Abstract
Control of colon cell fate in adenocarcinomas is disrupted, in part, due to aberrant Wnt/β-catenin signaling. The nuclear receptor peroxisome proliferator-activated receptor-γ (PPARγ) has been implicated in the development of colon cancers. In the adenomatous polyposis coli multiple intestinal neoplasia (APCMin) mouse cancer model, PPARγ expression in the colonic mucosa is markedly altered. In addition, PPARγ protein levels are elevated, possibly through sequestration by activated β-catenin in colon cancer cell lines. Induction of the Wnt/β-catenin pathway by LiCl also elevated PPARγ levels and induced PPARγ-dependent reporter and endogenous target genes. Mechanistically, PPARγ, through interactions with β-catenin and T cell transcription factor (Tcf)-4, may be a determinant of cell fate and is likely a target of the Wnt pathway in cancer cells.
Keywords: colon epithelium, Wnt signaling, nuclear receptors, APCMin mice, T cell transcription factor 4
The Wnt/β-catenin pathway plays a critical role in the development of the gastrointestinal tract. Central to this pathway is a multiprotein scaffold consisting of adenomatous polyposis coli (APC), glycogen synthase kinase (GSK)-3β, axin, and the transcriptional cofactor β-catenin. In the absence of Wnt ligands, β-catenin levels are kept low through constitutive phosphorylation by GSK-3β, which leads to the ubiquitination and degradation of β-catenin (1). Wnt signaling inhibits GSK-3β activity, reducing phosphorylation and subsequent degradation of β-catenin. β-catenin is stabilized and translocates to the nucleus to bind members of the T cell factor (Tcf)/lymphoidenhancing factor (LEF) family of transcription factors and induce target gene expression (1-3).
The downstream targets of the canonical Wnt signaling pathway comprise several genes important for cellular proliferation, such as c-myc and cyclin D1 (4, 5), underscoring the importance of Wnt signaling in the development of cancers. In the intestinal epithelium, Tcf-4 is the most prominently expressed Tcf family member, and Tcf-4 knockout mice fail to maintain a proliferative stem cell compartment in the crypts of the small intestine, due to G1 arrest and impairment of differentiation programs (6, 7).
Mutations in the APC gene are one of the initiating events in the development of sporadic (8) and hereditary familial colorectal tumors (9, 10). In the APCMin (multiple intestinal neoplasia) model, mice carry a nonsense mutation in the murine homolog of the APC gene and serve as a model of gastrointestinal neoplasia (11, 12). Homozygote mice die in utero, whereas heterozygote animals, although viable, develop multiple intestinal neoplasia at an early age and seldom survive beyond 3 months of age (2, 13).
Peroxisome Proliferator-Activated Receptor-γ (PPARγ) is a nuclear receptor that is ubiquitously expressed, with the highest levels observed in adipose tissue and the colon (14). PPARγ forms a heterodimer with the retinoic X receptor (RXR), and the resulting complex binds PPAR-responsive elements (PPRE) within target gene promoters (15, 16). Ligands for PPARγ include the endogenous prostaglandin derivate 15d-PGJ2 and the synthetic thiazolidinediones (17, 18). Although originally identified as a transcription factor essential for adipocyte differentiation (19), a role for PPARγ in epithelial cell differentiation cannot be excluded.
The expression of PPARγ in the normal colonic mucosa varies along the crypt axis, with highest levels in postmitotic cells facing the intestinal lumen (20). Furthermore, the exposure of cultured human colon cancer cells to PPARγ ligand induces growth inhibition and cellular differentiation (21). The role of PPARγ ligands, however, in regulating neoplastic transformation in vivo remains controversial. Aberrantly high levels of PPARγ have previously been seen in colon cancers (22). Troglitazone has been shown to inhibit tumor growth (21) and to reduce the formation of aberrant crypt foci (ACF) in mice (23, 24). In stark contrast, other studies showed that PPARγ ligands not only failed to suppress polyp formation, they also led to a small but significant increase in polyp numbers in APCMin mice (25, 26).
The observations that PPARγ expression in polyps from APCMin mice is increased (25) and that the integrity of APC is important for the tumor-suppressive activity of PPARγ (27) led us to determine PPARγ protein levels in APCMin mice. We found that protein levels were altered in colon tissue from the APCMin mouse. Furthermore, in transient transfection assays of epithelial cells, both the overexpression of β-catenin and LiCl activation of the Wnt signaling pathway led to augmentation of PPARγ protein levels. Signaling through the Wnt/β-catenin pathway activated a PPARγ-driven luciferase reporter gene and elevated expression of putative PPARγ target genes. PPARγ is also shown to physically interact with β-catenin and Tcf-4. In conclusion, the Wnt/β-catenin pathway likely regulates PPARγ function in colon epithelial cells.
Materials and Methods
Cell Lines and Mice. The colon carcinoma cell line SW480 (ATCC CCL-228) and the kidney epithelial cell line HEK293 (ATCC CRL-1573) were grown accordingly and used as stated in the transient transfections, coimmunoprecipitation, and RNA expression analyses. The C57BL/6J-ApcMin (APCMin) strain was purchased from The Jackson Laboratory.
Transient Transfections. Transfections were done by using either the FuGENE 6 (Roche Diagnostics) or Lipofectamine (Invitrogen) reagents, and they were harvested after 48 h. Luciferase assays were performed as described by the manufacturer (Promega). Rosiglitazone (BRL 49653) was purchased from Glaxo-SmithKline and used at 10 μM. Gifts of plasmids and expression vectors are gratefully acknowledged: PPARγ2 expression vector and the PPRE plasmid, (A. Berkenstam, Karo Bio, Huddinge, Sweden); the dnPPARγ2 plasmid (V. K. K. Chatterjee, University of Cambridge, Cambridge, U.K.); wild-type and mutated β-catenin-containing vectors (H. Clevers, University Medical Center Utrecht, Utrecht, The Netherlands); RXRα expression vector and its luciferase reporter gene (T. Perlmann, Ludwig Institute of Cancer Research, Stockholm); and the ERE and DRE reporter genes, full-length ERα, wild-type, and the constitutively active dioxin receptor expression vectors (mDRA1B) (L. Poellinger, Karolinska Institutet, Stockholm).
Immunoprecipitation and Western Blot. Cells were grown to confluence, treated as described, and then harvested and lysed according to ref. 28. Colonic epithelial cells were isolated and analyzed as described (28). Antibodies specific for actin (sc-1616), PPARγ (sc-7273), β-catenin (sc-1496), Jak1 (sc-513), Stat3 (sc-483), and Ku-70 (sc-1487) were purchased from Santa Cruz Biotechnology. The anti-Tcf-4 antibody (clone 6H5-3) was purchased from Upstate Biotechnology (Lake Placid, NY). Immunodetection with a goat secondary peroxidase-conjugated antibody (DAKO) and chemiluminescence were performed according to the manufacturers' protocols (ECL, Amersham Pharmacia). Immunoprecipitations were also performed with FLAG-conjugated Sepharose A beads (Sigma-Aldrich).
Immunofluorescence and Immunohistochemistry. Cells were seeded on coverslips and transfected as indicated. After 24 h, the cells were fixed in 4% formalin (Sigma Diagnostics), permeabilized with 0.1% Triton X-100, and subsequently stained with primary antibodies for PPARγ (sc-7273). Paraffin embedding of sections was done as described (28). Sections of specimens were incubated with antibodies for PPARγ (Biomol, Plymouth Meeting, PA) overnight at 4°C. Sections were then blocked in 2% goat preimmune serum, followed by 2% biotinylated goat anti-rabbit IgG. Thereafter, avidin biotin enzyme reagent was applied, followed by diaminobenzidine (DAB), and subsequently counterstained with hematoxylin. The goat anti-sera, DAB, and the avidin-biotin enzymes were obtained from Vector Laboratories.
RNase Protection and RT-PCR Assays. Total RNA was extracted by using TRIzol reagent (Invitrogen) and then analyzed by ribonuclease protection assay (described in ref. 28). PPARγ target gene expression levels were analyzed by the relative quantitative RT-PCR method. The total RNA was extracted by using the RNeasy Mini kit (Qiagen, Valencia, CA). The first-strand cDNA was synthesized from 1 μg of total RNA by using random hexamers and SuperScript II reverse transcriptase in a 20-μl reaction mixture (Invitrogen) according to the manufacturer's instructions. To normalize the signals from different RNA samples, β-actin was used as an internal standard. The following primers were used to amplify the PPARγ target genes: β-actin, sense primer 5′-CCTGGCACCCAGCACAAT-3′ and antisense primer 5′-GCCGATCCACACGGAGTACT-3′; keratin-20, sense primer 5′-CTGAATAAAGACCTAGCTCTCCTCAAA-3′ and antisense primer 5′-TGTTGCCCAGATGCTTGTGT-3′; ADFP, sense primer 5′-CTGTTCACCTGATTGAATTTGC-3′ and antisense primer 5′-AGAGCTTATCCTGAGCATCCTG-3′; and FABP2, sense primer 5′-AAATGGGTGTTAATATAGTGAAAA-3′ and antisense 5′-CCTTCTTGTGTAATTGTCAGCTTC-3′.
Real-time PCR was carried out in a 25-μl amplification mixture containing 0.5 μl of template cDNA, 12.5 μl of 2× SYBR Green I Master Mix, and 200 nM sense and antisense primers. The PCR conditions included a polymerase activation step at 95°C for 10 min followed by 40 cycles of 95°C for 15 s and 60°C for 60 s and run on an ABI PRISM 7500 Sequence Detector (Applied Biosystems). A nontemplate control was included for all of the primer pairs. The gene expression levels from the untreated control were set as 1 to compare the relative amounts of gene expression levels in the experimental groups. Statistical analysis of the results was performed in Microsoft excel. Significant difference was determined with P < 0.05 by the Student t test.
EMSA. Nuclear extracts from appropriately treated cells (see legends to Figs. 2 and 3) were made as described (29). The probe was derived from a single DR1 site in the rat CYP4A1 5′ flank with the following sequence: 5′-AACTAGGGTAAAGTTCAC-3′.
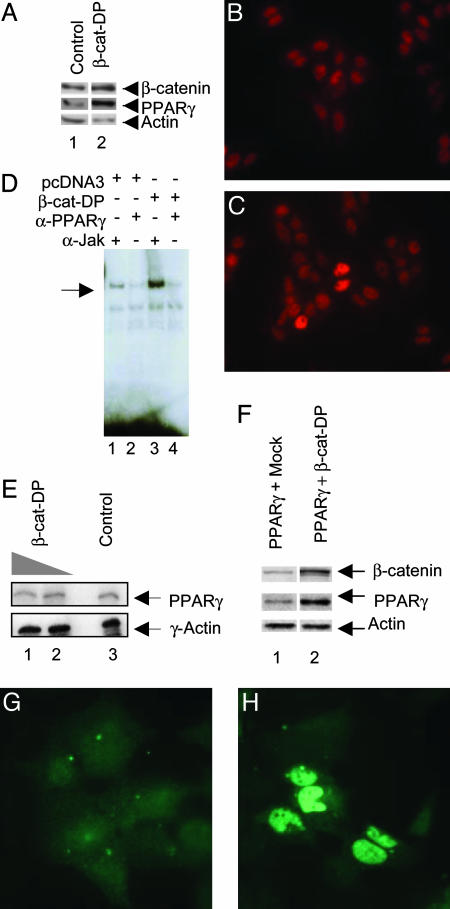
Fig. 2.
PPARγ protein levels are increased in β-catenin-transfected cells. (A) Western blot analysis of nuclear extracts from SW480 cells (lane 1) and cells transfected with constitutively active β-catenin (DP) (lane 2), sequentially probed with anti-PPARγ, anti-β-catenin, and anti-actin antibodies. (B and C) PPARγ immunostaining of SW480 cells in the presence (C) or absence (B) of transfected β-catenin-DP. The cells were stained with a primary monoclonal antibody directed against PPARγ and a secondary anti-mouse antibody conjugated to the fluorescent Cy3. (D) β-catenin-augmented PPARγ protein levels led to an increase in PPARγ-DNA complexes. Shown are EMSA with nuclear extracts from the SW480 cells transfected with 2 μg of either pcDNA3 or the β-catenin-DP expression vector. The extracts were incubated with the indicated antibodies before addition of a radiolabeled PPRE probe. The major PPARγ-containing complex is indicated by the arrow. (E) RNase protection assay showing the mRNA levels of PPARγ in SW480, after transfection with increasing amounts of β-catenin-DP (lane 1, 2 μg; lane 2, 1 μg; lane 3, nontransfected control SW480 cells). The transcript for human γ-actin was used as an internal control. (F) Western blot analysis of nuclear extracts from HEK293 cotransfected with PPARγ and either mock DNA (lane 1) or β-catenin-DP (lane 2). The membrane was sequentially probed with anti-PPARγ, anti-β-catenin, and anti-actin antibodies. (G and H) PPARγ immunostaining of HEK293 cells in the presence (H) or absence (G) of transfected β-catenin-DP.
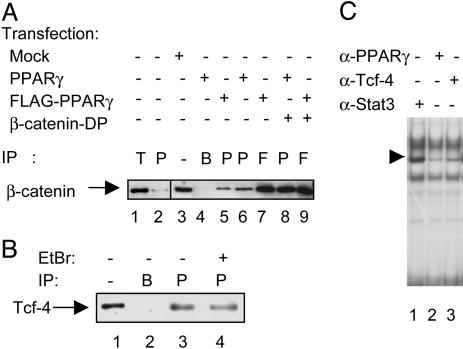
Fig. 3.
Physical interaction of PPARγ with β-catenin and Tcf-4. (A) Western blot analysis of β-catenin immunoprecipitations (IP) from the SW480 cell line. Whole-cell extracts were obtained from nontransfected cells (lanes 1 and 2), mock-transfected cells (lane 3), or cells transfected with PPARγ, FLAG-PPARγ, and/or β-catenin-DP as indicated (lanes 4-9). Lane 1 shows β-catenin coimmunoprecipitated by using an anti-Tcf-4 antibody (denoted T), whereas lane 2 shows β-catenin coimmunoprecipitated using an anti-PPARγ antibody. Lane 3 shows the endogenous β-catenin levels in SW480 cell lysates, whereas lane 4 represents a control IP with no antibody, only beads. In transfected cells, protein complexes containing β-catenin were immunoprecipitated (IP) with either a PPARγ-specific antibody (P, lanes 5, 6, and 8) or an anti-FLAG antibody (F, lanes 7 and 9). (B) Coimmunoprecipitations of endogenous Tcf-4 from the SW480 cell line. Lane 1 shows endogenous Tcf-4 levels in 25 μg of whole-cell extracts from SW480 cells, whereas lane 2 represents a mock-IP with no antibody, but with Sepharose A beads. Tcf-4 precipitated (IP) with a PPARγ-specific antibody (P) is shown in lane 3. Conditions for Tcf-4 IP in lane 4 are essentially as for lane 3, but with the addition of 10 μg/ml ethidium bromide during IP. (C) EMSA with nuclear extracts from the SW480 cells stimulated for 6 h with 20 mM LiCl. The extracts were incubated with different antibodies, as indicated, before addition of a radiolabeled PPRE probe. The major PPARγ-containing complex is indicated by the arrow.
Results
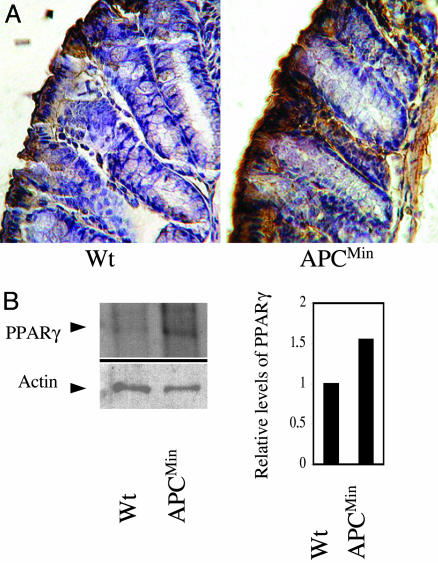
Altered PPARγ Protein Levels in APCMin Mice. Immunohistochemical analyses revealed differential distribution of PPARγ-positive cells in the colonic mucosa of APCMin, compared with wild-type mice (compare Fig. 1A Right and Left). We then determined PPARγ protein levels in colonic epithelial cells isolated from wild-type and APCMin mice. Western blot analysis indicated that levels of PPARγ in colonic epithelial cells isolated from APCMin mice were modestly elevated compared with cells from wild-type mice (Fig. 1B Left). A graphical representation of this difference is shown, using actin levels as a reference (Fig. 1B Right).
Fig. 1.
Levels of PPARγ protein are elevated in the APCMin mouse. (A) Immunohistochemical analysis depicting levels of PPARγ protein in sections from the distal colon of wild-type C57BL/6(Left) and C57BL/6J-APCMin (Right) mice by using a PPARγ-specific antibody. (B) Western blot analysis of protein extracts of colonic epithelial cells from wild-type and APCMin mice, sequentially probed for PPARγ and actin. (Right) A graphical representation of comparative PPARγ levels in relation to actin levels.
β-Catenin Overexpression Elevates PPARγ Protein Levels. Extending our observations made in the APCMin mice, we examined whether increased levels of constitutively active β-catenin could indeed result in elevated levels of PPARγ in human cell lines. To obtain increased levels of β-catenin, we took advantage of a mutant of β-catenin, termed β-catenin-DP, containing a Ser33Ala mutation that reduces phosphorylation by glycogen synthase kinase-3β and subsequent degradation (30). Expression of this protein in the colon carcinoma cell line SW480 resulted in elevated protein levels of endogenous PPARγ (Fig. 2A). These observations were also confirmed with immunocytochemistry. In control nontransfected SW480 cells, PPARγ is mainly in the nuclei, and the intensity of staining differs between the cells. In the presence of transfected β-catenin-DP, some cells demonstrate significantly higher levels of PPARγ staining in the nuclear compartment. The numbers of these intensively stained cells correlate with the transfection efficiency for this particular cell line, suggesting that the rise in intensity of PPARγ staining is a result of exogenous β-catenin-DP expression in these cells (Fig. 2 B and C). In addition, EMSA using a radiolabeled binding site for PPARγ and nuclear extracts from the SW480 cells transfected with β-catenin-DP showed a stronger PPARγ complex bound to DNA in the presence of transfected β-catenin-DP (Fig. 2D), consistent with an increase in total PPARγ protein levels.
Our next question was whether the β-catenin-dependent increase in PPARγ protein levels was mediated at the transcriptional level. Thus, we performed RNase protection assay (RPA) on RNA from SW480 colon cells transfected with either β-catenin or β-catenin-DP. No obvious alteration in levels of PPARγ transcripts was detected (Fig. 2E). Likewise, transfection experiments in HEK293 with a luciferase reporter gene driven by either the PPARγ-1 or γ-2 promoter, cotransfected with β-catenin-DP, showed that these constructs cannot be activated by overexpression of β-catenin (data not shown).
To circumvent the high basal levels of PPARγ expression as well as high levels of nuclear β-catenin in SW480 cells, we performed similar experiments in the HEK293 cell line. We found that protein levels of PPARγ were increased in cells transfected with β-catenin-DP, (Fig. 2F). Immunofluorescent stainings also confirmed this β-catenin-DP-induced increase in PPARγ levels (Fig. 2 G and H) in ≈30% of the cells.
Together, these data show that elevated levels of β-catenin result in increased PPARγ protein levels in two different epithelial cell lines. This increase in PPARγ protein levels seems to be a posttranscriptional event.
PPARγ Is Found in the Same Protein Complex as β-Catenin and Tcf-4. Protein stability modulated through protein-protein interaction would be one way to envisage the posttranscriptional increase in PPARγ protein levels. To examine this possibility, we explored physical interactions between PPARγ and components of the Wnt signaling pathway. We performed coimmunoprecipitation experiments using lysates from SW480 cells varyingly transfected with full-length PPARγ, flag-tagged PPARγ (FLAG-PPARγ), and/or β-catenin-DP constructs. Whole-cell extracts were prepared 24 h after the transfections, and protein complexes were immunoprecipitated with either anti-PPARγ or anti-FLAG antibodies and subsequently immunoblotted with an anti-β-catenin antibody. The results suggest that endogenous β-catenin and PPARγ are indeed found to interact, although weakly, in these cells (Fig. 3A, lane 2). By using extracts from cells transfected with wild-type PPARγ (lane 6) or FLAG-PPARγ (lanes 5 and 7), a significant increase was observed in levels of PPARγ-β-catenin complexes. This association was further enhanced through cotransfection with β-catenin-DP (lanes 8 and 9). The association between endogenous Tcf-4 and β-catenin is shown in lane 1 (Fig. 3A).
To determine whether Tcf-4 also associated with PPARγ, extracts from the SW480 cell line were immunoprecipitated with PPARγ antibodies and probed for Tcf-4. We found that endogenous PPARγ and Tcf-4 can complex in solution (Fig. 3B, lane 3). As a control to exclude any DNA-mediated nonspecific interaction between PPARγ and Tcf-4, we used ethidium bromide to intercalate with DNA. In Fig. 3B, lane 4, the coimmunoprecipitation of the PPARγ-Tcf-4 protein complex was retained even in the presence of ethidium bromide, corroborating that complex formation between the two proteins is DNA-independent. Endogenous Tcf-4 levels in SW480 extracts (lane 1) and a mock immunoprecipitation (lane 2) are also shown. These data correlate with EMSA results, which show that the protein complex binding to a radiolabeled PPRE is diminished after incubation with antibodies against Tcf-4 (Fig. 3C, lane 3). A PPARγ-specific antibody could also inhibit the complex formation (Fig. 3C, lane 2), whereas an unrelated antibody (α-Stat3) did not achieve this (Fig. 3C, lane 1). In conclusion, PPARγ seems to associate with both β-catenin and Tcf-4.
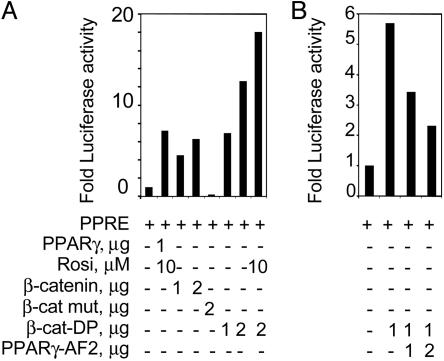
β-Catenin Activates a PPRE Linked to a Heterologous Reporter Gene. We next assessed whether overexpression of wild-type and mutated β-catenin influenced transcriptional activity of endogenous PPARγ. Cotransfections in HEK293 of either β-catenin or β-catenin-DP with a PPRE linked to the luciferase gene were performed. A clear dose-dependent elevation of the PPRE-coupled luciferase reporter gene activity was observed in the presence of transfected β-catenin compared with the PPRE vector alone (Fig. 4A). The synthetic ligand rosiglitazone was used as a positive control. Notably, the degradation-resistant β-catenin-DP enhanced the activation of the PPRE almost 2-fold compared with the wild-type β-catenin. This activation was further increased by ligand stimulation. In addition, the activation of the PPRE reporter gene by β-catenin was repressed by a dominant negative form of PPARγ (31) (Fig. 4B). Together, the data suggest that PPARγ is a central component of this transactivation complex regulated by β-catenin.
Fig. 4.
β-Catenin activates a PPRE-driven reporter gene. PPARγ activity was monitored in HEK293 cells, transiently transfected with a PPRE-containing luciferase reporter, in combination with either PPARγ expression vector or vectors expressing a variety of β-catenin constructs. Cells were harvested 48 h posttransfection and assayed for luciferase activity. Each transient transfection was performed in doublets of three independent experiments. The result depicts one such experiment. Luciferase readings were correlated to total protein content. The values show fold luciferase activation of each transfection relative to the activity of the PPRE reporter gene alone. (A) Cells were cotransfected with the PPRE, in the presence or absence of the PPARγ, β-catenin, a mutated β-catenin, or the β-catenin DP expression vectors, as indicated. Selected transfections were also treated with the PPARγ synthetic ligand rosiglitazone. (B) PPRE cotransfected with β-catenin DP and/or a dominant negative mutant of PPARγ, PPARγ-AF2.
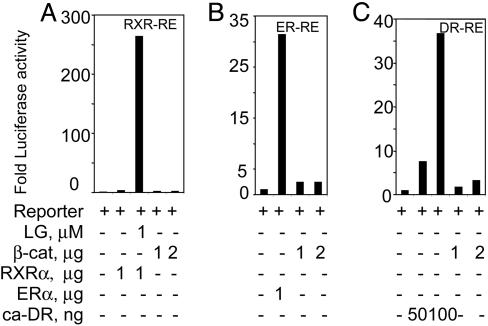
We then investigated whether β-catenin induced transactivation of other nuclear receptors in a way similar to that seen with PPARγ. PPARγ forms a transcriptionally active complex with RXR, to bind to and transactivate expression from the PPRE used here. To exclude activation of this PPRE through regulation of RXR by β-catenin, we examined whether β-catenin affected an RXR-dependent nuclear receptor response element (RXR-RE). We did not observe any significant transactivation of this reporter in the presence of transfected β-catenin, unlike the effect produced by the RXR synthetic ligand (Fig. 5A). Thus, the effect of β-catenin on the transactivation of PPRE is mediated through PPARγ, and not RXR. Notably, the dioxin receptor (DR)- and the estrogen receptor (ER)-driven reporter vectors showed only a modicum of induction by β-catenin, significantly less than that produced either in the presence of the ER (Fig. 5B), or when cotransfected with a constitutively active dioxin receptor (DR, Fig. 5C). In conclusion, it seems that β-catenin may have a preference for activation of PPARγ compared with the other nuclear receptors tested here.
Fig. 5.
β-Catenin-driven transactivation of other nuclear receptor response elements. Transient transfections were performed in HEK293 cells with luciferase reporter plasmids specific for a number of different nuclear receptors. Transfections and subsequent reporter gene assays were performed as described in the legend for Fig. 4. The amounts of expression vectors transfected are indicated. For each response element, the luciferase values reflect the fold luciferase activation relative to the activity of reporter gene alone. (A) RXR-dependent reporter (RXR-RE) was transfected in combination with either the β-catenin or the RXRα expression vectors in the presence or absence of 1 μM LG100268, a synthetic ligand for RXR. (B) ER-dependent reporter (ER-RE) was cotransfected with either the ERα or β-catenin expression plasmids. (C) DR-dependent reporter (DR-RE) was transfected together with either a plasmid expressing β-catenin or one expressing a constitutively active dioxin receptor (ca-DR).
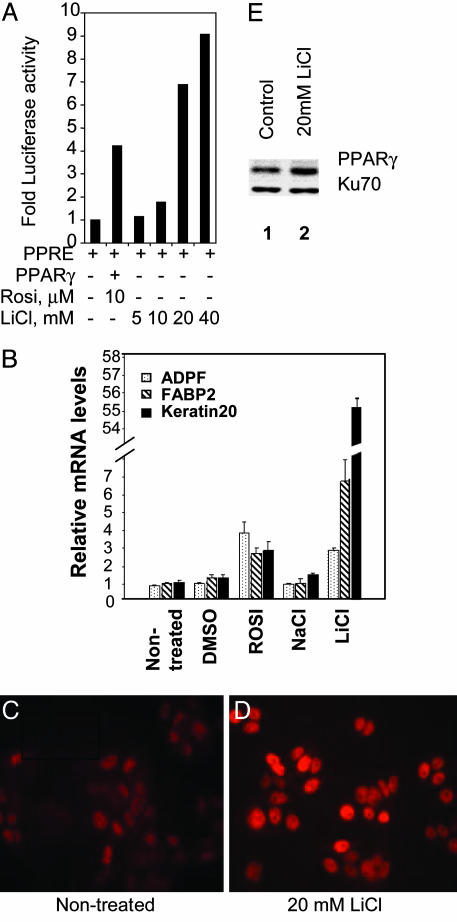
The Wnt Agonist LiCl Regulates PPARγ Activity. LiCl is an established agonist that mimics the Wnt-signaling pathway leading to the activation and stabilization of β-catenin (32). PPRE reporter-transfected HEK293 cells were incubated with different concentrations of LiCl for 6 h and subsequently analyzed for luciferase activity. As shown, increasing concentrations of LiCl activated transcription of the PPRE driven reporter gene (Fig. 6A) in a dose-dependent manner. In addition, the expression of three putative PPARγ target genes (33), FABP2 (fatty-acid binding protein-2), ADF2 (adipophilin), and Keratin 20 were elevated within 24 h after LiCl treatment, although differentially, as did rosiglitazone treatment (Fig. 6B).
Fig. 6.
LiCl treatment modulates protein levels as well as PPARγ transcriptional activity. LiCl, a Wnt-signaling agonist is able to transactivate PPRE in the HEK293 cells. (A) Stimulation of HEK293 cells with either rosiglitazone in the presence of a transfected PPARγ expression vector or with increasing amounts of LiCl in cells transiently transfected with PPRE. Rosiglitazone stimulation was performed for 24 h and LiCl for 6 h at indicated concentrations. (B) Real-time PCR analysis of PPARγ target gene expression in SW480 cells treated for 24 h with DMSO, 10 μM rosiglitazone, 20 mM NaCl, and 20 mM LiCl, or left untreated. RT-PCR was performed, as described in Materials and Methods,to determine relative mRNA levels of ADFP, FABP2, and Keratin 20 in comparison with β-actin expression. Three independent experiments were performed, and data from one representative analysis are depicted here. LiCl treatment of SW480 cells leads to an increase in PPARγ protein levels. Depicted is immunocytochemistry, showing PPARγ protein levels in SW480 cells treated with 20 mM LiCl for 24 h (D) or in untreated cells (C). (E) Western blot analysis of nuclear extracts from SW480 cells, comparing levels of PPARγ in cells untreated and treated with 20 mM of LiCl for 6 h. The blot was subsequently reprobed for Ku70 as a loading control.
Immunocytochemistry (Fig. 6 C and D) and Western blot analyses (Fig. 6E) of the SW480 cell line confirmed that LiCl stimulation led to increased levels of endogenous PPARγ. Thus, LiCl elevates PPARγ protein levels and induces expression of PPARγ target genes.
Discussion
Deregulation of Wnt/β-catenin plays a key role in the development of colorectal cancers (2). An increase in the constitutively active β-catenin/Tcf-4 complex is observed in the nuclei of colon cancer cells (34, 35). Intriguingly, PPARγ levels are increased in polyps from APCMin mice (25). Our observations suggest that β-catenin may play a role in the regulation of PPARγ function. Much controversy, though, has been generated by paradoxical studies that attribute both anticancer and tumor-promoting effects to PPARγ in colon cancers (36, 37). Although the effect of PPARγ on β-catenin has received some attention (38), the outcome of β-catenin/Tcf-4 on PPARγ activity in such cases has not been adequately investigated.
The data presented here show that PPARγ can form protein complexes with β-catenin and Tcf-4. Overexpression of stable β-catenin also results in elevated protein levels and enhanced transcriptional activity of PPARγ. Furthermore, mimicking Wnt signaling through LiCl resulted in the activation of PPARγ reporter and target genes. In our experiments, β-catenin-DP per se does not seem to exert a direct effect on PPARγ mRNA levels. These data, thus, establish a link between the activity of Wnt/β-catenin and PPARγ function in epithelial cells.
The data observed in experiments with the RXR reporter gene RXR-RE suggest that the β-catenin-dependent modulation of the PPARγ-dependent reporter gene PPRE is not mediated through RXR. Interestingly, two other nuclear receptors, DR and ER, can respond to β-catenin, although generating lower transactivation levels. This finding is concomitant with recent reports demonstrating that the androgen receptor (39, 40) and the vitamin D receptor (41) are subject to regulation by β-catenin.
Tcf-4 was found to complex with PPARγ in SW480 colon carcinoma cells, and recent observations of a direct link between Tcf-4 and the androgen receptor (42) support the possibility of a structural requirement for Tcf-4 for the formation of the PPARγ-β-catenin complex. However, a PPARγ-Tcf-4 complex occurring independently of β-catenin, and with divergent biological effects, cannot be formally excluded.
High levels of PPARγ correlate with highly differentiated cells at the top of the colonic crypts (20). In adenomas and carcinomas, both nuclear and cytoplasmic staining of β-catenin is observed, coinciding with the increased growth of the neoplastic cells (43, 44). The increased level of PPARγ protein observed in the colonic mucosa of APCMin mice could be the result of aberrant Wnt/β-catenin activity.
Our data show that PPARγ-β-catenin interaction seems to increase the stability of the PPARγ protein in epithelial cells. Furthermore, our results demonstrate a functional interaction between PPARγ and activated β-catenin in the nucleus. Either expression of a constitutively active Wnt/β-catenin or induction of the Wnt signaling by LiCl can equally transactivate a PPARγ-driven reporter gene and a selection of putative target genes. At present, both means of modulating PPARγ activity seem to depend on increasing PPARγ protein levels. Further studies on PPARγ, including posttranslational modifications and differential target gene expression, by normal and aberrant Wnt/β-catenin activity, are necessary to better understand the role of PPARγ in the colon.
Acknowledgments
This work was supported by Cancerfonden, Sweden; the Foundation for Knowledge and Competence Development, Sweden; and the Swedish Foundation for Strategic Research at the Strategic Research Center for Studies of Integrative Recognition in the Immune System, Karolinska Institute.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: APC, adenomatous polyposis coli; PPAR, peroxisome proliferator-activated receptor; PPRE, PPAR-responsive elements; RXR, retinoic X receptor; Tcf, T cell transcription factor; Min, multiple intestinal neoplasia; DR, dioxin receptor; ER, estrogen receptor.
References
- 1.Theodosiou, N. A. & Tabin, C. J. (2003) Dev. Biol. 259, 258-271. [DOI] [PubMed] [Google Scholar]
- 2.Giles, R. H., van Es, J. H. & Clevers, H. (2003) Biochim. Biophys. Acta 1653, 1-24. [DOI] [PubMed] [Google Scholar]
- 3.Barker, N., Morin, P. J. & Clevers, H. (2000) Adv. Cancer Res. 77, 1-24. [DOI] [PubMed] [Google Scholar]
- 4.He, T. C., Sparks, A. B., Rago, C., Hermeking, H., Zawel, L., da Costa, L. T., Morin, P. J., Vogelstein, B. & Kinzler, K. W. (1998) Science 281, 1509-1512. [DOI] [PubMed] [Google Scholar]
- 5.Shtutman, M., Zhurinsky, J., Simcha, I., Albanese, C., D'Amico, M., Pestell, R. & Ben-Ze'ev, A. (1999) Proc. Natl. Acad. Sci. USA 96, 5522-5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Korinek, V., Barker, N., Moerer, P., van Donselaar, E., Huls, G., Peters, P. J. & Clevers, H. (1998) Nat. Genet. 19, 379-383. [DOI] [PubMed] [Google Scholar]
- 7.van de Wetering, M., Sancho, E., Verweij, C., de Lau, W., Oving, I., Hurlstone, A., van der Horn, K., Batlle, E., Coudreuse, D., Haramis, A. P., et al. (2002) Cell 111, 241-250. [DOI] [PubMed] [Google Scholar]
- 8.Powell, S. M., Zilz, N., Beazer-Barclay, Y., Bryan, T. M., Hamilton, S. R., Thibodeau, S. N., Vogelstein, B. & Kinzler, K. W. (1992) Nature 359, 235-237. [DOI] [PubMed] [Google Scholar]
- 9.Joslyn, G., Carlson, M., Thliveris, A., Albertsen, H., Gelbert, L., Samowitz, W., Groden, J., Stevens, J., Spirio, L., Robertson, M., et al. (1991) Cell 66, 601-613. [DOI] [PubMed] [Google Scholar]
- 10.Groden, J., Thliveris, A., Samowitz, W., Carlson, M., Gelbert, L., Albertsen, H., Joslyn, G., Stevens, J., Spirio, L., Robertson, M., et al. (1991) Cell 66, 589-600. [DOI] [PubMed] [Google Scholar]
- 11.Su, L. K., Kinzler, K. W., Vogelstein, B., Preisinger, A. C., Moser, A. R., Luongo, C., Gould, K. A. & Dove, W. F. (1992) Science 256, 668-670. [DOI] [PubMed] [Google Scholar]
- 12.Moser, A. R., Pitot, H. C. & Dove, W. F. (1990) Science 247, 322-324. [DOI] [PubMed] [Google Scholar]
- 13.Gould, K. A. & Dove, W. F. (1997) Proc. Natl. Acad. Sci. USA 94, 5848-5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lefebvre, M., Paulweber, B., Fajas, L., Woods, J., McCrary, C., Colombel, J. F., Najib, J., Fruchart, J. C., Datz, C., Vidal, H., et al. (1999) J. Endocrinol. 162, 331-340. [DOI] [PubMed] [Google Scholar]
- 15.Debril, M. B., Renaud, J. P., Fajas, L. & Auwerx, J. (2001) J. Mol. Med. 79, 30-47. [DOI] [PubMed] [Google Scholar]
- 16.Mangelsdorf, D. J. & Evans, R. M. (1995) Cell 83, 841-850. [DOI] [PubMed] [Google Scholar]
- 17.Koutnikova, H. & Auwerx, J. (2002) Ann. N.Y. Acad. Sci. 967, 28-33. [DOI] [PubMed] [Google Scholar]
- 18.Lehmann, J. M., Moore, L. B., Smith-Oliver, T. A., Wilkison, W. O., Willson, T. M. & Kliewer, S. A. (1995) J. Biol. Chem. 270, 12953-12956. [DOI] [PubMed] [Google Scholar]
- 19.Tontonoz, P., Hu, E. & Spiegelman, B. M. (1994) Cell 79, 1147-1156. [DOI] [PubMed] [Google Scholar]
- 20.Mansen, A., Guardiola-Diaz, H., Rafter, J., Branting, C. & Gustafsson, J. A. (1996) Biochem. Biophys. Res. Commun. 222, 844-851. [DOI] [PubMed] [Google Scholar]
- 21.Sarraf, P., Mueller, E., Jones, D., King, F. J., DeAngelo, D. J., Partridge, J. B., Holden, S. A., Chen, L. B., Singer, S., Fletcher, C., et al. (1998) Nat. Med. 4, 1046-1052. [DOI] [PubMed] [Google Scholar]
- 22.DuBois, R. N., Gupta, R., Brockman, J., Reddy, B. S., Krakow, S. L. & Lazar, M. A. (1998) Carcinogenesis 19, 49-53. [DOI] [PubMed] [Google Scholar]
- 23.Tanaka, T., Kohno, H., Yoshitani, S., Takashima, S., Okumura, A., Murakami, A. & Hosokawa, M. (2001) Cancer Res. 61, 2424-2428. [PubMed] [Google Scholar]
- 24.Osawa, E., Nakajima, A., Wada, K., Ishimine, S., Fujisawa, N., Kawamori, T., Matsuhashi, N., Kadowaki, T., Ochiai, M., Sekihara, H., et al. (2003) Gastroenterology 124, 361-367. [DOI] [PubMed] [Google Scholar]
- 25.Saez, E., Tontonoz, P., Nelson, M. C., Alvarez, J. G., Ming, U. T., Baird, S. M., Thomazy, V. A. & Evans, R. M. (1998) Nat. Med. 4, 1058-1061. [DOI] [PubMed] [Google Scholar]
- 26.Lefebvre, A. M., Chen, I., Desreumaux, P., Najib, J., Fruchart, J. C., Geboes, K., Briggs, M., Heyman, R. & Auwerx, J. (1998) Nat. Med. 4, 1053-1057. [DOI] [PubMed] [Google Scholar]
- 27.Girnun, G. D., Smith, W. M., Drori, S., Sarraf, P., Mueller, E., Eng, C., Nambiar, P., Rosenberg, D. W., Bronson, R. T., Edelmann, et al. (2002) Proc. Natl. Acad. Sci. USA 99, 13771-13776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dubuquoy, L., Jansson, E. A., Deeb, S., Rakotobe, S., Karoui, M., Colombel, J. F., Auwerx, J., Pettersson, S. & Desreumaux, P. (2003) Gastroenterology 124, 1265-1276. [DOI] [PubMed] [Google Scholar]
- 29.Grant, P. A., Arulampalam, V., Ahrlund-Richter, L. & Pettersson, S. (1992) Nucleic Acids Res. 20, 4401-4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hsu, S. C., Galceran, J. & Grosschedl, R. (1998) Mol. Cell. Biol. 18, 4807-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gurnell, M., Wentworth, J. M., Agostini, M., Adams, M., Collinwood, T. N., Provenzano, C., Browe, P. O., Rajanayagam, O., Burris, T. P., Schwabe, et al. (2000) J. Immunol. 275, 5754-5759. [DOI] [PubMed] [Google Scholar]
- 32.Stambolic, V., Ruel, L. & Woodgett, J. R. (1996) Curr. Biol. 6, 1664-1668. [DOI] [PubMed] [Google Scholar]
- 33.Gupta, R. A., Brockman, J. A., Sarraf, P., Willson, T. M. & DuBois, R. N. (2001) J. Biol. Chem. 276, 29681-29687. [DOI] [PubMed] [Google Scholar]
- 34.Korinek, V., Barker, N., Morin, P. J., van Wichen, D., de Weger, R., Kinzler, K. W., Vogelstein, B. & Clevers, H. (1997) Science 275, 1784-1787. [DOI] [PubMed] [Google Scholar]
- 35.Morin, P. J., Sparks, A. B., Korinek, V., Barker, N., Clevers, H., Vogelstein, B. & Kinzler, K. W. (1997) Science 275, 1787-1790. [DOI] [PubMed] [Google Scholar]
- 36.Seed, B. (1998) Nat. Med. 4, 1004-1005. [DOI] [PubMed] [Google Scholar]
- 37.Peek, R. M., Jr. (2003) Gastroenterology 125, 619-621. [DOI] [PubMed] [Google Scholar]
- 38.Sharma, C., Pradeep, A., Wong, L., Rana, A. & Rana, B. (2004) J. Biol. Chem. 279, 35583-35594. [DOI] [PubMed] [Google Scholar]
- 39.Pawlowski, J. E., Ertel, J. R., Allen, M. P., Xu, M., Butler, C., Wilson, E. M. & Wierman, M. E. (2002) J. Biol. Chem. 277, 20702-20710. [DOI] [PubMed] [Google Scholar]
- 40.Truica, C. I., Byers, S. & Gelmann, E. P. (2000) Cancer Res. 60, 4709-4713. [PubMed] [Google Scholar]
- 41.Palmer, H. G., Gonzalez-Sancho, J. M., Espada, J., Berciano, M. T., Puig, I., Baulida, J., Quintanilla, M., Cano, A., de Herreros, A. G., Lafarga, M. & Munoz, A. (2001) J. Cell Biol. 154, 369-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Amir, A. L., Barua, M., McKnight, N. C., Cheng, S., Yuan, X. & Balk, S. P. (2003) J. Biol. Chem. 278, 30828-30834. [DOI] [PubMed] [Google Scholar]
- 43.Iwamoto, M., Ahnen, D. J., Franklin, W. A. & Maltzman, T. H. (2000) Carcinogenesis 21, 1935-1940. [DOI] [PubMed] [Google Scholar]
- 44.Hao, X. P., Pretlow, T. G., Rao, J. S. & Pretlow, T. P. (2001) Cancer Res. 61, 8085-8088. [PubMed] [Google Scholar]