Abstract
Dysregulation of microRNAs (miRNAs, miRs) and their putative target genes have been increasingly reported to contribute to colorectal cancer. However, miRNAs that directly target the mutated in colorectal cancer (MCC) gene, a tumor suppressor which is downregulated or inactivated in colorectal cancer, remain largely unknown. By using an array-based miRNA analysis, we identified a group of miRNAs that were dysregulated in human metastatic versus non-metastatic colorectal cancer tissues. One of these miRNAs, miR-4260, was predicted to target MCC in the miRDB database. Results using human HCT116 and HT29 colorectal cancer cell lines showed that miR-4260 mimic enhanced cell proliferation and migration and reduced apoptosis induced by the chemotherapeutic agent 5-fluorouracil while miR-4260 inhibitor had inverse effects. Furthermore, miR-4260 negatively regulated MCC as well as SMAD4 by directly binding to the 3'untranslational region (3'UTR). Using siRNAs targeting MCC or SMAD4, we showed that upregulation of MCC and SMAD4 was essential to mediate the functional roles of miR-4260 inhibitor in colorectal cancer cells. Our in vivo experiments indicated that inhibition of miR-4260 reduced colorectal tumor growth in nude mice subcutaneously implanted with HCT116 cells. Significantly, miR-4260 was increased in human colorectal cancer tissues with simultaneous downregulation of MCC and SMAD4, strongly suggesting the clinical relevance of targeting miR-4260 in the treatment of colorectal cancer. In summary, we identified miR-4260 as a novel oncomiR for colorectal cancer that targets MCC and SMAD4. Inhibition of miR-4260 can, therefore, be a potential therapeutic strategy for colorectal cancer.
Keywords: Colorectal cancer, miRNA-4260, MCC, SMAD4.
Introduction
Colorectal cancer is the third most common cancer and the fourth leading cause of cancer-related deaths worldwide 1. Biological agents that target vascular endothelial growth factor (VEGF) and epidermal growth factor receptor (EGFR), in combination with chemotherapy, have been shown to improve the clinical outcome for patients with colorectal cancer 2. However, the use of some molecularly targeted agents such as cetuximab and panitumumab is limited to metastatic colorectal cancer patients whose tumors have a wide-type Kirsten-ras (KRAS) gene 3. Development of novel biological agents that can target colorectal tumor growth and progression is critically needed 4.
The mutated in colorectal cancer (MCC) gene was originally isolated from the human chromosome 5q21 and has been identified as a potent tumor suppressor in colorectal cancer 5. Early evidence reveals that MCC negatively regulates the G1/S cell cycle transition and is highly expressed in well-differentiated cells, indicating its potential role as a tumor suppressor in both cell growth and differentiation 6, 7. Loss of MCC has been reported in different types of cancers, including colorectal cancer, hepatocellular carcinoma, and acute myeloid leukemia 8-10. In particularly, MCC promoter methylation is commonly present in colorectal cancer and is believed to be a frequent early event during colorectal carcinogenesis 11, 12. Based on a transposon-based genetic screen in mice with colorectal cancer, MCC was found to be dysregulated probably driving tumorigenesis 13. Furthermore, re-expression of MCC suppressed Wnt/β-catenin signal transduction and inhibited cell proliferation in various colorectal cancer cell lines 11. In addition to the promoter methylation, MCC loss during tumorigenesis could also be linked with LINE-1 retrotransposition events or microRNA (miRNA, miR) targeting as reported in liver cancer 8, 9. However, the upstream mechanisms leading to MCC loss in colorectal cancer remains largely unknown.
A class of small non-coding RNAs, miRNAs (18-25 nucleotides in length), can negatively regulate target genes at posttranscriptional level via directly binding to their 3'untranslated region (3'UTR) 14. To date, more than 1880 miRNAs have been identified in human (miRBase database, Release 21). One single miRNA can downregulate dozens to hundreds of target genes, and each miRNA can be regulated by a variety of upstream molecules, forming a complex gene regulatory system in the control of diverse biological processes 15. Increasing evidence indicates that miRNA dysregulation is involved in the initiation and development of colorectal cancer. Some of these miRNAs such as miR-17-5p 16, miR-21 17, miR-224 18, and miR-429 19 are upregulated while others including miR-126 20, miR-143 21, miR-185 22, miR-18-3p 23, miR-320a 24, and miR-330 25 are downregulated. Dysregulated patterns of expression of these miRNAs were implicated in the proliferation, apoptosis, angiogenesis, and metastasis of colorectal cancer cells 26. To our knowledge, there is no information on miRNA(s) directly targeting MCC, a tumor suppressor involved in colorectal tumorigenesis, as mentioned previously.
In this study, we performed miRNA array analysis in tumor tissues from human metastatic versus non-metastatic colorectal cancer. A total of 25 miRNAs were found to be dysregulated (24 upregulated and 1 downregulated) in human metastatic colorectal cancers compared to non-metastatic samples. Bioinformatics analysis predicted MCC to be the target of 221 miRNAs in miRDB (http://mirdb.org/miRDB/ index.html), among which only miR-4260 was confirmed to be upregulated in our miRNA arrays. Thus, we focused on an in-depth analysis of miR-4260 in colorectal cancer. Our results are highly suggestive of the clinical relevance of miR-4260 in colorectal carcinogenesis
Materials and Methods
Colorectal Cancer Tissue Specimens
Tissue specimens from tumor versus peritumoral region of colorectal cancer were collected from a total of 42 patients who signed informed consent. Complete clinicopathologic data were available from Tongji Hospital of Tongji University. The tumor tissue underwent macro-dissection to enhance the tumor content of the study material. All tissues were stored immediately in liquid nitrogen and conserved at -80oC for further use.
miRNA Arrays
Total RNA extracted from the Formalin-fixed, paraffin-embedded tumor tissue sections of 3 metastatic versus 3 non-metastatic colorectal cancer patients was used for miRNA array-based analysis using the Agilent Human miRNA 8x60K V18.0 Platform. The MIAME compliant data were submitted to Gene Expression Omnibus (GEO, platform ID: GSE93377).
Quantitative Real-time Polymerase Chain Reaction (qRT-PCR)
Total RNA was extracted from cells and tissues using Trizol (TaKaRa) and reverse transcribed to cDNA using Prime ScriptTM II 1st Strand cDNA Synthesis Kit (TaKaRa) according to the manufacture's manual. The expression levels of MCC and SMAD4 were analyzed by quantitative PCR with SYBR Green (TaKaRa) on 7900HT Fast Real-Time PCR System (Applied Biosystems, CA, USA). GAPDH was used as an internal control for normalization. The primer sequences were listed in Table 1. For miRNA analysis, total RNA was reverse transcribed to cDNA using iScriptTM cDNA Synthesis Kit (Bio-Rad). The Bulge-LoopTM miRNA qPCR Primer Set (RiboBio) was used to determine the expression level of miR-4260 with Takara SYBR on ABI 7900HT Fast Real-Time PCR System. 5s was used as an internal control for normalization.
Table 1.
The primer sequences used in this study
| Primer (human) | Sequences (5'-3') | |
|---|---|---|
| SMAD 4 | Forward | CTCATGTGATCTATGCCCGTC |
| Reverse | AGGTGATACAACTCGTTCGTAGT | |
| MCC | Forward | AATAAACGTCTCCAGCAAACAGA |
| Reverse | CGTTCCTCATAGCGAAGTGTC | |
| GAPDH | Forward | ATGACATCAAGAAGGTGGTG |
| Reverse | CATACCAGGAAATGAGCTTG | |
Colorectal Cancer Cell Line Culture and Transfection
HCT116 and HT29 cell lines were cultured in Dulbecco's Modified Eagle Medium (DMEM, Corning) and RPMI1640 (KeyGEN BioTECH), respectively, supplemented with 1% penicillin-streptomycin (Gibco) and 10% fetal bovine serum (FBS, HyClone) at 37 oC with 5% CO2. To investigate the functional roles of miR-4260 in colorectal cancer cells in vitro, HCT116 and HT29 cell lines were starved for 6 hrs and then transfected with miR-4260 mimic (50 nM, RiboBio, Guangzhou, China), inhibitor (100 nM, RiboBio, Guangzhou, China), or negative controls for 48 hrs. To investigate whether miR-4260 contributes to the regulation of colorectal cancer via targeting MCC and SMAD4, siRNA for these two genes (75 nM RiboBio, Guangzhou, China) was transfected in HCT116 and HT29 cells in combination with miR-4260 inhibitor for 48 hrs.
EdU Incorporation Assay
Cell proliferation was determined using EdU (5-ethynyl-2-deoxyuridine) assay (Ribobio). Briefly, 2×105/mL of HCT116 or HT29 cells were plated in 96-well plates and starved for 6 hrs before transfection. Cells were then transfected with an miR-4260 mimic, inhibitor, or negative controls for 48 hrs and exposed to EdU 2 hrs before the end of transfection. Subsequently, cells were fixed with 4% formaldehyde overnight at 4 oC. After neutralization with glycine and washing, cells were treated with 0.5% TritonX-100 for 30 min and reacted with Apollo® reaction cocktail for 30 min. Nuclei were stained with Hoechst 33342. The EdU-positive cells were visualized under a fluorescent microscope (Leica) and counted with Image J software.
Flow Cytometry for Cell Cycle and Apoptosis
After transfection with miR-4260 mimic, inhibitor, or the negative controls, HCT116 and HT29 cells were collected and fixed with cold absolute ethanol overnight. Cells were then treated with RNase A (KeyGEN BioTECH) and stained with propidium iodide (PI, Sigma) in the dark for 15 min. Cell cycle was determined by flow cytometry (Beckman) and analyzed by FlowJo software. At least 10,000 events were recorded for each sample.
To evaluate the role of miR-4260 in colorectal cancer cell apoptosis induced by the chemotherapeutic agent 5-fluorouracil (5-FU), HCT116 cells were transfected with miR-4260 mimic, inhibitor, or their negative controls for 48 hrs and incubated with 1 μg/mL of 5-FU in DMEM containing 10% FBS for 24 hrs before the end of transfection. Cells, as well as the culture medium, were collected and stained with Annexin V-FITC and PI (Bioworld). Cell apoptosis was determined by flow cytometry (Beckman). At least 10,000 events were recorded for each sample.
Transwell Assay
After transfection with miR-4260 mimic, inhibitor, or their negative controls for 24 hrs, HCT116 cells were resuspended at a density of 2×105/mL in DMEM without FBS. 200 μL of HCT116 cell suspension was applied onto polyvinyl/pyrrolidone-free polycarbonate membranes with 8.0 μm pores in the upper chamber of transwell system (Corning). The bottom wells were filled with 600 μL of DMEM containing 10% FBS. After incubation for 48 hrs, cells that migrated to the lower surface of the cabinet were fixed with 4% paraformaldehyde (PFA), stained with crystal violet (Beyotime), and photographed with Inverted Research Microscope (Leica).
Western Blotting
Total proteins were extracted from cells and tissues using RIPA lysis buffer (Beyotime) with 1% phenylmethanesulfonyl fluoride (PMSF, KeyGEN BioTECH). Protein concentration was determined by BCA Protein Assay Kit (TaKaRa) under 595 nm spectrophotometer (Bio-Rad). Equivalent amounts of protein samples were subjected to SDS-PAGE gels, transferred to polyvinylidene difluoride (PVDF) membranes (Millipore), and blotted with primary antibodies anti-SMAD4 (1:1000, Abcam) and anti-MCC (1:1000, Abcam). Proteins were then blotted with corresponding secondary antibodies and visualized by enhanced electrochemiluminescence (ECL) system (Tanon). GAPDH was used as a loading control.
Luciferase Reporter Assay
A fragment of the 3'untranslated region (3'UTR) of MCC or SMAD4 containing the target site of miR-4260 was obtained by PCR amplification and then cloned into the pGL3-Basic Vector (Promega). The primers used were listed in Table 2. 293T cells were co-transfected with PGL3-basic-3'UTR or PGL3-basic-3'UTR mut, Renila, miR-4260 mimic or negative control using Lipofectamine2000 Reagent in 24-well plates for 24h. Luciferase activity was measured by a dual-luciferase reporter assay Kit (Promega) according to the manufacturer's instructions.
Table 2.
The primer sequences used in luciferase reporter assay
| Primer | Sequences (5'-3') | |
|---|---|---|
| SMAD 4 3'UTR | Normal, Forward | TCTAGAGTCTGACATCCTGCCCCAA |
| Mutant, Forward | TCTAGAGTCTGACATCCTCGGGGTT | |
| Reverse | TCTAGAAGTATCCACATCAACAGAGAG | |
| MCC 3'UTR | Normal, Forward | TCTAGAGGACAATGGAGTGCCCCAAC |
| Mutant, Forward | TCTAGAGGACAATGGAGTCGGGGTTC | |
| Reverse | TCTAGACCTCTATCTTAAAGTTGAGACC | |
The bold is the seed sequence used in luciferase reporter assay.
In Vivo Xenograft Tumor Study
Six-week-old BALB/c nude mice were purchased from CAVENS Lab Animal Ltd (Changzhou, China) and bred under specific pathogen-free conditions in a 12h/12h light/dark circle. To form tumors, HCT116 cells were subcutaneously implanted into the right flank of nude mice (3×106 cells per mouse), and the tumor volume was measured every other day.
To examine the potential therapeutic role of miR-4260 inhibition in colorectal cancer development, a lentivirus-based miR-4260 sponge was established. Briefly, the sequences of miR-4260 sponge were designed and ligated into the Fugw. For lentivirus packaging, 293T cells were co-transfected with psPAX2, pMD2.G, and Fugw-miR-4260 sponge at the ratio of 3:1:4 using FuGene Transfection Reagent (Roche). After 48 to 72 hrs of transfection, the medium was harvested, centrifugated, and filtered for lentivirus collection. HCT116 cells were subcutaneously implanted into the right flank of 6-week old nude mice (3×106 cells per mouse) on day 0. Fourteen days after implantation, 50 μL of lentivirus-based miR-4260 sponge (108 PFU) or Fugw control was subcutaneously injected around the tumor, and the xenograft colorectal cancer tissues were harvested after another 14 days.
Immunohistochemical Staining for PCNA and Ki67
The xenograft colorectal cancer tissues were harvested, paraffin embedded, and cut into 5-μm-thick sections. Immunohistochemical staining was performed using SP Immunohistochemistry Kit (KeyGEN BioTECH) according to the manufacturer's instructions. Antigen retrieval was realized using pH 6.0 citrate buffer, and the endogenous peroxidase activity was blocked by 0.3% hydrogen peroxide. Sections were then blocked in 5% bovine serum albumin (BSA) and incubated with primary antibodies anti-PCNA (Abcam, 1:300) and anti-Ki67 (Abcam, 1:300) overnight at 4 oC. Sections were incubated with the secondary antibody and then stained with DAB substrate chromogen system (KeyGEN BioTECH). After counterstaining with hematoxylin, images were photographed with Inverted Research Microscope (Leica). The percentage of PCNA- or Ki67-positive cells to total cells was calculated to evaluate cell proliferation in the tumor region.
Statistical Analysis
All statistical analyses were performed using SPSS 20.0 software. Data were presented as the mean ± standard error of the mean (SEM). The quantitative data were analyzed using an independent-samples t-test for comparison between two groups. Comparisons among three or more than three groups were performed using one-way ANOVA test followed by Bonferroni's post hoc test. The paired-samples t-test was performed to analyze the expression level of miR-4260 in 42 pairs of matched tumor versus peri-tumoral region of human colorectal cancer tissues. The repeated-measures analyses of variance were used to analyze the tumor volume after miR-4260 sponge treatment. Statistical significance was identified with p<0.05.
Results
miR-4260 Is Upregulated in Colorectal Cancer
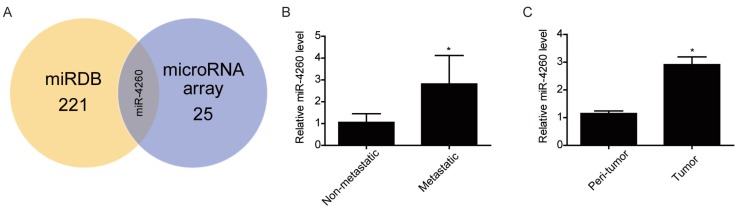
To screen miRNAs that might target MCC, which is involved in the regulation of colorectal cancer, we first performed miRNA arrays on FFPE tumor tissue sections from three human metastatic versus three non-metastatic colorectal cancers. As shown in Table 3, a total of 24 miRNAs were found to be upregulated while one miRNA was downregulated in the metastatic colorectal cancer tissues. Bioinformatics analysis by searching in the miRDB database (http://mirdb.org/miRDB/ index.html) predicted MCC to be targeted by 221 miRNAs, among which only miR-4260 was identified by miRNA arrays (Fig.1A) We validated the upregulation of miR-4260 in metastatic colorectal cancer tissues by qRT-PCRs (Fig. 1B). Furthermore, we examined the expression level of miR-4260 in 42 pairs of matched tumor versus peritumoral region of colorectal cancer specimens. The clinical characteristics of patients with colorectal cancer were listed in Table 4. As confirmed by qRT-PCRs, miR-4260 was significantly upregulated in tumor tissues compared to matched peritumoral regions (Fig. 1C), indicating that miR-4260 might be a potential regulator for colorectal cancer. Thus, miR-4260 was selected to be studied in-depth in the present study.
Table 3.
Dysregulated microRNAs in microarray analysis
| Name | Fold change | Regulation | p value |
|---|---|---|---|
| hsa-miR-1185-1-3p | 2.111893 | up | 0.044761 |
| hsa-miR-1291 | 2.110373 | up | 0.031324 |
| hsa-miR-198 | 2.029095 | up | 0.022614 |
| hsa-miR-4299 | 1.999009 | up | 0.010821 |
| hsa-miR-1185-2-3p | 1.878559 | up | 0.028431 |
| hsa-miR-652-5p | 1.826478 | up | 0.027472 |
| hsa-miR-370 | 1.822135 | up | 0.036486 |
| hsa-miR-4260 | 1.803656 | up | 0.029 |
| hsa-miR-663b | 1.801719 | up | 0.011135 |
| hsa-miR-3132 | 1.79591 | up | 0.032368 |
| hsa-miR-513a-5p | 1.785553 | up | 7.22E-04 |
| hsa-miR-2467-3p | 1.76938 | up | 0.04895 |
| hsa-miR-3621 | 1.740965 | up | 0.009596 |
| hsa-miR-4728-5p | 1.683318 | up | 0.026567 |
| hsa-miR-2278 | 1.636577 | down | 0.044191 |
| hsa-miR-514b-5p | 1.623918 | up | 0.036132 |
| hsa-miR-1202 | 1.619209 | up | 0.028448 |
| hsa-miR-3198 | 1.575268 | up | 0.026717 |
| hsa-miR-718 | 1.562195 | up | 0.032462 |
| hsa-miR-550b-2-5p | 1.559176 | up | 0.006162 |
| hsa-miR-4733-5p | 1.555993 | up | 0.008958 |
| hsa-miR-5096 | 1.553788 | up | 0.005777 |
| hsa-miR-4656 | 1.549922 | up | 0.021697 |
| hsa-miR-3196 | 1.519126 | up | 0.006716 |
| hsa-miR-575 | 1.517856 | up | 0.022388 |
Figure 1.
miR-4260 is increased in human colorectal cancer (A) MCC was predicted to be targeted by 221 miRNAs in miRDB and among them, miR-4260 was identified to be upregulated in human metastatic colorectal cancer compared to non-metastatic sample. (B) qRT-PCRs for miR-4260 expression in human metastatic versus non-metastatic colorectal cancer tissues (n=9 per group). (C) qRT-PCRs for miR-4260 expression in the tumor versus the peritumoral region of colorectal cancer from a total of 42 patients. *, p<0.05.
Table 4.
Clinical characteristics of colorectal cancer patients (n=42)
| Variable | Mean±SD or No.(%) |
|---|---|
| Age (X±S, years) | 58±10 |
| Sex | |
| Male | 27 (64.3%) |
| Female | 15 (35.7%) |
| Heart rate (X±S, times/min) | 76±7 |
| BMI (X±S, Kg/m2) | 24±3 |
| TNM stage | |
| I | 3 (7.1%) |
| II | 18 (42.9%) |
| III | 14 (33.3%) |
| IV | 7 (16.7%) |
| Tumor location | |
| Colon | 26 (61.9%) |
| Rectum | 16 (38.1%) |
| Comorbidity | |
| Hypertension | 11 (26.2%) |
| Diabetes Mellitus | 3 (7.1%) |
| Coronary heart disease | 5 (11.9%) |
miR-4260 Increases Colorectal Cancer Cell Proliferation and Migration and Reduces Apoptosis Induced by the Chemotherapeutic Agent 5-FU
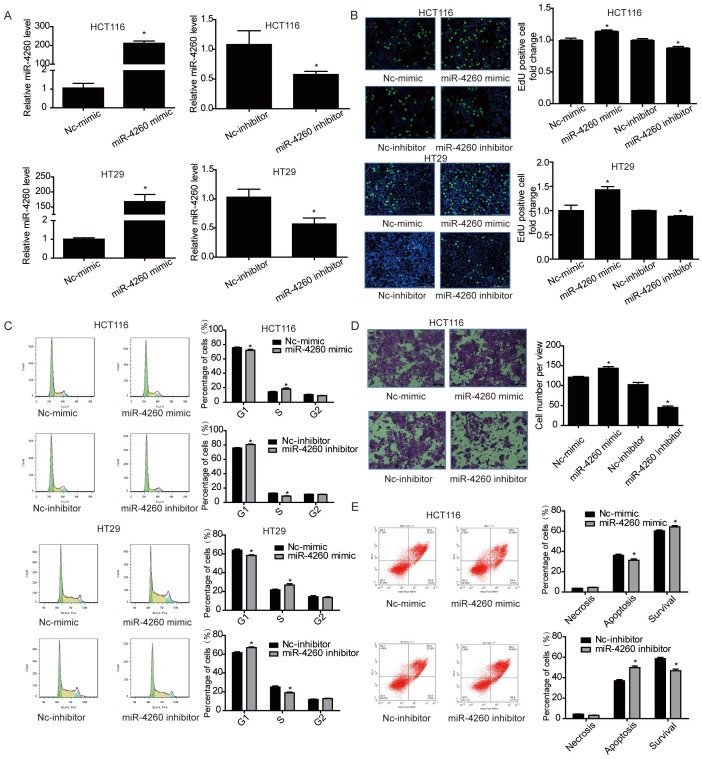
To evaluate the potential functional roles of miR-4260 in the biological behavior of colorectal cancer cells, HCT116 and HT29 cell lines were transfected with miR-4260 mimic, inhibitor, or negative controls. Subsequently, qRT-PCR was used to confirm their effects in increasing or decreasing miR-4260 (Fig. 2A). EdU incorporation assay and flow cytometry showed that the miR-4260 mimic increased cell proliferation and promoted G1/S cell cycle transition in both HCT116 and HT29 cell lines, while the miR-4260 inhibitor had inverse effects (Fig. 2B and 2C). Transwell assays further demonstrated that miR-4260 mimic increased the migration capacity of HCT116 cells, while miR-4260 inhibitor decreased it (Fig. 2D). Also, miR-4260 mimic reduced, while miR-4260 inhibitor enhanced the apoptosis of HCT116 cells treated by the chemotherapeutic agent 5-FU (Fig. 2E). Collectively, these results provide in vitro evidence that miR-4260 promoted colorectal cancer cell proliferation and migration and reduced apoptosis induced by 5-FU.
Figure 2.
miR-4260 promotes colorectal cancer cell proliferation and migration, and inhibits apoptosis induced by 5-FU (A) qRT-PCRs for miR-4260 expression in HCT116 and HT29 colorectal cancer cells transfected with miR-4260 mimic, inhibitor, or negative controls (n=3). (B) EdU incorporation assay for cell proliferation (n=4). EdU (green), DAPI (blue). Scale bar=50 μm. (C) Flow cytometry for cell cycle (n=4). (D) Transwell assay for cell migration capacity (n=3). Magnification=200x. (E) Flow cytometry for cell apoptosis induced by the chemotherapeutic agent 5-FU (n=4). *, p<0.05 versus relative negative control (nc).
MCC and SMAD4 Are Identified as Two Target Genes of miR-4260
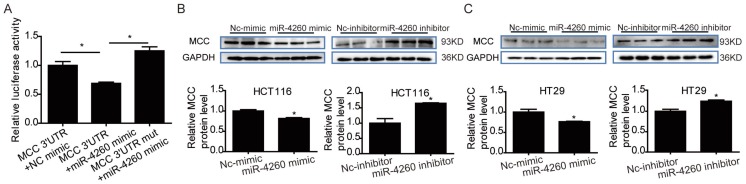
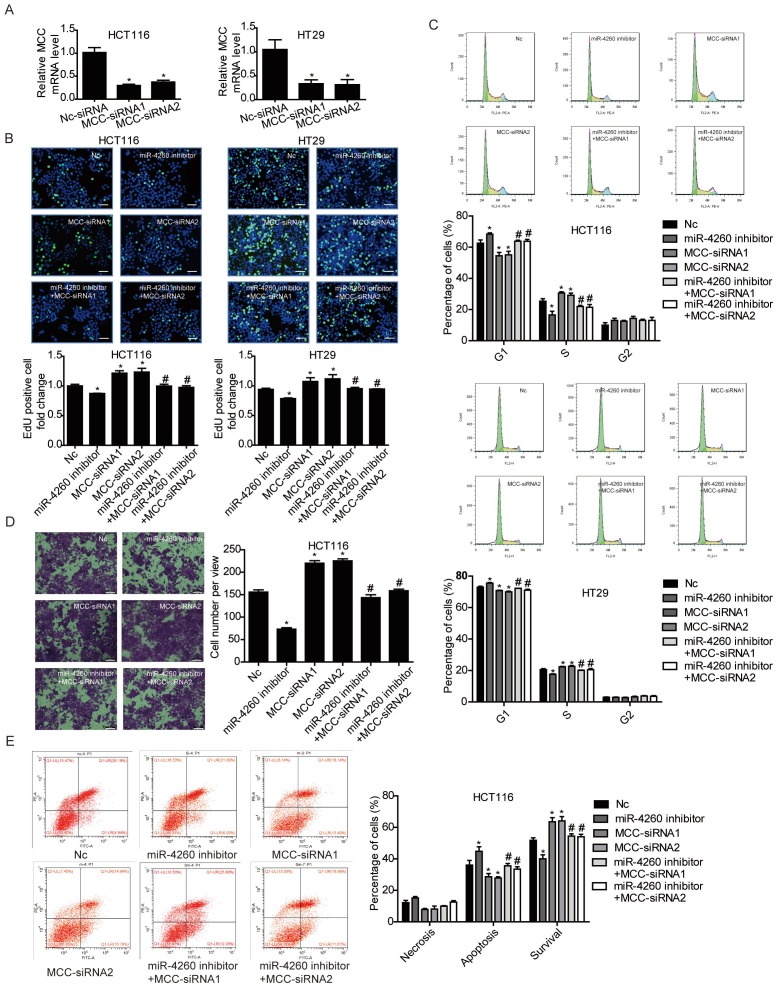
MCC is predicted to be directly targeted by miR-4260 according to the data in miRDB (http://mirdb.org/miRDB/index.html), As expected, luciferase assay showed that miR-4260 triggered a significant reduction in luciferase activity in cells transfected with the MCC 3'UTR construct. However, it had no effect when the putative miR-4260 binding site in MCC 3'UTR was mutated (Fig. 3A) indicating that MCC was a direct target of miR-4260. Also, Western blots showed that miR-4260 mimic reduced while miR-4260 inhibitor increased the protein level of MCC in HCT116 and HT29 cell lines. These results suggested that MCC could be negatively regulated by miR-4260 in colorectal cancer cells (Fig. 3B and 3C). A function-rescue assay was performed using siRNA targeting MCC to evaluate if the functional roles of miR-4260 in the proliferation, migration, and apoptosis of colorectal cancer cells were mediated through the MCC gene (Fig. 4A). Our results showed that miR-4260 inhibitor reduced the proliferation and G1/S cell cycle transition of HCT116 and HT29 cell lines, which could be totally abolished by MCC siRNAs (Fig. 4B and 4C). Silencing of MCC also abolished the effect of miR-4260 inhibitor on the migration and 5-FU-induced apoptosis in HCT116 cells (Fig. 4D and 4E). These data collectively confirmed MCC as a target gene of miR-4260 in colorectal cancer cells.
Figure 3.
MCC is a target gene of miR-4260 (A) Luciferase reporter assays demonstrated the MCC was a direct target of miR-4260 (n=6). (B) Western blot for MCC protein level in HCT116 cells transfected with miR-4260 mimic, inhibitor, or negative controls (n=3). (C) Western blot for MCC protein level in HT29 cells transfected with miR-4260 mimic, inhibitor, or negative controls (n=3). *, p<0.05.
Figure 4.
MCC is responsible for the effects of miR-4260 in human colorectal cancer cells (A) qRT-PCRs for MCC expression in HCT116 and HT29 cells transfected with siRNAs targeting MCC (n=3). (B) EdU incorporation assay for cell proliferation (n=4). EdU (green), DAPI (blue). Scale bar=50 μm. (C) Flow cytometry for cell cycle (n=4). (D) Transwell assay for cell migration capacity (n=3). Magnification=200x. (E) Flow cytometry for cell apoptosis induced by the chemotherapeutic agent 5-FU (n=4). *, p<0.05 versus negative control (nc); #, p<0.05 versus miR-4260 inhibitor.
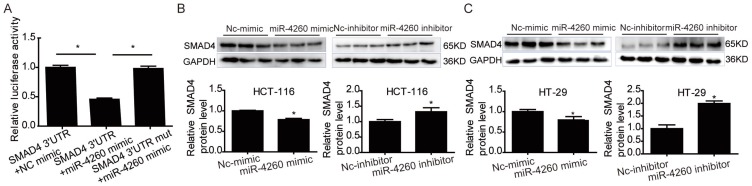
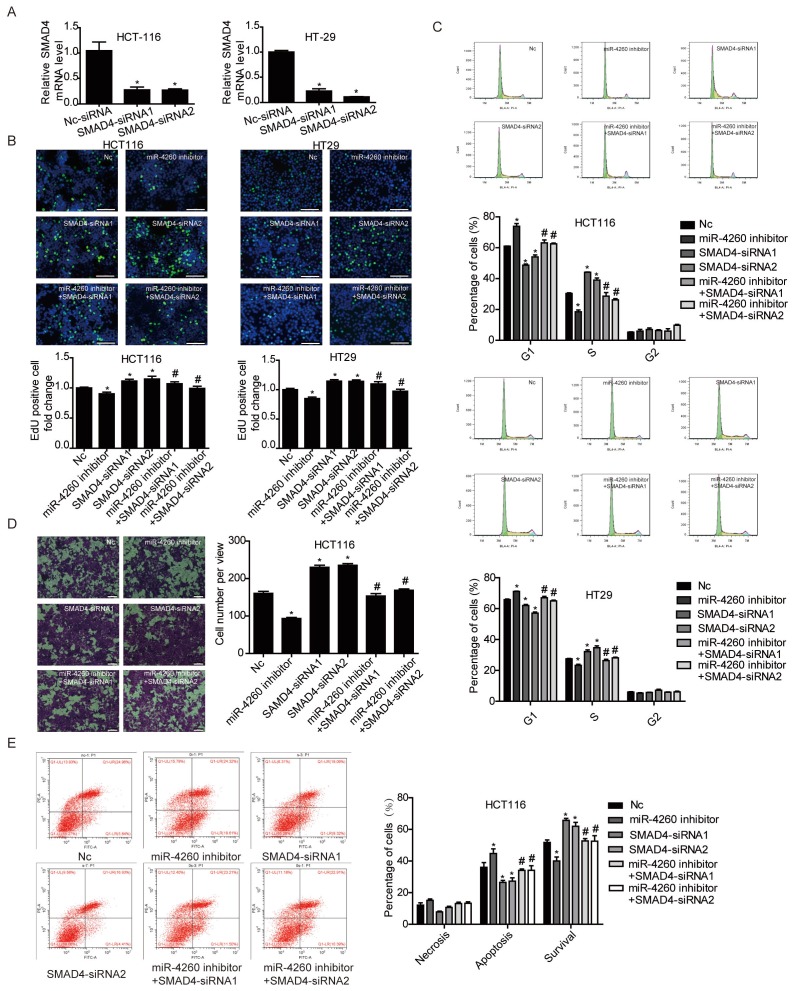
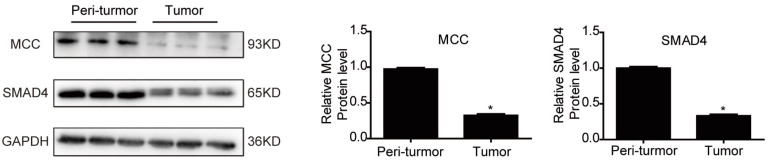
SMAD4 was previously reported to be downregulated by miRNA-130a/301a/454 family and miR-224, leading to cell proliferation and metastasis of colorectal cancer 27, 28. Based on TargetScan bioinformatics analysis (www.targetscan.org), SMAD4 is also predicted to be a target of miR-4260, though its functional role in relation to miR-4260 in colorectal cancer remains unclear. Here, we first demonstrated by the luciferase reporter assay that miR-4260 could directly target the 3'UTR of SMAD4 confirming it as a target gene of miR-4260 (Fig. 5A). Also, Western blot analysis showed that SMAD4 was negatively regulated at the protein expression level by miR-4260 in both HCT116 and HT29 cell lines (Fig. 5B and 5C). Similarly, silencing SMAD4 via siRNAs significantly abolished the protective effects of miR-4260 inhibitor in reducing cell proliferation and G1/S cell cycle transition in HCT116 and HT29 cell lines (Fig. 6A-C). The effect of miR-4260 inhibitor on cell migration and 5-FU-induced apoptosis could also be attenuated by SMAD4 siRNAs (Fig. 6D and 6E). Furthermore, concomitant to the increased expression level of miR-4260 in human colorectal cancer tissues, MCC and SMAD4 were found to be downregulated (Fig. 7). Thus, both SMAD4 and MCC were identified as target genes of miR-4260 mediating its critical roles in colorectal cancer.
Figure 5.
SMAD4 is a target gene of miR-4260 (A) Luciferase reporter assays demonstrated the SMAD4 was a direct target of miR-4260 (n=6). (B) Western blot for SMAD4 protein level in HCT116 cells transfected with miR-4260 mimic, inhibitor, or negative controls (n=3). (C) Western blot for SMAD4 protein level in HT29 cells transfected with miR-4260 mimic, inhibitor, or negative controls (n=3). *, p<0.05.
Figure 6.
SMAD4 is responsible for the effects of miR-4260 in human colorectal cancer cells (A) qRT-PCRs for SMAD4 expression in HCT116 and HT29 cells transfected with siRNAs targeting SMAD4 (n=3). (B) EdU incorporation assay for cell proliferation (n=4). EdU (green), DAPI (blue). Scale bar=50 μm. (C) Flow cytometry for cell cycle (n=4). (D) Transwell assay for cell migration capacity (n=3). Magnification=200x. (E) Flow cytometry for cell apoptosis induced by the chemotherapeutic agent 5-FU (n=4). *, p<0.05 versus negative control (nc); #, p<0.05 versus miR-4260 inhibitor.
Figure 7.
MCC and SMAD4 are downregulated in human colorectal cancer Western blot for MCC and SMAD4 protein levels in the tumor versus the peritumoral region of colorectal cancer from patients (n=3 per group). *, p<0.05.
Therapeutic Inhibition of miR-4260 Mitigates Tumor Growth of Implanted Colorectal Cancer
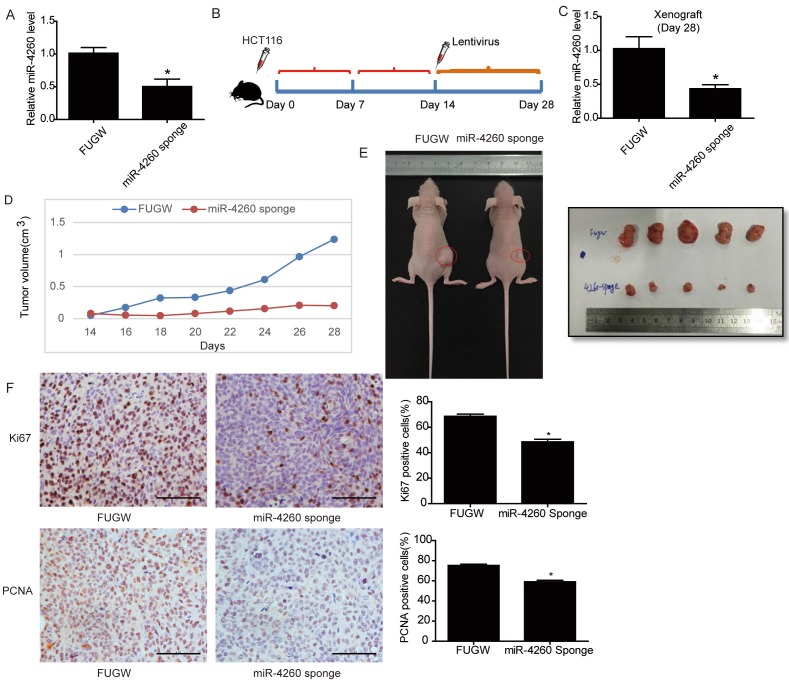
To further examine the potential therapeutic effect of miR-4260 inhibition in colorectal cancer development, a lentivirus-based miR-4260 sponge was established which was efficient in reducing miR-4260 expression level in HCT116 cells (Fig. 8A). First, HCT116 cells were subcutaneously implanted into the right flank of 6-week old nude mice (3×106 cells per mouse). Fourteen days after implantation, the lentivirus-based miR-4260 sponge was subcutaneously injected around the tumor, and the xenografted colorectal cancer tissues were harvested 14 days after the injection (Fig. 8B). Mice treated with miR-4260 sponge displayed reduced miR-4260 expression level in the xenograft tumor tissues harvested on day 28 after tumor implantation (Fig. 8C). Importantly, miR-4260 sponge efficiently reduced the tumor volume measured every other day and the tumor size on the day of sacrifice (p=0.016, Fig. 8D and 8E). Immunohistochemical staining also demonstrated that miR-4260 sponge reduced Ki67- and PCNA-positive cells in colorectal cancer tissues (Fig. 8F). These data suggested that therapeutic inhibition of miR-4260 can mitigate tumor growth of implanted colorectal cancer cells in vivo.
Figure 8.
Inhibition of miR-4260 mitigates tumor growth of implant colorectal cancer (A) qRT-PCRs for miR-4260 expression in HCT116 cells transfected with lentivirus-based miR-4260 sponge versus Fugw control (n=3). (B) Schema for subcutaneous implantation of HCT116 cells into the right flank of nude mice on day 0 and therapeutic injection of lentivirus-based miR-4260 sponge on day 14. (C) qRT-PCRs showed reduced miR-4260 expression level in the xenograft treated with miR-4260 sponge harvested on day 28 (n=5). (D) Tumor volume was measured every other day from the day of treatment with miR-4260 sponge (day 14) until the day of sacrifice (day 28) (n=5). (E) Representative images for general tumor morphology on day 28 (n=5). (F) Immunohistochemical staining for Ki67 and PCNA in the xenograft colorectal cancer tissues (n=5). Magnification=200x. *, p<0.05.
Discussion
Emerging evidence suggests that dysregulation of miRNAs is critically involved in the development and progression of colorectal cancer. Many target genes of these dysregulated miRNAs have been identified to regulate cell growth, proliferation, apoptosis, and metastasis in colorectal cancer 26. Although the MCC gene is implicated in a variety of cancers, including colorectal cancer 11-13, miRNAs that could directly target MCC in the regulation of colorectal cancer remain largely unknown. In one instance, downregulation of MCC targeted by miR-494 has been reported to contribute to cell proliferation and S-phase entry during liver tumorigenesis 9.
Based on miRNA arrays analysis, we identified a group of miRNAs that were dysregulated in colorectal cancer (24 upregulated and 1 downregulated in metastatic versus non-metastatic colorectal cancer). A shortcoming of this study was the small sample size of three pairs of metastatic and non-metastatic FFPE specimens used in miRNA arrays. It is possible that a larger sample size may have identified more miRNA candidates relevant for colon carcinogenesis. However, the focus of our study was to not only identify differentially expressed miRNAs but also to investigate their clinical relevance as well underlying potential molecular mechanisms. Among the 25 dysregulated miRNAs identified by the microarray platform, miR-4260 was predicted by the miRDB database to target MCC. We first validated upregulation of miR-4260 in the metastatic colorectal cancer tissues by qRT-PCR. To the best of our knowledge, any functional role of miR-4260 is currently not known. Thus, we set out to investigate miR-4260 as well as its potential target MCC in colorectal cancer. We showed that miR-4260 increased both cell proliferation and migration, while miR-4260 inhibitor had an opposite effect, indicating the potential growth-promoting effects of miR-4260 in colorectal cancer. Furthermore, miR-4260 overexpression reduced cell apoptosis after 5-FU treatment while miR-4260 inhibition augmented cell apoptosis in response to the chemotherapeutic agent 5-FU. This observation suggests that reducing miR-4260 level is an efficient way to diminish the anticancer drug resistance in colorectal cancer 27-29. More importantly, we demonstrated that therapeutic inhibition of miR-4260 efficiently mitigated tumor growth of the xenografted colorectal cancer in mice. Collectively, our in vitro and in vivo experiments provide strong evidence for the tumor-promoting effect of miR-4260 and a therapeutic effect of miR-4260 inhibition in colorectal cancer.
MCC is a tumor suppressor which was found to be downregulated or inactivated by promoter methylation in colorectal cancer pathology 8, 12, 13. Bioinformatics analysis, luciferase reporter assay, and Western blot validation collectively indicated that MCC was endogenously and negatively regulated by miR-4260 in both HCT116 and HT29 colorectal cancer cell lines. Furthermore, function-rescue assays revealed that MCC upregulation was necessary to mediate the suppressive effect of miR-4260 inhibition on cellular proliferation and migration, and resistance to 5-FU treatment in colorectal cancer cells. It has been reported that loss of MCC can lead to the activation of WNT/β-catenin signaling and thus promote cellular proliferation during colorectal tumorogenesis 11, 30, 31. Based on TargetScan bioinformatics analysis (www.targetscan.org), SMAD4 is also predicted to be a target of miR-4260. SMAD4 is an important tumor suppressor which was reported to be targeted by miR-224 and miR-130a/301a/454 in colorectal cancer 32, 33. Downregulation of SMAD4 promotes proliferation, migration, and invasion of colorectal cancer cells, however, the upstream mechanism of SMAD4 inactivation is poorly understood. Here, we showed that miR-4260 negatively regulated SMAD4 expression through directly binding to the 3'UTR. Using siRNAs targeting SMAD4, we further demonstrated that SMAD4 upregulation was essential to mediate the suppressive effect of miR-4260 inhibition on proliferation, migration, and resistance to 5-FU treatment in colorectal cancer cells. Taken together, our data clearly indicated that MCC and SMAD4, two key tumor suppressors, were direct targets of miR-4260 in colorectal cancer.
We also examined the expressions of miR-4260 and its targets in human colorectal cancer specimens. Significantly, miR-4260 was found to be increased in 42 pairs of matched tumor versus peritumoral region of colorectal cancer specimens strongly suggesting the clinical relevance of miR-4260 as a therapeutic target for colorectal cancer. Furthermore, both MCC and SMAD4 were downregulated in human colorectal cancer tissues. Lower expressions of MCC and SMAD4 have already been reported to be related to high tumorigenicity and poor prognosis in colorectal cancer patients 11, 12, 34, 35. Our data have shown that miR-4260 functions upstream of MCC and SMAD4 downregulating the expression of both genes. In the future, it is highly desirable to focus on the in vivo function of miR-4260 together with MCC or SMAD4 that appear to play a significant role in colorectal carcinogenesis.
In summary, we have identified, for the first time, miR-4260 as a novel oncomiR in colorectal cancer that promoted cell proliferation, migration, and reduces apoptosis induced by 5-FU via directly targeting MCC. We have also shown SMAD4 as another target gene of miR-4260. Significantly, therapeutic inhibition of miR-4260 inhibited colorectal cancer in vivo. Furthermore, we detected increased levels of miR-4260 in human colorectal cancer tissues together with downregulation of MCC and SMAD4. Our data provide compelling evidence that miR-4260 can potentially serve as a novel therapeutic target for colorectal cancer.
Acknowledgments
This work was supported by the grants from National Natural Science Foundation of China (81570362, 81200169 and 91639101 to JJ Xiao, 81400647 to Y Bei) and the development fund for Shanghai talents (to JJ Xiao).
Author Contributions
J.X. and Q.H. contributed equally to this work. J.X. and Q.H. designed the study, supervised all experiments and drafted the manuscript. D.L., J.Z., Y.B., T.C., M.H., Q.L. and S.F. performed the experiments and analyzed the data.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Moriarity A, O'Sullivan J, Kennedy J, Mehigan B, McCormick P. Current targeted therapies in the treatment of advanced colorectal cancer: a review. Ther Adv Med Oncol. 2016;8:276–93. doi: 10.1177/1758834016646734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhai Z, Yu X, Yang B, Zhang Y, Zhang L, Li X, Colorectal cancer heterogeneity and targeted therapy: Clinical implications, challenges and solutions for treatment resistance. Semin Cell Dev Biol; 2016. DOI: 10.1016/ j. semc db.2016.08.033. [DOI] [PubMed] [Google Scholar]
- 4.Henricks LM, Schellens JH, Huitema AD, Beijnen JH. The use of combinations of monoclonal antibodies in clinical oncology. Cancer Treat Rev. 2016;41:859–67. doi: 10.1016/j.ctrv.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 5.Kinzler KW, Nilbert MC, Vogelstein B, Bryan TM, Levy DB, Smith KJ. et al. Identification of a gene located at chromosome 5q21 that is mutated in colorectal cancers. Science. 1991;251:1366–70. doi: 10.1126/science.1848370. [DOI] [PubMed] [Google Scholar]
- 6.Matsumine A, Senda T, Baeg GH, Roy BC, Nakamura Y, Noda M. et al. MCC, a cytoplasmic protein that blocks cell cycle progression from the G0/G1 to S phase. J Biol Chem. 1996;271:10341–6. doi: 10.1074/jbc.271.17.10341. [DOI] [PubMed] [Google Scholar]
- 7.Senda T, Matsumine A, Yanai H, Akiyama T. Localization of MCC (mutated in colorectal cancer) in various tissues of mice and its involvement in cell differentiation. J Histochem Cytochem. 1999;47:1149–58. doi: 10.1177/002215549904700907. [DOI] [PubMed] [Google Scholar]
- 8.Shukla R, Upton KR, Munoz-Lopez M, Gerhardt DJ, Fisher ME, Nguyen T. et al. Endogenous retrotransposition activates oncogenic pathways in hepatocellular carcinoma. Cell. 2013;153:101–11. doi: 10.1016/j.cell.2013.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lim L, Balakrishnan A, Huskey N, Jones KD, Jodari M, Ng R. et al. MicroRNA-494 within an oncogenic microRNA megacluster regulates G1/S transition in liver tumorigenesis through suppression of mutated in colorectal cancer. Hepatology. 2014;59:202–15. doi: 10.1002/hep.26662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gamazon ER, Lamba JK, Pounds S, Stark AL, Wheeler HE, Cao X. et al. Comprehensive genetic analysis of cytarabine sensitivity in a cell-based model identifies polymorphisms associated with outcome in AML patients. Blood. 2013;121:4366–76. doi: 10.1182/blood-2012-10-464149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fukuyama R, Niculaita R, Ng KP, Obusez E, Sanchez J, Kalady M. et al. Mutated in colorectal cancer, a putative tumor suppressor for serrated colorectal cancer, selectively represses beta-catenin-dependent transcription. Oncogene. 2008;27:6044–55. doi: 10.1038/onc.2008.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kohonen-Corish MR, Sigglekow ND, Susanto J, Chapuis PH, Bokey EL, Dent OF. et al. Promoter methylation of the mutated in colorectal cancer gene is a frequent early event in colorectal cancer. Oncogene. 2007;26:4435–41. doi: 10.1038/sj.onc.1210210. [DOI] [PubMed] [Google Scholar]
- 13.Starr TK, Allaei R, Silverstein KA, Staggs RA, Sarver AL, Bergemann TL. et al. A transposon-based genetic screen in mice identifies genes altered in colorectal cancer. Science. 2009;323:1747–50. doi: 10.1126/science.1163040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 15.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J. et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–73. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 16.Ma Y, Zhang P, Wang F, Zhang H, Yang Y, Shi C. et al. Elevated oncofoetal miR-17-5p expression regulates colorectal cancer progression by repressing its target gene P130. Nat Commun. 2012;3:1291. doi: 10.1038/ncomms2276. [DOI] [PubMed] [Google Scholar]
- 17.Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S. et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–36. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 18.Liao WT, Li TT, Wang ZG, Wang SY, He MR, Ye YP. et al. microRNA-224 promotes cell proliferation and tumor growth in human colorectal cancer by repressing PHLPP1 and PHLPP2. Clin Cancer Res. 2013;19:4662–72. doi: 10.1158/1078-0432.CCR-13-0244. [DOI] [PubMed] [Google Scholar]
- 19.Li J, Du L, Yang Y, Wang C, Liu H, Wang L. et al. MiR-429 is an independent prognostic factor in colorectal cancer and exerts its anti-apoptotic function by targeting SOX2. Cancer Lett. 2013;329:84–90. doi: 10.1016/j.canlet.2012.10.019. [DOI] [PubMed] [Google Scholar]
- 20.Guo C, Sah JF, Beard L, Willson JK, Markowitz SD, Guda K. The noncoding RNA, miR-126, suppresses the growth of neoplastic cells by targeting phosphatidylinositol 3-kinase signaling and is frequently lost in colon cancers. Genes Chromosomes Cancer. 2008;47:939–46. doi: 10.1002/gcc.20596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen X, Guo X, Zhang H, Xiang Y, Chen J, Yin Y. et al. Role of miR-143 targeting KRAS in colorectal tumorigenesis. Oncogene. 2009;28:1385–92. doi: 10.1038/onc.2008.474. [DOI] [PubMed] [Google Scholar]
- 22.Liu M, Lang N, Chen X, Tang Q, Liu S, Huang J. et al. miR-185 targets RhoA and Cdc42 expression and inhibits the proliferation potential of human colorectal cells. Cancer Lett. 2011;301:151–60. doi: 10.1016/j.canlet.2010.11.009. [DOI] [PubMed] [Google Scholar]
- 23.Tsang WP, Kwok TT. The miR-18a* microRNA functions as a potential tumor suppressor by targeting on K-Ras. Carcinogenesis. 2009;30:953–9. doi: 10.1093/carcin/bgp094. [DOI] [PubMed] [Google Scholar]
- 24.Sun JY, Huang Y, Li JP, Zhang X, Wang L, Meng YL. et al. MicroRNA-320a suppresses human colon cancer cell proliferation by directly targeting beta-catenin. Biochem Biophys Res Commun. 2012;420:787–92. doi: 10.1016/j.bbrc.2012.03.075. [DOI] [PubMed] [Google Scholar]
- 25.Li Y, Zhu X, Xu W, Wang D, Yan J. miR-330 regulates the proliferation of colorectal cancer cells by targeting Cdc42. Biochem Biophys Res Commun. 2013;431:560–5. doi: 10.1016/j.bbrc.2013.01.016. [DOI] [PubMed] [Google Scholar]
- 26.Amirkhah R, Schmitz U, Linnebacher M, Wolkenhauer O, Farazmand A. MicroRNA-mRNA interactions in colorectal cancer and their role in tumor progression. Genes Chromosomes Cancer. 2015;54:129–41. doi: 10.1002/gcc.22231. [DOI] [PubMed] [Google Scholar]
- 27.Valeri N, Gasparini P, Braconi C, Paone A, Lovat F, Fabbri M. et al. MicroRNA-21 induces resistance to 5-fluorouracil by down-regulating human DNA MutS homolog 2 (hMSH2) Proc Natl Acad Sci U S A. 2010;107:21098–103. doi: 10.1073/pnas.1015541107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.To KK. MicroRNA: a prognostic biomarker and a possible druggable target for circumventing multidrug resistance in cancer chemotherapy. J Biomed Sci. 2013;20:99. doi: 10.1186/1423-0127-20-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nie J, Liu L, Zheng W, Chen L, Wu X, Xu Y. et al. microRNA-365, down-regulated in colon cancer, inhibits cell cycle progression and promotes apoptosis of colon cancer cells by probably targeting Cyclin D1 and Bcl-2. Carcinogenesis. 2012;33:220–5. doi: 10.1093/carcin/bgr245. [DOI] [PubMed] [Google Scholar]
- 30.Pangon L, Mladenova D, Watkins L, Van Kralingen C, Currey N, Al-Sohaily S. et al. MCC inhibits beta-catenin transcriptional activity by sequestering DBC1 in the cytoplasm. Int J Cancer. 2015;136:55–64. doi: 10.1002/ijc.28967. [DOI] [PubMed] [Google Scholar]
- 31.Murakami T, Mitomi H, Saito T, Takahashi M, Sakamoto N, Fukui N. et al. Distinct WNT/beta-catenin signaling activation in the serrated neoplasia pathway and the adenoma-carcinoma sequence of the colorectum. Mod Pathol. 2015;28:146–58. doi: 10.1038/modpathol.2014.41. [DOI] [PubMed] [Google Scholar]
- 32.Zhang GJ, Zhou H, Xiao HX, Li Y, Zhou T. Up-regulation of miR-224 promotes cancer cell proliferation and invasion and predicts relapse of colorectal cancer. Cancer Cell Int. 2013;13:104. doi: 10.1186/1475-2867-13-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu L, Nie J, Chen L, Dong G, Du X, Wu X. et al. The oncogenic role of microRNA-130a/301a/454 in human colorectal cancer via targeting Smad4 expression. PLoS One. 2013;8:e55532. doi: 10.1371/journal.pone.0055532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang B, Halder SK, Kashikar ND, Cho YJ, Datta A, Gorden DL. et al. Antimetastatic role of Smad4 signaling in colorectal cancer. Gastroenterology. 2010;138:969–80. doi: 10.1053/j.gastro.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alazzouzi H, Alhopuro P, Salovaara R, Sammalkorpi H, Jarvinen H, Mecklin JP. et al. SMAD4 as a prognostic marker in colorectal cancer. Clin Cancer Res. 2005;11:2606–11. doi: 10.1158/1078-0432.CCR-04-1458. [DOI] [PubMed] [Google Scholar]