Abstract
Cultural and linguistic groups are often expected to represent genetic populations. In this article, we tested the hypothesis that the hierarchical classification of languages proposed by J. Greenberg [(1987) Language in the Americas (Stanford Univ. Press, Stanford, CA)] also represents the genetic structure of Native North American populations. The genetic data are mtDNA sequences for 17 populations gleaned from literature sources and public databases. The hypothesis was rejected. Further analysis showed that departure of the genetic structure from the linguistic classification was pervasive and not due to an outlier population or a problematic language group. Therefore, Greenberg's language groups are at best an imperfect approximation to the genetic structure of these populations. Moreover, we show that the genetic structure among these Native North American populations departs significantly from the best-fitting hierarchical models. Analysis of median joining networks for mtDNA haplotypes provides strong evidence for gene flow across linguistic boundaries. In principle, the language of a population can be replaced more rapidly than its genes because language can be transmitted both vertically from parents to children and horizontally between unrelated people. However, languages are part of a cultural complex, and there may be strong pressure to maintain a language in place whereas genes are free to flow.
Keywords: language and genetics, mtDNA
Geneticists and anthropologists often expect that human language groups and gene pools will share a common structure (1–4). It is noted that both language and genes are passed from parents to children, mating tends to be endogamous with respect to linguistic groups, and splits in linguistic communities usually occur with splits in breeding populations (5, 6). Cavalli-Sforza et al. (7) have reported that genetic trees of major geographic populations correlate well with language families. They argue that a process consisting of population fissions, expansion into new territories, and isolation between ancestral and descendant groups will produced a tree-like structure common to both genes and languages. Linguists agree that population fissions and range expansions play an important role in the generation of linguistic diversity (8–10). The correlation between patterns of linguistic and genetic variation has been studied by many researchers in different world regions with mixed positive (11–14) and negative (15–17) findings. Regional differences in population history are likely to explain some of the discrepant results. However, it is difficult to draw firm conclusions because these studies are heterogeneous with respect to the genetic and population sampling units, definition of linguistic variables, and analytical methods. Researchers on this topic have identified the need for a method to directly compare language trees with population genetic trees (1, 3).
The potential correspondence between gene pools and language groups in Native North American populations is particularly interesting for several reasons. First, the initial colonization of the Americas involved a population radiation from an effectively small number of founders into an uninhabited region. Second, it occurred long enough ago to permit the accumulation of both linguistic and genetic differences, but not so long ago that the early history would necessarily have been erased by subsequent events. Third, there was a rich diversity of indigenous languages in North America at the time of European contact (9, 10, 18, 19). Early investigations of the correspondence between genetic groups and linguistic groups in Native North Americans produced equivocal results (5, 6). On the one hand, average genetic distances between populations in different language families were greater than average genetic distances between populations within language families. On the other hand, genetic distances were not significantly correlated with glottochronological distances. However, the methods and data available 30 years ago had limited power to resolve the question.
This article revisits the question of relationship between genetics and language in Native North American populations. Comparable mtDNA sequences are now available for many populations that represent diverse language groups. There are three advantages to the mtDNA data. First, they have high power to resolve population affinities. Second, it is easy to identify and remove non-Native American mtDNA sequences. Third, by contrasting gene and population trees, it is possible to discern patterns of gene flow. We begin our analyses by formally testing the hypothesis that a classification of languages proposed by Greenberg (19) represents the structure of Native North American gene pools. Although the majority of linguists do not endorse Greenberg's language classification (4, 10, 18, 20, 21), it is important to test this model because it remains one of the main tenets against which questions about Native American population genetics and physical anthropology are formulated (3). To preempt our results, this hypothesis is rejected. Because of this finding, we proceed to ask two questions. First, is the genetic structure of these populations hierarchical? Second, is there evidence from mtDNA sequences for exchange across the boundaries of widely accepted language families?
Materials and Methods
Sample. We obtained 1,056 mtDNA hypervariable segment (HVS) 1 sequences for 17 Native North American populations from the literature and public databases. No biological samples were handled. We included only sequences classified as founding Native American haplogroups A, B, C, D, or X (references in Table 1). The sequences were aligned and edited to 341 nucleotides covering the reference nucleotide positions 16024–16364 (22).
Table 1. Population sample size, estimated nucleotide diversity, sampling location, and language classification.
| Population | N | 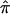 |
Family | Primary branch | Group | Subgroup | Branch | Subbranch | Ref(s). and accession nos. |
|---|---|---|---|---|---|---|---|---|---|
| Aleut | 164 | 0.0117 | 44 | ||||||
| Inupiaq | 15 | 0.0075 | Eskimo-Aleut | Eskimo | Inuit | 45; AF082222–AF082231 | |||
| Canadian Inuit | 46 | 0.0036 | Eskimo-Aleut | Eskimo | Inuit | AF186706–AF186751 | |||
| Greenland Inuit | 82 | 0.0049 | Eskimo-Aleut | Eskimo | Inuit | 46 | |||
| Central Yupik | 25 | 0.0033 | Eskimo-Aleut | Eskimo | Yupik | Alaskan | AF011645–AF011669 | ||
| Siberian Yupik | 52 | 0.0081 | Eskimo-Aleut | Eskimo | Yupik | Siberian | AF013684–AF013633 | ||
| Haida | 41 | 0.0062 | Na-Dene | Haida | 17 | ||||
| A. Athabascan | 40 | 0.0059 | Na-Dene | Continental | EA | Athabaskan | 45; AF184627–AF184647 | ||
| Navajo | 164 | 0.0166 | Na-Dene | Continental | EA | Athabaskan | Apachean | Navajo-Apache | 47; AF011670–AF011684 |
| Apache | 183 | 0.0146 | Na-Dene | Continental | EA | Athabaskan | Apachean | Navajo-Apache | 47 |
| Pima | 40 | 0.0202 | Amerind | Central | UA | Pimic | 48 | ||
| Cheyenne | 39 | 0.0204 | Amerind | Northern | AK | Almosan | Algic | Algonquin | 48 |
| Chippewa | 19 | 0.0109 | Amerind | Northern | AK | Almosan | Algic | Algonquin | 49 |
| Bella Coola | 41 | 0.0142 | Amerind | Northern | AK | Almosan | Mosan | Salish | 17 |
| Nuu-Chah-Nulth | 59 | 0.0143 | Amerind | Northern | AK | Almosan | Mosan | Wakashan | 17, 50 |
| Sioux | 16 | 0.0203 | Amerind | Northern | AK | Keresiouan | Siouan Yuchi | Siouan | 49 |
| Cherokee | 30 | 0.0147 | Amerind | Northern | AK | Keresiouan | Iroquoian | Southern | 49 |
EA, Eyak-Athabascan; UA, Uto-Aztecan; AK, Almosan-Keresiouan.
Table 1 provides each population's sample size, and placement in the Greenberg language classification (GLC). The terminology used for the language categories in this table is taken from Ruhlen (23). Fig. 1 presents the placement of these 17 populations within the GLC in tree form. Each of Greenberg's primary language families (Eskimo-Aleut, Na-Dene, and Amerind) is represented by at least four populations.
Fig. 1.
GLC for 17 populations
Statistical Analyses. The basic unit of analysis is a matrix composed of the average number of nucleotide substitutions between pairs of mtDNA sequences. The diagonal elements d̂ii pertain to averages for pairs drawn from the same population, whereas the off-diagonal elements d̂ij pertain to averages for pairs drawn from two different populations. Pairwise differences lead naturally to the net number of nucleotide substitutions d̂Aij = d̂ij - (d̂ii + d̂jj)/2 as a measure of genetic distance between populations (24).
Model trees (e.g., the GLC) were fitted to the matrix of observed, or “realized,” pairwise differences by using maximum likelihood (25–27). Each fitted tree produces a matrix of “expected” average pairwise nucleotide substitutions contingent on the assumption that the model tree accurately represents the true genetic relationships among populations. The lack-of-fit of the expected average pairwise differences relative to the realized pairwise differences is measured by a likelihood ratio statistic, Λ, which under the idealized circumstances of a large number of independent polymorphic sites is distributed as a χ2 variable (25). The degrees of freedom associated with this statistic are equal to r(r + 1)/2 minus the number of parameters specified by the fitted tree, where r is the number of populations sampled. To visualize the fit of a model tree, we plotted realized versus expected net nucleotide distances against one another. If a tree model fit the genetic data, the expected genetic distances for each population will be overestimated approximately as frequently as they are underestimated. The scattergram should assume a symmetric cigar-shaped distribution.
Either or both of two potential reasons can explain why the GLC model might fit poorly. The first is that the tree specified by the GLC is the wrong tree to describe the genetic relationships among these populations. The second is that the genetic data are not tree-like; in other words, no tree will fit the genetic data well. To distinguish among these alternatives, we applied the Fitch and neighbor-joining (NJ) algorithms to identify tree topologies that were optimized to the genetic data. We then used the maximum likelihood method to fit these topologies to the genetic data, applied the test for treeness, and examined plots of the realized versus expected genetic distances. The rationale underlying this test is that, if the genetic data are in fact tree-like, and only the language hierarchy in particular is incompatible with the genetic hierarchy, optimized trees will fit the genetic data. To determine whether there was evidence in the mtDNA sequences for local mate exchange across linguistic boundaries, we investigated the pattern of relationships among individual mtDNA sequences using median joining networks constructed for each haplogroup using the method of Bandelt et al. (28).
Results
Sequence Polymorphisms. The 1,056 mtDNA sequences contain 111 variable sites and 201 distinct haplotypes. The estimated nucleotide diversity (Table 1) shows an interesting trend within populations. With respect to Greenberg's three language families, the average nucleotide diversity within populations is low in Eskimo-Aleut populations and high in Amerind populations. However, nucleotide diversity varies considerably among the populations classified as Na-Dene-speaking. The Alaskan Athabascan and Haida populations, who reside in the North, have low nucleotide diversities, in the range of nucleotide diversities in the Eskimo-Aleut-speaking populations. The Navajo and Apache, who reside in the Southwest, have high nucleotide diversities, in the range of nucleotide diversities in populations classified as Amerind speaking. The A to G transition at nucleotide position (np) 16265 described by Starikovskaya et al. (29) appeared in all of the Eskimo samples, and none of the non-Eskimo samples (including Aleut). The A to G transitions at nps 16233 and 16331, identified by Torroni et al. (14), were observed in, and only in, the three Athabascan samples. Several sites were polymorphic only in populations classified as Amerind-speaking, but none occurred in all populations attributed to Amerind.
Statistical Analyses. Fig. 1 displays the GLC as a tree. Fig. 2A shows the result of fitting this tree to the genetic data. Fig. 2 A looks different from Fig. 1 because the branch lengths vary, and because the estimated length for many branches is zero. Notably, length zero was estimated for the branches leading to two of the three principal language families, Eskimo-Aleut and Na-Dene. The likelihood ratio statistic for the GLC model is high relative to its degrees of freedom, indicating a substantial lack of fit to the genetic data (Table 2). Note that the poor fit of the GLC does not indicate insufficient information; rather it shows that the genetic data contradict the structure postulated by the GLC model. Nevertheless, the GLC fits much better than an island model, which allows for genetic divergence, but assumes independent evolution, among populations (Table 2). To be certain that the poor fit of the GLC was not solely the result of one misplaced population, we repeated the analysis 17 times, each time leaving out a different sample from the analysis. The likelihood ratio statistic substantially exceeded its degrees of freedom in each of these 17 analyses (results not shown). We conclude that the poor fit of the GLC is systemic and not the result of a single outlier population.
Fig. 2.
Evalutation of GLC. (A) GLC fitted to mtDNA data. (B) Realized vs. expected genetic distances. The GLC overestimates distances for the Navajo (blue circles) and underestimates distances for the Canadian Inuit (pink circles).
Table 2. Treeness tests for five models.
| Model | Λ* | df |
|---|---|---|
| Island | 1776.98 | 135 |
| Greenberg language | 1112.92 | 139 |
| NJ tree | 795.74 | 122 |
| Fitch tree | 797.79 | 123 |
Ideally distributed as a χ2 variable
To identify the specific ways that the genetic data and GLC model depart, we examined the plot of expected vs. realized genetic distances (Fig. 2B). Several patterns that depart from the tree structure are apparent upon close examination. For example, the GLC expected distances consistently overestimate the realized genetic distances for several populations, including the Navajo, Aleut, and Siberian Yupik populations. This relationship means that these populations are genetically similar to populations with distantly related languages. Similarly, the GLC tree consistently underestimates the genetic distance between three Eskimo populations (Central Yupik, Canadian Inuit, and Inupaiq) and all other populations.
Both the Fitch and NJ trees fit the data substantially better than the GLC tree, and the NJ tree fits better than the Fitch tree (Table 2). This result indicates that the data are more tree-like than indicated by the GLC tree. Nonetheless, the likelihood ratio statistics for both the NJ and Fitch trees are high relative to their degrees of freedom (Table 2). This finding indicates a lack of fit to the genetic data for even the best-fitting trees. The scatter plot of realized versus expected genetic distances for the NJ tree (Fig. 3B) visually confirms the superior fit of the NJ tree over the GLC tree. The results are not shown for the Fitch tree because they are similar to the NJ tree. The lack of fit between the NJ tree and the genetic data is evident from the fact that there is a consistent trend for the realized genetic distances to exceed the expected genetic distances. This lop-sided pattern is difficult to explain and may be an artifact of applying the NJ algorithm to non-tree-like data.
Fig. 3.
Evaluation of NJ tree. (A) NJ tree. None of Greenberg's major language groups, Eskimo-Aleut (a), Na-Dene (b), or Amerind (c) forms a unique cluster. (B) Realized versus expected genetic distances.
A drawing of the NJ tree (Fig. 3A) reveals some interesting patterns. First, none of Greenberg's major language groups (Eskimo-Aleut, Na-Dene, or Amerind) forms a unique cluster. The most exclusive cluster that contains all Eskimo-Aleut populations (defined by branch a) also includes all four Na-Dene-speaking populations and the Amerind-speaking Cheyenne, Bella Coola, and Nuu Chah Nulth populations. The most exclusive cluster with all Na-Dene-speaking populations (defined by branch b) also includes six Eskimo-Aleut-speaking populations (Siberian Yupik, Greenland Inuit, Central Yupik, Canadian Inuit, and Inupiaq) and the Amerind-speaking Bella Coola. The most exclusive cluster with all Amerind-speaking populations (defined by branch c) includes the Eskimo-Aleut-speaking Aleuts and the Na-Dene-speaking Navajo. Second, there is a strong North-South geographic pattern to the clustering pattern. An Arctic-Pacific Northwest cluster that includes all Aleut-Eskimo populations, all Na-Dene populations, and the Amerind Nuu Chah Nulth and Bella Coola populations originates on one side of branch a, whereas a more Southern group includes the Pima, Cherokee, Sioux, and Chippewa Amerind-speaking population forms to the other side of branch a. The Southwestern Athabascan-speaking populations, Navajo and Apache, defy the geographic groupings, but this result is consistent with the archaeological record (30). Anthropologists agree that circa anno Domini 1400 the ancestors of Navajos and Apaches migrated from the Mackenzie Basin of Canada to the Southwest region, where they came into contact with Amerind-speaking populations who had been living there for thousands of years (31).
Fig. 4 shows the distribution of the canonical Native American mtDNA haplogroups in the 17 populations. The occurrence of haplogroup A differs markedly between the far Northern and the Southwestern samples. With only few exceptions, mtDNA lineages observed in the northern Na-Dene classified populations (Haida and Alaskan Athabascans) belong to haplogroup A (Fig. 4). Haplogroup A is also common in Eskimos and Aleuts. Outside of the far North, the only samples in which haplogroup A appears commonly are the Southwestern Athabascan-speaking populations (Navajo and Apache). mtDNA sequences belonging to haplogroups B and C are frequent primarily in the Amerind-classified populations (Fig. 4), including the Bella Coola, and Nuu Chah Nulth populations on the Northwest Coast. The Navajo and Apache are the only Na-Dene-classified populations with substantial frequencies of B- and C-group haplotypes, although haplogroup C is observed in the Haida and Alaskan Athabascan samples. Haplogroup D mtDNA sequences are common in the Eskimo and Aleut populations and the Bella Coola and Nuu Chah Nulth on the Northwest Coast. In summary, the distribution of the major mtDNA haplogroups exhibits a strong regional pattern in these samples. Haplogroups A and D are concentrated in the far North and Northwest Coast, whereas haplogroups B and C appear regularly on the Northwest Coast and further South. Recent surveys of Native North American mtDNA confirm the haplogroup distributions observed for the populations and regions represented here; however, broader distributions for the A, B, C, and D haplogroups are noted when samples from other regions are included (32, 33).
Fig. 4.
Median joining networks for mtDNA haplogroups A (A), B (B), C (C), and D (D).
Discussion
In this study, we adopt a rigorous approach to tree comparisons. First, a language classification is proposed as an a priori hypothesis for the genetic structure. Next, the language classification is fitted to the genetic data. And finally, the resulting tree is tested for treeness by using the method developed by Cavalli-Sforza and Piazza (25). The null hypothesis is that the pattern of relationships among languages is the same as the pattern of relationships among gene pools. We applied this approach to linguistic and genetic data for Native North Americans by fitting Greenberg's linguistic classification (19) to mitochondrial D-loop nucleotide sequence variations. The hypothesis of treeness was rejected. Therefore, a significant difference exists between Greenberg's linguistic classification and the genetic structures of these populations. Trees produced by the NJ and Fitch algorithms fit the genetic data better than did the Greenberg tree, but the treeness hypothesis was rejected even for these trees. Thus, we conclude that the genetic relationships among populations are not tree-like. The lack of treeness in the genetic data makes it unlikely that any hierarchical language classification will provide a close fit. The fact that not one of Greenberg's three major language groups (Amerind, Aleut-Eskimo, and Na Dene) formed an exclusive cluster in the better-fitting genetic trees produced by the NJ and Fitch algorithms shows that the problems with the GLC are pervasive and not due to a single problematic language group.
One possibility for the disagreement between the genetic data and the GLC is that the precontact pattern was disrupted by the entry of Europeans. We wish to emphasize why we do not feel that European contact is a likely explanation for our results. First, we restricted our analysis to mtDNA sequences that are established to be present in Native Americans and absent in Europeans. Second, population bottleneck and/or expansion events in the postcontact era would have changed relative branch lengths but would not have destroyed the treeness in the data. Third, 14 of the 17 populations are situated today in the same localities that they occupied at the time of first European contact. Fourth, mtDNA from ancient (precontact) Native American populations look similar to postcontact populations in the same region (34–36).
Another possibility for the disagreement between the genetic data and the GLC is that the GLC is a poor representation of the relationships among Native North American languages (4). In fact, most linguists do not accept the GLC (4, 10, 18, 20, 21). Greenberg's methods have been criticized (21, 37), and it is felt widely that there is not sufficient information to establish deep linguistic connections (10). Nevertheless, Greenberg included some undisputed language families within his classification. These undisputed families include the Eskimo-Aleut family and the Athabascan language group (10, 38, 39). We therefore find it quite interesting that the fitted branch to both Eskimo-Aleut and Athabascan was estimated to have zero length (Fig. 2). This finding suggests that the branch accounts for no increase in genetic relationship among its descendants. The NJ tree (Fig. 3) shows that the classifications within Aleut-Eskimo and Na Dene language families disagree with the population genetic relationships, regardless of whether or not they are analyzed within the context of the GLC. In fact, the NJ tree conflicts with generally accepted language groups presented by Campbell (10). Aleuts do not cluster with Eskimos. Neither the Yupik nor Inuit populations form distinct clusters within Eskimo. The two Algonquin-speaking populations do not form a cluster. The Navajo and Apache are separated in the NJ tree by the non-Athabascan-speaking Bella Coola. It is possible that a stronger relationship between language groups and gene pools would be found by directly assessing language areas that arise from diffusion of linguistic traits across phylogenetic boundaries of languages (4, 10).
The poor correspondence between the genetic data and language groups reflects the current population structure. Genetic and linguistic variation may have shared the same hierarchical relationships in the past. In fact, there are several features in the data that can be interpreted as remnants of such relationships. These features include the Eskimo-Aleut-specific transition polymorphism (29) and the two Athabascan-specific transition polymorphisms (14). Interestingly, these results are consistent with a linguistic model proposed by Nettle (9) that predicts that high diversity of language stocks is a transitory phase in linguistic evolution. According to Nettle's model, the entry of humans into a new region such as the Americas affords significant opportunity for the birth of new languages. Upon entry, population growth will be rapid and groups will fission and spread at high rate until the region is filled. New linguistic lineages will be founded, with population splits creating a tree-like structure. However, the rate of population fission will decline as open niches fill. Eventually, population dynamics will be governed by competition and efficiency of resource utilization. In this phase, linguistic diversity will decline as expanding communities absorb neighbors or as less successful communities fragment and members disperse.
Examination of the median joining networks for the canonical mtDNA haplogroups and population trees indicates a history of pervasive genetic exchange across linguistic boundaries. The distribution of mtDNA haplogroups in the Apache and Navajo presents the clearest example. As shown in Fig. 4, the distribution of the canonical Native American mtDNA haplogroups differs markedly between the far North and the Southwest. Notably, mtDNA sequences belonging to haplogroup B are not observed in the northern Na-Dene-attributed populations, and members of haplogroup C occur rarely (Fig. 4). By contrast, mtDNA sequences in Southwestern non-Athabascan speakers are characterized by the predominance of members of haplogroups B and C and the absence of members of haplogroup A. The haplogroup configuration for non-Athabascan speakers in the Southwest is exemplified in the present study by the Pima mtDNA sequences (Fig. 4) and has been established in surveys of haplogroups determined from diagnostic sites in many other Southwestern populations (33, 40). The Navajo and Apache possess many haplogroup A sequences typical of Northwestern populations with languages attributed to the Na Dene language family. However, DNA sequences belonging to haplogroups B and C are also common in the Navajo and Apache, and these are most likely due to immigrants from the local non-Athabascan-speaking populations. The elevated nucleotide diversity in the Navajo and Apache relative to their northern counterparts is the consequence of these haplogroup B and C mtDNA sequences.
Interestingly, the pattern of genetic exchange is not reciprocal. A-group haplotypes would have appeared in the Pima sample if they had absorbed a substantial number of Athabascan-speaking migrants. The pattern of asymmetrical genetic exchanges is all the more interesting given current mate exchange practices. Today, marriage practice in both the Western Apache and Navajo is strongly matrilineal. On this basis, we would not expect to see the inclusion of female lineages introduced from the surrounding non-Athabascan-speaking populations. However, the practice of matrilineality in these populations is likely to have begun after the Navajos and Apaches arrived in the Southwest (31). This practice makes it likely that the haplogroup B and C mtDNA sequences carried in the Navajo and Apache today were introduced early in their experience in the Southwest, and before the current cultural practices were initiated.
In addition to the presence of B- and C-group haplotypes in the Navajo and Apache, other features of the median-joining networks indicate genetic exchange between linguistically divergent neighboring populations in the Northwest and Arctic. For example, the Northwest Coast Bella Coola and Nuu Chah Nulth (attributed to Amerind) share A-group lineages with the Northwest Coast Haida (attributed to Na Dene), whereas the Eskimos in the far North share A-group lineages with interior dwelling Alaskan Athabascans. If language replacements predominated over gene replacements, we would expect good treeness for the genetic data but poor agreement between genetic and linguistic trees. In opposition to this expectation, even the best trees for the genetic data display a significant lack of treeness.
Many researchers have noted that either the language or the genes in a population can be replaced, and examples of both gene and language replacement have been identified (1, 2, 7). There has been a tendency to expect that language replacements will be more common (3, 41). In principle, language replacement can occur more rapidly than gene replacement because language can be transmitted both vertically from parents to children and horizontally between unrelated people, creating bilingual and multilingual individuals, whereas genes can be transmitted only vertically (1, 42). However, we found that the constraints imposed by mechanisms of transmission may be relatively weak as compared with other factors that will influence whether genes, languages, or both will be replaced.
Finally, we note that our findings are relevant to a multiple migration theory for the peopling of the Americas. According to this hypothesis, the ancestors of modern Amerind, Na-Dene, and Eskimo-Aleut speakers migrated into the Americas in three independent waves of migration, separated in time by thousands of years. Proponents have claimed support from a perceived parallel in patterns of variation in language, dental morphology, and classical genetic markers (43). Although it has been recognized that the original genetic data used in this argument lacked resolving power, our analyses of the currently available mtDNA sequences show that the patterns of variation in genes and languages actually disagree. Although our findings do not disprove that multiple migrations occurred historically, they dissipate a principal argument that was advanced to formulate the theory.
Acknowledgments
This work evaluates scientific hypotheses about the nature of correspondence between language groups and genetic populations. It is not a test of cultural practices or spiritual beliefs held by Native North Americans, and it should not be interpreted in that light. We thank Drs. D. (Weiss) Bolnick, J. Friedlaender, C. Lewis, R. Malhi, and S. Zegura for comments on earlier drafts of this paper. All interpretations and errors of omission or commission are those of the authors.
Abbreviations: GLC, Greenberg language classification; NJ, neighbor-joining.
References
- 1.Cavalli-Sforza, L. L. (1997) Proc. Natl. Acad. Sci. USA 94, 7719-7724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Diamond, J. & Bellwood, P. (2003) Science 300, 597-603. [DOI] [PubMed] [Google Scholar]
- 3.Renfrew, C. (2000) in America Past, America Present: Genes and Languages in the Americas and Beyond, ed. Renfrew, C. (McDonald Institute for Archaeological Research, Cambridge, U.K.), pp. 3-16.
- 4.Bolnick (Weiss), D. A., Shook (Shultz), B. A., Campbell, L. & Goddard, I. (2004) Am. J. Hum. Genet. 75, 519-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spuhler, J. N. (1979) in The First Americans: Origins, Affinities, and Adaptations, ed. Harper, A. B. (Bustav–Fischer, New York).
- 6.Spuhler, J. N. (1972) in The Assessment of Population Affinities in Man, ed. Huizinga, J. (Clarendon Press, Oxford).
- 7.Cavalli-Sforza, L. L., Piazza, A., Menozzi, P. & Mountain, J. (1988) Proc. Natl. Acad. Sci. USA 85, 6002-6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dixon, R. M. W. (1997) The Rise and Fall of Languages (Cambridge Univ. Press, Cambridge, U.K.).
- 9.Nettle, D. (1999) Proc. Natl. Acad. Sci. USA 96, 3325-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campbell, L. (1997) American Indian Languages: The Historical Linguistics of Native America (Oxford Univ. Press, New York).
- 11.Fagundes, N. J., Bonatto, S. L., Callegari-Jacques, S. M. & Salzano, F. M. (2002) Am. J. Phys. Anthropol. 117, 68-78. [DOI] [PubMed] [Google Scholar]
- 12.Merriwether, D. A., Friedlaender, J. S., Mediavilla, J., Mgone, C., Gentz, F. & Ferrell, R. E. (1999) Am. J. Phys. Anthropol. 110, 243-270. [DOI] [PubMed] [Google Scholar]
- 13.Piazza, A., Rendine, S., Minch, E., Menozzi, P., Mountain, J. & Cavalli-Sforza, L. L. (1995) in Proc. Natl. Acad. Sci. USA 92, 5836-5840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Torroni, A., Schurr, T. G., Cabell, M. F., Brown, M. D., Neel, J. V., Larsen, M., Smith, D. G., Vullo, C. M. & Wallace, D. C. (1993) Am. J. Hum. Genet. 53, 563-590. [PMC free article] [PubMed] [Google Scholar]
- 15.Bonatto, S. L. & Salzano, F. M. (1997) Proc. Natl. Acad. Sci. USA 94, 1866-1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Watson, E., Bauer, K., Aman, R., Weiss, G., von Haeseler, A. & Paabo, S. (1996) Am. J. Hum. Genet. 59, 437-444. [PMC free article] [PubMed] [Google Scholar]
- 17.Ward, R. H., Redd, A., Valencia, D., Frazier, B. & Paabo, S. (1993) Proc. Natl. Acad. Sci. USA 90, 10663-10667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nichols, J. (1990) Language 66, 475-521. [Google Scholar]
- 19.Greenberg, J. (1987) Language in the Americas (Stanford Univ. Press, Stanford, CA).
- 20.Mithun, M. (1990) Annu. Rev. Anthropol. 19, 309-330. [Google Scholar]
- 21.Ringe, D. (2000) in America Past, America Present: Genes and Languages in the Americas and Beyond, ed. Renfrew, C. (McDonald Institute for Archaeological Research, Cambridge, U.K.), pp. 139-162.
- 22.Anderson, S., Bankier, A. T., Barrell, B. G., de Bruijn, M. H., Coulson, A. R., Drouin, J., Eperon, I. C., Nierlich, D. P., Roe, B. A., Sanger, F., et al. (1981) Nature 290, 457-465. [DOI] [PubMed] [Google Scholar]
- 23.Ruhlen, M. (1991) Classification, A Guide to the World's Languages (Stanford Univ. Press, Stanford), Vol. 1, pp. 463. [Google Scholar]
- 24.Nei, M. (1987) Molecular Evolutionary Genetics (Columbia Univ. Press, New York).
- 25.Cavalli-Sforza, L. L. & Piazza, A. (1975) Theor. Popul. Biol. 8, 127-165. [DOI] [PubMed] [Google Scholar]
- 26.Urbanek, M., Goldman, D. & Long, J. C. (1996) Mol. Biol. Evol. 13, 943-953. [DOI] [PubMed] [Google Scholar]
- 27.Long, J. C. & Kittles, R. A. (2003) Hum. Biol. 75, 449-471. [DOI] [PubMed] [Google Scholar]
- 28.Bandelt, H. J., Forster, P. & Rohl, A. (1999) Mol. Biol. Evol. 16, 37-48. [DOI] [PubMed] [Google Scholar]
- 29.Starikovskaya, Y. B., Sukernik, R. I., Schurr, T. G., Kogelnik, A. M. & Wallace, D. C. (1998) Am. J. Hum. Genet. 63, 1473-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gunnerson, J. H. (1979) in Handbook of North American Indians: Southwest, ed. Ortiz, A. (Smithsonian Institution, Washington, D.C.), Vol. 9, pp. 162-169. [Google Scholar]
- 31.Opler, M. E. (1983) in Handbook of North American Indians: Southwest, ed. Ortiz, A. (Smithsonian Institution, Washington, D.C.), Vol. 10, pp. 368-392. [Google Scholar]
- 32.Malhi, R. S., Eshleman, J. A., Greenberg, J. A., Weiss, D. A., Schultz Shook, B. A., Kaestle, F. A., Lorenz, J. G., Kemp, B. M., Johnson, J. R. & Smith, D. G. (2002) Am. J. Hum. Genet. 70, 905-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malhi, R. S., Mortensen, H. M., Eshleman, J. A., Kemp, B. M., Lorenz, J. G., Kaestle, F. A., Johnson, J. R., Gorodezky, C. & Smith, D. G. (2003) Am. J. Phys. Anthropol. 120, 108-124. [DOI] [PubMed] [Google Scholar]
- 34.O'Rourke, D. H., Hayes, M. G. & Carlyle, S. W. (2000) Hum. Biol. 72, 15-34. [PubMed] [Google Scholar]
- 35.Stone, A. C. & Stoneking, M. (1993) Am. J. Phys. Anthropol. 92, 463-471. [DOI] [PubMed] [Google Scholar]
- 36.Carlyle, S. W., Parr, R. L., Hayes, M. G. & O'Rourke, D. H. (2000) Am. J. Phys. Anthropol. 113, 85-101. [DOI] [PubMed] [Google Scholar]
- 37.Ringe, D. (1996) Diachronica 13, 135-154. [Google Scholar]
- 38.Hill, J. H. & Hill, K. C. (2000) Am. Anthropol. 102, 161-163. [Google Scholar]
- 39.Golla, V. (2000) in America Past, America Present: Genes and Languages in the Americas and Beyond, ed. Renfrew, C. (McDonald Institute for Archaeological Research, Cambridge, U.K.), pp. 59-76.
- 40.Lorenz, J. G. & Smith, D. G. (1996) Am. J. Phys. Anthropol. 101, 307-323. [DOI] [PubMed] [Google Scholar]
- 41.Cavalli-Sforza, L. L. & Feldman, M. W. (2003) Nat. Genet. 33, Suppl., 266-275. [DOI] [PubMed] [Google Scholar]
- 42.Cavalli-Sforza, L. L. & Feldman, M. W. (1981) Cultural Transmission and Evolution: A Quantitative Approach (Princeton Univ. Press, Princeton). [PubMed]
- 43.Greenberg, J. H., Turner, C. G. & Zegura, S. L. (1986) Curr. Anthropol. 27, 477-497. [Google Scholar]
- 44.Rubicz, R., Melvin, K. L. & Crawford, M. H. (2002) Hum. Biol. 74, 743-760. [DOI] [PubMed] [Google Scholar]
- 45.Shields, G. F., Schmiechen, A. M., Frazier, B. L., Redd, A., Voevoda, M. I., Reed, J. K. & Ward, R. H. (1993) Am. J. Hum. Genet. 53, 549-562. [PMC free article] [PubMed] [Google Scholar]
- 46.Saillard, J., Forster, P., Lynnerup, N., Bandelt, H. J. & Norby, S. (2000) Am. J. Hum. Genet. 67, 718-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Budowle, B., Allard, M. W., Fisher, C. L., Isenberg, A. R., Monson, K. L., Stewart, J. E., Wilson, M. R. & Miller, K. W. (2002) Int. J. Legal Med. 116, 212-215. [DOI] [PubMed] [Google Scholar]
- 48.Kittles, R. A., Bergen, A. W., Urbanek, M., Virkkunen, M., Linnoila, M., Goldman, D. & Long, J. C. (1999) Am. J. Phys. Anthropol. 108, 381-399. [DOI] [PubMed] [Google Scholar]
- 49.Malhi, R. S., Schultz, B. A. & Smith, D. G. (2001) Hum. Biol. 73, 17-55. [DOI] [PubMed] [Google Scholar]
- 50.Ward, R. H., Frazier, B. L., Dew-Jager, K. & Paabo, S. (1991) in Proc. Natl. Acad. Sci. USA 88, 8720-8724. [DOI] [PMC free article] [PubMed] [Google Scholar]






