Abstract
The microbial dysbiosis associated with necrotizing enterocolitis (NEC) in preterm infants suggests that early exposure to probiotics may decrease and antibiotics may increase NEC risk. However, administration of Bifidobacterium breve strain BBG-001 to preterm infants did not affect NEC incidence in a multicenter randomised controlled phase 3 trial (PiPS trial). Using a subset of these subjects we compared the fecal microbiome of probiotic and placebo groups and assessed the impact of early antibiotic treatment. Extracted DNA from 103 fecal samples collected at 36 weeks post-menstrual age underwent PCR amplification of a fragment of the 16S rRNA gene. Heatmaps were constructed showing the proportions of sequences from bacterial families present at > 1% of the community. Stepwise logistic regression assessed the association between early antibiotic exposure and microbiome group. There was no difference in the microbial richness and diversity of the microbiome of preterm infants following treatment with probiotic or a placebo. Conversely, early antimicrobial exposure was associated with different patterns of colonisation, specifically a relative abundance of Proteobacteria.
These findings highlight that the potential influence of probiotics on the microbiome of preterm infants remains unclear whereas the modulatory effect of antibiotic exposure on microbial colonisation requires further research.
Keywords: Probiotic, Microbiome, Premature infant, Necrotizing enterocolitis
Abbreviations: AMR, antimicrobial resistant; NEC, necrotizing enterocolitis; NICU, Neonatal Intensive Care Unit
Highlights
-
•
Microbial dysbiosis has been associated with the development of neonatal necrotizing enterocolitis in premature infants.
-
•
Early administration of a probiotic compared to a placebo did not alter the microbiome of premature infants.
-
•
Early life exposure to antibiotics was associated with different patterns of colonisation in premature infants.
The association of altered microbial gastrointestinal colonisation (dysbiosis) with neonatal necrotizing enterocolitis suggests that, through their effects on microbial colonisation, probiotics and antimicrobials may modulate necrotizing enterocolitis risk. We compared microbial colonisation patterns of premature infants administered a probiotic or placebo in the early stages of life. We found no significant effect of probiotic administration on microbial colonisation, a finding which disputes the suggestion that prophylactic use of probiotics prevents the dysbiosis associated with necrotizing enterocolitis. In contrast, antibiotic exposure did alter the pattern of colonisation, indicating the need for further research in order to clearly ascertain the nature of this relationship.
1. Introduction
The bacteria that colonise newborn mammals contribute to health and disease in numerous ways (Deshmukh et al., 2014). The early pattern of development of the microbiome in infants nursed in intensive care has been linked with serious consequences including the development of neonatal necrotising enterocolitis (NEC) (Morowitz et al., 2010). NEC has a high associated mortality and morbidity and is the most common serious gastro-intestinal complication of prematurity with an incidence of between 6% to 10% in babies < 1500 g birthweight (Lin and Stoll, 2006).
Microbial diversity is characteristically reduced in infants at risk of developing NEC (Wang et al., 2009) leading to the proposition that the inflammatory response of the gut to an abnormal pattern of early microbial colonisation may underlie the pathogenesis of NEC (Neu and Walkerm, 2011). Consequently the administration of ‘beneficial’ live bacteria in the form of probiotics could help prevent NEC. The demonstration that probiotics reduce the risk of severe NEC in preterm infants supports this view, although such a reduction appears dependent on the infant's birthweight and the bacterial strain used (Aceti et al., 2015, AlFaleh and Anabrees, 2014). Mechanisms by which probiotics might prevent NEC include the possibility that they modify the composition, and increase the stability and diversity of gastrointestinal microbial populations (AlFaleh and Anabrees, 2014). However, the results of studies exploring the role of probiotics on the preterm gut and NEC incidence are confounded by small sample numbers and methodological inconsistencies (Aceti et al., 2015). Additionally, placebo controlled probiotic trials in infants have often omitted a comparison between the microbial composition of the two groups and subsequently do not allow a full understanding of the impact, if any, of probiotic administration on the developing microbiome.
The demonstration of intestinal dysbiosis following prolonged exposure to antimicrobials (Mshvildadze et al., 2010) and the associated increased risk of NEC (Greenwood et al., 2014) in preterm infants indicates a potential role for antimicrobial therapy in the modification of the microbiome and subsequent pathogenesis of NEC. Such dysbiosis is likely to depend on the antimicrobial used although, in the absence of robust trials, it is unclear as to which antimicrobial(s) confer the greatest NEC risk in preterm infants.
We have previously reported the results of a multicentre randomised placebo-controlled trial (PiPS trial) examining the impact of early administration of Bifidobacterium breve strain BBG-001 to 1315 babies born before 31 weeks of gestation and recruited within 48 h of birth (Costeloe et al., 2016). The primary endpoints were late onset blood stream infection, NEC and death. There was no evidence of differences in the primary endpoints when these outcomes in the probiotic and placebo groups were compared (Costeloe et al., 2016). To further explore the influence, if any, of probiotics on the preterm microbiome the present study carried out further microbial analysis on fecal samples collected at the end of the PiPS trial intervention period at 36 weeks post-menstrual age. In the light of previous reports demonstrating an effect of antimicrobials on the preterm gut, the impact of early antibiotic treatment was also examined.
2. Materials and Methods
2.1. PiPS Trial Protocol
The PiPS trial was a multicentre randomised masked placebo-controlled trial involving 1315 infants born at < 31 weeks gestational age randomised to receive either enteral Bifidobacterium breve strain BBG-001 or placebo. Infants were recruited between July 2010 and July 2013. Details of the trial have been published (Costeloe et al., 2016). Infants were recruited at 24 Neonatal Intensive Care Units (NICUs) within 48 h of birth. The interventions (probiotic and placebo preparations) were administered until 36 weeks post-menstrual age. Stool samples were collected from all infants at two weeks' postnatal age and 36 weeks' post-menstrual age. Samples included in this analysis were collected within the week following discontinuation of the trial intervention, coinciding with 36 weeks' post-menstrual age. Molecular analysis of stool samples was included within both the PiPS trial protocol and ethical approval documents (NRES Committee South Central: Oxford A, Reference 09-H0604-30).
Stool samples were transported to the laboratory using the Thermacor transportation system for diagnostic samples (Dyecor Ltd., Herefordshire, United Kingdom) and the Royal Mail. All samples were processed in the microbiology laboratory at Barts Health NHS Trust. Upon receipt in the laboratory, specimens were divided into two equal parts. One part was used for chemical, immunological and molecular analyses including the molecular detection of the trial strain (Bifidobacterium breve strain BBG). The other part was diluted 1:10 in a cryopreservative broth. Following processing of samples for the PiPS trial any residual sample was stored at − 80 °C.
2.2. Microbial Community Profiling
2.2.1. Selection of Samples
103 samples stored without preservative were selected from residual samples taken at 36 weeks' post-menstrual age and received by the laboratory between 23rd April 2012 and 12th February 2013. Only samples with > 0.5 g remaining were used. Selection was blinded to trial allocation status, information on individual characteristics and clinical outcomes. There were no duplicate samples analysed. The number of samples chosen for analysis was not based on a power calculation in large part because of the paucity of probiotic trial data from preterm infants which includes microbiome results. Bifodobacterium culture results for this group of samples were available from the PiPS study findings.
2.2.2. DNA Extraction and Partial 16S rRNA Gene Pyrosequencing
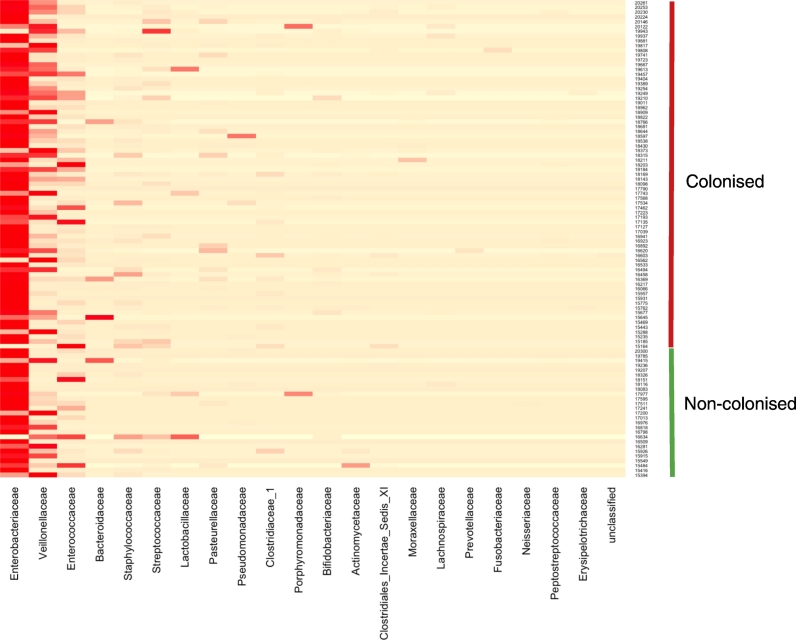
Samples were thawed and bacterial DNA was extracted from approximately 200 mg of fecal material following the QIAamp DNA Stool Minikit Protocol and using the QIAcube (Qiagen, Germany). A fragment of the 16S rRNA gene, encompassing the V1–V3 hypervariable regions, was amplified by PCR using the broad-range primers 27F-YM (Frank et al., 2008) and 519R (Lane et al., 1985). The Roche GS-FLX Titanium Series adapter sequence A and previously described unique 12-base error-correcting Golay barcodes (Fierer et al., 2008) were incorporated into the forward primers (5′-CCATCTCATCCCTGCGTGTCTCCGACTCAGNNNNNNNNNNNNAGAGTTTGATYMTGGCTCAG-3′) to enable multiplexing of multiple samples per run. The reverse primer (5′-CCTATCCCCTGTGTGCCTTGGCAGTCTCAGGWATTACCGCGGCKGCTG-3′) contained adapter sequence B at the 5′ end. PCR was performed using Extensor Hi-Fidelity PCR mastermix (Thermo-Scientific Abgene, UK) with the following conditions: 5 mins at 95 °C followed by 25 cycles of 95 °C for 45 s, 50 °C for 45 s, 72 °C for 90 s and a final extension of 72 °C for 5 mins. The amplicons were purified by means of a QIAquick PCR purification kit (Qiagen, Crawley, UK) following the manufacturer's instructions. The size and purity of the amplicons was assessed using the Agilent 2100 Bioanalyzer with the Agilent DNA 1000 kit (Agilent Technologies, Inc., Wokingham, UK), and quantified using the Qubit dsDNA HS Assay kit (Life Technologies, UK). The amplicons were then pooled together at equimolar concentrations (1 × 109 molecules/μl). Emulsion-PCR and unidirectional sequencing of the samples was performed using the Lib-L kit and the Roche 454 GS-FLX + Titanium series sequencer by the Department of Biochemistry, Cambridge University, Cambridge, UK. The primers used in this study had known mismatches with members of the family Bifidobacteriaceae (see Sim et al., 2012). We chose 16S rRNA primers consistent with previous published microbiome studies. For the PiPS study we included a specific PCR and cultured the intervention strain Bifidobacterium breve (strain BBG-001). The results of PCR analysis of the two week postnatal age samples, and of the 2 week postnatal age and the 36 week post menstrual age culture are shown in the original trial report (Costeloe et al., 2016). The Heatmap Fig. 2 shows the patterns of colonisation in infants by Bificobacterium breve colonisation status determined by culture. Specific PCR was not performed on the 36 week post-menstrual age samples.
Fig. 2.
Heatmap showing relative abundance of bacterial families amongst infant samples by trial intervention strain colonisation status. Two samples were excluded from the heatmap because the number of sequences was < 100. One sample dominated almost exclusively by Staphylococcaceae was also excluded.
2.2.3. Sequence Analysis
Signal processing of the raw data and generation of standard flowgram format (SFF) files was performed using the Long Amplicons 1 pipeline. The SFF files were processed using the mothur software suite (version 34.1) following the ‘454 standard operating procedure’ described at mothur.org (Schloss et al., 2009). The sequences were first deionised using the AmpliconNoise algorithm. Sequences < 440 bp in length with or without one of the following: > 2 mismatches to the forward primer sequence, > 1 mismatch to the barcode regions, and homopolymers > 8 bases in length, were discarded. The remaining sequences were trimmed to remove primers and barcodes and aligned to the SILVA 16S rRNA reference alignment. The UChime algorithm was used to identify chimeric sequences which were removed from the dataset. Sequences were clustered into operational taxonomic units (OTUs) at a genetic distance of (0.03) using the average neighbour algorithm and identified using a Naïve Bayesian classifier with the RDP reference set.
The sequences for each sample were randomly sub-sampled to 3250 sequences for statistical OTU-based diversity comparisons. The extent of sampling of the communities was assessed using Good's non-parametric coverage estimator (Good, 1953). The diversity of the communities was calculated using Simpson's inverse diversity index (Simpson, 1949). The community structure of the samples from treatment groups was compared using distance matrices generated with the thetaYC calculator (Yue and Clayton, 2005). The distance matrices were visualised using non-metric multidimensional scaling (NMDS) plots generated by means of the ggplot2 package in R (r-project.org). Analysis of molecular variance (AMOVA) (Excoffier et al., 1992) as implemented in mothur, was used to determine if there were statistically significant (p < 0.05) differences between treatments groups based on the thetaYC distance matrix.
2.2.4. Analysis of Colonisation Patterns
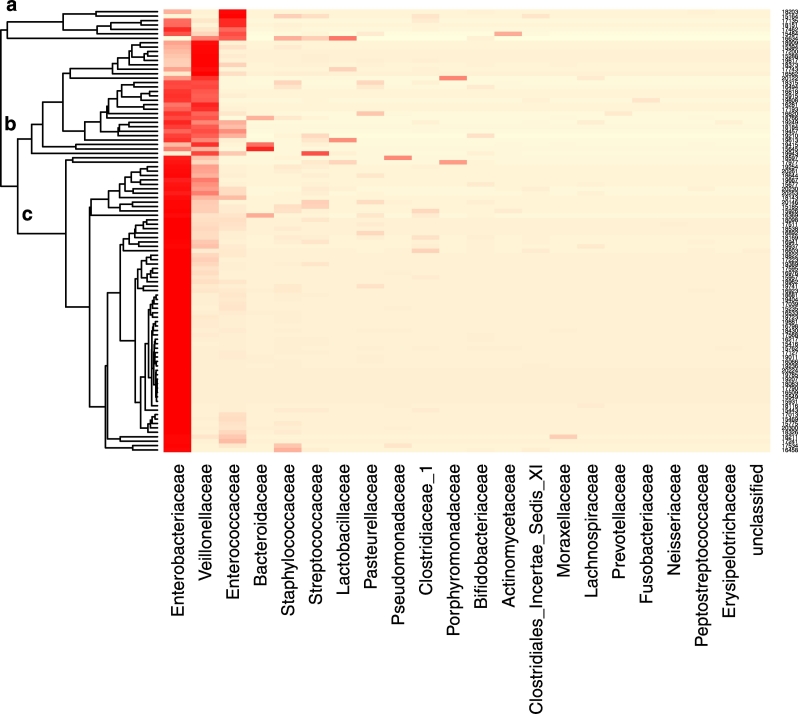
A heatmap showing the proportions of sequences from bacterial families present at > 1% of the community in infant samples was constructed using the vegan package in R. Samples were grouped by average linkage hierarchical clustering of a Bray Curtis dissimilarity matrix. Samples with > 100 OTUs were included in this heatmap and the logistical regression analysis of the relationship between antibiotic exposure and patterns of colonisation. This was the minimum number of OTUs considered to be sufficient for the data to be included in the derivation of the Heatmap shown in Fig. 4 (groups a, b, and c). We wanted to be inclusive in the derivation of the Heatmap and considered that 100 OTUs was sufficient to achieve groupings.
Fig. 4.
Heatmap showing relative abundance of predominant bacterial families and clustering of samples. Two samples were excluded from the heatmap because the number of sequences was < 100. One sample dominated almost exclusively by Staphylococcaceae was also excluded.
Microbiome groups were determined using the results of the heatmap. The association between early exposure to antibiotics and microbiome group was assessed using stepwise logistic regression, controlling for pre-specified covariates. Covariates included in the model selection procedure were: allocation (probiotic or placebo), baby's sex (male or female), mode of delivery (caesarean section or vaginal), gestational age at birth (weeks), birthweight (per 100 g), milk feeds from day 1 to day 14 postnatal age (formula and breast milk, formula only or breast milk only) and NEC confirmed by blinded endpoint review committee (yes or no). These covariates were defined in the PiPS trial protocol. The primary exposure was the total number of days on antibiotics from day 0 to day 14. Information was available from the PiPS daily log from day 0 to day 14 postnatal age for penicillin, aminoglycosides, cephalosporins, glycopeptides, carbapenems, β-lactam-inhibitor combinations, macrolides and other antibiotics.
Models were fitted using a backwards stepwise elimination approach. The significance level needed to enter the model was 0.5 and the significance level needed to stay in the model was 0.1. Using Stata SE for Windows (version 13.1) the likelihood-ratio test was used to decide which variables to enter into the model as it is more reliable for small sample sizes (Agresti, 1996). Correlation between independent variables was considered to determine if there were highly correlated variables which could be dropped before the model selection procedure. Adjusted logistic regression models were fitted to determine if there was a significant difference (p < 0.05) in the total number of days on antibiotics between day 0 to day 14 between the microbiome groups. Results are presented as adjusted odds ratios with 95% confidence intervals.
3. Results
Stool samples were received from 1235 infants alive at 36 weeks' post-menstrual age. We report the results of the microbiome analysis of fecal samples from 103 of these infants.
3.1. Comparison of Microbiome Composition in the Probiotic and Placebo Groups and by Colonisation Status
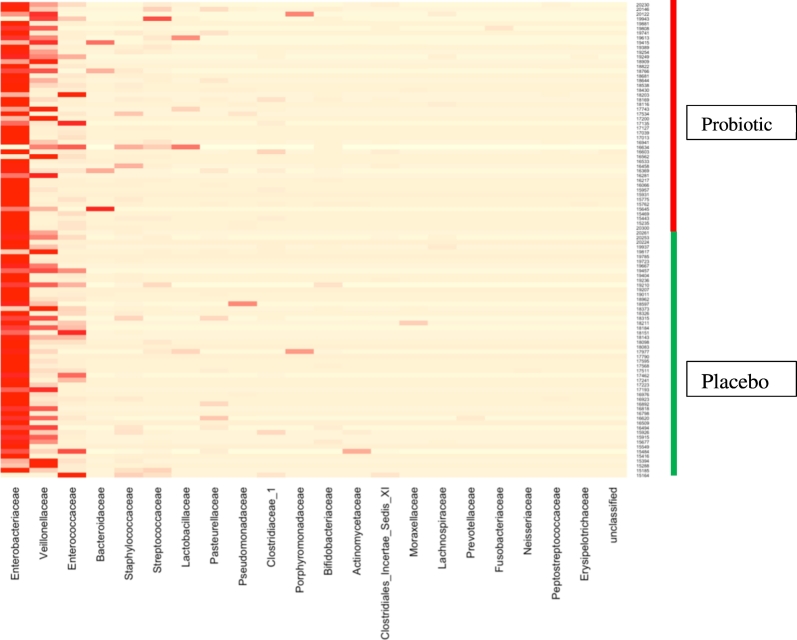
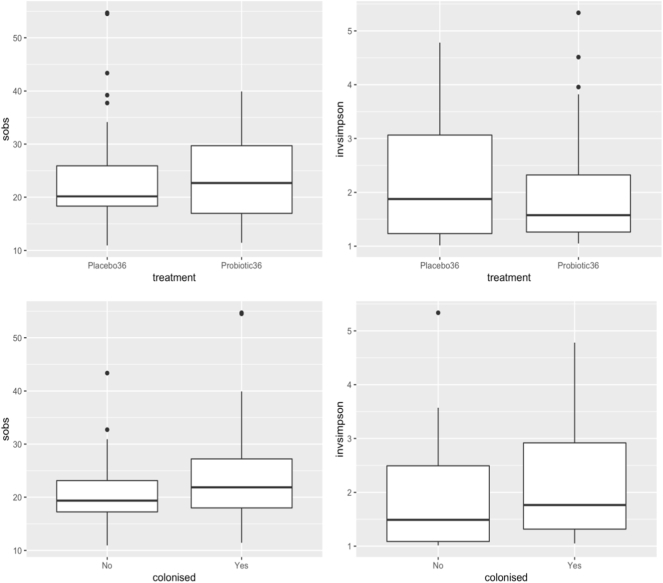
Heat maps showing the relative frequency of bacterial family sequences in individual samples are shown by allocation group in Fig. 1 and by colonisation status in Fig. 2. Box plots showing the richness (sobs) and diversity (invsimpson) of the microbiota in subjects by treatment and colonisation status are shown in Fig. 3. There were no statistically significant differences in richness and diversity between the subjects who received the probiotic or placebo nor between those colonised or not by the probiotic (Mann Whitney test). The analysis of richness and diversity was based on samples with > 3250 sequences and was therefore restricted to 88 samples (placebo 48 samples, probiotic 40, colonised 64, not colonised 24).
Fig. 1.
Heatmap showing relative abundance of predominant bacterial families amongst 100 infant samples by trial allocation group. Two samples were excluded from the heatmap because the number of sequences was < 100. One sample dominated almost exclusively by Staphylococcaceae was also excluded.
Fig. 3.
Box plots showing the richness (sobs) and diversity (invsimpson) of the microbiota in subjects treated with placebo (n = 48) or probiotic (n = 40) and colonised by the probiotic (n = 64) or not (n = 24).
Upper and lower edges of the boxes are the first and third quartiles; the line inside the box is the second quartile (median); individual dots are outliers.
3.2. Early Exposure to Antibiotics and Patterns of Microbial Colonisation
The heatmap shown in Fig. 4 of microbial colonisation in fecal microbiota collected at 36′ weeks post-menstrual age showed clustering of the samples into three groups designated a, b and c. Two infant samples gave < 100 OTUs and were excluded. In addition one sample dominated almost exclusively by Staphylococcaceae was excluded from the derivation of the heatmap. Group b included samples with a relative abundance of antibiotic sensitive microbes, particularly Veillonellaceae. Group c was dominated by Enterobacteriaceae and Group a by Enterococcaceae. Summary statistics for the groups a, b and c are shown in Table 1. The characteristics of the mothers and the infants from whom these samples were collected including details on mode of delivery, milk feeds, antibiotic administration and the development of NEC prior to sample collection were comparable by allocation status (probiotic and placebo groups) (Supplementary Table 1). The characteristics of this subset align broadly with those of the population of infants recruited to the PiPS trial (Costeloe et al., 2016). Each infant was the unit of randomisation in the PiPS trial. The Supplementary Table 1 shows details of multiple births. All of the proportions in the baseline characteristics used the total number of infants as a denominator.
Table 1.
Summary statistics of total number of days on antibiotics from day 0 to day 14 by microbiome group (SD, Standard Deviation; IQR, Interquartile Range).
| Microbiome group | Statistic | Total number of days on antibiotics from day 0 to day 14 |
|---|---|---|
| Group a | n (%) | 7 (7.0) |
| Mean (SD) | 9.3 (2.43) | |
| Median (IQR) | 10 (8 to 11) | |
| Group b | n (%) | 26 (26.0) |
| Mean (SD) | 6.4 (3.86) | |
| Median (IQR) | 6 (4 to 9) | |
| Group c | n (%) | 67 (67.0) |
| Mean (SD) | 8.5 (3.41) | |
| Median (IQR) | 8 (6 to 12) | |
| Groups a & c | n (%) | 74 (74.0) |
| Mean (SD) | 8.6 (3.32) | |
| Median (IQR) | 8 (6 to 11) |
We tested the hypothesis that antibiotic exposure in the first two weeks of life is associated with the colonisation of the gut by microbiome group by comparing colonisation group b with group a, and with groups a and c combined, adjusting for pre-specified covariates. ‘Gestational age’ was removed as it was found to be highly correlated with birthweight. ‘Milk feeds’ was also removed from the models as only one baby was fed on formula only in the first 14 days of life. The final models for both comparisons adjusted for mode of delivery (caesarean section or vaginal), NEC and birthweight (per 100 g). The results for models fitting the primary exposure of total number of days on antibiotics from day 0 to day 14 are shown in Table 2.
Table 2.
Statistical output for models fitting primary exposure of total number of days on antibiotics from day 0 to day 14.
| Model | N | Odds ratio | 95% CI | P-value |
|---|---|---|---|---|
| Unadjusted group b versus group c | 93 | |||
| Total number of days on antibiotics from day 0 to 14 | 0.84 | (0.74, 0.96) | 0.013 | |
| Adjusted group b versus group c | 93 | |||
| Total number of days on antibiotics from day 0 to 14 | 0.82 | (0.71, 0.95) | 0.009 | |
| Mode of delivery | 1.98 | (0.72, 5.41) | 0.185 | |
| NEC | 2.77 | (0.35, 21.68) | 0.332 | |
| Birthweight (per 100 g) | 0.93 | (0.79, 1.11) | 0.427 | |
| Unadjusted group b versus groups a&c | 100 | |||
| Total number of days on antibiotics from day 0 to 14 | 0.83 | (0.73, 0.95) | 0.008 | |
| Adjusted group b versus a&c | 100 | |||
| Total number of days on antibiotics from day 0 to 14 | 0.81 | (0.70, 0.94) | 0.007 | |
| Mode of delivery | 2.00 | (0.74, 5.42) | 0.173 | |
| NEC | 3.21 | (0.40, 25.46) | 0.269 | |
| Birthweight (per 100 g) | 0.94 | (0.79, 1.11) | 0.468 |
The odds of a baby being colonised by a microbiome from group b compared to group c decreased by 16% for every one day increase in the total number of days on antibiotics between days 0 and 14. This increased to 18% when adjusted for mode of delivery, NEC and birthweight (per 100 g). The odds of a baby being colonised by a microbiome from group b compared to groups a and c decreased by 17% for every one day increase in total number of days on antibiotics between days 0 and 14. This increased to 19% when adjusted for mode of delivery, NEC and birthweight (per 100 g). Days on antibiotics was highly statistically significant in all models.
4. Discussion
The microbiome of the gut of preterm infants has consistently been shown to have higher proportions of Proteobacteria compared to those of full term infants (Arboleya et al., 2016, Wang et al., 2009). These differences probably reflect the influence of a number of factors such as mode of delivery and postnatal exposures including the administration of antimicrobials (Mai et al., 2011, Wang et al., 2009). The use of probiotics has been postulated to represent a means by which the microbiome of the preterm infant may be altered such that it confers a protective effect against NEC. This study found no evidence of a difference in the patterns of colonisation or microbial diversity in the probiotic compared with placebo group by the end of the intervention period (36 weeks post-menstrual age), contrary to previous study findings (Ishizeki et al., 2013). By contrast, the number of days of antibiotics received in the first two weeks of life was statistically significantly associated with different patterns of colonisation.
The finding that antibiotic exposure in early life was associated with different patterns of colonisation and specifically a relative abundance of Proteobacteria in this study is consistent with studies in term and preterm infants (Tanaka et al., 2009). Use of antibiotics early in life has been associated both with increased levels of Enterobacteriaceae and an increased risk of NEC (Pammi et al., 2017, Greenwood et al., 2014). Warner et al. (2016) reported a relative abundance of Gammaproteobacteria and relative paucity of Negativicutes (includes Veillonella spp) preceding NEC in very low birthweight infants. Mai et al. (2011) and Morrow et al. (2013) reported that Proteobacteria were relatively dominant components of the microbiome (especially Enterobacteriaceae) preceding the onset of NEC in preterm infants in cases of NEC compared with controls. In addition to reduced microbial diversity, early antimicrobial administration may induce the development of a resistant population of intestinal bacteria harbouring antimicrobial resistant (AMR) genes (the resistome - Penders et al., 2013). The demonstration of increased levels of AMR genes in preterm infants receiving antibiotics (Gibson et al., 2016) supports this supposition. However, the detection of AMR genes in the preterm healthy gut (Rose et al., 2017) suggests that, even in the absence of antibiotics, AMR genes may be present perhaps reflecting the impact of gestational factors such as perinatal maternal antibiotics on the foetal microbiome (Francino, 2015). Large scale studies are required in this area to further characterise the evolution of the resistome.
The use of samples from a multicentre randomised placebo-controlled trial and the blinding of researchers to the treatment groups are strengths of the present study. However, the low sample number may prevent the detection of any small yet potentially important differences in the microbiomes of the probiotic and placebo groups. In addition only one sample from each infant was analysed at a single time point and yet the microbiome of the infant is in a constant state of flux especially during the early stages. For example, very low birthweight preterm infants have been shown to have a preponderance of Firmicutes at two days postpartum followed by Proteobacteria at 30 days (Arboleya et al., 2016). Consequently, determination of the microbiome at an isolated time point may fail to identify any differences that occur between the probiotic and placebo groups at an earlier or later date. Another weakness of this study is that the primers chosen (although consistent with those reported in previous relevant publications) were not optimised to detect Bifidobacteriaceae (Sim et al., 2012). The PiPS study included both a specific PCR for the intervention strain and culture and these results have already been reported (Costeloe et al., 2016). Differences in patterns of microbial colonisation have been ascribed to differences in the PCR primers used to target the 16S rRNA gene (Pammi et al., 2017).
The rapid progression of NEC in the preterm infant (Yang et al., 2014) highlights the importance of not only determining the most effective treatment options but also the prophylactic measures that can be taken to prevent its occurrence. The current study has not demonstrated an impact of probiotics on the microbiome of the preterm infant. Despite differences in microbial composition in response to antibiotics in the present study, the findings from this microbiome analysis of a subset of infants included in the PiPS trial and the outcomes of the trial itself neither refute nor support the hypothesis that NEC is a consequence of dysbiosis (Warner et al., 2016).
In conclusion, the possibility that specific bacteria or microbial factors play a protective role in determining the risk of NEC remains open and requires further investigation. In contrast, the altered microbiome in response to early antimicrobial administration demonstrates the importance of documenting and taking account of antibiotic exposure as a potential confounder in future probiotic studies.
Funding Sources
The PiPS Trial and subsequent research presented in this report was funded by the UK National Institute for Health Research Health Technology Assessment Programme (HTA 05/501/04).
Conflicts of Interests
All other authors declare no competing interests.
Author Contributions
Michael Millar conceived this study and prepared early manuscripts. Jo Seale contributed substantially to the microbiome analyses and the writing and preparation of the manuscript. Mark Wilks coordinated the laboratory work. Kate Costeloe & Edmund Juszczak coordinated the PiPS trial. Nicola Panton carried out the microbiome analyses with Jo Seale. William Wade oversaw the microbiome work, prepared the reports on the microbiome results and helped with the manuscript preparation. Pollyanna Hardy and Melanie Greenland carried out the statistical analyses. All authors read and approved the final manuscript.
Acknowledgements
We would like to acknowledge Angela Whiley who coordinated the storage and security of PiPS samples.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ebiom.2017.05.019.
Appendix A. Supplementary Data
Supplementary material
References
- Aceti A., Gori D., Barone G., Callegari M.L., Di Mauro A., Fantini M.P., Indrio F., Maggio L., Meneghin F., Morelli L., Zuccotti G., Corvaglia L., Italian Society of Neonatology Probiotics for prevention of necrotizing enterocolitis in preterm infants: systematic review and meta-analysis. Ital. J. Pediatr. 2015;41:89. doi: 10.1186/s13052-015-0199-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agresti A. vol. 135. Wiley; New York: 1996. An Introduction to Categorical Data Analysis. [Google Scholar]
- AlFaleh K., Anabrees J. Probiotics for prevention of necrotizing enterocolitis in preterm infants. Cochrane Database Syst. Rev. 2014;4:CD005496. doi: 10.1002/14651858.CD005496.pub4. [DOI] [PubMed] [Google Scholar]
- Arboleya S., Sánchez B., Solís G., Fernández N., Suárez M., Hernández-Barranco A.M., Milani C., Margolles A., de Los Reyes-Gavilán C.G., Ventura M., Gueimonde M. Impact of prematurity and perinatal antibiotics on the developing intestinal microbiota: a functional inference study. Int. J. Mol. Sci. 2016;17:E649. doi: 10.3390/ijms17050649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costeloe K., Hardy P., Juszczak E., Wilks M., Millar M.R. Probiotics in preterm infants study collaborative group. Bifidobacterium breve BBG-001 in very preterm infants: a randomised controlled phase 3 trial. Lancet. 2016;387:649–660. doi: 10.1016/S0140-6736(15)01027-2. [DOI] [PubMed] [Google Scholar]
- Deshmukh H.S., Liu Y., Menkiti O.R., Mei J., Dai N., O'Leary C.E., Oliver P.M., Kolls J.K., Weiser J.N., Worthen G.S. The microbiota regulates neutrophil homeostasis and host resistance to Escherichia coli K1 sepsis in neonatal mice. Nat. Med. 2014;20:524–530. doi: 10.1038/nm.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L., Smouse P.E., Quattro J.M. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N., Hamady M., Lauber C.L., Knight R. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc. Natl. Acad. Sci. U. S. A. 2008;105:17994–17999. doi: 10.1073/pnas.0807920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francino M.P. Antibiotics and the human gut microbiome: Dysbioses and accumulation of resistances. Front. Microbiol. 2015;6:1543. doi: 10.3389/fmicb.2015.01543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J.A., Reich C.I., Sharma S., Weisbaum J.S., Wilson B.A., Olsen G.J. Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl. Environ. Microbiol. 2008;74:2461–2470. doi: 10.1128/AEM.02272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson M.K., Wang B., Ahmadi S., Burnham C.A., Tarr P.I., Warner B.B., Dantas G. Developmental dynamics of the preterm infant gut microbiota and antibiotic resistome. Nat Microbiol. 2016;1:16024. doi: 10.1038/nmicrobiol.2016.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good I.J. The population frequencies of species and the estimation of population parameters. Biometrika. 1953;40:237–264. [Google Scholar]
- Greenwood C., Morrow A.L., Lagomarcino A.J., Altaye M., Taft D.H., Yu Z., Newburg D.S., Ward D.V., Schibler K.R. Early empiric antibiotic use in preterm infants is associated with lower bacterial diversity and higher relative abundance of Enterobacter. J. Pediatr. 2014;165:23–29. doi: 10.1016/j.jpeds.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizeki S., Sugita M., Takata M., Yaeshima T. Effect of administration of bifidobacteria on intestinal microbiota in low-birth-weight infants and transition of administered bifidobacteria: a comparison between one-species and three-species administration. Anaerobe. 2013;23:38–44. doi: 10.1016/j.anaerobe.2013.08.002. [DOI] [PubMed] [Google Scholar]
- Lane D.J., Pace B., Olsen G.J., Stahl D.A., Sogin M.L., Pace N.R. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc. Natl. Acad. Sci. U. S. A. 1985;82:6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin P.W., Stoll B.J. Necrotising enterocolitis. Lancet. 2006;368:1271–1283. doi: 10.1016/S0140-6736(06)69525-1. [DOI] [PubMed] [Google Scholar]
- Mai V., Young C.M., Ukhanova M., Wang X., Sun Y., Casella G., Theriaque D., Li N., Sharma R., Hudak M., Neu J. Fecal microbiota in premature infants prior to necrotizing enterocolitis. PLoS One. 2011;6:e20647. doi: 10.1371/journal.pone.0020647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morowitz M.J., Poroyko V., Caplan M., Alverdy J., Liu D.C. Redefining the role of intestinal microbes in the pathogenesis of necrotizing enterocolitis. Pediatrics. 2010;125:777–785. doi: 10.1542/peds.2009-3149. [DOI] [PubMed] [Google Scholar]
- Morrow A.L., Lagomarcino A.J., Schibler K.R., Taft D.H., Yu Z., Wang B., Altaye M., Wagner M., Gevers D., Ward D.V., Kennedy M.A., Huttenhower C., Newburg D.S. Early microbial and metabolomic signatures predict later onset of necrotizing enterocolitis in preterm infants. Microbiome. 2013;1:13. doi: 10.1186/2049-2618-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mshvildadze M., Neu J., Shuster J., Theriaque D., Li N., Mai V. Intestinal microbial ecology in preterm infants assessed with non-culture-based techniques. J. Pediatr. 2010;156:20–25. doi: 10.1016/j.jpeds.2009.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neu J., Walkerm W.A. Necrotizing enterocolitis. N. Engl. J. Med. 2011;364:255–264. doi: 10.1056/NEJMra1005408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pammi M., Cope J., Tarr P.I., Warner B.B., Morrow A.L., Mai V., Gregory E.G., Kroll S., McMurtry V., Ferris M.J., Engstrand L., Lilja H.E., Hollister E.B., Versalovic J., Neu J. Intestinal dysbiosis in preterm infants preceding necrotizing enterocolitis: a systematic review and meta-analysis. Microbiome. 2017;5:31. doi: 10.1186/s40168-017-0248-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penders J., Stobberingh E.E., Savelkoul P.H., Wolffs P.F. The human microbiome as a reservoir of antimicrobial resistance. Front. Microbiol. 2013;4:87. doi: 10.3389/fmicb.2013.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose G., Shaw A.G., Sim K., Wooldridge D.J., Li M.S., Gharbia S., Misra R., Kroll J.S. Antibiotic resistance potential of the healthy preterm infant gut microbiome. Peer J. 2017;5:e2928. doi: 10.7717/peerj.2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss P.D., Westcott S.L., Ryabin T., Hall J.R., Hartmann M., Hollister E.B., Lesniewski R.A., Oakley B.B., Parks D.H., Robinson C.J., Sahl J.W., Stres B., Thallinger G.G., Van Horn D.J., Weber C.F. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim K., Cox M.J., Wopereis H., Martin R., Knol J., Ming-Shi Li, Cookson W.O.C.M., Moffatt M.F., Kroll J.S. Improved detection of bifidobacteria with optimised 16S rRNA-gene based pyrosequencing. PLoS One. 2012;7:e32543. doi: 10.1371/journal.pone.0032543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson E.H. Measurement of diversity. Nature. 1949;163:688. [Google Scholar]
- Tanaka S., Kobayashi T., Songjinda P., Tateyama A., Tsubouchi M., Kiyohara C., Shirakawa T., Sonomoto K., Nakayama J. Influence of antibiotic exposure in the early postnatal period on the development of intestinal microbiota. FEMS Immunol. Med. Microbiol. 2009;56:80–87. doi: 10.1111/j.1574-695X.2009.00553.x. [DOI] [PubMed] [Google Scholar]
- Wang Y., Hoenig J.D., Malin K.J., Qamar S., Petrof E.O., Sun J., Antonopoulos D.A., Chang E.B., Claud E.C. 16S rRNA gene-based analysis of fecal microbiota from preterm infants with and without necrotizing enterocolitis. ISME J. 2009;3:944–954. doi: 10.1038/ismej.2009.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner B.B., Deych E., Zhou Y., Hall-Moore C., Weinstock G.M., Sodergren E., Shaikh N., Hoffmann J.A., Linneman L.A., Hamvas A., Khanna G., Rouggly-Nickless L.C., Ndao I.M., Shands B.A., Escobedo M., Sullivan J.E., Radmacher P.G., Shannon W.D., Tarr P.I. Gut bacteria dysbiosis and necrotising enterocolitis in very low birthweight infants: a prospective case-control study. Lancet. 2016;387:1928–1936. doi: 10.1016/S0140-6736(16)00081-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Guo Y., Kan Q., Zhou X.G., Zhou X.Y., Li Y. A meta-analysis of probiotics for preventing necrotizing enterocolitis in preterm neonates. Braz. J. Med. Biol. Res. 2014;47:804–810. doi: 10.1590/1414-431X20143857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue J.C., Clayton M.K. A similarity measure based on species proportions. Commun. Stat. Theory Methods. 2005;34:2123–2131. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material