SUMMARY
Bacterial skin infections represent some of the most common infectious diseases globally. Prevention and treatment of skin infections can involve application of a topical antimicrobial, which may be an antibiotic (such as mupirocin or fusidic acid) or an antiseptic (such as chlorhexidine or alcohol). However, there is limited evidence to support the widespread prophylactic or therapeutic use of topical agents. Challenges involved in the use of topical antimicrobials include increasing rates of bacterial resistance, local hypersensitivity reactions (particularly to older agents, such as bacitracin), and concerns about the indiscriminate use of antiseptics potentially coselecting for antibiotic resistance. We review the evidence for the major clinical uses of topical antibiotics and antiseptics. In addition, we review the mechanisms of action of common topical agents and define the clinical and molecular epidemiology of antimicrobial resistance in these agents. Moreover, we review the potential use of newer and emerging agents, such as retapamulin and ebselen, and discuss the role of antiseptic agents in preventing bacterial skin infections. A comprehensive understanding of the clinical efficacy and drivers of resistance to topical agents will inform the optimal use of these agents to preserve their activity in the future.
KEYWORDS: Staphylococcus aureus, antibiotic resistance, antiseptic, community-acquired infections, impetigo, skin infections
INTRODUCTION
The skin is one of the first lines of defense against microbial invasion (1). Healthy skin harbors a diverse range of bacteria, collectively known as the skin microbiome, and depending on host, bacterial, and environmental factors, this bacterial population may be protective or harmful (2, 3). Breaches in the skin, whether accidental (e.g., trauma or insect bite) or intentional (e.g., surgical incision), allow incursion of bacterial pathogens and can lead to skin and soft tissue infection (SSTI). SSTI is an extremely common infectious disease syndrome, with an estimated 14.2 million SSTI-related ambulatory care attendances in the United States in 2005 (4). Occasionally, treatment of SSTI may involve administration of a topical antibiotic agent, although supportive evidence for topical antibiotic use varies according to specific clinical manifestations. In addition to therapeutic indications, topical antibiotics and antiseptics are increasingly used in the prevention of skin infections, particularly to reduce surgical site infections (SSIs) in patients colonized with Staphylococcus aureus (5).
Theoretically, topical antibiotic use offers several advantages over systemic administration, including delivery of high concentrations of antimicrobial at the required site of action and a reduction in systemic toxicity (Table 1). However, the widespread use of commonly used topical antibiotics (particularly mupirocin and fusidic acid) has led to increasing bacterial resistance in some settings, limiting the potential efficacy of such agents. Moreover, there are recognized concerns about the possible deleterious ecological impact (so-called “collateral damage”) of increasingly widespread use of topical antiseptics, such as chlorhexidine and triclosan (6, 7). Given global concerns regarding antibiotic resistance and relatively limited therapeutic options, especially for some species, such as S. aureus, the appropriate use of topical agents and the prevention of further resistance are critical.
TABLE 1.
Theoretical advantages and disadvantages of topical antimicrobial therapy for bacterial skin infections
| Advantage/disadvantage |
|---|
| Advantages |
| May enable targeted delivery of a high concentration of antimicrobial to site of infection |
| Higher likelihood of adherence to treatment (e.g., in children) |
| Less potential for systemic side effects and toxicity |
| May avoid need for systemic antimicrobials |
| Ensures that site of infection is regularly inspected |
| Topical application allows use and development of agents that may not be able to be used systemically (e.g., neomycin or bacitracin) |
| Topical route of administration may be easier for patients and caregivers |
| Disadvantages |
| Limited evidence base for clinical effectiveness |
| Many agents associated with local allergy |
| Limited understanding of potentially deleterious effects on skin microbiota |
| Minimal depth of penetration, limiting use on intact skin |
| Unquantified effects on wound healing process |
| Widespread and unrestricted use is likely to select for bacterial resistance (e.g., fusidic acid and Staphylococcus aureus) |
| Potential for storage in patient homes, with possibility of recurrent use and contamination |
| Often combined with topical steroid therapy, meaning that primary prescribing indication may be for inflammation rather than infection |
| Potential perception by both patients and prescribers as more “benign” than systemic antimicrobials |
| May be difficult for some patients to apply to larger surface areas or skin folds |
Here we provide an overview of the major preventative and therapeutic uses for topical agents and a review of the clinical and molecular epidemiology of resistance to these agents. Specifically, we focus on common and emerging topical antibacterial and antiseptic agents. The scope of this review does not cover topical antifungal or antiviral agents, nor does it extensively cover skin and nasal decolonization, which was recently comprehensively reviewed (5). For convenience, we use the terms “topical antibiotic(s)” and “topical antiseptic(s)” throughout.
CLINICAL USAGE OF TOPICAL ANTIBIOTICS AND ANTISEPTICS
Topical antibiotics are among the most commonly prescribed antimicrobial agents. For example, in 2015 there were 4.7 million primary care prescriptions for topical antibiotics in the United Kingdom, at a cost of approximately $29.9 million (8). However, although they are widely used, evidence supports the prescription of topical antibiotics for only a small number of indications. To date, published antimicrobial stewardship guidelines have focused almost exclusively on systemically or intravenously administered antibiotics, and few studies have attempted to quantify the extent of topical antibiotic consumption or to assess the appropriateness of topical antibiotic prescribing practices. Using national data from outpatient physician practices, one study from the United States assessed topical antibiotic prescribing practices between 1993 and 2007 (9). The authors of that study found that approximately one-fourth of dermatology patient visits and approximately one-fifth of pediatric patient visits were associated with a topical antibiotic prescription. The most common diagnosis associated with a topical antibiotic prescription was skin neoplasm, and it was hypothesized that in such instances, topical antibiotics were being used as postoperative wound prophylaxis following minor surgery, a practice that is not supported by existing evidence (9). Similarly, a study from New Zealand assessed national trends and demographics in topical antibiotic prescribing practices between 2006 and 2014. Community usage of topical fusidic acid increased significantly over the study period, with the highest usage in children under 5 years of age (10). However, high rates of topical antibiotic usage were also observed in the >75-year-old age group, and it was suggested that given the limited evidence for prescribing topical antibiotics for older age groups, a proportion of this usage may be considered clinically inappropriate (10).
In the following sections, we provide an overview of the major clinical uses (both prophylactic and therapeutic) for topical antibiotic and antiseptic agents. Where possible, we review the evidence for and against their use.
Impetigo
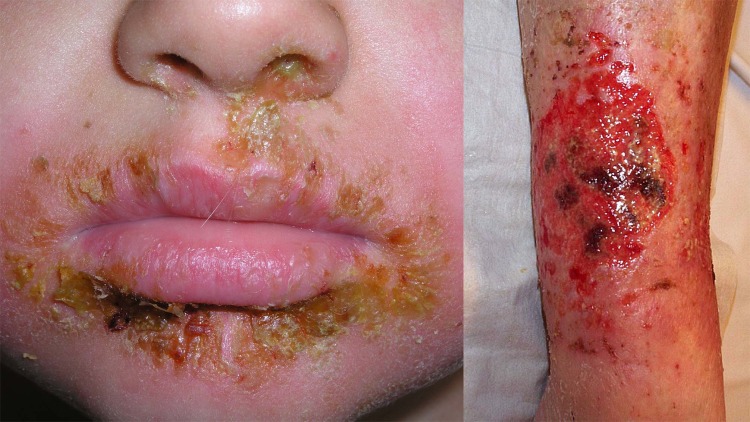
Impetigo is a common superficial and contagious bacterial skin infection that is usually caused by S. aureus and/or Streptococcus pyogenes. Clinically, there are two main syndromes: the more common nonbullous impetigo (impetigo contagiosa) and the less common bullous (blistering) impetigo (Fig. 1) (11, 12). Nonbullous impetigo typically manifests as small intraepidermal blisters, which subsequently form yellow-brown crusted lesions around the face, particularly the nose and mouth. It is most common in children aged 2 to 5 years old, and although self-limiting, it is generally treated with antibiotics to reduce symptom duration and to prevent further transmission of causative bacteria. Impetigo can be treated with topical antibiotics and/or systemic antibiotics, and the decision of which therapy to use is generally based on the number and extent of lesions, with minor disease usually treated with topical agents (11).
FIG 1.
Clinical presentations of impetigo. (Left) Typical crusting lesions of nonbullous impetigo. (Right) Blistering lesions characteristic of nonbullous impetigo. (Images are reproduced courtesy of Dermnet NZ under a Creative Commons agreement [CC BY-NC-ND 3.0 NZ] [the left panel was cropped minimally].)
Despite an estimated global prevalence in children of approximately 162 million (13), there is limited high-quality evidence to guide the appropriate empirical topical treatment of impetigo. A meta-analysis conducted in 2012 to assess interventions for impetigo included 24 randomized controlled trials (RCTs) that compared topical antibiotic therapy to placebo, another topical antibiotic, or a topical antiseptic (12). Overall, the authors of that analysis concluded that topical antibiotic therapy achieved significantly higher cure rates than those with placebo (risk ratio [RR], 2.24; 95% confidence interval [CI], 1.61 to 3.13), and they found no significant difference between the two main topical antibiotics used, i.e., mupirocin and fusidic acid. However, the quality of the included studies was variable, and most studies were conducted over a decade ago, when the prevalence of bacterial resistance to topical agents may have differed (14, 15). Importantly, there remain several knowledge gaps regarding the most appropriate topical treatment for impetigo (16). For example, given that impetigo is generally regarded as a self-limiting condition, there are relatively few trials comparing topical antibiotic treatment to treatment with placebo. Moreover, there are currently no studies comparing the use of topical antibiotic to that of placebo or topical antiseptic in settings with a high prevalence of resistance to commonly used topical agents. Finally, as laboratory testing is not part of the routine work-up for mild impetigo, true rates of resistance in causative pathogens are largely unknown. As such, caution should be exercised in extrapolating results of studies conducted in low-resistance settings, as it is possible that clinical and microbiological cure rates differ between settings. Given the current evidence, key points for practitioners to consider in prescribing topical antibiotics for impetigo include using the shortest possible duration of therapy and maintaining a close liaison with the microbiology laboratory regarding local resistance patterns.
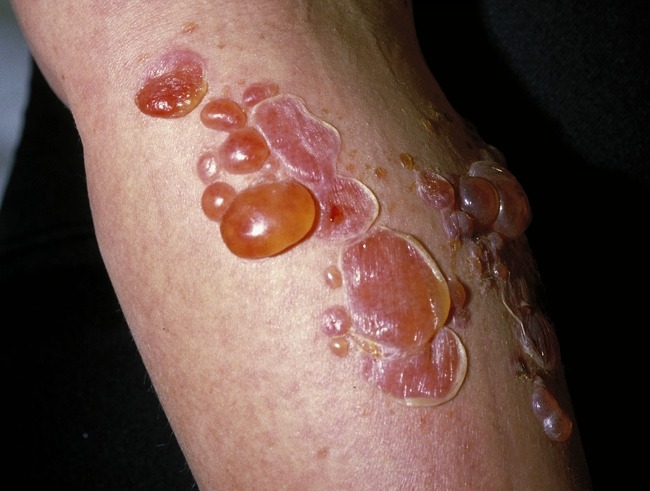
Chronic Wounds
Chronic (or “complex”) wounds include conditions such as venous leg ulceration and pressure ulcers (Fig. 2) (17). Both conditions are relatively common, with an estimated prevalence for venous leg ulceration of between 0.1% and 1% in high-income countries (18). For pressure ulceration, prevalence varies according to the setting (e.g., hospital versus community) and patient group (e.g., there is a higher prevalence in spinal injury patients) (19, 20). One population-based study in 2014 in the United Kingdom across community, health care, and residential care settings observed an overall prevalence of pressure ulceration of 4.6% (19). Similarly, studies from the United States and Europe show a range of prevalence values of approximately 4% to 43%, depending on the setting (21–23).
FIG 2.
Characteristic lower leg venous ulceration demonstrating shallow ulceration and surrounding reddened skin. (The image is reproduced courtesy of Dermnet NZ under a Creative Commons agreement [CC BY-NC-ND 3.0 NZ].)
One of the key factors in considering the use of topical antibiotics or antiseptics in the context of chronic wound management is deciding whether a wound is truly infected rather than asymptomatically colonized (24, 25). The vast majority of chronic wounds are colonized with at least one bacterial species, and accordingly, the decision on whether a wound is infected is based predominantly on clinical judgment rather than on microbiological analysis (26). However, specific pathogens (e.g., S. aureus, S. pyogenes, Enterobacteriaceae, and Pseudomonas aeruginosa) may have adverse impacts on wound healing, and along with clinical assessment and underlying pathology (e.g., diabetic neuropathy or vasculopathy), their isolation should be taken into account in considering antibiotic treatment (17, 27–30).
Either systemic or topical agents may be considered for treating infected chronic wounds, although, to date, evidence for using topical agents to effectively treat chronic wound infections is equivocal. In general, published studies do not recommend the use of topical agents for treating noninfected chronic wounds (31). For example, a 2014 Cochrane systematic review evaluated RCTs that involved antimicrobial therapy for promoting healing of venous leg ulceration (32). The authors identified 40 RCTs (recruiting 4,253 participants) that evaluated topical preparations. Although the RCTs were of variable quality, the authors of that review found statistical evidence to support the use of topical iodine in promoting wound healing compared to standard wound care (RR, 2.17; 95% CI, 1.30 to 3.60), although they could find no evidence to support the use of topical honey- or silver-based products (32). Based on the available data, they could not draw robust conclusions about the use of any other topical agents (including povidone-iodine, hydrogen peroxide, mupirocin, and chloramphenicol) in promoting healing of venous leg ulceration (32). Similarly, a 2016 systematic review evaluated 12 RCTs (including 576 participants) assessing the utility of topical agents in the healing of both infected and noninfected pressure ulcers (19). The authors concluded that there was insufficient evidence (based on the heterogeneity and quality of trials) to assess any benefit of topical agents on pressure ulcer healing (19). Interestingly, however, there was (limited) evidence to suggest that povidone-iodine may actually be detrimental for wound healing compared to nonantimicrobial alternatives, such as protease-modulating dressings (RR, 0.78; 95% CI, 0.62 to 0.98) (33) and hydrogel (RR, 0.64; 95% CI, 0.43 to 0.97) (34).
Wound Infection following Burn Injuries
Wound infections following burn injuries represent a major source of morbidity and mortality (35). The disruption of the skin epidermal barrier, coupled with avascular necrotic tissue and relative local and systemic immunosuppression, provides a “perfect storm” for colonization and proliferation of microbes (36, 37). It has been estimated that in patients with severe burns, up to three-fourths of all mortality is associated with sepsis, either from wound infections or from other infectious complications (e.g., pneumonia) (37–39). Numerous systemic and topical antibiotic and antiseptic agents have been used for the prevention and treatment of burn wound infections (BWI), with the rationale for application of topical therapy predominantly related to prophylaxis of BWI. It is postulated that topical therapy may reduce the microbial burden in the burn wound, thereby reducing the risk of infection and potentially promoting wound healing (36, 37, 40, 41).
To date, however, many of the studies assessing the efficacy of topical agents in BWI prophylaxis have been relatively small, with a variety of clinical endpoints and trial methodologies. A systematic review in 2013 attempted to assess the effectiveness of topical prophylaxis for BWI, and it included 26 RCTs (with 1,329 participants) evaluating a range of topical agents, such as silver sulfadiazine, neomycin, bacitracin, polymyxin B, and mafenide acetate (42). The authors found no evidence to support the use of topical antimicrobials (compared to either no intervention or any other intervention) for the prevention of BWI (42). Moreover, a subanalysis of 11 RCTs (with 645 participants) involving the use of 1% topical silver sulfadiazine found that patients treated with silver sulfadiazine actually had a higher risk of BWI and a longer hospital stay than those treated with either dressings or skin substitutes (odds ratio [OR], 1.87; 95% CI, 1.09 to 3.19), although the extent of bias within the included studies was unclear (42–51).
Prevention of Postsurgical Wound Infections
Infections following surgical procedures are a major cause of health care-associated infections (HCAI) and result in considerable clinical and economic burdens (52). Depending on the surgical procedure, preoperative intravenous antibiotics are often administered as part of bundles of interventions designed to prevent surgical site infections (SSIs) (53, 54). Prophylactic systemic antibiotic use has been studied extensively and is generally considered an effective and evidence-based contribution to SSI prevention (53). In addition to systemic antibiotics, contemporary SSI prevention strategies often involve the application of a topical antibiotic and/or antiseptic agent (5), with the two most common uses being antibiotic application (usually with mupirocin) to the nasal mucosa to eradicate preoperative S. aureus carriage and antiseptic body washes (usually with chlorhexidine) to reduce the bacterial load on the skin. The available evidence suggests that prophylactic nasal and skin decolonization is an effective strategy for preventing S. aureus SSI following some surgical procedures, predominantly orthopedic and cardiac surgeries (54). In particular, a large meta-analysis of 17 RCTs and quasi-experimental trials evaluated the protective effect of decolonization with nasal mupirocin on prevention of SSI post-cardiac or -orthopedic surgery (including studies that did and did not incorporate skin decolonization with chlorhexidine) (54). The authors of that study observed a significant protective effect of decolonization in preventing SSIs caused by S. aureus (pooled relative risk, 0.39; 95% CI, 0.31 to 0.50). Moreover, this reduction was observed for both methicillin-susceptible S. aureus (MSSA) (pooled relative risk, 0.50; 95% CI, 0.37 to 0.69) and methicillin-resistant S. aureus (MRSA) (pooled relative risk, 0.30; 95% CI, 0.15 to 0.62). The use of mupirocin and chlorhexidine is further discussed below, and additional evidence for their use in bacterial decolonization was recently thoroughly reviewed (5).
In contrast to the nasal application of topical antibiotics, there is comparatively limited evidence to support the administration of topical antibiotics (cf. antiseptics) directly at the surgical site (55). A previous review suggested that local application of topical antibiotics for surgical prophylaxis gave an unproven benefit for the majority of surgical procedures (55). Based on that analysis, it was concluded that locally applied topical antibiotics had a probable benefit in reducing SSI rates in joint arthroplasties (specifically when used in antibiotic-impregnated cement) and ophthalmic surgery and a possible benefit in reducing the rates in cosmetic breast augmentation and in obese patients undergoing abdominal surgery (55). In addition, a recent Cochrane review evaluated the use of locally applied topical antibiotics in the prevention of surgical wounds healing by primary intention (i.e., when the clean wound edges are actively held together with sutures, staples, or adhesive) (56). The authors identified 10 RCTs and four quasi-experimental studies that included a total of 6,466 patients. These studies covered a range of surgical procedures, from minor dermatological procedures conducted in an outpatient or emergency department to major surgeries conducted in an operating theater. When the use of topical antibiotics was compared to the use of no antibiotic, the authors observed a reduction in the risk of SSI (RR, 0.61; 95% CI, 0.42 to 0.87), with a number needed to treat to prevent one SSI (NNT) of 50. Similarly, the use of topical antibiotics was superior to the use of topical antiseptics in reducing the risk of SSI (RR, 0.49; 95% CI, 0.30 to 0.80; NNT, 24). The authors concluded that, overall, the use of topical antibiotics was associated with a probable reduced risk of SSI but that the included studies were of variable quality and heterogeneity. Moreover, due to a lack of statistical power and/or analyses, no conclusions could be drawn about whether topical antibiotics were associated with increased adverse outcomes, such as allergic contact dermatitis or increased antimicrobial resistance, or whether any specific antibiotic was superior to another (57).
In another recent systematic review, the use of topical antimicrobials (including both antibiotics and antiseptics) for the treatment of wound healing by secondary intention (i.e., open wounds that heal through new tissue growth, such as a perianal abscess) was assessed (58). The authors of the analysis evaluated 11 RCTs (including 886 patients) of variable quality that covered a range of surgical procedures. After synthesizing all studies, the authors concluded that there was insufficient evidence to establish the effectiveness of topical antimicrobials for promoting wound healing or reducing infection rates for wound healing by secondary intention. Therefore, at present, there is insufficient high-quality evidence for the routine use of topical antimicrobials for the treatment of postsurgical wound healing by either primary or secondary intention.
Prevention of Minor Traumatic Wound Infection
Minor traumatic skin wounds (e.g., abrasions or lacerations) are common presentations to primary care practices and emergency departments. To date, there is scarce evidence to support the adjunctive use of topical antibiotics in preventing infection or promoting wound healing following uncomplicated minor wounds. One previous narrative review was able to identify only two historical double-blinded placebo-controlled RCTs that compared infection rates in patients with minor wounds with and without the application of topical antibiotics (59). One study, from 1995, compared the use of petrolatum (placebo) and one of three topical antimicrobial preparations (bacitracin zinc ointment, silver sulfadiazine cream, and a triple antimicrobial ointment [TAO] containing bacitracin zinc, polymyxin B, and neomycin) for the prevention of infection following standard wound care (which included the use of sutures) in 465 patients with minor lacerations (60). The authors observed a significant difference in wound infection rates between the placebo group and the three groups who received topical therapy, although there was no significant difference in infection rates between the three treated groups. However, as noted previously (61), the majority of patients were graded as having “grade 1 infection” or a stitch abscess, which may have resolved with basic wound care regardless of whether topical antimicrobial therapy was used. The second placebo-controlled RCT was conducted in 1985 and compared the use of TAO ointment with that of placebo ointment (preparation not stated in the study) in preventing impetiginous lesions following minor wounds in 59 children aged 2 to 5 years (62). Although the authors observed a difference in infection rates between the TAO-treated group and the placebo-treated group (15% versus 47%), the study numbers were small, and the authors did not comment on the statistical significance of their findings or on whether the infections were clinically relevant (62). Future, larger RCTs to assess the efficacy of topical antibiotics in preventing posttraumatic wound infection are required to establish the utility of this approach.
CURRENTLY USED TOPICAL ANTIBACTERIAL AGENTS
Mupirocin
Mupirocin (first known as pseudomonic acid A) is produced naturally by Pseudomonas fluorescens (63). First used clinically in the 1980s, mupirocin is administered exclusively as a topical agent, as either a cream or an ointment (64–70). However, when it is administered systemically, mupirocin is rapidly degraded to an inactive metabolite, monic acid. To date, the main clinical uses for mupirocin have been treatment of minor staphylococcal skin infections and S. aureus nasal decolonization (12, 71, 72).
Mupirocin has a relatively broad range of antibacterial activity, covering all staphylococci (including MRSA), most streptococci (with the exception of Streptococcus bovis), and several Gram-negative bacteria, particularly Haemophilus influenzae, Neisseria gonorrhoeae, Neisseria meningitidis, and Moraxella catarrhalis (73). Mupirocin acts by reversibly binding to a bacterial enzyme called isoleucyl-tRNA synthetase. This enzyme catalyzes the conversion of isoleucine and tRNA to the isoleucyl-tRNA molecule (74, 75). Due to similarities between a moiety on the mupirocin structure and isoleucine, mupirocin can bind to isoleucyl-tRNA synthetase, specifically binding at a site called the Rossman fold and subsequently inhibiting bacterial RNA and protein synthesis (74, 76).
Because mupirocin has predominantly been used for the prevention and treatment of staphylococcal infections, to date, the majority of studies assessing the clinical and molecular epidemiology of mupirocin resistance have focused on staphylococci.
Phenotypic and genotypic characterization of mupirocin resistance.
For staphylococci, mupirocin resistance is described as either low level or high level, depending on the resistance phenotype and the molecular basis of resistance (see below). There are several laboratory methods for evaluating mupirocin resistance in staphylococci, including broth or agar dilution, disc diffusion, and Etest methods (77–81). However, there is currently no clear consensus on the most appropriate phenotypic differentiation of low- and high-level resistances, with different interpretative criteria used by the Clinical and Laboratory Standards Institute (CLSI) (82) and the European Committee for Antimicrobial Susceptibility Testing (EUCAST) (www.eucast.org). For example, for either broth microdilution or disc diffusion, the CLSI recommends differentiating only between “no high-level resistance” and “high-level resistance,” depending on whether there is a presence or absence of growth (82). In contrast, EUCAST defines MICs of ≤1 mg/ml as indicating susceptible strains and those of >256 mg/ml as indicating resistant strains, with an intermediate category of uncertain clinical significance (www.eucast.org).
At the molecular level, low-level resistance (indicated by MICs of 8 to 256 mg/ml) is determined by point mutations in the isoleucyl-tRNA synthetase gene (ileS) (76, 83–86), which is chromosomally borne (in contrast to plasmid borne). A range of point mutations have been associated with low-level resistance, with the most commonly reported mutations being V588F and V631F (74, 76, 87). These mutations are located in the Rossman fold, thereby limiting the ability of mupirocin to bind to the isoleucyl-tRNA synthetase enzyme (76). Importantly, the presence of these mutations has not been shown to confer a significant bacterial fitness cost in vitro, suggesting a possible fitness advantage for these low-level resistant strains in the context of ongoing mupirocin use (84, 85, 87). Interestingly, one recent study demonstrated the rapid development of mutations associated with low-level mupirocin resistance after only 14 days of exposure to subinhibitory mupirocin concentrations, with mutations being stably maintained in the absence of mupirocin (87).
In staphylococci, high-level resistance (MIC of >256 mg/ml) is most commonly mediated by the mupA gene (also known as ileS-2). This gene encodes a novel isoleucyl-tRNA synthetase (88, 89) and is primarily disseminated via plasmid-mediated horizontal gene transfer (74). A range of conjugative plasmids have been reported to harbor mupA, predominantly those related to the pSK1/pG01 family of plasmids (90–93). Moreover, within plasmids, mupA can be flanked by insertion sequences (IS), particularly IS257, which may promote recombination-mediated dissemination of mupA between plasmids (92, 94, 95). Importantly, the plasmids that harbor mupA may also contain additional resistance determinants, including those that encode resistance to aminoglycosides, macrolides, tetracycline, and clindamycin, raising the possibility that mupirocin use may select for coresistance to other antimicrobials (74). In addition, identical mupA-containing plasmids have been identified across a range of S. aureus lineages, including clonal complex 5 (CC5), CC8, CC22, and CC30, highlighting the ability of these plasmids to disseminate across major S. aureus clones (90, 91). Furthermore, studies have demonstrated in vitro transfer of mupA-containing plasmids between Staphylococcus epidermidis and S. aureus (96), and the presence of mupA-containing conjugative plasmids has also been documented for other coagulase-negative staphylococci (CoNS), such as Staphylococcus pseudintermedius (97), raising suggestions that CoNS may act as a possible reservoir of mupA.
Although the presence of mupA is almost always associated with high-level resistance, isolates harboring mupA and displaying low-level resistance were reported in two previous studies (98, 99). In one of these studies, mupA was found to be chromosomally integrated rather than plasmid borne, although the specific integration site and flanking elements were not characterized (99). Moreover, isolates that harbor mupA on a plasmid yet appear to be phenotypically susceptible to mupirocin have also been described. For some isolates, a frameshift mutation in mupA was described (100), although in other isolates, despite their appearing phenotypically susceptible, no apparent mupA mutation was detected, raising the possibility of mutations in a promoter or regulatory region (101).
More recently, an additional mechanism of high-level mupirocin resistance was described, thought to be determined by a novel, plasmid-borne gene, mupB, that was identified in a clinical MRSA strain in Canada (102). The mupB gene shares only 65.5% and 45.5% nucleotide similarities with mupA and ileS, respectively. To date, however, there are few studies that have systematically assessed staphylococci for the presence of mupB (103).
Prevalence of mupirocin resistance in staphylococci.
There have been numerous studies assessing the prevalence of mupirocin resistance in staphylococci, differing largely in the patient population studied (e.g., community, hospital, or intensive care unit [ICU]), bacterial species (e.g., S. aureus versus CoNS), associated resistance profile (e.g., MRSA versus MSSA), and nature of surveillance (e.g., active surveillance in patients previously treated with mupirocin versus passive surveillance of isolates from all patients). However, only subsets of these studies have assessed the molecular epidemiology and coresistance patterns of mupirocin-resistant isolates. Results of larger recent studies assessing mupirocin resistance in S. aureus are summarized in Table 2.
TABLE 2.
Recent published studies evaluating the prevalence of mupirocin resistance in Staphylococcus aureusa
| Country(ies) | Authors (reference) | Yr(s) | Study setting or population | Prevalence of phenotypic resistance (no. of resistant isolates/no. of isolates tested [%]) |
Prevalence of mupA (no. of isolates with mupA/total no. of resistant isolates) | ||
|---|---|---|---|---|---|---|---|
| MSSA | MRSA | Overall | |||||
| Germany, Switzerland, Austria | Kresken et al. (302) | 2001 | Hospital and outpatients | LLR, 1/624 (0.2) | LLR, 22/163 (13.5) | 30/787 (3.8) | NR |
| HLR, 2/624 (0.3) | HLR, 5/163 (3.1) | ||||||
| Greece | Petinaki et al. (303) | 1999–2002 | Hospital | NR | NR | LLR, 4/1,200 (0.3) | |
| HLR, 20/1,200 (1.7) | HLR, 20/20 | ||||||
| Ireland | Rossney and O'Connell (304) | 1999–2005 | Hospital (bloodstream isolates reported) | NR | 37/2,586 (1.4) | NR | NR |
| Netherlands | Donker et al. (305) | 2005 | Outpatients | NR | NR | 2/595 (0.3) | HLR, 2/2 |
| Spain | Perez-Roth et al. (91) | 2002–2009 | NR | NR | LLR, NR | NR | |
| HLR, 31/550 (5.6) | HLR, 31/31 | ||||||
| France | Trouillet-Assant et al. (103) | 2010 | Hospital | 0/615 | NR | ||
| France | Desroches et al. (104) | 2011–2012 | Hospital | NR | LLR, 5/367 (1.4) | 8/367 (2.2) | |
| HLR, 3/367 (0.8) | HLR, 3/3 | ||||||
| United States | Fritz et al. (101) | 2007–2009 | Community | NR | NR | 50/2,425 (2.1) | NR |
| United States | McDanel et al. (306) | 2008–2011 | Nursing homes | NR | LLR, 23/829 (2.8) | 101/829 (12.2) | HLR, 78/78 |
| HLR, 78/829 (9.4) | |||||||
| United States | Suwantarat et al. (307) | 2007–2013 | Neonatal intensive care unit | NR | LLR, 3/101 (3.0) | 3/101 (3.00) | NR |
| HLR, 0/101 | |||||||
| United States | Sciortino et al. (308) | NR | Hospital | NR | LLR, 3/82 (3.7) | 10/82 (12.2) | NR |
| HLR, 7/82 (8.5) | |||||||
| South Korea | Lee et al. (309) | 2006–2009 | Hospital | NR | LLR, 53/456 (11.6) | 62/456 (13.6) | |
| HLR, 9/456 (2.0) | HLR, 9/9 | ||||||
| United Kingdom | Horner et al. (310) | 2015 | Hospitals and community | NR | LLR, 0/520 | 4/520 (0.8) | NR |
| HLR, 4/520 (0.8) | |||||||
| Sweden | Fang et al. (311) | 2014 | Hospitals and community | NR | LLR, 2/743 (0.3) | 5/743 (0.7) | NR |
| HLR, 3/743 (0.4) | |||||||
| Malaysia | Ghasemzadeh-Moghaddam et al. (312) | 2011 | Hospitals | HLR, 2/164 (1.2) | HLR, 8/95 (8.4) | 10/259 (3.7) | HLR, 10/259 |
| United States | Warren et al. (313) | 2005–2012 | Intensive care units | NR | HLR, 35/504 (6.9) | 35/504 (6.9) | HLR, 35/35 |
| New Zealand | Williamson et al. (108) | 2013 | Community | NR | NR | 55/500 (11.0) | NR |
| Australia | Coombs et al. (314) | 2012 | Community | 68/2,334 | 8/510 | 76/2,844 (2.7) | NR |
| Australia | Coombs et al. (315) | 2011 | Hospital | 26/1,664 | 9/713 | 35/2,377 (1.5) | NR |
Abbreviations: MSSA, methicillin-susceptible Staphylococcus aureus; MRSA, methicillin-resistant S. aureus; NR, not reported; HLR, high-level resistance; LLR, low-level resistance.
In addition to that in S. aureus, mupirocin resistance is also found in CoNS, although this has been characterized less extensively. For example, one nationwide French study performed in 2011 and 2012 assessed mupirocin resistance in invasive CoNS isolates. The authors of that study described an overall mupirocin resistance rate of 10.3%, with a high-level mupA-mediated resistance rate of 5.6% (104). A higher resistance rate was demonstrated in a 2007 Irish study that observed a high-level mupirocin resistance prevalence of 22% among bloodstream isolates of CoNS (105). Similarly, a 2013 Belgian study described a mupA-mediated resistance rate of 20% for bloodstream isolates of S. epidermidis (106).
Clinical use of mupirocin and emergence of resistance in staphylococci.
Several studies have attempted to assess the association between clinical use of mupirocin and the emergence of resistance. Mupirocin resistance varies according to factors such as study setting (e.g., community versus hospital), availability of treatment (e.g., over-the-counter versus prescription), and patient populations (e.g., hemodialysis patients versus general patients).
Unsurprisingly, the emergence of resistance appears to be more common when there is unrestricted use of mupirocin in a large population of patients. For example, in New Zealand, mupirocin was available for purchase in community pharmacies between 1991 and 2000 (107). In 1999, approximately 28% of S. aureus isolates in one New Zealand study were resistant to mupirocin, with a higher resistance rate in community-associated isolates than in hospital-associated isolates (30.2% versus 19.8%, respectively) (107). Interestingly, a follow-up New Zealand study, conducted in 2013, demonstrated a reduction in the prevalence of mupirocin resistance, from 28% to 11% (108). This decline was concurrent with a decrease in the use of mupirocin in New Zealand following a regulatory change in 2000 restricting mupirocin use to “prescription only.” Similarly, in western Australia, widespread empirical use of mupirocin led to high rates of mupirocin resistance in MRSA during the early 1990s, peaking at 18% in 1993 (109). Subsequent regulatory changes resulted in a reduction in mupirocin resistance among MRSA isolates, to 0.3% in 1997 (110). In contrast, resistance development appears to be less frequent when community use of mupirocin is targeted rather than empirical. For example, in a large RCT involving 3,447 U.S. military trainees, a 5-day course of mupirocin was administered to those trainees found to be colonized with MRSA (111). Four months later, participants were assessed for MRSA colonization and the occurrence of skin infections. No mupirocin resistance was detected among MRSA isolates.
In the hospital setting, there are also notable differences in the emergence of mupirocin resistance depending on whether mupirocin use is widespread (e.g., universal decolonization of all preoperative patients) or restricted. For example, in one Brazilian study, all inpatients (between 1990 and 1995) found to be infected and/or colonized with MRSA were treated with a regimen that involved mupirocin and chlorhexidine to eradicate MRSA carriage (112). Subsequently, the rate of mupirocin resistance in MRSA was found to be 65% in 1995 (113). Following a restriction in mupirocin use to colonized patients only, mupirocin resistance decreased to 15% in 1999 and 2000 (112). Similarly, among renal dialysis patients, the routine and sustained use of prophylactic mupirocin to prevent peritoneal catheter site infections has been associated with subsequent isolation of resistant S. aureus (114, 115). Moreover, in one study, patients who were colonized with mupirocin-resistant strains were at an increased risk of exit site infection (114).
Conversely, when mupirocin use has been targeted to reducing nasal colonization with S. aureus in perioperative prophylaxis, the emergence of resistance has generally been less common (72, 116, 117). For example, in two large RCTs (with a combined total of 1,808 patients) assessing the effectiveness of mupirocin for preoperative S. aureus nasal decolonization for the prevention of surgical site infections, mupirocin-resistant S. aureus isolates were detected in only six patients (0.003%) (116, 118). Furthermore, of these six patients, only three were in the group that had received mupirocin (118). Similarly, in a Dutch study of cardiothoracic patients, mupirocin resistance was not detected in any S. aureus isolates from 868 patients following mupirocin treatment (119).
Like studies of S. aureus, some studies have also investigated the association between mupirocin use and resistance in CoNS. For example, a study from the Netherlands assessed trends in mupirocin resistance in both S. aureus and CoNS between 2006 and 2011 (120). The study demonstrated an increase in mupA-mediated mupirocin resistance in CoNS, from 8% in 2006 to 22% in 2011. Moreover, this increase was associated with a 3-fold increase in mupirocin usage over the study period (120). Similarly, in another study from the same Dutch institution, high-level mupirocin resistance among CoNS isolates was assessed for surgical inpatients before and after a universal decolonization regimen involving nasal mupirocin treatment and chlorhexidine body washes (121). Prior to decolonization, the rate of high-level mupirocin resistance was 21%, increasing to 43% after 5 days of mupirocin and chlorhexidine treatment, with 99.5% of resistance mediated by mupA (120).
Interestingly, a recent study by Hetem et al. (122) utilized a deterministic mathematical model to explore the emergence of mupirocin resistance in the context of hospital-based targeted or universal S. aureus decolonization regimens. They concluded that the risks of mupirocin resistance emergence were similar for both targeted and universal decolonization regimens and that, based on this finding, universal decolonization may be a more practical solution than targeted prophylaxis (122). Furthermore, based on analysis of published data assessing transmissibility rates of MRSA in hospitals (and incorporating a species-specific sensitivity analysis of transmissibility rates), they also concluded that high-level mupirocin resistance in CoNS was not a major risk for the emergence of high-level mupirocin-resistant S. aureus in the context of mupirocin use for decolonization (122).
In clinical practice, mupirocin is usually administered with chlorhexidine, a biguanide cationic antiseptic agent (see below). Interestingly, in one case-control study of an MRSA decolonization program, the presence of both low-level mupirocin-resistant isolates and the qacA/B genes (mediating reduced susceptibility to chlorhexidine) was associated with persistent MRSA carriage (98). The prevalence and relevance of chlorhexidine resistance are discussed separately below.
Fusidic Acid
Fusidic acid is a steroidal antibiotic derived from the fungus Fusidium coccineum. The most active derivative is the sodium salt (sodium fusidate), and this was first used clinically in the early 1960s for the treatment of staphylococcal infections. One of the important features of fusidic acid is the fact that it can be administered orally, intravenously, or topically. In particular, topical fusidic acid can be administered in a variety of preparations, including ointment, cream, lotion, and gel forms. Similar to those for mupirocin, the key clinical indications for topical fusidic acid are the treatment of superficial skin infections and eradication of nasal carriage of S. aureus.
Fusidic acid is primarily active against staphylococci (including most CoNS strains), with MICs for susceptible staphylococci ranging from 0.016 to 0.5 μg/ml (www.eucast.org). However, MICs for S. pyogenes are considerably higher, ranging from 1 to 16 μg/ml. Fusidic acid also has in vitro activity against several other Gram-positive bacteria, including corynebacteria and Gram-positive anaerobes. In general, Gram-negative bacteria are resistant to fusidic acid, with the exceptions of Neisseria and Moraxella species and some strains of the Bacteroides fragilis group.
Although the chemical structure of fusidic acid is similar to that of cephalosporin P, unlike cephalosporins, fusidic acid does not act on the cell wall but acts as a protein synthesis inhibitor, specifically at the translation phase. During bacterial protein synthesis, elongation of the polypeptide chain occurs as the ribosome moves along mRNA and accepts aminoacyl-tRNA units, in a reaction coupled to GTP hydrolysis. Elongation factor G (EF-G) is a GTPase involved in translocation of the mRNA-tRNA complex and is encoded by the chromosomal fusA gene. Binding of fusidic acid to EF-G produces a conformational change that prevents the dissociation of the EF-G–GDP complex from the ribosome, thus preventing binding of the next aminoacyl-tRNA unit and inhibiting further protein synthesis. This relatively unique mode of action means that there is currently no known cross-resistance with other antimicrobial classes.
Genotypic and phenotypic characterization of fusidic acid resistance.
Fusidic acid susceptibility testing can be performed using agar dilution, broth microdilution, disc diffusion, or Etest methods. Although there are no CLSI interpretive criteria for fusidic acid susceptibility, EUCAST (www.eucast.org) defines susceptibility in staphylococci for MICs of ≤1 mg/liter and resistance for MICs of >1 mg/liter (based on oral or intravenous doses of 500 mg twice daily or three times daily).
In staphylococci, there are a number of molecular mechanisms that mediate resistance to fusidic acid, both chromosomal and acquired. These mechanisms vary in their prevalence, mode of action, and impact on bacterial fitness. At the chromosomal level, resistance is most commonly associated with mutations in fusA, resulting in a structural change in EF-G and reduced fusidic acid binding to the EF-G ribosome complex. Although there are at least 30 fusA mutations, only a few result in phenotypic resistance to fusidic acid. Crystallographic analysis has demonstrated that the majority of mutations occur in structural domain III of EF-G, although they can also occur in domains I and V (123). Interestingly, mutations in domain V of EF-G have also been shown to be associated with the small colony variant (SCV) phenotype, with features such as reduced susceptibility to aminoglycosides and hemin auxotrophy (124). Within domain III (amino acids 404 to 483 in EF-G), the most commonly described mutations are L461K, H457K, and P406L, with the L461K mutation being the most common mutation associated with high-level resistance (125, 126). Site-directed mutagenesis and cloning of mutant fusA alleles demonstrated that these three mutations increase the fusidic acid MIC for S. aureus by at least 32-fold (126). Moreover, although some mutations in EF-G (e.g., P406L and H457Y) have been associated with an impairment of biological fitness in S. aureus (127), it has been demonstrated that secondary mutations in EF-G may compensate for this loss of fitness and potentially allow for the persistence of fusidic acid-resistant strains within a population (127).
In addition to chromosomal resistance-conferring mutations, there are several acquired genes that have been characterized in staphylococci that confer fusidic acid resistance. In particular, the fusB and fusC genes encode metalloproteins (FusB and FusC, respectively) that can bind to EF-G. Binding of these proteins enables the dissociation of the ribosomal EF-G–GDP complexes that form in the presence of fusidic acid (128, 129) and resumption of protein translation, despite the presence of fusidic acid.
Both fusB and fusC are located on mobile genetic elements; this enables transfer across S. aureus clones, and possibly across staphylococcal species (130, 131). fusB may be chromosomal or plasmid borne and typically confers low-level fusidic acid resistance. In S. aureus, fusB has been found on pUB101, a ubiquitous 21.9-kb plasmid that encodes β-lactamase and cadmium resistance (132–134), and recent work has described fusB in association with a highly clonal p11819-97 plasmid among isolates of the dominant European CA-MRSA CC80 clone (135). The fusB gene has also been described as being present on the chromosome of a European fusidic acid-resistant S. aureus impetigo clone (EEFIC) (136). In addition to that in S. aureus, fusB is also found in coagulase-negative staphylococci, and in one study assessing fusidic acid-resistant staphylococci from North America and Australia, fusB was more prevalent among CoNS strains than among S. aureus strains (65.0% versus 17.4%, respectively) (125). Furthermore, in another study from Europe, fusB was more prevalent among fusidic acid-resistant CoNS strains than among fusidic acid-resistant S. aureus strains (26.5% versus 10.1%, respectively) (137). Similar to that in S. aureus, fusB in CoNS can be either plasmid borne or chromosomal. For example, in one study, fusB was found to be harbored within chromosomally located pathogenicity islands (PIs) within S. epidermidis (138), and it was occasionally found to be colocated within these PIs with a putative virulence gene, vapE (138).
Subsequent to the characterization of fusB, a FusB homologue exhibiting 44% amino acid homology to FusB was reported for S. aureus (139, 140). In addition, a FusB homologue exhibiting 47% amino acid homology to FusB was reported for Staphylococcus saprophyticus (140). These two homologues were named FusC and FusD, respectively, and their encoding genes were designated fusC and fusD (140). More recently, another FusB homologue, FusF, encoded by the fusF gene, was described as a major resistance determinant in fusidic acid-resistant Staphylococcus cohnii (141).
The fusC gene is particularly prevalent among fusidic acid-resistant S. aureus and CoNS strains, and to date, it has always been identified within staphylococcal cassette chromosome (SCC) elements, with or without the mecA gene (139, 142–146). One recent study assessed the genetic context of fusC across different lineages of S. aureus, including ST5 and ST1 (147). In all lineages, fusC was always located within SCC elements, and it was hypothesized that SCC-mediated horizontal transfer was the main mechanism for fusC to be disseminated, both within S. aureus and across other staphylococcal species (147). Importantly, the genetic colocation of fusC with mecA may be important in driving the emergence of MRSA in settings where large amounts of fusidic acid are used. A particularly clear example of this is in New Zealand, a country with high (and increasing) usage of topical fusidic acid since the early 2000s (10, 108). Since 2005, a fusidic acid-resistant ST5 MRSA clone has emerged to become the dominant MRSA lineage in New Zealand, suggesting that in addition to fusidic acid use selecting solely for strains containing fusidic acid resistance determinants, there is also likely to be coselection of other resistance genes, such as mecA (108, 147).
Prevalence and clinical relevance of fusidic acid resistance.
The reported prevalence of fusidic acid resistance in staphylococci varies widely according to factors such as patient population, specimen type (e.g., clinical isolates versus carriage isolates), and geographic region. Importantly, many studies assessing fusidic acid resistance in staphylococci were conducted prior to the discovery of the fusC, fusD, and fusF genes and are unlikely to reflect the true prevalence of these mechanisms. Table 3 provides information on recent larger studies describing both the prevalence and mechanisms of fusidic acid resistance in staphylococci.
TABLE 3.
Recent large published studies evaluating the prevalence of fusidic acid resistance in Staphylococcus aureusa
| Country(ies) | Authors (reference) | Yr(s) | Study setting or population | Prevalence of phenotypic resistance (no. of resistant isolates/no. of isolates tested [%]) |
Acquired resistance (no. of isolates with detected mutations) |
No. of isolates with detected fusA mutations | |||
|---|---|---|---|---|---|---|---|---|---|
| MSSA | MRSA | Overall | fusB | fusC | |||||
| Nine European countries | den Heijer et al. (316) | 2010–2011 | Nasal carriage isolates from primary care | 194/6,814 (2.8) | 9/91 (9.9) | 203/6,905 (2.9) | 48 | 88 | 52 |
| United States | Farrell et al. (150) | 2014 | Hospitals | 4/956 (0.4) | 0/848 | 0 | 3 | 1 | |
| New Zealand | Williamson et al. (108) | 2013 | Community | NR | NR | 147/500 (29.4) | 0 | 147 | NR |
| United States, Australia, and Canada | Castanheira et al. (125) | 2007–2008 | Hospital | NR | NR | 25/4,167 (0.6) | 4 | 13 | 3 |
| Thirteen European countries | Castanheira et al. (137) | 2008 | Hospital | NR | NR | 288/2,700 (10.7) | 34 | 57 | 56 |
| Denmark | McLaws et al. (317) | 2003–2005 | NR | NR | 291/1,639 (17.8) | 196 (inferred by authors from previous work) | 57 | 39 | |
| China | Liu et al. (134) | 2008–2009 | Children's hospitals | 2/120 (1.2) | 2/66 (3.0) | 4/186 (2.2) | 2 | 2 | NR |
| Greece | Souli et al. (318) | 2012–2013 | Hospitals | 81/808 (10.0) | 212/372 (57.0) | 293/1,180 (24.8) | NR | NR | NR |
| United States | Jones et al. (319) | 2008–2009 | Hospitals | 15/3,463 (0.4) | 11/3,876 (0.3) | 26/7,339 (0.4) | 3 | 12 | 5 |
| Australia | Coombs et al. (320) | 2014 | Hospitals | 74/1,791 (4.1) | 17/414 (4.1) | 91/2,205 (4.1) | NR | NR | NR |
| Australia | Coombs et al. (314) | 2012 | Community | 135/2,334 (5.8) | 26/510 (5.1) | 161/2,844 (5.7) | NR | NR | NR |
Abbreviations: MSSA, methicillin-susceptible Staphylococcus aureus; MRSA, methicillin-resistant S. aureus; NR, not reported.
To date, several studies have identified potential associations between the use of topical fusidic acid and the emergence of fusidic acid resistance in staphylococci at both the patient and population levels. For example, in the United Kingdom, the use of topical fusidic acid approximately doubled over a 6-year period from 1995 to 2001, with a concomitant increase in fusidic acid resistance among S. aureus bloodstream isolates, from 2.0% in 1990 to 6.1% in 2001 (148). Moreover, a contemporaneous study from Wales identified a significant correlation between primary care practice level prescribing of fusidic acid and fusidic acid resistance in community MSSA isolates (149). Similarly, a more recent study from the United Kingdom analyzed trends in fusidic acid use and resistance in MRSA between 2002 and 2012 (142). In that study, although a slight decline in topical fusidic acid community use was observed, the percentage of fusidic acid resistance among contemporaneous MRSA bacteremia isolates increased markedly over the same period, from approximately 10% to 20% (142). In that study, fusidic acid was mainly mediated by fusC, encoded within SCC elements, again highlighting the potential for coselection of genetically linked resistance mechanisms, such as fusC and mecA. In addition, a recent study from New Zealand also assessed the trends of topical antimicrobial use and correlated these with resistance patterns (108). Between 1993 and 2012, there was a significant increase in topical fusidic acid use in the New Zealand community setting (108). Concurrent with this increase in usage, the prevalence of fusidic acid resistance in S. aureus in New Zealand increased from 17% in 1999 to 29% in 2013 (108). Although the authors of the study noted the “limitations in ecological correlations of antimicrobial prescribing and the development of resistance,” they hypothesized that the considerable increase in fusidic acid resistance was driven by high usage of fusidic acid in the New Zealand community (108). This hypothesis is supported by low rates of fusidic acid resistance in other countries that have low or negligible use of fusidic acid. In particular, fusidic acid resistance rates in S. aureus isolates are extremely low in the United States (0.3%), where fusidic acid is not yet widely used systemically and has not been used topically (125, 150).
In addition to population-level associations between fusidic acid use and resistance, several studies have assessed the development of fusidic acid resistance at the patient level, mainly in dermatology patients who have previously received topical fusidic acid therapy. For example, Shah and Mohanraj conducted a retrospective review of all dermatology patients with a positive S. aureus culture during a 4-month period in 2001 in a hospital in Yorkshire in the United Kingdom (151). They found that nearly two-thirds of all dermatology patients had used topical fusidic acid in the 6 months preceding the study period and that a significantly larger proportion of isolates from dermatology patients than that from other patients was fusidic acid resistant (51% versus 9.6%). Moreover, among patients who were infected with fusidic acid-resistant or -susceptible S. aureus isolates, 96% and 29%, respectively, had used topical fusidic acid therapy in the previous 6 months, suggesting an association between prior use and resistance. Similar findings were noted by Ravenscroft et al. in the United Kingdom (152) and Peeters et al. in the Netherlands (153). In particular, Peeters et al. observed an increase in fusidic acid resistance among S. aureus isolates from patients with atopic dermatitis, from 9.7% in 1995 to 23.4% in 2001, and they suggested that this increase was due to patients in this population receiving courses of topical fusidic acid to treat infected eczema lesions or to eradicate S. aureus carriage (153). Similarly, Sule et al. conducted a study of dermatology outpatients with atopic eczema and S. aureus colonization, and they observed a significant correlation between recent exposure to topical fusidic acid and the presence of fusidic acid-resistant S. aureus (154).
Although fusidic acid resistance in clinical isolates of S. aureus is well described, there is a scarcity of studies describing clinical failures of fusidic acid therapy (either systemic or topical). However, as discussed in a previous review (155), widespread and unregulated topical fusidic acid monotherapy should not be considered justifiable given the likelihood of resistance developing. This is particularly important given the (as yet) current utility of systemic fusidic acid in combination with other antimicrobials (e.g., rifampin) in the treatment of MRSA infections (156).
Neomycin
Neomycin is an aminoglycoside antimicrobial that is produced by Streptomyces fradiae and was first described in 1949 (157). Neomycin comprises three major chemically related compounds, namely, neomycin A (neamine), neomycin B (framycetin; also called neomycin sulfate), and neomycin C, with the quantity of each varying with the manufacturing process (158).
Due to its relative toxicity when administered systemically, neomycin is generally used only topically, either alone or in combination with other antimicrobials, particularly polymyxin B and/or bacitracin (see below). Topical preparations are available in a variety of formulations, such as gels, solutions, eye drops, and eardrops. Neomycin is active against staphylococci and most aerobic Gram-negative bacteria, although streptococci and Gram-positive bacilli are resistant.
Similar to other aminoglycosides, neomycin acts by binding to the 30S subunit of the bacterial ribosome to inhibit protein synthesis. Resistance is mediated through a number of mechanisms, with the most significant being enzymatic inactivation of the drug by chromosomally or plasmid-encoded aminoglycoside-modifying enzymes (159). Based on data from EUCAST, the MIC90 values of neomycin against S. aureus and CoNS isolates are 1 μg/ml and 0.25 μg/ml, respectively (www.eucast.org).
Topical neomycin-containing formulations are most commonly used for treatment of localized skin infections due to staphylococci and Gram-negative bacilli. In general, however, neomycin formulations perform less well than other topical antimicrobials, such as fusidic acid and mupirocin, for the treatment of common skin infections, such as impetigo (12). In addition, one of the major concerns regarding the use of topical neomycin is the apparently high prevalence of allergic contact dermatitis, which has been estimated at 1% to 6% but is thought to be higher in patients with a compromised skin barrier (160). Moreover, systemic complications, such as ototoxicity, can occur following topical neomycin therapy, including instances of severe ototoxicity in patients with eardrum perforation receiving neomycin eardrops (161).
Bacitracin
Bacitracin is a cyclic polypeptide antimicrobial, derived from the bacterium Bacillus subtilis, that is FDA approved for adults for the treatment of superficial bacterial skin infections. It is bactericidal by complexing with C55-isoprenyl pyrophosphate (IPP), which is a bacterial cell wall component that normally transports peptidoglycan across the bacterial cell membrane. Inhibition of IPP subsequently blocks cell wall formation (162).
Bacitracin is predominantly active against Gram-positive organisms, particularly S. aureus and S. pyogenes. In vitro studies suggest that other beta-hemolytic streptococci are either resistant or display reduced susceptibility (163), and this is occasionally used in the diagnostic microbiology laboratory as a distinguishing feature among certain beta-hemolytic streptococci (group A streptococcus usually appears to be susceptible to a 0.04-IU disc on blood agar). In general, Gram-negative bacteria are resistant, although the pathogenic Neisseria species (N. gonorrhoeae and N. meningitidis) and Haemophilus influenzae are usually susceptible (164).
Due to systemic toxicity (predominantly nephrotoxicity and thrombophlebitis), the use of bacitracin in humans is restricted to topical use. As such, there are no definitive pathogen-specific interpretive criteria for bacitracin susceptibility testing, although EUCAST data suggest that the wild-type distribution cutoff for Enterococcus faecalis and Enterococcus faecium is 32 μg/ml (www.eucast.org). The lack of definitive breakpoints means that there are few surveys of bacterial resistance to bacitracin, although resistance in human isolates of staphylococci and streptococci is thought to be low (165). Bacitracin can be administered alone or, more commonly, in combination with other topical antimicrobials, particularly polymyxin B (Polysporin) and/or neomycin (Neosporin) (163). It has previously been used for the treatment of minor skin, ear, or eye infections (165) but has strongly been associated with contact allergy, particularly in patients with preexisting skin conditions (165). As such, contemporary use of bacitracin has been superseded by use of other, safer topical agents.
Polymyxin B
Polymyxins are a group of antimicrobials that are products of Bacillus polymyxa and are active only against Gram-negative bacteria (166, 167). As cationic compounds, they are believed to interact with lipopolysaccharide, thereby disrupting the cell membrane, leading to cell death. Only polymyxin B and polymyxin E (colistin) are used clinically. According to the CLSI, the MIC cutoff for polymyxin B against Enterobacteriaceae is ≤2 μg/ml (82), although this is not commonly tested for in the clinical setting, as the limited Gram-positive spectrum of activity precludes the use of polymyxin B alone for bacterial skin infections. Polymyxin B is most often used in topical preparations alongside neomycin and bacitracin as a TAO (163). Although it has been available over-the-counter in the United States since the 1970s, a previous study demonstrated that rates of resistance to the three components of TAO among staphylococci, Enterobacteriaceae, and P. aeruginosa were generally low (163).
Retapamulin
Retapamulin belongs to the pleuromutilin class of antimicrobials, which are derived from Clitophilus scyphoides, an edible mushroom (168). Retapamulin is a semisynthetic member of this class, and similar to other members of this class, it has a novel mechanism of action in that it inhibits translation by binding to domain V of the 50S ribosomal subunit, acting at a site that is distinct from other agents, thus reducing the likelihood of cross-resistance (169). Pleuromutilin antibiotics (tiamulin and valnemulin) have been used in veterinary medicine for almost 30 years, predominantly in swine and poultry (170), but retapamulin has been registered for human use only since 2007 (171).
Retapamulin is licensed as a 1% ointment in the United States (Altabax) for the treatment of impetigo due to MSSA or S. pyogenes and in Europe (Altargo) for impetigo and infected minor wounds (172, 173). It has demonstrable in vitro activity against staphylococci and streptococci, with wild-type distribution cutoffs of 0.5 and 0.125 μg/ml for S. aureus and S. pyogenes, respectively. In vitro, development of resistance in multipassage studies can occur, with two studies reporting an increase in MIC values against S. aureus in serial passage studies (169, 174). Moreover, a small number of isolates with cross-resistance to linezolid or daptomycin were observed in one study (169). At the molecular level, retapamulin resistance in S. aureus has been associated with mutations in the rplC gene, which encodes ribosomal protein L3, and also with mutations in the 23S rRNA gene (175). In addition, resistance can be mediated by efflux pumps, such as VgaA or a variant, VgaAv (176), encoded by the vgaA or vgaAV gene, respectively. These genes are found on mobile genetic elements, and their products (belonging to the ATP-binding cassette protein family) are also associated with resistance to streptogramin A and lincosamide antibiotics (176). In addition, acquired retapamulin resistance can also be mediated by the cfr (chloramphenicol-florfenicol resistance)-encoded methyltransferase, which methylates the 23S rRNA subunit and prevents interaction with retapamulin (177).
To date, there are limited data on the global prevalence of retapamulin resistance among clinical isolates of S. aureus and S. pyogenes. One study from the United Kingdom in 2008 observed a resistance rate of <1% in S. aureus; notably, most of the strains included in this study were MRSA and were resistant to other topical agents, specifically mupirocin and/or fusidic acid (178). Similarly, a 2013 study from the United States evaluated retapamulin against 155 MRSA clinical isolates, including isolates resistant to vancomycin, linezolid, daptomycin, and mupirocin, and observed a resistance rate of 2.6% (179). More recently, however, a U.S. study evaluated S. aureus isolates from 400 children with SSTIs and identified 38 isolates (9.5%) that were resistant to retapamulin, with two isolates (0.5%) displaying cross-resistance to retapamulin and linezolid (180). In addition, four isolates were found to contain genes encoding the VgaA/Av efflux pumps, and all four of these isolates were also resistant to clindamycin, highlighting the potential for cross-resistance with acquired resistance mechanisms (180).
EMERGING TOPICAL ANTIBACTERIAL AGENTS
In response to rising rates of resistance to conventional antimicrobial agents, several compounds have emerged as potentially useful new topical agents, including some which have been repurposed from previously used agents. For example, ebselen is a synthetic organoselenium compound that was previously investigated for its anti-inflammatory and antioxidant activities (181). Previous in vitro preclinical work showed that ebselen displays bactericidal activity against multidrug-resistant clinical isolates of S. aureus, including MRSA and vancomycin-resistant S. aureus (VRSA) isolates (182, 183). Moreover, when it was evaluated in a mouse model of staphylococcal skin infection, topically applied ebselen significantly reduced bacterial loads and levels of proinflammatory cytokines (183). Ebselen is thought to act as a mimic of glutathione peroxidase, and potentially as an inhibitor of bacterial thioredoxin reductase (184); however, the exact mechanism of action remains unknown.
More recently, a number of antimicrobial peptides (AMPs) have been assessed for their antimicrobial efficacy. AMPs are naturally occurring, short (generally 5 to 15 amino acids) peptides that have attracted increasing attention as therapeutic agents for infections (185). AMPs are widely distributed in nature (185) and have broad-ranging antimicrobial activity in addition to potential host immunomodulatory effects (186). One promising topical compound is PXL150, a short, synthetic, broad-spectrum AMP that has activity against Gram-positive and Gram-negative pathogens (187, 188). An in vitro preclinical study suggested that PXL150 may have anti-inflammatory effects in a human monocytic cell line (187). Moreover, when it was used in a murine model of surgical wound infection, the application of topical PXL150 reduced the S. aureus load compared to that with placebo (189).
As topical antimicrobial agents, both ebselen and PXL150 are still in preclinical development. However, another emerging topical antimicrobial agent is pexiganan, a cationic peptide that is in phase 3 clinical development for topical use (190, 191). Clinically, pexiganan has been evaluated against systemic ofloxacin for the treatment of mildly infected diabetic foot ulcers, and it demonstrated clinical and microbiological equivalence in one study involving 835 patients (192). Pexiganan has a broad spectrum of activity against Gram-positive and Gram-negative pathogens (193), and it is thought to act by interacting with the negatively charged lipid bilayer, inducing toroid-like pore formation and subsequent disruption of the bacterial membrane (194). In vitro studies have shown good activity against S. aureus, CoNS, and S. pyogenes, with minimal development of resistance on serial passage at sub-MIC concentrations and no apparent cross-resistance to other commonly used topical agents, such as mupirocin and fusidic acid (190, 191).
TOPICAL BIOCIDES
Biocides are used extensively as disinfectants and antiseptics in topical and surface applications, including in the prevention of skin infections. They are key components of hospital infection control and cleaning programs and play an important role in the prevention of nosocomial infections. Biocides generally have a much broader spectrum of activity than those of antibiotics and typically have multiple nonspecific cellular targets. This broader target tropism may explain why resistance to biocides is far less prevalent than that to antibiotics, particularly at the high concentrations used in health care settings. Despite their widespread use, however, our understanding of the mechanisms of antimicrobial activity of these agents is often limited, and surveillance activities necessary for the identification and characterization of biocide-tolerant organisms are lacking. Importantly, the routine use of biocides is far less regulated than antibiotic use, leading to concern about the development of biocide resistance and the possible role that these agents may play in driving the emergence of multidrug-resistant pathogens (195). The following sections review the major topical biocides used in the prevention and treatment of bacterial skin infections.
Chlorhexidine
Chlorhexidine is a divalent cationic biguanide molecule that was first described in 1954 (196). Clinically, chlorhexidine has become the mainstay biocide in the prevention of health care-associated infections, and its role in decolonization regimens was recently extensively reviewed (5). Several different forms of chlorhexidine are used clinically, but the most common is the water-soluble form, chlorhexidine gluconate (197). Chlorhexidine can be incorporated into many products for use on the body, such as hand rubs, body washes, and antiseptic mouthwashes (198). Chlorhexidine can also be impregnated into wound dressings (199) and central line catheters (200) and is generally regarded as an extremely safe topical agent (201). Mild adverse effects include skin irritation and, more rarely, allergic reactions that include severe anaphylaxis (202, 203).
Chlorhexidine is most often used at concentrations of 0.5% to 4%, with the specific concentration dependent on the clinical indication. For example, hand disinfectants generally contain between 0.5% and 4% chlorhexidine (204), while MRSA decolonization is most often performed using a 1% chlorhexidine powder or with a 4% liquid (205). Presurgical skin disinfection often utilizes a 2% liquid suspension in 70% isopropyl alcohol (206), while bathing of ICU patients is normally performed using 4% liquid (207). There has been increasing adoption of the use of universal decolonization (using a combination of chlorhexidine bathing and intranasal mupirocin) of ICU patients to reduce health care-associated infections (208), raising concerns about the potential impact on bacterial resistance rates (209). To date, however, there is conflicting evidence regarding an increase in resistance to mupirocin and/or chlorhexidine as part of decolonization regimens (210, 211).
Mechanism and spectrum of action.
Chlorhexidine is a broad-spectrum biocide that also displays long-lasting residual activity in comparison to other biocides (212). It is most active against Gram-positive bacteria but also possesses activity against Gram-negative bacteria, some enveloped viruses, and fungi (213–215). However, it shows poor activity against nonenveloped viruses, and chlorhexidine is inactive against bacterial spores (201). Some bacterial species, such as mycobacteria (216), are intrinsically resistant to chlorhexidine because their outer membranes present an impermeable barrier that chlorhexidine cannot cross. Biofilm (217) and spore (197) formation also enables certain bacterial species to survive in the presence of chlorhexidine.
Chlorhexidine has both bacteriostatic and bactericidal activity, depending on the concentration used (218). Chlorhexidine is positively charged and, as such, binds to the negatively charged bacterial cell membrane and cell wall (197). At low concentrations, association of chlorhexidine with the cell membrane results in a loss of osmoregulatory and metabolic capacity, leading to the loss of cytosolic potassium ions, with subsequent inhibition of cellular respiration (197). At higher concentrations, chlorhexidine results in a complete loss of membrane integrity, with subsequent leakage of cellular contents from the cell and, ultimately, cell lysis and death.
Reduced susceptibility to chlorhexidine.
There are no standardized methods for chlorhexidine susceptibility testing, making comparability of data from published studies difficult. Numerous methods, including agar dilution, time-kill, and broth-based MIC and minimal bactericidal concentration (MBC) assays, have been used to phenotypically assess chlorhexidine susceptibility. Chlorhexidine has a low diffusion rate through solid agar, which precludes the use of disc diffusion susceptibility testing (205). Moreover, the antimicrobial effect of chlorhexidine may be overestimated if appropriate neutralization is not performed prior to susceptibility testing (219).
Efflux pumps are the most widely reported mechanism of resistance to chlorhexidine, and these protein complexes are able to actively pump chlorhexidine from the cell in an energy-dependent manner (220). In the majority of cases, efflux pumps have a broad range of substrates in addition to chlorhexidine (197). For example, Qac proteins, encoded by the quaternary ammonium compound (qac) genes, are multidrug efflux pumps that are distributed widely among Gram-positive and Gram-negative bacteria (221). They can broadly be split into two unrelated families, with QacA and QacB belonging to the major facilitator superfamily (MFS) and QacC (also referred to as Smr), QacE, QacEΔ1, QacF, QacG, QacH, QacJ, and QacZ belonging to the small multidrug resistance (SMR) family (221). However, apart from that of QacA, the role that Qac efflux systems play in chlorhexidine resistance has not yet been rigorously investigated.
The QacA protein is the most extensively studied of the Qac efflux systems and has been associated with increased tolerance to chlorhexidine (222, 223). It is usually found in Gram-positive bacteria, particularly staphylococci (221), although the qacA gene was also recently identified in clinical isolates of carbapenem-resistant Klebsiella pneumoniae displaying increased chlorhexidine tolerance (224). The qacA gene was first identified on S. aureus plasmid pSK1 (225) but has since been found on numerous other S. aureus plasmids, including pSK105, pSK107, pSK4032, pSK4769, pSK638, and pSK57 (205). Expression of qacA is under the regulatory control of a transcriptional regulator known as QacR (226). The gene encoding QacR (qacR) is located upstream of the qacA gene, and qacR and qacA are divergently transcribed (226). QacR acts as a repressor of qacA expression by binding to an operator sequence that overlaps the transcriptional start site of qacA, thus inhibiting expression (226). When an appropriate substrate (such as chlorhexidine) enters the cell, it binds to QacR, resulting in dissociation from DNA, which in turn relieves the repression of qacA gene expression (227).
Prevalence of chlorhexidine resistance.
In the absence of true clinical breakpoints, accurate determination of chlorhexidine susceptibility is challenging. Nevertheless, several studies have attempted to phenotypically assess the prevalence of reduced susceptibility to chlorhexidine among common skin pathogens, particularly staphylococci. For example, one U.S. study performed between 2010 and 2012 to assess clinical and colonizing MRSA isolates from the community found that only 1.6% of isolates were phenotypically nonsusceptible to chlorhexidine (228) by standard broth dilution assay. In contrast, some studies have reported relatively high rates of phenotypic resistance; for example, a study from Iranian hospitals conducted in 2012 and 2013 observed chlorhexidine MICs of 8 to 16 μg/ml in 30% of MSSA and 70% of MRSA isolates by broth dilution assay (229). Similarly, in a Taiwanese hospital study, 47.5% of MRSA isolates collected in 2005 were phenotypically nonsusceptible to chlorhexidine by agar dilution assay (230). Importantly, in the same study, the rate of chlorhexidine nonsusceptibility in isolates collected during 1990 was estimated to be only 1.7%, leading the authors of the study to suggest that chlorhexidine nonsusceptibility might be increasing (230). Like that in S. aureus, reduced chlorhexidine susceptibility in S. epidermidis has also been reported. One Swedish study using 143 isolates retrospectively collected between 1993 and 2011 from a single hospital used agar dilution and observed chlorhexidine tolerance in 37.5% of S. epidermidis isolates (231). A more recent Scottish study included 25 S. epidermidis isolates obtained between 2007 and 2014 from a single hospital intensive care unit, and it showed a chlorhexidine tolerance rate of 12% by agar dilution assay. In both studies, there was an association between increased chlorhexidine tolerance and the carriage of qacA/B genes (231, 232).
In addition to phenotypic resistance, numerous studies have assessed the rates of carriage of qac and smr genes in staphylococci (205). The prevalence of carriage is highly variable and is dependent on the geographical and epidemiological background of the study population. For example, in one U.S. study assessing MRSA isolates from a regional health care network in the District of Columbia, the carriage of the qacA/B genes was found to be <1% (5/493 isolates) (233). Similarly, another study assessing the prevalence of qacA/B in 86 MRSA isolates collected from prisoners of the Rikers Island jail system in the United States found an even lower prevalence, with no evidence of qacA/B in any of the collected isolates (234). In contrast, qacA/B genes were detected in 50/60 (83%) MRSA isolates in a 2009 study from a Malaysian hospital (235). Moreover, the prevalence of smr carriage can vary widely, with one Canadian study reporting a prevalence of approximately 7% for 334 MRSA isolates collected from two intensive care units between 2005 and 2009 (236), while another study, conducted between 2005 and 2008 in Chinese hospitals, observed an smr prevalence of approximately 77% (41/53 MRSA isolates) (237). Importantly, however, many studies that have assessed qacA and/or smr carriage have not performed parallel phenotypic susceptibility testing, precluding meaningful genotypic/phenotypic correlations with chlorhexidine nonsusceptibility. Previous work has demonstrated that carriage of these genes does not always result in phenotypic nonsusceptibility (238). As such, prevalence studies based on rates of gene carriage alone may overestimate rates of chlorhexidine tolerance, which may therefore limit the clinical usefulness of these studies.
Triclosan
Triclosan, a member of the bisphenol group of compounds, exhibits a broad spectrum of antimicrobial activity (201) and is found in numerous health care and hygiene products, including soaps, surgical scrubs, clinical hand washes, toothpastes, and mouthwashes (239). Clinically, triclosan has been used predominantly in MRSA decolonization regimens, although it has largely been superseded by chlorhexidine. Triclosan has also been incorporated into a range of fabrics and plastics, such as those used in surgical drapes, toothbrush handles, wound sutures, mop handles, and even children's toys (240). Much recent work has demonstrated the lack of efficacy of triclosan in household soap products, prompting the U.S. FDA to recently announce that, effective September 2017, the use of triclosan and 18 other biocidal chemicals would be prohibited in “consumer antiseptic products” (6).
Mechanism of action.
Triclosan exhibits broad-spectrum antimicrobial activity predominantly against bacteria, but it is also active against some viruses and fungi (241). For many years, triclosan was thought to target the cell membrane in a nonspecific manner, akin to some other biocides (242). However, several independent studies have demonstrated that triclosan acts on a defined target within the bacterial fatty acid biosynthetic pathway known as FabI (243–245), or InhA in Mycobacterium spp. (246). These essential proteins are NADH-dependent enoyl-acyl carrier protein reductases which are involved in the elongation cycle of fatty acids, an important step of lipid metabolism (247). Triclosan forms a stable complex with the amino acids of the FabI enzyme active site, where it acts as an inhibitor by mimicking the natural FabI substrate (244). The specific inhibition of FabI by triclosan results in the arrest of fatty acid biosynthesis within the cell. This in turn adversely affects a multitude of different cellular processes, including the synthesis of lipopolysaccharide, phospholipids, and lipoproteins (248), which may explain why triclosan was originally thought to target the cell membrane. However, it has also been suggested that triclosan may have nonspecific activity at higher concentrations, such as those used in topical antiseptics, where triclosan may cause cell lysis through effects on RNA and protein synthesis, leading to adverse effects on membrane integrity (240).
Mechanisms and prevalence of triclosan resistance.
Similar to the case for other biocides, there is no standardized method for triclosan susceptibility testing, and there are no defined susceptibility breakpoints. Nevertheless, resistance to triclosan is described in the literature, with a number of different mechanisms reported. Some organisms, such as P. aeruginosa, are inherently resistant to the biocide, while others, such as S. aureus, may become resistant following exposure (241). Resistance to triclosan is generally mediated by mutations within the fabI gene, encoding the enoyl-acyl carrier protein reductase. For example, in Escherichia coli, mutations that affect the active site of FabI are known to interfere with triclosan binding, leading to increased levels of triclosan resistance (244). Similarly, mutations in the fabI genes of S. aureus (249) and Acinetobacter baumannii (250) and the inhA gene of mycobacterial species (246) have been shown to confer increased tolerance to triclosan. In addition, mutations leading to increased FabI expression have also been shown to result in low and intermediate levels of triclosan resistance in A. baumannii (250) and S. aureus (251), respectively. In S. aureus, these mutations were found to arise most often in the upstream promoter region of the fabI gene (251). However, high-level resistance was observed only in strains that overexpressed mutant rather than wild-type FabI proteins, suggesting a synergistic effect of FabI mutation and overexpression in relation to triclosan resistance (251).
A recent study demonstrated that triclosan resistance in several staphylococcal species was mediated by the acquisition of a heterologous copy of the fabI gene known as sh-fabI, which is thought to have originated in Staphylococcus haemolyticus (252). Similarly to mutations that result in overexpression of fabI in other organisms, the acquisition of sh-fabI is thought to result in increased levels of FabI within the cell due to expression of both the native fabI gene and the acquired gene (252). Subsequent screening of available whole-genome sequences revealed the presence of variant sh-fabI alleles that contained single nucleotide polymorphisms (SNPs) predicted to confer triclosan resistance in a range of staphylococcal species (252). It was a concern that sh-fabI was located on a novel mobile genetic element, TnSha1 (253). The acquisition of sh-fabI in S. aureus was almost exclusively associated with a single copy of TnSha1 that was integrated into the chromosome, while in S. haemolyticus the sh-fabI gene was generally found to be part of a larger plasmid-borne IS1272-type element, TnSha2 (253). S. epidermidis was found to commonly carry both the TnSha1 and TnSha2 elements (253). IS-mediated transposition of TnSha1 and TnSha2 therefore represents a concerning example of horizontally acquired triclosan resistance determinants.
To date, there have been very few studies assessing the prevalence of triclosan resistance in clinical isolates of common skin pathogens. Although phenotypic testing for triclosan resistance was not performed, an in silico screen of >4,000 S. aureus and 300 S. epidermidis genomes revealed that the sh-fabI resistance gene was present in approximately 1.5% of S. aureus and 14% of S. epidermidis isolates (253). The finding of acquired triclosan resistance within staphylococcus isolates is of concern and, given the environmental ubiquity of triclosan, warrants ongoing investigation.
Povidone-Iodine
Iodine was originally discovered by Courtois in 1811, and although its potential as an antiseptic was recognized soon after its discovery, its use was limited by poor solubility, limited stability, and toxicity (254). Early medicinal iodine compounds, such as Lugol's solution and iodine tinctures, overcame some of these issues by combining free iodine with potassium iodide salts and alcohol, which greatly improved the solubility (255). However, a breakthrough occurred in the 1950s, when complexation of iodine with large organic polymers was found to result in the release of free iodine in aqueous systems. These iodophor complexes have since become the most common class of medicinal iodine compounds (254). Povidone-iodine is one such iodophor, which consists of complexed iodine and polyvinylpyrrolidone (PVP), or povidone (256). It is commonly used as a skin antiseptic, particularly prior to injections, surgery, and other invasive procedures. Although 10% povidone-iodine solutions are generally used for presurgical skin disinfection (257), lower concentrations, such as 5%, may be used for ophthalmic procedures (258). In addition, the use of more dilute solutions (less than 2%) may be effective for the prophylaxis and treatment of childhood conjunctivitis (259). Several studies have also proposed a use for dilute povidone-iodine in the management of chronic, nonhealing wounds (260, 261). However, the clinical efficacy of povidone-iodine in the management of such wounds remains controversial (262).
Mechanism of action and resistance.
In povidone-iodine preparations, PVP acts as a water-soluble carrier that releases the free iodine in solution (256). Although iodine is considered the bactericidal component, PVP is known to increase the antimicrobial efficiency of iodine as a consequence of its affinity for lipid membranes, which enables PVP to release free iodine in close proximity to the cell membranes of target microorganisms (257). Like chlorine, iodine is a powerful oxidizing agent that can quickly penetrate the cell membrane (201). Although the mechanism is not completely understood, once inside the cell, free iodine destabilizes membrane integrity, denatures nucleic acids, and can rapidly kill microorganisms by nonspecifically inhibiting essential cellular processes, including electron transport, cellular respiration, and protein synthesis (257).
Povidone-iodine has perhaps the broadest spectrum of activity of any antiseptic, with good activity against a range of bacteria, fungi, and protozoa, including Pseudomonas, Staphylococcus, Mycobacterium, Candida, and Trichophyton species (201). It also displays virucidal activity and can eradicate viruses such as influenza virus, HIV, and Ebola virus (263, 264). Some reports also suggest that, with increased exposure time, povidone-iodine may display sporicidal activity (265).
As with other biocides, there is no consensus on a standard method for povidone-iodine susceptibility testing. However, one previous study found that povidone-iodine activity was not detectable in susceptibility agar, suggesting that agar-based methodologies might not be appropriate for testing of this biocide (266). As with chlorhexidine, povidone-iodine must be neutralized appropriately before susceptibility is assessed in order to avoid overestimates of antimicrobial activity (267). Despite the widespread clinical use of povidone-iodine over many decades, as well as extensive testing of isolates, there have so far been no reports of resistance or increased tolerance to povidone-iodine in any laboratory-derived or clinical isolates (268). However, the clinical use of povidone-iodine in surgical antisepsis and decolonization regimens has generally been superseded by the use of chlorhexidine, particularly given the prolonged residual activity of chlorhexidine compared to that of povidone-iodine (269).
Alcohol
Alcohol-based biocides are widely used for surface disinfection and skin antisepsis and are commonly found in many antibacterial hand washes, being a mainstay of many hospital hand hygiene programs (270, 271). Alcohols are also often combined with other biocides, such as chlorhexidine (272, 273), which display residual activity following evaporation of the alcohol or with excipients that can lower the rate of alcohol evaporation and therefore increase the contact time and antimicrobial efficacy (274). Many different alcohols display potent antimicrobial activity; however, ethyl alcohol, isopropyl alcohol, and n-propanol are most commonly used as biocides (201). Alcohols are rapidly bactericidal and exhibit a broad spectrum of activity, particularly against vegetative bacteria. In addition, they are active against Mycobacterium, fungi, and viruses but have no activity against bacterial spores. The activities of both ethyl and isopropyl alcohols are highly dependent on the concentration used, with the bactericidal activities of both agents dropping sharply when the agents are diluted to below a 50% concentration. In general, optimal bactericidal activity is achieved at a concentration of 60% to 90% (275). Absolute alcohol is less bactericidal than alcohol which has been diluted to the optimal (60% to 90%) range with water (275). The specific mechanism by which alcohol achieves its antimicrobial activity is not well understood but may be related to protein denaturation (201) or to inhibition and uncoupling of mRNA and protein synthesis through direct effects on ribosomes and RNA polymerase (276). This process ultimately results in membrane damage, interference with essential metabolic pathways, and a loss of cellular integrity, leading to a loss of viability (201).
Resistance to alcohol.
There are no standardized susceptibility testing methods to assess alcohol tolerance, although the generation of enhanced biofuel-producing bacteria demonstrates that tolerance to alcohols, including ethanol and butanol, can readily be achieved (277–279). However, in the context of clinical usage, alcohol tolerance in bacteria such as staphylococci and streptococci has not been reported, and acquired resistance mechanisms to alcohol have not yet been identified. As with other biocides, this may be reflective of the nonspecific mode of bactericidal action. Alcohol exposure may also result in the increased production of biofilm for a number of clinically relevant species, including S. aureus (280), S. epidermidis (281), and A. baumannii (282). In addition, exposure of A. baumannii to low levels of alcohol can modulate the virulence response of the organism, leading to more severe disease in animal models of infection (282, 283). The clinical relevance of this observation to human disease, however, remains unknown.
Hydrogen Peroxide
Since its discovery in 1818 by Louis Thenard, hydrogen peroxide has become a widely used antiseptic agent that can be used in both liquid and gas forms (284). It is considered a potent and broad-spectrum antimicrobial that is active against all forms of microorganisms, including bacteria, viruses, and protozoa (285, 286). Importantly, it is also active against bacterial spores, protozoal cysts, and prions (287–289). In liquid form, hydrogen peroxide can be used on the skin as an antiseptic at concentrations of 3% to 6% (vol/vol) (284). It is also commonly used in dental disinfectants at concentrations between 0.4% and 1% and is often the active ingredient in contact lens solution, in which it is generally used at a concentration of 3% (290, 291). One recent study suggested that hydrogen peroxide and iodine may act synergistically against S. aureus, with lower concentrations of both agents required to effect killing (292).
The mechanism of action of hydrogen peroxide is not fully understood but is thought to be associated primarily with its oxidation activity. Hydrogen peroxide can rapidly cross the cell membrane; once it is inside the cell, the presence of trace metals, such as iron, catalyzes the production of highly reactive hydroxyl radicals, which results in the formation of lesions and nicks in cellular DNA, the cleavage of nucleic acid and protein backbones, and subsequent damage to the cell membrane (293, 294). This oxidative damage results in the impairment of many cellular processes, including RNA, DNA, and protein synthesis pathways, disruption of cellular homeostasis, and a loss of cellular integrity, which ultimately lead to a loss of viability (201).
Resistance to hydrogen peroxide.
There is no standard method for hydrogen peroxide susceptibility testing, nor any recognized breakpoints, although methods such as broth microdilution have been used to assess MIC and MBC values (295). Even though hydrogen peroxide has been used widely as a biocide for many years, the development of significant levels of tolerance among clinical isolates does not seem to have arisen. However, there are a small number of reports in the literature detailing hydrogen peroxide tolerance in several clinically relevant bacterial species, predominantly in organisms that produce catalase. Catalase is an enzyme that catalyzes the decomposition of hydrogen peroxide to oxygen and water (296). It is important in protecting cells from oxidative damage caused by reactive oxygen species (ROS) and is produced by almost all aerobic bacteria (297, 298). Although tolerance to exogenous hydrogen peroxide is rare, in some species the production of catalase may be associated with tolerance to hydrogen peroxide. For example, in one study, exposure of S. aureus to hydrogen peroxide was found to result in an increased frequency of SCV formation (299). Notably, the SCVs produced under these conditions expressed elevated levels of the KatA catalase enzyme, which rendered them tolerant to high levels of external hydrogen peroxide (299). Similarly, in E. faecalis, the production of the KatA catalase provided partial protection of the organism against environmental challenge with hydrogen peroxide (300), while in A. baumannii and Acinetobacter nosocomialis, the production of the KatG and KatE catalases is known to confer high-level tolerance to biocidal hydrogen peroxide (301). Although some organisms possess an intrinsic tolerance to hydrogen peroxide which can render the biocide ineffective, as described above, acquired resistance has not yet been reported.
CONCLUSIONS AND FUTURE CONSIDERATIONS
In summary, with the exception of impetigo and nasal decolonization of S. aureus, there are limited clinical data to support the widespread use of topical antimicrobials, including in the prevention and treatment of chronic wound infections. Importantly, widespread and indiscriminate use of topical agents, particularly mupirocin and fusidic acid, has led to the emergence of bacterial resistance, predominantly in staphylococci. In particular, the dissemination of acquired resistance mechanisms within and across staphylococcal species is of concern. Despite the fact that most topical agents are not used systemically, in the case of fusidic acid, resistance generated by topical use has largely precluded systemic use in some settings, such as New Zealand. Although agents such as retapamulin offer promise, resistance has already begun to emerge in staphylococci, with concerning reports of cross-resistance to systemic agents. In order to avoid further increases in resistance, consideration should be given to restricting the use of topical agents (e.g., to specialist-only prescribing).
Although antiseptics offer a possible alternative to the use of topical antimicrobials, robust clinical efficacy data are presently lacking, and the lack of standardized susceptibility testing limits monitoring of the development of tolerance and cross-resistance to other antimicrobial agents. Future work in this field should include trials undertaking head-to-head comparisons of topical antimicrobials and antiseptics in the treatment of impetigo, particularly in settings with a high prevalence of resistance to topical agents. Moreover, as universal decolonization regimens are increasingly implemented in health care settings, it is important that regular, systematic surveillance be conducted to identify any increase in resistance to mupirocin and/or chlorhexidine.
ACKNOWLEDGMENTS
The National Health and Medical Research Council, Australia, funded practitioner fellowship GNT1105905 to B.P.H. and early career fellowship GNT1123854 to D.A.W. The Microbiological Diagnostic Unit Public Health Laboratory is funded by the Victorian Government.
Biographies

Deborah A. Williamson is a clinical microbiologist and academic researcher and is Deputy Director of the Microbiological Diagnostic Unit Public Health Laboratory (MDU PHL). She is involved in the delivery of specialist public health laboratory services and in the diagnosis and surveillance of communicable diseases. Her research interests include the molecular epidemiology and pathogenesis of infections caused by antimicrobial-resistant pathogens and the translation of genomic technologies to questions of public health importance.

Glen P. Carter is the Senior Project Officer at the Doherty Centre for Applied Microbial Genomics (DAMG). Prior to this role, Dr. Carter received his Ph.D. in molecular microbiology from the University of Nottingham in 2005 and was then a postdoctoral researcher at Monash University for 10 years, where his research focused on the pathogenesis of Clostridium difficile and related pathogens and on the identification of small-molecule inhibitors active against these bacteria.

Benjamin P. Howden is a medical microbiologist, infectious disease physician, and molecular biologist. He is Director of the Microbiological Diagnostic Unit Public Health Laboratory (MDU PHL) and Medical Director of the Doherty Centre for Applied Microbial Genomics. He is responsible for the provision of public health laboratory services, the translation of microbial genomics into public health and clinical microbiology practice, and research investigating antimicrobial resistance, bacterial pathogenesis, bacterial evolution, and host-pathogen interactions.
REFERENCES
- 1.Wysocki AB. 1999. Skin anatomy, physiology, and pathophysiology. Nurs Clin North Am 34:777–797. [PubMed] [Google Scholar]
- 2.Findley K, Grice EA. 2014. The skin microbiome: a focus on pathogens and their association with skin disease. PLoS Pathog 10:e1004436. doi: 10.1371/journal.ppat.1004436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grice EA. 2015. The intersection of microbiome and host at the skin interface: genomic- and metagenomic-based insights. Genome Res 25:1514–1520. doi: 10.1101/gr.191320.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hersh AL, Chambers HF, Maselli JH, Gonzales R. 2008. National trends in ambulatory visits and antibiotic prescribing for skin and soft-tissue infections. Arch Intern Med 168:1585–1591. doi: 10.1001/archinte.168.14.1585. [DOI] [PubMed] [Google Scholar]
- 5.Septimus EJ, Schweizer ML. 2016. Decolonization in prevention of health care-associated infections. Clin Microbiol Rev 29:201–222. doi: 10.1128/CMR.00049-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McNamara PJ, Levy SB. 2016. Triclosan: an instructive tale. Antimicrob Agents Chemother 60:7015–7016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kampf G. 2016. Acquired resistance to chlorhexidine—is it time to establish an ‘antiseptic stewardship’ initiative? J Hosp Infect 94:213–227. doi: 10.1016/j.jhin.2016.08.018. [DOI] [PubMed] [Google Scholar]
- 8.Health and Social Care Information Centre. 2015. Prescription cost analysis, England—2015. http://content.digital.nhs.uk/catalogue/PUB20200.
- 9.Lapolla WJ, Levender MM, Davis SA, Yentzer BA, Williford PM, Feldman SR. 2011. Topical antibiotic trends from 1993 to 2007: use of topical antibiotics for non-evidence-based indications. Dermatol Surg 37:1427–1433. doi: 10.1111/j.1524-4725.2011.02122.x. [DOI] [PubMed] [Google Scholar]
- 10.Williamson D, Ritchie SR, Best E, Upton A, Leversha A, Smith A, Thomas MG. 2015. A bug in the ointment: topical antimicrobial usage and resistance in New Zealand. N Z Med J 128:103–109. [PubMed] [Google Scholar]
- 11.Hartman-Adams H, Banvard C, Juckett G. 2014. Impetigo: diagnosis and treatment. Am Fam Physician 90:229–235. [PubMed] [Google Scholar]
- 12.Koning S, van der Sande R, Verhagen AP, van Suijlekom-Smit LW, Morris AD, Butler CC, Berger M, van der Wouden JC. 2012. Interventions for impetigo. Cochrane Database Syst Rev 1:CD003261. doi: 10.1002/14651858.CD003261.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bowen AC, Mahe A, Hay RJ, Andrews RM, Steer AC, Tong SY, Carapetis JR. 2015. The global epidemiology of impetigo: a systematic review of the population prevalence of impetigo and pyoderma. PLoS One 10:e0136789. doi: 10.1371/journal.pone.0136789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koning S, van Suijlekom-Smit LW, Nouwen JL, Verduin CM, Bernsen RM, Oranje AP, Thomas S, van der Wouden JC. 2002. Fusidic acid cream in the treatment of impetigo in general practice: double blind randomised placebo controlled trial. BMJ 324:203–206. doi: 10.1136/bmj.324.7331.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Christensen OB, Anehus S. 1994. Hydrogen peroxide cream: an alternative to topical antibiotics in the treatment of impetigo contagiosa. Acta Derm Venereol 74:460–462. [DOI] [PubMed] [Google Scholar]
- 16.Vogel A, Lennon D, Best E, Leversha A. 2016. Where to from here? The treatment of impetigo in children as resistance to fusidic acid emerges N Z Med J 129:77–83. [PubMed] [Google Scholar]
- 17.Gould L, Abadir P, Brem H, Carter M, Conner-Kerr T, Davidson J, DiPietro L, Falanga V, Fife C, Gardner S, Grice E, Harmon J, Hazzard WR, High KP, Houghton P, Jacobson N, Kirsner RS, Kovacs EJ, Margolis D, McFarland Horne F, Reed MJ, Sullivan DH, Thom S, Tomic-Canic M, Walston J, Whitney J, Williams J, Zieman S, Schmader K. 2015. Chronic wound repair and healing in older adults: current status and future research. Wound Repair Regen 23:1–13. doi: 10.1111/wrr.12245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graham ID, Harrison MB, Nelson EA, Lorimer K, Fisher A. 2003. Prevalence of lower-limb ulceration: a systematic review of prevalence studies. Adv Skin Wound Care 16:305–316. doi: 10.1097/00129334-200311000-00013. [DOI] [PubMed] [Google Scholar]
- 19.Norman G, Dumville JC, Moore ZE, Tanner J, Christie J, Goto S. 2016. Antibiotics and antiseptics for pressure ulcers. Cochrane Database Syst Rev 4:CD011586. doi: 10.1002/14651858.CD011586.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raghavan P, Raza WA, Ahmed YS, Chamberlain MA. 2003. Prevalence of pressure sores in a community sample of spinal injury patients. Clin Rehabil 17:879–884. doi: 10.1191/0269215503cr692oa. [DOI] [PubMed] [Google Scholar]
- 21.VanGilder C, Amlung S, Harrison P, Meyer S. 2009. Results of the 2008-2009 International Pressure Ulcer Prevalence Survey and a 3-year, acute care, unit-specific analysis. Ostomy Wound Manage 55:39–45. [PubMed] [Google Scholar]
- 22.Moore Z, Johansen E, van Etten M. 2013. A review of PU risk assessment and prevention in Scandinavia, Iceland and Ireland (part II). J Wound Care 22:423–424, 426–428, 430–431. doi: 10.12968/jowc.2013.22.8.423. [DOI] [PubMed] [Google Scholar]
- 23.Moore Z, Johanssen E, van Etten M. 2013. A review of PU prevalence and incidence across Scandinavia, Iceland and Ireland (part I). J Wound Care 22:361–362, 364–368. doi: 10.12968/jowc.2013.22.7.361. [DOI] [PubMed] [Google Scholar]
- 24.Cutting KF, White RJ. 2005. Criteria for identifying wound infection—revisited. Ostomy Wound Manage 51:28–34. [PubMed] [Google Scholar]
- 25.Cutting KF, White RJ, Mahoney P. 2013. Wound infection, dressings and pain, is there a relationship in the chronic wound? Int Wound J 10:79–86. doi: 10.1111/j.1742-481X.2012.00947.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Howell-Jones RS, Wilson MJ, Hill KE, Howard AJ, Price PE, Thomas DW. 2005. A review of the microbiology, antibiotic usage and resistance in chronic skin wounds. J Antimicrob Chemother 55:143–149. doi: 10.1093/jac/dkh513. [DOI] [PubMed] [Google Scholar]
- 27.Schmidt K, Debus ES, St Jessberger, Ziegler U, Thiede A. 2000. Bacterial population of chronic crural ulcers: is there a difference between the diabetic, the venous, and the arterial ulcer? Vasa 29:62–70. doi: 10.1024/0301-1526.29.1.62. [DOI] [PubMed] [Google Scholar]
- 28.Schraibman IG. 1990. The significance of beta-haemolytic streptococci in chronic leg ulcers. Ann R Coll Surg Engl 72:123–124. [PMC free article] [PubMed] [Google Scholar]
- 29.Trengove NJ, Stacey MC, McGechie DF, Mata S. 1996. Qualitative bacteriology and leg ulcer healing. J Wound Care 5:277–280. doi: 10.12968/jowc.1996.5.6.277. [DOI] [PubMed] [Google Scholar]
- 30.Abbas M, Uckay I, Lipsky BA. 2015. In diabetic foot infections antibiotics are to treat infection, not to heal wounds. Expert Opin Pharmacother 16:821–832. doi: 10.1517/14656566.2015.1021780. [DOI] [PubMed] [Google Scholar]
- 31.National Institute of Health and Clinical Excellence (NICE). 2014. Pressure ulcers: prevention and management of pressure ulcers. Clinical guideline 179. NICE, London, United Kingdom: http://www.nice.org.uk/guidance/cg179/. [Google Scholar]
- 32.O'Meara S, Al-Kurdi D, Ologun Y, Ovington LG, Martyn-St James M, Richardson R. 2014. Antibiotics and antiseptics for venous leg ulcers. Cochrane Database Syst Rev 1:CD003557. doi: 10.1002/14651858.CD003557.pub5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nisi G, Brandi C, Grimaldi L, Calabro M, D'Aniello C. 2005. Use of a protease-modulating matrix in the treatment of pressure sores. Chir Ital 57:465–468. [PubMed] [Google Scholar]
- 34.Kaya AZ, Turani N, Akyuz M. 2005. The effectiveness of a hydrogel dressing compared with standard management of pressure ulcers. J Wound Care 14:42–44. doi: 10.12968/jowc.2005.14.1.26726. [DOI] [PubMed] [Google Scholar]
- 35.World Health Organization (WHO). 2006. Facts about injuries: burns. WHO and International Society for Burns Injuries, Geneva, Switzerland. [Google Scholar]
- 36.Gibran NS, Heimbach DM. 2000. Current status of burn wound pathophysiology. Clin Plast Surg 27:11–22. [PubMed] [Google Scholar]
- 37.Church D, Elsayed S, Reid O, Winston B, Lindsay R. 2006. Burn wound infections. Clin Microbiol Rev 19:403–434. doi: 10.1128/CMR.19.2.403-434.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dai T, Huang YY, Sharma SK, Hashmi JT, Kurup DB, Hamblin MR. 2010. Topical antimicrobials for burn wound infections. Recent Pat Antiinfect Drug Discov 5:124–151. doi: 10.2174/157489110791233522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharma BR. 2007. Infection in patients with severe burns: causes and prevention thereof. Infect Dis Clin North Am 21:745–759. doi: 10.1016/j.idc.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 40.Sevgi M, Toklu A, Vecchio D, Hamblin MR. 2013. Topical antimicrobials for burn infections—an update. Recent Pat Antiinfect Drug Discov 8:161–197. doi: 10.2174/15748898113089990001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Monafo WW, West MA. 1990. Current treatment recommendations for topical burn therapy. Drugs 40:364–373. doi: 10.2165/00003495-199040030-00004. [DOI] [PubMed] [Google Scholar]
- 42.Barajas-Nava LA, Lopez-Alcalde J, Roque i Figuls M, Sola I, Bonfill Cosp X. 2013. Antibiotic prophylaxis for preventing burn wound infection. Cochrane Database Syst Rev 6:CD008738. doi: 10.1002/14651858.CD008738.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barret JP, Dziewulski P, Ramzy PI, Wolf SE, Desai MH, Herndon DN. 2000. Biobrane versus 1% silver sulfadiazine in second-degree pediatric burns. Plast Reconstr Surg 105:62–65. doi: 10.1097/00006534-200001000-00010. [DOI] [PubMed] [Google Scholar]
- 44.Bugmann P, Taylor S, Gyger D, Lironi A, Genin B, Vunda A, La Scala G, Birraux J, Le Coultre C. 1998. A silicone-coated nylon dressing reduces healing time in burned paediatric patients in comparison with standard sulfadiazine treatment: a prospective randomized trial. Burns 24:609–612. doi: 10.1016/S0305-4179(98)00095-3. [DOI] [PubMed] [Google Scholar]
- 45.Caruso DM, Foster KN, Blome-Eberwein SA, Twomey JA, Herndon DN, Luterman A, Silverstein P, Antimarino JR, Bauer GJ. 2006. Randomized clinical study of Hydrofiber dressing with silver or silver sulfadiazine in the management of partial-thickness burns. J Burn Care Res 27:298–309. doi: 10.1097/01.BCR.0000216741.21433.66. [DOI] [PubMed] [Google Scholar]
- 46.Gerding RL, Emerman CL, Effron D, Lukens T, Imbembo AL, Fratianne RB. 1990. Outpatient management of partial-thickness burns: Biobrane versus 1% silver sulfadiazine. Ann Emerg Med 19:121–124. doi: 10.1016/S0196-0644(05)81793-7. [DOI] [PubMed] [Google Scholar]
- 47.Gerding RL, Imbembo AL, Fratianne RB. 1988. Biosynthetic skin substitute vs. 1% silver sulfadiazine for treatment of inpatient partial-thickness thermal burns. J Trauma 28:1265–1269. doi: 10.1097/00005373-198808000-00022. [DOI] [PubMed] [Google Scholar]
- 48.Gotschall CS, Morrison MI, Eichelberger MR. 1998. Prospective, randomized study of the efficacy of Mepitel on children with partial-thickness scalds. J Burn Care Rehabil 19:279–283. [DOI] [PubMed] [Google Scholar]
- 49.Hosseini SN, Karimian A, Mousavinasab SN, Rahmanpour H, Yamini M, Zahmatkesh SH. 2009. Xenoderm versus 1% silver sulfadiazine in partial-thickness burns. Asian J Surg 32:234–239. doi: 10.1016/S1015-9584(09)60400-0. [DOI] [PubMed] [Google Scholar]
- 50.Muangman P, Chuntrasakul C, Silthram S, Suvanchote S, Benjathanung R, Kittidacha S, Rueksomtawin S. 2006. Comparison of efficacy of 1% silver sulfadiazine and Acticoat for treatment of partial-thickness burn wounds. J Med Assoc Thai 89:953–958. [PubMed] [Google Scholar]
- 51.Noordenbos J, Dore C, Hansbrough JF. 1999. Safety and efficacy of TransCyte for the treatment of partial-thickness burns. J Burn Care Rehabil 20:275–281. [DOI] [PubMed] [Google Scholar]
- 52.Zimlichman E, Henderson D, Tamir O, Franz C, Song P, Yamin CK, Keohane C, Denham CR, Bates DW. 2013. Health care-associated infections: a meta-analysis of costs and financial impact on the US health care system. JAMA Intern Med 173:2039–2046. doi: 10.1001/jamainternmed.2013.9763. [DOI] [PubMed] [Google Scholar]
- 53.Anderson DJ, Podgorny K, Berrios-Torres SI, Bratzler DW, Dellinger EP, Greene L, Nyquist AC, Saiman L, Yokoe DS, Maragakis LL, Kaye KS. 2014. Strategies to prevent surgical site infections in acute care hospitals: 2014 update. Infect Control Hosp Epidemiol 35:605–627. doi: 10.1086/676022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schweizer ML, Chiang HY, Septimus E, Moody J, Braun B, Hafner J, Ward MA, Hickok J, Perencevich EN, Diekema DJ, Richards CL, Cavanaugh JE, Perlin JB, Herwaldt LA. 2015. Association of a bundled intervention with surgical site infections among patients undergoing cardiac, hip, or knee surgery. JAMA 313:2162–2171. doi: 10.1001/jama.2015.5387. [DOI] [PubMed] [Google Scholar]
- 55.McHugh SM, Collins CJ, Corrigan MA, Hill AD, Humphreys H. 2011. The role of topical antibiotics used as prophylaxis in surgical site infection prevention. J Antimicrob Chemother 66:693–701. doi: 10.1093/jac/dkr009. [DOI] [PubMed] [Google Scholar]
- 56.Heal CF, Banks JL, Lepper PD, Kontopantelis E, van Driel ML. 2016. Topical antibiotics for preventing surgical site infection in wounds healing by primary intention. Cochrane Database Syst Rev 11:CD011426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.OzFoodNet Working Group. 2010. OzFoodNet quarterly report, 1 July to 30 September 2010. Commun Dis Intell Q Rep 34:450–458. [PubMed] [Google Scholar]
- 58.Norman G, Dumville JC, Mohapatra DP, Owens GL, Crosbie EJ. 2016. Antibiotics and antiseptics for surgical wounds healing by secondary intention. Cochrane Database Syst Rev 3:CD011712. doi: 10.1002/14651858.CD011712.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Waterbrook AL, Hiller K, Hays DP, Berkman M. 2013. Do topical antibiotics help prevent infection in minor traumatic uncomplicated soft tissue wounds? Ann Emerg Med 61:86–88. doi: 10.1016/j.annemergmed.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 60.Dire DJ, Coppola M, Dwyer DA, Lorette JJ, Karr JL. 1995. Prospective evaluation of topical antibiotics for preventing infections in uncomplicated soft-tissue wounds repaired in the ED. Acad Emerg Med 2:4–10. doi: 10.1111/j.1553-2712.1995.tb03070.x. [DOI] [PubMed] [Google Scholar]
- 61.Rothrock SG. 2013. A single review article cannot define a standard of care for uncomplicated wounds. Ann Emerg Med 61:502. doi: 10.1016/j.annemergmed.2012.09.019. [DOI] [PubMed] [Google Scholar]
- 62.Maddox JS, Ware JC, Dillon HC Jr. 1985. The natural history of streptococcal skin infection: prevention with topical antibiotics. J Am Acad Dermatol 13:207–212. doi: 10.1016/S0190-9622(85)70160-0. [DOI] [PubMed] [Google Scholar]
- 63.Wuite J, Davies BI, Go M, Lambers J, Jackson D, Mellows G. 1983. Pseudomonic acid: a new topical antimicrobial agent. Lancet ii:394. [DOI] [PubMed] [Google Scholar]
- 64.Dacre JE, Emmerson AM, Jenner EA. 1983. Nasal carriage of gentamicin and methicillin resistant Staphylococcus aureus treated with topical pseudomonic acid. Lancet ii:1036. [DOI] [PubMed] [Google Scholar]
- 65.Reilly GD, Spencer RC. 1984. Pseudomonic acid—a new antibiotic for skin infections. J Antimicrob Chemother 13:295–298. doi: 10.1093/jac/13.3.295. [DOI] [PubMed] [Google Scholar]
- 66.Phillips LM, Yogev R, Esterly NB. 1985. The efficacy of mupirocin (pseudomonic acid) in the treatment of pyoderma in children. Pediatr Emerg Care 1:180–183. doi: 10.1097/00006565-198512000-00002. [DOI] [PubMed] [Google Scholar]
- 67.Rumsfield J, West DP, Aronson IK. 1986. Topical mupirocin in the treatment of bacterial skin infections. Drug Intell Clin Pharm 20:943–948. [DOI] [PubMed] [Google Scholar]
- 68.Ward A, Campoli-Richards DM. 1986. Mupirocin. A review of its antibacterial activity, pharmacokinetic properties and therapeutic use. Drugs 32:425–444. [DOI] [PubMed] [Google Scholar]
- 69.Eells LD, Mertz PM, Piovanetti Y, Pekoe GM, Eaglstein WH. 1986. Topical antibiotic treatment of impetigo with mupirocin. Arch Dermatol 122:1273–1276. [PubMed] [Google Scholar]
- 70.Gould PW, Villiger JW. 1986. Clinical and bacteriological efficacy of mupirocin (Bactroban): a new topical antibiotic. N Z Med J 99:516. [PubMed] [Google Scholar]
- 71.Coates T, Bax R, Coates A. 2009. Nasal decolonization of Staphylococcus aureus with mupirocin: strengths, weaknesses and future prospects. J Antimicrob Chemother 64:9–15. doi: 10.1093/jac/dkp159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.van Rijen MM, Bonten M, Wenzel RP, Kluytmans JA. 2008. Intranasal mupirocin for reduction of Staphylococcus aureus infections in surgical patients with nasal carriage: a systematic review. J Antimicrob Chemother 61:254–261. doi: 10.1093/jac/dkm480. [DOI] [PubMed] [Google Scholar]
- 73.Sutherland R, Boon RJ, Griffin KE, Masters PJ, Slocombe B, White AR. 1985. Antibacterial activity of mupirocin (pseudomonic acid), a new antibiotic for topical use. Antimicrob Agents Chemother 27:495–498. doi: 10.1128/AAC.27.4.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Thomas CM, Hothersall J, Willis CL, Simpson TJ. 2010. Resistance to and synthesis of the antibiotic mupirocin. Nat Rev Microbiol 8:281–289. doi: 10.1038/nrmicro2278. [DOI] [PubMed] [Google Scholar]
- 75.Thomas DG, Hann AC, Day MJ, Wilson JM, Russell AD. 1999. Structural changes induced by mupirocin in Staphylococcus aureus cells. Int J Antimicrob Agents 13:9–14. doi: 10.1016/S0924-8579(99)00090-4. [DOI] [PubMed] [Google Scholar]
- 76.Antonio M, McFerran N, Pallen MJ. 2002. Mutations affecting the Rossman fold of isoleucyl-tRNA synthetase are correlated with low-level mupirocin resistance in Staphylococcus aureus. Antimicrob Agents Chemother 46:438–442. doi: 10.1128/AAC.46.2.438-442.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Swenson JM, Wong B, Simor AE, Thomson RB, Ferraro MJ, Hardy DJ, Hindler J, Jorgensen J, Reller LB, Traczewski M, McDougal LK, Patel JB. 2010. Multicenter study to determine disk diffusion and broth microdilution criteria for prediction of high- and low-level mupirocin resistance in Staphylococcus aureus. J Clin Microbiol 48:2469–2475. doi: 10.1128/JCM.00340-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Palepou MF, Johnson AP, Cookson BD, Beattie H, Charlett A, Woodford N. 1998. Evaluation of disc diffusion and Etest for determining the susceptibility of Staphylococcus aureus to mupirocin. J Antimicrob Chemother 42:577–583. doi: 10.1093/jac/42.5.577. [DOI] [PubMed] [Google Scholar]
- 79.Finlay JE, Miller LA, Poupard JA. 1998. Comparison of the 5 microg disc and the Neo-Sensitab for determining the susceptibilities of Staphylococcus aureus isolates to mupirocin. J Antimicrob Chemother 42:403–405. doi: 10.1093/jac/42.3.403. [DOI] [PubMed] [Google Scholar]
- 80.Finlay JE, Miller LA, Poupard JA. 1997. Interpretive criteria for testing susceptibility of staphylococci to mupirocin. Antimicrob Agents Chemother 41:1137–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.de Oliveira NE, Cardozo AP, Marques Ede A, dos Santos KR, Giambiagi-deMarval M. 2007. Interpretive criteria to differentiate low- and high-level mupirocin resistance in Staphylococcus aureus. J Med Microbiol 56:937–939. doi: 10.1099/jmm.0.46965-0. [DOI] [PubMed] [Google Scholar]
- 82.Clinical and Laboratory Standards Institute. 2015. Performance standards for antimicrobial susceptibility testing, 26th ed CLSI, Wayne, PA. [Google Scholar]
- 83.Yun HJ, Lee SW, Yoon GM, Kim SY, Choi S, Lee YS, Choi EC, Kim S. 2003. Prevalence and mechanisms of low- and high-level mupirocin resistance in staphylococci isolated from a Korean hospital. J Antimicrob Chemother 51:619–623. doi: 10.1093/jac/dkg140. [DOI] [PubMed] [Google Scholar]
- 84.Hurdle JG, O'Neill AJ, Ingham E, Fishwick C, Chopra I. 2004. Analysis of mupirocin resistance and fitness in Staphylococcus aureus by molecular genetic and structural modeling techniques. Antimicrob Agents Chemother 48:4366–4376. doi: 10.1128/AAC.48.11.4366-4376.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hurdle JG, O'Neill AJ, Chopra I. 2004. The isoleucyl-tRNA synthetase mutation V588F conferring mupirocin resistance in glycopeptide-intermediate Staphylococcus aureus is not associated with a significant fitness burden. J Antimicrob Chemother 53:102–104. doi: 10.1093/jac/dkh020. [DOI] [PubMed] [Google Scholar]
- 86.Yang JA, Park DW, Sohn JW, Yang IS, Kim KH, Kim MJ. 2006. Molecular analysis of isoleucyl-tRNA synthetase mutations in clinical isolates of methicillin-resistant Staphylococcus aureus with low-level mupirocin resistance. J Korean Med Sci 21:827–832. doi: 10.3346/jkms.2006.21.5.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lee AS, Gizard Y, Empel J, Bonetti EJ, Harbarth S, Francois P. 2014. Mupirocin-induced mutations in ileS in various genetic backgrounds of methicillin-resistant Staphylococcus aureus. J Clin Microbiol 52:3749–3754. doi: 10.1128/JCM.01010-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hodgson JE, Curnock SP, Dyke KG, Morris R, Sylvester DR, Gross MS. 1994. Molecular characterization of the gene encoding high-level mupirocin resistance in Staphylococcus aureus J2870. Antimicrob Agents Chemother 38:1205–1208. doi: 10.1128/AAC.38.5.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nunes EL, dos Santos KR, Mondino PJ, Bastos MC, Giambiagi-deMarval M. 1999. Detection of ileS-2 gene encoding mupirocin resistance in methicillin-resistant Staphylococcus aureus by multiplex PCR. Diagn Microbiol Infect Dis 34:77–81. doi: 10.1016/S0732-8893(99)00021-8. [DOI] [PubMed] [Google Scholar]
- 90.Perez-Roth E, Lopez-Aguilar C, Alcoba-Florez J, Mendez-Alvarez S. 2006. High-level mupirocin resistance within methicillin-resistant Staphylococcus aureus pandemic lineages. Antimicrob Agents Chemother 50:3207–3211. doi: 10.1128/AAC.00059-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Perez-Roth E, Potel-Alvarellos C, Espartero X, Constela-Carames L, Mendez-Alvarez S, Alvarez-Fernandez M. 2013. Molecular epidemiology of plasmid-mediated high-level mupirocin resistance in methicillin-resistant Staphylococcus aureus in four Spanish health care settings. Int J Med Microbiol 303:201–204. doi: 10.1016/j.ijmm.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 92.Morton TM, Johnston JL, Patterson J, Archer GL. 1995. Characterization of a conjugative staphylococcal mupirocin resistance plasmid. Antimicrob Agents Chemother 39:1272–1280. doi: 10.1128/AAC.39.6.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Perez-Roth E, Kwong SM, Alcoba-Florez J, Firth N, Mendez-Alvarez S. 2010. Complete nucleotide sequence and comparative analysis of pPR9, a 41.7-kilobase conjugative staphylococcal multiresistance plasmid conferring high-level mupirocin resistance. Antimicrob Agents Chemother 54:2252–2257. doi: 10.1128/AAC.01074-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Needham C, Rahman M, Dyke KG, Noble WC. 1994. An investigation of plasmids from Staphylococcus aureus that mediate resistance to mupirocin and tetracycline. Microbiology 140:2577–2583. doi: 10.1099/00221287-140-10-2577. [DOI] [PubMed] [Google Scholar]
- 95.Perez-Roth E, Armas-Gonzalez E, Alcoba-Florez J, Mendez-Alvarez S. 2011. PCR-based amplification of heterogeneous IS257-ileS2 junctions for molecular monitoring of high-level mupirocin resistance in staphylococci. J Antimicrob Chemother 66:471–475. doi: 10.1093/jac/dkq493. [DOI] [PubMed] [Google Scholar]
- 96.Hurdle JG, O'Neill AJ, Mody L, Chopra I, Bradley SF. 2005. In vivo transfer of high-level mupirocin resistance from Staphylococcus epidermidis to methicillin-resistant Staphylococcus aureus associated with failure of mupirocin prophylaxis. J Antimicrob Chemother 56:1166–1168. doi: 10.1093/jac/dki387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Matanovic K, Perez-Roth E, Pintaric S, Seol Martinec B. 2013. Molecular characterization of high-level mupirocin resistance in Staphylococcus pseudintermedius. J Clin Microbiol 51:1005–1007. doi: 10.1128/JCM.02904-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lee AS, Macedo-Vinas M, Francois P, Renzi G, Schrenzel J, Vernaz N, Pittet D, Harbarth S. 2011. Impact of combined low-level mupirocin and genotypic chlorhexidine resistance on persistent methicillin-resistant Staphylococcus aureus carriage after decolonization therapy: a case-control study. Clin Infect Dis 52:1422–1430. doi: 10.1093/cid/cir233. [DOI] [PubMed] [Google Scholar]
- 99.Udo EE, Al-Sweih N, Noronha BC. 2003. A chromosomal location of the mupA gene in Staphylococcus aureus expressing high-level mupirocin resistance. J Antimicrob Chemother 51:1283–1286. doi: 10.1093/jac/dkg188. [DOI] [PubMed] [Google Scholar]
- 100.Driscoll DG, Young CL, Ochsner UA. 2007. Transient loss of high-level mupirocin resistance in Staphylococcus aureus due to MupA polymorphism. Antimicrob Agents Chemother 51:2247–2248. doi: 10.1128/AAC.00241-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fritz SA, Hogan PG, Camins BC, Ainsworth AJ, Patrick C, Martin MS, Krauss MJ, Rodriguez M, Burnham CA. 2013. Mupirocin and chlorhexidine resistance in Staphylococcus aureus in patients with community-onset skin and soft tissue infections. Antimicrob Agents Chemother 57:559–568. doi: 10.1128/AAC.01633-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Seah C, Alexander DC, Louie L, Simor A, Low DE, Longtin J, Melano RG. 2012. MupB, a new high-level mupirocin resistance mechanism in Staphylococcus aureus. Antimicrob Agents Chemother 56:1916–1920. doi: 10.1128/AAC.05325-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Trouillet-Assant S, Flammier S, Sapin A, Dupieux C, Dumitrescu O, Tristan A, Vandenesch F, Rasigade JP, Laurent F. 2015. Mupirocin resistance in isolates of Staphylococcus spp. from nasal swabs in a tertiary hospital in France. J Clin Microbiol 53:2713–2715. doi: 10.1128/JCM.00274-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Desroches M, Potier J, Laurent F, Bourrel AS, Doucet-Populaire F, Decousser JW, Microbs Study Group. 2013. Prevalence of mupirocin resistance among invasive coagulase-negative staphylococci and methicillin-resistant Staphylococcus aureus (MRSA) in France: emergence of a mupirocin-resistant MRSA clone harbouring mupA. J Antimicrob Chemother 68:1714–1717. doi: 10.1093/jac/dkt085. [DOI] [PubMed] [Google Scholar]
- 105.O'Shea S, Cotter L, Creagh S, Lydon S, Lucey B. 2009. Mupirocin resistance among staphylococci: trends in the southern region of Ireland. J Antimicrob Chemother 64:649–650. doi: 10.1093/jac/dkp227. [DOI] [PubMed] [Google Scholar]
- 106.Deplano A, Vandendriessche S, Nonhoff C, Dodemont M, Roisin S, Denis O. 2016. National surveillance of Staphylococcus epidermidis recovered from bloodstream infections in Belgian hospitals. J Antimicrob Chemother 71:1815–1819. doi: 10.1093/jac/dkw086. [DOI] [PubMed] [Google Scholar]
- 107.Upton A, Lang S, Heffernan H. 2003. Mupirocin and Staphylococcus aureus: a recent paradigm of emerging antibiotic resistance. J Antimicrob Chemother 51:613–617. doi: 10.1093/jac/dkg127. [DOI] [PubMed] [Google Scholar]
- 108.Williamson DA, Monecke S, Heffernan H, Ritchie SR, Roberts SA, Upton A, Thomas MG, Fraser JD. 2014. High usage of topical fusidic acid and rapid clonal expansion of fusidic acid-resistant Staphylococcus aureus: a cautionary tale. Clin Infect Dis 59:1451–1454. doi: 10.1093/cid/ciu658. [DOI] [PubMed] [Google Scholar]
- 109.Riley TV, Carson CF, Bowman RA, Mulgrave L, Golledge CL, Pearman JW, Grubb WB. 1994. Mupirocin-resistant methicillin-resistant Staphylococcus aureus in western Australia. Med J Aust 161:397–398. [DOI] [PubMed] [Google Scholar]
- 110.Torvaldsen S, Roberts C, Riley TV. 1999. The continuing evolution of methicillin-resistant Staphylococcus aureus in western Australia. Infect Control Hosp Epidemiol 20:133–135. doi: 10.1086/501594. [DOI] [PubMed] [Google Scholar]
- 111.Ellis MW, Griffith ME, Dooley DP, McLean JC, Jorgensen JH, Patterson JE, Davis KA, Hawley JS, Regules JA, Rivard RG, Gray PJ, Ceremuga JM, Dejoseph MA, Hospenthal DR. 2007. Targeted intranasal mupirocin to prevent colonization and infection by community-associated methicillin-resistant Staphylococcus aureus strains in soldiers: a cluster randomized controlled trial. Antimicrob Agents Chemother 51:3591–3598. doi: 10.1128/AAC.01086-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Vivoni AM, Santos KR, de-Oliveira MP, Giambiagi-deMarval M, Ferreira AL, Riley LW, Moreira BM. 2005. Mupirocin for controlling methicillin-resistant Staphylococcus aureus: lessons from a decade of use at a university hospital. Infect Control Hosp Epidemiol 26:662–667. doi: 10.1086/502599. [DOI] [PubMed] [Google Scholar]
- 113.Netto dos Santos KR, de Souza Fonseca L, Gontijo Filho PP. 1996. Emergence of high-level mupirocin resistance in methicillin-resistant Staphylococcus aureus isolated from Brazilian university hospitals. Infect Control Hosp Epidemiol 17:813–816. [PubMed] [Google Scholar]
- 114.Perez-Fontan M, Rosales M, Rodriguez-Carmona A, Falcon TG, Valdes F. 2002. Mupirocin resistance after long-term use for Staphylococcus aureus colonization in patients undergoing chronic peritoneal dialysis. Am J Kidney Dis 39:337–341. doi: 10.1053/ajkd.2002.30553. [DOI] [PubMed] [Google Scholar]
- 115.Annigeri R, Conly J, Vas S, Dedier H, Prakashan KP, Bargman JM, Jassal V, Oreopoulos D. 2001. Emergence of mupirocin-resistant Staphylococcus aureus in chronic peritoneal dialysis patients using mupirocin prophylaxis to prevent exit-site infection. Perit Dial Int 21:554–559. [PubMed] [Google Scholar]
- 116.Perl TM, Cullen JJ, Wenzel RP, Zimmerman MB, Pfaller MA, Sheppard D, Twombley J, French PP, Herwaldt LA, Mupirocin and the Risk of Staphylococcus aureus Study Team. 2002. Intranasal mupirocin to prevent postoperative Staphylococcus aureus infections. N Engl J Med 346:1871–1877. doi: 10.1056/NEJMoa003069. [DOI] [PubMed] [Google Scholar]
- 117.Fawley WN, Parnell P, Hall J, Wilcox MH. 2006. Surveillance for mupirocin resistance following introduction of routine peri-operative prophylaxis with nasal mupirocin. J Hosp Infect 62:327–332. doi: 10.1016/j.jhin.2005.09.022. [DOI] [PubMed] [Google Scholar]
- 118.Bode LG, Kluytmans JA, Wertheim HF, Bogaers D, Vandenbroucke-Grauls CM, Roosendaal R, Troelstra A, Box AT, Voss A, van der Tweel I, van Belkum A, Verbrugh HA, Vos MC. 2010. Preventing surgical-site infections in nasal carriers of Staphylococcus aureus. N Engl J Med 362:9–17. doi: 10.1056/NEJMoa0808939. [DOI] [PubMed] [Google Scholar]
- 119.Kluytmans JA, Mouton JW, VandenBergh MF, Manders MJ, Maat AP, Wagenvoort JH, Michel MF, Verbrugh HA. 1996. Reduction of surgical-site infections in cardiothoracic surgery by elimination of nasal carriage of Staphylococcus aureus. Infect Control Hosp Epidemiol 17:780–785. [DOI] [PubMed] [Google Scholar]
- 120.Bathoorn E, Hetem DJ, Alphenaar J, Kusters JG, Bonten MJ. 2012. Emergence of high-level mupirocin resistance in coagulase-negative staphylococci associated with increased short-term mupirocin use. J Clin Microbiol 50:2947–2950. doi: 10.1128/JCM.00302-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hetem DJ, Vogely HC, Severs TT, Troelstra A, Kusters JG, Bonten MJ. 2015. Acquisition of high-level mupirocin resistance in CoNS following nasal decolonization with mupirocin. J Antimicrob Chemother 70:1182–1184. doi: 10.1093/jac/dku522. [DOI] [PubMed] [Google Scholar]
- 122.Hetem DJ, Bootsma MC, Bonten MJ. 2016. Prevention of surgical site infections: decontamination with mupirocin based on preoperative screening for Staphylococcus aureus carriers or universal decontamination? Clin Infect Dis 62:631–636. doi: 10.1093/cid/civ990. [DOI] [PubMed] [Google Scholar]
- 123.Laurberg M, Kristensen O, Martemyanov K, Gudkov AT, Nagaev I, Hughes D, Liljas A. 2000. Structure of a mutant EF-G reveals domain III and possibly the fusidic acid binding site. J Mol Biol 303:593–603. doi: 10.1006/jmbi.2000.4168. [DOI] [PubMed] [Google Scholar]
- 124.Norstrom T, Lannergard J, Hughes D. 2007. Genetic and phenotypic identification of fusidic acid-resistant mutants with the small-colony-variant phenotype in Staphylococcus aureus. Antimicrob Agents Chemother 51:4438–4446. doi: 10.1128/AAC.00328-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Castanheira M, Watters AA, Bell JM, Turnidge JD, Jones RN. 2010. Fusidic acid resistance rates and prevalence of resistance mechanisms among Staphylococcus spp. isolated in North America and Australia, 2007–2008. Antimicrob Agents Chemother 54:3614–3617. doi: 10.1128/AAC.01390-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Besier S, Ludwig A, Brade V, Wichelhaus TA. 2003. Molecular analysis of fusidic acid resistance in Staphylococcus aureus. Mol Microbiol 47:463–469. doi: 10.1046/j.1365-2958.2003.03307.x. [DOI] [PubMed] [Google Scholar]
- 127.Besier S, Ludwig A, Brade V, Wichelhaus TA. 2005. Compensatory adaptation to the loss of biological fitness associated with acquisition of fusidic acid resistance in Staphylococcus aureus. Antimicrob Agents Chemother 49:1426–1431. doi: 10.1128/AAC.49.4.1426-1431.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Koripella RK, Chen Y, Peisker K, Koh CS, Selmer M, Sanyal S. 2012. Mechanism of elongation factor-G-mediated fusidic acid resistance and fitness compensation in Staphylococcus aureus. J Biol Chem 287:30257–30267. doi: 10.1074/jbc.M112.378521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Cox G, Thompson GS, Jenkins HT, Peske F, Savelsbergh A, Rodnina MV, Wintermeyer W, Homans SW, Edwards TA, O'Neill AJ. 2012. Ribosome clearance by FusB-type proteins mediates resistance to the antibiotic fusidic acid. Proc Natl Acad Sci U S A 109:2102–2107. doi: 10.1073/pnas.1117275109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Huang J, Ye M, Ding H, Guo Q, Ding B, Wang M. 2013. Prevalence of fusB in Staphylococcus aureus clinical isolates. J Med Microbiol 62:1199–1203. doi: 10.1099/jmm.0.058305-0. [DOI] [PubMed] [Google Scholar]
- 131.Farrell DJ, Castanheira M, Chopra I. 2011. Characterization of global patterns and the genetics of fusidic acid resistance. Clin Infect Dis 52(Suppl 7):S487–S492. doi: 10.1093/cid/cir164. [DOI] [PubMed] [Google Scholar]
- 132.O'Brien FG, Price C, Grubb WB, Gustafson JE. 2002. Genetic characterization of the fusidic acid and cadmium resistance determinants of Staphylococcus aureus plasmid pUB101. J Antimicrob Chemother 50:313–321. doi: 10.1093/jac/dkf153. [DOI] [PubMed] [Google Scholar]
- 133.Lannergard J, Norstrom T, Hughes D. 2009. Genetic determinants of resistance to fusidic acid among clinical bacteremia isolates of Staphylococcus aureus. Antimicrob Agents Chemother 53:2059–2065. doi: 10.1128/AAC.00871-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Liu Y, Geng W, Yang Y, Wang C, Zheng Y, Shang Y, Wu D, Li X, Wang L, Yu S, Yao K, Shen X. 2012. Susceptibility to and resistance determinants of fusidic acid in Staphylococcus aureus isolated from Chinese children with skin and soft tissue infections. FEMS Immunol Med Microbiol 64:212–218. doi: 10.1111/j.1574-695X.2011.00887.x. [DOI] [PubMed] [Google Scholar]
- 135.Stegger M, Wirth T, Andersen PS, Skov RL, De Grassi A, Simoes PM, Tristan A, Petersen A, Aziz M, Kiil K, Cirkovic I, Udo EE, del Campo R, Vuopio-Varkila J, Ahmad N, Tokajian S, Peters G, Schaumburg F, Olsson-Liljequist B, Givskov M, Driebe EE, Vigh HE, Shittu A, Ramdani-Bougessa N, Rasigade JP, Price LB, Vandenesch F, Larsen AR, Laurent F. 2014. Origin and evolution of European community-acquired methicillin-resistant Staphylococcus aureus. mBio 5:e01044-14. doi: 10.1128/mBio.01044-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.O'Neill AJ, Larsen AR, Skov R, Henriksen AS, Chopra I. 2007. Characterization of the epidemic European fusidic acid-resistant impetigo clone of Staphylococcus aureus. J Clin Microbiol 45:1505–1510. doi: 10.1128/JCM.01984-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Castanheira M, Watters AA, Mendes RE, Farrell DJ, Jones RN. 2010. Occurrence and molecular characterization of fusidic acid resistance mechanisms among Staphylococcus spp. from European countries (2008). J Antimicrob Chemother 65:1353–1358. doi: 10.1093/jac/dkq094. [DOI] [PubMed] [Google Scholar]
- 138.Chen HJ, Chang YC, Tsai JC, Hung WC, Lin YT, You SJ, Tseng SP, Teng LJ. 2013. New structure of phage-related islands carrying fusB and a virulence gene in fusidic acid-resistant Staphylococcus epidermidis. Antimicrob Agents Chemother 57:5737–5739. doi: 10.1128/AAC.01433-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Holden MT, Feil EJ, Lindsay JA, Peacock SJ, Day NP, Enright MC, Foster TJ, Moore CE, Hurst L, Atkin R, Barron A, Bason N, Bentley SD, Chillingworth C, Chillingworth T, Churcher C, Clark L, Corton C, Cronin A, Doggett J, Dowd L, Feltwell T, Hance Z, Harris B, Hauser H, Holroyd S, Jagels K, James KD, Lennard N, Line A, Mayes R, Moule S, Mungall K, Ormond D, Quail MA, Rabbinowitsch E, Rutherford K, Sanders M, Sharp S, Simmonds M, Stevens K, Whitehead S, Barrell BG, Spratt BG, Parkhill J. 2004. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc Natl Acad Sci U S A 101:9786–9791. doi: 10.1073/pnas.0402521101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.O'Neill AJ, McLaws F, Kahlmeter G, Henriksen AS, Chopra I. 2007. Genetic basis of resistance to fusidic acid in staphylococci. Antimicrob Agents Chemother 51:1737–1740. doi: 10.1128/AAC.01542-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Chen HJ, Hung WC, Lin YT, Tsai JC, Chiu HC, Hsueh PR, Teng LJ. 2015. A novel fusidic acid resistance determinant, fusF, in Staphylococcus cohnii. J Antimicrob Chemother 70:416–419. doi: 10.1093/jac/dku408. [DOI] [PubMed] [Google Scholar]
- 142.Ellington MJ, Reuter S, Harris SR, Holden MT, Cartwright EJ, Greaves D, Gerver SM, Hope R, Brown NM, Torok ME, Parkhill J, Koser CU, Peacock SJ. 2015. Emergent and evolving antimicrobial resistance cassettes in community-associated fusidic acid and meticillin-resistant Staphylococcus aureus. Int J Antimicrob Agents 45:477–484. doi: 10.1016/j.ijantimicag.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lin YT, Tsai JC, Chen HJ, Hung WC, Hsueh PR, Teng LJ. 2014. A novel staphylococcal cassette chromosomal element, SCCfusC, carrying fusC and speG in fusidic acid-resistant methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 58:1224–1227. doi: 10.1128/AAC.01772-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kinnevey PM, Shore AC, Brennan GI, Sullivan DJ, Ehricht R, Monecke S, Slickers P, Coleman DC. 2013. Emergence of sequence type 779 methicillin-resistant Staphylococcus aureus harboring a novel pseudo staphylococcal cassette chromosome mec (SCCmec)-SCC-SCCCRISPR composite element in Irish hospitals. Antimicrob Agents Chemother 57:524–531. doi: 10.1128/AAC.01689-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hung WC, Chen HJ, Lin YT, Tsai JC, Chen CW, Lu HH, Tseng SP, Jheng YY, Leong KH, Teng LJ. 2015. Skin commensal staphylococci may act as reservoir for fusidic acid resistance genes. PLoS One 10:e0143106. doi: 10.1371/journal.pone.0143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Chen HJ, Lin YT, Hung WC, Tsai JC, Hsueh PR, Teng LJ. 2016. Distribution of staphylococcal cassette chromosome (SCC) mec element types in fusidic acid-resistant Staphylococcus epidermidis and identification of a novel SCC7684 element. Antimicrob Agents Chemother 60:5006–5009. doi: 10.1128/AAC.00231-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Baines SL, Howden BP, Heffernan H, Stinear TP, Carter GP, Seemann T, Kwong JC, Ritchie SR, Williamson DA. 2016. Rapid emergence and evolution of Staphylococcus aureus clones harboring fusC-containing staphylococcal cassette chromosome elements. Antimicrob Agents Chemother 60:2359–2365. doi: 10.1128/AAC.03020-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Livermore D, James D, Duckworth G, Stephens P. 2002. Fusidic-acid use and resistance. Lancet 360:806. [DOI] [PubMed] [Google Scholar]
- 149.Mason BW, Howard AJ, Magee JT. 2003. Fusidic acid resistance in community isolates of methicillin-susceptible Staphylococcus aureus and fusidic acid prescribing. J Antimicrob Chemother 51:1033–1036. doi: 10.1093/jac/dkg190. [DOI] [PubMed] [Google Scholar]
- 150.Farrell DJ, Mendes RE, Castanheira M, Jones RN. 2016. Activity of fusidic acid tested against staphylococci isolated from patients in U.S. medical centers in 2014. Antimicrob Agents Chemother 60:3827–3831. doi: 10.1128/AAC.00238-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Shah M, Mohanraj M. 2003. High levels of fusidic acid-resistant Staphylococcus aureus in dermatology patients. Br J Dermatol 148:1018–1020. doi: 10.1046/j.1365-2133.2003.05291.x. [DOI] [PubMed] [Google Scholar]
- 152.Ravenscroft JC, Layton A, Barnham M. 2000. Observations on high levels of fusidic acid resistant Staphylococcus aureus in Harrogate, North Yorkshire, UK. Clin Exp Dermatol 25:327–330. doi: 10.1046/j.1365-2230.2000.00654.x. [DOI] [PubMed] [Google Scholar]
- 153.Peeters KA, Mascini EM, Sanders CJ. 2004. Resistance of Staphylococcus aureus to fusidic acid. Int J Dermatol 43:235–236. doi: 10.1111/j.1365-4632.2004.02172.x. [DOI] [PubMed] [Google Scholar]
- 154.Sule O, Brown NM, Willocks LJ, Day J, Shankar S, Palmer CR, Burrows NP. 2007. Fusidic acid-resistant Staphylococcus aureus (FRSA) carriage in patients with atopic eczema and pattern of prior topical fusidic acid use. Int J Antimicrob Agents 30:78–82. doi: 10.1016/j.ijantimicag.2007.02.015. [DOI] [PubMed] [Google Scholar]
- 155.Howden BP, Grayson ML. 2006. Dumb and dumber—the potential waste of a useful antistaphylococcal agent: emerging fusidic acid resistance in Staphylococcus aureus. Clin Infect Dis 42:394–400. doi: 10.1086/499365. [DOI] [PubMed] [Google Scholar]
- 156.Whitby M. 1999. Fusidic acid in the treatment of methicillin-resistant Staphylococcus aureus. Int J Antimicrob Agents 12(Suppl 2):S67–S71. doi: 10.1016/S0924-8579(98)00075-2. [DOI] [PubMed] [Google Scholar]
- 157.Waksman SA, Lechevalier HA. 1949. Neomycin, a new antibiotic active against Streptomycin-resistant bacteria, including tuberculosis organisms. Science 109:305–307. doi: 10.1126/science.109.2830.305. [DOI] [PubMed] [Google Scholar]
- 158.Tsuji K, Robertson JH, Baas R, McInnis DJ. 1969. Comparative study of responses to neomycins B and C by microbiological and gas-liquid chromatographic assay methods. Appl Microbiol 18:396–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Magnet S, Blanchard JS. 2005. Molecular insights into aminoglycoside action and resistance. Chem Rev 105:477–498. doi: 10.1021/cr0301088. [DOI] [PubMed] [Google Scholar]
- 160.Lio PA, Kaye ET. 2009. Topical antibacterial agents. Infect Dis Clin North Am 23:945–963. doi: 10.1016/j.idc.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 161.Yamasoba T, Tsukuda K. 2004. Ototoxicity after use of neomycin eardrops is unrelated to A1555G point mutation in mitochondrial DNA. J Laryngol Otol 118:546–550. doi: 10.1258/0022215041615245. [DOI] [PubMed] [Google Scholar]
- 162.Stone KJ, Strominger JL. 1971. Mechanism of action of bacitracin: complexation with metal ion and C55-isoprenyl pyrophosphate. Proc Natl Acad Sci U S A 68:3223–3227. doi: 10.1073/pnas.68.12.3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Jones RN, Li Q, Kohut B, Biedenbach DJ, Bell J, Turnidge JD. 2006. Contemporary antimicrobial activity of triple antibiotic ointment: a multiphased study of recent clinical isolates in the United States and Australia. Diagn Microbiol Infect Dis 54:63–71. doi: 10.1016/j.diagmicrobio.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 164.Evans FL. 1948. A note on the susceptibility of Hemophilus influenzae type B to bacitracin. J Bacteriol 56:507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Schalock PC, Zug KA. 2005. Bacitracin. Cutis 76:105–107. [PubMed] [Google Scholar]
- 166.Stansly PG, Schlosser ME. 1947. Studies on polymyxin: isolation and identification of Bacillus polymyxa and differentiation of polymyxin from certain known antibiotics. J Bacteriol 54:549–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Brownlee G. 1949. Antibiotics derived from bacillus polymyxa. Ann N Y Acad Sci 51:875–878. doi: 10.1111/j.1749-6632.1949.tb27313.x. [DOI] [PubMed] [Google Scholar]
- 168.Traczewski MM, Brown SD. 2008. Proposed MIC and disk diffusion microbiological cutoffs and spectrum of activity of retapamulin, a novel topical antimicrobial agent. Antimicrob Agents Chemother 52:3863–3867. doi: 10.1128/AAC.00399-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kosowska-Shick K, Clark C, Credito K, McGhee P, Dewasse B, Bogdanovich T, Appelbaum PC. 2006. Single- and multistep resistance selection studies on the activity of retapamulin compared to other agents against Staphylococcus aureus and Streptococcus pyogenes. Antimicrob Agents Chemother 50:765–769. doi: 10.1128/AAC.50.2.765-769.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.van Duijkeren E, Greko C, Pringle M, Baptiste KE, Catry B, Jukes H, Moreno MA, Pomba MC, Pyorala S, Rantala M, Ruzauskas M, Sanders P, Teale C, Threlfall EJ, Torren-Edo J, Torneke K. 2014. Pleuromutilins: use in food-producing animals in the European Union, development of resistance and impact on human and animal health. J Antimicrob Chemother 69:2022–2031. doi: 10.1093/jac/dku123. [DOI] [PubMed] [Google Scholar]
- 171.Paukner S, Riedl R. 2017. Pleuromutilins: potent drugs for resistant bugs—mode of action and resistance. Cold Spring Harb Perspect Med 7:a027110. doi: 10.1101/cshperspect.a027110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Scangarella-Oman NE, Shawar RM, Bouchillon S, Hoban D. 2009. Microbiological profile of a new topical antibacterial: retapamulin ointment 1%. Expert Rev Anti Infect Ther 7:269–279. doi: 10.1586/eri.09.7. [DOI] [PubMed] [Google Scholar]
- 173.Moody MN, Morrison LK, Tyring SK. 2010. Retapamulin: what is the role of this topical antimicrobial in the treatment of bacterial infections in atopic dermatitis? Skin Ther Lett 15:1–4. [PubMed] [Google Scholar]
- 174.Farrell DJ, Robbins M, Rhys-Williams W, Love WG. 2011. Investigation of the potential for mutational resistance to XF-73, retapamulin, mupirocin, fusidic acid, daptomycin, and vancomycin in methicillin-resistant Staphylococcus aureus isolates during a 55-passage study. Antimicrob Agents Chemother 55:1177–1181. doi: 10.1128/AAC.01285-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Gentry DR, Rittenhouse SF, McCloskey L, Holmes DJ. 2007. Stepwise exposure of Staphylococcus aureus to pleuromutilins is associated with stepwise acquisition of mutations in rplC and minimally affects susceptibility to retapamulin. Antimicrob Agents Chemother 51:2048–2052. doi: 10.1128/AAC.01066-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Gentry DR, McCloskey L, Gwynn MN, Rittenhouse SF, Scangarella N, Shawar R, Holmes DJ. 2008. Genetic characterization of Vga ABC proteins conferring reduced susceptibility to pleuromutilins in Staphylococcus aureus. Antimicrob Agents Chemother 52:4507–4509. doi: 10.1128/AAC.00915-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Candel FJ, Morales G, Picazo JJ. 2011. In vitro activity of retapamulin against linezolid and methicillin-resistant Staphylococcus aureus isolates. Rev Esp Quimioter 24:127–130. [PubMed] [Google Scholar]
- 178.Woodford N, Afzal-Shah M, Warner M, Livermore DM. 2008. In vitro activity of retapamulin against Staphylococcus aureus isolates resistant to fusidic acid and mupirocin. J Antimicrob Chemother 62:766–768. doi: 10.1093/jac/dkn266. [DOI] [PubMed] [Google Scholar]
- 179.Saravolatz LD, Pawlak J, Saravolatz SN, Johnson LB. 2013. In vitro activity of retapamulin against Staphylococcus aureus resistant to various antimicrobial agents. Antimicrob Agents Chemother 57:4547–4550. doi: 10.1128/AAC.00282-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.McNeil JC, Hulten KG, Kaplan SL, Mason EO. 2014. Decreased susceptibilities to retapamulin, mupirocin, and chlorhexidine among Staphylococcus aureus isolates causing skin and soft tissue infections in otherwise healthy children. Antimicrob Agents Chemother 58:2878–2883. doi: 10.1128/AAC.02707-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Parnham MJ, Sies H. 2013. The early research and development of ebselen. Biochem Pharmacol 86:1248–1253. doi: 10.1016/j.bcp.2013.08.028. [DOI] [PubMed] [Google Scholar]
- 182.Thangamani S, Younis W, Seleem MN. 2015. Repurposing clinical molecule ebselen to combat drug resistant pathogens. PLoS One 10:e0133877. doi: 10.1371/journal.pone.0133877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Thangamani S, Younis W, Seleem MN. 2015. Repurposing ebselen for treatment of multidrug-resistant staphylococcal infections. Sci Rep 5:11596. doi: 10.1038/srep11596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Lu J, Vlamis-Gardikas A, Kandasamy K, Zhao R, Gustafsson TN, Engstrand L, Hoffner S, Engman L, Holmgren A. 2013. Inhibition of bacterial thioredoxin reductase: an antibiotic mechanism targeting bacteria lacking glutathione. FASEB J 27:1394–1403. doi: 10.1096/fj.12-223305. [DOI] [PubMed] [Google Scholar]
- 185.Fox JL. 2013. Antimicrobial peptides stage a comeback. Nat Biotechnol 31:379–382. doi: 10.1038/nbt.2572. [DOI] [PubMed] [Google Scholar]
- 186.Yeung AT, Gellatly SL, Hancock RE. 2011. Multifunctional cationic host defence peptides and their clinical applications. Cell Mol Life Sci 68:2161–2176. doi: 10.1007/s00018-011-0710-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Myhrman E, Hakansson J, Lindgren K, Bjorn C, Sjostrand V, Mahlapuu M. 2013. The novel antimicrobial peptide PXL150 in the local treatment of skin and soft tissue infections. Appl Microbiol Biotechnol 97:3085–3096. doi: 10.1007/s00253-012-4439-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Bjorn C, Noppa L, Naslund Salomonsson E, Johansson AL, Nilsson E, Mahlapuu M, Hakansson J. 2015. Efficacy and safety profile of the novel antimicrobial peptide PXL150 in a mouse model of infected burn wounds. Int J Antimicrob Agents 45:519–524. doi: 10.1016/j.ijantimicag.2014.12.015. [DOI] [PubMed] [Google Scholar]
- 189.Hakansson J, Bjorn C, Lindgren K, Sjostrom E, Sjostrand V, Mahlapuu M. 2014. Efficacy of the novel topical antimicrobial agent PXL150 in a mouse model of surgical site infections. Antimicrob Agents Chemother 58:2982–2984. doi: 10.1128/AAC.00143-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Flamm RK, Rhomberg PR, Farrell DJ, Jones RN. 2016. In vitro spectrum of pexiganan activity; bactericidal action and resistance selection tested against pathogens with elevated MIC values to topical agents. Diagn Microbiol Infect Dis 86:66–69. doi: 10.1016/j.diagmicrobio.2016.06.012. [DOI] [PubMed] [Google Scholar]
- 191.Flamm RK, Rhomberg PR, Simpson KM, Farrell DJ, Sader HS, Jones RN. 2015. In vitro spectrum of pexiganan activity when tested against pathogens from diabetic foot infections and with selected resistance mechanisms. Antimicrob Agents Chemother 59:1751–1754. doi: 10.1128/AAC.04773-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Lipsky BA, Holroyd KJ, Zasloff M. 2008. Topical versus systemic antimicrobial therapy for treating mildly infected diabetic foot ulcers: a randomized, controlled, double-blinded, multicenter trial of pexiganan cream. Clin Infect Dis 47:1537–1545. doi: 10.1086/593185. [DOI] [PubMed] [Google Scholar]
- 193.Ge Y, MacDonald D, Henry MM, Hait HI, Nelson KA, Lipsky BA, Zasloff MA, Holroyd KJ. 1999. In vitro susceptibility to pexiganan of bacteria isolated from infected diabetic foot ulcers. Diagn Microbiol Infect Dis 35:45–53. doi: 10.1016/S0732-8893(99)00056-5. [DOI] [PubMed] [Google Scholar]
- 194.Gottler LM, Ramamoorthy A. 2009. Structure, membrane orientation, mechanism, and function of pexiganan—a highly potent antimicrobial peptide designed from magainin. Biochim Biophys Acta 1788:1680–1686. doi: 10.1016/j.bbamem.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Sheldon AT., Jr 2005. Antiseptic “resistance”: real or perceived threat? Clin Infect Dis 40:1650–1656. doi: 10.1086/430063. [DOI] [PubMed] [Google Scholar]
- 196.Davies GE, Francis J, Martin AR, Rose FL, Swain G. 1954. 1:6-Di-4′-chlorophenyldiguanidohexane (hibitane); laboratory investigation of a new antibacterial agent of high potency. Br J Pharmacol Chemother 9:192–196. doi: 10.1111/j.1476-5381.1954.tb00840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Russell AD. 1986. Chlorhexidine: antibacterial action and bacterial resistance. Infection 14:212–215. doi: 10.1007/BF01644264. [DOI] [PubMed] [Google Scholar]
- 198.Silvestri DL, McEnery-Stonelake M. 2013. Chlorhexidine: uses and adverse reactions. Dermatitis 24:112–118. doi: 10.1097/DER.0b013e3182905561. [DOI] [PubMed] [Google Scholar]
- 199.Loftus MJ, Florescu CJ, Stuart RL. 2014. Staphylococcus aureus bacteraemia associated with peripherally inserted central catheters: the role of chlorhexidine gluconate-impregnated sponge dressings. Med J Aust 200:317–318. doi: 10.5694/mja13.00092. [DOI] [PubMed] [Google Scholar]
- 200.Rupp ME, Lisco SJ, Lipsett PA, Perl TM, Keating K, Civetta JM, Mermel LA, Lee D, Dellinger EP, Donahoe M, Giles D, Pfaller MA, Maki DG, Sherertz R. 2005. Effect of a second-generation venous catheter impregnated with chlorhexidine and silver sulfadiazine on central catheter-related infections: a randomized, controlled trial. Ann Intern Med 143:570–580. doi: 10.7326/0003-4819-143-8-200510180-00007. [DOI] [PubMed] [Google Scholar]
- 201.McDonnell G, Russell AD. 1999. Antiseptics and disinfectants: activity, action, and resistance. Clin Microbiol Rev 12:147–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Sijbesma T, Rockmann H, van der Weegen W. 2011. Severe anaphylactic reaction to chlorhexidine during total hip arthroplasty surgery. A case report. Hip Int 21:630–632. doi: 10.5301/HIP.2011.8644. [DOI] [PubMed] [Google Scholar]
- 203.Parkes AW, Harper N, Herwadkar A, Pumphrey R. 2009. Anaphylaxis to the chlorhexidine component of Instillagel: a case series. Br J Anaesth 102:65–68. doi: 10.1093/bja/aen324. [DOI] [PubMed] [Google Scholar]
- 204.Maki DG. 1989. The use of antiseptics for handwashing by medical personnel. J Chemother 1(Suppl 1):S3–S11. [DOI] [PubMed] [Google Scholar]
- 205.Horner C, Mawer D, Wilcox M. 2012. Reduced susceptibility to chlorhexidine in staphylococci: is it increasing and does it matter? J Antimicrob Chemother 67:2547–2559. doi: 10.1093/jac/dks284. [DOI] [PubMed] [Google Scholar]
- 206.Mangram AJ, Horan TC, Pearson ML, Silver LC, Jarvis WR. 1999. Guideline for prevention of surgical site infection, 1999. Hospital Infection Control Practices Advisory Committee. Infect Control Hosp Epidemiol 20:250–278. [DOI] [PubMed] [Google Scholar]
- 207.Climo MW, Sepkowitz KA, Zuccotti G, Fraser VJ, Warren DK, Perl TM, Speck K, Jernigan JA, Robles JR, Wong ES. 2009. The effect of daily bathing with chlorhexidine on the acquisition of methicillin-resistant Staphylococcus aureus, vancomycin-resistant Enterococcus, and healthcare-associated bloodstream infections: results of a quasi-experimental multicenter trial. Crit Care Med 37:1858–1865. doi: 10.1097/CCM.0b013e31819ffe6d. [DOI] [PubMed] [Google Scholar]
- 208.Huang SS, Septimus E, Kleinman K, Moody J, Hickok J, Avery TR, Lankiewicz J, Gombosev A, Terpstra L, Hartford F, Hayden MK, Jernigan JA, Weinstein RA, Fraser VJ, Haffenreffer K, Cui E, Kaganov RE, Lolans K, Perlin JB, Platt R. 2013. Targeted versus universal decolonization to prevent ICU infection. N Engl J Med 368:2255–2265. doi: 10.1056/NEJMoa1207290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Batra R, Cooper BS, Whiteley C, Patel AK, Wyncoll D, Edgeworth JD. 2010. Efficacy and limitation of a chlorhexidine-based decolonization strategy in preventing transmission of methicillin-resistant Staphylococcus aureus in an intensive care unit. Clin Infect Dis 50:210–217. doi: 10.1086/648717. [DOI] [PubMed] [Google Scholar]
- 210.Hayden MK, Lolans K, Haffenreffer K, Avery TR, Kleinman K, Li H, Kaganov RE, Lankiewicz J, Moody J, Septimus E, Weinstein RA, Hickok J, Jernigan J, Perlin JB, Platt R, Huang SS. 2016. Chlorhexidine and mupirocin susceptibility of methicillin-resistant Staphylococcus aureus isolates in the REDUCE-MRSA Trial. J Clin Microbiol 54:2735–2742. doi: 10.1128/JCM.01444-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.McNeil JC, Kok EY, Vallejo JG, Campbell JR, Hulten KG, Mason EO, Kaplan SL. 2016. Clinical and molecular features of decreased chlorhexidine susceptibility among nosocomial Staphylococcus aureus isolates at Texas Children's Hospital. Antimicrob Agents Chemother 60:1121–1128. doi: 10.1128/AAC.02011-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Macias JH, Alvarez MF, Arreguin V, Munoz JM, Macias AE, Alvarez JA. 2016. Chlorhexidine avoids skin bacteria recolonization more than triclosan. Am J Infect Control 44:1530–1534. doi: 10.1016/j.ajic.2016.04.235. [DOI] [PubMed] [Google Scholar]
- 213.Gunther F, Kaiser SJ, Fries T, Frank U, Mutters NT. 2015. Susceptibility of multidrug resistant clinical pathogens to a chlorhexidine formulation. J Prev Med Hyg 56:E176–E179. [PMC free article] [PubMed] [Google Scholar]
- 214.Kawana R, Kitamura T, Nakagomi O, Matsumoto I, Arita M, Yoshihara N, Yanagi K, Yamada A, Morita O, Yoshida Y, Furuya Y, Chiba S. 1997. Inactivation of human viruses by povidone-iodine in comparison with other antiseptics. Dermatology 195(Suppl 2):S29–S35. [DOI] [PubMed] [Google Scholar]
- 215.Gantait S, Bhattacharyya J, Das S, Biswas S, Ghati A, Ghosh S, Goel P. 2016. Comparative assessment of the effectiveness of different cleaning methods on the growth of Candida albicans over acrylic surface. Contemp Clin Dent 7:336–342. doi: 10.4103/0976-237X.188554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Best M, Sattar SA, Springthorpe VS, Kennedy ME. 1990. Efficacies of selected disinfectants against Mycobacterium tuberculosis. J Clin Microbiol 28:2234–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Bonez PC, Dos Santos Alves CF, Dalmolin TV, Agertt VA, Mizdal CR, Flores VC, Marques JB, Santos RC, Anraku de Campos MM. 2013. Chlorhexidine activity against bacterial biofilms. Am J Infect Control 41:e119–e122. doi: 10.1016/j.ajic.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 218.Hugo WB, Longworth AR. 1964. Some aspects of the mode of action of chlorhexidine. J Pharm Pharmacol 16:655–662. doi: 10.1111/j.2042-7158.1964.tb07384.x. [DOI] [PubMed] [Google Scholar]
- 219.Kampf G. 2009. Effect of chlorhexidine probably overestimated because of lack of neutralization after sampling. Infect Control Hosp Epidemiol 30:811–812. doi: 10.1086/597522. [DOI] [PubMed] [Google Scholar]
- 220.Poole K. 2005. Efflux-mediated antimicrobial resistance. J Antimicrob Chemother 56:20–51. doi: 10.1093/jac/dki171. [DOI] [PubMed] [Google Scholar]
- 221.Wassenaar TM, Ussery D, Nielsen LN, Ingmer H. 2015. Review and phylogenetic analysis of qac genes that reduce susceptibility to quaternary ammonium compounds in Staphylococcus species. Eur J Microbiol Immunol 5:44–61. doi: 10.1556/EuJMI-D-14-00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Smith K, Gemmell CG, Hunter IS. 2008. The association between biocide tolerance and the presence or absence of qac genes among hospital-acquired and community-acquired MRSA isolates. J Antimicrob Chemother 61:78–84. doi: 10.1093/jac/dkm395. [DOI] [PubMed] [Google Scholar]
- 223.Lu Z, Chen Y, Chen W, Liu H, Song Q, Hu X, Zou Z, Liu Z, Duo L, Yang J, Gong Y, Wang Z, Wu X, Zhao J, Zhang C, Zhang M, Han L. 2015. Characteristics of qacA/B-positive Staphylococcus aureus isolated from patients and a hospital environment in China. J Antimicrob Chemother 70:653–657. doi: 10.1093/jac/dku456. [DOI] [PubMed] [Google Scholar]
- 224.Guo W, Shan K, Xu B, Li J. 2015. Determining the resistance of carbapenem-resistant Klebsiella pneumoniae to common disinfectants and elucidating the underlying resistance mechanisms. Pathog Glob Health 109:184–192. doi: 10.1179/2047773215Y.0000000022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Tennent JM, Lyon BR, Gillespie MT, May JW, Skurray RA. 1985. Cloning and expression of Staphylococcus aureus plasmid-mediated quaternary ammonium resistance in Escherichia coli. Antimicrob Agents Chemother 27:79–83. doi: 10.1128/AAC.27.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Grkovic S, Brown MH, Roberts NJ, Paulsen IT, Skurray RA. 1998. QacR is a repressor protein that regulates expression of the Staphylococcus aureus multidrug efflux pump QacA. J Biol Chem 273:18665–18673. doi: 10.1074/jbc.273.29.18665. [DOI] [PubMed] [Google Scholar]
- 227.Schumacher MA, Miller MC, Grkovic S, Brown MH, Skurray RA, Brennan RG. 2001. Structural mechanisms of QacR induction and multidrug recognition. Science 294:2158–2163. doi: 10.1126/science.1066020. [DOI] [PubMed] [Google Scholar]
- 228.Schlett CD, Millar EV, Crawford KB, Cui T, Lanier JB, Tribble DR, Ellis MW. 2014. Prevalence of chlorhexidine-resistant methicillin-resistant Staphylococcus aureus following prolonged exposure. Antimicrob Agents Chemother 58:4404–4410. doi: 10.1128/AAC.02419-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Hasanvand A, Ghafourian S, Taherikalani M, Jalilian FA, Sadeghifard N, Pakzad I. 2015. Antiseptic resistance in methicillin sensitive and methicillin resistant Staphylococcus aureus isolates from some major hospitals, Iran. Recent Pat Antiinfect Drug Discov 10:105–112. doi: 10.2174/1574891X10666150623093259. [DOI] [PubMed] [Google Scholar]
- 230.Wang JT, Sheng WH, Wang JL, Chen D, Chen ML, Chen YC, Chang SC. 2008. Longitudinal analysis of chlorhexidine susceptibilities of nosocomial methicillin-resistant Staphylococcus aureus isolates at a teaching hospital in Taiwan. J Antimicrob Chemother 62:514–517. doi: 10.1093/jac/dkn208. [DOI] [PubMed] [Google Scholar]
- 231.Prag G, Falk-Brynhildsen K, Jacobsson S, Hellmark B, Unemo M, Soderquist B. 2014. Decreased susceptibility to chlorhexidine and prevalence of disinfectant resistance genes among clinical isolates of Staphylococcus epidermidis. APMIS 122:961–967. doi: 10.1111/apm.12239. [DOI] [PubMed] [Google Scholar]
- 232.Hijazi K, Mukhopadhya I, Abbott F, Milne K, Al-Jabri ZJ, Oggioni MR, Gould IM. 2016. Susceptibility to chlorhexidine amongst multidrug-resistant clinical isolates of Staphylococcus epidermidis from bloodstream infections. Int J Antimicrob Agents 48:86–90. doi: 10.1016/j.ijantimicag.2016.04.015. [DOI] [PubMed] [Google Scholar]
- 233.McGann P, Kwak YI, Summers A, Cummings JF, Waterman PE, Lesho EP. 2011. Detection of qacA/B in clinical isolates of methicillin-resistant Staphylococcus aureus from a regional healthcare network in the eastern United States. Infect Control Hosp Epidemiol 32:1116–1119. doi: 10.1086/662380. [DOI] [PubMed] [Google Scholar]
- 234.Tanner J, Lin Y, Kornblum J, Herzig CT, Bystritsky R, Uhlemann AC, Lowy FD. 2014. Molecular characterization of methicillin-resistant Staphylococcus aureus clinical isolates obtained from the Rikers Island Jail System from 2009 to 2013. J Clin Microbiol 52:3091–3094. doi: 10.1128/JCM.01129-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Shamsudin MN, Alreshidi MA, Hamat RA, Alshrari AS, Atshan SS, Neela V. 2012. High prevalence of qacA/B carriage among clinical isolates of meticillin-resistant Staphylococcus aureus in Malaysia. J Hosp Infect 81:206–208. doi: 10.1016/j.jhin.2012.04.015. [DOI] [PubMed] [Google Scholar]
- 236.Longtin J, Seah C, Siebert K, McGeer A, Simor A, Longtin Y, Low DE, Melano RG. 2011. Distribution of antiseptic resistance genes qacA, qacB, and smr in methicillin-resistant Staphylococcus aureus isolated in Toronto, Canada, from 2005 to 2009. Antimicrob Agents Chemother 55:2999–3001. doi: 10.1128/AAC.01707-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Liu Q, Zhao H, Han L, Shu W, Wu Q, Ni Y. 2015. Frequency of biocide-resistant genes and susceptibility to chlorhexidine in high-level mupirocin-resistant, methicillin-resistant Staphylococcus aureus (MuH MRSA). Diagn Microbiol Infect Dis 82:278–283. doi: 10.1016/j.diagmicrobio.2015.03.023. [DOI] [PubMed] [Google Scholar]
- 238.Skovgaard S, Larsen MH, Nielsen LN, Skov RL, Wong C, Westh H, Ingmer H. 2013. Recently introduced qacA/B genes in Staphylococcus epidermidis do not increase chlorhexidine MIC/MBC. J Antimicrob Chemother 68:2226–2233. doi: 10.1093/jac/dkt182. [DOI] [PubMed] [Google Scholar]
- 239.Jones RD, Jampani HB, Newman JL, Lee AS. 2000. Triclosan: a review of effectiveness and safety in health care settings. Am J Infect Control 28:184–196. doi: 10.1067/mic.2000.102378. [DOI] [PubMed] [Google Scholar]
- 240.Schweizer HP. 2001. Triclosan: a widely used biocide and its link to antibiotics. FEMS Microbiol Lett 202:1–7. doi: 10.1111/j.1574-6968.2001.tb10772.x. [DOI] [PubMed] [Google Scholar]
- 241.Russell AD. 2004. Whither triclosan? J Antimicrob Chemother 53:693–695. doi: 10.1093/jac/dkh171. [DOI] [PubMed] [Google Scholar]
- 242.Escalada MG, Russell AD, Maillard JY, Ochs D. 2005. Triclosan-bacteria interactions: single or multiple target sites? Lett Appl Microbiol 41:476–481. doi: 10.1111/j.1472-765X.2005.01790.x. [DOI] [PubMed] [Google Scholar]
- 243.Stewart MJ, Parikh S, Xiao G, Tonge PJ, Kisker C. 1999. Structural basis and mechanism of enoyl reductase inhibition by triclosan. J Mol Biol 290:859–865. doi: 10.1006/jmbi.1999.2907. [DOI] [PubMed] [Google Scholar]
- 244.Heath RJ, Rubin JR, Holland DR, Zhang E, Snow ME, Rock CO. 1999. Mechanism of triclosan inhibition of bacterial fatty acid synthesis. J Biol Chem 274:11110–11114. doi: 10.1074/jbc.274.16.11110. [DOI] [PubMed] [Google Scholar]
- 245.McMurry LM, Oethinger M, Levy SB. 1998. Triclosan targets lipid synthesis. Nature 394:531–532. doi: 10.1038/28970. [DOI] [PubMed] [Google Scholar]
- 246.McMurry LM, McDermott PF, Levy SB. 1999. Genetic evidence that InhA of Mycobacterium smegmatis is a target for triclosan. Antimicrob Agents Chemother 43:711–713. doi: 10.1093/jac/43.5.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Heath RJ, Rock CO. 1995. Enoyl-acyl carrier protein reductase (fabI) plays a determinant role in completing cycles of fatty acid elongation in Escherichia coli. J Biol Chem 270:26538–26542. doi: 10.1074/jbc.270.44.26538. [DOI] [PubMed] [Google Scholar]
- 248.Heath RJ, White SW, Rock CO. 2001. Lipid biosynthesis as a target for antibacterial agents. Prog Lipid Res 40:467–497. doi: 10.1016/S0163-7827(01)00012-1. [DOI] [PubMed] [Google Scholar]
- 249.Brenwald NP, Fraise AP. 2003. Triclosan resistance in methicillin-resistant Staphylococcus aureus (MRSA). J Hosp Infect 55:141–144. doi: 10.1016/S0195-6701(03)00222-6. [DOI] [PubMed] [Google Scholar]
- 250.Chen Y, Pi B, Zhou H, Yu Y, Li L. 2009. Triclosan resistance in clinical isolates of Acinetobacter baumannii. J Med Microbiol 58:1086–1091. doi: 10.1099/jmm.0.008524-0. [DOI] [PubMed] [Google Scholar]
- 251.Grandgirard D, Furi L, Ciusa ML, Baldassarri L, Knight DR, Morrissey I, Largiader CR, Leib SL, Oggioni MR. 2015. Mutations upstream of fabI in triclosan resistant Staphylococcus aureus strains are associated with elevated fabI gene expression. BMC Genomics 16:345. doi: 10.1186/s12864-015-1544-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Ciusa ML, Furi L, Knight D, Decorosi F, Fondi M, Raggi C, Coelho JR, Aragones L, Moce L, Visa P, Freitas AT, Baldassarri L, Fani R, Viti C, Orefici G, Martinez JL, Morrissey I, Oggioni MR. 2012. A novel resistance mechanism to triclosan that suggests horizontal gene transfer and demonstrates a potential selective pressure for reduced biocide susceptibility in clinical strains of Staphylococcus aureus. Int J Antimicrob Agents 40:210–220. doi: 10.1016/j.ijantimicag.2012.04.021. [DOI] [PubMed] [Google Scholar]
- 253.Furi L, Haigh R, Al Jabri ZJ, Morrissey I, Ou HY, Leon-Sampedro R, Martinez JL, Coque TM, Oggioni MR. 2016. Dissemination of novel antimicrobial resistance mechanisms through the insertion sequence mediated spread of metabolic genes. Front Microbiol 7:1008. doi: 10.3389/fmicb.2016.01008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Durani P, Leaper D. 2008. Povidone-iodine: use in hand disinfection, skin preparation and antiseptic irrigation. Int Wound J 5:376–387. doi: 10.1111/j.1742-481X.2007.00405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Hugo WB. 1991. A brief history of heat and chemical preservation and disinfection. J Appl Bacteriol 71:9–18. [PubMed] [Google Scholar]
- 256.Shelanski HA, Shelanski MV. 1956. PVP-iodine: history, toxicity and therapeutic uses. J Int Coll Surg 25:727–734. [PubMed] [Google Scholar]
- 257.Zamora JL. 1986. Chemical and microbiologic characteristics and toxicity of povidone-iodine solutions. Am J Surg 151:400–406. doi: 10.1016/0002-9610(86)90477-0. [DOI] [PubMed] [Google Scholar]
- 258.Hosseini H, Ashraf MJ, Saleh M, Nowroozzadeh MH, Nowroozizadeh B, Abtahi MB, Nowroozizadeh S. 2012. Effect of povidone-iodine concentration and exposure time on bacteria isolated from endophthalmitis cases. J Cataract Refract Surg 38:92–96. doi: 10.1016/j.jcrs.2011.06.030. [DOI] [PubMed] [Google Scholar]
- 259.Khan FA, Hussain MA, Khan Niazi SP, Haq Z, Akhtar N. 2016. Efficacy of 2.5% and 1.25% povidone-iodine solution for prophylaxis of ophthalmia neonatorum. J Coll Physicians Surg Pak 26:121–124. doi: 10.2016/JCPSP.121124. [DOI] [PubMed] [Google Scholar]
- 260.Mitani O, Nishikawa A, Kurokawa I, Gabazza EC, Ikeda M, Mizutani H. 2016. Enhanced wound healing by topical application of ointment containing a low concentration of povidone-iodine. J Wound Care 25:521–529. doi: 10.12968/jowc.2016.25.9.521. [DOI] [PubMed] [Google Scholar]
- 261.Goldenheim PD. 1993. An appraisal of povidone-iodine and wound healing. Postgrad Med J 69(Suppl 3):S97–S105. [PubMed] [Google Scholar]
- 262.O'Meara S, Al-Kurdi D, Ologun Y, Ovington LG. 2010. Antibiotics and antiseptics for venous leg ulcers. Cochrane Database Syst Rev 2010:CD003557. doi: 10.1002/14651858.CD003557.pub3. [DOI] [PubMed] [Google Scholar]
- 263.Eggers M, Eickmann M, Kowalski K, Zorn J, Reimer K. 2015. Povidone-iodine hand wash and hand rub products demonstrated excellent in vitro virucidal efficacy against Ebola virus and modified vaccinia virus Ankara, the new European test virus for enveloped viruses. BMC Infect Dis 15:375. doi: 10.1186/s12879-015-1111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Sabracos L, Romanou S, Dontas I, Coulocheri S, Ploumidou K, Perrea D. 2007. The in vitro effective antiviral action of povidone-iodine (PVP-I) may also have therapeutic potential by its intravenous administration diluted with Ringer's solution. Med Hypotheses 68:272–274. doi: 10.1016/j.mehy.2006.07.039. [DOI] [PubMed] [Google Scholar]
- 265.Tennen R, Setlow B, Davis KL, Loshon CA, Setlow P. 2000. Mechanisms of killing of spores of Bacillus subtilis by iodine, glutaraldehyde and nitrous acid. J Appl Microbiol 89:330–338. doi: 10.1046/j.1365-2672.2000.01114.x. [DOI] [PubMed] [Google Scholar]
- 266.Hill RL, Casewell MW. 2000. The in-vitro activity of povidone-iodine cream against Staphylococcus aureus and its bioavailability in nasal secretions. J Hosp Infect 45:198–205. doi: 10.1053/jhin.2000.0733. [DOI] [PubMed] [Google Scholar]
- 267.Sheikh W. 1981. Development and validation of a neutralizer system for in vitro evaluation of some antiseptics. Antimicrob Agents Chemother 19:429–434. doi: 10.1128/AAC.19.3.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Capriotti K, Capriotti JA. 2012. Topical iodophor preparations: chemistry, microbiology, and clinical utility. Dermatol Online J 18:1. [PubMed] [Google Scholar]
- 269.Faoagali J, Fong J, George N, Mahoney P, O'Rourke V. 1995. Comparison of the immediate, residual, and cumulative antibacterial effects of Novaderm R,* Novascrub R,* Betadine surgical scrub, Hibiclens, and liquid soap. Am J Infect Control 23:337–343. doi: 10.1016/0196-6553(95)90263-5. [DOI] [PubMed] [Google Scholar]
- 270.Bolon M. 2011. Hand hygiene. Infect Dis Clin North Am 25:21–43. doi: 10.1016/j.idc.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 271.Sroka S, Gastmeier P, Meyer E. 2010. Impact of alcohol hand-rub use on meticillin-resistant Staphylococcus aureus: an analysis of the literature. J Hosp Infect 74:204–211. doi: 10.1016/j.jhin.2009.08.023. [DOI] [PubMed] [Google Scholar]
- 272.Rotter ML, Koller W. 1990. Surgical hand disinfection: effect of sequential use of two chlorhexidine preparations. J Hosp Infect 16:161–166. doi: 10.1016/0195-6701(90)90060-2. [DOI] [PubMed] [Google Scholar]
- 273.Larson EL, Eke PI, Laughon BE. 1986. Efficacy of alcohol-based hand rinses under frequent-use conditions. Antimicrob Agents Chemother 30:542–544. doi: 10.1128/AAC.30.4.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Bush LW, Benson LM, White JH. 1986. Pig skin as test substrate for evaluating topical antimicrobial activity. J Clin Microbiol 24:343–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Morton HE. 1950. The relationship of concentration and germicidal efficiency of ethyl alcohol. Ann N Y Acad Sci 53:191–196. doi: 10.1111/j.1749-6632.1950.tb31944.x. [DOI] [PubMed] [Google Scholar]
- 276.Haft RJ, Keating DH, Schwaegler T, Schwalbach MS, Vinokur J, Tremaine M, Peters JM, Kotlajich MV, Pohlmann EL, Ong IM, Grass JA, Kiley PJ, Landick R. 2014. Correcting direct effects of ethanol on translation and transcription machinery confers ethanol tolerance in bacteria. Proc Natl Acad Sci U S A 111:E2576–E2585. doi: 10.1073/pnas.1401853111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Woodruff LB, Pandhal J, Ow SY, Karimpour-Fard A, Weiss SJ, Wright PC, Gill RT. 2013. Genome-scale identification and characterization of ethanol tolerance genes in Escherichia coli. Metab Eng 15:124–133. doi: 10.1016/j.ymben.2012.10.007. [DOI] [PubMed] [Google Scholar]
- 278.Gerando HM, Fayolle-Guichard F, Rudant L, Millah SK, Monot F, Ferreira NL, Lopez-Contreras AM. 2016. Improving isopropanol tolerance and production of Clostridium beijerinckii DSM 6423 by random mutagenesis and genome shuffling. Appl Microbiol Biotechnol 100:5427–5436. doi: 10.1007/s00253-016-7302-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Akinosho H, Rydzak T, Borole A, Ragauskas A, Close D. 2015. Toxicological challenges to microbial bioethanol production and strategies for improved tolerance. Ecotoxicology 24:2156–2174. doi: 10.1007/s10646-015-1543-4. [DOI] [PubMed] [Google Scholar]
- 280.Luther MK, Bilida S, Mermel LA, LaPlante KL. 2015. Ethanol and isopropyl alcohol exposure increases biofilm formation in Staphylococcus aureus and Staphylococcus epidermidis. Infect Dis Ther 4:219–226. doi: 10.1007/s40121-015-0065-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Knobloch JK, Horstkotte MA, Rohde H, Kaulfers PM, Mack D. 2002. Alcoholic ingredients in skin disinfectants increase biofilm expression of Staphylococcus epidermidis. J Antimicrob Chemother 49:683–687. doi: 10.1093/jac/49.4.683. [DOI] [PubMed] [Google Scholar]
- 282.Nwugo CC, Arivett BA, Zimbler DL, Gaddy JA, Richards AM, Actis LA. 2012. Effect of ethanol on differential protein production and expression of potential virulence functions in the opportunistic pathogen Acinetobacter baumannii. PLoS One 7:e51936. doi: 10.1371/journal.pone.0051936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Fiester SE, Actis LA. 2013. Stress responses in the opportunistic pathogen Acinetobacter baumannii. Future Microbiol 8:353–365. doi: 10.2217/fmb.12.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Linley E, Denyer SP, McDonnell G, Simons C, Maillard JY. 2012. Use of hydrogen peroxide as a biocide: new consideration of its mechanisms of biocidal action. J Antimicrob Chemother 67:1589–1596. doi: 10.1093/jac/dks129. [DOI] [PubMed] [Google Scholar]
- 285.Omidbakhsh N, Sattar SA. 2006. Broad-spectrum microbicidal activity, toxicologic assessment, and materials compatibility of a new generation of accelerated hydrogen peroxide-based environmental surface disinfectant. Am J Infect Control 34:251–257. doi: 10.1016/j.ajic.2005.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Horn K, Otter JA. 2015. Hydrogen peroxide vapor room disinfection and hand hygiene improvements reduce Clostridium difficile infection, methicillin-resistant Staphylococcus aureus, vancomycin-resistant enterococci, and extended-spectrum beta-lactamase. Am J Infect Control 43:1354–1356. doi: 10.1016/j.ajic.2015.06.029. [DOI] [PubMed] [Google Scholar]
- 287.Barbut F, Menuet D, Verachten M, Girou E. 2009. Comparison of the efficacy of a hydrogen peroxide dry-mist disinfection system and sodium hypochlorite solution for eradication of Clostridium difficile spores. Infect Control Hosp Epidemiol 30:507–514. doi: 10.1086/597232. [DOI] [PubMed] [Google Scholar]
- 288.Hiti K, Walochnik J, Faschinger C, Haller-Schober EM, Aspock H. 2005. One- and two-step hydrogen peroxide contact lens disinfection solutions against Acanthamoeba: how effective are they? Eye (Lond) 19:1301–1305. doi: 10.1038/sj.eye.6701752. [DOI] [PubMed] [Google Scholar]
- 289.Rogez-Kreuz C, Yousfi R, Soufflet C, Quadrio I, Yan ZX, Huyot V, Aubenque C, Destrez P, Roth K, Roberts C, Favero M, Clayette P. 2009. Inactivation of animal and human prions by hydrogen peroxide gas plasma sterilization. Infect Control Hosp Epidemiol 30:769–777. doi: 10.1086/598342. [DOI] [PubMed] [Google Scholar]
- 290.Walker JT, Bradshaw DJ, Fulford MR, Marsh PD. 2003. Microbiological evaluation of a range of disinfectant products to control mixed-species biofilm contamination in a laboratory model of a dental unit water system. Appl Environ Microbiol 69:3327–3332. doi: 10.1128/AEM.69.6.3327-3332.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Hughes R, Kilvington S. 2001. Comparison of hydrogen peroxide contact lens disinfection systems and solutions against Acanthamoeba polyphaga. Antimicrob Agents Chemother 45:2038–2043. doi: 10.1128/AAC.45.7.2038-2043.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Zubko EI, Zubko MK. 2013. Co-operative inhibitory effects of hydrogen peroxide and iodine against bacterial and yeast species. BMC Res Notes 6:272. doi: 10.1186/1756-0500-6-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Repine JE, Fox RB, Berger EM. 1981. Hydrogen peroxide kills Staphylococcus aureus by reacting with staphylococcal iron to form hydroxyl radical. J Biol Chem 256:7094–7096. [PubMed] [Google Scholar]
- 294.Imlay JA, Chin SM, Linn S. 1988. Toxic DNA damage by hydrogen peroxide through the Fenton reaction in vivo and in vitro. Science 240:640–642. doi: 10.1126/science.2834821. [DOI] [PubMed] [Google Scholar]
- 295.Seier-Petersen MA, Nielsen LN, Ingmer H, Aarestrup FM, Agerso Y. 2015. Biocide susceptibility of Staphylococcus aureus CC398 and CC30 isolates from pigs and identification of the biocide resistance genes, qacG and qacC. Microb Drug Resist 21:527–536. doi: 10.1089/mdr.2014.0215. [DOI] [PubMed] [Google Scholar]
- 296.Deisseroth A, Dounce AL. 1970. Catalase: physical and chemical properties, mechanism of catalysis, and physiological role. Physiol Rev 50:319–375. [DOI] [PubMed] [Google Scholar]
- 297.Eason MM, Fan X. 2014. The role and regulation of catalase in respiratory tract opportunistic bacterial pathogens. Microb Pathog 74:50–58. doi: 10.1016/j.micpath.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 298.Binesse J, Lindgren H, Lindgren L, Conlan W, Sjostedt A. 2015. Roles of reactive oxygen species-degrading enzymes of Francisella tularensis SCHU S4. Infect Immun 83:2255–2263. doi: 10.1128/IAI.02488-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Painter KL, Strange E, Parkhill J, Bamford KB, Armstrong-James D, Edwards AM. 2015. Staphylococcus aureus adapts to oxidative stress by producing H2O2-resistant small-colony variants via the SOS response. Infect Immun 83:1830–1844. doi: 10.1128/IAI.03016-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Baureder M, Reimann R, Hederstedt L. 2012. Contribution of catalase to hydrogen peroxide resistance in Enterococcus faecalis. FEMS Microbiol Lett 331:160–164. doi: 10.1111/j.1574-6968.2012.02567.x. [DOI] [PubMed] [Google Scholar]
- 301.Sun D, Crowell SA, Harding CM, De Silva PM, Harrison A, Fernando DM, Mason KM, Santana E, Loewen PC, Kumar A, Liu Y. 2016. KatG and KatE confer Acinetobacter resistance to hydrogen peroxide but sensitize bacteria to killing by phagocytic respiratory burst. Life Sci 148:31–40. doi: 10.1016/j.lfs.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Kresken M, Hafner D, Schmitz FJ, Wichelhaus TA, Paul-Ehrlich-Society for Chemotherapy. 2004. Prevalence of mupirocin resistance in clinical isolates of Staphylococcus aureus and Staphylococcus epidermidis: results of the Antimicrobial Resistance Surveillance Study of the Paul-Ehrlich-Society for Chemotherapy, 2001. Int J Antimicrob Agents 23:577–581. doi: 10.1016/j.ijantimicag.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 303.Petinaki E, Spiliopoulou I, Kontos F, Maniati M, Bersos Z, Stakias N, Malamou-Lada H, Koutsia-Carouzou C, Maniatis AN. 2004. Clonal dissemination of mupirocin-resistant staphylococci in Greek hospitals. J Antimicrob Chemother 53:105–108. doi: 10.1093/jac/dkh028. [DOI] [PubMed] [Google Scholar]
- 304.Rossney A, O'Connell S. 2008. Emerging high-level mupirocin resistance among MRSA isolates in Ireland. Euro Surveill 13:8084. [PubMed] [Google Scholar]
- 305.Donker GA, Deurenberg RH, Driessen C, Sebastian S, Nys S, Stobberingh EE. 2009. The population structure of Staphylococcus aureus among general practice patients from The Netherlands. Clin Microbiol Infect 15:137–143. doi: 10.1111/J.1469-0691.2008.02662.X. [DOI] [PubMed] [Google Scholar]
- 306.McDanel JS, Murphy CR, Diekema DJ, Quan V, Kim DS, Peterson EM, Evans KD, Tan GL, Hayden MK, Huang SS. 2013. Chlorhexidine and mupirocin susceptibilities of methicillin-resistant Staphylococcus aureus from colonized nursing home residents. Antimicrob Agents Chemother 57:552–558. doi: 10.1128/AAC.01623-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Suwantarat N, Carroll KC, Tekle T, Ross T, Popoola VO, Milstone AM. 2015. Low prevalence of mupirocin resistance among hospital-acquired methicillin-resistant Staphylococcus aureus isolates in a neonatal intensive care unit with an active surveillance cultures and decolonization program. Infect Control Hosp Epidemiol 36:232–234. doi: 10.1017/ice.2014.17. [DOI] [PubMed] [Google Scholar]
- 308.Sciortino CV, Kemper M, Parthasarathy L, Lay J. 2015. Surveillance of methicillin-resistant Staphylococcus aureus mupirocin resistance in a Veterans Affairs Hospital. Infect Control Hosp Epidemiol 36:235–236. doi: 10.1017/ice.2014.10. [DOI] [PubMed] [Google Scholar]
- 309.Lee H, Lim H, Bae IK, Yong D, Jeong SH, Lee K, Chong Y. 2013. Coexistence of mupirocin and antiseptic resistance in methicillin-resistant Staphylococcus aureus isolates from Korea. Diagn Microbiol Infect Dis 75:308–312. doi: 10.1016/j.diagmicrobio.2012.11.025. [DOI] [PubMed] [Google Scholar]
- 310.Horner C, Utsi L, Coole L, Denton M. 2017. Epidemiology and microbiological characterization of clinical isolates of Staphylococcus aureus in a single healthcare region of the UK, 2015. Epidemiol Infect 145:386–396. doi: 10.1017/S0950268816002387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Fang H, Froding I, Gian B, Haeggman S, Tollstrom UB, Ullberg M, Nord CE. 2016. Methicillin-resistant Staphylococcus aureus in Stockholm, Sweden: molecular epidemiology and antimicrobial susceptibilities to ceftaroline, linezolid, mupirocin and vancomycin in 2014. J Glob Antimicrob Resist 5:31–35. doi: 10.1016/j.jgar.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 312.Ghasemzadeh-Moghaddam H, van Belkum A, Hamat RA, van Wamel W, Neela V. 2014. Methicillin-susceptible and -resistant Staphylococcus aureus with high-level antiseptic and low-level mupirocin resistance in Malaysia. Microb Drug Resist 20:472–477. doi: 10.1089/mdr.2013.0222. [DOI] [PubMed] [Google Scholar]
- 313.Warren DK, Prager M, Munigala S, Wallace MA, Kennedy CR, Bommarito KM, Mazuski JE, Burnham CA. 2016. Prevalence of qacA/B genes and mupirocin resistance among methicillin-resistant Staphylococcus aureus (MRSA) isolates in the setting of chlorhexidine bathing without mupirocin. Infect Control Hosp Epidemiol 37:590–597. doi: 10.1017/ice.2016.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Coombs GW, Daly DA, Pearson JC, Nimmo GR, Collignon PJ, McLaws ML, Robinson JO, Turnidge JD. 2014. Community-onset Staphylococcus aureus Surveillance Programme annual report, 2012. Commun Dis Intell Q Rep 38:E59–E69. [PubMed] [Google Scholar]
- 315.Coombs GW, Nimmo GR, Pearson JC, Collignon PJ, Bell JM, McLaws ML, Christiansen KJ, Turnidge JD. 2013. Australian Group on Antimicrobial Resistance Hospital-Onset Staphylococcus aureus Surveillance Programme annual report, 2011. Commun Dis Intell Q Rep 37:E210–E218. [PubMed] [Google Scholar]
- 316.den Heijer CD, van Bijnen EM, Paget WJ, Stobberingh EE. 2014. Fusidic acid resistance in Staphylococcus aureus nasal carriage strains in nine European countries. Future Microbiol 9:737–745. doi: 10.2217/fmb.14.36. [DOI] [PubMed] [Google Scholar]
- 317.McLaws FB, Larsen AR, Skov RL, Chopra I, O'Neill AJ. 2011. Distribution of fusidic acid resistance determinants in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 55:1173–1176. doi: 10.1128/AAC.00817-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Souli M, Karaiskos I, Galani L, Maraki S, Perivolioti E, Argyropoulou A, Charissiadou A, Zachariadou L, Tsiplakou S, Papaioannou V, Tsorlini H, Katsifa H, Baka V, Pantazi P, Paschali A, Kyratsa A, Trikka-Graphakos E, Giannopoulou P, Vogiatzakis E, Moraitou H, Papadogeorgaki H, Avgerinou H, Panagea T, Pantazatou A, Petinaki E, Stamatopoulou G, Toutouza M, Karatzoglou I, Kontopoulou K, Orfanidou M, Karantani I, Fytas P, Tzanetou K, Platsouka E, Kazila P, Chli A, Statiri N, Giamarellou H. 2016. Nationwide surveillance of resistance rates of Staphylococcus aureus clinical isolates from Greek hospitals, 2012–2013. Infect Dis (Lond) 48:287–292. doi: 10.3109/23744235.2015.1110858. [DOI] [PubMed] [Google Scholar]
- 319.Jones RN, Mendes RE, Sader HS, Castanheira M. 2011. In vitro antimicrobial findings for fusidic acid tested against contemporary (2008–2009) gram-positive organisms collected in the United States. Clin Infect Dis 52(Suppl 7):S477–S486. doi: 10.1093/cid/cir163. [DOI] [PubMed] [Google Scholar]
- 320.Coombs GW, Daley DA, Thin Lee Y, Pearson JC, Robinson JO, Nimmo GR, Collignon P, Howden BP, Bell JM, Turnidge JD, Australian Group on Antimicrobial Resistance. 2016. Australian Group on Antimicrobial Resistance Australian Staphylococcus aureus Sepsis Outcome Programme annual report, 2014. Commun Dis Intell Q Rep 40:E244–E254. [PubMed] [Google Scholar]