Significance
The causes of differentiation among populations are well circumscribed, but it remains unclear if they impact the proliferation of organisms over deep time. If, as some recent theory and observations suggest, population differentiation is not a rate-limiting control on species diversification, then the causes of population differentiation are unlikely to have macroevolutionary effects. We provide a large-scale test of the link between standardized estimates of rates of differentiation from population genetic data and speciation rates. Population differentiation rates predict speciation rates across New World birds, confirming the potential macroevolutionary importance of causes of differentiation. We also find that population differentiation and speciation rates are more tightly linked in the Tropics, which may help explain latitudinal differences in diversification dynamics.
Keywords: ephemeral speciation, trait-dependent diversification, comparative phylogeography, latitudinal diversity gradient
Abstract
An implicit assumption of speciation biology is that population differentiation is an important stage of evolutionary diversification, but its significance as a rate-limiting control on phylogenetic speciation dynamics remains largely untested. If population differentiation within a species is related to its speciation rate over evolutionary time, the causes of differentiation could also be driving dynamics of organismal diversity across time and space. Alternatively, geographic variants might be short-lived entities with rates of formation that are unlinked to speciation rates, in which case the causes of differentiation would have only ephemeral impacts. By pairing population genetics datasets from 173 New World bird species (>17,000 individuals) with phylogenetic estimates of speciation rate, we show that the population differentiation rates within species are positively correlated with their speciation rates over long timescales. Although population differentiation rate explains relatively little of the variation in speciation rate among lineages, the positive relationship between differentiation rate and speciation rate is robust to species-delimitation schemes and to alternative measures of both rates. Population differentiation occurs at least three times faster than speciation, which suggests that most populations are ephemeral. Speciation and population differentiation rates are more tightly linked in tropical species than in temperate species, consistent with a history of more stable diversification dynamics through time in the Tropics. Overall, our results suggest that the processes responsible for population differentiation are tied to those that underlie broad-scale patterns of diversity.
Speciation in most organisms is initiated via the geographic isolation and differentiation of populations. Many extrinsic and intrinsic factors determine the rate that populations differentiate within species, including the rate of geological and climatic change (1, 2), the dispersal ability of organisms (3), and the availability of ecological opportunities and strength of natural selection (4, 5). An implicit assumption of speciation biology is that these factors have an impact that percolates through to long evolutionary timescales and influences the proliferation of species. This connection, however, is not assured. The factors responsible for population differentiation can affect species diversification only if differentiation acts as a limiting control on diversification—for example, if the rate at which differentiated populations form within a lineage determines the rate at which that lineage can produce species—or if both differentiation and diversification are responses to the same causal processes. In either case, differentiation and diversification should be associated across evolutionary lineages.
Recent work, however, indicates that differentiation dynamics might be decoupled from those of diversification as measured over phylogenetic or paleontological timescales. Some macroevolutionary biologists suggest that geographic populations or variants are often ephemeral entities and that their rate of formation within a species might have little relation to speciation rates (6–8). Instead, speciation may be limited by other population-level processes, such as the persistence of differentiated populations (9) or the evolution of sufficient ecological divergence (5, 10) or reproductive isolation (11, 12) for differentiated populations to coexist in sympatry, or speciation may be random with respect to population-level processes. If differentiation and diversification are not associated, the factors responsible for differentiation cannot be expected to have macroevolutionary impacts.
The association between population differentiation and diversification can be tested by comparing present-day population differentiation within species to the speciation rate of those species’ lineages over deeper evolutionary time. A positive association between differentiation and speciation rate across lineages would support the hypothesis that differentiation is a rate-limiting control on speciation and contributes to large-scale diversity patterns. Few studies have attempted to make this comparison (13, 14). Kisel et al. (15) found no link between the magnitude of genetic divergence between populations and diversification rates in five sister clades of Costa Rican orchids. Haskell and Adhikari (16), however, found that the number of taxonomic subspecies within species predicted the number of species in avian genera, and Phillimore (17) found that the rate of avian subspecies formation in bird species was correlated with phylogenetic speciation rates. However, current subspecies taxonomy is often based on limited phenotypic data and appears to be an inconsistent indicator of intraspecific diversity in birds (18, 19). The examination of standardized, quantitative metrics of population genetic structure from large samples of species provides an important test of the relationship between differentiation and speciation independent of potentially arbitrary subspecific units that may not be comparable across diverse taxa.
Differences in the link between intraspecific diversity and speciation rates across studies may also stem from variation in the evolutionary importance of differentiation among organismal groups or across geographic areas. Population differentiation in temperate areas, for example, may be more ephemeral than in tropical regions (20–22), which could loosen its association with speciation rates. Alternatively, if differentiated populations form readily at low latitudes, but species formation is limited by the availability of vacant niches or the evolution of novel ecologies (23, 24), population differentiation may be a poorer predictor of speciation rate in the Tropics. Investigation into variation in the association between population differentiation and speciation rates could reveal differences in the temporal constancy of diversification rates that contribute to broad patterns of species richness among evolutionary lineages and geographic areas.
Here, we assess the association between population differentiation and speciation rates using quantitative estimates based on a large empirical dataset. We estimate population differentiation based on an application of the multispecies coalescent model to gene trees from population genetic and phylogeographic data from 173 species of New World birds. We compare population differentiation to speciation rates estimated for the lineages subtending the same 173 species using phylogenetic trees of all birds. We first test whether population differentiation and speciation rates are associated across all sampled species. Because the association between population differentiation and speciation rates may vary across geographic regions, we also test whether the association between differentiation and speciation rates in the tropics differs from that in the Temperate Zone. Finally, we perform a suite of supplementary tests to assess the robustness of recovered relationships to the inclusion of alternative predictors and to the approach used for sampling and analysis.
Results
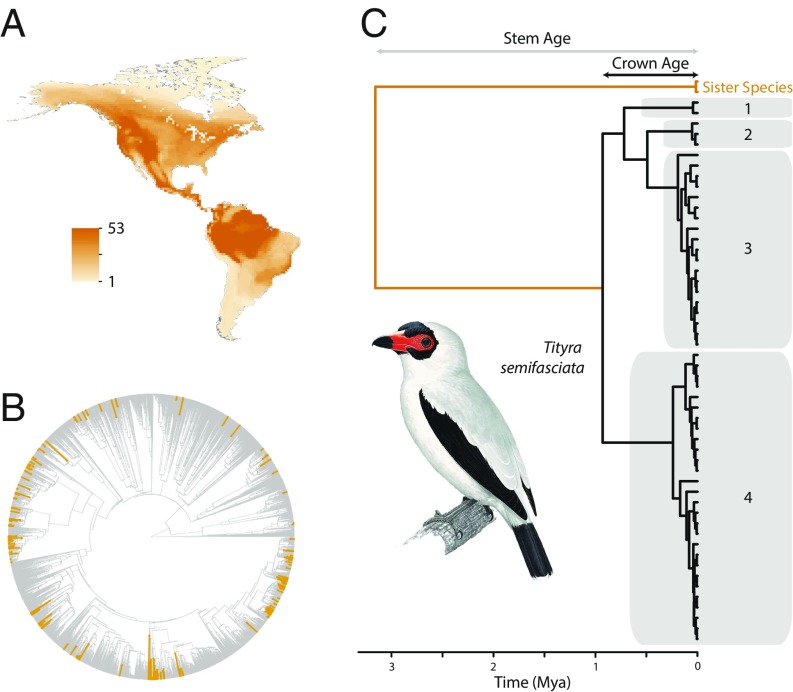
We estimated population differentiation based on genetic data using range-wide samples (n = 17,533) of 173 bird species from across the avian tree of life (Fig. 1A and SI Appendix) and inhabiting all biogeographic regions of the New World (Fig. 1B). We used a Bayesian implementation of the general mixed Yule-coalescent model (25, 26) to standardize population differentiation estimates within each species (Fig. 1C). The number of genetically distinguishable geographic populations within species varied from 1 to 24 with a median of 3 (Fig. 2). Because species might vary in the number of geographic populations simply due to differences in age, we calculated the rate of population formation since the crown age of each species (the age of the most recent common ancestor of extant haplotypes within the species) based on a time-calibrated gene tree. The rate at which geographic populations arose—hereafter the rate of population differentiation—varied from 0 to 6.64 divergences/million years (My) with a median of 0.54 divergences/My (Fig. 2).
Fig. 1.
Sampling strategy and approach to measuring population differentiation. (A) Overlaid distribution maps from the New World bird species used to estimate population genetic differentiation (n = 173). (B) The phylogenetic distribution of the study species within the tree of life of all birds (24). The orange branches indicate the species examined in this study. They are distributed throughout the tree and represent replicates with varying levels of phylogenetic independence for the purpose of comparative analysis. (C) An example of a mitochondrial gene tree used to estimate the rate of population genetic differentiation within one of the 173 study species, the Masked Tityra (Tityra semifasciata). The gray polygons represent population clusters for this species as inferred using bGMYC (19) based on a posterior probability threshold of shared population membership of 0.8. The stem age and crown age for this species, used to estimate rates of differentiation, are also depicted. The image was provided by Del Hoyo et al. (58).
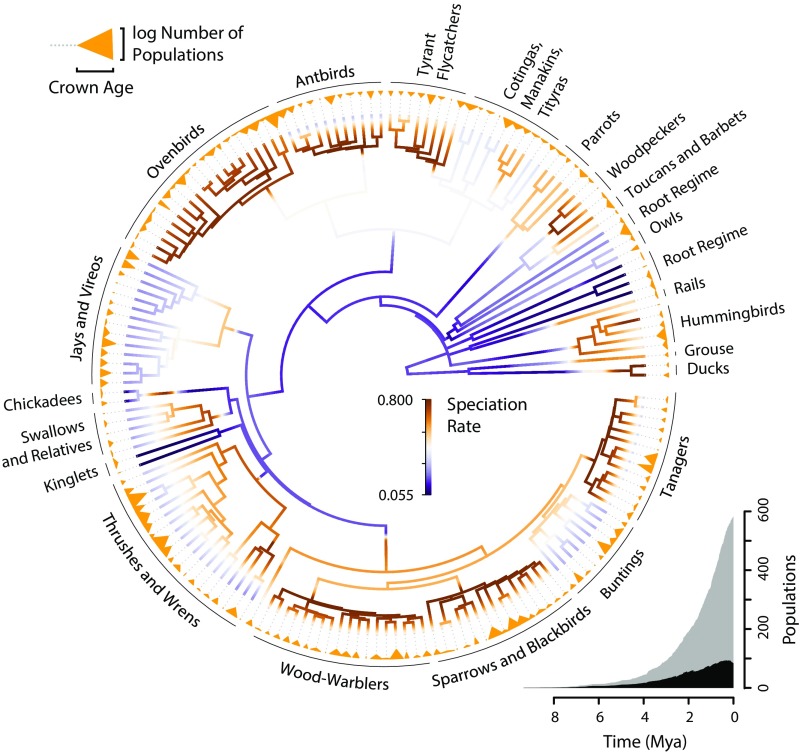
Fig. 2.
A circular phylogenetic tree of the 173 study species used to estimate rates of population genetic differentiation, colored with a gradient that depicts the BAMM speciation rate along each branch. Groups representing different evolutionary rate regimes based on the maximum credibility shift set are labeled. Encircling the phylogenetic tree are triangles representing each study species, the width of which represents the log number of populations inferred with bGMYC, and the height of which reflects the crown age of haplotypes in that species (crown height is not scaled to the timescale of the phylogeny). The color of the triangles does not reflect population differentiation rate. Inset at the bottom right is a plot of total accumulation of extant populations through time across all study species assuming no extinction (gray) and the number of those populations expected to ultimately form species based on the ratio of population differentiation to BAMM speciation rates of their respective lineages (black).
We estimated macroevolutionary speciation rates along the ancestral lineage leading to each of the 173 species in the population genetic datasets using two methods applied to an existing phylogenetic tree of all bird species (27). First, we computed a simple summary metric of speciation rate for each tip, called the diversification rate (DR) statistic (27), based on the weighted inverse of phylogenetic branch lengths. Because it does not explicitly account for extinction, the DR statistic is more tightly correlated with speciation rates at the tips of the tree than the net diversification rate (28). Speciation rates based on the DR statistic ranged from 0.03 to 3.35 species/My with a median of 0.16 species/My. Second, we obtained model-based speciation rate estimates using Bayesian analysis of macroevolutionary mixtures (BAMM) v.2.5, which jointly estimates the number of distinct evolutionary rate regimes across a phylogenetic tree and the speciation and extinction rates within each of the regimes (29–31). We extracted speciation rates for each study species based on the marginal posterior rate distribution at its tip on the phylogeny. Based on BAMM analysis, the marginal posterior speciation rates across the 173 study species varied from 0.05 to 0.66 species/My with a median of 0.14 species/My (Fig. 2 and SI Appendix, Figs. S1 and S2). Speciation rates from model-based analysis in BAMM and the summary DR statistic were correlated (R2 = 0.283, P < 0.001). Importantly, speciation rates inferred using both methods were slower than population differentiation rates. The median population differentiation rate was 3.35 times greater than the speciation rate using the DR statistic, or 3.70 times greater using the BAMM speciation rate (Fig. 2). These ratios are likely to be conservative because our population differentiation rate estimates do not account for population extinction within species. Although this result suggests that most geographic variants are ephemeral and do not persist to become reproductively isolated species, it does not preclude the possibility that variation among lineages in differentiation rate predicts variation in speciation rates.
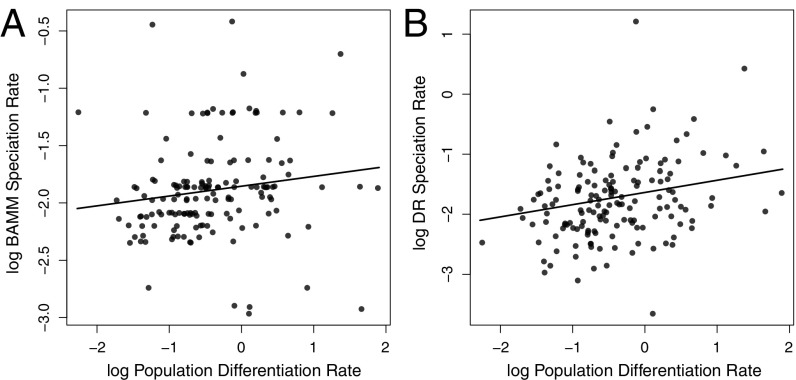
We tested whether population differentiation rates within species were associated with speciation rates inferred using both BAMM diversification analyses and the DR statistic. We tested for a relationship between population differentiation and BAMM speciation rates using structured rate permutations on phylogenies (STRAPP), a trait-dependent diversification test that avoids phylogenetic pseudoreplication while accounting for autocorrelation in evolutionary rates within evolutionary rate regimes (32). BAMM speciation rates were positively correlated with population genetic differentiation rates [Spearman’s correlation coefficient (r) = 0.264, P = 0.018, Fig. 3A]. We compared population differentiation and DR speciation rates using phylogenetic generalized least squares (PGLS) (33, 34). This test, pairing the DR statistic with PGLS analysis, is analogous to the trait-dependent diversification test proposed by Freckleton et al. (35). As with the BAMM speciation rate, the population differentiation rate predicted the DR speciation rate (PGLS slope = 0.203, P < 0.001, Fig. 3B). We simulated neutral change in population differentiation rates on the observed avian phylogenies to explore the propensity for false positives and found that it was minimal for both the STRAPP (P = 0.015) and PGLS (P = 0.055) tests. There were no correlations between the raw number of population clusters and speciation rate (STRAPP: r = 0.097, P = 0.287; PGLS slope = −0.070, P = 0.140), suggesting that the rate of population genetic differentiation rather than the level of standing differentiation is associated with speciation rate. These results provide quantitative evidence supporting the idea that population differentiation within species predicts macroevolutionary dynamics at a large spatial and taxonomic scale.
Fig. 3.
Plots showing the association between population differentiation and speciation rates across all 173 study species. Plots are presented based on speciation rates from (A) BAMM analysis [STRAPP correlation coefficient: (r) = 0.264, P = 0.018] and (B) the DR statistic (PGLS slope = 0.203, P < 0.001). The trend line for the plot using BAMM speciation rates is based on ordinary least-squares regression.
We conducted an additional series of tests to assess whether the association between population differentiation and speciation rates is an artifact of sampling or methodology. For brevity, we present results from STRAPP tests of BAMM speciation rates below, but results from PGLS of the DR statistic were similar and are presented in SI Appendix, SI Materials and Methods. The positive correlation between the population genetic differentiation rate and the speciation rate was robust to the taxonomy used to circumscribe species for the population-level analysis with a more finely subdivided taxonomy producing similar results to the primary taxonomy that we examined (r = 0.205, P = 0.038). Using a simple time threshold (9.501 My) instead of taxonomy to partition species diversity from intraspecific diversity also produced a similar result (r = 0.291, P = 0.011). Our result was robust to the use of lower (PP = 0.7; r = 0.256, P = 0.013) and higher (PP = 0.9; r = 0.267, P = 0.011) posterior probability thresholds for assigning individuals to population clusters, to whether the population differentiation rate was measured using the stem age rather than the crown age of a species (r = 0.319, P = 0.006), to the random removal of 20% (r = 0.244, P = 0.036) of samples from the dataset, to the removal of populations containing a single individual (r = 0.267, P = 0.012), and to models of population differentiation incorporating moderate [extinction/speciation ratio (eps) = 0.45; r = 0.262, P = 0.027] or high (eps = 0.9; r = 0.245, P = 0.048) population extinction rates. Population differentiation rate might be associated with speciation rate if clades with high speciation rates necessarily have shallower crown ages that result in elevated differentiation rates. However, crown age was not related to speciation rate (r = −0.104, P = 0.299).
Many other traits and environmental factors can potentially influence speciation rates in birds, including traits related to distribution, morphology, and behavior (e.g., 20, 36–38). We used multipredictor models to further explore the variation in speciation rates that was not explained by population differentiation alone; we considered a set of variables that represented range size, midpoint latitude, migratory distance, multivariate environmental niche, tarsus length (a proxy for body size), a wing shape metric (a proxy for dispersal ability), and presence or absence of sexual dichromatism (a proxy for strength of sexual selection) in each study species. Multivariate analyses were conducted using the DR statistic and PGLS, which readily accommodates multipredictor tests. We found that migratory distance (PGLS slope = 0.000, P = 0.041) and tarsus length (PGLS slope = 0.484, P = 0.024) were correlated with speciation rate in a multipredictor PGLS analysis, but population differentiation rate remained significant in the multivariate model (PGLS slope = 0.160, P = 0.003). In a comparison of PGLS models using the Akaike Information Criterion correcting for small sample sizes (AICc) scores, population differentiation rate was the variable responsible for the greatest model improvement (ΔAICc = 5.536, SI Appendix, Table S1). The model containing all nine variables explained less than twice as much variation in speciation rate overall (R2 = 0.180) compared with the population differentiation rate alone in one-way PGLS analysis (R2 = 0.098). This suggests that, although other variables may contribute to the noise observed in the relationship between population differentiation and speciation rates, population differentiation rate is the most important predictor among the predictors examined.
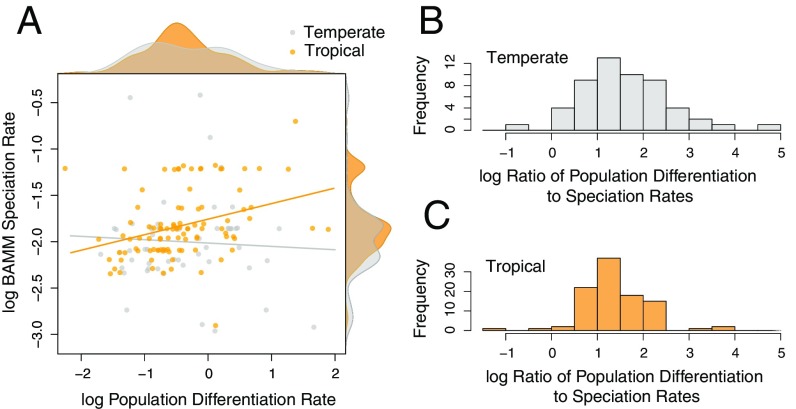
After dividing the species into temperate (n = 63) and tropical (n = 110) assemblages, we found no correlation between population differentiation and speciation rates in the temperate species (STRAPP: r = 0.106, P = 0.442; PGLS slope = 0.160, P = 0.086), but a strong positive correlation in the tropical species (STRAPP: r = 0.362, P = 0.014; PGLS slope = 0.229, P < 0.001; Fig. 4A and SI Appendix, Fig. S3). Resampling the tropical dataset to match the number of temperate species (n = 63) still produced stronger correlations than in the temperate dataset using STRAPP with BAMM speciation rates (P < 0.001), although not with PGLS of the DR statistic (P = 0.169). There was no latitudinal variation in our sample in either population differentiation rate (PGLS slope = −0.001, P = 0.888) or speciation rate (BAMM PGLS slope = 0.000, P = 0.866; DR statistic PGLS slope = 0.002, P = 0.537; Fig. 4A and SI Appendix, Fig. S3), nor in the ratio of population differentiation to speciation rate (BAMM PGLS slope = −0.001, P = 0.910; DR statistic PGLS slope = −0.003, P = 0.627). There was, however, a wide variance in the ratio of population differentiation to speciation rates at temperate latitudes, compared with a more peaked distribution among tropical species (F-test of equal variances: F = 1.837, P = 0.009 with BAMM speciation rates; F = 2.014, P = 0.003 with the DR statistic; Fig. 4 B and C and SI Appendix, Fig. S3). These results suggest that population differentiation leads to speciation at a relatively predictable rate in the tropics, but that this rate is less predictable in the Temperate Zone.
Fig. 4.
Plots showing differences in relative population differentiation and BAMM speciation rates between temperate (n = 63) and tropical (n = 110) species. (A) Tropical species show a relationship between population differentiation rates and speciation rates (r = 0.362, P = 0.014), whereas temperate species do not (r = 0.106, P = 0.442). Kernel density plots showing the relative distributions of rates between tropical and temperate species are plotted opposite the axis of the rate to which they correspond and show that neither differentiation or speciation rates differ markedly between temperate and tropical species. The ratio of population differentiation rate to speciation rate, however, is more variable in temperate species (B) than in tropical species (C). Similar results were observed using the DR statistic (SI Appendix, SI Materials and Methods).
Discussion
We found a robust association between population differentiation rate and speciation rate across New World birds, although considerable variance in speciation rate remained unexplained. Given the potential for estimation error in both population differentiation rates and speciation rates, detecting any association was remarkable. The rate of population differentiation within a species can be used, in part, to predict its speciation rate over longer timescales and vice versa. Statements of causality, however, would be misleading. Population differentiation may be a rate-limiting step in speciation or differentiation and speciation may be related through an unresolved causal structure involving other processes that affect rates at both timescales. In either case, our results support an implicit but largely untested assumption of speciation research, that the often-studied processes leading to population differentiation could also be responsible for elevated diversification rates over deep evolutionary time. Our results accord well with prior evidence that the number of taxonomic subspecies formed within a species is tied to species richness or speciation rate in the species’ higher taxonomic groups (16, 17). Moreover, our results provide an explicit timescale for population differentiation for direct comparison with diversification rates. Our results also bolster support for examples of links between speciation rate and traits thought to lead to population differentiation, such as limited dispersal ability or range fragmentation (36, 37, 39, 40).
Much of the variance that we observed in speciation rates is unexplained by population differentiation. This unexplained variance may be partly due to rate estimation error, but it also leaves room for other factors to contribute to speciation rate variation. Broad ecological traits of species and environmental variables may explain some speciation rate variation, and indeed we found evidence for weak associations with some of these variables (migratory distance and a proxy for body size). Aside from broad factors, population-level processes in addition to differentiation might also serve as proximate controls on the process of speciation. For allopatric speciation to be complete, geographically isolated populations must not only differentiate, but also persist until the evolution of reproductive isolation and ecological divergence permit the completion of speciation (6, 13). Variation among species in population persistence, time to reproductive isolation, or time to ecological differentiation therefore may explain some of the additional variance in speciation rate. However, extinction is notoriously difficult to estimate from data on extant lineages alone (9, 41), and measuring population persistence for exploration of this potential control using empirical data at the population level may be challenging. The rate of intrinsic postzygotic reproductive isolation does not predict speciation rate across birds (42), but premating reproductive isolation is also potentially important in birds (43) and may merit further investigation. Elevated ecological opportunities can be associated with increased speciation rates (44), and there is evidence that rates of ecological divergence vary regionally (45), but more data are needed to establish a link between rates of ecological divergence among populations and speciation rates. Regardless, although variation among lineages in factors like rates of population persistence, evolution of reproductive isolation, and ecological divergence may explain some variation in avian speciation rate, they are insufficient to erase the association between population differentiation and speciation observed in our datasets.
Population differentiation predicts speciation rate across all the New World birds examined, but the relationship appears to be stronger in tropical species and may be weak or absent in the Temperate Zone. Comparisons using larger samples of species, particularly from the Temperate Zone, are desirable to confirm this result. Even if additional research confirms that the association between population differentiation and speciation rates is an entirely tropical phenomenon, the association research would be of evolutionary importance given that most bird diversity is tropical and many temperate clades evolved from tropical ancestors (46, 47). In addition, the latitudinal difference in the association between population differentiation and speciation rate may provide information about geographical differences in how diversity accumulates. Coupled with the tighter relationship between population differentiation and speciation rates in tropical species, lineages in the tropics showed less variability in the ratio of population differentiation to speciation rates than temperate lineages. This pattern is consistent with a scenario in which the conversion of population differentiation to new species occurs predictably through time in the tropics, but is episodic or unpredictable at temperate latitudes. Climatic cycling over the past 420,000 y (48) suggests that major shifts in external environmental conditions may be the dominant driver of speciation rates at high latitudes, which could lead to cycles of differentiation and lineage loss that dampen the association between population differentiation and speciation in those regions. The tighter association between population splitting and speciation rates in the tropics may be due to the relative environmental stability in that region over recent timescales (49), which could relegate control of speciation rates to the population-level processes occurring constantly within lineages. Latitudinal differences in the correlation between population differentiation and speciation therefore support hypotheses that invoke greater tropical environmental stability as a cause of the latitudinal diversity gradient (20, 50) and suggest an underlying mechanism in the form of less episodic tropical diversification dynamics resulting from less dramatic climatic shifts.
To conclude, we predict that traits associated with processes that promote population differentiation will provide insights into attributes of organisms that predispose them to diversify. We also expect variation in those traits to help explain broader patterns of diversity among clades and regions. We anticipate that more and larger comparative, population-level datasets will allow investigation of additional processes, such as population persistence and ecological divergence, that might also contribute to the diversity of organisms worldwide.
Materials and Methods
Sampling and Taxonomy.
We examined population genetic data from 173 species from across the New World (SI Appendix). Species were defined as all nonsympatric monophyletic populations for which we had sampling, regardless of their current treatment by taxonomic authorities. Thus, metrics of population differentiation reflect geographic patterns of diversity among allopatric or parapatric groups, whereas metrics of speciation reflect deeper patterns among potentially sympatric and reproductively isolated groups. We alleviated the extent to which the inclusiveness of our species taxonomy could impact our results by focusing on rates of differentiation rather than standing levels of differentiation (see below). We expect differentiation rates to be similar in a species regardless of the taxonomic treatment used because a more inclusive treatment for a given species will generally result in an older species age in addition to more genetic structure. We also investigated the effect of taxonomic treatment on our results by applying a second taxonomy corresponding to the current taxonomy of the American Ornithological Society’s (AOS) North American (51, 52) and South American (53) Checklist committees. In situations where the North and South American committees differed in their treatment, we reverted to the North American committee’s treatment. The AOS taxonomy is more subdivided or “split” (260 species, 200 with sufficient samples to include in analyses) than the primary taxonomy (173 species), so examination of both provides an index of the impact of the level of taxonomic splitting on results. Finally, we examined a “taxonomy-free” approach in which we used a simple time threshold to distinguish between species diversity and intraspecific diversity. This threshold was based on the oldest crown age in the phylogeographic datasets (9.501 My) and was applied to every lineage in the dataset.
Molecular Data.
We examined previously published population-level mitochondrial datasets of New World birds, including a subset that we generated for this and related projects (22). We restricted our sampling to those datasets containing at least 10 samples (mean = 101) and range-wide coverage. We evaluated the robustness of our results to the level of sampling within species by randomly pruning 20% of the tips of the mitochondrial gene trees estimated from the full dataset and repeating analyses.
Population Differentiation Rate Estimation.
We estimated mitochondrial gene trees for each species using the Bayesian method implemented in BEAST v.1.7.5 (54). All trees were time-calibrated using an uncorrelated relaxed substitution rate based on published avian mitochondrial rates (SI Appendix, SI Materials and Methods). We included taxa deemed to be sister to study species based on prior phylogenetic work, and we extracted stem and crown age estimates for each species from maximum clade credibility (MCC) trees. We quantified phylogeographic structure using bGMYC (SI Appendix, SI Materials and Methods). We used the MCC tree from BEAST for each bGMYC run. bGMYC provides a posterior probability that two sequences belong to the same interbreeding population that can be used, along with a probability threshold, to determine the number of clusters present. For the primary analysis, we used a posterior probability threshold of 0.8 for clustering, but we also examined higher (0.9) and lower (0.7) thresholds.
To account for the fact that species might differ in the number of bGMYC clusters by virtue of differences in their age, we estimated the rate of bGMYC cluster formation, hereafter the population differentiation rate. We calculated rates using crown age, the age of the most recent common ancestor of extant haplotypes within the species. We used equation 6 from Magallón and Sanderson (55), which reduces to log(n)/t with no extinction (where n is the number of populations and t is the crown age). Although crown age is generally superior to stem age for rate estimation because it is positively correlated with diversity, which increases the comparability of rate estimates across species and taxonomic treatments (56), we also examined rates of population differentiation using the stem age. To account for population extinction, we also examined rates of population differentiation assuming different constant values for the relative rate of population extinction to population formation (SI Appendix, SI Materials and Methods). We did not control for area in our rate estimates because we expect population differentiation to have equivalent evolutionary importance regardless of the size of the area over which it is distributed (SI Appendix, SI Materials and Methods).
Speciation Rate Estimation.
We used time-calibrated MCC trees from a published phylogeny of all birds (27) for estimation of speciation rates. Tips in the phylogeny were collapsed in cases in which one of our study species was represented by multiple species in the taxonomy of the published phylogeny. That study placed species lacking genetic data using taxonomic constraints, but we removed these (leaving 6,670 species) from our analyses to reduce potential artifacts due to incorrect placement. We estimated speciation rates on the tree topology based on the Hackett et al. (57) backbone using the model implemented in the program BAMM v.2.5 (29, 30). BAMM was run assuming 67% sampling across the avian tree to account for species without genetic data and with a high value (100) for the prior on the expected number of macroevolutionary regime shifts. These settings, combined with the large size and heterogeneous diversification dynamics of the bird phylogeny, are expected to lead to accurate rate estimation using BAMM (31). Speciation rates for a given terminal branch on the tree were extracted from the marginal posterior distribution of rates, which is based on all processes sampled at that branch. We also estimated speciation rates using a simple summary statistic (the “DR statistic”) that reflects the number of splitting events subtending each tip on a phylogenetic tree (27). Specifically, it is the sum of the inverse of the branch lengths subtending a particular tip, down-weighted by half for each successive branch deeper in the tree. For this analysis we were unable to analytically account for incomplete sampling, so we used the phylogeny containing species with and without genetic data (n = 9,921) and calculated mean rates across 100 trees from the pseudoposterior of the published study (27).
Comparative Analyses.
We examined correlations between the population genetic differentiation rate and speciation rates inferred both using the DR statistic and diversification modeling in BAMM. We tested for correlations between log-transformed population differentiation and log-transformed BAMM speciation rates using STRAPP, a test that detects effects based on replicated associations between trait values and diversification rates from BAMM (28) (SI Appendix, SI Materials and Methods). This test accounts for covariance between species using permutations of trait values among species sharing the same evolutionary rate regime. For the DR statistic, we used PGLS (33, 34) to test for a correlation between log-transformed values and log-transformed population differentiation rates while accounting for relatedness between species based on phylogenetic distance in the avian tree (27).
We conducted multivariate analyses in which we tested whether any correlation between population differentiation and speciation rates persisted when other potentially important predictor variables were added to an analysis. The data for alternative predictor variables were gathered from existing databases of distribution and climatic data (distributional and environmental variables) and from museum specimens (ecomorphological variables) as detailed in SI Appendix, SI Materials and Methods. Finally, we conducted comparative analyses on both the full dataset and on datasets containing only species from either the Temperate Zone or the Tropical Zone. Species were assigned to the latitudinal zone based on the latitudinal midpoint of their breeding distribution relative to the tropics of Cancer and Capricorn (23.437° N and S). Curated datasets and scripts are available at https://github.com/mgharvey/differentiation_speciation.
Supplementary Material
Acknowledgments
We thank the field workers and museum staff who obtained and maintained the genetic resources and associated vouchers used in this study, in particular the staff of the Academy of Natural Sciences of Drexel University; the American Museum of Natural History; Colección Ornitologica Phelps; Cornell University Museum of Vertebrates; Field Museum of Natural History; Instituto Alexander von Humboldt; Instituto de Ciencias Naturales at the Universidad Nacional de Colombia; Laboratório de Genética e Evolução Molecular de Aves at the Universidade de São Paulo; Louisiana State University Museum of Natural Science; Museo de Historia Natural at the Universidad de los Andes; Museo de Zoología “Alfonso L. Herrera” at the Universidad Nacional Autónoma de Mexico; National Museum of Natural History; University of Kansas Natural History Museum; and University of Washington Burke Museum. We thank J. Klicka for providing data and feedback. We also thank J. V. Remsen, Jr., J. M. Brown, D. M. Baltz, S. Singhal, J. S. Mitchell, B. M. Winger, and P. O. Title for discussion and G. H. Thomas, A. J. Crawford, D. J. Futuyma, and six anonymous reviewers who provided comments that improved the manuscript. Funding was provided by National Science Foundation Grants IOS-1210556 (to M.G.H. and R.T.B.), DBI-1523893 (to M.G.H.), DEB-1146265 (to R.T.B.), IOS-0910285 (to R.T.B. and A.M.C.), DEB-1406932 (to R.T.B. and G.F.S.), and DEB-1256330 (to D.L.R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1617397114/-/DCSupplemental.
References
- 1.Hoorn C, et al. Amazonia through time: Andean uplift, climate change, landscape evolution, and biodiversity. Science. 2010;330:927–931. doi: 10.1126/science.1194585. [DOI] [PubMed] [Google Scholar]
- 2.Wade L. Cradle of life. Science. 2015;350:496–501. doi: 10.1126/science.350.6260.496. [DOI] [PubMed] [Google Scholar]
- 3.Smith BT, et al. The drivers of tropical speciation. Nature. 2014;515:406–409. doi: 10.1038/nature13687. [DOI] [PubMed] [Google Scholar]
- 4.Barreiro LB, Laval G, Quach H, Patin E, Quintana-Murci L. Natural selection has driven population differentiation in modern humans. Nat Genet. 2008;40:340–345. doi: 10.1038/ng.78. [DOI] [PubMed] [Google Scholar]
- 5.Price TD, et al. Niche filling slows the diversification of Himalayan songbirds. Nature. 2014;509:222–225. doi: 10.1038/nature13272. [DOI] [PubMed] [Google Scholar]
- 6.Mayr E. Animal Species and Evolution. Belknap Press; Cambridge, MA: 1963. [Google Scholar]
- 7.Stanley SM. Macroevolution: Pattern and Process. W. H. Freeman and Co.; San Francisco: 1979. [Google Scholar]
- 8.Rosenblum EB, et al. Goldilocks meets Santa Rosalia: An ephemeral speciation model explains patterns of diversification across time scales. Evol Biol. 2012;39:255–261. doi: 10.1007/s11692-012-9171-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dynesius M, Jansson R. Persistence of within-species lineages: A neglected control of speciation rates. Evolution. 2014;68:923–934. doi: 10.1111/evo.12316. [DOI] [PubMed] [Google Scholar]
- 10.Mayr E. Ecological factors in speciation. Evolution. 1947;1:263–288. [Google Scholar]
- 11.Dobzhansky T. Genetics and the Origin of Species. Columbia Univ Press; New York: 1937. [Google Scholar]
- 12.Mayr E. Systematics and the Origin of Species from the Viewpoint of a Zoologist. Columbia Univ Press; New York: 1942. [Google Scholar]
- 13.Allmon WD. A causal analysis of stages in allopatric speciation. Oxford Surv Evol Biol. 1992;8:219–257. [Google Scholar]
- 14.Barraclough TG, Nee S. Phylogenetics and speciation. Trends Ecol Evol. 2001;16:391–399. doi: 10.1016/s0169-5347(01)02161-9. [DOI] [PubMed] [Google Scholar]
- 15.Kisel Y, et al. Testing the link between population genetic differentiation and clade diversification in Costa Rican orchids. Evolution. 2012;66:3035–3052. doi: 10.1111/j.1558-5646.2012.01663.x. [DOI] [PubMed] [Google Scholar]
- 16.Haskell DG, Adhikari A. Darwin’s manufactory hypothesis is confirmed and predicts the extinction risk of extant birds. PLoS One. 2009;4:e5460. doi: 10.1371/journal.pone.0005460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Phillimore AB. Subspecies origination and extinction in birds. Ornithol Monogr. 2010;67:42–53. [Google Scholar]
- 18.Zink RM. The role of subspecies in obscuring avian biological diversity and misleading conservation policy. Proc Biol Sci. 2004;271:561–564. doi: 10.1098/rspb.2003.2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Remsen JV. Pattern, process, and rigor meet classification. Auk. 2005;122:403–413. [Google Scholar]
- 20.Weir JT, Schluter D. The latitudinal gradient in recent speciation and extinction rates of birds and mammals. Science. 2007;315:1574–1576. doi: 10.1126/science.1135590. [DOI] [PubMed] [Google Scholar]
- 21.Cutter AD, Gray JC. Ephemeral ecological speciation and the latitudinal biodiversity gradient. Evolution. 2016;70:2171–2185. doi: 10.1111/evo.13030. [DOI] [PubMed] [Google Scholar]
- 22.Smith BT, Seeholzer GF, Harvey MG, Cuervo AM, Brumfield RT. A latitudinal phylogeographic diversity gradient in birds. PLoS Biol. 2017;15:e2001073. doi: 10.1371/journal.pbio.2001073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rabosky DL. Ecological limits and diversification rate: Alternative paradigms to explain the variation in species richness among clades and regions. Ecol Lett. 2009;12:735–743. doi: 10.1111/j.1461-0248.2009.01333.x. [DOI] [PubMed] [Google Scholar]
- 24.Weir JT, Price TD. Limits to speciation inferred from times to secondary sympatry and ages of hybridizing species along a latitudinal gradient. Am Nat. 2011;177:462–469. doi: 10.1086/658910. [DOI] [PubMed] [Google Scholar]
- 25.Pons J, et al. Sequence-based species delimitation for the DNA taxonomy of undescribed insects. Syst Biol. 2006;55:595–609. doi: 10.1080/10635150600852011. [DOI] [PubMed] [Google Scholar]
- 26.Reid NM, Carstens BC. Phylogenetic estimation error can decrease the accuracy of species delimitation: A Bayesian implementation of the general mixed Yule-coalescent model. BMC Evol Biol. 2012;12:196. doi: 10.1186/1471-2148-12-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jetz W, Thomas GH, Joy JB, Hartmann K, Mooers AO. The global diversity of birds in space and time. Nature. 2012;491:444–448. doi: 10.1038/nature11631. [DOI] [PubMed] [Google Scholar]
- 28.Belmaker J, Jetz W. Relative roles of ecological and energetic constraints, diversification rates and region history on global species richness gradients. Ecol Lett. 2015;18:563–571. doi: 10.1111/ele.12438. [DOI] [PubMed] [Google Scholar]
- 29.Rabosky DL. Automatic detection of key innovations, rate shifts, and diversity-dependence on phylogenetic trees. PLoS One. 2014;9:e89543. doi: 10.1371/journal.pone.0089543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mitchell JS, Rabosky DL. Bayesian model selection with BAMM: Effects of the model prior on the inferred number of diversification shifts. Methods Ecol Evol. 2017;8:37–46. [Google Scholar]
- 31.Rabosky DL, Mitchell JS, Chang J. Is BAMM flawed? Theoretical and practical concerns in the analysis of multi-rate diversification models. Syst Biol. 2017 doi: 10.1093/sysbio/syx037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rabosky DL, Huang H. A robust semi-parametric test for detecting trait-dependent diversification. Syst Biol. 2016;65:181–193. doi: 10.1093/sysbio/syv066. [DOI] [PubMed] [Google Scholar]
- 33.Grafen A. The phylogenetic regression. Philos Trans R Soc Lond B Biol Sci. 1989;326:119–157. doi: 10.1098/rstb.1989.0106. [DOI] [PubMed] [Google Scholar]
- 34.Martins EP, Hansen TF. Phylogenies and the comparative method: A general approach to incorporating phylogenetic information into the analysis of interspecific data. Am Nat. 1997;149:646–667. [Google Scholar]
- 35.Freckleton RP, Phillimore AB, Pagel M. Relating traits to diversification: A simple test. Am Nat. 2008;172:102–115. doi: 10.1086/588076. [DOI] [PubMed] [Google Scholar]
- 36.Owens IPF, Bennett PM, Harvey PH. Species richness among birds: Body size, life history, sexual selection, or ecology? Proc Biol Sci. 1999;266:933–939. [Google Scholar]
- 37.Claramunt S, Derryberry EP, Remsen JV, Jr, Brumfield RT. High dispersal ability inhibits speciation in a continental radiation of passerine birds. Proc Biol Sci. 2012;279:1567–1574. doi: 10.1098/rspb.2011.1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang H, Rabosky DL. Sexual selection and diversification: Reexamining the correlation between dichromatism and speciation rate in birds. Am Nat. 2014;184:E101–E114. doi: 10.1086/678054. [DOI] [PubMed] [Google Scholar]
- 39.Riginos C, Buckley YM, Blomberg SP, Treml EA. Dispersal capacity predicts both population genetic structure and species richness in reef fishes. Am Nat. 2014;184:52–64. doi: 10.1086/676505. [DOI] [PubMed] [Google Scholar]
- 40.Jablonski D. Larval ecology and macroevolution in marine invertebrates. Bull Mar Sci. 1986;39:565–587. [Google Scholar]
- 41.Rabosky DL. Extinction rates should not be estimated from molecular phylogenies. Evolution. 2010;64:1816–1824. doi: 10.1111/j.1558-5646.2009.00926.x. [DOI] [PubMed] [Google Scholar]
- 42.Rabosky DL, Matute DR. Macroevolutionary speciation rates are decoupled from the evolution of intrinsic reproductive isolation in Drosophila and birds. Proc Natl Acad Sci USA. 2013;110:15354–15359. doi: 10.1073/pnas.1305529110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Price TD. Speciation in Birds. Roberts and Co.; Greenwood Village, CO: 2008. [Google Scholar]
- 44.Schluter D. The Ecology of Adaptive Radiation. Oxford Univ Press; Oxford: 2000. [Google Scholar]
- 45.Lawson AM, Weir JT. Latitudinal gradients in climatic-niche evolution accelerate trait evolution at high latitudes. Ecol Lett. 2014;17:1427–1436. doi: 10.1111/ele.12346. [DOI] [PubMed] [Google Scholar]
- 46.Hawkins BA, Diniz-Filho JAF, Jaramillo CA, Soeller SA. Post-Eocene climate change, niche conservatism, and the latitudinal diversity gradient of New World birds. J Biogeogr. 2006;33:770–780. [Google Scholar]
- 47.Diniz-Filho JAF, Rangel TFL, Bini LM, Hawkins BA. Macroevolutionary dynamics in environmental space and the latitudinal diversity gradient in New World birds. Proc Biol Sci. 2007;274:43–52. doi: 10.1098/rspb.2006.3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Petit JR, et al. Climate and atmospheric history of the past 420,000 years from the Vostok ice core, Antarctica. Nature. 1999;399:429–436. [Google Scholar]
- 49.Thomson LG, Mosley-Thomson E, Henderson KA. Ice-core palaeoclimate records in tropical South America since the Last Glacial Maximum. J Quat Sci. 2000;15:377–394. [Google Scholar]
- 50.Hewitt G. The genetic legacy of the Quaternary ice ages. Nature. 2000;405:907–913. doi: 10.1038/35016000. [DOI] [PubMed] [Google Scholar]
- 51.American Ornithologists’ Union . Check-List of North American Birds. 7th Ed. American Ornithologists’ Union; Washington, DC: 1998. [DOI] [PubMed] [Google Scholar]
- 52.Chesser RT, et al. Fifty-fourth supplement to the American Ornithologists’ Union Check-List of North American Birds. Auk. 2013;130:558–571. [Google Scholar]
- 53.Remsen JV, et al. 2014 A classification of the bird species of South America. AOU South American Classification Committee. Available at www.museum.lsu.edu/∼Remsen/SACCBaseline.htm. Accessed December 20, 2014.
- 54.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Magallón S, Sanderson MJ. Absolute diversification rates in angiosperm clades. Evolution. 2001;55:1762–1780. doi: 10.1111/j.0014-3820.2001.tb00826.x. [DOI] [PubMed] [Google Scholar]
- 56.Stadler T, Rabosky DL, Ricklefs RE, Bokma F. On age and species richness of higher taxa. Am Nat. 2014;184:447–455. doi: 10.1086/677676. [DOI] [PubMed] [Google Scholar]
- 57.Hackett SJ, et al. A phylogenomic study of birds reveals their evolutionary history. Science. 2008;320:1763–1768. doi: 10.1126/science.1157704. [DOI] [PubMed] [Google Scholar]
- 58.Del Hoyo J, Elliott A, Sargatal J, Christie DA, Juana E, editors. 2017 Handbook of the Birds of the World Alive (Lynx Edicions, Barcelona). Available at www.hbw.com. Accessed April 18, 2017.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.