Abstract
Pyrethroid resistance in malaria vector, An. funestus is increasingly reported across Africa, threatening the sustainability of pyrethroid-based control interventions, including long lasting insecticidal nets (LLINs). Managing this problem requires understanding of the molecular basis of the resistance from different regions of the continent, to establish whether it is being driven by a single or independent selective events. Here, using a genome-wide transcription profiling of pyrethroid resistant populations from southern (Malawi), East (Uganda), and West Africa (Benin), we investigated the molecular basis of resistance, revealing strong differences between the different African regions. The duplicated cytochrome P450 genes (CYP6P9a and CYP6P9b) which were highly overexpressed in southern Africa are not the most upregulated in other regions, where other genes are more overexpressed, including GSTe2 in West (Benin) and CYP9K1 in East (Uganda). The lack of directional selection on both CYP6P9a and CYP6P9b in Uganda in contrast to southern Africa further supports the limited role of these genes outside southern Africa. However, other genes such as the P450 CYP9J11 are commonly overexpressed in all countries across Africa. Here, CYP9J11 is functionally characterized and shown to confer resistance to pyrethroids and moderate cross-resistance to carbamates (bendiocarb). The consistent overexpression of GSTe2 in Benin is coupled with a role of allelic variation at this gene as GAL4-UAS transgenic expression in Drosophila flies showed that the resistant 119F allele is highly efficient in conferring both DDT and permethrin resistance than the L119. The heterogeneity in the molecular basis of resistance and cross-resistance to insecticides in An. funestus populations throughout sub-Saharan African should be taken into account in designing resistance management strategies.
Keywords: Malaria, Anopheles funestus, pyrethroids, insecticide resistance, cytochrome P450
Malaria remains one of the main causes of morbidity and mortality in Sub-Saharan Africa, predominantly in children under 5 yr and pregnant mothers (WHO 2015). Anopheles funestus s.s. is one of the major malaria vectors in Sub-Saharan Africa and is widely distributed across the continent (Gillies and De Meillon 1968). The important role of An. funestus in malaria transmission is supported by recent reports indicating high Plasmodium falciparum parasite infection rates in this vector in many Sub-Saharan countries (Coetzee and Koekemoer 2013; Dia et al. 2013). Malaria vector control relies heavily on the use of a single insecticide class—the pyrethroids. Pyrethroids are safe and fast acting (Zaim et al. 2000), and are the only class of insecticides approved for use on insecticide treated materials such as Long Lasting Insecticide Nets (LLINs) (http://www.who.int/whopes/en/). As in other malaria vectors, pyrethroid resistance in An. funestus has increasingly been reported in Sub-Saharan Africa from different regions, including southern [South Africa (Hargreaves et al. 2000; Brooke et al. 2001), Mozambique (Casimiro et al. 2006; Cuamba et al. 2010), Malawi (Hunt et al. 2010; Wondji et al. 2012)], East [Uganda and Kenya (Morgan et al. 2010; Mulamba et al. 2014) and Tanzania (Lwetoijera et al. 2014)], Central [Cameroon (Wondji et al. 2011; Menze et al. 2016)], or West Africa [Benin (Djouaka et al. 2011, R. Djouaka et al. 2016), Ghana (Okoye et al. 2008; Riveron et al. 2016), Senegal (Samb et al. 2016) and Nigeria (Ibrahim et al. 2014; R. J. Djouaka et al. 2016)]. These increasing reports of pyrethroid resistance in malaria vectors such as An. funestus is of great concern as it poses serious threats to the effectiveness of the malaria vector control tools across the continent (WHO 2012). Thus, the urgent calls to develop and implement suitable resistance management strategies against malaria vectors, to ensure sustainable effectiveness of malaria vector control interventions. Understanding the molecular basis of insecticide resistance in malaria vectors is critical for designing and implementing these resistance management strategies.
Cases of pyrethroids resistance reported so far in An. funestus populations are mainly caused by metabolic resistance mechanisms, with no evidence of target-site resistance through knockdown resistance (kdr) (Amenya et al. 2008; Okoye et al. 2008; Wondji et al. 2012; Riveron et al. 2013). Cytochrome P450s are known to be the primary enzyme family conferring resistance to pyrethroids. Molecular studies conducted in southern Africa, notably in Malawi and Mozambique, have revealed that the duplicated P450 genes, CYP6P9a, and CYP6P9b are the main genes driving pyrethroid resistance in this species in this region (Amenya et al. 2008; Wondji et al. 2009; Riveron et al. 2013). However, studies performed in Zambia suggested a diminishing role of these two duplicated P450s northward (Riveron et al. 2014a; Thomsen et al. 2014). Furthermore, a recent study has revealed a similar minor role of CYP6P9a and CYP6P9b across a south-north transect in Malawi, with low expression of these two genes in the north in contrast to high levels in the south, coupled with a nearly fixed resistant haplotype (Barnes et al. 2016). This variation of expression profiles in southern Africa suggests that there could also be significant differences in the underlying genetic drivers of pyrethroid resistance across African populations of An. funestus. However, the molecular basis of pyrethroid resistance in An. funestus in other African regions, such as in East or West Africa, remains poorly characterized despite the high level of pyrethroid resistance also reported in these regions (Okoye et al. 2008; Morgan et al. 2010; Djouaka et al. 2011; Mulamba et al. 2014).
Here, using a microarray genome-wide transcription analysis, we characterized the molecular basis of pyrethroid resistance in this major vector in West and East Africa and through a comparative analysis with southern African populations, we revealed sharp difference in the key genes driving resistance in each region. The P450 CYP9J11 commonly overexpressed in all countries was functionally characterized, and shown to confer resistance to pyrethroids and moderate cross-resistance to carbamates. In addition, allelic variation in the glutathione S-transferase gene, GSTe2, through the L119F mutation (Riveron et al. 2014b) was established to be playing a major role in both DDT and pyrethroid resistance in Benin.
Materials and Methods
Study sites and samples
Blood-fed (F0) females resting indoors were collected between 06.00 am and 12.00 pm in Tororo, Eastern Uganda (0.69° N, 34.18° E), in July 2012. Benin samples were collected in Pahou (6° 23′ N, 2° 13′ E) in southern Benin, West Africa in April 2011. The Malawian samples were collected in the Chikwawa District (0° 45′ N, 34° 5′E) in southern Malawi between July 2009 and April 2010. The F0 collection method and F1 rearing were conducted as described previously (Djouaka et al. 2011; Riveron et al. 2013; Mulamba et al. 2014). All F0 adults used for individual oviposition of the above F1 eggs were morphologically identified as belonging to the An. funestus group according to the key of Gillies and Coetzee (1987). A PCR assay was performed using the protocol of (Koekemoer et al. 2002) to confirm that collected F0 adults were An. funestus s.s. The study samples were 2- to 5-d-old F1 adult permethrin-resistant An. funestus s.s. mosquitoes.
Resistance profile of different populations
The resistance patterns of the three populations to various insecticides was determined as described previously (Djouaka et al. 2011; Riveron et al. 2013; Mulamba et al. 2014) following the World Health Organization (WHO) protocol (WHO 1998). The Pahou populations of An. funestus from Benin is highly resistant to DDT (0% mortality after 1 hr exposure), resistant to both Type I (permethrin; 66% mortality) and II (deltamethrin; 88% mortality) pyrethroid, resistant to carbamates (bendiocarb; 64% mortality), but fully susceptible to malathion (Djouaka et al. 2011). The Uganda population from Tororo is resistant to pyrethroids [permethrin (33% mortality), deltamethrin (20% mortality)] and DDT (61% mortality), but susceptible to other insecticide classes (Mulamba et al. 2014). The Malawi population from Chikwawa in 2010 was resistant to pyrethroid [permethrin (47.2% mortality), deltamethrin (42.3% mortality)] and carbamates (bendiocarb; 60% mortality), moderately resistant to DDT (87.8% mortality), and fully susceptible to organophosphates (Wondji et al. 2012; Riveron et al. 2013).
Detection of pyrethroid resistance genes using microarrays
The 8 × 60k Agilent microarray chip custom designed for An. funestus used for this study was previously described (Riveron et al. 2014a). Briefly, each chip contains 60mer probes designed from An. funestus published ESTs from transcriptome sequencing by 454 (8540) (Gregory et al. 2011), Illumina (15,527) (Crawford et al. 2010), or An. funestus cDNAs from GenBank (2850) (two probes for each EST). It also includes a set of P450 genes from the rp1 and rp2 QTL genomic regions (Wondji et al. 2009; Irving et al. 2012) (three probes for each gene), the complete set of Anopheles gambiae transcripts (13,000) (one probe each), and all of the An. gambiae detoxification genes (David et al. 2005) (three probes for each gene). In Benin, we also used the other 4 × 44k An. funestus chip (A-MEXP-2245), previously described (Riveron et al. 2013) in a triangular experimental design comparing resistant (R), control (C), and susceptible (S) samples.
The Picopure RNA Isolation Kit (Arcturus) was used to extract total RNA from three biological replicates, each made of batches of 10 2- to 5-d-old F1 An. funestus from each field sample that had survived exposure to 0.75% permethrin for 1 hr (R). The same was done also for the fully susceptible laboratory strain FANG (S). Mosquitoes from Benin not exposed to insecticide (C) were also extracted. The RNA extraction was performed as previously described (Riveron et al. 2014a). Complementary RNA (cRNA) was amplified from each sample using the Agilent Quick Amp Labeling Kit (two-color) following the manufacturer’s protocol. The cRNA samples from the susceptible strain FANG (S) were labeled with the cy3 dye, and cRNAs from the resistant samples (R) were labeled with cy5 dye. The cRNA quantity and quality were assessed before labeling using the NanoDrop and Bioanalyzer. Labeled cRNAs were hybridized to the arrays for 17 hr at 65° according to the manufacturer’s protocol. Five hybridizations were performed for each sample by swapping the biological replicates. Agilent GeneSpring GX 13.0 software was used to analyze the microarray data. The differentially expressed genes were identified using a threshold of twofold-change (FC) and a statistical significance of P < 0.01 with Benjamini-Hochberg correction for multiple testing. The BLAST2GO program was used to predict the functions of all the transcripts used to design the microarray chip (Conesa et al. 2005; Gotz et al. 2008). Gene Ontology (GO) enrichment analyses were preformed using BLAST2GO to detect the major GO terms over-represented among the sets of probes upregulated in various hybridizations and countries in comparison to the reference set made of the entire transcript set on the microarray chip. The Fisher’s test was used to assess the statistical significance of these tests.
Quantitative RT-PCR validation of the candidate resistance genes
Quantitative reverse transcription PCR (qRT-PCR) assays were performed to validate microarray results for the key candidate genes; 1 µg of RNA from each of the three biological replicates—the Resistant (R), Control (C), and FANG (S)—was used as a template for complementary (cDNA) synthesis using the superscript III (Invitrogen) following the manufacturer’s guide. The qRT-PCR was carried out as previously described (Kwiatkowska et al. 2013; Riveron et al. 2013) with the relative expression level and FC of each target gene in R and C relative to S calculated according to the 2−ΔΔCT method (Schmittgen and Livak 2008) after normalization with the housekeeping genes ribosomal protein S7 (RSP7; AFUN007153) and actin5C (AFUN006819). The primers are listed in Supplemental Material, Table S1 in File S1.
Heterologous expression of candidate genes in Escherichia coli
Cloning of CYP9J1 for expression in E. coli:
The full-length CYP9J11 was amplified from cDNAs (used for qRT-PCR) and cloned into the pJET1.2/blunt cloning vector (Thermo Scientific). The primers used are listed in Table S1 in File S1 as CYP9J11Full F and R. After sequence analysis, one clone predominant in the three countries was selected for functional characterization. This CYP9J11 allele was fused to a bacterial ompA+2 leader sequence, and expressed in E. coli JM109 cells using the pCW-ori+ vector as previously described (Pritchard et al. 1998; McLaughlin et al. 2008; Stevenson et al. 2011). Briefly, a DNA fragment containing the coding sequence for the ompA+2 signal peptide with a downstream alanine-proline linker, and the first ∼20 nt of CYP9J11 was first amplified using 50 ng of E. coli JM109 DNA as the template (Primer sets in Table S1 in File S1). Next, this CYP9J11 clone and the ompA+2 PCR fragment were used as templates in a fusion PCR under the same conditions described previously (Riveron et al. 2014a). The full-length sequence of CYP9J11 incorporating the ompA+2 leader was ligated into a modified pCW-ori+ vector plasmid, pB13 (Pritchard et al. 1998), via EcoRI and XBaI sites to produce pB13::ompA+2-CYP9J11. This construct was also sequenced to confirm the absence of PCR errors.
Membrane preparation:
Membranes containing CYP9J11 were obtained by cotransforming the E. coli cells JM109 with pB13::ompA+2-CYP9J11 with a plasmid containing the An. gambiae cytochrome P450 reductase, pACYC-AgCPR (McLaughlin et al. 2008). The expression of CYP9J11, membrane isolation, and determination of P450 content were carried out as previously described (McLaughlin et al. 2008; Stevenson et al. 2011). The membranes were stored in aliquots at −80°, and assayed for total protein concentration using NanoDrop spectrophotometer, P450 concentration (Omura and Sato 1964) and CPR activity by monitoring cytochrome c reduction (Strobel and Dignam 1978). The histidine-tagged An. gambiae cytochrome b5 was generated as previously described by Stevenson et al. (2011) and used for the metabolism assays.
Metabolism assays:
In vitro metabolism reactions between pyrethroids (deltamethrin and permethrin) and carbamates (bendiorcab and propuxur) and membranes expressing CYP9J11 were performed as previously described (Stevenson et al. 2011; Ibrahim et al. 2016a) in the presence of CPR with cytochrome b5. The reactions consisted of the following: 45 pmol of P450, 0.2 M Tris HCl pH 7.4, 0.25 mM MgCl2, 1 mM glucose-6-phosphate, 0.1 mM NADP+ (Melford), 1 unit/ml glucose-6-phosphate dehydrogenase (G6PDH), 0.8 µM cytochrome b5, and 0.2 mM of test insecticide in a final volume of 200 ml.
HPLC analysis:
Detection of the reaction outcome followed standard protocol (Stevenson et al. 2012), with the reactions stopped by addition of 0.1 ml ice-cold methanol and incubation for 5 min with shaking to dissolve all available pyrethroids. After a centrifugation of the samples, 150 µl of the supernatant was transferred into HPLC vials; 100 µl sample was loaded into an isocratic mobile phase of 90% methanol and 10% water with a flow-rate of 1 ml/min, and substrate peaks were separated on a 250 mm C18 column (Acclaim 120, Dionex) at 23°. The quantity of pyrethroid remaining in the samples was determined by reverse-phase HPLC with a monitoring absorbance wavelength of 226 nm (Agilent 1260 Infinity). Percentage depletion was calculated by comparing the area of the chromatogram from incubation with NADPH regeneration system to the tubes in which NADP+ was not added (NADP−). HPLC conditions for analysis of the nonpyrethroid insecticides was as described in a previous study (Ibrahim et al. 2016b).
Turnover and kinetic assays:
To determine the turnover of CYP9J11 with pyrethroids and bendiocarb, experiments with deltamethrin, permethrin, and bendiocarb were performed in with incubation time varied from 0 to 30 min. For kinetic constants, incubation was carried out with 20 µM each of deltamethrin, permethrin, and bendiocarb for 30 min. The turnover and steady-state kinetic parameters (Km and Vmax) were calculated as previously described (Ibrahim et al. 2015) using the enzyme kinetic module of GraphPad Prism 6.03 (GraphPad Software Inc., La Jolla, CA). Catalytic constants and efficiencies were determined from the steady-state parameters.
Transgenic expression of candidate genes in Drosophila strains and tests with insecticides
To functionally validate the role of An. funestus CYP9J11 (an ortholog of CYP9J5 in An. gambiae) in conferring pyrethroid resistance (CYP9J11 is consistently overexpressed in all three countries), transgenic Drosophila melanogaster flies expressing this gene were generated using the GAL4/UAS system. This is to establish whether CYP9J11 overexpression alone could confer resistance to pyrethroids. The construction of the transgenic strain followed the protocol we used successfully for the P450s CYP6P9a and CYP6P9b (Riveron et al. 2013), and CYP6M7 (Riveron et al. 2014a).
Briefly, the same predominant clone used for transgenic expression was selected to construct transgenic flies, and cloned into the pUASattB vector using primers containing restriction sites for bglII and XbaI (see Table S1 in File S1). The PhiC31 system was used to generate the transgenic line UAS-CYP9J11 by Genetic Services (Cambridge, MA). Ubiquitous expression of the transgene CYP9J11 in adult F1 progeny (experimental group) was obtained after crossing virgin females from the driver strain Act5C-GAL4 [“y[1] w[*]; P(Act5C-GAL4-w)E1/CyO,”“1;2”] (Bloomington Stock Center, Bloomington, IN) with homozygote UAS-CYP9J11 males. Similarly, adult F1 control progeny (control group) with the same genetic background as the experimental group but without expression of CYP9J11 were obtained by crossing virgin females from the driver strain Act5C-GAL4 and UAS recipient line males (which do not carry the pUASattb-CYP9J11 insertion).
Insecticide contact bioassays for both experimental and control F1 Drosophila melanogaster females were performed as previously described (Riveron et al. 2014a) using posteclosion females that were 2–5 d old for contact assay with the pyrethroids deltamethrin (0.15%) and permethrin (2%)-impregnated filter papers prepared in acetone and Dow Corning 556 Silicone Fluid (BHD/Merck, Germany). A total of 20–25 flies was placed in individual vial containing respective insecticide papers, and the mortality plus knockdown was scored after 1, 2, 3, 6, 12 24, 36, and 48 hr of exposure to the insecticide. For all assays, at least six replicates were performed. Student’s t-test was used to compare the mortality plus knockdown of the experimental group against the control group.
Investigating the role of allelic variation at GSTe2 in the permethrin resistance:
Due to the overexpression of GSTe2 in mosquitoes resistant to permethrin in Benin, we used the transgenic expression in Drosophila to assess whether the allelic variation observed at this gene with the L119F mutation was playing a role in the observed resistance. A transgenic line was generated using the susceptible L119-GSTe2 allele following the same protocol described previously for the resistant allele 119F-GSTe2 as well as for the bioassays with permethrin and deltamethrin, but also DDT (Riveron et al. 2014b). Student’s t-test was used to compare the mortality plus knockdown of the L119-GSTe2 group against the control group and 119F-GSTe2 group.
Genetic diversity of candidate resistance genes between different An. funestus populations from different regions of Africa
Genetic variability of CYP9J11:
The full-length coding region of CYP9J11 was amplified from cDNA of permethrin-resistant samples from Malawi, Uganda and Zambia to assess the polymorphism of this gene. The Zambia mosquitoes were collected in Katete district (14° 11′ 0′′ S, 31° 52′ 0′′ E) in 2010 as previously described (Riveron et al. 2014a). The amplification was performed using the same cDNA synthesized for qRT-PCR with the Phusion polymerase (Thermo Scientific), which was cloned and sequenced as described above.
Comparative genetic diversity of CYP6P9a and CYP6P9b between East and southern Africa:
To assess whether previously detected directional selection associated with high overexpression of CYP6P9a and CYP6P9b genes in southern Africa was also present in East Africa, mosquitoes from Tororo in Uganda were compared to those from Chikwawa in Malawi (Riveron et al. 2013). Genomic fragment of both genes spanning the full-length coding region and a portion of the 5′UTR region were amplified and sequenced directly in 10 susceptible (dead after 1 hr exposure) from Tororo and 10 resistant mosquitoes (alive after 1 hr exposure) to 0.75% permethrin. The primers used are listed in Table S1 in File S1. Polymorphic positions were detected through manual analysis of sequence traces using BioEdit, and as sequence differences in multiple alignments using ClustalW (Thompson et al. 1994). DnaSP 5.1 (Rozas et al. 2003) was used to define the haplotype phase (through the Phase program), and to assess genetic parameters of each gene such as nucleotide diversity (π) and haplotype diversity. A maximum likelihood phylogenetic tree of the haplotypes for each gene was constructed using MEGA 5.2 (Tamura et al. 2007) to assess the potential correlation between haplotypes and resistance phenotypes.
Data availability
The microarray data from this study were submitted to Array Express, accession numbers E-MTAB-5375, E-MTAB-5376, and E-MTAB-5424. The DNA sequences reported in this paper have been deposited in the GenBank database (accession numbers: KJ150626–KJ150674).
Results
Transcription profiling of the pyrethroid resistant population of Uganda
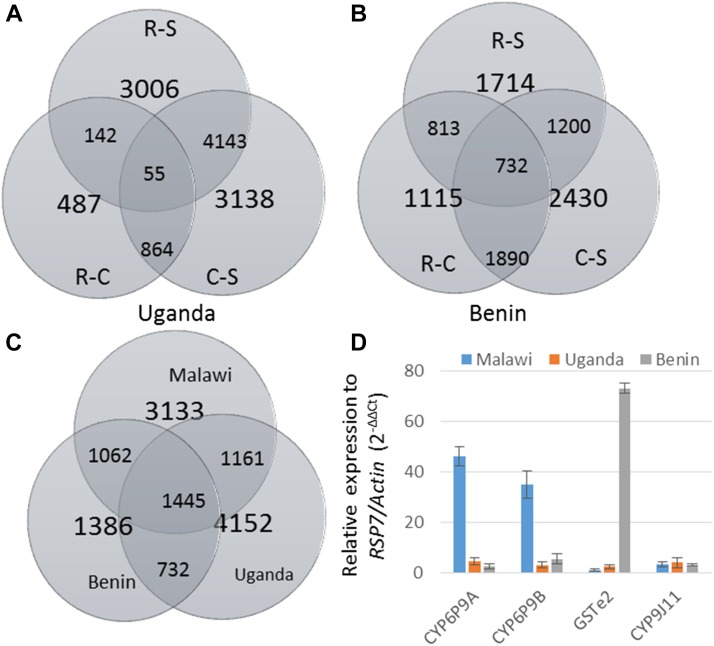
To detect the set of genes associated with permethrin resistance in Uganda, the 8 × 60k microarray chip was used to compare mosquitoes alive to permethrin exposure to control (nonexposed) (R-C) and to the fully susceptible laboratory strain FANG (R-S). The control mosquitoes were also compared to the susceptible FANG strain (C-S). High numbers of probes were significantly differentially expressed (P < 0.05) for the R-S (7346) and C-S (7479) comparisons (Figure 1A), most likely due to extensive genetic differences between the samples. In contrast, for the R-C comparison, a lower number (827) of differentially expressed probes was observed, as described previously in other similar studies (Riveron et al. 2013) due to the high level of resistance in the population. Consequently, only 55 probes were commonly differentially expressed in all three comparisons.
Figure 1.
Transcription profile of pyrethroid resistance across Benin, Uganda, and Malawi: (A, B) Venn diagram summarizing the number of probes differentially expressed in each and between comparisons in (A) Uganda and (B) in Benin at fold-change (FC) >2 and P < 0.05 in R-S, C-S and R-C comparisons, as well as the commonly expressed probes. (C) Venn-diagram of the comparison between Malawi, Uganda, and Benin for the R-S comparison only. (D) qRT-PCR expression of key resistance genes in the three countries when comparing the permethrin-resistant mosquitoes to FANG susceptible (R-S) mosquitoes.
R-S/C-S/R-C:
The cytochrome P450 CYP4C27 [Afun012777 using the ID system set by Crawford et al. (2010)] was the only detoxification gene commonly overexpressed in R-C, R-S and C-S, with highest fold change in R-S (FC10.3) (Table 1). Other genes had no annotation or no previous association with insecticide resistance such as an acyl-oxidase Afun004337 (AGAP011798-RA).
Table 1. List of top detoxification genes significantly overexpressed in pyrethroid resistant An. funestus in Uganda for all comparisons.
| Probe | Transcript | An. gambiae ID | R-C | R-S | C-S | Description |
|---|---|---|---|---|---|---|
| CUST_12777_PI426302897 | Afun012777 CYP4C27 | AGAP009246-PA | 3.6 | 10.3 | 6.8 | Cytochrome p450 |
| CUST_8293_PI426302897 | Afun008293 | AGAP008291-PA | 187.8 | 83.0 | Trypsin-related protease | |
| CUST_7663_PI426302897 | Afun007663 (CYP6M7) | AGAP008213-PA | 70.1 | 24.0 | Cytochrome p450 | |
| CUST_13921_PI426302897 | Afun013921 | AGAP006709-PA | 64.4 | 49.5 | Chymotrypsin 1 | |
| CUST_500_PI426302897 | Afun000500 | NA | 27.1 | 23.0 | Glycogenin | |
| CUST_9227_PI426302897 | Afun009227 | AGAP008141-PA | 22.8 | 21.2 | Argininosuccinate lyase | |
| CUST_7769_PI426302897 | Afun007769 (CYP9K1) | AGAP000818-PA | 21.5 | 16.1 | Cytochrome p450 | |
| CUST_15331_PI426302897 | Afun015331 (CYP307A1) | AGAP001039-PB | 16.6 | 3.9 | Cytochrome p450 | |
| CUST_11042_PI426302897 | Afun011042 | AGAP003321-PA | 13.3 | 8.1 | Glycine dehydrogenase | |
| CUST_13870_PI426302897 | Afun013870 | AGAP012697-PA | 11.9 | 14.8 | Sulfotransferase | |
| CUST_295_PI406199798 | AGAP000177-RA | AGAP000177-RA | 10.5 | 8.0 | Cuticle protein 7 | |
| CUST_4223_PI426302897 | Afun004223 (CYP4H17) | AGAP008358-PA | 10.1 | 7.5 | Cytochrome p450 | |
| CUST_15523_PI426302897 | Afun015523 | AGAP010581-PA | 8.1 | 5.7 | abc transporter | |
| CUST_4631_PI406199798 | AGAP005698-RA | AGAP005698-RA | 7.6 | 3.5 | Cuticular protein 4 | |
| CUST_1458_PI406199769 | combined_c738 | 6.7 | 4.9 | Short-chain dehydrogenase | ||
| CUST_13481_PI426302897 | Afun013481 (GSTe1) | AGAP009195-PA | 6.6 | 5.6 | Glutathione-s-transferase | |
| CUST_3246_PI426302897 | Afun003246 | AGAP006220-PA | 6.5 | 4.5 | Aldehyde oxidase | |
| CUST_8354_PI426302897 | Afun008354 (GSTD3) | AGAP004382-PA | 6.5 | 5.1 | Glutathione transferase | |
| CUST_12343_PI426302897 | Afun012343 (CYP4H17) | AGAP008358-PA | 6.0 | 4.4 | Cytochrome p450 4d1 | |
| CUST_11963_PI426302897 | Afun011963 | AGAP006220-PA | 5.7 | 4.0 | Aldehyde oxidase | |
| CUST_11037_PI426302897 | Afun011037 | AGAP003581-PA | 5.7 | 8.0 | Alcohol dehydrogenase | |
| CUST_376_PI406199788 | gb-CYP4H25 | 5.4 | 5.3 | Cytochrome p450 | ||
| CUST_12197_PI426302897 | Afun012197 (CYP304B1) | AGAP003066-PA | 5.2 | 3.3 | Cytochrome p450 | |
| CUST_7127_PI426302897 | Afun007127 (CYP4C36) | AGAP009241-PA | 5.2 | 2.6 | Cytochrome p450 | |
| CUST_6930_PI426302897 | Afun006930 (CYP6M2) | AGAP008212-PA | 5.1 | 5.3 | Cytochrome p450 | |
| CUST_7861_PI426302897 | Afun007861 (CYP6Z1) | AGAP008219-PA | 4.8 | 2.5 | Cytochrome p450 | |
| CUST_10949_PI426302897 | Afun010949 | AGAP010887-PA | 4.6 | 7.4 | Cuticular protein rr-1 family | |
| CUST_7696_PI406199798 | AGAP008141-RA | AGAP008141-RA | 4.6 | 2.2 | Argininosuccinate lyase | |
| CUST_3731_PI406199772 | CD577517.1 | 4.2 | 4.7 | Cuticle protein | ||
| CUST_7369_PI426302897 | Afun007369 (CYP6P9a) | AGAP002865-PA | 4.2 | 3.0 | Cytochrome p450 | |
| CUST_13871_PI426302897 | Afun013871 | AGAP012697-PA | 4.1 | 2.3 | Sulfotransferase | |
| CUST_13273_PI406199769 | combined_c6791 (CYP9J11) | AGAP012296-PA | 4.1 | 3.6 | Cytochrome p450 | |
| CUST_12461_PI426302897 | Afun012461 | AGAP000288-PA | 4.1 | 6.8 | Alcohol dehydrogenase | |
| CUST_7722_PI426302897 | Afun007722 | AGAP009850-PA | 4.0 | 3.6 | Abc transporter | |
| CUST_10360_PI426302897 | Afun010360 | AGAP006222-PA | 4.0 | 3.2 | UDP glucosyl transferases | |
| CUST_9866_PI426302897 | Afun009866 (GSTe5) | AGAP009192-PA | 3.9 | 2.7 | Glutathione-s-transferase | |
| CUST_9492_PI426302897 | Afun009492 | AGAP001722-PA | 3.8 | 8.8 | Carboxylesterase | |
| CUST_7469_PI426302897 | Afun007469 (CYP9J11) | AGAP012296-PA | 3.8 | 3.0 | Cytochrome p450 | |
| CUST_15244_PI426302897 | Afun015244 | AGAP000820-PA | 3.7 | 5.9 | Cuticular protein rr-1 family | |
| CUST_10836_PI426302897 | Afun010836 | AGAP006228-PA | 3.4 | 2.3 | Esterase b1 | |
| CUST_484_PI406199788 | gb-CYP9J3 | 3.3 | 2.1 | Cytochrome p450 | ||
| CUST_12666_PI426302897 | Afun012666 (CYP315A1) | AGAP002429-PA | 3.2 | 3.7 | Cytochrome p450 | |
| CUST_405_PI406199788 | gb-CYP6AD1 | 3.2 | 2.0 | Cytochrome p450 | ||
| CUST_9027_PI426302897 | Afun009027 | AGAP009463-PA | 3.1 | 2.1 | Abc transporter | |
| CUST_9335_PI426302897 | Afun009335 | AGAP003343-PA | 3.1 | 2.8 | Cytochrome p450 | |
| CUST_720_PI406199788 | gb-PX4B | 3.1 | 2.8 | Oxidase peroxidase | ||
| CUST_10630_PI426302897 | Afun010630 (CYP6P5) | AGAP002866-PA | 3.1 | 6.3 | Cytochrome p450 | |
| CUST_45_PI426302897 | Afun000045 (GSTe2) | AGAP009194-PA | 2.9 | 2.1 | Glutathione-s-transferase gst | |
| CUST_10994_PI426302897 | Afun010994 (CYP6P4) | AGAP002867-PA | 2.8 | 3.2 | Cytochrome p450 | |
| CUST_30_PI426302915 | CYP6Z3 | 2.8 | 2.4 | Cytochrome p450 | ||
| CUST_3315_PI406199769 | combined_c1675 | 2.7 | 2.6 | UDP glucosyl transferases | ||
| CUST_8909_PI426302897 | Afun008909 (CYP4K2) | AGAP002416-PA | 2.7 | 3.0 | Cytochrome p450 | |
| CUST_35_PI406199775 | COEAE6O | AGAP002863-PA | 2.6 | 3.1 | Carboxylesterase | |
| CUST_7499_PI426302897 | Afun007499 (GSTD1-5) | AGAP004164-PA | 2.5 | 2.1 | Glutathione transferase | |
| CUST_9584_PI426302897 | Afun009584 (CYP6M4) | AGAP008214-PA | 2.3 | 3.2 | Cytochrome p450 | |
| CUST_3394_PI426302897 | Afun003394 (CYP325A1) | AGAP000284-PA | 2.1 | 2.1 | Cytochrome p450 |
R-S/C-S only:
Among the most overexpressed genes commonly observed in R-S and C-S were proteases such as a trypsin-related protease (Afun008293), which was the top upregulated with FC187.8 in R-S and 83.07 in C-S. Other highly overexpressed proteases included chymotrypsin 1 (Afun013921), with FC64.4 in R-S and 49.5 in C-S. High overexpression of proteases is commonly reported in resistant mosquitoes either Anopheles (Kwiatkowska et al. 2013; Riveron et al. 2013), or Aedes albopictus (Ishak et al. 2016). Several detoxification genes were commonly upregulated in both comparisons, with a predominance of cytochrome P450s, notably CYP6M7 (Afun007663), which was the most overexpressed with FC70.1 in R-S and 24 in C-S. This P450 has previously been shown to metabolize pyrethroids (Riveron et al. 2014a). Other highly overexpressed P450 genes included CYP9K1 (Afun007769) with a higher fold change (FC21.5) in R-S than previously observed in southern Africa, suggesting a higher role played by this gene in Uganda. The CYP307A1 (Afun015331) exhibited a high FC in R-S (FC16.6). Other cytochrome P450s included CYP6 subfamily genes such as CYP6Z1, CYP6P5, CYP6P4, CYP6Z3, and, notably, the CYP6M8 (Afun006930), of which the ortholog from An. gambiae, CYP6M2 is responsible for pyrethroid resistance in this species (Stevenson et al. 2011; Mitchell et al. 2012) but was not previously associated with such resistance in An. funestus. A particular transcript (Afun07369) had a close hit to CYP6P9a, but none of the common probes for this gene highly overexpressed in southern Africa was observed in Uganda. CYP4 subfamily genes overexpressed included CYP4H17, CYP4C36, and CYP4K2, whereas CYP9J11, from the CYP9 subfamily, was also overexpressed in both comparisons. Glutathione S-transferases (GSTs) were also significantly overexpressed in pyrethroid resistant mosquitoes from Uganda compared to the susceptible FANG strain, notably genes of the epsilon class, including GSTe1 (Afun013481) (FC6.6 and 5.6, respectively, in R-S and C-S), GSTe5 (Afun009866) and GSTe2 (Afun000045), which, with FC of 2.9 and 2.1, exhibits a lower FC than the level observed in West Africa (Riveron et al. 2014b). GST genes from the Delta class were also overexpressed, including GSTD3 (Afun008354) (FC6.6 and 5.1) and GSTD1-5 (Afun007499). Other overexpressed detoxification gene families included sulfotransferases (with Afun013870 having a high FC of 11.9 and 14.8), carboxylesterases, aldehyde oxidases, ABC transporters, and other genes commonly associated with metabolic resistance to pyrethroid (Table 1). Cuticular protein genes were also among the overexpressed genes.
Transcription profiling of the pyrethroid resistant population of Benin
A similar approach was used in Benin using the 4 × 44k chip, as was carried out before the design of the 8 × 60k. High numbers of probes were significantly differentially expressed for the R-S (5617) and C-S (7735) comparisons (Figure 1B), most likely due to extensive genetic differences between the samples. Contrary to Uganda, the R-C comparison also showed a high number of probes differentially expressed (6033), leading to a higher number of probes commonly differentially expressed in all three comparisons (1890).
R-S/C-S/R-C:
The GST GSTe2 (Combined_c920) was the only detoxification gene commonly overexpressed in R-C, R-S, and C-S (Table 2). Three probes from this gene consistently had a higher overexpression in the R-S comparison from permethrin surviving mosquitoes vs. susceptible FANG than in the C-S comparison, supporting its association with permethrin resistance, in addition to its role as a main DDT metabolizer as previously established (Riveron et al. 2014b).
Table 2. List of top detoxification genes significantly overexpressed in pyrethroid resistant An. funestus in Benin for all comparisons.
| Probes | Transcript | C-S | R-C | R-S | Description |
|---|---|---|---|---|---|
| CUST_1822_PI406199769 | Combined_c920 | 11.9 | 2.6 | 35.5 | Glutathione-s-transferase gst |
| CUST_1822_PI406199769 | Combined_c920 | 8.8 | 2.0 | 25.2 | Glutathione-s-transferase gst |
| CUST_30_PI406199775 | Cyp6p9b | 3.9 | 2.9 | Cytochrome p450 | |
| CUST_25_PI406199775 | Cyp6p9a | 6.4 | 2.8 | Cytochrome p450 | |
| CUST_1616_PI406199772 | Ee589516.1 | 2.3 | 2.6 | D7-related 1 protein | |
| CUST_8241_PI406199769 | Combined_c4173 | 11.6 | 9.5 | Glycoprotein 93 | |
| CUST_1964_PI406199772 | Cd664227.1 | 2.4 | 2.0 | Alcohol dehydrogenase | |
| CUST_2550_PI406199769 | Combined_c1287 | 2.4 | 2.3 | Aldehyde dehydrogenase | |
| CUST_3110_PI406199772 | Cd577844.1 | 4.8 | 4.7 | Cuticle protein | |
| CUST_13273_PI406199769 | Combined_c6791 (CYP9J11) | 2.6 | 2.5 | Cytochrome p450 | |
| CUST_13272_PI406199769 | Combined_c6791 (CYP9J11) | 2.8 | 2.6 | Cytochrome p450 | |
| CUST_604_PI406199772 | Ee589610.1 | 3.2 | 2.8 | D7-related 1 protein | |
| CUST_1090_PI406199798 | Agap000881-ra | 2.1 | Aldehyde dehydrogenase | ||
| CUST_2068_PI406199798 | Agap002182-ra | 2.7 | ABC transporter | ||
| CUST_4410_PI406201128 | Agap001777-ra | 3.0 | ABC transporter | ||
| CUST_3109_PI406199772 | Cd577844.1 | 5.0 | Cuticle protein | ||
| CUST_4919_PI406199772 | Bu038983 | 4.7 | Cuticle protein | ||
| CUST_3398_PI406199772 | Cd577693.1 | 5.2 | Cuticle protein | ||
| CUST_48_PI406199775 | Cyp6z3 | 2.5 | Cytochrome p450 | ||
| CUST_10_PI406199775 | Cyp6p1 | 2.1 | Cytochrome p450 | ||
| CUST_43_PI406199775 | Cyp6z1 | 2.4 | Cytochrome p450 | ||
| CUST_45_PI406199775 | Cyp6z1 | 3.3 | Cytochrome p450 | ||
| CUST_27_PI406199775 | Cyp6p9a | 2.6 | Cytochrome p450 | ||
| CUST_44_PI406199775 | Cyp6z1 | 3.5 | Cytochrome p450 | ||
| CUST_717_PI406199772 | Ee589504.1 | 9.0 | D7-related 1 protein | ||
| CUST_359_PI406199772 | Ee589855.1 | 9.1 | D7-related 1 protein | ||
| CUST_1687_PI406199772 | Ee589439.1 | 8.2 | D7-related 1 protein | ||
| CUST_959_PI406199772 | Ee589285.1 | 3.6 | Gsg6 salivary protein | ||
| CUST_379_PI406199772 | Ee589823.1 | 2.2 | Gsg7 salivary protein | ||
| CUST_5934_PI406199769 | Combined_c3002 | 2.4 | Superoxide dismutase | ||
| CUST_21644_PI406201128 | Agap006867-ra | 5.2 | Adult-specific cuticular protein acp-20 | ||
| CUST_120_PI406199788 | Gb-COEAE1G | 5.1 | Alpha-esterase | ||
| CUST_21714_PI406201128 | Agap010906-ra | 3.6 | Cuticle protein | ||
| CUST_2401_PI406199769 | Combined_c1211 | 2.1 | Glucosyl glucuronosyl transferases | ||
| CUST_178_PI406199772 | Ee590018.1 | 2.2 | Gsg7 salivary protein | ||
| CUST_6_PI406199775 | Cyp6aa1 | 2.0 | Cytochrome p450 | ||
| CUST_11_PI406199775 | Cyp6p1 | 2.5 | Cytochrome p450 | ||
| CUST_24_PI406199775 | Cyp6p5 | 2.1 | Cytochrome p450 | ||
| CUST_26_PI406199775 | Cyp6p9a | 2.2 | Cytochrome p450 | ||
| CUST_1682_PI406199772 | Ee589442.1 | 2.5 | D7 protein | ||
| CUST_1182_PI406199772 | Ee589982.1 | 2.5 | D7-related 1 protein | ||
| CUST_892_PI406199772 | Ee589340.1 | 2.8 | D7-related 3 protein | ||
| CUST_3946_PI406199772 | Cd577403.1 | 3.2 | Glutathione s-transferase | ||
| CUST_14377_PI406199769 | Combined_c7513 | 2.6 | Glutathione transferase |
Common probes between two comparisons:
Among probes significantly overexpressed in at least two comparisons, the cytochrome P450 genes CYP6P9a and CYP6P9b were upregulated in both C-S and R-S, but with relatively low levels compared to previously reported levels in southern Africa (<6.4 FC). Two probes of the CYP9J11 were also upregulated, but between R-C and R-S only (Table 2). Other detoxification genes were upregulated, but only in one comparison. Those found in R-S included only the cytochrome P450s CYP6Z1 (three probes), CYP6Z3, CYP6P1, and another probe for CYP6P9a. It also included two ABC transporter genes (probes from An. gambiae transcripts AGAP002182 and AGAP001777, respectively), an aldehyde dehydrogenase, and cuticular protein genes (Table 2). Genes only present in the C-S comparison included an alpha-esterase (COEAE1G; FC5.1) and an UDP glycosyl transferase. Other detoxification genes, including the cytochrome P450s CYP6AA1, CYP6P5, and two GSTs, were upregulated only in the R-C comparison (Table 2).
GO enrichment analysis
Blast2go enrichment analysis for the set of probes upregulated in R-S and C-S comparisons did not detect many GO terms related to detoxification process in mosquitoes. In the case of the C-S comparison in Benin, for example, the major GO terms over-represented belong mainly to serine-type endopeptidase activity, odorant binding activity, protein DNA complex, and others (Figure S1). Similar results were obtained for other comparisons. The lack of GO terms associated with detoxification is similar to previous studies with this microarray chip in An. funestus (Riveron et al. 2013, 2014a). This is probably caused by the poor annotation of the set of Expressed Sequences Tags (ESTs) used for the microarray chip, and the composite nature of the microarray chip made of transcripts from different sources.
Regional comparison of expression profiles between West (Benin), East (Uganda) and southern (Malawi) Africa
The variation in the underlying resistance mechanisms to pyrethroid between geographical regions in Africa was analyzed by comparing the expression profiles from Benin and Uganda to that from Malawi in southern Africa using the 8 × 60k chip. The number of significantly differentially expressed probes is presented in Figure 1C.
Genes common in all regions:
Among the genes the most upregulated in all three regions were a trypsin-related protease gene (Afun008293), the P450 CYP6M7, the argininosuccinate lyase, and a glycogenin gene. However, although overexpressed in all regions, the expression levels vary significantly for some genes, such as for CYP6M7, which has a FC of 131.9 in Benin but only 12.5 in Malawi (Table 3). Among the detoxification genes commonly overexpressed in all three regions, cytochrome P450s were again dominant. Most of these P450 genes showed a similar level of expression in all the three countries, and included CYP4H17, CYP6Z1, CYP6M4, CYP6M2, CYP9J11, CYP9J13, CYP304B1, and a gene close to CYP6P9a (Afun007369). Another P450, CYP9K1, although commonly overexpressed in all three countries, was significantly more highly present in Uganda (FC16.1) than in Malawi (FC2.4) and Benin (FC6.2), suggesting a bigger role for this gene in Uganda. Other commonly expressed detoxification genes included an aldehyde oxidase (AGAP006220) and a UDP glucuronosyl transferase (AGAP006222).
Table 3. Detoxification genes commonly upregulated in Uganda (UG), Malawi (MAL), and Benin (BN) countries.
| Probe Name | An. funestus ID | An. gambiae ID | UG | MAL | BN | Description |
|---|---|---|---|---|---|---|
| CUST_8293_PI426302897 | Afun008293 | AGAP008291-PA | 83.0 | 32.5 | 74.4 | Trypsin-related protease |
| CUST_7663_PI426302897 | Afun007663 (CYP6M7) | AGAP008213-PA | 24.0 | 12.5 | 131.9 | Cytochrome p450 |
| CUST_500_PI426302897 | Afun000500 | 23.0 | 8.8 | 17.1 | Glycogenin | |
| CUST_9227_PI426302897 | Afun009227 | AGAP008141-PA | 21.2 | 66.3 | 29.2 | Argininosuccinate lyase |
| CUST_8887_PI426302897 | Afun008887 | AGAP011997-PA | 17.6 | 8.5 | 7.3 | Nucleotide binding protein 2 (nbp 2) |
| CUST_7769_PI426302897 | Afun007769 (CYP9K1) | AGAP000818-PA | 16.1 | 2.4 | 6.2 | Cytochrome p450 |
| CUST_1392_PI426302897 | Afun001392 | 10.7 | 8.8 | 6.0 | Glycine dehydrogenase | |
| CUST_11042_PI426302897 | Afun011042 | AGAP003321-PA | 8.1 | 18.5 | 6.4 | Glycine dehydrogenase |
| CUST_4223_PI426302897 | Afun004223 (CYP4H17) | AGAP008358-PA | 7.5 | 6.8 | 9.5 | Cytochrome p450 |
| CUST_10949_PI426302897 | Afun010949 | AGAP010887-PA | 7.4 | 3.0 | 4.2 | Cuticular protein rr-1 family |
| CUST_1459_PI406199769 | combined_c738 | 7.3 | 6.4 | 10.9 | Short-chain dehydrogenase | |
| CUST_6930_PI426302897 | Afun006930 (CYP6M2) | AGAP008212-PA | 5.3 | 4.3 | 5.0 | Cytochrome p450 |
| CUST_3246_PI426302897 | Afun003246 | AGAP006220-PA | 4.5 | 3.8 | 4.2 | Aldehyde oxidase |
| CUST_1563_PI406199772 | EE589574.1 | 4.4 | 2.1 | 2.8 | D7-related 1 protein | |
| CUST_12343_PI426302897 | Afun012343 (CYP4H17) | AGAP008358-PA | 4.4 | 2.9 | 4.2 | Cytochrome p450 4d1 |
| CUST_8347_PI426302897 | Afun008347 | AGAP009828-PA | 3.9 | 4.2 | 5.0 | Chymotrypsin 1 |
| CUST_9522_PI426302897 | Afun009522 (CYP9J13) | AGAP012292-PA | 3.5 | 4.5 | 2.8 | Cytochrome p450 |
| CUST_1710_PI406199772 | EE589412.1 | 3.3 | 2.2 | 2.6 | D7-related 1 protein | |
| CUST_12197_PI426302897 | Afun012197 (CYP304B1) | AGAP003066-PA | 3.3 | 2.8 | 4.1 | Cytochrome p450 |
| CUST_10360_PI426302897 | Afun010360 | AGAP006222-PA | 3.2 | 2.0 | 4.8 | Glucosyl glucuronosyl transferases |
| CUST_9584_PI426302897 | Afun009584 (CYP6M4) | AGAP008214-PA | 3.2 | 3.2 | 3.3 | Cytochrome p450 |
| CUST_27_PI426302915 | CYP6Z1 | 3.1 | 2.5 | 2.3 | Cytochrome p450 | |
| CUST_198_PI406199772 | EE590001.1 | 3.0 | 2.1 | 3.1 | D7-related 1 protein | |
| CUST_7369_PI426302897 | Afun007369 (CYP6P9a-like) | AGAP002865-PA | 3.0 | 2.5 | 4.4 | Cytochrome p450 |
| CUST_7469_PI426302897 | Afun007469 (CYP9J11) | AGAP012296-PA | 3.0 | 3.1 | 2.7 | Cytochrome p450 |
| CUST_3109_PI406199772 | CD577844.1 | 2.9 | 2.5 | 2.4 | Cuticle protein | |
| CUST_9335_PI426302897 | Afun009335 | AGAP003343-PA | 2.8 | 2.7 | 2.7 | Cytochrome p450 |
| CUST_2473_PI426302897 | Afun002473 | AGAP000553-PA | 2.5 | 4.5 | 2.5 | ATP-binding-cassette protein |
| CUST_7861_PI426302897 | Afun007861 | AGAP008219-PA | 2.5 | 3.1 | 2.2 | Cytochrome p450 |
| CUST_1097_PI406199769 | Combined_c557 | 2.5 | 6.4 | 5.1 | Trypsin | |
| CUST_10_PI426302915 | CYP6M4.seq | 2.4 | 2.6 | 3.2 | Cytochrome p450 | |
| CUST_798_PI426302897 | Afun000798 (CYP6M2) | AGAP008212-PA | 2.1 | 2.5 | 2.6 | Cytochrome p450 |
Genes common in only two regions:
Analysis of the list of genes commonly overexpressed in only two regions revealed that, for Uganda and Benin, the GSTs GSTe1 and GSTd3 were common to both countries, as were the P450s CYP307A, CYP314A1, and CYP315A. For those overexpressed only in Uganda and Malawi, the P450 CYP4C27 was detected, although with a higher expression in Uganda (FC10.3) than in Malawi (FC2.1). The CYP4C36 was also upregulated in both countries, similar to GSTd1-5. Other genes are also listed in Table S2 in File S1. The list of genes overexpressed only in Malawi and Benin is dominated by the CYP6P9a and CYP6P9b with several probes, but with a far higher overexpression in Malawi for both genes (e.g., FC39.4 for CYP6P9a in Malawi vs. only FC4.3 in Benin), suggesting that both genes are mainly driving resistance in southern Africa.
Some genes common to all countries were detected through different probes, such as GSTe2, which in Benin and Malawi was detected by probes against Combined_c920 transcript, whereas in Uganda and Malawi it was through probes for Afun000045 transcript, showing the impact of sequence polymorphism in the microarray results.
Quantitative RT-PCR:
Key genes exhibiting striking differences between regions (CYP6P9a, CYP6P9b, and GSTe2) or commonly overexpressed in all three countries (CYP9J11) were further validated by qRT-PCR. Analysis of the expression patterns confirmed the differences observed with microarrays, as both CYP6P9a and CYP6P9b were highly overexpressed only in Malawi, but just barely in Uganda and Benin, as seen with microarray (Figure 1D). Similarly, the GSTe2 was highly overexpressed in Benin permethrin resistant individuals (FC 73.1), but only very low-level expression of this gene was observed in Uganda and Malawi. The common overexpression of the CYP9J11 was also validated in all three countries, although at a lower fold-change compared to the other genes.
Functional characterization of key genes commonly overexpressed in all countries
Several genes (notably P450s) commonly overexpressed in the three geographical regions are located in the chromosomal regions spanning the three QTL (rp1, rp2, and rp3) previously detected for pyrethroid resistance in An. funestus (Wondji et al. 2009). Although the key genes driving resistance in rp1 and rp2 have already been characterized (Irving et al. 2012; Riveron et al. 2013), the genes driving resistance in rp3 remain uncharacterized. The CYP9J11 overexpressed in all three regions, and located in the 3L chromosomal region spanning the rp3 QTL, could be the pyrethroid metabolizer gene in this QTL. To validate this hypothesis, we performed a functional characterization of this gene.
Polymorphism analysis of CYP9J11:
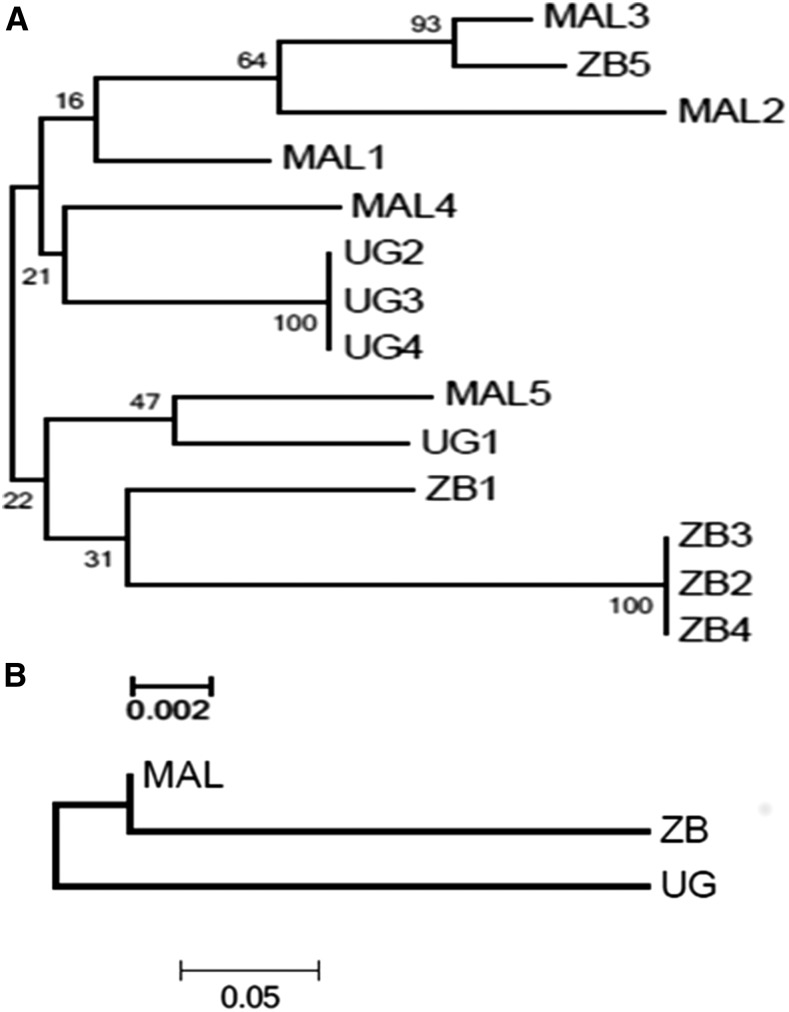
Analysis of the genetic variability of CYP9J11 full-length cDNA (1644 bp) for five clones each from Malawi and Zambia, and four from Uganda revealed a high polymorphism of this gene, with an average of 93 polymorphic sites observed for all combined 14 sequences and 17 amino acid changes observed in total (Table S3 in File S1). No evidence of directional selection was detected on CYP9J11, as shown by the lack of significant Tajima D and Fu and Li D’ estimates. No specific clades per the location of origin was observed between haplotypes, although the genetic distance tree revealed a closer genetic similarity between Malawi and Zambia than Uganda, as expected from geographical distance (Figure 2, A and B).
Figure 2.
Genetic diversity pattern of CYP9J11 in East (Uganda; UG) and southern [Malawi (MAL) and Zambia (ZB)] Africa. (A) Maximum likelihood tree of CYP9J11 from the cDNA haplotypes of the full-length 1644 bp sequence. (B) Genetic distances between African populations (Nst estimates) between the three countries.
Functional validation of CYP9J11 using heterologous expression in E. coli and metabolism assays
Pattern of expression of CYP9J11:
On average, CYP9J11 protein consistently expressed at low concentration (0.13 ± 0.007 nmol/mg protein) compared with previous estimates reported for CYP6M7 (0.15 ± 0.0 nmol/mg protein), and for CYP6P9a (0.42–1.0 nmol/mg) and CYP6P9b (0.35–0.42 nmol/mg), respectively.
Assessment of CYP9J11 pyrethroid activities and cross-resistance using metabolism assays:
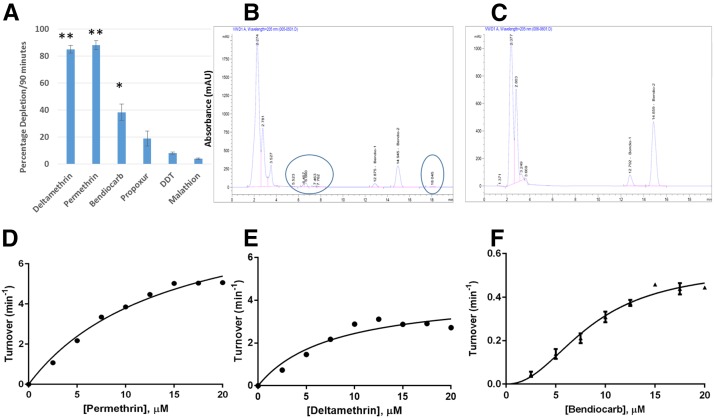
Disappearance of 20 µM insecticide substrates was determined after 90 min of incubation with the recombinant CYP9J11 in the presence of cytochrome b5 and NADPH regeneration system. CYP9J11 metabolized permethrin and deltamethrin with significant depletions of 88.05% ± 3.23 (P < 0.0001) and 95.05% + 0.74 (P < 0.0001), respectively (Figure 3A). These depletions were higher than obtained with both CYP6P9a and CYP6P9b alleles (Riveron et al. 2013, 2014a). Carbamates bendiocarb and propoxur, as well as the organophosphate malathion, were screened to investigate potential cross resistance by CYP9J11. Low and nonsignificant depletion was observed against DDT and malathion (Figure 3A), as observed previously from CYP6P9a, CYP6P9b, and CYP6M7. This result is consistent with malathion susceptibility across Africa so far. CYP9J11 significantly depleted bendiocarb; but with a lower depletion of 38.34% ± 7.01 (P < 0.05) than previously reported for CYP6Z1 (54.72% ± 0.45, P < 0.05) (Ibrahim et al. 2016a). In contrast to incubations with CYP6M7, CYP6P9a, and CYP6P9b (<10% depletions), CYPJ11-mediated metabolism of bendiocarb proceeded with polar metabolites eluting in the beginning of the HPLC chromatogram (Figure 3B). Initial reaction of carbamate metabolism has been described to produce very polar products that remain at the origin of the chromatogram (Kuhr 1970) and such highly polar metabolites have been recently described in metabolisms assay with bendiocarb and An. funestus CYP6Z1 protein (Ibrahim et al. 2016a).
Figure 3.
Functional validation of the role of CYP9J11 P450 gene in carbamate/pyrethroid resistance. (A) Percentage depletion of 20 µM carbamate and pyrethroid insecticides with CYP9J11. Results are an average of three replicates (n = 3) compared with negative control. * and ** Significantly different from negative control (−NADPH) at P < 0.05 and P < 0.01, respectively. (B) Polar metabolites with short retention time eluted at the beginning of chromatogram of CYP9J11 metabolism of bendiocarb (NADPH+). A third putative metabolite of bendiocarb metabolism eluted at 18.045 min. (C) Chromatogram of NADPH- incubation tubes devoid of polar metabolites with short retention indicating no metabolism of bendiocarb in the absence of NADPH regeneration agent. (D, E) Michaelis-Menten plot of CYP9J11 mediated metabolism of permethrin and deltamethrin, respectively. Results are an average of three replicates (n = 3) compared with negative control; (F) Allosteric sigmoidal curve of CYP9J11 metabolism of bendiocarb. Results are average of three replicates (n = 3) compared with negative control. Khalf = KM; h = 2.29.
Kinetics parameters of CYP9J11 metabolism of insecticides:
The CYP9J11-mediated metabolism of permethrin and deltamethrin follows a Michaelis-Menten pattern (Figure 3, A and B), but a decline in activity was observed with deltamethrin above 12.5 µM concentration, attributed to substrate or product inhibition. The turnover (Kcat) and KM obtained with permethrin was 9.260 min−1 ± 1.048 and 14.39 µM ± 3.12 leading to a catalytic efficiency of 0.643 min−1 µM−1 ± 0.157. The turnover for deltamethrin (4.338 min−1 ± 1.381) was, on average, half the value obtained with permethrin, but the affinity of CYP9J11 toward deltamethrin was surprising higher (KM of 7.957 ± 1.31). The catalytic efficiency of CYP9J11 for deltamethrin was calculated as 0.545 min−1 µM−1 ± 0.195, lower than obtained with permethrin. The catalytic efficiency of this enzyme toward permethrin is higher than obtained from An. funestus pyrethroid metabolizers CYP6P9a and CYP6P9b (Riveron et al. 2014a).
CYP9J11 was also tested with 20 µM bendiocarb and was shown to behave in allosteric fashion with this carbamate insecticide, with positive cooperativity (Hill coefficient, h = 2.29 ± 0.38) as described to be the case of some P450s (Atkins 2004). CYP9J11 portrayed sigmoidal curve with relatively low Khalf (lower than KM obtained with pyrethroids) and low maximal catalytic rate (Figure 3C). The dose-response curve was thus modeled using the GraphPad prism with relevant module as described (Copeland 2004). The Vmax and Khalf (KM) for bendiocarb were calculated as 0.04 min−1 ± 0.005 and 0.75 µM ± 0.2, respectively, leading to a very low catalytic efficiency of 0.053 min−1 µM−1 ± 0.0157, 12 times lower than compared with the values obtained with permethrin.
Transgenic expression of candidate genes in Drosophila flies
Validation of role of CYP9J11:
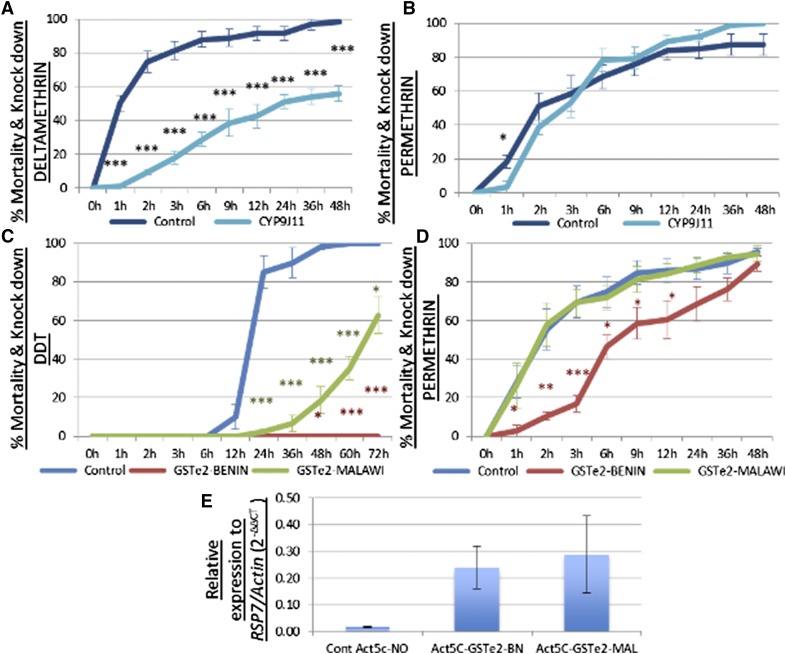
To confirm that CYP9J11 overtranscription alone can confer pyrethroid resistance, transgenic D. melanogaster individuals were generated expressing CYP9J11 (derived from permethrin resistant field mosquitoes from Uganda) under the control of the ubiquitous Act5C-GAL4 driver. Contact bioassays performed with 2% permethrin (type I pyrethroid) and 0.15% deltamethrin (type II) revealed that CYP9J11 overtranscription alone is sufficient to confer resistance to this class of insecticide. For deltamethrin, the flies overexpressing CYP9J11 were resistant with a significantly reduced mortality/knockdown rate compared to that observed for control flies (Figure 4A). Significantly reduced mortality/knockdown rates were recorded at all nine different exposure times for transgenic Act5C-CYP9J11 individuals when compared with the control group not expressing CYP9J11. For example, mortality rates were 1.04 ± 1% vs. 50.3 ± 4.4% (P < 0.001) at 1 hr, 9.5 ± 1.7% vs. 74.7 ± 6.32% (P < 0.001) at 2 hr and 56.03 ± 4.6% vs. 98.3 ± 3.3% (P < 0.001) at 24 hr (Figure 4A). These results demonstrate that CYP9J11 overtranscription alone is sufficient to confer resistance to deltamethrin. For permethrin, significantly reduced mortality/knockdown rate was recorded for transgenic Act5C-CYP9J11 flies when compared with the control after 1 hr exposure (3.33 ± 3.3% vs. 18.36 ± 3.8%; P < 0.05). However, similar mortality rates were recorded for both experimental and control samples at the rest of the exposure times, with no significant differences observed (Figure 4B).
Figure 4.
Functional validation of the role candidate resistance genes using transgenic expression in flies: (A) results of bioassay analysis of transgenic flies overexpressing CYP9J11 Act5C-CYP9J11 vs. control flies for deltamethrin. (B) The same bioassays with permethrin. (C) Functional validation of the role of allelic variation at the GSTe2 genes on the resistance phenotype using transgenic expression in flies through a comparative transgenic analysis of the 119F and L119-GSTe2 alleles using bioassay tests on transgenic Act5C-GSTe2-119F (GSTe2-Benin) and Act5C-GSTe2-L119 (GSTe2-Malawi) and flies (Exp-GSTe2), control strains [two parental (UAS-GSTe2 and GAL4-Actin) and F1 progeny that do not express the GSTe2 transgene (Cont-NO)]. (D) The same bioassays with permethrin. (E) Relative expression of the transgene GSTe2 alleles in the transgenic D. melanogaster strain (Act5C-GSTe2-MAL and Act5C-GSTe2-BN) and the control sample with no GSTe2 expression (Cont Act5c-NO). Data shown as mean ± SEM significantly different: *P < 0.05, **P < 0.01, and ***P < 0.001.
Confirmation of role of allelic variation of GSTe2 in both DDT and pyrethroid resistance:
Due to the consistent overexpression of GSTe2 in permethrin-resistant mosquitoes in Benin where the L119F mutation is fixed, the role of the allelic variation on this gene was investigated using transgenic expression. Comparative bioassays performed between a transgenic line expressing the susceptible L119 allele, and another one expressing the 119F resistant allele, revealed that the 119F mutation confers a higher resistance against both DDT and permethrin. For DDT, no mortality is observed in the flies expressing the resistant 119F allele for all the different exposure times, whereas significantly higher mortality rates were observed for the flies expressing the susceptible L119 allele from 24 to 72 hr exposure time (2.2–63%; P < 0.001) (Figure 4C). However, the fact that these mortality rates for flies expressing the susceptible L119 allele were lower than for flies not expressing the GSTe2 (2.22 ± 1.4, 18.65 ± 7.1, and 62.69 ± 9.6% vs. 85.12 ± 8.4, 98.33 ± 1.7 and 100%; P < 0.001; respectively at 24, 48, and 72 hr) suggests that even overexpression of the susceptible allele provide resistance against DDT in flies, but at a significantly lower level than with the 119F resistance allele. Bioassays with permethrin revealed that only flies expressing the resistant 119F allele had significantly lower mortality rate compared to control flies not expressing GSTe2 (2.78 ± 2.7, 10.52 ± 2.1, 16.74 ± 4.3, and 46.40 ± 6.07% vs. 27.98 ± 8.3, 55.44 ± 10.4, 69.60 ± 8.4, and 74.81 ± 7.8%; P < 0.01; respectively at 1, 2, 3, and 6 hr exposure time) (Figure 4D). Flies expressing the susceptible L119 allele showed the same high mortality rates as the control flies. Each GSTe2 allele was confirmed to be expressed only in the F1 progeny of the GAL4/UAS crosses by qRT-PCR (Figure 4E).
CYP6P9a and CYP6P9b polymorphisms in Uganda in comparison to southern Africa
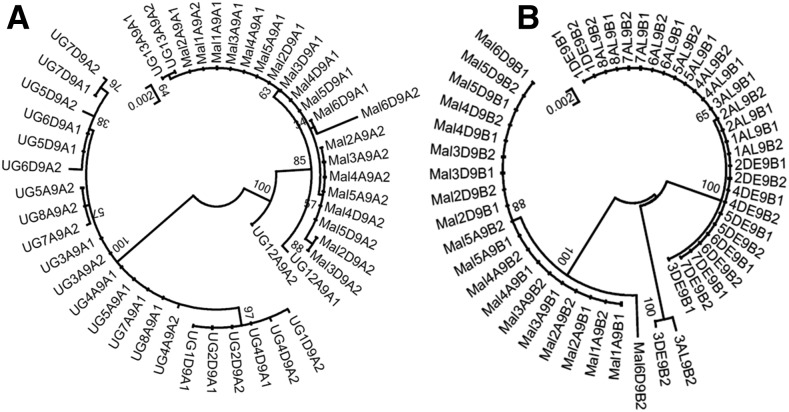
A comparative analysis of the polymorphism pattern of the duplicated P450 genes CYP6P9a and CYP6P9b was performed between permethrin-resistant and -susceptible mosquitoes from Uganda and those from Malawi. The aim was to assess whether the low expression of these genes in Uganda correlated with a higher genetic diversity of both genes in contrast to southern African, where a high overexpression was associated with a directional selection with reduced genetic diversity (Riveron et al. 2013, 2014a). Overall, both CYP6P9a and CYP6P9b genes exhibit a higher genetic diversity in Uganda than in Malawi as shown by the number of substitutions (50 vs. 13 for CYP6P9a; 45 vs. 12 for CYP6P9b), haplotypes (15 vs. 5 for CYP6P9a; 4 vs. 2 for CYP6P9b), and estimates of genetic diversity or number of nonsynonymous substitutions (Table S3 in File S1). This higher genetic diversity of both genes in Uganda correlates with their low overexpression and support a lower role of both genes in Uganda. However, the maximum likelihood trees of haplotypes of both genes for Uganda and Malawi (Figure 5, A and B) revealed that for CYP6P9a, four haplotypes from resistant mosquitoes clustered with haplotypes from Malawi. These Uganda haplotypes also exhibit the insertion of two AAs [CAAAAAA(AA)] in the promoter region characteristic of southern African resistant haplotypes (Ibrahim et al. 2015). For both genes, haplotypes of both countries cluster in separate clades (Figure 5, A and B).
Figure 5.
Molecular phylogenetic analysis of CYP6P9a (A) and CYP6P9b (B) in Uganda (UG) for both permethrin resistant and susceptible mosquitoes in comparison to Malawi (Mal) using the Maximum Likelihood method. The evolutionary history was inferred by using the Maximum Likelihood method based on the Tamura 3-parameter model. The tree with the highest log likelihood is shown. Initial tree(s) for the heuristic search were obtained by applying the Neighbor-Joining method to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 46 (CYP6P9a) and 50 (CYP6P9b) nucleotide sequences. All positions containing gaps and missing data were eliminated. There were a total of 1990 (CYP6P9a) and1757 (CYP6P9b) positions in the final dataset. Evolutionary analyses were conducted in MEGA6.
Discussion
Elucidation of resistance mechanisms to insecticide in mosquito vectors of tropical diseases such as malaria is a prerequisite for a better management of the growing problem of resistance to existing insecticide classes in public health sectors. If progress has been made in assessing the local transcription profiles associated with pyrethroid resistance in malaria vectors in Africa, generating a broader view of the molecular basis of resistance continent-wide has been limited. The regional comparison of the transcription profile of pyrethroid resistance in An. funestus across Africa revealed three main lessons discussed below.
The transcription profile of pyrethroid resistance is not uniform across the continent
The genome-wide analysis of the transcription profile associated with pyrethroid resistance highlighted a common trait, i.e., the predominant role of cytochrome P450 genes in the metabolic resistance observed in An. funestus population as previously reported in southern Africa (Riveron et al. 2013, 2014a) and in other mosquito species such as An. gambiae (Mitchell et al. 2012; Kwiatkowska et al. 2013) or in Aedes (Strode et al. 2008; Bariami et al. 2012; Saavedra-Rodriguez et al. 2012; Ishak et al. 2016). However, the drastic difference in the expression levels of key P450s suggests that the origin of resistance is not the same across the continent, and that there were independent selection events of resistance to pyrethroids in various populations. A clear example is that provided by the expression profile of the duplicated P450s CYP6P9a and CYP6P9b, the main pyrethroid resistance genes in southern African populations of An. funestus (Amenya et al. 2008; Riveron et al. 2013, 2014a), but which, from this study, seem to play no or little role in East Africa, as further supported by their higher genetic diversity in Uganda than in Malawi but also than in West (Benin and Ghana) and Central Africa (Cameroon) (Barnes et al. 2017). Such drastic variation is also in line with the gradual reduced expression of CYP6P9a and CYP6P9b in Zambia compared to Malawi and Mozambique (Riveron et al. 2014a; Thomsen et al. 2014; Barnes et al. 2016), and suggests barriers to gene flow previously detected between African populations of this mosquito species (Michel et al. 2005; Barnes et al. 2017). Variation in the transcription profiles of insecticide resistance genes are also reported in other mosquito species such as An. gambiae, where P450 genes such as CYP6P3 and CYP6M2 highly overexpressed in West (Mitchell et al. 2012; Kwiatkowska et al. 2013) and in Central (Fossog Tene et al. 2013; Antonio-Nkondjio et al. 2016) Africa are not significantly expressed in the southern populations in Zambia (Thomsen et al. 2014). Equally, in contrast, the P450 CYP6P4 from An. arabiensis has also been shown to be a major driver of pyrethroid resistance in populations from Chad (Ibrahim et al. 2016b) and Sudan (Abdalla et al. 2014). It is therefore important to avoid generalizing the underlying molecular basis of resistance across countries or the continent, but rather to determine as much as possible the main resistance genes in the different countries/regions, efforts that can impact the design of diagnostic tools or resistance management strategies. For example, the CYP6M2 in An. gambiae (Edi et al. 2014) and the CYP6Z1 (Ibrahim et al. 2016a) in An. funestus have been shown to confer cross-resistance between pyrethroids and carbamates, so their significant overexpression in a population should prevent using carbamates as alternative to pyrethroids in an IRS campaign.
The cytochrome P450 CYP9J11 is a common African pyrethroid resistance gene
If significant differences are observed between regions, there are also similarities with common genes observed across the continent such as the P450 CYP9J11 which was overexpressed in all three regions assessed here. However, because of its moderate level of expression, CYP9J11 may not be the primary resistance gene. Nevertheless, its significant catalytic efficiency in metabolising pyrethroid means it cannot be disregarded. CYP9J11 is the ortholog of CYP9J5 in An. gambiae, which was recently shown to also metabolize pyrethroids and pyriproxyfen, and to be overexpressed Africa-wide in An. gambiae field populations from West (Toe et al. 2015), Central (Fossog Tene et al. 2013), and East Africa (Nkya et al. 2014), suggesting that this gene could be important in providing protection to a wide range of xenobiotics in malaria vectors. CYP9J11 is also located on the 3L chromosome where the rp3 (resistance to pyrethroid 3) QTL had previously been detected, suggesting that it could be the main gene behind rp3 (Wondji et al. 2005, 2007, 2009). In addition to the ability to metabolize pyrethroids and confer resistance to An. funestus, CYP9J11 as previously shown for CYP6Z1 (Ibrahim et al. 2016a) is a cross-resistance gene, able to metabolize nonpyrethroid insecticides used in public health using noncanonical Michaelis-Menten kinetic mechanisms. Various P450s exhibit functional allostery using distributive catalysis to minimize toxicological effects of substrates (Atkins et al. 2002), for example, the promiscuous CYP3A4 (Wang et al. 2000), CYP2C9 (Tracy et al. 2002), and the recently characterized An. funestus CYP6Z1 (Ibrahim et al. 2016a). At low substrate concentrations, the slower substrate turnover afforded by cooperative CYPs compared with Michaelis-Menten enzymes can be a significant toxicological advantage, when toxic thresholds exist (Atkins et al. 2002). Possibly, bendiocarb is too “toxic” for CYP9J11, even though it can metabolize it, and this is why P450 employs distributive catalysis to effect its catalysis, as in the case of An. funestus CYP6Z1 (Ibrahim et al. 2016a).
3-Allelic variation of GST GSTe2 impacts pyrethroid resistance
The significant overexpression of GSTe2 in Benin in pyrethroid resistant mosquitoes (as seen by the FC > 2 in R-C comparing permethrin resistant to control, not-exposed, mosquitoes from Pahou) suggested that this gene could be involved in permethrin resistance. The significant lower mortality observed in transgenic Drosophila flies expressing the resistant 119F allele compared to those expressing the susceptible L119 allele supports the key role that allelic variation in this gene plays beside its overtranscription. As previously shown for DDT resistance, it is likely that the 119F also enlarging the GSTe2 binding cavity to facilitate access of pyrethroid, and allow either sequestration as suggested for GST action on pyrethroids (Vontas et al. 2005), or a direct metabolism as established by Riveron et al. (2014a). The ability of the transgenic expression in Drosophila flies to establish the phenotypic impact of the allelic variation of GSTe2 due to a single amino acid change highlights the robustness of this approach in functionally characterizing the role of candidate resistance genes in conferring resistance to insecticide. This shows that experimental results from transgenic Drosophila are very relevant to the phenotype obtained in mosquitoes, while providing the advantage that studies in Drosophila could be easily scaled up to hundreds of genes, with less work, cost and space required for storage of transgenic lines. Allelic variation was also recently shown to play an important role in the pyrethroid resistance conferred by the duplicated P450s CYP6P9a and CYP6P9b in southern African populations of An. funestus (Ibrahim et al. 2015), suggesting that, besides overtranscription of detoxification genes, amino acid changes in coding regions could also play a major role. Such cases will facilitate the design of DNA-based diagnostic tools to detect metabolic resistance in field populations, as done already for the L119F-GSTe2 mutation (Riveron et al. 2014a).
Conclusion
The comparative transcription analysis performed in this study between various African regions highlights that, although metabolic resistance is the common driving mechanism of pyrethroid resistance in An. funestus populations, there are significant special variations on the main genes associated with it, which could impact patterns of cross-resistance and resistance management strategies. The impact of many genes conferring resistance and cross-resistance to multiple resistant populations of An. funestus in sub-Saharan African is a challenge to resistance management. This phenomenon makes the resistance highly heterogeneous and complex, making the design of appropriate diagnostic tools operationally challenging. There is an overwhelming need for newer classes of insecticides that are safe but potent enough to control mosquito vectors of malaria and other diseases effectively. But caution must be exercised because of the presence of a number of detoxification enzymes that can confer cross-resistance, and a new insecticide may already be doomed before being deployed if resistance genes can already metabolize it.
Supplementary Material
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.117.040147/-/DC1.
Acknowledgments
This work was supported by a Wellcome Trust Master Fellowship in Public Health and Tropical Medicine to C.M. (092794/Z/10/Z), and a Wellcome Trust Research Career Development Fellowship to C.W. (083515/Z/07/Z).
Author contributions: C.S.W. conceived and designed the study; C.M., J.M.R., and R.D. carried out the sample collection and performed World Health Organization (WHO) bioassays; C.M., H.I., I.H.I., M.J.W., and C.S.W. performed the Microarray and qRT-PCR analyses; J.M.R., S.S.I., and H.I. performed the transgenic expression study; S.S.I. performed in vitro characterization work. C.M., H.I., and C.S.W. performed the sequencing of resistance genes; J.M.R., C.M., S.S.I., and C.S.W. analyzed the data and wrote the manuscript; All authors read and approved the manuscript. No conflict of interest statement declared by authors.
Footnotes
Communicating editor: J. A. Birchler
Literature Cited
- Abdalla H., Wilding C. S., Nardini L., Pignatelli P., Koekemoer L. L., et al. , 2014. Insecticide resistance in Anopheles arabiensis in Sudan: temporal trends and underlying mechanisms. Parasit. Vectors 7: 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amenya D. A., Naguran R., Lo T. C., Ranson H., Spillings B. L., et al. , 2008. Over expression of a cytochrome P450 (CYP6P9) in a major African malaria vector, Anopheles funestus, resistant to pyrethroids. Insect Mol. Biol. 17: 19–25. [DOI] [PubMed] [Google Scholar]
- Antonio-Nkondjio C., Poupardin R., Tene B. F., Kopya E., Costantini C., et al. , 2016. Investigation of mechanisms of bendiocarb resistance in Anopheles gambiae populations from the city of Yaounde, Cameroon. Malar. J. 15: 424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins W. M., 2004. Implications of the allosteric kinetics of cytochrome P450s. Drug Discov. Today 9: 478–484. [DOI] [PubMed] [Google Scholar]
- Atkins W. M., Lu W. D., Cook D. L., 2002. Is there a toxicological advantage for non-hyperbolic kinetics in cytochrome P450 catalysis? Functional allostery from “distributive catalysis”. J. Biol. Chem. 277: 33258–33266. [DOI] [PubMed] [Google Scholar]
- Bariami V., Jones C. M., Poupardin R., Vontas J., Ranson H., 2012. Gene amplification, ABC transporters and cytochrome P450s: unraveling the molecular basis of pyrethroid resistance in the dengue vector, Aedes aegypti. PLoS Negl. Trop. Dis. 6: e1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes K. G., Irving H., Chiumia M., Mzilahowa T., Coleman M., et al. , 2016. Restriction to gene flow is associated with changes in the molecular basis of pyrethroid resistance in the malaria vector Anopheles funestus. Proc. Natl. Acad. Sci. USA. 114: 286–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes K. G., Weedall G. D., Ndula M., Irving H., Mzilahowa T., et al. , 2017. Genomic footprints of selective sweeps from metabolic resistance to pyrethroids in African malaria vectors are driven by scale up of insecticide-based vector control. PLoS Genet. 13: e1006539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooke B. D., Kloke G., Hunt R. H., Koekemoer L. L., Temu E. A., et al. , 2001. Bioassay and biochemical analyses of insecticide resistance in southern African Anopheles funestus (Diptera: Culicidae). Bull. Entomol. Res. 91: 265–272. [DOI] [PubMed] [Google Scholar]
- Casimiro S., Coleman M., Mohloai P., Hemingway J., Sharp B., 2006. Insecticide resistance in Anopheles funestus (Diptera: Culicidae) from Mozambique. J. Med. Entomol. 43: 267–275. [DOI] [PubMed] [Google Scholar]
- Coetzee M., Koekemoer L. L., 2013. Molecular systematics and insecticide resistance in the major African malaria vector Anopheles funestus. Annu. Rev. Entomol. 58: 393–412. [DOI] [PubMed] [Google Scholar]
- Conesa A., Gotz S., Garcia-Gomez J. M., Terol J., Talon M., et al. , 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676. [DOI] [PubMed] [Google Scholar]
- Copeland R. A., 2004. Enzymes: A Practical Introduction to Structure, Mechanism, and Data Analysis. John Wiley & Sons, New York. [Google Scholar]
- Crawford J. E., Guelbeogo W. M., Sanou A., Traore A., Vernick K. D., et al. , 2010. De novo transcriptome sequencing in Anopheles funestus using Illumina RNA-seq technology. PLoS One 5: e14202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuamba N., Morgan J. C., Irving H., Steven A., Wondji C. S., 2010. High level of pyrethroid resistance in an Anopheles funestus population of the Chokwe district in Mozambique. PLoS One 5: e11010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David J. P., Strode C., Vontas J., Nikou D., Vaughan A., et al. , 2005. The Anopheles gambiae detoxification chip: a highly specific microarray to study metabolic-based insecticide resistance in malaria vectors. Proc. Natl. Acad. Sci. USA 102: 4080–4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dia I., Guelbeogo M. W., Ayala D., 2013. Advances and perspectives in the study of the malaria in Anopheles Mosquitoes—New Insights into Malaria Vectors, edited by Manguin S. InTech Publisher, Rijeka, Croatia. [Google Scholar]
- Djouaka R., Irving H., Tukur Z., Wondji C. S., 2011. Exploring mechanisms of multiple insecticide resistance in a population of the malaria vector Anopheles funestus in Benin. PLoS One 6: e27760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djouaka R., Riveron J. M., Yessoufou A., Tchigossou G., Akoton R., et al. , 2016. Multiple insecticide resistance in an infected population of the malaria vector Anopheles funestus in Benin. Parasit. Vectors 9: 453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djouaka R. J., Atoyebi S. M., Tchigossou G. M., Riveron J. M., Irving H., et al. , 2016. Evidence of a multiple insecticide resistance in the malaria vector Anopheles funestus in South West Nigeria. Malar. J. 15: 565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edi C. V., Djogbenou L., Jenkins A. M., Regna K., Muskavitch M. A., et al. , 2014. CYP6 P450 enzymes and ACE-1 duplication produce extreme and multiple insecticide resistance in the malaria mosquito Anopheles gambiae. PLoS Genet. 10: e1004236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fossog Tene B., Poupardin R., Costantini C., Awono-Ambene P., Wondji C. S., et al. , 2013. Resistance to DDT in an urban setting: common mechanisms implicated in both M and S forms of Anopheles gambiae in the city of Yaounde Cameroon. PLoS One 8: e61408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillies M. T., De Meillon B., 1968. Anophelinae of Africa South of Sahara (Ethiopian Zoogeographical region), Ed. 2. The South African Institute for Medical Research, Johannesburg, South Africa. [Google Scholar]
- Gillies M. T., Coetzee M., 1987. A Supplement to the Anophelinae of Africa South of the Sahara (Afrotropical Region). South African Institute for Medical Research, Johannesburg, South Africa. [Google Scholar]
- Gotz S., Garcia-Gomez J. M., Terol J., Williams T. D., Nagaraj S. H., et al. , 2008. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 36: 3420–3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory R., Darby A. C., Irving H., Coulibaly M. B., Hughes M., et al. , 2011. A de novo expression profiling of Anopheles funestus, malaria vector in Africa, using 454 pyrosequencing. PLoS One 6: e17418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves K., Koekemoer L. L., Brooke B. D., Hunt R. H., Mthembu J., et al. , 2000. Anopheles funestus resistant to pyrethroid insecticides in South Africa. J. Med. Vet. Entomol. 14: 181–189. [DOI] [PubMed] [Google Scholar]
- Hunt R., Edwardes M., Coetzee M., 2010. Pyrethroid resistance in southern African Anopheles funestus extends to Likoma Island in Lake Malawi. Parasit. Vectors 3: 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim S. S., Manu Y. A., Tukur Z., Irving H., Wondji C. S., 2014. High frequency of kdr L1014F is associated with pyrethroid resistance in Anopheles coluzzii in Sudan savannah of northern Nigeria. BMC Infect. Dis. 14: 441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim S. S., Riveron J. M., Bibby J., Irving H., Yunta C., et al. , 2015. Allelic variation of cytochrome P450s drives resistance to bednet insecticides in a major malaria vector. PLoS Genet. 11: e1005618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim S. S., Ndula M., Riveron J. M., Irving H., Wondji C. S., 2016a The P450 CYP6Z1 confers carbamate/pyrethroid cross-resistance in a major African malaria vector beside a novel carbamate-insensitive N485I acetylcholinesterase-1 mutation. Mol. Ecol. 25: 3436–3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim S. S., Riveron J. M., Stott R., Irving H., Wondji C. S., 2016b The cytochrome P450 CYP6P4 is responsible for the high pyrethroid resistance in knockdown resistance-free Anopheles arabiensis. Insect Biochem. Mol. Biol. 68: 23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irving H., Riveron J. M., Ibrahim S. S., Lobo N. F., Wondji C. S., 2012. Positional cloning of rp2 QTL associates the P450 genes CYP6Z1, CYP6Z3 and CYP6M7 with pyrethroid resistance in the malaria vector Anopheles funestus. Heredity (Edinb) 109: 383–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishak I. H., Riveron J. M., Ibrahim S. S., Stott R., Longbottom J., et al. , 2016. The Cytochrome P450 gene CYP6P12 confers pyrethroid resistance in kdr-free Malaysian populations of the dengue vector Aedes albopictus. Sci. Rep. 6: 24707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koekemoer L. L., Kamau L., Hunt R. H., Coetzee M., 2002. A cocktail polymerase chain reaction assay to identify members of the Anopheles funestus (Diptera: Culicidae) group. Am. J. Trop. Med. Hyg. 66: 804–811. [DOI] [PubMed] [Google Scholar]
- Kuhr R. J., 1970. Metabolism of carbamate insecticide chemicals in plants and insects. J. Agric. Food Chem. 18: 1023–1030. [Google Scholar]
- Kwiatkowska R. M., Platt N., Poupardin R., Irving H., Dabire R. K., et al. , 2013. Dissecting the mechanisms responsible for the multiple insecticide resistance phenotype in Anopheles gambiae s.s., M form, from Vallee du Kou, Burkina Faso. Gene 519: 98–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lwetoijera D. W., Harris C., Kiware S. S., Dongus S., Devine G. J., et al. , 2014. Increasing role of Anopheles funestus and Anopheles arabiensis in malaria transmission in the Kilombero Valley, Tanzania. Malar. J. 13: 331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin L. A., Niazi U., Bibby J., David J. P., Vontas J., et al. , 2008. Characterization of inhibitors and substrates of Anopheles gambiae CYP6Z2. Insect Mol. Biol. 17: 125–135. [DOI] [PubMed] [Google Scholar]
- Menze B. D., Riveron J. M., Ibrahim S. S., Irving H., Antonio-Nkondjio C., et al. , 2016. Multiple insecticide resistance in the malaria vector Anopheles funestus from Northern Cameroon is mediated by metabolic resistance alongside potential target site insensitivity mutations. PLoS One 11: e0163261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel A. P., Ingrasci M. J., Schemerhorn B. J., Kern M., Le Goff G., et al. , 2005. Rangewide population genetic structure of the African malaria vector Anopheles funestus. Mol. Ecol. 14: 4235–4248. [DOI] [PubMed] [Google Scholar]
- Mitchell S. N., Stevenson B. J., Muller P., Wilding C. S., Egyir-Yawson A., et al. , 2012. Identification and validation of a gene causing cross-resistance between insecticide classes in Anopheles gambiae from Ghana. Proc. Natl. Acad. Sci. USA 109: 6147–6152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan J. C., Irving H., Okedi L. M., Steven A., Wondji C. S., 2010. Pyrethroid resistance in an Anopheles funestus population from Uganda. PLoS One 5: e11872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulamba C., Riveron J. M., Ibrahim S. S., Irving H., Barnes K. G., et al. , 2014. Widespread pyrethroid and DDT resistance in the major malaria vector Anopheles funestus in East Africa is driven by metabolic resistance mechanisms. PLoS One 9: e110058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nkya T. E., Akhouayri I., Poupardin R., Batengana B., Mosha F., et al. , 2014. Insecticide resistance mechanisms associated with different environments in the malaria vector Anopheles gambiae: a case study in Tanzania. Malar. J. 13: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okoye P. N., Brooke B. D., Koekemoer L. L., Hunt R. H., Coetzee M., 2008. Characterisation of DDT, pyrethroid and carbamate resistance in Anopheles funestus from Obuasi, Ghana. Trans. R. Soc. Trop. Med. Hyg. 102: 591–598. [DOI] [PubMed] [Google Scholar]
- Omura T., Sato R., 1964. The carbon monoxide-binding pigment of liver microsomes. I. Evidence for its hemoprotein nature. J. Biol. Chem. 239: 2370–2378. [PubMed] [Google Scholar]
- Pritchard M. P., Glancey M. J., Blake J. A., Gilham D. E., Burchell B., et al. , 1998. Functional co-expression of CYP2D6 and human NADPH-cytochrome P450 reductase in Escherichia coli. Pharmacogenetics 8: 33–42. [DOI] [PubMed] [Google Scholar]
- Riveron J. M., Irving H., Ndula M., Barnes K. G., Ibrahim S. S., et al. , 2013. Directionally selected cytochrome P450 alleles are driving the spread of pyrethroid resistance in the major malaria vector Anopheles funestus. Proc. Natl. Acad. Sci. USA 110: 252–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riveron J. M., Ibrahim S. S., Chanda E., Mzilahowa T., Cuamba N., et al. , 2014a The highly polymorphic CYP6M7 cytochrome P450 gene partners with the directionally selected CYP6P9a and CYP6P9b genes to expand the pyrethroid resistance front in the malaria vector Anopheles funestus in Africa. BMC Genomics 15: 817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riveron J. M., Yunta C., Ibrahim S. S., Djouaka R., Irving H., et al. , 2014b A single mutation in the GSTe2 gene allows tracking of metabolically-based insecticide resistance in a major malaria vector. Genome Biol. 15: R27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riveron J. M., Osae M., Egyir-Yawson A., Irving H., Ibrahim S. S., et al. , 2016. Multiple insecticide resistance in the major malaria vector Anopheles funestus in southern Ghana: implications for malaria control. Parasit. Vectors 9: 504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas J., Sanchez-DelBarrio J. C., Messeguer X., Rozas R., 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19: 2496–2497. [DOI] [PubMed] [Google Scholar]
- Saavedra-Rodriguez K., Suarez A. F., Salas I. F., Strode C., Ranson H., et al. , 2012. Transcription of detoxification genes after permethrin selection in the mosquito Aedes aegypti. Insect Mol. Biol. 21: 61–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samb B., Konate L., Irving H., Riveron J. M., Dia I., et al. , 2016. Investigating molecular basis of lambda-cyhalothrin resistance in an Anopheles funestus population from Senegal. Parasit. Vectors 9: 449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen T. D., Livak K. J., 2008. Analyzing real-time PCR data by the comparative C-T method. Nat. Protoc. 3: 1101–1108. [DOI] [PubMed] [Google Scholar]
- Stevenson B. J., Bibby J., Pignatelli P., Muangnoicharoen S., O’Neill P. M., et al. , 2011. Cytochrome P450 6M2 from the malaria vector Anopheles gambiae metabolizes pyrethroids: sequential metabolism of deltamethrin revealed. Insect Biochem. Mol. Biol. 41: 492–502. [DOI] [PubMed] [Google Scholar]
- Stevenson B. J., Pignatelli P., Nikou D., Paine M. J., 2012. Pinpointing P450s associated with pyrethroid metabolism in the dengue vector, Aedes aegypti: developing new tools to combat insecticide resistance. PLoS Negl. Trop. Dis. 6: e1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel H. W., Dignam J. D., 1978. Purification and properties of NADPH-cytochrome P-450 reductase. Methods Enzymol. 52: 89–96. [DOI] [PubMed] [Google Scholar]
- Strode C., Wondji C. S., David J. P., Hawkes N. J., Lumjuan N., et al. , 2008. Genomic analysis of detoxification genes in the mosquito Aedes aegypti. Insect Biochem. Mol. Biol. 38: 113–123. [DOI] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S., 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599. [DOI] [PubMed] [Google Scholar]
- Thompson J. D., Higgins D. G., Gibson T. J., 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen E. K., Strode C., Hemmings K., Hughes A. J., Chanda E., et al. , 2014. Underpinning sustainable vector control through informed insecticide resistance management. PLoS One 9: e99822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toe K. H., N’Fale S., Dabire R. K., Ranson H., Jones C. M., 2015. The recent escalation in strength of pyrethroid resistance in Anopheles coluzzi in West Africa is linked to increased expression of multiple gene families. BMC Genomics 16: 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tracy T. S., Hutzler J. M., Haining R. L., Rettie A. E., Hummel M. A., et al. , 2002. Polymorphic variants (CYP2C9*3 and CYP2C9*5) and the F114L active site mutation of CYP2C9: effect on atypical kinetic metabolism profiles. Drug Metab. Dispos. 30: 385–390. [DOI] [PubMed] [Google Scholar]
- Vontas J., Blass C., Koutsos A. C., David J. P., Kafatos F. C., et al. , 2005. Gene expression in insecticide resistant and susceptible Anopheles gambiae strains constitutively or after insecticide exposure. Insect Mol. Biol. 14: 509–521. [DOI] [PubMed] [Google Scholar]
- Wang R. W., Newton D. J., Liu N., Atkins W. M., Lu A. Y., 2000. Human cytochrome P-450 3A4: in vitro drug-drug interaction patterns are substrate-dependent. Drug Metab. Dispos. 28: 360–366. [PubMed] [Google Scholar]
- WHO , 1998. Test Procedures for Insecticide Resistance Monitoring in Malaria Vectors, Bio-Efficacy and Persistence of Insecticides on Treated Surfaces. World Health Organization, Geneva, Switzerland. [Google Scholar]
- WHO , 2012. Global Plan for Insecticide Resistance Management (GPIRM) World Health Organization, Geneva, Switzerland. [Google Scholar]
- WHO , 2015. World Malaria Report 2015. World Health Organization, Geneva, Switzerland. [Google Scholar]
- Wondji C. S., Hunt R. H., Pignatelli P., Steen K., Coetzee M., et al. , 2005. An integrated genetic and physical map for the malaria vector Anopheles funestus. Genetics 171: 1779–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wondji C. S., Morgan J. C., Coetzee M., Hunt R., Steen K., et al. , 2007. Mapping a quantitative trait locus conferring pyrethroid resistance in the African malaria vector Anopheles funestus. BMC Genomics 8: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wondji C. S., Irving H., Morgan J., Lobo N. F., Collins F. H., et al. , 2009. Two duplicated P450 genes are associated with pyrethroid resistance in Anopheles funestus, a major malaria vector. Genome Res. 19: 452–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wondji C. S., Dabire R. K., Tukur Z., Irving H., Djouaka R., et al. , 2011. Identification and distribution of a GABA receptor mutation conferring dieldrin resistance in the malaria vector Anopheles funestus in Africa. Insect Biochem. Mol. Biol. 41: 484–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wondji C. S., Coleman M., Kleinschmidt I., Mzilahowa T., Irving H., et al. , 2012. Impact of pyrethroid resistance on operational malaria control in Malawi. Proc. Natl. Acad. Sci. USA 109: 19063–19070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaim M., Aitio A., Nakashima N., 2000. Safety of pyrethroid-treated mosquito nets. Med. Vet. Entomol. 14: 1–5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The microarray data from this study were submitted to Array Express, accession numbers E-MTAB-5375, E-MTAB-5376, and E-MTAB-5424. The DNA sequences reported in this paper have been deposited in the GenBank database (accession numbers: KJ150626–KJ150674).