Abstract
Experimentally restricting transgene expression exclusively to astrocytes has proven difficult. Using adeno-associated-virus-mediated gene transfer, we assessed two commonly used glial fibrillary acidic protein promoters: the full-length version gfa2 (2,210-bp human glial fibrillary acidic protein [GFAP] promoter) and the truncated variant gfaABC1D (681-bp GFAP promoter). The capacity to drive efficient, but also cell-type specific, expression of the EGFP in astrocytes was tested both in vitro in rat primary cortical cultures as well as in vivo in the rat striatum. We observed an efficient, but not entirely astrocyte-specific, gfa2-driven reporter expression. gfaABC1D exhibited a weaker activity, and most importantly, off-target, neuronal expression of the transgene occurred in a larger fraction of cells. Therefore, we explored the potential of a microRNA (miR)-specific target-sequence-based approach for abolishing off-target expression. When miR124 target sequences were incorporated into the 3′ UTR, neuronal gene expression was effectively silenced. However, unexpectedly, the insertion of an additional sequence in the 3′ UTR clearly diminished transgene expression. In conclusion, the gfaABC1D promoter on its own is not sufficient to specifically target transgene expression to astrocytes and is not well suited for AAV-based gene targeting, even if short promoter sequences are required. The combination with a miR de-targeting sequence represents a promising experimental strategy that eliminates off-target, neuronal expression.
Keywords: astrocyte, GFAP, promoter, transgene, targeting, gfaABC1D, gfa2, adeno-associated virus (AAV), gene therapy, microRNA
Introduction
Selectively targeting and genetically manipulating specific cell types of the nervous system represents a major challenge in neuroscience in general and particularly in the development of safe gene therapy strategies for the treatment of neurological disorders.1 Many studies have focused on astrocytes, the predominant glial cell type in the CNS that constitutes nearly half of human brain cells, because astroglial cells play a crucial role in neural development, for example, by coordinating synapse formation and axon guidance and supporting neuronal survival. They further regulate neuronal function by neurotransmitter recycling and extracellular potassium buffering, are involved in the energy metabolism of nervous tissue, and can modulate synaptic transmission.2 Targeted expression of transgenes in astrocytes has emerged as an important research tool to further advance our knowledge regarding neuron-glia interactions and to explore the role of astrocytic Ca2+ signaling. The ability to selectively and exclusively target astrocytes by means of astrocyte-specific promoter sequences is therefore crucial for experiments involving viral gene transfer.3
Astrocytes are also critically and actively involved in the pathogenesis of numerous neurodegenerative diseases such as amyotrophic lateral sclerosis,4, 5 Alzheimer’s disease,6 Huntington’s disease,7 and Parkinson’s disease (PD).8 A role for astrocytes has been demonstrated in the progression of Rett syndrome,9 and astrocytes have also been implicated in neurodevelopmental conditions like autism10 and schizophrenia (see Sloan and Barres11 for review). Experiments allowing targeted expression of transgenes in astrocytes not only improve our understanding of how astrocytes contribute to the pathogenesis of these disorders, but may also lead to potential targeted-gene therapies for pathways in which astrocytes participate. Astrocytes can serve as hosts for localized, and thus potentially safer, neurotrophic factor delivery, and neurotrophic-factor-based gene therapy targeting diseases like PD may be achieved.12 In conclusion, targeted gene expression in astrocytes is a key requirement for studying their contribution to physiological and pathophysiological processes, as well as for the development of safe gene therapy strategies that demand tightly targeted transgene expression.
The use of viral vectors for targeted genetic manipulation is a particularly successful and reliable approach permitting selective transgene expression in defined cell populations in adult animals. Adeno-associated viral (AAV) vectors are excellent gene delivery vehicles for sustained mammalian cell transduction. They offer many advantages for gene transfer, such as their ability to infect both dividing and non-diving cells, the capacity for long-term expression of transgenes, and a high degree of biosafety. An AAV-based gene therapy became the first viral vector system to be approved for clinical applications in Western nations in 2012.13 Despite the many positive aspects that recombinant AAV offer for gene therapy, their relatively small packaging capacity (4.7 kb) can be challenging. This problem can potentially be addressed by modifying the genetic cargo, and thus one strategy to circumvent the small AAV packaging capacity is the use of short promoter sequences. Unfortunately, cell-type-specific promoters tend to be bulky and can occupy considerable amounts of the viral genome. This also applies to the widely used human glial fibrillary acidic (gfa) protein promoter gfa2 (2,210-bp human glial fibrillary acidic protein [GFAP] promoter), which has a size of ∼2.2 kb. gfa2 has been extensively characterized and is most commonly used to induce astrocyte-specific gene expression.14 Truncated versions of the standard gfa2 promoter sequence have been developed to drive transgene expression in astrocytes.15 These shorter variants represent useful modifications for the AAV system because they accommodate the limited cloning capacity of the latter.
The utilization of microRNAs (miRs) that are specific for certain cell types is an additional promising approach for avoiding off-target gene expression. In contrast to transcriptional targeting using cell-specific promoters that positively regulate transgene expression, miR regulation operates at the post-transcriptional level and de-targets transgene expression.16 The remarkably small number of nucleotides required as binding sites on the mRNAs to induce target repression17 makes the miR-mediated de-targeting approach perfectly suited for use in AAV vectors. Neuron-specific miR124 target sequence (miR124),18 which is the most abundant miR in the brain,19 represses the translation of its target transcripts by binding to a specific target sequence and eliminates residual expression in neuronal cells.20 Thus, combined with a GFAP promoter to restrict expression of a transgene predominantly to GFAP-producing cells, the insertion of an miR124 target sequence for silencing off-target gene expression in neurons should greatly improve the specificity of astrocyte-targeting gene transfer.
Although previous approaches for achieving astrocyte-specific overexpression with AAV-mediated gene transfer relied on specific serotypes and GFAP promoters,12, 21, 22 the potential of miR-mediated regulation of transgene expression for providing an additional layer of regulation has not been explored yet.
In the present study, we re-evaluate the cell-type specificity of two commonly applied astrocyte-specific promoters for use in AAV vectors. We used the conventional 2,210-bp version of the gfa2 promoter and the truncated 681-bp sequence variant gfaABC1D (681-bp GFAP promoter), which was derived from the human gfa2 promoter.15 We tested the capacity of these two promoters to achieve efficient transgene expression in astroglial cells while ensuring the lowest possible off-target expression in neuronal cells both in vitro as well as in vivo. We further explored the potential of an miR124-specific target sequence with respect to abolishing neuronal expression of a transgene delivered by an AAV vector.
The principal aim of this study was to develop an AAV vector that exhibits a greatly improved specificity of astrocyte-targeted transgene expression by suppressing potential off-target expression in neurons while keeping the expression cassette still small in size.
Results
The Truncated gfa2 Promoter gfaABC1D Is Not Selective for Astrocytes In Vitro
We first evaluated the capacity of the astrocyte-specific full-length gfa2 promoter in comparison with that of the truncated gfaABC1D promoter for controlling expression of the EGFP reporter in primary rat cortical cultures containing a mixture of astrocytes and neurons. The genome layouts of vectors expressing the reporter EGFP are schematically shown in Figure 1. To allow a comparison of the promoter specificity, we selected AAV vectors of serotype 6 (AAV6) for packaging of the constructs for all in vitro experiments, because we never observed a clear cell type preference of this serotype. Rather, AAV6 was capable of transgene expression in both neurons and astrocytes with equally high efficacy that simply depended on transcriptional control elements incorporated into the viral genome.23 Astrocytic transduction has been observed with AAV6 in previous studies in vitro24, 25 and in vivo.25, 26
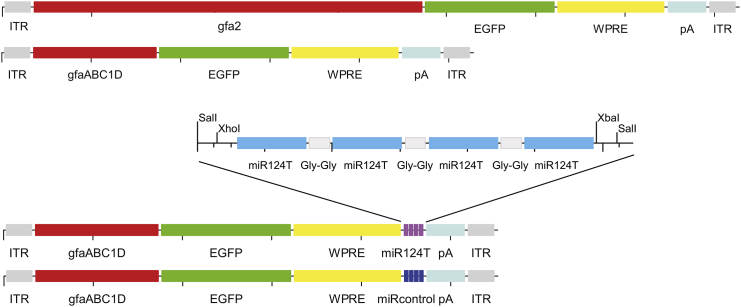
Figure 1.
Scheme of AAV Vector Genome Elements Used for Expression of EGFP in Rat Primary Cortical Cell Cultures and in the Rat Striatum
Expression cassettes with two different promoters driving EGFP reporter gene expression were used. For the de-targeting strategy, the vector was modified to carry four tandem copies of an artificial target sequence with complete complementarity for the endogenous neuronal miR124, which were inserted into the 3′ UTR region after the WPRE. A reporter construct containing the reversed strand of the 4× miR124T sequence was used as a negative control (miRcontrol). gfa2, 2,210-bp human GFAP promoter; gfaABC1D, 681-bp GFAP promoter; ITR, inverted terminal repeat; miR124T, neuron-specific miR124 target sequence; pA, bovine growth hormone polyadenylation site; WPRE, woodchuck hepatitis virus post-transcriptional regulatory element.
A total of 3 × 107, 5 × 107, or 6 × 107 transducing units (tu) of AAV6 vectors was added to each cell culture well of a 24-well plate containing approximately 250,000 cells at the day of seeding. To assess cell-type-specific transgene expression, we detected EGFP expression by direct fluorescence after immunocytochemical staining for cell-type-specific markers. To quantify EGFP expression among astrocytes, we performed immunostaining against the glial cell marker S100 to visualize astrocytes (Figure 2).
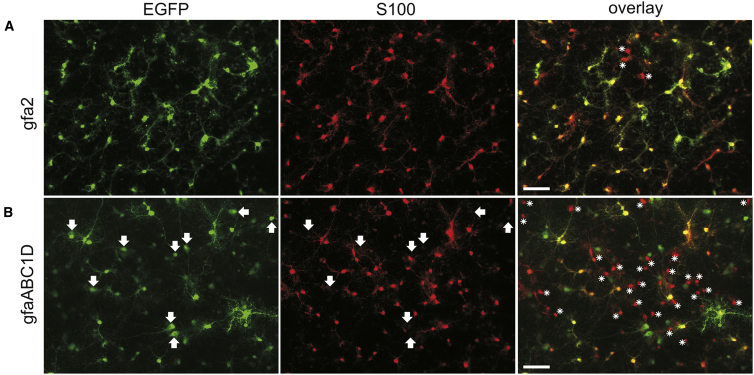
Figure 2.
In Vitro Application of Astroglial Targeting
(A and B) Using AAV6 vectors, the fluorescent reporter protein EGFP was expressed under the control of cell-type-specific promoters gfa2 (A) and gfaABC1D (B) in vitro. Rat primary cortical cultures were transduced at DIV 3 with 5 × 107 tu/well of AAV6-gfa2-EGFP or AAV6- gfaABC1D-EGFP. Fixation was performed 7 days posttransduction at DIV 10. Immunofluorescence staining against the glial cell marker S100 (red) was performed. Reporter transgene expression was detected by direct EGFP (green) fluorescence. Representative images from three independent experiments illustrate promoter efficacy and selectivity. Asterisks indicate S100-positive cells lacking EGFP expression. Arrows indicate EGFP+ cells lacking S100 immunoreactivity. Scale bars, 50 μm.
Readily detectable EGFP fluorescence following AAV delivery to primary cortical cultures emerged 2 days after transduction. Because the expression time course may differ between neurons and astrocytes, EGFP expression was assessed not before day 10 in vitro (DIV 10) (7 days posttransduction). In AAV-gfa2-EGFP-transduced primary cortical cultures, almost all cells labeled by anti-S100 immunofluorescence expressed EGFP at this time point. Vice versa, EGFP-positive (EGFP+) cells always showed S100 immunoreactivity (S100+) (Figure 2A). Thus, AAV-gfa2-EGFP transduction resulted in transgene expression restricted to cultured astrocytes.
Substituting gfa2 by the shorter gfaABC1D promoter variant resulted overall in a lower number of EGFP+ cells (Figure 2B), with many S100+ cells that did not show EGFP fluorescence. Surprisingly, EGFP+ cells were not always labeled by anti-S100 immunofluorescence (Figure 2B). Therefore, transgene expression driven by gfaABC1D appeared to be not limited to astrocytes.
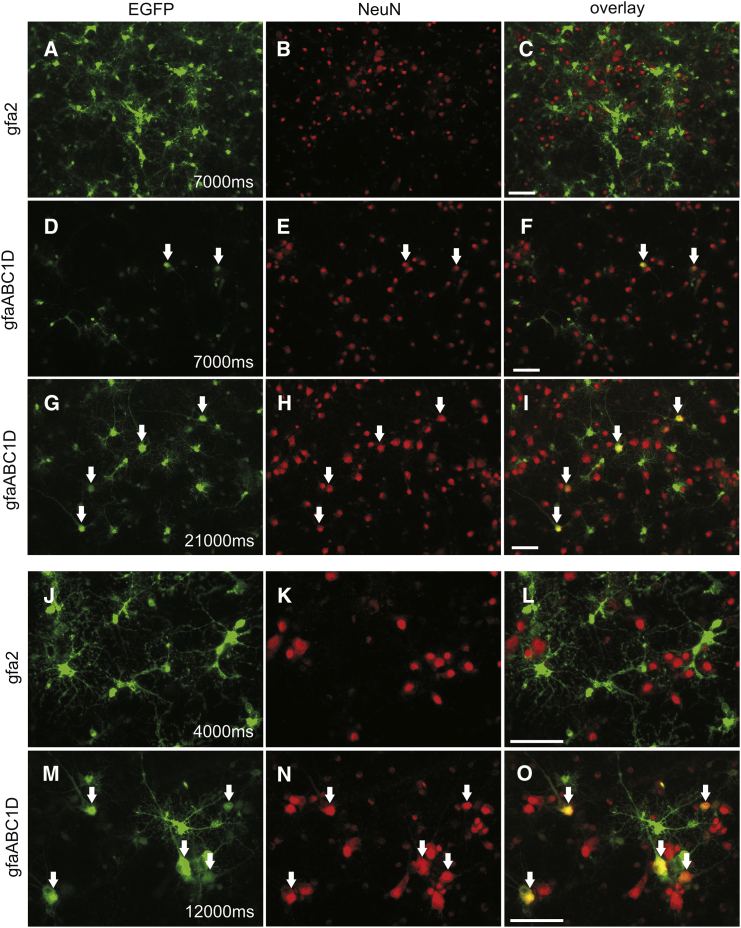
Next, the identity of EGFP+ cells in cortical cultures was examined by immunofluorescence with an antibody against the vertebrate neuron-specific nuclear protein (NeuN). EGFP and NeuN double-labeled cells were nearly absent after transfection with the gfa2 promoter (Figures 3A–3C and 3J–3L), indicating that off-target expression of EGFP in non-glial cell types was virtually nonexistent. In contrast, when driven by the gfaABC1D promoter, EGFP expression was also detected in NeuN-labeled (NeuN+) cells (Figures 3G–I and 3M–3O). We tried to limit the number of EGFP-expressing NeuN+ cells by significantly lowering the concentration of infectious particles (by 40%, from 5 × 107 tu/well [Figure 3] to 3 × 107 tu/well [Figure 4]). This reduction in virus titer induced an ∼50% decrease in expression level (Figure S1). However, reducing the concentration of infectious particles did not prevent neuronal reporter expression.
Figure 3.
Targeting of AAV to Rat Astrocytes In Vitro Using the Cell-Type-Specific Promoters gfa2 and gfaABC1D
Primary cortical cultures were transduced at DIV 3 with 5 × 107 tu/well of AAV6-gfa2-EGFP or AAV6- gfaABC1D-EGFP. Fixation was performed 7 days posttransduction at DIV 10. Immunofluorescence staining for the neuronal marker NeuN (red) was performed. EGFP (green) was visualized by direct fluorescence. Representative images from three independent experiments illustrate promoter efficacy (A–I) and selectivity (J–O). Numbers indicate exposure times. Arrows point to EGFP-expressing NeuN+ cells. (A–I) Images in (A) and (D) were acquired with an identical exposure time to compare the EGFP expression level between the gfa2 and gfaABC1D promoters. The level of expression of the reporter protein was clearly decreased in gfaABC1D-transduced cells. To be able to determine and compare the number of infected cells producing the reporter protein, we recorded the image in (G) with a 3-fold longer exposure time to increase the fluorescence intensity. EGFP+ cells were less abundant with the gfaABC1D promoter. (J–O) NeuN immunocytochemistry showed that the gfa2 promoter selectively targeted transgene expression to cells lacking NeuN (J–L). Driven by the gfaABC1D promoter, EGFP was expressed in a high number of NeuN+ cells (M–O). Scale bar, 50 μm.
Figure 4.
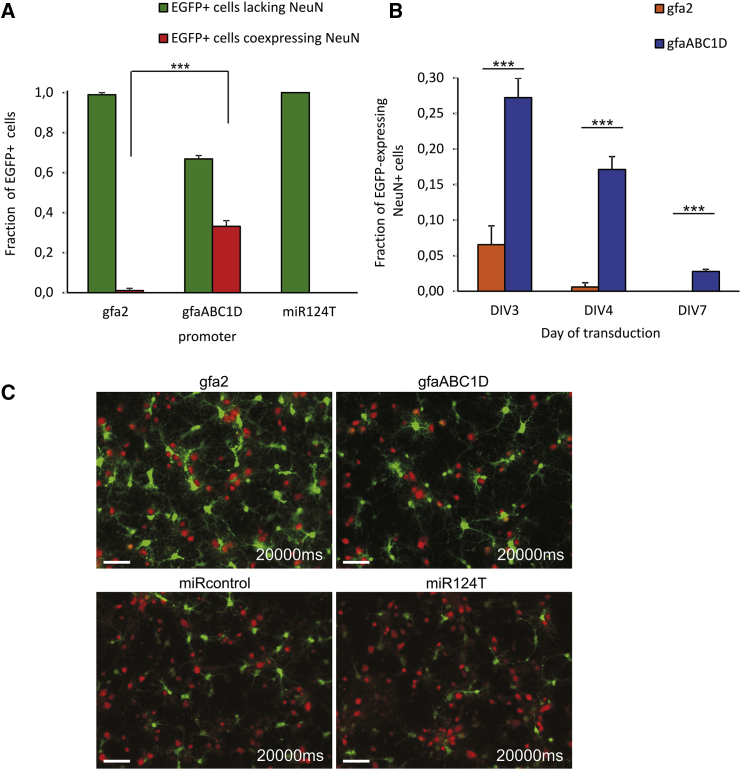
Endogenous miR124 Effectively De-targeted Transgene Expression from Rat Neurons In Vitro
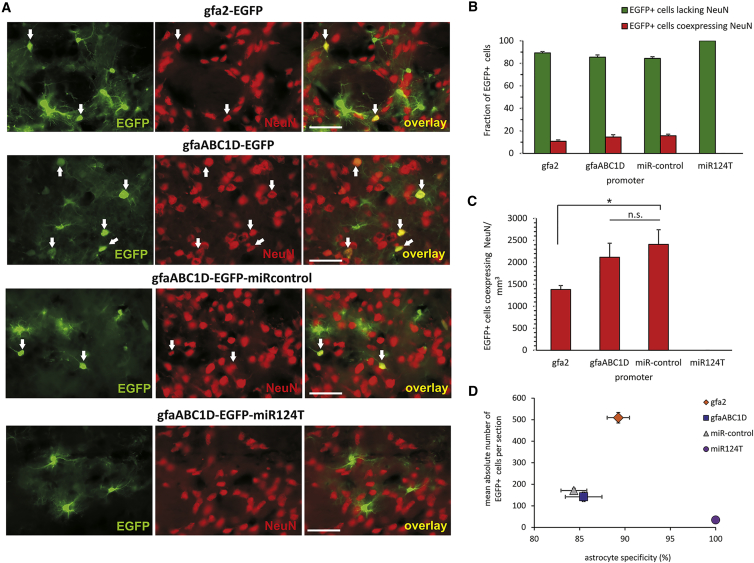
AAV vectors were used to express EGFP driven by the indicated promoters. The interaction of neuronal miR124 and its targeting sequence in the EGFP 3′ UTR was tested using constructs containing four repeats of the artificial target sites (miR124T) or reversed strand targeting sequence (miRcontrol) cloned into the 3′ UTR of the reporter gene. (A) Quantification of glial promoter selectivity following AAV delivery to primary cortical cultures. Depicted are the fractions of EGFP+ cells lacking NeuN or coexpressing NeuN. Primary cultures were transduced at DIV 4 with 3 × 107 tu/well of AAV6-gfa2-EGFP, AAV6-gfaABC1D-EGFP, or AAV6-gfaABC1D-EGFP-miR124T. Fixation was performed 10 days posttransduction at DIV 14. The gfa2 promoter drove EGFP expression almost exclusively in non-neuronal cells. The gfaABC1D promoter was significantly less cell specific. Insertion of miR124T prevented expression of EGFP in NeuN+ cells. (B) A decrease of the number of EGFP-expressing NeuN+ cells was achieved by transduction of the cell culture at later time points. However, when transgene expression was driven by gfaABC1D, a sizable number of NeuN+ cells was always expressing the reporter protein. Two-sided t test was applied for comparison. ***p < 0.0001. n = 7–15. Error bars represent SEM. (C) Cultures were transduced with the respective AAV vectors with 6 × 107 tu/well at DIV 4, and representative images were taken 11 days posttransduction. Cells were immunostained for neuronal cell marker NeuN (red). Transgene expression was detectable by direct EGFP fluorescence (green), which was acquired at a fixed exposure time. The gfa2 promoter drove a high level of EGFP expression, which is reduced with gfaABC1D. A further reduction in the EGFP expression level resulted from insertion of the miRcontrol and miR124T sequences. Scale bars, 50 μm.
Thus, in combination with an AAV vector, the gfaABC1D promoter was not selective for astrocytes, whereas the gfa2 promoter restricted EGFP expression to non-neuronal cells.
In order to compare EGFP-expression levels between the gfa2 and gfaABC1D promoter, we recorded images with an identical exposure time. The fluorescence intensity, and therefore the expression level of the reporter protein, was significantly lower in gfaABC1D-transduced cells (Figures 3A and 3D).
To determine and compare the total number of infected cells expressing the reporter protein, we recorded images with an automatically adjusted exposure time to increase the fluorescence intensity. The gfa2 promoter drove transgene expression noticeably with high efficacy (Figure 3A), whereas the use of the gfaABC1D promoter clearly resulted in a smaller number of EGFP+ astrocytes (Figure 3G).
Quantification of EGFP+ cells at DIV 14 (10 days posttransduction) confirmed that indeed the neuronal activity of gfa2 was significantly lower than that of gfaABC1D (Figure 4A). Extremely low levels of EGFP/NeuN colocalization were observed with gfa2. The percentage of EGFP+ cells coexpressing NeuN was 1.1% versus 98.9% ± 2.9% EGFP+ cells lacking NeuN. We conclude from these results that, in culture, the gfa2 promoter is highly astrocyte-specific when AAV6 is used as a transgene delivery vehicle. However, driven by the gfaABC1D promoter, EGFP was not exclusively expressed in non-neuronal cells. Although EGFP was expressed predominantly in astrocytes (66.9% ± 11.1%), a surprisingly high number of NeuN-positive (NeuN+) cells was also EGFP+ (33.1%).
In summary, a comparison of AAV6-gfa2-EGFP versus AAV6-gfaABC1D-EGFP expression showed that the gfaABC1D promoter performed less satisfyingly in several aspects compared with gfa2, driven by the gfaABC1D promoter: (1) EGFP expression levels were lower (Figures 3A and 3D); (2) the transduction efficacy was weaker (Figures 3A and 3G); and most importantly, (3) off-target expression of the EGFP transgene consistently occurred in a significantly larger fraction of cortical neurons when compared with the gfa2 promoter (Figures 3I and 3O).
miR124 Target Sequence Prevents Expression in Neuronal Cells In Vitro
Because the gfaABC1D promoter sequence did not restrict transgene expression to astroglial cells, we explored the potential of an miR124-specific target sequence as a means for abolishing neuronal expression of a transgene delivered by an AAV vector in primary cortical cultures. In order to silence off-target transgene synthesis in neurons, we constructed an miR-regulated AAV vector by inserting into the 3′ UTR of the EGFP expression cassette driven by the gfaABC1D promoter four identical tandem copies of a 21-bp sequence that was perfectly complementary to the miR124 (Figure 1). Multiple copies were used in order to optimize repression of the transgene.16 The sequence of the inhibitor used in our studies is the exact antisense copy of the mature miR124 sequence. A reporter construct containing the reversed strand of the 4× miR124T sequence was used as a negative control (miRcontrol).
Inserting 4× miR124T into AAV6-gfaABC1D-EGFP successfully eliminated neuronal expression of EGFP (Figure 4A). In contrast to the neuronal expression seen for gfaABC1D, no EGFP fluorescence was found to colocalize with the neuronal marker NeuN for gfaABC1D-miR124T. These results show that endogenous miR124 abolished transgene expression in neurons of primary cortical cultures.
The level of transgene expression when using either the gfaABC1D-miR124T vector or the gfaABC1D-miRcontrol vector was, however, lower than that when using the vector without the insertion at 3′ UTR (Figure 4C). Thus, the incorporation of an additional sequence after the woodchuck hepatitis virus post-transcriptional regulatory element (WPRE) apparently led to a reduction in the level of transgene expression that was unrelated to the specific miR target sequence. To exclude the possibility that a low endogenous expression of miR124 or miR506 (which exhibits the same seed sequence) in astrocytes may reduce expression of the EGFP transcript equipped with miR124T sequences in the 3′ UTR, we quantified the expression of these two miRs in our rat primary cortical cultures. We isolated total RNA and carried out real-time qPCR analysis on pure astrocytic and mixed glial-neuronal cultures (Supplemental Materials and Methods). The miR124 expression level was several orders of magnitude lower in astrocytes when compared with neurons, which is consistent with the finding that miR124 is specifically expressed in all neuronal populations, but absent in other CNS cell types.18, 27 Expression of miR506, if present, occurs at levels below the detection limit of our real-time qPCR assay (Figure S2). Thus, a specific interaction of endogenously expressed miR124 and miR506 is unlikely to account for the observed drop in expression level in astrocytic cells when using constructs with an inserted miR124T sequence in the 3′ UTR.
Transducing the primary cortical cultures at later time points, for example, at DIV 4 or DIV 7 instead of DIV 3, was another effective approach to lower the level of misexpression. Although weak neuronal EGFP expression still occurred with gfaABC1D, fewer cells (2.8%) were EGFP and NeuN double-labeled (Figure 4B). Possibly, this is a consequence of the larger total fraction of astrocytes in more mature cortical cultures and the lower responsiveness of older neurons to AAV-mediated transgene delivery. This is consistent with the observation that the number of EGFP+ neurons per culture dish depended on the density of astrocytes, i.e., a lower number of available astrocytes resulted in a higher number of transduced neurons. This correlation was particularly clear for the gfaABC1D promoter. Misexpression in neurons with gfa2 occurred only if the fraction of astrocytes in the cultures dropped to less than 25%. Although it thus appears to be possible to increase the level of cell specificity in cell culture simply by increasing the relative density of astrocytes, this is not a viable option for in vivo applications.
Targeting to Rat Astrocytes In Vivo
After in vitro characterization of our AAV6 vectors in primary rat cortical cells, we characterized gfa2- and gfaABC1D-driven transgene expression in the rat striatum. To this end, viral vectors were unilaterally injected into the striatum of adult rats, and immunohistochemistry for phenotypic marker NeuN followed by unbiased stereology was performed 3 weeks later. Low-power histological analysis revealed that the strongest EGFP expression in terms of transduced area (Figure 5A) and expression level (Figure 5B) was observed with the gfa2 promoter. Driven by gfaABC1D, a 4-fold smaller transduced area was obtained (Figure 7B).
Figure 5.
In Vivo Application of Astroglial Targeting
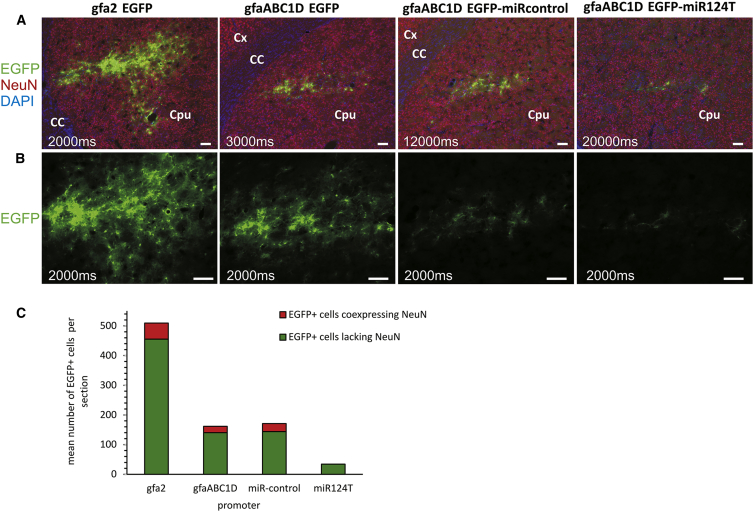
Expression pattern of the reporter gene EGFP. The indicated AAV6-EGFP vectors were administered with 1 × 108 tu to the striatum of adult rats, and brains were analyzed 3 weeks later to determine spread and selectivity of transgene expression. To determine whether the cell-type specificity was preserved in the gfa2 and gfaABC1D promoters, we performed immunohistochemical staining against NeuN (red) to visualize neurons. EGFP (green) was visualized by direct fluorescence. Astrocytes were identified by their typical shape and the absence of NeuN immunofluorescence. (A and B) Images are representative of three rats analyzed per vector type. (A) Low-magnification view of the transduced area. Images were acquired with an automatic exposure time. (B) Higher magnification of the injection site. Images were recorded with the same exposure time. Incorporation of miR sequences into the AAV vector had an apparent effect on transgene expression in astrocytes in the striatum. Expression levels of EGFP were very weak. (C) Stereological assessment of the number of transgene-expressing astrocytes and neurons per striatal section. For each vector, EGFP+ cells were counted and analyzed for costaining with NeuN (neuron). Quantification of the mean number of EGFP+ cells per section confirmed that the gfaABC1D promoter has a weaker activity compared with the gfa2 promoter. The miR-regulated AAV vector showed a further drop in efficiency. Scale bars, 100 μm. CC, corpus callosum; Cpu, caudate putamen (striatum); Cx, cortex.
Figure 7.
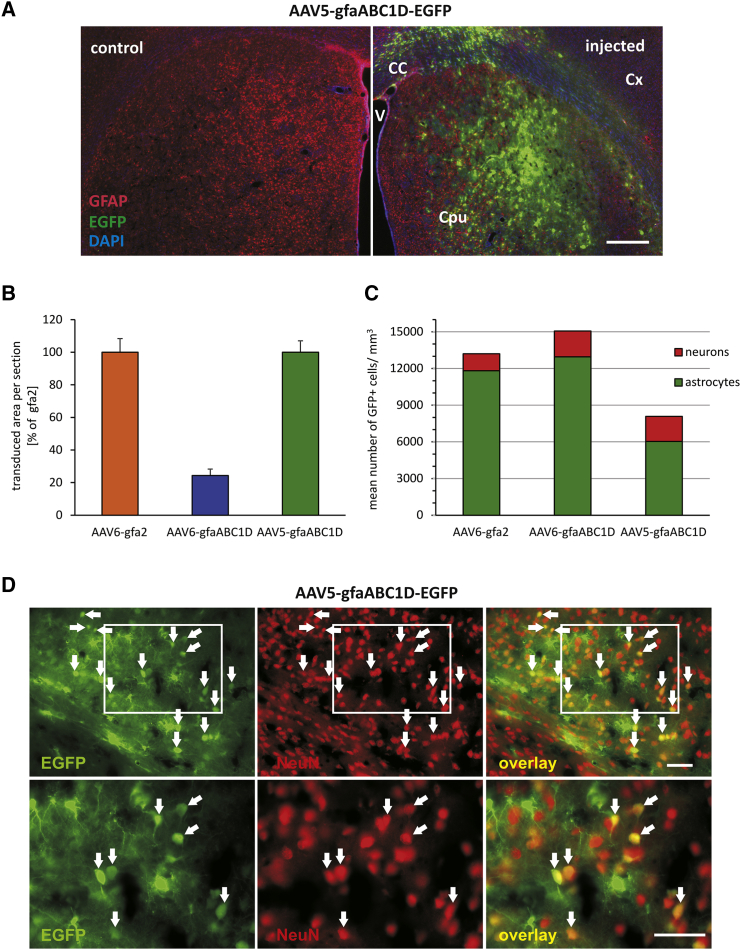
Specificity of Transgene Expression Was Not Improved by Application of AAV5 Serotype
(A) Using an AAV5 vector, EGFP was expressed under the control of the gfaABC1D promoter in the striatum of adult rats. Rats received one unilateral viral vector injection with AAV5-gfaABC1D-EGFP into the striatum. Shown is the expression of EGFP (green) in a GFAP (red)-stained striatal section. AAV5 allowed for widespread expression with the gfaABC1D promoter. Scale bar, 500 μm. (B) Stereological quantification of the transduced area per section. Expressing EGFP from the gfaABC1D promoter, the AAV5 vector transduced a four times larger area of striatal tissue when compared with AAV6 (n = 6 rats; error bars represent SEM). (C) Stereological quantification of the number of EGFP+ astrocytes and neurons per volume. AAV5-gfaABC1D caused off-target, neuronal expression with a number of EGFP+ neurons per cubic millimeter (mm3) that was comparable with that of AAV6-gfaABC1D. (D) Higher magnification images of NeuN-stained striatal sections from rats injected with AAV5-gfaABC1D-EGFP, illustrating that the viral vector transduced many neurons. Note that EGFP expression (green) was frequently colocalized (yellow) with the neuron-specific marker NeuN (red). Lower panels show higher magnification images of the areas indicated by the white boxes. Arrows indicate EGFP+ and NeuN+ neurons. EGFP/NeuN overlays are shown in the right column. Scale bars, 50 μm. CC, corpus callosum; Cpu, caudate putamen (striatum); Cx, cortex; V, ventricle.
Quantification of the mean number of EGFP+ cells per section confirmed that the gfaABC1D promoter had a weaker activity compared with the gfa2 promoter (Figure 5C). An average number of 455 EGFP+ astrocytes per striatal section was determined using gfa2, whereas only 140 EGFP+ astrocytes were counted after injection of gfaABC1D.
When analyzing the colocalization of EGFP fluorescence with immunohistochemically detected NeuN, we found that under in vivo conditions, neither the gfa2 nor the gfaABC1D promoter restricted EGFP expression exclusively to astrocytes, because both the gfa2 and the gfaABC1D promoter exhibited transgene expression in NeuN+ cells (Figure 5C). Figure 6A shows representative high-power images of the striatum after NeuN staining to identify neurons. The merged images reveal EGFP fluorescence in striatal neurons.
Figure 6.
Endogenous miR124 Effectively De-targeted Transgene Expression from Rat Neurons In Vivo
Immunohistochemical staining against NeuN was performed to test the interaction of neuronal miR124 and its targeting sequence in the EGFP 3′ UTR using constructs containing four copies of the artificial targeting sequence (miR124T) or reversed strand targeting sequence (miRcontrol) cloned into the 3′ UTR of the reporter gene. (A) Microscopy analysis of striatum of adult rats injected 3 weeks before with 1 × 108 tu of the indicated AAV6-EGFP vector. Counterstaining for NeuN (red) revealed EGFP fluorescence (green) in neurons. The merged images revealed some coexpressing cells. Shown are representative pictures from n = 3 rats per group. Arrows indicate EGFP+ neurons. Absence of any neuronal transgene expression by EGFP/NeuN overlay is shown for the AAV6-gfaABC1D-EGFP-miR124T vector. EGFP expression mediated by miR124T was limited exclusively to astrocytes. (B) Stereological quantification of transgene-expressing astrocytes and neurons. After insertion of miR124T sequences, EGFP+ neurons were not detected, and transgene expression was limited to astrocytes. (C) gfa2 and gfaABC1D caused a high level of neuronal expression. Only the gfaABC1D-miR124T viral vector was selective for striatal astrocytes. (D) The reporter transgene EGFP under control of the gfaABC1D promoter exhibited dramatic off-target, non-astrocytic expression patterns combined with a weak efficiency in vivo. Transgene expression restricted to astrocytes was restored by the incorporation of miR124T sequences. The gfa2 promoter was excellent in terms of activity, and EGFP immunoreactivity was mostly absent from neurons. Scale bars, 50 μm. *p = 0.038; p = 0.82, non-significant (n.s.). n = 5. One-way analysis of variance and post hoc Tukey’s test. Error bars represent SEM.
Because we did not obtain selective targeting for astrocytes but observed unwanted background expression in neurons, we combined the AAV-gfaABC1D targeting tool with the miR124 de-targeting strategy that proved to be successful in eliminating residual expression in neurons under in vitro conditions (see earlier). No expression of EGFP in NeuN+ cells was observed after incorporation of the neuron-specific miR target sequence miR124T into AAV-gfaABC1D vectors containing the EGFP reporter gene (Figure 5C). High-power images (Figure 6A) illustrate that EGFP fluorescence segregates from NeuN immunoreactivity, indicating astrocyte-specific transgene expression in the striatum.
The AAV-gfaABC1D vector and the gfaABC1D-miRcontrol vector performed equally well and produced similar results in terms of cell specificity and transduced area.
However, the insertion of the miR sequence (4× miR124T as well as miRcontrol) in the 3′ UTR strongly reduced the level of overall expression of the transgene as observed when EGFP fluorescence was recorded with identical exposure times (Figure 5B). As in cell culture, the incorporation of an additional sequence after the WPRE clearly diminished transgene expression. Although we were able to significantly improve the specificity of transgene expression and obtained virtually exclusive astrocyte-specific expression with the gfaABC1D-miR124T vector, the miR-de-targeting technique unfortunately caused a generally weaker total expression level in the context of AAV-mediated transgene delivery. It remains unclear why insertion of the miR124T or miRcontrol sequences in the 3′ UTR led to a non-specific reduction in transgene expression in astrocytes. The level of expression was for both sequences (either 4× miR124T or miRcontrol) lower than that obtained without insertion. This was observed in cell culture (Figure 4C) as well as in situ in the striatum of adult rats (Figure 5B). Thus, incorporating an additional sequence following the WPRE and not the actual design of the miR binding motif was responsible for the attenuated expression.
We quantified stereologically the fraction of EGFP+ striatal cells after injection of the AAV vectors (Figure 6B). Even though AAV-gfa2-promoter-controlled EGFP expression was predominantly found in astrocytes (89.3% ± 2.5%), a small amount of neuronal transgene expression was observed. Among all EGFP+ cells, 10.7% ± 2.5% were NeuN+ (n = 3 rats). Off-target, neuronal EGFP expression was higher using either the gfaABC1D promoter (14.6% ± 4.5% EGFP+ neurons) or the gfaABC1D-miRcontrol vector (15.6% ± 2.9% EGFP+ neurons).
In marked contrast to the other vectors, NeuN immunostaining of striatal sections from rats injected with AAV-gfaABC1D-EGFP-miR124T revealed that essentially all EGFP+ cells of all sections were NeuN-immunonegative, indicating exclusive astrocyte-specific expression (n = 3 rats; Figure 6).
The number of NeuN+ neurons that were also EGFP+ was calculated per area to further quantify neuronal EGFP expression in striatal sections (Figure 6C). gfa2, gfaABC1D, and gfaABC1D-miRcontrol viral vector caused a high level of neuronal expression. Only the gfaABC1D-miR124T vector effectively prevented EGFP expression in neuronal populations.
The capacity of the tested viral vectors to drive efficient but also cell-type-specific expression in vivo is summarized in Figure 6D. The gfa2 promoter performed excellently in terms of activity (high number of EGFP+ cells per section), and only a few EGFP+ neurons were detected. EGFP expression under control of the gfaABC1D promoter was weaker and occurred more frequently in non-astrocytic cells. Target specificity of EGFP expression was strongly improved by employing the miR-regulated AAV vector, albeit at the expense of a sizable drop in the number of EGFP+ cells per section. We therefore conclude that miR-mediated regulation of transgene expression is a viable and efficient method for improving target specificity of AAV-based transgene delivery in vivo.
Evaluation of an Alternative AAV Serotype
Because nervous cell tropism varies among AAV capsid serotypes, we tested whether packaging the gfaABC1D promoter construct into the serotype-5 capsid may reduce off-target expression in rat striatal neurons. We selected the AAV5 capsid for packaging because, similar to AAV6, this AAV serotype was reported to transduce both neurons and astrocytes in vivo.26, 28, 29, 30, 31
We determined: (1) spread (transduced area), (2) efficiency (transduced cells/volume), and (3) selectivity (fractional off-target expression) of transgene expression after injection of AAV5 in comparison with AAV6 (Figure 7).
The transduced areas obtained with the serotype-5 capsid in combination with gfaABC1D (Figure 7A) were on average four times larger when compared with the transduced area for the serotype-6 capsid. In fact, AAV5-gfaABC1D-EGFP injection resulted in a transduced area as large as with AAV6-gfa2-EGFP injection (Figure 7B).
Although AAV5-gfaABC1D-EGFP diffused over larger areas, the total number of EGFP+ cells per volume was ∼47% lower for AAV5 when compared with AAV6 when using the same promoter. With regard to the total number of transduced cells per volume, little difference was found between the two promotors when using serotype AAV6 (AAV6-gfa2 versus AAV6-gfaABC1D; Figure 7C).
We then quantified cell-type-specific EGFP expression (Figure 7C). Using AAV5, the mean number of EGFP+ neurons per volume was similar to that obtained with AAV6 (2,042 versus 2,117; red bars in Figure 7C), whereas the mean number of EGFP+ astrocytes per cubic millimeter (mm3) was 2-fold lower (green bars in Figure 7C). Therefore, the fraction of astrocytes among all EGFP+ cells decreased significantly, from 85% (AAV6-gfaABC1D) to 75% (AAV5-gfaABC1D). Thus, the fractional off-target, neuronal expression of the gfaABC1D promoter increased following AAV5 delivery. Using this latter serotype, we still frequently observed transgene-expressing NeuN+ cells in the target tissue (cf. Figures 7D and 6A, second row).
In conclusion, choosing AAV5 instead of an AAV6 serotype for application of the gfaABC1D vector did not improve astrocyte specificity in rat striatal neurons in situ. Thus, regardless of the specific serotype, the gfaABC1D promoter is unable to guarantee astrocyte-specific transgene expression in the context of AAV-mediated gene transfer.
Discussion
Studies that aim to selectively visualize or manipulate astroglial cells rely on an effective control of ectopic transgene expression and, therefore, require the identification of highly specific, but also efficient, and preferably small-sized recombinant promoters.
The mechanisms accounting for off-target, neuronal expression driven by glial promoters after gene transfer via viral vectors are largely unknown. In the case of AAV vectors, it remains possible that elements within the terminal repeat (TR) of AAV, which have been shown to have both enhancer and promoter function,32 are interacting with GFAP promoter elements to confer a more generalized expression of the transgene,33 thereby overriding the specificity of the promoter. Because the TRs are necessary for the encapsidation of the genome, and therefore constitute indispensable components of all recombinant AAV vectors, this TR transcriptional activity may interfere with all regulated expression cassettes32 and could represent a general problem for the design of AAV vectors for cell-specific targeting driven by cell-specific promoters.
The AAV vector genomes persist predominantly episomally upon transduction, but their genomes form concatemers.34 A low target specificity might be caused by this arrangement considering that transcriptional control elements from one genome can be brought to the proximity of elements from another genome and may interfere with each other.21
Although the gfa2 promoter was initially reported to consistently induce astrocyte-restricted expression in transgenic mice,35 a more recent study clearly showed significant off-target, neuronal expression of transgenes controlled by a constitutive gfa2 promoter in mouse lines.14 Unfortunately, later studies employing conditional manipulation of genes driven by the gfa2 promoter rarely re-evaluate cell specificity.3
When tested in AAV vectors in vivo, the gfa2 promoter showed inconsistent results with respect to directing transgene expression specifically to astrocytes.36 An early and rather unanticipated finding in astrocyte-specific AAV targeting was the widespread, robust, and predominant expression of the gfa2-promoter-driven EGFP transgene in neuronal cells in the adult rat spinal cord.33 In contrast, excellent and selective transgene expression in astrocytes was described with the gfa2 promoter following AAV injection into the mouse brain.22, 37
Nevertheless, in the context of AAV-mediated transgene delivery, the size of the 2,210-bp gfa2 promoter represents a major drawback, especially when driving large transgene expression. The same size limitation applies for other constructed gfa2-based promoters.38 Because of its compact size, the 681-bp GFAP promoter fragment, gfaABC1D, compares favorably with all other GFAP-derived promoters for AAV-based gene transfer. The gfaABC1D promoter was shown to be 100% astrocyte specific in transgenic mouse lines. gfaABC1D showed essentially the same expression pattern as the gfa2 promoter and an about 2-fold greater activity in transgenic mice.15 Therefore, it was recommended for gene targeting applications in which size matters. Our conclusion is, however, that the gfaABC1D promoter is not suitable for astrocyte-specific expression mediated by AAV. We demonstrate that the gfaABC1D promoter has a level of astrocyte specificity inferior to that of the gfa2 promoter and has a weaker activity, not recommending it for gene targeting use, not even if short promoter sequences are demanded.
The different extent of off-target neuronal expression of gfa2 and gfaABC1D indicates that promoter sequences required to silence expression in neurons reside in the segments removed from gfa2, and that negative regulatory elements are missing in gfaABC1D. The advantage of allowing larger transgenes to be inserted into the vector because of the shortening of the gfa2 promoter sequence is thus achieved at the expense of cell specificity and expression level.
In conclusion, due to its compact size and high activity in transgenic mouse lines, the gfaABC1D promoter was considered a better option than the traditional gfa2 promoter. However, because of its bad performance in terms of astrocyte specificity and promoter activity, gfaABC1D on its own is not suitable for targeted AAV-based gene transfer. Nevertheless, compared with the gfa2 promoter, gfaABC1D has the advantage of a 3-fold smaller size. That motivated us to exploit an miR-de-targeting strategy, which provides an additional layer of gene expression regulation when using cell-specific promoters, and to evaluate the ability of neuronal-expressed miR124 to restrict AAV-mediated transgene expression to astrocytic glial cells.
Our principal aim was to greatly improve the specificity of astrocyte-targeting gene transfer by abolishing or at least greatly minimizing off-target expression in neurons. Our experimental approach to achieve this goal, namely using multiple copies of a perfectly complementary target, aimed at optimizing repression of the transgene in the presence of miR124. The decision to use four complete complementary binding sites was based on the following experimental findings: (1) perfectly complementary miR target sequences allow per se higher suppression of the transgene than imperfectly complementary sequences; and (2) an increased number of miR target sites enhances miR-dependent transgene repression.39 Natural targets of miRs are often suppressed only to a fractional extent.40, 41 This is because most mRNAs have only one target site, which is not perfectly complementary, for a given miR.42 Moreover, the use of complete complementary miR target sequences, which enable rapid miR recycling, minimizes the risk of miR saturation.43 Because studies with lentiviral vectors20, 27 and a study using AAV in the retina44 demonstrated that integration of four repeats of an miR124 target sequence is sufficient for efficient transgene repression, we chose four tandem copies of an miR124 target sequence.
Because of the small size of the miR124T sequence (21 bp), a four-repeat insertion including small spacer sequences does not constitute a major capacity problem. Our 4× miR124T insert had a total size of 105 nt. The benefit of using less than four repeats of binding sites for minimizing the size of the transgene expression cassette would be small. However, because of the low packaging capacity of AAV vectors, a decrease of the total length of the miR124T insert could be advantageous. Deletion of up to 5 nt from the 5′ end of a miR122 target sequence was found to be well tolerated and did not influence transgene suppression.43 It remains to be tested whether shortening of the miR124 target sequence affects the miR124-mediated repression function.
Our results demonstrate that miR124 target sequences can efficiently restrict AAV-mediated transgene expression to astrocytes in vitro and in vivo. Because the sequence of mature miR124 and its expression in neuronal populations are completely conserved over species,45 this gene expression system based on AAV vectors, cell-specific promoters, and miR124-complementary sequences could be applied universally, in vertebrates and invertebrates.
We show that AAV vectors with astrocyte-specific promoters can be used for cell-type-specific targeting provided that miR-mediated regulation of transgene expression at the post-transcriptional level is applied to achieve an additional negative control of gene expression. Because astrocytes are potential therapeutic targets for the clinical treatment of neurological disorders, reliable astrocyte-specific targeting using cellular promoters in the context of AAV gene delivery is expected to be widely applicable. Selective introduction of transgenes into astrocytes to restore gene function might provide a therapeutic strategy that can prevent or delay neurodegeneration. Unfortunately, we observed a sizable drop in the level of astrocytic transgene expression after insertion of the miR sequence in the 3′ UTR, which could be disadvantageous for gene therapy applications. The attenuated expression can likely be tolerated in basic neurobiological research or disease modeling, but may be a concern for therapeutic approaches. Whether appropriate expression levels are achieved will thus depend on the particular transgene and the specific application, and needs to be carefully evaluated.
Because the miR124 and miR506 expression levels were below detection in astrocytes, it is unlikely that the respective miRs were responsible for reducing transgene expression in astrocytes. The miR expression level is known to be crucial for miR-mediated regulation because only the most abundant miRs within a cell mediate significant target suppression.46 Thus, a prerequisite for efficient miR-mediated repression of a miR target side-containing transcript is an miR expression above a minimum threshold.42
Reduced expression levels of different adenoviral genes following insertion of miR target sequences in the 3′ UTR of adenoviral vectors in spite of undetectable levels of the respective miRs have been observed before in vitro and in vivo.47 The 3′ UTR of an mRNA plays an important role for its stability.48 Insertion of additional sequences might affect mRNA stability for certain transgenes in an miR-independent manner.47
The WPRE is critical for strong expression and we generally see a WPRE-induced increase in protein expression of ∼5- to 15-fold. It is possible that the reduced transgene expression is a consequence of conformational changes in the WPRE induced by insertion at 3′ UTR close to the WPRE. The WPRE sequence containing RNA hairpin structures and three regulatory elements needs to assume a special conformation in order to stabilize the transcript. It is possible that the additional sequences, which were inserted immediately downstream the WPRE, form secondary structures preventing that conformation.
Because the known packaging limit of AAV constructs is <5.1 kb,49 and this in turn limits promoter length and hence selectivity, the challenge remains to develop short, yet highly astrocyte-specific and efficient, recombinant promoters. However, it is unlikely that promoter-only approaches will be sufficient to faithfully enable safe targeting of AAV vectors. On the other hand, miR-dependent regulation of transgene expression can effectively complement such strategies by providing an additional layer of transgene expression regulation and can further improve cell specificity when using AAV vectors.
Materials and Methods
Vector Construction
The AAV vectors were constructed to express EGFP as a reporter protein. The genome layout of the vectors used in this study is shown in Figure 1. The vector genomes consisted of inverted terminal repeats (ITRs), either the 2,210-bp human GFAP promoter gfa2 or the compact promoter version gfaABC1D (681 bp) derived from the conventional gfa2, the cDNA for EGFP, the WPRE for enhanced mRNA stability, and a bovine growth hormone polyadenylation site (pA).
For the validation of the de-targeting strategy, four tandem copies of a target sequence that is perfectly complementary to miR124 (4× miR124T) were inserted at a SalI site on 3′ UTR after the WPRE of the pAAV-gfaABC1D vector. At the 3′ end of the target sequence, we inserted a single adenosine residue to improve sensitivity to miR124.20, 50 A reporter construct containing the reversed strand of the 4× miR124T sequence was used as a negative control (miRcontrol). The final constructs were verified by DNA sequencing.
The miR124T (in capital letters) insert had the following sequence: 5′-gtcgacctcgagTGGCATTCACCGCGTGCCTTAattcgaaTGGCATTCACCGCGTGCCTTAaggtggaTGGCATTCACCGCGTGCCTTAaatgcatTGGCATTCACCGCGTGCCTTAtctagagtcgac-3′. The fragment was synthesized and cloned into pMA-RQ using SfiI and SfiI cloning sites (GeneArt; Life Technologies).
AAV Vector Production
The AAV constructs were packaged into AAV serotype-6-based viral vectors. The four different AAV6 vectors were produced by transient transfection of HEK293 cells using pDP6 as a helper plasmid. Cells were harvested at 2.5 days after transfection. Viral particles were purified by iodixanol step-gradient centrifugation, fast protein liquid chromatography (FPLC), and dialysis against PBS. Vector genome titers were determined by qPCR and ranged from 1 × 1010 to 2.5 × 1010 vector genomes (vg)/μL. Purity was confirmed by SDS-gel electrophoresis.
Vectors in Cell Culture
Primary cortical cells were prepared from embryonic day 18 rat pups according to standard protocols and were plated in 24-well plates.51 These preparations were seeded on poly-ornithine- and laminin-precoated glass coverslips at a density of 250,000 cells per well. Primary cultures were transduced by AAV vectors on days in vitro (DIV) 3, 4, or 7 by diluting the proper amount of viruses in 10 μL of PBS, which was then directly added to 750 μL of HCN culture medium in each well. A total of 3 × 107, 5 × 107, or 6 × 107 tu/well of the AAV6-EGFP vectors was used. The virus was not washed off. Cultures were fed by one-third media exchange on DIV 8 with fresh, equilibrated HCN medium.
Transgene expression was analyzed at 7, 10, or 11 days posttransduction. Cell counts were performed in at least five randomly selected fields per well and in at least six wells each of at least two independent replicates of the respective transduction.
Stereotaxic Vector Injections into the Rat Brain
All animal procedures were performed according to approved experimental animal licenses issued by the responsible animal welfare authority (Niedersächsisches Landesamt für Verbraucherschutz und Lebensmittelsicherheit [LAVES]) and supervised by local veterinarians.
Young adult female Wistar rats weighing between 220 and 280 g were housed in a 12 hr light-dark cycle and were provided with metamizole (1.5 mg/mL) in the drinking water 3 days before and for 1 week after surgery. Animals were anesthetized with a mixture of ketamine (100 mg/kg) and xylazine (5 mg/kg) and fixed in a stereotaxic frame (Kopf Instruments). Two injections were performed into the striatum, each with a total of 2 μL containing 1 × 108 tu at the following coordinates (in mm relative to bregma): mediolateral (ML) +0.2, anteroposterior (AP) +0.05, dorsoventral (DV) −0.5 and ML +0.38, AP −0.05, DV −0.5. Viruses were diluted in 2 μL of PBS and injected using a Nanoliter 2000 injector (World Precision Instruments) at 500 nL/min; the needle was left in place for 4 min both before and after the injection. Animals were sacrificed at 3 weeks postinjection. Rats were anesthetized and intracardially perfused with PBS followed by cold 4% paraformaldehyde (PFA) in PBS. Brains were then removed, postfixed in 4% PFA overnight, and cryoprotected in 30% sucrose in double-distilled water (ddH2O) for 48 hr. Thirty-micrometer-thick serial coronal sections from the striatum were collected on a cryostat (Leica) and processed for immunohistochemistry.
Immunostaining and Stereology
Cell phenotypes were identified by the primary antibodies mouse anti-NeuN (1:200; Chemicon) and rabbit anti-S100 (1:400; Dako). Corresponding Cy3-coupled secondary antibodies (Dianova) were used for detection. Reporter transgene expression was detected by direct EGFP fluorescence. Cell nuclei were visualized with DAPI (Sigma). For immunohistochemistry, primary antibodies were applied for 48 hr at 4°C followed by 2 hr incubation at room temperature (RT) with secondary antibodies on free-floating sections. For improved staining quality, antigen retrieval for 6 hr at 60°C in Tris-buffered saline (TBS [pH 9.0]) was applied beforehand. After immunostaining, the brain sections were mounted, and images were captured using AxioVision 4.6 software (Zeiss) on an Axioplan microscope equipped with a 16-bit grayscale charge-coupled device (CCD) camera.
To quantify the number of reporter-expressing cells in the striatum, we performed stereological counting by the optical fractionator method52 using the Stereo Investigator software (MicroBrightField Bioscience). Due to the limited AAV6-vector spread around the site of injection, we included all sections of the transduced striatum.
Statistical Analyses
Data were collected in a blinded manner. The unpaired two-sided t test was used for statistical comparison between two groups. Multiple comparisons were made by one-way analysis of variance followed by Tukey honestly significant difference test. Differences were considered significant at p < 0.05. Statistical tests were performed using the R software package. Values are shown as mean values with SEM.
Author Contributions
S.K. conceived the study. G.T. and J.T. conducted the experiments. All authors contributed to collection, analysis, and interpretation of experimental data. G.T. wrote the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Acknowledgments
This work was supported by the German Research Council-funded Center of Nanoscale Microscopy and Molecular Physiology of the Brain (CNMPB). We thank Sonja Heyroth, Monika Zebski, and Barbara Müller for excellent technical assistance. We are grateful to Uwe Michel for helpful discussion and to Mathias Bähr for continuous support.
Footnotes
Supplemental Information includes Supplemental Materials and Methods and two figures and can be found with this article online at http://dx.doi.org/10.1016/j.omtn.2017.03.009.
Supplemental Information
References
- 1.Lentz T.B., Gray S.J., Samulski R.J. Viral vectors for gene delivery to the central nervous system. Neurobiol. Dis. 2012;48:179–188. doi: 10.1016/j.nbd.2011.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sofroniew M.V., Vinters H.V. Astrocytes: biology and pathology. Acta Neuropathol. 2010;119:7–35. doi: 10.1007/s00401-009-0619-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sloan S.A., Barres B.A. Looks can be deceiving: reconsidering the evidence for gliotransmission. Neuron. 2014;84:1112–1115. doi: 10.1016/j.neuron.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nagai M., Re D.B., Nagata T., Chalazonitis A., Jessell T.M., Wichterle H., Przedborski S. Astrocytes expressing ALS-linked mutated SOD1 release factors selectively toxic to motor neurons. Nat. Neurosci. 2007;10:615–622. doi: 10.1038/nn1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamanaka K., Chun S.J., Boillee S., Fujimori-Tonou N., Yamashita H., Gutmann D.H., Takahashi R., Misawa H., Cleveland D.W. Astrocytes as determinants of disease progression in inherited amyotrophic lateral sclerosis. Nat. Neurosci. 2008;11:251–253. doi: 10.1038/nn2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nagele R.G., Wegiel J., Venkataraman V., Imaki H., Wang K.C., Wegiel J. Contribution of glial cells to the development of amyloid plaques in Alzheimer’s disease. Neurobiol. Aging. 2004;25:663–674. doi: 10.1016/j.neurobiolaging.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 7.Bradford J., Shin J.Y., Roberts M., Wang C.E., Sheng G., Li S., Li X.J. Mutant huntingtin in glial cells exacerbates neurological symptoms of Huntington disease mice. J. Biol. Chem. 2010;285:10653–10661. doi: 10.1074/jbc.M109.083287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Niranjan R. The role of inflammatory and oxidative stress mechanisms in the pathogenesis of Parkinson’s disease: focus on astrocytes. Mol. Neurobiol. 2014;49:28–38. doi: 10.1007/s12035-013-8483-x. [DOI] [PubMed] [Google Scholar]
- 9.Lioy D.T., Garg S.K., Monaghan C.E., Raber J., Foust K.D., Kaspar B.K., Hirrlinger P.G., Kirchhoff F., Bissonnette J.M., Ballas N., Mandel G. A role for glia in the progression of Rett’s syndrome. Nature. 2011;475:497–500. doi: 10.1038/nature10214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu T.W., Chahrour M.H., Coulter M.E., Jiralerspong S., Okamura-Ikeda K., Ataman B., Schmitz-Abe K., Harmin D.A., Adli M., Malik A.N. Using whole-exome sequencing to identify inherited causes of autism. Neuron. 2013;77:259–273. doi: 10.1016/j.neuron.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sloan S.A., Barres B.A. Mechanisms of astrocyte development and their contributions to neurodevelopmental disorders. Curr. Opin. Neurobiol. 2014;27:75–81. doi: 10.1016/j.conb.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drinkut A., Tereshchenko Y., Schulz J.B., Bähr M., Kügler S. Efficient gene therapy for Parkinson’s disease using astrocytes as hosts for localized neurotrophic factor delivery. Mol. Ther. 2012;20:534–543. doi: 10.1038/mt.2011.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bryant L.M., Christopher D.M., Giles A.R., Hinderer C., Rodriguez J.L., Smith J.B., Traxler E.A., Tycko J., Wojno A.P., Wilson J.M. Lessons learned from the clinical development and market authorization of Glybera. Hum. Gene Ther. Clin. Dev. 2013;24:55–64. doi: 10.1089/humc.2013.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Su M., Hu H., Lee Y., d’Azzo A., Messing A., Brenner M. Expression specificity of GFAP transgenes. Neurochem. Res. 2004;29:2075–2093. doi: 10.1007/s11064-004-6881-1. [DOI] [PubMed] [Google Scholar]
- 15.Lee Y., Messing A., Su M., Brenner M. GFAP promoter elements required for region-specific and astrocyte-specific expression. Glia. 2008;56:481–493. doi: 10.1002/glia.20622. [DOI] [PubMed] [Google Scholar]
- 16.Brown B.D., Venneri M.A., Zingale A., Sergi Sergi L., Naldini L. Endogenous microRNA regulation suppresses transgene expression in hematopoietic lineages and enables stable gene transfer. Nat. Med. 2006;12:585–591. doi: 10.1038/nm1398. [DOI] [PubMed] [Google Scholar]
- 17.Lim L.P., Lau N.C., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 18.Smirnova L., Gräfe A., Seiler A., Schumacher S., Nitsch R., Wulczyn F.G. Regulation of miRNA expression during neural cell specification. Eur. J. Neurosci. 2005;21:1469–1477. doi: 10.1111/j.1460-9568.2005.03978.x. [DOI] [PubMed] [Google Scholar]
- 19.Lagos-Quintana M., Rauhut R., Yalcin A., Meyer J., Lendeckel W., Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 20.Colin A., Faideau M., Dufour N., Auregan G., Hassig R., Andrieu T., Brouillet E., Hantraye P., Bonvento G., Déglon N. Engineered lentiviral vector targeting astrocytes in vivo. Glia. 2009;57:667–679. doi: 10.1002/glia.20795. [DOI] [PubMed] [Google Scholar]
- 21.Maddalena A., Tereshchenko J., Bähr M., Kügler S. Adeno-associated virus-mediated, mifepristone-regulated transgene expression in the brain. Mol. Ther. Nucleic Acids. 2013;2:e106. doi: 10.1038/mtna.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.von Jonquieres G., Mersmann N., Klugmann C.B., Harasta A.E., Lutz B., Teahan O., Housley G.D., Fröhlich D., Krämer-Albers E.M., Klugmann M. Glial promoter selectivity following AAV-delivery to the immature brain. PLoS ONE. 2013;8:e65646. doi: 10.1371/journal.pone.0065646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kügler S. Tissue-specific promoters in the CNS. Methods Mol. Biol. 2016;1382:81–91. doi: 10.1007/978-1-4939-3271-9_6. [DOI] [PubMed] [Google Scholar]
- 24.Howard D.B., Powers K., Wang Y., Harvey B.K. Tropism and toxicity of adeno-associated viral vector serotypes 1, 2, 5, 6, 7, 8, and 9 in rat neurons and glia in vitro. Virology. 2008;372:24–34. doi: 10.1016/j.virol.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koerber J.T., Klimczak R., Jang J.H., Dalkara D., Flannery J.G., Schaffer D.V. Molecular evolution of adeno-associated virus for enhanced glial gene delivery. Mol. Ther. 2009;17:2088–2095. doi: 10.1038/mt.2009.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hutson T.H., Verhaagen J., Yáñez-Muñoz R.J., Moon L.D. Corticospinal tract transduction: a comparison of seven adeno-associated viral vector serotypes and a non-integrating lentiviral vector. Gene Ther. 2012;19:49–60. doi: 10.1038/gt.2011.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Åkerblom M., Sachdeva R., Barde I., Verp S., Gentner B., Trono D., Jakobsson J. MicroRNA-124 is a subventricular zone neuronal fate determinant. J. Neurosci. 2012;32:8879–8889. doi: 10.1523/JNEUROSCI.0558-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davidson B.L., Stein C.S., Heth J.A., Martins I., Kotin R.M., Derksen T.A., Zabner J., Ghodsi A., Chiorini J.A. Recombinant adeno-associated virus type 2, 4, and 5 vectors: transduction of variant cell types and regions in the mammalian central nervous system. Proc. Natl. Acad. Sci. USA. 2000;97:3428–3432. doi: 10.1073/pnas.050581197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tenenbaum L., Chtarto A., Lehtonen E., Velu T., Brotchi J., Levivier M. Recombinant AAV-mediated gene delivery to the central nervous system. J. Gene Med. 2004;6(Suppl 1):S212–S222. doi: 10.1002/jgm.506. [DOI] [PubMed] [Google Scholar]
- 30.Shevtsova Z., Malik J.M., Michel U., Bähr M., Kügler S. Promoters and serotypes: targeting of adeno-associated virus vectors for gene transfer in the rat central nervous system in vitro and in vivo. Exp. Physiol. 2005;90:53–59. doi: 10.1113/expphysiol.2004.028159. [DOI] [PubMed] [Google Scholar]
- 31.Xie Y., Wang T., Sun G.Y., Ding S. Specific disruption of astrocytic Ca2+ signaling pathway in vivo by adeno-associated viral transduction. Neuroscience. 2010;170:992–1003. doi: 10.1016/j.neuroscience.2010.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haberman R.P., McCown T.J., Samulski R.J. Novel transcriptional regulatory signals in the adeno-associated virus terminal repeat A/D junction element. J. Virol. 2000;74:8732–8739. doi: 10.1128/jvi.74.18.8732-8739.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peel A.L., Klein R.L. Adeno-associated virus vectors: activity and applications in the CNS. J. Neurosci. Methods. 2000;98:95–104. doi: 10.1016/s0165-0270(00)00183-7. [DOI] [PubMed] [Google Scholar]
- 34.Clark K.R., Penaud-Budloo M. Evaluation of the fate of rAAV genomes following in vivo administration. Methods Mol. Biol. 2011;807:239–258. doi: 10.1007/978-1-61779-370-7_10. [DOI] [PubMed] [Google Scholar]
- 35.Brenner M., Kisseberth W.C., Su Y., Besnard F., Messing A. GFAP promoter directs astrocyte-specific expression in transgenic mice. J. Neurosci. 1994;14:1030–1037. doi: 10.1523/JNEUROSCI.14-03-01030.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mamber C., Verhaagen J., Hol E.M. In vivo targeting of subventricular zone astrocytes. Prog. Neurobiol. 2010;92:19–32. doi: 10.1016/j.pneurobio.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 37.Feng X., Eide F.F., Jiang H., Reder A.T. Adeno-associated viral vector-mediated ApoE expression in Alzheimer’s disease mice: low CNS immune response, long-term expression, and astrocyte specificity. Front. Biosci. 2004;9:1540–1546. doi: 10.2741/1323. [DOI] [PubMed] [Google Scholar]
- 38.de Leeuw B., Su M., ter Horst M., Iwata S., Rodijk M., Hoeben R.C., Messing A., Smitt P.S., Brenner M. Increased glia-specific transgene expression with glial fibrillary acidic protein promoters containing multiple enhancer elements. J. Neurosci. Res. 2006;83:744–753. doi: 10.1002/jnr.20776. [DOI] [PubMed] [Google Scholar]
- 39.Geisler A., Fechner H. MicroRNA-regulated viral vectors for gene therapy. World J. Exp. Med. 2016;6:37–54. doi: 10.5493/wjem.v6.i2.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poy M.N., Eliasson L., Krutzfeldt J., Kuwajima S., Ma X., Macdonald P.E., Pfeffer S., Tuschl T., Rajewsky N., Rorsman P., Stoffel M. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 41.Fazi F., Rosa A., Fatica A., Gelmetti V., De Marchis M.L., Nervi C., Bozzoni I. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell. 2005;123:819–831. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 42.Brown B.D., Gentner B., Cantore A., Colleoni S., Amendola M., Zingale A., Baccarini A., Lazzari G., Galli C., Naldini L. Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nat. Biotechnol. 2007;25:1457–1467. doi: 10.1038/nbt1372. [DOI] [PubMed] [Google Scholar]
- 43.Geisler A., Jungmann A., Kurreck J., Poller W., Katus H.A., Vetter R., Fechner H., Müller O.J. microRNA122-regulated transgene expression increases specificity of cardiac gene transfer upon intravenous delivery of AAV9 vectors. Gene Ther. 2011;18:199–209. doi: 10.1038/gt.2010.141. [DOI] [PubMed] [Google Scholar]
- 44.Karali M., Manfredi A., Puppo A., Marrocco E., Gargiulo A., Allocca M., Corte M.D., Rossi S., Giunti M., Bacci M.L. MicroRNA-restricted transgene expression in the retina. PLoS ONE. 2011;6:e22166. doi: 10.1371/journal.pone.0022166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cao X., Pfaff S.L., Gage F.H. A functional study of miR-124 in the developing neural tube. Genes Dev. 2007;21:531–536. doi: 10.1101/gad.1519207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mullokandov G., Baccarini A., Ruzo A., Jayaprakash A.D., Tung N., Israelow B., Evans M.J., Sachidanandam R., Brown B.D. High-throughput assessment of microRNA activity and function using microRNA sensor and decoy libraries. Nat. Methods. 2012;9:840–846. doi: 10.1038/nmeth.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shimizu K., Sakurai F., Tomita K., Nagamoto Y., Nakamura S., Katayama K., Tachibana M., Kawabata K., Mizuguchi H. Suppression of leaky expression of adenovirus genes by insertion of microRNA-targeted sequences in the replication-incompetent adenovirus vector genome. Mol. Ther. Methods Clin. Dev. 2014;1:14035. doi: 10.1038/mtm.2014.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilusz C.J., Wormington M., Peltz S.W. The cap-to-tail guide to mRNA turnover. Nat. Rev. Mol. Cell Biol. 2001;2:237–246. doi: 10.1038/35067025. [DOI] [PubMed] [Google Scholar]
- 49.Dong J.Y., Fan P.D., Frizzell R.A. Quantitative analysis of the packaging capacity of recombinant adeno-associated virus. Hum. Gene Ther. 1996;7:2101–2112. doi: 10.1089/hum.1996.7.17-2101. [DOI] [PubMed] [Google Scholar]
- 50.Wu L., Belasco J.G. Micro-RNA regulation of the mammalian lin-28 gene during neuronal differentiation of embryonal carcinoma cells. Mol. Cell. Biol. 2005;25:9198–9208. doi: 10.1128/MCB.25.21.9198-9208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taschenberger G., Toloe J., Tereshchenko J., Akerboom J., Wales P., Benz R., Becker S., Outeiro T.F., Looger L.L., Bähr M. β-synuclein aggregates and induces neurodegeneration in dopaminergic neurons. Ann. Neurol. 2013;74:109–118. doi: 10.1002/ana.23905. [DOI] [PubMed] [Google Scholar]
- 52.West M.J., Slomianka L., Gundersen H.J. Unbiased stereological estimation of the total number of neurons in thesubdivisions of the rat hippocampus using the optical fractionator. Anat. Rec. 1991;231:482–497. doi: 10.1002/ar.1092310411. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.