Summary
The blood-brain barrier (BBB) provides a constant homeostatic brain environment that is essential for proper neural function. An unusually low rate of vesicular transport (transcytosis) has been identified as one of the two unique properties of central nervous system (CNS) endothelial cells, relative to peripheral endothelial cells, that maintain the restrictive quality of the BBB. However, it is not known how this low rate of transcytosis is achieved. Here we provide a mechanism whereby the regulation of CNS endothelial cell lipid composition inhibits specifically the caveolae-mediated transcytotic route readily used in the periphery. An unbiased lipidomic analysis reveals significant differences in endothelial cell lipid signatures from the CNS and periphery, which underlie a suppression of caveolae vesicle formation and trafficking in brain endothelial cells. Furthermore, lipids transported by Mfsd2a establish a unique lipid environment that inhibits caveolae vesicle formation in CNS endothelial cells to suppress transcytosis and ensure BBB integrity.
Keywords: blood-brain barrier, transcytosis, CNS endothelial cells, caveolae, DHA, lipid transport, Mfsd2a, Cav-1, blood vessels, lipidomic mass spectrometry
Introduction
Vascular beds throughout the body have developed structural and functional heterogeneity to meet the demands of particular organs (Aird, 2007). The central nervous system (CNS) requires a constant homeostatic microenvironment that is free of blood-borne toxins, allowing for proper synaptic transmission and information processing. The endothelial cells that line the walls of CNS blood vessels form a continuous, non-fenestrated vasculature. These cells constitute the blood-brain barrier (BBB), which regulates solute flux between blood circulation and the brain parenchyma (Andreone et al., 2015; Zhao et al., 2015). BBB disruption has been implicated in the pathogenesis of neurodegenerative diseases (Zhao et al., 2015; Zlokovic, 2008). Since BBB functionality is essential for normal brain activity, the impermeable nature of the BBB prevents brain uptake of most neurotherapeutic agents (Pardridge, 2012). This signifies a major obstacle to drug treatment of CNS diseases.
Pioneering electron microscopy (EM) work by Brightman, Reese, and Karnovsky (Brightman and Reese, 1969; Reese and Karnovsky, 1967) identified two structural features of CNS endothelial cells that underlie the restrictive properties of the BBB. First, specialized tight junctions prevent the paracellular passage of water-soluble molecules from the circulatory system to the brain parenchyma. Second, CNS endothelial cells exhibit unusually low levels of vesicle trafficking, a phenomenon that has generally been assumed to limit transcellular transport (transcytosis). While conventional wisdom has emphasized the importance of tight junctions, recent advances have highlighted the regulation of transcytosis as a crucial factor in limiting BBB permeability under physiological conditions and after pathological assaults (Armulik et al., 2010; Ben-Zvi et al., 2014; Daneman et al., 2010b; Knowland et al., 2014). However, the mechanism by which the suppression of transcytosis is achieved in CNS endothelial cells is completely unknown. An understanding of how transcytosis is regulated to maintain BBB integrity is not only fundamental to understanding BBB function, but also to developing therapeutic strategies to open transiently the BBB for CNS drug delivery.
Transcytotic pathways in epithelial and endothelial cells fall into two categories: clathrin-mediated and non-clathrin-mediated (Mayor et al., 2014). The mechanisms of clathrin-mediated vesicle trafficking are the most-studied; these studies have identified the transferrin receptor-mediated transcytosis of iron as a key pathway that has been targeted for manipulation at the BBB for drug delivery (Bien-Ly et al., 2014; Niewoehner et al., 2014). Non-clathrin-mediated vesicle trafficking pathways utilize a broad range of large- and small-scale membrane invaginations that harness diverse cellular machinery, including dynamin, coat proteins, small GTPases, and RhoGAP proteins (Mayor et al., 2014). Among the many non-clathrin-mediated pathways, caveolae-mediated vesicle trafficking predominates as a transcytotic route used in the endothelial cells of peripheral organs, including heart and lung, to transport solutes from the blood to underlying tissue (Oh et al., 2007; Oh et al., 2014; Predescu et al., 1997; Schubert et al., 2001). Caveolae are 50–100 nm invaginations in the plasma membrane (Bastiani and Parton, 2010). The plasma membrane domains that form caveolae vesicles are highly ordered and enriched in saturated phospholipids, sphingolipids, ethanolamine plasmalogens (PE-p), and cholesterol (Parton and del Pozo, 2013; Pike et al., 2002). Additionally, the formation of caveolae vesicles requires the activity of both caveolin coat proteins and cytosolic adaptor proteins belonging to the cavin family (Bastiani and Parton, 2010). Although diverse functions have been identified for caveolae, their physiological role in different cell types and tissues is not fully understood (Cheng and Nichols, 2015).
Previously, we established that Major facilitator superfamily domain containing 2a (Mfsd2a) is a key regulator for BBB function (Ben-Zvi et al., 2014). Mfsd2a is specifically expressed in CNS endothelial cells. Genetic ablation of Mfsd2a in mice results in a leaky BBB, as extravasation of several intravenously injected tracers, including 10kD- and 70kD-dextrans, NHS-biotin, and horseradish peroxidase (HRP), into the brain parenchyma is observed in Mfsd2a−/− mice from embryonic stages through adulthood. EM examination of brain endothelial cells from Mfsd2a−/− mice reveals increased transcytosis, as the intracellular vesicle number significantly increases. Additionally, HRP-injected mutants exhibit HRP-filled vesicles invaginating from the luminal plasma membrane, within the endothelial cell cytoplasm, and exocytosing at the abluminal plasma membrane. While these Mfsd2a−/− mice have transcytotic defects, their tight junctions are normal. These findings demonstrate that Mfsd2a is specifically required to regulate transcytosis in CNS endothelial cells of the BBB and that the tracer extravasation observed in Mfsd2a−/− mice is solely due to increased levels of transcytotic vesicles within these cells. Therefore, Mfsd2a may serve as an ideal molecular handle to understand the mechanism by which transcytosis is regulated at the BBB.
There are several potential mechanisms whereby Mfsd2a regulates transcytosis in CNS endothelial cells, such as affecting transcytotic machinery via direct or indirect physical interactions or via controlling plasma membrane tension. Interestingly, in addition to its role in BBB function, Mfsd2a has been identified as a lipid transporter at the luminal plasma membrane of CNS endothelial cells to deliver the essential omega-3 fatty acid docosahexaenoic acid (DHA) into the brain (Nguyen et al., 2014). However, how DHA travels from the CNS endothelial cell plasma membrane to neurons is undetermined. DHA is heavily implicated in brain development (Innis, 2007), and indeed, Mfsd2a−/− mice have reduced total brain levels of DHA species and exhibit microcephaly (Nguyen et al., 2014). Therefore, although there is no existing example of lipids in endothelial cells playing a role in BBB function, examining if Mfsd2a’s lipid transport function is related to BBB integrity will help narrow down the possible mechanisms of Mfsd2a-mediated regulation of transcytosis.
In this study, we show how transcytosis is suppressed in CNS endothelial cells to ensure proper BBB function by focusing on Mfsd2a. Using a combination of mouse genetics, lipidomic mass spectrometry, and EM analysis, we elucidate a pathway underlying the cellular mechanism of BBB function: lipids transported by Mfsd2a create a unique lipid composition of CNS endothelial cells that inhibits specifically caveolae-mediated transcytosis to maintain BBB integrity. Moreover, unbiased lipidomic analyses reveal that the lipid signatures of endothelial cells from the CNS and the periphery exhibit significant differences, which underlie the suppression of caveolae vesicle formation and trafficking in brain endothelial cells. Thus, this study establishes that lipid composition of CNS endothelial cells serves as a key new player in the regulation of transcytosis and barrier permeability.
Results
Mfsd2a Cell-Autonomously Suppresses Vesicular Pit Formation and Cargo Uptake at the Plasma Membrane
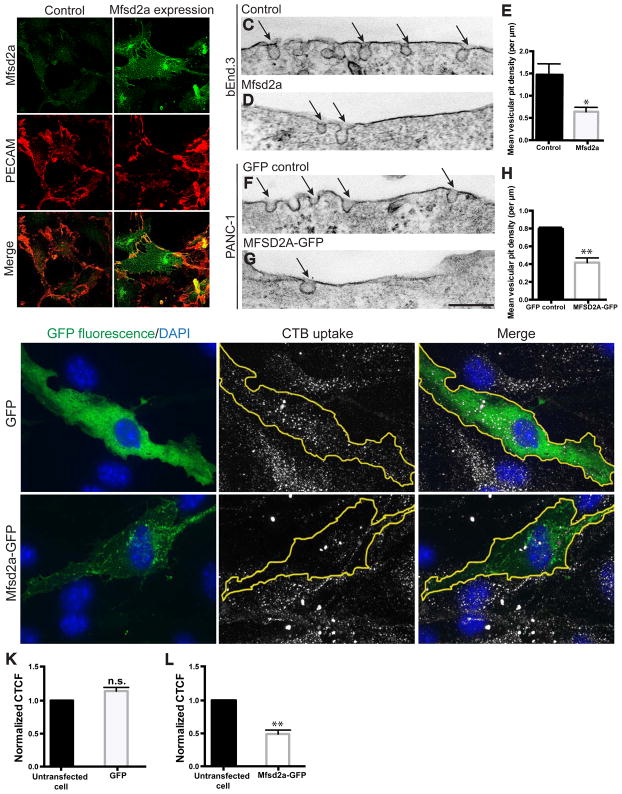
To understand how transcytosis is regulated at the BBB, we first asked whether Mfsd2a is sufficient to suppress endocytic vesicle formation, the first step of all transcytotic pathways. Mfsd2a was expressed using lentivirus with high efficiency in the immortalized mouse brain endothelial cell line bEnd.3, in which endogenous Mfsd2a expression was not detectable (Figure 1A). Exogenous expression of Mfsd2a displayed cell surface localization in bEnd.3 cells (Figure 1B), as well as cytoplasmic puncta, as seen in other cell lines (Nguyen et al., 2014; Reiling et al., 2011). Electron microscopy revealed a reduction in the number of apical (analogous to luminal) micro-invaginations (pits) in cells expressing Mfsd2a compared to mock infected control cells (Figures 1C–1E, Table S4A). A similar reduction in vesicular pit number was observed in a human pancreatic cancer cell line, PANC-1, stably transfected with human MFSD2A-GFP, compared to control cells stably transfected with GFP alone (Figures 1F–1H, Table S4A). These findings suggest that Mfsd2a expression is sufficient to inhibit vesicle formation.
Figure 1. Mfsd2a expression suppresses plasma membrane vesicular pit formation and cargo uptake in multiple cell types. See also Table S4.
(A–B) Expression of Mfsd2a via lentivirus infection in bEnd.3 cells, in which endogenous Mfsd2a expression was not detectable. Mfsd2a (green) in bEnd.3 cells displays cell surface localization and co-localization with PECAM (red), a cell surface marker (arrows). Scale bar, 50 μm. (C–H) Electron microscopy images showing the apical plasma membrane of bEnd.3 cells expressing mouse Mfsd2a (D) or PANC-1 cells expressing human MFSD2A-GFP (G). Mfsd2a-expressing cells show reduced vesicular pit density (arrows) in both cell types, compared to mock infected control bEnd.3 cells (C) or PANC-1 cells expressing GFP alone (F). n = 3, 25 image profiles from at least 15 cells per experiment. Scale bar, 150 nm. (I–J) Mfsd2a-expressing cells exhibit reduced cholera toxin subunit-B (CTB) uptake. Uptake of CTB (gray; middle panels) in bEnd.3 cells expressing either soluble GFP (I) or Mfsd2a-GFP (J) (green, GFP fluorescence; blue, DAPI; lefts panels.). Scale bar, 20 μm. (K–L) CTB uptake quantified as corrected total cell fluorescence (CTCF) per transfected cell compared to CTCF of at least 2 untransfected cells per image. n = 4, at least 15 transfected cells per experiment. All data are mean ± s.e.m.; n.s., not significant, * P < 0.05, ** P < 0.01 (Student’s t-test).
To further assess whether Mfsd2a can directly inhibit cargo uptake, we next loaded confluent bEnd.3 cell cultures with a fluorescent cholera toxin subunit-B (CTB) tracer (Figures 1I and 1J). Cells that express mouse Mfsd2a-GFP exhibited a 50% reduction in CTB uptake, as quantified by fluorescence intensity (Figure 1L), compared to cells that express GFP alone (Figure 1K). Use of the GFP-tagged constructs did not affect Mfsd2a protein localization (Reiling et al., 2011) (data not shown), and allowed for identification of positively transfected cells without quenching of the CTB fluorescence due to immunostaining protocol. These gain-of-function results are consistent with previous in vivo loss-of-function data (Ben-Zvi et al., 2014), and together demonstrate that Mfsd2a is necessary and sufficient for the suppression of vesicle formation and cargo uptake, cellular processes underlying transcytosis. Furthermore, they suggest that Mfsd2a acts via a direct mechanism to suppress CNS endothelial cell transcytosis in a cell-autonomous manner.
Mfsd2a Lipid Transport Function is Required for Suppression of Transcytosis and BBB Integrity
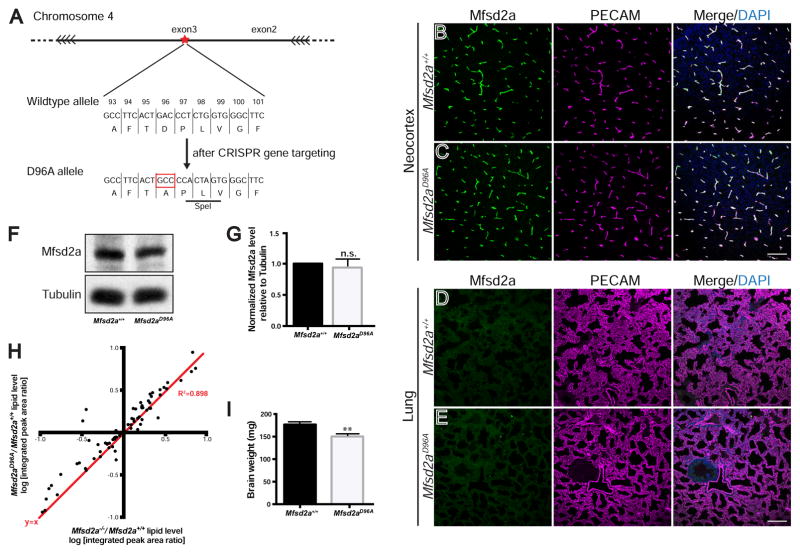
To investigate how Mfsd2a may act directly at the CNS endothelial cell plasma membrane to regulate vesicle trafficking, we first asked whether the reported lipid transport function of Mfsd2a is required to suppress transcytosis. Although there is no known role of lipids in CNS endothelial cells for BBB function, answering this question will help to narrow down and pinpoint Mfsd2a’s precise mechanism of action. Specifically to address this question, we generated a transporter-dead Mfsd2a knock-in mouse as an in vivo tool. Asp96 is a conserved amino acid required for the cation binding that is crucial for transporter activity in MFS family members (Ethayathulla et al., 2014), and the D96A aspartic acid to alanine point mutation abolishes Mfsd2a transport function in vitro (Nguyen et al., 2014; Reiling et al., 2011). A mouse line harboring the Mfsd2aD96A mutation in the endogenous Mfsd2a locus was generated via CRISPR/Cas9 gene editing. Specifically, one amino acid change resulting in the D96A missense mutation was introduced into exon 3 of Mfsd2a (Figures 2A, S1B, and S1C). Importantly, there was no observable difference in Mfsd2a expression pattern in Mfsd2aD96A/D96A (hereafter denoted as Mfsd2aD96A) mutants compared to Mfsd2a+/+ littermates by immunohistochemistry of neocortex and cerebellum (Figures 2B, 2C, S1D, and S1E). No difference in Mfsd2a expression was observed by Western blot analysis of brain lysate (Figures 2F and 2G), indicating that Mfsd2aD96A mutants express normal levels of Mfsd2a protein. Finally, Mfsd2a expression was restricted to BBB-containing CNS vasculature in Mfsd2aD96A mutants, and was absent in non-BBB-containing vasculature of peripheral organs, including the lung and liver (Figures 2E and S1G), similar to Mfsd2a+/+ littermates (Figures 2D and S1F).
Figure 2. A novel Mfsd2aD96A mouse abolishes the lipid transport function of Mfsd2a and displays microcephaly. See also Figures S1 and S2; Tables S1 and S2.
(A) Gene targeting strategy for the generation of Mfsd2aD96A mice. Magnified view of the genomic region that contains residue D96 in exon 3 of Mfsd2a. CRISPR/cas9 genome editing introduces the D96A point mutation (red box), as well as two additional silent mutations in codons 97/98 that generate a SpeI restriction site (black underline) for screening purposes. For additional details, see STAR methods. (B–C) Immunostaining at P4 of Mfsd2a protein (green) reveals vascular expression (purple, PECAM) in the neocortex of both Mfsd2a+/+ control (B) and Mfsd2aD96A mutants (C). n = 3 animals per genotype. Scale bar, 100 μm. (D–E) Immunostaining at P4 reveals absence of Mfsd2a protein expression in non-BBB-containing lung vasculature (purple, PECAM) of both Mfsd2a+/+ control (D) and Mfsd2aD96A mutants (E). n = 3 animals per genotype. Scale bar, 100 μm. (F) Western blot from P4 brain lysate of Mfsd2aD96A mutants and littermate Mfsd2a+/+ controls. (G) Quantification shows equal Mfsd2a protein levels between Mfsd2aD96A mutants and littermate Mfsd2a+/+ controls. Tubulin (loading control). n = 3 littermate pairs per genotype. Data are mean ± s.e.m.; n.s., not significant (Student’s t-test). (H) The lipid profile of Mfsd2aD96A mutant brains closely mimics that of Mfsd2a−/− mutant brains at P4. Dot plot representation of Mfsd2a−/−/Mfsd2a+/+ log-ratio (x-axis), compared to Mfsd2aD96A/Mfsd2a+/+ log-ratio (y-axis) of phospholipid levels measured by mass spectrometry. Each dot represents a single lipids species; red line: y=x; R2 = 0.898. Mfsd2aD96A experiment: n = 6 animals per genotype; Mfsd2a−/− experiment: n = 4 animals per genotype. (I) Mfsd2aD96A mutants exhibit microcephaly. Quantification of brain weight at P4. n = 5 animals per genotype. Data are mean ± s.e.m.; ** P < 0.01 (Student’s t-test).
To examine the effect of the Mfsd2aD96A mutation on the lipid transport function of Mfsd2a in vivo, we performed lipidomic mass spectrometry on brain lysate at postnatal stages. Lipidomic analysis of brain lysate revealed an altered lipid profile in Mfsd2aD96A mutants (Figure S2A), with the level of a broad range of phospholipid species changed at postnatal day 4 (P4), including but not limited to DHA-containing species. Importantly, the lipid profile of Mfsd2aD96A mutant brains closely mimicked that of Mfsd2a−/− mice, as the Mfsd2a−/−/Mfsd2a+/+ log-ratio was very similar to the Mfsd2aD96A/Mfsd2a+/+ log-ratio of phospholipid levels measured by mass spectrometry (Figures 2H, S2A, and S2B, Tables S1 and S2). Together, these data demonstrate that the Mfsd2aD96A mutation abolishes the lipid transport function of Mfsd2a in vivo without affecting protein expression, resulting in a lipid profile similar to that observed through global loss of Mfsd2a. Finally, Mfsd2aD96A mutants exhibited microcephaly at P4 (Figure 2I), recapitulating the phenotype observed in Mfsd2a−/− mice (Nguyen et al., 2014) and confirming that Mfsd2a lipid transport function is required for brain development.
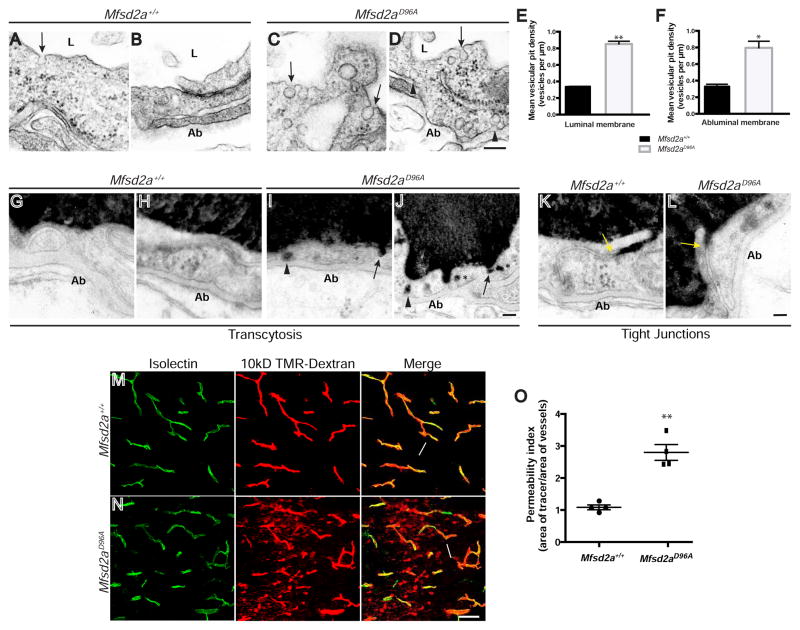
With this new genetic tool, we next asked if Mfsd2a lipid transport function is necessary for suppression of transcytosis and control of BBB permeability. Previously, it was shown that Mfsd2a−/− mice exhibit BBB leakage that is due to increased levels of transcytosis in CNS endothelial cells (Ben-Zvi et al., 2014). As an increased vesicle number in CNS endothelial cells is the hallmark of the increased transcytosis phenotype observed in Mfsd2a−/− mice, we examined vesicle number in Mfsd2aD96A mutants at P4 using EM (Figures 3A–3D). Mfsd2aD96A mutants exhibited a significant increase in density of both luminal and abluminal membrane-connected vesicles (Figures 3E, 3F, Table S4B), similar to the phenotype observed in Mfsd2a−/− mice, indicating an up-regulation of transcytosis. We next injected HRP into adult Mfsd2aD96A mutants and Mfsd2a+/+ littermates and examined sub-cellular tracer localization. While HRP tracer was confined to the vessel lumen in Mfsd2a+/+ mice (Figures 3G and 3H), tracer in Mfsd2aD96A mutants was observed being taken up into intracellular vesicles at the luminal plasma membrane, within the cytoplasm, and at the abluminal plasma membrane (Figures 3I and 3J), indicating that the upregulated vesicles in Mfsd2aD96A mutants act as transcytotic carriers. In both Mfsd2a+/+ wild-type littermates and Mfsd2aD96A mice, HRP tracer entered the space between CNS endothelial cells but was sharply halted at tight junction “kissing points” (Figures 3K and 3L), indicating that tight junctions are functional. Together, these data demonstrate that loss of Msfd2a transport function results in increased CNS endothelial cell transcytosis, without impacting tight junction integrity.
Figure 3. The lipid transport function of Mfsd2a is required for the suppression of transcytosis and maintenance of BBB integrity in vivo. See also Figures S3 and S6; Table S4.
(A–D) Increased vesicular density in CNS endothelial cells of Mfsd2aD96A mutants (C–D) compared to their littermate controls (A–B). Electron microscopy analysis at P4 reveals increased number of vesicles in Mfsd2aD96A mutants along the luminal (arrows) and abluminal (Ab; black arrowheads) plasma membranes. Scale bar, 150 nm. (E–F) Quantification of vesicular density along the luminal (E) and abluminal (F) plasma membranes. n = 3 animals per condition, 20 capillary cross-sections per animal. (G–J) Increased transcytosis is evident in CNS endothelial cells of HRP-injected Mfsd2aD96A mutants at P90. HRP tracer (black) is confined with the vessel lumen (L) of littermate control animals (G–H). Tracer-filled vesicles are observed invaginating from the luminal plasma membrane (arrows), within the cytoplasm (asterisks), and along to abluminal plasma membrane (arrowheads) in Mfsd2aD96A mutants (I–J). n = 3 animals per condition. Scale bar, 100 nm. (K–L) Tight junctions are functional in CNS endothelial cells of HRP-injected Mfsd2aD96A mutants. HRP tracer fills the vessel lumen (L) and enters intercellular clefts but is sharply halted at tight junction “kissing points” (yellow arrows) in both Mfsd2aD96A mutants (L) and Mfsd2a+/+ littermate control animals (K) at P90. n = 3 animals per condition. Scale bar, 100 nm. (M–N) Mfsd2aD96A mutants display BBB leakage. 10kD TMR-dextran tracer (red) is completely confined within vessels (green, Isolectin B4 (IB4)) (arrows) in Mfsd2a+/+ littermate control animals (M) at P4. Tracer-filled parenchyma cells (arrowheads) surround vessels in the brain of Mfsd2aD96A mutants (N). Scale bar, 50 μm. (O) Permeability index of tracer leakage in brain, as quantified by area of tracer divided by area of vessels per image (value = 1 signifying no leakage). n = 4 animals per genotype. All data are mean ± s.e.m.; * P < 0.05, ** P < 0.01 (Student’s t-test).
To determine whether the observed increased transcytosis results in BBB leakage in Mfsd2aD96A mutants, we performed intravenous injection of fluorescent tracer at P4 and examined tracer localization as a metric of BBB integrity. We chose 10kD-dextran tracer for analysis, as robust extravasation of this tracer was previously observed in Mfsd2a−/− mice (Ben-Zvi et al., 2014). While tracer was completely confined within the vasculature of Mfsd2a+/+ littermate control animals (Figure 3M), tracer extravasation was observed throughout the brain of Mfsd2aD96A mutants (Figure 3N), with tracer diffused into the brain parenchyma and taken up by non-vascular cells. Permeability index quantification (defined as area of tracer/area of vessels) revealed a statistically significant increase of BBB leakage in Mfsd2aD96A mutants compared to Mfsd2a+/+ controls (Figure 3O). BBB leakage in Mfsd2aD96A mutants persisted into adulthood, as HRP tracer extravasation into the forebrain parenchyma was observed at P90 (Figures S3A and S3B). Taken together, these data demonstrate that BBB leakage occurs in Mfsd2aD96A mice via increased transcytosis, similar to Mfsd2a−/− mice. Furthermore, they strongly implicate the lipid transport function of Mfsd2a in the suppression of vesicle trafficking.
Brain Capillaries Possess a Lipid Signature Underlying Suppression of Caveolae
Our results demonstrate that Mfsd2a cell-autonomously suppresses vesicle trafficking via a lipid transport-dependent mechanism. This raises the possibility that lipid transport directly affects the membrane lipid composition of CNS endothelial cells, which in turn may result in the suppression of transcytosis at the BBB. To further understand the link between lipids and vesicle trafficking, we next asked whether the lipid composition of CNS endothelial cells, a previously unexplored facet of BBB function, shows any differences compared to peripheral endothelial cells. To investigate this possibility, we performed lipidomic mass spectrometry analysis on acutely isolated capillaries from brain and lung of wild-type mice at P4. Lung vasculature does not possess the restrictive barrier properties of CNS vasculature, and lung endothelial cells are enriched in transcytotic vesicles, which have been shown to transport cargo from the circulation to the lung parenchyma (Ghitescu et al., 1986; Oh et al., 2007; Schubert et al., 2001). Western blot analysis showed that this capillary preparation is highly enriched in endothelial cells in both brain and lung, indicated by an abundance of VE-cadherin protein compared to total tissue lysate (Figure S4B). Residual pericytes were present in both brain and lung capillary preparations, and importantly, the brain capillary preparation was free of both neurons (NeuN) and astrocyte endfeet (Aquaporin-4) (Figures S4A and S4B).
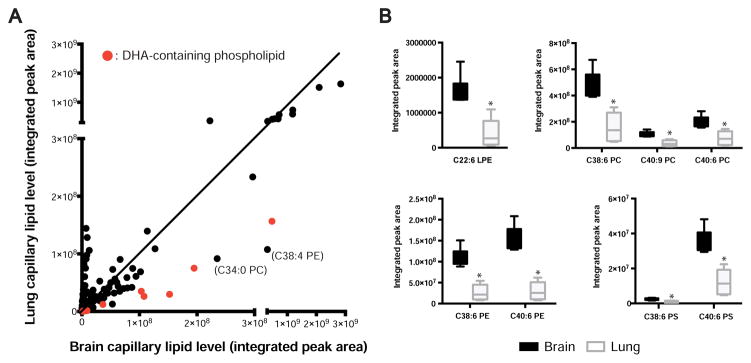
Lipidomic analysis revealed that although most lipid species were found at similar levels in isolated brain and lung capillaries, significant differences in several lipid species were observed (Figures 4A and S4C, Table S3). Remarkably, one of the most significantly enriched lipid populations in brain capillaries was DHA-containing phospholipid species in lysophosphatidylethanolamine (LPE), phosphatidylcholine (PC), phosphatidylethanolamine (PE), and phosphatidylserine (PS) subclasses, ranging from 1.9– to 5.1-fold enrichment (Figure 4B). Significant enrichment of a few non-DHA-containing lipids, such as C34:0 PC and C38:4 PE was also observed in brain capillaries (Figure 4A). On the other hand, modest decreases in several ethanolamine plasmalogen (PE-p) species were observed in brain capillaries compared to lung capillaries (Figure S5A). Although both the acutely isolated brain and lung capillary preparations retained residual pericytes, it unlikely that these cells contribute to the observed differences in lipid signature, as pericytes do not express Mfsd2a (Ben-Zvi et al., 2014), the only known DHA transporter in the brain, there is no evidence that pericytes are able to synthesize DHA de novo, and both brain and lung capillary preparations were highly enriched in endothelial cells.
Figure 4. Lipidomic profiling of brain versus lung capillaries reveals different lipid signatures, including changes in DHA-containing phospholipid species. See also Figures S4 and S5; Table S3.
(A) Dot plot representation of lipidomic profiling of brain versus lung capillaries from P4 wild-type mice. Dots represent average lipid level in the brain (x-axis) and lung (y-axis) for each metabolite tested, expressed as mass spectrometry integrated peak area. Red dots indicate DHA-containing phospholipid species. (B) DHA-containing phospholipid levels are increased in brain capillaries. Lipidomic analysis of DHA-containing phospholipid species from brain and lung capillaries of P4 wild-type mice. Value for each species is normalized to internal standard and expressed as integrated mass spectrometry peak area. (LPE, lyso-phosphatidylethanolamine; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PS, phosphatidylserine). * P < 0.004 (Student’s t-test, with Bonferroni correction for multiple comparisons). For all data, n = 6 groups per organ, 8 animals pooled per group.
Notably, lung endothelial cells have abundant caveolae vesicles that can act as transcytotic carriers (Ghitescu et al., 1986; Oh et al., 2007; Schubert et al., 2001). The formation and functionality of caveolae membrane domains can be affected by plasma membrane lipid composition (Lingwood and Simons, 2010; Parton and del Pozo, 2013). In particular, DHA is a highly unsaturated fatty acid that disrupts liquid-ordered caveolae domains, normally enriched in saturated phospholipids, sphingolipids, PE-p species, and cholesterol. DHA displaces Caveolin-1 (Cav-1), a coat protein of the caveolin family that is obligatory for caveolae vesicle formation, from the plasma membrane. Thus, enrichment of DHA in the plasma membrane renders caveolae membrane domains less capable of forming caveolae vesicles (Chen et al., 2007; Li et al., 2007; Ma et al., 2004). Therefore, the lipid composition of brain capillaries enriched in DHA species and low in PE-p species may reflect a plasma membrane microenvironment in CNS endothelial cells that is unfavorable for the assembly of functional caveolae domains and subsequent vesicle formation. Together, these analyses reveal significant differences in the lipid signature of BBB-containing brain capillaries versus non-BBB-containing lung capillaries, some of which may contribute to the suppression of transcytosis at the BBB by inhibiting caveolae vesicle formation and trafficking in CNS endothelial cells.
Significantly reduced levels of several triglycerides (TAG), and cholesteryl esters (CE) species were observed in brain capillaries versus lung capillaries (Figures S5B and S5C). TAG and CE are stored in intracellular lipid droplets (Guo et al., 2009), and both in vitro and in vivo evidence suggests that the storage of these lipids depends on caveolae vesicle trafficking (Cohen et al., 2004; Martin and Parton, 2005; Pol et al., 2004). Therefore, low levels of TAG and CE in brain capillaries may reflect a low level of caveolae trafficking. Together these observations raise the possibility that caveolae vesicles may be the subtype increased in CNS endothelial cells that accounts for the leaky BBB in Mfsd2a−/− mice.
Caveolae Vesicle Trafficking is Upregulated in Mfsd2a−/− Mice
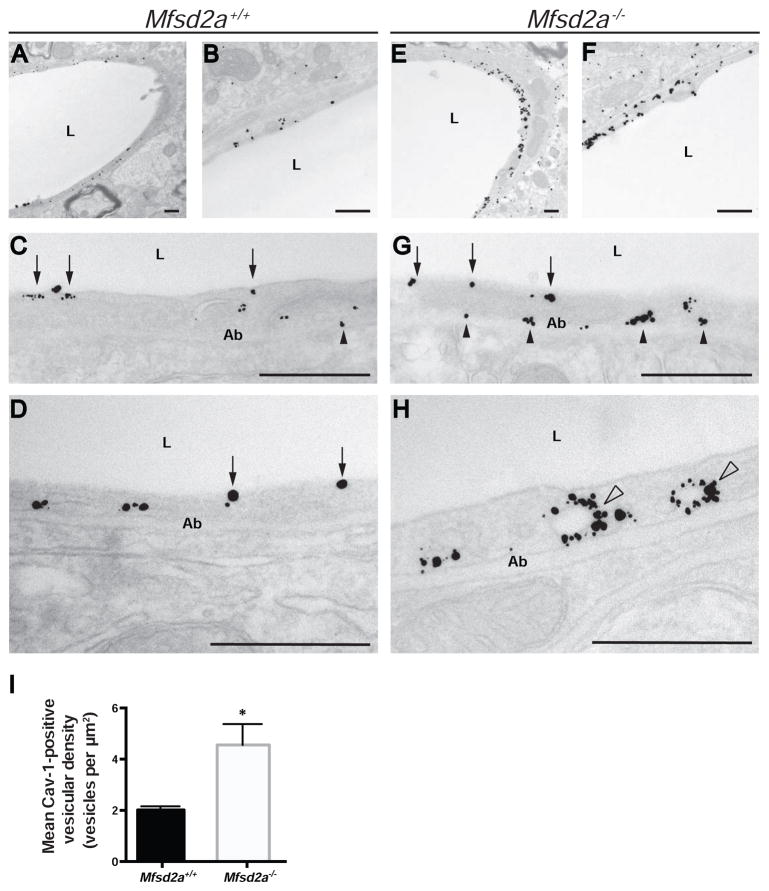
To test this hypothesis, we performed Cav-1 immuno-electron microscopy labeling in brains of Mfsd2a−/− mice and control littermates (Figures 5A–5H). Cav-1 immunoreactivity (gold particles) was seen along the luminal plasma membrane in CNS endothelial cells of Mfsd2a+/+ control mice, whereas immunoreactivity in Mfsd2a−/− mice was observed along the luminal and abluminal plasma membranes (Figure 5G), and surrounding vesicle structures in the cytoplasm (Figure 5H). Additionally, there was a significant increase in the number of Cav-1-positive vesicles in CNS endothelial cells of Mfsd2a−/− mice compared to controls (Figure 5I, Table S4C). These data suggest that caveolae vesicle trafficking is upregulated in mice lacking Mfsd2a, and that Mfsd2a may normally act to suppress caveolae-mediated transcytosis in CNS endothelial cells.
Figure 5. Mfsd2a−/− mice display an increased number of Cav-1-positive vesicles in CNS endothelial cells. See also Table S4.
(A–H) Cav-1 immunoreactivity in CNS endothelial cells of Mfsd2a+/+ and Mfsd2a−/− mice under electron microscopy. Electron microscopy analysis with immuno-gold labeling of Cav-1 protein in CNS endothelial cells of Mfsd2a+/+ (A–D) and Mfsd2a−/− mice (E–H) at P90. Gold particles (black puncta) are visualized along the luminal (arrows) and abluminal (Ab; black arrowheads) plasma membranes, as well as within the cytoplasm. Cytoplasmic particles are seen associated with vesicular structures (white arrowheads). All scale bars, 500 nm. (I) Quantification of Cav-1-positive vesicles in Mfsd2a+/+ and Mfsd2a−/− mice. n = 3 animals per genotype, 10 capillary cross-sections per animal. Data are mean ± s.e.m.; * P < 0.05 (Student’s t-test).
Caveolae-mediated Transcytosis is Specifically Suppressed Downstream of Mfsd2a Function at the BBB in vivo
To directly test the idea that the caveolae-mediated transcytosis pathway is suppressed downstream of Mfsd2a function at the BBB, we performed a genetic rescue experiment by crossing Mfsd2a−/− with Cav-1−/− mice, as Cav-1 is obligatory for caveolae formation. While Cav-1−/− mice are viable, they are unable to form caveolae vesicle structures and display defects in cargo uptake in vivo (Razani et al., 2001; Schubert et al., 2001). If the caveolae pathway is downstream of Mfsd2a in vivo, then crossing Cav-1−/− mice with Mfsd2a−/− mice should reverse the upregulated transcytosis seen in Mfsd2a−/− mice, as CNS endothelial cells of Mfsd2a−/−; Cav-1−/− double knockouts will no longer be able to form caveolae vesicles to act as transcytotic carriers. Alternatively, if Mfsd2a suppresses other transcytotic pathways, increased transcytosis will still be observed in Mfsd2a−/−; Cav-1−/− double knockouts.
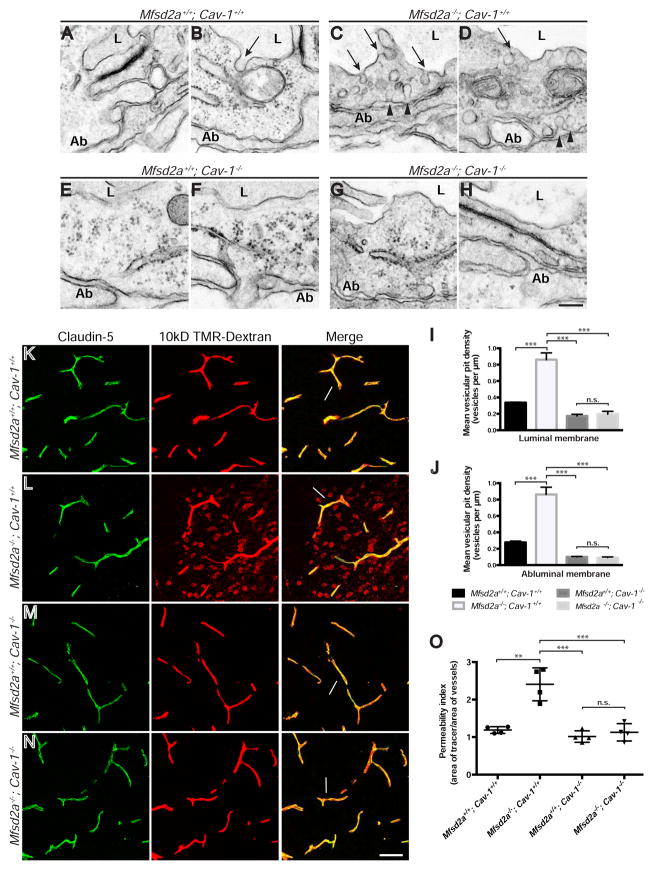
Similar to previous results (Ben-Zvi et al., 2014), EM analysis revealed occasional vesicles in Mfsd2a+/+; Cav-1+/+ wild-type CNS endothelial cells at P5 (Figures 6A and 6B), with an over 2.5-fold increase in total luminal and abluminal membrane-connected vesicle density in Mfsd2a−/−; Cav-1+/+ single knockouts (Figures 6C–6D and 6I–6J). Consistent with the finding that Cav-1−/− mice are unable to form caveolae vesicles, we observed a nearly 2-fold decrease in vesicle density in Mfsd2a+/+; Cav-1−/− single knockouts compared to wild-type levels (Figures 6E–6F and 6I–6J). However, in Mfsd2a−/−; Cav-1−/− double knockouts, a complete reversal of the Mfsd2a−/− single mutant phenotype was evident, with vesicle density mimicking that of Cav-1−/− single mutants (Figures 6G–6J, Table S4D). Therefore, the caveolae-mediated transcytosis pathway is the one that is normally suppressed by Mfsd2a. In contrast, no significant difference in the density of clathrin-coated pits, identified under EM by the presence of an electron dense clathrin coat (Figure S6A), was observed in Mfsd2a−/−, Cav-1−/−, Mfsd2a−/−; Cav-1−/−, or Mfsd2aD96A mutants compared to their littermate controls (Figure S6B–S6E, Tables S4E and S4F). Together, these data demonstrate that Mfsd2a acts upstream of Cav-1 specifically to suppress the caveolae vesicle trafficking pathway at the BBB.
Figure 6. Genetic inhibition of the caveolae pathway rescues both the increased vesicular density and BBB leakage phenotypes of Mfsd2a−/− mice. See also Figures S3, S6, and S7; Table S4.
(A–H) Genetic inhibition of the caveolae pathway rescues increased vesicular density in Mfsd2a−/− animals. Electron microscopy examination of BBB integrity in Mfsd2a+/+; Cav-1+/+ (A–B), Mfsd2a−/−; Cav-1+/+ (C–D), Mfsd2a+/+; Cav-1−/− (E–F), and Mfsd2a−/−; Cav-1−/− (G–H) mice at P5. Increased vesicular pit density in Mfsd2a−/−; Cav-1+/+ single mutant CNS endothelial cells is observed along the luminal (arrows) and abluminal (Ab; arrowheads) plasma membranes. Mfsd2a+/+; Cav-1+/+ wild-type, Mfsd2a+/+; Cav-1−/− single mutant and Mfsd2a−/−; Cav-1−/− double mutant cells exhibited very few vesicles. Scale bar, 150 nm. (I–J) Quantification of vesicular pit density at the luminal (I) and abluminal (J) plasma membranes. n = 4 animals per genotype, 20 capillary cross-sections per animal. (K–N) Genetic inhibition of the caveolae pathway rescues BBB leakage defects in Mfsd2a−/− animals. 10kD-dextran tracer (red) was injected into the circulation of Mfsd2a+/+; Cav-1+/+ (K), Mfsd2a−/−; Cav-1+/+ (L), Mfsd2a+/+; Cav-1−/− (M), and Mfsd2a−/−; Cav-1−/− (N) mice at P5. Tracer diffuses into the brain parenchyma in Mfsd2a−/−; Cav-1+/+ single mutants (L), with tracer-filled parenchyma cells (arrowheads) surrounding vessels (green, Claudin-5). Tracer is confined to the vasculature (arrows) in all other tested genotypes. Scale bar, 50 μm. (O) Permeability index of tracer leakage in brain, as quantified by area of tracer divided by area of vessels per image (value = 1 signifying no leakage). n = 4 animals per genotype. All data are mean ± s.e.m.; n.s., not significant, ** P < 0.01, *** P < 0.001 (one-way ANOVA, post-hoc Tukey test).
Finally, we asked if caveolae-mediated transcytosis is required for regulating BBB permeability by examining whether genetic loss of Cav-1 is able to rescue the BBB leakage observed in the brains of Mfsd2a−/− mice. To evaluate BBB permeability, we again examined 10kD-dextran tracer localization in the brain after intravenous injection at P5. Tracer was confined within the brain vasculature of Mfsd2a+/+; Cav-1+/+ wild-type and Mfsd2a+/+; Cav-1−/− single knockouts, indicating no BBB leakage in these mice (Figures 6K and 6M). Tracer extravasation was observed in Mfsd2a−/−; Cav-1+/+ single knockouts, as tracer diffused into the brain and was taken up by non-vascular parenchymal cells (Figure 6L). A significant increase in the permeability index quantification of Mfsd2a−/−; Cav-1+/+ single knockouts was observed, compared to both Mfsd2a+/+; Cav-1+/+ wild-type and Mfsd2a+/+; Cav-1−/− single knockouts (Figure 6O). However, no leakage was observed in Mfsd2a−/−; Cav-1−/− double knockouts, as tracer was confined within the brain vasculature and the permeability index quantification revealed no significant difference to that of both Mfsd2a+/+; Cav-1+/+ wild-type and Mfsd2a+/+; Cav-1−/− single knockouts (Figures 6N, 6O). This observed rescue of BBB leakage was also evidenced at adult stages, as extravasation of HRP tracer into the forebrain parenchyma was observed in Mfsd2a−/−; Cav-1+/+ single knockouts (Figure S3D), but not wild-type, Cav-1−/− single knockout, or Mfsd2a−/−; Cav-1−/− double knockout mice at P90 (Figures S3C, S3E, and S3F). Consistent with previous findings (Knowland et al., 2014), we found that tight junctions were functional in Cav-1−/− mice, as HRP tracer was halted at tight junction “kissing points” in both Cav-1 mutants (Figures S7C and S7D) and wild-type littermates (Figures S7A and S7B) under EM. Together with our vesicle density analysis, these data indicate that increased caveolae vesicle trafficking results in the BBB leakage observed in Mfsd2a−/− mice, as genetic loss of Cav-1 was able to rescue tracer extravasation. Furthermore, of the numerous vesicular trafficking pathways currently known, these data demonstrate that Mfsd2a maintains BBB integrity by specifically suppressing the caveolae-mediated transcytotic pathway.
Caveolae-mediated Transcytosis is Not Required for Mfsd2a-mediated Transport of DHA into the Brain Parenchyma for Brain Development
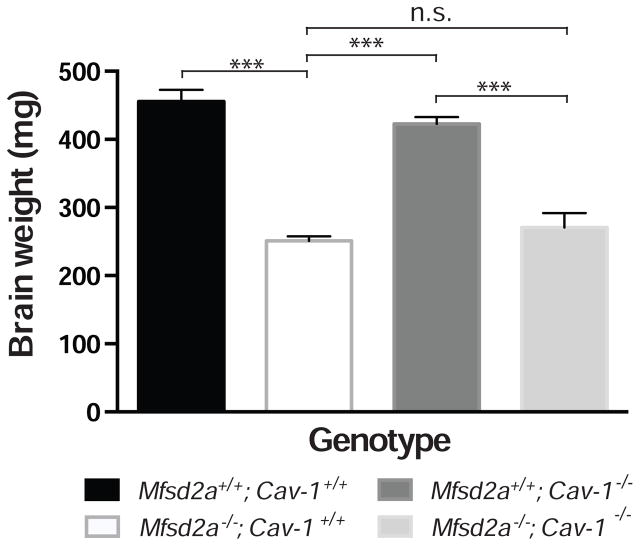
Interestingly, although Mfsd2a−/−; Cav-1−/− double knockouts were able to rescue the BBB defects seen in Mfsd2a−/− mice, they were unable to rescue the microcephaly phenotype (Figure 7). Thus, the caveolae pathway downstream of Mfsd2a in CNS endothelial cells specifically controls transcytosis and BBB function, but is not required for Mfsd2a-mediated transport of DHA into the brain parenchyma for proper brain development. Our analysis of the Mfsd2aD96A mouse together with the comparative brain and lung capillary lipidomic profiling indicate that the suppression of the caveolae pathway requires the transport of lipids, notably DHA-containing phospholipids, by Mfsd2a to regulate CNS endothelial cell plasma membrane composition and inhibit caveolae vesicle formation.
Figure 7. Genetic inhibition of the caveolae pathway fails to rescue the microcephaly defect seen in Mfsd2a−/− mice.
Quantification of brain weight at P90. n = 5 animals per genotype. Data are mean ± s.e.m.; n.s., not significant, *** P < 0.001 (one-way ANOVA, post-hoc Tukey test).
Discussion
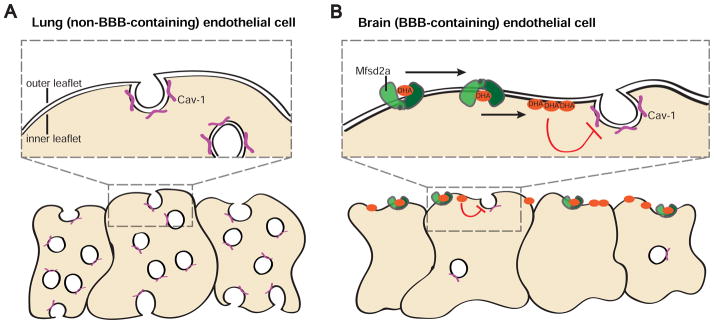
While it was observed nearly 50 years ago that CNS endothelial cells exhibit unusually low levels of transcytotic vesicles (Brightman and Reese, 1969; Reese and Karnovsky, 1967), it has remained poorly understood whether this low rate of transcytosis is important for BBB function, what type of vesicles are involved, and how vesicle formation and transcytosis are maintained at low levels in brain endothelial cells. In this study, we reveal that the unique membrane lipid composition of CNS endothelial cells regulates BBB permeability by suppressing caveolae-mediated transcytosis. Our findings support a model in which Mfsd2a-mediated transport of DHA controls the luminal plasma membrane DHA lipid composition of CNS endothelial cells such that it actively prevents the formation of functional caveolae membrane domains (Figures 8A and 8B). Thus, unlike lung endothelial cells, which have low levels of DHA and display high levels of transcytotic caveolae vesicles, Mfsd2a-mediated lipid transport into CNS endothelial cells renders them less capable of forming caveolae vesicles to act as transcytotic carriers. As a result, the BBB is functional. Together, these findings uncover a mechanism for the suppression of transcytosis in CNS endothelial cells to maintain BBB integrity, and reveal an essential role for the precise control of membrane lipid composition in the regulation of BBB permeability.
Figure 8. Model for the suppression of caveolae-mediated transcytosis via regulated membrane lipid composition at the BBB.
(A) Non-BBB containing lung endothelial cells do not express Mfsd2a, display low levels of DHA, and display high levels of transcytotic caveolae vesicles, characterized by the presence of Cav-1 coat protein (purple). (B) BBB-containing brain endothelial cells express Mfsd2a (green). Mfsd2a acts as a lipid flippase, transporting phospholipids, including DHA-containing species (orange), from the outer to inner plasma membrane leaflet. The increased levels of DHA, and presumably other lipid changes, alter the plasma membrane composition such that caveolae vesicles are unable to form and act as transcytotic carriers. Thus, brain endothelial cells display low levels of caveolae vesicles. This suppression of caveolae formation and trafficking subsequently ensures BBB integrity under normal conditions.
The Unique Lipid Composition of CNS Endothelial Cells Underlies BBB Function
Although the field has successfully identified several key gene and protein regulators for BBB function (Ben-Zvi et al., 2014; Chow and Gu, 2015; Daneman et al., 2010a; Junge et al., 2009; Liebner et al., 2008; Sohet et al., 2015; Tam et al., 2012; Zhou et al., 2014), the question of whether lipids can regulate BBB function has yet to be examined. Our demonstration of the in vivo requirement of the lipid transport function of Mfsd2a for the suppression of transcytosis and BBB permeability, coupled with our result of lipidomic profiling of capillaries from brain versus lung, reveal lipid composition of CNS endothelial cells as a key regulator for BBB function. Previous work on Mfsd2a has focused on the deleterious consequences of decreased DHA levels in brain parenchymal cells, such as behavioral abnormalities and microcephaly (Alakbarzade et al., 2015; Guemez-Gamboa et al., 2015; Nguyen et al., 2014). Our results demonstrate an additional direct functional role of DHA transport at the CNS endothelial cell luminal plasma membrane, the site of Mfsd2a expression in the brain, for suppression of caveolae transcytosis and maintenance of BBB integrity. Besides DHA species, C34:0 PC and C38:4 PE are also significantly enriched in brain capillaries relative to lung capillaries, and their function at the BBB remains to be determined. Together, our Mfsd2aD96A mouse model and brain versus lung lipidomic profiling illuminate a previously unknown role of CNS endothelial cell lipid composition in BBB integrity. These findings may usher in new lines of research into how the metabolomic, in addition to the proteomic, signature of CNS endothelial cells regulates BBB function.
Suppression of Caveolae-Mediated Transcytosis is Essential for BBB Function
Compared to the relatively well-studied mechanisms of clathrin-mediated vesicle trafficking, the physiology of caveolae is less understood. Caveolae have multiple putative functions at the plasma membrane, including cell signal transduction, mechanosensation, lipid metabolism, and plasma membrane organization, and their physiological role in different cell types and tissues is not fully clear (Cheng and Nichols, 2015; Parton and del Pozo, 2013). While endothelial cells in peripheral organs, such as the lung and heart, are enriched in caveolae, and previous studies have demonstrated that caveolae vesicles act as transcytotic carriers in these peripheral organ endothelial cells (Ghitescu et al., 1986; Oh et al., 2007; Oh et al., 2014; Schubert et al., 2001), there remains a long-standing debate as to what role, if any, caveolae play in transcytosis. In particular, recent in vitro studies have challenged the notion that caveolae significantly contribute to endocytic or transcytotic flux (Bitsikas et al., 2014; Shvets et al., 2015). Our genetic rescue experiments demonstrate that the presence of transcytotic caveolae vesicles account for BBB leakage in the absence of Mfsd2a, and thus assign a physiological role to caveolae-mediated transcytosis in CNS endothelial cells to regulate BBB permeability.
Regulated Lipid Composition Suppresses Caveolae-Mediated Transcytosis for BBB Function
The mutation of a single amino acid caused Mfsd2aD96A transporter-dead mice to phenocopy the increased transcytosis and BBB leakage defects observed in Mfsd2a−/− global knockout mice. Our unbiased lipidomic analysis showing DHA species as one of the most significantly enriched lipid populations in brain capillaries compared to lung capillaries further point to a direct role of DHA in the suppression of caveolae vesicles. How does DHA inhibit caveolae vesicles? Caveolae formation depends on both Cav-1 and cholesterol (Lingwood and Simons, 2010; Parton and del Pozo, 2013). Our findings support previous studies that have shown that DHA treatment of human retinal endothelial cells and human umbilical vein endothelial cells resulted in a significant depletion of cholesterol in the membrane as well as the displacement of Cav-1 and several signaling proteins resident in caveolae (Chen et al., 2007; Li et al., 2007). Moreover, in vivo studies show that feeding mice with omega 3 fatty acid enriched diet resulted in a concomitant reduction of cholesterol and Cav-1 in mouse colon epithelium (Ma et al., 2004). Therefore, it is likely that in CNS endothelial cells, Mfsd2a acts as a lipid flippase (Quek, et al., 2016), transporting phospholipids, including DHA-containing species, from the outer to the inner plasma membrane leaflet. The increased levels of DHA in the inner plasma membrane leaflet subsequently cause the displacement of cholesterol and Cav-1, thereby inhibiting caveolae formation. It is plausible that DHA enrichment in the plasma membranes results in higher membrane fluidity and low membrane curvature that is not favorable for caveolae formation.
In light of the emerging evidence in both the normal and diseased brain that transcytosis represents a major mechanism for BBB function (Armulik et al., 2010; Ben-Zvi et al., 2014; Chow and Gu, 2017; Daneman et al., 2010b; Haley and Lawrence, 2016; Knowland et al., 2014), our work provides a mechanistic understanding of how transcytotic pathways are suppressed in the CNS to ensure this function. Future studies, including examining the effect of acute removal of Mfsd2a function in adult mice and the effect of dietary manipulation of lipids on transcytosis and BBB permeability will shed new light on the development of new therapeutic strategies. It has been postulated that Mfsd2a may serve to modulate transcytotic mechanisms in CNS endothelial cells for therapeutic purposes (Betsholtz, 2014; Zhao and Zlokovic, 2014). By pinpointing the specific transcytotic route that Mfsd2a suppresses, and by illuminating a novel role of membrane lipid metabolism in BBB function, this work may prompt the development of new strategies to deliver drugs to the diseased CNS, potentially via the use of lipid-based biologics to “de-repress” the caveolae pathway.
STAR Methods
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Chenghua Gu (chenghua_gu@hms.harvard.edu)
Experimental Model and Subject Details
Animal Models
All mice were group housed in standard vivarium conditions, with ad libitum access to diet and water. Mfsd2a-null mice (MMRRC strain 032467-UCD; RRID: MGI:5755244) were maintained on a mixed B6/129 background and were used to test the involvement of Mfsd2a in transcytosis and BBB permeability. Cav-1-null mice (JAX: 004585; RRID:IMSR_JAX:004585, mixed B6/129 background) were used to test the involvement of caveolae-mediated transcytosis in BBB permeability.
Mfsd2aD96A mice were generated at HHMI Janelia Farm. The D96A mutation was generated using the following: gRNA sequence 5′-GCCTTCACTGACCCTCTGGTGGG-3′, donor oligonucleotide sequence 5′-TGCTTCTTCCCCATGGCAGGTGGAACCACTTCCTGCTTCCATTATCCTTTTTGT GGGCCGAGCCTGGGATGCCTTCACTGCCCCACTAGTGGGCTTCTGCATTAGC AAGTCCTCCTGGACCCGCCTGGGCCGCCTCATGCCCTGGTAAGTAGAGACGC TCCTTCGAAG-3′, and cas9 protein (Thermo Fisher Scientific, Cat #NC0789474 ), with an injection concentration: Cas9/gRNA/donor oligonucleotide = 100ng/100ng/20ng/μl. The mouse strain B6SJF1/J (JAX: 100012) was used for injection, with an injection efficiency of 62.5% (20/32). Founder Mfsd2aD96A mice were backcrossed to C57BL/6 mice (Charles River) for three generations prior to analysis. The open-access online tool, CCTop (Stemmer et al., 2015) was used to analyze the possibility of off-target DSBs in Mfsd2aD96A mice. The possibility of off-target mutations was experimentally assessed by Sanger sequencing of genomic DNA flanking top potential off-target sequences, with no mutations found in Mfsd2aD96A genomic DNA. A 250 bp PCR product containing Mfsd2a exon 3 was amplified using the following primers: 5′-TTGGTGGTTCAGCTGTGCCA-3′ and 5′-GACACCAGAAGCCTGCCTTT-3′, and Sanger sequenced with 5′-ATGCTCCGCTAAGGCAATGA-3′ to confirm generation of the D96A point mutation and SpeI restriction site. For genotyping, PCR products were digested with SpeI and run on agarose gels to test for the presence of the Mfsd2aD96A allele.
No animal randomization was performed and both male and female mice were used for experiments and no sex-based differences in phenotype were observed. Animals were genotyped via allele-specific PCR reactions and experiments were performed with observer not blinded to genotype. Information regarding the age of mice used for each specific experiment can be found in the Results and Figure Legends.
All animals were treated according to institutional and US National Institutes of Health (NIH) guidelines approved by the International Animal Care and Use Committee (IACUC) at Harvard Medical School.
Cell lines
HEK293T (ATCC, #CRL-3216) and bEnd.3 (ATCC, #CRL-2299) cells were cultured in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin in 5% CO2 at 37°C. Cell lines were obtained directly from ATCC, with no additional cell authentication performed. PANC-1 cells stably transfected with human MFSD2A-GFP (Reiling et al., 2011) we a obtained from Dr. David Sabatini and cultured in DMEM supplemented with 10% FBS, 1% penicillin/streptomycin, and 2μg/ml puromycin in 5% CO2 at 37°C.
Method Details
Lipid profiling
Lipids from whole brain, and brain and lung isolated capillary lysates were profiled using liquid chromatography tandem mass spectrometry (LC-MS) method operated on a Nexera X2 U-HPLC (Shimadzu Scientific Instruments) coupled to an Exactive Plus orbitrap MS (Thermo Fisher Scientific). Briefly, tissue samples were homogenized in a weight proportional volume of water (4μl/mg of tissue) using a TissueLyser II (QIAGEN). Equal starting amounts of tissue homogenate were used as starting material for all mass spectrometry. Lipids were extracted from homogenates (10 μl) using 190 μl of isopropanol containing 1,2-didodecanoyl-sn-glycero-3-phosphocholine as an internal standard (Avanti Polar Lipids). After centrifugation (10 min, 10,000 x g, ambient temperature), supernatants (2 μl) were injected directly onto a 100 × 2.1 mm ACQUITY BEH C8 column (1.7 μm; Waters). The column was eluted at a flow rate of 450 μL/min isocratically for 1 minute at 80% mobile phase A (95:5:0.1 vol/vol/vol 10 mM ammonium acetate/methanol/formic acid), followed by a linear gradient to 80% mobile-phase B (99.9:0.1 vol/vol methanol/formic acid) over 2 minutes, a linear gradient to 100% mobile phase B over 7 minutes, and then 3 minutes at 100% mobile-phase B. MS analyses were carried out using electrospray ionization in the positive ion mode using full scan analysis over m/z 200–1100 at 70,000 resolution and 3 Hz data acquisition rate. Additional MS settings were: ion spray voltage, 3.0 kV; capillary temperature, 300°C; probe heater temperature, 300 °C; sheath gas, 50; sweep gas, 5; and S-lens RF level 60. Raw data were processed using TraceFinder 3.2 software (Thermo Fisher Scientific) to integrate extracted ion chromatograms for approximately 240 lipids characterized by headgroup and total acyl chain carbon and double bound content. For data analysis, all individual lipid levels (integrated peak area) were normalized to internal standard values prior to statistical testing. Average values of integrated peak area were used for comparisons between conditions for each lipid species.
Immunohistochemistry and immunocytochemistry
Mouse tissues were fixed by immersion in 4% PFA/PBS overnight at 4°C, cryopreserved in 30% sucrose, and frozen in TissueTek OCT (Sakura). Tissue sections were blocked with 10% goat serum/5% BSA/PBST (0.5% Triton X-100) and stained overnight at 4°C with the following primary antibodies: α-Mfsd2a (1:200, Cell Signaling Technologies; RRID:AB_2617168), α-PECAM (1:200, BD Biosciences #553370; RRID:AB_394816), α-Claudin-5-488 conjugate (1:300, Thermo Fisher Scientific #352588; RRID:AB_2532189), isolectin B4 Alexa-488 conjugate (1:200, Thermo Fisher Scientific #I21411), followed by corresponding Alexa Fluor-conjugated secondary antibodies (1:1000, Thermo Fisher Scientific). Sections were mounted with ProLong Gold for imaging. bEnd.3 cells were fixed in 4% PFA/PBS for 15 minutes at room temperature as processed as described above.
BBB permeability assay
Postnatal
P4–P5 pups were anaesthetized with isoflurane and 10-kDa dextran tetramethylrhodamine (Thermo Fisher Scientific, Cat #D1817, 10 mg/ml, 10 μl per g body weight) was injected into the left heart ventricle with a Hamilton syringe. After five minutes of circulation, brains were dissected and fixed by immersion in 4% PFA/PBS at 4°C overnight, cryopreserved in 30% sucrose, and frozen in TissueTek OCT (Sakura). Sections of 14 μm were collected and co-stained with α-Claudin-5-488 conjugate (1:300, Thermo Fisher Scientific #352588; RRID:AB_2532189) or isolectin B4 Alexa-488 conjugate (1:200, Thermo Fisher Scientific #I21411) to visualize blood vessels. Adult: Adult P90 mice were briefly anaesthetized with isoflurane and HRP type II (Sigma Aldrich, Cat #P8250-50KU, 0.5 mg/g body weight dissolved in 0.4 ml PBS) was injected bi-laterally into the retro-orbital sinus. After 30 minutes of circulation, brains were dissected and fixed by immersion in 4%PFA/PBS at 4°C overnight. Following fixation, brains were washed in PBS and forebrain vibratome free-floating sections of 50 μm were collected, processed in a standard DAB reaction for 35 minutes, and mounted on 3% gelatin-coated slides. Slides were dehydrated through an ethanol series and mounted with Permount for brightfield imaging.
Transmission electron microscopy
In vivo
Brains from P4–P5 pups were dissected and fixed by immersion in 5% glutaraldehyde/4% PFA/0.1 M sodium-cacodylate for 10–14 days at room temperature. P90 mice were anaesthetized and HRP type II (Sigma Aldrich, Cat #P8250-50KU, 0.5 mg/g body weight dissolved in 0.4 ml PBS) was injected bilaterally into the retro-orbital sinus. After 30 minutes of circulation, brains were dissected and fixed by immersion, first in 5% glutaraldehyde/4% PFA/0.1 M sodium-cacodylate for 1 hour at room temperature, then overnight at 4°C in 4% PFA/0.1 M sodium-cacodylate. Following fixation, brains were washed overnight in 0.1 M sodium-cacodylate. Coronal vibratome free-floating sections of 50 μm were collected, post-fixed in 1% osmium tetroxide and 1.5% potassium ferrocyanide, dehydrated, and embedded in epoxy resin. Ultrathin sections of 80 nm were then cut from the block surface, collected on copper grids, and counter-stained with Reynold’s lead citrate. In vitro. Experiments on bEnd.3 cells were performed 5 days post-infection with lentivirus. bEnd.3 and PANC-1 confluent cell cultures on aclar coverslips were fixed in a 2.5% paraformaldehyde/5% glutaraldehyde/0.06% picric acid/0.2 M sodium-cacodylate mixer diluted 1:1 in culture media for 15 minutes at room temperature. Cultures were processed for TEM, as described above.
Immunogold labeling for electron microscopy
Adult P90 mice were anaesthetized and perfused through the heart with 30 ml PBS, followed by first 150 ml of a 5% glutaraldehyde/4% PFA/0.1 mM phosphate buffer fixative solution, then a 4% PFA/0.1 mM phosphate buffer fixative solution. Brains were dissected and post-fixed in 4% PFA/PBS for 30 minutes at 4°C. Coronal vibratome free-floating sections of 50 μm were collected and immersed in 0.1% sodium borohydride/PBS for 20 minutes at room temperature, blocked with 10% goat serum/0.5% gelatin/PBST (0.01% Triton X-100), and incubated with α-Cav-1 (1:100, Cell Signaling Technologies #3267; RRID:AB_2275453) overnight at room temperature. Sections were then incubated with gold-labeled goat α-rabbit IgG (1:50, Nanoprobes) overnight at room temperature, washed with PBS and sodium acetate, and silver-enhanced prior to processing for TEM.
Isolation of capillary fragments
Capillary isolation procedure was adapted from a combination of previously described protocols (Goldstein et al., 1984). P4 mouse brains were dissected and homogenized in PBS. Homogenates were centrifuged for 30 minutes on a density gradient of 16% dextran (Sigma Aldrich). The tissue pellet containing capillary fragments was resuspended in PBS and filtered through a 35 μm cell strainer to remove red blood cells and cell-type contaminates. P4 mouse lungs were processed as described above, with an additional tissue dissociation step using Collagenase/Dispase (Roche) for 30 minutes at 37°C prior to centrifugation on a dextran gradient. Isolated capillary fragments were either snap frozen in liquid nitrogen for lipid extraction or lysed and processed for Western blotting.
Western blotting
Protein lysate samples were boiled at 95°C, run on 6–12% polyacrylamide gels, and electrophoretically transferred onto PVDF membranes. Membranes were blocked with 5% BSA/TBST (0.1% Tween-20) or 5% non-fat milk/TBST and incubated overnight at 4°C with the following primary antibodies: α-Mfsd2a (1:1000, Cell Signaling Technologies; RRID:AB_2617168), α-NeuN (1:2000, Millipore #MAB377; RRID:AB_2298772), α-VE-Cadherin (1:1000, Abcam #ab33168; RRID:AB_870662), α-Aquaporin-4 (1:1000, Millipore #ABN411; RRID:AB_2637077), α-PDGFR-β (1:1000, Cell Signaling Technologies #3169; RRID:AB_2162497), α-β-Actin (1:5000, Sigma Aldrich #A2228; RRID:AB_476697), and α-Tubulin (1:8000, Sigma Aldrich #T5168; RRID:AB_477579). Membranes were then incubated with the appropriate HRP-conjugated secondary antibody (1:10000, Southern Biotech) and developed with regular or super ECL (Thermo Fisher Scientific). The intensity of individual bands was quantified using ImageJ.
Lentivirus production
HEK293T cells were transfected using Lipofectamine 2000 (Thermo Fisher Scientific) with the pHAGE-PGK_IZsGreen lentiviral plasmid containing full-length mouse Mfsd2a CDS, along with Δ-vpr and vsv-g packaging plasmids. Supernatant was harvested after three days and concentrated overnight at 4°C with Lenti-X Concentrator (Clontech). Lentivirus was stored fresh at 4°C for up to 24 hours prior to use.
In vitro cholera toxin subunit-B uptake assays
Uptake procedure was adapted from a combination of previously described protocols (Kirkham et al., 2005). Confluent bEnd.3 cells were transfected using Lipofectamine 2000 (Thermo Fisher Scientific) with either pCAG-GFP or pCAG-mMfsd2a-GFP for two days and serum-starved for two hours prior to experiments. Cultures were then incubated with cholera toxin subunit-B (CTB) Alexa-555 conjugate (Thermo Fisher Scientific, Cat #C34776, 150 μg/ml) for ten minutes at 37°C, washed several times with cold PBS, fixed in 4% PFA/PBS for 15 minutes at room temperature, washed with PBS, and mounted with ProLong Gold for imaging.
Imaging
An Olympus FluoView FV1000 or FV1200 laser scanning confocal microscope was used to image dextran tracer permeability assays and expression analyses. A Nikon Ti spinning disk confocal microscope was used to image the in vitro CTB uptake assay. An Olympus VS120-SL 5 slide scanner was used to image adult HRP tracer permeability assays. A 1200EX electron microscope (JOEL) equipped with a 2k CCD digital camera (AMT) was used for all TEM studies. Images were processed using ImageJ (NIH) and Adobe Photoshop.
Quantification and Statistical Analysis
Quantification of BBB leakage
Postnatal dextran tracer permeability assay
All quantification was performed blind using a custom ImageJ (NIH) macro (see Data S1). 6–8 images per animal were obtained from 14 μm thick coronal forebrain sections of the same rostrocaudal position using a 40x oil immersion objective. The same forebrain regions were imaged among littermate pairs. The same background subtraction and auto threshold parameters were applied to max z-projections of both Isolectin/Claudin-5 (endothelium vessel markers), and dextran (tracer) channels. Masks of both channels were generated and the permeability index of leakage was defined as the area of tracer mask divided by area of vessels mask. A leakage value of 1 signifies no tracer extravasation, while significantly higher values signify tracer extravasation.
TEM Quantification
For all TEM quantifications, mean vesicular pit/vesicular density values were calculated from the number of vesicles per μm of membrane for each image collected. All images were collected at 12000x magnification. Density values for each cell source/genotype reflect the average of mean density values obtained from each individual experiment/animal and are expressed as mean ± s.e.m.
Statistical Analysis
All statistical analyses were performed using Prism 7 (GraphPad Software). Two group comparisons were analyzed using an unpaired two-tailed Student’s t-test. Multiple group comparisons were analyzed using a one-way ANOVA, followed by a post-hoc Tukey’s test. Sample size for all experiments was determined empirically using standards generally employed by the field, and no data was excluded when performing statistical analysis. Standard error of the mean was calculated for all experiments and displayed as errors bars in graphs. Statistical details for specific experiments, including exact n values and what n represents, precision measures, statistical tests used, and definitions of significance can be found in the Figure Legends.
Data and Software Availability
A custom ImageJ macro was used to quantify postnatal dextran tracer permeability assays, as described in the Method Details. For script, see Data S1.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal α-Mfsd2a | Cell Signaling Technologies | Under Development; Cat#Mfsd2a14-87; RRID:AB_2617168 |
| Mouse monoclonal α-Claudin-5-488 conjugate | Thermo Fisher Scientific | Cat #352588; RRID:AB_2532189 |
| Rat polyclonal α-PECAM | BD Biosciences | Cat #553370; RRID:AB_394816 |
| Rabbit polyclonal α-Cav-1 | Cell Signaling Technologies | Cat #3267; RRID:AB_2275453 |
| Rabbit polyclonal α-NeuN | Millipore | Cat #MAB377; RRID:AB_2298772 |
| Rabbit polyclonal α-VE-cadherin | Abcam | Cat #ab33168; RRID:AB_870662 |
| Rabbit polyclonal α-PDGFR-β | Cell Signaling Technologies | Cat #3169; RRID:AB_2162497 |
| Rabbit polyclonal α-Aquaporin-4 | Millipore | Cat #ABN411; RRID:AB_2637077 |
| Mouse monoclonal α-β-Actin | Sigma | Cat #A2228; RRID:AB_476697 |
| Mouse monoclonal α-Tubulin | Sigma | Cat# T5168; RRID:AB_477579 |
| Bacterial and Virus Strains | ||
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| Isolectin GS-IB4, from Griffonia simplicifolia | Thermo Fisher Scientific | Cat #I21411 |
| 10-kDa Dextran-tetramethylrhodamine conjugate | Thermo Fisher Scientific | Cat #D1817 |
| Horseradish peroxidase, type II | Sigma-Aldrich | Cat #P8250-50KU |
| Cholera toxin subunit B, Alexa Fluor 555 conjugate | Thermo Fisher Scientific | Cat #C34776 |
| PNA Bio Cas9 protein | Thermo Fisher Scientific | Cat #NC0789474 |
| Critical Commercial Assays | ||
| Deposited Data | ||
| Experimental Models: Cell Lines | ||
| Human: HEK293T cells | ATCC | CRL-3216 |
| Mouse: bEnd.3 cells | ATCC | CRL-2299 |
| Human: PANC-1-MSFD2A-GFP | Dr. David Sabatini; (Reiling et al., 2011) | N/A |
| Experimental Models: Organisms/Strains | ||
| Mouse: Mfsd2a KO: B6;129S5-Mfsd2atm1Lex/Mmucd | Mouse Biology Program, University of California, Davis | MMRRC strain 032467-UCD; RRID: MGI:5755244 |
| Mouse: Cav-1 KO: STOCK Cav1tm1Mls/J | Jackson Laboratory | JAX: 004585; RRID:IMSR_JAX:004585 |
| Mouse: Mfsd2aD96A | This paper | N/A |
| Oligonucleotides | ||
| Mfsd2aD96A gRNA: GCCTTCACTGACCCTCTGGTGGG |
Integrated DNA Technologies | N/A |
| Mfsd2aD96A Donor Oligonucleotide: TGCTTCTTCCCCATGGCAGGTGGAACCACTTCCTGCT TCCATTATCCTTTTTGTGGGCCGAGCCTGGGATGCCT TCACTGCCCCACTAGTGGGCTTCTGCATTAGCAAGTC CTCCTGGACCCGCCTGGGCCGCCTCATGCCCTGGTAA GTAGAGACGCTCCTTCGAAG |
Integrated DNA Technologies | N/A |
| Genotyping Primers: Mfsd2aD96A: (Fwd): TTGGTGGTTCAGCTGTGCCA (Rev): GACACCAGAAGCCTGCCTTT (Sequencing): ATGCTCCGCTAAGGCAATGA |
Integrated DNA Technologies | N/A |
| Genotyping Primers: Mfsd2a: (WT Fwd): CCTGGTTTGCTAAGTGCTAGC (WT Rev): GTTCACTGGCTTGGAGGATGC (KO Fwd): CACTTCCTAAAGCCTTACTTC (KO Rev): GCAGCGCATCGCCTTCTATC |
Integrated DNA Technologies | N/A |
| Genotyping Primers: Cav1: (WT Fwd): GCACACCAAGGAGATTGACC (KO Fwd): CTCCAGACTGCCTTGGGAAAA (Rev): CTTGGCTGTCACCACACAC |
Integrated DNA Technologies | N/A |
| Recombinant DNA | ||
| pCAG-GFP | Matsuda and Cepko, 2004 | Addgene Plasmid #11150 |
| pHAGE-PGK_IZsGreen | Harvard Medical School, Jeng-Shin Lee | https://plasmid.med.harvard.edu/PLASMID/GetVectorDetail.do?vectorid=251 |
| Plasmid: pCAG-mMfsd2a-GFP | This paper | N/A |
| Plasmid: pHAGE-PGK_mMfsd2a-IZsGreen | This paper | N/A |
| Software and Algorithms | ||
| Prism 7 | GraphPad | https://www.graphpad.com/scientificsoftware/prism/ |
| CCTop – CRISPR/Cas9 target online predictor | Center for Organismal Studies Heidelberg; (Stemmer et al., 2015) | http://crispr.cos.uni-heidelberg.de/ |
| TraceFinder 3.2 | Thermo Fisher Scientific |
http://tools.thermofisher.com/content/sfs/manuals/ Man-XCALI-97598-TraceFinder-32-User-Clinical-ManXCALI97598-A-EN.pdf |
| Custom ImageJ macro for quantifying BBB permeability | This paper | Data S1 |
| Other | ||
Supplementary Material
Table S3. Lipidomic profiling of brain versus lung capillaries. Related to Figure 4.
Complete lipidomic analysis of lipid species from brain and lung capillaries of P4 wild-type mice (LPC, lyso-phosphatidylcholine; PC, phosphatidylcholine; LPE, lyso-phosphatidylethanolamine; PE, phosphatidylethanolamine; PS, phosphatidylserine; PI, phosphatidylinositol; p, plasmalogen; SM, sphingomyelin; CE, cholesteryl ester; MAG, monoacylglycerol; DAG, diacylglycerol; TAG, triacylglyceroyl). Value for each species is normalized to internal standard and expressed as integrated mass spectrometry peak area. n = 6 groups per organ, 8 animals pooled per group.
Acknowledgments
We thank Drs. Jonathan Cohen, Tom Schwartz, Christopher Harvey, Bob Datta, Chuck Weitz, and members of the Gu laboratory for comments on the manuscript; Dr. Radu Stan (Dartmouth University) for discussions; Dr. David Sabatini (MIT) for providing the PANC-1 cell line (Reiling et al., 2011); Dr. Christine Mössinger (Karolinska Institute) for advice regarding lipid biology; Roberto Rosa for technical assistance; the HMS Electron Microscopy Core Facility, the HMS Neurobiology Imaging Facility, and the Nikon Imaging Center at Harvard Medical School; Drs. Roberto Polakiewicz and Jianxin Xie (Cell Signaling Technologies) for generating the Mfsd2a antibody. This work was supported by NIH NRSA Award F31NS090669 (B.J.A.), the Mahoney postdoctoral fellowship (B.L.), the Howard Hughes Medical Institute (D.G), the Kaneb Fellowship (C.G.), Fidelity Biosciences Research Initiative (C.G.), and Harvard Blavatnik Biomedical Accelerator (C.G.), and the NIH DP1 NS092473 Pioneer Award (C.G.). The research of C.G. was also supported in part by a Faculty Scholar grant from the Howard Hughes Medical Institute.
Footnotes
Author Contributions
B.J.A. and C.G. conceived the project and designed experiments. K.B., A.A.D., and C.C. performed lipidomic mass spectrometry. B.J.A., A.T., and B.L. performed all other experiments. A.B.Z. initiated lipidomic experiments. D.D.G. provided experimental resources. B.J.A. and B.C. analyzed all data. B.J.A. and C.G. wrote the manuscript, with input from all authors.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aird WC. Phenotypic heterogeneity of the endothelium: I. Structure, function, and mechanisms. Circulation research. 2007;100:158–173. doi: 10.1161/01.RES.0000255691.76142.4a. [DOI] [PubMed] [Google Scholar]
- Alakbarzade V, Hameed A, Quek DQ, Chioza BA, Baple EL, Cazenave-Gassiot A, Nguyen LN, Wenk MR, Ahmad AQ, Sreekantan-Nair A, et al. A partially inactivating mutation in the sodium-dependent lysophosphatidylcholine transporter MFSD2A causes a non-lethal microcephaly syndrome. Nat Genet. 2015;47:814–817. doi: 10.1038/ng.3313. [DOI] [PubMed] [Google Scholar]
- Andreone BJ, Lacoste B, Gu C. Neuronal and vascular interactions. Annu Rev Neurosci. 2015;38:25–46. doi: 10.1146/annurev-neuro-071714-033835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armulik A, Genove G, Mae M, Nisancioglu MH, Wallgard E, Niaudet C, He L, Norlin J, Lindblom P, Strittmatter K, et al. Pericytes regulate the blood-brain barrier. Nature. 2010;468:557–561. doi: 10.1038/nature09522. [DOI] [PubMed] [Google Scholar]
- Bastiani M, Parton RG. Caveolae at a glance. Journal of cell science. 2010;123:3831–3836. doi: 10.1242/jcs.070102. [DOI] [PubMed] [Google Scholar]
- Ben-Zvi A, Lacoste B, Kur E, Andreone BJ, Mayshar Y, Yan H, Gu C. Mfsd2a is critical for the formation and function of the blood-brain barrier. Nature. 2014;509:507–511. doi: 10.1038/nature13324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betsholtz C. Physiology: Double function at the blood-brain barrier. Nature. 2014;509:432–433. doi: 10.1038/nature13339. [DOI] [PubMed] [Google Scholar]
- Bien-Ly N, Yu YJ, Bumbaca D, Elstrott J, Boswell CA, Zhang Y, Luk W, Lu Y, Dennis MS, Weimer RM, et al. Transferrin receptor (TfR) trafficking determines brain uptake of TfR antibody affinity variants. The Journal of experimental medicine. 2014;211:233–244. doi: 10.1084/jem.20131660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitsikas V, Correa IR, Jr, Nichols BJ. Clathrin-independent pathways do not contribute significantly to endocytic flux. eLife. 2014;3:e03970. doi: 10.7554/eLife.03970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brightman MW, Reese TS. Junctions between intimately apposed cell membranes in the vertebrate brain. The Journal of cell biology. 1969;40:648–677. doi: 10.1083/jcb.40.3.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Jump DB, Esselman WJ, Busik JV. Inhibition of cytokine signaling in human retinal endothelial cells through modification of caveolae/lipid rafts by docosahexaenoic acid. Investigative ophthalmology & visual science. 2007;48:18–26. doi: 10.1167/iovs.06-0619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng JP, Nichols BJ. Caveolae: One Function or Many? Trends Cell Biol. 2015 doi: 10.1016/j.tcb.2015.10.010. [DOI] [PubMed] [Google Scholar]
- Chow BW, Gu C. The molecular constituents of the blood-brain barrier. Trends in neurosciences. 2015;38:598–608. doi: 10.1016/j.tins.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow BW, Gu C. Gradual suppression of transcytosis governs functional blood-retinal barrier formation. Neuron. 2017;93:1325–1333. doi: 10.1016/j.neuron.2017.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen AW, Razani B, Schubert W, Williams TM, Wang XB, Iyengar P, Brasaemle DL, Scherer PE, Lisanti MP. Role of caveolin-1 in the modulation of lipolysis and lipid droplet formation. Diabetes. 2004;53:1261–1270. doi: 10.2337/diabetes.53.5.1261. [DOI] [PubMed] [Google Scholar]
- Daneman R, Zhou L, Agalliu D, Cahoy JD, Kaushal A, Barres BA. The mouse blood-brain barrier transcriptome: a new resource for understanding the development and function of brain endothelial cells. PloS one. 2010a;5:e13741. doi: 10.1371/journal.pone.0013741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daneman R, Zhou L, Kebede AA, Barres BA. Pericytes are required for blood-brain barrier integrity during embryogenesis. Nature. 2010b;468:562–566. doi: 10.1038/nature09513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ethayathulla AS, Yousef MS, Amin A, Leblanc G, Kaback HR, Guan L. Structure-based mechanism for Na(+)/melibiose symport by MelB. Nat Commun. 2014;5:3009. doi: 10.1038/ncomms4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghitescu L, Fixman A, Simionescu M, Simionescu N. Specific binding sites for albumin restricted to plasmalemmal vesicles of continuous capillary endothelium: receptor-mediated transcytosis. The Journal of cell biology. 1986;102:1304–1311. doi: 10.1083/jcb.102.4.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein GW, Betz AL, Bowman PD. Use of isolated brain capillaries and cultured endothelial cells to study the blood-brain barrier. Federation proceedings. 1984;43:191–195. [PubMed] [Google Scholar]
- Guemez-Gamboa A, Nguyen LN, Yang H, Zaki MS, Kara M, Ben-Omran T, Akizu N, Rosti RO, Rosti B, Scott E, et al. Inactivating mutations in MFSD2A, required for omega-3 fatty acid transport in brain, cause a lethal microcephaly syndrome. Nat Genet. 2015;47:809–813. doi: 10.1038/ng.3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Cordes KR, Farese RV, Jr, Walther TC. Lipid droplets at a glance. Journal of cell science. 2009;122:749–752. doi: 10.1242/jcs.037630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley MJ, Lawrence CB. The blood-brain barrier after stroke: Structural studies and the role of transcytotic vesicles. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 2016 doi: 10.1177/0271678X16629976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innis SM. Dietary (n-3) fatty acids and brain development. The Journal of nutrition. 2007;137:855–859. doi: 10.1093/jn/137.4.855. [DOI] [PubMed] [Google Scholar]
- Junge HJ, Yang S, Burton JB, Paes K, Shu X, French DM, Costa M, Rice DS, Ye W. TSPAN12 regulates retinal vascular development by promoting Norrin- but not Wnt-induced FZD4/beta-catenin signaling. Cell. 2009;139:299–311. doi: 10.1016/j.cell.2009.07.048. [DOI] [PubMed] [Google Scholar]
- Kirkham M, Fujita A, Chadda R, Nixon SJ, Kurzchalia TV, Sharma DK, Pagano RE, Hancock JF, Mayor S, Parton RG. Ultrastructural identification of uncoated caveolin-independent early endocytic vehicles. The Journal of cell biology. 2005;168:465–476. doi: 10.1083/jcb.200407078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowland D, Arac A, Sekiguchi KJ, Hsu M, Lutz SE, Perrino J, Steinberg GK, Barres BA, Nimmerjahn A, Agalliu D. Stepwise recruitment of transcellular and paracellular pathways underlies blood-brain barrier breakdown in stroke. Neuron. 2014;82:603–617. doi: 10.1016/j.neuron.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Zhang Q, Wang M, Liu F, Zhao S, Ma J, Luo N, Li N, Li Y, Xu G, et al. Docosahexaenoic acid affects endothelial nitric oxide synthase in caveolae. Archives of biochemistry and biophysics. 2007;466:250–259. doi: 10.1016/j.abb.2007.06.023. [DOI] [PubMed] [Google Scholar]
- Liebner S, Corada M, Bangsow T, Babbage J, Taddei A, Czupalla CJ, Reis M, Felici A, Wolburg H, Fruttiger M, et al. Wnt/beta-catenin signaling controls development of the blood-brain barrier. The Journal of cell biology. 2008;183:409–417. doi: 10.1083/jcb.200806024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- Ma DW, Seo J, Davidson LA, Callaway ES, Fan YY, Lupton JR, Chapkin RS. n-3 PUFA alter caveolae lipid composition and resident protein localization in mouse colon. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2004;18:1040–1042. doi: 10.1096/fj.03-1430fje. [DOI] [PubMed] [Google Scholar]
- Martin S, Parton RG. Caveolin, cholesterol, and lipid bodies. Seminars in cell & developmental biology. 2005;16:163–174. doi: 10.1016/j.semcdb.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Matsuda T, Cepko CL. Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc Natl Acad Sci U S A. 2004;101:16–22. doi: 10.1073/pnas.2235688100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor S, Parton RG, Donaldson JG. Clathrin-independent pathways of endocytosis. Cold Spring Harb Perspect Biol. 2014:6. doi: 10.1101/cshperspect.a016758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LN, Ma D, Shui G, Wong P, Cazenave-Gassiot A, Zhang X, Wenk MR, Goh EL, Silver DL. Mfsd2a is a transporter for the essential omega-3 fatty acid docosahexaenoic acid. Nature. 2014;509:503–506. doi: 10.1038/nature13241. [DOI] [PubMed] [Google Scholar]
- Niewoehner J, Bohrmann B, Collin L, Urich E, Sade H, Maier P, Rueger P, Stracke JO, Lau W, Tissot AC, et al. Increased brain penetration and potency of a therapeutic antibody using a monovalent molecular shuttle. Neuron. 2014;81:49–60. doi: 10.1016/j.neuron.2013.10.061. [DOI] [PubMed] [Google Scholar]
- Oh P, Borgstrom P, Witkiewicz H, Li Y, Borgstrom BJ, Chrastina A, Iwata K, Zinn KR, Baldwin R, Testa JE, et al. Live dynamic imaging of caveolae pumping targeted antibody rapidly and specifically across endothelium in the lung. Nat Biotechnol. 2007;25:327–337. doi: 10.1038/nbt1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh P, Testa JE, Borgstrom P, Witkiewicz H, Li Y, Schnitzer JE. In vivo proteomic imaging analysis of caveolae reveals pumping system to penetrate solid tumors. Nat Med. 2014;20:1062–1068. doi: 10.1038/nm.3623. [DOI] [PubMed] [Google Scholar]
- Pardridge WM. Drug transport across the blood-brain barrier. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 2012;32:1959–1972. doi: 10.1038/jcbfm.2012.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parton RG, del Pozo MA. Caveolae as plasma membrane sensors, protectors and organizers. Nature reviews Molecular cell biology. 2013;14:98–112. doi: 10.1038/nrm3512. [DOI] [PubMed] [Google Scholar]
- Pike LJ, Han X, Chung KN, Gross RW. Lipid rafts are enriched in arachidonic acid and plasmenylethanolamine and their composition is independent of caveolin-1 expression: a quantitative electrospray ionization/mass spectrometric analysis. Biochemistry. 2002;41:2075–2088. doi: 10.1021/bi0156557. [DOI] [PubMed] [Google Scholar]
- Pol A, Martin S, Fernandez MA, Ferguson C, Carozzi A, Luetterforst R, Enrich C, Parton RG. Dynamic and regulated association of caveolin with lipid bodies: modulation of lipid body motility and function by a dominant negative mutant. Molecular biology of the cell. 2004;15:99–110. doi: 10.1091/mbc.E03-06-0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Predescu SA, Predescu DN, Palade GE. Plasmalemmal vesicles function as transcytotic carriers for small proteins in the continuous endothelium. Am J Physiol. 1997;272:H937–949. doi: 10.1152/ajpheart.1997.272.2.H937. [DOI] [PubMed] [Google Scholar]
- Quek DQ, Nguyen LN, Fan H, Silver DL. Structural Insights into the Transport Mechanism of the Human Sodium-dependent Lysophosphatidylcholine Transporter MFSD2A. The Journal of biological chemistry. 2016;291:9383–9394. doi: 10.1074/jbc.M116.721035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razani B, Engelman JA, Wang XB, Schubert W, Zhang XL, Marks CB, Macaluso F, Russell RG, Li M, Pestell RG, et al. Caveolin-1 null mice are viable but show evidence of hyperproliferative and vascular abnormalities. The Journal of biological chemistry. 2001;276:38121–38138. doi: 10.1074/jbc.M105408200. [DOI] [PubMed] [Google Scholar]
- Reese TS, Karnovsky MJ. Fine structural localization of a blood-brain barrier to exogenous peroxidase. The Journal of cell biology. 1967;34:207–217. doi: 10.1083/jcb.34.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiling JH, Clish CB, Carette JE, Varadarajan M, Brummelkamp TR, Sabatini DM. A haploid genetic screen identifies the major facilitator domain containing 2A (MFSD2A) transporter as a key mediator in the response to tunicamycin. Proc Natl Acad Sci U S A. 2011;108:11756–11765. doi: 10.1073/pnas.1018098108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert W, Frank PG, Razani B, Park DS, Chow CW, Lisanti MP. Caveolae-deficient endothelial cells show defects in the uptake and transport of albumin in vivo. The Journal of biological chemistry. 2001;276:48619–48622. doi: 10.1074/jbc.C100613200. [DOI] [PubMed] [Google Scholar]
- Shvets E, Bitsikas V, Howard G, Hansen CG, Nichols BJ. Dynamic caveolae exclude bulk membrane proteins and are required for sorting of excess glycosphingolipids. Nat Commun. 2015;6:6867. doi: 10.1038/ncomms7867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohet F, Lin C, Munji RN, Lee SY, Ruderisch N, Soung A, Arnold TD, Derugin N, Vexler ZS, Yen FT, et al. LSR/angulin-1 is a tricellular tight junction protein involved in blood-brain barrier formation. The Journal of cell biology. 2015;208:703–711. doi: 10.1083/jcb.201410131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemmer M, Thumberger T, del Sol Keyer M, Wittbrodt J, Mateo JL. CCTop: an intuituve, flexible and reliable CRISPR/Cas9 target prediciton tool. PLOS ONE. 2015:10. doi: 10.1371/journal.pone.0124633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam SJ, Richmond DL, Kaminker JS, Modrusan Z, Martin-McNulty B, Cao TC, Weimer RM, Carano RA, van Bruggen N, Watts RJ. Death receptors DR6 and TROY regulate brain vascular development. Developmental cell. 2012;22:403–417. doi: 10.1016/j.devcel.2011.11.018. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Nelson AR, Betsholtz C, Zlokovic BV. Establishment and Dysfunction of the Blood-Brain Barrier. Cell. 2015;163:1064–1078. doi: 10.1016/j.cell.2015.10.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Zlokovic BV. Blood-brain barrier: a dual life of MFSD2A? Neuron. 2014;82:728–730. doi: 10.1016/j.neuron.2014.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Wang Y, Tischfield M, Williams J, Smallwood PM, Rattner A, Taketo MM, Nathans J. Canonical WNT signaling components in vascular development and barrier formation. The Journal of clinical investigation. 2014;124:3825–3846. doi: 10.1172/JCI76431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlokovic BV. The blood-brain barrier in health and chronic neurodegenerative disorders. Neuron. 2008;57:178–201. doi: 10.1016/j.neuron.2008.01.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S3. Lipidomic profiling of brain versus lung capillaries. Related to Figure 4.
Complete lipidomic analysis of lipid species from brain and lung capillaries of P4 wild-type mice (LPC, lyso-phosphatidylcholine; PC, phosphatidylcholine; LPE, lyso-phosphatidylethanolamine; PE, phosphatidylethanolamine; PS, phosphatidylserine; PI, phosphatidylinositol; p, plasmalogen; SM, sphingomyelin; CE, cholesteryl ester; MAG, monoacylglycerol; DAG, diacylglycerol; TAG, triacylglyceroyl). Value for each species is normalized to internal standard and expressed as integrated mass spectrometry peak area. n = 6 groups per organ, 8 animals pooled per group.