Abstract
Background
Biopsies from patients with high risk (HR) non-muscle invasive urothelial carcinoma (NMIUC), especially flat urothelial CIS frequently contain scant diagnostic material or denuded mucosa only, precluding further extensive genomic analysis. This study proposes to evaluate the use of Next Generation Sequencing (NGS) analysis of urine cytology material from patients with HR NMIUC in an attempt to identify genetic alterations that might correlate with clinical features and response to BCG treatment.
Methods
Forty-one cytology slides from patients with HR NMIUC and treated with intravesical BCG, were selected for this study. Histologic confirmation was available for all cases. The specimens were subjected to NGS analysis using a customized targeted exome capture assay comprised of 341 genes.
Results
In our cohort, genomic alterations were successfully identified and were able to detect mutations down to a 2% allele frequency and identify chromosomal rearrangements including copy number alterations and gene fusions. The most frequently altered genes included TERT, TP53, ERBB2 and chromatin remodeling genes, such as KDM6A and ARID1A. For patients with matched tumor tissue, cytology specimens revealed all mutations detected in tissue as well as additional mutations, suggesting that urine may more effectively capture the full genetic heterogeneity of disease than an individual cystectomy. Alterations in multiple genes correlated with clinical and histopathological features including response to BCG treatment, flat versus papillary architecture, and smoking history.
Conclusion
Urine specimens can replace tissue as substrate for NGS analysis. Several genomic alterations identified in urine specimens might be associated with histologic features and clinical characteristics.
Keywords: Bladder Cancer, BCG, Urine Cytology, Next-generation Sequencing
Introduction
Urothelial carcinoma of the bladder (UCB) is the fifth most common cancer in the United States, with an estimated 77,000 new cases diagnosed in the US in 2016.1–4 Among the newly diagnosed UCB patients, approximately 30% will present with high-risk (HR) non-muscle invasive urothelial carcinoma (NMIUC), a disease confined to the epithelium or subepithelial connective tissue of the bladder (stage Ta, Tis or T1).5–7 About 20% of these high–risk NMIUC will eventually progress to muscle invasive disease (stage T2, T3 or T4) and worse prognosis.8, 9 Disease management of high-risk NMIUC relies on the microscopic pathological examination of tissue or cells obtained from cystoscopic examinations. While the pathological evaluation of this material remains the standard of care, it is limited in its scope to predict individual recurrence, progression risk, and drug responsiveness.10, 11
In recent years, the clinical management of cancer has greatly benefitted from the rapid progression in the area of genomics, particularly next generation sequencing (NGS).12–15 Molecular diagnostics using NGS allows higher analytical sensitivity and the simultaneous analysis of numerous target genes and pathways for better tumor management and personalized drug therapies.10, 15–17 However, even with the ability to use smaller amounts of DNA and target technically challenging lesions, genomic profiling has been primarily restricted to tissue samples, including formalin-fixed, paraffin-embedded (FFPE) cell blocks. These samples are obtained from invasive surgical procedures, which can increase the risk of patient complications and cost of clinical management.18–20 Other disadvantages in the use of FFPE material is that DNA from FFPE is often moderately degraded with insufficient tumor cellularity and lack of subclonal tumor heterogeneity.21–25 These factors limit the ability to assess the association of different biomarkers with disease progression and drug response in specimens with limited tissue available for analysis. Due to these factors, other sample types such as effusion cytology and fine needle aspirate (FNA) specimens have been recognized as potential alternatives to FFPE for NGS analysis.18, 20
In this context, urine cytology specimens represent a potential source for genomic analysis of HR NMIUC, especially in patients with flat urothelial carcinoma in-situ (CIS). Biopsies from patients with flat urothelial CIS frequently contain scant diagnostic material or denuded mucosa only, precluding further extensive genomic analysis. On the other hand, urine cytology specimens obtained from patients with HR NMIUC are frequently cellular and contain enough DNA for genomic analysis. Therefore, urine cytology specimens could represent a platform for identification of biomarkers associated with clinical features, particularly biomarkers associated with response to BCG treatment in patients with HR NMIUC. Substratification of patients with HR NMIUC into responder and non-responder to BCG treatment represents a major need currently, as it is not possible to predict who will respond favorably to BCG and who might benefit from early aggressive treatment, such as cystectomy, to achieve cure.26 More recently, the European Organization for Research and Treatment of Cancer (EORTC) reiterated the need for molecular markers to better optimize patient selection and treatment strategy for patients with HR NMIUC treated with BCG.27
In this study, we propose to evaluate the use of NGS analysis of urine cytology material from patients with HR NMIUC. We analyzed the suitability of cytology specimens to replace FFPE material for genomic analysis and attempted to identify mutations or copy number alterations that might correlate with clinical features and response to BCG treatment.
Materials and Methods
Specimens
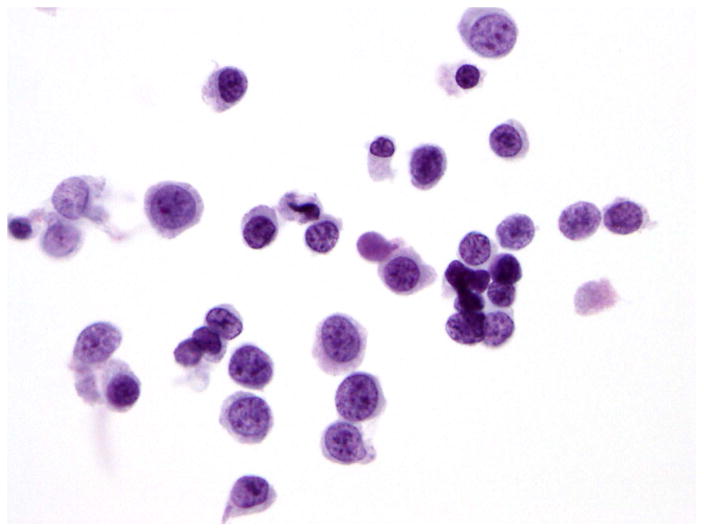
Fifty-two specimens from 41 patients with histologically proven HR NMIUC treated with intravesical BCG instillation were selected for this study. These samples included 41 ThinPrep or cytospin from urine cytology slides, 5 FFPE sections from matching cystectomy specimens and 6 FFPE germline (non-tumor) tissue samples from the same cohort of patients to use for genetic reference. The cytology slides had been CytoLyt® fixed, Papanicolaou stained, and archived for at least 10 years (range 10–12 years) and were obtained from both cystoscopic and voided urine specimens. All cytology slides were morphologically confirmed by a board-certified cytopathologist for the presence and percentage of tumor cells. All specimens satisfied the criteria of high grade urothelial carcinoma as defined in the Paris System for Reporting Urinary Cytology, including a minimum of 5 to 10 severely abnormal urothelial cells with and N/C ratio of 0.7 or greater, with cells showing moderate to severe hypochromasia, coarse chromatin, and markedly irregular nuclear membrane.28 Only cellular specimens containing at least 1,000 neoplastic cells with tumor cells representing over 50% of the cell population in the cytology preparation were included in this study (Figure 1). Thirty percent of the cases that matched our clinical and morphological criteria satisfied the cellularity criteria.
Figure 1.
Papanicolaou stained urine cytology specimen showing urothelial carcinoma.
Sample Preparation
Tumor DNA from cytology slides (1 slide per sample) was extracted by first dispersing 50 microliters of Qiagen ATL buffer onto the slide and then scraping the cells into 1.5-mL Eppendorf tubes with a sterile surgical blade. This was repeated twice for each slide, and the blade was rinsed with 30 μL of the buffer, resulting in a final volume of 180 μL of ATL Buffer. Then 20–40 μl of proteinase K was added and the mixture was then vortexed and incubated at 56°C. Lysates were then purified according to the Qiagen DNeasy protocol with an elution volume of 100 μl using Qiagen QIAamp DNA Tissue Kit (Qiagen Inc., Valencia, CA, USA) and the manufacturer’s protocol released April 2010.
DNA from FFPE tissue sections were extracted manually. Only tumor tissue sections with at least 50% neoplastic cells were selected. QIAamp DNA FFPE Tissue Kit (Qiagen Inc., Valencia, CA, USA) was used according to the manufacturer’s instructions. All extracted cytospin and FFPE DNA were quantified using dsQubit DNA assay (Life Technologies Corporation, Grand Island, NY).
Targeted Next-Generation Sequencing and Sequence Analysis
The extracted tumor and germline DNA samples from normal tissue were both prepared using the MSK-IMPACT (Memorial Sloan Kettering-Integrated Molecular Profiling of Actionable Cancer Targets) assay.29, 30 MSK-IMPACT is a targeted exon capture assay that uses target-specific probes to profile all protein-coding exons and select introns of 341 cancer-associated genes (Supplementary Table 1). It is followed by massive parallel sequencing performed on Illumina HiSeq 2500 (Illumina Inc., San Diego, CA, USA) (2 × 100 base paired reads). The genomic data obtained from the MSK-IMPACT assay includes somatic mutations, structural variants and copy number alterations from the 341-gene panel. For the purpose of this analysis, we manually reviewed all non-synonymous mutations, indels, and structural aberrations using Integrative Genomics Viewer.16, 19 Copy number variants were declared to be amplified if the fold change was ≥2 and deleted if the fold change was ≤-2. Further details on experimental methodology, algorithms used for mutation calling, and run statistics for MSK-IMPACT were described previously.29, 30
Data availability
All associated clinical data were obtained from prospectively maintained medical records. Genomic and clinical data are publicly available through the MSKCC cBio Portal for Cancer Genomics.21, 31
Statistical analysis
The patients were subdivided into 2 categories: responders (patients with no evidence of disease after one cycle of BCG for at least 2 years) and refractory (patients with no response to treatment). The criteria for inclusion in each category followed the European Organization for Research and Treatment of Cancer (EORTC) guidelines. We used Wilcoxon rank-sum test and Fisher’s exact test to analyze the two patient group’s distributions of demographic characteristics (Table 1). We performed univariate analyses to evaluate alteration frequencies and pathways within our cohort and compared our cohort against 125 patients with both copy number and mutation data from The Cancer Genome Atlas (TCGA) urothelial carcinoma of the bladder.32 Statistical analyses were also performed to identify differences associated with smoking history and histologic findings, particularly architecture and presence or absence of invasion.
Table 1.
Characteristics of Urothelial Carcinoma Patients (N=41).
| Characteristics | Overall | Disease Status | P-value‡ | |
|---|---|---|---|---|
| Refractory* n=29 (71%) | Response† n=12 (29%) | |||
| Age (years) | 66.0 (49.0–83.0) | 69.0 (47.5–82.5) | 65.0 (51.0–87.0) | 0.39§ |
| Gender | ||||
| Female | 16 (39.0) | 9 (31.0) | 7 (58.3) | |
| Male | 25 (61.0) | 20 (69.0) | 5 (41.7) | 0.16 |
| Tumor Stage | ||||
| pTa | 12 (29.3) | 8 (27.5) | 4 (33.3) | |
| pTis | 15 (36.6) | 10 (34.5) | 5 (41.7) | |
| pT1 | 13 (31.7) | 10 (34.5) | 3 (25.0) | |
| T1 | 1 (2.4) | 1 (3.5) | 0 (0) | 0.88 |
| Tumor Grade | ||||
| Non-invasive | 28 (68.3) | 18 (62.1) | 10 (83.3) | |
| Invasive | 13 (31.7) | 11 (37.9) | 2 (16.7) | 0.28 |
| Tumor Architecture | ||||
| Flat | 22 (53.7) | 16 (55.2) | 6 (50.0) | |
| Papillary | 19 (46.3) | 13 (44.8) | 6 (50.0) | 1.0 |
| Prior history of BCG therapy|| | ||||
| No | 22 (53.7) | 11 (37.9) | 11 (91.7) | |
| Yes | 19 (46.3) | 18 (62.1) | 1 (8.3) | 0.002 |
| Smoking | ||||
| Never | 13 (31.7) | 10 (34.5) | 3 (25.0) | |
| Ever | 28 (68.3) | 19 (65.5) | 9 (75.0) | 0.72 |
Refractory Disease Status: Patients with No response to therapy.
Response Disease Status: Patients with no evidence of disease for at least 2 years after one cycle of BCG.
Fishers exact test.
Wilcoxon rank sum test.
Prior history of BCG therapy: Bacillus Calmette-Guerin (BCG) treatment before admittance to Memorial Sloan Kettering Cancer Center (MSKCC).
Statistical analyses were conducted using SAS v.9.2 (SAS Institute, Cary, NC, USA) and R software version 3.2.0 (R Core Development Team, Vienna, Austria). All p-values <0.05 were considered statistically significant. Since this study is hypothesis generating, no adjustment for multiple comparisons was performed.
Results
Demographic and clinical characteristics of the 41 UCB patients included in this study are summarized in Table 1. Among the 41 patients, there were 29 patients (71%) with disease refractory to BCG treatment and 12 patients (29%) who responded to one cycle of BCG intravesical therapy. The patients who responded to BCG had either no recurrence or were disease free for over 2 years. Clinical follow up was available for all patients. Overall, our patient population had a median age of 66 years (interquartile range (IQR): 49–83) and was predominately male (61%), with a smoking history (68%).
The average amount of DNA extracted from 41 cytospin slides was 263 ng, ranging from 17 to 2080 ng. Targeted sequencing with MSK-IMPACT was performed for 41 cytospin tumor samples, 5 matched FFPE tumor samples from subsequent cystectomy specimens and 6 matched germline samples from 41 UCB patients. The mean sequencing coverage for all tumor and normal samples across 4980 exons and introns in the 341-gene IMPACT panel was 390x, with only 1% of exons covered below 100x (Figure 2). A total of 497 non-synonymous mutations and 137 gene-level copy number alterations were identified for all cytospin samples in our cohort. The mean number of somatic alterations (mutations and copy-number) per patient was 15.5 (ranging from 2–44 alterations).
Figure 2.
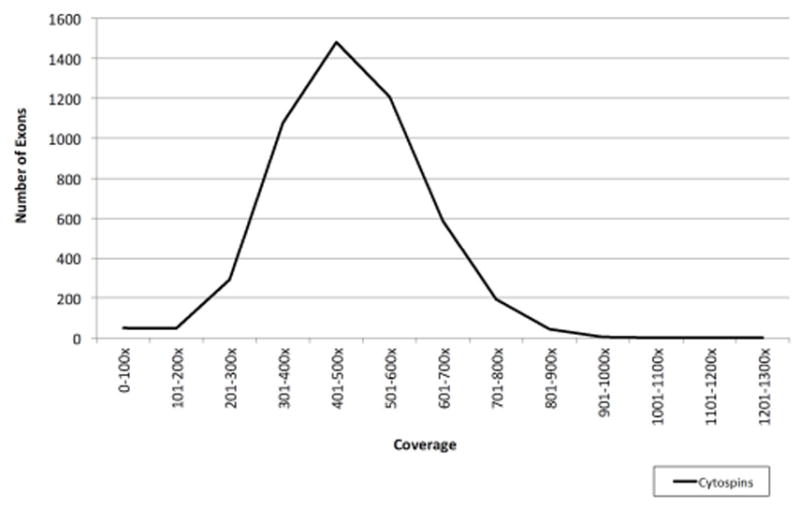
Range of coverage for cytospin samples in the 4,980 exons and introns targeted in MSK-IMPACT 341.
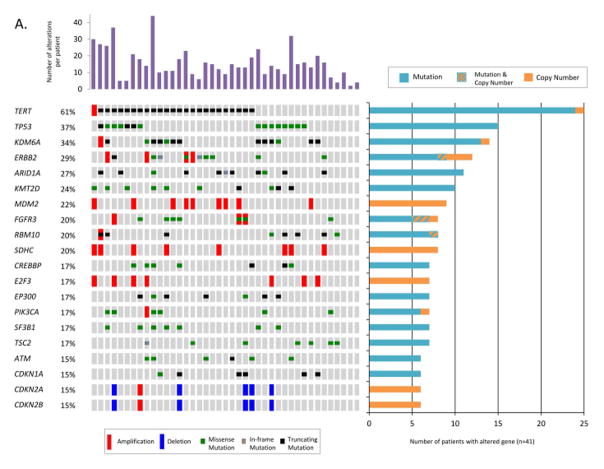
Altogether, 231/341 genes were altered in at least one patient. The most frequently altered genes across our patient cohort are displayed in Figure 3A. TERT was the single most frequently altered gene, with mutations observed in 61% (25/41) of cases. Of these 25 alterations, 24 were at one of two well-characterized mutational hotspots in the promoter region, and one represented an amplification (Figure 3B). The majority of the remaining commonly altered genes were chromatin remodeling genes, KDM6A (34%), ARID1A (27%), KMT2D (24%), FGFR3 (20%), CREBBP (17%), and EP300 (17%). In addition to mutations and copy number alterations, MSK-IMPACT was able to successfully detect chromosomal rearrangements in urine cytology specimens, including an intrachromosomal duplication producing the recurrent FGFR3-TACC3 gene fusion on chromosome 4 (Figure 3C).
Figure 3.
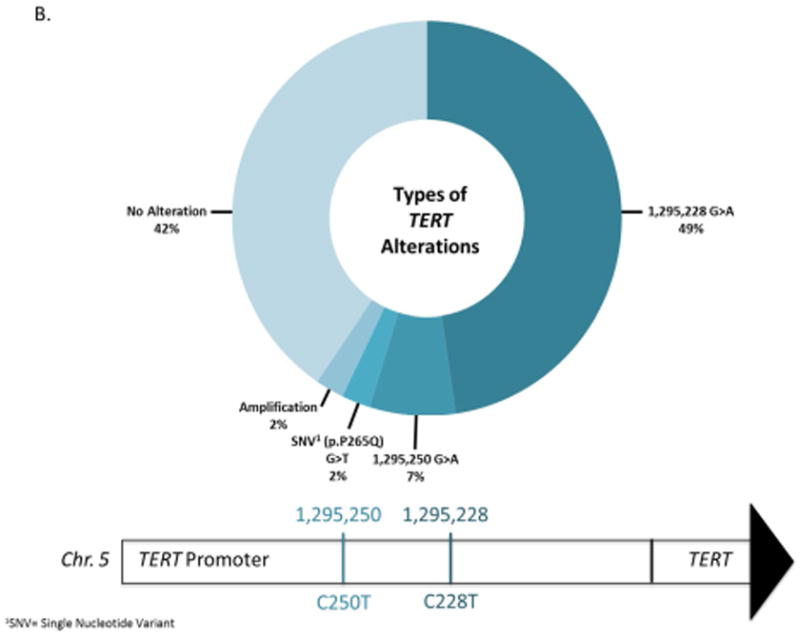
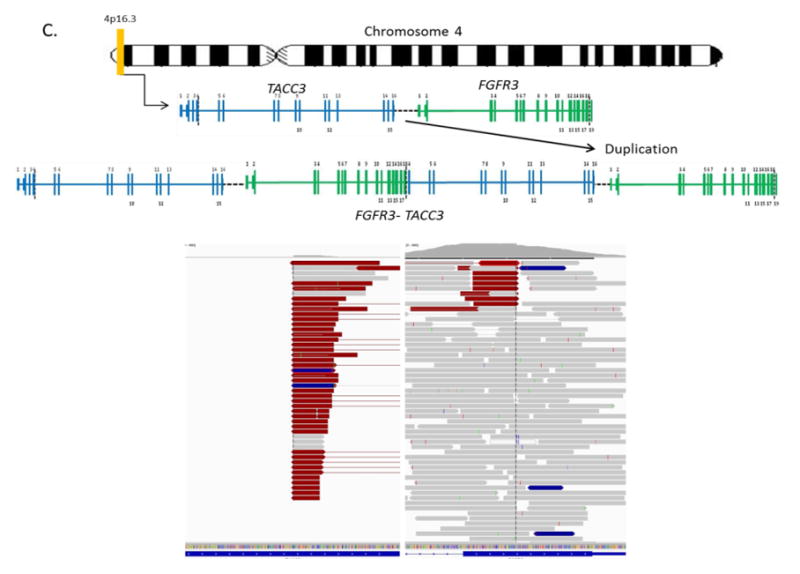
A) The oncoprint shows the most frequently altered genes for 41 high-grade urothelial carcinoma of the bladder patients analyzed by MSK-IMPACT. Alterations were categorized by type, as copy number amplifications (red bars) or deletions (blue bars), missense mutations (green), in-frame mutations (gray), truncating mutations (black) and promoter region mutations (yellow). The number of alterations per patient is depicted in the top bar graph. The number of patients with an alteration in a specified gene is shown in the right bar graph. B) Types of TERT alterations observed in 41 high-grade urothelial carcinoma of the bladder patients. C) FGFR3-TACC3 gene fusion on chromosome 4 (4p16.3). The breakpoints are located on exon 4 of TACC3 and exon 18 of FGFR3.
To confirm that the alterations we detected in urine were representative of bladder cancer, we compared our results to The Cancer Genome Atlas (TCGA) for urothelial carcinoma of the bladder (Figure 4).33 Our cohort and the urothelial carcinoma TCGA were highly concordant. Of the 20 genes mutated in at least 10% of patients in both cohorts, only four genes exhibited statistically significant differences in alteration prevalence between the two studies based on a univariate analysis. TERT (61% vs. 10%, p-value <0.001) and ERBB2 (29% vs. 12%, p-value=0.01) were altered more frequently in our cohort, while CDKN2A (15% vs 41%, p-value =0.02) and CDKN2B (15% vs 37%, p-value=0.02) were altered more frequently in TCGA (Figure 4). The underrepresentation of deletions of CDKN2A and CDKN2B, adjacent genes on chromosome 9, could indicate a reduced sensitivity of our targeted sequencing assay for identifying deletions in particular, due in part to conservative thresholds for calling copy number alterations. However, the enrichment of TERT mutations is completely explained by the inclusion of the TERT promoter in our sequencing panel and its omission from the whole exome sequencing assay utilized by TCGA. FGFR3-TACC3 gene fusions were observed in both TCGA and our cohort at a 2% frequency.
Figure 4.
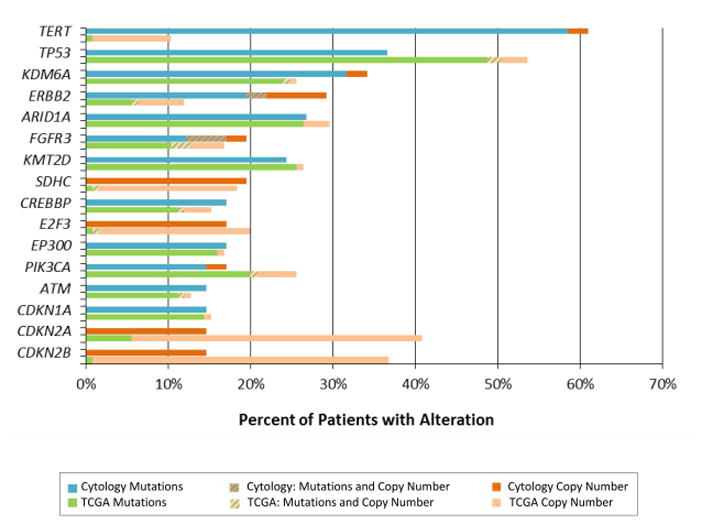
Prevalence of mutations and copy number alterations identified between our high-grade urothelial carcinoma of the bladder patients (cytology cohort) (n=41) and The Cancer Genome Atlas (TCGA) (n=125) high-grade urothelial carcinoma of the bladder cohort.
Correlation of genetic alterations with response to BCG treatment demonstrated differences among the 2 patient categories (Response and Refractory). Genetic alterations in RMB10 and EPHA3 were statistically more frequent in the responder category when compared patients refractory to BCG (42% vs. 10%, p-value=0.04 and 25% vs. 0%, p-value=0.02). No genes were detected to be statistically significantly more common in patients with refractory disease; however genes such as ARID1A, EP300, and CDKN1A were observed more often for these patient compared to responders (35% vs. 8%, 21% vs. 8%, 17% vs 8%).
We found that alterations in Switch/Sucrose Non-Fermentable (SWI/SNF) chromatin remodeling complex, which included genes ARID1A, ARID1B, and SMARCA4, were more common in flat tumor architecture compared to tumors with papillary architecture (59% vs. 21%, p-value=0.03). Other genes associated with flat architecture included TP53 (55% vs. 16%, p-value=0.02), SDHC (32% vs. 5%, p-value=0.05), BRIP1 (23% vs. 0%, p-value=0.05), and NF1 (23% vs. 0%, p-value=0.05). Conversely FGFR3 was altered more often in tumors with papillary architecture (5% vs. 37%, p-value=0.02). Analysis of different tumor stages (pTa, pTis, pT1, T1), demonstrated the presence of alterations in FOXL2 (p-value=0.03) and PTPRS (p-value=0.03) only in patients with pT1 tumor stage.
We also investigated alterations associated with smoking history and found that MDM2 amplifications (0% vs 32%, p-value=0.04) were exclusively found in patients who had a history of smoking. Meanwhile ATM and SMARCA4 were more commonly altered in patients with no history of smoking (39% vs. 4%, p-value=0.01 and 23% vs. 0%, p-value=0.03, respectively).
The comparison within the results between urine cytology specimens and matching FFPE tumor samples from 5 different patients are summarized in Figure 5. Every mutation identified in the FFPE samples was independently called in the corresponding urine cytospin with comparable allele frequency. However, not every mutation called in the urine cytospins was detectable in the corresponding FFPE sample. Only 2/5 patients exhibited 100% concordance between urine and FFPE. In 3/5 patients, at least 3 mutations present in urine were completely absent in FFPE tissue, including known oncogenic mutations in PIK3CA, TERT, and SMAD3 genes. Altogether, we detected 31% more mutations in urine than in FFPE tissue, indicating that urine may more effectively capture the full genetic heterogeneity of a patient’s cancer.
Figure 5.
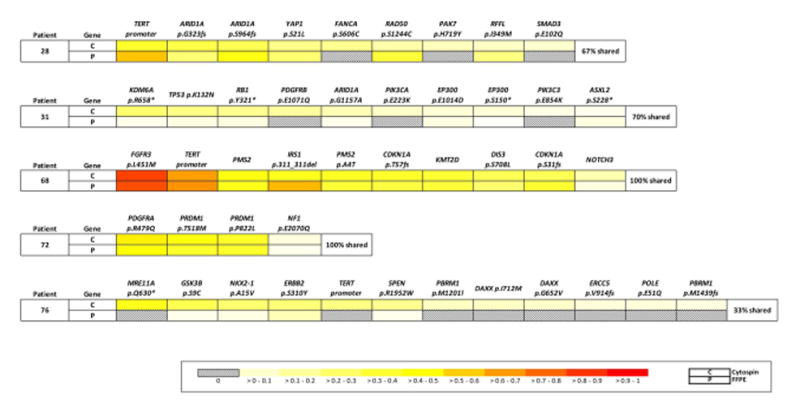
Comparison of mutations between paired urine cytology tumor samples (C) and FFPE tumor samples (P) from 5 patients. Alterations for each patient are ordered based on the variant frequencies of the urine cytology tumor in the first row of each patient row. Mutation frequencies are normalized within each tumor to allow for comparisons across samples, as illustrated through the color formatting key.
Discussion
The approach to the identification of UCB biomarkers in this study is unique as it uses urine cytology specimens for NGS analysis instead of the traditional FFPE or frozen specimens. Urine cytology specimens have several advantages over FFPE material. They are easier to obtain with minimal risk and discomfort to the patient than other conventional tumor collection methods, and they are inexpensive.18, 20 Also, they are more sensitive to detect flat CIS than cystoscopic bladder biopsies.22, 23 Also, urine specimens can, theoretically, capture the full heterogeneity of a patient’s disease more effectively than a single localized tissue biopsy. An important finding in this study was that urine specimens are not only suitable to detect genomic alterations with high accuracy and sensitivity, but seemed to be superior to FFPE. Only about 2% of urine samples in our cohort had an average coverage of below 200x compared to 20% of FFPE samples, which could indicate that FFPE degradation had an effect on the DNA yields and on sequencing coverage. Furthermore, our results demonstrated that urine cytology specimens detected an equal number or more genomic alterations than FFPE specimens in all 5 patients in which tumor tissue was available from the cystectomy specimens. No genomic alteration seen in the FFPE specimens was missed in the urine cytology specimen analysis and, overall, 31% more mutations were seen in the cytology specimens. These results were obtained despite the fact that the specimens used had been in storage for over 10 years due to regulatory reasons that required the cytology slides to remain intact for 10 years. These findings, in conjunction with the better coverage seen in the NGS analysis, confirm that urine cytology specimens can effectively replace FFPE in the genomic analysis of HR NMIUC. It fulfills a major need in the NGS analysis of urothelial CIS as no concurrent biopsy containing urothelial CIS contained sufficient material for NGS analysis.
We compared our cohort to the gene prevalence observed by TCGA.34 Similar to the TCGA findings, we found that the majority of the most commonly altered genes were associated with chromatin remodeling such as KDM6A, ARID1A, KMT2D, FGFR3, CREBBP, and EP300. Another similarity was the detection of FGFR3-TACC3 gene fusions in both TCGA and our cohort. Even though we found mostly similarities, we did observe some statistically significant differences between TCGA and our cohort. The largest difference between our study and the TCGA were that TERT mutations were more frequently found in our cohort, because of the inclusion of the TERT promoter region in MSK-IMPACT, whereas the promoter is absent in whole exome data from TCGA. The incidence of mutations in the TERT promoter region in our cohort is very similar to a prior report of 74% in noninvasive urothelial neoplasms.35 The lower frequencies of alterations in CDKN2A and CDKN2B observed in our cohort may be explained by differential sensitivity for detecting deletions by MSK-IMPACT and array based methods used by TCGA. We cannot explain why ERBB2 alterations are detected more often in our cytological samples, though it is notable that some of these mutations occur in the Furin-like cysteine rich region and the protein tyrosine kinase domain, which are commonly mutated codons in ERBB2 in UCB. Therefore, almost all variations in gene prevalence can be attributed to differences in the gene panels used or differences in pipeline calling criteria
One of the objectives of this study was to search for potential biomarkers that could predict response to BCG in high risk NMIUC. It is currently challenging to distinguish patients with HR NMIUC who will benefit from BCG treatment or early cystectomy. Cystectomies lead to excellent disease free survival rates, but they represent an overtreatment in over 50% of the cases, as not all patients will progress to muscle invasive disease. The current European Urological Association (EUA) guidelines suggest treating high risk NMIUC with BCG and proceed to cystectomy in case of recurrence or progression, although upfront cystectomy remains an option.7, 36 The problem with this approach is that patients who fail to achieve complete remission with BCG treatment have significantly lower survival rates than patients who were treated with upfront cystectomy.37 In our study, 2 genes (RMB10 and EPHA3) were shown to be associated with response to BCG in a univariate analysis. Unsurprisingly, RMB10 alterations have been previously associated with less aggressive urothelial carcinomas when compared to higher grade or invasive UCBs.38 Our analysis showed that the presence of RMB10 alterations was 6 times more likely in patients who responded to BCG treatment (Odds ratio=6). The other gene (EPHA3) associated with response to BCG (p-value=0.02) is part of the Eph receptor tyrosine kinases, which is an important family of signal transduction molecules that control many cellular processes, including cell adhesion and movement, cell shape and cell growth. EPHA3 alterations are found in 5–10% of lung adenocarcinomas,34 and 3% in the TCGA study of urothelial carcinomas. However, EPHA3’s association with other clinical features in UCB has not been previously reported.
Although no alterations were found to be statistically significantly associated with refractoriness to BCG treatment, alterations in ARID1A, EP300, and CDKN1A tended to be more altered in patients who did not respond to BCG treatment. ARID1A is a member of the Switch/Sucrose Non-Fermentable (SWI/SNF) chromatin remodeling complex. The incidence of ARID1A is variable in UCB with an incidence varying from 25–30% in samples evaluated by other investigators,33, 39, 40 except in the series reported by Gui et al., who reported a lower incidence of 13%.41 The low number of ARID1A in patients who responded to BCG (n=1, 8%) in comparison to 35% incidence in refractory patients suggests that ARID1A might be associated with worse prognosis, however statistical significance was not achieved. This finding would agree with the findings by Balbas-Martinez et al. who described an association of ARID1A mutation with higher stage and grade in urothelial bladder tumors leading to a worse prognosis.42 Another genetic alteration more frequently seen in patients’ refractory to BCG, EP300, is histone acetyltransferase gene that has been previously reported to be altered in 13% of patients with bladder cancer.41 The unaltered EP300 gene is believed to have a tumor suppressor gene role in bladder cancer, thus a mutation would silence its role as tumor suppressor gene. CDKN1A is a regulator of progression in G1 and S phases of the cell cycle,43 and is controlled by p53 in response to a variety of stress stimuli. Data reported by Cazier et al strongly suggest that loss of CDKN1A function promotes the growth of bladder carcinomas and may augment defects caused by inactivation of p53.44
The correlation of the genomic results with histological features support previous findings seen in papillary lesions, our results demonstrated a strong association of FGFR3 mutation with a papillary architecture (p-value=0.02). These findings are in line with prior studies using tissue sections that showed the association of FGFR3 mutations and papillary architecture in UCB. FGFR3 mutations in lesions with papillary architecture were first described by Billerey et al in tissue sections from low grade papillary UCB, but more recent studies in tissue sections have demonstrated FGFR3 mutations in both low grade and high grade papillary NMIUC.24, 25, 45–48 A flat (non-papillary) architecture was associated with significantly higher rate of mutations (p-value=0.03) in (SWI/SNF) chromatin remodeling complex genes. In addition to the association with chromatin remodeling genes, other genes such as TP53, SDHC, BRIP1, and NF1 were also found to be significantly more associated with a flat architecture. These findings provide new insight into the molecular classification of UCB with new markers and further support the existence of at least 2 molecular signatures in urothelial carcinomas.49, 50 One associated with FGFR3 alterations and papillary architecture and another one associated with flat architecture, alterations in the TP53 pathway and/or genetic alterations in the SWIF/SNF chromatin remodeling complex. The correlation of the genomic findings with histological findings also found an association between genomic alterations and invasion. Genetic alterations in FOXL2 and PTPRS were identified only in patients with pT1 disease and were absent in all non-invasive UCB in this cohort, suggesting that they might represent markers of progression in UCB. Although these genetic alterations have been previously described in urinary carcinoma, their relationship with stage has not been previously reported.51
Correlation of the smoking history with the genomic results showed that alterations in 3 genes, MDM2, ATM, and SMARC4, were associated with smoking status. MDM2 amplifications were significantly associated with a history of smoking (p-value=0.04), while ATM and SMARC4A, were significantly more associated with a non-smoking history with p values of 0.01 and 0.03, respectively. These findings in MDM2 are in contrast to findings in lung carcinoma in which MDM2 amplifications were not reported to correlate with a history of smoking in a meta-analysis reported by Bai et al.52 Conversely, ATM mutations were significantly more common in non-smoker patients, a finding similar to the ones seen in prior studies evaluating lung and breast carcinoma.53, 54 SMARC4A, another gene in the SWI/SNF chromatin remodeling complex, mutations have been reported in 75% of small cell carcinoma of the ovary; hypercalcemic type mutations but only 3.1% of carcinomas of the urinary tract (COSMIC).51 Its relationship with smoking has not been previously reported.
An area for potential future investigation is the use of targeted therapy in cases of high risk NMIUC as we identified several potentially actionable genetic alterations in ERBB2, CREBBP, and FGFR3 mutations among patients with refractory disease. These mutations were not solely found in patients with disease refractory to BCG, but can potentially play a therapeutic role in the event of refractoriness to BCG treatment. For instance, ERBB2 mutations or amplifications were found in 24% of patients with refractory disease in our series. It would be interesting to evaluate if NMIUC refractory to BCG can potentially be treated with an expanding number of anti-HER2 agents such as Trastuzumab, Lapatinib, Ado-trastuzumab emtansine, and Pertuzumab as suggested by Carneiro et al and Ross et al in cases of advanced UCB.55, 56 Similarly, tumors with activating mutations of FGFR3 might be sensitive to FGFR family inhibitors such as BGJ398.57, 58 Another gene that might be subject to targeted therapy is CREBBP. CREBBP mutations can potentially be treated with Mocetinostat, a potent selective inhibitor of histone acetyltransferases 1, 2, 3, and 11, which is currently in a phase 2 clinical trial for advanced bladder cancer. 59 Additionally, the presence of genomic alterations in the DNA repair-associated genes ATM and RB1, might predict response with cisplatin-based chemotherapy as demonstrated in muscle invasive bladder carcinomas by Plimack et al.60
Despite the promising results of this study, limitations of this study included small study size that compromised our power to detect small effect estimates. Future studies with larger sample sizes are needed to more effectively assess the association of BCG treatment and alterations in ARID1A, EP300 and CDKN1A. Additionally, not all samples were collected from treatment naïve patients as some patients had been treated with intravesical BCG prior to the collection of the specimen analyzed. At this moment, it is unknown what would be the impact of a prior BCG treatment on the genomic profile of the tumor cells.
In summary, the study shows that urine specimens represent a viable, powerful platform for NGS analysis of HR NMIUC. The use of urine specimens would eliminate a major limitation in the analysis of HR NMIUC, which is the lack of sufficient tissue for high coverage NGS analysis. Urine specimens can provide a genomic signature of HR NMIUC that can help the molecular classification of UCB and tailor the treatment of patients with HR NMIUC.
Supplementary Material
Acknowledgments
This research was funded by MSK Core Grant - NIH/NCI Cancer Center Support Grant P30 CA008748, Society of Memorial Sloan Kettering Cancer Center, and Marie Josée and Henry R. Kravis Center for Molecular Oncology.
Footnotes
There are no financial disclosures.
- Sasinya N. Scott: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Writing – original draft, Writing - review and editing, Visualization
- Irina Ostrovnaya: Software, Validation, Formal analysis, Data curation, Writing - review and editing
- Caroline M. Lin: Conceptualization, Formal analysis, Writing – original draft
- Nancy Bouvier: Resources
- Bernard Bochner: Conceptualization, Methodology, Validation, Investigation, Resources, Writing - review and editing, Supervision
- Gopakumar Iyer: Conceptualization, Writing – original draft, Writing – review and editing
- David Solit: Conceptualization, Methodology, Resources, Writing - review and editing, Supervision
- Michael F. Berger: Methodology, Formal analysis, Investigation, Writing - review and editing, Supervision, Project administration
- Oscar Lin: Conceptualization, Formal analysis, Writing – original draft, Writing - review and editing, Visualization, Supervision, Project administration
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Society AC. Cancer Facts and Figures 2016. Atlanta, Ga: 2016. [Google Scholar]
- 4.Surveillance E, and End Results Program. SEER. STAT FACTS SHEETS: BLADDER CANCER. 2013 Available from URL: http://seer.cancer.gov/statfacts/html/urinb.html.
- 5.Montironi R, Lopez-Beltran A. The 2004 WHO classification of bladder tumors: a summary and commentary. Int J Surg Pathol. 2005;13:143–153. doi: 10.1177/106689690501300203. [DOI] [PubMed] [Google Scholar]
- 6.Anastasiadis A, de Reijke TM. Best practice in the treatment of nonmuscle invasive bladder cancer. Ther Adv Urol. 2012;4:13–32. doi: 10.1177/1756287211431976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Babjuk M, Burger M, Zigeuner R, et al. EAU guidelines on non-muscle-invasive urothelial carcinoma of the bladder: update 2013. Eur Urol. 2013;64:639–653. doi: 10.1016/j.eururo.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 8.Sun M, Trinh QD. Diagnosis and staging of bladder cancer. Hematol Oncol Clin North Am. 2015;29:205–218. vii. doi: 10.1016/j.hoc.2014.10.013. [DOI] [PubMed] [Google Scholar]
- 9.Kirkali Z, Chan T, Manoharan M, et al. Bladder cancer: epidemiology, staging and grading, and diagnosis. Urology. 2005;66:4–34. doi: 10.1016/j.urology.2005.07.062. [DOI] [PubMed] [Google Scholar]
- 10.Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013;1303.3997. [Google Scholar]
- 14.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Talevich E, Shain AH, Botton T, et al. CNVkit: Copy number detection and visualization for targeted sequencing using off-target reads. 2014 doi: 10.1371/journal.pcbi.1004873. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robinson JT, Thorvaldsdottir H, Winckler W, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. journal. 2011;17:10–12. [Google Scholar]
- 19.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wagle N, Berger MF, Davis MJ, et al. High-throughput detection of actionable genomic alterations in clinical tumor samples by targeted, massively parallel sequencing. Cancer Discov. 2012;2:82–93. doi: 10.1158/2159-8290.CD-11-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fradet Y, Grossman HB, Gomella L, et al. A comparison of hexaminolevulinate fluorescence cystoscopy and white light cystoscopy for the detection of carcinoma in situ in patients with bladder cancer: a phase III, multicenter study. J Urol. 2007;178:68–73. doi: 10.1016/j.juro.2007.03.028. discussion 73. [DOI] [PubMed] [Google Scholar]
- 23.Yafi FA, Brimo F, Steinberg J, et al. Prospective analysis of sensitivity and specificity of urinary cytology and other urinary biomarkers for bladder cancer. Urol Oncol. 2015;33:66.e25–31. doi: 10.1016/j.urolonc.2014.06.008. [DOI] [PubMed] [Google Scholar]
- 24.Castillo-Martin M, Domingo-Domenech J, Karni-Schmidt O, et al. Molecular pathways of urothelial development and bladder tumorigenesis. Urol Oncol. 2010;28:401–408. doi: 10.1016/j.urolonc.2009.04.019. [DOI] [PubMed] [Google Scholar]
- 25.Diaz De Stahl T, Segersten U, Malmstrom PU. Molecular genetics of bladder cancer: an update. Minerva Urol Nefrol. 2008;60:205–216. [PubMed] [Google Scholar]
- 26.Saint F, Salomon L, Quintela R, et al. Do prognostic parameters of remission versus relapse after Bacillus Calmette-Guerin (BCG) immunotherapy exist? analysis of a quarter century of literature. Eur Urol. 2003;43:351–360. doi: 10.1016/s0302-2838(03)00048-4. discussion 360–351. [DOI] [PubMed] [Google Scholar]
- 27.Cambier S, Sylvester RJ, Collette L, et al. EORTC Nomograms and Risk Groups for Predicting Recurrence, Progression, and Disease-specific and Overall Survival in Non-Muscle-invasive Stage Ta-T1 Urothelial Bladder Cancer Patients Treated with 1–3 Years of Maintenance Bacillus Calmette-Guerin. Eur Urol. 2015 doi: 10.1016/j.eururo.2015.06.045. [DOI] [PubMed] [Google Scholar]
- 28.Barkan GA, Wojcik EM, Nayar R, et al. The Paris System for Reporting Urinary Cytology: The Quest to Develop a Standardized Terminology. Adv Anat Pathol. 2016;23:193–201. doi: 10.1097/PAP.0000000000000118. [DOI] [PubMed] [Google Scholar]
- 29.Won HH, Scott SN, Brannon AR, et al. Detecting somatic genetic alterations in tumor specimens by exon capture and massively parallel sequencing. J Vis Exp. 2013:e50710. doi: 10.3791/50710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J Mol Diagn. 2015;17:251–264. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Network CGAR. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507:315–322. doi: 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cancer Genome Atlas Research N. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507:315–322. doi: 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhuang G, Song W, Amato K, et al. Effects of cancer-associated EPHA3 mutations on lung cancer. J Natl Cancer Inst. 2012;104:1182–1197. doi: 10.1093/jnci/djs297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kinde I, Munari E, Faraj SF, et al. TERT promoter mutations occur early in urothelial neoplasia and are biomarkers of early disease and disease recurrence in urine. Cancer Res. 2013;73:7162–7167. doi: 10.1158/0008-5472.CAN-13-2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang SS, Dinney CP, Donat SM, et al. 12th Annual meeting of the Society of Urologic Oncology (SUO) bladder cancer sessions I and II summary report. Urol Oncol. 2012;30:944–947. doi: 10.1016/j.urolonc.2012.07.006. [DOI] [PubMed] [Google Scholar]
- 37.Lerner SP, Tangen CM, Sucharew H, et al. Failure to achieve a complete response to induction BCG therapy is associated with increased risk of disease worsening and death in patients with high risk non-muscle invasive bladder cancer. Urol Oncol. 2009;27:155–159. doi: 10.1016/j.urolonc.2007.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nordentoft I, Lamy P, Birkenkamp-Demtroder K, et al. Mutational context and diverse clonal development in early and late bladder cancer. Cell Rep. 2014;7:1649–1663. doi: 10.1016/j.celrep.2014.04.038. [DOI] [PubMed] [Google Scholar]
- 39.Wu JN, Roberts CW. ARID1A mutations in cancer: another epigenetic tumor suppressor? Cancer Discov. 2013;3:35–43. doi: 10.1158/2159-8290.CD-12-0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim PH, Cha EK, Sfakianos JP, et al. Genomic predictors of survival in patients with high-grade urothelial carcinoma of the bladder. Eur Urol. 2015;67:198–201. doi: 10.1016/j.eururo.2014.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gui Y, Guo G, Huang Y, et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat Genet. 2011;43:875–878. doi: 10.1038/ng.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Balbas-Martinez C, Rodriguez-Pinilla M, Casanova A, et al. ARID1A alterations are associated with FGFR3-wild type, poor-prognosis, urothelial bladder tumors. PLoS One. 2013;8:e62483. doi: 10.1371/journal.pone.0062483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gartel AL, Radhakrishnan SK. Lost in transcription: p21 repression, mechanisms, and consequences. Cancer Res. 2005;65:3980–3985. doi: 10.1158/0008-5472.CAN-04-3995. [DOI] [PubMed] [Google Scholar]
- 44.Cazier JB, Rao SR, McLean CM, et al. Whole-genome sequencing of bladder cancers reveals somatic CDKN1A mutations and clinicopathological associations with mutation burden. Nature Communications. 2014;5:3756. doi: 10.1038/ncomms4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lopez-Knowles E, Hernandez S, Malats N, et al. PIK3CA mutations are an early genetic alteration associated with FGFR3 mutations in superficial papillary bladder tumors. Cancer Res. 2006;66:7401–7404. doi: 10.1158/0008-5472.CAN-06-1182. [DOI] [PubMed] [Google Scholar]
- 46.Mhawech-Fauceglia P, Cheney RT, Schwaller J. Genetic alterations in urothelial bladder carcinoma: an updated review. Cancer. 2006;106:1205–1216. doi: 10.1002/cncr.21743. [DOI] [PubMed] [Google Scholar]
- 47.Zieger K, Dyrskjot L, Wiuf C, et al. Role of activating fibroblast growth factor receptor 3 mutations in the development of bladder tumors. Clin Cancer Res. 2005;11:7709–7719. doi: 10.1158/1078-0432.CCR-05-1130. [DOI] [PubMed] [Google Scholar]
- 48.Billerey C, Chopin D, Aubriot-Lorton MH, et al. Frequent FGFR3 mutations in papillary non-invasive bladder (pTa) tumors. Am J Pathol. 2001;158:1955–1959. doi: 10.1016/S0002-9440(10)64665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lindgren D, Frigyesi A, Gudjonsson S, et al. Combined gene expression and genomic profiling define two intrinsic molecular subtypes of urothelial carcinoma and gene signatures for molecular grading and outcome. Cancer Res. 2010;70:3463–3472. doi: 10.1158/0008-5472.CAN-09-4213. [DOI] [PubMed] [Google Scholar]
- 50.Goebell PJ, Knowles MA. Bladder cancer or bladder cancers? Genetically distinct malignant conditions of the urothelium. Urol Oncol. 2010;28:409–428. doi: 10.1016/j.urolonc.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 51.Wellcome Trust Sanger Institute. [accessed 2016]; Available from URL: available from: URL: www.cancer.sanger.ac.uk.
- 52.Bai J, Dai J, Yu H, et al. Cigarette smoking, MDM2 SNP309, gene-environment interactions, and lung cancer risk: a meta-analysis. J Toxicol Environ Health A. 2009;72:677–682. doi: 10.1080/15287390902840930. [DOI] [PubMed] [Google Scholar]
- 53.Huang S, Zhang Y, Zeng T. Effect of ATM-111 (G>A) Polymorphism on Cancer Risk: A Meta-Analysis. Genet Test Mol Biomarkers. 2016;20:359–366. doi: 10.1089/gtmb.2015.0320. [DOI] [PubMed] [Google Scholar]
- 54.Lo YL, Hsiao CF, Jou YS, et al. ATM polymorphisms and risk of lung cancer among never smokers. Lung Cancer. 2010;69:148–154. doi: 10.1016/j.lungcan.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 55.Carneiro BA, Meeks JJ, Kuzel TM, et al. Emerging therapeutic targets in bladder cancer. Cancer Treat Rev. 2015;41:170–178. doi: 10.1016/j.ctrv.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 56.Ross JS, Wang K, Al-Rohil RN, et al. Advanced urothelial carcinoma: next-generation sequencing reveals diverse genomic alterations and targets of therapy. Mod Pathol. 2014;27:271–280. doi: 10.1038/modpathol.2013.135. [DOI] [PubMed] [Google Scholar]
- 57.Abbosh PH, McConkey DJ, Plimack ER. Targeting Signaling Transduction Pathways in Bladder Cancer. Curr Oncol Rep. 2015;17:58. doi: 10.1007/s11912-015-0477-6. [DOI] [PubMed] [Google Scholar]
- 58.Palma N, Morris JC, Ali SM, et al. Exceptional Response to Pazopanib in a Patient with Urothelial Carcinoma Harboring FGFR3 Activating Mutation and Amplification. Eur Urol. 2015;68:168–170. doi: 10.1016/j.eururo.2015.02.023. [DOI] [PubMed] [Google Scholar]
- 59.De Souza C, Chatterji BP. HDAC Inhibitors as Novel Anti-Cancer Therapeutics. Recent Pat Anticancer Drug Discov. 2015;10:145–162. doi: 10.2174/1574892810666150317144511. [DOI] [PubMed] [Google Scholar]
- 60.Plimack ER, Dunbrack RL, Brennan TA, et al. Defects in DNA Repair Genes Predict Response to Neoadjuvant Cisplatin-based Chemotherapy in Muscle-invasive Bladder Cancer. Eur Urol. 2015;68:959–967. doi: 10.1016/j.eururo.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All associated clinical data were obtained from prospectively maintained medical records. Genomic and clinical data are publicly available through the MSKCC cBio Portal for Cancer Genomics.21, 31