Abstract
The main characteristics of ‘Khelaifiella massiliensis’ strain Mt13T (= CSUR P1935, = DSM100591), ‘Niameybacter massiliensis’ strain Mt14T (= CSUR P1909, = DSM100592), ‘Brachybacterium massiliense’ strain MT5T (= CSUR P2240, = DSM101766), ‘Enterobacter timonensis’ strain mt20T (= CSUR P2201, = DSM 101775) and ‘Massilibacillus massiliensis’ strain Marseille-P2411T (= CSUR P2411, = DSM102838), new species isolated from the gut of healthy African infants, are presented.
Keywords: Culturomics, gut microbiota, new species, taxono-genomic
Extensive knowledge of the gut microbiota composition has become essential for the understanding of many aspects of health and disease. For that purpose, stool samples were collected from a 7-month-old healthy girl with a weight-for-height z-score of 0.15 from Niger and a 38-month-old healthy girl with a weight-for-height z-score of – 0.12 from Senegal. The diversity of these samples was explored using the culturomics concept [1], [2]. Oral consent for this study was obtained from the children’s parents and the study received authorization from the local ethics committee of the Institut Federatif de Recherche IFR48 under number 09-022. As part of this study, strains Mt13T and Mt14T were isolated in the sample from Niger whereas strains mt20T, MT5T and Marseille-P2411T were isolated in the sample from Senegal.
Growth conditions and strains phenotypic characteristics
Strain Mt13T was first isolated after a 3-day pre-incubation in an anaerobic blood culture bottle supplemented with sheep blood and seeding on 5% sheep-blood-enriched Columbia agar (bioMérieux, Marcy l’Etoile, France) at 37°C in an anaerobic atmosphere. Large greyish and irregular colonies were obtained with a mean diameter of 8 mm. Cells were Gram-stain-positive bacilli with a mean diameter of 0.67 μm and a mean length of 2.99 μm. Catalase and oxidase activities were absent.
Strain Mt14T was first isolated after a 7-day pre-incubation in an anaerobic blood culture bottle supplemented with rumen and sheep blood and seeding on 5% sheep-blood-enriched Columbia agar at 37°C in an anaerobic atmosphere. Small white colonies were obtained with a mean diameter of 3 mm. These colonies were formed with Gram-stain-positive bacilli with a mean diameter of 0.61 μm and a mean length of 4.47 μm.
Strain MT5T and strain mt20T were first isolated after a 24-h and 3-day aerobic pre-incubation, respectively, in a liquid medium containing 37 g of Difco Marine Broth (Becton Dickinson, Le Pont de Claix, France) per litre of sterile water at 37°C and on 5% sheep-blood-enriched Columbia agar in aerobic conditions. Strain mt20T formed large brown colonies with a mean diameter of 8 mm. Cells were Gram-stain-negative bacilli with a mean diameter of 0.8 μm and a mean length of 1.3 μm. Strain mt20T exhibited oxidase and catalase activities. As for strain mt21T, it formed white full, circular colonies with a mean diameter of 3 mm, which were formed with Gram-stain-positive cooci. Cells had a mean diameter of 0.57 μm and exhibited catalase activity and no oxidase activity.
After a 7-day pre-incubation in an anaerobic blood culture bottle supplemented with sheep blood and rumen and seeding on 5% sheep-blood-enriched Colombia agar in an anaerobic atmosphere, strain Marseille-P2411T was isolated. Small translucent colonies with a white centre measuring a mean diameter of 2 mm were obtained. These Gram-stain-positive bacilli had a mean diameter of 0.6 μm and a mean length of 3.65 μm. Catalase and oxidase activities were not exhibited by strain Marseille-P2411T.
Strain identification
After a failed identification of all five strains using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) on a Microflex spectrometer (Bruker Daltonics, Bremen, Germany) [3], [4], the 16S rRNA gene was sequenced using fD1 and rP2 primers [5] as well as the rpoB gene for strain mt20T [6]. Kim et al. suggested a 98.65% and a 95% similarity level threshold to define a new species and a new genus, respectively, without performing DNA–DNA hybridization [7] using the 16S rRNA gene while a 97.7% threshold was set for identification using the rpoB gene [6].
The 16S rRNA gene of strain Mt13T (Accession number LN850733) showed a 94.23% similarity level with Hathewaya histolytica strain ATCC 19401T (Accession number AB566416) [8]. This classifies strain Mt13T as a putative new genus within the family Clostridiaceae (Fig. 1), for which we suggest the name Khelaifiella (Khe.lai.fi.el’la N.L. fem. n. composed of Khelaifia, in honour of the microbiologist Saber Khelaifia, and bacterium, meaning rod) with the type species being ‘Khelaifiella massiliensis’ (mas.si.li.en’sis; L. fem. adj. massiliensis for Massilia, the Roman name of Marseille, where strain Mt13T was first isolated). Strain Mt13T is the type strain of the species ‘Khelaifiella massiliensis’.
Fig 1.
Phylogenetic tree showing position of ‘Khelaifiella massiliensis’ strain Mt13T relative to other phylogenetically close species with validly published name. Sequences were aligned using CLUSTALW, and phylogenetic inferences were obtained using a maximum likelihood method within MEGA software [12]. Numbers at nodes are percentages of bootstrap values obtained by repeating the analysis 500 times to generate a majority consensus tree. Selenomonas sputigena was used as outgroup. Scale bar indicates 2% nucleotide sequence divergence.
The 16S rRNA gene sequences of strain Mt14T (Accession number LN850735) revealed a 92.18% similarity level with Cellulosilyticum lentocellum strain RHM5T (Accession number X71851) [9], confirming strain Mt14T as a new genus within the family Lachnospiraceae (Fig. 2). The name Niameybacter (Nia.mey.bac’ter for Niamey, the capital of Niger where the stool sample was collected and bacter for bacterium) is proposed for this new genus as well the type species ‘Niameybacter massiliensis’ (mas.si.li.en’sis; L. masc. adj. massiliensis for Massilia, the Roman name of Marseille, where strain Mt14T was first isolated). Strain Mt14T is the type strain of the species ‘Niameybacter massiliensis’.
Fig 2.
Phylogenetic tree showing position of ‘Niameybacter massiliensis’ strain Mt14T relative to other phylogenetically close species with validly published name. Sequences were aligned using CLUSTALW, and phylogenetic inferences were obtained using a maximum likelihood method within MEGA software [12]. Numbers at nodes are percentages of bootstrap values obtained by repeating the analysis 500 times to generate a majority consensus tree. Veillonella parvula was used as outgroup. Scale bar indicates 2% nucleotide sequence divergence.
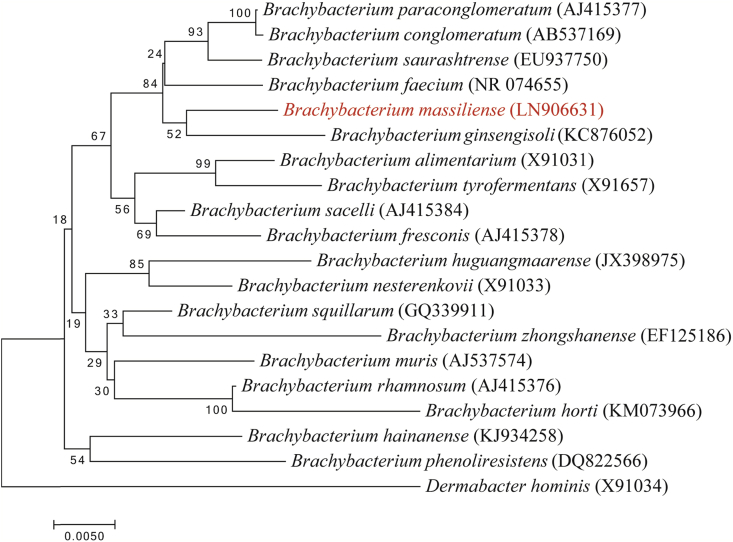
The 16S rRNA sequence of strain MT5T (Accession number LN906631) presented a 98.18% similarity level with Brachybacterium saurashtrense strain JG 06T (Accession number EU937750) [10]. Strain MT5T was therefore classified as a new species within the genus Brachybacterium (Fig. 3) for which we suggest the name ‘Brachybacterium massiliense’ (mas.si.li.en’se; L. masc. neut. adj. massiliense for Massilia, the Roman name of Marseille, where strain MT5T was first isolated). Strain MT5T is the type strain of the species ‘Brachybacterium massiliense’.
Fig 3.
Phylogenetic tree showing position of ‘Brachybacterium massiliensis’ strain MT5T relative to other phylogenetically close species with validly published name. Sequences were aligned using CLUSTALW, and phylogenetic inferences were obtained using a maximum likelihood method within MEGA software [12]. Numbers at nodes are percentages of bootstrap values obtained by repeating the analysis 500 times to generate a majority consensus tree. Dermabacter hominis was used as outgroup. Scale bar indicates 0.5% nucleotide sequence divergence.
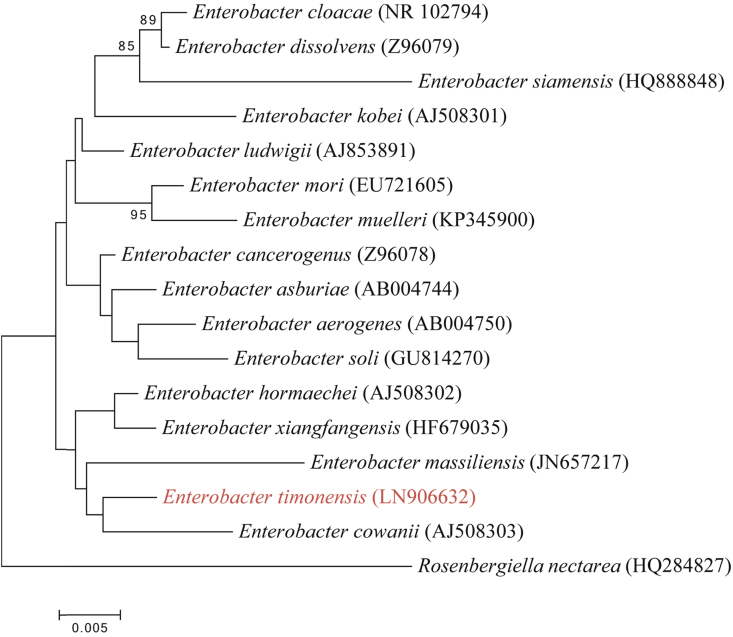
The 16S rRNA gene sequence of strain mt20T (accession number LN906632) showed a 98.4% similarity level with Enterobacter cloacae strain ATCC 13047T (Accession number NR_102794) [7], [8] so classifying strain mt20T as a new species within the genus Enterobacter (Fig. 4). The rpoB gene sequence (Accession number LN906633) showed a 95.12% similarity level with the rpoB gene of Enterobacter cloacae strain ATCC 13047T (Accession number AJ543726), confirming the status of strain mt20T as a putative new species [6]. Strain mt20T is the type strain of ‘Enterobacter timonensis’ sp. nov. (ti.mo.nen’sis. L. gen. masc. timonensis, of Timone, the name of the hospital where strain mt20T was first isolated).
Fig 4.
Phylogenetic tree showing position of ‘Enterobacter timonensis’ strain mt20T relative to other phylogenetically close species with validly published names. Sequences were aligned using CLUSTALW, and phylogenetic inferences were obtained using a maximum likelihood method within MEGA software [12]. Numbers at nodes are percentages of bootstrap values obtained by repeating the analysis 500 times to generate a majority consensus tree. Rosenbergiella nectarea was used as the outgroup. Scale bar indicates 0.5% nucleotide sequence divergence.
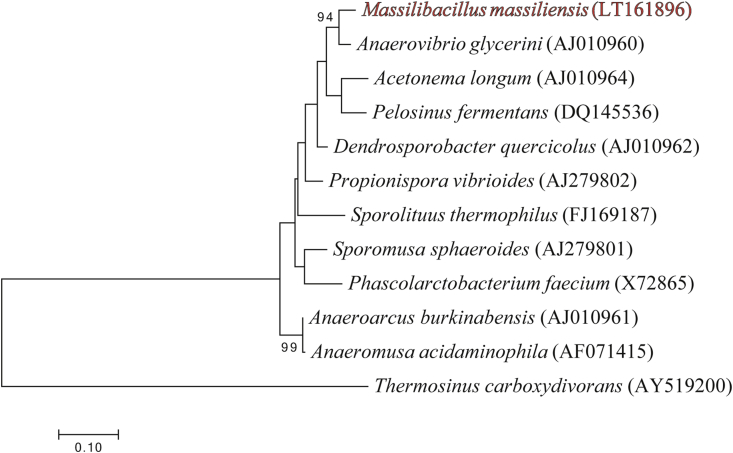
The 16S rRNA gene sequence of strain Marseille-P2411 (accession number LT161896) presented a similarity level of 94.2% with Anaerosinus glycerini strain DSM 5192 (GenBank Accession number NR_025297) [11], which is the phylogenetically closest species with a validly published name (Fig. 5). A new genus was created within the family Veillonellaceae named ‘Massilibacillus’ (mas.si’li, L., masc. adj., massili for Massilia, the old Roman name for Marseille where the genus was first isolated; bacillus as a reference to the rod shape of the cell). Strain Marseille-P2411T is the type species of ‘Massilibacillus massiliensis’ (mas.si.li.en’sis, L., masc. adj., massiliensis for Massilia, the old Roman name for Marseille where strain Marseille-P2411T was first isolated).
Fig 5.
Phylogenetic tree showing position of ‘Massilibacillus massiliensis’ strain Marseille-P2411T relative to other phylogenetically close species with validly published name. Sequences were aligned using CLUSTALW, and phylogenetic inferences were obtained using the maximum likelihood method within MEGA software [12]. Numbers at nodes are percentages of bootstrap values obtained by repeating the analysis 500 times to generate a majority consensus tree. Thermosinus carboxydivorans was used as outgroup. Scale bar indicates 1% nucleotide sequence divergence.
MALDI-TOF MS spectra
The MALDI-TOF MS spectra of ‘Khelaifiella massiliensis’, ‘Niameybacter massiliensis’, ‘Enterobacter massiliensis’, ‘Brachybacterium massiliense’ and ‘Massilibacillus massiliensis’ are available at http://www.mediterraneeinfection.com/article.php?laref=256&titre=urms-database.
Nucleotide sequence accession number
The 16S rRNA sequences of strains Mt13T, Mt14T, MT5T and Marseille-P2411T are deposited in the GenBank database under accession numbers LN850733, LN850735, LN906631 and LT161896, respectively.
The 16S rRNA and rpoB gene sequences for strain mt20T are also deposited in the GenBank database under accession numbers LN906632 and LN906633, respectively.
Deposit in a culture collection
Strains Mt13T, Mt14T, MT5T, mt20T and Marseille-P2411T were deposited in the Collection de Souches de l’Unité des Rickettsies (CSUR, WDCM 875) under numbers P1935, P1909, P2240, P2201 and P2411, respectively. Strains Mt14T, MT5T, mt20T and Marseille-P2411T were also deposited in the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (DSMZ) under numbers DSM100592, DSM101766, DSM101775 and DSM102838, respectively.
Transparency declaration
None to declare.
Acknowledgement
This study was funded by the Fondation Méditerranée Infection.
References
- 1.Lagier J.-C., Armougom F., Million M., Hugon P., Pagnier I., Robert C. Microbial culturomics: paradigm shift in the human gut microbiome study. Clin Microbiol Infect. 2012;18:1185–1193. doi: 10.1111/1469-0691.12023. [DOI] [PubMed] [Google Scholar]
- 2.Lagier J.-C., Hugon P., Khelaifia S., Fournier P.-E., La Scola B., Raoult D. The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota. Clin Microbiol Rev. 2015;28:237–264. doi: 10.1128/CMR.00014-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seng P., Abat C., Rolain J.M., Colson P., Lagier J.-C., Gouriet F. Identification of rare pathogenic bacteria in a clinical microbiology laboratory: impact of matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2013;51:2182–2194. doi: 10.1128/JCM.00492-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seng P., Drancourt M., Gouriet F., La Scola B., Fournier P.-E., Rolain J.M. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis. 2009;49:543–551. doi: 10.1086/600885. [DOI] [PubMed] [Google Scholar]
- 5.Drancourt M., Bollet C., Carlioz A., Martelin R., Gayral J.P., Raoult D. 16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates. J Clin Microbiol. 2000;38:3623–3630. doi: 10.1128/jcm.38.10.3623-3630.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adékambi T., Drancourt M., Raoult D. The rpoB gene as a tool for clinical microbiologists. Trends Microbiol. 2009;17:37–45. doi: 10.1016/j.tim.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 7.Kim M., Oh H.-S., Park S.-C., Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol. 2014;64:346–351. doi: 10.1099/ijs.0.059774-0. [DOI] [PubMed] [Google Scholar]
- 8.Lawson P.A., Rainey F.A. Proposal to restrict the genus Clostridium (Prazmowski) to Clostridium butyricum and related species. ResearchGate. 2015;35 doi: 10.1099/ijsem.0.000824. [DOI] [PubMed] [Google Scholar]
- 9.Cai S., Dong X. Cellulosilyticum ruminicola gen. nov., sp. nov., isolated from the rumen of yak, and reclassification of Clostridium lentocellum as Cellulosilyticum lentocellum comb. nov. Int J Syst Evol Microbiol. 2010;60:845–849. doi: 10.1099/ijs.0.014712-0. [DOI] [PubMed] [Google Scholar]
- 10.Gontia I., Kavita K., Schmid M., Hartmann A., Jha B. Brachybacterium saurashtrense sp. nov., a halotolerant root-associated bacterium with plant growth-promoting potential. Int J Syst Evol Microbiol. 2011;61:2799–2804. doi: 10.1099/ijs.0.023176-0. [DOI] [PubMed] [Google Scholar]
- 11.Strömpl C., Tindall B.J., Jarvis G.N., Lünsdorf H., Moore E.R.B., Hippe H. A re-evaluation of the taxonomy of the genus Anaerovibrio, with the reclassification of Anaerovibrio glycerini as Anaerosinus glycerini gen. nov., comb. nov., and Anaerovibrio burkinabensis as Anaeroarcus burkinensis [corrig.] gen. nov., comb. nov. Int J Syst Evol Microbiol. 1999;49:1861–1872. doi: 10.1099/00207713-49-4-1861. [DOI] [PubMed] [Google Scholar]
- 12.Kumar S., Stecher G., Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]