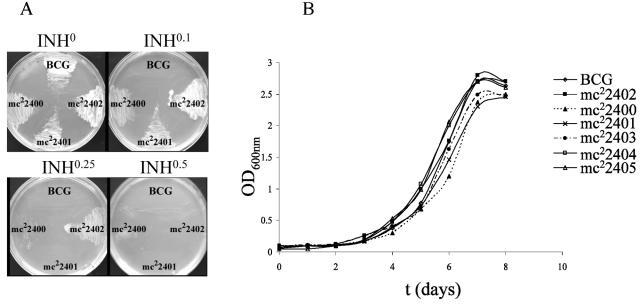
Since M. smegmatis is 100 times more resistant to INH than M. tuberculosis or M. bovis BCG is, we addressed whether ndh mutations could confer resistance to INH and ETH in slow-growing mycobacteria. Interestingly, a previous study identified mutations in ndh in INHr M. tuberculosis clinical isolates from Singapore, although no gene transfers were performed to prove that the resistance was directly mediated by the ndh mutations (27). Here, M. bovis BCG Pasteur was chosen with the intention of facilitating safety containment during French press procedures required for biochemical analyses. To enrich for ndh mutants and to eliminate the eventual selection of INHr strains carrying mutations within the katG gene, we isolated spontaneous mutants of M. bovis BCG resistant to both INH and ETH. The INHr and ETHr mutants were obtained by plating 10-fold serial dilutions of three independent M. bovis BCG cultures onto Middlebrook 7H10 plates containing 0.25 μg of INH/ml and 10 μg of ETH/ml. This double antibiotic selection led to the isolation of mutants at a frequency of 10−5 to 10−6. From 15 of the INHr ETHr strains sequenced for mutations in the ndh gene, 6 were found to possess mutations. Four of the six ndh mutants had a single point mutation in the ndh gene resulting in amino acid substitutions (Table 2; Fig. 1). The other two had acquired a single base pair insertion, theoretically causing a frameshift in the Ndh open reading frame (Table 2; Fig. 1). The base pair insertions were confirmed by sequencing ndh from two independent PCRs using two independent genomic DNA preparations as template. All six mutants displayed low-level resistance phenotypes to INH (2.5- to 6-fold), ETH (4- to 8-fold), and TRC (2- to 3-fold) (Table 2; Fig. 2A). Wild-type susceptibilities to INH, ETH, and TRC could be restored in each mutant by complementation with the wild-type M. bovis BCG ndh gene (data not shown). This suggests that resistance to all three drugs was directly mediated by mutations within the ndh gene in M. bovis BCG. Notably, all six ndh mutants had no growth defect in liquid medium without antibiotic (Fig. 2B).
TABLE 2.
Characterization of M. bovis BCG Pasteur ndh mutants
| Strain | ndh allele | Mutation
|
MIC (μg/ml)
|
|||
|---|---|---|---|---|---|---|
| DNA | Amino acid | INH | ETH | TRC | ||
| BCG Pasteur | ndh | Wild type | Wild type | 0.1 | 2.5 | 5 |
| mc22402 | ndh-3 | insbp272(a)a | Frameshift | 0.3 | 15 | 15 |
| mc22403 | ndh-443 | insbp439(t) | Frameshift | 0.6 | 15 | 10 |
| mc22404 | ndh-44 | C604T | R202W | 0.3 | 10 | 10 |
| mc22400 | ndh-33 | T716C | L239P | 0.25 | 12.5 | 12.5 |
| mc22401 | ndh-38 | T986C | V329A | 0.25 | 10 | 15 |
| mc22405 | ndh-55 | A1097G | D366G | 0.3 | 20 | 10 |
insbp272(a) refers to insertion base pair a at position 272.
FIG. 2.
Growth of M. bovis BCG Pasteur ndh mutants at 37°C. (A) Growth on solid media containing different concentrations (in micrograms per milliliter) of INH of three M. bovis BCG ndh mutants (mc22400, mc22401, and mc22402). (B) The ndh mutants and parent strain were grown in Middlebrook 7H9 broth supplemented with 10% OADC, 0.2% glycerol, and 0.05% Tween 80. The growth of the cultures was followed spectrophotometrically by measuring the OD600.
Coresistance to INH and ETH in ndh mutants is not due to InhA overexpression.
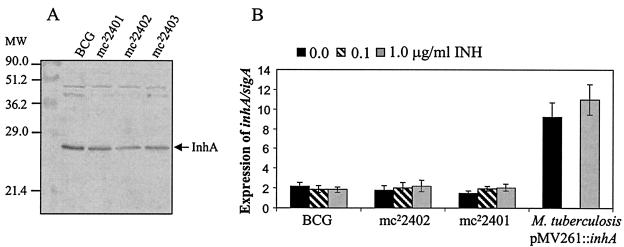
Since previous studies had demonstrated that overexpression of inhA conferred coresistance to INH and ETH in M. smegmatis, M. bovis BCG, and M. tuberculosis (1, 26), we hypothesized that the coresistance to INH and ETH could be mediated by the up-regulation of inhA. To test this hypothesis, the amounts of InhA protein in three M. bovis BCG ndh mutants (mc22401, mc22402, and mc22403) were compared to that of the parental M. bovis BCG strain by Western blot analysis using rabbit anti-InhA antibodies. This analysis showed no significant differences in InhA protein levels between the wild-type strain and the three M. bovis BCG ndh mutants (Fig. 3A). A further quantification of inhA was performed at a transcriptional level in two independent M. bovis BCG mutants (mc22401 and mc22402) using a molecular beacon mRNA assay (26) in the absence or presence of INH. Figure 3B shows that the level of inhA mRNA was similar in all strains tested, thus confirming the results obtained by immunoblotting. This rules out the possibility that coresistance to INH and ETH in the ndh M. bovis BCG mutants is mediated by higher InhA levels. Moreover, no increase in the levels of inhA mRNA was observed in the various strains, independent of the concentration of INH used (0, 0.1, or 1 μg/ml) (Fig. 3B). In contrast, M. tuberculosis transformed with pMV261::inhA, which has been previously shown to be coresistant to INH and ETH (26), showed a five- to sixfold increase in inhA mRNA levels. From these studies, we conclude that the coresistance to INH and ETH in the ndh mutants did not result from increased levels of inhA expression.
FIG. 3.
InhA expression in ndh mutants. (A) Western blot analysis. Detection of InhA in total protein extracts obtained from the different M. bovis BCG strains using rabbit anti-InhA antibodies raised against the M. tuberculosis InhA protein. (B) inhA mRNA levels in M. bovis BCG strains and the M. tuberculosis pMV261::inhA strain without addition of INH and after 4 h of incubation in the presence of 0.1 or 1.0 μg of INH/ml. inhA values were normalized using sigA levels as reference (26). Each value is the average of three culture replicates, each of which was evaluated three times. Error bars correspond to 95% confidence intervals. As a positive control of inhA overexpression, the M. tuberculosis strain harboring pMV261::inhA is shown (26), tested only without antibiotic and 1.0 μg of INH/ml.
The ndh mutants are defective in NdhII activity.
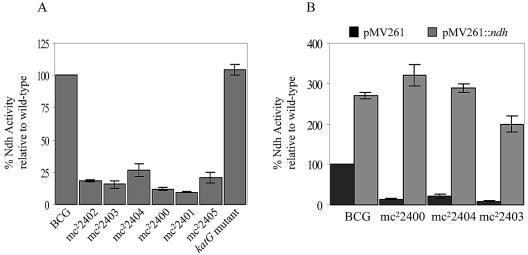
The identification of mutations in ndh suggested that the strains would be defective in NdhII activity. Therefore, the NdhII enzymatic activities were determined in the membrane fractions of the unique ndh mutants and six complemented mutants by measuring the rate of NADH oxidation in presence of menadione (Table 3; Fig. 4). In M. smegmatis, the level of NdhII activity of the different ndh mutants ranged from 5 to 48% of the activity of the wild-type strain, mc2155. In M. bovis BCG, all six M. bovis BCG ndh mutants lost between 75 and 90% of their NdhII activity (Fig. 4A). For comparison, we measured the NdhII activity in an INHr M. bovis BCG mutant having a single point mutation in katG (G985T, resulting in an Asp329Tyr amino acid change) and found it similar to that of the wild type (Fig. 4A). Complementation with pMV261::ndh restored NdhII activity in the mutants from both species to wild-type levels, suggesting that overexpression of a wild-type ndh gene could functionally restore the defect of NdhII activity in the mutants (Table 3; Fig. 4B).
TABLE 3.
Ndh activity and NADH/NAD+ ratios in M. smegmatis strains
| Strain | % Ndh activitya | [NADH] (nM)a | [NAD+] (nM)a | [NADH]/[NAD+] |
|---|---|---|---|---|
| mc2155 | 100 | 1,028 ± 22 | 1,379 ± 12 | 0.7 |
| mc22374 | 21.0 ± 1.0 | 1,742 ± 65 | 1,409 ± 19 | 1.2 |
| mc22375 | 44.8 ± 3.8 | 1,525 ± 31 | 1,132 ± 3 | 1.3 |
| mc22376 | 34.0 ± 3.0 | 2,322 ± 11 | 1,200 ± 2 | 1.9 |
| mc22377 | 4.8 ± 0.8 | 1,966 ± 90 | 1,222 ± 35 | 1.6 |
| mc22378 | 6.8 ± 0.5 | 1,508 ± 67 | 836 ± 19 | 1.8 |
| mc22379 | 29.2 ± 4.4 | 1,129 ± 9 | 1,100 ± 6 | 1.0 |
| mc22380 | 48.0 ± 2.0 | 1,760 ± 21 | 1,502 ± 17 | 1.2 |
| mc22381 | 8.0 ± 0.7 | 1,728 ± 41 | 1,116 ± 39 | 1.5 |
| mc22382 | 13.5 ± 0.7 | 1,845 ± 105 | 1,335 ± 45 | 1.4 |
| mc22383 | 13.8 ± 0.7 | 1,505 ± 2 | 1,113 ± 2 | 1.4 |
| mc22384 | 17.1 ± 2.2 | 1,268 ± 37 | 810 ± 5 | 1.6 |
| mc22385 | 15.9 ± 1.5 | 1,230 ± 105 | 938 ± 38 | 1.3 |
| mc22386 | 18.1 ± 1.8 | 1,525 ± 31 | 1,132 ± 3 | 1.4 |
| mc22387 | 18.2 ± 2.7 | 1,511 ± 0 | 1,072 ± 33 | 1.4 |
| mc22388 | 24.0 ± 1.1 | 1,316 ± 20 | 870 ± 30 | 1.5 |
| mc2155 pMV261 | 100 | 987 ± 23 | 1,372 ± 10 | 0.7 |
| mc2155 pMV261::ndh | 510.4 ± 55.7 | 362 ± 1 | 1,056 ± 14 | 0.3 |
| mc22378 pMV261 | 8.6 ± 0.4 | 3,196 ± 79 | 1,122 ± 3 | 2.8 |
| mc22378 pMV261::ndh | 841.3 ± 50 | 587 ± 29 | 1,098 ± 3 | 0.5 |
| mc22381 pMV261 | 7.9 ± 0.6 | 2,151 ± 30 | 1,271 ± 29 | 1.7 |
| mc22381 pMV261::ndh | 584.0 ± 75 | 577 ± 32 | 1,280 ± 17 | 0.4 |
| mc22377 pMV261 | 9.7 ± 1.4 | 2,203 ± 3 | 950 ± 22 | 2.3 |
| mc22377 pMV261::ndh | 587 ± 66 | 587 ± 1 | 1,267 ± 5 | 0.5 |
Values are means ± standard deviations.
FIG. 4.
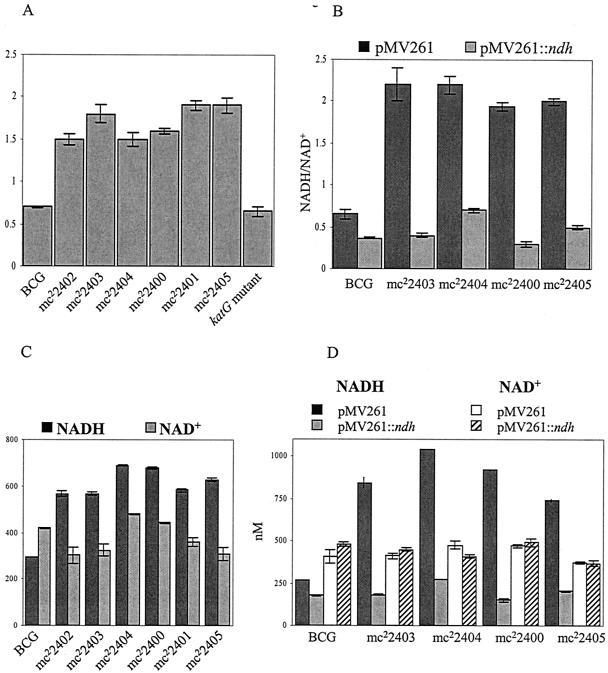
NADH dehydrogenase activity in M. bovis BCG ndh mutants and transformants. Ndh activity was measured as described in Materials and Methods and is given as a percentage relative to wild type (either M. bovis BCG Pasteur [A] or M. bovis BCG Pasteur transformed with pMV261 [B]). For comparison, the Ndh activity of an INHr M. bovis BCG strain having a single point mutation in katG (G985T, resulting in an Asp329Tyr amino acid change) is shown. The experiments were repeated three times, and the average is plotted with its standard deviation.
The NdhI activity was also assayed by measuring the rate of DCIP reduction in the presence of NHDH, as we were not able to measure the rate of NHDH oxidation in the presence of menadione. The NdhI activity was on average 95% lower than the NdhII activity (assayed by measuring the rate of DCIP reduction in the presence of NADH) in M. smegmatis mc2155 and 75% lower in wild-type M. bovis BCG. The M. smegmatis and M. bovis BCG ndh mutants had an NdhI activity comparable to that of the wild-type strains (data not shown).
ndh mutations result in altered NADH/NAD+ ratios in cells.
Since NdhII encoded by ndh oxidizes NADH into NAD+, we reasoned that the mutants with lowered NADH dehydrogenase activity would have an altered NADH/NAD+ ratio in the cells. To test this hypothesis, both intracellular NADH and NAD+ concentrations were measured using a sensitive cycling assay (30). Each mutant was grown at permissive temperature in broth containing neither INH nor ETH in a culture flask fully aerated. The NADH and NAD+ levels were found to be highly dependent on the density of the culture in that higher levels of NADH and NAD+ were measured in denser cultures (data not shown). Nevertheless, our results repeatedly showed that the NADH/NAD+ ratio was significantly higher in the ndh mutants than in wild-type M. smegmatis or M. bovis BCG (NADH/NAD+ ≈ 0.7 in parent strains and ≥1.0 in the ndh mutants; P < 0.002). The significance was determined using the Student t test (Table 3; Fig. 5A). As expected, in the M. bovis BCG INHr katG mutant which had no defect in NdhII, the NADH/NAD+ ratio was similar to that of the wild type (Fig. 5A). In M. bovis BCG, we observed that the NAD+ concentrations were comparable in both the wild-type and ndh mutants (Fig. 5C). The main difference in these mutants was the NADH concentration, which almost doubled in the ndh mutants compared to the wild-type strain (Fig. 5C).
FIG. 5.
NADH/NAD+ ratios, NADH concentrations, and NAD+ concentrations in M. bovis BCG ndh mutants and transformants. The M. bovis BCG strains were grown at 37°C to log phase (OD600 ≈ 0.8 to 1). The NADH and NAD+ concentrations were measured in triplicate as described in Materials and Methods. For comparison, the NADH/NAD+ ratio of an INHr M. bovis BCG strain having a single point mutation in katG (G985T, resulting in an Asp329Tyr amino acid change) is shown. The experiments were repeated three times, and the average is plotted with its standard deviation.
The NADH and NAD+ concentrations were also measured in ndh mutants transformed with the complementing pMV261::ndh plasmid (Table 3; Fig. 5B and D). The transformants had an NADH/NAD+ value (0.4 to 0.5) comparable to that of the wild-type strains transformed with their respective pMV261::ndh (0.3). Again, the main change between strains transformed with either pMV261 or pMV261::ndh was observed for the NADH concentrations, the NAD+ concentrations remaining comparable (Table 3; Fig. 5D). Altogether, these results suggest that coresistance to INH and ETH directly correlates with reduced NdhII activity and increased intracellular NADH/NAD+ ratios.
Increasing NADH concentrations prevent inactivation of InhA by the INH-NAD adduct.
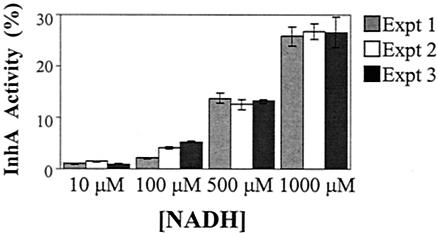
Previous studies have established that InhA is an NADH-specific enoyl-ACP reductase (11, 42) that is inhibited by a specific INH-NAD adduct (29, 40, 44, 45, 52). Since we have demonstrated that coresistance to INH and ETH in ndh mutants correlates with increased NADH/NAD+ ratios in the mutant cells, we hypothesized that the increased NADH concentrations mediate resistance by competitively inhibiting the binding of the INH-NAD adduct. Nguyen et al. (40) showed that high concentrations of NADH can prevent inhibition of InhA by the INH-NAD adduct. These authors reported that treatment of InhA with a 100 nM pool of INH-NAD adducts resulted in a 78% loss of InhA activity. However, when InhA was preincubated with 100 μM NADH and then treated with the pool of INH-NAD adducts, no InhA inhibition was observed, suggesting that high concentrations of NADH protect the adduct-mediated inhibition of InhA. To confirm the hypothesis that an excess of NADH could prevent inactivation of InhA by the INH-NAD adduct, the inhibition of InhA by the INH-NAD adduct was tested in the presence of increasing concentrations of NADH under three different conditions in vitro (Fig. 6). In the first two experiments (experiments 1 and 2 in Fig. 6), the enzyme was preincubated with NADH prior to the addition of the inhibitor [either as a preformed INH-NAD adduct or as a mixture of INH and Mn(III) pyrophosphate] and the substrate. In the third experiment, InhA was preincubated with the INH-NAD adduct for 3 h, and activity of the inhibited enzyme was then assayed in the presence of different concentrations of NADH. All three experiments yielded one consistent trend: increasing concentrations of NADH directly correlated with a decrease in the inhibition of InhA by the INH-NAD adduct. Therefore, we can conclude that high levels of NADH can protect InhA from the INH-NAD adduct, resulting in INH resistance.
FIG. 6.
InhA activity versus NADH concentration. In experiment 1, the INH-NAD adduct was first prepared by adding Mn(III) pyrophosphate (4 mM) into a mixture of INH (2 mM) and NAD+ (2 mM). InhA (60 nM) and NADH (10, 100, or 500 μM or 1 mM) were preincubated for 20 min prior to the addition of the INH-NAD adduct (10 μM). The reaction was initiated by the addition of 2-trans-dodecenoyl-CoA (50 μM). In experiment 2, InhA (60 nM) and NADH (10, 100, 500, or 1,000 μM) were preincubated for 20 min. INH (10 μM) and Mn(III) pyrophosphate (20 μM) were then added. The mixture was incubated for another 5 min before adding 2-trans-dodecenoyl-CoA (50 μM). In experiment 3, the INH-NAD adduct was prepared as in experiment 1 and incubated with InhA (5 μM) for 3 h. The inhibited InhA was separated from other components by using a desalting column, and the inhibited InhA (60 nM) was assayed under different concentrations of NADH (10, 100, or 500 μM or 1 mM). In each experiment, the InhA activity was assayed by monitoring the NADH absorption at 340 nm for 2 min, at room temperature, using 2-trans-dodecenoyl-CoA (50 μM) as a substrate.
Specific ndh mutations mediate a Ts bactericidal phenotype.
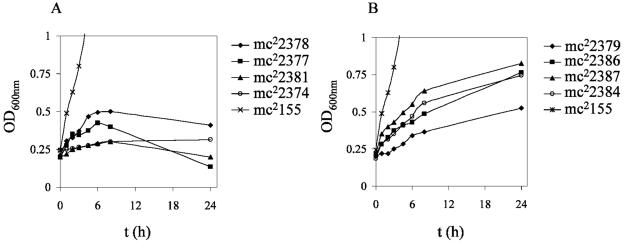
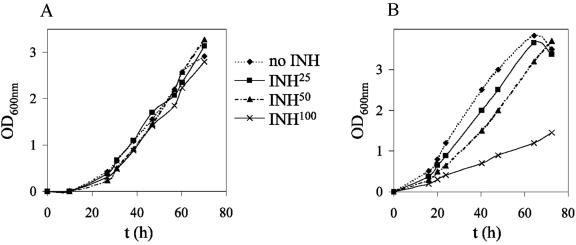
To further characterize the different ndh alleles, we tested whether incubation at 42°C was a bactericidal or static event in M. smegmatis ndh mutants. The mutants were grown at permissive temperature, titers were determined, and 10-fold serial dilutions were plated on plates containing no antibiotic and incubated at 42°C for 4 to 5 days. The plates incubated at 42°C were then shifted to the permissive temperature (30°C) and incubated for another 7 days. The phenotype was considered Ts bactericidal when less than 10% of cells could survive incubation at 42°C. Five ndh mutants (ndh-11 [Leu100Pro], ndh-32 [Arg145Cys], ndh-61 [Gly84Asp], ndh-63 [Arg145Cys], and ndh-93 [Arg145Cys]) did not survive incubation at 42°C, and the mutations were considered bactericidal. The other mutants did not grow at 42°C but were able to resume growth once the plates were shifted to permissive temperature (30°C). Interestingly, the lowest NdhII activity was observed for the mutants having a Ts bactericidal phenotype (mc22377, mc22378, and mc22381) (Table 3). In broth at nonpermissive temperature (42°C), growth ceased for all bactericidal mutants as well as five static mutants (Fig. 7A). In contrast, the seven other static mutants grew at the nonpermissive temperature, but at a much slower rate than wild-type M. smegmatis (Fig. 7B). After 24 h of incubation in Middlebrook 7H9 broth at nonpermissive temperature, the drop in the number of viable cells was less than 1 log for static ndh mutants (21 of 26), while the five bactericidal mutants had up to a 3-log drop in viable cells (data not shown). The reduction in viable cells was medium dependent, with maximal decreases in survival in minimal medium compared to results in tryptic soy or Mueller-Hinton broth (data not shown). Interestingly, most of the strains with mutations in the carboxy end of Ndh had no growth defect in medium containing up to 100 μg of INH/ml at the permissive temperature (Fig. 8A), whereas strains with mutations in the amino terminus of Ndh showed a slower growth rate with increasing concentration of INH (Fig. 8B).
FIG. 7.
Effect of nonpermissive temperature (42°C) on M. smegmatis ndh Ts mutants in liquid media. The ndh mutants were grown at 30°C to mid-log phase, diluted to an OD600 of ≈0.2, and shifted to 42°C (t = 0). The growth of the cultures was followed spectrophotometrically by measuring the OD600. To simplify the figures, only four mutants are shown per group. (A) Growth was ceased for the following M. smegmatis ndh Ts mutants: mc22374, mc22375, mc22376, mc22377, mc22378, mc22381, mc22383, and mc22385. (B) Shift to 42°C slows the growth of the following M. smegmatis ndh Ts mutants: mc22379, mc22380, mc22382, mc22384, mc22386, mc22387, and mc22388.
FIG. 8.
Growth of M. smegmatis ndh mutants at permissive temperature (30°C) with and without INH. The ndh mutants were grown at 30°C to mid-log phase and diluted to an OD600 of ≈0.05. The diluted cultures (10 ml) were added to four square bottles, and to each square bottle was added 0, 25, 50, or 100 μg of INH/ml. The cultures were incubated while shaking at 30°C. The growth of the cultures was followed spectrophotometrically by measuring the OD600. To simplify the figures, only one mutant is shown per group. (A) Typical growth curve for M. smegmatis ndh mutants not affected by INH, such as mc22374, mc22376, mc22378, mc22379, mc22380, mc22381, and mc22384 (mutant shown is mc22384). (B) Typical growth curves of M. smegmatis ndh mutants with a growth rate delayed by INH, such as mc22375, mc22377, mc22382, mc22383, mc22385, mc22386, mc22387, and mc22388 (mutant shown is mc22377).
DISCUSSION
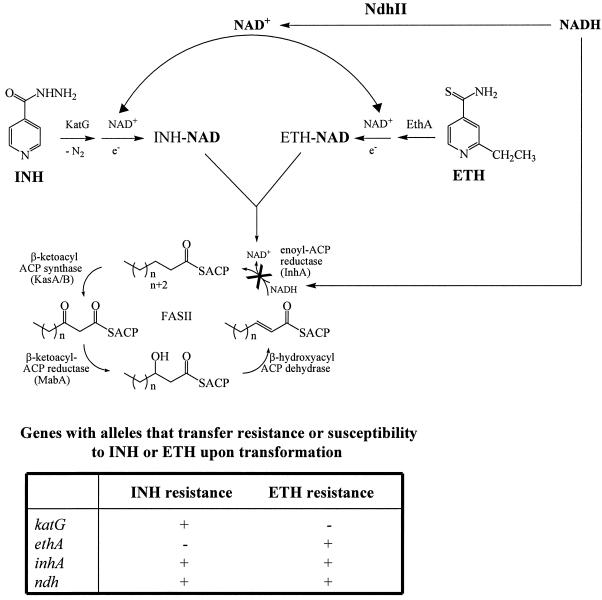
The analyses of mutations conferring resistance to drugs have been invaluable in elucidating the mechanisms of drug action and drug resistance. The discoveries of the phenotypes demonstrating that inactivation of the katG (20, 56, 57) and ethA (3, 10, 50) genes conferred resistance to INH and ETH, respectively, established that both INH and ETH are prodrugs with independent activation pathways. Resistance was shown to be mediated by recessive mutations causing a loss of prodrug-activating activities (Fig. 9). Genetic studies demonstrating target overexpression or target alteration revealed that INH and ETH shared a common target, InhA (1, 26). In contrast to the mutations in the INH or ETH activator genes, target mutations are dominant to the wild-type gene. Another mechanism of coresistance to INH and ETH was previously demonstrated to be mediated in M. smegmatis in a recessive fashion by mutations in ndh, a gene encoding a type II NADH dehydrogenase (36). It was unclear if a similar mechanism could function in slow-growing mycobacteria such as M. tuberculosis or M. bovis BCG, which are 100 times more sensitive to INH than M. smegmatis. Moreover, the precise mechanism by which mutations in ndh could mediate coresistance to both INH and ETH was also unclear.
FIG. 9.
Proposed mechanism of action of INH and ETH. INH and ETH are both prodrugs that are activated by the catalase-peroxidase KatG or the monooxygenase EthA, respectively. The activated forms react with NAD+ to form an INH-NAD or ETH-NAD adduct. These adducts inhibit the common target InhA, the NADH-dependent enoyl-ACP reductase of the fatty acid synthase type II system, resulting in mycolic acid biosynthesis inhibition and cell lysis. Resistance to INH or ETH is associated with recessive mutations in the genes encoding the activators of the drugs, katG and ethA, respectively, which prevent drug activation. Coresistance to INH and ETH is associated with dominant mutations in the gene encoding the common target of the drugs, inhA, which result in target amplification or target modification. A novel mechanism of coresistance to INH and ETH is by recessive mutations in ndh, which increase the NADH intracellular concentration and cause resistance by competitively inhibiting the binding of the INH-NAD or ETH-NAD adduct to InhA. This working model accounts for all known resistance phenotypes that have been transferred from drug-sensitive to drug-resistant strains to date.
To address these points, in this study 6 M. bovis BCG ndh mutants and 14 novel M. smegmatis ndh mutants coresistant to INH and ETH were isolated. Sequence analysis revealed the presence of specific mutations in the ndh gene. To date, no INHr ETHr M. tuberculosis mutants could be isolated in three independent attempts by directly plating M. tuberculosis H37Rv on INH and ETH plates, as done for M. bovis BCG (data not shown). We tried to obtain the ndh mutant strains found in INHr clinical isolates of M. tuberculosis described previously (27), but these strains were not saved. Our inability to isolate M. tuberculosis H37Rv ndh mutants might result from the concentrations of drugs used to do the screening for this particular strain, and it may reflect metabolic differences among M. tuberculosis strains. Interestingly, Middlebrook and Cohn had described the presence of INHr mutants that were catalase positive but failed to grow on Middlebrook minimal medium (35). It may be that ndh mutants of M. tuberculosis are auxotrophic for certain nutrients, as observed for some M. smegmatis mutants (36). Further analyses are under way to explore these possibilities. Nevertheless, complementation of the M. bovis BCG ndh mutations with the wild-type M. bovis BCG ndh gene restored full susceptibility to both INH and ETH, proving that these ndh mutations were sufficient to mediate the INH and ETH coresistance phenotypes in the M. tuberculosis complex.
The phenotype of coresistance mediated by mutations in inhA and ndh suggests that both INH and ETH target a common enzyme, InhA, and share a common mechanism of action. To address the molecular mechanism of resistance mediated by the ndh mutations, we first looked at the possibility that ndh mutations mediate coresistance to INH and ETH by overexpressing inhA. Using a quantitative PCR test, we were able to establish that the levels of inhA were not increased in the M. bovis BCG mutants analyzed. This was further confirmed at a translational level by Western blotting.
It was previously postulated that an NdhII defect in M. smegmatis would result in an altered NADH/NAD+ ratio due to an increase in the NADH concentration, which would prevent InhA inhibition (36, 45). To test this hypothesis, the NADH/NAD+ ratios were measured in all the M. bovis BCG and M. smegmatis ndh mutants and found to be greater than 1.0 in M. smegmatis and greater than 1.6 in M. bovis BCG, compared to 0.7 for the wild-type strains. In fact, the increase in the NADH/NAD+ ratios was mostly due to an increase in the NADH concentration. Nguyen et al. (40) first showed that preincubation of InhA with NADH prevented inhibition of InhA by the INH-NAD adduct. Our in vitro biochemical assays showed that increasing concentrations of NADH competitively inhibit binding of the INH-NAD adduct to InhA, irrespective of the order of addition of the different substrates or inhibitors to the InhA enzyme. Although the concentrations of NADH in these experiments are higher than the intracellular physiological concentrations present in the cells, it is unclear what the localized concentrations of NADH and INH-NAD adduct might be for InhA. The in vitro trend of protection from inhibition is fully consistent with the intracellular NADH increase observed for all ndh mutants. Therefore, we conclude that increased NADH/NAD+ ratios most likely competitively inhibit the binding of the INH-NAD or ETH-NAD adduct to InhA, thus resulting in INH and ETH resistance.
An alternative hypothesis was that the altered NADH/NAD+ ratios reduced or inhibited the activities of KatG and EthA, the activators of INH and ETH, respectively. However, Miesel et al. (36) showed that M. smegmatis ndh mutations causing INH resistance did not result in decreased KatG expression. Moreover, Fraaije et al. (14) reported that EthA, a FAD-containing monooxygenase, uses NADPH and not NADH as a cofactor. Thus, it appears that a high concentration of NADH does not prevent activation of INH or ETH.
We conclude that both INH and ETH are prodrugs which, when activated, form adducts with NAD, and the resulting INH-NAD and ETH-NAD adducts inactivate InhA (Fig. 9). We further conclude that InhA is the single primary target of action for both INH and ETH. Low-level resistance to these drugs is mediated by altering the NADH/NAD+ ratios. This alteration might be mediated by other metabolic enzymes than NdhII. Supporting this premise, the M. bovis BCG mutants isolated in our screen, which are coresistant to INH and ETH but have no mutations in ndh (see Results), have altered NADH/NAD+ ratios (C. Vilchèze and W. R. Jacobs, Jr., unpublished results). Efforts are under way to genetically identify the mutated genes in these strains. In addition, a number of clinical isolates of M. tuberculosis have been identified which are resistant to INH or coresistant to INH and ETH and possess no mutations in either inhA or ndh (38, 43). We postulate that such mutations could (i) mediate overexpression of inhA, (ii) alter ndh expression levels, or (iii) metabolically alter the NADH/NAD+ ratios. The mdh gene represents such a candidate (36), as well as the glf and ceoBC genes, which may contribute to INH resistance (8). Interestingly, the products of the glf and ceoBC genes are NAD+ binding proteins. Furthermore, mutations altering the NADH/NAD+ ratio might provide useful tools to elucidate key regulators of energy metabolism in this postgenomic era of biology.
The role that NdhII plays in cell metabolism has been unclear, but it has been proposed that NdhII might control the NADH pool to regulate cell energy metabolism (5, 16, 39). INHr ETHr M. smegmatis and M. bovis BCG ndh mutants have defective NdhII activity, resulting in altered NADH/NAD+ ratios. Complementation of the mutants with a wild-type ndh gene restored drug sensitivity, NdhII activity, and a NADH/NAD+ ratio similar to wild type. Therefore, it appears that NdhII is a major regulator of the NADH/NAD+ ratio in cells during exponential, aerobic growth for both fast- and slow-growing mycobacteria. The mutations that cause the reduced NdhII activity are distributed throughout the first 80% of the protein. Interestingly, according to the TransMembrane Hidden Markov model algorithm website (http://www.cbs.dtu.dk/services/TMHMM/), which predicts transmembrane domains, the NdhII protein of Mycobacterium species and related Corynebacterium contains a single transmembrane-spanning region, a region where no mutations have yet to be observed (Fig. 1). In contrast to the E. coli enzyme, which has been characterized as a membrane-bound protein, the mycobacterial NdhII enzyme appears to be membrane associated. Interestingly, all the mutations in ndh in M. smegmatis confer a Ts phenotype to the growth of M. smegmatis. The numbers of independent mutants isolated to date (Fig. 1) would argue that the Ts phenotype results not from Ts NdhII activity but rather from a conditional growth arrest caused by the altered NADH/NAD+ ratio, stabilizing activity. Certain mutants in ndh have been shown to lead to auxotrophies (36), suggesting that altered NADH/NAD+ ratios do affect overall metabolism, a reasonable expectation for the important electron donor NADH.
The mutations that alter NdhII activity from M. smegmatis could be readily classified based on their Ts phenotypes, which allowed us to test if thermal inactivation was either a bacteriostatic or bactericidal event. A set of mutations (Gly84Asp, Leu100Pro, and Arg145Cys), found in the region between the FAD and NAD binding sites, conferred a bactericidal effect upon inactivation. We believe that the development of compounds that target this NdhII region could lead to novel bactericidal drugs against mycobacterial pathogens. Further work to characterize these bactericidal biochemical events following thermal inactivation may provide insight into a novel mechanism by which mycobacterial cells die, thereby providing a rationale for designing better antimycobacterial drugs.
Acknowledgments
This work was supported by NIH grants AI43268 and AI46669. L.K. is supported by INSERM. J.C.S. is supported by the Welch foundation.
We thank Yossef Av-Gay for the critical reading of the manuscript and engaging discussion.
REFERENCES
- 1.Banerjee, A., E. Dubnau, A. Quemard, V. Balasubramanian, K. S. Um, T. Wilson, D. Collins, G. de Lisle, and W. R. Jacobs, Jr. 1994. inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science 263:227-230. [DOI] [PubMed] [Google Scholar]
- 2.Basso, L. A., R. Zheng, J. M. Musser, W. R. Jacobs, Jr., and J. S. Blanchard. 1998. Mechanisms of isoniazid resistance in Mycobacterium tuberculosis: enzymatic characterization of enoyl reductase mutants identified in isoniazid-resistant clinical isolates. J. Infect. Dis. 178:769-775. [DOI] [PubMed] [Google Scholar]
- 3.Baulard, A. R., J. C. Betts, J. Engohang-Ndong, S. Quan, R. A. McAdam, P. J. Brennan, C. Locht, and G. S. Besra. 2000. Activation of the pro-drug ethionamide is regulated in mycobacteria. J. Biol. Chem. 275:28326-28331. [DOI] [PubMed] [Google Scholar]
- 4.Bernstein, J. W., A. Lott, B. A. Steinberg, and H. L. Yale. 1952. Chemotherapy of experimental tuberculosis. Am. Rev. Tuberc. 65:357-374. [DOI] [PubMed] [Google Scholar]
- 5.Calhoun, M. W., K. L. Oden, R. B. Gennis, M. J. de Mattos, and O. M. Neijssel. 1993. Energetic efficiency of Escherichia coli: effects of mutations in components of the aerobic respiratory chain. J. Bacteriol. 175:3020-3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Canetti, G. 1965. Present aspects of bacterial resistance in tuberculosis. Am. Rev. Respir. Dis. 92:687-703. [DOI] [PubMed] [Google Scholar]
- 7.Canetti, G., B. Kreis, R. G. Thibier, and M. P. Le Lirzin. 1967. Current data on primary resistance in pulmonary tuberculosis in adults in France. 2d survey of the Centre d'Etudes sur la Resistance Primaire: 1965-1966. Rev. Tuberc. Pneumol. (Paris) 31:433-474. [PubMed] [Google Scholar]
- 8.Chen, P., and W. R. Bishai. 1998. Novel selection for isoniazid (INH) resistance genes supports a role for NAD+-binding proteins in mycobacterial INH resistance. Infect. Immun. 66:5099-5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corbett, E. L., C. J. Watt, N. Walker, D. Maher, B. G. Williams, M. C. Raviglione, and C. Dye. 2003. The growing burden of tuberculosis: global trends and interactions with the HIV epidemic. Arch. Intern. Med. 163:1009-1021. [DOI] [PubMed] [Google Scholar]
- 10.DeBarber, A. E., K. Mdluli, M. Bosman, L. G. Bekker, and C. E. Barry III. 2000. Ethionamide activation and sensitivity in multidrug-resistant Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 97:9677-9682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dessen, A., A. Quemard, J. S. Blanchard, W. R. Jacobs, Jr., and J. C. Sacchettini. 1995. Crystal structure and function of the isoniazid target of Mycobacterium tuberculosis. Science 267:1638-1641. [DOI] [PubMed] [Google Scholar]
- 12.Espinal, M. A. 2003. The global situation of MDR-TB. Tuberculosis 83:44-51. [DOI] [PubMed] [Google Scholar]
- 13.Fox, H. H. 1952. The chemical approach to the control of tuberculosis. Science 116:129-134. [DOI] [PubMed] [Google Scholar]
- 14.Fraaije, M. W., N. M. Kamerbeek, A. J. Heidekamp, R. Fortin, and D. B. Janssen. 2004. The prodrug activator EtaA from Mycobacterium tuberculosis is a Baeyer-Villiger monooxygenase. J. Biol. Chem. 279:3354-3360. [DOI] [PubMed] [Google Scholar]
- 15.Gennis, R. B., and V. Stewart. 1996. Respiration, p. 217-261. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, vol. 1. ASM Press, Washington, D.C.
- 16.Green, J., and J. R. Guest. 1994. Regulation of transcription at the ndh promoter of Escherichia coli by FNR and novel factors. Mol. Microbiol. 12:433-444. [DOI] [PubMed] [Google Scholar]
- 17.Heym, B., N. Honore, C. Truffot-Pernot, A. Banerjee, C. Schurra, W. R. Jacobs, Jr., J. D. van Embden, J. H. Grosset, and S. T. Cole. 1994. Implications of multidrug resistance for the future of short-course chemotherapy of tuberculosis: a molecular study. Lancet 344:293-298. [DOI] [PubMed] [Google Scholar]
- 18.Hok, T. T. 1964. A comparative study of the susceptibility to ethionamide, thiosemicarbazone, and isoniazid of tubercle bacilli from patients never treated with ethionamide or thiosemicarbazone. Am. Rev. Respir. Dis. 90:468-469. [DOI] [PubMed] [Google Scholar]
- 19.Jaworowski, A., G. Mayo, D. C. Shaw, H. D. Campbell, and I. G. Young. 1981. Characterization of the respiratory NADH dehydrogenase of Escherichia coli and reconstitution of NADH oxidase in ndh mutant membrane vesicles. Biochemistry 20:3621-3628. [DOI] [PubMed] [Google Scholar]
- 20.Johnsson, K., and P. G. Schultz. 1994. Mechanistic studies of the oxidation of isoniazid by the catalase peroxidase from Mycobacterium tuberculosis. J. Am. Chem. Soc. 116:7425-7426. [Google Scholar]
- 21.Kerscher, S. J. 2000. Diversity and origin of alternative NADH:ubiquinone oxidoreductases. Biochim. Biophys. Acta 1459:274-283. [DOI] [PubMed] [Google Scholar]
- 22.Kiepiela, P., K. S. Bishop, A. N. Smith, L. Roux, and D. F. York. 2000. Genomic mutations in the katG, inhA and aphC genes are useful for the prediction of isoniazid resistance in Mycobacterium tuberculosis isolates from Kwazulu Natal, South Africa. Tuberc. Lung Dis. 80:47-56. [DOI] [PubMed] [Google Scholar]
- 23.Kremer, L., L. G. Dover, H. R. Morbidoni, C. Vilcheze, W. N. Maughan, A. Baulard, S. Tu, N. Honore, V. Deretic, J. C. Sacchettini, C. Locht, W. R. J. Jacobs, and G. Besra. 2003. Inhibition of InhA activity, but not KasA activity, induces formation of a KasA-containing complex in mycobacteria. J. Biol. Chem. 278:20547-20554. [DOI] [PubMed] [Google Scholar]
- 24.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 25.Larsen, M. H. 2000. Some common methods in mycobacterial genetics, p. 313-320. In G. F. Hatfull and W. R. Jacobs, Jr. (ed.), Molecular genetics of mycobacteria. ASM Press, Washington, D.C.
- 26.Larsen, M. H., C. Vilcheze, L. Kremer, G. S. Besra, L. Parsons, M. Salfinger, L. Heifets, M. H. Hazbon, D. Alland, J. C. Sacchettini, and W. R. Jacobs, Jr. 2002. Overexpression of inhA, but not kasA, confers resistance to isoniazid and ethionamide in Mycobacterium smegmatis, M. bovis BCG and M. tuberculosis. Mol. Microbiol. 46:453-466. [DOI] [PubMed] [Google Scholar]
- 27.Lee, A. S., A. S. Teo, and S. Y. Wong. 2001. Novel mutations in ndh in isoniazid-resistant Mycobacterium tuberculosis isolates. Antimicrob. Agents Chemother. 45:2157-2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lefford, M. J. 1966. The ethionamide sensitivity of British pre-treatment strains of Mycobacterium tuberculosis. Tubercle 47:198-206. [DOI] [PubMed] [Google Scholar]
- 29.Lei, B., C. J. Wei, and S. C. Tu. 2000. Action mechanism of antitubercular isoniazid. Activation by Mycobacterium tuberculosis KatG, isolation, and characterization of inha inhibitor. J. Biol. Chem. 275:2520-2526. [DOI] [PubMed] [Google Scholar]
- 30.Leonardo, M. R., Y. Dailly, and D. P. Clark. 1996. Role of NAD in regulating the adhE gene of Escherichia coli. J. Bacteriol. 178:6013-6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Madison, B. M., S. H. Siddiqi, L. Heifets, W. Gross, M. Higgins, N. Warren, A. Thompson, G. Morlock, and J. C. Ridderhof. 2004. Identification of a Mycobacterium tuberculosis strain with stable, low-level resistance to isoniazid. J. Clin. Microbiol. 42:1294-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsushita, K., T. Ohnishi, and H. R. Kaback. 1987. NADH-ubiquinone oxidoreductases of the Escherichia coli aerobic respiratory chain. Biochemistry 26:7732-7737. [DOI] [PubMed] [Google Scholar]
- 33.McKie, J. H., and K. T. Douglas. 1991. Evidence for gene duplication forming similar binding folds for NAD(P)H and FAD in pyridine nucleotide-dependent flavoenzymes. FEBS Lett. 279:5-8. [DOI] [PubMed] [Google Scholar]
- 34.McMurry, L. M., P. F. McDermott, and S. B. Levy. 1999. Genetic evidence that InhA of Mycobacterium smegmatis is a target for triclosan. Antimicrob. Agents Chemother. 43:711-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Middlebrook, G., and M. L. Cohn. 1953. Some observations on the pathogenicity of isoniazid-resistant variants of tubercle bacilli. Science 118:297-299. [DOI] [PubMed] [Google Scholar]
- 36.Miesel, L., T. R. Weisbrod, J. A. Marcinkeviciene, R. Bittman, and W. R. Jacobs, Jr. 1998. NADH dehydrogenase defects confer isoniazid resistance and conditional lethality in Mycobacterium smegmatis. J. Bacteriol. 180:2459-2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Molenaar, D., M. E. van der Rest, A. Drysch, and R. Yucel. 2000. Functions of the membrane-associated and cytoplasmic malate dehydrogenases in the citric acid cycle of Corynebacterium glutamicum. J. Bacteriol. 182:6884-6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morlock, G. P., B. Metchock, D. Sikes, J. T. Crawford, and R. C. Cooksey. 2003. ethA, inhA, and katG loci of ethionamide-resistant clinical Mycobacterium tuberculosis isolates. Antimicrob. Agents Chemother. 47:3799-3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neijssel, O. M., and M. J. Teixeira de Mattos. 1994. The energetics of bacterial growth: a reassessment. Mol. Microbiol. 13:172-182. [DOI] [PubMed] [Google Scholar]
- 40.Nguyen, M., A. Quemard, S. Broussy, J. Bernadou, and B. Meunier. 2002. Mn(III) pyrophosphate as an efficient tool for studying the mode of action of isoniazid on the InhA protein of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 46:2137-2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Piatek, A. S., A. Telenti, M. R. Murray, H. El-Hajj, W. R. Jacobs, Jr., F. R. Kramer, and D. Alland. 2000. Genotypic analysis of Mycobacterium tuberculosis in two distinct populations using molecular beacons: implications for rapid susceptibility testing. Antimicrob. Agents Chemother. 44:103-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quemard, A., J. C. Sacchettini, A. Dessen, C. Vilcheze, R. Bittman, W. R. Jacobs, Jr., and J. S. Blanchard. 1995. Enzymatic characterization of the target for isoniazid in Mycobacterium tuberculosis. Biochemistry 34:8235-8241. [DOI] [PubMed] [Google Scholar]
- 43.Ramaswamy, S. V., R. Reich, S. J. Dou, L. Jasperse, X. Pan, A. Wanger, T. Quitugua, and E. A. Graviss. 2003. Single nucleotide polymorphisms in genes associated with isoniazid resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 47:1241-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rawat, R., A. Whitty, and P. J. Tonge. 2003. The isoniazid-NAD adduct is a slow, tight-binding inhibitor of InhA, the Mycobacterium tuberculosis enoyl reductase: adduct affinity and drug resistance. Proc. Natl. Acad. Sci. USA 100:13881-13886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rozwarski, D. A., G. A. Grant, D. H. Barton, W. R. Jacobs, Jr., and J. C. Sacchettini. 1998. Modification of the NADH of the isoniazid target (InhA) from Mycobacterium tuberculosis. Science 279:98-102. [DOI] [PubMed] [Google Scholar]
- 46.San, K. Y., G. N. Bennett, S. J. Berrios-Rivera, R. V. Vadali, Y. T. Yang, E. Horton, F. B. Rudolph, B. Sariyar, and K. Blackwood. 2002. Metabolic engineering through cofactor manipulation and its effects on metabolic flux redistribution in Escherichia coli. Metab. Eng. 4:182-192. [DOI] [PubMed] [Google Scholar]
- 47.Sonnhammer, E. L., G. von Heijne, and A. Krogh. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6:175-182. [PubMed] [Google Scholar]
- 48.Stover, C. K., V. F. de la Cruz, T. R. Fuerst, J. E. Burlein, L. A. Benson, L. T. Bennett, G. P. Bansal, J. F. Young, M. H. Lee, G. F. Hatfull, et al. 1991. New use of BCG for recombinant vaccines. Nature 351:456-460. [DOI] [PubMed] [Google Scholar]
- 49.Takayama, K., L. Wang, and H. L. David. 1972. Effect of isoniazid on the in vivo mycolic acid synthesis, cell growth, and viability of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2:29-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vannelli, T. A., A. Dykman, and P. R. Ortiz de Montellano. 2002. The antituberculosis drug ethionamide is activated by a flavoprotein monooxygenase. J. Biol. Chem. 277:12824-12829. [DOI] [PubMed] [Google Scholar]
- 51.Vilcheze, C., H. R. Morbidoni, T. R. Weisbrod, H. Iwamoto, M. Kuo, J. C. Sacchettini, and W. R. Jacobs, Jr. 2000. Inactivation of the inhA-encoded fatty acid synthase II (FASII) enoyl-acyl carrier protein reductase induces accumulation of the FASI end products and cell lysis of Mycobacterium smegmatis. J. Bacteriol. 182:4059-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilming, M., and K. Johnsson. 1999. Spontaneous formation of the bioactive form of the tuberculosis drug isoniazid. Angew Chem. Int. Ed. 38:2588-2590. [DOI] [PubMed] [Google Scholar]
- 53.World Health Organization. 2000. Anti-tuberculosis drug resistance in the world. Report no. 2: prevalence and trends. The W.H.O./IUATLD global project on anti-tuberculosis drug resistance surveillance. World Health Organization, Geneva, Switzerland.
- 54.Yagi, T. 1993. The bacterial energy-transducing NADH-quinone oxidoreductases. Biochim. Biophys. Acta 1141:1-17. [DOI] [PubMed] [Google Scholar]
- 55.Yagi, T., T. Yano, S. Di Bernardo, and A. Matsuno-Yagi. 1998. Procaryotic complex I (NDH-1), an overview. Biochim. Biophys. Acta 1364:125-133. [DOI] [PubMed] [Google Scholar]
- 56.Zhang, Y., T. Garbe, and D. Young. 1993. Transformation with katG restores isoniazid sensitivity in Mycobacterium tuberculosis isolates resistant to a range of drug concentrations. Mol. Microbiol. 8:521-524. [DOI] [PubMed] [Google Scholar]
- 57.Zhang, Y., B. Heym, B. Allen, D. Young, and S. Cole. 1992. The catalase-peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature 358:591-593. [DOI] [PubMed] [Google Scholar]