Summary
Onset of neurodegenerative disorders, including Huntington’s disease, is strongly influenced by aging. Hallmarks of aged cells include compromised nuclear envelope integrity, impaired nucleocytoplasmic transport, and accumulation of DNA double-strand breaks. We show that mutant huntingtin markedly accelerates all of these cellular phenotypes in a dose- and age-dependent manner in cortex and striatum of mice. Huntingtin-linked polyglutamine initially accumulates in nuclei, leading to disruption of nuclear envelope architecture, partial sequestration of factors essential for nucleocytoplasmic transport (Gle1 and RanGAP1), and intranuclear accumulation of mRNA. In aged mice, accumulation of RanGAP1 together with polyglutamine is shifted to perinuclear and cytoplasmic areas. Consistent with findings in mice, marked alterations in nuclear envelope morphology, abnormal localization of RanGAP1, and nuclear accumulation of mRNA were found in cortex of Huntington’s disease patients. Overall, our findings identify polyglutamine-dependent inhibition of nucleocytoplasmic transport and alteration of nuclear integrity as a central component of Huntington’s disease.
Introduction
Expansion of unstable repetitive sequences triggers various neurodegenerative diseases, including Huntington’s disease (HD), myotonic dystrophy, spinocerebellar ataxias, and instances of frontotemporal dementia (FTD) and/or amyotrophic lateral sclerosis (ALS) from hexanucleotide expansion in the C9ORF72 gene (Dejesus-Hernandez et al., 2011, Renton et al., 2011). Each of these diseases is characterized by abnormal nuclear and/or cytoplasmic aggregation of proteins.
Dynamic exchange of proteins and RNAs between nuclear and cytoplasmic compartments is essential for cellular functioning but is gradually impaired in the course of normal aging (Yannarell et al., 1977, Fifkova et al., 1987, Pujol et al., 2002, Scaffidi and Misteli, 2006, D’angelo et al., 2009). Long-living postmitotic cells, especially neurons, are particularly susceptible to such age-related damage. Indeed, impaired nucleocytoplasmic transport and nuclear integrity have been identified as hallmarks of aged neurons (Fifkova et al., 1987, Mertens et al., 2015). A flurry of recent reports has indicated that nucleocytoplasmic compartmentalization is also markedly impaired in several models of repeat expansion disorders (Chapple et al., 2008, Mapelli et al., 2012, Freibaum et al., 2015, Jovicic et al., 2015, Kaneb et al., 2015, Liu et al., 2015, Rodriguez et al., 2015, Zhang et al., 2015, Da Cruz and Cleveland, 2016, Lee et al., 2016, Nekrasov et al., 2016, Woerner et al., 2016, Zhang et al., 2016, Shi et al., 2017). Furthermore, apparent disruption of the nuclear envelope or disrupted nucleocytoplasmic transport has been reported in postmortem tissues from ALS (Kinoshita et al., 2009), Parkinson’s (Liu et al., 2012) and Alzheimer’s disease patients (Sheffield et al., 2006), and in cells expressing aggregation-prone proteins (Frost et al., 2016, Woerner et al., 2016).
To address the possible convergence of disease- and aging-related mechanisms in mediating nuclear dysfunction in the nervous system, we now focus on several mouse models, induced pluripotent stem cells (iPSC)-derived neuronal progenitors and autopsy samples from HD patients. In all of these, we identify a crucial role for mutant huntingtin in accelerating hallmarks of cellular aging by progressive, dose-dependent disruption of the nucleocytoplasmic import/export machinery associated with compromised nuclear architecture and genome instability.
Results
Dose and age-dependent disruption of the nuclear envelope in brains of HD mice and Huntington’s disease patients
We investigated nuclear envelope alterations in mice expressing physiological levels of a ~175 CAG trinucleotide repeat expansion within one (HttQ7/Q175) or both (HttQ175/Q175) endogenous huntingtin (Htt) alleles (Heikkinen et al., 2012, Menalled et al., 2012, Smith et al., 2014, Carty et al., 2015). We first tested whether wild-type mice with 7 glutamines (Q) in both endogenous Htt alleles (HttQ7/Q7) develop age-dependent nuclear envelope defects in cortex and striatum, two HD-vulnerable brain regions in which >80% of the cells are neurons detectable with the neuron-specific marker Neuronal Nuclear Antigen (NeuN) (Gusel’nikova and Korzhevskiy, 2015) (Figure S1A). For this, we followed lamin B1, a structural component of the nuclear lamina, as a marker of the nuclear envelope shape and an initial test of nuclear integrity. Consistent with aberrant nuclear architecture being a cellular hallmark of aging (Scaffidi and Misteli, 2006, Lopez-Otin et al., 2013), the number of abnormal nuclei increased two-fold in brains of 24 month-old compared to 6 month-old HttQ7/Q7 mice (from 21% to 39% in cortex and from 22% to 37% in striatum) (Figures 1A,C, S1B and S2A,B).
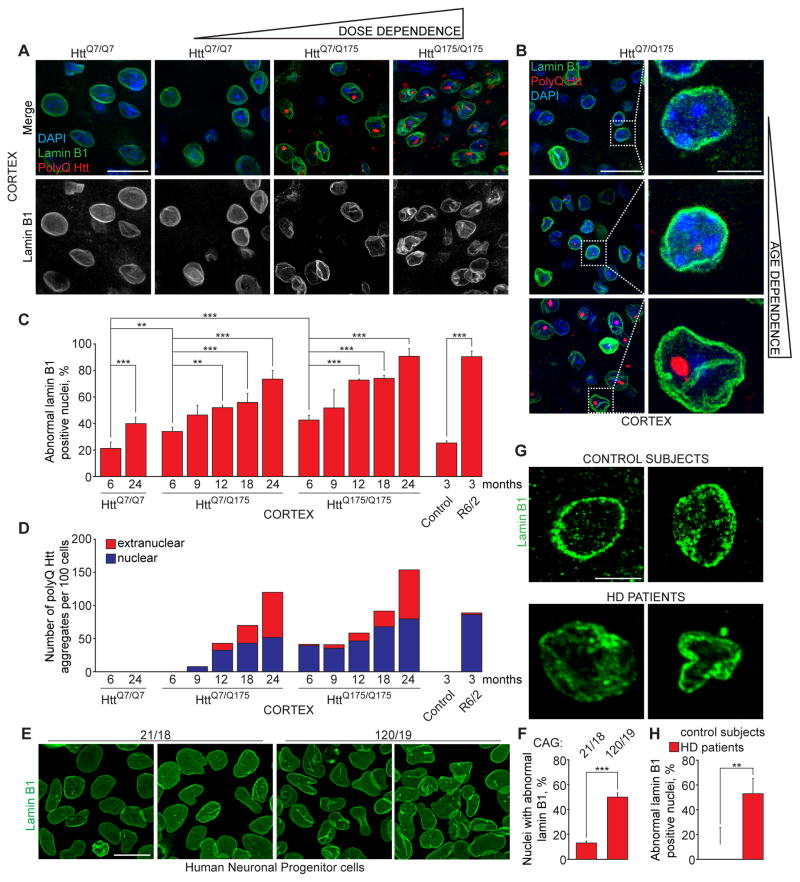
Figure 1. Expanded polyQ Huntingtin disrupts neuronal nuclear envelope morphology in a dose- and age-dependent manner.
(A) Immunofluorescence (IF) of lamin B1 (green) and polyQ Htt (red) in cortex of mice at indicated ages expressing one (HttQ7/Q175) or two (HttQ175/Q175) copies of expanded polyQ Htt and wild type littermates (HttQ7/Q7). (B) IF of lamin B1 (green) and polyQ Htt (red) in cortex of aging HttQ7/Q175 mice. A, B: Nuclei were stained with DAPI. (C) Percentage of altered nuclear envelopes in cortical sections of mice of indicated genotypes and ages, as determined by IF of lamin B1. 100–400 nuclei were counted per genotype and age. (D) Quantification of nuclear and extranuclear aggregates of polyQ Htt in cortex of HttQ175 and R6/2 mice. 200 cells were counted per genotype and age. (E, F) IF of lamin B1 (E) and percentage of altered nuclear envelopes (F) in human iPSC-derived neuronal progenitor cells with indicated number of CAG repeats. At least 600 cells were analyzed for each genotype. (G, H) IF of lamin B1 (G) and percentage of altered nuclear envelopes (H) in sections of motor cortex of two non-neurological disease control subjects and two Huntington’s disease patients of indicated ages. At least 200 nuclei were counted per each group. A, B, G: Z projections (3 μM) are shown. C, F, H: Data are shown as mean ± SEM. **: P<0.01, ***: P<0.001, chi-square test.
See also Figures S1 and S2.
HttQ175 mice closely mimic the human genetic context, express mutant huntingtin containing expanded polyglutamine (polyQ Htt), and gradually develop behavioral and pathological abnormalities similar to those observed in HD patients (Heikkinen et al., 2012, Menalled et al., 2012, Smith et al., 2014, Carty et al., 2015). Strikingly, expression of HttQ175 from both alleles markedly increased the number of cells with altered nuclear envelopes to 89% in cortex and 62% in striatum of 24 month-old HttQ175/Q175 mice (Figures 1A, C and S2A,B). Nuclear alterations triggered by expanded polyQ accumulation were dose dependent: by 24 months of age, cortex of heterozygous HttQ7/Q175 and homozygous HttQ175/Q175 mice contained 72% and 89% of misshapen nuclei, respectively (Figures 1A, C). Dose dependency was also evident in striatum with 38% and 62% aberrant nuclei in 24 month-old heterozygous HttQ175/Q7 and homozygous HttQ175/Q175 mice, respectively (Figures S2A,B).
Consistent with the polyQ Htt exacerbating aging-associated nuclear envelope alterations, longitudinal analyses of 6 to 24 month-old heterozygous HttQ7/Q175 and homozygous HttQ175/Q175 mice revealed gradual, age-dependent impairment of nuclear shape (Figures 1B,C and S2C). To precisely define the nuclear limits and determine the nuclear or extranuclear localization of polyQ Htt aggregates we used lamin B1 staining and DAPI. Intranuclear and extranuclear polyQ Htt aggregates increased in number and size with age in both heterozygous HttQ7/Q175 and homozygous HttQ175/Q175 mice, being the most abundant by 24 month-old (Figures 1A–D and S2A,C). Cytoplasmic aggregates were apparent only after 9 months of age and increasing 3-fold from 18 to 24 months of age in cortex of heterozygous and homozygous HttQ175 mice (Figure 1D). Both heterozygous and homozygous HttQ175 mice demonstrated age-related changes in shape of intranuclear aggregates. While nuclei of young mice contained mostly round-shaped aggregates, elongated egg-shaped aggregates were often detected in older mice (Figures 1A,B and S2C). Disruption of nuclear shape was already evident in 6 and 9 month-old heterozygous HttQ7/Q175 mice prior to appearance of extranuclear aggregates, with 33% and 46% of cortical cells demonstrating altered nuclear envelopes while only rare nuclear polyQ Htt aggregates are detected in cortex at this stage (Figures 1B–D). Consistently, cortex of 6 and 9 month-old homozygous HttQ175/Q175 mice contained 42% and 52% of cells with altered nuclear envelopes and predominantly nuclear polyQ Htt aggregates (Figures 1C–D, S2C).
In an additional HD model, the transgenic R6/2 mouse line (Mangiarini et al., 1996), severe alterations in nuclear lamina assembly were also present in 89% of cortical cells at much younger ages (3 months) (Figures 1C, S2D). R6/2 mice express a 23 kD human exon 1 fragment of huntingtin with a 120–125 polyQ expansion and develop severe, rapid onset of disease (Mangiarini et al., 1996). By 3 months of age nearly 100% of cortical cells with altered nuclear envelopes had nuclear polyQ aggregates (Figures 1C,D and S2D).
To test if the polyQ Htt-dependent changes in nuclear envelope morphology seen in mice reflected what occurs in human disease, we evaluated the nuclear envelope shape in iPSC-derived neuronal progenitors from HD and control individuals (Table S1). We observed that invaginations of the nuclear envelope were present in 50% of HD cells, compared to 13% in control cells, indicating that HD neuronal precursor cells are predisposed to nuclear defects (Figures 1E,F). We then confirmed the relevance of this cellular phenotype in human disease by analyzing the nuclear envelope shape within the motor cortex of 2 HD patients and 2 control individuals (Table S2). Strikingly, 55% of lamin B1-positive nuclei demonstrated misshapen nuclear envelopes as compared to 16 % in control individuals (Figure 1G,H).
Collectively, these data strongly indicate that expression of expanded polyQ-containing Htt exacerbates aging-associated alterations of the nuclear envelope architecture in cortex and striatum regions.
Expanded polyQ Huntingtin impairs nuclear integrity, inducing DNA damage
We next tested whether observed alterations in lamin assembly and nuclear envelope shape were associated with aging- and polyQ Htt-mediated nuclear envelope dysfunction. Envelope integrity is critically important for maintenance of genome stability and efficient DNA repair (Bukata et al., 2013). DNA double-strand breaks (DSBs) accumulate in aging tissues (Mandavilli and Rao, 1996a, Mandavilli and Rao, 1996b, Wang et al., 2009) and DNA fragmentation has been implicated in neurological disorders, including HD (Dragunow et al., 1995, Enokido et al., 2010, Lu et al., 2014). Sites of DSBs are marked by phosphorylated histone H2AX, designated as γH2AX (Mandavilli and Rao, 1996b, Rogakou et al., 1998). Nuclear γH2AX foci accumulated with age in cortex of wild type HttQ7/Q7 mice (Figures S3A, B) with 13% and 54% of cells containing γH2AX foci at 6- and 24 months of age, respectively. Expanded Htt expression led to increase in DSBs in a dose-dependent manner in 24 month-old HttQ175 mice relative to HttQ7/Q7 mice, with γH2AX foci observed in 65% and 80% of cells from heterozygous HttQ7/Q175 and homozygous HttQ175/Q175 mice, respectively (Figures S3A, B). Consistently, polyQ Htt expression increased the size of the γH2AX nuclear foci 3-fold in 24 month-old HttQ175/Q175 mice as compared to 24 month-old HttQ7/Q7 mice (Figure S3C). Additionally, DSBs accumulated in 73% of nuclei and increased in size in 3 month-old R6/2 mice, overall corroborating enhanced neuronal aging in polyQ Htt-expressing mice (Figure S3).
Intranuclear polyQ inhibits RNA export by sequestering RanGAP1 and Gle1
Alteration of nuclear envelopes during aging is associated with disruption of nucleocytoplasmic transport (Yannarell et al., 1977, D’angelo et al., 2009, Kelley et al., 2011). Since mutant huntingtin disrupted envelope integrity, we tested whether polyQ Htt also impaired nucleocytoplasmic transport of mRNA and proteins. We used fluorescence in situ hybridization (FISH) to identify polyadenylated (polyA) RNAs (mRNA), and measured the ratios of polyA RNA signal intensities in nucleus and cytoplasm of 24-month-old control HttQ7/Q7, heterozygous HttQ7/Q175 and homozygous HttQ175/Q175 mice. Expression of polyQ Htt was found to markedly exacerbate nuclear mRNA accumulation in a dose-dependent manner in HttQ175 mice (Figures 2A,B). In particular, 66% of cells in heterozygous HttQ7/Q175 and 93% of cells in homozygous HttQ175/Q175 displayed nuclear to cytoplasmic polyA ratios above the mean of control HttQ7/Q7 mice.
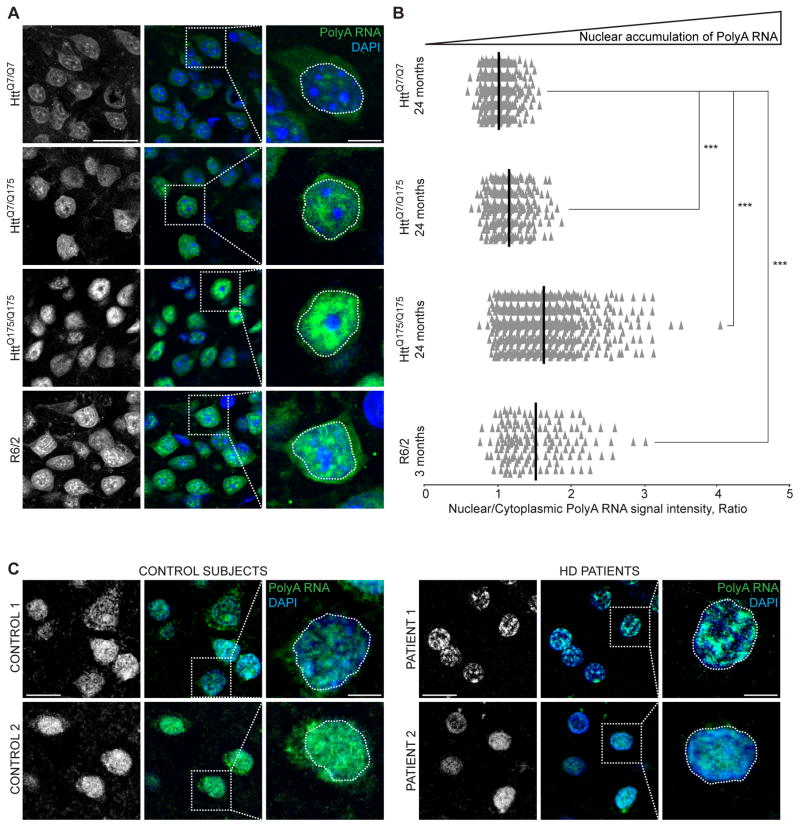
Figure 2. Expanded polyQ Huntingtin impairs mRNA export from nuclei.
(A) Fluorescence in situ hybridization (FISH) of polyA RNA (green) of cortex from mice of indicated genotypes and ages. Z projections of 30 consecutive slices (3 μM) are shown. (B) Quantification of (A). The ratio of nuclear to cytoplasmic signal intensities of PolyA RNA per cell in cortex from mice of indicated genotypes and ages normalized by 24 months HttQ7/Q7 mean. Each individual point represents a single cell. Mean is shown per each genotype and age. ***: P<0.001, unpaired t test, comparison with control 24 months HttQ7/Q7. 300–600 cells were counted per genotype and age. (C) FISH of polyA RNA (green) in sections of motor cortex of non-neurological disease control subjects and Huntington’s disease patients. Z projections of 30 consecutive slices (3 μM) are shown. A, C: nuclei were stained with DAPI.
See also Figure S3.
Compromised nuclear RNA export was evident in second HD model mice, as mRNA accumulated in nuclei of cortical cells of 3-month old R6/2 mice (Figures 2A,B). Despite a recent proposal that cytoplasmic-only polyQ aggregates drive nuclear export deficits in R6/2 mice (Woerner et al., 2016), nuclear mRNA accumulation strongly correlated with the presence of nuclear polyQ, as >99% of cells in the cortex of 3 month old R6/2 mice had intranuclear polyQ aggregates (in agreement with prior reports (Davies et al., 1997, Li et al., 2001)) (Figures 1D, S2D). To further address perturbed mRNA export in HD, we analyzed polyA RNA localization in motor cortex of Huntington’s disease patients (Table S2). Consistent with mouse models, the number of cells with marked nuclear mRNA accumulation increased two-fold in the cortex of Huntington’s disease patients as compared to control subjects (Figure 2C, Figure S3D).
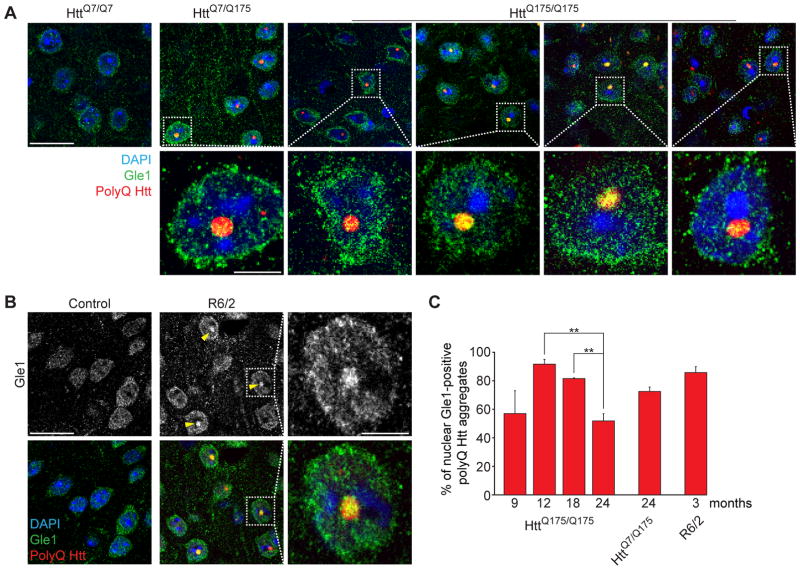
Moreover, in cortical regions of heterozygous HttQ7/Q175 and homozygous HttQ175/Q175 mice, nuclear expanded huntingtin co-aggregated with Gle1 (Figure 3A), a key component for mRNA export (Watkins et al., 1998) and with Ran GTPase-activating protein 1 (RanGAP1) (Figure 4A), a protein required for Ran-dependent nuclear import and export (Bischoff et al., 1995). Accordingly, nuclear Gle1/polyQ and RanGAP1/polyQ co-localizations were also observed in cortical regions of 3 month-old R6/2 mice (Figures 3B and 4A). Gle1 and Htt nuclear co-localization in HttQ175/Q175 mice was most evident at 12 and 18 months, and decreased at 24 months of age (Figures 3A,C). The decrease in nuclear Gle1 in 24 month-old mice is likely to be caused by reduced nuclear import of Gle1 due to chronic de-regulation of nuclear import machinery over the course of disease progression. Consistent with progressive impairment of nuclear import, RanGAP1/polyQ Htt co-localization within nuclei also peaked (in number and intensity) at 6 months of age and then decreased with age (Figure S4B, Movie S1), with the co-localization between RanGAP1 and polyQ Htt becoming perinuclear (Figures 4B,C and S4A,C and Movie S2). Prominent perinuclear RanGAP1/polyQ Htt co-localization was identified using Z-projections of 3 μM-thick optical stacks in cortex and striatum of older heterozygous HttQ7/Q175 and homozygous HttQ175/Q175 mice (Figures 4B–D and S4A,C). Consistently, in aged mice, intranuclear polyQ Htt inclusions were negative for RanGAP1 in cells with perinuclear RanGAP1/polyQ Htt aggregates (Figure 4B,C and Movie S2).
Figure 3. Nuclear polyQ Htt aggregates co-localize with the mRNA export factor Gle1.
(A, B) Immunofluorescence of polyQ Htt (red) and Gle1 (green) in cortical sections of (A) HttQ7/Q7, HttQ7/Q175 and HttQ175/Q175 mice of indicated ages and (B) 3-month-old control and R6/2 mice. Nuclei were stained with DAPI. (C) Percentage of nuclear polyQ aggregates positive for Gle1, in mice of indicated genotypes and ages. Data are shown as mean ± SEM. **: P<0.01, chi-square test. 100 cells were counted per genotype and age. A, B: Arrowheads point to Gle1/polyQ Htt nuclear co-localization.
See also Figure S4.
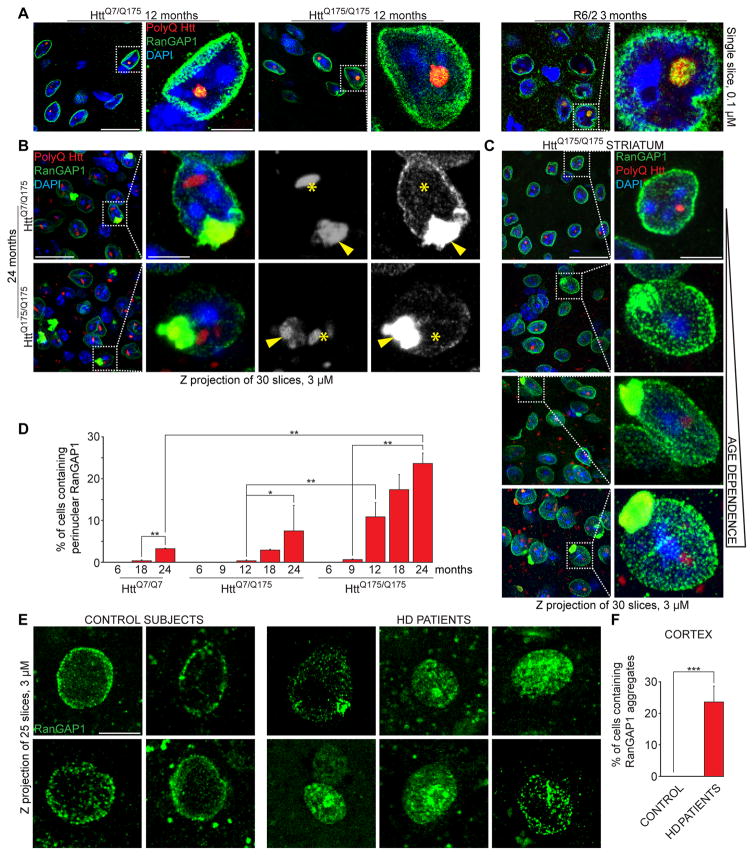
Figure 4. Expanded polyQ Huntingtin sequesters RanGAP1.
(A) Immunofluorescence (IF) of RanGAP1 (green) and polyQ Htt (red) in cortical sections of mice of indicated genotypes and ages. Arrowheads point to RanGap1/polyQ Htt nuclear co-localization. Single slices (0.1 μM) are shown. (B) IF of RanGAP1 (green) and polyQ Htt (red) in cortex of 24-month old mice of indicated genotypes. Localization of nuclear (asterisks) and perinuclear (arrowheads) polyQ Htt aggregates is shown. (C) IF of RanGAP1 in striatal sections of HttQ175/Q175 mice of indicated ages. (D) Percentage of cells containing perinuclear RanGAP1 in cortical sections of mice of indicated genotypes and ages. At least 600 cells per animal were analyzed. (E, F) IF of RanGAP1 (E) and percentage of cells containing mislocalized RanGAP1 (F) in sections of motor cortex of non-neurological disease control subjects and Huntington’s disease patients. At least 200 nuclei were counted per each group. B, C, E: Z projections (3 μM) are shown. D, F: Data are shown as mean ± SEM. *: P<0.05, **: P<0.01, ***: P<0.001, chi-square test.
See also Figure S4.
Accumulation of RanGAP1 and polyQ Htt in perinuclear area was age- and dose-dependent (Figures 4C,D and S4A), appearing initially in a few cells at 12 months of age in heterozygous HttQ7/Q175 mice and at 9 months of age in homozygous HttQ175/Q175. At later ages, both in cortex and striatum, the number of cells with aberrant perinuclear RanGAP1 increased, as did the size of the RanGAP1 aggregates in HttQQ7/Q175 and HttQ175/Q175 mice (Figures 4B–D and S4A,C). In addition, RanGAP1 (and more rarely Gle1) was observed inside cytoplasmic ring-shaped polyQ Htt aggregates at later ages in HttQ175 mice (Figures S4D,E). A gradual accumulation of RanGAP1, Gle1 and polyQ Htt in the cytoplasm – and/or reduced nuclear co-localization – further corroborated compromised nucleocytoplasmic transport of proteins in cells expressing polyQ Htt. Moreover, aberrant perinuclear accumulation of RanGAP1, strikingly similar to that observed in polyQ Htt-expressing mice, accompanied physiological neuronal aging as it was found in 2% of cells in the cortex of 24 month-old HttQ7/Q7 mice, but not in 6 month-old HttQ7/Q7 (Figure S4F).
Furthermore, examination of iPSC-derived neuronal precursor cells expressing mutant huntingtin revealed 40% of cells with altered RanGAP1 distribution characterized by nuclear envelope invaginations with focal increased staining intensity, as compared to 13% in control cells (Figure S4G,H and Table S1). Finally, human brain autopsies (Table S2) of control individuals revealed a primarily perinuclear RanGAP1 signal (Figures 4E,F), even at advanced ages (e.g., 92 or 94 years old). In contrast, RanGap1 in HD patient samples was heterogeneously distributed into perinuclear clusters in 25% of cells examined in motor cortex (Figures 4E,F), strongly supporting perturbed nucleocytoplasmic transport as a consequence of polyQ Htt expression and a new pathological hallmark in HD.
Discussion
Our findings provide in vivo support for repeat expansion-mediated acceleration of neuronal aging with altered nuclear integrity and nucleocytoplasmic transport in HD. Indeed, polyQ Htt dramatically exacerbates nuclear dysfunction in a dose- and age-related manner, including alterations in the shape of the nuclear envelope, accumulation of DNA double strand breaks and compromised nucleocytoplasmic transport of mRNA and proteins. Specifically, polyQ Htt initially forms intranuclear aggregates that sequester major regulators of nucleocytoplasmic transport Gle1 and RanGAP1, providing a likely explanation for polyQ Htt-mediated disruption of nucleocytoplasmic transport. The initial defects in nucleocytoplasmic transport are accompanied by extranuclear accumulation of co-aggregating polyQ Htt and RanGAP1. Perinuclear accumulation of RanGAP1 also occurs in a small fraction of cortical and striatal cells in normal aging of wild-type animals, representing a new hallmark of cellular aging associated with neurodegeneration. Notably, alterations of the nuclear envelope were already present in iPSC-derived neuronal progenitors from HD patients, and mutant Htt-dependent exacerbation of RanGAP1 mislocalization was confirmed in motor cortex of HD patients, demonstrating perturbed nucleocytoplasmic compartmentalization as a major component of HD.
Altogether, our data suggest common mechanisms underlying neuronal aging and pathogenesis. Indeed, several common hallmarks of aging, including mitochondria dysfunction, impaired protein homeostasis and degradation have previously been are associated with HD (Jenkins et al., 1993, Hay et al., 2004, Lin and Beal, 2006, Bennett et al., 2007). In addition, consistent with mutant Huntingtin accelerating cellular aging, a recent study reported an accumulation of epigenetic markers of aging in the brain of HD patients as compared to control subjects (Horvath et al., 2016). In line with this notion, several genetic, pharmacological and dietary anti-aging interventions have been reported to ameliorate aspects of HD in mouse models (Duan et al., 2003, Ma et al., 2007, Sadagurski et al., 2011, Tallaksen-Greene et al., 2014, Moreno et al., 2016).
PolyQ Htt-dependent accumulation of nuclear mRNA in HD patients and HD mouse models mice shown in this study adds to recent identification of similar mRNA export deficiency in cultured neurons following overexpression and aggregation of artificial β-sheet proteins, TDP-43, or Htt fragments (Woerner et al., 2016) and in iPS-derived neurons from ALS/FTD patients with C9ORF72 hexanucleotide expansion (Freibaum et al., 2015). Expanded RNA repeats and/or the polypeptides produced by translation of those repeats have recently been reported to interact with proteins that control nucleocytoplasmic compartmentalization, including RanGAP1 (Freibaum et al., 2015, Jovicic et al., 2015, Zhang et al., 2015, Lee et al., 2016, Woerner et al., 2016, Zhang et al., 2016, Shi et al., 2017). In addition, the crucial mRNA export factor Gle1 (Watkins et al., 1998) can affect toxicity in flies expressing C9ORF72 hexanucleotide-repeats (Freibaum et al., 2015), is mutated in rare cases of ALS (Kaneb et al., 2015), and has been associated with severity in a congenital motor neuron disease (Coyne et al., 2015). Hence, accumulating evidence supports an emerging disease mechanism, in which repeat expansion and cellular aging progressively impair nucleocytoplasmic compartmentalization.
STAR Methods
TABLE OF RESOURCES (Attached)
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Huntingtin Protein, clone mEM48 | Millipore | Cat# MAB5374, RRID:AB_10055116 |
| Lamin B1 (S-20) | Santa Cruz Biotechnology | Cat# sc-30264, RRID:AB_2136305 |
| Lamin B1 | Proteintech | Cat# 12987-1-AP, RRID:AB_2136290 |
| RanGAP1 (H-180) | Santa Cruz Biotechnology | Cat# sc-25630, RRID:AB_2176978 |
| GLE1 | Abcam | Cat# ab96007, RRID:AB_10678755 |
| Anti-phospho-Histone H2A.X (Ser139), clone JBW301 | Millipore | Cat# 05-636, RRID:AB_309864 |
| Biological Samples | ||
| Human post-mortem tissues | UCSD_HD Clinical Research Center | Supplementary Table 2 |
| Human post-mortem tissues | Massachusetts Alzheimer’s Disease Research Center | Supplementary Table 2 |
| Critical Commercial Assays | ||
| Sudan Black B | Fisher | BP-109-10 |
| Formamide, saline-sodium citrate | VWR International LLC | IB72020 |
| Experimental Models: Cell Lines | ||
| iPS | Coriell/Cedars Sinai | Supplementary Table 1 |
| CS001CTR21-n1 | Cedars Sinai | https://www.cedars-sinai.edu/Research/Research-Cores/Induced-Pluripotent-Stem-Cell-Core-/Stem-Cell-Lines.aspx#Control%20iPSC%20lines |
| CS09iHD109-n4 | Cedars Sinai | https://www.cedars-sinai.edu/Research/Research-Cores/Induced-Pluripotent-Stem-Cell-Core-/Stem-Cell-Lines.aspx#Huntingtons%20Disease%20(HD) |
| Experimental Models: Organisms/Strains | ||
| Mice | ||
| R6/2 (B6CBA-Tg(HDexon1)62Gpb/3J) | Jackson Laboratory | Stock # 006494 |
| F1 (B6CBAf1/J) | Jackson Laboratory | Stock # 100011 |
| Q175 Knock-in | Jackson Laboratory | Stock # 370437 |
| C57BL/6 (C57BL/6 NCrL) | Jackson Laboratory | Stock # 027 |
| Oligonucleotides | ||
| PolyT probe | IDT | 5′-/Cy3-TTT TTT TTT TTT TTT TTTTT-3′ |
| Software and Algorithms | ||
| Nikon EZ-C2 | Nikon | https://www.nikoninstruments.com/Products/Confocal-Microscopes/C2-Plus |
| Fiji | NIH | https://fiji.sc/#download |
| Acapella | Perkin Elmer | http://www.perkinelmer.com/content/relatedmaterials/brochures/bro_acapellasoftware |
| GraphPad Prism | GraphPad software | https://www.graphpad.com |
| IN Cell Analyzer 6000 Cell Imaging System | GE | http://www.gelifesciences.com/webapp/wcs/stores/servlet/productById/en/GELifeSciences-us/29043323 |
| Other | ||
TABLE WITH EXAMPLES FOR AUTHOR REFERENCE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-Snail | Cell Signaling Technology | Cat#3879S; RRID: AB_2255011 |
| Mouse monoclonal anti-Tubulin (clone DM1A) | Sigma-Aldrich | Cat#T9026; RRID: AB_477593 |
| Rabbit polyclonal anti-BMAL1 | This paper | N/A |
| Bacterial and Virus Strains | ||
| pAAV-hSyn-DIO-hM3D(Gq)-mCherry | Krashes et al., 2011 | Addgene AAV5; 44361-AAV5 |
| AAV5-EF1a-DIO-hChR2(H134R)-EYFP | Hope Center Viral Vectors Core | N/A |
| Cowpox virus Brighton Red | BEI Resources | NR-88 |
| Zika-SMGC-1, GENBANK: KX266255 | Isolated from patient (Wang et al., 2016) | N/A |
| Staphylococcus aureus | ATCC | ATCC 29213 |
| Streptococcus pyogenes: M1 serotype strain: strain SF370; M1 GAS | ATCC | ATCC 700294 |
| Biological Samples | ||
| Healthy adult BA9 brain tissue | University of Maryland Brain & Tissue Bank; http://medschool.umaryland.edu/btbank/ | Cat#UMB1455 |
| Human hippocampal brain blocks | New York Brain Bank | http://nybb.hs.columbia.edu/ |
| Patient-derived xenografts (PDX) | Children’s Oncology Group Cell Culture and Xenograft Repository | http://cogcell.org/ |
| Chemicals, Peptides, and Recombinant Proteins | ||
| MK-2206 AKT inhibitor | Selleck Chemicals | S1078; CAS: 1032350-13-2 |
| SB-505124 | Sigma-Aldrich | S4696; CAS: 694433-59-5 (free base) |
| Picrotoxin | Sigma-Aldrich | P1675; CAS: 124-87-8 |
| Human TGF-β | R&D | 240-B; GenPept: P01137 |
| Activated S6K1 | Millipore | Cat#14-486 |
| GST-BMAL1 | Novus | Cat#H00000406-P01 |
| Critical Commercial Assays | ||
| EasyTag EXPRESS 35S Protein Labeling Kit | Perkin-Elmer | NEG772014MC |
| CaspaseGlo 3/7 | Promega | G8090 |
| TruSeq ChIP Sample Prep Kit | Illumina | IP-202-1012 |
| Deposited Data | ||
| Raw and analyzed data | This paper | GEO: GSE63473 |
| B-RAF RBD (apo) structure | This paper | PDB: 5J17 |
| Human reference genome NCBI build 37, GRCh37 | Genome Reference Consortium | http://www.ncbi.nlm.nih.gov/projects/genome/assembly/grc/human/ |
| Nanog STILT inference | This paper; Mendeley Data | http://dx.doi.org/10.17632/wx6s4mj7s8.2 |
| Affinity-based mass spectrometry performed with 57 genes | This paper; and Mendeley Data | Table S8; http://dx.doi.org/10.17632/5hvpvspw82.1 |
| Experimental Models: Cell Lines | ||
| Hamster: CHO cells | ATCC | CRL-11268 |
| D. melanogaster: Cell line S2: S2-DRSC | Laboratory of Norbert Perrimon | FlyBase: FBtc0000181 |
| Human: Passage 40 H9 ES cells | MSKCC stem cell core facility | N/A |
| Human: HUES 8 hESC line (NIH approval number NIHhESC-09-0021) | HSCI iPS Core | hES Cell Line: HUES-8 |
| Experimental Models: Organisms/Strains | ||
| C. elegans: Strain BC4011: srl-1(s2500) II; dpy-18(e364) III; unc-46(e177)rol-3(s1040) V. | Caenorhabditis Genetics Center | WB Strain: BC4011; WormBase: WBVar00241916 |
| D. melanogaster: RNAi of Sxl: y[1] sc[*] v[1]; P{TRiP.HMS00609}attP2 | Bloomington Drosophila Stock Center | BDSC:34393; FlyBase: FBtp0064874 |
| S. cerevisiae: Strain background: W303 | ATCC | ATTC: 208353 |
| Mouse: R6/2: B6CBA-Tg(HDexon1)62Gpb/3J | The Jackson Laboratory | JAX: 006494 |
| Mouse: OXTRfl/fl: B6.129(SJL)-Oxtrtm1.1Wsy/J | The Jackson Laboratory | RRID: IMSR_JAX:008471 |
| Zebrafish: Tg(Shha:GFP)t10: t10Tg | Neumann and Nuesslein-Volhard, 2000 | ZFIN: ZDB-GENO-060207-1 |
| Arabidopsis: 35S::PIF4-YFP, BZR1-CFP | Wang et al., 2012 | N/A |
| Arabidopsis: JYB1021.2: pS24(AT5G58010)::cS24:GFP(-G):NOS #1 | NASC | NASC ID: N70450 |
| Oligonucleotides | ||
| siRNA targeting sequence: PIP5K I alpha #1: ACACAGUACUCAGUUGAUA | This paper | N/A |
| Primers for XX, see Table SX | This paper | N/A |
| Primer: GFP/YFP/CFP Forward: GCACGACTTCTTCAAGTCCGCCATGCC | This paper | N/A |
| Morpholino: MO-pax2a GGTCTGCTTTGCAGTGAATATCCAT | Gene Tools | ZFIN: ZDB-MRPHLNO-061106-5 |
| ACTB (hs01060665_g1) | Life Technologies | Cat#4331182 |
| RNA sequence: hnRNPA1_ligand: UAGGGACUUAGGGUUCUCUCUAGGGACUUAGGGUUCUCUCUAGGGA | This paper | N/A |
| Recombinant DNA | ||
| pLVX-Tight-Puro (TetOn) | Clonetech | Cat#632162 |
| Plasmid: GFP-Nito | This paper | N/A |
| cDNA GH111110 | Drosophila Genomics Resource Center | DGRC:5666; FlyBase:FBcl0130415 |
| AAV2/1-hsyn-GCaMP6-WPRE | Chen et al., 2013 | N/A |
| Mouse raptor: pLKO mouse shRNA 1 raptor | Thoreen et al., 2009 | Addgene Plasmid #21339 |
| Software and Algorithms | ||
| Bowtie2 | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| Samtools | Li et al., 2009 | http://samtools.sourceforge.net/ |
| Weighted Maximal Information Component Analysis v0.9 | Rau et al., 2013 | https://github.com/ChristophRau/wMICA |
| ICS algorithm | This paper; Mendeley Data | http://dx.doi.org/10.17632/5hvpvspw82.1 |
| Other | ||
| Sequence data, analyses, and resources related to the ultra-deep sequencing of the AML31 tumor, relapse, and matched normal. | This paper | http://aml31.genome.wustl.edu |
| Resource website for the AML31 publication | This paper | https://github.com/chrisamiller/aml31SuppSite |
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contacts Don Cleveland (dcleveland@ucsd.edu)
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
R6/2 and HttQ175
HttQ175 mice were described earlier (Menalled et al., 2012) and were provided by CHDI foundation (CHDI-81003003) and maintained on C57BL/6 background. Males and females heterozygous for the expanded allele were bred together to obtain a colony of wild-type, heterozygous and homozygous animals; CAG repeats were maintained between 160–190.
R6/2 mice (Mangiarini et al., 1996) were obtained from Jackson laboratories and maintained by crossing transgene positive males with F1 (CBA x C57BL/6) females; CAG repeats were maintained between 120–145.
For both strains, males were perfused in a 20% paraformaldehyde solution and the tissue collected for imaging. Maintenance and experimental procedures were approved by the Institutional Animal Care and Use Committee of the University of California, San Diego.
Human
Human post-mortem tissues
Information of human samples is provided in Table S2.
Cell lines
Human Neuronal Progenitors
Human induced pluripotent stem cells (Consortium, 2012) were cultured on Matrigel coated (BD Biosciences) 6-well plates in mTeSR 1 media (STEMCELL Technologies) in a 37°C incubator at 5% CO2 with fresh-media changes (2.5mL/well) every day. To obtain human neuronal progenitor cell (NPC) populations, human iPS lines were differentiated following standardized STEMdiff™ NIM method (STEMCELL Technologies) following manufacturer’s instructions. In short, single cell suspensions of human iPS cells were prepared by harvesting individual iPS colonies using ACCUTASE™ (STEMCELL Technologies) and re-suspending in STEMdiff™ neural induction media (supplemented with 10 μM Y-27632). Total cell counts and viability were determined using the Countess Automated Cell Counter (ThermoFisher Scientific).
For initiation of uniform sized embryoid body formations, 3 million cells/mL in a single cell suspension was plated for each human iPS population in independent wells of an AggreWell800 plate (STEMCELL Technologies), resulting in approximately 10,000 single cells/micro-well. The resulting neural aggregates were incubated in neural induction media for four days with daily 1mL/well media changes. Mature embryoid bodies were harvested on day 5 and plated on PLO/Laminin coated 6-well plates and cultured for additional 7 days in neural induction media. The fidelity of the neural rosette formation from each embryoid body was checked over the course of the next 7 days. On day 12, high fidelity neural rosettes were harvested through individual neural rosette selection and replated onto Matrigel coated 6-well plates for further neuronal progenitor population expansion in NPC media. The neuronal progenitor populations were maintained in NPC media.
Additional information is provided in Table S1.
METHOD DETAILS
Mice surgery and tissue preservation
Anesthetized animals were subjected to transcardial perfusion with ice-cold Sorenson’s phosphate buffer (SPB), and fixed with 4% paraformaldehyde (PFA) in phosphate buffer (PBS). Brains were removed, trimmed with coronal cuts immediately rostral to the forebrain, post-fixed in 4% PFA for 23h, cryoprotected in 30% sucrose, embedded in HistoPrep (TM) (SH75-125D, Fisher Chemical) followed by a rapid incubation in −40°C 2-Methylbutane (Fisher Scientific), and kept at −80°C. Fixed brains were cut using the Leica 2800E Frigocut cryostat at −20°C. 35 μm thick free-floating sections were preserved in PBS containing Sodium Azide (0.02%) at 4°C.
For RNA FISH experiments, the 10-μm thick sections were immediately mounted on Fisherbrand Superfrost Plus Microscope Slides (Thermo Fisher Scientific) after sectioning, and preserved in dry ice.
For an experiment shown in Figure 4A (R6/2), paraffin-embedded sections of 10 μm thickness prepared by UCSD histology core were used.
Immunofluorescence (free-floating OCT-embedded sections)
The sections were washed in PBS (3 times, 5 minutes each), permeabilized and blocked in a PBS/0.3% Tween 20/1.5% BSA for one hour at room temperature, followed by an overnight incubation with the primary antibody diluted in a PBS/0.2% TritonX100 solution at room temperature. Next, sections were washed in PBS (3 times, 10 minutes each) and incubated with the secondary antibody (diluted in PBS) for one hour, washed twice with PBS (10 minutes each), and then incubated for 10 minutes with PBS/DAPI (Thermo Fisher Scientific, 100ng/ml) solution. To quench autofluorescence, sections were incubated with 0.1% Sudan Black/70% EtOH for 1 minute, and then washed twice briefly with PBS. Sections were mounted on Fisherbrand Superfrost Plus Microscope Slides (Thermo Fisher Scientific) with ProLong Gold antifade reagent with DAPI (Thermo Fisher Scientific).
Immunofluorescence (paraffin-embedded sections)
Immunofluorescence of paraffin-embedded sections for an experiment shown in Figure 4A (R6/2) and Figures 1G, 2C and 4E (Human) was performed as following: CitriSol (Thermo Fisher Scientific)-treated sections were incubated in 100% EtOH, 90% EtOH and dH2O, permeabilized in 0.2% TritonX100/PBS, followed by antigen retrieval and blocking with 2% FBS/0.2% TritonX100/PBS. Then sections were incubated with Anti-Huntingtin Protein Antibody, clone mEM48 (1:500), RanGAP1 Antibody (H-180, sc-25630) (1:500) and lamin B1 Antibody (S-20 sc-30264) (1:5000) diluted in 2% FBS/PBS overnight at 4°C and, after additional washing and blocking steps, incubated with Goat anti Rabbit Alexa488, Invitrogen, A11008, (1:500) antibody, diluted in PBS for 1h at room temperature. Following incubations with 0.01% DAPI and 0.1% Sudan Black/70% EtOH, sections were washed with PBS and mounted with ProLong Gold antifade reagent with DAPI (Thermo Fisher Scientific).
Immunofluorescence (human neuronal progenitors)
For immunofluorescence, human neuronal progenitors were grown on Matrigel coated 6-well plates in a 37°C incubator at 5% CO2 with fresh media changes every third day. NPCs were harvested using TrypLE Express (GIBCO), re-suspended in fresh NPC media, and plated onto Matrigel coated 96-well glass bottom plate (PerkinElmer) at a total cell density of 20,000 cells/well. One day later, cells were washed once with PBS to remove residual media and fixed using 4% paraformaldehyde (Tousimis) in PBS for 20 minute at room temperature. After washing twice with PBS, cells were permeabilized with 0.1% Triton X-100 for 10 minute at room temperature. Cells were washed twice again with PBS and blocked with 1% bovine albumin in PBS for 1 hour at room temperature. Cells were incubated with primary antibodies (anti-laminB1 antibody (Proteintech 12987-1-AP) at 1:500, anti-RanGAP1 (H-180, sc-25630) at 1:500), at the indicated dilution in the blocking solution supplemented with 0.02% Tween-20 and incubated at room temperature on a rocking platform (BellCo Technologies) for 2 hours. Appropriate fluorescently tagged secondary antibodies conjugated to Alexa488/Alexa595 (ThermoFisher Scientific) were incubated for 30 minute at room temperature in the blocking buffer. The nuclei were stained sequentially using Hoechst 33342 dye (Life Technologies) at 1:1000 dilution in PBS for 15 minute at room temperature. The cells were washed finally with PBS twice and kept in PBS for imaging analysis.
Antibodies
The following primary antibodies were used for immunofluorescence experiments: Anti-Huntingtin Protein Antibody, clone mEM48, MAB-5374, Millipore (1:500); lamin B1 Antibody (S-20), sc-30264, Santa Cruz (1:5000); lamin B1 Antibody, 12987-1-AP, Proteintech (1:500); RanGAP1 Antibody (H-180), sc-25630, Santa Cruz (1:500); Anti-GLE1 antibody (ab96007), Abcam (1:200); Anti-phospho-Histone H2A.X (Ser139) Antibody, clone JBW301, Millipore (1:500). Secondary antibodies include: Donkey anti Mouse Cy3, Jackson immunology, 715-165-151, (1:500); Goat anti Rabbit Alexa488, Invitrogen, A11008, (1:500); Donkey anti Goat Alexa488, Invitrogen, A11055, (1:500).
RNA Fluorescence in situ hybridization (FISH)
All hybridization steps were performed under RNase-free conditions. Brain OCT frozen sections (10 μm) were dried for 10 min at 60°C. Paraffin-embedded sections (from human autopsies) were washed with xylene and EtOH (100% and 70%) for deparaffinization and with water for hydration, followed by antigen retrieval. Then, samples were permeabilized in 0.2% Triton X100/PBS for 10 min at room temperature and blocked (at 60°C for 1 h) with hybridization buffer (50% formamide, saline-sodium citrate (SSC, 2x), 50 mM sodium phosphate (pH 7), 10% dextran sulfate, and 2 mM vanadyl sulfate ribonucleosides). The sections were then hybridized with 5’-labelled Cy3-(d)T20 oligonucleotides (Integrated DNA Technologies) (200 mM) for 1h/overnight at 37°C or 60°C. After hybridization, slides were washed in 0.1% Tween-20/2xSCC for 5 min (at room temperature) and three times (10 min each) in 0.1x SSC at 65°C. Autofluorescence was quenched by 0.25% SudanBlack B/70% ethanol and cell nuclei were stained with DAPI.
Image acquisition
Images were acquired with Nikon Eclipse laser scanning confocal microscope with Nikon C2 power device, using the Nikon EZ-C2 software. Brain sections were imaged with 100X, 60X and 20X objectives.
For Z-stacks acquired at 100X, 3 μm thick layers were imaged by taking 30 consecutive images (0.1μm Z-step). Z-projections were generated with the standard deviation Z-projection algorithm using ImageJ-based open-source Fiji software package.
For Z-stacks, acquired at 20X, 15 μm thick layers were imaged by taking 10 consecutive images (1.5 μm Z-step), and Z-projections were generated as mentioned above.
For human iPS-derived neuronal progenitors immunofluorescence analysis, 40X images were acquired on IN Cell Analyzer 6000 Cell Imaging System (laser based confocal imaging platform from GE Healthcare Life Sciences). For lamin B1 and RanGAP1 based nuclear morphology analysis, 10 random Fields of View (FOVs) spread evenly across the entire surface of each well of a 96-well plate were captured for each cell line. Multiple independent wells for each cell lines were analyzed.
QUANTIFICATION AND STATISTICAL ANALYSIS
All quantitative analyses relied on systematic uniform random sampling.
A. Nuclear Envelope shape (lamin B1 staining)
We defined 2 types of NE morphology (Figure S1B):
normal, where nuclear lamina is characterized by a predominantly round and regular shape;
altered, with moderate or severe alterations, an irregular shape of nuclear lamina with several invaginations or protrusions.
100–400 cells (cortex) and 300–700 cells (striatum) from at least 2 animals per each genotype and age were analyzed.
B. PolyQ Huntingtin aggregates
PolyQ Htt aggregates were quantified manually per cell. Lamin B1 and DAPI staining were used to address nuclear/extranuclear polyQ Htt localization on images acquired with a 100X objective. 200 cells for each genotype and age were counted.
C. DNA Double Strand breaks
Thresholds for γH2AX and DAPI signals were defined using Nikon Elements software, and new binary layers containing both signals were generated. γH2AX foci number and areas were quantified using Nikon Elements or FIJI IMAGEJ software. 100–200 cells per animal were counted.
D. Gle1/PolyQ Htt and RanGAP1/PolyQ Htt co-localization in nuclei
Nuclear PolyQ Htt aggregates within 3 μm Z stacks (100x magnification, 30 consecutive images, 0.1μm Z-step) were analyzed for the presence of Gle1 or RanGAP1 signals. Aggregates were considered “co-localized”, if co-localizing Gle1 and PolyQ Htt signals or RanGAP1 and PolyQ Htt signals formed distinct spots, observed in any of corresponding channels: PolyQ Htt and Gle1, or PolyQ Htt and RanGAP1 respectively. Nuclear localization was defined by DAPI. At least 100 cells from at least 2 animals were analyzed for each genotype and age.
E. RanGAP1 perinuclear accumulation
20x images were analyzed. Cells, positive or negative for perinuclear accumulated RanGAP1 were counted manually (at least 600 cells per animal were analyzed).
RanGAP1 alterations in the human neuronal progenitor cells were determined by the detection of RanGAP1 positive nuclear invaginations. Nuclear RanGAP1 was first defined by overlaying Hoechst 33342 dye based nuclear area masks to segment individual nuclei. Each segmented nucleus was then scored for the presence or absence of RanGAP1 intensity based nuclear invagination. For automated detection, nuclear invaginations were defined as unilateral spikes in per pixel intensity forming a high intensity protrusion extending inwards from the periphery (outlined by Hoechst 33342 dye based individual nuclear perimeter) of a given nucleus. For obtaining percent of cells with nuclear invagination, cells from each Field of View were combined to take a ratio of total number of nuclei scored positive over the total number of nuclei measured. Multiple wells with 10 fields of views per well were analyzed for each cell line. At least 5000 cells per cell lines were analyzed.
F. PolyA RNA distribution
60x-100x images were analyzed. Using Acapella software (Perkin Elmer), cells were segmented by first identifying nuclei based on Dapi, and then cytoplasm based on PolyA RNA. Mean intensities of nuclear and cytoplasmic PolyA RNA signals were determined for each cell. Ratio of nuclear to cytoplasmic signal intensities is shown in Figure 2B. 300–600 cells were analyzed from mice of following genotypes and ages: 24 month-old HttQ7/Q7 (n=3), 24 month-old HttQ7/Q175 (n=3), 24 month-old HttQ175/Q175 (n=3), 3 month-old R6/2 (n=1).
Quantification of cells with nuclear or nuclear/cytoplasmic RNA localization in human autopsy samples was performed manually by a blinded investigator; counting the cells that presented polyA signal only in the nucleus (nuclear mRNA accumulation) or in the nucleus & cytoplasm (100x magnification, 30 consecutive images, 0.1μm Z-step). 150 cells were counted.
G. Human Nuclear Envelope shape (lamin B1 staining)
Nuclear envelope morphology was examined manually in lamin B1 positive cells. 3 μm Z stacks images (100x magnification, 30 consecutive images, 0.1μm Z-step) were analyzed. 100 cells were counted per subject.
Statistical Analysis
Statistical analyses were performed using GraphPad Prism. All data are shown as the mean ± standard error of the mean (SEM). The statistical significance of the differences between two groups was investigated by unpaired t test or by chi-square test with Yates’ correction. Statistical tests are indicated in each figure legend along with the corresponding significance level (p value). The number of cells analyzed per experiment is provided in the corresponding figure legends.
Supplementary Material
Immunofluorescence of RanGAP1 (green) and polyQ Htt (red) in cortex of 6 month-old HttQ175/Q175 mice. 30 consecutive slices (3 μM) are shown. Arrowheads indicate polyQ/RanGAP1 nuclear co-localization. Nuclei were stained with DAPI.
Immunofluorescence of RanGAP1 (green) and polyQ Htt (red) in cortex of 18 month-old HttQ175/Q175 mice. 30 consecutive slices (3 μM) are shown. Arrowheads indicate polyQ/RanGAP1 nuclear co-localization. Asterisks indicate polyQ/RanGAP1 perinuclear co-localization. Nuclei were stained with DAPI.
Acknowledgments
We would like to express a special thanks to Dr. Douglas MacDonald from CHDI Inc. for constructive discussions and continuous support. We are grateful to Drs. Jody Corey-Bloom, Eliezer Masliah and Ivy Trinh (UCSD ADCR Brain Bank), and Dr. Matthew Frosch and Jose McLean at the Massachusetts Alzheimer’s Disease Research Center (PG50 AG005134) for providing us with the human tissue samples. We also thank John Ravits and Jennifer Stauffer for the technical support on FISH methodology. This work was supported by CHDI Inc. F.G.R., C.C.M. and J.A. received salary support from CHDI Inc. A.G. and D.W.C. received salary support from the Ludwig Institute for Cancer Research. C.L.-T received salary support from the Ludwig Institute for Cancer Research, the Massachusetts General Hospital and NINDS/NIH (R01NS087227).
Footnotes
Author Contributions:
F.G.R., C.C.M., A.G., D.W.C. and C.L.T. designed research; F.G.R., C.C.M., A.G., R.S.A., V.C.W., A.G.B, D.W.C. and C.L.T analyzed the data; F.G.R., C.C.M., R.S.A., J.A., and R.T. performed research; F.G.R., A.G., D.W.C. and C.L.T wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bennett EJ, Shaler TA, Woodman B, Ryu KY, Zaitseva TS, Becker CH, Bates GP, Schulman H, Kopito RR. Global changes to the ubiquitin system in Huntington’s disease. Nature. 2007;448:704–708. doi: 10.1038/nature06022. [DOI] [PubMed] [Google Scholar]
- Bischoff FR, Krebber H, Kempf T, Hermes I, Ponstingl H. Human RanGTPase-activating protein RanGAP1 is a homologue of yeast Rna1p involved in mRNA processing and transport. Proc Natl Acad Sci U S A. 1995;92:1749–1753. doi: 10.1073/pnas.92.5.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukata L, Parker SL, D’angelo MA. Nuclear pore complexes in the maintenance of genome integrity. Curr Opin Cell Biol. 2013;25:378–386. doi: 10.1016/j.ceb.2013.03.002. [DOI] [PubMed] [Google Scholar]
- Carty N, Berson N, Tillack K, Thiede C, Scholz D, Kottig K, Sedaghat Y, Gabrysiak C, Yohrling G, Von Der Kammer H, et al. Characterization of HTT inclusion size, location, and timing in the zQ175 mouse model of Huntington’s disease: an in vivo high-content imaging study. PLoS One. 2015;10:e0123527. doi: 10.1371/journal.pone.0123527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapple JP, Bros-Facer V, Butler R, Gallo JM. Focal distortion of the nuclear envelope by huntingtin aggregates revealed by lamin immunostaining. Neurosci Lett. 2008;447:172–174. doi: 10.1016/j.neulet.2008.09.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium HDI. Induced pluripotent stem cells from patients with Huntington’s disease show CAG-repeat-expansion-associated phenotypes. Cell Stem Cell. 2012;11:264–278. doi: 10.1016/j.stem.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne AN, Yamada SB, Siddegowda BB, Estes PS, Zaepfel BL, Johannesmeyer JS, Lockwood DB, Pham LT, Hart MP, Cassel JA, et al. Fragile X protein mitigates TDP-43 toxicity by remodeling RNA granules and restoring translation. Hum Mol Genet. 2015;24:6886–6898. doi: 10.1093/hmg/ddv389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’angelo MA, Raices M, Panowski SH, Hetzer MW. Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell. 2009;136:284–295. doi: 10.1016/j.cell.2008.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Cruz S, Cleveland DW. CELL BIOLOGY. Disrupted nuclear import-export in neurodegeneration. Science. 2016;351:125–126. doi: 10.1126/science.aad9872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies SW, Turmaine M, Cozens BA, Difiglia M, Sharp AH, Ross CA, Scherzinger E, Wanker EE, Mangiarini L, Bates GP. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell. 1997;90:537–548. doi: 10.1016/s0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- Dejesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9plinked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragunow M, Faull RL, Lawlor P, Beilharz EJ, Singleton K, Walker EB, Mee E. In situ evidence for DNA fragmentation in Huntington’s disease striatum and Alzheimer’s disease temporal lobes. Neuroreport. 1995;6:1053–1057. doi: 10.1097/00001756-199505090-00026. [DOI] [PubMed] [Google Scholar]
- Duan W, Guo Z, Jiang H, Ware M, Li XJ, Mattson MP. Dietary restriction normalizes glucose metabolism and BDNF levels, slows disease progression, and increases survival in huntingtin mutant mice. Proc Natl Acad Sci U S A. 2003;100:2911–2916. doi: 10.1073/pnas.0536856100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enokido Y, Tamura T, Ito H, Arumughan A, Komuro A, Shiwaku H, Sone M, Foulle R, Sawada H, Ishiguro H, et al. Mutant huntingtin impairs Ku70-mediated DNA repair. J Cell Biol. 2010;189:425–443. doi: 10.1083/jcb.200905138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fifkova E, Tonks M, Cullen-Dockstader K. Changes in the nuclear pore complexes of the dentate granule cells in aged rats. Exp Neurol. 1987;95:755–762. doi: 10.1016/0014-4886(87)90314-1. [DOI] [PubMed] [Google Scholar]
- Freibaum BD, Lu Y, Lopez-Gonzalez R, Kim NC, Almeida S, Lee KH, Badders N, Valentine M, Miller BL, Wong PC, et al. GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature. 2015;525:129–133. doi: 10.1038/nature14974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost B, Bardai FH, Feany MB. Lamin Dysfunction Mediates Neurodegeneration in Tauopathies. Curr Biol. 2016;26:129–136. doi: 10.1016/j.cub.2015.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusel’nikova VV, Korzhevskiy DE. NeuN As a Neuronal Nuclear Antigen and Neuron Differentiation Marker. Acta Naturae. 2015;7:42–47. [PMC free article] [PubMed] [Google Scholar]
- Hay DG, Sathasivam K, Tobaben S, Stahl B, Marber M, Mestril R, Mahal A, Smith DL, Woodman B, Bates GP. Progressive decrease in chaperone protein levels in a mouse model of Huntington’s disease and induction of stress proteins as a therapeutic approach. Hum Mol Genet. 2004;13:1389–1405. doi: 10.1093/hmg/ddh144. [DOI] [PubMed] [Google Scholar]
- Heikkinen T, Lehtimaki K, Vartiainen N, Puolivali J, Hendricks SJ, Glaser JR, Bradaia A, Wadel K, Touller C, Kontkanen O, et al. Characterization of neurophysiological and behavioral changes, MRI brain volumetry and 1H MRS in zQ175 knock-in mouse model of Huntington’s disease. PLoS One. 2012;7:e50717. doi: 10.1371/journal.pone.0050717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, Langfelder P, Kwak S, Aaronson J, Rosinski J, Vogt TF, Eszes M, Faull RL, Curtis MA, Waldvogel HJ, et al. Huntington’s disease accelerates epigenetic aging of human brain and disrupts DNA methylation levels. Aging (Albany NY) 2016;8:1485–1512. doi: 10.18632/aging.101005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins BG, Koroshetz WJ, Beal MF, Rosen BR. Evidence for impairment of energy metabolism in vivo in Huntington’s disease using localized 1H NMR spectroscopy. Neurology. 1993;43:2689–2695. doi: 10.1212/wnl.43.12.2689. [DOI] [PubMed] [Google Scholar]
- Jovicic A, Mertens J, Boeynaems S, Bogaert E, Chai N, Yamada SB, Paul JW, 3rd, Sun S, Herdy JR, Bieri G, et al. Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat Neurosci. 2015;18:1226–1229. doi: 10.1038/nn.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneb HM, Folkmann AW, Belzil VV, Jao LE, Leblond CS, Girard SL, Daoud H, Noreau A, Rochefort D, Hince P, et al. Deleterious mutations in the essential mRNA metabolism factor, hGle1, in amyotrophic lateral sclerosis. Hum Mol Genet. 2015;24:1363–1373. doi: 10.1093/hmg/ddu545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley JB, Datta S, Snow CJ, Chatterjee M, Ni L, Spencer A, Yang CS, Cubenas-Potts C, Matunis MJ, Paschal BM. The defective nuclear lamina in Hutchinson-gilford progeria syndrome disrupts the nucleocytoplasmic Ran gradient and inhibits nuclear localization of Ubc9. Mol Cell Biol. 2011;31:3378–3395. doi: 10.1128/MCB.05087-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita Y, Ito H, Hirano A, Fujita K, Wate R, Nakamura M, Kaneko S, Nakano S, Kusaka H. Nuclear contour irregularity and abnormal transporter protein distribution in anterior horn cells in amyotrophic lateral sclerosis. J Neuropathol Exp Neurol. 2009;68:1184–1192. doi: 10.1097/NEN.0b013e3181bc3bec. [DOI] [PubMed] [Google Scholar]
- Lee KH, Zhang P, Kim HJ, Mitrea DM, Sarkar M, Freibaum BD, Cika J, Coughlin M, Messing J, Molliex A, et al. C9orf72 Dipeptide Repeats Impair the Assembly, Dynamics, and Function of Membrane-Less Organelles. Cell. 2016;167:774–788 e717. doi: 10.1016/j.cell.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Li SH, Yu ZX, Shelbourne P, Li XJ. Huntingtin aggregate-associated axonal degeneration is an early pathological event in Huntington’s disease mice. J Neurosci. 2001;21:8473–8481. doi: 10.1523/JNEUROSCI.21-21-08473.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- Liu GH, Qu J, Suzuki K, Nivet E, Li M, Montserrat N, Yi F, Xu X, Ruiz S, Zhang W, et al. Progressive degeneration of human neural stem cells caused by pathogenic LRRK2. Nature. 2012;491:603–607. doi: 10.1038/nature11557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu KY, Shyu YC, Barbaro BA, Lin YT, Chern Y, Thompson LM, James Shen CK, Marsh JL. Disruption of the nuclear membrane by perinuclear inclusions of mutant huntingtin causes cell-cycle re-entry and striatal cell death in mouse and cell models of Huntington’s disease. Hum Mol Genet. 2015;24:1602–1616. doi: 10.1093/hmg/ddu574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu XH, Mattis VB, Wang N, Al-Ramahi I, Van Den Berg N, Fratantoni SA, Waldvogel H, Greiner E, Osmand A, Elzein K, et al. Targeting ATM ameliorates mutant Huntingtin toxicity in cell and animal models of Huntington’s disease. Sci Transl Med. 2014;6:268ra178. doi: 10.1126/scitranslmed.3010523. [DOI] [PubMed] [Google Scholar]
- Ma TC, Buescher JL, Oatis B, Funk JA, Nash AJ, Carrier RL, Hoyt KR. Metformin therapy in a transgenic mouse model of Huntington’s disease. Neurosci Lett. 2007;411:98–103. doi: 10.1016/j.neulet.2006.10.039. [DOI] [PubMed] [Google Scholar]
- Mandavilli BS, Rao KS. Accumulation of DNA damage in aging neurons occurs through a mechanism other than apoptosis. J Neurochem. 1996a;67:1559–1565. doi: 10.1046/j.1471-4159.1996.67041559.x. [DOI] [PubMed] [Google Scholar]
- Mandavilli BS, Rao KS. Neurons in the cerebral cortex are most susceptible to DNA-damage in aging rat brain. Biochem Mol Biol Int. 1996b;40:507–514. doi: 10.1080/15216549600201073. [DOI] [PubMed] [Google Scholar]
- Mangiarini L, Sathasivam K, Seller M, Cozens B, Harper A, Hetherington C, Lawton M, Trottier Y, Lehrach H, Davies SW, et al. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87:493–506. doi: 10.1016/s0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- Mapelli L, Canale C, Pesci D, Averaimo S, Guizzardi F, Fortunati V, Falasca L, Piacentini M, Gliozzi A, Relini A, et al. Toxic effects of expanded ataxin-1 involve mechanical instability of the nuclear membrane. Biochim Biophys Acta. 2012;1822:906–917. doi: 10.1016/j.bbadis.2012.01.016. [DOI] [PubMed] [Google Scholar]
- Menalled LB, Kudwa AE, Miller S, Fitzpatrick J, Watson-Johnson J, Keating N, Ruiz M, Mushlin R, Alosio W, Mcconnell K, et al. Comprehensive behavioral and molecular characterization of a new knock-in mouse model of Huntington’s disease: zQ175. PLoS One. 2012;7:e49838. doi: 10.1371/journal.pone.0049838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens J, Paquola AC, Ku M, Hatch E, Bohnke L, Ladjevardi S, Mcgrath S, Campbell B, Lee H, Herdy JR, et al. Directly Reprogrammed Human Neurons Retain Aging-Associated Transcriptomic Signatures and Reveal Age-Related Nucleocytoplasmic Defects. Cell Stem Cell. 2015;17:705–718. doi: 10.1016/j.stem.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno CL, Ehrlich ME, Mobbs CV. Protection by dietary restriction in the YAC128 mouse model of Huntington’s disease: Relation to genes regulating histone acetylation and HTT. Neurobiol Dis. 2016;85:25–34. doi: 10.1016/j.nbd.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov ED, Vigont VA, Klyushnikov SA, Lebedeva OS, Vassina EM, Bogomazova AN, Chestkov IV, Semashko TA, Kiseleva E, Suldina LA, et al. Manifestation of Huntington’s disease pathology in human induced pluripotent stem cell-derived neurons. Mol Neurodegener. 2016;11:27. doi: 10.1186/s13024-016-0092-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujol G, Soderqvist H, Radu A. Age-associated reduction of nuclear protein import in human fibroblasts. Biochem Biophys Res Commun. 2002;294:354–358. doi: 10.1016/S0006-291X(02)00492-8. [DOI] [PubMed] [Google Scholar]
- Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, Van Swieten JC, Myllykangas L, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez R, Hernandez-Hernandez O, Magana JJ, Gonzalez-Ramirez R, Garcia-Lopez ES, Cisneros B. Altered nuclear structure in myotonic dystrophy type 1-derived fibroblasts. Mol Biol Rep. 2015;42:479–488. doi: 10.1007/s11033-014-3791-4. [DOI] [PubMed] [Google Scholar]
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA doublestranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- Sadagurski M, Cheng Z, Rozzo A, Palazzolo I, Kelley GR, Dong X, Krainc D, White MF. IRS2 increases mitochondrial dysfunction and oxidative stress in a mouse model of Huntington disease. J Clin Invest. 2011;121:4070–4081. doi: 10.1172/JCI46305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaffidi P, Misteli T. Lamin A-dependent nuclear defects in human aging. Science. 2006;312:1059–1063. doi: 10.1126/science.1127168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield LG, Miskiewicz HB, Tannenbaum LB, Mirra SS. Nuclear pore complex proteins in Alzheimer disease. J Neuropathol Exp Neurol. 2006;65:45–54. doi: 10.1097/01.jnen.0000195939.40410.08. [DOI] [PubMed] [Google Scholar]
- Shi KY, Mori E, Nizami ZF, Lin Y, Kato M, Xiang S, Wu LC, Ding M, Yu Y, Gall JG, et al. Toxic PRn poly-dipeptides encoded by the C9orf72 repeat expansion block nuclear import and export. Proc Natl Acad Sci U S A. 2017;114:E1111–E1117. doi: 10.1073/pnas.1620293114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GA, Rocha EM, Mclean JR, Hayes MA, Izen SC, Isacson O, Hallett PJ. Progressive axonal transport and synaptic protein changes correlate with behavioral and neuropathological abnormalities in the heterozygous Q175 KI mouse model of Huntington’s disease. Hum Mol Genet. 2014;23:4510–4527. doi: 10.1093/hmg/ddu166. [DOI] [PubMed] [Google Scholar]
- Tallaksen-Greene SJ, Sadagurski M, Zeng L, Mauch R, Perkins M, Banduseela VC, Lieberman AP, Miller RA, Paulson HL, Albin RL. Differential effects of delayed aging on phenotype and striatal pathology in a murine model of Huntington disease. J Neurosci. 2014;34:15658–15668. doi: 10.1523/JNEUROSCI.1830-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Jurk D, Maddick M, Nelson G, Martin-Ruiz C, Von Zglinicki T. DNA damage response and cellular senescence in tissues of aging mice. Aging Cell. 2009;8:311–323. doi: 10.1111/j.1474-9726.2009.00481.x. [DOI] [PubMed] [Google Scholar]
- Watkins JL, Murphy R, Emtage JL, Wente SR. The human homologue of Saccharomyces cerevisiae Gle1p is required for poly(A)+ RNA export. Proc Natl Acad Sci U S A. 1998;95:6779–6784. doi: 10.1073/pnas.95.12.6779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woerner AC, Frottin F, Hornburg D, Feng LR, Meissner F, Patra M, Tatzelt J, Mann M, Winklhofer KF, Hartl FU, et al. Cytoplasmic protein aggregates interfere with nucleocytoplasmic transport of protein and RNA. Science. 2016;351:173–176. doi: 10.1126/science.aad2033. [DOI] [PubMed] [Google Scholar]
- Yannarell A, Schumm DE, Webb TE. Age-dependence of nuclear RNA processing. Mech Ageing Dev. 1977;6:259–264. doi: 10.1016/0047-6374(77)90026-4. [DOI] [PubMed] [Google Scholar]
- Zhang K, Donnelly CJ, Haeusler AR, Grima JC, Machamer JB, Steinwald P, Daley EL, Miller SJ, Cunningham KM, Vidensky S, et al. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature. 2015;525:56–61. doi: 10.1038/nature14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YJ, Gendron TF, Grima JC, Sasaguri H, Jansen-West K, Xu YF, Katzman RB, Gass J, Murray ME, Shinohara M, et al. C9ORF72 poly(GA) aggregates sequester and impair HR23 and nucleocytoplasmic transport proteins. Nat Neurosci. 2016;19:668–677. doi: 10.1038/nn.4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Immunofluorescence of RanGAP1 (green) and polyQ Htt (red) in cortex of 6 month-old HttQ175/Q175 mice. 30 consecutive slices (3 μM) are shown. Arrowheads indicate polyQ/RanGAP1 nuclear co-localization. Nuclei were stained with DAPI.
Immunofluorescence of RanGAP1 (green) and polyQ Htt (red) in cortex of 18 month-old HttQ175/Q175 mice. 30 consecutive slices (3 μM) are shown. Arrowheads indicate polyQ/RanGAP1 nuclear co-localization. Asterisks indicate polyQ/RanGAP1 perinuclear co-localization. Nuclei were stained with DAPI.