Abstract
Hybrid zones have yielded considerable insight into many evolutionary processes, including speciation and the maintenance of species boundaries. Presented here are analyses from a hybrid zone that occurs among three salamanders – Plethodon jordani, Plethodon metcalfi and Plethodon teyahalee – from the southern Appalachian Mountains. Using a novel statistical approach for analysis of non-clinal, multispecies hybrid zones, we examined spatial patterns of variation at four markers: single-nucleotide polymorphisms (SNPs) located in the mtDNA ND2 gene and the nuclear DNA ILF3 gene, and the morphological markers of red cheek pigmentation and white flecks. Concordance of the ILF3 marker and both morphological markers across four transects is observed. In three of the four transects, however, the pattern of mtDNA is discordant from all other markers, with a higher representation of P. metcalfi mtDNA in the northern and lower elevation localities than is expected given the ILF3 marker and morphology. To explore whether climate plays a role in the position of the hybrid zone, we created ecological niche models for P. jordani and P. metcalfi. Modelling results suggest that hybrid zone position is not determined by steep gradients in climatic suitability for either species. Instead, the hybrid zone lies in a climatically homogenous region that is broadly suitable for both P. jordani and P. metcalfi. We discuss various selective (natural selection associated with climate) and behavioural processes (sex-biased dispersal, asymmetric reproductive isolation) that might explain the discordance in the extent to which mtDNA and nuclear DNA and colour-pattern traits have moved across this hybrid zone.
Keywords: climate change, differential introgression, ecological niche modelling, hybrid zone, Plethodon, salamander
Introduction
Hybrid zones, which may form after secondary contact between two partially reproductively isolated populations, have long been utilized in studies of speciation and the maintenance of species boundaries (Barton & Hewitt 1985). One important aspect of hybrid zone studies is that different markers, either molecular or morphological, may exhibit different patterns in frequency change across a hybrid zone. These differences may indicate important ecological and evolutionary dynamics in the gene or gene regions under study (Teeter et al. 2008, 2010) or between the interacting species (e.g. Arntzen & Wallis 1991; Brito 2007).
One pattern that may emerge in hybrid zone analyses is that of differential introgression. Numerous studies have documented differential introgression of mtDNA relative to nuclear DNA and morphology (e.g. Funk & Omland 2003; Chan & Levin 2005). Many of these are phylogenetic studies between closely related species that share mtDNA haplotypes, suggesting a pattern of current or historic mtDNA gene flow (e.g. Weisrock & Larson 2006; Linnen & Farrell 2007). Many other cases documenting mtDNA introgression come from studies of naturally occurring hybrid zones showing shifts in clines of mtDNA relative to nuclear DNA and morphology (e.g. Arntzen & Wallis 1991; Sequeira et al. 2005; Vörös et al. 2006; Brito 2007; Hofman & Szymura 2007; Leaché & Cole 2007; Kawakami et al. 2008).
A pattern of differential introgression may indicate hybrid zone movement. Recent empirical work suggests that hybrid zone movement may be more common than once thought (Buggs 2007). This has led to inferences on the ecological and evolutionary dynamics among participating species (e.g. Arntzen & Wallis 1991; Hairston et al. 1992; Rohwer et al. 2001; García-París et al. 2003). In a recent review, Buggs (2007) identified 23 studies which documented hybrid zone movement and another 16 studies which had patterns consistent with movement. These studies utilize two different approaches: first, long-term monitoring of molecular and morphological markers across a hybrid zone is a reliable method for detecting movement. Second, analysing differential patterns of introgression across a suite of markers at a single point in time may allow inference of movement. One cause of hybrid zone movement is range expansion or contraction as a result of climate change. Although evidence for this phenomenon is limited (but see Britch et al. 2001 and Walls 2009), there is widespread evidence that species’ ranges have shifted in response to Pleistocene cooling and warming (e.g. Davis & Shaw 2001; Peterson et al. 2004; Brito 2007). Furthermore, shifts in hybrid zone position resulting from climate change are expected to be greatest for montane species, as geographically proximate locations may experience considerable climatic differences (Hewitt 1996; Guralnick 2007; but see Peterson 2003).
Alternatively, a pattern of differential introgression may indicate differential selection on the markers under study or regions that are linked to those markers (Barton 1979). For example, such a pattern might be interpreted as selection acting differentially on different mtDNA alleles owing to the important metabolic functions of the mitochondrion (Boutilier 2001; Nouette-Gaulain et al. 2005, Lynn et al. 2007; Tattersall & Ultsch 2008) or to nuclear DNA-encoded phenotypic traits exhibited by one, but not the other, parental species. Lastly, noncoincident, biparentally inherited nuclear and maternally inherited mtDNA clines may be because of mating or dispersal asymmetries (Dakin 2006).
In this study, we present an analysis of a naturally occurring hybrid zone among three species of salamanders in the genus Plethodon. The hybrid zone occurs at the boundary between the Great Smoky and Balsam Mountains in the southern Appalachians of North Carolina and Tennessee. The high elevation species P. jordani and P. metcalfi are known to hybridize along two ridgelines, Balsam Mountain and Hyatt Ridge (Hairston 1950; Highton 1970; Hairston et al. 1992). These ridge-lines are high elevation corridors that connect the Great Smoky to the Balsam Mountains, encompassing the range of P. jordani and a portion of the range of P. metcalfi, respectively. A third species, P. teyahalee, inhabits lower elevations throughout much of the southern Appalachians and hybridizes with the former species at intermediate elevations (Peabody 1978; Manzo 1988; Reagan 1992). The most complete study of this system was conducted by Hairston et al. (1992). These authors sampled salamanders from the same five localities along the southern portion of Balsam Mountain each year for an 18-year period and recorded the amount of red cheek pigmentation, which is present in P. jordani but absent in P. metcalfi. That study documented extensive hybridization along the ridgeline, and because no movement was detected, yielded some insight into the short-term stability of the hybrid zone.
The analyses presented here expand on this study in four ways: (i) patterns of spatial variation are examined for mtDNA, nuclear DNA and morphological markers; (ii) localities with hybrids among P. jordani, P. metcalfi and P. teyahalee are analysed across four transects, two along high elevation ridgelines predominately connecting the ranges of P. jordani and P. metcalfi, and two elevational transects between hybrid localities of the former species with that of P. teyahalee; (iii) sampling in this study was performed at a spatially fine scale, allowing for increased resolution in the detection of introgression; and (iv) ecological niche models are created for P. jordani and P. metcalfi to explore whether ecological factors play a role in determining the position of this hybrid zone. It is only recently that the GIS-based method of ecological niche modelling (ENM) has been used in hybrid zone studies (Cicero 2004; Swenson 2006, 2008; Martínez-Freiría et al. 2008; Swenson et al. 2008), although earlier studies have incorporated habitat-genotype associations (Bridle et al. 2001). Moreover, as the width of this hybrid zone is narrow, the fine scale at which ENM is utilized in this study represents a novel application to the study of hybrid zones and highlights the utility of ENM, in concert with genetic and morphological data, for understanding hybrid zone dynamics.
Methods
Study species
Salamanders in the genus Plethodon (family Plethodontidae) comprise a monophyletic group that is distributed throughout the eastern and western United States (Petranka 1998). Although under some debate, the estimated number of species is around 55 (Collins & Taggart 2009). All members of this group are fully terrestrial and undergo direct development; therefore, population density is generally diffuse and uniform throughout the environment. Plethodon jordani, P. metcalfi, and P. teyahalee are primarily forest inhabitants, and all three species (especially P. jordani and P. metcalfi) occur at high densities (Highton 1970; Merchant 1972; Hairston 1980a,b).
Sampling
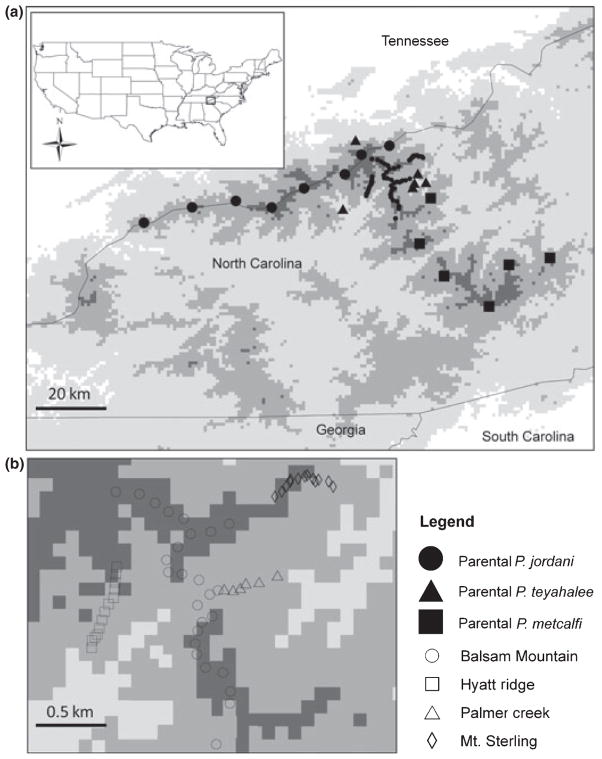
Salamanders were sampled along transects from discrete localities with each locality having a radius of <50 m. Two high elevation transects were established along Balsam Mountain and Hyatt Ridge (Fig. 1 and Supporting information Table S1). Salamanders were collected from 24 localities along the 24-km-long Balsam Mountain, with a minimum of five salamanders sampled at each locality; and 13 localities along 6 km of the southernmost portion of Hyatt Ridge, with a minimum of 10 samples per locality. Two elevational transects were also created that connect high and low elevation populations (Fig. 1 and Supporting information Table S1). Salamanders were collected from eight localities, with a minimum of five salamanders per locality, from the Palmer Creek transect beginning at 1390 m in elevation and extending 8 km to 951 m. The Mt Sterling transect follows Mt Sterling Ridge for 1.5 km to Mt Sterling summit (at 1768 m elevation) before descending for 2 km to 1134 m. Salamanders were collected from 13 localities, with a minimum of five salamanders sampled at each locality.
Fig. 1.
(a) Map of study area showing collection localities of parental (Plethodon jordani, Plethodon metcalfi, and Plethodon teyahalee) and hybrid samples. Inset depicts location of study within the continental United States. (b) Expanded map of hybrid zones from A showing collection localities of hybrid samples. In both a and b, darkened areas = high elevation and light areas = low elevation.
Parental animals from each of the three species were also sampled at locations distant from known areas of hybridization (Fig. 1 and Supporting information Table S2). These included eight localities in the Great Smoky Mountains (the range of P. jordani), six from the Balsam Mountains (within the range of P. metcalfi) and six from low elevations in the Great Smoky Mountains (within the range of P. teyahalee). It should be noted that Highton & Peabody (2000) and Weisrock & Larson (2006) uncovered two genetically divergent lineages of P. metcalfi corresponding to the Balsam and Blue Ridge Mountains. As only populations from the Balsam Mountains are known to hybridize with P. jordani, only these populations of P. metcalfi were sampled.
Tissue collection and DNA extraction
Samples were collected in the field from 2004 to 2007 during the months of May–July. Salamanders were captured by hand and 10–20 mm of the tail tip removed for genetic analysis. In the field, vials containing tissue samples were immediately placed in an ice-salt mixture (approximately −20 °C) until they could be transferred to liquid nitrogen 3–48 h later. At the end of each field season, samples were removed from the liquid nitrogen and stored at −80 °C. All samples have been catalogued into the tissue collection of the Division of Reptiles and Amphibians, Museum of Zoology at the University of Michigan (under accession number 2008–09 no. 3, and uniquely identified by field number; Supporting information Tables S3 and S4).
DNA was extracted from tail tissue using a standard phenol-chloroform protocol (Museum of Vertebrate Zoology, University of California, Berkeley, CA, USA). Briefly, 0.5–20 mg of frozen tissue was washed in 1 mL of cold STE buffer and then incubated in a mixture of lysis buffer, proteinase K and RNase A. Samples were centrifuged and pellets discarded. The resultant supernatant was subjected to three rounds of purification with a phenol-chloroform mixture and centrifugation. DNA was precipitated in approximately 900 mL of cold 95% ethanol, centrifuged and the supernatant discarded. The resultant pellet was washed twice with 70% ethanol, allowed to dry and resuspended in 100 μL of TE buffer.
Morphological markers
Animals were scored in the field for two morphological markers. The first, red cheek pigmentation, is present in all the animals captured within the range of P. jordani and absent in parental populations of P. metcalfi and P. teyahalee. Animals were scored on a 14-point scale, with 0 indicating the complete absence of red cheek pigmentation and 13 indicating bright and pervasive red cheek pigmentation, extending onto the throat, shoulders and forelimbs. Assigned scores accounted for both extent and intensity of red pigmentation, as well as both the right and left cheeks. The second marker, white flecking, is present in pure populations of P. teyahalee, and is absent in populations of P. jordani and P. metcalfi. The amount and pattern of white flecking is variable in P. teyahalee, but lateral flecks are generally abundant and large, while dorsal flecks are sparse and small. White flecks were scored as being either present or absent. To achieve consistency in scoring the morphological markers, all animals were scored by MWHC. Digital photographs of the right and left sides of the head were taken of each animal. These images have been catalogued into the digital image collection of the Division of Reptiles and Amphibians, Museum of Zoology at the University of Michigan (under accession number 2008–09 no. 3, image numbers 49–960, and uniquely identified by field number; Supporting information Tables S3 and S4).
mtDNA marker
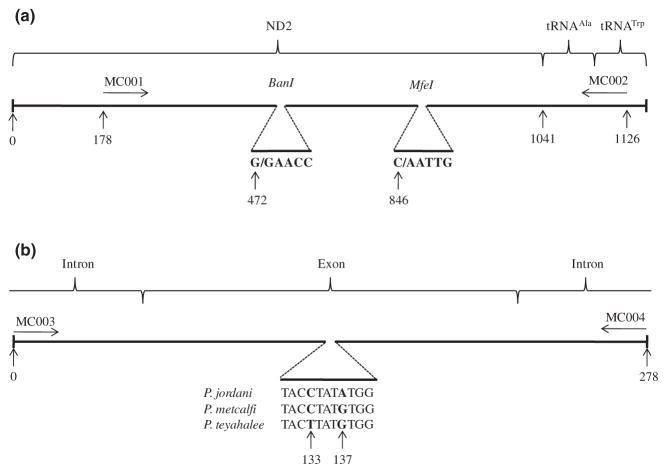
A series of two single-nucleotide polymorphisms (SNPs) were identified in the mtDNA gene NADH subunit II (ND2): the first distinguishes P. jordani from P. metcalfi and P. teyahalee, and the second distinguishes P. teyahalee from P. jordani and P. metcalfi. Thus, when used in tandem, the SNPs were diagnostic for each species. The panel used to identify these single-nucleotide differences consisted of the following pure parental samples: 18 animals from eight populations within the range of P. jordani, 16 animals from five populations within the range of P. metcalfi and 13 animals from six populations within the range of P. teyahalee (Fig. 1 and Supporting information Table S3). Approximately 950 base pairs of the ND2 gene, the entire tRNAAla gene and a portion of the tRNATrp gene were amplified with the forward primer MC001 (5′-TTTCTAACCCAATCTATAGCATCC-3′) and the reverse primer MC002 (5′-GTCTTGCAAGTTC-GAGTCAGA-3′), designed using the online software Primer3 (Fig. 2a). Representative ND2 sequence data have been deposited in GenBank under accession numbers HM775317–HM775319. Polymerase chain reaction (PCR) protocols followed Weisrock et al. (2001), with the inclusion of a 5-min hot start at 95 °C and a 7-min final extension at 72 °C. Sequencing was performed on an ABI 3730 XL automated DNA sequencer through the University of Michigan DNA Sequencing Core facility. Resulting sequences were aligned using Sequencher 4.8.
Fig. 2.
(a) Map of the mtDNA ND2 gene with adjacent tRNA genes and relative positions of forward (MC001) and reverse (MC002) primers. Expanded sections of the gene show the six-base-pair restriction enzyme (BanI and MfeI) recognition sites and cut sites (indicated by ‘ / ’) that, when used in tandem, are diagnostic for Plethodon jordani, Plethodon metcalfi, and Plethodon teyahalee. (b) Partial map of nuclear ILF3 gene showing relative positions of the middle exon and two introns and the forward (MC003) and reverse (MC004) primers. Expanded section depicts diagnostic SNPs (in bold). Base pairs are indicated below arrows in both A and B. Maps are not drawn to scale.
Scoring samples at the mtDNA locus was carried out using restriction fragment length polymorphism (RFLP) digests. The PCR product for each sample was divided into two equal aliquots and digested with BanI and MfeI restriction enzymes. When used in tandem, RFLP digestions were unambiguous when scoring the ND2 gene. Digestion with BanI cut the PCR product of P. jordani into fragments with approximate lengths of 300 and 650 base pairs, while leaving the products of P. metcalfi and P. teyahalee whole. Similarly, digestion with MfeI cut the PCR product of P. teyahalee into fragments with approximate lengths of 280 and 670 base pairs, while leaving the products of P. jordani and P. metcalfi whole (Fig. 2a). On rare occasions, P. jordani samples (i.e. those cut with BanI) shared the P. teyahalee allele and were cut with MfeI; however, because P. teyahalee was never cut with BanI, resulting fragments remained diagnostic. Banding patterns from the RFLP digestions were visualized and scored on 2% NuSieve gels.
Nuclear DNA markers
As with the mtDNA marker, a panel was developed to identify diagnostic nuclear SNPs. The panel consisted of 12 animals from six localities in the range of P. jordani, 11 animals from five localities in the range of P. metcalfi and eight animals from five localities in the range of P. teyahalee (Fig. 1 and Supporting information Table S3). Two SNPs (separated by four base pairs) were identified in the nuclear gene interleukin enhancer binding factor 3 (ILF3) that, when used in tandem, could distinguish the three parental species. Samples were scored at the nuclear gene markers by sequencing each PCR product and scoring the sequences by eye. Approximately 280 base pairs of the middle exon (and partial sequences of the surrounding introns) of ILF3 were amplified with the forward primer MC003 (5′-CCAGGCATTTATGCATCCTT-3′) and the reverse primer MC004 (5′-CGTGCTAGCCTCGGTAACAT-3′), designed using Oligo 6.71 (Fig. 2b). PCR was performed using a hot start of 94 °C for 3 min, 20 cycles at 94°C for 30 s, 65 °C minus 0.5 °C / cycle for 30 s and 72°C for 1 min, followed by 20 cycles at 94 °C for 30 s, 55 °C for 30 s and 72 °C for 1 min, with a final extension at 72 °C for 8 min (E. Jockusch pers. comm.). Sequencing and alignments were performed as described earlier.
Hybrid zone analysis
To address questions about concordance or discordance between markers, we could not use cline-fitting methods typically applied to two-species hybrid zones (e.g. Barton & Baird 1998) or traditional cytonuclear disequilibrium analyses for the two-species case (Asmussen et al. 1987). Instead, two different types of analyses were performed to explore differential patterns of introgression. The first consisted of two chi-square contingency table tests on the entire data set that were used specifically to test for differential introgression of mtDNA relative to the ILF3 marker. The first test was performed on individual samples and tested the null hypothesis of no difference in relative abundance of P. jordani, P. metcalfi and P. teyahalee genotypes between the two markers. The second test compared sample localities that were classified by their most common genotype at the nuclear and mtDNA markers and tested the null hypothesis that the number of sites dominated by each species’ genotype was the same for the two markers. In both tests, significance was assigned at the P ≤ 0.05 level.
Second, we used generalized log-linear models (GLMs) to fit specific genetic models to the genotypic contingency table for the entire data set (Table 1). Genotypes were grouped according to whether they were (i) ‘parental’ (homozygous at the nuclear marker and had homospecific mtDNA), (ii) ‘homozygous hybrid’ (homozygous at the nuclear marker and had heterospecific mtDNA), (iii) ‘2-way heterozygote’ (heterozygous at the nuclear marker and had mtDNA from one of the nuclear parents) or (iv) ‘3-way heterozygote’ (heterozygous at the nuclear marker and had mtDNA from a third parental species). Although these designations apply only to the two genetic markers assayed (‘parentals’ might be genetically mixed at other parts of the genome), they allow us to specify a symmetrical model in which the genotypic contingency table is predicted only by the marginal genotype frequencies and these four hybrid categories. In addition to this symmetrical model and the null model (in which each possible 2-locus genotype is predicted only by the frequencies of the single-locus genotypes), we tested nine asymmetrical models constructed by including interaction terms between single-locus genotypes and the generic hybrid categories. These asymmetrical models incorporate variation within hybrid categories according to the specific ancestry of genotypes. These analyses test for patterns of association (linkage disequilibrium) between mtDNA and ILF3 genotypes that might be caused by spatial structure, mating behaviour and / or selection. Given the spatial structure of sampling, we expected to reject the statistical null model therefore what is most interesting is the biological interpretation of symmetrical versus asymmetrical models. Models were fitted using Poisson GLMs in the stats package of R 2.6.2 (http://www.r-project.org).
Table 1.
Contingency table depicting genotype groupings1 used in the generalized log-linear models
| mtDNA Genotypes2 | Nuclear Genotypes3 | |||||
|---|---|---|---|---|---|---|
|
| ||||||
| J / J | M / M | T / T | J / M | J / T | M / T | |
| J | P | HoH | HoH | He2 | He2 | He3 |
| M | HoH | P | HoH | He2 | He3 | He2 |
| T | HoH | HoH | P | He3 | He2 | He2 |
Genotype groupings of parental (P), homozygous hybrids (HoH), 2-way heterozygotes (He2), and 3-way heterozygotes (He3) are as described in the text.
mtDNA genotypes are as follows: J = P. jordani, M = P. metcalfi, and T = P. teyahalee.
Each allele in the nuclear genotype is given for homozygotes (J / J, M / M, and T / T) and heterozygotes (J / M, J / T, and M / T).
Two methods were used to assess the suitability of each model. The first is the Akaike information criterion (AIC). AIC can be used to decide on the best of a given set of models, or as a multimodel inference tool that assesses the suitability of the full set of models given the data (Burnham & Anderson 2004). Models are ranked according to their AIC values, with lower values indicating higher suitability. Often, as is done here, AIC values are given as ΔAIC, which is the difference of each AIC value from that of the best model. The second method uses the residual deviance as a measure of the goodness-of-fit of a model relative to a saturated model, which acts as a baseline (Agresti 2007). Higher values indicate more variation is unaccounted for by the model and, therefore, lower values indicate models with a better fit. Significance levels are determined by comparison of each model to the saturated model. Therefore, an insignificant residual deviance (P > 0.05) means the saturated model does not fit significantly better than the tested model, which we would infer is an adequate description of the data because the residual variance is adequately explained as sampling error.
Ecological niche modelling
Hybrid zones that are maintained by a balance between dispersal of hybrids and selection against those hybrids are termed tension zones and may move so as to coincide with geographic barriers, population density troughs (i.e. regions of poor quality habitat for one or both parental lineages) or ecotonal regions (Barton & Hewitt 1985). As such, environmental factors (e.g. climate) might play an important role in maintaining the spatial locations of hybrid zones. For example, many hybrid zones are associated with ecotones where lineages that are adapted to different climatic conditions come into geographic contact and interbreed (e.g. Cicero 2004; Swenson 2006). Alternatively, hybrid zones are also common in environmentally homogeneous regions where previously isolated lineages that share similar climatic requirements come into geographic contact. The latter type of hybrid zone may be maintained primarily by dispersal of parental forms and endogenous selection against hybrids (Barton & Hewitt 1985), or they may correspond to locations where previously isolated lineages are in the process of merging (e.g. Pereira & Wake 2009).
To explore whether climatic factors might influence the structure of the hybrid zone, we used ecological niche modelling (ENM) to predict the geographic distribution of climatically suitable habitats for P. jordani and P. metcalfi, and the extent to which such locations geographically overlap. We used Maxent version 3.2 to model the potential geographic distributions of P. jordani and P. metcalfi. Briefly, Maxent predicts the expected distribution of a species using data on the environmental conditions where it is known to occur and randomly selected background locations in the study area. Maxent is a general approach for characterizing probability distributions from incomplete information and computes a probability distribution that describes the relative suitability of each grid cell as a function of the environmental variables at all the known occurrence locations (Phillips et al. 2006). When the model is projected into geographic space, it produces a map of the species’ potential geographic distribution.
To construct the models, we used 19 temperature and precipitation variables from the WordClim data set with 30-second spatial resolution (Hijmans et al. 2005a) and georeferenced occurrence locations for P. jordani (n = 374) and P. metcalfi (n = 289) obtained from the U.S. National Museum of Natural History. Models for P. teyahalee are not included as the model resolution is not fine enough to permit meaningful interpretation in the narrow area of hybridization between this species and P. jordani and P. metcalfi. Most of the records used in the analysis were collected by R. Highton and assigned to species using morphology and allozymes. Locality records occurring within the same map pixel were removed to avoid pseudoreplication. To calibrate the model, we used quadratic features, and default parameters for the number of background pixels, regularization, the convergence threshold and the maximum number of iterations (following Phillips et al. 2006). We randomly selected 75% of the occurrence locations to construct the model; the remaining 25% were set aside to test the model. We calculated the area under the receiving operator characteristic (AUC) to test whether the model could discriminate between the test localities and 10 000 localities randomly selected from across the study region (defined as the United States east of the Mississippi River).
Maxent assigns a continuous suitability score to each grid cell in the study area (referred to as the cumulative probability). Thus, to map locations where suitable habitats for both species come into geographic contact or overlap, a threshold value for presence–absence must be employed. For each species, we recorded the cumulative probability associated with each georeferenced occurrence location. We then classified as climatically unsuitable, any grid cell falling in the lower 5th percentile of this empirical distribution of suitability scores. Finally, given this threshold for suitability, we used the grid overlay function in DIVA GIS 5.2 (Hijmans et al. 2005b) to map the locations of habitats that were suitable for both P. jordani and P. metcalfi. Our threshold adequately captures patterns of climatic suitability as it results in very few presence locations (5 of 663) incorrectly being classified as occurring in unsuitable habitats.
As predicted by tension zone theory, an association might exist between allele frequency and climate as a result of the hybrid zone settling at a geographic barrier, population density trough or ecotonal area (Barton & Hewitt 1985). If selection associated with climatic factors influences the position of the hybrid zone, one might expect an association to exist between climatic suitability and gene frequencies. For example, if the nuclear DNA of P. jordani confers greater fitness to the climatic conditions in the hybrid zone, then one would expect the P. jordani ILF allele to be present in greater frequency at sites that have higher suitability for P. jordani than for P. metcalfi. To explore this possibility, we employed a hybrid index to score the proportion of P. jordani and P. metcalfi alleles at each site. Our index ranged from 1 (only P. jordani alleles present) to −1 (only P. mecalfi alleles present). Positive scores indicated a greater frequency of the P. jordani allele; negative scores indicated a greater frequency of the P. metcalfi allele. Sites in which P. jordani and P. metcalfi alleles were present in equal frequencies received a score of 0. Similarly, we scored whether each site was more climatically suitable for P. jordani or P. metcalfi by calculating the difference in climatic suitability (i.e. cumulative probabilities from Maxent) between the two species. This climatic suitability index ranged from 100 to −100, with the former score indicating that a site has the maximum suitability for P. jordani and is completely unsuitable for P. metcalfi, and the latter score indicating that a site has the maximum suitability for P. metcalfi and is completely unsuitable for P. jordani. Similar to the allele frequency index, scores of zero indicated that a site is equally suitable for both species. Positive scores indicated that a site was more suitable P. jordani; negative scores that a site was more suitable for P. metcalfi. We then used Spearman’s rank correlation to test for a significant association between the allele frequency- and habitat-suitability scores, and the habitat-suitability scores and cheek colouration.
We caution that distribution-based niche modelling makes the implicit assumption that biotic factors (i.e. competition) do not prevent species from occupying the full extent of climatic conditions in which they can survive and successfully reproduce. If competitive interactions strongly influence a species distribution, then the geographic extent of climatically suitable habitats could be drastically underestimated (Pearson & Dawson 2003; Kozak et al. 2008). However, given that the niche models of both species predict large areas of suitable habitat beyond their empirical range limits (and in the hybrid zone), it does not appear that biotic interactions have strongly biased our results in such a way (see Results).
Finally, some ENM algorithms have been criticized for overfitting climatic variables to species’ presence records and climatic variables (Peterson et al. 2007). Given that models of P. jordani and P. metcalfi (and many other species of Plethodon, see Kozak & Wiens 2006) predict large areas of highly suitable habitat outside of their empirical distributions, overfitting does not appear to have strongly influenced the predicted distributions of these species. Nevertheless, if the climatic niche breadth of either species has been underestimated either because of biotic interactions or because of over-fitting, then the overlapping zone of suitable habitats for P. jordani and P. metcalfi in the hybrid zone would in reality be even wider. Thus, our conclusion that location and dynamics of the hybrid zone are not associated with steep gradients in climatic suitability for either species (see Results) is robust to these potential shortcomings of ENM algorithms.
Results
General patterns
Every salamander captured within the range of pure Plethodon jordani had at least some red cheek pigmentation (mean = 8.02, range = 1–13, n = 90), while red pigmentation was entirely absent in pure P. metcalfi localities (n = 55) and P. teyahalee (n = 21). Similarly, the presence of white flecks was found to be diagnostic for pure P. teyahalee (n = 11) and was completely absent in pure P. jordani and P. metcalfi localities (Supporting information Table S3). Within the hybrid zone, the number of animals collected per transect is as follows: Balsam Mountain, 5–16 animals per locality, 264 animals total; Hyatt Ridge, 10–15 per locality, 155 total; Palmer Creek, 5–16, 86 total; and Mt Sterling, 5–13 per locality, 140 total (Supporting information Table S4). Salamanders captured within the hybrid zone show a wide range of cheek pigmentation scores (range = 0–13, n = 645), and 14 individuals showed the presence of both red cheek pigmentation and white flecks.
Hybrid zone analysis
Most combinations of genotypic classes are represented in the hybrid zone (Table 2). Curiously, no P. metcalfi / P. teyahalee heterozygotes were found at the nuclear marker. Similarly, no individuals were found that were homozygous for P. teyahalee at the nuclear marker while having P. jordani mtDNA. Other genotypic classes are also uncommon. For example, only a single individual was sampled that was heterozygous for P. jordani and P. metcalfi at the nuclear marker but had P. teyahalee mtDNA. Similarly, only two individuals were sampled that were homozygous for the P. metcalfi nuclear allele but had P. teyahalee mtDNA. Most other hybrid genotype combinations are moderately well represented. For example, P. jordani / P. metcalfi heterozygotes at the nuclear marker with P. jordani mtDNA were found in 28 individuals; P. jordani / P. teyahalee heterozygotes at the nuclear marker with P. metcalfi mtDNA were found in 15 individuals; and P. jordani / P. teyahalee heterozygotes with P. jordani mtDNA were found in 8 individuals. Results from the contingency table test on individuals show that P. metcalfi mtDNA is most common, while the P. jordani ILF3 genotype is most common (χ2 = 193.1835, d.f. = 2, P < 0.0001; Table 3). Similarly, results from the contingency table test on localities show that P. metcalfi mtDNA is most common and the P. jordani ILF3 genotype is most common at the majority of localities (χ2 = 9.0195, d.f. = 2, P = 0.0110; Table 4). This suggests P. metcalfi mtDNA is more widespread than the P. metcalfi ILF3 genotype, and the P. jordani ILF3 genotype is more widespread than P. jordani mtDNA.
Table 2.
Summary of salamander samples from the hybrid zone that are classified by their nuclear and mtDNA genotypes. The number of animals with at least some red on their cheeks and with at least some white flecks is given in parentheses, respectively
| Nuclear DNA | mtDNA | ||
|---|---|---|---|
|
| |||
| P. jordani | P. metcalfi | P. teyahalee | |
| P. jordani / P. jordani | 180 (177.1) | 168 (142.2) | 15 (10.6) |
| P. metcalfi / P. metcalfi | 4 (3.0) | 96 (12.3) | 2 (0.2) |
| P. teyahalee / P. teyahalee | 0 (0.0) | 5 (3.1) | 47 (2.42) |
| P. jordani / P. metcalfi | 28 (27.0) | 67 (25.3) | 1 (0.0) |
| P. jordan / P. teyahalee | 8 (8.1) | 15 (10.2) | 9 (5.5) |
| P. metcalfi / P. teyahalee | 0 (0.0) | 0 (0.0) | 0 (0.0) |
Table 3.
Contingency table showing individual sample data used to test for differential patterns of introgression
| Species | mtDNA | Nuclear DNA | Total | ||
|---|---|---|---|---|---|
|
|
|
||||
| Obs. | Exp. | Obs. | Exp. | ||
| Plethodon jordani | 227 | 365 | 868 | 730 | 1095 |
| P. metcalfi | 369 | 235 | 336 | 470 | 705 |
| P. teyahalee | 82 | 78 | 152 | 156 | 234 |
| Total | 678 | 678 | 1356 | 1356 | 2034 |
Table 4.
Contingency table showing sample localities classified by their most common genotype
| Species | mtDNA | Nuclear DNA | Total | ||
|---|---|---|---|---|---|
|
|
|
||||
| Obs. | Exp. | Obs. | Exp. | ||
| Plethodon jordani | 27 | 36 | 45 | 36 | 72 |
| P. metcalfi | 35 | 27.5 | 20 | 27.5 | 55 |
| P. teyahalee | 12 | 10.5 | 9 | 10.5 | 21 |
| Total | 74 | 74 | 74 | 74 | 148 |
The general linear models depict general patterns of association that might be functions of the population spatial structure, nonrandom mating, natural selection, or any combination thereof. Based on resulting ΔAIC values and residual deviances, the models may be categorized into three groups (Table 5). The first group contains those models with large ΔAIC values and significant residual deviances, meaning these models are not suitable given the data. This group includes the null model, the symmetrical model and the following asymmetrical models: P. metcalfi / P. teyahalee heterozygotes, P. metcalfi homozygotes, P. jordani / P. metcalfi heterozygotes and P. jordani / P. teyahalee heterozygotes (all are ILF3 genotypes). The second group contains those models with moderate ΔAIC values and smaller, although still significant, residual deviances. This group includes P. jordani homozygotes (ILF3 genotype), and the models varying P. jordani and P. metcalfi mtDNA. One interesting trend observed in this group is the overrepresentation of P. jordani homozygotes at the ILF3 marker, which suggests P. jordani hybrids are more likely to backcross with P. jordani than with P. metcalfi. Another trend is the atypical patterns of P. jordani and P. metcalfi mtDNA, which reflects an overrepresentation of P. metcalfi mtDNA and a subsequent underrepresentation of P. jordani mtDNA. The third group contains two models that have low ΔAIC values and insignificant residual deviances. These models – P. teyahalee homozygotes (at the ILF3 marker) and P. teyahalee mtDNA – are the only two models not rejected by the goodness-of-fit test. Overall, these results demonstrate associations among genotypes (rejection of the null model), with a disproportionate underrepresentation of P. teyahalee genotypes in hybrids (for the symmetrical model, the residual deviance for ‘parental’ P. teyahalee genotypes is positive, and the residual deviances of all hybrid genotypes with P. teyahalee mtDNA or ILF3 alleles are negative). This finding is consistent with observations that P. teyahalee is the most ecologically (based on elevation differences in species ranges) and morphologically (Highton & Peabody 2000) divergent of the three species.
Table 5.
Results of general linear models showing the Akaike information criterion values (given as ΔAIC), residual deviances (and associated degrees of freedom) and significance levels
| Model | Residual | |||
|---|---|---|---|---|
|
| ||||
| ΔAIC1 | Deviance2 | d.f.3 | P-value4 | |
| Null | 387.3449 | 405.4704 | 10 | 6.4467 × 10−81 |
| Symmetrical | 55.6383 | 69.7638 | 8 | 5.4763 × 10−12 |
| Asymmetrical5 | ||||
| P. metcalfi / P. teyahalee | – | – | – | – |
| P. metcalfi / P. metcalfi | 57.0183 | 69.1439 | 7 | 2.2001 × 10−12 |
| P. jordani / P. metcalfi | 54.0597 | 66.1852 | 7 | 8.6866 × 10−12 |
| P. jordani / P. teyahalee | 54.0597 | 66.1852 | 7 | 8.6866 × 10−12 |
| P. jordani | 15.8975 | 24.0230 | 5 | 2.1491 × 10−4 |
| P. metcalfi | 13.2398 | 21.3654 | 5 | 6.9090 × 10−4 |
| P. jordani / P. jordani | 12.8195 | 24.9450 | 7 | 7.7603 × 10−4 |
| P. teyahalee / P. teyahalee | 1.8222 | 13.9477 | 7 | 0.052120 |
| P. teyahalee | 0.0000 | 8.1255 | 5 | 0.14950 |
Akaike information criterion values given as the difference from the best model.
Residual deviance is a measure of the goodness-of-fit of a model to the data. Higher values indicate that more variation is unaccounted for by the model and, therefore, lower values indicate models with a better fit.
Degrees of freedom of the residual deviance.
Insignificant residual deviance (P > 0.05) means the residual variance is adequately explained as sampling error and, therefore, the model is an adequate description of the data. Insignificant residual P-values are given in bold.
Results for all nine possible asymmetrical models are given (except for P. metcalfi / P. teyahalee heterozygotes because none were found at the nuclear marker). Each asymmetrical model was constructed by adding interaction terms involving the listed marker genotype and all relevant genotypic categories. Genotypes with ‘ / ’ are diploid ILF3 genotypes and all others are mtDNA genotypes.
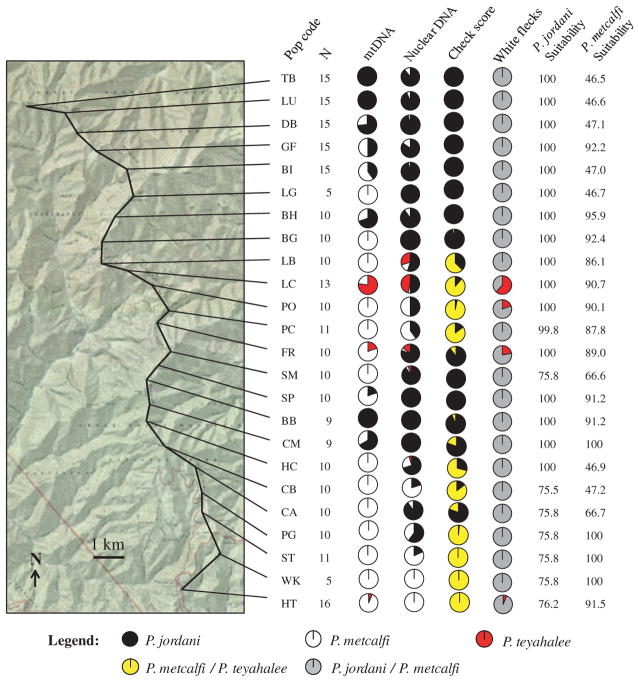
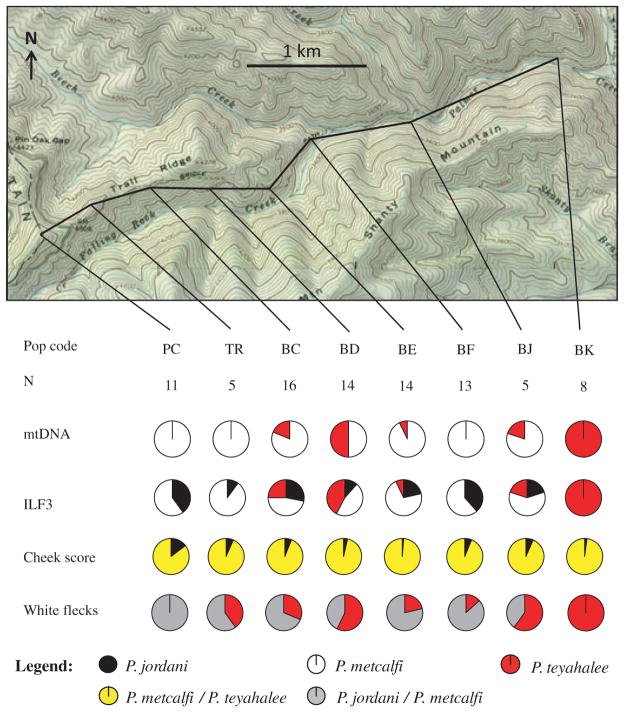
Three patterns emerge from the Balsam Mountain transect: first, at four localities near the centre of the transect (LB, LC, PO, and FR), the presence of P. teyahalee was detected both morphologically and genetically (Fig. 3). This occurs at Pin Oak Gap, a low point in the otherwise high elevation of the Balsam Mountain ridgeline. Second, frequencies of the P. jordani nuclear allele and the incidence of red cheek pigmentation are in close agreement. That is, populations from localities along the ridgeline that show a predominance of the P. jordani nuclear allele also have a high cheek pigmentation score (e.g. localities LG, BH, BG, and SP; Fig. 3). Third, frequencies of P. jordani mtDNA are largely discordant with respect to those of the nuclear allele and red cheek pigmentation. This is most clearly seen in localities GF, BI, LG, BG, LB, SM, SP, HC, CB, and CA (Fig. 3). Notably, this represents a shift of P. metcalfi mtDNA northwards relative to the P. jordani ILF3 allele and morphology.
Fig. 3.
Map of Balsam Mountain showing transect (bold line), collection localities, sample sizes, marker scores and habitat-suitability values. Pie charts are interpreted as follows: mtDNA = proportion of samples at a given locality that are diagnostic for each parental species, nuclear DNA = proportion of alleles at a given locality that are diagnostic for each parental species, cheek score = the average scaled cheek pigmentation score (i.e. average score divided by the average for pure parental Plethodon jordani), and white flecks = the proportion of animals that have at least some white flecks. Habitat-suitability values are extracted from the ecological niche models presented in Fig. 7 and are given as a percentage from 0 to 100. Note that more than one collection locality may lie within a single grid cell; therefore, identical values may not be independent from one another.
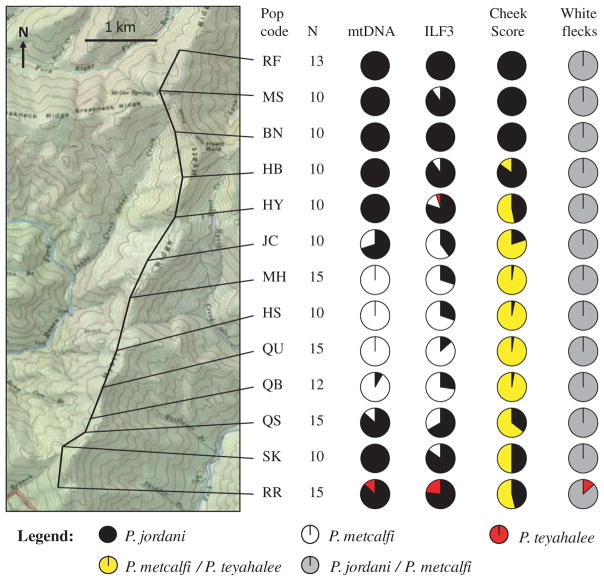
The pattern of differential introgression is not seen in the Hyatt Ridge transect (Fig. 4). Rather, there is coincidence among all markers. Also unlike Balsam Mountain, the P. teyahalee allele does not appear on Hyatt Ridge, except for one individual that is heterozygous at the ILF3 marker (with P. jordani mtDNA and no white flecks) and one apparently pure P. teyahalee from the southernmost collection locality (which has a lower elevation than most other sites on the ridgeline).
Fig. 4.
Map of Hyatt Ridge showing transect (bold line), collection localities, sample sizes and marker scores. Interpretation of marker scores is as given in Fig. 3.
The pattern of differential introgression is strongly apparent in the Palmer Creek transect (Fig. 5). The upslope (western) end of the transect begins in a region containing P. jordani and P. metcalfi hybrids (locality PC); however, the ILF3 and mtDNA markers show a prevalence of P. metcalfi alleles. The average cheek pigmentation score is 0.9, with 2 of 11 animals having some red pigmentation and a considerable number of P. jordani ILF3 alleles (8 of 20 alleles). There is, however, a complete absence of P. jordani mtDNA at this site. The presence of the P. jordani ILF3 allele extends downslope to 1000-m elevation, almost to the valley floor. The presence of some red cheek pigmentation extends to the lowest elevation sampled (locality BK at 951 m with an average cheek pigmentation score of 0.125). This pattern is in contrast to the complete absence of P. jordani mtDNA along the transect. Patterns in white flecks are consistent with those of red cheek pigmentation and nuclear DNA. Thus, there is a clear discordance with P. jordani mtDNA being restricted to the highest elevations sampled despite the presence of the P. jordani ILF3 allele and morphology extending much of the way to the valley below.
Fig. 5.
Map of Palmer Creek showing transect (bold line), collection localities, sample sizes and marker scores. Interpretation of marker scores is as given in Fig. 3.
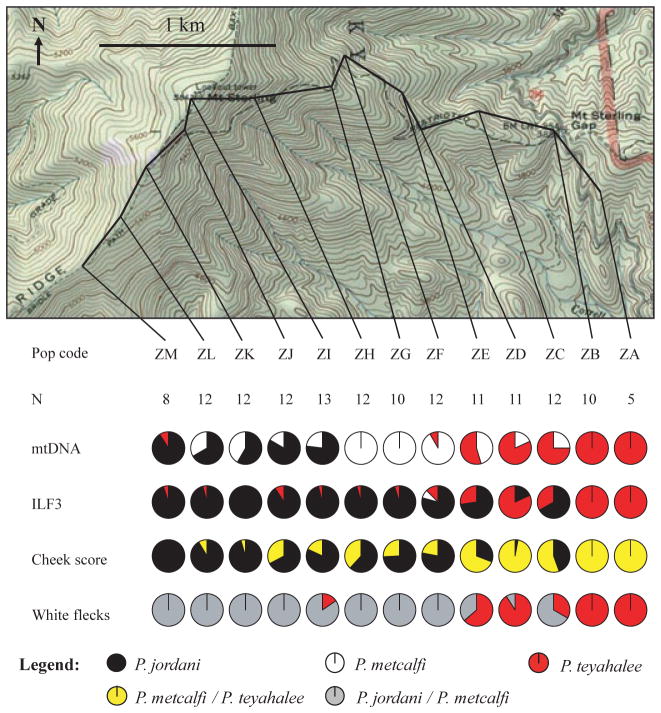
Patterns of marker frequencies are more complicated along the Mt Sterling transect (Fig. 6). Along the ridge-line of the western portion of the transect (localities ZM-ZI; 1561–1768 m elevation), there is a predominance of the P. jordani ILF3 allele, mtDNA and morphology, although P. metcalfi mtDNA is slightly overrepresented. At localities beginning immediately off the ridge top, there is a complete absence of P. jordani mtDNA. This is in sharp contrast to the patterns of the P. jordani ILF3 allele and red cheek pigmentation, both of which predominate down to an elevation of 1280 m. As in the Palmer Creek and Balsam Mountain transects, patterns in P. teyahalee (white flecks) are largely coincident with those of red cheek pigmentation and nuclear DNA. Thus, the same pattern of discordance between P. metcalfi mtDNA and the P. jordani ILF3 allele and morphology that is found in the Palmer Creek transect is also seen in the Mt Sterling transect.
Fig. 6.
Map of Mt Sterling showing transect (bold line), collection localities, sample sizes and marker scores. Interpretation of marker scores is as given in Fig. 3.
Ecological niche models
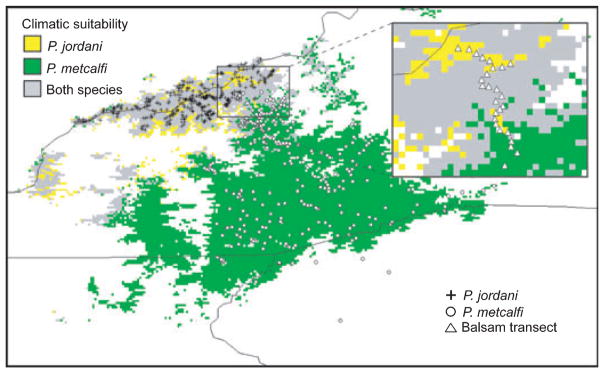
The predicted geographic distributions for P. jordani and P. metcalfi are shown in Fig. 7. The area under the receiving characteristic (AUC) shows that the ecological niche models strongly discriminate between randomly selected locations across the study region and the training (AUCP. jordani = 0.99, AUCP. metcalfi = 0.97) and the test localities (AUCP. jordani = 1.0, AUCP. metcalfi = 0.99).
Fig. 7.
Ecological niche modelling results showing present-day predicted geographic distributions for Plethodon jordani and Plethodon metcalfi. For comparison, collection localities are shown for both Plethodon jordani (+) and Plethodon metcalfi (O). The inner box encompasses the hybrid zone and is expanded to show sampling localities (Δ) along the Balsam Mountain transect. Colours indicate habitat-suitability values as assigned by Maxent and are given as percentages (cumulative probability × 100; see text for more detail).
The geographic distributions of climatically suitable habitats for P. jordani and P. metcalfi are not entirely overlapping. Nevertheless, it seems unlikely that divergent climatic adaptation or the presence of population density troughs influences the position of the hybrid zone. The climatic conditions are highly suitable for both species at many of the sampling locations in the hybrid zone (Fig. 7). Across the hybrid zone, there is no relationship between allele frequency scores and climatic suitability scores (mtDNA × climatic suitability: ρ = −0.197, P > 0.35; ILF × climatic suitability: ρ = −0.107; P > 0.618). Furthermore, there is also no relationship between cheek colouration scores and habitat-suitability scores (ρ = −0.145; P > 0.496). Thus, it seems unlikely that natural selection associated with climate and / or gradients in population density influences the dynamics of the hybrid zone.
Discussion
In this study, we document strong discordance in the extent to which mtDNA and nuclear DNA and colour-pattern traits have moved across a salamander hybrid zone in the Great Smoky Mountains. A variety of selective (natural selection associated with climate) and behavioural processes (sex-biased dispersal, asymmetric reproductive isolation) acting alone or together might explain this discordance. Here, we discuss the evidence favouring each of these explanations.
Unlike some hybrid zones (e.g. Cicero 2004; Swenson 2006; Martínez-Freiría et al. 2008; Swenson et al. 2008), we found no evidence that the location of the hybrid zone between P. jordani and P. metcalfi is associated with steep gradients in climate or habitat suitability. The ecological niche models suggest that the climatic conditions are suitable for both species across much of the hybrid zone. Similarly, we found no relationship between the climatic suitability scores for P. jordani and P. metcalfi across the transect and the proportion of samples having alleles that were diagnostic for either species. Thus, it seems unlikely that exogenous selection associated with climate could promote the movement of either P. metcalfi mtDNA (e.g. Boutilier 2001; Nouette-Gaulain et al. 2005; Lynn et al. 2007; Tattersall & Ultsch 2008) or P. jordani nuclear DNA across the hybrid zone (e.g. Doiron et al. 2002; Bachtrog et al. 2006). The ecological niche models do suggest that the hybrid zone lies in a region containing suitable habitat for both P. jordani and P. metcalfi. Thus, while exogenous, climate-associated selection may not be acting to maintain hybrid zone position, the hybrid zone may nonetheless be constrained by this region of overlapping, suitable habitats. Furthermore, if this hybrid zone is a tension zone (i.e. maintained by a balance between dispersal and selection), then the zone may have settled into this region of overlapping habitats without actually being maintained by climate-associated selection.
Male-biased dispersal is often invoked as a mechanism by which nuclear DNA may move across a hybrid zone more readily than mtDNA (e.g. Jockusch & Wake 2002; García-París et al. 2003). However, the available evidence does not support the idea that greater dispersal of male Plethodon underlies the discordant patterns of genetic and morphological variation in the hybrid zone. The Hyatt Ridge transect is particularly informative in this regard. Along Hyatt Ridge, dispersal of pure P. metcalfi from the south is impossible, and no pattern of asymmetrical introgression is observed. If greater dispersal of male P. jordani were responsible for the pattern of differential introgression observed in the Balsam Mountain, Palmer Creek and Mt Sterling transects, then differential introgression should also be observed in the Hyatt Ridge transect. In addition, a recent study that directly tested for male-biased dispersal in a related species with similar behaviour and ecology to P. jordani and P. metcalfi (Plethodon cinereus) found that dispersal is equally restricted in both sexes (Cabe et al. 2007).
An alternative explanation for the pattern of discordance of genetic and morphological variation is that the hybrid zone is moving. The underrepresentation of P. jordani mtDNA that is seen in the Balsam Mountain, Palmer Creek, and Mt Sterling transects may have resulted from a shift in hybrid zone position southward towards the range of P. metcalfi and downslope towards the range of P. teyahalee. Support for this hypothesis comes from laboratory-staged mating trials between P. jordani and P. metcalfi as reported by Reagan (1992). Specifically, she found that heterospecific crosses between female P. metcalfi and male P. jordani yielded about a 10% mating success rate (11 deposited spermatophores and eight inseminations of 100 staged crosses), while the reverse cross yielded only a 1% success rate (1 spermatophore and 1 insemination of 100 crosses). Under this scenario, the front of the expanding P. jordani distribution could not effectively remove the P. metcalfi mtDNA left in its wake because of mating asymmetry. Consequently, the extensive occurrence of P. metcalfi mtDNA may best be viewed as a relict, i.e. a ‘footprint’, of the historic range of P. metcalfi.
Lastly, it is not possible to rule out that positive selection for P. jordani cheek colouration (along with linked nuclear genes) contributes to the discordant patterns seen across the hybrid zone. Early studies on P. jordani morphology show that red cheek pigmentation has an aposematic function, serving as a warning to potential predators (Huheey 1960; Brodie & Howard 1973; Hensel & Brodie 1976). While not toxic, all three species are noxious, and when disturbed, such as during a predator attack, copious amounts of glandular secretions are released from the skin (Huheey 1960; Brodie & Howard 1973; Hensel & Brodie 1976; Brodie et al. 1979). Another plethodontid salamander, Desmognathus imitator, is likely a Batesian mimic of P. jordani, with about 25% of the population possessing red cheeks like their noxious model (Orr 1968; Brodie & Howard 1973). Aposematism and mimicry of red cheek pigmentation suggest this trait is under selection, and hybrids possessing at least some red pigmentation may have a selective advantage when the red pigmentation and the associated noxious secretions are common, but a disadvantage when rare. If this is the case, increased fitness of red-cheeked populations may be an explanation for the patterns of introgression observed in this study. Additional work on the introgression of this morphological trait and possible differences in defensive chemistry between species is needed.
The dynamics of hybridization seen in the Palmer Creek and Mt Sterling transects, may differ somewhat from those found between P. jordani and P. metcalfi along Balsam Mountain and Hyatt Ridge. First, there is likely to be differential adaptation of the parental species. Plethodon teyahalee is a large species that is restricted to low elevations, whereas P. jordani and P. metcalfi are smaller species that are only found in cool, moist, high elevation habitats. The latter two species may be restricted to high elevations as a result of smaller body size, which leads to greater rates of evaporative water loss (Spotila & Berman 1976). Second, previous studies (Hairston 1980a,b, 1983; Hairston et al. 1987; Adams 2004) have demonstrated that competitive interactions may play a role in the distributions of P. jordani and P. teyahalee. Lastly, P. jordani and P. metcalfi occur at a greater density than P. teyahalee (Highton 1970; Merchant 1972; Hairston 1980a,b). Population density is one determinant of hybrid zone structure and, when asymmetrical, may result in the movement of the hybrid zone towards the less dense parental species (Barton & Hewitt 1985).
Hypotheses of hybrid zone movement based on patterns of differential introgression of genes and traits have been proposed in other systems. For example, in fire salamanders (Salamandra) on the Iberian Peninsula, García-París et al. (2003) found strong discordance between mtDNA on the one hand and allozymes, morphology and life history on the other, which they attributed to male-biased dispersal. In another study, a hybrid zone among lizards in the genus Sceloporus of the western United States appears to have shifted 1.5 km as a result of anthropogenic changes to the habitat (Leaché & Cole 2007). Lastly, Rohwer et al. (2001) examined hybridization between warbler species of the genus Dendroica from the Pacific Northwest. Patterns of differential introgression between mtDNA and a suite of morphological markers led the researchers to conclude that the hybrid zone is moving. Independent observations on behaviour and inferences from historical data suggest mating asymmetry may be the cause of hybrid zone movement in that system.
The findings presented here have considerable potential to explain the patterns observed in other hybrid zones among Plethodon in the southern Appalachians. A rapid radiation leading to high species richness (Highton 1995; Kozak et al. 2006; Wiens et al. 2006) has resulted in myriad hybrid zones among Plethodon in the southeastern United States (Highton & Peabody 2000). Given the mountainous terrain encompassing the ranges of many of these species, the seemingly narrow climatic specificity of many high elevation Plethodon species (Kozak & Wiens 2006, 2010), and past oscillations in climate, we may reasonably expect many of these hybrid zones to be dynamic. One such example occurs between P. shermani and P. teyahalee. In a 20-year study, Hairston et al. (1992) documented a shift in hybrid zone position, which they attributed to changes in land use early last century. More recent analyses, however, suggest that modern climate change may actually be driving hybrid zone movement in that system (Walls 2009).
Hairston et al. (1992) made the assumption that red cheek pigmentation is neutrally diffusing across the P. jordani-P. metcalfi hybrid zone. Based on the most recent warming period during the Hypsithermal Interval (approximately 5000 years ago; see Pielou 1991), the authors hypothesized that populations of P. jordani and P. metcalfi migrated along the Balsam Mountain ridgeline from their mountain top refuges and met near the centre of the hybrid zone as inferred by the authors. However, the expanded and finer-scale sampling performed in this study uncovered extensive hybridization farther north than that documented by Hairston et al. (1992), thus nearly doubling the width of the hybrid zone. Furthermore, this study documents a nonclinal transition between the parental species, which suggests a much more complex biogeographic history than the one outlined by Hairston et al. (1992). A more likely scenario is that repeated bouts of isolation and secondary contact (perhaps as early as eight million years ago, i.e. shortly after molecular clock estimates place the date of divergence; Highton 1995; Kozak et al. 2006; Wiens et al. 2006) have left a complex, mosaic pattern of hybridization. In addition, severely limited dispersal abilities (as suggested by small home range sizes; Madison & Shoop 1970; Merchant 1972; Nishikawa 1990), long generation times (every other year beginning at 4 years of age for P. jordani and P. metcalfi; Hairston 1983) and stasis during the 18-year study period of Hairston et al. (1992) suggest this hybrid zone, if moving, may be doing so very slowly. Hairston et al.’s (1992) hypothesis of neutral diffusion gives way to one of differential introgression, and possibly hybrid zone movement, on an evolutionary time scale not readily measured by ecological studies.
Supplementary Material
Acknowledgments
We especially thank the crew at the Appalachian Highlands Science Learning Center and R. Highton for help with field logistics. We also thank M. Vance, V. Chatfield and K. Hamed for help with field work, and K. Luzynski and A. Conti for help with laboratory work. This research was funded through awards to MWHC from the following institutions: the Department of Ecology and Evolutionary Biology, the Museum of Zoology, and the Horace H. Rackham School of Graduate Studies at the University of Michigan; the Society for the Study of Amphibians and Reptiles; and the North Carolina Herpetological Society. This manuscript was greatly improved through the help of three anonymous reviewers.
Footnotes
This work forms part of M.W.H.C.’s Ph.D. thesis on the evolutionary dynamics of salamanders in the Plethodon glutinosus group. He is currently studying interactions between the amphibian chytrid fungus and frogs of the eastern United States. K.H.K. is interested in the evolutionary processes of southern Appalachian salamanders, especially as revealed by ecological niche modelling and physiological limits. B.M.F. studies conservation, population genetics, and patterns of hybridization across a variety of salamander taxa. P.K.T.’s research focuses on speciation genetics, as informed by an extensive house mouse hybrid zone in Eastern Europe.
Additional supporting information may be found in the online version of this article.
Table S1 Transects, sites codes, elevation, and latitude and longitude for animals captured in the hybrid zone
Table S2 Sites codes, elevation, and latitude and longitude for parental taxa
Table S3 Pure parental individuals (Plethodon jordani, P. metcalfi, and P. teyahalee) and marker scores used in panel for marker development
Table S4 Samples (arranged by transect) and marker scores for all samples used in analyses
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Adams DC. Character displacement via aggressive interference in Appalachian salamanders. Ecology. 2004;85:2664–2670. [Google Scholar]
- Agresti A. An Introduction to Categorical Data Analysis. John Wiley and Sons Inc; New Jersey: 2007. [Google Scholar]
- Arntzen JW, Wallis GP. Restricted gene flow in a moving hybrid zone of the newts Triturus cristatus and T. marmoratus in western France. Evolution. 1991;4:805–826. doi: 10.1111/j.1558-5646.1991.tb04352.x. [DOI] [PubMed] [Google Scholar]
- Asmussen MA, Arnold J, Avise JC. Definition and properties of disequilibrium statistics for association between nuclear and cytoplasmic genotypes. Genetics. 1987;115:755–768. doi: 10.1093/genetics/115.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D, Thornton K, Clark A, Andolfatto P. Extensive introgression of mitochondrial DNA relative to nuclear genes in the Drosophila yakuba species group. Evolution. 2006;60:292–302. [PubMed] [Google Scholar]
- Barton NH. Gene flow past a cline. Heredity. 1979;43:333–339. [Google Scholar]
- Barton NH, Baird SJE. Analyse v 1.3. Edinburgh: 1998. http://www.biology.ed.ac.uk/research/institutes/evolution/software/Mac/Analyse/ [Google Scholar]
- Barton NH, Hewitt GM. Analysis of hybrid zones. Annual Review of Ecology and Systematics. 1985;16:113–148. [Google Scholar]
- Boutilier RG. Mechanisms of metabolic defense against hypoxia in hibernating frogs. Respiration Physiology. 2001;128:365–377. doi: 10.1016/s0034-5687(01)00312-7. [DOI] [PubMed] [Google Scholar]
- Bridle JR, Baird SJE, Butlin RK. Spatial structure and habitat variation in a grasshopper hybrid zone. Evolution. 2001;55:1832–1843. doi: 10.1111/j.0014-3820.2001.tb00832.x. [DOI] [PubMed] [Google Scholar]
- Britch SC, Cain ML, Howard DJ. Spatio-temporal dynamics of the Allenomobius fasciatus-A. socius mosaic hybrid zone: a 14-year perspective. Molecular Ecology. 2001;10:627–638. doi: 10.1046/j.1365-294x.2001.01215.x. [DOI] [PubMed] [Google Scholar]
- Brito PH. Contrasting patterns of mitochondrial and microsatellite genetic structure among Western European populations of tawny owls (Strix aluco) Molecular Ecology. 2007;16:3423–3437. doi: 10.1111/j.1365-294X.2007.03401.x. [DOI] [PubMed] [Google Scholar]
- Brodie ED, Jr, Howard RR. Experimental study of Batesian mimicry in the salamanders Plethodon jordani and Desmognathus ochrophaeus. American Midland Naturalist. 1973;90:38–46. [Google Scholar]
- Brodie ED, Jr, Nowak RT, Harvey WR. The effectiveness of antipredator secretions and behavior of selected salamanders against shrews. Copeia. 1979;1979:270–274. [Google Scholar]
- Buggs RJA. Empirical study of hybrid zone movement. Heredity. 2007;99:301–312. doi: 10.1038/sj.hdy.6800997. [DOI] [PubMed] [Google Scholar]
- Burnham KP, Anderson DR. Multimodel inference: understanding AIC and BIC in model inference. Sociological Methods Research. 2004;33:261–304. [Google Scholar]
- Cabe PR, Hanlon TJ, Aldrich ME, Connors L, Marsh DM. Fine-scale population differentiation and gene flow in a terrestrial salamander (Plethodon cinereus) living in continuous habitat. Heredity. 2007;98:1–8. doi: 10.1038/sj.hdy.6800905. [DOI] [PubMed] [Google Scholar]
- Chan KMA, Levin SA. Leaky prezygotic isolation and porous genomes: rapid introgression of maternally inherited DNA. Evolution. 2005;59:720–729. [PubMed] [Google Scholar]
- Cicero C. Barriers to sympatry between avian sibling species (Paridae: Baeolophus) in local secondary contact. Evolution. 2004;58:1573–1587. doi: 10.1111/j.0014-3820.2004.tb01737.x. [DOI] [PubMed] [Google Scholar]
- Collins JT, Taggart TW. Standard Common and Current Scientific Names for north American Amphibians, Turtles, Reptiles, and Crocodilians. 6. Publication of The Center for North American Herpetology; Lawrence, Kansas: 2009. [Google Scholar]
- Dakin E. Cytonuclear disequilibria in a spatially structured hybrid zone. Theoretical Population Biology. 2006;70:82–91. doi: 10.1016/j.tpb.2005.12.005. [DOI] [PubMed] [Google Scholar]
- Davis MB, Shaw RG. Range shifts and adaptive responses to Quaternary climate change. Science. 2001;292:673–679. doi: 10.1126/science.292.5517.673. [DOI] [PubMed] [Google Scholar]
- Doiron S, Bernatchez L, Blier PU. A comparative mitogenomic analysis of the potential adaptive value of arctic charr mtDNA introgression in brook charr populations (Salvelinus fontinalis Mitchill) Molecular Biology and Evolution. 2002;19:1902–1909. doi: 10.1093/oxfordjournals.molbev.a004014. [DOI] [PubMed] [Google Scholar]
- Funk DJ, Omland KE. Species-level paraphyly and polyphyly: Frequency, causes, and consequences, with insights from animal mitochondrial DNA. Annual Review of Ecology and Systematics. 2003;34:397–423. [Google Scholar]
- García-París M, Alcobendas M, Buckley D, Wake DB. Dispersal of viviparity across contact zones in Iberian populations of fire salamanders (Salamandra) inferred from discordance of genetic and morphological traits. Evolution. 2003;57:129–143. doi: 10.1111/j.0014-3820.2003.tb00221.x. [DOI] [PubMed] [Google Scholar]
- Guralnick R. Differential effects of past climate warming on mountain and flatland Species distributions: a multispecies North American mammal assessment. Global Ecology and Biogeography. 2007;16:14–23. [Google Scholar]
- Hairston NG. Intergradation in Appalachian salamanders of the genus Plethodon. Copeia. 1950;1950:262–273. [Google Scholar]
- Hairston NG. The experimental test of an analysis of field distributions: competition in terrestrial salamanders. Ecology. 1980a;61:817–826. [Google Scholar]
- Hairston NG. Evolution under interspecific competition: field experiments on terrestrial salamanders. Evolution. 1980b;34:409–420. doi: 10.1111/j.1558-5646.1980.tb04829.x. [DOI] [PubMed] [Google Scholar]
- Hairston NG. Growth, survival and reproduction of Plethodon jordani: trade-offs between selective pressures. Copeia. 1983;1983:1024–1035. [Google Scholar]
- Hairston NG, Sr, Nishikawa KC, Stenhouse SL. The evolution of competing species of terrestrial salamanders: niche partitioning or interference? Evolutionary Ecology. 1987;1:247–262. [Google Scholar]
- Hairston NG, Wiley RH, Smith CK, Kneidel KA. The dynamics of two hybrid zones in Appalachian salamanders of the genus Plethodon. Evolution. 1992;46:930–938. doi: 10.1111/j.1558-5646.1992.tb00610.x. [DOI] [PubMed] [Google Scholar]
- Hensel JL, Jr, Brodie ED., Jr An experimental study of aposematic coloration in the salamanders Plethodon jordani. Copeia. 1976;1976:59–65. [Google Scholar]
- Hewitt GM. Some genetic consequences of ice ages, and their role in divergence and speciation. Biological Journal of the Linnean Society. 1996;58:247–276. [Google Scholar]
- Highton R. Evolutionary interactions between species of North American salamanders of the genus Plethodon. Part I. Genetic and ecological relationships of Plethodon jordani and P. glutinosus in the southern Appalachian Mountains. Evolutionary Biology. 1970;4:211–241. [Google Scholar]
- Highton R. Speciation in eastern North American salamanders of the genus Plethodon. Annual Review of Ecology and Systematics. 1995;26:579–600. [Google Scholar]
- Highton R, Peabody RB. Geographic protein variation and speciation in salamanders of the Plethodon jordani and Plethodon glutinosus complexes in the southern Appalachian mountains with the description of four new species. In: Bruce RC, Jaeger R, Houck LD, editors. The Biology of Plethodontid Salamander. Kluwer Academic / Plenum; New York: 2000. pp. 31–94. [Google Scholar]
- Hijmans RJ, Guarino L, Jarvis A, et al. DIVA-GIS version 5.2. 2005a http://divagis.org.
- Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. Very high resolution interpolated climate surfaces for global land areas. International Journal of Climatology. 2005b;25:1965–1978. [Google Scholar]
- Hofman S, Szymura JM. Limited mitochondrial DNA introgression in a Bombina hybrid zone. Biological Journal of the Linnean Society. 2007;91:295–306. [Google Scholar]
- Huheey JE. Mimicry in the color pattern of certain Appalachian salamanders. Journal of the Elisha Mitchell Scientific Society. 1960;76:246–251. [Google Scholar]
- Jockusch EL, Wake DB. Falling apart and merging: diversification of slender salamanders (Plethodontidae: Batrachoseps) in the American West. Biological Journal of the Linnean Society. 2002;76:361–391. [Google Scholar]
- Kawakami T, Butlin RK, Adams M, Paull DJ, Cooper SJB. Genetic analysis of a chromosomal hybrid zone in the Australian morabine grasshoppers (Vandiemenella viatica species group) Evolution. 2008;63:139–152. doi: 10.1111/j.1558-5646.2008.00526.x. [DOI] [PubMed] [Google Scholar]
- Kozak KH, Wiens JJ. Does niche conservatism promote speciation? A case study in North American salamanders. Evolution. 2006;60:2604–2621. [PubMed] [Google Scholar]
- Kozak KH, Wiens JJ. Niche conservatism drives elevational diversity patterns in Appalachian salamanders. American Naturalist. 2010;176:40–54. doi: 10.1086/653031. [DOI] [PubMed] [Google Scholar]
- Kozak KH, Weisrock DW, Larson A. Rapid lineage accumulation in a non-adaptive radiation: phylogenetic analysis of diversification rates in eastern North American woodland salamanders (Plethodontidae: Plethodon) Proceedings of the Royal Society Royal Society B-Biological Sciences. 2006;273:539–546. doi: 10.1098/rspb.2005.3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak KH, Graham CH, Wiens JJ. Integrating GIS-based environmental data into evolutionary biology. Trends in Ecology and Evolution. 2008;23:141–148. doi: 10.1016/j.tree.2008.02.001. [DOI] [PubMed] [Google Scholar]
- Leaché AD, Cole CJ. Hybridization between multiple fence lizard lineages in an ecotone: locally discordant variation in mitochondrial DNA, chromosomes, and morphology. Molecular Ecology. 2007;16:1035–1054. doi: 10.1111/j.1365-294X.2006.03194.x. [DOI] [PubMed] [Google Scholar]
- Linnen CR, Farrell BD. Mitonuclear discordance is caused by rampant mitochondrial introgression in Neodiprion (Hymenoptera: Diprionidae) sawflies. Evolution. 2007;61:1417–1438. doi: 10.1111/j.1558-5646.2007.00114.x. [DOI] [PubMed] [Google Scholar]
- Lynn EG, Zhongping L, Minerbi D, Sack MN. The regulation, control, and consequences of mitochondrial oxygen utilization and disposition in the heart and skeletal muscle during hypoxia. Antioxidants and Redox Signaling. 2007;9:1353–1361. doi: 10.1089/ars.2007.1700. [DOI] [PubMed] [Google Scholar]
- Madison DM, Shoop CR. Homing behavior, orientation, and home range of salamanders tagged with tantalum-182. Science. 1970;168:1484–1487. doi: 10.1126/science.168.3938.1484. [DOI] [PubMed] [Google Scholar]
- Manzo PA. PhD thesis. University of Maryland; College Park, Maryland: 1988. A Morphometric Analysis of Plethodon jordani Blatchley and its Hybrids. [Google Scholar]
- Martínez-Freiría F, Sillero N, Lizana M, Brito JC. GIS-based niche models identify environmental correlates sustaining a contact zone between three species of European vipers. Diversity and Distributions. 2008;14:452–461. [Google Scholar]
- Merchant H. Estimated population size and home range of the salamanders Plethodon jordani and Plethodon glutinosus. Journal of the Washington Academy of Sciences. 1972;62:248–257. [Google Scholar]
- Nishikawa KC. Intraspecific spatial relationships of two species of terrestrial salamanders. Copeia. 1990;1990:418–426. [Google Scholar]
- Nouette-Gaulain K, Malgat M, Rocher C, et al. Time course of differential mitochondrial energy metabolism adaptation to chronic hypoxia in right and left ventricles. Cardiovascular Research. 2005;66:132–140. doi: 10.1016/j.cardiores.2004.12.023. [DOI] [PubMed] [Google Scholar]
- Orr LP. The relative abundance of mimics and models in a supposed mimetic complex in salamanders. Journal of the Elisha Mitchell Scientific Society. 1968;84:303–304. [Google Scholar]
- Peabody RB. PhD thesis. University of Maryland; College Park, Maryland: 1978. Electrophoretic Analysis of Geographic Variation and Hybridization of Two Appalachian Salamanders, Plethodon jordani and Plethodon glutinosus. [Google Scholar]
- Pearson RG, Dawson TP. Predicting the impact of climate change on the distribution of species: are bioclimate envelope models useful? Global Ecology and Biogeography. 2003;12:361–371. [Google Scholar]
- Pereira RJ, Wake DB. Genetic leakage after adaptive and nonadaptive divergence in the Ensatina eschscholtzii ring species. Evolution. 2009;63:2288–2301. doi: 10.1111/j.1558-5646.2009.00722.x. [DOI] [PubMed] [Google Scholar]
- Peterson AT. Projected climate change effects on Rocky Mountain and Great Plains birds: generalities of biodiversity consequences. Global Change Biology. 2003;9:647–655. [Google Scholar]
- Peterson AT, Martínez-Meyer E, González-Salazar C. Reconstructing the Pleistocene geography of the Aphelocoma jays (Corvidae) Diversity and Distributions. 2004;10:237–246. [Google Scholar]
- Peterson AT, Papes M, Eaton M. Transferability and model evaluation in ecological niche modeling: a comparison of GARP and Maxent. Ecography. 2007;30:550–560. [Google Scholar]
- Petranka JW. Salamanders of the United States and Canada. Smithsonian Institution; Washington DC: 1998. [Google Scholar]
- Phillips SJ, Anderson RP, Schapire RE. Maximum entropy modeling of species geographic distributions. Ecological Modelling. 2006;190:231–259. [Google Scholar]
- Pielou EC. After the Ice Age: The Return of Life to Glaciated North America. The University of Chicago Press; Illinois: 1991. [Google Scholar]
- Reagan NL. PhD thesis. The University of Chicago; Chicago, Illinois: 1992. Evolution of Sexual Isolation in Salamanders in the Genus Plethodon. [Google Scholar]
- Rohwer S, Bermingham E, Wood C. Plumage and mitochondrial DNA haplotype variation across a moving hybrid zone. Evolution. 2001;55:405–422. doi: 10.1111/j.0014-3820.2001.tb01303.x. [DOI] [PubMed] [Google Scholar]
- Sequeira F, Alexandrino F, Rocha S, Arntzen JW, Ferrand N. Genetic exchange across a hybrid zone within the Iberian endemic golden-striped salamander. Chioglossa lusitanica Molecular Ecology. 2005;14:245–254. doi: 10.1111/j.1365-294X.2004.02390.x. [DOI] [PubMed] [Google Scholar]
- Spotila JR, Berman EN. Determination of skin resistance and the role of the skin in controlling water loss in amphibians and reptiles. Comparative Biochemistry and Physiology. 1976;55:407–411. doi: 10.1016/0300-9629(76)90069-4. [DOI] [PubMed] [Google Scholar]
- Swenson NG. Gis-based niche models reveal unifying climatic mechanisms that maintain the location of avian hybrid zones in a North American suture zone. Evolutionary Biology. 2006;19:717–725. doi: 10.1111/j.1420-9101.2005.01066.x. [DOI] [PubMed] [Google Scholar]
- Swenson NG. The past and future influence of geographic information systems on hybrid zone, phylogeographic and speciation research. Evolutionary Biology. 2008;21:421–434. doi: 10.1111/j.1420-9101.2007.01487.x. [DOI] [PubMed] [Google Scholar]
- Swenson NG, Fair JM, Heikoop J. Water stress and hybridization between Quercus gambelii and Quercus grisea. Western North American Naturalist. 2008;68:498–507. [Google Scholar]
- Tattersall GJ, Ultsch GR. Physiological ecology of aquatic overwintering in ranid frogs. Biological Review. 2008;83:119–140. doi: 10.1111/j.1469-185X.2008.00035.x. [DOI] [PubMed] [Google Scholar]
- Teeter KC, Payseur BA, Harris LW, et al. Genome-wide patterns of gene flow across a house mouse hybrid zone. Genome Research. 2008;18:67–76. doi: 10.1101/gr.6757907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeter KC, Thibodeau LM, Gompert Z, Buerkle AC, Nachman MW, Tucker PK. The variable genomic architecture of isolation between hybridizing species of house mice. Evolution. 2010;64:472–485. doi: 10.1111/j.1558-5646.2009.00846.x. [DOI] [PubMed] [Google Scholar]
- Vörös J, Alcobendas M, Martínez-Solano I, García-París M. Evolution of Bombina bombina and Bombina variegata (Anura: Discoglossidae) in the Carpathian Basin: a history of repeated mt-DNA introgression across species. Molecular Phylogenetics and Evolution. 2006;38:705–718. doi: 10.1016/j.ympev.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Walls S. The role of climate in the dynamics of a hybrid zone in Appalachian salamanders. Global Change Biology. 2009;15:1903–1910. [Google Scholar]
- Weisrock DW, Larson A. Testing hypotheses of speciation in the Plethodon jordani species complex with allozymes and mitochondrial DNA sequences. Biological Journal of the Linnean Society. 2006;89:25–51. [Google Scholar]
- Weisrock DW, Macey JR, Ugurtas IH, Larson A, Panenfuss TJ. Molecular phylogenetics and historical biogeography among salamandrids of the “true” salamander clade: rapid branching of numerous highly divergent lineages in Mertensiella luschani associated with the rise of Anatolia. Molecular Phylogenetics and Evolution. 2001;18:434–448. doi: 10.1006/mpev.2000.0905. [DOI] [PubMed] [Google Scholar]
- Wiens JJ, Engstrom TN, Chippindale PT. Rapid diversification, incomplete isolation, and the “speciation clock” in North American salamanders (genus Plethodon): testing the hybrid swarm hypothesis of rapid radiation. Evolution. 2006;60:2585–2603. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.