Abstract
Claudins provide tight junction barrier selectivity. The human CLDN5 gene contains a high-frequency single-nucleotide polymorphism (rs885985), where the G allele encodes for glutamine (Q) and the A allele encodes for an amber stop codon. Thus, these different CLDN5 alleles define nested open reading frames (ORFs) encoding claudin-5 proteins that are 303 or 218 amino acids in length. Interestingly, human claudin-16 and claudin-23 also have long ORFs. The long form of claudin-5 contrasts with the majority of claudin-5 proteins in the National Center for Biotechnology Information protein database, which are less than 220 amino acids in length. Screening of genotyped human lung tissue by immunoblot revealed only the 218–amino acid form of claudin-5 protein; the long-form claudin-5 protein was not detected. Moreover, when forcibly expressed in transfected cells, the long form of human claudin-5 was retained in intracellular compartments and did not localize to the plasma membrane, in contrast to the 218–amino acid form, which localized to intercellular junctions. This suggests that the 303–amino acid claudin-5 protein is rarely expressed, and, if so, is predicted to adversely affect cell function. Potential roles for upstream ORFs in regulating claudin-5 expression are also discussed.
Keywords: claudin, epithelium, endothelium, single nucleotide polymorphism, upstream open reading frame
Introduction
Tight junctions enable epithelial barriers to form by regulating the paracellular space between cells in direct contact.1,2 Although a multitude of proteins are required to form fully functional tight junctions, claudin family transmembrane proteins provide the structural basis to control paracellular ion and solute flux through tight junctional barriers.3,4 There are roughly two dozen different claudins, which differ in permeability as well as compatibility of interaction.3,5 By expressing multiple different claudin proteins, a tissue can fine-tune barrier function to match the permeability requirements for specific organs.6–8
Claudins span the bilayer four times, with both the N- and C-termini oriented towards the cytoplasm and two extracellular domains that interact with adjacent claudins on neighboring cells to form paracellular channels.9–11 By and large, each claudin mRNA contains an intron-less open reading frame (ORF). However there are some exceptions. For instance, there are two human claudin-18 splice variants, one expressed predominantly in the lung and the other expressed as a stomach-specific isoform.6,12,13 There are also two classes of claudin-10 splice variants that differ in permselectivity, with claudin-10b being more chloride permeable than claudin-10a, along with several subvariants that do not form tight junctions.14,15 There is also a claudin-7 splice variant with a C-terminal tail truncation.16 In addition to differential splicing, single-nucleotide polymorphisms (SNPs) have the potential to alter claudin structure and function and thus harbor the potential to be unrecognized disease modifiers.17,18 However, SNPs in general tend to have a low frequency of penetrance.
Claudin-5 is prominently expressed by the vascular endothelium and plays critical roles in the blood–brain barrier and pulmonary endothelial barrier as well as acting as a modulator for epithelial function.6,19,20 In investigating the amino acid sequence of human claudin-5 in the National Center for Biotechnology Information (NCBI) database, we found that there were two ORFs represented, one 303–amino acid ORF and a second nested ORF that codes for 218 amino acids. The longer gene product was surprising, since claudin-5 proteins produced by most other species are ~ 218 amino acids in length. In fact, the average size of a human claudin ORF reflects a gene product that is typically ~ 220 amino acids, although two notable exceptions are claudin-16 (305 amino acids)21,22 and claudin-23 (292 amino acids). Moreover, in both the NCBI and Uniprot databases, there are two entries for human claudin-5 protein that reflect gene products that are 218 and 303 amino acids in length, respectively. Given these apparent discrepancies, we further examined the human CLDN5 coding sequence and found a documented SNP with near-Mendelian penetrance in the general population, dbSNP cluster ID rs885985. The rs885985 A allele of CLDN5 contains a nonsense codon and thus only codes for the 218–amino acid claudin-5 ORF; however, the G allele contains an overlapping ORF that could theoretically encode for both the 303– and 218–amino acid gene products. Thus, we examined human lung tissue by immunoblot to measure the size(s) of claudin-5 protein produced. Regardless of genotype, only the 218–amino acid claudin-5 was detectable. This has implications for other mammalian CLDN5 genes present in the NCBI database where ORFs were adjusted to encode for 303–amino acid gene products. Moreover, a synthetic 303–amino acid form of human claudin-5 was not properly processed by transfected human cell lines and did not localize to sites of cell–cell junctions. These data suggest that normal human claudin-5 is 218 amino acids in length and that upstream start codons in claudin-5 are more likely to have a role in regulating gene expression.23 Furthermore, we hypothesize that any conditions where the 303–amino acid form of claudin-5 is expressed may have pathological consequences.
Materials and methods
Sequence acquisition and analysis
The human CLDN5 sequence (Chromosome 22 – NC_000022.1, Gene ID: 7122) came from the National Institutes of Health, United States National Library of Medicine, (NCBI) database (https://www.ncbi.nlm.nih.gov/gene/7122). Predicted human protein sequence NP_003268.2 corresponds to the 303–amino acid claudin-5 ORF and was used for an NCBI Basic Local Alignment Tool (BLASTp) homology search (https://blast.ncbi.nlm.nih.gov/). The rs885985 SNP alleles, G and A, were identified from the human CLDN5 sequence, as annotated in dbSNP Geneview database Build 149 (https://www.ncbi.nlm.nih.gov/SNP/snp_ref.cgi?locusId=7122). All databases were accessed on February 14, 2017.
Claudin-5 cDNA constructs, cell transfection, and immunofluorescence
Untagged human claudin-5 (218–amino acid ORF) in pcDNA3.1 and N-terminal YFP–tagged human claudin-5 in pEYFP C1 using a CMV promoter were produced as previously described.24,25 Human claudin-5 (303–amino acid ORF; G allele) was obtained from Origene (SC325743, transcript version 1 in a pcmv6-XL4 vector). The claudin-5 insert was polymerase chain reaction (PCR) amplified using the primers 5′-GATTGGCTCTCGAGCGATGACCGGCG-3′ and 5′-CGTGCCCAAGCTTCTCAGACGTAGTTC-3′ and inserted into pcDNA3.1 or pYFP C1 using HindIII and XbaI. Plasmid sequences were verified by sequencing (Eurofins MWG).
HeLa cells or Caco2 cells were maintained in Dulbecco’s modified Eagle media (DMEM; Sigma) containing 10% FBS, 0.25 μg/mL amphotericin B (ThermoFisher), 100 U/mL penicillin, and 10 mg/mL streptomycin (Sigma). For transfection, cells were plated on rat tail collagen type I–coated glass coverslips, Transwells, or tissue culture plastic (100-mm dish). When cells were 80–90% confluent, the media was replaced with Opti-Mem. Cells were transiently transfected with the different claudin-5 cDNAs using XtremeGene HP DNA-transfection reagent (Roche). Expression of the different claudin-5 constructs was analyzed 48 h after transfection by immunofluorescence or immunoblot.
For immunofluorescence microscopy, cells transduced with YFP-tagged claudin-5 were washed three times with Dulbecco’s phosphate-buffered saline containing Ca2+ and Mg2+ (DPBS++), fixed with 1:1 methanol/acetone for 2 min at room temperature, and then washed again three times with DPBS++ and mounted in Mowiol on a glass slide. Fluorescence images were taken using an Olympus IX70 microscope with a U-MWIBA filter pack (BP460–490, DM505, BA515–550). Minimum and maximum intensities were adjusted for images in parallel so that the intensity scale remained linear to maximize dynamic range.
Genotyping and immunoblot
De-identified tissue from posttransplant human donor lungs was collected by the Emory Transplant Center under an Institutional Review Board–approved tissue-acquisition protocol.
Genotyping was designed and performed on lung tissue samples by the GenoTyping Center of America (Ellsworth, ME). SNP rs885985 was detected using SYBR Green melting-curve analysis followed by gel electrophoresis. The locus was amplified using SYBR Green PCR Mix (primers: 5′-GTCACGATGTTGTGGTCCAG-3′ and 5′-CAAGAGCCGTTGTTTCCCTA-3′). The reaction was run on a Roche LightCycler 480 with conditions suggested for SYBR Green amplification. The PCR product was digested overnight with AvrII, and products were resolved by electrophoresis on a 2% agarose gel. The A allele had expected band sizes of 269 and 166 bp. The G allele had an expected band size of 439 bp. Heterozygous samples had 439-, 269-, and 166-bp bands. All genotypes were detectable.
For immunoblot, total lung tissue was resuspended in 100 μL buffer per 100 mg in RIPA buffer (Cell Signaling). The tissue was minced using a tissue homogenizer and incubated on ice for 30 min before samples were sonicated for 3 × 15-s pulses. Debris was pelleted by centrifugation for 10 min at 13,200 × g at 4 °C. Protein concentration of the supernatant was determined by bicinchoninic acid (BCA) assay (ThermoFisher Pierce #23225). Reducing SDS sample buffer (10% glycerol, 1.25% SDS, 50 mM Tris pH 6.7, 8.3 mg/mL dithiothreitol (DTT)) was added to the supernatant. HeLa cells transfected with either the 303–or 218–amino acid form of untagged human claudin-5 and solubilized in reducing SDS sample buffer were used as standards. Protein samples were heated for 10 min at 65 °C and then resolved by SDS-PAGE using 4–20% Mini-PROTEAN TGX stain-free gradient SDS polyacrylamide gels, transferred to nitrocellulose membranes (BioRad, Hercules, CA), and immunostained using primary mouse anti-claudin-5 (4C3C2, ThermoFisher #35-2500) in combination with Goat anti-mouse IgG IRDye680RD (Licor #92668070). For band detection, fluorescence imaging was performed using the Odyssey Classic imager (LI-COR) and analyzed with Image Studio (LI-COR).
Results
Two common CLDN5 alleles encode different predicted ORFs
As shown in Figure 1A, SNP rs885985 within the human CLDN5 gene encodes a missense mutation, where the G allele is glutamine (Q) and the A allele is an amber (TAG) stop codon. The full predicted ORF for claudin-5 in the G allele is thus a gene product of 303 amino acids that also encodes a 218–amino acid protein (Fig. 1B). By contrast, the stop codon in the A allele means that it only supports the 218–amino acid version of claudin-5.
Figure 1.
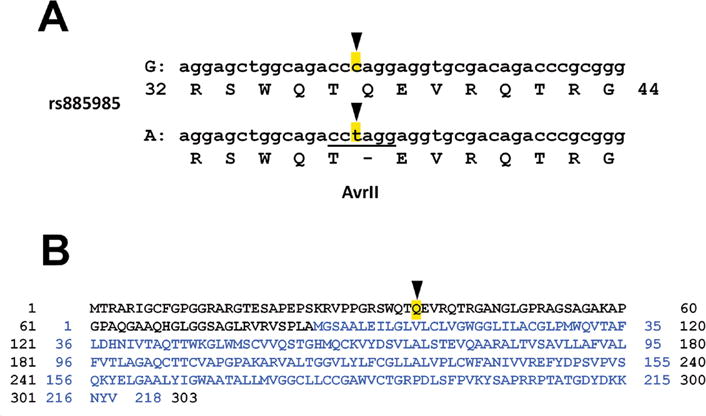
Two human claudin-5 alleles encode distinct overlapping open reading frames. (A) SNP rs885985 has two major alleles, G and A, which encode for glutamine (Q) or a stop signal, respectively. The A allele has an AvrII site not present in the G allele, which was used for genotyping (Fig. 3C). (B) Translation of the two ORFs for the G allele (1–303) and the A allele (1–218; blue). The Q37 codon in the G allele that is replaced with a stop codon in the A allele is highlighted.
The penetrance of both the G and A alleles is extremely high, with net frequencies of G = 0.5032 and A = 0.4968 in the general population, as sampled in the 1000genomes project database (Table 1).26 Interestingly, different subpopulations differ in the relative frequency of the G and A alleles; African populations tended to have higher G allele frequency, whereas Indian populations tended to favor the A allele (Table 1). Whether this is linked to any physiologic consequence in either of these subpopulations remains to be determined.
Table 1.
Frequency of CLDN5 rs885985 G/A alleles
| G | A | G + A | Freq G | Freq A | ||
|---|---|---|---|---|---|---|
| Total | 2520 | 2488 | 5008 | 0.503 | 0.497 | |
| YRI | Yoruba in Ibadan, Nigeria | 182 | 34 | 216 | 0.843 | 0.157 |
| ESN | Esan in Nigeria | 172 | 26 | 198 | 0.869 | 0.131 |
| GWD | Gambian in Western Divisions in Gambia | 164 | 62 | 226 | 0.726 | 0.274 |
| LWK | Luhya in Webuye, Kenya | 162 | 36 | 198 | 0.818 | 0.182 |
| ACB | African Caribbeans in Barbados | 157 | 35 | 192 | 0.818 | 0.182 |
| MSL | Mende in Sierra Leone | 123 | 47 | 170 | 0.724 | 0.276 |
| FIN | Finnish in Finland | 112 | 86 | 198 | 0.566 | 0.434 |
| JPT | Japanese in Tokyo, Japan | 110 | 98 | 208 | 0.529 | 0.471 |
| CHB | Han Chinese in Bejing, China | 107 | 99 | 206 | 0.519 | 0.481 |
| CHS | Southern Han Chinese | 97 | 113 | 210 | 0.462 | 0.538 |
| IBS | Iberian population in Spain | 95 | 119 | 214 | 0.444 | 0.556 |
| TSI | Toscani in Italia | 93 | 121 | 214 | 0.435 | 0.565 |
| PUR | Puerto Ricans from Puerto Rico | 91 | 117 | 208 | 0.438 | 0.563 |
| ASW | Americans of African Ancestry in Southwest United States | 88 | 34 | 122 | 0.721 | 0.279 |
| CLM | Colombians from Medellin, Colombia | 82 | 106 | 188 | 0.436 | 0.564 |
| CDX | Chinese Dai in Xishuangbanna, China | 81 | 105 | 186 | 0.435 | 0.565 |
| KHV | Kinh in Ho Chi Minh City, Vietnam | 79 | 119 | 198 | 0.399 | 0.601 |
| CEU | Utah Residents (CEPH) Northwestern European | 78 | 120 | 198 | 0.394 | 0.606 |
| GBR | British in England and Scotland | 68 | 114 | 182 | 0.374 | 0.626 |
| STU | Sri Lankan Tamil from the United Kingdom | 65 | 139 | 204 | 0.319 | 0.681 |
| MXL | Mexican Ancestry from Los Angeles, California | 57 | 71 | 128 | 0.445 | 0.555 |
| PEL | Peruvians from Lima, Peru | 56 | 114 | 170 | 0.329 | 0.671 |
| GIH | Gujarati Indian from Houston, Texas | 55 | 151 | 206 | 0.267 | 0.733 |
| ITU | Indian Telugu from the United Kingdom | 55 | 149 | 204 | 0.270 | 0.730 |
| BEB | Bengali from Bangladesh | 49 | 123 | 172 | 0.285 | 0.715 |
| PJL | Punjabi from Lahore, Pakistan | 42 | 150 | 192 | 0.219 | 0.781 |
NOTE: Genotypes of different subpopulations were obtained from the 1000genomes project database.26 G and A are the number of each allele detected in each subpopulation; G + A is the total number of alleles measured; Freq G and Freq A are the frequency for each allele. Total is the sum total of all the subpopulations represented in the database. Except for the total values, the table is sorted from high frequency to low frequency of the G allele.
Homology with CLDN5 in other species
The 303–amino acid ORF of human CLDN5 was surprising considering that, as of February, 2017, of the 276 claudin-5 protein sequences in the NCBI protein database, 215 of these are 220 amino acids or shorter in length. Given this, we performed a BLAST search to identify ORFs present in CLDN5 genes in other species that were recognized using the human 303–amino acid claudin-5 sequence (Fig. 2).27
Figure 2.
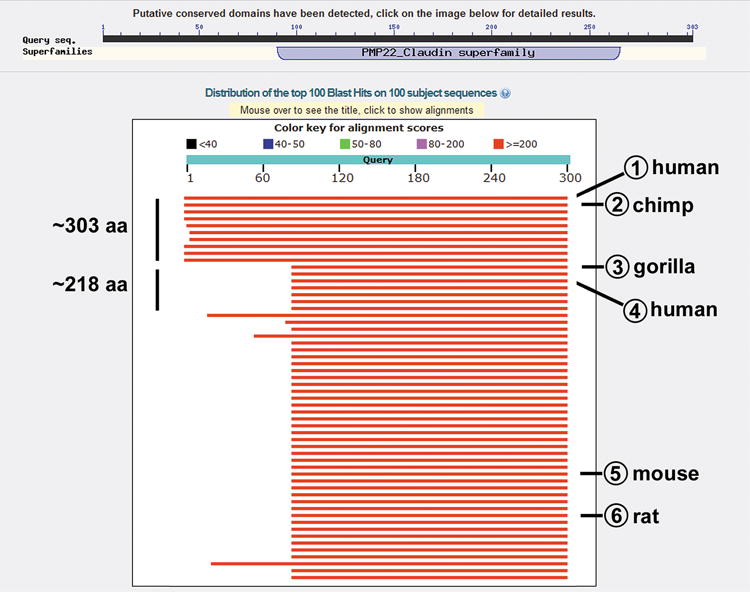
Annotated screen capture of BLAST search using the 303–amino acid human claudin-5 sequence. The 303–amino acid sequence in Fig. 1 (accession number NP_003268.2) was used in a BLASTp search accessed February 14, 2017, and a screen image was saved. Approximate sizes of putative proteins are indicated on the left. Shown are Blast hits corresponding to (1) the original human query sequence, (2) chimpanzee (Pan troglodytes; XP_009436065.1), (3) gorilla (Gorilla gorilla gorilla; XP_004063074.1), (4) a human 218–amino acid form (Homo sapiens; AAH19290.2/BAD96186.1), (5) mouse (Mus musculus; NP_038833.2) and 6) (rat (Rattus norvegicus; NP_113889.1). Note that 2 and 3 represent predicted sequences based on mRNA ORFs.
A BLASTp homology search using human CLDN5 sequence revealed eight hits in the 303–amino acid size range. These were chimpanzee (Pan troglodytes; XP_009436065.1), rhesus macaque (Macaca mulatta; XP_015005195.1), crab-eating macaque (Macaca fascicularis; XP_005596069.1), olive baboon (Papio Anubis; XP_017800620.1), black-capped squirrel monkey (Saimiri boliviensis; XP_003942723.1), common marmoset (Callithrix jacchus; XP_002743588.1), Weddell seal (Leptonychotes weddellii; XP_006752396.1), and Pacific walrus (Odobenus rosmarus divergens; XP_004417759.1). Sequence alignments between the human and chimpanzee claudin-5 proteins and representative mRNAs are shown in Figures S1 and S2, respectively. While these predicted peptide sequences are based on mRNA and expressed sequence tag (EST) data, all contain codon corrections that have not been experimentally validated. Of note were amber mutations that were presumed to encode for amino acids as opposed to acting as bona fide stop codons.
All of the mRNAs mentioned above that encode for a putative 303–amino acid ORF also contain an AUG initiation codon and a continuous ORF needed to produce the more typically sized 218–amino acid claudin-5 protein. The olive baboon, black-capped squirrel monkey, and common marmoset mRNAs lack the AUG start codon necessary to initiate translation for a 303–amino acid gene product. Additionally, chimpanzee, rhesus macaque, crab-eating macaque, olive baboon, Weddell seal, and Pacific walrus claudin-5 sequences contain an amber stop codon upstream of the AUG start site that could be used to encode for a more commonly sized 218–amino acid claudin-5 gene product. The only other homologous ORF approaching 303 amino acids in this BLAST search (Fig. 2) was from the gray mouse lemur (Microcebus murinus; XP_012632046.1). This ORF contains a predicted sequence that encodes for a 218–amino acid version of claudin-5 that is 91.7% identical to human claudin-5. However, the upstream predicted peptide motif for the gray mouse lemur shows considerable divergence from the comparable human sequence (< 55% similarity, < 30% identity).
The gorilla CLDN5 gene (Gorilla gorilla gorilla; XP_004063074.1) unequivocally encodes for a 218–amino acid form of claudin-5, as it lacks an AUG site with the ability to encode for a 303–amino acid ORF and also has an amber stop codon upstream of the AUG initiation site for the 218–amino acid form of claudin-5 (Figs. S1 and S2). This length is consistent with the CLDN5 gene products found in ~ 75% of the other species represented in the NCBI database, including rodent and canine claudin-5 as well those found in several non-mammalian genomes (Fig. 2).
Both the G and A alleles of human CLDN5 produce a 218–amino acid protein
We then examined de-identified, posttransplant human lung tissue and a human colon cancer cell line (Caco2) by immunoblot to determine whether one or both isoforms of claudin-5 protein were expressed. HeLa cells, which we previously established to be claudin null,24 were transfected with cDNAs coding for either the 303–amino acid form or the 218–amino acid form of human claudin-5 using the cytomegalovirus (CMV) promoter as a strong driver of transcription and used as standards. As shown in Figure 3A, human lung and Caco2 cells both only showed expression of the 218–amino acid form of claudin-5.
Figure 3.
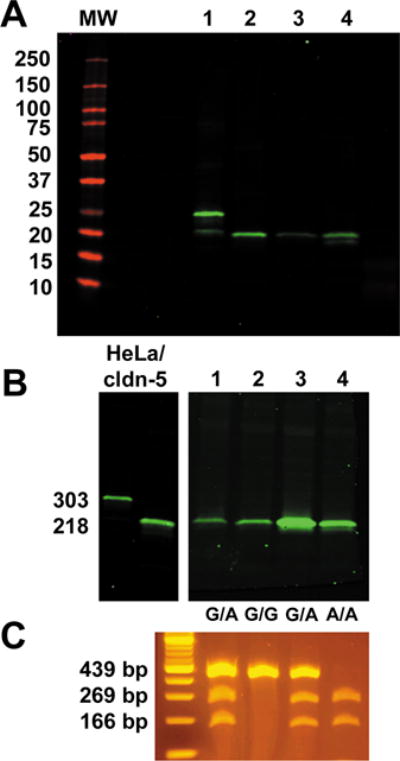
Normal human claudin-5 is 218 amino acids regardless of genotype. (A) Claudin-5 immunoblot showing (1) HeLa cells transfected with untagged human claudin-5 (303), (2) HeLa cells transfected with untagged human claudin-5 (218), (3) human lung tissue, and (4) Caco2 cells. (B) Immunoblot of HeLa cells transfected with either the 303– or 218–amino acid form of claudin-5. Lanes 1–4 correspond to total lung samples from different donors with different genotypes. (C) DNA isolated from the samples in B was amplified by PCR and analyzed for AvrII cleavage as described in the methods section. Genotypes for the lanes in B were 1: G/A, 2: G/G, 3: G/A; and 4: A/A. In all cases, only the 218–amino acid form of claudin-5 was detected.
To determine whether tissues of different genotypes produced differently sized claudin-5 proteins, we examined claudin-5 protein present in four different human lung tissues that were representative of G/G, A/A and G/A genotypes by immunoblot (Fig. 3B and 3C). As shown in Figure 3C, all three different genotypes were represented in this set of samples. Despite the differences in genotype, all four tissues only exhibited the 218–amino acid form of human claudin-5. Although this does not rule out the possibility that the longer form of human claudin-5 may be expressed under certain circumstances, it demonstrates that the 218–amino acid form of claudin-5 is most likely to be expressed under normal conditions, with the caveat that we examined a relatively limited set of tissues in this study. Moreover, an analysis of the regions surrounding the two initiation codons for Kozak sequences28,29 (using http://www.cbs.dtu.dk/services/NetStart/) revealed that the initiation codon for the 218–amino acid protein (CCUCUAGCCAUGGGGUCC; score = 0.762) was a stronger initiation sequence than that of the 303 –amino acid protein (UGCGGCACGAUGACCCGC; score = 0.649),30 consistent with it being more likely to be used to initiate translation. These sites are highly conserved among human, chimpanzee, and gorilla claudin-5 mRNAs, although the gorilla mRNA lacks the upstream AUG and instead has an ACG sequence (Fig. S2).
The long form of claudin-5 is not transported to the plasma membrane
Since we were able to force expression of the long form of human claudin-5 in claudin-null HeLa cells24 with the CMV promoter, we examined the intracellular distribution of this protein by fluorescence microscopy. As shown in Figure 4A and as previously described,24 the 218–amino acid form of claudin-5 was transported to the plasma membrane of HeLa cells and enriched at sites of cell–cell contact. By contrast, the 303–amino acid form was not and was instead retained in intracellular compartments. On the basis of nuclear envelope labeling, this most likely included the endoplasmic reticulum, suggesting improper processing of the long form of claudin-5. Although this could potentially be due to overexpression, HeLa cells transfected with the 218–amino acid form of claudin-5 expressed 3.1 ± 0.5 fold (n = 3) more claudin-5 protein than HeLa cells transfected with the 303–amino acid form of claudin-5, suggesting that mistargeting of the long form was not the result of overexpression.
Figure 4.
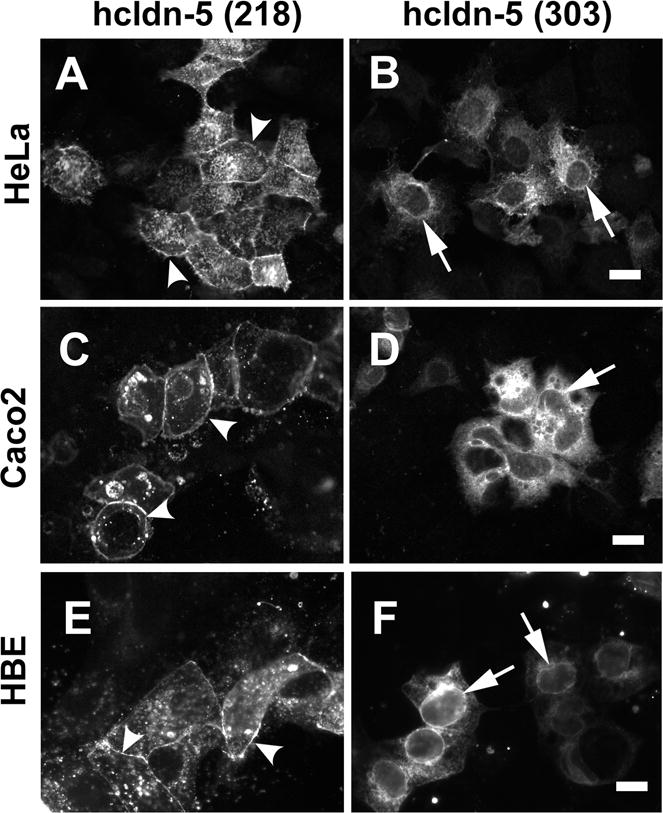
The long form of claudin-5 is not transported to the plasma membrane. HeLa cells (A,B) were transfected with untagged variants of either the 218 amino acid form (A) or the 303 amino acid form of claudin-5 (B), then fixed, immunostained using anti-claudin-5 and then imaged by fluorescence microscopy. Caco2 cells (C,D) or HBE cells (E,F) were transfected with N-terminal YFP-tagged variants of either the 218 amino acid form (C,E) or the 303 amino acid form of claudin-5 (D,F), then fixed and imaged by fluorescence microscopy. In each case, the 218 amino acid form of claudin-5 was transported to the plasma membrane where it localizes to intercellular junctions (arrowheads). By contrast, the 303 amino acid, long form of claudin-5 is retained in an intracellular compartment that most likely is the endoplasmic reticulum, based on nuclear envelope labeling (arrows). Bar, 10 μM.
Since HeLa cells are claudin-null, it was possible that the long form of claudin-5 might be properly transported in epithelial cells expressing other claudins and producing fully formed, native tight junctions. To distinguish the transfected claudin-5 construct from endogenous claudin-5, we used N-terminally tagged versions of the 218– and 303–amino acid forms of claudin-5, since we had previously shown that EYFP–claudin-5 (218 amino acids) was properly transported to the plasma membrane of transfected HeLa cells and that the tag does not interfere with tight junction localization.24 When transfected into either human Caco2 intestinal epithelial cells (Fig. 4C) or human bronchiolar epithelial (HBE) cells (Fig. 4E), the 218–amino acid form of EYFP–claudin-5 showed junctional localization. By contrast, the 303–amino acid form of EYFP–claudin-5 was retained in intracellular compartments in both cell lines (Fig. 4D and 4F), despite that fact that both Caco2 and HBE cells express endogenous claudins, including claudin-5, that are properly trafficked to tight junctions.31 Taken together, these results are consistent with the 218–amino acid form of human claudin-5 being normally expressed in healthy human cells.
Discussion
In this study, we found that, while two different human CLDN5 genotypes encode for two different gene products, only one gene product was expressed in the samples we examined. A human claudin-5 that is 218 amino acids in size is consistent with the vast majority of other claudin-5 proteins found in the protein database, most notably the gorilla CLDN5 gene, which is incapable of producing a long version of claudin-5. Our data also suggest that other long-form claudin-5 ORFs present in the database for other species owing to the database curation of stop codons into amino acid–coding codons either represent bona fide alleles or may be improperly represented in the database. Whether this is the case will require higher-quality sequencing of multiple different individuals from each species.
Although we did not detect any endogenous long claudin-5 proteins in the samples we examined, we were able to induce synthesis of this protein using a CMV-based cDNA construct. Among the ramifications for this finding, it means that caution should be used in obtaining cDNAs from different sources and that it is critical to ensure which isoform is being expressed, particularly since the long form of claudin-5 is not properly processed when expressed by transfected cells.
In our analysis of human claudin gene products, claudin-16 and claudin-23 are the only claudins besides claudin-5 that have an ORF able to support production of an atypically long protein and that also contain an in-frame internal AUG site with the capacity to produce a more standard-sized claudin protein (Table 2). Claudin-16 has been studied in considerable detail, and it was found that endogenous claudin-16 is a 235–amino acid protein instead of the full-length 305–amino acid form encoded by the entire ORF.21,22 Additionally, there is a SNP (rs386669518) at amino acid 55 of the long form of claudin-16 expressed at 16.7% frequency that results in a frame shift leading to a premature translation stop at position 90.22 This is consistent with a lack of selective pressure to maintain the ability to produce the 305–amino acid version of claudin-16. Less is known about claudin-23; however, it has an ORF that could support a protein that is 292 amino acids long. However, by immunoblot, the claudin-23 protein is more consistent with a 213–amino acid gene product.32
Table 2.
Upstream AUG codons in human claudin mRNAs
| Gene product | mRNA | Upstream AUGs
|
Upstream stop in frame | |
|---|---|---|---|---|
| In frame | Out of frame | |||
| Claudin-1 | 0 | 0 | y | |
| Claudin-2 | 1 | 1 | 2 | y |
| 2 | 1 | 7 | y | |
| 3 | 0 | 2 | y | |
| Claudin-3 | 0 | 1 | n | |
| Claudin-4 | 0 | 0 | y | |
| Claudin-5 | 1 | 3 | 4 | allelica |
| 2 | 1 | 1 | allelic | |
| Claudin-6 | 0 | 0 | y | |
| Claudin-7 | 1 | 0 | 0 | y |
| 2 | 1 | 0 | y | |
| 3b | 0 | 0 | y | |
| Claudin-8 | 0 | 0 | y | |
| Claudin-9 | 1 | 4 | y | |
| Claudin-10ac | a | 0 | 3 | y |
| Claudin-10b | b | 0 | 0 | y |
| Claudin-11 | 1 | 0 | 0 | y |
| 2d | 2 | 1 | y | |
| Claudin-12 | 1 | 1 | 3 | y |
| 2 | 1 | 2 | y | |
| 3 | 1 | 2 | y | |
| Claudin-14 | 1/α | 7 | 6 | y |
| 5/β | 2 | 7 | y | |
| γ | 1 | 3 | y | |
| 3/δ | 2 | 2 | y | |
| ε | 0 | 1 | y | |
| Claudin-15 | 1 | 3 | 5 | y |
| 2 | 0 | 0 | y | |
| Claudin-16 | 2 | 2 | n | |
| Claudin-17 | 0 | 0 | n | |
| Claudin-18.1 | 1 | 0 | 0 | n |
| Claudin-18.2 | 2 | 0 | 0 | n |
| Claudin-19 | 1 | 0 | 0 | n |
| 2 | 0 | 0 | n | |
| 3 | 0 | 0 | n | |
| Claudin-20 | 2 | 4 | y | |
| Claudin-22 | 2 | 5 | y | |
| Claudin-23e | 5 | 0 | n | |
| Claudin-24 | 0 | 0 | n | |
NOTE: Listed are validated mRNAs from the NCBI website as accessed on 2/14/2017 (https://www.ncbi.nlm.nih.gov/gene/). The number of in-frame and out-of-frame AUG sites upstream of the initiation site used for translation of a 218– to 230–amino acid gene product are listed. Whether there were any in-frame stop codons between the upstream AUG sites and the site of initiation that would prevent continuous translation is denoted by “y.” Messenger RNAs containing ORFs with the capacity to produce long claudin gene products are highlighted in blue.
One human claudin-5 allele encodes for an amino acid, while another is a stop codon.
This transcript produces a tail-truncated gene product.
Includes all splicing isoforms listed in Ref. 15.
This transcript produces a truncated gene product lacking the first transmembrane domain.
The size of the claudin-23 protein has not been rigorously examined, but the 292–amino acid ORF has several internal start codons, one of which would encode for a 213–amino acid gene product that is compatible with the size of the protein expressed, as measured by immunoblot.32
When expressed by transfected cells, the long form of claudin-16 is not properly trafficked to tight junctions.21 This is reminiscent of what we observed for claudin-5. However, in contrast to claudin-5, which we found was largely retained in the endoplasmic reticulum, claudin-16 appears to accumulate in lysosomes.21 This suggests that claudin-16 is transported to the cell surface and rapidly internalized, in contrast to claudin-5, which appears unlikely to exit the secretory compartment. Whether the long form of claudin-5 is transported to the cell surface remains to be determined, although it clearly is impaired in comparison with the 218–amino acid form. The inability of the long form of claudin-5 to be properly processed raises the intriguing hypothesis that there may be situations where the 303–amino acid form of claudin-5 is expressed, leading to pathologic outcomes. For instance, there are seven SNPs in human claudin-5 mRNA near the start site of the 218–amino acid form, as well as 24 SNPs upstream from the start site, that might affect the initiation of translation at that site. There also are nearly four dozen SNPs in the 5′ untranslated region (UTR) of human claudin-5. Although most of these SNPs are in the frequency range of at most 2%, the 5′ UTR includes a high-frequency SNP, rs739370 that has a minor allele frequency of 0.298. However, to date, endogenous expression of the 303–amino acid form of human claudin-5 has not been detected.
Given the potential for expression of a claudin-5 protein prone to misprocessing, we speculate that there may be human diseases in which the 303–amino acid form of claudin-5 is expressed. Although the long form of claudin-5 is not likely to have a dominant-negative effect on other claudins,5,33 it could still impair barrier function by not being properly integrated into functional tight junctions. Moreover, high levels of ER-localized misfolded proteins could render cells susceptible to ER-associated stress and unfolded protein responses that predispose cells to apoptosis and/or increased sensitivity to proinflammatory stimuli.34,35
The potential for expression of the long form of claudin-5 may be particularly significant in individuals with a G/G genotype, which is highly represented in some human subpopulations (Table 1), and could be relevant to vascular or pulmonary disease in tissues where claudin-5 is highly expressed. Whether the long form of claudin-5 is associated with human disease will require screening of larger tissue sample sets by immunoblot to determine if and when the long form of claudin-5 is expressed. The existence of AUG motifs in the 5′ untranslated regions of mRNAs is quite common, present in over 50% of human mRNAs. Some of these AUGs define upstream ORFs (uORFs) that encode for peptides varying in size from 10 to 100 amino acids.23,36–38 Upstream ORFs have been found to act as repressors of translation, since ribosomes binding to AUGs in the 5′ untranslated region can stall and prevent access to the primary ORF, thus inhibiting translation. Mutations affecting uORFs have been implicated in several human diseases, including craniofrontonasal syndrome,39 cystic fibrosis,40 and Marie-Unna hereditary hypotrichosis.41 Of the 22 human claudins, 14 have AUGs that are either in or out of frame with the main ORF of the gene product that could regulate translation in this manner, although whether these influence claudin translation remains to be determined. Moreover, only claudin-5, claudin-16, and, potentially, claudin-23 have upstream AUGs that are contiguous with the main ORF and have the potential to produce a larger protein.
Evidence also has emerged that small peptides encoded by uORFs can be produced, are stable, and have roles in regulating cell function.37,42 Of particular relevance to human claudin-5, the A allele of rs885985 breaks up the large ORF, resulting in a uORF that encodes for a 36–amino acid peptide. It is tempting to speculate that this peptide may have a functional role in cells harboring this claudin-5 allele that is lacking in cells with a G/G genotype. Although this remains to be determined, it seems likely that there are genetic variations in the upstream regions of claudin mRNAs that will ultimately be linked to human disease susceptibility.
Supplementary Material
Figure S1. Alignment of claudin-5 amino acid sequences from human, chimpanzee, and gorilla.
Figure S2. Alignment of claudin-5 mRNA sequences from human, chimpanzee, and gorilla.
Acknowledgments
We thank Samuel A. Molina, PhD for critical reading of the manuscript. This work was supported by Emory Alcohol and Lung Biology Center/National Institutes of Health (NIH) Grant P50-AA013757 (M.K.), R01-HL116958 (M.K.), R01- AA025854 (M.K.), the German Academic Exchange Service (DAAD) (B.S.), R25-GM099644 (R.M.L.), Emory+Children’s Center of Excellence for Cystic Fibrosis Research (M.K.), and Emory University Integrated Cellular Imaging Microscopy Core of the Winship Cancer Institute Comprehensive Cancer Center Grant P30CA138292. We thank Oskar Laur, PhD, of the Emory Custom Cloning Core Facility for production of claudin-5 expression vectors. This study was supported in part by the Emory Integrated Genomics Core (EIGC), which is subsidized by the Emory University School of Medicine and is one of the Emory Integrated Core Facilities.
Footnotes
Author contributions were as follows: conception and design: B.S., J.D.C., M.K.; acquisition of data R.M.C., B.S., W.S.S., M.K.; analysis and interpretation of data: R.M.C., B.S., J.D.C., D.C.N., M.K.; participated in drafting the manuscript R.M.C., B.S., J.D.C., D.C.N., M.K.; approved the final version of the submitted manuscript: R.M.C., B.S., W.S.S., J.D.C., D.C.N., M.K.
Competing interests
The authors declare no competing interests.
References
- 1.Van Itallie CM, Anderson JM. Architecture of tight junctions and principles of molecular composition. Semin Cell Dev Biol. 2014;36:157–165. doi: 10.1016/j.semcdb.2014.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krug SM, Schulzke JD, Fromm M. Tight junction, selective permeability, and related diseases. Semin Cell Dev Biol. 2014;36:166–176. doi: 10.1016/j.semcdb.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 3.Gunzel D. Claudins: vital partners in transcellular and paracellular transport coupling. Pflugers Arch. 2016;469:35–44. doi: 10.1007/s00424-016-1909-3. [DOI] [PubMed] [Google Scholar]
- 4.Furuse M, Tsukita S. Claudins in occluding junctions of humans and flies. Trends Cell Biol. 2006;16:181–188. doi: 10.1016/j.tcb.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 5.Overgaard CE, Daugherty BL, Mitchell LA, Koval M. Claudins: control of barrier function and regulation in response to oxidant stress. Antioxid Redox Signal. 2011;15:1179–1193. doi: 10.1089/ars.2011.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schlingmann B, Molina SA, Koval M. Claudins: Gatekeepers of lung epithelial function. Semin Cell Dev Biol. 2015;42:47–57. doi: 10.1016/j.semcdb.2015.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turner JR, Buschmann MM, Romero-Calvo I, Sailer A, Shen L. The role of molecular remodeling in differential regulation of tight junction permeability. Semin Cell Dev Biol. 2014;36:204–212. doi: 10.1016/j.semcdb.2014.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Capaldo CT, Nusrat A. Claudin switching: Physiological plasticity of the Tight Junction. Semin Cell Dev Biol. 2015;42:22–29. doi: 10.1016/j.semcdb.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 9.Krause G, Protze J, Piontek J. Assembly and function of claudins: Structure-function relationships based on homology models and crystal structures. Semin Cell Dev Biol. 2015;42:3–12. doi: 10.1016/j.semcdb.2015.04.010. [DOI] [PubMed] [Google Scholar]
- 10.Suzuki H, Tani K, Tamura A, Tsukita S, Fujiyoshi Y. Model for the architecture of claudin-based paracellular ion channels through tight junctions. J Mol Biol. 2015;427:291–297. doi: 10.1016/j.jmb.2014.10.020. [DOI] [PubMed] [Google Scholar]
- 11.Suzuki H, Nishizawa T, Tani K, Yamazaki Y, Tamura A, Ishitani R, Dohmae N, Tsukita S, Nureki O, Fujiyoshi Y. Crystal structure of a claudin provides insight into the architecture of tight junctions. Science. 2014;344:304–307. doi: 10.1126/science.1248571. [DOI] [PubMed] [Google Scholar]
- 12.Niimi T, Nagashima K, Ward JM, Minoo P, Zimonjic DB, Popescu NC, Kimura S. claudin-18, a novel downstream target gene for the T/EBP/NKX2.1 homeodomain transcription factor, encodes lung- and stomach-specific isoforms through alternative splicing. Mol Cell Biol. 2001;21:7380–7390. doi: 10.1128/MCB.21.21.7380-7390.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jovov B, Van Itallie CM, Shaheen NJ, Carson JL, Gambling TM, Anderson JM, Orlando RC. Claudin-18: a dominant tight junction protein in Barrett’s esophagus and likely contributor to its acid resistance. Am J Physiol Gastrointest Liver Physiol. 2007;293:G1106–1113. doi: 10.1152/ajpgi.00158.2007. [DOI] [PubMed] [Google Scholar]
- 14.Van Itallie CM, Rogan S, Yu A, Vidal LS, Holmes J, Anderson JM. Two splice variants of claudin-10 in the kidney create paracellular pores with different ion selectivities. Am J Physiol Renal Physiol. 2006;291:F1288–1299. doi: 10.1152/ajprenal.00138.2006. [DOI] [PubMed] [Google Scholar]
- 15.Gunzel D, Stuiver M, Kausalya PJ, Haisch L, Krug SM, Rosenthal R, Meij IC, Hunziker W, Fromm M, Muller D. Claudin-10 exists in six alternatively spliced isoforms that exhibit distinct localization and function. J Cell Sci. 2009;122:1507–1517. doi: 10.1242/jcs.040113. [DOI] [PubMed] [Google Scholar]
- 16.Zheng JY, Yu D, Foroohar M, Ko E, Chan J, Kim N, Chiu R, Pang S. Regulation of the expression of the prostate-specific antigen by claudin-7. J Membr Biol. 2003;194:187–197. doi: 10.1007/s00232-003-2038-4. [DOI] [PubMed] [Google Scholar]
- 17.Koval M. Claudin heterogeneity and control of lung tight junctions. Annu Rev Physiol. 2013;75:551–567. doi: 10.1146/annurev-physiol-030212-183809. [DOI] [PubMed] [Google Scholar]
- 18.Gong Y, Hou J. Claudins in barrier and transport function-the kidney. Pflugers Arch. 2016 doi: 10.1007/s00424-016-1906-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nitta T, Hata M, Gotoh S, Seo Y, Sasaki H, Hashimoto N, Furuse M, Tsukita S. Size-selective loosening of the blood-brain barrier in claudin-5-deficient mice. J Cell Biol. 2003;161:653–660. doi: 10.1083/jcb.200302070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morita K, Sasaki H, Furuse M, Tsukita S. Endothelial claudin: claudin-5/TMVCF constitutes tight junction strands in endothelial cells. J Cell Biol. 1999;147:185–194. doi: 10.1083/jcb.147.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hou J, Paul DL, Goodenough DA. Paracellin-1 and the modulation of ion selectivity of tight junctions. J Cell Sci. 2005;118:5109–5118. doi: 10.1242/jcs.02631. [DOI] [PubMed] [Google Scholar]
- 22.Weber S, Schlingmann KP, Peters M, Nejsum LN, Nielsen S, Engel H, Grzeschik KH, Seyberth HW, Grone HJ, Nusing R, Konrad M. Primary gene structure and expression studies of rodent paracellin-1. J Am Soc Nephrol. 2001;12:2664–2672. doi: 10.1681/ASN.V12122664. [DOI] [PubMed] [Google Scholar]
- 23.Morris DR, Geballe AP. Upstream open reading frames as regulators of mRNA translation. Mol Cell Biol. 2000;20:8635–8642. doi: 10.1128/mcb.20.23.8635-8642.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Daugherty BL, Ward C, Smith T, Ritzenthaler JD, Koval M. Regulation of heterotypic claudin compatibility. J Biol Chem. 2007;282:30005–30013. doi: 10.1074/jbc.M703547200. [DOI] [PubMed] [Google Scholar]
- 25.Schlingmann B, Overgaard CE, Molina SA, Lynn KS, Mitchell LA, Dorsainvil White S, Mattheyses AL, Guidot DM, Capaldo CT, Koval M. Regulation of claudin/zonula occludens-1 complexes by hetero-claudin interactions. Nat Commun. 2016;(7):12276. doi: 10.1038/ncomms12276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Genomes Project C. Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 28.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 29.Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pedersen AG, Nielsen H. Neural network prediction of translation initiation sites in eukaryotes: perspectives for EST and genome analysis. Proc Int Conf Intell Syst Mol Biol. 1997;5:226–233. [PubMed] [Google Scholar]
- 31.Overgaard CE, Schlingmann B, Dorsainvil White S, Ward C, Fan X, Swarnakar S, Brown LA, Guidot DM, Koval M. The relative balance of GM-CSF and TGFbeta1 regulates lung epithelial barrier function. Am J Physiol Lung Cell Mol Physiol. 2015;308:L1212–L1223. doi: 10.1152/ajplung.00042.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maryan N, Statkiewicz M, Mikula M, Goryca K, Paziewska A, Strzalkowska A, Dabrowska M, Bujko M, Ostrowski J. Regulation of the expression of claudin 23 by the enhancer of zeste 2 polycomb group protein in colorectal cancer. Mol Med Rep. 2015;12:728–736. doi: 10.3892/mmr.2015.3378. [DOI] [PubMed] [Google Scholar]
- 33.Koval M. Differential pathways of claudin oligomerization and integration into tight junctions. Tissue Barriers. 2013;1:e24518. doi: 10.4161/tisb.24518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang M, Kaufman RJ. Protein misfolding in the endoplasmic reticulum as a conduit to human disease. Nature. 2016;529:326–335. doi: 10.1038/nature17041. [DOI] [PubMed] [Google Scholar]
- 35.Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 36.Wethmar K, Barbosa-Silva A, Andrade-Navarro MA, Leutz A. uORFdb–a comprehensive literature database on eukaryotic uORF biology. Nucleic Acids Res. 2014;42:D60–67. doi: 10.1093/nar/gkt952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bazzini AA, Johnstone TG, Christiano R, Mackowiak SD, Obermayer B, Fleming ES, Vejnar CE, Lee MT, Rajewsky N, Walther TC, Giraldez AJ. Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. Embo J. 2014;33:981–993. doi: 10.1002/embj.201488411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnstone TG, Bazzini AA, Giraldez AJ. Upstream ORFs are prevalent translational repressors in vertebrates. Embo J. 2016;35:706–723. doi: 10.15252/embj.201592759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Twigg SR, Babbs C, van den Elzen ME, Goriely A, Taylor S, McGowan SJ, Giannoulatou E, Lonie L, Ragoussis J, Sadighi Akha E, Knight SJ, Zechi-Ceide RM, Hoogeboom JA, Pober BR, Toriello HV, Wall SA, Rita Passos-Bueno M, Brunner HG, Mathijssen IM, Wilkie AO. Cellular interference in craniofrontonasal syndrome: males mosaic for mutations in the X-linked EFNB1 gene are more severely affected than true hemizygotes. Hum Mol Genet. 2013;22:1654–1662. doi: 10.1093/hmg/ddt015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lukowski SW, Bombieri C, Trezise AE. Disrupted post-transcriptional regulation of the cystic fibrosis transmembrane conductance regulator (CFTR) by a 5′UTR mutation is associated with a CFTR-related disease. Hum Mutat. 2011;32:E2266–2282. doi: 10.1002/humu.21545. [DOI] [PubMed] [Google Scholar]
- 41.Wen Y, Liu Y, Xu Y, Zhao Y, Hua R, Wang K, Sun M, Li Y, Yang S, Zhang XJ, Kruse R, Cichon S, Betz RC, Nothen MM, van Steensel MA, van Geel M, Steijlen PM, Hohl D, Huber M, Dunnill GS, Kennedy C, Messenger A, Munro CS, Terrinoni A, Hovnanian A, Bodemer C, de Prost Y, Paller AS, Irvine AD, Sinclair R, Green J, Shang D, Liu Q, Luo Y, Jiang L, Chen HD, Lo WH, McLean WH, He CD, Zhang X. Loss-of-function mutations of an inhibitory upstream ORF in the human hairless transcript cause Marie Unna hereditary hypotrichosis. Nat Genet. 2009;41:228–233. doi: 10.1038/ng.276. [DOI] [PubMed] [Google Scholar]
- 42.Magny EG, Pueyo JI, Pearl FM, Cespedes MA, Niven JE, Bishop SA, Couso JP. Conserved regulation of cardiac calcium uptake by peptides encoded in small open reading frames. Science. 2013;341:1116–1120. doi: 10.1126/science.1238802. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Alignment of claudin-5 amino acid sequences from human, chimpanzee, and gorilla.
Figure S2. Alignment of claudin-5 mRNA sequences from human, chimpanzee, and gorilla.


