Abstract
It has long been suspected that sensory signal transmission is inhibited in the mammalian brain during sleep. We hypothesized that Cav3.1 T-type Ca2+ channel currents inhibit thalamic sensory transmission to promote sleep. We found that T-type Ca2+ channel activation caused prolonged inhibition (>9 s) of action-potential firing in thalamic projection neurons of WT but not Cav3.1 knockout mice. Inhibition occurred with synaptic transmission blocked and required an increase of intracellular Ca2+. Furthermore, focal deletion of the gene encoding Cav3.1 from the rostral–midline thalamus by using Cre/loxP recombination led to frequent and prolonged arousal, which fragmented and reduced sleep. Interestingly, sleep was not disturbed when Cav3.1 was deleted from cortical pyramidal neurons. These findings support the hypothesis that thalamic T-type Ca2+ channels are required to block transmission of arousal signals through the thalamus and to stabilize sleep.
Keywords: insomnia, α1G, thalamus, arousal, Cre recombinase
Each year, 60 million Americans experience frequent or extended periods of insomnia characterized by difficulty initiating or maintaining sleep. Understanding how sleep is produced should provide insight into insomnia and identify targets for its treatment.
Sensory perceptions and motor responses are blunted during sleep. Because sensory and motor signals are relayed to and from the cerebral cortex through the thalamus, inhibition of thalamocortical signal transmission could play an important role in achieving stable sleep. Consistent with this notion, the firing response of thalamic projection neurons to visual stimulation is suppressed during sleep in primates (1). Although not yet shown, visceral sensory signals (e.g., respiration), which relay through midline thalamus, represent a potent arousal signal and, therefore, may also be suppressed to facilitate sleep (2). The molecular and cellular basis for sensory suppression during sleep and its potential role in preventing arousal have not been well studied.
One potential mechanism for sensory inhibition is the marked increase of T-type Ca2+ channel-mediated Ca2+ transients observed in thalamus during sleep (3, 4). Thalamic neurons hyperpolarize during sleep, causing T-type Ca2+ channels to deinactivate and enabling their activation with excitatory input (4–6). Cav3.1 is the major T-type Ca2+ channel transcript in thalamic projection neurons, where it is required for low-threshold Ca2+ spikes and action-potential bursts (7–9). Interestingly, in animals with Cav3.1 T-type Ca2+ channel globally deleted pain-evoked action-potential firing in thalamic neurons of the ventrobasal nucleus is increased (9). The cellular mechanism of this Cav3.1 control of action-potential firing is difficult to deduce based on data from a global knockout (KO) because Cav3.1 is very broadly expressed throughout the brain. Nevertheless, the finding led us to hypothesize that thalamic Cav3.1 may inhibit action-potential firing directly and that this inhibition may serve to stabilize sleep.
To assess the role of Cav3.1 in thalamic projection neurons, we examined its effect on action-potential firing in vitro. To determine whether this thalamic function of Cav3.1 plays a role in sleep, we selectively deleted the gene from thalamic projection neurons and examined vigilance states by recording the electroencephalogram (EEG) and electromyogram (EMG). We found that Cav3.1 activation initiates a prolonged inhibition of action-potential firing in thalamic projection neurons in vitro. We also found that targeted deletion of Cav3.1 gene in thalamocortical neurons destabilizes sleep.
Materials and Methods
General Animal Handling. Mice were housed in a pathogen-free environment regulated at 21–23°C under a 12:12-h light/dark schedule (lights on from 7 a.m. to 7 p.m.). All procedures relating to animal care and treatment were approved by the Massachusetts Institute of Technology and Harvard Medical School Institutional Animal Care and Use Committees, and they conformed to institutional and National Institutes of Health guidelines.
Generation of Thalamic-Cav3.1 KO Mice. A 120-kb-long bacterial artificial chromosome (BAC) clone, containing the complete genomic sequence of mouse Kv3.2, was isolated by screening a high-density colony array of mouse C57BL/6 genomic BAC clones (Genome Systems, St. Louis) with a 339-bp cDNA fragment, which overlaps the 5′ translational start site of Kv3.2. Sequence from a 3.5-kb subcloned fragment of Kv3.2 BAC-matched mouse Kv3.2 cDNA. Using BAC recombination methods developed by Yang et al. (10), a 2.6-kb NotI fragment of pZQCRE (11) was inserted into the translational start site of Kv3.2, replacing the expression of Kv3.2 with Cre recombinase. This fragment included Cre recombinase (plasmid pBS317; a gift from Brian Sauer, Stowers Institute for Medical Research, Kansas City, MO), a nuclear-localization signal (12), and exon–intron splicing and a polyadenylation signals. Cesium-gradient-purified, supercoiled BAC DNA containing the Kv3.2 promoter and the Cre gene was injected into the pronucleus of C57BL/6 zygotes as described (13). Candidate founders were tested by PCR and Southern blot analysis. Positive lines were crossed to lacZ reporter line Rosa26 (14) or alkaline phosphatase reporter line Z/AP (15). Transgenic line, K128-Cre, recombined lacZ reporter to produce strong β-galactosidase staining in rostral and midline thalamic nuclei (xiphoid, reunions, rhomboid, centromedial, intermediodorsal, paraventricular, paracentral, centrolateral, submedius, mediodorsal, lateral dorsal, anteromedial, suprageniculate, and paratenial). Moderate staining was observed in other thalamic nuclei: anteroventral, anterodorsal, ventral lateral, ventral anterior, parafascicular, and posterior thalamic group. Weak staining was observed in lateral posterior, dorsal lateral geniculate, medial geniculate, and ventral posterior thalamic nuclei. Strong recombination was observed also in ventral medial and dorsal medial hypothalamic nuclei, pontine reticular nucleus, mesencephalic fifth, and 12th cranial nerve nuclei. Moderate recombination was also present in the superficial piriform cortex, indusium griseum, dorsal endopiriform nucleus, medial and median preoptic hypothalamic nuclei, medial vestibular nucleus, ventral cochlear nucleus, nucleus of trapezoid body, and abducens nucleus. Very small numbers of neurons expressed in the lateral septum, locus coeruleus, substantia nigra pars reticulata, and parabrachial nucleus.
Construction of Floxed T-Type Calcium Channel Cav3.1. A BAC clone containing the Cav3.1 gene was isolated from a mouse C57BL/6 genomic BAC clone library (Genome Systems) and confirmed by PCR, T/A cloning, and sequencing. Restriction mapping, Southern blot hybridization, and subcloning and ligation were used to create a replacement vector for homologous recombination in ES cells. Briefly, a 7-kb EcoRV–PstI fragment containing exons 9–13 was used. A loxP sequence was placed into a SpeI site 5′ of exon 9. An LFNT cassette (containing a loxP sequence) was placed into a SpeI site 3′ of exon 12 to produce an ≈2.5 kb of loxP-flanked genomic DNA and a 1.5-kb 5′ and 3-kb 3′ flanking sequence. A diptheria toxin A gene (DT-A) was placed at a site flanking the targeting construct for negative selection (16). An LFNT cassette (K. Nakazawa and S.T., unpublished data) was modified to delete a TK cassette (LFNT-TK). A pair of FRT sites (recognition sequences for a yeast-derived, site-specific Flp recombinase) that flank PGK-Neo (for positive selection) and a 3′ loxP sequence remained after deletion of TK. The PGK-Neo cassette was deleted after germline transmission by crossing to a Flp deleter transgenic mouse (17). The “floxed” Cav3.1 mice (fCav3.1) were crossed to a global Cre deleter (K9-Cre; M.P.A. and S.T., unpublished data), a thalamic Cre deleter (K128-Cre), or a cortical Cre deleter (CW2-Cre, CAMKIIα promoter driving cortical and hippocampal CA1 pyramidal neuron expression) (18).
Histocytochemistry. Mice were perfused with ice-cold 4% paraformaldehyde in PBS buffer. For X-gal staining, brains were removed and postfixed at 4°C for 30 min. Coronal sections (50 μm thick) were cut on a Vibratome in PBS. Sections were incubated in 0.1 M PBS containing 0.01% SDS, 0.02% Nonidet P-40, and 2 mM MgCl2 at 4°C for 15 min, followed by β-galactosidase reaction in 1 PBS (pH 8.0) containing 0.5 mg/ml X-gal, 5 mM K4Fe(CN)6/3H2O, 5 mM K3Fe(CN)6, and 2 mM MgCl2 at 37°C for 24 h. For alkaline phosphatase staining, sections were heat-inactivated in PBS at 70°C for 1 h, equilibrated with a reaction buffer (100 mM Tris·HCl, pH 9.5/100 mM NaCl/50 mM MgCl2/0.1% Tween 20/2 mM levamisole; Vector Laboratories) for 15 min at room temperature and incubated with 0.4 mg/ml nitroblue tetrazolium chloride/0.19 mg/ml 5-bromo-4-chloro-3-indolyl-phosphate/100 mM Tris, pH 9.5/50 mM MgSO4 (NBT/BCIP tablets; Roche Molecular Biochemicals) at room temperature for 10–20 min. Section were postfixed in 10% formalin for 2 h and stained with nuclear fast red (Polysciences).
In Situ Hybridization. Terminal deoxyribonucleotidyl transferase (Invitrogen) and ATP [33P] (New England Nuclear) labeled the following set of 33-bp antisense oligonucleotides: 5′-AGAGCAGCCCAAGATGACGTGGAGGCCATGCCG-3′, 5′-TCTGAAAGACAGTGACAATGGCCCAGAGCAGGG-3′, 5′-GCCGA AGGGACCGTAGACGAGCAGCT TCAGCAG-3′, and 5′-ATTCTTCCGGTCTGGCAACGTGTCCCCATCCCG-3′. Probes were designed to hybridize sequences flanked by loxP sequences and to be minimally homologous to other channels. Unincorporated nucleotides were removed on Sephadex G-50 spin columns (Pharmacia). Hybridization buffers, temperatures, and wash stringency were performed as described by Talley et al. (19). Slides were exposed to film (Hyperfilm β-MAX, Amersham Biosciences) for 1 week and analyzed for relative intensity by using image-analysis software (mcid, Imaging Research, St. Catherine's, ON, Canada). For resolution of cellular labeling, slides were dipped in liquid autoradiography emulsion (NTB2, Kodak, Sigma–Aldrich), exposed for 5–9 weeks, and examined by dark-field and bright-field microscopy. Nissl counterstain was used.
Implantation of EEG/EMG Electrodes. Under anesthesia with ketamine/xylazine (100/10 mg/kg, injected i.p.), screw electrodes were implanted into the skull (1-mm anterior to bregma and 1-mm anterior to lambda, 1.5-mm lateral to midline). EMG electrodes (AS633; Cooner Wire, Chatsworth, CA) were inserted into neck extensor muscles. Leads were attached to a 2 × 2-pin header secured to the skull by using dental acrylic. An activity transmitter (TA-F20; Data Sciences International, St. Paul, MN) was placed into the peritoneal cavity.
Sleep Recordings. Recordings were performed under a 12:12-h light/dark schedule (lights on from 7 a.m. to 7 p.m.). Mice were habituated for 10 days to a counterbalanced, light-weight cable connected to a low-torque commutator (4-TBC-9-S; Crist Instrument, Hagerstown, MD), affixed to the center of the top of the cage. Signals from EEG/EMG electrodes were amplified and bandpass-filtered by using model 12 amplifiers (Grass Instruments, Quincy, MA). EEG signals were amplified ×5,000 and bandpass-filtered at 0.3–30 Hz. EMG signals were amplified ×5,000 and bandpass filtered at 2–100 Hz. Both signals were acquired digitally at 128 Hz by using Sleep Sign (Kissei Comtec, Matsumoto, Japan). Behavioral state was scored in 10-s epochs as wake, rapid-eye-movement (REM), or non-REM (NR) sleep by using automated methods, followed by visual screening by a blinded single examiner (20).
Submerged Slice Patch Clamp Recording. Mice were anesthetized by using isoflurane, and the brains were submerged into 4°C sucrose solution (250 mM sucrose/5 mM KCl/10 mM glucose/25 mM sodium bicarbonate/5 mM KCl/1.25 mM NaH2PO4/1 mM CaCl2/5 mM MgSO4/95% O2/5% CO2 (carbogen gas). Coronal sections (250 μm) were prepared on a Vibratome Plus 3000 (Vibratome, St. Louis, MO) in 4°C sucrose. Slices were incubated at 35°C for 30 min and then at room temperature for <8 h in an interface chamber containing ACSF buffer (125 mM NaCl/25 mM glucose/25 mM sodium bicarbonate/2.5 mM KCl/1.25 mM NaH2PO4/2 mM CaCl2/1 mM MgCl2/95% O2/5% CO2. Recordings were performed at room temperature by using infrared-guided whole-cell patch-clamp technique. Data were acquired with an EPC10 triple patch clamp amplifier and acquisition system (HEKA, Lambrecht, Germany). Slices were recorded in rapidly flowing (1 ml/min), carbogen-perfused ACSF on the stage of an upright microscope fitted with differential interference contrast (DIC) optics and camera (Optical Analysis, Nashua, NH).
T-Type Calcium Channel Current Measurements. Whole-cell, voltage clamp was performed with 1 μM tetrodotoxin/30 μM bicuculline/1 mM 4-aminopyridine/10 mM tetraethylammonium (TEA) chloride/2 mM cesium chloride in the ACSF. The pipette contained 90 mM cesium methanesulfonate, 11 mM cesium chloride, 0.5 mM magnesium dichloride, 0.5 mM CaCl2, 10 mM Hepes, 11 mM EGTA, 4 mM magnesium·ATP, 0.3 mM Tris·GTP, and 7 mM Tris·phosphocreatine (pH 7.3; Tris base).
Ca2+ Spike and Action-Potential Measurements. Current clamp studies were performed with 10 μM CNQX and 30 μM bicuculline in ACSF. The pipette contained 135 mM potassium gluconate, 4 mM KCl, 2 mM NaCl, 10 mM Hepes, 4 mM magnesium·ATP, 0.3 mM Tris·GTP, and 7 mM Tris·phosphocreatine (pH 7.25; Tris base). In some experiments, the following drugs were added to the bath: cAMP agonist forskolin (10 μM) or TEA chloride (4 mM). BAPTA (15 mM) was substituted for equimolar potassium gluconate in the pipette.
Statistical Analysis. Statistical analysis was conducted by using statview (SAS Institute, Cary, NC). Data were analyzed by two-tailed t test, ANOVA, or repeated-measures ANOVA. Values in graphs were expressed as mean ± SEM.
Results
Generation of Mice for Gene Manipulations in Thalamus. Cav3.1 T-type calcium channels were focally deleted from a subset of adult neurons by using the bacteriophage P1-derived Cre/loxP-recombination system (11) (Figs. 1A and 2 A). To delete loxP-flanked sequences selectively in thalamic projection neurons, we placed the Cre recombinase gene under control of the promoter sequences for the potassium channel Kv3.2 (Kv3.2-Cre; Fig. 1 A). The Kv3.2 promoter was chosen because the transcript is first expressed ≈12 days after birth and is highly restricted (>90% of transcript) to thalamic projection neurons (21).
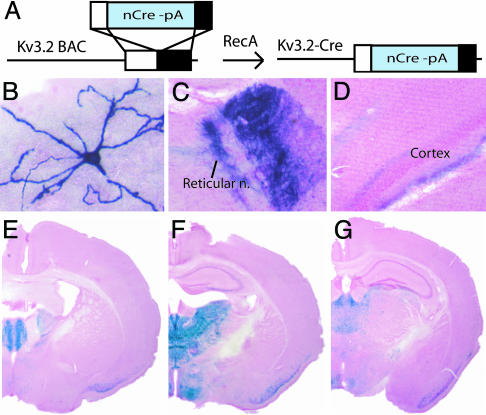
Fig. 1.
Generation of K128-Cre transgenic mice that delete loxP-flanked sequence in rostral–midline thalamic projection neurons. (A) Schematic diagram of the procedure for creating a transgenic construct in which nuclear-targeted Cre recombinase gene with polyadenylation sequence (nCre-pA) is placed under control of the Kv3.2 promoter in a 120-kb genomic BAC DNA fragment. (B–D) K128-Cre transgene deletes loxP-flanked sequence to label neuron processes by alkaline phosphatase staining in 1-week-old (anterior ventral thalamic nucleus, representative example, n = 15 cells) (B) and 1-month-old (anterior ventral thalamic nucleus, C; presubicular cortex, D) K128-Cre/+; Z/AP/+ double-transgenic male mouse. (E–G) K128-Cre transgene deletes loxP-flanked sequence to label neuron nuclei blue by X-gal staining in a 4-month-old male K128-Cre/+; Rosa26/+, double-transgenic male mouse. Coronal brain sections were counterstained with nuclear fast red.
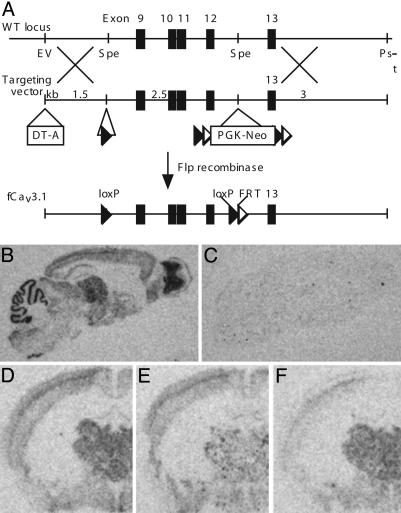
Fig. 2.
Generation of mice carrying a loxP-flanked Cav3.1 gene (fCav3.1/fCav3.1) for postnatal, region-restricted deletion of Cav3.1 T-type Ca2+ channel. (A) Schematic representation of the procedure for inserting loxP sequences around exons 9–12 of the gene encoding T-type Ca2+ channel, Cav3.1 (fCav3.1; see Supporting Materials and Methods, which is published as supporting information on the PNAS web site, for details). (B and C) Distribution of Cav3.1 mRNA in brains of 4-month-old WT (B) and global Cav3.1 deleted (C) male mice. Parasagittal brain sections are shown. (D–F) Distribution of Cav3.1 mRNA in brains of 4-month-old WT (D), thalamic (E), or cortical (F) Cav3.1 KO male mice. The genotypes are fCav3.1/fCav3.1; fCav3.1/fCav3.1, K128-Cre/+; and fCav3.1/fCav3.1, CW2-Cre/+, respectively. Coronal brain sections are shown. DNA flanked by LoxP and FRT sequences is deleted by Cre and Flp recombinase, respectively. DT-A and PGK-Neo are negative and positive selection markers, respectively. See Materials and Methods for more details.
K128-Cre, a mouse line containing the Kv3.2-Cre transgene, was found to express Cre recombinase activity strongly in rostral–midline thalamic projection neurons. By crossing K128-Cre to two Cre recombinase-dependent reporters [Z/AP, which labels cellular membranes with alkaline phosphatase (Fig. 1 B–D), and Rosa26, which labels cellular nuclei with β-galactosidase (Fig. 1 E–G)], we established the cell type and regional pattern of Cre recombinase activity. At an early age (1 week old), only rare, scattered neurons expressed the Z/AP reporter, consistent with the postnatal expression of Kv3.2 (Fig. 1B). The neurons displayed a multipolar dendritic arbor (Fig. 1B), and a dense axonal projection to reticular thalamic nucleus (Fig. 1C), and superficial and deep layers of cortex (Fig. 1D). Using the Rosa26 reporter (Fig. 1 E–G), the K128-Cre transgene was found to recombine selectively in rostral and midline thalamic nuclei despite the broad thalamic expression of native Kv3.2 (see Materials and Methods for details). Recombination was also observed in superficial layers of piriform cortex, ventromedial and dorsomedial hypothalamic nuclei (Fig. 1 E–G). The complete pattern of recombination is described in Materials and Methods. The results demonstrate that K128-Cre transgene deletes loxP-flanked sequences strongly in rostral–midline thalamic projection neurons.
Generation of Mice for Focal Deletion of T-Type Ca2+ Channel Cav3.1. To focally delete Cav3.1, loxP sequences were placed into introns flanking exons 9–12 of the gene encoding Cav3.1 in C57/BL6 mouse embryonic stem cells by using homologous recombination (Fig. 2A). Exons 9–12 were deleted by crossing homozygous loxP-flanked mice (fCav3.1/fCav3.1) to Cre transgenic mice including a global Cre deleter (K9-Cre), a thalamic Cre deleter (K128-Cre), and a cortical Cre deleter (CW2-Cre, targets cortical pyramidal neurons, 18). In situ hybridization to Cav3.1 mRNA showed a WT pattern of expression in homozygous fCav3.1 mice (Fig. 2 B and D) (19) and a complete loss of transcript with global deletion (Fig. 2C). Mice from the thalamic Cre deleter cross showed a marked reduction of Cav3.1 mRNA in rostral–midline thalamus but normal expression in cortex (Fig. 2E). Mice from the cortical Cre deleter cross showed decreased Cav3.1 mRNA transcript in cerebral cortex but no change in the thalamus (Fig. 2F). This genetic approach allows cell-type- and region-restricted deletion of Cav3.1 from the thalamus by using K128-Cre and from the cortex by using CW2-Cre.
T-type Ca2+ channel-mediated Ca2+ transients increase in thalamic projection neurons during sleep (22), but the purpose of this increase is unknown. To define the function of these currents, we first confirmed that this current in thalamic projection neurons depends on Cav3.1 (Fig. 5, which is published as supporting information on the PNAS web site), as reported (8). We then examined the effects of Cav3.1 activation on action-potential firing.
T-Type Ca2+ Channel Cav3.1 Activation Inhibits Action-Potential Firing. Transient hyperpolarization, which activates Cav3.1, inhibited action-potential firing in neurons of WT mice. By contrast, this stimulus had no effect on neurons from global Cav3.1 KO mice. Nonadapting action-potentials were observed under baseline conditions, in some cases, or with depolarizing current injections (Fig. 3 A and B). These action potentials were inhibited by a transient (1 s) hyperpolarizing current injection for >9 s (Fig. 3A a and b) in neurons of WT mice. Repeated hyperpolarization every 9 s revealed that inhibition duration depended on current-injection size (Fig. 3A b–d) and was reversible (Fig. 3Ad). In neurons from global Cav3.1 KO mice (Fig. 3 B and C), inhibition was abolished.
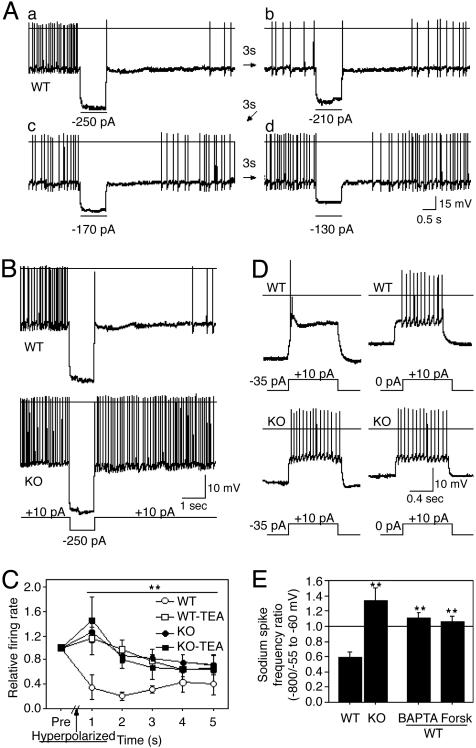
Fig. 3.
Cav3.1 T-type calcium channel activation inhibits Na+ action-potential firing. A hyperpolarizing current injection (1 s) caused prolonged inhibition of Na+ action potentials in neurons of WT (A–C)(n = 13), but not global Cav3.1 KO (B and C)(n = 16), mice. Cells were obtained from lateral dorsal thalamic nucleus, and they displayed a multipolar dendritic arbor. (C) TEA (4 mM) blocks inhibition in WT neurons (P < 0.002, repeated-measures ANOVA, n = 5 each). (D) Depolarizing current injections, in hyperpolarized neurons (–80 mV, left traces), evoked a Ca2+ potential and brief Na+ action-potential burst in WT (n = 12/12) but no Ca2+ potential (n = 0/15) and persistent Na+ action-potential firing in KO (n = 15/15). Ca2+ potential activation and Na+ action-potential inhibition are missing with current injections to neurons held at –55 to –60 mV (right traces). (E) The ratio of Na+ action-potential firing frequencies evoked by current injections from –80 vs. –55 to –60 mV revealed inhibition in WT (n = 7), but not KO (n = 7), neurons. Inhibition was blocked in WT neurons perfused with intracellular BAPTA (15 mM, n = 9) or extracellular forskolin (10 μM, n = 6). Current pulses are shown below the traces. *, P < 0.02; **, P < 0.01, unpaired t test.
Tonic hyperpolarization, which also deinactivates Cav3.1, inhibited evoked action-potential firing in thalamic projection neurons of WT but not global Cav3.1 KO mice. Depolarizing current injections in hyperpolarized WT neurons (Vm = –80 mV; Fig. 3D, left traces), evoked a low-threshold Ca2+ potential, a brief action-potential burst, and a plateau of membrane-potential depolarization that evoked no further action potentials. In contrast, the same conditions evoked a continuous train of action potentials in global Cav3.1 KO neurons. Depolarizing current injections evoked a continuous train of action potentials in neurons of both genotypes when left at resting membrane potential (Vm = –55 to –60 mV; Fig. 3D, right traces). Comparing the action-potential firing rates evoked from a holding potential of –80 mV and –55 to –60 mV, we observed inhibition in WT but not global Cav3.1 KO mice (Fig. 3E). The results indicate that one of the major Cav3.1 functions is to inhibit action-potential firing.
This Cav3.1-mediated inhibition of action-potential firing does not occur through GABAergic feedback (23, 24). First, AMPA and GABAA synaptic transmissions were blocked (10 μM CNQX and 300 μM bicuculline) in our experiments. Second, no inhibitory synaptic currents were observed in the tracings. Third, action-potential inhibition was blocked, but action-potential bursts were preserved in neurons exposed to an intracellular calcium chelator (BAPTA; Fig. 3E), a nonselective potassium channel blocker (TEA chloride; Fig. 3C), and an agent that increases intracellular cAMP (forskolin; Fig. 3E). The data suggest that Cav3.1 activation inhibits action-potential firing through a Ca2+-dependent, TEA-inhibited, and cAMP-regulated pathway.
Based on our current findings that thalamic Cav3.1 T-type Ca2+ channel activation inhibits action-potential firing and the evidence for increased thalamic T-type Ca2+ channel activity during sleep (22), we hypothesized that thalamic Cav3.1 may be necessary for well maintained sleep. Consistent with this hypothesis, mice with global or thalamic Cav3.1 deletion displayed fragmented sleep. The vigilance-state hypnograms during the rest (light) period showed frequent brief awakenings (W) that interrupted NR sleep in KO mice (Fig. 4A). During the light period, global and thalamic KO mice had more total wake bouts (Fig. 4B), mainly because of an increase of brief (20–40 s) wake bouts (bout histogram analysis, data not shown). Because of these frequent awakenings, NR bouts were much shorter (Fig. 4B, right graph). Mice with cortical Cav3.1 deletion, by contrast, showed no disruption of sleep (Fig. 4 A and B). All mice showed normal amounts and durations of REM sleep (Fig. 4B). The results demonstrate that loss of thalamic Cav3.1 causes frequent brief arousals that fragment sleep.
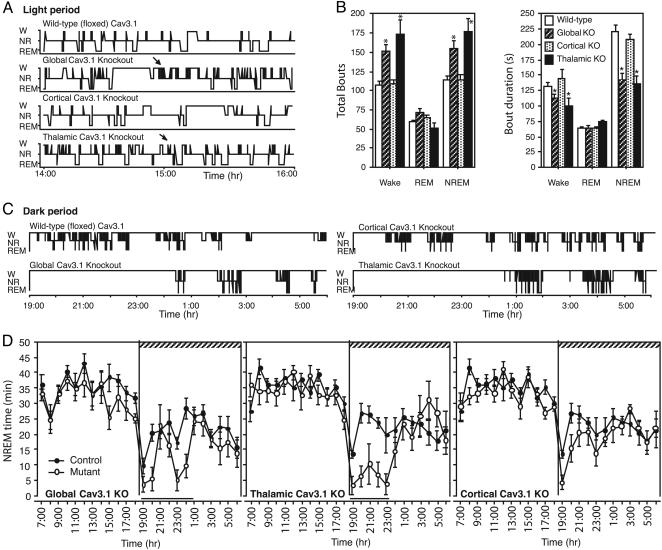
Fig. 4.
Global and thalamic Cav3.1 deletion disrupts sleep. (A) Vigilance-state hypnograms, during the 12-h light (rest) period, revealed NR sleep interrupted by frequent brief wake bouts (W) in global and thalamic, but not cortical Cav3.1, KO mice. REM was unchanged. (B) Global and thalamic Cav3.1 KO mice displayed more brief and numerous wake and NR bouts. Bar graphs of bout number (Left) and duration (Right) for wake, REM, and NR are shown for WT (floxed), global, cortical, and thalamic Cav3.1 KO mice. Data are from the 12-h light period. *, P < 0.02, unpaired t test. (C) Vigilance-state hypnograms during the 12-h dark (active) period revealed delayed sleep onset after lights out in global and thalamic Cav3.1 KO. (D) NR sleep time over the 24-h light/dark period showed decreased NR sleep during the 12-h dark (hatched bar) period in global and thalamic Cav3.1 KO mice. Black bar indicates P < 0.02 (repeated-measures ANOVA; n = 8 mice each genotype).
Mice with global or thalamic Cav3.1 deletion also had difficulty sleeping during the dark period. All mice displayed decreased NR sleep immediately after the lights were turned off (Fig. 4 C and D). However, after this ≈30-min wake period, WT mice slept, but global and thalamic KO mice remained awake for up to 6 h (Fig. 4C and Fig. 6, which is published as supporting information on the PNAS web site). Consequently, sleep onset was delayed (latency to sleep: WT, 28 ± 28 min; thalamic Cav3.1 deletion, 226 ± 58 min; mean ± SE; P < 0.003, unpaired t test), and total sleep was reduced (Fig. 4D, total NR sleep: WT, 11 h 24 ± 16 min; thalamic Cav3.1 deletion, 9 h 54 min ± 19 min; mean ± SE; P = 0.003, unpaired t test). In contrast, cortical Cav3.1 KO mice had only a mild delay in sleep onset (latency to sleep: cortical Cav3.1 deletion, 85 ± 19 min; mean ± SE; P < 0.02, unpaired t test, compared with WT) and had normal amounts of total sleep (Fig. 4B, P > 0.05, unpaired t test).
Animals with excessive locomotor activity (e.g., global dopamine transporter KO mice; ref. 25) can experience fragmented and decreased sleep. However, in contrast to the dopamine transporter KO, global Cav3.1 KO mice had mildly reduced locomotor activity during the 12-h dark period (WT, 7.9 ± 1.19; KO, 5.34 ± 0.46; mean ± SE; P < 0.01, unpaired t test; n = 8 mice each genotype). Animals with a markedly reduced locomotor activity can have sleep fragmentation because of poor consolidation (e.g., global albumin D-binding protein KO; ref. 26). However, the albumin D-binding protein KO mouse displays decreased delta wave power suggesting decreased homeostatic sleep drive. In contrast, global and thalamic Cav3.1 KO mice display increased delta wave power during NR sleep (data not shown). Interestingly, hypersynchronous delta waves have been described in patients with sleep disorders characterized by frequent arousal (27).
Discussion
In conclusion, the results suggest Cav3.1 is required in the thalamus, but not the cortex, to stabilize sleep. Despite their reduced activity and possible increased homeostatic sleep drive, Cav3.1 KO mice have difficulty initiating and maintaining sleep.
Sensory gating may help ensure that sleep is not disrupted. We have demonstrated that Cav3.1 T-type Ca2+ channel activation inhibits action potentials through an intracellular pathway involving increases of intracellular Ca2+. This gating of thalamic action potentials is defective in global Cav3.1 KO mice. We have also shown that Cav3.1 T-type Ca2+ channel function is required in the thalamus, but not in the cerebral cortex to achieve normal patterns of sleep. Mice with thalamic Cav3.1 deletion experience frequent arousals from sleep. During the dark period, these mice also experience sleep loss because of a delayed sleep onset. Based on these findings, we suggest Cav3.1 T-type Ca2+ channels inhibit the transmission of thalamic action potentials to prevent feedforward excitation, which could destabilize activity within the neural circuitry maintaining the sleep state (28).
Inhibition of thalamic transmission may occur during various conditions that activate Cav3.1. A feedback-inhibitory pathway has been identified (23, 24) that interconnects neurons of the thalamic relay and reticular nuclei. Based on the >9 s inhibition that we observed, GABAergic synaptic inputs from the reticular nuclei could be transformed into a prolonged inhibition by Cav3.1. Thalamic projection neurons also display various spontaneous rhythmic activities that are thought to involve T-type Ca2+ channels (29, 30). One purpose of these rhythmic activities may be to activate Cav3.1 and, thereby, provide a continuous inhibition of transmission independent of GABAergic signaling.
Cav3.1 inhibits action potentials through a TEA-sensitive, Ca2+-dependent and cAMP-inhibited intracellular pathway. The sensitivity to a potassium channel blocker, TEA, the slow kinetics of inhibition, and the dependence on Ca2+ suggest the slow after hyperpolarization current (sAHP) may be responsible. This potassium current is activated slowly by Ca2+ and inhibited by the cAMP-dependent protein kinase, consistent with our observations (31).
Although some recombination occurred outside the thalamus, the intense pattern of Cav3.1 deletion observed in rostral–midline thalamus in K128-Cre suggests that this region may normally carry arousal signals that disrupt sleep (2, 32). Alternatively, the midline (intralaminar) thalamic nuclei may produce Cav3.1-mediated action-potential bursts during sleep that excite reticular thalamic neurons to provide feedforward inhibition to other thalamic nuclei (33).
Some researchers have speculated that thalamic bursts produced by the T-type Ca2+ channel may underlie the cortical EEG waves observed during sleep (4, 5). We were surprised to find that cortical EEG power within the delta frequency range (1–4 Hz) was not significantly reduced but, rather, was moderately increased (data not shown). The observation suggests that other systems (e.g., intracortical circuits or basal forebrain GABAergic projections; ref. 34) drive the delta waves of the cortical EEG during sleep. We speculate that this delta-wave-generating system may be critical to the maintenance of sleep when thalamic sensory filtering by Cav3.1 is defective.
The pattern of brief arousals in Cav3.1 KO mice resembles the sleep-microstructure alterations recently recognized in patients with insomnia and other sleep disorders of arousal (35). It remains to be established whether these mice rouse from sleep because of inappropriate transmission of normal sensations or because of pathologic sensations like those in sleep apnea or periodic limb movement disorder. If these brief arousals from sleep in Cav3.1 KO mice were caused by inadequate gating of sensory stimuli, it would possibly provide a perspective on human insomnia in which relatively mild sensations can disrupt sleep.
Supplementary Material
Acknowledgments
We thank Lorene Leiter, Xiaoning Zhou, and Dennis King for technical assistance. M.P.A. is supported by a Howard Hughes Fellowship for Physicians, a Mentored Clinical Scientist Development Award from the National Institute of Mental Health, and a Career Award in the Biomedical Sciences from the Burroughs Wellcome Fund. This research was also supported by National Institutes of Health Grants R01-NS32925 and P50-MH58880 (to S.T.) and R01-MH62589 and P01-HL60292 (to T.E.S.), as well as a grant from the RIKEN Brain Research Institute (to S.T.).
Abbreviations: REM, rapid eye movement; KO, knockout; BAC, bacterial artificial chromosome; EEC, electroencephalogram; EMG, electromyogram; TEA, tetraethylammonium; NR, non-REM.
References
- 1.Livingstone, M. S. & Hubel, D. H. (1981) Nature 291, 554–561. [DOI] [PubMed] [Google Scholar]
- 2.Saper, C. B. (2002) Annu. Rev. Neurosci. 25, 433–469. [DOI] [PubMed] [Google Scholar]
- 3.Jahnsen, H. & Llinas, R. (1984) J. Physiol. 349, 227–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Steriade, M., McCormick, D. A. & Sejnowski, T. J. (1993) Science 262, 679–685. [DOI] [PubMed] [Google Scholar]
- 5.McCormick, D. A. & Bal, T. (1997) Annu. Rev. Neurosci. 20, 185–215. [DOI] [PubMed] [Google Scholar]
- 6.Huguenard, J. R. (1996) Annu. Rev. Physiol. 58, 329–348. [DOI] [PubMed] [Google Scholar]
- 7.Perez-Reyes, E., Cribbs, L. L., Daud, A., Lacerda, A. E., Barclay, J., Williamson, M. P., Fox, M., Rees, M. & Lee, J. H. (1998) Nature 391, 896–900. [DOI] [PubMed] [Google Scholar]
- 8.Kim, D., Song, I., Keum, S., Lee, T., Jeong, M. J., Kim, S. S., McEnery, M. W. & Shin, H. S. (2001) Neuron 31, 35–45. [DOI] [PubMed] [Google Scholar]
- 9.Kim, D., Park, D., Choi, S., Lee, S., Sun, M., Kim, C. & Shin, H. S. (2003) Science 302, 117–119. [DOI] [PubMed] [Google Scholar]
- 10.Yang, X. W., Model, P. & Heintz, N. (1997) Nat. Biotechnol. 15, 859–865. [DOI] [PubMed] [Google Scholar]
- 11.Tsien, J. Z., Huerta, P. T. & Tonegawa, S. (1996) Cell 87, 1317–1338. [DOI] [PubMed] [Google Scholar]
- 12.Kalderon, D., Roberts, B. L., Richardson, W. D. & Smith, A. E. (1984) Cell 39, 499–509. [DOI] [PubMed] [Google Scholar]
- 13.Hogan, B., Beddington, R., Constantini, F. & Lacy, E. (1994) in Manipulating the Mouse Embryo: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, New York), 2nd Ed..
- 14.Soriano, P. (1999) Nat. Genet. 21, 70–71. [DOI] [PubMed] [Google Scholar]
- 15.Lobe, C. G., Koop, K. E., Kreppner, W., Lomeli, H., Gertsenstein, M. & Nagy, A. (1999) Dev. Biol. 208, 281–292. [DOI] [PubMed] [Google Scholar]
- 16.Yagi, T., Ikawa, Y., Yoshida, K., Shigetani, Y., Takeda, N., Mabuchi, I., Yamamoto, T. & Aizawa, S. (1990) Proc. Natl. Acad. Sci. USA 87, 9918–9922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farley, F. W., Soriano, P., Steffen, L. S. & Dymecki, S. M. (2000) Genesis 28, 106–110. [PubMed] [Google Scholar]
- 18.Zeng, H., Chattarji, S., Barbarosie, M., Rondi-Reig, L., Philpot, B. D., Miyakawa, T., Bear, M. F. & Tonegawa, S. (2001) Cell 107, 617–629. [DOI] [PubMed] [Google Scholar]
- 19.Talley, E. M., Cribbs, L. L., Lee, J. H., Daud, A., Perez-Reyes, E. & Bayliss, D. A. (1999) J. Neurosci. 19, 1895–1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mochizuki, T., Crocker, A., McCormack, S., Yanagisawa, M., Sakurai, T. & Scammell, T. E. (2004) J. Neurosci. 24, 6291–6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rudy, B., Kentros, C., Weiser, M., Fruhling, D., Serodio, P., Vega-Saenz de Miera, E., Ellisman, M. H., Pollock, J. A. & Baker, H. (1992) Proc. Natl. Acad. Sci. USA 89, 4603–4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCarley, R. W., Benoit, O. & Barrionuevo, G. (1983) J. Neurophysiol. 50, 798–818. [DOI] [PubMed] [Google Scholar]
- 23.Le Masson, G., Renaud-Le Masson, S., Debay, D. & Bal, T. (2002) Nature 417, 854–858. [DOI] [PubMed] [Google Scholar]
- 24.Von Krosigk, M., Bal, T. & McCormick, D.A. (1993) Science 261, 361–364. [DOI] [PubMed] [Google Scholar]
- 25.Wisor, J. P., Nishino, S., Ichiro, S., Uhl, G. H., Mignot, E. & Edgar, D. M. (2001) J. Neurosci. 21, 1787–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Franken, P., Lopez-Molina, L., Marcacci, L., Schibler, U. & Tafti, M. J. Neurosci. 15, 617–625. [DOI] [PMC free article] [PubMed]
- 27.Pressman, M. R. (2004) Sleep 27, 706–710. [DOI] [PubMed] [Google Scholar]
- 28.Saper, C. B., Chou, T. C. & Scammell, T. E. (2001) Trends Neurosci. 24, 726–731. [DOI] [PubMed] [Google Scholar]
- 29.Hughes, S. W., Cope, D. W., Blethyn, K. L. & Crunelli, V. (2002) Neuron 33, 947–958. [DOI] [PubMed] [Google Scholar]
- 30.Hughes, S. W., Lorincz, M., Cope, D. W., Blethyn, K. L., Kekesi, K. A., Parri, H. R., Juhasz, G. & Crunelli, V. (2004) Neuron 22, 253–268. [DOI] [PubMed] [Google Scholar]
- 31.Vogalis, F., Storm, J. F. & Lancaster B. (2003) Eur. J. Neurosci. 18, 3155–3166. [DOI] [PubMed] [Google Scholar]
- 32.Van der Werf, Y. D., Witter, M. P. & Groenewegen, H. J. (2002) Brain Res. Rev. 39, 107–140. [DOI] [PubMed] [Google Scholar]
- 33.Crabtree, J. W., Collingridge, G. L. & Isaac, J. T. (1998) Nat. Neurosci. 1, 389–394. [DOI] [PubMed] [Google Scholar]
- 34.Modirrousta M., Mainville L. & Jones B.E. (2004) Neuroscience 129, 803–810. [DOI] [PubMed] [Google Scholar]
- 35.Terzano, M. G., Parrino, L. & Smerieri, A. (2001) Rev. Neurol. (Paris) 157, 62–66. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.