Abstract
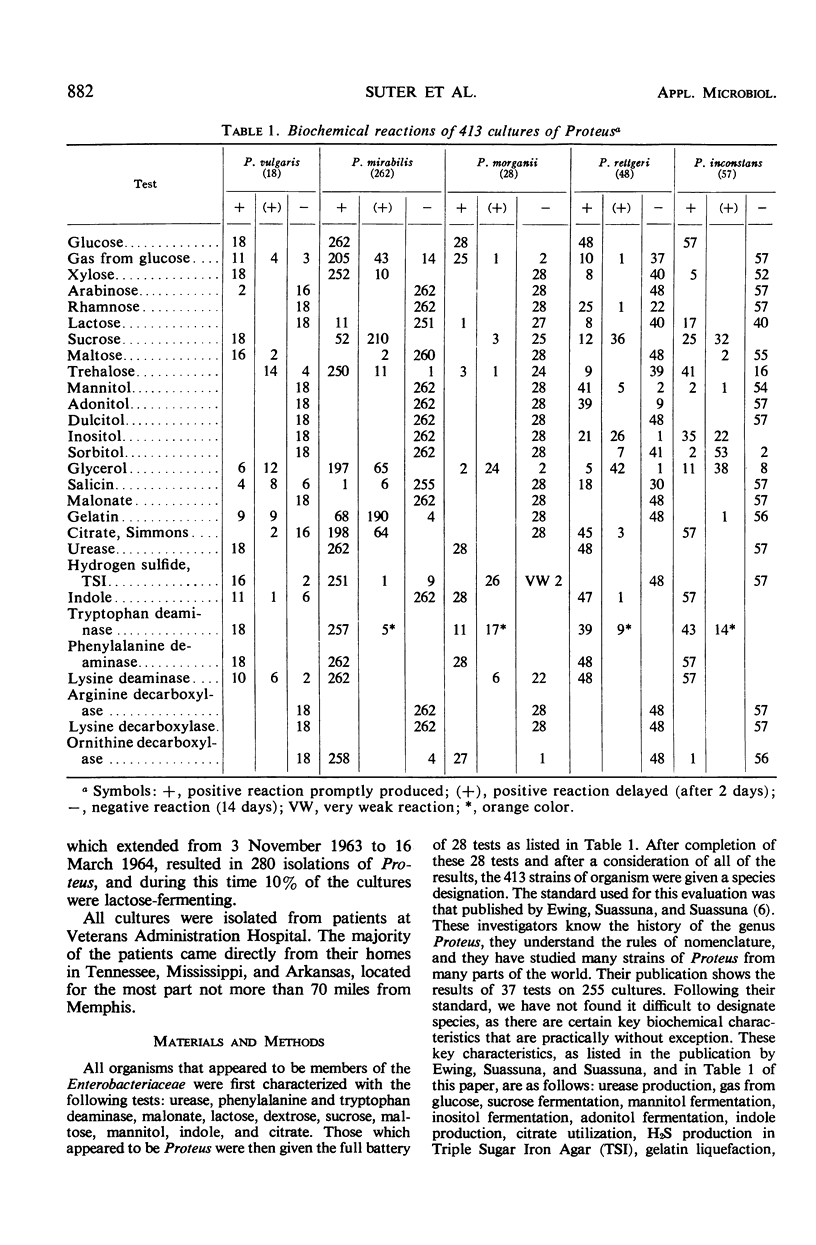
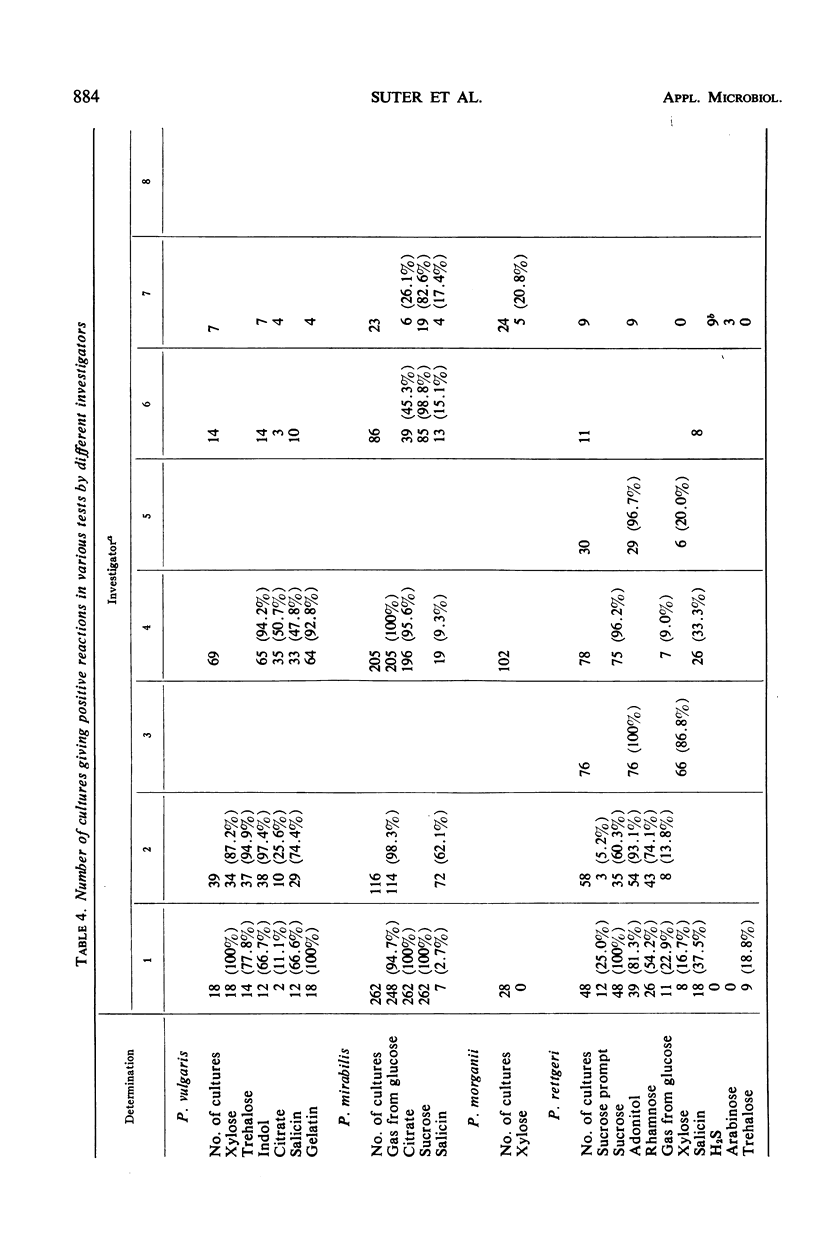
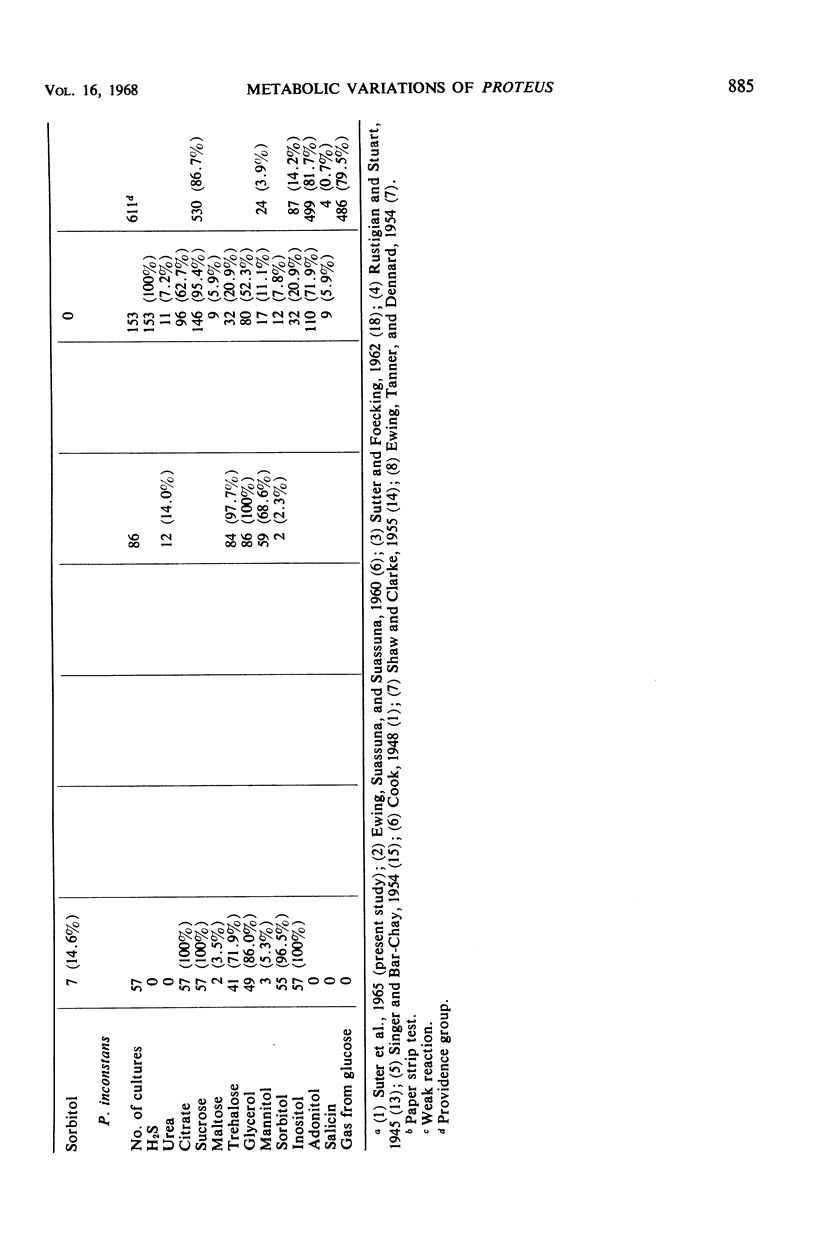
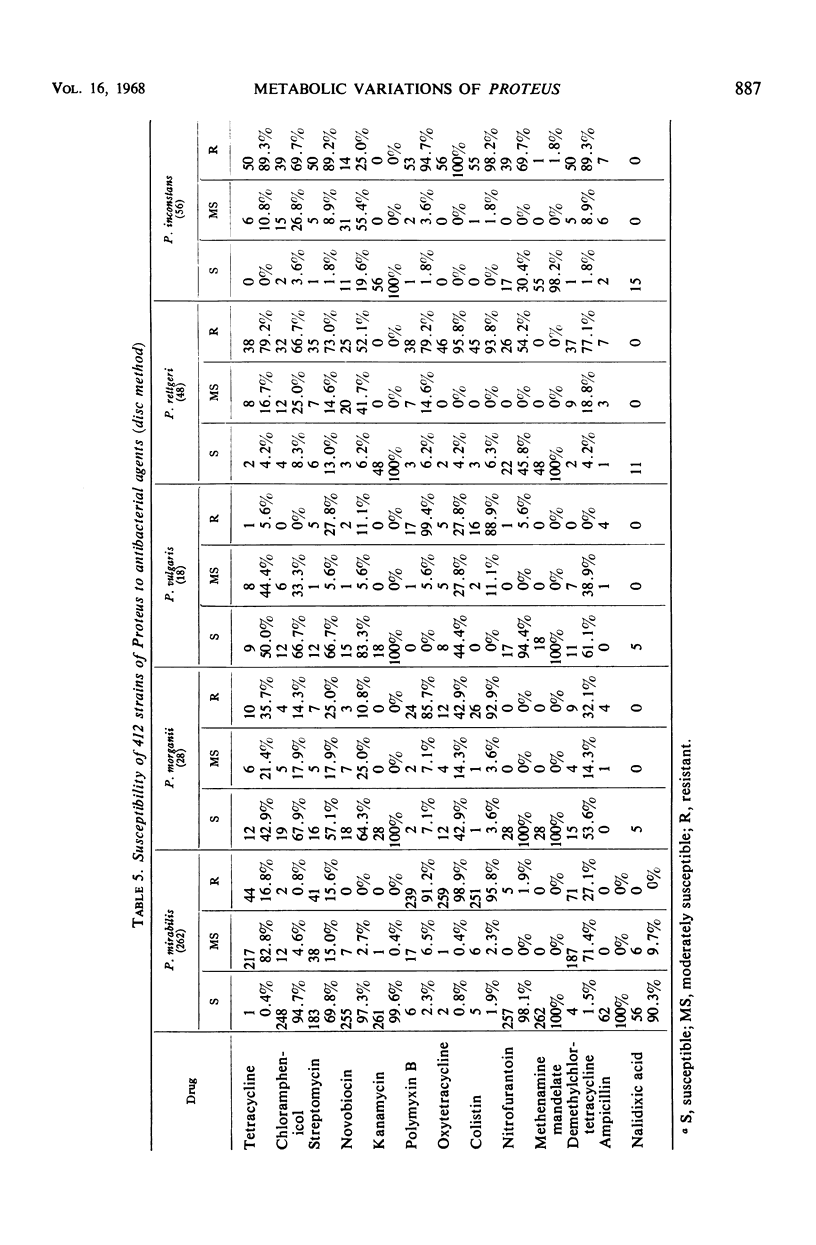
The number of strains of Proteus studied was 413, and these were obtained from all clinical materials with the exception of fecal specimens. Lactose was fermented by 37 strains (P. inconstans, 29%; P. rettgeri, 16%; P. mirabilis, 4.2%; P. morganii, 3.6%; and P. vulgaris, 0%) of which 33 were from the genitourinary system. These 33 strains constituted 12.7% of the 260 strains isolated from this source. Biochemically, P. mirabilis was the least variable, and P. rettgeri was the most variable of the five species of Proteus tested. P. inconstans and P. rettgeri resembled each other more closely than any of the other species of Proteus. Comparison of results obtained in the Memphis area with those found in other locations showed that biochemical characteristics varied most with the substances citrate, salicin, xylose, trehalose, and mannitol. In contrast to earlier reports from Israel and England, none of the strains of P. inconstans in the present study was able to attack urea. All five species of Proteus tested (by the disc method) were highly susceptible to methenamine mandelate. P. mirabilis, P. morganii, and P. vulgaris were also highly susceptible to nitrofurantoin. All strains of P. mirabilis were susceptible to ampicillin. P. inconstans was the most resistant species of Proteus. Of the other 356 urease-positive strains tested, 79% were susceptible to chloramphenicol, whereas only 3.8% of the 56 urease-negative strains (P. inconstans) were susceptible. When tested with streptomycin, 61% of urease-positive strains were susceptible and 1.8% of the urease-negative strains were susceptible. Of 36 lactose-positive strains, 33.8% were susceptible to chloramphenicol, whereas 72.8% of all lactose-negative strains were susceptible. Again, of the lactose-positive strains, 17% were susceptible to streptomycin, whereas 56.3% of all lactose-negative strains were susceptible.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- EDWARDS P. R., FIFE M. A. Lysine-iron agar in the detection of Arizona cultures. Appl Microbiol. 1961 Nov;9:478–480. doi: 10.1128/am.9.6.478-480.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EWING W. H., TANNER K. E., DENNARD D. A. The Providence group: an intermediate group of enteric bacteria. J Infect Dis. 1954 Mar-Apr;94(2):134–140. doi: 10.1093/infdis/94.2.134. [DOI] [PubMed] [Google Scholar]
- FALKOW S. Activity of lysine decarboxlase as an aid in the identification of Salmonellae and Shigellae. Am J Clin Pathol. 1958 Jun;29(6):598–600. doi: 10.1093/ajcp/29.6_ts.598. [DOI] [PubMed] [Google Scholar]
- Jeljaszewicz J., Hawiger J. The resistance to antibiotics of strains of Streptococcus viridans, Streptococcus faecalis, Escherichia coli, Pseudomonas aeruginosa, Proteus and Klebsiella isolated in Poland. Bull World Health Organ. 1966;35(2):243–246. [PMC free article] [PubMed] [Google Scholar]
- POTEE K. G., WRIGHT S. S., FINLAND M. In vitro susceptibility of recently isolated strains of Proteus to ten antibiotics. J Lab Clin Med. 1954 Sep;44(3):463–477. [PubMed] [Google Scholar]
- PROOM H. Amine production and nutrition in the Providence group. J Gen Microbiol. 1955 Aug;13(1):170–175. doi: 10.1099/00221287-13-1-170. [DOI] [PubMed] [Google Scholar]
- RANTZ L. A., RANTZ H. H. Antibiotic sensitivity of clinically important bacteria. A nine-year study. Arch Intern Med. 1962 Nov;110:739–743. doi: 10.1001/archinte.1962.03620230185025. [DOI] [PubMed] [Google Scholar]
- Rustigian R., Stuart C. A. The Biochemical and Serological Relationships of the Organisms of the Genus Proteus. J Bacteriol. 1945 May;49(5):419–436. doi: 10.1128/jb.49.5.419-436.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHAW C., CLARKE P. H. Biochemical classification of Proteus and Providence cultures. J Gen Microbiol. 1955 Aug;13(1):155–161. doi: 10.1099/00221287-13-1-155. [DOI] [PubMed] [Google Scholar]
- SINGER J., BAR-CHAY J. Biochemical investigation of Providence strains and their relationship to the Proteus group. J Hyg (Lond) 1954 Mar;52(1):1–8. doi: 10.1017/s0022172400027194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SINGER J., VOLCANI B. E. An improved ferric chloride test for differentiating Proteus-Providence group from other Enterobacteriaceae. J Bacteriol. 1955 Mar;69(3):303–306. doi: 10.1128/jb.69.3.303-306.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SUTTER V. L., FOECKING F. J. Biochemical characteristics of lactose-fermenting Proteus rettgeri from clinical specimens. J Bacteriol. 1962 Apr;83:933–935. doi: 10.1128/jb.83.4.933-935.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TURCK M., LINDEMEYER R. I., PETERSDORF R. G. Comparison of single-disc and tube-dilution techniques in determining antibiotic sensitivities of gram-negative pathogens. Ann Intern Med. 1963 Jan;58:56–65. doi: 10.7326/0003-4819-58-1-56. [DOI] [PubMed] [Google Scholar]
- Welch H., Poole A. K. The Pleo-antigenicity of a Variant of Proteus X19. J Bacteriol. 1934 Nov;28(5):523–540. doi: 10.1128/jb.28.5.523-540.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]


