Abstract
The structural integrity of tissue proteins is damaged in processes ranging from remodeling of the extracellular matrix to destruction by microbial pathogens. Leukocytes play a prominent role in tissue surveillance and repair. However, it remains enigmatic what features of structurally decayed proteins prompt recognition by leukocyte cell-surface receptors. Here, we report that adhesion of human neutrophil granulocytes to fibrinogen is greatly increased by plasmin digestion in a mode where αXβ2 dominates the integrin-dependent binding. The bacterial protease subtilisin also enhances binding by αXβ2. The αX ligand binding domain has an unusually high affinity for carboxyl groups, with KD at ≈100 μM. Our findings implicate enhanced accessibility of negatively charged residues in structurally decayed proteins as a pattern recognition motif for αXβ2 integrin. Comparisons among integrins show relevance of these findings to the large number of ligands recognized by αMβ2 and αXβ2 but not αLβ2. The observations suggest that the pericellular proteolysis at the leading edge of neutrophils not only facilitates passage through the extracellular matrix but also manufactures binding sites for αXβ2.
Keywords: plasmin, scavenger receptor
The detrimental influence of unfolded or denatured proteins and the importance of regulating removal are underscored for the intracellular environment in eukaryotes by the high elaboration of the ubiquitination pathway. By contrast, little is known about how damaged proteins are recognized in the extracellular compartment, particularly given the broad range of cleavages catalyzed by the vast array of proteolytic enzymes to which the extracellular environment is exposed. Infectious agents, inflammatory cells, and processes including coagulation, wound healing, angiogenesis, and tissue remodeling in development engender structural decay of proteins. Evidence from murine leukocytes implicates so-called macrophage scavenger receptors in binding denatured collagen (1). However, the best characterized scavenger receptor ligands are a diverse range of polyanionic species such as modified low-density lipoprotein and lipopolysaccharides (2). Myeloid leukocytes bind to an exceedingly large number of protein ligands as well as denatured protein (3) through the structurally and functionally similar αXβ2 (p150,95, CD18/CD11c) and αMβ2 (Mac-1, CD18/CD11b) integrins, but the molecular basis for recognition of multiple ligands and selectivity for denatured proteins remains obscure.
Fibrinogen (Fg) is one of the best studied ligands of αMβ2 and αXβ2 (4, 5). Plasma Fg, derived from hepatic synthesis, assembles after cleavage by thrombin into fibrin in hemostasis. However, it has recently been established that Fg is also secreted by epithelial cells in inflammation and is assembled together with fibronectin independently of thrombin into fibrils in the extracellular matrix during wound repair (6, 7). The receptor for urokinase-type plasminogen activator closely associates with both αXβ2 and αMβ2 in the cell membranes of neutrophils, and the complex with αXβ2 remains stable on leading edge lamellapodia during migration (8, 9). The recruitment of activated urokinase-type plasminogen activator to the neutrophil leading edge clears a migratory path through pericellular proteolysis by plasmin of its well known substrate Fg as well as other extracellular matrix components (10, 11). However, the influence on leukocyte adhesion of proteolysis of integrin ligands is not well understood. Indeed, extracellular matrix degradation by proteases released from injured tissue or microbes could also establish a danger signal (12). Fg is a target for multiple proteases, in consequence of its labile structure (13) and large flexible and disordered regions (14). Binding of Fg to leukocyte receptors, its incorporation into the extracellular matrix, and its susceptibility to loss of structural integrity would suit Fg to act as a sentinel of tissue damage. Here, we investigate the recognition of chemically denatured or proteolyzed Fg by leukocyte cell-surface receptors and identify aberrant exposure of negatively charged residues in structurally decayed protein as a pattern recognition motif for αXβ2 integrin.
Materials and Methods
Neutrophil Adhesion Assays. Human neutrophils were isolated from freshly drawn blood as described (15) and diluted to a final concentration of 106 cells per ml in cold neutrophil dilution buffer [1 mM MgCl2/1 mM CaCl2/5 mM d-glucose/1% (vol/vol) FCS (Omega Scientific, Tarzana, CA)/150 mM NaCl/10 mM Hepes, pH 7.4]. The adhesion assay was carried out essentially as described in ref. 5. Sixty-well Terasaki plates (Nalge–Nunc International) were coated with Fg (Enzyme Research Laboratories, South Bend, IN) in coating buffer (150 mM NaCl/20 mM Tris·HCl, pH 9.0) for 1 h at 37°C in a humidified incubator. Residual binding sites were blocked by incubation of the wells with 0.05% (wt/vol) polyvinylpyrrolidone in PBS for 1 h at room temperature. Human plasma-derived plasmin (Calbiochem) and subtilisin Carlsberg (Calbiochem) in 10 μl of 10 mM CaCl2/150 mM NaCl/0.05% (vol/vol) polyoxyethylenesorbitan monolaureate/20 mM Tris·HCl, pH 7.4, were applied to the wells for 30 min at 37°C, followed by three washes in neutrophil dilution buffer. The Terasaki plates were then chilled on ice. Neutrophils were pretreated for 15 min on ice with 10 μg/ml isotypic control Ab, αX ligand binding-blocking mAb 3.9 (Biosource International, Camarillo, CA), αM-blocking mAb CBRM1/29, a combination of the αX and αM blocking mAbs, excess (100 μg/ml) β2 blocking mAb YCF5, 2 ng/ml recombinant TNF-α (Sigma), or 10 mM EDTA in neutrophil dilution buffer. The Terasaki plate wells were aspirated, and plates were placed back on ice. Cells were immediately added (5 μl in each of six wells), and, after 30 min at 4°C, the plates were transferred to 37°C for 15 min and then washed 10 times by being manually dipped into a container with 155 mM NaCl/1 mM potassium phosphate/2 mM sodium phosphate, pH 7.4. The Terasaki plates were inverted for 30 min (retaining the well contents by surface tension), and the washing was repeated; the cells were then fixed with formaldehyde. For each experimental condition, phase-contrast images representing 80% of the surface of each of six separate wells were obtained, and the number of cells in each well was scored from printed micrographs.
K562 Transfectant Cell Adhesion Assays. V-well microtiter plates (Corning, Corning, NY) were coated with Fg and treated with proteases as described above. iC3b (Calbiochem) was diluted in coating buffer and incubated as described for Fg. In addition to plasmin and subtilisin, the plastic-immobilized Fg was treated with human neutrophil elastase (Sigma) and bovine thrombin (Calbiochem). The digestion was stopped by washing the wells four times in L15 medium (Sigma) supplemented with 10% (vol/vol) FBS and 10 mM Hepes, pH 7.4 (L15/FBS). Denaturation of the plastic-immobilized Fg was induced by applying to each well for 1.5 h at room temperature 100 μl of 6 M guanidine hydrochloride dissolved in Fg coating buffer, followed by blocking with polyvinylpyrrolidone as above. The denaturation was tested by ELISA (Zymed) in the plastic wells with conformation-sensitive mAb 313 (American Diagnostica, Greenwich, CT) and mAb 2C2-G7 (BD Biosciences, San Diego) insensitive to Fg. αMβ2- and αXβ2-expressing K562 cells (16, 17) were cultured in RPMI medium 1640 with 2 mM Gln, 10 units/ml penicillin, 10 μg/ml streptomycin, and 10% (vol/vol) FCS in the presence of 4 μg/ml puromycin (Sigma) or 200 μg/ml hygromycin B (Invitrogen), respectively, and fluorescently labeled as described in ref. 18. The cells were resuspended in L15/FBS to 106 cells/ml. To activate integrins, the Mg2+- and Ca2+-containing medium was supplemented with 1.5 mM MnCl2 followed by incubation for 15–20 min at 37°C. Cell suspension (100 μl) was added to each well, incubated at 37°C for 30 min, followed by centrifugation at room temperature for 5 min at 378 × g. Fluorescence signals were read in a fluorescence concentration analyzer (Idexx Laboratories, Westbrook, MA), and the fraction of binding cells in each well was estimated by comparison with the signal from uncoated wells blocked with polyvinylpyrrolidone as described (19). The amount of protein remaining in the well after proteolytic digestion was determined by colorimetric detection with a bicinchoninic acid protein assay kit (Pierce).
Analysis of Recombinant I Domain Binding. Wild-type and Ile-314→Gly mutated αX I domains were produced and applied in surface plasmon resonance (SPR) assays as described in ref. 20. For probing the influence of proteolysis on ligand properties, ≈10,000 response units of Fg (corresponding to 10 ng of Fg per mm2 of flow cell surface) was immobilized and digested with 1.9 milliunits/ml plasma-derived human plasmin (Calbiochem) diluted as described for the cell adhesion assays. The digestion was monitored in real time by observing the decline in resonance units and stopped with injection of a mixture of covalent and noncovalent protease inhibitors (Complete; Roche) to obtain a digestion of ≈50% (wt/wt). Denaturation of Fg was induced by flowing in 300 μl over 30 min of 6 M guanidine dissolved in running buffer (20). For determining the affinity of the αX I domain to amino acids, other small molecules, poly-l-Glu (catalog no. P-4761, Sigma), porcine heparin (catalog no. H-8537, Sigma), and shark cartilage chondroitin 6-sulfate, type C (catalog no. C-4384, Sigma), the compounds were dissolved in SPR running buffer, and the stock solution was adjusted to pH 6.0, with the Mg2+ concentration raised to 10 mM to avoid any influence from divalent cation chelation by the compounds. Various concentrations of each compound were mixed with a fixed concentration of 537 nM αX I domain. The binding properties of the high-affinity αX I domain were compared with the equivalent αM I domain, i.e., comprising the amino acid sequence from Glu-123 to Gly-321, with a Cys-128→Ser substitution, a C-terminal His-6 tag, and an Ile-316→Gly mutation to activate the domain. The bacterial expression and purification were carried out as described in ref. 20 for the αX I domain, except that we employed a 150 mM NaCl/20 mM Tris·HCl, pH 7.4, buffer for the final gel permeation chromatography purification. This buffer supplemented with 1 mM MgCl2 was also used as SPR running buffer. The interaction between intercellular adhesion molecule-1 (ICAM-1) and the disulfide-linked high-affinity αLβ2 I domain was tested as described in ref. 21. Denaturation of the ICAM was carried out as described above, supplementing, however, the guanidine buffer with 1 mM 1,4-DTT. The affinity of the αM and αL I domains for compounds was tested as for the αX I domain in Tris buffer with 10 mM MgCl2 with domain concentrations of 3 μM and 500 nM, respectively. Direct binding of the αX I domains was tested by incubating either wild-type or mutated I domain with Glu or Lys coupled through their α-amino groups and a one-carbon atom spacer to cyanogen bromide-activated 4% beaded agarose (catalog nos. G-2759 and L-5631, Sigma). The I domains were applied at concentrations of 300 μg/ml running buffer with 1 mM Mg2+ or 1 mM EDTA and incubated for 30 min, followed by two brief washes in running buffer. The beads were eluted in buffer with 25 mM EDTA, and the eluates were subjected to SDS/PAGE and Coomassie staining. The SPR data were analyzed by fitting the single-species Langmuir equation to the sensorgrams with the biaevaluation software (Biacore, Uppsala) or as described by Svitel et al. (22) with a correction for transport effects in the matrix (unpublished observations).
Results
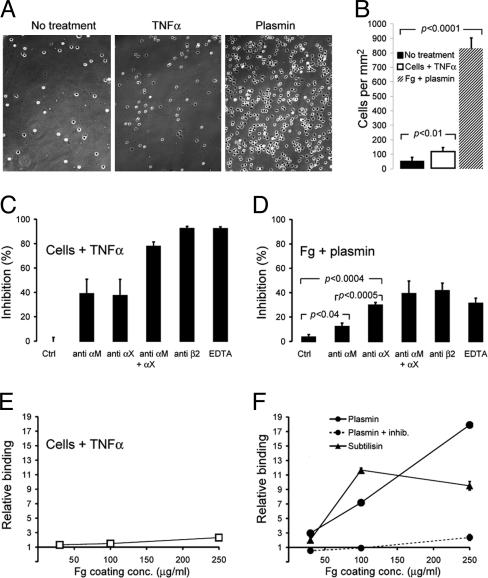
Neutrophil Adhesion to Protease-Digested Fg. To analyze how neutrophils respond to digestion of substrate for integrin-dependent adhesion, binding to proteolyzed Fg was tested (Fig. 1). Contrary to the hypothesis that proteolysis would remove binding sites, plasmin digestion of Fg enhanced neutrophil adhesion many fold (Fig. 1 A and B). Furthermore, a bacterial protease with markedly less substrate specificity, subtilisin (23), also strongly enhanced adhesion (Fig. 1F). Binding correlated with the Fg-coating concentration, indicating that other proteins, either released from the cells or contributed by 1% serum added to the medium to quench residual proteolytic activity, did not contribute significantly to the adhesion (Fig. 1F). Omission of serum from the neutrophil medium did not affect the enhanced binding to plasmin-treated Fg (data not shown). Blocking mAbs and use of EDTA demonstrated that a substantial portion of the adhesion to the plasmin-treated Fg was integrin-dependent and demonstrated the primary role of integrin αXβ2 in this adhesion (Fig. 1D). This is particularly striking, given that αXβ2 is expressed in ≈4-fold lesser amounts on the neutrophil surface than αMβ2 (24). For comparison, we also examined TNF-α-stimulated adhesion to Fg-coated surfaces (Fig. 1 A and B). As reported earlier (5), the binding is entirely mediated by β2 integrins with a significant contribution by αXβ2 (Fig. 1 C and E), in some cases equaling or exceeding the contribution by αMβ2 (5). Nonetheless, TNF-α is a weaker agent than proteolyzed/denatured Fg in the extent to which it stimulates overall neutrophil adhesion (Fig. 1 A and B) and, more specifically, in the total amount of αXβ2-dependent adhesion (Fig. 1 C–F) it promotes.
Fig. 1.
Neutrophil adhesion to immobilized Fg. (A) Micrographs of wells with unstimulated (Left) or TNF-stimulated (Center) human neutrophils binding to Fg, or unstimulated neutrophils binding to plasmin-treated Fg (Right). (B) Average density of neutrophil binding from six different experiments with five different donors (mean value ± SEM). (C) The contribution of integrins to adhesion by TNF-α-stimulated neutrophils was analyzed by addition of an isotypic control Ab, a function blocking anti-αM mAb (CBRM1/29), a function blocking anti-αX mAb (3.9), a combination of these Abs, a function blocking β2 Ab (YCF5.1), or 10 mM EDTA. The percentage of inhibition was calculated from comparison with neutrophil binding in the absence of any addition. (D) Adhesion of unstimulated neutrophils to plasmin-treated Fg. Abs or EDTA were applied as in C. Experiments in A–D were carried out in parallel with wells coated with Fg at a concentration of 250 μg/ml. (E and F) The influence of Fg coating on neutrophil adhesion. (E) Binding of TNF-α-stimulated neutrophils. The binding under stimulating conditions at each coating concentration was divided by the binding by unstimulated neutrophils applied in parallel to give relative binding. (F) Binding of unstimulated neutrophils to protease-treated Fg surfaces. In all panels, surfaces were either untreated or treated with 1.3 μM subtilisin, 24 nM plasmin, or 24 nM plasmin in the presence of protease inhibitors. The binding at each coating concentration was divided by the binding in parallel to untreated surfaces to give the relative binding.
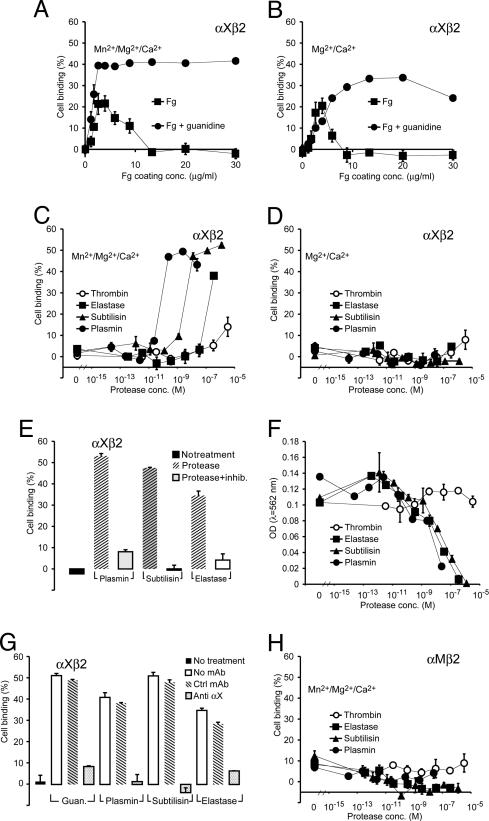
Adherence by αMβ2 and αXβ2 Transfectants to Proteolyzed or Guanidine-Treated Fg. A more detailed picture of αMβ2 and αXβ2 integrin ligand preferences was obtained from cell adhesion studies with K562 cell transfectants (Fig. 2). Binding of αXβ2/K562 transfectants increased and then decreased to baseline at higher Fg coating concentration (Fig. 2 A and B), whereas the amount of Fg immobilized steadily increased with coating concentration (see Fig. 5A, which is published as supporting information on the PNAS web site). The sharp peak in cell binding at a coating concentration of ≈3 μg Fg/ml (Fig. 2 A and B) is in quantitative agreement with the finding that adsorption to surfaces at concentrations <10 μg of Fg/ml promotes loss in structure as shown by spectroscopy (13) and binding by conformation-specific mAbs (25). Guanidine treatment converted the native, nonbinding form adsorbed at higher Fg concentrations to a maximally adhesive form (Fig. 2 A and B), whereas it decreased binding by an Fg Ab preferentially recognizing the native conformation (Fig. 5B). In agreement with the results for neutrophils described above, protease treatment of native Fg enabled it to efficiently support adhesion of activated αXβ2/K562 transfectants (Fig. 2 C–E, G). Although the efficiency of activating binding differed between plasmin, subtilisin, and human neutrophil elastase (Fig. 2C), they removed similar amounts of adsorbed Fg (Fig. 2F). Furthermore, plasmin and subtilisin activated adhesiveness at protease concentrations that removed little overall Fg. These findings rule out an alteration in protein sorption on the plastic surface as the mechanism for substrate activation. Naked plastic wells incubated with the serum-containing K562 cell medium did not support any binding by the αXβ2 transfectants (data not shown). Importantly, because the native structure of Fg was retained by coating of the molecule at high protein densities the observed proteolysis and its consequences on ligand activation is likely to be possible in a physiologic presentation of the substrate. The most specific protease, thrombin, removed little Fg and activated little binding. By contrast to the results with αXβ2, activated αMβ2/K562 transfectants bound weakly to Fg, and this binding was not enhanced with proteolysis (Fig. 2H), even though the αMβ2/K562 cells were as active as αXβ2/K562 cells in binding to the cleaved complement component iC3b (Fig. 5C).
Fig. 2.
Binding assays with K562 cells expressing recombinant αXβ2 or αMβ2 integrin. For each condition, binding was measured in triplicate wells and stated as mean ± SEM. (A and B) Binding of αXβ2/K562 cells in wells incubated with various concentrations of Fg with or without subsequent treatment by 6 M guanidine. (C and D) Protease induction of αXβ2/K562 cell binding. Wells were coated with 100 μg/ml Fg and treated with plasmin, subtilisin, or neutrophil elastase. All cell adhesion studies were carried out in the presence of 10% (vol/vol) FCS to avoid any effect of residual enzymatic activity in the wells on cellular function. (E) Wells were coated as in C and treated with 1.9 milliunit/ml plasmin (24 nM), 1 milliunit/ml subtilisin (1.3 μM), or 1.6 unit/ml human neutrophil elastase (354 nM) with or without protease inhibitors. (F) Bicinchoninic acid assay of the amount of Fg remaining in the wells after incubation with proteases. (G) Specificity of interaction with αXβ2/K562 cells. A blocking (3.9) or isotypic control Ab to αX was mixed with cells before application to wells either treated with guanidine or proteases. (H) Interaction between proteolyzed Fg and αMβ2/K562 expressing cells was tested in the presence of Mn2+ as for the αXβ2/K562 cells.
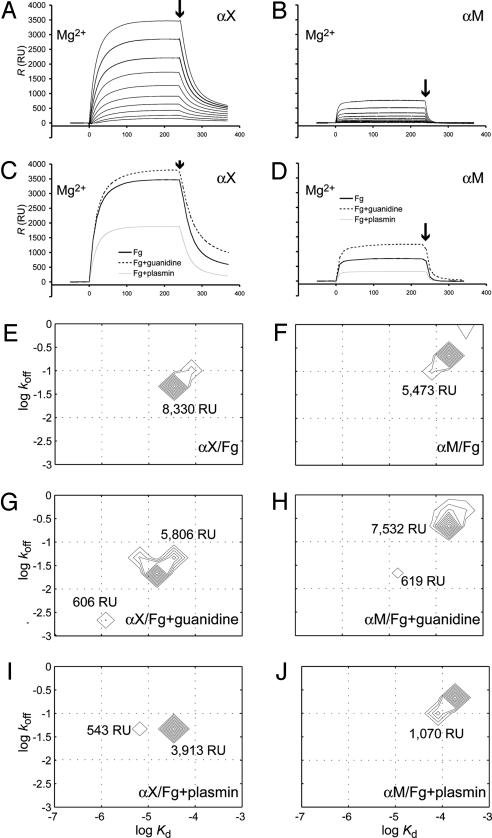
Analysis by SPR of αM and αX I Domain Binding to Native, Proteolyzed, or Guanidine-Treated Fg. The differences between αMβ2 and αXβ2 were mirrored in SPR assays with αM and αX I domains stabilized in their active conformations (20, 26), where the αX I domain generated a severalfold higher response than αM I domain in the binding to Fg (Fig. 3 A and B). With the SPR assays, it is possible to quantify the absolute mass of Fg and I domain bound to the surfaces because this is linearly related to change in refractive index at the surface (see Fig. 6G, which is published as supporting information on the PNAS web site). Remarkably, the number of bound αX I domains per immobilized Fg molecule at saturation was ≈12.5 as estimated by the single-species Langmuir equation (Fig. 6G). This is likely an underestimate of the ratio for soluble Fg, as seen with the αL I domain binding to ICAM-1, where saturation was reached at 0.4 αL I domain per ICAM-1, consistent with binding to a single site and the inaccessibility of this site on a fraction of the immobilized ICAM-1 molecules (Fig. 6G). To make a robust estimate of affinities and saturating levels of αX I domain binding sites on the immobilized Fg and to resolve the potentially important issue of heterogeneity in the ensemble of interactions, the widely used single-species Langmuir equation is insufficient. Recently, a methodology was developed, which extracts the distribution of affinities and kinetic properties for heterogeneous ligand interactions from the SPR sensorgrams without a priori assumptions concerning the number of different classes of interactions or constraints on the shape of the distribution (22). In this setting, the observed SPR signal from the I domains binding to the immobilized Fg is considered as a sum of multiple classes of interactions, each class of sites after individual Langmuir adsorption isotherms. By analyzing the observed kinetic properties of I domain binding, it is possible to acquire the distribution of kon and koff values, and hence the KD, together with the abundance of the classes of binding sites. This approach allowed us rigorously to test the conversion of Fg ligand properties on exposure to guanidine or plasmin (Figs. 3 and 6 A and B). Native Fg bound both the αX and αM I domains in a largely homogenous mode with affinities (KD) of ≈50 μM for the αX I domain and 200 μM for the αM I domain (Fig. 3 E and F) with the associated saturating amounts of I domain estimated to be 18.2 and 11.0 per Fg molecule, respectively. As a control, we also tested the αL I domain binding to ICAM-1 (Fig. 6 E and F), which estimated binding of 0.56 I domains per ICAM-1 molecule. Upon treatment of Fg with guanidine, two subpopulations of binding sites were observed (Fig. 3 G and H). The major subpopulation was shifted to higher affinity for αX (Fig. 3G) but was essentially unchanged for αM (Fig. 3H), in agreement with single species Langmuir estimates (Fig. 6G). The minor subpopulation of αX binding sites has a higher affinity with KD of 2 μM (Fig. 3G). The number of high-affinity binding sites (606 response units) corresponds to 1.3 I domains bound per Fg dimer. The ratio of 0.65 I domains per guanidine-treated Fg monomer suggests partial exposure of a single, high-affinity binding site. Similarly, the minor subpopulation of αM binding sites (619 response units) binds 0.65 I domain/Fg monomer with a KD of 12 μM (Fig. 3H). These observations agree with mapping of a linear binding epitope for the αX and αM I domain in the C-terminal part of the Fg γ-chain (27). In transgenic mice, mutation of this segment ablated αMβ2-mediated binding by primary human neutrophils to murine Fg (28), and unmasking of this site enhanced binding by the αMβ2 integrin to human Fg (29). The specificity of these results for αM and αX is emphasized by comparison to the αL I domain, where unmasking is not required for binding to ICAM-1, and guanidine treatment indeed weakened binding (Fig. 6G).
Fig. 3.
The interaction between Fg and high-affinity αX and αM I domains measured by SPR. The I domains were applied in parallel to flow cells coupled with native (A and B), proteolyzed, or guanidine-treated (C and D)Fginthe presence of 1 mM MgCl2. The affinity for native, proteolyzed, or guanidine-treated Fg for the I domains was measured with a range of 10 concentrations of I domains from 0.28 to 10.6 μM (as shown for native Fg in A and B). For comparison, sensorgrams are shown in C and D from injections of the I domains at the highest applied concentration of 10.6 μM over the surfaces with native, proteolyzed, or guanidine-treated Fg. The end of injection phase is indicated with arrows. (E–J) Two-dimensional off-rate-constant and affinity distribution analyses for heterogeneous surface sites. The calculation was carried out for surfaces with native (E and F), guanidine-treated (G and H), or plasmin-treated (I and J) Fg for measurements either with the αX or αM I domain. The distribution of species at different rate and equilibrium constants is indicated by contour lines, and the total abundance of binding sites in each peak was obtained by integration of the peaks and labeled in response units (RU).
Plasmin treatment of Fg also exposed a high-affinity binding site for the αX I domain. About 10% (543 response units) of the total binding was shifted to a KD of 6 μM, i.e., an ≈10-fold higher affinity than the bulk of the interactions (Fig. 3I). Similarly to the results with guanidine-treated Fg, this shows a qualitative difference between the native and plasmin-digested Fg as ligands for the αX I domain. This finding is significant in the comparison with the cell adhesion studies because it explains why, despite net loss of protein, the surfaces treated with proteases were able to support strong cell adhesion through the αXβ2 integrin. A similar mode of on-rate-driven affinity increase was discernible with guanidine-treated Fg (Fig. 3G), suggesting that partial unfolding occurs as a consequence of proteolysis in agreement with earlier spectroscopic studies (30). However, by contrast to the guanidine-treated Fg, no koff-driven affinity increase for the αX I domain was observed (Fig. 3 G and I). This offers a simple explanation for the requirement of integrin activation with manganese to obtain cell adhesion through αXβ2 to protease-treated Fg (Fig. 2 C and D), whereas denatured Fg supported binding in the absence of exogenous integrin activation as a quantitatively more potent ligand (Fig. 2 A and B). Furthermore, plasmin treatment did not enhance binding of the αM I domain to Fg (Fig. 3J), in agreement with the cell adhesion studies (Fig. 2H).
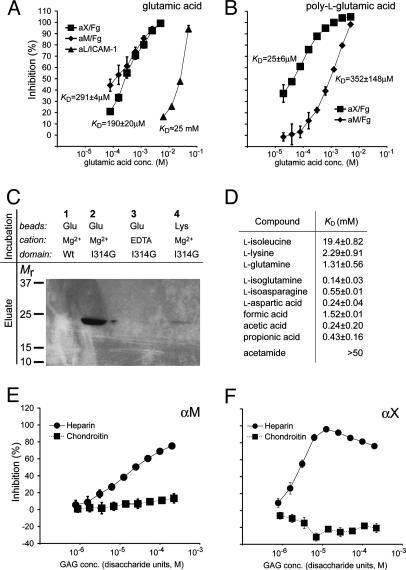
Anionic Species Are Potent Ligands for the αX and αM I Domains. Ligand binding by integrin I domains is Mg2+-dependent (Fig. 6 C and D), and crystal structures show binding of the Mg2+ to acidic side chains in ligands (31–33). Considering the diversity of αX and αM ligands, and the presence of multiple recognition sites within a single protein ligand, we considered the possibility that the key ligand binding motif might reduce to a single acidic side chain. Indeed, the KD for Glu binding to both the αX and αM I domains determined by SPR inhibition assays (34) of binding to Fg was ≈200 μM (Fig. 4A). By contrast, the KD for inhibition of the much more specific binding of the αL I domain to ICAM-1 was 100-fold weaker, at 25 mM. Compared to binding to Asp and Glu with side chain carboxyl groups, acetic and propionic acids also showed good potency, whereas binding to amino acids with only free α-carboxyl groups was 10- to 100-fold less potent (Fig. 4D). This suggests preferential binding in cleaved proteins to side chain rather than α carboxyl groups. Interestingly, l-isoglutamine, which has amidated α-carboxyl and free γ-carboxyl groups, was the most potent inhibitor (KD = 140 μM, Fig. 4D) and also the closest analogue of a Glu residue in a peptide backbone. Direct binding was demonstrated by a conformation-dependent and Mg2+-dependent association between glutamate-coupled beads and the high-affinity αX I domain (Fig. 4C). Although specific residues around the metal ion-dependant adhesion site must contribute some specificity to the wide range of ligands recognized (35–37), the findings suggest that the promiscuous ligand recognition by αXβ2 and αMβ2 is primarily because of high affinity for acidic side chains. Other cell adhesion receptors such as CD2 rely on ligand affinities in the order of 100 μM (38), and therefore both the low- and high-affinity sites on Fg for the αM and αX I domains are in a physiologically meaningful range.
Fig. 4.
Interaction between I domain and charged compounds. (A and B) The interaction between the high-affinity αX, αM, and αL I domains and acidic molecules was measured by SPR inhibition assays as described by Karlsson (34). A standard curve for the interaction between various concentrations of each I domain and immobilized ligand (Fg for αX and αM, or ICAM-1 for αL) was established. (A) The I domains were mixed with Glu (inhibitor) in concentrations as indicated, and the response level was converted by use of the standard curve to an estimate of the free amount of I domain (not bound to inhibitor) at each concentration of inhibitor. The amount of inhibitor-bound and free I domain was used to calculate KD. (B) Inhibition of the αX and αM I domains to Fg with poly-l-Glu is shown together with the KD values calculated as in A, with the concentration of inhibitor corresponding to the total Glu concentration. (C) Direct binding of the αX I domain to Glu. Either the wild-type or high-affinity I314G αX I domain was incubated with Glu-coupled beads in the presence of Mg2+ or EDTA as indicated, followed by elution with EDTA, SDS/PAGE, and Coomassie staining. (D) KD values for binding of the αX I domain to various small compounds determined as in A. (E and F) Inhibition of the αM (E) and αX (F) I domain binding to Fg in the presence of heparin or chondroitin sulfate C. Concentrations are given as the molar concentrations of the sulfated disaccharide units of each glycosaminoglycan (GAG) polymer, for comparison to other inhibitors. The Mr of these units was estimated to be ≈500 and 445 for heparin and chondroitin sulfate, respectively.
Poly-l-Glu was examined as a model for interaction with unfolded, acidic regions in proteins. Interestingly, the affinity of the αX I domain for poly-l-Glu (KD = 25 μM) was ≈10-fold higher than for monomeric Glu, whereas the affinity of the αM I domain for monomeric and polymeric Glu was similar (Fig. 4B). This correlates with the higher affinity of αX but not αM I domain for proteolyzed Fg (Fig. 3 I and J). Furthermore, the affinity of αX I domain for poly-Glu of 25 μM is in the same range as the high-affinity site on plasmin-digested Fg (6 μM). Overall, the results suggest that the αX I domain recognizes proteolyzed and denatured linear polypeptide segments where the polyanionic character is no longer restricted by higher-order protein structure.
Heparin is another linear, polyanionic molecule. αMβ2 and αXβ2 bind to heparin and not chondroitin sulfate (39), and to the glucuronic acid derivative glucuronoxylomannan (40). In agreement, we found inhibition by heparin and not chondroitin sulfate of I domain binding to Fg (Fig. 4 E and F). These glycosaminoglycans each contain 50% uronic acids and sulfates, but in different stereochemistries. The stereochemistry of the anionic sulfate and carboxyl groups is fixed by the covalent structure of these glycosaminoglycans, in contrast to the stereochemistry of carboxyl groups in proteins, which depends highly on overall protein conformation. Heparin is released by mast cell degranulation, and thus also functions as a danger signal.
Discussion
The stronger interaction between the αX I domain and heparin compared with the αM I domain recapitulates the findings on the binding of polyglutamate and proteolyzed Fg. There is a most interesting structural correlate. In the αX I domain, a groove containing positively charged residues runs through the metal ion-dependent adhesion site (20), resembling heparin binding sites in other proteins (41). However, in αM this positively charged groove is interrupted by the substitution of a Glu for Lys-242 in αX (see figure 1 of ref. 20).
We find that acidic residues that are exposed in denatured or proteolyzed Fg function as a damage tag for recognition by the integrin αXβ2. The similarity with scavenger receptor recognition of polyanions is striking. Intriguingly, as seems to be a general characteristic of scavenger receptors (2), αXβ2 converges functions in homeostasis and innate immune defense. The binding sites in extracellular matrix manufactured through plasmin proteolysis provide a functional explanation for the stable association between αXβ2 and urokinase-type plasminogen activator receptor on the lamellipodium of migrating neutrophils (8, 9). In scenarios where unchecked proteolysis is inflicted by pathogens or tissue destruction, the structural decay of Fg may set off neutrophil accumulation through αXβ2-mediated adhesion in cooperation with other proinflammatory stimuli. The aberrant negative charge exposure by proteolyzed Fg is in this sense a danger signal akin to the release of cytoplasmic constituents such as uric acid from dying cells (42) with the origin linked with damage or injury to tissues.
Supplementary Material
Acknowledgments
We thank Barry S. Coller and Samuel C. Silverstein for reviewing the manuscript. This work was supported by National Institutes of Health Grant CA31799 and a fellowship from the Carlsberg Foundation (to T.V.-J.).
Abbreviations: Fg, fibrinogen; SPR, surface plasmon resonance; ICAM-1, intercellular adhesion molecule-1.
References
- 1.Gowen, B. B., Borg, T. K., Ghaffar, A. & Mayer, E. P. (2000) Matrix Biol. 19, 61–71. [DOI] [PubMed] [Google Scholar]
- 2.Gordon, S. (2002) Cell 111, 927–930. [DOI] [PubMed] [Google Scholar]
- 3.Davis, G. E. (1992) Exp. Cell Res. 200, 242–252. [DOI] [PubMed] [Google Scholar]
- 4.Wright, S. D., Weitz, J. I., Huang, A. D., Levin, S. M., Silverstein, S. C. & Loike, J. D. (1988) Proc. Natl. Acad. Sci. USA 85, 7734–7738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loike, J. D., Sodeik, B., Cao, L., Leucona, S., Weitz, J. I., Detmers, P. A., Wright, S. D. & Silverstein, S. C. (1991) Proc. Natl. Acad. Sci. USA 88, 1044–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pereira, M., Rybarczyk, B. J., Odrljin, T. M., Hocking, D. C., Sottile, J. & Simpson-Haidaris, P. J. (2002) J. Cell Sci. 115, 609–617. [DOI] [PubMed] [Google Scholar]
- 7.Rybarczyk, B. J., Lawrence, S. O. & Simpson-Haidaris, P. J. (2003) Blood 102, 4035–4043. [DOI] [PubMed] [Google Scholar]
- 8.Kindzelskii, A. L., Laska, Z. O., Todd, R. F., III, & Petty, H. R. (1996) J. Immunol. 156, 297–309. [PubMed] [Google Scholar]
- 9.Kindzelskii, A. L., Eszes, M. M., Todd, R. F., III, & Petty, H. R. (1997) Biophys. J. 73, 1777–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murphy, G. & Gavrilovic, J. (1999) Curr. Opin. Cell Biol. 11, 614–621. [DOI] [PubMed] [Google Scholar]
- 11.Petty, H. R., Worth, R. G. & Todd, R. F., III (2002) Immunol. Res. 25, 75–95. [DOI] [PubMed] [Google Scholar]
- 12.Gallucci, S. & Matzinger, P. (2001) Curr. Opin. Immunol. 13, 114–119. [DOI] [PubMed] [Google Scholar]
- 13.Kim, J. & Somorjai, G. A. (2003) J. Am. Chem. Soc. 125, 3150–3158. [DOI] [PubMed] [Google Scholar]
- 14.Yang, Z., Mochalkin, I., Veerapandian, L., Riley, M. & Doolittle, R. F. (2000) Proc. Natl. Acad. Sci. USA 97, 3907–3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carman, C. V., Jun, C.-D., Salas, A. & Springer, T. A. (2003) J. Immunol. 171, 6135–6144. [DOI] [PubMed] [Google Scholar]
- 16.Petruzzelli, L., Luk, J. & Springer, T. A. (1995) in Leucocyte Typing V: White Cell Differentiation Antigens, eds. Schlossman, S. F., Boumsell, L., Gilks, W., Harlan, J., Kishimoto, T., Morimoto, T., Ritz, J., Shaw, S., Silverstein, R., Springer, T., et al. (Oxford Univ. Press, New York), pp. 1581–1585.
- 17.Lu, C., Ferzly, M., Takagi, J. & Springer, T. A. (2001) J. Immunol. 166, 5629–5637. [DOI] [PubMed] [Google Scholar]
- 18.Lu, C. & Springer, T. A. (1997) J. Immunol. 159, 268–278. [PubMed] [Google Scholar]
- 19.Weetall, M., Hugo, R., Friedman, C., Maida, S., West, S., Wattanasin, S., Bouhel, R., Weitz-Schmidt, G. & Lake, P. (2001) Anal. Biochem. 293, 277–287. [DOI] [PubMed] [Google Scholar]
- 20.Vorup-Jensen, T., Ostermeier, C., Shimaoka, M., Hommel, U. & Springer, T. A. (2003) Proc. Natl. Acad. Sci. USA 100, 1873–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shimaoka, M., Lu, C., Palframan, R., von Andrian, U. H., Takagi, J. & Springer, T. A. (2001) Proc. Natl. Acad. Sci. USA 98, 6009–6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Svitel, J., Balbo, A., Mariuzza, R. A., Gonzales, N. R. & Schuck, P. (2003) Biophys. J. 84, 4062–4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meldal, M., Svendsen, I., Breddam, K. & Auzanneau, F. I. (1994) Proc. Natl. Acad. Sci. USA 91, 3314–3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lanier, L. L., Arnaout, M. A., Schwarting, R., Warner, N. L. & Ross, G. D. (1985) Eur. J. Immunol. 15, 713–718. [DOI] [PubMed] [Google Scholar]
- 25.Moskowitz, K. A., Kudryk, B. & Coller, B. S. (1998) Thromb. Haemostasis 79, 824–831. [PubMed] [Google Scholar]
- 26.Xiong, J.-P., Li, R., Essafi, M., Stehle, T. & Arnaout, M. A. (2000) J. Biol. Chem. 275, 38762–38767. [DOI] [PubMed] [Google Scholar]
- 27.Ugarova, T. P. & Yakubenko, V. P. (2001) Ann. N.Y. Acad. Sci. 936, 365–385. [DOI] [PubMed] [Google Scholar]
- 28.Flick, M. J., Du, X., Witte, D. P., Jirouskova, M., Soloviev, D. A., Busuttil, S. J., Plow, E. F. & Degen, J. L. (2004) J. Clin. Invest. 113, 1596–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lishko, V. K., Kudryk, B., Yakubenko, V. P., Yee, V. C. & Ugarova, T. P. (2002) Biochemistry 41, 12942–12951. [DOI] [PubMed] [Google Scholar]
- 30.Azpiazu, I. & Chapman, D. (1992) Biochim. Biophys. Acta 1119, 268–274. [DOI] [PubMed] [Google Scholar]
- 31.Lee, J.-O., Rieu, P., Arnaout, M. A. & Liddington, R. (1995) Cell 80, 631–638. [DOI] [PubMed] [Google Scholar]
- 32.Emsley, J., Knight, C. G., Farndale, R. W., Barnes, M. J. & Liddington, R. C. (2000) Cell 101, 47–56. [DOI] [PubMed] [Google Scholar]
- 33.Shimaoka, M., Xiao, T., Liu, J.-H., Yang, Y., Dong, Y., Jun, C.-D., McCormack, A., Zhang, R., Joachimiak, A., Takagi, J., et al. (2003) Cell 112, 99–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karlsson, R. (1994) Anal. Biochem. 221, 142–151. [DOI] [PubMed] [Google Scholar]
- 35.Yakubenko, V. P., Lishko, V. K., Lam, S. C. & Ugarova, T. P. (2002) J. Biol. Chem. 277, 48635–48642. [DOI] [PubMed] [Google Scholar]
- 36.Li, R., Rieu, P., Griffith, D. L., Scott, D. & Arnaout, M. A. (1998) J. Cell Biol. 143, 1523–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rieu, P., Sugimori, T., Griffith, D. L. & Arnaout, M. A. (1996) J. Biol. Chem. 271, 15858–15861. [DOI] [PubMed] [Google Scholar]
- 38.Davis, S. J., Ikemizu, S., Wild, M. K. & van der Merwe, P. A. (1998) Immunol. Rev. 163, 217–236. [DOI] [PubMed] [Google Scholar]
- 39.Diamond, M. S., Alon, R., Parkos, C. A., Quinn, M. T. & Springer, T. A. (1995) J. Cell Biol. 130, 1473–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taborda, C. P. & Casadevall, A. (2002) Immunity 16, 791–802. [DOI] [PubMed] [Google Scholar]
- 41.Mulloy, B. & Linhardt, R. J. (2001) Curr. Opin. Struct. Biol. 11, 623–628. [DOI] [PubMed] [Google Scholar]
- 42.Shi, Y., Evans, J. E. & Rock, K. L. (2003) Nature 425, 516–521. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.