Abstract
The constitutive androstane receptor (CAR/Nr1i3) is an important xenobiotic-sensing nuclear receptor that is highly expressed in the liver and is well known to have species differences. During development, age-specific activation of CAR may lead to modified pharmacokinetics and toxicokinetics of drugs and environmental chemicals, leading to higher risks for adverse drug reactions in newborns and children. The goal of this study was to systematically investigate the age- and species-specific regulation of various drug-processing genes (DPGs) after neonatal or adult CAR activation in the livers of wild-type, CAR-null, and humanized CAR transgenic mice. At either 5 or 60 days of age, the three genotypes of mice were administered a species-appropriate CAR ligand or vehicle once daily for 4 days (i.p.). The majority of DPGs were differentially regulated by age and/or CAR activation. Thirty-six DPGs were commonly upregulated by CAR activation regardless of age or species of CAR. Although the cumulative mRNAs of uptake transporters were not readily altered by CAR, the cumulative phase I and phase II enzymes as well as efflux transporters were all increased after CAR activation in both species. In general, mouse CAR activation produced comparable or even greater fold increases of many DPGs in newborns than in adults; conversely, humanized CAR activation produced weaker induction in newborns than in adults. Western blotting and enzyme activity assays confirmed the age and species specificities of selected CAR-targeted DPGs. In conclusion, this study systematically compared the effect of age and species of CAR proteins on the regulation of DPGs in the liver and demonstrated that the regulation of xenobiotic biotransformation by CAR is profoundly modified by age and species.
Introduction
The liver is a major organ for the biotransformation of various drugs and environmental chemicals. Many phase I and phase II drug-metabolizing enzymes as well as uptake and efflux transporters, together called drug-processing gene (DPGs), are highly expressed in hepatocytes (Klaassen and Aleksunes, 2010; Fu et al., 2016; Almazroo et al., 2017). Xenobiotics are taken up into the hepatocytes by various uptake transporters in the solute carrier and the solute carrier organic anion transporter (SLCO) families (Klaassen and Aleksunes, 2010), where they are frequently modified by various phase I enzymes, including those that catalyze 1) oxidation reactions, such as cytochrome P450s (P450s), alcohol dehydrogenases, aldehyde dehydrogenases (ALDHs), molybdenum hydroxylases, aldehyde oxidases, dimeric dihydrodiol dehydrogenases, amine oxidases, flavin-containing monooxygenases (FMOs), and peroxidases; 2) reduction reactions, such as azo- and nitro-reduction, carbonyl reduction, disulfide reduction, sulfoxide reduction, quinone reduction by NAD(P)H quinone oxidoreductases (NQOs), dehalogenation, and dehydroxylation; and 3) hydrolysis reactions, as catalyzed by carboxyesterases (CESs), cholinesterases, paraoxonases, epoxide hydrolases (EPHXs), peptidases, and β-glucuronidases (Parkinson et al., 2013; Cui and Li, 2016). Phase II reactions, including glucuronidation by UDP-glucuronosyltransferases (UGTs), sulfonation by sulfotransferases (SULTs), glutathione conjugation by glutathione-S-transferases (GSTs), and amino acid conjugation by various enzymes such as medium-chain CoA ligases and acyl-CoA [glycine N-acetyltransferases (NATs), seryl-tRNA synthetases, and bile acid conjugation enzymes], increase the water solubility of the substrates and favor their excretion. Other phase II reactions including acetylation (by NATs), methylation (by various methyltransferases), and fatty acid conjugation (by acyltransferases and cholesterol ester hydrolases) result in decreased water solubility of the substrates (Parkinson et al., 2013; Cui and Li, 2016). Many conjugated metabolites are substrates for efflux transporters in the ATP-binding cassette (ABC) family and some solute carrier families (Klaassen and Aleksunes, 2010).
The constitutive androstane receptor (CAR) is an important xenobiotic-sensing transcription factor that is highly expressed in the liver (Bookout et al., 2006; Yan and Xie, 2016). Prototypical CAR activators include the anticonvulsant phenobarbital (indirect activator) in both rodents and humans, 1,4-bis-[2-(3,5- dichloropyridyloxy)]benzene,3,3′,5,5′-tetrachloro-1,4-bis(pyridyloxy)benzene (TCPOBOP, direct ligand) in rodents only (Tzameli et al., 2000), as well as 6-(4-chlorophenyl)imidazo[2,1-b][1,3]thiazole-5-carbaldehyde-O-(3,4-dichlorobenzyl)oxime (CITCO, direct ligand) in humans only (Maglich et al., 2003; Windshügel and Poso, 2011). Upon activation, it is well known that CAR upregulates the expression of a wide spectrum of DPGs in the adult liver and in liver-derived cell cultures in a CAR-dependent manner (Wortham et al., 2007; Aleksunes and Klaassen, 2012; Li et al., 2015; Cui and Klaassen, 2016). In addition to the prototypical CAR activators, many other human-relevant xenobiotics, such as certain herbal medicines and environmental chemicals, can also activate CAR (Huang et al., 2004; Ross et al., 2010; Sueyoshi et al., 2014; Zhang et al., 2015). Therefore, CAR is partially involved in adverse drug reactions. Species differences between CAR proteins in humans and rodents are one potential problem in investigating CAR in animal models (Holsapple et al., 2006; Ross et al., 2010; Yang and Wang, 2014). For example, activation of the mouse constitutive androstane receptor (mCAR) promotes tumors in the liver (Yamamoto et al., 2004; Huang et al., 2005). Conversely, activation of the human constitutive androstane receptor (hCAR) is associated with cell cycle arrest and enhanced apoptosis, suggesting anticancer potential (Chakraborty et al., 2011). Although it is generally recognized that CAR is important in regulating drug metabolism and transport in the livers of both species, there have been no systematic studies to date specifically comparing the effects of CAR activation on all the hepatic DPG expression between mice and humans. Human livers and in vitro human primary hepatocyte cultures are the gold standards to investigate the effect of CAR activation in humans. However, these approaches are limited by a scarcity of sources, large biologic and/or technical variations, and ethical concerns. To understand the species differences in CAR-mediated regulation of hepatic target genes in vivo, various hCAR transgenic (TG) mice have been generated (Scheer et al., 2008; Zhang et al., 2013). The hCAR-TG mice used in our study include not only the exons and promoters but also 73-kb upstream and 91-kb downstream regulatory DNA into the mouse genome utilizing bacterial artificial chromosome TG technology, allowing investigation of the regulation of CAR target genes as well as the transcription of CAR itself (Zhang et al., 2013). This hCAR-TG mouse model expresses major hCAR splicing variants (SVs) in humans, and it is functional in terms of upregulating the prototypical CAR target gene Cyp2b10 (which is the human CYP2B6 homolog), allowing an in vivo investigation on hCAR function in response to chemical activation.
Another important yet understudied area is the chemical-mediated regulation of CAR-targeted genes during development. At birth, profound changes occur in the liver transition from a hematopoietic organ to the major organ for xenobiotic biotransformation. Although the basal ontogenic expression of various DPGs has been characterized in wild-type (WT) mouse livers (O'Sullivan et al., 1989; Cui et al., 2010, 2012a; Peng et al., 2012, 2013; Lu et al., 2013; Gunewardena et al., 2015) and in human liver samples (Hines and McCarver, 2002; McCarver and Hines, 2002; Hines, 2007, 2013; Mooij et al., 2014, 2016; Thomson et al., 2016), very little is known regarding the age-specific transcriptional responses of hepatic DPGs after exposure to foreign chemicals such as therapeutic drugs, dietary factors, and environmental toxicants (Lee et al., 2008; Xiao et al., 2012). Age-specific expression of chromatin epigenetic marks in the liver suggests that the accessibility of transcription factors to target genes in the genome and the subsequent changes in target gene expression are highly regulated by age (Lu et al., 2012). Among various xenobiotics to which pediatric patients are exposed, a considerable portion may activate the CAR (Huang et al., 2004; Ross et al., 2010; Sueyoshi et al., 2014; Zhang et al., 2015), placing newborns and children at a higher risk for adverse drug reactions and chemical-induced liver injuries. Neonatal activation of mCAR in the mouse liver leads to persistent increase in certain drug-metabolizing enzymes in the liver in adult age (Chen et al., 2012; Li et al., 2016a); however, to date, there have been no systematic studies conducted to determine to what extent the acute pharmacological activation of CAR leads to age- and species-specific regulation of hepatic DPGs.
Therefore, the purpose of this study was to use RNA sequencing (RNA-seq) to systematically characterize and compare the regulation of DPGs by CAR in an age- and species-specific manner, using WT, hCAR-TG, and CAR-null mice of neonatal and adult age. The selection of 393 important DPGs was based on previous studies in the literature (Parkinson et al., 2013; Cui and Li, 2016).
Materials and Methods
Chemicals and Reagents.
Corn oil, TCPOBOP, and CITCO were purchased from Sigma-Aldrich (St. Louis, MO). A liquid alanine aminotransferase (ALT) assay kit was purchased from Pointe Scientific Inc. (Canton, MI). RNA-Bee reagent was purchased from Tel-Test Inc. (Friendswood, TX). The primers were designed using Primer3 (http://bioinfo.ut.ee/primer3-0.4.0/) and were purchased from Integrated DNA Technologies (Coralville, IA). High-capacity cDNA reverse transcription kits, Power SYBR Green PCR Master Mix, Bolt 4%–12% Bis-Tris Plus gel, Bolt antioxidant, 10× phosphate-buffered saline (pH 7.4), and UltraPure DNase/RNase-Free Distilled Water were purchased from Life Technologies (Carlsbad, CA). P450-Glo screening systems for CYP1A2, CYP2B6, CYP3A4, and the NADPH regeneration system were purchased from Promega (Madison, WA).
Animal Procedures.
The University of Washington Office of Animal Welfare approved all experiments. Eight-week-old male C57BL/6J mouse breeders were purchased from the Jackson Laboratory (Bar Harbor, ME). Breeders of CAR-null mice in the C57BL/6 background were obtained from Amgen (Thousand Oaks, CA). Breeders of hCAR-TG mice in the C57BL/6 background were provided by the University of Kansas Medical Center (Kansas City, KS) (Zhang and Klaassen, 2013). Mice were housed and bred according to the standard conditions in the animal facilities of the University of Washington, as described by the American Animal Association Laboratory Animal Care Guidelines. All animals were given ad libitum access to purified water and irradiated Picolab Rodent Diet 20 number 5053 (PMI Nutrition International, Brentwood, MO) and were housed under relative humility of 40%–60% on a 12-hour light/dark cycle. Mice were randomly assigned to various treatment groups. To avoid variations due to the estrous cycle, only male pups were used in this study. A dose-response study of TCPOBOP and CITCO was performed at neonatal age 5 days, whereas a single optimal dose was selected at adult age 60 days based on the literature. As described in Fig. 1A, at neonatal age 5 days, WT and CAR-null mice were administered various doses of the mCAR ligand TCPOBOP (0.3 mg/kg, 1 mg/kg, or 3 mg/kg, i.p.) or vehicle (corn oil, 10 ml/kg, i.p.), whereas hCAR-TG mice were administered various doses of the hCAR ligand CITCO (2.5 mg/kg, 10 mg/kg, or 30 mg/kg, i.p.) or vehicle (corn oil, 10 ml/kg, i.p.), once daily for 4 consecutive days (n = 5 per group). At adult age 60 days, WT and CAR-null mice were administered the mCAR ligand TCPOBOP (3 mg/kg, i.p.) or vehicle (corn oil, 10 ml/kg, i.p.), whereas hCAR-TG mice were administered the hCAR ligand CITCO (30 mg/kg, i.p.) or vehicle (corn oil, 10 ml/kg, i.p.) once daily for 4 consecutive days (n = 5 per group). Livers were removed 24 hours after the final dose, immediately frozen in liquid nitrogen, and stored at −80°C until further analysis.
Fig. 1.
(A) Animal dosing regimen: WT, CAR-null, and hCAR-TG mice at two developmental ages were administered vehicle (corn oil, 10 ml/kg, i.p.) or a species-appropriate CAR ligand (i.p.) once daily for 4 consecutive days. Specifically, at neonatal age 5 days, WT and CAR-null mice were administered corn oil or the mCAR ligand TCPOBOP daily at one of the three doses: 0.3, 1, or 3 mg/kg; hCAR-TG mice were administered corn oil or the hCAR ligand CITCO daily at one of the three doses: 2.5, 10, or 30 mg/kg. At adult age 60 days, WT (n = 5 per group) and CAR-null (n = 4 per group) mice were administered corn oil or TCPOBOP daily (3 mg/kg), whereas hCAR-TG mice (n = 5 per group) were administered corn oil or CITCO daily (30 mg/kg). (B) mRNA expression of mCAR in the livers of control and chemical-treated mice of the three genotypes at the two developmental ages. (C) Expression of various hCAR SVs in the livers of vehicle- and CITCO-treated hCAR-TG mice at days 5 and 60. Data were generated by RT-qPCR and are expressed as the mean percent of the housekeeping gene β-actin ± S.E.M. Asterisks indicate statistically significant differences compared with vehicle-treated groups of the same age in WT mice.
RNA Isolation.
Total RNA was isolated from livers of WT, CAR-null, and hCAR-TG mice using RNA-Bee reagent (Tel-Test Inc.) following the manufacturer’s instructions. RNA was dissolved in DNase/RNase-Free water, and the concentrations were determined spectrophotometrically at 260 nm using a Nano-Drop 1000 spectrophotometer (Thermo Scientific, Waltham, MA). Integrity of the total RNA was evaluated using an Agilent 2100 Bioanalyzer (Agilent Technologies Inc., Santa Clara, CA). Samples with RNA integrity values larger than 8.0 were used for further analysis.
Real-Time Quantitative Polymerase Chain Reaction Analysis.
The total RNA samples were reverse transcribed into cDNAs using a high-capacity cDNA kit (Life Technologies) according to the manufacturer’s instructions. A real-time quantitative polymerase chain reaction (RT-qPCR) assay was performed using Power SYBR Green PCR Master Mix (Life Technologies) under a Bio-Rad CFX384 Real-Time Detection System (Bio-Rad, Hercules, CA). Primer sequences for real-time PCR reactions are listed in Supplemental Table 3. Expression values were analyzed by the following equation: relative % = 2−dCt target gene/2−dCt β-actin ×100%, where dCt represents the differences in cycle threshold numbers between each of the target genes and the reference gene average.
cDNA Library Construction and RNA-Seq.
Three representative liver samples from WT (corn oil and TCPOBOP treated) and hCAR-TG mice (corn oil and CITCO treated) were selected for RNA-seq at the University of Washington Genome Sciences Sequencing Facilities. The cDNA libraries from all samples were prepared from 1.25 µg total RNA in an automated and high-throughput format using the TruSeq Stranded mRNA kit (Illumina, San Diego, CA). All steps required for the sequence library construction were performed in an automated workflow on a Sciclone NGSx Workstation (Perkin Elmer, Waltham, MA). During library construction, ribosomal RNA was depleted by means of a poly(A) enrichment. RNA fragmentation and first- and second-strand cDNA syntheses were performed, and each library was technically barcoded using the Illumina adapters and was amplified using a total of 13 cycles of PCR. After amplification and cleanup, library concentrations were quantified using the Quant-iT dsDNA assay (Life Technologies). Libraries were subsequently normalized and pooled based on the Agilent 2100 Bioanalyzer results. Sequencing was performed on an Illumina HiSEq 2000 sequencer using a 50-bp paired-end multiplexing strategy with five samples per lane.
RNA-Seq Data Analysis.
After the sequencing images were generated by the sequencing platform, raw data collection, image analysis, and base calls were generated in Illumina Real-Time Analysis software. Illumina Real-Time Analysis output base calling files were converted to qseq files using the Illumina BCL Converter software and were subsequently converted to FASTQ files for downstream analysis. Custom scripts developed in house were used to process the FASTQ files and to output de-multiplexed FASTQ files by lane and index sequence. Quality of the reads was examined using the FASTX toolkit (http://hannonlab.cshl.edu/fastx_toolkit/; Cold Spring Harbor Laboratory, Cold Spring Harbor, NY) and FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/; Babraham Institute Bioinformatics Group, Cambridge, UK). RNA-seq reads from the FASTQ files were mapped to the UCSC Genome Browser mm10 mouse reference genome (University of California, Santa Cruz, CA) using HISAT (Johns Hopkins University, Baltimore, MD). Aligned data were sorted and converted to BAM format using SAMtools (version 0.1.9), and then Cufflinks (version 2.2.1) was used for isoform assembly and quantitation analysis. mRNA abundance was expressed as fragments per kilobase of exons per million reads mapped (FPKM). Differential expression analysis was performed using Cuffdiff, with a Benjamini–Hochberg false-discovery rate (FDR-BH) < 0.05. The RNA sequencing data is deposited in the NCBI Gene Expression Omnibus database (GSE98666).
Western Blot Analysis.
Liver samples were weighed and transferred into a Dounce homogenizer with 5 volumes (w/v) of ST buffer (10 mM Tris base and 250 mM sucrose, pH 7.5) containing a protease inhibitor cocktail (1:200) (Sigma-Aldrich). The livers were homogenized in an ice-cold condition and then transferred to polycarbonate tubes (Beckman Coulter Inc., Brea, CA). Crude membranes were prepared by ultraspeed centrifugation of the liver homogenates at 10,000g for 10 minutes at 4°C using an L8-70 M Ultracentrifuge (Beckman Coulter Inc.). Protein concentrations were determined using a Qubit Protein Assay Kit (Life Technologies) per the manufacturer’s instructions. Equal amounts of protein samples with loading buffer were loaded and electrophoretically separated on a NuPAGE Novex 4%–12% Bis-Tris gel and then transferred onto a polyvinylidene difluoride blotting membrane at 20 V for 5 hours. Subsequently, membranes were blocked with 5% nonfat milk dissolved in phosphate-buffered saline solution containing 0.05% Tween 20 (PBST) for 1 hour and were then probed with primary antibodies diluted in PBST with 1% nonfat milk at 4°C overnight. The following primary antibodies were used at the stated dilution: mouse anti-rat CYP1A2 (1:1000, sc-53241; Santa Cruz Biotechnology, Inc., Dallas, TX), rabbit anti-mouse Cyp2b10 monoclonal antibody (1:5000, AB9916; EMD Millipore, Billerica, MA), mouse anti-rat CYP3A1 monoclonal antibody (1:500, sc-53246; Santa Cruz Biotechnology, Inc.), and rat anti-mouse multidrug resistance-associated protein Mrp4 (1:2000, M4I-10; University of Kansas Medical Center). The membranes were then incubated with a species-appropriate secondary antibody conjugated with horseradish peroxidase (Sigma-Aldrich) diluted 1% nonfat milk in PBST. Protein bands were developed using a Novex ECL Chemiluminescent Substrate Reagent Kit (Life Technologies). Membranes were stripped and reincubated with primary antibodies for β-actin or histone H3 (Abcam, Cambridge, MA) as loading controls.
Enzyme Activity Quantifications.
Crude membranes were prepared as described in the section above on western blot analysis. Enzyme activities for Cyp1a, Cyp2b, and Cyp3a were quantified using the P450-Glo CYPA1, CYP2B, and CYP3A assay systems (Promega) following the manufacturer’s protocol. Briefly, 20 µg protein and a P450-Glo substrate (100 µM Luciferin-ME for Cyp1a, 3 µM Luciferin-2B6 for Cyp2b, and 3 µM IPA [2-(4-(diisopropoxymethyl)-4,5-dihydrothiazol-2-yl)benzo[d]thiazol-6-ol] for Cyp3a) were combined in potassium phosphate buffer in a 2× concentrated mixture. The reaction was initiated by adding 1 volume of the 2× concentrated NADPH regenerating system (25 µl added for a final volume of 50 µl in an opaque 96-well plate). The products produced in the P450 reaction were detected as a luminescent signal from a luciferase reaction using a Platelumino biomedical system (Promega). Signals were allowed to stabilize for 20 minutes at room temperature before they were read on a luminometer.
Serum ALT Quantification.
Serum samples from WT (corn oil, TCPOBOP), CAR-null (corn oil, TCPOBOP), and hCAR-TG (corn oil, CITCO) mice were analyzed by standard enzyme-colorimetric assays using a liquid ALT kit according to the manufacturer’s protocol (Pointe Scientific). Absorbance was quantified spectrophotometrically at wavelengths of 340 and 540 nm.
Statistical Analysis.
A total of 393 DPGs with known functions for xenobiotic biotransformation were retrieved from the Cufflinks output for further analysis. Genes that were expressed with an average FPKM > 1 in at least one group were considered to be significantly expressed in the liver, and this criterion was selected based on the literature (Peng et al., 2016). Differential expression of the RNA-seq data was considered at FDR-BH < 0.05. A Venn diagram was generated using the VennDiagram package in R software (R Project for Statistical Computing, Vienna, Austria) utilizing the draw.quad.venn function. Heatmaps were plotted using JMP software (Ward method, standardized data; SAS, Cary, NC). For RT-qPCR and protein assays, two-way analysis of variance was used for multiple-group comparisons (SPSS Statistics 19; IBM, Armonk, NY) (Tukey’s post hoc test, P < 0.05).
Results
Regulation of mCAR and hCAR Signaling in the Developing Livers of WT, CAR-Null, and hCAR-TG Mice.
The animal dosing regimen is shown in Fig. 1A. Because the dose response of CAR ligands in adult mice and primary hepatocyte culture is well established (Honkakoski et al., 1992; Honkakoski and Negishi, 1998; Maglich et al., 2003; Scheer et al., 2008), a single optimal dose at adult age was selected in our study. Because relatively less is known regarding the dose-response relationship between CAR activation and targeted DPG expression in newborns, a dose response of TCPOBOP and CITCO was performed at neonatal age 5 days. Serum ALT levels were low in all treatment groups, except for a moderate increase in the livers of WT mice at the medium dose of TCPOBOP. The highest average serum ALT value was 33.54 U/l, which was within the normal range (Supplemental Fig. 1). To confirm the mouse genotypes and the regulation of CAR in the liver by age and CAR ligands, RT-qPCR was performed in the livers of WT, CAR-null, and hCAR-TG mice (Fig. 1B). As expected, mCAR mRNA was only detected in the livers of WT mice but not in the livers of CAR-null or hCAR-TG mice. During development, mCAR mRNA increased from 5 to 60 days of age in the livers of control WT mice. Interestingly, the mCAR ligand TCPOBOP at the highest dose markedly decreased mCAR mRNA at both ages (Fig. 1B; Supplemental Fig. 2A). These data suggest a negative feedback mechanism upon pharmacological activation of CAR to reduce CAR signaling at the transcription level. Regarding the regulation of hCAR mRNA, five hCAR SVs (namely, SV0, SV1, SV2, SV3, and SV4; Lamba et al., 2004; Zhang et al., 2013) were quantified as shown in Fig. 1C. This nomenclature is the same as used in a previous study (Jinno et al., 2004), in that SV0 refers to NM_005122, SV1 has an in-frame 12-bp insertion, SV2 has an in-frame 15-bp insertion, and SV3 has both 12-bp and 15-bp insertions, whereas SV4 has an in-frame 117-bp deletion. However, this nomenclature is slightly different from another study (Arnold et al., 2004), in that SV0 (our study) is termed SV3, SV3 (our study) is termed SV6 by Arnold et al., and SV2 and SV4 are the same for both articles. SV0 and SV2 were the major hCAR transcripts, followed by SV1 and SV3, which were expressed at lower levels; SV4 was minimally expressed throughout liver development. All four expressed transcripts (SV0–SV3) were increased from 5 to 60 days of age. Unlike mCAR, the hCAR ligand CITCO did not alter the expression of any hCAR SVs at either age. The prototypical CAR target gene Cyp2b10 was upregulated by both mCAR and hCAR activation at day 60 in a CAR-dependent manner; however, at day 5, Cyp2b10 was upregulated by mCAR at all three doses of TCPOBOP in the livers of WT mice and by hCAR at the medium and high doses of CITCO in the livers of hCAR-TG mice (Supplemental Fig. 2B). In summary, these observations confirmed the three genotypes of mice and demonstrated that both the mCAR and hCAR transcripts were upregulated from neonatal to adult age. However, only the mCAR ligand downregulated hepatic mCAR expression, whereas the hCAR ligand did not alter the expression of hCAR transcripts at either developmental age. In newborns, both mCAR and hCAR are functionally active and respond to chemical induction of target genes.
Regulation of Hepatic DPGs by Pharmacological Activation of mCAR and hCAR during Development.
To determine the effect of age, species, and pharmacological activation of CAR on the regulation of DPGs in the liver, RNA-seq was performed in the livers of WT and hCAR-TG mice treated with vehicle or a species-appropriate CAR ligand at 5 and 60 days of age.
A dose response of the mCAR ligand TCPOBOP and the hCAR ligand CITCO was established previously in adult mice and primary hepatocyte cultures (Honkakoski et al., 1992; Honkakoski and Negishi, 1998; Maglich et al., 2003; Scheer et al., 2008); however, very little is known regarding the optimal dose range of CAR activation in newborns. Therefore, a dose-response study of TCPOBOP and CITCO was performed at neonatal age 5 days in the livers of WT, CAR-null, and hCAR-TG mice (Supplemental Figs. 2 and 3). mCAR mRNA was downregulated, whereas the prototypical CAR target gene Cyp2b10 was upregulated by TCPOBOP at all doses in the livers of WT mice at 5 days of age in a CAR-dependent manner (Supplemental Fig. 2). Interestingly, at neonatal age 5 days, many other DPGs were dose-dependently regulated by CAR activation. For example, phase I enzymes Cyp1a2, Cyp3a11, Nqo1, and Aldh1a1; phase II enzymes Ugt2b35, Gstm1, Gstm3, and Sult5a1; and efflux transporters Mrp3 and Mrp4 were all upregulated by both mCAR and hCAR activation in a CAR-dependent manner (Supplemental Fig. 3). Based on the dose-response studies on DPG expression in WT and hCAR-TG mice at age 5 days (Supplemental Figs. 2 and 3), the highest doses were selected for RNA-seq for both TCPOBOP- and CITCO-treated groups, whereas a single optimal dose was selected at adult age 60 days based on the literature.
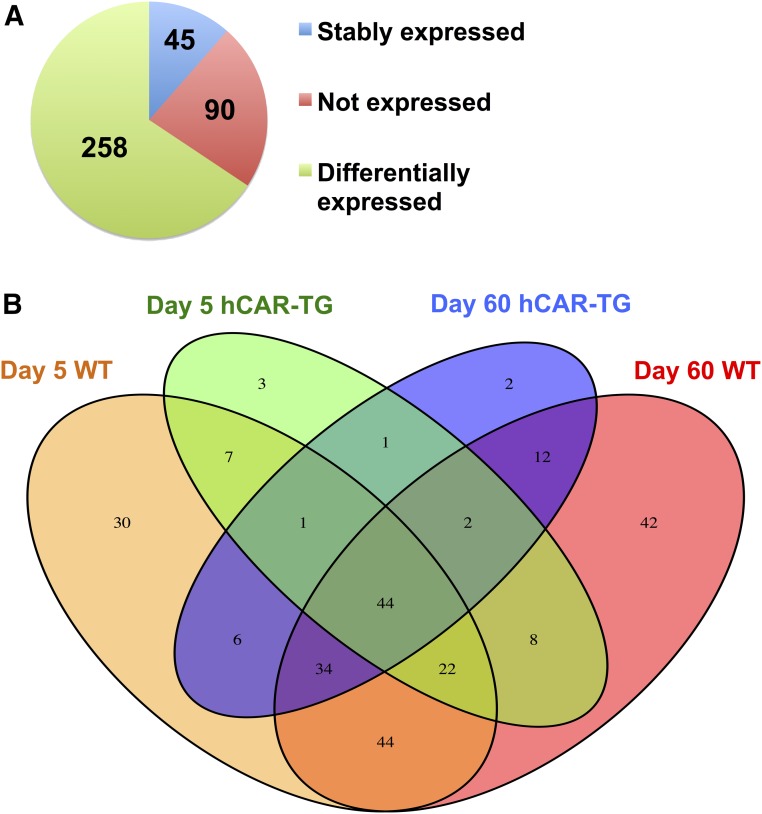
RNA-seq generated approximately 47–68 million reads per sample, among which approximately 40–60 million reads were uniquely mapped to the mouse reference genome (National Center for Biotechnology Information no. GRCm/38/mm10). As shown in Fig. 2A, among all 393 DPGs with known important functions in xenobiotic biotransformation (Supplemental Table 1) (Parkinson et al., 2013), 90 DPGs were not expressed in the livers of any groups (threshold: average FPKM < 1 in all treatment groups). However, 303 genes were expressed in the livers of at least one group, among which 258 DPGs were differentially regulated by mCAR or hCAR activation on either day 5 or day 60 (FDR-BH < 0.05), and 45 genes were stably expressed among all treatment groups.
Fig. 2.
RNA-seq was conducted in the livers of control and CAR ligand–treated male WT and hCAR-TG mice at day 5 and day 60 of age (n = 3 per group). (A) Pie chart showing DPGs that are not expressed in any groups (red), stably expressed in all groups (blue), and differentially expressed by the CAR ligand in at least one of the four comparisons (i.e., day 5 WT vehicle versus day 5 WT TCPOBOP; day 60 WT vehicle versus day 60 WT TCPOBOP; day 5 hCAR-TG vehicle versus day 5 hCAR-TG CITCO; or day 60 hCAR-TG vehicle versus day 60 hCAR-TG CITCO, FDR-BH < 0.05). (B) Venn diagram showing the commonly and uniquely regulated DPGs among the four comparisons as described in (A). The Venn diagram was generated using the VennDiagram package in R software utilizing the draw.quad.venn function.
A Venn diagram (Fig. 2B) of the 258 differentially regulated DPGs displayed a clear separation by age and species of CAR (threshold: average FPKM > 1 in at least one group; FDR-BH < 0.05 in at least one comparison). Regarding age specificity, between day 5 WT (orange) and day 60 WT (red), there were 144 DPGs commonly regulated by mCAR at both ages, whereas 44 DPGs were uniquely regulated by mCAR activation only at neonatal age day 5 and 64 DPGs were uniquely regulated by mCAR activation only at day 60. Similarly, hCAR activation also displayed age specificity, in that between day 5 hCAR-TG (green) and day 60 hCAR-TG (blue), there were 48 DPGs differentially regulated at both ages by hCAR activation, whereas 40 DPGs were uniquely regulated by hCAR activation only at neonatal age day 5 and 54 DPGs were uniquely regulated by hCAR activation at adult age day 60.
Regarding species specificity, at the same developmental age, mCAR and hCAR produced overlapping but not identical effects on the expression of DPGs (Fig. 2B). At neonatal age day 5, 74 DPGs were differentially regulated by both mCAR and hCAR activation, whereas more DPGs (n = 114) were uniquely regulated only by mCAR and fewer DPGs (n = 14) were uniquely regulated only by hCAR. A similar trend was observed at adult age day 60, in that 92 DPGs were differentially regulated by both mCAR and hCAR activation, whereas 116 were uniquely regulated only by mCAR and fewer DPGs (n = 10) were uniquely regulated only by hCAR.
In summary, pharmacological activation of CAR displayed both age and species specificities in modulating the expression of DPGs in the liver. Although age profoundly alters the CAR-targeted DPG profiles in both genotypes, mCAR appears to target more DPGs than hCAR at both ages.
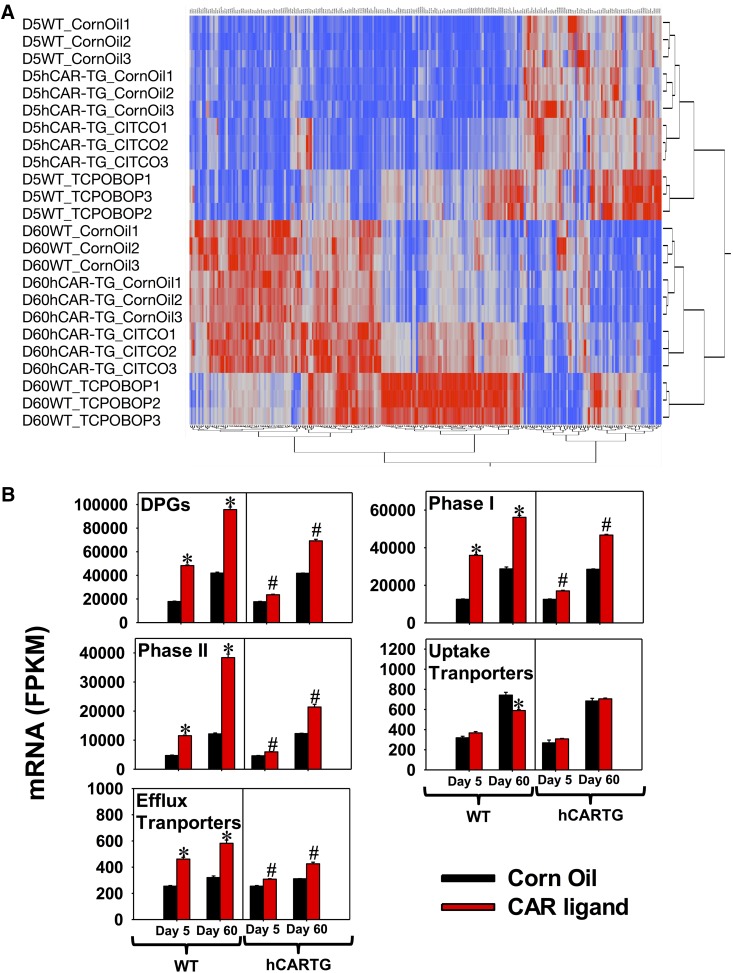
A two-way hierarchical clustering dendrogram of individual FPKMs of the 258 differentially regulated DPGs is shown in Fig. 3A. As expected, samples within the same treatment group were clustered together. Age appeared to be a more predominant separation factor than other factors such as chemical treatment and genotypes, evidenced by the first two major branches splitting the two ages in the dendrogram. DPGs in the upper right corner were neonatally enriched, whereas DPGs in the lower left corners were adult enriched. At the same developmental age, mCAR activation generally resulted in greater alterations in targeted DPG expression than hCAR activation, evidenced by the further separation between the WT corn oil–treated group and the WT TCPOBOP-treated group within the same age. hCAR activation appeared to upregulate more DPGs at adult age day 60 compared with those at neonatal age day 5.
Fig. 3.
(A) Two-way hierarchical clustering dendrogram of all 258 differentially regulated DPGs by pharmacological activation of CAR in the livers of WT and hCAR-TG mice at day 5 and day 60 of age (FDR-BH < 0.05) (JMP version 12.0, using the Ward method and standardized data option). Each row represents an individual mouse with FPKM values of all differentially regulated DPGs in the liver, whereas each column shows the mRNA expression of an individual DPG among all of the mice. Red represents relatively high expression, and blue represents relatively low expression. (B) Cumulative FPKM of differentially regulated DPGs, phase I enzymes, phase II enzymes, uptake transporters, and efflux transporters in the livers of control and CAR ligand–treated WT and hCAR-TG mice at day 5 and day 60 of age. Individual FPKM values of genes of the same category were added together within the same treatment group, and results are expressed as the mean FPKM ± S.E.M. of three individual biologic replicates per group. Asterisks indicate statistically significant differences compared with vehicle-treated groups of the same age in WT mice; pound signs indicate statistically significant differences compared with vehicle-treated groups of the same age in hCAR-TG mice.
To estimate the overall capacity of all DPGs in the liver from the transcriptomic data, cumulative FPKM values of the 258 differentially regulated DPGs were plotted as shown in Fig. 3B (threshold: average FPKM > 1 in at least one group; FDR-BH < 0.05 in at least one comparison). Both mCAR and hCAR activation upregulated the cumulative expression of DPGs at both days 5 and 60; however, hCAR activation resulted in a much weaker fold increase compared with mCAR activation, and hCAR activation at day 5 produced an even weaker inducible effect than at day 60. Regarding the regulation of specific categories of DPGs, in control livers of both genotypes, the cumulative mRNAs of all DPGs, phase I enzymes, phase II enzymes, uptake transporters, and efflux transporters were all increased from 5 to 60 days of age. After CAR activation, cumulative expression of phase I enzymes, phase II enzymes, and efflux transporters was upregulated by both mCAR and hCAR at both developmental ages, with mCAR producing greater mRNA fold increases than hCAR at both ages, whereas day 60 was a more inducible age than day 5 for hCAR activation. In contrast, cumulative mRNAs of uptake transporters were not readily inducible by CAR at either age or genotype; mCAR activation downregulated cumulative mRNAs of uptake transporters at 60 days of age.
In summary, the majority of liver-expressed DPGs were differentially regulated by pharmacological activation of CAR in an age- and species-specific manner. In general, DPGs in the phase I enzyme, phase II enzyme, and efflux transporter categories were more responsive to CAR activation than uptake transporters. Although mCAR activation increased the cumulative expression of enzymes and efflux transporters at both ages, hCAR activation preferably upregulated the cumulative expression of these genes in adult age.
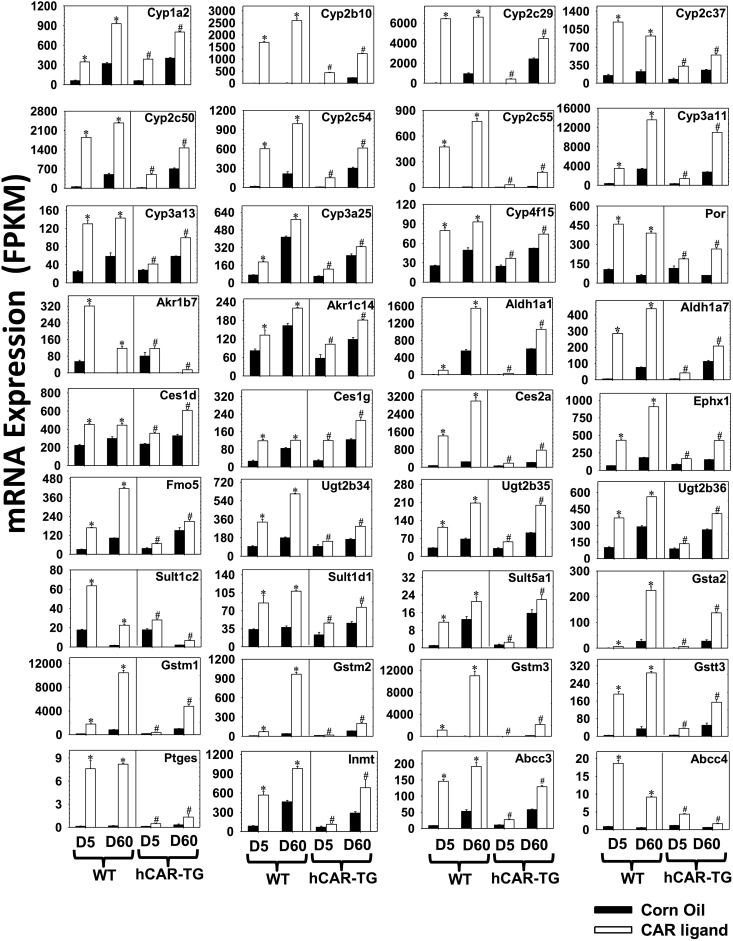
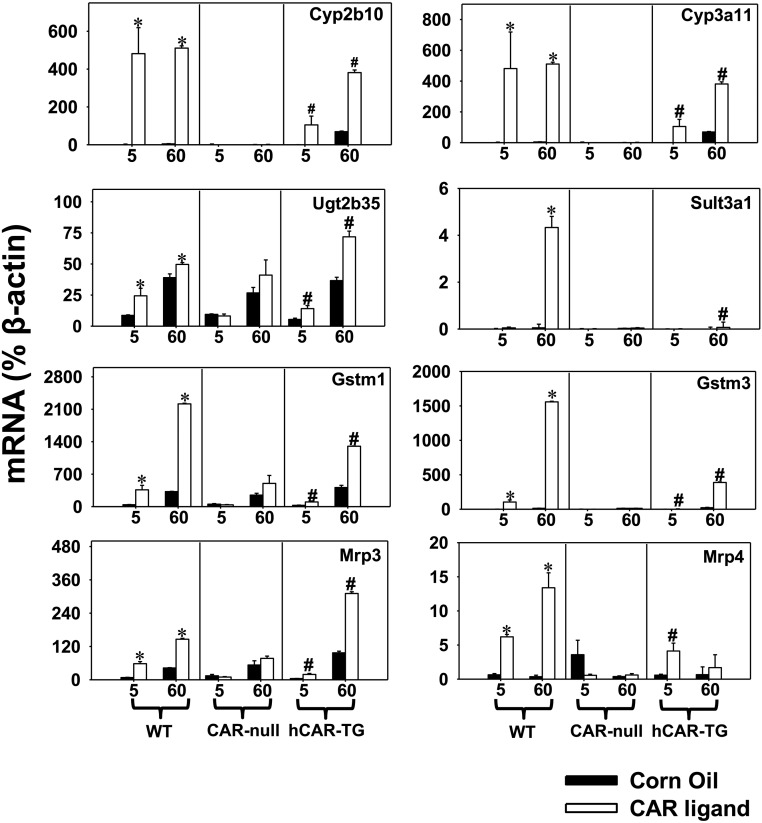
DPGs That Were Common Targets of mCAR and hCAR at Both Developmental Ages 5 Days and 60 Days.
Among the 44 differentially regulated DPGs by both mCAR and hCAR activation at both ages, DPGs (n = 36 in total) that were altered at least 30% (average FPKM > 1 in at least one group, FDR-BH < 0.05 in all groups) are shown in Fig. 4. All 36 DPGs were upregulated by CAR activation across species and age, indicating that the effect of CAR on common DPG targets across species and development is inducible rather than suppressive. Many commonly regulated DPGs had a greater mRNA fold increase on neonatal age day 5 compared with adult age day 60 of the same genotype (Table 1). Specifically, this “neonatal preference” was manifested by a greater fold increase at day 5 by both mCAR and hCAR activation for the mRNAs of P450s, including Cyp1a2, Cyp2b10, Cyp2c29, Cyp2c37, Cyp2c50, Cyp2c54, and Cyp3a25; other phase I enzymes, such as Aldh1a1 and Aldh1a7, Ces1g, and Fmo5; phase II enzymes Gstt3 and Sult5a1; and basolateral efflux transporters Abcc3 (Mrp3) and Abcc4 (Mrp4) [threshold: (fold increase at day 5) – (fold increase at day 60) > 30%] (Fig. 4; Table 1). In addition, mCAR activation resulted in a greater fold increase at day 5 in the mRNAs of the following: P450s (Cyp2c55, Cyp3a11, Cyp3a13, and Cyp4f15), carboxyesterases (Ces1d and Ces2a), epoxide hydrolase Ephx1, glucuronidation enzymes Ugt2b35 and Ugt2b36, glutathione conjugation enzyme prostaglandin E synthase Ptges, and methyltransferase indolethylamine N-methyltransferase Inmt, compared with day 60 WT livers. hCAR activation resulted in a moderately greater fold increase at day 5 in the mRNA of Sult1d1 compared with hCAR-TG livers at day 60. Although most commonly regulated DPGs were preferentially upregulated in newborns, seven DPGs (P450 oxidoreductase Por, aldo-keto reductase Akr1b7, Sult1c2, Gsta2, Gstm1, Gstm2, and Gstm3) were preferentially upregulated by both mCAR and hCAR at adult age day 60, and four DPGs (Cyp2c55, Ces2a, Ephx1, and Ugt2b35) were preferentially upregulated by hCAR at adult age day 60 (Table 1). Table 1 also shows DPGs that were upregulated by CAR ligands to a similar extent between day 5 and day 60 of the same genotype.
Fig. 4.
DPGs that were commonly regulated by both mCAR and hCAR and at both developmental ages (FDR-BH < 0.05, and with at least a 30% increase or decrease in mRNA expression). There are three individual biologic replicates per group. Asterisks indicate statistically significant differences compared with vehicle-treated groups of the same age in WT mice; pound signs indicate statistically significant differences compared with vehicle-treated groups of the same age in hCAR-TG mice.
TABLE 1.
DPGs commonly regulated by both mCAR and hCAR activation and at both developmental ages
Data are expressed as ratios of FPKM in the CAR ligand–treated group over the vehicle-treated group of the same age and genotype.
| Gene Symbol | WT (TCPOBOP/Corn Oil) |
hCAR-TG (CITCO/Corn Oil) |
||
|---|---|---|---|---|
| Day 5 | Day 60 | Day 5 | Day 60 | |
| Cyp1a2 | 5.72 | 2.90 | 6.48 | 1.99 |
| Cyp2b10 | 397.45 | 201.52 | 154.93 | 5.44 |
| Cyp2c29 | 215.63 | 6.98 | 73.99 | 1.83 |
| Cyp2c37 | 7.98 | 4.09 | 4.37 | 2.16 |
| Cyp2c50 | 33.58 | 4.74 | 22.84 | 2.07 |
| Cyp2c54 | 35.16 | 4.66 | 20.73 | 2.04 |
| Cyp2c55 | 284.18 | 124.01 | 7.98 | 14.05 |
| Cyp3a11 | 9.48 | 4.03 | 4.24 | 4.00 |
| Cyp3a13 | 5.24 | 2.44 | 1.48 | 1.70 |
| Cyp3a25 | 2.67 | 1.39 | 2.09 | 1.33 |
| Cyp4f15 | 3.14 | 1.89 | 1.50 | 1.42 |
| Por | 4.34 | 6.46 | 1.64 | 4.42 |
| Akr1b7 | 5.93 | 371.33 | 1.45 | 17.68 |
| Akr1c14 | 1.62 | 1.35 | 1.80 | 1.53 |
| Aldh1a1 | 23.59 | 2.80 | 5.96 | 1.76 |
| Aldh1a7 | 57.03 | 5.77 | 7.37 | 1.84 |
| Ces1d | 2.04 | 1.50 | 1.51 | 1.85 |
| Ces1g | 4.55 | 1.44 | 4.17 | 1.70 |
| Ces2a | 18.97 | 12.18 | 2.94 | 3.59 |
| Ephx1 | 6.37 | 5.00 | 1.97 | 2.78 |
| Fmo5 | 5.53 | 4.07 | 1.84 | 1.39 |
| Ugt2b34 | 3.50 | 3.39 | 1.52 | 1.77 |
| Ugt2b35 | 3.45 | 3.08 | 1.85 | 2.18 |
| Ugt2b36 | 3.66 | 1.94 | 1.53 | 1.56 |
| Sult1c2 | 3.59 | 13.44 | 1.57 | 3.28 |
| Sult1d1 | 2.54 | 2.87 | 2.00 | 1.68 |
| Sult5a1 | 10.98 | 1.63 | 1.86 | 1.40 |
| Gsta2 | 7.09 | 8.54 | 4.72 | 5.09 |
| Gstm1 | 12.39 | 12.74 | 2.05 | 4.81 |
| Gstm2 | 8.01 | 23.52 | 1.50 | 2.40 |
| Gstm3 | 127.97 | 192.51 | 3.09 | 16.06 |
| Gstt3 | 43.17 | 8.47 | 6.79 | 3.09 |
| Ptges | 53.61 | 42.97 | 3.74 | 4.12 |
| Inmt | 6.71 | 2.12 | 1.65 | 2.38 |
| Abcc3 | 16.53 | 3.62 | 2.56 | 2.22 |
| Abcc4 | 21.09 | 16.46 | 3.68 | 2.65 |
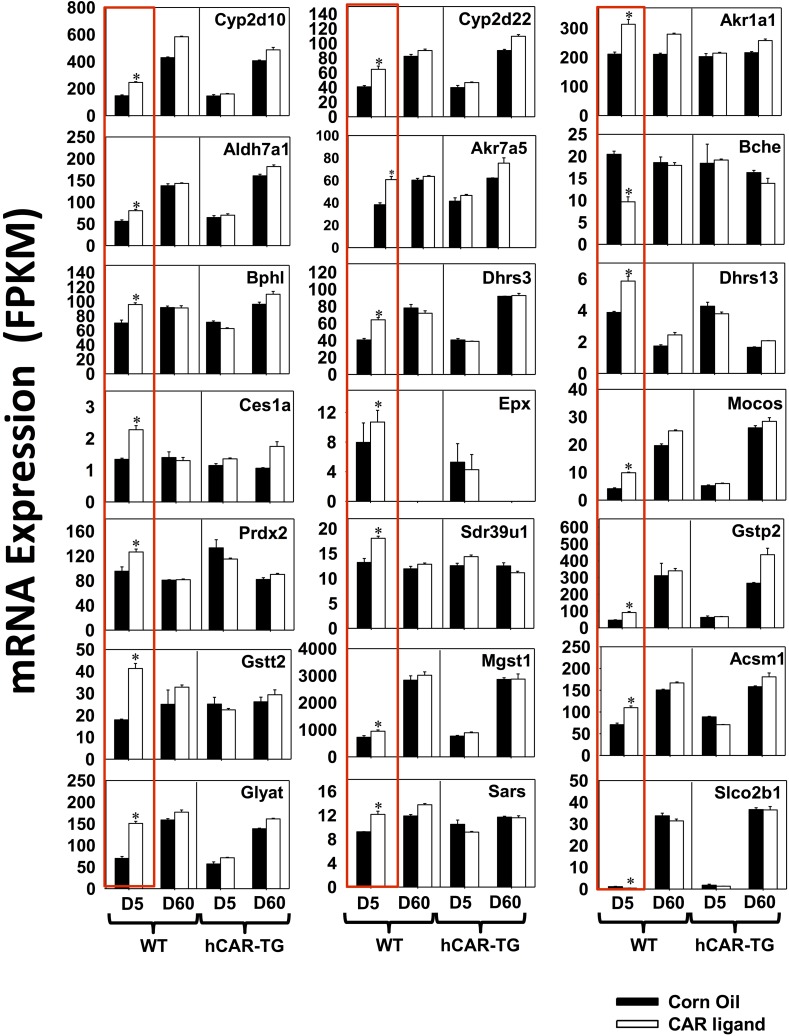
DPGs That Were Uniquely Regulated by mCAR at Neonatal Age Day 5.
Among the 30 DPGs uniquely regulated by mCAR at neonatal age day 5 (Fig. 2B), there were 21 DPGs for which the mRNAs were altered at least 30%. These genes are shown in Fig. 5, highlighted in red boxes (threshold: average FPKM > 1 in at least one group, FDR-BH < 0.05 only in the day 5 WT group and fold change above 30%). These DPGs include the following: phase I P450 enzymes Cyp2d10 (67% increase) and Cyp2d22 (58% increase); other phase I enzymes Akr1a1 (49% increase), Akr7a5 (58%), Aldh7a1 (44% increase), butyrylcholinesterase Bche (53% decrease), biphenyl hydrolase-like (serine hydrolase, breast epithelial mucin-associated antigen) Bphl (36% increase), Ces1a (70% increase), dehydrogenase/reductase (SDR family) Dhrs3 (58% increase), Dhrs13 (51% increase), eosinophil peroxidase Epx (35% increase), molybdenum cofactor sulfurase Mocos (1.4-fold increase), peroxiredoxin Prdx2 (33% increase), and Sdr39u1 (37% increase); phase II glutathione conjugation enzymes Gstp2 (100% increase), Gstt2 (1.31-fold increase), and microsomal glutathione S-transferase (Mgst1) (31% increase); phase II amino acid conjugation enzymes acyl-CoA synthetase medium-chain family member Acsm1 (55% increase), glycine-N-acyltransferase Glyat (1.16-fold increase), and seryl-aminoacyl-tRNA synthetase Sars (31% increase); as well as the basolateral uptake transporter Slco2b1 (Oatp2b1) (62% decrease, although the basal expression was very low). At adult age day 60 in WT mice and at both developmental ages in hCAR-TG mice, these genes were not readily altered by CAR activation.
Fig. 5.
DPGs that were differentially regulated only by mCAR activation at day 5 (FDR-BH < 0.05, and with at least a 30% increase or decrease in mRNA expression). There are three individual biologic replicates per group. Asterisks indicate statistically significant differences compared with vehicle-treated groups of the same age in WT mice.
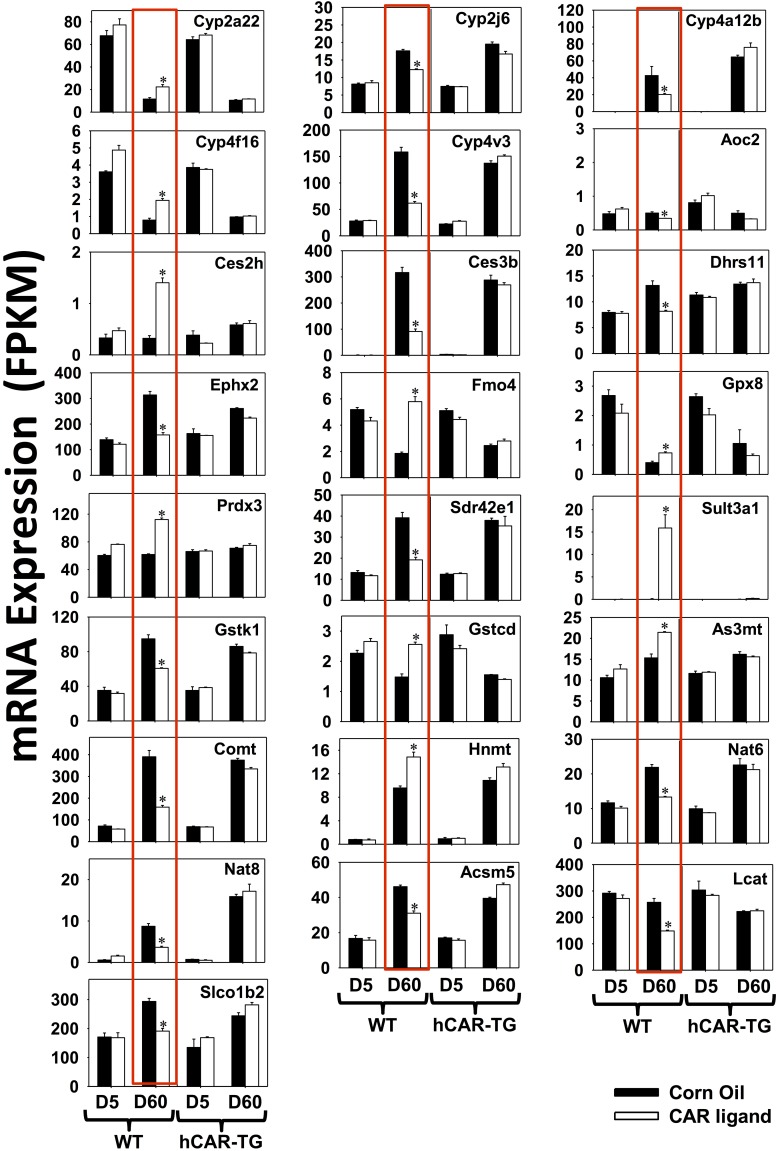
DPGs That Were Uniquely Regulated by mCAR at Adult Age Day 60.
Among the 42 DPGs uniquely regulated by mCAR at day 60 of age (Fig. 2B), there were 25 DPGs for which the mRNAs were altered at least 30%. These genes are shown in Fig. 6 and highlighted in red boxes (threshold: average FPKM > 1 in at least one group, FDR-BH < 0.05 only in day 60 WT group and fold change above 30%). These DPGs include the following: phase I P450s Cyp2a22 (93% increase), Cyp2j6 (30% decrease), Cyp4a12b (52% decrease), Cyp4f16 (1.45-fold increase), and Cyp4v3 (61% decrease); other phase I enzymes, including copper-containing amine oxidase Aoc2 (31% decrease), Ces2h (3.36-fold increase), Ces3b (71% decrease), Dhrs11 (38% decrease), Ephx2 (50% decrease), Fmo4 (2.13-fold increase), glutathione peroxidase Gpx8 (82% increase), peroxiredoxin Prdx3 (82% increase), and short chain dehydrogenase/reductase family Sdr42e1 (51% decrease); phase II sulfotransferase Sult3a1 (185.20-fold increase); glutathione transferases Gstk1 (36% decrease) and Gstcd (73% increase); methyltransferases arsenic (+3 oxidation state) methyltransferase As3mt (40% increase), catechol-O-methyltransferase Comt (59% decrease), and histamine N-methyltransferase Hnmt (55% increase); acetyltransferases Nat6 (39% decrease) and Nat8 (58% decrease); amino acid conjugation enzyme Acm5 (33% decrease); fatty acid conjugation enzyme lecithin cholesterol acyltransferase Lcat (42% decrease); and the basolateral uptake transporter Slco1b2 (Oatp1b2) (35% decrease).
Fig. 6.
DPGs that were differentially regulated only by mCAR activation at day 60 (FDR-BH < 0.05, and with at least a 30% increase or decrease in mRNA expression). There are three individual biologic replicates per group. Asterisks indicate statistically significant differences compared with vehicle-treated groups of the same age in WT mice.
DPGs That Were Uniquely Regulated by hCAR at Developmental Ages Day 5 or Day 60.
Compared with mCAR, unique hCAR-targeted DPGs were fewer at both developmental ages (three at day 5 and two at day 60) (Fig. 2B). Specifically, as highlighted in red boxes, only Fmo1 (35% decrease), monoamine oxidase Maoa (30% decrease), and Nat2 (58% increase) mRNAs were uniquely regulated by hCAR activation at neonatal age day 5 (Fig. 7A) (average FPKM > 1 in at least one group, FDR-BH < 0.05, and fold change at least 30%), and only Ugt2b5 mRNA (38% increase) was uniquely regulated by hCAR activation at adult age day 60 (Fig. 7B).
Fig. 7.
DPGs that were differentially regulated only by hCAR activation at day 5 (A) and at day 60 (B) (FDR-BH < 0.05, and with at least a 30% increase or decrease in mRNA expression). There are three individual biologic replicates per group. Pound signs indicate statistically significant differences compared with vehicle-treated groups of the same age in hCAR-TG mice.
Age Affecting the CAR-Mediated Regulatory Patterns of Certain DPGs in the Liver.
Interestingly, in the livers of WT mice, between day 5 and day 60, mCAR activation produced opposite effects in the regulation of certain DPGs, including P450s (Cyp2c38, Cyp2c40, Cyp2c69, Cyp2e1, Cyp2f2, and Cyp2j5), alcohol dehydrogenase Adh7, aldehyde oxidase Aox2, Ugt2a3, and Nnmt, as shown in Fig. 8A (highlighted in red boxes). Specifically, these DPGs were all upregulated by mCAR at neonatal age day 5 but were all downregulated by mCAR activation at adult age day 60. Among these DPGs, Cyp2c40 and Cyp2c69 were also downregulated by hCAR activation at adult age day 60. Similarly, age also reversed the CAR-mediated regulatory patterns in the livers of hCAR-TG mice (Fig. 8B), in that Cyp4a14 and Cyp4a32 mRNAs were both upregulated by hCAR activation at neonatal age day 5 but were both downregulated by hCAR activation at adult age day 60 (highlighted in red boxes).
Fig. 8.
Age specificity in CAR-mediated regulation of DPGs in WT (A) and hCAR-TG (B) mice. The CAR-mediated regulatory patterns between day 5 and day 60 were opposite in the livers of the same genotype (FDR-BH < 0.05). There are three individual biologic replicates per group. Data are shown as the mean FPKM ± S.E.M. Asterisks indicate statistically significant differences compared with vehicle-treated groups in WT mice; pound signs indicate statistically significant differences compared with vehicle-treated groups in hCAR-TG mice.
In summary, age was a critical modifying factor in CAR signaling and subsequent regulatory patterns of DPGs in the liver. In general, neonatal age provided a more permissive environment for upregulation of DPGs commonly targeted by CAR of both the mouse and human origins. However, unique age- and/or species-specific regulations of DPGs were also observed, and in certain scenarios, age completely reversed the CAR-mediated regulatory patterns of certain DPGs.
RT-qPCR Confirmation of Selected DPGs in the Livers of WT, CAR-Null, and hCAR-TG Mice.
RT-qPCR was performed to validate selected DPGs targeted by CAR in the livers of WT, CAR-null, and hCAR-TG mice. A shown in Fig. 9, RT-qPCR not only confirmed the RNA-seq data of the “universally regulated” DPGs by CAR activation (namely, Cyp2b10, Cyp3a11, Ugt2b35, Gstm1, Gstm3, Mrp3, and Mrp4) (Fig. 4) as well as Sult3a1 that was uniquely regulated in day 60 WT livers (Fig. 6), but it also demonstrated that these differential regulations were CAR dependent (data from CAR-null mice). In summary, RT-qPCR results confirmed the regulation of these DPGs by CAR in an age- and species-specific manner.
Fig. 9.
Validation of selected DPGs in the livers of WT, CAR-null, and hCAR-TG mice treated with corn oil or a species-appropriate CAR ligand using RT-qPCR (n = 5 biologic replicates per group). Data were normalized to the expression of the housekeeping gene β-actin and are presented as the mean ± S.E.M. Asterisks indicate statistically significant differences compared with vehicle-treated groups of the same age in WT mice; pound signs indicate statistically significant differences compared with vehicle-treated groups of the same age in hCAR-TG mice.
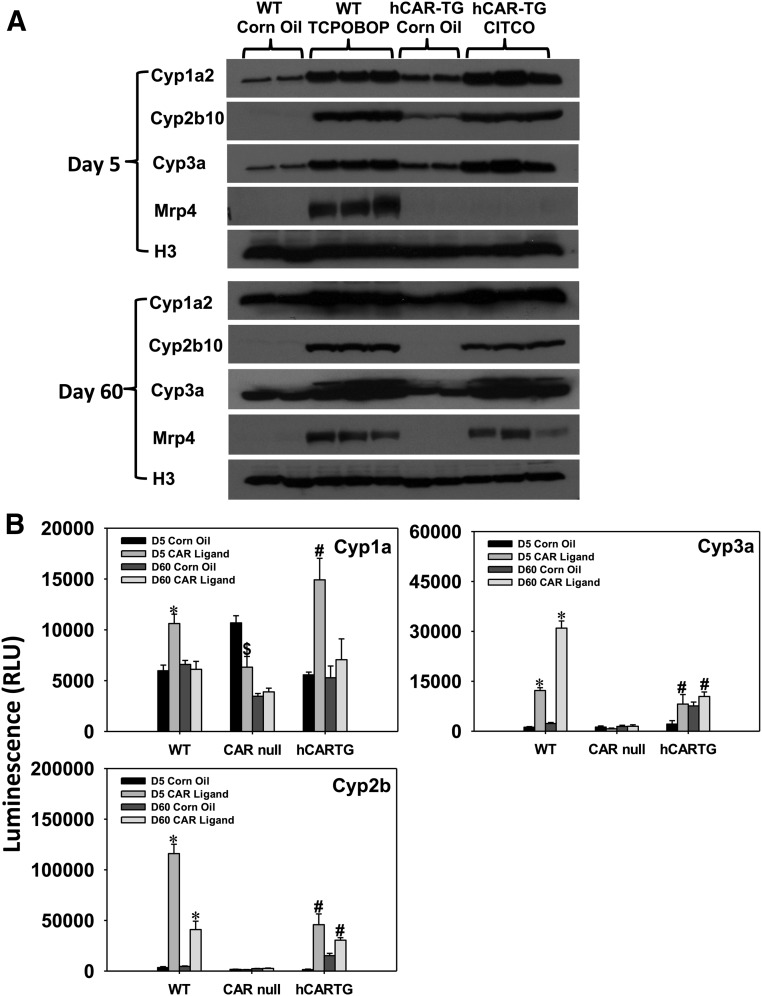
Protein Expression and Enzyme Activities of Selected CAR-Targeted DPGs in the Liver.
Western blot experiments were performed to determine protein expression of Cyp1a2, Cyp2b10, Cyp3a, and basolateral efflux transporter Mrp4 (Fig. 10) in the livers of control and CAR ligand–treated WT or hCAR-TG mice at days 5 and 60 of developmental age. Cyp1a2 protein was upregulated by CAR ligands in the livers of both WT and hCAR-TG mice more prominently at 5 days of age (5-fold by TCPOBOP and 3.14-fold by CITCO, respectively; quantification with ImageJ software [data not shown]; National Institutes of Health, Bethesda, MD). In contrast, although there was also an apparent increase in Cyp1a2 protein by CAR ligands at day 60, the fold increase was very moderate. As shown in Fig. 10B, consistent with the protein data, Cyp1a enzyme activities were preferably increased by activation of CAR of both species at neonatal age day 5 but not adult age day 60. Considering that Cyp1a2 mRNA was marked upregulated at adult age day 60 (Fig. 4), these observations suggest that there is a suppressive post-transcriptional mechanism to prevent the upregulation of a functional protein. The day 5 specific upregulation of Cyp1a enzyme activity was CAR dependent; in fact, , there was even a constitutive decrease in Cyp1a enzyme activity in the livers of 5-day-old CAR-null mice (Fig. 10B).
Fig. 10.
(A) Western blotting analysis of the proteins Cyp1a2, Cyp2b10, Cyp3a, Mrp4, and histone H3 (loading control) in the livers of WT and hCAR-TG mice treated with corn oil or a species-appropriate CAR ligand at day 5 (top) and day 60 (bottom). Due to space limitations on the gel, two representative control samples and three CAR ligand–treated samples were loaded. (B) Enzyme activities of Cyp1a, Cyp2b, and Cyp3a in crude membranes of the livers from WT, CAR-null, and hCAR-TG mice treated with corn oil or a species-appropriate CAR ligand (n = 5 per group). Promega P450-Glo enzyme activity assay systems were used as described in the Materials and Methods. Data are presented as mean ± S.E.M. Asterisks indicate statistically significant differences compared with vehicle-treated groups of the same age in WT mice; dollar signs indicate statistically significant differences compared with vehicle-treated groups of the same age in CAR-null mice; and pound signs indicate statistically significant differences compared with vehicle-treated groups of the same age in hCAR-TG mice.
CAR activation in the livers of WT and hCAR-TG mice upregulated protein expression of Cyp2b10 and Cyp3a11 at both day 5 and day 60 of age, consistent with mRNA data (Fig. 10A). The fold induction of Cyp2b enzyme activity was most prominent by TCPOBOP and CITCO at 5 days of age (16.92-fold and 8.26-fold, respectively), followed by upregulation at 60 days of age (7.35-fold and 112%, respectively) (Fig. 10B). Cyp3a activity was induced in TCPOBOP-treated mouse livers at ages 5 days and 60 days (9.34-fold and 12.16-fold, respectively), whereas this induction was observed in the CITCO-treated group to a much lesser extent (Fig. 10B).
In the liver of WT mice, Mrp4 protein was upregulated at day 5 by TCPOBOP and to a lesser extent at day 60; however, in the liver of hCAR-TG mice, Mrp4 protein was only upregulated by CITCO at day 60 but not at day 5 (Fig. 10A). To note, although Mrp4 mRNA was upregulated by CITCO at day 60 in CAR-TG livers, both the basal and induced levels of this gene were very low compared with at neonatal age, suggesting that the apparent mRNA increase from low levels does not translate into biologic significance.
Discussion
Taken together, this study used a transcriptomic approach to compare age and species specificities of mCAR and hCAR in modulating in vivo pharmacological regulation of drug-metabolizing enzymes and transporters in the livers of mice. This study also validated the necessity of CAR in regulating the expression of selected DPGs in the liver of CAR-null mice. Three regulatory patterns have been identified: DPGs in pattern 1 (upregulated with age) and pattern 2 (downregulated with age) were predominantly regulated by age, whereas DPGs in pattern 3 were predominantly regulated by CAR activation. Results from this study also demonstrated that although several DPGs are upregulated by CAR in both species and in both ages after ligand activation, there were also distinct age and species effects of CAR in modulating the expression of many other DPGs. These distinct effects can be summarized into the following four categories: 1) pharmacological activation of CAR has both inducible effects and suppressive effects in the expression of many DPGs, 2) both age and species differences may alter CAR-mediated regulatory patterns of certain DPGs, 3) the transcription of some DPGs only responds to a specific age or a specific species, and 4) pharmacological activation of CAR may alter expression of DPGs that are known to be prototypical targets of other important transcription factors in an age- and species-specific manner, indicating that there are age- and species-specific cross-talks between CAR and other receptor signaling pathways in the liver.
During postnatal liver development, the maturation of DPGs is a pivotal factor contributing to the developmental regulation of metabolic drug clearance in children. Under basal conditions, previous studies have summarized the ontogenic gene expression profiles of some DPGs including phase I enzymes, phase II enzymes, and transporters in the mouse or rat liver by CAR pharmacological activation (Cheng et al., 2005; Alnouti and Klaassen, 2006; Alnouti and Klaassen, 2008; Cui et al., 2010, 2012a,b; Peng et al., 2012, 2013; Pratt-Hyatt et al., 2013). Understanding the regulation of DPGs in newborns and children is important, so as to improve the safety and efficacy of drugs in pediatric patients. However, relatively less is known regarding the inducibility of DPGs by chemical inducers in newborns. Because newborns are exposed to various types of xenobiotics, including therapeutic drugs and environmental toxicants, it is important to characterize the potential age-specific modulation of DPGs after xenobiotic exposure. This study fills this critical knowledge gap regarding CAR-targeted DPGs during development.
Because many CAR activators are frequently used as therapeutic drugs in pediatric patients, understanding the common and unique CAR-targeted DPGs during liver development is critical. For example, targeting CAR agonists has been suggested as one of the ultimate goals for the management and treatment of total parenteral nutrition cholestasis, which is common in infants (Hendaus, 2013). The traditional herbal medicine Yin Zhi Huang from Artemisia capillaris is a CAR activator that has been widely used in Asia to prevent and treat neonatal jaundice (Huang et al., 2004), which is a clinical disorder of reduced bile flow. Herbal extract of Phyllanthus amarus root has been shown to decrease bilirubin and oxidative stress in drug-induced neonatal jaundice in mice, and this correlates with upregulated hepatic CAR and Cyp3a expression that account for enhanced bilirubin clearance (Maity et al., 2013). Given the wide application of CAR activators in pediatric patients, the age-specific regulation of hepatic DPGs by CAR may serve as a major risk factor for adverse drug reactions in newborns and children and may alter the pharmacokinetics of drugs and other environmental chemicals. For example, phenobarbital, an indirect CAR activator that has been used as an effective and safe anticonvulsant drug in infants with seizures, is primarily metabolized in the liver by CYP2C9 in humans; the half-life of this drug is longer in adults (100 hours) compared with 4-week-old infants (67 hours) (Pacifici, 2016). Interestingly, our study showed that most of the differentially regulated Cyp2c family members were more induced in newborns than in adults by both mCAR and hCAR (Table 1). The greater inducibility of Cyp2c enzymes may at least in part explain the lower half-life of Cyp2c substrates (e.g., phenobarbital) in newborns. The persistent environmental toxicants polybrominated diphenyl ethers (PBDEs), which cause a wide spectrum of toxicities including neurodevelopmental disorders (Costa and Giordano, 2007; Hoffman et al., 2012), have been detected in human breast milk in U.S. women at worrisome levels (Daniels et al., 2010). Several breast milk–enriched PBDE congeners were shown to be primarily metabolized by the human CYP2B6 enzyme (Erratico et al., 2012; Gross et al., 2015), generating hydroxylated metabolites that are generally considered more toxic than the parent compounds. The mouse homolog Cyp2b10 was upregulated to a much greater extent in the livers of both WT and hCAR-TG mice in newborns than in adults (Table 1), and this may in part explain why newborns are more vulnerable to PBDE-mediated toxicities than adults. Regarding human CYP3A4, which is the most abundant hepatic phase I P450 enzyme that metabolizes approximately 50% marketed drugs (Zhou, 2008), our shows that its mouse homolog Cyp3a11 is equally inducible by hCAR between newborns and adults but is more inducible by mCAR between newborns and adults (Table 1). Although CYP3A4 is generally considered to be lowly expressed in human livers in newborns (Ince et al., 2013), our study suggests that, after xenobiotic exposure, the contribution of this enzyme in potential adverse drug reactions in pediatric patients should not be underestimated. In addition, it is also logical to predict that therapeutic drugs that are CAR activators may predispose pediatric patients to environmental chemical-induced toxicities by inducing certain DPGs.
Previously, a systematic study was conducted using multiplex suspension branched DNA amplification technology to evaluate the inducibility of various DPGs in the livers of adult mice after treatment with CAR ligand TCPOBOP (Aleksunes and Klaassen, 2012). In the livers of adult WT mice, consistent with the previous study, all of the DPGs that were shown to be differentially regulated by TCPOBOP by Aleksunes and Klaassen (2012) were also shown to be differentially regulated in the same direction by CAR in our study (Supplemental Table 2). This includes the TCPOBOP-mediated upregulation of the following: phase I enzymes including P450s (Cyp1a2, Cyp2b10, Cyp3a11) and Nqo1; phase II enzymes including GSTs (Gsta1, Gsta4, Gstm1, Gstm3, Gstm4, Gstt1), SULTs (Sult1e1, Sult3a1, Sult5a1), 3′-phosphoadenosine 5′-phosphosulfate synthase Papss2, and UGTs (Ugt1a1, Ugt1a9, Ugt2b34–Ugt2b36); and transporters Slco1a4, Abcc2, and Abcc3, as well as the TCPOBOP-mediated downregulation of Ugt2a3 and Slco1a1. This study also demonstrated that most of these DPGs were hCAR targets at 60 days of adult age, except for Nqo1, Sult3a1, Papss2, and Ugt2a3, for which the mRNAs remained unchanged in the livers of adult hCAR-TG mice after CITCO treatment (Supplemental Table 2). In newborns, although most of these DPGs were still differentially regulated by TCPOBOP in the livers of WT mice, four DPGs (namely, Gsta1, Sult1e1, Sult3a1, and Slco1a1) were not readily regulated by TCPOBOP at day 5, and many more CAR-targeted DPGs shown in the previous study (Aleksunes and Klaassen, 2012) were unresponsive to CITCO in the livers of hCAR-TG mice at day 5 (Supplemental Table 2). In summary, although many DPGs are commonly regulated by CAR between newborns and adults and between mice and humans, age and species specificities do exist.
In a study utilizing microarray profiling in the livers of WT and CAR-null mice after exposure to various CAR activators, Oshida et al. (2015) constructed a CAR-dependent biomarker signature after xenobiotic insult. Specifically, there are 83 CAR-dependent genes identified by Oshida et al. (2015) from the two genotypes of mice after exposure to three structurally diverse CAR activators (CITCO, phenobarbital, and TCPOBOP). Compared with the previous study, our study confirmed the bona fide CAR target genes, such as Cyp2b10, Cyp2c55, Gstm1, Ugt2b34, Cyp1a2, Cyp4f15, and Aldh1a1.
Another recent study used WT and CAR-null HepaRG cells, which are human liver cancer–derived cell lines, to determine transcriptome-wide regulation of human CAR in vitro (Li et al., 2015). The investigators found that CITCO differentially regulated 135 genes, whereas phenobarbital differentially regulated 133 genes in an hCAR-dependent manner (Li et al., 2015). Interestingly, compared with the hCAR-TG data in our study (Supplemental Table 2), many hCAR-targeted DPGs in HepaRG cells were also differentially regulated in hCAR knockout conditions after CITCO or phenobarbital treatment (Li et al., 2015). In addition, hCAR activation by CITCO differentially regulated many more DPGs in our study than in HepaRG cells in the previous study by Li et al. (2015). The differences in the observations indicate that CITCO may have certain off-target effects in modulating DPG expression in human hepatocytes; furthermore, hCAR activation in mouse hepatocytes in vivo may differ from hCAR activation in human hepatocytes in vitro, likely due to species differences in the enhancers of DPGs and/or chromatin epigenetic differences between the two model systems. Future studies are needed to determine the epigenetic mechanisms of the age and species specificities of CAR in modulating DPGs in the liver.
Acknowledgments
The authors thank members of Dr. Debbie Nickerson’s laboratory at the University of Washington Genome Science Department and all members of the University of Washington EDGE Functional Genomics and Proteomics Facility Core for technical assistance with sequencing. The authors also thank members of Dr. Cui’s laboratory for help with tissue collection and manuscript proofreading.
Abbreviations
- ABC
ATP-binding cassette
- AKR
aldo-keto reductase
- ALDH
aldehyde dehydrogenase
- ALT
alanine aminotransferase
- CAR
constitutive androstane receptor
- CES
carboxyesterase
- CITCO
6-(4-chlorophenyl)imidazo[2,1-b][1,3]thiazole-5-carbaldehyde-O-(3,4-dichlorobenzyl)oxime
- DPG
drug-processing gene
- EPHX
epoxide hydrolase
- FDR-BH
Benjamini–Hochberg false-discovery rate
- FPKM
fragments per kilobase of exons per million reads mapped
- GST
glutathione-S-transferase
- hCAR
human constitutive androstane receptor
- IPA
2-(4-(diisopropoxymethyl)-4,5-dihydrothiazol-2-yl)benzo[d]thiazol-6-ol
- mCAR
mouse constitutive androstane receptor
- MRP
multidrug resistance-associated protein
- NAT
N-acetyltransferase
- NQO
NAD(P)H quinone oxidoreductase
- P450
cytochrome P450
- PBDE
polybrominated diphenyl ether
- PBST
phosphate-buffered saline solution containing 0.05% Tween 20
- RNA-seq
RNA sequencing
- RT-qPCR
real-time quantitative polymerase chain reaction
- SLCO
solute carrier organic anion transporter
- SULT
sulfotransferase
- SV
splicing variant
- TCPOBOP
1,4-bis-[2-(3,5-dichloropyridyloxy)]benzene,3,3′,5,5′-tetrachloro-1,4-bis(pyridyloxy)benzene
- TG
transgenic
- UGT
UDP-glucuronosyltransferase
- WT
wild type
Authorship Contributions
Participated in research design: Cheng, Cui.
Conducted experiments: Cheng, Cui.
Performed data analysis: Cheng, Bammler, Cui.
Wrote or contribute to the writing of the manuscript: Cheng, Bammler, Cui.
Footnotes
This research was supported by the National Institutes of Health National Institute of Environmental Health Sciences [Grants R01-ES025708, R01-ES019487, and P30-ES007033 (to the University of Washington Interdisciplinary Center for Exposures, Diseases, Genomics, and Environment)], the National Institutes of Health National Institute of General Medical Sciences [Grant R01-GM111381], and the University of Washington Department of Environmental and Occupational Health Sciences Murphy Endowment.
 This article has supplemental material available at dmd.aspetjournals.org.
This article has supplemental material available at dmd.aspetjournals.org.
References
- Aleksunes LM, Klaassen CD. (2012) Coordinated regulation of hepatic phase I and II drug-metabolizing genes and transporters using AhR-, CAR-, PXR-, PPARα-, and Nrf2-null mice. Drug Metab Dispos 40:1366–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almazroo OA, Miah MK, Venkataramanan R. (2017) Drug metabolism in the liver. Clin Liver Dis 21:1–20. [DOI] [PubMed] [Google Scholar]
- Alnouti Y, Klaassen CD. (2006) Tissue distribution and ontogeny of sulfotransferase enzymes in mice. Toxicol Sci 93:242–255. [DOI] [PubMed] [Google Scholar]
- Alnouti Y, Klaassen CD. (2008) Tissue distribution, ontogeny, and regulation of aldehyde dehydrogenase (Aldh) enzymes mRNA by prototypical microsomal enzyme inducers in mice. Toxicol Sci 101:51–64. [DOI] [PubMed] [Google Scholar]
- Arnold KA, Eichelbaum M, Burk O. (2004) Alternative splicing affects the function and tissue-specific expression of the human constitutive androstane receptor. Nucl Recept 2:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bookout AL, Jeong Y, Downes M, Yu RT, Evans RM, Mangelsdorf DJ. (2006) Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell 126:789–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty S, Kanakasabai S, Bright JJ. (2011) Constitutive androstane receptor agonist CITCO inhibits growth and expansion of brain tumour stem cells. Br J Cancer 104:448–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WD, Fu X, Dong B, Wang YD, Shiah S, Moore DD, Huang W. (2012) Neonatal activation of the nuclear receptor CAR results in epigenetic memory and permanent change of drug metabolism in mouse liver. Hepatology 56:1499–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Maher J, Chen C, Klaassen CD. (2005) Tissue distribution and ontogeny of mouse organic anion transporting polypeptides (Oatps). Drug Metab Dispos 33:1062–1073. [DOI] [PubMed] [Google Scholar]
- Costa LG, Giordano G. (2007) Developmental neurotoxicity of polybrominated diphenyl ether (PBDE) flame retardants. Neurotoxicology 28:1047–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui JY, Choudhuri S, Knight TR, Klaassen CD. (2010) Genetic and epigenetic regulation and expression signatures of glutathione S-transferases in developing mouse liver. Toxicol Sci 116:32–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui JY, Gunewardena SS, Yoo B, Liu J, Renaud HJ, Lu H, Zhong XB, Klaassen CD. (2012a) RNA-Seq reveals different mRNA abundance of transporters and their alternative transcript isoforms during liver development. Toxicol Sci 127:592–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui JY, Klaassen CD. (2016) RNA-Seq reveals common and unique PXR- and CAR-target gene signatures in the mouse liver transcriptome. Biochim Biophys Acta 1859:1198–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui JY, Li CY. (2016) Regulation of xenobiotic metabolism in the liver, in Comprehensive Toxicology, 3rd ed, chap 9.05. [Google Scholar]
- Cui JY, Renaud HJ, Klaassen CD. (2012b) Ontogeny of novel cytochrome P450 gene isoforms during postnatal liver maturation in mice. Drug Metab Dispos 40:1226–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels JL, Pan IJ, Jones R, Anderson S, Patterson DG, Jr, Needham LL, Sjödin A. (2010) Individual characteristics associated with PBDE levels in U.S. human milk samples. Environ Health Perspect 118:155–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erratico CA, Szeitz A, Bandiera SM. (2012) Oxidative metabolism of BDE-99 by human liver microsomes: predominant role of CYP2B6. Toxicol Sci 129:280–292. [DOI] [PubMed] [Google Scholar]
- Fu ZD, Selwyn FP, Cui JY, Klaassen CD. (2016) RNA sequencing quantification of xenobiotic-processing genes in various sections of the intestine in comparison to the liver of male mice. Drug Metab Dispos 44:842–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross MS, Butryn DM, McGarrigle BP, Aga DS, Olson JR. (2015) Primary role of cytochrome P450 2B6 in the oxidative metabolism of 2,2′,4,4′,6-pentabromodiphenyl ether (BDE-100) to hydroxylated BDEs. Chem Res Toxicol 28:672–681. [DOI] [PubMed] [Google Scholar]
- Gunewardena SS, Yoo B, Peng L, Lu H, Zhong X, Klaassen CD, Cui JY. (2015) Deciphering the developmental dynamics of the mouse liver transcriptome. PLoS One 10:e0141220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendaus M. (2013) TPN cholestasis in infants: what do we know? Review. Georgian Med News 216:32–39. [PubMed] [Google Scholar]
- Hines RN. (2007) Ontogeny of human hepatic cytochromes P450. J Biochem Mol Toxicol 21:169–175. [DOI] [PubMed] [Google Scholar]
- Hines RN. (2013) Developmental expression of drug metabolizing enzymes: impact on disposition in neonates and young children. Int J Pharm 452:3–7. [DOI] [PubMed] [Google Scholar]
- Hines RN, McCarver DG. (2002) The ontogeny of human drug-metabolizing enzymes: phase I oxidative enzymes. J Pharmacol Exp Ther 300:355–360. [DOI] [PubMed] [Google Scholar]
- Hoffman K, Adgent M, Goldman BD, Sjödin A, Daniels JL. (2012) Lactational exposure to polybrominated diphenyl ethers and its relation to social and emotional development among toddlers. Environ Health Perspect 120:1438–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holsapple MP, Pitot HC, Cohen SM, Boobis AR, Klaunig JE, Pastoor T, Dellarco VL, Dragan YP. (2006) Mode of action in relevance of rodent liver tumors to human cancer risk. Toxicol Sci 89:51–56. [DOI] [PubMed] [Google Scholar]
- Honkakoski P, Auriola S, Lang MA. (1992) Distinct induction profiles of three phenobarbital-responsive mouse liver cytochrome P450 isozymes. Biochem Pharmacol 43:2121–2128. [DOI] [PubMed] [Google Scholar]
- Honkakoski P, Negishi M. (1998) Protein serine/threonine phosphatase inhibitors suppress phenobarbital-induced Cyp2b10 gene transcription in mouse primary hepatocytes. Biochem J 330:889–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Zhang J, Moore DD. (2004) A traditional herbal medicine enhances bilirubin clearance by activating the nuclear receptor CAR. J Clin Invest 113:137–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Zhang J, Washington M, Liu J, Parant JM, Lozano G, Moore DD. (2005) Xenobiotic stress induces hepatomegaly and liver tumors via the nuclear receptor constitutive androstane receptor. Mol Endocrinol 19:1646–1653. [DOI] [PubMed] [Google Scholar]
- Ince I, Knibbe CA, Danhof M, de Wildt SN. (2013) Developmental changes in the expression and function of cytochrome P450 3A isoforms: evidence from in vitro and in vivo investigations. Clin Pharmacokinet 52:333–345. [DOI] [PubMed] [Google Scholar]
- Jinno H, Tanaka-Kagawa T, Hanioka N, Ishida S, Saeki M, Soyama A, Itoda M, Nishimura T, Saito Y, Ozawa S, et al. (2004) Identification of novel alternative splice variants of human constitutive androstane receptor and characterization of their expression in the liver. Mol Pharmacol 65:496–502. [DOI] [PubMed] [Google Scholar]
- Klaassen CD, Aleksunes LM. (2010) Xenobiotic, bile acid, and cholesterol transporters: function and regulation. Pharmacol Rev 62:1–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamba JK, Lamba V, Yasuda K, Lin YS, Assem M, Thompson E, Strom S, Schuetz E. (2004) Expression of constitutive androstane receptor splice variants in human tissues and their functional consequences. J Pharmacol Exp Ther 311:811–821. [DOI] [PubMed] [Google Scholar]
- Lee JS, Ward WO, Wolf DC, Allen JW, Mills C, DeVito MJ, Corton JC. (2008) Coordinated changes in xenobiotic metabolizing enzyme gene expression in aging male rats. Toxicol Sci 106:263–283. [DOI] [PubMed] [Google Scholar]
- Li CY, Cheng SL, Bammler TK, Cui JY. (2016a) Editor’s highlight: neonatal activation of the xenobiotic-sensors PXR and CAR results in acute and persistent down-regulation of PPARα-Signaling in mouse liver. Toxicol Sci 153:282–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Mackowiak B, Brayman TG, Mitchell M, Zhang L, Huang SM, Wang H. (2015) Genome-wide analysis of human constitutive androstane receptor (CAR) transcriptome in wild-type and CAR-knockout HepaRG cells. Biochem Pharmacol 98:190–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Cui JY, Gunewardena S, Yoo B, Zhong XB, Klaassen CD. (2012) Hepatic ontogeny and tissue distribution of mRNAs of epigenetic modifiers in mice using RNA-sequencing. Epigenetics 7:914–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Gunewardena S, Cui JY, Yoo B, Zhong XB, Klaassen CD. (2013) RNA-sequencing quantification of hepatic ontogeny and tissue distribution of mRNAs of phase II enzymes in mice. Drug Metab Dispos 41:844–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maglich JM, Parks DJ, Moore LB, Collins JL, Goodwin B, Billin AN, Stoltz CA, Kliewer SA, Lambert MH, Willson TM, et al. (2003) Identification of a novel human constitutive androstane receptor (CAR) agonist and its use in the identification of CAR target genes. J Biol Chem 278:17277–17283. [DOI] [PubMed] [Google Scholar]
- Maity S, Nag N, Chatterjee S, Adhikari S, Mazumder S. (2013) Bilirubin clearance and antioxidant activities of ethanol extract of Phyllanthus amarus root in phenylhydrazine-induced neonatal jaundice in mice. J Physiol Biochem 69:467–476. [DOI] [PubMed] [Google Scholar]
- McCarver DG, Hines RN. (2002) The ontogeny of human drug-metabolizing enzymes: phase II conjugation enzymes and regulatory mechanisms. J Pharmacol Exp Ther 300:361–366. [DOI] [PubMed] [Google Scholar]
- Mooij MG, Schwarz UI, de Koning BA, Leeder JS, Gaedigk R, Samsom JN, Spaans E, van Goudoever JB, Tibboel D, Kim RB, et al. (2014) Ontogeny of human hepatic and intestinal transporter gene expression during childhood: age matters. Drug Metab Dispos 42:1268–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooij MG, van de Steeg E, van Rosmalen J, Windster JD, de Koning BA, Vaes WH, van Groen BD, Tibboel D, Wortelboer HM, de Wildt SN. (2016) Proteomic analysis of the developmental trajectory of human hepatic membrane transporter proteins in the first three months of life. Drug Metab Dispos 44:1005–1013. [DOI] [PubMed] [Google Scholar]
- O’Sullivan JL, Haedicke GJ, Bevivino J. (1989) Spontaneous postpartum factor VIII inhibitor development with bleeding into the face and neck. Plast Reconstr Surg 84:802–805. [DOI] [PubMed] [Google Scholar]
- Oshida K, Vasani N, Jones C, Moore T, Hester S, Nesnow S, Auerbach S, Geter DR, Aleksunes LM, Thomas RS, et al. (2015) Identification of chemical modulators of the constitutive activated receptor (CAR) in a gene expression compendium. Nucl Recept Signal 13:e002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacifici GM. (2016) Clinical pharmacology of phenobarbital in neonates: effects, metabolism and pharmacokinetics. Curr Pediatr Rev 12:48–54. [DOI] [PubMed] [Google Scholar]
- Parkinson AOB, Buckley DB, Kazmi F, Czerwinski M, Parkinson O. (2013) Biotransformation of xenobiotics, in Casarett and Doull’s Toxicology: The Basic Science of Poisons (Casarett LJ, Doull J, Klaassen CD. eds) pp xv, 1310, 1311 (leaf of plates), McGraw-Hill, New York. [Google Scholar]
- Peng L, Cui JY, Yoo B, Gunewardena SS, Lu H, Klaassen CD, Zhong XB. (2013) RNA-sequencing quantification of hepatic ontogeny of phase-I enzymes in mice. Drug Metab Dispos 41:2175–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L, Piekos S, Guo GL, Zhong XB. (2016) Role of farnesoid X receptor in establishment of ontogeny of phase-I drug metabolizing enzyme genes in mouse liver. Acta Pharm Sin B 6:453–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L, Yoo B, Gunewardena SS, Lu H, Klaassen CD, Zhong XB. (2012) RNA sequencing reveals dynamic changes of mRNA abundance of cytochromes P450 and their alternative transcripts during mouse liver development. Drug Metab Dispos 40:1198–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt-Hyatt M, Lickteig AJ, Klaassen CD. (2013) Tissue distribution, ontogeny, and chemical induction of aldo-keto reductases in mice. Drug Metab Dispos 41:1480–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J, Plummer SM, Rode A, Scheer N, Bower CC, Vogel O, Henderson CJ, Wolf CR, Elcombe CR. (2010) Human constitutive androstane receptor (CAR) and pregnane X receptor (PXR) support the hypertrophic but not the hyperplastic response to the murine nongenotoxic hepatocarcinogens phenobarbital and chlordane in vivo. Toxicol Sci 116:452–466. [DOI] [PubMed] [Google Scholar]
- Scheer N, Ross J, Rode A, Zevnik B, Niehaves S, Faust N, Wolf CR. (2008) A novel panel of mouse models to evaluate the role of human pregnane X receptor and constitutive androstane receptor in drug response. J Clin Invest 118:3228–3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sueyoshi T, Li L, Wang H, Moore R, Kodavanti PR, Lehmler HJ, Negishi M, Birnbaum LS. (2014) Flame retardant BDE-47 effectively activates nuclear receptor CAR in human primary hepatocytes. Toxicol Sci 137:292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson MM, Hines RN, Schuetz EG, Meibohm B. (2016) Expression patterns of organic anion transporting polypeptides 1B1 and 1B3 protein in human pediatric liver. Drug Metab Dispos 44:999–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzameli I, Pissios P, Schuetz EG, Moore DD. (2000) The xenobiotic compound 1,4-bis[2-(3,5-dichloropyridyloxy)]benzene is an agonist ligand for the nuclear receptor CAR. Mol Cell Biol 20:2951–2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windshügel B, Poso A. (2011) Constitutive activity and ligand-dependent activation of the nuclear receptor CAR-insights from molecular dynamics simulations. J Mol Recognit 24:875–882. [DOI] [PubMed] [Google Scholar]
- Wortham M, Czerwinski M, He L, Parkinson A, Wan YJ. (2007) Expression of constitutive androstane receptor, hepatic nuclear factor 4 alpha, and P450 oxidoreductase genes determines interindividual variability in basal expression and activity of a broad scope of xenobiotic metabolism genes in the human liver. Drug Metab Dispos 35:1700–1710. [DOI] [PubMed] [Google Scholar]
- Xiao D, Chen YT, Yang D, Yan B. (2012) Age-related inducibility of carboxylesterases by the antiepileptic agent phenobarbital and implications in drug metabolism and lipid accumulation. Biochem Pharmacol 84:232–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Moore R, Goldsworthy TL, Negishi M, Maronpot RR. (2004) The orphan nuclear receptor constitutive active/androstane receptor is essential for liver tumor promotion by phenobarbital in mice. Cancer Res 64:7197–7200. [DOI] [PubMed] [Google Scholar]
- Yan J, Xie W. (2016) A brief history of the discovery of PXR and CAR as xenobiotic receptors. Acta Pharm Sin B 6:450–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Wang H. (2014) Signaling control of the constitutive androstane receptor (CAR). Protein Cell 5:113–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XJ, Shi Z, Lyv JX, He X, Englert NA, Zhang SY. (2015) Pyrene is a novel constitutive androstane receptor (CAR) activator and causes hepatotoxicity by CAR. Toxicol Sci 147:436–445. [DOI] [PubMed] [Google Scholar]
- Zhang YK, Lu H, Klaassen CD. (2013) Expression of human CAR splicing variants in BAC-transgenic mice. Toxicol Sci 132:142–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou SF. (2008) Drugs behave as substrates, inhibitors and inducers of human cytochrome P450 3A4. Curr Drug Metab 9:310–322. [DOI] [PubMed] [Google Scholar]