Abstract
Waterlogged paddy soils possess anoxic zones in which microbes actively induce reductive nitrogen transformation (RNT). In the present study, a shotgun RNA sequencing analysis (metatranscriptomics) of paddy soil samples revealed that most RNT gene transcripts in paddy soils were derived from Deltaproteobacteria, particularly the genera Anaeromyxobacter and Geobacter. Despite the frequent detection of the rRNA of these microbes in paddy soils, their RNT-associated genes have rarely been identified in previous PCR-based studies. This metatranscriptomic analysis provides novel insights into the diversity of RNT microbes present in paddy soils and the ecological function of Deltaproteobacteria predominating in these soils.
Keywords: paddy soils, metatranscriptomics, denitrification, dissimilatory nitrate reduction to ammonium, nitrogen fixation
Paddy soils are characterized by temporal anaerobic conditions caused by waterlogging, and the active occurrence of anaerobic biogeochemical processes (9). Among these active processes, biological reductive nitrogen transformation (RNT), i.e., denitrification (NO3−→NO2−→NO→N2O→N2), dissimilatory nitrate reduction to ammonium (DNRA; NO3−→NO2−→NH4+), and nitrogen fixation (N2→NH4+) contribute to less leaching of nitrogen pollutants (NO3−, NO2−, and N2O) into the environment and the greater retention of nitrogen-based nutrients (NH4+) for rice plants in waterlogged paddy soils than in upland soils (8, 22). Therefore, the identification of microbial drivers of RNT in paddy soils is important for successful rice production with minimal environmental nitrogen burden.
However, a comprehensive understanding of the RNT microbial community has not yet been achieved. In order to investigate RNT microbes in paddy soils, genes encoding the enzymes that catalyze each reaction have been assessed via PCR-based culture-independent methods, as represented by a clone library analysis (13, 24). Recent studies based on bacterial genomics reported that the diversity of microbes harboring RNT genes is greater than previously considered; PCR-based methods have underrepresented this diversity because of mismatches in the sequences of the primers used (5, 10, 21), indicating the need for alternative methods without a PCR bias. Furthermore, simultaneous assessments of microbes involved in denitrification, DNRA, and nitrogen fixation in a single paddy field have not yet been performed. Moreover, limited information is available on the transcriptional profiles in situ of RNT microbes in paddy soils because of the small number of field studies conducted based on soil RNA, which directly implicates RNT microbial activity. In the present study, we investigated RNT-associated microbial diversity in paddy soils via a shotgun RNA sequencing analysis without any prior PCR preparation (metatranscriptomics).
In order to obtain a more complete understanding of paddy soils with various biogeochemical properties spatially and seasonally (9, 12), soil RNA extracted from paddy soils in shallow (S1, S3) and deep (S2, S4) layers under waterlogged (S1, S2) and drained (S3, S4) conditions (Fig. S1) were subjected to a metatranscriptomic analysis using an Illumina MiSeq sequencer (Illumina, San Diego, CA, USA). The sequences of RNT genes were retrieved from the metatranscriptomic libraries obtained and taxonomically annotated through a tandem similarity search with the blat and blast programs (full methods in Supplementary information).
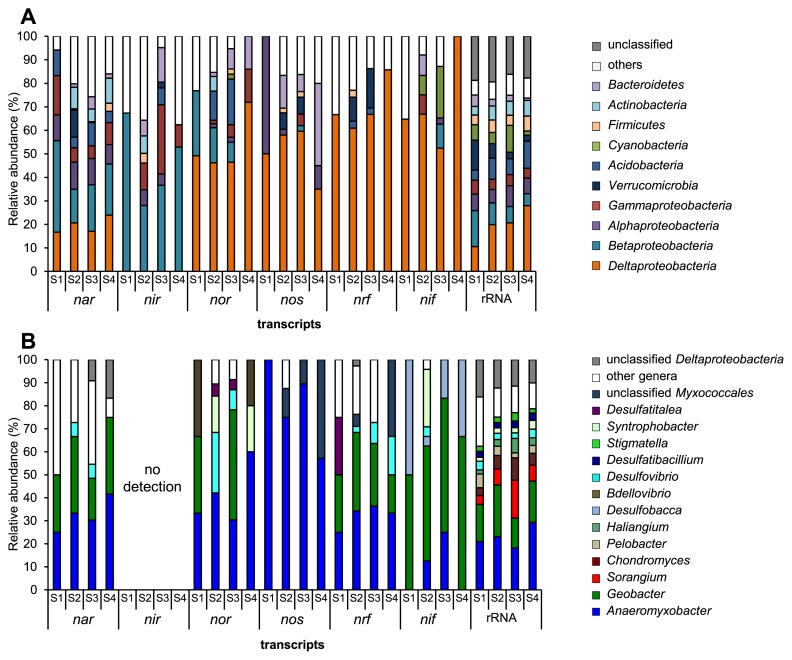
Four reactions crucial to denitrification are catalyzed by the following enzymes: NO3− reductase (Nar), NO2− reductase (Nir), NO reductase (Nor), and N2O reductase (Nos). The nar transcripts detected in all soil samples using metatranscriptomics were related to those of Deltaproteobacteria, Betaproteobacteria, Alphaproteobacteria, Gammaproteobacteria, and Acidobacteria (Fig. 1A), suggesting that these bacterial groups are involved in the reduction of NO3− to NO2−. The nir transcripts were mostly derived from Betaproteobacteria, Gammaproteobacteria, and Alphaproteobacteria (Fig. 1A), the members of which include common denitrifiers (5); these were also frequently detected in the same paddy soils in our previous PCR-based survey (24). Furthermore, nor and nos transcripts were predominantly detected in Deltaproteobacteria (Fig. 1A), the transcripts of which were rarely detected via previous PCR assays (2, 24). Successive denitrification steps were considered to be associated with common denitrifiers, such as Betaproteobacteria, Gammaproteobacteria, Alphaproteobacteria, and Actinobacteria, which harbor nir, nor, and/or nos (18). However, the metatranscriptomic data obtained in the present study suggested that the reduction of NO2− into NO was driven by these denitrifiers, and that the reduction of NO and N2O was mainly progressed by non-denitrifiers such as Deltaproteobacteria, Bacteroidetes, Acidobacteria, and Verrucomicrobia, which harbor nor and/or nos, but not nir (5). Thus, paddy soil denitrification appears to be a cooperative process by each nitrogen oxide reducer, i.e., NO2− reducers (denitrifiers) and NO/N2O reducers (non-denitrifiers), similar to nitrification (NH4+→NO2−→NO3−) orchestrated by NH4+-oxidizing bacteria/archaea and NO2−-oxidizing bacteria (6). These inferences in the denitrification process may be verified using co-culture experiments on denitrifiers and non-denitrifiers.
Fig. 1.
Microbial diversity of RNT gene transcripts and rRNA. Taxonomic distribution of nar, nir, nor, nos, nrf, and nif transcripts, and rRNA at the phylum and proteobacterial class level (A), and deltaproteobacterial genus level (B). Sample IDs indicate data derived from paddy soils in shallow (S1, S3) and deep (S2, S4) layers under waterlogged (S1, S2) and drained (S3, S4) conditions. Data represent the mean of triplicates.
DNRA, another NO3− reduction process, is catalyzed by Nar and NH4+-forming NO2− reductase (Nrf). Most of the nrf transcripts belong to Deltaproteobacteria, while some belong to Verrucomicrobia (Fig. 1A). Together with the frequent detection of nar transcripts derived from Deltaproteobacteria as described above, Deltaproteobacteria appear to mainly contribute to DNRA dynamics in paddy soils. Although DNRA has been geochemically detected in paddy soils (1, 23), limited information is available on DNRA microbial diversity. To the best of our knowledge, the present study is the first to attempt to identify the key player groups in DNRA in paddy soils.
Diazotrophs harboring nitrogenase (Nif) drive nitrogen fixation. The taxonomic composition of nif transcripts was dominated by Deltaproteobacteria (Fig. 1A), indicating that Deltaproteobacteria represents a key player group in nitrogen fixation. Rhizospheric Alphaproteobacteria, Betaproteobacteria, and Gammaproteobacteria and phototrophic Cyanobacteria were considered to be key diazotrophs in paddy soils (13, 18). However, we detected more nif transcripts in Deltaproteobacteria than in these well-known diazotrophs; our results were consistent with a recent metatranscriptomic analysis based on a microcosm study on Italian paddy soils (11).
The abundance of RNT genes derived from Deltaproteobacteria, as described above, was also demonstrated in a shotgun DNA sequencing analysis (metagenomics) (Fig. S2A). Additionally, the microbial community structure based on rRNA gene/transcript sequences showed that Deltaproteobacteria is a major group in paddy soil microbes (Fig. 1A, S2A). These results support Deltaproteobacteria being a key player group driving RNT in paddy soils.
Further analyses on Deltaproteobacteria at the genus level revealed the consistent detection of RNT gene transcripts in metatranscriptomic data derived from the genera Anaeromyxobacter and Geobacter (Fig. 1B), as well as their RNT genes in metagenomic data (Fig. S2B). These genera represent obligate anaerobes and metal reducers predominating in paddy soils (7, 20) and exhibit some RNT activities in vitro (summarized in Table S1). Although the nitrogen fixation activity of Anaeromyxobacter has yet to be characterized, the genomes of some Anaeromyxobacter spp. conserve a similar nif cluster to that of Geobacter spp. exhibiting nitrogen fixation activity (Fig. S3). Together with the detection of the nif transcripts of Anaeromyxobacter in this study, it is plausible that Anaeromyxobacter spp. perform nitrogen fixation. However, in contrast to Anaeromyxobacter and Geobacter rRNA genes, their RNT genes have rarely been detected in paddy soil samples using PCR-based techniques (4, 24). Thus, the putative role of these genera in the RNT process has received little attention despite their predominance in paddy soils. The limited coverage of RNT gene-specific PCR primers used in previous studies may have led to the oversight of these genera (10, 17); additionally, the GC content may be implicated because the nor/nos/nrf/nif of Anaeromyxobacter spp. showed markedly higher GC contents than the rRNA genes and nor/nos/nrf/nif of other bacteria (Table S2). Even improved nos universal primers, which have enabled lower rates of sequence mismatches, were unable to amplify Anaeromyxobacter nos (10). Therefore, a metatranscriptomic analysis represents a more effective approach to examine the diversity of functional microbes, without any PCR bias arising from the high GC content of target genes as well as primer limitations.
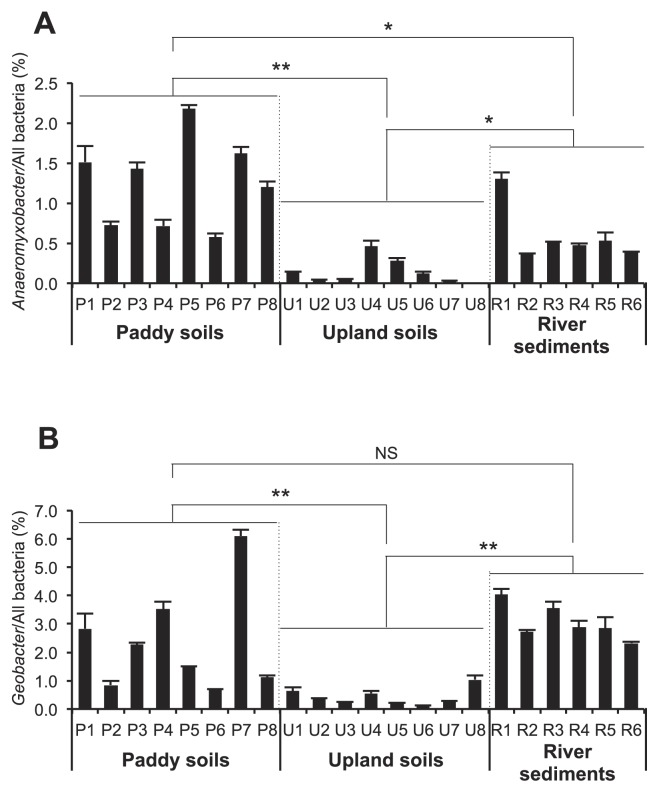
Anaeromyxobacter and Geobacter, which have frequently been detected in Japanese, Chinese, and Italian paddy soils (4, 11, rRNA data in Fig. 1B), predominate more in paddy soils than in upland soils, as confirmed by the present study (Fig. 2A, B; Table S3). Their universal distribution and predominance in paddy soils support Anaeromyxobacter and Geobacter being the key RNT players in paddy soils. Furthermore, the predominance of these genera was found in river sediments (Fig. 2A, B; Table S3); the RNT genes of Anaeromyxobacter were frequently and globally detected in upland soil environments in recent shotgun metagenomics studies (14, 15), indicating the contribution of these bacteria to RNT not only in paddy soils, but also in other environments. The further application of metatranscriptomics across different environments will expand our knowledge on the diversity of RNT microbes in nature as well as the ecological function of Deltaproteobacteria in soil environments.
Fig. 2.
Distribution of Anaeromyxobacter and Geobacter in various soil environments. Proportions of Anaeromyxobacter (A) and Geobacter (B) against all bacteria estimated by a quantitative PCR method. The mean±SD is shown (n=3). The paddy soil sample P3 was collected from the same paddy field used for the metatranscriptomic analysis in this study. Asterisks indicate significant differences (Mann-Whitney U test; *, p<0.01; **, p<0.001); NS, not significant. Details of soil samples and qPCR data are summarized in Table S3.
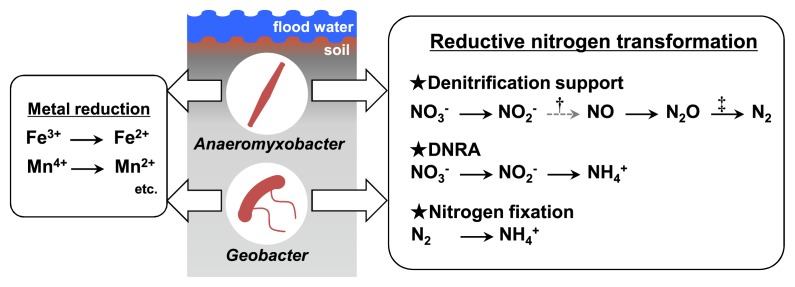
Previous studies on paddy soils identified the predominance of Deltaproteobacteria and their ecological roles in dissimilatory metal reduction, sulfur/sulfate reduction, and hydrogen production (7, 9, 19). Although genomic studies showed the ubiquitous possession of the RNT genes of Deltaproteobacteria, the association of RNT with Deltaproteobacteria has not been considered because of the rare detection of their RNT genes in soil environments through PCR-based analyses. The present study revealed the novel ecological functions of Anaeromyxobacter and Geobacter within Deltaproteobacteria dominating in paddy soils, namely, RNT, denitrification support, and NH4+ production via DNRA and nitrogen fixation (Fig. 3).
Fig. 3.
Ecological functions of Anaeromyxobacter and Geobacter, belonging to Deltaproteobacteria, predominant in paddy soils, expanded by metatranscriptomics in this study. Anaeromyxobacter and Geobacter, ubiquitously predominant in paddy soils, are key player groups in the reduction of iron and manganese, which actively progresses in paddy soils soon after waterlogging (3, 4, 7, 9, 11, 20, Fig. 1B, S2B). Metatranscriptomics in this study suggested that these genera also associate with reductive nitrogen transformation, i.e., denitrification, DNRA, and nitrogen fixation. Sketches of Anaeromyxobacter and Geobacter are based on electron microscope photographs published elsewhere (3, 16). †, NO2− reduction to NO is driven by common denitrifiers within Alphaproteobacteria, Betaproteobacteria, and Gammaproteobacteria (5), not by Anaeromyxobacter and Geobacter within Deltaproteobacteria; ‡, Anaeromyxobacter reduce N2O to N2, whereas Geobacter do not (Table S1).
Supplementary information
Acknowledgements
We thank the technical staff of Niigata Agricultural Research Institute for their assistance with field management, and T. Shinano (Hokkaido Agricultural Research Center, NARO), T. Hasegawa, H. Akiyama (Institute for Agro-Environmental Science, NARO), S. Asakawa (Nagoya University), S. Obata (Emeritus Professor of Mie University), and Y. Ono (Fukushima Agricultural Technology Center) for collecting and providing soil samples. This study was supported by JSPS KAKENHI Grant Numbers JP22248038, JP25252013, JP15KT0014, and JP15K14675, and the Science and Technology Research Promotion Program for Agriculture, Forestry, Fisheries and Food Industry (27004C), Japan.
References
- 1.Buresh R.J., Patrick W.H. Nitrate reduction to ammonium in anaerobic soil. Soil Sci Soc Am J. 1978;42:913–918. [Google Scholar]
- 2.Chen Z., Liu J., Wu M., Xie X., Wu J., Wei W. Differentiated response of denitrifying communities to fertilization regime in paddy soil. Microb Ecol. 2012;63:446–459. doi: 10.1007/s00248-011-9909-5. [DOI] [PubMed] [Google Scholar]
- 3.Childers S.E., Ciufo S., Lovley D.R. Geobacter metallireducens accesses insoluble Fe (III) oxide by chemotaxis. Nature. 2002;416:767–769. doi: 10.1038/416767a. [DOI] [PubMed] [Google Scholar]
- 4.Ding L.J., Su J.Q., Xu H.J., Jia Z.J., Zhu Y.G. Long-term nitrogen fertilization of paddy soil shifts iron-reducing microbial community revealed by RNA-13C-acetate probing coupled with pyrosequencing. ISME J. 2015;9:721–734. doi: 10.1038/ismej.2014.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Graf D.R., Jones C.M., Hallin S. Intergenomic comparisons highlight modularity of the denitrification pathway and underpin the importance of community structure for N2O emissions. PLoS One. 2014;9:e114118. doi: 10.1371/journal.pone.0114118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hirsch P.R., Mauchline T.H. The importance of the microbial N cycle in soil for crop plant nutrition. Adv Appl Microbiol. 2015;93:45–71. doi: 10.1016/bs.aambs.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 7.Hori T., Muller A., Igarashi Y., Conrad R., Friedrich M.W. Identification of iron-reducing microorganisms in anoxic rice paddy soil by 13C-acetate probing. ISME J. 2010;4:267–278. doi: 10.1038/ismej.2009.100. [DOI] [PubMed] [Google Scholar]
- 8.Ishii S., Ikeda S., Minamisawa K., Senoo K. Nitrogen cycling in rice paddy environments: past achievements and future challenges. Microbes Environ. 2011;26:282–292. doi: 10.1264/jsme2.me11293. [DOI] [PubMed] [Google Scholar]
- 9.Itoh H., Ishii S., Shiratori Y., Oshima K., Otsuka S., Hattori M., Senoo K. Seasonal transition of active bacterial and archaeal communities in relation to water management in paddy soils. Microbes Environ. 2013;28:370–380. doi: 10.1264/jsme2.ME13030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones C.M., Graf D.R., Bru D., Philippot L., Hallin S. The unaccounted yet abundant nitrous oxide-reducing microbial community: a potential nitrous oxide sink. ISME J. 2013;7:417–426. doi: 10.1038/ismej.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim Y., Liesack W. Differential assemblage of functional units in paddy soil microbiomes. PLoS One. 2015;10:e0122221. doi: 10.1371/journal.pone.0122221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kögel-Knabner I., Amelung W., Cao Z., Fiedler S., Frenzel P., Jahn R., Kalbitz K., Kölbel A., Schloter M. Biogeochemistry of paddy soils. Geoderma. 2010;157:1–14. [Google Scholar]
- 13.Mårtensson L., Díez B., Wartiainen I., Zheng W., El-Shehawy R., Rasmussen U. Diazotrophic diversity, nifH gene expression and nitrogenase activity in a rice paddy field in Fujian, China. Plant and Soil. 2009;325:207–218. [Google Scholar]
- 14.Nelson M.B., Martiny A.C., Martiny J.B. Global biogeography of microbial nitrogen-cycling traits in soil. Proc Natl Acad Sci USA. 2016;113:8033–8040. doi: 10.1073/pnas.1601070113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Orellana L.H., Rodriguez-R L.M., Higgins S., Chee-Sanford J.C., Sanford R.A., Ritalahti K.M., Löffler F.E., Konstantinidis K.T. Detecting nitrous oxide reductase (nosZ) genes in soil metagenomes: method development and implications for the nitrogen cycle. MBio. 2014;5:e01193–14. doi: 10.1128/mBio.01193-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanford R.A., Cole J.R., Tiedje J.M. Characterization and description of Anaeromyxobacter dehalogenans gen. nov., sp. nov., an aryl-halorespiring facultative anaerobic myxobacterium. Appl Environ Microbiol. 2002;68:893–900. doi: 10.1128/AEM.68.2.893-900.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanford R.A., Wagner D.D., Wu Q., et al. Unexpected nondenitrifier nitrous oxide reductase gene diversity and abundance in soils. Proc Natl Acad Sci USA. 2012;109:19709–19714. doi: 10.1073/pnas.1211238109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shapleigh J.P. The denitrifying prokaryotes. In: Dworkin M., Falkow S., Rosenberg E., Schleifer K.H., Stackebrandt E., editors. The Prokaryotes. Springer; New York: 2006. pp. 769–792. [Google Scholar]
- 19.Shrestha P.M., Kube M., Reinhardt R., Liesack W. Transcriptional activity of paddy soil bacterial communities. Environ Microbiol. 2009;11:960–970. doi: 10.1111/j.1462-2920.2008.01821.x. [DOI] [PubMed] [Google Scholar]
- 20.Weber K.A., Achenbach L.A., Coates J.D. Microorganisms pumping iron: anaerobic microbial iron oxidation and reduction. Nat Rev Microbiol. 2006;4:752–764. doi: 10.1038/nrmicro1490. [DOI] [PubMed] [Google Scholar]
- 21.Wei W., Isobe K., Nishizawa T., Zhu L., Shiratori Y., Ohte N., Koba K., Otsuka S., Senoo K. Higher diversity and abundance of denitrifying microorganisms in environments than considered previously. ISME J. 2015;9:1954–1965. doi: 10.1038/ismej.2015.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiong Z.Q., Huang T.Q., Ma Y.C., Xing G.X., Zhu Z.L. Nitrate and ammonium leaching in variable- and permanent-charge paddy soils. Pedosphere. 2010;20:209–216. [Google Scholar]
- 23.Yin S.X., Chen D., Chen L.M., Edis R. Dissimilatory nitrate reduction to ammonium and responsible microorganisms in two Chinese and Australian paddy soils. Soil Biol Biochem. 2002;34(8):1131–1137. [Google Scholar]
- 24.Yoshida M., Ishii S., Fujii D., Otsuka S., Senoo K. Identification of active denitrifiers in rice paddy soil by DNA- and RNA-based analyses. Microbes Environ. 2012;27:456–461. doi: 10.1264/jsme2.ME12076. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.