Abstract
Spatiotemporal control of the cAMP signaling pathway is governed by both hormonal stimulation of cAMP generation by adenylyl cyclases (activation phase) and cAMP hydrolysis by phosphodiesterases (PDEs) (termination phase). The termination phase is initiated by PDEs actively targeting the protein kinase A (PKA) R-subunit through formation of a PDE-PKAR-cyclic adenosine monophosphate (cAMP) complex (the termination complex). Our results using PDE8 as a model PDE, reveal that PDEs mediate active hydrolysis of cAMP bound to its receptor RIα by enhancing the enzymatic activity. This accelerated cAMP turnover occurs via formation of a stable PDE8-RIα complex, where the protein-protein interface forms peripheral contacts and the central ligand cements this ternary interaction. The basis for enhanced catalysis is active translocation of cAMP from its binding site on RIα to the hydrolysis site on PDE8 through direct “channeling.” Our results reveal cAMP channeling in the PDE8-RIα complex and a molecular description of how this channel facilitates processive hydrolysis of unbound cAMP. Thus, unbound cAMP maintains the PDE8-RIα complex while being hydrolyzed, revealing an undiscovered mode for amplification of PKA activity by cAMP-mediated sequestration of the R-subunit by PDEs. This novel regulatory mode explains the paradox of cAMP signal amplification by accelerated PDE-mediated cAMP turnover. This highlights how target effector proteins of small-molecule ligands can promote enzyme-mediated ligand hydrolysis by scaffolding effects. Enhanced activity of the PDE8-RIα complex facilitates robust desensitization, allowing the cell to respond to dynamic levels of cAMP rather than steady-state levels. The PDE8-RIα complex represents a new class of PDE-based complexes for specific drug discovery targeting the cAMP signaling pathway.
Introduction
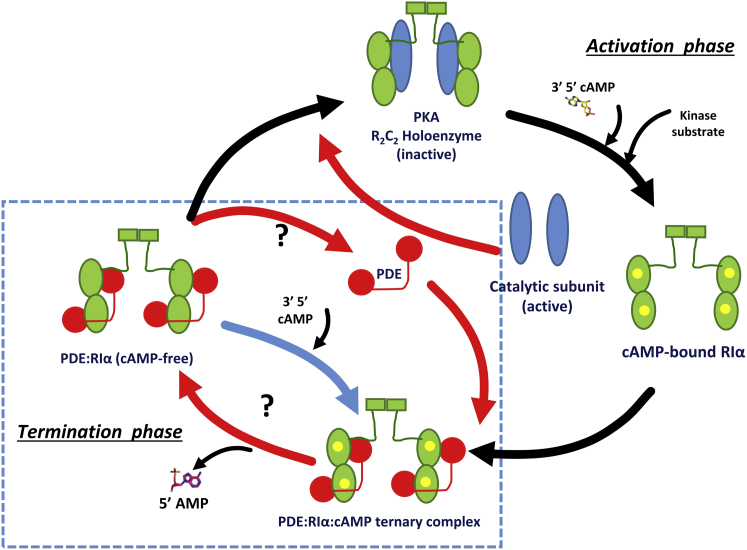
Cyclic 3′, 5′-adenosine monophosphate (cAMP) is an important second messenger that mediates a broad range of responses in the cell, with enormous implications for cellular metabolism (1). The cAMP signaling pathway is governed by two phases: 1) an activation phase, where, catalyzed by adenylyl cyclases, hormones stimulate cAMP synthesis through specific G-protein-coupled receptors (GPCRs), after which cAMP binding activates cellular targets such as protein kinase A (PKA); and 2) a termination phase that operates through phosphodiesterases (PDEs) to hydrolyze cAMP to 5′-AMP (Fig. 1). The activation phase is well characterized, where sequential cAMP binding mediates conformational changes in the inactive PKA holoenzyme, dissociating it into active catalytic subunits (C) and a dimer of regulatory subunits (R) (2, 3, 4). The dissociated/active PKA C-subunit functions to phosphorylate a large number of intracellular target substrates in the cell. The R-subunit exists as a stable dimer, and four non-redundant isoforms of the R-subunit are known (RIα, RIIα, RIβ, and RIIβ) (5). Each monomer of R consists of a C-subunit inhibitory pseudosubstrate site, a dimerization/docking domain at the N-terminus, and a tandem array of two cyclic nucleotide binding domains (CNB:A and CNB:B) for cAMP binding (6, 7). Structure and dynamics of the PKA holoenzyme and of free C- and R-subunits bound to cAMP and various analogs provide atomic resolution details of the effects of cAMP-mediated activation of PKA in the cell (activation phase). However, far less is known about how cAMP tightly bound to the R-subunit is released to facilitate reassociation with the C-subunit (termination phase), thus resetting the PKA holoenzyme to resume a new cycle of activation-termination. Signal termination is critical in cAMP signaling, as constitutively active C- or aberrant R-subunits result in metabolic diseases, some of which are lethal (8, 9, 10).
Figure 1.
Overview of the activation and termination phases in cAMP signaling. The activation phase is initiated by cAMP and substrate-mediated dissociation of the inactive PKA holoenzyme, a complex of a dimeric regulatory subunit (R, green) bound to two catalytic subunits (C, blue) (R2C2), to release free, active C-subunit. The termination phase begins with the targeting of the cAMP-bound R-subunit by dimeric PDEs (red spheres), which hydrolyze bound cAMP (yellow spheres) to generate the cAMP-free R-subunit. How PDEs preferentially hydrolyze bound cAMP is unknown. Further, it is unclear how adaptation to steady-state levels of cAMP is achieved. To see this figure in color, go online.
An important aspect of cAMP signaling is that the signaling output response resulting from cAMP-dependent PKA activation has been observed to be enhanced through increases in both signal activation and inactivation via increasing rates of cAMP synthesis or cAMP hydrolysis, respectively (11, 12). Although it is self-evident how increased cAMP synthesis through hormonal stimulation of GPCRs would increase PKA activity, how enhanced cAMP degradation by PDEs might activate PKA remains a paradox (13). Regulated cAMP degradation by PDEs results in uneven clustering of cAMP pools inside the cells, referred to as cAMP compartmentalization, where PKA and PDEs are shown to coexist as macromolecular assemblies (14). The reported ∼70% reduction in the apparent activation constant for PKA due to rapid PDE-mediated turnover of cAMP suggested that cAMP signaling output was amplified in the presence of dynamic changes in cAMP levels (13). Further, computer simulations suggested an as yet undiscovered consequence of compartmentalization in explaining the enigma of how the increased activity of PDEs mediated amplification of cAMP signaling output (12, 15, 16).
The current assumption of competitive displacement of bound cAMP by the C-subunit (17) followed by PDE-mediated hydrolysis of displaced cAMP is insufficient to account for the rapid hydrolysis of cAMP observed under a broad range of cAMP concentrations (18). Moreover, at physiological cAMP concentrations, activation of the C-subunit is achieved upon substrate-mediated dissociation from R-cAMP-C ternary complex, subsequently resulting in the phosphorylation of numerous cytoplasmic and nuclear targets (19, 20). Therefore, C-induced cAMP release and PDE-mediated passive hydrolysis of displaced cAMP would only contribute minimally toward cAMP signal termination. Hence, the only efficient route to control the catalytic activity of the PKA C-subunit necessitates its reassociation with the “cAMP-free R-subunit,” generated by direct hydrolysis of bound-cAMP by PDEs through the active-site coupling model proposed earlier (21). Although all PDEs, including PDE8, are assumed to solely hydrolyze unbound cAMP freely diffusing in cells (bulk cAMP), our recent studies indicate an important role for PDEs in actively catalyzing hydrolysis of cAMP bound to its primary receptor, the R-subunit (22, 23, 24), thereby offering a mechanism for the dynamic turnover of cAMP levels.
The concept of substrate channeling has been primarily described in metabolic enzymes, where supramolecular complexes function to restrict diffusion of reaction intermediates through the formation of specific channels between the associated constituent proteins (25). Channeling ensures that the enzyme reaction operates in a single coordinated step without release of ligands/reaction intermediates into the solvent. In tryptophan synthase and pyruvate dehydrogenase, for instance (26, 27), substrate channeling occurs by translocation of the product from one enzyme directly into the other active site within the same supramolecular assembly. Although channels have only rarely been described in signaling pathways, our recent studies point to substrate channeling as a mechanism to explain the calibrated rapid hydrolysis of cAMP bound to its target PKA through formation of a PDE-PKAR complex (23, 28). This is particularly important as cAMP binds the R-subunit with high affinity and does not readily dissociate in solution (29). In this study, we describe how this channel functions to directly access cAMP tightly bound to its target, the regulatory subunit of PKA, and enables steering toward the active site of PDE8. Furthermore, this complex would contribute to robustness of the cAMP signaling response. Properties common to all biological signaling systems include a broad dynamic range (robustness), sensitivity to small changes in stimulus levels, and adaptation to different baseline stimulus levels (30). This would ensure that the cAMP-PKA pathway is activated only under large fluxes of cAMP observable either with hormonal stimulation (13) or through PDE-mediated hydrolysis leading to adaptation to steady-state levels of cAMP (12). Therefore, the PDE8-PKAR complex not only mediates robust desensitization but is also essential in maintaining optimal PKA activity. The consequent sequestration of PKA R-subunits by PDEs even under low concentrations of cAMP through stable maintenance of the PDE8-PKAR complex keeps the C-subunit active and offers an entirely new paradigm for signaling-output amplification by cAMP degradation. This further highlights the significance of channeling in both the activation and termination phases of the cAMP signaling pathway.
To describe the functioning of the PDE8-PKA-RIα complex for rapid cAMP-PKA signal termination, we have used a combination of amide hydrogen-deuterium exchange mass spectrometry (HDXMS), enzyme assays, and fluorescence polarization (FP) spectroscopy. These powerful complementary techniques together provide peptide-level protein dynamics information and probe ligand association-dissociation kinetics, thereby providing unique insights into complex formation and cAMP dissociation. Our results show that the PDE8-PKA-RIα complex is stable in the presence of cAMP and serves to catalyze rapid turnover of cAMP through both CNB:A and CNB:B sites. We postulate that the PDE8-PKA-RIα channel functions to mediate active and rapid hydrolysis of cAMP while sequestering the R-subunit away from the PKA C-subunit. Based on our results, we describe the steps in cAMP hydrolysis mediated by the PDE8-PKA-RIα complex. This provides an explanation for how dynamic cAMP fluxes form the basis for molecular adaptation (12) and how PKA is poised to respond preferentially to large changes in cAMP either through adenylyl cyclase stimulation or activation of PDE upon complexation with the PKA R-subunit. This further underscores the importance of the PDE8-PKA-RIα complex as a novel target for drug discovery with important implications for regulation of cAMP signaling.
Materials and Methods
Materials
Chemically ultra-competent Escherichia coli BL 21 (DE3) bacterial strains used for protein expression were obtained from Life Technologies (Carlsbad, CA). TALON cobalt resin for affinity purification was from ClonTech (Mountain View, CA) and BioGel HTP hydroxyapatite beads were from BioRad laboratories (Hercules, CA). The fluorescent analogs of cAMP were 8-(2-(fluoresceinyl) aminoethylthio) adenosine-3′,5′-cyclic monophosphate (8-(fluo)-cAMP) and 2′-(6-(fluoresceinyl) aminohexylcarbamoyl) adenosine-3′,5′-cyclic monophosphate (2′-fluo-AHC-cAMP) were acquired from Biolog Life Science Institute (Bremen, Germany). The AMP-Glo assay kit was obtained from Promega (Madison, WI). LC/MS grade acetonitrile, methanol, and water were from Fisher Scientific (Waltham, MA), and trifluoroacetic acid (TFA), sequence-analysis grade, was from Fluka BioChemika (Buchs, Switzerland). Deuterium oxide was from Cambridge Isotope Laboratories (Tewksbury, MA). All other reagents and chemicals were research grade or higher from Sigma-Aldrich (St. Louis, MO).
Expression and purification of RIα
A deletion construct of the R-subunit (RIαAB, residues 75–380), cloned in the pRSETa expression vector, was transformed into the E. coli BL21 (DE3) strain for protein expression and purification as described previously (23). The full-length construct of the R-subunit (RIα), cloned in the pRSETa vector encoding ampicillin resistance, was transformed into the E. coli BL21 (DE3) strain for protein expression and purification. The harvested bacterial pellet (10 × g) was resuspended in lysis buffer (20 mM Tris, pH 7.5, 150 mM NaCl, and 1 mM β-mercaptoethanol) supplemented with protease inhibitor cocktail and subjected to lysis by sonication for 20 min. The lysate was centrifuged at 17,000 × g for 30 min and the supernatant was then incubated with TALON cobalt resin for 3 h at 277 K with gentle shaking. Proteins non-specifically bound to cobalt resin were washed out using wash buffer (lysis buffer + 5 mM imidazole), and RIα was eluted with elution buffer (lysis buffer + 250 mM imidazole, pH 7.5). The eluted protein was subsequently loaded onto a size-exclusion chromatograph using HiLoad 16/60 superdex 200 prep grade with lysis buffer on an AKTA FPLC system (GE Healthcare, Marlborough, MA). The quality of the purified RIα was confirmed using denaturing sodium dodecyl sulfate polyacrylamide gel electrophoresis and quantified by Bradford colorimetric assay.
Expression and purification of PDE8AC
The catalytic domain of PDE8A1 (residues 472–829) was cloned into pETDuet-1 plasmid and purified as described (23, 31). In short, PDE8AC was expressed and purified from the E. coli BL21 (DE3) strain via an unfolding (his-tagged purification) and refolding (BioGel HTP hydroxyapatite bead purification) process. Refolded PDE8AC protein was purified by ion-exchange chromatography (MonoQ 5/50 GL column) followed by size-exclusion chromatography (HiLoad 16/60 Superdex 200 pg column) on an AKTA FPLC system (GE Healthcare). The quality of the purified PDE8AC fractions was determined by denaturing sodium dodecyl sulfate polyacrylamide gel electrophoresis and quantified by Bradford colorimetric assay. It has been previously shown that a deletion construct of PDE8A1 spanning the catalytic domain is enzymatically active like the full-length protein and does not require any modification for activation (23, 31, 32). We therefore used the catalytic domain construct for the findings presented here. The phosphodiesterase activity of refolded PDE8AC protein was confirmed using the malachite green phosphate assay kit (BioAssay systems, Hayward, CA).
Phosphodiesterase activity assay
To monitor the enzymatic activity of PDE8A complexed to PKA-RIα and simultaneously compare it with the kinetics of free PDE8A, we used a linked AMP-Glo assay (Promega, Madison, WI) for monitoring cAMP hydrolysis. This assay measures the amount of AMP generated as a result of phosphodiesterase activity by converting the AMP formed to ATP, which is coupled to a luminescence readout using an in-built luciferase-luciferin detection system (33). The activity of free- and RIα-bound PDE8AC (1 nM) was determined over a range of cAMP concentrations (0.1, 0.25, 0.5, 1, 5, 10, and 25 μM) by incubating the reactions for 1, 2, and 5 min at room temperature. This range of cAMP concentrations was determined based on previously reported values for the KM of PDE8A (31). The amount of AMP formed by enzymatic activity of PDE8A in the two states was detected by following the manufacturer’s instructions, and the luminescence was detected by a GloMax Discover multimode plate reader (Promega, Madison, WI). An average of three independent measurements was calculated and fitted onto a curve for the Michaelis-Menten equation using GraphPad Prism 6.0 (San Diego, CA).
Fluorescence polarization spectroscopy
Fluorescence polarization assays were performed using two fluorescent analogs of cAMP: 1) a PDE-resistant analog, 8-(fluo)-cAMP (17), henceforth referred to as “8fl-cAMP,” and 2) an analog susceptible to PDE hydrolysis, 2′-fluo-AHC-cAMP (34), henceforth referred to as “2′fl-cAMP” (23, 35). Like cAMP, these analogs have been previously shown to bind to the R-subunit with very high affinity (17). Purified full-length RIα was saturated with the fluorescent cAMP analogs (20 μM) individually for 24 h at 277 K by slow mixing. Unbound ligands were removed by size-exclusion chromatography on a HiLoad 16/60 Superdex 200 pg column on an AKTA FPLC system. Fluorescence polarization (FP) assays were performed in 96-well opaque black plates from Greiner Bio-One (Kremsmünster, Austria) using a Synergy 4 multi-detection microplate reader (BioTek, Winooski, VT). For both ligands, the excitation wavelength, λex = 485 nm, and emission wavelength, λem =524 nm, were used with a bandwidth of 20 nm and an instrument G-factor of 0.87.
All FP experiments were carried out in dark with fluorescent cAMP-bound RIα at a final concentration of 5 μM and at 298 K. In the first set of experiments, unlabeled cAMP (330 μM) and PDE8AC (10 μM) at a 2:1 molar ratio were added separately to fluorescent-cAMP saturated RIα at t = 0 min and t = 20 min as shown in Fig. 3. To monitor PDE-mediated unbinding of cAMP, PDE8AC was added at time t = 20 min to fluorescent cAMP-saturated RIα with unlabeled cAMP (330 μM, t = 0 min). Next, the complex was pre-formed by addition of PDE8AC to fluorescent-cAMP-bound RIα at t = 0 min followed by unlabeled cAMP (330 μM) or AMP (2 mM) at 20 min. FP of 8fl-cAMP (5 μM), 2′fl-cAMP (5 μM), and PDE8AC (10 μM) were also obtained as control experiments. All experiments were performed three different times, each as technical triplicates. The mean ± SD values were calculated and the graphs were plotted using GraphPad Prism 6.0.
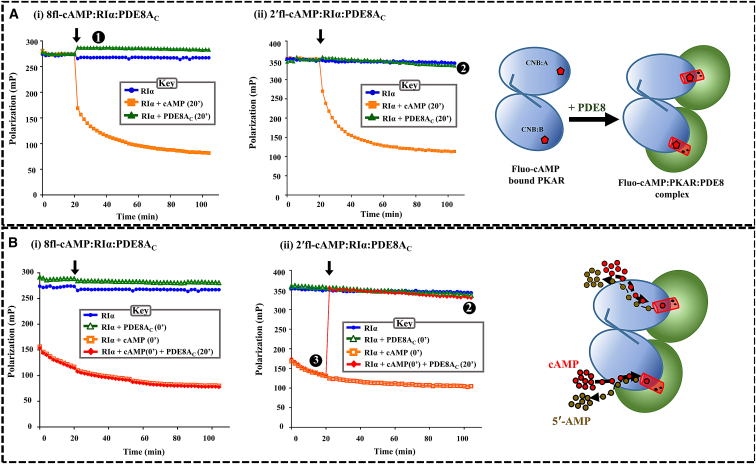
Figure 3.
Complexation of PDE8-PKA-RIα facilitates hydrolysis of cAMP by direct channeling. (A) To full-length RIα (solid circles) saturated with either 8fl-cAMP (A i) or 2′fl-cAMP (A ii), 330 μM cAMP (solid squares) and 10 μM PDE8AC (solid triangles) were added at 20 min, and polarization values were recorded for 100 min. Higher polarization indicates reduced mobility of the fluorescent ligand and, hence, the complexation of PDE8AC and RIα (step 1). Differences between the properties of the two fluorescent cAMPs is demonstrated by PDE8-mediated hydrolysis of cAMP (step 2) and, hence, decreased polarization by the end of 100 min. (B) To fluorescent cAMP, saturated RIα (solid circles), cAMP (open squares), and PDE8AC (open triangles) were added at 0 min; cAMP (0 min) was followed by PDE8AC at time t = 20 min to reactions with 8′fl-cAMP (B i) and 2′fl-cAMP (B ii), which depicts processive cAMP hydrolysis by the PDE8AC-RIα complex (step 3). Time plots of polarization were generated using GraphPad Prism 6.0. Each point represents an average value obtained from triplicate measurements from two independent experiments. Error bars (0–3 mP units) are too small to be readily visible in the plots. The cartoon represents complexation of PKA-RIα saturated with the fluorescent ligand (red pentagon) with PDE8 (top) and hydrolysis of cAMP by PKA-RIα (blue circles) (bottom), with PDE8AC represented as green circles and the channel as a red cylinder. To see this figure in color, go online.
Amide hydrogen-deuterium exchange mass spectrometry
Deuterium labeling experiments were performed in 30 μL reaction volumes with D2O at a final concentration of 90%. For HDXMS reactions of deletion construct RIαAB (1.5 μM), PDE8AC (4.5 μM) was added to cAMP-sepharose affinity purified RIαAB, followed by hydrogen-deuterium exchange reactions at 0.5, 1, 2, 5, 10, 15, 30, and 60 min. HDXMS experiments on the PDE8AC-RIα complex were carried out 1) to map the changes in RIα upon PDE8 complexation (1.5 μM), where saturating amounts of PDE8AC (4.5 μM) were maintained (a 3:1 molar ratio of PDE8AC-RIα); and 2) to map the changes on PDE8AC (1.5 μM), where saturating amounts of RIα (4.5 μM) were used (a 1:3 molar ratio of PDE8AC-RIα) to maintain all of PDE8AC in complex form. Each sample set was tested in the absence and in the presence of cAMP (330 μM). Control experiments of PDE8AC and PDE8AC-cAMP were also carried out. Complexation was first carried out by mixing PDE8AC to RIα. Deuterium exchange was then carried out by addition deuterated (20 mM Tris pH 7.5, 50 mM NaCl, 5 mM MgCl2, 20 μM ZnSO4, 5 mM β-mercaptoethanol) buffer.
All reactions were done in triplicate at 298 K with labeling times of 0.5, 1, 5, 10, 30, 60, and 100 min. The exchange reaction was quenched by lowering the pHread to 2.5 using chilled 0.1% TFA. Quenched protein samples were then injected onto nano-UPLC sample manager and subjected to cleavage by immobilized pepsin column (Poroszyme, ABI, Foster City, CA) with continuous flow of 0.1% formic acid in LC-MS grade water (pH 2.5) at 100 μL/min flow rate. The pepsin digested peptides were then trapped on to VanGuard followed by separation through reverse-phase 2.1 × 5 mm C-18 trap (ACQUITY BEH, Waters, Milford, MA) liquid chromatography column. Peptides were eluted using 8–40% gradient of acetonitrile in 0.1% formic acid (pH 2.5) at 40 μL/min flow rate, pumped by nano-ACQUITY binary solvent manager and analyzed on a SYNAPT G2-Si mass spectrometer (Waters, Milford, MA), acquiring in MSE mode to detect the peptides and measure their masses. Mass spectrometer was continuously calibrated with 200 fmol/μL of Glu-fibrinopeptide B (Glu-Fib) as a standard, at a flow rate of 5 μL/min. Same time scale parameters were followed for quench (3 s), pepsin digestion (3 min), chromatographic separation coupled to mass spectrometer (10 min) as described previously (21, 36).
The digested peptides from undeuterated controls were then identified using Protein Lynx Global Server (PLGS v3.0) software (Waters, Milford, MA). Peptides were matched and identified from primary sequence database of the individual proteins and considered only if they appeared twice among the triplicates with a minimum of four fragment ion digests. The peptides identified from undeuterated controls were then used to map the deuteration profiles of all other experimental samples using DynamX v2.0 (Waters, Milford, MA). Each peptide was then analyzed at every time point for the different states, and non-overlapping peptides with intensities at high signal-to-noise ratio were considered for quantitative analysis and data interpretation. The isotopic envelope of each peptide was used to determine the centroid mass. Subtraction of the centroid of the undeuterated mass spectra for each peptide from the deuterium exchanged states was used to calculate the average number of deuterons exchanged in each peptide by the program DynamX (Waters, Milford, MA). All values reported are not corrected for back-exchange and each value is an average of three independent deuterium exchange experiments.
Results
Activation of PDE8 hydrolysis of cAMP by RIα
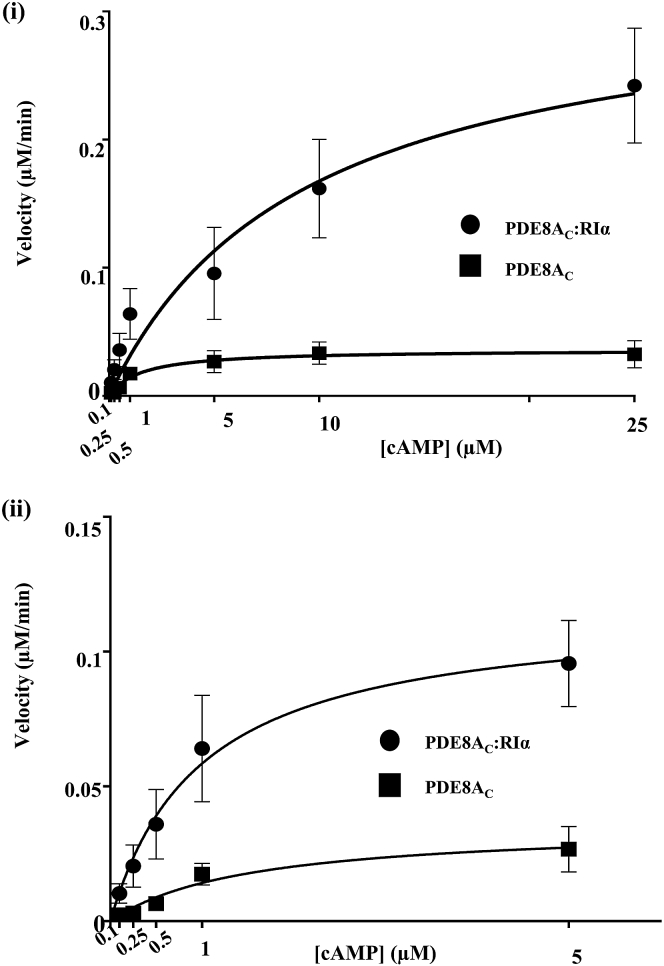
Substrates of intracellular PDEs are assumed to predominantly be free, unbound cyclic nucleotides in solution. cAMP-specific PDEs, such as PDE8, however, show hydrolytic activity for unbound cAMP in solution (11) as well as cAMP bound to their protein targets (22). We previously reported RIα-mediated activation (13-fold) of RegA, a PDE from D. discoideum (21). Here, we set out to measure rates of PDE8-mediated hydrolysis of unbound cAMP with cAMP pre-bound to its receptor, full-length PKA-RIα, using a linked assay where the AMP generated was measured by conversion to ATP detected by an increase in luciferin luminescence using an AMP-Glo assay (Materials and Methods). Enhanced AMP synthesis (cAMP hydrolysis) was observed in PDE8AC in the presence of PKA-RIα (Fig. 2). Because RIα encompasses two cAMP binding sites, a two-site binding equation (Fig. S1) was used to fit the plot for hydrolysis of bound-cAMP by PDE8AC. Significantly, plots of AMP synthesis (cAMP hydrolysis) with time for PDE8AC in the presence of RIα revealed an interesting burst-and-lag-phase kinetics profile, suggesting two catalytic rates of cAMP hydrolysis. This can be explained by the different affinities of the two distinct cAMP binding sites in the RIα, CNB:A and CNB:B, each coupled to the active site of each PDE8AC monomer in the PDE8AC-RIα complex. This would result in two distinct composite active sites, each saturable with cAMP at different concentrations. One active site was saturable at concentrations up to 5 μM cAMP, which was hydrolyzed at high initial velocity, whereas the second active site was saturable with cAMP at concentrations >10 μM (see the Supporting Material).
Figure 2.
Activation of cAMP hydrolysis by PDE8A:RIα complex. (i) Plot of AMP generation (μM/min), reflecting cAMP hydrolysis, versus substrate cAMP concentrations (μM) by PDE8AC (squares), with curve fitting onto Michaelis-Menten kinetics, and the PDE8AC:RIα complex (circles), with curve fitting to a curve describing the sum of the two-site binding of the ligand. The plot for PDE8AC hydrolysis of cAMP bound to RIα showed biphasic kinetics of cAMP hydrolysis, where an initial burst phase of rapid cAMP hydrolysis was followed by a lag phase of slower cAMP hydrolysis. (ii) We resolved the burst-phase kinetics by fitting the curve to the Michaelis-Menten equation for calculating the hydrolysis rate. The values obtained were an average of triplicate measurements, with error bars shown, and the graph was plotted by fitting the curve to the Michaelis-Menten equation in GraphPad Prism 6.0 (San Diego, CA).
Consequently, the plot for hydrolysis of bound cAMP was fit only for the fast-hydrolyzing site (Fig. 2 ii) using the Michaelis-Menten equation. The Vmax and KM values for hydrolysis of varying concentrations of cAMP by free PDE8AC were calculated to be Vmax = 0.04 ± 0.001 μM/min (per nmol of enzyme) and KM = 1.5 ± 0.3 μM, and Vmax = 0.16 ± 0.009 μM/min (per nmol of enzyme) and KM = 0.98 ± 0.05 μM for hydrolysis of different concentrations of cAMP by PDE8AC in the presence of RIα. Enhanced cAMP hydrolysis rates were observed for the PDE8AC reaction in the presence of stoichiometric amounts of RIα, indicating that PDE8AC preferentially hydrolyzed cAMP bound to RIα, resulting in faster cAMP hydrolysis by the resultant PDE8AC-RIα complex. This preference of PDE8AC for cAMP bound to its effector target was observed despite unbound cAMP being in vast excess in the reaction and the strong cAMP binding affinity for the RIα (KD ∼ 2 nM) (17, 37). Considering the higher affinity of RIα (>1000 times) for cAMP relative to that of PDE8A for cAMP and the slightly lower cAMP affinity for PDE8A-RIα (0.98 μM vs. 1.5 μM for free PDE8A), RIα-mediated activation of PDE8AC must result from formation of a stable PDE8AC-RIα complex with enhanced catalytic properties.
Processive cAMP hydrolysis by PDE8-PKA-RIα complex
Activation of cAMP hydrolysis by the PDE8-PKA-RIα complex can be modeled into the following three steps: 1) PDE8 binding to cAMP-bound RIα; 2) Hydrolysis of bound cAMP; and 3) Processive cAMP hydrolysis by PDE8-RIα complex. To monitor each of these steps we used fluorescent analogs of cAMP as reporters of PDE8AC-RIα-cAMP ternary complexation, by measuring real-time changes in their polarization intensities using FP spectroscopy (35). The recognition and complexation of PDE8 to cAMP-bound RIα (step 1), was probed using the PDE-resistant fluorescent analog 8fl-cAMP. On the other hand, binding and hydrolysis events (steps 2 and 3) were simultaneously monitored using the PDE-hydrolyzable fluorescent analog 2′fl-cAMP. Full-length RIα was saturated with these two fluorescent cAMP analogs and the resultant complexes were referred to as “8fl-cAMP-RIα” and “2′fl-cAMP-RIα” for RIα bound to 8fl-cAMP and 2′fl-cAMP, respectively. First, FP values of 8fl-cAMP-RIα and 2′fl-cAMP-RIα remained unchanged throughout the timescale of the experiment (100 min) (Fig. 3, A and B, blue), indicative of a strong association of the fluorescent cAMP analogs to RIα, with no detectable dissociation. Next, addition of a molar excess of unlabeled cAMP at 0 or 20 min resulted in competitive displacement of 8fl-cAMP and 2′fl-cAMP, respectively, from RIα, as seen by the decrease in fluorescence polarization (Fig. 3, A and B, respectively; orange).
Step 1
To monitor formation of a ternary complex of PDE8AC with cAMP-bound RIα, equimolar PDE8AC was added to 8fl-cAMP-RIα and 2′fl-cAMP-RIα individually. The increase in polarization for PDE8AC complexes with 8fl-cAMP-RIα was observed (Fig. 3, A and B, green). These polarization values were greater than that for free 8fl-cAMP-RIα, reflecting that PDE8AC bound to cAMP-bound RIα led to ternary complexation. We did not observe any significant changes in polarization for PDE8AC alone with the two fluorescent cAMP analogs (Fig. S2 i), and hence, the increased polarization observed for the above reactions can be attributed only to complexation (Fig. 3, A and B, cartoons). These results confirm that PDE8AC preferentially interacted with cAMP-bound RIα.
Step 2
Although increased polarization was observed upon addition of PDE8AC, differences in FP between the 8fl-cAMP-RIα-PDE8AC and 2′fl-cAMP-RIα-PDE8AC complexes highlight the unique properties of the fluorescent cAMP analogs. Constant FP values in (green triangles) indicate an inactive initiation 8fl-cAMP-RIα-PDE8AC complex, whereas reduced FP values in Fig. 3 B ii (green triangles) at the end of the experiment (100 min) indicates an endpoint complex bound to product, i.e., 2′fl-AMP-RIα-PDE8AC. A small increase in FP observed for 2′fl-cAMP-RIα-PDE8AC (Fig. 3, A ii and B ii) at the instant of addition of PDE8Ac reflects the rapid hydrolysis of 2′fl-cAMP to 2′fl-AMP by the PDE8AC-RIα complex. The rate of hydrolysis of cAMP to AMP is fast, and the fluorescent analog bound to the proteins gets completely hydrolyzed within a minute. It is to be noted that the decrease in FP values over a period of 100 min is indicative of a slow dissociation of the PDE8AC-RIα complex. Importantly, these results also suggest that the cAMP was directly translocated to PDE8AC (without dissociation of the PDE8-PKA-RIα complex), as neither an abrupt decrease in polarization during the reaction nor a progressive increase in polarization for PDE8AC alone or with fluorescent cAMP analogs was detected (Fig. S2 i).
Step 3
We next set out to monitor enhanced hydrolysis of cAMP by the ternary complex by adding PDE8AC at 20 min to both 8fl-cAMP-RIα and 2′fl-cAMP-RIα samples initially (0 min) incubated with a molar excess of cAMP. This led to a sharp increase in FP for the 2′fl-cAMP-RIα reaction (Fig. 3 B ii, red), indicating that addition of PDE8AC mediated a rapid hydrolysis of cAMP followed by reassociation of 2′fl-cAMP (or 2′fl-AMP) to the PDE8AC-RIα complex. Estimated rates of hydrolysis based on this experiment are consistent with the catalytic rates of PDE8AC (32). Surprisingly, no increase in FP was observed upon addition of PDE to 8fl-cAMP-RIα (Fig. 3 B i, red), signifying that this particular analog, once competitively displaced, was unable to reassociate to the PDE8AC-RIα complex. This supports a model where the two active sites are tightly coupled in the PDE8-RIα complex in the presence of an excess cAMP or 5′-AMP product.
The lack of reassociation of a PDE-resistant cAMP analog suggested that PDE8AC-RIα is a catalytic complex, stably maintained in the presence of either an excess cAMP “substrate” or 5′-AMP (henceforth referred to as “AMP”) product. We tested this by adding PDE-substrate (330 μM cAMP) or PDE-product (2 mM AMP) in excess to the preformed PDE8AC-RIα-cAMP ternary complex. No significant change in polarization was observed, indicating that the presence of extraneous substrate/product did not competitively displace “pre-formed” PDE8AC-RIα complex interactions (Fig. S2, ii and iii). These results prove that the larger fluorophore cAMP-AMP ligand stays stably bound to the ternary complex.
The FP results reveal complexation of RIα and PDE8AC, where 8fl-cAMP is unable to bind the PDE8AC-RIα complex, but 2′fl-cAMP does bind it, suggesting that the PDE8AC-RIα complex acted as a novel catalytic enzyme core. Pre-bound 2′fl-cAMP-RIα-PDE8AC and 8fl-cAMP-RIα-PDE8AC complexes do not result in dissociation of the fluorescent cAMP analog from the complex. This can be attributed to substitutions at the 2′- and 8-positions of cAMP that enhanced binding through the bulky fluorescein moiety, thereby increasing the dwell times of the fluorescent analogs with the complex. On the other hand, the bulky fluorescein substitution at the 8-carbon position of the adenine ring prevents re-entry of 8fl-cAMP to the PDE8-RIα complex, indicating an orderly association of PDE8 to RIα. Together, these results confirmed the formation of a stable PDE8AC-RIα complex in the presence of substrate/product that facilitates hydrolysis of cAMP through direct translocation of cAMP from RIα to PDE8. This could be interpreted as an activation of PDE8 catalysis by allosteric activation upon binding to RIα or through substrate channeling mediated by the coupling of the two active sites. That free fluorophores do not associate to PDE8 in solution (Fig. S2) and that excess cAMP is unable to dissociate a preformed complex point to channeling as a mechanism for RIα-dependent enhancement of PDE8 catalysis.
Bimodal kinetics of deuterium exchange for monitoring bound cAMP hydrolysis
Our FP results indicated stable complexation of cAMP-bound RIα with PDE8AC, mediating enhanced hydrolysis (Fig. 2). Further, excess cAMP was important for stably maintaining the dynamic ternary complex of PDE8A, PKA-RIα, and cAMP (Fig. 3). To map the intermolecular interactions of the ternary complex, we used amide hydrogen-deuterium exchange mass spectrometry (HDXMS) to monitor conformational dynamics of the ternary RIα-PDE8-cAMP interactions leading to hydrolysis of cAMP to AMP. To delineate interdimer interactions in the native dimeric RIα from cooperativity effects between the two CNB domains (intradimeric interactions), we chose two constructs of RIα for HDXMS analysis. These included 1) a deletion construct expressed as a monomer (referred to as “RIαAB”), which represents a simpler system for mapping the interaction of RIα-PDE8 in limited- and excess-cAMP conditions; and 2) a full-length construct expressed as a dimer (referred to as “RIα”), to obtain interactions for the native full-length protein.
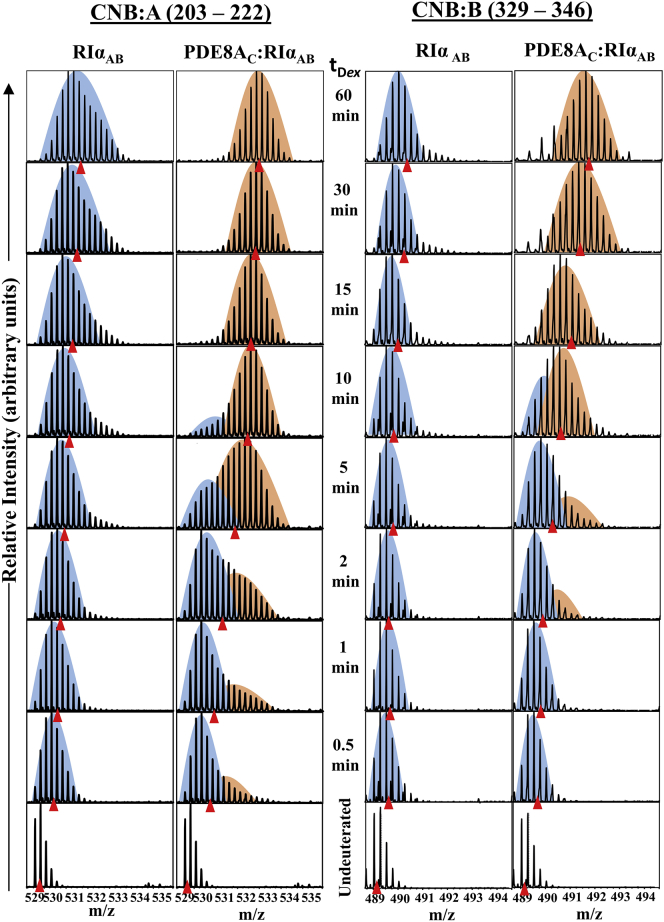
Protein-wide time-dependent increases in deuterium exchange were observed for PDE8AC-RIαAB complex relative to free RIαAB (Fig. S3 A). Both cAMP binding loci exhibited increased deuterium uptake, which upon closer examination showed a characteristic bimodal pattern of deuterium exchange at early exchange times, as shown in Fig. 4. We observed bimodal kinetics for both cAMP binding domains, CNB-A (residues 203–222; left column) and CNB:B (residues 329–346; right column) of RIαAB only in the presence of PDE8AC. Bimodal distributions are reflective of an ensemble of multiple conformations of proteins or peptides in solution (28, 38). In this case, we observed two conformations, a ligand-free conformation (Fig. 4, orange) and a ligand-bound conformation (Fig. 4, blue) of RIαAB present in the complex. With increasing deuteration times, the bimodal characteristic is not observable (time >10 min) and the increased unimodal exchange likely reflects release of ligand. Correspondingly, decreased deuterium exchange was observed in peptides spanning the active site of PDE8AC in the presence of RIαAB (Fig. S3 B). This showed that the hydrolysis of cAMP from RIα occurred only upon active association with PDE8AC.
Figure 4.
Bimodal kinetics of CNB domains monitor PDE8-mediated cAMP release from RIα. Mass spectra of peptides CNB:A (residues 203–222, left column) and CNB:B (residues 329–346, right column), for RIαAB and PDE8AC-RIαAB at different deuterium labeling (tDex) times are shown. Centroids are indicated by red triangles. The bimodal kinetics represent at least two populations—a lower-exchanging population of ligand-bound RIαAB, highlighted in blue, and a higher-exchanging population of ligand-free RIαAB, highlighted in orange. To see this figure in color, go online.
We compared the isotopic distribution profiles of the mass spectra of the peptide spanning the cAMP binding sites (residues 203–222) for different states of RIαAB-apo (without cAMP), RIαAB-cAMP (with excess cAMP), PDE8AC-RIαAB, and PDE8AC-RIαAB-cAMP (with excess cAMP) after 10 min labeling time (Fig. S3 D). The deuterium exchange profile for PDE8AC-RIαAB-cAMP after 10 min uniquely showed a broader mass spectral width, with ion sticks in the middle of the spectrum having equivalent intensities (Fig. S3 D, right). This broadening of the mass spectral envelope without a dip in intensities represents more than a simple sum of apo and cAMP-bound conformations and highlights at least one additional conformation that represents an unbroken channel that moves cAMP from the RIα binding site to the PDE8 active site. This, together with the FP, is indicative of “channeling” in the PDE8AC-RIα complex in the presence of an excess of cAMP.
PDE8-PKA-RIα interactions by HDXMS
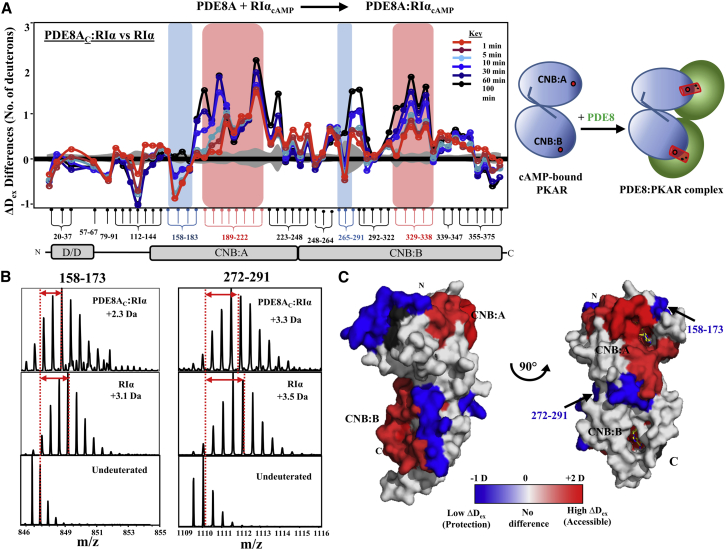
We next extended these studies to complexes of full-length dimeric RIα with PDE8AC. Pepsin proteolysis generated 56 peptides corresponding to a sequence coverage of ∼85% (Fig. S4) of RIα. Comparison of HDXMS results of the PDE8AC-RIα complex with cAMP-bound RIα, shown as a “difference plot” in Fig. 5 A, showed major differences in deuterium exchange in peptides spanning the following regions: 1) residues 112–144; 2) the αB:C helix (residues 227–244); 3) residues 157–161 and 271–275; and 4) both cAMP binding pockets (CNB:A and CNB:B). First, the interdomain linker αB:C helix showed decreased exchange upon interaction with PDE8AC. Second, across different states of RIα, subtractive peptide analysis identified two regions, residues 157–161 and 271–275, that showed decreased exchange upon interaction with PDE8AC (Fig. 5, blue boxes). These span residues from the β2-3 loop identified as allosteric sites for cAMP binding, and together with the β4-5 loop, they have been identified as putative sites for binding additional partner proteins (6). Significantly, compared to free RIα, these regions showed decreased exchange (Fig. 5 B) in the PDE8AC-RIα complex and are therefore denoted as core interaction sites with PDE8AC. These differences in deuterium exchange (10 min) are mapped onto the structure of RIα (PDB: 1RGS (39)) in Fig. 5 C, with the two potential PDE8-binding sites highlighted in blue.
Figure 5.
PDE8-PKA-RIα interaction interface on cAMP-bound RIα. (A) Differences in average deuterons exchanged (Y-axis) in PDE8AC-RIα relative to cAMP-bound RIα for each pepsin fragment peptide listed from N to C terminus (X-axis). Peptides spanning continuous regions are grouped by brace brackets. Peptides spanning the cAMP binding regions in CNB:A (189–222) and CNB:B (329–338) are highlighted in red and peptides specific to PDE8AC binding are in blue. Positive and negative differences in deuterium exchange represent increased and decreased exchange respectively in the PDE8AC-RIα complex. Deuterium exchange times for every peptide are depicted and colored according to key. Standard deviations are shaded gray. Plots were generated using DynamX 2.0 software (Waters, Milford, MA). Cartoon representation of two domains of RIα in blue, connected by a linker; cAMP as red spheres; PDE8 dimer in green, forming a PDE8AC-RIα complex (right). (B) Stacked mass spectra for RIα and PDE8AC-RIα binary complex after 10 min deuterium labeling are shown for the two putative PDE8-binding peptides. Average deuterons exchanged indicated by centroid obtained by calculating the differences between the deuterated and undeuterated peptide centroids (red arrows). (C) Significant differences in deuterium exchange in the binary complex compared to RIα are mapped onto the structure of RIα (PDB: 1RGS (39)) represented in surface in two different orientations. Regions showing increased exchange after 10 min of deuterium labeling are colored in shades of red and regions with decreased exchange in blue. cAMP molecules are shown as yellow sticks. No-coverage regions are in gray.
cAMP stabilizes PDE8-PKA-RIα complex
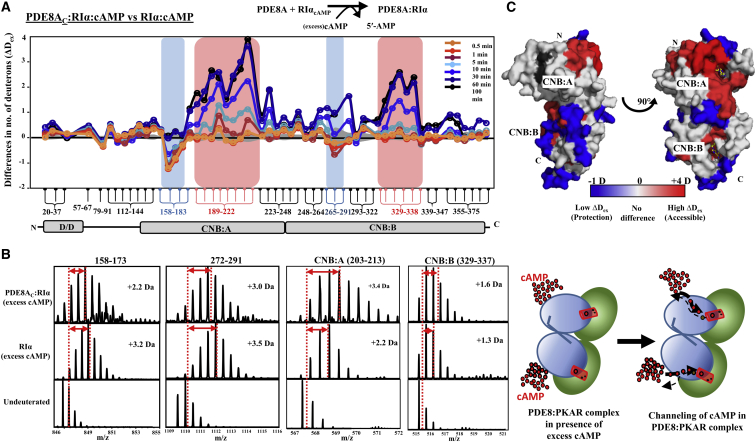
To monitor how the PDE8-PKA-RIα complex hydrolyzed excess cAMP, we measured HDXMS of the ternary complex in excess cAMP (330 μM) and compared it to that of RIα under the same conditions. In contrast to the experiments with limiting cAMP concentrations, we observed sustained reduction in exchange over longer time points (1–30 min) of deuterium labeling at the two PDE8 interaction sites in the complex (Fig. 6 A, blue boxes). This indicated that a molar excess of cAMP positively contributed to stability of the complex. Further, the magnitude of reduction and time dependence at the two PDE8AC binding sites reflected a slower dissociation at the PDE8AC interaction site flanking the CNB:A site compared to the CNB:B site. Gradual increases in exchange with time across both cAMP binding sites indicated that PDE8AC binding to RIα mediated release of cAMP (Fig. 6, A and B). These differences, mapped onto the structure of the RIα monomer (Fig. 6 C), suggest a tube-like complex with the PDE8-binding regions highlighted in blue forming a peripheral contacts that straddle a region showing increased deuterium exchange associated with cAMP hydrolysis (Fig. 6, cartoon). Deuterium uptake plots for representative peptides spanning RIα, based on deuterium uptake values at 1 and 30 min for the R-subunit in different conditions (Table ST1), are shown in Fig. S5.
Figure 6.
Mapping PDE8-PKA-RIα interactions in the presence of excess cAMP. (A) Differences in the average number of deuterons exchanged (Y axis) in PDE8AC-RIα-cAMP relative to RIα-cAMP for each pepsin-fragment peptide listed from the N- to the C-terminus (X axis). Peptides spanning continuous regions are grouped by brace brackets. Peptides spanning the cAMP binding regions, CNB:A (189–222) and CNB:B (329–338), are highlighted in red and peptides specific to PDE8AC binding in blue. Positive differences in deuterium exchange indicate increased exchange and negative differences indicate decreased exchange in the PDE8AC-RIα complex in excess cAMP. The deuterium labeling time for every peptide is depicted and color-coded according to the key. Standard deviations are shaded gray. (B) Stacked mass spectra for RIα with excess cAMP and PDE8AC-RIα-cAMP ternary complex after 10 min deuterium labeling are shown for representative peptides highlighted for PDE8-binding sites and cAMP binding pockets. Differences in their masses, i.e., the centroids are represented by double-headed arrows, compared to that of the undeuterated control (vertical dashed line). (C) Differences in deuteration levels were mapped on the structure of the monomer of RIα, with regions showing increased deuterium exchange in shades of red and decreased deuterium exchange in blue. cAMP is shown in yellow sticks. A cartoon representation of the processive hydrolysis of cAMP by the activated PDE8AC-RIα complex is shown. To see this figure in color, go online.
Enhanced hydrolysis of cAMP from PKA-RIα to PDE8
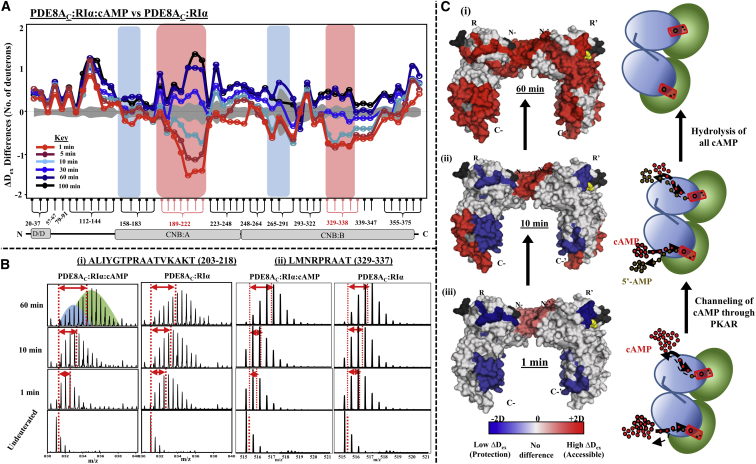
A comparison of deuterium exchange of ternary complexes of PDE8AC-RIα-cAMP in limiting and excess cAMP (Fig. 7 A) showed no differences in deuterium exchange at the putative PDE8AC interaction site, suggesting that the complex remained intact throughout the timescale of the experiment and that processive cAMP binding and hydrolysis resulted in a larger decrease in deuterium exchange in both CNB:A and CNB:B (Fig. 7 A). Based on our results from fluorescence polarization (Fig. 3), we conclude that differences in deuterium exchange observed are due to the PDE8AC-RIα-cAMP ternary complex rapidly hydrolyzing all cAMP molecules to AMP to eventually generate a ligand-free end-state complex.
Figure 7.
Channeling and turnover of cAMP through RIα in the PDE8-PKA-RIα complex as seen in the inversion of HDXMS kinetics at CNB:A and CNB:B. (A) Differences in average deuterons exchanged (Y axis) between PDE8AC-RIα-cAMP (the ternary complex) and PDE8AC-RIα complex for pepsin fragment peptides of RIα as listed from the N- to the C-terminus (X axis). Peptides spanning contiguous regions are grouped by brace brackets. Negative differences indicate decreased deuterium exchange, and positive differences denote increased deuterium exchange in the ternary complex. The two domains CNB:A and CNB:B and the PDE8-binding sites are highlighted by pink and blue boxes, respectively. The deuterium labeling times for the peptides are depicted and colored according to the key. Standard deviations are shaded gray. (B) Cyclic nucleotide binding domains CNB:A and CNB:B are indicated by stacked mass spectra of a representative peptide spanning residues 203–218 (i) and 329–338 (ii), respectively, including spectra from the ternary complex PDE8AC-RIα-cAMP (left) and the binary complex PDE8AC-RIα (right) at different labeling times. The shift in centroids with deuterium exchange is represented by double-headed red arrows, and the reference point for undeuterated RIα is shown as a vertical dashed line. (C) Surface representation of the crystal structure of RIα (PDB: 4MX3), with regions showing significant differences in deuterium exchange in the ternary complex, are mapped with color coding as in (A). Relative differences between the two states at three different labeling times—60 min (iii), 10 min (ii), and 1 min (i)—are displayed. Regions showing decreased exchange are colored in shades of blue and regions showing increased exchange in shades of red. The two RIα monomers are indicated as R (left) and R′ (right). cAMP molecules are shown as yellow spheres. No-coverage regions are in gray. A cartoon schematic is also shown of processive hydrolysis by the PDE8-PKAR complex by channeling of cAMP through PKAR resulting in release of 5′-AMP.
Peptides from the cAMP pocket in CNB:A in the ternary complex exhibited characteristic bimodal distributions of the mass spectral isotopic envelope (Fig. 7 B i). Bimodal distributions in HDXMS indicate sample heterogeneity (38), in this instance representative of a mixture of higher-exchanging cAMP-free and lower-exchanging cAMP-bound conformations (t = 60 min, Fig. 7 B). Transition of RIα from the cAMP-bound to cAMP-free conformations in real time is mapped onto its structure in Fig. 7 C, which shows protein-wide increase in deuterium exchange accompanying cAMP hydrolysis. The transitions in deuterium exchange corresponding to changes from protection to increased exchange at CNB regions in the complex with time suggests direct translocation of cAMP from RIα to PDE8AC. The distinct time-dependent HDXMS kinetics for the PDE8AC-RIα complex in the presence of excess cAMP is thus suggestive of a channel stabilizing the PDE8AC-RIα complex and accelerating cAMP turnover.
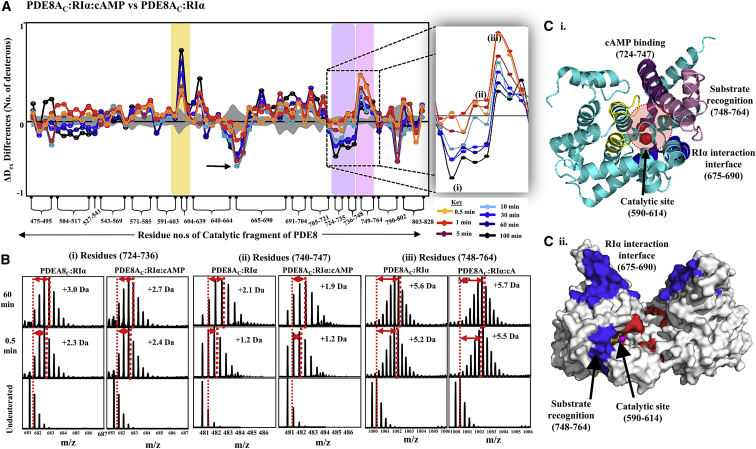
We then set out to generate a map of the complementary interface on PDE8AC for RIα interactions. Saturating concentrations of RIα were added to PDE8 in the absence or presence of cAMP, and a total of 66 peptides were obtained, corresponding to ∼87% sequence coverage of PDE8AC (Fig. S6). Residues associated with cAMP binding and catalysis showed significant changes in deuterium exchange (Fig. 8). The catalytic site (residues 604–639; Fig. 8, A–C, yellow) showed increased exchange with time in the ternary complex of PDE8AC-RIα-cAMP as compared to the binary complex of PDE8AC-RIα (limited cAMP), indicating rapid hydrolysis of cAMP by the ternary complex. Two peptides (residues 724–736 and 740–747; Fig. 8, A–C, lilac) mediate stacking interactions with the adenine ring of cAMP, whereas the third peptide spanning the M-loop (residues 748–764; Fig. 8, A–C) is obligatory for cAMP binding (32).
Figure 8.
Enhanced hydrolysis by cAMP translocation in the PDE8-PKA-RIα complex. (A) Plot of the average differences in deuterium uptake (Y axis) between PDE8AC-RIα-cAMP and PDE8AC-RIα, with residue numbers for pepsin fragment peptides of the PDE8A catalytic domain listed from the N- to the C-terminus (X axis). Positive changes denote increased deuterium exchange and negative changes denote decreased exchange in the PDE8AC-RIα complex in excess cAMP. Peptides spanning the catalytic site of PDE8A are highlighted in yellow, substrate binding sites in purple, and the PDE substrate recognition site peptides in pink. Standard deviations are shaded gray. The inset shows a magnification of the three peptides that interact with cAMP, spanning residues 724–736 (i), 740–747 (ii), and 748–764 (iii). (B) Stacked spectral plots for the three peptides (i–iii) are shown for the PDE8AC-RIα complex without and with cAMP, as indicated for different deuterium labeling times. The shifts in centroid values are represented by double-headed arrows. (C) (i) Crystal structure of the monomer of the PDE8A catalytic domain (PDB: 3ECN (32)) depicting the catalytic site in yellow (highlighted with two metal ions in red), cAMP binding sites in purple, the substrate recognition site in pink, and the RIα binding site in blue. (ii) Differences in deuterium exchange observed for PDE8AC-RIα are mapped onto the surface representation of the monomer of PDE8AC, with regions showing decreased exchange in blue and regions with increased exchange in red. To see this figure in color, go online.
Complexation of RIα and PDE8 induces conformational changes leading to translocation of cAMP to the PDE8 active site, as observed by the decreased deuterium uptake at peptides when comparing PDE8AC-RIα complex in excess cAMP with PDE8AC-RIα (Fig. 8 B). The putative RIα interaction sites (Fig. 8 C, black arrows) in PDE8AC-RIα-cAMP complex exchanged fewer deuterons throughout the labeling times, signifying complexation. To validate that these observed changes reflect the effects of complexation with RIα in the presence of cAMP, we performed HDXMS and compared the results for PDE8AC-RIα with excess cAMP to those for PDE8AC incubated with excess cAMP (Fig. S7). In excess cAMP, increased deuterium exchange was observed at the catalytic site in PDE8AC-RIα complex compared to PDE8AC alone. Thus, these results indicate RIα-mediated rapid hydrolysis of cAMP by PDE8A upon formation of the ternary complex. (Refer to Table ST2 for deuterium uptake at 1 and 30 min for PDE8AC).
Discussion
This study builds upon a previously proposed mechanism for termination of the cAMP-PKA signaling pathway through coupling of the binding sites of cAMP-target RIα with the PDE8 active site (23). This has important implications for understanding how cAMP signaling output is controlled through the termination phase of cAMP signaling by modulating PDE action. More importantly, this study highlights a unique mechanism wherein the enzymatic activity of PDE8 in the complex accelerates cAMP hydrolysis. Our results highlight a major advancement in cAMP signaling regulation, where the PDE8-PKAR complex represents a new enzyme core that mediates accelerated cAMP hydrolysis by steering cAMP directly from the binding site to the catalytic core of the PDE. This ensures that the preferential output response in cAMP signaling through increases in PKA C-subunit activation would be toward large fluxes rather than steady-state levels of cAMP. This would enable cellular adaptation to hormonal input signals. In this context, our model highlights a critical biological role for the PDE8-PKAR complex in promoting rapid cAMP hydrolysis and robustness in response to dynamic levels of cAMP in the cAMP-PKA signaling pathway. Although large fluxes in cAMP levels through hormonal modulation of adenylyl cyclase are well understood, we describe here for the first time to our knowledge, how a cAMP receptor, PKA-RIα, enhances cAMP hydrolysis rates through dynamic interactions with a PDE. In addition to its well-recognized function of hydrolyzing free cAMP (bulk cAMP) inside the cell, we show that PDEs additionally directly target cAMP-bound PKA R-subunits and catalyze hydrolysis to 5′-AMP. PDEs are thus exquisitely poised to carry out hydrolysis on both bulk and bound cAMP at different rates, resulting in cAMP compartmentalization. Active hydrolysis of cAMP in PDE8-PKAR-subunit complexes appears to be essential to effective regulation of cAMP signaling. Patients with acrodysostosis showed aberrant cAMP responses to hormonal stimuli that were attributed to mutations in PKA-RIα (Y173, T207, D267, and F291) as well as PDE4D (40, 41). The sites of mutation mapped onto residues in RIα and equivalent residues in PDE8A form the interaction interface in the PDE8-RIα complex.
Our HDXMS and FP assays also describe how cAMP stabilizes the PDE8A-RIα complex by cementing protein interactions at the periphery through the formation of a channel between the active sites of PDE8 and the cAMP binding site on PKA-RIα (Fig. 9). In proposing a “channeling” model, we considered several of the possible explanations for the enhancement of cAMP hydrolysis by the PDE8-PKAR complex. 1) cAMP-bound RIα interacted with PDE8 to facilitate rapid cAMP binding and dissociation cycles. The FP results preclude such a model, as the complex once formed is inaccessible to 8fl-cAMP (Fig. 3 B i). 2) The PDE8-PKAR complex is stabilized by the ligand cAMP, which tethers the PDE8 active site and the RIα binding site using different functional groups and mediates parallel interactions. Although such a model would promote colocalization of the two proteins, akin to a heterobifunctional cross-linker, the inability of 8fl-cAMP to bind a preformed PDE8-PKAR complex suggests that the cAMP ligand alone is insufficient for stabilizing the complex. 3) A third model relies on “channeling” to couple the two active sites in the PDE8-PKAR complex to promote enhanced cAMP hydrolysis and signal termination, and this is the model most consistent with the FP and HDXMS results.
Figure 9.
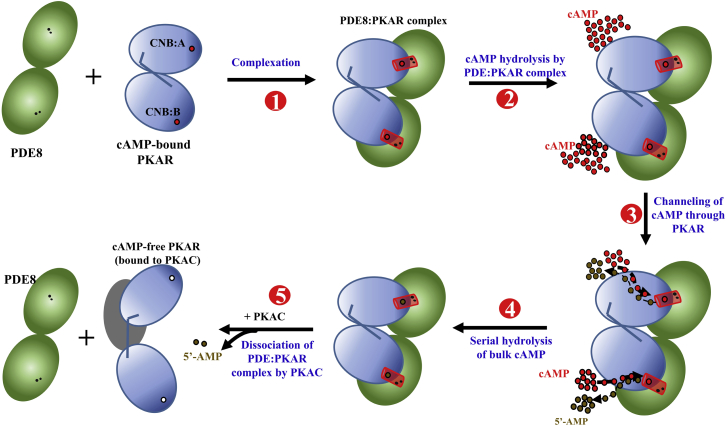
Processive hydrolysis of cAMP by channeling through the PDE8-PKAR complex. A PDE8 dimer (green circles) binds the cAMP (red circles)-bound PKA regulatory subunit (blue), forming a PDE8-PKAR complex (step 1), bringing the cAMP binding site of PKAR in close proximity to the PDE8 active site (two catalytic metal cations (black dots)) to form a “channel” (red cylinder). This channel drives hydrolysis of the cAMP flux by facilitating translocation of cAMP via PKAR (step 2), resulting in enhanced serial hydrolysis of cAMP (step 3) followed by release of the 5′-AMP product (step 4), until all cAMP molecules are hydrolyzed by the PDE8-PKAR complex. Entry of the C-subunit at this stage (step 5) promotes PKA holoenzyme formation by PDE dissociation and resets cAMP signaling for a new activation/termination cycle. To see this figure in color, go online.
We believe that channeling, described for the first time in cAMP signaling, enhances the robustness of the cAMP response. The PDE-PKAR interactions and complexation would be further enhanced in the presence of AKAP proteins, which mediate colocalization and formation of larger assemblies or cAMP signalosomes (42).
The PDE8-PKA-RIα complex is stabilized by ligand binding
Our FP results showed competitive displacement of fluorescent cAMP analogs by cAMP from free RIα, but not from PDE8-bound RIα. Higher polarization reflects decreased ligand mobility with concomitant increases in the relative molecular mass of the complex; therefore, the increased polarization observed for fluorescent cAMP-saturated RIα and PDE8 can only occur as a result of stable complexation (Fig. 9, step 1). Interestingly, the lack of difference in polarization observed in the ternary complexes mediated by the two separate fluorescent analogs of cAMP (8fl-cAMP-RIα-PDE8AC or 2′fl-cAMP-RIα-PDE8AC) supports a cAMP channeling model wherein both the PDE-non-hydrolyzable and PDE-susceptible analogs promote the stability of the ternary complex. This enhanced stability is contributed by the fluorescein moiety, which mediates additional stacking contacts with the hydrophobic residues at the opening of the active site of PDE8.
Channeling model for PDE-mediated cAMP dissociation from PKAR
On the basis of our assays and HDXMS results, we propose a cAMP channeling model in the PDE8-PKA-RIα terminator complex for accelerated cAMP turnover in cAMP signal termination. The steps are as follows: 1) PDE8 binds to the cAMP-bound R-subunit, inducing large-scale protein-wide conformational changes, including in the αB:C helix of RIα, which spans the CNB:A capping residue W260. This weakens the π-π stacking interactions between the aromatic ring of W260 and the adenine ring of cAMP. This would also occur in parallel at CNB:B with the capping residue Y371 and would weaken the equivalent π-π stacking interactions with cAMP. 2) PDE8 binding mediates disruption of bonds between the cyclic phosphate of cAMP and the critical anchoring residues in both cAMP binding pockets (CNB:A and CNB:B), greatly weakening cAMP binding. 3) Conserved hydrophobic amino acids on the surface of PDE8, spanning the substrate recognition M-loop, present a shell of strong aromatic (π-ring) contacts to draw the adenine ring of cAMP away from RIα to PDE8 while maintaining the integrity of the ternary complex (43). 4) The active site of enzyme PDE8 ringed by positively charged residues would further draw out the negatively charged cyclic phosphate of cAMP to the PDE8 active site for active hydrolysis.
The PDE8-PKA R-subunit channel would thus catalyze active cAMP hydrolysis without diffusion of any unhydrolyzed cAMP into solution. Processive hydrolysis of all cAMP molecules would thus take place through repeated cycles of binding of cAMP to the R-subunit, channeling, and PDE-mediated hydrolysis to AMP (Fig. 9, steps 2–4).
Implications of receptor-enzyme cross talk in cAMP signaling
Accelerated cAMP hydrolysis by the PDE8-RIα complex without allowing diffusion of ligand into the cytoplasm would lead to the formation of cAMP-free RIα (Fig. 9, step 5) through a rapid reduction in cAMP flux. This would enable robust desensitization of the cAMP signal and raise the response threshold for subsequent stimulus cycles. Importantly, restricted diffusion of cAMP into the cytoplasm spatially localizes the signaling response and prevents nonspecific or secondary elicitation of the non-canonical signaling pathway associated with second messengers. Lastly, the channel in the PDE8-PKA-RIα terminator complex offers, to our knowledge, a new target with improved specificity for small-molecule disruptors of cAMP signaling and PDE function.
Author Contributions
N.K.T. and S.K. carried out the experiments. N.K.T., S.K., and G.S.A. designed the experiments, analyzed and interpreted the results, and wrote the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
This work was supported by grants from the Singapore Ministry of Education Academic Research Fund—Tier 1 (MOE2016-T1-A18-114) awarded to G.S.A., National University of Singapore, Singapore.
Editor: Elizabeth Komives.
Footnotes
Seven figures and two tables are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)30461-7.
Supporting Material
References
- 1.Sassone-Corsi P. The cyclic AMP pathway. Cold Spring Harb. Perspect. Biol. 2012;4:4. doi: 10.1101/cshperspect.a011148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Das R., Esposito V., Melacini G. cAMP activation of PKA defines an ancient signaling mechanism. Proc. Natl. Acad. Sci. USA. 2007;104:93–98. doi: 10.1073/pnas.0609033103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim C., Xuong N.H., Taylor S.S. Crystal structure of a complex between the catalytic and regulatory (RIα) subunits of PKA. Science. 2005;307:690–696. doi: 10.1126/science.1104607. [DOI] [PubMed] [Google Scholar]
- 4.Taylor S.S., Zhang P., Kornev A.P. PKA: lessons learned after twenty years. Biochim. Biophys. Acta. 2013;1834:1271–1278. doi: 10.1016/j.bbapap.2013.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Skalhegg B.S., Tasken K. Specificity in the cAMP/PKA signaling pathway. Differential expression, regulation, and subcellular localization of subunits of PKA. Front. Biosci. 2000;5:D678–D693. doi: 10.2741/skalhegg. [DOI] [PubMed] [Google Scholar]
- 6.Kornev A.P., Taylor S.S., Ten Eyck L.F. A generalized allosteric mechanism for cis-regulated cyclic nucleotide binding domains. PLOS Comput. Biol. 2008;4:e1000056. doi: 10.1371/journal.pcbi.1000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malmstrom R.D., Kornev A.P., Amaro R.E. Allostery through the computational microscope: cAMP activation of a canonical signalling domain. Nat. Commun. 2015;6:7588. doi: 10.1038/ncomms8588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amieux P.S., McKnight G.S. The essential role of RI alpha in the maintenance of regulated PKA activity. Ann. N. Y. Acad. Sci. 2002;968:75–95. doi: 10.1111/j.1749-6632.2002.tb04328.x. [DOI] [PubMed] [Google Scholar]
- 9.Berthon A.S., Szarek E., Stratakis C.A. PRKACA: the catalytic subunit of protein kinase A and adrenocortical tumors. Front. Cell Dev. Biol. 2015;3:26. doi: 10.3389/fcell.2015.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bruystens J.G., Wu J., Taylor S.S. PKA RIα homodimer structure reveals an intermolecular interface with implications for cooperative cAMP binding and Carney complex disease. Structure. 2014;22:59–69. doi: 10.1016/j.str.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conti M., Beavo J. Biochemistry and physiology of cyclic nucleotide phosphodiesterases: essential components in cyclic nucleotide signaling. Annu. Rev. Biochem. 2007;76:481–511. doi: 10.1146/annurev.biochem.76.060305.150444. [DOI] [PubMed] [Google Scholar]
- 12.Houslay M.D. Adaptation in cyclic AMP signalling processes: a central role for cyclic AMP phosphodiesterases. Semin. Cell Dev. Biol. 1998;9:161–167. doi: 10.1006/scdb.1997.0221. [DOI] [PubMed] [Google Scholar]
- 13.Leiser M., Fleischer N., Erlichman J. Enhanced activation of cAMP-dependent protein kinase by rapid synthesis and degradation of cAMP. J. Biol. Chem. 1986;261:15486–15490. [PubMed] [Google Scholar]
- 14.Conti M., Mika D., Richter W. Cyclic AMP compartments and signaling specificity: role of cyclic nucleotide phosphodiesterases. J. Gen. Physiol. 2014;143:29–38. doi: 10.1085/jgp.201311083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedrich P., Aszódi A. Cyclic AMP turnover and signal amplification. Biochem. J. 1989;257:621–623. doi: 10.1042/bj2570621b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leiser M., Fleischer N., Erlichman J. Cyclic AMP turnover and signal amplification: a reply. Biochem. J. 1989;257:623–624. doi: 10.1042/bj2570621b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ogreid D., Ekanger R., Døskeland S.O. Comparison of the two classes of binding sites (A and B) of type I and type II cyclic-AMP-dependent protein kinases by using cyclic nucleotide analogs. Eur. J. Biochem. 1989;181:19–31. doi: 10.1111/j.1432-1033.1989.tb14689.x. [DOI] [PubMed] [Google Scholar]
- 18.Houslay M.D. Underpinning compartmentalised cAMP signalling through targeted cAMP breakdown. Trends Biochem. Sci. 2010;35:91–100. doi: 10.1016/j.tibs.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 19.Viste K., Kopperud R.K., Døskeland S.O. Substrate enhances the sensitivity of type I protein kinase A to cAMP. J. Biol. Chem. 2005;280:13279–13284. doi: 10.1074/jbc.M413065200. [DOI] [PubMed] [Google Scholar]
- 20.Kopperud R., Christensen A.E., Døskeland S.O. Formation of inactive cAMP-saturated holoenzyme of cAMP-dependent protein kinase under physiological conditions. J. Biol. Chem. 2002;277:13443–13448. doi: 10.1074/jbc.M109869200. [DOI] [PubMed] [Google Scholar]
- 21.Moorthy B.S., Gao Y., Anand G.S. Phosphodiesterases catalyze hydrolysis of cAMP-bound to regulatory subunit of protein kinase A and mediate signal termination. Mol. Cell. Proteomics. 2011;10:002295. doi: 10.1074/mcp.M110.002295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krishnamurthy S., Moorthy B.S., Anand G.S. Dynamics of phosphodiesterase-induced cAMP dissociation from protein kinase A: capturing transient ternary complexes by HDXMS. Biochim. Biophys. Acta. 2013;1834:1215–1221. doi: 10.1016/j.bbapap.2013.02.028. [DOI] [PubMed] [Google Scholar]
- 23.Krishnamurthy S., Moorthy B.S., Anand G.S. Active site coupling in PDE:PKA complexes promotes resetting of mammalian cAMP signaling. Biophys. J. 2014;107:1426–1440. doi: 10.1016/j.bpj.2014.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moorthy B.S., Gao Y., Anand G.S. Phosphodiesterases catalyze hydrolysis of cAMP-bound to regulatory subunit of protein kinase A and mediate signal termination. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.002295. M110 002295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spivey H.O., Ovádi J. Substrate channeling. Methods. 1999;19:306–321. doi: 10.1006/meth.1999.0858. [DOI] [PubMed] [Google Scholar]
- 26.Miles E.W. Tryptophan synthase: a multienzyme complex with an intramolecular tunnel. Chem. Rec. 2001;1:140–151. doi: 10.1002/tcr.4. [DOI] [PubMed] [Google Scholar]
- 27.Wang W., Baker P., Seah S.Y. Comparison of two metal-dependent pyruvate aldolases related by convergent evolution: substrate specificity, kinetic mechanism, and substrate channeling. Biochemistry. 2010;49:3774–3782. doi: 10.1021/bi100251u. [DOI] [PubMed] [Google Scholar]
- 28.Krishnamurthy S., Tulsian N.K., Anand G.S. Parallel allostery by cAMP and PDE coordinates activation and termination phases in cAMP signaling. Biophys. J. 2015;109:1251–1263. doi: 10.1016/j.bpj.2015.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herberg F.W., Dostmann W.R., Taylor S.S. Crosstalk between domains in the regulatory subunit of cAMP-dependent protein kinase: influence of amino terminus on cAMP binding and holoenzyme formation. Biochemistry. 1994;33:7485–7494. doi: 10.1021/bi00189a057. [DOI] [PubMed] [Google Scholar]
- 30.Yuan J., Branch R.W., Berg H.C. Adaptation at the output of the chemotaxis signalling pathway. Nature. 2012;484:233–236. doi: 10.1038/nature10964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan Z., Wang H., Ke H. Refolding and kinetic characterization of the phosphodiesterase-8A catalytic domain. Protein Expr. Purif. 2009;64:82–88. doi: 10.1016/j.pep.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang H., Yan Z., Ke H. Kinetic and structural studies of phosphodiesterase-8A and implication on the inhibitor selectivity. Biochemistry. 2008;47:12760–12768. doi: 10.1021/bi801487x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mondal S., Hsiao K., Goueli S.A. A bioluminescent assay for monitoring conjugation of ubiquitin and ubiquitin-like proteins. Anal. Biochem. 2016;510:41–51. doi: 10.1016/j.ab.2016.06.016. [DOI] [PubMed] [Google Scholar]
- 34.Huang W., Zhang Y., Sportsman J.R. A fluorescence polarization assay for cyclic nucleotide phosphodiesterases. J. Biomol. Screen. 2002;7:215–222. doi: 10.1177/108705710200700305. [DOI] [PubMed] [Google Scholar]
- 35.Rossi A.M., Taylor C.W. Analysis of protein-ligand interactions by fluorescence polarization. Nat. Protoc. 2011;6:365–387. doi: 10.1038/nprot.2011.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wales T.E., Fadgen K.E., Engen J.R. High-speed and high-resolution UPLC separation at zero degrees Celsius. Anal. Chem. 2008;80:6815–6820. doi: 10.1021/ac8008862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ogreid D., Døskeland S.O. Cyclic nucleotides modulate the release of [3H] adenosine cyclic 3′,5′-phosphate bound to the regulatory moiety of protein kinase I by the catalytic subunit of the kinase. Biochemistry. 1983;22:1686–1696. doi: 10.1021/bi00276a026. [DOI] [PubMed] [Google Scholar]
- 38.Wang L.C., Morgan L.K., Anand G.S. The inner membrane histidine kinase EnvZ senses osmolality via helix-coil transitions in the cytoplasm. EMBO J. 2012;31:2648–2659. doi: 10.1038/emboj.2012.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Su Y., Dostmann W.R.G., Varughese K.I. Regulatory subunit of protein kinase A: structure of deletion mutant with cAMP binding domains. Science. 1995;269:807–813. doi: 10.1126/science.7638597. [DOI] [PubMed] [Google Scholar]
- 40.Elli F.M., Bordogna P., Mantovani G. Screening of PRKAR1A and PDE4D in a large Italian series of patients clinically diagnosed with Albright hereditary osteodystrophy and/or pseudohypoparathyroidism. J. Bone Miner. Res. 2016;31:1215–1224. doi: 10.1002/jbmr.2785. [DOI] [PubMed] [Google Scholar]
- 41.Rhayem Y., Le Stunff C., Clauser E. Functional characterization of PRKAR1A mutations reveals a unique molecular mechanism causing acrodysostosis but multiple mechanisms causing Carney complex. J. Biol. Chem. 2015;290:27816–27828. doi: 10.1074/jbc.M115.656553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lefkimmiatis K., Zaccolo M. cAMP signaling in subcellular compartments. Pharmacol. Ther. 2014;143:295–304. doi: 10.1016/j.pharmthera.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang Y.M., Huber G., McCammon J.A. Electrostatic steering enhances the rate of cAMP binding to phosphodiesterase: Brownian dynamics modeling. Protein Sci. 2015;24:1884–1889. doi: 10.1002/pro.2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.