ABSTRACT
AMPK-related protein kinases (ARKs) coordinate cell growth, proliferation, and migration with environmental status. It is unclear how specific ARKs are activated at specific times. In the fission yeast Schizosaccharomyces pombe, the CaMKK-like protein kinase Ssp1 promotes cell cycle progression by activating the ARK Cdr2 according to cell growth signals. Here, we demonstrate that Ssp1 activates a second ARK, Ssp2/AMPKα, for cell proliferation in low environmental glucose. Ssp1 activates these two related targets by the same biochemical mechanism: direct phosphorylation of a conserved residue in the activation loop (Cdr2-T166 and Ssp2-T189). Despite a shared upstream kinase and similar phosphorylation sites, Cdr2 and Ssp2 have distinct regulatory input cues and distinct functional outputs. We investigated this specificity and found that distinct protein phosphatases counteract Ssp1 activity toward its different substrates. We identified the PP6 family phosphatase Ppe1 as the primary phosphatase for Ssp2-T189 dephosphorylation. The phosphatase inhibitor Sds23 acts upstream of PP6 to regulate Ssp2-T189 phosphorylation in a manner that depends on energy but not on the intact AMPK heterotrimer. In contrast, Cdr2-T166 phosphorylation is regulated by protein phosphatase 2A but not by the Sds23-PP6 pathway. Thus, our study provides a phosphatase-driven mechanism to induce specific physiological responses downstream of a master protein kinase.
KEYWORDS: AMPK, Cdr2, Ssp1, fission yeast, kinase, phosphatase, pombe
INTRODUCTION
Cells control proliferation, polarized growth, and migration according to nutrient and environmental status. These processes must be tightly regulated during normal growth and development, and loss of this coordination is coupled with distinct steps in the initiation and maturation of tumors (1, 2). The AMP-activated protein kinase (AMPK)-related kinase (ARK) subfamily of protein kinases is essential for coordination of cell growth, proliferation, and migration with environmental status (3). Human cells express at least 13 members of this conserved kinase subfamily, including the metabolic sensor AMPK, the cytoskeletal kinase MARK1/Par-1, and the SAD kinases that regulate cell growth and polarity (4). Given their functions in diverse cell biological processes, it is remarkable that all ARKs can be activated by a shared upstream activating kinase, which phosphorylates the activation loop of ARK kinase domains to turn them “on” (4). To meet the needs of a rapidly changing environment, different ARKs need to be turned on and off at different times. This leads to a simple question: how does a common activation mechanism activate different ARKs at different times?
The defining member of the ARK subfamily is AMPK. This conserved heterotrimeric complex is activated by low cellular energy status, typically the result of low levels of environmental nutrients (5). AMPK activation shifts cells toward catabolism and promotes the Warburg effect in cancer cells (5–7). The AMPK heterotrimer consists of a catalytic α subunit, a scaffolding β subunit, and an energy-sensing γ subunit, which contains nucleotide-binding CBS domains. AMPK catalytic activity depends on phosphorylation of the α subunit activation loop by upstream kinases. When cellular energy levels are low, the CBS domains in the γ subunit are bound to AMP, leading to high levels of phosphorylation within the α subunit activation loop. Under high-cellular-energy conditions, the CBS domains are bound to ATP, and the α subunit is dephosphorylated and thus inactive. The β subunit mediates conformational changes to couple the γ subunit energy sensing to activation of the catalytic α subunit (5, 7). Current models suggest that these conformational changes alter the access of inactivating phosphatases to the AMPK α subunit activation loop (8). In budding yeast, the catalytic α subunit can also be activated in an energy-dependent manner in the absence of the β and γ subunits (9, 10). This noncanonical mechanism represents heterotrimer-independent regulation of the α subunit, but the molecular component(s) for heterotrimer-independent energy sensing remains unknown.
All ARKs are activated by phosphorylation of the activation loop in their kinase domain (3, 4), and at least three activating kinases have been discovered in humans. Most notably, the tumor suppressor LKB1 was shown to phosphorylate and activate all 13 human ARKs in vitro (4). Similarly, budding yeast ARKs have three upstream kinases, and all three must be deleted simultaneously to prevent activation loop phosphorylation of AMPK (11, 12). The mechanisms that regulate ARK activation by upstream kinases have been well studied, but the counteracting phosphatases that remove this modification to inactivate ARKs are less understood. In humans, protein phosphatase 2C (PP2C) is thought to act as the primary phosphatase for AMPK, based primarily on in vitro studies (13–15). Budding yeast AMPK activation highlights the importance of phosphatases, as the upstream activating kinases do not appear to be regulated (8, 16). Rather, dynamic control may come from protein phosphatases, and PP2C, PP1, and PP6 have been implicated in deactivating budding yeast AMPK (10, 17, 18).
ARKs in the fission yeast Schizosaccharomyces pombe may represent a simplified version of this signaling network. A single upstream kinase, Ssp1, appears to be the sole ARK-activating kinase in fission yeast, unlike the functionally overlapping and redundant activating kinases in humans and budding yeast. Mutations in ssp1+ lead to pleiotropic defects, including energy stress, cell cycle progression, and cell polarity (19–23), consistent with activation of fission yeast ARKs for diverse cellular functions. Genetic experiments place Ssp1 upstream of Ssp2/AMPKα for growth in low glucose (22, 24, 25), and Ssp1 is required for phosphorylation of Ssp2 under low-nitrogen or low-glucose conditions (21, 26). Further, we recently identified Ssp1 as the sole upstream kinase for the activation loop of Cdr2 (27), an ARK that regulates cell size at division. Ssp1 progressively phosphorylates Cdr2-T166 as cells increase in size due to the inhibitory action of Pom1, a protein kinase that localizes to cell tips. Specifically, Pom1 directly phosphorylates the carboxyl terminus of Cdr2 (27, 28), and this modification antagonizes Cdr2 activation loop phosphorylation by Ssp1. The dynamic control of Cdr2 activation loop phosphorylation suggests a role for an inactivating protein phosphatase. More generally, dephosphorylation of ARK activation loops has the potential to generate dynamic control of these diverse enzymes but remains poorly understood, especially in the fission yeast model system.
In this study, we found that Ssp1 phosphorylates and activates Cdr2 and Ssp2 under distinct growth conditions for distinct physiological functions. This raises the question of where specificity arises in the face of constitutive Ssp1 activity. We show that different phosphatases counter Ssp1 activity on these two ARKs, indicating that phosphatases can generate specificity in this ARK network downstream of the master activator, Ssp1.
RESULTS
Ssp1 activates Ssp2 under low glucose.
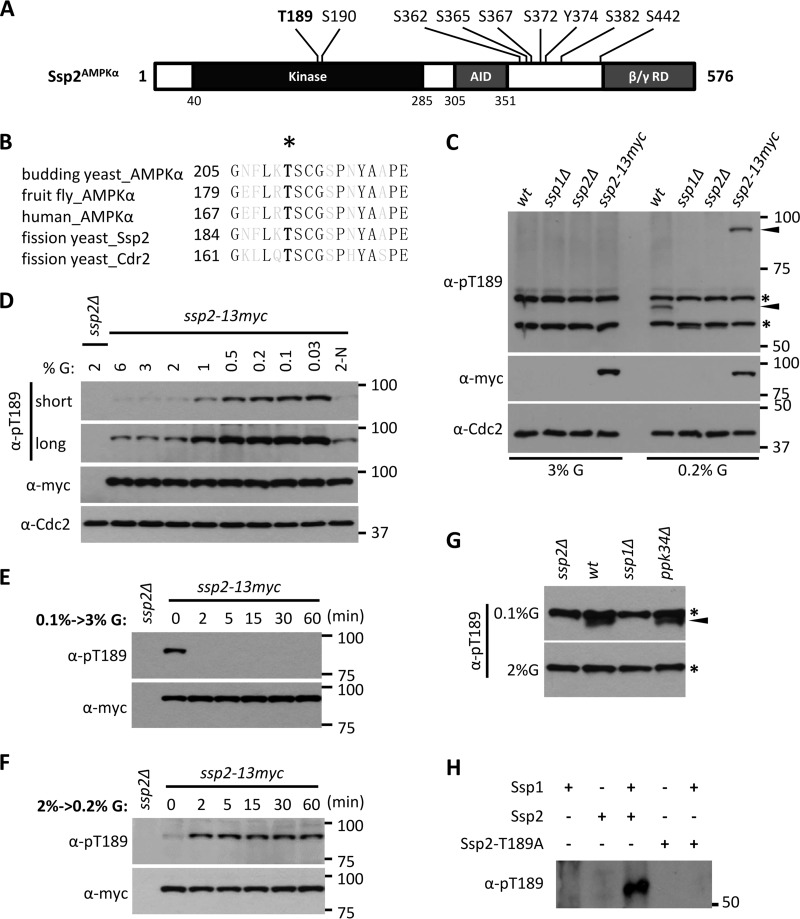
Our recent phosphoproteomics study revealed in vivo phosphorylation of Ssp2 (29), the catalytic α subunit of the fission yeast AMPK heterotrimer (Fig. 1A). In particular, we identified phosphorylation at the conserved threonine residue that is phosphorylated in the activation loops of AMPKs from diverse organisms (Ssp2-T189) (Fig. 1B). To investigate regulation of Ssp2, we generated a phosphospecific antibody against Ssp2-T189 phosphorylation (Ssp2-pT189). In whole-cell extracts, we detected a specific band for wild-type cells grown in low (0.2%) glucose but not in high (3%) glucose (Fig. 1C), consistent with previous studies (21, 26). This band was absent in ssp2Δ cells and migrated at the predicted higher molecular weight in lysates from cells expressing the fusion protein Ssp2-13–myc (Fig. 1C). Ssp2-T189 phosphorylation in cells was exquisitely sensitive to the glucose concentration. We detected low levels of Ssp2-pT189 under standard high-glucose conditions (2% to 6%) and increasing phosphorylation as glucose became limiting (Fig. 1D). Upon changes in the glucose concentration, the Ssp2-T189 phosphorylation state changed rapidly. Specifically, Ssp2-pT189 was induced within 2 min of glucose removal and was lost with similar speed when glucose was restored (Fig. 1E and F). The total protein level of Ssp2 was unchanged in these experiments (Fig. 1E and F). These results reveal the presence of a kinase-phosphatase control system that acts on AMPK with remarkably rapid kinetics.
FIG 1.
Ssp1 phosphorylates Ssp2-T189 according to environmental glucose. (A) Ssp2/AMPKα phosphorylation sites identified in our recent phosphoproteomics study (29). T189 and S190 reside in the activation loop (T loop). AID, autoinhibitory domain; β/γ RD, regulatory domain of AMPK β and γ subunits. (B) Sequence alignment of activation loops of AMPKα from different species, as well as Cdr2. The black letters represent invariant residues; the asterisk denotes phosphorylated threonine. (C) Glucose-dependent detection of Ssp2-pT189. The indicated strains (wt, wild type) were grown in high (3%) or low (0.2%) glucose, and whole-cell extracts were blotted with the indicated antibodies. Anti-Cdc2 was used as a loading control. For anti-Ssp2-pT189, the asterisks denote background bands, and the arrowheads mark Ssp2-pT189 bands. (D) Ssp2-pT189 monitors glucose concentrations. Cells were shifted from 2% glucose (G) to the indicated glucose concentrations for 6 h, and then whole-cell extracts were blotted. 2-N, medium containing 2% glucose but lacking a nitrogen source. Both short and long exposures are shown for Ssp2-pT189. Anti-Cdc2 was used as a loading control. (E) Rapid dephosphorylation of Ssp2-pT189 upon shift from low (0.1%) to high (3%) glucose. The samples were whole-cell extracts. (F) Rapid phosphorylation of Ssp2-T189 upon shift from high (2%) to low (0.2%) glucose. The samples were whole-cell extracts. (G) Detection of Ssp2-pT189 in whole-cell extracts from the indicated strains grown under low (0.1%) or high (2%) glucose. The asterisks indicate background bands, and the arrowhead marks a specific Ssp2-pT189 band. (H) In vitro kinase assay using the indicated recombinant full-length proteins, which were all expressed and purified from bacteria.
We noted that the sequence surrounding Ssp2-T189 is nearly identical to the activation loop of Cdr2 (Fig. 1B), an AMPK-related protein kinase that functions in fission yeast cell cycle progression (30, 31). Cell growth induces Cdr2 activation through phosphorylation at this site (Cdr2-T166) (27). The sequence identity between the Cdr2 and Ssp2 activation loops suggests possible connections in the regulatory mechanisms of the two protein kinases, though they appear to function in distinct cellular processes. Cdr2-T166 is phosphorylated by the CaMKK-related protein kinase Ssp1 (27). Several lines of evidence identify Ssp1 as the upstream protein kinase for Ssp2-pT189. First, we found that Ssp2-pT189 was absent in ssp1Δ cells grown under low glucose (Fig. 1C and G), consistent with previous reports for Ssp1-Ssp2 phosphorylation under low nitrogen and low glucose (21, 26). Phosphorylation of Ssp2-T189 did not require Ppk34 (Fig. 1G), the other fission yeast CaMKK that has similarity to Ssp1 (32). To confirm and extend these results, we purified recombinant Ssp1 and Ssp2 proteins from bacteria and then performed in vitro kinase assays. We detected Ssp2-pT189 in the presence but not in the absence of Ssp1, and this signal was lost in the nonphosphorylatable mutant Ssp2-T189A (Fig. 1H). We conclude that Ssp1 phosphorylates Ssp2-T189 both in vitro and in cells. Thus, Ssp1 has two known substrates: Ssp2/AMPKα and Cdr2.
Distinct regulatory inputs and functional outputs for Ssp2 and Cdr2.
Ssp1 phosphorylates highly related motifs in Ssp2 and Cdr2 (Fig. 1B), raising the possibility that the two substrates are coregulated by similar physiological stimuli. In contrast to this prediction, Cdr2-T166 was phosphorylated to the same degree under both high- and low-glucose conditions (Fig. 2A), which dramatically changed Ssp2-pT189 levels. This indicates that changes in Ssp1 kinase activity do not underlie glucose-regulated phosphorylation of Ssp2-T189. Further, physical tethering of Ssp1 and Ssp2 using the green fluorescent protein (GFP) binding peptide (GBP) system (33) induced Ssp2-T189 phosphorylation under high-glucose conditions (Fig. 2B), confirming that Ssp1 remains active in high glucose. We previously showed that Cdr2-pT166 levels depend on the protein kinase Pom1, which directly phosphorylates residues outside the Cdr2 kinase domain. These modifications by Pom1 inhibit phosphorylation of Cdr2-T166 by Ssp1 and connect Ssp1-Cdr2 signaling with cell size at division (27). Unlike Cdr2-pT166, the levels of Ssp2-pT189 were the same in wild-type and pom1Δ cells (Fig. 2C). Therefore, Cdr2 and Ssp2 are activated by distinct physiological input signals, despite sharing the same upstream kinase and a highly conserved activation mechanism.
FIG 2.
Distinct regulation and functions of two substrates of Ssp1. (A) Comparison of Cdr2-pT166 and Ssp2-pT189 at different glucose levels. Cells were shifted from 3% glucose to different glucose levels for 10 min or 3 h, and then whole-cell extracts were blotted. No tag, strain without integrated epitope tags. (B) Physical tethering of Ssp1 and Ssp2 induces Ssp2-pT189 in high glucose. GBP binds to GFP with nanomolar affinity. The indicated strains were grown in high (2%) or low (0.1%) glucose, and whole-cell extracts were probed with the indicated antibodies. (C) Comparison of Ssp2-pT189 levels in whole-cell extracts from wild-type and pom1Δ cells grown under 2% or 0.1% glucose levels. (D) Phosphorylation of Ssp2-T189 but not Cdr2-T166 is required for cell growth under low-glucose conditions. Tenfold serial dilutions of the indicated strains were spotted on EMM4S with 2% or 0.1% glucose. Amk2 and Cbs2 are β and γ subunits of the AMPK heterotrimer, respectively. (E) Detection of Ssp2-pT189 in whole-cell extracts from the indicated strains grown in 0.1% or 2% glucose. The asterisks indicate background bands, and the arrowhead marks a specific Ssp2-pT189 band. (F) Cell lengths of dividing, septated cells of the indicated strains (means ± standard deviations [SD]; n > 50 for each value). The P values from two-tailed Student's t tests are indicated with brackets for specific pairs.
We next addressed the connection between these different “input” signals and the functional “outputs” of Ssp1-ARK signaling. Consistent with Ssp2-T189 phosphorylation by Ssp1 under low glucose, we found that ssp1Δ, ssp2Δ, and ssp2-T189A mutant strains were unable to grow like the wild type under low-glucose conditions (Fig. 2D). Further, deletion of the β (Amk2) and γ (Cbs2) subunits of the AMPK heterotrimer dramatically reduced both cell proliferation (Fig. 2D) (24, 25) and Ssp2-pT189 levels (Fig. 2E) under low glucose. These results confirm that Ssp1 phosphorylates Ssp2-T189 to activate AMPK for cell proliferation under low-glucose conditions. In contrast, cdr2Δ and cdr2-T166A mutants showed no defects in proliferation under low-glucose conditions (Fig. 2D). Mutations that impair Ssp1-Cdr2 signaling alter cell cycle progression (27), leading to increased cell size at division. We found that ssp2Δ and ssp2-T189A mutants had no effect on cell size at division, even when combined with cdr2-T166A mutation (Fig. 2F). We conclude that Ssp1-Ssp2 signaling modulates cell proliferation under low-glucose conditions, while Ssp1-Cdr2 signaling controls cell cycle progression.
Our combined results reveal striking specificity in the signaling networks controlled by the master regulatory protein kinase, Ssp1. Extracellular glucose tightly controls activation of one substrate (Ssp2) but not another (Cdr2) for a defined functional output: cell proliferation under low glucose. Ssp1 signaling to Cdr2 is modulated by cell growth signals for timely cell cycle progression, and this process is independent of Ssp2. This specificity occurs despite phosphorylation at a site that is highly conserved between the two substrates, raising a simple question: how does a shared activation mechanism turn on different substrates at different times?
Phosphatases actively counter Ssp1 kinase.
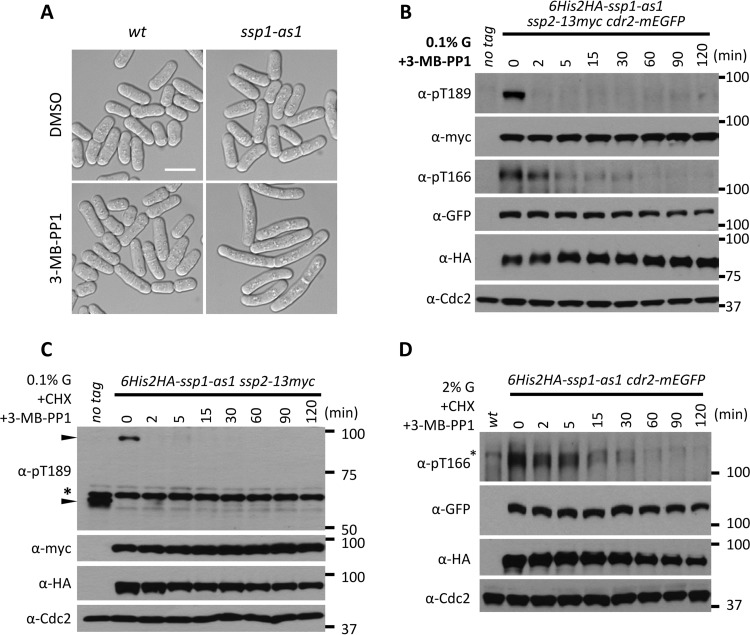
The balance of Ssp1 kinase versus a counteracting phosphatase dictates the level of activating phosphorylation for Cdr2 and Ssp2 at any given time. This suggests the possibility that phosphatases contribute to specificity in the Ssp1 signaling network. As a first step toward understanding this balance, we sought to determine if Cdr2-T166 and Ssp2-T189 are actively dephosphorylated in cells. We generated an analog-sensitive mutation in the Ssp1 kinase domain that confers rapid and specific inhibition by the ATP analog 3-MB-PP1 (34, 35). This ssp1-as1 (analog-sensitive) mutant (ssp1-L221G) did not exhibit defects when grown in the presence of dimethyl sulfoxide (DMSO) control but phenocopied ssp1Δ upon addition of 3-MB-PP1 (Fig. 3A). We integrated epitope tags at endogenous genomic loci to generate the 6His-2HA–ssp1-as1 ssp2-13–myc cdr2-mEGFP strain. When grown under low-glucose conditions, both Ssp2-T189 and Cdr2-T166 were phosphorylated in the strain, as expected. Upon addition of the inhibitor 3-MB-PP1, Ssp2-T189 phosphorylation was abolished within 2 min (Fig. 3B). This result indicates that phosphatases rapidly target Ssp2-T189 even under conditions that promote high steady-state levels of phosphorylation. Similarly, Cdr2-T166 was rapidly dephosphorylated upon Ssp1 inhibition, although the dephosphorylation rate was lower than for Ssp2 (Fig. 3B). These results are not due to protein turnover, as similar results were obtained in the presence of the translation inhibitor cycloheximide (CHX) (Fig. 3C and D). Further, Cdr2-pT166 was dephosphorylated similarly under high- and low-glucose conditions (Fig. 3D). We conclude that Ssp2-T189 and Cdr2-T166 are actively dephosphorylated by protein phosphatases in cells, even under conditions that promote steady-state phosphorylation. This raises the possibility that dephosphorylation plays an active role in dynamic control of the Ssp1 signaling network.
FIG 3.
Phosphatases actively counter Ssp1 kinase activity. (A) Phenotypes of wild-type ssp1+ and ssp1-as1 mutant cells treated with DMSO or 15 μM 3-MB-PP1 for 10 h at 37°C. ssp1-as1 harbors an L221G mutation; note the elongation of ssp1-as1 cells treated with 3-MB-PP1. Scale bar, 10 μm. (B) Ssp2-pT189 and Cdr2-pT166 are rapidly dephosphorylated upon inhibition of Ssp1. Cells were grown in YE4S with 0.1% glucose to mid-log phase and treated with 15 μM 3-MB-PP1. Whole-cell extracts were generated at the indicated time points and then probed using the indicated antibodies. Anti-Cdc2 was used as a loading control. (C) Cells were grown as for panel B and then treated with both 15 μM 3-MB-PP1 and 200 mg/ml CHX. Whole-cell extracts were generated at the indicated time points and then probed using the indicated antibodies. Anti-Cdc2 was used as a loading control. The asterisk marks a background band, and the arrowheads mark specific Ssp2-pT189 bands. (D) Cells were grown in YE4S with 2% glucose to mid-log phase and treated with 15 μM 3-MB-PP1 and 200 mg/ml CHX. Whole-cell extracts were generated at the indicated time points and then probed using the indicated antibodies. Anti-Cdc2 was used as a loading control. The asterisk marks a background band. Note that Cdr2-pT166 dephosphorylation is not glucose regulated.
An energy-sensing PP6 pathway inactivates Ssp2/AMPK.
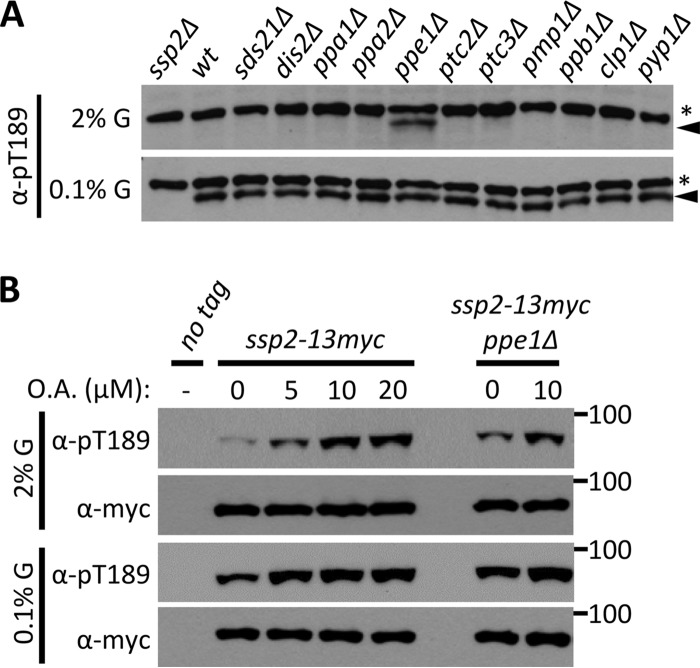
We next sought to identify the protein phosphatase pathways that counter Ssp1 kinase. We exploited the glucose-regulated phosphorylation of Ssp2-T189 to screen a collection of over 30 protein phosphatases (PP), including PP1 (sds21+ and dis2+), PP2A (ppa1+ and ppa2+), PP2C (ptc2+ and ptc3+), and PP6 (ppe1+). Among the mutants tested, only deletion of the PP6 catalytic subunit Ppe1 (36) led to Ssp2-T189 phosphorylation in high glucose (Fig. 4A), similar to the role of PP6 in budding yeast (17). This suggests that the PP2A-like protein phosphatase PP6 counteracts the activation of Ssp2 by Ssp1 through dephosphorylation of Ssp2-pT189 under high-glucose conditions. Consistent with the possibility of ectopic Ssp1-Ssp2 activation in ppe1 mutants, the ssp1 and ssp2 genes were originally identified in a screen for mutants that suppress loss of ppe1 (19). The level of Ssp2-pT189 in the ppe1Δ mutant is further increased by treatment with okadaic acid (OA) (Fig. 4B), which inhibits the PP1/PP2A/PP4/PP6 subfamilies (37, 38), indicating that additional phosphatases are likely to act directly or indirectly on Ssp2 under high glucose. We conclude that PP6 is the primary phosphatase that inactivates fission yeast AMPK under high-glucose conditions by dephosphorylating Ssp2-T189.
FIG 4.
PP6 is the primary phosphatase that keeps AMPK inactive in high glucose. (A) Ppe1 is required for dephosphorylation of Ssp2-pT189 under high-glucose conditions. Phosphatase mutant strains were grown in high (2%) or low (0.1%) glucose, and whole-cell lysates were blotted with anti-Ssp2-pT189 antibody. The asterisks denote background bands, and the arrowheads mark Ssp2-pT189 bands. (B) Effects of the indicated concentrations of okadaic acid (OA) on Ssp2-pT189 in ppe1+ and ppe1Δ strains. Whole-cell extracts were blotted.
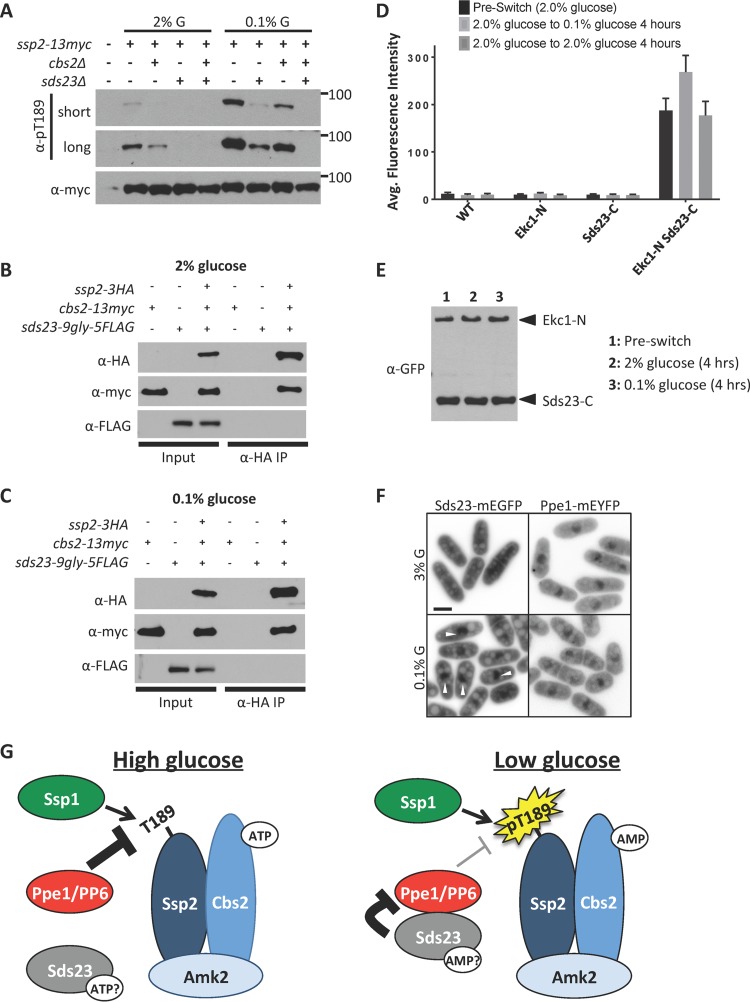
We investigated the regulation of PP6-AMPK by testing the role of Sds23, which inhibits PP6 activity by binding to the PP6 regulatory subunit Ekc1 (22). Much like the AMPK γ subunit Cbs2, Sds23 is comprised of four nucleotide-binding CBS domains. Ssp2-pT189 was nearly absent in sds23Δ cells grown in low glucose (Fig. 5A), which typically induces this phosphorylation. This suggests that Sds23 inhibits PP6 under low-glucose conditions to allow AMPK activation. Canonical energy sensing by the AMPK heterotrimer requires the γ subunit Cbs2. In cbs2Δ mutants, we observed low levels of Ssp2-pT189 (Fig. 5A) that were nonetheless regulated by glucose. This likely reflects the so-called “heterotrimer-independent” phosphorylation of the AMPK activation loop previously seen in budding yeast (9, 10). Interestingly, the sds23Δ cbs2Δ double mutant abolished detectable Ssp2-pT189 in low glucose (Fig. 5A). We conclude that Sds23 is required for heterotrimer-independent phosphorylation of Ssp2-T189.
FIG 5.
Sds23-PP6 signaling controls heterotrimer-independent phosphorylation of Ssp2-T189. (A) Detection of Ssp2-pT189 levels in the indicated mutants grown in 2% or 0.1% glucose. Whole-cell extracts were blotted. Note that Ssp2-pT189 is not detected in long exposures for a cbs2Δ sds23Δ double mutant. (B) Coimmunoprecipitation of Ssp2 with Cbs2 but not Sds23 under high (2%) glucose conditions. (C) Coimmunoprecipitation of Ssp2 with Cbs2 but not Sds23 under low (0.1%) glucose conditions. (D) Quantification of intracellular YFP fluorescence for the indicated strains from BiFC experiments. The values (arbitrary units) represent mean cytoplasmic fluorescence (n = 100 cells each; the error bars represent standard deviations). (E) Detection of Ekc1-NYFP and Sds23-CYFP protein levels before and after shift to 2% or 0.1% glucose. Equal amounts of whole-cell extracts were blotted. (F) Localization of Sds23-mEGFP and Ppe1-mEYFP in 3% glucose or 0.1% glucose. The arrowheads mark nuclear Sds23. The images are inverted single focal planes. Scale bar, 5 μm. (G) Schematic diagrams of pathways that control phosphoactivation of Ssp2 according to the glucose concentration. Nucleotide levels impact AMPK activation through canonical energy sensing mediated by Cbs2, and also through the Sds23-PP6 pathway.
We considered two possible mechanisms for the role of Sds23 in this pathway. First, Sds23 could act as an additional γ subunit for energy sensing by AMPK, based on the similar domain layouts of Cbs2 and Sds23. However, Ssp2 consistently coimmunoprecipitated with Cbs2 but not with Sds23 (Fig. 5B and C). This stable interaction was independent of cellular energy states (Fig. 5B and C) and confirms that Sds23 is not a subunit of AMPK (22). A second possible mechanism is that Sds23 differentially inhibits PP6 under high- versus low-glucose conditions due to energy sensing by its CBS domains. Sds23 binds to Ekc1, the regulatory subunit of PP6 (22). We investigated Sds23-Ekc1 interactions using bimolecular fluorescence complementation (BiFC) (also called “split yellow fluorescent protein [YFP]”) in living cells. We integrated functional tags at the endogenous loci of Ekc1 and Sds23 and found that cells expressing either Ekc1-NYFP (the N-terminal fragment of YFP) or Sds23-CYFP (the C-terminal fragment of YFP) alone did not fluoresce. However, cells expressing both Ekc1-NYFP and Sds23-CYFP generated strong YFP fluorescence, consistent with the physical interaction of Sds23 and Ekc1 (Fig. 5D). Importantly, the fluorescence intensity increased when we switched cells from high glucose (2%) to low glucose (0.1%) but was unchanged in control switches (2% to 2%) (Fig. 5D). This increase in fluorescence was not due to changes in Sds23 or Ekc1 protein levels (Fig. 5E). These experiments indicate that low cellular energy increases the interaction between Sds23 and PP6.
We extended this finding by testing spatial regulation within the Sds23-PP6 pathway. Ssp2 localizes in the cytoplasm in high glucose but shifts to the nucleus upon activation in low glucose (21). We integrated functional fluorescent tags at the endogenous loci for Sds23 and Ppe1 and imaged their localization under high versus low glucose. Ppe1-monomeric enhanced YFP (mEYFP) was concentrated in the nucleus, but also in the cytoplasm, under both high- and low-glucose conditions (Fig. 5F). Interestingly, Sds23-monomeric enhanced GFP (mEGFP) localized evenly in the cytoplasm and nucleus under high glucose, but the Sds23-mEGFP concentration in the nucleus increased by 60% upon shift to low-glucose conditions (Fig. 5F) (P < 0.0001 by two-tailed Student's t test; n = 20 cells for each condition). Translocation of Sds23 into the nucleus increases spatial overlap with its inhibitory target Ppe1 under low glucose. This spatial control step is consistent with both increased Sds23-PP6 interactions by BiFC and the additive/synergistic effects of sds23Δ and cbs2Δ on Ssp2-pT189 under low glucose. We conclude that AMPK activation is sensitive to environmental glucose because of two independent mechanisms. First, the Cbs2 γ subunit mediates canonical energy sensing by binding to ATP versus AMP and thereby controlling activation through allostery within the heterotrimer. Second, we have uncovered a novel glucose-regulated Sds23-PP6 pathway for AMPK activation. These combined experiments have revealed the dynamic kinase-phosphatase control system that coordinates fission yeast AMPK activation with glucose availability (Fig. 5G).
A distinct phosphatase pathway targets Cdr2.
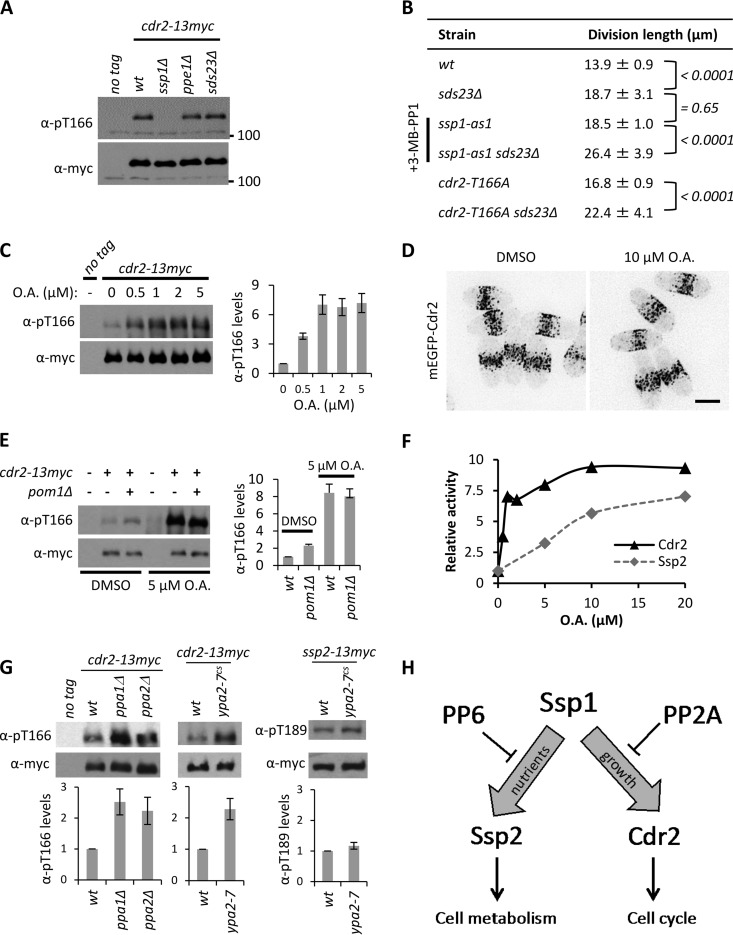
We next tested if the Sds23-PP6 phosphatase pathway also targets Cdr2-T166, which shares sequence homology with Ssp2-T189 and is also phosphorylated and activated by Ssp1 (27). Remarkably, Cdr2-pT166 levels were completely unaffected by the ppe1Δ and sds23Δ mutations (Fig. 6A), which dramatically altered Ssp2-pT189 levels. sds23Δ mutant cells are elongated at division (Fig. 6B), indicative of a cell cycle defect. We used genetic epistasis to test if this phenotype reflects a role for Sds23 in regulating Cdr2-T166 phosphorylation. The cell elongation phenotypes of sds23Δ and cdr2-T166A mutants were additive (Fig. 6B). This contrasts with the nonadditive phenotype when combining mutations in Cdr2 and Ssp1, which act in a linear pathway (27). Further, sds23Δ ssp1-as mutants displayed additive defects when Ssp1 kinase activity was inhibited by 3-MB-PP1 (Fig. 6B). We conclude that the Sds23-PP6 phosphatase pathway specifically regulates Ssp2-pT189 and does not dephosphorylate Cdr2-pT166.
FIG 6.
Cdr2-pT166 dephosphorylation requires PP2A but not PP6. (A) Detection of Cdr2-pT166 in the indicated strains; whole-cell extracts were blotted. The unit for 100 is kDa. (B) Lengths of dividing, septated cells of the indicated strains (means ± SD; n > 50 for each value). The P values of two-tailed Student t tests are indicated with brackets for the respective pairs. (C) (Left) Detection of Cdr2-pT166 after treatment with the indicated concentrations of OA for 30 min; whole-cell extracts were blotted. (Right) Quantification of Cdr2-pT166 levels with values normalized to DMSO control treatment. The error bars indicate standard deviations from two biological replicates. (D) Localization of mEGFP-Cdr2 with control DMSO or 10 μM okadaic acid treatment for 30 min. The images are inverted maximum projections from deconvolved z-series. Scale bar, 5 μm. (E) (Left) Detection of Cdr2-pT166 in the indicated strains with control DMSO or 5 μM okadaic acid treatment; whole-cell extracts were blotted. (Right) Quantification of Cdr2-pT166 levels; the values were normalized to the wild type with DMSO treatment after subtracting background from untagged strains. The error bars indicate standard deviations from two biological replicates. (F) Comparison of Cdr2-pT166 and Ssp2-pT189 sensitivity to okadaic acid. Cells of the ssp2-13–myc cdr2-13–myc strain were grown in 2% glucose, and the indicated concentrations of OA were added for 30 min. The levels of Ssp2-pT189 and Cdr2-pT166 were detected in whole-cell extracts and normalized to total levels of Ssp2 and Cdr2. For both proteins, “relative activity” refers to the phosphorylation level (arbitrary units) normalized to 0 μM OA. (G) (Top) Detection of Cdr2-pT166 or Ssp2-pT189 in whole-cell extracts from the indicated strains. (Bottom) Quantification of Cdr2-pT166 or Ssp2-pT189 levels, with relative values (arbitrary units) normalized to the wild type. The error bars indicate standard deviations from three biological replicates. (H) Schematic diagram for the role of PP6 and PP2A phosphatases in countering Ssp1 activity.
To investigate Cdr2-pT166 dephosphorylation, we used the phosphatase inhibitor OA. We found that low levels of OA induced an almost 10-fold increase in cellular Cdr2-pT166 levels (Fig. 6C). This change in Cdr2-pT166 levels did not affect Cdr2 localization (Fig. 6D), supporting a localization mechanism that is independent of Cdr2 kinase activity (39). We next compared the effects of okadaic acid and pom1Δ, which increases Cdr2-pT166 2- to 3-fold (27). OA had a much stronger effect than pom1Δ on Cdr2-pT166, and their effects were nonadditive (Fig. 6E). These data indicate that Cdr2 is inactivated by an OA-sensitive protein phosphatase. Further, regulation of Cdr2 by Pom1 requires this phosphatase to be active.
OA inhibits PP1/PP2A/PP4/PP6 family protein phosphatases but acts selectively on PP2A at low concentrations (37, 38). A concentration series experiment revealed that Cdr2-pT166—but not the PP6 target, Ssp2-pT189—was strongly affected by low concentrations of OA (Fig. 6F). PP2A is a heterotrimeric protein phosphatase complex (40), and fission yeast expresses two catalytic subunits, Ppa1 and Ppa2 (41). We found that Cdr2-pT166 levels were increased approximately 2.5-fold in ppa1Δ and ppa2Δ single-mutant cells (Fig. 6G), consistent with PP2A-dependent dephosphorylation. PP2A activity is essential for cell viability, so we were unable to test a ppa1Δ ppa2Δ double mutant due to synthetic lethality (41). Importantly, these single mutants did not increase Ssp2-pT189 levels in our AMPK phosphatase screens (Fig. 4A). Fission yeast PP2A activity requires the conserved activator Ypa2 (42, 43). We shifted the cold-sensitive ypa2-7 mutant to the nonpermissive temperature and observed a 2.5-fold increase in Cdr2-pT166 levels but not in Ssp2-pT189 levels (Fig. 6G). These combined experiments reveal a PP2A-dependent pathway for dephosphorylation of Cdr2-pT166, which may represent a conserved regulatory mechanism. PP2A has long been connected with cell cycle regulation (41, 44–46), and was recently linked to dephosphorylation of the Cdr2-like protein kinase Hsl1 in budding yeast (47). We conclude that distinct phosphatase pathways target the Ssp1 substrates Cdr2 and Ssp2 to promote different cellular functions (Fig. 6H).
DISCUSSION
Our current and previous findings have shown that Ssp1 is necessary for Cdr2-pT166 and Ssp2-pT189 in cells and is sufficient to phosphorylate their activation loops directly in vitro. These combined results identify Ssp1 as the sole upstream kinase for activation loop phosphorylation of the fission yeast ARKs Cdr2 and Ssp2. The presence of a single activating kinase for fission yeast ARKs contrasts with other cell types, such as budding yeast and human cells, where redundant activating kinases must be inactivated in combination to ablate phosphorylation of ARK activation loops (11, 12, 48–52). Thus, fission yeast presents a unique experimental system to study ARK activation by a single upstream kinase. ssp1Δ cells exhibit pleiotropic defects (19–22), including increased cell size at division and growth defects in limiting glucose, which are explained by the substrates Cdr2 and Ssp2. However, additional phenotypes in ssp1Δ cells, including defects in cell polarity and stress responses (19, 20), indicate the presence of additional, unidentified targets. We note that other putative ARKs in the fission yeast genome show strong sequence similarity to the activation loops of Cdr2 and Ssp1. These kinases, including Kin1/MARK, Ppk1, and Ppk16, fit as potential Ssp1 targets due to their similar activation loop sequences and their connections to ssp1Δ phenotypes and represent key targets for future work. Defining this simplified fission yeast ARK system has the potential to elucidate signaling principles that apply to human ARK-activating kinases, such as the tumor suppressor LKB1.
Ssp1 phosphorylates identical sequences (pTSCGSP) in the activation loops of Cdr2 and Ssp2, raising the question of how these substrates are differentially phosphorylated under different growth conditions. We exploited the simplicity of the fission yeast ARK activation system to detect rapid dephosphorylation of both Cdr2-pT166 and Ssp2-pT189. The dephosphorylation rates were different for Cdr2 and Ssp2, and indeed, we identified distinct counteracting phosphatases for these two Ssp1 substrates. Kinases are traditionally thought to confer specificity on complex phosphorylation networks, but we propose that phosphatases can generate specificity downstream of a master regulatory kinase, such as Ssp1. Phosphatase-driven specificity is not mutually exclusive with other potential mechanisms, such as regulation of the upstream kinase or even the substrate itself, as seen for both Cdr2 and Ssp2 (5, 27). We anticipate that this concept could expand as additional ARK-inactivating phosphatases are identified in fission yeast and other organisms.
By manipulating the kinase-phosphatase balance for Cdr2 and Ssp2 activation loops, we made several unexpected observations. First, when phosphatases were inhibited by okadaic acid, the levels of Cdr2-pT166 increased approximately 10-fold. This means that only 10% of Cdr2 molecules in a cell are active, at least in an asynchronous population of cells. Cdr2 protein is concentrated in large cortical-node structures at the plasma membrane (Fig. 6D) (39), but our results suggest that a significant fraction of the protein is not phosphorylated in its activation loop and is therefore inactive. This suggests that all nodes are not necessarily catalytically active structures, consistent with some kinase-independent functions for Cdr2 (39, 53–55). Future studies should consider the dynamic Cdr2 activation system as a critical regulatory layer that operates in addition to node formation. Second, we observed rapid dephosphorylation of Ssp2-pT189 when Ssp1 was inhibited under low-glucose conditions, which promote high steady-state levels of Ssp2-pT189. AMPK bound to AMP has been suggested to adopt a conformational state that precludes access of inactivating phosphatases (8). Our observation means that phosphatases fully dephosphorylate Ssp2-pT189 under low cellular energy as quickly as we can generate whole-cell extracts, despite conditions that promote high steady-state levels of phosphorylation. This suggests the existence of additional allosteric mechanisms beyond access of phosphatases that contribute to energy-dependent regulation of fission yeast AMPK.
Importantly, we found that the PP6 inhibitor Sds23 is required for heterotrimer-independent regulation of fission yeast AMPK. The mechanism that enables AMPK energy sensing in the absence of the canonical energy-sensing subunit has been mysterious. Our data suggest that low cellular energy, which we experimentally induced by growing cells in low glucose, increases the physical interaction and spatial overlap of Sds23 with its inhibitory target, PP6. Because PP6 dephosphorylates Ssp2-pT189, this increased Sds23-PP6 interaction would promote Ssp2-pT189 levels even in the absence of the intact AMPK heterotrimer. This model is supported by the previous finding that Sds23-PP6 regulates 3-hydroxy-3-methyl-glutaryl (HMG)–coenzyme A (CoA) reductase in a manner that depends on glucose but is independent of AMPK (56). CBS domains, such as those found in both Sds23 and Cbs2/AMPKγ, have been shown to bind nucleotides and act as cellular energy sensors (57). Binding of the Sds23 CBS domains to ATP versus AMP has the potential to mediate binding affinities to PP6 and potentially other factors. This represents an intriguing future direction because Sds23 binds to other protein phosphatases and appears to regulate diverse signaling events (22). We anticipate that Sds23-like proteins play a similar role in heterotrimer-independent AMPK activation in other organisms. For example, budding yeast has two Sds23 homologs (Sds23 and Sds24) that arose from genome duplication. It will be interesting to determine if they regulate budding yeast AMPK-PP6 connections in a similar fashion and act redundantly in such a mechanism. Humans do not have an obvious Sds23 homolog, but other proteins containing CBS domains might have evolved such a function. Our work points to phosphatase-regulatory pathways as critical features of dynamic signaling networks, such as ARK activation in yeast and human cells.
MATERIALS AND METHODS
Yeast strains and growth.
Standard S. pombe media and methods were used (58). The strains used in this study are listed in Table S1 in the supplemental material; plasmids are listed in Table S2. Gene tagging and deletion were performed using PCR and homologous recombination (59), and integrations were verified by colony PCR, fluorescence, or backcross as needed. The nonphosphorylatable ssp2-T189A and analog-sensitive ssp1-as1 (ssp1-L221G) alleles were generated by QuikChange II mutagenesis (Stratagene) and integrated at the leu1+ locus using pJK148 (60) or at the endogenous loci through 5-fluoroorotic acid (5-FOA) counterselection. Tetrad dissections were performed to generate double mutants. To cross the sterile sds23Δ mutant, a pJK148 plasmid with sds23+ (pJM1035) was integrated into the leu1+ locus of JM3467 (sds23Δ::natR ULA− h+), and leu1-32 natR progeny were selected from subsequent crosses. Additional details for construction of all the strains and plasmids are available upon request. For the growth assay shown in Fig. 2D, cells were grown in Edinburgh minimal medium with four supplements (EMM4S) plus 2% glucose to mid-log phase at 32°C and centrifuged. The cells were then washed 3 times and resuspended in EMM4S plus 0.1% glucose and spotted in 10-fold serial dilutions (starting at an optical density at 595 nm [OD595] of 0.1) on EMM4S plates with 2% glucose or 0.1% glucose. The plates were incubated at 32°C for 3 to 4 days before scanning. Cold-sensitive ypa2-7 mutants (42) were grown to mid-log phase at 32°C and then shifted to 19°C for 8 h before sampling.
Glucose limitation and drug treatments.
For glucose concentration assays (Fig. 1C and D), cells were grown in EMM4S plus 2% glucose to mid-log phase; the cells were pelleted, washed three times, and then resuspended in EMM4S with different glucose concentrations for 6 h before sampling. For other glucose shift experiments, cells were grown in yeast extract medium with four supplements (YE4S) plus 2% glucose and then washed twice and resuspended in YE4S plus 2% glucose or YE4S plus 0.1% glucose for 10 min before sampling. To inhibit Ssp1 kinase activity, ssp1-as1 mutant cells were treated with 15 μM the nonhydrolyzable ATP analog 3-MB-PP1 {1-(tert-butyl)-3-(3-methylbenzyl)-1H-pyrazolo [3,4-d] pyrimidin-4-amine; catalog no. 529582; Millipore}. For okadaic acid treatments, cells in mid-log phase were centrifuged, washed, and resuspended in 500 μl of fresh medium before addition of okadaic acid (Santa Cruz Biotechnology) from 1 mM stock in DMSO. The cells were treated for 30 min at room temperature before imaging or sampling.
In vitro kinase assays and Western blots.
Full-length Ssp1, Ssp2, and Ssp2-T189A were cloned into pGEX6P1 (glutathione S-transferase [GST] tag) vector (GE Healthcare), expressed in E. coli strain BL21 (Rosetta), and purified with glutathione-agarose resin (Sigma). The purified proteins were released from the resin by overnight incubation with 3C protease at 4°C. For in vitro kinase assays, the purified proteins were incubated in kinase assay buffer (30 mM Tris-HCl, 100 mM NaCl, 10 mM MgCl2, 1 mM EGTA, 10% glycerol) supplemented with 20 μM ATP in 40-μl reaction mixtures at 30°C for 30 min. The reactions were stopped by boiling in SDS-PAGE sample buffer for 5 min at 95°C before SDS-PAGE analysis. Whole-cell extracts were prepared as described previously (61). For coimmunoprecipitations, logarithmic-phase cells at an OD595 of 6 were collected and washed into medium containing either 2% glucose or 0.1% glucose. The cells were resuspended in 200 μl immunoprecipitation (IP) buffer [20 mM 4-(2-hydroxyethyl-1-piperazineethanesulfonic acid), pH 7.4, 1 mM EDTA, 150 mM NaCl, 0.2% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, complete EDTA-free protease inhibitor tablets (Roche)] and 400 μl glass beads and lysed using a Mini-Beadbeater-16 (BioSpec, Bartlesville, OK; two cycles of 2 min each at maximum speed). The lysates were then spun at 16,000 × g for 5 min at 4°C, and the supernatants were recovered and incubated with antihemagglutinin (anti-HA) magnetic beads (Thermo Scientific Pierce) for 1.5 h at 4°C. The beads were then washed five times with IP buffer, resuspended in SDS-PAGE sample buffer, and boiled for 5 min at 99°C. Western blots were probed with anti-Cdc2 (SC-53; Santa Cruz), anti-GFP (53), anti-myc (SC-40; Santa Cruz), anti-Cdr2-pT166 (27), anti-FLAG (F7425; Sigma), anti-mCherry (ab183628; Abcam), and anti-HA (clone 16B12; Covance) antibodies. Rabbit anti-Ssp2-pT189 antibody was generated against the phosphopeptide GNFLK(pT)SCGSPNY (21st Century Biochemicals, Inc.).
Imaging and data analysis.
Microscopy was performed at room temperature with a DeltaVision imaging system (Applied Precision) equipped with an Olympus IX-71 inverted wide-field microscope, a Photometrics CoolSnap HQ2 camera, and an Insight solid-state illumination unit. For Fig. 6D, a z-series with a 0.5-μm step size was acquired and then iteratively deconvolved in SoftWoRx software (Applied Precision). Maximum-intensity projection images were generated with ImageJ 1.49 (National Institutes of Health). Other images were single focal planes positioned in the cell middle. For cell length measurements, more than 50 septated cells were grown in EMM4S at 32°C to mid-log phase and then stained with Blankophor (MP Biomedicals) and measured. Two-tailed Student's t tests were performed for statistical comparison of cell lengths at division. To quantify Cdr2-pT166 or Ssp2-pT189 levels, inverted images of Western blots were analyzed in ImageJ 1.49. The ratios of Cdr2-pT166 or Ssp2-pT189 to the total Cdr2 or Ssp2 were calculated after background (intensity in a blank region) subtraction. The values were normalized to those from the control experiments to show the relative changes. For BiFC experiments, wild-type (no fluorescent tag), Sds23C (Sds23-VC155), Ekc1N (Ekc1-VN173), and Sds23C Ekc1N (Sds23-VC155 Ekc1-VN173) strains were grown in EMM4S at 25°C and then shifted to EMM4S containing either 2% glucose or 0.1% glucose for 4 h. All the strains were imaged under identical parameters. Average YFP intensity in the cytoplasm was measured for 100 cells by drawing a box (22 pixels by 18 pixels) and correcting for background signal.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Moseley laboratory and the Biochemistry Department for discussions, T. Gauvin for technical help, M. Balasubramanian, P. Espenshade, and V. Simanis for strains, E. Griffin, A. Gladfelter, H. Opalko, and C. Allard for comments on the manuscript, and S. MacNeill for BiFC plasmids.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00494-16.
REFERENCES
- 1.Hanahan D, Weinberg RA. 2000. The hallmarks of cancer. Cell 100:57–70. doi: 10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. 2011. Hallmarks of cancer: the next generation. Cell 144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Bright NJ, Thornton C, Carling D. 2009. The regulation and function of mammalian AMPK-related kinases. Acta Physiol (Oxford) 196:15–26. doi: 10.1111/j.1748-1716.2009.01971.x. [DOI] [PubMed] [Google Scholar]
- 4.Lizcano JM, Goransson O, Toth R, Deak M, Morrice NA, Boudeau J, Hawley SA, Udd L, Makela TP, Hardie DG, Alessi DR. 2004. LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. EMBO J 23:833–843. doi: 10.1038/sj.emboj.7600110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hardie DG. 2007. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat Rev Mol Cell Biol 8:774–785. doi: 10.1038/nrm2249. [DOI] [PubMed] [Google Scholar]
- 6.Hardie DG, Alessi DR. 2013. LKB1 and AMPK and the cancer-metabolism link—ten years after. BMC Biol 11:36. doi: 10.1186/1741-7007-11-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shackelford DB, Shaw RJ. 2009. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat Rev Cancer 9:563–575. doi: 10.1038/nrc2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rubenstein EM, McCartney RR, Zhang C, Shokat KM, Shirra MK, Arndt KM, Schmidt MC. 2008. Access denied: Snf1 activation loop phosphorylation is controlled by availability of the phosphorylated threonine 210 to the PP1 phosphatase. J Biol Chem 283:222–230. doi: 10.1074/jbc.M707957200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elbing K, Rubenstein EM, McCartney RR, Schmidt MC. 2006. Subunits of the Snf1 kinase heterotrimer show interdependence for association and activity. J Biol Chem 281:26170–26180. doi: 10.1074/jbc.M603811200. [DOI] [PubMed] [Google Scholar]
- 10.Ruiz A, Liu Y, Xu X, Carlson M. 2012. Heterotrimer-independent regulation of activation-loop phosphorylation of Snf1 protein kinase involves two protein phosphatases. Proc Natl Acad Sci U S A 109:8652–8657. doi: 10.1073/pnas.1206280109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hong SP, Leiper FC, Woods A, Carling D, Carlson M. 2003. Activation of yeast Snf1 and mammalian AMP-activated protein kinase by upstream kinases. Proc Natl Acad Sci U S A 100:8839–8843. doi: 10.1073/pnas.1533136100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sutherland CM, Hawley SA, McCartney RR, Leech A, Stark MJ, Schmidt MC, Hardie DG. 2003. Elm1p is one of three upstream kinases for the Saccharomyces cerevisiae SNF1 complex. Curr Biol 13:1299–1305. doi: 10.1016/S0960-9822(03)00459-7. [DOI] [PubMed] [Google Scholar]
- 13.Davies SP, Helps NR, Cohen PT, Hardie DG. 1995. 5′-AMP inhibits dephosphorylation, as well as promoting phosphorylation, of the AMP-activated protein kinase. Studies using bacterially expressed human protein phosphatase-2C alpha and native bovine protein phosphatase-2AC. FEBS Lett 377:421–425. [DOI] [PubMed] [Google Scholar]
- 14.Sanders MJ, Grondin PO, Hegarty BD, Snowden MA, Carling D. 2007. Investigating the mechanism for AMP activation of the AMP-activated protein kinase cascade. Biochem J 403:139–148. doi: 10.1042/BJ20061520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moore F, Weekes J, Hardie DG. 1991. Evidence that AMP triggers phosphorylation as well as direct allosteric activation of rat liver AMP-activated protein kinase. A sensitive mechanism to protect the cell against ATP depletion. Eur J Biochem 199:691–697. [DOI] [PubMed] [Google Scholar]
- 16.Hong SP, Momcilovic M, Carlson M. 2005. Function of mammalian LKB1 and Ca2+/calmodulin-dependent protein kinase kinase alpha as Snf1-activating kinases in yeast. J Biol Chem 280:21804–21809. doi: 10.1074/jbc.M501887200. [DOI] [PubMed] [Google Scholar]
- 17.Ruiz A, Xu X, Carlson M. 2011. Roles of two protein phosphatases, Reg1-Glc7 and Sit4, and glycogen synthesis in regulation of SNF1 protein kinase. Proc Natl Acad Sci U S A 108:6349–6354. doi: 10.1073/pnas.1102758108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruiz A, Xu X, Carlson M. 2013. Ptc1 protein phosphatase 2C contributes to glucose regulation of SNF1/AMP-activated protein kinase (AMPK) in Saccharomyces cerevisiae. J Biol Chem 288:31052–31058. doi: 10.1074/jbc.M113.503763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsusaka T, Hirata D, Yanagida M, Toda T. 1995. A novel protein kinase gene ssp1+ is required for alteration of growth polarity and actin localization in fission yeast. EMBO J 14:3325–3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rupes I, Jia Z, Young PG. 1999. Ssp1 promotes actin depolymerization and is involved in stress response and new end take-off control in fission yeast. Mol Biol Cell 10:1495–1510. doi: 10.1091/mbc.10.5.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Valbuena N, Moreno S. 2012. AMPK phosphorylation by Ssp1 is required for proper sexual differentiation in fission yeast. J Cell Sci 125:2655–2664. doi: 10.1242/jcs.098533. [DOI] [PubMed] [Google Scholar]
- 22.Hanyu Y, Imai KK, Kawasaki Y, Nakamura T, Nakaseko Y, Nagao K, Kokubu A, Ebe M, Fujisawa A, Hayashi T, Obuse C, Yanagida M. 2009. Schizosaccharomyces pombe cell division cycle under limited glucose requires Ssp1 kinase, the putative CaMKK, and Sds23, a PP2A-related phosphatase inhibitor. Genes Cells 14:539–554. doi: 10.1111/j.1365-2443.2009.01290.x. [DOI] [PubMed] [Google Scholar]
- 23.Freitag SI, Wong J, Young PG. 2014. Genetic and physical interaction of Ssp1 CaMKK and Rad24 14-3-3 during low pH and osmotic stress in fission yeast. Open Biol 4:130127. doi: 10.1098/rsob.130127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsuzawa T, Fujita Y, Tohda H, Takegawa K. 2012. Snf1-like protein kinase Ssp2 regulates glucose derepression in Schizosaccharomyces pombe. Eukaryot Cell 11:159–167. doi: 10.1128/EC.05268-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saitoh S, Mori A, Uehara L, Masuda F, Soejima S, Yanagida M. 2015. Mechanisms of expression and translocation of major fission yeast glucose transporters regulated by CaMKK/phosphatases, nuclear shuttling, and TOR. Mol Biol Cell 26:373–386. doi: 10.1091/mbc.E14-11-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davie E, Forte GM, Petersen J. 2015. Nitrogen regulates AMPK to control TORC1 signaling. Curr Biol 25:445–454. doi: 10.1016/j.cub.2014.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deng L, Baldissard S, Kettenbach AN, Gerber SA, Moseley JB. 2014. Dueling kinases regulate cell size at division through the SAD kinase Cdr2. Curr Biol 24:428–433. doi: 10.1016/j.cub.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhatia P, Hachet O, Hersch M, Rincon SA, Berthelot-Grosjean M, Dalessi S, Basterra L, Bergmann S, Paoletti A, Martin SG. 2014. Distinct levels in Pom1 gradients limit Cdr2 activity and localization to time and position division. Cell Cycle 13:538–552. doi: 10.4161/cc.27411. [DOI] [PubMed] [Google Scholar]
- 29.Kettenbach AN, Deng L, Wu Y, Baldissard S, Adamo ME, Gerber SA, Moseley JB. 2015. Quantitative phosphoproteomics reveals pathways for coordination of cell growth and division by the conserved fission yeast kinase pom1. Mol Cell Proteomics 14:1275–1287. doi: 10.1074/mcp.M114.045245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Breeding CS, Hudson J, Balasubramanian MK, Hemmingsen SM, Young PG, Gould KL. 1998. The cdr2(+) gene encodes a regulator of G2/M progression and cytokinesis in Schizosaccharomyces pombe. Mol Biol Cell 9:3399–3415. doi: 10.1091/mbc.9.12.3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanoh J, Russell P. 1998. The protein kinase Cdr2, related to Nim1/Cdr1 mitotic inducer, regulates the onset of mitosis in fission yeast. Mol Biol Cell 9:3321–3334. doi: 10.1091/mbc.9.12.3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cisneros-Barroso E, Yance-Chavez T, Kito A, Sugiura R, Gomez-Hierro A, Gimenez-Zaragoza D, Aligue R. 2014. Negative feedback regulation of calcineurin-dependent Prz1 transcription factor by the CaMKK-CaMK1 axis in fission yeast. Nucleic Acids Res 42:9573–9587. doi: 10.1093/nar/gku684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rothbauer U, Zolghadr K, Tillib S, Nowak D, Schermelleh L, Gahl A, Backmann N, Conrath K, Muyldermans S, Cardoso MC, Leonhardt H. 2006. Targeting and tracing antigens in live cells with fluorescent nanobodies. Nat Methods 3:887–889. doi: 10.1038/nmeth953. [DOI] [PubMed] [Google Scholar]
- 34.Bishop AC, Ubersax JA, Petsch DT, Matheos DP, Gray NS, Blethrow J, Shimizu E, Tsien JZ, Schultz PG, Rose MD, Wood JL, Morgan DO, Shokat KM. 2000. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature 407:395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- 35.Bishop AC, Buzko O, Shokat KM. 2001. Magic bullets for protein kinases. Trends Cell Biol 11:167–172. doi: 10.1016/S0962-8924(01)01928-6. [DOI] [PubMed] [Google Scholar]
- 36.Shimanuki M, Kinoshita N, Ohkura H, Yoshida T, Toda T, Yanagida M. 1993. Isolation and characterization of the fission yeast protein phosphatase gene ppe1+ involved in cell shape control and mitosis. Mol Biol Cell 4:303–313. doi: 10.1091/mbc.4.3.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hardie DG. 1999. Protein phosphorylation: a practical approach, 2nd ed Oxford University Press, London, United Kingdom. [Google Scholar]
- 38.Swingle M, Ni L, Honkanen RE. 2007. Small-molecule inhibitors of ser/thr protein phosphatases: specificity, use and common forms of abuse. Methods Mol Biol 365:23–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morrell JL, Nichols CB, Gould KL. 2004. The GIN4 family kinase, Cdr2p, acts independently of septins in fission yeast. J Cell Sci 117:5293–5302. doi: 10.1242/jcs.01409. [DOI] [PubMed] [Google Scholar]
- 40.Shi Y. 2009. Serine/threonine phosphatases: mechanism through structure. Cell 139:468–484. doi: 10.1016/j.cell.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 41.Kinoshita N, Ohkura H, Yanagida M. 1990. Distinct, essential roles of type 1 and 2A protein phosphatases in the control of the fission yeast cell division cycle. Cell 63:405–415. doi: 10.1016/0092-8674(90)90173-C. [DOI] [PubMed] [Google Scholar]
- 42.Goyal A, Simanis V. 2012. Characterization of ypa1 and ypa2, the Schizosaccharomyces pombe orthologs of the peptidyl prolyl isomerases that activate PP2A, reveals a role for Ypa2p in the regulation of cytokinesis. Genetics 190:1235–1250. doi: 10.1534/genetics.111.138040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bernal M, Sanchez-Romero MA, Salas-Pino S, Daga RR. 2012. Regulation of fission yeast morphogenesis by PP2A activator pta2. PLoS One 7:e32823. doi: 10.1371/journal.pone.0032823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Wulf P, Montani F, Visintin R. 2009. Protein phosphatases take the mitotic stage. Curr Opin Cell Biol 21:806–815. doi: 10.1016/j.ceb.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 45.Jiang Y. 2006. Regulation of the cell cycle by protein phosphatase 2A in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 70:440–449. doi: 10.1128/MMBR.00049-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Felix MA, Cohen P, Karsenti E. 1990. Cdc2 H1 kinase is negatively regulated by a type 2A phosphatase in the Xenopus early embryonic cell cycle: evidence from the effects of okadaic acid. EMBO J 9:675–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zapata J, Dephoure N, Macdonough T, Yu Y, Parnell EJ, Mooring M, Gygi SP, Stillman DJ, Kellogg DR. 2014. PP2ARts1 is a master regulator of pathways that control cell size. J Cell Biol 204:359–376. doi: 10.1083/jcb.201309119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hawley SA, Pan DA, Mustard KJ, Ross L, Bain J, Edelman AM, Frenguelli BG, Hardie DG. 2005. Calmodulin-dependent protein kinase kinase-beta is an alternative upstream kinase for AMP-activated protein kinase. Cell Metab 2:9–19. doi: 10.1016/j.cmet.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 49.Hurley RL, Anderson KA, Franzone JM, Kemp BE, Means AR, Witters LA. 2005. The Ca2+/calmodulin-dependent protein kinase kinases are AMP-activated protein kinase kinases. J Biol Chem 280:29060–29066. doi: 10.1074/jbc.M503824200. [DOI] [PubMed] [Google Scholar]
- 50.Momcilovic M, Hong SP, Carlson M. 2006. Mammalian TAK1 activates Snf1 protein kinase in yeast and phosphorylates AMP-activated protein kinase in vitro. J Biol Chem 281:25336–25343. doi: 10.1074/jbc.M604399200. [DOI] [PubMed] [Google Scholar]
- 51.Woods A, Dickerson K, Heath R, Hong SP, Momcilovic M, Johnstone SR, Carlson M, Carling D. 2005. Ca2+/calmodulin-dependent protein kinase kinase-beta acts upstream of AMP-activated protein kinase in mammalian cells. Cell Metab 2:21–33. doi: 10.1016/j.cmet.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 52.Woods A, Johnstone SR, Dickerson K, Leiper FC, Fryer LG, Neumann D, Schlattner U, Wallimann T, Carlson M, Carling D. 2003. LKB1 is the upstream kinase in the AMP-activated protein kinase cascade. Curr Biol 13:2004–2008. doi: 10.1016/j.cub.2003.10.031. [DOI] [PubMed] [Google Scholar]
- 53.Moseley JB, Mayeux A, Paoletti A, Nurse P. 2009. A spatial gradient coordinates cell size and mitotic entry in fission yeast. Nature 459:857–860. doi: 10.1038/nature08074. [DOI] [PubMed] [Google Scholar]
- 54.Guzman-Vendrell M, Rincon SA, Dingli F, Loew D, Paoletti A. 2015. Molecular control of the Wee1 regulatory pathway by the SAD kinase Cdr2. J Cell Sci 128:2842–2853. doi: 10.1242/jcs.173146. [DOI] [PubMed] [Google Scholar]
- 55.Martin SG, Berthelot-Grosjean M. 2009. Polar gradients of the DYRK-family kinase Pom1 couple cell length with the cell cycle. Nature 459:852–856. doi: 10.1038/nature08054. [DOI] [PubMed] [Google Scholar]
- 56.Burg JS, Espenshade PJ. 2011. Glucose controls phosphoregulation of hydroxymethylglutaryl coenzyme A reductase through the protein phosphatase 2A-related phosphatase protein, Ppe1, and Insig in fission yeast. J Biol Chem 286:27139–27146. doi: 10.1074/jbc.M111.233452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Scott JW, Hawley SA, Green KA, Anis M, Stewart G, Scullion GA, Norman DG, Hardie DG. 2004. CBS domains form energy-sensing modules whose binding of adenosine ligands is disrupted by disease mutations. J Clin Invest 113:274–284. doi: 10.1172/JCI19874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moreno S, Klar A, Nurse P. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol 194:795–823. doi: 10.1016/0076-6879(91)94059-L. [DOI] [PubMed] [Google Scholar]
- 59.Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A III, Steever AB, Wach A, Philippsen P, Pringle JR. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14:943–951. [DOI] [PubMed] [Google Scholar]
- 60.Keeney JB, Boeke JD. 1994. Efficient targeted integration at leu1-32 and ura4-294 in Schizosaccharomyces pombe. Genetics 136:849–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sreenivasan A, Kellogg D. 1999. The elm1 kinase functions in a mitotic signaling network in budding yeast. Mol Cell Biol 19:7983–7994. doi: 10.1128/MCB.19.12.7983. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.