Abstract
RNA polymerase (RNAP) is the central motor of gene expression since it governs the process of transcription. In prokaryotes, this holoenzyme is formed by the RNAP core and a sigma factor. After approaching and binding the specific promoter site on the DNA, the holoenzyme‐promoter complex undergoes several conformational transitions that allow unwinding and opening of the DNA duplex. Once the first DNA basepairs (∼10 bp) are transcribed in an initial transcription process, the enzyme unbinds from the promoter and proceeds downstream along the DNA while continuously opening the helix and polymerizing the ribonucleotides in correspondence with the template DNA sequence. When the gene is transcribed into RNA, the process generally is terminated and RNAP unbinds from the DNA. The first step of transcription–initiation, is considered the rate‐limiting step of the entire process. This review focuses on the single‐molecule studies that try to reveal the key steps in the initiation phase of bacterial transcription. Such single‐molecule studies have, for example, allowed real‐time observations of the RNAP target search mechanism, a mechanism still under debate. Moreover, single‐molecule studies using Förster Resonance Energy Transfer (FRET) revealed the conformational changes that the enzyme undergoes during initiation. Force‐based techniques such as scanning force microscopy and magnetic tweezers allowed quantification of the energy that drives the RNAP translocation along DNA and its dynamics. In addition to these in vitro experiments, single particle tracking in vivo has provided a direct quantification of the relative populations in each phase of transcription and their locations within the cell.
Keywords: RNAP, transcription initiation, single‐molecule studies, promoter search, closed to open complex, abortive initiation, promoter escape, intermediates, conformational changes
Introduction
RNA polymerase (RNAP) is the biological motor responsible for the fundamental catalytic activity of RNA synthesis. RNAP is a DNA–binding enzyme that targets promoter sequences within the DNA and at these locations it is capable of locally melting the DNA (initiation phase; Fig. 1). After these initial steps, RNAP can start to continuously convert chemical energy [in the form of nucleoside triphosphates (NTPs) bond hydrolysis] into direct motion accompanied with continuous synthesis of RNA (elongation phase). RNAP translocates along the DNA to read the gene until the corresponding RNA chain is transcribed, and eventually it releases the products and unbinds from the DNA (termination phase). These steps of initiation, elongation, and termination describe the general working mechanism of RNAP. The different RNAPs from the three domains of life however do exhibit substantial variations within specific steps of this general working mechanism.1 Here, we focus on prokaryotic RNAP. The bacterial RNAP has a known structure formed by a core, which in all bacteria is made of five subunits, αΙαΙββ’ω. The core forms a complex with one of seven different σ factors,2 resulting in the “holoenzyme” complex, which is the active form capable of transcription in the cell. Each σ factor directs the RNAP toward a specific subset of promoters. Regulation of the pool of σ factors thus allows expression of specific genes relevant to the environmental condition.3 Among these factors, σ70 has the highest affinity for the RNAP core. It is the most abundant (>5‐fold respect to the other factors),4 recognizes the majority of the genes and is therefore known as the “housekeeping” σ factor. Moreover, σ70 represents the family of factors that are able to recognize promoters and initiate transcription without any cofactors or energy source. The σ54 family members, on the other hand, require the presence of enhancers (e.g., ATPase) that provide the complex with energy from hydrolysis of ATP/GTP. σ54 is responsible for expression of the genes related to nitrogen metabolism and stress response.5 In addition, other binding proteins also regulate the complex formation and its activity, as well as fluctuations of the initial availability of NTPs.6 Note that in the text we will indicate clearly what results applies to σ54 and σ70. RNAP activity is furthermore modulated by cellular features such as the local structure of the nucleoid (e.g., supercoiled DNA) and the presence of nonspecific DNA‐binding proteins (by e.g., macromolecular crowding).7
Figure 1.
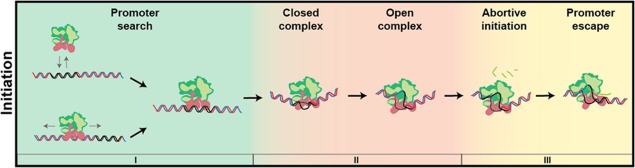
Transcription initiation scheme: This review focuses on the initiation process and how its intermediates have been recently unraveled by single‐molecule techniques. RNAP initiation is divided into three substeps that correspond to the three subsections of this review: I: the promoter search process (green background), II: the transition from the closed complex to the open complex (red background), and III: the mechanisms of initial transcription and promoter escape (yellow background). Based on ref 11.
The main steps of the bacterial transcription process have been revealed thanks to ensemble averaging data from biochemical studies.8 In parallel, structural studies yielded insight in the shape and configuration of RNAP as well as conformational transitions.9 With the advent of single‐molecules techniques, such as single‐molecule FRET, colocalization single‐molecule spectroscopy, magnetic tweezers combined with microscopy, optical tweezers combined with fluorescence microscopy, DNA curtains, and scanning force microscopy, the dynamics and mechanics of transcription have been elucidated in great detail. These methods allowed the unraveling of transient intermediates and rare events, their real‐time kinetics and possible concurrent reaction pathways. Furthermore, these methods allow direct analysis and control of RNAP activity by manipulating the DNA and monitoring in real‐time its behavior under different chemical conditions.
This review will provide an overview of such single‐molecule studies on transcription initiation of the E. coli RNAP. The three subsections of this review correspond to the three main steps of the initiation process that RNAP undergoes before processive RNA transcription can occur (see scheme in Fig. 1): (1) promoter search, (2) transition from the closed to the open complex, and (3) from abortive initiation to promoter escape. For insights into the resulting elongation process observed with single‐molecule experiments, we refer to excellent dedicated reviews such as that by Larson et al.10
Promoter search
The initiation process begins with nonspecific DNA binding, search for a promoter site and sequence recognition. The initial complex between the RNAP and the promoter has limited stability. The holoenzyme contacts the DNA double helix and when it recognizes the two hexamers of consensus DNA sequence, it forms the closed complex (RPc).11 The RPc can interact with promoters owing to the binding sites present at three specific σ factor regions.11 In general, there are five promoter elements upstream the gene coding region (indicated as +1bp element) that a σ factor can distinguish, depending on its structure.12 The main recognized promoter regions vary in length from 2 to 6 bps (e.g., TATAAT‐ or TTGACA‐sequence at −10 and −35 element, respectively). Since the length of the RNAP target site is 5 orders of magnitude shorter than the complete bacterial genome, the probability of direct binding (within 1bp from the promoter) seems to be limited. It is generally acknowledged that specific DNA‐binding proteins can accelerate the rate of target sequence binding thanks to a “facilitated diffusion” (FD) mechanism: initial binding at a random non‐specific DNA site, followed by sliding (1D diffusive excursions) along the DNA and/or translocation through hopping/jumping/intersegmental transfer.13 However, the RNAP promoter search mechanism is still under debate. For example, recent single‐molecule observations have reported that bacterial RNAP locates its target through direct binding from solution (3D diffusion), in contradiction with previous bulk studies and single‐molecule observations that favor the concept of FD.
The first visual examination of single RNAP molecules moving over DNA was performed on DNA stretched by dielectrophoresis and anchored to the surface at both ends by Kabata et al.14 Fluorescently labeled RNAP was observed moving both along λ‐phage DNA and T7 DNA. The observation of 40% of the molecules that deviate from the direction of the bulk flow was classified as RNAP moving on the extended DNA. Direct high resolution evidence of RNAP sliding along DNA was obtained by sequentially imaging with atomic force microscopy (AFM) in liquid by Guthold et al.15 Single σ70 RNAPs were observed both moving (D1D=1·10−13 cm2/s) [Fig. 2(A)], with a non‐specific binding lifetime of 600 s, and transferring between DNA fragments. The large lifetimes (∼2–3 order of magnitudes higher than was previously reported) was explained as a possible effect of surface interactions. Such surface effects can lead to a higher activation energy for conformational changes before dissociation as well as a presumed rapid re‐association induced by the vicinity of the two molecules on the surface. These surface interaction effects preclude direct comparisons to bulk kinetics studies.
Figure 2.
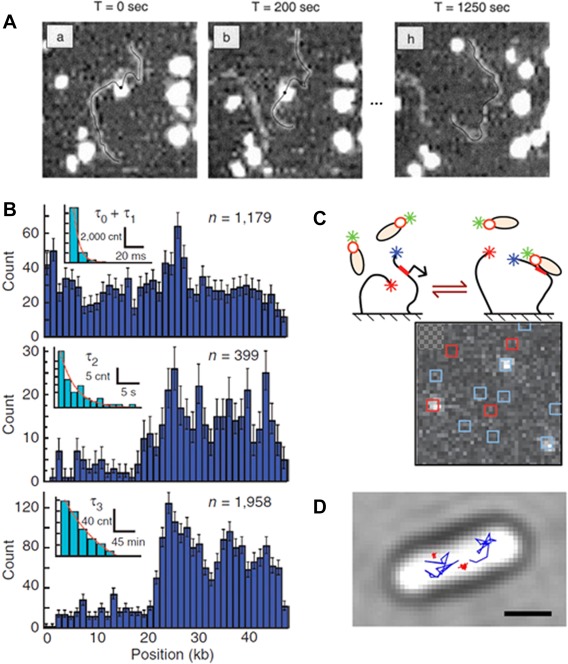
The promoter search mechanism. A: AFM was applied to observe one‐dimensional diffusion of RNAP along DNA. Consecutive images (here at t = 0, 200, 1250 s) were acquired after initial observation of the RNAP‐DNA complex formation. RNAP molecules (white spot) appeared to slide back and forth on the DNA (black line). Reproduced from Ref 15. B: DNA curtains combined with fluorescence microscopy allowed to quantify the distribution of binding positions along λ‐DNA (dark blue histograms) and their binding lifetime (light blue, insets) for three kinetically distinct populations: fast and sequence nonspecific (top), intermediate at λ AT‐rich regions (middle), and long at promoter regions (bottom). Reproduced from Ref 17. C: Schematic of the single‐molecule constructs used for CoSMos experiments. Surface‐anchored DNAs (black lines) and σ54‐RNAP complexes are shown before and after binding. The labeling was set on the RNAP (green star), promoter‐containing DNA (blue star on DNA) and promoter‐free DNA (red star on DNA). The colocalization of the fluorophores was examined in consecutive images allowing to follow the RNAP complex binding kinetics. Reproduced from Ref 18. D: Single‐molecule tracking of individual RNAPs enables distinction of mobile (blue) and DNA‐bound molecules (red). The image shows sample trajectories observed in vivo. Reproduced from Ref 20.
In the same year, Harada et al. showed σ70 RNAP (Cy3‐labeled) sliding along DNA in the absence of surface interactions by combining total internal reflection fluorescence microscopy (TIRFM) with optical manipulation of DNA.16 Individual binding and sliding events were quantified on single λ‐phage DNA molecules suspended in solution between optically trapped beads. The inhomogeneity of binding along the DNA revealed that RNAP binds more frequently to regions rich in adenine and thymidine. At these AT‐rich sequences, RNAP dissociation exhibits a fast and a slow dissociation rate. Experiments showed that RNAP mostly slides along DNA within the region characterized by fast dissociation (with D = 10−10cm2/s). RNAP molecules dissociated slowly from the positions of the promoters (and sequences common to promoters) at a rate of 0.66 s−1. This rate is several times slower than the binding lifetimes at nonspecific locations. Fluorescence observations were performed under low but significant DNA tension (∼5 pN). When DNA was relaxed, in a more physiological condition, the specific binding at promoter sequences was enhanced.
In contrast, a more recent study by Wang et al. revealed no evidence for microscopic 1D diffusion (<0.5% of molecules). σ70‐RNAPs tagged with 25 nm diameter quantum dots were visualized by TIRFM on DNA curtains that were stretched over the surface of a lipid bilayer.17 Three populations of DNA‐binding events by RNAP were found with different binding lifetimes (∼0.03 s, ∼3 s, ∼6000 s) associated, respectively, with nonspecific interactions and, at promoters regions, closed and open complexes [Fig. 2(B)]. The authors suggest that prior reports of extensive sliding may have been confounded by aggregates of RNAPs. It was concluded that in this assay promoter search is dominated by 3D diffusion, the effect of which is further enhanced at high RNAP concentration when the probability of a direct collision increases.
In line with the findings by Wang et al. for Qdot‐tagged σ70‐RNAP, Friedman et al. found that the enhancer dependent σ54‐RNAP also approaches its target primarily through a 3D diffusion process.18 The RNAP complex was kept in conditions such that it stays in the closed complex state, and the experiments were performed using colocalization single‐molecule spectroscopy (CoSMoS) multiwavelength fluorescence microscopy [Fig. 2(C)] on short surface‐tethered DNA molecules of 800–3500 bp. The study compares the binding kinetics on promoter and promoter‐free DNAs, showing that RNAP binds more frequently and 10‐fold faster on promoter DNA (with 90% of these events as promoter‐specific binding events). This observation is in conflict with the FD model, which predicts approximately identical rates of binding to promoter‐containing and promoter‐free DNA. Furthermore, the impact of the length of non‐specific DNA flanking the promoter sequence was tested. This experiment revealed a significant increase of short‐lived (nonspecific) binding events on DNA that was increased in length up to 3kbp, but no enhanced frequency of closed‐complex formation (i.e., long‐lived events). These results suggest FD does not play a significant role for σ54‐RNAP in the range of tested DNA lengths near physiological salt conditions. The authors further noted that previous studies, that showed evidence for sliding and that were conducted with σ70 at nonphysiological low ionic strength, did not directly show that sliding is followed by promoter capture.
A recent single‐molecule method allows imaging labeled RNAPs in live cells by photoactivation and subsequent localization of single fluorophores. Bakshi et al. used superresolution fluorescence microscopy for localization and real‐time tracking of single RNAPs in the cell.19 Following each localization over a sequence of frames visualizes, the trajectory of each complex. Observations revealed four RNAP populations associated with different activity states. Subdiffusive trajectories were related to “slow” RNAPs that are stably bound to DNA. These events (∼50% of the observed proteins) are RNAPs engaged in transcription (at one of the stages among initiation, elongation, pausing, or termination). The other half of the observed trajectories described as ‘mixed‐state, shows the interconversion of the RNAP between two states. The fast transition between the nonspecifically bound (∼30%) and the free (∼10%) RNAPs occurred with an effective diffusion constant of 0.21 um2/s. These transitions happen on the 100 ms‐timescale, suggesting that the time RNAPs spend on nonspecific sequences at physiological salt conditions is shorter than what was observed in in vitro studies. Imaging conditions for observation of the longer‐lived events (7 s) showed transitions between the slow and the mixed‐state (stop‐go or go‐stop, double transitions are not clearly observed). This work provides the first direct partitioning estimation of RNAP fractional populations in vivo.
More recently, RNAPs were observed in a homogenous distribution across the nucleoid and interacting with the DNA through random non‐specific binding during the promoter search process20 [Fig. 2(D)]. Stracy et al. found that the more densely packed is the DNA region, the higher is the concentration of mobile RNAPs. RNAP can thus find promoters located throughout the nucleoid: two populations were detected with 48% of bound and 52% of mobile RNAPs, in agreement with Bakshi et al.19 Inducing chromosome decompaction results in an increase of RNAPs mobility (with a 2‐fold higher diffusion constant), probably due to the movement between more distant available DNA strands. Since the apparent diffusion constant is one order of magnitude smaller than the expected free diffusion constant, RNAPs transiently bind to DNA and subsequently unbind back to solution. The bound RNAP diffusion constant was measured to be 0.11 um2/s and they estimated that RNAP spends 85% of its promoter search time (30–120s) nonspecifically bound to DNA.
The mechanism by which RNAP finds its promoter sites remains a highly debated issue. In vivo recent studies followed RNAPs motion in the nucleoid where RNAP has to find promoters on a 4.6Mbp circular chromosome that is ∼104‐fold compacted in comparison to the linear DNA used in in vitro experiments. In the cell, RNAP was found to be homogenously distributed across the nucleoid, with half of the molecules statically bound to the DNA, and the other half mobile (free in solution or non‐specifically bound to DNA). The first in vitro single‐molecule experiments could directly observe the movement of bound RNAPs, providing evidences of sliding along DNA (however, promoter engage after diffusing has not been demonstrated yet). However, some later studies claim the 3D diffusion as the main (and only relevant) mechanism involved in the promoter search process. Thus, at this point, without more experimental results, no definitive conclusion can be drawn about the nature of the search, at least for σ70.
From closed to open complex
After binding to a promoter and forming the closed complex (RPc), RNAP unwinds one turn (∼14 bp) of DNA at the transcription start site, resulting in a ssDNA “bubble” enclosed inside the core. Subsequently it forms the catalytically active open complex (RPo). For this transition to occur, the RPc needs to undergo several conformational changes that leads to the DNA relocation, unwinding and bending within the RNAP core pocket.11 The holoenzyme is a stretchable structure, especially the two pincers (β and β′ domains) of the clamp. The transcription bubble was recently found to be flexible as well: it expands upon scrunching of the DNA inside the catalytic center as the DNA is pulled in the core, and later closes upon relaxation when it is released from the core during the promoter escape phase.21
The first single‐molecule evidence of the DNA bending during RPo formation was reported by AFM experiments by Rees et al. for σ70 RNAP22 (and by Rippe et al. for σ54 RNAP23), where a reduction in the DNA contour length suggested the DNA wrapping. High‐resolution AFM, together with biochemical experiments, was employed by Rivetti et al. to study the open complex of σ70 holoenzyme at the λPR promoter.24 The AFM images showed a shortening (∼30 nm, ∼90 bp) of the DNA contour length (in respect to free DNA) together with a bending of ∼60°–70° when the open complex is formed. According to these observations, the DNA wraps ∼300° around the RNAP surface [Fig. 3(A)]. Moreover, the inactive complexes did not show such variations, suggesting that these conformational changes are necessary for transcription activation. Analysis of the RPo structure quantified the DNA wrapping to 2/3 upstream the transcription site; in this configuration, the DNA opening would happen near the active cleft. Authors hypothesized that the DNA wrapping has several advantages: it maximizes the contact area between DNA and the protein (since the DNA‐RNAP contact is twice the length of the protein), therefore it could facilitate the DNA untwisting and introduce energetic benefits to induce the promoter escape.
Figure 3.
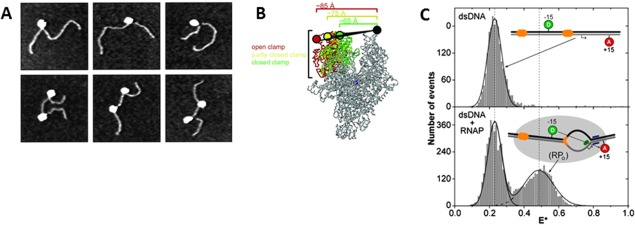
From the closed complex state to the formation of the open complex. A: AFM images of open complexes on DNA templates with one RNAP bound (upper row) or two complexes bound to promoters (bottom row). The impact of RNAP binding on DNA shape and length suggested the DNA wrapping around the enzyme. Reproduced from Ref 24. B: smFRET measurements to determine the RNAP clamp conformations during the initiation phase. The fluorescent probes were incorporated at the tips of β and β' subunits. The picture shows conformational differences between the open (red), closed (yellow) and “collapsed” (green) clamp states. Reproduced from Ref 26. C: Detection of the open complex formation with smFRET. The donor and acceptor fluorophores were placed on both sites of the transcription bubble (at −15 and +15 position). The ratio E* represents the measured FRET that occurred upon formation of the open complex. The bottom plot shows that upon DNA opening another conformational state is introduced in the FRET signal. Reproduced from Ref 28.
Revyakin et al. observed the same DNA compaction by stretching torsionally constrained DNA with magnetic tweezers while monitoring in real‐time the DNA end‐to‐end distance.25 The magnetic‐induced bead rotations allow to introduce negative/positive superhelical turns that correspond to a linear change in the end‐to‐end distance of the 4‐kbp DNA tether, containing a single promoter. Local structural rearrangements at the RNAP‐DNA complex can be in turn converted into a tens‐of‐nanometer detectable bead movement, allowing a direct observation of DNA unwinding and compaction due to the RNAP action in wrapping and/or bending the DNA (with a space resolution of ∼1 bp and ∼5 nm for the unwinding and compaction observations, respectively). They quantified a 13 bp‐unwinding that corresponds to the values extrapolated from DNA footprinting, while the DNA compaction correspond to what previously observed with AFM by Rivetti et al.24 Moreover, the real‐time measurements allowed to quantify the rate of formation of the unwound complex (0.3–1 s) and the rate between unwinding and rewinding events (0.03–1 s) (both consistent with conventional experiments). The RNAP unwounding rate and the stability of the formed complex are found to be strongly affected by the DNA supercoiling degree. The presented DNA nanomanipulation mimic the in vivo DNA supercoiled condition and it directly showed how RNAP activity is profoundly affected by the DNA mechanical state.
Ten years later Chakraborty et al. monitored in real‐time the conformational changes of the σ70 RNAP clamp during DNA opening by employing smFRET.26 Donor and acceptor were placed on the two pincers of the RNAP clamp (β and β′). The RNAP core and the holoenzyme were found to have three states when free in solution [Fig. 3(B)]: the majority of molecules belong to an open clamp with the active‐center cleft wide enough to accommodate dsDNA. The two other subpopulations were identified as a closed clamp (with a cleft able to accommodate only ssDNA) and a “collapsed” closed clamp (insufficiently wide for any DNA). The clamp stays open when the RNAP is in the closed complex state and the subsequent intermediates states. At the RPo stage, RNAP unwinds DNA in the active‐center cleft and the clamp is now found in one of the closed conformations. Direct interaction between the DNA inside the active‐center cleft and the clamp seems to induce the closure process. With NTPs in solution, the initial transcribing complex (ITC) is followed by the promoter‐escape and the complex did not reveal any structural rearrangement of the clamp during this transition. Importantly, the stability of the open complex and of the following elongation complex are directly related to the pincers closure stability.
In the open complex state, the RNAP enzymatic activity changes depending on the stage of the process, possibly due to different interactions within the promoter. This variation is connected with a flexibility in the position of the DNA bubble in respect to the active center of the RPo (mechanistic model).27 Robb et al. also used smFRET on freely diffusing RNAP in solution to investigate the DNA flexibility in respect to the RNAP active site during the open complex stage.28 This was the first experimental observation of the conformational dynamics in the transcription bubble of the RPo [Fig. 3(C)]. In this study, the FRET signal between donor and acceptor probes was quantified for different promoter regions: the conformational fluctuations (ms timescale) in the bubble's apparent length and width showed the formation of the bubble itself and a consequent start‐site variation. The flexibility of the promoter sites was tested by introducing sequence modifications and could hence confirm the mechanistic model where the bubble relocation in the RPo proceeds through a DNA scrunching mechanism.
The conformational changes of the σ54 holoenzyme in the initiation steps were characterized by Friedman et al.29 and this study provided the first quantitative visualization of its initiation pathway. The RNAP‐σ54 complex stays in the RPc conformation until ATPase or NtrC are added to trigger the RPo formation. As a consequence, these two conformational stages are easily controlled and separated. The two previously unknown intermediates of unstable closed complexes (short‐lived and long‐lived RPc with the latter containing melted DNA) were found to preceed the rate‐limiting step at which an enhancer triggers the conformational change to open complex. The short‐lived closed complex is obligatory for the long‐lived closed complex formation, which is, in turn, a required forerunner of the open complex. This last conformational change requires, on average, 30 attempts of promoter binding, so most of the time the long‐lived closed complex does not mature into an open‐complex. When RNAP climbs the necessary energetic barrier (ΔG 0>+8.4 kJ/mol from ATP/GTP hydrolysis), and the NTPs are available, the open complex starts to transcribe, after ∼8 s.
Experimental analyses at the structural and single‐molecule level have identified conformational intermediates of RNAP in presence and absence of DNA. When in solution, RNAP exists in different conformational states. Once bound to DNA, clamp closure results from a direct interaction with the DNA inside the active‐center cleft. Stable closure of clamp and pincers is necessary for transition to the open complex. In addition, DNA‐bubble dynamics can directly affect gene regulation by impacting initiation frequencies and transcription starting point locations.
From abortive initiation to promoter escape
Once the RNAP opens the DNA, the complex is ready to catalyze the formation of the first RNA bonds, and formation of the ITC. The NTPs enter the catalytic center of the enzyme through the secondary channel of RNA. Once a RNA chain is synthesized, it is extruded through the RNA exit channel. This operation requires a conformational change consisting of displacing the 3.2 region of the σ factor and its C‐terminal region. The primary transcription occurs while the RNAP is still bound to the promoter, and therefore requires scrunching of the DNA within the core.30 Interestingly, often the process does not proceed directly to elongation, but the synthesis aborts after forming short RNA products (∼2–10 nt). The short RNA transcripts are released from the core and the polymerase translocates backwards to recreate again the open complex. The process of abortive initiation can be interrupted by pauses, the duration of which depends on the length of the initial transcript.31 Finally, after multiple attempts, the RNAP synthesizes an RNA product of 8 nt, or longer, and successfully enters the elongation phase. The motor unbinds from the promoter site and proceeds to move downstream the coding sequence while continuously unwinding and opening the DNA duplex, and simultaneous polymerizing NTPs. The number of abortive initiation cycles before promoter escape depends on sequences of the promoter and the initial transcribed region.32 After promoter clearance, the σ factor is no longer necessary and therefore might be released from the complex. However, the σ factor can also be retained in the early elongation complex and induce promoter‐proximal pausing events.33
The initial synthesis of RNA has been extensively studied in various bulk assays to determine the major catalytic sites, structure or mechanism.9, 32, 34, 35, 36, 37, 38 Unfortunately, these studies did not resolve the timescale and productivity of the abortive initiation process, the moment of promoter escape, or the occurrence of pausing events. FRET has been the most common single‐molecule approach to study the abortive initiation to promoter escape phase. The method employed relies on the observation of the RNAP translocations with respect to DNA.39 The first single‐molecule results were published by Margeat et al.40 Alternating‐laser excitation and TIRFM were used to image the abortive complexes and the process of promoter escape of immobilized complexes. This approach allowed real‐time observations of single transcription complexes in many iterations of abortive initiation. At saturating concentrations of NTPs, the complex spends the majority of its time in the state in which the RNAP leading edge translocates forward relative to DNA. The RNA product synthesis and the forward translocation of the edge was found to be fast. In stark contrast, the dissociation of the abortive products and reverse translocation of RNAP were relatively slow, implying this step is rate‐limiting to the whole initiation phase.
In the same year Kapanidis et al. used the similar experimental methods to observe the RNAP‐active‐center translocation mechanism.30 Different locations of the FRET pairs were used to validate and/or distinguish between the three possible mechanisms of early transcription: inch‐worming (detachment and translocation of the active center only), transient excursions (translocation by 1 bp per added NTP) and scrunching (contraction of the DNA inside the active center). smFRET experiments provided evidence for the latter model. The scrunching model postulates net changes in the DNA unwinding within the core: the RNAP pulls downstream DNA into itself, which requires expansion of the transcription bubble. The authors suggest that the evidence in favor of the scrunching model is a proof of the previously hypothesized stressed intermediate.41 Such unstable intermediate state has an elevated internal energy that could serve as the driving force for initiation.
A parallel study published by Revyakin et al.,21 used magnetic tweezers to also look into this process. The same group had previously proven that magnetic tweezers can detect changes in DNA end‐to‐end distance that correspond to unwinding of 1bp or compaction of 5nm of DNA25 [Fig. 4(A)]. Again, the model of translocation that corresponded to the observed results was the scrunching mechanism. Observation of positively and negatively supercoiled DNA allowed evaluation of the compacting and unwinding forces; the results demonstrated the existence of an obligatory stressed intermediate with approximately one turn of DNA unwinding. The scrunching of the DNA requires breakage of a base‐pair interaction and the synthesis of an RNA product of >2 nt long. The free energy from one hydrogen bond is ∼2 kcal/mol per bp,42 thus it is possible that at a typical promoter a total of ∼14 to 18 kcal/mol of energy is accumulated in the stressed intermediate. This energy is relatively high in respect to the free energy involved in the RNAP‐promoter interaction (∼7–9 kcal/mol) and in the RNAP‐σ70 complex formation (∼13 kcal/mol), therefore it could be the energy needed for transition to the elongating complex.43, 44 Interestingly, scrunching was also observed for productive initiation, when the more stable elongation complex has been formed: 80% of the RNAPs scrunched DNA for >1s. The compaction probably occurs in all or nearly all transcription cycles and may be obligatory for the promoter escape.
Figure 4.
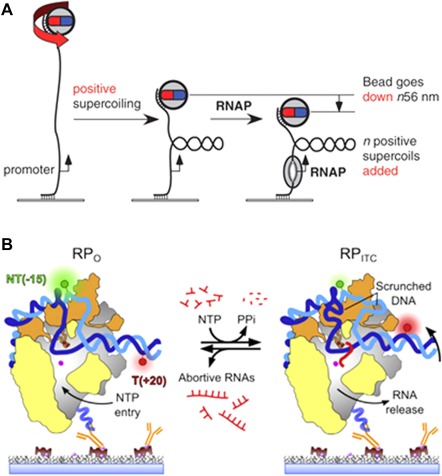
Experimental approaches to study the abortive initiation and promoter escape processes. A: Magnetic tweezers experiments allow real‐time manipulation of single DNA molecules. By rotating the beads attached at one end of the DNA tether a promoter‐containing DNA was supercoiled. Subsequently, the unwinding by RNAP at the promoter could be monitored since it induces gain or loss of supercoils (depending on the coiling direction). A quantification of the end‐to‐end distance of the DNA allows detection of single unwinding turns (56 nm). Reproduced from Ref 21. B: smFRET study of surface‐immobilized RNAP complexes. The initial FRET efficiency in the open complex (left) is low; when NTPs are added, the DNA scrunching within the core moves the acceptor (red) towards the donor (green) corresponding to a FRET efficiency increase. Reproduced from Ref 45.
A recent study using smFRET was published by Duchi et al.45 Contrarily to the previous FRET studies that were limited by low temporal resolution and short observation times, this study allowed the observation of RNAP pausing during initial transcription [Fig. 4(B)]. Authors report highly stable scrunched states and pausing, and evidence for relaxation of the complex by RNA backtracking during abortive initiation. The major determinant that stabilizes the complex is σ70 region 3.2 that partially occupies the RNA exit channel. The RNA clashes with this sigma loop once the first 5–6 nt are transcribed, while ∼20 s pauses occur when RNA is already 6–7 nt long. The backtracked RNAP cannot perform synthesis since the active site is blocked by the 3' end of the RNA (which can later lead to loss of the RNA). Interestingly, the results also revealed another pause, right before the promoter escape. At that point the maximal scrunching is expected to be reached, therefore the pause could be related to the destabilization of contacts between sigma region 4 and −35 promoter element, and complete displacement of the 3.2 region from the RNA exit channel. The proposed initiation model assumes that pausing is controlled by modulation of the 6 to 7 nt RNA transition. The formation of a 7‐mer transcript stabilizes the ITC and triggers translocation.
Finally, a recent article by Lerner et al.31 reports the occurrence of backtracked pausing as well. The study used smFRET kinetic assays and magnetic tweezers to investigate backtracking pausing that can occur at different stages of initial transcription. The smFRET study revealed diverse kinetics of pausing dependent of the RNA length: the exit kinetics of RNAP for 4,6 or 7‐mer was slower than for 2nt long RNA, which was in turn similar to the promoter clearance kinetics (RNA of 11 nt). The magnetic tweezers experiments focused on the pause‐backtracked intermediate and the effect of stress‐like conditions. The population of paused complexes decreases in response to an induced imbalance in NTPs concentration. Authors proposed a modified transcription model: with the addition of a “slow abortive release” pathway that can occur when the seventh nucleotide is incorporated. After the complex enters the paused‐backtracked state it can either proceed to translocate to the open complex with slow backtracking steps, or cleave off the RNA chain within the secondary channel. This can result in ITC complex with 5 nt, that can either release the abortive product or restart the polymerization.
The dynamics of promoter escape has been elucidated by a range of single‐molecule studies: smFRET studies confirmed the DNA scrunching mechanism that results in a stressed intermediate that serves as potential driving force for abortive initiation. Mechanical manipulation and supercoiling measurements, however, have quantified relevant forces and DNA unwinding rates. While single‐molecule analyses revealed how abortive initiation is interrupted by backtracked‐pauses and that release of the abortive products depends on transcript lengths.
Discussion
Single molecule methods have contributed significantly to our understanding of bacterial transcription. Here, we reviewed in particular those studies that have addressed the initiation process of transcription, with focus on (1) promoter search, (2) closed to open complex formation, and (3) abortive initiation and promoter escape.
In promoter search studies, conflicting in vitro datasets continue to fuel controversy regarding the impact of facilitated diffusion (FD) on the rate of promoter binding. Despite exquisite abilities of single‐molecule methods to qualify and quantify 1D and 3D search processes,14, 15, 16, 17, 18 the required artificial in vitro conditions are a potential source of controversy. A key question is to what extent FD and 1D‐sliding along DNA contribute to promoter approach processes in vivo. In vivo (superresolution) fluorescence microscopy studies now start tackling these questions, yielding quantitative clues, such as the relative cellular populations of RNAP of different mobilities,19, 20 that link our in vitro understanding to the realistic situation in the cell.
Several experiments have aimed to analyze the conformational dynamics during transcription, which is of particular significance during the series of events that make up transcription initiation. Conformational intermediate states, both of RNAP and of DNA, free in solution or in complex, have been observed and quantified directly.24, 25, 26, 29 A key challenge is to now connect these conformational states and their dynamic interconversion to motions and activity of the binding partners during promoter approach. What are the conformational dynamics while binding, unbinding, and/or sliding along nonspecific DNA? Can we resolve these conformational intermediates during transitions from closed to open complex?
The last phase of initiation can be considered to be abortive initiation and promoter escape. Here, evidence for stressed intermediates and scrunching of DNA inside the RNAP were found, as well as for paused, backtracked states during the initial RNAP synthesis.30, 31, 40, 45 Traditionally, the primary role of σ factors is considered to be in promoter recognition. Recent single‐molecule analysis however revealed that σ factors can be retained after promoter escape, with possible regulatory consequences for the elongation phase of transcription.
In summary, single‐molecule studies have exposed molecular mechanisms, conformational intermediates, and dynamic activities during the initial phases of transcription initiation as reviewed here for bacteria, as well as for eukaryotic46, 47, 48, 49, 50 and phage51, 52, 53, 54, 55, 56 systems. More work is now needed to connect knowledge of conformational dynamics to knowledge of molecular motions and activities, perhaps even observing conformation and molecular location and activity concurrently on long DNA molecules representative of genomic DNA. Such integrated knowledge is functional to enhance our fundamental understanding of transcription and for continued studies on how transcription is affected and regulated by the many biophysical and biochemical cues in the cell. Furthermore, this understanding is accelerating the discovery of new drugs by redefining diseases at the molecular level.57 A final hurdle is to link the detailed in vitro datasets to those acquired in the complex and crowded environment in vivo. The field is now seeing encouraging steps toward this goal as exemplified by the exploitation of quantitative super‐resolution analyses of transcription in living cells. The fields of single‐molecule analysis and cell biology are thus well on their way to jointly resolve how to switch the motor on during transcription initiation.
Acknowledgment
This work was supported by a STW HTSM prject grant (G.J.L.W. & W.H.R), a VIDI grant (W.H.R.) and a VENI grant (I.H.) all from the Nederlandse Organisatie voor Wetenschappelijk Onderzoek, and a H2020 FETopen grant (G.J.L.W.).
References
- 1. Decker KB, Hinton DM (2013) Transcription regulation at the core: similarities among bacterial, Archaeal, and eukaryotic RNA polymerases. Annu Rev Microbiol 67:113–139. [DOI] [PubMed] [Google Scholar]
- 2. Murakami KS (2013) X‐ray crystal structure of Escherichia coli RNA polymerase σ70 holoenzyme. J Biol Chem 288:9126–9134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Browning D, Busby S (2004) The regulation of bacterial transcription initiation. Nat Rev Microbiol 2:57–65. [DOI] [PubMed] [Google Scholar]
- 4. Ishihama A (2000) Functional modulation of Escherichia coli RNA polymerase. Annu Rev Microbiol 54:499–518. [DOI] [PubMed] [Google Scholar]
- 5. Feklistov A, Sharon BD, Darst SA, Gross CA (2014) Bacterial sigma factors: a historical, structural, and genomic perspective. Annu Rev Microbiol 68:357–376. [DOI] [PubMed] [Google Scholar]
- 6. Browning DF, Busby SJ (2016) Local and global regulation of transcription initiation in bacteria. Nat Rev Microbiol 14:638–650. [DOI] [PubMed] [Google Scholar]
- 7. Li G‐W, Berg OG, Elf J (2009) Effects of macromolecular crowding and DNA looping on gene regulation kinetics. Nat Phys 5:294–297. [Google Scholar]
- 8. Burgess RR (1971) RNA polymerase. Annu Rev Biochem 40:1:711–740. [DOI] [PubMed] [Google Scholar]
- 9. Zhang Y, Feng Y, Chatterjee S, Tuske S, Ho M, Arnold E, Ebright R (2012) Structural basis of transcription initiation. Science 338:1076–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Larson MH, Landick R, Block SM (2011) Single‐molecule studies of RNA polymerase: one singular sensation, every little step it takes. Mol Cell 41:249–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Murakami KS, Masuda S, Darst SA (2002) Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 Å resolution. Science 296. [PAGE #S]. [DOI] [PubMed] [Google Scholar]
- 12. Haugen SP, Ross W, Gourse RL (2008) Advances in bacterial promoter recognition and its control by factors that do not bind DNA. Nat Rev Microbiol 6:507–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mirny L, Slutsky M, Wunderlich Z, Tafvizi A, Leith J, Kosmrlj A (2009) How a protein searches for its site on DNA: the mechanism of facilitated diffusion. J Phys A: Math Theory 42:434013–434023. [Google Scholar]
- 14. Kabata H, Kurosawa O, Arai I, Washizu M, Margarson S, Glass R, Shimamoto N (1993) Visualization of single molecules of RNA polymerase sliding along DNA. Science 262:1561–1563. [DOI] [PubMed] [Google Scholar]
- 15. Guthold M, Zhu X, Rivetti C, Yang G, Thomson N, Kasas S, Hansma H, Smith B, Hansma P, Bustamante C (1999) Direct observation of one‐dimensional diffusion and transcription by Escherichia coli RNA polymerase. Biophys J 77:2284–2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Harada Y, Funatsu T, Murakami K, Nonoyama Y, Ishihama A, Yanagida T (1999) Single‐molecule imaging of RNA polymerase‐DNA interactions in real time. Biophys J 76:709–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang F, Redding S, Finkelstein I, Gorman J, Reichman D, Greene E (2013) The promoter‐search mechanism of Escherichia coli RNA polymerase is dominated by three‐dimensional diffusion. Nat Struct Mol Biol 20:174–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Friedman LJ, Mumm JP, Gelles J (2013) RNA polymerase approaches its promoter without long‐range sliding along DNA. Proc Natl Acad Sci USA 110:9740–9745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bakshi S, Dalrymple RM, Li W, Choi H, Weisshaar JC (2013) Partitioning of RNA polymerase activity in live Escherichia coli from analysis of single‐molecule diffusive trajectories. Biophys J 105:2676–2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Stracy M, Lesterlin C, Garza de Leon F, Uphoff S, Zawadzki P, Kapanidis A (2015) Live‐cell superresolution microscopy reveals the organization of RNA polymerase in the bacterial nucleoid. Proc Natl Acad Sci U S A 112:E4390–E4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Revyakin A, Liu C, Ebright RH, Strick TR (2006) Abortive initiation and productive initiation by RNA polymerase involve DNA scrunching. Science 314:1139–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rees W, Keller R, Vesenka J, Yang G, Bustamante C (1993) Evidence of DNA bending in transcription complexes imaged by scanning force microscopy. Science 260:1646–1649. [DOI] [PubMed] [Google Scholar]
- 23. Rippe K, Guthold M, von Hippel PH, Bustamante C (1997) Transcriptional activation via DNA‐looping: visualization of intermediates in the activation pathway of E. coli RNA polymerase·σ54 holoenzyme by scanning force microscopy. J Mol Biol 270:125–138. [DOI] [PubMed] [Google Scholar]
- 24. Rivetti C, Guthold M, Bustamante C (1999) Wrapping of DNA around the E.coli RNA polymerase open promoter complex. EMBO J 18:4464–4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Revyakin A, Ebright RH, Strick TR (2004) Promoter unwinding and promoter clearance by RNA polymerase: detection by single‐molecule DNA nanomanipulation. Proc Natl Acad Sci U S A 101:4776–4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chakraborty A, Wang D, Ebright Y, Korlann Y, Kortkhonjia E, Kim T, Chowdhury S, Wigneshweraraj S, Irschik H, Jansen R, Nixon B, Knight J, Weiss S, Ebright R (2012) Opening and closing of the bacterial RNA polymerase clamp. Science 337:591–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Carpousis AJ, Stefano JE, Gralla JD (1982) 5′ Nucleotide heterogeneity and altered initiation of transcription at mutant lac promoters. J Mol Biol 157:619–633. [DOI] [PubMed] [Google Scholar]
- 28. Robb N, Cordes T, Hwang L, Gryte K, Duchi D, Craggs T, Santoso Y, Weiss S, Ebright R, Kapanidis A (2013) The transcription bubble of the RNA polymerase–promoter open complex exhibits conformational heterogeneity and millisecond‐scale dynamics: implications for transcription start‐site selection. J Mol Biol 425:875–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Friedman LJ, Gelles J (2012) Mechanism of transcription initiation at an activator‐dependent promoter defined by single‐molecule observation. Cell 148:679–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kapanidis A, Margeat E, Ho S, Kortkhonjia E, Weiss S, Ebright R (2006) Initial transcription by RNA polymerase proceeds through a DNA‐scrunching mechanism. Science 314:1144–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lerner E, Chung S, Allen B, Wang S, Lee J, Lu S, Grimaud L, Ingargiola A, Michalet X, Alhadid Y, Borukhov S, Strick T, Taatjes D, Weiss S (2016) Backtracked and paused transcription initiation intermediate of Escherichia coli RNA polymerase. Proc Natl Acad Sci U S A 113:E6562–E6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hsu LM (2009) Monitoring abortive initiation. Methods 47:25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ring B, Yarnell W, Roberts J (1996) Function of E. coli RNA polymerase sigma factor sigma 70 in promoter‐proximal pausing. Cell 86:485–493. [DOI] [PubMed] [Google Scholar]
- 34. Ko J, Heyduk T (2014) Kinetics of promoter escape by bacterial RNA polymerase: effects of promoter contacts and transcription bubble collapse. Biochem J 463. [PAGE #S]. [DOI] [PubMed] [Google Scholar]
- 35. Vahia AV, Martin CT (2011) Direct tests of the energetic basis of abortive cycling in transcription. Biochemistry 50:7015–7022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Borukhov S, Severinov K (2002) Role of the RNA polymerase sigma subunit in transcription initiation. Res Microbiol 153:557–562. [DOI] [PubMed] [Google Scholar]
- 37. Murakami KS, Masuda S, Campbell EA, Muzzin O, Darst SA (2002) Structural basis of transcription initiation: an RNA polymerase holoenzyme‐DNA complex. Science 296:1285–1290. [DOI] [PubMed] [Google Scholar]
- 38. Basu R, Warner B, Molodtsov V, Pupov D, Esyunina D, Fernández‐Tornero C, Kulbachinskiy A, Murakami K (2014) Structural basis of transcription initiation by bacterial RNA polymerase holoenzyme. J Biol Chem 289:24549–24559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lee N, Kapanidis A, Wang Y, Michalet X, Mukhopadhyay J, Ebright R, Weiss S (1992) Accurate FRET measurements within single diffusing biomolecules using alternating‐laser excitation. Biophys J 88:2939–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Margeat E, Kapanidis A, Tinnefeld P, Wang Y, Mukhopadhyay J, Ebright R, Weiss S (2006) Direct observation of abortive initiation and promoter escape within single immobilized transcription complexes. Biophys J 90:1419–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Straney DC, Crothers DM (1987) Comparison of the open complexes formed by RNA polymerase at the Escherichia coli Zac UV5 promoter. J Mol Biol 193:879–202. [DOI] [PubMed] [Google Scholar]
- 42. Breslauer KJ, Frank R, Blöcker H, Marky LA (1986) Predicting DNA duplex stability from the base sequence. Proc Natl Acad Sci U S A 83:3746–3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Record MT, Reznikoff WS, Craig ML, McQuade KL, Schlax PJ (1996) Escherichia coli RNA polymerase (Esigma70), promoters, and the kinetics of the steps of transcription initiation In: Neidhardt FC, et al., editors. Escherichia coli and Salmonella Cellular and Molecular Biology. Washington DC: ASM Press; pp. 792–821. [Google Scholar]
- 44. Gill SC, Weitzel SE, von Hippel PH (1991) Escherichia coli sigma 70 and NusA proteins. I. Binding interactions with core RNA polymerase in solution and within the transcription complex. J Mol Biol 220:307–324. [DOI] [PubMed] [Google Scholar]
- 45. Duchi D, Bauer D, Fernandez L, Evans G, Robb N, Hwang L, Gryte K, Tomescu A, Zawadzki P, Morichaud Z, Brodolin K, Kapanidis A (2016) RNA polymerase pausing during initial transcription. Mol Cell 63:939–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Larson DR, Zenklusen D, Wu B, Chao JA, Singer RH (2011) Real‐time observation of transcription initiation and elongation on an endogenous yeast gene. Science 332. [PAGE #S]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fazal FM, Meng CA, Murakami K, Kornberg RD, Block SM (2015) Real‐time observation of the initiation of RNA polymerase II transcription. Nature 525:274–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Revyakin A, Zhang Z, Coleman R, Li Y, Inouye C, Lucas J, Park S, Chu S, Tjian R (2012) Transcription initiation by human RNA polymerase II visualized at single‐molecule resolution. Genes Dev 26:1691–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Izeddin I, Récamier V, Bosanac L, Cissé I, Boudarene L, Dugast‐Darzacq C, Proux F, Bénichou O, Voituriez R, Bensaude O, Dahan M, Darzacq X, Bender M, Groudine M, Gnirke A, Stamatoyannopoulos J, Mirny L, Lander E, Dekker J (2014) Single‐molecule tracking in live cells reveals distinct target‐search strategies of transcription factors in the nucleus. Elife 3:23352–23373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Horn AE, Kugel JF, Goodrich JA (2016) Single molecule microscopy reveals mechanistic insight into RNA polymerase II preinitiation complex assembly and transcriptional activity. Nucleic Acids Res 44:7132–7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Skinner GM, Baumann CG, Quinn DM, Molloy JE, Hoggett JG (2004) Promoter binding, initiation, and elongation by bacteriophage T7 RNA polymerase. A single‐molecule view of the transcription cycle. J Biol Chem 279:3239–3244. [DOI] [PubMed] [Google Scholar]
- 52. Kim JH, Larson RG (2007) Single‐molecule analysis of 1D diffusion and transcription elongation of T7 RNA polymerase along individual stretched DNA molecules. Nucleic Acids Res 35:3848–3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Endo M, Tatsumi K, Terushima K, Katsuda Y, Hidaka K, Harada Y, Sugiyama H (2012) Direct visualization of the movement of a single T7 RNA polymerase and transcription on a DNA nanostructure. Angew Chem 124:8908–8912. [DOI] [PubMed] [Google Scholar]
- 54. Skinner GM, Kalafut BS, Visscher K (2011) Downstream DNA tension regulates the stability of the T7 RNA polymerase initiation complex. Biophys J 100:1034–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhang Z, Revyakin A, Grimm J, Lavis L, Tjian R (2014) Single‐molecule tracking of the transcription cycle by sub‐second RNA detection. Elife 3:e01775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tang G‐Q, Roy R, Bandwar RP, Ha T, Patel SS (2009) Real‐time observation of the transition from transcription initiation to elongation of the RNA polymerase. Proc Natl Acad Sci U S A 106:22175–22180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bates S (2011) The role of gene expression profiling in drug discovery. Curr Opin Pharmacol 11:549–556. [DOI] [PubMed] [Google Scholar]


