ABSTRACT
Adaptive laboratory evolution (ALE) experiments are often designed to maintain a static culturing environment to minimize confounding variables that could influence the adaptive process, but dynamic nutrient conditions occur frequently in natural and bioprocessing settings. To study the nature of carbon substrate fitness tradeoffs, we evolved batch cultures of Escherichia coli via serial propagation into tubes alternating between glucose and either xylose, glycerol, or acetate. Genome sequencing of evolved cultures revealed several genetic changes preferentially selected for under dynamic conditions and different adaptation strategies depending on the substrates being switched between; in some environments, a persistent “generalist” strain developed, while in another, two “specialist” subpopulations arose that alternated dominance. Diauxic lag phenotype varied across the generalists and specialists, in one case being completely abolished, while gene expression data distinguished the transcriptional strategies implemented by strains in pursuit of growth optimality. Genome-scale metabolic modeling techniques were then used to help explain the inherent substrate differences giving rise to the observed distinct adaptive strategies. This study gives insight into the population dynamics of adaptation in an alternating environment and into the underlying metabolic and genetic mechanisms. Furthermore, ALE-generated optimized strains have phenotypes with potential industrial bioprocessing applications.
IMPORTANCE Evolution and natural selection inexorably lead to an organism's improved fitness in a given environment, whether in a laboratory or natural setting. However, despite the frequent natural occurrence of complex and dynamic growth environments, laboratory evolution experiments typically maintain simple, static culturing environments so as to reduce selection pressure complexity. In this study, we investigated the adaptive strategies underlying evolution to fluctuating environments by evolving Escherichia coli to conditions of frequently switching growth substrate. Characterization of evolved strains via a number of different data types revealed the various genetic and phenotypic changes implemented in pursuit of growth optimality and how these differed across the different growth substrates and switching protocols. This work not only helps to establish general principles of adaptation to complex environments but also suggests strategies for experimental design to achieve desired evolutionary outcomes.
KEYWORDS: adaptive laboratory evolution, Escherichia coli, adaptive mutations, phenotypic variation
INTRODUCTION
In heterotrophs such as Escherichia coli, catabolism of carbon substrates is the driving force behind the energy generation and chemical synthesis necessary for homeostasis and anabolism (1). Although glucose is the most readily metabolized carbohydrate (2), the frequent environmental availability of other carbon compounds long ago led most organisms to evolve the ability to subsist on a range of nutritional sources. These alternative compounds can vary greatly in energy content, the point at which they enter into the metabolic network, and their impact on cellular phenotype (3). The ability of alternative compounds to sustain growth and proliferation in the absence of glucose makes it almost essential for a robust bacterium to be able to switch between carbon growth substrates as environmental circumstances dictate. For example, an enteric E. coli bacterium depends on its ability to switch to metabolizing different carbohydrates as it passes through the digestive tract (4). Such switching between carbon sources has relevance to more than just natural environments; the use of genetically engineered microbes to produce commercially valuable chemicals frequently relies on batch growth, which can include a stage at which the cells run out of the preferred nutrient (e.g., glucose) and have to switch to a less-than-optimal alternative (e.g., xylose) (5).
Understanding diauxic shifts has been a long-standing effort of the scientific community (6). Diauxic lag has traditionally been understood to result from catabolite repression, wherein the depletion of the preferred substrate relieves the repression on genes for metabolizing the remaining substrate(s). However, recent work has shown that cells in a multisubstrate environment can display divergent bet-hedging behaviors, which can result in subpopulations that grow differently on the substrates (7). Moreover, slightly different microbial strains can have notably different lag-phase durations and behaviors, which can be targets for natural selection in a competitive environment (8). Adaptive laboratory evolution, or ALE, serves as a technique that harnesses natural selection to arrive at genetic and phenotypic outcomes that are difficult to predict a priori (9). ALE work so far has examined E. coli adaptation to a number of environments characterized by temporal heterogeneity; fluctuations in temperature (10, 11), pH (12), UV irradiation (13), and random stressors (14) have all been studied. It is well established that homogenous environments tend to develop narrow niche width “specialists” while heterogeneous environments usually lead to broader niche width “generalists” (15), to the extent that the failure of a generalist to develop in certain fluctuating environments is seen as surprising (16). However, evolution of E. coli on a glucose-acetate mixture (in which the glucose is first depleted before a diauxic shift to acetate occurs, creating temporal variability) has been shown to lead repeatedly to coexisting specialists rather than generalists (17), due to competition for limited resources and the fitness trade-offs of glucose versus acetate specialization (18). While several cases of E. coli evolution to alternating growth substrates have been studied, few substrates have been examined (19) and analyses have been limited to fitness assays (20) or in-depth study of a single operon (21).
In this study, we sought to investigate carbon source switching with different compounds of industrial relevance, and to examine evolutionary outcomes via a number of different data types. ALE was used to adapt E. coli cultures for ∼1,000 generations to a dynamic, nutrient-excess environment in which the available carbon substrate alternated with every tube of growth and cultures were serially propagated while still growing exponentially. Although such resource-abundant laboratory environments have few natural counterparts, the conditions of excess help to ensure that selection occurs for growth rate, without the complicating factors of resource competition or changing growth phases that introduce new stressors (22). Populations evolved to these switching environments had substrate-specific fitnesses comparable to those reached by single-substrate-evolved control cultures. The adaptive mechanism used to achieve this fitness improvement varied based on substrate, and evolved strains likewise exhibited phenotypic, genetic, and transcriptomic dynamics that varied across compounds. Genome-scale metabolic models were used to help interpret substrate-specific results and to explain the propensity for specialist or generalist development, given different switching setups.
RESULTS
Evolved population fitness.
Adaptive laboratory evolution (ALE) was employed to adapt Escherichia coli to an environment with constantly alternating carbon growth substrates. Wild-type E. coli K-12 MG1655 was used, and substrate switching was examined on four different carbon compounds, glucose, xylose, glycerol, and acetate (ac) (E. coli wild-type growth rates at 37°C were 0.73, 0.55, 0.45, and 0.27 h−1, respectively). ALE replicates were each assigned a substrate in addition to glucose, and at the end of each growth tube a culture was passed to a fresh tube of M9 minimal medium containing the next carbon source, such that substrate procession was glucose → [substrate] → glucose → [substrate] →, etc. (Fig. 1A). Three tubes switched between glucose and xylose (1,180 generations of growth; 164 tubes of growth; 3.1 × 1012 cumulative cell divisions), three between glucose and glycerol (1,170 generations; 162 tubes; 3.0 × 1012 cumulative cell divisions [CCD]), and three between glucose and acetate (650 generations; 91 tubes; 1.8 × 1012 CCD). Because each tube underwent serial passage at the same optical density (at a point at which nutrients were still in excess and cultures were still growing exponentially), the generations, tubes, and CCD of growth upon each individual substrate were half that of the total. As controls, cultures were also evolved to each of the four different compounds on their own with no switching.
FIG 1.
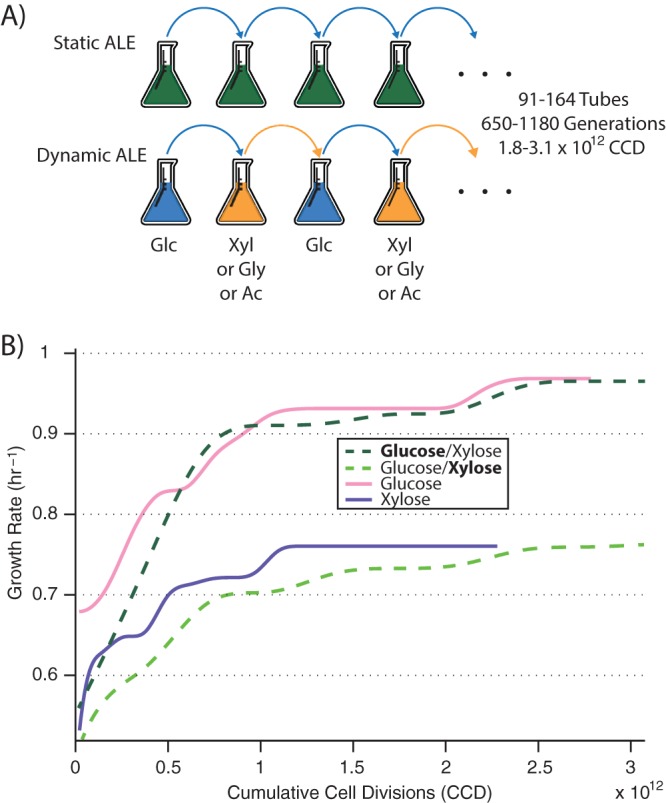
Experimental setup and evolutionary trajectories. (A) “Static ALE” exposes cultures to a constant environment, while a “dynamic ALE” introduces temporal variability in carbon growth substrate. (B) Example plot of fitness trajectories (i.e., growth rates) for statically evolved (pink/purple) and switching (dark green/light green) cultures on glucose and xylose over the course of the ALE experiment. Similar fitnesses are reached, although under switching conditions cultures take longer to get there. CCD; cumulative cell divisions; Glc, glucose; Xyl, xylose; Gly, glycerol; Ac, acetate.
Comparison of evolutionary trajectories between the “dynamic” (switching) and “static” (nonswitching) populations gives the first indication of consequences of the substrate switching. As a general trend across all compounds, replicates undergoing switching adapted at a lower rate, but were still able to reach final fitness values comparable to those of control populations exposed solely to a single compound (Fig. 1B). Although establishing a statistically significant difference between the final growth rates was made more difficult by the small number of samples, average final population growth rates on a compound failed to differentiate dynamic and static ALE setups (one-way ANOVA, P > 0.05). However, cultures under static conditions reached their half-maximal final growth rates, on average, 43% faster than switching cultures (see File S1 in the supplemental material). Thus, it seems that adaptation proceeds at a higher rate when the selection pressure is more sustained (in this case, greater “time under selection” leads to a greater selective force), but fitness plateaus toward a similar value since the same objective is still being optimized for (namely, exponential-phase growth rate on a compound).
Genetic analysis.
Population sequencing was performed on the cultures evolved under dynamic conditions to examine whether specialists (coexisting subpopulations optimized for the different substrates) or generalists (one main dominant strain capable of good growth on both substrates) had developed. The endpoint populations for each dynamic ALE replicate were grown up on each of the different carbon sources they had been switching between and population sequencing results were compared (see Materials and Methods). Consider, as an example, a population switching between glucose and acetate. If specialist subpopulations are present, then growing the culture on glucose will preferentially enrich for the glucose specialist, and likewise for the acetate specialist with acetate growth. Population sequencing would then show a significant change in mutational frequency dependent upon which growth enrichment was performed. If there were no specialist subpopulations, but instead one dominant generalist, then the mutational frequencies would be roughly identical regardless of the substrate used for growth enrichment. Applying this sequencing analysis to all evolved replicates revealed the propensities of various compounds to elicit subpopulation development (see File S2 in the supplemental material). All three glucose/glycerol replicates were isogenic, all three glucose/acetate replicates were subpopulations, and two of three glucose/xylose replicates were clearly isogenic, while the third replicate developed hypermutability, which complicated the genetic analysis. As examples for each case, the glucose/acetate switcher Glc/Ac 3 developed two distinct specialist populations (a sapA rpoC mutant for glucose growth and an nrd ptsP rpoC mutant for acetate growth, in addition to two mutations which swept both subpopulations), while the glucose/glycerol switcher Glc/Gly 2 was a generalist, having the same population composition regardless of growth substrate (Table 1).
TABLE 1.
Substrate-induced mutational frequency differences or lack thereof
| Mutation | Gene | Population frequency (%) when grown on: |
||
|---|---|---|---|---|
| Glucose | Acetate | Glycerol | ||
| Apparent specialists (Glc/Ac 3) | ||||
| L14R (CTT→CGT) | sapA | 85 | 0 | |
| Δ88 bp | nrdA-nrdB | 0 | 68 | |
| Δ1 bp (708/2,247 nta) | ptsP | 0 | 100 | |
| R98H (CGC→CAC) | rpoC | 95 | 11 | |
| K398 M (AAG→ATG) | rpoC | 7 | 79 | |
| IS5 (141–144/144 nt) | yobF | 100 | 100 | |
| Δ1 bp intergenic (−33/+33) | pyrE-rph | 100 | 100 | |
| Apparent generalists (Glc/Gly 2) | ||||
| IS5 (258–261/491 nt) | ybbD | 35 | 42 | |
| Δ82 bp | pyrE-rph | 84 | 90 | |
| M272I (ATG→ATT) | glpK | 100 | 100 | |
| E672K (GAA→AAA) | rpoB | 100 | 100 | |
nt, nucleotides.
Comparing mutational frequencies across the switching-adapted cultures and single-substrate-evolved controls revealed key genes under selective pressure under these conditions (Table 2). Many of the repeatedly mutated genes are seen frequently in other evolution experiments, with the fitness benefit already either known or inferred—pyrE-rph mutations for improved minimal medium growth (23), pykF and hns-tdk mutations for glucose growth (24, 25), glpK mutations for glycerol growth (26), and rpoB and rpoC mutations to serve as large-scale transcriptional rewirings (27, 28). In addition to these oft seen mutational targets, several genes stood out as indicators of differing adaptive strategies between dynamic and static conditions. Most striking are mutations in the cspC-yobF region, in which a number of distinct mutations were seen across five out of six glucose/xylose and glucose/acetate populations (with the sixth having a mutated cspE rather than cspC), but only in a single acetate-evolved replicate. These dynamically favored mutations may be altering the role of CspC as a transcript stabilizer in stressful environments, a functionality more relevant as a target for adaptation when the carbon source is frequently changing (29). Similarly, mutations in ptsP (3 unique dynamic mutations, with 2 in one lineage), relA (3 unique dynamic mutations), and sapB (2 unique dynamic mutations) appeared to be dynamically favored, while mutations in rho (4 unique static mutations) and xylR (2 unique static mutations) appeared statically favored. Although, as a whole, these data indicate differing adaptive strategies for dynamic and static growth environments, the explicit biochemical mechanisms through which such mutations enable fitness improvements remain unclear without detailed follow-up analyses (26, 28).
TABLE 2.
Genetic regions mutated repeatedly during selection
| Gene | No. of unique mutations | Dynamic occurrences (fraction of replicates) | Static occurrences (fraction of replicates) |
|---|---|---|---|
| rpoC | 8 | Glc/Xyl (3/3) | Glc (1/3) |
| Glc/Gly (1/3) | Xyl (2/4) | ||
| Glc/Ac (3/3) | Gly (2/2) | ||
| cspC-yobF | 6 | Glc/Xyl (2/3) | Ac (1/3) |
| Glc/Ac (3/3) | |||
| rho | 5 | Glc/Ac (1/3) | Xyl (2/4) |
| Ac (2/3) | |||
| glpK | 5 | Glc/Gly (3/3) | Gly (2/2) |
| Glc/Xyl (1/3) | |||
| ptsP | 4 | Glc/Ac (2/3) | Ac (1/3) |
| rpoB | 4 | Glc/Gly (2/3) | Glc (2/3) |
| Glc/Ac (1/3) | |||
| pyrE-rph | 4 | Glc/Gly (2/3) | Glc (2/3) |
| Glc/Ac (3/3) | Xyl (4/4) | ||
| Ac (1/3) | |||
| relA | 3 | Glc/Xyl (3/3) | NAa |
| pykF | 3 | Glc/Xyl (1/3) | NA |
| Glc/Ac (2/3) | |||
| xylR | 2 | NA | Xyl (2/4) |
| sapB | 2 | Glc/Xyl (1/3) | NA |
| Glc/Ac (1/3) | |||
| hns-tdk | 2 | NA | Glc (2/3) |
NA, not applicable.
Physiological analysis of evolved strains.
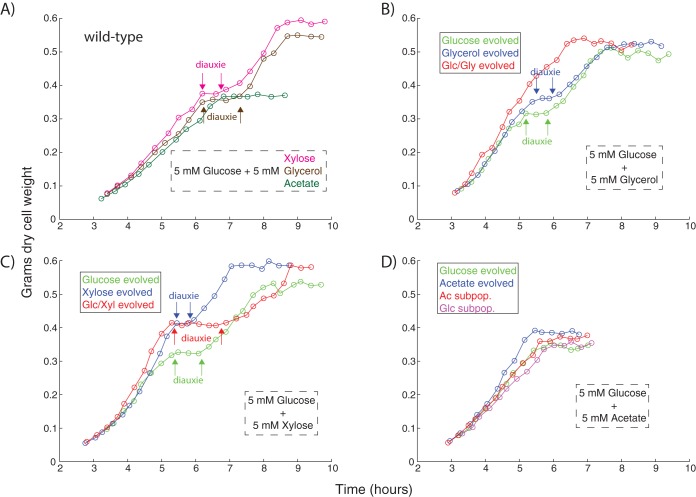
Representative clones were isolated from evolved cultures for purposes of physiological characterization. For cultures that evolved substrate specialists, this involved isolating a clone for each of the observed subpopulations. Clones were sequenced to ensure that they were representative of the evolved cultures, containing the same key mutations revealed from population sequencing (see File S2). The substrate-switching phenotypes of the clones were characterized via a series of diauxic growth curves. Clones were grown in the presence of 5 mM glucose and 5 mM of an additional carbon substrate, depending on the environment the clone had been exposed to over the course of the evolution. The 5 mM concentrations were chosen such that sufficiently dense final optical densities (ODs) could be obtained, but not without utilizing both of the available carbon compounds.
Unsurprisingly, the different carbon substrates led to different growth phenotypes for the various evolved strains (Fig. 2). The wild type achieved the highest density on glucose/xylose, an intermediate density on glucose/glycerol, and the lowest density on glucose/acetate. While xylose and glycerol both caused diauxic lag phases, there was no such lag when the wild type was grown on glc/ac, likely due to the frequent presence of acetate as an overflow metabolite in regular batch culturing (30) and inability to trigger anabolism following glucose exhaustion (31). Nevertheless, overall growth rate on glc/ac improved in all evolved strains, with the static acetate-evolved strain growing most robustly. Similarly, the static xylose-evolved strain outperformed the other strains under conditions of glc/xyl diauxie. The dynamically evolved glc/xyl generalist's increased lag phase duration appears counterintuitive, but a simultaneous multisubstrate environment is not something the cells were ever exposed to during the ALE; in this case, intertube substrate switching adaptation did not extend to intratube switching. In contrast, the clone that evolved to an environment switching between glucose and glycerol was able to completely abolish the lag phase that typically occurs midway through growth on both substrates. Moreover, this lag phase was not abolished in either of the statically evolved glucose or glycerol controls, leading to much different performance in the diauxic growth test. Overall, the variable diauxic growth phenotypes of the evolved strains across substrate conditions highlight the complexity of adaptation to dynamic environments.
FIG 2.
Diauxic growth curves. (A) Wild type performance on all substrate mixtures. (B) Evolved strains on glucose plus glycerol. (C) Evolved strains on glucose plus xylose. (D) Evolved strains on glucose plus acetate.
Transcriptomic analysis of evolved strains.
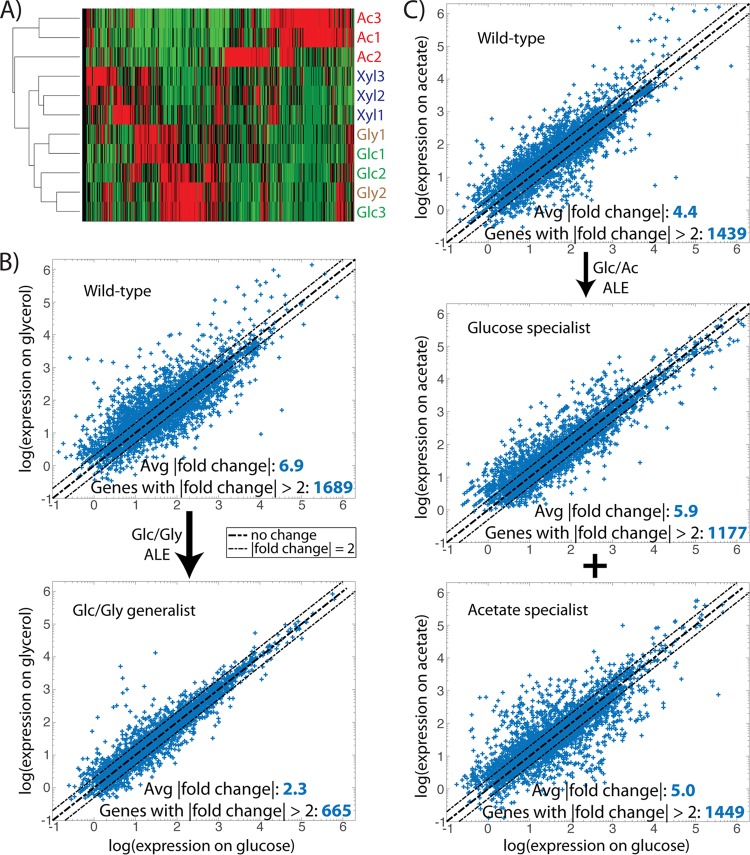
Transcriptome sequencing (RNA-seq) was performed on both statically and dynamically evolved clones to probe the transcriptional states of the strains under relevant substrate growth conditions. For a given growth environment, principal-component analysis (PCA) was expected to cluster statically evolved strains together in a region corresponding to the “optimal” expression state for fast growth on that substrate. Dynamically evolved strains, however, would be expected to cluster apart from the static controls, given that their expression state evolved in response to multisubstrate exposure. This transcriptomic “distance” serves as an indicator of dissimilarity that should mirror substrate differences; for example, if [substrate 1] is more similar to glucose than [substrate 2], then a strain evolved dynamically to glucose/[substrate 1] should fall closer to glucose-evolved controls than a glucose/[substrate 2] strain. Indeed, PCA reinforced the conclusion drawn from population sequencing that growth of evolved strains on glycerol and xylose creates cellular states more similar to glucose than acetate; that is, glc/xyl and glc/gly generalists clustered closer to glucose optimality than did the glc/ac specialist strains (Fig. 3A). Similar logic implies that the dynamically evolved strains have moved closer to transcript optimality, represented by the statically evolved controls, than the wild-type starting strain, which is what is seen for glycerol (Mahalanobis distance to static strains = 4.89 for wild type, 2.63 for dynamic) and xylose (1.88 for wild type, 1.35 for dynamic) (Fig. 3B and C). However, in the case of glc/ac switching, the specialist strains appear to have adopted a different transcriptional strategy than acetate-evolved controls, falling further away from the optimum expression state of the static strains than does the wild type (Fig. 3D). This specialist discrepancy may result from the nature of the strains as coexisting subpopulations and the resultant transcriptional adjustments necessary for alternating dominance between tubes during the ALE experiment. Although it is possible that the two specialists interact and exhibit different phenotypes when cultured together versus independently, this appears unlikely given the precipitous drop in frequency of acetate-characteristic mutations after glucose enrichment, and likewise for glucose-characteristic mutations after acetate enrichment (Table 1; see also File S2).
FIG 3.
Principal component analysis (PCA) of RNA-seq on various growth substrates. PCA plots clustering expression states of different strains grown on glucose (A), glycerol (B), xylose (C), and acetate (D). Overall, the statically evolved strains grouped together. In addition, for growth on glucose, the generalist strains grouped together in closer proximity to the static strains than did the specialist strains. glc/xyl, glucose/xylose generalist; glc/gly, glucose/glycerol generalist; GLC/ac, glucose specialist subpopulation; glc/AC, acetate specialist subpopulation; WT, wild type.
Hierarchical clustering of expression data for the statically evolved strains further highlights the differing transcriptional strategies adopted in pursuit of substrate optimality (Fig. 4A). Glucose and glycerol strains clustered most closely together, followed by xylose, with acetate adaptation resulting in the most distinct pattern of relative gene expression levels. For dynamically evolved strains, these substrate-optimal transcriptional patterns cannot be adopted without leaving the cells in a state where widespread expression changes must be made between every growth tube, an adaptation strategy unlikely to prove optimal, given the time and energy it takes to alter expression levels (32). It would be expected that for the rapidly switching ALE environment utilized herein, the transcriptome will adapt, such that shifts in expression level needed between the two relevant substrates become minimized. To test this, RNA-seq data for individual strains (wild type or evolved) were compared to determine how expression levels changed when a strain was grown on glucose versus when it was grown on [alternative substrate] (Fig. 4B and C). For glucose/glycerol switching, the wild-type starting strain had 1,689 genes with greater than 2-fold changes in gene expression across the two substrates and an average gene fold change magnitude of 6.9, while the evolved glucose/glycerol generalist significantly decreased (paired t test, P = 0.018) to 665 genes and a 2.3-fold average expression change, respectively (Fig. 4B). Similarly, the wild type on glucose/xylose had 1,676 genes with a |fold change| >2 and an average fold change of 4.9, which significantly decreased (P = 0.034) in the glucose/xylose generalist to 842 genes and a 2.9-fold average change. This stands in stark contrast with the phenotype of the dynamically evolved glucose/acetate specialist strains, which did not approach any closer to transcriptional parity across the substrates than the wild-type strain (Fig. 4C). Unlike the generalists, the coexisting glucose and acetate specialist subpopulations obviated the need for a single genotype capable of reconciling the dissimilar optimal expression states between the two substrates. Thus, it becomes clear that global transcriptional analyses across substrates can be used to interpret the differential development of specialists or generalists under various substrate switching regimes.
FIG 4.
Global transcriptome changes in statically and dynamically evolved strains. (A) Hierarchical clustering of strains statically evolved to each of the four studied substrates. Strains optimized for glucose and glycerol growth have the most similarity in expression state, followed by xylose and then acetate. (B and C) Scatter plots of expression data for the wild-type ancestor and dynamically evolved strains when grown on glucose versus glycerol (B) or glucose versus acetate (C). Points falling on the diagonal line represent genes whose expression does not vary, while those falling outside the dashed lines are genes with more than 2-fold changes in expression across the two substrates. (B) The number and magnitude of transcriptional shifts between glucose and glycerol growth conditions significantly decreased in the dynamically evolved generalist strain. (C) The number and magnitude of transcriptional shifts between glucose and acetate growth conditions did not significantly change in the dynamically evolved specialist strains.
Metabolic modeling.
To better understand the mechanisms underlying growth-substrate-driven adaptive responses, genome-scale metabolic modeling techniques were applied. By modeling the metabolic reaction network of an organism with a stoichiometric matrix (33) and applying relevant physiological constraints, predictions can be made of optimal growth behavior and metabolic flux states in a particular environment (34). Here, the relevant environments are identical except for the different carbon growth substrates.
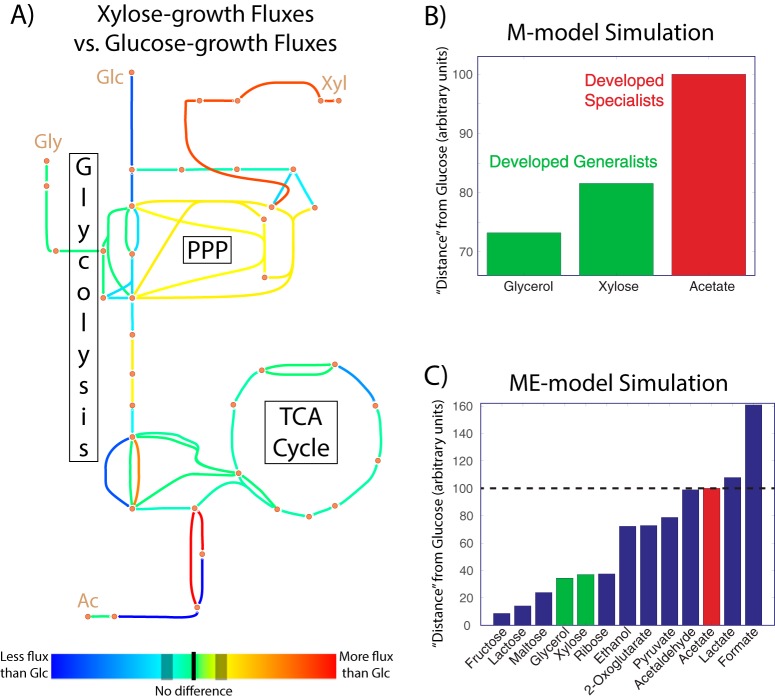
The experimental analyses performed thus far indicate that ALE cultures adapted differently to different switching schemes, becoming either generalists or specialists in response to different substrate combinations. Robust growth under glucose/xylose and glucose/glycerol switching was achieved by versatile generalist strains, whereas glucose/acetate switching failed to select for such strains, instead opting for coexisting specialist subpopulations as the strategy for growth improvement. One potential explanation for this discrepancy is that glucose and acetate are too metabolically dissimilar (requiring conflicting or disparate pathways for metabolism) for a single strain to easily evolve for robust growth on both, while xylose and glycerol are both similar enough to glucose to avoid this. To test this, optimal growth of the wild-type strain was modeled on the four different relevant substrates individually, and the flux states necessary for optimal growth were inferred from Monte Carlo sampling of a genome-scale M-Model (35). This sampling yielded flux distributions for each chemical reaction in the model; the three nonstandard substrates were pairwise compared with glucose (Fig. 5). As an example case, consider how metabolic fluxes resulting from growth on xylose compare with glucose-growth fluxes (Fig. 5A). Glucose import and conversion reactions have lower flux, while xylose import/conversion reactions have higher flux, and these differences impact the metabolic flux network as a whole, e.g., via increased nonoxidative pentose phosphate pathway (PPP) activity from the conversion of xylose to xylulose 5-phosphate. Expression profiling could be used to perform such analyses or support modeling-derived results (e.g., RNA-seq data show that the glucose phosphotransferase system [PTS] permease ptsG decreased 2.4-fold in expression upon xylose growth, while the xylulokinase xylB increased expression 120-fold), but scalar gene expression values do not give insight into reaction directionality, which can cause important network features to be overlooked. Expression of pgi, for example, changed by less than 2% in glucose versus xylose growth, but modeling showed that, although flux magnitude did not appreciably change, the direction did; increased nonoxidative PPP activity from xylose growth leads to increased fructose 6-phosphate levels, which drive flux “backward” through pgi to glucose 6-phosphate in the direction of gluconeogenesis. A 2-sample Kolmogorov-Smirnov test was applied to the modeling-determined fluxes to quantify a metabolic distance between glucose and each of the substrates, and this scaled as expected—glycerol and xylose are more similar to glucose than acetate is (Fig. 5B).
FIG 5.
Metabolic flux analysis of substrate differences. (A) Flux map of central carbon metabolism, with colors indicating the extent of difference in reaction fluxes between optimal growth on, as an example, glucose and xylose. Lines closer to the red end of the spectrum represent reactions where flux increases under xylose growth, while lines closer to the blue end of the spectrum represent reactions with decreased flux under xylose growth. (B) Substrate distance from glucose based on Monte Carlo sampling of the M-model network, with coloration indicating whether E. coli evolved to glucose/[substrate] switching developed into specialists or generalists. (C) Same as for panel B but using Euclidean distance between ME-model-predicted expression levels as the distance metric, and including simulation results for substrates not experimentally tested.
M-Models, although powerful tools for predicting and analyzing physiology, do not quantitatively predict gene expression, which can in certain circumstances lead to inaccurate predictions (36). Monte Carlo sampling is one way to skirt this issue, but genome-scale models of metabolism that factor in gene expression and its concomitant energy costs, dubbed ME-Models, have recently been developed (37). This additional model content allows for quantitative predictions of the optimal gene expression and flux state in a given environment without the need for random sampling. Taking the Euclidean distance between the expression state on glucose and on the other substrates is another way to quantify the extent of dissimilarity, and the results are in excellent agreement with M-Model sampling (Fig. 5C). Moreover, ME-Models do not require physiological data on substrate uptake rates, so many compounds can be computationally tested. Dynamic ALEs on compounds with a lower glucose distance than glycerol (from these simulations, mono- and disaccharides) would likely lead to generalists, and compounds more distant than acetate (other carboxylic acids) would likely lead to specialists, but it is as of yet unclear at what intermediate substrate distance a dynamic evolution would begin to favor specialists over generalists. This study thus helps to establish the explanatory and predictive power of metabolic modeling for understanding why and under what circumstances generalists or specialists arise, but future experimental work will be needed to determine the extent and limitations of this efficacy.
DISCUSSION
In this study, E. coli cultures were evolved for upwards of 1,000 generations under environmental conditions in which the available carbon source alternated repeatedly between glucose and either glycerol, xylose, or acetate. These dynamically evolved cultures reached fitnesses comparable to those of statically evolved controls grown only on a single compound. Genetic analysis revealed that glucose/glycerol and glucose/xylose switchers adopted a generalist strategy, while glucose/acetate switching resulted in the development of specialist subpopulations for each of the two carbon sources. Mutational comparison between static and dynamic strains highlighted genes important for robust growth on the various substrates, as well as ones targeted differentially, depending on the static or dynamic nature of the evolutionary environment. The diauxic phenotype of dynamically evolved strains varied across the substrates, but in one case the lag phase was completely abolished, whereas in statically evolved controls it was not. Transcriptional analysis further highlighted the divergence in substrate-optimal expression states and resultant evolved expression shifts, and genome-scale metabolic modeling provided insight into the metabolic basis underlying substrate differences and generalist versus specialist development.
Several important conclusions can be drawn from this study. Although the genetic basis for fitness improvement between static and dynamic conditions was mostly similar, several genetic regions stood out as being differentially targeted based on the temporal nature of the evolution environment. This is most noticeable in the cspC-yobF region, which acquired 5 unique mutations across the 6 total replicates of glc/xyl and glc/ac strains but only 1 mutation in a single acetate static strain. That such mutations occur disproportionately in dynamic conditions, and are not constrained to a single substrate pair, implies that this region influences substrate switching in general, perhaps through the interaction of CspC interactions with RNA polymerase complexes in response to stress (29). Further analysis could establish the underlying mechanism for this influence and find ways to leverage this knowledge for genetic engineering to produce strains with robust growth under dynamic conditions. However, simply looking at genes that mutate repeatedly does not provide the whole picture. Another striking finding from this study is the evolved diauxic behavior of a glucose/glycerol generalist, which eliminated its lag phase where glucose and glycerol statically evolved strains did not. In both the generalist and the glycerol-evolved strain the key mutations (Table 1) were in glpK and rpoC, genes known to be targeted under glycerol evolution (26). Despite this genetic similarity, the different evolutionary histories selected for different mutations within the same genes, with a resultant phenotypic difference that would not be deduced from genotype alone. Additionally, diauxic lag phase elimination or duration reduction can significantly increase bioprocess fermentation efficiency (38). The successful improvement in diauxic phenotype from both dynamic (e.g., glc/gly generalist, Fig. 2B) and static (e.g., xylose-evolved strain, Fig. 2C) ALE environments highlights the importance of utilizing both methods. ALE studies such as this one can thus help to expand the genetic knowledge base and indicate promising directions for genetic engineering of a desired phenotype, as well as naturally generating strains with industrially valuable phenotypes.
In addition to phenotypic and genetic analysis, transcriptomics and metabolic modeling helped to explain the observed evolutionary outcomes. Hierarchical clustering of static strain transcriptomes established the ordinality of substrate similarity (glucose → glycerol → xylose → acetate) that had been hinted at by the genetic and phenotypic ALE results. Generalist strains were found to shift their expression from the wild-type starting state closer to, but still distinct from, statically evolved strains, while specialists adopted a different transcriptional strategy that moved them further from static strains than from the wild type. Where the generalists evolved to minimize the number and magnitude of gene expression shifts across two substrates, the specialists were not subject to this adaptive constraint, given their tactic of coexistence and niche partitioning. Genome-scale modeling was also performed to examine the metabolic differences between the relevant substrates, and perfectly recapitulated the experimentally determined substrate similarity hierarchy. These results indicate promising avenues of investigation for future studies. First, evolving to static conditions can yield insight into the results of dynamic evolutions. The amount to which expression states differed among statically evolved strains explained the development of subpopulations under dynamic environments, and as ALE studies increase in number there are more and more available data researchers can reference when designing studies of their own (24, 25). However, when such data are lacking or it is prudent to avoid the time and resource costs of performing a static ALE, genome-scale metabolic modeling serves as a way to make these predictions without requiring starting data. The propensity for specialist subpopulation development can be deduced from the modeling-quantified substrate distances, and ALE studies can be designed accordingly, depending on the desired outcome. If a single dominant genotype were desired, such as when optimizing a genetically engineered strain (39), an environment favoring generalist development could be selected; if overall culture performance were instead the important factor, then an environment favoring specialists would not need to be avoided, allowing naturally evolved specialists to substitute for artificially engineered microbial consortia that would have the same collective phenotype (40, 41).
Overall, the data presented herein provide insight into adaptive strategies and evolutionary outcomes in dynamic environments, and demonstrate the efficacy of various data types for analyzing or designing such studies. Dynamic environments present a much more complicated selection pressure than static alternatives, with increased environmental heterogeneity known to lead to greater population heterogeneity (20). As dynamic ALE studies increase in number (11, 14) it is essential that appropriate experimental tools are in place to properly guide analyses and assess outcomes. Moreover, the dynamically driven development of generalists or specialists is of clinical importance in regard to antibiotic resistance and treatment regimes (42). The genome-scale models used in this study to quantify metabolic variation due to growth on different substrates can also model the network perturbations caused by antibiotics targeting specific biochemical reactions (43). With modeling-driven predictions and omics data follow-up characterizations after ALE experiments, strides can be made in both basic evolutionary research and applied clinical and biotechnological studies.
MATERIALS AND METHODS
Adaptive laboratory evolution and phenotypic profiling.
Strains were evolved in an automated system that tracked growth rates and propagated cultures in constant exponential growth phase, as described previously (44). Starting with wild-type Escherichia coli K-12 MG1655 (ATCC 4706), cultures were serially propagated (100 μl passage volume) in 15 ml (working volume) tubes of M9 minimal medium kept at 37°C and well-mixed for full aeration. Cultures were propagated upon reaching an optical density at 600 nm (OD600) of 0.3 (Tecan Sunrise plate reader; equivalent to an OD600 of ∼1 on a typical instrument with 1 cm path length), a point at which nutrients were still in excess and exponential growth was still occurring (confirmed with growth curves and high-pressure liquid chromatography [HPLC] measurements). The M9 medium contained either 4 g/liter glucose, 4 g/liter xylose, 4 g/liter acetate, or 0.2% (by volume) glycerol. Dynamic cultures were alternately passed between glucose medium and one of the three alternative medium types, while static cultures were ever grown on only a single type of medium. For dynamic cultures, average growth period in glucose tubes (time from inoculation to passage) decreased from 8.2 to 5.3 h over the course of the ALE experiment, while average xylose growth period decreased from 9.1 to 6.0 h, glycerol growth period decreased from 10.4 to 5.8 h, and acetate growth period decreased from 22.5 to 12.9 h. Lag phases were not evident when passing cultures between alternating substrates, likely because lag would occur immediately after passage and before OD600 values were detectable. Diauxic growth tests were performed under identical conditions to the ALE experiment, but M9 medium (5 mM glucose plus 5 mM xylose or acetate or glycerol) was used.
DNA and RNA sequencing.
Genomic DNA, either clonal or population, was isolated using the Macherey-Nagel NucleoSpin tissue kit, following the manufacturer's protocol for use with bacterial cells. The quality of isolated genomic DNA was assessed using Nanodrop UV absorbance ratios. DNA was quantified using the Qubit double-stranded DNA (dsDNA) high-sensitivity assay. Paired-end whole genome DNA sequencing libraries were generated using Illumina's Kappa kit and run on an Illumina MiSeq platform with a PE600v3 kit. The generated DNA sequencing FASTQ files were processed with the breseq computational pipeline (45) and aligned to the E. coli genome (NCBI accession no. NC_000913.3) to identify mutations. For population sequencing, evolved endpoint populations were used to inoculate a tube of medium with the desired carbon source and DNA was then harvested following growth enrichment overnight. Mean read depth for each population was at least 110 times, and only mutations with greater than 15% population frequency in at least one growth condition were examined. Mutations differing by more than 1.5× in frequency across different carbon source growth enrichments were taken as evidence of subpopulations (see File S2). DNA sequencing data from this study are available from the Sequence Read Archive database (accession no. SRP103966).
RNA sequencing data were generated under conditions of aerobic, exponential-phase growth on M9 minimal medium plus the relevant carbon substrate at the concentrations used in the ALE experiment. Cells were harvested using the Qiagen RNAprotect bacterial reagent according to the manufacturer's specifications. Prior to RNA extraction, pelleted cells were stored at −80°C, then thawed and incubated with lysozyme, SuperaseIn, protease K, and 20% sodium dodecyl sulfate for 20 min at 37°C. Total RNA was isolated and purified using a Qiagen RNeasy minikit column according to the manufacturer's specifications. rRNA was removed using Epicentre's Ribo-Zero rRNA-removal kit for Gram-negative bacteria. A KAPA Stranded RNA-Seq kit was used to generate paired-end strand-specific RNA sequencing libraries, which were then run on an Illumina HiSeq sequencer. RNA-seq reads were aligned to the E. coli genome using Bowtie 2 (46), and values (fragments per kilobase per million [FPKM]) were calculated with cufflinks (47). Each sample had at least 80× mean read depth coverage. Sample normalization was performed with cuffnorm, and differential expression levels were quantified via cuffdiff (48). RNA-sequencing data from this study are available from the Gene Expression Omnibus database (accession no. GSE97944).
In silico modeling.
Monte Carlo sampling of M-Model flux distributions under different substrate growth conditions was performed on the most current genome-scale model of E. coli metabolism, iJO1366 (49), using the Matlab COBRA Toolbox (50), as described previously (35). Substrate uptake rates for the different carbon compounds were set to wild-type values (see File S3 in the supplemental material) and the allowable growth rate to within 10% of model-determined optimum, and Monte Carlo sampling was performed with the sampleCbModel function using default parameters. A 2-sample Kolmogorov-Smirnov test statistic was used to pairwise compare, for every reaction in the model, the difference in flux distribution between xylose- or glycerol- or acetate-growth and glucose-growth. The cumulative sum of test statistics for every reaction led to a quantitative value of metabolic distance from glucose, and values were normalized such that the distance of acetate from glucose was 100. Flux differences were visualized by mapping model outputs to a metabolic pathway map (Fig. 5A) via the Escher tool (51).
A genome-scale model of E. coli metabolism and gene expression (ME-Model), iLE1678-ME (52), was used to simulate growth on each of the four carbon substrates used in this study, as well as several other common growth substrates. To model each condition, the uptake of the growth substrate was unconstrained and the uptakes of other carbon substrates were set to 0. The remaining default ME-Model parameters are optimized to model the growth of a laboratory-evolved E. coli K-12 MG1655 strain, so they remained set to their default values. The ME-Model was simulated by computing the maximum feasible growth rate of the model under the imposed in silico conditions via a bisection procedure (53) that uses a quadruple-precision version of the MINOS optimizer (54). A single ME-Model simulation provides predictions of the transcriptome, proteome, and metabolic flux state required for the cell to grow optimally. Using these values, the metabolic distance of a compound from glucose was quantified by calculating the Euclidean distance between ME-Model-predicted translation reaction fluxes (proteome), and values were normalized such that the distance of acetate was 100.
Accession number(s).
RNA sequencing data from this study are available from the Gene Expression Omnibus (GEO) database under the accession number GSE97944. DNA sequencing data from this study are available from the Sequence Read Archive database (accession no. SRP103966).
Supplementary Material
ACKNOWLEDGMENTS
We thank Richard Szubin, Ying Hefner, and Connor Olson for their assistance with conducting experiments. This work was funded under Novo Nordisk Foundation Center for Biosustainability grant no. NNF10CC1016517.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00410-17.
REFERENCES
- 1.Noor E, Eden E, Milo R, Alon U. 2010. Central carbon metabolism as a minimal biochemical walk between precursors for biomass and energy. Mol Cell 39:809–820. doi: 10.1016/j.molcel.2010.08.031. [DOI] [PubMed] [Google Scholar]
- 2.Gorke B, Stulke J. 2008. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat Rev Microbiol 6:613–624. doi: 10.1038/nrmicro1932. [DOI] [PubMed] [Google Scholar]
- 3.Hempfling WP, Mainzer SE. 1975. Effects of varying the carbon source limiting growth on yield and maintenance characteristics of Escherichia coli in continuous culture. J Bacteriol 123:1076–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Savageau MA. 1998. Demand theory of gene regulation. II. Quantitative application to the lactose and maltose operons of Escherichia coli. Genetics 149:1677–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zaldivar J, Nielsen J, Olsson L. 2001. Fuel ethanol production from lignocellulose: a challenge for metabolic engineering and process integration. Appl Microbiol Biotechnol 56:17–34. doi: 10.1007/s002530100624. [DOI] [PubMed] [Google Scholar]
- 6.Siegal ML. 2015. Shifting sugars and shifting paradigms. PLoS Biol 13:e1002068. doi: 10.1371/journal.pbio.1002068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Solopova A, van Gestel J, Weissing FJ, Bachmann H, Teusink B, Kok J, Kuipers OP. 2014. Bet-hedging during bacterial diauxic shift. Proc Natl Acad Sci U S A 111:7427–7432. doi: 10.1073/pnas.1320063111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.New AM, Cerulus B, Govers SK, Perez-Samper G, Zhu B, Boogmans S, Xavier JB, Verstrepen KJ. 2014. Different levels of catabolite repression optimize growth in stable and variable environments. PLoS Biol 12:e1001764. doi: 10.1371/journal.pbio.1001764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dragosits M, Mattanovich D. 2013. Adaptive laboratory evolution—principles and applications for biotechnology. Microb Cell Fact 12:64. doi: 10.1186/1475-2859-12-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bennett AF, Lenski RE, Mittler JE. 1992. Evolutionary adaptation to temperature. I. Fitness responses of Escherichia coli to changes in its thermal environment. Evolution 46:16–30. [DOI] [PubMed] [Google Scholar]
- 11.Ketola T, Mikonranta L, Zhang J, Saarinen K, Ormala AM, Friman VP, Mappes J, Laakso J. 2013. Fluctuating temperature leads to evolution of thermal generalism and preadaptation to novel environments. Evolution 67:2936–2944. doi: 10.1111/evo.12148. [DOI] [PubMed] [Google Scholar]
- 12.Hughes BS, Cullum AJ, Bennett AF. 2007. An experimental evolutionary study on adaptation to temporally fluctuating pH in Escherichia coli. Physiol Biochem Zool 80:406–421. doi: 10.1086/518353. [DOI] [PubMed] [Google Scholar]
- 13.Alcantara-Diaz D, Brena-Valle M, Serment-Guerrero J. 2004. Divergent adaptation of Escherichia coli to cyclic ultraviolet light exposures. Mutagenesis 19:349–354. doi: 10.1093/mutage/geh039. [DOI] [PubMed] [Google Scholar]
- 14.Karve SM, Daniel S, Chavhan YD, Anand A, Kharola SS, Dey S. 2015. Escherichia coli populations in unpredictably fluctuating environments evolve to face novel stresses through enhanced efflux activity. J Evol Biol 28:1131–1143. doi: 10.1111/jeb.12640. [DOI] [PubMed] [Google Scholar]
- 15.Kassen R. 2002. The experimental evolution of specialists, generalists, and the maintenance of diversity. J Evol Biol 15:173–190. doi: 10.1046/j.1420-9101.2002.00377.x. [DOI] [Google Scholar]
- 16.Jasmin JN, Kassen R. 2007. Evolution of a single niche specialist in variable environments. Proc Biol Sci 274:2761–2767. doi: 10.1098/rspb.2007.0936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saxer G, Doebeli M, Travisano M. 2010. The repeatability of adaptive radiation during long-term experimental evolution of Escherichia coli in a multiple nutrient environment. PLoS One 5:e14184. doi: 10.1371/journal.pone.0014184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friesen ML, Saxer G, Travisano M, Doebeli M. 2004. Experimental evidence for sympatric ecological diversification due to frequency-dependent competition in Escherichia coli. Evolution 58:245–260. [PubMed] [Google Scholar]
- 19.Mitchell A, Romano GH, Groisman B, Yona A, Dekel E, Kupiec M, Dahan O, Pilpel Y. 2009. Adaptive prediction of environmental changes by microorganisms. Nature 460:220–224. doi: 10.1038/nature08112. [DOI] [PubMed] [Google Scholar]
- 20.Cooper TF, Lenski RE. 2010. Experimental evolution with E. coli in diverse resource environments. I. Fluctuating environments promote divergence of replicate populations. BMC Evol Biol 10:11. doi: 10.1186/1471-2148-10-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quan S, Ray JC, Kwota Z, Duong T, Balazsi G, Cooper TF, Monds RD. 2012. Adaptive evolution of the lactose utilization network in experimentally evolved populations of Escherichia coli. PLoS Genet 8:e1002444. doi: 10.1371/journal.pgen.1002444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kram KE, Geiger C, Ismail WM, Lee H, Tang H, Foster PL, Finkel SE. 2017. Adaptation of Escherichia coli to long-term serial passage in complex medium: evidence of parallel evolution. mSystems 2:e00192-16. doi: 10.1128/mSystems.00192-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conrad TM, Joyce AR, Applebee MK, Barrett CL, Xie B, Gao Y, Palsson BO. 2009. Whole-genome resequencing of Escherichia coli K-12 MG1655 undergoing short-term laboratory evolution in lactate minimal medium reveals flexible selection of adaptive mutations. Genome Biol 10:R118. doi: 10.1186/gb-2009-10-10-r118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sandberg TE, Pedersen M, LaCroix RA, Ebrahim A, Bonde M, Herrgard MJ, Palsson BO, Sommer M, Feist AM. 2014. Evolution of Escherichia coli to 42 degrees C and subsequent genetic engineering reveals adaptive mechanisms and novel mutations. Mol Biol Evol 31:2647–2662. doi: 10.1093/molbev/msu209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.LaCroix RA, Sandberg TE, O'Brien EJ, Utrilla J, Ebrahim A, Guzman GI, Szubin R, Palsson BO, Feist AM. 2015. Use of adaptive laboratory evolution to discover key mutations enabling rapid growth of Escherichia coli K-12 MG1655 on glucose minimal medium. Appl Environ Microbiol 81:17–30. doi: 10.1128/AEM.02246-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng KK, Lee BS, Masuda T, Ito T, Ikeda K, Hirayama A, Deng L, Dong J, Shimizu K, Soga T, Tomita M, Palsson BO, Robert M. 2014. Global metabolic network reorganization by adaptive mutations allows fast growth of Escherichia coli on glycerol. Nat Commun 5:3233. doi: 10.1038/ncomms4233. [DOI] [PubMed] [Google Scholar]
- 27.Conrad TM, Frazier M, Joyce AR, Cho BK, Knight EM, Lewis NE, Landick R, Palsson BO. 2010. RNA polymerase mutants found through adaptive evolution reprogram Escherichia coli for optimal growth in minimal media. Proc Natl Acad Sci U S A 107:20500–20505. doi: 10.1073/pnas.0911253107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Utrilla J, O'Brien EJ, Chen K, McCloskey D, Cheung J, Wang H, Armenta-Medina D, Feist AM, Palsson BO. 2016. Global rebalancing of cellular resources by pleiotropic point mutations illustrates a multi-scale mechanism of adaptive evolution. Cell Syst 2:260–271. doi: 10.1016/j.cels.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shenhar Y, Biran D, Ron EZ. 2012. Resistance to environmental stress requires the RNA chaperones CspC and CspE. Environ Microbiol Rep 4:532–539. doi: 10.1111/j.1758-2229.2012.00358.x. [DOI] [PubMed] [Google Scholar]
- 30.Basan M, Hui S, Okano H, Zhang Z, Shen Y, Williamson JR, Hwa T. 2015. Overflow metabolism in Escherichia coli results from efficient proteome allocation. Nature 528:99–104. doi: 10.1038/nature15765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Enjalbert B, Cocaign-Bousquet M, Portais JC, Letisse F. 2015. Acetate exposure determines the diauxic behavior of Escherichia coli during the glucose-acetate transition. J Bacteriol 197:3173–3181. doi: 10.1128/JB.00128-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosenfeld N, Young JW, Alon U, Swain PS, Elowitz MB. 2005. Gene regulation at the single-cell level. Science 307:1962–1965. doi: 10.1126/science.1106914. [DOI] [PubMed] [Google Scholar]
- 33.Feist AM, Herrgard MJ, Thiele I, Reed JL, Palsson BO. 2009. Reconstruction of biochemical networks in microorganisms. Nat Rev Microbiol 7:129–143. doi: 10.1038/nrmicro1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orth JD, Thiele I, Palsson BO. 2010. What is flux balance analysis? Nat Biotechnol 28:245–248. doi: 10.1038/nbt.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schellenberger J, Palsson BO. 2009. Use of randomized sampling for analysis of metabolic networks. J Biol Chem 284:5457–5461. doi: 10.1074/jbc.R800048200. [DOI] [PubMed] [Google Scholar]
- 36.Lerman JA, Hyduke DR, Latif H, Portnoy VA, Lewis NE, Orth JD, Schrimpe-Rutledge AC, Smith RD, Adkins JN, Zengler K, Palsson BO. 2012. In silico method for modelling metabolism and gene product expression at genome scale. Nat Commun 3:929. doi: 10.1038/ncomms1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Brien EJ, Lerman JA, Chang RL, Hyduke DR, Palsson BO. 2013. Genome-scale models of metabolism and gene expression extend and refine growth phenotype prediction. Mol Syst Biol 9:693. doi: 10.1038/msb.2013.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Slininger PJ, Thompson SR, Weber S, Liu ZL, Moon J. 2011. Repression of xylose-specific enzymes by ethanol in Scheffersomyces (Pichia) stipitis and utility of repitching xylose-grown populations to eliminate diauxic lag. Biotechnol Bioeng 108:1801–1815. doi: 10.1002/bit.23119. [DOI] [PubMed] [Google Scholar]
- 39.Mundhada H, Seoane JM, Schneider K, Koza A, Christensen HB, Klein T, Phaneuf PV, Herrgard M, Feist AM, Nielsen AT. 2017. Increased production of L-serine in Escherichia coli through adaptive laboratory evolution. Metab Eng 39:141–150. doi: 10.1016/j.ymben.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 40.Xia T, Eiteman MA, Altman E. 2012. Simultaneous utilization of glucose, xylose and arabinose in the presence of acetate by a consortium of Escherichia coli strains. Microb Cell Fact 11:77. doi: 10.1186/1475-2859-11-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Erbilgin O, Bowen BP, Kosina SM, Jenkins S, Lau RK, Northen TR. 2017. Dynamic substrate preferences predict metabolic properties of a simple microbial consortium. BMC Bioinformatics 18:57. doi: 10.1186/s12859-017-1478-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roemhild R, Barbosa C, Beardmore RE, Jansen G, Schulenburg H. 2015. Temporal variation in antibiotic environments slows down resistance evolution in pathogenic Pseudomonas aeruginosa. Evol Appl 8:945–955. doi: 10.1111/eva.12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murima P, McKinney JD, Pethe K. 2014. Targeting bacterial central metabolism for drug development. Chem Biol 21:1423–1432. doi: 10.1016/j.chembiol.2014.08.020. [DOI] [PubMed] [Google Scholar]
- 44.Sandberg TE, Long CP, Gonzalez JE, Feist AM, Antoniewicz MR, Palsson BO. 2016. Evolution of E. coli on [U-13C]glucose reveals a negligible isotopic influence on metabolism and physiology. PLoS One 11:e0151130. doi: 10.1371/journal.pone.0151130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deatherage DE, Barrick JE. 2014. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol Biol 1151:165–188. doi: 10.1007/978-1-4939-0554-6_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. 2013. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Orth JD, Conrad TM, Na J, Lerman JA, Nam H, Feist AM, Palsson BO. 2011. A comprehensive genome-scale reconstruction of Escherichia coli metabolism—2011. Mol Syst Biol 7:535. doi: 10.1038/msb.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Becker SA, Feist AM, Mo ML, Hannum G, Palsson BO, Herrgard MJ. 2007. Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox. Nat Protoc 2:727–738. doi: 10.1038/nprot.2007.99. [DOI] [PubMed] [Google Scholar]
- 51.King ZA, Drager A, Ebrahim A, Sonnenschein N, Lewis NE, Palsson BO. 2015. Escher: a web application for building, sharing, and embedding data-rich visualizations of biological pathways. PLoS Comput Biol 11:e1004321. doi: 10.1371/journal.pcbi.1004321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lloyd CJ, Ebrahim A, Yang L, King ZA, Catoiu E, O'Brien EJ, Liu JK, Palsson BO. 2017. COBRAme: a computational framework for building and manipulating models of metabolism and gene expression. bioRxiv doi: 10.1101/106559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang L, Ma D, Ebrahim A, Lloyd CJ, Saunders MA, Palsson BO. 2016. solveME: fast and reliable solution of nonlinear ME models. BMC Bioinformatics 17:391. doi: 10.1186/s12859-016-1240-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ma D, Yang L, Fleming RM, Thiele I, Palsson BO, Saunders MA. 2017. Reliable and efficient solution of genome-scale models of metabolism and macromolecular expression. Sci Rep 7:40863. doi: 10.1038/srep40863. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.