ABSTRACT
During macrophage development, myeloid progenitor cells undergo terminal differentiation coordinated with reduced cell cycle progression. Differentiation of macrophages from myeloid progenitors is accompanied by increased expression of the E26 transformation-specific transcription factor PU.1. Reduced PU.1 expression leads to increased proliferation and impaired differentiation of myeloid progenitor cells. It is not understood how PU.1 coordinates macrophage differentiation with reduced cell cycle progression. In this study, we utilized cultured PU.1-inducible myeloid cells to perform genome-wide chromatin immunoprecipitation sequencing (ChIP-seq) analysis coupled with gene expression analysis to determine targets of PU.1 that may be involved in regulating cell cycle progression. We found that genes encoding cell cycle regulators and enzymes involved in lipid anabolism were directly and inducibly bound by PU.1 although their steady-state mRNA transcript levels were reduced. Inhibition of lipid anabolism was sufficient to reduce cell cycle progression in these cells. Induction of PU.1 reduced expression of E2f1, an important activator of genes involved in cell cycle and lipid anabolism, indirectly through microRNA 223. Next-generation sequencing identified microRNAs validated as targeting cell cycle and lipid anabolism for downregulation. These results suggest that PU.1 coordinates cell cycle progression with differentiation through induction of microRNAs targeting cell cycle regulators and lipid anabolism.
KEYWORDS: E2F1, microRNA, PU.1, cell cycle, differentiation, lipid synthesis, myeloid cells
INTRODUCTION
A central problem in biology is understanding how cell division is regulated in response to developmental and environmental cues. During macrophage development, proliferating myeloid progenitor cells undergo terminal differentiation that is coordinated with reduced cell cycle progression (1). However, terminally differentiated macrophages are not necessarily permanently cell cycle arrested, as shown by evidence that epidermal Langerhans cells and brain microglia can reenter the cell cycle to self-renew (2, 3). Thus, cell cycle arrest and terminal differentiation, while normally coincident, can be independently and actively regulated. Much remains to be learned about the mechanisms by which cell-type-specific transcription factors coordinate cell cycle arrest with differentiation.
PU.1 (encoded by the Spi1 gene) is an E26 transformation-specific (ETS) family transcription factor that is a concentration-dependent regulator of blood cell differentiation (4, 5). Differentiation of macrophages from myeloid progenitors is accompanied by a 5-fold upregulation of PU.1 concentration that is required for differentiation (5–7). Reduced PU.1 expression caused by mutation or repression leads to increased proliferation and impaired differentiation of myeloid progenitor cells (7). Inactivating mutations of the SPI1 gene encoding human PU.1 are associated with acute myeloid leukemia (AML) (8), and mutation of Spi1 is sufficient to induce AML in mouse models (7, 9). Reduced PU.1 expression promotes increased cell cycle entry in hematopoietic stem cells (10). Minimal reductions in PU.1 levels are sufficient to induce a preleukemic state with increased proliferation of myeloid progenitors, leading to AML (11). Spi1BN/BN mice (also called BN mice) express PU.1 at ∼20% of normal levels, do not generate mature macrophages, and have increased proliferation of myeloid progenitors in the spleen (6, 12). Myeloid progenitor cells from Spi1BN/BN fetal liver (BN cells) proliferate indefinitely in culture in response to granulocyte-macrophage colony-stimulating factor (GM-CSF) without undergoing differentiation (13). Induction of PU.1 in BN cells causes reduced cell cycle progression and myeloid differentiation (12). Therefore, PU.1 is a transcription factor that is essential for the coordination of myeloid terminal differentiation with cell cycle arrest. However, it is not clear how PU.1 accomplishes these important functions.
E2F1 to E2F3 are transcription factors that are required for cell cycle progression in the myeloid lineage by activating genes critical for G1-to-S-phase cell cycle progression (14, 15). E2F1 regulates the metabolic requirements of cell division by activating multiple genes involved in lipid anabolism (16). Proliferating cells increase their rate of synthesis of fatty acids during S and G2/M phases of the cell cycle, and this increase at G2/M is required for cell division (17). Therefore, E2F1 is an important determinant of cell cycle progression in mammalian cells since it coordinates lipid anabolism with the cell cycle.
In this study, we utilized cultured PU.1-inducible BN (iBN) cells to discover mechanisms of cell cycle regulation by PU.1. We performed genome-wide chromatin immunoprecipitation sequencing (ChIP-seq) analysis coupled with gene expression analysis to determine direct targets of PU.1 that may be involved in the regulation of cell cycle progression. We found that genes encoding cell cycle regulators, as well as enzymes in lipid anabolic pathways, were directly and inducibly bound by PU.1. The gene encoding E2F1 was directly activated by PU.1; however, mature E2f1 mRNA transcripts were downregulated upon PU.1 induction. Analysis of microRNAs (miRNAs) induced by PU.1 revealed upregulation of multiple miRNAs targeting E2f1 as well as mRNAs of genes involved in lipid anabolism such as Acly encoding ATP citrate lyase (ACL). Pharmacologic inhibition of ACL was sufficient to induce cell cycle arrest and differentiation in BN cells. Our results suggest that PU.1 coordinates cell cycle progression with differentiation through induction of microRNAs targeting cell cycle regulators and lipid anabolism.
RESULTS
Increased PU.1 concentration reduces cell cycle progression and induces myeloid differentiation.
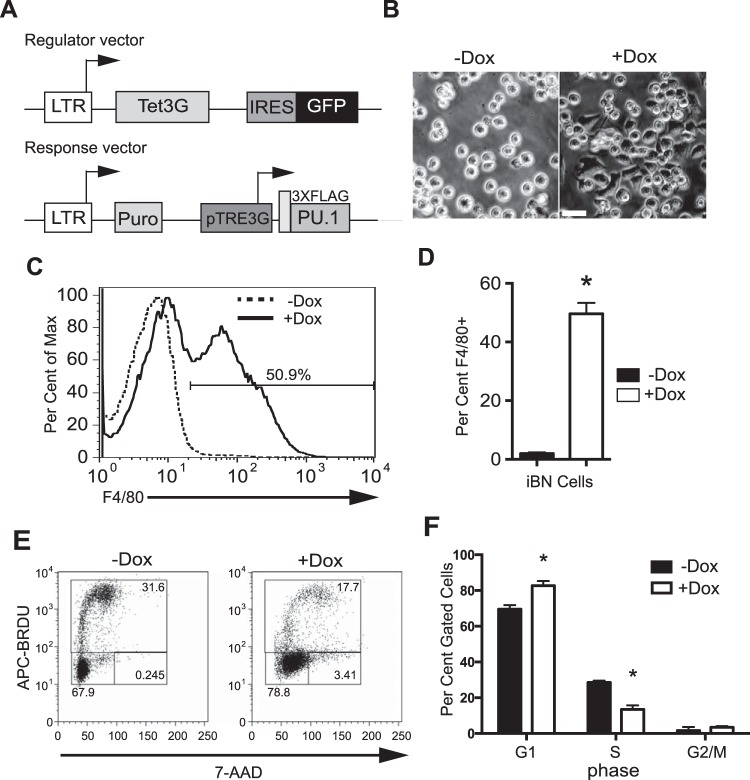
Spi1BN/BN cells (BN cells) are fetal liver-derived myeloid progenitor-like cells that proliferate in response to granulocyte-macrophage colony-stimulating factor (GM-CSF) (12, 13). BN cells express PU.1 at 20% of normal levels, below a threshold required to induce cell cycle arrest and myeloid differentiation, permitting the cells to proliferate indefinitely (13). The effects of PU.1 concentration were studied using PU.1-inducible BN (iBN) cells, in which PU.1 expression can be induced in response to doxycycline (Dox) (12) (Fig. 1A). Induction of PU.1 expression resulted in macrophage-like cellular morphology (Fig. 1B) and cell surface expression of F4/80 (Fig. 1C and D). F4/80 is a marker of macrophage differentiation encoded by Adgre1, a gene activated directly by PU.1 (18). PU.1 induction resulted in reduced cell cycle progression, as determined by an increase in G1 and a reduction in S-phase frequencies (Fig. 1E and F). Therefore, increased concentrations of PU.1 in iBN cells reduced cell cycle progression and induced myeloid differentiation.
FIG 1.
PU.1 reduces cell cycle progression and induces myeloid differentiation in iBN cells. (A) Schematic of the doxycycline (Dox)-inducible system. The top panel shows the regulator vector. The bottom panel shows the response vector. LTR, long terminal repeat; Tet3G, third-generation tetracycline-responsive transcription factor; IRES, internal ribosomal entry site; GFP, green fluorescent protein; Puro, puromycin resistance gene; pTRE3G, third-generation tetracycline-regulatable element promoter; 3XFLAG-PU.1, cDNA encoding epitope-tagged PU.1. (B) Acquisition of adherent macrophage-like cellular morphology after 72 h of induction with 1,000 ng/ml Dox. Scale bar, 20 μm. (C) Acquisition of cell surface F4/80 expression after PU.1 induction. Inducible BN (iBN) cells were untreated or treated with 1,000 ng/ml Dox for 72 h, followed by flow cytometric analysis. The arrow indicates the frequency of F4/80-positive cells. (D) Quantification of results shown in panel C for three biological replicates. (E) Reduction of cell cycle progression after PU.1 induction. iBN cells were untreated or treated with 1,000 ng/ml Dox for 72 h and then analyzed for cell cycle. The y axis indicates bromodeoxyuridine (BRDU) incorporation. The x axis represents DNA content as determined by 7-amino-actinomycin D (7-AAD) staining. Boxes show G1 (lower left), S (upper), and G2/M (lower right) phases of the cell cycle. (F) Quantification of results shown in panel E for four biological replicates. *, P < 0.05.
Peak-to-gene association after induction of PU.1 expression in iBN cells.
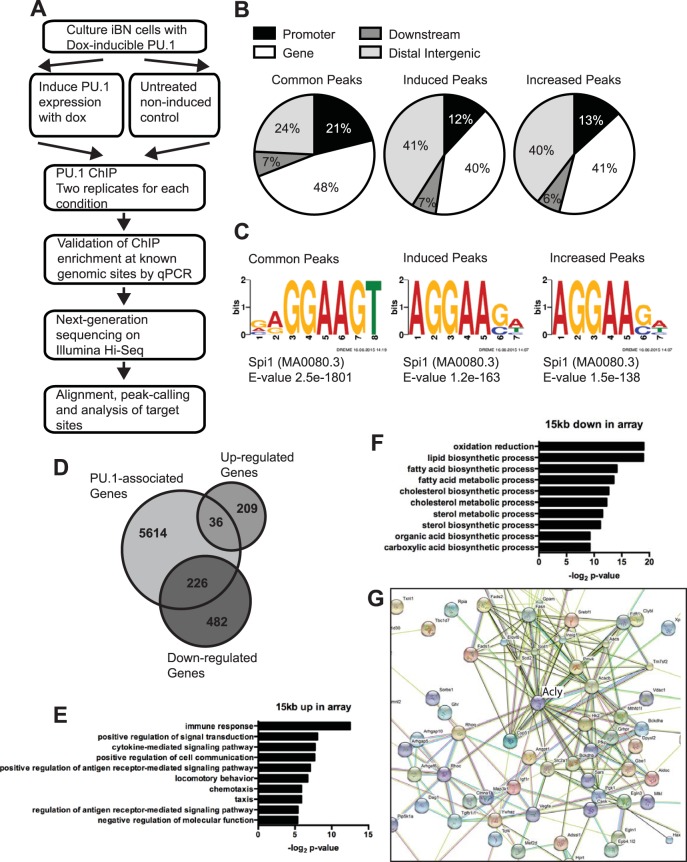
The iBN cell line is an opportune system to determine the mechanism(s) by which increased PU.1 concentration coordinates reduced cell cycle progression with terminal myeloid differentiation. Changes in gene expression after PU.1 induction in iBN cells were determined using Affymetrix Mouse Gene 2.0 ST arrays as previously described (12). To determine changes in target gene binding by PU.1, anti-PU.1 chromatin immunoprecipitation sequencing (ChIP-seq) was performed on iBN cells with or without PU.1 induction (Fig. 2A). Single-end reads were generated by Illumina sequencing and aligned to mouse genome reference mm9 using Bowtie. Peaks were called using model-based analysis of ChIP-seq (MACS). A total of 44,233 significantly enriched regions of PU.1 binding were observed in noninduced cells, and upon induction of PU.1 this number increased 1.6-fold to 71,582 regions. DiffBind was used to stringently compare PU.1-associated genomic regions (peaks) between uninduced and PU.1-induced iBN cells. DiffBind reported 5,602 peaks to be increased in induced iBN cells compared to levels in uninduced cells. Of these 5,602 induced regions, 2,779 did not have a peak detectable above background in the noninduced cells and were therefore considered induced peaks. The remaining 2,823 regions were observed in noninduced cells and were increased 2-fold or higher upon induction; they were therefore considered increased peaks. All other peaks were unchanged (common peaks). Common peaks, induced peaks, and increased peaks had similar patterns of location relative to annotated transcription start sites (Fig. 2B). Induced peaks had a decreased frequency within 10 kb of transcription start sites (12% compared to 18% for all regions) and an increased frequency at distal intergenic regions (40% compared to 29% for all regions) (Fig. 2B, center panel). The most common DNA sequence motif identified for all peaks was the canonical PU.1-binding motif 5′-GGAA-3′ at 75% for common peaks, 55% for induced peaks, and 38% for increased peaks (Fig. 2C).
FIG 2.
Genome-wide analysis of induced PU.1 binding sites in iBN cells. (A) Workflow for generating ChIP sequencing data. (B) Distribution of ChIP-seq peaks within the genome. PU.1 peaks were divided into common peaks (PU.1-associated genomic regions that did not change upon PU.1 induction), induced peaks (genomic regions that acquired PU.1 association upon PU.1 induction), and increased peaks (genomic regions that were more highly associated with PU.1 upon induction). (C) DNA sequence motifs associated with genomic regions containing common peaks, induced peaks, or increased peaks, as indicated. (D) Venn diagram showing overlap of PU.1-associated genes with genes whose mRNA levels were either up- or downregulated as determined by microarray analysis. (E) Biological pathway analysis of 36 upregulated mRNAs associated with PU.1 peaks. (F) Biological pathway analysis of 226 downregulated mRNAs associated with PU.1 peaks. (G) Functional protein association analysis of PU.1-downregulated genes. STRING was used to integrate proteins encoded by 226 downregulated mRNAs associated with PU.1 peaks.
Next, we determined the relationship between PU.1 binding regions and changes in gene expression upon induction of PU.1 in iBN cells. GREAT was used to identify 5,840 genes associated with the 5,602 PU.1-increased or -induced peaks. Analysis of microarray data (12) indicated that 245 genes were upregulated and that 708 genes were downregulated more than 1.5-fold. Of 5,840 genes directly bound by PU.1, 36 genes were upregulated, and 226 genes were downregulated (see Data Set S1 in the supplemental material) (Fig. 2D). This short list of genes is likely to be directly regulated by PU.1.
DAVID was used to assign to biological pathways PU.1-upregulated or PU.1-downregulated genes that were directly associated with induced or increased peaks. The 36 PU.1-upregulated genes were highly associated with the immune response, positive regulation of signal transduction, and cytokine-mediated signaling pathway, as expected based on previous studies (Fig. 2E) (19). This upregulated list included known targets of PU.1 direct binding, including Cybb encoding gp91phox and Adgre1 encoding F4/80 (Fig. 1C and D). Unexpectedly, the 226 PU.1-downregulated genes were highly associated with pathways such as oxidation reduction, lipid biosynthetic process, and fatty acid metabolic process (Fig. 2F). To explore this further, proteins encoded by PU.1-downregulated genes were analyzed for interactions using STRING. An interaction cluster was observed for enzymes involved in fatty acid anabolism that was centered on Acly encoding ATP citrate lyase (ACL) (Fig. 2G). This enzyme plays a key role in de novo lipid biosynthesis by generation of acetyl coenzyme A (acetyl-CoA) from citrate (20, 21). Taken together, these results suggest that myeloid differentiation and reduced cell cycle progression induced by PU.1 are associated with downregulation of genes involved in fatty acid anabolism.
Direct upregulation of Cybb transcription by PU.1.
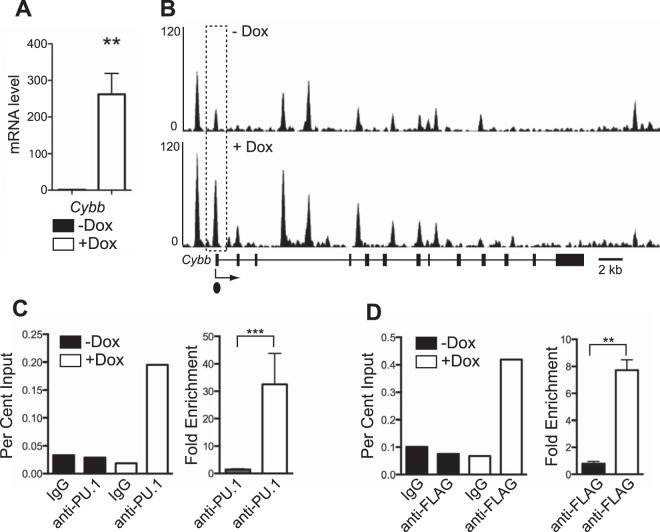
One of the most highly induced genes from microarray data that was associated with increased PU.1 binding was Cybb encoding cytochrome b-245 beta, also known as gp91phox. gp91phox is essential to generate superoxide in myeloid cells and is essential for the innate immune response to pathogens (22). Induction of Cybb by PU.1 was confirmed by reverse transcription-quantitative PCR (RT-qPCR) analysis (Fig. 3A). PU.1 binding was associated with several regions within the Cybb locus in uninduced and induced iBN cells (Fig. 3B). A region located near the Cybb transcription start site increased its PU.1 association by approximately 30-fold upon PU.1 induction (Fig. 3B to D). Taken together, these data suggest that Cybb is a direct target gene of PU.1 activation.
FIG 3.
Activation of Cybb is associated with direct binding of PU.1. (A) Induction of PU.1 expression in iBN cells increases Cybb mRNA transcript levels. Cybb mRNA transcript levels were determined using RT-qPCR of RNA prepared 72 h after induction with 1,000 ng/ml Dox. (B) PU.1 association with the Cybb promoter is inducible. Shown are ChIP-seq tracks without (−) and with (+) Dox at 72 h after PU.1 induction. The Cybb gene structure is shown below the bottom panel; boxes represent exons. The dotted box indicates an increased site of PU.1 association. The dot underneath the bottom panel indicates the location of the primer pair used for ChIP-qPCR analysis. (C) Quantification of PU.1 association with the Cybb promoter. Anti-PU.1 ChIP was performed 72 h after Dox induction. The left panel shows percent input determined by qPCR using primers recognizing the Cybb promoter. The right panel shows fold enrichment comparing percent input from anti-PU.1 to that from IgG ChIP for three biological replicates. (D) Quantification of PU.1 association with Cybb promoter using anti-FLAG ChIP performed 72 h after Dox induction. The left panel shows percent input determined by qPCR using primers recognizing the Cybb promoter. The right panel shows fold enrichment comparing percent input from anti-FLAG to that of IgG ChIP for three biological replicates. **, P < 0.005; ***, P < 0.001.
ACL inhibition induces cell cycle arrest and differentiation.
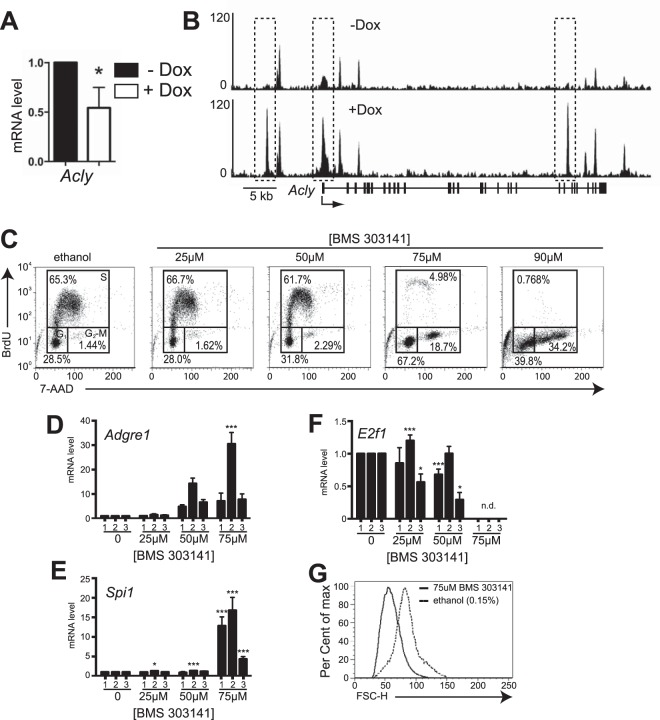
As described above, genes involved in lipid anabolism, such as Acly, were associated with increased PU.1 binding upon PU.1 induction even though steady-state mRNA levels were decreased, as determined by microarray analysis. RT-qPCR analysis confirmed that Acly mRNA transcript levels were reduced upon PU.1 induction (Fig. 4A). ChIP-seq analysis showed that PU.1 induction resulted in increased PU.1 binding at three sites within the Acly locus (Fig. 4B). Pharmacologic inhibition of ACL (encoded by Acly) has been shown to inhibit the growth of cancer cells in vitro and in vivo by limiting the cellular pool of acetyl-CoA (23, 24). We reasoned that downregulation of Acly in iBN cells might be involved in cell cycle regulation. To test this idea, BN cells were cultured with the ACL inhibitor BMS303141 (25). Treatment of BN cells with concentrations of BMS303141 above 50 μM induced cell cycle arrest (Fig. 4C). Cell cycle arrest induced by BMS303141 was coordinated with increased steady-state levels of mRNA transcripts for Adgre1 as well as Spi1 (Fig. 4D and E), decreased steady-state mRNA transcript levels of E2f1 (Fig. 4F), and reduced cellular size as measured by forward light scatter (Fig. 4G). In summary, inhibition of ACL activity in BN cells by BMS303141 was sufficient to induce cell cycle arrest, reduce cell size, and change gene expression, which is suggestive of myeloid differentiation.
FIG 4.
Inhibition of ATP citrate lyase activity induces cell cycle arrest and differentiation in BN cells. (A) Induction of PU.1 expression in iBN cells reduces Acly mRNA transcript levels. Acly mRNA transcript levels were determined using RT-qPCR of RNA prepared 72 h after induction of PU.1 with 1,000 ng/ml Dox. (B) PU.1 association with the Acly locus is inducible. Shown are ChIP-seq tracks without (−) and with (+) Dox 72 h after PU.1 induction. The Acly gene structure is shown below the bottom panel; vertical boxes represent exons. The dotted box indicates the site of increased PU.1 association. (C) Inhibition of ATP citrate lyase (ACL) activity induces cell cycle arrest in BN cells. BN cells were treated with the ACL inhibitor BMS303141 at the indicated concentrations for 48 h before cell cycle analysis using flow cytometry. The control was 0.01% ethanol. One representative experiment is shown. (D to F) Induction of myeloid differentiation in BN cells by ACL inhibition. Shown are fold changes in mRNA transcript levels of Adgre1, Spi1, and E2f1 normalized to the B2m transcript level in three independent experiments, indicated by numbers 1 to 3. mRNA transcript levels were measured 48 h after treatment with the indicated concentrations of BMS303141. n.d., not detected. (G) ACL inhibition reduces cell size. Flow cytometry was used to determine relative cell size as measured by forward scatter (FSC-H) 48 h after inhibition of ACL activity by 75 μM BMS303141 in BN cells. *, P < 0.05; ***, P < 0.001.
Indirect downregulation of E2f1 transcription by PU.1.
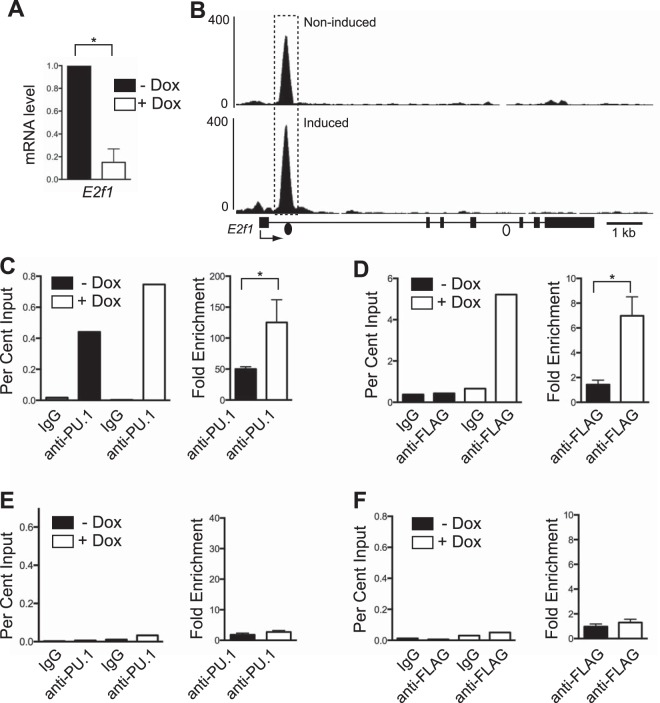
E2F1 has been described to regulate hundreds of genes to promote cell cycle progression, of which prototypic examples include cyclins and cyclin-dependent kinases (26). E2F1 also activates genes encoding enzymes involved in lipid anabolism to promote the synthesis of fatty acids required for cell growth (16). Previous studies showed that increased PU.1 was associated with reduced E2F1 expression in iBN cells (12). RT-qPCR analysis confirmed that steady-state levels of mature E2f1 mRNA transcripts were reduced in iBN cells 72 h after PU.1 induction (Fig. 5A). Examination of ChIP-seq data revealed a single site of PU.1 interaction in intron 1 of E2f1 in uninduced iBN cells that was increased upon PU.1 induction (Fig. 5B). Analysis using either anti-PU.1 ChIP (Fig. 5C) or anti-FLAG ChIP (Fig. 5D) showed that PU.1 occupancy of this site increased upon PU.1 induction. After PU.1 induction, there was no enrichment of PU.1 binding to a negative-control site located in intron 4 of the E2f1 gene (Fig. 5E and F). Therefore, in the iBN cell line, PU.1 interacts inducibly with a site in intron 1 of the E2f1 gene.
FIG 5.
Reduction of E2f1 mRNA transcripts is associated with increased PU.1 binding. (A) Induction of PU.1 expression in iBN cells decreases E2f1 mRNA transcript levels. E2f1 mRNA transcript levels were determined using RT-qPCR of RNA prepared 72 h after induction with 1,000 ng/ml Dox. (B) PU.1 association with a site in E2f1 intron 1 is inducible. Shown are ChIP-seq tracks without (−) and with (+) Dox 72 h after PU.1 induction. The E2f1 gene structure is shown below the bottom panel; boxes represent exons. The dashed box indicates a site of increased PU.1 association. Filled and open dots below the bottom panel indicate locations of primer pairs used for ChIP-qPCR analysis. (C) Quantification of PU.1 association with E2f1 intron 1. Anti-PU.1 ChIP was performed 72 h after Dox induction. The left panel shows percent input determined by qPCR using primers (represented by the filled dot in panel B) recognizing the E2f1 intron 1. The right panel shows fold enrichment comparing percent input from anti-PU.1 to that of IgG ChIP for three biological replicates. (D) Quantification of PU.1 association with E2f1 intron 1 using anti-FLAG ChIP performed 72 h after Dox induction. The left panel shows percent input determined by qPCR using primers recognizing E2f1 intron 1. The right panel shows fold enrichment comparing percent input from anti-FLAG to that of IgG ChIP for three biological replicates. (E and F) Quantification of PU.1 association with a negative-control site within E2f1 intron 4. Anti-PU.1 ChIP or anti-FLAG ChIP was performed 72 h after Dox induction. The left panels show percent input determined by qPCR using primers (represented by the open dot in panel B) recognizing E2f1 intron 4. The right panels show fold enrichment comparing percent input from anti-PU.1 to that of IgG ChIP for three biological replicates. *, P < 0.05.
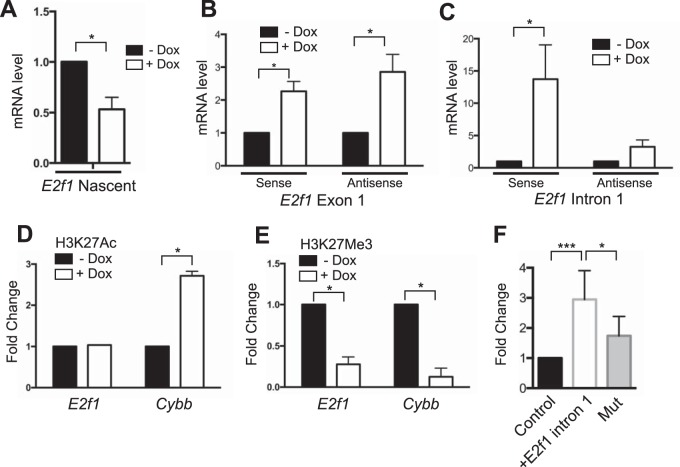
Next, we set out to determine if E2f1 mRNA transcript levels are reduced by PU.1 directly or indirectly. First, we determined whether nascent E2f1 mRNA transcripts were reduced by PU.1 induction. Nascent RNA was isolated 24 h after PU.1 induction in iBN cells and used to determine levels of E2f1 mRNA transcripts. RT-qPCR analysis showed that nascent E2f1 mRNA transcript levels were reduced upon PU.1 induction (Fig. 6A). Next, we determined if immature, unspliced mRNA transcripts in the E2f1 locus were affected by PU.1 induction. Primers were designed to recognize exon 1 or intron 1 of the E2f1 gene, detecting immature RNA transcripts. cDNA was synthesized using strand-specific E2f1 primers that did not prime cDNA synthesis for the reference gene B2m (data not shown). Interestingly, both sense and antisense immature E2f1 nascent mRNA transcripts were increased in iBN cells upon PU.1 induction (Fig. 6B and C). This result suggests that PU.1 activates rather than represses E2f1 transcription.
FIG 6.
Activation of E2f1 transcription by PU.1. (A) Reduction of E2f1 mature nascent mRNA transcript levels by PU.1 induction. E2f1 mature mRNA transcript levels were determined using RT-qPCR of nascent mRNA prepared 72 h after induction with 1,000 ng/ml Dox. (B) Induction of immature E2f1 nascent mRNA transcript levels by PU.1 induction. cDNA was synthesized using strand-specific primers. Immature E2f1 mRNA transcripts levels were determined using a primer set specific for E2f1 exon 1. (C) Induction of E2f1 immature nascent mRNA transcript levels by PU.1 induction. Immature E2f1 mRNA transcripts levels were determined using a primer set specific for E2f1 intron 1. (D) Induction of H3K27 acetylation by PU.1 at the Cybb promoter. Chromatin was prepared from iBN cells 72 h after induction with 1,000 ng/ml Dox. ChIP was performed using anti-H3K27Ac antibody. qPCR analysis was performed using primers recognizing the E2f1 or Cybb promoter. (E) Reduction of H3K27 trimethylation by PU.1 at the E2f1 and Cybb promoters. Chromatin was prepared from iBN cells 72 h after induction with 1,000 ng/ml Dox. ChIP was performed using anti-H3K27Me3 antibody. qPCR analysis was performed using primers recognizing the E2f1 or Cybb promoters. (F) A regulatory region in intron 1 of E2f1 behaves as a transcriptional activator. Plasmid DNA for pGL3-promoter (control), pGL3-promoter-E2F1 intron 1, or pGL3-promoter-E2F1 intron 1 with a mutated PU.1 binding site (Mut) was transiently transfected into WEHI-3B cells. Normalized luciferase activity was determined 24 h after transfection and is expressed as fold change relative to the level of the control. *, P < 0.05; ***, P < 0.001.
To further investigate if PU.1 acts as a transcriptional activator for E2f1, we used chromatin immunoprecipitation (ChIP) followed by qPCR to determine if induction of PU.1 in iBN cells induces changes in histone modifications near the PU.1 binding site in intron 1 of E2f1. ChIP-qPCR was performed using antibodies for the histone tail modification H3K27Ac (acetylation of histone H3 at K27), which is associated with transcriptional activation, and H3K27Me3 (trimethylation of histone H3 at K27), which is associated with transcriptional repression. Primers were designed to amplify a region containing the PU.1 binding site in E2f1 (Fig. 5B, filled dot). As a control, PCR primers were designed to flank the inducible PU.1 binding site at the transcription start site of Cybb (Fig. 3B, dot). Using this assay, H3K27Ac association was increased at the Cybb promoter upon PU.1 induction although it was not significantly increased at the E2f1 PU.1 binding site (Fig. 6D). In contrast, H3K27Me3 association was reduced upon PU.1 induction for both the E2f1 and Cybb genes (Fig. 6E). These results are most consistent with PU.1 functioning as a transcriptional activator at the E2f1 intron 1 binding site. To further test this idea, a 400-bp fragment of E2f1 intron 1 centered on the PU.1 binding site was cloned and ligated into a luciferase reporter vector containing a minimal promoter. The PU.1 binding site was mutated by changing the core interaction motif from GGAA to GGAC (mutation underlined), a sequence with which PU.1 cannot interact (27). Upon transient transfection into the WEHI-3B myeloid cell line, the reporter containing the E2f1 regulatory region showed transcriptional activation that was reduced upon mutation of the predicted PU.1 binding site (Fig. 6F). Taken together, these data suggest that PU.1 acts as a transcriptional activator for the E2f1 gene by interacting with a site in intron 1. However, mature E2f1 transcript levels were reduced upon PU.1 induction, suggesting a posttranscriptional mechanism of mRNA downregulation.
To determine potential consequences of changes in E2F1 expression, we constructed a retroviral vector encoding full-length murine E2F1 and Thy1.1 as a marker (Fig. S1A). E2F1 retrovirus was generated and used to infect cultured BN cells. After 96 h of cell culture, few Thy1.1-expressing (Thy1.1+) cells were present in cultures of cells infected with the E2F1 retroviral vectors, whereas cells infected with the control vector proliferated normally (Fig. S1B and C). To determine if cells infected with E2F1 were undergoing apoptosis, infected cells were analyzed using flow cytometry for terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) staining. This analysis showed a high degree of TUNEL staining in Thy1.1+ cells infected with E2F1 in contrast to few TUNEL-positive cells in Thy1.1+ cells infected with control retrovirus (Fig. S1D and E). Therefore, increased E2F1 expression promoted apoptosis in cultured BN cells. Taken together, these results suggest that BN cells are sensitive to changes in E2F1 levels. Furthermore, these results suggest that PU.1 reduces E2f1 mRNA transcript levels indirectly rather than directly.
PU.1 regulates E2f1 through miR-223.
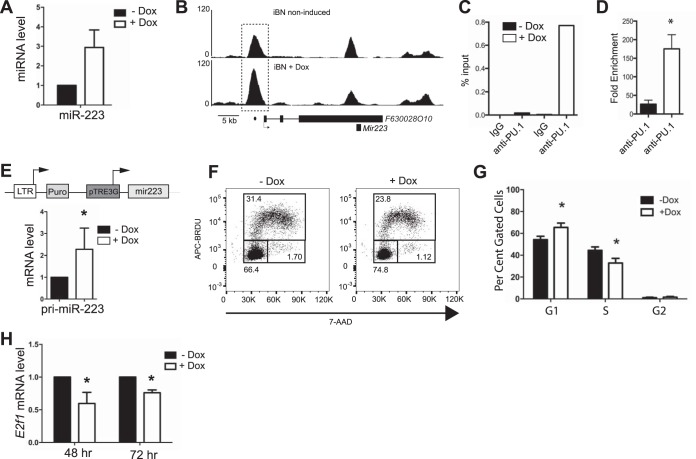
MicroRNAs (miRNAs) are small noncoding RNAs that target mRNAs for degradation or translational inhibition and are an important mechanism for fine-tuning mRNA transcript levels during blood cell development (28). E2f1 mRNA transcript levels were previously suggested to be negatively regulated by microRNA 223 (miR-223) (29). The gene encoding miR-223 was shown to be directly activated by PU.1 (30). qPCR analysis showed that miR-223 was upregulated by PU.1 upon doxycycline induction in iBN cells (Fig. 7A). ChIP analysis showed that the F630028O10 gene that encodes miR-223 (30) was associated with increased PU.1 binding upon PU.1 induction (Fig. 7B to D). To determine whether induction of miR-223 is sufficient to reduce cell cycle progression in iBN cells, a 447-bp genomic DNA fragment encoding miR-223 was cloned and ligated into the MIG-TRE3G response vector (where MIG is a murine stem cell virus/internal ribosome entry site/green fluorescent protein retroviral expression vector) (Fig. 7E, upper panel). BN cells were infected with an MIG-Tet3G regulator vector (where Tet3G is a third-generation tetracycline-responsive transcription factor) and a MIG-TRE3G response vector encoding miR-223 and selected using puromycin and cell sorting to generate an miR-223-inducible BN cell line. miR-223 was inducible by doxycycline in this cell line, as detected using primers recognizing primary miR-223 transcripts (Fig. 7E, lower panel). Upon doxycycline induction of miR-223, cell cycle progression was reduced (Fig. 7F and G). Induction of miR-223 by doxycycline was sufficient to reduce steady-state E2f1 mRNA transcript levels (Fig. 7H). In summary, PU.1 induction increased miR-223 levels in BN cells, and increased miR-223 expression was sufficient to reduce E2f1 mRNA transcript levels, leading to reduced cell cycle progression.
FIG 7.
Regulation of E2f1 mRNA levels and cell cycle progression by microRNA 223. (A) miR-223 levels were determined using a TaqMan miRNA assay 72 h after PU.1 induction in iBN cells using 1,000 ng/ml Dox. (B) PU.1 association with a site in the F630028O10 promoter is inducible. Shown are ChIP-seq tracks without (−) and with (+) Dox 72 h after PU.1 induction. The F630028O10 gene structure is shown below the bottom panel; boxes represent exons. The dashed box indicates a site of increased PU.1 association. The dot below the bottom panel indicates the location of the primer pair used for ChIP-qPCR analysis. (C) Quantification of PU.1 association with the F630028O10 promoter. Anti-PU.1 ChIP was performed 72 h after Dox induction. The panel shows percent input determined by qPCR using primers (represented by the dot in panel B) recognizing the F630028O10 promoter. (D) Fold enrichment comparing percent input from anti-PU.1 to that of IgG ChIP for three biological replicates. (E) Construction of an inducible system for miR-223. A genomic segment encoding miR-223 was ligated into the pTRE3G retroviral response vector as shown in the top panel. BN cells were infected with regulator and response vectors and selected to generate miR-223 BN cells. Induction of miR-223 BN cells with 1,000 ng/ml Dox for 48 h resulted in induction of primary miR-223 (pri-miR-223) transcripts as detected by qPCR analysis. (F) Reduction of cell cycle progression after miR-223 induction. Inducible cells were untreated or treated with 1,000 ng/ml Dox for 72 h and then analyzed for cell cycle. The y axis indicates bromodeoxyuridine (BRDU) incorporation. The x axis represents DNA content as determined by 7-amino-actinomycin D (7-AAD) staining. Boxes show G1 (lower left), S (upper), and G2/M (lower right) phases of the cell cycle. (G) Quantification of results shown in panel F for four biological replicates. (H) Induction of miR-223 expression in inducible BN cells results in reduced steady-state E2f1 mRNA transcripts. miR-223 BN cells were induced with 1,000 ng/ml Dox for 48 or 72 h. E2f1 mRNA transcript levels were determined by RT-qPCR. *, P < 0.05.
PU.1 induces microRNAs that target cell cycle regulation and lipid anabolism.
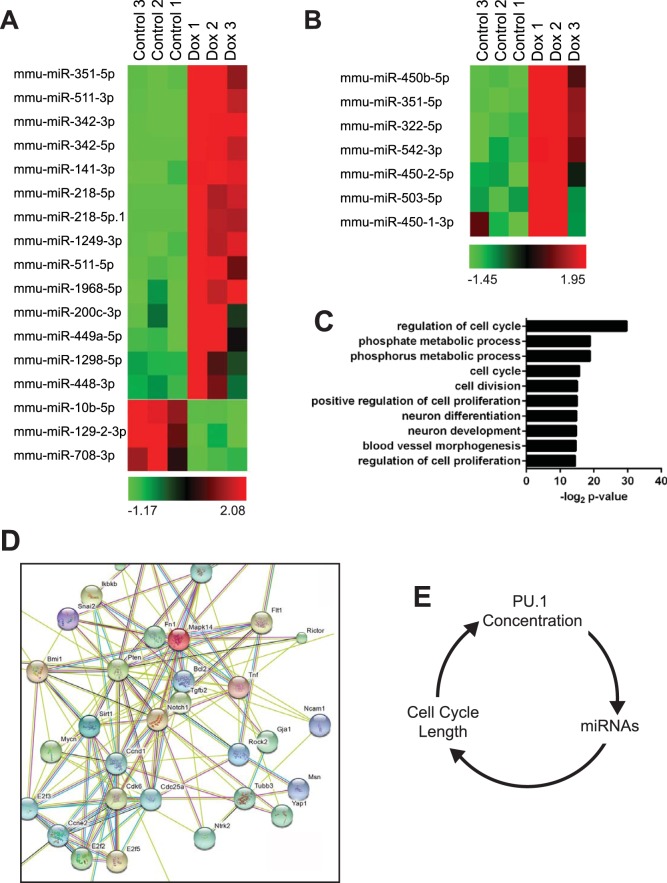
To determine if PU.1 induces microRNAs capable of inducing the pattern of changes in gene expression shown in Fig. 2, microRNA sequencing (miR-seq) was conducted using small RNAs enriched from uninduced or induced iBN cells. Illumina TruSeq small RNA libraries were generated from three biological replicates of 72-h-induced or uninduced cells and sequenced, generating 29 to 36 million reads per sample. Sequenced miRNAs were aligned to mouse genome mm10 and miRBase, version 21, and miRDeep2 was used to identify known and novel miRNAs. In all samples, the most abundant miRNA was miR-146b-5p, representing 11.6% of control reads and 12.2% of induced reads. DESeq2 was used to determine differentially expressed miRNAs. Seventeen miRNAs exhibited a greater than 2-fold change after PU.1 induction (Fig. 8A and B). Of these, 14 were significantly increased in response to increased PU.1, and 3 demonstrated decreased expression. Among the most highly induced miRNAs was miR-342, which demonstrated 19-fold induction of the miR-342-3p and 2-fold induction of the miR-342-5 p sequences. miR-342 is encoded in the third intron of the Evl gene, and upregulation of miR-342 was associated with increased PU.1 binding in the first intron (Fig. S2A). Changes in expression of miR-342-3p were confirmed using quantitative real-time PCR using a TaqMan small RNA assay (Fig. S2B). DESeq2 also indicated significant upregulation of miR-351 and increased expression of miRNAs in the adjacent locus containing miR-322 (miR-424 in Homo sapiens), a known regulator of macrophage differentiation (31). This locus contained a PU.1 binding site that demonstrated significant enrichment in PU.1 binding following induction (Fig. S2C).
FIG 8.
Analysis of microRNAs induced by PU.1 using miRNA-seq (A) Expression of miRNAs induced or repressed ≥2-fold 72 h after PU.1 induction in iBN cells using 1,000 ng/ml Dox. (B) Heat map showing quantification of miRNAs in the miR-322 cluster. (C) Biological pathway analysis of validated miRNA targets. Validated targets of significantly induced miRNAs were associated with the biological pathways shown on the y axis. (D) Functional protein association analysis of validated genes regulated by PU.1-induced miRNAs. STRING was used to integrate proteins encoded by 49 genes regulated by PU.1-induced miRNAs. (E) Model for the interaction between PU.1 concentration, expression of miRNAs, and cell cycle length to regulate cell cycle progression in the myeloid lineage. mmu, Mus musculus.
Next, miRGate was used to identify validated gene targets of the 14 upregulated miRNAs. This analysis identified 49 validated gene targets that were analyzed for Gene Ontology (GO) using DAVID (Data Set S2). Interestingly, the top biological process predicted to be regulated by validated miRNA targets was regulation of cell cycle (GO:0051726) followed by phosphate metabolic process and phosphorus metabolic process (Fig. 8C). The first process included E2f1 that was regulated by miR-342-3p, miR-342-5p, miR-449a-5p, and miR-1298-5p. The other two processes included enzymes involved in phospholipid metabolism, including the product of Acly, which is a validated target of miR-141-3p, miR-1968-5p, and miR-511-3p. STRING analysis of the protein products of the 49 genes revealed a regulatory network centered on CCND1, CCNE2, CCDK6, E2F3, and E2F2, all key regulators of cell cycle progression (Fig. 8D). In summary, PU.1 induced expression of miRNAs that target mRNAs encoding key regulators of cell cycle and lipid anabolism for downregulation.
DISCUSSION
In this study, we used the inducible BN cell line system to investigate the mechanism by which PU.1 coordinates differentiation with reduced cell cycle progression in the myeloid lineage. We performed genome-wide ChIP-seq analysis coupled with gene expression analysis to determine target genes of PU.1 that may be involved in cell cycle regulation. Unexpectedly, we found that downregulated genes associated with direct PU.1 binding encoded enzymes that function in lipid anabolism. Inhibition of ACL activity was sufficient to induce cell cycle arrest and differentiation in BN cells. E2f1, an important transcriptional regulator of the G1-to-S-phase transition and metabolism, was downregulated by PU.1 indirectly through miR-223. MicroRNAs upregulated by PU.1 were predicted to downregulate a network of cell cycle-regulatory and lipid anabolic genes. These results suggest that PU.1 coordinates cell cycle progression with differentiation through induction of microRNAs targeting cell cycle regulators and lipid anabolism.
We found that induction of PU.1 in iBN cells resulted in reduced expression of E2F1. E2F1 to E2F3 activate genes in myeloid cells to induce progression of G1 to S phase (14). We found that downregulation of E2F1 expression by PU.1 in iBN cells was mediated indirectly through miR-223, and this downregulation was associated with reduced cell cycle progression. MicroRNA-sequencing analysis identified 14 miRNAs that were significantly increased by PU.1 induction. These 14 miRNAs were validated as targeting 49 genes that were downregulated in our gene expression analysis. Notably, E2f1 is targeted by miR-342-3p, miR-342-5p, miR-449a-5p, and miR-1298-5p. E2f1 has miR-342-3p binding sites at positions 262 to 268 and at positions 284 to 290 of the 3′ untranslated region (UTR) (32). STRING analysis also suggested that E2F2 and E2F3 were targeted by induced miRNAs (Fig. 8). E2F2 and E2F3 are validated targets of miR-342-3p. E2F1 is an important activator of lipid anabolism at the G1-to-S-phase transition. Genes encoding lipid anabolic enzymes are downregulated in E2f1−/− mouse hepatocytes, and, conversely, in obese mice E2F1 is upregulated (16). Reanalysis of published E2F1 ChIP-seq data (33) showed that E2F1 interacts with the Acly promoter (data not shown). Thus, reduced E2F1 expression levels, induced by PU.1 through miR-223, would be expected to impair cell cycle progression by a variety of mechanisms including reduced lipid anabolism. Further work will need to be performed to determine the identity of genes repressed by PU.1 through miR-223 as opposed to other microRNAs. Experiments could include the generation of miR-223 knockout iBN cells for such analysis.
Although there are reports in the literature suggesting that PU.1 can function as a transcriptional repressor, there are few descriptions of a mechanism for transcriptional repression by PU.1 (10, 13). Indeed, genome-wide ChIP-seq studies demonstrated that PU.1 binding in the genome of multiple blood cell types is always associated with histone marks of transcriptional activation (34, 35). We found that PU.1 binding was associated with transcriptional activation of the E2f1 gene as determined by qPCR analysis of immature unspliced mRNA transcripts and ChIP analysis of H3K27Me3. Genes such as Acly were downregulated by PU.1 even as they were associated with induced peaks of PU.1 association. Therefore, we expect that genes such as Acly are transcriptionally activated by PU.1 but regulated posttranscriptionally by PU.1-activated miRNAs. We speculate that induction of miRNAs by PU.1 is an important mechanism for gene repression upon increased PU.1 concentration. We cannot rule out additional mechanisms for gene repression by PU.1, such as transcriptional pausing, but investigation of such mechanisms will require additional work.
An interesting question is why there might be a need for PU.1 to function in a concentration-dependent fashion as both a direct activator and indirect repressor of gene expression in myeloid cells. We speculate that genes such as E2f1 and Acly need to have finely tuned levels of expression at different phases of the cell cycle. In support of this idea, we found that overexpression of E2F1 induced apoptosis in BN cells (see Fig. S1 in the supplemental material) and that treatment of BN cells with the ACL inhibitor BMS303141 resulted in cell cycle arrest (Fig. 4). We speculate that expression of genes encoding E2f1 and Acly is transcriptionally activated by PU.1 and poised for rapid up- or downregulation by miRNAs in response to environmental cues.
Previous studies suggested a link between PU.1 levels and regulation of cell cycle progression. Small reductions in PU.1 levels lead to increased proliferation and reduced differentiation of hematopoietic stem cells and myeloid progenitor cells (6, 7, 10, 11). Reintroduction of PU.1 into proliferating cells expressing low levels of PU.1 rapidly induces cell cycle arrest and differentiation (5, 7, 12). Although PU.1 is expressed throughout myeloid differentiation, PU.1 protein levels increase during terminal differentiation into macrophages as a consequence of protein accumulation during a lengthening cell cycle (36). A positive feedback loop between PU.1 protein levels and the length of the cell cycle reinforces myeloid differentiation (36). Our results showed that inhibition of cell cycle progression with the ACL inhibitor BMS303141 led to increased levels of Spi1 and Adgre1 mRNA transcripts (Fig. 4D and E). Adgre1 is a direct target gene of PU.1 in macrophages that encodes the cell surface protein F4/80 (18). PU.1 regulates its own gene expression in myeloid cells through binding sites in an upstream regulatory region and the promoter region (10). We speculate that inhibition of ACL by BMS303141 results in cell cycle lengthening, resulting in accumulation of PU.1 protein. Increased PU.1 protein levels would be expected to lead to increased levels of Spi1 and Adgre1 mRNAs and would also be expected to further reinforce cell cycle arrest (Fig. 8E).
Nutrient status is an important input into the cell cycle clock, and it has long been appreciated that a reduction in the concentration of key nutrients will induce cell cycle arrest in cultured cells (37, 38). Lipid anabolism is a potentially important input into the cell cycle clock in developing myeloid cells (17). There is little known about how lipid levels are sensed by the cell cycle machinery, and it will be important to investigate this mechanism in future studies (37, 38). The results presented in this paper suggest that PU.1 coordinates cell cycle progression with differentiation through induction of microRNAs targeting cell cycle regulators and lipid anabolism.
MATERIALS AND METHODS
Cell culture.
PU.1-inducible BN cells (iBN cells) were generated as previously described (12). iBN cells were cultured in Iscove's modified Dulbecco's medium (IMDM) (Wisent, St-Bruno, Quebec, Canada) supplemented with 1 ng/ml granulocyte-macrophage colony-stimulating factor (GM-CSF; Peprotech, Quebec, Canada), 10% fetal bovine serum (Wisent), 100 U/ml penicillin-streptomycin, 2 mM l-glutamine (Wisent), and 5 × 10−5 mol/liter 2-mercaptoethanol (Sigma-Aldrich, St. Louis, MO). PU.1 or miR-223 induction experiments were performed by culture of inducible BN cells in 1.0 ng/ml GM-CSF in the presence or absence of 1,000 ng/ml doxycycline for 24 to 72 h. WEHI-3B cells were cultured in complete RPMI 1640 medium (Wisent) with the supplements described above. Platinum-E (Plat-E) retroviral packaging cells were cultured in complete low-glucose Dulbecco's modified Eagle's medium (DMEM; Wisent) with the supplements described above.
Flow cytometry.
Flow cytometric analysis was performed on cells washed in Dulbecco's phosphate-buffered saline (PBS; Wisent) containing 0.05 mol/liter EDTA and 0.5% bovine serum albumin (BSA; Sigma-Aldrich). F4/80 surface expression was determined using allophycocyanin (APC)-conjugated mouse anti-F4/80 antibody (Caltag Laboratories/Invitrogen, Carlsbad, CA) at a 1:200 dilution. Cell cycle analysis was performed using an APC-bromodeoxyuridine (BrdU) flow kit according to the manufacturer's protocol (BD Biosciences, Mississauga, Canada). TUNEL analysis for apoptosis was performed using an ApoBrdU Red DNA fragmentation kit according to the manufacturer's instructions (Biovision, Inc., Milpitas, CA). Flow cytometric analysis and sorting were performed using a FACSCalibur and FACSARia III instrument, respectively (BD Immunocytometry Systems, San Jose, CA) at the London Regional Flow Cytometry Facility. Flow cytometry data were analyzed using FlowJo, version 10 (Tree Star, Ashland, OR).
RNA preparation.
Total RNA was isolated using TRIzol (Life Technologies, Grand Island, NY) and reverse transcribed using an iScript cDNA synthesis kit (Bio-Rad, Hercules, CA). Nascent RNA enrichment was performed according to the manufacturer's instructions using a Click-iT nascent RNA capture kit (Life Technologies). Small RNAs were isolated using an miRNeasy minikit (Qiagen, Toronto, ON, Canada) and eluted in RNase-free water.
RT-qPCR.
Oligonucleotide primers were ordered from Integrated DNA Technologies (Coralville, IA). Primer sequences for reverse transcription-quantitative PCR (RT-qPCR) are described in Table S1 in the supplemental material. Relative mRNA levels were determined using β2-microglobulin (B2m) as a reference gene. Quantitative PCR was performed using PerfeCTa SYBR green supermix (Quanta, Gaithersburg, MD) with a Rotor-Gene 6000 instrument (Qiagen) for mRNA analysis. For miRNA analysis, cDNAs were synthesized using a TaqMan miRNA reverse transcription kit (ThermoFisher Scientific, Mississauga, Ontario, Canada). Quantitative PCR was performed using SensiFast Probe Hi-Rox (Bioline, Taunton, MA) using an Applied Biosystems StepOne real-time PCR system for miRNA analysis (ThermoFisher). Abundance of mature miRNAs was normalized to U6 spliceosomal RNA amplified in triplicate. Fold change in expression was calculated using the delta CT (where CT is threshold cycle) method (39).
Plasmid construction.
Primer sequences for plasmid construction are listed in Table S1 in the supplemental material. PCR products were cloned using a StrataClone PCR cloning kit (Agilent Technologies, La Jolla, CA). cDNA encoding mouse E2F1 was purchased from Origene (Rockville, MD). NotI sites were added by PCR. E2F1 cDNA was ligated into the NotI site of murine stem cell virus I (MSCV-I) Thy1.1 retroviral vector (17442; Addgene, Cambridge, MA) using T4 DNA ligase (New England BioLabs, Ipswich, MA). For luciferase assays, a 400-bp fragment of the first intron of the E2f1 gene was amplified from mouse genomic DNA using LA Taq (TaKaRa Bio, Mountain View, CA). BglII sites were added by PCR. The E2F1 intron 1 regulatory region was ligated into the BglII site of the pGL3-promoter luciferase reporter vector (Promega, Madison, WI). Site-directed mutagenesis of the predicted PU.1 binding site from GGAA to GGAC (the mutation is underlined) was performed using a Q5 site-directed mutagenesis kit and protocol (New England BioLabs). A 447-bp genomic fragment encoding miR-223 was amplified from mouse genomic DNA using LA Taq (TaKaRa Bio). This fragment was ligated into pRetroX-TRE3G (TaKaRa). All plasmids were confirmed by DNA sequencing. Plasmid DNA was prepared using a Maxi Plasmid kit (Geneaid, New Taipei City, Taiwan). To generate retroviral particles, platinum E (Plat-E) retroviral packaging cells were transfected with plasmids using polyethylenimine (PEIpro; PolyPlus, Illkirch, France). Cell-free supernatant containing virus was collected 48 h after transfection.
Transient transfection.
Electroporation was used to introduce the luciferase vectors into WEHI-3B cells. Renilla luciferase (pRL-TK) was used as a control (Promega). Electroporation was performed using a Gene Pulser II with settings of 950 μF and 220 V (Bio-Rad, Mississauga, Ontario, Canada). Luciferase activity was measured 24 h after electroporation using a Dual-Luciferase kit (Promega). Luminescence was measured using a plate reader (BioTek, Winooski VT).
ChIP and ChIP-seq.
Doxycycline-induced and noninduced iBN cells were cross-linked with 1% formaldehyde for 10 min at room temperature before being treated with 125 mM glycine to terminate cross-linking. Fixed cells were lysed in lysis buffer (50 mM Tris-HCl [pH 8.1], 10 mM EDTA, 1% SDS) containing Halt protease inhibitor mixture (ThermoFisher Scientific). Chromatin solutions were sonicated to yield DNA fragments in the range of 150 to 300 bp using a Bioruptor-300 sonicator (Diagenode, Sparta, NJ). Antibodies used for chromatin immunoprecipitation (ChIP) included anti-H3K27Ac (Abcam, Toronto, Ontario, Canada), anti-H3K27Me3 (Millipore Canada, Etobicoke, Ontario, Canada), anti-FLAG M2 (Sigma-Aldrich, St. Louis, MO), or anti-PU.1 T-21 (Santa Cruz Biotechnology, Santa Cruz, CA). Sonicated chromatin was incubated with antibodies conjugated to protein G Dynabeads (Life Technologies) overnight at 4°C. Bound beads were washed once with low-salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl [pH 8.0], 150 mM NaCl), once with high-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl [pH 8.0], 500 mM NaCl), once with LiCl buffer (0.25 M LiCl, 1% Nonidet P-40, 1% Na-deoxycholate, 1 mM EDTA, 10 mM Tris-HCl [pH 8.0]), and twice with Tris-EDTA buffer at pH 8. Immunocomplexes were eluted with elution buffer (1% SDS, 0.1 M NaHCO3). Cross-links were reversed in a final volume of 300 μl, containing 200 mM NaCl, overnight at 65°C. DNA was purified using a QiaQuick PCR cleanup system (Qiagen). qPCR was performed using Bio-Rad SYBR green supermix on a Rotor-Gene 6000 instrument (Qiagen) with primers listed in Table S1. Threshold cycle values obtained from qPCR were used to determine the percent input of each ChIP reaction product using the comparative threshold cycle method. Single-end reads were generated on an Illumina HiSeq instrument using the services of Genome Quebec.
Sequencing of small RNAs.
Libraries were generated from 1,000 ng of RNA using a TruSeq small RNA sample prep kit (Illumina, San Diego, CA), according to the manufacturer's recommendations. cDNA purification was performed on a Pippin Prep instrument (Sage Science, Beverly, MA). Final libraries were quantified using a Quant-iT PicoGreen double-stranded DNA (dsDNA) assay kit (Life Technologies) and a Kapa Illumina Genome Analyzer with revised primers-SYBR Fast Universal kit (D-Mark Biosciences, Toronto, Ontario, Canada). Average fragment size was determined using an LaChip GX (PerkinElmer, Waltham MA) instrument. Libraries were multiplexed and sequenced on an Illumina HiSeq 2000 SR50 using the services of Genome Quebec.
Computational and bioinformatic methods.
Detailed bioinformatic methods are described in supplemental bioinformatic methods. Gene lists are provided as Data Sets S1 and S2. ChIP-seq analysis was performed using Galaxy Suite. Sequencing adapters were removed from sequencing reads using Trim Galore (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/). DNA sequences generated by ChIP-seq were aligned to mouse genome reference mm9 using Bowtie (40). Peaks were called using MACS (41). Peak-to-gene associations were performed using GREAT (42). Biological pathway analysis was performed using DAVID (43). Gene interactions were determined using STRING (44). For microRNA-seq, trimmed reads were processed and mapped to the mm10 mouse genome using miRDeep2 Mapper, version 2.0.0. Collapsed reads were mapped to mirBase release 21 (45) using miRDeep2 for identification of novel and known miRNAs (46). Known miRBase mouse miRNAs were quantified using miRDeep2 quantifier, version 2.0.0, and differential analysis of fold change was conducted using DESeq2 (47). Differentially expressed miRNAs were considered significant if they had a log fold change greater than 1 and a P value of <0.05 (48). Significantly overexpressed miRNAs determined using DESeq2 were analyzed for predicted and validated targets using miRGate (49). Computational predictions of 3′ UTRs were determined using TargetScan and Miranda.
Statistical analysis.
All statistical analysis was performed on results of biological replicate experiments unless otherwise indicated. Significance was determined by Student's t test or one-way analysis of variance (ANOVA) using Prism, version 5 (GraphPad Software, La Jolla, CA). RT-qPCR analysis was performed using REST software (39) (Qiagen). Differences were considered significant with a P value of <0.05.
Accession number(s).
ChIP-seq and microRNA-seq data are available from the Gene Expression Omnibus (GEO) under accession number GSE79193. Microarray data are available under GEO accession number GSE52577.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the contribution of the staff of the Genome Quebec Innovation Centre, Montreal, Canada, for next-generation sequencing services. We acknowledge access to the Shared Hierarchical Academic Research Computing Network (SHARCNET) and Compute/Calcul Canada. We thank Peng Shao for performing RT-qPCR experiments. We thank Kristen Chadwick of the London Regional Flow Cytometry Core for assistance with cell sorting and analysis. We thank Fred Possmeyer and David Carter for helpful discussions.
This work was supported by grant 386046 from the Natural Sciences and Engineering Research Council of Canada.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00013-17.
REFERENCES
- 1.Passegue E, Wagers AJ, Giuriato S, Anderson WC, Weissman IL. 2005. Global analysis of proliferation and cell cycle gene expression in the regulation of hematopoietic stem and progenitor cell fates. J Exp Med 202:1599–1611. doi: 10.1084/jem.20050967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perdiguero EG, Geissmann F. 2016. The development and maintenance of resident macrophages. Nat Immunol 17:2–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sieweke MH, Allen JE. 2013. Beyond stem cells: self-renewal of differentiated macrophages. Science 342:1242974. doi: 10.1126/science.1242974. [DOI] [PubMed] [Google Scholar]
- 4.Iwasaki H, Somoza C, Shigematsu H, Duprez EA, Iwasaki-Arai J, Mizuno S, Arinobu Y, Geary K, Zhang P, Dayaram T, Fenyus ML, Elf S, Chan S, Kastner P, Huettner CS, Murray R, Tenen DG, Akashi K. 2005. Distinctive and indispensable roles of PU.1 in maintenance of hematopoietic stem cells and their differentiation. Blood 106:1590–1600. doi: 10.1182/blood-2005-03-0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeKoter RP, Singh H. 2000. Regulation of B lymphocyte and macrophage development by graded expression of PU.1. Science 288:1439–1441. doi: 10.1126/science.288.5470.1439. [DOI] [PubMed] [Google Scholar]
- 6.Houston IB, Kamath MB, Schweitzer BL, Chlon TM, DeKoter RP. 2007. Reduction in PU.1 activity results in a block to B-cell development, abnormal myeloid proliferation, and neonatal lethality. Exp Hematol 35:1056–1068. doi: 10.1016/j.exphem.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosenbauer F, Wagner K, Kutok JL, Iwasaki H, Le Beau MM, Okuno Y, Akashi K, Fiering S, Tenen DG. 2004. Acute myeloid leukemia induced by graded reduction of a lineage-specific transcription factor, PU1. Nat Genet 36:624–630. doi: 10.1038/ng1361. [DOI] [PubMed] [Google Scholar]
- 8.Lavallee VP, Baccelli I, Krosl J, Wilhelm B, Barabe F, Gendron P, Boucher G, Lemieux S, Marinier A, Meloche S, Hebert J, Sauvageau G. 2015. The transcriptomic landscape and directed chemical interrogation of MLL-rearranged acute myeloid leukemias. Nat Genet 47:1030–1037. doi: 10.1038/ng.3371. [DOI] [PubMed] [Google Scholar]
- 9.Cook WD, McCaw BJ, Herring C, John DL, Foote SJ, Nutt SL, Adams JM. 2004. PU.1 is a suppressor of myeloid leukemia, inactivated in mice by gene deletion and mutation of its DNA binding domain. Blood 104:3437–3444. doi: 10.1182/blood-2004-06-2234. [DOI] [PubMed] [Google Scholar]
- 10.Staber PB, Zhang P, Ye M, Welner RS, Nombela-Arrieta C, Bach C, Kerenyi M, Bartholdy BA, Zhang H, Alberich-Jorda M, Lee S, Yang H, Ng F, Zhang J, Leddin M, Silberstein LE, Hoefler G, Orkin SH, Gottgens B, Rosenbauer F, Huang G, Tenen DG. 2013. Sustained PU.1 levels balance cell-cycle regulators to prevent exhaustion of adult hematopoietic stem cells. Mol Cell 49:934–946. doi: 10.1016/j.molcel.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Will B, Vogler TO, Narayanagari S, Bartholdy B, Todorova TI, da Silva Ferreira M, Chen J, Yu Y, Mayer J, Barreyro L, Carvajal L, Neriah DB, Roth M, van Oers J, Schaetzlein S, McMahon C, Edelmann W, Verma A, Steidl U. 2015. Minimal PU.1 reduction induces a preleukemic state and promotes development of acute myeloid leukemia. Nat Med 21:1172–1181. doi: 10.1038/nm.3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ziliotto R, Gruca MR, Podder S, Noel G, Ogle CK, Hess DA, DeKoter RP. 2014. PU.1 promotes cell cycle exit in the murine myeloid lineage associated with downregulation of E2F1. Exp Hematol 42:204–217.e1. doi: 10.1016/j.exphem.2013.11.011. [DOI] [PubMed] [Google Scholar]
- 13.Kamath MB, Houston IB, Janovski AJ, Zhu X, Gowrisankar S, Jegga AG, DeKoter RP. 2008. Dose-dependent repression of T-cell and natural killer cell genes by PU.1 enforces myeloid and B-cell identity. Leukemia 22:1214–1225. doi: 10.1038/leu.2008.67. [DOI] [PubMed] [Google Scholar]
- 14.Trikha P, Sharma N, Opavsky R, Reyes A, Pena C, Ostrowski MC, Roussel MF, Leone G. 2011. E2f1–3 are critical for myeloid development. J Biol Chem 286:4783–4795. doi: 10.1074/jbc.M110.182733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishida S, Huang E, Zuzan H, Spang R, Leone G, West M, Nevins JR. 2001. Role for E2F in control of both DNA replication and mitotic functions as revealed from DNA microarray analysis. Mol Cell Biol 21:4684–4699. doi: 10.1128/MCB.21.14.4684-4699.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Denechaud PD, Lopez-Mejia IC, Giralt A, Lai Q, Blanchet E, Delacuisine B, Nicolay BN, Dyson NJ, Bonner C, Pattou F, Annicotte JS, Fajas L. 2016. E2F1 mediates sustained lipogenesis and contributes to hepatic steatosis. J Clin Invest 126:137–150. doi: 10.1172/JCI81542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scaglia N, Tyekucheva S, Zadra G, Photopoulos C, Loda M. 2014. De novo fatty acid synthesis at the mitotic exit is required to complete cellular division. Cell Cycle 13:859–868. doi: 10.4161/cc.27767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Reilly D, Addley M, Quinn C, MacFarlane AJ, Gordon S, McKnight AJ, Greaves DR. 2004. Functional analysis of the murine Emr1 promoter identifies a novel purine-rich regulatory motif required for high-level gene expression in macrophages. Genomics 84:1030–1040. doi: 10.1016/j.ygeno.2004.08.016. [DOI] [PubMed] [Google Scholar]
- 19.Turkistany SA, DeKoter RP. 2011. The transcription factor PU.1 is a critical regulator of cellular communication in the immune system. Arch Immunol Ther Exp (Warsz) 59:431–440. doi: 10.1007/s00005-011-0147-9. [DOI] [PubMed] [Google Scholar]
- 20.Zaidi N, Swinnen JV, Smans K. 2012. ATP-citrate lyase: a key player in cancer metabolism. Cancer Res 72:3709–3714. doi: 10.1158/0008-5472.CAN-11-4112. [DOI] [PubMed] [Google Scholar]
- 21.Srere PA. 1972. The citrate enzymes: their structures, mechanisms, and biological functions. Curr Top Cell Regul 5:229–283. doi: 10.1016/B978-0-12-152805-8.50013-7. [DOI] [PubMed] [Google Scholar]
- 22.Holland SM. 2010. Chronic granulomatous disease. Clin Rev Allergy Immunol 38:3–10. doi: 10.1007/s12016-009-8136-z. [DOI] [PubMed] [Google Scholar]
- 23.Bauer DE, Hatzivassiliou G, Zhao F, Andreadis C, Thompson CB. 2005. ATP citrate lyase is an important component of cell growth and transformation. Oncogene 24:6314–6322. doi: 10.1038/sj.onc.1208773. [DOI] [PubMed] [Google Scholar]
- 24.Hatzivassiliou G, Zhao F, Bauer DE, Andreadis C, Shaw AN, Dhanak D, Hingorani SR, Tuveson DA, Thompson CB. 2005. ATP citrate lyase inhibition can suppress tumor cell growth. Cancer Cell 8:311–321. doi: 10.1016/j.ccr.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 25.Li JJ, Wang H, Tino JA, Robl JA, Herpin TF, Lawrence RM, Biller S, Jamil H, Ponticiello R, Chen L, Chu CH, Flynn N, Cheng D, Zhao R, Chen B, Schnur D, Obermeier MT, Sasseville V, Padmanabha R, Pike K, Harrity T. 2007. 2-Hydroxy-N-arylbenzenesulfonamides as ATP-citrate lyase inhibitors. Bioorg Med Chem Lett 17:3208–3211. doi: 10.1016/j.bmcl.2007.03.017. [DOI] [PubMed] [Google Scholar]
- 26.DeGregori J, Leone G, Miron A, Jakoi L, Nevins JR. 1997. Distinct roles for E2F proteins in cell growth control and apoptosis. Proc Natl Acad Sci U S A 94:7245–7250. doi: 10.1073/pnas.94.14.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li SK, Abbas AK, Solomon LA, Groux GM, DeKoter RP. 2015. Nfkb1 activation by the E26 transformation-specific transcription factors PU.1 and Spi-B promotes Toll-like receptor-mediated splenic B cell proliferation. Mol Cell Biol 35:1619–1632. doi: 10.1128/MCB.00117-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hong SH, Kim KS, Oh IH. 2015. Concise review: Exploring miRNAs—toward a better understanding of hematopoiesis. Stem Cells 33:1–7. doi: 10.1002/stem.1810. [DOI] [PubMed] [Google Scholar]
- 29.Pulikkan JA, Dengler V, Peramangalam PS, Peer Zada AA, Muller-Tidow C, Bohlander SK, Tenen DG, Behre G. 2010. Cell-cycle regulator E2F1 and microRNA-223 comprise an autoregulatory negative feedback loop in acute myeloid leukemia. Blood 115:1768–1778. doi: 10.1182/blood-2009-08-240101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fukao T, Fukuda Y, Kiga K, Sharif J, Hino K, Enomoto Y, Kawamura A, Nakamura K, Takeuchi T, Tanabe M. 2007. An evolutionarily conserved mechanism for microRNA-223 expression revealed by microRNA gene profiling. Cell 129:617–631. doi: 10.1016/j.cell.2007.02.048. [DOI] [PubMed] [Google Scholar]
- 31.Rosa A, Ballarino M, Sorrentino A, Sthandier O, De Angelis FG, Marchioni M, Masella B, Guarini A, Fatica A, Peschle C, Bozzoni I. 2007. The interplay between the master transcription factor PU.1 and miR-424 regulates human monocyte/macrophage differentiation. Proc Natl Acad Sci U S A 104:19849–19854. doi: 10.1073/pnas.0706963104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tai MC, Kajino T, Nakatochi M, Arima C, Shimada Y, Suzuki M, Miyoshi H, Yatabe Y, Yanagisawa K, Takahashi T. 2015. miR-342-3p regulates MYC transcriptional activity via direct repression of E2F1 in human lung cancer. Carcinogenesis 36:1464–1473. doi: 10.1093/carcin/bgv152. [DOI] [PubMed] [Google Scholar]
- 33.Cao AR, Rabinovich R, Xu M, Xu X, Jin VX, Farnham PJ. 2011. Genome-wide analysis of transcription factor E2F1 mutant proteins reveals that N- and C-terminal protein interaction domains do not participate in targeting E2F1 to the human genome. J Biol Chem 286:11985–11996. doi: 10.1074/jbc.M110.217158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang JA, Mortazavi A, Williams BA, Wold BJ, Rothenberg EV. 2012. Dynamic transformations of genome-wide epigenetic marking and transcriptional control establish T cell identity. Cell 149:467–482. doi: 10.1016/j.cell.2012.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lara-Astiaso D, Weiner A, Lorenzo-Vivas E, Zaretsky I, Jaitin DA, David E, Keren-Shaul H, Mildner A, Winter D, Jung S, Friedman N, Amit I. 2014. Immunogenetics. Chromatin state dynamics during blood formation. Science 345:943–949. doi: 10.1126/science.1256271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kueh HY, Champhekar A, Nutt SL, Elowitz MB, Rothenberg EV. 2013. Positive feedback between PU.1 and the cell cycle controls myeloid differentiation. Science 341:670–673. doi: 10.1126/science.1240831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee IH, Finkel T. 2013. Metabolic regulation of the cell cycle. Curr Opin Cell Biol 25:724–729. doi: 10.1016/j.ceb.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aguilar V, Fajas L. 2010. Cycling through metabolism. EMBO Mol Med 2:338–348. doi: 10.1002/emmm.201000089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pfaffl MW, Horgan GW, Dempfle L. 2002. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. 2010. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang DW, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 44.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C. 2015. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kozomara A, Griffiths-Jones S. 2014. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42:D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Friedlander MR, Chen W, Adamidi C, Maaskola J, Einspanier R, Knespel S, Rajewsky N. 2008. Discovering microRNAs from deep sequencing data using miRDeep. Nat Biotechnol 26:407–415. doi: 10.1038/nbt1394. [DOI] [PubMed] [Google Scholar]
- 47.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Abbas AK, Le K, Pimmett VL, Bell DA, Cairns E, DeKoter RP. 2014. Negative regulation of the peptidylarginine deiminase type IV promoter by NF-κB in human myeloid cells. Gene 533:123–131. doi: 10.1016/j.gene.2013.09.108. [DOI] [PubMed] [Google Scholar]
- 49.Andrés-León E, González Peña D, Gómez-López G, Pisano DG. 2015. miRGate: a curated database of human, mouse and rat miRNA-mRNA targets. Database (Oxford) 2015:bav035. doi: 10.1093/database/bav035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.