Abstract
The paramyxo- and pneumoviruses are members of the order Mononegavirales, a group of viruses with non-segmented, negative strand RNA genomes. The polymerases of these viruses are multi-functional complexes, capable of transcribing subgenomic capped and polyadenylated mRNAs and replicating the genome. Although there is no native structure available for any complete paramyxo- or pneumovirus polymerase, functional and structural studies of a fragment of a pneumovirus polymerase protein and mutation analyses and resistance profiling of small-molecule inhibitors have generated a wealth of mechanistic information. This review integrates these data with the structure of a related polymerase, identifying similarities, differences, gaps in knowledge, and avenues for antiviral drug development.
Keywords: Paramyxovirus, pneumovirus, Mononegavirales, polymerase, capping, inhibitor
Paramyxo- and pneumovirus taxonomy and genome organization
The families Paramyxoviridae and Pneumoviridae are members of the order Mononegavirales, the non-segmented, negative strand RNA viruses (nsNSVs). Until 2016, they were classified as two subfamilies within the same family, but distinctions between them, and similarities between the pneumoviruses and filoviruses (another family within the nsNSVs) warranted their separation (1). The paramyxovirus and pneumovirus families collectively include a number of major human pathogens, as well as viruses infecting other mammals, birds, fish and reptiles. The paramyxovirus family consists of seven genera. The best-studied representative of this family in terms of polymerase activity is Sendai virus, but some information is also available for the parainfluenza viruses (PIV) 1–5, measles, rinderpest, mumps, and Newcastle disease viruses. The pneumovirus family includes two genera, with respiratory syncytial virus (RSV) and human metapneumovirus (HMPV) being the most extensively investigated representatives of each.
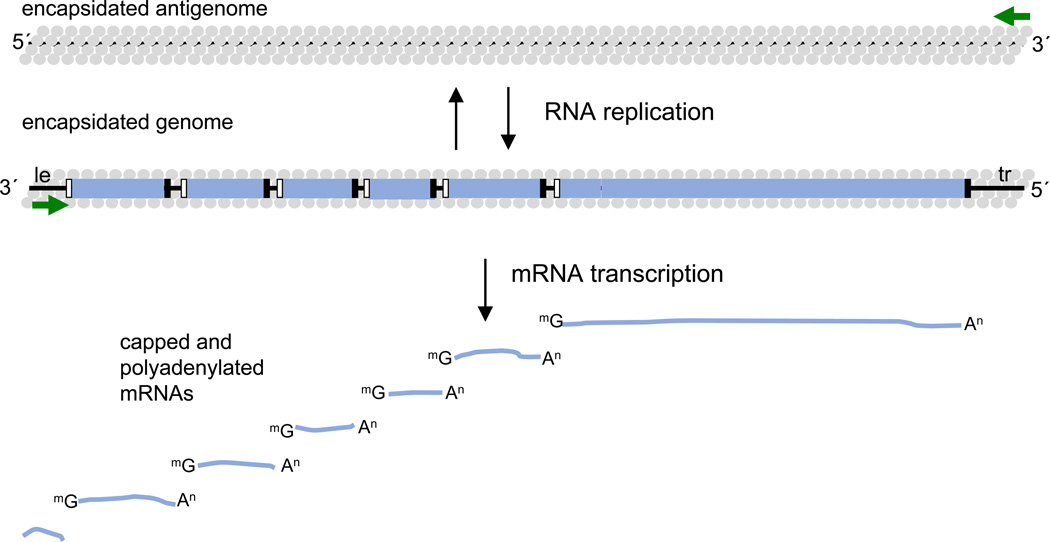
The paramyxo- and pneumoviruses differ in the number of genes they possess, but their genome structures and mechanisms of gene expression and genome replication are very similar (Figure 1) (2). Their genomes are approximately 15 kb in length and encode between six and ten mRNAs. The genes are arranged in the same orientation along the genome, and are delineated with conserved elements, known as gene start and gene end sequences, which flank each of the genes. At the 3´ and 5´ ends of the genome are short extragenic regions, a 3´ leader region, of ~ 50 nt and a 5´ trailer region, of variable length. The genome is transcribed into subgenomic, capped and polyadenylated mRNAs that correspond to each gene, and replicated to yield a genomic-length, positive sense antigenome, which in turn acts as a template for genome RNA synthesis. The accidental generation of bi- and multicistronic mRNAs was demonstrated, but para- and pneumoviruses lack a mechanism to translate downstream open reading frames of multicistronic mRNAs. Transcription and replication take place in the cytoplasm of the host cell and are performed by the viral polymerase complex without involvement of nuclear enzymes. Transcription is initiated at a single promoter within the 3´ leader region. The polymerase generates subgenomic mRNAs by responding to the gene start and gene end signals on the genome: at a gene start signal it initiates RNA synthesis, it then modifies the 5´ end of the RNA to add a methylated cap, and elongates the mRNA and when it reaches the gene end signal it polyadenylates and releases it. Having released the transcript, the polymerase by default remains attached to the template and scans to locate the next gene start signal, where it reinitiates RNA synthesis (Figure 1). To replicate the genome, the polymerase again initiates at the leader promoter, but in this case it is highly processive and disregards the gene junction signals to generate a positive sense antigenome RNA. The antigenome subsequently acts as a template for the polymerase to synthesize progeny genome RNAs. The replication products differ from mRNAs by being encapsidated along their length with nucleoprotein (N) as they are synthesized. Thus the RNA template for the polymerase is encased within N protein at all times of infection, and in fact only encapsidated genomic and antigenomic RNA can serve as template for the polymerase complex (Figure 1).
Figure 1. Schematic diagram showing the typical paramyxovirus or pneumovirus genome organization.
The genes are shown as blue rectangles with gene start and gene end signals indicated with black and white rectangles, respectively. The leader (le) and trailer (tr) regions at the 3´ and 5´ ends of the genome, respectively, are indicated. The promoters at the 3´ ends of the genome and antigenome are shown with green arrows. The products of transcription and genome replication are represented below and above the genome, respectively. Transcription yields a short uncapped and non-polyadenylated RNA complementary to the leader, and mRNAs with 5´ caps and 3´ poly A tails, corresponding to each of the genes. Replication yields antigenome and genome RNAs that are encapsidated with N protein (grey circles).
Paramyxo- and pneumovirus polymerase structure and enzymatic activities
Overview of the nsNSV polymerase structure
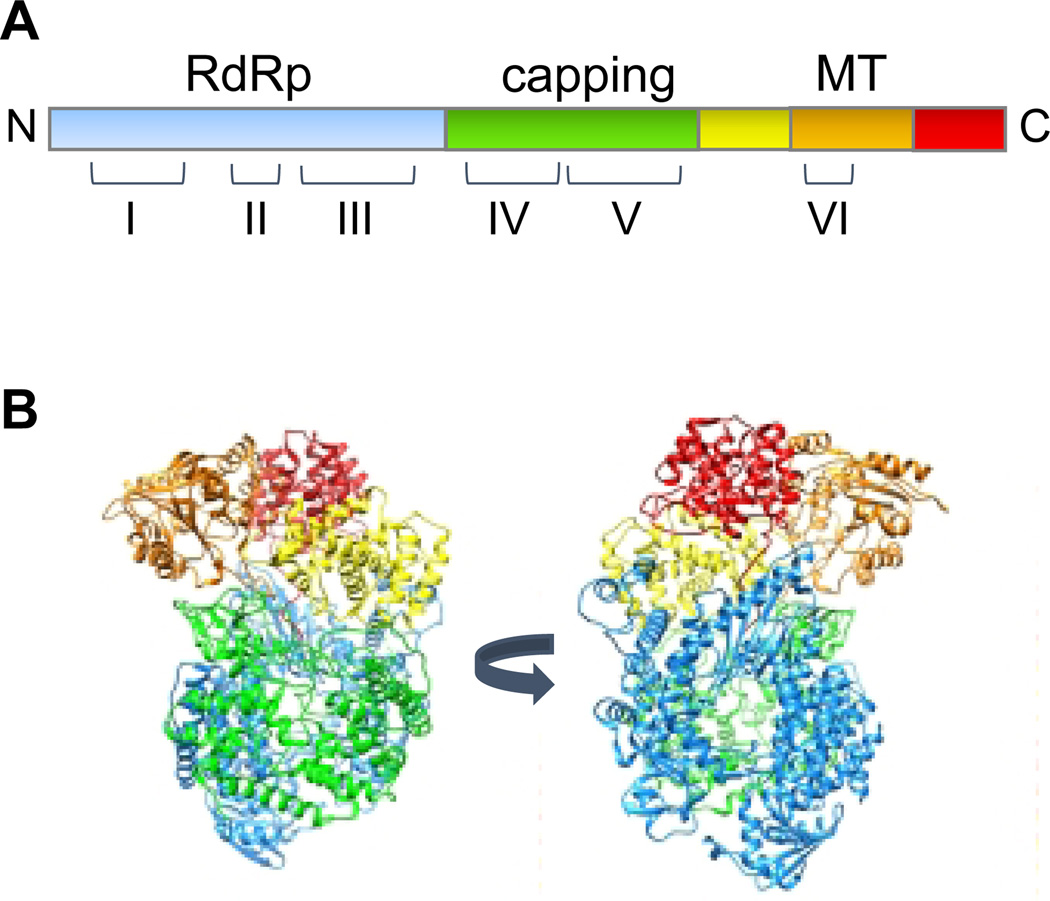
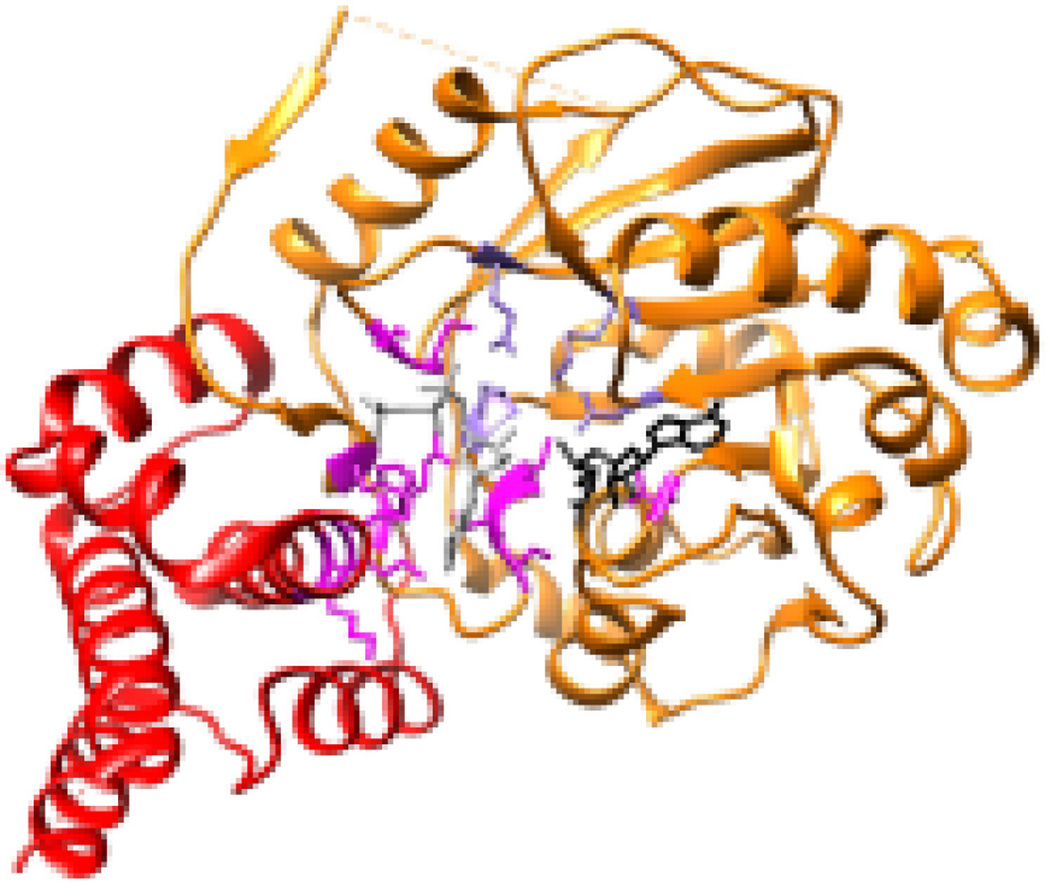
The polymerase of the paramyxo- and pneumoviruses is a multi-functional complex, with RNA dependent RNA polymerase (RdRp) activity, and enzymatic activities that add and methylate the mRNA caps. It consists of a complex of two proteins, the large polymerase subunit (L) and a phosphoprotein (P) (3, 4). The L proteins contain the catalytic domains required for RNA synthesis, cap addition, and cap methylation. They are over 2,000 amino acids in length and approximately 250 kDa in molecular weight. The P proteins are co-factors that assist polymerase expression and association with the nucleocapsid template (3, 5–7). The P proteins are highly divergent in size and sequence, whereas the L proteins are more conserved. There is no high-resolution structure available for a complete L protein of any paramyxo- or pneumovirus. However, in 2015, the structure of almost the entire L protein of another nsNSV, vesicular stomatitis virus (VSV) was solved in complex with a fragment of P by cryo-EM to 3.8 Angstrom, allowing construction of a near-atomic model of the protein (8). The VSV L structure is shown in Figure 2. It resembles a doughnut-ring in shape, which is comprised of the N-terminal portion of the protein, associated with globular appendages that are formed by the C-terminal portion. Specific functions could be ascribed to the VSV L domains by alignment with other enzymes and detailed mutational analysis. The doughnut-ring (shown in blue) is responsible for RNA polymerization. It resembles a right-hand structure with a palm and connecting finger and thumb sub-domains, similar to that of other polymerases (8, 9). This domain has two channels, which are likely to be the template and NTP entry channels, based on analogy with other RdRps (8, 10). The domain responsible for adding the guanosine cap is shown in green. In the reconstructed conformation of the L protein, which likely represents a pre-initiation state, the capping domain is folded over the polymerization domain creating a cage-like structure, and a loop protrudes into the polymerization active site. By analogy with other RdRps, this loop is likely a priming loop that functions to prevent hairpin formation of the template and position the initiating NTP to enable de novo initiation of RNA synthesis (11–15). The methyltransferase domain (shown in orange) is adjacent to the capping domain. Because the latter is folded over the polymerization domain, there is no obvious exit channel for the nascent strand, leading to the conclusion that there must be a significant reorganization of the polymerase that swings the capping domain away from the polymerization domain and probably into closer proximity to the methyltransferase domain. This is supported by negative stain electron microscopy studies that showed that the lobes can adopt different conformations (8, 16). The remaining globular domains, a connector, and C-terminal domains may be involved in structural reconfiguration and methyltransferase function.
Figure 2. Structure of an nsNSV polymerase.
(A) Linear representation of the VSV L protein, indicating the positions of conserved regions I-VI relative to the different domains. (B) Views of the VSV L protein structure (PDB 5a22) (8). The domains identified by Liang et al are as follows: RdRP (blue), capping (green), connector domain (yellow), methyltransferase domain (orange) and C-terminal domain (red).
Relationship of the VSV L protein structure with those of the paramyxo and pneumoviruses
Although there are no complete L structures available for the paramyxo- and pneumoviruses, all nsNSV L proteins share six regions of strong amino acid similarity, known as conserved regions I to VI (17), that correspond to regions within the RdRp, capping, and methyltransferase domains (Figure 2). This shared overall organization allows meaningful alignment of the paramyxo- and pneumovirus L proteins with that of VSV. The Sendai virus L protein, in particular, has been subjected to extensive mutation analysis, using a variety of assays to examine P protein and template binding, leader RNA synthesis, mRNA transcription and genome replication (18–24). Additional information is available from more focused mutation analysis of other viruses, and from experiments with a small molecule inhibitor of RSV capping (25). Thus, the VSV L structure provides a framework for integrating this information and drawing conclusions regarding the structure-function relationships of paramyxo- and pneumovirus polymerases. In addition, the structure of a C-terminal fragment of the HMPV L protein in the presence or absence of substrate has been resolved by X-ray crystallography and analyzed by mutation analysis (26), which provides further information regarding the spatial organization of the methyltransferase and C-terminal domain that helps inform our understanding of the paramyxo- and pneumovirus polymerases.
RNA synthesis
RNA synthesis is believed to follow the “two-metal-ion” mechanism of catalysis proposed by Steitz (27, 28). This is indicated by the fact that conserved regions II and III of nsNSV L proteins possess a set of conserved motifs (A to D) found in all RdRps (9, 29). The functions of these motifs has been reviewed extensively previously (29). Motif A is involved in Mg2+ coordination and possibly sugar selection. In poliovirus it is a β-turn, α-helix followed by a longer α-helix. This motif contains a conserved aspartate residue that is thought to play a role in coordinating the divalent cation during nucleotidyl transfer. Motif B is thought to be involved in selection of ribose versus deoxyribose. In HIV reverse transcriptase, this motif is a β-strand, loop, α-helix. Motif C is a β-strand, turn, β-strand that contains a GDN motif, analogous to GDD found in other polymerases, at the turn. The aspartate residue in motif C is in close proximity to the aspartate of motif A and is thought to be involved in coordinating the second divalent cation required for catalysis. Motif D is part of the palm core. The locations of these motifs in the VSV L protein are indicated in gold in Figure 3A. Substitutions in residues flanking the aspartate and aromatic residue of motif A and the loop and α-helix of motif B inhibited all Sendai virus RNA synthesis (24, 30), and several studies with paramyxo- and pneumovirus polymerases have confirmed that the GDN motif is essential for phosphodiester bond formation (31–35). The priming loop has not been subjected to mutation analysis, but substitutions in the adjacent residues also inhibited all Sendai virus RNA synthesis activity (19). However, MeV and canine distemper virus adaptation to growth in the presence of a pan-morbillivirus polymerase inhibitor has yielded substitutions in these typically fully sequence conserved adjacent residues that did not affect polymerase function in cell culture but mediated robust viral escape from inhibition (36, 37). In the VSV L protein structure, the N-terminal domain of the protein forms a buttress to the polymerization domain. This region of the protein is overall less well conserved than that of the remainder of the polymerization domain. For many paramyxoviruses, the N-terminal region of L has also been shown to interact with the respective P protein (18, 38–43)(44), and as the P proteins are highly variable, this might account for the relatively low level of conservation in this part of L. Nonetheless, the N terminal domain contains some stretches of amino acid similarity and identity which make up conserved region I (17), which lies within the polymerization cavity and adjacent to the template entry channel. Substitutions close to this region have been shown to inhibit all Sendai virus RNA synthesis as have substitutions in a loop in conserved region II, which also flanks this channel (18, 24) (Figure 3A and B). These data indicate that the template entry channel and the core RNA polymerization domain are probably highly similar between Sendai virus and VSV, and thus many aspects of this structure are likely shared by all paramyxo- and pneumoviruses.
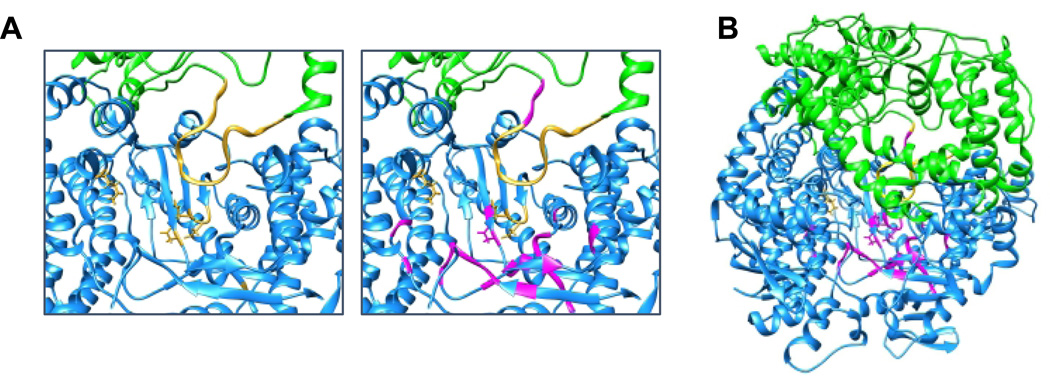
Figure 3. The RdRp domain.
(A) In the left panel, the RdRp active site within the VSV L protein is shown, with the motifs A-D and the putative priming loop colored gold. The right panel shows the same image with residues that correspond to inhibitory substitutions in Sendai virus L shown in magenta. (B) A wider view of the image shown in panel A, from the perspective of looking through the template entry site. Corresponding amino acids were made by aligning Sendai and measles virus L protein sequences and referring to the alignment presented by Liang et al (8).
Cap addition and methylation
The capping mechanism of VSV has been shown to occur by a mechanism that is biochemically distinct from the conventional capping mechanism used by eukaryotes, DNA, and dsRNA viruses (45). It occurs through a RNA:GDP polyribonucleotidyltransferase (PRNTase), rather than guanylyltransferase activity. Beginning with GTP and triphosphorylated RNA substrates, the polymerase converts GTP to GDP through a guanosine 5´ triphosphatase activity, and a histidine within the capping domain becomes covalently bound to mono-phosphorylated RNA (pRNA). Nucleophilic attack of the 5´ α-phosphate of pRNA by the β-phosphate of GDP results in formation and release of capped GpppRNA from the L protein (45–48). The VSV cap is methylated at the guanine N-7 and ribose 2´-O positions using S-adenosyl methionine as a substrate. In VSV, this occurs through a distinctive mechanism, in which the ribose 2´-O methylation precedes that of guanine N-7 (49–51). The final product is an mRNA with a cap 1 structure, equivalent to that of cellular mRNAs. The motifs on the VSV polymerase required for cap addition and methylation are well defined. Cap addition depends on a number of residues within conserved region V, which cluster close to the histidine required to generate the covalent attachment to RNA (8, 52, 53). While we have a good understanding of which specific residues are involved in the different steps of cap formation (53), to date no single amino acid substitution has been identified that completely inhibits the GTPase activity. Both methylation reactions involve motifs in conserved region VI, a single S-adenosyl methionine binding site, conferred by a GxGxG motif (49), and methylase activity conferred by a K-D-K-E catalytic tetrad (54). In the three-dimensional structure, these motifs are in close proximity to each other and the domain has strong similarity with other enzymes that perform similar reactions (8, 55, 56).
Relatively little is known regarding the capping reaction in paramyxo- and pneumoviruses. In large part this is due to the lack of a trans-capping assay, used to study VSV PRNTase activity in isolation from other polymerase activities, for any paramyxo- or pneumovirus polymerase. Regarding paramyxovirus capping activity, one paper presents data suggesting alternatively that rinderpest virus L protein forms phosphoamide bond with GMP, consistent with a host cell-like guanylyltransferase (rather than PRNTase) mechanism for capping (57). It remains puzzling, however, that residues essential for the PRNTase reaction in VSV are conserved in all paramyxoviruses including rinderpest virus (52, 53). Furthermore, substitutions in conserved region V of Sendai virus in a stretch of amino acids immediately N-terminal to a conserved glycine, shown in VSV to be required for formation of the L-pRNA intermediate (53), inhibit transcription without blocking replication (19), which is consistent with the notion that residues in this region are involved a role in capping (Figure 4). In the case of the pneumoviruses, it has been shown that RSV polymerase transfers GDP, rather than GMP, to the 5´ end of the RNA, consistent with the polymerase having a PRNTase activity (58). Confirmation that capping activity is located within the same domain in RSV as in VSV comes from work with small molecule inhibitors of RSV. A series of compounds was identified that inhibited production of full-length mRNAs, but instead resulted in accumulation of short ~50 nt RNAs containing a 5´ triphosphate, most likely generated as a consequence of capping failure (25). Resistance to these inhibitors mapped to three residues surrounding essential capping motifs (8, 25). Thus, the preponderance of data is consistent with the paramyxo- and pneumoviruses having a PRNTase activity similar to that of VSV and other rhabdoviruses, but more extensive analysis is required to fully characterize their capping domains and the reactions that they perform.
Figure 4. The capping domain.
The capping domain of the VSV L protein is shown with residues essential for capping activity indicated in gold. Residues that correspond to positions of capping inhibitor resistance mutations are shown in red; residues that correspond to substitutions that inhibit Sendai virus transcription, but not replication, are shown in magenta.
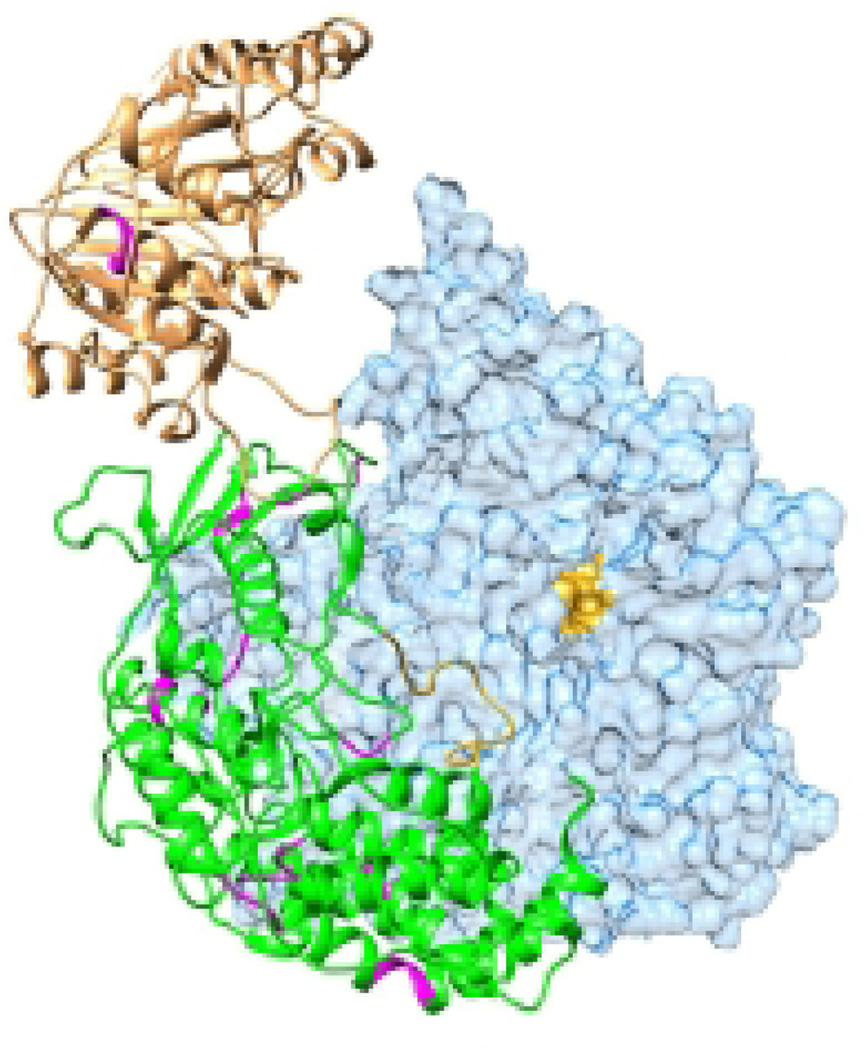
The methyltransferase activity of the paramyxo- and pneumoviruses is much better characterized. The methylation reactions are performed by conserved region VI of the paramyxo- and pneumoviruses, which contain the GxGxG and K-D-K-E motifs. L fragments containing conserved region VI of Sendai virus, rinderpest virus and HMPV are able to methylate RNAs provided in trans (26, 59) (60), and substitutions in conserved region VI of Sendai virus inhibit cap methylation (20, 22, 23). The structure of the C-terminus of the HMPV L protein, containing conserved region VI and the C-terminal domain was recently resolved to 2.2 Angstroms in the presence and absence of S-adenosyl methionine and NTPs, and the enzymatic properties of this region were analyzed using biochemical assays (26). The crystal structure predicted that the C-terminal domain of HMPV L folds over the active site cleft of the methyltransferase domain in conserved region VI and creates a substrate binding pocket that presumably accommodates the nucleotide that is being methylated (Figure 5). This region of the C-terminal domain contains a K-K-G motif, that is conserved in paramyxo- and pneumoviruses and has been shown to be required for mRNA production by parainfluenza virus type 2 (42). This motif positions the nucleotide close to the K-D-K-E catalytic motif. A single S-adenosyl methionine binding pocket was identified capable of adopting an open or closed conformation, which would allow uptake of the S-adenosyl methionine substrate and release of the S-adenosyl homocysteine by-product for both methylation reactions. Finally, this fragment of the HMPV L protein was shown to have NTPase activity, which might contribute to the elusive GTPase activity required for cap addition, but so far the exact site that performs this activity is not known. Although the active sites for methylation are similar between the viruses, there are some differences compared to VSV. Whereas methylation events by VSV polymerase are obligatorily sequential (50), this does not seem to apply to some paramyxo- and pneumoviruses. For example, Newcastle disease virus and RSV mRNAs have been described that lack 2´ O methylation (58, 61), the Sendai virus L fragment described above performed guanosine N7 methylation exclusively (59), and although the fragment containing wild type HMPV L sequence performed 2´ O and N7 methylations in sequential order, some variants were more inhibited in 2´ O methylation than N7, in toto indicating that this order is not obligatory (26). In further contrast to VSV L, the HMPV L fragment could methylate uncapped RNA (26). Finally, the HMPV structure contains a hydrophobic nucleoside binding pocket that is not apparent in the VSV L structure (26). Although the function of this pocket is not known at present, it was speculated that it may accommodate the guanosine cap of the RNA to allow methylation at the 2´ O ribose site. However, substitutions in this pocket did not interfere with either N7 or 2´ O methylation and it is not well conserved beyond the pneumoviruses.
Figure 5. The methyltransferase domain.
The methyltransferase and C-terminal domain of the HMPV L protein (PDB 4UCI) is shown, with domains colored as in Figure 2. Residues that are required for methyltransferase activity are shown in magenta with the K-D-K-E residues in a lighter shade. S-adenosyl methionine is shown in black, and GTP (which represents the RNA substrate) in grey.
Integrating polymerase structure with different events in transcription and replication
Promoter recognition and access by the polymerase
In the paramyxoviruses and pneumoviruses, both transcription and replication are initiated at the 3´ promoter, but the organization of the N-RNA template and the promoters are distinct (62). As noted above, the template for the polymerase is not naked RNA, but rather is encased along its length with nucleoprotein (N). Therefore, before the polymerase can form a complex with the promoter RNA, it needs to locate the promoter and displace the N protein to unveil the RNA and allow it to enter the template channel. The mechanism by which promoter recognition occurs is apparently different for the paramyxo- versus pneumoviruses. The paramyxovirus promoters are bipartite, with one element located at the 3´ end of the leader region and the second internally between positions 77 and 96, depending on the virus (63–69). The two promoter elements must be in the correct phase relative to the N protein surrounding them to be functional, and so they must be positioned appropriately relative to one another. Mapping of the two elements onto the N-RNA structure indicates that they are positioned on the same face of the helix (66, 67, 70). Together, these data suggest that both promoter elements are contacted simultaneously during initial interactions of the polymerase with the template. Whether polymerase loading onto the RNP involves specific interactions between L and/or P and/or other factors and the exposed RNA bases is not known. However, the initial contact between the polymerase complex and the bipartite promoter, rather than the 3´ end of the N-RNA, appears to be the key factor that allows the paramyxovirus polymerase to locate its binding site in the promoter, since studies with Sendai virus have shown that the bipartite promoter does not need to be at the 3´ end of the RNA to be functional. Provided that the promoter is intact, it can even be recognized even if it is displaced 100 nucleotides from the 3´ terminus by heterologous sequence (70), although the exact mechanism by which this occurs is not known.
In contrast to the bipartite promoter of the paramyxoviruses, the pneumovirus promoter is a monopartite promoter in which all the signals required for RNA replication are contained within the first 36 nt of the template RNA (71). Also in contrast to the paramyxoviruses, there is no requirement for the pneumovirus RNA to be in a particular phase with respect to N protein (72), and whereas the paramyxovirus promoter shows some tolerance to be recognized when located internally, the pneumovirus promoter can only be placed up to 6 additional nucleotides downstream from the 3´ end (73). Thus, the 3´ end of the pneumovirus template appears to play a more restrictive role in aiding the polymerase to locate the promoter, and the N protein, while present and almost certainly important, seems less critical for positioning the bases of the promoter to contact with the polymerase.
Structures of a variety of nsNSV N proteins were solved and all share an overall organization of the core structure into N-terminal (NTD) and C-terminal (CTD) folding domains (74–78). When assembled into the corresponding polymeric nucleocapsids, a linear alignment creates a groove between the string of parallel core domains that harbors the genomic or antigenomic RNA. Having contacted the promoter, the polymerase complex therefore needs to unveil the RNA in order to route it through the template channel of the RdRp domain. While the molecular mechanism of RNA release is not fully understood yet, alternative hypotheses have been proposed recently. Based on insight gained from the crystal structure of RSV N, a hinge-like movement was suggested upon binding of the RSV P protein to the N-protein NTD, resulting in reorientation of the CTD and opening of the RNA groove (76). This model is supported by the recent comparison of parainfluenza virus 5 (PIV5)-derived N ring assemblies (78) with monomeric N of the related Nipah virus (79), which revealed a more open conformation between the NTD and CTD in monomeric N than in the PIV5 N assembly. These distinct N conformations were thought to resemble different stages of CTD rotation relative to NTD, allowing RNA encapsidation and release (78). Alternatively, a cryo-EM reconstruction of mumps virus nucleocapsids suggested that a more conservative local conformational change induced by interaction of the polymerase complex with an NTD α-helix adjacent to the RNA groove is sufficient to release the RNA (80), eliminating the request of the first model for deep-seated conformational changes in the highly stable N protein core when assembled in the nucleocapsid. Independent of the mechanism of local RNA release from the nucleocapsid, however, the N protein chain must be routed over the surface of the P-L polymerase complex while the RNA strand is directed through the template channel (81), followed by reuniting of the RNA strand and N protein core as the polymerase advances along the template.
Initiation of transcription and replication
The 3´ promoter elements of the paramyxo- and pneumoviruses show similarity to one another, being highly pyrimidine-rich, particularly with uracil residues, in each case (Figure 6). However, there are differences in initiation site selection and the mechanism of initiation. Studies with RSV have shown that two initiation sites are present in the promoter: a replication initiation site at position 1U and a transcription initiation site at position 3C (62, 82). The promoter sequence of HMPV is similar to that of RSV, suggesting that it follows a similar initiation mechanism. How the polymerase accurately initiates from two sites on the promoter is not clear, but experiments with templates containing substitutions or a deletion at position 1 of the promoter indicated that the initiating ATP, and possibly the second NTP, CTP, is selected independently of the template (83, 84). These findings suggest the possibility that initiation with GTP at position 3 is the default, and that initiation at position 1 occurs when the polymerase is preloaded with ATP and CTP.
Figure 6. Comparison of paramyxo- and pneumovirus promoters.
The sequences at the 3´ end of the representative paramyxo- and pneumovirus leader promoters are shown, with uracil residues colored in red. The sequences are aligned with the likely initiation site in the paramyxovirus promoters (position 1) opposite the dominant initiation site in the pneumovirus promoters (position 3) to illustrate the alignment of uracil residues in most of the promoters. Abbreviations are Atlantic salmon paramyxovirus (ASPV), Newcastle disease virus (NDV), Fer de Lance virus (FDLV), measles virus (MeV), and mumps virus (MuV).
In VSV, RNA synthesis initiation occurs opposite position 1 of the promoter only, and, unlike the situation with RSV, this initiation opposite position 1 is templated (85). Whether the paramyxoviruses have two initiation sites within the promoter, similar to RSV, or a single initiation site at position 1, similar to VSV, is not known. However, promoter sequences are more consistent with a model in which the only initiation site on the promoter is at position 1, since RdRps have a preference to initiate with purine (29). Because paramyxoviruses feature G residues at positions 2, 3 and in some cases position 4 of their promoters (Figure 6), it is unlikely that the polymerase initiates opposite any of these positions. Furthermore, primer extension analysis of abortive RNAs from the Sendai virus promoter failed to detect an internal initiation site (86). These and other data support that in the paramyxoviruses, both transcription and RNA replication are initiated from position 1 (62). However, a conserved feature between the paramyxovirus and pneumovirus families is polymerase initiation from position 1 with pppApC, suggesting that ATP and CTP might play an important role in positioning the 3´ end of the template appropriately for 3´ terminal initiation (83).
Also the interactions between the template, incoming NTPs, and polymerase are not well understood for any nsNSV, but a model can be proposed based on the promoter sequences discussed above. Inspection of VSV, pneumovirus, and paramyxovirus promoters suggests alignment of the uridine tract if the pneumovirus sequences overhang by two nucleotides at the 3´ end (Figure 6). Thus, we propose that the uridine (or pyrimidine) tract plays an identical role in paramyxo- and pneumoviruses, namely positioning the RNA to locate the polymerase active site opposite positions 3 and 4 in case of the pneumoviruses, and positions 1 and 2 in case of the paramyxoviruses. In the case of the pneumoviruses, the polymerase could recruit GTP and initiate directly opposite position 3 if ATP and CTP are not bound, or ratchet back towards the 3´ end should they be bound. For the paramyxoviruses, ATP and CTP might help stabilize interactions to allow the polymerase to initiate opposite position 1 with high efficiency.
As noted above, a key polymerase determinant in initiation of de novo RNA synthesis is a priming loop, which in the VSV polymerase structure appears to protrude from the capping domain into the polymerization active site (8). Priming loops typically contain an aromatic residue that forms stacking interactions with the incoming NTP (11, 12, 15). In influenza virus, a conserved aromatic residue was shown to be required for de novo initiation of replication, but not primer mediated initiation of transcription (15). There is little primary sequence conservation of the region of the paramyxo- and pneumovirus L proteins that corresponds to the VSV priming loop, but in the pneumoviruses and most (although not all) paramyxoviruses a conserved tyrosine residue, corresponding to a tryptophan in VSV, is positioned at the tip of the priming loop and close to the polymerization active site. This residue presents as a strong candidate for priming loop activity since an aromatic side chain is required, although it remains unclear what the counterpart is in some paramyxoviruses. It is also unclear how the pneumovirus priming loop manages to enable two different initiation events.
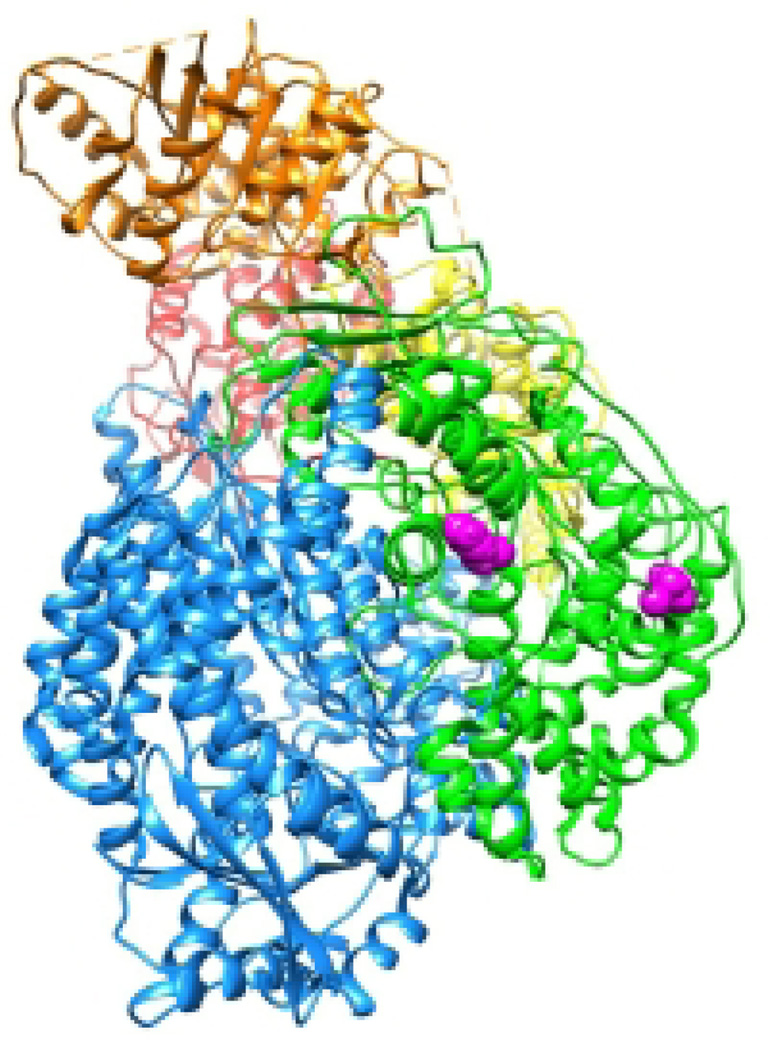
Having initiated RNA synthesis, the polymerase must undergo significant reorganization, creating exit channels for the template and nascent RNA transcript (8). This reorganization likely opens a channel for the RNA template, such that it exits on the same face at it enters to allow easy repackaging into the nucleocapsid structure, as is the case for bunyavirus, another virus with an encapsidated RNA genome (10, 87). The transcript RNA presumably exits following movement of the capping domain. Substitutions on the exterior of the Sendai virus capping domain (in conserved regions IV and V) and in the methyltransferase domain inhibit RNA synthesis without affecting L protein accumulation or P binding (19, 20)(23). As these regions of the polymerase are distant from the RNA synthesis active site (Figure 7) and would not be expected to be directly involved in RNA synthesis, these substitutions might be preventing the domains from undergoing the necessary structural rearrangement required for continued RNA synthesis. This hypothesis is consistent with the fact that some Sendai virus L variants had a temperature sensitive phenotype, suggesting altered protein organization or interactions (20).
Figure 7. Substitutions that may inhibit polymerase rearrangement.
The VSV L protein with RdRP, capping and methyltransferase domains is shown. The RdRp is in pale blue with the active site (viewed through the template entry channel) and putative priming loop colored gold. The capping domain is colored green and the methyltransferase in orange. Residues corresponding to substitutions that inhibit both transcription and replication in Sendai virus, that are distant from the RdRp active site are shown in magenta.
Transcription reinitiation at the gene start signals
After initiation of RNA synthesis at the 3´ promoter element, the polymerase is non-processive, unless in replicase mode, and releases the RNA transcript stochastically, usually before the end of the leader region (82, 86, 88–91). Having released the RNA, the polymerase scans the template to locate the gene start signal for the first gene and reinitiates RNA synthesis, again using a de novo initiation mechanism (82, 86). Re-initiation also occurs at each of the remaining gene start signals on the genome. Since these signals share strong similarity throughout each viral genome, the mechanism of re-initiation is likely to be identical at each gene (2, 82, 86). However, the re-initiation events differ from the inaugural initiation at the 3´ end of the template, since during gene start re-initiation the template is threaded through both the entry and exit channels. As noted above, there is no obvious template exit channel in the existing VSV polymerase structure, and so we can only speculate as to the structure of the polymerase during re-initiation. The structural differences between the polymerase initiating at the promoter versus the gene start signals might only be slight, as release of the initial transcript might allow the capping domain and priming loop to refold back into place on the polymerization domain. Notably, the gene start signals of the paramyxo- and pneumoviruses show strong similarities to their promoters (62) and the initiating NTP is the same in the gene start signals as at the promoter: ATP for the paramyxoviruses, and GTP (the NTP required for position 3 initiation within the leader) for the pneumoviruses. These observations suggest that the polymerase makes similar contacts with the gene start signal and initiating NTP as with the promoter, except that perhaps the changes incurred by the formation of the template exit channel result in slightly different optimal initiation sequences at the promoter versus the internal gene start signals.
Capping, methylation and elongation of the mRNA
Having re-initiated RNA synthesis at the gene start signal, the polymerase caps and methylates the nascent mRNA as it is being extruded from the active site. The proximity of the capping domain to the polymerization active site could allow an RNA exit channel to be formed that positions the 5´ end of the RNA in the capping active site. In VSV, capping and methylation occur within 31 nt (92) and capping is necessary for the RNA to be efficiently elongated beyond 40 nt (93)(94). This restriction also appears to apply to RSV, based on studies with the putative capping inhibitor described above, which caused accumulation of abortive transcripts approximately 40–50 nt in length (25). In VSV, the sequence at the 5´ end of the mRNA transcripts is important for efficient capping and methylation (95)(50). Similarly, fragments of the Sendai virus and HMPV L proteins could methylate a capped RNA in trans, but required a specific RNA sequence for this activity (26, 59), which suggests specific interactions between the 5´ end of the nascent mRNA transcript and the polymerase. The differences between the gene start and promoter sequences could account for why mRNA are elongated, capped, and methylated, whereas RNAs synthesized from the promoter are not. When substitutions were made in the RSV leader promoter to increase its similarity to a gene start signal, the RNA was elongated more efficiently (82). It is not known, however, whether this more efficient elongation was due to cap addition or to the sequence favoring polymerase processivity. Once the polymerase has capped the mRNA, it can continue elongation until the end of the gene. In the P/V gene of the paramyxoviruses, the polymerase can perform transcriptional editing to yield mRNA transcripts containing additional inserted nucleotides (and thus altered open reading frames), to expand the coding capacity of the gene (96–99). This is thought to occur through a mechanism of controlled polymerase backsliding triggered by a sequence in the RNA template (99, 100).
Polyadenylation and release of the mRNA
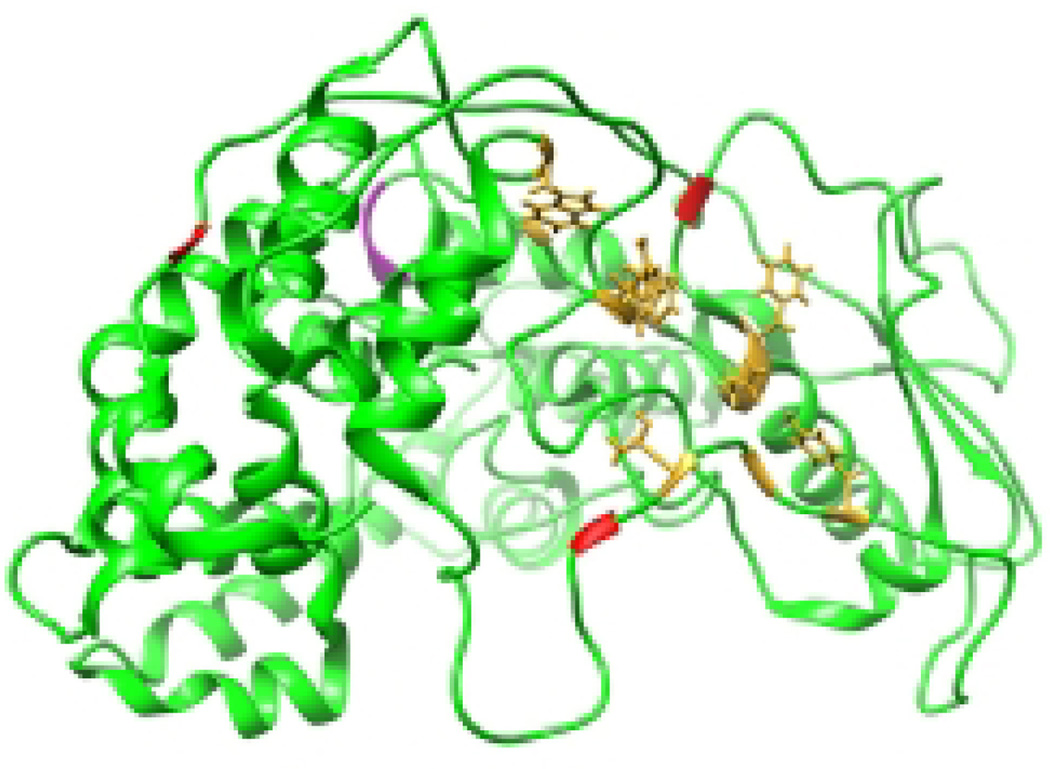
When the polymerase reaches a gene end signal, it polyadenylates and releases the mRNA. The gene end signals for the paramyxo- and pneumoviruses are 10–13 nt in length. Most notably, they contain a U-tract of 4 to 5 nucleotides, and it is believed that reiterative stuttering on the U tract creates the ~ 300–400 nt poly A tail through non-templated backsliding resembling in principle the mRNA editing process. The mRNA is released from the polymerase following polyadenylation by an unknown mechanism. Little is known about the interactions between the polymerase, the RNA template, and the transcript that enable reiterative stuttering and mRNA release. The stuttering occurs through repeated slippage of the transcript relative to the template within the polymerase active site, but it is not clear what precipitates mRNA release after 300–400 adenylate additions. Two variants of the RSV polymerase have been described that show a reduced ability to recognize the gene end signal and instead create multi-cistronic mRNAs (101, 102). Mapping of these amino acid substitutions onto the VSV L protein indicates that they lie close to one another on the exterior of the capping domain, suggesting that this domain might also be involved in mRNA release (Figure 8).
Figure 8. Substitutions that inhibit transcription termination at the gene end signal.
The VSV L protein is shown, colored as in Figure 2. Residues corresponding to substitutions in RSV L protein that inhibit mRNA release at a gene end signal are shown as magenta spheres.
Negotiation of the intergenic region
The nature of the intergenic regions located between the gene end signal of an upstream gene and the gene start signal of the next gene varies between paramyxo- and pneumovirueses. In the former, they are either a highly conserved trinucleotide or variable in length and sequence (2). In the latter, they are variable and in the case of RSV, one of the gene junctions is even overlapping, such that the gene start signal of the downstream gene lies upstream of the gene end signal for the preceding gene (103). Having released the mRNA of the upstream gene, the polymerase scans the gene junctions to locate the next gene start signal (104, 105). As noted above, this can reasonably only occur with the template RNA continuously threaded through the polymerase active site and the N proteins encapsidated the template being displaced as the polymerase moves along, since the polymerase is not able to initiate at internal gene start signals independently of initiation at the 3´ leader promoter. Interestingly, in some paramyxoviruses the gene start signals must be in correct phase relative to the N protein (81). A possible explanation put forward suggests that the polymerase might receive signals from the N proteins being displaced in addition to the sequence motifs within the RNA (81).
Encapsidation and commitment to replication
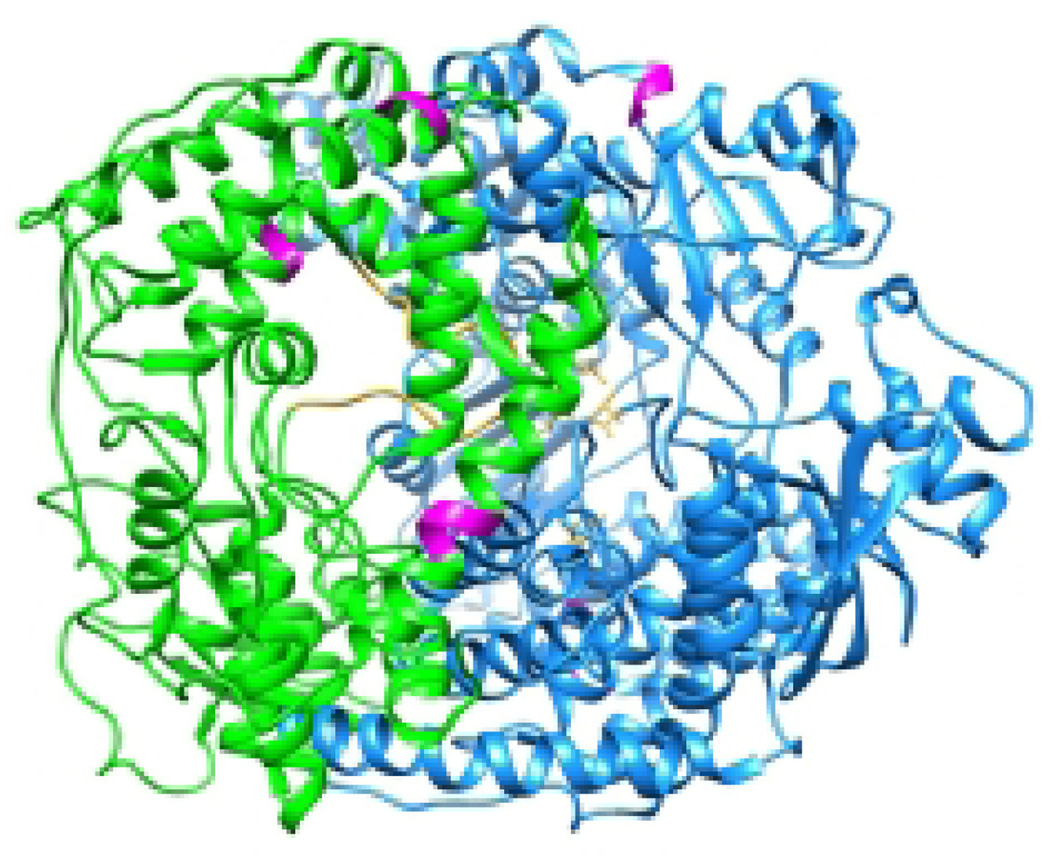
The ability of the polymerase to be fully processive in replicase mode and to continue through the leader, the first gene, and all remaining gene end signals without triggering polyadenylation and strand release depends on concurrent encapsidation of the replication product with N protein (71, 86, 106). N is maintained in soluble monomeric form through interaction with P in N0-P complexes (107). As P also interacts with L, it is thought that this dual binding specificity is instrumental for the nascent RNA to become associated with N protein as it is being synthesized (107). Biochemically studies have indicated that the L and N proteins do not interact directly with each other. However, the Moyer group identified several amino acid substitutions in the Sendai virus L protein that inhibited replication without significantly inhibiting transcription (20, 24). Mapping of these residues onto the VSV L protein structure shows that four of the five sets of mutated residues lie on the same face of the L protein, within conserved regions II and IV, and thus span the capping and polymerization domains (Figure 9). Conceivably, these residues are located close to the transcript exit site, and it is therefore tantalizing to speculate that this face of the polymerase might represent an N or N-P binding site, guiding correct delivery of N proteins to the nascent replication product for co-transcriptional encapsidation.
Figure 9. Substitutions that inhibit RNA replication specifically.
The VSV L protein is shown, colored as in Figure 2. Residues corresponding to substitutions in Sendai virus L protein that inhibit replication, but not transcription, are shown in magenta.
Antiviral strategies targeting pneumo- and paramyxovirus polymerase activity
Therapeutic targeting of the pneumo- and paramyxovirus polymerase machinery has emerged as a promising antiviral approach for four major reasons: i) each of the different enzymatic functions carried out by the polymerase complex is essential for successful virus replication; ii) because the polymerase complexes act both as replicase and transcriptase of the viral genome, interference with polymerase activity not only affects the synthesis of progeny viral genomes but also reduces the expression of viral proteins counteracting the host antiviral response, which are among the first and often most abundantly expressed after infection. This two-pronged effect has high potential for antiviral synergies, since a virus population with pharmaceutically retarded growth rate will encounter a potent host innate immune response that cannot be contained through interfering viral proteins (37); iii) while several enzymatic activities carried out by the polymerase complex such as mRNA capping, methylation, and polyadenylation are also found in mammalian cells, the structural organization of the host cell protein complexes involved and the viral machinery are starkly distinct as discussed above, reducing the risk of undesirable cross-inhibition; and iv) further contributing to a low potential for off-target effects, mammalian cells lack signature activities of the viral polymerase complex, RNA synthesis based on RNA templates and PRNTase activity for mRNA capping as opposed to host cell guanylyltransferase activity.
Multiple catalytic centers present in the L protein and a number of essential protein-protein interactions between viral proteins such as L and P and P-L and the encapsidated RNP template and viral polymerase components and mandatory host co-factors such as the human translation elongation factor eEF1A (108) provide a multitude of candidate druggable targets for small-molecule inhibition. A frequent problem especially of allosteric polymerase blockers is a narrow indication spectrum, however, restricting potent antiviral activity to a specific family member or pathogens of a single genus within the family (109, 110).
Host-directed antiviral therapy, targeting host factors required for virus replication rather than viral proteins directly, has enjoyed a renaissance in the past decade due to its promise to broaden the indication range, since host factors such as effectors of cellular signaling pathways (111, 112) or polymerase cofactors such as eEF1A (113, 114) are recruited by a variety of viral pathogens. However, the benefits of host-directed antivirals – the expanded indication range and, in addition, a lowered frequency of viral escape from inhibition – are typically offset by unacceptable side effects resulting from pharmaceutical interference with key cellular components. While it may be possible to generate pathogen target-specificity, for instance through blockage of host factor-viral protein-protein interfaces (PPI), small-molecule inhibition of PPI formation is challenging per se (115) and a major advantage of the host-directed approach, expanding the indication spectrum, would likely be lost through this strategy. We therefore believe that the clinical application of host-directed antivirals may likely remain limited to some very specific indications such as chemokine co-receptor targeting for antiretroviral therapy (116). However, widespread use for the treatment of seasonal virus infections may remain elusive, especially when considering a predominantly pediatric patient population as envisioned for anti-pneumo- and anti-paramyxovirus therapeutics (110, 117). Rather, allosteric and competitive inhibitors of the polymerase complex hold in our opinion the highest promise to best combine two key elements of the desirable drug profile, potent antiviral activity and high tolerability, despite the likely restriction to a narrow indication range. Combination therapies, for instance consisting of a nucleoside analog and an allosteric polymerase inhibitor, may address possible resistance issues experienced with virus-directed monotherapies and even allow to further reduce dose levels of each individual drug if synergistic effects are achieved.
In the following, we will discuss strategies and druggable target candidates for pneumo- and paramyxovirus polymerase inhibition by example of a panel of RSV polymerase blockers that have completed advanced development and were tested in clinical trials.
Inhibitors of RSV polymerase activity
Nucleoside analogs
The clinical potential of nucleoside analogs that feature atypical bases and compete with natural nucleotides for incorporation into the nascent strand is best illustrated by their game-changing role in modern antiretroviral therapy (118). In fact, the current standard of care for treatment of inexperienced HIV patients still consists of a backbone of two distinct nucleoside analogs that are combined with a non-nucleoside reverse transcriptase blocker or a protease or integrase inhibitor (119). The canonical mechanism of inhibition of nucleoside analogs is through chain termination after intracellular conversion to the corresponding nucleotide through the cellular phosphorylation machinery and incorporation by the advancing polymerase (120–123).
Unlike the structurally diverse spectrum of distinct nucleoside inhibitors available for antiretroviral therapy, only one nucleoside variant, ribavirin, is licensed for the treatment of RSV infection to date and a second substrate analog, ALS-8176, was advanced to phase II clinical trials. Of these, only ALS-8176 acts as a chain terminator, while the mechanism of antiviral activity of ribavirin, showing a broadened indication spectrum that spans in addition to RSV a variety of paramyxovirus family members, hepatitis C virus and others, is still controversially discussed (124). Chain termination was proposed (125, 126) but potency of ribavirin against hepatitis C polymerase is low in vitro, strongly arguing against direct inhibition of the polymerase complex (127, 128). According to an alternative hypothesis, the compound may rather permit continued polymerization after incorporation into a newly synthesized strand, but subsequently supports equally efficient base-pairing with either cytosine or uracil (129). The resulting hypermutagenesis of the viral genome ultimately interrupts the replication cycle through error catastrophe (130, 131). Independent of the molecular mechanism of activity, however, the drug was essentially abandoned for RSV treatment due to limited efficacy combined with the induction of strong adverse effects. In particular hemolytic anemia is a hallmark for ribavirin toxicity, since the compound is efficiently phosphorylated to the nucleotide form, but cannot be reconverted to the inactive and membrane-permeable nucleoside in erythrocytes, due to lack of the required phosphatase activity (132). As a consequence, ribavirin accumulates to intracellular concentrations of up to 100-fold that of serum levels in red blood cells.
The second nucleoside analog currently under consideration for RSV treatment, ALS-8176, was well tolerated in a phase II clinical trial and significantly reduced RSV burden when adult volunteers were treated at the onset of infection (133). In contrast to the unclear mechanism of action of ribavirin, cytidine analogs of the ALS-8176 class immediately blocked polymerase activity in in vitro assays of RSV RdRp function, consistent with chain termination as the underlying molecular basis for inhibition (134). Resistance profiles of ALS-8176 were determined experimentally through viral adaptation and identified four point mutations in the L protein (M628L, A789V, L795I, and I796V) that in combination mediate robust escape from inhibition (134). Three of these substitutions are positioned in immediate linear proximity to the highly conserved GDN motif (residues 810 to 813) that is considered to form the catalytic center for phosphodiester bond formation (35, 135). This finding was underscored by localization of the residues in a 3D-homology model of RSV L that was generated based on the electron density maps released for related VSV L (8). This model positioned all four residues as lining the substrate channel walls in close proximity to the GDN catalytic center (136). Remarkably, residues in the corresponding microdomain were implicated in mediating morbillivirus resistance to inhibition by a novel class of allosteric RdRp inhibitors (36, 37), spotlighting the pneumo- and paramyxovirus polymerase substrate entry channel as a promising druggable target not only for nucleoside analog blockers but equally allosteric inhibitors. It remains to be evaluated whether allosteric inhibitors binding to this target plug the channel and thus prevent substrate entry into the catalytic center, and whether analogs of the morbillivirus inhibitor class can be engineered with high affinity for the RSV L target.
Non-nucleoside RSV polymerase inhibitors in clinical trials
To date, only one structural class of allosteric RSV RdRp inhibitors was advanced to clinical testing. The clinical candidate of this class, RSV604 (137), is orally bioavailable and reduced viral load and disease symptoms in some stem cell transplant recipients with RSV disease, but only in those patients in which drug plasma concentrations reached EC90 concentrations (138). Mechanistic characterization revealed that the compound blocks de novo synthesis of viral RNA but does not interfere with synthesis from assembled viral ribonucleocapsid complexes (139), arguing against direct inhibition of the viral P-L polymerase complex. This finding was consistent with resistance profiling of RSV604, which identified escape mutations exclusively in the viral N protein (137), and label-free positive target identification studies through surface plasmon resonance that suggested direct docking of the compound to the N protein (139). Mapping of resistance sites (N105D, K107N, I129L, and L139I) onto a crystal structure of RSV N (76) positioned all resistance sites into a poorly sequence conserved β-hairpin region and adjacent α-helix located at the distal end of the NTD. This end was suggested to serve as a docking site for the P-L polymerase complex to the assembled N-RNA, supposedly allowing polymerase access to the RNA template by inducing the hinge movement of the CTD relative to the NTD upon P-L binding as discussed above as a possible mechanism for template RNA unveiling (76). Subsequently solved co-crystal structures of the N NTD domain with a C-terminal portion of the viral P protein revealed that the α-helix carrying resistance residues 129 and 139 forms part of a hydrophobic pocket that transiently accommodates the C-terminal most phenylalanine residue of the P protein, as the P-L polymerase complex advances along the ribonucleoprotein template (140).
While this P-N interface represents an attractive target for small-molecule inhibition due to the transient nature of the interaction and the low binding affinity (KD values are in the 50 µM range), direct competition of RSV604 with P docking into the pocket is unlikely, considering that the compound only prevents de novo RNP assembly but not transcription from preassembled N:RNA complexes (137). A possible explanation may be that the drug binding site is only accessible in N monomers, but not after incorporation of N into nucleocapsids, thus demanding preloading of viral genomes with the drug during replication and N-RNA assembly for efficient inhibition. Alternatively, although buoyed by a structurally attractive hypothesis, we lack at present experimental confirmation that the identified escape mutations mediate primary resistance to RSV604. The actual drug binding site could be physically distinct from the NTD region carrying the resistance mutations and escape be mediated through long-range structural effects. Independent of the molecular mechanisms of inhibition and viral escape, however, RSV604 has established fundamental proof-of-concept for the feasibility of successful therapeutic targeting of an nsNSV N protein.
We believe that the recent advances in the structural and mechanistic understanding of the pneumo- and paramyxovirus polymerase complex provides groundbreaking new opportunity for the development of much needed next generation antivirals. For instance, an appropriately designed drug screening campaign can directly target the low-affinity P-N:RNA interface, having high potential for successful structure-informed hit discovery.
HIGHLIGHTS.
Paramyxoviruses and pneumoviruses are non-segmented negative strand RNA viruses
Their polymerases can transcribe and replicate the viral genome
Residues essential for different steps in these processes have been identified
These residues can be mapped onto the structure of a related polymerase
Small molecule inhibitors that target the polymerase complex are described
Acknowledgments
We thank Molly Braun for assistance preparing the figures in this article. This work was supported, in part, by funding from The Hartwell Foundation and public health service grant AI113321 from NIH/ NIAID (to R.F.) and public health service grants AI011002 and HD079327 from the NIH/NIAID and NIH/NICHD (to R.K.P.).
ABBREVIATIONS
- nsNSV
non-segmented negative strand RNA virus
- RSV
respiratory syncytial virus
- HMPV
human metapneumovirus
- VSV
vesicular stomatitis virus
- PIV
parainfluenza virus
- MeV
measles virus
- L
large polymerase subunit
- P
phosphoprotein
- N
nucleoprotein
- RdRp
RNA dependent RNA polymerase
- PRNTase
RNA:GDP polyribonucleotidyl transferase
- NTD
N-terminal domain
- CTD
C-terminal domain
Footnotes
Conflicts of Interest
R.F.’s research is supported in part by sponsored research agreements with Alios Biopharma and Merck. R.K.P. is an inventor on US patent 8729059 “Paramyxovirus Family Inhibitors and Methods of Use Thereof”. This study could affect his personal financial status.
REFERENCES
- 1.Afonso CL, Amarasinghe GK, Banyai K, Bao Y, Basler CF, Bavari S, Bejerman N, Blasdell KR, Briand FX, Briese T, Bukreyev A, Calisher CH, Chandran K, Cheng J, Clawson AN, Collins PL, Dietzgen RG, Dolnik O, Domier LL, Durrwald R, Dye JM, Easton AJ, Ebihara H, Farkas SL, Freitas-Astua J, Formenty P, Fouchier RA, Fu Y, Ghedin E, Goodin MM, Hewson R, Horie M, Hyndman TH, Jiang D, Kitajima EW, Kobinger GP, Kondo H, Kurath G, Lamb RA, Lenardon S, Leroy EM, Li CX, Lin XD, Liu L, Longdon B, Marton S, Maisner A, Muhlberger E, Netesov SV, Nowotny N, Patterson JL, Payne SL, Paweska JT, Randall RE, Rima BK, Rota P, Rubbenstroth D, Schwemmle M, Shi M, Smither SJ, Stenglein MD, Stone DM, Takada A, Terregino C, Tesh RB, Tian JH, Tomonaga K, Tordo N, Towner JS, Vasilakis N, Verbeek M, Volchkov VE, Wahl-Jensen V, Walsh JA, Walker PJ, Wang D, Wang LF, Wetzel T, Whitfield AE, Xie JT, Yuen KY, Zhang YZ, Kuhn JH. Taxonomy of the order Mononegavirales: update 2016. Archives of virology. 2016;161:2351–2360. doi: 10.1007/s00705-016-2880-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lamb RA, Parks GD. Paramyxoviridae: the viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. Fifth. Lippincott Williams and Wilkins; pp. 1449–1496. [Google Scholar]
- 3.Horikami SM, Curran J, Kolakofsky D, Moyer SA. Complexes of Sendai virus NP-P and P-L proteins are required for defective interfering particle genome replication in vitro. J Virol. 1992;66:4901–4908. doi: 10.1128/jvi.66.8.4901-4908.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mazumder B, Adhikary G, Barik S. Bacterial expression of human respiratory syncytial viral phosphoprotein P and identification of Ser237 as the site of phosphorylation by cellular casein kinase II. Virology. 1994;205:93–103. doi: 10.1006/viro.1994.1623. [DOI] [PubMed] [Google Scholar]
- 5.Bloyet LM, Welsch J, Enchery F, Mathieu C, de Breyne S, Horvat B, Grigorov B, Gerlier D. HSP90 Chaperoning in Addition to Phosphoprotein Required for Folding but Not for Supporting Enzymatic Activities of Measles and Nipah Virus L Polymerases. J Virol. 2016;90:6642–6656. doi: 10.1128/JVI.00602-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bousse T, Takimoto T, Matrosovich T, Portner A. Two regions of the P protein are required to be active with the L protein for human parainfluenza virus type 1 RNA polymerase activity. Virology. 2001;283:306–314. doi: 10.1006/viro.2001.0881. [DOI] [PubMed] [Google Scholar]
- 7.Sourimant J, Rameix-Welti MA, Gaillard AL, Chevret D, Galloux M, Gault E, Eleouet JF. Fine mapping and characterization of the L-polymerase-binding domain of the respiratory syncytial virus phosphoprotein. J Virol. 2015;89:4421–4433. doi: 10.1128/JVI.03619-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liang B, Li Z, Jenni S, Rahmeh AA, Morin BM, Grant T, Grigorieff N, Harrison SC, Whelan SP. Structure of the L Protein of Vesicular Stomatitis Virus from Electron Cryomicroscopy. Cell. 2015;162:314–327. doi: 10.1016/j.cell.2015.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poch O, Sauvaget I, Delarue M, Tordo N. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. Embo J. 1989;8:3867–3874. doi: 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reguera J, Gerlach P, Cusack S. Towards a structural understanding of RNA synthesis by negative strand RNA viral polymerases. Current opinion in structural biology. 2016;36:75–84. doi: 10.1016/j.sbi.2016.01.002. [DOI] [PubMed] [Google Scholar]
- 11.Appleby TC, Perry JK, Murakami E, Barauskas O, Feng J, Cho A, Fox D, 3rd, Wetmore DR, McGrath ME, Ray AS, Sofia MJ, Swaminathan S, Edwards TE. Viral replication. Structural basis for RNA replication by the hepatitis C virus polymerase. Science. 2015;347:771–775. doi: 10.1126/science.1259210. [DOI] [PubMed] [Google Scholar]
- 12.Butcher SJ, Grimes JM, Makeyev EV, Bamford DH, Stuart DI. A mechanism for initiating RNA-dependent RNA polymerization. Nature. 2001;410:235–240. doi: 10.1038/35065653. [DOI] [PubMed] [Google Scholar]
- 13.Hong Z, Cameron CE, Walker MP, Castro C, Yao N, Lau JY, Zhong W. A novel mechanism to ensure terminal initiation by hepatitis C virus NS5B polymerase. Virology. 2001;285:6–11. doi: 10.1006/viro.2001.0948. [DOI] [PubMed] [Google Scholar]
- 14.Mosley RT, Edwards TE, Murakami E, Lam AM, Grice RL, Du J, Sofia MJ, Furman PA, Otto MJ. Structure of hepatitis C virus polymerase in complex with primer-template RNA. J Virol. 2012;86:6503–6511. doi: 10.1128/JVI.00386-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Te Velthuis AJ, Robb NC, Kapanidis AN, Fodor E. The role of the priming loop in influenza A virus RNA synthesis. Nat Microbiol. 2016;1:16029. doi: 10.1038/nmicrobiol.2016.29. [DOI] [PubMed] [Google Scholar]
- 16.Rahmeh AA, Schenk AD, Danek EI, Kranzusch PJ, Liang B, Walz T, Whelan SP. Molecular architecture of the vesicular stomatitis virus RNA polymerase. Proc Natl Acad Sci U S A. 2010;107:20075–20080. doi: 10.1073/pnas.1013559107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poch O, Blumberg BM, Bougueleret L, Tordo N. Sequence comparison of five polymerases (L proteins) of unsegmented negative-strand RNA viruses: theoretical assignment of functional domains. J Gen Virol. 1990;71(Pt 5):1153–1162. doi: 10.1099/0022-1317-71-5-1153. [DOI] [PubMed] [Google Scholar]
- 18.Chandrika R, Horikami SM, Smallwood S, Moyer SA. Mutations in conserved domain I of the Sendai virus L polymerase protein uncouple transcription and replication. Virology. 1995;213:352–363. doi: 10.1006/viro.1995.0008. [DOI] [PubMed] [Google Scholar]
- 19.Cortese CK, Feller JA, Moyer SA. Mutations in domain V of the Sendai virus L polymerase protein uncouple transcription and replication and differentially affect replication in vitro and in vivo. Virology. 2000;277:387–396. doi: 10.1006/viro.2000.0615. [DOI] [PubMed] [Google Scholar]
- 20.Feller JA, Smallwood S, Horikami SM, Moyer SA. Mutations in conserved domains IV and VI of the large (L) subunit of the sendai virus RNA polymerase give a spectrum of defective RNA synthesis phenotypes. Virology. 2000;269:426–439. doi: 10.1006/viro.2000.0234. [DOI] [PubMed] [Google Scholar]
- 21.Horikami SM, Moyer SA. Alternative amino acids at a single site in the Sendai virus L protein produce multiple defects in RNA synthesis in vitro. Virology. 1995;211:577–582. doi: 10.1006/viro.1995.1440. [DOI] [PubMed] [Google Scholar]
- 22.Murphy AM, Grdzelishvili VZ. Identification of sendai virus L protein amino acid residues affecting viral mRNA cap methylation. J Virol. 2009;83:1669–1681. doi: 10.1128/JVI.01438-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murphy AM, Moerdyk-Schauwecker M, Mushegian A, Grdzelishvili VZ. Sequence-function analysis of the Sendai virus L protein domain VI. Virology. 2010;405:370–382. doi: 10.1016/j.virol.2010.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smallwood S, Easson CD, Feller JA, Horikami SM, Moyer SA. Mutations in conserved domain II of the large (L) subunit of the Sendai virus RNA polymerase abolish RNA synthesis. Virology. 1999;262:375–383. doi: 10.1006/viro.1999.9933. [DOI] [PubMed] [Google Scholar]
- 25.Liuzzi M, Mason SW, Cartier M, Lawetz C, McCollum RS, Dansereau N, Bolger G, Lapeyre N, Gaudette Y, Lagace L, Massariol MJ, Do F, Whitehead P, Lamarre L, Scouten E, Bordeleau J, Landry S, Rancourt J, Fazal G, Simoneau B. Inhibitors of respiratory syncytial virus replication target cotranscriptional mRNA guanylylation by viral RNA-dependent RNA polymerase. J Virol. 2005;79:13105–13115. doi: 10.1128/JVI.79.20.13105-13115.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paesen GC, Collet A, Sallamand C, Debart F, Vasseur JJ, Canard B, Decroly E, Grimes JM. X-ray structure and activities of an essential Mononegavirales L-protein domain. Nature communications. 2015;6:8749. doi: 10.1038/ncomms9749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steitz TA. A mechanism for all polymerases. Nature. 1998;391:231–232. doi: 10.1038/34542. [DOI] [PubMed] [Google Scholar]
- 28.Steitz TA, Steitz JA. A general two-metal-ion mechanism for catalytic RNA. Proc Natl Acad Sci U S A. 1993;90:6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Reilly EK, Kao CC. Analysis of RNA-dependent RNA polymerase structure and function as guided by known polymerase structures and computer predictions of secondary structure. Virology. 1998;252:287–303. doi: 10.1006/viro.1998.9463. [DOI] [PubMed] [Google Scholar]
- 30.Smallwood S, Hovel T, Neubert WJ, Moyer SA. Different substitutions at conserved amino acids in domains II and III in the Sendai L RNA polymerase protein inactivate viral RNA synthesis. Virology. 2002;304:135–145. doi: 10.1006/viro.2002.1644. [DOI] [PubMed] [Google Scholar]
- 31.Chattopadhyay A, Raha T, Shaila MS. Effect of single amino acid mutations in the conserved GDNQ motif of L protein of Rinderpest virus on RNA synthesis in vitro and in vivo. Virus Res. 2004;99:139–145. doi: 10.1016/j.virusres.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 32.Fix J, Galloux M, Blondot ML, Eleouet JF. The insertion of fluorescent proteins in a variable region of respiratory syncytial virus L polymerase results in fluorescent and functional enzymes but with reduced activities. The open virology journal. 2011;5:103–108. doi: 10.2174/1874357901105010103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Magoffin DE, Halpin K, Rota PA, Wang LF. Effects of single amino acid substitutions at the E residue in the conserved GDNE motif of the Nipah virus polymerase (L) protein. Arch Virol. 2007;152:827–832. doi: 10.1007/s00705-006-0881-1. [DOI] [PubMed] [Google Scholar]
- 34.Noton SL, Deflube LR, Tremaglio CZ, Fearns R. The respiratory syncytial virus polymerase has multiple RNA synthesis activities at the promoter. PLoS pathogens. 2012;8:e1002980. doi: 10.1371/journal.ppat.1002980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sleat DE, Banerjee AK. Transcriptional activity and mutational analysis of recombinant vesicular stomatitis virus RNA polymerase. J Virol. 1993;67:1334–1339. doi: 10.1128/jvi.67.3.1334-1339.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoon JJ, Krumm SA, Ndungu JM, Hoffman V, Bankamp B, Rota PA, Sun A, Snyder JP, Plemper RK. Target analysis of the experimental measles therapeutic AS-136A. Antimicrob Agents Chemother. 2009;53:3860–3870. doi: 10.1128/AAC.00503-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krumm SA, Yan D, Hovingh ES, Evers TJ, Enkirch T, Reddy GP, Sun A, Saindane MT, Arrendale RF, Painter G, Liotta DC, Natchus MG, von Messling V, Plemper RK. An orally available, small-molecule polymerase inhibitor shows efficacy against a lethal morbillivirus infection in a large animal model. Sci Transl Med. 2014;6 doi: 10.1126/scitranslmed.3008517. 232ra252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cevik B, Holmes DE, Vrotsos E, Feller JA, Smallwood S, Moyer SA. The phosphoprotein (P) and L binding sites reside in the N-terminus of the L subunit of the measles virus RNA polymerase. Virology. 2004;327:297–306. doi: 10.1016/j.virol.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 39.Chattopadhyay A, Shaila MS. Rinderpest virus RNA polymerase subunits: mapping of mutual interacting domains on the large protein L and phosphoprotein p. Virus Genes. 2004;28:169–178. doi: 10.1023/B:VIRU.0000016855.25662.95. [DOI] [PubMed] [Google Scholar]
- 40.Holmes DE, Moyer SA. The phosphoprotein (P) binding site resides in the N terminus of the L polymerase subunit of sendai virus. J Virol. 2002;76:3078–3083. doi: 10.1128/JVI.76.6.3078-3083.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Horikami SM, Smallwood S, Bankamp B, Moyer SA. An amino-proximal domain of the L protein binds to the P protein in the measles virus RNA polymerase complex. Virology. 1994;205:540–545. doi: 10.1006/viro.1994.1676. [DOI] [PubMed] [Google Scholar]
- 42.Nishio M, Tsurudome M, Garcin D, Komada H, Ito M, Le Mercier P, Nosaka T, Kolakofsky D. Human parainfluenza virus type 2 L protein regions required for interaction with other viral proteins and mRNA capping. J Virol. 2011;85:725–732. doi: 10.1128/JVI.01226-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parks GD. Mapping of a region of the paramyxovirus L protein required for the formation of a stable complex with the viral phosphoprotein P. J Virol. 1994;68:4862–4872. doi: 10.1128/jvi.68.8.4862-4872.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smallwood S, Moyer SA. The L polymerase protein of parainfluenza virus 3 forms an oligomer and can interact with the heterologous Sendai virus L, P and C proteins. Virology. 2004;318:439–450. doi: 10.1016/j.virol.2003.09.045. [DOI] [PubMed] [Google Scholar]
- 45.Ogino T, Banerjee AK. An unconventional pathway of mRNA cap formation by vesiculoviruses. Virus research. 2011;162:100–109. doi: 10.1016/j.virusres.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ogino T, Banerjee AK. Unconventional mechanism of mRNA capping by the RNA-dependent RNA polymerase of vesicular stomatitis virus. Molecular cell. 2007;25:85–97. doi: 10.1016/j.molcel.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 47.Ogino T, Banerjee AK. Formation of guanosine(5’)tetraphospho(5’)adenosine cap structure by an unconventional mRNA capping enzyme of vesicular stomatitis virus. J Virol. 2008;82:7729–7734. doi: 10.1128/JVI.00326-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ogino T, Banerjee AK. The HR motif in the RNA-dependent RNA polymerase L protein of Chandipura virus is required for unconventional mRNA-capping activity. J Gen Virol. 2010;91:1311–1314. doi: 10.1099/vir.0.019307-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li J, Wang JT, Whelan SP. A unique strategy for mRNA cap methylation used by vesicular stomatitis virus. Proc Natl Acad Sci U S A. 2006;103:8493–8498. doi: 10.1073/pnas.0509821103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rahmeh AA, Li J, Kranzusch PJ, Whelan SP. Ribose 2’-O methylation of the vesicular stomatitis virus mRNA cap precedes and facilitates subsequent guanine-N-7 methylation by the large polymerase protein. J Virol. 2009;83:11043–11050. doi: 10.1128/JVI.01426-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Testa D, Banerjee AK. Two methyltransferase activities in the purified virions of vesicular stomatitis virus. J Virol. 1977;24:786–793. doi: 10.1128/jvi.24.3.786-793.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li J, Rahmeh A, Morelli M, Whelan SP. A conserved motif in region v of the large polymerase proteins of nonsegmented negative-sense RNA viruses that is essential for mRNA capping. J Virol. 2008;82:775–784. doi: 10.1128/JVI.02107-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Neubauer J, Ogino M, Green TJ, Ogino T. Signature motifs of GDP polyribonucleotidyltransferase, a non-segmented negative strand RNA viral mRNA capping enzyme, domain in the L protein are required for covalent enzyme-pRNA intermediate formation. Nucleic acids research. 2016;44:330–341. doi: 10.1093/nar/gkv1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li J, Fontaine-Rodriguez EC, Whelan SP. Amino acid residues within conserved domain VI of the vesicular stomatitis virus large polymerase protein essential for mRNA cap methyltransferase activity. J Virol. 2005;79:13373–13384. doi: 10.1128/JVI.79.21.13373-13384.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bujnicki JM, Rychlewski L. In silico identification, structure prediction and phylogenetic analysis of the 2’-O-ribose (cap 1) methyltransferase domain in the large structural protein of ssRNA negative-strand viruses. Protein engineering. 2002;15:101–108. doi: 10.1093/protein/15.2.101. [DOI] [PubMed] [Google Scholar]
- 56.Ferron F, Longhi S, Henrissat B, Canard B. Viral RNA-polymerases -- a predicted 2’-O-ribose methyltransferase domain shared by all Mononegavirales. Trends Biochem Sci. 2002;27:222–224. doi: 10.1016/s0968-0004(02)02091-1. [DOI] [PubMed] [Google Scholar]
- 57.Gopinath M, Shaila MS. RNA triphosphatase and guanylyl transferase activities are associated with the RNA polymerase protein L of rinderpest virus. J Gen Virol. 2009;90:1748–1756. doi: 10.1099/vir.0.010975-0. [DOI] [PubMed] [Google Scholar]
- 58.Barik S. The structure of the 5’ terminal cap of the respiratory syncytial virus mRNA. J Gen Virol. 1993;74(Pt 3):485–490. doi: 10.1099/0022-1317-74-3-485. [DOI] [PubMed] [Google Scholar]
- 59.Ogino T, Kobayashi M, Iwama M, Mizumoto K. Sendai virus RNA-dependent RNA polymerase L protein catalyzes cap methylation of virus-specific mRNA. J Biol Chem. 2005;280:4429–4435. doi: 10.1074/jbc.M411167200. [DOI] [PubMed] [Google Scholar]
- 60.Gopinath M, Shaila MS. Evidence for N(7) guanine methyl transferase activity encoded within the modular domain of RNA-dependent RNA polymerase L of a Morbillivirus. Virus Genes. 2015;51:356–360. doi: 10.1007/s11262-015-1252-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Colonno RJ, Stone HO. Newcastle disease virus mRNA lacks 2’-O-methylated nucleotides. Nature. 1976;261:611–614. doi: 10.1038/261611a0. [DOI] [PubMed] [Google Scholar]
- 62.Noton SL, Fearns R. Initiation and regulation of paramyxovirus transcription and replication. Virology. 2015 doi: 10.1016/j.virol.2015.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hoffman MA, Banerjee AK. Precise mapping of the replication and transcription promoters of human parainfluenza virus type 3. Virology. 2000;269:201–211. doi: 10.1006/viro.2000.0223. [DOI] [PubMed] [Google Scholar]
- 64.Marcos F, Ferreira L, Cros J, Park MS, Nakaya T, Garcia-Sastre A, Villar E. Mapping of the RNA promoter of Newcastle disease virus. Virology. 2005;331:396–406. doi: 10.1016/j.virol.2004.10.040. [DOI] [PubMed] [Google Scholar]
- 65.Mioulet V, Barrett T, Baron MD. Scanning mutagenesis identifies critical residues in the rinderpest virus genome promoter. J Gen Virol. 2001;82:2905–2911. doi: 10.1099/0022-1317-82-12-2905. [DOI] [PubMed] [Google Scholar]
- 66.Murphy SK, Ito Y, Parks GD. A functional antigenomic promoter for the paramyxovirus simian virus 5 requires proper spacing between an essential internal segment and the 3’ terminus. J Virol. 1998;72:10–19. doi: 10.1128/jvi.72.1.10-19.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tapparel C, Maurice D, Roux L. The activity of Sendai virus genomic and antigenomic promoters requires a second element past the leader template regions: a motif (GNNNNN)3 is essential for replication. J Virol. 1998;72:3117–3128. doi: 10.1128/jvi.72.4.3117-3128.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Walpita P. An internal element of the measles virus antigenome promoter modulates replication efficiency. Virus research. 2004;100:199–211. doi: 10.1016/j.virusres.2003.12.025. [DOI] [PubMed] [Google Scholar]
- 69.Walpita P, Peters CJ. Cis-acting elements in the antigenomic promoter of Nipah virus. J Gen Virol. 2007;88:2542–2551. doi: 10.1099/vir.0.83035-0. [DOI] [PubMed] [Google Scholar]
- 70.Vulliemoz D, Roux L. “Rule of six”: how does the Sendai virus RNA polymerase keep count? J Virol. 2001;75:4506–4518. doi: 10.1128/JVI.75.10.4506-4518.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McGivern DR, Collins PL, Fearns R. Identification of internal sequences in the 3’ leader region of human respiratory syncytial virus that enhance transcription and confer replication processivity. J Virol. 2005;79:2449–2460. doi: 10.1128/JVI.79.4.2449-2460.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Samal SK, Collins PL. RNA replication by a respiratory syncytial virus RNA analog does not obey the rule of six and retains a nonviral trinucleotide extension at the leader end. J Virol. 1996;70:5075–5082. doi: 10.1128/jvi.70.8.5075-5082.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cowton VM, Fearns R. Evidence that the respiratory syncytial virus polymerase is recruited to nucleotides 1 to 11 at the 3’ end of the nucleocapsid and can scan to access internal signals. J Virol. 2005;79:11311–11322. doi: 10.1128/JVI.79.17.11311-11322.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Green TJ, Zhang X, Wertz GW, Luo M. Structure of the vesicular stomatitis virus nucleoprotein-RNA complex. Science. 2006;313:357–360. doi: 10.1126/science.1126953. [DOI] [PubMed] [Google Scholar]
- 75.Albertini AA, Wernimont AK, Muziol T, Ravelli RB, Clapier CR, Schoehn G, Weissenhorn W, Ruigrok RW. Crystal structure of the rabies virus nucleoprotein-RNA complex. Science. 2006;313:360–363. doi: 10.1126/science.1125280. [DOI] [PubMed] [Google Scholar]
- 76.Tawar RG, Duquerroy S, Vonrhein C, Varela PF, Damier-Piolle L, Castagne N, MacLellan K, Bedouelle H, Bricogne G, Bhella D, Eleouet JF, Rey FA. Crystal structure of a nucleocapsid-like nucleoprotein-RNA complex of respiratory syncytial virus. Science. 2009;326:1279–1283. doi: 10.1126/science.1177634. [DOI] [PubMed] [Google Scholar]
- 77.Gutsche I, Desfosses A, Effantin G, Ling WL, Haupt M, Ruigrok RW, Sachse C, Schoehn G. Structural virology. Near-atomic cryo-EM structure of the helical measles virus nucleocapsid. Science. 2015;348:704–707. doi: 10.1126/science.aaa5137. [DOI] [PubMed] [Google Scholar]
- 78.Alayyoubi M, Leser GP, Kors CA, Lamb RA. Structure of the paramyxovirus parainfluenza virus 5 nucleoprotein-RNA complex. Proc Natl Acad Sci U S A. 2015;112:E1792–E1799. doi: 10.1073/pnas.1503941112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yabukarski F, Lawrence P, Tarbouriech N, Bourhis JM, Delaforge E, Jensen MR, Ruigrok RW, Blackledge M, Volchkov V, Jamin M. Structure of Nipah virus unassembled nucleoprotein in complex with its viral chaperone. Nature structural & molecular biology. 2014;21:754–759. doi: 10.1038/nsmb.2868. [DOI] [PubMed] [Google Scholar]
- 80.Severin C, Terrell JR, Zengel JR, Cox R, Plemper RK, He B, Luo M. Releasing the genomic RNA sequestered in the mumps virus nucleocapsid. J Virol. 2016 doi: 10.1128/JVI.01422-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kolakofsky D. Paramyxovirus RNA synthesis, mRNA editing, and genome hexamer phase: A review. Virology. 2016;498:94–98. doi: 10.1016/j.virol.2016.08.018. [DOI] [PubMed] [Google Scholar]
- 82.Tremaglio CZ, Noton SL, Deflube LR, Fearns R. Respiratory syncytial virus polymerase can initiate transcription from position 3 of the leader promoter. J Virol. 2013;87:3196–3207. doi: 10.1128/JVI.02862-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Noton SL, Cowton VM, Zack CR, McGivern DR, Fearns R. Evidence that the polymerase of respiratory syncytial virus initiates RNA replication in a nontemplated fashion. Proc Natl Acad Sci U S A. 2010;107:10226–10231. doi: 10.1073/pnas.0913065107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Noton SL, Fearns R. The first two nucleotides of the respiratory syncytial virus antigenome RNA replication product can be selected independently of the promoter terminus. RNA. 2011;17:1895–1906. doi: 10.1261/rna.2813411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Morin B, Rahmeh AA, Whelan SP. Mechanism of RNA synthesis initiation by the vesicular stomatitis virus polymerase. The EMBO journal. 2012;31:1320–1329. doi: 10.1038/emboj.2011.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vidal S, Kolakofsky D. Modified model for the switch from Sendai virus transcription to replication. J Virol. 1989;63:1951–1958. doi: 10.1128/jvi.63.5.1951-1958.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gerlach P, Malet H, Cusack S, Reguera J. Structural Insights into Bunyavirus Replication and Its Regulation by the vRNA Promoter. Cell. 2015;161:1267–1279. doi: 10.1016/j.cell.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ghosh A, Nayak R, Shaila MS. Synthesis of leader RNA and editing of P mRNA during transcription by rinderpest virus. Virus research. 1996;41:69–76. doi: 10.1016/0168-1702(95)01276-1. [DOI] [PubMed] [Google Scholar]
- 89.Horikami SM, Moyer SA. Synthesis of leader RNA and editing of the P mRNA during transcription by purified measles virus. J Virol. 1991;65:5342–5347. doi: 10.1128/jvi.65.10.5342-5347.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kurilla MG, Stone HO, Keene JD. RNA sequence and transcriptional properties of the 3’ end of the Newcastle disease virus genome. Virology. 1985;145:203–212. doi: 10.1016/0042-6822(85)90154-0. [DOI] [PubMed] [Google Scholar]
- 91.Leppert M, Rittenhouse L, Perrault J, Summers DF, Kolakofsky D. Plus and minus strand leader RNAs in negative strand virus-infected cells. Cell. 1979;18:735–747. doi: 10.1016/0092-8674(79)90127-2. [DOI] [PubMed] [Google Scholar]
- 92.Tekes G, Rahmeh AA, Whelan SP. A freeze frame view of vesicular stomatitis virus transcription defines a minimal length of RNA for 5’ processing. PLoS pathogens. 2011;7:e1002073. doi: 10.1371/journal.ppat.1002073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ogino T. Capping of vesicular stomatitis virus pre-mRNA is required for accurate selection of transcription stop-start sites and virus propagation. Nucleic acids research. 2014;42:12112–12125. doi: 10.1093/nar/gku901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Stillman EA, Whitt MA. Transcript initiation and 5’-end modifications are separable events during vesicular stomatitis virus transcription. J Virol. 1999;73:7199–7209. doi: 10.1128/jvi.73.9.7199-7209.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang JT, McElvain LE, Whelan SP. Vesicular Stomatitis Virus mRNA Capping Machinery Requires Specific cis-Acting Signals in the RNA. J Virol. 2007;81:11499–11506. doi: 10.1128/JVI.01057-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cattaneo R, Kaelin K, Baczko K, Billeter MA. Measles virus editing provides an additional cysteine-rich protein. Cell. 1989;56:759–764. doi: 10.1016/0092-8674(89)90679-x. [DOI] [PubMed] [Google Scholar]
- 97.Paterson RG, Lamb RA. RNA editing by G-nucleotide insertion in mumps virus P-gene mRNA transcripts. J Virol. 1990;64:4137–4145. doi: 10.1128/jvi.64.9.4137-4145.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Thomas SM, Lamb RA, Paterson RG. Two mRNAs that differ by two nontemplated nucleotides encode the amino coterminal proteins P and V of the paramyxovirus SV5. Cell. 1988;54:891–902. doi: 10.1016/S0092-8674(88)91285-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Vidal S, Curran J, Kolakofsky D. Editing of the Sendai virus P/C mRNA by G insertion occurs during mRNA synthesis via a virus-encoded activity. J Virol. 1990;64:239–246. doi: 10.1128/jvi.64.1.239-246.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hausmann S, Garcin D, Morel AS, Kolakofsky D. Two nucleotides immediately upstream of the essential A6G3 slippery sequence modulate the pattern of G insertions during Sendai virus mRNA editing. J Virol. 1999;73:343–351. doi: 10.1128/jvi.73.1.343-351.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cartee TL, Megaw AG, Oomens AG, Wertz GW. Identification of a single amino acid change in the human respiratory syncytial virus L protein that affects transcriptional termination. J Virol. 2003;77:7352–7360. doi: 10.1128/JVI.77.13.7352-7360.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Juhasz K, Murphy BR, Collins PL. The major attenuating mutations of the respiratory syncytial virus vaccine candidate cpts530/1009 specify temperature-sensitive defects in transcription and replication and a non-temperature-sensitive alteration in mRNA termination. J Virol. 1999;73:5176–5180. doi: 10.1128/jvi.73.6.5176-5180.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Collins PL, Olmsted RA, Spriggs MK, Johnson PR, Buckler-White AJ. Gene overlap and site-specific attenuation of transcription of the viral polymerase L gene of human respiratory syncytial virus. Proc Natl Acad Sci U S A. 1987;84:5134–5138. doi: 10.1073/pnas.84.15.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Fearns R, Collins PL. Model for polymerase access to the overlapped L gene of respiratory syncytial virus. J Virol. 1999;73:388–397. doi: 10.1128/jvi.73.1.388-397.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Plattet P, Strahle L, le Mercier P, Hausmann S, Garcin D, Kolakofsky D. Sendai virus RNA polymerase scanning for mRNA start sites at gene junctions. Virology. 2007;362:411–420. doi: 10.1016/j.virol.2006.12.033. [DOI] [PubMed] [Google Scholar]
- 106.Gubbay O, Curran J, Kolakofsky D. Sendai virus genome synthesis and assembly are coupled: a possible mechanism to promote viral RNA polymerase processivity. J Gen Virol. 2001;82:2895–2903. doi: 10.1099/0022-1317-82-12-2895. [DOI] [PubMed] [Google Scholar]
- 107.Bourhis JM, Canard B, Longhi S. Structural disorder within the replicative complex of measles virus: functional implications. Virology. 2006;344:94–110. doi: 10.1016/j.virol.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 108.Wei T, Li D, Marcial D, Khan M, Lin MH, Snape N, Ghildyal R, Harrich D, Spann K. The eukaryotic elongation factor 1A is critical for genome replication of the paramyxovirus respiratory syncytial virus. PLoS One. 2014;9:e114447. doi: 10.1371/journal.pone.0114447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Krumm SA, Ndungu JM, Yoon JJ, Dochow M, Sun A, Natchus M, Snyder JP, Plemper RK. Potent host-directed small-molecule inhibitors of myxovirus RNA-dependent RNA-polymerases. PLoS ONE. 2011;6:e20069. doi: 10.1371/journal.pone.0020069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cox R, Plemper RK. Structure-guided design of small-molecule therapeutics against RSV disease. Expert opinion on drug discovery. 2016:1–14. doi: 10.1517/17460441.2016.1174212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Schwegmann A, Brombacher F. Host-directed drug targeting of factors hijacked by pathogens. Sci Signal. 2008;1 doi: 10.1126/scisignal.129re8. re8. [DOI] [PubMed] [Google Scholar]
- 112.Prussia A, Thepchatri P, Snyder JP, Plemper RK. Systematic Approaches towards the Development of Host-Directed Antiviral Therapeutics. Int J Mol Sci. 2011;12:4027–4052. doi: 10.3390/ijms12064027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Das T, Mathur M, Gupta AK, Janssen GM, Banerjee AK. RNA polymerase of vesicular stomatitis virus specifically associates with translation elongation factor-1 alphabetagamma for its activity. Proc Natl Acad Sci U S A. 1998;95:1449–1454. doi: 10.1073/pnas.95.4.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Qanungo KR, Shaji D, Mathur M, Banerjee AK. Two RNA polymerase complexes from vesicular stomatitis virus-infected cells that carry out transcription and replication of genome RNA. Proc Natl Acad Sci U S A. 2004;101:5952–5957. doi: 10.1073/pnas.0401449101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Scott DE, Bayly AR, Abell C, Skidmore J. Small molecules, big targets: drug discovery faces the protein-protein interaction challenge. Nature reviews. 2016;15:533–550. doi: 10.1038/nrd.2016.29. [DOI] [PubMed] [Google Scholar]
- 116.MacArthur RD, Novak RM. Reviews of anti-infective agents: maraviroc: the first of a new class of antiretroviral agents. Clin Infect Dis. 2008;47:236–241. doi: 10.1086/589289. [DOI] [PubMed] [Google Scholar]
- 117.Plemper RK, Hammond AL. Synergizing vaccinations with therapeutics for measles eradication. Expert opinion on drug discovery. 2014;9:201–214. doi: 10.1517/17460441.2014.867324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Arts EJ, Hazuda DJ. HIV-1 antiretroviral drug therapy. Cold Spring Harb Perspect Med. 2012;2:a007161. doi: 10.1101/cshperspect.a007161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.posting date. Guidelines: HIV. World Health Organization; 2016. http://www.who.int/hiv/pub/guidelines/en/. [Online.] [Google Scholar]
- 120.Waqar MA, Evans MJ, Manly KF, Hughes RG, Huberman JA. Effects of 2’,3’-dideoxynucleosides on mammalian cells and viruses. J Cell Physiol. 1984;121:402–408. doi: 10.1002/jcp.1041210218. [DOI] [PubMed] [Google Scholar]
- 121.Huang P, Farquhar D, Plunkett W. Selective action of 3’-azido-3’-deoxythymidine 5’-triphosphate on viral reverse transcriptases and human DNA polymerases. J Biol Chem. 1990;265:11914–11918. [PubMed] [Google Scholar]
- 122.Barditch-Crovo P, Toole J, Hendrix CW, Cundy KC, Ebeling D, Jaffe HS, Lietman PS. Anti-human immunodeficiency virus (HIV) activity, safety, and pharmacokinetics of adefovir dipivoxil (9-[2-(bis-pivaloyloxymethyl)-phosphonylmethoxyethyl]adenine) in HIV-infected patients. J Infect Dis. 1997;176:406–413. doi: 10.1086/514057. [DOI] [PubMed] [Google Scholar]
- 123.Balzarini J, Naesens L, Herdewijn P, Rosenberg I, Holy A, Pauwels R, Baba M, Johns DG, De Clercq E. Marked in vivo antiretrovirus activity of 9-(2-phosphonylmethoxyethyl)adenine, a selective anti-human immunodeficiency virus agent. Proc Natl Acad Sci U S A. 1989;86:332–336. doi: 10.1073/pnas.86.1.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Thomas E, Ghany MG, Liang TJ. The application and mechanism of action of ribavirin in therapy of hepatitis C. Antivir Chem Chemother. 2012;23:1–12. doi: 10.3851/IMP2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bougie I, Bisaillon M. Initial binding of the broad spectrum antiviral nucleoside ribavirin to the hepatitis C virus RNA polymerase. J Biol Chem. 2003;278:52471–52478. doi: 10.1074/jbc.M308917200. [DOI] [PubMed] [Google Scholar]
- 126.Fernandez-Larsson R, O’Connell K, Koumans E, Patterson JL. Molecular analysis of the inhibitory effect of phosphorylated ribavirin on the vesicular stomatitis virus in vitro polymerase reaction. Antimicrob Agents Chemother. 1989;33:1668–1673. doi: 10.1128/aac.33.10.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Howe AY, Cheng H, Johann S, Mullen S, Chunduru SK, Young DC, Bard J, Chopra R, Krishnamurthy G, Mansour T, O’Connell J. Molecular mechanism of hepatitis C virus replicon variants with reduced susceptibility to a benzofuran inhibitor, HCV-796. Antimicrob Agents Chemother. 2008;52:3327–3338. doi: 10.1128/AAC.00238-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Nakamura M, Saito H, Ikeda M, Tada S, Kumagai N, Kato N, Shimotohno K, Hibi T. Possible molecular mechanism of the relationship between NS5B polymorphisms and early clearance of hepatitis C virus during interferon plus ribavirin treatment. J Med Virol. 2008;80:632–639. doi: 10.1002/jmv.21125. [DOI] [PubMed] [Google Scholar]
- 129.Wright PJ, Crameri G, Eaton BT. RNA synthesis during infection by Hendra virus: an examination by quantitative real-time PCR of RNA accumulation, the effect of ribavirin and the attenuation of transcription. Arch Virol. 2005;150:521–532. doi: 10.1007/s00705-004-0417-5. [DOI] [PubMed] [Google Scholar]
- 130.Crotty S, Andino R. Implications of high RNA virus mutation rates: lethal mutagenesis and the antiviral drug ribavirin. Microbes Infect. 2002;4:1301–1307. doi: 10.1016/s1286-4579(02)00008-4. [DOI] [PubMed] [Google Scholar]
- 131.Aljabr W, Touzelet O, Pollakis G, Wu W, Munday DC, Hughes M, Hertz-Fowler C, Kenny J, Fearns R, Barr JN, Matthews DA, Hiscox JA. Investigating the Influence of Ribavirin on Human Respiratory Syncytial Virus RNA Synthesis by Using a High-Resolution Transcriptome Sequencing Approach. J Virol. 2016;90:4876–4888. doi: 10.1128/JVI.02349-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Page T, Connor JD. The metabolism of ribavirin in erythrocytes and nucleated cells. Int J Biochem. 1990;22:379–383. doi: 10.1016/0020-711x(90)90140-x. [DOI] [PubMed] [Google Scholar]
- 133.DeVincenzo J, Fathi H, Mcclure M, Westland C, Chanda S, Lambkin-Williams R, Smith P, Harrison L, Symons J, Scaglioni-Weinlich C, Zhang Q, Nieforth K, Beigelman L, Blatt L, Fry J. Treatment with Oral ALS-008176, a Nucleoside Analog, Rapidly Reduces RSV Viral Load and Clinical Disease Severity in a Healthy Volunteer Challenge Study. Open Forum Infectious Diseases. 2014;Volume 1:S66–S69. [Google Scholar]
- 134.Deval J, Hong J, Wang G, Taylor J, Smith LK, Fung A, Stevens SK, Liu H, Jin Z, Dyatkina N, Prhavc M, Stoycheva AD, Serebryany V, Liu J, Smith DB, Tam Y, Zhang Q, Moore ML, Fearns R, Chanda SM, Blatt LM, Symons JA, Beigelman L. Molecular Basis for the Selective Inhibition of Respiratory Syncytial Virus RNA Polymerase by 2’-Fluoro-4’-Chloromethyl-Cytidine Triphosphate. PLoS pathogens. 2015;11:e1004995. doi: 10.1371/journal.ppat.1004995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Malur AG, Gupta NK, De Bishnu P, Banerjee AK. Analysis of the mutations in the active site of the RNA-dependent RNA polymerase of human parainfluenza virus type 3 (HPIV3) Gene Expr. 2002;10:93–100. [PMC free article] [PubMed] [Google Scholar]
- 136.Fearns R, Deval J. New antiviral approaches for respiratory syncytial virus and other mononegaviruses: Inhibiting the RNA polymerase. Antiviral Res. 2016;134:63–76. doi: 10.1016/j.antiviral.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 137.Chapman J, Abbott E, Alber DG, Baxter RC, Bithell SK, Henderson EA, Carter MC, Chambers P, Chubb A, Cockerill GS, Collins PL, Dowdell VC, Keegan SJ, Kelsey RD, Lockyer MJ, Luongo C, Najarro P, Pickles RJ, Simmonds M, Taylor D, Tyms S, Wilson LJ, Powell KL. RSV604, a novel inhibitor of respiratory syncytial virus replication. Antimicrob Agents Chemother. 2007;51:3346–3353. doi: 10.1128/AAC.00211-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Marty FM, Chemaly RF, Liakopoulou E, Dent JC. A doubleblind, randomised, placebo-controlled study to evaluate the safety and efficacy of RSV604 in adults with respiratory syncytial virus (RSV) infection following stem cell transplantation. Hong Kong, China: IX International Symposium on Respiratory Viral Infections; 2007. [Google Scholar]
- 139.Challa S, Scott AD, Yuzhakov O, Zhou Y, Tiong-Yip CL, Gao N, Thresher J, Yu Q. Mechanism of action for respiratory syncytial virus inhibitor RSV604. Antimicrob Agents Chemother. 2015;59:1080–1087. doi: 10.1128/AAC.04119-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ouizougun-Oubari M, Pereira N, Tarus B, Galloux M, Lassoued S, Fix J, Tortorici MA, Hoos S, Baron B, England P, Desmaele D, Couvreur P, Bontems F, Rey FA, Eleouet JF, Sizun C, Slama-Schwok A, Duquerroy S. A Druggable Pocket at the Nucleocapsid/Phosphoprotein Interaction Site of Human Respiratory Syncytial Virus. J Virol. 2015;89:11129–11143. doi: 10.1128/JVI.01612-15. [DOI] [PMC free article] [PubMed] [Google Scholar]